Abstract
Kinases are annotated in many nucleoside biosynthetic gene clusters (BGCs) but generally are considered responsible only for self-resistance. Here, we report an unexpected 2’-phosphorylation of nucleoside biosynthetic intermediates in the nikkomycin and polyoxin pathways. This phosphorylation is a unique cryptic modification as it is introduced in the third of seven steps during aminohexuronic acid (AHA) nucleoside biosynthesis, retained throughout the pathway’s duration, and is removed in the last step of the pathway. Bioinformatic analysis of reported nucleoside BGCs suggests the presence of cryptic phosphorylation in other pathways and the importance of functional characterization of kinases in nucleoside biosynthetic pathways in general. This study also functionally characterized all of the enzymes responsible for AHA biosynthesis and revealed that AHA is constructed via a unique oxidative C-C bond cleavage reaction. The results suggest a divergent biosynthetic mechanism for three classes of antifungal nucleoside natural products.
INTRODUCTION
Nucleoside natural products constitute an important class of molecules with diverse structures and biological activities useful in medicine, agriculture, and research1,2. While approximately 29 BGCs have been reported3–5, characterization of these pathways is frequently challenging because hydrophilic biosynthetic intermediates do not accumulate or are difficult to detect in gene disruption experiments. Notably, these challenges limited our understanding of how the conserved nucleoside moiety in antifungal peptidyl nucleosides (PNs) is biosynthesized, even though the BGCs have been known for more than a decade6,7.
PNs have wide-ranging applications in medicine (nikkomycin Z, 1)8 and agriculture (polyoxin D, 2)9 (Figure 1a), and these compounds exhibit antifungal activities through selective inhibition of chitin synthase10. In contrast to many antifungal compounds, PNs exhibited no side effects in humans during dose-finding clinical trials8 and the United States Environmental Protection Agency does not regulate polyoxin residue on agricultural products (EPA Reg. No. 68173-1). Thus, PNs represent an attractive platform for the development of novel antifungals.
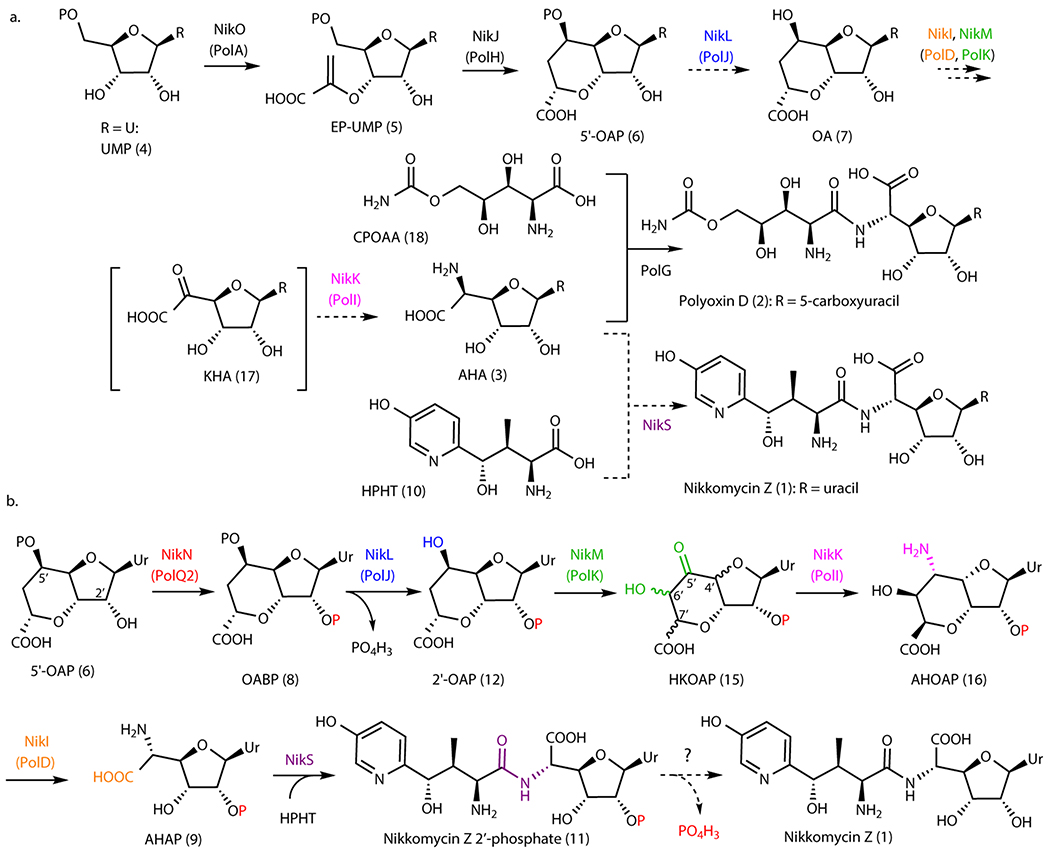
Figure 1. Nikkomycin and polyoxin pathways.
a. Overview of PN biosynthetic pathways. Steps shown in dashed arrows represent previous proposals that are revised in this study. Nucleobase (R) may be uracil (Ur), 5-methyluracil, 5-hydroxymethyluracil, or 5-carboxyuracil for polyoxins, and uracil, 5-methyluracil, imidazolone, or pseudouracil for nikkomycins. b. Revised mechanism of nikkomycin Z biosynthesis. The enzyme responsible for dephosphorylating nikkomycin Z 2’-phosphate has not been identified.
PNs are characterized by a non-proteinogenic amino acid ligated to a conserved six-carbon nucleoside, aminohexuronic acid (AHA, 3). The biosynthesis of PNs has been studied for more than three decades, and the BGCs for nikkomycins6,11 and polyoxins7 were reported in the 1990’s and 2000’s, respectively. Still, our understanding in the biosynthesis of AHA is limited to the first two steps: (i) coupling of uracil 5’-monophosphate (UMP, 4) and phosphoenol pyruvate to form enolpyruvyl UMP (EP-UMP, 5) by NikO/PolA (Figure 1a, Supplementary Table 1)7,12 and (ii) a radical-mediated cyclization of EP-UMP into octosyl acid 5’-phosphate (5’-OAP, 6)13,14. Subsequent steps in AHA biosynthesis remain uncharacterized.
Four enzymes (PolJ/NikL, PolK/NikM, PolD/NikI, and PolI/NikK) were previously proposed to convert 5’-OAP into AHA based on their conservation between the nikkomycin and polyoxin pathways (Figure 1a)7,14. While a previous study reported14 that PolJ dephosphorylates 5’-OAP to yield octosyl acid (OA, 7), PolJ also catalyzed dephosphorylation of EP-UMP at apparently comparable efficiency, which left ambiguity about the relevance of these observations to AHA biosynthesis. Furthermore, while gene knockout strains for all four enzymes have been reported14–17, no biosynthetic intermediates were described. Therefore, the mechanism for 5’-OAP’s transformation into AHA remained unknown.
Here, we report that AHA biosynthesis (Figure 1b) proceeds through an unexpected and cryptic 2’-phosphorylation catalyzed by a kinase (PolQ2). Previous proposals suggested that PolQ2 and its homologs only participated in the export of the final metabolite. However, our characterizations revealed that PolQ2 catalyzes an ATP-dependent 2’-phosphorylation of 5’-OAP to yield octosyl acid 2’,5’-bisphosphate (OABP, 8). This finding permitted the in vitro reconstitution of AHA biosynthesis, where OABP is dephosphorylated at the 5’-position, followed by conversion of the octosyl acid backbone into AHA backbone via hydroxylation, epimerization, transamination, and oxidative C-C bond cleavage (Figure 1b). Finally, the amide ligase, NikS, couples AHA 2’-phosphate (AHAP, 9) to 4’-hydroxy-2’-pyridinyl homothreonine (HPHT, 10), the N-terminal amino acid of nikkomycin Z, to yield nikkomycin Z 2’-phosphate (11). Gene knockout studies verified the physiological relevance of these in vitro characterizations. This unexpected 2’-phosphorylation explains why biosynthetic intermediates may have been overlooked in previous gene disruption studies. A comparison of nucleoside BGCs suggests the conservation of kinases in ~60% of reported nucleoside natural product biosynthetic pathways. While most of these kinases are uncharacterized, cryptic phosphorylation may be relevant to the biosynthesis of many other nucleoside natural products.
RESULTS
Search for the enzyme that acts on 5’-OAP
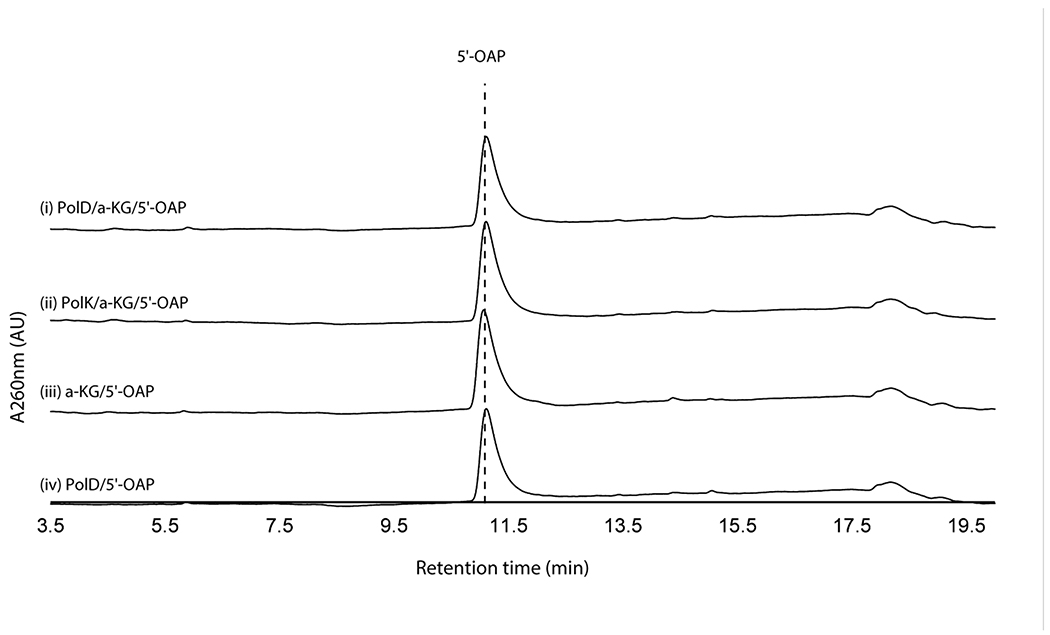
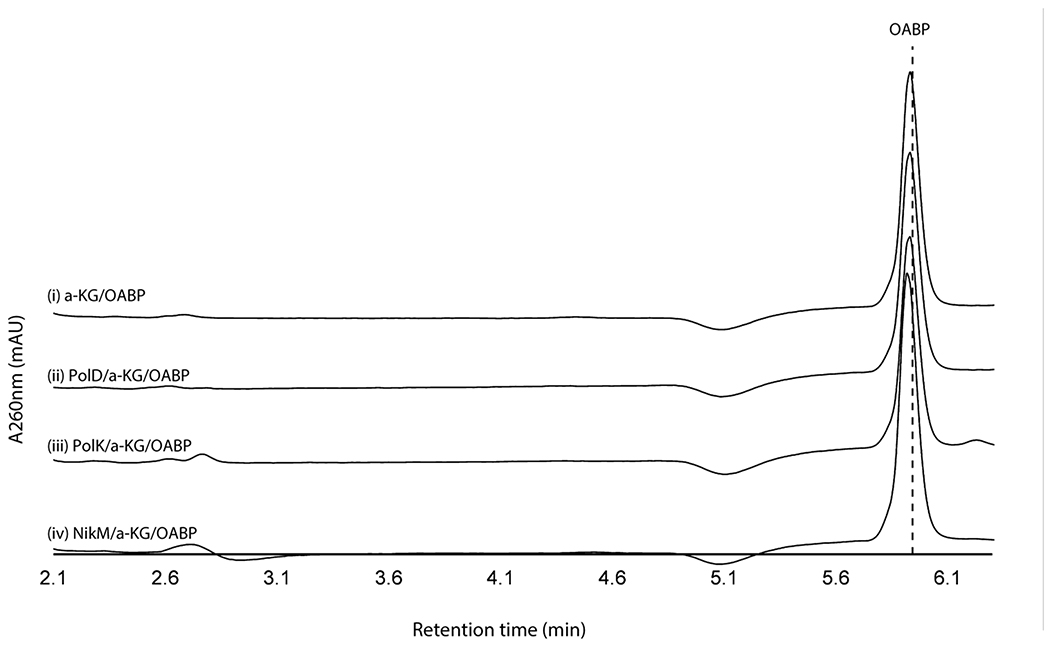
We initially hypothesized that the four enzymes (PolJ/NikL phosphatase, PolI/NikK aminotransferase, PolD/NikI α-ketoglutarate (α-KG) dependent oxygenase, and PolK/NikM α-KG dependent oxygenase; Supplementary Table 1) conserved between the nikkomycin and polyoxin pathways may be responsible for the transformation of 5’-OAP into AHA. To test this hypothesis, we initially focused on in vitro characterization of primarily the polyoxin homologs (PolJ phosphatase, PolK oxygenase, and PolD oxygenase, see Supplementary Figure 1 for SDS-PAGE) since they generally were more soluble than the nikkomycin homologs. The only exception was NikK, whose aminotransferase activity had been reported for proteinogenic amino acids, although the nucleoside substrate was not identified18. NikK was expressed and purified as an N-terminal fusion with maltose binding protein (MBP) to improve its solubility and stability. PolD and PolK were expressed with an N-terminal His-tag. Both an N-terminal His-tagged protein and an MBP fusion protein were investigated for PolJ. Of these, PolJ, PolK, and PolD did not exhibit activities towards 5’-OAP (Extended Data Figure 1, see below for PolJ). NikK was not tested, since 5’-OAP lacks an amine or ketone and cannot be a substrate of an aminotransferase.
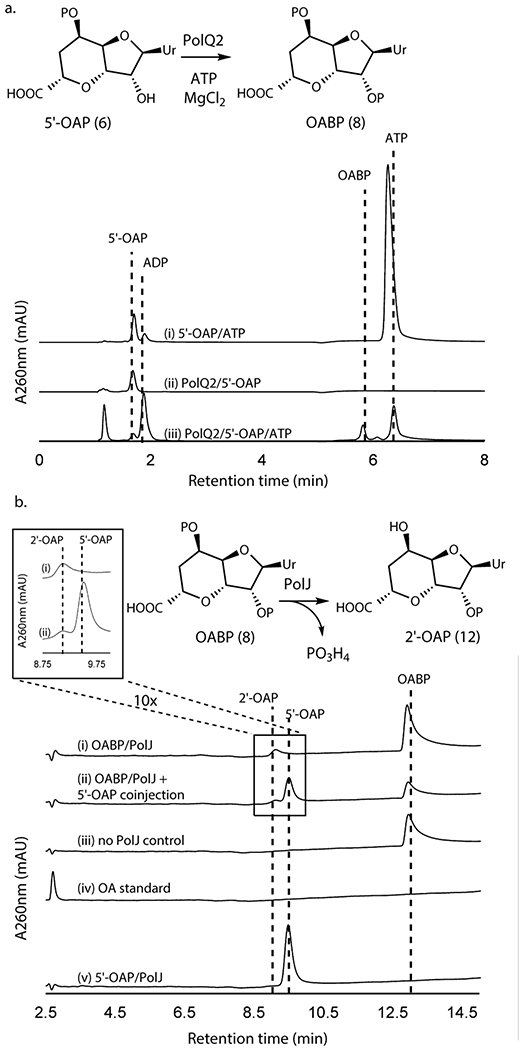
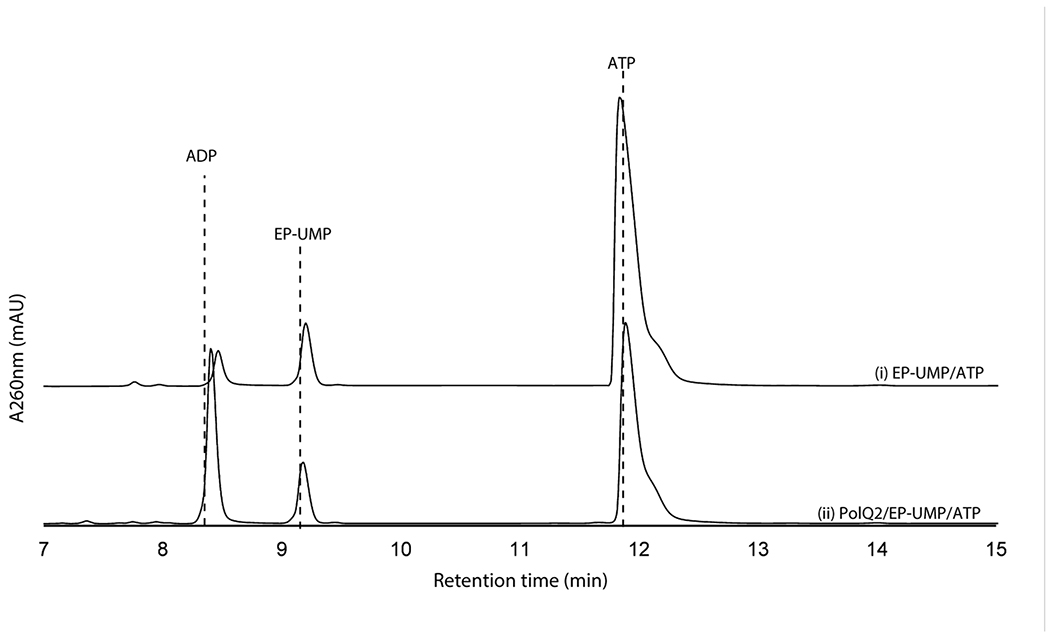
We subsequently extended our study to another conserved enzyme, PolQ2/NikN (Supplementary Table 1). NikN contains two domains: an N-terminal major facilitator superfamily19 (MFS) transporter domain and a C-terminal P-loop NTPase superfamily domain. In the polyoxin pathway, these domains are encoded by two independent but co-transcribed genes, polQ1 and polQ2, respectively. Because of this genetic organization, the P-loop NTPase was proposed as a regulatory subunit/domain of the MFS transporter and only responsible for PN export to the extracellular space7. For our studies, we focused on PolQ2 to avoid purification of NikN with a transmembrane MFS domain. PolQ2 was expressed and purified as an N-terminal fusion with MBP, and incubated with 5’-OAP in the presence of ATP and MgCl2. Analysis of the reaction by high performance anion exchange chromatography (HPAEC) revealed the formation of a uracil-derived molecule (Figure 2a). Notably, the compound’s retention time (RT, 5.9 min) was consistent with the addition of a phosphate to 5’-OAP, and product formation was concomitant with ATP consumption. We observed formation of both ADP and AMP, where ADP is likely the by-product of 5’-OAP’s phosphorylation, and AMP is likely an artifact of promiscuous phosphatase activity. PolQ2’s product was structurally characterized by HRMS and NMR (Supplementary Note), which were consistent with OABP. PolQ2 demonstrated a turnover rate of ~0.1-0.21 min−1 with 5’-OAP as the substrate (Supplementary Figure 2), which may be compared to 6.8 min−1 reported for NikO12 and ~0.3 min−1 for NikJ/PolH13. No activity was observed with EP-UMP despite prolong incubation (12 h, Extended Data Figure 2). Thus, PolQ2 specifically acts on 5’-OAP, and the observed turnover rate suggested its relevance to AHA biosynthesis.
Figure 2. Transformation of 5’-OAP into 2’-OAP by PolQ2 and PolJ.
a. PolQ2 assays with 5’-OAP. Shown are HPAEC chromatograms at 260 nm for a control without the enzyme (i), a control without ATP (ii), and an assay in the complete condition (iii). b. PolJ assays with OABP or 5’-OAP. Shown are HPLC chromatograms at 260 nm for an assay with OABP (i), co-injection of a 5’-OAP standard and the assay with OABP (ii), a control with OABP without the enzyme (iii), an OA standard (iv), and an assay with 5’-OAP (v). All enzyme activities were qualitatively reproducible in at least three independent assays in multiple different enzyme preparations.
Selective 5’-dephosphrylation of OABP by PolJ
Of the remaining conserved enzymes, only the phosphatase PolJ produced a new compound from OABP (Figure 2b, Extended Data Figure 3), which migrated closely to 5’-OAP on HPAEC. The same product was formed by His-tagged and MBP fusion forms of PolJ. PolJ’s product was isolated and structurally characterized by HRMS and NMR (Supplementary Note), which were consistent with 2’-OAP (12). Phosphorylation at the 2’-OH was unambiguously assigned based upon the 3JP-H coupling of the 3’-H signal and the downfield shift of the 2’-H resonance at 4.89 ppm, compared to 4.26 ppm for 5’-OAP. PolJ catalyzed the 5’-dephosphorylation of OABP at the rate of 0.016 min−1 (Supplementary Figure 3), and did not dephosphorylate 5’-OAP (Figure 2b).
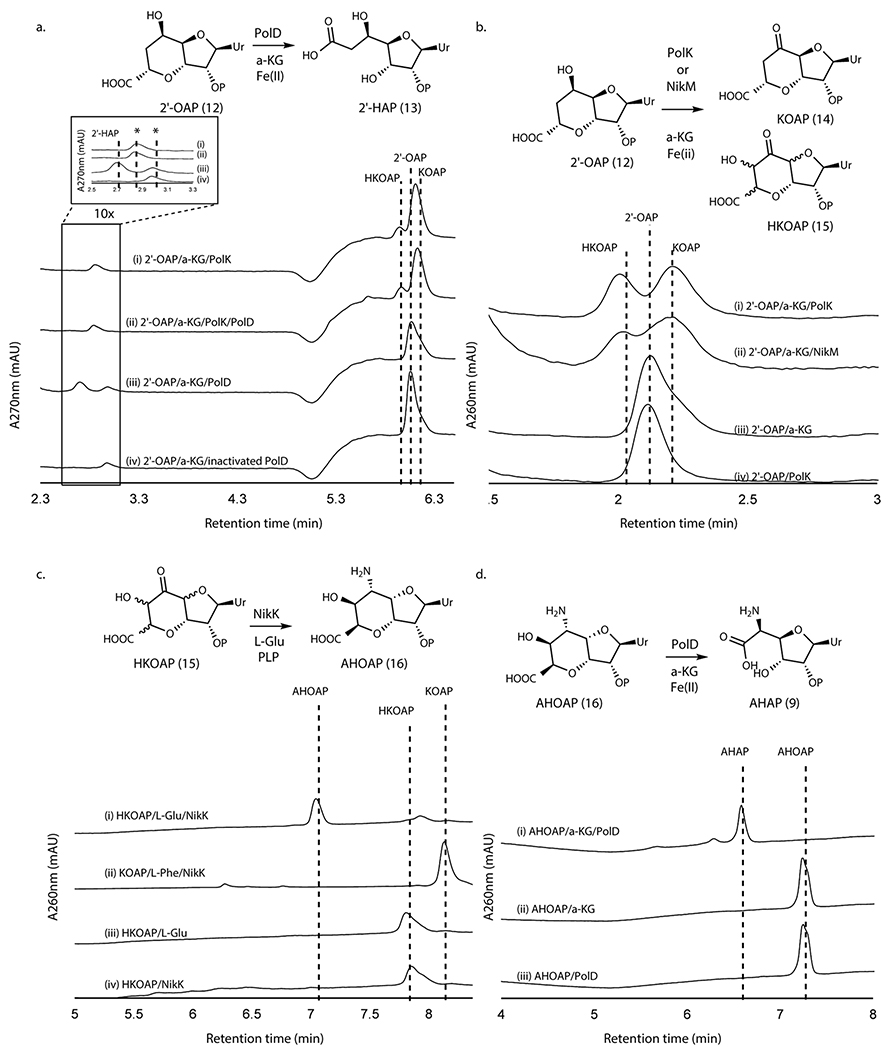
Oxidation of 2’-OAP by PolK
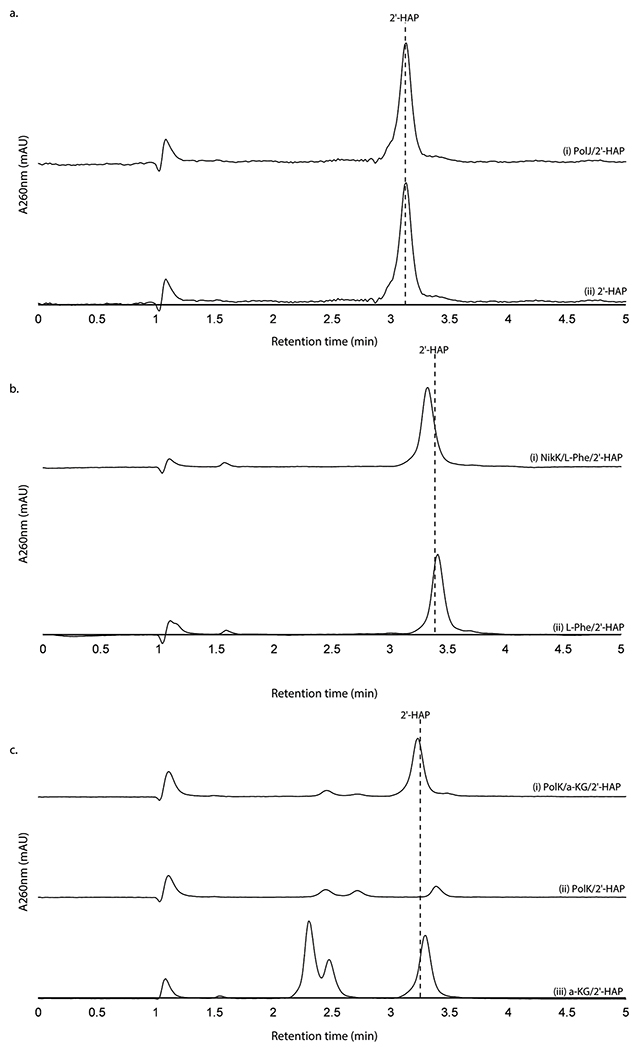
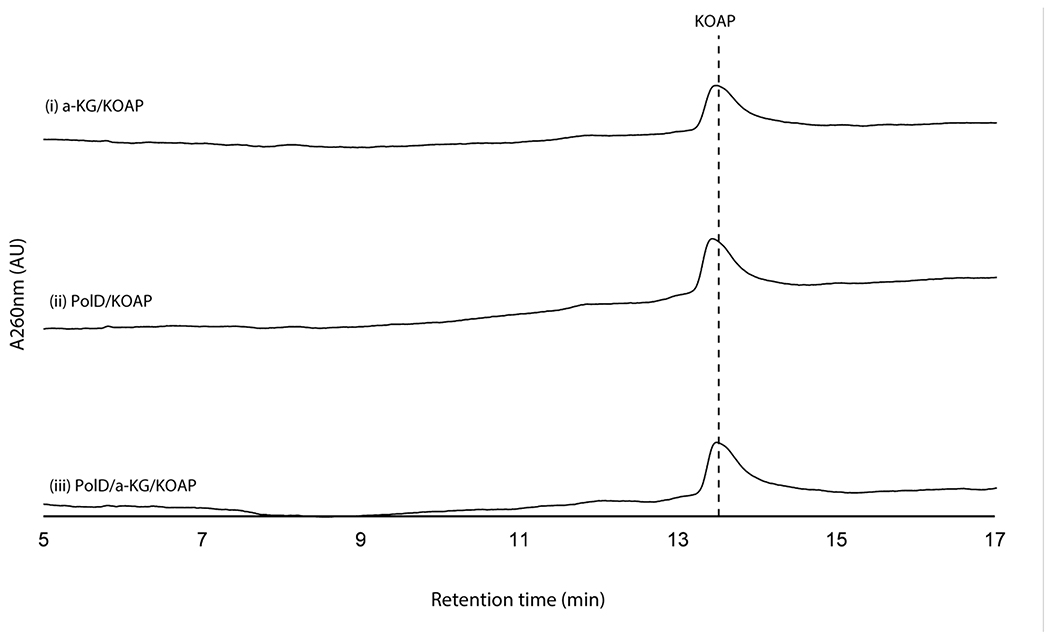
When assayed with 2’-OAP, both oxygenases, PolD and PolK, yielded products in an α-KG dependent fashion. PolD yielded a single product that migrated very early on HPAEC (Figure 3a), while PolK assay yielded two identical products in a ~1:1 ratio which migrated very closely to 2’-OAP (Figure 3b). PolD’s product was isolated and characterized by LC-HRMS/MS, which was consistent with heptosyl acid 2’-phosphate (2’-HAP, 13; Supplementary Note). However, this compound was not converted to any advanced intermediates by either phosphatase PolJ, aminotransferase NikK, or oxygenase PolK (Extended Data Figure 4). When 2’-OAP was assayed in the presence of both oxygenases PolK and PolD, 2’-HAP was not formed, and only PolK’s products were observed (Figure 3a). These observations suggest that 2’-HAP is likely an off-pathway product that is irrelevant to AHA biosynthesis.
Figure 3. Transformation of 2’-OAP into AHAP.
a. PolK and PolD assays with 2’-OAP. Shown are HPAEC chromatograms at 270 nm for an assay with PolK (trace i), an assay with PolD and PolK (ii), an assay with PolD (iii), and a control with heat-inactivated PolD (iv). The peaks shown with asterisks are an unknown contaminant associated with both PolD and PolK, which has λmax at 247 nm and is not a uracil-related compound. b. PolK and NikM assays with 2’-OAP. Shown are HPAEC chromatograms at 260 nm for an assay with PolK (trace i), an assay with NikM (ii), a control without the enzyme (iii), and a control PolK assay without α-KG (iv). c. NikK assays with HKOAP or KOAP. Shown are HPAEC chromatograms at 260 nm for a NikK assay with HKOAP (trace i), a NikK assay with KOAP (scaled 1/6x) (ii), a control with HKOAP without NikK (iii), a control with HKOAP without l-Glu (iv). d. PolD assays with AHOAP. Shown are HPAEC chromatograms at 260 nm for the PolD assay in a complete condition (trace i), a control without the enzyme (ii), and a control without α-KG (iii). All enzyme activities were qualitatively reproducible in at least three independent assays in multiple different enzyme preparations.
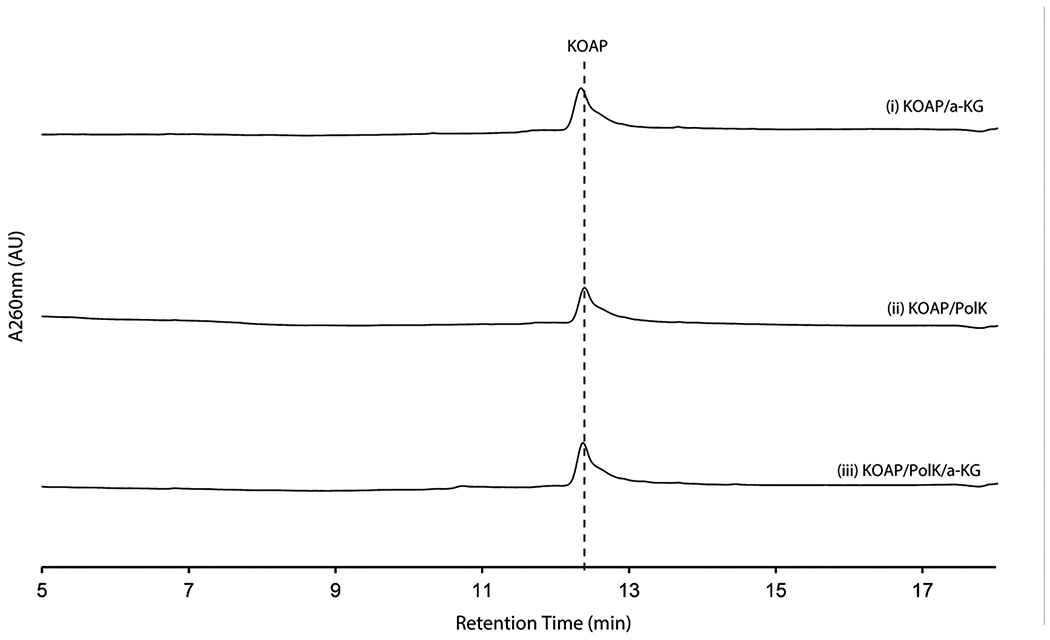
PolK’s two products were isolated for further characterization. The product with later RT on HPAEC (2.2 min in Figure 3b) was stable and characterized by LC-HRMS and NMR analyses, which were consistent with 5’-keto-octosyl acid 2’-phosphate (KOAP, 14; Supplementary Note). In contrast, the product with an earlier RT (2.0 min in Figure 3b) exhibited limited chemical stability. Interpretation of the 1H NMR spectra of a mixture of this product and its degraded compounds were challenging, but the absence of 1H signals at 2.0 – 2.5 ppm suggested an oxidation of C6’ (Supplementary Note). Based on the exact mass of this compound determined by LC-HRMS (+ 31.989 Da from 2’-OAP, Supplementary Note) and its acceptance by the PLP-dependent aminotransferase NikK (Figure 3c; see below for details), which requires the presence of a ketone, the structure of this compound was inferred as a hydrate form of 6’-hydroxy-5’-keto-octosyl acid 2’-phosphate (HKOAP, 15). The stereochemistry of C4’, C6’ and C7’ of HKOAP cannot be assigned. As described below, it is possible that the C4’ and C7’ of HKOAP have opposite stereochemistry from 2’-OAP. Recombinant NikM, the homolog of PolK in the nikkomycin pathway, also catalyzed the formation of HKOAP and KOAP (Figure 3b), suggesting that the mechanism of AHA biosynthesis is likely conserved between the two pathways.
No substantial amount of HKOAP was formed when isolated KOAP was incubated with oxygenase PolK for 20 hrs (Extended Data Figure 5), suggesting that KOAP is not an intermediate of HKOAP formation. When HKOAP and KOAP were individually incubated with NikK aminotransferase in the presence of l-Glu or l-Phe as an amino donor, only HKOAP was converted to another compound (Figure 3c). Neither HKOAP nor KOAP was a substrate of the other oxygenase, PolD (Figure 3a, Extended Data Figure 6). These observations suggested that HKOAP is an on-pathway intermediate that is aminated by NikK. Intriguingly, a C4’-epimer of dephosphorylated KOAP (octosyl acid C; Supplementary Figure 4)20 and a 6’-hydroxylated and dephosphorylated 2’-OAP (nikkomycin SOZ; Supplementary Figure 4)21 were previously isolated from culture media of Streptomyces cacaoi and Streptomyces tendae, respectively, suggesting the relevance of our in vitro observations to both polyoxin and nikkomycin biosynthesis and prior literature in the field.
HKOAP is selectively aminated by NikK
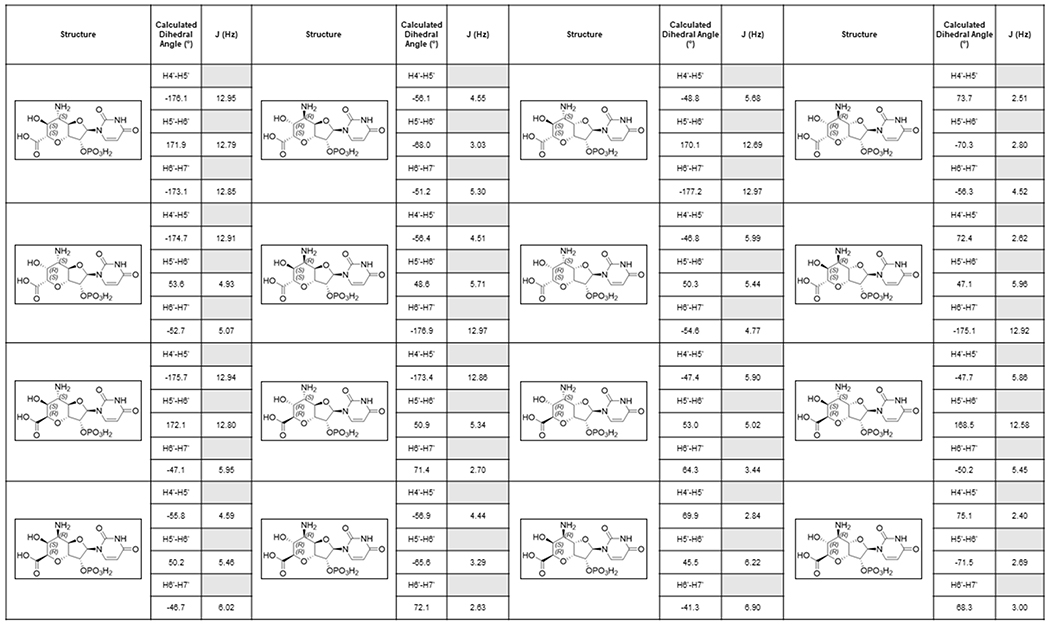
As described above, the aminotransferase NikK selectively accepts HKOAP to form a less anionic molecule. LC-HRMS and FTIR analyses of this compound (Supplementary Note) were consistent with the conversion of a ketone of HKOAP into an amine. However, characterization by NMR was challenging due to broadened 1H NMR signals and a slow T1 relaxation (0.5-1.0 s; Supplementary Note), likely from a slow conformational equilibrium or extended H-bond interactions in solution. We consequently characterized a dephosphorylated analog. Upon dephosphorylation using calf intestinal alkaline phosphatase (CIP) as confirmed by LC-HRMS (Supplementary Note), the resulting compound exhibited sharper NMR signals that were consistent with 5’-amino-6’-hydroxy-octosyl acid (AHOA, Supplementary Note). In particular, the absence of proton signals at 2.0 - 2.5 ppm was consistent with the presence of 6’-OH, and JH4’-H5’ = 3.5 Hz, JH5’-H6’ = 10.5 Hz and JH6’-H7’ = 4.9 Hz were most consistent with 4’S, 5’S, 6’S, 7’R stereochemistry (Extended Data Figure 7). Based on these characterizations, we propose the structure of NikK’s product as (4’S, 5’S, 6’S, 7’R)-5’-amino-6’-hydroxy-octosyl acid 2’-phosphate (AHOAP, 16). The 4’S and 7’R stereochemistry was unexpected and suggests an epimerization of C4’ and C7’ by either oxygenase PolK or aminotransferase NikK.
NikK was previously proposed to catalyze a transamination of ketohexuronic acid (KHA, 17; Figure 1a) to AHA7. Thus, we tested this hypothesis by incubating AHA with NikK in the presence of α-KG. We did not detect KHA formation (Supplementary Figure 5), suggesting that NikK does not catalyze the transamination between AHA and KHA. These observations suggest that the physiological function of NikK is likely the transformation of HKOAP into AHOAP.
PolD catalyzes oxidative C-C bond cleavage of AHOAP
When AHOAP was assayed with oxygenase PolD in the presence of α-KG and Fe2+, PolD efficiently converted AHOAP into another compound that migrated earlier on HPAEC (Figure 3d). NMR and LC-HRMS characterization of this product (Supplementary Note) was consistent with AHA 2’-phosphate (AHAP, 9), and treatment of this product with CIP yielded AHA. PolD catalyzed the conversion of AHOAP into AHAP at least 150-fold faster than the conversion of 2’-OAP into 2’-HAP under similar conditions (Extended Data Figure 8), suggesting that the former is likely relevant to AHA biosynthesis. These observations support that PolD catalyzes a unique oxidative C-C bond cleavage reaction in AHA biosynthesis.
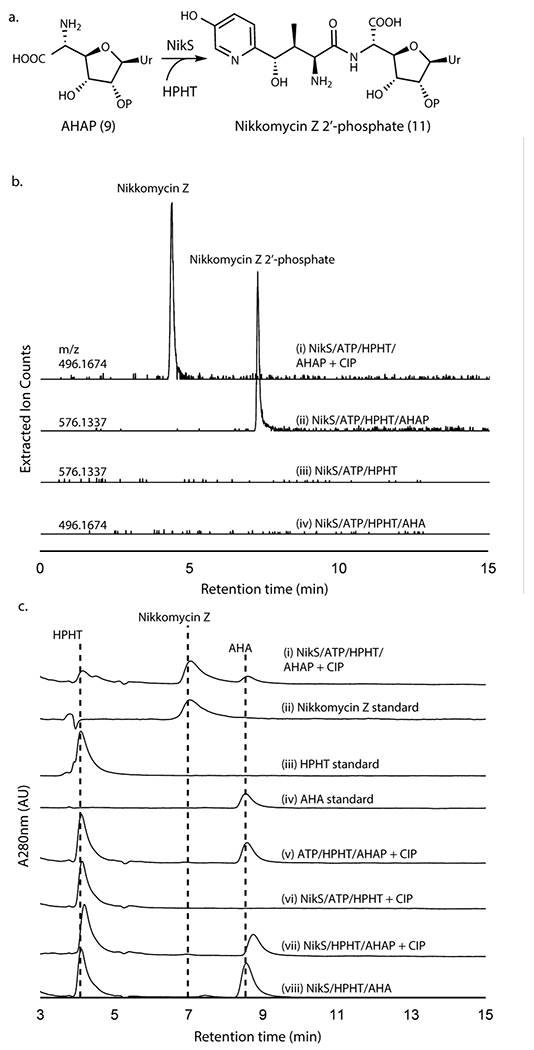
NikS amide ligase requires 2’-phosphorylation
With the in vitro reconstitution of AHAP biosynthesis, we investigated the coupling of AHAP with HPHT by NikS. When AHAP and HPHT were incubated with NikS in the presence of ATP and MgCl2, we observed the formation of a new peak with the molecular weight consistent with phosphorylated nikkomycin Z (Figure 4a,b; Supplementary Figure 6a). The NikS product was converted to nikkomycin Z upon treatment with CIP (Figure 4b,c; Supplementary Figure 6b). Intriguingly, NikS did not catalyze the ligation of AHA with HPHT even after prolonged incubation (Figure 4b,c). These observations provide strong evidence that NikS selectively conjugates HPHT with AHAP, suggesting the importance of 2’-phosphate for substrate recognition. Furthermore, these data suggest that at least in nikkomycin biosynthesis, AHA is likely a shunt metabolite.
Figure 4. NikS activity assays with AHA or AHAP as the amine substrate.
a. Amide ligation catalyzed by NikS. b. LC-HRMS analysis of NikS assays with AHA or AHAP. Shown are extracted ion chromatograms (EIC) at the indicated m/z with a 5 ppm window for an assay with AHAP and HPHT followed by CIP treatment (trace i), an assay with AHAP and HPHT (ii), a control without AHAP (iii), and an assay with AHA and HPHT (iv). Traces ii and iii were monitored at m/z for nikkomycin Z 2’-phosphate ([M+H]+ = 576.1337) and traces i and iv were monitored at m/z for nikkomycin Z ([M+H]+ = 496.1674). c. HPLC analysis of NikS assays with AHAP or AHA. Shown are HPLC UV chromatograms at 280 nm for an assay with NikS with AHAP and HPHT after a CIP treatment (trace i), a nikkomycin Z standard (scaled 3x) (ii), an HPHT standard (iii), an AHA standard (iv), a no enzyme control after a CIP treatment (v), a control without AHAP after a CIP treatment (vi), a control without ATP after a CIP treatment (vii), and a NikS assay with AHA and HPHT (viii). All enzymatic activities as determined by HPLC analyses were reproducible in at least three independent assays and all LC-HRMS analyses were reproducible in two assays.
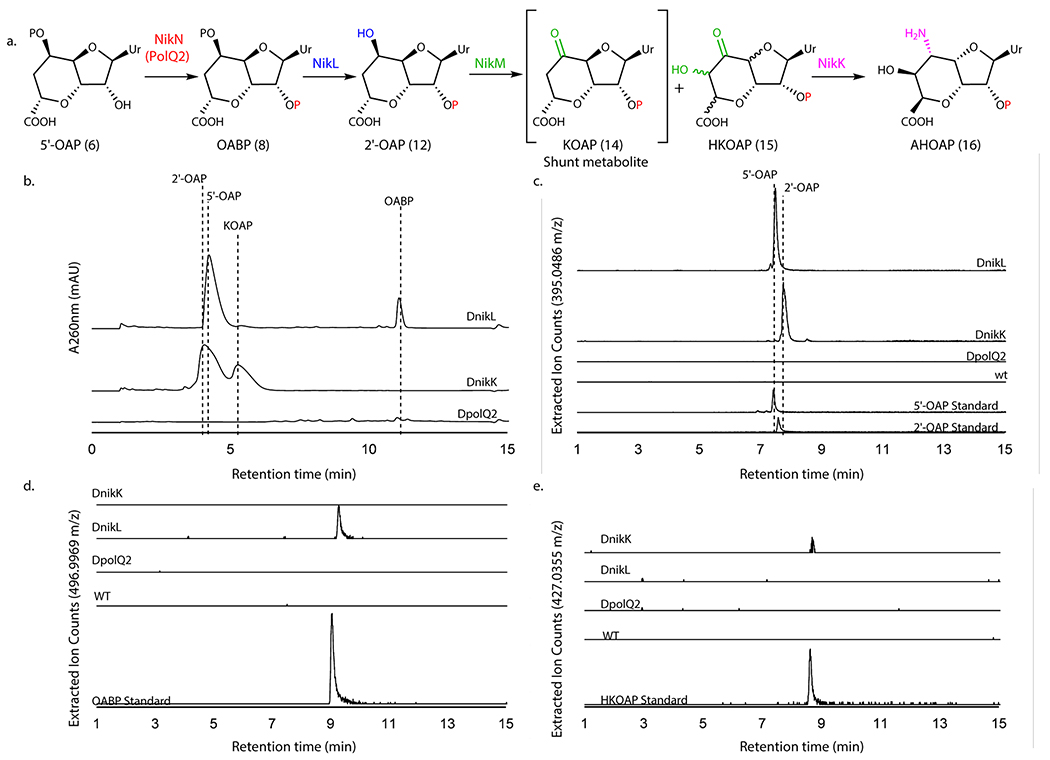
Genetic characterization of polQ2, nikL, and nikK
The in vitro characterization of these seven enzymes (PolQ2, PolJ, PolK/NikM, NikK, PolD, and NikS) suggested the presence of an unexpected cryptic phosphorylation during AHA biosynthesis. To test the physiological relevance of these observations, we performed gene knockout studies for polQ2, nikL, and nikK. Due to the high titer of nikkomycin Z (~0.15 mg/mL, isolation yield), we primarily focused on the nikkomycin pathway, except for the polQ2 kinase (Figure 5a). A selective knockout of kinase activity would be challenging in the nikkomycin pathway because the corresponding kinase (NikN) is fused with a putative MFS transporter.
Figure 5. LCMS characterization of metabolites produced by WT and mutant S. cacaoi and S. tendae.
a. The transformation of 5’-OAP into AHOAP. b. HPAEC chromatograms at 260 nm of nucleotide fraction of culture media of S. tendae ΔnikL::kanR and ΔnikK::kanR, and S. cacaoi ΔpolQ2. The identities of 5’-OAP, 2’-OAP, and KOAP were determined by isolation and characterization with NMR. OABP was identified by co-injection with an authentic standard. c-e. LC-HRMS analysis of S. tendae wt, ΔnikK::kanR, and ΔnikL::kanR and S. cacaoi ΔpolQ2 culture media. Shown are EIC for 2’-OAP and 5’-OAP (m/z 395.0486 ± 0.0020; c), OABP (m/z 496.9969 ± 0.0025, d), and HKOAP (m/z 427.0355 ± 0.0021, e). The observations were reproducible for 2-3 different clones for each mutant strain. The culture was repeated twice for each clone.
The polQ2 gene was disrupted in-frame by homologous recombination in S. cacaoi. The resulting polQ2 disruptant (ΔpolQ2) was then characterized for polyoxin production using LC-HRMS. Under conditions that the S. cacaoi WT strain produced polyoxins, S. cacaoi ΔpolQ2 did not (Figure 5b; Extended Data Figure 9). Complementation of S. cacaoi ΔpolQ2 with the wt polQ2 gene restored polyoxin production (Supplementary Figure 7a, Extended Data Figure 9). These observations are consistent with the essential function of PolQ2 in polyoxin biosynthesis.
The nikL and nikK genes were individually disrupted in S. tendae with a kanamycin resistance gene (kanR) replacement. The resulting disruptants S. tendae ΔnikL::kanR, and ΔnikK::kanR did not produce detectable amounts of nikkomycin Z (Supplementary Figure 7b), confirming that these genes are essential for nikkomycin biosynthesis. Genetic complementation of ΔnikK::kanR with the wt nikK gene restored nikkomycin Z production (Supplementary Figure 7b).
We searched for the characterized biosynthetic intermediates in the culture media of the gene disruptants, and observed accumulation of 5’-OAP (~0.25 mg/mL isolated yield) and OABP (~0.03 mg/mL, LCMS quantitation) in ΔnikL::kanR, and 2’-OAP (~0.2 mg/mL isolated yield) and KOAP (~0.1 mg/mL isolated yield) in ΔnikK::kanR mutants (Figure 5c). These accumulated metabolites were isolated and characterized by LC-HRMS (Figure 5d,e) and NMR (Supplementary Note). OABP accumulation in ΔnikL is consistent with the proposed function of NikL (Figure 5a), and its relatively low accumulation is consistent with OABP’s inability to passively diffuse through the membrane due to its strong negative charge. LCMS analysis also revealed that the ΔnikK::kanR strain accumulated trace amounts of HKOAP (~0.01 mg/mL, LCMS quantitation, Figure 5e), which is consistent with the in vitro function of NikK (Figure 5a). The trace amount of HKOAP accumulation likely derives from HKOAP’s limited stability as described above. The accumulation of HKOAP and KOAP is also consistent with the in vitro function of NikM, which promiscuously catalyzed the conversion of 2’-OAP into HKOAP and KOAP (Figure 5a). Therefore, the results from the gene knockout studies were consistent with the in vitro assays and provide in vivo support for the proposed AHA biosynthetic mechanism involving unexpected cryptic phosphorylation.
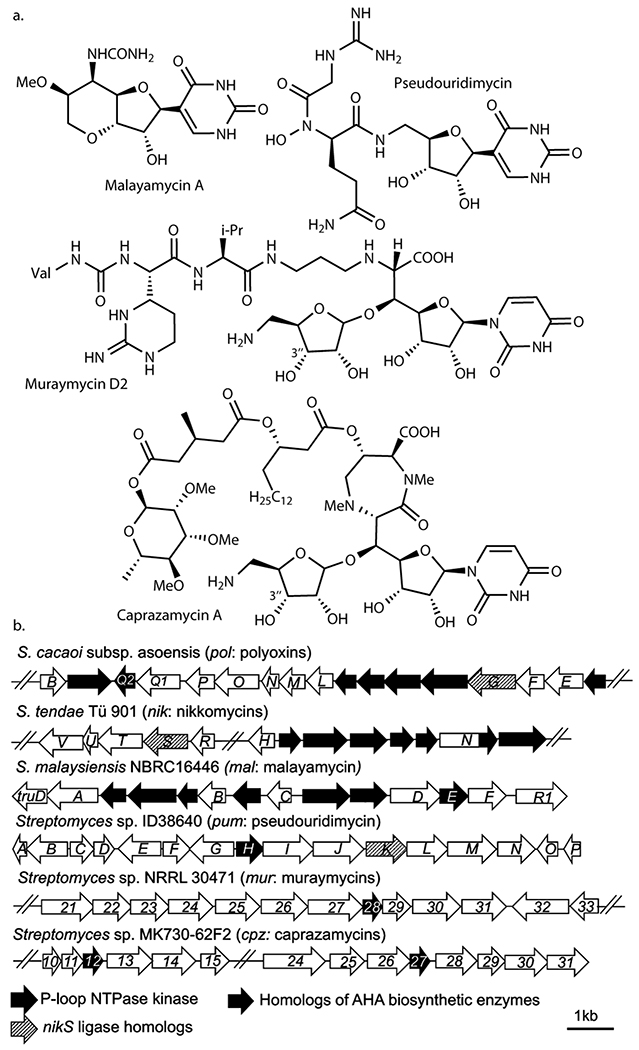
Roles of kinases in nucleoside BGCs
To investigate the potential generality of cryptic phosphorylation, we surveyed reported nucleoside natural product BGCs and identified kinases in 17 of 29 reported BGCs (Supplementary Figure 8). Nine of these kinases belong to the P-loop NTPase superfamily, including MalE (malayamycin; Figure 6a)22, PumH (pseudouridimycin)23, Mur28 (muraymycins)24, and Cpz12 and Cpz27 (caprazamycins)25. The malayamycin22 and pseudouridimycin23 BGCs contain other AHA biosynthetic enzymes or NikS homologs (Figure 6b). Mur28 kinase from the muraymycin pathway catalyzes the 3”-OH phosphorylation of a GlyU-ADR disaccharide biosynthetic intermediate24, and homologous kinases are conserved among GlyU-ADR containing nucleoside BGCs (caprazamycin25, A-9028926, and muraminomycin27). A 3”-phosphorylated caprazamycin precursor has been isolated28. More recently, while this manuscript was in revision, one of the muraymycin biosynthetic enzymes (Mur24) was reported to act specifically on 3”-phosphorylated GlyU-ADR29. Therefore, these pathways likely involve cryptic phosphorylation. Eight other nucleoside BGCs contain putative kinases in families related to phosphorylation of nucleosides and carbohydrates (Supplementary Figure 8), and thus it is still conceivable that these kinases catalyze phosphorylation of carbohydrates or nucleoside/nucleotide biosynthetic intermediates. While some of these kinases may have differing roles, such as self-resistance as previously proposed30,31, these analyses and the PolQ2’s essential role in AHA biosynthesis suggest the importance of the functional characterization of kinases in nucleoside BGCs to understand their biosynthetic mechanisms in general.
Figure 6. Nucleoside natural products predicted to be biosynthesized through cryptic phosphorylation.
a. Structures of nucleoside natural products whose biosynthesis may involve cryptic phosphorylation. b. BGCs of the natural products shown in a.
DISCUSSION
This report describes the mechanism of AHA biosynthesis in PN biosynthetic pathways and the in vitro preparation of nikkomycin Z 2’-phosphate (Figure 1b). In particular, we demonstrate that the 2’-phosphorylation is introduced by a P-loop NTPase kinase PolQ2 (NikN) and is retained for the pathway’s duration. Previously, PolQ2/NikN kinases were thought to be involved exclusively in the export of the final metabolites since no additional phosphorylation beyond 5’-OAP was immediately apparent, and these kinases are associated with the putative MFS transporter. In contrast, our in vitro and in vivo characterizations of PolQ2 and the downstream enzymes reveal that the kinase is essential for AHA biosynthesis and is responsible for cryptic phosphorylation at the 2’-OH.
The discovery of 2’-phosphorylation in AHA biosynthesis has important implications on our general understanding of nucleoside natural product biosynthesis. Phosphorylated intermediates behave differently from nucleosides and can be easily missed when characterizing biosynthetic pathways. AHAP and its precursors were not reported in previous gene knockout studies14–17 likely because phosphorylation was not expected. Based upon our bioinformatics analysis suggesting the potential abundance of such cryptic phosphorylation, this may be the case for other nucleoside natural product pathways as well. Seventeen of the 29 reported nucleoside BGCs contained kinases, of which nine belonged to the P-loop kinase superfamily. In particular, cryptic phosphorylation is likely involved in the biosynthetic pathways of malayamycin, pseudouridimycin, and GlyU-ADR-containing nucleosides (e.g. caprazamycins). Therefore, these findings suggest the importance of the functional characterization of kinases in nucleoside BGCs in general.
Previously reported cryptic modifications of biosynthetic intermediates usually serve chemical roles, e.g. cryptic chlorination installs a leaving group for subsequent cyclopropanation32. Cryptic acylation is found in the biosyntheses of arginine33, butirosin34,35, and the polyoxin N-terminal amino acid, carbamoylpolyoxamic acid36 (CPOAA, 18), and is thought to function as a protecting group. Transient phosphorylation has been reported to act as a leaving group for dehydration in the biosynthesis of ribosomally synthesized post-translationally modified peptides (RiPPs)37. Compared to these precedents, cryptic phosphorylation in AHA biosynthesis is unique since phosphorylation is introduced early in the pathway, retained for the pathway’s duration, and has no apparent chemical role.
Consequently, cryptic phosphorylation in AHA biosynthesis is more similar to the recent reports of loading biosynthetic intermediates to an acyl carrier protein (ACP) during mitomycin biosynthesis38,39, where the function of ACP was not immediately apparent. We propose two possible biological functions for phosphorylation in AHA biosynthesis. The first is a specific recognition by downstream enzymes. With the exception of nicotinamide adenine dinucleotide 2’-phosphate (NADP), nucleoside 2’-phosphate is rare in nature, suggesting that 2’-phosphorylation may serve as a specific recognition tag. Since PNs have substantialvariations in their nucleobase structures that the AHA biosynthetic machinery must accommodate, 2’-phosphorylation may be a mechanism to produce structurally diverse natural products while maintaining the crucial specificity for efficient metabolic flux. A second possible function is to prevent the diffusion of intermediates to the extracellular space. In fact, no phosphorylated intermediates were observed in our MS analysis of WT S. tendae. Even in gene knockout strains, the amount of OABP in the culture media was ~ 25-fold less than 5’-OAP, suggesting that the additional phosphate group minimizes its diffusion/export to the extracellular media. Thus, 2’-phosphorylation may allow efficient metabolic flux during PN biosynthesis.
While the current study revealed a majority of the steps in nikkomycin biosynthesis, the mechanism of dephosphorylation of nikkomycin Z 2’-phosphate remains unknown. Nikkomycin Z 2’-phosphate is not detectable in WT S. tendae, suggesting the presence of a mechanism of dephosphorylation. Two possibilities are conceivable: (i) promiscuous activity of phosphatase NikL or (ii) a reverse reaction of the NikN kinase domain. In the latter case, it is possible that dephosphorylation is coupled to the export of nikkomycin Z through the MFS transporter. Such a mechanism may explain the absence of detectable accumulation of nikkomycin Z 2’-phosphate and the genetic association of the kinase and MFS transporter.
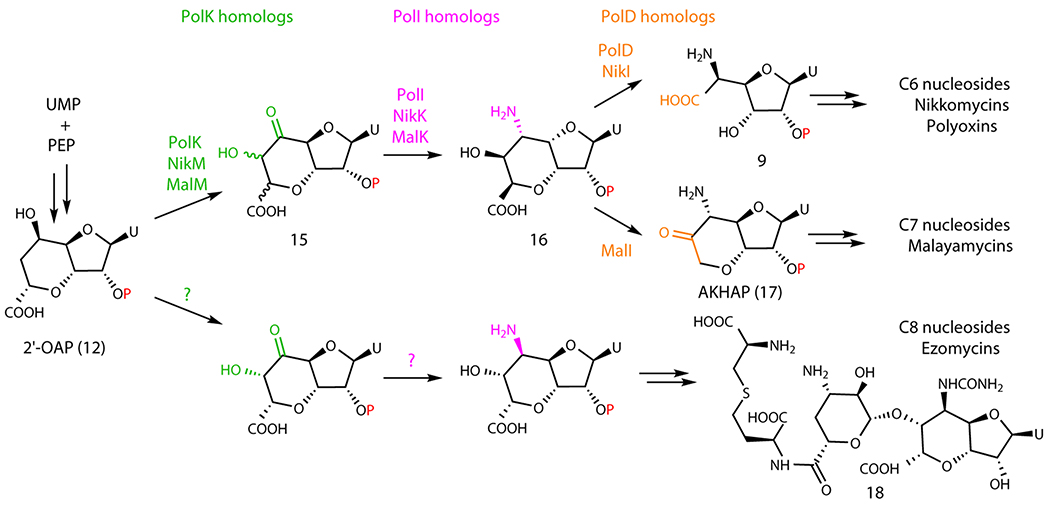
This study also revealed the functions of the two α-KG-dependent dioxygenases, PolK and PolD. PolK catalyzes the dual hydroxylation, while PolD catalyzes an oxidative C-C bond cleavage. Based on these observations, we propose a divergent mechanism for the biosynthesis of the structurally related antifungal nucleosides malayamycins and ezomycins. Ezomycin A1 (Extended Data Figure 10) contains a C8 bicyclic sugar, while malayamycins contain a C7 bicyclic sugar (Figure 6a); AHA in the PNs contains a C6 sugar. We propose that the size of the sugars of AHA and malayamycins is controlled by the PolD homologs. The malayamycin BGC carries both PolK and PolD homologs (MalM and MalI, respectively)22. It is likely that MalI catalyzes the C-C bond cleavage, but instead of cleaving between C6’-C7’ as observed for PolD, MalI cleaves the C7’-C8’ bond. As for the ezomycins, their nucleoside structure suggests that the pathway likely diverges earlier from either 2’-OAP or HKOAP.
Three mechanisms are conceivable for the PolD-catalyzed C-C bond cleavage reaction. The first mechanism involves H-5’ atom abstraction by Fe(IV)=O followed by C6’-C7’ bond cleavage and C7’ hydroxylation by Fe(III)-OH (Supplementary Figure 9a). Subsequent release of glyoxalate and further oxidation of C6’ into a carboxylate will yield AHAP. Alternatively, a processive oxidative decarboxylation could transform AHOAP into AHAP (Supplementary Figure 9b). Finally, although unprecedented, it is possible that PolD generates ketoacid intermediates, followed by their oxidative decarboxylation by a mechanism analogous to the oxidative cleavage of α-KG into succinate (Supplementary Figure 9c). Mechanistic studies are currently underway to distinguish these possibilities.
Our study also revises the previously proposed function of NikS40. Initial proposals suggested that NikS loads HPHT or its precursor onto NikT’s acyl carrier domain40. More recently, NikS was proposed to catalyze an ATP-dependent amide ligation between AHA and HPHT41. However, no in vitro functional characterization was reported. Our results demonstrate that NikS does not catalyze ligation between HPHT and AHA, and instead ligates HPHT and AHAP to form nikkomycin Z 2’-phosphate. Therefore, AHAP is likely the physiological substrate of NikS, and AHA is an off-pathway shunt metabolite. Intriguingly, in a recent report for PolG42, the NikS homolog in the polyoxin pathway, recombinant PolG ligates AHA and CPOAA (Figure 1a). Together with our studies, this earlier report suggests that PolG may promiscuously accept both AHA and AHAP, while only AHAP is physiologically relevant. Alternatively, AHAP may be dephosphorylated to AHA prior to its ligation with CPOAA in the polyoxin pathway. Nevertheless, our study resolves a long-standing mystery regarding NikS’s catalytic function, and identifies a potential difference in substrate specificity between the amide ligases of the nikkomycin and polyoxin pathways.
In conclusion, the identification of unexpected cryptic 2’-phosphorylation catalyzed by a P-loop NTPase kinase, PolQ2/NikN, resolved long-standing mysteries regarding AHA biosynthesis. The presence of related kinases in numerous nucleoside biosynthetic pathways suggests the relevance of cryptic phosphorylation beyond the nikkomycin/polyoxin pathways and the importance of functionally characterizing these kinases for our understanding of nucleoside natural product biosynthesis in general.
METHODS
General chemicals, reagents and analytical methods
All reagents were purchased from commercial sources without further purification. NMR spectra were collected on a 500 and 800 MHz Varian NMRs and on 500 and 700 MHz Bruker NMRs housed within the Duke University Nuclear Magnetic Resonance Spectroscopy facility. NMR spectra were processed and analyzed with SpinWorks 4.2.8.0. HPLC analyses was conducted on a Hitachi D-2000 Elite system consisting of a L-2130 pump, L-2300 column oven, L-2200 autosampler, L-2455 diode array equipped with a ODS Hypersil column (Thermo Fischer Scientific) or an Xbridge Amide column (3.5 μm, 4.6 x 250 mm, Waters). High performance anion exchange chromatography (HPAEC) analyses was performed on a Thermo Scientific ICS-5000+ system consisting of a Dionex ICS-5000+ SP pump, Dionex ICS 5000+ DC column oven, Dionex AS-AP autosampler, and Dionex PDA diode array with a DNAPac PA-100 4x250 mm column (Thermo Scientific). Liquid chromatography high resolution mass spectrometry (LC-HRMS) data was collected with an Agilent Technologies 1200 Series LC and an Agilent Technologies 6224 ESI-TOF with dual ESI source and an Agilent Technologies HILIC Plus column or on an RPLC-ESI/MS/MS was performed using a Shimadzu LC system (comprising a solvent degasser, two LC-10A pumps and a SCL-10A system controller using a Zorbax SB-C8 reversed-phase column (5 μm, 2.1 x 50 mm) from Agilent (Palo Alto, CA)) coupled to a high-resolution TripleTOF5600 mass spectrometer (Sciex, Framingham, MA). For LC-ESI/MS/MS, data acquisition and analysis were performed using the Analyst TF1.5 software (Sciex, Framingham, MA). Size exclusion column chromatography analyses were conducted on an AKTA purifier (GE Healthcare Life Sciences) with a Superdex 200 10/300 GL column (GE Healthcare Life Sciences, part number 17-5175-01). IR spectra were acquired in transmission mode (4 scans and 4 cm−1 resolution) using CaF2 disks on a PerkinElmer Frontier FTIR equipped with an MCT detector. The spectrum of the compound was baselined in the Spectrum 10.5.2.636 (PerkinElmer) using the multi-point baselining function, and plotted using Origin 2017 (64-bit) SR2 b9.4.2.380 (OriginLab). pET28-NikS and pLacI-GroELES were provided as gifts by the labs of Dr. Steven Van Lanen (University of Kentucky), and Dr. Dewey McCafferty (Duke University), respectively.
General microbiology and cloning methods
The strains and plasmids used in this study are listed in Supplementary Tables 2 and 3, respectively. E. coli and Streptomyces strains were grown and manipulated following standard methods 43,44. Spore stocks were generated from S. cacaoi var. asosensis and S. tendae Tü901 grown on MS agar 44. Selection of Streptomyces mutants was performed using MS and TSB (Tryptic Soy Agar, CRITERION™ Dehydrated Culture Media). The spores of Streptomyces strains were prepared on MS agar, collected, and stored at −80 °C in 20% glycerol for further analysis. Antibiotics were added to the media at the following final concentrations when appropriate: 25 μg/ml apramycin, 50 μg/ml hygromycin, 30 μg/ml thiostrepton , 25 μg/ml kanamycin, 25 μg/ml chloramphenicol and 25 μg/ml nalidixic acid. DNA extraction from E. coli was carried out using a QIAprep Spin Miniprep Kit (Qiagen). Restriction enzymes (NEB) were used according to the instructions provided by the manufacturers. PCR amplifications of DNA fragments used in the expression constructs and verification of the constructs were performed using Q5 High-Fidelity DNA polymerase (NEB). E. coli and Streptomyces colony PCR was performed as previously described45.
General methods of protein expression and purification.
In general, polyoxin homologs (PolQ2 kinase, PolJ phosphatase, PolD oxygenase, and PolK oxygenase) were characterized as they were more soluble and stable than nikkomycin homologs. The exceptions were NikK aminotransferase and NikS amide ligase. NikK was chosen as its aminotransferase activity towards proteinogenic amino acids was previously reported18. NikS was chosen as its carboxylate substrate (HPHT) was readily available to us by hydrolysis of nikkomycin Z. To obtain soluble protein expressions, we screened for expression conditions and tags. As a consequence, PolQ2, NikK, and NikM were expressed as N-terminal fusion with His-tagged MBP. PolD, PolK, and NikS were expressed as N-terminal His-tagged proteins. PolJ was expressed as both an N-terminal His-tagged MBP fusion protein as well as an N-terminal His-tagged protein. His-tagged PolK was co-expressed with the GroES GroEL molecular chaperone system to facilitate its folding, while all the other proteins were expressed without the chaperone system. The resulting His-tagged or MBP fusion proteins were used in the assays without removal of the His-tag or MBP fusion. To be clear about the procedure, we distinguish the MBP fusion and His-tagged proteins in the Methods by adding the prefixes “MBP-” or “His-”, respectively, to each enzyme name. However, we do not distinguish between the MBP fusion and His-tagged proteins in the main text to avoid confusion. Measurable effects on our functional characterization were not detected with either tag.
Cloning of His-PolK, MBP-NikM, His-PolD, His-PolJ, MBP-PolJ, MBP-PolQ2, and MBP-NikK
The clonings of polK and polD were previously reported46. For the His-tagged PolJ expression, the polJ gene was PCR amplified under standard conditions from the gDNA of S. cacaoi using polJ-F/R primers in Supplementary Table 4 and was cloned into pCR2.1-TOPO (Invitrogen) by following the manufacturer’s protocol. The plasmid was digested with HindIII and NdeI restriction enzymes and the polJ gene was subcloned into pET28b to yield pET-HisPolJ. For the MBP fusion constructs, polQ2, polJ, nikM and nikK were PCR amplified under standard conditions from gDNA of S. cacaoi or S. tendae using LIC-MBP-polQ2-F/R, LIC-MBP-nikM-F/R, or LIC-MBP-nik-F/R primers (Supplementary Table 4), respetively. The PCR products were gel purified, treated with T4 DNA Polymerase (New England Biolabs) in the presence of dGTP and were cloned into pMCSG9 following published procedures47, yielding pMCSG9-MBPPolQ2, pMCSG9-PolJ, pMCSG9-MBPNikM, and pMCSG9-MBPNikK.
Expression of His-PolD, His-PolK, MBP-PolQ2, His-PolJ, MBP-PolJ, MBP-NikM, MBP-NikK, and His-NikS
His-PolD was expressed as previously reported13. His-PolK was expressed in E. coli BL21(DE3) harboring a pLacI-GroELES chaperone plasmid and pET28-PolK. A single colony was grown in LB medium (8 mL) with 50 mg/L kanamycin and 25 mg/L chloramphenicol, and incubated at 37 °C, 200 rpm overnight until saturation. The entire overnight culture then was used to inoculate 1.5 L of LB medium with the same antibiotics, which was grown at 37 °C, 200 rpm until OD600 = 0.6 – 0.8. Protein expression was induced with 0.25 mM IPTG and 8.6 mg/L FeCl3, and the culture was incubated at 15 °C, 180 rpm for 20 h. The cells were harvested by centrifugation, washed with Buffer A (50 mM Tris pH 7.6, 150 mM NaCl, 10% glycerol), frozen with liquid nitrogen, and stored at −20 °C. Typically, 1.0-3.0 g of wet cell paste was obtained per liter. Identical conditions were used for the expression of His-PolJ, MBP-PolJ, MBP-NikM, MBP-PolQ2, MBP-NikK, and His-NikS in E. coli BL21(DE3) cells (without the pLacI-GroELES plasmid or addition of FeCl3) harboring either pET28b-HisPolJ, pMCSG9-MBPPolJ, pMCSG9-MBPNikM, pMCSG9-MBPPolQ2, pMCSG9-MBPNikK, or pET28-NikS in the presence of appropriate antibiotics. Approximately 2.5 – 3.0 g of wet cell paste was obtained per liter of culture.
Purification of His-PolD, His-PolK, MBP-PolQ2, His-PolJ, MBP-PolJ, MBP-NikM, His-NikS, and MBP-NikK
His-PolD, His-PolK, and MBP-NikM were purified, and reconstituted as previously reported (Supplementary Figures 10 – 12)13. For MBP-PolQ2 purification, the cell pellet of E. coli BL21(DE3)/pMCSG9-MBPPolQ2 was suspended in Buffer B (50 mM Tris pH 7.6, 500 mM NaCl, 10% glycerol), homogenized by a dounce homogenizer, and lysed by two passages through French pressure cell operated at 14,000 psi. The lysate was clarified by centrifugation (21,000 x g, 20 min, 4 °C), and the supernatant was incubated with Ni-NTA agarose resin (Genesee Scientific, 20 mL equilibrated in Buffer A supplemented with 40 mM imidazole) for 1 h at 4 °C. The resin was subsequently packed into a column, and the column was washed with 10 column volumes (CV) of Buffer B supplemented with 40 mM imidazole. The bound proteins were eluted with Buffer B supplemented with 200 mM imidazole. Fractions containing MBP-PolQ2 were confirmed by 12.5% SDS-PAGE, combined, and exchanged into Buffer A with Sephadex G25 resin (GE Healthcare Life Sciences). MBP-PolQ2 was kept at 4 °C for short-term use (<1 week) or flash frozen in liquid nitrogen and stored at −80 °C for long-term storage. His-PolJ, MBP-PolJ, MBP-NikK, and His-NikS were purified identically except for following differences: For His-PolJ and MBP-PolJ, the cell pellet of E. coli BL21(DE3) with pET-HisPolJ or pMCSG9-MBPPolJ was suspended in Buffer C (50 mM Tris pH 7.6, 300 mM NaCl, 10% glycerol), and Ni-NTA agarose resin was equilibrated and washed with Buffer C supplemented with 40 mM imidazole. His-PolJ precipitated at 4 °C in ~6 h, and thus was frozen in liquid nitrogen immediately after purification and stored at −80 °C. For MBP-NikK, the cell pellet of E. coli BL21(DE3)/pMCSG9-MBPNikK was suspended in Buffer C supplemented with 2 mg/mL PLP, and Ni-NTA agarose resin was equilibrated and washed with Buffer C supplemented with 10 mM imidazole. Binding of PLP to MBP-NikK was confirmed by the yellow color of the purified protein and by UV absorbance features of PLP and PMP at 425 nm and 330 nm, respectively. Typically, ~0.9 eq. of PLP was copurified (Supplementary Figure 13). For His-NikS purification, the cell pellet was resuspended in Buffer D (25 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol), and Ni-NTA agarose resin was equilibrated and washed with Buffer D supplement with 30 mM imidazole. His-NikS was eluted with Buffer D supplemented with 200 mM imidazole. The typical yields of purification were 3-5 mg/g cell paste (MBP-PolQ2), 1.5-4 mg/g cell paste (His-PolJ), 10-12 mg/g cell paste (MBP-NikK), and 10 mg/g cell paste (His-NikS). In all cases, the His-tagged and MBP fusion proteins were used in the assays without removal of the His-tag or MBP.
PolQ2 Activity Assay
MBP-PolQ2 (40 μM) was incubated with 500 μM of EP-UMP or 5’-OAP in Buffer A supplemented with 4 mM MgCl2 and 2 mM ATP at 25 °C. After 15, 30, 60, 120, 240 min, aliquots (10 μL) were diluted 4x with 100 mM NH4OAc pH 4.7, boil quenched for 2 min at 95 °C, and were clarified with centrifugation. Then 12 μL of the supernatant was injected into and analyzed by HPAEC using a DNAPac PA100 4 x 250 mm column (Thermo Scientific) equilibrated in 11% 1 M NH4OAc pH 4.7 (solvent B) in water (solvent A). Elution of the product was performed at a flow rate of 1.5 mL/min with 11% solvent B for 3.5 min followed by 18% solvent B for 4.5 min. Chromatography was monitored by UV absorbance at 260 nm.
PolJ Activity Assays
His-PolJ or MBP-PolJ (10 μM) was incubated with 100 μM of 5’-OAP or OABP in Buffer A supplemented with 20 mM MgCl2 at 25 °C. After 15, 50, 120, and 240 min, aliquots (10 μL) were diluted 4x in water, boil quenched at 95 °C for 2 min, and were clarified by centrifugation. Then, 12 μL of the supernatant was injected into and analyzed by HPAEC using a DNAPac PA100 4x250 mm column (Thermo Scientific) under identical conditions to those described for PolQ2. Chromatography was monitored by UV absorbance at 260 nm.
PolD, PolK, and NikM Activity Assay
His-PolD (10 μM), His-PolK (40 μM), or MBP-NikM (40 μM) was incubated with 100 μM of substrate (2’-OAP, OABP, AHOAP, or KOAP), 100 μM (NH4)2Fe(SO4)2·6H2O, 1 mM ascorbate, and 2 mM α-KG in oxygen saturated Buffer A at 25 °C. After 15, 60, 120, and 1080 min, aliquots (10 μL) were collected, diluted 5x in dH2O, boil quenched for 2 min at 95 °C, and were clarified by centrifugation. Then, 12 μL of the supernatant was injected into and analyzed by HPAEC (for AHOAP, 2’OAP and OABP) using a DNAPac PA100 4 x 250 mm column (Thermo Scientific). KOAP assays were analyzed by HPLC using an Xbridge Amide (Waters) column. Chromatography was monitored by UV absorbance at 260 nm.
NikK Activity Assay
MBP-NikK (10 μM) was incubated with HKOAP (100 μM) or KOAP (500 μM) in the presence of l-Glu (3 mM) or l-Phe (2 mM) at 25 °C. After 15, 60, 120, and 1080 min, aliquots (10 μL) were collected, diluted 5x in dH2O, boil quenched for 2 min at 95 °C, and were clarified by centrifugation. Then, 12 μL of the supernatant was injected into and analyzed by HPAEC using a DNAPac PA100 4 x 250 mm column (Thermo Scientific). Completion of the reaction with HKOAP was observed after 15 minutes or less, while no detectable product was observed for MBP-NikK with KOAP after 1080 minutes.
NikS Activity Assay
His-NikS (100 μM) was incubated with HPHT (150 μM) and either AHAP (150 μM) or AHA (150 μM) in 100 mM Tris-HCl pH 8.0 buffer supplemented with 3 mM MgCl2, and 2 mM ATP. After 15, 60, 120 min, aliquots (30 μL) were withdrawn and boil quenched at 95 °C for 2 min. To the quenched reactions, CutSmart 10x buffer (3 μL; NEB) and 5 units of alkaline calf intestine phosphatase (CIP; CalBioTech) were added. The reactions were incubated at 37 °C for 30 min, boil quenched at 95 °C for 2 min, and allowed to cool to room temperature. Samples were diluted in 80 μL of MeCN, clarified by centrifugation, and analyzed by HPLC equipped with an Xridge Amide (Waters) column under isocratic conditions: 25% 100 mM ammonium acetate pH 4.8/ 75% MeCN. UV absorbance was monitored at 280 nm. Identical samples were submitted for LC-HRMS analysis with or without CIP treatment, and the MS data were analyzed by MassHunter B.07.00 (Agilent Technologies).
Construction of polQ2 in-frame deletion mutants
PCR-based homologous recombination and temperature sensitive selection were used to create Streptomyces deletion mutants (Supplementary Figure 14). Gene deletion was accomplished using pKC113948, a temperature sensitive Streptomyces-E.coli shuttle vector that was modified from a previously reported protocol49. For polQ2 deletion, markerless in-frame deletion was performed. The 5’ flanking region of polQ2 (5’-polQ2) was amplified with primers KA16F and KA16R (Supplementary Table 4) to generate a 1,294 bp DNA fragment with terminal EcoRI and NdeI sites. The 3’ flanking region of polQ2 (3’-polQ2) was amplified with primers KA17F1 and KA17R1 to generate a 1,431 bp DNA fragment with terminal NdeI and HindIII sites. Then, the PCR products of 5’-polQ2 and 3’-polQ2 were individually cloned into pJET 1.2 blunt cloning vector (Supplementary Table 4) following the manufacturer’s instructions, to generate pJET/5’-polQ2 and pJET/3’-polQ2, respectively. The recombinant plasmids were amplified in E. coli DH10β at 30 °C, and extracted using QIAprep Spin Miniprep kit (QIAGEN, USA). The DNA sequences were verified using pJET universal primers. The flanking fragments were digested from pJET/5’-polQ2 and pJET/3’-polQ2, at their restriction sites described above, and subcloned into pKC1139 digested with EcoRI and HindIII to create pKC1139/ΔpolQ2. The resulting plasmid was introduced into E. coli ET12567/pUZ800250,51 by a CaCl2 transformation and was grown at 30 °C for 2 days. The conjugation between S. cacaoi wt with E. coli ET12567/pUZ8002 carrying pKC1139/ΔpolQ2 was carried out as described in the general Streptomyces genetics protocol44. 0.5 mL of overnight culture of E. coli ET12567/pUZ8002/pKC1139/ΔpolQ2 was transferred to 50 ml of fresh LB containing 25 μg/ml apramycin, 25 μg/ml kanamycin and 25 μg/ml chloramphenicol, incubated with shaking at 30°C, 220 rpm until the OD600nm reach 0.4. The cells were harvested by centrifugation (4,000 rpm for 5 min at 4 °C), washed twice with LB at 4 °C, and resuspended with 5 ml of LB. Meanwhile, 10 μl of S. cacaoi spore suspension (108-109 CFU) was added to a mixture of 250 μL of 2xYT and 250 μL TSB:YEME (a mixed media containing all the ingredients for TSB and YEME media with additions of 5 mM MgCl2 and 0.5% w/v glycine) at 45°C for 30 min, then mixed with 500 μl of E. coli ET12567/pUZ8002/pKC1139/polQ2 once the spore suspension has cooled down and let it stand at room temperature for 10 min. The mixture was spun down at 8,000 rpm for 3 min. The resulting pellet was resuspended with 200-250 μl of the supernatant and plated on MS agar and incubated at 28 °C for 20 hr. The conjugation plates were then overlaid with 25 μg/ml apramycin and 25 μg/ml nalidixic acid and were incubated at 28 °C for 3-5 days until the ex-conjugants were observed. Then, the plates were transferred to 37 °C for 2 days, and the ex-conjugants were streaked on fresh MS agar containing 25 μg/ml apramycin and 25 μg/ml nalidixic acid and incubated at 37 °C for 5 days to promote the loss of the temperature sensitive plasmid. The resulting single crossover ex-conjugants were re-streaked for 2 rounds on MS agar containing 25 μg/ml nalidixic acid, incubated at 37 °C for 5 days to initiate the double crossover during the elimination of E. coli. Then single colonies were grown in TSB overnight and spread on the MS agar for 7 days until they fully sporulated. The spores were then collected, diluted, and plated on the MS agar to select for the double crossover ex-conjugants, which were tested for apramycin sensitivity by patching the colonies, in order, on TSB agar with and without apramycin, respectively, and incubated at 37 °C for 5 days. Mycelia of apramycin sensitive ex-conjugants were scraped and used for Streptomyces colony PCR to screen for the deletion genotype using primers KA23F and KA23R (Supplementary Figure 15a). The amplified PCR fragments correspond to the size of polQ2 deletion genotype were subjected for sequencing using primers KA18F, KA18R, KA23F and KA23R (Supplementary Table 4).
Genetic complementation of S. cacaoi ΔpolQ2 with wt polQ2 gene
The polQ2 gene was subcloned from pET28b/polQ2 into the NdeI/HindIII site of pUWL201PWT (Streptomyces multi-copy expression vector, pUWL201PW52, with oriT inserted into the PstI site) creating pUWL201PWT/polQ2. The NdeI/HindIII site is immediately downstream of the constitutive ermE* promoter, allowing constitutive expression of PolQ2. The pUWL201PWT/polQ2 plasmid was then introduced into conjugative E. coli ET12567/pUZ8002. The conjugal transfer of pUWL201PWT/polQ2 from E. coli ET12567/pUZ8002 into S. cacaoi ΔpolQ2 was carried out as described above. S. cacaoi ex-conjugants were selected on MS agar overlaid with 30 μg/ml thiostrepton and 25 μg/ml nalidixic acid, grown at 28°C for 5-7 days. The resulting recombinant strain, S. cacaoi ΔpolQ2+pUWL201PWT/polQ2 was streaked on the same media containing 30 μg/ml thiostrepton and 25 μg/ml nalidixic acid until the strains were free from E. coli cells. The recombinant strains were confirmed by colony PCR using primers LIC-MBP-polQ2-F and LIC-MBP-polQ2-R (Supplementary Table 4).
Construction of nikK and nikL disruption mutants
ΔnikK and ΔnikL mutants were created by replacing the target gene with a kanamycin resistant gene (Supplementary Figure 16). The replacement fragments contained the target’s 5’ flanking region, a kanamycin resistant gene, and the target’s 3’ flanking region, respectively, and were amplified using overlapping PCR. Primers and fragments amplified for ΔnikK and ΔnikL knockout mutants are shown in Supplementary Table 4 and Supplementary Figure 16. Cloning and conjugation were performed as described above. Selection of double crossover ex-conjugants was performed by screening for apramycin sensitivity and kanamycin resistance. Verification of the resulting mutants was performed by Streptomyces colony PCR using primer pairs HSU-Nik11 and HSU-Nik15 for S. tendae ΔnikK::kanR, and HSU-Nik16 and HSU-Nik20 for S. tendae ΔnikL::kanR (Supplementary Table 4, Supplementary Figure 15b).
Genetic complementation of S. tendae ΔnikK::kanR with wt nikK gene
The nikK gene was amplified from pMCSG9-MBPNikK by PCR using primers KA09F and KA09R (Supplementary Table 4) and cloned into pJET1.2 blunt cloning vector (Thermo Fisher Scientific) following the manufacturer’s instructions. After sequence confirmation using pJET universal primers, the pJET1.2-nikK was digested with NdeI and HindIII and the nikK fragment was subcloned into the NdeI and HindIII site of pIJ10257 53 to yield pIJ10257/nikK. The NdeI and HindIII site is downstream of the constitutive ermE* promoter, allowing constitutive expression of nikK. Intergeneric conjugation of E. coli ET12567/pUZ8002 carrying the pIJ10257/nikK with S. tendae ΔnikK::kanR was carried out as described above using the spore suspension that was heat shocked at 37°C for 30 min instead . S. tendae ex-conjugants were selected on MS agar overlaid with 50 μg/ml hygromycin and 25 μg/ml nalidixic acid, grown at 28°C for 5-7 days. The resulting recombinant strain, S. tendae ΔnikK::pIJ10257/nikK was streaked on the same media containing 50 μg/ml hygromycin and 25 μg/ml nalidixic acid until the strains were free from E. coli cells. Streptomyces colony PCR was used to verify the recombinant strains using primers KA09F and KA09R (Supplementary Table 4).
HPAEC and LC-HRMS analysis of S. tendae and S. cacaoi culture supernatants
S. cacaoi and S. tendae strains were grown in fermentation medium P (40 g/L mannitol, 10 ml/L glycerol, 10 g/L soluble starch, 20 g/L soy peptone, 10 g/L yeast extract, 5 g/L ammonium sulfate, 3.75 mg/l FeSo4•7H2O, 6.9 g/L NaH2PO4•7H2O, 8.7 g/L K2HPO4) or medium N (43 g/L mannitol, 12 g/L soluble starch, 20 g/L soy peptone, 10 g/L yeast extract, 3.75 mg/l FeSO4•7H2O), respectively54. A 1:1 mixture of TSB and the fermentation medium P or N (total 10 mL) in 50 ml tube with 1-2 cm of spring coil was inoculated with 10 μl (108-109 CFU) of spores of S. cacaoi or S. tendae, respectively, and incubated at 28°C with shaking at 225 rpm for 2-3 days. The resulting Streptomyces liquid cultures (0.5-2 mL) were then diluted in 50 mL of the fermentation medium P or N containing 100 mM PIPES buffer pH 6.0 in 250 ml baffled flasks containing stainless steel springs with the starting OD600nm = 0.1. The cultures were grown at 28°C with shaking at 225 rpm. During the fermentation, the pH was monitored. The pH was at ~6.5 between days 1-5, and increased to ~7.0 on day 6-7. After 5-7 days of culture, the culture media were harvested, cleared by centrifugation at 14,000 rpm, 4 °C for 20 min to remove mycelia, and analyzed by LC-HRMS. Nikkomycins were chromatographed on an Agilent HILIC plus column at 35 °C with solvents A (10 mM NH4OAc pH 6.8) and B (acetonitrile): 5%-55% A 10 min, 55% A 4 min and 5% A 4 min with a flow rate of 0.22 mL/min. Polyoxins and AHA biosynthetic intermediates were chromatographed on a Waters Xbridge Amide column at 40 °C using solvents A (10mM NH4OAc pH 10.0) and B (acetonitrile): Linear gradient 15-50% A for 20 min at a flow rate of 0.5 mL/min. The elution was monitored by UV absorption at 254 nm as well as ESI-TOF MS. The MS data were analyzed by MassHunter (Agilent Technologies).
For the HPAEC Analysis of 5’-OAP, 2’-OAP, OABP and KOAP in the culture media, 20 ml of 7 day culture supernatant from S. cacaoi ΔpolQ2, S. tendae ΔnikK::kanR and S. tendae ΔnikL::kanR were loaded onto a QAE Sephadex A25 column (10 mL; bicarbonate form). Compounds were eluted with a linear gradient from 20-400 mM ammonium bicarbonate pH 7.6 over 10 CV. Fractions eluted between 250 – 400 mM ammonium bicarbonate contained all the nucleotide intermediates, which were combined, lyophilized, resuspended in water (200 uL), and analyzed by HPAEC monitored at 260 nm.
Data Availability Statement
The authors declare that all the data supporting the findings of this study are available within the paper, Extended Data, and Supplementary Information. All plasmids, analytical amounts of reported compounds, and raw data are available upon request. Sequences are deposited at National Center for Biotechnology Information under accession nos.: PolQ2, ABX24486; PolJ, ABX24494; PolK, ABX24493; PolD, ABX24500; NikL, CAC80910; NikM, CAC80911; NikK, CAC80909; NikS, CAC11141.
Extended Data
Extended Data Fig. 1. Activity of PolD and PolK with 5’-OAP.
Shown are HPLC chromatograms at 260 nm for a full reaction with PolK, a full reaction with PolD, a control without enzyme, and a control without α-KG. No consumption of 5’-OAP was detected. The results were reproducible in at least two independent assays.
Extended Data Fig. 2. Activity of PolQ2 with EP-UMP.
Shown are HPAEC chromatograms at 260 nm for a control without the enzyme (trace i) and the complete reaction (trace ii). No consumption of EP-UMP is detected. The results were reproducible in at least two independent assays.
Extended Data Fig. 3. Activity of PolD, PolK, and NikM with OABP.
No consumption of OABP was observed after 24 hours. ATPase activity was observed in all the reactions except for no enzyme control, suggesting that it is likely due to a trace contamination of an endogeneous E. coli phosphatase. The results were reproducible in at least two independent assays.
Extended Data Fig. 4. Activity assays of PolJ, NikK, and PolK with 2’-HAP.
a. PolJ phosphatase (10 μM) was incubated with 2’-HAP (100 μM) in 50 mM Tris pH 8.0 supplemented with 10 mM MgCl2 at 25 °C for 18 hrs. b. NikK aminotransferase (30 μM) was incubated with 2’-HAP (200 μM) in 200 mM Tris pH 9.0 supplemented with 1 mM MgCl2 and 10 mM l-Glu at 25 °C for 2 hrs. c. PolK oxygenase (25 μM) was incubated with 2’-HAP (200 μM) in 150 mM NaCl, 50 mM Tris pH 7.5 supplemented with 1 mM Fe2+, 1 mM ascorbate, 200 μM α-KG at 25 °C for 18 hrs. No product formation was detected within typical reaction timescales for the enzymes with their on-pathway substrates. The results were reproducible in at least two independent assays.
Extended Data Fig. 5. Activity assays of PolK with KOAP.
PolK (40 μM) was incubated with KOAP (100 μM) in the presence of 100 μM Fe2+, 2 mM ascorbate, and 1 mM α-KG for 20 hours at 25 °C. Reaction traces for PolK with KOAP under typical conditions. Even after prolonged incubation (20 hours), no detectable product is observed. The results were reproducible in at least two independent assays.
Extended Data Fig. 6. Activity of PolD with KOAP.
PolD (40 μM) was incubated with KOAP (100 μM) in the presence of 100 μM Fe(II), 1 mM ascorbate, and 2 mM α-KG for 2 hours at 25 °C. No product formation or KOAP consumption. For comparison, under similar conditions, PolD completed the conversion of AHOAP to AHAP in <15 minutes. The results were reproducible in at least two independent assays.
Extended Data Fig. 7. Stereochemistry of AHOAP.
Coupling constants for possible stereochemistry of the 5’-amino, 6’-hydroxyl, 7’ carboxylic acid groups. Experimental evidence of JH4’-H5’ = 3.5 Hz, JH5’-H6’ = 10.5 Hz and JH6’-H7’ = 5.6 Hz for AHOA is most consistent with the stereochemistry of 4’-S, 5’-S, 6’-S, 7’-R indicating that AHOA and AHOAP have 4’-S, 5’-S, 6’-S, 7’-R stereochemistry. Dihedral angles were calculated with ChemDraw Professional v19.0 and ChemDraw 3D v19.0 (PerkinElmer Informatics).
Extended Data Fig. 8. Comparison of the rates of reactions between PolD + AHOAP vs. PolD + 2’-OAP.
The PolD assays with AHOAP were performed with 10 μM PolD, 0.1mM Fe2+, 2 mM ascorbate, 100 μM AHOAP in 150 mM NaCl, 50 mM Tris pH 7.6, 10% glycerol. The PolD assays with 2’-OAP were performed with 30μM PolD, 1mM ascorbate, 0.5 mM Fe2+, 200 μM 2’-OAP in 150 mM NaCl, 50 mM Tris pH 7.6.
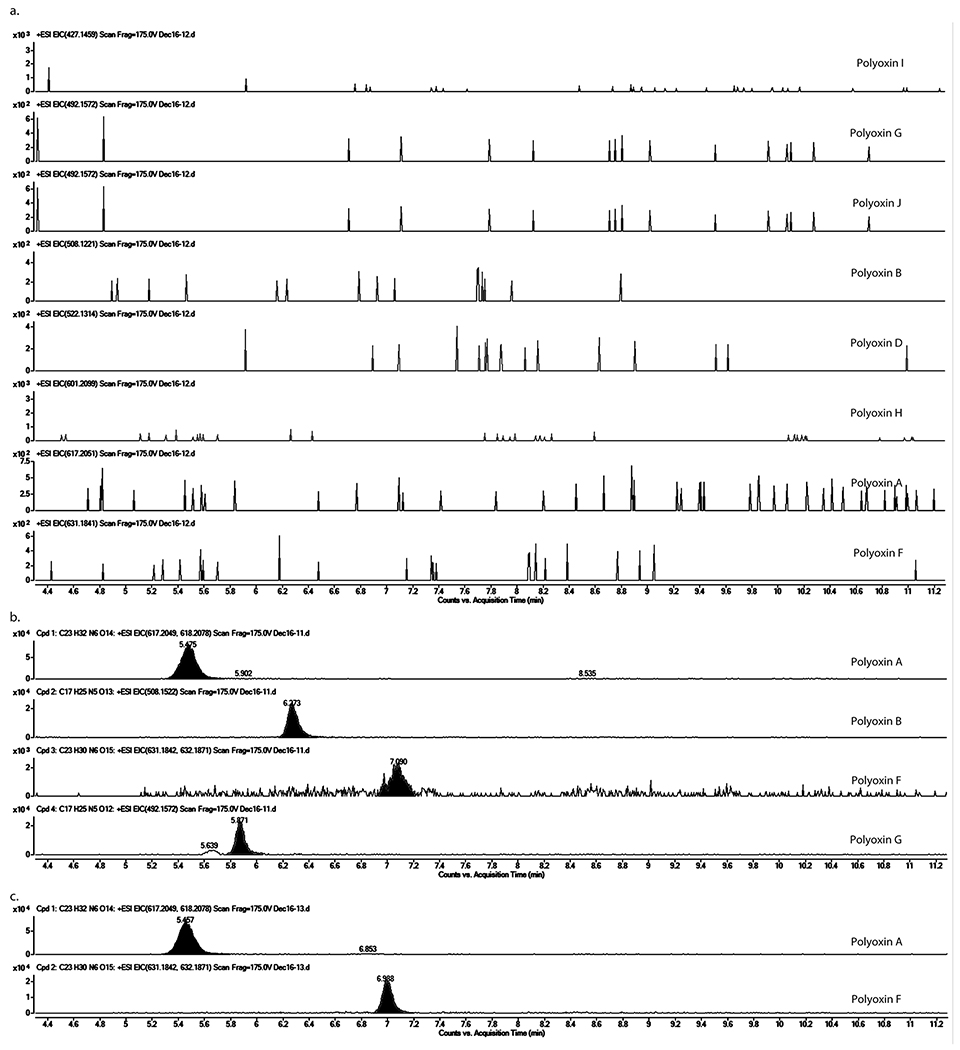
Extended Data Fig. 9. LC-HRMS analysis of culture media of S. cacaoi ΔpolQ2, wt, and ΔpolQ2 + polQ2.
Shown are EICs (calculated m/z for [M+H]+ ± 5 ppm) for polyoxins A, B, D, F, G, H, I, and J. No polyoxin production was detected in ΔpolQ2 (a), polyoxin A, B, F, and G were found in the wt strain (b), and polyoxin A and F were detected in the ΔpolQ2 + polQ2 strain (c). The observations were reproducible for 2-3 different clones for each mutant strain. The culture was repeated twice for each clone.
Extended Data Fig. 10. Proposed divergent biosynthesis of antifungal nucleoside natural products.
Proposed tailoring of sugar size by oxidative C-C bond cleavage by PolD homologs.
Supplementary Material
Acknowledgement
The authors thank the Duke University Shared Instruments Core Facility (Dr. Peter Silinski) and the Duke University Lipidomics Core Facility (Dr. Ziqiang Guan) for assistance with the LC-HRMS and LC HRMS/MS analyses, respectively. We thank the Duke University NMR Spectroscopy Core Facility (Dr. Benjamin Bobay, Dr. Ronald Venters, and Don Mika) for their assistance with the NMR analyses. We thank Dr. Allison Stelling for assistance with the FTIR analyses. This work was supported by the Duke University School of Medicine, and National Institute of General Medical Sciences R01 GM115729 (to K.Y.). M.M.D. was supported in part by the Duke Medical Scientist Training Program (T32 GM007171).
Footnotes
The authors declare no competing financial interest.
REFERENCE
- 1.Winn M, Goss RJ, Kimura K & Bugg TD Antimicrobial nucleoside antibiotics targeting cell wall assembly: recent advances in structure-function studies and nucleoside biosynthesis. Nat Prod Rep 27, 279–304 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Isono K Nucleoside Antibiotics - Structure, Biological-Activity, and Biosynthesis. J Antibiot 41, 1711–1739 (1988). [DOI] [PubMed] [Google Scholar]
- 3.Niu G & Tan H Nucleoside antibiotics: biosynthesis, regulation, and biotechnology. Trends Microbiol 23, 110–119 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Chen WQ et al. Natural and engineered biosynthesis of nucleoside antibiotics in Actinomycetes. J Ind Microbiol Biot 43, 401–417 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Shiraishi T & Kuzuyama T Recent advances in the biosynthesis of nucleoside antibiotics. J Antibiot (Tokyo) 72, 913–923 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Bruntner C, Lauer B, Schwarz W, Mohrle V & Bormann C Molecular characterization of co-transcribed genes from Streptomyces tendae Tu901 involved in the biosynthesis of the peptidyl moiety of the peptidyl nucleoside antibiotic nikkomycin. Mol Gen Genet 262, 102–114 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Chen W et al. Characterization of the polyoxin biosynthetic gene cluster from Streptomyces cacaoi and engineered production of polyoxin H. J Biol Chem 284, 10627–10638 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nix DE, Swezey RR, Hector R & Galgiani JN Pharmacokinetics of nikkomycin Z after single rising oral doses. Antimicrob Agents Ch 53, 2517–2521 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osada H Discovery and applications of nucleoside antibiotics beyond polyoxin. J Antibiot (Tokyo) 72, 855–864 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Hector RF Compounds active against cell walls of medically important fungi. Clin Microbiol Rev 6, 1–21 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohrle V, Roos U & Bormann C Identification of cellular proteins involved in nikkomycin production in Streptomyces tendae Tu901. Mol Microbiol 15, 561–571 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Ginj C, Ruegger H, Amrhein N & Macheroux P 3’-Enolpyruvyl-UMP, a novel and unexpected metabolite in nikkomycin biosynthesis. Chembiochem 6, 1974–1976 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Lilla EA & Yokoyama K Carbon extension in peptidylnucleoside biosynthesis by radical SAM enzymes. Nat Chem Biol 12, 905–907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He N et al. Construction of an octosyl acid backbone catalyzed by a radical S-adenosylmethionine enzyme and a phosphatase in the biosynthesis of high-carbon sugar nucleoside antibiotics. Chem Sci 8, 444–451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lauer B, Sussmuth R, Kaiser D, Jung G & Bormann C A putative enolpyruvyl transferase gene involved in nikkomycin biosynthesis. J Antibiot 53, 385–392 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Chen W, Zeng HM & Tan HR Cloning, sequencing, and function of sanF: A gene involved in nikkomycin biosynthesis of Streptomyces ansochromogenes. Curr Microbiol 41, 312–316 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Xu J, Liu G & Tan H sanC- a novel gene involved in nikkomycin biosynthesis in Streptomyces ansochromogenes. Lett Appl Microbiol 36, 234–238 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Binter A et al. Characterization of the PLP-dependent aminotransferase NikK from Streptomyces tendae and its putative role in nikkomycin biosynthesis. Febs J 278, 4122–4135 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Pao SS, Paulsen IT & Saier MH Jr. Major facilitator superfamily. Microbiol Mol Biol Rev 62, 1–34 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isono K, Crain PF & Mccloskey JA Isolation and Structure of Octosyl Acids - Anhydrooctose Uronic Acid Nucleosides. J Am Chem Soc 97, 943–945 (1975). [Google Scholar]
- 21.Schuz TC, Fiedler HP, Zahner H, Rieck M & Konig WA Metabolic Products of Microorganisms .263. Nikkomycins Sz, Sx, Soz and Sox, New Intermediates Associated to the Nikkomycin Biosynthesis of Streptomyces-Tendae. J Antibiot 45, 199–206 (1992). [DOI] [PubMed] [Google Scholar]
- 22.Hong H, Samborskyy M, Zhou Y & Leadlay PF C-Nucleoside Formation in the Biosynthesis of the Antifungal Malayamycin A. Cell Chem Biol 26, 493–501 e495 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Sosio M et al. Analysis of the Pseudouridimycin Biosynthetic Pathway Provides Insights into the Formation of C-nucleoside Antibiotics. Cell Chem Biol 25, 540–549 e544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui Z et al. Self-Resistance during Muraymycin Biosynthesis: a Complementary Nucleotidyltransferase and Phosphotransferase with Identical Modification Sites and Distinct Temporal Order. Antimicrob Agents Chemother 62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaysser L et al. Identification and manipulation of the caprazamycin gene cluster lead to new simplified liponucleoside antibiotics and give insights into the biosynthetic pathway. J Biol Chem 284, 14987–14996 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Funabashi M et al. The biosynthesis of liposidomycin-like A-90289 antibiotics featuring a new type of sulfotransferase. Chembiochem 11, 184–190 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Chi X et al. The muraminomicin biosynthetic gene cluster and enzymatic formation of the 2-deoxyaminoribosyl appendage. Medchemcomm 4, 239–243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiraishi T, Hiro N, Igarashi M, Nishiyama M & Kuzuyama T Biosynthesis of the antituberculous agent caprazamycin: Identification of caprazol-3”-phosphate, an unprecedented caprazamycin-related metabolite. J Gen Appl Microbiol 62, 164–166 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Cui Z et al. Pyridoxal-5’-phosphate-dependent alkyl transfer in nucleoside antibiotic biosynthesis. Nat Chem Biol 16, 904–911 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y et al. Discovery and characterization of the tubercidin biosynthetic pathway from Streptomyces tubercidicus NBRC 13090. Microb Cell Fact 17, 131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z et al. Functional and kinetic analysis of the phosphotransferase CapP conferring selective self-resistance to capuramycin antibiotics. J Biol Chem 285, 12899–12905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaillancourt FH, Yeh E, Vosburg DA, O’Connor SE & Walsh CT Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosynthesis. Nature 436, 1191–1194 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Cunin R, Glansdorff N, Pierard A & Stalon V Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev 50, 314–352 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Llewellyn NM, Giri R, Huang F & Spencer JB Biosynthesis of the unique amino acid side chain of butirosin: possible protective-group chemistry in an acyl carrier protein-mediated pathway. Chem Biol 12, 665–675 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Llewellyn NM, Li Y & Spencer JB Biosynthesis of butirosin: transfer and deprotection of the unique amino acid side chain. Chem Biol 14, 379–386 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Qi J et al. Deciphering Carbamoylpolyoxamic Acid Biosynthesis Reveals Unusual Acetylation Cycle Associated with Tandem Reduction and Sequential Hydroxylation. Cell Chem Biol 23, 935–944 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Arnison PG et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30, 108–160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen HP & Yokoyama K Characterization of Acyl Carrier Protein-Dependent Glycosyltransferase in Mitomycin C Biosynthesis. Biochemistry 58, 2804–2808 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogasawara Y, Nakagawa Y, Maruyama C, Hamano Y & Dairi T In vitro characterization of MitE and MitB: Formation of N-acetylglucosaminyl-3-amino-5-hydroxybenzoyl-MmcB as a key intermediate in the biosynthesis of antitumor antibiotic mitomycins. Bioorg Med Chem Lett 29, 2076–2078 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Lauer B et al. Molecular characterization of co-transcribed genes from Streptomyces tendae Tu901 involved in the biosynthesis of the peptidyl moiety and assembly of the peptidyl nucleoside antibiotic nikkomycin. Mol Gen Genet 264, 662–673 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Li Y, Zeng H & Tan H Cloning, function, and expression of sanS: a gene essential for nikkomycin biosynthesis of Streptomyces ansochromogenes. Curr Microbiol 49, 128–132 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Gong R et al. An ATP-Dependent Ligase with Substrate Flexibility Involved in Assembly of the Peptidyl Nucleoside Antibiotic Polyoxin. Appl Environ Microbiol 84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS REFERENCES
- 43.Sambrook J & Russell DW Molecular Cloning: A laboratory manual. (Cold Spring Harbor Laboratory Press, 2001). [Google Scholar]
- 44.Kieser T, Bibb MJ, Buttner MJ, Chater KF & Hopwood DA Practical Streptomyces Genetics. (John Innes Foundation, 2000). [Google Scholar]
- 45.Thanapipatsiri A, Claesen J, Gomez-Escribano JP, Bibb M & Thamchaipenet A A Streptomyces coelicolor host for the heterologous expression of Type III polyketide synthase genes. Microb Cell Fact 14, 145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hover BM, Lilla EA & Yokoyama K Mechanistic Investigation of cPMP Synthase in Molybdenum Cofactor Biosynthesis Using an Uncleavable Substrate Analogue. Biochemistry 54, 7229–7236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eschenfeldt WH, Lucy S, Millard CS, Joachimiak A & Mark ID A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol Biol 498, 105–115 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bierman M et al. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116, 43–49 (1992). [DOI] [PubMed] [Google Scholar]
- 49.Mo S et al. Roles of fkbN in positive regulation and tcs7 in negative regulation of FK506 biosynthesis in Streptomyces sp. strain KCTC 11604BP. Applied and environmental microbiology 78, 2249–2255 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paget MS, Chamberlin L, Atrih A, Foster SJ & Buttner MJ Evidence that the extracytoplasmic function sigma factor sigmaE is required for normal cell wall structure in Streptomyces coelicolor A3(2). Journal of bacteriology 181, 204–211 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacNeil DJ et al. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111, 61–68 (1992). [DOI] [PubMed] [Google Scholar]
- 52.Doumith M et al. Analysis of genes involved in 6-deoxyhexose biosynthesis and transfer in Saccharopolyspora erythraea. Molecular & general genetics : MGG 264, 477–485 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Hong HJ, Hutchings MI, Hill LM & Buttner MJ The role of the novel Fem protein VanK in vancomycin resistance in Streptomyces coelicolor. The Journal of biological chemistry 280, 13055–13061 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Schuz TC, Fiedler HP, Zahner H, Rieck M & Konig WA Metabolic products of microorganisms. 263. Nikkomycins SZ, SX, SoZ and SoX, new intermediates associated to the nikkomycin biosynthesis of Streptomyces tendae. The Journal of antibiotics 45, 199–206 (1992). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all the data supporting the findings of this study are available within the paper, Extended Data, and Supplementary Information. All plasmids, analytical amounts of reported compounds, and raw data are available upon request. Sequences are deposited at National Center for Biotechnology Information under accession nos.: PolQ2, ABX24486; PolJ, ABX24494; PolK, ABX24493; PolD, ABX24500; NikL, CAC80910; NikM, CAC80911; NikK, CAC80909; NikS, CAC11141.