Extended Data Fig.2|. HIST1H1 mutations are genetic drivers in lymphoma and confer loss of function.
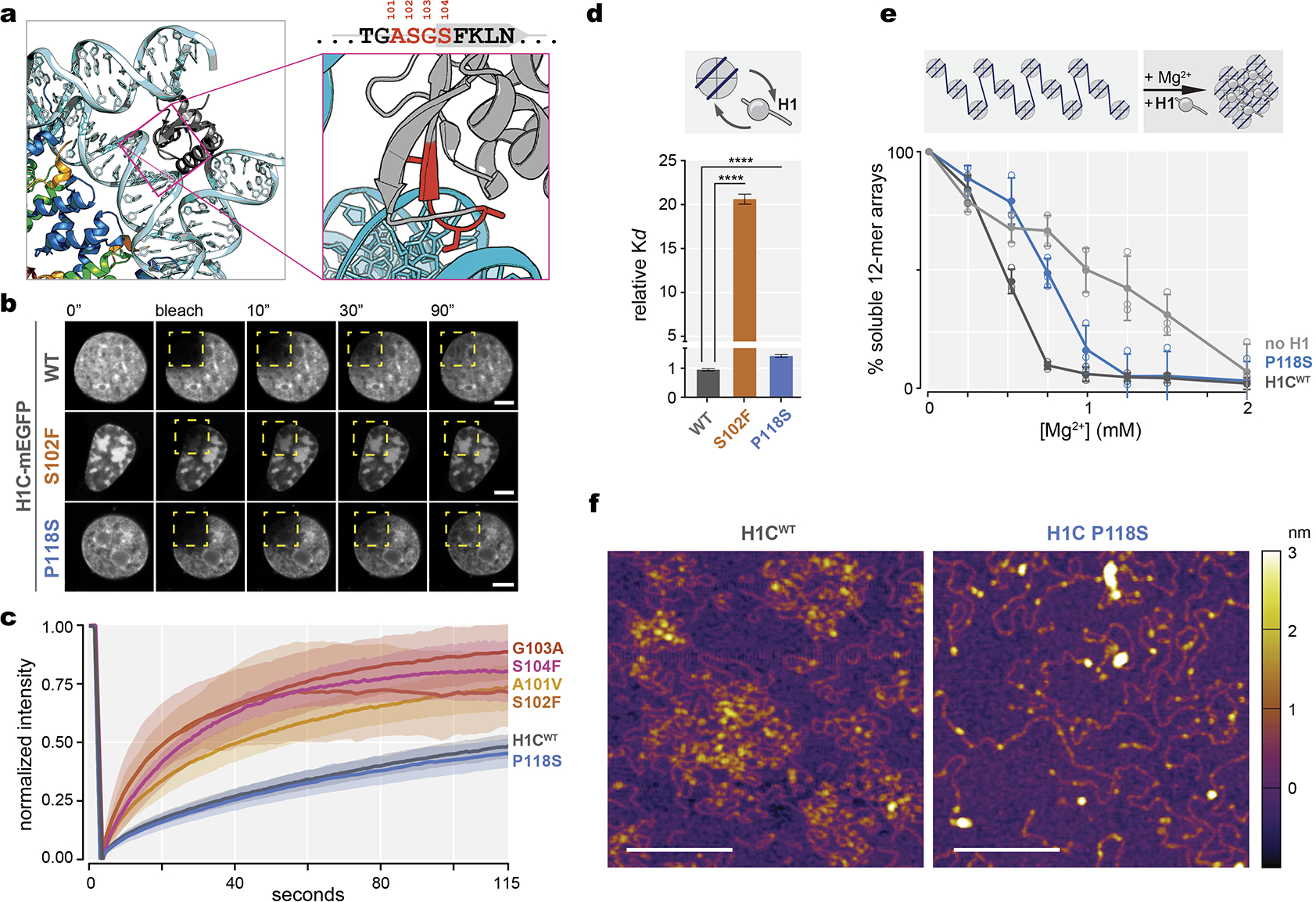
a, Crystal structure of the linker histone globular domain (gray) bound to nucleosome [PDB, 4QLC], with zoomed-in view of ASGS amino acid residues highlighted in red. b, Representative images of fluorescence recovery after photobleaching of ectopically expressed, meGFP-tagged wild-type H1C, S102F, and P118S mutants in 3T3 cells prior to, immediately after, and at 10, 30 and 90 seconds after bleaching the area (yellow dashed square). Scale bars = 5 μm. c, Quantification of normalized intensity as representation of turnover kinetics from (E) for wild-type H1C (n=18), and mutants A101V (n=15), S102F (n=9), S104F (n=10), G103A (n=10), and P118S (n=10) cell measurements, shaded area indicates 95% C.I. Data are pooled from two independent biological experiments.d, Dissociation constant (Kd) of recombinant mutant H1C S102F and P118S compared to WT H1C binding to mononucleosomes determined by biolayer interferometry. Data are mean ± s.e.m (two-sided unpaired t-test, ****P<0.0001). Data are global fit from five concentration measurements. e, Chromatin fiber oligomerization upon serial precipitation by Mg2+ as percent soluble 12-mer arrays was determined for no H1, WT H1C and C-terminal domain P118S mutant. Data are mean ± SD. Data are pooled from three independent biological experiments. f, Atomic force microscopy imaging of chromatin arrays in presence of wild-type H1 and C-terminal domain P118S mutant, scale bars, 200 nm. Images are representative from two independent biological experiments.
