Abstract
Objective
Most women with epithelial ovarian cancer (EOC) are diagnosed after the disease has metastasized and survival in this group remains poor. Circulating proteins associated with the risk of developing EOC have the potential to serve as biomarkers for early detection and diagnosis. We integrated large-scale genomic and proteomic data to identify novel plasma proteins associated with EOC risk.
Methods
We used the germline genetic variants most strongly associated (P<1.5×10−11) with plasma levels of 1,329 proteins in 3,301 healthy individuals from the INTERVAL study to predict circulating levels of these proteins in 22,406 EOC cases and 40,941 controls from the Ovarian Cancer Association Consortium (OCAC). Association testing was performed by weighting the beta coefficients and standard errors for EOC risk from the OCAC study by the inverse of the beta coefficients from INTERVAL.
Results
We identified 26 proteins whose genetically predicted circulating levels were associated with EOC risk at false discovery rate<0.05. The 26 proteins included MFAP2, SEMG2, DLK1, and NTNG1 and a group of 22 proteins whose plasma levels were predicted by variants at chromosome 9q34.2. All 26 protein association signals identified were driven by association with the high-grade serous histotype that comprised 58% of the EOC cases in OCAC. Regional genomic plots confirmed overlap of the genetic association signal underlying both plasma protein level and EOC risk for the 26 proteins. Pathway analysis identified enrichment of seven biological pathways among the 26 proteins (Padjusted<0.05), highlighting roles for Focal Adhesion-PI3K-Akt-mTOR and Notch signaling.
Conclusion
The identified proteins further illuminate the etiology of EOC and represent promising new EOC biomarkers for targeted validation by studies involving direct measurement of plasma proteins in EOC patient cohorts.
Keywords: epithelial ovarian cancer, risk, circulating proteins, circulating biomarkers, genome-wide association study
Introduction
Ovarian cancer is the most common cause of death from gynecological malignancy in the United States and accounted for an estimated 295,000 incident cases and 184,000 deaths globally in 2018 [1,2]. Despite advances in treatment, survival rates in ovarian cancer continue to remain low, in part, due to the late detection of most cases [3]. Nearly four decades after its discovery [4], circulating levels of the protein cancer antigen 125 (CA-125) continue to be used to screen women at high risk of developing ovarian cancer, such as those with a hereditary cancer syndrome, and women with abnormal findings on examination and/or ultrasound. However, CA-125 has limited sensitivity and specificity in these settings [5]. Furthermore, screening asymptomatic women for CA125 level, despite the use of serial measurements and algorithmic approaches to the interpretation of these levels [6] – and even in combination with transvaginal ultrasound – does not reduce ovarian cancer mortality and is not recommended by the US Preventive Services Task Force [7]. Human epididymis secretory protein E4 (HE4) has been developed in recent years as a blood-based protein biomarker for the diagnosis of ovarian carcinoma [8], and the combination of CA-125 and HE4 is a more accurate predictor of ovarian malignancy than either biomarker alone [9]. However, there remains an urgent unmet need to identify novel circulating protein biomarkers that will be more useful for the early detection of this aggressive disease.
Studies in search of new plasma protein biomarkers in ovarian cancer have been restricted to small sample sizes and evaluated limited protein panels [10,11]. In the current study, we adopted a different approach to the identification of circulating protein biomarkers of ovarian cancer risk using large-scale data from two genome-wide association studies (GWAS). The first data set was a GWAS of healthy blood donors in the INTERVAL study that has identified robust associations between inherited genetic variants and plasma protein levels [12]. The second data set was the the largest and latest published GWAS meta-analysis from the Ovarian Cancer Association Consortium (OCAC) [13]. While epithelial ovarian cancer (EOC) accounts for approximately 90% of all ovarian cancer cases, EOC itself is a diverse entity with distinct histological subtypes: high-grade serous (the most common and lethal histotype), low-grade serous, clear cell, mucinous, endometrioid, and low malignant potential (serous or mucinous) tumors. The OCAC GWAS included associations with all invasive and histotype-specific EOC susceptibility. We used the inherited genetic variants robustly associated with plasma protein levels in the INTERVAL GWAS to predict these levels in the OCAC GWAS where plasma protein levels have not actually been measured but the variants have been genotyped. Such predictions are likely to suffer from less selection bias and confounding because the genetic variants on which they are based are randomly allocated at gametogenesis and fixed after conception. Our study design enabled a comprehensive appraisal of the role of the levels of over 1,300 plasma proteins in more than 22,000 ovarian cancer cases and over 40,000 controls.
Methods
Circulating (plasma) protein data set
We used effect size estimates (beta coefficients) from genome-wide association analyses linking 667 single nucleotide polymorphisms (SNPs) to the circulating (plasma) levels of 1,329 proteins in 3,301 healthy participants from the INTERVAL study [12], a bioresource of blood donors in England who were recruited into a multi-center randomized trial of blood donation frequency [14]. Each of these SNPs was associated with at least one of the plasma proteins at genome-wide significance (defined as P < 1.5 × 10−11 in the INTERVAL analysis [12]) and was the SNP most strongly associated with the circulating levels of that protein. Five hundred and eight-five SNPs were associated with the levels of only one circulating protein each while 82 SNPs were associated with multiple proteins (ranging from 36 SNPs that were associated with two proteins each to one SNP that was associated with 95 proteins; Table S1). We restricted analysis to SNPs that had minor allele frequency (MAF) > 1% and had either been genotyped or imputed with quality score > 0.8 – both in the INTERVAL analytic sample and in the OCAC data set (described below). These SNP-protein associations included 908 trans-associations where the top SNP associated with the protein was > 1 Mb away from the gene encoding the protein and 421 cis-associations where the top SNP associated with the protein was < 1 Mb away from the gene encoding the protein. Plasma protein levels in the INTERVAL study were quantified using an expanded aptamer-based multiplex protein assay called SOMAscan [12,15] and germline genotypes were measured on Affymetrix Axiom UK Biobank array with imputation into a combined combined 1000 Genomes Phase 3-UK10K reference panel. We used the same protein names and identifiers, including UniProt and SOMAmer IDs (Table S1), as used in the original INTERVAL genetic report [12] for consistency. That report contains additional details of sample and genotype quality control, imputation, and association analysis in the INTERVAL study.
Epithelial ovarian cancer data set
Summary statistics (beta coefficients and standard errors) from a GWAS meta-analysis for EOC susceptibility in women of European ancestry were obtained from OCAC [13]. The GWAS meta-analysis included 22,406 invasive EOC cases overall and 40,941 controls and this “all invasive EOC” case-control set was the focus of the primary analysis in the current study. EOC histotype-specific summary statistics from the same GWAS meta-analysis were also evaluated for seven histological subtypes as a secondary analysis in the current study. This included high-grade serous (13,037 cases), low-grade serous (1,012 cases), low malignant potential serous (1,954 cases), invasive mucinous (1,417 cases), low malignant potential mucinous (1,149), clear cell (1,366), and endometrioid (2,810 cases) EOC cases and 40,941 controls. Additional details of sample and genotype quality control, imputation, and association analytic procedures for the OCAC GWAS meta-analysis have been previously published [13].
Statistical analysis
We used the Wald ratio to estimate the effect of genetically predicted circulating protein levels on ovarian cancer risk. The Wald estimator in this context is the ratio of the beta coefficient for a SNP from the ovarian cancer GWAS meta-analysis to the beta coefficient for the same SNP from the plasma protein genome-wide association analysis. The SNP most strongly associated with the circulating level of each protein in the INTERVAL data set was used. The standard error of the Wald estimator is the ratio of the standard error for the SNP from the ovarian cancer GWAS meta-analysis to the absolute value of the beta coefficient for the SNP from the plasma protein genome-wide association analysis. These analyses were performed using the R (version 3.6.2) statistical computing language. P-values were calculated using the formula: “pnorm(abs(Wald_estimator)/standard_error_of_Wald_estimator, lower.tail=FALSE) * 2” and the multiple comparisons burden for testing 1,329 SNP-protein-ovarian cancer associations was accounted for using false discovery rate (FDR) control by the method of Benjamini and Hochberg as implemented in the “p.adjust” function. The Wald estimator allowed for incorporation of the beta coefficient for the SNP from the plasma protein analysis and allowed easy inference of the direction of the association (whether positive or inverse) between plasma protein level and ovarian cancer risk. Therefore, we preferred the Wald estimator over directly testing for the genetic association between the top plasma protein level-associated SNP and ovarian cancer risk (although in practice both approaches provided almost identical P-values). As noted above, our primary analysis was for all invasive EOC risk, given that this combined phenotype had the largest sample size, while in secondary analyses we evaluated histotype-specific risk.
We followed up genetically predicted circulating levels of proteins that were found to be associated (FDR < 0.05) in our study with ovarian cancer risk to assess whether the top plasma protein level-associated SNP was part of the top ovarian cancer genetic association signal in the same genomic region – a positional overlap that would reinforce the role of the SNP as a driver of both circulating protein levels and ovarian cancer risk. We did this by visualizing ovarian cancer genetic associations for all SNPs with MAF > 1% and imputation quality > 0.8 in the OCAC data set in the 500 kb window centered on the top protein-associated SNP (i.e., +/- 250 kb on either side) using two-way scatter plots generated in Stata (version 14, StataCorp LP, College Station, TX). For SNPs with stronger P-values for association with ovarian cancer risk in OCAC as compared to the top protein-associated SNP, the correlation between the stronger P-value SNPs and the top protein-associated SNP was calculated using the LDlink online tool and data from the 1000 Genomes European ancestry populations [16]. If the same SNP association signal drives both plasma protein level and ovarian cancer risk, we expected one of the following three scenarios to be true: (i) the top protein-associated SNP is also the top ovarian cancer associated SNP or (ii) it is strongly correlated (r2 > 0.9) with the top ovarian cancer associated SNP(s) or (iii) there are multiple independent (r2 < 0.01) genetic association signals in OCAC in the same region and the top protein-associated SNP is one of these associations. A second follow-up analysis of proteins that achieved FDR < 0.05 in our study involved mapping these to the genes encoding them and evaluating the genes for enrichment of pathways (at P < 0.05 after adjustment for testing multiple pathways) using the Enrichr online tool [17] and the “WikiPathways Human 2019” database [18] that contains annotations for 472 known biological pathways. A final follow-up analysis involved searching for genome-wide signficant associations (P < 5 × 10−8) between the top plasma protein level-associated SNP for each of the proteins that achieved FDR < 0.05 in our study and other diseases and traits in the published (i.e., MEDLINE indexed) literature. This search was performed using the PhenoScanner (version 2) online tool [19], querying published European-ancestry GWAS. The aim was to identify pleiotropic diseases and traits that may provide an alternative explanation for the plasma protein-EOC risk associations identified, stemming from their associations with the same top SNPs. Such pleiotropic diseases and traits associated with the same SNPs may also be the cause or consequence of plasma protein level changes that in turn are associated with EOC risk.
Results
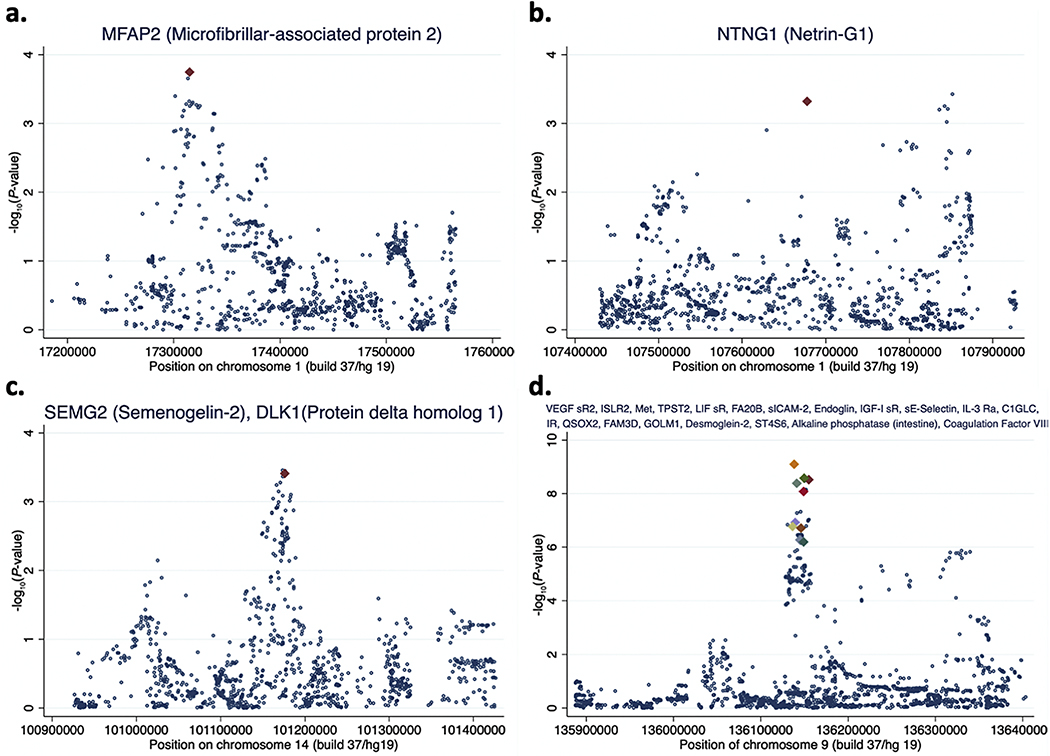
Genetically predicted circulating levels of 26 proteins were associated with all invasive EOC risk at FDR < 0.05 (13 positive and 13 inverse associations; Table 1 and Table S1). First, this included a positive association between MFAP2 encoded by MFAP2 on chromosome 1 and all invasive EOC risk (Microfibrillar-associated protein 2; PWald =1.8 × 10−4, FDR= 0.01).The top MFAP2 plasma protein-associated SNP rs4920605 (POCAC-GWAS-all = 1.82 × 10−4) in the INTERVAL study was ~8 kb from the transcription start site (TSS) of MFAP2. There was only one SNP in the same region, rs143483351 (POCAC-GWAS-all = 1.76 × 10−4),a multi-allelic variant 2 kb from rs4920605, with a slightly stronger association with all invasive EOC risk (Fig. 1 (a) and Table S2). SNP rs143483351 could not be evaluated in LDlink [16] for correlation with rs4920605 because it was a multi-allelic variant. Second, our FDR < 0.05 results also included an inverse association between NTNG1 encoded by NTNG1 on chromosome 1 (in a genomic region distinct from MFAP2) and all invasive EOC risk (Netrin-G1; PWald = 4.9 × 10−4, FDR= 0.03). The top NTNG1 plasma protein-associated SNP rs115668827 (POCAC-GWAS-all = 4.9 × 10−4) in the INTERVAL study was ~4 kb from the TSS of NTNG1. There was only one SNP in the same region, rs11185086 (POCAC-GWAS-all = 3.8 × 10−4), 173 kb from rs115668827, with a stronger association with all invasive EOC risk (Fig. 1 (b) and Table S2). However, rs11185086 and rs115668827 represented independent signals in the same region (r2 = 7 × 10−4). Third, the list of 26 plasma proteins identified included positive associations between SEMG2 (Semenogelin-2) and ovarian cancer risk and DLK1 (Protein delta homolog 1) and ovarian cancer risk (for both associations – PWald = 4.0 × 10−4, FDR = 0.02). The top SEMG2 plasma protein-associated SNP and the top DLK1 plasma protein-associated SNP in the INTERVAL study was the same SNP, rs12881760 (POCAC-GWAS-all = 3.96 × 10−4), which is ~16 kb from the TSS of DLK1 on chromosome 14. SEMG2 is encoded by SEMG2 on chromosome 20 and rs12881760 is associated with its circulating level by acting in trans. SNP rs12881760 is part of a cluster of three SNPs that includes rs10144381 (POCAC-GWAS-all = 3.50 × 10−4) and rs12881545(POCAC-GWAS-all = 3.56 × 10−4), which are within 3 kb of each other and strongly correlated (r2 > 0.93), and together mark the strongest association signal with all invasive EOC risk in the DLK1 region (Fig. 1 (c) and Table S2).
Table 1.
Associations identified between genetically predicted circulating (plasma) protein levels and all invasive epithelial ovarian cancer risk.
| Protein | Protein full name | TopSNP associated with plasma level of protein | Chr | Posa | Cis/transb | Gene encoding protein | Gene mapped to SNP | ORc | 195 %CL | U95 %CL | P | FDR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FA20B | Glycosaminoglycan xylosylkinase | rs587729126 | 9 | 136138765 | trans | FAM20B | ABO | 1.47 | 1.30 | 1.66 | 8.1x10−10 | 4.5x10−7 |
| slCAM-2 | Intercellular adhesion molecule 2 | rs587729126 | 9 | 136138765 | trans | ICAM2 | ABO | 0.65 | 0.57 | 0.75 | 8.1x10−10 | 4.5x10−7 |
| VEGF sR2 | Vascular endothelial growth factor receptor 2 | rs635634 | 9 | 136155000 | trans | KDR | ABO | 0.84 | 0.80 | 0.89 | 3.0x10−9 | 4.5x10−7 |
| ISLR2 | Immunoglobulin superfamily containing leucine-rich repeat protein 2 | rs115478735 | 9 | 136149711 | trans | ISLR2 | ABO | 0.80 | 0.75 | 0.86 | 2.8x10−9 | 4.5x10−7 |
| Met | Flepatocyte growth factor receptor | rs635634 | 9 | 136155000 | trans | MET | ABO | 0.77 | 0.71 | 0.84 | 3.0x10−9 | 4.5x10−7 |
| TPST2 | Protein-tyrosine sulfotransferase 2 | rs115478735 | 9 | 136149711 | trans | TPST2 | ABO | 1.36 | 1.23 | 1.51 | 2.8x10−9 | 4.5x10−7 |
| LIFsR | Leukemia inhibitory factor receptor | rs635634 | 9 | 136155000 | trans | LIFR | ABO | 0.72 | 0.64 | 0.80 | 3.0x10−9 | 4.5x10−7 |
| Endoglin | Endoglin | rs635634 | 9 | 136155000 | trans | ENG | ABO | 0.66 | 0.57 | 0.76 | 3.0x10−9 | 4.5x10−7 |
| IGF-I sR | Insulin-like growth factor 1 receptor | rs635634 | 9 | 136155000 | trans | IGF1R | ABO | 0.64 | 0.55 | 0.74 | 3.0x10−9 | 4.5x10−7 |
| sE-Selectin | E-selectin | rs2519093 | 9 | 136141870 | trans | SELE | ABO | 0.92 | 0.89 | 0.95 | 4.2x10−9 | 4.7x10−7 |
| IL-3 Ra | Interleukin-3 receptor subunit alpha | rs2519093 | 9 | 136141870 | trans | IL3RA | ABO | 0.89 | 0.86 | 0.93 | 4.2x10−9 | 4.7x10−7 |
| C1GLC | ClGALTl-specific chaperone 1 | rs2519093 | 9 | 136141870 | trans | C1GALT1C1 | ABO | 1.17 | 1.11 | 1.24 | 4.2x10−9 | 4.7x10−7 |
| IR | Insulin receptor | rs507666 | 9 | 136149399 | trans | INSR | ABO | 0.85 | 0.80 | 0.90 | 8.4x10−9 | 8.6x10−7 |
| QSOX2 | Sulfhydryl oxidase 2 | rs149092047 | 9 | 136139907 | trans | QSOX2 | ABO | 1.09 | 1.06 | 1.13 | 1.2x10−7 | 1.0x10−5 |
| FAM3D | Protein FAM3D | rs149092047 | 9 | 136139907 | trans | FAM3D | ABO | 1.12 | 1.07 | 1.16 | 1.2x10−7 | 1.0x10−5 |
| GOLM1 | Golgi membrane protein 1 | rs149092047 | 9 | 136139907 | trans | GOLM1 | ABO | 1.15 | 1.09 | 1.22 | 1.2x10−7 | 1.0x10−5 |
| Desmoglein-2 | Desmoglein-2 | rs687621 | 9 | 136137065 | trans | DSG2 | ABO | 1.39 | 1.23 | 1.57 | 1.7x10−7 | 1.3x10−5 |
| ST4S6 | Carbohydrate sulfotransferase 15 | rs550057 | 9 | 136146597 | trans | CHST15 | ABO | 0.79 | 0.72 | 0.86 | 2.0x10−7 | 1.4x10−5 |
| Alkaline phosphatase, intestine | Intestinal-type alkaline phosphatase | rs550057 | 9 | 136146597 | trans | ALPI | ABO | 0.74 | 0.66 | 0.83 | 2.0x10−7 | 1.4x10−5 |
| Coagulation Factor VIII | Coagulation Factor VIII | rs9411377 | 9 | 136145404 | trans | F8 | ABO | 1.16 | 1.09 | 1.22 | 5.5x10−7 | 3.7x10−5 |
| BGAT | Flisto-blood group ABO System transferase | rs505922 | 9 | 136149229 | cis | ABO | ABO | 1.05 | 1.03 | 1.08 | 6.7x10−7 | 4.0x10−5 |
| DC-SIGN | CD209 antigen | rs505922 | 9 | 136149229 | trans | CD209 | ABO | 1.09 | 1.05 | 1.12 | 6.7x10−7 | 4.0x10−5 |
| MFAP2 | Microfibrillar-associated protein 2 | rs4920605 | 1 | 17315425 | cis | MFAP2 | MFAP2 | 1.27 | 1.12 | 1.45 | 1.8x10−4 | 0.011 |
| SEMG2 | Semenogelin-2 | rs12881760 | 14 | 101176335 | trans | SEMG2 | DLK1 | 1.10 | 1.04 | 1.15 | 4.0x10−4 | 0.021 |
| DLK1 | Protein delta homolog 1 | rs12881760 | 14 | 101176335 | cis | DLK1 | DLK1 | 1.10 | 1.04 | 1.16 | 4.0x10−4 | 0.021 |
| IMTIMG1 | Netrin-G1 | rsll5668827 | 1 | 107678268 | cis | NTNG1 | NTNG1 | 0.89 | 0.84 | 0.95 | 4.9x10−4 | 0.025 |
Build 37/hl9 position.
Cis if thetop SNP is < 1 Mbfrom thè gene encoding protein and trans if thè top SNP is > 1 Mb from thè gene encoding thè protein.
Odds ratio (OR), lower 95% confidence limit (L95%CL), upper 95% confidence limit (U95%CL), P-value, and false discovery rate (FDR) from thè current study. OR is in terms of all invasive EOC risk per standard deviation increase in circulating (plasma) protein level
Fig. 1.
Regional genetic association plots. Genetic association with all invasive epithelial ovarian cancer risk (negative logarithm base 10 P-value) from the Ovarian Cancer Association Consortium study is plotted on the Y-axis and chromosomal position (build 37/hg 19) is plotted on the X-axis. SNPs are marked with blue dots or colored diamonds. SNPs marked with colored diamonds are the SNPs most strongly associated in the INTERVAL study with circulating (plasma) levels of the proteins named in the titles of the plots.
The remaining 22 of the 26 all invasive EOC risk-associated circulating proteins identified were proxied by 10 correlated SNPs (r2 > 0.38) spanning a ~10 kb interval on chromosome 9 (Table 1). Three of these SNPs were the top SNP for one protein each, four for two proteins each, two for three proteins each, and one SNP was the top SNP for five proteins in the INTERVAL data set (Table 1). Ten proteins demonstrated a positive association and 12 showed an inverse association with all invasive EOC risk. Twenty-one of the 22 proteins were encoded by genes > 1 Mb away from this chromosome 9 interval (trans-associations) and most were in fact encoded by genes located on other chromosomes. The only exception to this was the plasma protein BGAT (Histo-blood group ABO system transferase) encoded by ABO and the top BGAT plasma level-associated SNP, rs505922, is ~2 kb from the TSS of ABO. The ABO locus (chromosome 9q34.2) is a known genome-wide significant (P < 5 × 10−8) locus for all invasive and high-grade serous ovarian cancer risk [13,20]. The ten protein level-associated SNPs spanned the ABO locus and were among the top 50 all invasive EOC risk SNPs in the 500 kb region (Table S2 and Fig. 1 (d)). The top all invasive EOC risk SNP in the region, rs587729126 (POCAC-GWAS-all = 8.3 × 10−10),was the top SNP in the INTERVAL study for association with circulating levels of FA20B (Glycosaminoglycan xylosylkinase) and sICAM-2 (Intercellular adhesion molecule 2). This overlap of top associations led to these two proteins emerging as the plasma proteins whose genetically predicted levels were most strongly associated with all invasive EOC risk in our analysis (for both associations – PWald = 8.1 × 10−10, FDR = 4.5 × 10−7; Table 1). The PhenoScanner search indicated that eight of the ten protein level-associated SNPs that spanned the ABO locus were associated with 62 traits (Table S3). Overall, for all 26 proteins identified (associated with SNPs in the regions presented in Fig. 1 and discussed above), we observed a clear overlap between the top circulating protein level-associated SNP and the top all invasive EOC risk association, lending further confidence to the association between plasma protein levels predicted by these SNPs and disease risk. We did not identify any additional proteins at FDR < 0.05 in any of the histotype-specific analyses (Table S1). An inspection of the high-grade serous EOC results (Table S1) confirmed that all 26 FDR < 0.05 protein associations with all invasive EOC risk were driven by associations in the high-grade serous EOC sample, which contributed the largest number of cases to the all invasive EOC sample. Pathway enrichment analysis of the genes encoding the 26 proteins identified seven pathways at Padjusted < 0.05 (Table 2).
Table 2.
Pathways enriched among the genes encoding the 26 all invasive epithelial ovarian cancer risk-associated circulating protein biomarkers identified.
| Pathwaya | Overlapb | P-value | Adjusted Pc | Genesd |
|---|---|---|---|---|
| Pathways Regulating Hippo Signaling WP4540 | 4/98 | 7.5 × 10−6 | 0.004 | INSR; KDR; MET; IGF1R |
| Hippo-Merlin Signaling Dysregulation WP4541 | 4/120 | 1.7 × 10−5 | 0.004 | INSR; KDR; MET; IGF1R |
| Focal Adhesion-PI3K-Akt-mTOR-signaling pathway WP3932 | 5/303 | 3.9 × 10−5 | 0.006 | INSR; IL3RA; KDR; MET; IGF1R |
| PI3K-Akt Signaling Pathway WP4172 | 5/340 | 6.8 × 10−5 | 0.008 | INSR; IL3RA; KDR; MET; IGF1R |
| Ras Signaling WP4223 | 4/184 | 8.9 × 10−5 | 0.008 | INSR; KDR; MET; IGF1R |
| Ebola Virus Pathway on Flost WP4217 | 3/129 | 6.1 × 10−4 | 0.041 | CD209; ICAM2; IGF1R |
| Canonical and Non-canonical Notch signaling WP3845 | 2/27 | 5.6 × 10−4 | 0.044 | MFAP2; DLK1 |
From the “WikiPathways 2019 Fluman” pathway database (with associated WP identifier number).
The number of genes out of the 26 genes evaluated/the total number of genes annotated to the pathway.
Adjusted for testing 472 pathways.
The genes (out of the 26 genes evaluated) that are annotated to the pathway.
Discussion
By combining genome-wide association data from 22,406 all invasive EOC cases and 40,941 controls and plasma proteome-wide genetic association data from 3,301 healthy individuals, we identified 26 proteins whose genetically inferred circulating levels were associated with EOC risk after false discovery rate control (FDR < 0.05). The combination of these data sets offered unprecedented scale to evaluate the role of over 1,300 plasma proteins in the development of EOC and identified circulating protein biomarkers with the potential for clinical translational in the early detection and diagnosis of EOC.
We observed that the top plasma protein level-associated SNP was either the top all invasive EOC risk SNP in the 500 kb region centred on the SNP (Fig. 1 (a) and (d)) or it was the top SNP of one of two independent (r2 < 0.01) all invasive EOC risk associations in the region (Fig. 1 (b)) or it was part of a cluster of highly correlated (r2 > 0.9) SNPs that together marked the top all invasive EOC risk association in the region (Fig. 1 (c)). This suggests that our results are unlikely to be due to linkage disequilibrium contamination, i.e., regional genetic architecture where the top plasma protein level-associated SNP is weakly correlated with the top all invasive EOC risk SNP and results in a spurious association underpinned by two distinct SNP signals (one for protein and another for EOC). While the focus of our analysis was the use of SNPs associated with plasma protein levels to evaluate the association between plasma protein levels and EOC risk and not the direct genetic association between SNPs and EOC risk, we note that there were 667 unique SNPs used in the analysis (Table S1) and the 13 unique SNPs underpinning the 26 proteins identified (Table 1) were all associated with all invasive EOC risk at P < 0.05/667, which would be the conventional threshold for statistical significance if this was a SNP-based association study of 667 SNPs. Ten of the 13 SNPs were genome-wide significant (P < 5 × 10−8) as they are located at a previously reported all invasive and high-grade serous EOC risk locus at or near ABO on chromosome 9q34.2 [13,20]. The three remaining SNPs (spanning three distinct genomic regions; Fig. 1 (a), (b), and (c)) may well represent as yet unidentified genetic susceptibility loci for all invasive EOC that we are presently underpowered to detect at GWAS levels of significance (P < 5 × 10−8). Thus, loci known to be associated with the plasma proteome may aid in the discovery of sub-threshold GWAS loci for disease susceptibility in much the same way as previously demonstrated for other biological information integrated into genetic association studies [21].
Pathway analysis highlighted that five of the 26 proteins whose genetically predicted plasma levels were associated with all invasive EOC risk at FDR < 0.05 belonged to the “Focal Adhesion-PI3KAkt-mTOR-signaling pathway (Ppathway (adjusted) = 0.006; Table 2), which was the maximum overlap seen between any established biomolecular pathway and the 26 proteins. The genes encoding these proteins were located across different chromosomes, but the SNPs associated most strongly with their plasma levels were all located at the 9q34.2 locus. The PI3K-Akt-mTOR intracellular signaling cascade is a major regulator of the cell cycle and has key roles in cellular quiescence, growth and proliferation, and cancer cell survival and metastasis [22]. Somatic aberrations in this pathway are found in the majority of high-grade serous ovarian tumors [23]. Another pathway identified at Ppathway(adjusted) < 0.05 was Notch signaling and this association was driven, in turn, by associations between genetically predicted circulating levels of two Notch proteins, MFAP2 (chromosome 1p36.13) and DLK1 (chromosome 14q32.2), and all invasive EOC risk. It is noteworthy that these two plasma proteins associated with EOC risk at FDR < 0.05 were encoded by genes located on distinct chromosomes but the genes/proteins were members of the same biological pathway. DLK1, a noncanonical Notch ligand, has a demonstrated role in promoting ovarian carcinogenesis via Notch activation and epithelial-mesenchymal transition [24]. The microfibrillar-associated protein 2 (MFAP2), previously named microfibril-associated glycoprotein 1 (MAGP1), activates integrin signaling and is a potential oncogene [25–27]. Another protein identified at FDR < 0.05, Netrin-G1 or NTNG1, is involved in apoptosis and known to be dysregulated particularly in endocrine-related tumors [28,29]. Further, the gene that encodes NTNG1 has been shown to be overexpressed in malignant ovarian tumors [30].
Larger genetic association studies of the circulating proteome as well as for EOC susceptibility may identify additional candidate biomarkers for EOC. Moreover, such studies may profile additional proteins (including CA-125, which was not profiled in the INTERVAL study) and include individuals of non-European ancestries, offering new opportunities for plasma protein biomarker discovery. The present study was unable to identify associations for EOC histotypes other than for the most common high-grade serous hisotype and this is another area where larger sample sizes might help. The INTERVAL and OCAC data sets used in this analysis were based on participants of European ancestry and there is a compelling need for similar trans-ancestry analyses. Smaller GWAS of ovarian cancer risk in women of African and East Asian ancestry have been reported by OCAC but there is no circulating protein level GWAS comparable to the INTERVAL study as yet in a cohort that is not of European ancestry [31,32]. A major strength of the current analysis was the ability to appraise the roles of over 1,300 proteins. A vital next step in assessing the role of the plasma proteome in EOC risk and validating our findings will involve directly measuring the 26 proteins shortlisted by our study in EOC case and control sample collections that have pre-diagnostic and longitudinal follow-up biospecimens available such as the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial [33]. The pleiotropic associations observed at the 9q34.2 locus where eight of the ten SNPs associated with plasma protein levels were also associated with 62 other traits leaves open the possibility that some of these traits, rather than the protein levels, may underlie the association with EOC risk. Alternatively, some of these traits may lie up- or downstream of the protein levels and mediate the association with EOC risk as part of the same causal pathway. Further studies will be required to dissect these possibilities. For example, five of the eight SNPs at the 9q34.2 (ABO) locus are associated with low density lipoprotein-cholesterol (LDL-C) levels with the SNP alleles predicting lower LDL-C levels associating with reduced EOC risk (Table S3). This is consistent with a recent analysis based on the OCAC data set which showed that lower LDL-C level genetically predicted by SNPs in or near HMGCR, which encodes the enzyme inhibited by statins, was associated with reduced EOC risk [34].
In conclusion, our integrative analysis of large-scale proteomic and genomic data sets identified several associations between genetically predicted circulating protein levels and EOC risk that were statistically significant after FDR control and biologically plausible. These plasma proteins are candidate biomarkers with the potential for application in the early diagnosis of this aggressive gynecological cancer. The associations shed new light on EOC biology and should inform a range of follow-up laboratory-based studies and targeted biomarker validation projects wherein the 26 identified plasma proteins are directly tracked in incident EOC cases and controls over time.
Supplementary Material
Research Highlights.
This study analyzed 667 germline genetic variants known to be associated with circulating (plasma) levels of 1,329 proteins
These variants were used to predict plasma protein levels in 22,406 epithelial ovarian cancer cases and 40,941 controls
Genetically predicted levels of 26 proteins were associated with all invasive epithelial ovarian cancer risk
The identified proteins were enriched for the Focal Adhesion-PI3K-Akt-mTOR signaling, Notch signaling, and other pathways
The identified proteins have the potential to serve as circulating biomarkers particularly for high-grade serous epithelial ovarian cancer risk
Acknowledgments
This work was supported by a grant (R01 CA211574) from the US National Institutes of Health (National Cancer Institute). We thank the participants of the many studies contributing to the Ovarian Cancer Association Consortium (OCAC) and the INTERVAL study as well as the many researchers involved in OCAC and INTERVAL who have made this work possible. A full list of grant funding for OCAC is available in PMID:28346442/PMCID:PMC5612337/DOI:10.1038/ng.3826 and for INTERVAL is available in PMID:29875488/PMCID:PMC6697541/DOI:10.1038/s41586018-0175-2.
Footnotes
Conflict of Interest statement
The authors have no conflict of interest to declare.
Availability of data and code
The Ovarian Cancer Association Consortium (OCAC) data set can be downloaded from: ftp://ftp.ebi.ac.uk/pub/databases/gwas/summary_statistics/PhelanCM_28346442_GCST004462
The genetic associations with circulating protein biomarkers were obtained from the INTERVAL study and can be downloaded from: https://static-content.springer.com/esm/art%3A10.1038%2Fs41586-018-0175-2/MediaObjects/41586_2018_175_MOESM4_ESM.xlsx
Code used to perform the analysis reported in this paper is available at: https://github.com/siddhartha-kar/circulating-proteins-and-ovarian-cancer
Other online tools used –
LDlink: https://ldlink.nci.nih.gov
Enrichr: https://amp.pharm.mssm.edu/Enrichr
WikiPathways: https://www.wikipathways.org
PhenoScanner: http://www.phenoscanner.medschl.cam.ac.uk/
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2020, CA Cancer J Clin. 70 (2020) 7–30. [DOI] [PubMed] [Google Scholar]
- [2].Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A, Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA Cancer J Clin. 68 (2018) 394–424. [DOI] [PubMed] [Google Scholar]
- [3].Matulonis UA, Sood AK, Fallowfield L, Howitt BE, Sehouli J, Karlan BY, Ovarian cancer, Nat Rev Dis Primers. 2 (2016) 16061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bast RC, Feeney M, Lazarus H, Nadler LM, Colvin RB, Knapp RC, Reactivity of a monoclonal antibody with human ovarian carcinoma, J. Clin. Invest. 68 (1981) 1331–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Moss EL, Hollingworth J, Reynolds TM, The role of CA125 in clinical practice, J. Clin. Pathol. 58 (2005) 308–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dochez V, Caillon H, Vaucel E, Dimet J, Winer N, Ducarme G, Biomarkers and algorithms for diagnosis of ovarian cancer: CA125, HE4, RMI and ROMA, a review, J Ovarian Res. 12 (2019) 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].US Preventive Services Task Force, Grossman DC, Curry SJ, Owens DK, Barry MJ, Davidson KW, Doubeni CA, Epling JW, Kemper AR, Krist AH, Kurth AE, Landefeld CS, Mangione CM, Phipps MG, Silverstein M, Simon MA, Tseng C-W, Screening for Ovarian Cancer: US Preventive Services Task Force Recommendation Statement, JAMA. 319 (2018) 588–594. [DOI] [PubMed] [Google Scholar]
- [8].Moore RG, McMeekin DS, Brown AK, DiSilvestro P, Miller MC, Allard WJ, Gajewski W, Kurman R, Bast RC, Skates SJ, A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass, Gynecol Oncol. 112 (2009) 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Moore RG, Brown AK, Miller MC, Skates S, Allard WJ, Verch T, Steinhoff M, Messerlian G, DiSilvestro P, Granai CO, Bast RC, The use of multiple novel tumor biomarkers for the detection of ovarian carcinoma in patients with a pelvic mass, Gynecol Oncol. 108 (2008) 402–408. [DOI] [PubMed] [Google Scholar]
- [10].Enroth S, Berggrund M, Lycke M, Broberg J, Lundberg M, Assarsson E, Olovsson M, Stålberg K, Sundfeldt K, Gyllensten U, High throughput proteomics identifies a high-accuracy 11 plasma protein biomarker signature for ovarian cancer, Commun Biol. 2 (2019) 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Enroth S, Berggrund M, Lycke M, Lundberg M, Assarsson E, Olovsson M, Stålberg K, Sundfeldt K, Gyllensten U, A two-step strategy for identification of plasma protein biomarkers for endometrial and ovarian cancer, Clin Proteomics. 15 (2018) 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, Burgess S, Jiang T, Paige E, Surendran P, Oliver-Williams C, Kamat MA, Prins BP, Wilcox SK, Zimmerman ES, Chi A, Bansal N, Spain SL, Wood AM, Morrell NW, Bradley JR, Janjic N, Roberts DJ, Ouwehand WH, Todd JA, Soranzo N, Suhre K, Paul DS, Fox CS, Plenge RM, Danesh J, Runz H, Butterworth AS, Genomic atlas of the human plasma proteome, Nature. 558 (2018) 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Phelan CM, Kuchenbaecker KB, Tyrer JP, Kar SP, Lawrenson K, Winham SJ, Dennis J, Pirie A, Riggan MJ, Chornokur G, Earp MA, Lyra PC, Lee JM, Coetzee S, Beesley J, McGuffog L, Soucy P, Dicks E, Lee A, Barrowdale D, Lecarpentier J, Leslie G, Aalfs CM, Aben KKH, Adams M, Adlard J, Andrulis IL, Anton-Culver H, Antonenkova N, AOCS study group, Aravantinos G, Arnold N, Arun BK, Arver B, Azzollini J, Balmaña J, Banerjee SN, Barjhoux L, Barkardottir RB, Bean Y, Beckmann MW, Beeghly-Fadiel A, Benitez J, Bermisheva M, Bernardini MQ, Birrer MJ, Bjorge L, Black A, Blankstein K, Blok MJ, Bodelon C, Bogdanova N, Bojesen A, Bonanni B, Borg Å, Bradbury AR, Brenton JD, Brewer C, Brinton L, Broberg P, Brooks-Wilson A, Bruinsma F, Brunet J, Buecher B, Butzow R, Buys SS, Caldes T, Caligo MA, Campbell I, Cannioto R, Carney ME, Cescon T, Chan SB, Chang-Claude J, Chanock S, Chen XQ, Chiew Y-E, Chiquette J, Chung WK, Claes KBM, Conner T, Cook LS, Cook J, Cramer DW, Cunningham JM, D’Aloisio AA, Daly MB, Damiola F, Damirovna SD, Dansonka-Mieszkowska A, Dao F, Davidson R, DeFazio A, Delnatte C, Doheny KF, Diez O, Ding YC, Doherty JA, Domchek SM, Dorfling CM, Dörk T, Dossus L, Duran M, Dürst M, Dworniczak B, Eccles D, Edwards T, Eeles R, Eilber U, Ejlertsen B, Ekici AB, Ellis S, Elvira M, EMBRACE Study, Eng KH, Engel C, Evans DG, Fasching PA, Ferguson S, Ferrer SF, Flanagan JM, Fogarty ZC, Fortner RT, Fostira F, Foulkes WD, Fountzilas G, Fridley BL, Friebel TM, Friedman E, Frost D, Ganz PA, Garber J, García MJ, Garcia-Barberan V, Gehrig A, GEMO Study Collaborators, Gentry-Maharaj A, Gerdes A-M, Giles GG, Glasspool R, Glendon G, Godwin AK, Goldgar DE, Goranova T, Gore M, Greene MH, Gronwald J, Gruber S, Hahnen E, Haiman CA, Håkansson N, Hamann U, Hansen TVO, Harrington PA, Harris HR, Hauke J, HEBON Study, Hein A, Henderson A, Hildebrandt MAT, Hillemanns P, Hodgson S, Høgdall CK, Høgdall E, Hogervorst FBL, Holland H, Hooning MJ, Hosking K, Huang R-Y, Hulick PJ, Hung J, Hunter DJ, Huntsman DG, Huzarski T, Imyanitov EN, Isaacs C, Iversen ES, Izatt L, Izquierdo A, Jakubowska A, James P, Janavicius R, Jernetz M, Jensen A, Jensen UB, John EM, Johnatty S, Jones ME, Kannisto P, Karlan BY, Karnezis A, Kast K, KConFab Investigators, Kennedy CJ, Khusnutdinova E, Kiemeney LA, Kiiski JI, Kim S-W, Kjaer SK, Köbel M, Kopperud RK, Kruse TA, Kupryjanczyk J, Kwong A, Laitman Y, Lambrechts D, Larrañaga N, Larson MC, Lazaro C, Le ND, Le Marchand L, Lee JW, Lele SB, Leminen A, Leroux D, Lester J, Lesueur F, Levine DA, Liang D, Liebrich C, Lilyquist J, Lipworth L, Lissowska J, Lu KH, Lubinoski J, Luccarini C, Lundvall L, Mai PL, Mendoza-Fandiño G, Manoukian S, Massuger LFAG, May T, Mazoyer S, McAlpine JN, McGuire V, McLaughlin JR, McNeish I, Meijers-Heijboer H, Meindl A, Menon U, Mensenkamp AR, Merritt MA, Milne RL, Mitchell G, Modugno F, Moes-Sosnowska J, Moffitt M, Montagna M, Moysich KB, Mulligan AM, Musinsky J, Nathanson KL, Nedergaard L, Ness RB, Neuhausen SL, Nevanlinna H, Niederacher D, Nussbaum RL, Odunsi K, Olah E, Olopade OI, Olsson H, Olswold C, O’Malley DM, Ong K-R, Onland-Moret NC, OPAL study group, Orr N, Orsulic S, Osorio A, Palli D, Papi L, Park-Simon T-W, Paul J, Pearce CL, Pedersen IS, Peeters PHM, Peissel B, Peixoto A, Pejovic T, Pelttari LM, Permuth JB, Peterlongo P, Pezzani L, Pfeiler G, Phillips K-A, Piedmonte M, Pike MC, Piskorz AM, Poblete SR, Pocza T, Poole EM, Poppe B, Porteous ME, Prieur F, Prokofyeva D, Pugh E, Pujana MA, Pujol P, Radice P, Rantala J, Rappaport-Fuerhauser C, Rennert G, Rhiem K, Rice P, Richardson A, Robson M, Rodriguez GC, Rodríguez-Antona C, Romm J, Rookus MA, Rossing MA, Rothstein JH, Rudolph A, Runnebaum IB, Salvesen HB, Sandler DP, Schoemaker MJ, Senter L, Setiawan VW, Severi G, Sharma P, Shelford T, Siddiqui N, Side LE, Sieh W, Singer CF, Sobol H, Song H, Southey MC, Spurdle AB, Stadler Z, Steinemann D, Stoppa-Lyonnet D, Sucheston-Campbell LE, Sukiennicki G, Sutphen R, Sutter C, Swerdlow AJ, Szabo CI, Szafron L, Tan YY, Taylor JA, Tea M-K, Teixeira MR, Teo S-H, Terry KL, Thompson PJ, Thomsen LCV, Thull DL, Tihomirova L, Tinker AV, Tischkowitz M, Tognazzo S, Toland AE, Tone A, Trabert B, Travis RC, Trichopoulou A, Tung N, Tworoger SS, van Altena AM, Van Den Berg D, van der Hout AH, van der Luijt RB, Van Heetvelde M, Van Nieuwenhuysen E, van Rensburg EJ, Vanderstichele A, Varon-Mateeva R, Vega A, Edwards DV, Vergote I, Vierkant RA, Vijai J, Vratimos A, Walker L, Walsh C, Wand D, Wang-Gohrke S, Wappenschmidt B, Webb PM, Weinberg CR, Weitzel JN, Wentzensen N, Whittemore AS, Wijnen JT, Wilkens LR, Wolk A, Woo M, Wu X, Wu AH, Yang H, Yannoukakos D, Ziogas A, Zorn KK, Narod SA, Easton DF, Amos CI, Schildkraut JM, Ramus SJ, Ottini L, Goodman MT, Park SK, Kelemen LE, Risch HA, Thomassen M, Offit K, Simard J, Schmutzler RK, Hazelett D, Monteiro AN, Couch FJ, Berchuck A, Chenevix-Trench G, Goode EL, Sellers TA, Gayther SA, Antoniou AC, Pharoah PDP, Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer, Nat. Genet. 49 (2017) 680–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Di Angelantonio E, Thompson SG, Kaptoge S, Moore C, Walker M, Armitage J, Ouwehand WH, Roberts DJ, Danesh J, INTERVAL Trial Group, Efficiency and safety of varying the frequency of whole blood donation (INTERVAL): a randomised trial of 45000donors,Lancet. 390 (2017) 2360–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rohloff JC, Gelinas AD, Jarvis TC, Ochsner UA, Schneider DJ, Gold L, Janjic N, Nucleic Acid Ligands With Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents, Mol Ther Nucleic Acids. 3 (2014) e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Machiela MJ, Chanock SJ, LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants, Bioinformatics. 31 (2015) 3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A, Enrichr: a comprehensive gene set enrichment analysis web server 2016 update, Nucleic Acids Res. 44 (2016) W90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Slenter DN, Kutmon M, Hanspers K, Riutta A, Windsor J, Nunes N, Mélius J, Cirillo E, Coort SL, Digles D, Ehrhart F, Giesbertz P, Kalafati M, Martens M, Miller R, Nishida K, Rieswijk L, Waagmeester A, Eijssen LMT, Evelo CT, Pico AR, Willighagen EL, WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research, Nucleic Acids Res. 46 (2018) D661–D667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, Butterworth AS, Staley JR, PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations, Bioinformatics. 35 (2019) 4851–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kuchenbaecker KB, Ramus SJ, Tyrer J, Lee A, Shen HC, Beesley J, Lawrenson K, McGuffog L, Healey S, Lee JM, Spindler TJ, Lin YG, Pejovic T, Bean Y, Li Q, Coetzee S, Hazelett D, Miron A, Southey M, Terry MB, Goldgar DE, Buys SS, Janavicius R, Dorfling CM, van Rensburg EJ, Neuhausen SL, Ding YC, Hansen TVO, Jønson L, Gerdes A-M, Ejlertsen B, Barrowdale D, Dennis J, Benitez J, Osorio A, Garcia MJ, Komenaka I, Weitzel JN, Ganschow P, Peterlongo P, Bernard L, Viel A, Bonanni B, Peissel B, Manoukian S, Radice P, Papi L, Ottini L, Fostira F, Konstantopoulou I, Garber J, Frost D, Perkins J, Platte R, Ellis S, EMBRACE, Godwin AK, Schmutzler RK, Meindl A, Engel C, Sutter C, Sinilnikova OM, GEMO Study Collaborators, Damiola F, Mazoyer S, Stoppa-Lyonnet D, Claes K, De Leeneer K, Kirk J, Rodriguez GC, Piedmonte M, O’Malley DM, de la Hoya M, Caldes T, Aittomäki K, Nevanlinna H, Collée JM, Rookus MA, Oosterwijk JC, Breast Cancer Family Registry, Tihomirova L, Tung N, Hamann U, Isaccs C, Tischkowitz M, Imyanitov EN, Caligo MA, Campbell IG, Hogervorst FBL, HEBON, Olah E, Diez O, Blanco I, Brunet J, Lazaro C, Pujana MA, Jakubowska A, Gronwald J, Lubinski J, Sukiennicki G, Barkardottir RB, Plante M, Simard J, Soucy P, Montagna M, Tognazzo S, Teixeira MR, KConFab Investigators, Pankratz VS, Wang X, Lindor N, Szabo CI, Kauff N, Vijai J, Aghajanian CA, Pfeiler G, Berger A, Singer CF, Tea M-K, Phelan CM, Greene MH, Mai PL, Rennert G, Mulligan AM, Tchatchou S, Andrulis IL, Glendon G, Toland AE, Jensen UB, Kruse TA, Thomassen M, Bojesen A, Zidan J, Friedman E, Laitman Y, Soller M, Liljegren A, Arver B, Einbeigi Z, Stenmark-Askmalm M, Olopade OI, Nussbaum RL, Rebbeck TR, Nathanson KL, Domchek SM, Lu KH, Karlan BY, Walsh C, Lester J, Australian Cancer Study (Ovarian Cancer Investigators), Australian Ovarian Cancer Study Group, Hein A, Ekici AB, Beckmann MW, Fasching PA, Lambrechts D, Van Nieuwenhuysen E, Vergote I, Lambrechts S, Dicks E, Doherty JA, Wicklund KG, Rossing MA, Rudolph A, Chang-Claude J, Wang-Gohrke S, Eilber U, Moysich KB, Odunsi K, Sucheston L, Lele S, Wilkens LR, Goodman MT, Thompson PJ, Shvetsov YB, Runnebaum IB, Dürst M, Hillemanns P, Dörk T, Antonenkova N, Bogdanova N, Leminen A, Pelttari LM, Butzow R, Modugno F, Kelley JL, Edwards RP, Ness RB, du Bois A, Heitz F, Schwaab I, Harter P, Matsuo K, Hosono S, Orsulic S, Jensen A, Kjaer SK, Hogdall E, Hasmad HN, Azmi MAN, Teo S-H, Woo Y-L, Fridley BL, Goode EL, Cunningham JM, Vierkant RA, Bruinsma F, Giles GG, Liang D, Hildebrandt MAT, Wu X, Levine DA, Bisogna M, Berchuck A, Iversen ES, Schildkraut JM, Concannon P, Weber RP, Cramer DW, Terry KL, Poole EM, Tworoger SS, Bandera EV, Orlow I, Olson SH, Krakstad C, Salvesen HB, Tangen IL, Bjorge L, van Altena AM, Aben KKH, Kiemeney LA, Massuger LFAG, Kellar M, Brooks-Wilson A, Kelemen LE, Cook LS, Le ND, Cybulski C, Yang H, Lissowska J, Brinton LA, Wentzensen N, Hogdall C, Lundvall L, Nedergaard L, Baker H, Song H, Eccles D, McNeish I, Paul J, Carty K, Siddiqui N, Glasspool R, Whittemore AS, Rothstein JH, McGuire V, Sieh W, Ji B-T, Zheng W, Shu X-O, Gao Y-T, Rosen B, Risch HA, McLaughlin JR, Narod SA, Monteiro AN, Chen A, Lin H-Y, Permuth-Wey J, Sellers TA, Tsai Y-Y, Chen Z, Ziogas A, Anton-Culver, Gentry-Maharaj A, Menon U, Harrington P, Lee AW, Wu AH, Pearce CL, Coetzee G, Pike MC, Dansonka-Mieszkowska A, Timorek A, Rzepecka IK, Kupryjanczyk J, Freedman M, Noushmehr H, Easton DF, Offit K, Couch FJ, Gayther S, Pharoah PP, Antoniou AC, Chenevix-Trench G, Consortium of Investigators of Modifiers of BRCA1 and BRCA2, Identification of six new susceptibility loci for invasive epithelial ovarian cancer, Nat. Genet. 47 (2015) 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang X, Tucker NR, Rizki G, Mills R, Krijger PH, de Wit E, Subramanian V, Bartell E, Nguyen X-X, Ye J, Leyton-Mange J, Dolmatova EV, van der Harst P, de Laat W, Ellinor PT, Newton-Cheh C, Milan DJ, Kellis M, Boyer LA, Discovery and validation of sub-threshold genome-wide association study loci using epigenomic signatures, Elife. 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Porta C, Paglino C, Mosca A, Targeting PI3K/Akt/mTOR Signaling in Cancer, Front Oncol. 4 (2014) 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ediriweera MK, Tennekoon KH, Samarakoon SR, Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer: Biological and therapeutic significance, Semin. Cancer Biol. 59 (2019) 147–160. [DOI] [PubMed] [Google Scholar]
- [24].Huang C-C, Cheng S-H, Wu C-H, Li W-Y, Wang J-S, Kung M-L, Chu T-H, Huang S-T, Feng C-T, Huang S-C, Tai M-H, Delta-like 1 homologue promotes tumorigenesis and epithelial-mesenchymal transition of ovarian high-grade serous carcinoma through activation of Notch signaling, Oncogene. 38 (2019) 3201–3215. [DOI] [PubMed] [Google Scholar]
- [25].Yao L-W, Wu L-L, Zhang L-H, Zhou W, Wu L, He K, Ren J-C, Deng Y-C, Yang D-M, Wang J, Mu G-G, Xu M, Zhou J, Xiang G-A, Ding Q-S, Yang Y-N, Yu H-G, MFAP2 is overexpressed in gastric cancer and promotes motility via the MFAP2/integrin a5ß1/FAK/ERK pathway, Oncogenesis. 9 (2020) 17 10.1038/s41389-020-0198-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gong X, Dong T, Niu M, Liang X, Sun S, Zhang Y, Li Y, Li D, lncRNA LCPAT1 Upregulation Promotes Breast Cancer Progression via Enhancing MFAP2 Transcription, Mol Ther Nucleic Acids. 21 (2020) 804–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wu M, Ding Y, Jiang X, Chen Y, Wu N, Li L, Wang H, Huang Y, Xu N, Teng L, Overexpressed MAGP1 Is Associated With a Poor Prognosis and Promotes Cell Migration and Invasion in Gastric Cancer, Front Oncol. 9 (2019) 1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Arakawa H, Netrin-1 and its receptors in tumorigenesis, Nat. Rev. Cancer. 4 (2004) 978–987. [DOI] [PubMed] [Google Scholar]
- [29].Hao W, Yu M, Lin J, Liu B, Xing H, Yang J, Sun D, Chen F, Jiang M, Tang C, Zhang X, Zhao Y, Zhu Y, The pan-cancer landscape of netrin family reveals potential oncogenic biomarkers, Sci Rep. 10 (2020) 5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Papanastasiou AD, Pampalakis G, Katsaros D, Sotiropoulou G, Netrin-1 overexpression is predictive of ovarian malignancies, Oncotarget. 2 (2011) 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Manichaikul A, Peres LC, Wang X-Q, Barnard ME, Chyn D, Sheng X, Du Z, Tyrer J, Dennis J, Schwartz AG, Cote ML, Peters E, Moorman PG, Bondy M, Barnholtz-Sloan JS, Terry P, Alberg AJ, Bandera EV, Funkhouser E, Wu AH, Pearce CL, Pike M, Setiawan VW, Haiman CA, African American Breast Cancer Consortium (AABC), African Ancestry Prostate Cancer Consortium (AAPC), Palmer JR, LeMarchand L, Wilkens LR, Berchuck A, Doherty JA, Modugno F, Ness R, Moysich K, Karlan BY, Whittemore AS, McGuire V, Sieh W, Lawrenson K, Gayther S, Sellers TA, Pharoah P, Schildkraut JM, African American Cancer Epidemiology Study (AACES) and the Ovarian Cancer Association Consortium (OCAC), Identification of novel epithelial ovarian cancer loci in women of African ancestry, Int J Cancer. 146 (2020) 2987–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lawrenson K, Song F, Hazelett DJ, Kar SP, Tyrer J, Phelan CM, Corona RI, Rodríguez-Malavé NI, Seo J-H, Adler E, Coetzee SG, Segato F, Fonseca MAS, Amos CI, Carney ME, Chenevix-Trench G, Choi J, Doherty JA, Jia W, Jin GJ, Kim B-G, Le ND, Lee J, Li L, Lim BK, Adenan NA, Mizuno M, Park B, Pearce CL, Shan K, Shi Y, Shu X-O, Sieh W, Australian Ovarian Cancer Study Group, Thompson PJ, Wilkens LR, Wei Q, Woo YL, Yan L, Karlan BY, Freedman ML, Noushmehr H, Goode EL, Berchuck A, Sellers TA, Teo S-H, Zheng W, Matsuo K, Park S, Chen K, Pharoah PDP, Gayther SA, Goodman MT, Genome-wide association studies identify susceptibility loci for epithelial ovarian cancer in east Asian women, Gynecol. Oncol. 153 (2019) 343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhu CS, Huang W-Y, Pinsky PF, Berg CD, Sherman M, Yu KJ, Carrick DM, Black A, Hoover R, Lenz P, Williams C, Hawkins L, Chaloux M, Yurgalevitch S, Mathew S, Miller A, Olivo V, Khan A, Pretzel SM, Multerer D, Beckmann P, Broski KG, Freedman ND, The Prostate, Lung, Colorectal and Ovarian Cancer (PLCO) Screening Trial Pathology Tissue Resource, Cancer Epidemiol. Biomarkers Prev. 25 (2016) 1635–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yarmolinsky J, Bull CJ, Vincent EE, Robinson J, Walther A, Smith GD, Lewis SJ, Relton CL, Martin RM, Association Between Genetically Proxied Inhibition of HMG-CoA Reductase and Epithelial Ovarian Cancer, JAMA. 323 (2020) 646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.