Abstract
We have successfully generated induced pluripotent stem cells (iPSC) from dermal fibroblasts of a patient with a homozygous p.Leu272Pro mutation in the gene encoding the linear deubiquitinase OTULIN. Biallelic loss of function mutations in this gene are responsible for the OTULIN deficiency termed Otulipenia or OTULIN-related autoinflammatory syndrome (ORAS). The iPSC carrying homozygous L272P OTULIN gene mutations are phenotypically normal and they have capacity to differentiate toward the three germ layers. These iPSC have great potential to study the role of linear ubiquitination in the regulation of immune responses and other cellular pathways.
1. -25167335560Resource Table
| Unique stem cell lines identifier | NIHTVBi014-A |
| Alternative names of stem cell lines | iPSC p342 |
| Institution | National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health (NIH), Bethesda, Maryland, USA |
| Contact information of distributor | Manfred Boehm; boehmm@nhlbi.nih.gov |
| Type of cell lines | iPSC |
| Origin | Human |
| Cell Source | Dermal fibroblasts |
| Clonality | Clonal cell lines |
| Method of reprogramming | Sendai-virus vectors containing the transcription factors Oct-4, Klf4, Sox2 and c-MYC |
| Multiline rationale | Lines derived from the individual |
| Gene modification | Yes |
| Type of modification | Hereditary |
| Associated disease | OTULIN-related autoinflammatory syndrome (ORAS) |
| Gene/locus | OTULIN, 5p15.2 |
| Method of modification | N/A |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | 2015 |
| Cell line repository/bank | N/A |
| Ethical approval | National Institutes of Health Ethics Committee (Approval Number: 15H0190) |
2. Resource utility
The human induced pluripotent stem cells (hiPSC) carrying homozygous L272P OTULIN gene mutations possess the potential of differentiating into variety of cell types including immune cells. The derivatives sustaining the mutation could be a powerful platform allowing for investigating the molecular mechanisms of disease, and for drug screening to search the targets for treatment of these patients.
3. Resource details
The OUTLIN is the key deubiquitinase (DUB) with the role to remove linear also known as Methionine-1 (M1)-linked ubiquitin chains from various signaling pathways, including the canonical NF-κB pathway, which regulate immune homeostasis and responses to infection (Keusekotten et al., 2013). It has been reported that M1-specific deubiquitinase OTULIN is essential for preventing TNF-associated systemic inflammation in humans and mice. Biallelic hypomorphic mutation in human OTULIN gene causes a potentially fatal autoinflammatory condition variably termed Otulipenia, OTULIN-related autoinflammatory syndrome (ORAS), or Autoinflammation, panniculitis, and dermatosis syndrome (AIPDS). This monogenic autoinflammatory disease is characterized by neonatal-onset of recurrent fever, erythematous rash with painful nodules and lipodystrophy, joint and gastrointestinal inflammation (Damgaard et al., 2016, Zhou et al., 2016). OTULIN deficiency is a rare and potentially lethal disease and very few patients have been reported in the literature. Patients present with a severe inflammatory phenotype that is responsive to treatment with corticosteroids or cytokine inhibitors. The hematopoietic stem cell transplantation (HSCT) could rescue the phenotype and has been successfully performed in one patient (Damgaard et al., 2019). However, the precise molecular mechanism of dysregulated M1-linked polyubiquitin signaling resulting in the disease is largely unknown.
We identified a male patient carrying the homozygous missense mutation p.Leu272Pro (614712.0001) in the OTULIN gene by exome sequencing. The patient was enrolled into our NHLBI clinical protocols for further investigations. Information regarding clinical features of this patient were obtained using the standard clinical questionnaire (Table 1). Skin punch biopsy samples were collected at the NIH Clinical Center. Using a Sendai-OKSM delivery system expressing four transcription factors (OCT4, SOX2, KLF4, and C-MYC), we have successfully generated iPSC (p342) from skin fibroblasts from the patient with the homozygous mutation in the OTULIN gene. We have also derived iPSC lines from healthy volunteers (Control) who did not have the mutation (data did not shown). The iPSC (p342) maintained typical morphologies and expressed phenotypically the pluripotency markers OCT4, NANOG, TRA-160, SSEA4, and SOX2, as shown by immunocytochemistry (Fig. 1A) and/or FACS (Fig. 1B), and/or real-time (RT)-qPCR (Fig. 1E). The iPSC (p342) were free of Sendai virus confirmed by RT-PCR at 15th passage (Fig. 1C). Genotyping of the generated iPSC (p342) showed the point mutation L272P in the OTULIN gene that were the same as their parental fibroblasts (Fig. 1D). To test the differentiation potential of the cells, we performed a monolayer differentiation assay to drive the cells towards the three germ layers in vitro. We determined the marker gene expression for the mesoderm (RUNX1), endoderm (AFP), and ectoderm (NESTIN) by RT-qPCR, which showed comparable expression levels between the iPSC (p342) and control lines (Control) (Fig. 1E). Short tandem repeat (STR) profiles indicated that the iPSC (p342) matched with its parental fibroblast completely in 15 amplified STR loci (see Supplementary file 1). All cultures were routinely tested for Mycoplasma contamination and were found to be Mycoplasma free as shown in Supplementary file 2. The iPSC (p342) demonstrated chromosomal stability and a normal karyotype with G-banding (Fig. 1F). In conclusion, the hiPSC (p342) exhibited the pluripotent potential for self-renew and differentiation, suggesting the successful generation of iPSCs from ORAS patient. To the best of our knowledge, this is the first report of human iPSC that were generated from a patient with OTULIN deficiency (see Table 2).
Table 1.
Summary of a patient with a homozygous L272P mutation in the OTULIN gene.
| iPSC line names | Abbreviation in figures | Gender | Age (years) | Ethnicity | Genotype of locus | Disease |
|---|---|---|---|---|---|---|
| NIHTVBi014-A | p342 | M | 11 | Pakistani | OTULIN, 5p15.2 | ORAS |
Fig. 1. Validation of human induced pluripotent stem cells (iPSC) generated from a patient with a homozygous L272P mutation in the OTULIN gene.
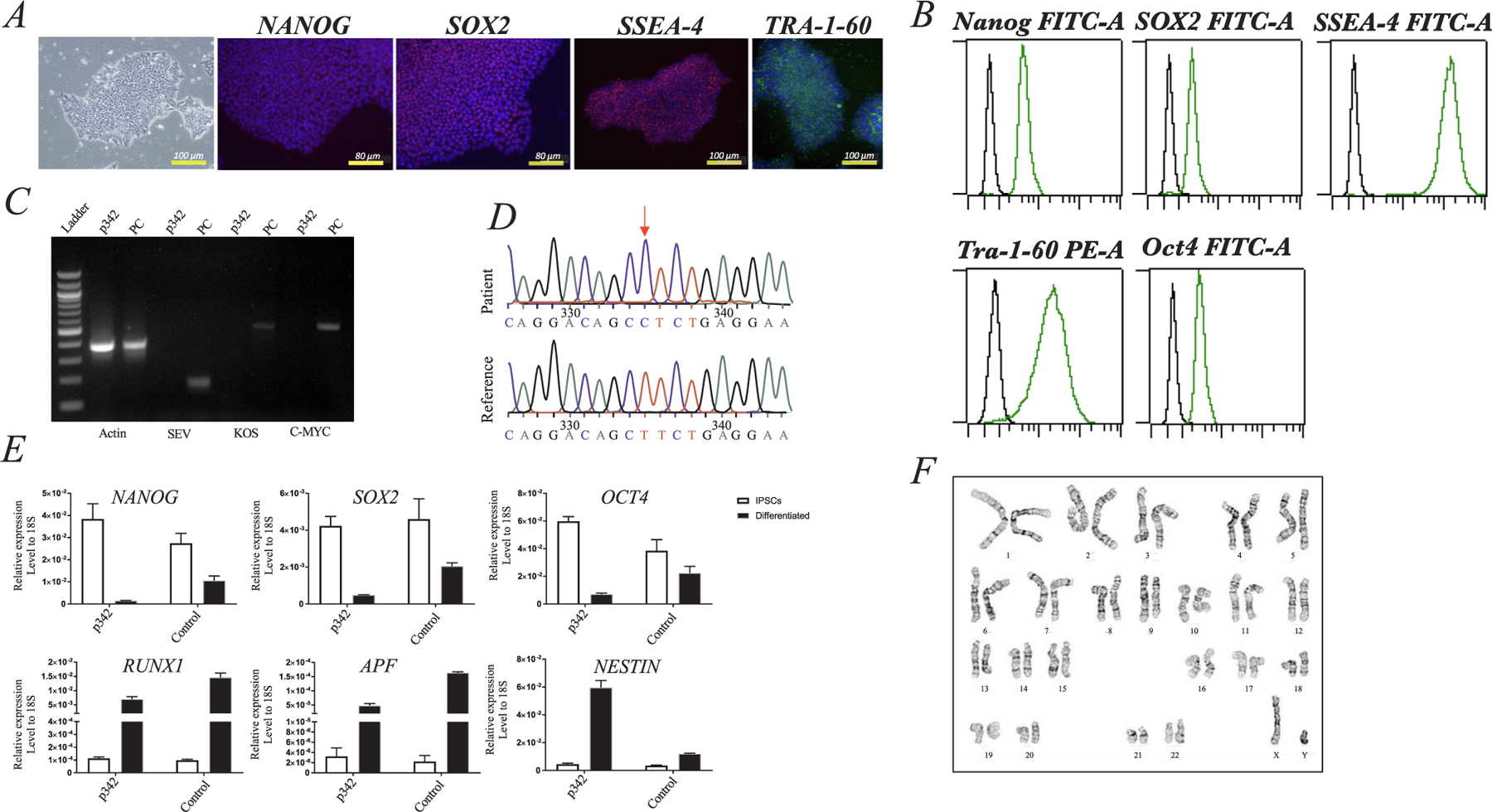
(A) Phase contrast images of iPSC clones were growing on passage 15 on a feeder-free plate. Expression of pluripotent markers (Nanog, Sox2, SSEA4, and Tra-1–60) were analyzed by immunofluorescence assay; DAPI staining of cell nuclei in blue. (B) The expression level of pluripotent markers (Nanog, Sox2, SSEA4, TRA-1–60, and Oct4) was quantitative analysis by Flow Cytometry Analysis. (C) qPCR demonstrated Sendai Virus free in iPSC at 15th passage. (D) PCR and DNA sequencing identified L272P mutation in the Otulin gene (red arrows). (E) Expression of pluripotent genes (NANOG, SOX2, and OCT3/4) was confirmed in iPSC derived from a ORAS patient as assessed by RT-qPCR (open bar). The iPSC from a ORAS patient were able to differentiate into three germ layers using monolayer differentiation in vitro at day 7, as shown by gene expression of AFP, NESTIN and RUNX1 (black bar). The All genes tested shown comparable expression levels between the iPSC from a ORAS patient and control lines (Control). Data are represented as means ± SEM relative to mRNA levels. (F) G-Banding assay showed normal chromosomal stability in a ORAS patient derived iPSC.
Table 2.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Phase-contrast microscope | Normal | Fig. 1A |
| Phenotype | Qualitative analysis (immunofluorescence staining) | Expression of pluripotency markers: OCT4, NANOG, SSEA4 and TRA-1–60 | Fig. 1A |
| Quantitative analysis (RT-qPCR) | Expression of pluripotency markers: SOX2 and NANOG | Fig. 1E | |
| Qualitative analysis (FACS) | Expression of pluripotency markers: NANOG, SOX2, SSEA4, TRA-1–60, OCT4 | Fig. 1B | |
| Genotype | Karyotype (G-banding) and resolution | 46, XY; resolution 450–500 bands | Fig. 1F |
| Identity | Microsatellite PCR OR STR analysis | Not performed | N/A |
| 15 sites tested, 100% match | Supplementary file 1 | ||
| Mutation analysis (IF APPLICABLE) | DNA sequencing | Homozygous, OTULIN gene point mutation | Fig. 1D |
| Southern blot OR WGS | Not performed | N/A | |
| Microbiology and virology | Mycoplasma testing by luminescence | Negative | Supplementary file 2 |
| Differentiation potential | Monolayer differentiation assay | Differentiating cells are expression of RUNX1, AFP, and NESTIN; iPSC were able to differentiate into three germ layers | Fig. 1E |
| Donor screening (OPTIONAL) | HIV1 + HIV2, hepatitis B virus, hepatitis C virus | Not performed | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | Not performed | N/A |
| HLA tissue typing | Not performed | N/A |
4. Materials and methods
4.1. Subjects and derivation of fibroblasts
The fibroblasts were derived from skin punch biopsy samples obtained from the ORAS patient who carries homozygous OTULIN gene mutation. Those cells were propagated in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum and 1% penicillin–-streptomycin, as previously described (Jin et al., 2016). This study was approved by the NHLBI’s institutional review board, and samples were collected after obtaining informed written consents.
4.2. Generation and culture of human iPSC from fibroblasts
Fibroblasts from the ORAS patient were reprogrammed for generating iPSC lines by using CytoTune™-iPS 2.0 Sendai Reprogramming Kit (Invitrogen). The iPSC colonies were picked up at 21 days posttransduction, and expanded in a typical hESC/iPSC culture condition (Jin et al., 2016).
4.3. Immunofluorescent staining and Flow cytometry analysis (FACS)
iPSC colonies were fixed with 4% paraformaldehyde and stained following the previous protocol (Jin et al., 2016). In brief, cells were incubated with primary antibodies against NANOG, SOX2, SSEA4, and TRA-1–60 (Table 3) at 4 °C overnight. Following washing with PBS, they were incubated with appropriate fluorophore-tagged secondary antibodies at room temperature for 1 h. After washing with PBS, nuclei were stained with DAPI. Images were captured using a fluorescence microscope (Zeiss).
Table 3.
Reagents.
| Antibodies used for immunocytochemistry | ||||||
|---|---|---|---|---|---|---|
| Antibody | Dilution | Company | Cat# | RRID | ||
| Primary antibodies | Rabbit anti-SOX2 | 1:100 | Cell Signaling Technology | 3579 | AB_2722343 | |
| Mouse anti-NANOG | 1:100 | Cell Signaling Technology | 4893 | AB_10548762 | ||
| Mouse anti-SSEA4 | 1:100 | MilliporeSigma | MAB4304 | AB_177629 | ||
| Mouse anti-TRA-1–60 | 1:150 | MilliporeSigma | MAB4360 | AB_2119183 | ||
| Alexa Fluor 488 anti-SSEA4 Antibody | 1:10 | BioLegend | 330412 | AB_1089198 | ||
| PE anti-TRA-1–60 Antibody | 1:10 | BioLegend | 330610 | AB_2119065 | ||
| Alexa Fluor 488 anti-SOX2 Antibody | 1:10 | BioLegend | 656110 | AB_2563957 | ||
| Alexa Fluor 488 anti-OCT4 Antibody | 1:10 | BioLegend | 653708 | AB_2563184 | ||
| Alexa Fluor 647 anti-NANOG Antibody | 1:10 | BioLegend | 674210 | AB_2650619 | ||
| Secondary antibodies | Alexa Fluor 594 Donkey anti-rabbit | 1:300 | Life Technologies | A21207 | AB_141637 | |
| Alexa Fluor 594 Donkey anti-mouse | 1:300 | Life Technologies | A21203 | AB_141633 | ||
| Alexa Fluor 488 Donkey anti-mouse | 1:300 | Life Technologies | A21202 | AB_141607 | ||
| Alexa Fluor 555 Goat anti-mouse | 1:300 | Life Technologies | A21426 | AB_2535847 | ||
| Primers used for RT-qPCR and PCR | ||||||
| Target | Forward/reverse primer (5′−3′) | |||||
|
| ||||||
| NANOG | AGG GAA ACA ACC CAC TTC T/CCT TCT GCG TCA CAC CAT T | |||||
| SOX2 | CCC AGC AGA CTT CAC ATG T/CCT CCC ATT TCC CTC GTT TT | |||||
| AFP | AGC TTG GTG GAT GAA AC/CCC TCT TCA GCA AAG CAG AC | |||||
| NESTIN | GCG TTG GAA CAG AGG TTG GA/TGG GAG CAA AGA TCC AAG AC | |||||
| RUNX1 | CTG CCC ATC GCT TTC AAG GT/GCC GAG TAG TTT TCA TTG CC | |||||
| OTULIN | TGT AAA ACG ACG GCC AGT GGA AAC CTG AAT GTT GTG AGC/AGG AAA CAG CTA TGA CCA TTA GAT CTT CCA GCC CCA GTC | |||||
| Sev | GGA TCA CTA GGT GAT ATC GAG C/ACC AGA CAA GAG TTT AAG AGA TAT GTA TC | |||||
| Sev c-MYC | TAA CTG ACT AGC AGG CTT GTC G/TCC ACA TAC AGT CCT GGA TGA TGA TG | |||||
| Sev Kos | ATG CAC CGC TAC GAC GTG AGC GC/ACC TTG ACA ATC CTG ATG TGG | |||||
| β-ACTIN | GAG AAG ATG ACC CAG ATC ATG TTT/GGC AGC TCG TAG CTC TTC TCC A | |||||
For FACS analysis, iPSC were dissociated into a single cell by using Trypsin-EDTA (Invitrogen). Following by fixation and permeabilization, the cells were stained with antibodies designed (Table 3). The data acquisition was performed on a MACSQuant Flow Cytometer (Miltenyi) and the results were analyzed with FlowJo software (FlowJo, LLC).
4.4. Monolayer differentiation assay
To assess differentiation ability of iPSC in vitro, cells were dissociated into small clumps with 0.5 μM EDTA and then cultured on Matrigel Precoated Plates (Corning) with differentiation medium consisting of 90% KnockOut DMEM, 10% FBS, 2 mM L-glutamine, 0.1 mM non-essential amino acids, and 0.1 mM 2-mercaptoethanol (Invitrogen). After seven days, cells were harvested for further analysis.
4.5. Gene expression analysis by RT-PCR
The total RNA was isolated by using RNeasy Mini Kits (Qiagen). cDNA was synthesized by reverse transcription (RT) using Super script™ III (Invitrogen).
After 15 passages, iPSC was tested for Sendai virus (SeV) residues. PCR was performed using the primers indicated in Table 3 and following the instructions as recommended by the manufacturer. The iPSC obtained at passage 1 served as positive control (PC) shown in Fig. 1C.
Endogenous mRNA expression levels of NANOG, SOX2, AFP, NESTIN, and RUNX1 were determined in iPSC (iPSC) and in differentiating cells at day 7 (differentiated) shown in Fig. 1E. For this, RT-qPCR was performed by using SYBR Green Premix on a Real-Time PCR Detection System (Bio-Rad). Assays were run in triplicate and the results were normalized to 18S ribosomal RNA expression. Primers used for RT-qPCR are shown in Table 3.
4.6. Karyotyping assay
The karyotype of the iPSC was evaluated by the WiCell Research Institute using G-banding metaphase karyotype analysis.
4.7. DNA sequencing and STR
Genomic DNA was extracted by using DNeasy Blood & Tissue Kit (Qiagen). To amplify the corresponding deletion position in OTULIN gene, PCR was performed with specific primers (Table 3). Following purification, the PCR products were sent to Eurofins Scientific for sequencing.
STR analysis was performed by WiCell Research Institute, which generated a STR profile via the Promega Powerplex® 16 System to verify STR polymorphisms for 15 loci plus amelogenin in genomic DNA extracted from iPSCs and their parental fibroblasts.
4.8. Mycoplasma detection
To validate the cultures of iPSC were Mycoplasma free, supernatant were collected after culturing for 48 h while the confluency of cells was at last 80%. The mycoplasma analysis was performed using the MycoAlert™ Mycoplasma Detection Kit (Lonza, LT27–224). The absence of mycoplasma contamination was confirmed in the culture tested (Supplement file 2).
Supplementary Material
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2020.101921.
References
- Keusekotten K, Elliott PR, Glockner L, Fiil B, Damgaard RB, Kulathu Y, Wauer T, Hospenthal MK, Gyrd-Hansen M, Krappmann D, Hofmann K, Komander D, 2013. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell 153, 1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard RB, Walker JA, Marco-Casanova P, Morgan NV, Titheradge HL, Elliott PR, McHale D, Maher ER, McKenzie ANJ, Komander D, 2016. The deubiquitinase OTULIN is an essential negative regulator of inflammation and autoimmunity. Cell 166, 1215–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Yu X, Demirkaya E, Deuitch N, Stone D, Tsai W-L, Kuehn HS, Wang H, Yang D, Park YH, Ombrello AK, Blake M, et al. , 2016. Biallelic hypomorphic mutations in a linear deubiquitinase define otulipenia, an early-onset autoinflammatory disease. Proc. Nat. Acad. Sci 113, 10127–10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard RB, Elliott PR, Swatek KN, Maher ER, Stepensky P, Elpeleg O, Komander D, Berkun Y, 2019. OTULIN deficiency in ORAS causes cell type-specific LUBAC degradation, dysregulated TNF signalling and cell death. EMBO Mol Med 11, e9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, St Hilaire C, Huang Y, Yang D, Dmitrieva NI, Negro A, Schwartzbeck R, Liu Y, Yu Z, Walts A, Davaine JM, Lee DY, Donahue D, Hsu KS, Chen J, Cheng T, Gahl W, Chen G, Boehm M, 2016. Increased activity of TNAP compensates for reduced adenosine production and promotes ectopic calcification in the genetic disease ACDC. Sci. Signal 9, ra121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


