Abstract
Background:
Substantial research has investigated the adverse effects of traffic-related air pollutants (TRAP) on human health. Convincing associations between TRAP and respiratory and cardiovascular diseases are known, but the underlying biological mechanisms are not well established. High-resolution metabolomics (HRM) is a promising platform for untargeted characterization of molecular mechanisms between TRAP and health indexes.
Objectives:
We examined metabolic perturbations associated with short-term exposures to TRAP, including carbon monoxide (CO), nitrogen dioxide (NO2), ozone (O3), fine particulate matter (PM2.5), organic carbon (OC), and elemental carbon (EC) among 180 participants of the Center for Health Discovery and Well-Being (CHDWB), a cohort of Emory University-affiliated employees.
Methods:
A cross-sectional study was conducted on baseline visits of 180 CHDWB participants enrolled during 2008-2012, in whom HRM profiling was determined in plasma samples using liquid chromatography-high-resolution mass spectrometry with positive and negative electrospray ionization (ESI) modes. Ambient pollution concentrations were measured at an ambient monitor near downtown Atlanta. Metabolic perturbations associated with TRAP exposures were assessed following an untargeted metabolome-wide association study (MWAS) framework using feature-specific Tobit regression models, followed by enriched pathway analysis and chemical annotation.
Results:
Subjects were predominantly white (76.1%) and non-smokers (95.6%), and all had at least a high school education. In total, 7,821 and 4,123 metabolic features were extracted from the plasma samples by the negative and positive ESI runs, respectively. There are 3421 features significantly associated with at least one air pollutant by negative ion mode, and 1691 features by positive ion mode. Biological pathways enriched by features associated with the pollutants are primarily involved in nucleic acids damage/repair (e.g., pyrimidine metabolism), nutrient metabolism (e.g., fatty acid metabolism), and acute inflammation (e.g., histidine metabolism and tyrosine metabolism). NO2 and EC were associated most consistently with these pathways. We confirmed the chemical identity of 8 metabolic features in negative ESI and 2 features in positive ESI, including metabolites closely linked to oxidative stress and inflammation, such as histamine, tyrosine, tryptophan, and proline.
Conclusions:
We identified a range of ambient pollutants, including components of TRAP, associated with differences in the metabolic phenotype among the cohort of 180 subjects. We found Tobit models to be a robust approach to handle missing data among the metabolic features. The results were encouraging of further use of HRM and MWAS approaches for characterizing molecular mechanisms underlying exposure to TRAP.
Keywords: Traffic-related air pollution, high-resolution Metabolomics, metabolomics-wide association study, pathway analysis
1. INTRODUCTION
Outdoor air pollution is an important environmental risk factor for human health all over the world (Lelieveld et al. 2015). With the fast development of urbanization and increasing number of vehicles on the roads, traffic-related air pollution (TRAP) have become a major source of ambient air pollution in urban areas, which contribute 25–40% of the ambient levels of major air pollutants (Greenbaum 2013; Health Effects Institute 2010). These TRAP pollutants include carbon monoxide (CO), nitrogen dioxide (NO2), fine particulate matter [PM2.5, with components such as elemental carbon (EC), organic carbon (OC), and metals] that are emitted directly from vehicles via combustion processes and tire and brake wear, along with ozone (O3), a secondary by-product (Greenbaum 2013).
There has been substantial research investigating the effects of TRAP on respiratory and cardiovascular diseases, where TRAP and its components were found to be associated with cardiopulmonary morbidity and mortality, with impacts on stroke, asthma exacerbation, impaired lung function, and non-asthmatic respiratory allergy (Health Effects Institute 2010). Over the last decade, more studies have further characterized health effects associated with TRAP and ambient air pollution for susceptible populations in a broader view of clinical outcomes. These epidemiological literature pointed to detrimental effects beyond the cardiovascular and respiratory systems, with observed associations of TRAP and its components with outcomes such as diabetes mellitus, hypertension disorder of pregnancy, preterm birth, low birth weight, and neurotoxicity (Brauer et al. 2008; Costa et al. 2017; Eze et al. 2015; National Toxicology Program 2019; Wilhelm et al. 2011).
Given the abundance of studies reporting TRAP-related adverse health outcomes, it is critical to identify biological mechanisms underlying the effects of TRAP. Systemic inflammatory markers, oxidative stress factors, and cell counts in blood have been the main endpoints measured in the more recent studies, which were considered as proxies of TRAP-induced internal perturbation (Carvalho et al. 2018; Chiu et al. 2016; Golan et al. 2018; Jacobs et al. 2010; Krishnan et al. 2013; Kubesch et al. 2015; Sarnat et al. 2014; Zuurbier et al. 2011). However, controversies exist among these studies, which are likely largely due to the lack of robust and specific biomarkers that accurately reflect TRAP exposure or the corresponding effects. In addition, some researchers have employed plasma circulating microRNAs, whole blood RNA, or mitochondrial abundance to identify changes in gene expression following exposure to TRAP; the findings indicate some molecular mechanisms involved in the pathogenesis of multiple diseases, such as breast and lung cancers, and cardiovascular diseases (Chu et al. 2016; Krauskopf et al. 2018; Zhong et al. 2016). Overall, research is in progress to provide insights that TRAP can induce diverse biological responses in the human body, while the underlying molecular mechanisms remain largely inconclusive. Exploration of novel internal biomarkers which could sensitively reflect the direct or indirect health responses to exposure to TRAP is warranted.
High-resolution metabolomics (HRM) is an advanced analytical method for identification of internal metabolites (e.g., present in a given biological media, such as blood or saliva). This method provides new opportunities for epidemiologists to investigate the associations of external exposures with endogenous processes at the molecular level (Jones et al. 2012; Uppal et al. 2016). In most prior work, targeted methods have been used to identify and quantify a defined set of metabolites (e.g., targeted biomarkers of inflammation or oxidative stress) in a single sample run. In recent years, untargeted HRM has emerged as a powerful platform to improve internal exposure estimation to complex environmental mixtures by providing identification and quantitation of thousands of metabolites in biological samples associated with endogenous and exogenous processes (Bundy et al. 2009; Lankadurai et al. 2013; Miller and Jones 2014; Uppal et al. 2016). Several previous studies have demonstrated the applicability of using high-resolution metabolomics as a central platform linking TRAP exposures to internal dose and biological responses (Chen et al. 2019; Ladva et al. 2018; Liang et al. 2018b; Liang et al. 2019; van Veldhoven et al. 2019).
To expand on this growing body of research and further explore the biological pathways perturbed by TRAP exposures, we performed a cross-sectional study nested in a cohort with a sample size of 180 participants. In the present study, we followed an untargeted HRM MWAS workflow to identify the biological pathways perturbed by TRAP among participants at baseline in the Center for Health Discovery and Wellbeing (CHDWB), a cohort of Emory University-affiliated employees in Atlanta, Georgia, USA. The CHDWB cohort was an observational study designed to investigate deep clinical and metabolic phenotyping and the effects of clinical self-knowledge and health partner counseling (Tabassum et al. 2014). We applied untargeted HRM to 180 plasma samples collected at participants’ baseline visit to obtain HRM profiles and applied these data in epidemiologic analyses using Tobit regression model to identify metabolic features associated with short-term exposures to ambient CO, NO2, O3, PM2.5, EC, and OC. Pathway enrichment analysis and chemical annotation were conducted to identify biological pathways enriched by significant metabolic features and validate the observed untargeted metabolic associations.
2. MATERIAL AND METHODS
2.1. Study Design
The present study was a cross-sectional design that included the baseline visits of 180 participants in the CHDWB cohort. Details of the cohort can be found elsewhere (Brigham 2010; Rask et al. 2011; Tabassum et al. 2014). Briefly, the CHDWB cohort was initiated in May 2008 and randomly recruited employees affiliated with Emory University from 2008 to 2012. The participants were free of poorly controlled chronic diseases or acute illness at the time of recruitment. Basic demographics and plasma samples were collected during the clinical visit, along with tobacco and alcohol usage. A total of 180 participants completed the baseline visits and had their plasma samples analyzed by untargeted HRM profiling at the Clinical Biomarkers Laboratory at Emory University (Bellissimo et al. 2019). All participants provided informed consent, and the study was approved by the Emory University Institutional Review Board.
2.2. Exposure Assessment
For the study period (2008-2012), continuous measurements of CO, NO2, O3, PM2.5, EC, and OC were made at Jefferson St. (JST), an ambient monitoring site near downtown Atlanta. Measurements from the JST site have been used previously to generate population exposure estimates in analyses examining short-term health effects associated with ambient air pollution exposures (Darrow et al. 2009; Metzger et al. 2003; Strickland et al. 2010; Tolbert et al. 2000) and is generally considered to be representative of daily variation in Atlanta urban background pollutant concentrations and composition (Edgerton et al. 2005; Liang et al. 2018a; Solomon et al. 2002). The details of site information and measure methods can be found elsewhere (Hansen et al. 2006). Briefly, CO was measured continuously with 1-min resolution using a TEI Model 48S NDIR analyzer. NO2 was not directly measured but converted photolytically from nitrogen monoxide (NO), and NO was measured continuously using a TEI Model 42ctl analyzer via chemiluminescence. O3 was measured by UV-based ozone analyzer. All trace gases were aggregated to the daily level and reported as daily 1-hr maximum values (for CO and NO2) or daily 8-hr maximum values (for O3). PM2.5 was measured continuously with an R&P Model 1400 a/b tapered element oscillating microbalance (TEOM). OC and EC were measured using an R&P Model 5400 ambient particulate carbon monitor with 60-min resolution. PM2.5 and its components were reported as daily 24-hr averages. The daily concentration of each pollutant was assigned to each participant according to the date of their baseline visit. Daily meteorological data were obtained from the Atlanta Hartsfield-Jackson International Airport.
2.3. High-Resolution Metabolomics
In total, we collected and analyzed 180 plasma samples using established protocols (Ladva et al. 2017; Liang et al. 2018b; Liang et al. 2019). Briefly, the biological samples were randomized into blocks of 20 for de-identifying and blinding. A pooled plasma sample had been referenced against the National Institute of Standards and Technology (NIST) 1950 standard reference material and added to each analytical batch. Samples underwent the following process before the analysis: 65 μL of plasma stored at −80°C was added to 130 μL of acetonitrile which contains 3.5 μL mixture of 14 stable isotope standards (Soltow et al. 2013). Precipitated proteins were pelleted via centrifugation for 10 min at 4°C and 14,100 × g after mixing and incubation at 4°C for 30 min. Following protein precipitation, triplicate 10 mL aliquots were analyzed by reverse-phase C18 liquid chromatography with Fourier transform mass spectrometry (Dionex Ultimate 3000, Q-Exactive, Thermo Scientific, Waltham, MA). The analysis was operated in either negative or positive electrospray ionization (ESI) mode, since having data from both modes should provide a more comprehensive profile of metabolism than data extracted from only one mode; compounds are ionized with different ionization efficiency between the positive and negative ion mode, which results in varying sensitivity and detection limits (Liigand et al. 2017). Each compound would have a mass-to-charge (m/z) ratio with the within 85 to 1275 and a retention time that the compound needed to pass through the chromatography column. Data from each analytical run was saved in both the .RAW format and converted to the .mzMl format using ProteoWizard (v.3). Then, metabolic profiles were extracted by apLCMS with modifications using the R package xMSanalyzer (Uppal et al. 2013). The detailed codes for xMSanalyzer are provided in the Supplemental Material. Briefly, two sets of minimum length of elution time and minimum proportion of scans were employed, as xMSanalyzer is able to merge features depending on the correlation (0.7) between two features with same m/z and retention time (defined tolerance levels) detected at these two settings. Only features with median coefficient of variance or percent intensity difference among all samples within technical replicates < 70% were included to maintain feature consistency within technical replicates.
2.4. Data Analysis
Associations of air pollution with metabolic features and associated pathways were assessed following an untargeted MWAS workflow which comprises data-driven significant feature detection using regression models, knowledge-driven methods for pathway analysis, and feature annotation/identification (Uppal et al. 2016). This workflow facilitates the detection of metabolite and metabolic pathways that are associated with the exposure of interest among high-throughput metabolomics data. To prepare the HRM data for data analysis, extracted metabolic features present in less than 10% of the participants were excluded. This was done in order to reduce the background noise generated by the measurement equipment (Alonso et al. 2015), and also as a means of focusing the analytic dataset to more common metabolites (i.e., endogenous metabolites that are common across people) in order to facilitate detection of biological pathways later in the analysis. The missing values of features was primarily the result of detection limitations of the analytical method. In other words, we did not know the “true” signal intensity below the method’s limit of detection (LOD); i.e., the data were left-censored. The Tobit model is designed to estimate linear relationships when there is censoring in the dependent variable, and assumes that the dependent variable follows a censored normal distribution. In the case of censoring, the Tobit model can yield an unbiased coefficient estimate for the independent variable whereas the coefficient estimate may be biased when using a usual linear regression model (McBee 2010). We considered the minimum value of feature intensity among the full metabolomics dataset as LOD in the present study.
Tobit regression models were conducted for each air pollutant-feature pair, controlling for potential temporal confounders. We also assessed a range of demographic and lifestyle factors as potential (non-temporal) confounders, including age, race/ethnicity, gender, marital status, annual household income ($/year), education, smoking status, consumption of alcoholic beverages, and body mass index; Since no variables were consistently significantly associated with 6 air pollutants (Table S2), we did not include demographic or lifestyle factors in the regression models. The basic form of the model was:
where refers to the observed intensity of metabolic feature m for individual i; refers to the latent log-transformed feature intensity; refers to the coefficient of metabolic feature m for the air pollutant, indicating the change in feature intensity for a one unit increase in pollution. Lag 1-day (i.e., previous day) pollutant concentrations were fitted; since the lag effect of air pollution was reported in our previous study, we also considered the moving average of lag 1-2 day concentrations (Strickland et al. 2010). These two exposure windows were compared to each other to better capture the distribution of air pollution effects over time. In the regression model, variables Year (3-level: 2008, 2009, >2009), season (4-level: spring, summer, fall, winter), and weekday (5-level) were determined by the date of baseline visit. Apparent_temp was included in the model with the lag or moving average corresponding to that of the pollutant exposures included in the model, and was computed using the daily mean air temperature in combination with the daily mean dew point (Steadman 1984). We introduced a linear term and a quadratic term of apparent temperature into the model to account for potential nonlinear relationships between apparent temperature and metabolic features. Associations between pollutant and individual metabolic feature were visualized using Manhattan plots that displayed negative log10 of p-values, with the retention time of metabolic feature i on the x-axis against the −log10(p) for on the y-axis. Multiple comparison correction was conducted using the Benjamini-Hochberg false discovery rate (FDRB-H) procedure, a widely used procedure in MWAS study, at a 5% false positive threshold. All analyses were performed in R (v.3.5.1).
2.5. Pathway Analysis and Chemical Annotation
To predict biological functions and molecular mechanisms associated with these significant features, pathway analysis was conducted using mummichog (v.2.0.1), a novel bioinformatics platform that infers and categorizes functional biological activity directly from mass spectrometry output, without prior metabolite validation (Li et al. 2013). Briefly, metabolic features were separated into two groups based on their statistical significance in regression models with pollutant concentrations, and mummichog made putative annotations of each feature by mapping them to its metabolite database based on their m/z. The insignificant metabolic features were selected at random and mapped to known metabolite pathways, repeating the process 1000 times to estimate the null distribution of pathway activities. The significant group of annotated features were also mapped to the known pathways, and over-represented pathways were detected by Fisher’s exact test (FET) using the Gaussian hypergeometric probability distribution. An adjusted p-value for each pathway was generated based on all p-values of FET for all pathways and the null distribution calculated in the previous step, which weights significance in favor of pathways enriched by more significant features. We applied two strategies to select eligible metabolic features for pathway analysis: (i) at raw p-values < 0.05; (ii) at adjusted p-values < 0.05 using the Benjamini-Hochberg method for multiple comparison correction. For the first approach, to control for false positive discovery rate, we excluded pathways identified by mummichog with a p-value higher than 0.05 and those containing less than 3 significant metabolic features that were matched with known compounds by m/z. To further reduce the possibility of false positive findings, each of the pollutant-driven metabolic features was screened for spectrum peak quality and purity by manual examination of their respective extracted ion chromatographs (EICs). Finally, we confirmed a selected number of annotated metabolites by comparison of m/z, retention time and ion dissociation patterns to authentic chemical reference standards analyzed in our lab using the identical method and instrument parameters via tandem mass spectrometry.
2.6. Sensitivity Analysis
Feature-specific regression analyses, pathway enrichment analysis, and chemical annotation were also performed using regular multiple linear regression (MLR) models as a sensitivity analysis. For MLR analyses, missing values (i.e., metabolic features missing for a given participant) were assigned the half of the minimum feature intensity (also defined as the limit of detection) observed across all metabolic features in the dataset (note that the Tobit model, given its design, does not require such imputation). Otherwise, the MLR model was constructed similar to the Tobit model, with the same covariate control. The performance of MLR models was compared to that of Tobit models based on the total number of significant metabolic features detected for each pollutant, and based on comparisons of the p-values calculated by either the Tobit or MLR models relative to each feature’s percent presence among participants given the assumption that the significance of features with fewer missing values should be more robust across different statistical methods.
3. RESULTS
Baseline information for the 180 participants is shown in Table 1. Three-quarters of the participants were over the age of 42, and 76.1% of them were white. Over half of participants had completed graduate school. They predominantly were not current smokers (95.6%). Most baseline visits (95.0%) were conducted between 2008 and 2009, and all visits occurred on weekdays (none on the weekend).
Table 1.
Participant characteristics and the temporal characteristics of baseline visits.
| Characteristics | Number | Proportion (%) |
|---|---|---|
| Age, years [median (Q1-Q3)] | 51.0 (42-57) | |
| BMI, kg/m2 [median (Q1-Q3)] | 26.4 (23.6-29.7) | |
| Race/ethnicity | ||
| White | 137 | 76.1 |
| Black | 34 | 18.9 |
| Other racesa | 9 | 5.0 |
| Gender | ||
| Female | 113 | 62.8 |
| Male | 67 | 37.2 |
| Marital status | ||
| Married | 117 | 65.0 |
| Other statusesb | 63 | 35.0 |
| Annual household income ($/year) | ||
| 0-50,000 | 17 | 10.1 |
| 50,000-100,000 | 44 | 26.0 |
| 100,000-200,000 | 58 | 34.3 |
| 200,000+ | 50 | 29.6 |
| Missing | 11 | |
| Education | ||
| College and high school | 78 | 43.3 |
| Graduate school and above | 102 | 56.7 |
| Smoking status | ||
| Non-smoker | 172 | 95.6 |
| Current smoker | 8 | 4.4 |
| Consumption of alcoholic beverages | ||
| Yes | 139 | 77.2 |
| No | 41 | 22.8 |
| Year of visit | ||
| 2008 | 76 | 42.2 |
| 2009 | 95 | 52.8 |
| Over 2009 | 9 | 5.0 |
| Weekday of visit | ||
| Monday | 39 | 21.7 |
| Tuesday | 37 | 20.6 |
| Wednesday | 40 | 22.2 |
| Thursday | 30 | 16.7 |
| Friday | 34 | 18.9 |
| Season of visit | ||
| Spring | 29 | 16.1 |
| Summer | 63 | 35.0 |
| Autumn | 44 | 24.4 |
| Winter | 44 | 24.4 |
Other races includes American Indian or Alaskan Native and Asian.
Other statuses includes single, divorced, widowed, separated, and partnered.
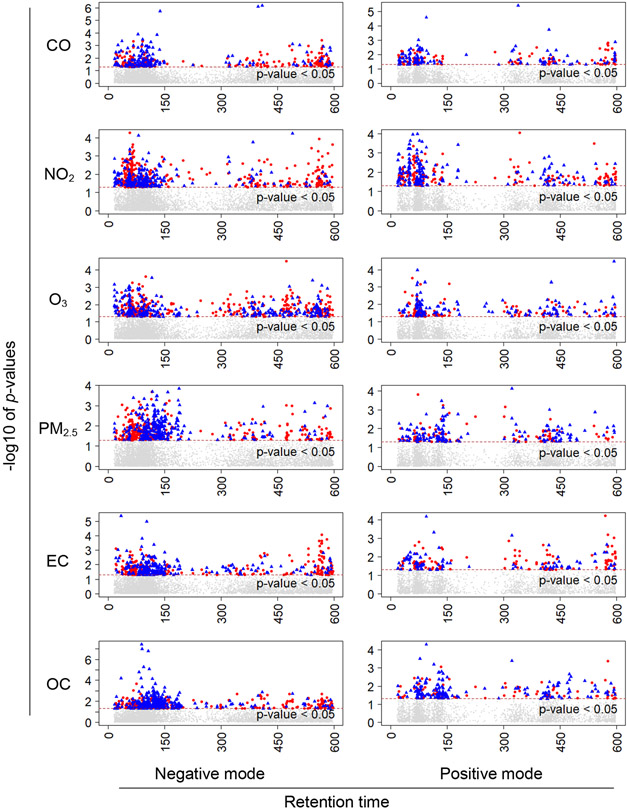
We reliably (i.e., using two combinations of relatively stringent parameters for feature detection in xMSanalyzer (Uppal et al. 2013)) extracted 7,821 metabolic features by negative ion mode and 4,123 by positive ion mode in plasma samples with relatively good data quality. After data filtering for removing features that were present in less than 10% of participants, the data contained 7,106 features from negative ESI and 3,628 features from positive ESI, respectively. We performed Tobit models for all pollutants of interest. Numbers of significant features (p-value ≤ 0.05) for the pollutants are summarized in Table 2. There are 3421 features significantly associated with at least one air pollutant by either exposure window by negative ion mode, and 1691 features by positive ion mode. Figure 1 and Figure S1 depict the results for each pollutant in Manhattan plots displaying the −log10 p values of each metabolic feature against its retention time, by exposure time window lag 1 and moving average of lag 1-2 respectively.
Table 2.
Number of significant metabolic features by negative and positive electrospray ion mode associated with lag 1 day (i.e., previous day) and the moving average of lag 1-2 days pollution from Tobit models.
| Lag* | Negative mode a | Positive mode a | |
|---|---|---|---|
| CO | L1 | 400 | 216 |
| MA | 465 | 173 | |
| NO2 | L1 | 618 | 338 |
| MA | 526 | 280 | |
| O3 | L1 | 603 | 245 |
| MA | 376 | 247 | |
| PM2.5 | L1 | 603 | 259 |
| MA | 545 | 244 | |
| EC | L1 | 498 | 208 |
| MA | 606 | 230 | |
| OC | L1 | 581 | 251 |
| MA | 473 | 254 |
L1, the exposure at lag 1 day; MA, the moving average of exposure at lag 1-2 days; EC, elemental carbon, OC, organic carbon.
Metabolic features were statistically significant with p-values less than 0.05.
Figure 1. Manhattan plots of associations between log-transformed metabolic feature intensity and 1-day lag concentration of air pollutants from Tobit models.
X-axis denotes the retention time (in seconds), Y-axis denotes the negative log10 of the p-values calculated from the Tobit model. Significant features with p-values less than 0.05 are colored. Blue triangles and red circles denote negative and positive associations, respectively.
3.1. Pathway Enrichment Analysis
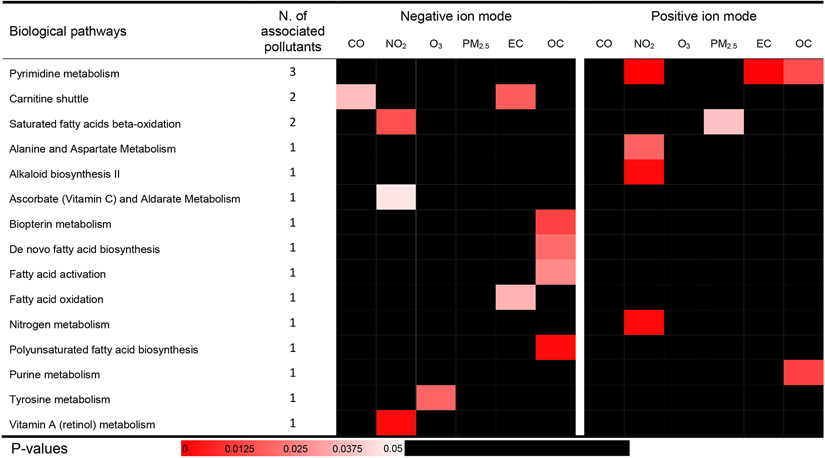
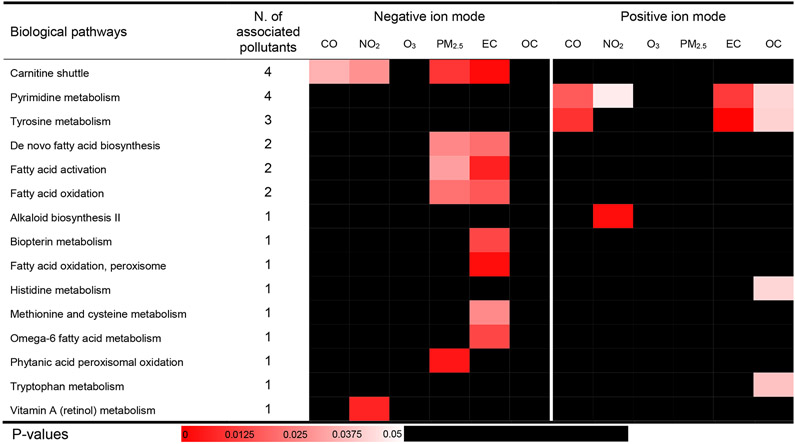
We performed pathway enrichment analyses and examined whether the features associated with the pollutants of interest co-occurred as enriched metabolites within specific metabolic pathways. Twenty-one biological pathways were associated with at least one pollutant (Figures 2 and 3) by either exposure time window. The significant pathways mainly pertain to nucleic acids damage and repair pathways (i.e., purine metabolism, pyrimidine metabolism), nutrient metabolism (i.e., fatty acid metabolism, vitamin A metabolism), and acute inflammation (i.e., histidine metabolism, tyrosine metabolism, alanine and aspartate metabolism). The 1-day lag and 1-2 day MA results share most biological pathways, especially fatty acid metabolism and nucleotide metabolism. There was no overlap in identified pathways between features detected in negative ESI mode and those detected in positive ESI mode, which could be due to the differences in features identified by each mode. Although thousands of metabolic features were detected in the negative and positive ESI modes, only 71 features were detected in both modes based on matching m/z with a tolerance of 10 ppm and retention time with a tolerance of 10 s. The majority of these matched features were weakly correlated, which indicated that they were not the same compounds, and the few matched features with a correlation coefficient > 0.5 were not significantly associated any air pollutants. Pyrimidine metabolism was associated with the most pollutants, including 1-day lag concentrations of NO2, EC, and OC, and the moving average of 1-2 day lag concentrations of CO, NO2, PM2.5, and EC. The 1-day lag concentration of NO2 was associated with seven biological pathways. The moving average concentration of EC was associated with ten pathways, five of which are involved in lipid metabolism. More than half of the biological pathways identified were observed for both lag periods assessed (i.e., Figure 2 & 3; pyrimidine metabolism, carnitine shuttle, alkaloid biosynthesis II, biopterin metabolism, de novo fatty acid biosynthesis, fatty acid activation, fatty acid oxidation, tyrosine metabolism and vitamin A metabolism), which could be explained partly by the high correlations between the 1-day lag and 2-day moving average concentrations (Figure S8). However, several biological pathways were exclusively associated with the 2-day moving averages, such as PM2.5, which may be explained by the considerable lagged effect of PM2.5 (Lim et al. 2016). Given the number of significant metabolic features after FDR correction was too low to conduct the pathway analysis, no biological pathways were detected in mummichog based on the standard (ii), since the input number of significant metabolic features was too low.
Figure 2. Metabolic pathways associated with 1-day lag pollution from Tobit models.
Cells are shaded according to the magnitude of p-values generated by mummichog for each pathway. Pathways enriched by more than 2 annotated significant metabolic features were included and ordered alphabetically and following the total number of the significant associations between pathways and pollutants by either negative or positive ion modes.
Figure 3. Metabolic pathways associated with the moving average of lag 1-2 days pollution from Tobit models.
Cells are shaded according to the magnitude of p-values generated by mummichog for each pathway. Pathways enriched by more than 2 annotated significant metabolic features were included and ordered alphabetically and following the total number of the significant associations between pathways and pollutants by either negative or positive ion modes.
3.2. Feature annotation
We confirmed the chemical identity of 8 metabolic features (features matched to multiple metabolites are excluded) from negative ESI (Table 3) and 2 metabolic features from positive ESI (Table 4) showing significant associations with one or more air pollutants with level one evidence (i.e., the proposed metabolite has been confirmed via appropriate measurement of an authentic reference standard with the same experimental conditions as the present plasma samples) (Schymanski et al. 2014). Their corresponding parameter estimates from Tobit models are also shown, indicating whether the metabolites were positively or negatively associated with the pollutants (Table 3).
Table 3.
Metabolites matched with the metabolic features extracted via negative ion mode and significantly associated with air pollutants in Tobit models.
| Experimental mass, m/z |
Retention time (s) |
Metabolite matched | Associated pollutants * |
Coefficients |
|---|---|---|---|---|
| 105.0178 | 146.7 | Glycerate | CO MA EC MA |
−0.35 −0.28 |
| 110.0709 | 434.4 | Histamine | CO MA NO2 L1 NO2 MA EC L1 EC MA |
−9.97 −0.14 −0.32 −7.56 −7.64 |
| 118.0495 | 112.3 | Allothreonine Threonine Homoserine |
CO MA PM2.5 L1 EC L1 EC MA |
0.19 0.01 0.18 0.16 |
| 129.0543 | 110.3 | Alpha-Ketoisocaproic acid | EC MA | 0.15 |
| 130.0859 | 114.0 | Norleucine Isoleucine Leucine |
EC MA | 0.20 |
| 132.029 | 119.7 | Aspartate | OC MA | −0.07 |
| 148.0425 | 102.5 | Methionine | EC MA | 0.16 |
| 164.0706 | 101.2 | Phenylalanine | EC L1 EC MA OC L1 OC MA |
0.18 0.19 0.06 0.06 |
| 180.0655 | 86.8 | Tyrosine | EC L1 EC MA |
0.15 0.21 |
| 191.0188 | 98.3 | Citrate Isocitric acid |
EC L1 EC MA |
−0.78 −0.77 |
| 203.082 | 94.7 | Tryptophan | EC L1 EC MA |
0.17 0.15 |
L1, the exposure at lag 1 day; MA, the moving average of exposure at lag 1-2 days; EC, elemental carbon, OC, organic carbon.
Significance at alpha = 0.05.
Table 4.
Metabolites matched with the metabolic features extracted via positive ion mode and significantly associated with air pollutants by Tobit models.
| Experimental mass, m/z |
Retention time (s) |
Metabolite matched | Associated pollutants * |
Coefficients |
|---|---|---|---|---|
| 90.0555 | 116.2 | Sarcosine Alanine |
EC MA | 0.15 |
| 113.0350 | 330.7 | Uracil | CO MA NO2 L1 NO2 MA EC L1 EC MA OC L1 |
3.86 0.10 0.13 3.79 3.25 1.16 |
| 116.0709 | 122.5 | Proline | OC L1 OC MA |
0.05 0.06 |
| 118.0865 | 93.3 | Betaine Valine Norvaline Aminopentanoate |
EC L1 EC MA |
0.10 0.11 |
| 120.0657 | 130.1 | Threonine Homoserine Allothreonine |
PM2.5 L1 EC L1 |
0.01 0.11 |
| 123.0452 | 106.4 | Hydroxybenzaldehyde Benzoate |
PM2.5 L1 OC L1 OC MA |
−0.01 −0.05 −0.81 |
L1, the exposure at lag 1 day; MA, the moving average of exposure at lag 1-2 days; EC, elemental carbon, OC, organic carbon.
Significance at alpha = 0.05.
3.3. Sensitivity Analysis
Feature-specific MLR models were performed in place of Tobit models as a sensitivity analysis. The results from MLR models are shown in Table S3 and Figures S2-S3 in the Supplemental Material. The significant pathways identified using MLR model results (Figures S4 and S5) were mostly contained within the results observed using Tobit models (Figures 2 and 3). We further compared the performance of these two modeling approaches based on their p-values of the same feature, and the distribution of those features with respect to their % presence among participants. As shown in Table S3, Tobit models identified more significant features than did MLR models. While p-values resulting from the two regression approaches were highly correlated for CO and PM2.5 (Figures S6-S7, the rest air pollutants had the same pattern as CO), there were instances of dramatic differences in p-values for the same feature, in particular several features that were significant (i.e., p-value < 0.05) in MLR models but had a p-value of 1.00 in Tobit models.
4. DISCUSSION
In the present study, we applied HRM to identify metabolic alterations associated with exposures to ambient CO, NO2, O3, PM2.5, OC, and EC in 180 adults from an Emory University-based employee cohort using a cross-sectional design. We identified several biological pathways that were associated with one or more air pollutants. The identified pathways are involved in various biochemical processes in the human body, including nucleic acids damage and repair (pyrimidine metabolism and purine metabolism), nutrient metabolism (e.g., fatty acid beta oxidation, tryptophan metabolism, vitamin A metabolism), and acute inflammation (e.g., histidine metabolism, tyrosine metabolism, alanine and aspartate metabolism). These biochemical processes are responsible for maintaining homeostasis and wellbeing in humans.
We found that pyrimidine metabolism was specifically associated with short-term exposures to traffic-related pollutants, including 1-day lag concentrations of NO2, EC, and OC, and the moving average of 1-2 day lag concentrations of CO, NO2, EC, and OC, respectively. Purine metabolism was associated with 1-day lag concentration of OC. One of the metabolic features detected in the positive ion mode was identified as uracil with level 1 evidence (i.e. chemical annotation using reference standards); this feature was significantly associated with CO, NO2, EC, and OC, and was putatively annotated by mummichog as uracil as well. Uracil, a key component of RNA, serves as an essential reactant in pyrimidine and purine metabolisms. Our findings indicated that DNA damage might be a potential mechanism of the adverse effects of TRAP exposure, which was reported in previous studies as well (Carvalho et al. 2018; Huang et al. 2012). In addition to DNA damage, gene expression could be altered due to TRAP exposure. Krauskopf et al. identified changes of gene express profiles after exposure to TRAP by plasma circulating miRNA (a regulator for gene expression), and Chu et al. detected differentially expressed genes that have implicated a range of cellular responses and pathways such as oxidative stress among trucking industry workers with regular exposures to TRAP (Chu et al. 2016; Krauskopf et al. 2018). Pyrimidine and purine metabolism were also reported as significant pathways with TRAP exposure in previous studies using untargeted metabolomics (Jeong et al. 2018; Liang et al. 2018b; Liang et al. 2019; Walker et al. 2018). Walker et al. found that pyrimidine metabolism and purine metabolism were associated with the long-term exposure to ultrafine particles (UFP) in a community-based participatory cross-sectional study (Walker et al. 2018). Annual average exposure to NO2 was also associated with pyrimidine metabolism among patients with adult-onset asthma or cardio-cerebrovascular diseases compared with healthy controls (Jeong et al. 2018). Liang et al. reported multiple ambient air pollutants, including black carbon, nitric oxide, and PM2.5 to be associated with purine metabolism among a panel of 54 college students living in dormitories located either near or far from a major highway (Liang et al. 2018b). The associations of PM2.5, black carbon, OC, and water-soluble OC with pyrimidine metabolism were observed as well in a semi-controlled crossover study of car commuters (Liang et al. 2019). Our current results provide further evidence of exposure to TRAP inducing variation in nucleotide metabolism as a mechanism of action responding to nucleic acid damage and repair.
We found that the moving average of 1-2 day lagged EC was associated with several pathways involved in fatty acid biosynthesis and metabolism, including carnitine shuttle, de novo fatty acid biosynthesis, fatty acid activation, fatty acid oxidation, and omega-6 fatty acid metabolism. Metabolic features that were annotated by mummichog as octadecenoyl-CoA, tetracosanoyl-CoA, and (4R,8R,12R)-trimethyl-2E-tridecenoyl-CoA were enriched in these pathways consistently. One-day lag concentrations of CO was associated with the carnitine shuttle pathway, and for NO2 and PM2.5, 1-day lag concentrations were associated with saturated fatty acid beta-oxidation. Multiple metabolic features enriched in the two pathways were annotated as fatty-acyl-CoAs, which plays a pivotal role in fatty acid metabolism. However, we failed to match these metabolic features in our in-house list of identified metabolites by authentic chemical reference standards, as the list did not contain these short-lived compounds. A few MWAS studies also reported several lipid metabolism pathways (including fatty acid activation, de novo fatty acid biosynthesis, and carnitine shuttle) associated with air pollutants (Jeong et al. 2018; Miller et al. 2016; Walker et al. 2018). These findings implicated an essential role played by fatty acids (FAs) in the health effect of exposure to TRAP, potentially supplying energy and signaling (Gropper and Smith 2012), while human studies that investigated the impact of TRAP on blood lipid profiles are scarce.
Five significant pathways of amino acid metabolism were identified in our study: methionine and cysteine metabolism, tyrosine metabolism, histidine metabolism, tryptophan metabolism, and alanine and aspartate metabolism. Correspondingly, mummichog annotated a metabolic feature as methionine associated with the moving average of EC and a group of short-lived compounds involved in these biological pathways. We also confirmed the identify of methionine, tyrosine, histamine, and tryptophan, which were each associated with EC (and for histamine, other TRAPs) in negative ESI mode. Methionine can regulate multiple metabolic processes, including lipid metabolism and oxidative stress (Martinez et al. 2017), and the detrimental effect of PM2.5 on heart rate variability could by modified by genetic variations in the methionine pathway or differences in methionine intake (Baccarelli et al. 2008). Tyrosine, histidine, and tryptophan are aromatic amino acid and susceptible to the attack of reactive oxidative species (ROS) (Stadtman 2006). ROS can mediate the conversion process of tyrosine residues to hydroxyl derivatives (for example, 3-nitrotyrosine), histidine residues to 2-oxohistidine and asparagine, and tryptophan residues to formyl-kynurenine and kynurenine (Stadtman 2006). The oxidative modification of amino acid residues of proteins is involved in the etiology or progression of many diseases (Stadtman and Berlett 1998). 3-nitrotyrosine is used as a marker of oxidative stress, and Rossner et al. reported a significantly higher level of 3-nitrotyrosine in plasma among bus drivers compared with healthy volunteers spending most daily times indoors (Khan et al. 1998; Rossner et al. 2007). Histamine is the core component of histidine metabolism and a well-known inflammatory agent involved in airway hyper-responsiveness (Juniper et al. 1981). Nasal challenge with allergen plus diesel exhaust particles (DEPs) induced a higher level of histamine in nasal lavage of nonsmoking volunteers compared to allergen alone within a single-blind, randomized, and placebo-controlled crossover study, and an in-vitro experiment with a mouse mast cell line (activated by Immunoglobulin E in advance) confirmed the release of histamine responding to the DEPs in a dose-dependent manner (Diaz-Sanchez et al. 2000). In an animal study using hamsters, histamine elevation was considerably slower in plasma than that in bronchoalveolar lavage fluid, and plasma histamine reached the climax after the intratracheal instillation of DEP at six hours (Nemmar et al. 2003). In the current study, the feature annotated by histamine was negatively associated with CO, NO2, and EC, and similar to the findings reported by Liang et al. of a negative association between histidine, a precursor of histamine, and EC among a panel of commuters (Liang et al. 2019). Few studies have employed markers of protein oxidation to investigate the effect of TRAP on the human body, and our current findings motivate future work in this area.
To the best of our knowledge, Tobit models have not been used in addressing missing values in mass spectrometry-based metabolomics data. As such, we used MLR models with missing value imputation of half the minimum feature intensity as a sensitivity analysis. Replacing missing values by the half of the minimum of non-missing values is a commonly used approach and is an approach suggested by almost all statistical packages and online toolkits, although limitations exist (e.g., distorting distributions) (Wei et al. 2018). Compared to the MLR model with imputation by a constant, which might underestimate the slopes, the Tobit model assumes a censored normal distribution for , with the distribution censored on the left at log(LOD). In other words, the probability density of is the same as that for for and is equal to the probability of observing if (i.e., missing values). Thus, the coefficient estimates from the Tobit model are potentially less biased than those from the MLR model. To confirm this, we further compared the results derived by the two models. As shown in Table S3, in general, Tobit models identified more significant features than MLR models. Figures S6-S7 suggest that features present in more samples had more similar and robust estimated associations with pollution among the two regression approaches, Tobit and MLR, with similar p-values (e.g., darker color points along the 1:1 line in these plots). We also observed that some features were significantly associated with pollution in MLR models, but had p-values close to 1 in Tobit models; there were no features only associated with pollution in Tobit and not MLR models, suggesting that the Tobit model approach is more conservative than MLR model approach. We used half of the minimum feature intensity of the whole dataset for imputation of missing values among features in MLR models. For features with high abundance and a high percent presence among participants, the relatively low number of imputed values may have been outliers which would severely distort the association between these features and exposures in MLR models.
There are several limitations in the present study. First, metabolism exhibits diurnal variation. Diurnal changes of energy expenditure and intake distribution are associated with many factors, including time of day, amount of sleep, timing of meals, and light-dark cycles (Walker et al. 2016). Principal component analysis on averaged metabolic profiles according to time of sampling shows three time-of-day patterns: morning, afternoon, and night (Park et al. 2009). Diurnal changes may influence intra-individual variation in response to TRAP, so ideally the time of day of sampling should be considered in the interpretation of results; this level of information was missing from our study, which may have added to between-participant variability in feature intensities. Second, the exposure assessment in the current study was limited to measurements made at one ambient monitoring site located near downtown Atlanta, and assigned to participants living across the city. Therefore, the spatial gradients in day-to-day pollutant concentrations were not captured. Exposures of participants living near highways may have been underestimated. Moreover, due to the lack of information on daily activities, we were not able to account for factors that may have affect participants’ daily exposures to ambient concentrations, such as time spent inside or outside. Finally, we used raw p-values to select significant features in order to conduct the pathway analysis by mummichog, false discoveries might exist regarding significant metabolic features given the multiple evaluations in a single experiment. In addition, since mummichog use putative annotation of metabolic features based on m/z, it may introduce errors when predicting metabolic functional activity and therefore, conventional chemical identification process is needed to validate findings from these pathway enrichment analyses. The results should be interpreted with caution, because considering the high chance of false positive findings, the tentative nature of our findings should not be underrecognized, which requires validation in much larger cohorts.
5. CONCLUSIONS
Using an untargeted MWAS approach among the Emory CHDWB cohort, we identified associations between a range of ambient pollutants, including components of TRAP, and perturbations to the metabolic phenotype. The results demonstrate the use of HRM as a viable platform for untargeted characterization of molecular mechanisms underlying exposures to TRAP. The biological pathways identified are primarily involved in nucleic acid damage and repair, nutrient metabolism, and acute inflammation. These results add evidence for the hypothesis that exposure to TRAP can induce biological effects on the human body via a range of mechanisms manifested by perturbed biological pathways.. Future work on HRM and air pollution in the CHDWB cohort will utilize information from the repeated measures over the set of annual follow-up visits for each participant in the cohort to validate the observed findings and examine how metabolic perturbations are associated with TRAP change over time. We anticipate that these data will provide a rich resource for validating the underlying mechanisms associated with air pollution, when used in combination with more comprehensive exposure assessment (e.g., air pollution data from Community Multiscale Air Quality Modeling System of the U.S. Environmental Pollution Association) or feature identification (e.g., verifying compounds with authentic chemical standards analyzed under the same experimental conditions) (Sumner et al. 2007).
Supplementary Material
Highlights.
Traffic-related air pollutants (TRAPs) associated with nucleic acids damage and repair, nutrient metabolism, and acute inflammation pathways.
Pyrimidine metabolism and carnitine shuttle consistently associated with TRAPs.
Histamine and uracil associated with carbon monoxide, nitrogen dioxide, and elemental carbon.
Tobit model performed as well as multiple linear regression models in metabolomics application
ACKNOWLEDGEMENTS
This work was based on information from the Emory Predictive Health Institute and Center for Health Discovery and Well Being Database, supported by Emory University, the National Institutes of Health (UL1TR000454], and the National Heart Lung and Blood Institute (P20 HL113451). The study was also supported by HERCULES Exposome Research Center through the National Institute of Environmental Health Sciences (P30 ES019776). We are grateful to Jane Clark and ViLinh Tran for assisting with access to the CHDWB database and supporting the efforts of the TRAPHIC study. We also thank our ongoing collaborations with Jim Mulholland and Ted Russell at the Georgia Institute of Technology and their assistance with use of Atlanta air quality data.
All participants provided informed consent, and the study was approved by the Emory University Institutional Review Board.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alonso A; Marsal S; Julia A Analytical methods in untargeted metabolomics: state of the art in 2015. Front Bioeng Biotechnol 2015;3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A; Cassano PA; Litonjua A; Park SK; Suh H; Sparrow D; Vokonas P; Schwartz J Cardiac autonomic dysfunction - Effects from particulate air pollution and protection by dietary methyl nutrients and metabolic Polymorphisms. Circulation 2008;117:1802–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A; Wright RO; Bollati V; Tarantini L; Litonjua AA; Suh HH; Zanobetti A; Sparrow D; Vokonas PS; Schwartz J Rapid DNA methylation changes after exposure to traffic particles. American journal of respiratory and critical care medicine 2009;179:572–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellissimo MP; Cai Q; Ziegler TR; Liu KH; Tran PH; Vos MB; Martin GS; Jones DP; Yu T; Alvarez JA Plasma high-resolution metabolomics differentiates adults with normal weight obesity from lean individuals. Obesity (Silver Spring) 2019;27:1729–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer M; Lencar C; Tamburic L; Koehoorn M; Demers P; Karr C A cohort study of traffic-related air pollution impacts on birth outcomes. Environ Health Perspect 2008;116:680–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigham KL Predictive health: the imminent revolution in health care. J Am Geriatr Soc 2010;58 Suppl 2:S298–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundy JG; Davey MP; Viant MR Environmental metabolomics: a critical review and future perspectives. Metabolomics 2009;5:3 [Google Scholar]
- Carvalho RB; Carneiro MFH; Barbosa F; Batista BL; Simonetti J; Amantea SL; Rhoden CR The impact of occupational exposure to traffic-related air pollution among professional motorcyclists from Porto Alegre, Brazil, and its association with genetic and oxidative damage. Environ Sci Pollut R 2018;25:18620–18631 [DOI] [PubMed] [Google Scholar]
- Chen C; Li H; Niu Y; Liu C; Lin Z; Cai J; Li W; Ge W; Chen R; Kan H Impact of short-term exposure to fine particulate matter air pollution on urinary metabolome: A randomized, double-blind, crossover trial. Environment international 2019;130:104878. [DOI] [PubMed] [Google Scholar]
- Chen Z; Salam MT; Toledo-Corral C; Watanabe RM; Xiang AH; Buchanan TA; Habre R; Bastain TM; Lurmann F; Wilson JP; Trigo E; Gilliland FD Ambient air pollutants have adverse effects on insulin and glucose homeostasis in Mexican Americans. Diabetes care 2016;39:547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH; Garshick E; Hart JE; Spiegelman D; Dockery DW; Smith TJ; Laden F Occupational vehicle-related particulate exposure and inflammatory markers in trucking industry workers. Environmental research 2016;148:310–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu JH; Hart JE; Chhabra D; Garshick E; Raby BA; Laden F Gene expression network analyses in response to air pollution exposures in the trucking industry. Environ Health 2016;15:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa LG; Cole TB; Coburn J; Chang YC; Dao K; Roque PJ Neurotoxicity of traffic-related air pollution. Neurotoxicology 2017;59:133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrow LA; Klein M; Flanders WD; Waller LA; Correa A; Marcus M; Mulholland JA; Russell AG; Tolbert PE Ambient air pollution and preterm birth: a time-series analysis. Epidemiology 2009;20:689–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Sanchez D; Penichet-Garcia M; Saxon A Diesel exhaust particles directly induce activated mast cells to degranulate and increase histamine levels and symptom severity. J Allergy Clin Immunol 2000; 106:1140–1146 [DOI] [PubMed] [Google Scholar]
- Edgerton ES; Hartsell BE; Saylor RD; Jansen JJ; Hansen DA; Hidy GM The Southeastern Aerosol Research and Characterization Study: Part II. Filter-based measurements of fine and coarse particulate matter mass and composition. J Air Waste Manag Assoc 2005;55:1527–1542 [DOI] [PubMed] [Google Scholar]
- Eze IC; Hemkens LG; Bucher HC; Hoffmann B; Schindler C; Kunzli N; Schikowski T; Probst-Hensch NM Association between ambient air pollution and diabetes mellitus in Europe and North America: systematic review and meta-analysis. Environ Health Persp 2015;123:381–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan R; Ladva C; Greenwald R; Krall JR; Raysoni AU; Kewada P; Winquist A; Flanders WD; Liang DH; Sarnat J Acute pulmonary and inflammatory response in young adults following a scripted car commute. Air Qual Atmos Hlth 2018;11:123–136 [Google Scholar]
- Gouveia-Figueira S; Karimpour M; Bosson JA; Blomberg A; Unosson J; Sehlstedt M; Pourazar J; Sandstrom T; Behndig AF; Nording ML Mass spectrometry profiling reveals altered plasma levels of monohydroxy fatty acids and related lipids in healthy humans after controlled exposure to biodiesel exhaust. Analytica chimica acta 2018;1018:62–69 [DOI] [PubMed] [Google Scholar]
- Greenbaum DS Sources of air pollution: gasoline and diesel engines. in: Straif K, Cohen A, Samet J, eds. Air Pollution and Cancer: the International Agency for Research on Cancer; 2013 [Google Scholar]
- Gropper SS; Smith JL Advanced nutrition and human metabolism ed^eds: Cengage Learning; 2012 [Google Scholar]
- Hansen DA; Edgerton E; Hartsell B; Jansen J; Burge H; Koutrakis P; Rogers C; Suh H; Chow J; Zielinska B; McMurry P; Mulholland J; Russell A; Rasmussen R Air quality measurements for the aerosol research and inhalation epidemiology study. J Air Waste Manag Assoc 2006;56:1445–1458 [DOI] [PubMed] [Google Scholar]
- Health Effects Institute. Traffic-Related Air Pollution: A Critical Review of the Literature on Emissions, Exposure, and Health Effects. A Special Report of the Institute's Panel on the Health Effects of Traffic-Related Air Pollution; 2010
- Huang HB; Lai CH; Chen GW; Lin YY; Jaakkola JJ; Liou SH; Wang SL Traffic-related air pollution and DNA damage: a longitudinal study in Taiwanese traffic conductors. PloS one 2012;7:e37412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q; Hu D; Wang X; Chen Y; Wu Y; Pan L; Li H; Zhang J; Deng F; Guo X; Shen H The modification of indoor PM2.5 exposure to chronic obstructive pulmonary disease in Chinese elderly people: A meet-in-metabolite analysis. Environment international 2018;121:1243–1252 [DOI] [PubMed] [Google Scholar]
- Jacobs L; Nawrot TS; de Geus B; Meeusen R; Degraeuwe B; Bernard A; Sughis M; Nemery B; Panis LI Subclinical responses in healthy cyclists briefly exposed to traffic-related air pollution: an intervention study. Environ Health 2010;9:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong A; Fiorito G; Keski-Rahkonen P; Imboden M; Kiss A; Robinot N; Gmuender H; Vlaanderen J; Vermeulen R; Kyrtopoulos S; Herceg Z; Ghantous A; Lovison G; Galassi C; Ranzi A; Krogh V; Grioni S; Agnoli C; Sacerdote C; Mostafavi N; Naccarati A; Scalbert A; Vineis P; Probst-Hensch N Perturbation of metabolic pathways mediates the association of air pollutants with asthma and cardiovascular diseases. Environment international 2018;119:334–345 [DOI] [PubMed] [Google Scholar]
- Jiang S; Bo L; Gong CY; Du XH; Kan HD; Xie YQ; Song WM; Zhao JZ Traffic-related air pollution is associated with cardio-metabolic biomarkers in general residents. Int Arch Occ Env Hea 2016;89:911–921 [DOI] [PubMed] [Google Scholar]
- Jones DP; Park Y; Ziegler TR Nutritional metabolomics: progress in addressing complexity in diet and health. Annu Rev Nutr 2012;32:183–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juniper E; Frith P; Hargreave FJT Airway responsiveness to histamine and methacholine: relationship to minimum treatment to control symptoms of asthma. 1981;36:575–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan J; Brennand DM; Bradley N; Gao B; Bruckdorfer R; Jacobs M 3-Nitrotyrosine in the proteins of human plasma determined by an ELISA method. Biochem J 1998;332 (Pt 3):807–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauskopf J; Caiment F; van Veldhoven K; Chadeau-Hyam M; Sinharay R; Chung KF; Cullinan P; Collins P; Barratt B; Kelly FJ; Vermeulen R; Vineis P; de Kok TM; Kleinjans JC The human circulating miRNome reflects multiple organ disease risks in association with short-term exposure to traffic-related air pollution. Environment international 2018;113:26–34 [DOI] [PubMed] [Google Scholar]
- Krishnan RM; Sullivan JH; Carlsten C; Wilkerson HW; Beyer RP; Bammler T; Farin F; Peretz A; Kaufman JD A randomized cross-over study of inhalation of diesel exhaust, hematological indices, and endothelial markers in humans. Particle and fibre toxicology 2013;10:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubesch NJ; de Nazelle A; Westerdahl D; Martinez D; Carrasco-Turigas G; Bouso L; Guerra S; Nieuwenhuijsen MJ Respiratory and inflammatory responses to short-term exposure to traffic-related air pollution with and without moderate physical activity. Occupational and environmental medicine 2015;72:284–293 [DOI] [PubMed] [Google Scholar]
- Ladva CN; Golan R; Greenwald R; Yu T; Sarnat SE; Flanders WD; Uppal K; Walker DI; Tran V; Liang D; Jones DP; Sarnat JA Metabolomic profiles of plasma, exhaled breath condensate, and saliva are correlated with potential for air toxics detection. Journal of breath research 2017;12:016008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladva CN; Golan R; Liang D; Greenwald R; Walker DI; Uppal K; Raysoni AU; Tran V; Yu T; Flanders WD; Miller GW; Jones DP; Sarnat JA Particulate metal exposures induce plasma metabolome changes in a commuter panel study. PloS one 2018;13:e0203468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankadurai BP; Nagato EG; Simpson MJ Environmental metabolomics: an emerging approach to study organism responses to environmental stressors. Environmental Reviews 2013;21:180–205 [Google Scholar]
- Lelieveld J; Evans JS; Fnais M; Giannadaki D; Pozzer A The contribution of outdoor air pollution sources to premature mortality on a global scale. Nature 2015;525:367–371 [DOI] [PubMed] [Google Scholar]
- Li SZ; Park Y; Duraisingham S; Strobel FH; Khan N; Soltow QA; Jones DP; Pulendran B Predicting Network Activity from High Throughput Metabolomics. Plos Comput Biol 2013;9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D; Golan R; Moutinho JL; Chang HH; Greenwald R; Sarnat SE; Russell AG; Sarnat JA Errors associated with the use of roadside monitoring in the estimation of acute traffic pollutant-related health effects. Environmental research 2018a;165:210–219 [DOI] [PubMed] [Google Scholar]
- Liang D; Moutinho JL; Golan R; Yu T; Ladva CN; Niedzwiecki M; Walker DI; Sarnat SE; Chang HH; Greenwald R; Jones DP; Russell AG; Sarnat JA Use of high-resolution metabolomics for the identification of metabolic signals associated with traffic-related air pollution. Environment international 2018b;120:145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang DH; Ladva CN; Golan R; Yu TW; Walker DI; Sarnat SE; Greenwald R; Uppal K; Tran V; Jones DP; Russell AG; Sarnat JA Perturbations of the arginine metabolome following exposures to traffic-related air pollution in a panel of commuters with and without asthma. Environment international 2019;127:503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liigand P; Kaupmees K; Haav K; Liigand J; Leito I; Girod M; Antoine R; Kruve A Think negative: finding the best electrospray ionization/MS mode for your analyte. Analytical chemistry 2017;89:5665–5668 [DOI] [PubMed] [Google Scholar]
- Lim H; Kwon HJ; Lim JA; Choi JH; Ha M; Hwang SS; Choi WJ Short-term Effect of Fine Particulate Matter on Children's Hospital Admissions and Emergency Department Visits for Asthma: A Systematic Review and Meta-analysis. J Prev Med Public Health 2016;49:205–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton MF; Yancey PG; Davies SS; Jerome WG; Linton EF; Song WL; Doran AC; Vickers KC The role of lipids and lipoproteins in atherosclerosis Endotext [Internet]: MDText. com, Inc; 2019 [Google Scholar]
- Martinez Y; Li X; Liu G; Bin P; Yan WX; Mas D; Valdivie M; Hu CAA; Ren WK; Yin YL The role of methionine on metabolism, oxidative stress, and diseases. Amino acids 2017;49:2091–2098 [DOI] [PubMed] [Google Scholar]
- McBee M Modeling outcomes with floor or ceiling effects: an introduction to the Tobit model. Gifted Child Quart 2010;54:314–320 [Google Scholar]
- Metzger K; Tolbert P; Klein M; Peel J; Flanders W Case-crossover analyses of cardiovascular emergency department visits and ambient air quality, Atlanta, Georgia, 1993–2000: ISEE-333. Epidemiology 2003;14:S66 [Google Scholar]
- Miller DB; Ghio AJ; Karoly ED; Bell LN; Snow SJ; Madden MC; Soukup J; Cascio WE; Gilmour MI; Kodavanti UP Ozone exposure increases circulating stress hormones and lipid metabolites in humans. American journal of respiratory and critical care medicine 2016;193:1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GW; Jones DP The nature of nurture: refining the definition of the exposome. Toxicological sciences : an official journal of the Society of Toxicology 2014;137:1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Toxicology Program. NTP monograph on the systematic review of traffic-related air pollution and hypertensive disorders of pregnancy. Research Triangle Park, NC: National Toxicology Program,; 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemmar A; Nemery B; Hoet PH; Vermylen J; Hoylaerts MF Pulmonary inflammation and thrombogenicity caused by diesel particles in hamsters: role of histamine. American journal of respiratory and critical care medicine 2003;168:1366–1372 [DOI] [PubMed] [Google Scholar]
- Park Y; Kim SB; Wang B; Blanco RA; Le NA; Wu S; Accardi CJ; Alexander RW; Ziegler TR; Jones DP Individual variation in macronutrient regulation measured by proton magnetic resonance spectroscopy of human plasma. Am J Physiol Regul Integr Comp Physiol 2009;297:R202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask KJ; Brigham KL; Johns MME Integrating comparative effectiveness research programs into predictive health: a unique role for academic health centers. Acad Med 2011;86:718–723 [DOI] [PubMed] [Google Scholar]
- Rossner P Jr.; Svecova V; Milcova A; Lnenickova Z; Solansky I; Santella RM; Sram RJ Oxidative and nitrosative stress markers in bus drivers. Mutat Res 2007;617:23–32 [DOI] [PubMed] [Google Scholar]
- Sarnat JA; Golan R; Greenwald R; Raysoni AU; Kewada P; Winquist A; Sarnat SE; Dana Flanders W; Mirabelli MC; Zora JE; Bergin MH; Yip F Exposure to traffic pollution, acute inflammation and autonomic response in a panel of car commuters. Environmental research 2014;133:66–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymanski EL; Jeon J; Gulde R; Fenner K; Ruff M; Singer HP; Hollender J Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environmental science & technology 2014;48:2097–2098 [DOI] [PubMed] [Google Scholar]
- Solomon PA; Chameides W; Weber R; Middlebrook A; Kiang CS; Russell AG; Butler A; Turpin B; Mikel D; Scheffe R; Cowling E; Edgerton E; John JS; Jansen J; McMurry P; Hering S; Bahadori T Overview of the 1999 Atlanta Supersites Project. 2002;
- Soltow QA; Strobel FH; Mansfield KG; Wachtman L; Park Y; Jones DP High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics 2013;9:S132–S143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman ER Protein oxidation and aging. Free Radic Res 2006;40:1250–1258 [DOI] [PubMed] [Google Scholar]
- Stadtman ER; Berlett BS Reactive oxygen-mediated protein oxidation in aging and disease. Drug Metab Rev 1998;30:225–243 [DOI] [PubMed] [Google Scholar]
- Steadman RG A universal scale of apparent temperature. J Clim Appl Meteorol 1984;23:1674–1687 [Google Scholar]
- Strickland MJ; Darrow LA; Klein M; Flanders WD; Sarnat JA; Waller LA; Sarnat SE; Mulholland JA; Tolbert PE Short-term associations between ambient air pollutants and pediatric asthma emergency department visits. American journal of respiratory and critical care medicine 2010;182:307–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW; Amberg A; Barrett D; Beale MH; Beger R; Daykin CA; Fan TW; Fiehn O; Goodacre R; Griffin JL; Hankemeier T; Hardy N; Harnly J; Higashi R; Kopka J; Lane AN; Lindon JC; Marriott P; Nicholls AW; Reily MD; Thaden JJ; Viant MR Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007;3:211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surowiec I; Karimpour M; Gouveia-Figueira S; Wu J; Unosson J; Bosson JA; Blomberg A; Pourazar J; Sandstrom T; Behndig AF; Trygg J; Nording ML Multi-platform metabolomics assays for human lung lavage fluids in an air pollution exposure study. Analytical and bioanalytical chemistry 2016;408:4751–4764 [DOI] [PubMed] [Google Scholar]
- Tabassum R; Cunningham L; Stephens EH; Sturdivant K; Martin GS; Brigham KL; Gibson G A Longitudinal study of health improvement in the Atlanta CHDWB Wellness Cohort. J Pers Med 2014;4:489–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert PE; Mulholland JA; MacIntosh DL; Xu F; Daniels D; Devine OJ; Carlin BP; Klein M; Dorley J; Butler AJ; Nordenberg DF; Frumkin H; Ryan PB; White MC Air quality and pediatric emergency room visits for asthma in Atlanta, Georgia, USA. Am J Epidemiol 2000;151:798–810 [DOI] [PubMed] [Google Scholar]
- Uppal K; Soltow QA; Strobel FH; Pittard WS; Gernert KM; Yu T; Jones DP xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics 2013;14:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal K; Walker DI; Liu K; Li S; Go YM; Jones DP Computational metabolomics: a framework for the million metabolome. Chemical research in toxicology 2016;29:1956–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veldhoven K; Kiss A; Keski-Rahkonen P; Robinot N; Scalbert A; Cullinan P; Chung KF; Collins P; Sinharay R; Barratt BM; Nieuwenhuijsen M; Rodoreda AA; Carrasco-Turigas G; Vlaanderen J; Vermeulen R; Portengen L; Kyrtopoulos SA; Ponzi E; Chadeau-Hyam M; Vineis P Impact of short-term traffic-related air pollution on the metabolome - Results from two metabolome-wide experimental studies. Environment international 2019;123:124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlaanderen JJ; Janssen NA; Hoek G; Keski-Rahkonen P; Barupal DK; Cassee FR; Gosens I; Strak M; Steenhof M; Lan Q; Brunekreef B; Scalbert A; Vermeulen RCH The impact of ambient air pollution on the human blood metabolome. Environmental research 2017;156:341–348 [DOI] [PubMed] [Google Scholar]
- Walker DI; Go Y-M; Liu K; Pennell KD; Jones DP Population screening for biological and environmental properties of the human metabolic phenotype: implications for personalized medicine Metabolic Phenotyping in Personalized and Public Healthcare: Elsevier; 2016 [Google Scholar]
- Walker DI; Lane KJ; Liu K; Uppal K; Patton AP; Durant JL; Jones DP; Brugge D; Pennell KD Metabolomic assessment of exposure to near-highway ultrafine particles. J Expo Sci Environ Epidemiol 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R; Wang J; Su M; Jia E; Chen S; Chen T; Ni Y Missing value imputation approach for mass spectrometry-based metabolomics data. Sci Rep 2018;8:663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M; Ghosh JK; Su J; Cockburn M; Jerrett M; Ritz B Traffic-related air toxics and preterm birth: a population-based case-control study in Los Angeles County, California. Environ Health 2011;10:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J; Cayir A; Trevisi L; Sanchez-Guerra M; Lin X; Peng C; Bind MA; Prada D; Laue H; Brennan KJ; Dereix A; Sparrow D; Vokonas P; Schwartz J; Baccarelli AA Traffic-related air pollution, blood pressure, and adaptive response of mitochondrial abundance. Circulation 2016;133:378–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuurbier M; Hoek G; Oldenwening M; Meliefste K; Krop E; van den Hazel P; Brunekreef A In-traffic air pollution exposure and CC16, blood coagulation, and inflammation markers in healthy adults. Environ Health Perspect 2011;119:1384–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.