Abstract
Background and Objectives:
Previous studies have revealed that sulfation, as mediated by the estrogen-sulfating cytosolic sulfotransferase (SULT) SULT1E1, is involved in the metabolism of 17β-estradiol (E2), 4-hydroxytamoxifen (4OH-tamoxifen) and diethylstilbestrol in humans. It is an interesting question whether the genetic polymorphisms of SULT1E1, the gene that encodes the SULT1E1 enzyme, may impact on the metabolism of E2 and these two drug compounds through sulfation.
Methods:
In this study, five missense coding single nucleotide polymorphisms (SNPs) of the SULT1E1 gene were selected for investigating the sulfating activity of the coded SULT1E1 allozymes toward E2, 4OH-tamoxifen and diethylstilbestrol. Corresponding cDNAs were generated by site-directed mutagenesis and recombinant SULT1E1 allozymes were bacterially expressed, affinity-purified, and characterized using enzymatic assays.
Results:
Purified SULT1E1 allozymes were shown to display differential sulfating activities toward E2, 4OH-tamoxifen and diethylstilbestrol. Kinetic analysis revealed further distinct Km (reflecting substrate affinity) and Vmax (reflecting catalytic activity) values of the five SULT1E1 allozymes with E2, 4OH-tamoxifen and diethylstilbestrol as substrates.
Conclusions:
Taken together, these findings highlighted the significant differences in E2- as well as drug-sulfating activities of SULT1E1 allozymes, which may have implications in the differential metabolism of E2, 4OH-tamoxifen and diethylstilbestrol in individuals with different SULT1E1 genotypes.
Keywords: Cytosolic sulfotransferase, sulfate conjugation, SULT1E1, single nucleotide polymorphisms, E2, 4OH-tamoxifen, diethylstilbestrol
1. Introduction
Tamoxifen, a selective estrogen receptor modulator (SERM), is widely used to treat metastatic breast cancer in pre- as well as post-menopausal women [1]. 4OH-tamoxifen has been shown to be a Phase I metabolite of tamoxifen capable of suppressing the cell proliferation of breast cancer [2–4]. Despite the common use of tamoxifen as a first-line therapy, breast cancer in tamoxifen-treated patients may eventually relapse due to the development of de novo or acquired resistance, which remains an obstacle in breast cancer therapy [5]. To cope with this challenge, diethylstilbestrol has been investigated for use as an adjuvant therapy to enhance the quality of life and improve the survival rate of patients with advanced breast cancer [6–8]. Tamoxifen and diethylstilbestrol, however, may cause serious side effects that include the induction of endometrial cancer, ocular toxicity, and venous thromboembolic events (associated with tamoxifen) and cardiovascular complications [9, 10], [11–15]. In view of their adverse effects, it is important to understand in greater detail the metabolism and deactivation mechanisms of 4OH-tamoxifen as an active metabolite of tamoxifen, as well as diethylstilbestrol. In relation to this latter notion, previous studies have demonstrated the involvement of sulfation in the metabolism of 4OH-tamoxifen and diethylstilbestrol [16–18].
Sulfation, which is catalyzed by the cytosolic sulfotransferase (SULT) enzymes, provides a major pathway for the biotransformation and excretion of a wide range of xenobiotics including drugs [19–21]. In humans, thirteen distinct SULTs have been identified [22, 23]. Besides endogenous estrogens such as E2, estrogen sulfotransferase (SULT1E1) has been recognized as one of the major SULTs capable of sulfating 4OH-tamoxifen and diethylstilbestrol [16–18], [24–26]. Like many other genes, single nucleotide polymorphisms (SNPs) of the SULT1E1 gene have been reported [27–33]. It is an important question whether SULT1E1 non-synonymous coding SNPs (cSNPs) may affect the sulfating activity of the coded SULT1E1 allozymes toward E2, 4OH-tamoxifen and diethylstilbestrol, thereby influencing their metabolism in individuals with different SULT1E1 genotypes.
In this study, five SULT1E1 allozymes coded by selected missense cSNPs identified in major SNP databases were generated, expressed and purified. The sulfating activity of purified SULT1E1 allozymes toward E2, 4OH-tamoxifen and diethylstilbestrol were evaluated. In addition, kinetic experiments were conducted to determine their kinetic parameters for evaluating their substrate affinity and catalytic efficiency in mediating the sulfation of E2, 4OH-tamoxifen and diethylstilbestrol.
2. Materials and Methods
2.1. Materials.
E2 and 4OH-tamoxifen were products of Cayman Chemical Company (Ann Arbor, MI, USA). Diethylstilbestrol, adenosine 5’-triphosphate (ATP), dimethyl sulfoxide (DMSO), dithiothreitol (DTT), 3’-phosphoadenosine-5’-phosphosulfate (PAPS), isopropyl-1-thio-β-D-galactopyranoside (IPTG), and N-2-hydroxylpiperazine-N’-2-ethanesulfonic acid (HEPES) were from Sigma-Aldrich Chemical Company (St. Louis, MO, USA). Ecolume scintillation cocktail was obtained from MP Biomedicals (Solon, OH, USA). Carrier-free sodium [35S]sulfate was from American Radiolabeled Chemicals, Inc. (St. Louis, MO, USA). Recombinant human bifunctional ATP sulfulyase/adenosine 5’-phosphosulfate kinase was prepared as previously described [34], and used to synthesize PAP[35S] from ATP and [35S]sulfate based on an established procedure [34]. Cellulose thin-layer chromatography (TLC) plates were products of Macherey-Nagel (Düren, Germany). Dpn I was purchased from New England BioLabs (Ipswich, MA, USA). PrimeStar Max DNA polymerase was a product of Takara Bio Inc. (Mountain View, CA, USA). All blue prestained protein markers were obtained from BioLand Scientific LLC. (Paramount, CA, USA). Oligonucleotide primers were synthesized by Eurofins Genomics (Louisville, KY, USA). All other reagents and chemicals used were of the highest grades commercially available.
2.2. Identification and analysis of the human SULT1E1 SNPs.
Three genomic databases, including the U.S National Center for Biotechnology Information (NCBI), the Pharmacogenomics Knowledge Base (PharmGKB), and the Universal Protein Resource (UniProt), were comprehensively searched for the non-synonymous cSNPs of the human SULT1E1 gene.
2.3. Generation of cDNAs encoding different human SULT1E1 allozymes.
A PCR-based site-directed mutagenesis procedure was used to generate cDNAs encoding different SULT1E1 allozymes. Each pair of mutagenic oligonucleotide primers that is listed in Table 1 was used in conjugation with the template (the wild-type SULT1E1 cDNA) packaged in the pGEX-2TK prokaryotic expression vector. PCR-amplification conditions were an initial denaturation for 30 s at 94°C, followed by 12 cycles of template denaturation for 30 s at 95°C, mutagenic primer annealing for 1 min at 55°C, and extension for 15 min at 72°C. Upon completion of PCR amplification, reaction mixtures were supplemented with Dpn I endonuclease to digest the wild-type SULT1E1 cDNA/pGEX-2TK. The “mutated” SULT1E1 cDNA/pGEX-2TK plasmids were individually transformed into competent DH5α E. coli cells. SULT1E1 cDNA/pGEX-2TK plasmids isolated from the transformed cells were analyzed by nucleotide sequencing to verify the desired “mutations”.
Table 1.
Primer sets used in the site-directed mutagenesis of the cDNA encoding human SULT1E1 allozymes
| SULT1E1 Allozyme and Corresponding Amino Acid Substitution | Mutagenic Primer Set | Minor Allele Frequency |
|---|---|---|
| SULT1E1-A43D | 5’-GATGATCTTGTCATTGACACCTACCCTAAATCTGGT-3’ 5’-ACCAGATTTAGGGTAGGTGTCAATGACAAGATCATC-3’ |
|
| SULT1E1-A131P | 5’-TATCTTTGCCGGAATCCAAAGGATGTGGCTGTTTCC-3’ 5’-GGAAACAGCCACATCCTTTGGATTCCGGCAAAGATA-3’ |
(0.00007–0.0002) |
| SULT1E1-R186L | 5’-AAGGGAAAGAGTCCACTTGTACTATTTCTTTTCTAC-3’ 5’-GTAGAAAAGAAATAGTACAAGTGGACTCTTTCCCTT-3’ |
(0.000008–0.00002) |
| SULT1E1-P214T | 5’-TTCCTGGAAAGGAAGACATCAGAGGAGCTTGTGGAC-3’ 5’-GTCCACAAGCTCCTCTGATGTCTTCCTTTCCAGGAA-3’ |
(0.000008–0.00008) |
| SULT1E1-D220V | 5’-TCAGAGGAGCTTGTGGTCAGGATTATACATCATACT-3’ 5’-AGTATGATGTATAATCCTGACCACAAGCTCCTCTGA-3’ |
(0.000008) |
2.4. Expression and purification of recombinant SULT1E1 allozymes.
To express SULT1E1 allozymes, each pGEX-2TK harboring a “mutated” SULT1E1 cDNA was individually transformed into competent BL21 E. coli cells. The transformed cells were grown at 37°C in 1 liter of LB medium containing 100 μg/ml ampicillin to OD600 nm =~0.5. Afterwards, the cells were induced with 0.1 mM IPTG at 25°C for 8 hours, collected by centrifugation, and resuspended in 20 ml of an ice-cold lysis buffer containing 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, and 150 mm NaCl. After the addition of 100 μl of a protease inhibitor mixture, the cells were homogenized using an Aminco French press apparatus. The crude homogenate was centrifuged at 10,000 × g for 30 min at 4°C, and the collected supernatant was fractionated using 0.5 ml glutathione-Sepharose. The bound fusion protein was treated with 2 ml of a thrombin digestion buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 2.5 mM CaCl2) that contained 0.5 unit/ml bovine thrombin. After incubation at room temperature for 15 min with constant agitation, the preparation was subjected to centrifugation. SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was performed to analyze the purity of recombinant SULT1E1 allozyme present in the collected supernatant.
2.5. Sulfotransferase assay.
The sulfating activity of the recombinant SULT1E1 allozymes toward E2, 4OH-tamoxifen or diethylstilbestrol was determined using radiolabeled PAP[35S] as a sulfate donor. The reaction mixture, with a final volume of 20 μl, contained 50 mM HEPES buffer at pH 7.4, 1 mM DTT, 14 μM PAP[35S], and a substrate dissolved in DMSO. The final concentration of DMSO in the reaction mixture was 10% (v/v). A control with DMSO replacing the substrate was analyzed in parallel. The reaction was initiated by adding 2 μl (containing 0.5 μg) of a SULT1E1 allozyme to the reaction mixture and incubated at 37°C for 10 min. Afterwards, the reaction was terminated by heating at 100°C for 3 min. After centrifugation of reaction mixture at 13,000 rpm for 3 min, 2 μl of the cleared supernatant was applied onto a cellulose TLC plate. TLC separation was conducted using a solvent system containing n-butanol/isopropanol/formic acid/water in a ratio of 3:1:1:1 (by volume). Upon completion of TLC, the plate was air-dried and then subjected to autoradiography to localize the radiolabeled sulfated product. The detected [35S] sulfated product spot was cut out and eluted with 500 μl of water in a vial for 45 min. Afterwards, 2 ml of the Ecolume scintillation liquid was added to each vial and mixed thoroughly with the eluate, and the [35S] radioactivity therein was determined using a liquid scintillation counter [35]. Based on the measured count per minute (cpm), the specific activity was calculated in the unit of nmol of sulfated product produced per min per mg enzyme.
2.6. Kinetics studies.
In the kinetic experiments, varying concentrations for each of the three substrates were used in the SULT assays. All enzymatic assays were performed in triplicate. The apparent kinetic constants were determined using the GraphPad Prism® 7.0 software that aligned with the Michaelis-Menten kinetics with non-linear regression. One-way analysis of variance (ANOVA) was implemented for inter-group comparison, followed by Tukey’s post hoc analysis. Statistical significance was set at P-values < 0.05.
2.7. Rotamer analysis of single point mutations.
Side-chain conformation of a mutated amino acid residue was simulated using the Dunbrack backbone-dependent rotamer library [36]. The structure of SULT1E1, resolved with E2 and PAP, (Protein Data Bank code: 4JVL), was referred to as the wild-type. Hydrophobic and hydrogen-binding interactions of the point-mutated amino acid residue with other residues, E2, or PAP, were also simulated by Find Clashes/Contacts tool in USCF Chimera software [37].
3. Results
3.1. Identification and analysis of different SULT1E1 SNPs.
Three online SNP databases, located at the websites of the U.S. National Center for Biotechnology Information (NCBI), the Pharmacogenomics Knowledge Base (PharmGKB), and the Universal Protein Resource (UniProt), were systematically searched for SULT1E1 genotypes. A total of 5,607 SULT1E1 SNPs were identified. The identified SULT1E1 genotypes were analyzed and categorized into coding including (synonymous, non-synonymous (missense), and nonsense SNPs) and non-coding SNPs including (introns, 3’-untranslated region (3’UTR), and 5’-untranslated region (5’UTR) SNPs). Of the 220 identified SULT1E1 missense coding SNPs (cSNPs), five were selected for further investigation based on the locations (e.g., proximity to substrate-binding- and PAPS-binding site) and changes in physiochemical properties (e.g., acidic to/from basic, polar to/from non-polar, turn inducing to/from non-turn inducing residues) of the altered amino acid residues. Table 1 shows the amino acid variations and locations, the documented allelic frequency of these five SULT1E1 cSNPs, as well as the mutagenic primers sets designed for PCR-amplification. Fig. 1 shows a ribbon diagram of the structure of human SULT1E1-E2-PAP complex indicating the locations of the amino acid residues involved in the SULT1E1 cSNPs as depicted using Structure Comparison Analysis in USCF Chimera, a molecular modeling software.
Fig. 1. Ribbon diagram of the structure of human SULT1E1-17β-estradiol (E2)-3’-phosphoadenosine 5-phosphate (PAP) complex showing the locations of amino acid residues involved in the SULT1E1 cSNPs studied.
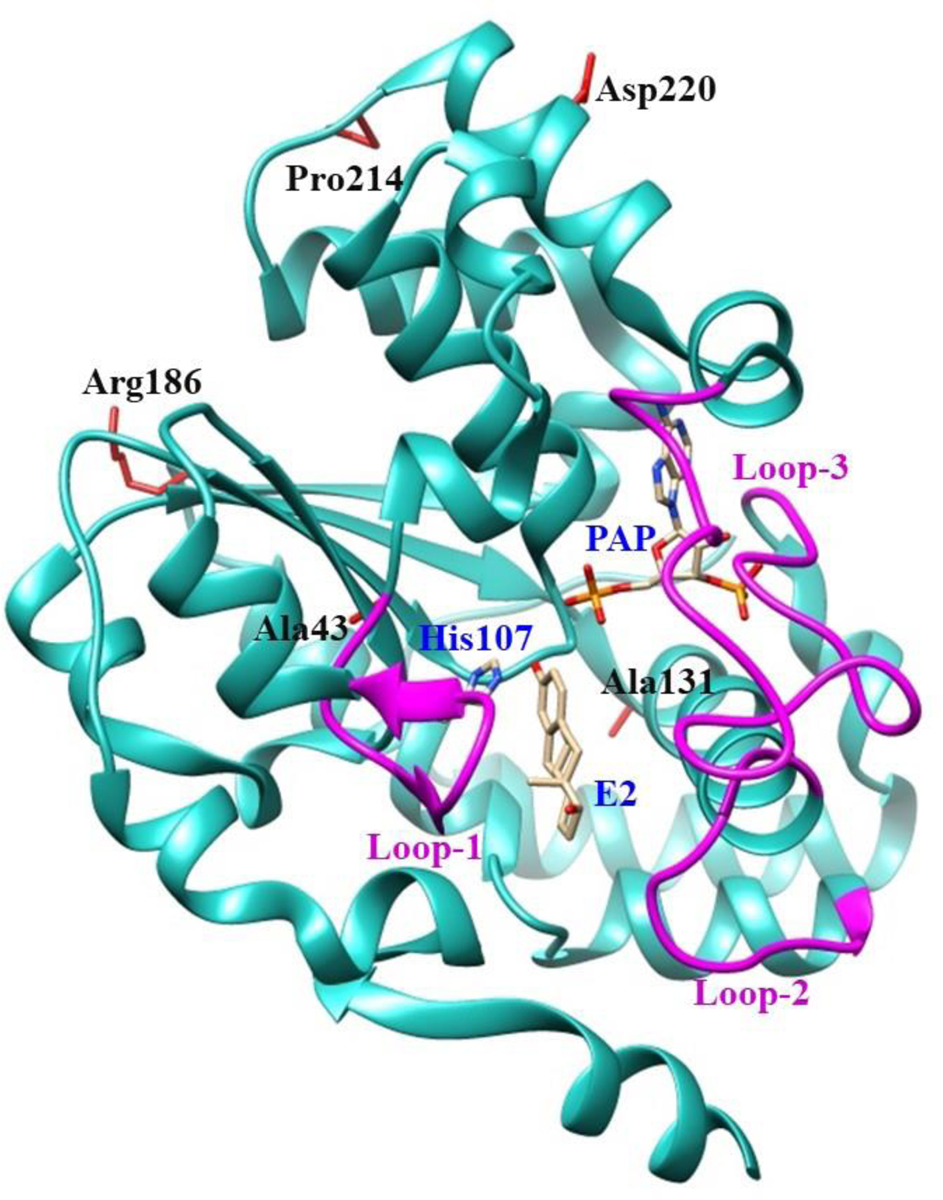
The structure of SULT1E1 (Protein Data Bank code: 4JVL), containing the polypeptide from Asp2 (N-terminus) to Glu293 (C-terminus), was depicted using Structure Comparison Analysis in USCF Chimera, a molecular modeling software [37]. E2 and PAP molecules in the structure are shown by bond structures. Loop-1 (Phe80-Asp90), loop-2 (Met144-Ser153), and loop-3 (Pro235-Gly25), shown by magenta color, form a gate for substrate entry [57, 58]. Side chains of the amino acid residues involved in the SULT1E1 cSNPs Ala43, Ala131, Arg186, Pro214 and Asp220 are indicated by bond structures (red color). PAP, (3′-phosphoadenosine 5′-phosphate); E2, (17β-Estradiol).
3.2. Preparations of recombinant human SULT1E1 allozymes.
pGEX-2TK prokaryotic expression vector carrying individual cDNAs encoding different SULT1E1 allozymes were transformed into BL21 E. coli cells. Following induction of recombinant protein expression by IPTG, the glutathione-Sepharose affinity chromatography was performed to fractionate the recombinant SULT1E1 allozymes from the homogenates of E. coli cells. The untagged recombinant SULT1E1 allozymes were released from the bound GST fusion proteins upon treatment with bovine thrombin. As shown in Fig. 2, SDS-polyacrylamide gel electrophoretic pattern confirmed that the apparent molecular weights of the purified SULT1E1 allozymes were consistent with the reported molecular weight (35,126kDa) of the wild-type SULT1E1.
Fig. 2. SDS gel electrophoretic pattern of the purified human SULT1E1 allozymes.
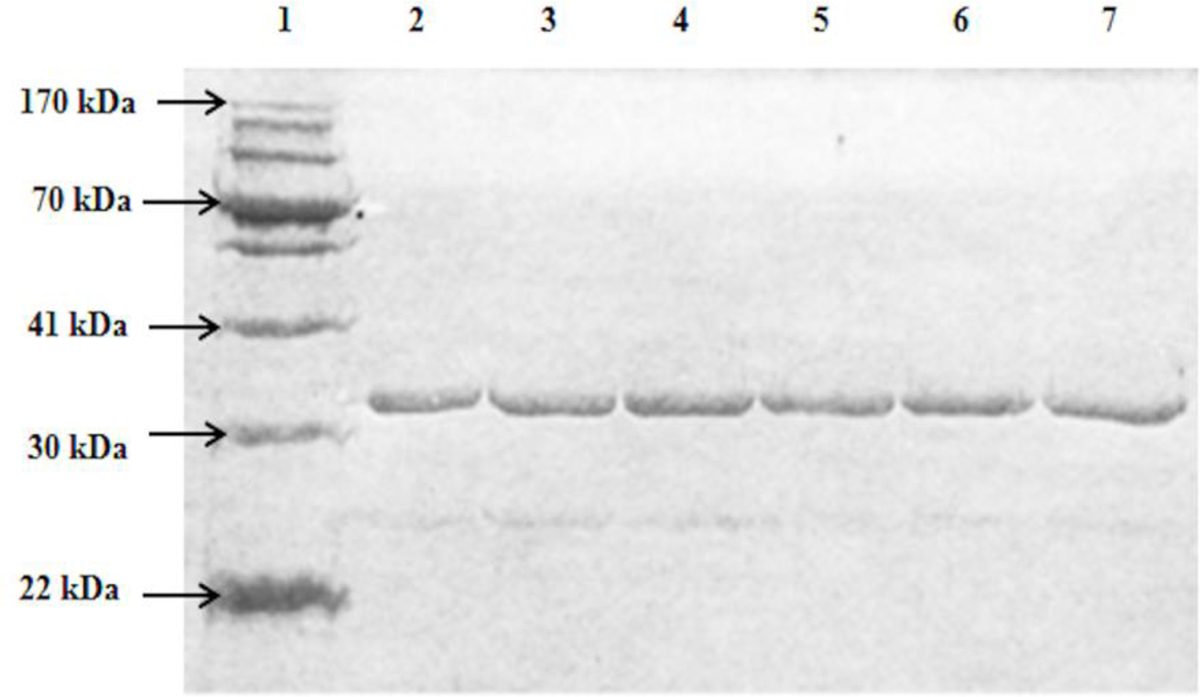
SDS-PAGE was performed on a 12% gel, followed by Coomassie blue staining. Samples analyzed in lanes 1–7. Lane 1 corresponds to the migrating positions of protein molecular weight markers co-electrophoresed. Samples analyzed in lanes 2 through 7 correspond to SULT1E1-WT, SULT1E-A43D, SULT1E1-A131P, SULT1E1-R186L, SULT1E1-P214T, and SULT1E1-D220V.
3.3. Characterization of the E2-sulfating activity of human SULT1E1 allozymes.
The concentration dependence of the sulfation of E2 by wild-type SULT1E1 was first examined. As shown in Fig. 3, the rate of reaction continued to proportionately increase up to a substrate concentration of 4 μM. At higher concentration, substrate inhibition was observed. Based on these results, three substrate concentrations, 0.5 μM, 2 μM, and 4 μM, were selected for screening the E2-sulfating activity of SULT1E1 allozymes. Results obtained are shown in Fig. 4.
Fig. 3. Concentration dependence of the sulfation of E2 by human wild-type SULT1E1.
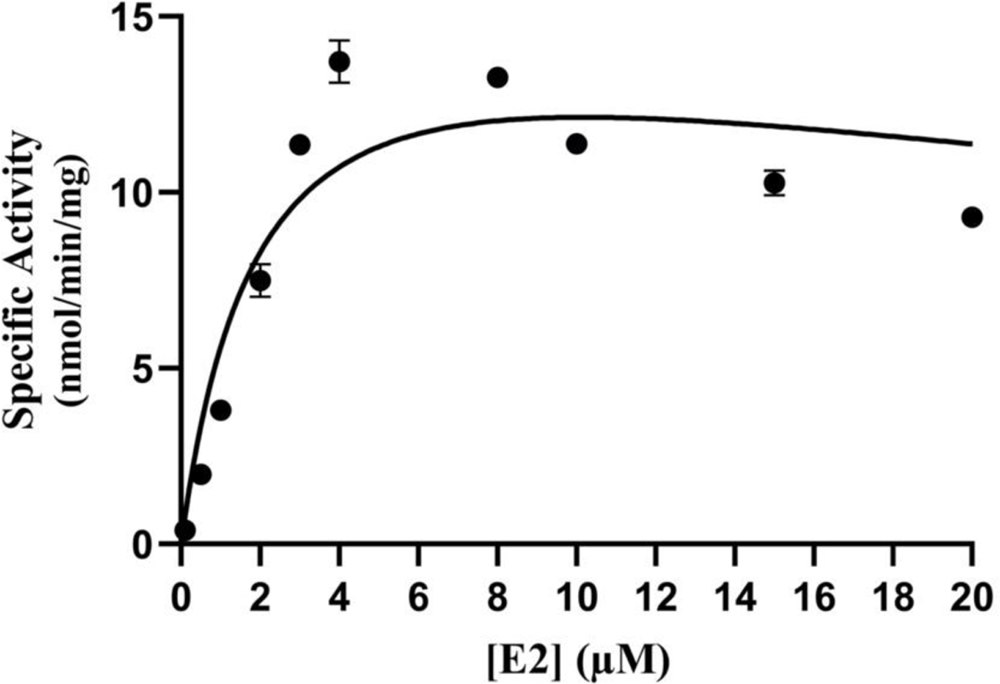
The fitting curve was generated based on Michaelis-Menten kinetics. Data shown represent calculated mean ± standard deviation derived from three experiments.
Fig. 4. Specific activities of the human SULT1E1 allozymes toward E2.
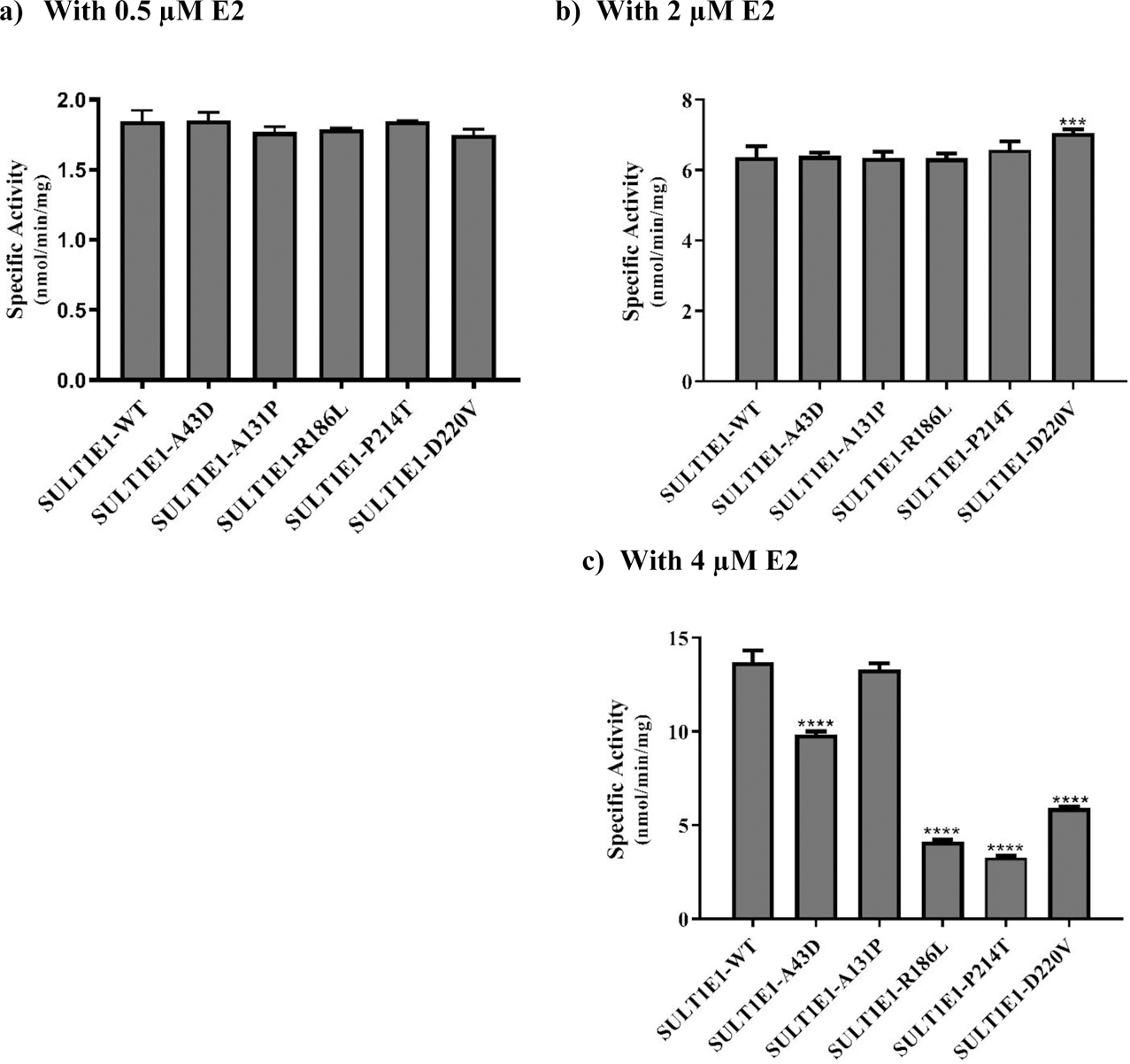
(a) Using 0.5 μM E2. (b) Using 2 μM E2. c) Using 4 μM E2. Data shown represent mean ± standard deviation derived from three independent determinations. One-way ANOVA was performed followed by Tukey’s post hoc analysis. **** Statistical significant p<0.0001 from SULT1E1-WT.
With 0.5 μM E2 as a substrate, the sulfating activities of all five SULT1E1 allozymes (SULT1E1-A43D, SULT1E1-A131P, SULT1E1-R186L, SULT1E1-P214T, and SULT1E1-D220V) were comparable to that of the wild-type enzyme (Fig. 4a). Similarly, with 2 μM E2 as a substrate, activities comparable to that of the wild-type were found for four allozymes (SULT1E1-A43D, SULT1E1-A131P, SULT1E1-R186L, and SULT1E1-P214T), while the sulfating activity of SULT1E1-D220V was 11% higher than the wild-type (Fig. 4b). With 4 μM E2 as a substrate (Fig. 4c), while the sulfating activity of SULT1E1-A131P was comparable to that of the wild-type, the activities of the other four allozymes (SULT1E1-A43D, SULT1E1-R186L, SULT1E1-P214T, and SULT1E1-D220V) were significantly lower than the wild-type enzyme. Of these four allozymes, SULT1E1-P214T showed the lowest E2-sulfating activity (3.27 ± 0.10 nmol/min/mg), being only 24% of that determined for the wild-type. The other three (SULT1E1-A43D, SULT1E1-R186L, and SULT1E1-D220V) showed sulfating activities that were, respectively, 28%, 70%, and 60% lower than the wild-type enzyme.
Kinetic assays were performed using different concentrations of E2 to investigate further the effect of genetic polymorphisms on the E2-sulfating activity of SULT1E1 allozymes. The kinetic parameters Vmax (reflecting the catalytic activity), Km (reflecting the substrate affinity), and Vmax/Km (reflecting the catalytic efficiency) determined for the wild-type and SULT1E1 allozymes are compiled in Table 2. Among the five SULT1E1 allozymes, the Km values of SULT1E1-R186L and SULT1E1-P214T were at least 25% lower than that of the wild-type enzyme. In contract, the Km values for the other three allozymes (SULT1E1-A43D, SULT1E1-A131P, and SULT1E1-D220V) were all more than 2 times higher than the wild-type enzyme. Regarding Vmax, the values determined for two allozymes (SULT1E1-R186L and SULT1E1-P214T) were, respectively, 47% and 60% lower than that of the wild-type. On the other hand, the Vmax values for the remaining three (SULT1E1-A43D, SULT1E1-A131P, and SULT1E1-D220V) were, respectively, 30%, 17%, and 19% higher than that of the wild-type. Based on these results, the calculated parameters that reflect the catalytic efficiency (Vmax/Km) values were significantly lower for all five SULT1E1 allozymes, compared with the wild-type. Of the five, SULT1E1-A43D and SULT1E1-D220V exhibited, respectively, values 42% and 47% lower than the wild-type enzyme.
Table 2.
Kinetic constants of the human SULT1E1 allozymes in catalyzing the sulfation of E2
| SULT1E1 allozyme | Vmax (nmol/min/mg) | Km (μM) | Vmax/Km (ml/min/mg) |
|---|---|---|---|
| SULT1E1-WT1 | 19.30 ± 1.2 | 2.07 ± 0.42 | 9.51 ± 1.38 |
| SULT1E1-A43D | 24.98 ±2.92**** | 5.12 ± 0.9**** | 4.91 ± 0.23**** |
| SULT1E1-A131P | 22.48 ± 2.66**** | 3.99 ± 0.94**** | 5.74 ± 0.71**** |
| SULT1E1-R186L | 10.31 ± 1.29**** | 1.55 ± 0.42**** | 6.84 ± 1.06**** |
| SULT1E1-P214T | 7.79 ± 0.89**** | 1.53 ± 0.41**** | 5.24 ± 0.85**** |
| SULT1E1-D220V | 23.15 ± 4.35**** | 5.27 ± 1.4**** | 4.49 ± 1.99**** |
Data shown represent mean ± SD derived from 3 independent experiments. One-way ANOVA was performed followed by Tukey’s post hoc analysis.
Statistical significant p<0.0001 from SULT1E1-WT.
Wild-type SULT1E1
3.4. Characterization of the 4OH-tamoxifen-sulfating activity of human SULT1E1 allozymes.
The concentration dependence of the sulfation of 4OH-tamoxifen by wild-type SULT1E1 was first examined. As shown in Fig. 5, the rate of reaction continued to increase up to a substrate concentration of 300 μM. At higher 4OH-tamoxifen concentration, substrate inhibition was observed. Based on these results, three substrate concentrations, 10 μM, 50 μM, and 200 μM, were selected for screening the 4OH-tamoxifen-sulfating activity of SULT1E1 allozymes. Results obtained are shown in Fig. 6.
Fig. 5. Concentration dependence of the sulfation of 4OH-tamoxifen by human wild-type SULT1E1.
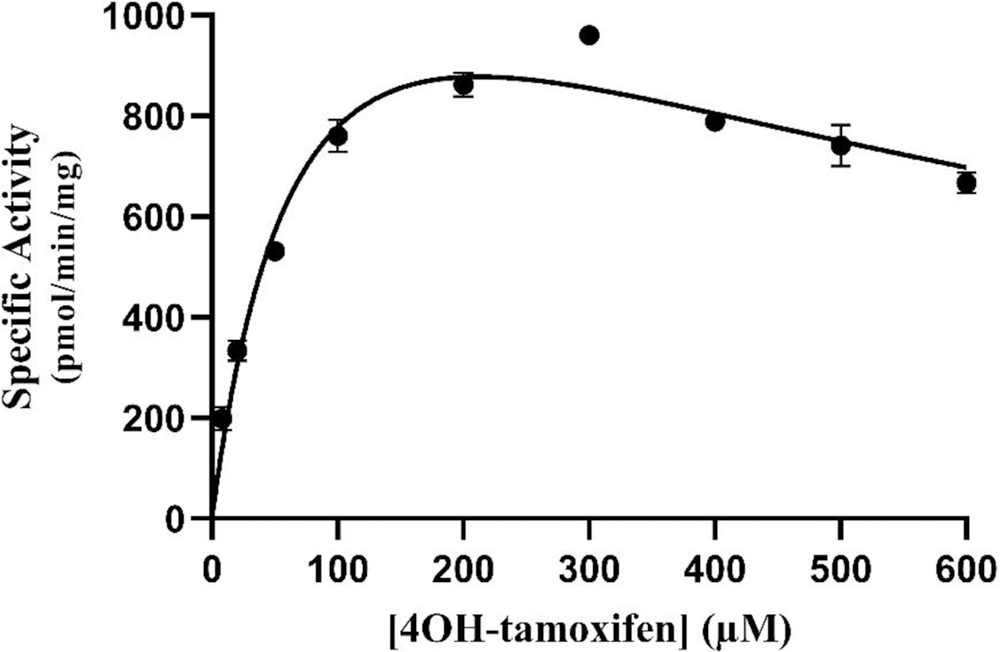
The fitting curve was generated based on Michaelis-Menten kinetics. Data shown represent calculated mean ± standard deviation derived from three experiments.
Fig. 6. Specific activities of the human SULT1E1 allozymes toward 4OH-tamoxifen.
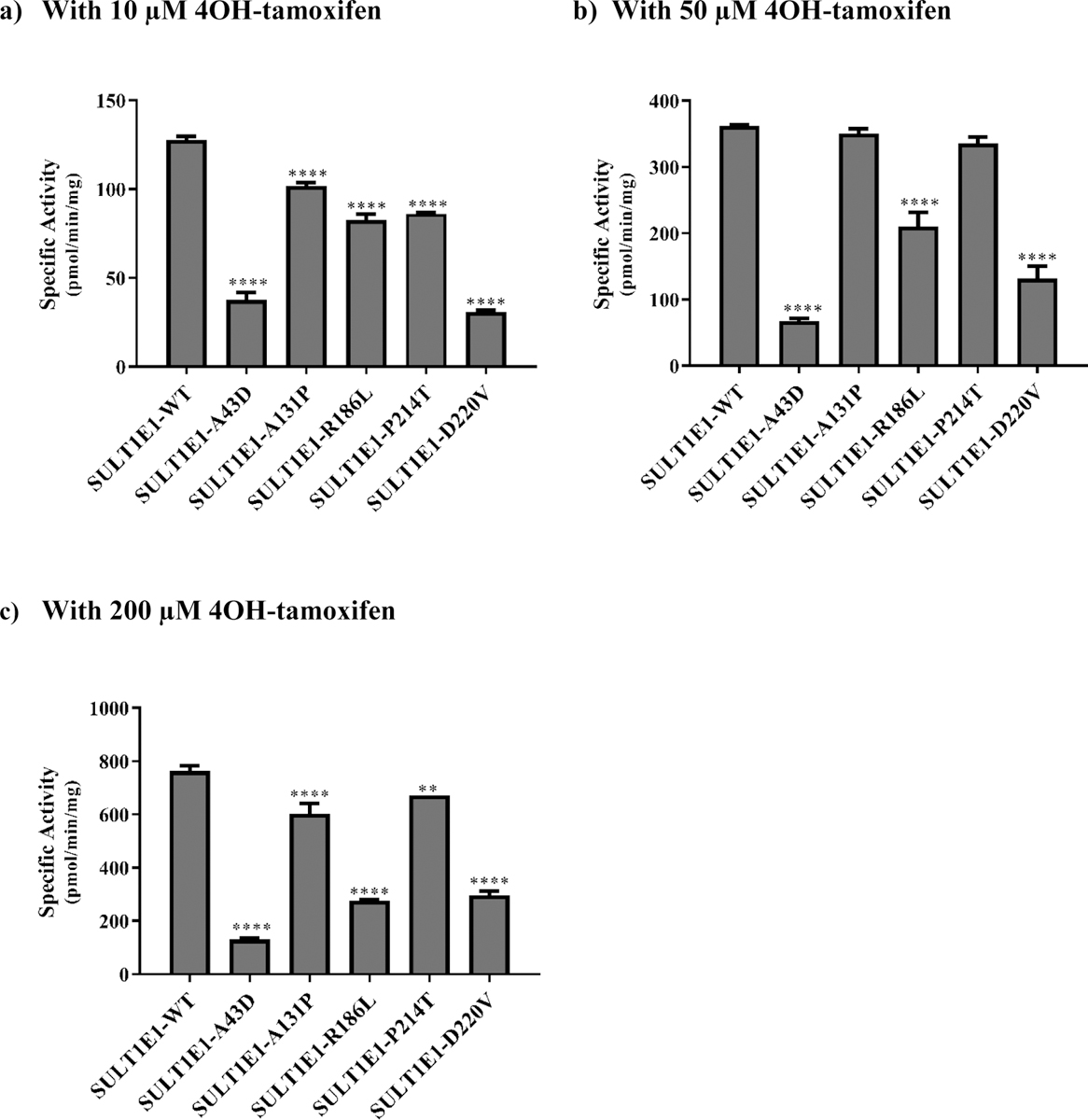
(a) Using 10 μM 4OH-tamoxifen. (b) Using 50 μM 4OH-tamoxifen. (c) Using 200 μM 4OH-tamoxifen. Data shown represent mean ± standard deviation derived from three independent determinations. One-way ANOVA was performed followed by Tukey’s post hoc analysis. **** Statistical significant p<0.0001 from SULT1E1-WT.
With 10 μM 4OH-tamoxifen as a substrate (Fig. 6a), all five SULT1E1 allozymes (SULT1E1-A43D, SULT1E1-A131P, SULT1E1-R186L, SULT1E1-P214T, and SULT1E1-D220V) showed lower sulfating activities compared with the wild-type enzyme. Of the five, SULT1E1-D220V displayed the lowest 4OH-tamoxifen-sulfating activity (30.70 ± 0.001 pmol/min/mg), being only 24% of that determined for the wild-type. Of the remaining four allozymes, the sulfating activities of three (SULT1E1-A131P, SULT1E1-R186L, and SULT1E1-P214T) were at least 20% lower than the wild-type, while that of SULT1E1-A43D was over two-thirds lower than SULT1E1-WT. With 50 μM 4OH-tamoxifen as a substrate (Fig. 6b), the sulfating activities of SULT1E1-A131P and SULT1E1-P214T were comparable to that of the wild-type enzyme, while the other three SULT1E1 allozymes (SULT1E1-A43D, SULT1E1-R186L, and SULT1E1-D220V) displayed significantly lower sulfating activities. Among them, SULT1E1-A43D showed the lowest 4OH-tamoxifen-sulfating activity, being only 18% of that determined for the wild-type, while the other two (SULT1E1-R186L, and SULT1E1-D220V) showed sulfating activities that were approximately 42% and 64%, respectively, lower than the wild-type enzyme. With 200 μM 4OH-tamoxifen as a substrate (Fig. 6c), all five allozymes (SULT1E1-A43D, SULT1E1-A131P, SULT1E1-R186L, SULT1E1-P214T, and SULT1E1-D220V) displayed significantly lower sulfating activities than the wild-type enzyme. Of them, SULT1E1-A43D showed the lowest 4OH-tamoxifen-sulfating activity (130.30 ± 0.01 pmol/min/mg), being only 17% of that determined for the wild-type. Of the other four allozymes, the sulfating activities of two (SULT1E1-A131P and SULT1E1-P214) were at least 12% lower than the wild-type, while the sulfating activities of the other two (SULT1E1-R186L and SULT1E1-D220V) were approximately 61% lower than the wild-type enzyme.
To examine further the effects of genetic polymorphisms on the 4OH-tamoxifen-sulfating activity of SULT1E1 allozymes, kinetic experiments were conducted using varying concentrations of 4OH-tamoxifen as substrates. The kinetic parameters (Vmax, Km, and Vmax /Km) determined for the wild-type and SULT1E1 allozymes are compiled in Table 3. Among the five SULT1E1 allozymes, SULT1E1-R186L displayed a Km value close to that of the wild-type enzyme, while the Km values for the remaining allozymes (SULT1E1-A43D, SULT1E1-A131P, SULT1E1-P214T, and SULT1E1-D220V) were all significantly higher. Of them, the Km values of SULT1E-A43D and SULT1E1-P214T were at least 45% higher than that of the wild-type, while the Km values of SULT1E1-A131P and SULT1E1-D220V were 71% and 79% higher, respectively. In regard to Vmax, the values determined for all SULT1E1 allozymes, except SULT1E1-A131P, were all significantly lower than that of the wild-type, with SULT1E1-A43D showing the lowest Vmax value, being only 19% of that determined for the wild-type. Based on the determined Vmax and Km values, the calculated Vmax/Km values were significantly lower for all five SULT1E1 allozymes, compared with the wild-type. Of them, SULT1E1-D220V and SULT1E1-A43D showed much lower values (~ 7.5 and 10 times, respectively, lower than the wild-type enzymes) than the other three allozymes.
Table 3.
Kinetic constants of the human SULT1E1 allozymes in catalyzing the sulfation of 4OH-tamoxifen
| SULT1E1 allozyme | Vmax (pmol/min/mg) | Km (μM) | Vmax/Km (ml/min/mg) |
|---|---|---|---|
| SULT1E1-WT1 | 907.20 ± 28.43 | 29.42 ± 3.17 | 0.031 ± 0.002 |
| SULT1E1-A43D | 178.90 ± 8.494**** | 54 ± 8.18 | 0.003 ± 0.000**** |
| SULT1E1-A131P | 944.05 ± 37.16 | 101.79 ± 10.90**** | 0.009 ± 0.000**** |
| SULT1E1-R186L | 324.80 ± 10.23**** | 29.43 ± 6.93 | 0.011 ± 0.002**** |
| SULT1E1-P214T | 770.25 ± 17.58*** | 56.18 ± 4.52 | 0.014 ± 0.000**** |
| SULT1E1-D220V | 510.60 ± 42.01**** | 144.80 ± 24.10**** | 0.004 ± 0.000**** |
Data shown represent mean ± SD derived from 3 independent experiments. One-way ANOVA was performed followed by Tukey’s post hoc analysis.
Statistical significant p<0.0001 from SULT1E1-WT
Wild-type human SULT1E1
3.5. Characterization of diethylstilbestrol-sulfating activity of human SULT1E1 allozymes.
The concentration dependence of the sulfation of diethylstilbestrol by wild-type SULT1E1 was first examined. As shown in Fig. 7, the rate of reaction continued to increase up to a substrate concentration of 6 μM. At higher diethylstilbestrol concentration, substrate inhibition was observed. Based on these results, three substrate concentrations, 0.5 μM, 3 μM, and 6 μM, were selected for screening the diethylstilbestrol-sulfating activity of SULT1E1 allozymes. Results obtained are shown in Fig. 8.
Fig. 7. Concentration dependence of the sulfation of diethylstilbestrol by human wild-type SULT1E1.
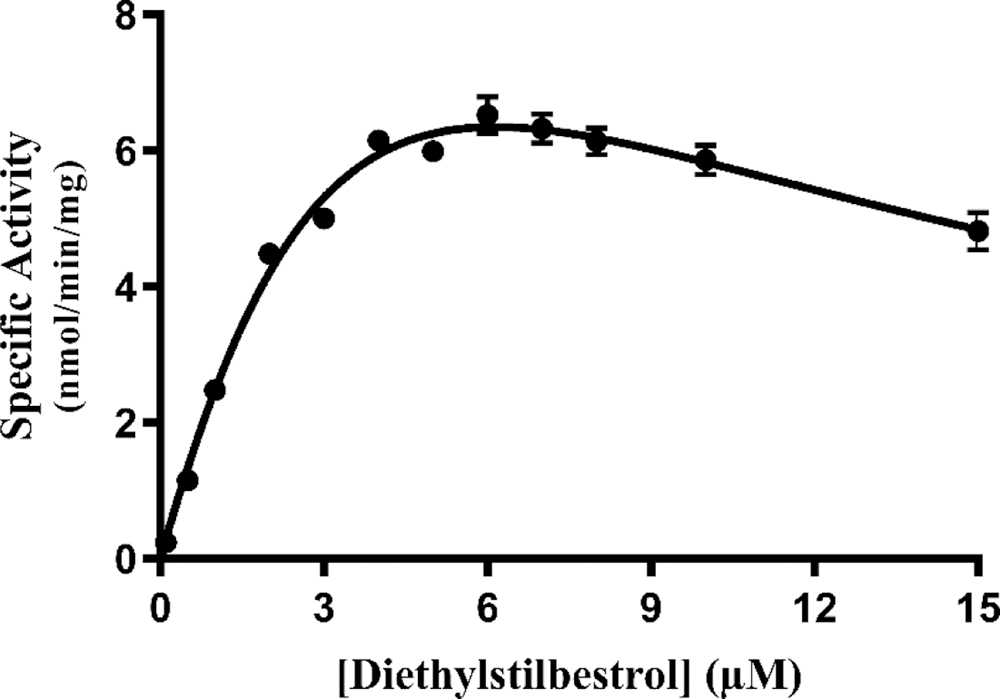
The fitting curve was generated based on Michaelis-Menten kinetics. Data shown represent calculated mean ± standard deviation derived from three experiments.
Fig. 8. Specific activities of the human SULT1E1 allozymes toward diethylstilbestrol.
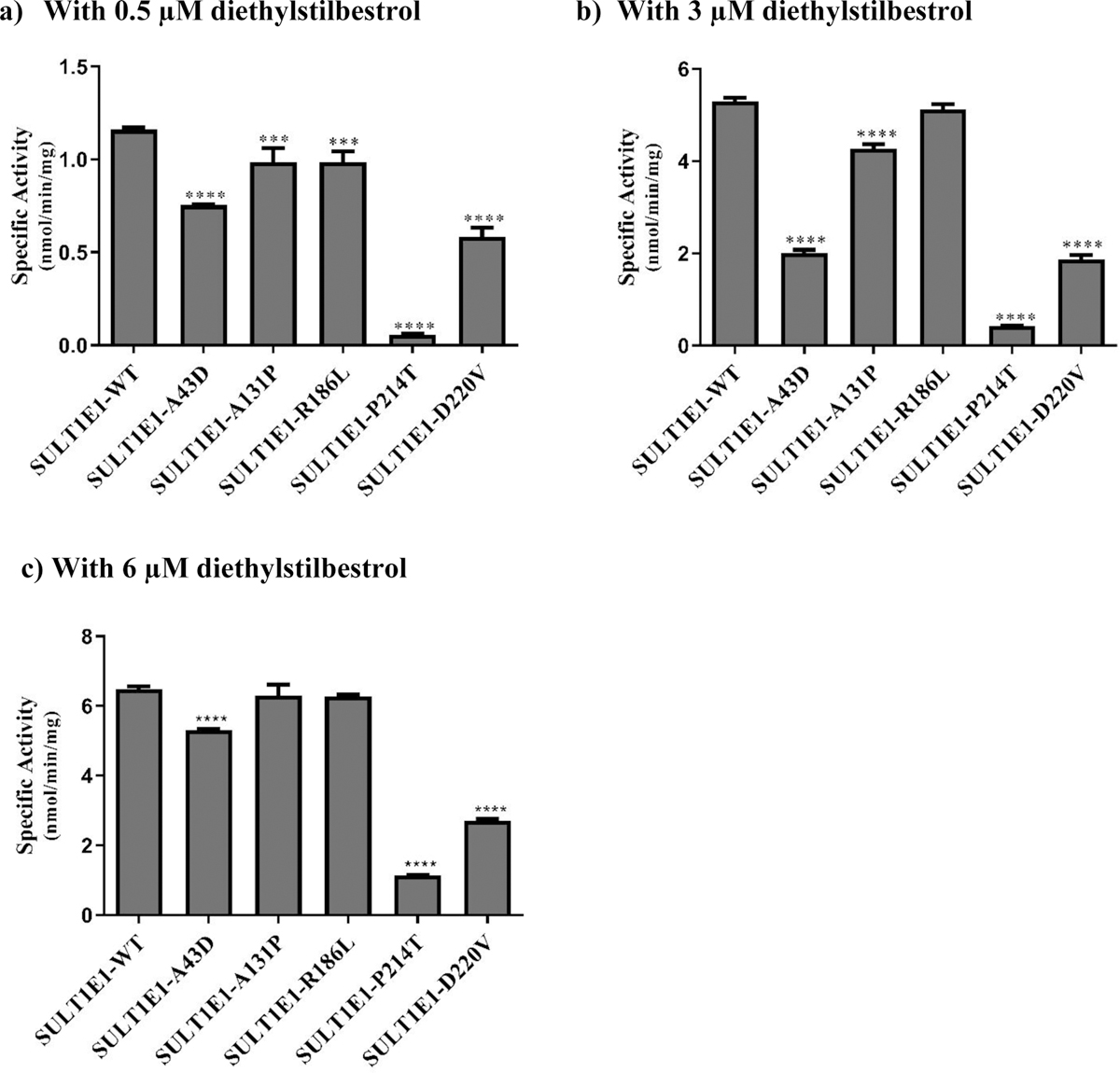
(a) Using 0.5 μM diethylstilbestrol. (b) Using 3 μM diethylstilbestrol. c) Using 6 μM diethylstilbestrol. Data shown represent mean ± standard deviation derived from three independent determinations. One-way ANOVA was performed followed by Tukey’s post hoc analysis. **** Statistical significant p<0.0001 from SULT1E1-WT.
With 0.5 μM diethylstilbestrol as a substrate (Fig. 8a), all five SULT1E1 allozymes (SULT1E1-A43D, SULT1E1-A131P, SULT1E1-R186L, SULT1E1-P214T, and SULT1E1-D220V) exhibited significantly lower sulfating activities compared with the wild-type enzyme. Among them, SULT1E1-P214T displayed the lowest diethylstilbestrol-sulfating activity, being 20 times lower than that of the SULT1E1-WT. Of the remaining four allozymes, the sulfating activities of two (SULT1E1-A131P and SULT1E1-R186L) were approximately 15% lower than the wild-type, while the activities of the other two (SULT1E1-A43D and SULT1E1-D220V) were at least 35% lower than the wild-type enzyme. With 3 μM of diethylstilbestrol as a substrate (Fig. 8b), the sulfating activity of SULT1E1-R186L was comparable to that of the wild-type, while the other four allozymes (SULT1E1-A43D, SULT1E1-A131P, SULT1E1-P214T, and SULT1E1-D220V) displayed significantly lower sulfating activities compared to the wild-type enzyme. The sulfating activity of SULT1E1-A131P was 19% lower than the wild-type, whereas the remaining three SULT1E1 allozymes showed much lower activities than the wild-type enzyme. Among the three, SULT1E1-P214T displayed the lowest diethylstilbestrol-sulfating activity, being 12 times lower than that of the wild-type, while SULT1E1-A43D and SULT1E1-D220V showed sulfating activities that were approximately two-thirds lower than the wild-type enzyme. With 6 μM of diethylstilbestrol as a substrate (Fig. 8c), the sulfating activities of SULT1E1-A131P and SULT1E1-R186L were comparable to that of the wild-type enzyme, while three other SULT1E1 allozymes (SULT1E1-A43D, SULT1E1-P214T, and SULT1E1-D220V) displayed significantly lower sulfating activities compared to the wild-type enzyme. Of the three, the sulfating activity of SULT1E1-A43D was 18% lower than the wild-type enzyme, while the other two showed much lower sulfating activities than the wild-type, with SULT1E1-D220V showing a sulfating activity approximately 41% lower and SULT1E1-P214T displaying a sulfating activity 6 times lower than the wild-type enzyme.
To investigate further the impact of genetic polymorphisms on the diethylstilbestrol-sulfating activity of SULT1E1 allozymes, kinetic experiments were performed using varying concentrations of diethylstilbestrol. The kinetic parameters (Vmax, Km, and Vmax /Km) determined for the wild-type and SULT1E1 allozymes are compiled in Table 4. Of the five SULT1E1 allozymes tested, SULT1E1-P214T showed a Km value close to that of the wild-type enzyme, while the Km values of SULT1E-A43D and SULT1E1-D220V were more than 30% lower and those of SULT1E1-A131P and SULT1E1-R186L were 40% and 79%, respectively, higher than the wild-type. In regard to Vmax, SULT1E1-A43D, SULT1E1-D220V, and SULT1E1-P214T showed values 27%, 58%, and 81%, respectively, lower than the wild-type. In contrast, SULT1E1-A131P and SULT1E1-R186L displayed Vmax values 8% and 32% higher values than the wild-type enzyme. Consequently, SULT1E1-P214T showed the lowest Vmax /Km value, being approximately 6-fold lower than the wild-type. SULT1E1-A131P, SULT1E1-R186L, and SULT1E1-D220 showed Vmax /Km values that were over 21% lower than the wild-type. In contrast, the Vmax/Km value for SULT1E1-A43D was nearly 25% higher than the wild-type enzyme.
Table 4.
Kinetic constants of the human SULT1E1 allozymes in catalyzing the sulfation of diethylstilbestrol
| SULT1E1 allozyme | Vmax (nmol/min/mg) | Km (μM) | Vmax/Km (ml/min/mg) |
|---|---|---|---|
| SULT1E1-WT1 | 8.54 ± 0.30 | 2.27 ± 0.19 | 3.77 ± 0.18 |
| SULT1E1-A43D | 6.10 ± 0.12**** | 1.00 ± 0.08**** | 4.70 ± 0.19*** |
| SULT1E1-A131P | 9.32 ± 0.65**** | 3.17± 0.23**** | 2.94 ± 0.01*** |
| SULT1E1-R186L | 11.18 ± 0.38**** | 4.07± 0.36**** | 2.76 ± 0.15*** |
| SULT1E1-P214T | 1.56 ± 0.04**** | 2.32 ± 0.15 | 0.68 ± 0.03*** |
| SULT1E1-D220V | 3.56 ± 0.16**** | 1.53 ± 0.24**** | 2.35 ± 0.27*** |
Data shown represent mean ± SD derived from 3 independent experiments. One-way ANOVA was performed followed by Tukey’s post hoc analysis.
Statistical significant p<0.0001 from SULT1E1-WT.
Wild-type human SULT1E1
3.6. Structure simulation analyses of the single amino acid mutations.
Rotamer analyses were performed in order to investigate the effect of amino acid substitution on the conformation of respective SULT1E1 allozymes (Fig. 9). Of the five amino acid substitutions, A43D appeared to affect the interaction with Thr106 and Leu108 adjacent to the catalytic His residue (His107), implying an altered conformation of the His107-containing loop. A43D substitution, therefore, may affect the catalytic activity of the resulting SULT1E1 allozyme. R186L was also found to alter the surrounding conformation since the hydrogen bond between Arg186 and Cys122 in the wild-type enzyme is lost in the R186L allozyme. Similarly, D220V was also found to alter the surrounding conformation since the hydrogen bond between Asp220 and Arg200 in the wild-type enzyme is lost in the D220V allozyme. On the other hand, A131P appeared to have limited effect on the conformation of the corresponding SULT1E1 allozyme.
Fig. 9. Hydrophobic interaction and hydrogen bond analyses of the SULT1E1 allozymes.
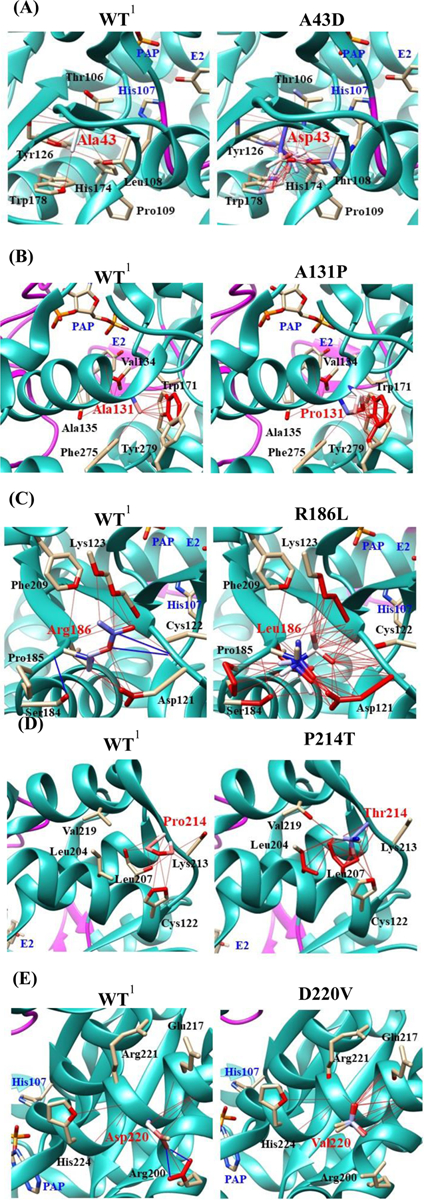
Atoms interacted with Ala43 (A), Ala131 (B), Arg186 (C), Pro214 (D), and Asp220 (E) are colored by the blue-white-red gradient (left panels; WT). Estimated interaction formed with Asp43 in A43D (A), Pro131 in A131P (B), Leu186 in R186L (C), Thr214 in P214T (D), and Val220 in D220V (E) are colored by the blue-white-red gradient (right panels; substituted). Side-chain conformation of a substituted residue was simulated using the Dunbrack backbone-dependent rotamer library [36]. Hydrophobic and hydrogen bond interactions of the substituted residues were also simulated by Find Clashes/Contacts tool in USCF Chimera software [37]. Top five-ranked rotamers of each substituted residue are modeled using the Dunbrack backbone-dependent rotamer library [36] and interaction was analyzed by Find Clashes/Contacts tool in USCF Chimera software. Hydrogen bonds formed with Cys122 (C) and Arg200 (E) are shown by blue slid lines (left panels). 1Wild-type human SULT1E1; PAP, (3’-phosphoadenosine 5’-phosphate); E2, (17β-Estradiol).
4. Discussion
The hepatic cytochrome P450, CYP2D6, mediates the metabolism of tamoxifen to form 4OH-tamoxifen, which is highly potent in antagonizing the estrogenic effects in mammalian tissues and may effectively suppress the proliferation of breast cancer cells [2–4]. It is under investigation for use in a topical gel formulation to alleviate the symptoms of cyclical mastalgia [38]. Diethylstilbestrol, an established treatment for prostate cancer in some European countries [39], has been proposed for use in an adjunctive estrogen treatment, together with other endocrine agents such as tamoxifen, to improve the beneficial effects of the combination therapy for metastatic breast cancer in postmenopausal patients [6–8]. SULT1E1, which is known to be expressed in estrogen-responsive tissues, including breast and prostate tissues [40–42], has been shown to be capable of sulfating both 4OH-tamoxifen and diethylstilbestrol [16–18]. It is noted that while SULT1A1 displayed higher sulfating activity toward 4OH-tamoxifen and diethylstilbestrol [16–18, 43], SULT1E1 has been shown to be more abundantly expressed in estrogen-responsive tissues and thus is likely a major responsible enzyme that metabolizes these drugs in breast tissue [40–42]. Variations in tamoxifen metabolism have been reported [33], [44–50]. Interestingly, a recent clinical study has demonstrated a correlation between genetic polymorphisms of tamoxifen-metabolizing enzymes including SULT1E1 and inter-individual variations of plasma concentrations of 4OH-tamoxifen in breast cancer patients treated with tamoxifen [33]. SULT1E1 is known to be the most efficient sulfotransferase enzyme in catalyzing the sulfation of endogenous estrogens, particularly E2 [24–26]. Considering its pivotal role in mediating estrogen sulfation, SULT1E1 may act as a key regulator of estrogen hemostasis. Additionally, previous epidemiological studies have demonstrated the correlation between genetic polymorphisms of SULT1E1 and the risk for breast cancer in Korean and Jewish women, as well as endometrial cancer in Caucasian women [28, 29, 31]. Another study has demonstrated that genetic variations in the human SULT1E1 gene may affect the sulfating activity of the encoded enzyme toward E2 [51]. The current study was carried out to investigate how genetic polymorphisms of the SULT1E1 gene, leading to amino acid variation, may affect the sulfating activity of coded SULT1E1 allozymes toward E2, 4OH-tamoxifen and diethylstilbestrol. Following a comprehensive database search for human SULT1E1 gene, five missense cSNPs of SULT1E1 were selected and their corresponding cDNAs were generated by site-directed mutagenesis. Recombinant SULT1E1 allozymes were expressed, purified, and characterized for their sulfating activity toward E2, 4OH-tamoxifen and diethylstilbestrol.
Initial experiments showed differential E2-sulfating activities among the five SULT1E1 allozymes (Fig. 4c). Kinetic analyses demonstrated further the distinct kinetic parameters Vmax (reflecting the catalytic activity), Km (reflecting the substrate affinity), and Vmax/Km (reflecting the catalytic efficiency) for these SULT1E1 allozymes in mediating E2 sulfation (Table 2). With 4OH-tamoxifen as a substrate, an initial investigation also demonstrated differential 4OH-tamoxifen-sulfating activities among the five SULT1E1 allozymes (Fig. 6). Kinetic analyses revealed the distinct kinetic parameters Vmax, Km, and Vmax/Km for these SULT1E1 allozymes in mediating 4OH-tamoxifen sulfation (Table 3). With diethylstilbestrol as a substrate, the sulfating activities, as shown in Fig. 8 also revealed significant variations in diethylstilbestrol-sulfating activities among the five examined SULT1E1 allozymes. Likewise, the kinetic constants compiled in Table 4 revealed significant changes in Vmax, Km, and Vmax/Km values of the five SULT1E1 allozymes with diethylstilbestrol as a substrate. Overall, the findings of this study showed clearly an impact of SULT1E1 missense coding SNPs (cSNPs) on the sulfation of E2, 4OH-tamoxifen and diethylstilbestrol by coded SULT1E1 allozymes.
Several crystal structures of human SULT1E1 have been reported [52–54]. Some key elements/ residues revealed in these structures include a 5’-phosphosulphate-binding (PSB) loop (45TYPKSGT51), a PAP adenine-binding region (W52, Y192 and T226), a 3’-phosphate-binding region (R129, S137, and 256RKG258), substrate-binding residues (Y20, F23, P46, F75, F80, C83, K85, M89, K105, H107, F138, F141, 145VAGH148, Y168, Y239, L242, and 246IM247), and a C-terminal dimerization motif (265KNHFTVALNE274) [52–56]. It is interesting to note that many of the amino acid residues associated with the SULT1E1 cSNPs examined in the current study fall within or are in close proximity to these important structural elements. To investigate the effect of amino acid substitution on the conformation of respective SULT1E1 allozymes, rotamer analyses were performed. Fig. 9 depicts the estimated hydrophobic interaction and hydrogen bonding between the variant residues with surrounding residues in respective SULT1E1 allozymes, in comparison with wild-type SULT1E1. For example, SULT1E1-A43D was shown to display differential catalytic efficiencies toward E2, 4OH-tamoxifen and diethylstilbestrol. It is noted that in wild-type SULT1E1 molecule, A43 is close to proline (Pro46) and lysine (Lys47) that are involved in, respectively, the substrate- and cofactor (PAPS)-binding [52, 54]. Rotamer analysis showed that A43D substitution may alter the conformation of the His107-containing loop, thereby changing the position of the catalytic residue, His107 (Fig. 9). The substitution of alanine (a non-polar and hydrophobic residue) with aspartic acid (an acidic residue) in SULT1E1-A43D may lead to an increased binding affinity for diethylstilbestrol and a decreased binding affinity for E2 and 4OH-tamoxifen, and thus the differential catalytic efficiencies toward these three substrates. Interestingly, P214T also showed differential catalytic efficiencies toward E2, 4OH-tamoxifen and diethylstilbestrol. The substitution of proline (a turn-inducing residue) with threonine (a non-turn-inducing residue) may introduce a kink in the alpha-helix region of the SULT-P214T molecule. Rotamer analysis indicated that the substitution may alter the interaction between Pro214 and Leu207/Cys122, which may lead to altered conformation of Pro214-containing PAP adenine-binding region and the adjacent loop-3. Conformation change of loop-3 may result in differential catalytic efficiencies of SULT-P214T toward E2, 4OH-tamoxifen and diethylstilbestrol. Similar to P214T, substitutions R186L and D220V may also cause the conformational changes in PAP adenine-binding region and the adjacent loop-3, leading to the disruption of the capability of the PAPS adenine moiety to form hydrogen bonds to tyrosine (Tyr192) and threonine (Thr226) residues that are located downstream from the R186L and D220V amino acid substitutions [54]. On the other hand, the replacement of alanine with proline in SULT1E1-A131P may disrupt the hydrogen bond interactions of the 3’-phosphate of the PAPS molecule with both Arg129 and Ser137 [52, 54]. Nevertheless, the effect of A131P substitution may not affect dramatically the catalytic activity of SULT1E1-A131P. Finally, it should be pointed out that there may be other effects of the amino acids substitutions in allozymes studied. Particularly, some of these amino acids substitutions that occur in the interior of the enzyme molecule may affect proteins turnover as well as steady-state levels. Additional studies are warranted in order to clarify this interesting issue.
5. Conclusion
In summary, the current study represented a first attempt aiming to clarify the impact of the genetic polymorphisms of the SULT1E1 gene on the enzymatic properties of the coded SULT1E1 protein products. Results obtained demonstrated clearly the differential sulfating activities of coded SULT1E1 allozymes toward E2, 4OH-tamoxifen and diethylstilbestrol. Pending additional studies, such information may have implications in assessing the risk for breast, ovarian, and endometrial cancers, as well as in designing individualized regimens of 4OH-tamoxifen and diethylstilbestrol for patients with distinct SULT1E1 genotypes, thereby enhancing efficacy and minimizing risk for toxicity of these two drugs.
Supplementary Material
Fig. S1. Time-dependent of the sulfation of 17β-Estradiol by human wild-type SULT1E1.
Key Points:
The results obtained from this study imply that individuals with different SULT1E1 genotypes may have differential capacity in sulfating E2, as well as 4OH-tamoxifen and diethylstilbestrol. Pending additional studies, the results may in the future help predict the risk for estrogen-related diseases and optimize the therapeutic uses for 4OH-tamoxifen and diethylstilbestrol in order to improve their efficacy and reduce risk for side effects in patients with different SULT1E1 genotypes.
Funding:
This work was supported in part by a grant from National Institutes of Health (Grant # R03HD071146).
Footnotes
Declarations
Conflicts of interest: The authors (A.E., F. A., M.A., A.B., M.R., K.K., and M.L.) declare no conflicts of interest.
Ethics Approval: N/A
Availability of data and material: Data and materials will be made available upon request.
Code availability: N/A
Consent to participate: N/A
Consent for publication: All authors have consented to the publication of this manuscript. No permissions from other parties are required.
References
- [1].Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med 1998;339:1609–18. 10.1056/NEJM199811263392207 [DOI] [PubMed] [Google Scholar]
- [2].Crewe HK, Ellis SW, Lennard MS, Tucker GT. Variable contribution of cytochromes P450 2D6, 2C9 and 3A4 to the 4-hydroxylation of tamoxifen by human liver microsomes. Biochem Pharmacol 1997;53(2):171–8. 10.1016/s0006-2952(96)00650-8 [DOI] [PubMed] [Google Scholar]
- [3].Crewe HK, Notley LM, Wunsch RM, Lennard MS, Gillam EM. Metabolism of tamoxifen by recombinant human cytochrome P450 enzymes: formation of the 4-hydroxy, 4’-hydroxy and N-desmethyl metabolites and isomerization of trans-4-hydroxytamoxifen. Drug Metab Dispos 2002;30(8):869–74. 10.1124/dmd.30.8.869 [DOI] [PubMed] [Google Scholar]
- [4].Jordan VC, Collins MM, Rowsby L, Prestwich G. A monohydroxylated metabolite of tamoxifen with potent antioestrogenic activity. J Endocrinol 1977;75(2):305–16. 10.1677/joe.0.0750305 [DOI] [PubMed] [Google Scholar]
- [5].Chang M Tamoxifen resistance in breast cancer. Biomol Ther (Seoul) 2012;20(3):256–67. 10.4062/biomolther.2012.20.3.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Iwase H, Yamamoto Y. Clinical benefit of sequential use of endocrine therapies for metastatic breast cancer. Int J Clin Oncol 2015;20:253–61. 10.1007/s10147-015-0793-8 [DOI] [PubMed] [Google Scholar]
- [7].Lonning PE, Taylor PD, Anker G, Iddon J, Wie L, Jorgensen LM, Mella O, Howell A. High-dose estrogen treatment in postmenopausal breast cancer patients heavily exposed to endocrine therapy. Breast Cancer Res Treat 2001;67:111–6. 10.1023/a:1010619225209 [DOI] [PubMed] [Google Scholar]
- [8].Mahtani RL, Stein A, Vogel CL. High-dose estrogen as salvage hormonal therapy for highly refractory metastatic breast cancer: a retrospective chart review. Clin Ther 2009;31:2371–78. 10.1016/j.clinthera.2009.11.002 [DOI] [PubMed] [Google Scholar]
- [9].de Voogt HJ, Smith PH, Pavone-Macaluso M, de Pauw M, Suciu S. Cardiovascular side effects of diethylstilbestrol, cyproterone acetate, medroxyprogesterone acetate and estramustine phosphate used for the treatment of advanced prostatic cancer: results from European Organization for Research on Treatment of Cancer trials 30761 and 30762. J Urol 1986;135(2):303–7. 10.1016/s0022-5347(17)45620-5 [DOI] [PubMed] [Google Scholar]
- [10].Fisher B, Costantino JP, Redmond CK, Fisher ER, Wickerham DL, Cronin WM. Endometrial cancer in tamoxifen-treated breast cancer patients: findings from the National Surgical Adjuvant Breast and Bowel Project (NSABP) B-14. J Natl Cancer Inst 1994;86(7):527–37. 10.1093/jnci/86.7.527 [DOI] [PubMed] [Google Scholar]
- [11].Latifyan S, Vansteelandt C, Lecomte S, Efira A. Tamoxifen induced thromboembolic events in breast cancer. Some possible mechanisms. Rev Med Brux 2017;38(6):494–500. [PubMed] [Google Scholar]
- [12].Manikandan R, Srirangam SJ, Pearson E, Brown SCW, O’Reilly P, Collins GN. Diethylstilboestrol versus bicalutamide in hormone refractory prostate carcinoma: a prospective randomized trial. Urol Int 2005;75(3):217–21. 10.1159/000087797 [DOI] [PubMed] [Google Scholar]
- [13].Nayfield SG, Gorin MB. Tamoxifen-associated eye disease. A review. J Clin Oncol 1996;14(3):1018–26. 10.1200/JCO.1996.14.3.1018 [DOI] [PubMed] [Google Scholar]
- [14].Salomao SR, Watanabe SE, Berezovsky A, Motono M. Multifocal electroretinography, color discrimination and ocular toxicity in tamoxifen use. Curr Eye Res 2007;32(4):345–52. 10.1080/02713680701229638 [DOI] [PubMed] [Google Scholar]
- [15].Smith DC, Redman BG, Flaherty LE, Li L, Strawderman M, Pienta KJ. A phase II trial of oral diethylstilbesterol as a second-line hormonal agent in advanced prostate cancer. Urology. 1998;52(2):257–60. 10.1016/s0090-4295(98)00173-3 [DOI] [PubMed] [Google Scholar]
- [16].Falany JL, Pilloff DE, Leyh TS, Falany CN. Sulfation of raloxifene and 4-hydroxytamoxifen by human cytosolic sulfotransferases. Drug Metab Dispos 2006;34(3):361–68. 10.1124/dmd.105.006551 [DOI] [PubMed] [Google Scholar]
- [17].Nishiyama T, Ogura K, Nakano H, Ohnuma T, Kaku T, Hiratsuka A, Muro K, Watabe T. Reverse geometrical selectivity in glucuronidation and sulfation of cis- and trans-4-hydroxytamoxifens by human liver UDP-glucuronosyltransferases and sulfotransferases. Biochem Pharmacol 2002;63(10):1817–30. 10.1016/s0006-2952(02)00994-2 [DOI] [PubMed] [Google Scholar]
- [18].Suiko M, Sakakibara Y, Liu MC. Sulfation of environmental estrogen-like chemicals by human cytosolic sulfotransferases. Biochem Biophys Res Commun 2000;267(1):80–4. 10.1006/bbrc.1999.1935 [DOI] [PubMed] [Google Scholar]
- [19].Falany C, Roth JA. Properties of human cytosolic sulfotransferases involved in drug metabolism In: Jeffery EH, editors. Human Drug Metabolism; From Molecular Biology to Man. FL: CRC Press; 1993. pp. 101–15. [Google Scholar]
- [20].Mulder GJ, Jakoby WB. Sulfation in conjugation reactions In: Mulder GJ, Jakoby WB, editors. Drug Metabolism. London: Taylor and Francis; 1990. pp. 107–61. [Google Scholar]
- [21].Weinshilboum R, Otterness D. Sulfotransferase enzymes In: Kaufmann FC, editors. Conjugation-Deconjugation Reactions in Drug Metabolism and Toxicity. Berlin: Springer-Verlag; 1994. pp. 45–78. [Google Scholar]
- [22].Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MW. A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics. 2004;14(3):199–211. 10.1097/00008571-200403000-00009 [DOI] [PubMed] [Google Scholar]
- [23].Freimuth RR, Wiepert M, Chute CG, Wieben ED, Weinshilboum RM. Human cytosolic sulfotransferase database mining: identification of seven novel genes and pseudogenes. Pharmacogenomics J 2004;4(1):54–65. 10.1038/sj.tpj.6500223 [DOI] [PubMed] [Google Scholar]
- [24].Falany CN, Krasnykh V, Falany JL. Bacterial expression and characterization of a cDNA for human liver estrogen sulfotransferase. J Steroid Biochem Mol Biol 1995;52(6):529–39. [DOI] [PubMed] [Google Scholar]
- [25].Zhang H, Varlamova O, Vargas FM, et al. Sulfuryl transfer: the catalytic mechanism of human estrogen sulfotransferase. J Biol Chem 1998;273(18):10888–92. [DOI] [PubMed] [Google Scholar]
- [26].Petrotchenko EV, Doerflein ME, Kakuta Y, et al. Substrate gating confers steroid specificity to estrogen sulfotransferase. J Biol Chem 1999;274(42):30019–22. [DOI] [PubMed] [Google Scholar]
- [27].Agarwal N, Alex AB, Farnham JM, Patel S, Gill D, Buckley TH, Stephenson RA, Cannon-Albright L. Inherited Variants in SULT1E1 and Response to Abiraterone Acetate by Men with Metastatic Castration Refractory Prostate Cancer. J Urol 2016;196(4):1112–6. 10.1016/j.juro.2016.04.079 [DOI] [PubMed] [Google Scholar]
- [28].Choi JY, Lee KM, Park SK, Noh DY, Ahn SH, Chung HW, Han W, Kim JS, Shin SG, Jang IJ, Yoo KY, Hirvonen A, Kang D. Genetic polymorphisms of SULT1A1 and SULT1E1 and the risk and survival of breast cancer. Cancer Epidemiol Biomarkers Prev 2005;14(5):1090–5. 10.1158/1055-9965.EPI-04-0688 [DOI] [PubMed] [Google Scholar]
- [29].Cohen S, Laitman Y, Kaufman B, Milgrom R, Nir U. Friedman E. SULT1E1 and ID2 genes as candidates for inherited predisposition to breast and ovarian cancer in Jewish women. Fam Cancer. 2009;8(2):135–44. 10.1007/s10689-008-9218-4 [DOI] [PubMed] [Google Scholar]
- [30].Daniels J, Kadlubar S. Sulfotransferase genetic variation: from cancer risk to treatment response. Drug Metab Rev 2013;45(4):415–22. 10.3109/03602532.2013.835621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hirata H, Hinoda Y, Okayama N, Suehiro Y, Kawamoto K, Kikuno N, Rabban JT, Chen LM, Dahiya R. CYP1A1, SULT1A1, and SULT1E1 polymorphisms are risk factors for endometrial cancer susceptibility. Cancer. 2008;112(9):1964–73. 10.1002/cncr.23392 [DOI] [PubMed] [Google Scholar]
- [32].Rebbeck TR, Troxel AB, Wang Y, Walker AH, Panossian S, Gallagher S, Shatalova EG, Blanchard R, Bunin G, DeMichele A, Rubin SC, Baumgarten M, Berlin M, Schinnar R, Berlin JA, Strom BL. Estrogen sulfation genes, hormone replacement therapy, and endometrial cancer risk. J Natl Cancer Inst 2006;98(18):1311–20. 10.1093/jnci/djj360 [DOI] [PubMed] [Google Scholar]
- [33].Woo HI, Lee SK, Kim J, Kim SW, Yu J, Bae SY, Lee JE, Nam SJ, Lee SY. Variations in plasma concentrations of tamoxifen metabolites and the effects of genetic polymorphisms on tamoxifen metabolism in Korean patients with breast cancer. Oncotarget 2017;8(59):100296–311. 10.18632/oncotarget.22220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yanagisawa K, Sakakibara Y, Suiko M, Takami Y, Nakayama T, Nakajima H, Takayanagi K, Natori Y, Liu MC. cDNA cloning, expression, and characterization of the human bifunctional ATP sulfurylase/adenosine 5’-phosphosulfate kinase enzyme. Biosci Biotechnol Biochem 1998;62(5):1037–40. 10.1271/bbb.62.1037 [DOI] [PubMed] [Google Scholar]
- [35].Hui Y, Liu MC. Sulfation of ritodrine by the human cytosolic sulfotransferases (SULTs): Effects of SULT1A3 genetic polymorphism. Eur J Pharmacol 2015;761:125–9. 10.1016/j.ejphar.2015.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dunbrack RL Jr. Rotamer libraries in the 21st century. Curr Opin Struct Biol 2002;12(4):431–40. 10.1016/S0959-440X(02)00344-5 [DOI] [PubMed] [Google Scholar]
- [37].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 2004;25(13):1605–12. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- [38].Mansel R, Goyal A, Nestour EL, Masini-Eteve V, O’Connell K. Afimoxifene Breast Pain Research Group, A phase II trial of Afimoxifene (4-hydroxytamoxifen gel) for cyclical mastalgia in premenopausal women. Breast Cancer Res Treat 2007;106(3):389–97. 10.1007/s10549-007-9507-x [DOI] [PubMed] [Google Scholar]
- [39].Turo R, Tan K, Thygesen H, Sundaram SK, Chahal R, Prescott S, Cross WR. Diethylstilboestrol (1 mg) in the management of castration-resistant prostate cancer. Urol Int 2015;94(3):307–12. 10.1159/000365198 [DOI] [PubMed] [Google Scholar]
- [40].Falany JL, Falany CN. Expression of cytosolic sulfotransferases in normal mammary epithelial cells and breast cancer cell lines. Cancer Res 1996;56(7):1551–55. [PubMed] [Google Scholar]
- [41].Nakamura Y, Suzuki T, Fukuda T, Ito A, Endo M, Moriya T, Arai Y, Sasano H. Steroid sulfatase and estrogen sulfotransferase in human prostate cancer. Prostate 2006;66(9):1005–12. 10.1002/pros.20426 [DOI] [PubMed] [Google Scholar]
- [42].Qian Y, Deng C, Song WC. Expression of estrogen sulfotransferase in MCF-7 cells by cDNA transfection suppresses the estrogen response: potential role of the enzyme in regulating estrogen-dependent growth of breast epithelial cells. J Pharmacol Exp Ther 1998;286(1):555–60 [PubMed] [Google Scholar]
- [43].Hui Y, Luo L, Zhang L, Kurogi K, Zhou C, Sakakibara Y, Suiko M, Liu MC. Sulfation of afimoxifene, endoxifen, raloxifene, and fulvestrant by the human cytosolic sulfotransferases (SULTs): A systematic analysis. J Pharmacol Sci 2015;128(3):144–9. 10.1016/j.jphs.2015.06.004 [DOI] [PubMed] [Google Scholar]
- [44].Areepium N, Panomvana D, Rungwanonchai P, Sathaporn S, Voravud N. Effects of CYP2D6 and UGT2B7 polymorphisms on pharmacokinetics of tamoxifen in Thai breast cancer patients. Breast Cancer (Dove Med Press). 2013;5:73–8. 10.2147/BCTT.S47172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].de Vries Schultink AH, Zwart W, Linn SC, Beijnen JH, Huitema AD. Effects of Pharmacogenetics on the Pharmacokinetics and Pharmacodynamics of Tamoxifen. Clin Pharmacokinet. 2015;54(8):797–810. 10.1007/s40262-015-0273-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Fernandez-Santander A, Gaibar M, Novillo A, Romero-Lorca A, Rubio M, Chicharro LM, Tejerina A, Bandres F. Relationship between genotypes Sult1a2 and Cyp2d6 and tamoxifen metabolism in breast cancer patients. PLoS One. 2013;8(7):e70183 10.1371/journal.pone.0070183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gjerde J, Hauglid M, Breilid H, Lundgren S, Varhaug JE, Kisanga ER, Mellgren G, Steen VM, Lien EA. Effects of CYP2D6 and SULT1A1 genotypes including SULT1A1 gene copy number on tamoxifen metabolism. Ann Oncol 2008;19(1):56–61. 10.1093/annonc/mdm434 [DOI] [PubMed] [Google Scholar]
- [48].Lim JS, Chen XA, Singh O, Yap YS, Ng RC, Wong NS, Wong M, Lee EJ, Chowbay B. Impact of CYP2D6, CYP3A5, CYP2C9 and CYP2C19 polymorphisms on tamoxifen pharmacokinetics in Asian breast cancer patients. Br J Clin Pharmacol 2011;71(5):737–50. 10.1111/j.1365-2125.2011.03905.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Murdter TE, Schroth W, Bacchus-Gerybadze L, Winter S, Heinkele G, Simon W, Fasching PA, Fehm T. German Tamoxifen and AI Clinician Group, Eichelbaum M, Schwab M, Brauch H. Activity levels of tamoxifen metabolites at the estrogen receptor and the impact of genetic polymorphisms of phase I and II enzymes on their concentration levels in plasma. Clin Pharmacol Ther 2011;89(5):708–17. 10.1038/clpt.2011.27 [DOI] [PubMed] [Google Scholar]
- [50].Zafra-Ceres M, de Haro T, Farez-Vidal E, Blancas I, Bandres F, de Duenas EM, Ochoa-Aranda E, Gomez-Capilla JA, Gomez-Llorente C. Influence of CYP2D6 polymorphisms on serum levels of tamoxifen metabolites in Spanish women with breast cancer. Int J Med Sci 2013;10(7):932–7. 10.7150/ijms.5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Adjei AA, Thomae BA, Prondzinski JL, Eckloff BW, Wieben ED, Weinshilboum RM. Human estrogen sulfotransferase (SULT1E1) pharmacogenomics: gene resequencing and functional genomics. Br J Pharmacol 2003;139(8):1373–82. 10.1038/sj.bjp.0705369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Pedersen LC, Petrotchenko E, Shevtsov S, Negishi M. Crystal structure of the human estrogen sulfotransferase-PAPS complex: evidence for catalytic role of Ser137 in the sulfuryl transfer reaction. J Biol Chem 2002;277(20):17928–32. 10.1074/jbc.M111651200 [DOI] [PubMed] [Google Scholar]
- [53].Shevtsov S, Petrotchenko EV, Pedersen LC, Negishi M. Crystallographic analysis of a hydroxylated polychlorinated biphenyl (OH-PCB) bound to the catalytic estrogen binding site of human estrogen sulfotransferase. Environ Health Perspect 2003;111(7):884–8. 10.1289/ehp.6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Thomas MP, Potter BV. The structural biology of oestrogen metabolism. J Steroid Biochem Mol Biol 2013;137:27–49. 10.1016/j.jsbmb.2012.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kakuta Y, Pedersen LG, Carter CW, Negishi M, Pedersen LC. Crystal structure of estrogen sulphotransferase. Nat Struct Biol 1997;4(11):904–8. 10.1038/nsb1197-904 [DOI] [PubMed] [Google Scholar]
- [56].Petrotchenko EV, Pedersen LC, Borchers CH, Tomer KB, Negishi M. The dimerization motif of cytosolic sulfotransferases. FEBS Lett 2001;490:39–43. 10.1016/s0014-5793(01)02129-9 [DOI] [PubMed] [Google Scholar]
- [57].Cook I, Wang T, Almo SC, Kim J, Falany CN, Leyh TS. The gate that governs sulfotransferase selectivity. Biochemistry. 2012;52(2):415–24. 10.1021/bi301492j [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Tibbs ZE, Rohn-Glowacki KJ, Crittenden F, Guidry AL, Falany CN. Structural plasticity in the human cytosolic sulfotransferase dimer and its role in substrate selectivity and catalysis. Drug Metab Pharmacokinet. 2015;30(1):3–20. 10.1016/j.dmpk.2014.10.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Time-dependent of the sulfation of 17β-Estradiol by human wild-type SULT1E1.


