Abstract
Current literature implicates arachidonic acid-derived leukotrienes and prostaglandins in the pathogenesis of chronic rhinosinusitis. However, other omega-3 and omega-6 derived lipid mediators, such as specialized pro-resolving mediators (SPMs), may also be important in chronic inflammatory disorders of the upper airway. We hypothesize that SPMs differ among CRS subtypes compared to controls and in relation to sinonasal microbiota. Ethmoid sinus tissue and middle meatal swabs were collected from a convenience sample of 66 subjects, including non-CRS controls, CRS with polyps (CRSwNP), and CRS without polyps (CRSsNP). Lipid mediator pathways were analyzed by liquid chromatography/tandem mass spectrometry. Bacterial taxa were profiled in parallel by 16S rRNA gene sequencing. Resolvin D2 was elevated in both CRSwNP (p=0.00076) and CRSsNP (p=0.030) compared with non-CRS controls. Lipoxin A4 was significantly increased in CRSwNP compared with CRSsNP (p=0.000033) and controls (p=0.044). Cigarette smoking was associated with significantly lower concentrations of several 15-lipoxygenase metabolites including resolvin D1 (p=0.0091) and resolvin D2 (p=0.0097), compared with never-smokers. Several of the lipid compounds also correlated with components of the sinonasal mucosal microbiota, including bacterial pathogens such as Pseudomonas aeruginosa. These data suggest that dysfunctional lipid mediator pathways in CRS extend beyond the traditional descriptions of leukotrienes and prostaglandins and include SPMs. Furthermore, dysregulated SPM signaling may contribute to persistent inflammation and bacterial colonization in CRS.
Keywords: Chronic Rhinosinusitis, Airway inflammation, Lipidomics, Microbiome
SUMMARY
We tested the hypothesis that lipidomic pathways differ among chronic rhinosinusitis (CRS) subtypes compared to controls and in relation to sinonasal microbiota. We examined surgical sinus tissue from 66 patients, including non-CRS controls, CRS with polyps (CRSwNP), and CRS without polyps (CRSsNP). Lipidomic pathways were analyzed by liquid chromatography/tandem mass spectrometry and bacterial taxa were profiled in parallel by 16S rRNA gene sequencing. We observed significantly elevated levels of Resolvin D2 in both CRSwNP and CRSsNP compared with non-CRS controls. Lipoxin A4 was significantly increased in CRSwNP compared with CRSsNP and controls. Cigarette smoking was associated with significantly lower concentrations of several 15-lipoxygenase metabolites including resolvin D1 and resolvin D2, compared with never-smokers. Several lipids also correlated with components of the sinonasal microbiota, including bacterial pathogens such as Pseudomonas aeruginosa. These data suggest that CRS subtypes have distinct lipidomic and microbial signatures which include specialized pro-resolving mediators.
1.0. INTRODUCTION
Chronic rhinosinusitis (CRS) is a chronic inflammatory disease characterized by nasal congestion, rhinorrhea, facial pain, and hyposmia.[1] The paranasal sinuses in CRS demonstrate varying accumulations of innate immune cells (e.g., neutrophils, eosinophils, fibroblasts, macrophages, goblet cells) and may be accompanied by the formation of nasal polyps (CRS with nasal polyps [CRSwNP], in contrast to CRS without polyps [CRSsNP]).[2, 3] Past efforts to characterize CRS endotypes have used variations in the predominant innate immune cell types observed within the inflammatory milieu, either neutrophils (CRSsNP) or eosinophils (CRSwNP), along with corresponding cytokines and primary helper T-cells (Th1 or Th2, respectively).[4] These cellular/molecular phenotypes are highly variable within and across patient populations worldwide.[5] CRSwNP is also associated with several other clinical conditions, including bronchial asthma and aspirin sensitivity (i.e., aspirin-exacerbated respiratory disease [CRSwNP-AERD]).[2–4]
Much attention in CRS has been devoted to understanding the origins of clinically apparent persistent inflammation. Initiation of acute inflammation typically results from an adaptive signaling cascade involving prostaglandins and leukotrienes, which together result in increased vascular permeability, transudative tissue edema, production of pro-inflammatory cytokines, and neutrophilic infiltration, followed by macrophage driven phagocytosis.[6] Similarly, resolution of acute inflammation entails a highly-choreographed and active process that is dependent on specialized pro-resolving lipid mediators (SPMs) derived from omega-3 and omega-6 polyunsaturated fatty acids.[7] These endogenously produced autacoids consist of arachidonic acid (AA) derived lipoxins (LXA4, LXB4), eicosapentanoic acid (EPA) derived resolvins (RvE1, RvE2), and docosahexaenoic acid (DHA) derived protectins (PD1), resolvins (RvD1, RvD2, RvD3), and maresins (MaR1, MaR2). Each of these compounds has spatiotemporally controlled and tissue-specific actions, which include halting neutrophil chemotaxis, increasing clearance of apoptotic neutrophils from the inflammatory milieu (efferocytosis), and directing the local chemokine and cytokine environment towards homeostasis.[8] Dysregulation of these pathways contributes to the pathogenesis of several chronic disease states such as bronchial asthma.[9]
Although earlier studies reported lipoxin deficiency in CRS-AERD,[10] a detailed examination of eicosanoid and docosanoid derivatives, particularly SPMs, is lacking in CRS. Here we report targeted liquid chromatography mass spectrometry (LC-MS/MS) profiling of 32 DHA and AA metabolites (Figure 1) in surgical tissue from patients with and without CRS. Prior studies suggest that nasal polyp fibroblasts are enriched in DHA when compared to non-polyp nasal mucosa.[11] This suggests that DHA pathway derivatives may elevated in nasal polyp tissue. We hypothesized that CRS subtypes (CRSsNP, CRSwNP, CRSwNP-AERD) and non-CRS controls would be characterized by unique lipid mediator profiles, allowing for a more detailed understanding of the molecular aberrations leading to and perpetuating CRS. In parallel, we examined the local microbiota to determine if specific bacteria corresponded with lipid mediator signatures and inflammatory phenotypes. This exploratory analysis of lipidome-microbiome associations was motivated by reports that lipid mediators modify the host innate immune response to bacterial pathogens,[12–16] suggesting that sinonasal bioactive lipids might directly influence, and be influenced by, commensal and pathogenic microbiota of the upper airways.
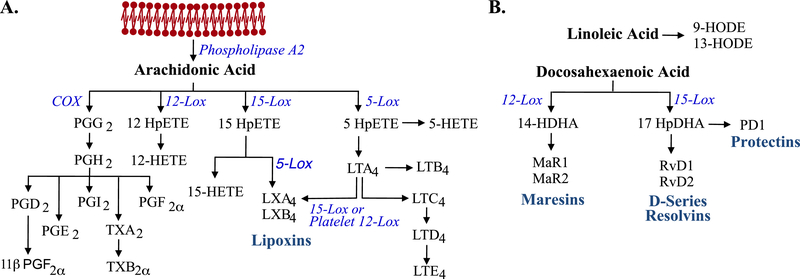
Figure 1. Simplified metabolic pathways of arachidonic acid and docosahexaenoic acid.
A. Arachidonic acid pathway metabolism. Membrane phospholipids are mobilized by phospholipase A2. Arachidonic acid is metabolized by either cyclooxygenase enzymes to form prostaglandins, prostacyclins, and thromboxanes or lipoxygenase pathways to form leukotrienes and lipoxins. B. Docosahexaenoic acid pathway metabolism. Docosahexaenoic acid is obtained the diet and oxidized by specific lipoxygenases to form precursors (14-HpDHA and 17-HpDHA) which are further metabolized to form resolvins, protectins, and maresins. Adapted from Harizi et al.[60] and Serhan and Petasis.[61]
2.0. MATERIALS AND METHODS
2.1. Patient selection and tissue collection
This cross-sectional study was approved by the Institutional Review Board of the University of Colorado (COMIRB protocol number 15–0574) and all study participants provided informed consent. The diagnosis of CRS was made according to the updated Adult Sinusitis Clinical Practice Guidelines, and accordingly, CRS patients were initially managed medically with a minimum trial of saline rinses, oral antibiotics and topical intranasal steroids.[1] Those with continued evidence of disease who elected to undergo endoscopic sinus surgery were enrolled in the study. Patients less than 18 years of age, with antibiotic use within 1 month of surgery (systemic or topical), or with cystic fibrosis, immunodeficiency or autoimmune diseases were excluded from the study. Control sinus tissue was obtained from patients undergoing nasal surgery for non-sinus related disease including anterior skull-base approaches and orbital decompressions, where no hormonal abnormalities were present, and normal sinuses were noted radiographically and endoscopically. After consent was obtained, additional information was collected including general demographic data, smoking history, allergy and asthma history, presence of aspirin intolerance with a respiratory reaction, oral aspirin challenge, prior surgical history, recent oral corticosteroid use, and SNOT-22 symptom survey. Sinus samples were collected at surgery from the ethmoid region, including the uncinate process, ethmoid bulla and nasal polyps emanating from this area. At the time of sample collection, the surgeon noted the presence or absence of purulence and transferred the tissue to a sterile sample collection cup and placed the sample on ice. Samples were then transferred to a 1.5 mL microcentrifuge tube and stored at −80°C until further processing. In total, sinus epithelial tissue from 66 patients met inclusion criteria and were included in the study (Table 1).
Table 1.
Tissue sample information and patient demographic data
| CON | CRSsNP | CRSwNP | |
|---|---|---|---|
| N | 13 | 20 | 33 |
| Age (Yrs)1 | 57 (30.0–59.0) | 50 (36.8–61.5) | 50 (37.8–55.5) |
| Sex (F/M)2 | 69.2% (9/13) | 65.0% (13/20) | 39.4% (13/33) |
| Allergy (Y/N)2 | 15.4% (2/13) | 35.0% (7/20) | 65.6% (21/32) |
| Asthma (Y/N)2 | 7.7% (1/13) | 25.0% (5/20) | 75.8% (25/33) |
| Aspirin Sensitivity (Y/N)2 | 0.0% (0/13) | 0.0% (0/20) | 36.4% (12/33) |
| Prior Nasal Surgery (Y/N)2 | 15.4% (2/11) | 25.0% (5/20) | 54.5% (18/33) |
| Purulence (Y/N)2 | 0.0% (0/13) | 45.0% (9/20) | 50.0% (16/32) |
| Smoking (Ever/Never)2 | 53.8% (7/13) | 30.0% (6/20) | 42.4% (14.33) |
| Saline Wash (Y/N)2 | 15.4% (2/13) | 95% (19/20) | 93.9% (31/33) |
| Topical Steroids (Y/N)2 | 23.1% (3/13) | 85% (17/20) | 90.9% (30/33) |
| Systemic Steroids (Y/N)2 | 0.0% (0/13) | 5.0% (1/20) | 40.6% (13/32) |
| SNOT221 | 38.5 (17–44.8) | 42.5 (27–58) | 49.0 (38.8–60) |
Median (IQR)
Proportion: First option / Total. Denominators may not equal N since values for some variables were not available for all subjects.
2.2. Lipid mediator extraction and metabolomic profiling
All standards and internal standards used for LC-MS/MS analysis of AA, DHA, and LA derived lipid mediators were purchased from Cayman Chemical (Ann Arbor, Michigan, USA). All HPLC and extraction solvents were HPLC grade or better. Lipid mediators in sinus epithelium samples were isolated as described by Yang et al.[17] The following procedural steps were performed on each of the sinus tissue samples separately. Tissue samples were pre-weighed and transferred into 1.5mL microcentrifuge tubes. 1mL of methanol and 10uL of internal standard solution (100pg total/each of 5(S)-HETE-d8, 8-iso-PGF2α-d4, 9(S)-HODE-d4, LTB4-d4, LTD4-d5, LTE4-d5, PGE2-d4, PGF2α-d9, RvD2-d5 and RvD1-d5 in ethanol) was added and then the sample was vortexed and stored overnight at −20°C. The sample was then transferred to a DUALL all glass size 21 tissue homogenizer (Kimball Chase, Vineland, NJ) and ground until homogenized, resting frequently on ice. The homogenate was transferred to a 1.5mL centrifuge tube and centrifuged at 14,000 RPM for 10 minutes at 4°C. The supernatant was diluted to 10mL with water and the sample rapidly acidified to pH 3.5. The samples were then immediately applied to Hypersep C18 500mg/6mL SPE columns (Thermo-Fisher, Fairlawn, NJ) that were prewashed with 20mL of methanol followed by 20mL of water. The SPE columns were then washed with 10mL of water followed by 10mL of hexane. Lipid mediators were eluted with 8mL of methyl formate (eicosanoids and docosanoid fraction) followed by 10mL of methanol (cysteinyl leukotriene fraction). Both fractions were taken down under a stream of nitrogen and reconstituted with 100% ethanol and combined into a reduced surface activity/maximum recovery glass autosampler vial analyzed immediately or stored at −70°C until analysis for no more than 1 week.
Quantitation of lipid mediators was performed using 2-dimensional reverse phase HPLC tandem mass spectrometry as previously described by Armstrong et al. [18] The HPLC system consisted of an Agilent 1260 autosampler (Agilent Technologies, Santa Clara, CA), an Agilent 1260 binary loading pump (pump 1), an Agilent 1260 binary analytical pump (pump 2) and a 6-port switching valve. Pump 1 buffers consisted of 0.1% formic acid in water (solvent A) and 9:1 v:v acetonitrile:water with 0.1% formic acid (solvent B). Pump 2 buffers consisted of 0.01% formic acid in water (solvent C) and 1:1 v:v acetonitrile:isopropanol (solvent D). 10uL of each extracted sample was injected onto an Agilent SB-C18 2.1X5mm 1.8um trapping column using pump 1 at 2mL/min for 30 seconds with a solvent composition of 97% solvent A:3% solvent B. At 31 seconds the switching valve changed the flow to the trapping column from pump 1 to pump 2. The flow was reversed and the trapped lipid mediators were eluted onto an Agilent Eclipse Plus C-18 2.1X150mm 1.8um analytical column using the following gradient at a flow rate of 0.3mL/min: hold at 75% solvent A:25% solvent D from 0–30 seconds, then a linear gradient from 25–75% D over 20 minutes followed by an increase from 75–100% D from 20–21 minutes, then holding at 100% D for 2 minutes. During the analytical gradient pump 1 washed the injection loop with 100% B for 22.5 minutes at 0.2mL/min. Both the trapping column and the analytical column were re-equilibrated at starting conditions for 5 minutes before the next injection.
Mass spectrometric analysis was performed on an Agilent 6490 triple quadruple mass spectrometer in negative ionization mode. The drying gas was 250°C at a flow rate of 15mL/min and the sheath gas was 350°C at 12mL/min. The nebulizer pressure was 35psi and the capillary voltage was 3500V.
Data for lipid mediators was acquired in dynamic MRM mode using experimentally optimized collision energies obtained by flow injection analysis of authentic standards (Supplemental Table 1). Calibration standards for each lipid mediator were analyzed over a range of concentrations from 0.25–250pg on column. Calibration curves for each lipid mediator were constructed using Agilent Masshunter Quantitative Analysis software. Tissue samples were quantified using the calibration curves to obtain the on-column concentration, followed by multiplication of the results by the appropriate dilution factor and normalization to input tissue weight to obtain the concentration in pg/mg of tissue (tissue homogenates).
2.3. Microbiome analysis
Middle meatus swabs were obtained during the procedure in which sinus tissues were collected for lipidomics. DNA was prepared from swabs using a phenol:choloroform bead-beating protocol.[19, 20] Broad-range bacterial 16S rRNA gene amplicons were generated using barcoded primers targeting approximately 300 base pairs of the V1V2 variable region of the 16S rRNA gene: 27F-YM (5′ - AGAGTTTGATYMTGGCTCAG) and 338R (5′ - TGCTGCCTCCCGTAGGAGT).[21–23] Pooled amplicons were diluted to 15pM and spiked with 25% of the Illumina PhiX control DNA prior to loading the Illumina MiSeq platform with a 600 cycle version 3 reagent kit. Quality-filtering, chimera removal, demultiplexing, and sequencing classification using SINA/Silva[24, 25] used our previously described methods.[19, 20, 26] This process generated 2,073,120 sequences for 56 samples (median of 23,015 sequences/sample; interquartile range: 13,484 – 49,729). The minimum Goods coverage score (a measure of depth of sequence coverage) across all sequence libraries was 99.2 % at the rarefaction point of 5,000 sequences, indicating excellent depth of sequence coverage. Demultiplexed 16S rRNA gene sequences and associated metadata were deposited in the NCBI Sequence Read Archive under accession number PRJNA678776.
2.4. Statistical analysis:
The Explicet and R software packages [27, 28] were used for data analysis and figure generation. Overall differences in lipid mediator levels between groups were assessed by multivariate analysis of variance (MANOVA) tests, using log10-transformed values as outcome variables and the Pillai-Bartlett test statistic. MANOVA tests were first conducted for each variable in Table 2, with experimental batch included as a covariate. Four variables with p<0.1 (CRS/polyps, use of saline washes, use of topical steroids, smoking history [never vs ever]) were selected for a MANOVA with multiple predictor variables. Between-group differences in individual lipid mediator levels were assessed by analysis of variance (ANOVA) tests, using log10-transformed values as outcome variables. Because the MANOVA tests indicated that CRS subtype and smoking history were the dominant determinants of lipid profiles, ANOVA tests of CRS subtype were first adjusted for smoking history and vice versa; batch was also included as a covariate in these analyses. For analysis of SNOT-22 scores, subjects were dichotomized using a cutoff score of 50.[29]
Table 2.
Multivariate Analysis of Variance Results.
| P-Value | Notes | |
|---|---|---|
| Age | 0.35 | Decade |
| Sex | 0.40 | F, M |
| CRS/Polyps | 0.00045 | CON, CRSsNP, CRSwNP |
| Allergies | 0.75 | Y, N |
| Asthma | 0.76 | Y, N |
| Aspirin Sensitivity | 0.17 | Y, N |
| Previous Surgery | 0.18 | Y, N |
| Purulence | 0.45 | Y, N |
| Smoking | 0.036 | NVR, EVR |
| Saline Wash | 0.089 | Y, N |
| Topical Steroids | 0.051 | Y, N |
| Systemic Steroids | 0.22 | Y, N |
| SNOT22 >= 501 | 0.48 | Y, N |
Dichotomozed at cutoff of 50 [29]
Associations between overall microbiota composition (beta-diversity) and individual lipid species were evaluated by the Microbiome Regression-Based Kernel Association Test (MiRKAT)[30] with individual lipid species as outcome variables and microbiome, CRS status (CRSwNP vs. CRSsNP vs. Control), smoking (ever vs. never smoker), and lipidomic batch included as covariates. In this analysis, kernel matrices were constructed using weighted (Bray-Curtis) and unweighted (Jaccard) beta-diversity indices calculated at the genus, family, order, and phylum levels. Omnibus p-values considering multiple kernels are reported for each lipid species. Associations between relative abundances of bacterial taxa and lipids were assessed by linear modeling of center-log transformed [31] taxon abundances as outcome variables, individual lipids (log10-transformed) as main predictor variables, first adjusting for CRS status (CRSwNP vs. CRSsNP vs. Control), smoking (ever vs. never smoker), and lipidomic batch. To visualize results, heatmaps were constructed based on t-statistics obtained from pairwise linear models of bacteria vs. lipids; rows and columns in which no associations were significant (p > 0.05) were excluded from the figure. Heatmaps were drawn with the R heatmap.2 function, with complete-linkage hierarchical clustering (hclust R function) of Euclidean distances (dist R function).
3.0. RESULTS
Targeted lipidomics was performed on tissue specimens collected during endoscopic sinus surgery from 53 adult CRS patients and 13 control subjects without signs or symptoms of CRS (Table 1). All 41 compounds in the standard calibration mix (32 analytes plus 9 deuterium labeled internal standards) were detected and confirmed by MS/MS spectral analysis (Supplemental Figure 1). All analytes were identified in the tissue samples except for the maresins (MaR1 and MaR2).
3.1. Clinical/demographic variables
Because CRS is characterized by a complex set of overlapping co-morbid conditions, we first assessed the extent to which these and other clinical/demographic variables were associated with the panel of lipid mediators through MANOVA tests of one or multiple predictor variables (Table 2). The most significant association with the lipid panel involved the 3-group comparison of CRSsNP vs. CRSwNP vs. controls (p = 0.00045). Cigarette smoking history (never vs. ever smokers) was the only other variable that reached significance (p = 0.036), while use of saline washes (p = 0.089) and topical steroids (p = 0.051) trended towards significance. A MANOVA analysis including CRS subtype, smoking history, saline washes, and topical steroids as predictor variables found that only CRS subtype (p = 0.00069) and smoking history (p = 0.046) remained significant; no interaction between CRS subtype and smoking history was evident (p = 0.29). Consequently, the following analyses (Fig. 2) were directed primarily towards identifying associations of individual lipid species with CRS subtype and smoking history (controlling for each). Because of the exploratory nature of this study, we focus on results with nominal p-values less than 0.05, but also note associations that remained significant after correcting p-values for multiple comparisons (i.e., fdr < 0.10).
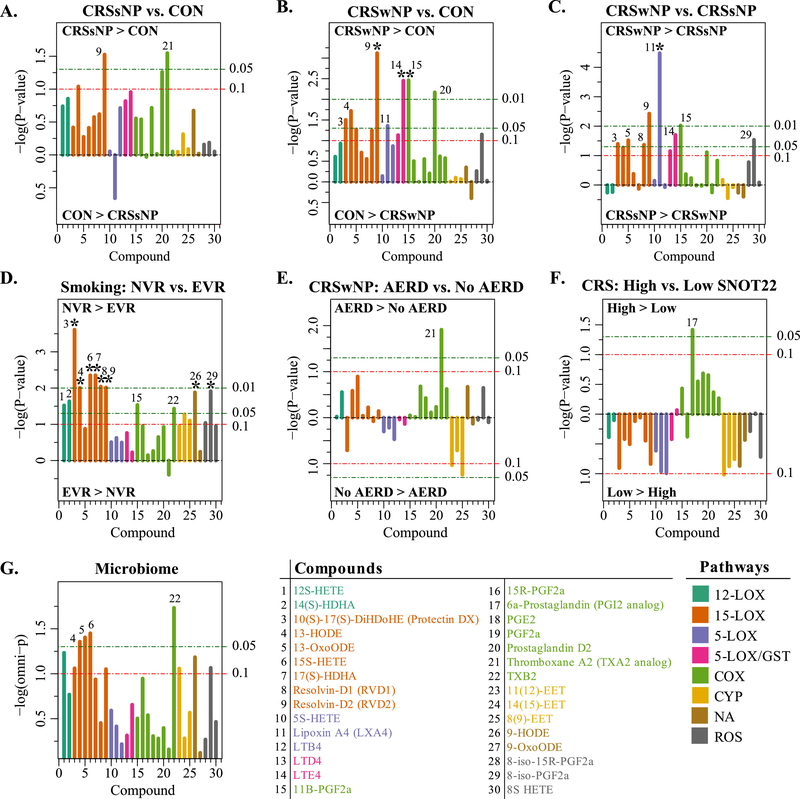
Figure 2. Differences in tissue concentration of lipid mediators between different pairs of groups.
Panels illustrate generalized increase in lipid mediators of inflammation in CRS compared to controls, in particular in the CRSwNP subtype. A. Compound differences between CRSsNP and Controls. Length of each line is proportional to −1*log10(p-value), such that longer lines indicate lower p-values. The horizontal lines show the cutoffs for p<0.1, p<0.05, and p<0.01. Lines pointing upward are for compounds with mean abundance greater in the first listed group (i.e. CRSsNP in panel A), compared with the second group (i.e. CON in Panel A). B. Compound differences between CRSwNP and Controls. C. Compound differences between CRSwNP and CRSsNP. D. Compares compound differences between never to ever smokers. E. Compares CRSwNP-AERD to aspirin tolerant CRSwNP (No AERD). F. Compares CRS patients with SNOT22 ≥50 to those with SNOT22 <50. G. Associations between composition of sinonasal microbiota and lipids. Individual compounds are listed at the bottom and the lines are color-coded by the predominant enzymatic pathways that produce the compounds. Compounds with nominal p-values <0.05 are enumerated in each panel. Asterisks denote compounds with false discovery rate (FDR) corrected p-values <0.05.
3.2. 15-lipoxygenase metabolites
DHA-derived specialized pro-resolving mediators resolvin D1 and D2 (RvD1 and RvD2) were detected in human sinus tissue for the first time here. RvD2 was significantly elevated in both CRSsNP (p = 0.030) and CRSwNP (p = 0.00076; fdr = 0.023) compared with control subjects (Fig. 2A and 2B). Furthermore, RvD2 was enriched in CRSwNP compared with CRSsNP (p = 0.0038; fdr = 0.056). RvD1 was elevated in CRSwNP compared with controls (p = 0.056). Higher concentrations of RvD1 (p=0.0091; fdr = 0.049) and RvD2 (p=0.0097; fdr = 0.049) also were observed in never-smokers compared to ever-smokers (Fig. 2D). Similarly, never smokers demonstrated increased levels of 15-lipoxygenase metabolites including the RvD1 precursor 17(s)-HDHA (p = 0.0044; fdr = 0.045) and protectin DX (p = 0.00025; fdr = 0.0073) (Fig. 2D). Protectin DX is 10S,17S-diHDHA with trans, cis, trans triene geometry but has distinct actions and arises from a different enzymatic pathway from the bioactive neuroprotectin D1 (10R,17S-diHDHA with trans, trans, cis triene geometry) which was not evaluated in the current study.
3.3. 5-lipoxygenase pathway mediators
Several AA derivative compounds that depend on 5-lipoxygenase activity in addition to 15-lipoxygenase were also increased in CRSwNP compared with controls (Fig. 2B). For example, LXA4 (p = 0.044) and the cysteinyl leukotrienes LTD4 (p = 0.074) and LTE4 (p = 0.0035; fdr = 0.035) all were increased in CRSwNP compared with controls. Similarly, LXA4 was significantly higher in CRSwNP compared with CRSsNP (p = 0.000033; fdr = 0.00099). Lower levels of LXA4 were observed in CRSsNP compared to controls though this relationship was not significant (p = 0.22).
3.4. Cyclooxygenase pathway mediators
Compared to controls, Prostaglandin D2 (PGD2) was increased in both CRSwNP (p = 0.0068; fdr = 0.051) and CRSsNP (p = 0.05) (Fig. 2B). CRSwNP specimens also had higher levels of 11β PGF2α than both CRSsNP (p = 0.0095; fdr = 0.095) and control tissue (p =0.0034; fdr = 0.035). Similarly, higher levels of 11β PGF2α were observed in never- vs. ever-smokers (p = 0.029; fdr = 0.082). No significant differences in prostaglandin E2 (PGE2) levels were observed across groups. Thromboxane A2 (TXA2) was significantly higher in CRSsNP vs. controls (p = 0.028) and in CRSwNP-AERD vs. CRSwNP without AERD (p = 0.012) (Fig. 2E).
3.5. 12-lipoxygenase mediators
Although recently identified macrophage-derived maresins (MaR1 and MaR2)[32] were not identified, 14(s) hydroxy-DHA (14(S)-HDHA) serves as a marker of maresin pathway activation (the true precursor molecule 14-hydroperoxy DHA (14 HpDHA) is not captured in the present study). 14(S)-HDHA was elevated in never smokers (p = 0.023; fdr = 0.077) suggesting that loss of the maresin pathway may contribute to pathogenicity related to cigarette smoke.
3.6. Associations with CRS symptoms
The MANOVA tests described above did not detect a significant association between lipid mediator levels and scores obtained through the SNOT-22 symptom-based survey (p = 0.48; dichotomized at a cutoff of 50).[29] Nonetheless, we sought to determine whether any individual lipid mediator pathway or metabolite species differed by SNOT-22 score, in order to identify potential biomarkers of greater disease severity. Because SNOT-22 scores were expected a priori to be higher for CRS than for non-CRS subjects, we restricted this analysis to CRS patients. Although only a few compounds approached significance (Fig. 2F), COX pathway mediators were generally enriched in subjects with high SNOT-22 scores (≥50), with 6-α-prostaglandin reaching significance (p = 0.038). In contrast, most other compounds, were diminished in subjects with higher SNOT-22 scores (≥50).
3.7. Microbiome associations
Middle meatal swabs from 62 of the 66 subjects were available for bacterial community profiling and 56 produced sufficient DNA for 16S rRNA amplicon sequencing. Using the Microbiome Regression-Based Kernel Association Test (MiRKAT)[30], we found that differences in overall microbiome composition between subjects (i.e., beta-diversity) were associated with between-individual differences in several lipid compounds, especially the 15-lipoxygenase metabolites (Fig. 2G).
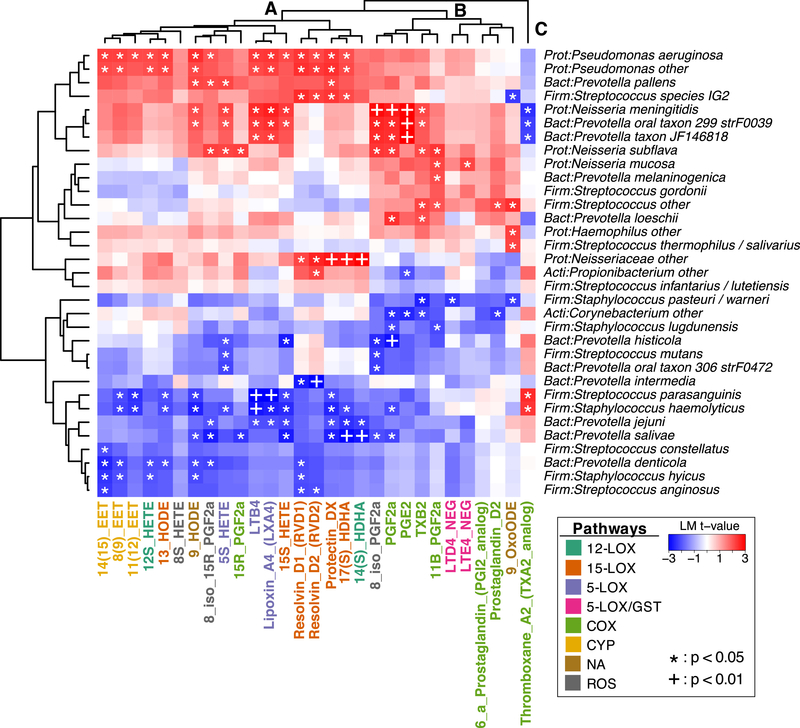
We next identified associations between individual lipids and selected bacterial genera/species belonging to noteworthy upper airway taxa (e.g., Corynebacterium, Haemophilus, Moraxella, Neisseria, Prevotella, Propionibacterium, Pseudomonas, Staphylococcus, Streptococcus). After correcting for CRS subtype and smoking history, multiple significant positive and negative associations (p < 0.05) were noted between lipids and bacterial taxa (Fig. 3). Hierarchical clustering resulted in three high-level groupings of lipids (labeled A-C in Fig. 3). Cluster A included all lipids belonging to the 12-lipoxygenase, 15-lipoxygenase, 5-lipoxygenase, and CYP pathways including the SPMs RvD1, RvD2, Protectin DX, and LXA4. Most of the lipids in this cluster were positively associated with Pseudomonas aeruginosa, unclassified Pseudomonas spp., and Prevotella pallens, and negatively associated with diverse members of the genera Staphylococcus, Streptococcus, and Prevotella. Cluster B comprised all of the 5-LOX/GST and COX lipids, including LTD4, LTE4, PGF2α, PGD2, and PGE2. In contrast to Cluster A, the lipids of Cluster B lipids were not significantly associated with Pseudomonas spp. and were associated with different species of Staphylococcus, Streptococcus, and Prevotella. Both Cluster A and Cluster B were positively associated with several species of Neisseria. Finally, Cluster C consisted solely of Thromboxane A2, which had a unique pattern of bacterial associations that was the inverse of the other lipids.
Figure 3. Associations between bacterial genera/species and lipid mediators.
Heatmaps summarize strength of association between pairs of taxa and lipids, as measured by t-statistics obtained through linear modeling (results were adjusted for CRS subtype and smoking history). Positive associations are in red while negative associations are in blue. Dendrograms show the results of hierarchical clustering along both rows and columns. Three primary clusters of lipids, denoted by “A”, “B”, and “C” are discussed in the text. Taxa that could be annotated only to the genus level are denoted “other”. *: p < 0.05. +: p< 0.01.
4.0. DISCUSSION AND CONCLUSIONS
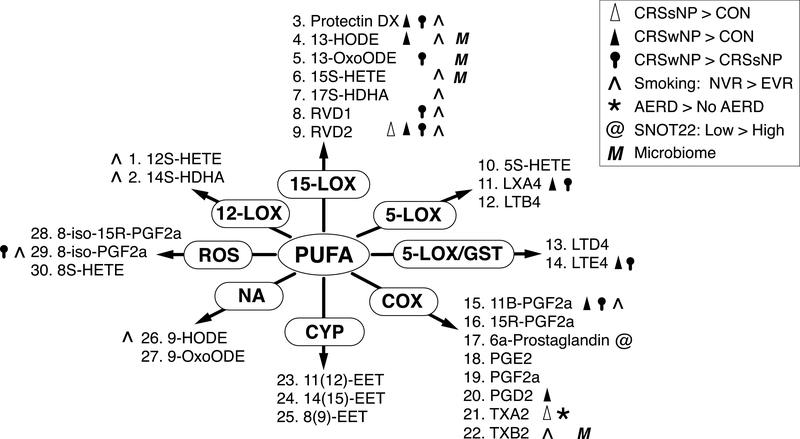
Biological disease processes in CRS are complex and multifaceted, requiring new, integrative approaches to fully understand the host and environmental factors driving disease initiation and chronic tissue inflammation. In this study, we applied a novel approach combining targeted metabolomics and microbiome profiling to identify associations between host CRS phenotypes, sinonasal lipid mediators, and key environmental factors (i.e., smoking history and sinonasal microbiota) in human subjects. Our combined analysis identifies lipidomic signatures of CRS subtypes in the setting of specific comorbidities and environmental factors such as smoking and mucosa-associated microbiota (Table 2, Fig. 4). In particular, we provide evidence supporting the hypothesis that SPMs, such as lipoxins, resolvins, and protectins, contribute to sinus mucosal homeostasis in addition to their protective effects in the lower airway.[33]
Figure 4. Summary of Primary Results.
Symbols denote lipid compounds (enumerated as in Fig. 2) that differed in abundance between particular comparison groups. These are organized by enzymatic pathways operating on poly-unsaturated fatty acid (PUFA) precursors.
Prior reports have identified elevations in cysteinyl leukotrienes (LTC4 and LTE4) and the cysteinyl leukotriene receptor CysLT1 in patients with nasal polyps using ELISA and real time PCR. [34–37] We similarly observed higher levels of Cys-LTs in CRSwNP patients compared to controls. Current understanding of AA-pathway dynamics in CRSwNP focuses on decreased COX2 expression, which is thought to shunt AA metabolism to lipoxygenase pathways, thus leading to high levels of leukotrienes and lower levels of prostaglandins. However, we also observed high levels of PGD2 in CRSwNP patients compared with controls, suggesting a larger role for arachidonic acid metabolism in polyp development. PGD2 and Cys-LTs regulate type II respiratory inflammatory disease and nasal polyps, in particular, though activation of type II innate lymphoid (ILC2) cells by binding specific receptors including the prostaglandin D2 receptor (CRTH2) and cysteinyl leukotriene receptors (CysLTr). [38–40]
These findings highlight the utility of a broad-based lipidomics approach that can capture complex changes in metabolism. Earlier reports demonstrated decreased levels of prostaglandin E2 in epithelial cells cultured from polyp patients,[35, 41] however in our samples, which comprised multiple cell types, PGE2 was low across all patients. In fact, we observed similar levels of PGE2 and LTB4 in all sampled tissues. These compounds are likely not the most informative for characterizing CRS subtypes since they facilitate both acute inflammation and pro-resolving actions though lipid mediator class-switching.[42, 43]
Recent data document increased 5-lipoxygenase expression and lipoxin A4 production in CRSwNP tissue compared with tissue from CRSsNP patients and healthy controls.[35] We similarly observed a surplus of LXA4 in CRSwNP compared with both CRSsNP (p = 0.000033) and control patients (p = 0.044). CRSsNP is often characterized by neutrophilic inflammation with Th1-predominant infiltrates enriched in IFNγ and TNFα.[44] LXA4 in addition to many other SPMs (RvD1, RvD2, MaR1) decrease IFNγ and TNFα production in CD4+ Th1 and Th17 cells. [45, 46] Thus in the healthy state, LXA4 attenuates the neutrophilic inflammatory response and a deficiency in lipoxin pathway products may contribute to the non-resolving inflammatory phenotype seen in CRSsNP which warrants further study.
Significantly higher levels of RvD1 and RvD2 were found in never smokers tissue compared with ever smoker sinus tissue. RvD1 reduces eosinophil and neutrophil infiltration, decreases smoking-induced emphysema and promotes tissue regeneration and Th2 differentiation in models of cigarette smoke-induced lung inflammation in mice.[47, 48] Decreased levels of DHA-derived resolvins in smokers’ sinus tissue may provide a possible mechanism for the observed poor outcomes among smokers with CRS and warrants further investigation.[49]
Whereas smoking and CRSsNP are characterized by robust type 1 inflammation, CRSwNP is more often associated with eosinophilia or type 2 inflammation.[50] We observed increased levels of RvD1 and RvD2 in CRSwNP patients. Interestingly, D-series resolvins are known to regulate type 2 inflammation. For example, RvD1 triggers polarization of macrophages toward the alternatively activated pro-resolving M2 phenotype via activation of the RvD1 receptor GPR32,[51] and CRSwNP patients have a more immunotolerant inflammatory milieu with elevated levels of M2 macrophages.[52–54] In methacholine mouse model of allergic airway disease, RvD1 decreased lower airway eosinophilia, mucous secretion, and promoted faster clearance of inflammatory infiltrates (efferocytosis).[55] SPMs and resolvins in particular also possess distinct roles in the adaptive immune response, which may also play a role in eosinophilic sinonasal polyps.[53] Phipps et al. demonstrated that RvD1 and its precursor 17-hydroxydocosahexaenoic acid (17-HDHA) suppress differentiation of naïve B cells into IgE secreting plasma cells and decrease IgE production by human B cells in vitro.[56] Taken together, the abundance of resolvins we observed in CRSwNP tissue may reflect an exuberant response to persistent tissue eosinophilia in an effort to promote homeostasis. Or, an interesting alternative is the possibility of SPM resistance in CRS, allowing for the propagation of continued inflammation that is a hallmark of the disease.
Our observation that tissue RvD2 is elevated in the chronic phase of CRSwNP is novel and introduces a role for resolution pharmacology in CRS. Since these compounds are differentially expressed in CRS subtypes, these pathways deserve further exploration in CRS pathophysiology and may offer unique molecular targets for novel therapeutics in the future. Although not statistically significant, DHA-derived resolvins were minimally elevated in CRSsNP relative to controls, indicative of the chronic inflammatory state in the airway. These observations, coupled with the lipoxin findings discussed above, lead us to speculate that a primary deficiency in SPMs may facilitate persistent inflammation in CRSsNP.
We observed distinct associations between lipid mediators and bacterial taxa that were more pronounced in CRSsNP and CRSwNP compared with controls, suggesting that activation of lipid mediator pathways in occurs concomitantly with shifts in microbiota. The interactions of microbes and SPMs on epithelial surfaces is an area of active research and most studies to date have utilized intestinal epithelia as a model system. For example, intestinal SPMs, including LXA4 and RvE1, increase expression of bactericidal permeability-increasing protein, resulting in antimicrobial effects against Gram-negative pathogens and mitigating the harmful effects of LPS.[14, 57] In murine models of acute peritonitis infections with E. coli and S. aureus, RvD2 decreases neutrophil infiltration, increases phagocytosis of bacteria, and expedites resolution.[58] Furthermore, SPMs such as RvD1, RvD5, and PD1 have been shown to lower the antibiotic requirements needed to clear infection in a murine model E. coli peritonitis. Despite the clear role of SPMs for mucosal homeostasis in the setting of pathogenic microbes in the gut, no studies have examined the role of SPMs in sinus mucosa. Here we identify both positive and negative associations between SPMs and sinus commensals and pathogens (e.g., Pseudomonas aeruginosa) that indicate a need for mechanistic follow up studies.
Several limitations in the current study must be noted. Most of the enrolled CRS patients (47/53) were treated with topical steroids, systemic steroids, or saline nasal irrigation at some point prior to surgery, consistent with clinical treatment guidelines, although relatively few subjects were on systemic corticosteroids in the month before surgery (Table 1).[1] Glucocorticoids can inhibit phospholipase A2 and skew downstream lipid mediator production from arachidonic acid. Both steroid exposure and saline irrigation could potentially influence lipidome and microbiome results, as evidenced by the results of the MANOVA analysis (Table 2, p = 0.089 and p = 0.051 for saline wash and topical steroids, respectively). Prescribed topical corticosteroid use and saline irrigation in CRS patients limited our ability to tease out their effects in this study, a common challenge in CRS research. However, that pathway and analyte differences are observed within the CRS cohort, supports the representative nature of these findings to endogenous tissue processes, rather than treatment per se. Additionally, this study utilized human surgical tissues, which invariably reflect different timepoints in each patient’s cycle of inflammatory disease and wound healing. The choreography and tight regulation of resolution pathways create challenges to examining chronically inflamed tissue in a cross-sectional fashion. Similarly, temporal and causal relationships between altered lipid and microbial profiles and disease phenotypes are difficult to ascertain through a cross-sectional design. Additionally, this study analyzed whole tissue rather than cell-specific roles. The contribution of myeloid cells within the inflammatory milieu can affect the synthesis of lipid mediators. For example, recent application of single-cell RNA sequencing indicates that prostaglandin D2 gene expression is found in sinonasal mast cells, whereas 15-lipoxygenase activity predominates in ciliated epithelial cells.[59]
In conclusion, lipid mediator pathway dysfunction in CRS, CRSwNP, and CRSwNP-AERD extends beyond traditional descriptions of leukotrienes and prostaglandins. Disease state and comorbidities such as cigarette smoke exposure play important roles in determining an individual’s sinonasal microbiota and host-specific immune function.[26] Specifically, DHA-derived SPMs RvD1 and RvD2 are differentially expressed in CRSwNP and individuals with prior smoking exposure, indicating that active resolution of inflammation may be disrupted in CRS, and thus should be given attention in the CRS pathophysiology paradigm that currently focuses primarily on pro-inflammatory activities. The apparent deficiency of SPMs in CRS offers new therapeutic targets that warrant further exploration. These potent lipid mediators are endogenously produced and possess anti-inflammatory and pro-resolving actions that lack the immunosuppressive properties of long-term corticosteroids that are often relied upon in CRS. For instance, particular endotypes might be responsive to 5- or 12-lipoxygenase inhibitors or monoclonal antibodies targeting eicosanoid receptors, or even probiotic strategies to correct LM-SPM imbalances. Finally, future studies applying multi-omic techniques to both sinus mucosa and its surface microbiota hold great promise to clarifying CRS endotypes and providing mechanistic insight into the effects of external factors such as smoking and microbiota on mucosal immunity. Ultimately this type of analysis could prove clinically useful by establishing signature metabolomic and microbiome profiles that can reliably distinguish CRS endotypes and suggest endotype-specific targeted therapies.
Supplementary Material
Supplemental Figure 1. Comparison of MRM extracted ion chromatograms for 25 selected analytes and internal standards in a 250pg calibration standard mix with identification of internal standard and select target analyte peaks (A) and Resolvin D2 in a 250pg calibration standard (B) and a sinus tissue sample (C).
Highlights:
Distinct lipidomic signatures in CRS offer new therapeutic targets for each subtype
Pro-resolving mediators RvD2 and LXA4 are elevated in CRSwNP compared to CRSsNP
Sinonasal RVD1 and RvD2 are decreased in cigarette smokers compared with never-smokers
Upper airway microbiota are associated with lipid mediators in underlying tissue
5.0. ACKNOWLEDGEMENTS
The authors thank all study participants and the study coordinator, Elyse Handley, BA for their support.
Support: Research reported in this publication was supported by the National Institute On Deafness and Other Communication Disorders (NIDCD) of the National Institutes of Health under a training grant to the Department of Otolaryngology at the University of Colorado (T32-DC012280) and grant number K23-DC014747 (VRR), as well as the Flight Attendants Medical Research Institute grants CIA130066 (DNF, VRR) and CIA160014 (DNF, VRR). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. VRR has served as a consultant for Medtronic, Inc., and Optinose US, which are unaffiliated with the current study.
Abbreviations:
- 16S rRNA
Small-subunit ribosomal ribonucleic acid gene
- CRS
Chronic rhinosinusitis
- CRSw/sNP
CRS with/without nasal polyps
- CRSwNP-AERD
CRSwNP with aspirin exacerbated respiratory disease
- CRSwNP-AT
Aspirin tolerant CRSwNP
- LC-MS/MS
Liquid chromatography tandem mass spectrometry
- SNOT-22
Sinonasal outcome test – 22 questions
- SPM
Specialized pro-resolving lipid mediator(s)
- Th1/Th2
T-helper lymphocyte cell, subtype 1/ subtype 2
- V1V2
16S rRNA gene variable regions 1 and 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Rosenfeld RM, Piccirillo JF, Chandrasekhar SS, Brook I, Kumar KA, Kramper M, Orlandi RR, Palmer JN, Patel ZM, Peters A, Walsh SA, Corrigan MD, Clinical practice guideline (update): Adult Sinusitis Executive Summary, Otolaryngol Head Neck Surg, 152 (2015) 598–609. [DOI] [PubMed] [Google Scholar]
- [2].Rajan JP, Wineinger NE, Stevenson DD, White AA, Prevalence of aspirin-exacerbated respiratory disease among asthmatic patients: A meta-analysis of the literature, J Allergy Clin Immunol, 135 (2015) 676–681 e671. [DOI] [PubMed] [Google Scholar]
- [3].Orlandi RR, Kingdom TT, Hwang PH, Smith TL, Alt JA, Baroody FM, Batra PS, Bernal-Sprekelsen M, Bhattacharyya N, Chandra RK, Chiu A, Citardi MJ, Cohen NA, DelGaudio J, Desrosiers M, Dhong HJ, Douglas R, Ferguson B, Fokkens WJ, Georgalas C, Goldberg A, Gosepath J, Hamilos DL, Han JK, Harvey R, Hellings P, Hopkins C, Jankowski R, Javer AR, Kern R, Kountakis S, Kowalski ML, Lane A, Lanza DC, Lebowitz R, Lee HM, Lin SY, Lund V, Luong A, Mann W, Marple BF, McMains KC, Metson R, Naclerio R, Nayak JV, Otori N, Palmer JN, Parikh SR, Passali D, Peters A, Piccirillo J, Poetker DM, Psaltis AJ, Ramadan HH, Ramakrishnan VR, Riechelmann H, Roh HJ, Rudmik L, Sacks R, Schlosser RJ, Senior BA, Sindwani R, Stankiewicz JA, Stewart M, Tan BK, Toskala E, Voegels R, Wang de Y, Weitzel EK, Wise S, Woodworth BA, Wormald PJ, Wright ED, Zhou B, Kennedy DW, International Consensus Statement on Allergy and Rhinology: Rhinosinusitis, Int Forum Allergy Rhinol, 6 Suppl 1 (2016) S22–S209. [DOI] [PubMed] [Google Scholar]
- [4].Akdis CA, Bachert C, Cingi C, Dykewicz MS, Hellings PW, Naclerio RM, Schleimer RP, Ledford D, Endotypes and phenotypes of chronic rhinosinusitis: a PRACTALL document of the European Academy of Allergy and Clinical Immunology and the American Academy of Allergy, Asthma & Immunology, J Allergy Clin Immunol, 131 (2013) 1479–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhang N, Van Zele T, Perez-Novo C, Van Bruaene N, Holtappels G, DeRuyck N, Van Cauwenberge P, Bachert C, Different types of T-effector cells orchestrate mucosal inflammation in chronic sinus disease, J Allergy Clin Immunol, 122 (2008) 961–968. [DOI] [PubMed] [Google Scholar]
- [6].Tabas I, Glass CK, Anti-inflammatory therapy in chronic disease: challenges and opportunities, Science, 339 (2013) 166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Serhan CN, Pro-resolving lipid mediators are leads for resolution physiology, Nature, 510 (2014) 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Buckley CD, Gilroy DW, Serhan CN, Proresolving lipid mediators and mechanisms in the resolution of acute inflammation, Immunity, 40 (2014) 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Levy BD, Vachier I, Serhan CN, Resolution of inflammation in asthma, Clin Chest Med, 33 (2012) 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Perez-Novo CA, Claeys C, Van Zele T, Holtapples G, Van Cauwenberge P, Bachert C, Eicosanoid metabolism and eosinophilic inflammation in nasal polyp patients with immune response to Staphylococcus aureus enterotoxins, Am J Rhinol, 20 (2006) 456–460. [DOI] [PubMed] [Google Scholar]
- [11].Ayyad SJ, Roca-Ferrer J, Picado C, Fatty Acid Composition of Cultured Fibroblasts Derived from Healthy Nasal Mucosa and Nasal Polyps, Sinusitis, 1 (2016) 55–64. [Google Scholar]
- [12].Campbell EL, Serhan CN, Colgan SP, Antimicrobial aspects of inflammatory resolution in the mucosa: a role for proresolving mediators, J Immunol, 187 (2011) 3475–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Canny G, Levy O, Furuta GT, Narravula-Alipati S, Sisson RB, Serhan CN, Colgan SP, Lipid mediator-induced expression of bactericidal/ permeability-increasing protein (BPI) in human mucosal epithelia, Proc Natl Acad Sci U S A, 99 (2002) 3902–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Campbell EL, MacManus CF, Kominsky DJ, Keely S, Glover LE, Bowers BE, Scully M, Bruyninckx WJ, Colgan SP, Resolvin E1-induced intestinal alkaline phosphatase promotes resolution of inflammation through LPS detoxification, Proc Natl Acad Sci U S A, 107 (2010) 14298–14303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Okano M, Fujiwara T, Haruna T, Kariya S, Makihara S, Higaki T, Nishizaki K, Prostaglandin E(2) suppresses staphylococcal enterotoxin-induced eosinophilia-associated cellular responses dominantly through an E-prostanoid 2-mediated pathway in nasal polyps, J Allergy Clin Immunol, 123 (2009) 868–874 e813. [DOI] [PubMed] [Google Scholar]
- [16].Perez-Novo CA, Waeytens A, Claeys C, Cauwenberge PV, Bachert C, Staphylococcus aureus enterotoxin B regulates prostaglandin E2 synthesis, growth, and migration in nasal tissue fibroblasts, J Infect Dis, 197 (2008) 1036–1043. [DOI] [PubMed] [Google Scholar]
- [17].Yang R, Chiang N, Oh SF, Serhan CN, Metabolomics-lipidomics of eicosanoids and docosanoids generated by phagocytes, Curr Protoc Immunol, Chapter 14 (2011) Unit 14 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Armstrong M, Manke J, Nkrumah-Elie Y, Shaikh SR, Reisdorph N, Improved quantification of lipid mediators in plasma and tissues by liquid chromatography tandem mass spectrometry demonstrates mouse strain specific differences, Prostaglandins Other Lipid Mediat, 151 (2020) 106483. [DOI] [PubMed] [Google Scholar]
- [19].Feazel LM, Robertson CE, Ramakrishnan VR, Frank DN, Microbiome complexity and Staphylococcus aureus in chronic rhinosinusitis, Laryngoscope, 122 (2012) 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ramakrishnan VR, Gitomer S, Kofonow JM, Robertson CE, Frank DN, Investigation of sinonasal microbiome spatial organization in chronic rhinosinusitis, Int Forum Allergy Rhinol, 7 (2017) 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Frank DN, BARCRAWL and BARTAB: Software tools for the design and implementation of barcoded primers for highly multiplexed DNA sequencing, BMC bioinformatics, 10 (2009) 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ, Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes, Appl Environ Microbiol, 74 (2008) 2461–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lane DJ, 16S/23S rRNA sequencing, in: Stackebrandt E, Goodfellow M (Eds.) Nucleic acid techniques in bacterial systematics, Wiley, New York, 1991, pp. 115–175. [Google Scholar]
- [24].Pruesse E, Peplies J, Glockner FO, SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes, Bioinformatics, 28 (2012) 1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO, The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucleic Acids Res, 41 (2013) D590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ramakrishnan VR, Frank DN, Impact of cigarette smoking on the middle meatus microbiome in health and chronic rhinosinusitis, Int Forum Allergy Rhinol, 5 (2015) 981–989. [DOI] [PubMed] [Google Scholar]
- [27].Robertson CE, Harris JK, Wagner BD, Granger D, Browne K, Tatem B, Feazel LM, Park K, Pace NR, Frank DN, Explicet: graphical user interface software for metadata-driven management, analysis and visualization of microbiome data, Bioinformatics, 29 (2013) 3100–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].R.C. Team, R: A Language and Environment for Statistical Computing, Vienna, Austria, R Foundation for Statistical Computing, Vienna, Austria, 2019. [Google Scholar]
- [29].Toma S, Hopkins C, Stratification of SNOT-22 scores into mild, moderate or severe and relationship with other subjective instruments, Rhinology, 54 (2016) 129–133. [DOI] [PubMed] [Google Scholar]
- [30].Zhao N, Chen J, Carroll IM, Ringel-Kulka T, Epstein MP, Zhou H, Zhou JJ, Ringel Y, Li H, Wu MC, Testing in Microbiome-Profiling Studies with MiRKAT, the Microbiome Regression-Based Kernel Association Test, Am J Hum Genet, 96 (2015) 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Aitchison j., The Statistical Analysis of Compositional data, Chapman & Hall Ltd., London, 1986. [Google Scholar]
- [32].Serhan CN, Dalli J, Karamnov S, Choi A, Park CK, Xu ZZ, Ji RR, Zhu M, Petasis NA, Macrophage proresolving mediator maresin 1 stimulates tissue regeneration and controls pain, FASEB J, 26 (2012) 1755–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Serhan CN, Levy BD, Resolvins in inflammation: emergence of the pro-resolving superfamily of mediators, J Clin Invest, 128 (2018) 2657–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Choi JH, Kim MA, Park HS, An update on the pathogenesis of the upper airways in aspirin-exacerbated respiratory disease, Curr Opin Allergy Clin Immunol, 14 (2014) 1–6. [DOI] [PubMed] [Google Scholar]
- [35].Perez-Novo CA, Watelet JB, Claeys C, Van Cauwenberge P, Bachert C, Prostaglandin, leukotriene, and lipoxin balance in chronic rhinosinusitis with and without nasal polyposis, J Allergy Clin Immunol, 115 (2005) 1189–1196. [DOI] [PubMed] [Google Scholar]
- [36].Steinke JW, Bradley D, Arango P, Crouse CD, Frierson H, Kountakis SE, Kraft M, Borish L, Cysteinyl leukotriene expression in chronic hyperplastic sinusitis-nasal polyposis: importance to eosinophilia and asthma, J Allergy Clin Immunol, 111 (2003) 342–349. [DOI] [PubMed] [Google Scholar]
- [37].Sousa AR, Parikh A, Scadding G, Corrigan CJ, Lee TH, Leukotriene-receptor expression on nasal mucosal inflammatory cells in aspirin-sensitive rhinosinusitis, N Engl J Med, 347 (2002) 1493–1499. [DOI] [PubMed] [Google Scholar]
- [38].Mjosberg JM, Trifari S, Crellin NK, Peters CP, van Drunen CM, Piet B, Fokkens WJ, Cupedo T, Spits H, Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161, Nat Immunol, 12 (2011) 1055–1062. [DOI] [PubMed] [Google Scholar]
- [39].Poposki JA, Klingler AI, Tan BK, Soroosh P, Banie H, Lewis G, Hulse KE, Stevens WW, Peters AT, Grammer LC, Schleimer RP, Welch KC, Smith SS, Conley DB, Raviv JR, Karras JG, Akbari O, Kern RC, Kato A, Group 2 innate lymphoid cells are elevated and activated in chronic rhinosinusitis with nasal polyps, Immun Inflamm Dis, 5 (2017) 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Barrett NA, Boyce JA, Activation of group 2 innate lymphoid cells: a new role for cysteinyl leukotrienes, J Allergy Clin Immunol, 132 (2013) 214–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kowalski ML, Pawliczak R, Wozniak J, Siuda K, Poniatowska M, Iwaszkiewicz J, Kornatowski T, Kaliner MA, Differential metabolism of arachidonic acid in nasal polyp epithelial cells cultured from aspirin-sensitive and aspirin-tolerant patients, Am J Respir Crit Care Med, 161 (2000) 391–398. [DOI] [PubMed] [Google Scholar]
- [42].Levy BD, Clish CB, Schmidt B, Gronert K, Serhan CN, Lipid mediator class switching during acute inflammation: signals in resolution, Nat Immunol, 2 (2001) 612–619. [DOI] [PubMed] [Google Scholar]
- [43].Basil MC, Levy BD, Specialized pro-resolving mediators: endogenous regulators of infection and inflammation, Nat Rev Immunol, 16 (2016) 51–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fokkens WJ, Lund VJ, Mullol J, Bachert C, Alobid I, Baroody F, Cohen N, Cervin A, Douglas R, Gevaert P, Georgalas C, Goossens H, Harvey R, Hellings P, Hopkins C, Jones N, Joos G, Kalogjera L, Kern B, Kowalski M, Price D, Riechelmann H, Schlosser R, Senior B, Thomas M, Toskala E, Voegels R, Wang de Y, Wormald PJ, European Position Paper on Rhinosinusitis and Nasal Polyps 2012, Rhinology. Supplement, (2012) 3 p preceding table of contents, 1–298. [PubMed] [Google Scholar]
- [45].Ariel A, Chiang N, Arita M, Petasis NA, Serhan CN, Aspirin-triggered lipoxin A4 and B4 analogs block extracellular signal-regulated kinase-dependent TNF-alpha secretion from human T cells, J Immunol, 170 (2003) 6266–6272. [DOI] [PubMed] [Google Scholar]
- [46].Chiurchiu V, Leuti A, Dalli J, Jacobsson A, Battistini L, Maccarrone M, Serhan CN, Proresolving lipid mediators resolvin D1, resolvin D2, and maresin 1 are critical in modulating T cell responses, Sci Transl Med, 8 (2016) 353ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hsiao HM, Sapinoro RE, Thatcher TH, Croasdell A, Levy EP, Fulton RA, Olsen KC, Pollock SJ, Serhan CN, Phipps RP, Sime PJ, A novel anti-inflammatory and pro-resolving role for resolvin D1 in acute cigarette smoke-induced lung inflammation, PloS one, 8 (2013) e58258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kim KH, Park TS, Kim YS, Lee JS, Oh YM, Lee SD, Lee SW, Resolvin D1 prevents smoking-induced emphysema and promotes lung tissue regeneration, International journal of chronic obstructive pulmonary disease, 11 (2016) 1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Christensen DN, Franks ZG, McCrary HC, Saleh AA, Chang EH, A Systematic Review of the Association between Cigarette Smoke Exposure and Chronic Rhinosinusitis, Otolaryngol Head Neck Surg, 158 (2018) 801–816. [DOI] [PubMed] [Google Scholar]
- [50].Mehta H, Nazzal K, Sadikot RT, Cigarette smoking and innate immunity, Inflamm Res, 57 (2008) 497–503. [DOI] [PubMed] [Google Scholar]
- [51].Schmid M, Gemperle C, Rimann N, Hersberger M, Resolvin D1 Polarizes Primary Human Macrophages toward a Proresolution Phenotype through GPR32, J Immunol, 196 (2016) 3429–3437. [DOI] [PubMed] [Google Scholar]
- [52].Krysko O, Holtappels G, Zhang N, Kubica M, Deswarte K, Derycke L, Claeys S, Hammad H, Brusselle GG, Vandenabeele P, Krysko DV, Bachert C, Alternatively activated macrophages and impaired phagocytosis of S. aureus in chronic rhinosinusitis, Allergy, 66 (2011) 396–403. [DOI] [PubMed] [Google Scholar]
- [53].Duffney PF, Falsetta ML, Rackow AR, Thatcher TH, Phipps RP, Sime PJ, Key roles for lipid mediators in the adaptive immune response, J Clin Invest, 128 (2018) 2724–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Croasdell A, Lacy SH, Thatcher TH, Sime PJ, Phipps RP, Resolvin D1 Dampens Pulmonary Inflammation and Promotes Clearance of Nontypeable Haemophilus influenzae, J Immunol, 196 (2016) 2742–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rogerio AP, Haworth O, Croze R, Oh SF, Uddin M, Carlo T, Pfeffer MA, Priluck R, Serhan CN, Levy BD, Resolvin D1 and aspirin-triggered resolvin D1 promote resolution of allergic airways responses, J Immunol, 189 (2012) 1983–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kim N, Ramon S, Thatcher TH, Woeller CF, Sime PJ, Phipps RP, Specialized proresolving mediators (SPMs) inhibit human B-cell IgE production, European journal of immunology, 46 (2016) 81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Levy O, Canny G, Serhan CN, Colgan SP, Expression of BPI (bactericidal/permeability-increasing protein) in human mucosal epithelia, Biochem Soc Trans, 31 (2003) 795–800. [DOI] [PubMed] [Google Scholar]
- [58].Chiang N, Dalli J, Colas RA, Serhan CN, Identification of resolvin D2 receptor mediating resolution of infections and organ protection, J Exp Med, 212 (2015) 1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ordovas-Montanes J, Dwyer DF, Nyquist SK, Buchheit KM, Vukovic M, Deb C, Wadsworth MH 2nd, Hughes TK, Kazer SW, Yoshimoto E, Cahill KN, Bhattacharyya N, Katz HR, Berger B, Laidlaw TM, Boyce JA, Barrett NA, Shalek AK, Allergic inflammatory memory in human respiratory epithelial progenitor cells, Nature, 560 (2018) 649–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Chiang N, Fredman G, Backhed F, Oh SF, Vickery T, Schmidt BA, Serhan CN, Infection regulates pro-resolving mediators that lower antibiotic requirements, Nature, 484 (2012) 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Serhan CN, Petasis NA, Resolvins and protectins in inflammation resolution, Chem Rev, 111 (2011) 5922–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Comparison of MRM extracted ion chromatograms for 25 selected analytes and internal standards in a 250pg calibration standard mix with identification of internal standard and select target analyte peaks (A) and Resolvin D2 in a 250pg calibration standard (B) and a sinus tissue sample (C).