Abstract
Polycystic ovarian syndrome and its associated endocrine abnormalities comprise one of the most common metabolic spectrum disorders within the human race. Due to the variance in phenotypic expression among individuals and within family lineages, attention has been turned to genetic and epigenetic changes in which the root cause of the disorder may lie. Further understanding of DNA/histone methylation and microRNA patterns may help to improve the accuracy of diagnosis and lead to future treatment options.
Keywords: PCOS, Genetics, Epigenetics, DNA, Methylation, MicroRNA
Introduction
Polycystic ovarian syndrome (PCOS) and its associated endocrine abnormalities comprise one of the most common metabolic spectrum disorders within the human race[1]. Based on the frequency in which PCOS in seen clinically and the ramifications the disease process can have on reproductive and metabolic health, research is underway with the hopes of finding not only the underlying etiology of the disorder, but also better tools for diagnosis and treatment. Due to the variance in phenotypic expression among individuals and within family lineages, attention has been turned to genetic and epigenetic changes in which the root cause of the disorder may lie.
Genetics
PCOS is known to be a complex disorder in which heritable genetics has long been suspected due to familial clustering of symptoms. Sisters often share hyperandrogenic and menstrual similarities with their mother while their male siblings may demonstrate hyperandrogenism via symptoms such as early balding[2]. Initial studies into a truly genetic etiology revealed a possible autosomal dominant mode of inheritance however further investigation has driven the revelation that PCOS is multigenic in origin. Work continues within this investigative arena however challenges exist due to lack of consistent diagnostic criteria and similarities in phenotypic expression across different ethnic groups[3].
Multiple population-wide genome-wide association studies (GWAS) have been leveraged to investigate PCOS with results revealing numerous pathogenic variants. Conservation has been noted across many promoter genes, including the following major contributors: LHCGR, THADA and DENND1A[3–6]. Luteinizing hormone/chorionic gonadotropic receptors (LHCGR) are found in theca and mature granulosa cells of the adult ovary with associated polymorphisms driving excess androgen production[7]. DENND1A encodes for DENN proteins with overexpression also driving excess ovarian steroidogenesis[8]. Changes in expression of the thyroid adenoma-associated gene (THADA) alter insulin secretion and subsequent insulin resistance by modifying pancreatic beta cell function. Further associated genes and their respective association with PCOS pathophysiology can be seen in figure 1.
Figure 1.
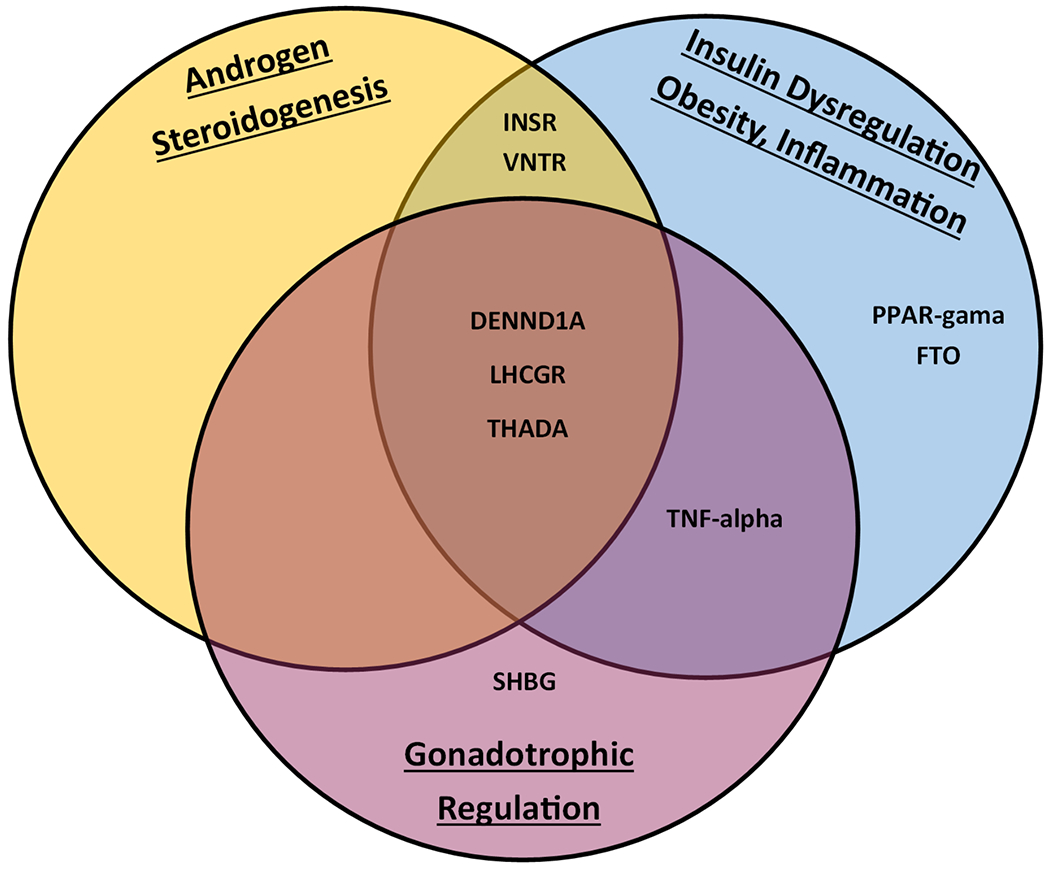
Promoter Genes identified by GWAS. (VNTR – Variable Number Tandem Repeats Insulin Gene; INSR – Insulin Receptor Gene; PPAR-Gama – Peroxisome Proliferator-Activated Receptor; FTO – Fat Mass/Obesity Gene; DENND1A – DENN Domain-containing Protein 1; LHCGR – Luteinizing Hormone/Chorionic Gonadotropin Receptor; THADA – Thyroid Adenoma-Associated Protein; TNF-Alpha – Tumor Necrosis Factor-Alpha; SHBG – Sex Hormone Binding Globulin).
The large volume of data generated by GWAS and conclusions drawn from its analysis were not without controversy. Ultimately, no single gene loci was identified as the common etiology for PCOS. The discovery of these loci did however open the door for further study into their DNA foundations and the changes which cause associated differences in expression[3, 4].
Mitochondrial DNA heritability may be another contributing factor to the genetic lineage of PCOS[9]. This topic will be covered in further detail at the conclusion of this chapter.
Epigenetics
Given the lack of specific, heritable findings as a single etiology for PCOS, investigators began looking at other factors which could change gene expression characteristics. Environmental exposures, specifically increased exposure to in-utero androgens, has become a leading theory in fetal programming for PCOS. Dumesic et al. proposed maternal insulin resistance leads to fetal exposure to hyperinsulinemia, which in turn drives fetal ovarian steroidogenesis with excess androgen production. In a normal pregnancy, this excess androgen should be resolved by placental aromatase activity however placental dysfunction in PCOS pregnancies has been previously reported. This two-hit mechanism of hyperinsulinemia driving excess androgen production, with decreased placental aromatase activity to counterbalance the system, likely results in a hyperandrogenic fetal environment[5, 10]. Proven by elevated testosterone levels in second trimester amniocentesis and umbilical vein sampling of female infants born to mothers with PCOS, the Barker hypothesis correlates this exposure with re-programming of offending genes, such as those identified via GWAS[4, 5, 11]. Animal models using sheep, mice and monkeys have confirmed this hypothesis with the reported development of PCOS phenotypes and associated DNA methylation changes following exposure to increased androgen levels in-utero[3, 10].
This epigenetic re-programming is facilitated by modifications to chromatin without additions nor deletions to the existing DNA and can be accomplished via one of two mechanisms, or both in combination. The first includes methylation, hydroxymethylation, formylation or carboxylation modifications of cytosine at its 5th carbon in the pyrimidine ring followed by a guanine, also known as CpG islands[3, 10]. Methylation of DNA is generally thought to inhibit gene expression while hydroxymethylation increases expression[12]. The second mechanism is histone modification by acetylation, methylation, phosphorylation, ubiquitylation or sumoylation. Epigenetic re-programing can occur in both somatic and germ cells however only germ cells have the potential to pass these changes to offspring[3].
Once epigenetic changes occur, DNA transcription is either down-regulated or up-regulated. This is followed by a correlated change in translation and protein production which ultimately affects gene expression. Confirmation of these methylated/hypomethylated changes to genome-wide DNA have been studied through peripheral blood sampling via next-generation sequencing in women with PCOS, revealing numerous genes which differ in methylation status between women with and without the disease. Similar to previous GWAS studies, the LHCGR gene encoding for LH receptor presence was again identified as having modified expression levels in PCOS. Others include FST – encodes for follistatin, LMNA – encodes for Lamin A/C, PPARGC1A – encodes for peroxisome proliferation, and EPHX1 – encodes for epoxide hydrolase. As demonstrated when searching for a single, global genetic cause for PCOS, none of these individual gene methylation alterations are responsible for the disease process. They are instead building blocks which encode for the physiologic processes of follicular development, steroidogenesis, glucose metabolism, insulin regulation and inflammatory mediation[10]. Changes in their methylation status results in the derangement of multiple systems leading to syndromic outcomes.
While initially promising, concerns for lack of statistical significance in these serum genome-wide methylation findings were presented due the inability to specify which cell line or tissue type produced detectable levels of epigenetic changes. This led to further study of tissue-specific cell lines, such as those within granulosa cells of PCOS women[10, 12, 13]. Again found with decreased levels of methylation and subsequent over-expression were LHCGR and PPAR-gama genes[14].
Xu et al. took this analysis perspective further by examining the differences between obese versus non-obese PCOS patients in regard to DNA methylation levels. Their findings indicate that obese PCOS patients have higher levels of serum global methylation present than no-obese PCOS patients. This indicates obesity may play a role within the disease process, not only as a phenotypic expression but also an epigenetic modifier itself[12]. Further evaluation of adipose tissue by this same group in 2011 demonstrated the presence of elevated DNA methylation levels, contributing to the above findings. Interestingly, different adipose-specific gene sets were methylated in infant versus adult rhesus monkey models who were exposed to androgens in-utero[15]. This alludes to the constant change present within epigenetic modifications and furthers the notion that while PCOS may originate from pre-programming in-utero, the disease process continues to be driven by epigenetic changes resulting from internal environmental changes such as body habitus.
Histone epigenetic modifications, most commonly via acetylation or methylation, complements the direct methylation changes in DNA. As mentioned previously, these modifications may be paired and potentiate one another or appear in opposition. Proper gene expression requires the presence of the correct pattern of both DNA and histone epigenetic modifiers[10, 16]. Hosseini et al. confirmed the pathophysiologic link between histone acetylation and PCOS while studying CYP19A1 in ovarian cumulus cells when compared to controls. Increased serum levels of acetylation in histone H3 and methylation of H3K9 were found in PCOS patients, reducing the expression of CYP19A1, ultimately reducing cytochrome P450 aromatase activity[16]. This loss in aromatase function is likely a contributing factor to the hyperandrogenic phenotype and subsequent pathology in PCOS.
Another facet of epigenetic modifications influencing gene expression involves the presence of microRNA (miRNA). Previously studied in other adult disease states, changes in miRNA expression are known to be associated with diabetes, insulin resistance, inflammation and cancer[6]. These non-coding single-stranded RNA molecules regulate gene expression following translation and are capable of modulating DNA methyltransferases and histone deacetylases[17]. While present in large, stable quantities within serum, tissue origins are difficult to trace, as is the case with previously discussed correlation of serum DNA methylation levels. In regards to PCOS, a multitude of miRNAs have been associated with disease pathogenesis to include metabolism/insulin and androgen regulation as well as adipose generation and inflammation[18]. These are listed in figure 2 with their pathogenesis correlate.
Figure 2.
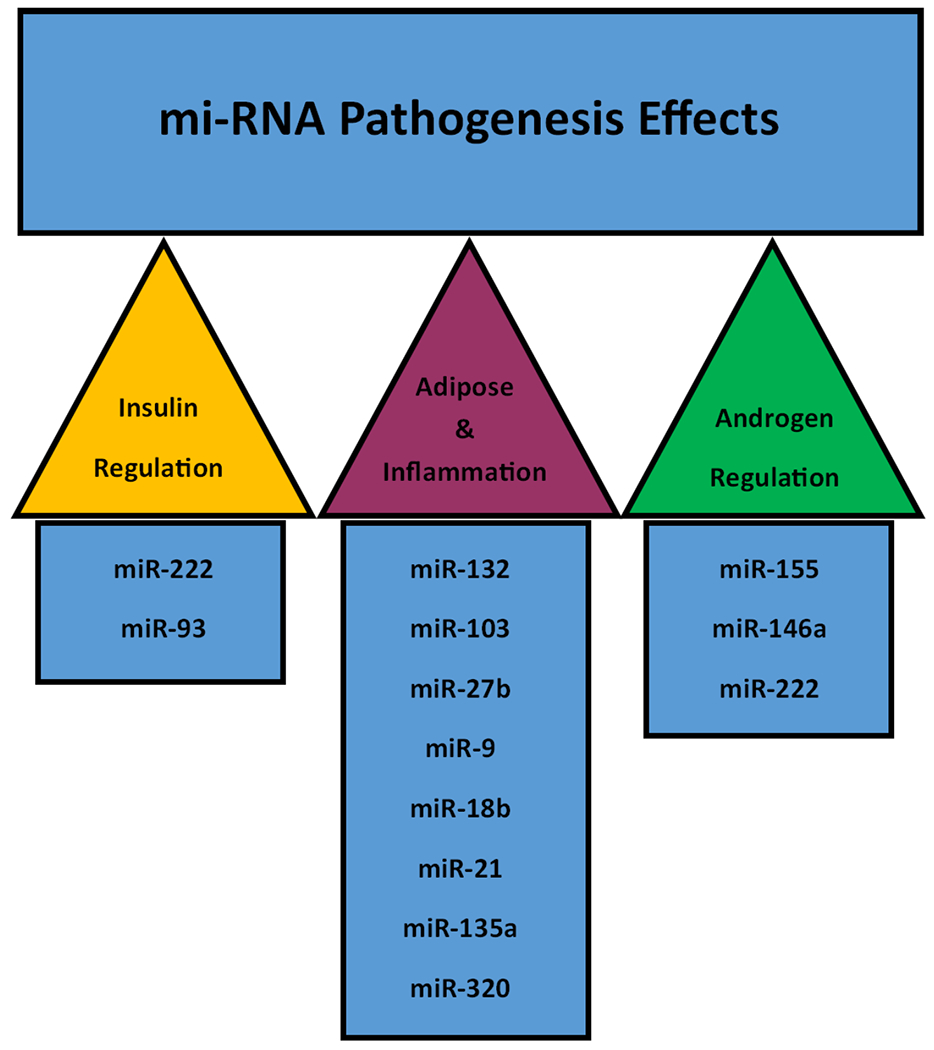
MiRNAs and their associated pathophysiologic effects.
Perhaps one of the most well-studied of these miRNAs is miR-222. Previously linked to type 2 and gestational diabetes, miR-222 has consistently been found to be elevated in serum quantities across multiple studies[6]. MiR-93 is also known to correlate with cellular lipid and glucose metabolism and is directly linked to GLUT4 expression. GLUT4 acts at the major insulin-mediated transporter of glucose into adipocytes and decreased expression has been demonstrated in PCOS patients with inversely proportional MiR-93 levels in serum[19]. These miRNA correlations may contribute to insulin resistance via over-expression, thus establishing one of the major pathophysiologic pillars of PCOS.
While much is still be learned regarding miRNA and its role within the foundation for PCOS, its stable presence within the serum may be useful for future diagnostic testing as opposed subjective choice between multiple organization-defined diagnostic criteria[6]. This standardization would allow for more defined research into the etiology of PCOS when studying a cohort meeting the same inclusion threshold, research-wide.
Finally, mitochondrial DNA (mtDNA) may be another frontier in the research of PCOS and its heritability through genetic and epigenetic mechanisms. Studies have identified 16 variants in the mt-tRNA of PCOS women, contributing to inflammation via reactive oxygen species formation and insulin resistance. As mitochondrial DNA is maternally inherited, linkages could exist between family members with similar PCOS phenotypes[9]. Epigenetic control is also suspected through the methylated regulation of genes such as PARK2, ESR1 and INS, all of which have demonstrated control of mitochondrial activity[20]. Known controversy exists regarding the above mitochondrial associations with PCOS and more studies are need for further validation.
Conclusion
Much work has been done in attempt to identify the cause of PCOS and its associated co-morbidities. While genetic linkages have been found, these appear to be just as complex at the disease process itself. The continuing growth of knowledge regarding the multitude of genes associated with the PCOS, and their epigenetic regulators, provides promise for improvement in future understanding of the disease, its diagnosis and treatment modalities. Continued research into the epigenetic root cause of PCOS is needed to not only validate what has been recently discovered but also to continue to grow the common knowledge base.
References
- 1.Azziz R, Introduction. Fertility and Sterility, 2016. 106(1): p. 4–5. [DOI] [PubMed] [Google Scholar]
- 2.Trikudanathan S, Polycystic Ovarian Syndrome. Medical Clinics of North America, 2015. 99(1): p. 221–235. [DOI] [PubMed] [Google Scholar]
- 3.Mukherjee S, Pathomechanisms of polycystic ovary syndrome Multidimensional approaches. Frontiers in Bioscience, 2018. 10(3): p. 384–422. [DOI] [PubMed] [Google Scholar]
- 4.Filippou P and Homburg R, Is foetal hyperexposure to androgens a cause of PCOS? Human Reproduction Update, 2017. 23(4): p. 421–432. [DOI] [PubMed] [Google Scholar]
- 5.Dumesic DA, et al. , Mechanisms of intergenerational transmission of polycystic ovary syndrome. Reproduction, 2020. 159(1): p. R1–R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ilie IR and Georgescu CE, Polycystic Ovary Syndrome-Epigenetic Mechanisms and Aberrant MicroRNA. 2015, Elsevier; p. 25–45. [DOI] [PubMed] [Google Scholar]
- 7.Narayan P, Genetic Models for the Study of Luteinizing Hormone Receptor Function. Front Endocrinol (Lausanne), 2015. 6: p. 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McAllister JM, et al. , Overexpression of a DENND1A isoform produces a polycystic ovary syndrome theca phenotype. Proc Natl Acad Sci U S A, 2014. 111(15): p. E1519–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shukla P and Mukherjee S, Mitochondrial dysfunction: An emerging link in the pathophysiology of polycystic ovary syndrome. Mitochondrion, 2020. 52: p. 24–39. [DOI] [PubMed] [Google Scholar]
- 10.Vázquez-Martínez ER, et al. , DNA methylation in the pathogenesis of polycystic ovary syndrome. Reproduction, 2019. 158(1): p. R27–R40. [DOI] [PubMed] [Google Scholar]
- 11.Barker DJ, The fetal and infant origins of adult disease. BMJ, 1990. 301(6761): p. 1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, et al. , Comprehensive analysis of genome-wide DNA methylation across human polycystic ovary syndrome ovary granulosa cell. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan J-X, et al. , Aberrant expression and DNA methylation of lipid metabolism genes in PCOS: a new insight into its pathogenesis. Clinical Epigenetics, 2018. 10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang P, et al. , Hypomethylation of the LH/choriogonadotropin receptor promoter region is a potential mechanism underlying susceptibility to polycystic ovary syndrome. Endocrinology, 2014. 155(4): p. 1445–52. [DOI] [PubMed] [Google Scholar]
- 15.Xu N, et al. , Epigenetic mechanism underlying the development of polycystic ovary syndrome (PCOS)-like phenotypes in prenatally androgenized rhesus monkeys. PLoS One, 2011. 6(11): p. e27286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosseini E, et al. , Role of epigenetic modifications in the aberrant CYP19A1 gene expression in polycystic ovary syndrome. Archives of Medical Science, 2019. 15(4): p. 887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ambros V, microRNAs: tiny regulators with great potential. Cell, 2001. 107(7): p. 823–6. [DOI] [PubMed] [Google Scholar]
- 18.Schroen B and Heymans S, Small but smart--microRNAs in the centre of inflammatory processes during cardiovascular diseases, the metabolic syndrome, and ageing. Cardiovasc Res, 2012. 93(4): p. 605–13. [DOI] [PubMed] [Google Scholar]
- 19.Chen YH, et al. , miRNA-93 Inhibits GLUT4 and Is Overexpressed in Adipose Tissue of Polycystic Ovary Syndrome Patients and Women With Insulin Resistance. Diabetes, 2013. 62(7): p. 2278–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou S, Tang X, and Chen HZ, Sirtuins and Insulin Resistance. Front Endocrinol (Lausanne), 2018. 9: p. 748. [DOI] [PMC free article] [PubMed] [Google Scholar]


