Abstract
Objective:
Our aim was to elucidate the role of diet in type 1 diabetes (T1D) by examining combinations of nutrient intake in the progression from islet autoimmunity (IA) to T1D.
Methods:
We measured 2457 metabolites and dietary intake at the time of seroconversion in 132 IA-positive children in the prospective Diabetes Autoimmunity Study in the Young. IA was defined as the first of two consecutive visits positive for at least one autoantibody (insulin, GAD, IA-2, or ZnT8). By December 2018, 40 children progressed to T1D. Intakes of 38 nutrients were estimated from semiquantitative food frequency questionnaires. We tested the association of each metabolite with progression to T1D using multivariable Cox regression. Nutrient patterns that best explained variation in candidate metabolites were identified using reduced rank regression (RRR), and their association with progression to T1D was tested using Cox regression adjusting for age at seroconversion and high-risk HLA genotype.
Results:
In stepwise selection, 22 nutrients significantly predicted at least two of the 13 most significant metabolites associated with progression to T1D, and were included in RRR. A nutrient pattern corresponding to intake lower in linoleic acid, niacin, and riboflavin, and higher in total sugars, explained 18% of metabolite variability. Children scoring higher on this metabolite-related nutrient pattern at seroconversion had increased risk for progressing to T1D (HR = 3.17, 95%CI = 1.42-7.05).
Conclusions:
Combinations of nutrient intake reflecting candidate metabolites are associated with increased risk of T1D, and may help focus dietary prevention efforts.
Keywords: dietary intake, metabolomics, nutrient patterns, progression, T1D
1. ∣ INTRODUCTION
Type 1 diabetes (T1D) is characterized by destruction of the insulin-producing beta cells in the pancreas, and affects over 100 000 children and adolescents in the United States.1 Clinical T1D is preceded by a period of detectable and persistent autoimmunity to islet antigens, called islet autoimmunity (IA). However, not all children with IA progress to T1D. Therefore, it is important to identify factors influencing progression from IA to T1D.
Non-genetic factors are involved in the development of T1D, which also has a well-defined genetic risk component.2 Dietary intake in early life and throughout childhood has been implicated in T1D risk, though without conclusive findings of a single responsible risk factor.3 Higher intake of some foods or nutrients at seroconversion has been associated with increased risk of progression to T1D, including: glycemic load,4 total sugars and sugar-sweetened beverages,5 and cow's milk protein.6 Other dietary factors, including vitamin D intake and status,7,8 and omega-3 fatty acid intake and status,9 were not associated with progression. These traditional investigations of individual dietary risk factors may oversimplify true dietary exposure since multiple nutrients are contained in foods, and nutrients may work syner-gistically or antagonistically in the body. We sought to improve upon previous investigation into diet and progression from IA to T1D by considering combinations of nutrient intake, or nutrient patterns.
Our aim was to identify nutrient patterns of intake at seroconversion associated with risk of progression to T1D in the Diabetes Autoimmunity Study in the Young (DAISY), as shown in Figure 1A. To capture disease-related variation in nutrient intake, first we conducted a metabolome-wide association study to identify metabolites and lipids associated with progression from IA to T1D. As markers of biological processing and response to nutrition, metabolites have been used together with reported dietary intake to elucidate the role of diet in the development or prevention of other outcomes, such as cardiovascular disease.10 Next, we used reduced rank regression (RRR) to create nutrient patterns best explaining those candidate metabolites. Finally, we tested the nutrient pattern with the risk of progression from IA to T1D. The nutrient pattern identified by our study was more strongly associated with T1D than any of the constituent metabolites or nutrients, suggesting joint consideration of multiple dietary factors may be important for elucidating the role of diet in T1D development.
FIGURE 1.
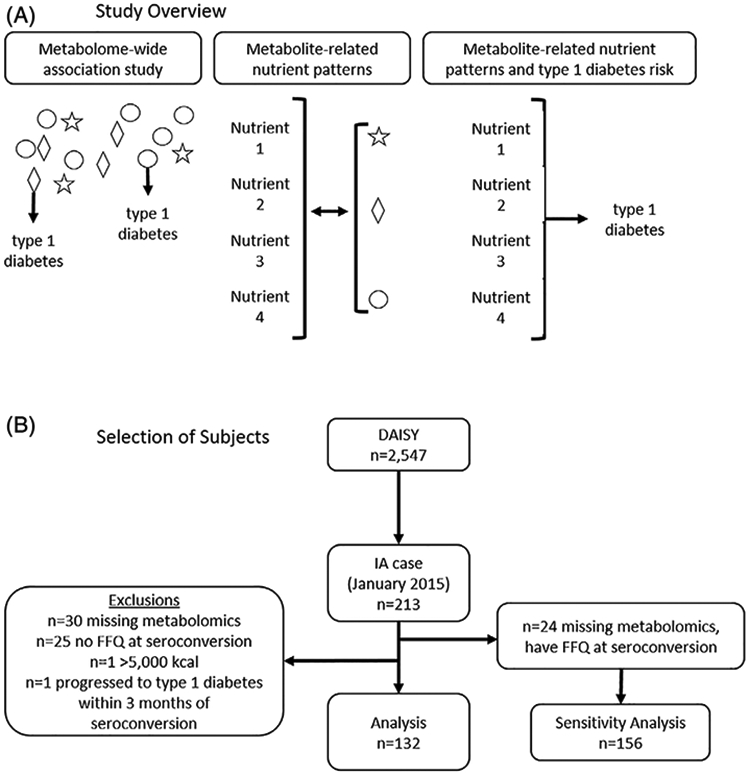
Study design for identifying metabolite-related nutrient patterns associated with progression from islet autoimmunity (IA) to T1D. A, We conducted a metabolome-wide association study to identify seroconversion lipidomics and metabolomics associated with progression to T1D by December 2018. Significant metabolites and nutrient patterns that best summarized them were identified using reduced rank regression. Metabolite-related nutrient pattern scores were tested for the risk of T1D. B, Of the 213 IA cases identified in DAISY by January 2015, 132 had untargeted metabolomics measures and dietary intake available at the time of seroconversion. DAISY, Diabetes Autoimmunity Study in the Young; T1D, type 1 diabetes
2. ∣ MATERIALS AND METHODS
2.1. ∣ Study design
From 1993 to 2004, DAISY recruited 2547 children in Colorado who were at high risk for developing T1D.11 Prospective follow-up for the development of islet autoantibodies and T1D is ongoing, and has included clinic visits at 9, 15, and 24 months, and annually thereafter.12 An IA case is defined by the presence of one or more confirmed autoantibody to insulin, GAD65, IA-2, or ZnT8 on two or more consecutive clinic visits. Seroconversion is defined as the first visit at which IA was detected. IA cases follow an accelerated protocol with clinic visits every 3 to 6 months until T1D is diagnosed by a physician following standard criteria, including typical symptoms of polyuria, polydipsia and/or weight loss and a random glucose > 11.1 mmoL/L or an OGTT with a fasting plasma glucose ≥ 7.0 mmoL/L or 2-hours glucose > 11.1 mmoL/L.13 The Colorado Multiple Institutional Review Board approved all DAISY study protocols (COMIRB 92-080). Informed consent was obtained from the parents/legal guardians of all children. Assent was obtained from children age 7 years and older. All research was performed in accordance with relevant guidelines/regulations.
To identify metabolomics and nutrient patterns present at seroconversion and associated with progression from IA to T1D, we examined IA cases who seroconverted prior to January 2015 and their prospective follow-up for T1D through December 2018. Plasma samples from the first of the two consecutive autoantibody positive visits were used for metabolomics. Where the first sample was unavailable, we used plasma from the second of the two consecutive autoantibody positive visits that defined an IA case.
2.2. ∣ Metabolomics data acquisition, normalization, and preprocessing
Untargeted metabolomics measures were acquired from non-fasting plasma samples using GC-TOF MS (primary metabolism), CSH-QTOF MS (complex lipids), and HILIC-QTOF MS (biogenic amines), at the NIH West Coast Metabolomics Center at the University of California, Davis. Samples were kept at −80°C prior to analysis. Samples were allowed to thaw on wet-ice and kept cold during extraction, once thawed samples were inverted multiple times to ensure serum homogeneity. Samples were extracted by taking 30 μL of serum and performing a liquid-liquid extraction first previously described14 with modification, including addition of labeled internal standards for quality control and retention time correction.15 The aqueous phase, containing polar metabolites, was split into two equal volumes and dried then re-suspended in 1:1 acetonitrile: water to precipitate any remaining lipids. One polar aliquot was then prepared for and analyzed by HILIC-QTOF MS/MS.16 The second polar aliquot was analyzed by GC-TOF-MS.17 The non-polar phase was analyzed by CSH-QTOF MS/MS for identification and relative quantification of complex lipids and free fatty acids.15
Peak picking, integration, alignment, and annotation of GC-TOF-MS data was performed using BinBase.18 Liquid chromatography (LC) data (CSH-QTOF-MS and HILIC-QTOF-MS) were processed with MS-Dial, 19 and complex lipids were annotated with LipidBlast20 and Massbank of North America (http://mona.fiehnlab.ucdavis.edu/). MS-FLO was used to remove erroneous peaks and reduce the false discovery rate in LC datasets.21
After data were collected, annotated and postprocessed they were normalized to account for any instrument drift, variation, or batch effects, which may have occurred. Data were normalized by our in-house normalization algorithm; systematic error removal using random forest (SERRF).22 SERRF is a QC-based normalization method using the random forest algorithm.23 Briefly, SERRF was designed to be used in place of compound independent normalization methods such as locally estimated scatterplot smoothing (LOESS). While LOESS normalizes compounds independently, SERRF takes the metabolites' correlation into consideration and automatically assigns higher weight to important compounds. SERRF is nonparametric, non-linear, less prone to overfitting, robust to outliers and noise, and fast to train.
After normalization, we performed quality control checks at the sample and metabolite level. Samples with a high proportion of features estimated to be zero were considered low abundance (n = 2) and excluded from analyses. Each metabolite was transformed using box-cox transformation analysis, to improve heteroscedasticity and normality.24 Metabolites with a coefficient of variation more than two absolute deviations from the median were excluded from analyses (metabolites = 344), as they may be unreliable or produce unstable estimates. Coefficient of variation median and median absolute deviations was calculated separately by panel. After quality checks, there were 2457 untargeted metabolites for statistical analyses, including 1905 (77%) unannotated metabolites.
2.3. ∣ Dietary intake assessment
Dietary intake was measured annually using the Willett semiquantitative food frequency questionnaire (FFQ) starting at age 2 years. Parents completed an FFQ representing their child's average intake over the previous year until age 10 years, after which study participants self-reported using the Youth Adolescent Questionnaire (YAQ). The FFQ has been previously validated,25 and the FFQ and YAQ shown to be comparable in the DAISY study population.26
The average amounts of daily nutrient intakes were estimated from the FFQ and YAQ at the Channing Laboratory, Harvard, MA. Nutrients that were measured throughout the DAISY study were considered for inclusion in nutrient pattern analyses. Where available, nutrient variables included the intake from both foods and supplements. We used the residual method to calculate the energy-adjusted nutrient intakes.27 For each nutrient, residuals from the linear regression of total calories on nutrient intake are saved and scaled by adding the population mean nutrient intake as a constant. There were 39 energy-adjusted nutrients available for nutrient pattern analysis (Table S1).
2.4. ∣ Statistical methods
IA cases missing metabolomics and FFQ at seroconversion (n = 30), those without a FFQ at seroconversion (n = 25), those reporting unreasonable total calories (>5000 kcal, n = 1), or whose seroconversion visit occurred less than 3 months prior to T1D diagnosis (n = 1) were excluded from all analyses (Figure 1B). The 24 IA cases with FFQ available at seroconversion but no metabolomics measures were included in a sensitivity analysis (described below). All metabolite variables were standardized to the population mean and SD prior to analyses to allow for direct comparison of the magnitude of association with T1D. We used Cox regression to identify seroconversion metabolomics associated with progression from IA to T1D, adjusting for age at seroconversion and high-risk HLA (DR3/4) genotype. The time-to-event was defined as the time from seroconversion (the first of two consecutive positive visits) to diagnosis with T1D. We adjusted for multiple comparisons using the Benjamini-Hochberg false discovery rate.28
From the most significant metabolites identified in discovery (minimum FDR q-value), we used RRR to identify nutrient patterns that best explained metabolite variation. RRR helps summarize highdimensional data by creating linear combinations of nutrients (nutrient patterns) that maximize the variance explained in a set of response variables (metabolites).29 The number of patterns retained for analyses was determined using Hotellings T2, which minimizes the predicted residual sum of squares. We reduced the number of nutrients included in RRR by first applying a stepwise regression selection procedure for each metabolite.30 Nutrients that significantly predicted at least two metabolites (P-value < .1, Table S1) were included in nutrient pattern analysis. Energy-adjusted nutrient intakes were standardized to the mean and SD prior to the RRR. Results of RRR included metabolite weights, nutrient loadings, and a nutrient pattern score for each participant, indicating how similar their intake was to the nutrient pattern. Weights and loadings more extreme than ±0.2 were used to interpret the nutrient pattern.
We tested the association of metabolite-related nutrient pattern scores with progression from IA to T1D using Cox regression. For comparison with the nutrient pattern, we included an additional analysis testing individual nutrient associations with progression from IA to T1D for nutrients that loaded strongly (more extreme than ±0.2) on the nutrient pattern. While there were insufficient additional IA cases with FFQ at seroconversion to test for replication of the association between nutrient pattern score and progression to T1D, we conducted a sensitivity analysis of all IA cases with FFQ at seroconversion (n = 132 + 24 = 156) using Cox regression. All multivariable Cox regression models were adjusted for age at seroconversion and high-risk HLA (DR3/4) genotype. BMI is not associated with progression to T1D in DAISY,31 and therefore was not included in the multivariable models. SAS 9.4 (SAS Institute Inc., Cary, North Carolina) was used for all statistical analyses.
3. ∣ RESULTS
Of 132 IA cases with metabolomics and dietary intake measured at the time of seroconversion, 40 progressed to T1D by December 2018. Characteristics of the study population are shown in Table 1. T1D progressors were 48% male, mostly non-Hispanic white, and 63% had a first degree relative with T1D. These characteristics were similar in the non-progressor group. However, compared to non-progressors, those who progressed to T1D had higher-risk HLA genotypes (43% vs 28%) and seroconverted to IA positivity at younger ages (5 years vs 7.8 years). Residual-adjusted nutrient intakes were comparable across ages at seroconversion (Figure S1).
TABLE 1.
Characteristics of 132 IA cases at seroconversion
| Progressed to type 1 diabetes (n = 40) |
Non-progressors (n = 92) |
||||
|---|---|---|---|---|---|
| n | % | n | % | P-valuea | |
| Male | 19 | 47.5 | 45 | 48.9 | .883 |
| Non-Hispanic white | 36 | 90.0 | 68 | 73.9 | .079 |
| High-risk HLA (DR3/4) | 17 | 42.5 | 26 | 28.3 | .046 |
| First degree relative with type 1 diabetes | 25 | 62.5 | 52 | 56.5 | .595 |
| - | Mean | SD | Mean | SD | - |
| Age at seroconversion (years) | 5.0 | 3.8 | 7.8 | 4.3 | .025 |
| Time from seroconversion to T1D or last follow-up (years) | 5.5 | 2.8 | 9.8 | 4.1 | <.0001 |
| Total calories | 2146 | 492 | 2078 | 739 | .811 |
Abbreviations: IA, islet autoimmunity; T1D, type 1 diabetes.
From Cox regression.
From individual multivariable Cox regression models, 201 metabolites (8%) had nominal associations (P-value < .05) with risk of progression to T1D (Figure 2). None of these associations reached significance after adjustment for multiple comparisons (q-value < 0.05). Thirteen metabolites were tied at the minimum q-value of 0.3451, including: threonine, histidine, choline, and 10 unannotated compounds from the biogenic amines and complex lipids panels. All 13 of these metabolites were used in the nutrient pattern analysis. Mass spectra for the compounds are included in Figure S2. The majority of nominally significant metabolites were associated with increased risk of progression to T1D (Figure 2).
FIGURE 2.
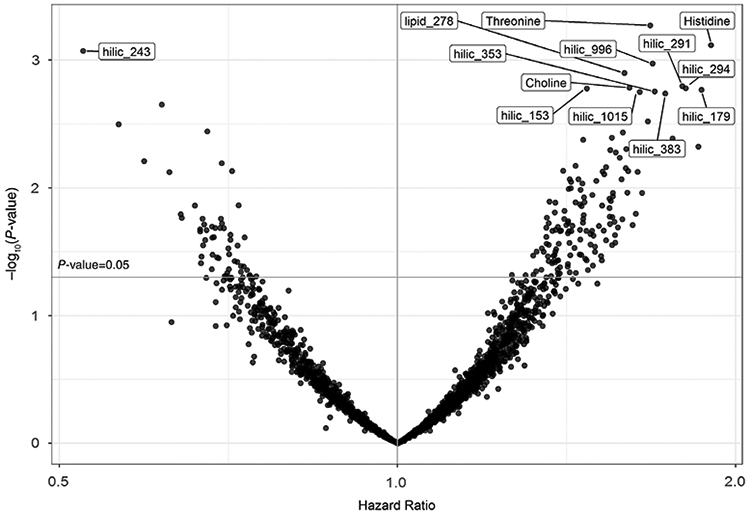
Cox regression results showing the association of 2457 metabolites with progression from IA to T1D. 201 metabolites (8%) had nominal P-value < .05 (above the guideline), and none were significant at q-value < 0.05. The 13 labeled metabolites had the minimum q-value of 0.3451 and were selected for nutrient pattern analyses. These included threonine, histidine, choline, and 10 unannotated compounds. Unannotated compounds were labeled by the untargeted platform on which they were measured (ie, HILIC_243 was measured using HILIC-QTOF-MS)
To identify nutrient patterns using RRR, we first selected nutrients that were significant predictors of the 13 metabolites identified in discovery. Stepwise selection results indicated that 22 of 39 nutrients predicted at least two metabolites (Table S1). From RRR, one nutrient pattern was identified that explained 18% of metabolite variation and 5% of nutrient variation. The metabolite weights and nutrient loadings that define the nutrient pattern are shown in Figure 3. A higher score on the nutrient pattern corresponded to a diet with higher intake of total sugars, vitamin C, and monounsaturated fat, and lower in linoleic acid, niacin, riboflavin, vitamin K, vitamin B12, and caffeine (Figure 3A). Higher scores on the nutrient pattern also corresponded to higher amounts of unknown metabolites hilic_291, hilic_294, hilic_179, hilic_353, hilic_1015, hilic_383, and lower amounts of hilic_243 (Figure 3B). Known metabolites threonine, histidine, and choline were poorly explained by the nutrient pattern, as indicated by weights closer to O.
FIGURE 3.
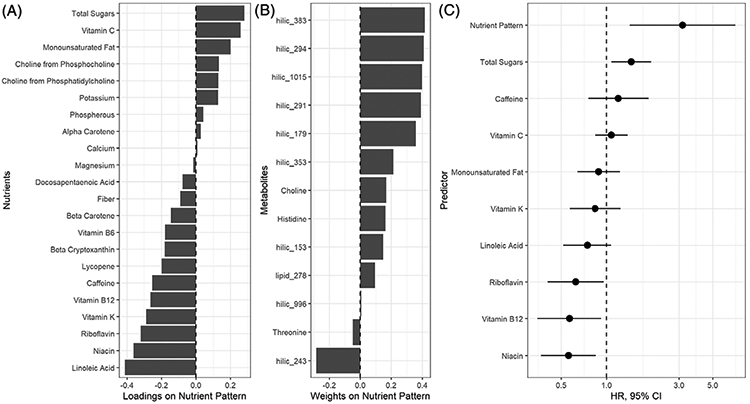
A seroconversion metabolite-related nutrient pattern and its association with progression from IA to T1D. One dietary pattern explained 5% of nutrient variation and 18% of metabolite variation. A, The loadings indicate which combinations of nutrients defined the nutrient pattern scores. B, The weights indicate which metabolites were best explained by the nutrient pattern. C, Multivariable Cox regression results showing the associations of the nutrient pattern score and individual nutrients in progression from IA to T1D. Hazard ratios represent one SD change in each nutrient, metabolite, or the nutrient pattern score
In multivariable Cox regression, nutrient pattern scores at seroconversion were associated with increased risk of progression from IA to T1D (Figure 3C, HR = 3.17, 95%CI = 1.42-7.05). In analyses of the nine nutrients that loaded high on the nutrient pattern, only vitamin B12, riboflavin, niacin, and total sugars had significant individual associations with progression from IA to T1D (Figure 3C). The association between nutrient pattern score and risk of progression was robust in sensitivity analysis (HR = 2.49, 95%CI = 1.23-5.05).
4. ∣ DISCUSSION
We identified a nutrient pattern of intake at seroconversion that was significantly associated with increased risk of progression to T1D. Higher nutrient pattern scores (and increased risk of progression) corresponded to higher intake of total sugars, vitamin C, and monounsaturated fat, and lower in linoleic acid, niacin, riboflavin, vitamin K, vitamin B12, and caffeine. Accounting for combinations of nutrient intake in the diet using a nutrient pattern score resulted in a stronger association with progression to T1D (HR = 3.17) than the individual nutrients highly contributing to the pattern (HR range = 0.56-1.45) and the metabolites used to inform the pattern (HR range = 0.52-1.9), despite a wider confidence interval.
Several of the nutrients that highly contributed to the nutrient pattern have been investigated in the development of IA or T1D for years, often resulting in small or inconsistent associations with disease endpoints. For example, higher intake of total sugars was previously associated with increased risk of progression in DAISY.5 Dietary intake of the omega-6 fatty acid, linoleic acid, was not associated with progression from IA to T1D.9 However, linoleic acid status has been inconsistently identified as protective for development of IA.32 Similarly, the niacin derivative nicotinamide was protective for T1D in mice, but shown to be ineffective in a prevention trial.33 The failure to account for the inter-relationship between nutrients may explain the inconsistencies surrounding the role of dietary intake in T1D development. Using a nutrient pattern rather than individual nutrients may better reflect etiologically relevant exposures, and help clarify the role of nutrition in T1D.
Other nutrients contributing to the pattern have not been directly studied but have plausible connection to T1D. For example, riboflavin34 and vitamin K (K1, phylloquinone)35 have antioxidant properties that may protect islet cells from destruction by free radicals and oxidative stress. Through its role as a cofactor in one-carbon metabolism, vitamin B12 affects DNA methylation, which is also implicated in the disease process.36 Notably, vitamin C contributes in an unexpected direction to the nutrient pattern. Due to its antioxidant and anti-inflammatory properties, vitamin C would be expected to have an inverse relationship with risk of progression to T1D. Results from the TEDDY study indicated higher vitamin C was associated with lower risk of IA.37 However, these findings are not directly comparable, since they measured plasma ascorbic acid rather than diet and did not investigate the role of plasma ascorbic acid in progression from IA to T1D. Foods high in sugar may also contain higher amounts of vitamin C (eg, fruits, fruit juices, juice drinks with added vitamin C)—the tendency to receive both of these nutrients from ingestion of the same foods might explain the unexpected direction of effect we found for vitamin C in the covariation-based pattern analysis.
The metabolite discovery was primarily used as a means to capture meaningful, disease-related variation in nutrient pattern intake. However, a broad conclusion of our metabolomics findings indicate that metabolic differences at seroconversion distinguish which IA cases will go on to T1D, similar to previous studies comparing IA cases and T1D cases to autoantibody negative controls.38-40 Though conducted in different disease stages, each study has identified various amino acids as dysregulated prior to the development of different disease endpoints. In our study, however, threonine and histidine were poorly correlated with diet and therefore less influential in the nutrient pattern. The other study comparing IA cases to T1D cases investigated the lipidome in cord blood.41
The majority of our candidate metabolites were unknown compounds measured on the HILIC panel, making comparison to previous metabolomics studies infeasible. As metabolomics databases grow, annotation and identification of these compounds may become possible. We have included mass spectra for the compounds in the supplement to facilitate potential future comparisons. Future work should identify and annotate these unknown compounds, and consider the strong correlation between metabolites, which makes the use of typical multiple comparison procedures too strict. Despite the limitations of including unknown compounds in our analyses, they were useful for identifying combinations of nutrient intake associated with T1D progression.
The 13 candidate metabolites were mostly associated with increased risk of progression to T1D. While these associations were only nominal, our nutrient pattern explained over 18% of metabolite variability—over three times the amount of variability explained in previous dietary (food) pattern studies,42,43 and studies of individual dietary factors and targeted metabolites.44 This suggests that the metabolites used as response variables for identifying the nutrient pattern are related to nutrition. The novel use of nutrients rather than food groups in pattern analysis likely contributes to the increased variability explained by the nutrient pattern—nutrient intakes have more continuous distributions and may perform better in covariation-based analyses such as RRR. Nutrient patterns may be harder to interpret than dietary patterns of food. However, the great complexity of harmonizing foods across countries may lead to difficulties creating generalizable dietary food patterns associated with outcomes.43 Nutrients are more comparable across different populations and therefore may be more amenable to the development and cross-cultural implementation of dietary interventions to prevent T1D.
Studies investigating complex dietary intake in disease development are important, particularly for diseases such as T1D where dietary factors are among the leading hypothesized environmental risk factors. However, multivariate methods used to accomplish this (such as RRR) are sensitive to the selection of variables used in the analyses.45 To minimize the chances of random findings we have practiced judicious selection of response variables for pattern analysis. Modeling energy-adjusted, standardized nutrient intakes rather than unstandardized, unadjusted, or food groups improves the likelihood that our results will generalize to new populations, as they will be less affected by the variability of intake across age and geographic settings. Furthermore, we accounted for relevant confounders. DAISY's small sample size inhibited our ability to test for replication of this nutrient pattern, but our sensitivity analysis including an additional 24 IA cases was consistent with our primary findings. Replication is an important next step for establishing the robustness of our findings and should be conducted in other T1D studies collecting nutrient intake.
As hypothesized, examining nutrient intakes in combinations, to account for their correlation in intake and in biological processing, resulted in a stronger magnitude of association than any of the individual constituent nutrients. The relationship of highly contributing nutrients to the nutrient pattern is consistent with existing literature and hypotheses, suggesting that the contents of the diet may work in combination to affect the development of T1D, and should be considered jointly in future work.
Supplementary Material
ACKNOWLEDGEMENTS
Thank you to the participants and families of the DAISY study and the clinical research staff at the Barbara Davis Center for Diabetes, whose continued commitment make such research possible.
Funding information
National Institutes of Health, Grant/Award Numbers: R01-DK104351, R01-DK32493
Footnotes
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/pedi.13085.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.Mayer-Davis EJ, Lawrence JM, Dabelea D, et al. Incidence trends of type 1 and type 2 diabetes among youths, 2002–2012. N Engl J Med. 2017;376(15):1419–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett JC, Clayton DG, Concannon P, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41(6):703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Virtanen SM. Dietary factors in the development of type 1 diabetes. Pediatr Diabet. 2016;17(S22):49–55. [DOI] [PubMed] [Google Scholar]
- 4.Lamb MM, Yin X, Barriga K, et al. Dietary Glycemic index, development of islet autoimmunity, and subsequent progression to type 1 diabetes in young children. J Clin Endocrinol Metab. 2008;93(10):3936–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamb MM, Frederiksen B, Seifert JA, Kroehl M, Rewers M, Norris JM. Sugar intake is associated with progression from islet autoimmunity to type 1 diabetes: the diabetes autoimmunity study in the young. Diabetologia. 2015;58(9):2027–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamb MM, Miller M, Seifert JA, et al. The effect of childhood cow's milk intake and HLA-DR genotype on risk of islet autoimmunity and type 1 diabetes: the diabetes autoimmunity study in the young. Pediatr Diabet. 2015;16(1):31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raab J, Giannopoulou EZ, Schneider S, et al. Prevalence of vitamin D deficiency in pre-type 1 diabetes and its association with disease progression. Diabetologia. 2014;57(5):902–908. [DOI] [PubMed] [Google Scholar]
- 8.Simpson M, Brady H, Yin X, et al. No association of vitamin D intake or 25-hydroxyvitamin D levels in childhood with risk of islet autoimmunity and type 1 diabetes: the diabetes autoimmunity study in the young (DAISY). Diabetologia. 2011;54(11):2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller MR, Yin X, Seifert J, et al. Erythrocyte membrane omega-3 fatty acid levels and omega-3 fatty acid intake are not associated with conversion to type 1 diabetes in children with islet autoimmunity: the diabetes autoimmunity study in the young (DAISY). Pediatr Diabet. 2011;12(8):669–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Gonzalez MA, Bes-Rastrollo M. Dietary patterns, Mediterranean diet, and cardiovascular disease. Curr Opin Lipidol. 2014;25(1): 20–26. [DOI] [PubMed] [Google Scholar]
- 11.Rewers M, Bugawan TL, Norris JM, et al. Newborn screening for HLA markers associated with IDDM: diabetes autoimmunity study in the young (DAISY). Diabetologia. 1996;39(7):807–812. [DOI] [PubMed] [Google Scholar]
- 12.Norris JM, Barriga K, Klingensmith G, et al. Timing of initial cereal exposure in infancy and risk of islet autoimmunity. JAMA. 2003;290 (13):1713–1720. [DOI] [PubMed] [Google Scholar]
- 13.Association AD. 2. Classification and diagnosis of diabetes. Diabet Care. 2017;40(suppl 1):S11–S24. [DOI] [PubMed] [Google Scholar]
- 14.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49(5):1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cajka T, Smilowitz JT, Fiehn O. Validating quantitative untargeted lipidomics across nine liquid chromatography-high-resolution mass spectrometry platforms. Anal Chem. 2017;89(22):12360–12368. [DOI] [PubMed] [Google Scholar]
- 16.Showalter MR, Nonnecke EB, Linderholm AL, et al. Obesogenic diets alter metabolism in mice. PLoS One. 2018;13(1):e0190632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiehn O, Wohlgemuth G, Scholz M, et al. Quality control for plant metabolomics: reporting MSI-compliant studies. Plant J Cell Mol Biol. 2008;53(4):691–704. [DOI] [PubMed] [Google Scholar]
- 18.Skogerson K, Wohlgemuth G, Barupal DK, Fiehn O. The volatile compound BinBase mass spectral database. BMC Bioinform. 2011;12:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsugawa H, Cajka T, Kind T, et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods. 2015;12(6):523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kind T, Liu K-H, Yup Lee D, DeFelice B, Meissen JK, Fiehn O. LipidBlast-in-silico tandem mass spectrometry database for lipid identification. Nat Methods. 2013;10(8):755–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeFelice BC, Mehta SS, Samra S, et al. Mass spectral feature list optimizer (MS-FLO): a tool to minimize false positive peak reports in untargeted liquid chromatography-mass spectroscopy (LC-MS) data processing. Anal Chem. 2017;89(6):3250–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan S, Kind T, Cajka T, et al. Systematic error removal using random forest for normalizing large-scale untargeted lipidomics data. Anal Chem. 2019;91:3590–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breiman L Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 24.Box GE, Cox DR. An analysis of transformations. J R Stat Soc Ser B Methodol. 1964;26(2):211–252. [Google Scholar]
- 25.Parrish LA, Marshall JA, Krebs NF, Rewers M, Norris JM. Validation of a food frequency questionnaire in preschool children. Epidemiology. 2003;14(2):213–217. [DOI] [PubMed] [Google Scholar]
- 26.Lamb MM, Ross CA, Brady HL, Norris JM. Comparison of children's diets as reported by the child via the youth/adolescent questionnaire and the parent via the Willett food-frequency questionnaire. Public Health Nutr. 2007;10(07):663–670. [DOI] [PubMed] [Google Scholar]
- 27.Willett W Nutritional Epidemiology OUP USA. 2012;547. [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57(1):289–300. [Google Scholar]
- 29.Hoffmann K, Schulze MB, Schienkiewitz A, Nöthlings U, Boeing H. Application of a new statistical method to derive dietary patterns in nutritional epidemiology. Am J Epidemiol. 2004;159(10):935–944. [DOI] [PubMed] [Google Scholar]
- 30.Schulze MB, Hoffmann K, Manson JE, et al. Dietary pattern, inflammation, and incidence of type 2 diabetes in women. Am J Clin Nutr. 2005;82(3):675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamb MM, Yin X, Zerbe GO, et al. Height growth velocity, islet autoimmunity and type 1 diabetes development: the diabetes autoimmunity study in the young. Diabetologia. 2009;52(10):2064–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niinistö S, Takkinen H-M, Erlund I, et al. Fatty acid status in infancy is associated with the risk of type 1 diabetes-associated autoimmunity. Diabetologia. 2017;60(7):1223–1233. [DOI] [PubMed] [Google Scholar]
- 33.Gale E a M, Bingley PJ, Emmett CL, Collier T. European nicotinamide diabetes intervention trial (ENDIT) group. European nicotinamide diabetes intervention trial (ENDIT): a randomised controlled trial of intervention before the onset of type 1 diabetes. Lancet Lond Engl. 2004; 363(9413):925–931. [DOI] [PubMed] [Google Scholar]
- 34.Cobianchi L, Fornoni A, Pileggi A, et al. Riboflavin inhibits IL-6 expression and p38 activation in islet cells. Cell Transplant. 2008;17(5):559–566. [DOI] [PubMed] [Google Scholar]
- 35.Varsha MKNS, Thiagarajan R, Manikandan R, Dhanasekaran G. Vitamin K1 alleviates streptozotocin-induced type 1 diabetes by mitigating free radical stress, as well as inhibiting NF-κB activation and iNOS expression in rat pancreas. Nutr Burbank Los Angel Cty Calif. 2015;31(1):214–222. [DOI] [PubMed] [Google Scholar]
- 36.Johnson RK, Vanderlinden LA, Dong F, et al. Longitudinal DNA methylation differences precede type 1 diabetes. Sci Rep. 2020;10(1):3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattila M, Erlund I, Lee H-S, et al. Plasma ascorbic acid and the risk of islet autoimmunity and type 1 diabetes: the TEDDY study. Diabetologia. 2020;63(2):278–286. 10.1007/s00125-019-05028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orešič; M, Simell S, Sysi-Aho M, et al. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J Exp Med. 2008;205(13):2975–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pflueger M, Seppänen-Laakso T, Suortti T, et al. Age- and islet autoimmunity-associated differences in amino acid and lipid metabolites in children at risk for type 1 diabetes. Diabetes. 2011;60 (11):2740–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Q, Parikh H, Butterworth MD, et al. Longitudinal metabolome-wide signals prior to the appearance of a first islet autoantibody in children participating in the TEDDY study. Diabetes. 2020;69(3): 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamichhane S, Ahonen L, Sparholt Dyrlund T, et al. Cord-blood lipidome in progression to islet autoimmunity and type 1 diabetes. Biomolecules. 2019;9(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer J, Döring A, Herder C, Roden M, Koenig W, Thorand B. Dietary patterns, subclinical inflammation, incident coronary heart disease and mortality in middle-aged men from the MONICA/KORA Augsburg cohort study. Eur J Clin Nutr. 2011;65(7):800–807. [DOI] [PubMed] [Google Scholar]
- 43.Johnson RK, Vanderlinden L, DeFelice BC, et al. Metabolite-related dietary patterns and the development of islet autoimmunity. Sci Rep. 2019;9(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Floegel A, von Ruesten A, Drogan D, et al. Variation of serum metabolites related to habitual diet: a targeted metabolomic approach in EPIC-Potsdam. Eur J Clin Nutr. 2013;67(10):1100–1108. [DOI] [PubMed] [Google Scholar]
- 45.Weikert C, Schulze MB. Evaluating dietary patterns: the role of reduced rank regression. Curr Opin Clin Nutr Metab Care. 2016;19(5): 341–346. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


