Abstract
Ras activates its effectors at the membrane. Active PI3Kα and its associated kinases/phosphatases assemble at membrane regions enriched in signaling lipids. By contrast, the Raf kinase domain extends into the cytoplasm and its assembly is away from the crowded membrane surface. Our structural membrane-centric outlook underscores the spatiotemporal principles of membrane and signaling lipids which helps clarify PI3Kα activation. Here we focus on mechanisms of activation driven by PI3Kα driver mutations, spotlighting the PI3Kα double (multiple) activating mutations. Single mutations can be potent, but double mutations are stronger: their combination is specific, a single strong driver cannot fully activate PI3K, and two weak drivers may or may not do so. By contrast, two strong drivers may successfully activate PI3K, where one, e.g. H1047R, modulates membrane interactions facilitating substrate binding at the active site (km) and the other, e.g. E542K and E545K, reduces the transition state barrier (ka), releasing autoinhibition by nSH2. Although mostly unidentified, weak drivers are expected to be common, so we ask here how common double mutations are likely to be and why PI3Kα with double mutations responds effectively to inhibitors. We provide a structural view of hotspot and weak driver mutations in PI3Kα activation, explain their mechanisms, compare these with mechanisms of Raf activation, and point to targeting cell-specific, chromatin-accessible, and parallel (or redundant) pathways to thwart the expected emergence of drug resistance. Collectively, our biophysical outlook delineates activation and highlights the challenges of drug resistance.
Keywords: Ras, PI3Kα, drug resistance, activating mutations, K-Ras, inhibitors, B-Raf, PI3Kα/Akt signaling
The Membrane and Signaling Lipids in Ras Signaling
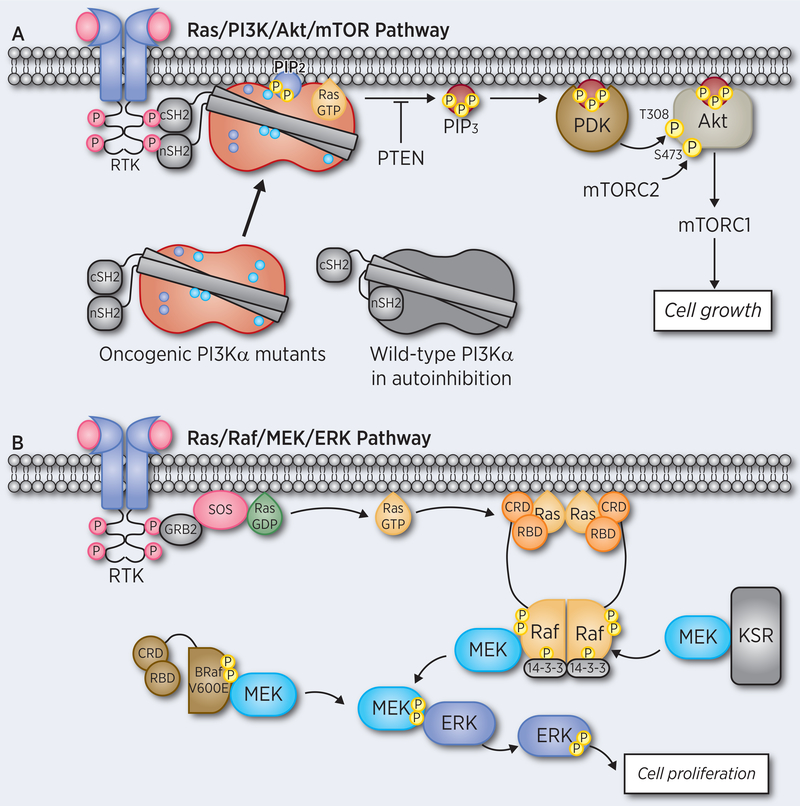
Signal transduction originates at the membrane (1). Kinases and phosphatases in the Ras pathways, such as phosphatidylinositol 3-kinase α (PI3Kα), Akt (also known as protein kinase B), 3-phosphoinositide-dependent protein kinase 1 (PDK1), and phosphatase and tensin homologue (PTEN) receive their incoming cues at the membrane in regions enriched by signaling lipids such as phosphatidylinositol-3,4,5-bisphosphate (PIP3) or phosphatidylinositol-3,4-bisphosphate (PIP2) (Figure 1A). Those activated in the cytoplasm, can receive mediated signals, as in the case of B-Raf activation of mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (ERK), with B-Raf itself mediated by receptor tyrosine kinase (RTK) (1) and Ras nanoclustering. Membrane attributes include curvature (2), preferred phospholipid composition and membrane order/disorder as key factors in anchoring and differentiating among Ras isoforms (3), and isoforms of other Ras family GTPases (4). The cardinal role of these factors in nanoclustering has been discussed (5,6) and recently, Ras assemblies at the membrane (7) have also been explored from this standpoint.
Figure 1.
Ras activates its effectors and signaling pathways at the membrane. (A) Active PI3Kα and its associated kinases/phosphatases, Akt, PDK1, PTEN, assemble at and are confined to membrane regions enriched in signaling lipids. The figure portrays activation of oncogenic PI3Kα by RTK signaling and Ras where the driver mutations mimic physiological events, as proposed in the seminal paper of Roger Williams and his co-workers (87), and the chain of events initiating at the plasma membrane down to the nucleus via the PI3K/Akt pathway. (B) The activation of Ras/Raf/MEK/ERK pathway mainly occurs in the cytoplasm away from the membrane. The dimerized Raf or monomeric Raf mutant (V600E) phosphorylates the downstream MEK and ERK for cell proliferation.
Physical principles underlie signaling landscapes. Among these, (i) substrate phosphorylation (dephosphorylation) cannot be efficiently executed by activated kinases (phosphatases) that diffuse freely in the cytoplasm. Effective regulation requires coupling to external cues, which necessitates membrane attachment (8). Membrane interaction can be direct as in the case of PI3K or mediated through another molecule (9). (ii) Equilibrium among the conformational species in the ensemble, for example between the closed autoinhibited and the open conformations, is a key factor in activation. Relieving Raf autoinhibition – which is driven by the high affinity binding of Ras to Raf’s Ras binding domain (RBD) at the membrane – will increase the population of the open state versus the autoinhibited state due to its higher stability (10). Finally, (iii) the membrane and its surface are densely crowded. Since function involves large dynamic multimolecular assemblies, which include large scaffolding proteins (11–13) not all can contact the membrane; activated Ras/PI3K/Akt/PDK1/PTEN proteins need to be at the membrane surface. However, that is not the case for Raf, where only two small Raf domains, RBD and cysteine-rich domain (CRD) must be there. The remainder of Raf is in the cytoplasm (Figure 1B). Further, there are water layers adjacent to biological membranes (14). The water at the charged membrane surface pushes proteins away from the crowded membrane, increasing their effective concentrations, thus clustering in the cytoplasm. At the same time, the depletion of proteins at the surface allows for accelerated diffusion of those proteins that contact lipid head groups (15). Since wild type kinases typically populate the inactive state, the interactions of PI3K, Akt and PDK1 with the membrane can be short-lived.
Below, we clarify the mechanisms of PI3Kα driver mutations, assembly and signaling at the membrane. As we discuss below, the remarkable recent experimental observations of PI3Kα driver mutations, single, double and multiple (16,17), can be understood within the framework of these principles. In a clinical setting, the mutations promote a membrane-attached, appropriately oriented and exposed active site, enabling recruitment of PIP2 in the absence of an external cue, stabilizing its binding, and reducing the kinetic barrier of phosphorylating it to PIP3. These scenarios are contrasted by those of driver mutations in Raf, whose activation is in the cytoplasm. Collectively, our membrane-centric outlook helps unravel signaling by Ras’s effectors and downstream pathways in drug resistance.
PI3Kα: Single Mutations Can Be Potent; Double Mutations Stronger
PI3Ks are a family of lipid kinases that mediate the PI3K/Akt/mTOR signaling pathway in the cell (18–25). Class I PI3K phosphorylates PIP2 to PIP3 at the membrane to deliver a cell growth signal, a necessary component of proliferation (Figure 1A) (26). They are involved in disease (20,27–30) and they, and their pathways, are major drug targets (26,31–39). PI3Kα, the key member of the PI3K lipid kinase family, is one of the most highly mutated proteins in cancer and an important drug target (40–47). PI3Kα is an obligate heterodimer, containing the regulatory p85α and the catalytic p110α subunits (Figure 2A). It is recruited to the membrane by Ras and the RTK, such as platelet-derived growth factor receptor (PDGFR) (48,49) (Figure 1A). The structural basis for its activation and inhibition has been explored (20,50–52). Recently we studied the structural features in the activation mechanism of PI3Kα upon nSH2 release (53,54). The mechanism clarifies the structural rearrangements that take place at the membrane upon interaction of the p85α subunit with the phosphorylated tyrosine (pY) motif at the C-terminal of an RTK, which release PI3Kα’s autoinhibition by the nSH2 domain. This interaction increases the population of PI3Kα catalytically-competent state, with the kinase domain (KD) preorganized for membrane interaction and PIP2 phosphorylation (28,55,56). The conformational changes involving the movements of the iSH2 domain in the p85α relative to KD have also been captured in the activated PI3Kα with the E545K mutation (57). The mechanistic scenario helps to understand how single and double PI3Kα driver mutations exert their actions and why PI3K inhibitors can potently target them (58).
Figure 2.
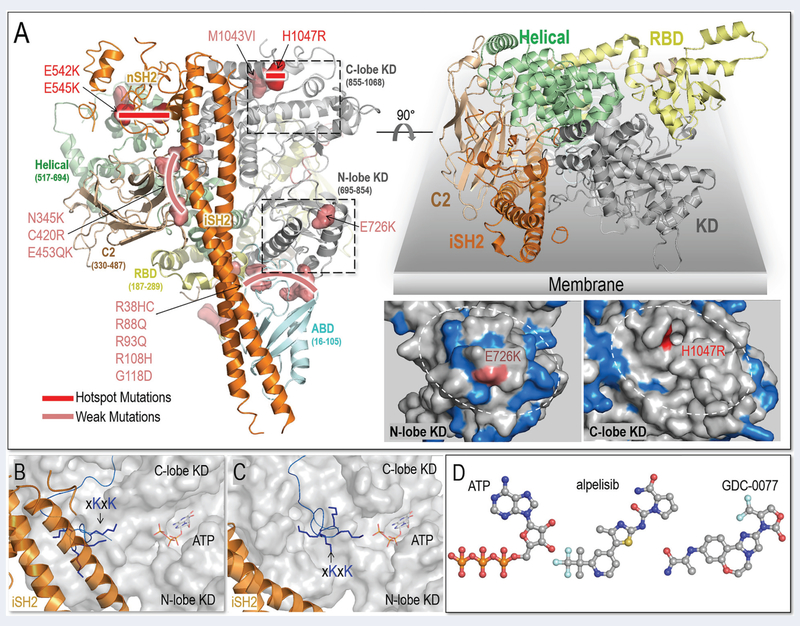
Structural insight into PI3Kα activation and inhibitors. (A) PI3Kα structure (PDB ID: 4OVV) and oncogenic mutations (left panel). The top right panel illustrates the membrane interactions of PI3Kα. The C-lobe of the kinase domain (KD) contains fewer basic residues on the surface than its N-lobe. The basic residues are highlighted in marine color and the plausible membrane binding area denoted by the white dash circle (right bottom panel). The snapshots of the (B) PI3Kα (PDBID: 4OVV) and (C) PI3Kα with nSH2 released (constructed from MD simulations) suggest the conformational changes of PI3Kα. (D) PI3Kα inhibitors structurally resemble ATP, competing with ATP to interact with the kinase domain (The drug structures are from DrugBank).
Single potent activating hotspot mutations in PI3Kα are common in tumor samples (16,59–62). The three hotspot mutations, E542K, E545K and H1047R are in the helical domain and KD of the p110α subunit (63–65) (Figure 2A). E542K and E545K activate PI3Kα by substituting the effect of p85α-RTK binding, reducing the barrier height of the transition state (ka) via a reorganization of the active site; H1047R can substitute for the action of active Ras binding on the membrane, enhancing the population time of the PIP2 substrate at the active site (km) (66–68). This mechanism is in line with earlier single-molecule imaging experiments which indicate that simultaneous activation by a receptor activation loop (from PDGFR, an RTK) and H-Ras leads to strong, synergistic activation of PI3Kα, resulting in a large increase in net kinase activity via the membrane recruitment mechanism (69) as well as additional experimental data on activation by membrane-localized H-Ras (70). These gain-of-function mutations promote phosphorylation of downstream proteins, aberrant cell growth and tumor development in vivo (59).
In addition to frequent strong hotspot mutations, PI3Kα has rare and weak activating mutations; some promote proliferation in vivo and in vitro, but with lower oncogenicity (63,71–81) (Figure 2A). Recently, Vasan et al. observed double and multiple mutations on the same allele (in cis): one hotspot and one (or several) weak (16). They observed that combinations of the weak mutations (E453K/Q, E726K, and M1043V/I) with the hotspots (E542K, E545K, and H1047R) are frequent in breast cancer (Figure 2A). Double/multiple mutations result in additive signaling potency in tumor proliferation and growth (16), the outcome of the ‘double-trouble’ highlighted in Toker’s Commentary (82). The activation mechanism suggests how the mutations work.
During fetal development weak driver mutants arise in PI3K (CLOVES, congenital lipomatous overgrowth, vascular malformations, and epidermal nevi syndrome) (83,84) and in Ras family members (rasopathies). Shvartsman and his colleagues (85) reasoned that because cancer mutations would be embryonic lethal when inherited through the germ line, it might be expected that mutations that cause developmental disorders, but allow survival of the carrier such as those in rasopathy, are weaker than those that cause cancer (86).
A Structural View of Hotspot and Weak Mutations in PI3Kα Activation
The occurrence and strong transforming potential of double/multiple mutations suggest a sum of multiple contributions. Full activation involves two events: release of the nSH2 domain of the p85α to relieve the autoinhibition, and membrane association for catalysis (87–89). Hotspot mutations promote PI3Kα activation by mimicking the two events. p85α has five domains. The interaction of its nSH2 domain with p110α is responsible for PI3Kα autoinhibition (90). RTK’s pY motif has high affinity to nSH2 (91–93). It competes with p110α for nSH2, releasing the autoinhibition (56). Hotspot mutations E542K and E545K are at the surface of the helical domain, where they mediate the interactions with nSH2. Their mutations to lysine residues with opposite charges disrupt the interfacial salt bridges, relieving the autoinhibition (59,94,95). PI3Kα is a lipid kinase. Its interaction with the membrane is required for catalysis. Active membrane-latched Ras promotes PI3Kα-membrane interaction. H1047 is on the surface of the KD (94). Its mutation to arginine (H1047R) increases the positive charge, thus promoting membrane interaction (59) (Figure 2B). Its local interactions may also enhance the stability of the catalytically competent PI3Kα-substrate interactions. Collectively, the hotspot mutations produce active catalytic site (ka) and promote the population of PIP2 in the active site (km).
Most of the weak mutations are far away from both the nSH2 and the catalytic site (96). The majority locate at p110α C2, adaptor binding domain (ABD) and ABD-RBD linker. C2 and ABD accommodate the p85α iSH2 domain, implying the involvement of iSH2 in PI3Kα activation (Figure 2A). The PIP2 substrate is negatively charged. PI3Kα activation loop contains the conserved basic boxes (xKxK and KRER), controlling the substrate specificity (97). In the crystal structures of PI3Kα with the bound nSH2 (PDB ID: 4OVV), the iSH2 interacts with the first basic box in the activation loop (Figure 2B). The conformational changes spurred by nSH2 release explain how weak mutations in C2 and ABD can collaborate with hotspot mutations leading (mainly via ka) to more potent PI3Kα activation (53). Upon nSH2 release, iSH2 frees the basic box in the activation loop on the KD’s surface for catalysis and promotes membrane interactions (Figure 2C) (53). In protein kinases, when the activation loop is in the extended conformation, the catalytic residues are ‘correctly’ positioned and oriented, enhancing activation in correspondence to their ka. The movements of ABD, C2 and iSH2 are linked (57). The weak mutations in C2 (N345K, C420R, and E453K/Q) and ABD domains (R38H/C, R88Q, R93Q, R108H, and G118/D) amplify the single hotspot via the coupled iSH2.
In the KD, the two weak mutations that are frequently observed in PI3Kα double mutants are E726K and M1043V/I. E726K is on the surface of the N-lobe, and M1043V/I is in the C-lobe. E726K can promote PI3Kα membrane interaction by increasing the positive charge. In that sense it resembles the H1047R hotspot, and since both N- and C-lobes interact with the membrane for catalysis, the collaboration of E726K in the N-lobe and H1047R in the C-lobe are expected. However, unlike H1047 which provides positive charge for the less charged surface on the C-lobe, there are several positively charged residues around E726, making it a weaker mutation (Figure 2A). H1047 and M1043 are at the regulatory arch of KD’s C-lobe. M1043 is buried, contributing to hydrophobic core interactions of the regulatory arch. Its mutation to valine or isoleucine with shorter hydrophobic side chains may alter the local hydrophobic interactions, promoting activation. Its minor effects might suggest that it is a ‘latent’ driver (98), that is, a mutation which behaves as a passenger, and does not confer a cancer hallmark. However, when coupled with other evolving mutations, can drive cancer and drug resistance. The actions of the hotspot and weak mutations can be coupled in PI3Kα activation, collectively leading to the enhanced substrate binding and membrane interactions.
PI3Kα with Double Mutations Responds Effectively to Inhibitors
PI3Kα with double/multiple mutations is more responsive at least to certain tested inhibitors as compared to single hotspot mutants (16). A population of breast cancer patients with double/multiple mutations displayed deep and prolonged clinical benefit from alpelisib, which was recently approved by the Food and Drug Administration (FDA). GDC0077 is undergoing clinical evaluation for breast cancer. Both drugs documented increased PI3K inhibition in patients.
Structurally, the inhibitors resemble ATP, thus can quench catalysis by interfering with ATP loading (Figure 2D). The ATP pocket in KD targeted by inhibitors are conserved among PI3K isoforms. The crystal structures (PDB ID: 4JPS) indicate the structural details of alpelisib interacting with PI3Kα. The alpelisib, as well as many other PI3K inhibitors, are loaded by the ATP pocket (58). Their interactions with the specificity pocket, affinity pocket and hinge region help in defining the isoform specificity and potency. PI3Kα activation features the structural and dynamic changes of KD in the active conformation (16,97). The additive activation of PI3Kα by double-mutations is expected to modulate the dynamics of ATP pockets for inhibitor response.
With the universal use of ATP in the cell, ATP-competitive drugs, especially the less specific ones like alpelisib, can elicit multiple side-effects. Even a PI3Kα inhibitor like GDC0077 is likely to retain certain affinity to other ATP binding sites, and concentration-dependent side-effects may take place. Cells with multiple mutations in the PIK3CA and NOTCH1 genes exhibit stronger dependencies on the mutated genes, enhanced downstream signaling, and (or) greater sensitivity to inhibitory drugs than those with single mutations (17).
Why Double Mutations in PI3Kα?
If single mutations are potent, why has PI3Kα evolved double/multiple mutations as observed in some cancer cells? Do some cells demand higher PI3Kα activity for proliferation via double/multiple mutations as supported by single hotspots promoting tumor growth in some systems, but failing in others (16)? In line with this, in breast cancer, PI3Kα mutations may co-exist with dysfunctional human epidermal growth factor receptor 2 (HER2) upstream and downstream proteins (99,100). Here we offer an alternative possible explanation.
Oncogenic mutations are commonly observed to be differentially expressed in tumors. Weak mutations in PI3Kα double/multiple mutations in breast cancer include E453K/Q, E726K, and M1043V/I; in other cancers R88Q and R93Q (16). The patterns of Ras hot spots and weaker mutations in lung, pancreatic, colorectal or skin cancers provide another well-known cell-specific example, even though those are documented for single, not double/multiple mutations (101). The preferred mutational distributions in tumors can be interpreted in the framework of distinct pre-existing cellular networks (102), which reflect tissue (tumor)-specific locally accessible chromatin states (103,104). Double mutations in PI3Kα comprise one hotspot and one weak activation mutation. Significantly, Vasan et al. did not observe double hotspot mutations, even though in principle they may generate ~1,000-fold higher downstream Akt phosphorylation than the single mutations (16,66). Notably, a recent more comprehensive analysis (17) did observe some, pointing to their rarity. To date, no other proteins were observed to carry them.
So why double (multiple) mutations and why one hotspot and the other(s) weak? And why double strong mutations are still possible for PI3K?
The activity threshold determines the pathological consequences of oncogenic PIK3CA (105) and single mutations cannot fully activate PI3Kα to reach the necessary threshold. In principle, two strong hot spots can – by acting together, they can lead to higher rate of replication. This can however result in breaks in the double-stranded DNA and cell senescence. That is, if the extent of proliferation is too high, the balance between senescence and proliferation can be tipped, leading to cell cycle arrest. Thus, since single hotspots cannot fully activate PI3K, tumor development cannot rely on single mutations for growth and proliferation. PI3K can achieve full activation through cooperation of two drivers, where one, e.g. H1047R facilitates the substrate binding at the active site through its positive charge interaction with the membrane (km) and the other, e.g. E545K, reduces the transition state barrier (ka) by reorganizing the active site as observed recently. The arguments above hold for co-occurring hotspot mutations in PI3Kα, and in PI3Kα and PTEN phosphatase.
A combination of additive contributions of strong and weaker drivers can enhance PI3K activity and proliferation at a level that can be sustained. We expect weak drivers to be common, albeit to date mostly unidentified (98). Like latent drivers, together they effectively shift the protein ensemble from the inactive to the active state (89). They can be statistically rare since for an observable functional change, they need to cooperate with additional mutations, and these are not considered in the cancer-specific protein sequence analysis. In line with this, a recent pan-cancer analysis observed multiple driver mutations to be common, occurring more frequently than expected in the same oncogene, with overrepresentation of functionally weak, rare mutations, which confer enhanced oncogenicity and sensitivity to drugs in combination as compared to single mutations (17).
The next obvious question to ask is are such double (multiple) hotspot/weak mutations the rule, or is PI3Kα an exception? Autoinhibition is common. Mutations that abolish it can be powerful drivers (106), as indeed now shown by Vasan et al. (16). Statistical analyses pairing them with additional tumor-specific mutations in patient samples can uncover those mutations (89). Nonetheless, such combinations may not be commonplace. Their occurrence and combined actions are likely to be structure- and activation mechanism (ka and km)-dependent and need to be explored on a case-by-case basis. PI3K is large and its activation at the membrane is complex. Smaller proteins may be fully activated by a single hotspot. Finally, in those PI3Kα cases where one hotspot is enough, cell-specific downstream signaling protein nodes could be expressed at higher levels or mutated to compensate for the absence of a second compensatory mutation in the same allele. The clinical correlation with the PI3Kα double mutations testifies to their significance. All mutants enhance or retain their interaction with the membrane. In those cases where the mutations are not at the membrane-interacting surface, as in those cases which involve only release of the nSH2 autoinhibition, the outcome is rearrangement exposing the active site at the membrane and enhancing the population of the PIP2 at the substrate binding site. As we discuss below in the context of Ras effectors and their assemblies, activating mutations often work by releasing autoinhibition (89).
Taken together, cell growth depends on the PI3Kα/Akt/mTOR pathway and PTEN which regulates it (Figure 1A). Double mutations may enhance PI3Kα activation, making its output enough for cell growth. This also explains why double mutations have higher response rate to PI3Kα inhibitors, such as alpelisib. We hypothesize that PI3K inhibitors have higher efficacy in the double mutation context since cancers driven by these multiple mutations are more specifically dependent on this particular signaling node. For a more comprehensive description of PI3K inhibitors and their mechanisms see Zhang et al. (58).
Assemblies of Ras Effectors: At the Membrane and Away from the Membrane
The assembly including Ras, PI3Kα, Akt, PDK1 and PTEN is at the membrane.
PI3Kα is activated as a functional heterodimer by RTK and a monomeric Ras molecule. Above, we discussed the role of RTK and Ras in activating it at the membrane and activating mutations. Downstream, signaling proceeds by PIP3 acting as a cofactor recruiting Akt1 amino-terminal pleckstrin homology (PH) domain. Active Akt1 is confined to membranes enriched in either PIP3 or PIP2 (3, 4), which maintain spatial and temporal control (107,108); even though numerous Akt1 phosphorylation substrates were identified (109), only few were carefully confirmed (110), suggesting that activation and substrate phosphorylation may be tightly coupled to membrane attachment (107). At the same time, genetic studies in several model organisms and human cancers suggested a role for Akt in the regulation of FOXO family transcription factors (111). Thus, while kinase intermediates may be involved, it is unlikely that these PI3K targets are regulated at the membrane. Full Akt activation requires phosphorylation of T308 in the activation loop by PDK1 and S473 in the carboxy-terminus by mTORC2. Autoinhibitory intramolecular PH–KD interactions maintain Akt1 in an inactive state (112). Activation involves a conformational change that loosens the PH‒KD domain interaction. In line with this, human tumors carry driver mutations at the PH–KD interface. In the closed PH‒KD interacting state (PH-in), PDK1 is unable to access and phosphorylate T308; in the open PH-out state it can. In an alternative pS473-dependent mechanism (113), Akt activation is driven by interaction between the C-terminal tail and the PH‒KD linker that relieves Akt1 autoinhibition. In another allosteric mechanism, activating Akt1 could involve dual Ser477/Thr479 phosphorylation.
PDK1 (Figure 1A) also binds PIP3 through the PH domain, which is recruited to the membrane via binding of phosphatidylserine at a site distinct from the phosphoinositide-binding site (114). Binding of the PH domains of both PDK1 and Akt1 to PIP3 results in their colocalization at the plasma membrane, where PDK1 can phosphorylate and activate Akt1 (115,116). Autoinhibitory PDK1 homodimer conformations have been proposed, although the exact scenario is yet to be worked out (117). PTEN tumor suppressor binds the plasma membrane and hydrolyzes PIP3 to form PIP2. Its loss-of-function mutations block this activity. Modeling obtained its membrane-associated state consistent with experimental results for triple mutant R161E/K163E/K164E (118). The activation mechanism is still unclear, and neither are the mechanisms of PTEN activating mutations (119). Scaffolding proteins such as IQ motif containing GTPase activating protein 1 (IQGAP1) (11) and adaptor protein such as Grb2-associated-binding protein 2 (GAB2) (120) also associate with the PI3K/Akt assembly, albeit not interact with the membrane. Altogether, this large dynamic multimolecular assembly is anchored at the membrane, with signaling lipids acting either as substrates or cofactors.
The mitogen-activated protein kinase (MAPK) assembly is not at the membrane.
Different from PI3Kα’s KD, there is no significant population of Raf’s KD interacting neither with Ras nor with the membrane. The different environments and activation scenarios clarify the altered patterns of signaling and activating mutations. Raf’s activation is away from the membrane, with a long (~150 residues) intrinsically disordered linker connecting the KD and the CRD that attaches Raf to the membrane (121) and the nearby RBD, positioning Raf KD in the cytoplasm. As we discussed above, from the physical standpoint, this is understandable: the surface of the plasma membrane is crowded, and proteins act in very large complexes that include multiple kinases, scaffolding and adaptor proteins such as galectin. Raf’s activation requires dimerization of its KD. MEK and ERK are dimers as well. Kinase suppressor of Ras (KSR) is also in the complex (13), and 14–3-3 is attached, and critically involved in the autoinhibition and activation, as the recent cryo-EM and crystal structures indicate (122,123). KSR and 14–3-3 are dimers as well. IQGAP1 scaffolding protein is large and binds to and regulates cell signaling also through the Raf/MEK/ERK pathway (124) and tethers ERK to actin filaments (125). It can attach to numerous partners (126–128), but not Ras (129). The sheer physical size and coordinated activation, function and complexity is likely a prime reason for Raf’s KD positioning away from the brimming membrane surface, albeit regulated by it through attachment of other domains (e.g. CRD, RBD). Crowding, slow diffusion rates, and temporal spatial nanoclusters (130), with epidermal growth factor receptor (EGFR, i.e. ErbB1) also directly binding actin, and both ErbB receptors and Met interacting with integrins (1), also increase the MAPK assembly sizes, and govern the numbers and locations of complexes (131). The water layer at the membrane surface further contributes to protein molecules diffusing away from the crowded membrane and assembling in the cytoplasm (15).
Ras contributes to Raf activation by shifting Raf’s conformational ensembles, which under physiological conditions are dominated by the closed, autoinhibited inactive states, to the open, active state. In the inactive state, Raf’s RBD and CRD can interact with the KD at a surface partly overlapping the dimerization interface. The assembly is further stabilized by the 14–3-3 as the recent structures indicate (122,123) (Figure 1B). While not measured, the stability of the KD–RBD/CRD is likely to be higher than the dimeric KD association. However, the affinity of the RBD–Ras interaction is in the low nanomolar range scale, much stronger than its interaction with the KD. Taken together, this suggests the following activation scenario (89). Raf populates three states. In the inactive state, which is the one where Raf spends most of its time, Ras is GDP-bound and Raf mostly populates its autoinhibited state, with minor populations in the open ‘free’ state and the Ras-bound state. However, in the presence of active GTP-bound Ras, the equilibrium shifts; the RBD of the open, free Raf state, binds Ras depleting the open free state population. The population of the autoinhibited states then shifts toward this open free state, maintaining the equilibrium, and concomitantly exposing the dimerization surface resulting in KD dimerization and activation. Phosphorylation plays a cardinal regulatory role in the relative populations of the states. In B-Raf, Ser446 phosphorylation weakens the autoinhibition; 14–3-3 proteins bind to pSer259 in C-Raf (Raf-1) and pSer365 in B-Raf (132–135), stabilizing the autoinhibition. Protein phosphatase 2A (PP2A) and protein phosphatase 1 (PP1) relieve it, with the equilibrium trending to the free open state (136–140). 14–3-3 also binds pSer621 in C-Raf and Ser729 in B-Raf (132,134,135). Even though the interaction of the KD with the RBD-CRD appears weak, binding of the KD at both sites can stabilize the autoinhibited state (133,136,141–144).
Different from PI3Kα, under physiological conditions Raf is activated by Ras dimers or nanoclusters, where Ras monomers are in spatial proximity (5), which promote Raf KD dimerization (Figure 1B). Hotspot mutations in the linker can relieve the autoinhibition (3,89); if occurring in the catalytic KD they can activate monomeric Raf, without Ras involvement as observed in drug resistance (3,145–147) in the classical case of V600E missense mutation in 80–90% of B-Raf mutants (My Cancer Genome: https://www.mycancergenome.org/content/alteration/braf-v600e/). To the best of our knowledge, to date no activating hotspot mutations were identified in the RBD or CRD. Whether double co-acting mutations are present in the same allele is unclear; again, to date none were found. The key requirement for full Raf activation is dimerization which requires a high effective local concentration (thus proximity) and availability of the interacting KD surface. Raf’s KD activation was explored in atomistic detail (148). However, conformational details related to the long-disordered linker and the recently uncovered KD–CRD/RBD interactions (122,123), are still missing due to high flexibility and the exact location of RBD in the assembly is still unclear. Our on-going modeling and simulations aim to help resolve this conundrum and obtain the full activation scenario.
The Biological Significance of the Two Modes of Activation, At and Away from the Membrane
Ras-mediated PI3Kα and Raf activation exemplify two different cellular modes of activation – at the membrane and away from the membrane. They epitomize different mechanisms of autoinhibition, patterns of driver mutations and phosphorylation. The efficient, dynamic, cascading kinases in the MAPK pathway are all dimers, occupy massive space, are better off away from the membrane, especially when considering also the scaffolding proteins which are involved in their assembly (149). In contrast, in the PI3K pathway, both Ras and RTK activators, are located at/in the membrane, and after activation the PIP3 product is in the membrane, to be exploited for further signal transduction. The distinct autoinhibition mechanisms adopted by PI3Kα and Raf evolved to fit their cellular location and activation scenarios: in Raf, the CRD and RBD block access to the kinase domain dimerization interface; in PI3Kα the p85α and p110α interdomain interfaces hinder the active site exposure to the membrane and substrate PIP2 access. Phosphorylation events tighten the autoinhibition in both PI3Kα and Raf, but in different ways. In Raf, at the cytoplasm, phosphorylated residues in the conserved region 2 (CR2, pS365 in B-Raf) and at the C-terminal CR3 (pS729 in B-Raf) serve as 14–3-3 binding sites (123); in PI3K, at the membrane, no additional protein is involved. Phosphorylated Ser608 (and Ser807) in p85α (150) may interact with p110α at the C-lobe, with the acidic residues around it interacting with the surrounding basic residues as well, increasing the stability of the autoinhibited state. With respect to driver mutations (151), no drivers have been identified in the RBD or CRD at the membrane. Instead, the vast majority of B-Raf’s 300 mutations (152) are in the activation loop near V600E, the major mutation in melanoma, or in the GSGSFG phosphate binding loop (P-loop) at residues 464–469 (152,153). This is not the case in PI3Kα, where most, including p85α truncation, are involved in relieving the autoinhibition or enhancing and stabilizing lipid substrate docking. This is critical, since the cavity is relatively small, with part of the PIP2 is still in the membrane. These two chores – shifting the ensemble away from the autoinhibited state and increasing the population of PIP2 at its pocket – are respectively taken up by two sets of mutations. For the first, E542K and E545K in the helical domain with charge reversal, mimicking the physiological process, as does Q546K/R. Cys420 (C2) and Asn345 (C2) at the iSH2–C2 interface are also mutated to basic residues (Arg or Lys), and may disrupt the iSH2–C2 interface through a repulsive force. p85α frequent truncation mutations starting from iSH2, destroy the iSH2–C2 interface, and via a conformational change involving iSH2 rotation promote exposure to the membrane and activation. Several are in linkers at the ABD-kinase domain interface; others at the ABD-RBD linker. Some involved in key contacts are mutated to uncharged amino acids (Cys or Gln). For the second, some, are at the surface of the kinase domain including H1047R are all mutated into Lys or Arg in cancer, promoting PI3Kα membrane interaction. A more complete list along with their mechanisms are described in Zhang et al. (53).
Thus, at the membrane, with limited space and the needed proximity to the small signaling lipid PIP2, occluding the active site through the p85α regulatory subunit and membrane attachment are critical. For Raf, at the cytoplasm with activation involving dimerization, and the presence of a huge signalosome assembly (149), 14–3-3 is called upon to help, with barely any mutations away from the kinase domain to disengage autoinhibition. This huge assembly is still dynamically anchored – to the cytoskeleton, where e.g. ERK controls protrusion initiation and protrusion speed (that emerge via the WRC (WAVE2 regulatory complex, which activates the Arp2/3 actin nucleator for actin assembly (154)); migration which is enabled by MAPK-induced cell softening via actin cytoskeleton re-organization (155), and more (e.g. (156)). IQGAP1 scaffolding protein is one way through which the interactions can be mediated, as it binds Raf, MEK, ERK and actin (127,128,157).
Conclusions
Lipid kinases mediate and transduce signals via lipids such as sphingolipids and phosphoinositides (158). Their dynamic assemblies include membrane-attached protein kinases that bind signaling lipids as cofactors, and a phosphatase that hydrolyzes the phosphorylated lipids to keep cell growth signaling in check. This is not the case for Raf’s KD and its assembly in the cytoplasm. Here we underscore some spatiotemporal physical principles regulating the assemblies at, and away from, the membrane. This helps clarify the activation mechanisms and delineates their driver mutations, with the recently identified PI3Kα double and multiple activating mutations providing an excellent example. Effective inhibition of kinases, especially lipid kinases, still faces immense hurdles with limited progress. Even though the so-called isoform specific PI3Kα inhibitors are becoming available at the bedside, these ATP-competitive drugs are toxic at effective concentrations. Drug resistance via parallel or redundant pathways is also expected. Eventually, coupling allosteric drugs (159,160) with combinatorial drug regimens and creative new twists (161) may forge a way forward (58).
The mechanism of activation of single and double co-occurring mutations led us to suggest new pharmacological strategies for PI3Kα: a combination of allosteric and orthosteric inhibitors targeting the same molecule; and following mother nature a rescue mutation strategy to guide drug discovery (58). In the first, the allosteric inhibitor may modulate the active site conformation to now favorably bind an inhibitor to overcome drug resistance; in the second, the allosteric site may be optimally located. Cancer appears to rely on the PI3K node and signaling pathway more than on other pathways; thus, PI3Kα inhibitors may have higher efficacy in the double and multiple mutation context. Notwithstanding, the expected emergence of drug resistance argues for targeting cell-type specific parallel (or redundant) chromatin-accessible proliferation pathways. The challenge in drug regimen is how to identify the proliferation pathway likely to emerge next (162).
Here we emphasized that biological regulation is cell-context dependent. PIK3α lipid kinase is activated at the crowded membrane surface and this is where its assembly locates; Raf protein kinase is in the cytoplasm. Their differing physical environments underlie their distinct modes of action, oncogenic mutations and inhibition. Future biophysical challenges could consider their broader framework including the cytoskeleton; they could also take up identification of their cell-specific proliferation pathways which are linked to their chromatin accessibility(162).
Acknowledgements
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Trenker R, Jura N. Receptor tyrosine kinase activation: From the ligand perspective. Curr Opin Cell Biol 2020;63:174–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, Phelps C, Huang T, Mostofian B, Wu L, Zhang Y, et al. High-throughput, single-particle tracking reveals nested membrane domains that dictate KRas(G12D) diffusion and trafficking. Elife 2019;8:e46393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai CJ, Nussinov R. Allosteric activation of RAF in the MAPK signaling pathway. Curr Opin Struct Biol 2018;53:100–6 [DOI] [PubMed] [Google Scholar]
- 4.Sartorel E, Unlu C, Jose M, Massoni-Laporte A, Meca J, Sibarita JB, et al. Phosphatidylserine and GTPase activation control Cdc42 nanoclustering to counter dissipative diffusion. Mol Biol Cell 2018;29:1299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nussinov R, Tsai CJ, Jang H. Is Nanoclustering essential for all oncogenic KRas pathways? Can it explain why wild-type KRas can inhibit its oncogenic variant? Semin Cancer Biol 2019;54:114–20 [DOI] [PubMed] [Google Scholar]
- 6.Maxwell KN, Zhou Y, Hancock JF. Rac1 Nanoscale Organization on the Plasma Membrane Is Driven by Lipid Binding Specificity Encoded in the Membrane Anchor. Mol Cell Biol 2018;38:e00186–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nussinov R, Tsai CJ, Jang H. Ras assemblies and signaling at the membrane. Curr Opin Struct Biol 2020;62:140–8 [DOI] [PubMed] [Google Scholar]
- 8.Kolch W, Kiel C. From oncogenic mutation to dynamic code. Science 2018;361:844–5 [DOI] [PubMed] [Google Scholar]
- 9.Bugaj LJ, Sabnis AJ, Mitchell A, Garbarino JE, Toettcher JE, Bivona TG, et al. Cancer mutations and targeted drugs can disrupt dynamic signal encoding by the Ras-Erk pathway. Science 2018;361:892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nussinov R, Tsai CJ, Jang H. Does Ras Activate Raf and PI3K Allosterically? Front Oncol 2019;9:1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei T, Choi SY, Buehler D, Anderson RA, Lambert PF. A PI3K/AKT Scaffolding Protein, IQ Motif-Containing GTPase Associating Protein 1 (IQGAP1), Promotes Head and Neck Carcinogenesis. Clin Cancer Res 2020;26:301–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorisse L, Li Z, Wagner CD, Worthylake DK, Zappacosta F, Hedman AC, et al. Ubiquitination of the scaffold protein IQGAP1 diminishes its interaction with and activation of the Rho GTPase CDC42. J Biol Chem 2020:doi: 10.1074/jbc.RA119.011491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavoie H, Sahmi M, Maisonneuve P, Marullo SA, Thevakumaran N, Jin T, et al. MEK drives BRAF activation through allosteric control of KSR proteins. Nature 2018;554:549–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins MJ, Polcik M, Fukuma T, Sader JE, Nakayama Y, Jarvis SP. Structured water layers adjacent to biological membranes. Biophys J 2006;91:2532–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nawrocki G, Im W, Sugita Y, Feig M. Clustering and dynamics of crowded proteins near membranes and their influence on membrane bending. P Natl Acad Sci USA 2019;116:24562–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasan N, Razavi P, Johnson JL, Shao H, Shah H, Antoine A, et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kalpha inhibitors. Science 2019;366:714–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito Y, Koya J, Araki M, Kogure Y, Shingaki S, Tabata M, et al. Landscape and function of multiple mutations within individual oncogenes. Nature 2020;582:95–9 [DOI] [PubMed] [Google Scholar]
- 18.Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K Pathway in Human Disease. Cell 2017;170:605–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bilanges B, Posor Y, Vanhaesebroeck B. PI3K isoforms in cell signalling and vesicle trafficking. Nat Rev Mol Cell Biol 2019;20:515–34 [DOI] [PubMed] [Google Scholar]
- 20.Burke JE. Structural Basis for Regulation of Phosphoinositide Kinases and Their Involvement in Human Disease. Mol Cell 2018;71:653–73 [DOI] [PubMed] [Google Scholar]
- 21.Hawkins PT, Stephens LR. PI3K signalling in inflammation. Bba-Mol Cell Biol L 2015;1851:882–97 [DOI] [PubMed] [Google Scholar]
- 22.Fritsch R, Downward J. SnapShot: Class I PI3 K Isoform Signaling. Cell 2013;154:940–1 [DOI] [PubMed] [Google Scholar]
- 23.De Santis MC, Sala V, Martini M, Ferrero GB, Hirsch E. PI3K Signaling in Tissue Hyper-Proliferation: From Overgrowth Syndromes to Kidney Cysts. Cancers (Basel) 2017;9:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanhaesebroeck B, Stephens L, Hawkins P. PI3K signalling: the path to discovery and understanding. Nat Rev Mol Cell Biol 2012;13:195–203 [DOI] [PubMed] [Google Scholar]
- 25.Nussinov R, Tsai CJ, Muratcioglu S, Jang H, Gursoy A, Keskin O. Principles of K-Ras effector organization and the role of oncogenic K-Ras in cancer initiation through G1 cell cycle deregulation. Expert Rev Proteomics 2015;12:669–82 [DOI] [PubMed] [Google Scholar]
- 26.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer 2015;15:7–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanhaesebroeck B, Whitehead MA, Pineiro R. Molecules in medicine mini-review: isoforms of PI3K in biology and disease. J Mol Med 2016;94:5–11 [DOI] [PubMed] [Google Scholar]
- 28.Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov 2014;13:140–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Getz G, Gabriel SB, Cibulskis K, Lander E, Sivachenko A, Sougnez C, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pereira B, Chin SF, Rueda OM, Vollan HKM, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun 2016;7:11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verret B, Cortes J, Bachelot T, Andre F, Arnedos M. Efficacy of PI3K inhibitors in advanced breast cancer. Ann Oncol 2019;30 Suppl 10:x12–x20 [DOI] [PubMed] [Google Scholar]
- 32.Vasan N, Toska E, Scaltriti M. Overview of the relevance of PI3K pathway in HR-positive breast cancer. Ann Oncol 2019;30 Suppl 10:x3–x11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stark AK, Sriskantharajah S, Hessel EM, Okkenhaug K. PI3K inhibitors in inflammation, autoimmunity and cancer. Curr Opin Pharmacol 2015;23:82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janku F, Yap TA, Meric-Bernstam F. Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol 2018;15:273–91 [DOI] [PubMed] [Google Scholar]
- 35.Mayer IA, Arteaga CL. The PI3K/AKT Pathway as a Target for Cancer Treatment. Annu Rev Med 2016;67:11–28 [DOI] [PubMed] [Google Scholar]
- 36.Ellis H, Ma CX. PI3K Inhibitors in Breast Cancer Therapy. Curr Oncol Rep 2019;21:110. [DOI] [PubMed] [Google Scholar]
- 37.Nussinov R, Tsai CJ, Mattos C. ‘Pathway drug cocktail’: targeting Ras signaling based on structural pathways. Trends Mol Med 2013;19:695–704 [DOI] [PubMed] [Google Scholar]
- 38.Marshall JDS, Whitecross DE, Mellor P, Anderson DH. Impact of p85 alpha Alterations in Cancer. Biomolecules 2019;9:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bheemanaboina RRY. Isoform-Selective PI3K Inhibitors for Various Diseases. Curr Top Med Chem 2020;20:1. [DOI] [PubMed] [Google Scholar]
- 40.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014;505:495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mukohara T PI3K mutations in breast cancer: prognostic and therapeutic implications. Breast Cancer (Dove Med Press) 2015;7:111–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burke JE, Williams RL. Synergy in activating class I PI3Ks. Trends Biochem Sci 2015;40:88–100 [DOI] [PubMed] [Google Scholar]
- 43.Dornan GL, Burke JE. Molecular Mechanisms of Human Disease Mediated by Oncogenic and Primary Immunodeficiency Mutations in Class IA Phosphoinositide 3-Kinases. Front Immunol 2018;9:575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ito Y, Hart JR, Vogt PK. Isoform-specific activities of the regulatory subunits of phosphatidylinositol 3-kinases - potentially novel therapeutic targets. Expert Opin Ther Tar 2018;22:869–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu S, Jang H, Muratcioglu S, Gursoy A, Keskin O, Nussinov R, et al. Ras Conformational Ensembles, Allostery, and Signaling. Chem Rev 2016;116:6607–65 [DOI] [PubMed] [Google Scholar]
- 46.Nussinov R, Wang GQ, Tsai CJ, Jang H, Lu SY, Banerjee A, et al. Calmodulin and PI3K Signaling in KRAS Cancers. Trends Cancer 2017;3:214–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venot Q, Blanc T, Rabia SH, Berteloot L, Ladraa S, Duong JP, et al. Targeted therapy in patients with PIK3CA-related overgrowth syndrome. Nature 2018;558:540–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Mao Y, Bouaziz M, Yu H, Qu X, Wang F, et al. Lens differentiation is controlled by the balance between PDGF and FGF signaling. PLoS Biol 2019;17:e3000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsolakos N, Durrant TN, Chessa T, Suire SM, Oxley D, Kulkarni S, et al. Quantitation of class IA PI3Ks in mice reveals p110-free-p85s and isoform-selective subunit associations and recruitment to receptors. Proc Natl Acad Sci U S A 2018;115:12176–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vadas O, Burke JE, Zhang XX, Berndt A, Williams RL. Structural Basis for Activation and Inhibition of Class I Phosphoinositide 3-Kinases. Sci Signal 2011;4:re2, 1–12 [DOI] [PubMed] [Google Scholar]
- 51.Wu H, Shekar SC, Flinn RJ, El-Sibai M, Jaiswal BS, Sen KI, et al. Regulation of Class IA PI 3-kinases: C2 domain-iSH2 domain contacts inhibit p85/p110alpha and are disrupted in oncogenic p85 mutants. Proc Natl Acad Sci U S A 2009;106:20258–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X, Dai J, Ni D, He X, Zhang H, Zhang J, et al. Insight into the mechanism of allosteric activation of PI3Kalpha by oncoprotein K-Ras4B. Int J Biol Macromol 2019;144:643–55 [DOI] [PubMed] [Google Scholar]
- 53.Zhang MZ, Jang H, Nussinov R. The mechanism of PI3K activation at the atomic level. Chem Sci 2019;10:3671–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang M, Jang H, Nussinov R. Structural Features that Distinguish Inactive and Active PI3K Lipid Kinases. Journal of Molecular Biology 2020:doi.org/10.1016/j.jmb.2020.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang M, Jang H, Nussinov R. The structural basis for Ras activation of PI3Kalpha lipid kinase. Phys Chem Chem Phys 2019;21:12021–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nolte RT, Eck MJ, Schlessinger J, Shoelson SE, Harrison SC. Crystal structure of the PI 3-kinase p85 amino-terminal SH2 domain and its phosphopeptide complexes. Nat Struct Biol 1996;3:364–74 [DOI] [PubMed] [Google Scholar]
- 57.Ioannis Galdadas FLG, Zoe Cournia. Unravelling the effect of the E545K mutation on PI3Kα kinase Chem Sci 2020:DOI: 10.1039/c9sc05903b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang MZ, Jang H, Nussinov R. PI3K inhibitors: review and new strategies. Chem Sci 2020;11:5855–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci U S A 2006;103:1475–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams R, Berndt A, Miller S, Hon WC, Zhang X. Form and flexibility in phosphoinositide 3-kinases. Biochem Soc Trans 2009;37:615–26 [DOI] [PubMed] [Google Scholar]
- 61.Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, Hooi CS, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res 2004;64:7678–81 [DOI] [PubMed] [Google Scholar]
- 62.Madsen RR, Vanhaesebroeck B, Semple RK. Cancer-Associated PIK3CA Mutations in Overgrowth Disorders. Trends Mol Med 2018;24:856–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004;304:554. [DOI] [PubMed] [Google Scholar]
- 64.Arafeh R, Samuels Y. PIK3CA in cancer: The past 30 years. Semin Cancer Biol 2019;59:36–49 [DOI] [PubMed] [Google Scholar]
- 65.Samuels Y, Waldman T. Oncogenic mutations of PIK3CA in human cancers. Curr Top Microbiol Immunol 2010;347:21–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A 2008;105:2652–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pang H, Flinn R, Patsialou A, Wyckoff J, Roussos ET, Wu H, et al. Differential enhancement of breast cancer cell motility and metastasis by helical and kinase domain mutations of class IA phosphoinositide 3-kinase. Cancer Res 2009;69:8868–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vatte C, Al Amri AM, Cyrus C, Chathoth S, Alsayyah A, Ahmad A, et al. Helical and kinase domain mutations of PIK3CA, and their association with hormone receptor expression in breast cancer. Oncol Lett 2019;18:2427–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buckles TC, Ziemba BP, Masson GR, Williams RL, Falke JJ. Single-Molecule Study Reveals How Receptor and Ras Synergistically Activate PI3K alpha and PIP3 Signaling. Biophys J 2017;113:2396–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siempelkamp BD, Rathinaswamy MK, Jenkins ML, Burke JE. Molecular mechanism of activation of class IA phosphoinositide 3-kinases (PI3Ks) by membrane-localized HRas. Journal of Biological Chemistry 2017;292:12256–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gymnopoulos M, Elsliger MA, Vogt PK. Rare cancer-specific mutations in PIK3CA show gain of function. Proc Natl Acad Sci U S A 2007;104:5569–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun MH, Hillmann P, Hofmann BT, Hart JR, Vogt PK. Cancer-derived mutations in the regulatory subunit p85 alpha of phosphoinositide 3-kinase function through the catalytic subunit p110 alpha. P Natl Acad Sci USA 2010;107:15547–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dornan GL, Stariha JTB, Rathinaswamy MK, Powell CJ, Boulanger MJ, Burke JE. Defining How Oncogenic and Developmental Mutations of PIK3R1 Alter the Regulation of Class IA Phosphoinositide 3-Kinases. Structure 2019;28:145–56 [DOI] [PubMed] [Google Scholar]
- 74.Cheung LW, Hennessy BT, Li J, Yu S, Myers AP, Djordjevic B, et al. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov 2011;1:170–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lindhurst MJ, Parker VER, Payne F, Sapp JC, Rudge S, Harris J, et al. Mosaic overgrowth with fibroadipose hyperplasia is caused by somatic activating mutations in PIK3CA. Nat Genet 2012;44:928–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lucas CL, Zhang Y, Venida A, Wang Y, Hughes J, McElwee J, et al. Heterozygous splice mutation in PIK3R1 causes human immunodeficiency with lymphoproliferation due to dominant activation of PI3K. J Exp Med 2014;211:2537–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mandelker D, Gabelli SB, Schmidt-Kittler O, Zhu JX, Cheong I, Huang CH, et al. A frequent kinase domain mutation that changes the interaction between PI3K alpha and the membrane. P Natl Acad Sci USA 2009;106:16996–7001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Y, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science 2007;317:239–42 [DOI] [PubMed] [Google Scholar]
- 79.Schroeder C, Riess A, Bonin M, Bauer P, Riess O, Dobler-Neumann M, et al. PIK3R1 mutations in SHORT syndrome. Clin Genet 2014;86:292–4 [DOI] [PubMed] [Google Scholar]
- 80.Shekar SC, Wu HY, Fu Z, Yip SC, Nagajyothi, Cahill SM, et al. Mechanism of constitutive phosphoinositide 3-kinase activation by oncogenic mutants of the p85 regulatory subunit. Journal of Biological Chemistry 2005;280:27850–5 [DOI] [PubMed] [Google Scholar]
- 81.Urick ME, Rudd ML, Godwin AK, Sgroi D, Merino M, Bell DW. PIK3R1 (p85 alpha) Is Somatically Mutated at High Frequency in Primary Endometrial Cancer. Cancer Research 2011;71:4061–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Toker A Double trouble for cancer gene. Science 2019;366:685–6 [DOI] [PubMed] [Google Scholar]
- 83.Hanker AB, Kaklamani V, Arteaga CL. Challenges for the Clinical Development of PI3K Inhibitors: Strategies to Improve Their Impact in Solid Tumors. Cancer Discov 2019;9:482–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kurek KC, Luks VL, Ayturk UM, Alomari AI, Fishman SJ, Spencer SA, et al. Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am J Hum Genet 2012;90:1108–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jindal GA, Goyal Y, Yamaya K, Futran AS, Kountouridis I, Balgobin CA, et al. In vivo severity ranking of Ras pathway mutations associated with developmental disorders. Proc Natl Acad Sci U S A 2017;114:510–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kiel C, Serrano L. Structure-energy-based predictions and network modelling of RASopathy and cancer missense mutations. Mol Syst Biol 2014;10:727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burke JE, Perisic O, Masson GR, Vadas O, Williams RL. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110alpha (PIK3CA). Proc Natl Acad Sci U S A 2012;109:15259–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miller MS, Schmidt-Kittler O, Bolduc DM, Brower ET, Chaves-Moreira D, Allaire M, et al. Structural basis of nSH2 regulation and lipid binding in PI3K alpha. Oncotarget 2014;5:5198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nussinov R, Tsai CJ, Jang H. Autoinhibition can identify rare driver mutations and advise pharmacology. FASEB J 2020;34:16–29 [DOI] [PubMed] [Google Scholar]
- 90.Thorpe LM, Spangle JM, Ohlson CE, Cheng H, Roberts TM, Cantley LC, et al. PI3K-p110alpha mediates the oncogenic activity induced by loss of the novel tumor suppressor PI3K-p85alpha. Proc Natl Acad Sci U S A 2017;114:7095–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Panayotou G, Gish G, End P, Troung O, Gout I, Dhand R, et al. Interactions between Sh2 Domains and Tyrosine-Phosphorylated Platelet-Derived Growth-Factor Beta-Receptor Sequences - Analysis of Kinetic-Parameters by a Novel Biosensors-Based Approach. Mol Cell Biol 1993;13:3567–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Piccione E, Case RD, Domchek SM, Hu P, Chaudhuri M, Backer JM, et al. Phosphatidylinositol 3-Kinase P85 Sh2 Domain Specificity Defined by Direct Phosphopeptide Sh2 Domain Binding. Biochemistry-Us 1993;32:3197–202 [DOI] [PubMed] [Google Scholar]
- 93.Rordorfnikolic T, Vanhorn DJ, Chen DX, White MF, Backer JM. Regulation of Phosphatidylinositol 3’-Kinase by Tyrosyl Phosphoproteins - Full Activation Requires Occupancy of Both Sh2 Domains in the 85-Kda Regulatory Subunit. Journal of Biological Chemistry 1995;270:3662–6 [DOI] [PubMed] [Google Scholar]
- 94.Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, et al. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science 2007;318:1744–8 [DOI] [PubMed] [Google Scholar]
- 95.Leontiadou H, Galdadas I, Athanasiou C, Cournia Z. Insights into the mechanism of the PIK3CA E545K activating mutation using MD simulations. Sci Rep 2018;8:15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Burke JE, Perisic O, Williams RL. Allosteric activation of PI3Ka by oncogenic mutations. Oncotarget 2013;4:180–1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pirola L, Zvelebil MJ, Bulgarelli-Leva G, Van Obberghen E, Waterfield MD, Wymann MP. Activation loop sequences confer substrate specificity to phosphoinositide 3-kinase alpha (PI3Kalpha ). Functions of lipid kinase-deficient PI3Kalpha in signaling. J Biol Chem 2001;276:21544–54 [DOI] [PubMed] [Google Scholar]
- 98.Nussinov R, Tsai CJ. ‘Latent drivers’ expand the cancer mutational landscape. Curr Opin Struct Biol 2015;32:25–32 [DOI] [PubMed] [Google Scholar]
- 99.Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res 2005;65:2554–9 [DOI] [PubMed] [Google Scholar]
- 100.Perez-Tenorio G, Alkhori L, Olsson B, Waltersson MA, Nordenskjold B, Rutqvist LE, et al. PIK3CA mutations and PTEN loss correlate with similar prognostic factors and are not mutually exclusive in breast cancer. Clin Cancer Res 2007;13:3577–84 [DOI] [PubMed] [Google Scholar]
- 101.Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. J Cell Sci 2016;129:1287–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ibanez Gaspar V, Catozzi S, Ternet C, Luthert PJ, Kiel C. Analysis of Ras-effector interaction competition in large intestine and colorectal cancer context. Small GTPases 2020:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sack LM, Davoli T, Li MZ, Li YY, Xu QK, Naxerova K, et al. Profound Tissue Specificity in Proliferation Control Underlies Cancer Drivers and Aneuploidy Patterns. Cell 2018;173:499–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Haigis KM, Cichowski K, Elledge SJ. Tissue-specificity in cancer: The rule, not the exception. Science 2019;363:1150–1 [DOI] [PubMed] [Google Scholar]
- 105.Madsen RR, Knox RG, Pearce W, Lopez S, Mahler-Araujo B, McGranahan N, et al. Oncogenic PIK3CA promotes cellular stemness in an allele dose-dependent manner. P Natl Acad Sci USA 2019;116:8380–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nussinov R, Tsai C, Jang H. Why Are Some Driver Mutations Rare? Trends in pharmacological sciences 2019;40:919–29 [DOI] [PubMed] [Google Scholar]
- 107.Lucic I, Rathinaswamy MK, Truebestein L, Hamelin DJ, Burke JE, Leonard TA. Conformational sampling of membranes by Akt controls its activation and inactivation. Proc Natl Acad Sci U S A 2018;115:E3940–E9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Leonard TA. Reply to Agarwal: Activity against nuclear substrates is not necessarily mediated by nuclear Akt. Proc Natl Acad Sci U S A 2018;115:E6101–E2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Manning BD, Toker A. AKT/PKB Signaling: Navigating the Network. Cell 2017;169:381–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.George S, Rochford JJ, Wolfrum C, Gray SL, Schinner S, Wilson JC, et al. A family with severe insulin resistance and diabetes due to a mutation in AKT2. Science 2004;304:1325–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tzivion G, Dobson M, Ramakrishnan G. FoxO transcription factors; Regulation by AKT and 14–3-3 proteins. Biochim Biophys Acta 2011;1813:1938–45 [DOI] [PubMed] [Google Scholar]
- 112.Parikh C, Janakiraman V, Wu WI, Foo CK, Kljavin NM, Chaudhuri S, et al. Disruption of PH-kinase domain interactions leads to oncogenic activation of AKT in human cancers. Proc Natl Acad Sci U S A 2012;109:19368–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chu N, Salguero AL, Liu AZ, Chen Z, Dempsey DR, Ficarro SB, et al. Akt Kinase Activation Mechanisms Revealed Using Protein Semisynthesis. Cell 2018;174:897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lucas N, Cho W. Phosphatidylserine binding is essential for plasma membrane recruitment and signaling function of 3-phosphoinositide-dependent kinase-1. J Biol Chem 2011;286:41265–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PRJ, Reese CB, et al. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase B alpha. Curr Biol 1997;7:261–9 [DOI] [PubMed] [Google Scholar]
- 116.Cordon-Barris L, Pascual-Guiral S, Yang S, Gimenez-Llort L, Lope-Piedrafita S, Niemeyer C, et al. Mutation of the 3-Phosphoinositide-Dependent Protein Kinase 1 (PDK1) Substrate-Docking Site in the Developing Brain Causes Microcephaly with Abnormal Brain Morphogenesis Independently of Akt, Leading to Impaired Cognition and Disruptive Behaviors. Mol Cell Biol 2016;36:2967–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Masters TA, Calleja V, Armoogum DA, Marsh RJ, Applebee CJ, Laguerre M, et al. Regulation of 3-phosphoinositide-dependent protein kinase 1 activity by homodimerization in live cells. Sci Signal 2010;3:ra78. [DOI] [PubMed] [Google Scholar]
- 118.Lumb CN, Sansom MSP. Defining the Membrane-Associated State of the PTEN Tumor Suppressor Protein. Biophys J 2013;104:613–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Smith IN, Thacker S, Seyfi M, Cheng FX, Eng C. Conformational Dynamics and Allosteric Regulation Landscapes of Germline PTEN Mutations Associated with Autism Compared to Those Associated with Cancer. American Journal of Human Genetics 2019;104:861–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Adams SJ, Aydin IT, Celebi JT. GAB2--a scaffolding protein in cancer. Mol Cancer Res 2012;10:1265–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Travers T, Lopez CA, Van QN, Neale C, Tonelli M, Stephen AG, et al. Molecular recognition of RAS/RAF complex at the membrane: Role of RAF cysteine-rich domain. Sci Rep-Uk 2018;8:8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kondo Y, Ognjenovic J, Banerjee S, Karandur D, Merk A, Kulhanek K, et al. Cryo-EM structure of a dimeric B-Raf:14–3-3 complex reveals asymmetry in the active sites of B-Raf kinases. Science 2019;366:109–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Park E, Rawson S, Li KH, Kim BW, Ficarro SB, Gonzalez-Del Pino G, et al. Architecture of autoinhibited and active BRAF-MEK1–14-3–3 complexes. Nature 2019;575:545–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sanchez-Laorden B, Viros A, Marais R. Mind the IQGAP. Cancer Cell 2013;23:715–7 [DOI] [PubMed] [Google Scholar]
- 125.Vetterkind S, Poythress RH, Lin QQ, Morgan KG. Hierarchical scaffolding of an ERK1/2 activation pathway. Cell Commun Signal 2013;11:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sayedyahossein S, Li ZG, Hedman AC, Morgan CJ, Sacks DB. IQGAP1 Binds to Yes-associated Protein (YAP) and Modulates Its Transcriptional Activity. Journal of Biological Chemistry 2016;291:19261–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Smith JM, Hedman AC, Sacks DB. IQGAPs choreograph cellular signaling from the membrane to the nucleus. Trends Cell Biol 2015;25:171–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nussinov R, Jang H. Dynamic multiprotein assemblies shape the spatial structure of cell signaling. Prog Biophys Mol Bio 2014;116:158–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Morgan CJ, Hedman AC, Li Z, Sacks DB. Endogenous IQGAP1 and IQGAP3 do not functionally interact with Ras. Sci Rep 2019;9:11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nussinov R, Tsai CJ, Jang H. Oncogenic KRas mobility in the membrane and signaling response. Semin Cancer Biol 2019;54:109–13 [DOI] [PubMed] [Google Scholar]
- 131.Bray D Signaling complexes: Biophysical constraints on intracellular communication. Annu Rev Bioph Biom 1998;27:59–75 [DOI] [PubMed] [Google Scholar]
- 132.Michaud NR, Fabian JR, Mathes KD, Morrison DK. 14–3-3 Is Not Essential for Raf-1 Function - Identification of Raf-1 Proteins That Are Biologically Activated in a 14–3-3-Independent and Ras-Independent Manner. Mol Cell Biol 1995;15:3390–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tzivion G, Luo ZJ, Avruch J. A dimeric 14–3-3 protein is an essential cofactor for Raf kinase activity. Nature 1998;394:88–92 [DOI] [PubMed] [Google Scholar]
- 134.Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14–3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 1996;84:889–97 [DOI] [PubMed] [Google Scholar]
- 135.Rommel C, Radziwill G, Lovric J, Noeldeke J, Heinicke T, Jones D, et al. Activated Ras displaces 14–3-3 protein from the amino terminus of c-Raf-1. Oncogene 1996;12:609–19 [PubMed] [Google Scholar]
- 136.Lavoie H, Therrien M. Regulation of RAF protein kinases in ERK signalling. Nat Rev Mol Cell Bio 2015;16:281–98 [DOI] [PubMed] [Google Scholar]
- 137.Dhillon AS, Meikle S, Yazici Z, Eulitz M, Kolch W. Regulation of Raf-1 activation and signalling by dephosphorylation. Embo J 2002;21:64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Abraham D, Podar K, Pacher M, Kubicek M, Welzel N, Hemmings BA, et al. Raf-1-associated protein phosphatase 2A as a positive regulator of kinase activation. Journal of Biological Chemistry 2000;275:22300–4 [DOI] [PubMed] [Google Scholar]
- 139.Jaumot M, Hancock JF. Protein phosphatases 1 and 2A promote Raf-1 activation by regulating 14–3-3 interactions. Oncogene 2001;20:3949–58 [DOI] [PubMed] [Google Scholar]
- 140.Ory S, Zhou M, Conrads TP, Veenstra TD, Morrison DK. Protein phosphatase 2A positively regulates ras signaling by dephosphorylating KSR1 and Raf-1 on critical 14–3-3 binding sites. Curr Biol 2003;13:1356–64 [DOI] [PubMed] [Google Scholar]
- 141.Tran NH, Wu X, Frost JA. B-Raf and Raf-1 are regulated by distinct autoregulatory mechanisms. J Biol Chem 2005;280:16244–53 [DOI] [PubMed] [Google Scholar]
- 142.Matallanas D, Birtwistle M, Romano D, Zebisch A, Rauch J, von Kriegsheim A, et al. Raf family kinases: old dogs have learned new tricks. Genes Cancer 2011;2:232–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dumaz N, Marais R. Protein kinase A blocks Raf-1 activity by stimulating 14–3-3 binding and blocking Raf-1 interaction with Ras. J Biol Chem 2003;278:29819–23 [DOI] [PubMed] [Google Scholar]
- 144.Molzan M, Ottmann C. Synergistic binding of the phosphorylated S233- and S259-binding sites of C-RAF to one 14–3-3zeta dimer. J Mol Biol 2012;423:486–95 [DOI] [PubMed] [Google Scholar]
- 145.Freeman AK, Ritt DA, Morrison DK. Effects of Raf dimerization and its inhibition on normal and disease-associated Raf signaling. Mol Cell 2013;49:751–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Durrant DE, Morrison DK. Targeting the Raf kinases in human cancer: the Raf dimer dilemma. Br J Cancer 2018;118:3–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Thevakumaran N, Lavoie H, Critton DA, Tebben A, Marinier A, Sicheri F, et al. Crystal structure of a BRAF kinase domain monomer explains basis for allosteric regulation. Nat Struct Mol Biol 2015;22:37–43 [DOI] [PubMed] [Google Scholar]
- 148.Jambrina PG, Rauch N, Pilkington R, Rybakova K, Nguyen LK, Kholodenko BN, et al. Phosphorylation of RAF Kinase Dimers Drives Conformational Changes that Facilitate Transactivation. Angew Chem Int Edit 2016;55:983–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mysore VP, Zhou Z-W, Ambrogio C, Li L, Kapp JN, Lu C, et al. A structural model of a Ras-Raf signalosome. bioRxiv 2020:DOI: 10.1101/2020.07.15.165266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Layton MJ, Saad M, Church NL, Pearson RB, Mitchell CA, Phillips WA. Autophosphorylation of serine 608 in the p85 regulatory subunit of wild type or cancer-associated mutants of phosphoinositide 3-kinase does not affect its lipid kinase activity. BMC Biochem 2012;13:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer 2014;14:455–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 2011;39:D945–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell 2004;6:313–9 [DOI] [PubMed] [Google Scholar]
- 154.Mendoza MC, Er EE, Zhang W, Ballif BA, Elliott HL, Danuser G, et al. ERK-MAPK drives lamellipodia protrusion by activating the WAVE2 regulatory complex. Mol Cell 2011;41:661–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rudzka DA, Spennati G, McGarry DJ, Chim YH, Neilson M, Ptak A, et al. Migration through physical constraints is enabled by MAPK-induced cell softening via actin cytoskeleton re-organization. J Cell Sci 2019;132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Descot A, Hoffmann R, Shaposhnikov D, Reschke M, Ullrich A, Posern G. Negative regulation of the EGFR-MAPK cascade by actin-MAL-mediated Mig6/Errfi-1 induction. Mol Cell 2009;35:291–304 [DOI] [PubMed] [Google Scholar]
- 157.Roy M, Li Z, Sacks DB. IQGAP1 binds ERK2 and modulates its activity. J Biol Chem 2004;279:17329–37 [DOI] [PubMed] [Google Scholar]
- 158.Carrera AC, Anderson R. The cell biology behind the oncogenic PIP3 lipids. J Cell Sci 2019;132:jcs228395. [DOI] [PubMed] [Google Scholar]
- 159.Nussinov R, Tsai CJ. Allostery in Disease and in Drug Discovery. Cell 2013;153:293–305 [DOI] [PubMed] [Google Scholar]
- 160.Nussinov R, Tsai CJ. Unraveling structural mechanisms of allosteric drug action. Trends in Pharmacological Sciences 2014;35:256–64 [DOI] [PubMed] [Google Scholar]
- 161.Zhang Z, Shokat KM. Bifunctional Small-Molecule Ligands of K-Ras Induce Its Association with Immunophilin Proteins. Angew Chem Int Ed Engl 2019;58:16314–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nussinov R, Tsai CJ, Jang H. Are Parallel Proliferation Pathways Redundant? Trends Biochem Sci 2020;45:554–63 [DOI] [PubMed] [Google Scholar]