Abstract
A case-control design determined whether konzo, an upper motoneuron disease linked to food (cassava) toxicity was associated with protein carbamoylation and genetic variations. Exon sequences of thiosulfate sulfurtransferase (TST) or mercaptopyruvate sulfurtransferase (MPST), plasma cyanide detoxification rates, and 2D-LC-MS/MS albumin carbamoylation were assessed in 40 children [21 konzo-affected and 19 putatively healthy controls, mean (SD) age: 9.2 (3.0) years] subjected to cognition and motor testing using the Kaufman Assessment Battery and the Bruininks/Oseretsky Test, respectively. Konzo was significantly associated with higher levels of carbamoylated peptides 206–219 (LDELRDEGKASSAK, pep1) after adjusting for age, gender, albumin concentrations and BUN [regression coefficient: 0.03 (95%CI:0.02 – 0.05), p = 0.01]. Levels of pep1 negatively correlated with performance scores at all modalities of motor proficiency (r = 0.38 to 0.61; all p < 0.01) or sequential processing (memory)(r = − 0.59, p = 0.00) and overall cognitive performance ( r = − 0.48, p = 0.00) but positively with time needed for cyanide detoxification in plasma (r = 0.33, p = 0.04). Rare potentially damaging TST p.Arg206Cys (rs61742280) and MPST p.His317Tyr (rs1038542246) heterozygous variants were identified but with no impact on subject phenotypes. Protein carbamoylation appears to be a reliable marker for cassava related neurodegeneration.
I. INTRODUCTION
Protein carbamoylation has been associated with a host of human conditions, including but not limited to cardiovascular diseases, kidney failure, diabetes, and aging1–8. The post-translational modification (PTM) occurs when isocyanic acid covalently adducts sulfhydryl or α and/or ε-amino groups of amino acids or proteins in living organisms. Isocyanic acid may derive from cyanate (OCN), a spontaneous metabolite urea in aqueous solutions or from peroxide-catalyzed oxidation of thiocyanate (SCN), a metabolite of cyanide1,9–11.
PTM including carbamoylation are of significant interest to the mechanisms of neurodegeneration as they may cause conformational changes in protein structure and function leading to cell death or modifications that can serve as biomarkers of pathogenic states12,13. We and others have shown that OCN toxicity may be associated with protein carbamoylation, changes in enzyme functions, memory deficits and motor system degeneration in laboratory animals13–16. We have proposed that similar events may occur in konzo, a distinct upper motor neuron disease associated with chronic dietary reliance on food from improperly processed cyanogenic cassava, which contains high amounts of β-glucosides notably linamarin14,17. Upon ingestion of poorly processed cyanogenic cassava, linamarin is hydrolyzed to via the gut flora linamarase-detoxifying mechanisms and cyanide release ensues. Under normal conditions, cyanide is in turn metabolized into the purportedly less toxic SCN via sulfurtransferase-regulated pathways. However, under chronic malnutrition and, perhaps, genetic variations in cyanide detoxifying enzymes, sulfurtransferase detoxification capabilities are compromised and oxidative mechanisms are triggered resulting in an increase production of OCN, a motor system toxicant known to induce protein carbamoylation14,18,19.
A genetic component of disease susceptibility to konzo has not been ruled out though plausible as familial clusters of disease exist18,20. In the present study, we searched for genetic variations in genes coding for the two main host cyanide detoxifying enzymes, thiosulfate sulfurtransferase (TST a.k.a rhodanese) and mercapto pyruvate sulfurtransferase (MPST) and assessed levels of protein (serum albumin) carbamoylation in children from the konzo heavily affected district of Kahemba in the Democratic Republic of Congo.
II. METHODS
II.1. Study Area
Ongoing research on cassava related motor and cognitive deficits takes place in the district of Kahemba, Bandundu province, Democratic Republic of Congo. Kahemba residents rely on cassava farming and limited livestock (goat, pigs, chicken) for subsistence. During the last decade, the population of Kahemba has faced a severe agroecological crisis due to population displacement with the end of war in Angola. Because of food shortages, residents of Kahemba were forced to adopt shortcuts in cassava processing methods, a phenomenon that has led to food (cassava) cyanogenic exposure. Shortly after, a sudden rise in the number of cases with konzo was documented with prevalence up to 10% in certain villages21. We have reported a sustained deterioration in motor and cognitive functions in association with the aforementioned exposure in children from a large (N = 200) 4-year prospective cohort of children20,22.
II.2. Study Subjects
Forty children [21 boys, 19 girls, overall mean (SD) age: 9.2 (3.0)] from Kahemba were included in the present study. Of these, 17 had a severe form of konzo i.e. unable to walk, 2 pairs of twins with a mild form of the disease i.e. using walking sticks, and 19 close neighbours had no visible neurological abnormalities, thus considered as controls. All the 17 severely affected subjects from the abovementioned cohort study were included in the present to better contrast the study findings. The pairs of twins were included for the sole purpose of internal genetic validation. Children with konzo fulfilled the WHO definition criteria for the disease, notably: (1) a visible symmetric spastic abnormality of gait while walking or running; (2) a history of onset of less than 1 week followed by a non-progressive course in an otherwise formerly healthy person; and (3) bilaterally exaggerated knee or ankle jerks without symptoms of spinal disease23.
All study subjects had their cognition and motor performance scores measured on the Kaufman Assessment Battery for Children, 2nd edition (KABC-II) for cognition and the Bruininks/Oseretsky Test, 2nd Edition (BOT-2), for motor proficiency, respectively. Routine clinical chemistry, urinary and plasma concentrations of thiocyanate as markers of cyanogenic exposure as well the plasma cyanide detoxification rates [expressed as the number of milliseconds required to detoxify cyanide to yield one μmol of thiocyanate per mg of protein ms/(μmol/mg protein] were readily available from the aforementioned cohort study20,24.
Ethics statement
Informed consent and child assent were obtained verbally by investigators who were fluent in Lingala and/or Kikongo, the local spoken languages. Ethical approval of research activities including informed consent and assent procedures was obtained from the Oregon Health & Science University (OHSU) Institutional Review Board FWA00000161 and from the Ministry of Health of the Democratic Republic of Congo (DRC).
II.3. Exon sequencing and protein modeling of cyanide detoxifying enzymes
II.3.1. Exon sequencing of cyanide detoxifying candidate genes
Genomic DNA was extracted from the buffy coat using a Maxwell 16 Blood DNA Purification kit (Promega): AS1010 according to manufacturer’s instruction. DNA samples were immediately frozen to −80 °C and shipped on dry ice to OHSU for storage and analysis. Prior to sequencing, DNA concentration was determined using a Nanodrop (ND-1000, Thermo Fisher Scientific, Waltham, MA) and DNA primers for exon sequencing were designed using Primer3 V.0.4.0 software (http://frodo.wi.mit.edu/). The primer design was based on the nucleotide sequences of TST and MPST as published in GenBank (Accession No. NT_011520.12 and NT_011520, respectively). TST and MPST exons amplifications were performed in a volume of 25 μl with final concentrations being 10 mM QNTPs, 10xQ Taq buffer, GC-rich/Q solution, 5 units Taq polymerase, 10 μM primers, and 10ng/μl of genomic DNA. We used Taq PCR Core Kit DNA from Qiagen (Germantown, MD) and the amplification by polymerase chain reaction (PCR) was carried out in Peltier Thermal cycler (PTC-200) from MJ Research (Ramsey, MN). Cycling conditions were as follows: DNA was denatured at 94°C for 15 seconds, annealing temperature was 70°C for 30 seconds with an extension step at 72°C for 1 minute. The extension step of 70°C for 30 seconds was followed by a temperature gradient with an increment of 1°C decrease per cycle for 15 cycles, 30 seconds each cycle from 70°C to 55°C. The temperature was brought back to 94°C for 15 seconds, and decreased to 55°C for 30 seconds followed by one cycle at 72°C for 1 minute. Additional 25 cycles of 94°C for 15 seconds; 55°C for 30 seconds, and 72°C for 1 minute were added. Finally, a cycle of 72°C for 10 minutes and the reaction was set to end at 15°C. Excess primer and nucleotides were removed by ExoSAP-IT enzyme according to the manufacturer’s instruction (Affymetrix, Santa Clara, CA). A mixture of 160 μl dye, 240 μl of 5x buffer, and 240 μl deionized water was prepared and 4 μl of the mix was added to 6 μl of the amplicon. Samples were run in the thermocycler set at 96°C for 1 minute and 50 seconds, 96 °C for 10 seconds, 52 for 5 seconds and 60°C for 4 minutes. The Sequencing was performed on ABI 3100XL Genetic Analyzer from Applied Biosystem (Foster City, California) using BigDye 3.1 sequencing chemistry reagents. The chromatograms were read with FinchTV 1.4.0, and sequences were blasted using NCBI blast engine. The Human genome (Hg38) reference was used for the genomic coordinate’s and the dbSNP database was used to determine allele frequency as found in gnomad and topmed data bases.
II.3.2. In silico protein modeling of mutated cyanide detoxifying enzymes
TST, MPST, and corresponding potentially damaging point-mutant proteins were modeled using the SWISS-model server via EXPASY (https://swissmodel.expasy.org). All models’ GMQE scores are 0.98 or higher, indicating high-quality models. To assess the change in the Gibb’s free energy of folding, SWISS-model PDB files were submitted to both STRUM6(https://zhanglab.ccmb.med.umich.edu/STRUM/about.html) and MAESTROweb7 (https://biwww.che.sbg.ac.at/maestro/web) servers25–28.
II.4. Protein (serum albumin) carbamoylation
II.4.1. Identification of carbamoylated peptides
Carbamoylation studies were performed on albumin, the most abundant protein in serum. To first identify carbamoylated peptides, serum was pooled from 20 control and 19 case subjects and 100 μg portions of each were trypsinized overnight as previously described14. The resulting peptides were then solid phase extracted using Sep Pak Light C18 cartridges (Waters Corp., Milford, MA), and separated into 8 fractions by loading onto a 2.1 × 5mm Reliasil cation exchange cartridge (Opti-Lynx, 11–02870-ES, Optimize Technologies, Portland, OR) and elution by injection of 300 μl of 17, 21, 25, 37.5, 56.5, 75, 150, and 375 mM ammonium formate solution containing 25% acetonitrile and 0.4% formic acid. The 8 cation exchange fractions, including the flow through, were then dried by vacuum concentration, re-dissolved in 5% formic acid and analyzed by LC-MS/MS using an Agilent 1100 series capillary LC system (Agilent Technologies) and an LTQ linear ion trap mass spectrometer (Thermo Scientific). Data-dependent MS/MS data was collected as previously described14, except a 95 min acetonitrile gradient was used for the 9 fractions from each digest (14 hours of data collection per sample). Peak lists from raw instrument files and matching of MS/MS data to peptide sequences was also as previously described14 except a Uniprot database (Swiss Bioinformatics Institute, Geneva, Switzerland) containing human sequences and common contaminants (20,433 entries) was used. Peptide false discovery was controlled by amending the database with sequence-reversed entries as previously described29, except unmodified and modified peptides containing a +43 mass increase at lysines, indicating carbamoylation, were independently filtered to maintain an approximately 1.5% peptide false discovery rate in both peptide classes. Confidently filtered results were then loaded into Scaffold software (Version 4.4.1, Proteome Software, Portland, OR) to assist visualization of annotated MS/MS spectra.
II.4.2. Quantification of carbamoylated serum albumin peptides
Carbamoylated albumin peptides 206–219 (LDELRDEGKASSAK, pep1) and 438–452 (KVPQVSTPTLVEVSR, pep2) identified during the 2-dimensional LC-MS/MS analysis described above, as well as unmodified peptides 66–75 (LVNEVTEFAK), 397–413 (VFDEFKPLVEEPQNLIK), and 599–609 (LVAASQAALGL) used for normalization of carbamoylated peptides 206–219 and 438–452, were subjected to a targeted mass spectrometric analysis to measure differences in levels of albumin carbamoylation in individual subjects. Serum was diluted 10-fold in 50 mM ammonium bicarbonate (AB), and 8.4 μl (~50 μg) was used for trypsin digestion. The digestion mixture was prepared by adding 2 μl of 1% ProteaseMax surfactant (ProMega), 83.1 μl of 50 mM AB, 1 μl 0.5 M dithiothreitol (DTT), and incubated at 56°C for 20 minutes. Samples were then alkylated by adding 2.7 μl of 0.55 M iodoacetamide and incubation at room temperature in the dark for 15 minutes. An additional 1 μl 1% ProteaseMax was then added and digestion initiated by addition of 1.8 μl of proteomics grade trypsin (Sigma) dissolved at 1 μg/μl concentration in 1 mM HCl. Following incubation at 37°C for 3 hrs, samples were acidified by addition of 5 μl 10% TFA, incubated at room temp for 5 minutes, centrifuged at 14,000 g for 10 min, and the supernatant used for targeted LC-MS analysis. Ten μl samples containing approximately 2.5 μg of plasma digest were injected onto a 1 mm × 8 mm peptide Opti-Trap cartridge (Optimize Technologies) at a 20 μl/minute flow rate in a mobile phase containing 0.1% formic acid (mobile phase A). After 5 min, the trap cartridge was placed in-line with a 0.5 mm × 250 mm column containing 5 μm Zorbax SB-C18 stationary phase (Agilent Technologies), and peptides separated by a 2–7.5 % gradient of mobile phase B (100% acetonitrile, 0.1% formic acid) for 1 min, 7.5–18.75% B for 30 min, 18.75–45% B for 30 min, 45–100% B for 4 minutes at a 10 μl/minute flow rate using an 1100 series capillary HPLC (Agilent Technologies). Peptides were analyzed using an LTQ Velos Pro linear ion trap (Thermo Fisher Scientific, Waltham, MA) by sequentially isolating the major charge states of the five albumin peptides listed above using an isolation window of 2 Da and collecting full MS/MS spectra of fragment ions in centroid mode using a normalized collision energy of 35. Sequest search results were used to create a spectral library for each targeted peptide. Quantification of peptides was then performed using version 2.5 of Skyline software30 by summing the integrated peaks of the 4–5 most intense fragment ions from each of the 5 peptides. Peaks for pep1 and pep2 were normalized using the summed peak intensities for unmodified albumin peptides 66–75, 397–413, and 599–609.
II.5. Statistical analysis
Biochemical data and performance scores at neuropsychological testing were compared across study groups using the Mann-Whitney test. Spearman correlation was used to explore relationships between plasma cyanide detoxification rates (CDR), plasma and urinary concentrations of SCN, blood urea nitrogen (BUN, an endogenous precursor of cyanate), serum albumin concentrations, motor and cognitive performance scores, and levels of pep1 and pep2. CDR were expressed as the number of milliseconds required to detoxify cyanide to yield one μmol of SCN per mg of protein [ms/(μmol/mg protein)] i.e. slower cyanide detoxifier had higher CDR. We also performed statistical analyses on the 40 subjects as a whole as previous studies have indicated that subclinical forms of the disease may exist and non-konzo children from konzo affected areas may perform worse than those from a non-konzo area at neuropsychological testing20,31. Logistic regression models assessed the relationships between konzo and levels of pep1 and/or pep2 before and after adjusting for age, gender, concentrations of serum albumin, and BUN. STATA software (updated version 11.2) was used for all analyses at the statistical significance set at p ≤ 0.05.
III. RESULTS
III.1. Genetic variations in TST and MPST in study subjects
A total of seven TST and four MPST single nucleotide polymorphisms (SNPs) were identified among those affected with konzo and/or their respective controls (Table 1). All TST SNPs were considered rare alleles in both the general and African populations based on statistics from the aggregate data bases gnome and topmed. Two of the variants, rs1049280 and rs974658299 were located in the 3’-UTR. Variants rs35156365, rs142340242, rs142340242, rs148893857, and rs375907649 were determined to be likely silent/benign changes based on SIFT/CONDEL predictions. One TST variant (rs61742280), causing a likely deleterious C>T mutation was identified that resulted in an amino acid change at p.Arg206Cys within exon 3. Four variants in a directly upstream gene MPST were also identified in individuals with and without konzo. Three of these variants were located in the 3’UTR or Intronic regions, likely causing minimal influence on the protein structure and function. However, a C>T variant resulting in a p.His317Tyr amino acid change of isoform1 was predicted to be possibly damaging, but this same variant when expressed in isoform 2 was determined to be benign. Possibly deleterious heterozygous TST variant rs61742280 was found in 2/21 children with konzo and 3/19 children with no konzo whereas possibly damaging heterozygous MPST rs1038542246 was found in only one child with konzo.
Table 1.
TST and MPST SNPs and nucleotide or amino acid changes
| Gene | Coordinate | SNP rs# | Reference Seq | Gene Location | Nucleotide Change | AA Change | Predicted Pathogenicity | General Allele Frequency | African Allele Frequency |
|---|---|---|---|---|---|---|---|---|---|
| TST | 37011023 | rs1049280 | NM_001270483 | 3’-UTR | G>A | N/A | N/A | 0.0205 | 0.0076 |
| TST | 37018427 | rs35156365 | NM_001270483 | Exon 2 | A>C | p.Glu102Asp | Benign | 0.0113 | 0.1104 |
| TST | 37011305 | rs61742280 | NM_001270483 | Exon 3 | C>T | p.Arg206Cys | Deleterious | 0.0009 | 0.0094 |
| TST | 37011129 | rs142340242 | NM_001270483 | Exon 3 | C>T | p.Cys264Cys | Benign | 0.0005 | 0.0054 |
| TST | 37011180 | rs148893857 | NM_001270483 | Exon 3 | G>A | p.Thr247Thr | Benign | 0.0001 | 0.0009 |
| TST | 37011290 | rs375907649 | NM_001270483 | Exon 3 | A>C | p.Met211Leu | Benign | 0.00009 | 0.0003 |
| TST | 37010819 | rs974658299 | NM_001270483 | 3’-UTR | A>G | N/A | N/A | 0.0002 | 0.0010 |
| MPST | 37024123 | rs112260704 | XM_011530196 | Intronic | C>G | N/A | N/A | 0.0316 | 0.1116 |
| MPST | 37029550 | rs59832951 | NM_001013436 | 3’-UTR | G>A | N/A | N/A | 0.0138 | 0.1387 |
| MPST | 37024849 | rs60296118 | XM_011530196 | Intronic | C>T | N/A | N/A | 0.0821 | 0.3922 |
| MPST | 37029509 | rs1038542246 | NM_001013436 | Exon 3 | C>T (Isoform 1) | p.His317Tyr | Possibly Damaging | 0.000003 | 0.00004 |
| MPST | 37029509 | rs1038542246 | NM_001013436 | Exon 3 | C>T (Isoform 2) | p.His297Tyr | Benign | 0.000003 | 0.00004 |
TST = Thiosulfate Sulfur Transferase; MPST = Mercapto Pyruvate Sulfur Transferase
III.2. In silico predictions of SNP effects on TST/MPST functions
To predict consequences of SNP TST rs61742280 or MPST rs1038542246 on protein function, we examined structures of the TST and MPST proteins using homology-modeled structures (Figure 1). In TST, Arg206 is partially exposed to the aqueous environment; however, the Arg206 guanidinium group appears to be engaged in hydrogen bonding with the adjacent side chain of Glu173 and, perhaps more importantly, the backbone carbonyl of Glu286. To better understand these interactions, we analyzed the change in stability due to the point-mutation using servers that predict the change in Gibbs free energy (ΔΔGpred) of folding due to a point-mutation. According to both STRUM and MAESTRO web servers, the Arg206Cys point-mutation is predicted to destabilize the protein with ΔΔGpred values of −0.34 (negative values are deemed unfavorable by STRUM) and 1.448 (positive values are deemed unfavorable by MAESTROweb), respectively. The MPST His317Tyr mutation was less conspicuous. His317 is the terminal residue of what is predicted to be an unordered coil, and of the entirety of the this MPST isoform. According to structural prediction, His317 does not engage in any important contacts with the protein, though we acknowledge that the slightly bulkier Tyr may affect the packing of the C-terminal polypeptide against the protein.
Figure 1. In silico predictions of SNP effects on TST/MPST functions.
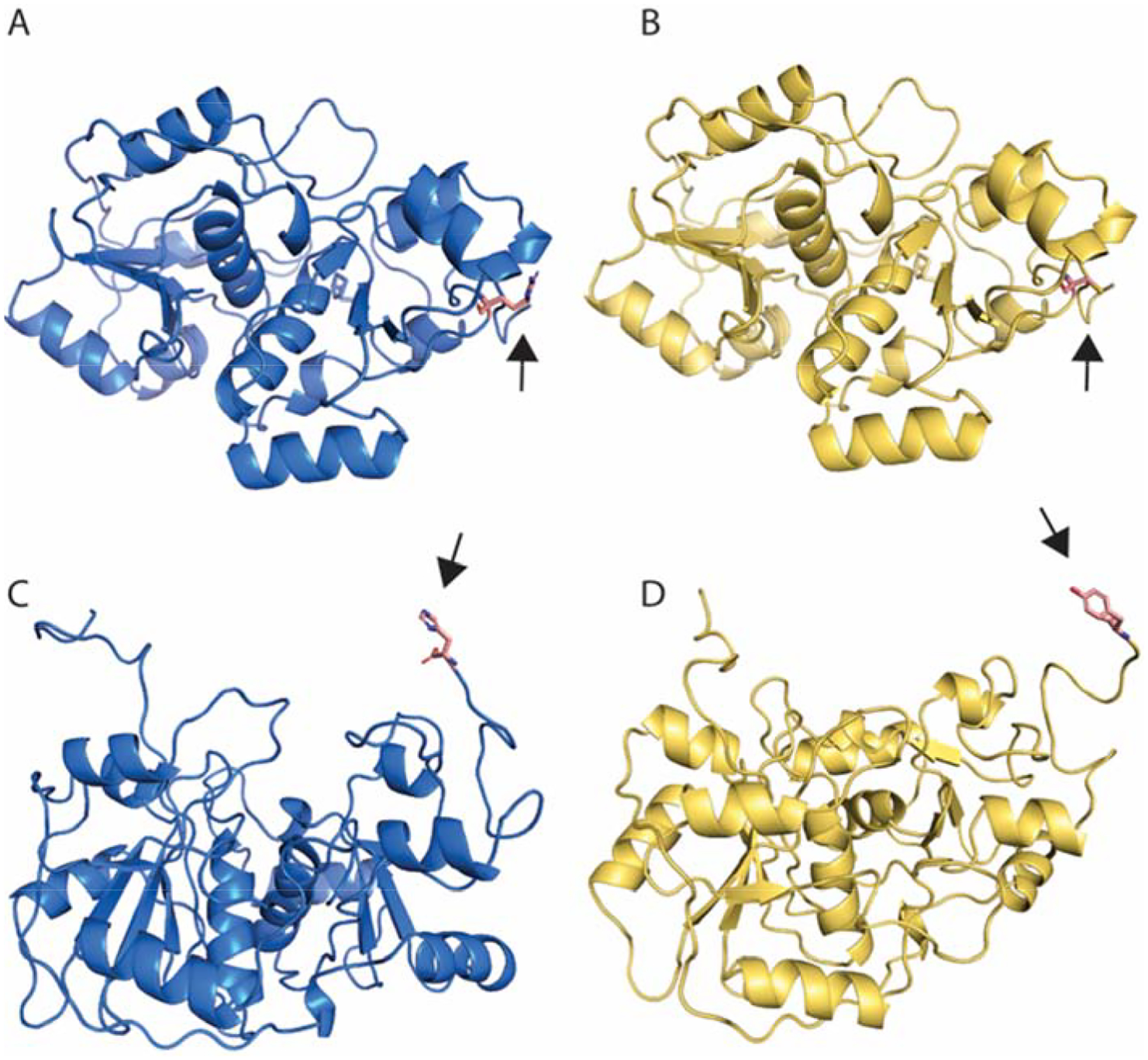
A) Predicted wild-type TST structure harboring an arginine amino acid at position 206. B) The predicted TST mutant, harboring a likely deleterious SNP resulting in a cysteine substitution at position 206. C) The predicted wild-type MPST protein containing a histidine at position 317 of the isoform 1 transcript. D) Predicted MPST mutant resulting in a tyrosine substitution at the terminal amino acid 317. According to both STRUM and MAESTRO web servers, the Arg206Cys point-mutation is predicted to destabilize the protein with ΔΔGpred values of −0.34 (negative values are deemed unfavorable by STRUM). According to structural prediction, His317 does not engage in any important contacts with the protein, though we acknowledge that the slightly bulkier Tyr may affect the packing of the C-terminal polypeptide against the protein.
III.3. Biomarkers of cyanogenic exposure, carbamoylation, and motor and cognitive performance
Overall, median (IQR) concentrations of plasma and urinary SCN were 197 (118 – 301) and 344 (172 – 688) μmol/l, respectively. Blood urea nitrogen (BUN) was within normal ranges for all subjects. In general, levels of pep1 or pep2 negatively correlated with concentrations of serum albumin (r = − 0.55, p = 0.01 or − 0.61, p < 0.01) for pep1 or pep2, respectively). A similar trend was seen for pep1 in children with konzo (p = − 0.47, p = 0.05). These children had lower levels of serum albumin and plasma SCN (PSCN), an indication of poor nutrition and sulfur-driven cyanide detoxification. They also performed worse at neuropsychological testing and had significantly higher pep1 206–219 (LDELRDEGKASSAK) and pep2 438–452 (KVPQVSTPTLVEVSR) compared to those with no konzo (Table 2 and Figures 2 and 3). In addition, their serum albumin concentrations positively correlate with performances scores at neuropsychological testing for sequential processing (r = 0.52, p = 0.05), learning (r = 0.52, p = 0.04), planning (r = 0.75, p = 0.01), delayed recognition (r = 0.56, p = 0.03), overall mental processing (r = 0.59, p = 0.02); or fine motor control (r = 0.73, p < 0.01), body coordination (r = 0.86, p < 0.01), and agility (r = 0.89, p < 0.01).
Table 2.
Median (IQR) exposure levels, biochemical characteristics, and performance scores at neuropsychological testing
| Domains | Konzo | Non-Konzo | *p |
|---|---|---|---|
| Plasma SCN in μmol/l | 162 (150 – 190) | 228 (192 – 272) | 0.05 |
| Urinary SCN in μmol/l | 344 (344 – 688) | 344 (258 – 516) | NS |
| Serum Albumin in g/dl | 3.2 (2.8 – 3.6) | 4.2 (3.7 – 4.6) | 0.00 |
| BUN in mmol/l | 1.8 (1.6 – 2.3) | 2.0 (1.9 – 2.6) | NS |
| CDR in ms/(μmol/mg protein) | 442.9 (390.2 – 555.9) | 403.4 (274.4 – 484.7) | NS |
| Fine Motor Control | 25 (21–32) | 31.5 (26–39) | 0.03 |
| Manual Coordination | 22 (20–25) | 36 (32–47.5) | 0.00 |
| Strength and Agility | 20 (20–21) | 40.5 (36.5–43.5) | 0.00 |
| Body Coordination | 20 (20–25) | 41.5 (33–48.5) | 0.00 |
| Overall Motor Proficiency | 20 (20–21) | 33.5 (31.5–41.5) | 0.00 |
| Sequential Processing | 63 (54–68) | 72.5 (66–89.5) | 0.01 |
| Simultaneous Processing | 55 (43–58) | 66 (52–72) | 0.03 |
| Learning | 64 (48–73) | 67 (64–76.5) | 0.18 |
| Planning | 51 (51–57) | 57 (51–57) | 0.30 |
| Overall Mental Processing Index | 52 (49–57) | 57.5 (54.5–68) | 0.01 |
SCN = Thiocyanate; BUN = Blood Urea Nitrogen; CDR = Cyanide Detoxification Rate. Higher CDR i.e. longer time to detoxify cyanide reflects slower (poor) cyanide detoxification. Higher score at neuropsychological testing domains reflects better proficiency.
*Mann-Whitney test
Figure 2.
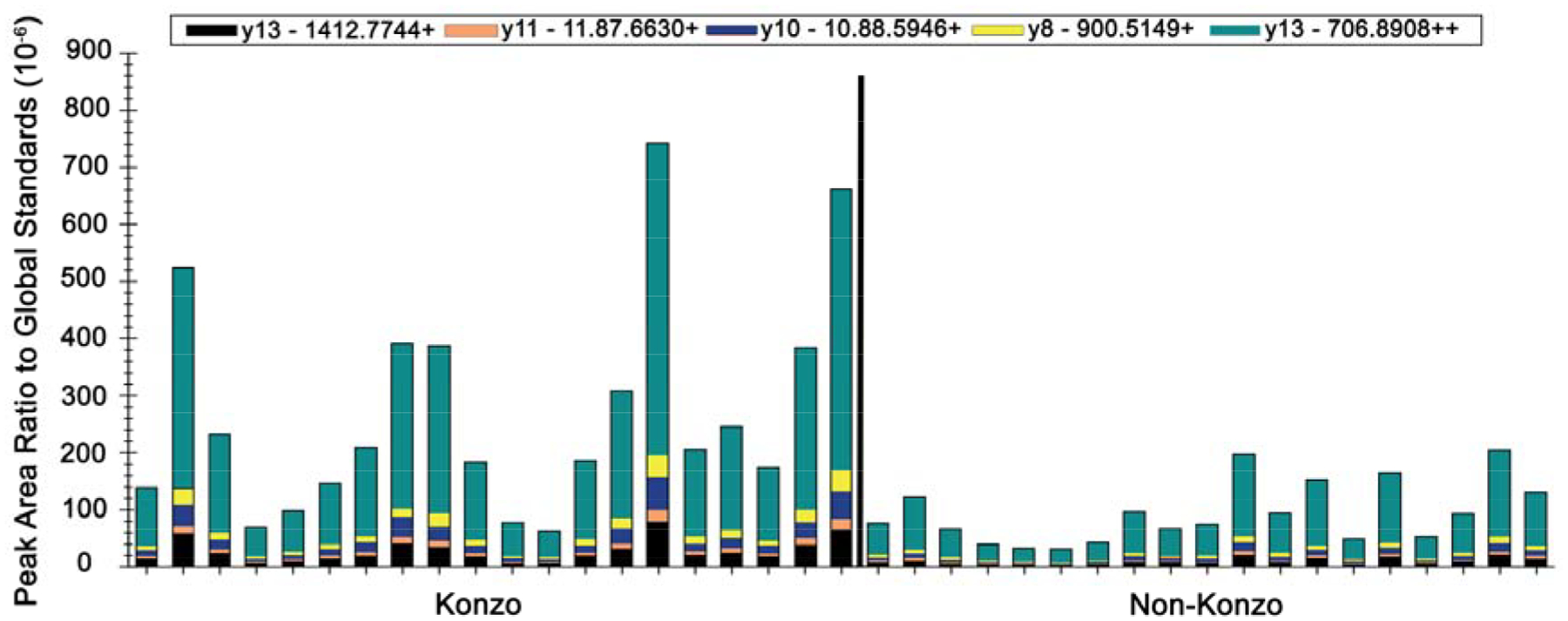
Relative abundance of pep1 206–219 (LDELRDEGKASSAK, pep1) peptide in children with konzo (N = 20) relative to those with no konzo (N = 19). Differences in abundance of fragment ions and pep 1 were noticeable (p < 0.01, Mann-Whitney test).
Figure 3.
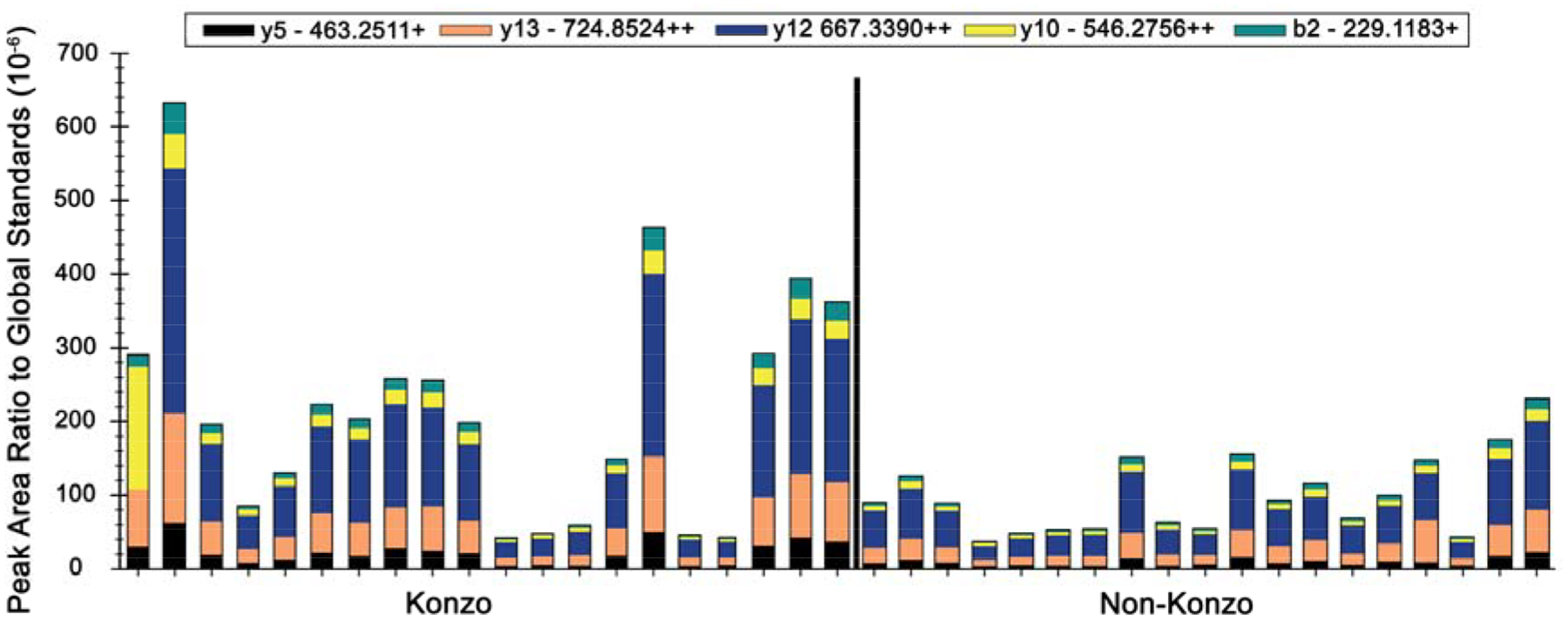
Relative abundance of pep2 438–452 (KVPQVSTPTLVEVSR peptide in children with konzo (N = 20) relative to those with no konzo (N = 19). Differences in abundance of fragment ions and pep2 were noticeable (p < 0.01, Mann-Whitney test).
Overall, a significant and positive correlation was found between levels of pep1 and pep2 (r = 0.71, p < 0.01). Levels of Pep1 negatively but positively correlated with PSCN (r = − 0.50, p = 0.02) and CDR (r = 0.45, p = 0.04), respectively. Correlations between pep1 and pep2 remained significant within study groups. PSCN remained negatively correlated with pep1 (r = − 0.67, p = 0.05) or pep2 (r = − 0.73, p = 0.02) in healthy children. Negative correlations were found between levels of pep1 and performance scores at testing for fine motor control (r = − 0.38, p = 0.02), manual coordination (r = − 0.47, p = 0.00), strength and agility (r = − 0.51, p = 0.00), body coordination (r = − 0.58, p = 0.00), and overall motor proficiency (r = − 0.61, p = 0.00); or sequential processing ( r = − 0.59, p = 0.00) and overall cognitive performance ( r = − 0.48, p = 0.00). The correlation between levels of pep1 and performance scores for fine motor control remain significant in children with overt paralysis (r = − 0.47, p = 0.04).
Logistic regression showed that konzo was significantly associated with higher levels of carbamoylated peptides pep1 or pep2 [(regression coefficient: 0.02 (95%CI: 0.01 – 0.03), p = 0.004 for pep1; regression coefficient: 0.01 (95%CI: 0.00 – 0.02), p = 0.01 for pep2)]. After adjusting for age, gender, levels of serum albumin and BUN, the associations remain significant for pep 1 [regression coefficient: 0.03 (95%CI:0.02 – 0.05), p = 0.01]. Neurocognitive performance and carbamoylation levels in the few children with possibly deleterious heterozygous TST rs61742280 or MPST rs1038542246 variants were unremarkably changed.
IV. DISCUSSION
We report for the first time higher albumin carbamoylation in children with the cassava-related paralytic disease known as konzo. Motor (fine motor control, manual and body coordination, along with strength and agility, and overall motor proficiency) and cognitive (sequential processing and overall mental processing index) deficits were associated with lower concentrations in serum albumin and higher carbamoylation after controlling for BUN, an endogenous precursor of cyanate. These particular findings indicate that the levels of carbamoylation seen in our study subjects may be driven by poor nutrition and an external source of isocyanic acid most likely associated with cyanogenic compounds from an almost exclusive dietary reliance on food from improperly processed cassava19,32. In addition, they suggest that serum PTM may serve as peripheral and surrogate markers of distant neurological insults and protein carbamoylation may have been the longtime missing link between cassava toxicity and neurocognition deficits seen in konzo18,33.
In contrast to plasma and/or urinary SCN, which mostly reflects acute exposure to cassava cyanogens, protein (serum) albumin carbamoylation appears to be a more attractive marker candidate for cassava related neurological diseases degeneration because of the aforementioned associations and the relatively extended half-life of carbamoylated products5.
In this small group of children, targeted exon sequencing has revealed a number of benign and two possibly damaging SNPs. While the TST Arg206Cys mutation may be sufficiently deleterious to prevent proper folding or to inactivate TST, it is also possible that the surface-exposed cysteine reacts (e.g. through carbamoylation) to cause protein modifications and, subsequently, prevent substrate-binding or catalysis. We have predicted that the His317Tyr mutation in MPST may not play key roles in protein structure and function, however additional biochemical analysis is needed. The identification of extremely rare SNPs in these two important enzymes does raise the possibility that these variants are likely be found in a homozygous state within the study population. If individuals harbor such mutations in a homozygous pattern, then it is likely that functionality of both TST and MPST could be affected, thereby reducing the efficiency of host cyanide detoxification pathways and increasing risks for neurotoxicity.
The overall burden of cassava related neurological disease needs to be determined accordingly and attention be placed on other cassava-related conditions, including tropical ataxic neuropathy, tropical diabetes, and a poorly documented cerebellar-parkinsonian-dementia syndrome (CPDS) reported many years ago in Nigeria. Whether cassava-related neurodegeneration is on a continuum of subtle and subclinical phenotypes that can evolved into overt phenotypes such as konzo, TAN, and CPDS needs to be determined. It is possible that rates of cyanide exposure, detoxification, protein carbamoylation, and genomic variations, contribute to the many possible phenotypes33,34.
Interpretation of our study findings requires some caution because of a relatively small sample size, lack of direct measurements of cyanate and/or isocyanic acid whether in plasma or brain tissues, and lack of a non-cassava dietarily reliant controls as community-wide exposure occurs in Kahemba21. Pathways that lead to higher carbamoylation and potential targets for treatment still need to be established. Apart from nutritional deficiencies and protein post-translational modifications that can negatively influence enzyme detoxifying capabilities, a search for deleterious SNPs using high-throughput sequencing will prove useful. Functional metagenomic analysis of the gut flora is of paramount importance to capture the likely potential role of the gut microbiome, since bacterial β-glycosidases are at the forefront of initial linamarin (main cassava cyanogen) detoxification. The toxicity potential of cassava crop varieties should also be addressed as risks for neurotoxicity may be modulated at the level of plant-microbiota-host interactions in constantly changing environments34. Studies on cassava-related neurotoxicity offers invaluable opportunities to provide insights into mechanisms of human disease across numerous disciplines, in particular those addressing issues of molecular ageing and neurodegeneration among millions who rely on cyanogenic cassava as the main source of food mostly under the tropics5,12,35.
Poor nutrition promotes carbamoylation and brain deficits under cassava toxicity
Slower sulfur-mediated cyanide detoxification in plasma promotes carbamoylation
Serum carbamoylation is a marker of brain deficits associated with cassava toxicity
ACKNOWLEDGMENTS
Supported by the NIH grant NIEHS/FIC R01ES019841. We are grateful to the Kahemba community and participating families for their support in this study. This paper is dedicated to co-author Jean-Pierre Banea Mayambu, a research pioneer on konzo who suddenly passed away during the revision process.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- 1.Delporte C, Zouaoui Boudjeltia K, Furtmuller PG, et al. Myeloperoxidase-catalyzed oxidation of cyanide to cyanate: A potential carbamylation route involved in the formation of atherosclerotic plaques? J Biol Chem 2018;293:6374–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mori D, Matsui I, Shimomura A, et al. Protein carbamylation exacerbates vascular calcification. Kidney Int 2018;94:72–90. [DOI] [PubMed] [Google Scholar]
- 3.Monhemi H, Tabaee SS. The effects of mutation and modification on the structure and stability of human lysozyme: A molecular link between carbamylation and atherosclerosis. J Mol Graph Model 2020;100:107703. [DOI] [PubMed] [Google Scholar]
- 4.Carracedo J, Ramirez-Carracedo R, Martinez de Toda I, et al. Protein Carbamylation: A Marker Reflecting Increased Age-Related Cell Oxidation. Int J Mol Sci 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorisse L, Pietrement C, Vuiblet V, et al. Protein carbamylation is a hallmark of aging. Proc Natl Acad Sci U S A 2016;113:1191–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delanghe S, Delanghe JR, Speeckaert R, Van Biesen W, Speeckaert MM. Mechanisms and consequences of carbamoylation. Nat Rev Nephrol 2017;13:580–93. [DOI] [PubMed] [Google Scholar]
- 7.Hawkins CL. Protein carbamylation: a key driver of vascular calcification during chronic kidney disease. Kidney Int 2018;94:12–4. [DOI] [PubMed] [Google Scholar]
- 8.Long J, Vela Parada X, Kalim S. Protein Carbamylation in Chronic Kidney Disease and Dialysis. Adv Clin Chem 2018;87:37–67. [DOI] [PubMed] [Google Scholar]
- 9.Holzer M, Zangger K, El-Gamal D, et al. Myeloperoxidase-derived chlorinating species induce protein carbamylation through decomposition of thiocyanate and urea: novel pathways generating dysfunctional high-density lipoprotein. Antioxid Redox Signal 2012;17:1043–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velasquez MT, Ramezani A, Raj DS. Urea and protein carbamylation in ESRD: surrogate markers or partners in crime? Kidney Int 2015;87:1092–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Nicholls SJ, Rodriguez ER, et al. Protein carbamylation links inflammation, smoking, uremia and atherogenesis. Nat Med 2007;13:1176–84. [DOI] [PubMed] [Google Scholar]
- 12.Jaisson S, Pietrement C, Gillery P. Protein Carbamylation: Chemistry, Pathophysiological Involvement, and Biomarkers. Adv Clin Chem 2018;84:1–38. [DOI] [PubMed] [Google Scholar]
- 13.Guru KrishnaKumar V, Baweja L, Ralhan K, Gupta S. Carbamylation promotes amyloidogenesis and induces structural changes in Tau-core hexapeptide fibrils. Biochim Biophys Acta Gen Subj 2018;1862:2590–604. [DOI] [PubMed] [Google Scholar]
- 14.Kimani S, Moterroso V, Lasarev M, et al. Carbamoylation correlates of cyanate neuropathy and cyanide poisoning: relevance to the biomarkers of cassava cyanogenesis and motor system toxicity. Springerplus 2013;2:647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crist RD, Grisolia S, Bettis CJ, Grisolia J. Carbamoylation of proteins following administration to rats of carbamoyl phosphate and cyanate and effects on memory. European journal of biochemistry / FEBS 1973;32:109–16. [DOI] [PubMed] [Google Scholar]
- 16.Tellez I, Johnson D, Nagel RL, Cerami A. Neurotoxicity of sodium cyanate. New pathological and ultrastructural observations in Maccaca nemestrina. Acta neuropathologica 1979;47:75–9. [DOI] [PubMed] [Google Scholar]
- 17.Kassa RM, Kasensa NL, Monterroso VH, et al. On the biomarkers and mechanisms of konzo, a distinct upper motor neuron disease associated with food (cassava) cyanogenic exposure. Food and chemical toxicology : an international journal published for the British Industrial Biological Research Association 2011;49:571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tshala-Katumbay D, Mumba N, Okitundu L, et al. Cassava food toxins, konzo disease, and neurodegeneration in sub-Sahara Africans. Neurology 2013;80:949–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tor-Agbidye J, Palmer VS, Lasarev MR, et al. Bioactivation of cyanide to cyanate in sulfur amino acid deficiency: relevance to neurological disease in humans subsisting on cassava. Toxicological sciences : an official journal of the Society of Toxicology 1999;50:228–35. [DOI] [PubMed] [Google Scholar]
- 20.Boivin MJ, Okitundu D, Makila-Mabe Bumoko G, et al. Neuropsychological effects of konzo: a neuromotor disease associated with poorly processed cassava. Pediatrics 2013;131:e1231–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okitundu Luwa EAD, Bumoko Makila-Mabe G, Ayanne MT, et al. [Persistence of konzo epidemics in Kahemba, Democratic Republic of Congo: phenomenological and socio-economic aspects]. The Pan African medical journal 2014;18:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boivin MJ, Okitundu D, Makila-Mabe B, et al. Cognitive and motor performance in Congolese children with konzo during 4 years of follow-up: a longitudinal analysis. Lancet Glob Health 2017;5:e936–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.WHO. Konzo: a distinct type of upper motor neuron disease. Weekly Epidemiological Record 1996;71:225–32.8718826 [Google Scholar]
- 24.Kambale KJ, Ali ER, Sadiki NH, et al. Lower sulfurtransferase detoxification rates of cyanide in konzo-A tropical spastic paralysis linked to cassava cyanogenic poisoning. Neurotoxicology 2017;59:256–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 2018;46:W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bienert S, Waterhouse A, de Beer TA, et al. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res 2017;45:D313–D9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011;27:343–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 2009;30 Suppl 1:S162–73. [DOI] [PubMed] [Google Scholar]
- 29.Wilmarth PA, Riviere MA, David LL. Techniques for accurate protein identification in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. Journal of ocular biology, diseases, and informatics 2009;2:223–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacLean B, Tomazela DM, Shulman N, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010;26:966–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tshala-Katumbay D, Eeg-Olofsson KE, Tylleskar T, Kazadi-Kayembe T. Impairments, disabilities and handicap pattern in konzo--a non-progressive spastic para/tetraparesis of acute onset. Disabil Rehabil 2001;23:731–6. [DOI] [PubMed] [Google Scholar]
- 32.Tylleskar T, Banea M, Bikangi N, Nahimana G, Persson LA, Rosling H. Dietary determinants of a non-progressive spastic paraparesis (Konzo): a case-referent study in a high incidence area of Zaire. Int J Epidemiol 1995;24:949–56. [DOI] [PubMed] [Google Scholar]
- 33.Simsek B, Cakatay U. Could cyanogenic glycoside rich diet cause increased risk for carbamylation-induced protein damage in individuals with chronic inflammatory diseases? Med Hypotheses 2019;130:109275. [DOI] [PubMed] [Google Scholar]
- 34.Tshala-Katumbay D, Mwanza JC, Rohlman DS, Maestre G, Oria RB. A global perspective on the influence of environmental exposures on the nervous system. Nature 2015;527:S187–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorisse L, Jaisson S, Pietrement C, Gillery P. [Protein carbamylation: when protein and chronologic ageings meet]. Med Sci (Paris) 2016;32:684–6. [DOI] [PubMed] [Google Scholar]


