Abstract
Three-dimensional (3D) bioprinting of patient-specific auricular cartilage constructs would aid in the reconstruction process of traumatically injured or congenitally deformed ear cartilage. To achieve this, a hydrogel-based bioink is required that recapitulates the complex cartilage microenvironment. Tissue-derived decellularized extracellular matrix (dECM)-based hydrogels have been used as bioinks for cell-based 3D bioprinting because they contain tissue-specific ECM components that play a vital role in cell adhesion, growth, and differentiation. In this study, porcine auricular cartilage tissues were isolated and decellularized, and the decellularized cartilage tissues were characterized by histology, biochemical assay, and proteomics. This cartilage-derived dECM (cdECM) was subsequently processed into a photo-crosslinkable hydrogel using methacrylation (cdECMMA) and mixed with chondrocytes to create a printable bioink. The rheological properties, printability, and in vitro biological properties of the cdECMMA bioink were examined. The results showed cdCEM was obtained with complete removal of cellular components while preserving major ECM proteins. After methacrylation, the cdECMMA bioinks were printed in anatomical ear shape and exhibited adequate mechanical properties and structural integrity. Specifically, auricular chondrocytes in the printed cdECMMA hydrogel constructs maintained their viability and proliferation capacity and eventually produced cartilage ECM components, including collagen and glycosaminoglycans (GAGs). The potential of cell-based bioprinting using this cartilage-specific dECMMA bioink is demonstrated as an alternative option for auricular cartilage reconstruction.
Keywords: Bioink, bioprinting, decellularization, extracellular matrix, methacrylation, cartilage tissue engineering
1. Introduction
Elastic cartilage is a unique type of cartilage tissue found exclusively in the head and neck area. Due to the abundance of elastic fibers, this cartilage is highly flexible and functionally and morphologically distinct from hyaline and fibrocartilage [1]. Like other cartilage tissues, elastic cartilage is avascular and has no self-regenerative capacity. Therefore, surgical reconstructive procedures are required to restore shape and function of the affected anatomical part when the cartilage tissue is damaged (i.e. in burns) or congenitally deformed (i.e. in microtia). Current reconstruction techniques utilize autologous tissue grafts such as rib cartilage [2, 3] or synthetic implants made from nonabsorbable polymers (i.e. MedPor®) [4–6]. Autologous reconstruction using rib cartilage has a high donor site morbidity [7] and requires a complex surgical procedure. Synthetic implants, on the other hand, have higher infection rates and complications and can eventually protrude [8].
Tissue engineering has made significant advances as an alternative option in cartilage reconstruction. A first-in-human trial has been performed for the reconstruction of nasal [9] and ear [10] cartilage, and several tissue-engineered cartilage tissues are currently being investigated in clinical trials [11]. Nevertheless, the bioengineering of auricular cartilage tissues remains extremely challenging. Firstly, creating a complex shape such as the auricle or nostril has proven difficult [12, 13]. Secondly, keeping this shape both in vitro and in vivo provides a major challenge [14–17]. Because of these limitations, several strategies have been examined. Recently, cell-based 3D bioprinting strategies have enabled bioengineering of clinically relevant tissue constructs by imitating the tissue complexity [18–20]. Additionally, extracellular matrix (ECM)-derived bioinks have been utilized to accelerate the tissue maturation and formation in the bioprinted tissue constructs [21, 22]. These ECM-derived bioinks may retain tissue-specific structural and functional molecules that regulate cell behavior and tissue homeostasis [21, 23, 24].
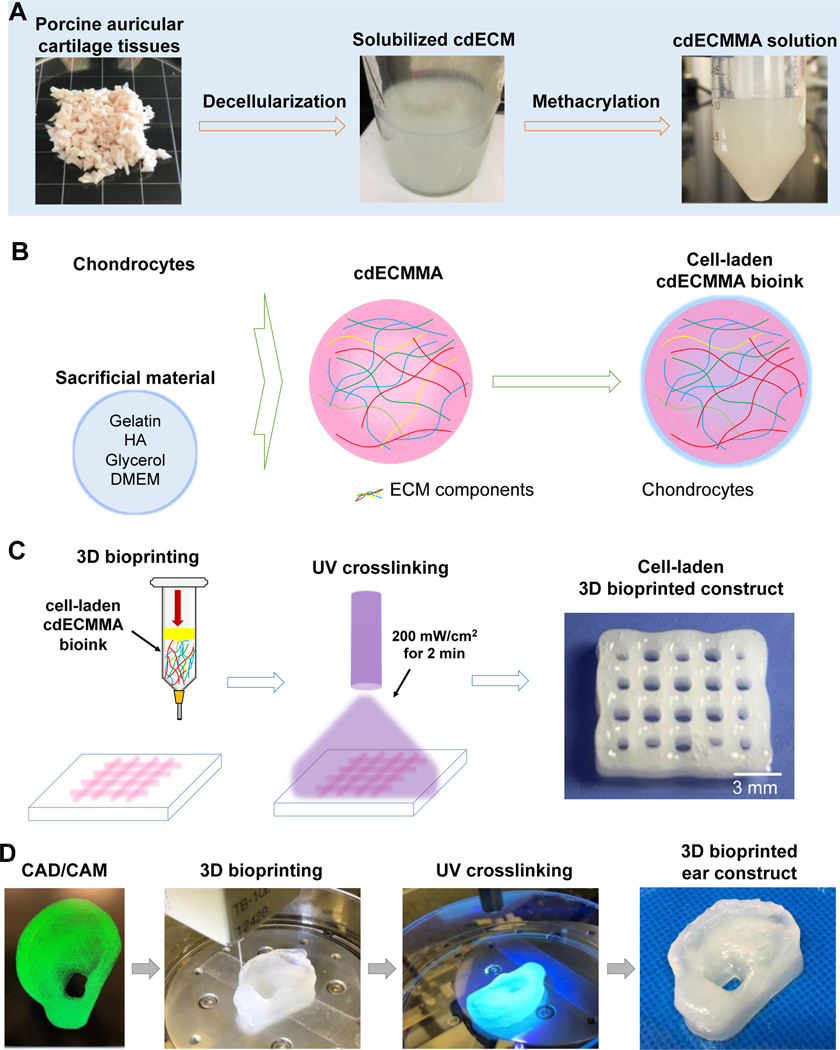
In this study, we hypothesize that cartilage-derived ECM components could provide cartilage-specific structural and biochemical signals to promote cellular activities and function, as well as tissue maturation and formation. We aim to develop a photo-crosslinkable cartilage-derived hydrogel bioink for bioprinting of patient-specific cartilage constructs. This chemical modification of cartilage-derived decellularized ECM (cdECM) by methacrylate reaction offers the rapid structural integrity of cdECM-based constructs after the printing process [21, 22, 24]. We also analyze the composition of decellularized elastic cartilage tissue using a proteomics approach. The process for formulation of cdECM methacrylate (cdECMMA) hydrogel, including tissue preparation, decellularization, solubilization, and methacrylation is shown in Figure 1.
Figure 1.
Schematic diagram of the auricular cartilage-derived ECM (cdECM) bioink development. (A) Methacrylation of cdECM obtained by decellularization and solubilization process. (B) Illustration of cdECMMA bioink formulation containing cells. (C) 3D bioprinting process using cell-laden cdECMMA bioink. (D) 3D bioprinting workflow from 3D CAD/CAM model to bioprinted ear construct for personalized auricular reconstruction.
2. Materials and methods
2.1. Decellularization of auricular cartilage tissue
Ear cartilage was isolated from Yorkshire pigs (female, 10–15 months old) according to the guidelines of the Wake Forest University Institutional Animal Care and Use Committee (IACUC). Under the aseptic condition, the skin was carefully removed leaving remnants of the perichondrium intact, and the remaining ear cartilage tissues were cut into small pieces (3–8 mm2). The obtained tissue pieces were rinsed using deionized water and subjected to three freezing-thawing cycles (−80 to 37°C). Following the freezing-thawing process, the cartilage tissues were agitated in 1% Triton X-100 solution containing a protease Inhibitor (Pierce Protease Inhibitor Mini Tablet, Thermo Fisher Scientific, Waltham, MA) for 24 h at 4°C, followed by three washes in phosphate-buffered solution (PBS). Subsequently, the sample was transferred to a 50 ml tube containing Hanks Buffered Salt Solution (HBSS) supplemented with 200 U/ml DNase and continuously agitated for 12 h at room temperature. Finally, the sample was washed 6 times in PBS, frozen at −80°C, lyophilized for 3 days, and cryomilled (SPEX Certiprep Cryogenic Mill 6870, Metuchen, NJ) into a powder form. The prepared decellularized cartilage tissue powder was stored at −20°C until the use.
2.2. Characterizations of decellularized auricular cartilage tissue
To assess the cellularity of cartilage tissue after decellularization, double-stranded DNA (dsDNA) content was measured using the Quant-iT™ Picogreen® dsDNA assay (Life Technologies, Carlsbad, CA) according to the manufacturer’s instruction. Briefly, 10 mg/ml of tissue sample was dissolved in the TE buffer solution (pH 7.5, 10 mM Tris-HCL and 1 mM EDTA), followed by the addition of Quant-iT™ Picogreen® reagent. The solution was incubated in triplicate in a 96-well plate for 5 min at room temperature. Subsequently, the absorbance was read at 520 nm using a CytoFluor microplate reader (MTX Lab Systems Inc., Vienna, VA). The supplied Lambda DNA standard was serially diluted to provide a standard curve ranging from 0 to 1 μg/ml.
Collagenous fiber bundles in both native and decellularized cartilage tissues were observed by second-harmonic generation and two-photon excited autofluorescence (SHG/2PF) microscopy, as described previously [11]. Briefly, the samples were placed on an inverted petri dish and covered with a glass slide to generate a flat interface. A commercial two-photon laser-scanning microscope (TrimScope I, Lavision BioTec, Bielefeld, Germany) and a femtosecond Ti-sapphire laser source (Chameleon Ultra II, Coherent, Palo Alto, CA) were used for SHG/2PF imaging. Data acquisition was performed with TriMScope I software (Impector Pro, Lavision BioTec). Image stacks were stored in 16-bit tiff-format and processed in Image J (NIH, Bethesda, MD).
For histological analysis, native and decellularized cartilage tissues were fixed with 10% neutral buffered formalin (NBF) for 3 days at room temperature. The samples were then paraffin-embedded and sectioned a thickness of 5-μm using a microtome. The sections were deparaffinized and stained with hematoxylin and eosin (H&E) for general morphology, Masson’s trichrome (MTS) for collagen, and Alcian Blue (pH 1.0) for sulfated glycosaminoglycans (GAGs), respectively. The stained samples were imaged by a brightfield/fluorescent microscope (Leica Microsystems, Inc., Buffalo Grove, IL).
For quantification of ECM components in native and decellularized cartilage tissues, collagen and GAGs were analyzed. Briefly, collagen was extracted by 0.5M acetic acid/pepsin at 4°C overnight. The solubilized collagen contents were quantified using the Sircol Soluble Collagen Assay (Biocolor Ltd., Carrickfergus, Northern Ireland). The collagen content of each sample was measured using the spectrophotometer at 555 nm. Sulfated GAGs were extracted by pepsin digestion buffer for 3 h at 65°C. The extracted GAG contents were quantified using the Blyscan sGAG Assay (Biocolor Ltd.). The GAG content of each sample was measured using the spectrophotometer at 656 nm.
2.3. Nano LC-MS/MS for proteomic analysis
To prepare the cartilage samples for proteomic analysis, both native and decellularized cartilage powders were dissolved in NuPAGE™ lithium dodecyl sulfate (LDS) buffer (Invitrogen, Carlsbad, CA) containing 10 mM dithiothreitol (DTT) in 50 mM ammonium bicarbonate at 10 mg/ml, vortexed three times, and heated to 70°C for 4 h. The solution was then sonicated three times and centrifuged to remove the remaining insoluble components. The supernatant was stored at −20°C until the use. The following day, after thawing, samples were heated to 99°C for 3 min. Twenty-five μl of each sample was applied to a freshly prepared 12.5% acrylamide SDS-PAGE gel and run for 15 min at 80V. Next, gels were washed with deionized water and fixed in 50% EtOH/1% phosphoric acid for 10 min and stained overnight with 1% Coomassie brilliant blue R250 (Thermo Fisher Scientific) in 40% MeOH/1% phosphoric acid containing 1.5 M ammonium sulfate. Finally, the gels were destained with deionized water and scanned using a digital scanner (Hewlett-Packard, Palo Alto, CA). In-gel digestion and LC-MS/MS were performed as described previously [25]. All chemicals were obtained from Millipore Sigma (St Louis, MO) unless stated otherwise.
2.4. Protein identification
MS/MS spectra were searched against a Uniprot Sus scrofa (porcine) reference proteome FASTA file (last modified March 2018, 40,710 entries) using MaxQuant 1.5.2.8 software (Max Planck Institute of Biochemistry, Planegg, Germany). Enzyme specificity was set to trypsin and up to two missed cleavages were allowed. Cysteine carbamidomethylation was treated as a fixed modification, and methionine oxidation and N-terminal acetylation as variable modifications. Peptide precursor ions were searched with a maximum mass deviation of 4.5 ppm and fragment ions with a maximum mass deviation of 20 ppm. Peptide and protein identifications were filtered at an FDR of 1% using the decoy database strategy. The minimal peptide length was 7 amino acids, the minimum Andromeda score for modified peptides was 40, and the minimum delta score was 6. Proteins that could not be differentiated based on MS/MS spectra alone were grouped into protein groups (default MaxQuant setting). Searches were performed with the label-free quantification option selected. Spectral counts, i.e. the number of identified MS/MS spectra for a given protein, were used as a proxy for protein abundance [26]. Raw counts were normalized based on the sum of spectral counts for all identified proteins in a particular sample, relative to the average sample sum determined with all samples. To find statistically significant differences in normalized counts between groups, we applied the beta-binomial test, which considers within-sample and between-sample variation using an alpha level of 0.05 [27]. For hierarchical clustering analysis, protein abundance data were normalized to zero mean and unit variance, and Euclidian distance and Ward linkage were used for clustering. All analyses were performed using dedicated R scripts.
Gene symbols for identified proteins were uploaded to the web-based STRING tool (version 10.0, http://string.embl.de, String Consortium 2020) to retrieve protein-protein association data [28]. The data were imported, visualized, and annotated in Cytoscape version 3.6.1 [29]. Gene ontology analysis was performed using the BiNGO app for Cytoscape to retrieve overrepresented biological process terms (corrected p-value <0.05) [30].
2.5. Methacrylation of cartilage-derived dECM (cdECMMA)
Cartilage-derived dECM methacrylate (cdECMMA) was prepared following the decellularization process. Briefly, 10 mg/ml of the pulverized decellularized cartilage tissue was enzymatically digested in a solution of 1 mg/ml porcine pepsin in 5 M acetic acid for 48 h at room temperature under continuous stirring. To precipitate the dissolved ECM components, 20% w/v sodium chloride (NaCl) was added to the solution and centrifuged at 3000 rpm for 15 min at 4°C. The resulting cartilage-derived ECM precipitate (cdECM) was dialyzed for 2 days against deionized water at 4°C using 3,500 MWCO dialysis tubing to completely remove all low molecular weight chemicals with changes in deionized water twice daily. For methacrylate, 300 mg of cdECM was dissolved in 80 mL of 5 M acetic acid, vortexed until dissolved, and pH adjusted to 8–9 using NaOH. Then, 0.6 mL methacrylic anhydride (MAA) was added dropwise under vigorous stirring at 4°C for 2 days. A 4-fold of acetone was added to the cdECM-MA solution and centrifuged at 3000 rpm for 15 min at 4°C. After disposal of the supernatant, the precipitated cdECM-MA was dialyzed for 7 days against deionized water at 4°C using 3,500 MWCO dialysis tubing (Spectrum Laboratories, Inc., Los Angeles, CA) with changes in deionized water twice daily. After Lyophilization, the product was stored at −20°C until the use. Gelatin methacrylate (GelMA) as a control bioink was prepared according to the previous protocol [21].
The degree of methacrylate was confirmed using a 2,4,6-trinitrobenzene sulfonic acid (TNBS) assay. The TNBS reagent can be quantified by reacting with the free lysine amines to form a chromogenic TNP derivative with absorbance at 346 nm. Briefly, lyophilized GelMA and cdECMMA were dissolved in a 0.1 M sodium bicarbonate buffer (pH 8.5) at a density of 0.2 mg/ml, and 250 μl of TNBS reagent was added. After incubation for 2 h, the reaction was stopped by adding 125 ml of 1M hydrochloric acid (HCl) and 250 μl of 10% SDS. The optical density (OD) of the solution was measured using a spectrophotometer (SpectraMax M5 microplate reader; Molecular Devices, San Jose) at 346 nm. The degree of methacrylation was calculated using the following equation (1).
| (1) |
The cdECMMA hydrogels after photo-crosslinking were carefully blotted to remove excess surface liquid, and the total swelled weight was measured (Ws). The samples were then fully dried using a freeze-dryer overnight, and total dry weight was measured (Wd). Swelling ratios were calculated using the following equation (2).
| (2) |
The gel stiffness of the cdECMMA hydrogels after photo-crosslinking was analyzed using a Discovery Hybrid Rheometer DHR-2 (TA Instruments, New Castle, DE). The cdECMMA hydrogels were cast and crosslinked using a mold to obtain discs of 1 cm in diameter and 2 mm in height. The gel stiffness was measured using a parallel plate contact mode at 18°C using 8-mm geometry.
2.6. Preparation of cdECMMA-based bioinks
For the cdECMMA-based bioink, a solution containing 37.5 mg/ml gelatin (type A) with 3 mg/ml hyaluronic acid (HA) in phenol-red free DMEM was prepared at 37°C for 1 h according to our previous studies [18, 21]. Then, the lyophilized cdECMMA (20, 30, and 40 mg/mL) was added to the solution together with 10% v/v glycerol and a photoinitiator (0.1% v/v 2-hydroxy-4’-(2-hydroxyethoxy)-2-methylpropriophenone) (Irgacure 2959, CIBA Chemicals, Tarrytown, NY). The cdECMMA-based formulations were vigorously mixed on ice using two 3 mL syringes connected to a 3-way stopcock. As a control, a GelMA-based formulation was prepared by the procedure described above. Photocrosslinking of these bioink formulations was induced by exposure to UV light (200 mW/cm2) for 2 min.
2.7. Rabbit chondrocyte culture and expansion
Auricular chondrocytes were isolated from ear cartilage biopsies from rabbits (New Zealand White, Charles River Laboratories, Wilmington, MA) according to our previous work [31]. All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at Wake Forest University. Auricular cartilage was dissected from the external ear after removing the perichondrium. The harvested cartilage was cut to 1-mm3 pieces and treated with 0.2% type II collagenase (Thermo Fisher Scientific) under gentle agitation at 37°C for 3 h. The digested tissue was then filtered using a 100-μm filter to collect cells, and the collected cells were pelleted by centrifugation at 1000 rpm for 5 min and washed three times with sterile phosphate-buffered saline (PBS). The resulting cells were cultured and expanded in Dulbecco’s Modified Eagle Medium/F12 (DMEM/F12) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotic/antimycotic at 37°C, 5% CO2. The cells were expanded up to passage 4 for further experiments.
2.8. Bioprinting of cell-laden constructs using cdECMMA-based bioinks
The cdECMMA-based bioinks with different concentrations of cdECMMA (20, 30, and 40 mg/mL) were prepared as described above. A bioink formulation containing GelMA (30 mg/mL) instead of cdECMMA was used as a control. The prepared bioink formulations were mixed with chondrocytes (20 ×106 cells/mL), and the cell-laden bioinks were loaded into a sterile plastic syringe at 37°C and then cooled on ice for 10 min. To fabricate 3D bioprinted constructs, our integrated tissue-organ printing (ITOP) system was used, which contains a X, Y, Z-axis stage/controller and multiple dispensing modules. The cell-laden bioink was printed at 45–50 kPa pressure and 180 mm/min. Finally, the bioprinted cell-laden cdECMMA constructs were photo-crosslinked by UV light (200 mW/cm2) for 2 min. The bioprinted cell-laden GelMA constructs were also fabricated for the comparison.
2.9. In vitro cell viability and proliferation
LIVE/DEAD™ viability/cytotoxicity kit (Thermo Fisher Scientific) was used to assess the cell viability in the bioprinted constructs. At 1, 3, and 7 days in culture, the constructs were washed in PBS, followed by incubation with 1 μl/ml Cal-AM and 3 μl/ml Eth-D in PBS at 37°C for 30 min. The cell-laden constructs were imaged using a confocal microscope (TCS LSI Macro Confocal; Leica Microsystems, Inc.). The quantification for % cell viability was performed using ImageJ software.
Cell proliferation was analyzed at 1, 3, and 7 days in culture using the AlamarBlue™ Assay kit (Thermo Fisher Scientific). Briefly, the bioprinted cell-laden constructs were washed three times with DPBS and incubated in a fresh medium containing 10% v/v AlamarBlue™ dye solution at 37°C. After 4 h incubation, 200 μL of the medium was collected and measured the fluorescence with excitation at 560 nm and emission at 590 nm using a spectrophotometer.
2.10. In vitro cartilage tissue formation in the bioprinted cdECMMA constructs
The bioprinted cdECMMA constructs (10 × 10 × 3 mm3) containing primary chondrocytes were cultured in DMEM/F12 supplemented with 10% FBS and 1% antibiotic/antimycotic. After 4 weeks in culture, the constructs were fixed in 10% NBF overnight, dehydrated, embedded in paraffin, and sectioned to 5-μm thickness for the histological analyses. The sections were rehydrated and stained with H&E, Safranin O/Fast Green, and Alcian Blue (pH 1.0)/Sirius Red, which confirmed the general morphology and GAG production.
For the quantification of collagen and GAG assays, the collagen in the bioprinted constructs was extracted by 0.5M acetic acid/pepsin at 4°C overnight. The solubilized collagen was quantified using the Sircol Soluble Collagen Assay and measured using the spectrophotometer at 555 nm. Sulfated GAGs in the bioprinted constructs were extracted by pepsin digestion buffer at 65°C for 3 h and quantified using the Blyscan sGAG Assay. The GAG content of each sample was measured using the spectrophotometer at 656 nm.
2.11. Statistical analysis
Data were analyzed with Student’s t-test or one-way analysis of variance (ANOVA) using Graphpad Prism software (GraphPad Software, Inc., La Jolla, CA). Variables are expressed as a mean ± standard deviation (SD), and p < 0.05 was considered statistically significant.
3. Results
3.1. Characterization of decellularized auricular cartilage tissue
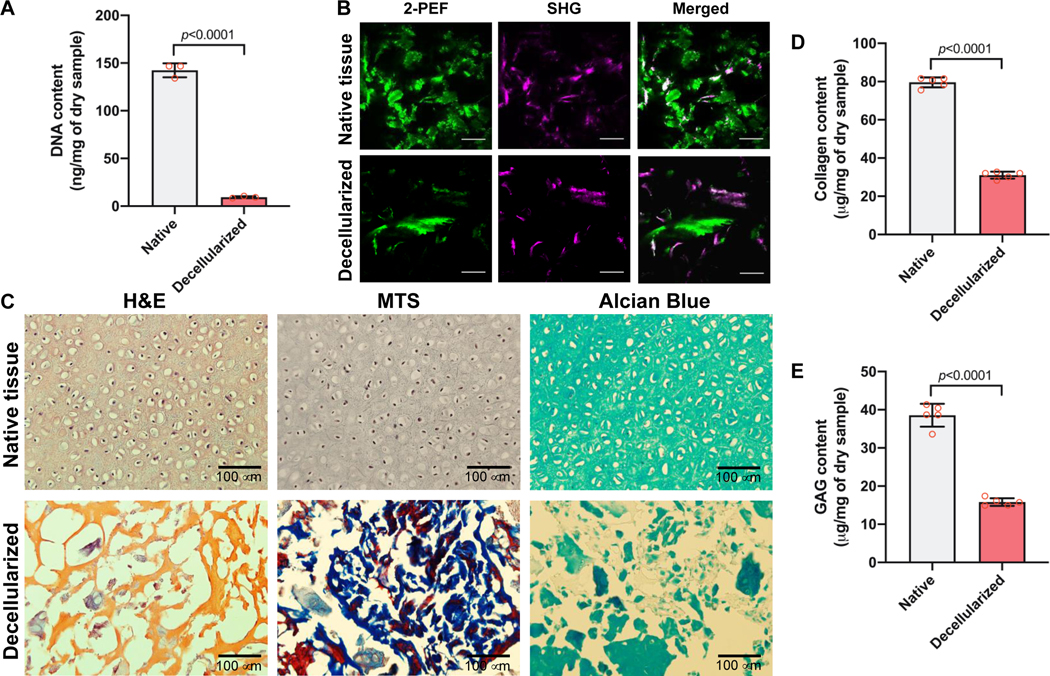
The porcine ear cartilage tissue was decellularized by the freezing-thawing method followed by Triton X-100 and DNase treatment. After decellularization, DNA content was quantified to validate the cellular components in the decellularized cartilage tissue. The cellular components of the decellularized cartilage tissue revealed a significant decrease compared with the native cartilage, as measured by the dsDNA content (9.4 ± 0.8 ng/mg for decellularized and 142.5 ± 6.0 ng/mg for native cartilage tissue, p<0.001; Fig. 2A).
Figure 2.
Characterizations of cdECM. (A) DNA contents before and after decellularization of cartilage tissue (n=3). Data are represented mean ± SD. The p-value by Student t-test is indicated. (B) Second-harmonic generation and two-photon autofluorescence (SHG/2-PEF) images of collagenous fiber bundles of native and decellularized auricular cartilage tissues. Scale = 100 μm. (C) Histological analyses of native and decellularized auricular cartilage tissues by H&E, MTE, and Alcian Blue staining. (D) Collagen contents (n=5) and (E) GAG contents (n=5) of native and decellularized auricular cartilage tissues. Data are represented mean ± SD. The p-value by Student t-test is indicated.
To confirm the ECM preservation after decellularization, the collagen bundles in both native and decellularized cartilage tissues were observed by SHG/2PF imaging. The SHG signal revealed no difference between native and decellularized cartilage tissues (Fig. 2B). Histologically, the cellular components were completely removed by the decellularization process, as confirmed by H&E staining. Masson’s trichrome staining indicated the collagenous matrix in the decellularized cartilage tissue, and the glycosaminoglycans (GAGs) presented in the tissues were detected by Alcian Blue staining (Fig. 2C). In addition, collagen and GAGs were detected in the native and decellularized cartilage tissues (Fig. 2D,E). However, the contents of both collagen and GAGs after decellularization was significantly lower than those of the native tissue.
3.2. Proteomic analysis of decellularized cartilage tissue
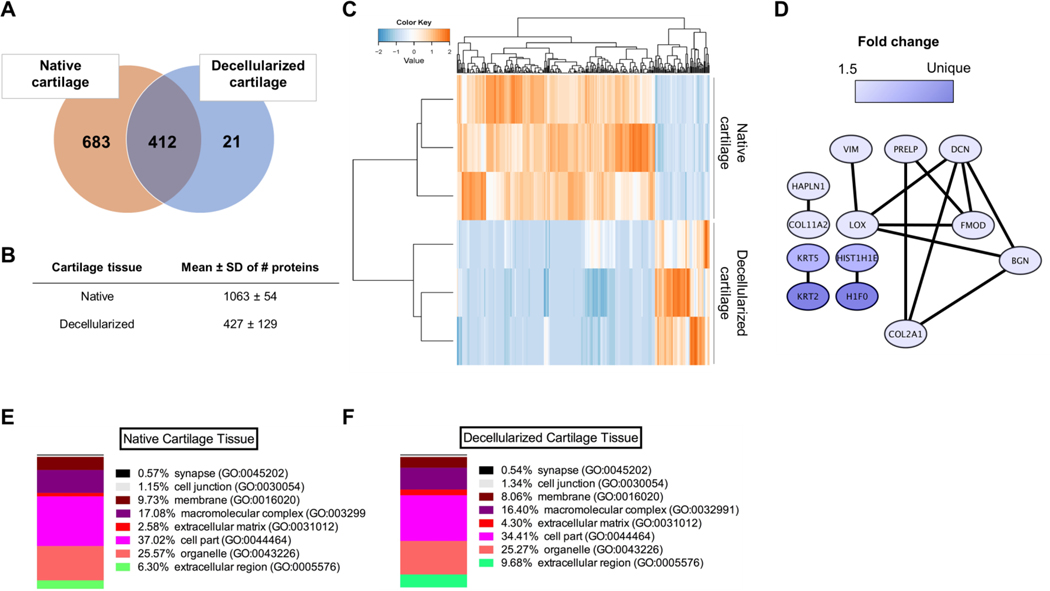
The tissue samples were processed for proteomics involving protein gel prefractionation, followed by combined fractionation and analysis on a nano LC-MS/MS platform, including an orbitrap mass spectrometer. The analysis revealed 683 unique proteins found solely in native cartilage, 21 unique proteins found solely in decellularized cartilage, and 412 proteins found in both native and decellularized tissues (Fig. 3A). The total number of proteins identified in both tissues was 1063 ± 54 for native and 427 ± 129 for decellularized cartilage tissues (Fig. 3B, n=3). Unsupervised hierarchical cluster analysis using normalized spectral count data for all identified proteins showed clear clustering of proteins found in native versus decellularized tissues (Fig. 3C). Among the identified database entries for decellularized cartilage tissue, 13 entries exhibited a highly significant difference in abundance (at least 1.5-fold, p<0.05) (Fig. 3D). Importantly, supervised cluster analysis revealed separate clustering of native and decellularized cartilage tissues, indicating substantial changes in the decellularized cartilage proteome compared to native cartilage (Fig. 3C). This difference was related mostly to proteins found in the ECM (Fig. 3D). In addition, analysis of the percentage (%) of gene hits against a total number of genes associated with a certain cellular location in both native (Fig. 3E) and decellularized (Fig. 3F) cartilage revealed that decellularized cartilage tissue contained a higher percentage of proteins associated with the gene ontology (GO) terms: extracellular matrix and extracellular region (4.3% and 9.7% for decellularized versus 2.6% and 6.3% for native cartilage). All ECM proteins (GO term extracellular matrix) both present and absent in decellularized cartilage tissue are listed in Supplementary Table 1. Noticeable absent ECM proteins in decellularized cartilage tissue were elastin, cartilage matrix protein, cartilage-associated protein, chondroadherin-like protein, versican core protein, and some collagens.
Figure 3.
Proteomic analysis of native and decellularized auricular cartilage tissues. (A) Venn diagram showing numbers of unique proteins found in either native (683) or decellularized (21), and numbers of proteins found in both tissues (412) (in at least 2 out of 3 samples). (B) Data are represented mean ± SD of numbers of proteins identified using mass spectrometry (MS/MS) of native cartilage tissue and decellularized cartilage tissue (n=3). (C) Heat map and supervised cluster analysis using protein expression data from native and decellularized cartilage tissue. Notice biological variation in both native and decellularized tissue samples (orange). Only proteins found in 2 out of 3 samples were processed for further analysis. (D) Protein-protein network of significantly upregulated proteins in decellularized cartilage (p<0.05, fold change ≥1.5, detected in at least 2 out of 3 samples). The networks were generated using default settings in String and visualized using Cytoscape. (E) Percentage (%) of gene hits against total number of genes associated with a certain cellular location in native cartilage tissue and (F) decellularized cartilage tissue (Panther GO - Slim cellular component, detected in at least 2 out of 3 samples).
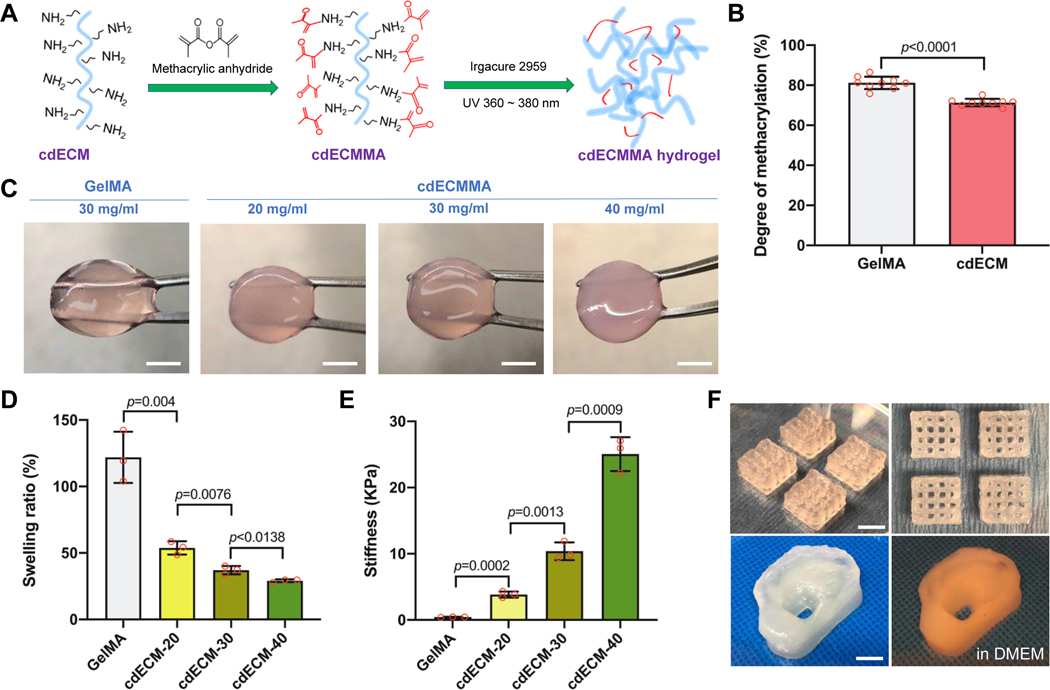
3.3. Characterization of cartilage-derived dECMMA hydrogels
Figure 4A shows the illustration of the methacrylation of the solubilized cdECM and photo-crosslinking of cdECMMA. After methacrylation, the degree of substitution of amino groups in the cdECM before and after methacrylation estimated by TNBS assay presented 71 ± 2% of methacrylation of cdECMMA, while the GelMA showed 81 ± 3% of methacrylation (Fig. 4B). Under the UV light, the cdECMMA hydrogels were produced with different concentrations (20, 30, and 40 mg/ml) (Fig 4C). The swelling ratio of the cdECMMA hydrogels decreased with the increase of the concentration of cdECMMA. When compared with the GelMa hydrogel (30 mg/mL, 122 ± 19%), all concentrations of the cdECMMA hydrogels (20 mg/ml, 54 ± 5; 30 mg/ml, 37 ± 3; and 40 mg/ml, 29 ± 1) showed significantly lower swelling ratio (Fig. 4D). The stiffness of the cdECMMA hydrogels significantly increased with increasing the cdECMMA concentration (20 mg/ml, 3837 ± 462 Pa; 30 mg/ml, 10381 ± 1339 Pa; and 25050 ± 2573 Pa) (Fig. 4E). The GelMA hydrogel at 30 mg/ml obtained 436 ± 40 Pa of the gel stiffness.
Figure 4.
Synthesis and characterization of photo-crosslinkable cdECM bioinks. (A) Schematic illustration of methacrylation and UV crosslinking of cdECM and (B) degree of methacrylation (%) of GelMA and cdECM-MA (n=9). (C) Gross appearance of cdECM-MA bioink constructs with different concentrations (scale bar: 5 mm). (D) Swelling ratios and (E) gel stiffness of cdECMMA hydrogels with different concentrations. GelMA hydrogel served as a control. All data are represented mean ± SD. The p-values by Student t-test are indicated. (F) Lattice structure (top, scale bar: 5 mm) and ear-shape of 3D printed cdECM-MA bioink constructs (30 mg/ml) (bottom, scale bar: 10 mm).
For the bioprinting process, the cdECMMA hydrogels were reconstituted with gelatin, HA, and glycerol as bioink formulations [18, 21]. The cdECMMA-based bioinks were printed to fabricate lattice-shaped and ear-shaped cell-laden constructs (Fig. 4F). After photo-crosslinking of cdECMMA, the un-crosslinked components (gelatin, HA, and glycerol) can be rapidly removed for the bioprinted constructs in the culture condition. The bioprinted cdECMMA constructs provided proper structural integrity and mechanical stability after photo-crosslinking (Supplementary Movie 1).
3.4. In vitro cell viability and proliferation
To determine whether the cdECMMA could support the cell viability and proliferation, the bioprinted cell-laden cdECMMA constructs with different concentrations were cultured at 1, 3, and 7 days. The Live/Dead staining assay indicated over 90% cell viability with all concentrations of the cdECMMA constructs (Fig. 5A,B). The GelMA constructs exhibited significantly lower cell viability (88 ± 3% at 1 day and 89 ± 4% at 3 days in culture). After 7 days in culture, all bioprinted constructs showed over 98% cell viability. The cell proliferation in the bioprinted constructs containing auricular chondrocytes was also determined. The results indicated that the increase of the cdECMMA concentration improved cell proliferation in the bioprinted constructs (Fig. 5C). The higher concentration of the cdECMMA (40 mg/ml) showed significantly higher cell proliferation compared with the GelMA constructs at 1, 3, and 7 days in culture.
Figure 5.
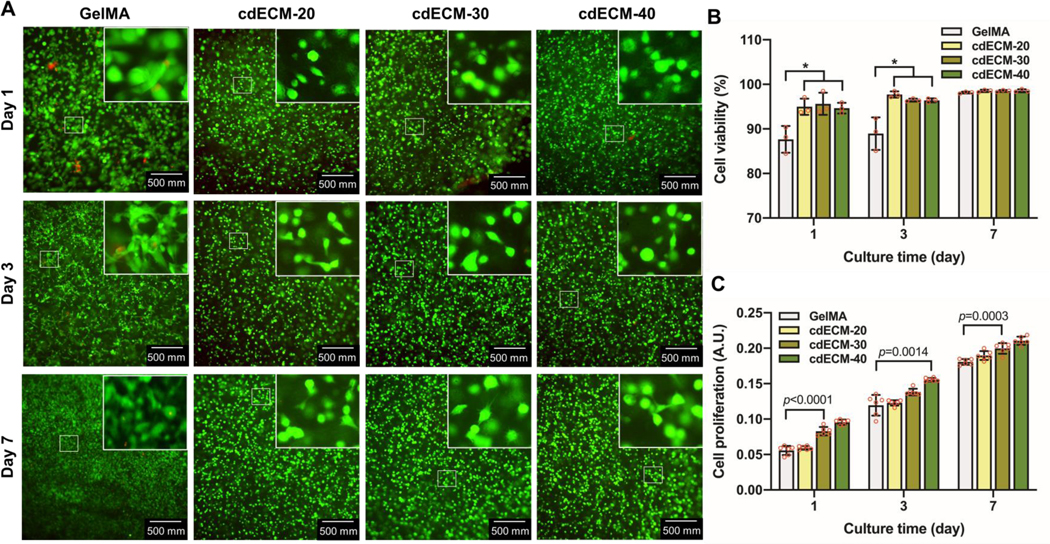
Cell viability and proliferation in the bioprinted cdECMMA constructs at 1, 3, and 7 days in culture. (A) Live/Dead™ stained images and (B) quantification (n=3, *p<0.05). (C) AlamarBlue™ assay for cell proliferation (n=6). GelMA construct served as a control. All data are represented mean ± SD. The p-values by one-way ANOVA followed by Tukey’s test are indicated.
3.5. In vitro cartilage tissue formation in the bioprinted cdECMMA constructs
To demonstrate the tissue-specific effects of the cartilage-derived dECMMA on cartilage tissue formation, the bioprinted constructs containing chondrocytes were examined histologically and biochemically. After 4 weeks in culture, H&E stained images revealed the presence of typical triangular and ovoid-shaped chondrocytes that settled normal-appearing lacunae. However, chondrocytes in the GelMA constructs did not show the typical cellular morphology. Moreover, Safranin-O and Alcian Blue staining confirmed the presence of sulfated GAGs in the bioprinted cdECMMA constructs (Fig. 6A). Quantitatively, the ECM production, total collagen (Fig. 6B) and GAGs (Fig. 6C), in the bioprinted cdECMMA constructs was significantly higher than in the GelMA constructs. And the increase of the cdECMMA concentration accelerated both collagen and GAG production. This indicates the cartilage-specific dECMMA bioinks can improve the cellular functions and accelerate the tissue maturation in the bioprinted constructs.
Figure 6.
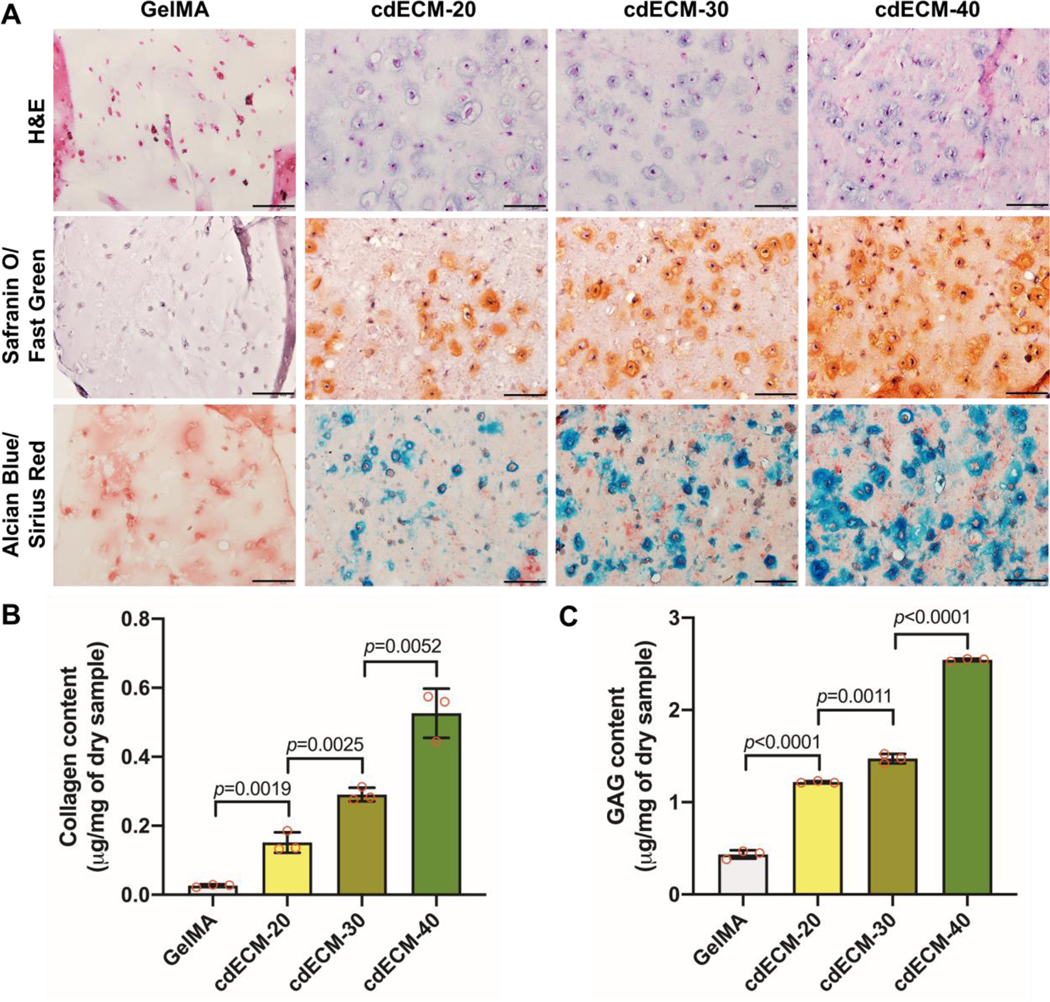
In vitro cartilage tissue formation of 3D bioprinted cdECMMA constructs containing rabbit auricular chondrocytes after 28 days in culture. (A) Histological evaluations by H&E, Safranin O/Fast Green, and Alcian Blue/Sirius Red staining. Scale bar: 100 μm. Biochemical assay for (B) collagen (n=3) and (C) GAG (n=3) production in the 3D printed constructs. GelMA construct served as a control. All data are represented mean ± SD. The p-values by one-way ANOVA followed by Tukey’s test are indicated.
4. Discussion
This study investigated whether the cartilage-derived dECM could be utilized for cell-based 3D bioprinting strategies and further auricular reconstruction [32]. For the bioprinting process, the cdECM was and formulated with gelatin, HA, and glycerol based on our previous works [18, 21, 33, 34]. These supporting components in the bioink formulation provided a proper printability and initial structural integrity. Also, the cdECM was chemically modified as a photo-crosslinkable hydrogel (cdECMMA) that could provide the improved structural integrity of the printed dECM-based constructs [21, 22, 24]. This, this cdECMMA-based bioink was printable, and the bioprinted cdECMMA constructs were mechanically and structurally stable and provided a proper 3D microenvironment for cellular activities in the construct. These results are an important step towards bioengineered auricular cartilage tissue construct that can be used for patient-specific facial reconstruction.
Decellularization of native tissues or organs can be achieved through a variety of methods, all of which affect and alter the native tissue architecture [35]. For elastic cartilage decellularization, a wide range of techniques have been used to remove cellular components and preserve the ECM components [36–39]. Unlike other soft tissues or solid organs, removing the cellular components of the avascular cartilage tissue is extremely difficult [40]. To overcome this, we utilized a mixture of physical, chemical, and enzymatic processes to remove the cellular components thoroughly. After decellularization, cdECM revealed low levels of dsDNA (9.4 ±0.8 ng/mg dry weight), which is an important criterium for decellularization because DNA is directly correlated to adverse host reactions [41, 42]. In addition, following decellularization, collagen architecture was preserved as observed using two-photon laser-scanning microscopy. Furthermore, histological and biochemical analyses confirmed the presence of collagens and GAGs in the dECM.
The resulting ECM proteome was analyzed using mass spectrometry. Many ECM proteins such as collagens I, II, III, and VI, growth factors (transforming growth factors (TGFs)), proteoglycans (keratocan, decorin, aggrecan core protein, lumican, mimecan, and biglycan) and cartilage-specific proteins (chondroadherin, CILP-2, and COMP) were identified and presented following the decellularization process. However, elastin was not found in the cdECM, which is a main ECM component in elastic cartilage. This result could be explained by the fact that elastin is a highly crosslinked insoluble protein that cannot be solubilized into a hydrogel [43]. Nevertheless, the absence of elastin did not alter the mechanical properties of the cdECMMA constructs when compared to the GelMA construct. Although the elastin could be removed by the decellularization, the cdECMMA contained many elastin-related proteins, including elastin microfibril interface-located protein (EMILIN)-1 and 3, which play a role in elastic fiber formation [44], and fibrillin 1 and 2, which play an important role in tissue mechanical properties [45]. In addition, microfibrillar-associated protein 2 (MFAP2), and fibulin 5 and 7 were present in the cdECMMA, of which fibulin-5 is an essential protein in elastic fiber development [46]. Based on these results, further studies will investigate whether the cdECMMA could stimulate the chondrocytes to produce the elastin fibers both in vitro and in vivo.
The increase in mechanical properties of the cdECMMA hydrogels can be accomplished by increased concentration [47, 48]. However, the cdECMMA hydrogels exhibited approximately 25-fold higher gel stiffness when compared with the same concentration (30 mg/ml) of GelMA hydrogel. Although this increase in the gel stiffness cannot be clarified by hydrogel concentration or UV polymerization (as these remained constant), the starting gel stiffness or degree of functionalization may play a role in cell behavior [49]. In addition, the chondrocytes in the bioprinted cdECMMA constructs maintained higher cell viability and proliferation capacity than in the GelMA constructs. Increasing the concentration of cdECMMA had a positive effect on cell proliferation. More importantly, the cdECMMA could maintain the morphological phenotype of auricular chondrocytes and promote ECM production in the bioprinted constructs, as confirmed histologically and biochemically.
Although this study is promising to develop personalized auricular constructs for reconstruction, several concerns need to be addressed. For instance, we observed a large protein heterogeneity between samples in the mass spectrometric analyses, which makes it difficult to draw a specific conclusion on protein abundance in cdECM samples. This could be explained by donor [50, 51], tissue [52, 53], or cell heterogeneity [54]. Although this is common in tissue engineering, future pooling of samples may decrease heterogeneity between the samples allowing a more accurate representation of the decellularized cartilage proteome. Further in vivo studies using the cdECMMA-based bioprinting are currently being performed in an animal model to demonstrate cartilage tissue development and formation.
5. Conclusion
A photo-crosslinkable cartilage-derived ECM-based bioink was successfully developed for auricular cartilage reconstruction. This bioink provided the desired printability, structural and mechanical stability, and the cartilage-specific microenvironment that could promote cellular activities and maturation of the chondrocytes in the bioprinted constructs. The cdECMMA-based bioprinted constructs may be an effective therapeutic approach for personalized auricular cartilage reconstruction.
Supplementary Material
Acknowledgments
This study was supported by the National Institutes of Health (1P41EB023833). We would like to acknowledge Prof. C.R. Jimenez and Dr. S.R. Piersma from the OncoProteomics laboratory of the Amsterdam UMC for their help with tissue workup and interpretation of proteomics data and Dr. L. van Haasterecht for help with SHG imaging.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found in the online version, at doi: 10.1016/j.actbio.2020.11.029.
References
- [1].Cole A, A review of diversity in the evolution and development of cartilage: the search for the origin of the chondrocyte, Eur Cell Mater 21(122) (2011) 9. [DOI] [PubMed] [Google Scholar]
- [2].Soukup B, Mashhadi SA, Bulstrode NW, Health-related quality-of-life assessment and surgical outcomes for auricular reconstruction using autologous costal cartilage, Plastic and reconstructive surgery 129(3) (2012) 632–640. [DOI] [PubMed] [Google Scholar]
- [3].Chauhan DS, Guruprasad Y, Auricular reconstruction of congenital microtia using autogenous costal cartilage: report of 27 cases, Journal of maxillofacial and oral surgery 11(1) (2012) 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kludt NA, Vu H, Auricular reconstruction with prolonged tissue expansion and porous polyethylene implants, Annals of plastic surgery 72 (2014) S14–S17. [DOI] [PubMed] [Google Scholar]
- [5].Fernandes JR, Driscoll DN, Burn ear reconstruction using porous polyethylene implants and tissue expansion, Journal of Burn Care & Research 37(4) (2016) e348–e352. [DOI] [PubMed] [Google Scholar]
- [6].Hwang CM, Lee BK, Green D, Jeong SY, Khang G, Jackson JD, Atala A, Lee SJ, Yoo JJ, Auricular reconstruction using tissue-engineered alloplastic implants for improved clinical outcomes, Plast Reconstr Surg 133(3) (2014) 360e–369e. [DOI] [PubMed] [Google Scholar]
- [7].Nayyer L, Patel KH, Esmaeili A, Rippel RA, Birchall M, O’Toole G, Butler PE, Seifalian AM, Tissue engineering: revolution and challenge in auricular cartilage reconstruction, Plastic and reconstructive surgery 129(5) (2012) 1123–1137. [DOI] [PubMed] [Google Scholar]
- [8].Lewin S, Complications after Total porous implant ear reconstruction and their management, Facial Plastic Surgery 31(06) (2015) 617–625. [DOI] [PubMed] [Google Scholar]
- [9].Fulco I, Miot S, Haug MD, Barbero A, Wixmerten A, Feliciano S, Wolf F, Jundt G, Marsano A, Farhadi J, Engineered autologous cartilage tissue for nasal reconstruction after tumour resection: an observational first-in-human trial, The Lancet 384(9940) (2014) 337–346. [DOI] [PubMed] [Google Scholar]
- [10].Zhou G, Jiang H, Yin Z, Liu Y, Zhang Q, Zhang C, Pan B, Zhou J, Zhou X, Sun H, In vitro regeneration of patient-specific ear-shaped cartilage and its first clinical application for auricular reconstruction, EBioMedicine 28 (2018) 287–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu Y, Zhou G, Cao Y, Recent progress in cartilage tissue engineering—our experience and future directions, Engineering 3(1) (2017) 28–35. [Google Scholar]
- [12].Visscher DO, Bos EJ, Peeters M, Kuzmin NV, Groot ML, Helder MN, van Zuijlen PP, Cartilage tissue engineering: preventing tissue scaffold contraction using a 3D-printed polymeric cage, Tissue Engineering Part C: Methods 22(6) (2016) 573–584. [DOI] [PubMed] [Google Scholar]
- [13].Visscher DO, Van Eijnatten M, Liberton NP, Wolff J, Hofman MB, Helder MN, Griot JPWD, Van Zuijlen PP, MRI and additive manufacturing of nasal alar constructs for patient-specific reconstruction, Scientific reports 7(1) (2017) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liao HT, Zheng R, Liu W, Zhang WJ, Cao Y, Zhou G, Prefabricated, ear-shaped cartilage tissue engineering by scaffold-free porcine chondrocyte membrane, Plastic and reconstructive surgery 135(2) (2015) 313e–321e. [DOI] [PubMed] [Google Scholar]
- [15].Cohen BP, Bernstein JL, Morrison KA, Spector JA, Bonassar LJ, Tissue engineering the human auricle by auricular chondrocyte-mesenchymal stem cell co-implantation, PloS one 13(10) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Reiffel AJ, Kafka C, Hernandez KA, Popa S, Perez JL, Zhou S, Pramanik S, Brown BN, Ryu WS, Bonassar LJ, High-fidelity tissue engineering of patient-specific auricles for reconstruction of pediatric microtia and other auricular deformities, PloS one 8(2) (2013) e56506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cervantes TM, Bassett EK, Tseng A, Kimura A, Roscioli N, Randolph MA, Vacanti JP, Hadlock TA, Gupta R, Pomerantseva I, Design of composite scaffolds and three-dimensional shape analysis for tissue-engineered ear, Journal of the Royal Society Interface 10(87) (2013) 20130413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kang HW, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A, A 3D bioprinting system to produce human-scale tissue constructs with structural integrity, Nat Biotechnol 34(3) (2016) 312–9. [DOI] [PubMed] [Google Scholar]
- [19].Seol YJ, Lee H, Copus JS, Kang HW, Cho DW, Atala A, Lee SJ, Yoo JJ, 3D Bioprinted BioMask for Facial Skin Reconstruction, Bioprinting 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Merceron TK, Burt M, Seol YJ, Kang HW, Lee SJ, Yoo JJ, Atala A, A 3D bioprinted complex structure for engineering the muscle-tendon unit, Biofabrication 7(3) (2015) 035003. [DOI] [PubMed] [Google Scholar]
- [21].Ali M, Pr AK, Yoo JJ, Zahran F, Atala A, Lee SJ, A Photo-Crosslinkable Kidney ECM-Derived Bioink Accelerates Renal Tissue Formation, Adv Healthc Mater 8(7) (2019) e1800992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kim W, Lee H, Lee J, Atala A, Yoo JJ, Lee SJ, Kim GH, Efficient myotube formation in 3D bioprinted tissue construct by biochemical and topographical cues, Biomaterials 230 (2020) 119632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].West-Livingston LN, Park J, Lee SJ, Atala A, Yoo JJ, The Role of the Microenvironment in Controlling the Fate of Bioprinted Stem Cells, Chem Rev 120(19) (2020) 11056–11092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lee H, Kim W, Lee J, Yoo JJ, Kim GH, Lee SJ, Effect of Hierarchical Scaffold Consisting of Aligned dECM Nanofibers and Poly(lactide-co-glycolide) Struts on the Orientation and Maturation of Human Muscle Progenitor Cells, ACS Appl Mater Interfaces 11(43) (2019) 39449–39458. [DOI] [PubMed] [Google Scholar]
- [25].Piersma SR, Warmoes MO, de Wit M, de Reus I, Knol JC, Jimenez CR, Whole gel processing procedure for GeLC-MS/MS based proteomics, Proteome Sci 11(1) (2013) 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liu H, Sadygov RG, Yates JR 3rd, A model for random sampling and estimation of relative protein abundance in shotgun proteomics, Anal Chem 76(14) (2004) 4193–201. [DOI] [PubMed] [Google Scholar]
- [27].Pham TV, Piersma SR, Warmoes M, Jimenez CR, On the beta-binomial model for analysis of spectral count data in label-free tandem mass spectrometry-based proteomics, Bioinformatics 26(3) (2010) 363–9. [DOI] [PubMed] [Google Scholar]
- [28].Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C, STRING v10: protein-protein interaction networks, integrated over the tree of life, Nucleic Acids Res 43(Database issue) (2015) D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T, Cytoscape: a software environment for integrated models of biomolecular interaction networks, Genome Res 13(11) (2003) 2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Maere S, Heymans K, Kuiper M, BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks, Bioinformatics 21(16) (2005) 3448–9. [DOI] [PubMed] [Google Scholar]
- [31].Lee SJ, Broda C, Atala A, Yoo JJ, Engineered cartilage covered ear implants for auricular cartilage reconstruction, Biomacromolecules 12(2) (2011) 306–13. [DOI] [PubMed] [Google Scholar]
- [32].Lee SC, Gillispie G, Prim P, Lee SJ, Physical and Chemical Factors Influencing the Printability of Hydrogel-based Extrusion Bioinks, Chem Rev 120(19) (2020) 10834–10886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kim JH, Kim I, Seol YJ, Ko IK, Yoo JJ, Atala A, Lee SJ, Neural cell integration into 3D bioprinted skeletal muscle constructs accelerates restoration of muscle function, Nat Commun 11(1) (2020) 1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Park JH, Gillispie GJ, Copus JS, Zhang W, Atala A, Yoo JJ, Yelick PC, Lee SJ, The effect of BMP-mimetic peptide tethering bioinks on the differentiation of dental pulp stem cells (DPSCs) in 3D bioprinted dental constructs, Biofabrication 12(3) (2020) 035029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gilbert TW, Sellaro TL, Badylak SF, Decellularization of tissues and organs, Biomaterials 27(19) (2006) 3675–3683. [DOI] [PubMed] [Google Scholar]
- [36].Utomo L, Pleumeekers MM, Nimeskern L, Nürnberger S, Stok KS, Hildner F, van Osch GJ, Preparation and characterization of a decellularized cartilage scaffold for ear cartilage reconstruction, Biomedical materials 10(1) (2015) 015010. [DOI] [PubMed] [Google Scholar]
- [37].Rahman S, Griffin M, Naik A, Szarko M, Butler PE, Optimising the decellularization of human elastic cartilage with trypsin for future use in ear reconstruction, Scientific reports 8(1) (2018) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Dimou Z, Michalopoulos E, Katsimpoulas M, Dimitroulis D, Kouraklis G, Stavropoulos-Giokas C, Tomos P, Evaluation of a Decellularization Protocol for the Development of a Decellularized Tracheal Scaffold, Anticancer research 39(1) (2019) 145–150. [DOI] [PubMed] [Google Scholar]
- [39].Gong YY, Xue JX, Zhang WJ, Zhou GD, Liu W, Cao Y, A sandwich model for engineering cartilage with acellular cartilage sheets and chondrocytes, Biomaterials 32(9) (2011) 2265–2273. [DOI] [PubMed] [Google Scholar]
- [40].Gong YY, Xue JX, Zhang WJ, Zhou GD, Liu W, Cao Y, A sandwich model for engineering cartilage with acellular cartilage sheets and chondrocytes, Biomaterials 32(9) (2011) 2265–73. [DOI] [PubMed] [Google Scholar]
- [41].Nagata S, Hanayama R, Kawane K, Autoimmunity and the clearance of dead cells, Cell 140(5) (2010) 619–630. [DOI] [PubMed] [Google Scholar]
- [42].Zheng MH, Chen J, Kirilak Y, Willers C, Xu J, Wood D, Porcine small intestine submucosa (SIS) is not an acellular collagenous matrix and contains porcine DNA: possible implications in human implantation, Journal of Biomedical Materials Research Part B: Applied Biomaterials: An Official Journal of The Society for Biomaterials, The Japanese Society for Biomaterials, and The Australian Society for Biomaterials and the Korean Society for Biomaterials 73(1) (2005) 61–67. [DOI] [PubMed] [Google Scholar]
- [43].Mecham RP, Methods in elastic tissue biology: elastin isolation and purification, Methods 45(1) (2008) 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bressan GM, Daga-Gordini D, Colombatti A, Castellani I, Marigo V, Volpin D, Emilin, a component of elastic fibers preferentially located at the elastin-microfibrils interface, The Journal of cell biology 121(1) (1993) 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Green EM, Mansfield JC, Bell JS, Winlove CP, The structure and micromechanics of elastic tissue, Interface focus 4(2) (2014) 20130058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Yanagisawa H, Davis EC, Starcher BC, Ouchi T, Yanagisawa M, Richardson JA, Olson EN, Fibulin-5 is an elastin-binding protein essential for elastic fibre development in vivo, Nature 415(6868) (2002) 168–171. [DOI] [PubMed] [Google Scholar]
- [47].Sawkins MJ, Bowen W, Dhadda P, Markides H, Sidney LE, Taylor AJ, Rose FR, Badylak SF, Shakesheff KM, White LJ, Hydrogels derived from demineralized and decellularized bone extracellular matrix, Acta biomaterialia 9(8) (2013) 7865–7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wu J, Ding Q, Dutta A, Wang Y, Huang Y.-h., Weng H, Tang L, Hong Y, An injectable extracellular matrix derived hydrogel for meniscus repair and regeneration, Acta biomaterialia 16 (2015) 49–59. [DOI] [PubMed] [Google Scholar]
- [49].Pepelanova I, Kruppa K, Scheper T, Lavrentieva A, Gelatin-Methacryloyl (GelMA) hydrogels with defined degree of functionalization as a versatile toolkit for 3D cell culture and extrusion bioprinting, Bioengineering 5(3) (2018) 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Andrews SH, Kunze M, Mulet-Sierra A, Williams L, Ansari K, Osswald M, Adesida AB, Strategies to mitigate variability in engineering human nasal cartilage, Scientific reports 7(1) (2017) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hellingman CA, Verwiel ET, Slagt I, Koevoet W, Poublon RM, Nolst-Trenité GJ, De Jong RJB, Jahr H, Van Osch GJ, Differences in cartilage-forming capacity of expanded human chondrocytes from ear and nose and their gene expression profiles, Cell transplantation 20(6) (2011) 925–940. [DOI] [PubMed] [Google Scholar]
- [52].Hsueh M-F, Khabut A, Kjellströ S, Önnerfjord P, Kraus VB, Elucidating the molecular composition of cartilage by proteomics, Journal of proteome research 15(2) (2016) 374–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Albro M, Bergholt M, St-Pierre J, Guitart AV, Zlotnick H, Evita E, Stevens M, Raman spectroscopic imaging for quantification of depth-dependent and local heterogeneities in native and engineered cartilage, NPJ Regenerative medicine 3(1) (2018) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Salter DM, Godolphin JL, Gourlay MS, Chondrocyte heterogeneity: immunohistologically defined variation of integrin expression at different sites in human fetal knees, Journal of Histochemistry & Cytochemistry 43(4) (1995) 447–457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.