Abstract
Dynamically tunable biomaterials are of particular interest in biomedical engineering because of potential utility for shape-change materials, drug and cell delivery and tissue regeneration. Stimuli-responsive proteins formed into hydrogels are potential candidates for such systems, due to the genetic tailorability and control over structure-function. Here we report the synthesis of genetically engineered Silk Elastin-Like Protein (SELP) photoresponsive hydrogels. Polymerization of the SELPs and monomeric adenosylcobalamin (AdoB12)-dependent photoreceptor C-terminal adenosylcobalamin binding domain (CarHC) was achieved using genetically encoded SpyTag-SpyCatcher peptide-protein pairs under mild physiological conditions. The hydrogels exhibited a rapid gel to sol phase transition upon exposure to visible light. The materials were also evaluated for cytotoxicity and the encapsulation and release of L929 murine fibroblasts from 3D cultures. The design of these photo-responsible proteins provides new stimuli-responsive SELP-CarHc hydrogels for dynamically tunable protein-based materials.
Keywords: silk, elastin, stimuli-responsive, biomaterials, silk-elastin-like proteins, bioengineering
Graphical abstracts
1. Introduction
Silk-Elastin-Like Proteins (SELPs) are well-studied genetically engineered biopolymers noted for elastic properties and utilized in biomedical applications for controlled drug delivery [1]. Traditional biomaterials have functioned as passive scaffolds for cells, tissues and biomolecules and often consist of synthetic polymers. These polymers tend to present limitations in the ability to respond to different types of signals or stimuli involved in biological processes, or in some cases may impose cytocompatibility challenges such as with acrylamide-based systems [2, 3]. Thus, advanced, dynamic and stimuli-responsive materials that can mimic the complexity of biological systems would be advantageous, with retention of biological compatibility. These next-generation materials should have the capability to respond to artificial and biological signals and dynamically adjust or tune to a desired outcome [4, 5].
Silk-elastin-like protein polymers (SELPs) are genetically engineered chimeric proteins consisting of repeat units of silk and elastin peptide motifs [1, 6]. The GXGVP sequence of the elastin domain provides elasticity and dynamic features, as well as solubility. The GAGAGS sequence of the silk domain provides mechanical stiffness and physical crosslinks, with more limited solubility. The resulting SELPs have a combination of useful biological and mechanical properties including elasticity, stiffness and tunable stimuli-responsive features [7]. The silk to elastin ratio in SELPs impacts assembly, informing the design of materials with predictable mechanical stiffness [8, 9]. These types of SELP materials have been fabricated into various structures which are useful in tissue engineering and drug delivery [9].
Along these lines, the fabrication of light-responsive biomaterials are of interest as a facile approach to regulate molecule, cell and tissue dynamics with high spatiotemporal accuracy, as most light sources are cytocompatible [10-15]. Further, with advances in synthetic chemistry, progress has been made in the synthesis of dynamically tunable photoresponsive biomaterials [16-18]. For 3-D and 4-D cell culture, some chemically-decorated synthetic hydrogels have been prepared with a combination of photochemistry and other orthogonal click reactions [19-22]. For controlled release from these matrices, photoresponsive devices have been prepared [10, 17, 23-25]. However, despite these successes, many challenges remain. Conventional hydrogels are often limiting in desired dynamic signals, such as spatial or temporal control of formation and degradation, response to biological stimuli such as enzyme catalysis or ligand receptor binding, and mechanical strength. Thus, the dynamic features of materials utilized to date are mostly limited to simple forms of physical and chemical properties which do not mimic the complex conditions found in vivo. Thus, advanced engineering of mechanics and spatiotemporal presentation of bioactive moieties, as well as the manipulation of multiscale shape, structure, and architecture are desired to achieve more tunable and relevant features.
The assembly of genetically engineered peptides into molecular networks having regulated and controlled properties can be a useful strategy to prepare light-responsive biomaterials [26-29]. Nature has evolved many proteins and functional domains which can sense and respond to various environmental and biological stimuli, including light, as well as metal ions, pH, oxidative stress and specific molecules [30]. These protein domains have also been used in the synthesis of dynamically tunable and stimuli-responsive biomaterials. For example, calmodulin-based protein biomaterials have been synthesized and are responsive to calcium ions and trifluoperazine, with dynamic properties [30, 31].
Recently, the light-responsive transcriptional regulator protein of bacterial carotenoid synthesis (CarH) was reported that can sense and respond to visible light. The C-terminal domain of CarH protein (CarHc) is an adenosylcobalamin (AdoB12) binding domain that responds to light in the presence of AdoB12 [32-35]. The AdoB12-dependent protein is involved in the biosynthesis of carotenoids in response to light. CarH has four monomeric domains and the monomeric apoprotein forms tetramers upon binding AdoB12 in the dark [32]. Upon exposure of this tetramerized CarHc domain complexed with AdoB12 in the dark to white or green light (522 nm), the complex dissociates into monomers due to the cleavage of the carbon-cobalt bond in the CarHc tetramer domain, along with a conformational change in protein structure. The dissociation of the CarHc tetramer proceeds through a cob(III)alamin intermediate which forms a stable adduct with the protein [33, 34].
SpyTag/SpyCatcher chemistry is a suitable system to generate long-chain protein polymers [36]. Genetically encoded SpyTag and SpyCatcher consist of a peptide and protein pair that spontaneously form a strong and irreversible isopeptide bond under physiological conditions [36-38]. The SpyTag-SpyCatcher chemistry is suitable for immobilization, labeling, binding and generating new types of protein architectures. Genetically encoded SpyTag can react and form a spontaneous amide bond with its partner SpyCatcher under a wide range of conditions. The system is biologically compatible and the product of this reaction is stable to Sodium Dodecyl Sulfate (SDS) detergent even at 100°C. The chemistry is efficient and modular, thus useful in biomolecular imaging, synthesis of biologically active materials and the control of biomacromolecular topology [26, 28, 38-43]. These same features and utility suggest options to generate complex and responsive biomaterials.
The purpose of this study was the construction of bioactive photoresponsive SELPs using light-triggered assembly-disassembly features of the CarHc protein. The proteins were prepared with SpyTag-SpyCatcher chemistry towards tunable and light stimuli-responsive features. The assembly of the protein components into supramolecular architectures as light-responsive and multipurpose biopolymers was pursued.
2. Materials and methods
2.1. Design, construction, cloning, expression and protein purification
The SELPs monomer (amino acid sequence = (GVGVP)4(GYGVP)1(GVGVP)3(GAGAGS)1) was designed with one silk domain (GAGAGS) and eight elastin domains (GVGVP) to generate SE8Y with silk to elastin domain ratio of 1:8. (Fig. 1A). The short oligonucleotides encoding the SELP monomer (GGTGTAGGAGTACCCGGTGTAGGCGTTCCGGGTGTTGGAGTTCCGGGCGTTGGTGT ACCAGGGTATGGCGTACCGGGCGTTGGTGTGCCTGGTGTTGGAGTTCCGGGAGTTGG TGTCCCAGGAGCTGGTGCGGGGTCC) was commercially synthesized (GeneScript, NJ, USA). SE8Y monomer was cloned into the pET30a expression vector between BseRI and AcuI (both these sites were inserted into the pET30a vector by modifying these vector restriction sites) (Supplementary Fig. S1). Dimers of SE8Y [(SE8Y)2] were prepared by using recursive directional ligation (RDL) for the seamless cloning of monomeric genes in the pET30a vector. The RDL cloning strategy is provided in the supplementary information (Supplementary Fig. S9). SpyTag, SpyCatcher and CarHc were cloned individually into the pUC57 cloning vector between BamHI/EcoRI restriction sites. To make a final recombinant protein cassette of ACA, SpyTag, (SE8)2, CarHc domain, (SE8)2 and SpyTag were sub-cloned between NdeI/KpnI, KpnI/BamHI, BamHI/EcoRI, EcoRI/SacI, and SacI/XhoI restriction sites of pET30a vector. Similarly, the final recombinant protein cassette of BCB was prepared by sub-cloning SpyCatcher, (SE8)2, CarHc domain, (SE8)2 and SpyCatcher between NdeI/KpnI, KpnI/BamHI, BamHI/EcoRI, EcoRI/SacI, and SacI/XhoI restriction sites of pET30a expression vector. The detailed cloning plan is provided in the supplementary information (Supplementary Fig. S10) Cloning was carried out with the help of gene-specific primers listed in supplementary materials (Supplementary Table 1). All clones were confirmed by restriction digestion and sequencing (Genewiz, Boston lab, Cambridge, MA, USA). pUC57 cloning vector and pET30a expression vector was used for cloning and expression. E. coli strain 10-β was used for cloning and plasmid amplification. Plasmids were isolated with the help of QIAprep spin miniprep kits (Qiagen, USA). For protein expression, E. coli strain BL21 (DE3) was used and the cells were grown at 37°C and 250 rpm in Hyper Broth medium supplemented with glucose and the selection marker kanamycin to mid-log phase. To overexpress proteins, cells were induced by adding 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) when the optical density, OD600, was between 0.6 to 0.8 at 37°C. Induction was continued until 4 h. SELP-Fusion Proteins (ACA and BCB) were purified following standard Inversion Transition Cycling (ITC) method (Supplementary Fig. S11)[7, 44, 45] . Cells were harvested using by centrifugation at 6,000 rpm and 4°C for 30 minutes. The pellet was resuspended in 1X PBS buffer (3g pellet in 45ml) with 10 mg lysozyme. Cells were lysed by sonication at 40% power for 10s, then paused for 5 minutes, and then repeated 3 times. Lysed cells were centrifuged at 8,000 rpm, 4°C for 15 minutes to pellet the cell debris. Clear lysate (supernatant) was saved and diluted with an equal volume of 2X TN buffer (300 mM NaCl, 20 mm Tris-HCl, pH8). Samples were incubated in a 40°C water bath for 1h and then centrifuged at 5,000 rpm, 40°C for 3 minutes. The supernatant was discarded and the pellet resuspended with 10 ml of deionized (DI) water. Target proteins were recovered in DI water by shaking at 4°C/25°C overnight. Samples were centrifuged at 8,000 rpm, 4°C for 15 minutes, and the supernatant was dialyzed against DI water for 3 days at 4°C using 10,000 Da MWCO dialysis tubing on a stir plate. Lyophilization was run at −100°C and lyophilized proteins were stored at −80°C. Eluted purified proteins were analyzed by SDS-PAGE on NuPage 4-20% bis-Tris gels. The gels were stained with blue safe stain and visualized with a gel documentation system (Syngene, MD, USA).
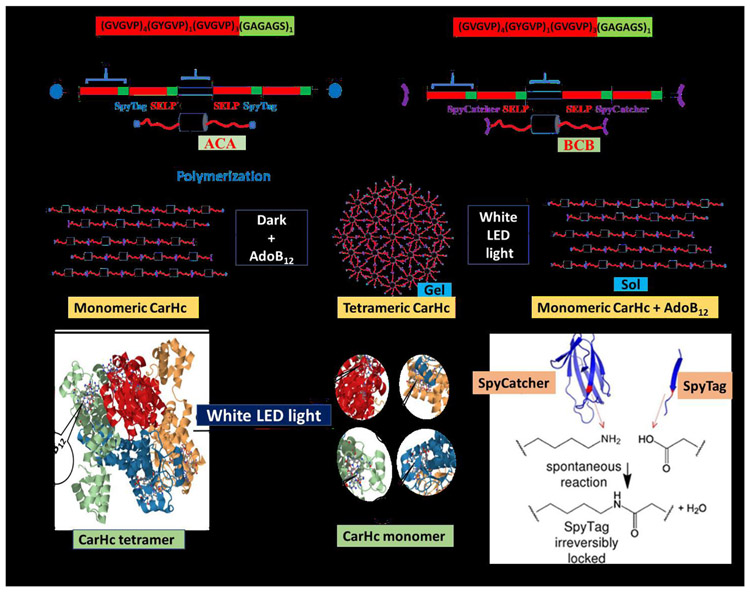
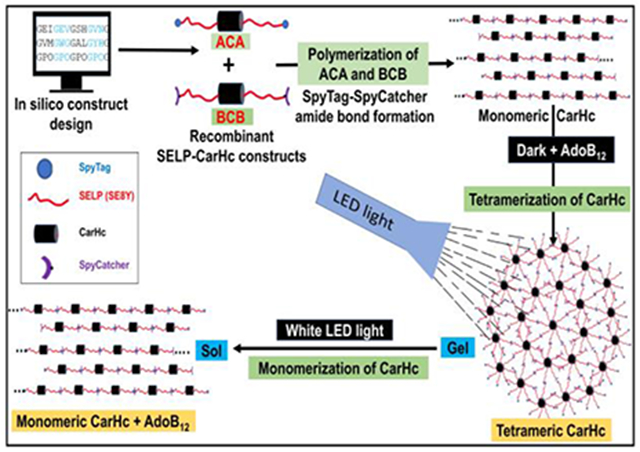
Fig. 1. Diagrammatic representation of the assembly of recombinant SELPs incorporated with photoresponsive CarHc peptide.
The recombinant protein (SELPs with CarHc) assembled into a molecular network through adenosylcobalamin (AdoB12) induced CarHc tetramerization in the dark and disassembled upon exposure to light (white light). Color code; Black block- CarHc domain; Red-green blocks- SELP peptide. A: Block diagram of SELP monomer B: Diagrammatic representation of domain arrangement in the recombinant fusion protein ACA. Red color: elastin domain; green color: silk domain; black color: CarHc domain. At both ends, SpyTag is attached (blue color). C: Diagrammatic representation of domain arrangement in the recombinant fusion proteins BCB. Red color: elastin domain; green color: silk domain; black color: CarHc domain. At both ends, SpyCatcher is attached (purple color). D: Diagrammatic representation of proteins oligomerized/polymerized through SpyTag-SpyCatcher chemistry. The resulting polymers can further be assembled into a molecular network. Red color indicates separate silk and elastin proteins incorporated with photoresponsive CarHc protein in between. E: The resulting polymers can further be assembled into a molecular network through AdoB12-induced CarHc tetramerization in the dark F: Dissociation of CarHc tetramer into monomers resulting in a gel sol transformation under white light exposure G: Structures of CarHc protein. (a) CarHc dark state tetramer, showing the four identical subunits in four colors, and four molecules of AdoB12 one bound to each subunit. (b) CarHc light-state monomer with AdoB12 one bound to each subunit. Light exposure disassembles tetrameric CarHc accompanied by the degradation of AdoB12. Panel H adapted and modified from Figure 4 in reference [49]. H: SpyTag-SpyCatcher chemistry. The amide bond formation between SpyTag and SpyCatcher peptides.
2.2. Polymerization and hydrogel preparation
To check polymerization of ACA and BCB, purified and lyophilized proteins were dissolved in PBS to yield 10 wt% solution and mixed in equimolar ratio and allowed to polymerize at normal physiological conditions, room temperature, and at 4°C. The integrity of the polymerized proteins was checked with 4-20% SDS-PAGE. AdoB12 (Sigma Aldrich, USA) was dissolved in PBS to a final concentration of 10 mM. In the case of the two separate recombinant proteins, they were mixed at an equimolar ratio and allowed to polymerize for 5 minutes, followed by the addition of a stoichiometric amount of AdoB12 in the dark at room temperature. For example, to prepare SELP-CarHc hydrogels, 20μl of 10 wt % of 40 kDa protein was mixed with 31.25μl of 10 wt% of 62 kDa protein, followed by the addition of 2.5μl of 10mM of AdoB12 and allowed to polymerize in the dark for 12 h. 4:1 CarHc to AdoB12 ratio was maintained because one molecule of AdoB12 reacts with four CarHc domain to form a tetramer of CarHc. To prepare enzymatically cross-linked SELP-CarHc hydrogels, lyophilized protein powder was dissolved in PBS at 4°C for 4 h to form a stock solution. Horseradish peroxidase (HRP) type VI lyophilized powder (Sigma Aldrich, St. Louis, MO) was mixed with deionized water to form a stock solution of HRP of 40 mg/mL with a concentration of 10,000 U/ml. To fabricate enzymatically cross-linked 10% SELP-CarHc hydrogels, 6 μL of HRP stock solution was added to 100 μL 10% SELP-CarHc-Adob12 solution, and then the crosslinking reaction of SELP was initiated by adding 0.2 μL of 30 wt% H2O2 solution to the SELP and HRP mixture with a final H2O2 concentration of 18 mM. The reaction mixture was gently mixed and incubated at 4°C overnight to allowed to polymerize.
2.3. Scanning Electron Microscopy
The recombinant proteins ACA and BCB were formed into hydrogels at 4°C upon the addition of 10 mM of AdoB12, as described above. The hydrogels were lyophilized with a Labconco freeze dryer. The dried hydrogel samples were fractured in liquid nitrogen to expose cross-sections, followed by attachment to a carbon tape on a stub and coating with Pt/Pd to a thickness of 10 nm with an EMS 200T D dual-head sputter coater. SEM images were taken with a field emission scanning electron microscope (Ultra55, Zeiss) using an SE2 detector at 5.00 kV.
2.4. Dynamic shear rheology measurements
Dynamic time, strain, and frequency sweep experiments were run on a strain-controlled rheometer (ARES-RFS, TA Instruments, USA) with a standard transparent parallel-plate geometry (8 mm diameter). For test materials, hydrogels were prepared from the recombinant proteins by mixing 10 wt% of both proteins in PBS and 10 mM of AdoB12 in PBS. The real-time gelation process and gel-sol transition were monitored by dynamic time-sweep with the fixed frequency of 1 rad/s and the strain of 5% at 25°C for 12 h. For gelation, all ingredients were mixed as described in the Methods section and placed on parallel plates of the rheometer and tested as per the set parameter (all experiments performed in dark). Dynamic stress sweep of protein gel was performed at a fixed frequency of 6.28 rad/s at 25°C. To prevent light exposure and to maintain dark conditions, all samples were covered with aluminum foil. For the light-dependent degradation rheological tests, the gel form of material was utilized. Photolysis was conducted by exposing the SELP-CarHc hydrogels to an LED light (30 klux) (Electrix™, New Haven, USA). The light source was focused on the material between two parallel plates of the rheometer and dynamic time sweeps were used to monitor the gel-sol transition.
2.5. Light response
The 10 wt% solution of each protein and the 10 mM solution of AdoB12 were prepared in PBS. To prepare SELP-CarHc hydrogels, AdoB12 and solutions of both proteins were mixed and cured in the dark for 12 h at room temperature. To examine the effect of light on protein release, the gels were either exposed to white LED light (Electrix™, New Haven, USA) (30 klux) for 20 min to 1 h or kept in the dark as controls. Both types of samples were immersed with 500 μL PBS and transferred to the dark.
2.6. Cytocompatibility
Cytocompatibility of the hydrogels was investigated by culturing L929 murine fibroblasts from mouse subcutaneous connective tissues (ATCC, Manassas, VA) on the hydrogel surfaces or within the hydrogel matrices for 7 days. Lyophilized protein components were sterilized by ethyleneoxide (ETO) and then dissolved in ultrapure water, while AdoB12 prepared in 1X PBS was sterile filtered using 0.22 μm Polyvinyl Difluoride (PVDF) syringe filters. For surface-seeding, a 10% w/v prehydrogel solution was mixed with AdoB12 at a final concentration of 10 mM, pipetted into 48 well plates and incubated at 4°C in the dark for 4h. L929 murine fibroblasts were seeded on the hydrogels at a density of 8,000 cells/cm2 and cultured for 7 days in an incubator at 37°C with 5% CO2. Cell viability was analyzed by live/dead assay (Invitrogen, Carlsbad, CA) following the cell suspension protocol without washing steps provided by the manufacturer to preserve dead or loosely attached cells. Briefly, 2 μL of 50 μM calcein and 4 μL of 2 mM ethidium homodimer-1 solutions were added directly into 1 mL of culture media. Samples were incubated for 20 min at room temperature in the dark and imaged under a fluorescence microscope (Keyence, IL, USA). Metabolic activity of the cells was monitored using alamarBlue viability assay (Invitrogen, Carlsbad, CA) at 1, 3, 5, and 7 days, and percent dye reduction was calculated according to the manufacturer’s instructions. For cell encapsulation, cells were mixed with 200μL pre-hydrogel solution in 1X PBS at a density of 1x106 cells.mL−1 and gelation was induced with AdoB12 at 4°C in the dark. After 4 h, the hydrogels were dipped in growth media and cultured at 37°C. Metabolic activity of the encapsulated cells was determined by alamarBlue assay at days 1, 3, 5, and 7. For the cell release studies, the pre-hydrogel solution and cell suspension were mixed in Eppendorf tubes. After 4 h of gelation at 4°C, cell-laden gels were transferred into 48-well plates and flushed with growth media for 24 h culture in an incubator at 37°C with 5% CO2. After exposing the samples to white LED light (30 klux) for 20 min, released cells were collected by centrifugation at 1,400 rpm for 5 min and seeded in 48 well plates at a density of 10,000 cells.cm−2. After 12 h of incubation in growth media, cells were live/dead stained and cell viability was estimated from 5 random fluorescent micrographs of each sample (n=3) using the live/dead staining macro of the software ImageJ.
2.7. Statistical Analysis
All data are indicated as mean ±S.D. (standard deviation) for n ≥ 3. GraphPad Prism (GraphPad Software, La Jolla, CA) was used to perform a two-way analysis of variance (ANOVA) with Tukey's post hoc multiple comparison test to determine statistical significance (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001).
3. Results
3.1. Protein construct design
To prepare SELP-CarHc polymers, two gene constructs were designed: 1. SypTag-(SE8Y)2 - CarHc-(SE8Y)2-SpyTag (ACA; 40 kDa) and 2. SpyCatcher-(SE8Y)2-CarHc-(SE8Y)2-SpyCatcher (BCB; 62 kDa). The SE8Y monomer consists of a 46 amino acid sequence with 8 repeat units of GVGVP elastin peptide and 1 unit of GAGAGS silk peptide at a ratio of 8:1((GVGVP)4(GYGVP)1(GVGVP)3(GAGAGS)) (Fig. 1A-C). The complete nucleotide and amino acid sequences for both constructs are given in the supplementary materials (Supplementary Figs. S2-8). Detail cloning steps necessary to access fusion of ACA and BCB proteins are provided in the supplementary information (Supplementary Figs. S9-10). Both fusion proteins were purified by ITC method at a transition temperature of 40°C with an average yield of purified protein of about 2 g/L. ITC method steps are also given in supplementary information (Supplementary Fig. S11). We proposed that the formation of protein-polymers would be based on the interaction and covalent bond formation between SpyTag and SpyCatcher placed at the ends of both constructs. These SELP-CarHc polymers would be dominated by AdoB12-induced tetramerization of the CarHc domain to form hydrogels (Fig. 1B-F). The purity and molecular size of proteins were analyzed on 4-20% SDS-PAGE and MALDI-TOF TOF mass spectrum (Supplementary Figs. S12-15). Purity of ABA was high quality, however, we observed some impurities in BCB. We attempted to separate the impurities by size exclusion chromatography but yield was low and insufficient for further experimentation.
3.2. Synthesis of SELP-CarHc hydrogels
Purified and lyophilized protein powder was dissolved in Phosphate Buffer Saline (PBS) to achieve a 10 wt% solution in PBS. The 10 wt% solution of ACA and BCB proteins were mixed at an equimolar ratio and allowed to polymerize and followed by the addition of a stoichiometric amount of AdoB12 to initiate gelation in the dark at room temperature. Gelation started within 5 minutes and continued typically for at least 2 h. A red gel-like material formed that was sensitive to light. Before the synthesis of a hydrogel, the protein polymers ACA and BCB were assessed by SDS-PAGE. (Supplementary Fig. S16). Proteins polymers of higher molecular weight (ranging from ~175 kDa and above) than either of ACA and BCB were observed, indicating polymerization of both proteins. The ACA and BCB conjugates formed porous and entangled networks at the microscale, revealed by scanning electron microscopy (SEM) (Supplementary Fig. S17), which confirmed the formation of hydrogels. The SEM images showed that freeze-dried SELP hydrogels were porous and the average pore size was about 7 μm.
3.3. Light-induced gel-sol transition of SELP- CarHc hydrogel
The synthesized gels were exposed to light and the gel converted to liquid with about 20 min of exposure to white LED light (30 klux) (Fig. 2). This gel-sol transition can be explained by the light-induced disassembly of CarHc tetramers (Fig. 3A). The polymer integrity was also analyzed before gel formation and after light exposure of the hydrogels by SDS-PAGE (Supplementary Fig. S18). This result confirmed the polymerization of the ABA and BCB proteins aided by the SpyTag SpyCatcher reaction during gelation. For control, we enzymatically crosslinked the gel using tyrosine of elastin block in SELP-CarHc polymer and this gel was not responding to the light because dityrosine crosslinking of elastin block cannot disrupt by white light (Fig. 3B).
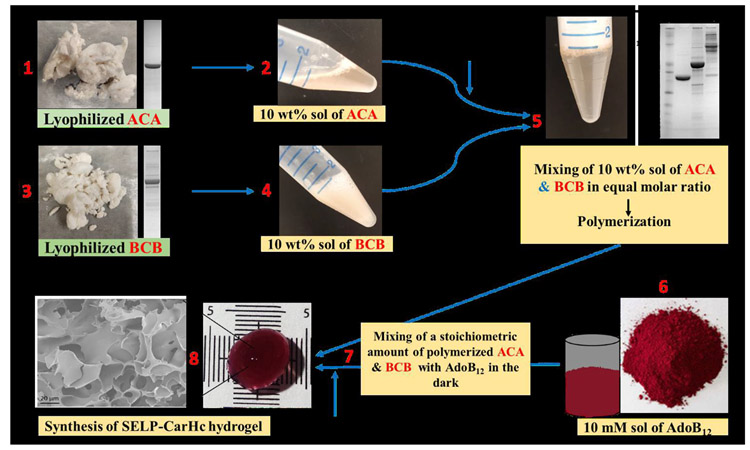
Fig. 2. Biosynthesis scheme of SELP-CarHc hydrogels.
Steps indicating the biosynthesis of a hydrogel. Proteins ACA and BCB were dissolved in PBS to yield 10 wt% solutions. AdoB12 was dissolved in PBS to a final concentration of 10 mM. Protein A and B were mixed at an equimolar ratio and polymerized, followed by the addition of a stoichiometric amount of AdoB12 in the dark at 25°C temperature and incubated overnight. Structure of AdoB12 reproduced from Figure 1 in Chemaly (2016). Scale bars are 10 mm. SDS-PAGE gel picture is indicating the purification of ACA (step 1), BCB (step 3), and polymerization of ACA and BCB (step 5). SEM image of a hydrogel is attached in step 8. Detailed description is given in supplementary information.
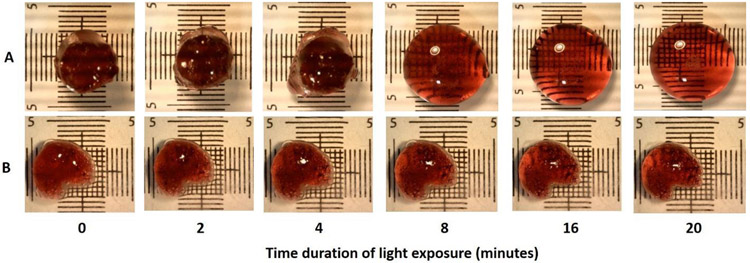
Fig. 3. Light-induced gel-sol phase change of SELP-CarHc hydrogels.
A. A red-colored 10% ACA and BCB hydrogel exposed to 30 klux of white light for 20 minutes showing gradual phase transition from gel to sol. B. A control gel; enzymatically crosslinked SELP-CarHc hydrogel was exposed to same light source for same time period. Grids are in mm.
3.4. Rheological properties of SELP-CarHc hydrogel
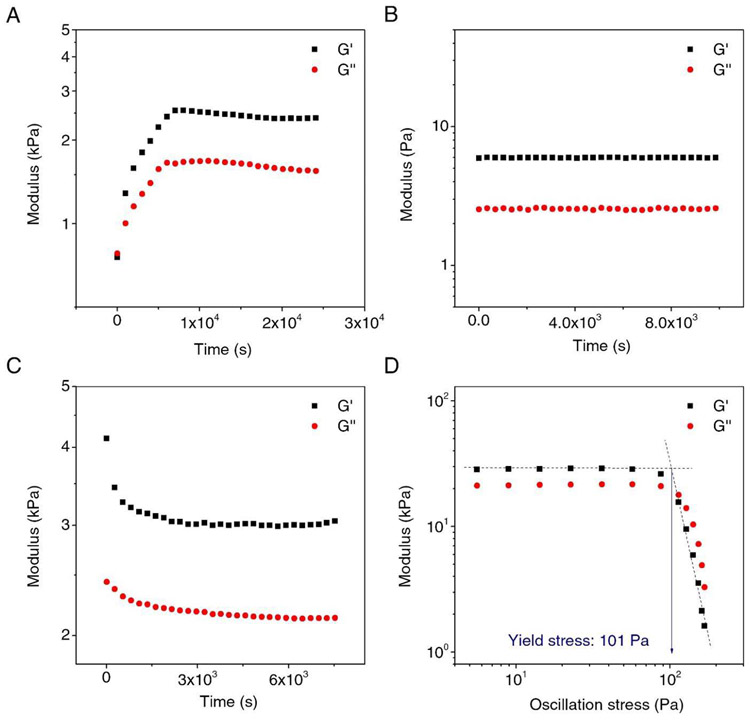
The gelation and dissociation process of SELP samples were analyzed by dynamic time and stress sweeps in rheometry. The mixture of ACA, BCB, and AdoB12 in the dark showed a gradual increase in storage modulus G’, indicating the building up of molecular network (Fig. 4A). After 2 h, the modulus reached ~2.5 kPa for G’ and ~1.5 kPa for G”. In the control experiment, where only the ACA and BCB were mixed in the absence of the AdoB12. the G’ was lower by three-orders of magnitude and was time-independent within at least 2 h (Fig. 4B). Conventionally, G’ indicates the crosslinking densities and a larger G’ than G” indicated that the sample was a gel. The results suggested that AdoB12 was essential to form CarHc tetramerization that led to a higher crosslinking density in comparison with the linear polymerization by only ACA and BCB. When the hydrogel, formed by mixing ACA, BCB, and AdoB12, was exposed to the white light (30 klux), the G’ dropped from 4.13 kPa to 3.15 kPa in about 20 min and remained largely constant afterward (Fig. 4C). During the light exposure, the G’ also remained larger than the G”, suggesting gel softening due to partial dissociation of the molecular network. The rest of the molecular network may be associated with the silk fibroin domain, requiring further investigation. . These results suggested that the white light was responsible for the dissociation of the gel, mainly via the photo-cleavage of the C-Co bond. The yield stress of the gel of ACA, BCB, and AdoB12 was also examined, which was ~101 Pa (Fig. 4D). The yield stress indicated the minimum stress to initiate flow and the mechanical strength of the gel. Of note, G” became larger than G’ after the point of yield stress, indicating a stress-driven gel-sol conversion.
Fig. 4. Rheological test for SELP-CarHC hydrogels formation and response to light.
A. Evaluation of the storage modulus (G′) and loss modulus (G″) of SELP-CarHC hydrogels at room temperature (25°C) in the dark as a function of time. B. The control experiment of A without adding AdoB12. C. The G′ and G″ of the SELP-CarHC hydrogel under white light indicating a gel-sol transition D. Determination of the yield stress of the SELP-CarHc hydrogel.
3.5. Cytocompatibility and cell encapsulation of SELP-CarHc Hydrogels
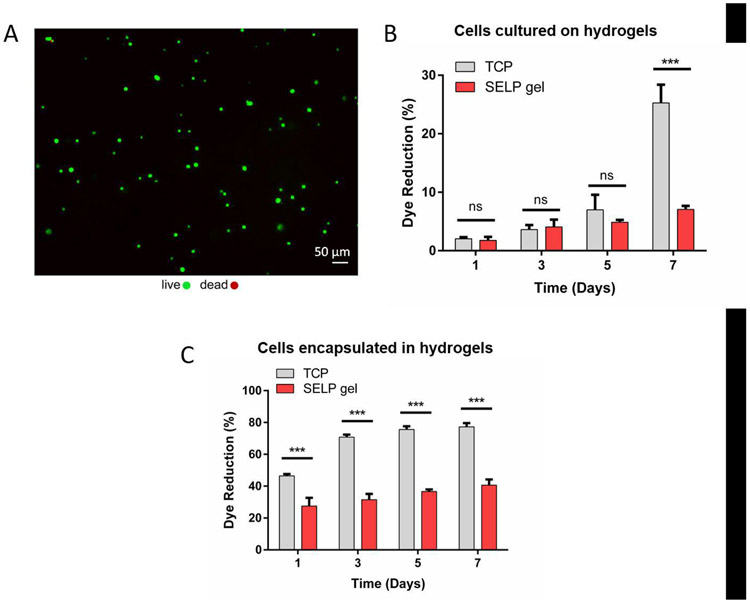
Cytotoxicity of the synthesized SELP-CarHc hydrogels was analyzed using L929 murine fibroblasts cultured on hydrogel surfaces or encapsulated within the hydrogels. Cells that were seeded onto the hydrogels attached to the substrate surface and their viability was determined by live/dead staining, with no significant cell death observed at day 1 (Fig. 5A). This result suggested that the SELP-CarHc hydrogels were nontoxic to the L929 murine fibroblasts. Moreover, the gradual increase in the metabolic activity of the cells grown on the hydrogel surfaces over 7 days (Fig. 5B) indicated that the hydrogels supported cell growth. Metabolic activity of the cells encapsulated and cultured within the hydrogels increased slightly over 7 days of culture, suggesting that the cells were viable but did not proliferate extensively (Fig. 5C).
Fig. 5. Cell viability.
Cytocompatibility test of L929 murine fibroblasts grown on or encapsulated within the SELP-CarHc hydrogels. A. Live/dead fluorescent micrograph of the cells seeded onto the hydrogel surface at day 1. Green: calcein (live), red: EthD-1 (dead). (B-C) % dye reduction by the cells grown grown on the hydrogel surfaces (B) or encapsulated within the hydrogels (C) over 7 days of culture as an indicator of metabolic activity. TCP: Tissue Culture Plastic control. (n = 3, *p < 0.05, **p < 0.01 and ***p < 0.001). Statistical analysis (two-way ANOVA with Tukey’s post-hoc test) is provided as a Table in the supplementary information (Tables S2 and S3).
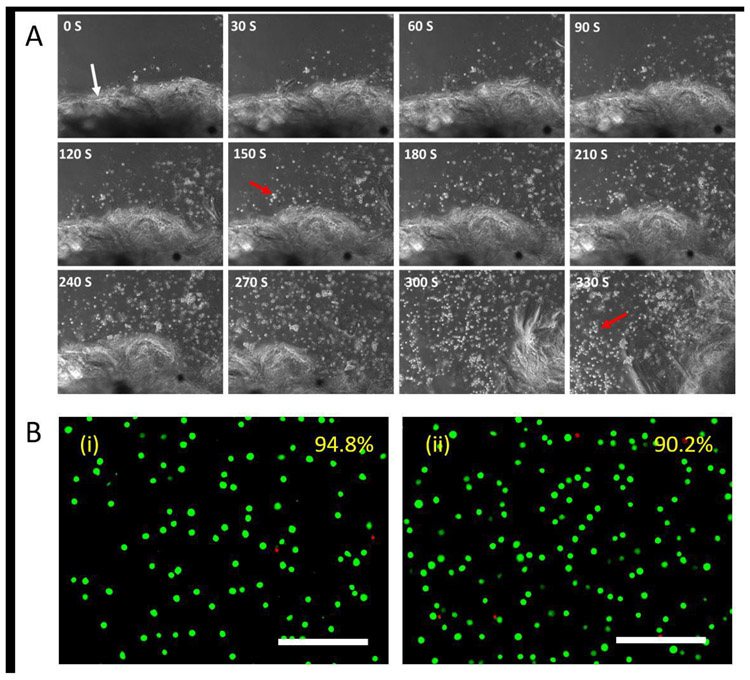
The on-demand release of the encapsulated cells was also investigated through the exposure of the hydrogels to light. Rapid dissociation of the hydrogels was observed upon shining white light of 30 klux onto the samples, and the encapsulated cells were released from the matrix into solution (Fig. 6A). Digital analysis of the Live/dead micrographs of the released cells revealed cell viability of around 90% compared to ~95% determined for the untreated control cells (Fig. 6B), indicating that the encapsulation and on-demand release were cytocompatible with the L929 murine fibroblasts.
Fig. 6. Release of encapsulated cells from hydrogels upon light exposure.
A. L929 murine fibroblasts were encapsulated in SELP-CarHc hydrogels (gel boundary is indicated by white arrow) and cultured for 24 h. Gel was exposed to the white light of 30klux and to initiate cell release along with the dissociation of the hydrogel. Micrograph of light exposure and cell release is indicated at every 30 seconds. The red arrow indicates released cells from the hydrogel. B. Live/dead staining micrographs of (i) untreated control and (ii) encapsulated cells 12 h after release. Average cell viabilities estimated from 5 random images are shown in the upper right corner. Scale bars: 200 μm.
4. Discussion
Stimuli-responsive biomaterials are of interest in biomedical engineering for drug delivery, cell delivery and tissue regeneration. Protein polymers, in particular, can provide tight control of sequence chemistry and molecular weight, thus affording tunable biomaterials features. However, the formation of dynamically tunable and stimuli-responsive protein hydrogels remains challenging. The limitations include the understanding of the relationships between protein sequence and function for the rational development of new protein polymers. There are methods available to modify proteins to make them dynamically tunable, such as azobenzene-based protein modulation. However, UV light is needed to trigger azobenzene photo switches [46] and this can be harmful to DNA and cells. To overcome these problems, genetically engineered proteins can be utilized to develop both fundamental insights into sequence-function designs, as well as to develop functional biomaterials for medical utility. Biocompatibility of silk-based biomaterials has been demonstrated with Food and Drug Administration, USA (FDA) approvals. The long-term biocompatibility of various silk formats has been demonstrated in animal models and in humans and an absence of toxicity, pyrogenicity and allergenicity [47]. The present work demonstrated the design and fabrication of a new family of light-responsive protein hydrogels by exploiting the CarHc domain and silk and elastin peptide motifs as building blocks.
The SpyTag-SpyCatcher reaction initiates polymer formation and AdoB12-dependent CarHc self-assembly into tetramer was responsible for gelation. The gelation time depends on the kinetics of the SpyTag-SpyCatcher and AdoB12-dependent CarHc tetramerization. Upon exposure of the hydrogels to white light, the CarHc tetramer dissociates into monomers based on the conformational change produced by C-Co bond cleavage [33, 48, 49]. This light-induced conformational change results in the gel to sol transition. Photodegradation of AdoB12 in the SELP-CarHc hydrogel is not a typical radical mechanism as dissociation occurs into 4′,5′-anhydroadenosine instead of the more common 5′-dAdo radicals [35]. AdoB12 dependent CarHc tetramer formation in the dark and light-controlled AdoB12 dependent photo regulation and degradation of CarHc into monomers was demonstrated previously as a fundamental aspect of light-controlled assembly-disassembly [32, 34, 50].
In the present study, SpyTag and SpyCatcher were able to form SELP-CarHc protein polymers in which interchain interactions were mediated by the AdoB12 induced tetramerization of the CarHc domain. This tetramerization was through interchain interactions for SELP-CarHc hydrogels in the dark. Polymer integrity before and after light exposure showed no degradation of the polymer and suggested that the material phase transition was caused by tetramerization and monomerization of CarHc and not because of alteration of SpyTag SpyCatcher bond formation. Dynamic shear rheology showed a gradual increase in storage modulus with time in the dark and a rapid drop upon exposure to light to confirm the gelation process in the dark and light-induced dissociation of the hydrogel mediated by AboB12. Moreover, the yield stress was comparable with other proteinaceous hydrogels [51] and a widely-used commercial hydrogel, Carbopol [52], indicating the potential utility of the SELP-CarHc hydrogels for injections and 3D printing [53, 54].
Increase in metabolic activity of L929 murine fibroblasts was observed over 7 days suggesting cytocompatibility when grown on the SELP-CarHc hydrogels. 3D cell culture system also supported the encapsulation of cells similar to the enzymatically-crosslinked silk-based hydrogels previously reported [55]. Moreover, subsequent release of the encapsulated cells was achieved by exposure to white light without having to use proteolytic degradation or complicated chemical treatments [56, 57]. The recovery of viable encapsulated cells suggested that the SELP-CarHc hydrogel is potential as a cell delivery system. Previous studies also show that silk and SELP- based materials were suitable for cell culture and in vivo studies [1, 58-62].
In the future, light-responsive biomaterials could be utilized for cell culture systems and more broadly for many areas of biomedical research. The ability to trigger changes in biomaterial properties on-demand and with spatiotemporal control, using safe visible light, can be helpful for many areas of cell culture (positioning in 3D, controlled release), cell delivery (to support cell protection from shear forces during injection, with release on-demand once in situ), and many other areas of cell delivery, tissue systems and treatments. For example, photo-responsive stiffening biomaterials can be useful in studies of disease modeling, while materials that soften can be used in cell and therapeutic delivery. Furthermore, light-driven protein-based biomaterials can be used to direct cell proliferation, migration, and differentiation through suitable modifications in photosensitive material control over mechanics, signal presentation, and biomolecule release. These types of materials could be used to study the impact of stiffness on immune responses and cell health, with such dynamically compliant materials providing additional insights into cell mechanobiological responses.
5. Conclusion
In summary, recombinant protein-based light-sensitive SELP-CarHc hydrogels were synthesized using SpyTag-SpyCatcher chemistry and were successfully demonstrated to undergo a dynamic phase transition upon exposure to white light. AdoB12-dependent CarHc tetramerization was essential for the formation of the hydrogels in the dark, while upon exposure to white light a gel to sol phase transition was demonstrated due to the conversation of CarHc tetramer back to monomer. This photo-induced hydrogel dissociation supported the facile release of encapsulated L929 murine fibroblasts from 3D cultures. These results offer an important first step towards light-reversible photo responsive hydrogels for utility in cell encapsulation and release in dynamic control conditions.
Supplementary Material
Statement of significance.
Dynamically tunable stimuli-responsive biomaterials provide potential utility in biomedical engineering as candidates for soft robotics and biomimetic devices, as well as vehicles for cell and drug delivery/release. Photoresponsive systems are of particular interest as the activation mode, due to the simple input required and avoidance of additions of chemical catalysts or other exogenous components to drive the process. Here, SELP-based light responding hydrogels prepared here self-assembled into a dynamic hydrogels in the presence of AdoB12 in the dark and disassembled upon exposure to light to release encapsulated cells. This SELP-based stimuli-responsive hydrogel represents a useful strategy for designing smart biomaterials for cell, protein and molecule delivery, here using light as the triggering mechanism.
Acknowledgments
We thank the National Institute of Health (NIH) (P41EB027062, U01EB014976) and the Army Research Office W911NF-17-1-0384) for financial support for this study. The authors acknowledge the Harvard MRSEC (DMR-1420570) and the Harvard University Center for Nanoscale Systems (CNS) for providing rheological and SEM measurements, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interest
The authors declare no conflict of interest
References
- [1].Huang W, Rollett A, Kaplan DL, Silk-elastin-like protein biomaterials for the controlled delivery of therapeutics, Expert opinion on drug delivery 12(5) (2015) 779–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kopeček J, Yang J, Hydrogels as smart biomaterials, Polymer international 56(9) (2007) 1078–1098. [Google Scholar]
- [3].Krishna OD, Kiick KL, Protein-and peptide-modified synthetic polymeric biomaterials, Peptide Science: Original Research on Biomolecules 94(1) (2010) 32–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Langer R, Tirrell DA, Designing materials for biology and medicine, Nature 428(6982) (2004) 487. [DOI] [PubMed] [Google Scholar]
- [5].Burdick JA, Murphy WL, Moving from static to dynamic complexity in hydrogel design, Nature communications 3 (2012) 1269. [DOI] [PubMed] [Google Scholar]
- [6].Roberts EG, Rim NG, Huang W, Tarakanova A, Yeo J, Buehler MJ, Kaplan DL, Wong JY, Fabrication and Characterization of Recombinant Silk-Elastin-Like-Protein (SELP) Fiber, Macromolecular bioscience 18(12) (2018) 1800265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Huang W, Tarakanova A, Dinjaski N, Wang Q, Xia X, Chen Y, Wong JY, Buehler MJ, Kaplan DL, Design of Multistimuli Responsive Hydrogels Using Integrated Modeling and Genetically Engineered Silk–Elastin-Like Proteins, Advanced functional materials 26(23) (2016) 4113–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wang Q, Xia X, Huang W, Lin Y, Xu Q, Kaplan DL, High Throughput Screening of Dynamic Silk-Elastin-Like Protein Biomaterials, Advanced functional materials 24(27) (2014) 4303–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Xia X-X, Xu Q, Hu X, Qin G, Kaplan DL, Tunable self-assembly of genetically engineered silk–elastin-like protein polymers, Biomacromolecules 12(11) (2011) 3844–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].DeForest CA, Anseth KS, Cytocompatible click-based hydrogels with dynamically tunable properties through orthogonal photoconjugation and photocleavage reactions, Nature chemistry 3(12) (2011) 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tomatsu I, Peng K, Kros A, Photoresponsive hydrogels for biomedical applications, Advanced drug delivery reviews 63(14-15) (2011) 1257–1266. [DOI] [PubMed] [Google Scholar]
- [12].Katz JS, Burdick JA, Light-responsive biomaterials: Development and applications, Macromolecular bioscience 10(4) (2010) 339–348. [DOI] [PubMed] [Google Scholar]
- [13].Yagai S, Kitamura A, Recent advances in photoresponsive supramolecular self-assemblies, Chemical Society Reviews 37(8) (2008) 1520–1529. [DOI] [PubMed] [Google Scholar]
- [14].Chang VY, Fedele C, Priimagi A, Shishido A, Barrett CJ, Photoreversible Soft Azo Dye Materials: Toward Optical Control of Bio-Interfaces, Advanced Optical Materials 7(16) (2019) 1900091. [Google Scholar]
- [15].Pianowski ZL, Recent Implementations of Molecular Photoswitches into Smart Materials and Biological Systems, Chemistry–A European Journal 25(20) (2019) 5128–5144. [DOI] [PubMed] [Google Scholar]
- [16].Ercole F, Davis TP, Evans RA, Photo-responsive systems and biomaterials: photochromic polymers, light-triggered self-assembly, surface modification, fluorescence modulation and beyond, Polymer Chemistry 1(1) (2010) 37–54. [Google Scholar]
- [17].Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS, A versatile synthetic extracellular matrix mimic via thiol-norbornene photopolymerization, Advanced Materials 21(48) (2009) 5005–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Boelke J, Hecht S, Designing molecular photoswitches for soft materials applications, Advanced Optical Materials 7(16) (2019) 1900404. [Google Scholar]
- [19].DeForest CA, Polizzotti BD, Anseth KS, Sequential click reactions for synthesizing and patterning three-dimensional cell microenvironments, Nature materials 8(8) (2009) 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].DeForest CA, Tirrell DA, A photoreversible protein-patterning approach for guiding stem cell fate in three-dimensional gels, Nature materials 14(5) (2015) 523. [DOI] [PubMed] [Google Scholar]
- [21].Li L, Scheiger JM, Levkin PA, Design and applications of photoresponsive hydrogels, Advanced Materials 31(26) (2019) 1807333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ruskowitz ER, DeForest CA, Photoresponsive biomaterials for targeted drug delivery and 4D cell culture, Nature Reviews Materials 3(2) (2018) 1–17. [Google Scholar]
- [23].Alvarez-Lorenzo C, Bromberg L, Concheiro A, Light-sensitive intelligent drug delivery systems, Photochemistry and photobiology 85(4) (2009) 848–860. [DOI] [PubMed] [Google Scholar]
- [24].Zhang YS, Khademhosseini A, Advances in engineering hydrogels, Science 356(6337) (2017) eaaf3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].LeValley PJ, Neelarapu R, Sutherland B, Dasgupta S, Kloxin CJ, Kloxin AM, Photolabile linkers: exploiting labile bond chemistry to control mode and rate of hydrogel degradation and protein release, Journal of the American Chemical Society (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sun F, Zhang W-B, Mahdavi A, Arnold FH, Tirrell DA, Synthesis of bioactive protein hydrogels by genetically encoded SpyTag-SpyCatcher chemistry, Proceedings of the National Academy of Sciences 111(31) (2014) 11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Banta S, Wheeldon IR, Blenner M, Protein engineering in the development of functional hydrogels, Annual review of biomedical engineering 12 (2010) 167–186. [DOI] [PubMed] [Google Scholar]
- [28].Gao X, Fang J, Xue B, Fu L, Li H, Engineering protein hydrogels using SpyCatcher-SpyTag chemistry, Biomacromolecules 17(9) (2016) 2812–2819. [DOI] [PubMed] [Google Scholar]
- [29].Fang J, Mehlich A, Koga N, Huang J, Koga R, Gao X, Hu C, Jin C, Rief M, Kast J, Forced protein unfolding leads to highly elastic and tough protein hydrogels, Nature communications 4 (2013) 2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sui Z, King WJ, Murphy WL, Protein-Based Hydrogels with Tunable Dynamic Responses, Advanced Functional Materials 18(12) (2008) 1824–1831. [Google Scholar]
- [31].Murphy WL, Dillmore WS, Modica J, Mrksich M, Dynamic hydrogels: translating a protein conformational change into macroscopic motion, Angewandte Chemie International Edition 46(17) (2007) 3066–3069. [DOI] [PubMed] [Google Scholar]
- [32].Kutta RJ, Hardman SJ, Johannissen LO, Beilina B, Messiha HL, Ortiz-Guerrero JM, Elías-Arnanz M, Padmanabhan S, Barran P, Scrutton NS, The photochemical mechanism of a B 12-dependent photoreceptor protein, Nature communications 6 (2015) 7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jost M, Fernández-Zapata J, Polanco MC, Ortiz-Guerrero JM, Chen PY-T, Kang G, Padmanabhan S, Elías-Arnanz M, Drennan CL, Structural basis for gene regulation by a B 12-dependent photoreceptor, Nature 526(7574) (2015) 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ortiz-Guerrero JM, Polanco MC, Murillo FJ, Padmanabhan S, Elías-Arnanz M, Light-dependent gene regulation by a coenzyme B12-based photoreceptor, Proceedings of the National Academy of Sciences 108(18) (2011) 7565–7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jost M, Simpson JH, Drennan CL, The transcription factor CarH safeguards use of adenosylcobalamin as a light sensor by altering the photolysis products, Biochemistry 54(21) (2015) 3231–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, Howarth M, Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin, Proceedings of the National Academy of Sciences 109(12) (2012) E690–E697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Reddington SC, Howarth M, Secrets of a covalent interaction for biomaterials and biotechnology: SpyTag and SpyCatcher, Current opinion in chemical biology 29 (2015) 94–99. [DOI] [PubMed] [Google Scholar]
- [38].Zhang W-B, Sun F, Tirrell DA, Arnold FH, Controlling macromolecular topology with genetically encoded SpyTag–SpyCatcher chemistry, Journal of the American Chemical Society 135(37) (2013) 13988–13997. [DOI] [PubMed] [Google Scholar]
- [39].Schoene C, Fierer JO, Bennett SP, Howarth M, SpyTag/SpyCatcher cyclization confers resilience to boiling on a mesophilic enzyme, Angewandte Chemie International Edition 53(24) (2014) 6101–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bedbrook CN, Kato M, Kumar SR, Lakshmanan A, Nath RD, Sun F, Sternberg PW, Arnold FH, Gradinaru V, Genetically encoded spy peptide fusion system to detect plasma membrane-localized proteins in vivo, Chemistry & biology 22(8) (2015) 1108–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Matsunaga R, Yanaka S, Nagatoishi S, Tsumoto K, Hyperthin nanochains composed of self-polymerizing protein shackles, Nature communications 4 (2013) 2211. [DOI] [PubMed] [Google Scholar]
- [42].Chen AY, Deng Z, Billings AN, Seker UO, Lu MY, Citorik RJ, Zakeri B, Lu TK, Synthesis and patterning of tunable multiscale materials with engineered cells, Nature materials 13(5) (2014) 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Veggiani G, Nakamura T, Brenner MD, Gayet RV, Yan J, Robinson CV, Howarth M, Programmable polyproteams built using twin peptide superglues, Proceedings of the National Academy of Sciences 113(5) (2016) 1202–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hassouneh W, Christensen T, Chilkoti A, Elastin-like polypeptides as a purification tag for recombinant proteins, Current protocols in protein science 61(1) (2010) 6.11. 1–6.11. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Meyer DE, Chilkoti A, 18 Protein Purification by Inverse Transition Cycling, Protein-protein interactions: A molecular cloning manual (2002) 329–344. [Google Scholar]
- [46].Zhu M, Zhou H, Azobenzene-based small molecular photoswitches for protein modulation, Organic & biomolecular chemistry 16(44) (2018) 8434–8445. [DOI] [PubMed] [Google Scholar]
- [47].Yucel T, Lovett ML, Kaplan DL, Silk-based biomaterials for sustained drug delivery, Journal of Controlled Release 190 (2014) 381–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jones AR, The photochemistry and photobiology of vitamin B 12, Photochemical & Photobiological Sciences 16(6) (2017) 820–834. [DOI] [PubMed] [Google Scholar]
- [49].Chemaly SM, New light on vitamin B12: The adenosylcobalamin-dependent photoreceptor protein CarH, South African Journal of Science 112(9-10) (2016) 1–9. [Google Scholar]
- [50].Gruber K, Kräutler B, Coenzyme B12 repurposed for photoregulation of gene expression, Angewandte Chemie International Edition 55(19) (2016) 5638–5640. [DOI] [PubMed] [Google Scholar]
- [51].Olsen BD, Kornfield JA, Tirrell DA, Yielding behavior in injectable hydrogels from telechelic proteins, Macromolecules 43(21) (2010) 9094–9099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].R Varges P, M Costa C, S Fonseca B, F Naccache M, de Souza Mendes PR, Rheological characterization of carbopol® dispersions in water and in water/glycerol solutions, Fluids 4(1) (2019) 3. [Google Scholar]
- [53].Mouser VH, Melchels FP, Visser J, Dhert WJ, Gawlitta D, Malda J, Yield stress determines bioprintability of hydrogels based on gelatin-methacryloyl and gellan gum for cartilage bioprinting, Biofabrication 8(3) (2016) 035003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Mu X, Fitzpatrick V, Kaplan DL, From Silk Spinning to 3D Printing: Polymer Manufacturing using Directed Hierarchical Molecular Assembly, Advanced Healthcare Materials (2020) 1901552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hasturk O, Jordan KE, Choi J, Kaplan DL, Enzymatically crosslinked silk and silk-gelatin hydrogels with tunable gelation kinetics, mechanical properties and bioactivity for cell culture and encapsulation, Biomaterials 232 (2020) 119720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Santoro M, Tatara AM, Mikos AG, Gelatin carriers for drug and cell delivery in tissue engineering, Journal of controlled release 190 (2014) 210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Vo TN, Kasper FK, Mikos AG, Strategies for controlled delivery of growth factors and cells for bone regeneration, Advanced drug delivery reviews 64(12) (2012) 1292–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ding Z, Zhou M, Zhou Z, Zhang W, Jiang X, Lu X, Zuo B, Lu Q, Kaplan DL, Injectable Silk Nanofiber Hydrogels for Sustained Release of Small-Molecule Drugs and Vascularization, ACS Biomaterials Science & Engineering 5(8) (2019) 4077–4088. [DOI] [PubMed] [Google Scholar]
- [59].Zhu C, Ding Z, Lu Q, Lu G, Xiao L, Zhang X, Dong X, Ru C, Kaplan DL, Injectable Silk–Vaterite Composite Hydrogels with Tunable Sustained Drug Release Capacity, ACS Biomaterials Science & Engineering 5(12) (2019) 6602–6609. [DOI] [PubMed] [Google Scholar]
- [60].Parker RN, Cairns DM, Wu WA, Jordan K, Guo C, Huang W, Martin-Moldes Z, Kaplan DL, Smart Material Hydrogel Transfer Devices Fabricated with Stimuli-Responsive Silk-Elastin-Like Proteins, Advanced Healthcare Materials 9(11) (2020) 2000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Price R, Poursaid A, Cappello J, Ghandehari H, In vivo evaluation of matrix metalloproteinase responsive silk–elastinlike protein polymers for cancer gene therapy, Journal of Controlled Release 213 (2015) 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Poursaid A, Price R, Tiede A, Olson E, Huo E, McGill L, Ghandehari H, Cappello J, In situ gelling silk-elastinlike protein polymer for transarterial chemoembolization, Biomaterials 57 (2015) 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.