Abstract
Background
There has been increasing interest in machine learning based natural language processing (NLP) methods in radiology; however, models have often used word embeddings trained on general web corpora due to lack of a radiology-specific corpus.
Purpose
We examined the potential of Radiopaedia to serve as a general radiology corpus to produce radiology specific word embeddings that could be used to enhance performance on a NLP task on radiological text.
Materials and Methods
Embeddings of dimension 50, 100, 200, and 300 were trained on articles collected from Radiopaedia using a GloVe algorithm and evaluated on analogy completion. A shallow neural network using input from either our trained embeddings or pre-trained Wikipedia2014+Gigaword5 (WG) embeddings was used to label the Radiopaedia articles. Labeling performance was evaluated based on exact match accuracy and Hamming loss. The McNemar’s test with continuity and the Benjamini-Hochberg correction and a 5×2 cross validation paired two-tailed t-test were used to assess statistical significance.
Results
For accuracy in the analogy task, 50-dimensional (50-D) Radiopaedia embeddings outperformed WG embeddings on tumor origin analogies (p < 0.05) and organ adjectives (p < 0.01) whereas WG embeddings tended to outperform on inflammation location and bone vs. muscle analogies (p < 0.01). The two embeddings had comparable performance on other subcategories. In the labeling task, the Radiopaedia-based model outperformed the WG based model at 50, 100, 200, and 300-D for exact match accuracy (p < 0.001, p < 0.001, p< 0.01, and p < 0.05, respectively) and Hamming loss (p < 0.001, p < 0.001, p < 0.01, and p < 0.05, respectively).
Conclusion
We have developed a set of word embeddings from Radiopaedia and shown that they can preserve relevant medical semantics and augment performance on a radiology NLP task. Our results suggest that the cultivation of a radiology-specific corpus can benefit radiology NLP models in the future.
Keywords: natural language processing, word embeddings, analogy completion, multi-label classification
Graphical Abstract
Introduction
Natural language processing1 (NLP) techniques are a broad category of methods that have been utilized to extract useful information from free texts. NLP may play an important role due to the nature of a radiologist’s core task of converting information in medical images into a text format (i.e. radiology report). Radiology reports represent a vast corpus of medical information and a form of annotation for the associated medical images; however, their unstructured, free text nature often makes it difficult to convert them into a computer-friendly representation. Language standards such as RadLex and SNOMED CT were created to help give more structure to text (1,2). Further parsing and structuring of this vast amount of free-text information has created an important niche for NLP applications in radiology.
Traditional NLP systems in radiology were constructed using a grammatical rule system to give structure to free narrative text (3,4). However, rule-based approaches take a great deal of effort to develop due to the need to create domain-specific tasks and are unadaptable to variation in individual and institutional practices. In recent years, machine learning approaches have been gaining popularity as they do not require laborious hand-engineered rules (5,6). Convolutional neural network (7,8) and recurrent neural network (9,10) approaches have been widely used and were applied to text-classification tasks such as flagging the presence of pulmonary embolism (7,11) or pneumonia (12,13) within a radiology report.
An important step in many machine learning based NLP models is encoding words into a numerical vector representation known as a word embedding (6). GloVe (Global Vectors for Word Representation) (14) and word2vec (15) are popular methods of creating word embeddings that have seen use in radiology NLP models (7,16). Initialization of the embedding layer with medical domain data has been shown to improve results (17,18); however, radiology NLP models have often relied on embeddings trained on a general corpus or trained from scratch to the specific training set (7,11,19). We hypothesize that embeddings trained on a radiology-focused corpus can capture underlying medical semantics which can then be used to enhance a model’s performance on a radiology NLP task. Radiopaedia.org (20) is a readily available resource under a Creative Commons license (CC BY-NC-SA 3.0) that can act as a radiology corpus. In this study, we develop a set of GloVe word embeddings trained on Radiopaedia and compare them against a set of pre-trained embeddings on an analogy completion task and a multi-label text classification task.
Materials and Methods
Data Curation
Main text from Radiopaedia articles on May 14, 2020 were collected along with their respective “System” label(s) using Python’s BeautifulSoup (21). No articles were excluded. Text from Radiopaedia cases were not included. Text was preprocessed using sentence and word tokenization from the NLTK package (22). English stop-words (e.g. “the”), punctuation, capitalization, and special characters were removed. Words containing punctuation or special characters were split into separate tokens wherever a character was removed. Embeddings trained on Wikipedia 2014 and Gigaword 5 (WG), a corpus of 6 billion tokens, were obtained from Stanford NLP (14). The article characteristics of the dataset and the first 50–50 split are summarized in Table 1.
Table 1.
Summary of Article Characteristics
| Features | Seed 1 Set A (n=6950) | Seed 1 Set B (n=6950) | Overall (n=13,900) |
|---|---|---|---|
| Max tokens in article | 2704 | 3493 | 3493 |
| Minimum tokens in article | 7 | 7 | 7 |
| Average article token count | 163.29 | 163.74 | 163.52 |
| Label Cardinality | 1.45 | 1.44 | 1.44 |
| Label Density | 0.072 | 0.072 | 0.072 |
| Label Categories | |||
| Musculoskeletal | 1663 | 1614 | 3277 |
| CNS | 1296 | 1251 | 2547 |
| Chest | 865 | 861 | 1726 |
| Head and Neck | 800 | 840 | 1640 |
| Gastrointestinal | 620 | 595 | 1215 |
| Paediatrics | 596 | 594 | 1190 |
| Vascular | 578 | 592 | 1170 |
| Miscellaneous | 573 | 587 | 1160 |
| Urogenital | 465 | 455 | 920 |
| Oncology | 379 | 400 | 779 |
| Obstetrics | 380 | 375 | 755 |
| Hepatobiliary | 294 | 309 | 603 |
| Spine | 308 | 288 | 596 |
| Gynaecology | 292 | 277 | 569 |
| Cardiac | 268 | 287 | 555 |
| Trauma | 280 | 262 | 542 |
| Breast | 152 | 184 | 336 |
| Haematology | 133 | 135 | 268 |
| Interventional | 94 | 86 | 180 |
| Forensic | 21 | 14 | 35 |
CNS = Central Nervous System
Label cardinality is defined as the average labels per article
Label density is defined as the label cardinality divided by the total number of possible labels
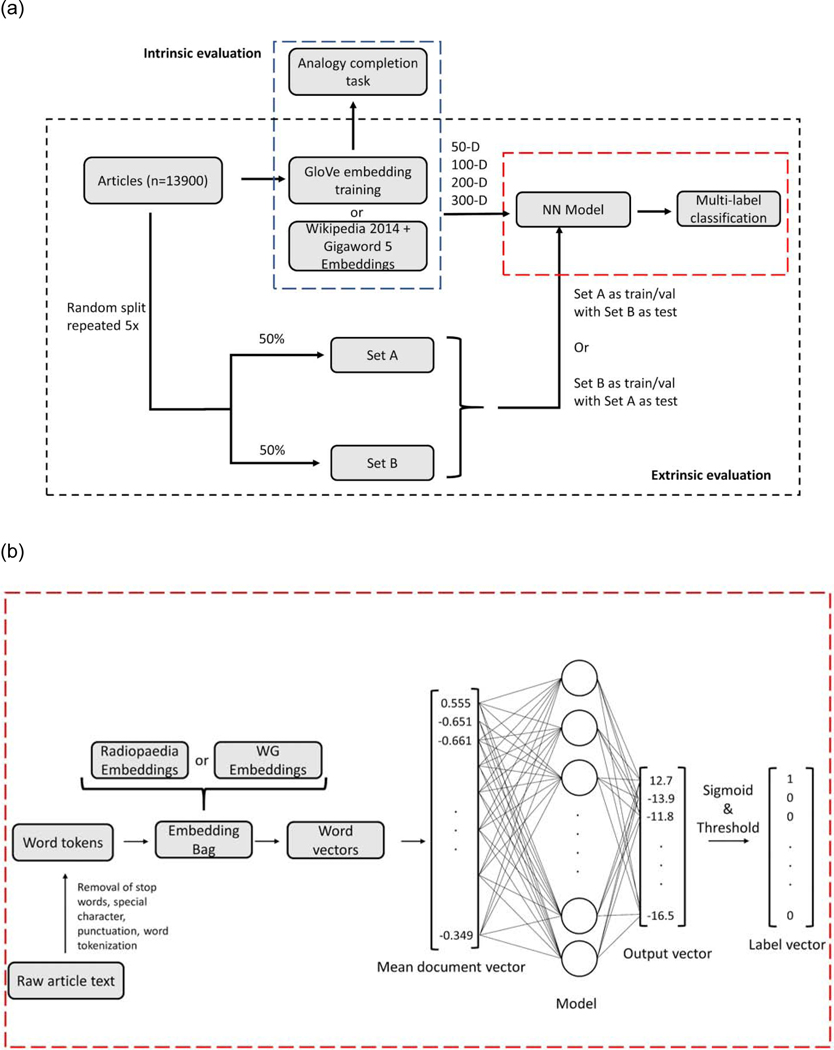
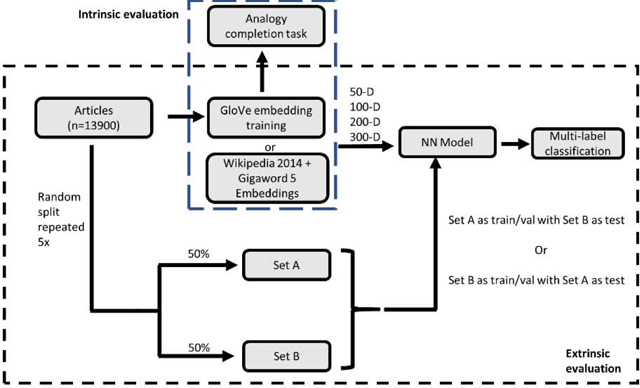
Word Embedding Training
Figure 1a depicts the process in which articles were split for embedding training. A Python based GloVe model (23) was used to train word embeddings on Radiopaedia.org text (approx. 2.2 million tokens), henceforth referred to as Radiopaedia embeddings. Hyperparameters for word embedding training were obtained from a previously published GloVe model (14) except using 25 training epochs for all dimensions: 50, 100, 200, and 300.
Figure 1.
(a) Schematic of intrinsic and extrinsic evaluation. The set of articles were randomly split 50–50 five times for extrinsic evaluation. Set A was used as the train/validation set and Set B as the test set for one instance and vice versa for a second instance. For 10 training sets and two embeddings types with 4 different dimensions (50,100,200,300) each, a total of 80 models were trained. (b) shows an expanded view of the model training pipeline for multi-label classification for a single model. Not all elements in vectors or neurons in the model are shown. Abbreviations: NN (Neural Network), WG (Wikipedia 2014 + Gigaword 5)
Intrinsic Evaluation
Intrinsic evaluation quantifies a word embedding’s ability to understand the relationship between words in a language domain (24). Existing datasets of embeddings such as the Google analogy test set (15) tend to evaluate non-medical relationships. There is currently no state-of-the-art medical analogy dataset, so a custom set of 1754 analogies was created. The creation of this set was done without referencing the embedding vocabularies or word frequencies. This set included 8 semantic categories. These categories were chosen by TC and JHS based on their relevance to radiology and the ability to create single word analogies with a concrete ground truth. The complete analogy dataset is publicly released (https://bit.ly/3hp4Vrg). Table 2 shows sample analogies from each category. A 3CosAdd method was implemented for analogy completion in which the word embedding model was tasked with determining word “d” given words a, b, and c in an analogy “a is to b as c is to d” (25,26).
Table 2.
Selected Sample Medical Analogies from Each Semantic Category
| Semantic Category | Analogy Examples† |
|---|---|
| Benign vs. Malignant | benign:malignant :: fibroma:fibrosarcoma benign:malignant :: leiomyoma:leiomyosarcoma myxoma:myxosarcoma :: adenoma:adenocarcinoma |
| Body Cavity | heart:thorax :: uterus:pelvis lung:thorax :: stomach:abdomen colon:abdomen :: bladder:pelvis |
| Bone Group | ulna:forearm :: tibia:leg radius:forearm :: ilium:hip fibula:leg :: scaphoid:wrist |
| Bone vs. Muscle | radius:bone :: gracilis:muscle humerus:bone :: deltoid:muscle fibula:bone :: trapezius:muscle |
| Organ Adjective | gastric:stomach :: hepatic:liver hepatic:liver :: ovarian:ovary pulmonary:lung :: thymic:thymus |
| Inflammation Location | pneumonia:lung :: encephalitis:brain pneumonitis:lung :: prostatitis:prostate myocarditis:heart :: endometritis:uterus |
| Surgical Procedures | mastectomy:breast :: colectomy:colon cholecystectomy:gallbladder :: hysterectomy:uterus pneumonectomy:lung :: cystectomy:bladder |
| Tumor Origin | astrocystoma:brain :: osteosarcoma:bone nephroblastoma:kidney :: hepatoblastoma:liver myxoma:heart :: thymoma:thymus |
a is to b as c is to d represented as a:b::c:d
Top 1 and top 3 analogy completion accuracy was compared between Radiopaedia embeddings and WG pre-trained GloVe embeddings. The accuracy was defined as correct analogies divided by total analogies. Input words to the analogy were suppressed in the output. In addition, Radiopaedia embeddings also suppressed output of words with occurrence frequency of one in the corpus in order to remove rare, misspelled words from the data curation process. This word filtering was not applied to WG embeddings as the vocabulary of these embeddings have already been sifted through a word-frequency filter prior to their release (14). Outputs were marked as correct only if the top output (top 1 accuracy) or top 3 outputs (top 3 accuracy) exactly matched the ground truth.
Extrinsic Evaluation
Extrinsic evaluations assess an embedding’s ability to facilitate downstream NLP tasks such as text classification and sentiment analysis (24). We evaluated the embeddings on a multi-label classification task on Radiopaedia articles. Article labels were derived from Radiopaedia.org’s 19 “System” labels that were already assigned to each article (20). A 20th “Miscellaneous” label was created and assigned to articles with no default label. Articles were randomly subdivided following a 5×2 cross-validation method which consists of 5 randomized 50–50 train-test splits (27). We randomly reserved 5% of the train subset for internal validation.
A shallow neural network was created in PyTorch using an EmbeddingBag layer with mean reduction and a Linear layer (28). The EmbeddingBag layer converts incoming word tokens into a word vector. All word vectors from a given document are averaged element-wise in mean reduction. The mean document vector is then used as input into a neural net for classification. Figure 1b depicts the model training pipeline. Model training froze the embedding layer and trained using the Adam optimizer to minimize PyTorch’s BCEWithLogitsLoss criterion. The BCEWithLogitsLoss function from PyTorch combines a sigmoid layer followed by binary cross-entropy loss together in its implementation which is done to take advantage of the log-sum-exp trick for numerical stability. If a token was not in the embedding vocabulary, the word was assigned a randomized vector constructed element-wise by randomly sampling from a Gaussian distribution that had a mean and standard deviation equal to the mean and standard deviation of that element across the training set of word embedding vectors. Two versions of the model were trained: one using Radiopaedia embeddings as input and the other using WG. Model output was interpreted using a threshold of 0.5. Performance was evaluated using an exact match accuracy and the Hamming loss (29). The Hamming loss can be interpreted as the proportion of incorrect labels relative to the total number of labels, thus a perfect Hamming loss value is 0.
Error analysis
Incorrect analogies and incorrectly labeled articles were manually reviewed for systematic errors. Incorrect analogies completed with proposed words were tabulated along with the corresponding cosine similarity. Top 3 outputs were examined for all incorrect analogies for 50-D RAD and 300-D WG which were the respective best performing models. The text of incorrectly labeled articles were manually examined for ambiguity or incorrect ground truth labels. Systematic errors identified from the process were recorded for further discussion. Error analysis was conducted by a radiology fellow (PGY-6, JHS), medical student (MS-4, TC), and undergraduate engineering student (ME) under the supervision of attending radiologist (15 years of experience, THV).
To further examine the errors produced by the models, 100 articles were randomly chosen from the pool of incorrectly labeled articles for each of the 8 models. Incorrectly labeled articles were defined as article labels that did not have an exact match. The labels produced by the model were compared with the ground truth and categorized into four categories by TC. Errors were categorized as no label, labels that were close to the ground truth, labels that were far off the ground truth, and questionable ground truth labels.
Data Visualization
We implemented t-Distributed Stochastic Neighbor Embedding (t-SNE) using Python’s scikit-learn package (30) to visualize the high-dimensional separation of predictions in a 2-dimensional space. t-SNE is a dimension reduction technique using Gaussian kernels to map high dimensional data to a lower dimension with the goal of preserving relative positions of data points (31). Input dimensions to t-SNE were first reduced to 30 using principal component analysis. Additionally, the high dimension embedding data was organized using a k-means algorithm to fit 20 clusters over 1000 iterations.
Statistical Analysis
McNemar tests with continuity correction were used to evaluate analogy performance. For each analogy subcategory, the Radiopaedia and WG embeddings of the same dimension were compared. Statistical significance was determined by applying a Benjamini-Hochberg correction on the p-values with false discovery rate of 0.05. Model performance was assessed using a 5×2 cross-validation paired t-test (27). A two-tailed test with p-value less than 0.05 was considered significant. All statistical analysis was carried out using R statistical software (v4.0.0) (32). All code for embedding training, intrinsic evaluation, extrinsic evaluation, and error analysis results can be found hosted at our GitHub repository (https://bit.ly/3hp4Vrg).
Results
Dataset Summary
A total of 13,900 articles (~2.2 million tokens) were collected. There were 9,191 articles with 1 system label, 3,673 with 2 labels, 807 with 3 labels, 144 with 4 labels, 48 with 5 labels, and 37 with 6 or more labels. The most common labels were “Musculoskeletal” and “CNS”. The “Forensic” and “Interventional” labels had the fewest associated articles.
Intrinsic Evaluation
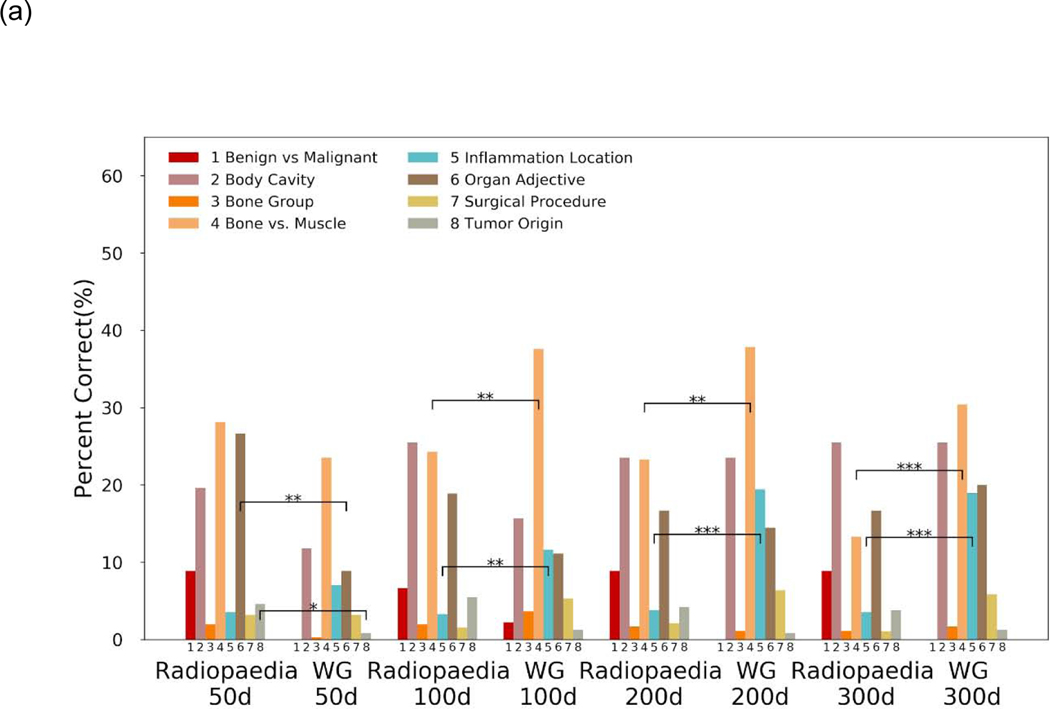
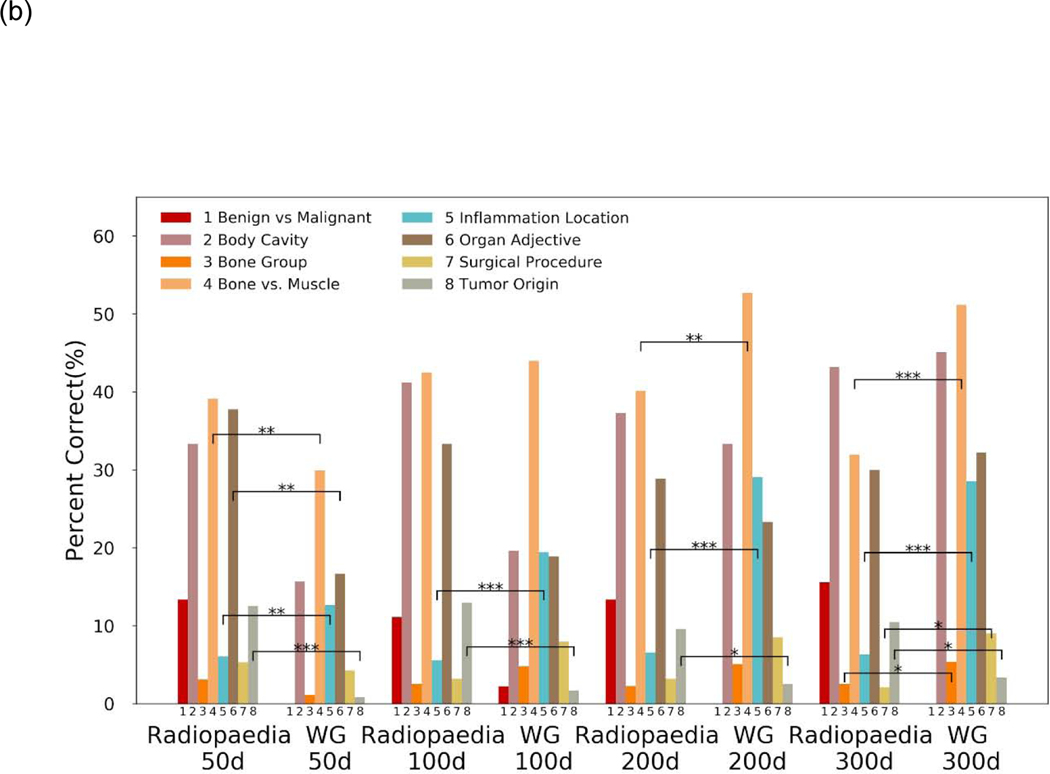
A total of 1,754 medico-radiological analogies were evaluated. Radiopaedia embeddings performed quite similarly to the Wikipedia-Gigaword embeddings for top 1 accuracy (Figure 2a). The 50-dimensional Radiopaedia embeddings performed the best of the Radiopaedia embeddings. Compared to the 50-D WG embeddings, the 50-D Radiopaedia embeddings had significantly better performance on tumor origin analogies (4.6% vs. 0.8% adjusted-p < 0.05) and organ adjectives (26.7% vs. 9.3% adjusted-p < 0.01). At 100-D and greater WG embeddings had better top 1 accuracy on inflammation and bone vs. muscle analogies (adjusted p < 0.01) (Figure 2a). Performance for top 3 accuracy reveals some additional underlying patterns (Figure 2b). While accuracy expectedly increased across the board due to the less stringent requirement, Radiopaedia embeddings saw greater improvements in top 3 accuracy performance in organ adjective, bone vs. muscle, and tumor origin analogies than WG embeddings (Supplementary Table 1 and 2). WG embeddings continued to outperform on inflammation location analogies.
Figure 2.
Analogy completion task performance by various embeddings for (a) top 1 accuracy results and (b) top 3 accuracy results. Performance is separated by semantic categories. Significant differences in category performance according to McNemar’s test with continuity correction between Radiopaedia and WG embeddings of a given dimension. Significance is denoted * for BH adjusted-p <0.05, ** for BH adjusted-p< 0.01 and *** for BH adjusted-p < 0.001. No marking means no statistical significance. Abbreviations: WG = Wikipedia 2014 + Gigaword 5 embeddings, d = embedding dimensions, BH = Benjamini-Hochberg
Extrinsic Evaluation
A 5×2 cross-validation method was used to compare models that used different pre-trained embeddings. Models were evaluated on a multi-label task of Radiopaedia articles. In each randomized 50–50 split, 6,950 articles were used as a testing set. The models that used embeddings trained on Radiopaedia outperformed models that used WG embeddings in exact match accuracy (50-D: by 0.100 p< 0.001; 100-D: 0.060, p<0.001; 200-D: 0.038, p<0.01;300-D: 0.020, p<0.05; Table 3) and Hamming loss for all dimensions (50-D: by −0.103, p<0.001;100-D:−0.0058, p<0.001,200-D:−0.0040, p<0.01;300-D:−0.0032,p<0.05; Table 3).
Table 3.
Model Performance on Multi-Labeling Task
| Parameter | 50-D | 100-D | 200-D | 300-D |
|---|---|---|---|---|
| Exact Match Accuracy (EMA) | ||||
| EMARadiopaedia | 0.358 | 0.440 | 0.476 | 0.487 |
| EMAWG | 0.258 | 0.380 | 0.438 | 0.467 |
| EMARadiopaedia - EMAWG‡ | 0.100 ±0.00546 |
0.060 ±0.00745 |
0.038 ±0.00740 |
0.020 ±0.00538 |
| p-value | < 0.001* | < 0.001* | 0.003* | 0.013* |
| Hamming Loss (HL) | ||||
| HLRadiopaedia | 0.0495 | 0.0436 | 0.0402 | 0.0393 |
| HLWG | 0.0598 | 0.0494 | 0.0442 | 0.0425 |
| HLRadiopaedia- HLWG‡ | −0.0103 ±0.000579 |
−0.0058 ±0.000693 |
−0.0040 ±0.000595 |
−0.0032 ±0.00110 |
| p-value | < 0.001* | < 0.001* | 0.001* | 0.032* |
Displayed exact match accuracy and Hamming loss correspond to the first permutation of the first seed.
WG = Wikipedia 2014 + Gigaword 5 Embeddings
errors are given as one standard deviation derived from the averaged 5×2 cross validation variance
indicates significant difference between Radiopaedia and WG
Data Visualization
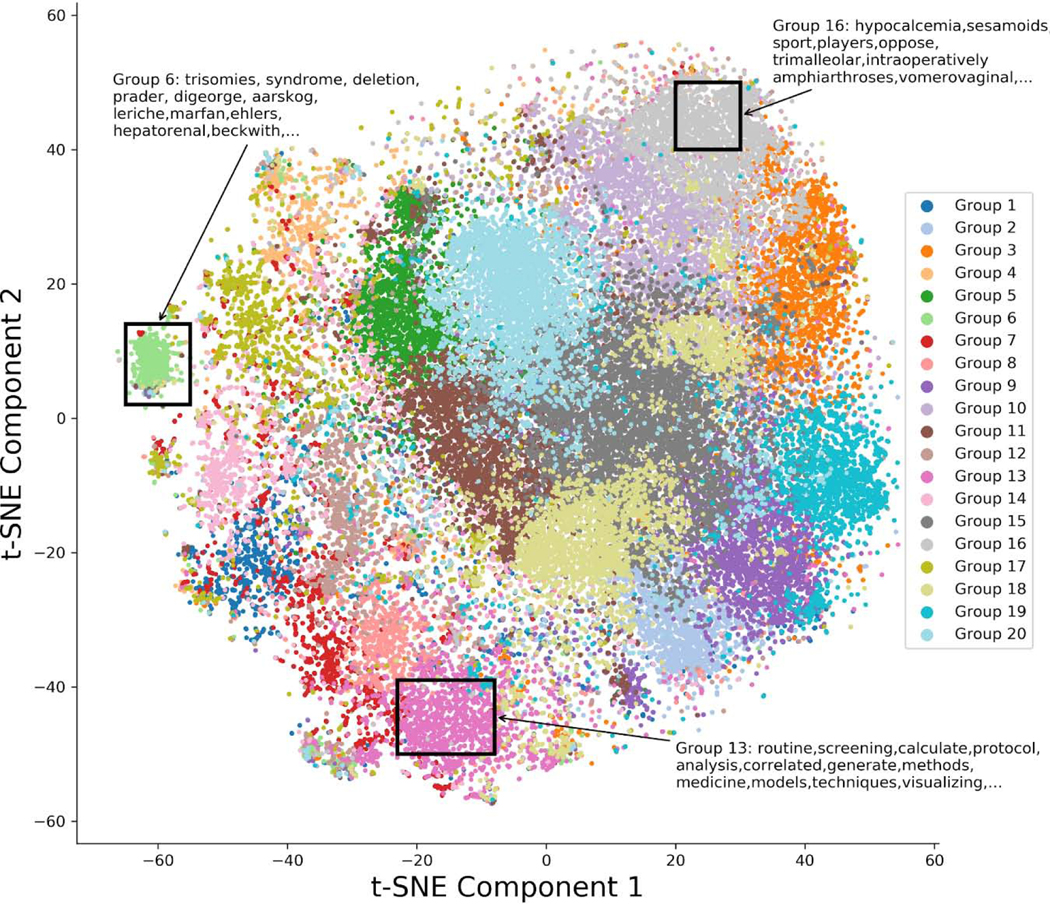
Manual examination of k-means clusters of the word-embedding revealed GloVe was able to group semantically similar words together. Figure 3 shows the t-SNE plot and sample words from selected regions. Some clusters, such as the Group 18 cluster, appear to have less well-defined boundaries for their points which may be due to the relative limitations of a single t-SNE map to visualize non-metric relationships. The t-SNE plot shows that the GloVE model was able to capture some of the semantic meaning between words as similar groups of terms are appropriately clustered close together.
Figure 3.
Two-dimensional t-distributed stochastic neighbor embedding plot of 50 dimensional Radiopaedia embeddings. Each point is colored according to each 2-D point’s respective 50-D k-means cluster assignment. Sample words from selected regions are shown. Similar words are grouped together indicating that the word embeddings were able to preserve the semantic meanings of words.
Error Analysis
Radiopaedia embeddings had 75 analogies marked incorrect due to an out of vocabulary word whereas WG embeddings had 649. Even when incorrect, the output generated from Radiopaedia embeddings tended to be closer to the ground truth than the WG embeddings (Table 5). Some of the analogies marked incorrect were due to Radiopaedia reporting the plural form of the ground truth. Radiopaedia embedding analogy completion output was subjectively more similar to ground truth than the WG embeddings which may explain why it saw much higher increase in performance from top 1 to top 3 accuracy.
Table 5.
Error Analysis for Intrinsic and Extrinsic Evaluations
| Model | Query | Ground Truth | Output | Interpretation |
|---|---|---|---|---|
| 50-D-RAD† | gastric:stomach::renal:? | kidney | (pelvis, 0.636) (kidneys, 0.623) (calyces,0.558) | Output is reasonably close to ground truth. Model output plural form of ground truth and thus was marked incorrect |
| 50-D-RAD | pubis:hip::trapezium:? | wrist | (replacements, 0.622) (scaphoid, 0.569) (arthroplasty, 0.565) | Output correctly identifies that the analogies are dealing with bones and even offers an analogous wrist bone. |
| 300-D-WG‡ | pubis:hip::trapezium:? | wrist | (hop,0.436) (pop,0.376) (rap,0.362) | Uses incorrect association of hip with hip-hop and other music related terms |
| 50-D-RAD | medulloblastoma:brain::myxoma:? | heart | (cardiac 0.700) (parenchyma, 0.623) (shocks, 0.619) | Output is reasonably close to ground truth but did not give the correct term |
| 300-D-WG | medulloblastoma:brain::myxoma:? | heart | (virus, 0.451) (muscles,0.418) (nerve,0.374) | Unclear association between output words and query |
| 300-D-RAD | “Triangulation is a technique for localizing lesions seen on at least two views on 2D mammography…” Ref. (20) | Breast | Breast, MSK | Close prediction – The model correctly identifies the major article label which is breast but additionally adds an erroneous MSK label likely due to all of the anatomical terms such as lateral and oblique stated throughout the article. |
| 300-D-RAD | “the four branches of the thoracoacromial artery are... A: acromial B: breast (pectoral) C: clavicular D: deltoid …” Ref. (20) | Chest, HN, Vascular | MSK | Distant prediction - The shallow neural net is unable to capture long term dependencies to understand the context of these otherwise MSK terms. |
| 300-D-RAD | “The sphenopetrosal suture is the cranial suture connecting the greater wing of sphenoid...” Ref. (20) | CNS,HN, MSK | CNS,HN | Questionable Ground Truth - A very reasonable prediction by the model that did not count towards its exact match accuracy due to a questionable ground truth label. There were some inconsistencies in what fell under MSK and what was HN in the ground truth labels. |
CNS = Central Nervous System, HN = head and neck, MSK = musculoskeletal, ONC = oncology For analogies, the top 3 outputs are shown along with associated cosine similarity
RAD = Radiopaedia embedding
WG = Wikipedia 2014 + Gigaword 5 embedding
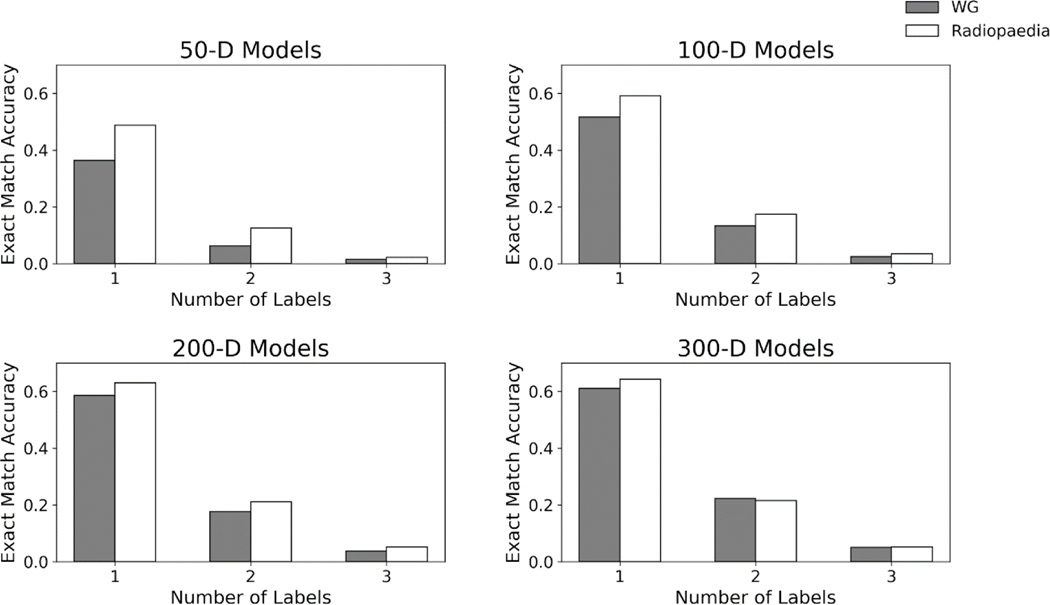
With a threshold of 0.5, the 50-D WG model had 3176 articles unlabeled out of the test set of 6950 articles; 100-D had 2034; 200-D had 1333, and 300-D had 840. For Radiopaedia embedding based models, 50-D had 2318; 100-D had 1641; 200-D had 1641, and 300-D had 1243 articles unlabeled. This was a major source for lowered exact match accuracy for all models. In addition, there was a sharp drop off in accuracy as the number of ground truth labels of an article increased (Figure 4). Table 4 shows the distribution of error types for the 100 randomly selected incorrectly labeled articles for each model. A chi-square test of homogeneity was performed between each of pair of models with the same dimensions. The distribution of error types were not significantly different between Radiopaedia and WG embeddings for 50-D: (χ2 = 0.59 p=0.90), 100-D: (χ2 = 2.57, p = 0.46), 200-D: (χ2 = 6.52, p = 0.09), and 300-D: (χ2 = 1.65, p = 0.65). Table 5 shows selected examples of erroneous predictions for intrinsic and extrinsic evaluations.
Figure 4.
Model performance stratified by the number of labels an article had. Dramatic drop in exact match accuracy was seen as the number of article labels increased. For articles with four or more ground truth labels, exact match accuracy was zero for all articles.
Table 4.
Classification of Article Label Errors
| Model | No Label | Close Prediction | Distant Prediction | Questionable Ground Truth |
|---|---|---|---|---|
| 50-D-Rad† | 60 | 19 | 10 | 11 |
| 50-D-WG‡ | 59 | 19 | 8 | 14 |
| 100-D-Rad | 44 | 32 | 8 | 16 |
| 100-D-WG | 55 | 24 | 7 | 14 |
| 200-D-Rad | 31 | 39 | 19 | 11 |
| 200-D-WG | 39 | 34 | 9 | 18 |
| 300-D-Rad | 31 | 37 | 13 | 19 |
| 300-D-WG | 25 | 44 | 15 | 16 |
RAD = Radiopaedia embedding
WG = Wikipedia 2014 + Gigaword 5 embedding
Discussion
We have shown that articles from Radiopaedia.org (20) can act as a radiology corpus that can train word embeddings that have a strong intrinsic understanding of medical language and facilitate the performance of a model on a radiology oriented natural language processing (NLP) task. The relatively larger increase in analogy completion performance from top 1 to top 3 accuracy seen with Radiopaedia trained embeddings supports the idea that its outputs are closer to the ground truth. Models that used Radiopaedia embeddings saw a 5–10% increase over Wikipedia 2014 + Gigaword 5 (WG) embeddings (14) in exact match accuracy at 50- and 100-Dimensions and approximately 3% increase at 200 and 300 dimensions which were statistically significant improvements. In both intrinsic and extrinsic evaluation, Radiopaedia embeddings had stronger relative performance at lower dimensions, suggesting that there are diminishing returns at dimensions beyond 100 for this version of a Radiopaedia GloVe model.
Deep learning NLP approaches have increased in popularity for automating extraction of information from radiological texts. However, NLP is still a relatively unexplored tool in radiology. For instance, transfer learning from ImageNet is an established approach in training computer vision models (33,34), yet similar approaches in radiology NLP models have been sparse. Some models do not use transfer learning at all and instead train their embeddings on their own domain specific corpus (19) while others have employed transfer learning by using pre-trained word embeddings from general web corpora (7,8). We have presented one use case where word embedding pretraining on a radiology corpus is useful, but additional research is required to precisely identify the most effective scenarios for using radiology domain specific word embeddings.
General web corpora present some issues when trying to use them for medical tasks. In the case of the popular pre-trained WG embeddings, there is an inherent issue with comprehensive coverage of radiological language and more importantly enormous distractions from non-medical terms, as was highlighted in the example of “hip” being associated with “hop”. Furthermore, the WG embedding vocabulary is truncated based on word frequency which will naturally remove the esoteric words of medicine. A drawback of non-context dependent word embeddings, such as those generated by GloVe, is that a single vector is used to represent all the meanings of a word. Words such as “film” have very different every day and medical implications. When derived from a general corpus, embeddings for such words can have their colloquial meanings dominate their semantics which may decrease their applicability to radiology.
Wikipedia represents an intriguing corpus, having both general and radiology specific articles. Some previous studies used Pennington and coworker’s GloVe embeddings that were pretrained on Wikipedia (7,8). We aimed to show that embeddings trained on a radiology specific corpus could offer similar or better performance than a general corpus. Combined with its vast token size, Wikipedia trained embeddings could be expected to outperform Radiopaedia embeddings, but this was not the case in our study. Despite being trained on a corpus about 2700 times smaller, the Radiopaedia embeddings offered performance comparable to and sometimes better than the WG embeddings on a medico-radiological analogy completion task, showing that text relevancy may be as important as quantity in creating optimal medical embeddings. This performance may be attributed to the highly succinct nature of Radiopaedia articles which allows for word relationships to form without noisy associations with non-medical words; although, this succinctness also makes it more difficult for a model to obtain grammatical understanding of the language.
The Radiopaedia embeddings had encouraging performance on analogies involving semantically difficult analogies such as body cavity and tumor origin analogies. Even for analogies that were incorrect, the Radiopaedia embeddings choices tended to be closer to the underlying semantic meaning than the pre-trained WG embeddings. The overall performance of Radiopaedia embeddings in this study was likely underestimated. Error analysis of the intrinsic evaluation revealed numerous cases where the answer was off only by plural or different forms of adjectives. For extrinsic evaluation, exact match accuracy was expectedly low due to the stringent requirements of the metric. However, error analysis revealed that in numerous instances the ground truth labels were somewhat arbitrary.
This word embeddings set, released as open source, has not reached its full potential as it only integrates a single resource. Radiopaedia does not have to be used in isolation but can rather act as a cornerstone for creating a more comprehensive radiology corpus similar to MIMIC-III which acts as a comprehensive critical care clinical corpus (35). There has already been some work in developing large-scale standardized corpora for radiology. For instance, the MIMIC-CXR dataset (36) was developed to help address the need for a standardized radiology dataset. Creating large clinical report datasets like MIMIC-CXR or RadCore (37) requires extensive processing to de-identify reports. Our results suggest that general radiology text, which are more accessible than clinical data, can also train models that can capture semantics relevant to radiology. In the future, assuming protected health information concerns are addressed and licensing issues are overcome, a more versatile corpus can be created by using not only clinical radiology reports but also general radiology text such as commercial radiology textbooks, and open resources such as the radiology articles in Radiopaedia, Wikipedia, and PubMed abstracts. Future NLP work in radiology can take advantage of this corpus by using it to train word embeddings or using it as a benchmarking standard for various radiology NLP tasks.
Our study had several limitations. The first is the potential bias of Radiopaedia trained embeddings being used to classify Radiopaedia articles. However, the word embedding training process was blind to the ground truth labels of the articles, though bias from the word choices of Radiopaedia articles indeed remains. A second limitation of our study is the limited number of analogies we could create. Due to limitations on word embedding creation, terms that spanned multiple words (e.g. squamous cell carcinoma) could not be used. Strict analogy completion is also a difficult task given the many similar meaning words in medical vocabulary, and thus analogies needed to be limited to those with as concrete of a ground truth as possible. The set of analogies evaluated was not a true random sample from the set of all possible medical analogies, so statistical significance should be interpreted with caution. We also did not investigate syntactic analogies as we were more interested in the semantic knowledge that Radiopaedia offers. The classification of errors is inherently subjective, and thus there may be variability in classification and interpretation of the errors. Therefore, we have released the full data used in our error analysis. Finally, while we have shown that our embeddings can enhance a model’s performance on an NLP task regarding text relevant to radiology, an equivalent enhancement may not be seen when using these embeddings on a separate corpus (e.g. radiology reports).
Radiopaedia.org is a valuable resource that contains many domain-specific word relationships relevant to developing NLP tools for radiology. We have created a set of medical analogies and shown that embeddings trained on Radiopaedia are able to capture difficult medical semantics at the same level or sometimes better than a comprehensive corpus order of magnitudes larger. These embeddings have shown potential to enhance model performance on a radiology article multi-label classification task. Further work can characterize how these embeddings can improve performance on clinical NLP tasks or identify when certain types of pre-trained embeddings may be more appropriate than others. Our results suggest that the cultivation of a radiology-specific corpus can benefit radiology NLP models in the future.
Supplementary Material
Highlights.
Radiopaedia can be used as a domain-specific corpus in radiology NLP tasks
Domain specific embeddings offer comparable performance on analogy completion
Domain specific embeddings did significantly better on multi-label classification
The source code, embeddings, and analogy dataset are publicly released
Acknowledgments:
JHS was supported by the National Institute of Biomedical Imaging and Bioengineering (NIBIB) 5T32EB001631 Grant.
Footnotes
NLP - Natural Language Processing ; GloVe - Global Vectors for Word Representation; WG - WIkipedia 2014 + Gigaword 5; t-SNE - t-Distributed Stochastic Neighbor Embedding; CNS - Central Nervous System; EMA - Exact Match Accuracy; HL - Hamming Loss
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Langlotz CP. RadLex: A New Method for Indexing Online Educational Materials. RadioGraphics. Radiological Society of North America; 2006;26(6):1595–1597. [DOI] [PubMed] [Google Scholar]
- 2.SNOMED CT. U.S. National Library of Medicine; https://www.nlm.nih.gov/healthit/snomedct/index.html. Accessed August 2, 2020. [Google Scholar]
- 3.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1(2):161–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pons E, Braun LMM, Hunink MGM, Kors JA. Natural Language Processing in Radiology: A Systematic Review. Radiology. 2016;279(2):329–343. [DOI] [PubMed] [Google Scholar]
- 5.Kreimeyer K, Foster M, Pandey A, et al. Natural language processing systems for capturing and standardizing unstructured clinical information: A systematic review. J Biomed Inform. 2017;73:14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorin V, Barash Y, Konen E, Klang E. Deep Learning for Natural Language Processing in Radiology-Fundamentals and a Systematic Review. J Am Coll Radiol JACR. 2020;17(5):639–648. [DOI] [PubMed] [Google Scholar]
- 7.Chen MC, Ball RL, Yang L, et al. Deep Learning to Classify Radiology Free-Text Reports. Radiology. Radiological Society of North America; 2017;286(3):845–852. [DOI] [PubMed] [Google Scholar]
- 8.Banerjee I, Ling Y, Chen MC, et al. Comparative effectiveness of convolutional neural network (CNN) and recurrent neural network (RNN) architectures for radiology text report classification. Artif Intell Med. 2019;97:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miao S, Xu T, Wu Y, et al. Extraction of BI-RADS findings from breast ultrasound reports in Chinese using deep learning approaches. Int J Med Inf. 2018;119:17–21. [DOI] [PubMed] [Google Scholar]
- 10.Lee C, Kim Y, Kim YS, Jang J. Automatic Disease Annotation From Radiology Reports Using Artificial Intelligence Implemented by a Recurrent Neural Network. AJR Am J Roentgenol. 2019;212(4):734–740. [DOI] [PubMed] [Google Scholar]
- 11.Banerjee I, Chen MC, Lungren MP, Rubin DL. Radiology report annotation using intelligent word embeddings: Applied to multi-institutional chest CT cohort. J Biomed Inform. 2018;77:11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendonça EA, Haas J, Shagina L, Larson E, Friedman C. Extracting information on pneumonia in infants using natural language processing of radiology reports. J Biomed Inform. 2005;38(4):314–321. [DOI] [PubMed] [Google Scholar]
- 13.Dublin S, Baldwin E, Walker RL, et al. Natural Language Processing to identify pneumonia from radiology reports. Pharmacoepidemiol Drug Saf. 2013;22(8):834–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pennington J, Socher R, Manning C. Glove: Global Vectors for Word Representation. Proc 2014 Conf Empir Methods Nat Lang Process EMNLP Doha, Qatar: Association for Computational Linguistics; 2014. p. 1532–543 https://www.aclweb.org/anthology/D14-1162. Accessed May 10, 2020. [Google Scholar]
- 15.Mikolov T, Chen K, Corrado G, Dean J. Efficient Estimation of Word Representations in Vector Space. ArXiv13013781 Cs. 2013;http://arxiv.org/abs/1301.3781. Accessed June 1, 2020. [Google Scholar]
- 16.Banerjee I, Madhavan S, Goldman RE, Rubin DL. Intelligent Word Embeddings of Free-Text Radiology Reports. ArXiv171106968 Cs. 2017;http://arxiv.org/abs/1711.06968. Accessed June 1, 2020. [PMC free article] [PubMed] [Google Scholar]
- 17.Jagannatha AN, Yu H. Bidirectional RNN for Medical Event Detection in Electronic Health Records. Proc Conf Assoc Comput Linguist North Am Chapter Meet. 2016;2016:473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Y. Recurrent neural networks for classifying relations in clinical notes. J Biomed Inform. 2017;72:85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan J, Zhu H, Tahmasebi A. Classification of Pulmonary Nodular Findings based on Characterization of Change using Radiology Reports. AMIA Summits Transl Sci Proc. 2019;2019:285–294. [PMC free article] [PubMed] [Google Scholar]
- 20.Radiopaedia.org, the wiki-based collaborative Radiology resource. Radiopaedia. https://radiopaedia.org/?lang=us. Accessed June 1, 2020.
- 21.Richardson, Leonard. Beautiful soup documentation. 2007.
- 22.Bird S, Loper E, Klein E. Natural Language Processing with Python. O’Reilly Media Inc; 2009. [Google Scholar]
- 23.Kula M. maciejkula/glove-python. 2020https://github.com/maciejkula/glove-python. Accessed June 1, 2020. [Google Scholar]
- 24.Schnabel T, Labutov I, Mimno D, Joachims T. Evaluation methods for unsupervised word embeddings. Proc 2015 Conf Empir Methods Nat Lang Process Lisbon, Portugal: Association for Computational Linguistics; 2015. p. 298–307 https://www.aclweb.org/anthology/D15-1036. Accessed June 1, 2020. [Google Scholar]
- 25.Levy O, Goldberg Y. Linguistic Regularities in Sparse and Explicit Word Representations. Proc Eighteenth Conf Comput Nat Lang Learn Ann Arbor, Michigan: Association for Computational Linguistics; 2014. p. 171–180https://www.aclweb.org/anthology/W14-1618. Accessed June 1, 2020. [Google Scholar]
- 26.Mikolov T, Yih W, Zweig G. Linguistic Regularities in Continuous Space Word Representations. Proc 2013 Conf North Am Chapter Assoc Comput Linguist Hum Lang Technol Atlanta, Georgia: Association for Computational Linguistics; 2013. p. 746–751https://www.aclweb.org/anthology/N13-1090. Accessed June 9, 2020. [Google Scholar]
- 27.Dietterich TG. Approximate statistical tests for comparing supervised classification learning algorithms. Neural Comput. 1998;10(7):1895–1923. [DOI] [PubMed] [Google Scholar]
- 28.Paszke A, Gross S, Massa F, et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library In: Wallach H, Larochelle H, Beygelzimer A, Alché-Buc F, Fox E, Garnett R, editors. Adv Neural Inf Process Syst 32. Curran Associates, Inc; 2019. p. 8026–8037http://papers.nips.cc/paper/9015-pytorch-an-imperative-style-high-performance-deep-learning-library.pdf. Accessed June 3, 2020. [Google Scholar]
- 29.Tsoumakas G, Katakis I. Multi-Label Classification: An Overview. Int J Data Warehous Min. 2009;3:1–13. [Google Scholar]
- 30.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12(85):2825–2830. [Google Scholar]
- 31.van der Maaten L, Hinton G. Visualizing Data using t-SNE. J Mach Learn Res. 2008;9(Nov):2579–2605. [Google Scholar]
- 32.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 33.Deng J, Dong W, Socher R, Li L-J, Kai Li, Li Fei-Fe. ImageNet: A large-scale hierarchical image database. 2009 IEEE Conf Comput Vis Pattern Recognit 2009. p. 248–255. [Google Scholar]
- 34.Shin H-C, Roth HR, Gao M, et al. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans Med Imaging. 2016;35(5):1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data. Nature Publishing Group; 2016;3(1):160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson AEW, Pollard TJ, Berkowitz SJ, et al. MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Sci Data. Nature Publishing Group; 2019;6(1):317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hassanpour S, Langlotz CP. Information extraction from multi-institutional radiology reports. Artif Intell Med. 2016;66:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.