Abstract
Borrelia burgdorferi is the causative agent of Lyme disease, the leading tick-borne illness in the United States. However, due to, in part, to the significant number of proteins of unknown function encoded across the complex fragmented genome, the molecular mechanisms of B. burgdorferi infection remain largely undefined. Previous work identified the virulence determinant gene, bbk13, which is critical for B. burgdorferi’s ability to establish a productive disseminated infection. BBK13 is an immunogenic, non-surface exposed protein of unknown function predicted to harbor an N-terminal transmembrane domain and annotated as a member of the SIMPL domain protein superfamily (PF04402). In eukaryotes, SIMPL domain proteins have been shown to contribute to NF-kappa-B signaling but have no known functions in prokaryotes. Herein we investigated the biochemical and biophysical properties of BBK13 toward elucidation of its function. Bioinformatics analysis revealed secondary and tertiary structural homology between BBK13 and two other prokaryotic SIMPL domain proteins for which the crystal structures have been solved, Brucella abortus BP26 and Campylobacter jejuni cjSLP. Furthermore, comparable to BP26, recombinant BBK13 self-assembled into multimeric complexes in vitro and endogenous BBK13 was found in large oligomeric complexes in the spirochete membrane. Together these data suggest that the oligomeric structure of BBK13 may be important for the molecular function of this critical infection protein.
Keywords: Borrelia burgdorferi, Lyme disease, bbk13, lp36, SIMPL domain protein, blue native gel, size exclusion chromatography, far western blot
INTRODUCTION
The causative agent of Lyme disease in North America is the spirochete Borrelia burgdorferi [1, 2], which is transmitted to humans through the bite of an infected Ixodes scapularis tick [3]. Through the initial infected tick bite site, B. burgdorferi disseminates through the blood of its mammalian host, leading to colonization of distal tissues. B. burgdorferi colonization leads to multiple pathologies that occur in the joints (Lyme arthritis), the heart (Lyme carditis), and the central nervous system (Lyme neuroborreliosis) [4].
We recently identified the crucial role of gene bbk13, located on virulence-associated linear plasmid 36 (lp36), in the establishment of early B. burgdorferi infection [5]. Borrelia burgdorferi lacking bbk13 demonstrated a significant reduction in population expansion within the skin inoculation site, resulting in attenuated disseminated infection. Furthermore, we demonstrated that BBK13 is an immunogenic protein that localizes to the B. burgdorferi membrane but is likely not surface exposed [5]. These findings are consistent with the association of BBK13 with B. burgdorferi inner membrane lipid rafts [6]. BBK13 is a 232 amino acid protein, which is predicted to harbor an N-terminal transmembrane domain and is annotated as a member of the SIMPL domain protein superfamily (PF04402) [7]. In eukaryotes, SIMPL domain proteins have been shown to function in NF-kappa-B signaling regulation [8]. Although SIMPL domain-containing proteins are present across pathogenic and non-pathogenic bacterial species [5, 9], the function(s) of these proteins in prokaryotes remain unknown. The crystal structures of two bacterial SIMPL domain proteins, BP26 from Brucella abortus and cjSLP fromCampylobacter jejuni, have been solved [9, 10]. Although these two proteins share little primary sequence identity, both consist of two domains that adopt an α-helix-decorated β-sheet secondary structure and a boomerang-shaped tertiary structure [9, 10]. BP26 is found in a hexadecameric complex that forms a channel-like structure [10]. In contrast, cjSLP is monomeric in solution [9]. It has been suggested that BP26 and cjSLP represent distinct groups of bacterial SIMPL domain proteins each with unique local structural features and oligomeric states.
Herein we investigated the biochemical and biophysical properties of BBK13. We determined that BBK13 likely shares a similar secondary and tertiary structure with the other SIMPL domain proteins, particularly BP26. Further, like BP26, recombinant BBK13 self-assembled into multimeric complexes in vitro. Moreover, endogenous BBK13 was found in large oligomeric complexes in the spirochete membrane, suggesting that the oligomeric structure is important for the molecular function of this protein.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The B. burgdorferi clones used in this study were derived from the low-passage infectious clone B31 A3-68 Δbbe02, which lacks the plasmids cp9 and lp56, along with the gene bbe02 on lp25 [11]. The generation of the mutant Δbbk13/pBSV2G (Δbbk13) and Δbbk13/ pBSV2G bbk13+ (Δbbk13/bbk13+) complement B. burgdorferi clones was described previously [5]. Spirochetes were cultured at 35°C in Barbour-Stoenner-Kelly II (BSKII) growth medium containing gelatin and 6% rabbit serum. As needed for B. burgdorferi growth, the following antibiotics were used: kanamycin (200 μg/mL); streptomycin (50 μg/mL); gentamycin (40 μg/mL). Cloning steps were performed using Escherichia coli DH5α and recombinant protein was produced in Escherichia coli NiCo21(DE3) (New England Biolabs), grown in LB broth liquid medium or on LB agar plates. Kanamycin was used for E. coli cultures at 50 μg/ml or ampicillin at 100 μg/ml, as appropriate.
BBK13 sequence alignment and modeling
Amino acid sequence alignment of BBK13 (AAC66132.2), BP26 (AAO39767.1) and cjSLP (A0A3Z9IF87) was performed using Clustal Omega (MegAlign Pro, DNASTAR Lasergene 16). Secondary and tertiary structural modeling was performed with I-TASSER [12–14] and Phyre2 [15].
Expression and purification of recombinant BBK13 proteins
Expression and purification of recombinant 6xHis tagged BBK1325-232 (rBBK13-HIS) was performed as previously described [5]. Briefly, the bbk13 DNA sequence minus the transmembrane domain was cloned into the His tag protein expression vector pET28A. Protein production in E. coli was induced with 1mM isopropyl-β-D-thiogalactopyranoside (IPTG) and purified using Ni Sepharose high performance histidine tagged protein purification resin (Cytiva). Following histidine purification, eluted protein was further purified by cation exchange using strong cation-exchange minispin columns (Pierce). The rBBK13-HIS purity was confirmed by Coomassie stained SDS-PAGE gels and quantified using Bradford protein reagent (Thermo Scientific). Generation of the rGST-BBK13 fusion protein was executed by amplifying the BBK13 DNA sequence, excluding the transmembrane domain, from B31 A3 genomic DNA using Phusion DNA polymerase (Thermo Scientific) and primers 2336 (5’ gttgttGGATCCggaataaaaaacattggcacc 3’) and 2459 (5’ ggccgCTCGAGctaatccaaataataagaaacgg 3’). The BBK13 fragment was cloned into the glutathione S-transferase (GST) fusion protein expression vector pGEX-6p1 using restriction enzymes BamHI and XhoI and verified by DNA sequence analysis. Protein production was carried out in E. coli NiCo21(DE3) cells grown in LB at 37°C with shaking to an optical density at 600nm of ~1.3. The induction of protein expression was achieved by the addition of 1mM IPTG per culture and incubated at 18°C with shaking for 16 hours. Cells were harvested by centrifugation and lysed by sonication. Recombinant GST-BBK13 was purified using glutathione sepharose 4B according to the manufacturer’s protocol (Cytiva). Purity of rGST-BBK13 was determined by Coomassie stained SDS-PAGE gel. The relative rGST-BBK13 protein concentration was determined using a Bradford assay (Thermo Scientific).
Far-western blot
Two μg of rBBK13-HIS were separated by SDS-PAGE on 4-12% Tris/glycine polyacrylamide gels (Invitrogen) and transferred to nitrocellulose membranes. Membranes were then blocked and probed overnight at 4°C in Tris-buffered saline-Tween (TBST; 25 mM Tris, 150 mM NaCl, 0.1% Tween) + 2% skim milk, supplemented with either 2 μg/ml purified rGST or rGST-BBK13. Following overnight incubation membranes were washed and probed with primary antibody anti-GST (1:10,000) followed by goat anti-mouse secondary antibodies conjugated to HRP. Loading control of rBBK13-HIS was performed by blotting membranes with anti-BBK13 (1:10,000) [5] followed by secondary goat anti-rabbit antibodies conjugated to HRP.
Blue native-PAGE analysis
Analysis of native rGST-BBK13 by BN-PAGE was performed as previously described [16]. BN-PAGE gels were visualized by incubation in Imperial protein stain (Coomassie R-250) (Thermo Scientific). To evaluate rGST-BBK13 complex migration, the relative migration distance (Rf) of the native protein standards, size range 20-1,200 kDa, (Invitrogen) and were calculated and plotted against molecular mass as previously described [16] and the molecular masses of rGST-BBK13 complex species were estimated.
Analytical Size Exclusion Chromatography
Fast protein liquid chromatography with High Prep 16/60 Sephacryl S-300 resin (Cytiva) was used to analyze the oligomeric states of 500 μg rGST-BBK13. Protein standards used to calibrate the SEC resin: Thyroglobulin (669 kDa), Ferritin (440 kDa), Aldolase (158 kDa), Conalbumin (75 kDa), Ovalbumin (43 kDa) (Cytiva). The corresponding plot of the log of the molecular mass of the protein standards as a function of their elution volume (r2 = 0.9638) was used to estimate the apparent molecular mass of the native rGST-BBK13 complex species. Immunoblot validation of BBK13 containing fractions was performed using anti-BBK13 antibodies, as described above.
Isolation of Borrelia burgdorferi membrane fraction
Subcellular fractionation of B. burgdorferi was performed as described previously [5]. Briefly, WT, Δbbk13 and Δbbk13/bbk13+ B. burgdorferi were grown to mid-log phase (5 × 107 to 7 × 107/ml) in BSKII growth medium, pelleted at 3,210 × g for 10 minutes, washed twice with cold HN buffer (50 mM HEPES, 50 mM NaCl, pH 7.5) and resuspended in 1 ml of HN buffer and 10 μl of Halt protease inhibitor cocktail (Thermo Scientific). Samples were lysed via sonication and spun at 125,000 × g to separate the membrane and soluble components. The membrane fraction was immediately used for membrane protein solubilization/extraction (see below).
Two dimensional Blue native-PAGE
Blue native polyacrylamide gel electrophoresis (BN-PAGE) of native membrane proteins was performed as previously described [17, 18]. Membrane proteins were solubilized with protein extraction buffer consisting of β-dodecyl maltoside (DM) (1.5% w/v) and 750 mM aminocaproic acid dissolved in 50 mM Bis-Tris pH 7.0. Samples were incubated on ice for 30 minutes and mixed by pipetting every 10 minutes. Solubilized membrane proteins were separated from cellular debris by centrifugation at 14,000 × g for 30 minutes at 4°C and supernatant was transferred and diluted to 1×107 spirochete/μl in BNG sample buffer (3% sucrose, 50 mM 6-aminocaproic acid, 10 mM Bis-Tris pH 7.0, and 0.5% Coomassie G250). For 2-dimensional studies, BN-PAGE lanes were excised and incubated in SDS-PAGE equilibration buffer (25 mM Tris pH 7.5, 1% SDS w/v, 1% DTT w/v) for 30 minutes. Second dimension SDS-PAGE separation of individual proteins was performed using 10% polyacrylamide gels containing 0.1% SDS. Following second dimension separation, proteins were transferred onto nitrocellulose membrane and blocked in 5% skim milk dissolved in TBST. Immunoblotting was performed with antibodies against BBK13 [5] (1:10,000) or OspA [19] (H5332, 1:1,000).
Immunofluorescence
1.5 ml of mid-log B. burgdorferi culture was pelleted at 1000 Δ g for 3 minutes and resuspended in PBS by gentle pipetting. B. burgdorferi were adhered to poly-L-lysine-coated 8-well chamber slides and fixed with 2% methanol-free formaldehyde in PBS with 0.01% v/v Triton-X-100 (PBT). Blocking was performed 300 μl of PBT with 5% w/v bovine serum albumin (PBT-BSA) followed by an endogenous biotin blocking kit (Invitrogen). Rabbit polyclonal antibodies against BBK13 [5] were biotinylated using the APEX™ Biotin-XX antibody labeling kit, according to the manufacturer’s instructions (Invitrogen). Spirochetes were incubated for 48 hours at 4°C without agitation in 1:1000 biotinylated anti-BBK13 antibodies in PBT-BSA followed by incubation with 1:400 Alexa 488-conjugated streptavidin and 1:1000 DAPI (Sigma) in PBT-BSA for 1 hour.
Surface expression of BBK13 was examined as described [20]. Membrane permeabilization was achieved by incubating slides in PBS with 0.1% Triton-X-100 for 30 minutes at room temperature. Incubation of a duplicate slide in PBS-MgCl2 served as a non-permeabilized control. Samples were blocked with PBS-MgCL2 with 0.75% w/v BSA followed by an endogenous biotin blocking kit (Invitrogen). Samples were incubation with undiluted biotinylated anti-BBK13 antibodies at 37°C for 4 hours followed by incubation with 1:400 Alexa 488-conjugated streptavidin in blocking solution for 30 minutes at 37°C.
Fluorescent and phase contrast images were acquired on a Zeiss LSM 710 confocal microscope using either a Plan-Apochromat 100X/1.46 oil objective or a 20X Plan-Apochromat 20X/0.8 air objective. Image acquisition settings and minor adjustments of brightness and contrast after image acquisition were identical across all B. burgdorferi clones within the same experiment.
RESULTS
BBK13 demonstrates strong structural homology to BP26.
Comparative analysis of the crystal structures of BP26 and cjSLP suggests that these two proteins may represent different subgroups of bacterial SIMPL domain containing proteins [9]. Alignment of the primary amino acid sequences of BBK13, BP26 and cjSLP (Figure 1A) revealed low sequence similarity between the three proteins. Secondary and tertiary structural modeling of BBK13 was performed using I-TASSER [12–14] (Figure 1). The predicted secondary structure for BBK13 was comprised of 3-4 α-helices and 12 β-strands (Figure 1A, B). Structural alignment using TM-align demonstrated high structural similarity between the BBK13 model and BP26 (PDB: 4hvz) and resulted in a root-mean-square deviation (RMSD) of 1.662 Å. The RMSD value represents the average distance between the atoms of the structures of two proteins after optimal superposition. The lower the RMSD, the more similar the two protein structures [21]. Less structural similarity was found between the BBK13 model and cjSLP (PBD: 7c50), with an RMSD of 4.276 Å (Figure 1B). Together these data suggest that BBK13 is more closely related to BP26.
Figure 1. BBK13 demonstrates structural similarity to known SIMPL domain proteins.
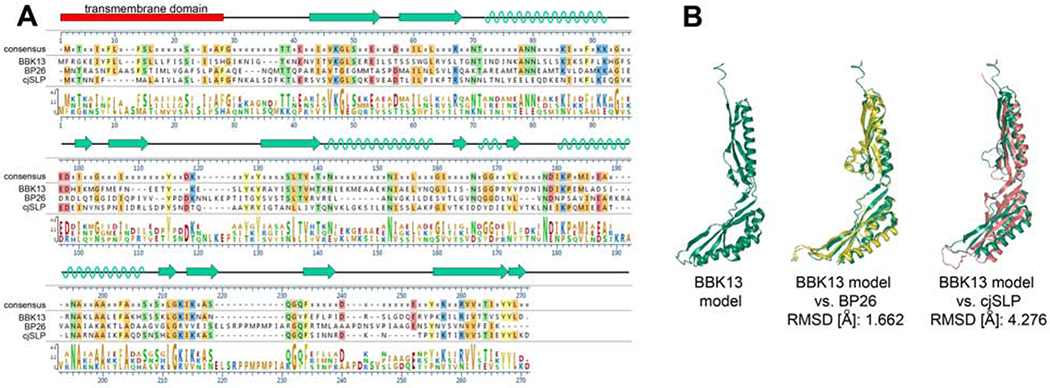
(A) Amino acid sequence alignment of BBK13, BP26 and cjSLP. Amino acids identical to the consensus sequence are highlighted according to their side chain chemistry. Amino acid conservation is also displayed graphically. The I-TASSER predicted alpha helix (green helix) and beta sheet (green arrows) secondary structure of BBK13 is shown. (B) The I-TASSER tertiary structure model of BBK13 (green) alone and the BBK13 model aligned with BP26 structure (yellow) or cjSLP structure (pink). The root-mean-square deviation (RMSD) values for the aligned structures are shown.
Recombinant BBK13 demonstrates protein-protein interaction with itself.
Given the finding that BBK13 is predicted to have strong structural homology to BP26 we sought to examine the ability of BBK13 to associate with itself to form multimers. To begin, we performed a far-western blot to assess BBK13-BBK13 interaction. Two versions of recombinant BBK13 with unique epitope tags, one with a C-terminal histidine tag (rBBK13-HIS) and another with an N-terminal fusion with glutathione S-transferase (rGST-BBK13), were used. The rBBK13-HIS protein was resolved by SDS-PAGE and transferred to a nitrocellulose membrane. The rBBK13-HIS containing membrane was probed with rGST-BBK13 or rGST alone and an immunoblot using anti-GST primary antibodies was performed (Figure 2A). The far-western blot probed with rGST-BBK13 resulted in detection of a protein of ~24 kDa, the theoretical molecular mass of rBBK13-HIS, whereas the far-western blot probed with rGST alone did not (Figure 2B), together indicating that BBK13 interacts with itself. Equal loading of the rBBK13-HIS “bait” was confirmed by traditional immunoblot with anti-BBK13 antibodies (Figure 2C).
Figure 2. BBK13 to BBK13 interactions are detected by far-western blot.
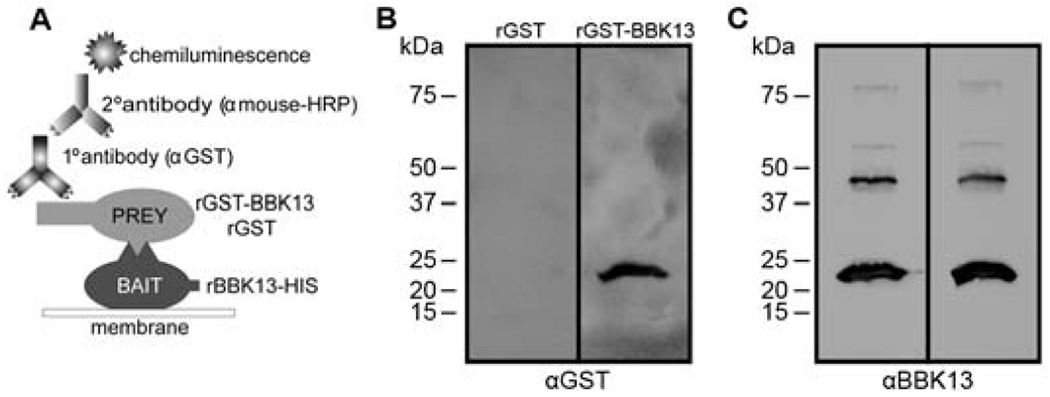
(A) Schematic representation of the far-western blot method used to identify recombinant BBK13 interactions. (B) rBBK13-HIS was resolved by SDS-PAGE and transferred to a nitrocellulose membrane. The membranes were probed with either rGST-BBK13 or rGST (negative control). Protein-protein interactions were detected by immunoblot with antibodies against GST (αGST). (C) The nitrocellulose membranes were stripped and reprobed with antibodies against BBK13 (αBBK13). Molecular mass standards are shown in kilodaltons (kDa).
Recombinant BBK13 forms multimeric complexes in solution.
To examine the native state of rGST-BBK13 in solution, the purified protein, along with recombinant GST (rGST) alone as a negative control, was subjected to blue native-PAGE (Figure 3A). Four major species were observed for rGST-BBK13, complexes I, II, III and IV, with relative migration distance (Rf) values of 0.86, 0.70, 0.53 and 0.17, respectively (Figure 3). The Rf values for the molecular mass standards were calculated and plotted versus molecular mass in kilodaltons (kDa) (Figure 3B). The second-order polynomial best fit standard curve was used to estimate the molecular masses of the rGST-BBK13 complex species. rGST-BBK13 complex I had an estimated molecular mass of 45 kDa. This size is consistent with theoretical molecular mass for rGST-BBK13 and the monomeric size of the recombinant protein observed by SDS-PAGE (Figure 3B). The molecular masses of rGST-BBK13 complexes II, III and IV were estimated at 156, 352 and 962 kDa, respectively. Although native GST is known to dimerize [22], higher-order complexes were not observed for rGST alone, suggesting that rGST-BBK13 complex formation was independent of the GST tag. The multimeric states of rGST-BBK13 were also analyzed by size exclusion chromatography using High Prep 16/60 Sephacryl S-300 resin. Four major elution peaks were detected (Figure S1A). Western blot analysis of elution fractions representative of these peaks demonstrated that rGST-BBK13 comprised peaks 1-3 (Figure S1B). Consistent with the BN-PAGE results, we detected both estimated monomeric and multimeric species of rGST-BBK13 with estimated molecular masses of 42 kDa, 232 kDa and >781 kDa, respectively (Figure S1C). Together these data indicate the ability of rGST-BBK13 to oligomerize in solution.
Figure 3. Recombinant BBK13 oligomerizes in solution.
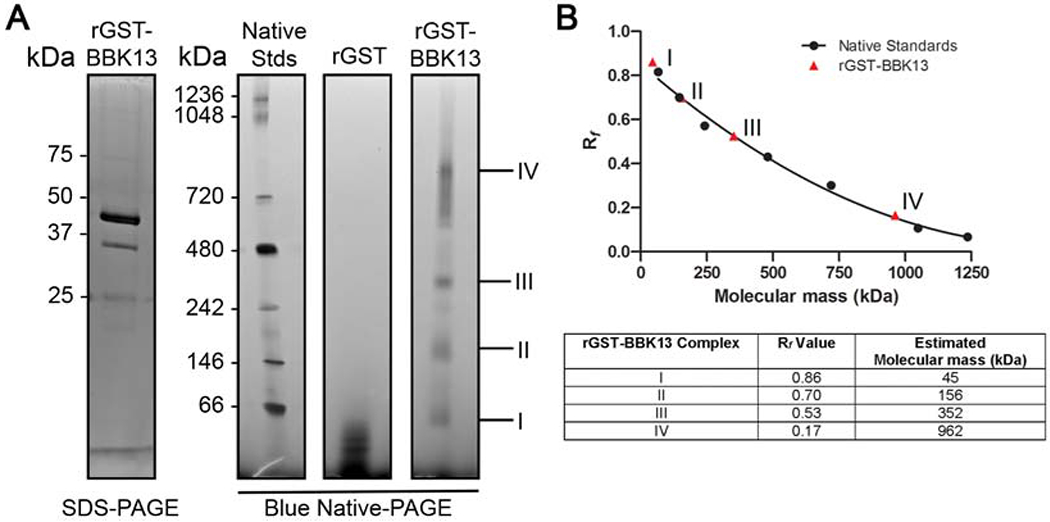
(A) SDS-PAGE analysis of recombinant fusion protein GST-BBK13 (rGST-BBK13) and blue native-PAGE (BN-PAGE) resolution of native rGST-BBK13 or recombinant GST alone (rGST). Molecular masses are shown in kilodaltons (kDa). Major rGST-BBK13 complexes are indicated (I-IV). (B) Relative migration values (Rf) of native protein standards in the BN-PAGE were calculated and plotted versus molecular mass (kDa). A second order polynomial line of best fit standard curve (r2=0.9912) was used to approximate the molecular masses of the rGST-BBK13 complexes based on their Rf values in the BN-PAGE. The Rf value and estimated molecular mass of each rGST-BBK13 complex are shown in the table.
Endogenous BBK13 is found in large oligomeric complexes in the spirochete membrane.
Next, we sought to determine the oligomeric state of endogenous BBK13 in B. burgdorferi. Spirochete membranes were isolated from wild type, Δbbk13 mutant and Δbbk13 complement B. burgdorferi [5] under native conditions. The membrane proteins were resolved in the first dimension by BN-PAGE (Figure 4A). Each lane of the BN-PAGE was excised and subsequently resolved in the second dimension by SDS-PAGE followed by immunoblot analysis using anti-BBK13 (Figure 4B) or anti-OspA (Figure S2) antibodies. BN-PAGE revealed a number of large protein complexes in the B. burgdorferi membrane (Figure 4A), as has been reported previously [18]. BBK13 was detected in protein complexes of 480 kDa up to great than 1000 kDa. The size range of the OspA-associated membrane complexes was consistent with previous findings [18], providing validation for our 2D BN-PAGE method. Furthermore, we found that these complexes were distinct from those of BBK13 and were independent of the presence or absence of BBK13 (Figure S2).
Figure 4. BBK13 is found within large complexes in the spirochete membrane.
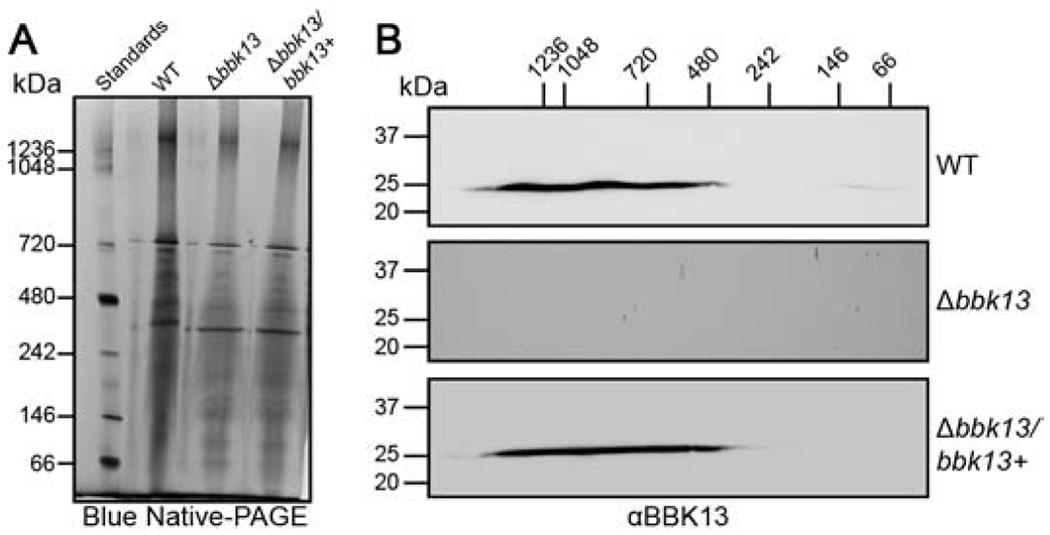
(A) One dimensional blue native-PAGE (BN-PAGE) of Borrelia burgdorferi total membrane lysates. Total membrane protein complexes isolated from wild type (WT), Δbbk13 and Δbbk13/bbk13+ B. burgdorferi, were resolved by BN-PAGE. Native protein standards (Standards) are shown in kilodaltons (kDa). (B) Each BN-PAGE sample lane was excised and subjected to second dimension 10% SDS-PAGE, followed by immunoblot analysis for BBK13 (αBBK13). Molecular mass standards for the second dimension SDS-PAGE (left) and the first dimension BN-PAGE (top) are shown in kilodaltons (kDa).
BBK13 is detected homogeneously across the spirochete.
Given the findings that BBK13 was in large protein complexes in the spirochete membrane we investigated whether this resulted in a detectable staining pattern by fluorescence microscopy. BBK13-specific staining was detected for WT and Δbbk13/bbk13+ spirochetes (Figure S3A). A largely homogenous staining pattern for BBK13 was observed. At the level of resolution of confocal fluorescence microscopy no significant sites of enrichment of BBK13 were detected (Figure S3A). Furthermore, consistent with previous reports by us and others of the non-surface exposed membrane localization of BBK13 [5, 6], membrane permeabilization was required for BBK13 detection (Figure S3B).
DISCUSSION
BBK13 is critical for B. burgdorferi to mount a productive disseminated infection [5], yet the function of BBK13 is not known. BBK13 is annotated as a SIMPL domain containing protein. Bioinformatics and structural modeling analyses revealed a boomerang-like structure for BBK13 comprised of four alpha-helices and twelve beta-sheets similar to that of the elucidated structures of SIMPL domain containing proteins cjSLP and BP26 [9, 10]. Alignment of the modeled structure of BBK13 with the known structures of cjSLP and BP26 indicated strong structural homology between BBK13 and BP26. In congruence with BP26, but in contrast to cjSLP [9, 10], BBK13 oligomerized both in vitro and in vivo. We demonstrated by far-western blot, blue native-PAGE and size exclusion chromatography (SEC) that recombinant BBK13 undergoes homomeric protein-protein interactions to form oligomeric complexes. Blue native-PAGE and SEC provided consistent molecular mass estimations for the rBBK13 complexes. These data suggested that rBBK13 oligomers may be comprised of units of trimers, including the possibilities for a single trimer, a hexamer and a trimer of hexamers. Two dimensional blue native-PAGE of the B. burgdorferi membrane proteins demonstrated that endogenous BBK13 is in large complexes. Interestingly, BBK13 monomer was not detected, suggesting that in vivo BBK13 is maintained in a higher-order oligomeric complex within the spirochete membrane, yet site of BBK13 enrichment were not observable by confocal fluorescence microscopy. The presence or absence of BBK13 did not appear to affect the overall profile of the native membrane protein complexes. Yet, our data do not rule out the possibility that the BBK13 complexes also include other B. burgdorferi proteins.
The function(s) of SIMPL domain containing protein in prokaryotes remains unknown. Recently, it was proposed that BP26 may function as a Brucella adhesin due to the ability of the recombinant protein to bind to select extracellular matrix proteins, including type I collagen and vitronectin [23]. However, this function has not been established in vivo and the mechanism of this function is unclear given the periplasmic localization of BP26. Like BP26, BBK13 appears to localize to the spirochete periplasm [5] and immunofluorescence assay of intact spirochetes confirmed that the protein is not surface exposed, likely ruling out a role for BBK13 as an adhesin. The bbk13 gene is important for B. burgdorferi to establish a productive disseminated infection [5]. Furthermore, B. burgdorferi lacking bbk13 are not recovered from heart tissue up to 3 weeks post-inoculation [5]. B. burgdorferi must overcome significant physiological and immune barriers in the mammalian host in order to establish a disseminated infection. We recently observed that the infection defect of the bbk13 mutant B. burgdorferi was ameliorated in severely immunocompromised Nod SCID gamma (NSG) mice, resulting in near wild type levels of infection in distal tissues, including the heart, 3 weeks post-inoculation (unpublished data). These findings suggest that BBK13 may contribute to immune evasion by the spirochete and it is tempting to predict that the oligomeric structure of the protein could be important for this. Future studies are focused on elucidation of the molecular mechanism of BBK13-dependent infectivity and how BBK13 oligomerization contributes to this mechanism.
Supplementary Material
Highlights.
BBK13 is SIMPL domain protein critical for B. burgdorferi infection
BBK13 demonstrates structural homology with Brucella BP26
Recombinant BBK13 self-associates and forms multimers in solution
Endogenous BBK13 forms higher order oligomers in the spirochete membrane
ACKNOWLEDGEMENTS
We would like to thank Kyle Mamounis and Victor Davidson for technical assistance with the size exclusion chromatography studies. We greatly appreciate the significant technical and intellectual contributions of Travis Jewett.
FUNDING SOURCES
This work was supported by the National Institutes of Health [R01AI099094 to M.W.J.] and the Deborah and Mark Blackman-Global Lyme Alliance postdoctoral fellowship [G.F.A].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- [1].Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP, Lyme disease-a tick-borne spirochetosis?, Science, 216 (1982) 1317–1319. [DOI] [PubMed] [Google Scholar]
- [2].Steere AC, Grodzicki RL, Kornblatt AN, Craft JE, Barbour AG, Burgdorfer W, Schmid GP, Johnson E, Malawista SE, The spirochetal etiology of Lyme disease, N Engl J Med, 308 (1983) 733–740. [DOI] [PubMed] [Google Scholar]
- [3].Burgdorfer W, Keirans JE, Ticks and Lyme disease in the United States, Ann Intern Med, 99 (1983) 121. [DOI] [PubMed] [Google Scholar]
- [4].Hu LT, Lyme Disease, Ann Intern Med, 165 (2016) 677. [DOI] [PubMed] [Google Scholar]
- [5].Aranjuez GF, Kuhn HW, Adams PP, Jewett MW, Borrelia burgdorferi bbk13 Is Critical for Spirochete Population Expansion in the Skin during Early Infection, Infection and immunity, 87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Toledo A, Huang Z, Coleman JL, London E, Benach JL, Lipid rafts can form in the inner and outer membranes of Borrelia burgdorferi and have different properties and associated proteins, Molecular microbiology, 108 (2018) 63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, Thanki N, Yamashita RA, Yang M, Zhang D, Zheng C, Lanczycki CJ, Marchler-Bauer A, CDD/SPARCLE: the conserved domain database in 2020, Nucleic acids research, 48 (2020) D265–D268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Vig E, Green M, Liu Y, Yu KY, Kwon HJ, Tian J, Goebl MG, Harrington MA, SIMPL is a tumor necrosis factor-specific regulator of nuclear factor-kappaB activity, The Journal of biological chemistry, 276 (2001) 7859–7866. [DOI] [PubMed] [Google Scholar]
- [9].Oh HB, Yoon SI, Structural analysis of a Simpl-like protein from Campylobacter jejuni, Biochemical and biophysical research communications, 529 (2020) 270–276. [DOI] [PubMed] [Google Scholar]
- [10].Kim D, Park J, Kim SJ, Soh YM, Kim HM, Oh BH, Song JJ, Brucella immunogenic BP26 forms a channel-like structure, J Mol Biol, 425 (2013) 1119–1126. [DOI] [PubMed] [Google Scholar]
- [11].Rego RO, Bestor A, Rosa PA, Defining the plasmid-borne restriction-modification systems of the Lyme disease spirochete Borrelia burgdorferi, Journal of bacteriology, 193 (2011) 1161–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Roy A, Kucukural A, Zhang Y, I-TASSER: a unified platform for automated protein structure and function prediction, Nature protocols, 5 (2010) 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y, The I-TASSER Suite: protein structure and function prediction, Nat Methods, 12 (2015) 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang Y, I-TASSER server for protein 3D structure prediction, BMC bioinformatics, 9 (2008) 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ, The Phyre2 web portal for protein modeling, prediction and analysis, Nature protocols, 10 (2015) 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kenedy MR, Luthra A, Anand A, Dunn JP, Radolf JD, Akins DR, Structural modeling and physicochemical characterization provide evidence that P66 forms a beta-barrel in the Borrelia burgdorferi outer membrane, Journal of bacteriology, 196 (2014) 859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Schagger H, von Jagow G, Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form, Anal Biochem, 199 (1991) 223–231. [DOI] [PubMed] [Google Scholar]
- [18].Yang X, Promnares K, Qin J, He M, Shroder DY, Kariu T, Wang Y, Pal U, Characterization of multiprotein complexes of the Borrelia burgdorferi outer membrane vesicles, J Proteome Res, 10 (2011) 4556–4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Barbour AG, Tessier SL, Todd WJ, Lyme disease spirochetes and ixodid tick spirochetes share a common surface antigenic determinant defined by a monoclonal antibody, Infection and immunity, 41 (1983) 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bestor A, Rego RO, Tilly K, Rosa PA, Competitive advantage of Borrelia burgdorferi with outer surface protein BBA03 during tick-mediated infection of the mammalian host, Infection and immunity, 80 (2012) 3501–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chothia C, Lesk AM, The relation between the divergence of sequence and structure in proteins, EMBO J, 5 (1986) 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wilce MC, Parker MW, Structure and function of glutathione S-transferases, Biochim Biophys Acta, 1205 (1994) 1–18. [DOI] [PubMed] [Google Scholar]
- [23].ElTahir Y, Al-Araimi A, Nair RR, Autio KJ, Tu H, Leo JC, Al-Marzooqi W, Johnson EH, Binding of Brucella protein, Bp26, to select extracellular matrix molecules, BMC Mol Cell Biol, 20 (2019) 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


