Abstract
Intrinsic antiviral host factors confer cellular defense by limiting virus replication and are often counteracted by viral countermeasures. We reasoned that host factors that inhibit viral gene expression could be identified by determining proteins bound to viral DNA (vDNA) in the absence of key viral antagonists. Herpes simplex virus 1 (HSV-1) expresses ICP0, which functions as an E3 ubiquitin ligase required to promote infection. Cellular substrates of ICP0 have been discovered as host barriers to infection, but mechanisms for inhibition of viral gene expression are not fully understood. To identify restriction factors antagonized by ICP0, we compared proteomes associated with vDNA during HSV-1 infection with wild-type (WT) virus and mutant lacking functional ICP0 (ΔICP0). We identified the cellular protein Schlafen 5 (SLFN5) as an ICP0 target that binds vDNA during HSV-1 ΔICP0 infection. We demonstrated that ICP0 mediates ubiquitination of SLFN5 which leads to its proteasomal degradation. In the absence of ICP0, SLFN5 binds vDNA to repress HSV-1 transcription by limiting accessibility of RNA polymerase II to viral promoters. These results highlight how comparative proteomics of proteins associated with viral genomes can identify host restriction factors, and reveal that viral countermeasure can overcome SLFN antiviral activity.
Intrinsic host defenses function cooperatively to limit replication and spread of viral pathogens from the outset of nuclear infection1–4. Conversely, evolution provides viruses with elegant strategies to subvert these host defenses, often through binding and inducing degradation of the cellular restriction factors1,5–12. Although there is rapidly expanding knowledge of restriction factors for RNA viruses, there is a critical need to identify and better understand intrinsic cellular defenses against viruses with DNA genomes. Since viruses hijack the cellular ubiquitin machinery to modify the host cell proteome and subvert these inhibitory defenses, identifying substrates for viral-induced ubiquitination can reveal cellular restriction factors5–7. The immediate early ICP0 viral protein of herpes simplex virus type 1 (HSV-1) promotes transactivation of viral genes and regulates reactivation from latency13–15. ICP0 contains an E3 ubiquitin ligase domain that antagonizes host defenses through proteasomal degradation of intrinsic antiviral factors in infected cells13,16–36. Cellular substrates of ICP0 have been discovered as host barriers to infection, but their mechanisms for inhibition of viral gene expression are not fully understood33–36. We sought to discover antiviral host factors that limit HSV-1 infection by identifying proteins bound to viral DNA genomes (vDNA) in the absence of ICP0. The Isolation of Proteins On Nascent DNA (iPOND) technique was developed as a way of identifying proteins associated with newly-synthesized DNA during replication37. This approach involves metabolic incorporation of 5-ethynyl-2´-deoxyuridine (EdU) into newly-synthesized DNA, which can be biotinylated by click chemistry to allow affinity purification and determination of proteins bound to DNA by mass spectrometry-based proteomics. It has recently been adapted to isolate and define proteins accumulated on nuclear-replicating vDNA genomes38–40. We reasoned that antiviral factors that are targeted by ICP0 to overcome inhibition of viral transcription and replication would be associated with the viral genome in its absence and could be identified by iPOND and mass spectrometry. By comparing vDNA-associated proteomes for wild-type HSV-1 and a ΔICP0 mutant, we identified the cellular SLFN5 protein as a target for ICP0-mediated degradation. SLFN5 is a nuclear member of the Schlafen family of proteins that have been implicated in immune cell proliferation, differentiation and antiviral retsriction41–50. Employing iPOND in a comparative proteomics approach presents an approach to identify substrates of viral antagonists and reveal host factors that act on viral DNA genomes to restrict infection.
Results
Comparative proteomics identifies SLFN5 on vDNA in the absence of ICP0.
Here we employed iPOND to identify proteins differentially associated with vDNA during WT or ΔICP0 HSV-1 infection of human foreskin fibroblasts (HFFs) at 8 hours post-infection (hpi) (Fig. 1a). Host factors known to be substrates of ICP0-mediated degradation were isolated from viral genomes during ΔICP0, but not WT, HSV-1 infection (Fig. 1b). We performed three-dimensional principal component analysis (PCA) clustering to identify cellular factors that showed binding profiles similar to known ICP0 substrates DNA-PKcs, IFI16, and PML (Fig. 1c; Extended Data Fig. 1a). Clustered proteins enriched on ΔICP0 relative to WT HSV-1 genomes were considered putative ICP0 substrates that are targeted for degradation to overcome cellular antiviral restriction (Extended Data Fig. 1b,c). Among these putative substrates, we identified the SLFN5 protein (Fig. 1c). Identification of SLFN5 in iPOND-MS from uninfected cells may reflect functions on cellular DNA41–45. Although the cytoplasmic SLFN11 protein has been reported to inhibit virus infection by controlling protein synthesis46, no antiviral function has been ascribed to nuclear SLFN proteins, and no viral countermeasures to SLFN proteins have been identified.
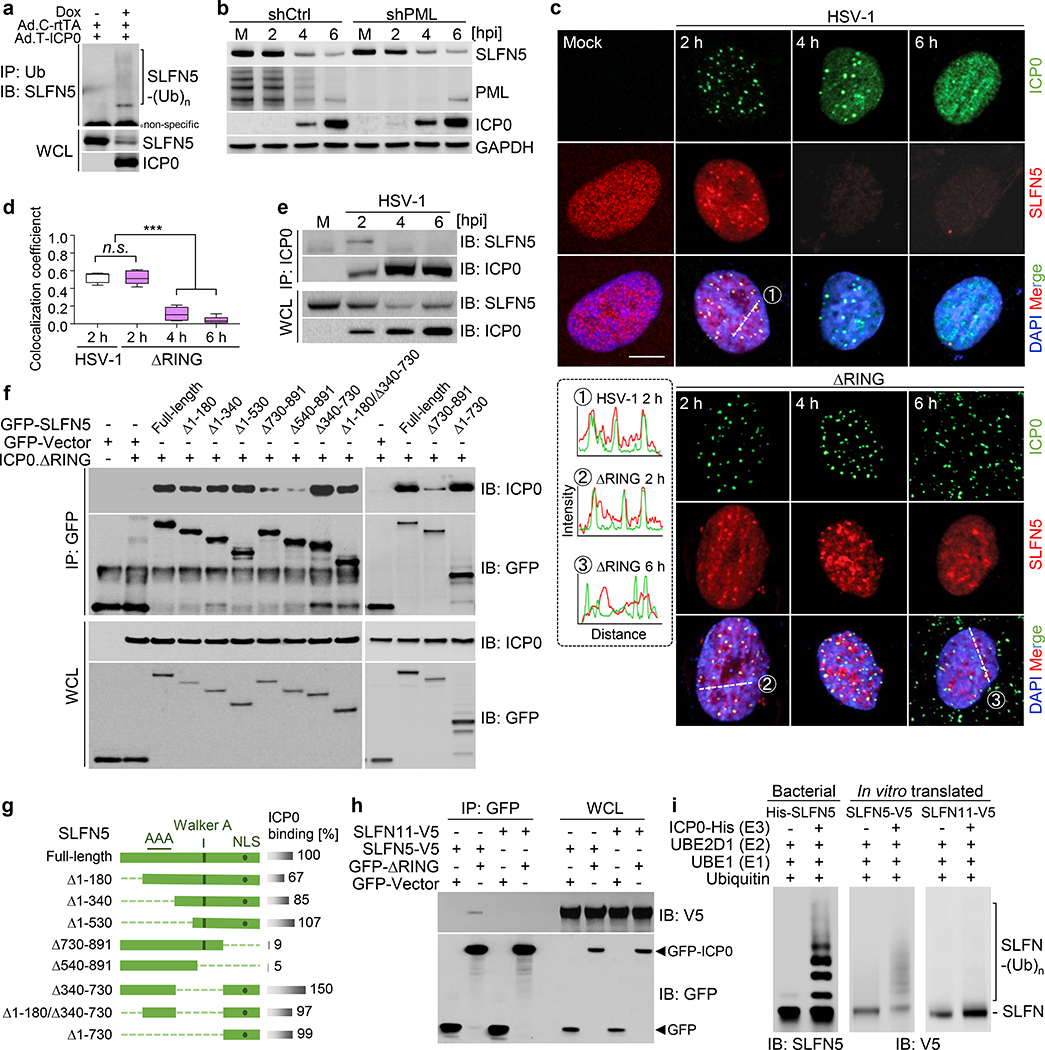
Fig. 1 |. ICP0 targets SLFN5 for degradation.
a, Workflow schematic showing how iPOND-MS was combined with PCA-based clustering to identify cellular substrates of ICP0. b, Plot highlights a trend in differential abundance for known ICP0 substrates across iPOND proteomes at 8 hpi. c, PCA-based clustering of proteins identified in iPOND-MS proteomes by abundance similarity to known ICP0 substrates. Proteins are projected onto a 3D PCA space. Indicated known restriction factors are assigned as cluster centers. Proteins were clustered based on their proximity to cluster centers in 3D PCA space. d, HFF cells were infected with HSV-1 WT or ΔICP0 mutant virus at an MOI of 3 and iPOND isolation of SLFN5 was examined at 8 hpi. e, Comparison of changes in protein abundance of ICP0 targets during HSV-1 infection. Whole cell proteome analysis reveals SLFN5 protein reduction during HSV-1 infection. f, Comparison with known ICP0 substrate proteins reveals SLFN5 degradation is dependent on the ICP0 RING finger domain and that SLFN11 is not degraded. g, SLFN5 turnover is proteasome-dependent. The proteasome inhibitor MG132 was added at 2 hpi and lysates were prepared for immunoblotting at indicated time points. h, The turnover rate of SLFN5 in WT or ΔRING mutant-infected HFF cells was compared by cycloheximide (CHX) chase followed by densitometric analysis of immunoblots. Data are the mean ± SD. Comparisons between groups were performed using the two-tailed unpaired Student’s t-test. **, p < 0.005. n = 3 biologically independent experiments. Immunoblots show representative data from n = 3 biologically independent experiments.
ICP0 targets SLFN5 for ubiquitination and poteasomal degradation.
To explore SLFN5 during HSV-1 infection, we first confirmed by immunoblotting that SLFN5 was differentially bound to ΔICP0 EdU-labelled vDNA during HSV-1 infection (Fig. 1d). The absence of SLFN5 in the proteome isolated on vDNA by iPOND-MS from WT HSV-1 infections suggests that it is depleted by ICP0. Proteins isolated by iPOND were therefore also examined in whole cell proteome abundance data generated over a time-course of HSV-1 infection51. SLFN5 decreased in protein abundance in a similar fashion to known ICP0 substrates DNA-PKcs, PML, IFI16, USP7, and ATRX (Fig. 1e; Extended Data Fig. 1d). These SLFN5 proteomics results were further validated by immunoblot analysis over a time-course of infection, where SLFN5 levels decreased similarly to other known substrates of ICP0 during WT HSV-1 infection (Fig. 1f). The decrease in SLFN5 expression was specific, since the levels of other family members were not decreased, including SLFN11 (Fig. 1f) which has been assigned antiviral functions46. Expression of a functionally inactive ICP0 RING domain (ΔRING) did not decrease SLFN5 protein levels during HSV-1 infection (Fig. 1f). Furthermore, the reduction in SLFN5 in HSV-infected cells was abolished by treatment with the proteasome inhibitor MG132 (Fig. 1g). When protein synthesis was inhibited by cycloheximide, SLFN5 protein was rapidly turned over during WT, but not ΔRING, HSV-1 infection (Fig. 1h). Together, these data identify SLFN5 as a target of ICP0-mediated proteasomal degradation. Furthermore, the observation that SLFN5 accumulates on viral genomes in the absence of ICP0, similar to other antiviral substrates of ICP0, suggests that SLFN5 is a putative host restriction factor.
We next investigated the requirement for SLFN5 reduction during HSV-1 infection. To confirm that ICP0 is sufficient to induce SFLN5 degradation independently of other viral factors, we employed an adenovirus transduction vector with doxycycline-inducible expression of ICP0 (Fig. 2a). Expression of ICP0 reduced SLFN5 protein abundance, with detectable levels of SLFN5 polyubiquitination following immunoprecipitation. This was further supported by co-transfection of tagged-SLFN5 with ICP0 in HEK293T cells, which lack endogenous SLFN546 (Extended Data Fig. 2a,b), confirming ICP0 RING-dependent degradation of SLFN5. We next examined whether targeting of SLFN5 is due to direct interaction with ICP0 or is mediated indirectly through the degradation of PML nuclear bodies (NBs)52–55. In PML depleted cells, SLFN5 was degraded by ICP0, suggesting PML-independent direct targeting (Fig. 2b). The RING-dependent degradation of endogenous SLFN5 was also consistently observed in other cell lines (Extended Data Fig. 2c,g). Since ICP0 has SUMO-targeting properties56,57 and SLFN5 has been predicted as a SUMO-dependent substrate for ubiquitination58, we examined SLFN5 degradation in cells depleted for the SUMO-conjugating enzyme UBC9 (Extended Data Fig. 2h). SLFN5 degradation was unaffected by UBC9 depletion, suggesting a SUMO-independent mechanism of ICP0 degradation. We therefore examined whether ICP0 interacts directly with SLFN5. While some human and mouse SLFN proteins have been detected in the cytoplasm, human SLFN5 is predominantly nuclear. Immunofluorescence demonstrated SLFN5 to be diffusely nuclear in uninfected cells but to colocalize in nuclear puncta with WT and ΔRING mutant ICP0 at 2 hpi (Fig. 2c,d). SLFN5 signal decreased by 4 hpi in WT HSV-infected cells due to ICP0-mediated degradation, and relocalized with ΔRING into larger structures that are presumably sites of vDNA accumulation (Fig. 2d). Endogenous SLFN5 co-immunoprecipitated with ICP0 from WT HSV-1 infected cells at 2 hpi (Fig. 2e) but not at 4 hpi, a time point at which SLFN5 levels were significantly diminished. To identify the domain of SLFN5 that interacts with ICP0 we constructed a series of GFP-tagged SLFN5 deletion mutants (Fig. 2f,g). Full-length SLFN5 and truncation mutants mainly localized to the nucleus (Extended Data Fig. 3a), except for the Δ730–891 and Δ540–891 mutants which lack a predicted nuclear localization sequence (NLS) in the carboxyl (C)-terminal region (aa812–815, RKRK)59. We mapped the ICP0-binding region of SLFN5 by co-immunoprecipitation from co-transfected HEK293T cells. ICP0 interacted with full-length SLFN5 and mutants that retain residues 730–891, but their interaction was diminished in the absence of the C-terminal region (Fig. 2f,g). Since C-terminal mutants lack the SLFN5 NLS, we also tested interaction with a cytoplasmic ICP0 mutant (cICP0)60, but no interaction was detected (Extended Data Fig. 3b). These data suggest ICP0 binds the C-terminal region of nuclear SLFN5. To verify the specificity of the ICP0-SLFN5 interaction among other SLFN proteins, we assessed ICP0 interaction with SLFN11 via co-immunoprecipitation. Under conditions where ICP0 interacted with SLFN5, we did not detect an interaction with SLFN11 (Fig. 2h). Proteins that interact with viral factors such as ICP0 often contain intrinsically disordered regions61 which we observed in the C-terminal region of SLFN5 but not SLFN11 (Extended Data Fig. 3c) and may facilitate association with ICP0. As final evidence of targeted ubiquitination, we performed in vitro ubiquitination assays using recombinant proteins. These assays showed that full-length SLFN5 produced by bacteria or in vitro translation could be ubiquitinated by recombinant ICP0 (Fig. 2i; Extended Data Fig. 2i). In contrast, ICP0 did not ubiquitinate either SLFN11 or the SLFN5 Δ730–891 truncation that lacks ICP0 binding. Together, these data demonstrate that ICP0 binds SLFN5 and is sufficient for ubiquitination and proteasomal degradation independently of other viral cofactors.
Fig. 2 |. ICP0 interacts with SLFN5.
a, Ubiquitination of endogenous SLFN5 by ICP0 expressed by inducible recombinant adenoviruses vector transduction. Denaturing immunoprecipitation (IP) was carried out with anti-Ub antibody and SLFN5 detected by immunoblot (IB). b, SLFN5 degradation is PML-independent and is detected in cells with PML depleted. c, HFF cells infected with HSV-1 WT or ΔRING virus at MOI of 3 were subject to immunofluorescence with antibodies to ICP0 and SLFN5. Scale bar, 10 μm. The fluorescence plot profiles (at dashed lines) were analyzed by ImageJ. d, Quantification of the colocalization coefficient showed colocalization of ICP0 with SLFN5 at early infection times of both viruses. Each box plot represents five cells per condition with whiskers from minimum to maximum, median indicated by horizontal bar in the box, and box limits extending from quartile 1 to quartile 3. Data are the mean ± SD. Comparisons between groups were performed using the two-tailed unpaired Student’s t-test. ***, p < 0.0005. n.s., not significant. e, ICP0 was immunoprecipitated from HSV-1 infected HFF cells and immunoblot detected interaction with endogenous SLFN5 at 2 hpi. f, ICP0 interacts through the C-terminal region of SLFN5. HEK293T cells were co-transfected with plasmids encoding ICP0 ΔRING and full-length or mutants of GFP-SLFN5. Proteins were immunoprecipitated with anti-GFP Ab, followed by immunoblotting. g, Schematic structure of full-length and mutant SLFN5 proteins. The relative strength [%] of each interaction compared to full-length was determined by densitometric analysis of co-purified ICP0 protein bands. h, ICP0 interacts with SLFN5, but not with SLFN11. Co-IP were carried out with plasmids co-transfected cells in the presence of benzonase. i, In vitro ubiquitination assays with bacterially purified or in vitro translated proteins reveal that ICP0 ubiquitinates SLFN5 directly, but not SLFN11. Immunoblots show representative data from n = 3 biologically independent experiments.
SLFN5 associates with vDNA in the absence of ICP0.
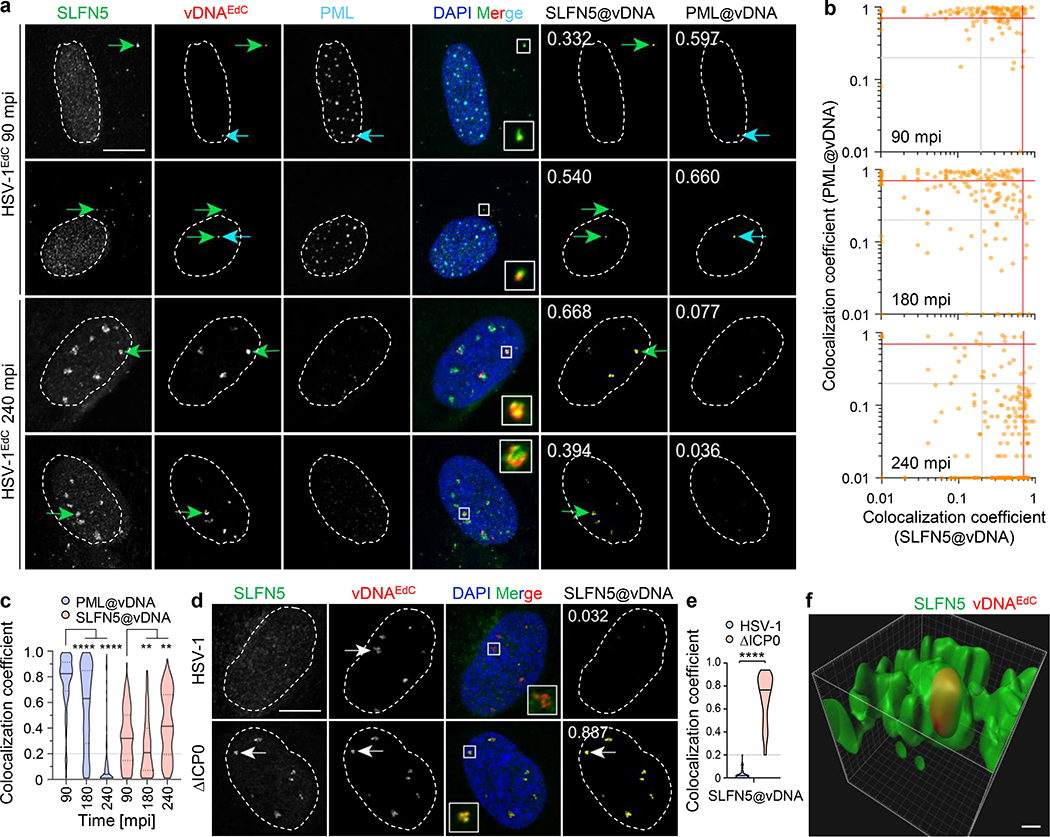
We used immunofluorescence and confocal microscopy to localize SLFN5 during HSV-1 infection (Fig. 3). HFF cells were infected with HSV-1 grown in the presence of ethynyl-tagged deoxycytidine (EdC) to label vDNA that can be detected by click chemistry62. SLFN5 co-localized with input HSV-1 genomes in puncta during early times of infection, including prematurely uncoated cytosolic genomes and incoming genomes in the nucleus (Fig. 3a–c). As previously reported62, at early times input vDNA in the nucleus colocalizes with PML nuclear bodies (NBs) before PML is degraded by ICP0 (Fig. 3a–c). Some PML NBs also colocalize with SLFN5 foci at sites of input vDNA (Fig. 3b). By 4 hpi the PML has been decreased via ICP0 degradation but remaining SLFN5 colocalized with vDNA as pre-replication foci are formed. At early times of infection, the viral transcriptional activator ICP4 marks site of gene expression from incoming viral genomes (Extended Data Fig. 4a). SLFN5 partially localized with these virus-induced structures, further supporting association with incoming vDNA genomes. We then examined SLFN5 localization at later times when vDNA synthesis had been initiated at viral replication compartments (VRCs). We labeled sites of vDNA synthesis by EdC incorporation and detection via click chemistry (Fig. 3d). To allow comparison between WT and ΔICP0 mutant HSV-1 infection, we added acycloguanosine (ACG) to limit VRC expansion. During WT HSV-1 infection, the staining for SLFN5 decreased and did not colocalize with vDNA (Fig. 3d,e) or with VRCs located by staining for the DNA binding protein ICP8 (Extended Data Fig. 4b). In contrast, SLFN5 colocalized with vDNA and VRCs during infection with ΔICP0 mutant HSV-1 (Fig. 3d; Extended Data Fig. 4b). Quantification showed a high colocalization coefficient for vDNA and SLFN5 in cells infected with HSV-1 ΔICP0 (Fig. 3e), with high-resolution Z-series imaging suggesting vDNA to be entrapped by a shell of SLFN5 in the nucleus (Fig. 3f). These observations are similar to patterns detected for the antiviral protein PML (Extended Data Fig. 4b), which was previously reported to entrap nuclear HSV-1 DNA62. Together with the iPOND data, these results support the conclusion that SLFN5 binds vDNA in the absence of ICP0.
Fig. 3 |. SLFN5 colocalizes with HSV-1 DNA.
HFF cells were infected at an MOI of 1 with HSV-1EdC and samples fix at the indicated times post-infection. Input viral DNA (vDNA) was detected by click chemistry (red). SLFN5 (green) and PML (cyan) were detected by indirect immunofluorescence. Nuclei were stained with DAPI (blue). a, Representative confocal microscopy images showing colocalization of SLFN5 and vDNA. Green and cyan arrows highlight examples of SLFN5 and PML colocalization at vDNA, respectively. Inserts; magnified regions of interest (boxes). Cut mask images (yellow) show regions of SLFN5 or PML (as indicated) colocalization at vDNA. W. colocalization coefficients shown (0 = no colocalization; 1 = perfect colocalization). Scale bar, 10 μm b, Scatter plots showing paired weighted (w.) colocalization coefficients of SLFN5 and PML at vDNA per nuclei. Grey and red lines indicate arbitrary weighted colocalization coefficient gating at 0.2 and 0.7, respectively. c, Violin plots showing weighted colocalization coefficients of SLFN5 or PML at vDNA. Black line; median: Dotted lines; 5 and 95th percentile range: Grey line; threshold of detection (0.2). ** p < 0.01, **** p < 0.0001; Dunn’s multiple comparison test (n90mpi = 126, n180mpi = 155, n240mpi = 193 over 3 biological replicates). d, Immunofluorescence for SLFN5 and viral DNA (vDNA). HFF cells were infected with WT HSV-1 or ΔICP0 mutant virus at an MOI of 3 prior to overlay with medium containing 0.5 μM EdC and 50 μM ACG. Samples were fixed at 6 hpi. De novo nascent vDNA synthesis was detected by click chemistry (red) and SLFN5 (green) by indirect immunofluorescence. Representative confocal microscopy images showing nuclear localization of SLFN5 at vDNA. White arrows highlight examples of SLFN5 localization at vDNA. Cut mask images (yellow) show region of SLFN5 colocalization at vDNA. Colocalization coefficients shown (0 = no colocalization; 1 = perfect colocalization). Scale bar, 10 μm e, Violin plots showing weighted colocalization coefficients of SLFN5 at vDNA. Black line; median; Dotted lines; 5 and 95th percentile range; Grey line; threshold of detection (0.2). n ≥ 100 infected cells per sample population derived from 3 independent experiments. **** p < 0.0001; Mann-Whitney U-test (nHSV-1 = 122, nΔICP0 = 114 over three biological replicates). f, 3D reconstruction of high-resolution Z-series confocal image showing SLFN5 (green) entrapment of HSV-1 DNA EdC vDNA (red). Scale bar, 0.4 μm.
ICP0 counteracts SLFN5-mediated inhibition of HSV-1 infection.
To assess the functional relevance of SLFN5 degradation, we used a lentiviral vector to generate cells expressing shRNAs to SLFN5 (Extended Data Fig. 5a). We compared vDNA replication in the presence and absence of SLFN5 during WT and ΔRING HSV-1 infection (Fig. 4a, Extended Data Fig. 5b). Accumulation of vDNA for WT HSV-1 was moderately increased in the absence of SLFN5 (~2-fold). However, vDNA abundance of ΔRING HSV-1 was significantly increased (>10-fold) in SLFN5-depleted cells relative to infected control cells (Fig. 4a). Consistent with increased vDNA replication, SLFN5 depletion resulted in a 3- and 13-fold increase in progeny production of ΔRING virus at 24 and 48 hpi, respectively (Fig. 4b). SLFN5 depletion also increased accumulation of viral proteins for ΔRING virus, with the most pronounced effect at lower virus MOIs (Extended Data Fig. 5c). Since SLFN5 has been implicated in control of cell growth43 and morphology44, we examined the effect of SLFN5 on cell proliferation. We did not observe any change in morphology and growth rate when SLFN5 was depleted in HeLa cells (Extended Data Fig. 5d–f), suggesting a direct antiviral effect on HSV-1 replication rather than an indirect effect on cell growth. We confirmed that the effects of SLFN5 depletion on HSV-1 replication were achieved when both siRNA-mediated knockdown and single guide RNA-mediated CRISPR/Cas9 knockout (KO) were employed (Fig. 4c; Extended Data Fig. 5g). To complement the knockdown approach, we also examined whether ectopic SLFN5 overexpression impacted HSV-1 replication. We examined vDNA replication in SLFN5 KO HeLa cells which had been transduced with lentiviral vector to express HA-tagged SLFN5 under a tetracycline-inducible promoter. While vDNA levels of WT virus were not significantly altered, levels of vDNA during ΔRING infection were significantly decreased in the presence of SLFN5 when compared to empty vector control cells (Fig. 4d). The impact of SLFN5 depletion and reconstitution on ΔRING virus supports our conclusion that SLFN5 restricts HSV-1 infection and this restriction is alleviated by ICP0 during WT HSV-1 infection. One possible way that SLFN5 could affect viral replication is via the putative helicase activity63. To determine whether the Walker A helicase motif of SLFN5 affects HSV-1 replication, SLFN5 KO cells were transduced with adenoviral vectors expressing WT or Walker A mutant (K584A) SLFN5. Expression of both WT and mutant SLFN5 reduced HSV-1 protein expression and progeny production compared to the control, demonstrating helicase-independent antiviral activity (Fig. 4e,f). We also tested the impact of SLFN5 overexpression on HSV-1 replication for full-length SLFN5 compared to the Δ730–891 truncation that lacks the ICP0 binding domain (Extended Data Fig. 6a). When SLFN5 proteins were induced in cells that were then infected with HSV-1, only full-length SLFN5 was able to inhibit ΔRING HSV-1 replication (Extended Data Fig. 6b,c).
Fig. 4 |. ICP0 counteracts SLFN5-mediated suppression of HSV-1 replication.
HeLa cells stably depleted for SLFN5 by shRNA were infected with HSV-1 at an MOI of 0.1. HSV-1 replication was monitored by qPCR (a) and plaque assays (b). n = 3 biologically independent experiments. c, HSV-1 replication in siRNA-transfected HFF cells was monitored by viral DNA qPCR. n = 3 biologically independent experiments. d, HeLa cells were stably transduced with control lentivirus (Tet.Vector-HA) or lentivirus containing tetracycline-inducible SLFN5-HA gene (Tet.SLFN5-HA). SLFN5-HA was induced with doxycycline (0.5 μg/ml) for 48 h. Cells were infected with HSV-1 WT or ΔRING virus at an MOI of 0.1. At 24 hpi, viral DNA was measured by qPCR. n = 4 biologically independent experiments. SLFN5 expression was confirmed by immunoblotting. e, f, Walker A motif mutant (K584A) showed similar restriction as WT SLFN5. Recombinant adenoviral transduction system was used for ectopic expression of SLFN5 proteins. Transduced cells for 24 h were infected with ΔRING at an MOI of 3 (e) or an MOI of 0.1 (f) and HSV-1 infectivity was analyzed by immunoblotting (e) and plaque assays (f). n = 3 biologically independent experiments. g, h, Transient knockdown of SLFN5 or SLFN11. The cells were infected ΔRING at an MOI of 3 (g) or an MOI of 0.1 (h). Viral protein at 9 hpi was analyzed by immunoblotting (g) and viral DNA at 24 hpi was monitored by qPCR (h). n = 6 biologically independent experiments. Data are the mean ±SD. Comparisons between groups were performed using the two-tailed unpaired Student’s t-test. *, p < 0.05, **, p < 0.005, ***, p < 0.0005. n.s., not significant. Immunoblots show representative data from n = 3 biologically independent experiments.
We further addressed the specificity of the inhibitory effect of SLFN5 by analyzing additional SLFN family proteins and other viruses. It has been reported that cytoplasmic SLFN11 inhibits protein synthesis during HIV infection46. Depletion of SLFN11 using siRNA did not affect HSV-1 protein expression or vDNA replication (Fig. 4g,h). We found that HSV-2 was also able to decrease SLFN5 levels (Extended Data Fig. 7a) and showed increased viral protein expression when SFLN5 was depleted (Extended Data Fig. 8b). In contrast, other DNA viruses (HCMV or Ad5) neither degraded SLFN5 (Extended Data Fig. 7b,c) nor were restricted by SLFN5 (Extended Data Fig. 8b,c). These results suggest virus specificity in SLFN family-mediated antiviral restriction.
SLFN5 suppresses HSV-1 gene transcription by limiting RNA polymerase accessibility.
We next explored whether SLFN5 impacts HSV-1 gene transcription. We analyzed accumulation of viral gene transcripts in the presence of viral DNA polymerase inhibitor phosphonoacetic acid (PAA) in either control or SLFN5 knockdown cells. Viral replication was completely blocked by PAA (Fig. 5a), enabling transcript levels to be measured from incoming vDNA. Viral transcription was enhanced for the immediate-early gene ICP27 and early gene TK in SLFN5-depleted cells compared to controls (Fig. 5a). The increase in gene expression was further supported by concomitant increases in early viral protein levels during infection in the presence of PAA (Fig. 5b). Late protein production is dependent on vDNA replication, and therefore proteins such as VP21 were not detected. Since RNA levels are determined by the balance between synthesis and decay, we analyzed de novo transcription and mRNA stability by 4sU metabolic pulse labelling (Fig. 5c and 5d). In SLFN5 knockdown cells, both total mRNA and nascent mRNA for viral genes were increased by 3-fold (Fig. 5c). We calculated similar RNA decay rates for both ICP27 and TK transcripts when compared to SLFN5 knockdown cells (Fig. 5d). These results indicate that SLFN5 inhibits viral gene transcription but does not impact mRNA stability.
Fig. 5 |. SLFN5 associates with vDNA to suppress HSV-1 gene transcription.
a, Control and SLFN5-depleted HeLa cells were infected with ΔRING virus at an MOI of 3 in the absence or presence of phosphonoacetic acid (PAA, 200 μg/ml). Accumulation of viral DNA and mRNA (ICP27 and TK) was measured by qPCR and RT-qPCR, respectively. n = 3 biologically independent experiments. b, Accumulation of viral proteins were analyzed by immunoblot analysis (representative data from n = 3 biologically independent experiments). c, 4sU metabolic pulsing reveals SLFN5-mediated suppression of viral gene transcription. Schematic of 4sU experiments (top). Cells were pulsed for 30 min at 5.5 hpi with ΔRING virus at an MOI of 3. Total RNA was isolated and 4sU-labeled RNA was then conjugated to biotin and isolated by use of streptavidin beads. Total mRNA or nascent mRNA of ICP27 and TK were analyzed by RT-qPCR. n = 3 biologically independent experiments. d, RT-qPCR analysis of mRNA half-lives from 4sU experiments. Half-lives were normalized to that for GAPDH. n = 3 biologically independent experiments. e, ChIP assays with anti-RNAP II specific antibody was performed in control or SLFN5-silenced HeLa cells infected with ΔRING at 3 hpi. n = 3 biologically independent experiments. f, ChIP assays with anti-RNAP II specific antibody was performed in control or SLFN5-silenced HeLa cells infected with ΔRING at an MOI of 1 or 10 for 3 h. n = 3 biologically independent experiments. g, ΔRING virus replication was monitored by plaque assays at 15 hpi. n = 3 biologically independent experiments. Data are the mean ± SD. Comparisons between groups were performed using the two-tailed unpaired Student’s t-test. *, p < 0.05, **, p < 0.005, n.s., not significant. h, Model for SLFN5 association with the viral genome and inhibition of transcription by minimizing RNAP II accessibility in the absence of ICP0. ICP0 degrades SLFN5 to promote transcription of viral genes.
To assess whether SLFN5 effects were specific to HSV-1, we examined infection with the HCMV. SLFN5 depletion enabled increased viral protein expression of viral proteins such as IE1/IE2 and UL44 within the first 24 hours of infection but was decreased at later times (Extended Data Fig. 8b). We saw similar effects when examining mRNA levels for immediate-early and late viral transcripts (Extended Data Fig. 9a,b). HCMV replication yield was also not significantly changed in SLFN5-deleted HFF cells (Extended Data Fig. 9c). One difference between HSV-1 and HCMV is the time course of infection, with HSV-1 replication being much quicker than HCMV. Since SLFN5 has been shown to regulate STAT1-mediated gene transcription and interferon-stimulated genes (ISG) expression, depletion of SLFN5 may generate higher levels of ISG signaling that results in lower levels of HCMV replication. We observed that SLFN5 knockdown led to higher levels of ISG15 expression, and this was further increased during HCMV infection (Extended Data Fig. 8b,d). Therefore, we propose that SLFN5 has a direct role in the cellular restriction of HSV-1 infection prior to the induction of innate immune defences and ISG expression.
Finally, we employed chromatin immunoprecipitation (ChIP) assays to detect association of cellular proteins with the HSV-1 genome. When we used an antibody to HA-tagged SLFN5, ChIP pulldown from ΔRING virus infection recovered increasing yields of vDNA over time (Extended Data Fig. 10a). Although WT HSV-1 replicates to a higher level than the ΔRING mutant, we observed less vDNA isolated by HA ChIP during WT HSV-1 infection, consistent with SLFN5 degradation by ICP0. We found that SLFN5 bound indiscriminately along the length of the HSV-1 genome, suggesting that it does not recognize a specific DNA sequence (Extended Data Fig. 10b). Since SLFN5 inhibits viral gene expression (Fig. 5a,c), we examined whether it impacts RNA polymerase II (RNAP II) binding to transcription start sites (TSS) on viral promoters. ChIP assays showed that RNAP II binding at viral promoters was significantly higher in SLFN5 knockdown cells than in controls (Fig. 5e). Since the defects of an ICP0 mutant can be overcome at high MOIs, we also examined RNAP II ChIP with increasing MOI of HSV-1 (Fig. 5f). As expected, the effect of SLFN5 loss on the degree of RNAP II binding to the viral genome decreased as the MOI of ΔRING virus increased from MOI 1 to 10. In addition, this point was further strengthened by examining the effect of SLFN5 depletion on virus yield as ΔRING virus MOI increases (Fig. 5g). The difference in virus particles produced in the absence of SLFN5 decreased as MOI increased from 1 to 10. Together these data demonstrate MOI-dependent restriction of ICP0 mutant virus replication by SLFN5. These results suggest that SLFN5 restricts HSV-1 infection by binding vDNA to prevent access to RNAP II, and thus inhibiting transcription of viral promoters.
Discussion
Our study presents an innovative approach to identify host restriction factors by defining proteins associated with virus genomes in the absence of a key viral immune antagonist. We found the human SLFN5 protein associates with HSV-1 genomes in the absence of the viral ICP0 protein, a critical regulator of host intrinsic immune defenses to HSV-1 infection. The SLFN proteins have been implicated in multiple functions41,42, which include suppression of retrovirus replication via inhibition of protein synthesis by SLFN1146,64 and inhibition of influenza virus by SLFN1447. The SLFN proteins have also been indirectly linked to virus infection via interferon signaling45,47. Our results present the first example of direct inhibition of virus gene expression through SLFN5 binding to vDNA in the nucleus. Our model for antiviral activity proposes that SLFN5 binding to vDNA inhibits transcription from viral genomes by limiting accessibility to RNAP II (Fig. 5h). We demonstrate that the E3 ubiquitin ligase activity of ICP0 marks SLFN5 for proteasome-mediated degradation during WT HSV-1 replication to promote the efficient initiation of viral transcription. This represents the first report of a viral countermeasure targeting a member of the SLFN family, highlighting the importance of intrinsically expressed host factors in the inhibition of vDNA pathogens. Our comparative proteomics approach demonstrates how antiviral host restriction factors can be revealed by identifying proteomes associated with vDNA genomes in the absence of viral anatagonists. This approach could be extended to other DNA viruses where targets of viral proteins that promote infection are unknown.
Methods
Cell culture.
All cells were obtained from the American Type Culture Collection (ATCC) without independent authentication and grown in a 5% CO2 humidified incubator at 37°C. Primary human foreskin fibroblasts (HFFs; ATCC SCRC-1041), HEK293 (ATCC CRL-1573), HEK293T (ATCC CRL-3216), HeLa (ATCC CCL-2), U2OS (ATCC HTB-96), Vero (ATCC CCL-81), and retinal pigmented epithelial (RPE-1; ATCC, CRL-4000) cells were grown in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS) (VWR) and penicillin (100 U/ml)/streptomycin (100 μg/ml) (Invitrogen). Cells were transfected using Lipofectamine 2000 or RNAiMAX (Invitrogen) following the manufacturer’s instructions.
Plasmids.
The pcDNA6.2 plasmids expressing C-terminal V5-tagged SLFN5 and SLFN11 were kindly provided by Sara L. Sawyer (University of Colorado Boulder). Plasmids expressing GFP-SLFN5 (pcDNA6.2-/N-EmGFP-DEST), His-SLFN5 (pDEST17), Adenoviral SLFN5-V5 (pAd/CMV/V5-DEST), and SLFN5-HA under a tetracycline-inducible promoter (pLIX_402, gifted from David Root, Addgene plasmid # 41394) were created using Gateway recombination technology (Invitrogen). Point and deletion mutants of SLFN5 were generated using the Stratagene QuickChange Site-Directed Mutagenesis protocol. pLKO.1-shSLFN5 plasmids (RHS4533-EG162394) were purchased from Dharmacon. pX330-GFP-Cas9 plasmid for human SLFN5 knockout was constructed with single guide RNA (5’-GATGCAGGAAAAGTCACCCT-3’).
Viruses and Titration.
Parental virus HSV-1 strain was 17syn+ and the matched ICP0 deletion mutant ΔICP0 was dl140365. The ICP0 ΔRING domain deletion mutant was FXE66. Viruses were kindly provided by Roger Everett (Glasgow, Scotland) propagated in Vero cells and titrated in U2OS cells. For 5-ethynyl-2’-deoxycytidine (EdC) labelling of HSV-1 genomes, RPE cells were infected with HSV-1 (MOI 0.001) or ΔICP0 (MOI 0.5). At 24 hpi, EdC was added at a final concentration of 0.5 μM. Fresh EdC was pulsed into infected cultures at 24 h intervals until extensive cytopathic effect was observed. Supernatants containing labelled viruses were clarified by centrifugation (423 xg for 10 min) and filtered through a 0.45 μm sterile filter and pelleted using a Beckman TLA100 Ultracentrifuge (33,800 xg for 3h at 4°C). Virion pellets were resuspended and pooled in 500 μl complete DMEM medium. For HSV-1 plaque assays, U2OS cells in 12-well plates were infected with ten-fold serial dilutions of viruses. After virus adsorption for 1h, the cells were overlaid with medium containing 0.5% carboxylmethylcellulose. Plaques were stained with crystal violet at 3 days post infection.
iPOND-MS.
We followed the iPOND protocol in virus infection previously described40,67. Per condition, eight 15 cm cell culture dishes containing HFF (1.0 × 107 cells) were mock-infected or infected with HSV-1 17syn+ or dl1403 at an MOI 3. Cells were pulsed with 10 μM EdU (Invitrogen) for 15 min at 8 hpi. Cells were fixed with 1% paraformaldehyde in PBS for 20 min at room temperature, crosslinking was quenched with 125 mM glycine and cells were scraped and harvested. All conditions were performed in triplicate. Samples were processed for iPOND as described previously40,67, with the following adaptations: after click chemistry reaction, cell pellets were resuspended in 0.5 ml of lysis buffer (20 mM HEPES pH 7.9, 400 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Triton X-100) supplemented with 1 mM dithiothreitol (DTT) and cOmplete™ Protease Inhibitor Cocktail (Roche) and 1 mM PMSF and sonicated with a Bioruptor (Diagenode) for 20 min in 30 sec on/off cycles at a high intensity. Capture of DNA-protein complexes was carried out by incubating lysates with 120 μl streptavidin Dynabeads M-280 (Invitrogen) for 16 h at 4 °C in the dark. Beads were washed once in lysis buffer, once in 1 M NaCl, four times in wash buffer (20 mM HEPES pH 7.4, 110 mM KOAc, 2 mM MgCl2, 0.1% Tween 20, 0.1% Triton X-100, 150 mM NaCl) and once in PBS. Then, 60 μl of 1X LDS sample buffer (Invitrogen) containing 10% DTT was used to elute proteins from the beads. Eluted proteins were boiled at 95 °C for 45 min to reverse crosslinks. iPOND isolates were separated on ~0.8 cm on a 10% Bis-Tris Novex mini-gel (Invitrogen) using the MOPS buffer electrophoresis system. The gel was stained with Coomassie Brilliant Blue and the band excised. Gel segments were destained with 50% methanol/1.25% acetic acid, reduced with 5 mM DTT, and alkylated with 40 mM iodoacetamide (Sigma). Gel pieces were then washed with 20 mM ammonium bicarbonate (Sigma) and dehydrated with acetonitrile (Thermo Scientific). Trypsin (Promega) (5 ng/μl in 20 mM ammonium bicarbonate) was added to the gel pieces and proteolysis was allowed to proceed overnight at 37 °C. Peptides were extracted with 0.3% trifluoroacetic acid (J.T.Baker), followed by 50% acetonitrile. Extracts were combined and the volume was reduced by vacuum centrifugation. Tryptic digests were analyzed by LC-MS/MS on a hybrid LTQ Orbitrap Elite mass spectrometer (Thermo) coupled with a nanoLC Ultra (Eksigent Technologies). Peptides were separated by reverse phase (RP)-HPLC on a nanocapillary column, 75μm id x 15 cm Reprosil-pur 3μM, 120 A (Dr. Maisch, HPLC GmbH) in a Nanoflex chip system (Eksigent). Mobile phase A consisted of 1% methanol (Fisher)/0.1% formic acid (Thermo Fisher Scientific) and mobile phase B of 1% methanol/0.1% formic acid/80% acetonitrile. Peptides were eluted into the mass spectrometer at 300 nl/min with each RP-LC run comprising a 120-min gradient from 5 to 35% B. The mass spectrometer was set to repetitively scan m/z from 300 to 1800 (r = 240,000 for LTQ-Orbitrap Elite) followed by data-dependent MS/MS scans on the twenty most abundant ions, with a minimum signal of 1500, dynamic exclusion with a repeat count of 1, repeat duration of 30 s, exclusion size of 500 and duration of 60 s, isolation width of 2.0, normalized collision energy of 35, and waveform injection and dynamic exclusion enabled. FTMS full scan AGC target value was 1×106, whereas MSn AGC was 1×104, respectively. FTMS full scan maximum fill time was 500 ms, whereas ion trap MSn fill time was 50 ms; microscans were set at one. FT preview mode, charge state screening, and monoisotopic precursor selection were all enabled with rejection of unassigned and 1+ charge states.
MS Data Processing and Database Searching.
MS raw files were analyzed by MaxQuant software version 1.5.2.8. MS/MS spectra were searched by the Andromeda search engine against the Human UniProt FASTA database (9606; 136,251 entries) (version July 2014). The database included 247 common contaminants, discarded during data analysis. The search included variable modifications of methionine oxidation and amino-terminal acetylation, and fixed modification of carbamidomethyl cysteine. Trypsin was specified as the digestive enzyme. Minimal peptide length was set to six amino acids and a maximum of two missed cleavages was allowed. The false discovery rate (FDR) was set to 0.01 for peptide-spectrum matches (PSMs) and protein identifications. Protein grouping was enabled. Peptide identification was performed with an allowed precursor mass deviation up to 4.5 ppm after time-dependent mass calibration and an allowed fragment mass deviation of 20 ppm. Protein identification required at least one unique or razor peptide per protein group. Label-free quantification in MaxQuant was performed using the intensity-based absolute quantification (iBAQ) algorithm. The human proteome was searched using the match-between-runs functionality with the retention time alignment window set to 20 min and the match time window to 1 min. Proteins were filtered to eliminate the identifications from the reverse database, only identified by site and common contaminants. Proteomics data are supplied in Supplementary Table 1.
Data normalization and analysis.
MaxQuant output was filtered to remove identified common contaminants, proteins identified in the reverse protein database, and proteins that were quantified with 0 MS/MS counts. Missing data were imputed using the BPCA method in the pcaMethods68 R package. SLFN5 did not contain missing quantification data. iBAQ intensities were transformed to log2 values, with unidentified values assigned as “NA”. Data were normalized by subtracting the sample medians from log2 transformed iBAQ values within the respective samples. The log2 fold changes of protein iBAQ quantification, used to compare protein abundance across samples, were calculated by comparing the averaged the log2 transformed and normalized iBAQ values for each replicate within the samples. Hypothesis testing was performed using unpaired, two-tailed student’s t-tests comparing the log2 transformed, normalized iBAQ values within the compared samples. Multiple testing correction was not performed69. Z-scores based on log2, normalized average iBAQ values were used to measure relative abundance of a protein within a sample.
3D PCA Clustering Analysis.
PCA analysis was performed on the log2 transformed, normalized protein iBAQ data. The protein abundance data from each replicate, within the mock, WT HSV and ΔICP0 HSV infections, were considered discretely within the PCA analysis (3 replicates x 3 samples = 9 total replicates). PCA was performed using the “prcomp” function, within the R statistical software package. Cluster grouping was evaluated by quantifying “loading” data distances with the first three dimensions of PCA space. A specified protein was assigned as the cluster center and proteins within a sphere of 0.0125 units from the cluster center were clustered with the selected protein. The sphere size was selected based on the range of the distribution of “loading” values for each of the first three PCA dimensions. This approach does not assign proteins to unique clusters if a protein can be grouped with multiple distinct cluster centers. Clusters for known ICP0 substrates (ATRX, IFI16, PML, and DNA-PKcs) were generated and compared. The intersection of the clusters contained the highest confidence predictions of ICP0 substrates within the iPOND dataset.
siRNA.
Gene knockdown experiments by siRNA were carried out using Lipofectamine RNAiMAX transfection reagent (Invitrogen). siGENOME non-targeting control, SLFN5, and SLFN11 SMARTpool siRNAs were purchased from Dharmacon. Cells in 12-well plates were transfected with 20 pmol/ml of siRNA and 2 μl of RNAiMAX.
CRISPR/Cas9 KO.
For the HeLa SLFN5 KO generation, HeLa cells were transfected with pX330, which contains a GFP cassette and Cas9 nuclease, harboring sgSLFN5. At 24h post transfection, GFP expressing cells were sorted by fluorescence-activated cell sorting (FACS) into 96-well plates for clonal expansion. Immunoblot analysis of SLFN5 was used to validate the KO cell lines.
Viral vector production.
Recombinant adenoviruses expressing ICP0 was obtained from P. Schaffer70. Recombinant adenoviruses expressing SLFN5 and K584A mutant were produced using pAd/CMV/V5-DEST vector from Gateway technology (Invitrogen). Then the plasmid was digested with PacI restriction enzyme and transfected into HEK293 cells and adenoviruses were collected at 7 days after transfection, according to the manufacturer’s protocol.
Antibodies.
The following antibodies were used: anti-SLFN5 (Sigma-Aldrich; Cat.HPA017760; Lot.B96361), anti-SLFN11 (Novus Biologicals; Cat.NBP1–92368; Lot.H96783), anti-PML (Bethyl Laboratory; Cat.A301–167A, Santa Cruz; Cat.sc-966; Lot.H1413), anti-IFI16 (Santa Cruz; Cat.sc-8023; Lot.C1312), anti-ATRX (Santa Cruz; Cat.sc-15408), anti-DNA-PKcs (Santa Cruz; Cat.sc-5282; Lot.G280), anti-SUMO2+3 (Abcam; Cat.ab3742; Lot.GR8249–1), anti-RNAP II (Santa Cruz; Cat.sc-56767), anti-GFP rabbit (Abcam; Cat.ab290; Lot.GR3251545–1), anti-GFP mouse (Millipore; Cat.MAB2510; Lot.2512480), anti-RAD50 (GeneTex; Cat.GTX70228; Lot.40186), anti-V5 (Santa Cruz; Cat.sc-271944; Lot.E2217), anti-HA (Abcam; Cat.ab9110; Lot.GR3217183–2), anti-GAPDH (GeneTex; Cat.GTX100118; Lot.42158), anti-α-Tubulin (Santa Cruz; Cat.sc-69969; Lot.DO412), anti-β-Actin (Sigma-Aldrich; Cat.a5441; Lot.064M4789V), anti-KU70 (Abcam; Cat.ab83501; Lot.GR3176811–2), anti-Histone H3 (Abcam; Cat.ab1791; Lot.GR3198176–1), anti-ICP0 (Santa Cruz; Cat.sc-53070; Lot. A0313), anti-ICP8 (gifted from David M. Knipe), anti-TK (Santa Cruz; Cat.sc-28037; Lot.K1813), anti-VP21 and anti-gD (gifted from Gary H. Cohen), anti-IE1/IE2 (Virusys; Cat.P1215; Lot.A1345070), anti-UL44 (Virusys; Cat.ca006–100; Lot.C1034151), adenovirus late protein antibody staining Hexon, Penton and Fiber (gift from James M. Wilson), and anti-DBP (gift from Arnold J. Levine).
Immunoblot Analysis.
Cells were washed with PBS, and total cell extracts were prepared by boiling the cell pellets in NuPAGE 1X LDS Sample Buffer (Invitrogen). Proteins were separated via SDS-PAGE and visualized using SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Scientific) and G:Box imaging system (Syngene). For SLFN5 half-life analysis in HSV-1 infection, HFF cells infected as indicated were incubated with 100 μg/ml of the protein synthesis inhibitor cycloheximide and collected at indicated time points. Protein bands were quantified by densitometric analysis using ImageJ software.
Immunofluorescence.
Cells on glass coverslips were infected with HSV-1 at an MOI 3. Cells were grown on glass coverslips in 24-well plates and either mock infected or infected with the indicated virus. Cells were washed in PBS, fixed in 4% paraformaldehyde for 10 min, permeabilized with 0.5% Triton X-100 in PBS for 10 min or pre-extracted to enhance immunofluorescence signal of DNA-protein complex with a buffer containing 20 mM HEPES pH7.9, 20 mM NaCl, 5 mM MgCl2, and 0.5 % NP40 for 1 min, and then fixed with 4% PFA prior to permeabilization with Triton-X100, and blocked with 3% bovine serum albumin for 1h. Cells were incubated for 1h with primary antibodies followed by Alexa Fluor 488 anti-mouse or Alexa Fluor 647 anti-rabbit antibodies (Invitrogen) for 1h at room temperature. Nuclei were visualized by staining with 4’,6-diamidino-2-phenylindol (DAPI). Coverslips were mounted using ProLong Gold Antifade Reagent (Life Technologies) and immunofluorescence was visualized using a Zeiss LSM 710 Confocal microscope (Cell and Developmental Microscopy Core at UPenn) and ZEN 2011 software. Images were processed using ImageJ. For click chemistry imaging of HSV-1 DNA, cells were infected at an MOI of 3 with WT or ΔICP0 mutant HSV-1 prior to overlay with medium containing 0.5 μM EdC and 50 μM ACG on coverslips. At 6 hpi, cells were washed twice in CSK buffer (10 mM HEPES, 100 mM NaCl, 300 mM Sucrose, 3 mM MgCl2, 5 mM EGTA), fixed in 1.8% formaldehyde, permeabilized with 0.5% Triton X-100 in CSK buffer for 10 min, and blocked with 2% human serum (MP Biomedicals) in PBS for 30 min. EdC labelled vDNA was detected using Alexa Fluor 555 picolyl azide (C10638; Thermo Fisher Scientific) as per manufacturer’s guidelines. Zen black software (Zeiss) was used for image capture, generating cut mask channels, and calculating weighted colocalization coefficients. High-resolution Z-series images were captured under LSM 880 Airy scan deconvolution settings using 1:1:1 capture conditions at 0.035 μm intervals. Images were processed using Imaris (Bitplane) software to produce rendered 3D image reconstructions.
Co-immunoprecipitation (Co-IP).
HEK293T cells (8 × 105 in 6-well plate) were transfected with the indicated plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. At 24 h post-transfection, cells were harvested in 500 μl of ice-cold co-IP buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1% Triton X-100, 50 mM NaF, 1mM Na3VO4) with protease inhibitors and sonicated with a Bioruptor for 5 min in 30 sec on/off cycles at a high intensity. The lysates were cleared and incubated with 2 μg of anti-GFP antibody (Abcam) per sample for 4 h at 4°C with constant rotation. The 20 μl of Dynabeads Protein G (Novex) were added to the samples and rotated at 4°C for 1 h. The beads were washed four times with ice-cold co-IP buffer and resuspended in 1X LDS sample buffer. For endogenous co-IPs, HFF cells (3 × 106 in 100-mm dish) were mock infected or infected with HSV-1 at an MOI of 3. Cells were harvested at indicated time points and sonicated in 500 μl of co-IP buffer. The clarified cell lysates were incubated with 2 μg of anti-ICP0 antibody (Santa Cruz) per sample for 4 h at 4°C. The 20 μl of Dynabeads Protein G were incubated for 1 h. The beads were then washed with co-IP buffer and resuspended in 1X LSD sample buffer.
Chromatin Immunoprecipitation (ChIP).
HeLa cells (8 × 106 per sample) were used for ChIP assays. Cells were fixed with 1% formaldehyde for 15 min and then quenched with 125 mM glycine for 5 min at room temperature. The cell pellet was washed with cold PBS, resuspended in 1 ml ChIP buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.5, 5 mM EDTA, 0.5% NP-40, 1% Triton X-100) supplemented with protease inhibitor and 1 mM PMSF. The nuclei were collected by centrifugation at 2,000 xg for 5 min at 4°C and sonicated with a Bioruptor (Diagenode) for 10 min in 30 sec on/off cycles at a high intensity. The cleared lysate was used for IP with anti-Histone H3 (Abcam; ab1791), anti-HA (Abcam; ab9110) antibodies, and the rabbit or mouse control IgG (Invitrogen; 31884; 31880). Antibodies (5 μg) were incubated with the lysate for 4 h at 4°C with constant rotation. Protein G Dynabeads (40 μl) were added to the samples and rotated at 4°C for 1 h. Each immune complex was washed five times in 1 ml of cold ChIP buffer, eluted by addition of 100 μl of Elution buffer (10 mM Tris-HCl pH 8.0, 5 mM EDTA, 1% SDS, 20 μg of RNase A) via boiling for 10 min at 95°C. After elution of precipitated DNA, qPCRs were performed, the IgG background was subtracted, and the obtained data were normalized to input DNA and expressed as percent input.
Reverse Transcription Polymerase Chain Reaction (RT-PCR) and Quantitative PCR (qPCR).
To measure mRNA levels, total RNA was isolated (RNeasy Mini Kit, Qiagen) and was reverse transcribed using the high-capacity RNA-to-cDNA Kit (Applied Biosystems). The successful removal of DNA contamination was confirmed by amplifying the RNA in each sample without the reverse transcription reaction. For DNA, total DNA was extracted using PureLink genomic DNA Mini Kit (Invitrogen). Amplifying sequences were detected using Power SYBR green (Applied Biosystems) PCR reporter dye in a ViiA 7 real-time PCR system (Applied Biosystems). Relative levels were normalized to RPLP0.
Ubiquitination Assays.
To assess endogenous ubiquitination of SLFN5, cells were harvested in PBS containing 10 mM NEM and lysed in 1% SDS by boiling for 10 min. The lysates were diluted to 0.1% SDS by adding TBST, and immunoprecipitated with anti-ubiquitin (P4D1, Santa Cruz) antibody, followed by immunoblotting. For in vitro ubiquitination reactions, bacterially purified His-SLFN5 (500 ng) was incubated with 50 ng of UBE1 (UBPBio; B1100), 250 ng of UBE2D1 (UBPBio; C1400), 5 μg of ubiquitin (UBPBio, E1100), and 2 mM ATP in the absence or presence of 100 ng of ICP0-His in 40 mM Tris-HCl pH 7.6, 50 mM NaCl, and 1 mM DTT at for 2 h at 4°C. The reaction was stopped by boiling in LDS sample buffer and analyzed by immunoblotting using anti-SLFN5 antibody.
RNA Transcription and Stability Profiling.
To assess relative RNA transcription rate and RNA half-life, cells were treated with 200 μM 4-thiouridine (4sU; Sigma T4509) for exactly 30 min. Infection was stopped and RNA harvested using 1 ml TRIzol (Thermo Fisher Scientific), following manufacturer’s instructions. A fraction of the total RNA was reserved as input, and the remaining 4sU-labeled nascent RNA was biotinylated using MTSEA-Biotin-XX (Biotium; 90066) as previously described71,72. Nascent RNA was separated from unlabeled RNA using MyOne C1 Streptavidin Dynabeads (Thermo Fisher Scientific; 65–001), biotin was removed from nascent RNA using 100 mM dithiothreitol (DTT), and RNA was isopropanol precipitated. Total RNA (1 μg) and an equivalent volume of nascent RNA were converted to cDNA and qPCR was performed as described above. Relative transcription rates were determined by the ΔΔCt method to compare nascent transcript levels between control and siRNA treated cells normalized to nascent GAPDH RNA. RNA half-life was determined using the previously described formula t1/2 = −t × [ln(2)/DR] where t is the 4sU labeling time (0.5 h) and DR is the decay rate defined as Nascent/Total RNA73. Half-lives were normalized to the half-life of GAPDH set at 8 h as previously determined74.
Statistics and Reproducibility.
Biological replicate information is indicated in the figure legends. All results are given as mean ± SD and analyzed by using statistical tools implemented in GraphPad Prism 7.0 software. Statistical analyses were performed using the standard two-tailed unpaired Student’s t test with the assumption of normality for analysis or Mann-Whitney U-test of two groups. Multiple test correction was not implemented. Differences with p < 0.05 were considered to be significant and p-value ranges are provided in each figure. Details regarding statistical analysis are reported in each figure legend, and exact p-values for each analysis are provided as source data.
Data Availability.
Source data for figures and associated statistical analysis are provided with this manuscript. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE75 partner repository with the dataset identifier PXD018773 (http://www.ebi.ac.uk/pride/archive/projects/PXD018773).
Code availability.
The scripts used to analyze the iPOND proteomics data are available from the corresponding author upon request or can be accessed via GitHub. https://github.com/JosephDybas/HSV_iPOND.
Extended Data
Extended Data Fig. 1: Comparative analysis of protein abundance between PCA-based clustering of HSV iPOND data and whole cell proteome data identifies potential substrates of ICP0.
a, Venn diagram of PML, ATRX, IFI16, and DNA-PKcs PCA-based clusters identifies proteins that are clustered with each known substrate. b, Volcano plots showing log2 fold change (x-axis) and associated statistical significance (y-axis) for proteins identified by iPOND proteomics, quantifying proteins associated with HSV genomes. Known ICP0 substrates (green) and clustered proteins (blue and red) are indicated. iPOND data comparing HSV WT and ΔICP0 infection shows known substrates and clustered proteins enriched on the ΔICP0 genome. P-values were calculated using the two-tailed unpaired Student’s t-test. n = 3 biologically independent experiments. c, Heatmap showing iPOND abundances as z-scores for mock, WT HSV-1, and ΔICP0 during infection. This list is proteins that are more than 2-fold enriched (dashed gray box in b) on the ΔICP0 virus genome compared to WT HSV-1 genome. These are clustered using a hierarchical clustering algorithm that analyzes the abundance for each condition. Therefore, the proteins with the same trends for mock-HSV-ΔICP0 are close together on the heatmap. d, Whole cell proteome abundance data over a time course of WT HSV-1 infection show decreases in known ICP0 substrates PML, IFI16, and DNA-PKcs, as well as SLFN5. Additional proteins (gray line) showing similar trends in the heat map (c) are not decreased during HSV infection.
Extended Data Fig. 2: Comparison of SLFN5 degradation in different cells.
a, HEK293T cells were transfected with plasmids encoding SLFN5-V5 and GFP-ICP0 (WT or ΔRING), as indicated. At 24 h after transfection, total cell lysates were prepared, and immunoblot analysis was carried out with indicated antibodies. GAPDH was used as a loading control. b, HEK293T cells were transfected with plasmids encoding GFP-SLFN5 and ICP0 (WT or ΔRING), and the cells were monitored for the GFP signals and the fluorescence intensity was measured by ImageJ software. Scale bar, 300 μm. c, HEK293 cells were infected with WT HSV-1 or ΔRING virus at an MOI of 3. Cell lysates were prepared at the indicated time points and subjected to immunoblot analysis. d, HeLa cells were infected with WT HSV-1 or ΔRING virus at an MOI of 3. Cell lysates were prepared at the indicated time points and subjected to immunoblot analysis. e, HeLa cells were transduced with recombinant adenoviral vectors encoding ICP0 together with Ad.C-rtTA, an adenovirus expressing the doxycycline regulated transactivator. At 24 h after transduction in the absence or presence of doxycycline (1 μg/ml), immunoblot analysis was performed with indicated antibodies. f, g, Similar experimental setup as in (d and e); U2OS cells were used. h, SLFN5 degradation is independent of SUMOylation, and is detected in cells depleted of the SUMO E2 conjugating enzyme UBC9. i, In vitro ubiquitination assay with bacterially purified SLFN5 Δ730–891 truncation mutant which lacks the ICP0 interaction region demonstrates it is not ubiquitinated. Immunoblot data shows representative data from n = 3 biologically independent experiments.
Extended Data Fig. 3: ICP0 interacts SLFN5 via the carboxy terminal region of SLFN5.
a, Subcellular localization of SLFN5 mutants was analyzed. HEK293T cells were transfected with plasmids encoding GFP-SLFN5 (full-length or various deletion mutants). At 24 hours after transfection, the cells monitored for the GFP signals. Scale bar, 50 μm. b, HEK293T cells were co-transfected with plasmids encoding various version of GFP-SLFN5 and ICP0 ΔRING or cytoplasmic ICP0 mutant (cICP0, also called D8) ΔRING, as indicated. The cells were subject to co-IP with anti-GFP Ab, followed by immunoblotting. Immunoblot and immunofluorescence images show representative data from n = 3 biologically independent experiments. c, SLFN5 interaction with ICP0 maps to a part of the intrinsicly disordered region of SLFN5. The SLFN5 and SLFN11 protein sequences were analyzed for disorder tendency using IUPred2A. The domain structure of SLFN is shown above. The green bar indicates an interaction region of ICP0 or a homolgous region of SLFN11 with SLFN5. Red lines indicate unique disorder region of SLFN5.
Extended Data Fig. 4: Colocalization of SLFN5 with replication compartments of HSV-1.
a, HFF cells were infected with WT HSV-1 at an MOI of 10 and immunofluorescence was examined at 2 hpi. ICP4 marks viral pre-replication foci from incoming viral genomes. b, HFF cells were infected with WT HSV or ΔICP0 mutant virus at an MOI of 3 and immunofluorescence was examined at 8 hpi. ICP8 marks viral replication compartments and DAPI marks cellular DNA. Scale bar, 10 μm. Fluorescence plot profile analyzed by ImageJ showed colocalization of SLFN5 and viral proteins (at dashed line). Immunofluorescence images show representative data from n = 3 biologically independent experiments.
Extended Data Fig. 5: SLFN5 suppresses HSV-1 replication.
a, Efficiency of SLFN5 knockdown in HFF and HeLa cells. Immunoblot analysis indicating SLFN5 and β-actin levels following transduction with lentiviruses encoding shRNAs that are nontargeting control (Ctrl) or specific for SLFN5 (#1 to #5). b, Viral DNA replication yields from shRNA-transduced HFF cells infected with ΔRING virus at an MOI of 0.1 and harvested at 24 h after infection. Quantification of viral DNA was carried out by qPCR. n = 5 biologically independent experiments. c, HeLa cells transduced with shRNA were mock infected or infected with HSV-1 (WT or ΔRING) at different MOIs (0.5 to 3) as indicated. Cell lysates were prepared at 9 h after infection and immunoblot nalysis was performed with antibodies to ICP0, VP21, and β-actin. d, Phase micrograph of cell morphology under subconfluent or confluent culture conditions. Scale bar, 100 μm. e, Growth of shRNA transduced cells over time. n = 3 biologically independent experiments. f, 2,000 cells were seeded in a 96-well plate, and cell proliferation was measured over 6 days by colorimetric analysis using the water-soluble tetrazolium salt, WST-8 (Cell Counting Kit-8, Dojindo) at the indicated time point. n = 3 biologically independent experiments. g, CRISPR/Cas9-mediated SLFN5 gene editing and permanent depletion from HeLa cells. Guide RNA transfected cells were selected and cloned for a month. Immunoblot analysis of SLFN5 in control cells or SLFN5 knockout cells. Viral DNA replication yields from cells infected with ΔRING at an MOI of 0.1 were analyzed by qPCR. HeLa cells were infected with ΔRING at an MOI of 0.1 for 24 h. Viral DNA was measured by qPCR. n = 3 biologically independent experiments. Data are the mean ± SD. Comparisons between groups were performed using the two-tailed unpaired Student’s t-test. **, p < 0.005, ***, p < 0.0005, n.s., not significant. Immunoblots show representative data from n = 3 biologically independent experiments.
Extended Data Fig. 6: SLFN5 Δ730–891 mutant does not inhibit HSV-1 replication.
a, HeLa cells were stably transduced with lentivirus containing tetracycline-inducible SLFN5-HA genes (WT and Δ730–891). SLFN5-HA was induced with doxycycline (0.5 μg/ml) for 48 h. SLFN5 expression was confirmed by immunoblot analysis. b, c, Cells were infected with HSV-1 (b) and ΔRING virus (c) at an MOI of 0.1. At 24 hpi, viral DNA was measured by qPCR. n = 3 biologically independent experiments. Data are the mean ± SD. Comparisons between groups were performed using the two-tailed unpaired Student’s t-test. **, p < 0.005. n.s., not significant.
Extended Data Fig. 7: Degradation of SLFN5 by other DNA viruses.
a, HFF cells were mock infected or infected with HSV-2 at an MOI of 3, and MG132 (5 μM) was added at 2 h after virus inoculation as indicated. Total cell lysates were prepared at indicated time points and examined by immunoblot analysis with indicated antibodies. ICP8 serves as an infection control. b, HFF cells were mock infected or infected with HCMV (TB40/E) at an MOI of 3 and total cell lysates were prepared at indicated time points and examined by immunoblot analysis with indicated antibodies. UL44 serves as an infection control. Circle marks cross-reacting viral protein. c, A549 cells were mock infected or infected with adenovirus (Ad5) at an MOI of 20 and MG132 (5 μM) was added at 2 h after virus inoculation, as indicated. Cell lysates were prepared at the indicated time points and examined by immunoblot analysis with indicated antibodies. RAD50 served as a degradation control and DBP as a control for infection. Immunoblots show representative data from n = 3 biologically independent experiments.
Extended Data Fig. 8: Restriction of other DNA viruses by SLFN5.
HeLa cells were mock infected or infected with other DNA viruses as indicated. a, Immunoblot analysis demonstrating increased HSV-2 protein levels in SLFN5-depleted cells. b, HCMV proteins are slightly increased in SLFN5-depleted cells at immediate-early time point (24 hpi), but decreased at late times (72 hpi) as compared to control cells. The ISG15 protein was increased in the absence of SLFN5 and further increased upon HCMV infection. c, SLFN5 does not affect adenovirus protein levels. d, SLFN5 knockdown results in higher level of ISG15 expression in HFF and HeLa cells. Immunoblots show representative data from n = 3 biologically independent experiments.
Extended Data Fig. 9: Effects of SLFN5 on HCMV infection.
a, b, HCMV mRNA (UL123, an immediate-early gene and UL94, a late gene) expression from shRNA-transduced HFF cells infected with HCMV at an MOI of 3 and harvested at 12 h (a) and 72 h (b) after infection. Quantification of viral transcripts was carried out by RT-qPCR. Viral DNA replication yields from shRNA-transduced HFF cells infected with ΔRING virus at an MOI of 0.1 and harvested at 24 h after infection. Quantification of viral DNA was carried out by qPCR. c, Progeny production of HCMV was monitored by infectious center assays using anti-IE1 antibody. n = 3 biologically independent experiments. Data are the mean ± SD. Comparisons between groups were performed using the two-tailed unpaired Student’s t-test. *, p < 0.05, **, p < 0.005.
Extended Data Fig. 10: Interaction of SLFN5 with HSV-1 DNA.
a. ChIP assay with anti-HA antibody was performed in SLFN5-HA-expressed HeLa cells. ΔRING virus was infected at an MOI of 3 for indicated time points in the absence or presence of PAA. ChIP DNA was assayed by qPCR using primers specific for US11 TSS. n = 3 biologically independent experiments. b. ΔRING virus was infected at an MOI of 3 for 3 h. ChIP DNA was assayed by qPCR using primers specific for the HSV-1 DNA or human DNA loci. n = 3 biologically independent experiments. Data are the mean ± SD. Comparisons between groups were performed using the two-tailed unpaired Student’s t-test. *, p < 0.05, **, p < 0.005.
Supplementary Material
Acknowledgements
We thank members of the Weitzman Lab for insightful discussions and input. We are grateful to Gary H. Cohen, Roger D. Everett, David M. Knipe, Sara L. Sawyer, and Priscilla A. Schaffer for generous gifts of reagents. C. B. was supported by the Medical Research Council (http://mrc.ukri.org) grantMC_UU_12014/5. Work in the Weitzman lab was supported in part by grants from the National Institutes of Health (AI115104 and NS082240 to M.D.W.) and funds from the Children’s Hospital of Philadelphia. A.M.P. was supported by the NCI T32 Training Grant in Tumor Virology T32-CA115299 and Individual National Research Service Award F32-AI138432. J.M.D. was supported by Individual National Research Service Award F32-AI147587.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Malim MH & Bieniasz PD HIV Restriction Factors and Mechanisms of Evasion. Cold Spring Harb. Perspect. Med. 2, a006940 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu H & Zheng Y Role of ND10 nuclear bodies in the chromatin repression of HSV-1. Virol. J 13, 62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tavalai N & Stamminger T Intrinsic cellular defense mechanisms targeting human cytomegalovirus. Virus Res. 157, 128–33 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Glass M & Everett RD Components of Promyelocytic Leukemia Nuclear Bodies (ND10) Act Cooperatively To Repress Herpesvirus Infection. J. Virol. 87, 2174–2185 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crow MS, Lum KK, Sheng X, Song B & Cristea IM Diverse mechanisms evolved by DNA viruses to inhibit early host defenses. Crit. Rev. Biochem. Mol. Biol. 51, 452–481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajsbaum R & García-Sastre A Viral evasion mechanisms of early antiviral responses involving regulation of ubiquitin pathways. Trends Microbiol. 21, 421–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dybas JM, Herrmann C & Weitzman MD Ubiquitination at the interface of tumor viruses and DNA damage responses. Curr. Opin. Virol. 32, 40–47 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colomer-Lluch M, Ruiz A, Moris A & Prado JG Restriction Factors: From Intrinsic Viral Restriction to Shaping Cellular Immunity Against HIV-1. Front. Immunol 9, 2876 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simon V, Bloch N & Landau NR Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat. Immunol. 16, 546–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komatsu T, Nagata K & Wodrich H The Role of Nuclear Antiviral Factors against Invading DNA Viruses: The Immediate Fate of Incoming Viral Genomes. Viruses 8, 1–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bieniasz PD Restriction factors: A defense against retroviral infection. Trends Microbiol. 11, 286–291 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Harris RS, Hultquist JF & Evans DT The restriction factors of human immunodeficiency virus. J. Biol. Chem. 287, 40875–40883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boutell C & Everett RD Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J. Gen. Virol. 94, 465–481 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Knipe DM & Cliffe A Chromatin control of herpes simplex virus lytic and latent infection. Nature Reviews Microbiology vol. 6 211–221 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Cai W & Schaffer PA Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J. Virol. 66, 2904–2915 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodríguez MC, Dybas JM, Hughes J, Weitzman MD & Boutell C The HSV-1 ubiquitin ligase ICP0: Modifying the cellular proteome to promote infection. Virus Res. 285, 198015 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boutell C & Everett RD The herpes simplex virus type 1 (HSV-1) regulatory protein ICP0 interacts with and ubiquitinates p53. J. Biol. Chem. 278, 36596–36602 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Everett RD, Orr A & Preston CM A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 17, 7161–7169 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lium EK & Silverstein S Mutational analysis of the herpes simplex virus type 1 ICP0 C3HC4 zinc ring finger reveals a requirement for ICP0 in the expression of the essential alpha27 gene. J. Virol. 71, 8602–8614 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkinson J, Lees-Miller SP & Everett RD Herpes Simplex Virus Type 1 Immediate-Early Protein Vmw110 Induces the Proteasome-Dependent Degradation of the Catalytic Subunit of DNA-Dependent Protein Kinase. J. Virol. 73, 650–657 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lees-Miller SP et al. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J. Virol. 70, 7471–7477 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaurushiya MS et al. Viral E3 Ubiquitin Ligase-Mediated Degradation of a Cellular E3: Viral Mimicry of a Cellular Phosphorylation Mark Targets the RNF8 FHA Domain. Mol. Cell 46, 79–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lilley CE et al. A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses. EMBO J. 29, 943–55 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chelbi-Alix MK & De Thé H Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18, 935–941 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Everett RD et al. The Disruption of ND10 during Herpes Simplex Virus Infection Correlates with the Vmw110- and Proteasome-Dependent Loss of Several PML Isoforms. J. Virol. 72, 6581–6591 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perusina Lanfranca M, Mostafa HH & Davido DJ Two Overlapping Regions within the N-Terminal Half of the Herpes Simplex Virus 1 E3 Ubiquitin Ligase ICP0 Facilitate the Degradation and Dissociation of PML and Dissociation of Sp100 from ND10. J. Virol. 87, 13287–13296 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukashchuk V & Everett RD Regulation of ICP0-Null Mutant Herpes Simplex Virus Type 1 Infection by ND10 Components ATRX and hDaxx. J. Virol. 84, 4026–4040 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orzalli MH, DeLuca NA & Knipe DM Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc. Natl. Acad. Sci. U. S. A. 109, E3008–17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Everett RD, Earnshaw WC, Findlay J & Lomonte P Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 18, 1526–1538 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lomonte P, Sullivan KF & Everett RD Degradation of Nucleosome-associated Centromeric Histone H3-like Protein CENP-A Induced by Herpes Simplex Virus Type 1 Protein ICP0. J. Biol. Chem. 276, 5829–5835 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Lomonte P & Morency E Centromeric protein CENP-B proteasomal degradation induced by the viral protein ICP0. FEBS Lett. 581, 658–662 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Perusina Lanfranca M, Mostafa HH & Davido DJ HSV-1 ICP0: An E3 Ubiquitin Ligase That Counteracts Host Intrinsic and Innate Immunity. Cells 3, 438–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alandijany T Host Intrinsic and Innate Intracellular Immunity During Herpes Simplex Virus Type 1 (HSV-1) Infection. Frontiers in Microbiology vol. 10 2611 (Frontiers Media S.A., 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merkl PE, Orzalli MH & Knipe DM Mechanisms of Host IFI16, PML, and Daxx Protein Restriction of Herpes Simplex Virus 1 Replication. J. Virol. 92, 1–19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lilley CE, Chaurushiya MS, Boutell C, Everett RD & Weitzman MD The intrinsic antiviral defense to incoming HSV-1 genomes includes specific DNA repair proteins and is counteracted by the viral protein ICP0. PLoS Pathog 7, e1002084 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuchet-Lourenço D et al. SUMO pathway dependent recruitment of cellular repressors to herpes simplex virus type 1 genomes. PLoS Pathog. 7, e1002123 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sirbu BM et al. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 25, 1320–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dembowski JA & DeLuca NA Selective recruitment of nuclear factors to productively replicating herpes simplex virus genomes. PLoS Pathog. 11, e1004939–e1004939 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dembowski JA & DeLuca NA Temporal Viral Genome-Protein Interactions Define Distinct Stages of Productive Herpesviral Infection. MBio 9, e01182–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reyes ED et al. Identifying Host Factors Associated with DNA Replicated During Virus Infection. Mol. Cell. Proteomics 16, 2079–2097 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu F, Zhou P, Wang Q, Zhang M & Li D The Schlafen family: complex roles in different cell types and virus replication. Cell Biol. Int. 42, 2–8 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Mavrommatis E, Fish EN & Platanias LC The schlafen family of proteins and their regulation by interferons. J. Interferon Cytokine Res. 33, 206–210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katsoulidis E et al. Role of Interferon α (IFNα)-inducible Schlafen-5 in Regulation of Anchorage-independent Growth and Invasion of Malignant Melanoma Cells. J. Biol. Chem. 285, 40333–41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sassano A et al. Human Schlafen 5 (SLFN5) Is a Regulator of Motility and Invasiveness of Renal Cell Carcinoma Cells. Mol. Cell. Biol. 35, 2684–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arslan AD et al. Human SLFN5 is a transcriptional co-repressor of STAT1-mediated interferon responses and promotes the malignant phenotype in glioblastoma. Oncogene 36, 6006–6019 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li M et al. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 491, 125–128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seong RK et al. Schlafen 14 (SLFN14) is a novel antiviral factor involved in the control of viral replication. Immunobiology 222, 979–988 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J-Y et al. Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control. Nat. Commun 9, 1165 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valdez F et al. Schlafen 11 restricts flavivirus replication. J. Virol. 93, 104–123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De La Casa-Espero’n E From mammals to viruses: The Schlafen genes in developmental, proliferative and immune processes. Biomol. Concepts 2, 159–169 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Kulej K et al. Time-resolved Global and Chromatin Proteomics during Herpes Simplex Virus Type 1 (HSV-1) Infection. Mol. Cell. Proteomics 16, S92–S107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schreiner S et al. Proteasome-Dependent Degradation of Daxx by the Viral E1B-55K Protein in Human Adenovirus-Infected Cells. J. Virol. 84, 7029–7038 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dutrieux J et al. PML/TRIM19-Dependent Inhibition of Retroviral Reverse-Transcription by Daxx. PLoS Pathog. 11, 1–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernassola F et al. Ubiquitin-dependent Degradation of p73 Is Inhibited by PML. J. Exp. Med. 199, 1545–1557 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Louria-Hayon I et al. The Promyelocytic Leukemia Protein Protects p53 from Mdm2-mediated Inhibition and Degradation. J. Biol. Chem. 278, 33134–33141 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Boutell C et al. A viral ubiquitin ligase has substrate preferential SUMO targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. PLoS Pathog. 7, e1002245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sloan E et al. Analysis of the SUMO2 Proteome during HSV-1 Infection. PLoS Pathog. 11, e1005059 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar R, González-Prieto R, Xiao Z, Verlaan-de Vries M & Vertegaal ACO The STUbL RNF4 regulates protein group SUMOylation by targeting the SUMO conjugation machinery. Nat. Commun. 8, 1809 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neumann B, Zhao L, Murphy K & Gonda TJ Subcellular localization of the Schlafen protein family. Biochem. Biophys. Res. Commun. 370, 62–66 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Taylor KE, Chew MV, Ashkar AA & Mossman KL Novel roles of cytoplasmic ICP0: proteasome-independent functions of the RING finger are required to block interferon-stimulated gene production but not to promote viral replication. J. Virol. 88, 8091–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lou DI et al. An Intrinsically Disordered Region of the DNA Repair Protein Nbs1 Is a Species-Specific Barrier to Herpes Simplex Virus 1 in Primates. Cell Host Microbe 20, 178–188 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alandijany T et al. Distinct temporal roles for the promyelocytic leukaemia (PML) protein in the sequential regulation of intracellular host immunity to HSV-1 infection. PLoS Pathog. 14, e1006769–e1006769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Geserick P, Kaiser F, Klemm U, Kaufmann SHE & Zerrahn J Modulation of T cell development and activation by novel members of the Schlafen (slfn) gene family harbouring an RNA helicase-like motif. Int. Immunol. 16, 1535–1548 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Stabell AC et al. Non-human primate schlafen11 inhibits production of both host and viral proteins. PLOS Pathog. 12, e1006066 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stow ND & Stow EC Isolation and Characterization of a Herpes Simplex Virus Type 1 Mutant Containing a Deletion within the Gene Encoding the Immediate Early Polypeptide Vmw110. J. Gen. Virol. 67, 2571–2585 (1986). [DOI] [PubMed] [Google Scholar]
- 66.Everett RD Construction and Characterization of Herpes Simplex Virus Type 1 Mutants with Defined Lesions in Immediate Early Gene 1. J. Gen. Virol. 70, 1185–1202 (1989). [DOI] [PubMed] [Google Scholar]
- 67.Sirbu BM, Couch FB & Cortez D Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat. Protoc. 7, 594–605 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stacklies W, Redestig H, Scholz M, Walther D & Selbig J pcaMethods - A bioconductor package providing PCA methods for incomplete data. Bioinformatics 23, 1164–1167 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Pascovici D, Handler DCL, Wu JX & Haynes PA Multiple testing corrections in quantitative proteomics: A useful but blunt tool. Proteomics 16, 2448–2453 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Halford WP, Kemp CD, Isler JA, Davido DJ & Schaffer PA ICP0, ICP4, or VP16 Expressed from Adenovirus Vectors Induces Reactivation of Latent Herpes Simplex Virus Type 1 in Primary Cultures of Latently Infected Trigeminal Ganglion Cells. J. Virol. 75, 6143–6153 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Poling BC, Price AM, Luftig MA & Cullen BR The Epstein-Barr virus miR-BHRF1 microRNAs regulate viral gene expression in cis. Virology 512, 113–123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Russo J, Heck AM, Wilusz J & Wilusz CJ Metabolic labeling and recovery of nascent RNA to accurately quantify mRNA stability. Methods 120, 39–48 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Price AM, Messinger JE & Luftig MA c-Myc Represses Transcription of Epstein-Barr Virus Latent Membrane Protein 1 Early after Primary B Cell Infection. J. Virol. 92, e01178–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dölken L et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 14, 1959–1972 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perez-Riverol Y et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for figures and associated statistical analysis are provided with this manuscript. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE75 partner repository with the dataset identifier PXD018773 (http://www.ebi.ac.uk/pride/archive/projects/PXD018773).