Abstract
Colorectal cancer (CRC) remains one of the most commonly diagnosed cancers and a leading cause of cancer-related deaths worldwide. The stepwise accumulation of epigenetic alterations in the normal colorectal epithelium has been reported to act as a driving force for the initiation and promotion of tumorigenesis in CRC. From a mechanistic standpoint, emerging evidence indicates that within the colonic epithelium, the diverse gut microbiota can interact with host cells to regulate multiple physiological processes. In fact, recent studies have found that the gut microbiota represents a potential cause of carcinogenesis, invasion, and metastasis via DNA methylation, histone modifications, and non-coding RNAs - providing an epigenetic perspective for the connection between the gut microbiota and CRC. Herein, we comprehensively review the recent research that provides a comprehensive yet succinct evidence connecting the gut microbiota to CRC at an epigenetic level, including carcinogenic mechanisms of cancer-related microbiota, and the potential for utilizing the gut microbiota as CRC biomarkers. These scientific findings highlight a promising future for manipulating the gut microbiota to improve clinical outcomes in patients suffering from CRC.
Keywords: Colorectal cancer, Gut microbiota, Diagnostic biomarkers, Epigenetic modifications, non-coding RNAs
1. INTRODUCTION
Colorectal cancer (CRC) remains the third leading cause of cancer-related deaths among men and women in the United States, with an estimated 147,950 new cases and 53,200 deaths projected in 2020 [1]. Even with advances in CRC screenings and therapeutic strategies, CRC still remains one of the most devastating malignancies. Patients with metastatic CRC have a median overall survival of only ~30 months [2]. Comprehensive studies on disease pathogenesis at the molecular level have provided a unique perspective for its detection, surveillance, and therapeutic approaches for improving outcomes and survival in CRC patients [3]. The driving force for CRC tumorigenesis is a combination of multiple factors, with various host and environmental factors involved in tumor formation and growth [4]. To some extent, CRC has been described as a hereditary susceptibility syndrome [5], in which genetic predisposition or familial influences, such as Lynch Syndrome [6], familial adenomatous polyposis [7], and Peutz–Jeghers syndrome [8], influence the overall risk of developing CRC. However, it is estimated that only a minority of CRC cases develop through germline transmission of genetic alterations; while the majority of cases are believed to result from host-environmental interactions [9]. Environmental factors thought to act as either carcinogens or tumor-promoting agents can manifest in the accumulation of epigenetic variations in host cells, among which the gut microbiota as emerged as an important player for its pathogenic role in various cancers, including CRC [10, 11]. Therefore, a better understanding of the epigenetic connection between the gut microbiota and CRC pathogenesis will likely yield novel insights into the impact of environmental exposure in CRC. In this review article, using comprehensive search terms related to the gut microbiota, CRC and epigenetics, we conducted a literature search through the database searches in PubMed, Embase, and Web of Science to search for all relevant articles and abstracts up until Novermber 2020, including both clinical trials and basic research. We highlight the carcinogenic mechanisms of CRC-related microbiota and discuss the potential for utilizing the gut microbiota as biomarkers in this malignancy. Such a knowledge will provide a theoretical basis for the potential use of gut microbiota as biomarkers for cancer screening, diagnosis and risk prediction.
2. THE GUT MICROBIOTA AND CRC
For human beings, from birth, microbiota colonizes the skin, digestive tract, respiratory tract, reproductive tract, and other parts in contact with the external environment, among which the colon and rectum represent ideal habitats for containing the largest number and variety of microbiota [12]. With the most expansive mucosal surface area in contact with the outside environment, the human colorectal epithelium comprises of a considerable gut microbiota ecosystem, comprising of more than 1014 microorganisms, that primarily include Bacteroidetes, Proteobacteria, Actinobacteria, Firmicutes, and Fusobacteria [13]. In this context, the intestinal environment, dietary regulation, and drug metabolism have a rapid and major impact on the communal characteristics of the gut microbiota [14, 15]. The gut microbiota, in turn, plays a fundamental role in overall human health, by assisting in host digestion and absorption, regulating the development and function of the mucosal barrier, promoting the maturation of immune tissues, and affecting tolerance to gastrointestinal antigens [16, 17]. However, at the same time, metabolites of the gut microbiota that have toxic effects on the human body, including but not limited to phenol, p-cresol and indole are recognized to be involved in the occurrence and development of CRC [18]. In general, the gut microbiota likely acts as a common denominator during cancer pathogenesis by acting as bridge for various tumor stages, from environmental effects and inflammation of the gastrointestinal mucosa to colorectal mucosal cancerization.
2.1. ADVANCED APPROACHES FOR IDENTIFICATION OF GUT MICROBIOTA
Polymerase chain reaction (PCR), which can identify the presence and abundance of specific bacterial strains among the complex community of microbiota through specific primer design, has been considered to be the most attractive approach for studying the gut microbiota profiles. However, PCR testing is limited to detecting a particular strain using specifically designed primers, and thus cannot fully provide a comprehensive description of the gut microbiome [19]. The emergence of next-generation sequencing (NGS) technologies have enabled large-scale studies of the gut microbiota to identify previously unknown bacterial species and strains, and has revealed high variability in microbiome composition between individuals and between different body sites within the same person [20, 21]. To date, several types of large-scale analyses based on NGS techniques have been used to assess the gut microbiota, including 16S amplicon sequencing, which is based on sequencing hypervariable regions of the 16S ribosomal RNA (rRNA) [22], and shotgun analysis, which is based on direct sequencing of the total DNA (metagenome) and/or total RNA (metatranscriptome) [23]. Such techniques have greatly expanded our knowledge of the diversity and multi-functionality of the gut microbiota (Figure 1).
Fig. 1.
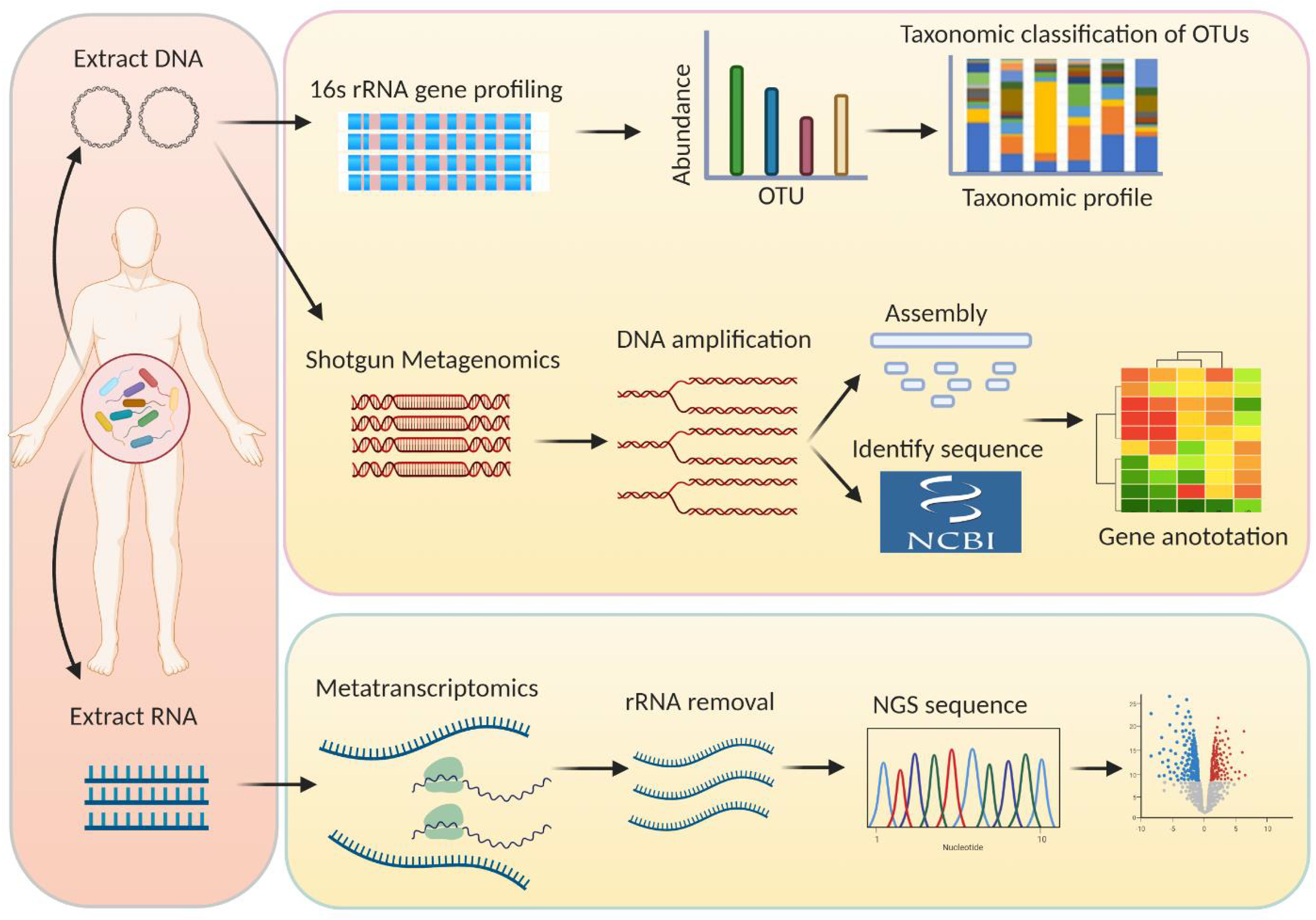
General pipeline of different sequencing and bioinformatic strategies for the gut microbiota analysis. Both methods starting from DNA and RNA extraction of the gut microbiota samples. The extracted DNA is either subjected to 16S rRNA gene profiling or sheared into small DNA fragments to perform shotgun metagenomics, and after rRNA removal, the extracted RNA is subjected to metatranscriptomic sequencing. rRNA, ribosomal RNA; OUT, operational taxonomic units; NGS, next-generation sequencing.
In this context, 16S amplicon sequencing is a relatively simple, low-cost method to obtain a broad overview of microbiome composition [24], in which specific regions common to all microbiota genomes are first amplified and sequenced. Sequencing reads are clustered into operational taxonomic units (OTUs) based on sequence homology against known 16S rRNA gene databases, which is a commonly used measure of microbial diversity. Population abundance is quantified and the diversity of the microbiome is identified by the resulting OTUs in various specimens [25]. However, data generated by 16S amplicon sequencing is limited in several aspects, due to the lack of information on microbiota function and limited “taxonomic resolution.” For instance, in most cases, microbiota cannot be identified precisely at the species level [26]. To overcome these limitations, metagenomic shotgun sequencing—sequencing all genes presented in the microbiome rather than just a single taxonomic marker gene—can provide information about the abundances of genes at all taxonomic levels by mapping reference genomes/genes. Metagenomic shotgun sequencing can also identify gene content and infer its functional potential of proteins encoding in the microbiome using functional annotated databases [27]. Both 16S amplicon sequencing and metagenomic shotgun sequencing are based on genomic DNA sequences from microbiota samples. RNA-expressing genes express their functional activity better than DNA because many genes are only conditionally expressed. RNA transcripts from the microbiome provide a more comprehensive description of gene expression of the microbiota. Therefore, metatranscriptomic sequencing allows the identification of expressed transcripts, providing different insights from DNA‐based microbiome sequencing methods. Transcriptome abundance can also be used to compare gene expression profiles between various microbiota. However, it is more expensive to obtain comparable profiles of the microbiota using metagenomic and metatranscriptomic sequencing compared to using 16S amplicon sequencing [25]. In addition, data generated from metagenomic and metatranscriptomic sequencing has been challenging to analyze, owing to its substantial complexity compared to data generated by 16S amplicon sequencing.
2.2. COMPOSITION OF GUT MICROBIOTA IN HEALTHY PEOPLE
The structure and physiological activity of the gut microbiota are closely related to the health of an organism [28]. Gut microbiota are combined in a certain proportion; wherein, each strain is interdependent and competes to maintain a critical ecological balance [29]. In the normal human gastrointestinal tract, the relationship between the host and the gut microbiota is mutually beneficial and symbiotic: the digestive tract provides an environment for the colonization and survival of specific microbiota, and the microbiota in turn plays unique functions in maintaining intestinal health, such as nutrient metabolism and intestinal protection [29]. In addition, the gut microbiota participates in catabolism and synthesis of substances in the intestinal tract and decomposes macromolecular compounds that cannot be otherwise digested by the host into final metabolic products, providing energy for the host as well as nutrition for its own growth and reproduction [30]. For example, gram-positive microbiota, such as Lactobacillus spp., Streptococcus spp., and Faecalibacterium spp. can synthesize B vitamins, vitamin K, niacin, and a variety of amino acids with anti-inflammatory, anti-tumor and anti-bacterial effects [31–33]. Furthermore, epithelial cells, mucosal layers, and intestinal microbiota secretions can form an effective intestinal barrier to prevent the invasion of harmful bacteria and other pathogens [34].
The rapid development of NGS technologies is deepening our understanding of the origin of the gut microbiota and the landscape of its evolution. Due to differences in oxygen content, pH values, antimicrobial peptide levels and intestinal motility at different anatomical sites within the gut, the overall composition of the gut microbiota varies greatly [35, 36]. The gut microbiota colonizes the entire length of the intestine tract, and the load of microbiota generally increases from the duodenum to the distal colon, ranging from 103 to 104 mL−1 content in the stomach, duodenum, and jejunum, to 108 mL−1 in the ileum, and up to 1011 mL−1 in the colon [35, 36]. Gut-commensal microbiota can be anatomically defined as: (i) lumen-commensal, (ii) mucus‐resident, (iii) epithelium‐resident, and (iv) lymphoid tissue-resident populations (summarized in Table 1). The translocation and ecological imbalance of gut commensal microbiota are closely related to carcinogenesis and development of CRC.
Table 1.
The Composition of Microbiota in Various Anatomical Regions of the Gut
| Location | Major microbiota | REFs |
|---|---|---|
| Lumen-commensal microbiota | Bacteroides spp., Prevotella spp., Mucispirillum spp., Lactobacillus spp., Ruminococcus spp., Oscillospira spp., Sutterella.spp, Desulfovibrio spp., Fusobacterium spp. | [126] |
| Mucus‐resident microbiota | Streptococcaceae, Actinomycinaeae, Corynebacteriaceae, Mucispirillum spp., Lachnospiraceae, Lactobacillus spp., Veillonella spp., Helicobacter spp. | [127, 128] |
| Epithelium‐resident microbiota | Adherent-invasive E. coli, segmented filamentous bacteria, B. fragilis, Clostridium spp. | [76, 129] |
| Lymphoid tissue-resident microbiota | Achromobacter spp., Alcaligenes spp., Bordetella spp. and Ochrobactrum spp. | [130] |
2.3. COMPOSITION OF GUT MICROBIOTA IN PATIENTS WITH CRC
The microbial diversity and balance are the key characteristic features of a healthy gut, as a rich gastrointestinal ecosystem can cope with the challenges of various factors that promote disease occurrence [37]. By comparing the gut microbiota of younger individuals (20–39 years old) vs. elderly (>60 years old), the results have revealed that younger individuals tend to gain more microbial taxa, while elderly individuals tend to lose microbiota diversity in a healthy gut[38]. Although CRC incidence and mortality trends have declined overall, these trends in early-onset CRC (EOCRC; in patients <50 years old) are actually on a rise, worldwide[39]. By analyzing healthcare claims data from all geographic areas of the United States, it is confirmed that the increased risk of EOCRC is associated with metabolic dysregulation, which is often accompanied by the presence of the gut microbiota dysbiosis [40]. Nowadays, due to various risk factors such as antibiotic use, diet, obesity and stress, the gut microbiota dysbiosis often occurs in younger generations, which may in part explain the increasing risk of EOCRC[41]. Even in the individuals with genetic predisposition to CRC, the gut microbiota still contributes to CRC risk substantially. For example, a significant increase of Bacteroidetes and Proteobacteria as well as a reduction of Firmicutes were observed in Lynch syndrome fecal samples [42]. Leveraging stool meta-transcriptomes, another study showed that the progression toward carcinogenesis of lynch syndrome can be predicted in modest power by gut microbial transcription[43]. Accumulated studies to detect gut microbiota in experimental animal models and in patients have indicated that various stages of CRC disease progression are often associated with significant ecological disorders in CRC tissues and with microorganisms in the adjacent mucosa [44]. [42, 43]Key characteristics of major changes in the gut microbiota associated with CRC are summarized in Table 2. For example, in patients with adenomas, the abundance of Bilophilia, Desulfovibrio, Corynebacterium and Phascolarctobacterium in the fecal matter was significantly higher, whereas in patients with serrated polyps, Erysipelotrichia and Fusobacteria were predominantly more, compared with their relatively abundance in healthy people [45]. Both adenomas and serrated polyps may develop into CRC. As adenomas and serrated polyps develop into CRC, toxins produced by pathogenic microbiota present an increasing trend in intestinal mucosal tissues, such as cytotoxic necrosis factor and cycle suppressor produced by Fusobacterium nucleatum and Bacteroidetes fragilis. In addition, as illustrated in Figure 2, the abundance of some of the invasive microbiota also illustrates a similar upward trend, which includes enteroinvasive Escherichia coli (EIEC) [46].
Table 2.
Major Gut Microbiota Associated with the Occurrence and Development of CRC
| Microbiota | Model | Association with CRC | REFs |
|---|---|---|---|
| F. nucleatum | CRC xenograft mouse model | Promotes metastasis by activating autophagy signaling. | [108] |
| ApcMin/+ mice | Modulates the tumor immune microenvironment via FadA that activates the β-catenin pathway. | [131] | |
| Human umbilical vein endothelial cells | Secretes FadA that binds endothelial cell surface cadherins. | [132] | |
| Enterococcus Faecalis | IL10−/− mice | Secretes the metalloprotease gelatinase that compromises the epithelial barrier. | [133] |
| Rat intestinal colonization model | Produces the reactive oxygen species extracellular superoxide and hydrogen peroxide that cause epithelial cell DNA damage. | [134] | |
| B. fragilis | ApcMin/+ mice | Mediates colitis and tumorigenesis. | [135] |
| HT29/C1 cells | Secretes the BFT toxin that stimulates E-cadherin cleavage and facilitates CRC metastasis. | [136] | |
| E. coli | ApcMin/+ mice | Triggers a pro-carcinogenic multi-step inflammatory cascade. | [137] |
| Mice intestinal loop model | Induces DNA damage in vivo and triggers genomic instability in mammalian epithelial cells. | [138] | |
| ApcMin/+ mice | Secretes colibactin that induces autophagy. | [78] | |
| Streptococcus gallolyticus | AOM CRC model | Increases β-catenin nuclear localization and c-Myc and cyclin D1 expression. | [139] |
| Escherichia coli NC101 | AOM/IL10−/− mice | Produces colibactin that induces CRC. | [140] |
| Clostridium nexile | Not yet identified | Contributes to the anticancer effect of pseudomonas aeruginosa. | [141] |
| Clostridium septicum | Not yet identified | Produces alpha toxin that binds GPI-anchored cell surface receptors. | [142] |
| Lactobacillus casei BL23 | AOM/DSS CRC model | Downregulates IL-22 that mediates an immunomodulatory effect. | [143] |
| Faecalibacterium prausnitzii | Colitis mouse model | Induces IL-10 that protects against cancer formation. | [144] |
| Streptococcus bovis | AOM CRC model | Produces inflammatory cytokines that promotes colon lesions. | [145] |
| Eubacterium rectale | Colitis-induced CRC mice model | Produces butyrate to induce the anti-inflammatory cytokine IL-10. | [146, 147] |
Fig. 2.
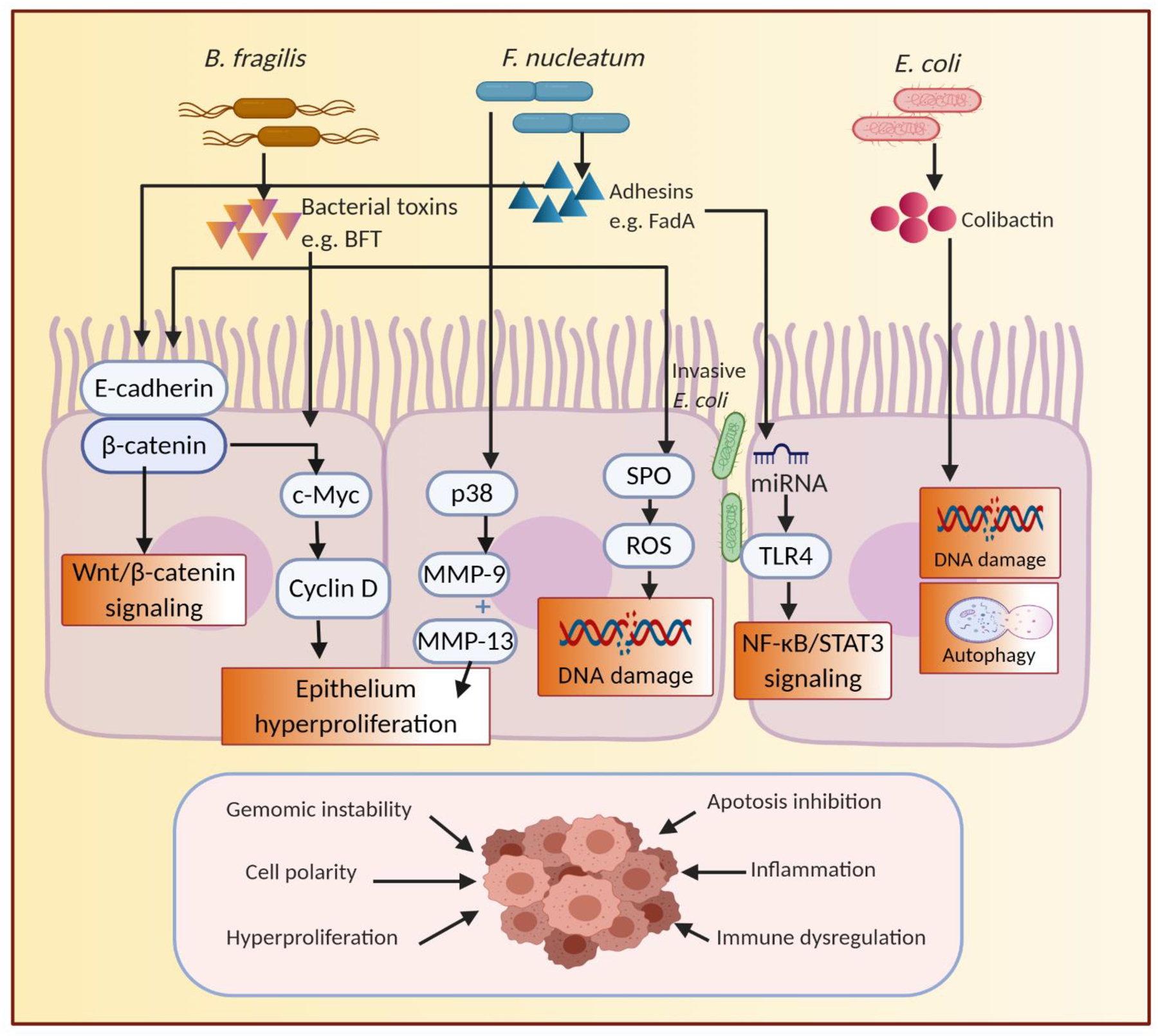
Mechanistic links involved in the gut microbiota promoting carcinogenesis of CRC. Interactions between the gut microbiota and host contribute to the alterations at the molecular level, such as the biosynthesis of toxins interfering with the regulation of cell proliferation and apoptosis by Wnt/β-Catenin and NF-κB/STAT3 signaling pathway, or damaging DNA by producing SPO, or regulating autophagy, that ultimately lead to the onset and progression of CRC. BFT, B. fragilis toxin; FadA, F. nucleatum adhesin A; MMP, matrix metalloproteinases; SPO, spermine oxidase; ROS, reactive oxygen species; TLR4, toll-like receptors 4; NF-κB, nuclear factor kappa B; STAT3, signal transducer and activator of transcription 3.
F. nucleatum
F. nucleatum, a gram-negative anaerobic bacterium commonly found in the mouth, which was first found to be associated with CRC incidence in 2012 [47, 48]. Fluorescence quantitative PCR (qPCR), fluorescence in situ hybridization [49], and droplet digital PCR [50] were used to demonstrate that F. nucleatum was highly enriched in CRC tissues. The qPCR results revealed that the average total abundance of F. nucleatum was 415 times higher in 99 CRC tissue specimens compared to the corresponding normal mucosal tissues [49]. Subsequent studies have used metatranscriptomic sequencing and metagenomic analysis to confirm the increased abundance of F. nucleatum in CRC tissues vis-à-vis healthy tissues [51–53]. Furthermore, the matched F. nucleatum strain was detected in both saliva specimens and CRC tissues in 75% of the CRC-positive patients, suggesting that this bacterium in CRC tissues likely originates in the oral cavity, and digestive tract transmission may be one mechanisms underlying its diffusion [54]. The correlation between F. nucleatum and CRC has been widely confirmed; while the potential mechanisms for its mechanistic role in cancer pathogenesis remain unclear but are an area of active investigation.
One mechanism connecting F. nucleatum and CRC may be epithelial-to-mesenchymal transition (EMT). Microscopic approaches were used to observe epithelial cells incubated with F. nucleatum transdifferentiating into mesenchymal-like cells, showing an enhanced ability to invade [55]. However, western blot analysis of total E-cadherin protein revealed an equal expression of this epithelial cell adhesion protein before and after incubation with F. nucleatum. Accumulating evidence suggests that the potential toxicity of F. nucleatum and its ability to destroy intestinal epithelial cells are primarily due to F. nucleatum adhesin A (FadA), a virulence factor that regulates bacterial adhesion and invasion [56, 57]. The FadA gene expression was significantly higher in CRC specimens than in adjacent normal tissues [58]. In another study, F. nucleatum emerged as a high-risk factor for CRC metastasis and was found to be bound to E-cadherin-expressing cells via FadA [59]. This is of biological importance, as E-cadherin activates the β-catenin pathway, which promotes CRC cell growth. Recently, fadA was shown to up-regulate the expression of the Wnt/β-catenin modulator Annexin A1 through E-cadherin [60]. Dysregulation of the E-cadherin/β-catenin complex leads to CRC cell metastasis. Combined with the regulatory effect of F. nucleatum on E-cadherin/β-catenin, this strain may promote occurrence and progression of CRC by reducing E-cadherin-dependent cell–cell adhesion. Additionally, F. nucleatum can promote the secretion of the matrix metalloproteinases MMP-9 and MMP-13 by activating the mitogen-activated protein kinase p38, increasing the proliferation and survival of infected epithelial cells [61]. Furthermore, recent studies reported that the microRNA (miRNA) miR-21 plays a key regulatory role in the direct correlation between F. nucleatum and proliferation, invasive activity, and xenograft tumor formation in mice [62, 63].
B. fragilis
B. fragilis, a common gram-negative obligate anaerobe, is more abundant in fecal samples from patients with CRC compared to controls [64]. Another study used pyrosequencing analysis to confirm that although the absolute abundance of bacteria per gram of feces was similar between patients with CRC and healthy people, B. fragilis abundance was significantly higher in patients with CRC than in healthy people [65]. However, it has also been shown that B. fragilis is less abundant in CRC tissues than in adjacent non-cancer tissues [66, 67]. These differences may be due to the distinct species of B. fragilis existing in the gastrointestinal tracts of patients with CRC. Two main classes of B. fragilis that colonize most humans have been described: enterotoxigenic B. fragilis (ETBF), which secrete B. fragilis toxin (BFT), causing diarrhea, peritonitis, and intra-abdominal abscesses in humans; and non-toxigenic B. fragilis (NTBF), which do not secrete BFT [68].
Some investigations have suggested that ETBF may operate as a pathogenic bacteria, triggering an immediate and robust inflammatory response, causing an ecological imbalance in the intestinal microbial community [64, 69]. The key factor for ETBF virulence in CRC is attributed to BFT, a secreted 20-kDa zinc-dependent metalloprotease toxin [70]. Expression of the BFT gene is more common in the intestinal mucosa of patients with CRC compared to healthy individuals, especially in patients with advanced CRC [64]. BFT expression induces cleavage of the extracellular domain of E-cadherin in colonic epithelial cells, leading to increased epithelial cell permeability. E-cadherin stimulates cellular signaling through the β-catenin/Wnt pathway, which is active in certain CRC cases [71]. Another study showed that after BFT treatment of CRC cells, loss of membrane-associated E-cadherin activated the nuclear localization of ß-catenin and induced c-Myc translation, leading to continuous cell proliferation [7]. An additional study revealed that BFT can stimulate the production of spermine oxidase (SPO) in intestinal epithelial cell lines, suggesting that enterotoxin has a direct effect on the production of abundant reactive oxygen species (ROS) and thereby causing DNA damage [72].
Escherichia coli
E. coli, a member of the Enterobacteriaceae family, is the most common symbiotic, gram-negative anaerobe in the gastrointestinal tract, which causes diverse effects on gut health by different biological components [73]. For example, some strains are known for their probiotic properties, such as the Nissle 1917 strain of E. coli, which prevents invasion of human intestinal epithelial cells by various pathogens and has been used as a probiotic to treat gastrointestinal disorders for more than a century [74]. In contrast, some E. coli strains show genotoxic activity, which has deleterious effects on host DNA and might ultimately cause colon cancer. Numerous studies have reported elevated colonization levels of colonic mucosa-associated E. coli in patients with CRC compared to healthy individuals. For instance, mucosal specimens from patients with CRC contain more than 70% microbiota, generally E. coli [75]. One study reported that mucosa-associated or internalized E. coli increased significantly in CRC tissues compared to corresponding non-tumor normal tissues [76]. Another study reported that E. coli was detected in and accounts for 62% and 77% of patients presenting with adenomas and carcinomas, respectively [77]. In addition, the levels of pathogenic E. coli strains that produce the toxin cyclomodulin were more prevalent in stage III and IV CRC tissues than in stage I cancers, indicating that the abundance of pathogenic E. coli may be related to CRC stage and prognosis [76].
Various studies have demonstrated a clear link between mucosa-adherent E. coli and CRC. Importantly, E. coli promotes colon tumorigenesis in CRC mouse models after microbiota transplantation in various CRC mouse models, including ApcMin/+ mice [78], azoxymethane (AOM)-treated IL10−/−mice [79], AOM/dextran sodium sulfate (DSS)-treated mice [80], and ApcMin/+ /IL10−/− mice [81]. The biological roles for E. coli in CRC etiology have also been demonstrated. In particular, colibactin, a bacterial toxin synthesized by E. coli carrying the polyketide synthase (pks) gene, can cause DNA damage and genomic alterations and instability, involved in colorectal carcinogenesis [82]. This conclusion was supported by another study, in which mammalian epithelial cells exposed to pks-positive E. coli exhibited transient DNA damage, dysfunctional DNA repair, and an increased frequency of gene mutations [83]. Moreover, pks-positive E. coli induce autophagy and DNA damage repair in intestinal epithelial cells; inhibition of this protective process increases the inflammatory and carcinogenic effects of E. coli in susceptible mice [78].
3. EFFECTS OF GUT MICROBIOTA AND THEIR METABOLITES ON EPIGENETIC REGULATION OF CRC
Most CRC cases begin with the growth of polyps in the inner lining of the colon and rectum, which subsequently develop into dysplastic adenomas, and eventually cancer. In addition to multiple genetic mutations, epigenetic modifications also contribute to the pathogenesis of this disease during its initiation and progression, through processes such as tissue invasion and metastasis [84]. Epigenetic modifications broadly refer to phenotypic changes secondary to changes in gene expression that do not involve permanent changes in the DNA sequence. Accumulating data indicate that the gut microbiota can regulate epigenetic modifications in the host, thus enabling manipulation of the host’s chromatin configuration and functionality. And these modifications can last from one cell division to the next, and hence can be inherited even if the gene sequence is not altered [85]. Therefore, in some sense, epigenetic modifications provide a potentially important interface linking the dynamic interactions between the microbiota and the host genome.
The link between epigenetic modifications and the gut microbiota has been known to be involved in the crosstalk of microbiota-derived metabolites [86]. Microbiota-derived metabolites have received widespread attention for beneficial effects on both cellular energy metabolism and intestinal homeostasis [58]. Short-chain fatty acids (SCFAs) are the most important metabolites of the gut microbiota, as they act as a direct energy source for host cells, stimulate the production of hormones in the body, and play a role in regulating food intake in the brain [87]. Other microbial metabolites, such as bile acids, branched-chain amino acids, indole propionic acid, and endocannabinoids, affect the body’s energy expenditure by influencing thermogenesis and adipose tissue browning [88]. In addition to these direct metabolic effects, it is becoming increasingly apparent that microbiota-derived metabolites can be important but indirect regulators of the epigenetic mechanisms. The epigenetic mechanisms that have a role in cancer development include DNA methylation, histone modifications and non-coding RNAs. Subsequently, from these perspectives, we will discuss the impact of the gut microbiota and their metabolites on epigenetic modifications during colorectal carcinogenesis. Studies about gut microbiota involved in CRC epigenomic modifications are summarized in Table 3.
Table 3.
Gut Microbiota Involved with Epigenetic Alterations in CRC
| Epigenetic modification | Microbiota studied | Model | Key findings | REFs |
|---|---|---|---|---|
| Methylation | Murine gut microbiota | GF mice | The methylation of WIF1, PENK, and NPY were associated with CRC dysbiosis. | [94] |
| Murine gut microbiota | Lgr5-EGFP-CreER mice | Gene methylation was increased by microbiota transplantation. | [148] | |
| L. acidophilus, B. infantis, and Klebsiella species | Human intestinal epithelial cells (IECs) | Microbiota treatment resulted in differential methylation changes in 200 regions of DNA. | [149] | |
| Murine gut microbiota | GF mice | The number of changes in the methylation status of genes increased with age of GF mice. | [150] | |
| ETBF | MinApcΔ716+/− mice | ETBF-induced tumors contained methylated tumor suppressor genes. | [151] | |
| Histone modifications | Murine gut microbiota | wild-type mice | Histone marks H3K4me1 and H3K27ac were enriched at poised or active enhancers. | [152] |
| Antibiotic-treated murine microbiota | GF mice | De novo generation of oscillating histone marks and rhythmically expressed genes. | [153] | |
| Murine gut microbiota | wild-type mice | Bacterial presence resulted in numerous changes in histone acetylation in the proximal colon tissue. | [154] | |
| Murine gut microbiota | GF mice | The location of H3K4 methylation marks was modified when gut microbes colonized. | [155] | |
| Antibiotic-treated murine microbiota | GF mice | Derived SCFAs promoted H3K18 crotonylation by inhibiting HDACs. | [156] | |
| Non-coding RNAs | E. coli strains or fecal-derived murine microbiota | GF mice | Distinct changes in lncRNA signatures occurred after GF mice were reconstituted with normal mouse microbiota or with E. coli alone. | [105] |
| F. nucleatum | CRC xenograft model | F. nucleatum caused resistance to oxaliplatin and 5-FU via downregulation of miR-4802 and miR-18a. | [108] | |
| Murine gut microbiota | GF mice | A total of 19 miRNAs in IESCs significantly differed in expression depending on microbial status. | [157] | |
| Murine gut microbiota | GF mice | Microbiota-dependent miR-21–5p expression in IECs regulated intestinal epithelial permeability via ARF4. | [107] | |
| Antibiotic-treated murine microbiota | GF mice | miRNAs let-7b, miR-141, and miR-200a expression were significantly reduced in GF mice. | [106] | |
| Murine gut microbiota | IEC-miRNA-deficient mice | The presence of gut microbes was associated with decreased production of miRNAs. | [158] |
3.1. DNA METHYLATION
DNA methylation is an epigenetic biological process that predominantly occurs through covalent addition of a methyl group (CH3) to the 5-carbon of a cytosine residue, resulting in 5-methylcytosine (5-mC), which subsequently affects the function and inhibits the transcription of the target gene [89]. Numerous reports have shown that the gut microbiota can modify the methylation pattern of cancer-related genes, thus providing insight into new directions for identifying relationships among the gut microbiota, the epigenetic landscape of host cells, and the occurrence and development of CRC. For example, in a study comparing the methylation profiles of ETBF-induced and spontaneous tumors in the distal colon of ApcMin/+ mice, greater hypermethylation and reduced hypomethylation of differentially methylated regions were observed in ETBF-induced tumors than in spontaneous tumors [90]. Ecological dysbiosis of the gut microbiota can induce host gene methylation, but to date, studies on specific bacteria regulating CRC methylation and its regulatory mechanisms are very limited. A population-based study reported that a high load of F. nucleatum in CRC tissues was associated with high microsatellite instability and a CpG island methylator phenotype [91]. In another study, F. nucleatum was correlated with wild-type tumor suppressor TP53, methylation of the mismatch repair gene hMLH1, genomic hypermutation, and mutation of the chromatin remodelers CHD7/8 [92]. Similarly, comparison of gastric biopsy results between patients infected with Helicobacter pylori and healthy individuals revealed that chronic gastritis was associated with hypermethylation of the promoter region of E-cadherin (CDH1), the DNA methyltransferase (DNMT) MGMT, the Wnt inhibitor WIF1, and the MLH1 gene [93]. Furthermore, fecal bacterial species, including several Parvimonas species, were recently identified in individuals with a higher cumulative methylation index in the blood [94]. During DNA methylation, S-adenosyl methionine (SAM) acts as a methyl donor to enable DNMTs to form 5-mC [95]. Microbial metabolites such as folate are potentially involved in the synthesis of 5-methyltetrahydrofolate, which is itself a methyl group donor for SAM [96]. Indeed, in human colonic cells, global DNA and p53 region-specific hypomethylation were induced by folate depletion, and the common microbial genera bifidobacteria and lactobacillus produce folate [97]. Another intriguing study highlighted that volunteers given bifidobacteria exhibited high concentrations of folate in the feces [98], which suggests that this probiotic may affect DNA methylation patterns via folate production.
3.2. HISTONE MODIFICATIONS
The acetylation of histone lysine residues is involved in transcription, translation, and DNA repair, and is regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). HATs catalyze the transfer of acetyl groups from acetyl coenzyme A (Ac-CoA) to amino-terminal lysine residues on histones, whereas HDACs reverse this process. Consequently, HATs and HDACs collaboratively maintain the balance of histone acetylation in vivo to achieve homeostasis [99]. In addition to stabilization of acetylated histones, HDACs can also regulate CRC cell proliferation and apoptosis by modulating the acetylation status of p53 and tubulin protein [100]. In turn, gut microbiota can regulate the activity of HDACs by producing epigenetic metabolites such as SCFAs [86], as illustrated in Figure 3. Many studies surrounding the gut microbiota, CRC, and histone modifications have focused on the roles of SCFAs as HDAC inhibitors. Several different mechanisms by which SCFAs enter colonic intestinal cells have been proposed, including passive diffusion, counter-transport with bicarbonate, transporter monocarboxylic acid transporter 1, and sodium-coupled monocarboxylic acid transporter 1 [101]. The most important SCFA-producing bacteria are Firmicute, Bacteroidetes, Proteobacteria, and Actinobacteria [102]. How SCFAs regulate tumor formation and histone deacetylation remains unclear and is an active area of investigation. Chromatin maps of colorectal epithelial cells isolated from conventional and germ-free (GF) mice showed a decrease in diacetylated lysine on the histone subunit H3 in GF mice [102]. Supplementation with several SCFAs (acetate, propionate, and butyrate) resulted in a histone profile closer to that of conventional mice, suggesting that these metabolic by-products have the potential to induce histone modifications [103].
Fig. 3.
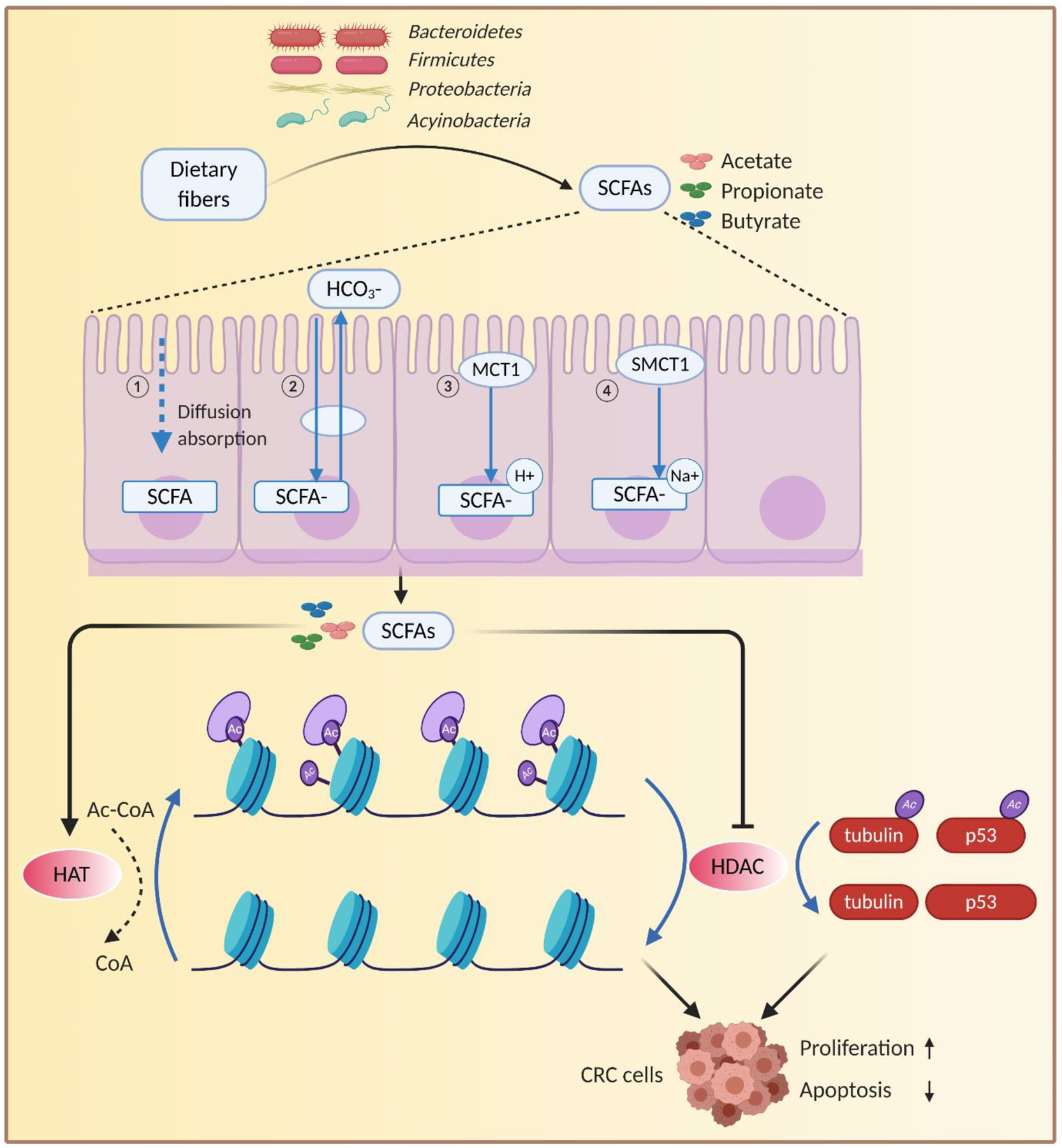
Epigenomic interactions between the gut microbiota and CRC via SCFAs. Gut bacteria in the colon or rectum produce a number of low molecular weight SCFAs, which can be absorbed by epithelial cells, and cause epigenetic modifications in DNA methylation and histone acetylation via activation or inhibition of certain enzymes such as DNMTs, HDACs. SCFAs, short-chain fatty acids; MCT1, monocarboxylate transporter 1; SMCT1, sodium-dependent monocarboxylate transporter 1; HATs, histone acetyltransferases; HDACs, histone deacetylases; CoA, coenzyme A; Ac-CoA, acetyl coenzyme A.
3.3. NON-CODING RNAs (NcRNAs)
NcRNAs are important regulators of epigenetic status, and have functional significance in modulating gene expression during CRC tumorigenesis [104]. Whether the gut microbiota modulates ncRNA expression in the host to impact CRC is under investigation. Most studies have used GF and conventional mice to detect differences in long noncoding RNAs (lncRNAs) and miRNA expression in the presence of gut microbiota. Exon microarrays were used to compare lncRNA expression profiles between GF and conventional mice, and distinct changes in lncRNA signatures were observed after GF mice were reconstituted with normal mouse microbiota or with E. coli alone [105]. Reduced expression of three miRNAs known to be expressed in intestinal epithelial cells (let-7b, miR-141, and miR-200a) was observed in the feces of GF mice compared to the conventional mice. Following a 6-week treatment with antibiotics to deplete the gut microbiota in rats, miRNA expression was also reduced [106]. Other studies have shown that gut microbiota can alter the expression of CRC-related miRNAs; for example, commensal bacteria induce the expression of miR-21–5p in intestinal epithelial cells [107]. To demonstrate the functional impact of microbially mediated miRNA changes on CRC development, global miRNA expression profiling was used to identify several differentially expressed miRNAs between F. nucleatum-rich samples from patients with recurrent CRC and low amount of F. nucleatum samples from non-recurrent patients. Among the downregulated miRNAs, F. nucleatum targeted miR-18a and miR-4802 to alter CRC chemotherapeutic response by activating the autophagy pathway [108].
4. THE VIABILITY OF THE GUT MICROBIOTA AS A BIOMARKER FOR CRC
The progression from normal colonic mucosa to adenoma and eventually to CRC orchestrates in several stages, over at least 10 years, and appropriate measures can be taken to avoid the occurrence of CRC at any stage during this period [109]. Even if cancer is diagnosed, the 5-year survival of patients treated during the early stages of CRC is encouraging, ranging from 72% to 93% [110]. Therefore, early detection of precancerous lesions and CRC is the key to improving the chance of a cure.
Non-invasive screening strategies for early CRC involve either checking the presence of minute quantities of blood in the stool, such as with a fecal immunohistochemistry test (FIT) or a guaiac-based fecal occult blood test, or performing multi-target molecular detection of DNA, RNA, and protein markers of neoplasia in the feces [111, 112]. Single-sample FIT tests are approximately 20%–30% sensitive for advanced neoplasia detection and approximately 80% sensitive for CRC detection [113]. Multi-target molecular detection for high-grade precancerous lesions is approximately 46% sensitive [114]. Therefore, it is necessary to explore novel non-invasive screening methods. Due to differences in the gut microbiota between patients with colon adenoma or CRC and healthy people, characteristic changes in fecal microbiota and metabolites are promising biomarkers for early CRC screening. The possibility of using gut microbiota evaluation as a CRC screening tool via Bayesian statistics was first proposed in 2014 [115]. Targeted detection of CRC-related dysbiosis and microbiota species in fecal samples may represent a promising non-invasive screening method to overcome the limitations and the poor performance of current CRC early diagnosis tools.
Notable advances in the development of non-invasive, early screening methods have been made through the identification of relevant and specific microbial characteristics in patients with CRC. Many studies have assessed the diagnostic accuracy of fecal F. nucleatum as a potential non-invasive biomarker for CRC. In a meta‐analysis included ten studies comprising 1450 patients with CRC and 1421 healthy individuals, fecal F. nucleatum performed well as a diagnostic biomarker for CRC, with 71% sensitivity and 76% specificity [116]. Indeed, in one study of 439 subjects (203 colorectal cancer and 236 healthy subjects), fecal F. nucleatum performed well as a diagnostic biomarker, with sensitivity better than FIT for the detection of CRC [117]. By using qPCR to analyze 103 patients with advanced adenoma, 104 patients with CRC, and 102 healthy controls, it is identified that the combination of F. nucleatum and FIT has a high sensitivity (92.3%) and an area under the curve (AUC) of 0.95 in detecting CRC [118]. The expression levels of the FadA gene is significantly higher in the colon tissue of patients with adenoma and adenocarcinoma than in that of normal individuals; thus, this unique adhesin of F. nucleatum may be an ideal diagnostic marker to identify individuals with CRC risk [58]. After metagenomic profiling of fecal microbiota, a non-invasive microbial biomarker panel (F. nucleatum, Peptostreptococcus stomatis, Parvimonas micra, and Solobacterium moorei) for CRC was established and validated from ethnically different cohorts in China, Denmark, France, and Austria [119]. Another study also performed fecal metagenomic and metabolomic profiling on fecal samples from a large cohort of 616 participants, providing that the shifts of F. nucleatum and its metabolites (branched-chain amino acids, phenylalanine, and bile acids) occurred from the very early stages of CRC development, which is of significance for etiology and diagnosis [120]. Based on an analysis of metagenomic samples from a 526-case CRC cohort, a panel of seven CRC-enriched bacteria (B. fragilis, F. nucleatum, Porphyromonas asaccharolytica, P. micra, Prevotella intermedia, Alistipes finegoldii, and Thermanaerovibrio acidaminovorans) was identified with an AUC value of 0.80 [121]. Moreover, high abundance of F. nucleatum and B. fragilis has been identified as independent indicators of poor survival in patients with CRC [122]. Various studies (summarized in Table 4) have used characteristic gut microbiota to construct CRC diagnostic models, which can be divided into the following four categories: (i) individual strains dominated by F. nucleatum; (ii) combinations of more than three kinds of microbiota as biomarkers; (iii) inclusion of viruses, fungi, or metabolites as biomarkers; and (iv) prediction models jointly constructed by combination of the above microbiota and FIT or other conventional tests.
Table 4.
Fecal Gut Microbiota as A potential Biomarker for Non-invasive Diagnosis of CRC
| Microbiota and Other Tests | Cohort Size | Sample Type | Detection Method | REFs | |
|---|---|---|---|---|---|
| Noncancer Controls | CRC | ||||
| F. nucleatum | 60 | 188 | Fecal samples | 16S rRNA gene sequencing and qPCR | [50] |
| F. nucleatum | 370 | 120 | Fecal samples | 16S rRNA gene sequencing and qPCR | [159] |
| Clostridium symbiosum, F. nucleatum, FIT and carcinoembryonic antigen | 454 | 327 | Fecal samples | qPCR | [160] |
| F. nucleatum and FIT | 205 | 104 | Fecal samples | Metagenomics and qPCR | [118] |
| F. nucleatum, clbA-positive bacteria | 962 | 174 | Fecal samples | 16S rRNA gene sequencing and qPCR | [161] |
| F. nucleatum, Atopobium parvulum, Actinomyces odontolyticus, Metabolites: branched-chain amino acids, phenylalanine, bile acids | 251 | 365 | Fecal sample | Metagenomics and qPCR | [120] |
| F. nucleatum, Peptostreptococcus stomatis, Parvimonas micra, Solobacterium moorei | 187 | 137 | Fecal sample | Metagenomics and qPCR | [119] |
| B. fragilis, F. nucleatum, Porphyromonas asaccharolytica, Parvimonas micra, Prevotella intermedia, Alistipes finegoldii, Thermanaerovibrio acidaminovorans | 271 | 255 | Fecal sample | Metagenomics and qPCR | [121] |
| F. nucleatum, Enterococcus faecalis, Streptococcus bovis, Enterotoxigenic B. fragilis, Porphyromonas spp | 73 | 20 | Fecal samples | 16S rRNA gene sequencing and qPCR | [162] |
| F. nucleatum, Bifidobacterium, Faecalibacterium prausnitzii | 367 | 536 | Fecal samples | 16S rRNA gene sequencing and qPCR | [163] |
| F. nucleatum, Bacteroides clarus, Roseburia intestinalis, Clostridium hathewayi | 236 | 203 | Fecal samples | 16S rRNA gene sequencing and qPCR | [117] |
In order to translate basic research findings for their clinic utility, clinical trials are currently being performed to determine the translational potential of the gut microbiota as biomarker of CRC. A clinical trial (NCT01778595) was recently conducted to identify a useful diagnostic biomarker Proteobacteria whose median relative abundance was 3-fold higher in colorectal adenoma patients than in healthy individuals. Another trial (NCT02845973) enrolled 1325 participants and identified that one or more gut bacterial species in feces may contribute to early diagnosis of CRC. The results showed that fecal Clostridium symbiosum, maybe a promising noninvasive biomarker for early diagnosis of CRC, with a higher diagnostic power than F. nucleatum, FIT and CEA. In addition to evaluating the gut microbiota as diagnostic biomarker for CRC, investigators have also studied bacterial diversity for its effectiveness as a prognostic predictor and recurrence monitor for CRC. A clinical trial (NCT04223102) is currently recruiting patients to evaluate the association between the gut microbiota and response to neoadjuvant therapy. Another trial (NCT03385213) is currently underway to discover whether there are differences in the gut microbiota between patients with recurrent CRC and those without. All these data suggest that exploration of the gut microbiota can be used not only for non-invasive and accurate diagnostic approaches for CRC but may also serve as prognostic tools for CRC treatment in the future.
5. CONCLUSIONS AND PERSPECTIVE
Affected by various factors such as diet, age, and health status, there are natural differences in the composition of the gut microbiota among individuals, but the overall complexity of the human gut microbiota is still relatively conservative. Therefore, it is of universal significance to study the microbiota that play a major role in terms of quantity and function. Specific microbiota (e.g., B. fragilis, F. nucleatum, and E. coli) have been increasingly found to be carcinogenic in CRC. To conclude, CRC may be the result of enhanced interaction between pathogenic microbiota and a disordered host response at both the genomic and epigenomic level.
Although great progress has been made in investigating the gut microbiota, there are still some unresolved issues. To date, annotations from metagenomic data of microbial taxa mostly reach the level of genus or species, which is not detailed enough to understand which specific strains participate in the pathogenesis of human CRC or the mechanisms by which they do so. Further research identifying microbial strains based on multi-omics and data mining algorithm is urgently needed. In addition, the current state of microbial research on CRC has potential limitations: because most CRC cohorts are cross-sectional, it is difficult to reveal the dynamic balance of gut microbiota and to infer the causal relationship between gut microbiota disorders and CRC. Thus, it is urgent that we integrate multi-omics data across populations for longitudinal microbial profiling to further understand the exact role of the gut microbiota in CRC tumorigenesis.
Along with the development of advanced techniques and the deepening of mechanism research, a more comprehensive and in-depth understanding of the gut microbiota in CRC will undoubtedly contribute to the emergence of novel and precise diagnostic strategies in the foreseeable future. Of note, fecal samples because of their easy availability, repeatable sampling, non-invasive and inexpensive nature, can be used as a suitable substitute to reflect the gut microbiota. However, if contamination occurs or the time interval between sample deposition and collection is too long, the fecal microbiota may become altered[123]. In addition, fecal collection kits, storage conditions, transportation conditions and DNA extraction methods all influence detection results, which makes it difficult to guide clinical diagnosis and treatment decisions [124]. Thus far, it has been challenging to get an accurate result from a somewhat improper specimen. The accuracy of samples has a significant influence on the value of research on the gut microbiota. The current gold standard for fecal sampling is instant freezing of fecal materials at −80°C and without preservatives to preserve microbial integrity[125]. However, in some cases, the ideal conditions for storage of fecal specimens at −80°C are not met, especially for large-scale population studies. Therefore, to ensure the reliability of research, more optimized and precise sampling methods are urgent needed. Overall, it is expected that wider range of standardized specimen collection and preservation, more standardized and rational clinical design, and more powerful database sharing and sequencing analysis techniques will make clinical application of the gut microbiota as a CRC biomarker possible.
ACKNOWLEDGEMENTS
We would like to thank Dr. Sarah Wilkinson for her important insights, useful suggestions and careful editing of this manuscript.
FUNDING
The present work was supported by the grants CA72851, CA181572, CA184792, CA187956 and CA202797 from the National Institute of Health (NIH) to AG, as well as the China Scholarship Council [grant numbers 201906220156] to YZ.
Abbreviations:
- 5-mc
5-methylcytosine
- Ac-CoA
acetyl coenzyme A
- AOM
azoxymethane
- AUC
area under the curve
- BFT
B. fragilis toxin
- CRC
colorectal cancer
- DNMTs
DNA methyltransferases
- DSS
dextran sodium sulfate
- EIEC
enteroinvasive Escherichia coli
- ETBF
enterotoxigenic B. fragilis
- FadA
F. nucleatum adhesin A
- FIT
fecal immunohistochemistry test
- GF
germ-free
- HATs
histone acetyltransferases
- HDACs
histone deacetylases
- lncRNA
long noncoding RNA
- ncRNAs
noncoding RNAs
- NGS
next-generation sequencing
- NTBF
non-toxigenic B. fragilis
- OTUs
operational taxonomic units
- PCR
polymerase chain reaction
- pks
polyketide synthase
- ROS
reactive oxygen species
- rRNA
ribosomal RNA
- SAM
S-Adenosyl methionine
- SCFAs
short-chain fatty acids
- SPO
spermine oxidase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: None of the authors has any potential conflicts to disclose.
References:
- [1].Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A, Colorectal cancer statistics, 2020, CA Cancer J Clin 70(3) (2020) 145–164. [DOI] [PubMed] [Google Scholar]
- [2].Jeon J, Du M, Schoen RE, Hoffmeister M, Newcomb PA, Berndt SI, Caan B, Campbell PT, Chan AT, Chang-Claude J, Giles GG, Gong J, Harrison TA, Huyghe JR, Jacobs EJ, Li L, Lin Y, Le Marchand L, Potter JD, Qu C, Bien SA, Zubair N, Macinnis RJ, Buchanan DD, Hopper JL, Cao Y, Nishihara R, Rennert G, Slattery ML, Thomas DC, Woods MO, Prentice RL, Gruber SB, Zheng Y, Brenner H, Hayes RB, White E, Peters U, Hsu L, Colorectal Transdisciplinary S, Genetics C Epidemiology of Colorectal Cancer, Determining Risk of Colorectal Cancer and Starting Age of Screening Based on Lifestyle, Environmental, and Genetic Factors, Gastroenterology 154(8) (2018) 2152–2164 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fillon M, Study aims to improve colorectal cancer screening rates, CA Cancer J Clin 69(3) (2019) 161–163. [DOI] [PubMed] [Google Scholar]
- [4].Dow LE, O’Rourke KP, Simon J, Tschaharganeh DF, van Es JH, Clevers H, Lowe SW, Apc Restoration Promotes Cellular Differentiation and Reestablishes Crypt Homeostasis in Colorectal Cancer, Cell 161(7) (2015) 1539–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yurgelun MB, Kulke MH, Fuchs CS, Allen BA, Uno H, Hornick JL, Ukaegbu CI, Brais LK, McNamara PG, Mayer RJ, Schrag D, Meyerhardt JA, Ng K, Kidd J, Singh N, Hartman AR, Wenstrup RJ, Syngal S, Cancer Susceptibility Gene Mutations in Individuals With Colorectal Cancer, J Clin Oncol 35(10) (2017) 1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Espenschied CR, LaDuca H, Li S, McFarland R, Gau CL, Hampel H, Multigene Panel Testing Provides a New Perspective on Lynch Syndrome, J Clin Oncol 35(22) (2017) 2568–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shah RS, Plesec T, Bhatt A, Abnormal Biliary Mucosa Uncovered in a Familial Adenomatous Polyposis Patient, Gastroenterology 158(5) (2020) e1–e2. [DOI] [PubMed] [Google Scholar]
- [8].Sengupta S, Bose S, Peutz-Jeghers Syndrome, N Engl J Med 380(5) (2019) 472. [DOI] [PubMed] [Google Scholar]
- [9].Keum N, Giovannucci E, Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies, Nat Rev Gastroenterol Hepatol 16(12) (2019) 713–732. [DOI] [PubMed] [Google Scholar]
- [10].Nunez O, Fernandez-Navarro P, Martin-Mendez I, Bel-Lan A, Locutura JF, Lopez-Abente G, Arsenic and chromium topsoil levels and cancer mortality in Spain, Environ Sci Pollut Res Int 23(17) (2016) 17664–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wild CP, Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology, Cancer Epidemiol Biomarkers Prev 14(8) (2005) 1847–50. [DOI] [PubMed] [Google Scholar]
- [12].Hillman ET, Lu H, Yao T, Nakatsu CH, Microbial Ecology along the Gastrointestinal Tract, Microbes Environ 32(4) (2017) 300–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, Mele MC, What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases, Microorganisms 7(1) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lukens JR, Gurung P, Vogel P, Johnson GR, Carter RA, McGoldrick DJ, Bandi SR, Calabrese CR, Vande Walle L, Lamkanfi M, Kanneganti TD, Dietary modulation of the microbiome affects autoinflammatory disease, Nature 516(7530) (2014) 246–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL, Mapping human microbiome drug metabolism by gut bacteria and their genes, Nature 570(7762) (2019) 462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, Tuohy K, Gut microbiota functions: metabolism of nutrients and other food components, Eur J Nutr 57(1) (2018) 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Iacob S, Iacob DG, Luminos LM, Intestinal Microbiota as a Host Defense Mechanism to Infectious Threats, Front Microbiol 9 (2018) 3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nowak A, Libudzisz Z, Influence of phenol, p-cresol and indole on growth and survival of intestinal lactic acid bacteria, Anaerobe 12(2) (2006) 80–4. [DOI] [PubMed] [Google Scholar]
- [19].Tsuji H, Matsuda K, Nomoto K, Counting the Countless: Bacterial Quantification by Targeting rRNA Molecules to Explore the Human Gut Microbiota in Health and Disease, Front Microbiol 9 (2018) 1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, Knight R, Experimental and analytical tools for studying the human microbiome, Nat Rev Genet 13(1) (2011) 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Weinstock GM, Genomic approaches to studying the human microbiota, Nature 489(7415) (2012) 250–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Callahan BJ, Wong J, Heiner C, Oh S, Theriot CM, Gulati AS, McGill SK, Dougherty MK, High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution, Nucleic Acids Res 47(18) (2019) e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bikel S, Valdez-Lara A, Cornejo-Granados F, Rico K, Canizales-Quinteros S, Soberon X, Del Pozo-Yauner L, Ochoa-Leyva A, Combining metagenomics, metatranscriptomics and viromics to explore novel microbial interactions: towards a systems-level understanding of human microbiome, Comput Struct Biotechnol J 13 (2015) 390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Youssef N, Sheik CS, Krumholz LR, Najar FZ, Roe BA, Elshahed MS, Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys, Appl Environ Microbiol 75(16) (2009) 5227–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ranjan R, Rani A, Metwally A, McGee HS, Perkins DL, Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing, Biochem Biophys Res Commun 469(4) (2016) 967–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Johnson JS, Spakowicz DJ, Hong BY, Petersen LM, Demkowicz P, Chen L, Leopold SR, Hanson BM, Agresta HO, Gerstein M, Sodergren E, Weinstock GM, Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis, Nat Commun 10(1) (2019) 5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chu W, Han Q, Xu J, Wang J, Sun Y, Li W, Chen ZJ, Du Y, Metagenomic analysis identified microbiome alterations and pathological association between intestinal microbiota and polycystic ovary syndrome, Fertil Steril 113(6) (2020) 1286–1298 e4. [DOI] [PubMed] [Google Scholar]
- [28].Ding T, Schloss PD, Dynamics and associations of microbial community types across the human body, Nature 509(7500) (2014) 357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sears CL, Garrett WS, Microbes, microbiota, and colon cancer, Cell Host Microbe 15(3) (2014) 317–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Irrazabal T, Belcheva A, Girardin SE, Martin A, Philpott DJ, The multifaceted role of the intestinal microbiota in colon cancer, Mol Cell 54(2) (2014) 309–20. [DOI] [PubMed] [Google Scholar]
- [31].Yamamoto Y, Nakanishi Y, Murakami S, Aw W, Tsukimi T, Nozu R, Ueno M, Hioki K, Nakahigashi K, Hirayama A, Sugimoto M, Soga T, Ito M, Tomita M, Fukuda S, A Metabolomic-Based Evaluation of the Role of Commensal Microbiota throughout the Gastrointestinal Tract in Mice, Microorganisms 6(4) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Schroeder BO, Fight them or feed them: how the intestinal mucus layer manages the gut microbiota, Gastroenterol Rep (Oxf) 7(1) (2019) 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Saputo S, Faustoferri RC, Quivey RG Jr., Vitamin D Compounds Are Bactericidal against Streptococcus mutans and Target the Bacitracin-Associated Efflux System, Antimicrob Agents Chemother 62(1) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gouyer V, Dubuquoy L, Robbe-Masselot C, Neut C, Singer E, Plet S, Geboes K, Desreumaux P, Gottrand F, Desseyn JL, Delivery of a mucin domain enriched in cysteine residues strengthens the intestinal mucous barrier, Sci Rep 5 (2015) 9577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wlodarska M, Kostic AD, Xavier RJ, An integrative view of microbiome-host interactions in inflammatory bowel diseases, Cell Host Microbe 17(5) (2015) 577–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mowat AM, Agace WW, Regional specialization within the intestinal immune system, Nat Rev Immunol 14(10) (2014) 667–85. [DOI] [PubMed] [Google Scholar]
- [37].Galloway-Pena JR, Smith DP, Sahasrabhojane P, Wadsworth WD, Fellman BM, Ajami NJ, Shpall EJ, Daver N, Guindani M, Petrosino JF, Kontoyiannis DP, Shelburne SA, Characterization of oral and gut microbiome temporal variability in hospitalized cancer patients, Genome Med 9(1) (2017) 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ghosh TS, Das M, Jeffery IB, O’Toole PW, Adjusting for age improves identification of gut microbiome alterations in multiple diseases, Elife 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Low EE, Demb J, Liu L, Earles A, Bustamante R, Williams CD, Provenzale D, Kaltenbach T, Gawron AJ, Martinez ME, Gupta S, Risk Factors for Early-Onset Colorectal Cancer, Gastroenterology 159(2) (2020) 492–501 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chen H, Zheng X, Zong X, Li Z, Li N, Hur J, Fritz CD, Chapman W Jr., Nickel KB, Tipping A, Colditz GA, Giovannucci EL, Olsen MA, Fields RC, Cao Y, Metabolic syndrome, metabolic comorbid conditions and risk of early-onset colorectal cancer, Gut (2020). [DOI] [PMC free article] [PubMed]
- [41].Hofseth LJ, Hebert JR, Chanda A, Chen H, Love BL, Pena MM, Murphy EA, Sajish M, Sheth A, Buckhaults PJ, Berger FG, Early-onset colorectal cancer: initial clues and current views, Nat Rev Gastroenterol Hepatol 17(6) (2020) 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ferrarese R, Zuppardo RA, Puzzono M, Mannucci A, Amato V, Ditonno I, Patricelli MG, Raucci AR, Clementi M, Elmore U, Rosati R, Testoni PA, Mancini N, Cavestro GM, Oral and Fecal Microbiota in Lynch Syndrome, J Clin Med 9(9) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yan Y, Drew DA, Markowitz A, Lloyd-Price J, Abu-Ali G, Nguyen LH, Tran C, Chung DC, Gilpin KK, Meixell D, Parziale M, Schuck M, Patel Z, Richter JM, Kelsey PB, Garrett WS, Chan AT, Stadler ZK, Huttenhower C, Structure of the Mucosal and Stool Microbiome in Lynch Syndrome, Cell Host Microbe 27(4) (2020) 585–600 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Park CH, Han DS, Oh YH, Lee AR, Lee YR, Eun CS, Role of Fusobacteria in the serrated pathway of colorectal carcinogenesis, Sci Rep 6 (2016) 25271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hale VL, Chen J, Johnson S, Harrington SC, Yab TC, Smyrk TC, Nelson H, Boardman LA, Druliner BR, Levin TR, Rex DK, Ahnen DJ, Lance P, Ahlquist DA, Chia N, Shifts in the Fecal Microbiota Associated with Adenomatous Polyps, Cancer Epidemiol Biomarkers Prev 26(1) (2017) 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Drewes JL, Corona A, Sanchez U, Fan Y, Hourigan SK, Weidner M, Sidhu SD, Simner PJ, Wang H, Timp W, Oliva-Hemker M, Sears CL, Transmission and clearance of potential procarcinogenic bacteria during fecal microbiota transplantation for recurrent Clostridioides difficile, JCI Insight 4(19) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M, Genomic analysis identifies association of Fusobacterium with colorectal carcinoma, Genome Res 22(2) (2012) 292–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA, Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma, Genome Res 22(2) (2012) 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Li YY, Ge QX, Cao J, Zhou YJ, Du YL, Shen B, Wan YJ, Nie YQ, Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients, World J Gastroenterol 22(11) (2016) 3227–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Suehiro Y, Sakai K, Nishioka M, Hashimoto S, Takami T, Higaki S, Shindo Y, Hazama S, Oka M, Nagano H, Sakaida I, Yamasaki T, Highly sensitive stool DNA testing of Fusobacterium nucleatum as a marker for detection of colorectal tumours in a Japanese population, Ann Clin Biochem 54(1) (2017) 86–91. [DOI] [PubMed] [Google Scholar]
- [51].Sun CH, Li BB, Wang B, Zhao J, Zhang XY, Li TT, Li WB, Tang D, Qiu MJ, Wang XC, Zhu CM, Qian ZR, The role of Fusobacterium nucleatum in colorectal cancer: from carcinogenesis to clinical management, Chronic Dis Transl Med 5(3) (2019) 178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Oh HJ, Kim JH, Bae JM, Kim HJ, Cho NY, Kang GH, Prognostic Impact of Fusobacterium nucleatum Depends on Combined Tumor Location and Microsatellite Instability Status in Stage II/III Colorectal Cancers Treated with Adjuvant Chemotherapy, J Pathol Transl Med 53(1) (2019) 40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].de Carvalho AC, de Mattos Pereira L, Datorre JG, Dos Santos W, Berardinelli GN, Matsushita MM, Oliveira MA, Duraes RO, Guimaraes DP, Reis RM, Microbiota Profile and Impact of Fusobacterium nucleatum in Colorectal Cancer Patients of Barretos Cancer Hospital, Front Oncol 9 (2019) 813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Komiya Y, Shimomura Y, Higurashi T, Sugi Y, Arimoto J, Umezawa S, Uchiyama S, Matsumoto M, Nakajima A, Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity, Gut 68(7) (2019) 1335–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ma CT, Luo HS, Gao F, Tang QC, Chen W, Fusobacterium nucleatum promotes the progression of colorectal cancer by interacting with E-cadherin, Oncol Lett 16(2) (2018) 2606–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yeoh YK, Chen Z, Wong MCS, Hui M, Yu J, Ng SC, Sung JJY, Chan FKL, Chan PKS, Southern Chinese populations harbour non-nucleatum Fusobacteria possessing homologues of the colorectal cancer-associated FadA virulence factor, Gut (2020). [DOI] [PMC free article] [PubMed]
- [57].Li Q, Tan L, Wang H, Kou Y, Shi X, Zhang S, Pan Y, Fusobacterium nucleatum Interaction with Pseudomonas aeruginosa Induces Biofilm-Associated Antibiotic Tolerance via Fusobacterium Adhesin A, ACS Infect Dis (2020). [DOI] [PubMed]
- [58].Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW, Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin, Cell Host Microbe 14(2) (2013) 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zhang S, Li C, Liu J, Geng F, Shi X, Li Q, Lu Z, Pan Y, Fusobacterium nucleatum promotes epithelial-mesenchymal transiton through regulation of the lncRNA MIR4435–2HG/miR-296–5p/Akt2/SNAI1 signaling pathway, FEBS J (2020). [DOI] [PMC free article] [PubMed]
- [60].Rubinstein MR, Baik JE, Lagana SM, Han RP, Raab WJ, Sahoo D, Dalerba P, Wang TC, Han YW, Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/beta-catenin modulator Annexin A1, EMBO Rep 20(4) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Uitto VJ, Baillie D, Wu Q, Gendron R, Grenier D, Putnins EE, Kanervo A, Firth JD, Fusobacterium nucleatum increases collagenase 3 production and migration of epithelial cells, Infect Immun 73(2) (2005) 1171–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, Li H, Guo B, Zhu Q, Wei Q, Moyer MP, Wang P, Cai S, Goel A, Qin H, Ma Y, Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-kappaB, and Up-regulating Expression of MicroRNA-21, Gastroenterology 152(4) (2017) 851–866 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Proenca MA, Biselli JM, Succi M, Severino FE, Berardinelli GN, Caetano A, Reis RM, Hughes DJ, Silva AE, Relationship between Fusobacterium nucleatum, inflammatory mediators and microRNAs in colorectal carcinogenesis, World J Gastroenterol 24(47) (2018) 5351–5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, Ellis B, Carroll KC, Albesiano E, Wick EC, Platz EA, Pardoll DM, Sears CL, The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients, Clin Infect Dis 60(2) (2015) 208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Haghi F, Goli E, Mirzaei B, Zeighami H, The association between fecal enterotoxigenic B. fragilis with colorectal cancer, BMC Cancer 19(1) (2019) 879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S, Zhao L, Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers, ISME J 6(2) (2012) 320–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP, Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults, PLoS One 8(8) (2013) e70803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sears CL, Geis AL, Housseau F, Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis, J Clin Invest 124(10) (2014) 4166–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Bao Y, Tang J, Qian Y, Sun T, Chen H, Chen Z, Sun D, Zhong M, Chen H, Hong J, Chen Y, Fang JY, Long noncoding RNA BFAL1 mediates enterotoxigenic Bacteroides fragilis-related carcinogenesis in colorectal cancer via the RHEB/mTOR pathway, Cell Death Dis 10(9) (2019) 675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wu S, Dreyfus LA, Tzianabos AO, Hayashi C, Sears CL, Diversity of the metalloprotease toxin produced by enterotoxigenic Bacteroides fragilis, Infect Immun 70(5) (2002) 2463–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Devaux CA, Mezouar S, Mege JL, The E-Cadherin Cleavage Associated to Pathogenic Bacteria Infections Can Favor Bacterial Invasion and Transmigration, Dysregulation of the Immune Response and Cancer Induction in Humans, Front Microbiol 10 (2019) 2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Goodwin AC, Destefano Shields CE, Wu S, Huso DL, Wu X, Murray-Stewart TR, Hacker-Prietz A, Rabizadeh S, Woster PM, Sears CL, Casero RA Jr., Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis, Proc Natl Acad Sci U S A 108(37) (2011) 15354–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, Popova IE, Parikh SJ, Adams LG, Tsolis RM, Stewart VJ, Baumler AJ, Host-derived nitrate boosts growth of E. coli in the inflamed gut, Science 339(6120) (2013) 708–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Altenhoefer A, Oswald S, Sonnenborn U, Enders C, Schulze J, Hacker J, Oelschlaeger TA, The probiotic Escherichia coli strain Nissle 1917 interferes with invasion of human intestinal epithelial cells by different enteroinvasive bacterial pathogens, FEMS Immunol Med Microbiol 40(3) (2004) 223–9. [DOI] [PubMed] [Google Scholar]
- [75].Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, Englyst H, Williams HF, Rhodes JM, Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer, Gastroenterology 127(1) (2004) 80–93. [DOI] [PubMed] [Google Scholar]
- [76].Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, Dechelotte P, Bonnet R, Pezet D, Darfeuille-Michaud A, Colonization of the human gut by E. coli and colorectal cancer risk, Clin Cancer Res 20(4) (2014) 859–67. [DOI] [PubMed] [Google Scholar]
- [77].Swidsinski A, Khilkin M, Kerjaschki D, Schreiber S, Ortner M, Weber J, Lochs H, Association between intraepithelial Escherichia coli and colorectal cancer, Gastroenterology 115(2) (1998) 281–6. [DOI] [PubMed] [Google Scholar]
- [78].Lucas C, Salesse L, Hoang MHT, Bonnet M, Sauvanet P, Larabi A, Godfraind C, Gagniere J, Pezet D, Rosenstiel P, Barnich N, Bonnet R, Dalmasso G, Nguyen HTT, Autophagy of Intestinal Epithelial Cells Inhibits Colorectal Carcinogenesis Induced by Colibactin-Producing Escherichia coli in Apc(Min/+) Mice, Gastroenterology 158(5) (2020) 1373–1388. [DOI] [PubMed] [Google Scholar]
- [79].Metzger R, Maruskova M, Krebs S, Janssen KP, Krug AB, Increased Incidence of Colon Tumors in AOM-Treated Apc (1638N/+) Mice Reveals Higher Frequency of Tumor Associated Neutrophils in Colon Than Small Intestine, Front Oncol 9 (2019) 1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Srivatsa S, Paul MC, Cardone C, Holcmann M, Amberg N, Pathria P, Diamanti MA, Linder M, Timelthaler G, Dienes HP, Kenner L, Wrba F, Prager GW, Rose-John S, Eferl R, Liguori G, Botti G, Martinelli E, Greten FR, Ciardiello F, Sibilia M, EGFR in Tumor-Associated Myeloid Cells Promotes Development of Colorectal Cancer in Mice and Associates With Outcomes of Patients, Gastroenterology 153(1) (2017) 178–190 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Tomkovich S, Yang Y, Winglee K, Gauthier J, Muhlbauer M, Sun X, Mohamadzadeh M, Liu X, Martin P, Wang GP, Oswald E, Fodor AA, Jobin C, Locoregional Effects of Microbiota in a Preclinical Model of Colon Carcinogenesis, Cancer Res 77(10) (2017) 2620–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Fais T, Delmas J, Barnich N, Bonnet R, Dalmasso G, Colibactin: More Than a New Bacterial Toxin, Toxins (Basel) 10(4) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Bossuet-Greif N, Vignard J, Taieb F, Mirey G, Dubois D, Petit C, Oswald E, Nougayrede JP, The Colibactin Genotoxin Generates DNA Interstrand Cross-Links in Infected Cells, mBio 9(2) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Hong SN, Genetic and epigenetic alterations of colorectal cancer, Intest Res 16(3) (2018) 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Weyrich A, Lenz D, Fickel J, Environmental Change-Dependent Inherited Epigenetic Response, Genes (Basel) 10(1) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Miro-Blanch J, Yanes O, Epigenetic Regulation at the Interplay Between Gut Microbiota and Host Metabolism, Front Genet 10 (2019) 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM, The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism, J Lipid Res 54(9) (2013) 2325–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Nawa N, Hirata K, Kawatani K, Nambara T, Omori S, Banno K, Kokubu C, Takeda J, Nishimura K, Ohtaka M, Nakanishi M, Okuzaki D, Taniguchi H, Arahori H, Wada K, Kitabatake Y, Ozono K, Elimination of protein aggregates prevents premature senescence in human trisomy 21 fibroblasts, PLoS One 14(7) (2019) e0219592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Jones PA, Functions of DNA methylation: islands, start sites, gene bodies and beyond, Nat Rev Genet 13(7) (2012) 484–92. [DOI] [PubMed] [Google Scholar]
- [90].Cheng AS, Li MS, Kang W, Cheng VY, Chou JL, Lau SS, Go MY, Lee CC, Ling TK, Ng EK, Yu J, Huang TH, To KF, Chan MW, Sung JJ, Chan FK, Helicobacter pylori causes epigenetic dysregulation of FOXD3 to promote gastric carcinogenesis, Gastroenterology 144(1) (2013) 122–133 e9. [DOI] [PubMed] [Google Scholar]
- [91].Koi M, Okita Y, Carethers JM, Fusobacterium nucleatum Infection in Colorectal Cancer: Linking Inflammation, DNA Mismatch Repair and Genetic and Epigenetic Alterations, J Anus Rectum Colon 2(2) (2018) 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, Yamano HO, Sugai T, An B, Shureiqi I, Toyota M, Kondo Y, Estecio MR, Issa JP, Fusobacterium in colonic flora and molecular features of colorectal carcinoma, Cancer Res 74(5) (2014) 1311–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Kawanaka M, Watari J, Kamiya N, Yamasaki T, Kondo T, Toyoshima F, Ikehara H, Tomita T, Oshima T, Fukui H, Daimon T, Das KM, Miwa H, Effects of Helicobacter pylori eradication on the development of metachronous gastric cancer after endoscopic treatment: analysis of molecular alterations by a randomised controlled trial, Br J Cancer 114(1) (2016) 21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Sobhani I, Bergsten E, Couffin S, Amiot A, Nebbad B, Barau C, de’Angelis N, Rabot S, Canoui-Poitrine F, Mestivier D, Pedron T, Khazaie K, Sansonetti PJ, Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures, Proc Natl Acad Sci U S A 116(48) (2019) 24285–24295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Moore LD, Le T, Fan G, DNA methylation and its basic function, Neuropsychopharmacology 38(1) (2013) 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Abbasi IHR, Abbasi F, Wang L, Abd El Hack ME, Swelum AA, Hao R, Yao J, Cao Y, Folate promotes S-adenosyl methionine reactions and the microbial methylation cycle and boosts ruminants production and reproduction, AMB Express 8(1) (2018) 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Wasson GR, McGlynn AP, McNulty H, O’Reilly SL, McKelvey-Martin VJ, McKerr G, Strain JJ, Scott J, Downes CS, Global DNA and p53 region-specific hypomethylation in human colonic cells is induced by folate depletion and reversed by folate supplementation, J Nutr 136(11) (2006) 2748–53. [DOI] [PubMed] [Google Scholar]
- [98].Pompei A, Cordisco L, Amaretti A, Zanoni S, Matteuzzi D, Rossi M, Folate production by bifidobacteria as a potential probiotic property, Appl Environ Microbiol 73(1) (2007) 179–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Legube G, Trouche D, Regulating histone acetyltransferases and deacetylases, EMBO Rep 4(10) (2003) 944–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Liu S, Chang W, Jin Y, Feng C, Wu S, He J, Xu T, The function of histone acetylation in cervical cancer development, Biosci Rep 39(4) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Sivaprakasam S, Bhutia YD, Yang S, Ganapathy V, Short-Chain Fatty Acid Transporters: Role in Colonic Homeostasis, Compr Physiol 8(1) (2017) 299–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Vital M, Howe AC, Tiedje JM, Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data, mBio 5(2) (2014) e00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Wichmann A, Allahyar A, Greiner TU, Plovier H, Lunden GO, Larsson T, Drucker DJ, Delzenne NM, Cani PD, Backhed F, Microbial modulation of energy availability in the colon regulates intestinal transit, Cell Host Microbe 14(5) (2013) 582–90. [DOI] [PubMed] [Google Scholar]
- [104].Morlando M, Fatica A, Alteration of Epigenetic Regulation by Long Noncoding RNAs in Cancer, Int J Mol Sci 19(2) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Liang L, Ai L, Qian J, Fang JY, Xu J, Long noncoding RNA expression profiles in gut tissues constitute molecular signatures that reflect the types of microbes, Sci Rep 5 (2015) 11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Moloney GM, Viola MF, Hoban AE, Dinan TG, Cryan JF, Faecal microRNAs: indicators of imbalance at the host-microbe interface?, Benef Microbes 9(2) (2018) 175–183. [DOI] [PubMed] [Google Scholar]
- [107].Nakata K, Sugi Y, Narabayashi H, Kobayakawa T, Nakanishi Y, Tsuda M, Hosono A, Kaminogawa S, Hanazawa S, Takahashi K, Commensal microbiota-induced microRNA modulates intestinal epithelial permeability through the small GTPase ARF4, J Biol Chem 292(37) (2017) 15426–15433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang JY, Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy, Cell 170(3) (2017) 548–563 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Sievers CK, Grady WM, Halberg RB, Pickhardt PJ, New insights into the earliest stages of colorectal tumorigenesis, Expert Rev Gastroenterol Hepatol 11(8) (2017) 723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].O’Connell JB, Maggard MA, Ko CY, Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging, J Natl Cancer Inst 96(19) (2004) 1420–5. [DOI] [PubMed] [Google Scholar]
- [111].Brenner H, Chen H, Fecal occult blood versus DNA testing: indirect comparison in a colorectal cancer screening population, Clin Epidemiol 9 (2017) 377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Schreuders EH, Grobbee EJ, Spaander MC, Kuipers EJ, Advances in Fecal Tests for Colorectal Cancer Screening, Curr Treat Options Gastroenterol 14(1) (2016) 152–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Robertson DJ, Lee JK, Boland CR, Dominitz JA, Giardiello FM, Johnson DA, Kaltenbach T, Lieberman D, Levin TR, Rex DK, Recommendations on Fecal Immunochemical Testing to Screen for Colorectal Neoplasia: A Consensus Statement by the US Multi-Society Task Force on Colorectal Cancer, Gastroenterology 152(5) (2017) 1217–1237 e3. [DOI] [PubMed] [Google Scholar]
- [114].Bosch LJW, Melotte V, Mongera S, Daenen KLJ, Coupe VMH, van Turenhout ST, Stoop EM, de Wijkerslooth TR, Mulder CJJ, Rausch C, Kuipers EJ, Dekker E, Domanico MJ, Lidgard GP, Berger BM, van Engeland M, Carvalho B, Meijer GA, Multitarget Stool DNA Test Performance in an Average-Risk Colorectal Cancer Screening Population, Am J Gastroenterol 114(12) (2019) 1909–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Zackular JP, Rogers MA, Ruffin M.T.t., Schloss PD, The human gut microbiome as a screening tool for colorectal cancer, Cancer Prev Res (Phila) 7(11) (2014) 1112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Zhang X, Zhu X, Cao Y, Fang JY, Hong J, Chen H, Fecal Fusobacterium nucleatum for the diagnosis of colorectal tumor: A systematic review and meta-analysis, Cancer Med 8(2) (2019) 480–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J, Brim H, Ashktorab H, Ng SC, Ng SSM, Zheng S, Chan FKL, Sung JJY, Yu J, Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer, Clin Cancer Res 23(8) (2017) 2061–2070. [DOI] [PubMed] [Google Scholar]
- [118].Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G, Lam TYT, Zhang L, Wu JCY, Chan FKL, Ng SSM, Wong MCS, Ng SC, Wu WKK, Yu J, Sung JJY, Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia, Gut 66(8) (2017) 1441–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, Tang L, Zhao H, Stenvang J, Li Y, Wang X, Xu X, Chen N, Wu WK, Al-Aama J, Nielsen HJ, Kiilerich P, Jensen BA, Yau TO, Lan Z, Jia H, Li J, Xiao L, Lam TY, Ng SC, Cheng AS, Wong VW, Chan FK, Xu X, Yang H, Madsen L, Datz C, Tilg H, Wang J, Brunner N, Kristiansen K, Arumugam M, Sung JJ, Wang J, Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer, Gut 66(1) (2017) 70–78. [DOI] [PubMed] [Google Scholar]
- [120].Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, Hosoda F, Rokutan H, Matsumoto M, Takamaru H, Yamada M, Matsuda T, Iwasaki M, Yamaji T, Yachida T, Soga T, Kurokawa K, Toyoda A, Ogura Y, Hayashi T, Hatakeyama M, Nakagama H, Saito Y, Fukuda S, Shibata T, Yamada T, Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer, Nat Med 25(6) (2019) 968–976. [DOI] [PubMed] [Google Scholar]
- [121].Dai Z, Coker OO, Nakatsu G, Wu WKK, Zhao L, Chen Z, Chan FKL, Kristiansen K, Sung JJY, Wong SH, Yu J, Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers, Microbiome 6(1) (2018) 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Wei Z, Cao S, Liu S, Yao Z, Sun T, Li Y, Li J, Zhang D, Zhou Y, Could gut microbiota serve as prognostic biomarker associated with colorectal cancer patients’ survival? A pilot study on relevant mechanism, Oncotarget 7(29) (2016) 46158–46172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Tang Q, Jin G, Wang G, Liu T, Liu X, Wang B, Cao H, Current Sampling Methods for Gut Microbiota: A Call for More Precise Devices, Front Cell Infect Microbiol 10 (2020) 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Antosca K, Hoen AG, Palys T, Hilliard M, Morrison HG, Coker M, Madan J, Karagas MR, Reliability of stool microbiome methods for DNA yields and sequencing among infants and young children, Microbiologyopen 9(5) (2020) e1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Tap J, Cools-Portier S, Pavan S, Druesne A, Ohman L, Tornblom H, Simren M, Derrien M, Effects of the long-term storage of human fecal microbiota samples collected in RNAlater, Sci Rep 9(1) (2019) 601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Li H, Limenitakis JP, Fuhrer T, Geuking MB, Lawson MA, Wyss M, Brugiroux S, Keller I, Macpherson JA, Rupp S, Stolp B, Stein JV, Stecher B, Sauer U, McCoy KD, Macpherson AJ, The outer mucus layer hosts a distinct intestinal microbial niche, Nat Commun 6 (2015) 8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Dieterich W, Schink M, Zopf Y, Microbiota in the Gastrointestinal Tract, Med Sci (Basel) 6(4) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Robertson BR, O’Rourke JL, Neilan BA, Vandamme P, On SLW, Fox JG, Lee A, Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents, Int J Syst Evol Microbiol 55(Pt 3) (2005) 1199–1204. [DOI] [PubMed] [Google Scholar]
- [129].Ladinsky MS, Araujo LP, Zhang X, Veltri J, Galan-Diez M, Soualhi S, Lee C, Irie K, Pinker EY, Narushima S, Bandyopadhyay S, Nagayama M, Elhenawy W, Coombes BK, Ferraris RP, Honda K, Iliev ID, Gao N, Bjorkman PJ, Ivanov II, Endocytosis of commensal antigens by intestinal epithelial cells regulates mucosal T cell homeostasis, Science 363(6431) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Fung TC, Bessman NJ, Hepworth MR, Kumar N, Shibata N, Kobuley D, Wang K, Ziegler CGK, Goc J, Shima T, Umesaki Y, Sartor RB, Sullivan KV, Lawley TD, Kunisawa J, Kiyono H, Sonnenberg GF, Lymphoid-Tissue-Resident Commensal Bacteria Promote Members of the IL-10 Cytokine Family to Establish Mutualism, Immunity 44(3) (2016) 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS, Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment, Cell Host Microbe 14(2) (2013) 207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Wu J, Li Q, Fu X, Fusobacterium nucleatum Contributes to the Carcinogenesis of Colorectal Cancer by Inducing Inflammation and Suppressing Host Immunity, Transl Oncol 12(6) (2019) 846–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Steck N, Hoffmann M, Sava IG, Kim SC, Hahne H, Tonkonogy SL, Mair K, Krueger D, Pruteanu M, Shanahan F, Vogelmann R, Schemann M, Kuster B, Sartor RB, Haller D, Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation, Gastroenterology 141(3) (2011) 959–71. [DOI] [PubMed] [Google Scholar]
- [134].Huycke MM, Abrams V, Moore DR, Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA, Carcinogenesis 23(3) (2002) 529–36. [DOI] [PubMed] [Google Scholar]
- [135].Housseau F, Sears CL, Enterotoxigenic Bacteroides fragilis (ETBF)-mediated colitis in Min (Apc+/−) mice: a human commensal-based murine model of colon carcinogenesis, Cell Cycle 9(1) (2010) 3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Wu S, Powell J, Mathioudakis N, Kane S, Fernandez E, Sears CL, Bacteroides fragilis enterotoxin induces intestinal epithelial cell secretion of interleukin-8 through mitogen-activated protein kinases and a tyrosine kinase-regulated nuclear factor-kappaB pathway, Infect Immun 72(10) (2004) 5832–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, Dejea CM, Fathi P, Chen J, Finard BB, Tam AJ, McAllister F, Fan H, Wu X, Ganguly S, Lebid A, Metz P, Van Meerbeke SW, Huso DL, Wick EC, Pardoll DM, Wan F, Wu S, Sears CL, Housseau F, Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells, Cell Host Microbe 23(2) (2018) 203–214 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrede JP, Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells, Proc Natl Acad Sci U S A 107(25) (2010) 11537–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Kumar R, Herold JL, Schady D, Davis J, Kopetz S, Martinez-Moczygemba M, Murray BE, Han F, Li Y, Callaway E, Chapkin RS, Dashwood WM, Dashwood RH, Berry T, Mackenzie C, Xu Y, Streptococcus gallolyticus subsp. gallolyticus promotes colorectal tumor development, PLoS Pathog 13(7) (2017) e1006440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Kahrstrom CT, Bacterial pathogenesis: E. coli claims the driving seat for cancer, Nat Rev Cancer 12(10) (2012) 658–9. [DOI] [PubMed] [Google Scholar]
- [141].Chen L, Tai WC, Brar MS, Leung FC, Hsiao WL, Tumor grafting induces changes of gut microbiota in athymic nude mice in the presence and absence of medicinal Gynostemma saponins, PLoS One 10(5) (2015) e0126807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Melton-Witt JA, Bentsen LM, Tweten RK, Identification of functional domains of Clostridium septicum alpha toxin, Biochemistry 45(48) (2006) 14347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Jacouton E, Chain F, Sokol H, Langella P, Bermudez-Humaran LG, Probiotic Strain Lactobacillus casei BL23 Prevents Colitis-Associated Colorectal Cancer, Front Immunol 8 (2017) 1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Martin R, Miquel S, Benevides L, Bridonneau C, Robert V, Hudault S, Chain F, Berteau O, Azevedo V, Chatel JM, Sokol H, Bermudez-Humaran LG, Thomas M, Langella P, Functional Characterization of Novel Faecalibacterium prausnitzii Strains Isolated from Healthy Volunteers: A Step Forward in the Use of F. prausnitzii as a Next-Generation Probiotic, Front Microbiol 8 (2017) 1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Ellmerich S, Scholler M, Duranton B, Gosse F, Galluser M, Klein JP, Raul F, Promotion of intestinal carcinogenesis by Streptococcus bovis, Carcinogenesis 21(4) (2000) 753–6. [DOI] [PubMed] [Google Scholar]
- [146].Chen L, Jiang B, Zhong C, Guo J, Zhang L, Mu T, Zhang Q, Bi X, Chemoprevention of colorectal cancer by black raspberry anthocyanins involved the modulation of gut microbiota and SFRP2 demethylation, Carcinogenesis 39(3) (2018) 471–481. [DOI] [PubMed] [Google Scholar]
- [147].Riviere A, Selak M, Lantin D, Leroy F, De Vuyst L, Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut, Front Microbiol 7 (2016) 979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Yu DH, Gadkari M, Zhou Q, Yu S, Gao N, Guan Y, Schady D, Roshan TN, Chen MH, Laritsky E, Ge Z, Wang H, Chen R, Westwater C, Bry L, Waterland RA, Moriarty C, Hwang C, Swennes AG, Moore SR, Shen L, Postnatal epigenetic regulation of intestinal stem cells requires DNA methylation and is guided by the microbiome, Genome Biol 16 (2015) 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Cortese R, Lu L, Yu Y, Ruden D, Claud EC, Epigenome-Microbiome crosstalk: A potential new paradigm influencing neonatal susceptibility to disease, Epigenetics 11(3) (2016) 205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Pan WH, Sommer F, Falk-Paulsen M, Ulas T, Best P, Fazio A, Kachroo P, Luzius A, Jentzsch M, Rehman A, Muller F, Lengauer T, Walter J, Kunzel S, Baines JF, Schreiber S, Franke A, Schultze JL, Backhed F, Rosenstiel P, Exposure to the gut microbiota drives distinct methylome and transcriptome changes in intestinal epithelial cells during postnatal development, Genome Med 10(1) (2018) 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Maiuri AR, Peng M, Podicheti R, Sriramkumar S, Kamplain CM, Rusch DB, DeStefano Shields CE, Sears CL, O’Hagan HM, Mismatch Repair Proteins Initiate Epigenetic Alterations during Inflammation-Driven Tumorigenesis, Cancer Res 77(13) (2017) 3467–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Davison JM, Lickwar CR, Song L, Breton G, Crawford GE, Rawls JF, Microbiota regulate intestinal epithelial gene expression by suppressing the transcription factor Hepatocyte nuclear factor 4 alpha, Genome Res 27(7) (2017) 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Thaiss CA, Levy M, Korem T, Dohnalova L, Shapiro H, Jaitin DA, David E, Winter DR, Gury-BenAri M, Tatirovsky E, Tuganbaev T, Federici S, Zmora N, Zeevi D, Dori-Bachash M, Pevsner-Fischer M, Kartvelishvily E, Brandis A, Harmelin A, Shibolet O, Halpern Z, Honda K, Amit I, Segal E, Elinav E, Microbiota Diurnal Rhythmicity Programs Host Transcriptome Oscillations, Cell 167(6) (2016) 1495–1510 e12. [DOI] [PubMed] [Google Scholar]
- [154].Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-Wilt GA, Rabaglia ME, Keller MP, Attie AD, Rey FE, Denu JM, Diet-Microbiota Interactions Mediate Global Epigenetic Programming in Multiple Host Tissues, Mol Cell 64(5) (2016) 982–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Kelly D, Kotliar M, Woo V, Jagannathan S, Whitt J, Moncivaiz J, Aronow BJ, Dubinsky MC, Hyams JS, Markowitz JF, Baldassano RN, Stephens MC, Walters TD, Kugathasan S, Haberman Y, Sundaram N, Rosen MJ, Helmrath M, Karns R, Barski A, Denson LA, Alenghat T, Microbiota-sensitive epigenetic signature predicts inflammation in Crohn’s disease, JCI Insight 3(18) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Fellows R, Denizot J, Stellato C, Cuomo A, Jain P, Stoyanova E, Balazsi S, Hajnady Z, Liebert A, Kazakevych J, Blackburn H, Correa RO, Fachi JL, Sato FT, Ribeiro WR, Ferreira CM, Peree H, Spagnuolo M, Mattiuz R, Matolcsi C, Guedes J, Clark J, Veldhoen M, Bonaldi T, Vinolo MAR, Varga-Weisz P, Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases, Nat Commun 9(1) (2018) 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Peck BC, Mah AT, Pitman WA, Ding S, Lund PK, Sethupathy P, Functional Transcriptomics in Diverse Intestinal Epithelial Cell Types Reveals Robust MicroRNA Sensitivity in Intestinal Stem Cells to Microbial Status, J Biol Chem 292(7) (2017) 2586–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Liu S, da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L, Comstock LE, Gandhi R, Weiner HL, The Host Shapes the Gut Microbiota via Fecal MicroRNA, Cell Host Microbe 19(1) (2016) 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Baxter NT, Ruffin M.T.t., Rogers MA, Schloss PD, Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions, Genome Med 8(1) (2016) 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Xie YH, Gao QY, Cai GX, Sun XM, Sun XM, Zou TH, Chen HM, Yu SY, Qiu YW, Gu WQ, Chen XY, Cui Y, Sun D, Liu ZJ, Cai SJ, Xu J, Chen YX, Fang JY, Fecal Clostridium symbiosum for Noninvasive Detection of Early and Advanced Colorectal Cancer: Test and Validation Studies, EBioMedicine 25 (2017) 32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Eklof V, Lofgren-Burstrom A, Zingmark C, Edin S, Larsson P, Karling P, Alexeyev O, Rutegard J, Wikberg ML, Palmqvist R, Cancer-associated fecal microbial markers in colorectal cancer detection, Int J Cancer 141(12) (2017) 2528–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Rezasoltani S, Sharafkhah M, Asadzadeh Aghdaei H, Nazemalhosseini Mojarad E, Dabiri H, Akhavan Sepahi A, Modarressi MH, Feizabadi MM, Zali MR, Applying simple linear combination, multiple logistic and factor analysis methods for candidate fecal bacteria as novel biomarkers for early detection of adenomatous polyps and colon cancer, J Microbiol Methods 155 (2018) 82–88. [DOI] [PubMed] [Google Scholar]
- [163].Guo S, Li L, Xu B, Li M, Zeng Q, Xiao H, Xue Y, Wu Y, Wang Y, Liu W, Zhang G, A Simple and Novel Fecal Biomarker for Colorectal Cancer: Ratio of Fusobacterium Nucleatum to Probiotics Populations, Based on Their Antagonistic Effect, Clin Chem 64(9) (2018) 1327–1337. [DOI] [PubMed] [Google Scholar]


