Abstract
Haematopoietic stem cells (HSCs) reside in specialized microenvironments in the bone marrow (BM), often referred to as “niches”, which represent complex regulatory milieux influenced by multiple cellular constituents, including nerves1,2. Although sympathetic nerves regulate the HSC niche3-6, the contribution of nociceptive neurons in the BM remains unclear. Here we show that nociceptive nerves are required for enforced HSC mobilization and collaborate with sympathetic nerves for HSC maintenance in BM. Nociceptor neurons drive granulocyte colony-stimulating factor (G-CSF)-induced HSC mobilization via the secretion of calcitonin gene-related peptide (CGRP). Unlike sympathetic nerves which regulate HSCs indirectly via the niche3,4,6, CGRP acts directly on HSCs via the receptor activity modifying protein-1 (RAMP1) and calcitonin-receptor-like receptor (CALCRL), to promote egress by activating Gαs/Adenylyl cyclase/cAMP pathway. Remarkably, the ingestion of food containing capsaicin (a natural component of chili peppers), significantly enhanced HSC mobilization. Thus, targeting the nociceptive nervous system is a novel strategy to improve the HSC yield for stem cell-based therapeutics.
The ability to migrate is a hallmark of HSCs which travel to different haematopoietic sites during development and continue to be released from the adult bone marrow throughout life. The migratory property of HSCs has facilitated their harvest in blood and their transplantation for curative anti-cancer therapies. The detailed mechanisms by which HSCs migrate out of the bone marrow, however, remain largely enigmatic. Additionally, the mobilized HSC yield can be insufficient in some donors7, indicating that improved methods are needed.
In tissues exposed to the external environment (e.g. skin, lung, gut), nociceptor sensory neurons detect damaging stimuli, and can regulate the ensuing immune response by releasing neurotransmitters and other regulatory molecules8-10. However, the role of nociceptors in a deep tissue like the bone marrow (BM) — other than perception of pain — is not well understood.
Neural collaboration to maintain HSCs
We surveyed the total nerve densities in the BM by analysing the pan-neuronal marker class III β tubulin (TUBB3) expression using immunofluorescence. While adrenergic fibres from the SNS comprised a meaningful fraction of the nerve staining area identified by anti-tyrosine hydroxylase (TH), the vast majority of the staining area (~77%) was by nociceptive nerves marked by CGRP (Fig.1a,b; Extended Data Fig.1a). Analysis of total nerve length per BM area revealed similar length densities of the CGRP+ and TH+ nerves (Extended Data Fig.1b), suggesting that CGRP+ and TH+ nerves exhibit similar projections in BM and that the increased nociceptive nerve area may reflect their larger bundles.
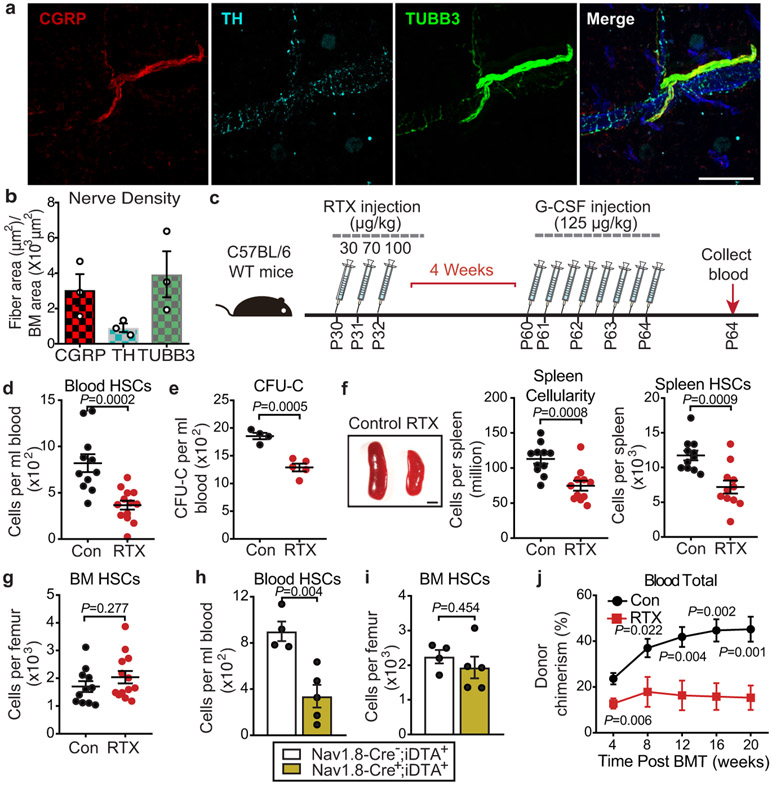
Figure 1. Nociceptive neurons promote HSC mobilization.
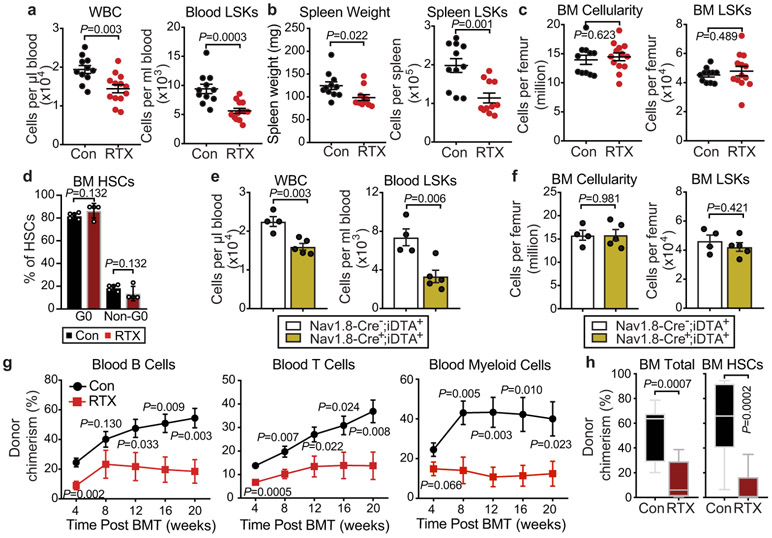
a, b, Representative confocal z-stack projection of C57BL/6 mouse femur stained for CGRP (red), TH (light blue), and TUBB3 (green) nerve fibres. Scale bar, 100μm. Femoral sensory and sympathetic innervation quantified by CGRP+, TH+ or TUBB3+ stained area divided by total BM area. n=3 mice. c, Schematic illustration of G-CSF-induced HSC mobilization and analyses. d, Absolute numbers of HSCs (Lin−Sca-1+cKit+CD150+CD48−) per mL of peripheral blood following G-CSF-induced mobilization in saline- or RTX-treated mice. n=11,13 mice, respectively. e, Colony-forming units (BFU-E, CFU-GM, and CFU-GEMM) per mL of G-CSF-mobilized blood from saline- or RTX-treated mice. n=4,5 mice, respectively. f, Left, representative images of the spleen following G-CSF administration in saline- or RTX-treated mice. Scale bar, 2mm; middle and right, cellularity and absolute numbers of HSCs in the spleen of saline and RTX-treated mice after G-CSF. n=11 mice per group. g, HSC numbers per femur from saline- or RTX-treated mice after G-CSF. n=11, 13 mice, respectively. h, i, HSC numbers per mL of peripheral blood or per femur following G-CSF mobilization in Nav1.8-Cre−;iDTA+ and Nav1.8-Cre+;iDTA+. n=4,5 mice, respectively. j, Peripheral blood donor chimerism (CD45.2+) in CD45.1-recipient mice transplanted with mobilized blood (CD45.2) derived from saline or RTX-treated mice mixed with CD45.1 competitor BM cells at the indicated time points post-transplantation. n=9,8 mice, respectively. Error bars represent s.e.m. Two-tailed unpaired Student’s t test.
Given their afferent functions, it is possible that nociceptor neurons sense the BM environment and relay these signals to the central nervous system to circuit back via efferent fibres from the SNS. Alternatively, nociceptors may exert efferent influences and directly alter organ physiology9. To distinguish between these possibilities, we treated wild-type C57BL/6 mice systemically with either vehicle, resiniferatoxin (RTX), 6-hydroxydopamine (6OHDA), or both to differentially ablate nociceptive or sympathetic nerve fibres, respectively (Extended Data Fig.1c-e). While sympathetic denervation significantly suppressed B lymphopoiesis—in line with previous studies11, neither sympathetic nor sensory denervation alone altered HSC numbers in the BM (Extended Data Fig.2a-d). However, the dual denervation induced significant HSC expansion and led to a myeloid bias of haematopoiesis (Extended Data Fig.2c,d). Competitive transplantation experiments revealed a marked reduction in long-term reconstitution after dual denervation compared with the other groups (Extended Data Fig.2e,f). However, the homing efficiency of HSCs and Lin− Sca-1+ c-Kit+ (LSK) cells was similar between the two groups, suggesting that the reduced BM reconstitution is independent of HSC homing (Extended Data Fig.2g,h). To investigate further the dual denervation in a genetic model, we intercrossed Nav1.8-Cre transgenic mice with Cre-inducible diphtheria toxin A (iDTA) animals to specifically ablate Nav1.8-expressing sensory neurons (Extended Data Fig.3a-d). Confocal immunofluorescence microscopy analysis of BM from Nav1.8-Cre;iTdTomato mice revealed that all CGRP+ nerves were iTdTomato+ and that the majority (>70%) of Nav1.8+ nociceptive nerves were CGRP+ (Extended Data Fig.3a). Dual depletion using Nav1.8-Cre/iDTA mice combined with 6OHDA completely recapitulated the haematopoietic phenotype observed with RTX/6OHDA dual treatment (Extended Data Fig.3e,f). Thus, nociceptive and sympathetic nerves do not act as a circuit but collaborate as parallel efferent regulators to maintain HSCs in BM niches.
As nociceptor neurons secrete two major neurotransmitters, CGRP and substance P (SP)10, we evaluated their specific contributions in regulating HSCs in concert with adrenergic signals. We found that the continuous delivery of CGRP, but not SP, returned HSC numbers, B cells and myeloid cells to control levels (Extended Data Fig.3g). Additionally, HSC numbers were normalized by administration of a pan-β-adrenergic receptor agonist (Extended Data Fig.3g), further indicating that nociceptive and sympathetic nerves form an integrated efferent innervation network regulating steady-state HSC function.
Nociceptors regulate HSC mobilization
Granulocyte colony-stimulating factor (G-CSF)-enforced HSC mobilization is controlled in part by peripheral sympathetic nerve-derived norepinephrine (NE) signalling via regulation of the BM niche3. We explored the possibility that nociceptor nerve-derived neurotransmitter release in the BM microenvironment might also influence G-CSF-induced HSC mobilization. Remarkably, RTX-induced nociceptor neuron ablation alone led to a marked reduction of mobilized phenotypic HSCs and colony-forming progenitors (Fig.1c-e), and LSKs cells (Extended Data Fig.4a) compared to vehicle-treated controls in response to G-CSF. Consistent with the reduced mobilization, the spleen size and its HSC content were significantly reduced in RTX-treated mice compared to nerve-intact mice (Fig.1f; Extended Data Fig.4b). However, we observed no haematopoietic deficits in the BM (Fig.1g; Extended Data Fig.4c,d), suggesting that the absence of nociceptor neurons selectively affected HSC egress. We obtained similar results with G-CSF-mobilized Nav1.8-Cre;iDTA genetic model (Fig.1h,i; Extended Data Fig.4e,f). To assess the blood content in functional HSCs, we transfused blood from G-CSF-, vehicle- or RTX-treated mice mixed with competitor BM cells into lethally irradiated recipients. Mice reconstituted with the mobilized blood from the RTX-treated mice consistently exhibited lower donor multi-lineage reconstitution 20 weeks post-transplantation (Fig.1j; Extended Data Fig.4g,h). These data indicate that nociceptor neurons are required to drive robust G-CSF-elicited HSC mobilization from the BM into the blood.
The neuropeptide CGRP promotes HSC egress
As CGRP collaborated with adrenergic signals in HSC maintenance, we investigated its role in HSC mobilization (Fig.2a). Treatment with CGRP significantly increased the neuropeptide levels in the BM extracellular fluid (Fig.2b) and dramatically increased the HSC numbers mobilized by G-CSF (Fig.2c). By contrast, no statistically significant improvement in mobilization was observed with SP administration (Extended Data Fig.5a,b). Moreover, CGRP administration rescued the mobilization deficit of mice lacking nociceptors to levels similar to those observed in saline treated wild-type controls (Fig.2c). While administration of CGRP did not alter the BM cellularity, it significantly reduced the BM HSC numbers (Extended Data Fig.5c), consistent with previous studies suggesting that sustained CGRP treatment may suppress the proliferation of HSPCs12. To exclude further the possibility that nociceptive neurons may act via a sympathetic efferent circuit, we depleted sympathetic nerves using 6OHDA and evaluated whether CGRP could still induce HSC mobilization. We found that CGRP treatment led to a significant and proportional (~3-fold) increase in HSC mobilization in the absence of sympathetic neurons (Extended Data Fig.5d). These results suggest that G-CSF-induced HSC mobilization is regulated by the nociceptor nerve-derived neuropeptide CGRP.
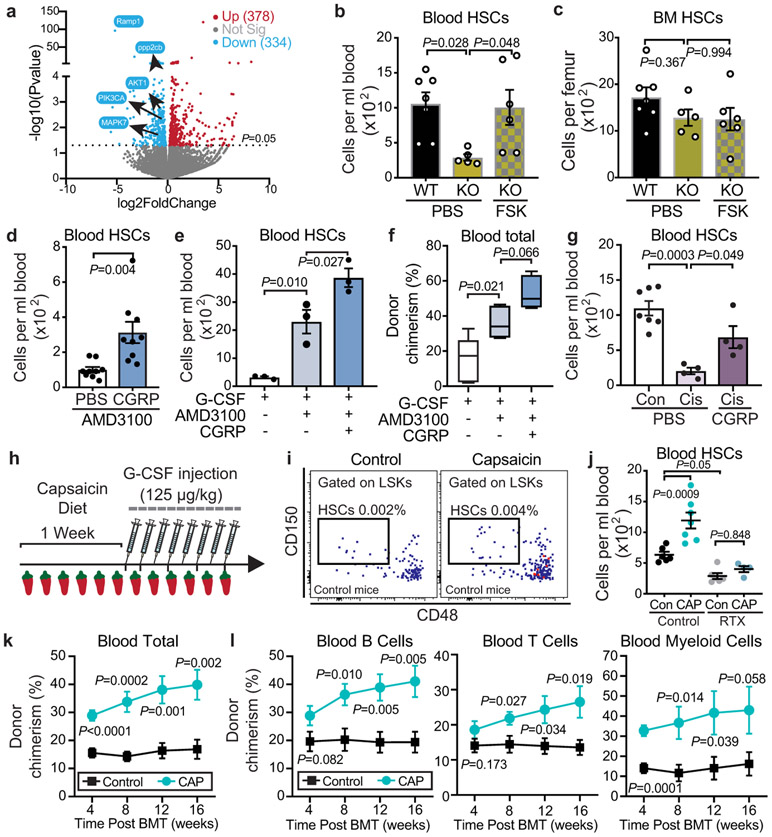
Figure 2. Nociceptor-derived CGRP induces HSC mobilization via its cognate receptor CALCRL/ RAMP1.
a, Schematic illustration of the rescue experiment. b, Bone marrow extracellular fluid (BMEF) CGRP levels measured by ELISA (n=11,6,3,4 mice, respectively). c, Absolute numbers of HSCs (Lin−Sca-1+cKit+CD150+CD48−) per mL of peripheral blood following G-CSF mobilization in saline- or RTX-treated mice implanted with osmotic pumps containing saline or CGRP. n=18,9,7,7 mice, respectively. d, e, HSC numbers per mL of peripheral blood or per femur from Ramp1+/+ (WT) or Ramp1−/− (KO) mice with or without G-CSF. n=5 mice per group. f, Ramp1 mRNA levels in sorted HSCs, endothelial and stromal compartments (from RNA-seq datasets5,28). g, Schematic illustration of reciprocal transplantation of BM cells from Ramp1−/− (KO) or Ramp1+/+ (WT) mice into lethally irradiated Ramp1+/+ (WT) or Ramp1−/− (KO) recipients and G-CSF was injected 8 weeks post-transplantation. h, HSCs per mL of peripheral blood following G-CSF treatment after reciprocal transplantation. n=6,7,5,6 mice, respectively. i, Schematic illustration of mobilization experiment with Vav1-iCre+;Calcrlf/f mice. j, k, HSCs per mL of blood or per femur following G-CSF treatment in Vav1-iCre+;Calcrlw/w or Vav1-iCre+;Calcrlf/f mice. n=6 mice per group. Error bars represent s.e.m. Two-tailed unpaired Student’s t test (b,c,d,e,j,k) or one-way ANOVA (h).
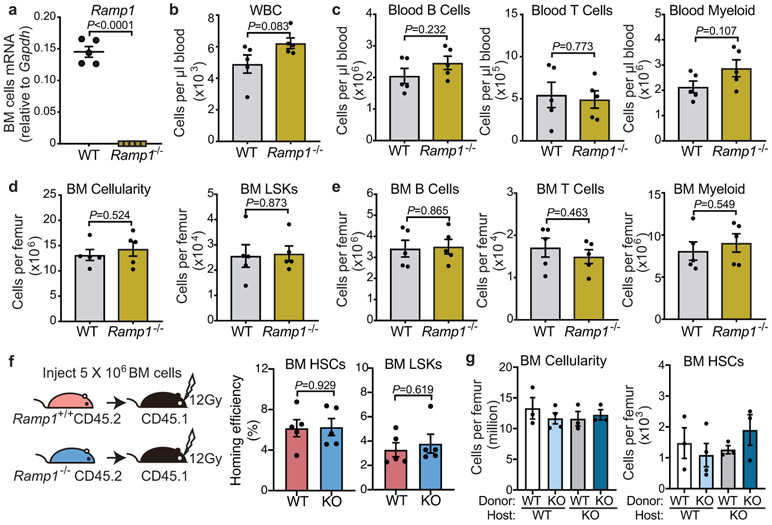
To investigate how CGRP promoted HSC egress, we evaluated the function of its receptors formed by heterodimers of CALCRL and RAMP1, each of which is required to initiate a CGRP response. We examined G-CSF-induced HSC mobilization in Ramp1−/− mice compared with Ramp1+/+ littermates (Extended Data Fig.6a), and found marked reduction in G-CSF-induced HSC mobilization in mice lacking Ramp1 (Fig.2d). The reduction of mobilized HSCs in Ramp1−/− mice was not due to lower HSC reserves in BM (Fig.2e). Additionally, we found no haematopoietic deficit at steady state and Ramp1 deficiency did not affect the homing ability of HSCs and LSKs to the BM (Extended Data Fig.6b-f), indicating that Ramp1 deletion selectively impairs HSC egress into the circulation.
CGRP acts on haematopoietic cells
To identify the cell target of nociceptors, we first surveyed Ramp1 expression levels among various stromal and haematopoietic elements. While Ramp1 was expressed at low levels in stromal cells, robust expression was detected in HSCs (Fig.2f). To ascertain whether Ramp1 was required on haematopoietic or stromal compartment, we generated chimeric mice by reciprocal transplantation of BM cells from Ramp1−/− or Ramp1+/+ mice into lethally irradiated Ramp1+/+ or Ramp1−/− recipients (Fig.2g). We found that the mobilization defect in the Ramp1−/− recipients was rescued when donor HSCs were wild type (Fig.2h). By contrast, chimeras in which Ramp1−/− donor HSCs engrafted wild-type recipients exhibited the same reduction in HSC mobilization (Fig.2h) without alteration of cellularity or HSC numbers in BM (Extended Data Fig.6g). To confirm the haematopoietic origin of the defect, we intercrossed Calcrlf/f mice with Vav1-iCre to conditionally delete Calcrl in haematopoietic cells (Fig.2i). We found similar HSC mobilization defect in Vav1-iCre+; Calcrlf/f mice while BM HSCs and cellularity were not altered (Fig.2j,k). In addition, we did not observe any significant changes in the number of niche-associated cells or CXCL12 expression within distinct niche compartments (Extended Data Fig.7a-c). Thus, CGRP acts via its heterodimeric receptor RAMP1/CALCRL on haematopoietic cells to promote HSC egress.
Next, we sought to define the CGRP-targeted haematopoietic cell type regulating HSC mobilization. Quantitative RT-PCR analyses revealed that Ramp1 was expressed at highest levels in HSCs, and at lower levels in monocytes and macrophages (Fig.3a). We investigated the role of monocytes/macrophages which are known to regulate HSC mobilization13-15. Depletion of BM monocytes/macrophages, using clodronate liposome13-15, significantly reduced BM CXCL12 levels, leading to a significant increase of HSPCs in blood (Fig.3b-f). Strikingly, CGRP administration to mononuclear-cell-depleted mice markedly enhanced HSC mobilization without affecting the BM CXCL12 levels (Fig.3e,f). Their synergistic effect excludes the possibility that CGRP is acting on mononuclear phagocytes to promote HSC mobilization.
Figure 3. RAMP1 signals directly in HSCs.
a. Quantification of Ramp1 mRNA levels in sorted cell populations from the BM by qRT-PCR. n=3-4 biological samples. b, Schematic illustration of the macrophage depletion experiment. c, Macrophage numbers (Gr-1−F4/80+CD115intSSCint/lo) per femur from PBS or clodronate-liposome-treated mice implanted with osmotic pumps containing saline or CGRP. n=7 mice per group. d, CXCL12 levels in BMEF measured by ELISA. n=7 biological samples. e, f, HSCs (Lin−Sca-1+cKit+CD150+CD48−) and LSKs (Lin−Sca-1+cKit+) per mL of peripheral blood. n=7 mice per group. g, Experimental design of the transwell assay. h, Colony-forming units per well of migrated stem progenitor cells from Ramp1+/+ (WT) or Ramp1−/− (KO) mice. n=5, 4 mice, respectively. i, Schematic illustration of the mixed chimerism experiment. j, Chimerism of mobilized HSC in blood normalized to the chimerism of BM HSCs. n=4,4,3,3 mice, respectively. Error bars represent s.e.m. Two-tailed unpaired Student’s t test (c-e, h) or one-way ANOVA (j).
To address whether CGRP/RAMP1 signals were HSPC autonomous, we analysed the migration capacity of haematopoietic progenitors toward the chemoattractant CXCL12 (Fig.3g). These analyses revealed that the transmigration of lineage-depleted progenitor cells from Ramp1−/− mice was significantly reduced (Fig.3h). To confirm the HSPC autonomous effect in vivo, we generated mixed chimeras in which CD45.1+ recipient mice were transplanted with a mix of CD45.1+ wild-type and CD45.2+ Ramp1−/− or Ramp1+/+ BM at 1:1 ratio (Fig.3i). These analyses revealed significant impairments of Ramp1−/− HSC mobilization (Fig.3j), further suggesting that CGRP acts directly on HSPCs.
CGRP activates Gαs/AC/cAMP pathways
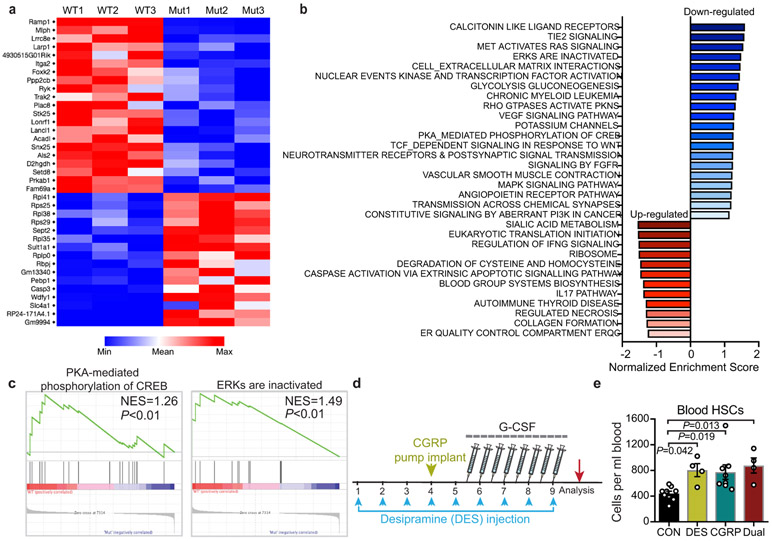
We found no alterations in the expression of receptors involved in HSC trafficking (CXCR4, VLA4 or CD44) after CGRP administration, invoking an independent mechanism (Extended Data Fig.7d). To obtain further mechanistic insight, we analysed the genome-wide changes in gene expression of purified HSCs from Ramp+/+ and Ramp1−/− mice by RNA-seq (Fig.4a; Extended Data Fig.8a). Gene set enrichment analysis revealed downregulation of genes associated with Gαs/AC/cAMP pathways, including cAMP-dependent PKA-mediated phosphorylation of CREB and activation of ERKs (Extended Data Fig.8b,c). Since Gαs can activate adenylyl cyclase (AC) to generate cAMP, we tested whether stimulation of AC with forskolin could rescue the mobilization defect of Ramp1-deficient HSCs. We found that AC activation completely restored the numbers of G-CSF-mobilized HSCs in Ramp1−/− mice (Fig.4b), without affecting the BM HSC numbers (Fig.4c). These results indicate that the CGRP/RAMP1 pathway promotes HSC mobilization via activation of downstream Gαs/AC/cAMP signalling.
Figure. 4. CGRP amplifies HSC mobilization via the Gαs/AC/cAMP pathway.
a, Volcano plot showing differentially expressed genes (P<0.05) in Ramp1−/− compared to Ramp1+/+ HSCs. b, c, Absolute numbers of HSCs per mL of blood or per femur following G-CSF administration in Ramp1+/+ and Ramp1−/− mice treated with vehicle or forskolin. n=7,5,6 mice, respectively. d, HSC numbers in plerixafor-mobilized blood from saline or CGRP-treated mice. n=9 mice per group. e, HSCs per mL of blood from mice treated with G-CSF, G-CSF+AMD3100 or G-CSF+AMD3100+CGRP. n=3 mice per group. f, Blood donor chimerism (CD45.2) in CD45.1-recipient mice transplanted with mobilized blood (CD45.2) mixed with CD45.1 competitor BM cells, 16 weeks post-transplantation. n=5 mice per group. g, HSC numbers in G-CSF-mobilized blood of saline or cisplatin-treated mice with or without CGRP administration. n=7,4,4 mice, respectively. h, Experimental design to determine the effect of capsaicin diet on HSC mobilization. i, j, Representative gating strategy and quantification of HSCs per mL of blood following G-CSF administration in saline or RTX-treated mice fed on control or capsaicin-containing diet. n=6,7,6,4 mice, respectively. k, l, Blood leukocyte chimerism in CD45.1-recipient mice transplanted with mobilized blood (CD45.2) derived from mice fed on control or capsaicin diet mixed with CD45.1 competitor BM cells. n=9 mice per group. Error bars represent s.e.m. Two-tailed unpaired Student’s t test (d,k,l) or one-way ANOVA for (b,c,e,f,g,j). For box plots, the box spans from the 25th to 75th percentiles and the centerline shows the median. Whiskers represent minimum to maximum range.
The CGRP-induced Gαs/AC/cAMP pathway activation in HSCs raised the possible collaboration with plerixafor (AMD3100), an FDA-approved drug antagonist of CXCR4 used with G-CSF to elicit HSPCs in patients that mobilize poorly or in patients in which G-CSF is contraindicated7,16. Plerixafor administration with CGRP markedly increased HSC mobilization over plerixafor alone (Fig.4d). Remarkably, CGRP administration further increased the mobilization induced by concurrent plerixafor and G-CSF treatment (Fig.4e), and this was confirmed by long-term competitive repopulation of mobilized blood (Fig.4f). To investigate if adrenergic stimulation could enhance the mobilization by CGRP, we treated mice with the noradrenaline reuptake inhibitor desipramine which enhances HSC mobilization by increasing the adrenergic tone in the BM17,18. However, we did not observe further mobilization by combining desipramine and CGRP stimulation (Extended Data Fig.8d,e), suggesting that they may be acting on the same mobilizable pool. To assess whether CGRP administration could rescue the defective HSC mobilization from diseased BM, we evaluated animals that had received multiple cycles of chemotherapy, which is a common cause for insufficient HSC yield in the clinic19. As expected4, G-CSF-induced HSPC mobilization was significantly impaired in the cisplatin-treated mice, and we found that this deficit was significantly rescued by CGRP administration (Fig.4g). These data suggest broad contributions of CGRP-induced signals in improving HSC mobilization.
Spicy food enhances HSC mobilization
The role of nociceptors in promoting HSC mobilization led us to hypothesize that the ingestion of spicy food—which can trigger nociceptive nerve activation—could influence HSC mobilization from the BM. To explore this possibility, we fed C57BL/6 mice with food containing capsaicin, a chili pepper extract and natural ligand activating TRPV1+ neurons (Fig.4h, Extended Data Fig.9a). Although when given the choice, mice preferred regular chow over spicy chow, their daily food intake and body weight were not altered on a spicy food diet (Extended Data Fig.9b,c). Capsaicin diet significantly increased CGRP levels in the BMEF without altering HSC number or their proliferation (Extended Data Fig.9d,e). Remarkably, G-CSF-induced HSPC egress in mice that consumed spicy food was significantly higher than those fed with standard chow (Fig.4i,j; Extended Data Fig.9f). This was confirmed by competitive repopulation assays of mobilized blood (Fig.4k,l). By contrast, capsaicin did not enhance HSC mobilization when nociceptors were depleted by RTX (Fig.4j), indicating that capsaicin diet most likely acted via nociceptors, although additional mechanisms remain possible. To determine whether the increased repopulation activity with capsaicin diet was due to the increased numbers or an increased engraftment capacity of HSCs, we carried out competitive repopulation experiments with sorted HSCs and found no difference in the repopulation activities between the capsaicin-treated and control groups (Extended Data Fig.9g-j). Similar results were obtained with sorted HSCs from mice stimulated with CGRP (Extended Data Fig.9k-m). These results suggest that the increased BM repopulation activity elicited by the capsaicin diet or CGRP administration was not due to enhanced HSC competitiveness but from a greater number of mobilized functional HSCs.
Discussion
Nociceptors allow rapid detection of external insults such as pain, cold, or heat to avoid organ damage8,10. While nociceptors are integral to the generation of an immune response protecting the body’s integrity, our results uncover an important role of nociceptor neurons in an organ isolated from the external environment where nerve-derived CGRP release regulates HSC maintenance and egress from the BM (Extended Data Fig.10). CGRP acts via CALCRL/RAMP1 expressed on HSPCs, leading to Gαs-mediated cAMP elevations that promote their egress. These results are in line with previous studies suggesting that Gαs promotes HSC mobilization via a then unidentified GPCR20. Of note, prostaglandin E2 (PGE2) interaction with the Gαs-coupled E-prostanoid receptor-4 (EP4) also induces cAMP elevations that regulate HSPC homing and mobilization21,22. PGE2 enhances homing in part by increasing CXCR4 expression on HSPCs, whereas the inhibition of EP4 signalling in stromal cells (not HSPCs) promotes HSPC egress21,22. By contrast, CGRP alters neither CXCR4 expression nor homing to bone marrow. The differing responses to cAMP elevation by different GPCRs may involve different cAMP-response element binding (CREB)-dependent or independent mechanisms23 and/or the activation of co-signalling pathways that may specify the response.
It is interesting to point out that G-CSF administration is commonly associated with bone pain, often reported after a single dose of G-CSF24, which is consistent with the present results suggesting that pain likely arises from nociceptive neuron activation rather than from the rapid haematopoietic cell expansion in the confined BM space. G-CSF has indeed been reported to activate directly receptors expressed on sensory neurons, release CGRP and cause pain25. Three antibody drugs targeting CGRP have recently been approved by the US Food and Drug administration and the European Medicines Agency to treat migraine26. Based on the present results, CGRP inhibition would interfere with HSC mobilization. On the other hand, TRPV1 receptor agonists have been formulated for pain relief27 and, if administered at relatively low doses, could substitute to the ingestion of the 10 Jalapeno peppers/day for 4 days predicted to enhance mobilization in humans. Enhancing signals from the nociceptive nervous system may thus provide new avenues toward improved HSC collection.
METHODS
Mice.
Nav1.8-Cre mice were a gift from Dr. John Wood (University College London). Nav1.8-Cre mice were bred with iDTA mice (from Jackson Laboratory) and iTdTomato mice (from Jackson Laboratory) to generate Nav1.8-Cre/iDTA nociceptor-deficient mice for functional studies and Nav1.8-Cre/iTdTomato mice for imaging experiments, respectively. Ramp1−/− mice were generated in the 129S6/SvEv background29. All experiments with Ramp1+/+ and Ramp1−/− mice were carried out using littermates by intercrossing Ramp1+/− heterozygous mice. Carcrlf/f mice were generated as previously described30 and were bred with Vav1-iCre mice from Jackson Laboratory to conditionally delete Calcrl in haematopoietic cells. C57BL/6 CD45.1 and CD45.2 congenic mice were purchased from the Jackson Laboratory. Unless indicated otherwise, eight- to ten-week old mice of both genders were used for experiments. All mice were maintained in pathogen-free conditions under a 12h:12h light/dark cycle, temperature 70 ± 2°F, humidity 40-70% and fed with autoclaved food. This study complied with all ethical regulations involving experiments with mice, and all experimental procedures performed on mice were approved by the Animal Care and Use Committee of Albert Einstein College of Medicine.
Drug and chemical treatments.
G-CSF was administered subcutaneously (s.c.) at a dose of 125 μg/kg twice a day (8 divided doses) beginning in the evening of the first day and blood was harvested 3h after the final morning dose. Newborn mice (2-4 day old) were s.c. injected with resiniferatoxin (RTX, Sigma-Aldrich, 50 μg/kg) and then three RTX escalating doses (30 μg/kg, 70 μg/kg and 100 μg/kg) were s.c. injected into 4-week old mice on 3 consecutive days. Control littermates were injected with vehicle solution on the same days. For sympathetic nerve disruption in adults, 2 doses of 6OHDA or vehicle, 100 mg/kg on day 0, 250 mg/kg on day 2, were injected i.p. and analyses were performed on day 5. For steady-state experiments, CGRP (Sigma-Aldrich), substance P (GenScript) and isoproterenol (Sigma-Aldrich) were dosed at 14 μg/day, 5 μg/day and 40 μg/day, respectively, using s.c. implanted Alzet osmotic pumps (model 1007D). For mobilization experiments, CGRP (Tocris) was delivered into mice at 2.4 μg/day using Alzet osmotic pumps (model 1007D). Control animals were implanted with Alzet pumps containing saline (PBS; Gibco). Forskolin (Cayman Chemical) was given i.p. at a dose of 2.5 mg/kg twice a day (8 divided doses) at the same time with G-CSF treatment. For in vivo depletion of macrophages, clodronate liposomes or PBS liposome as control (250 μL) were infused i.v. one week before analysis. For chemotherapy treatment, mice were injected with cisplatin (10 mg/kg once per week; Teva) for 5 weeks, and were recovered for 6-8 weeks before experiment. Desipramine (Tocris, 10 mg/kg i.p.) or saline (as control) treatment was started 4 days before G-CSF injection and continued throughout the experiment. For AMD3100-induced mobilization, mice were received a single dose of AMD3100 (Sigma, 5 mg/kg, i.p.) 3 hour before blood collection. C57BL/6 wild-type mice were fed with capsaicin containing diet (100 ppm, 100 mg/kg) or control diet from a week before G-CSF administration and until analysis.
Bone marrow transplantation assays.
Competitive repopulation assays were performed using the CD45.1/CD45.2 congenic system. CD45.1 recipient mice were lethally irradiated (12 Gy, two split doses) in a Cesium Mark 1 irradiator (JL Shepherd & Associates). For competitive repopulation assays, 0.5 x 106 donor BM cells (CD45.2) or 50 μL of mobilized blood (CD45.2) were transplanted together with 0.5 x 106 CD45.1 competitor BM cells into lethally irradiated CD45.1 recipients. For competitive HSC repopulation assays, 250 donor HSCs (CD45.2) were sorted from mobilized blood and transplanted together with 0.5 x 106 CD45.1 competitor BM cells into lethally irradiated CD45.1 recipients. CD45.1/CD45.2 chimerism of recipient blood and BM was analysed up to 16 or 20 weeks after transplantation by FACS. For RAMP1 reciprocal BM transplantation, 2 x 106 donor BM cells from Ramp1+/+ or Ramp1−/− mice were transplanted into lethally irradiated Ramp1+/+ or Ramp1−/− recipient mice (12 Gy, two split doses). For mixed chimera experiment, 106 CD45.1 BM cells from C57BL/6 wild-type mice were mixed with 106 CD45.2 BM cells (1:1 ratio) from Ramp1+/+ or Ramp1−/− mice and transplanted into lethally irradiated CD45.1 recipient mice (12 Gy, two split doses). For G-CSF-induced mobilization experiment with Vav1-iCre mice, 2 x 106 donor BM cells from Vav1-iCre+; Calcrlw/w or Vav1-iCre+; Calcrlf/f into lethally irradiated CD45.1 recipient mice (12 Gy, two split doses). G-CSF was administrated to induce mobilization in recipient mice at least 8 weeks post transplantation.
Bone marrow homing assay.
Donor BM cells were harvested from PBS-, RTX/6OHDA-treated mice (CD45.2), Ramp1+/+ or Ramp1−/− mice (CD45.2) and injected (5 x 106 cells) into lethally irradiated wild-type mice (CD45.1, 12 Gy, single dose). Sixteen hours after irradiation and injection, the recipient BM was harvested (femurs, tibiae, humeri and pelvis) to determine the number of homed donor HSCs and LSKs using FACS analysis. The homing efficiency was determined by dividing the absolute numbers of homed HSCs or LSK cells in BM by the absolute numbers of injected HSCs or LSKs (input).
CFU-C assay.
The haematopoietic progenitor colony-forming units in culture (CFU-C) assay was assayed by plating RBC-lysed peripheral blood into methylcellulose (Stem Cell Technologies, Cat#:3434). Cultures were plated in 35mm culture dishes and incubated for 7 days in humid chambers before colonies were scored.
Transwell migration assay.
Femurs, tibiae and humeri from C57BL/6 wild-type mice were flushed by 1mL of phosphate-buffered saline (PBS, Corning) in a syringe with a 21-gauge needle, erythrocytes were lysed, and lineage-positive cells were immunomagnetically depleted from the BM cells using a biotinylated lineage antibody cocktail (CD3e, B220, CD11b, Ter119 and Gr-1, at 1:100 dilution; BD Bioscience 559971) and subsequent streptavidin magnetic beads (Miltenyi Biotech 130-48-101). Lineage-depleted BM cell were transferred into the 6.5mm transwell plates (Corning 3421) containing alpha MEM (Gibco) supplemented with 10% FBS (Stem Cell Technologies) and the bottoms of transwells were filled with media containing 100 ng/mL CXCL12 (Peprotech). Transwell plates were incubated at 37°C for 3 h and migrated cells in the bottom wells were then plated in CFU-C as described above.
Flow cytometry and sorting.
Peripheral blood was harvested by retro-orbital bleeding of mice anesthetized with isoflurane and collected in polypropylene tubes containing EDTA. Blood parameters were determined with the Advia120 Hematology System (Siemens). BM cells were obtained by flushing and dissociating using a 1-mL syringe with phosphate-buffered saline (PBS, Corning) via a 21-gauge needle. For analysis of stromal and endothelial cell populations, intact flushed BM plugs were digested at 37°C for 30min in 1 mg/mL collagenase type IV (Gibco), 2 mg/mL Dispase (Gibco) and 500 μg/mL DNase I (Sigma-Aldrich) in Hank’s balanced salt solution (HBSS, Gibco). For FACS analysis and sorting, red blood cells were lysed and washed in ice-cold PEB (PBS containing 0.5% BSA and 2 mM EDTA) before staining with antibodies in PEB. The following antibodies were used for FACS: the anti-lineage panel cocktail (CD3e, B220, CD11b, Ter119 and Gr-1, at 1:100 dilution) was from BD Bioscience (559971), anti-Sca-1-FITC (D7; 11-5981-85), anti-CD117(c-Kit)-PE/Cy7 (2B8; 105814), anti-CD48-PerCP-eFluor710 (HM48-1; 46-0481-85), anti-CD150-PE (TC15-12F12.2; 115904), anti-B220-APC-eFluor780 (RA3-6B2; 47-0452-82), anti-CD3e-PerCP-Cy5.5 (145-2C11; 45-0031-82), anti-Gr-1-FITC (RB6-8C5; 11-5931-85), anti-CD11b-PE (M1/17; 12-0112-83), anti-CD45.1-PE/Cy (A20; 25-0453-82), anti-CD45.2-FITC (104; 109806), anti-Sca-1-APC (D7; 17-5981-83), anti-CD117-BV421 (2B8; 105828), anti-CD45-PerCP-Cy5.5 (30-F11, 45-0451-82), anti-Ter119-PerCP-Cy5.5 (TER-119; 45-5921-82), anti-CD31-PE/Cy7 (390; 25-0311-82), anti-CD51-PE (RMV-7; 12-0512-83), anti-PDGFRα(CD140α)-APC (APA5; 17-1401-81), anti-Gr-1-APC (RB6-8C5; 17-5931-82), anti-F4/80-PE (BM8; 123110), anti-CD115-PE/C7 (AFS98; 25-1152-82), streptavidin APC-eFluor 780 (47-4317-82), anti-CXCR4-AF647 (L276F12, 146504), anti-CD49d-APC (R1-2, 17-0492-82), anti-CD44-APC (IM7, 17-0441-82), all purchased from BioLegend or eBioscience. Unless otherwise specified, all antibodies were used at a 1:100 dilution. Cell cycle was analysed with Ki67 and Hoechst 33342 (Sigma). FACS analyses were carried out using BD LSRII flow cytometry (BD Biosciences) and cell sorting experiments were performed using a MoFlo Astrios (Beckman Coulter). Data were analysed with FlowJo 10.4.0 (LCC) and FACS Diva 6.1 software (BD Biosciences).
Immunofluorescence imaging.
Anti-CD31-Alexa Fluor 647 (MEC13.3; 102516; BioLegend) and anti-CD144 (VE-Cadherin)- Alexa Fluor 647 (BV13; 138006; BioLegend) (7.5 μg each) were injected intravenously to mice and mice were perfusion fixed with 4% paraformaldehyde (PFA) 10min after the injection. Bones were post-fixed with 4% PFA for another 15min at room temperature (RT). For cryopreservation, the bones were incubated sequentially in 15% sucrose/PBS at 4°C overnight and 30% sucrose/PBS at RT for 2h, and embedded and flash frozen in SCEM embedding medium (SECTION-LAB). Frozen sections were prepared 20μM thick with a Cryostat (CM3050, Leica) using Kawamoto’s tape transfer method. For immunofluorescent staining, sections were rinsed with PBS, post-fixed with 4% PFA for 30 min following by blocking with 20% donkey serum (Sigma) in 0.5% Triton X-100/PBS for 2h at RT. The following primary antibodies were used for nerve staining, anti-tyrosine hydroxylase (TH) antibody (AB152, Millipore), anti-CGRP (ab36001, Abcam) and anti-TUBB3-Alexa Fluor 488 (657404), used at 1:100 (TH and TUBB3) or 1:1000 (CGRP) dilution in 2% donkey serum with 0.1% Triton X-100/PBS for 48-72 hr at 4°C. Primary antibody staining was washed thrice with PBS, and incubated with secondary antibodies, used at 1:200 dilution in 2% donkey serum with 0.1% Triton X-100/PBS at RT for 1 h. Secondary antibodies: donkey anti-rabbit IgG BV421 (406410, BioLegend), donkey anti-goat IgG AF568 (A11057, Invitrogen), donkey anti-rabbit IgG AF488 (A21206, Invitrogen), donkey anti-goat IgG AF488 (A11055, Invitrogen). All images were acquired at room temperature using a Zeiss Axio examiner D1 microscope (Zeiss) with confocal scanner unit (Yokogawa). For quantification of nerve fibers, Z-stack images were stitched together and reconstructed using Slide Book Software 6.0 (Intelligent Imaging Innovations). Nerve densities were quantified by total length or total area (as indicated in the figure legends) of nerves fibres divided by the bone marrow area from 8-20 z-stack projections per mouse and 3-5 mice per group (each dot in the graph represents one mouse).
RNA isolation and quantitative real time PCR (q-PCR).
RNA was isolated as previously described using Dynabeads® mRNA DIRECT™ Purification Kit (61012, Invitrogen). Conventional reverse transcription, using the RNA to cDNA EcoDry Premix (639549, Takara), was performed in accordance with the manufacturer’s instructions. Q-PCR was performed with SYBR GREEN on QuantStudio 6 Flex Real-Time PCR System (Applied Biosystem, Thermo Fisher). The sequences of the oligonucleotides used can be provided upon reasonable request.
RNA-Seq analysis.
Total RNA from 2,000 sorted HSCs was extracted using the RNeasy Plus Micro kit (Qiagen), and assayed for integrity and purity using an Aligent 2100 Bioanalyzer (Agilent Technologies). RNA-seq data generated from Illumina Platform PE150 were processed using the following pipeline. Briefly, paired-end sequencing reads were aligned to the mouse genome using Spliced Transcripts Alignment to a Reference (STAR) v2.6.1. Alignments were parsed using Tophat program. HTSeq v0.6.1 was used to count the read numbers mapped of each gene and then FPKM (Fragments Per Kilobase of exon model per Million mapped reads) of each gene was calculated based on the length of the gene and reads count mapped to this gene. Differential expression analysis between two groups was performed using the DESeq2 R package, which provide statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting p values were adjusted using the Benjamini and Hochberg’s approach for controlling the False Discovery Rate (FDR).
ELISA.
Mouse CXCL12/SDF-1 alpha Quantikine ELISA Kit was purchased from R&D (Cat# MCX120) and used according to the manufacturer’s instructions. CGRP EIA Kit was purchased from Cayman Chemical (Cat# 589001) and used according to the manufacturer’s instructions. All BMEF used for ELISA were collected by flushing the BM of one or two femurs into 500 μL PBS and subsequently pelleting the cells by centrifugation. The resulting supernatant was removed from the cell pellet and frozen at −80°C for analysis.
Quantification and statistical analysis.
All data are presented as mean ± s.e.m. n represents mouse number in each experiment, as detailed in the figure legends. Statistical significance was determined by unpaired, two-tailed Student t test to compare two groups or one-way ANOVA for multiple group comparisons. Data statistical analyses and presentations were performed using GraphPad Prism 8 (GraphPad Software), FACS Diva 6.1 software (BD Biosciences, FlowJo 10.4.0 (LLC), Slide Book Software 6.0 (Intelligent Imaging Innovations), and QuantStudio 6 Real-Time PCR Software (Applied Biosystem, Thermo Fisher).
Extended Data
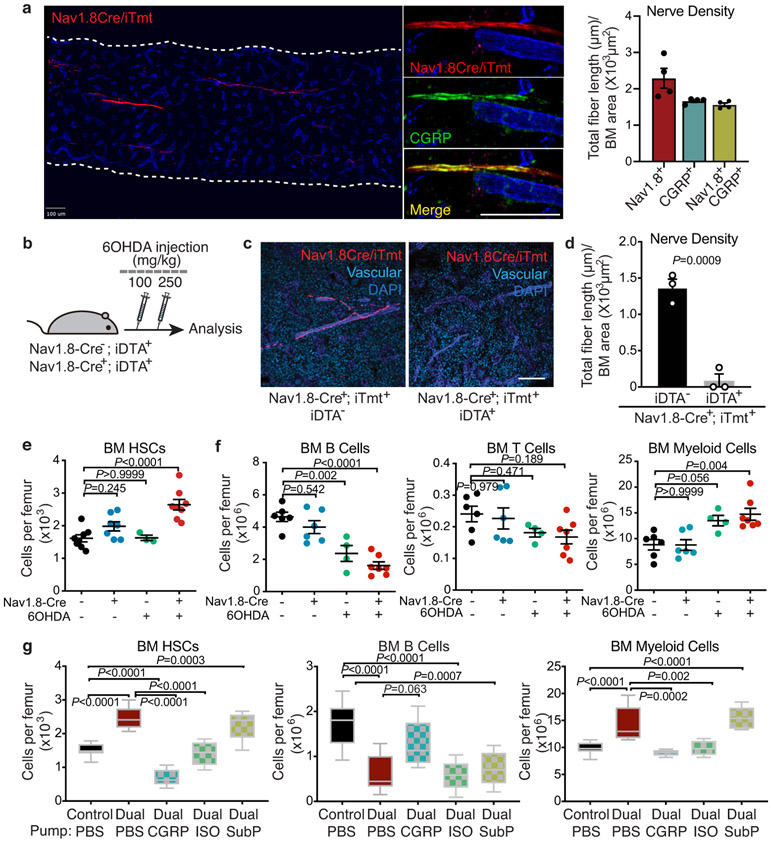
Extended Data Fig. 1. Characterization of nociceptive and sympathetic innervation in the bone marrow.
a, b, Representative confocal z-stack projection montages of C57BL/6 mouse femur stained for CGRP (nociceptive nerves), TH (sympathetic nerves), and TUBB3 (all peripheral nerves), CD31+ CD144+ double-positive vasculature. Scale bar, 100 μm. Femoral sensory and sympathetic innervation quantified by the total length of all CGRP+, TH+, or TUBB+ nerve fibres divided by the bone marrow area. n=3 mice. c, Schematic illustration of the pharmacological denervation experiment using RTX and 6OHDA. d, e, Representative images of confocal z-stack projections from femurs of control-, RTX-, 6OHDA-, dual-denervated mice stained for CGRP+ nociceptive nerve fibres, TH+ sympathetic nerve fibres, CD31+CD144+ blood vessels and DAPI. Scale bar, 100 μm. Femoral sensory and sympathetic innervation quantified by the total length of CGRP+ or TH+ nerves divided by total bone marrow area. Data from n=4, 5, 4, 4 mice, respectively. Error bars represent s.e.m. one-way ANOVA.
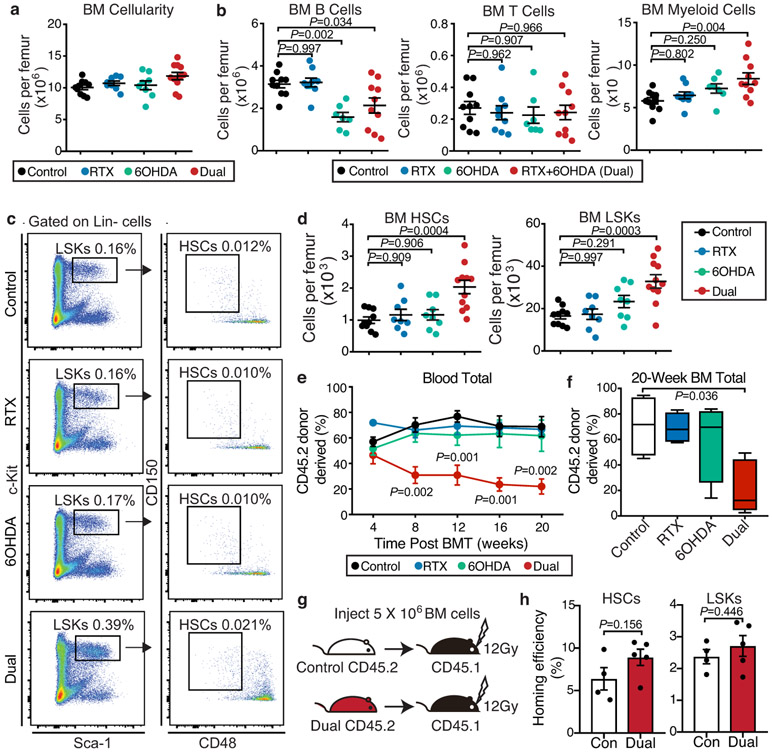
Extended Data Fig. 2. Nociceptive or sympathetic nerves are dispensable for HSC maintenance, whereas the depletion of both systems expands poorly functional HSCs.
a, b, BM cellularity and absolute numbers of B cells (B220+), T cells (CD3e+) and myeloid cells (Mac-1+) per femur from control, RTX, 6OHDA, or dual-denervated mice. n=10, 8, 8, 11 (a) and 10, 9, 7, 10 (b) mice, respectively. c, Representative FACS plots showing the gating strategy for Lin− Sca-1+ c-Kit+ CD150+ CD48− HSCs. The same gating strategy was used throughout. d, Absolute number of HSCs and LSK cells (Lin− Sca-1+ c-Kit+) per femur from control, RTX, 6OHDA, or dual-denervated mice. n=10, 8, 8, 11 mice, respectively. e, Peripheral blood chimerism (CD45.2+) in CD45.1-recipient mice transplanted with 0.5 x 106 CD45.1 competitor BM cells mixed with 0.5 x 106 donor BM cells (CD45.2) from control, RTX, 6OHDA, or dual-denervated mice at the indicated time points post-transplantation. n=4, 4, 4, 5 mice, respectively. f, Bone marrow chimerism (CD45.2+) 20 weeks after transplantation. g. Experimental design to determine the homing efficiency of HSCs and LSK cells (CD45.2) from control- or dual-denervated mice to the bone marrow of recipients (CD45.1). h, Homing efficiency of donor CD45.2 HSCs and LSK cells detected in the recipient bone marrow. n=4,5 mice, respectively. Error bars represent s.e.m. Two-tailed unpaired Student’s t test (h) or one-way ANOVA (a, b, d-f). For box plots, the box spans from the 25th to 75th percentiles and the centerline was plotted at the median. Whiskers represent minimum to maximum range.
Extended Data Fig. 3. Expansion of phenotypic HSCs following dual sympathetic and nociceptive denervation is normalized by administration of CGRP or a β-adrenergic agonist.
a, Representative confocal z-stack projections from femurs of Nav1.8-Cre+, iTdTomato+ mice stained for CD31+ CD144+ vasculature and CGRP+ nociceptive nerves. Nociceptive innervation quantified by the total length of all Nav1.8+ (red), CGRP+ (green), and Nav1.8+ CGRP+ double positive (yellow) nerves divided by the bone marrow area. n=4 mice. b, Schematic illustration of the dual denervation experiment with Nav1.8-Cre; iDTA mice. c, d, Representative images of confocal z-stack projections from femurs of Nav1.8-Cre+; iTdTomato+; iDTA− or Nav1.8-Cre+; iTdTomato+; iDTA+ mice stained for blood vessels (blue). Quantification of TdTomato+ nociceptive nerves in the femurs. n=3 mice per group. e, f, Absolute numbers of HSCs (Lin− Sca-1+ c-Kit+ CD150+ CD48−), B cells (B220+), T cells (CD3e+) and myeloid cells (Mac-1+) per femur from Nav1.8-Cre−; iDTA+ or Nav1.8-Cre+; iDTA+ mice with or without 6OHDA treatment. Each dot represents data from individual mice. n=8, 7, 3, 8 (e) and 6, 6, 4, 7 (f) mice, respectively. g, BM cellularity and absolute numbers of HSCs, B cells and myeloid cells per femur from control and dual-denervated mice implanted with osmotic pumps containing saline as control, CGRP, isoproterenol or substance P. n=14, 9, 5, 5, 5 mice respectively. Error bars represent s.e.m. Two-tailed unpaired Student’s t test (d) or one-way ANOVA (e-g). For box plots, the box spans from the 25th to 75th percentiles and the centerline was plotted at the median. Whiskers represent minimum to maximum range.
Extended Data Fig. 4. G-CSF-induced HSPC mobilization is impaired in mice lacking nociceptor neurons.
a, White blood cell counts and absolute numbers of LSK (Lin− Sca-1+ c-Kit+) cells per mL of peripheral blood following G-CSF mobilization in vehicle-treated control and RTX-treated mice. n=11, 13 mice, respectively. b, Spleen weight and the absolute numbers of LSK cells in the spleen following G-CSF mobilization in vehicle-treated control and RTX-treated mice n=11 mice per group. c, Bone marrow cellularity and the absolute numbers of LSK cells in the bone marrow following G-CSF-induced mobilization in vehicle-treated control and RTX-treated mice. n=11, 13 mice, respectively. d, Cell cycle analysis of BM HSCs (Lin− Sca-1+ c-Kit+ CD150+ CD48−) from control or RTX-treated animals determined by FACS using anti-Ki67 and Hoechst 33342 staining. n=6, 4 mice, respectively. e, White blood cell counts and absolute numbers of LSK cells per mL of peripheral blood following G-CSF-induced mobilization in Nav1.8-Cre−; iDTA+ and Nav1.8-Cre+; iDTA+. n=4, 5 mice, respectively. f, Bone marrow cellularity and the absolute numbers of LSK cells in the bone marrow following G-CSF-induced mobilization in Nav1.8-Cre−; iDTA+ and Nav1.8-Cre+; iDTA+. n=4, 5 mice, respectively. g, Peripheral blood B-cell (B220+CD45.2+), blood T-cell (CD3e+CD45.2+), and myeloid-cell (Mac-1+CD45.2+) donor chimerism in CD45.1-recipient mice transplanted with mobilized blood (CD45.2) derived from saline or RTX-treated mice mixed with CD45.1 competitor BM cells at the indicated time points post-transplantation. n=9,8 mice, respectively. h, Total bone marrow chimerism (CD45.2+) and bone marrow HSC chimerism (Lin− Sca-1+ c-Kit+ CD150+ CD48− CD45.2+) 20 weeks after transplantation. n=9, 8 mice, respectively. Error bars represent s.e.m. Two-tailed unpaired Student’s t test. For box plots, the box spans from the 25th to 75th percentiles and the centerline was plotted at the median. Whiskers represent minimum to maximum range.
Extended Data Fig. 5. The neuropeptide CGRP, but not substance P, promotes G-CSF-induced HSC mobilization.
a, Absolute numbers of HSCs (Lin− Sca-1+ c-Kit+ CD150+ CD48−) and LSK cells (Lin− Sca-1+ c-Kit+) per mL of peripheral blood following G-CSF-induced mobilization in C57BL/6 mice implanted with osmotic pumps containing saline or substance P. n=5 mice per group. b, Bone marrow cellularity and the absolute numbers of LSK cells and HSCs in the bone marrow following G-CSF administration in C57BL/6 mice implanted with osmotic pumps containing saline or substance P. n=5 mice per group. c, Bone marrow cellularity and the absolute numbers of HSCs per femur from mice described in Fig. 2a. n=18, 9, 7, 7 mice, respectively. d, Experimental design to determine the effect of CGRP on HSC mobilization of 6OHDA-denervated mice (left). Absolute number of HSCs per mL of peripheral blood following G-CSF administration in saline-or 6OHDA- treated C57BL/6 mice implanted with osmotic pumps containing saline or CGRP. n=6 mice per group. Error bars represent s.e.m. Two-tailed unpaired Student’s t test.
Extended Data Fig. 6. Ramp1-deficient mice exhibit no haematopoietic defect at steady state.
a, Ramp1 mRNA expression levels determined by quantitative PCR in total bone marrow cells derived from Ramp1+/+ or Ramp1−/− mice. n=5 biological sample. b, c, White blood cell counts, and the absolute numbers of B cells (B220+), T cell (CD3e+) and myeloid cells (Mac-1+) per mL of peripheral blood from Ramp1+/+ or Ramp1−/− mice at steady state. n=5 mice per group. d, e, Bone marrow cellularity, and the absolute numbers of LSK (Lin− Sca-1+ c-Kit +) cells, B cells (B220+), T cell (CD3e+) and myeloid cells (Mac-1+) per femur from Ramp1+/+ or Ramp1−/− mice at steady state. n=5 mice per group. f, Experimental design to determine the homing efficiency of HSCs (Lin− Sca-1+ c-Kit+ CD150+ CD48−) and LSK cells from Ramp1+/+ and Ramp1−/− mice (CD45.2) to the bone marrow of lethally irradiated recipients (CD45.1) (left panel). Percentage of donor CD45.2+ HSCs and LSK cells detected in the recipient bone marrow (right panel). n=5 mice per group. g, BM cellularity and the absolute number of HSCs in the BM. n=3, 4, 3, 3 mice, respectively. Error bars represent s.e.m. Two-tailed unpaired Student’s t test (a-f) or one-way ANOVA (g).
Extended Data Fig. 7. Nociceptive nerve-deficient mice exhibit no alteration of HSC niche components following G-CSF treatment.
a, Absolute numbers of MSCs (CD45− Ter119− CD31− CD51+ PDGFRα+), ECs (CD45− Ter119− CD31high), and macrophages (Gr-1− F4/80+ CD115int SSCint/lo) per femur from saline- or RTX-treated mice following G-CSF treatment. n=4, 4 (left and middle), 6, 8 (right) mice, respectively. b, qRT-PCR quantification of Cxcl12 mRNA levels in total bone marrow cells, sorted MSCs and ECs from saline- or RTX-treated mice following G-CSF treatment. n=5 mice per group. c, Bone marrow extracellular fluid (BMEF) CXCL12 levels measured by ELISA. n=3, 5, 4 (left), 3, 4, 5 (middle), 4, 5 (right) mice, respectively. d. Mean fluorescence intensities (MFI) in the expression of CXCR4, VLA4 and CD44 on HSCs (Lin− Sca-1+ c-Kit+ CD150+ CD48−). n=4 mice per group Error bars represent s.e.m. Two-tailed unpaired Student’s t test (a, b, d) or one-way ANOVA (c).
Extended Data Fig. 8. Transcriptome analysis by RNA-seq of HSCs from Ramp1+/+ and Ramp1−/− mice.
a, Heatmap shows differentially expressed genes between wild-type- and Ramp1−/−-sorted HSCs (adjusted P value <0.05). b, Gene set enrichment analyses showing up-regulated and down-regulated pathways in Ramp1−/− HSCs compared to wild-type HSCs (P<0.01, n=3 biological replicates per group). c, Gene set enrichment analyses showing significant alterations of gene sets involved in the Gαs/AC/cAMP pathway. d, e, Schematic illustration of the dual stimulation experiment (d). Absolute number of HSCs (Lin− Sca-1+ c-Kit+ CD150+ CD48−) in the mobilized peripheral blood of mice treated with control saline, desipramine (DES), CGRP, or both DES and CGRP (e). n=9, 4, 8, 4 mice, respectively. Error bars represent s.e.m. One-way ANOVA.
Extended Data Fig. 9. Spicy food ingestion enhances HSC mobilization.
a, Scoville heat scale for chili peppers. 100 ppm=100 mg/kg. b. Three pellets of standard chow (brown) and spicy chow (red) were provided to 2 mice (6:00 pm on day 1). Sixteen hours later (10:00 am on day 2), two of the three standard pellets were consumed while spicy food pellets remained untouched. c, Daily food intake and the body weight of mice fed with standard or capsaicin-containing diet. n=5 (left), 4 mice (right) per group. d, CGRP levels in the BMEF from mice fed with control- or capsaicin- diet. n=9 mice per group. e, Absolute numbers of HSCs (Lin− Sca-1+ c-Kit+ CD150+ CD48−) in the bone marrow following G-CSF-induced mobilization in mice fed with standard or capsaicin-containing chow (left). n=6, 7 mice, respectively. Cell cycle analysis of BM HSCs was determined by FACS using anti-Ki67 and Hoechst 33342 staining. n=6 mice. f, White blood cell counts and absolute numbers of LSK cells (Lin− Sca-1+ c-Kit+) per mL of peripheral blood following G-CSF-induced mobilization in mice fed with standard or capsaicin-containing diet. n=6, 7 mice, respectively. g, i, Peripheral total blood donor chimerism (CD45.2+) in CD45.1-recipient mice transplanted with 0.5 x 106 CD45.1 competitor BM cells and 250 HSCs sorted from mobilized blood (g) or BM (i) from mice fed with standard or capsaicin chow. n=9, 8 (g), 8, 8 (i) mice, respectively. h, j. Peripheral blood B cell (B220+CD45.2+), T cell (CD3e+CD45.2+) and myeloid cell (Mac-1+CD45.2+) donor chimerism in CD45.1-recipient mice transplanted with 250 blood HSCs (h) or BM HSCs (j) with competitor bone marrow cells at 16 weeks post-transplantation. n=9, 8 (h), 8, 8 (j) mice, respectively. k, Experimental design to determine the effects of CGRP administration on HSC competitiveness. l, Peripheral total blood donor chimerism (CD45.2+) in CD45.1-recipient mice transplanted with 0.5 x 106 CD45.1 competitor BM cells and 250 HSCs sorted from mobilized blood from PBS- or CGRP- treated mice. n=7, 9 mice, respectively. m, Peripheral blood B cell, T cell and myeloid cell donor chimerism in CD45.1-recipient mice transplanted with 250 blood HSCs with competitor bone marrow cells at 16 weeks post-transplantation. n=7, 9 mice, respectively. Error bars represent s.e.m. Two-tailed unpaired Student’s t test. For box plots, the box spans from the 25th to 75th percentiles and the centerline was plotted at the median. Whiskers represent minimum to maximum range.
Extended Data Fig. 10. Nociceptor-mediated HSC mobilization.
Schematic representation showing that nociceptive nerve-derived CGRP, but not substance P, acts via CGRP receptors on HSCs to enhance their mobilization via cAMP-medicated signalling pathway.
Acknowledgements
We thank Dr. John Wood for providing Nav1.8-Cre mice. We are grateful to C. Prophete for technical assistance, Drs A. Birbrair and N. Asada for advice with the initial experiments, and Dr. D. Sun from Human Stem Cell FACS and Xenotransplantation Facility for assistance with cell sorting. This work was supported by U01 and R01 grants from the National Institutes of Health (NIH) (DK116312, DK056638, DK112976 and to P.S.F). HHL is the recipient of a F32 Ruth L. Kirschstein Postdoctoral Individual National Research Service Award (HL142243-01).
Footnotes
Competing interests P.S.F. serves as consultant for Pfizer, has received research funding from Ironwood Pharmaceuticals and is shareholder of Cygnal Therapeutics. The rest of the authors declare no competing interests.
Data availability.
RNA sequencing data have been deposited in the Gene Expression Omnibus under accession number GSE156449 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE156449). Source data behind Figures 1-4 and Extended Data Figures 1-10 are available within the manuscript files.
REFERENCES
- 1.Gao X, Xu C, Asada N & Frenette PS The hematopoietic stem cell niche: from embryo to adult. Development 145, doi: 10.1242/dev.139691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoggatt J, Kfoury Y & Scadden DT Hematopoietic Stem Cell Niche in Health and Disease. Annu Rev Pathol 11, 555–581, doi: 10.1146/annurev-pathol-012615-044414 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Katayama Y et al. Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell 124, 407–421, doi: 10.1016/j.cell.2005.10.041 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Lucas D et al. Chemotherapy-induced bone marrow nerve injury impairs hematopoietic regeneration. Nat Med 19, 695–703, doi: 10.1038/nm.3155 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maryanovich M et al. Adrenergic nerve degeneration in bone marrow drives aging of the hematopoietic stem cell niche. Nat Med 24, 782–791, doi: 10.1038/s41591-018-0030-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mendez-Ferrer S et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 466, 829–834, doi: 10.1038/nature09262 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bensinger W, DiPersio JF & McCarty JM Improving stem cell mobilization strategies: future directions. Bone Marrow Transplant 43, 181–195, doi: 10.1038/bmt.2008.410 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Ordovas-Montanes J et al. The Regulation of Immunological Processes by Peripheral Neurons in Homeostasis and Disease. Trends Immunol 36, 578–604, doi: 10.1016/j.it.2015.08.007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavlov VA, Chavan SS & Tracey KJ Molecular and Functional Neuroscience in Immunity. Annual Review of Immunology 36, 783–812, doi: 10.1146/annurev-immunol-042617-053158 PMID - 29677475 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinho-Ribeiro FA, Verri WA & Chiu IM Nociceptor Sensory Neuron–Immune Interactions in Pain and Inflammation. Trends in Immunology 38, 5–19, doi: 10.1016/j.it.2016.10.001 PMID - 27793571 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsunokuma N et al. Depletion of Neural Crest-Derived Cells Leads to Reduction in Plasma Noradrenaline and Alters B Lymphopoiesis. J Immunol 198, 156–169, doi: 10.4049/jimmunol.1502592 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Suekane A et al. CGRP-CRLR/RAMP1 signal is important for stress-induced hematopoiesis. Sci Rep 9, 429, doi: 10.1038/s41598-018-36796-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chow A et al. Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J Exp Med 208, 261–271, doi: 10.1084/jem.20101688 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christopher MJ, Rao M, Liu F, Woloszynek JR & Link DC Expression of the G-CSF receptor in monocytic cells is sufficient to mediate hematopoietic progenitor mobilization by G-CSF in mice. J Exp Med 208, 251–260, doi: 10.1084/jem.20101700 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winkler IG et al. Bone marrow macrophages maintain hematopoietic stem cell (HSC) niches and their depletion mobilizes HSCs. Blood 116, 4815–4828, doi: 10.1182/blood-2009-11-253534 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Broxmeyer HE et al. Rapid mobilization of murine and human hematopoietic stem and progenitor cells with AMD3100, a CXCR4 antagonist. J Exp Med 201, 1307–1318, doi: 10.1084/jem.20041385 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucas D et al. Norepinephrine reuptake inhibition promotes mobilization in mice: potential impact to rescue low stem cell yields. Blood 119, 3962–3965, doi: 10.1182/blood-2011-07-367102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shastri A et al. Stimulation of adrenergic activity by desipramine enhances hematopoietic stem and progenitor cell mobilization along with G-CSF in multiple myeloma: A pilot study. Am J Hematol 92, 1047–1051, doi: 10.1002/ajh.24843 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Ford CD, Green W, Warenski S & Petersen FB Effect of prior chemotherapy on hematopoietic stem cell mobilization. Bone Marrow Transplant 33, 901–905, doi: 10.1038/sj.bmt.1704455 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Adams GB et al. Haematopoietic stem cells depend on Galpha(s)-mediated signalling to engraft bone marrow. Nature 459, 103–107, doi: 10.1038/nature07859 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagedorn EJ, Durand EM, Fast EM & Zon LI Getting more for your marrow: Boosting hematopoietic stem cell numbers with PGE2. Exp Cell Res 329, 220–226, doi: 10.1016/j.yexcr.2014.07.030 PMID - 25094063 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoggatt J et al. Differential stem- and progenitor-cell trafficking by prostaglandin E2. Nature 495, 365–369, doi: 10.1038/nature11929 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sands WA & Palmer TM Regulating gene transcription in response to cyclic AMP elevation. Cell Signal 20, 460–466, doi: 10.1016/j.cellsig.2007.10.005 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Pulsipher MA et al. Adverse events among 2408 unrelated donors of peripheral blood stem cells: results of a prospective trial from the National Marrow Donor Program. Blood 113, 3604–3611, doi: 10.1182/blood-2008-08-175323 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schweizerhof M et al. Hematopoietic colony-stimulating factors mediate tumor-nerve interactions and bone cancer pain. Nat Med 15, 802–807, doi: 10.1038/nm.1976 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Charles A & Pozo-Rosich P Targeting calcitonin gene-related peptide: a new era in migraine therapy. Lancet, doi: 10.1016/S0140-6736(19)32504-8 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Cui M, Gosu V, Basith S, Hong S & Choi S Polymodal Transient Receptor Potential Vanilloid Type 1 Nocisensor: Structure, Modulators, and Therapeutic Applications. Adv Protein Chem Struct Biol 104, 81–125, doi: 10.1016/bs.apcsb.2015.11.005 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Asada N et al. Differential cytokine contributions of perivascular haematopoietic stem cell niches. Nat Cell Biol 19, 214–223, doi: 10.1038/ncb3475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li M et al. Deficiency of RAMP1 attenuates antigen-induced airway hyperresponsiveness in mice. PLoS One 9, e102356, doi: 10.1371/journal.pone.0102356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fritz-Six KL, Dunworth WP, Li M & Caron KM Adrenomedullin signaling is necessary for murine lymphatic vascular development. J Clin Invest 118, 40–50, doi: 10.1172/JCI33302 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA sequencing data have been deposited in the Gene Expression Omnibus under accession number GSE156449 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE156449). Source data behind Figures 1-4 and Extended Data Figures 1-10 are available within the manuscript files.