Abstract
The first important step in a structure-based virtual screening is the judicious selection of a receptor protein. In cases where holo protein receptor structure is unavailable, significant reduction in virtual screening performance has been reported. In this work, we present a robust method to generate reliable holo protein structure conformations from apo structures using molecular dynamics simulation with restraints derived from holo structure binding site templates. We perform benchmark tests on two different datasets: 40 structures from a directory of useful decoy-enhanced (DUD-E) and 84 structures from Gunasekaran dataset. Our results show successful refinement of apo binding site structures toward holo conformations in 82% of the test cases. In addition, virtual screening performance on 40 DUD-E structures are significantly improved using our MD-refined structures as receptors with an average enrichment factor EF1% value of 6.2 compared to apo structures with 3.5. Docking of native ligands to the refined structures shows an average ligand RMSD of 1.97 Å (DUD-E dataset and Gunasekaran dataset) relative to ligands in the holo crystal structures, which is comparable to the self-docking (i.e., docking of native ligand back to its crystal structure receptor) average, 1.34 Å (DUD-E dataset) and 1.36 Å (Gunasekaran dataset). On the other hand, docking to the apo structures yields an average ligand RMSD of 3.65 Å (DUD-E) and 2.90 Å (Gunasekaran). These results indicate that our method is robust and can be useful to improve virtual screening performance in apo structures.
Graphical Abstract

INTRODUCTION
Protein-ligand binding is facilitated by physicochemical interactions between binding site residues of the protein and the ligand for specificity and/or relay of communication between active and allosteric sites. These interactions tend to change the shape of the binding pocket going from apo (ligand unbound) to holo (ligand bound) structure of a protein. The extent to which apo structures are different from holo structures have been explored systematically by many research groups. In 2005, Gutteridge and Thornton analyzed 60 enzymes and showed that apo enzymes were structurally different compared to holo enzymes.1 But, the majority of enzymes had small Cα root mean square deviation (RMSD) of ≤ 1 Å between apo and holo structures. In 2007, Gunasekaran and Nussinov analyzed 98 apo-holo pairs that were categorized into rigid (≤ 0.5 Å), moderate (0.5 Å ≤ and ≤ 2.0 Å), and flexible (> 2 Å), based on apo-holo Cα RMSD differences.2 They showed that rigid proteins had more polar-polar interactions at the binding site, whereas flexible proteins preferred hydrophobic interactions. In 2008, Brylinski and Skolnick analyzed a larger set of 521 apo-holo protein pairs and showed that 80% had RMSD of ≤ 1 Å.3 Recently, Clark et al analyzed over 4,000 crystal structures (of 305 proteins) and reported that induced backbone changes across apo-holo pairs are generally small and within 0.5 Å.4 However, they pointed out that larger differences are seen when analyzing sidechain orientations across apo-holo pairs.
Investigations on sidechain orientations across apo-holo pairs reveal more flexibility at the binding site, which is usually missing in typical analysis where only backbones are considered. In 1999, Heringa and Argos reported that binding site side chains underwent rotameric changes upon ligand binding.5 Gaudreault et al further explored sidechain rearrangements upon ligand binding using 188 apo-holo structure pairs and found that 90% of them had at least one residue with significant rotameric change.6 Recently, we have shown that sidechain orientations (chemical feature points) are more informative than backbone conformations in determining protein-carbohydrate interactions.7 The general consensus across analysis from several research groups is that backbones tend to undergo small conformational changes, but side chains sample larger conformational space upon ligand binding.
It is widely accepted that protein structure with a holo conformation is the suitable target structure for ligand docking. Due to the differences of the binding site structures between apo and holo structures, apo structures are seldom used for docking. McGovern and Shoichet analyzed docking results of 95,000 ligands to 10 protein targets and showed that holo structures were significantly better than their apo counterparts in discriminating binders from nonbinders.8 They described that most apo structures had binding site conformations that were inadequate for ligand binding, because of incorrect sidechain orientations or loops that blocked proper docking. Similarly, Lee et al analyzed docking results of 8 receptors and showed that their enrichment levels were significantly lowered in apo structures compared to their holo counterparts.9
In order to solve this problem, Zavodszky and Kuhn introduced flexible SLIDE docking to 20 proteins having negligible apo and holo backbone differences.10 They reported that while rigid docking failed in half of the cases, flexible docking by manipulating protein’s sidechain orientations of the apo structures was able to dock all ligands within 2.5 Å of their crystal structure pose. Incorrect sidechain rotations of 60° or greater were shown to be detrimental in rigid dockings of apo structures in many instances.10-12 While flexible docking can be a solution to this docking problem, some apo proteins with significant backbone differences to their holo counterparts remain a problem. In addition, judicious choice of flexible residues is challenging and can complicate the docking process. Another approach that considers receptor flexibility is ensemble docking, where molecular dynamics (MD) simulations are used to obtain different receptor conformations for docking.13, 14 Ogrizek and colleagues showed that short MD simulations helped obtained relevant receptor conformations that improved ligand enrichment in docking.15 Physics-based MD simulation provides different conformational states of the binding site that can facilitate rigid docking. At the same time, this approach lacks direction or specificity to obtain relevant holo conformations for docking. To solve this problem, we have developed a robust method that uses MD simulation with restraints derived from binding site templates to get holo conformations from apo structures. Predicted binding site templates are in their holo states, and they are obtained using our local structure alignment tool, G-LoSA (graph-based local structure alignment, https://compbio.lehigh.edu/GLoSA).16-19 We have successfully conducted ligand-binding-site structure refinement of modeled or predicted structures from Astex dataset using this approach.20 Our method selected proper binding site templates to derive Cα-distance restraint potentials for MD simulations and reported consistently better refinement of 37 out of 40 targets with an average Cα RMSD improvement of 0.9 Å.
In this work, motivated by our success in ligand-binding-site structure refinement, we extend its application to generate holo structures from 40 apo structures in DUD-E (a directory of useful decoys, enhanced) dataset and 84 apo structures in Gunasekaran dataset.2, 21 Recognizing previous findings that apo-holo pairs tend to have similar backbone conformations but different sidechain orientations, we add sidechain center-of-mass (SC-COM) distance restraints to properly direct their orientations during MD simulations. Our results show consistently better refinement of apo binding site structures toward holo conformations with all-heavy-atom average RMSD improvement of 0.63 Å for DUD-E dataset and 0.52 Å for Gunasekaran dataset. In addition, virtual screening on 40 DUD-E targets are consistently improved in refined structures with enrichment factors EF1% value of 6.2 compared to apo structures with 3.5. Docking of native ligands to both the refined and initial apo structures also shows consistent improvements in binding modes. An average ligand RMSD relative to ligands in the holo crystal structures improves by 1.70 Å (DUD-E) and 0.94 Å (Gunasekaran dataset), respectively. Together, these results indicate a robust method of generating reliable holo structures from apo structures that can be useful to improve structure-based virtual screening docking accuracy for apo structures.
METHODS
In our workflow (Figure S1), an input is an apo structure on which G-LoSA search is performed to obtain binding site templates, to derive Cα and SC-COM distance restraint potentials, and to run restrained MD simulations. This workflow is similar in nature to our previously published ligand-binding-site structure refinement method.20 In this work, for benchmark test, we selected apo structures from two different datasets, DUD-E and Gunasekaran datasets.2, 21 The DUD-E dataset consists of 102 holo protein target with bound ligands that have pharmacological precedence. For each holo structure, we searched for its apo counterpart in the Protein Data Bank (PDB) with the same sequence identity and without a ligand at the binding site.22 Gunasekaran dataset consists of apo-holo pairs, and we simply used the apo structures for our test. Transmembrane proteins were excluded from our test. We only kept one apo structure when a protein happened to be in both datasets. Altogether, our benchmark datasets consisted of 40 structures from DUD-E and 84 structures from Gunasekaran dataset (Table S1, S2).
The binding site residues are defined as protein residues that are within 4.5 Å of the bound ligand in the holo crystal structures. Following classification criteria used in Gunasekaran dataset, we first calculated the apo-holo RMSD using the ligand-binding-site residues and classified our test cases into 3 groups: group 1 having negligible backbone conformational change with Cα RMSD of < 0.5 Å; group 2 with intermediate backbone motions where Cα RMSD is between 0.5 Å < and ≤ 1.5 Å; and group 3 having large backbone movements with Cα RMSD of > 1.5 Å (Table S1, S2).
G-LoSA is a robust local structure alignment tool that aligns protein local structures based on their geometry and physicochemical features in a sequence order independent manner.16-19 A comparative performance evaluation study by Govindaraj et al showed that G-LoSA outperformed other widely used local structure alignment tools.23 We ran G-LoSA search on each apo structure in our datasets to predict its binding site and select the appropriate binding site template. Through G-LoSA search, all available binding site templates in our PDB library were aligned onto each apo query protein (without prior knowledge of its binding site) and the templates were ranked based on their GA-scores. The GA-score is a normalized chemical feature-based and size-independent structure similarity score that ranges between 0 and 1, where 1 represents a perfect alignment. GA-score for all templates are provided in Table S1 and S2. Templates with more than 10 residues and 0.6 GA-scores were selected. Small templates with less than 10 binding residues have been suggested to contain invalid ligands or crystal reagents that have no biological relevance.24
Using the shortest augmenting path algorithm to solve the linear sum assignment problem, we identified equivalent aligned residues.25 One top template was selected for each query apo structure (Table S1, S2). From this template and the identified equivalent aligned residues, we derived two different distance restraint potentials applied to each apo structure: method 1 using only Cα distance restraints and method 2 using both Cα and SC-COM distance restraints. We included SC-COM distance restraints because previously published analysis of apo-holo pairs has revealed that sidechain orientations showed more conformational variations than the backbones and the changes in sidechain conformations directly influenced docking results.4-6,10-12
In method 1, we calculated a distance matrix (M) between Cα atoms of the selected template residues that were equivalent to the query binding site residues, and derived a harmonic distance restraint potential for the query protein using equation 1. For method 2, in addition to the Cα distance restraints, we also calculated each residue’s SC-COM and obtained a distance matrix (M) between SC-COM points to derive a similar harmonic distance restraint potential using equation 1.
| (1) |
where k is the force constant, rij is the distance between ith and jth Cα atoms in the target protein, and r0,ij is the distance between the equivalent atoms in the template. K was set to 1.5 kcal/(mol·Å2) for method 1 and 2, based on parameter optimization conducted in our previous paper.20 In addition, we added weak positional restraints (with a force constant of 0.5 kcal/(mol·Å2)) to Cα atoms of residues that were not part of the binding sites. We have shown previously that this approach is effective in preventing the overall protein structure from drifting from the query structure.20
For each target apo structure, we performed all-atom MD simulations in explicit solvents using its distance restraint potentials from method 1 or method 2. Simulation inputs were prepared using CHARMM-GUI Solution Builder.26, 27 Each apo structure was solvated in a cubic box with TIP3P water models with 10 Å padding in each direction.28 Using short 2,000 steps of Monte Carlo simulations for ion placement, each system was neutralized with Na+ and Cl− ions. We then applied periodic boundary conditions in the NPT (constant particle number, pressure, and temperature) ensemble using Langevin thermostat.29 Electrostatic interactions were handled by the particle-mesh Ewald summation and the force-based switching was used between 10 and 12 Å to truncate van der Waals interactions.30, 31 Covalently bound hydrogen atoms were constrained using the SHAKE algorithm.32 All simulations used the CHARMM36m force field.33 The systems were minimized using the steepest descent method for 5,000 steps followed by 1-ns equilibration. We conducted 3 x 50-ns production runs (with method 1), 3 x 50-ns production runs (with method 2), and 3 x 50-ns production runs (without distance restraints) as a control group. Each replica started with the same apo structure but using different initial velocity random seeds. All MD simulations were carried out using the OpenMM package.34
The final refined structure for each target is the average structure from the final frames of three replicas. This structure was resolvated in a TIP3P water box and neutralized using Na+ and Cl− ions and then followed by short minimization using the steepest descent method for 5,000 steps and 25-ps equilibration to remove potentially distorted geometries from structural averaging. Structure averaging had been shown to dampen divergence and amplifying improvements when used in structure refinement protocols.35
For visualization of binding pockets, we used surface representations with electrostatic potentials calculated using PBEQ Solver (Poisson-Boltzmann equation solver) in CHARMM-GUI.26, 36, 37 The native ligands were overlaid onto the holo, apo, and refined structures to assess structural changes at the binding site.
In order to test the usefulness of our MD-refined apo structures, we ran virtual screening using AutoDock Vina.38 For the 40 receptor structures from DUD-E, we retrieved their active and decoy ligands from DUD-E database.21 The number of active ligands were 10,308 and decoys were 616,619 ligands, which showed about 60 decoys per active ligand. For each target, virtual screening was conducted on all three types of receptors, apo, holo, and refined structures. Receptors and ligands were prepared using default vina protocols.38 Search space was determined to be a cubic grid with an edge dimension of 25 Å. The center of the mass of the crystal ligand was chosen as the center of the box. Exhaustiveness was set to 8 and only one output (the top scoring) of binding pose/binding energy per ligand was considered. Evaluation metric that we used for virtual screening results was enrichment factor (EF) at 1% and 2%. EF measures the concentration of active ligands among the x% ranked compounds as compared to active compounds concentration in the entire database.39 The enrichment value is computed as following:
| (2) |
Additionally, we evaluated the binding pose of native ligands when docked into all three receptor types, holo, apo and refined structures. The holo structures from DUD-E and Gunasekaran datasets were used as control for self-docking (i.e., docking of native ligand back to its crystal structure receptor). Receptor structures were rigid during docking. To avoid binding pose bias, the ligand conformations were randomized prior to docking. Each search space was located at the binding site based on the coordinate of the native ligand from the holo crystal structure. We added 5.0 Å padding in each direction from the ligand to ensure that ligand docking is close to the binding site. Using the same parameters, we docked the native ligand to the initial apo structures and the refined holo structures. To compare ligand binding modes, we aligned the protein binding site residues and calculated ligand RMSD (without aligning the ligands) relative to ligands in the holo crystal structures.
RESULTS AND DISCUSSION
G-LoSA search yields proper binding site templates for MD simulations with restraints
For all 124 apo structures in DUD-E and Gunasekaran datasets, we ran local structure alignments using G-LoSA to obtain a top template as described in Methods. The apo structures and their holo counterparts, as well as selected binding site template PDB IDs are listed in Table S1 and S2. Using our method, we excluded homologous proteins in our library whose sequence identity is >30% to our benchmark target protein. To assess similarities between our selected G-LoSA templates and the apo target structures, we ran sequence identity and TM-score calculations.40-42 Our data show that the average sequence identity between apo and template proteins is 25% (DUD-E) and 25% (Gunasekaran) (Table S1, S2). Average TM-scores are 0.56 (DUD-E) and 0.48 (Gunasekaran), indicating that many template structures do not have very similar global folds to the query structures. Because G-LoSA functions in a sequence order independent way at the local structure level (i.e., the binding site), the template selection from similar protein is not necessarily the case for any given target protein. Additionally, 53/124 (43%) targets have TM-scores less than 0.5, indicating that the two protein structures do not have similar global folds either (Table S1, S2).42
Our test cases were classified into 3 groups based on apo-holo Cα RMSD of the ligand-binding-site residues (Table S1, S2): group 1 (RMSD of < 0.5 Å), group 2 (0.5 Å < RMSD ≤ 1.5 Å), and group 3 (RMSD > 1.5 Å). Dissimilar structures between the target proteins and the selected templates can be found in every group. Specifically, there are 44% (15/43 in group 1), 48% (20/61 in group 2), 25% (6/20 in group 3) structures with different global folds.
Overall, we demonstrate that our method of identifying binding site template structures is robust, because it selects proper templates without having to necessarily rely on similar sequence and structure to the query protein. Identification of appropriate template binding site is a critical step in our workflow to derive restraint potentials for MD simulations (Figure S1).
Adding sidechain center-of-mass distance restraints improves sidechain orientations
Following the optimized parameters for distance restraints described in our previous paper (see Methods), we performed all-atom MD simulations of 40 DUD-E apo proteins to generate holo protein structures.20 For each target, we conducted three independent 50-ns simulations with Cα distance restraints (method 1), Cα and SC-COM distance restraints (method 2), or without distance restraints (control group).
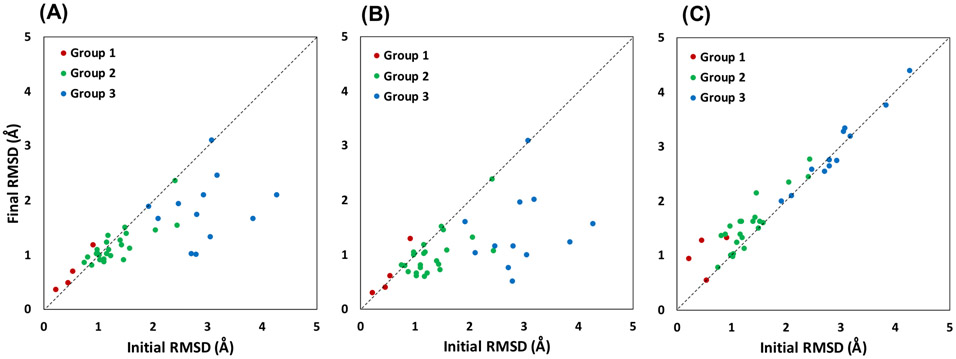
Our overall results of DUD-E proteins show consistently successful generation of holo conformations for 30 out of 40 apo structures (Table S3, Figure 1). In Gunasekaran set, 72 out of 84 structures undergo improvement toward their holo structure conformations (Table S4, Figure 2). Our assessment is based on all-heavy-atom RMSD of binding site residues relative to the PDB holo structure counterparts. For 40 DUD-E proteins, using method 1 an average RMSD change from the apo structure toward their holo conformation is 0.40 Å (initial 1.71 Å, final 1.31 Å), and the method 2 shows an average RMSD change of 0.63 Å (initial 1.71 to final 1.09 Å). Similarly, in Gunasekaran dataset, method 2 shows better improvement of 0.52 Å as compared to method 1 with 0.29 Å average improvement. Clearly, the addition of SC-COM distance restraints improves sidechain orientations toward the correct holo conformations. In addition, our results indicate that successful refinements originate from the distance restraints, because the control group simulations without distance restraints show worse results (Table S3, S4, Figure1C, 2C).
Figure 1.
Ligand-binding-site all-heavy-atom RMSD values of the initial apo structures and MD-derived structures against their holo crystal structures for 40 targets in DUD-E dataset. (A) Method 1 using Cα distance restraints. The average improvement is 0.40 Å. (B) Method 2 using Cα and SC-COM distance restraints. The average improvement is 0.63 Å. (C) Control group without distance restraints and no improvement in RMSD. The structures are separated into three groups based on their initial binding site Cα RMSD compared to their holo structures: group 1 (< 0.5 Å, red), group 2 (0.5-1.5 Å, green), and group 3 (> 1.5 Å, blue).
Figure 2.
Ligand-binding-site all-heavy-atom RMSD values of the initial apo structures and MD-derived structures against their holo crystal structures for 84 targets in Gunasekaran dataset. (A) Method 1 using Cα distance restraints. The average improvement is 0.29 Å. (B) Method 2 using Cα and SC-COM distance restraints. The average improvement is 0.52 Å. (C) Control group without distance restraints and no improvement in RMSD. The structures are separated into three groups based on their initial binding site Cα RMSD compared to their holo structures: group 1 (< 0.5 Å, red), group 2 (0.5-1.5 Å, green), and group 3 (> 1.5 Å, blue).
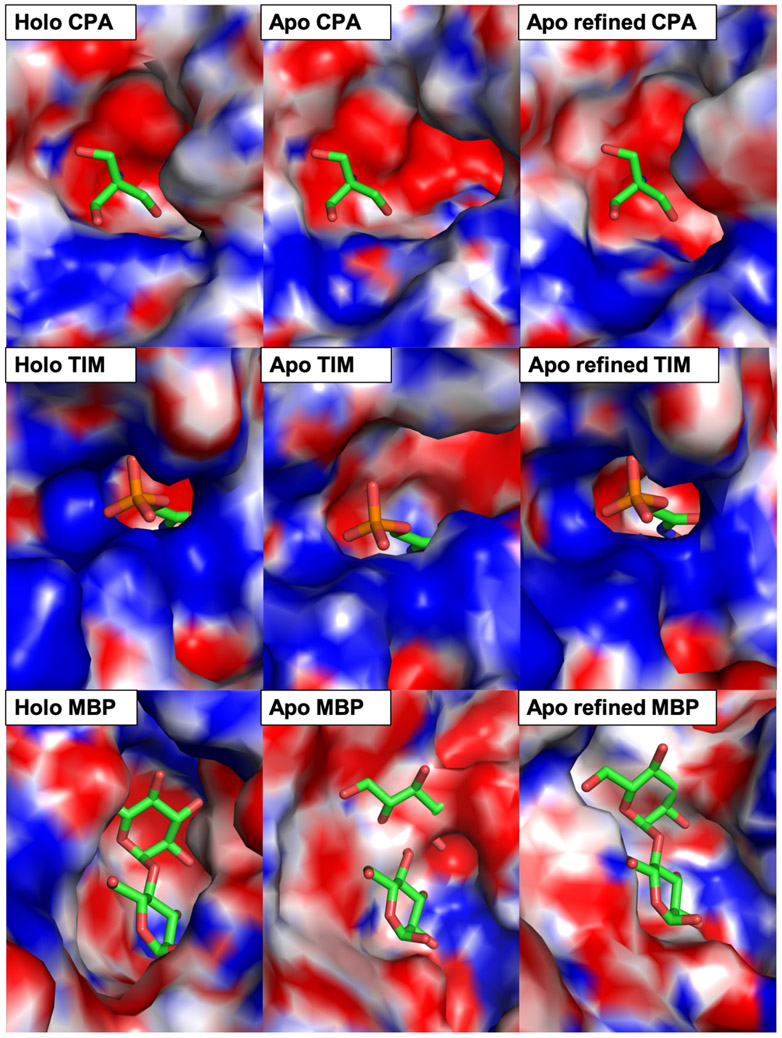
Group 1 contains 4 structures in our DUD-E set and their binding sites are rigid, i.e., the apo structures have almost identical conformations to their holo counterparts. It is generally accepted that RMSD changes of < 0.5 Å can be attributed to positional uncertainty from crystal structures solved at 2.0 Å resolution.43 Therefore, little changes are observed after MD simulations with distance restraints regardless of using method 1 or method 2 (Table S3, Figure 1). In DUD-E set, using method 1, the average all-heavy-atom RMSD of MD-derived holo structures relative to PDB holo structures increase by 0.15 Å (initial 0.53, final 0.68), and using method 2, the RMSD increase with an average of 0.12 Å. Similarly, in Gunasekaran set with 39 proteins in group 1, using method 1, we observe little changes in the overall shape of the binding pocket with only 0.02 Å RMSD improvement on average. A slightly better improvement in all-heavy-atom RMSD is observed using method 2 with an average of 0.14 Å (Initial 0.58, final 0.44) (Table S4, Figure 2). Despite the small changes, It is worth noting that small improvements in sidechain orientations have been reported to significantly improved docking results.10-12 One such structure in this group (no. 13) is carboxypeptidase A (CPA) bound to tromethamine, where the apo-holo pairs contain one sidechain (Tyr248) with big conformational difference.44 In its holo structure, Tyr248 faces the ligand, whereas in the apo form, Tyr248 faces away from the ligand. Accordingly, apo structure contains a larger binding pocket that eliminates specific interactions between Tyr248 and the ligand (Figure 3). Refinement with method 2 corrects the sidechain orientation of Tyr248, resulting in a smaller and more specific binding pocket for tromethamine (Figure 3).
Figure 3.
Representative structure no. 13 (carboxypeptidase A: holo PDB 1arm and apo PDB 1yme), no. 79 (triosephosphate isomerase: holo PDB 7tim and apo PDB 1ypi), and no. 81 (maltodextrin binding protein: holo PDB 1anf and apo PDB 1omp) from Gunasekaran dataset. Protein structures are shown in electrostatic potential surface representation and native ligands are overlaid at their binding site through protein structure alignments. All-heavy-atom RMSD of the binding sites relative to their holo counterparts are: CPA (initial 1.95 Å, refined 0.46 Å), TIM (initial 2.41 Å, refined 0.52 Å), and MBP (initial 3.47 Å, refined 3.51 Å).
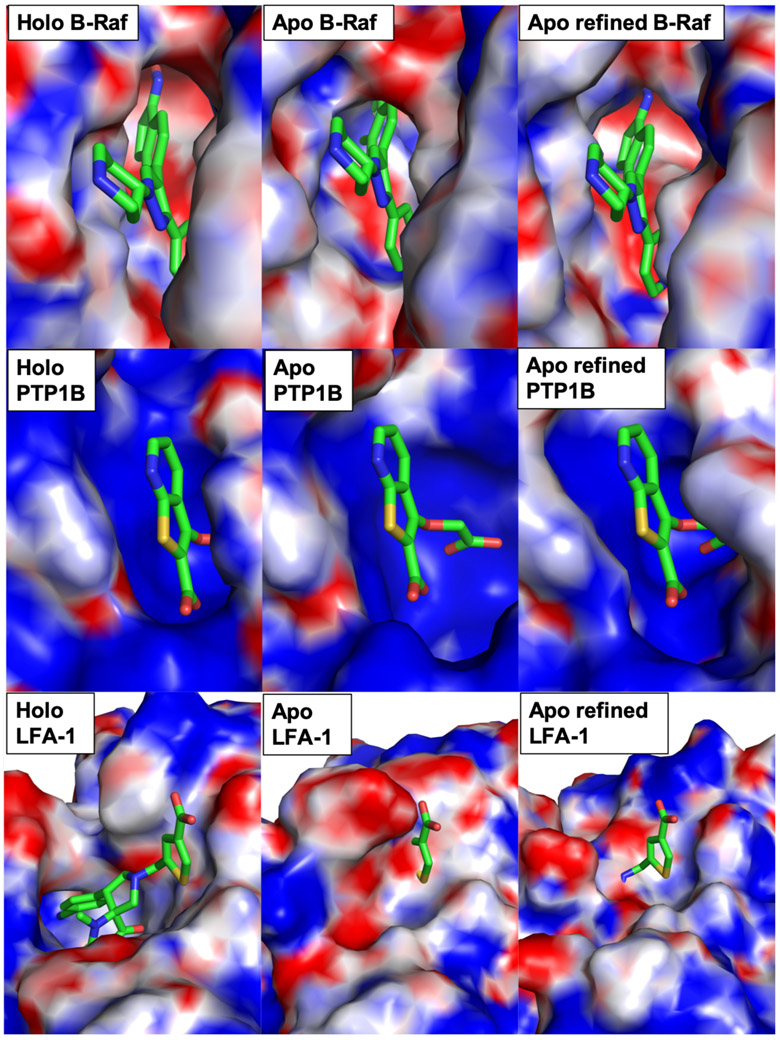
Apo-holo pairs with moderate conformational changes are categorized in group 2. On average our results show that qualities of MD-derived holo structures are improved using either method 1 or method 2 (Table S3, S4, Figure 1, 2). In DUD-E set, using method 1, the changes are small with improvement of 0.15 Å. This improvement is enhanced with method 2 showing an average RMSD improvement of 0.31 Å (initial 1.31, final 0.99). In Gunasekaran set, method 2 also show better improvement of 0.53 Å as compared to method 1 with 0.23 Å average improvement. A representative target for this group is B-Raf kinase in complex with a pyrazole-based inhibitor (SM5), and its apo counterpart is PDB ID 6uan (Table S3, no. 22). The protein-ligand complex crystal structure reveals a deep binding pocket that allows for the oxime moiety of SM5 to form a hydrogen bond with the side chain of Glu501 at the base of the pocket (Figure 4).45 However, in its apo form, sidechain orientations of Lys483 and Glu594 obstruct the binding site, resulting in a smaller pocket size that does not properly accommodate SM5’s oxime moiety. Through our refinement method, the conformations of these two side chains are corrected (RMSD initial 1.43 Å, final 0.81 Å, Table S3, no. 22) and the overlaid of SM5 appears to fit correctly (Figure 4).
Figure 4.
Representative structure no. 22 (B-Raf kinase: holo PDB 3d4q and apo PDB 6uan), no. 35 (PTP1B: holo PDB 2azr and apo PDB 2hnp), and no. 38 (LFA-1: holo PDB 2ica and apo PDB 1zon) from DUD-E dataset. Protein structures are shown in electrostatic potential surface representation and native ligands are overlaid at their binding site through protein structure alignments. All-heavy-atom RMSD of the binding sites relative to their holo counterparts are: B-Raf kinase (initial 1.43 Å, refined 0.81 Å), PTP1B (initial 2.71 Å, refined 0.75 Å), and LFA-1 (initial 3.08, refined 3.09).
Group 3 contains 20 structures with large conformational changes. In DUD-E set, we show the most significant changes in an average RMSD improvement of 1.51 Å toward their holo conformations (initial 2.93, final 1.42) using method 2 (Table S3). Using method 1, we see slightly less improvement of 1.10 Å. Similarly, in Gunasekaran set, we also observe significant changes in both methods 1 and 2 with 1.90 and 2.33 Å improvements, respectively (Table S4). Big changes in this group indicate that our method of generating holo conformations from apo structure is robust. One of the protein targets in this group is protein-tyrosine phosphatase 1B (PTP1B) from DUD-E set that is well-known to adopt an open conformation in its apo form and a closed conformation in its holo form.46, 47 More specifically, in its holo form, bound to a bicyclic thiophene inhibitor, the WPD loop adopts a closed conformation where the ligand’s thiophene ring is sandwiched between Phe182 and Tyr46 (Figure 4).48 Whereas, in its apo form, this WPD loop is open and Phe182 is located 13 Å away from the thiophene ring of the ligand in the overlaid structure (Figure 4). As a result, the binding pocket becomes larger, and specific interactions between the ligand and binding residues are diminished. Our refinement method correctly identifies the WPD loop and positions it in the holo closed conformation through MD simulations with restraints (Figure 4). The all-heavy-atom RMSD changes from an initial value of 2.71 Å to a refined 0.75 Å relative to the holo crystal structure (Table S3, no. 35). An example structure from Gunasekaran set from group 3 is triosephosphate isomerase (TIM, no. 79). In its apo form, loop 6 (residues 168-177) adopts an open conformation and the catalytic residue Glu165 is pointing away from the binding pocket.49 In contrast, its holo form bound to phosphoglycohydroxamate has a closed conformation of loop 6 and Glu165 pointing into the binding site.50 As a result, the holo structure has a smaller and more specific binding pocket than its apo counterpart (Figure 3). Our refinement method shows a successful result for TIM, where its holo structure conformation was generated from its apo structure through MD simulations with restraints (initial 2.41, final 0.52 Å RMSD).
Despite many successful cases with ligand-binding-site RMSD improvements (102 out of 124 targets), there are 22 unsuccessful cases. The unsuccessful cases suggest that G-LoSA could not find good templates for refinement. To better understand the relationship between the RMSD improvement and GA-score, we plot the RMSD improvement as a function of GA-score. Figure 5 shows that there is no correlation between GA-score and RMSD improvement. The average GA-scores from the unsuccessful and the successful cases are of the same value of 0.75. It suggests that GA-score alone is not a good predictor of whether or not a target apo protein can be successfully refined. Also shown in Figure 5, the changes in ligand-binding-site structures are small for the 22 unsuccessful cases, with an average RMSD change of 0.05 Å. Separating the unsuccessful cases into their respective groups, there are 12/43 (28%) cases in group 1, 8/61 (13%) in group 2, and 2/20 (10%) in group 3. Not surprisingly, group 1, which contains rigid binding sites (i.e., very similar apo and holo binding site conformations) show more unsuccessful cases with no RMSD improvement and with very small structural changes (Figure 5). Moreover, we look into the two structures in group 3 that show unsuccessful result in generating a holo conformation from an apo structure (Table S3, no. 38, Table S4, no. 81). From DUD-E set, the protein is leukocyte function-associated antigen-1 I-domain (LFA-1), and it adopts a largely open conformation of its C-terminal helix in the holo conformation bound to a spirocyclic hydantoin antagonist (Figure 4).51 In its apo form, this helix obstructs the binding site in a closed conformation, and Leu302, Gln303, Lys305, and Ile306 occupy the binding pocket.52 Our method did not identify a proper binding site template to generate the holo conformation for this apo structure (Table S1). From Gunasekaran set, it is a maltodextrin binding protein (MBP) that undergoes a relatively large conformational change at the binding pocket upon ligand binding (Figure 3). Unfortunately, binding templates obtained from G-LoSA search could not find a proper template (Table S2). As a result, MD simulations with restraints did not generate the correct holo structure conformation.
Figure 5.
Ligand-binding-site all-heavy-atom RMSD improvement (RMSD(apo) – RMSD(refined)) as a function of GA-score, where RMSD(apo) represents the RMSD of a PDB apo structure with respect to its holo structure in the PDB, and RMSD(refined) is the RMSD of an MD-refined structure with respect to the PDB holo structure. The circles represent all 124 targets. The structures are separated into three groups based on their initial binding site Cα RMSD compared to their holo structures: group 1 (< 0.5 Å, red), group 2 (0.5-1.5 Å, green), and group 3 (> 1.5 Å, blue). Clearly, the 22 targets with unsuccessful refinements show little deviations from their initial structures.
Overall, when comparing all-heavy-atom RMSD from method 1 to method 2, we see clear improvements in sidechain orientations in method 2 (Figure 1, 2). As mentioned above, changes in sidechain orientations at the binding site can have significant impacts on the overall shape of the binding pocket (Figure 3, 4), and correct shape of the binding pocket is ultimately important for proper docking for virtual screening.
Virtual screening and docking poses evaluation show that the MD-derived holo structures perform consistently better than the initial apo structures
In order to evaluate the quality of our refined structures, we conducted virtual screening for 40 DUD-E targets with their known active and decoy ligands using Vina.21, 38 As a control group, we also conducted virtual screening for the crystal holo structures of DUD-E targets to compare their results with the apo and the MD-refined holo cases. The enrichment level for enrichment factors at 1% (EF1%) and 2% (EF2%) attained during virtual screening for each target is reported in Table S5. Additionally, we compare the EF1% and EF2% of apo structures against refined structures and holo structures in Figure 6. Overall, when 1% of the top ranked compounds are selected (EF1%), on average, refined structures have a value of 6.2, which is almost two times better than the apo average of 3.5. Similarly, looking at 2% top ranked compounds (EF2%), the refined structures have an average value of 5.3 compared to 3.3 average from apo structures. The control group with holo crystal structures shows the best enrichment levels with EF1% of 10.0 and EF2% of 7.3. These enrichment values are comparable to the previously published data using DUD-E dataset from Wojcikowski et al and Pereira et al.53, 54 Comparing the results from apo and holo crystal structures, our results further confirm the widely accepted notion that holo structures are superior to apo structures in virtual screening campaigns.8, 9
Figure 6.
Comparison of EF1% and EF2% results from apo structures to refined structures and holo structures. The circles represent each of the receptors from 40 DUD-E targets. (A) EF1% comparing apo vs. refined structures. The average improvement is 2.7. (B) EF1% from the control group with PDB holo structures. The average improvement is 6.5. (C) EF2% comparing apo vs. refined structures. The average improvement is 2.0. (D) EF2% from the control group with PDB holo structures. The average improvement is 4.0.
In group 1, all of the 4 refined structures show improvements in EF1%, despite the fact that only 1 of them show improvement in ligand-binding-site RMSD (Table S5, S3). This suggests that short MD simulations with minimization and equilibration procedures could help optimize the conformations of side chains in the binding site. At the same time, it is worth noting that the difference in average EF1% is quite small: 4.9 for refined structures and 3.7 for apo structures.
In group 2, 17/24 receptors in the refined structures perform better than apo structure at EF1% and similarly 19/24 at EF2%. This is comparable to 18/24 structures that undergo improvement in their ligand-binding-site RMSD (Table S3). The average improvements in enrichment values are also bigger in this group. At EF1%, the refined structures have 6.4 compared to 3.7 in apo structures, and at EF2%, the values are 5.4 and 3.5 for refined and apo structures, respectively.
In group 3, 11/12 refined structures have better EF1% than the apo structures and all of the refined structures have better EF2% than the apo structures. During refinement, 11/12 structures show better binding site RMSD, which is consistent with the virtual screening results. The largest difference in enrichment values are seen in this group. At EF1%, the refined receptors have 6.2 compared to apo receptors with 2.9, and at EF2%, the values are 5.3 and 2.8 for refined and apo structures, respectively. It suggests that large improvement in binding site RMSD for receptors in group 3 are really useful to improve virtual screening results.
In addition, we performed ligand docking of native ligands onto the initial apo protein structures and MD-refined structures, as well as to the crystal holo structures (i.e., self-docking). AutoDock Vina was used for rigid docking.38 Ligand poses from docking were assessed based on their all-heavy-atom ligand RMSD relative to the crystal structure after aligning the binding site residues of the proteins, but not the ligand. Our results show consistently better ligand poses in the refined structures compared to the initial unrefined apo structures. For 40 DUD-E structures, the refined structures improve docking poses of native ligands by 1.68 Å on average (Table S6, Figure 7A). For 84 Gunasekaran structures, the refined structures improve docking poses of native ligands by 0.94 Å on average (Table S7, Figure 7C). A general consensus about correct binding pose is a docking pose with ligand RMSD ≤ 2 Å.38, 55 We show that our method of generating holo structures from apo structures yield significant improvements in docking binding modes of native ligands. The average results from our refined structures are 1.97 Å ligand RMSD (DUD-E dataset) and 1.96 Å (Gunasekaran dataset), which are comparable to the self-docking averages, 1.34 Å (DUD-E dataset) and 1.36 Å (Gunasekaran dataset). On the other hand, the unrefined apo structures yield an average of 3.65 Å ligand RMSD (DUD-E) and 2.90 Å (Gunasekaran) (Table S6, S7, Figure 7).
Figure 7.
RMSD values of the native ligand binding poses on the initial apo structures and MD-derived structures against their holo crystal structures. (A) 40 targets in DUD-E dataset. The average improvement is 1.68 Å. (B) Control group, PDB holo structures. The average improvement is 2.31 Å. (C) 84 targets in Gunasekaran dataset. The average improvement is 0.94 Å. (D) Control group, PDB holo structures. The average improvement is 1.54 Å.
In DUD-E dataset, group 1 does not show improvement in ligand poses (apo 1.71 Å, refined holo 1.83 Å) (Table S6). Since both ligand RMSDs are below 2 Å, they are considered successful docking poses. Similarly, small average improvement of 0.46 Å is seen in group 1 from Gunasekaran dataset (apo 2.25 Å, refined holo 1.79 Å) (Table S7). However, there are 39 proteins in this group as compared to only 4 in DUD-E dataset. In the refined group, 31 out of 39 structures have ligand RMSD ≤ 2 Å, while the apo structures only contain 20 out of 39 structures with ligand RMSD ≤ 2 Å (Table S7). This result clearly indicates that small improvements in sidechain orientations (in rigid proteins) have significant effects in the ligand binding modes from docking. An example is CPA, where a difference in the sidechain orientation of Tyr248 significantly changes the shape of the binding pocket (Figure 3). As a result, docking of tromethamine to the apo structure results in a ligand RMSD of 3.09 Å, while docking to the refined structure improves the ligand RMSD to 1.25 Å (Table S7).
Group 2 has an average ligand pose improvement of 1.58 Å (DUD-E dataset) and 1.43 Å (Gunasekaran dataset). Changes in ligand RMSD for 24 proteins in DUD-E dataset are significant, because the apo-based results contain only 2 successful binding poses (≤ 2 Å) and the refined holo-based results have 15 successful docking poses (Table S6). The structure of B-Raf kinase (no. 22) is a good example that shows differences in sidechain orientations of the binding pocket (Figure 4). Docking to the obstructed apo binding pocket has a highly incorrect binding pose with a ligand RMSD of 5.99 Å. In contrast, a correct ligand pose is seen when docking to the refined holo structure with a ligand RMSD of 1.73 Å (Table S6, no. 22). Group 2 (37 proteins) in Gunasekaran dataset also shows significant improvements in ligand poses, where docking to the apo structures have 5/37 correct poses, whereas results from refined holo structures have 24/37 correct poses (Table S7).
Group 3 from both datasets also show significant improvements in ligand poses with 2.46 Å (12 proteins in DUD-E) and 0.99 Å (8 proteins Gunasekaran). Proteins in this group undergo the largest changes in their binding site structures, which can explain their big improvements in docking results. From DUD-E dataset, PTP1B has an open conformation in its apo form that leaves out specific interactions between Phe182 and the ligand, bicyclic thiophene inhibitor (Figure 4). As a result, incorrect ligand pose was obtained when docked to the apo structure (ligand RMSD 2.60 Å), and this binding mode was corrected to 1.38 Å, when docked to the refined holo structure (Table S6, no. 35). Similarly, in Gunasekaran dataset, TIM has an open conformation in its apo form that hinders proper docking of the ligand, phosphoglycohydroxamate (Figure 3). Docking to the apo structure yields an incorrect binding pose with a ligand RMSD of 2.98 Å, and this binding pose is corrected when docked to the refined structure with a ligand RMSD of 1.76 Å (Table S7, no. 79).
In the cases where apo structures are not refined properly, we see no significant improvement in docking poses. In DUD-E dataset, ligand docking to LFA-1 shows similar binding modes in apo and refined structures with 4.84 Å and 4.45 Å, respectively (Table S6, no. 38). The ligand is blocked from accessing the binding site by a few residues in both apo and refined structures (Figure 4). Similarly, in Gunasekaran dataset, the binding site of MBP did not undergo proper refinement, leaving a few residues blocking the binding pocket (Figure 3). As a result, docking of maltose shows unsuccessful ligand poses in both apo and refined structures with 5.12 Å and 5.16 Å, respectively (Table S7, no. 81).
CONCLUSIONS
One of the most important aspects in a structure based virtual screening campaign is the proper selection of a protein receptor. The conformation of the receptor binding site needs to be in a specific state to correctly accommodate ligand docking. Several studies have demonstrated that a holo (ligand bound) receptor conformation is superior to an apo (ligand free) structure to get meaningful outcomes in virtual screening.8, 9, 11 Therefore, it is necessary to obtain a holo structure conformation that is conducive for ligand docking. In here, we present a method that generates reliable holo protein conformations from apo structures. We use our local structure alignment tool, G-LoSA, to obtain a holo binding site template for an input, apo structure. We run MD simulation with restraints derived from a holo template to obtain a holo structure conformation from the apo protein. Our results show consistently successful generation of holo conformations from apo structures in 82% (102/124) of our test cases. Using two different datasets, DUD-E and Gunasekaran, we show good results across easy, medium, and hard apo targets. Moreover, we show significant improvements in virtual screening performance for 40 DUD-E targets using our MD-derived holo structures.
The protocol presented here is inspired by our previously published ligand-binding-site structure refinement protocol to refine homology model structures.20 This protocol is further optimized to generate reliable holo protein conformations from apo structures by adding SC-COM distance restraints. Adding sidechain restraints is rationalized by several studies that have reported small differences in backbone RMSD, but larger conformational varieties in sidechain orientations between apo-holo pairs.4-6 Furthermore, small differences in sidechain orientations between apo-holo pairs have been shown to negatively affect ligand docking to apo structures.8-12
We expect that our protocol will serve as a useful computational tool to properly utilize apo protein receptor for virtual screening whenever holo structures are not available. A well-prepared receptor structure can be quickly tested by docking known binders and using our high-throughput MD simulations to estimate the quality of the binding pocket.56
In addition, we observe that our method can also detect and identify sidechain residues that undergo conformational changes upon ligand binding. These conformational changes are often slow in the context of standard ligand binding free energy simulation, hindering the simulation convergence. Going forward, we plan to extend our method to automatically identify residues with such slow degrees of freedom. It has been shown with T4 lysozyme that the sidechain of Val111 undergoes conformational reorientation during ligand binding, resulting in a kinetic trap that complicates the convergence of free energy calculations.57 Jiang and Roux have shown that they could overcome the kinetically trapped sidechain conformations at the protein receptor using free energy perturbation with Hamiltonian replica exchange MD.58 Mobley et al also showed that their confine-and-release method improved their free energy calculations and resulted in better agreement with experimental data.59 Similarly, we plan to connect the identification of residues with slow degrees of freedom for enhanced sampling to improve the convergence of ligand binding free energy calculations in CHARMM-GUI Free Energy Calculator.60, 61
Supplementary Material
Table S1, List of 40 apo targets and their PDB holo structures from DUD-E dataset
Table S2, List 84 apo targets and their PDB holo structures from Gunasekaran dataset
Table S3, Results of 40 targets from DUD-E dataset
Table S4, Results of 84 targets from Gunasekaran dataset
Table S5, Virtual screening enrichment factor (EF) results for 40 DUD-E targets
Table S6, Ligand RMSD relative to experimental holo structures from 40 DUD-E targets
Table S7, Ligand RMSD relative to experimental holo structures from 84 Gunasekaran targets
Figure S1, Schematic workflow of ligand binding site refinement to generate holo protein structure conformations from apo structures.
Acknowledgement
This work has been supported by NIH GM126140 and XSEDE MCB070009 (to WI) and NIH P41-GM104601 and the Advanced Scientific Computing Research program, DOE Office of Science (to WJ). We thank to Hui Sun Lee for a helpful discussion.
Footnotes
Supporting Information
The Supporting Information is available free of charge.
Methods
The authors declare no competing financial interests.
REFERENCES
- 1.Gutteridge A; Thornton J, Conformational Changes Observed in Enzyme Crystal Structures upon Substrate Binding. J Mol Biol 2005, 346, 21–28. [DOI] [PubMed] [Google Scholar]
- 2.Gunasekaran K; Nussinov R, How Different are Structurally Flexible and Rigid Binding Sites? Sequence and Structural Features Discriminating Proteins That Do and Do Not Undergo Conformational Change upon Ligand Binding. J Mol Biol 2007, 365, 257–273. [DOI] [PubMed] [Google Scholar]
- 3.Brylinski M; Skolnick J, What Is the Relationship Between the Global Structures of Apo and Holo Proteins? Proteins Struct Funct Genet 2008, 70, 363–377. [DOI] [PubMed] [Google Scholar]
- 4.Clark JJ; Benson ML; Smith RD; Carlson HA, Inherent Versus Induced Protein Flexibility: Comparisons Within and Between Apo and Holo Structures. PLoS Comput Biol 2019, 15, e1006705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heringa J; Argos P, Strain in Protein Structures as Viewed Through Nonrotameric Side Chains: II. Effects upon Ligand Binding. Proteins Struct Funct Bioinf 1999, 37, 44–55. [PubMed] [Google Scholar]
- 6.Gaudreault F; Chartier M; Najmanovich R, Side-chain Rotamer Changes upon Ligand Binding: Common, Crucial, Correlate with Entropy and Rearrange Hydrogen Bonding. Bioinformatics 2012, 28, i423–i430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Y; Park SJ; Im W, A Systematic Analysis of Protein-Carbohydrate Interactions in the PDB. Glycobiology 2020. [DOI] [PubMed] [Google Scholar]
- 8.McGovern SL; Shoichet BK, Information Decay in Molecular Docking Screens Against Holo, Apo, and Modeled Conformations of Enzymes. J Med Chem 2003, 46, 2895–2907. [DOI] [PubMed] [Google Scholar]
- 9.Lee HS; Lee CS; Kim JS; Kim DH; Choe H, Improving Virtual Screening Performance Against Conformational Variations of Receptors by Shape Matching with Ligand Binding Pocket. J Chem Inf Model 2009, 49, 2419–2428. [DOI] [PubMed] [Google Scholar]
- 10.Zavodszky MI; Kuhn LA, Side-chain Flexibility in Protein-ligand Binding: The Minimal Rotation Hypothesis. Protein Sci 2005, 14, 1104–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erickson JA; Jalaie M; Robertson DH; Lewis RA; Vieth M, Lessons in Molecular Recognition: The Effects of Ligand and Protein Flexibility on Molecular Docking Accuracy. J Med Chem 2004, 47, 45–55. [DOI] [PubMed] [Google Scholar]
- 12.Halperin I; Ma B; Wolfson H; Nussinov R, Principles of Docking: An Overview of Search Algorithms and a Guide to Scoring Functions. Proteins Struct Funct Bioinf 2002, 47, 409–443. [DOI] [PubMed] [Google Scholar]
- 13.Wong CF, Flexible Receptor Docking for Drug Discovery. Expert Opin Drug Discovery 2015, 10, 1189–1200. [DOI] [PubMed] [Google Scholar]
- 14.Osguthorpe DJ; Sherman W; Hagler AT, Generation of Receptor Structural Ensembles for Virtual Screening Using Binding Site Shape Analysis and Clustering. Chem Biol Drug Des 2012, 80, 182–193. [DOI] [PubMed] [Google Scholar]
- 15.Ogrizek M; Turk S; Lesnik S; Sosic I; Hodoscek M; Mirkovic B; Kos J; Janezic D; Gobec S; Konc J, Molecular Dynamics to Enhance Structure-based Virtual Screening on Cathepsin B. J Comput Aided Mol Des 2015, 29, 707–712. [DOI] [PubMed] [Google Scholar]
- 16.Lee HS; Im W, G-LoSA for Prediction of Protein-Ligand Binding Sites and Structures. Methods Mol Biol 2017, 1611, 97–108. [DOI] [PubMed] [Google Scholar]
- 17.Lee HS; Im W, G-LoSA: An Efficient Computational Tool for Local Structure-centric Biological Studies and Drug Design. Protein Sci 2016, 25, 865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee HS; Im W, Ligand Binding Site Detection by Local Structure Alignment and Its Performance Complementarity. J Chem Inf Model 2013, 53, 2462–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee HS; Im W, Identification of Ligand Templates using Local Structure Alignment for Structure-Based Drug Design. J Chem Inf Model 2012, 52, 2784–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guterres H; Lee HS; Im W, Ligand-Binding-Site Structure Refinement Using Molecular Dynamics with Restraints Derived from Predicted Binding Site Templates. J Chem Theory Comput 2019, 15, 6524–6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mysinger MM; Carchia M; Irwin JJ; Shoichet BK, Directory of Useful Decoys, Enhanced (DUD-E): Better Ligands and Decoys for Better Benchmarking. J Med Chem 2012, 55, 6582–6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berman HM; Westbrook J; Feng Z; Gilliland G; Bhat TN; Weissig H; Shindyalov IN; Bourne PE, The Protein Data Bank. Nucleic Acids Res 2000, 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Govindaraj RG; Brylinski M, Comparative Assessment of Strategies to Identify Similar Ligand-binding Pockets in Proteins. BMC Bioinf 2018, 19, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khazanov NA; Carlson HA, Exploring the Composition of Protein-ligand Binding Sites on a Large Scale. PLoS Comput Biol 2013, 9, e1003321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derigs U, The Shortest Augmenting Path Method for Solving Assignment Problems - Motivation and Computational Experience. Ann Oper Res 1985, 4, 57–102. [Google Scholar]
- 26.Jo S; Kim T; Iyer VG; Im W, CHARMM-GUI: A Web-based Graphical User Interface for CHARMM. J Comput Chem 2008, 29, 1859–1865. [DOI] [PubMed] [Google Scholar]
- 27.Lee J; Cheng X; Swails JM; Yeom MS; Eastman PK; Lemkul JA; Wei S; Buckner J; Jeong JC; Qi Y; Jo S; Pande VS; Case DA; Brooks CL 3rd; MacKerell AD Jr.; Klauda JB; Im W, CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J Chem Theory Comput 2016, 12, 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jorgensen WL; Chandrasekhar J; Madura JD; Impey RW; Klein ML, Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys 1983, 79, 926–935. [Google Scholar]
- 29.Allen MP, Tildesley DJ, Computer Simulations of Liquids. Clarendon Press: Oxford: 1987. [Google Scholar]
- 30.Essmann U; Perera L; Berkowitz ML; Darden T; Lee H; Pedersen LG, A Smooth Particle Mesh Ewald Method. J Chem Phys 1995, 103, 8577–8593. [Google Scholar]
- 31.Steinbach PJ; Brooks BR, New Spherical-Cutoff Methods for Long-Range Forces in Macromolecular Simulation. J Comput Chem 1994, 15, 667–683. [Google Scholar]
- 32.Barth E; Kuczera K; Leimkuhler B; Skeel RD, Algorithms for Constrained Molecular Dynamics. J Comput Chem 1995, 16, 1192–1209. [Google Scholar]
- 33.Huang J; Rauscher S; Nawrocki G; Ran T; Feig M; de Groot BL; Grubmuller H; MacKerell AD Jr., CHARMM36m: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat Methods 2017, 14, 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eastman P; Swails J; Chodera JD; McGibbon RT; Zhao Y; Beauchamp KA; Wang LP; Simmonett AC; Harrigan MP; Stern CD; Wiewiora RP; Brooks BR; Pande VS, OpenMM 7: Rapid Development of High Performance Algorithms for Molecular Dynamics. PLoS Comput Biol 2017, 13, e1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park H; DiMaio F; Baker D, The Origin of Consistent Protein Structure Refinement from Structural Averaging. Structure 2015, 23, 1123–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jo S; Vargyas M; Vasko-Szedlar J; Roux B; Im W, PBEQ-Solver for Online Visualization of Electrostatic Potential of Biomolecules. Nucleic Acids Res 2008, 36, W270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Im W; Beglov D; Roux B, Continuum Solvation Model: Computation of Electrostatic Forces from Numerical Solutions to the Poisson-Boltzmann Equation. Comput Phys Commun 1998, 111, 59–75. [Google Scholar]
- 38.Trott O; Olson AJ, AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J Comput Chem 2010, 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang N; Shoichet BK; Irwin JJ, Benchmarking Sets for Molecular Docking. J Med Chem 2006, 49, 6789–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang X, Miller W , A Time-efficient, Linear-space Local Similarity Algorithm. Adv Appl Math 1991, 12, 337–357. [Google Scholar]
- 41.Zhang Y; Skolnick J, Scoring Function for Automated Assessment of Protein Structure Template Quality. Proteins Struct Funct Bioinf 2004, 57, 702–710. [DOI] [PubMed] [Google Scholar]
- 42.Xu J; Zhang Y, How Significant is a Protein Structure Similarity with TM-score = 0.5? Bioinformatics 2010, 26, 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daopin S; Davies DR; Schlunegger MP; Grutter MG, Comparison of Two Crystal Structures of TGF-beta2: The Accuracy of Refined Protein Structures. Acta Crystallogr sect D Biol Crystallogr 1994, 50, 85–92. [DOI] [PubMed] [Google Scholar]
- 44.Greenblatt HM; Feinberg H; Tucker PA; Shoham G, Carboxypeptidase A: Native, Zinc-removed and Mercury-replaced Forms. Acta Crystallogr sect D Biol Crystallogr 1998, 54, 289–305. [DOI] [PubMed] [Google Scholar]
- 45.Hansen JD; Grina J; Newhouse B; Welch M; Topalov G; Littman N; Callejo M; Gloor S; Martinson M; Laird E; Brandhuber BJ; Vigers G; Morales T; Woessner R; Randolph N; Lyssikatos J; Olivero A, Potent and Selective Pyrazole-based Inhibitors of B-Raf Kinase. Bioorg Med Chem Lett 2008, 18, 4692–4695. [DOI] [PubMed] [Google Scholar]
- 46.Schubert HL; Fauman EB; Stuckey JA; Dixon JE; Saper MA, A Ligand-induced Conformational Change in the Yersinia Protein Tyrosine Phosphatase. Protein Sci 1995, 4, 1904–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brandao TA; Johnson SJ; Hengge AC, The Molecular Details of WPD-loop Movement Differ in the Protein-tyrosine Phosphatases YopH and PTP1B. Arch Biochem Biophys 2012, 525, 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moretto AF; Kirincich SJ; Xu WX; Smith MJ; Wan ZK; Wilson DP; Follows BC; Binnun E; Joseph-McCarthy D; Foreman K; Erbe DV; Zhang YL; Tam SK; Tam SY; Lee J, Bicyclic and Tricyclic Thiophenes as Protein Tyrosine Phosphatase 1B Inhibitors. Bioorg Med Chem 2006, 14, 2162–2177. [DOI] [PubMed] [Google Scholar]
- 49.Lolis E; Alber T; Davenport RC; Rose D; Hartman FC; Petsko GA, Structure of Yeast Triosephosphate Isomerase at 1.9-A Resolution. Biochemistry 1990, 29, 6609–6618. [DOI] [PubMed] [Google Scholar]
- 50.Davenport RC; Bash PA; Seaton BA; Karplus M; Petsko GA; Ringe D, Structure of the Triosephosphate Isomerase-phosphoglycolohydroxamate Complex: An Analogue of the Intermediate on the Reaction Pathway. Biochemistry 1991, 30, 5821–5826. [DOI] [PubMed] [Google Scholar]
- 51.Potin D; Launay M; Monatlik F; Malabre P; Fabreguettes M; Fouquet A; Maillet M; Nicolai E; Dorgeret L; Chevallier F; Besse D; Dufort M; Caussade F; Ahmad SZ; Stetsko DK; Skala S; Davis PM; Balimane P; Patel K; Yang Z; Marathe P; Postelneck J; Townsend RM; Goldfarb V; Sheriff S; Einspahr H; Kish K; Malley MF; DiMarco JD; Gougoutas JZ; Kadiyala P; Cheney DL; Tejwani RW; Murphy DK; McIntyre KW; Yang X; Chao S; Leith L; Xiao Z; Mathur A; Chen BC; Wu DR; Traeger SC; McKinnon M; Barrish JC; Robl JA; Iwanowicz EJ; Suchard SJ; Dhar TG, Discovery and Development of 5-[(5S,9R)-9-(4-cyanophenyl)-3-(3,5-dichlorophenyl)-1-methyl-2,4-dioxo-1,3,7-tria zaspiro[4.4]non-7-yl-methyl]-3-thiophenecarboxylic acid (BMS-587101)--A Small Molecule Antagonist of Leukocyte Function Associated Antigen-1. J Med Chem 2006, 49, 6946–6949. [DOI] [PubMed] [Google Scholar]
- 52.Qu A; Leahy DJ, The Role of the Divalent Cation in the Structure of the I Domain from the CD11a/CD18 Integrin. Structure 1996, 4, 931–942. [DOI] [PubMed] [Google Scholar]
- 53.Wojcikowski M; Ballester PJ; Siedlecki P, Performance of Machine-learning Scoring Functions in Structure-based Virtual Screening. Sci Rep 2017, 7, 46710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pereira JC; Caffarena ER; Dos Santos CN, Boosting Docking-Based Virtual Screening with Deep Learning. J Chem Inf Model 2016, 56, 2495–2506. [DOI] [PubMed] [Google Scholar]
- 55.Bursulaya BD; Totrov M; Abagyan R; Brooks CL 3rd, Comparative Study of Several Algorithms for Flexible Ligand Docking. J Comput Aided Mol Des 2003, 17, 755–763. [DOI] [PubMed] [Google Scholar]
- 56.Guterres H; Im W, Improving Protein-Ligand Docking Results with High-Throughput Molecular Dynamics Simulations. J Chem Inf Model 2020, 60, 2189–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morton A; Matthews BW, Specificity of Ligand Binding in a Buried Nonpolar Cavity of T4 lysozyme: Linkage of Dynamics and Structural Plasticity. Biochemistry 1995, 34, 8576–8588. [DOI] [PubMed] [Google Scholar]
- 58.Jiang W; Roux B, Free Energy Perturbation Hamiltonian Replica-Exchange Molecular Dynamics (FEP/H-REMD) for Absolute Ligand Binding Free Energy Calculations. J Chem Theory Comput 2010, 6, 2559–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mobley DL; Chodera JD; Dill KA, The Confine-and-Release Method: Obtaining Correct Binding Free Energies in the Presence of Protein Conformational Change. J Chem Theory Comput 2007, 3, 1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jo S; Jiang W; Lee HS; Roux B; Im W, CHARMM-GUI Ligand Binder for Absolute Binding Free Energy Calculations and Its Application. J Chem Inf Model 2013, 53, 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim S; Oshima H; Zhang H; Kern NR; Re S; Lee J; Roux B; Sugita Y; Jiang W; Im W, CHARMM-GUI Free Energy Calculator for Absolute and Relative Ligand Solvation and Binding Free Energy Simulations. J Chem Theory Comput 2020, 16, 7207–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, List of 40 apo targets and their PDB holo structures from DUD-E dataset
Table S2, List 84 apo targets and their PDB holo structures from Gunasekaran dataset
Table S3, Results of 40 targets from DUD-E dataset
Table S4, Results of 84 targets from Gunasekaran dataset
Table S5, Virtual screening enrichment factor (EF) results for 40 DUD-E targets
Table S6, Ligand RMSD relative to experimental holo structures from 40 DUD-E targets
Table S7, Ligand RMSD relative to experimental holo structures from 84 Gunasekaran targets
Figure S1, Schematic workflow of ligand binding site refinement to generate holo protein structure conformations from apo structures.