Abstract
Centriole amplification in multiciliated cells occurs in a pseudo-cell cycle regulated process that typically utilizes a poorly characterized molecularly dense structure called the deuterosome. We identified the centrosomal protein Cep70 as a novel deuterosome-associated protein that forms a complex with other deuterosome proteins, CCDC78 and Deup1. Cep70 dynamically associates with deuterosomes during centriole amplification in the ciliated epithelia of Xenopus embryos. Cep70 is not found in nascent deuterosomes prior to amplification. However, it becomes localized at deuterosomes at the onset of centriole biogenesis and remains there after the completion of centriole amplification. Deuterosome localization requires a conserved C-terminal “Cep70” motif. Depletion of Cep70 using morpholino oligos or CRISPR/Cas9 editing in F0 embryos leads to a severe decrease in centriole formation in both endogenous MCCs, as well as ectopically induced MCCs. Consistent with a decrease in centrioles, endogenous MCCs have defects in the process of radial intercalation. We propose that Cep70 represents a novel regulator of centriole biogenesis in MCCs.
Introduction:
Centriole duplication in cycling cells has been studied in great detail where it is known that daughter centrioles are born from pre-existing mother centrioles in a cell cycle-dependent manner (Nigg and Holland, 2018). Multiciliated cells (MCCs) represent a unique post-mitotic cell type that utilizes a pseudo-cell cycle state in order to generate over 100 centrioles that will become the basal bodies for their motile cilia (Al Jord et al., 2017). While daughter centrioles can template off of the existing parental centrioles, it is generally appreciated that the vast majority of nascent centrioles in MCCs template from an MCC-specific structure called the deuterosome (Spassky and Meunier, 2017). Interestingly, centrioles are also known to be able to nucleate de novo without a preexisting template. In cell culture systems laser ablation of the existing centrioles leads to de novo centriole formation as the cells enter the cell cycle (Khodjakov et al., 2002). Similarly, it has been shown that daughter centrioles can nucleate de novo in MCC precursors that are devoid of both parental centrioles and deuterosomes (Mercey et al., 2019). In MCCs, this likely occurs via an electron dense cloud of material identified in classic TEM studies as the fibrogranular material (Chang et al., 1979). The molecular composition of this material remains poorly characterized.
While much of the centriole biogenesis machinery is common between cell cycle centriole duplication and MCC centriole amplification, there are regulatory features unique to each system. One of the critical regulators of centriole biogenesis found in both cycling cells and MCCs is Cep152 (Blachon et al., 2008; Klos Dehring et al., 2013). In Xenopus MCCs, overexpression of Cep152 leads to a significant increase in centriole amplification, and conversely, loss of functional Cep152 at the deuterosome leads to a decrease in centriole biogenesis (Klos Dehring et al., 2013). Additionally, CCDC78 and Deup1 have been identified as deuterosome proteins. In Xenopus, fluorescently tagged versions of both Deup1 and CCDC78 localize to both centrioles and deuterosomes. However, an N-terminal tagged GFP-CCDC78 preferentially localizes to the deuterosome. CCDC78 has been implicated in centriole biogenesis in MCCs in Xenopus but CCDC78 depletion has no effect on centriole duplication in other cell types (Klos Dehring et al., 2013). The initial report of Deup1, using RNAi knockdown approaches, suggested that Deup1 was involved in centriole biogenesis in MCCs. However, more recent work analyzing multiple tissues in mouse genetic knock-out models and morpholino (MO) depletion in Xenopus has determined that Deup1 is not required for centriole amplification (Mercey et al., 2019; Zhao et al., 2013). The transcription factor E2F4 forms a complex with DP1 and GemC1 or MCIDAS to regulate transcription of genes associated with centriole amplification in MCCs (Kim et al., 2018; Terre et al., 2016). Despite these recent advances, there is much that remains unknown about the regulation of centriole amplification in MCCs.
Cep70 is a centrosomal-associated protein that has been connected with the regulation of microtubule nucleation in mammalian systems (Shi et al., 2011; Shi et al., 2012; Shi et al., 2015; Yang et al., 2014). Cep70 physically interacts with gamma tubulin and has been implicated in the formation of proper mitotic spindles (Shi et al., 2011). Furthermore, perturbations in Cep70 levels has been associated with numerous cancers. Importantly, these mutations have been implicated in paclitaxel sensitivity, further linking Cep70 to microtubule function (Lazo, 2017; Shi et al., 2017a; Shi et al., 2017b). However, genome wide RNAi screens of human cells assaying for either broad mitotic defects or specific defects in centriole duplication have not identified Cep70 as an essential regulator of these processes (Balestra et al., 2013; Neumann et al., 2010). Additionally, MO depletion of Cep70 in zebrafish does not result in mitotic or centriole duplication defects, although it does result in defective ciliogenesis (Wilkinson et al., 2009). Of particular relevance, CRC70, the Chlamydomonas orthologue of Cep70, has been proposed to act as a scaffold during centriole duplication (Shiratsuchi et al., 2011). It localizes to nascent procentrioles and, when depleted, key centriolar components such as Sas6 are not recruited. Additionally, depletion leads to phenotypes consistent with loss of centriole duplication and furthermore a loss of cilia. Importantly, overexpression of Cep70 in both Chlamydomonas and in human NIH3T3 cells results in the formation of ectopic aggregates that contain key centriolar proteins such as Sas6 (Shiratsuchi et al., 2011). These results imply that Cep70 could act as a critical scaffold for recruiting proteins required for centriole biogenesis. Here we present evidence that in Xenopus, Cep70 is largely dispensable for cell cycle regulated centriole duplication but is essential for MCC centriole amplification.
Results:
CCDC78 was previously identified as one of the first molecular components of the deuterosome. In the epidermis of Xenopus embryos, CCDC78 localizes to both centrioles and deuterosomes in MCCs, however an N-terminal GFP fusion preferentially localizes to deuterosomes. Both MO knockdown and F0 CRISPR/Cas9 based gene editing of CCDC78 results in a 50% reduction in the number of centrioles in MCCs (Collins et al., 2020; Klos Dehring et al., 2013). While this reduction is significant, it also implies that CCDC78 is not essential for centriole biogenesis but instead is part of the regulatory machinery that determines proper centriole numbers. To gain further insight into deuterosome function, we commissioned Hybrigenics Inc. to perform a yeast two hybrid screen of human fetal brain tissue using human CCDC78 as a bait (Hybrigenics Inc). Using this approach, we identified Cep70 as a potential CCDC78 interacting protein. Consistent with its known role as a centriolar protein, Cep70 tagged with fluorescent proteins (FPs) localizes strongly to all centrioles including the basal bodies of MCCs marked with Centrin4-RFP (Figure 1A–B). Importantly, we also find FP-Cep70 below the apical surface of MCCs at deuterosomes marked with GFP-CCDC78 and GFP-Deup1 (Figure 1C–D). To validate the yeast two hybrid and immunofluorescence co-localization results, we performed a biochemical analysis, employing a co-immunoprecipitation (co-IP) assay using anti-FLAG beads. Embryos were injected with FLAG-tagged CCDC78 and GFP-tagged Cep70 mRNAs, and co-IPs were performed at the early tailbud stage. Biochemical analysis confirmed that GFP-Cep70 co-immunoprecipitated with FLAG-CCDC78 but not with FLAG beads alone, implying an interaction between CCDC78 and Cep70 (Figure 1E and Supplemental Figure 1A). Similarly, GFP-CCDC78 co-immunoprecipitated with FLAG-Cep70 but not with FLAG beads alone (Figure 1F and Supplemental Figure 1B). Since GFP-CCDC78 resides primarily at the deuterosome this result further implicates Cep70 at the deuterosome. We next tested if Cep70 interacted with another deuterosome protein, Deup1. GFP-Deup1 pulled down with both FLAG-CCDC78 as well as FLAG-Cep70 (Figure 1G and Supplemental Figure 1C). These results suggest that Cep70, CCDC78 and Deup1 are part of a deuterosome complex. Collectively, our co-IP data, co-localization studies and the yeast two hybrid experiments demonstrate that Cep70 is capable of biochemically interacting with CCDC78, suggesting that Cep70 is a deuterosome-associated protein likely involved in centriole biogenesis in MCCs.
Figure 1. Cep70 is a deuterosome associated protein.
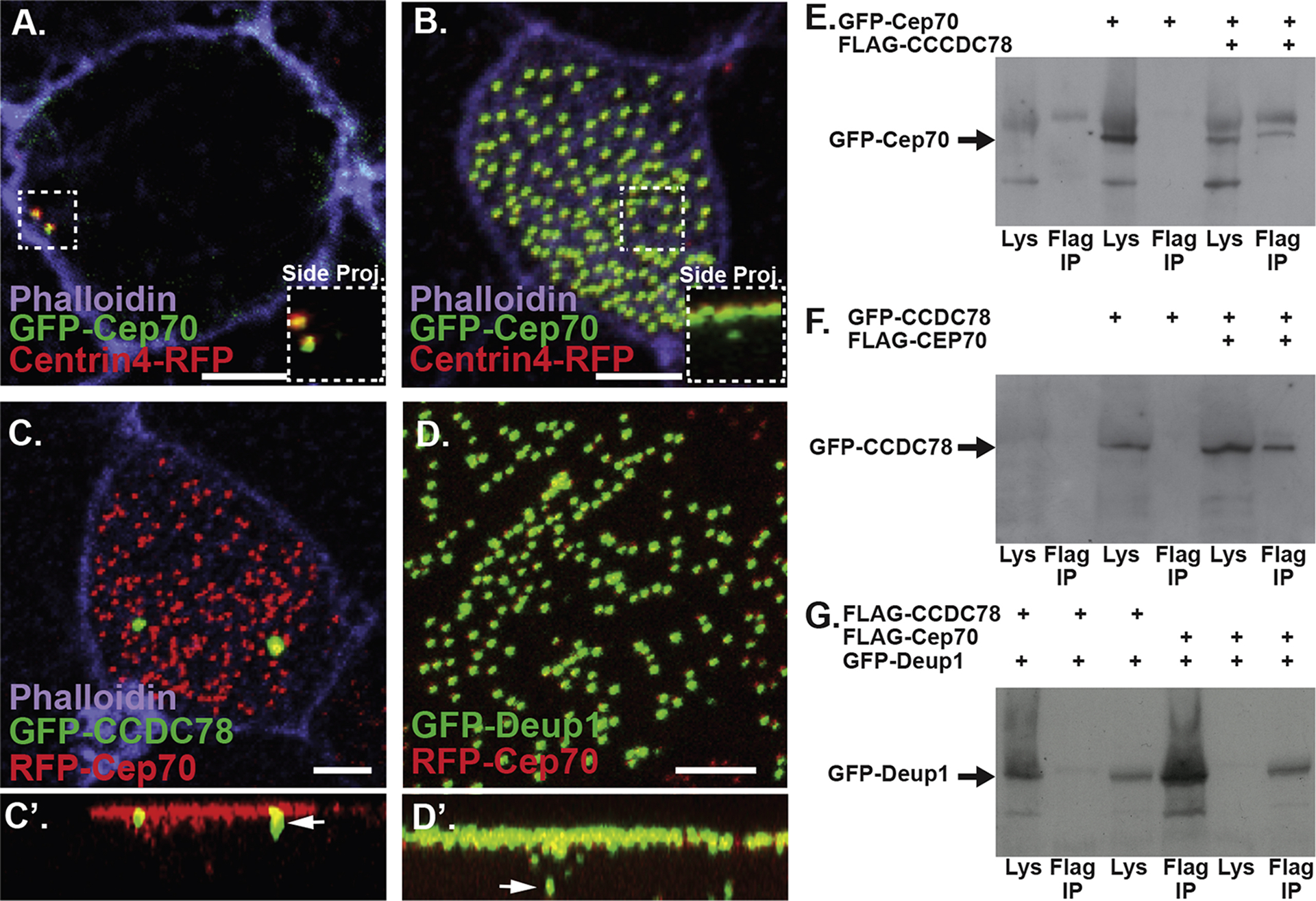
A–B, Z-projections of a non-MCC (A) and an MCC (B) labeled with the centriole marker Centrin4-RFP (red) and GFP-Cep70 (green), inlays are side projections. Scale bar if 5μm. C–D, Mature MCCs in embryos injected with RFP-Cep70 and GFP-CCDC78 (C) and GFP-Deup1 (D). C’–D’, side projections of C–D with arrows identifying colocalization below the apical surface. Scale bar is 5μm. E, Co-IP using FLAG beads to pull down FLAG-CCDC78 followed by an anti-GFP western blot to identify GFP-Cep70. F, Co-IP using FLAG beads to pull down FLAG-Cep70 followed by anti-GFP western blot to identify GFP-CCDC78. G, Co-IP using FLAG beads to pull down either FLAG-CCDC78 (lanes 1–3) or FLAG-Cep70 (lanes 4–6) followed by anti-GFP western blot to identify GFP-Deup1.
The over-expression (OE) of FP-Cep70 can lead to non-specific protein aggregates similar to what has been seen in other systems, however, using centriole and deuterosome markers independently we were able to characterize a dynamic localization of Cep70. In Xenopus, centriole amplification occurs below the apical surface prior to radial intercalation making imaging challenging (Zhang and Mitchell, 2015). To circumvent this issue we ectopically expressed the MCC-inducing factor Multicilin (MCIDAS) which converts outer epithelial cells into MCCs (Stubbs et al., 2012). Injection of MCIDAS mRNA containing a 3’ fusion of human Glucocorticoid Receptor (hGR-MCIDAS) creates a Dexamethasone (Dex)-inducible MCIDAS that allows for temporal control of MCC induction in the outer epithelia (Stubbs et al., 2012). As previously shown MCIDAS induction converts epithelial cells in MCCs within 10Hrs after Dex treatment (Stubbs et al., 2012). However, we see considerable cell to cell variation in the timing of both deuterosome and centriole formation. The earliest identifiable step in MCC specification is the formation of nascent deuterosomes, marked with GFP-CCDC78 which can occur within 2Hrs of Dex treatment (Figure 2A). Interestingly, FP-Cep70 does not co-localize with these early deuterosomes but remains at the parental centrioles (Figure 2A–B). However, in cells that have accumulated a large number of deuterosomes (~10) we begin to find FP-Cep70 co-localizing with FP-CCDC78 while it is also maintained at parental centrioles (Figure 2C–D). Finally, approximately 6HRs after Dex treatment we begin to see nascent centriole formation with FP-Cep70 localizing at deuterosomes marked with FP-CCDC78 (yellow foci in Figure 2E) as well as what we interpret as nascent centrioles (red foci in Figure 2E). Similarly, we find FP-Cep70 co-localizing with Centrin4-FP at nascent centrioles (yellow foci in Figure 2F) budding off of what we interpret as deuterosomes marked with FP-Cep70 (green foci in Figure 2F). After centriole biogenesis, deuterosomes coalesce from 15–20 deuterosomes into several structures that reside below the apical surface and we continue to see co-localization of FP-Cep70 and GFP-CCDC78 (Figure 1C–D). The key take away from these localization studies is that while Cep70 consistently co-localizes with centrioles it does not co-localize at nascent deuterosomes but is instead recruited to deuterosomes prior to centriole amplification. The recruitment of Cep70 to the deuterosome prior to centriole nucleation led us to hypothesize that Cep70 could be critically involved in the regulation centriole amplification.
Figure 2. Dynamic localization of Cep70 to the deuterosome.
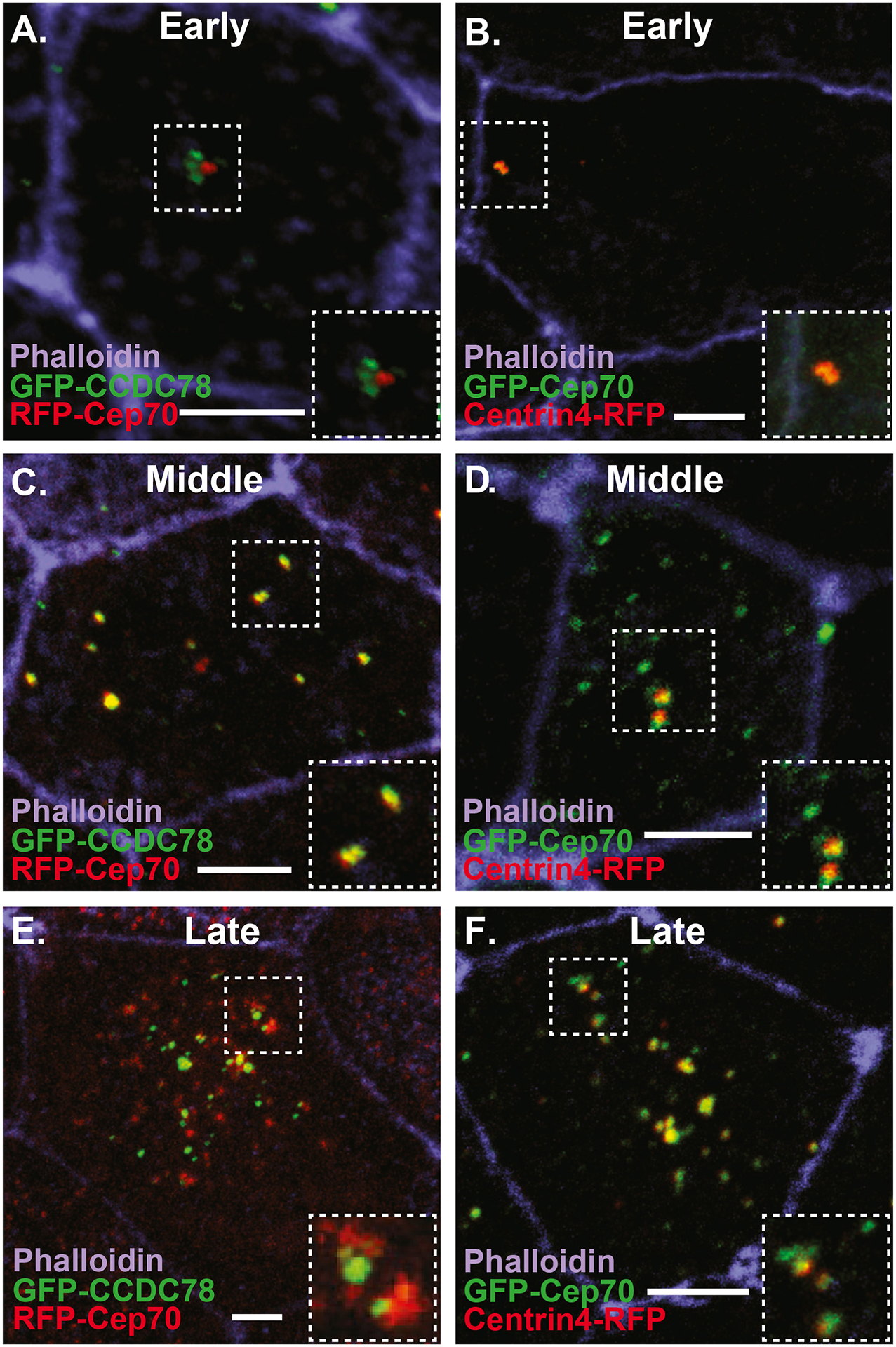
A–F Images of ectopic MCCs from embryos injected with the MCC inducing factor hGR-MCIDAS, treated with Dex and co-injected with RFP-Cep70 and the deuterosome marker GFP-CCDC78 (A, C, E) or GFP-Cep70 and the centriole marker Centrin4-RFP (B, D, F) stained with phalloidin. A–B, Early phase (2–3Hrs) of MCC development with Cep70 co-localizing to parental centrioles (B) but not nascent deuterosomes marked by CCDC78 (A). C–D, Middle phase (4–6Hrs) of MCC development showing FP-Cep70 co-localizing with GFP-CCDC78 at deuterosomes (yellow foci in E) but not at parental centrioles (red foci in C) and co-localizing with centrin4-RFP at parental centrioles (yellow foci in D) but not deuterosomes (green foci in D). Late phase (> 6Hrs) of MCC development showing FP-Cep70 co-localizing with GFP-CCDC78 at deuterosomes (yellow foci in E) but not at presumptive nascent centrioles (red foci in E) and co-localizing with Centrin4-RFP at nascent centrioles (yellow foci in F) but not presumptive deuterosomes (green foci on F). Scale bar is 5μm.
The Cep70 protein, like many centrosomal proteins, contains multiple coiled-coil domains (Truebestein and Leonard, 2016). Additionally, at the C-terminal there is a conserved domain that was initially called the Cep70 motif (Cep70M) based on conservation between CRC70 in Chlamydomonas and the vertebrate Cep70 (AA 505–562 in Xenopus, Supplemental Figure 2.) (Shiratsuchi et al., 2011). This region has some relation to a protein-protein interaction domain called the tetratricopeptide repeat (TPR), although this is only reliably identified as such in the human protein (Supplemental Figure 2.). To better understand the regulation of Cep70 localization we generated four Cep70 truncation constructs fused to RFP (Figure 3A). These include protein fragments that contain the first coiled-coil domain (C1), the second coiled-coil domain (C2), the combined coiled-coil domains (CC) and the C terminal Cep70M (Figure 3A). Neither the C1 nor the C2 fragments reliably co-localize with GFP-CCDC78 at the deuterosome (17.1% and 4.9% of cells respectively) but both were frequently found at the apical surface localized to centrioles (54.3% and 78.1%) (Figure 3A, B, C). Additionally, both fragments were observed enriched in cytoplasmic aggregates (88.6% and 87.8%) (Figure 3A, B, C). Similarly, the CC fragment containing both coiled-coil domains primarily localized to cytoplasmic aggregates (75.7% of cells), but also occasionally localized either at the deuterosome (41.4% of cells) or apically with centrioles (44.4% of cells) (Figure 3A, D). In contrast to the coiled-coil data, the Cep70M domain rarely localized to aggregates (8.3%), was enriched at deuterosomes (47.2%) and was reliably but very weakly localized to the centrioles (~100%) (Figure 3A, E). These data indicate that the regulation of Cep70 localization is likely complex, but surprisingly establishes the Cep70M domain as important for both centriolar and deuterosome localization. Both coiled-coil domains and TPR domains are well established mediators of protein-protein interactions (Perez-Riba and Itzhaki, 2019; Truebestein and Leonard, 2016). The coiled-coil domain of Cep70 has previously been implicated in its interactions with gamma tubulin and during the stabilization of the mitotic spindle. While the Cep70M domain is evolutionarily conserved, a clear function for it is still unknown. Our localization data suggest that part of that function could be recruitment to the deuterosome during centriole amplification.
Figure 3. Different domains of Cep70 contribute to different cellular localizations.
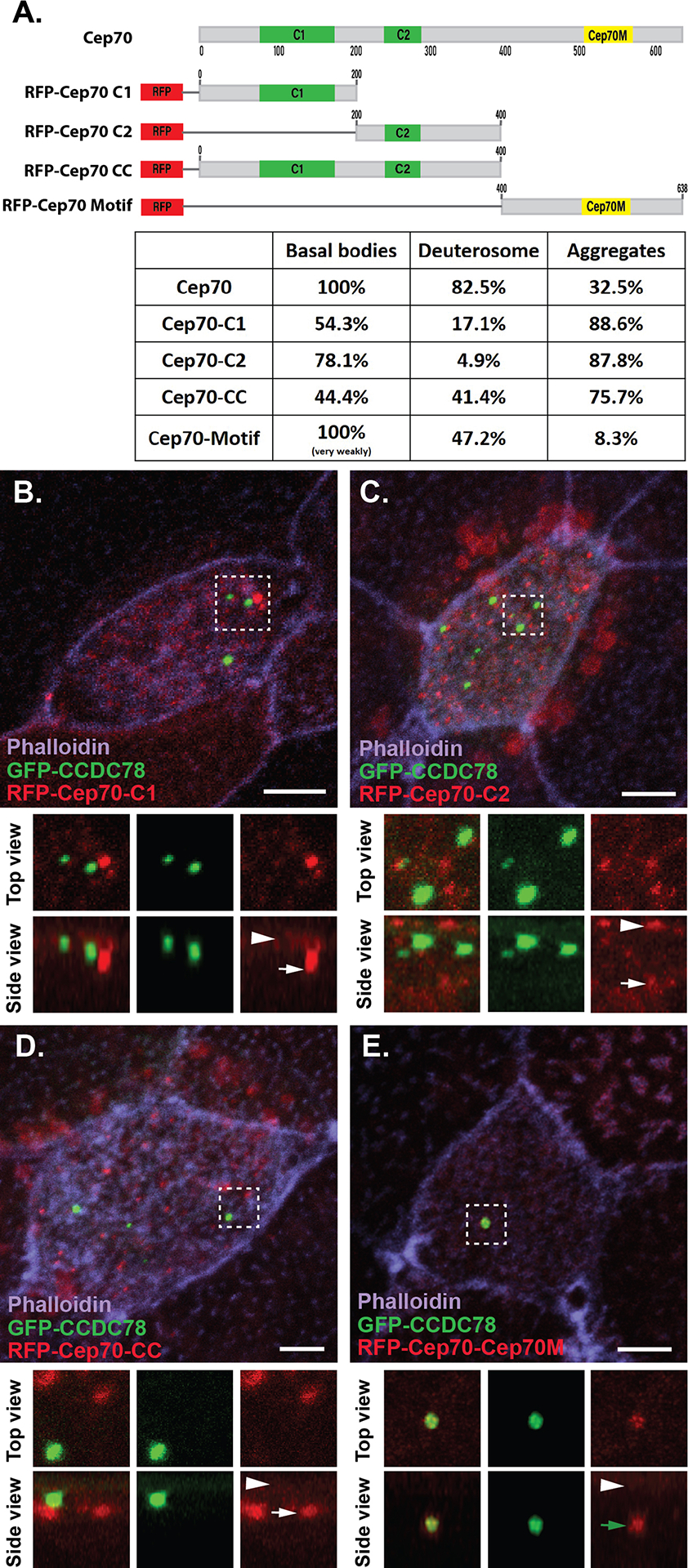
A, Domain map of Cep70 and description of the 4 individual domain constructs used in B-E along with quantification of localization for each construct (n > 30 cells for each condition). Scale bars are 5μm. B–E, Localization of GFP-CCDC78 (green) and RFP-Cep70 (red) domain constructs in mature MCCs stained with phalloidin (purple) with boxed areas blown up and shown without phalloidin both top and side projections. B, RFP-Cep70-C1 localizing weakly at the level of basal bodies (arrowhead) and in cytoplasmic aggregates (white arrow). C, RFP-Cep70-C2 localizing strongly at the level of basal bodies (arrowhead) and in cytoplasmic aggregates (white arrow). D, RFP-Cep70-CC localizing weakly at the level of basal bodies (arrowhead) and in cytoplasmic aggregates (white arrow). E, RFP-Cep70-Cep70M localizing very weakly at the level of basal bodies (arrowhead) and at the deuterosome (green arrow).
The localization of Cep70 to the deuterosome during centriole biogenesis led us to ask if Cep70 is important for centriole formation in MCCs. First, we generated a MO targeting the translational start site of Cep70. Injection into all four blastomeres at the 4 cells stage resulted in only 2% survival (n=135 from 5 individual experiments). Importantly, we achieved a partial rescue of 22% survival (n=160 from 5 individual experiments) in embryos where we co-injected GFP-Cep70 mRNA not targeted by the MO. To avoid lethality, we injected MOs mosacially into 1 cell at the 4 cell stage together with a fluorescent marker (Dextran blue or membrane-RFP) to label morphant cells. In mosaic embryos, MCCs containing a control MO generate 124 +/− 43 centrioles per cell whereas Cep70 MO containing cells only generate 41 +/− 26 centrioles (Figure 4A, C and E). Importantly, this decrease in centrioles can be partially rescued by the injection of mRNA encoding a Cep70 construct not targeted by the MO and results in 94 +/− 42 centrioles per cell (Figure 4D–E). While there is some experiment to experiment variation in overall centriole numbers based on the size of the embryos, we do not see a significant differences within experiments when we compare WT to control MO or WT to GFP-Cep70 expressing embryos (Supplemental Figure 3). These results indicate that depletion of Cep70 adversely affects centriole amplification. Additionally, we assessed the ability of morphant MCCs to generate cilia and found that control morphants cells in mosaic embryos generated proper cilia numbers whereas Cep70 morphant cells had few discernable cilia (Figure 4F–G). Finally, we assayed centriole number in non-MCCs and found a slight defect in centriole duplication with Control MOs containing 1.79 +/− 1.03 whereas Cep70 MO cells had 1.51 +/− 0.95 (p=0.054). However, the low survival rates of more complete knockdowns suggests that there is likely an important non-MCC function for Cep70.
Figure 4. Cep70 is required for centriole amplification.
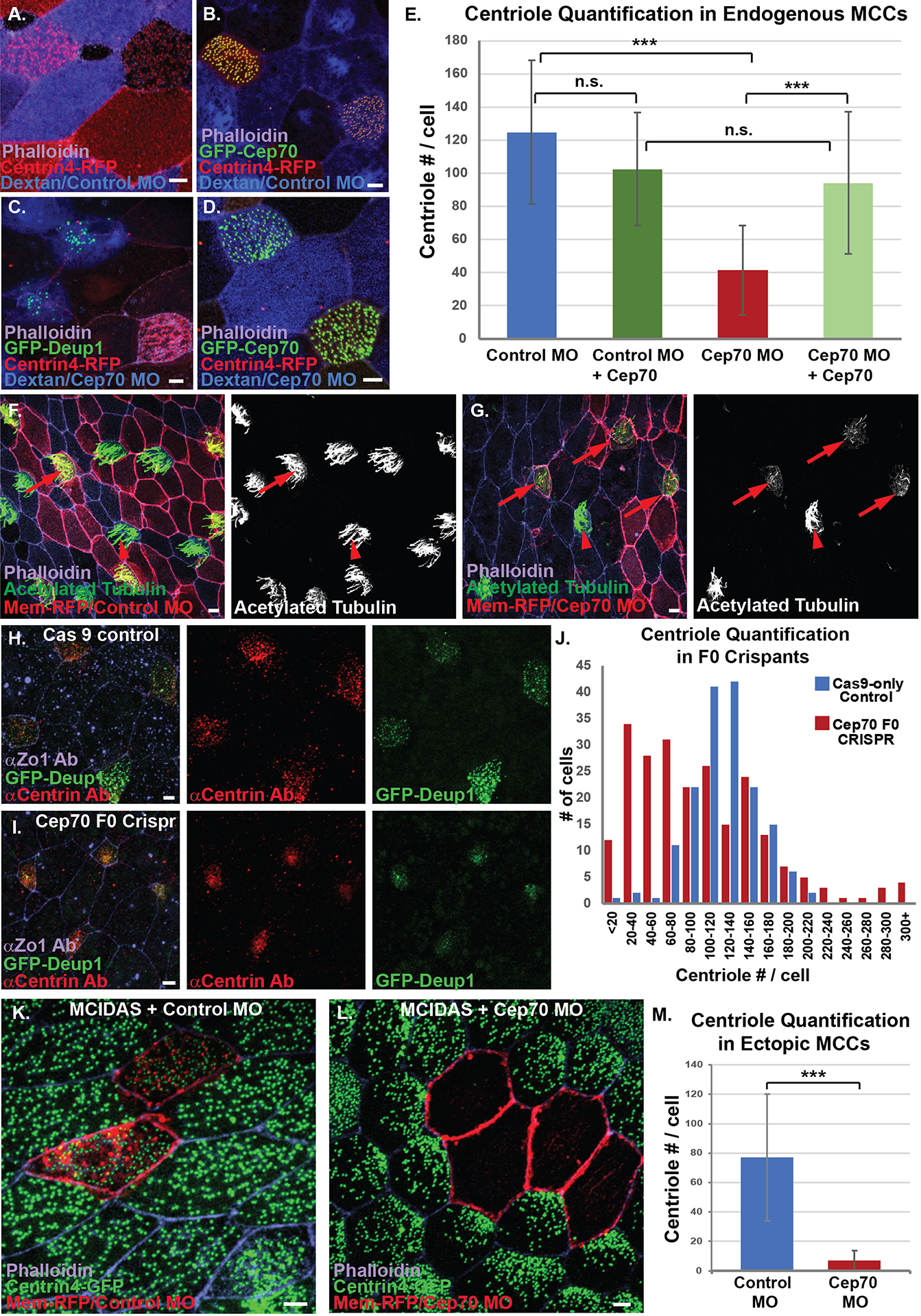
A–E, MO analysis in endogenous MCCs. A, MCCs from embryos injected with Centrin-RFP (red) and mosaically with Control MO marked with Dextran (blue) stained with phalloidin (purple). B, MCCs from embryos injected with Centrin-RFP (red) and mosaically with Control MO marked with Dextran (blue) together with GFP-Cep70 and stained with phalloidin (purple). C, MCCs from embryos injected with Centrin-RFP (red), GFP-Deup1 (green) and mosaically with Cep70 MO marked with Dextran (Blue) stained with phalloidin (purple). D, MCCs from embryos injected with Centrin-RFP (red) and GFP-Cep70 (green) mosaically with Cep70 MO marked with Dextran (blue) stained with phalloidin (purple). E, Quantification of centriole number in embryos from conditions imaged in A–D (*** p<0.001, n.s. not significant, n > 85 cells). F, Mosaic embryo showing strong cilia staining with acetylated tubulin (green/white) in both WT cells (arrowhead, not red) and in control MO cells (arrow, red), stained with phalloidin (purple). G, Mosaic embryo showing strong cilia staining (green/white) in WT cells (arrowhead, not red) but not in Cep70 MO cells (arrow, red), stained with phalloidin. H–J, CRISPR analysis in transgenic Tub-GFP-Deup1 (green) embryos. H, Cas9 alone control embryo stained with anti-centrin mAb (red) and phalloidin (purple). I, MCCs (green) from embryos injected with Cas9 and Cep70 sgRNA stained with anti-centrin mAb (red) and phalloidin (purple). J, Quantification of centriole number in MCCs from F0 crispant embryos; data is presented as a histogram of the number of cells observed in each bin (as labeled, n > 167 cells). K–M, MO analysis in hGR-MCIDAS induced ectopic MCCs. K, MCCs from embryos uniformly injected with hGR-MCIDAS and centrin-GFP (green) and mosaically with control MO together with mem-RFP as a marker (red) and stained with phalloidin (purple). L, MCCs from embryos uniformly injected with hGR-MCIDAS and centrin-GFP (green) and mosaically with Cep70 MO together with mem-RFP as a marker (red) and stained with phalloidin (purple). K, Quantification of the average centriole number from MCCs from conditions in I–J(***p<0.001, n > 107 cells). Scale bars are 5μm.
To further validate the Cep70 depletion data, we performed F0 CRISPR/Cas9 gene editing of Cep70. In contrast to our Cep70 MO mosaic embryos in which morphant cells are labeled, our F0 CRISPR experiments result in blindly mosaic embryos. Regardless of this limitation, we can still observe a severe phenotype in centriole formation in cells devoid of Cep70. A histogram of centriole number in control embryos (Cas9 with no sgRNA) shows a tight peak for centriole quantification, with a majority of cells containing between 100 and 140 centrioles (Figure 4H and J). In stark contrast, the histogram of Cep70 crispants shows a much broader distribution with the peak at cells containing between 20–40 centrioles (Figure 4I and J). We interpret the broad distribution to reflect the likelihood that the F0 animals are mosaic such that cells with low numbers of centrioles are mutant whereas cell with normal centriole number are wild type. However, we also see a small increase in the number of cells that have massive increases in centriole number which could reflect a rare defect in successful cell division (Figure 4J). Regardless, the overall decrease in centriole numbers is consistent with our MO data and indicates an important role for Cep70 in MCC centriole amplification.
We have previously published a role for both centrioles and microtubules in the process of radial intercalation, whereby MCCs apically insert into the outer epithelium and expand their apical domain (Collins et al., 2020; Werner et al., 2014). Consistent with this, MCCs containing Cep70 MO often completely fail to intercalate (Figure 5A). Additionally, one metric for proper intercalation is the average apical surface area. In morphant MCCs we observe a significant decrease in apical size from 223 +/− 121 μm2 in control MO to 77 +/− 80 μm2 in Cep70 MO (Figure 5B). Furthermore, in Cep70 crispant embryos, we see a dramatic shift in apical size towards smaller sized cells (Figure 5C). Due to these intercalation defects, we believe that our quantifications of the effect of Cep70 depletion on centriole decrease, are likely an underestimate of the true phenotype, as we could miss quantifying cells that have completely failed intercalation. To address this concern, we generated ectopic MCCs by expressing hGR-MCIDAS with Dex induction to generate ectopic MCCs in the outer epithelium. This allows centriole amplification to occur in the outer epithelial cells, eliminating the requirement for radial intercalation. Importantly, in this context there is no statistical difference in the apical size of ectopic MCCs containing either the control or Cep70 MO (Figure 5D). However, while the ectopic MCCs containing a control MO generated on average 77 +/− 43 centrioles, ones containing a Cep70 MO generated only 6.8 +/− 6.9 centrioles per cell (Figure 4K, L and M). These results indicate that Cep70 is a critical regulator of centriole biogenesis and furthermore that the defects in apical size seen in endogenous MCCs was in fact a failure of proper intercalation rather than an overall cell size defect.
Figure 5. Cep70 affects radial intercalation of MCCs.
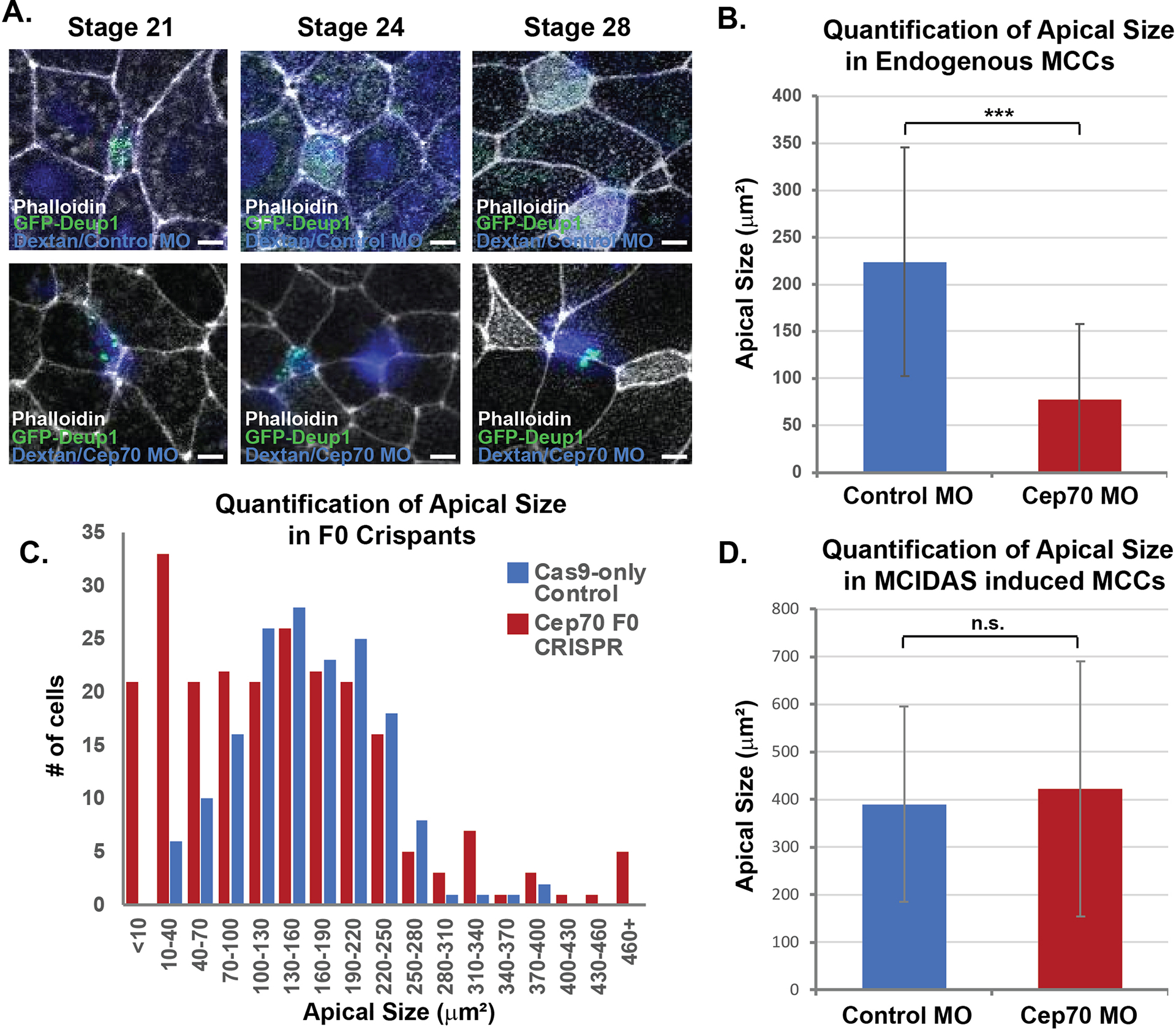
A, Time course of MCCs (marked with transgenic Tub-GFP-Deup1) undergoing intercalation in embryos injected with Dextran and a control MO (blue, top) or Cep70 MO (blue, bottom) showing examples of MCCs that have failed to intercalate (blue, bottom). Scale Bar is 5 μm. B, Quantification of MCC apical size in morphant embryos from experiments in Figure 4 A, C, and E (*** p<0.001, n > 85 cells). C, Quantification of apical size in F0 crispant embryos; data is presented as a histogram of the number of cells observed in each bin (as labeled, n > 167 cells). D, Quantification of ectopic MCC apical size in morphant embryos from experiments in Figure 4 I–K (n.s. not significant, n > 107 cells).
Discussion
Cep70 co-localizes and biochemically interacts with 2 known deuterosome proteins, CCDC78 and Deup1. The regulation of Cep70 localization appears complex such that multiple domains appear to contribute to both centriolar and deuterosome localization. Of particular significance is the role for the Cep70 motif in deuterosome localization. This domain is broadly conserved across evolution and yet its function remains unclear. Our data provides evidence that one role for this domain is to facilitate deuterosome localization which is presumably a factor for its role in centriole biogenesis in MCCs.
The depletion of Cep70 results in a significant decrease in the number of centrioles generated in both endogenous MCCs and ectopically-induced MCCs. However, while cell cycle-regulated centriole duplication defects are subtle, we demonstrate that this is likely a result of incomplete knockdown as non-mosaic MO experiments and CRISPR experiments resulted in low survival rates. Importantly, Cep70 does not appear to be involved in deuterosome nucleation as it does not localize to nascent deuterosomes. Consistent with a role of Cep70 in centriole biogenesis, deuterosomes do not appear competent to nucleate centrioles until Cep70 is recruited to the deuterosome. Overall, our results implicate Cep70 in the process of centriole amplification in MCCs. Recent work has indicated that deuterosmes and indeed parental centrioles are both dispensable for centriole biogenesis in MCCs (Mercey et al., 2019). This suggests that the centriole biogenesis machinery can function without condensing into a deuterosome. While Cep70 localizes to the deuterosome during amplification the strong phenotype associated with its loss of function (stronger than the loss of deuterosomes), would suggests that its function goes beyond simply localizing to the deuterosome and that it is in fact a critical regulator of centriole biogenesis independent of that localization.
The loss of centrioles in Cep70 morphant and crispant MCCs leads to a failure to properly undergo radial intercalation. We recently showed that MCCs with a 50% reduction in centrioles were delayed by several hours in intercalation; however, they eventually intercalated (Collins et al., 2020). The more severe failure of some Cep70 morphant and crispant MCCs could reflect a more severe delay that would eventually resolve itself or it could reflect an additional role in regulating MT dynamics. The MT stabilizing protein CLAMP is essential for radial intercalation independent of centriole number indicating that MT regulation is an essential aspect of intercalation (Werner et al., 2014). Consistent with this, MCCs treated with the MT depolymerizing drug nocodazole also fail to properly intercalate. Therefore, the intercalation failure of Cep70 depleted cells could be the result of a loss of centrioles or could be the combination of centriolar loss coupled to a separate defect in the MT network. Given the considerable literature linking Cep70 to MT function this scenario is particularly appealing. Future work will be required to distinguish between these possibilities.
Cep70 has an evolutionarily conserved role in centriole biogenesis. One hypothesis is that Cep70 was ancestrally important as a scaffold protein during centriole biogenesis as reflected in the Chlamydomonas data but that this requirement was co-opted by other proteins in the cycling cells of vertebrates. We propose that the massive centriole amplification that occurs in MCCs puts considerable stress on the biogenesis machinery revealing a previously unidentified role for Cep70 during centriole biogenesis. The deutersome and centriole amplification in general remain a poorly understood feature of MCCs. This work adds Cep70 to the short list of proteins critically required for proper centriole amplification.
Methods:
All experiments in Xenopus were performed using previously described techniques (Werner and Mitchell, 2013). Embryos were obtained via in vitro fertilization using standard protocols (Sive et al., 2007) approved by the Northwestern University Institutional Animal Care and Use Committee.
Plasmids and mRNAs:
Cep70 (Xl.15467) of Xenopus laevis (Thermo, MXL1736–202797545) was amplified with PCR and cloned into pCS2+ containing N-terminal GFP or RFP or C-terminal FLAG with XhoI site by Gibson Assembly. For deuterosome and centriole markers, GFP-CCDC78 (Xl.4890) (Klos Dehring et al., 2013) and Centrin4-RFP (Xl.50473) were used as previously described (Werner et al., 2011). CCDC78 (Xl.4890) was cloned into N-terminal FLAG tagged pCS2+ with StuI and XhoI sites (Klos Dehring et al., 2013). hGR-MCIDAS was a generous gift from Chris Kintner (Stubbs et al., 2012). The coiled-coil and Cep70M domains of Cep70 were assigned using Cluster Omega and BOXSHADE. Each domain was amplified by PCR and cloned into pCS2+ containing an N-terminal RFP with Gibson Assembly using the following primers.
Cep70_C1_F) GGA TCC CAT CGA TTC atg tca gag cag ctg cca tc (Tm:60.5’C),
Cep70_C1_R) GTTCTAGAGGCTCGAGAtctggggaggcggctca (Tm:59.8’C)
Cep70_C2_F) GGA TCC CAT CGA TTC gac aat aat gga aca gat cag ca (Tm:59.2’C)
Cep70_C2_R) GTTCTAGAGGCTCGAGAagggctgctgagtactgag (Tm:59.5’C)
Cep70_Cep70M_F) GGA TCC CAT CGA TTC cgg gcc cct cag ctc ct (Tm:61.8’C)
Cep70_Cepp70M_R) GTTCTAGAGGCTCGAGA ctaatgcgacgctcgtgcct (Tm:62.5’C)
Deup1 (XM_018247783) was amplified from Xenopus laevis cDNA at NF stage 20–22 then cloned into GFP or RFP tagged pCS2+ vector with StuI + Xbal. For generating the Deup1 Xenopus laevis transgenic line, the α-tubulin promoter (TUBA1A-B on Scaffold 127187) was cloned in front of GFP-Deup1 in the IsceI vector. The Tub-GFP-Deup1 transgenic was generated and maintained at the National Xenopus Resource Center (NXR). mRNAs of each of the DNA constructs were synthesized with the Sp6 mMessage Machine kit (Life Technologies, AM1340) and purified by RNeasy MinElute Cleanup Kit (QIAGEN, 74204).
Co-Immunoprecipitation/Western Blot:
Embryos were injected in each blastomere at the four-cell stage with 200pg of mRNAs for each condition and animal caps were isolated at NF stage 10.5–12. The animal caps were reared in 0.5X Marc’s Modified Ringer’s (MMR) media until the desired stage (stage 22–24). For co-IP, the standard protocol was performed with FLAG beads (Sigma, A2220) and binding with the beads was done at 4°C for 4–6hours. Following IP we performed a western blot using anti-GFP Ab (1:2000, Aves Labs Inc, GFP-1020) followed by either donkey anti-chicken HRP secondary Ab (1:5000, Fisher) or goat anti-chicken HRP Ab (1:2000, Thermo). The membrane was developed with Pierce ECL Western Blotting Substrate (Thermo, 32109) following the manufacture’s protocol.
MO analysis:
Tub-GFP-Deup1 Transgenic Xenopus laevis embryos (Transgenic female × WT male, whose yield was approximately 50% transgenic) were injected at the 2-cell stage with centrin-RFP RNA in all four quadrants. At the 4-cell stage, one blastomere was injected with either Cep70 MO (5’ GCTGCTCTGACATCTCCCCTTCTTC 3’, Gene-Tools, Inc.) or Control MO (5’ CCTCTTACCTCAGTTACAATTTATA 3’, Gene-Tools. Inc.) with Dextran-blue dye. Embryos were grown until stage 28 and fixed in 3% PFA/PBS solution and stained with 1:500 phalloidin 670 and then imaged. hGR-MCIDAS MO experiments were performed similarly except for the addition of hGR-MCIDAS and the treatment with Dexamethasone at Stage 10.5.
CRISPR analysis:
Tub-GFP-Deup1 embryos were injected at the 1-cell stage with Cas9 protein and a mixture of guides at 1:10 (500 ng/μL). Guides were synthesized from an oligo guide template (obtained by PCR) with a T7 site and universal reverse template and using purified PCR product as a T7 template. The T7 transcription reaction went overnight and was then purified using GE Illumina columns and EtOH precipitation, then diluted and apportioned into 500 ng/μL aliquots and stored at −80°C until use. Single guide RNA (sgRNA) was designed using CHOPCHOP. sg-RNAs were used as a mix to increase likelihood of editing. The following guides were used: Exon 2 (−strand GTTCTTCCTTTGCAGCTGAAGG), Exon 5 (+strand GCCAAAATCCGGGATCTAGAGG), Exon8 (+strand GAGATATAAGGTGGCTGATGTGG) and Exon9 (+strand TGCAGCAGGAGTTGGAGGGCAGG). Embryos were grown to stage 28 and fixed in ice-cold Methanol at −20°C for 48 hours, then rehydrated in Methanol series and incubated in 10% HIGS, then 1:100 anti-centrin Ab (Millipore 04–1624) and 1:150–1:200 anti-ZO1 Ab (Thermo 61–7300) overnight at 4°C, incubated with Cy3 anti-mouse secondary Ab and Cy5 anti-rabbit secondary Ab.
Imaging and analysis:
All microscopy was performed on a Nikon A1R laser scanning confocal microscope using a 60X oil Plan-Apo objective (1.4 N.A.). Data analysis was performed using either Nikon NIS Elements or using Fiji (Schindelin et al., 2012). Apical area and centriole number were measured manually.
Supplementary Material
Supplemental Figure 1. Full Western Blots from Figure 1. A, Full Western Blot of Co-IP data presented in Figure 1C with lanes as labeled. B, Full Western Blot of Co-IP data presented in Figure 1D with lanes as labeled. C, Full Western Blot of Co-IP data presented in Figure 1E with lanes as labeled.
Supplemental Figure 2. Alignment of the conserved C terminal of Cep70 proteins. BOXSHADE analysis of CRC70 from C. reinhadtii and Cep70 from X. laevis, D. rerio, M. musculus, H. sapiens. The TPR domain identified in the human protein is outlined in red and the conserved Cep70 motif is identified in blue.
Supplemental Figure 3. Non-MO and Cep70 OE controls for centriole quantification. Quantification of centriole numbers in Non-MO embryos, with or without GFP-Cep70 as well as Control MO with or without GFP-Cep70 along with statistical comparisons (n > 40 cells).
Highlights.
Cep70 is a novel component of the deuterosome that biochemically interacts with CCDC78 and Deup1.
Cep70 dynamically associates with the deuterosome during development of multiciliated cells.
Morpholino or CRISPR depletion of Cep70 dramatically decreases centriole biogenesis in multiciliated cells.
Acknowledgements:
This work was supported by funding from NIH-NIGMS (GM089970). We would like to thanks Chris Kintner for generously sharing reagents. We would especially like to thank the National Xenopus Resource Center and the Marine Biological Laboratory (MBL) for technical support and reagents. Additionally, aspects of this work were supported by a Whitman Fellowship from the MBL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Al Jord A, Shihavuddin A, Servignat d’Aout R, Faucourt M, Genovesio A, Karaiskou A, Sobczak-Thepot J, Spassky N, Meunier A, 2017. Calibrated mitotic oscillator drives motile ciliogenesis. Science 358, 803–806. [DOI] [PubMed] [Google Scholar]
- Balestra FR, Strnad P, Fluckiger I, Gonczy P, 2013. Discovering regulators of centriole biogenesis through siRNA-based functional genomics in human cells. Dev Cell 25, 555–571. [DOI] [PubMed] [Google Scholar]
- Blachon S, Gopalakrishnan J, Omori Y, Polyanovsky A, Church A, Nicastro D, Malicki J, Avidor-Reiss T, 2008. Drosophila asterless and vertebrate Cep152 Are orthologs essential for centriole duplication. Genetics 180, 2081–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JP, Mayahara H, Yoloyama M, Ubukata A, Moller PC, 1979. An ultrastructural study of morphogenesis of fibrogranular complex and centriole in ductuli efferentes of Chinese hamster. Tissue Cell 11, 401–412. [DOI] [PubMed] [Google Scholar]
- Collins C, Majekodunmi A, Mitchell B, 2020. Centriole Number and the Accumulation of Microtubules Modulate the Timing of Apical Insertion during Radial Intercalation. Curr Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL, Sluder G, Cassels G, Sibon O, Wang CL, 2002. De novo formation of centrosomes in vertebrate cells arrested during S phase. J Cell Biol 158, 1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Ma L, Shokhirev MN, Quigley I, Kintner C, 2018. Multicilin and activated E2f4 induce multiciliated cell differentiation in primary fibroblasts. Sci Rep 8, 12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klos Dehring DA, Vladar EK, Werner ME, Mitchell JW, Hwang P, Mitchell BJ, 2013. Deuterosome-mediated centriole biogenesis. Dev Cell 27, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo PA, 2017. Is Centrosomal Protein 70, a Centrosomal Protein with New Roles in Breast Cancer Dissemination and Metastasis, a Facilitator of Epithelial-Mesenchymal Transition? Am J Pathol 187, 494–497. [DOI] [PubMed] [Google Scholar]
- Mercey O, Levine MS, LoMastro GM, Rostaing P, Brotslaw E, Gomez V, Kumar A, Spassky N, Mitchell BJ, Meunier A, Holland AJ, 2019. Massive centriole production can occur in the absence of deuterosomes in multiciliated cells. Nat Cell Biol 21, 1544–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, Cetin C, Sieckmann F, Pau G, Kabbe R, Wunsche A, Satagopam V, Schmitz MH, Chapuis C, Gerlich DW, Schneider R, Eils R, Huber W, Peters JM, Hyman AA, Durbin R, Pepperkok R, Ellenberg J, 2010. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464, 721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Holland AJ, 2018. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat Rev Mol Cell Biol 19, 297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riba A, Itzhaki LS, 2019. The tetratricopeptide-repeat motif is a versatile platform that enables diverse modes of molecular recognition. Curr Opin Struct Biol 54, 43–49. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Li D, Wang Y, Liu S, Qin J, Wang J, Ran J, Zhang Y, Huang Q, Liu X, Zhou J, Liu M, 2017a. Discovery of Centrosomal Protein 70 as an Important Player in the Development and Progression of Breast Cancer. Am J Pathol 187, 679–688. [DOI] [PubMed] [Google Scholar]
- Shi X, Sun X, Liu M, Li D, Aneja R, Zhou J, 2011. CEP70 protein interacts with gamma-tubulin to localize at the centrosome and is critical for mitotic spindle assembly. J Biol Chem 286, 33401–33408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Wang J, Yang Y, Ren Y, Zhou J, Li D, 2012. Cep70 promotes microtubule assembly in vitro by increasing microtubule elongation. Acta Biochim Biophys Sin (Shanghai) 44, 450–454. [DOI] [PubMed] [Google Scholar]
- Shi X, Wang Y, Sun X, Wang C, Jiang P, Zhang Y, Huang Q, Liu X, Li D, Zhou J, Liu M, 2017b. Centrosomal Protein 70 Is a Mediator of Paclitaxel Sensitivity. Int J Mol Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Yao Y, Wang Y, Zhang Y, Huang Q, Zhou J, Liu M, Li D, 2015. Cep70 regulates microtubule stability by interacting with HDAC6. FEBS Lett 589, 1771–1777. [DOI] [PubMed] [Google Scholar]
- Shiratsuchi G, Kamiya R, Hirono M, 2011. Scaffolding function of the Chlamydomonas procentriole protein CRC70, a member of the conserved Cep70 family. J Cell Sci 124, 2964–2975. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM, 2007. Xenopus laevis In Vitro Fertilization and Natural Mating Methods. CSH protocols 2007, pdb prot4737. [DOI] [PubMed] [Google Scholar]
- Spassky N, Meunier A, 2017. The development and functions of multiciliated epithelia. Nat Rev Mol Cell Biol 18, 423–436. [DOI] [PubMed] [Google Scholar]
- Stubbs JL, Vladar EK, Axelrod JD, Kintner C, 2012. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat Cell Biol 14, 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terre B, Piergiovanni G, Segura-Bayona S, Gil-Gomez G, Youssef SA, Attolini CS, Wilsch-Brauninger M, Jung C, Rojas AM, Marjanovic M, Knobel PA, Palenzuela L, Lopez-Rovira T, Forrow S, Huttner WB, Valverde MA, de Bruin A, Costanzo V, Stracker TH, 2016. GEMC1 is a critical regulator of multiciliated cell differentiation. EMBO J 35, 942–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truebestein L, Leonard TA, 2016. Coiled-coils: The long and short of it. Bioessays 38, 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner ME, Hwang P, Huisman F, Taborek P, Yu CC, Mitchell BJ, 2011. Actin and microtubules drive differential aspects of planar cell polarity in multiciliated cells. J Cell Biol 195, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner ME, Mitchell BJ, 2013. Using Xenopus Skin to Study Cilia Development and Function, in: Marshall WF (Ed.), Methods in Enzymology, Volume 525: Cilia, Part B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner ME, Mitchell JW, Putzbach W, Bacon E, Kim SK, Mitchell BJ, 2014. Radial intercalation is regulated by the Par complex and the microtubule-stabilizing protein CLAMP/Spef1. J Cell Biol 206, 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson CJ, Carl M, Harris WA, 2009. Cep70 and Cep131 contribute to ciliogenesis in zebrafish embryos. BMC Cell Biol 10, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ran J, Liu M, Li D, Li Y, Shi X, Meng D, Pan J, Ou G, Aneja R, Sun SC, Zhou J, 2014. CYLD mediates ciliogenesis in multiple organs by deubiquitinating Cep70 and inactivating HDAC6. Cell Res 24, 1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Mitchell BJ, 2015. Centriole biogenesis and function in multiciliated cells. Methods Cell Biol 129, 103–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Zhu L, Zhu Y, Cao J, Li S, Huang Q, Xu T, Huang X, Yan X, Zhu X, 2013. The Cep63 paralogue Deup1 enables massive de novo centriole biogenesis for vertebrate multiciliogenesis. Nat Cell Biol 15, 1434–1444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Full Western Blots from Figure 1. A, Full Western Blot of Co-IP data presented in Figure 1C with lanes as labeled. B, Full Western Blot of Co-IP data presented in Figure 1D with lanes as labeled. C, Full Western Blot of Co-IP data presented in Figure 1E with lanes as labeled.
Supplemental Figure 2. Alignment of the conserved C terminal of Cep70 proteins. BOXSHADE analysis of CRC70 from C. reinhadtii and Cep70 from X. laevis, D. rerio, M. musculus, H. sapiens. The TPR domain identified in the human protein is outlined in red and the conserved Cep70 motif is identified in blue.
Supplemental Figure 3. Non-MO and Cep70 OE controls for centriole quantification. Quantification of centriole numbers in Non-MO embryos, with or without GFP-Cep70 as well as Control MO with or without GFP-Cep70 along with statistical comparisons (n > 40 cells).


