Abstract
Recent advances have begun to clarify the physiological and pathological roles of non-coding RNAs (ncRNAs) in various diseases, including cancer. Among these, microRNAs (miRNAs) have been the most studied and have emerged as key players that are involved in the regulation of important growth regulatory pathways in cancer pathogenesis. The ability of a single ncRNA to modulate the expression of multiple downstream gene targets and associated pathways, have provided a rationale to pursue them for therapeutic drug development in cancer. In this context, early data from pre-clinical studies have demonstrated that synthetic miRNA-based therapeutic molecules, along with various protective coating approaches, has allowed for their efficient delivery and anti-tumor activity. In fact, some of the miRNA-based cancer therapeutic strategies have shown promising results even in early-phase human clinical trials. While the enthusiasm for ncRNA-based cancer therapeutics continue to evolve, the field is still in the midst of unraveling a more precise understanding of the molecular mechanisms and specific downstream therapeutic targets of other lesser studied ncRNAs such as the long-non-coding RNAs, transfer RNAs, circular RNAs, small nucleolar RNAs, and piwi-interacting RNAs. This review article provides the current state of knowledge and the evolving principles for ncRNA-based therapeutic approaches in cancer, and specifically highlights the importance of data to date and the approaches that are being developed to overcome the challenges associated with their delivery and mitigating the off-target effects in human cancers.
Keywords: non-coding RNAs, microRNAs, long non-coding RNAs, piRNAs, snoRNAs, cancer, therapy
1. INTRODUCTION
Decades of accumulating research indicates that dysregulated expression of certain genes in critical growth regulatory pathways is a major driver of oncogenesis in human malignancies. Although the prevailing consensus is that altered gene expression is causally related to cancer pathogenesis, the underlying mechanisms driving the neoplastic growth of cancer cells are far more complex. Extensive investigations in the context of genetic causes of cancer have revealed that aberrant gene expression is not only a consequence of protein-coding genes, but to a large extent is also mediated by the regulatory actions of non-coding genomic elements in the human genome. The Encyclopedia of DNA Elements (ENCODE) transcriptome project concluded that only ~1.2% of the genome comprises protein-coding genes, whereas ~80% of it is actively transcribed into a variety of non-coding RNAs (ncRNAs), some of which have been characterized, and some of which are under active interrogation [1]. Although ncRNAs were initially deemed as “transcriptional noise,” “junk DNA,” or “dark genomic matter,” research in the past two decades has provided convincing and irrefutable evidence favoring biological roles for various types of ncRNAs in various diseases, including cancer [2]. Broadly speaking, all ncRNAs can be divided into two categories based on size: small ncRNAs (sncRNAs), which are shorter than 200 nucleotides and long ncRNAs (lncRNAs) that are longer than 200 nucleotides. The sncRNA category includes microRNAs (miRNAs), transfer RNAs (tRNAs), piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNA) [3]. Although specific biological functions for some sncRNAs continue to be realized and appreciated, a fascinating theme that has emerged to date is that hierarchically, ncRNAs represent a higher-level gene regulatory domain, and a single ncRNA is theoretically capable of controlling the expression of many downstream gene (mRNA) targets.
In view of the increased recognition of the biological roles of ncRNAs in various diseases, it is not surprising that recent years have seen a concerted effort to evaluate the translational and clinical significance of sncRNAs in cancer and other diseases [4–10]. Given that a single sncRNA can control the expression of several mRNA targets in distinct cancer-associated pathways, an early hypothesis was that using a ncRNA-based therapeutic approach would address the issue of the multi-faceted nature of cancer pathogenesis and resultant tumor heterogeneity present in various cancers. Indeed, numerous studies and clinical trials have already been initiated to leverage this aspect of ncRNAs, and ncRNA-based anti-cancer drug development has gained significant momentum and is potentially ripe for breakthroughs [11]. Based upon the evidence gathered to date, this review evaluates the advantages and challenges associated with ncRNA-based cancer therapy, and summarizes the knowledge surrounding emerging therapeutic strategies for the application of ncRNAs in cancer treatment.
2. MICRORNA (MIRNA)-BASED THERAPY IN CANCER
While the evidence for miRNA-based cancer therapy in cancer is still in relative early stages, burgeoning body of literature and scientific evidence indicates that this concept has merit, while additional research is needed to overcome existing challenges as we improve our understanding for their functional downstream targets.
2.1. miRNAs in Cancer
miRNAs are endogenous single-stranded sncRNAs that are 18–25 nucleotides in length and are present in animals, plants, and some viruses. miRNAs bind to the 3’-untranslated regions of target genes that regulate cellular processes including the cell cycle, apoptosis, cellular development, differentiation, and metabolism. miRNAs were first discovered in 1993 in C. elegans [12, 13], but the first evidence for aberrant miRNA expression and its biological consequences in human cancer were not revealed until 2002, when Calin and colleagues identified genomic alterations in the miR-15a/16 cluster in leukemia [14]. Publication of this seminal study paved the way for a plethora of investigations describing miRNA dysregulation in cancer. Moreover, microarray profiling has since demonstrated dysregulated expression of various miRNAs in a variety of tumor types, highlighting the functional relevance of these sncRNAs in oncogenesis [15]. Today, miRNAs are by far the most studied ncRNAs in cancer, with more than 80,000 publications catalogued in PubMed involving miRNAs and cancer.
A single miRNA has the potential to modulate the expression of multiple genes and influence various biological systems in a specific manner. Cancer-associated miRNAs are generally classified into one of two subcategories: tumor suppressor miRNAs (or “tumor-suppressor-miRs”) and oncogenic miRNAs (or “onco-miRs”). Examples of well-characterized tumor-suppressor-miRs include miR-34a, miR-145, and the let-7 family; well-established onco-miRs include miR-21 and miR-155. Interestingly, several miRNAs appear to possess dual functionality, acting as both as a tumor suppressor and as an oncogene. For instance, although miR-200c inhibits epithelial-to-mesenchymal transition (EMT) and blocks initiation of cancer metastasis, it is also frequently overexpressed in late-stage cancers and involved in promoting distant metastasis [8, 16, 17]. Therefore, miR-200c appears to have both tumor suppressor and oncogenic functions, which manifest in a context-dependent manner, and relate to specific stages of carcinogenesis. While the enthusiasm continues to grow, presently, the ncRNA-based cancer research still is not as mature and well-established as classical protein-encoding genes. This highlights the need for continued research in developing a better understanding of the role of ncRNAs in cancer, identification of signaling pathways and the specific downstream genetic targets, as this will all be pivotal in a better realization of their therapeutic potential.
2.2. Clinical Application of miRNAs in Cancer Therapy
Many miRNAs inhibit cellular signaling pathways by suppressing the expression of multiple genes within a single growth regulatory pathway. Additionally, due to crosstalk between various signaling pathways, miRNAs can theoretically affect the functionality of multiple interconnected growth regulatory pathways all at once – which, provides an attractive rationale for the development of miRNA-based cancer therapeutics. Cancer development is a heterogeneous process that evolves by negating the impact of various environmental stressors. Most cancer cells acquire resistance to therapeutic drugs by activating survival signals, evading immune responses, and blocking programmed cell death or apoptotic pathways [18]. Not surprisingly, majority of therapeutic agents that target a single gene or pathway are effective for only for a limited time period, until the cancer cells figure out alternate mechanisms to evade the efficacy of a specific drug – a concept that has been the Achilles heel of most anti-cancer modalities to date. Cancer therapeutic strategies that modulate the expression of miRNAs in tumors act by restoring miRNA expression, replenishing endogenously depleted miRNAs, or inhibiting overexpressed miRNAs through the application of small-molecule antagonists. Because a number of miRNAs are epigenetically silenced due to hypermethylation of their promoter regions, clinically-approved hypomethylating agents such as 5-azacytidine and decitabine were initially considered as potential therapeutic options [19]. However, these drugs are non-specific and cause global demethylation of all genomic DNA, which leads to the upregulation of many other epigenetically silenced miRNAs and protein-coding genes. To overcome this challenge, recent technological advancements have allowed for the development of novel miRNA mimics and inhibitors that can regulate the expression of specific miRNAs for precise therapeutic application in cancer.
2.3. miRNA Mimics in Cancer Therapy
To date, preclinical studies using ectopically overexpressed miRNAs far outnumber those that suppress miRNA expression. Expression vectors and oligonucleotide-based mimics are currently the most commonly used methods to synthetically overexpress target miRNAs. The miRNA mimics are typically double-stranded synthetic oligonucleotides that, upon entering cancer cells, are processed by the cellular machinery to form active, single-stranded molecules. These synthetic oligonucleotides subsequently incorporate into the RNA-induced silencing complex (RISC) and acquire phosphorothioate backbone modifications. However, during this process, a significant portion of backbone modifications are lost before the molecules can reach their target locations, which presents a challenge for their effective therapeutic clinical application [20]. Furthermore, because double-stranded RNAs that are not formulated for stable durability within the body, degrade quickly within biological fluids, emphasizing the need for unique protective delivery systems that must be designed to maintain a sufficiently effective dosage [21]. Such well-recognized inefficiencies of synthetic oligonucleotides are not unique to miRNA-based therapeutics, and essentially overlap with those of other conventional molecular therapeutic drugs. To overcome these technical impediments for effective drug delivery, chemical modifications for a newer generation of molecular mimics are generally designed to optimize RNA oligonucleotide stability, while limiting potential off-target effects. Collectively, these recent technological advancements have significantly advanced the use of miRNA-based molecules in cancer therapeutics, but more research is needed—and is currently underway—to address the enduring challenges associated with their delivery.
Nonetheless, a number of well-characterized miRNAs have already passed the initial litmus test and are now in early-phase clinical testing. One such example is MRX34, a synthetic mimic of the highly studied tumor suppressor miR-34a that suppresses metastasis and stemness in various cancers, which was tested in lung cancer patients [22]. While the clinical trial with MRX34 was halted due to immune-related adverse effects in a small group of patients, it did exhibit antitumor activity in a subset of patients with refractory advanced solid tumors [23]. Considering that a single miRNA modulates the expression of multiple genes, side effects observed during the MRX34 trial were not entirely surprising. Regardless, this study suggested that some miRNA mimics might require combinatorial administration along with another drug to minimize their side effects and provided an important proof-of-concept, confirming findings from previous preclinical experiments and demonstrating promise for clinical application of miRNA mimics as cancer therapeutics.
2.4. miRNA Inhibitors as Cancer Therapeutic Agents
As for miRNA mimics, suppression of onco-miRs through the use of miRNA inhibitors is of significant research interest. In contrast to miRNA mimics, which are predominantly created using oligonucleotide-based technologies, several distinct methods are used to generate miRNA inhibitors. It is also easier and more practical to design miRNA inhibitors compared to miRNA mimics, due to the specific structural and pharmacokinetic properties of miRNA inhibitors [24]. The Figure 1 illustrates various strategies to inhibit, as well as promote, the expression of miRNAs in cancers, hence providing a rationale for their clinical application as cancer therapeutic agents.
Figure 1.
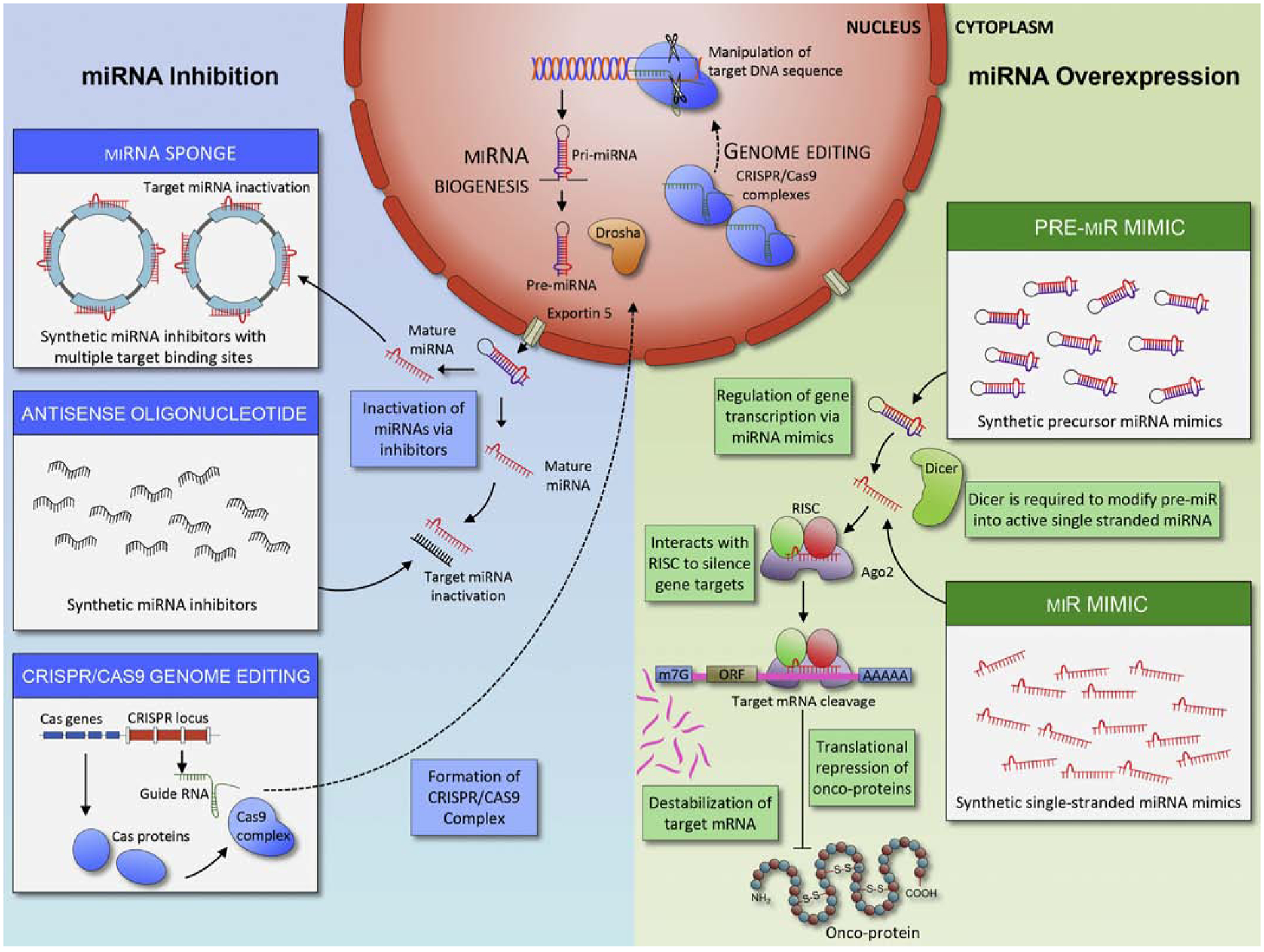
Modulation of miRNA expression in a tumor. Modulation of the expression of miRNAs can occurs via several distinct strategies. To achieve overexpression of miRNAs (green), miRNA mimics enter tumor cells as “pre” (Top right) or “mature” (Bottom right) miRNA forms. Dicer cleaves pre-miRNAs into short RNA fragments. Subsequently, miRNA forms complex with RISC and Ago2 proteins and binds to DNA to suppress RNA transcription. In contrast, the suppression of miRNA expression in a tumor (blue) can occur via several methodologies, including miR-sponges (top left), and antisense oligonucleotides (middle left) and CRISPR/Cas9 genome editing (bottom left). For genome editing, CRISPR/Cas9 complex enters nucleus and changes DNA sequence of target miRNAs. Both miRNA sponge and antisense oligonucleotides bind to miRNAs and inhibit their functions.
2.4.1. Oligonucleotide-based Approach:
Antisense oligonucleotides (ASOs) are single-stranded RNAs that are complementary to sense segments of target mRNA sequences [25] and function as competitive inhibitors to suppress the expression of target miRNAs. Locked nucleic acids (LNAs) are ASOs substituted with several bicyclic RNA analogues that form a “locked” conformation, which provides high intrinsic target affinity and suitability for inhibiting short RNA and DNA segments [26]. The locked ribose conformation ideally targets Watson-Crick binding and significantly improves hybridization to oligonucleotides for targeting miRNAs [27]. Currently, ASO is the most commonly used method for targeting specific miRNAs. In addition, recently heteroduplex oligonucleotide antimir has been developed and demonstrated that structural modification resulted in substantial improvement of target suppression efficiency [28].
2.4.2. miRNA Sponges:
Similar to ASO, miRNA sponges are typically DNA plasmids or transcribed RNAs, that contain multiple miRNA binding motifs [29]. miRNA sponges are designed to contain binding sites complementary to heptamers in the seed sequences of target miRNAs; thus, a single miRNA sponge can block an entire family of miRNAs containing the same target binding sequence [30]. Although in vitro studies have demonstrated that miRNA sponges inhibit specific miRNAs and downregulate their respective downstream targets, significantly higher concentrations (i.e. much higher than for ASO-based inhibitors) are required to achieve effective target mRNA inhibition, which can potentially increase unwanted off-target effects [31].
2.4.3. CRISPR/Cas9 Genome Editing:
Clustered regularly interspaced short palindromic repeats (CRISPR) is a novel adaptive immune system found in many prokaryotes that encounter invading foreign genetic elements. These unique sequences in the genomic loci, discovered 30 years ago in Escherichia coli, serve as a defense mechanism against viruses. CRISPR sequences, along with associated enzymes such as CRISPR-associated protein 9 (Cas9), can cut nucleic acids and disable viruses. In the type II CRISPR system, the Cas9 protein partially complexes with repeat sequences in a spacer-containing RNA and transactivating CRISPR RNA (tracrRNA). The CRISPR/Cas9 complex cleaves foreign genetic material upon recognition of target gene sequences. The use of this system of cutting and inserting nucleic acids has recently become a popular method for editing the genome [32]. To date, the CRISPR/Cas9 system has been exploited to knock out various ncRNAs, including miRNAs, lncRNAs, and small nucleolar RNAs (snoRNAs) [33–35]. CRISPR is efficient, specific, and cost-effective. However, because liposome or nanoparticle-based systems are not very proficient at delivering CRISPR/Cas9 machinery to cells, this system requires viral vector delivery to achieve high delivery efficacy and therefore is not currently very popular in therapeutic settings [36]. Nevertheless, as CRISPR becomes the foremost genome-editing strategy, its integration into clinical applications may eventually replace the use of oligonucleotide-based inhibitors.
2.5. Delivery Strategies
Despite the availability of various mimics and inhibitors, it is difficult to modulate the expression of ncRNAs in tumors without first improving specific pharmacological barriers. Synthetic ncRNA mimics and inhibitors generally degrade rapidly in biological fluids, absorb poorly into the intracellular space, and often may fail to reach specific target locations [37]. Chemically modifications of RNA backbones prolongs the half-life of ncRNAs in vivo and could significantly alter intracellular interaction with target ncRNAs [38]. Furthermore, artificial protective coatings, such as nanoparticles and polymer-based formulations, are popular methods that protect fragile synthetic ncRNAs [39]. In the following section, we describe various delivery systems and their potential advantages and disadvantages.
2.5.1. Liposomes
Liposomes are lipid-bilayer compounds composed of aqueous cores surrounded by hydrophobic membranes. They are commonly used in various manufactured products, from dietary supplements to cosmetics and recognized as a relatively safe delivery method. Therefore, it is not surprising that liposomes are the most commonly used delivery vehicles for miRNA mimics and inhibitors [21]. Through years of compositional and structural optimization, the current generation of liposome-based delivery systems offer highly efficient delivery to a target cell while maintaining high loading capacity [40].
2.5.2. Cationic polymers
The cationic polymer polyethylene imine (PEI) is another popular delivery technology commonly used for the transfer of plasmid DNA and small interfering RNA (siRNA) [41, 42]. Polymer PEI coating provides a net cationic charge, which shields molecules from enzymatic degradation, while enhancing the interaction with anionic cell membrane polysaccharides [31]. Although the delivery efficiency of polymer PEI coating is high, a cationic-based delivery system is often hampered by excessive interactions with serum proteins [43].
2.5.3. Peptide based transmembrane structures
Certain structural modifications can stabilize miRNA mimics and inhibitors for improved delivery. One standard structural modification used for drug delivery attaches a peptide with a low pH-induced transmembrane structure (pHLIP) to miRNA inhibitors [44]. This peptide forms a transmembrane alpha-helix when it is exposed to acidic conditions, such as those normally found in tumors. The pHLIP structure can then translocate molecules such as anti-miRs across the cell membrane through a non-endocytic route, resulting in high drug delivery efficiency [45, 46].
2.5.4. Adeno-associated viral vectors
Use of an adeno-associated virus (AAV) system is another intriguing technology for miRNA delivery. For example, an AAV was successfully used to deliver miRNA mimics through tail vein injections in a murine model of hepatocellular carcinoma [47]. However, safety concerns associated with any viral delivery system complicate potential clinical applications.
2.5.5. Exosomes
Exosomes were initially discovered three decades ago as cellular fragments attributed as a waste disposal mechanism; however, these small extracellular vehicles are now a topic of active interrogation due to their recognition as important mediators of cellular signalling in cancer [9, 48, 49]. Exosomes are small, 40–140 nm in size, membrane vesicles that inherit molecular cargo of their cell of origin [50–52], and contribute to cancer pathogenesis [53–55]. Intriguingly, a recent study showed that cancer exosomes contain miRNA-processing complexes that convert pre-miRNAs to mature-miRNAs, enriching cancer exosomes with miRNAs – a mechanism that is absent in normosomes (exosomes, originating from non-cancerous cells) [56]. Furthermore, knockdown of Vps4a, an exosome packaging protein frequently dysregulated in hepatocellular carcinoma, was shown to alter exosomal miRNA expression profiles [57]. This led to several studies validating the functional role of exosomal miRNAs in oncogenesis [58, 59]. By virtue of their small size, encapsulating miRNAs in exosomes is relatively simple [60]. Standard transfection methods can be used to introduce synthetic oligonucleotides into exosomes and surface ligands can be genetically modified to improve the target specificity of the exosomes. The biological origin of exosomes ensures high biocompatibility with their target tumor types and minimizes toxicity. Therefore, these tiny vesicles show significant potential as alternative delivery vehicles for ncRNAs.
2.6. Preclinical miRNA-based Cancer Therapy
As our understanding for the molecular mechanisms involving miRNAs has improved over the past decades, scientific evidence continues to build with regards to their efficacy as anti-cancer therapeutics in pre-clinical model systems.
2.6.1. Tumor-suppressor-miRs:
During the last decade, use of short interfering RNA (siRNA) and short hairpin RNA (shRNA)-based pre-clinical animal studies have resulted in an improved understanding and a better characterization of the functional roles of miRNAs in cancer [61]. The results obtained from these efforts have prompted the use of miRNA mimic-based cancer therapeutics in pre-clinical settings, as summarized in Table 1.
Table 1.
A list of miRNA-mimics examined for their cancer therapeutic potential in pre-clinical studies*
| miRNA | Year | Delivery system | Cancer | Mechanisms (Target genes) | Target genes | Model | Methodology | Ref |
|---|---|---|---|---|---|---|---|---|
| miR-26 | 2009 | Adeno-associated virus | Liver | G1 cell cycle arrest | CCND2, CCNE2 | In vivo | tet-oMYC;LAP-tTA | (36) |
| miR-34a | 2010 | Nanoparticles | Lung | Tumor load reduction | Survivin | In vivo | Intravenous | (60) |
| miR-34a & 143/145 | 2011 | Nanoparticles | Pancreatic | Apoptosis enhancement | SIRT1, CD44, ALDH1, KRAS2, RREB1 | In vivo | Xenograft | [73] |
| miR-34a | 2012 | Nanoparticles | Neuroblastoma | Vascularization reduction | TIMP2 | In vivo | Xenograft | (58) |
| miR-107 | 2012 | Nanoparticles | Head and neck | Cancer initiation cell reduction | NANOG, OCT3/4, SOX4 | In vivo | Xenograft | (68) |
| miR-155 | 2012 | Nanoparticles | Ovarian | Immunosuppressive mediator suppression | - | In vivo | Orthotopic | (126) |
| miR-5, 10, 7 | 2012 | Adeno-associated virus | Retinal | Endogenous VEGF suppression | VEGF | In vivo | Intramuscular | (70) |
| Let-7a | 2013 | Nanoparticles | Lung | Proliferation reduction | RAS | In vitro | 3D tumor growth assay | (62) |
| miR-29b | 2013 | Nanoparticles | Leukemia | Growth reduction | DNMTs, CDK6, SP1, KIT, FLT3 | In vivo | Intravenous | (125) |
| miR-200c | 2013 | Nanoparticles | Ovarian, Lung, Real Breast | Tumor angiogenesis reduction | IL-8 and CXCL1 | In vivo | Intrapulmonary | (10) |
| miR-203 | 2013 | Nanoparticles | Esophageal | Proliferation and migration reduction | Ran, ΔNp63 | In vitro | Cell lines | (65) |
| miR-1 | 2014 | Nanoparticles | Glioblastoma | GBM sphere reduction | MET, EGFR | In vitro | Patient derived stem cells | (69) |
| miR-16 | 2014 | Nanoparticles | Gastric | Apoptosis enhancement | - | In vivo | Xenograft | (123) |
| miR-34a | 2014 | Stable lipid particles | Multiple myeloma | Apoptosis enhancement | ERK2, AKT | In vivo | Orthotopic | (59) |
| miR-34a | 2014 | Nanoparticles | Breast | Migration reduction | NOTCH 1 | In vivo | Xenograft | (57) |
| Let-7b | 2015 | Neutral lipid emulsion | Lung | Tumor growth reduction | MYC | In vivo | Kras; P53 model | (55) |
| miR-34a | 2015 | Neutral lipid emulsion | Lung | Proliferation reduction | MET, MYC | In vivo | Kras; P53 model | [69] |
| miR-34a | 2015 | Nanoparticles | Prostate | Non-canonical autophagy enhancement | MET, AXL, MYC | In vivo | Femur | (56) |
| miR-145 | 2015 | Lipofectamine RNAi max | Bladder | Apoptosis enhancement | MYC, SOCS7, FSCN1, CDH1 | In vivo | Xenograft | [79] |
| miR-145 | 2015 | PEI nanocarrier | Prostate | Proliferation reduction | FSCN1, MYC, IGFIR | In vivo | Xenograft | [78] |
| miR-495 | 2015 | Neutral lipid emulsion | Lung | Tumor burden and Proliferation reduction | - | In vivo | Xenograft | (122) |
| miR-514-3p | 2015 | Nanoparticles | Neuroblastoma | Apoptosis enhancement | Survivin | In vivo | Xenograft | (124) |
| Let-7g | 2016 | Nanoparticles | Liver | Cell survival enhancement | MYC | In vivo | Orthotopic | [175] |
| miR-542 | 2016 | Nanoparticles | Breast | Apoptosis enhancement | P53 activation | In vitro | Cell lines | [176] |
| miR-34a | 2017 | Nanoparticles | Colon | Apoptosis enhancement | BCL-2 | In vivo | Xenograft | [177] |
| miR-542 | 2017 | Nanoparticles | Gastric | Apoptosis enhancement | P53 activation | In vivo | Xenograft | [178] |
| miR-7 | 2018 | Nanoparticles | Ovary | Chemosensitization enhancement | EGFR/ERK pathway | In vivo | Xenograft | [179] |
| miR-20a | 2018 | Nanoparticles | Colon | Liver metastasis reduction | ARHGA P1 and E2F1 | In vivo | Liver metastasis model | [180] |
| miR-27a | 2018 | Nanoparticles | Liver | Proliferation reduction | FOXO1, PPAP-γ | In vivo | Xenograft | [181] |
| miR-375 | 2018 | Nanoparticles | Liver | Autophagy and Proliferation reduction | - | In vivo | Xenograft | [182] |
| miR-27 | 2019 | Nanoparticles | Liver | Apoptosis enhancement | FOXO1, PPAR | In vivo | Xenograft | [183] |
| miR-122 | 2019 | Nanoparticle | Liver | Apoptosis enhancement | ADAM17 | In vivo | Xenograft | [184] |
| miR-125b | 2019 | Nanoparticles | Ovarian | Increased Macrophage repolarization | - | In vivo | Orthotopic | [185] |
| miR-139-5p | 2019 | Nanoparticles | Colon | Proliferation and migration reduction | - | In vivo | Xenograft | [186] |
| miR-143 | 2019 | Nanoparticles | Prostate | Proliferation reduction | UPAR | In vivo | Xenograft | [187] |
| miR-212 | 2019 | Nanoparticles | Pancreas | Chemosensitization enhancement | USP9X | In vivo | PDX | [188] |
| miR-660 | 2019 | Nanoparticles | Lung | Growth reduction | MDM2, p-53 | In vivo | PDX | [189] |
| miR-873 | 2019 | Nanoparticles | Pancreas, breast | Growth reduction | KRAS | In vivo | Xenograft | [190] |
| miR-34a | 2020 | Nanoparticles | Breast | Tumor growth reduction | - | In vivo | Xenograft | [191] |
Chronologically listed
Among various miRNAs, miR-34a is by far the most investigated miRNA in preclinical studies. miR-34a has multiple tumor suppressor-like features: it forms a positive feedback loop with tumor suppressor TP53; targets several key oncogenes including BCL2, MYC, NOTCH1, CCND1, SNAIL1, and SIRT1 [62, 63]; and functions as a switch to suppress the cancer self-renewal and EMT-inducing transcription factor, Snail [64, 65]. However, miR-34a is frequently downregulated in multiple cancer types through aberrant transcriptional regulation, genomic deletions, and promoter hypermethylation [66–68]. Therefore, in order to increase its expression, miR-34a mimics were examined in several preclinical animal models of cancer including lung, breast, pancreatic, and prostate cancer, as well as neuroblastoma – all of which, demonstrated a successful increase in its expression following introduction of miRNA mimics and eventual decrease in tumor growth [69–73]. In a multiple myeloma xenograft model, addition of lipid-particle-coated miR-34a inhibited apoptosis by suppressing Erk2 and Akt, which in turn attenuated tumor growth [74]. Another study in a mouse model of lung cancer revealed that liposome-encapsulated miR-34a suppressed an alternative apoptosis inhibitor, Birc1, and reduced tumor burden [75]. Intriguingly, results from a recent study in lung adenocarcinoma cell lines demonstrated that the miR-34a mimic MRX34 improved efficacy of radiotherapy in p53-proficient cancer through suppression of the immune response inhibitor PD-L1 [76], suggesting that miR-34a has immune-boosting potential.
Likewise, miR-143 and miR-145, a well-established putative tumor-suppressor-miR cluster, has been a focus of many studies for its role as a therapeutic target. The expression of these two miRNAs is frequently downregulated in cancer [77], and mechanistically, the miR-143/miR-145 cluster regulates the expression of key oncogenes, including the MYC and FSCN1. First investigated in 2011 in a pancreatic cancer xenograft model, treatment with miR-143 and miR-145 mimics in lipid-based nanoparticles resulted in the overexpression of the tumor suppressor gene, KRAS2, resulting in an overall attenuated tumor growth [73]. The therapeutic efficacy of a miR-145 mimic was subsequently also confirmed several years later in other mouse models of prostate and bladder cancer [78, 79]. Collectively, these results demonstrate that miR-34a, miR-143 and miR-145 mimics have effective tumor suppressor activity across multiple types of cancers.
Given that miRNAs are critical in regulating key oncogenic pathways, several miRNA mimics have the ability to block specific growth signaling pathways. EMT is a well-established mechanism in cancer, which has important implications for tumor metastasis and acquisition of drug resistance [80] – a molecular processes by which epithelial cells lose adhesion and gain mesenchymal-like migratory and invasive properties essential for the ensuing process of metastasis. miRNAs such as the miR-200 family, let-7 family and miR-34a are well-known inhibitors of EMT-inducing transcription factors [80, 81]. In particular, the miR-200 family members and miR-203 form negative feedback loops with the ZEB1 and ZEB2 transcription factors, which act to suppress the epithelial cell adhesion molecule E-cadherin, thereby potently inhibiting the process of EMT [82, 83]. A nanoparticle formulation comprising of miR-203 suppressed cellular proliferation and migration in a number of cell lines including esophageal, ovarian, lung, renal, and breast cancer [84]. Furthermore, liposomal nanoparticle delivery of miR-200c inhibited tumor angiogenesis in renal and breast cancers [17]. Collectively these studies support therapeutic targeting of oncogenic pathways using miRNAs.
Despite well-established association of some miRNAs with oncogenic pathways, an obvious challenge for developing novel miRNA-based therapeutic drugs is the requirement to replenish constitutively downregulated tumor-suppressor-miRs. Unfortunately, to date, only a handful of miRNAs, such as members of the Let-7 and miR-200 families, are accepted as bona fide tumor-suppressor-miRs. However, their reported upregulation in some cancers raises concerns about their mechanisms of action and whether they truly are tumor-suppressor-miRs [8, 85]. Nevertheless, mimics based upon these miRNAs have exhibited significant and effective anti-tumorigenic potential in mouse models of lung, renal, and breast cancers [17, 65, 69, 86].
2.6.3. miRNA Inhibitors:
Similar to tumor-suppressor-miRs, few miRNAs have now been recognized to be bona fide oncogenic miRNAs or onco-miRs. Most miRNAs have expression patterns that differ depending on the organ in which they are expressed; thus, identifying specific miRNA inhibitors that lack or have minimal off-target effects have largely proven to be a challenging task in preclinical studies. Nevertheless, a considerable number of miRNA inhibitors have been evaluated in preclinical animal models (Table 2).
Table 2.
A list of miRNA-inhibitors examined for their cancer therapeutic potential in pre-clinical studies*
| miRNA | Year | Delivery system | Cancer | Mechanism | Target | Model | Ref | |
|---|---|---|---|---|---|---|---|---|
| miR-10b | 2012 | Nanoparticles | Breast | Wound healing reduction | RHOC | In vitro | Cell line | [192] |
| miR-21 | 2012 | Solid lipid nanoparticles | Lung | Proliferation and invasion reduction | - | In vitro | Cell line | [94] |
| miR-155 | 2012 | Nanoparticles | Lymphoma | Tumor growth reduction | - | In vivo | Xenograft | [89] |
| miR-21 | 2013 | Lentiviral vector | Pancreatic | Proliferation and angiogenesis reduction | RhB | In vivo | Xenograft | [93] |
| miR-155 | 2013 | Polymer carrier | Leukemia | LIN28 increased and RUNX1 inhibited | LIN28, RUNX 1 | In vivo | Cell line | [193] |
| miR-520d-3p | 2013 | Nanoparticles | Ovarian | Proliferation and migration suppression | EPHB2 | In vivo | Orthotopic | [194] |
| miR-211 | 2014 | LNA | Multiple myeloma | Tumor growth suppression | p27Kip 1 | In vivo | Orthotopic | [195] |
| miR-31 | 2015 | LNA | Esophageal | Tumor formation and proliferation suppression | STK40 | In vivo | Zinc-deficient model | [97] |
| miR-126 | 2015 | Nanoparticles | Leukemia | Targeting leukemia stem cells | - | In vivo | Tail vein | [196] |
| miR-214 | 2015 | LNA | Colorectal cancer | Inflammation and tumor formation suppression | PDLM 2, PTEN | In vivo | DSS-AOM | [98] |
| miR-155 | 2015 | pHLIP | Lymphoma | Tumor growth suppression | BACH1 | In vivo | miR-155 transgenic mouse | [44] |
| miR-17 | 2017 | Nanoparticles | Liver | Tumor growth suppression | TGFBR 2 | In vivo | Xenograft | [197] |
| miR-21 | 2017 | Nanoparticles | Breast | Drug resistance reduction | - | In vivo | Xenograft | [198] |
| miR-21 | 2018 | Nanocarriers | Breast | Proliferation suppression | - | In vivo | Xenograft | [199] |
| miR-21 | 2018 | Nanoparticles | Ovarian | Drug resistance reduction | - | In vitro | Cell line | [200] |
| miR-21 | 2018 | Nanoparticles | Liver | Drug resistance reduction | - | In vivo | Xenograft | [201] |
| miR-21 and 10b | 2018 | Nanoparticles | Brain | Cell cycle arrest enhancement | - | In vivo | Xenograft | [202] |
| miR-214 | 2018 | Exosomes | Stomach | Drug resistance reduction | - | In vivo | Tail vein | [203] |
| miR-221 | 2018 | Nanoparticles | Leukemia | Tumorigenicity suppression | p27Kip | In vivo | Metastasis model | [204] |
| miR-21 | 2019 | Nanoparticles/LNA | Breast | Migration suppression | PTEN, PDCD4 | In vivo | Xenograft | [205] |
| miR-204 | 2019 | Nanoparticles | Ovarian | Angiogenesis suppression | THBS1 | In vivo | Orthotopic tumor | [206] |
Chronologically listed
For instance, miR-155 is a highly prominent, putative onco-miR involved in metastasis and tumor progression, as well as participates in fundamental biological processes such as stem cell development and inflammation [87]. miR-155 overexpression in a transgenic mice model provided the first evidence that a single dysregulated miRNA could cause a B-cell malignancy [88]. This discovery became the basis for initiating therapeutic evaluation of various miR-155 inhibitors in animal models of lymphoma and leukemia [44, 89]. Anti-miR-155 delivered in a pHLIP-based miRNA-inhibitor exhibited a significant anti-tumorigenic efficacy when overexpressed in a lymphoma mouse model [44]. Not surprisingly, therapeutic use of miR-155 inhibitor is currently being tested for hematological malignancies (Cobomarsen [MRG-106]) phase II clinical trial for cutaneous T-cell lymphoma and phase I clinical trial for adult T-cell lymphoma/leukemia [90]
Likewise, miR-21 is another well-recognized and well-established onco-miR frequently dysregulated in many cancers [91]. The overexpression of miR-21 enhances proliferation, metastasis, invasion, and drug resistance [92]. Lentiviral vector knockdown of miR-21 inhibited tumor growth in a xenograft pancreatic cancer model [93]. Similarly, miR-21 inhibitors delivered in solid lipid nanoparticles significantly suppressed cellular proliferation, migration, and invasion in human lung cancer cells [94]. Currently, anti-miR-21 oligonucleotides are being clinically tested for Alport Syndrome [95]. Considering that miR-21 is a well-established oncogene, this oligonucleotide could be used for cancer treatment in the near future.
Inflamed tumor microenvironments are well-recognized as critical components of tumor progression, cellular proliferation, survival and migration, and inflammation in the tumor microenvironment has long been considered a key pathway in oncogenesis [96]. Recently, several inflammation-associated miRNA inhibitors have been interrogated for their therapeutic efficacy in a variety of cancers. For example, in esophageal cancer, zinc deficiency activates the miR-31 promoter region and NF-κB binding sites. NF-κB is a gene which is known to regulate inflammation and a LNA-based miR-31 inhibitor attenuated the serine-threonine kinase 40 (STK40) and NF-κB-controlled inflammation, which subsequently resulted in suppression of esophageal pre-neoplasia in zinc-deficient rats [97]. High-throughput functional screening of miRNAs in human colorectal cancer also identified miR-214 as a regulator of NF-κB [98]. In a colitis-associated colorectal cancer mouse model, inhibition of miR-214 significantly reduced the severity of chemically induced colitis, thereby decreasing the number and size of lesions. In both of these studies, miRNA inhibitors displayed remarkable efficacy in rodent models of inflammation-associated cancers, highlighting their clinical potential.
3. Long non-coding RNA (lncRNA)-based Cancer Therapy
3.1. lncRNAs in Cancer
The lncRNAs are a class of ncRNAs that are typically longer than 200 nucleotides; despite lacking protein-coding capability, they are involved in the pathogenesis of cancer. Of the approximately 60,000 lncRNAs identified from human tumor tissues and cancer cell lines, a significant majority (more than 70%) are still awaiting appropriate annotations [99]. Nonetheless, the functional roles of numerous lncRNAs whose expression is often dysregulated in various cancers have been investigated (Table 3). Although it remains largely unclear whether dysregulation of lncRNAs is the cause or the consequence for cancer pathogenesis, this group of ncRNAs have added a new dimension to the already complex molecular architecture of carcinogenesis. Mechanistically, the majority of well-studied lncRNAs display functional similarity to typical protein-coding oncogenes and tumor suppressors involved in tumor initiation, progression, and metastasis. lncRNAs appear to play critical roles in oncogenesis and have significant potential as cancer therapeutic targets. Interestingly, a recent comprehensive genomic characterization of lncRNAs across a number of human cancers have discovered that although both lncRNAs and protein-coding genes have similar frequencies of tumor-associated dysregulation, a significantly higher proportion of altered lncRNAs were deemed cancer type-specific [100]. Furthermore, some lncRNAs display organ-specific dualities, serving as both tumor suppressors and oncogenes. In addition, recently a highly efficient RNA-seq-based method for lncRNA profiling of highly degraded FFPE fixed cancer samples has been established [101], which could accelerate lncRNA profiling in various cancers. Collectively, a thorough functional investigation of lncRNAs is needed to determine their true role in cancer, prior to their evaluation in preclinical studies as potential therapeutic targets.
Table 3.
A list of potential lncRNAs that can be therapeutic targeted in cancer
| lncRNA | Function | Reported cancer type | Mechanism | REF |
|---|---|---|---|---|
| ANRIL | Oncogene | Bladder, lung, liver, cervical, stomach | Interacts with PRC2 and CBX7 | [207] |
| BANCER | Oncogene | Stomach, skin | Promotes proliferation and metastasis via regulation of NF-kB1, p21, MAPK pathways | [208, 209] |
| CCAT1-L | Oncogene | Colon | Transcriptionally regulates MYC and promotes long-range chromatin looping | [210] |
| CCAT2 | Oncogene | Esophagus, stomach, breast, colon | Enhances Wnt signaling pathway via TCF7L2 interaction | [211–213] |
| CRNDE | Oncogene | Colon | Negatively regulated by insulin and insulin-like growth factors and may regulate the expression of genes involved in metabolism | [214] |
| HCP5 | Oncogene | Stomach | Sequesters miR-3619-5p and upregulates PPARGC1A, which Induces stemness and drug resistance | [215] |
| HOTAIR | Oncogene | Esophagus, stomach, colon, liver, lung, breast, ovary, bladder, prostate, glioma, melanoma | Serves as a scaffold to assemble PRC2 and LSD1 complexes to the HOXD gene cluster. Epigenetic silencing of HOXD genes in multiple tissues | [216] |
| HULC | Oncogene | Liver, pancreas | Modulates abnormal lipid metabolism through miR-9-mediated RXRA signaling pathway | [217, 218] |
| lincRNA-ATB | Oncogene | Liver, breast, colon, pancreas | miR-200 family sponge. Upregulates ZEB1 and ZEB2. | [219] |
| lincRNA-ROR | Oncogene | Breast | Competitive endogenous RNA for miR-145 | [220, 221] |
| MALAT 1 | Oncogene | Lung, prostate colon, liver | Forms molecular scaffolds for ribonucleoprotein complexes in the nucleus. Transcriptional regulator for genes involved in cell cycle regulation, cancer metastasis, and cell migration. | [222–224] |
| MCF2L-AS1 | Oncogene | Colon | Enhances cell proliferation and invasion through crosstalk with miR-874-3p/FOXM1 signaling axis | [133] |
| PCA3 | Oncogene | Prostate | Enhances cell proliferation through regulation of PRUNE2 | [225] |
| PRNCR 1 (PCAT8) | Oncogene | Colon, prostate | Binds to the androgen receptor and enhances ligand-dependent and ligand-independent androgen-receptor-mediated gene activation and increases proliferation | [226, 227] |
| PVT-1 | Oncogene | Breast, bladder, colon, kidney, pancreas | Regulates MYC oncogene | [228] |
| UCA1 | Oncogene | Colon, bladder, breast, esophagus, stomach, liver, skin | Regulates CREB | [229] |
| GAS5 | Tumor suppressor | Breast, prostate, lung | Encodes glucocorticoid response element, which binds to the DNA binding domain and blocks the activity of glucocorticoid receptor, androgen, progesterone, and mineralocorticoid. | [230] |
| H19 | Tumor suppressor | Breast, lung, pancreas, stomach, bladder, prostate, colon, skin | Cancer metastasis tumor suppressor and generates miR-675 | [231] |
| lincRNA-p21 | Tumor suppressor | Colon | Enhances p21 activity | [232] |
| MEG3 | Tumor suppressor | Brain, bladder, bone marrow, breast, colon, liver, lung, prostate | Interacts with the tumor suppressor p53 and regulates its target gene expression. | [233] |
| ncRuPAR | Tumor suppressor | Colon, stomach | Inhibits tumor progression by downregulation of protease-activated receptor-1(PAR-1) | [210] |
| TUG1 | Tumor suppressor | Bladder, esophagus | Regulates miR-145. Suppresses epithelial-to-mesenchymal transition and radio-resistance. | [229, 234] |
3.2. lncRNAs as Potential Therapeutic Targets
Hypoxia is a major factor that contributes in a multitude of ways to cancer progression and acquisition of chemotherapeutic resistance. Therefore, targeting signaling pathways associated with hypoxia are deemed attractive for achieving tumor suppression and progression, as well as an escape mechanism from chemoresistance. Accumulating evidence indicates that NEAT1 [102], UCA1 [103], and H19 [104] are functionally relevant lncRNAs that are modulated by hypoxia during oncogenesis. Furthermore, long intergenic noncoding RNA (lincRNA)-p21 is a hypoxia-responsive lncRNA that forms a positive feedback loop with HIF-1α to drive glycolysis and promote tumor growth [105]. In breast cancer, upregulation of the HIF-1α-inducing lncRNA EFNA3 facilitates Ephrin-A3 accumulation at cell surface to promote extravasation, and eventual metastasis [106]. Moreover, linc-RoR overexpression in the extracellular tumor environment during hypoxia in hepatocellular cancer cells has been shown to act as a miR-sponge for the tumor suppressive miR-145, which permits the self-renewal of cancer cells [107]. linc-RoR-induced upregulation of miR-145 downstream targets p70S6K1, PDK1, and HIF-1α, genes associated with hypoxia, and resulted in the acceleration of cellular proliferation [108].
DNA damage repair (DDR) is an evolutionarily conserved process that maintains genomic integrity but is frequently dysregulated in cancer. Although this system normally protects healthy cells from tumorigenic DNA damage and replication errors, most cancer cells acquire some form of enhanced DDR that eventually results in radiotherapeutic or chemotherapeutic resistance [109–111]. Recently, several studies demonstrated that DDR dysregulation alters the expression of various lncRNAs [112, 113]. Two studies showed that a DDR-associated lncRNA, p21-associated ncRNA DNA damage-activated (PANDA), suppressed apoptosis via inhibition of p53-associated downstream genes FAS, PUMA, and CCNB1 [114, 115]. Furthermore, another oncogenic lncRNA, the antisense ncRNA in the INK4 locus (ANRIL), is upregulated in an ATM-dependent manner following DNA damage [116]. ANRIL epigenetically represses tumor suppressor gene, INK4B, expression by recruiting the polycomb repressor complex (PRC) [117, 118]. Because these lncRNAs play critical roles in the DDR process, their expression can modulate the response of cancer cells to radio or chemotherapy.
Various treatment options, such as radiotherapy, chemotherapy, hormone therapy, and biological therapies exist for patients with metastatic cancer; however, responses to these treatments may differ significantly among patients. Therefore, identifying ncRNAs that become dysregulated in the metastatic process may result in the discovery of potential therapeutic targets that could be personalized to render otherwise resistant tumors responsive to conventional therapies. In particular, emerging evidence indicates that several lncRNAs are involved in the regulation of EMT, a key metastasis pathway, through epigenetic, transcriptional and post transcriptional regulation of RNA, DNA or proteins [119]. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a lncRNA that is consistently upregulated in multiple cancers [120] and its expression is associated with poor clinical outcomes in various cancers [121]. MALAT1 induces EMT through the activation of the Wnt signaling pathway and may therefore serve as a promising independent prognostic factor for early-stage metastatic non-small cell lung cancer [122]. Similarly, several other recognized oncogenic lncRNAs are dysregulated in EMT, suggesting that these lncRNAs could affect metastasis [121]. Expression of the lncRNA H19 is greater in bladder cancer tissues compared to adjacent normal mucosa, and it is even higher in metastatic tissues [123]. Mechanistically, H19 is upregulated during EMT in a positive feedforward loop, resulting in ablation of E-cadherin expression [124]. HOX antisense intergenic RNA (HOTAIR) is one of the most well-studied oncogenic lncRNAs that is involved in the EMT process in breast and gastric cancer cells, and is frequently upregulated in lymph node metastasis [125, 126]. Elevated HOTAIR expression in several cancer types is associated with resistance to chemotherapeutics, suggesting that inhibitors of HOTAIR could potentially resensitize a patient’s tumor to a specific chemotherapy [127–129]. Consistent with its involvement in EMT, HOTAIR is also known to promote cancer stem cell like properties in breast and colon cancers [130, 131]. Emerging evidence indicates that lncRNAs are aberrantly expressed in diverse cancer stem cells and they appear to be involved in the regulation of cancer stem cells at different molecular levels [132]. Moreover, lncRNA MCF2L-AS1 has been recently identified as an oncogene which regulate cell migration and invasion through sponging of miR-874–3p and its downstream target FOXM1 [133], a postulated master regulator of cancer metastasis [134]. As additional studies define the biological roles of lncRNAs in cancer, mainstream clinical use of molecular inhibitors targeting unique lncRNAs to prevent metastasis might be closer to becoming a reality for the personalized treatment of cancer patients.
4. Other ncRNA-based Cancer Therapeutic Targets
Successful miRNA-based cancer research has prompted further investigations to identify other ncRNA families that might serve as potential therapeutic targets in cancer. High-throughput sequencing approaches have resulted in the identification of many new classes of ncRNAs that contribute to the pathogenesis of various diseases, including cancer. We list and illustrate several families of ncRNAs known to be involved in oncogenesis with a promising therapeutic potential (Figure 2).
Figure 2.
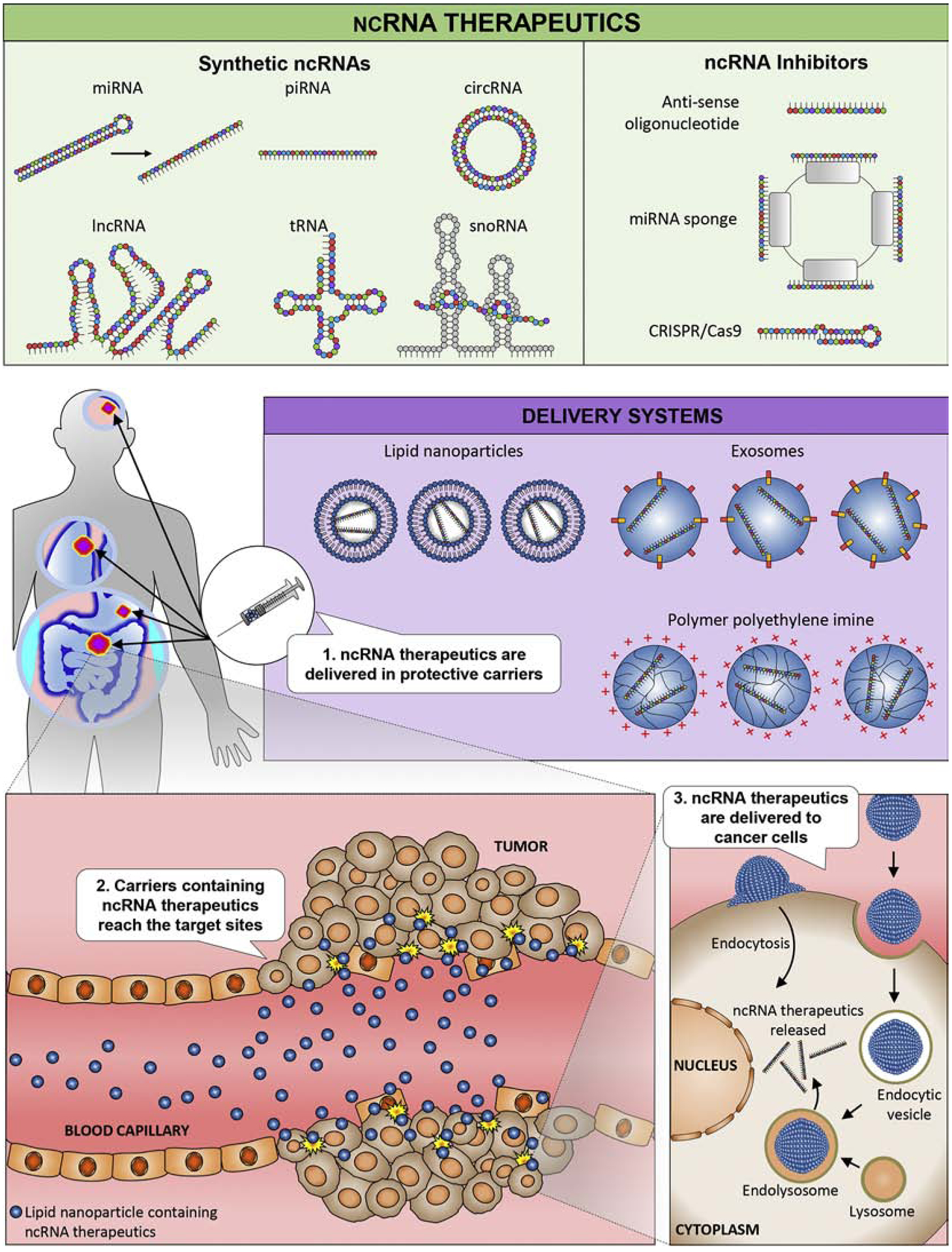
Concepts in ncRNA-based therapy. (Top) Two major strategies to modulate the expression of ncRNAs in tumors. ncRNAs include miRNAs, lncRNAs, tRNAs, piRNAs, circRNAs, and snoRNAs. Synthetic generation of these ncRNAs for use in overexpression and ncRNA inhibitors such as antisense oligonucleotides, CRISPR/Cas9, or miRNA sponges can inhibit the expression of target ncRNAs. (Middle) Delivery of the mimics/inhibitors to the tumor can occur via protective delivery mechanisms including lipid nanoparticles, exosomes, or polymer polyethylene imine. (Bottom) Subsequently, the mimics/inhibitors reach the tumor site and modulate the expression of the target ncRNAs in the tumor.
4.1. tRNAs:
The transfer RNA (tRNA) fragments (tRFs) are present in most organisms; are heterogeneous in size, nucleotide composition, biogenesis, and function; and are the second most abundant family of sncRNAs after miRNAs [135]. tRNAs have higher turnover rates in cancer vis-à-vis normal cells [136], become frequently overexpressed in cancers under stress [137, 138], correlate with clinical stages of cancer, and their expression levels are elevated in the serum and urine of cancer patients [139, 140]. The high abundance of tRNAs in blood makes them legitimate candidates to serve as diagnostic markers in cancer. Several tRFs bind YBX1, an RNA-binding protein, and in this manner stabilize multiple oncogenic transcripts that suppress cell growth and invasion [141]. While tRNAs have important roles in cancer, yet the elucidation of their functional roles in oncogenesis is in early stages; hence, we must wait for data to accumulate, before we further consider their therapeutic applications in the clinic.
4.2. circRNAs:
Circular RNAs (circRNAs) are another novel class of endogenous ncRNAs that are emerging as important molecular modulators in cancer. Initially discovered in RNA viruses in the 1970s, circRNAs, unlike conventional linear RNAs, form covalently closed loops with neither 5’ or 3’ polarities nor polyadenylated tails [142, 143]. Although the functional role of circRNAs is evolving each day, several studies have to date demonstrated that circRNAs harbor specific binding sites and primarily act as miRNA sponges or gene regulators [144]. For example, a recently identified circRNA, ciRS-7, was shown to function as a miR-7 sponge [145]. Given that miR-7 acts as a tumor-suppressor-miRNA in various cancers and regulates the expression of several oncogenes such as EGFR, RAF1, PAK1, and PIK3CD [146–148], ciRS-7 appears to be an oncogenic circRNA based on its modulation of the miRNA activity [10]. Furthermore, a recent examination of spatial expression of ciRS-7 in tumors showed that ciRS-7 is completely absent in the cancer cells, but highly expressed in stromal cells within the tumor microenvironment [149], highlighting the complex role of circular RNAs in cancer. In addition, recently functional roles of several circRNAs that are dysregulated in cancers have been clarified. circHIPK3, circWDR77 and circZFR have been shown to regulate cancer cell proliferations [150–152]. Moreover, circMYLK and circIRAK3 were shown to regulate EMT in prostate cancer and non-small cell lung carcinoma respectively [153, 154]. The structural stability of circRNAs is particularly appealing for designing efficiently deliverable therapeutic drugs, further underscoring their therapeutic potential.
4.3. piRNAs:
The piwi-interacting RNAs (piRNAs) are sncRNAs expressed in eukaryotic cells [155]. Initially believed to be entirely absent in cancer, a growing body of evidence indicates that not only piRNAs are aberrantly expressed in cancers, but they are also involved in tumor pathogenesis [156]. Although they are 26–31 nucleotides in length, which is similar in size to miRNAs, piRNAs are distinctly different because they lack the sequence conservation present in miRNAs. Functionally, piRNAs interact with piwi regulatory proteins to form RNA-protein complexes that induce epigenetic and post-transcriptional gene silencing. Most of the mechanisms underlying piRNA function remain in their infancy but are an active area of investigation. For example, several piRNAs appear to act as tumor suppressors through the degradation of downstream messenger RNAs [157, 158]. In contrast, piR-651 [159], piR-823 [160], and piR-Hep1[161] are upregulated in gastric and hepatocellular cancers and appear to function as oncogenes. Although piRNAs appear to be functionally relevant sncRNAs in cancer, and targeting select piRNAs may have therapeutic effects, more investigations are required to clarify their fundamental roles in oncogenesis.
4.4. snoRNAs:
snoRNAs are well-conserved, metabolically stable RNAs that are 60–300 nucleotides in length and highly abundant in human and other organisms [162]. Considering that most snoRNAs localize primarily to the nucleus, it appears unlikely that their function is similar to that of other ncRNAs. For years, it was assumed that snoRNAs functioned as housekeeping genes [163]. However, accumulating evidence indicates that snoRNAs have oncogenic roles as well [164]. In 2000, dysregulation of the C/D box type snoRNA U50 was discovered in B-cell lymphoma [165], which prompted the investigation of snoRNAs in other types of cancers. These investigations led to the discovery that amplification of SNORA42 is common occurrence in many human cancers [166–168]. In lung cancer, mechanistic gain and loss-of-function experiments demonstrated that SNORA42 enhances tumor growth [166]. In colorectal cancer, high SNORA42 expression was associated with poor prognosis of patients [167]. Furthermore, SNORA50A and SNORA50B were found to be frequently deleted in cancers [33]. In a mouse model, deleting SNORA50A/B using CRISPR/Cas9 enhanced tumorigenicity by increasing the amount of active oncogene, KRAS, thereby hyperactivating the RAS-ERK1/ERK2 signaling pathway. Given that snoRNAs are highly abundant and easily detectable in solid tumors and blood, and may prove functionally relevant in oncogenesis, they may become a major target for cancer therapy.
5. Potential Clinical Implications
The clinical application of ncRNAs as potential therapeutic targets in cancer can manifest in two scenarios: using ncRNAs to “replenish” suppressed or missing RNAs (replacement therapy) or to “block” the effects of over-active oncogenic RNAs. The ncRNA-based replacement therapy primarily benefits patients with reduced tumor-suppressor-miR expression or those with an overexpression of the downstream targets of these miRNAs. Replenishing downregulated miRNAs (or the use of miRNA mimics) that have multiple gene targets critical in oncogenesis could be an attractive treatment modality in cancer patients with low expression of tumor-suppressor-miRs. Furthermore, RNA-sequencing-based miRNA profiling of cancer samples could provide further insights on which specific miRNAs are dysregulated in patients and thus provide a rationale for the restoration of expression of these targets either through replacement therapy or the use of inhibitors. In addition, it is important to recognize that molecular characteristics and functional roles of miRNAs vary between tumor types. Therefore, the effectiveness of miRNA therapeutics must be tested in individual tumor types and tumor specific efficacy of these therapeutics needs to be clarified. Collectively, further understanding of the functional roles of target miRNAs and tumor specific efficacy of miRNA therapeutics will be key factors for the development of successful miRNA therapeutics.
Beyond cancer cells, the immune environment plays an important role in promoting or preventing the process of carcinogenesis. Several miRNAs appear central to various immunological responses. Therefore, administering miRNA mimics that target critical tumor-promoting immune cells may protect patients at high risk for cancer. For example, miR-568 mimics inhibit the production of the tumor-promoting cytokines TGF-β and IL-10, and induce proliferation of tumor-protecting regulatory T cells [169]. Tissue resident macrophages and stromal cells drive propagation of gastrointestinal neoplasms through inflammatory mediators. In this regard, miR-155 knockdown represses pro-inflammatory mediators (TNFα, IL-1b, and IL-6) in macrophages [170]. Therefore, inhibitors that target immune-directing miRNAs could complement existing and future immunotherapies, such as adaptive T-cell therapy and cancer vaccines.
miRNAs that counter tumor formation and progression by neutralizing tumor-promoting mutations are also enticing entities to explore for use in cancer treatment. For example, somatic KRAS mutations are frequently found in patients with blood and solid tumor cancers, including leukemia and colorectal cancer [171, 172]. Thus, administering a synthetic tumor-suppressor-miR that targets KRAS, such as Let-7, may benefit a large number of cancer patients. Indeed, in mice with KRAS-active non-small cell lung cancer, therapeutic delivery of synthetic Let-7 mimics reduced tumor burden [173]. As another example, approximately one-third of tumors harbor β-catenin activating mutations, which in turn upregulate miR-34a levels. In a mouse model in which β-catenin was overactivated in the liver, using miR-34a-LNA to inhibit miRNA-34a inhibited proliferation of hepatocellular carcinoma [174]. Collectively, these studies highlight the potential that miRNAs remain an attractive therapeutic targeting in cancer patients.
6. Opportunities and Challenges
Although our understanding for the roles of ncRNAs in various cancers continues to improve, there is still much to learn. miRNAs are the most well studied among the family of ncRNAs; therefore, their presence at the forefront of ncRNA-based cancer therapeutics is not surprising. Unfortunately, to date, it has been difficult to definitively categorize the majority of miRNAs as either tumor suppressors or oncogenes. However, recent technological advancements and the increased affordability of RNA-sequencing-based profiling technologies will allow a comprehensive assessment of their functional roles in multiple organ systems. Similar approaches can elucidate the roles of other ncRNAs in cancer as well. To put it simply, the major obstacle for using ncRNA-based therapeutic strategies in cancer patients have largely been due to the lack of a clear knowledge regarding their functional roles in oncogenesis, and the specific downstream genetic targets they regulate. On the other hand, enough technological advances have been made to synthesize and manufacture most ncRNA mimics and inhibitors for utilization in pre-clinical studies and eventually in human clinical trials. In conclusion, while this field may seem to be in infancy, we are currently witnessing the bourgeoning potential of ncRNAs in cancer therapy - and, it is only a matter of time in the near future, when we may begin to successfully exploit their potential for therapeutic targeting in cancer as we embark on the journey for precision oncological treatments in cancer patients.
Acknowledgements:
The authors would like to thank Dr. Wenhao Weng for his useful insights, and Dr. Sarah Wilkinson, City of Hope, Beckman Research Institute, Duarte, CA and Dr. Margaret Hinshelwood, manager of the Office of Scientific Publications, Baylor Charles A. Sammons Cancer Center, Dallas, for critical suggestions and editing to further improve the quality of this article.
Funding: The present work was supported by the grants CA72851, CA181572, CA184792, and CA202797 from the National Cancer Institute, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: No conflicts of interest exist for any of the authors.
REFERENCES
- [1].Consortium EP, An integrated encyclopedia of DNA elements in the human genome, Nature, 489 (2012) 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Costa FF, Non-coding RNAs: new players in eukaryotic biology, Gene, 357 (2005) 83–94. [DOI] [PubMed] [Google Scholar]
- [3].Pauli A, Rinn JL, Schier AF, Non-coding RNAs as regulators of embryogenesis, Nature reviews. Genetics, 12 (2011) 136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Crichton DJ, Altinok A, Amos CI, Anton K, Cinquini L, Colbert M, Feng Z, Goel A, Kelly S, Kincaid H, Liu D, Lombeyda S, Mahabal A, Mishra A, Patriotis C, Srivastava S, Cancer Biomarkers and Big Data: A Planetary Science Approach, Cancer cell, (2020). [DOI] [PubMed] [Google Scholar]
- [5].Jung G, Hernandez-Illan E, Moreira L, Balaguer F, Goel A, Epigenetics of colorectal cancer: biomarker and therapeutic potential, Nat Rev Gastroenterol Hepatol, 17 (2020) 111–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lane JS, Hoff DV, Cridebring D, Goel A, Extracellular Vesicles in Diagnosis and Treatment of Pancreatic Cancer: Current State and Future Perspectives, Cancers (Basel), 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Matsuyama T, Kandimalla R, Ishikawa T, Takahashi N, Yamada Y, Yasuno M, Kinugasa Y, Hansen TF, Fakih M, Uetake H, Gyorffy B, Goel A, A novel mesenchymal-associated transcriptomic signature for risk-stratification and therapeutic response prediction in colorectal cancer, Int J Cancer, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Toiyama Y, Hur K, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A, Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer, Annals of surgery, 259 (2014) 735–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tovar-Camargo OA, Toden S, Goel A, Exosomal microRNA Biomarkers: Emerging Frontiers in Colorectal and Other Human Cancers, Expert Rev Mol Diagn, 16 (2016) 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, Cai S, Qin H, Ma Y, Goel A, Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer, Clinical cancer research : an official journal of the American Association for Cancer Research, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wen D, Danquah M, Chaudhary AK, Mahato RI, Small molecules targeting microRNA for cancer therapy: Promises and obstacles, Journal of controlled release : official journal of the Controlled Release Society, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lee RC, Feinbaum RL, Ambros V, The C elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14, Cell, 75 (1993) 843–854. [DOI] [PubMed] [Google Scholar]
- [13].Wightman B, Ha I, Ruvkun G, Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans, Cell, 75 (1993) 855–862. [DOI] [PubMed] [Google Scholar]
- [14].Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM, Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia, Proceedings of the National Academy of Sciences of the United States of America, 99 (2002) 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Calin GA, Croce CM, MicroRNA signatures in human cancers, Nature reviews. Cancer, 6 (2006) 857–866. [DOI] [PubMed] [Google Scholar]
- [16].Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celia-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua Y, Wei Y, Hu G, Garcia BA, Ragoussis J, Amadori D, Harris AL, Kang Y, Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization, Nature medicine, 17 (2011) 1101–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pecot CV, Rupaimoole R, Yang D, Akbani R, Ivan C, Lu C, Wu S, Han HD, Shah MY, Rodriguez-Aguayo C, Bottsford-Miller J, Liu Y, Kim SB, Unruh A, Gonzalez-Villasana V, Huang L, Zand B, Moreno-Smith M, Mangala LS, Taylor M, Dalton HJ, Sehgal V, Wen Y, Kang Y, Baggerly KA, Lee JS, Ram PT, Ravoori MK, Kundra V, Zhang X, Ali-Fehmi R, Gonzalez-Angulo AM, Massion PP, Calin GA, Lopez-Berestein G, Zhang W, Sood AK, Tumour angiogenesis regulation by the miR-200 family, Nature communications, 4 (2013) 2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gottesman MM, Mechanisms of cancer drug resistance, Annual review of medicine, 53 (2002) 615–627. [DOI] [PubMed] [Google Scholar]
- [19].Galm O, Herman JG, Baylin SB, The fundamental role of epigenetics in hematopoietic malignancies, Blood reviews, 20 (2006) 1–13. [DOI] [PubMed] [Google Scholar]
- [20].Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjaer N, Babu BR, Hojland T, Abramov M, Van Aerschot A, Odadzic D, Smicius R, Haas J, Andree C, Barman J, Wenska M, Srivastava P, Zhou C, Honcharenko D, Hess S, Muller E, Bobkov GV, Mikhailov SN, Fava E, Meyer TF, Chattopadhyaya J, Zerial M, Engels JW, Herdewijn P, Wengel J, Kjems J, A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity, Nucleic acids research, 37 (2009) 2867–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhang Y, Wang Z, Gemeinhart RA, Progress in microRNA delivery, Journal of controlled release : official journal of the Controlled Release Society, 172 (2013) 962–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Misso G, Di Martino MT, De Rosa G, Farooqi AA, Lombardi A, Campani V, Zarone MR, Gulla A, Tagliaferri P, Tassone P, Caraglia M, Mir-34: a new weapon against cancer?, Molecular therapy. Nucleic acids, 3 (2014) e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bader AG, miR-34 - a microRNA replacement therapy is headed to the clinic, Frontiers in genetics, 3 (2012) 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dirin M, Winkler J, Influence of diverse chemical modifications on the ADME characteristics and toxicology of antisense oligonucleotides, Expert opinion on biological therapy, 13 (2013) 875–888. [DOI] [PubMed] [Google Scholar]
- [25].Dias N, Stein CA, Antisense oligonucleotides: basic concepts and mechanisms, Molecular cancer therapeutics, 1 (2002) 347–355. [PubMed] [Google Scholar]
- [26].Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, Hansen HF, Koch T, Pappin D, Hannon GJ, Kauppinen S, Silencing of microRNA families by seed-targeting tiny LNAs, Nature genetics, 43 (2011) 371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lennox KA, Behlke MA, Chemical modification and design of anti-miRNA oligonucleotides, Gene therapy, 18 (2011) 1111–1120. [DOI] [PubMed] [Google Scholar]
- [28].Yoshioka K, Kunieda T, Asami Y, Guo H, Miyata H, Yoshida-Tanaka K, Sujino Y, Piao W, Kuwahara H, Nishina K, Hara RI, Nagata T, Wada T, Obika S, Yokota T, Highly efficient silencing of microRNA by heteroduplex oligonucleotides, Nucleic Acids Res, 47 (2019) 7321–7332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ebert MS, Sharp PA, MicroRNA sponges: progress and possibilities, Rna, 16 (2010) 2043–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ling H, Fabbri M, Calin GA, MicroRNAs and other non-coding RNAs as targets for anticancer drug development, Nature reviews. Drug discovery, 12 (2013) 847–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Baumann V, Winkler J, miRNA-based therapies: strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents, Future medicinal chemistry, 6 (2014) 1967–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sánchez-Rivera FJ, Jacks T, Applications of the CRISPR-Cas9 system in cancer biology, Nature reviews. Cancer, 15 (2015) 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Siprashvili Z, Webster DE, Johnston D, Shenoy RM, Ungewickell AJ, Bhaduri A, Flockhart R, Zarnegar BJ, Che Y, Meschi F, Puglisi JD, Khavari PA, The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer, Nature genetics, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhao Y, Dai Z, Liang Y, Yin M, Ma K, He M, Ouyang H, Teng CB, Sequence-specific inhibition of microRNA via CRISPR/CRISPRi system, Scientific reports, 4 (2014) 3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ho TT, Zhou N, Huang J, Koirala P, Xu M, Fung R, Wu F, Mo YY, Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines, Nucleic acids research, 43 (2015) e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Li L, He ZY, Wei XW, Gao GP, Wei YQ, Challenges in CRISPR/CAS9 Delivery: Potential Roles of Nonviral Vectors, Human gene therapy, 26 (2015) 452–462. [DOI] [PubMed] [Google Scholar]
- [37].Tabernero J, Shapiro GI, LoRusso PM, Cervantes A, Schwartz GK, Weiss GJ, Paz-Ares L, Cho DC, Infante JR, Alsina M, Gounder MM, Falzone R, Harrop J, White AC, Toudjarska I, Bumcrot D, Meyers RE, Hinkle G, Svrzikapa N, Hutabarat RM, Clausen VA, Cehelsky J, Nochur SV, Gamba-Vitalo C, Vaishnaw AK, Sah DW, Gollob JA, Burris HA 3rd, First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement, Cancer discovery, 3 (2013) 406–417. [DOI] [PubMed] [Google Scholar]
- [38].Garzon R, Marcucci G, Croce CM, Targeting microRNAs in cancer: rationale, strategies and challenges, Nature reviews. Drug discovery, 9 (2010) 775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Malik R, Roy I, Design and development of antisense drugs, Expert opinion on drug discovery, 3 (2008) 1189–1207. [DOI] [PubMed] [Google Scholar]
- [40].Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, Borodovsky A, Borland T, Constien R, de Fougerolles A, Dorkin JR, Narayanannair Jayaprakash K, Jayaraman M, John M, Koteliansky V, Manoharan M, Nechev L, Qin J, Racie T, Raitcheva D, Rajeev KG, Sah DW, Soutschek J, Toudjarska I, Vornlocher HP, Zimmermann TS, Langer R, Anderson DG, A combinatorial library of lipid-like materials for delivery of RNAi therapeutics, Nature biotechnology, 26 (2008) 561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bolcato-Bellemin AL, Bonnet ME, Creusat G, Erbacher P, Behr JP, Sticky overhangs enhance siRNA-mediated gene silencing, Proceedings of the National Academy of Sciences of the United States of America, 104 (2007) 16050–16055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Liu X, Chen H, Laurini E, Wang Y, Dal Col V, Posocco P, Ziarelli F, Fermeglia M, Zhang CC, Pricl S, Peng L, 2-difluoromethylene-4-methylenepentanoic acid, a paradoxical probe able to mimic the signaling role of 2-oxoglutaric acid in cyanobacteria, Organic letters, 13 (2011) 2924–2927. [DOI] [PubMed] [Google Scholar]
- [43].Goula D, Benoist C, Mantero S, Merlo G, Levi G, Demeneix BA, Polyethylenimine-based intravenous delivery of transgenes to mouse lung, Gene therapy, 5 (1998) 1291–1295. [DOI] [PubMed] [Google Scholar]
- [44].Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM, Saltzman WM, Slack FJ, MicroRNA silencing for cancer therapy targeted to the tumour microenvironment, Nature, 518 (2015) 107–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Thevenin D, An M, Engelman DM, pHLIP-mediated translocation of membrane-impermeable molecules into cells, Chem Biol, 16 (2009) 754–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Reshetnyak YK, Andreev OA, Lehnert U, Engelman DM, Translocation of molecules into cells by pH-dependent insertion of a transmembrane helix, Proceedings of the National Academy of Sciences of the United States of America, 103 (2006) 6460–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT, Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model, Cell, 137 (2009) 1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Trams EG, Lauter CJ, Salem N Jr., Heine U, Exfoliation of membrane ecto-enzymes in the form of micro-vesicles, Biochimica et biophysica acta, 645 (1981) 63–70. [DOI] [PubMed] [Google Scholar]
- [49].Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C, Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes), The Journal of biological chemistry, 262 (1987) 9412–9420. [PubMed] [Google Scholar]
- [50].Tauro BJ, Greening DW, Mathias RA, Mathivanan S, Ji H, Simpson RJ, Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids, Molecular & cellular proteomics : MCP, 12 (2013) 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, Simpson RJ, Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature, Molecular & cellular proteomics : MCP, 9 (2010) 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Simpson RJ, Jensen SS, Lim JW, Proteomic profiling of exosomes: current perspectives, Proteomics, 8 (2008) 4083–4099. [DOI] [PubMed] [Google Scholar]
- [53].Wei JX, Lv LH, Wan YL, Cao Y, Li GL, Lin HM, Zhou R, Shang CZ, Cao J, He H, Han QF, Liu PQ, Zhou G, Min J, Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells, Hepatology, 61 (2015) 1284–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Taylor DD, Gercel-Taylor C, MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer, Gynecologic oncology, 110 (2008) 13–21. [DOI] [PubMed] [Google Scholar]
- [55].Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr., Carter BS, Krichevsky AM, Breakefield XO, Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers, Nature cell biology, 10 (2008) 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Melo SA, Sugimoto H, O’Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, Lucci A, Ivan C, Calin GA, Kalluri R, Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis, Cancer cell, 26 (2014) 707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wei JX, Lv LH, Wan YL, Cao Y, Li GL, Lin HM, Zhou R, Shang CZ, Cao J, He H, Han QF, Liu PQ, Zhou G, Min J, Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells, Hepatology, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhang L, Zhang S, Yao J, Lowery FJ, Zhang Q, Huang WC, Li P, Li M, Wang X, Zhang C, Wang H, Ellis K, Cheerathodi M, McCarty JH, Palmieri D, Saunus J, Lakhani S, Huang S, Sahin AA, Aldape KD, Steeg PS, Yu D, Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth, Nature, 527 (2015) 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Challagundla KB, Wise PM, Neviani P, Chava H, Murtadha M, Xu T, Kennedy R, Ivan C, Zhang X, Vannini I, Fanini F, Amadori D, Calin GA, Hadjidaniel M, Shimada H, Jong A, Seeger RC, Asgharzadeh S, Goldkorn A, Fabbri M, Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy, Journal of the National Cancer Institute, 107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Li SP, Lin ZX, Jiang XY, Yu XY, Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools, Acta Pharmacol Sin, 39 (2018) 542–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Lynam-Lennon N, Maher SG, Reynolds JV, The roles of microRNA in cancer and apoptosis, Biol Rev Camb Philos Soc, 84 (2009) 55–71. [DOI] [PubMed] [Google Scholar]
- [62].Hermeking H, The miR-34 family in cancer and apoptosis, Cell death and differentiation, 17 (2010) 193–199. [DOI] [PubMed] [Google Scholar]
- [63].Okada N, Lin CP, Ribeiro MC, Biton A, Lai G, He X, Bu P, Vogel H, Jablons DM, Keller AC, Wilkinson JE, He B, Speed TP, He L, A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression, Genes & development, 28 (2014) 438–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Siemens H, Jackstadt R, Hunten S, Kaller M, Menssen A, Gotz U, Hermeking H, miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions, Cell cycle, 10 (2011) 4256–4271. [DOI] [PubMed] [Google Scholar]
- [65].Bu P, Chen KY, Chen JH, Wang L, Walters J, Shin YJ, Goerger JP, Sun J, Witherspoon M, Rakhilin N, Li J, Yang H, Milsom J, Lee S, Zipfel W, Jin MM, Gumus ZH, Lipkin SM, Shen X, A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells, Cell stem cell, 12 (2013) 602–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, Ferrara P, McKeon F, Caput D, Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers, Cell, 90 (1997) 809–819. [DOI] [PubMed] [Google Scholar]
- [67].Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Korner H, Knyazev P, Diebold J, Hermeking H, Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer, Cell cycle, 7 (2008) 2591–2600. [DOI] [PubMed] [Google Scholar]
- [68].Tanaka N, Toyooka S, Soh J, Kubo T, Yamamoto H, Maki Y, Muraoka T, Shien K, Furukawa M, Ueno T, Asano H, Tsukuda K, Aoe K, Miyoshi S, Frequent methylation and oncogenic role of microRNA-34b/c in small-cell lung cancer, Lung cancer, 76 (2012) 32–38. [DOI] [PubMed] [Google Scholar]
- [69].Kasinski AL, Kelnar K, Stahlhut C, Orellana E, Zhao J, Shimer E, Dysart S, Chen X, Bader AG, Slack FJ, A combinatorial microRNA therapeutics approach to suppressing non-small cell lung cancer, Oncogene, 34 (2015) 3547–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Gaur S, Wen Y, Song JH, Parikh NU, Mangala LS, Blessing AM, Ivan C, Wu SY, Varkaris A, Shi Y, Lopez-Berestein G, Frigo DE, Sood AK, Gallick GE, Chitosan nanoparticle-mediated delivery of MiRNA-34a decreases prostate tumor growth in the bone and its expression induces non-canonical autophagy, Oncotarget, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Deng X, Cao M, Zhang J, Hu K, Yin Z, Zhou Z, Xiao X, Yang Y, Sheng W, Wu Y, Zeng Y, Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer, Biomaterials, 35 (2014) 4333–4344. [DOI] [PubMed] [Google Scholar]
- [72].Tivnan A, Orr WS, Gubala V, Nooney R, Williams DE, McDonagh C, Prenter S, Harvey H, Domingo-Fernandez R, Bray IM, Piskareva O, Ng CY, Lode HN, Davidoff AM, Stallings RL, Inhibition of neuroblastoma tumor growth by targeted delivery of microRNA-34a using anti-disialoganglioside GD2 coated nanoparticles, PloS one, 7 (2012) e38129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Pramanik D, Campbell NR, Karikari C, Chivukula R, Kent OA, Mendell JT, Maitra A, Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice, Molecular cancer therapeutics, 10 (2011) 1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Di Martino MT, Campani V, Misso G, Gallo Cantafio ME, Gulla A, Foresta U, Guzzi PH, Castellano M, Grimaldi A, Gigantino V, Franco R, Lusa S, Cannataro M, Tagliaferri P, De Rosa G, Tassone P, Caraglia M, In vivo activity of miR-34a mimics delivered by stable nucleic acid lipid particles (SNALPs) against multiple myeloma, PloS one, 9 (2014) e90005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Chen Y, Zhu X, Zhang X, Liu B, Huang L, Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy, Molecular therapy : the journal of the American Society of Gene Therapy, 18 (2010) 1650–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Cortez MA, Ivan C, Valdecanas D, Wang X, Peltier HJ, Ye Y, Araujo L, Carbone DP, Shilo K, Giri DK, Kelnar K, Martin D, Komaki R, Gomez DR, Krishnan S, Calin GA, Bader AG, Welsh JW, PDL1 Regulation by p53 via miR-34, Journal of the National Cancer Institute, 108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Kent OA, McCall MN, Cornish TC, Halushka MK, Lessons from miR-143/145: the importance of cell-type localization of miRNAs, Nucleic acids research, 42 (2014) 7528–7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Zhang T, Xue X, He D, Hsieh JT, A prostate cancer-targeted polyarginine-disulfide linked PEI nanocarrier for delivery of microRNA, Cancer letters, 365 (2015) 156–165. [DOI] [PubMed] [Google Scholar]
- [79].Inamoto T, Taniguchi K, Takahara K, Iwatsuki A, Takai T, Komura K, Yoshikawa Y, Uchimoto T, Saito K, Tanda N, Kouno J, Minami K, Uehara H, Hirano H, Nomi H, Kiyama S, Akao Y, Azuma H, Intravesical administration of exogenous microRNA-145 as a therapy for mouse orthotopic human bladder cancer xenograft, Oncotarget, 6 (2015) 21628–21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Brabletz T, EMT and MET in metastasis: where are the cancer stem cells?, Cancer cell, 22 (2012) 699–701. [DOI] [PubMed] [Google Scholar]
- [81].Hermeking H, MicroRNAs in the p53 network: micromanagement of tumour suppression, Nature reviews. Cancer, 12 (2012) 613–626. [DOI] [PubMed] [Google Scholar]
- [82].Park SM, Gaur AB, Lengyel E, Peter ME, The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2, Genes & development, 22 (2008) 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, Brunton VG, Morton J, Sansom O, Schuler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T, The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs, Nature cell biology, 11 (2009) 1487–1495. [DOI] [PubMed] [Google Scholar]
- [84].Cao M, Deng X, Su S, Zhang F, Xiao X, Hu Q, Fu Y, Yang BB, Wu Y, Sheng W, Zeng Y, Protamine sulfate-nanodiamond hybrid nanoparticles as a vector for MiR-203 restoration in esophageal carcinoma cells, Nanoscale, 5 (2013) 12120–12125. [DOI] [PubMed] [Google Scholar]
- [85].Liu WJ, Xu Q, Sun LP, Dong QG, He CY, Yuan Y, Expression of serum let-7c, let-7i, and let-7f microRNA with its target gene, pepsinogen C, in gastric cancer and precancerous disease, Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine, 36 (2015) 3337–3343. [DOI] [PubMed] [Google Scholar]
- [86].Lee HY, Mohammed KA, Kaye F, Sharma P, Moudgil BM, Clapp WL, Nasreen N, Targeted delivery of let-7a microRNA encapsulated ephrin-A1 conjugated liposomal nanoparticles inhibit tumor growth in lung cancer, International journal of nanomedicine, 8 (2013) 4481–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Tili E, Croce CM, Michaille JJ, miR-155: on the crosstalk between inflammation and cancer, International reviews of immunology, 28 (2009) 264–284. [DOI] [PubMed] [Google Scholar]
- [88].Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM, Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice, Proceedings of the National Academy of Sciences of the United States of America, 103 (2006) 7024–7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM, Slack FJ, Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma, Proceedings of the National Academy of Sciences of the United States of America, 109 (2012) E1695–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Witten L, Slack FJ, miR-155 as a novel clinical target for hematological malignancies, Carcinogenesis, 41 (2020) 2–7. [DOI] [PubMed] [Google Scholar]
- [91].Pan X, Wang ZX, Wang R, MicroRNA-21: a novel therapeutic target in human cancer, Cancer biology & therapy, 10 (2010) 1224–1232. [DOI] [PubMed] [Google Scholar]
- [92].Hong L, Han Y, Zhang Y, Zhang H, Zhao Q, Wu K, Fan D, MicroRNA-21: a therapeutic target for reversing drug resistance in cancer, Expert opinion on therapeutic targets, 17 (2013) 1073–1080. [DOI] [PubMed] [Google Scholar]
- [93].Sicard F, Gayral M, Lulka H, Buscail L, Cordelier P, Targeting miR-21 for the therapy of pancreatic cancer, Molecular therapy : the journal of the American Society of Gene Therapy, 21 (2013) 986–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Shi SJ, Zhong ZR, Liu J, Zhang ZR, Sun X, Gong T, Solid lipid nanoparticles loaded with anti-microRNA oligonucleotides (AMOs) for suppression of microRNA-21 functions in human lung cancer cells, Pharmaceutical research, 29 (2012) 97–109. [DOI] [PubMed] [Google Scholar]
- [95].Gomez IG, MacKenna DA, Johnson BG, Kaimal V, Roach AM, Ren S, Nakagawa N, Xin C, Newitt R, Pandya S, Xia TH, Liu X, Borza DB, Grafals M, Shankland SJ, Himmelfarb J, Portilla D, Liu S, Chau BN, Duffield JS, Anti-microRNA-21 oligonucleotides prevent Alport nephropathy progression by stimulating metabolic pathways, J Clin Invest, 125 (2015) 141–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Coussens LM, Werb Z, Inflammation and cancer, Nature, 420 (2002) 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Taccioli C, Garofalo M, Chen H, Jiang Y, Tagliazucchi GM, Di Leva G, Alder H, Fadda P, Middleton J, Smalley KJ, Selmi T, Naidu S, Farber JL, Croce CM, Fong LY, Repression of Esophageal Neoplasia and Inflammatory Signaling by Anti-miR-31 Delivery In Vivo, Journal of the National Cancer Institute, 107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Polytarchou C, Hommes DW, Palumbo T, Hatziapostolou M, Koutsioumpa M, Koukos G, van der Meulen-de Jong AE, Oikonomopoulos A, van Deen WK, Vorvis C, Serebrennikova OB, Birli E, Choi J, Chang L, Anton PA, Tsichlis PN, Pothoulakis C, Verspaget HW, Iliopoulos D, MicroRNA214 Is Associated With Progression of Ulcerative Colitis, and Inhibition Reduces Development of Colitis and Colitis-Associated Cancer in Mice, Gastroenterology, 149 (2015) 981–992 e911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM, The landscape of long noncoding RNAs in the human transcriptome, Nature genetics, 47 (2015) 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, Fan L, Kandalaft LE, Tanyi JL, Li C, Yuan CX, Zhang D, Yuan H, Hua K, Lu Y, Katsaros D, Huang Q, Montone K, Fan Y, Coukos G, Boyd J, Sood AK, Rebbeck T, Mills GB, Dang CV, Zhang L, Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers, Cancer cell, 28 (2015) 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Iraola-Guzmán S, Brunet-Vega A, Pegueroles C, Saus E, Hovhannisyan H, Casalots A, Pericay C, Gabaldón T, Target Enrichment Enables the Discovery of lncRNAs with Somatic Mutations or Altered Expression in Paraffin-Embedded Colorectal Cancer Samples, Cancers (Basel), 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Choudhry H, Schodel J, Oikonomopoulos S, Camps C, Grampp S, Harris AL, Ratcliffe PJ, Ragoussis J, Mole DR, Extensive regulation of the non-coding transcriptome by hypoxia: role of HIF in releasing paused RNApol2, EMBO Rep, 15 (2014) 70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Xue M, Li X, Li Z, Chen W, Urothelial carcinoma associated 1 is a hypoxia-inducible factor-1alpha-targeted long noncoding RNA that enhances hypoxic bladder cancer cell proliferation, migration, and invasion, Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine, 35 (2014) 6901–6912. [DOI] [PubMed] [Google Scholar]
- [104].Matouk IJ, Mezan S, Mizrahi A, Ohana P, Abu-Lail R, Fellig Y, Degroot N, Galun E, Hochberg A, The oncofetal H19 RNA connection: hypoxia, p53 and cancer, Biochimica et biophysica acta, 1803 (2010) 443–451. [DOI] [PubMed] [Google Scholar]
- [105].Yang F, Zhang H, Mei Y, Wu M, Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect, Mol Cell, 53 (2014) 88–100. [DOI] [PubMed] [Google Scholar]
- [106].Gomez-Maldonado L, Tiana M, Roche O, Prado-Cabrero A, Jensen L, Fernandez-Barral A, Guijarro-Munoz I, Favaro E, Moreno-Bueno G, Sanz L, Aragones J, Harris A, Volpert O, Jimenez B, del Peso L, EFNA3 long noncoding RNAs induced by hypoxia promote metastatic dissemination, Oncogene, 34 (2015) 2609–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, Wu M, Xiong J, Guo X, Liu H, Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal, Dev Cell, 25 (2013) 69–80. [DOI] [PubMed] [Google Scholar]
- [108].Takahashi K, Yan IK, Haga H, Patel T, Modulation of hypoxia-signaling pathways by extracellular linc-RoR, J Cell Sci, 127 (2014) 1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Srivastava AK, Han C, Zhao R, Cui T, Dai Y, Mao C, Zhao W, Zhang X, Yu J, Wang QE, Enhanced expression of DNA polymerase eta contributes to cisplatin resistance of ovarian cancer stem cells, Proceedings of the National Academy of Sciences of the United States of America, 112 (2015) 4411–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Banerjee R, Russo N, Liu M, Basrur V, Bellile E, Palanisamy N, Scanlon CS, van Tubergen E, Inglehart RC, Metwally T, Mani RS, Yocum A, Nyati MK, Castilho RM, Varambally S, Chinnaiyan AM, D’Silva NJ, TRIP13 promotes error-prone nonhomologous end joining and induces chemoresistance in head and neck cancer, Nature communications, 5 (2014) 4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Bandey I, Chiou SH, Huang AP, Tsai JC, Tu PH, Progranulin promotes Temozolomide resistance of glioblastoma by orchestrating DNA repair and tumor stemness, Oncogene, 34 (2015) 1853–1864. [DOI] [PubMed] [Google Scholar]
- [112].Zhang C, Peng G, Non-coding RNAs: an emerging player in DNA damage response, Mutat Res Rev Mutat Res, 763 (2015) 202–211. [DOI] [PubMed] [Google Scholar]
- [113].Lukas J, Altmeyer M, A lncRNA to repair DNA, EMBO Rep, 16 (2015) 1413–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL, A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response, Cell, 142 (2010) 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Kong B, Langerod A, Borresen-Dale AL, Kim SK, van de Vijver M, Sukumar S, Whitfield ML, Kellis M, Xiong Y, Wong DJ, Chang HY, Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters, Nature genetics, 43 (2011) 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Wan G, Mathur R, Hu X, Liu Y, Zhang X, Peng G, Lu X, Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway, Cell Signal, 25 (2013) 1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H, Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA, Nature, 451 (2008) 202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y, Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene, Oncogene, 30 (2011) 1956–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Cheng JT, Wang L, Wang H, Tang FR, Cai WQ, Sethi G, Xin HW, Ma Z, Insights into Biological Role of LncRNAs in Epithelial-Mesenchymal Transition, Cells, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Lin R, Maeda S, Liu C, Karin M, Edgington TS, A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas, Oncogene, 26 (2007) 851–858. [DOI] [PubMed] [Google Scholar]
- [121].Wu Y, Lu W, Xu J, Shi Y, Zhang H, Xia D, Prognostic value of long non-coding RNA MALAT1 in cancer patients, Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine, (2015). [DOI] [PubMed] [Google Scholar]
- [122].Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F, Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition, Mol Biosyst, 8 (2012) 2289–2294. [DOI] [PubMed] [Google Scholar]
- [123].Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J, Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression, Cancer letters, 333 (2013) 213–221. [DOI] [PubMed] [Google Scholar]
- [124].Matouk IJ, Halle D, Raveh E, Gilon M, Sorin V, Hochberg A, The role of the oncofetal H19 lncRNA in tumor metastasis: orchestrating the EMT-MET decision, Oncotarget, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Cai B, Wu Z, Liao K, Zhang S, Long noncoding RNA HOTAIR can serve as a common molecular marker for lymph node metastasis: a meta-analysis, Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine, 35 (2014) 8445–8450. [DOI] [PubMed] [Google Scholar]
- [126].Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR, Goel A, Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis, Carcinogenesis, 35 (2014) 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Liu Z, Sun M, Lu K, Liu J, Zhang M, Wu W, De W, Wang Z, Wang R, The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression, PloS one, 8 (2013) e77293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, Li S, Zhao JC, Yu J, LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer, Oncogene, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Wang Y, Wang H, Song T, Zou Y, Jiang J, Fang L, Li P, HOTAIR is a potential target for the treatment of cisplatinresistant ovarian cancer, Mol Med Rep, 12 (2015) 2211–2216. [DOI] [PubMed] [Google Scholar]
- [130].Deng J, Yang M, Jiang R, An N, Wang X, Liu B, Long Non-Coding RNA HOTAIR Regulates the Proliferation, Self-Renewal Capacity, Tumor Formation and Migration of the Cancer Stem-Like Cell (CSC) Subpopulation Enriched from Breast Cancer Cells, PLoS One, 12 (2017) e0170860. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [131].Pádua Alves C, Fonseca AS, Muys BR, de Barros E Lima Bueno R, Bürger MC, de Souza JE, Valente V, Zago MA, Silva WA, Brief report: The lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines, Stem Cells, 31 (2013) 2827–2832. [DOI] [PubMed] [Google Scholar]
- [132].Ma Z, Wang YY, Xin HW, Wang L, Arfuso F, Dharmarajan A, Kumar AP, Wang H, Tang FR, Warrier S, Tergaonkar V, Sethi G, The expanding roles of long non-coding RNAs in the regulation of cancer stem cells, Int J Biochem Cell Biol, 108 (2019) 17–20. [DOI] [PubMed] [Google Scholar]
- [133].Zhang Z, Yang W, Li N, Chen X, Ma F, Yang J, Zhang Y, Chai X, Zhang B, Hou X, Luo S, Hua Y, LncRNA MCF2L-AS1 aggravates proliferation, invasion and glycolysis of colorectal cancer cells via the crosstalk with miR-874–3p/FOXM1 signaling axis, Carcinogenesis, (2020). [DOI] [PubMed] [Google Scholar]
- [134].Raychaudhuri P, Park HJ, FoxM1: a master regulator of tumor metastasis, Cancer Res, 71 (2011) 4329–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Lee YS, Shibata Y, Malhotra A, Dutta A, A novel class of small RNAs: tRNA-derived RNA fragments (tRFs), Genes & development, 23 (2009) 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Borek E, Baliga BS, Gehrke CW, Kuo CW, Belman S, Troll W, Waalkes TP, High turnover rate of transfer RNA in tumor tissue, Cancer Res, 37 (1977) 3362–3366. [PubMed] [Google Scholar]
- [137].Gebetsberger J, Polacek N, Slicing tRNAs to boost functional ncRNA diversity, RNA Biol, 10 (2013) 1798–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X, Stress induces tRNA cleavage by angiogenin in mammalian cells, FEBS Lett, 583 (2009) 437–442. [DOI] [PubMed] [Google Scholar]
- [139].Gehrke CW, Kuo KC, Waalkes TP, Borek E, Patterns of urinary excretion of modified nucleosides, Cancer Res, 39 (1979) 1150–1153. [PubMed] [Google Scholar]
- [140].Lakings DB, Waalkes TP, Borek E, Gehrke CW, Mrochek JE, Longmore J, Adamson RH, Composition, associated tissue methyltransferase activity, and catabolic end products of transfer RNA from carcinogen-induced hepatoma and normal monkey livers, Cancer Res, 37 (1977) 285–292. [PubMed] [Google Scholar]
- [141].Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF, Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement, Cell, 161 (2015) 790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Chen LL, Yang L, Regulation of circRNA biogenesis, RNA Biol, 12 (2015) 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK, Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures, Proceedings of the National Academy of Sciences of the United States of America, 73 (1976) 3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H, Circular RNA: A new star of noncoding RNAs, Cancer letters, 365 (2015) 141–148. [DOI] [PubMed] [Google Scholar]
- [145].Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J, Natural RNA circles function as efficient microRNA sponges, Nature, 495 (2013) 384–388. [DOI] [PubMed] [Google Scholar]
- [146].Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S, Purow B, microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma, Cancer Res, 68 (2008) 3566–3572. [DOI] [PubMed] [Google Scholar]
- [147].Fang Y, Xue JL, Shen Q, Chen J, Tian L, MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma, Hepatology, 55 (2012) 1852–1862. [DOI] [PubMed] [Google Scholar]
- [148].Reddy SD, Ohshiro K, Rayala SK, Kumar R, MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions, Cancer Res, 68 (2008) 8195–8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Kristensen LS, Ebbesen KK, Sokol M, Jakobsen T, Korsgaard U, Eriksen AC, Hansen TB, Kjems J, Hager H, Spatial expression analyses of the putative oncogene ciRS-7 in cancer reshape the microRNA sponge theory, Nat Commun, 11 (2020) 4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, Sun H, Pan Y, He B, Wang S, CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7, Cell Death Dis, 9 (2018) 417. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [151].Chen J, Cui L, Yuan J, Zhang Y, Sang H, Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124, Biochem Biophys Res Commun, 494 (2017) 126–132. [DOI] [PubMed] [Google Scholar]
- [152].Tan A, Li Q, Chen L, CircZFR promotes hepatocellular carcinoma progression through regulating miR-3619–5p/CTNNB1 axis and activating Wnt/β-catenin pathway, Arch Biochem Biophys, 661 (2019) 196–202. [DOI] [PubMed] [Google Scholar]
- [153].Dai Y, Li D, Chen X, Tan X, Gu J, Chen M, Zhang X, Circular RNA Myosin Light Chain Kinase (MYLK) Promotes Prostate Cancer Progression through Modulating Mir-29a Expression, Med Sci Monit, 24 (2018) 3462–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Wu J, Jiang Z, Chen C, Hu Q, Fu Z, Chen J, Wang Z, Wang Q, Li A, Marks JR, Guo C, Chen Y, Zhou J, Yang L, Lin C, Wang S, CircIRAK3 sponges miR-3607 to facilitate breast cancer metastasis, Cancer letters, 430 (2018) 179–192. [DOI] [PubMed] [Google Scholar]
- [155].Seto AG, Kingston RE, Lau NC, The coming of age for Piwi proteins, Mol Cell, 26 (2007) 603–609. [DOI] [PubMed] [Google Scholar]
- [156].Siddiqi S, Matushansky I, Piwis and piwi-interacting RNAs in the epigenetics of cancer, J Cell Biochem, 113 (2012) 373–380. [DOI] [PubMed] [Google Scholar]
- [157].Ross RJ, Weiner MM, Lin H, PIWI proteins and PIWI-interacting RNAs in the soma, Nature, 505 (2014) 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Watanabe T, Lin H, Posttranscriptional regulation of gene expression by Piwi proteins and piRNAs, Mol Cell, 56 (2014) 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z, Zhou H, Li QN, piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells, Clin Chim Acta, 412 (2011) 1621–1625. [DOI] [PubMed] [Google Scholar]
- [160].Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z, Guo J, piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells, Cancer letters, 315 (2012) 12–17. [DOI] [PubMed] [Google Scholar]
- [161].Law PT, Qin H, Ching AK, Lai KP, Co NN, He M, Lung RW, Chan AW, Chan TF, Wong N, Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma, J Hepatol, 58 (2013) 1165–1173. [DOI] [PubMed] [Google Scholar]
- [162].Kiss T, Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions, Cell, 109 (2002) 145–148. [DOI] [PubMed] [Google Scholar]
- [163].Gee HE, Buffa FM, Camps C, Ramachandran A, Leek R, Taylor M, Patil M, Sheldon H, Betts G, Homer J, West C, Ragoussis J, Harris AL, The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis, Br J Cancer, 104 (2011) 1168–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Thorenoor N, Slaby O, Small nucleolar RNAs functioning and potential roles in cancer, Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine, 36 (2015) 41–53. [DOI] [PubMed] [Google Scholar]
- [165].Tanaka R, Satoh H, Moriyama M, Satoh K, Morishita Y, Yoshida S, Watanabe T, Nakamura Y, Mori S, Intronic U50 small-nucleolar-RNA (snoRNA) host gene of no protein-coding potential is mapped at the chromosome breakpoint t(3;6)(q27;q15) of human B-cell lymphoma, Genes to cells : devoted to molecular & cellular mechanisms, 5 (2000) 277–287. [DOI] [PubMed] [Google Scholar]
- [166].Mei YP, Liao JP, Shen J, Yu L, Liu BL, Liu L, Li RY, Ji L, Dorsey SG, Jiang ZR, Katz RL, Wang JY, Jiang F, Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis, Oncogene, 31 (2012) 2794–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Okugawa Y, Toiyama Y, Toden S, Mitoma H, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A, Clinical significance of SNORA42 as an oncogene and a prognostic biomarker in colorectal cancer, Gut, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Goeze A, Schluns K, Wolf G, Thasler Z, Petersen S, Petersen I, Chromosomal imbalances of primary and metastatic lung adenocarcinomas, The Journal of pathology, 196 (2002) 8–16. [DOI] [PubMed] [Google Scholar]
- [169].Li W, Kong LB, Li JT, Guo ZY, Xue Q, Yang T, Meng YL, Jin BQ, Wen WH, Yang AG, MiR-568 inhibits the activation and function of CD4(+) T cells and Treg cells by targeting NFAT5, International immunology, 26 (2014) 269–281. [DOI] [PubMed] [Google Scholar]
- [170].Du F, Yu F, Wang Y, Hui Y, Carnevale K, Fu M, Lu H, Fan D, MicroRNA-155 deficiency results in decreased macrophage inflammation and attenuated atherogenesis in apolipoprotein E-deficient mice, Arteriosclerosis, thrombosis, and vascular biology, 34 (2014) 759–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Russo M, Siravegna G, Blaszkowsky LS, Corti G, Crisafulli G, Ahronian LG, Mussolin B, Kwak EL, Buscarino M, Lazzari L, Valtorta E, Truini M, Jessop NA, Robinson HE, Hong TS, Mino-Kenudson M, Di Nicolantonio F, Thabet A, Sartore-Bianchi A, Siena S, Iafrate AJ, Bardelli A, Corcoran RB, Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer, Cancer discovery, 6 (2016) 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Mithraprabhu S, Khong T, Ramachandran M, Chow A, Klarica D, Mai L, Walsh S, Broemeling D, Marziali A, Wiggin M, Hocking J, Kalff A, Durie B, Spencer A, Circulating tumour DNA analysis demonstrates spatial mutational heterogeneity that coincides with disease relapse in myeloma, Leukemia, (2017). [DOI] [PubMed] [Google Scholar]
- [173].Trang P, Wiggins JF, Daige CL, Cho C, Omotola M, Brown D, Weidhaas JB, Bader AG, Slack FJ, Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice, Molecular therapy : the journal of the American Society of Gene Therapy, 19 (2011) 1116–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Gougelet A, Sartor C, Bachelot L, Godard C, Marchiol C, Renault G, Tores F, Nitschke P, Cavard C, Terris B, Perret C, Colnot S, Antitumour activity of an inhibitor of miR-34a in liver cancer with beta-catenin-mutations, Gut, (2015). [DOI] [PubMed] [Google Scholar]
- [175].Zhou K, Nguyen LH, Miller JB, Yan Y, Kos P, Xiong H, Li L, Hao J, Minnig JT, Zhu H, Siegwart DJ, Modular degradable dendrimers enable small RNAs to extend survival in an aggressive liver cancer model, Proceedings of the National Academy of Sciences of the United States of America, 113 (2016) 520–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Wang S, Zhang J, Wang Y, Chen M, Hyaluronic acid-coated PEI-PLGA nanoparticles mediated co-delivery of doxorubicin and miR-542–3p for triple negative breast cancer therapy, Nanomedicine, 12 (2016) 411–420. [DOI] [PubMed] [Google Scholar]
- [177].Xie Y, Murray-Stewart T, Wang Y, Yu F, Li J, Marton LJ, Casero RA Jr., Oupicky D, Self-immolative nanoparticles for simultaneous delivery of microRNA and targeting of polyamine metabolism in combination cancer therapy, Journal of controlled release : official journal of the Controlled Release Society, 246 (2017) 110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Li T, Zhang Y, Meng YP, Bo LS, Ke WB, miR-542–3p Appended Sorafenib/All-trans Retinoic Acid (ATRA)-Loaded Lipid Nanoparticles to Enhance the Anticancer Efficacy in Gastric Cancers, Pharmaceutical research, 34 (2017) 2710–2719. [DOI] [PubMed] [Google Scholar]
- [179].Cui X, Sun Y, Shen M, Song K, Yin X, Di W, Duan Y, Enhanced Chemotherapeutic Efficacy of Paclitaxel Nanoparticles Co-delivered with MicroRNA-7 by Inhibiting Paclitaxel-Induced EGFR/ERK pathway Activation for Ovarian Cancer Therapy, ACS Appl Mater Interfaces, 10 (2018) 7821–7831. [DOI] [PubMed] [Google Scholar]
- [180].Marquez J, Fernandez-Pineiro I, Arauzo-Bravo MJ, Poschmann G, Stuhler K, Khatib AM, Sanchez A, Unda F, Ibarretxe G, Bernales I, Badiola I, Targeting liver sinusoidal endothelial cells with miR-20a-loaded nanoparticles reduces murine colon cancer metastasis to the liver, Int J Cancer, (2018). [DOI] [PubMed] [Google Scholar]
- [181].Zheng X, Zhang F, Zhao Y, Zhang J, Dawulieti J, Pan Y, Cui L, Sun M, Shao D, Li M, He K, Zhang M, Li J, Chen L, Self-assembled dual fluorescence nanoparticles for CD44-targeted delivery of anti-miR-27a in liver cancer theranostics, Theranostics, 8 (2018) 3808–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Zhao P, Li M, Wang Y, Chen Y, He C, Zhang X, Yang T, Lu Y, You J, Lee RJ, Xiang G, Enhancing anti-tumor efficiency in hepatocellular carcinoma through the autophagy inhibition by miR-375/sorafenib in lipid-coated calcium carbonate nanoparticles, Acta Biomater, 72 (2018) 248–255. [DOI] [PubMed] [Google Scholar]
- [183].Wang Z, Zhao K, Zhang Y, Duan X, Zhao Y, Anti-GPC3 Antibody Tagged Cationic Switchable Lipid-Based Nanoparticles for the Co-Delivery of Anti-miRNA27a And Sorafenib in Liver Cancers, Pharmaceutical research, 36 (2019) 145. [DOI] [PubMed] [Google Scholar]
- [184].Ning Q, Liu YF, Ye PJ, Gao P, Li ZP, Tang SY, He DX, Tang SS, Wei H, Yu CY, Delivery of Liver-Specific miRNA-122 Using a Targeted Macromolecular Prodrug toward Synergistic Therapy for Hepatocellular Carcinoma, ACS Appl Mater Interfaces, 11 (2019) 10578–10588. [DOI] [PubMed] [Google Scholar]
- [185].Parayath NN, Gandham SK, Leslie F, Amiji MM, Improved anti-tumor efficacy of paclitaxel in combination with MicroRNA-125b-based tumor-associated macrophage repolarization in epithelial ovarian cancer, Cancer letters, 461 (2019) 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Zhao Y, Xu J, Le VM, Gong Q, Li S, Gao F, Ni L, Liu J, Liang X, EpCAM Aptamer-Functionalized Cationic Liposome-Based Nanoparticles Loaded with miR-139–5p for Targeted Therapy in Colorectal Cancer, Mol Pharm, (2019). [DOI] [PubMed] [Google Scholar]
- [187].Wach S, Brandl M, Borchardt H, Weigelt K, Lukat S, Nolte E, Al-Janabi O, Hart M, Grässer F, Giedl J, Jung R, Stöhr R, Hartmann A, Lieb V, Höbel S, Peters A, Stäubert C, Wullich B, Taubert H, Aigner A, Exploring the MIR143-UPAR Axis for the Inhibition of Human Prostate Cancer Cells In Vitro and In Vivo, Molecular therapy. Nucleic acids, 16 (2019) 272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Chen W, Zhou Y, Zhi X, Ma T, Liu H, Chen BW, Zheng X, Xie S, Zhao B, Feng X, Dang X, Liang T, Delivery of miR-212 by chimeric peptide-condensed supramolecular nanoparticles enhances the sensitivity of pancreatic ductal adenocarcinoma to doxorubicin, Biomaterials, 192 (2019) 590–600. [DOI] [PubMed] [Google Scholar]
- [189].Moro M, Di Paolo D, Milione M, Centonze G, Bornaghi V, Borzi C, Gandellini P, Perri P, Pastorino U, Ponzoni M, Sozzi G, Fortunato O, Coated cationic lipid-nanoparticles entrapping miR-660 inhibit tumor growth in patient-derived xenografts lung cancer models, Journal of controlled release : official journal of the Controlled Release Society, 308 (2019) 44–56. [DOI] [PubMed] [Google Scholar]
- [190].Mokhlis HA, Bayraktar R, Kabil NN, Caner A, Kahraman N, Rodriguez-Aguayo C, Zambalde EP, Sheng J, Karagoz K, Kanlikilicer P, Abdel Aziz AAH, Abdelghany TM, Ashour AA, Wong S, Gatza ML, Calin GA, Lopez-Berestein G, Ozpolat B, The Modulatory Role of MicroRNA-873 in the Progression of KRAS-Driven Cancers, Molecular therapy. Nucleic acids, 14 (2019) 301–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Ahir M, Upadhyay P, Ghosh A, Sarker S, Bhattacharya S, Gupta P, Ghosh S, Chattopadhyay S, Adhikary A, Delivery of dual miRNA through CD44-targeted mesoporous silica nanoparticles for enhanced and effective triple-negative breast cancer therapy, Biomater Sci, (2020). [DOI] [PubMed] [Google Scholar]
- [192].Jin H, Yu Y, Chrisler WB, Xiong Y, Hu D, Lei C, Delivery of MicroRNA-10b with Polylysine Nanoparticles for Inhibition of Breast Cancer Cell Wound Healing, Breast cancer : basic and clinical research, 6 (2012) 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Klimenko OV, Shtilman MI, Transfection of Kasumi-1 cells with a new type of polymer carriers loaded with miR-155 and antago-miR-155, Cancer gene therapy, 20 (2013) 237–241. [DOI] [PubMed] [Google Scholar]
- [194].Nishimura M, Jung EJ, Shah MY, Lu C, Spizzo R, Shimizu M, Han HD, Ivan C, Rossi S, Zhang X, Nicoloso MS, Wu SY, Almeida MI, Bottsford-Miller J, Pecot CV, Zand B, Matsuo K, Shahzad MM, Jennings NB, Rodriguez-Aguayo C, Lopez-Berestein G, Sood AK, Calin GA, Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment, Cancer discovery, 3 (2013) 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Di Martino MT, Gulla A, Gallo Cantafio ME, Altomare E, Amodio N, Leone E, Morelli E, Lio SG, Caracciolo D, Rossi M, Frandsen NM, Tagliaferri P, Tassone P, In vitro and in vivo activity of a novel locked nucleic acid (LNA)-inhibitor-miR-221 against multiple myeloma cells, PloS one, 9 (2014) e89659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Dorrance AM, Neviani P, Ferenchak GJ, Huang X, Nicolet D, Maharry KS, Ozer HG, Hoellarbauer P, Khalife J, Hill EB, Yadav M, Bolon BN, Lee RJ, Lee LJ, Croce CM, Garzon R, Caligiuri MA, Bloomfield CD, Marcucci G, Targeting leukemia stem cells in vivo with antagomiR-126 nanoparticles in acute myeloid leukemia, Leukemia, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Huang X, Magnus J, Kaimal V, Karmali P, Li J, Walls M, Prudente R, Sung E, Sorourian M, Lee R, Davis S, Yang X, Estrella H, Lee EC, Chau BN, Pavlicek A, Zabludoff S, Lipid Nanoparticle-Mediated Delivery of Anti-miR-17 Family Oligonucleotide Suppresses Hepatocellular Carcinoma Growth, Molecular cancer therapeutics, 16 (2017) 905–913. [DOI] [PubMed] [Google Scholar]
- [198].Rui M, Qu Y, Gao T, Ge Y, Feng C, Xu X, Simultaneous delivery of anti-miR21 with doxorubicin prodrug by mimetic lipoprotein nanoparticles for synergistic effect against drug resistance in cancer cells, International journal of nanomedicine, 12 (2017) 217–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Bose RJC, Uday Kumar S, Zeng Y, Afjei R, Robinson E, Lau K, Bermudez A, Habte F, Pitteri SJ, Sinclair R, Willmann JK, Massoud TF, Gambhir SS, Paulmurugan R, Tumor Cell-Derived Extracellular Vesicle-Coated Nanocarriers: An Efficient Theranostic Platform for the Cancer-Specific Delivery of Anti-miR-21 and Imaging Agents, ACS Nano, 12 (2018) 10817–10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Vandghanooni S, Eskandani M, Barar J, Omidi Y, AS1411 aptamer-decorated cisplatin-loaded poly(lactic-co-glycolic acid) nanoparticles for targeted therapy of miR-21-inhibited ovarian cancer cells, Nanomedicine (Lond), 13 (2018) 2729–2758. [DOI] [PubMed] [Google Scholar]
- [201].Li M, Su Y, Zhang F, Chen K, Xu X, Xu L, Zhou J, Wang W, A dual-targeting reconstituted high density lipoprotein leveraging the synergy of sorafenib and antimiRNA21 for enhanced hepatocellular carcinoma therapy, Acta Biomater, 75 (2018) 413–426. [DOI] [PubMed] [Google Scholar]
- [202].Malhotra M, Sekar TV, Ananta JS, Devulapally R, Afjei R, Babikir HA, Paulmurugan R, Massoud TF, Targeted nanoparticle delivery of therapeutic antisense microRNAs presensitizes glioblastoma cells to lower effective doses of temozolomide, Oncotarget, 9 (2018) 21478–21494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Wang X, Zhang H, Bai M, Ning T, Ge S, Deng T, Liu R, Zhang L, Ying G, Ba Y, Exosomes Serve as Nanoparticles to Deliver Anti-miR-214 to Reverse Chemoresistance to Cisplatin in Gastric Cancer, Molecular therapy : the journal of the American Society of Gene Therapy, 26 (2018) 774–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Deng R, Shen N, Yang Y, Yu H, Xu S, Yang YW, Liu S, Meguellati K, Yan F, Targeting epigenetic pathway with gold nanoparticles for acute myeloid leukemia therapy, Biomaterials, 167 (2018) 80–90. [DOI] [PubMed] [Google Scholar]
- [205].Yin H, Xiong G, Guo S, Xu C, Xu R, Guo P, Shu D, Delivery of Anti-miRNA for Triple-Negative Breast Cancer Therapy Using RNA Nanoparticles Targeting Stem Cell Marker CD133, Molecular therapy : the journal of the American Society of Gene Therapy, 27 (2019) 1252–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Chen X, Mangala LS, Mooberry L, Bayraktar E, Dasari SK, Ma S, Ivan C, Court KA, Rodriguez-Aguayo C, Bayraktar R, Raut S, Sabnis N, Kong X, Yang X, Lopez-Berestein G, Lacko AG, Sood AK, Identifying and targeting angiogenesis-related microRNAs in ovarian cancer, Oncogene, 38 (2019) 6095–6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Aguilo F, Zhou MM, Walsh MJ, Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression, Cancer Res, 71 (2011) 5365–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Zhang ZX, Liu ZQ, Jiang B, Lu XY, Ning XF, Yuan CT, Wang AL, BRAF activated non-coding RNA (BANCR) promoting gastric cancer cells proliferation via regulation of NF-kappaB1, Biochem Biophys Res Commun, 465 (2015) 225–231. [DOI] [PubMed] [Google Scholar]
- [209].Sun X, Jiao X, Pestell TG, Fan C, Qin S, Mirabelli E, Ren H, Pestell RG, MicroRNAs and cancer stem cells: the sword and the shield, Oncogene, 33 (2014) 4967–4977. [DOI] [PubMed] [Google Scholar]
- [210].Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, Yang L, Chen LL, Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus, Cell Res, 24 (2014) 513–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Zhang X, Xu Y, He C, Guo X, Zhang J, Zhang L, Kong M, Chen B, Zhu C, Elevated expression of CCAT2 is associated with poor prognosis in esophageal squamous cell carcinoma, J Surg Oncol, 111 (2015) 834–839. [DOI] [PubMed] [Google Scholar]
- [212].Wang CY, Hua L, Yao KH, Chen JT, Zhang JJ, Hu JH, Long non-coding RNA CCAT2 is up-regulated in gastric cancer and associated with poor prognosis, Int J Clin Exp Pathol, 8 (2015) 779–785. [PMC free article] [PubMed] [Google Scholar]
- [213].Redis RS, Sieuwerts AM, Look MP, Tudoran O, Ivan C, Spizzo R, Zhang X, de Weerd V, Shimizu M, Ling H, Buiga R, Pop V, Irimie A, Fodde R, Bedrosian I, Martens JW, Foekens JA, Berindan-Neagoe I, Calin GA, CCAT2, a novel long non-coding RNA in breast cancer: expression study and clinical correlations, Oncotarget, 4 (2013) 1748–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Graham LD, Pedersen SK, Brown GS, Ho T, Kassir Z, Moynihan AT, Vizgoft EK, Dunne R, Pimlott L, Young GP, Lapointe LC, Molloy PL, Colorectal Neoplasia Differentially Expressed (CRNDE), a Novel Gene with Elevated Expression in Colorectal Adenomas and Adenocarcinomas, Genes Cancer, 2 (2011) 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Wu H, Liu B, Chen Z, Li G, Zhang Z, MSC-induced lncRNA HCP5 drove fatty acid oxidation through miR-3619–5p/AMPK/PGC1α/CEBPB axis to promote stemness and chemoresistance of gastric cancer, Cell Death Dis, 11 (2020) 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Bhan A, Mandal SS, LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer, Biochimica et biophysica acta, 1856 (2015) 151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Yang X, Xie X, Xiao YF, Xie R, Hu CJ, Tang B, Li BS, Yang SM, The emergence of long non-coding RNAs in the tumorigenesis of hepatocellular carcinoma, Cancer letters, 360 (2015) 119–124. [DOI] [PubMed] [Google Scholar]
- [218].Peng W, Gao W, Feng J, Long noncoding RNA HULC is a novel biomarker of poor prognosis in patients with pancreatic cancer, Med Oncol, 31 (2014) 346. [DOI] [PubMed] [Google Scholar]
- [219].Li W, Kang Y, A new Lnc in metastasis: long noncoding RNA mediates the prometastatic functions of TGF-beta, Cancer cell, 25 (2014) 557–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Eades G, Wolfson B, Zhang Y, Li Q, Yao Y, Zhou Q, lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6, Mol Cancer Res, 13 (2015) 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Josephy PD, Chiu AL, Eling TE, Prostaglandin H synthase-dependent mutagenic activation of benzidine in a Salmonella typhimurium Ames tester strain possessing elevated N-acetyltransferase levels, Cancer Res, 49 (1989) 853–856. [PubMed] [Google Scholar]
- [222].Wei Y, Niu B, Role of MALAT1 as a Prognostic Factor for Survival in Various Cancers: A Systematic Review of the Literature with Meta-Analysis, Dis Markers, 2015 (2015) 164635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Tian X, Xu G, Clinical value of lncRNA MALAT1 as a prognostic marker in human cancer: systematic review and meta-analysis, BMJ Open, 5 (2015) e008653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Yoshimoto R, Mayeda A, Yoshida M, Nakagawa S, MALAT1 long non-coding RNA in cancer, Biochimica et biophysica acta, (2015). [DOI] [PubMed] [Google Scholar]
- [225].Salameh A, Lee AK, Cardó-Vila M, Nunes DN, Efstathiou E, Staquicini FI, Dobroff AS, Marchiò S, Navone NM, Hosoya H, Lauer RC, Wen S, Salmeron CC, Hoang A, Newsham I, Lima LA, Carraro DM, Oliviero S, Kolonin MG, Sidman RL, Do KA, Troncoso P, Logothetis CJ, Brentani RR, Calin GA, Cavenee WK, Dias-Neto E, Pasqualini R, Arap W, PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3, Proc Natl Acad Sci U S A, 112 (2015) 8403–8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Yang L, Qiu M, Xu Y, Wang J, Zheng Y, Li M, Xu L, Yin R, Upregulation of long non-coding RNA PRNCR1 in colorectal cancer promotes cell proliferation and cell cycle progression, Oncol Rep, 35 (2016) 318–324. [DOI] [PubMed] [Google Scholar]
- [227].Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, Evans CP, Rosenfeld MG, lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs, Nature, 500 (2013) 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Colombo T, Farina L, Macino G, Paci P, PVT1: a rising star among oncogenic long noncoding RNAs, Biomed Res Int, 2015 (2015) 304208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Xue M, Chen W, Li X, Urothelial cancer associated 1: a long noncoding RNA with a crucial role in cancer, J Cancer Res Clin Oncol, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Ma C, Shi X, Zhu Q, Li Q, Liu Y, Yao Y, Song Y, The growth arrest-specific transcript 5 (GAS5): a pivotal tumor suppressor long noncoding RNA in human cancers, Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine, (2015). [DOI] [PubMed] [Google Scholar]
- [231].Yu X, Li Z, Long non-coding RNA growth arrest-specific transcript 5 in tumor biology, Oncol Lett, 10 (2015) 1953–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Zhai H, Fesler A, Schee K, Fodstad O, Flatmark K, Ju J, Clinical significance of long intergenic noncoding RNA-p21 in colorectal cancer, Clin Colorectal Cancer, 12 (2013) 261–266. [DOI] [PubMed] [Google Scholar]
- [233].Zhou Y, Zhang X, Klibanski A, MEG3 noncoding RNA: a tumor suppressor, J Mol Endocrinol, 48 (2012) R45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Huang MD, Chen WM, Qi FZ, Sun M, Xu TP, Ma P, Shu YQ, Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2, Mol Cancer, 14 (2015) 165. [DOI] [PMC free article] [PubMed] [Google Scholar]


