Abstract
Magnetic resonance imaging (MRI) is an increasingly important tool for the diagnosis and treatment of prostate cancer. However, interpretation of MRI suffers from high inter-observer variability across radiologists, thereby contributing to missed clinically significant cancers, overdiagnosed low-risk cancers, and frequent false positives. Interpretation of MRI could be greatly improved by providing radiologists with an answer key that clearly shows cancer locations on MRI. Registration of histopathology images from patients who had radical prostatectomy to pre-operative MRI allows such mapping of ground truth cancer labels onto MRI. However, traditional MRI-histopathology registration approaches are computationally expensive and require careful choices of the cost function and registration hyperparameters. This paper presents ProsRegNet, a deep learning-based pipeline to accelerate and simplify MRI-histopathology image registration in prostate cancer. Our pipeline consists of image preprocessing, estimation of affine and deformable transformations by deep neural networks, and mapping cancer labels from histopathology images onto MRI using estimated transformations. We trained our neural network using MR and histopathology images of 99 patients from our internal cohort (Cohort 1) and evaluated its performance using 53 patients from three different cohorts (an additional 12 from Cohort 1 and 41 from two public cohorts). Results show that our deep learning pipeline has achieved more accurate registration results and is at least 20 times faster than a state-of-the-art registration algorithm. This important advance will provide radiologists with highly accurate prostate MRI answer keys, thereby facilitating improvements in the detection of prostate cancer on MRI. Our code is freely available at https://github.com/pimed//ProsRegNet.
Keywords: Image registration, radiology-pathology fusion, MRI, Histopathology, prostate cancer, deep learning
1. Introduction
Prostate cancer is the second leading cause of cancer death and the most diagnosed cancer in men in the United States, with an estimated 33,330 deaths and 191,930 new cases in 2020 (American Cancer Society 2020). Diagnosis, staging, and treatment planning of prostate cancer is increasingly assisted by magnetic resonance imaging (MRI) (Turkbey et al., 2012); Verma et al. (2012). The Prostate Imaging Reporting and Data System (PI-RADS) Weinreb et al. (2016) was developed to standardize the acquisition, interpretation, and reporting of prostate MRI. Despite the widespread adoption of PIRADS, the performance of MRI still suffers from high levels of variation across radiologists Sonn et al. (2019), reduced positive predictive value (27-58%) Westphalen et al. (2020), low inter-reader agreement (k = 0.46-0.78) Ahmed et al. (2017), and large variations in reported sensitivity (58-96%) and specificity (23-87%) Ahmed et al. (2017). It also has been shown that high interobserver disagreement in prostate MRI significantly affects prostate biopsy practice including aborting planned biopsy and reduced number of region of interest samples Rosenzweig et al. (2020). One major barrier to improvement in MRI interpretation is the lack of a pathologic reference standard to provide radiologists detailed feedback about their performance. Image registration Shao et al. (2016) of the pre-surgical MRI with histopathology images after surgical resection of the prostate (radical prostatectomy) addresses this issue by enabling mapping of the extent of cancer from the ground-truth histopathology images onto the MRI. Such mapping allows side-by-side comparison of the histopathology and MRI images, which can be use in the training of radiologists to improve their interpretation of MRI. Furthermore, accurate cancer labels achieved by image registration may facilitate the development of radiomic and deep learning approaches for early prostate cancer detection and risk stratification on pre-operative MRI Cao et al. (2019); Lovegrove et al. (2016); Wang et al. (2018); (Bhattacharya et al., 2020).
Several MRI-histopathology image registration approaches have been developed to account for elastic tissue deformation occurred during histological preparation inducing tissue fixation, sectioning, and mounting on histologic slides. Turkbey et al. developed patient-specific 3D printed molds for the resected prostate that are designed based on the pre-operative MRI and allow sectioning of the prostate in-plane with the same slice thickness as MRI Turkbey et al. (2011). A radiologist will first carefully segment the edge of the prostate gland as well as indicate areas of suspected prostate cancer. From this segmentation, a 3D volume will be created, which is then imported into a modeling software to create a personalized mold such that the orientation of the prostate specimen is aligned with the original MRI to guide the gross sectioning of the ex vivo prostate to have exact slice correspondences with the MRI Priester et al. (2014). Several approaches Losnegård et al. (2018); Rusu et al. (2019); Wu et al. (2019) rely on patient-specific 3D printed molds to establish better histopathology and MRI slice correspondences in order to improve the registration of MRI and histopathology images. While some approaches work directly with MR and histopathology images alone, others require additional steps including a separate ex vivo MRI of the prostate Wu et al. (2019), fiducial markers Ward et al. (2012), or advanced image similarity metrics Chappelow et al. (2011); Li et al. (2017). Several pipelines have been developed for direct integration of MR and histopathology images by 3D histopathology volume reconstruction Losnegård et al. (2018); Rusu et al. (2019); Samavati et al. (2011); Stille et al. (2013), but most are time-consuming, computationally expensive, and can suffer from partial volume effects and artifacts due to large spacing between images.
Typically, a geometric transformation can be parameterized by either a few (affine) or a large number of (deformable) variables. Previous automated MRI-histopathology registration approaches estimate variables that encode geometric transformations by optimizing a cost function for tens or hundreds of iterations (Goubran et al., 2013, 2015); Rusu et al. (2017). Therefore, this optimization process is computationally expensive and can take several minutes to finish. Moreover, the estimated transformation is often sensitive to the choice of hyperparameters (e.g., the number of iterations and the cost function), making traditional registration approaches complex to set up and reducing their generalization.
To address this important gap, this paper presents a deep learning based pipeline for efficient MRI-histopathology registration. In the past few years, deep learning has been successfully used in many medical image registration problems. A deep learning based registration network can be considered as a function that takes two images, a fixed image and a moving image, as the input and directly outputs a unique transformation without requiring additional optimization. Many deep learning approaches (Balakrishnan et al., 2018, 2019); Dalca et al. (2018); Ghosal and Ray (2017); Krebs et al. (2019); Yang et al. (2017); Zhang (2018) assume that the fixed and moving images have already been aligned by affine registration and only focus on the deformable registration. However, the affine registration of MRI and histopathology images of the prostate is challenging since they are considerably different modalities while having different contents. Therefore, prior deformable registration approaches cannot be directly used for MRI histopathology registration where affine registration is a necessity due to large geometric changes of the prostate during histological preparation. Rocco et al. proposed a multi-stage deep learning framework (CNNGeometric) that can handle both affine and deformable deformations of natural images Rocco et al. (2017). Inspired by their study, we developed the ProsRegNet registration pipeline for affine and deformable registration of the MRI and histopathology images. Our registration pipeline includes preprocessing and postprocessing modules, and the registration network that estimates an affine transformation at the first stage and a more accurate thin-plate-spline transformation at the second stage. Some other deep learning registration approaches can also jointly estimate the affine and deformable transformations de Vos et al. (2019); Shen et al. (2019). Similar to our ProsRegNet approach, the approach developed in de Vos et al. (2019) used a feature extraction network followed by a parameter estimation network. Unlike our approach, their model lacked the feature matching component which has been shown to increase the generalization capabilities of registration networks to unseen images Rocco et al. (2017). In our study, we will show that our ProsRegNet registration network trained with images from one cohort generalizes well to unseen images from other cohorts. Moreover, the models developed by (de Vos et al., 2019); Shen et al. (2019) were trained in an unsupervised manner using the normalized cross correlation, which can be unsuitable for MRI-histopathology registration as the intensities are not correlated. To our knowledge, we are the first to apply deep learning to the problem of MRI-histopathology registration of the prostate. We will demonstrate that our deep learning registration pipeline can achieve better registration accuracy than the state-of-the-art RAPSODI approach Rusu et al. (2020) while being much more computationally efficient and easier to use for non-experts users.
This paper has the following major contributions:
We are the first to use deep learning to solve the challenging problem of registering MRI and histopathology images of the prostate.
We avoid the shortcomings of multi-modal similarity measures for MRI-histopathology registration by training our registration network with mono-modal synthetic image pairs in an unsupervised manner using a mono-modal dissimilarity measure. During the testing, we applied our network to multi-modal image registration as the network has learned how to solve image registration problems irrespective of the image modalities.
We improved the stability of the training by parameterizing the transformations using the sum of an identity transform and the estimated parameter vector scaled by a small weight.
We trained our network with a large set of MRI and histopathology prostate images and evaluated our approach relative to the state-of-the-art traditional and deep learning registration methods.
Our code is one of the very few freely available MRI-histopathology registration codes.
2. Materials and methods
2.1. Data acquisition
This study approved by the Institutional Review Board at Stanford University included 152 subjects with biopsy-confirmed prostate cancer from three cohorts at different institutions. The first cohort consists of 111 patients who had a pre-operative MRI scan and underwent radical prostatectomy at Stanford University. The excised prostate was submitted for histological preparation and we used a patient-specific 3D printed mold to generate whole-mount histopathology images that had slice-to-slice correspondences with the MRI. Experts determined the correspondences between T2-weighted (T2-w) MRI and histopathology slices. The prostate, cancer, urethra, and other anatomic landmarks on histopathology images were manually segmented by an expert genitourinary pathologist. Two hundred fifty-seven anatomic landmarks visible on both MRI and histopathology images, e.g., benign prostate hyperplasia nodules and ejaculatory ducts were chosen for a subset of 12 subjects from the first cohort. The second cohort consisted of 16 patients from the publicly available “Prostate Fused-MRI Pathology” dataset in The Cancer Imaging Archive (TCIA) [dataset] Madabhushi and Feldman (2016). Each patient had an MRI along with digitized histopathology images of the corresponding radical prostatectomy specimen. Each surgically excised prostate specimen was originally sectioned and quartered resulting in four images for each section. The four images were then digitally stitched together to produce a pseudowhole mount section. Annotations of cancer presence on the pseudo-whole mount sections were made by an expert pathologist. Slice correspondences were established between the individual T2-w MRI and stitched pseudo-whole mount sections by the program in Toth et al. (2014) and checked for accuracy by an expert pathologist and radiologist. The third cohort consisted of 25 patients from the publicly available TCIA “Prostate-MRI” dataset [dataset] Choyke et al. (2016). Each patient had a preoperative MRI and underwent a radical prostatectomy. A mold was generated from each MRI, and the prostatectomy specimen was first placed in the mold, then cut in the same plane as the MRI. The data was generated at the National Cancer Institute, Bethesda, Maryland, USA between 2008-2010. For all of the three cohorts, the prostate on each MRI scan was manually segmented and used in the registration procedure. The prostate segmentation serves to drive the alignment while the urethra and other anatomic landmarks were only used to evaluate the registration. We summarized details of datasets from the above three cohorts in Table 1.
Table 1.
Summary of datasets. We used 152 = 111 + 16 + 25 patients from three cohorts. T2-w MRI: T2-weighted MRI, H&E: Hematoxylin and Eosin, TR: repetition time, TE: echo time, H: in-plane image height, W: in-plane image width, D: through-plane image depth.
| Cohort 1 (Stanford) | Cohort 2 (TCIA) | Cohort 3 (TCIA) | ||||
|---|---|---|---|---|---|---|
| Number of patients | 111 |
16 |
25 |
|||
| Modality | MRI | Histology | MRI | Histology | MRI | Histology |
| Manufacturer | GE | - | Siemens | - | Philips | - |
| Coil type | Surface | - | Endorectal | - | Endorectal | - |
| Sequence | T2-w MRI | Whole-mount | T2-w MRI | Pseudo-whole mount | T2-w MRI | Blockface-whole mount |
| Acquisition characteristics | TR: [3.9s, 6.3s], TE: [122ms, 130ms] | H&E stained, 3D-printed mold | TR: [3.7s, 7.0s], TE: 107ms | H&E stained | TR: 8.9s, TE: 120 ms | H&E stained, mold |
| Image size | H,W: {256, 512}, D: [24, 43] | H,W:[1663, 7556] | H,W: 320x320, D: [21, 31] | H,W:[2368, 6324] | H,W: 512, D: 26 | H,W: [496, 2881] |
| In-plane resolution (mm) | [0.27, 0.94] | {0.0081, 0.0162} | [0.41, 0.43] | 0.0072 | 0.27 | {0.0846, 0.0216} |
| Distance between slices | [3mm, 5.2mm] | [3mm, 5.2mm] | 4mm | Free hand | 3mm | 3mm |
2.2. State-of-the-art RAPSODI registration framework
We briefly summarize the state-of-the-art RAPSODI (Radiology pathology spatial open-source multi-dimensional integration) framework for the registration of MRI and histopathology images Rusu et al. (2020). The RAPSODI approach assumes known slice correspondences between MRI and histopathology images, and starts with 3D reconstruction of the histopathology specimen by registering each histopathology slice to its adjacent slice. The purpose of the 3D reconstruction of the histopathology volume is to initialize the histopathology slices in the registration with the MRI. Then 2D rigid, affine and deformable transformations between each histopathology image and the corresponding T2-w MRI slice are estimated iteratively using gradient descent. The rigid and affine registrations use the prostate masks as the input and the sum of squared differences as the cost function. The deformable registration uses the images masked by the prostate segmentation as the input, free-from deformations as the deformation model and the Mattes mutual information as the cost function. Early stopping is used in the deformable registration to prevent overfitting. Compared to our deep learning registration approach, RAPSODI requires significant user input including careful choice of similarity metric and registration hyperparameters such as step size, and the number of iterations. The RAPSODI approach has been shown to be highly accurate and we will compare it with our deep learning registration pipeline.
2.3. Deep learning ProsRegNet pipeline
We propose the ProsRegNet (Prostate Registration Network) pipeline to register T2-w MRI and histopathology images, which consists of image preprocessing, transformation estimation by deep neural networks, and postprocessing, as shown in Fig. 1.
Fig. 1.
Proposed pipeline for registration of MRI and histopathology images. The yellow rectangle highlights the prostate in the MRI slice. The preprocessed images IA and IB represent the moving and the fixed images, respectively. Images IA and IB are fed into the image registration neural network to estimate θ that represents the affine and nonrigid transformation parameters. Cancer labels (the red outlines) in the histopathology slice are then deformed into the MRI slice using the estimated transformations.
2.3.1. Preprocessing
Mounting of tissue sections on glass slides can produce several significant artifacts, including tissue shrinkage, in-plane rotation and horizontal flipping, that will affect alignment with the corresponding MR images. We manually corrected for the gross rotation angle and determined whether horizontal flipping was present for each histopathology slice, as shown in IA in Fig. 1. We applied the same rotation and flip transformations to the binary mask of the prostate, cancer regions, urethra, and other regions of the prostate in the histopathology slice. A bounding box around the prostate mask was applied to extract prostate slices from the T2-w MRI, as shown in IB in Fig. 1. We normalized the intensity of each cropped MRI slice from 0 to 255. The histopathology and MRI images were multiplied by the corresponding prostate masks to facilitate the registration process. The resulting images IA and IB were then resampled to 240×240 before feeding into the registration neural networks. This preprocessing procedure has been applied to images going through the CNNGeometric and ProsRegNet networks.
2.3.2. Image registration neural networks
Both ProsRegNet and CNNGeometric registration networks consisted of feature extraction, feature matching, and transformation parameter estimation and utilized a two-stage registration architecture (see Fig. 2). In the first stage, an affine transformation was estimated to align the two images globally. In the second stage, the affine transformation is used as an initial transform to facilitate the estimation of a more accurate thin-plate spline (TPS) transformation. There are two major differences between our ProgRegNet model and the prior CNN Geometric model. First, our ProgRegNet model used image intensity differences to train the registration networks in an unsupervised manner, while CNNGeometric used a loss based on point location differences in a supervised training. Second, our ProsRegNet model improved the stability of the training by parameterizing the transformations using the sum of an identity transform and the estimated parameter vector scaled by a small weight, while CNNGeometric directly used the estimated parameter vector.
Fig. 2.
Two-stage registration framework using deep neural networks (Rocco et al., 2017). The first stage estimates an affine transform that globally aligns the two images. The second stage uses the affine transform as initialization to determine a thin-plate spline transform. Composing the two transforms gives the resulting correspondence map between IA and IB.
We use the same feature extraction and regression networks as in Rocco et al. (2017). The inputs to the geometric matching networks are a moving image IA and a fixed image IB. Those two images were passed through the same pre-trained feature extraction convolutional neural network (ResNet-101 He et al. (2016) network cropped at the third layer) to produce the corresponding feature maps fA and fB, respectively. Each feature map is an image of size (w, h) whose value at each voxel is a d-dimensional vector, where d is the number of features. The feature maps fA and fB were fed into a correlation layer followed by normalization. The correlation layer combines fA and fB into a single correlation map cAB of the same size. At each voxel location (i, j), cAB(i, j) is a vector of length wh whose k-th element is given by:
| (1) |
where k = h( jk 1)+ik. The correlation map cAB was normalized using a rectified linear unit (ReLU) followed by a channel-wise L2-normalization. The resulting tentative correspondence map fAB was passed through a regression network to estimate parameters of the geometric transformation between IA and IB. The regression network consisted of two stacked layers, where each layer begins with a convolutional unit and is followed by batch normalization and ReLU. A final fully connected (FC) layer regresses the parameters of the geometric transform, as shown in Fig. 3.
Fig. 3.
Regression network for estimating transformation parameters from the correspondence map fAB Rocco et al. (2017).
The output of the regression network (θ) is a vector of 6 elements when performing affine registration. Unlike Rocco et al. (2017) that directly use θ = (θ1 , … ,θ6) as the affine matrix, we propose to use , where α is a small number and is the parameter vector for identity affine transform. To be more specific, the affine transformation associated with θ is given by:
| (2) |
where (x, y) is any spatial location, and we choose α = 0.1 in this paper.
Using instead of α as the affine matrix guarantees that the initial estimate of ϕθ during the network training is close to the identity map and thus improves the stability of our registration network. We parameterize the nonrigid transformations using a thin-plate spline grid of size 6x6 instead of 3 × 3 in Rocco et al. (2017) for more accurate registration. This requires θ to be a vector of 2 × 6 × 6 = 72 elements. Similarly, we use instead of θ to parameterize the nonrigid transforms, where is the parameter vector for the identity thin-plate spline transform.
Unlike Rocco et al. (2017) that uses the differences between the original and deformed coordinate locations (location matching error) as the loss function, we define the loss function as the sum of squared differences (SSD) between the fixed and the deformed image (since IA and IB are from the same modality during the training):
| (3) |
where ϕθ is the transformation parameterized by θ, and W and H are the width and height of the images. Since all MR and histopathology images have been masked during the preprocessing, our SSD cost can quickly drive the registration process during the training.
2.3.3. Postprocessing
After affine and deformable image registrations, the histopathology images and the prostate, urethra, anatomic landmarks, and cancer labels on the histopathology images were mapped to the corresponding MRI slices using the estimated composite (affine + deformable) transformation. Although the histopathology images were resampled to have a size of 240×240, the deformed histopathology images still have the same size as the original histopathology images since we applied the estimated composite transformation directly to the original high-resolution histopathology images. For visualization purposes, sampling artifacts in the deformed images were removed by binary thresholding to set the intensity of pixels outside the prostate to be zero.
2.4. Training dataset
Since the ground truth spatial correspondences between the MRI and histopathology images are lacking, we trained our neural networks using uni-modal image pairs generated by synthetic transformations (Fig. 4). For each 2D image IA, we applied a simulated transformation ϕ to deform it into the image IB. The 3-tuple (IA, IB, ϕ) will be used as one training example. The transformation ϕ can be either an affine transformor a thin-plate spline transform. To guarantee the plausibility of the simulated transformations, the variables used to parameterize the transformations were randomly sampled from bounded intervals. When simulating the affine transformations, the rotation angle ranged from −10 degrees to +10 degrees, the scaling coefficients ranged from 0.8 to 1.2, the shifting coefficients were within 5% of the image size, and the shearing coefficients were within 5%. When simulating the thin-plate-spline transformations, the movement of each control point was within 5% of the image size. We chose these intervals as they represent typical transformation ranges we observed when using RAPSODI and were shown to be sufficiently wide to cover the transformations observed in our diverse patient cohorts. For the training, we used 1,390 MRI and histopathology images and the corresponding prostate masks of 99 patients from Cohort 1. Prostate masks were used to train the affine registration network and masked MRI and histopathology images were used to train the deformable registration network. Although our registration neural network was trained with image pairs of the same modality, we will show that it can be generalized to the multi-modal registration of MRI and histopathology images for all three cohorts.
Fig. 4.
Generating training dataset by applying known transformations. IA is the original image, ϕ is either an affine or thin-plate spline transform, and IB is the deformed image by applying ϕ to IA. Each tuple (IA, IB, ϕ) is considered as one training example.
2.5. Experiments
We trained the neural networks on the NVIDIA GeForce RTX 2080 GPU (8GB memory, 14000 MHz clock speed). We used an initial learning rate of 0.001, a learning rate decay of 0.95, a batch size of 64, and the Adam optimizer Kingma and Ba (2017), for which both the affine and deformable registration networks were trained with 50 epochs. For each deformation model, the network with the minimum validation loss during the training was used in the testing.
In total, we experimented with three different approaches for registration of MRI and the corresponding histopathology images: the traditional RAPSODI registration framework Rusu et al. (2020) (RAPSODI), a prior deep learning registration framework developed by Rocco et al. Rocco et al. (2017) (CNNGeometric), and our deep learning ProsRegNet pipeline (ProsRegNet), We tested the RAPSODI approach on the Intel Core i9-9900K CPU (8-Core, 16-Thread, 3.6 GHz (5.0 GHz Turbo)) and tested the CNNGeometric and ProsRegNet approaches on the GeForce RTX 2080 GPU. In total, we used datasets of 53 prostate cancer patients (12 from Cohort 1, 16 from Cohort 2, and 25 from Cohort 3) to evaluate the performance of the above three registration approaches.
2.6. Evaluation metrics
The Dice coefficient, the Hausdorff distance, and the mean landmark error were used to evaluate the alignment accuracy for the deformed histopathology and the corresponding MRI images. The Dice coefficient measures the relative overlap between MA and MB, which is given by:
| (4) |
where MA denotes the deformed histopathology prostate mask, MB denotes corresponding MRI prostate mask, and denotes the cardinality (number of elements) of a set.
The Hausdorff distance measures how close the prostate boundaries are defined in A and B, which is given by:
| (5) |
where ∥·∥ is the standard L2 metric, sup represents the supremum, and inf represents the infimum.
The mean landmark error measures the accuracy of point-to-point correspondences found by image registration. Let ϕ denote the resulting transformation from image registration. Our experts labeled N landmark pairs in the fixed T2w MRI and the moving histopathology image, denoted by (p1 , p’1 ),…, (pN , p’N). Then the mean landmark error for the resulting transformation ϕ from image registration is given by:
| (6) |
We used an identical approach to evaluate the distance between urethra segmentation on MRI and the corresponding deformed urethra segmentation on histopathology images (urethra deviations). All evaluation measures were computed on a slice by slice basis in 2D and averaged across several slices to obtain per patient measures.
3. Results
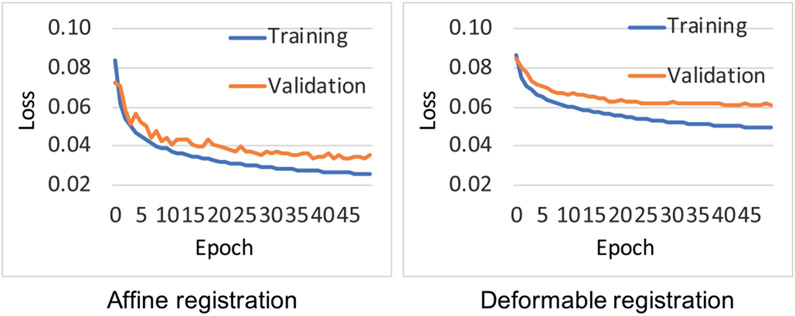
Fig. 5 shows the training loss and validation loss curves of the ProsRegNet affine and deformable registration networks. From this Fig., we can see that the validation loss has converged at 50 epochs for both networks and there is no issue of overfitting. We also notice that we had a slight unrepresentative sample for the training and we except better performance if the networks were trained with a larger dataset.
Fig. 5.
Training loss and validation loss curves of ProsRegNet affine and deformable registration networks.
To evaluate the plausibility of the estimated geometric transformations, we used each of them to deform a 2D grid image. By investigating all deformed grid images, we conclude that the composite transformations estimated by our ProsRegNet network are smooth and biologically plausible. Fig. 6 shows a typical deformed grid image from each cohort.
Fig. 6.
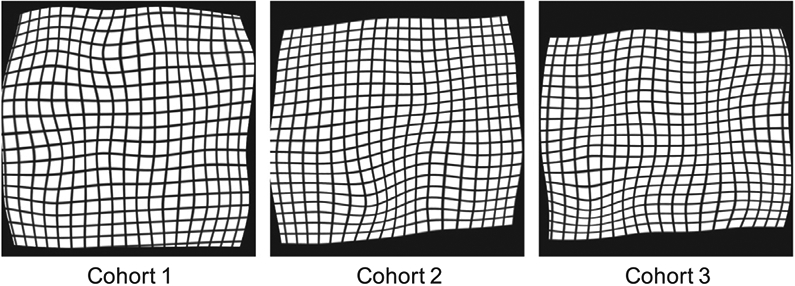
Typical deformed grid images from ProsRegNet registration.
3.1. Qualitative alignment accuracy
Fig. 7 shows the registration results of three patients with large cancerous regions (one from each cohort). The prostate boundaries on the MRI and the histopathology sections appeared well aligned for all three subjects, suggesting that the ProsRegNet pipeline achieved accurate global alignment of the prostate. Anatomic regions of the prostate on the MR and the histopathology images were also well aligned. Accurate alignment of anatomic regions indicates that the ProsRegNet pipeline has achieved promising alignment of local prostate features. The results in Fig. 7 demonstrate that our ProsRegNet pipeline generalizes across cohorts even if they were not part of the training, showing accurate registration for images from different cohorts acquired by different protocols. Our accurate alignment of the histopathology and MRI images suggests that we can carefully map the cancer labels in the histopathology images to the corresponding MRI slices using the estimated transformations.
Fig. 7.
Registration results for three different subjects (one from each cohort) using the proposed ProsRegNet deep learning registration pipeline. The MRI slices were chosen as the fixed images. (Left) MRI, (Middle) registered histopathology image, (Right) MRI overlaid with registered histopathology image. Cancer labels from the histopathology images were mapped onto MRI using estimated transformations from image registration.
3.2. Quantitative results
We evaluated various measures to assess the quality of alignment between the histopathology images and corresponding MRI slices. Those measures assess the overall alignment of the prostate (Dice coefficient), the distance between the prostate boundaries (Hausdorff Distance), and anatomic landmark deviation. Moreover, we also evaluated the execution time of the RAPSODI, CNNGeometric, and ProsRegNet approaches. Fig. 8 shows the box plots of the Dice Coefficient, Hausdorff distance, urethra deviation, and computation time of different approaches for all three cohorts. The results show that there is no significant difference (p-value > 0.05) between the Dice coefficient and the urethra deviation of the RAPSODI and ProsRegNet approaches for all three cohorts. Our ProsRegNet approach achieved significantly lower (p ≤ 0.05) Hausdorff distance than the RAPSODI approach for the second and the third cohorts. Our ProsRegNet approach has achieved significantly higher Dice coefficient and lower Hausdorff distance than the deep learning CNNGeometric approach for all three cohorts. Also, there is no significant difference between the urethra deviation of all three approaches for all cohorts. Notice that both our ProsRegNet and the CNNGeometric deep learning approaches were at least 20x faster to register the images than the iterative optimization performed by RAPSODI. In summary, the ProsRegNet pipeline has achieved better alignment near the prostate boundary than the RAPSODI approach while being several orders of magnitude faster, and it has also achieved better alignment of the overall shape and boundary of the prostate than prior CNNGeometric deep learning approach.
Fig. 8.
Box plots of different measures for the RAPSODI, CNNGeometric, and ProRegNet registration approaches of three cohorts. SS: statistically significant (p ≤ 0.05), NS: not significant (p > 0.05).
Table 2 summarizes the Dice coefficients for the whole prostate, Hausdorff distances for the prostate boundary, urethra deviations, and anatomic landmark errors after registration for the three cohorts. The results show that both the ProsRegNet and RAPSODI approaches have achieved a higher Dice Coefficient than the prior CNNGeometric approach. The high Dice coefficient indicates that our ProsRegNet pipeline can accurately align the overall shape and edges of the prostate for all of the three cohorts. The results also show that the ProsRegNet pipeline achieved a lower Hausdorff distance than both the RAPSODI and CNNGeometric approaches. The low Hausdorff distance implies that our ProsRegNet pipeline can have a small registration error of no more than 2mm near the prostate boundary. No significant differences were found between the RAPSODI, ProsRegNet, and CNNGeometric approaches in terms of urethra deviation and landmark error. The urethra deviation and landmark error indicate that our ProsRegNet pipeline has an average registration error of no more than 3mm inside the prostate. It is notable that the average running time of the ProsRegNet and CNNGeometric approaches was 1-4 seconds, compared to 31-264 seconds of the state-of-the-art RAPSODI approach and compared to running times of 120-750 seconds reported for other traditional approaches Li et al. (2017); (Losnegård et al., 2018).
Table 2.
Registration results of the RAPSODI, CNNGeometric, ProsRegNet approaches of three cohorts.
| Dataset | Registration Approach |
Dice Coefficient | Hausdorff Distance (mm) |
Urethra Deviation (mm) |
Landmark Error (mm) |
Computation Time (second) |
|---|---|---|---|---|---|---|
| Cohort 1 | RAPSODI | 0.979 (± 0.01) | 1.83 (± 0.50) | 2.48 (± 0.78) | 2.88 (± 0.73) | 264 (± 150) |
| CNNGeometric | 0.962 (± 0.01) | 2.43 (± 0.83) | 2.62 (± 0.86) | 2.72 (± 0.75) | 4 (±2) | |
| ProsRegNet | 0.975 (± 0.01) | 1.72 (± 0.42) | 2.37 (± 0.76) | 2.68 (± 0.68) | 4 (±2) | |
| Cohort 2 | RAPSODI | 0.965 (± 0.01) | 2.58 (± 1.05) | 2.96 (± 1.23) | NA | 60 (± 47) |
| CNNGeometric | 0.948 (± 0.01) | 3.05 (± 0.69) | 2.78 (± 2.03) | NA | 3 (±1) | |
| ProsRegNet | 0.961 (± 0.01) | 1.98 (± 0.28) | 2.51 (± 0.82) | NA | 3 (±1) | |
| Cohort 3 | RAPSODI | 0.966 (± 0.01) | 2.62 (± 1.32) | 3.3 (± 1.90) | NA | 31 (± 11) |
| CNNGeometric | 0.946 (± 0.01) | 2.68 (± 0.33) | 2.83 (± 1.2) | NA | 1 (±1) | |
| ProsRegNet | 0.967 (± 0.01) | 1.96 (± 0.29) | 2.91 (± 1.99) | NA | 1 (±1) |
3.3. Alignment of prostate cancers
One major goal of MRI-histopathology registration is to map the ground truth cancer labels from the histopathology images onto MRI. Here, we evaluate the accuracy of different approaches for registering cancerous regions using patients from the first cohort and the second cohorts. For the first cohort, two body imaging radiologists with more than five years of experience manually labeled regions of clinically significant prostate cancer on T2-w MRI of 35 patients. The following exclusion criterion was applied to handle inconsistency between the radiologists’ and pathologists’ annotations: (1) the size of two cancer labels of the same region differs by more than 100%, (2) there is no overlap between two cancer labels of the same region, (3) cancer labels are too tiny (less than 25 pixels). For the second cohort, the authors of the dataset have provided cancer labels on MRI by performing landmark-based registration of MRI and histopathology images. Table 3 shows the Dice coefficient and Hausdorff distance between cancer labels from the radiologists’ or landmark-based registration and cancer labels achieved by each of the registration approaches. The results show that ProsRegNet achieved better alignment of the prostate cancer boundaries (Hausdorff distance) than RAPSODI and CNNGeometric for both cohorts. Although CNNGeometric achieved slightly higher Dice coefficient than RAPSODI and ProsRegNet for the second cohort, our ProsRegNet approach achieved the highest Dice coefficient for the first cohort. In summary, our ProsRegNet approach has achieved comparable or better alignments of cancerous regions relative to CNNGeometric and RAPSODI. Notice that the accuracy of our analysis may be compromised by inconsistency between the radiologist’s cancer labels and the pathologists’ cancer labels (first cohort), and also errors in landmark-based registration (second cohort).
Table 3.
Accuracy of the RAPSODI, CNNGeometric, ProsRegNet approaches for aligning cancerous regions.
| Dataset | Registration Approach |
Dice Coefficient |
Hausdorff Distance (mm) |
|---|---|---|---|
| Cohort 1 | RAPSODI | 0.624 (± 0.12) | 6.02 (± 2.78) |
| CNNGeometric | 0.610 (± 0.11) | 5.70 (± 2.22) | |
| ProsRegNet | 0.628 ( ± 0.10) | 5.42 (± 2.32) | |
| Cohort 2 | RAPSODI | 0.573 (± 0.13) | 5.42 (± 2.00) |
| CNNGeometric | 0.575 ( ± 0.12) | 5.34 (± 2.14) | |
| ProsRegNet | 0.563 (±0.14) | 4.87 (±1.53) |
3.4. Other training schemes
In this section, we investigate two additional training schemes, one for ProsRegNet and the other one for CNNGeometric. For the first training scheme, we trained both the affine and deformable registration networks of ProsRegNet directly by the prostate masks of 99 patients from the first cohort and tested the performance on 53 patients from three cohorts (see Table 4). Compared to results presented in Table 2, training and testing ProsRegNet with only prostate masks has improved the alignment of prostate boundaries, with Dice coefficient increased by 0.4%-1.0% and Hausdorff distance decreased by 13.4%-18.7%. However, this training scheme has also deteriorated the registration results inside the prostate, with urethra deviation increased by 13.5%-25.7% and landmark error increased by 24.6%. Those results show that training the ProsRegNet model with masked MRI and histopathology images facilitates the alignment of features inside the prostate. Since alignment of features inside the prostate is more important than alignment of the prostate boundaries, we do not recommend training and testing ProsRegNet only on the prostate masks.
Table 4.
Registration results of ProsRegNet trained with only prostate masks and CNNGeometric trained with multi-modal image pairs.
| Dataset | Registration Approach | Dice Coefficient | Hausdorff Distance (mm) | Urethra Deviation (mm) | Landmark Error (mm) |
|---|---|---|---|---|---|
| Cohort 1 | ProsRegNet (masks only) | 0.979 (± 0.01) | 1.49 (± 0.44) | 2.98 (± 0.82) | 3.39 (± 0.68) |
| CNNGeometric (multi-modal) | 0.960 (± 0.01) | 2.42 (± 0.55) | 2.55 (± 0.73) | 2.79 (± 0.74) | |
| Cohort 2 | ProsRegNet (masks only) | 0.971 (± 0.01) | 1.61 (± 0.33) | 2.85 (± 1.34) | NA |
| CNNGeometric (multi-modal) | 0.910 (± 0.03) | 4.08 (± 1.14) | 2.82 (± 1.34) | NA | |
| Cohort 3 | ProsRegNet (masks only) | 0.976 (± 0.01) | 1.60 (± 0.38) | 3.57 (± 2.28) | NA |
| CNNGeometric (multi-modal) | 0.947 (± 0.01) | 3.00 (± 0.82) | 3.17 (± 2.07) | NA |
For the second training scheme, we investigated the efficacy of training a multi-modal deep learning network on MRI-histopathology image pairs pre-aligned by RAPSODI. We chose the CNNGeometric model over the ProsRegNet model since the SSD loss function used by the ProsRegNet model cannot be directly used for multi-modal registration. Again, we trained the CNNGeometric model on MRI-histopathology image pairs of 99 patients from the first cohort and evaluated its performance using 53 patients from three cohorts (see Table 4). shows the registration results of the multi-modal CNNGeometric network for the three cohorts. Compared to results in Table 2, the performance of the multi-modal CNNGeometric model is worse than the uni-modal ProsRegNet model for both the global and local alignment of the MRI and histopathology images. One factor that compromised the performance of the multi-modal CNNGeometric is that the MRI-histopathology image pairs used in the training are from RAPSODI registration and therefore do not have perfect spatial correspondences.
4. Discussion
Accurately aligning MRI with histopathology images provides a detailed answer key regarding precise cancer locations on MRI. As such, it has tremendous potential for improving the interpretation of prostate MRI and providing labeled imaging data to establish and validate prostate cancer detection models based on radiomics or machine learning methods Metzger et al. (2016). In this paper, we have developed the novel ProsRegNet deep learning approach for 2D registration of MRI and histopathology images. It is challenging to directly train a multi-modal network for registering MR and histopathology images due to the lack of either an effective loss function for unsupervised learning or MRI-histopathology image pairs with accurate spatial correspondences for supervised learning. We tackled this problem from a different perspective by training a uni-modal ProsRegNet network which learns how to combine high-level features in the MRI and histopathology images to solve image registration problems. The trained ProsRegNet network has the capabilities to solve uni-modal registration problems in the context of MRI and histopathology images and thus can be used to register the two modalities in a multi-modal manner. Our experiments and results provide empirical evidence that, although our ProsRegNet was trained with pairs of images from the same modality, it can be generalized to achieve very accurate MRI-histopathology registration. This paper is the first attempt to apply deep learning to the registration of MRI and histopathology images of the prostate.
Our study is the largest prostate MRI-histopathology registration study, using 654 of pairs of histopathology and MRI slices of 152 prostate cancer patients from three different institutions and MRIs from three different manufactures. The wide range of parameters of synthetic transformations used during the training allowed ProgRegNet to accurately cover large affine and deformable transformations observed in three different cohorts, which include MR images acquired with or without using an endorectal coil, as well as histopathology images acquired as whole mounts, quadrants or at low resolution. We showed that our ProsRegNet pipeline achieved a very high Dice coefficient (0.96-0.98), a very low Hausdorff distance (1.7-2.0mm), a relatively low urethra deviation (2.4-2.9mm) and a relatively low landmark error (2.7mm) compared to results reported in previous studies Chappelow et al. (2011); (Kalavagunta et al., 2015); Li et al. (2017); Losnegård et al. (2018); Park et al. (2008); Reynolds et al. (2015); Ward et al. (2012); Wu et al. (2019). Moreover, in a direct comparison of the state-of-the-art RAPSODI pipeline Rusu et al. (2020), we showed that ProsRegNet achieved slightly better performance while being 20x-60x faster. This allows our ProsRegNet approach to execute the histopathology-MRI registration in real-time interactive software, otherwise not possible with any previous method. By significantly speeding up the registration process, our approach can help to create a large dataset of labeled MRI using ground-truth histopathology images which is crucial for the training of prostate cancer detection methods on pre-operative MRI using machine learning.
Even recent deep learning cancer prediction studies Cao et al. (2019); Sumathipala et al. (2018) that use histopathology images as the reference, rely on cognitive alignment (mentally projecting the histopathology images onto the MRI) to create cancer labels on MRI. This time-consuming labeling is inaccurate and biases the labels towards visible extent of cancer on MRI (known to underestimate the real size of cancer Piert et al. (2018) and missing MRI-invisible lesions). Our ProsRegNet pipeline allows the efficient creation of labels on MRI with accurate borders, including MRI invisible lesions. In addition, once trained, our deep learning network is parameter-free when registering unseen pairs of MRI and histopathology images, alleviating the need of modifying registration hyperparameters, e.g. step size, number of iterations. By making the registration set up less complicated, our approach is more accessible to non-expert users than the traditional methods.
Although this study demonstrates promising results for MRI-histopathology registration, there are some limitations related to human input: prostate segmentation on MRI and histopathology images, gross rotation and flip of the histopathology images and identifying slice-to-slice correspondences. Our team is working on developing methods to automate these steps, yet they are beyond the scope of the current study. Nonetheless, our proposed work simplifies the registration step without requiring manual picking of landmarks or complex selection of features for multifeature scoring functions Chappelow et al. (2011); Li et al. (2017); Ward et al. (2012).
We have shown that our deep learning pipeline can achieve fast and accurate registration of the histopathology and MRI images. Accurate registration could improve radiologists interpretation of MRI by allowing side-by-side comparison of MR and histopathology images. Indeed, we use these side-by-side comparisons in a multidisciplinary prostate MRI tumor board at our institution. Accurate registration also allows mapping of the ground truth extent and grade of prostate cancer from histopathology images onto the corresponding preoperative MRI. Such accurate labels mapped from histopathology images on MRI will help develop and validate radiomic and machine learning approaches for detecting cancer locations within the prostate based on MRI to guide biopsies and focal treatment.
5. Conclusion
We have developed a deep learning pipeline for efficient registration of MRI and histopathology images of the prostate for patients that underwent radical prostatectomy. The performance of the deep neural networks for aligning the MRI and histology is promising and slightly better than state-of-the-art registration approaches. Compared to traditional approaches that require significant user input (e.g., careful choice of registration parameters) and considerable computing time, our pipeline achieved very accurate and efficient alignment with less user input. The ease of use and speed make our pipeline attractive for clinical implementation to allow direct comparison of MR and histological images to improve radiologist accuracy in reading MRI. Furthermore, this pipeline could serve as a useful tool for image alignment in developing radiomic and deep learning approaches for early detection of prostate cancer.
Acknowledgments
This work was supported by the Department of Radiology at Stanford University, Radiology Science Laboratory (Neuro) from the Department of Radiology at Stanford University, and the National Cancer Institute (U01CA196387 to James D. Brooks)
Footnotes
Declaration of Competing Interest
The authors declare that they do not have any competing financial interests or personal relationships that could have influenced the work in this paper.
References
- Ahmed HU, El-Shater Bosaily A, Brown LC, Gabe R, Kaplan R, Parmar MK, et al. , 2017. Diagnostic accuracy of multi-parametric mri and trus biopsy in prostate cancer (promis): a paired validating confirmatory study. Lancet North Am. Ed 389, 815–822. URL https://www.ncbi.nlm.nih.gov/pubmed/28110982. [DOI] [PubMed] [Google Scholar]
- American Cancer Society, 2020. Facts & Fig.s 2020. American Cancer Society, Atlanta, GA: URL: https://www.cancer.org/cancer/prostate-cancer/about/key-statistics.html. [Google Scholar]
- Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV, 2018. An unsupervised learning model for deformable medical image registration. Proc. IEEE Conf. Comput. Vision Pattern Recognit 9252–9260. URL https://ieeexplore.ieee.org/document/8579062. [Google Scholar]
- Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV, 2019. Voxelmorph: a learning framework for deformable medical image registration. IEEE Trans. Med. Imaging 38, 1788–1800. URL https://arxiv.org/abs/1809.05231. [DOI] [PubMed] [Google Scholar]
- Bhattacharya I, Seetharaman A, Shao W, Sood R, Kunder CA, Fan RE, et al. , 2020. Corrsignet: Learning correlated prostate cancer signatures from radiology and pathology images for improved computer aided diagnosis arXiv preprint arXiv:2008.00119 URL https://arxiv.org/abs/2008.00119. [Google Scholar]
- Cao R, Mohammadian Bajgiran A, Afshari Mirak S, Shakeri S, Zhong X, Enzmann D, et al. , 2019. Joint prostate cancer detection and gleason score prediction in mp-mri via focalnet. IEEE Trans. Med. Imaging 38, 2496–2506. URL https://ieeexplore.ieee.org/document/8653866. [DOI] [PubMed] [Google Scholar]
- Chappelow J, Bloch BN, Rofsky N, Genega E, Lenkinski R, Dewolf W, et al. , 2011. Elastic registration of multimodal prostate MRI and histology via multi-attribute combined mutual information. Med. Phys 38, 2005–2018. URL https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3078156/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choyke P, Turkbey B, Pinto P, Merino M, Wood B, 2016. Data From PROSTATE-MRI. The Cancer Imaging Archive URL: https://wiki.cancerimagin-garchive.net/display/Public/PROSTATE-MRI. [Google Scholar]
- Dalca AV, Balakrishnan G, Guttag J, Sabuncu MR, 2018. Unsupervised learning for fast probabilistic diffeomorphic registration. In: International Conference on Medical Image Computing and Computer-Assisted Intervention Springer, pp. 729–738 URL: http://www.mit.edu/~adalca/files/papers/miccai2018voxel-morph-pd.pdf. [Google Scholar]
- Ghosal S, Ray N, 2017. Deep deformable registration: enhancing accuracy by fully convolutional neural net. Pattern Recognit. Lett 94, 81–86. URL https://arxiv.org/abs/1611.08796. [Google Scholar]
- Goubran M, Crukley C, de Ribaupierre S, Peters TM, Khan AR, 2013. Image registration of ex-vivo mri to sparsely sectioned histology of hippocampal and neocortical temporal lobe specimens. Neuroimage 83, 770–781. URL https://www.ncbi.nlm.nih.gov/pubmed/23891884. [DOI] [PubMed] [Google Scholar]
- Goubran M, de Ribaupierre S, Hammond RR, Currie C, Burneo JG, Parrent AG, et al. , 2015. Registration of in-vivo to ex-vivo mri of surgically resected specimens: a pipeline for histology to in-vivo registration. J. Neurosci. Methods 241, 53–65 URL: https://www.ncbi.nlm.nih.gov/pubmed/25514760. [DOI] [PubMed] [Google Scholar]
- He K, Zhang X, Ren S, Sun J, 2016. Deep residual learning for image recognition. Proc. CVPR 770–778 URL: https://arxiv.org/abs/1512.03385. [Google Scholar]
- Kalavagunta C, Zhou X, Schmechel SC, Metzger GJ, 2015. Registration of in vivo prostate MRI and pseudo-whole mount histology using Local Affine Transformations guided by Internal Structures (LATIS). J. Magn. Reson. Imaging : JMRI 41, 1104 URL https://www.ncbi.nlm.nih.gov/pubmed/24700476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma D, Ba J, 2017. Adam: A method for stochastic optimization arXiv.org URL: https://arxiv.org/abs/1412.6980. [Google Scholar]
- Krebs J, Delingette H, Mailhé B, Ayache N, Mansi T, 2019. Learning a probabilistic model for diffeomorphic registration. IEEE Trans. Med. Imaging 38, 2165–2176 URL: https://ieeexplore.ieee.org/document/8633848. [DOI] [PubMed] [Google Scholar]
- Li L, Pahwa S, Penzias G, Rusu M, Gollamudi J, Viswanath S, et al. 2017. Co-Registration of ex vivo Surgical Histopathology and in vivo T2 weighted MRI of the Prostate via multi-scale spectral embedding representation. Sci Rep 7, 8717–8717. URL: https://www.ncbi.nlm.nih.gov/pubmed/28821786.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losnegård A, Reister L, Halvorsen OJ, Beisland C, Castilho A, Muren LP, et al. , 2018. Intensity-based volumetric registration of magnetic resonance images and whole-mount sections of the prostate. Comput. Med. Imaging Graph 63, 24–30 URL: https://www.ncbi.nlm.nih.gov/pubmed/29276002. [DOI] [PubMed] [Google Scholar]
- Lovegrove CE, Matanhelia M, Randeva J, Eldred-Evans D, Miah HTS, Winkler M, et al. , 2016. The Role of Pathology Correlation Approach in Prostate Cancer Index Lesion Detection and Quantitative Analysis with Multiparametric MRI NIH URL: https://www.ncbi.nlm.nih.gov/pubmed/25683501. [DOI] [PMC free article] [PubMed]
- Madabhushi A, Feldman M, 2016. Fused Radiology-Pathology Prostate Dataset. The Cancer Imaging Archive URL: https://wiki.cancerimagingarchive.net/display/Public/Prostate+Fused-MRI-Pathology. [Google Scholar]
- Metzger GJ, Kalavagunta C, Spilseth B, Bolan PJ, Li X, Hutter D, et al. , 2016. Detection of prostate cancer: quantitative multiparametric mr imaging models developed using registered correlative histopathology. Radiology 279, 805–816. URL https://www.ncbi.nlm.nih.gov/pubmed/26761720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Piert MR, Khan A, Shah R, Hussain H, Siddiqui J, et al. , 2008. Registration methodology for histological sections and in vivo imaging of human prostate. Acad. Radiol 15, 1027–1039 URL: https://www.ncbi.nlm.nih.gov/pubmed/18620123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piert M, Shankar P, Montgomery J, Kunju L, Rogers V, Siddiqui J, Rajendiran T, Hearn J, George A, Shao X, Davenport M, 2018. Accuracy of tumor segmentation from multi-parametric prostate mri and 18 f-choline pet/ct for focal prostate cancer therapy applications. EJNMMI Res. 8, 1–14. URL https://www.ncbi.nlm.nih.gov/pubmed/29589155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priester A, Natarajan S, Le JD, Garritano J, Radosavcev B, Grundfest W, et al. , 2014. A system for evaluating magnetic resonance imaging of prostate cancer using patient-specific 3d printed molds. Am. J. Clin. Exp. Urology 2, 127. [PMC free article] [PubMed] [Google Scholar]
- Reynolds HM, Williams S, Zhang A, Chakravorty R, Rawlinson D, Ong CS, et al. , 2015. Development of a registration framework to validate MRI with histology for prostate focal therapy. Med. Phys 42, 7078–7089. URL https://www.ncbi.nlm.nih.gov/pubmed/26632061. [DOI] [PubMed] [Google Scholar]
- Rocco I, Arandjelovic R, Sivic J, 2017. Convolutional neural network architecture for geometric matching. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition URL: https://arxiv.org/abs/1703.05593. [DOI] [PubMed] [Google Scholar]
- Rosenzweig B, Laitman Y, D.E. Z, Raz O, Ramon J, Dotan Z, et al. , 2020. Effects of ”real life” prostate mri inter-observer variability on total needle samples and indication for biopsy. Urological Oncol.. URL https://www.ncbi.nlm.nih.gov/pubmed/32303407. [DOI] [PubMed] [Google Scholar]
- Rusu M, Kunder C, Fan R, Ghanouni P, West R, Sonn G, et al. , 2019. Framework for the co-registration of MRI and histology images in prostate cancer patients with radical prostatectomy, in: Medical Imaging 2019: Image Processing, Proc. SPIE; URL: 10.1117/12.2513099. [DOI] [Google Scholar]
- Rusu M, Rajiah P, Gilkeson R, Yang M, Donatelli C, Thawani R, et al. , 2017. Co-registration of pre-operative ct with ex vivo surgically excised ground glass nodules to define spatial extent of invasive adenocarcinoma on in vivo imaging: a proof-of-concept study. Eur. Radiol 27, 4209–4217. URL https://www.ncbi.nlm.nih.gov/pubmed/28386717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusu M, Shao W, Kunder CA, Wang JB, Soerensen SJ, Teslovich NC, Sood RR, Chen LC, Fan RE, Ghanouni P, et al. , 2020. Registration of pre-surgical mri and histopathology images from radical prostatectomy via rapsodi. Medical Physics URL: https://aapm.onlinelibrary.wiley.com/doi/full/10.1002/mp.14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavati N, Mcgrath DM, Lee J, van Kwast T, Jewett M, Mnard C, Brock KK, 2011. Biomechanical model-based deformable registration of mri and histopathology for clinical prostatectomy. J. Pathol. Informatics 2 URL: http://search.proquest.com/docview/1034797427.S10-S10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, Christensen GE, Johnson HJ, Hyun Song J, Durumeric OC, Johnson CP, et al. , 2016. Population shape collapse in large deformation registration of mr brain images. In: The IEEE Conference on Computer Vision and Pattern Recognition (CVPR) Workshops, pp. 109–117 URL. [Google Scholar]
- Shen Z, Han X, Xu Z, Niethammer M, 2019. Networks for joint affine and non-parametric image registration. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4224–4233 URL: https://openaccess.thecvf.com/contentCVPR2019/papers/ Shen Networks for Joint Affine and Non-Parametric Image Registration CVPR 2019 paper.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonn GA, Fan RE, Ghanouni P, Wang NN, Brooks JD, Loening AM, et al. , 2019. Prostate magnetic resonance imaging interpretation varies substantially across radiologists. Eur. Urology Focus 5, 592–599. URL https://www.ncbi.nlm.nih.gov/pubmed/29226826. [DOI] [PubMed] [Google Scholar]
- Stille M, Smith EJ, Crum WR, Modo M, 2013. 3d reconstruction of 2d fluorescence histology images and registration with in vivo mr images: Application in a rodent stroke model. J. Neurosci. Methods 219, 27–40. URL https://www.ncbi.nlm.nih.gov/pubmed/23816399. [DOI] [PubMed] [Google Scholar]
- Sumathipala Y, Lay N, Turkbey B, Smith C, Choyke PL, Summers RM, 2018. Prostate cancer detection from multi-institution multiparametric mris using deep convolutional neural networks. J. Med. Imaging 5, 044507 URL https://www.ncbi.nlm.nih.gov/pubmed/30840728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth R, Shih N, Tomaszewski J, Feldman M, Kutter O, Yu D, et al. , 2014. Histostitcherc: an informatics software platform for reconstructing whole-mount prostate histology using the extensible imaging platform framework. J. Pathol. Informatics 5 8–8, URL http://www.jpathinformatics.org/article.asp?issn=2153-3539. year=2014;volume=5;issue=1;spage=8;epage=8; aulast=Toth;type=0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkbey B, Mani H, Shah V, Rastinehad AR, Bernardo M, Pohida T, et al. , 2011. Multiparametric 3T prostate magnetic resonance imaging to detect cancer: histopathological correlation using prostatectomy specimens processed in customized magnetic resonance imaging based molds. J. Urol 186, 1818–1824 URL: https://www.ncbi.nlm.nih.gov/pubmed/21944089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkbey L,B, Choyke L,P, 2012. Multiparametric MRI and prostate cancer diagnosis and risk stratification. Curr. Opin. Urol 22, 310–315. URL https://www.ncbi.nlm.nih.gov/pubmed/22617060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Turkbey B, Muradyan N, Rajesh A, Cornud F, Haider MA, et al. , 2012. Overview of dynamic contrast-enhanced MRI in prostate cancer diagnosis and management. AJR. Am. J. Roentgenol 198, 1277–1288. URL http://search.proquest.com/docview/1016673380/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos BD, Berendsen FF, Viergever MA, Sokooti H, Staring M, Išgum I, 2019. A deep learning framework for unsupervised affine and deformable image registration. Med. Image Anal 52, 128–143 URL: https://www.sciencedirect.com/science/article/abs/pii/S1361841518300495. [DOI] [PubMed] [Google Scholar]
- Wang Z, Liu C, Cheng D, Wang L, Yang X, Cheng KT, 2018. Automated detection of clinically significant prostate cancer in mp-mri images based on an end-to-end deep neural network. IEEE Trans. Med. Imaging 37, 1127–1139. URL https://www.ncbi.nlm.nih.gov/pubmed/29727276. [DOI] [PubMed] [Google Scholar]
- Ward AD, Crukley C, Mckenzie CA, Montreuil J, Gibson E, Romagnoli C, et al. , 2012. Prostate: registration of digital histopathologic images to in vivo mr images acquired by using endorectal receive coil. Radiology 263 URL https://www.ncbi.nlm.nih.gov/pubmed/22474671. [DOI] [PubMed] [Google Scholar]
- Weinreb JC, Barentsz JO, Choyke PL, Cornud F, Haider MA, Macura KJ, et al. , 2016. Pi-rads prostate imaging reporting and data system: 2015, version 2. Eur. Urol 69, 16–40 URL: https://www.ncbi.nlm.nih.gov/pubmed/26427566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphalen AC, McCulloch CE, Anaokar JM, Arora S, Barashi NS, Barentsz JO, et al. , 2020. Variability of the positive predictive value of pirads for prostate mri across 26 centers: experience of the society of abdominal radiology prostate cancer disease-focused panel. Urological Oncol.. URL https://pubs.rsna.org/doi/10.1148/radiol.2020190646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HH, Priester A, Khoshnoodi P, Zhang Z, Shakeri S, Afshari Mirak S, et al. , 2019. A system using patient-specific 3D-printed molds to spatially align in vivo MRI with ex vivo MRI and whole-mount histopathology for prostate cancer research. J. Magn. Reson. Imaging 49 URL https://www.ncbi.nlm.nih.gov/pubmed/30069968. [DOI] [PubMed] [Google Scholar]
- Yang X, Kwitt R, Styner M, Niethammer M, 2017. Quicksilver: Fast predictive image registration-a deep learning approach. NeuroImage 158, 378–396 URL-https://www.sciencedirect.com/science/article/pii/S1053811917305761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, 2018. Inverse-consistent deep networks for unsupervised deformable image registration arXiv.org URL https://arxiv.org/abs/1809.03443. [Google Scholar]