Abstract
Complement hyperactivation, angiopathic thrombosis, and protein-losing enteropathy (CHAPLE disease) is a lethal disease caused by genetic loss of the complement regulatory protein CD55 leading to overactivation of complement and innate immunity together with immunodeficiency due to immunoglobulin (Ig) wasting in the intestine. We report in vivo human data that we accumulated using the complement C5 inhibitor eculizumab for the medical treatment of CHAPLE patients and observed cessation of gastrointestinal pathology together with restoration of normal immunity and metabolism. We found that patients rapidly renormalized Ig concentrations and other serum proteins as revealed by aptamer profiling, re-established a healthy gut microbiome, discontinued Ig replacement and other treatments, and exhibited catch-up growth. Thus, we show blockade of C5 by eculizumab effectively re-establishes the regulation of the innate immune complement system to substantially reduce the pathophysiological manifestations of CD55 deficiency in humans.
In 1961, T.A. Waldmann described serum hypoproteinemia associated with protein-losing enteropathy (PLE)1. The disease pathogenesis was unknown, and temporizing measures such as albumin infusions and immunoglobulin (Ig) replacement therapy (IgRT) became the conventional therapies. In 2017, the discovery of “CD55 deficiency with hyperactivation of complement, angiopathic thrombosis, and PLE (CHAPLE disease, OMIM #226300) revealed that complement and innate immunity hyperactivation caused by CD55 (also known as decay acceleration factor DAF) gene deficiency can cause this disorder2, 3. The cardinal features are severe PLE due to primary intestinal lymphangiectasia (PIL) due to the inflammatory attack on intestinal lymphatic vessels by complement and innate immune overactivation2, 4. CHAPLE leads to severe pathophysiology including diarrhea with protein wasting, vomiting, abdominal pain, and edema that cause a metabolic starvation state; recurrent infections due to hypogammaglobulinemia; and severe, often fatal, thromboembolic complications1, 2, 4. The disease occurs globally but prevails in areas with high consanguinity, such as the Igdir region of eastern Turkey, where there is a high carrier frequency of CD55 loss of function (LOF) alleles. Lethal CHAPLE disease, called “tedirgin” in the local language (meaning ‘agitated’), is prevalent there, and desperate parents of affected children resort to folk remedies since conventional therapies do not improve or extend life. Thus, understanding the immune and metabolic derangements due to the CD55 loss and how they change with complement interventions is critical.
The complement system is a cascade of proteins coordinated with innate and adaptive immunity to destroy pathogens and clear immune complexes, apoptotic cells, and debris5, 6. Complement activation produces bioactive peptides, anaphylatoxins, that can alter both innate and adaptive immune responses and ultimately lead to the assembly of a membrane attack complex (MAC) that can lyse targets such as pathogens or cells7. Unwanted complement activation on host/self cells is regulated by the cell surface glycoproteins CD55 (also called decay accelerating factor; DAF), CD46, and CD59, which protect normal hematopoietic, endothelial, and epithelial cells from complement-mediated damage8. In the gastrointestinal (GI) tract, lymph recirculation through lymph vessels called lacteals return serum proteins such as albumin and Ig to the venous circulation. The genetic loss of CD55 induces local complement hyperactivation that deposits MAC on GI lymphatics causing PLE2. Other severe diseases, such as paroxysmal nocturnal hemoglobinuria (PNH) and atypical hemolytic uremic syndrome (aHUS), result from the loss of complement inhibitors and uncontrolled complement activation on erythrocytes and kidney basal membrane cells, respectively2, 9, 10, 11, 12. Both conditions are effectively treated with the complement inhibitor, eculizumab (Soliris). Eculizumab is a monoclonal antibody that binds to and inhibits the activation of C5, which occurs normally as consequence of the activation of the central complement component C3. CD55 is a negative regulator of the so-called C3 and C5 convertases that mediate cleavage activation of C3 and C5. We found that eculizumab successfully abrogated complement activation in vitro in CHAPLE patient T cells2.
Previous studies reported that eculizumab could improve the condition of 3 members of a CD55-deficient family3, 13. These promising results raised several important questions. Would eculizumab have broad efficacy in families with different genetic backgrounds and CD55 mutations? What physiological manifestations of disease would be alleviated, and would healthy immunity and metabolism be re-established. What are the drug pharmacokinetics and pharmacodynamics for complement control? Are there pharmacogenomic variants that determine treatment efficacy and dosing? Because PLE causes a starvation state, what are the specific metabolic effects of the disease and treatment? Multiplexed proteomic platforms have identified novel biomarkers and new disease mechanisms. For example, the investigation of inflammatory bowel disease using “slow off-rate modified aptamers” (SOMAmers) revealed key serum protein changes independent of transcriptome changes suggesting this could help elucidate CHAPLE disease mechanisms14. Finally, despite ubiquitous CD55 expression in the body, the severe complement hyperactivation in CHAPLE disease mainly affects the GI tract. Could microbiome studies yield insights into the GI pathogenesis?15, 16 We, therefore, comprehensively investigated eculizumab as a medical treatment in CHAPLE patients with different CD55 gene mutations.
Results
Natural history of a case series of CHAPLE disease
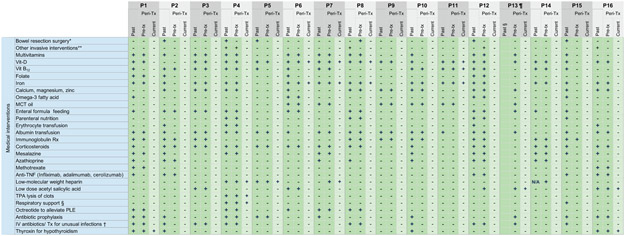
We evaluated 16 CHAPLE patients from 14 families diagnosed with recessive biallelic CD55 gene mutations causing decreased CD55 expression and complement overactivation (Extended Data Fig. 1 and Supplementary Fig. 1)2. All patients manifested severe PLE leading to hypoproteinemia, low immunoglobulin (Ig) concentrations and recurrent infections, abdominal pain, nausea, vomiting, diarrhea, loss of appetite, weight loss, and edema (Extended Data Fig. 1,2 and Supplementary Information)2, 3. Patients progressed to three life-threatening conditions: hypoproteinemia causing metabolic derangements, starvation, and infections; debilitating GI inflammation, ulceration, and obstruction; and severe, often fatal, thromboembolic disease (Fig. 1a and Extended Data Fig. 2, and Supplementary Information, case histories). Patients required frequent hospitalizations and many medical interventions, including albumin infusions and Ig replacement therapy (IgRT) that failed to alleviate disease (Fig. 1b and Extended Data Fig. 3). In the extended families, we found 32 probable CHAPLE patients, of whom eight died in childhood (25%) and many others severely ill.
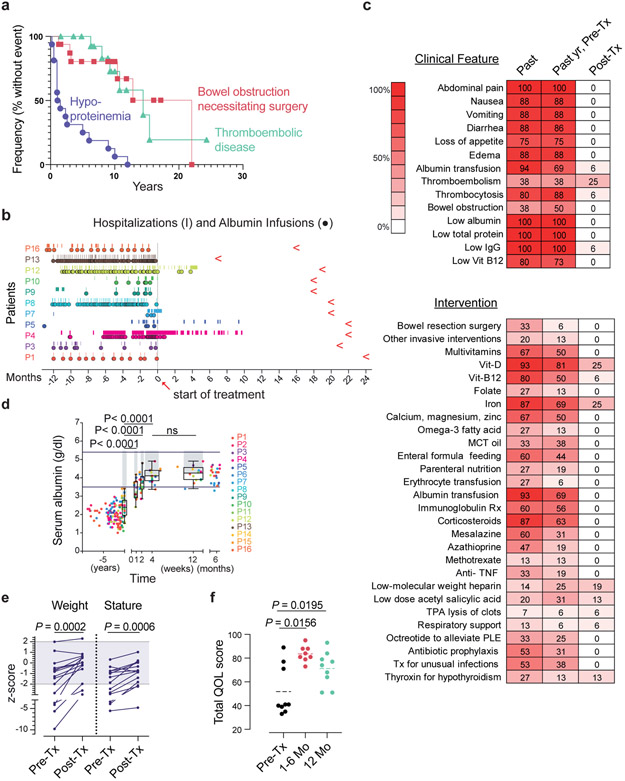
Fig. 1 ∣. The treatment effect of eculizumab in CHAPLE disease.
a, A Kaplan- Meier plot showing cumulative frequency of being without hypoproteinemia (purple), bowel obstruction (red), and thromboembolic disease (green) vs. time (years)(n = 16). b, Timeline of hospitalizations (vertical line) and albumin infusions (circles) for 11 patients pretreatment (pre-Tx)(−) and post-treatment (post-Tx)(+) time (months). Red leftward arrowheads indicate the end of the observation period. c, Heatmaps of the prevalence (% of patients, n = 16) in the study population of the indicated clinical parameter or therapeutic intervention, respectively, over the patient lifetime (past), past year pre-Tx, and the post-Tx observation period when a particular patient has been on regular eculizumab treatment. d, Serum albumin concentration before and after treatment beginning at t = 0. Horizontal bars show normal range; gray bars indicate statistical comparison range using mixed-effects analysis with Tukey’s multiple comparisons correction (n = 16). The statistical comparison of 4 vs 12 weeks revealed an adjusted P value of 0.3905 (ns = not significant). For each time point, the box plot shows the median, interquartile range, minimum and maximum values. e, Plots of weight and height (stature) z-scores compared to population averages pre-Tx and after 20 months of treatment using a paired t-test (n = 12). Gray region indicates the normal range. Z-scores were calculated using an online calculator (https://peditools.org/growthpedi/index.php) that uses CDC data tables as chart source. f, Total quality of life (QOL) score of patients using the KINDLR questionnaire (n = 9 for pre-Tx and 12 months’ assessments; n = 8 at 1-6 months). The statistical comparisons were made by Wilcoxon matched- pairs signed rank test. All P values are two-sided.
Eculizumab causes rapid improvement in most symptoms and overall health
During medical care, we filed appeals to the Turkish Medicines and Medical Devices Agency (TMMDA) to provide eculizumab not as a clinical trial but for off-label use in the care of each of the 15 patients. The 16th patient was treated in the Netherlands using eculizumab provided by medical insurance. We observed treatment effects over a median of 20 months (interquartile range 18 to 22 months) totaling 309 patient-months of data. Strikingly, most patients no longer required hospitalization or transfusions of albumin or Ig following eculizumab therapy (Fig. 1b). Three patients did not receive the full regimen due to inaccessibility (Patient 5) or medical reasons (Patients 12), or there was only a partial drug response (Patient 4, and Supplementary Information); however, all three responded to treatment adjustments. Long-standing pathophysiological signs and symptoms were eliminated by eculizumab (Fig. 1c top and Extended Data Fig. 1,2). Most previous treatment interventions became unnecessary (Fig. 1c bottom, Extended Data Fig. 3). However, supplementation of iron and vitamin D, thyroxin, anticoagulants, and thrombolytic medications continued to be needed by some patients. Within months, the children exhibited a healthier appearance and function. Serum albumin increased into the normal range within 2-4 weeks and remained normal, at least 6 months or longer (Fig. 1d and Extended Data Fig. 1). The patients’ heights and weights improved, and the total quality-of-life (QOL) scores increased (Fig. 1e,f and Extended Data Fig. 2; Supplementary Fig. 2,3). However, thrombocytosis, thrombosis, and pulmonary embolism persisted in some patients (Fig. 1c, Extended Data Fig. 2). Overall, eculizumab treatment substantially restored normal physiology and immunity.
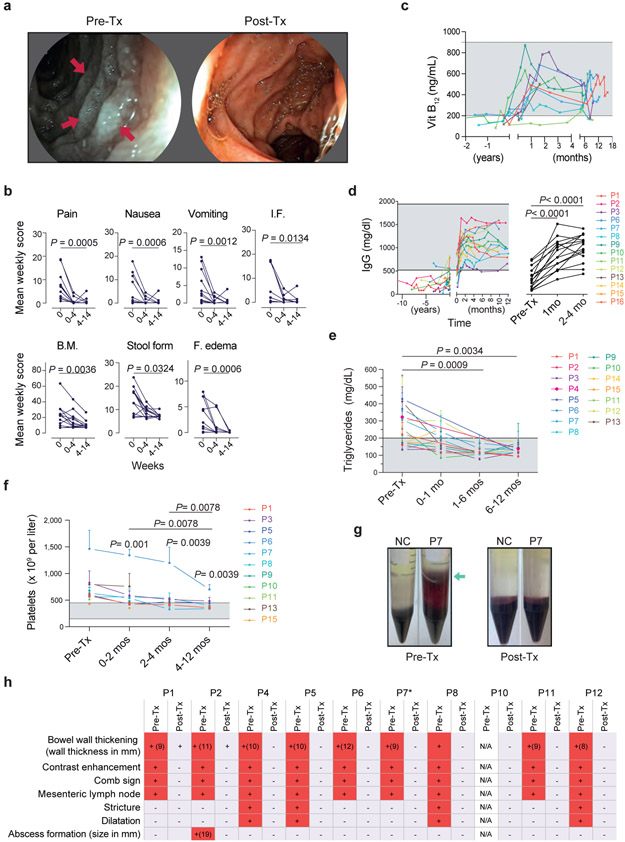
CHAPLE disease involves complement damage to GI lymphatics, so we evaluated the intestinal mucosa by endoscopy2. We found that lymphangiectasia, reflected by the dense white aggregates imparting a grayish cast on the mucosa, was eliminated by treatment (Fig. 2a). Eculizumab also promoted the healing of mucosal ulcers (Supplementary Fig. 4). Prospective symptom diaries revealed that the treatment reduced all GI symptoms and edema within 4 weeks (Fig. 2b, Extended Data Fig. 2 and Supplementary Fig. 5). Restoration of normal alimentary function was illustrated by increases in vitamin B12 and serum Ig, making IgRT and vitamin supplements unnecessary (Fig. 2c,d; Extended Data Fig. 1). We found markedly improved immune function with reduced and less severe infections. CHAPLE patients also have abnormal increases in triglycerides and platelets that were reversed by eculizumab (Fig. 2e,f). Moreover, a blood cell aggregation abnormality was also eliminated by treatment (Fig. 2g). Radiological abnormalities, including bowel wall thickening, contrast enhancement, intestinal stricture, and proximal dilatation, free fluid collection, and abscess formation, also resolved with treatment (Fig. 2h). However, eculizumab therapy did not correct thromboembolic disease in Patients −4, −7, and −14, or ischemic gliotic foci in P12. Thus, eculizumab effectively ameliorated most disease findings except for thromboembolic disease.
Fig. 2 ∣. The gastrointestinal, circulatory, hematologic and metabolic manifestations of CHAPLE disease with or without eculizumab.
a, Duodenal endoscopy images pretreatment (pre-Tx) showing white lymph globules due to lymphangiectasia (red arrows) and lymph leakage imparting a grayish color to the mucosa and after 14 months of treatment showing lymphangiectasia replaced by normal mucosa. b, Mean total weekly scores as defined in the Methods for the indicated parameters in each patient during the pre-Tx (0), 0-4 weeks and 4-14 weeks post-treatment (post-Tx) are plotted. Statistics used the Friedman test and Dunn’s multiple comparisons test (two-sided P value; n = 13 patients at each time point). I.F. = inability to feed, B.M. = number of bowel movements. c, Vitamin B12 concentration in serum before and after treatment beginning at t = 0 (n = 9). d, Serum immunoglobulin (IgG) concentration before and after treatment beginning at t = 0 (n = 15 for pre-Tx and 2-4 months’ assessments; n = 13 for 1-month assessment). e, Calculated mean and standard deviation for intra-patient repeated measurements of fasting blood triglyceride concentrations during pre-Tx periods or the indicated number of months Post-Tx for each patient (n = 15). In d and e statistical comparisons were made by mixed effects analysis and Dunnett’s multiple comparisons test (two-sided P value). f, Mean and standard deviation values for multiple platelet count measurements obtained in each patient during the pre-Tx period or the indicated number of months Post-Tx. Two-sided P values are calculated from Wilcoxon matched-pairs signed-rank test, based on the calculated means of multiple measurements for a given interval (n = 11 patients). g, Photographs of representative pre-Tx samples from P7 or normal control (NC) showing erythrocytes abnormally infiltrating the supernatant (green arrows) during Ficoll gradient separation of PBMCs and disappearance of this phenotype post-Tx. h, Summary of radiological features before and after treatment. Red (+) shows presence of the sign and light blue (−) indicates absence. N/A means radiological studies not available. *P7 had voluminous abdominal fluid before therapy that resolved following treatment (not shown). Shaded areas in c-f show normal range.
Dosing intervals, interrupted therapy, and pharmacokinetics
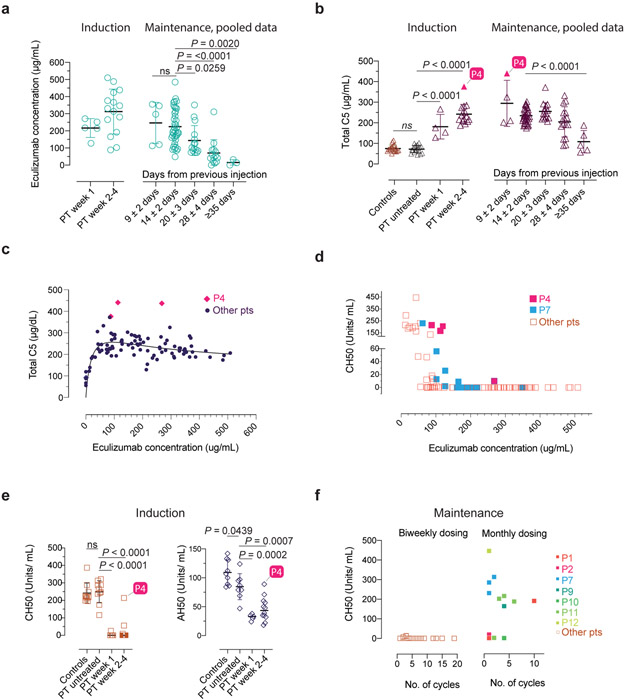
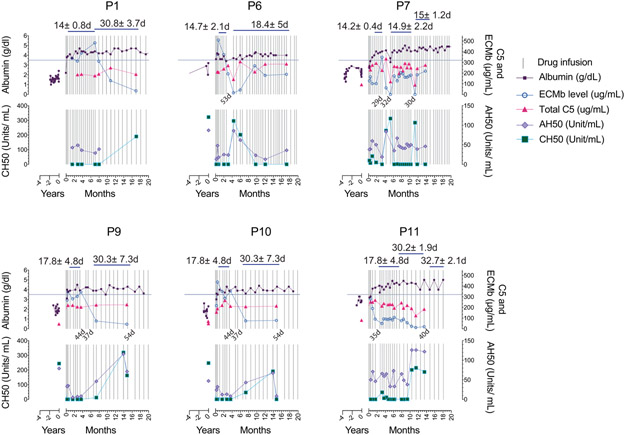
We next studied the pharmacokinetics and pharmacodynamics of eculizumab and total C5 (free + eculizumab-bound). Kurolap et al. In a previous report, an augmented induction regimen was used to account for GI protein wasting with twice-weekly injections13. However, we found that once-weekly induction doses achieved a median trough concentration of eculizumab of 217 and 330 μg/ml, at week 1 and weeks 2-4, respectively, which were much greater than 100 μg/mL, the minimum recommended concentration (Fig. 3a). Eculizumab complexed with C5 to increase the total C5 (free + eculizumab-bound) in the blood to a median of 219 μg/ml (Fig. 3b). Even with the first dose, the eculizumab and total C5 concentrations reached ~200 μg/ml and were stable with the maintenance doses (Fig. 3a,b). We also observed that if dosing was delayed, eculizumab and total C5 concentrations fell slowly and reached baseline at 35 days (Fig. 3a,b). Total C5 for all patients reached a plateau when eculizumab reached 60-100 μg/ml and the ratio of total C5 to eculizumab dropped below 2:1 (the theoretical ratio at which C5 becomes saturated by eculizumab) at 100 μg/ml eculizumab (Fig. 3c, Supplementary Fig. 6a). We, therefore, tested the activity of the classic complement pathway, employing the total hemolytic complement (CH50) test, and found it was completely inhibited by eculizumab > 100 μg/ml (Fig. 3d). Both the AH50 and CH50, measuring the target lysis-inducing activity of the alternative and classical pathways, respectively, were correlated with eculizumab concentrations and strongly suppressed at 1 week (Fig. 3e; Supplementary Fig. 6b,c). If dosing was delayed, eculizumab and total C5 concentrations progressively decreased, and AH50 and CH50 test value outcomes progressively increased with time (Fig. 3f, Extended Data Fig. 4). Thus, eculizumab rapidly and potently inhibits uncontrolled complement activation and reverses PLE in vivo.
Fig. 3 ∣. Serum eculizumab concentration, total C5 and functional complement inhibition during the induction and maintenance phases of treatment.
a-b, Eculizumab and total C5 concentrations with respect to timing of therapy during the indicated periods. The respective exact P values comparing 9±2 vs. 14±2 days was 0.9803 (ns = not significant) in a, and controls vs untreated patients was 0.9926 (ns = not significant) in b. c-d, Pooled analyses of patient samples showing total C5 and CH50 levels across the range of eculizumab concentrations. e, CH50 and AH50 levels with respect to timing of therapy during the indicated periods. f, CH50 values with respect to number of cycles on the standard biweekly dosing intervals or the modified monthly-dosing regimen in selected cases. Error bars indicate mean and S.D. values. Multiple group comparisons were made by ordinary one-way ANOVA test. During post hoc analyses, Dunnett's multiple comparisons test analyzed differences between the standard 14 ± 2 days group with others in the maintenance pooled analyses in a and b, and patient (PT) untreated values with other groups in b and e, during the induction period, respectively. Adjusted P values for multiple testing are indicated. The P value comparing the controls vs untreated patients was 0.9795 (ns = not significant) in e. The number of samples investigated for each parameter were as follows: Total C5: 19 healthy controls and 114 samples from 15 CHAPLE patients; eculizumab concentrations: 96 samples from 15 patients; CH50: 14 healthy controls and 121 samples from 15 patients; AH50: 9 healthy controls and 25 samples from 13 patients. ECMb: eculizumab.
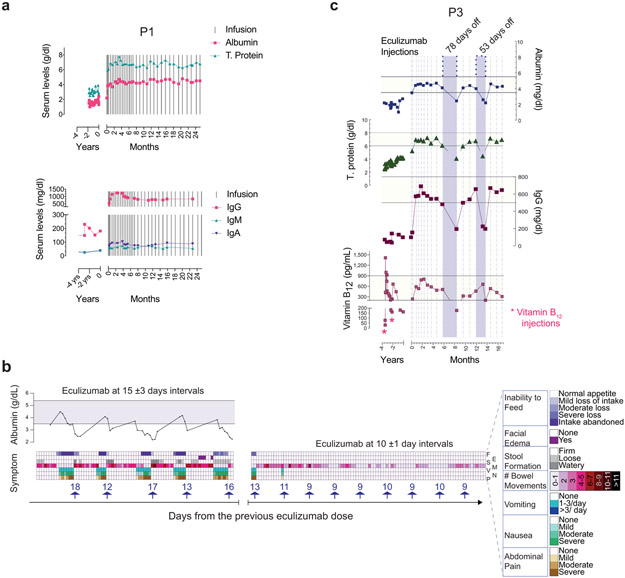
We also tried extended dosing intervals11, 12. Using 4-week dosing, we found no disease relapse in P1, P3, P5, P8, P9, and P10, and P11 (Extended Data Fig. 4,5a). By contrast, P4, the most severely affected in our cohort, showed disease relapse unless we used a 10-day dosing interval (Extended Data Fig. 5b). This finding was not explained by the CD55 mutation in P4, since the identical amino acid change is present in P2 and P5, who responded normally to eculizumab (Extended Data Fig. 1). In P4, we observed less CH50 and AH50 inhibition at comparable concentrations of serum eculizumab than other patients (Fig. 3d, e). Also, total C5 accumulation was much higher in P4, suggesting eculizumab bound to C5 (Fig. 3b,c). We, therefore, performed whole-genome sequencing on P4 and his affected sister, P5, who had responded better to treatment. We detected no novel or extremely rare variants but a single-nucleotide polymorphism (SNP) (rs17611) encoding a V802I amino acid substitution in C5 that was homozygous in P4 and heterozygous in P5. This SNP is common (MAF=0.4590 in gnomAD database) with global homozygosity of about 23% (32516 homozygous in 141333 total individuals, https://gnomad.broadinstitute.org/variant/9-123769200-C-T?dataset=gnomad_r2_1) and previously associated with inflammatory diseases, including liver fibrosis and rheumatoid arthritis17, 18. The substituted residue is outside of the eculizumab binding site but alters the structure so that 5 new amino acid clashes occur in the complex (Supplementary Fig. 7a-c)19. Further genotyping showed that P8, P13, and P14 were also homozygous for I802 (Supplementary Fig. 7d). These patients had comparatively higher antibody-C5 complex concentrations, reduced C5a, and slightly higher soluble C5b-9 complex (sC5b-9) but did not require increased administration of eculizumab (Supplementary Fig. 7e-g). Previous in vitro work showed that eculizumab blocks C5I802 activity, but the variant protein is less efficiently cleaved by human neutrophil elastase17, 20. Taken together, the C5802I/802I variant appears to alter the antibody-C5 complex and possibly its turnover21; however, more complicated factors are likely involved in the poor response of P4 to eculizumab therapy.
Eculizumab therapy was interrupted due to medication inaccessibility (P3 and P8) or a medical decision because of myocarditis (P12), and all experienced disease relapse (Extended Data Fig. 5c, Supplementary Fig. 8, and Supplementary Information). Thus, ongoing eculizumab therapy is necessary to sustain remission. Hence, CHAPLE disease differs from previous diseases in which the treatment can be tapered and discontinued22.
Complement markers before and during eculizumab treatment
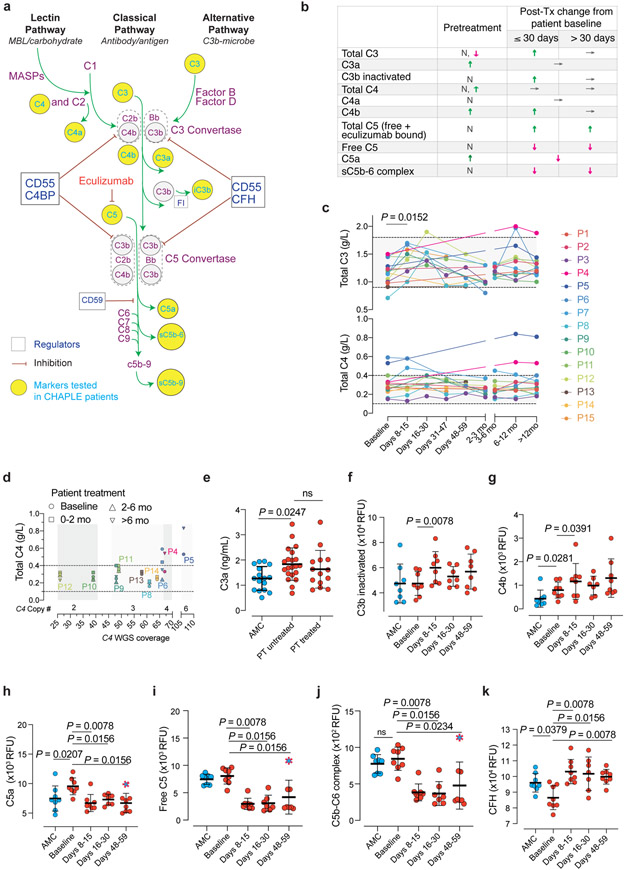
Because complement variations contribute to disease variability, we assessed blood concentrations of C3, C4, and C5, and their activation products together with copy number variations (CNV) in the C4 gene (Fig. 4a-j)23. We observed C3a, C4b, C5a, and soluble terminal complex (sC5b-9) were increased and complement Factor H (CFH) was decreased in baseline disease compared to age-matched healthy controls (AMCs) (Fig. 4 and Supplementary Fig. 7f-g). After treatment, free C5, C5a, sC5b-6, and sC5b-9 were swiftly reduced. Also, we found increased C4b, and CFH but no reduction in C3a with therapy (Fig. 4b,e,f,g, k). Intriguingly, we detected a temporary increase in total C3 and inactive C3b but not C3b with treatment (Fig. 4c). Perhaps the active C3b is rapidly inactivated by the inhibitory proteins, including CFH and Factor I. These changes may be secondary to dissociation of the C5-convertase complexes (by the regulators other than CD55) upon abrupt removal of their substrate, the C5 molecule, from the circulation since eculizumab-bound form is inaccessible to the enzyme. Overall, eculizumab blocked the generation of the terminal complement activation mediators in CHAPLE patients and caused secondary upstream effects.
Fig. 4 ∣. Blood concentrations of complement proteins and their activated products before and during eculizumab treatment.
a, Pathway schematic of the complement system. iC3b = inactivated C3b, FI = complement factor I. CFH = Complement factor H. MASP = mannose-associated serine protease. b, Summary of alterations in selected circulating complement markers in CHAPLE patients. Arrows indicate the direction of change, if any: ↓(decrease), ↑(increase), or →(no change). N = Normal. c, Blood concentration in grams/liter (g/L) of intact C3 and C4, at baseline and during eculizumab treatment. Mean of repeated measurements at each time interval was plotted for individual patients. A mixed-effects model assessed the significance between values at different time points, with Tukey’s multiple comparisons test calculating adjusted P values for each pair (n = 15 patients). d, Serum C4 levels in relation to estimated Complement C4 gene copy numbers based on C4 WGS coverage. WGS: Whole-genome sequencing. In c and d, horizontal dashed lines show reference ranges. e-j, Blood concentration of complement products generated during complement activation at baseline and after eculizumab treatment. k, The soluble phase inhibitor CFH levels at baseline and during treatment. Statistics used to compare C3a between the three groups included an ordinary one-way ANOVA and the Tukey’s multiple comparisons test. Adjusted P values are indicated. Mann Whitney (for comparing AMC vs patient baseline values), and Wilcoxon matched-pairs signed rank tests (between patient baseline vs different post-treatment time points) analyzed the differences between groups in f-k (n = 8 control subjects and n = 8 patients, for each analysis). Red dashed symbols filled with blue in h-j indicate values that correspond to dropped eculizumab concentrations after dose skipping. All P values are two-sided. Sample size for C3a: 16 AMC, 21 untreated-, and 13 treated- patients. Error bars indicate mean and standard deviation. AMC= age-matched control.
C4 concentrations remained stable during treatment, but four subjects, P4, P5, and P7, with a history of thrombosis, and P6 with thrombocytosis, showed increased C4 (Fig. 4c). We evaluated C4 CNV and found two patients had two C4 copies, six with three C4 copies, one with four C4 copies, and one with six C4 copies determined by estimation by whole-genome DNA sequencing (WGS) coverage (Supplementary Fig. 7h). The most common copy number of C4 genes, present between 50% and 60% of healthy human subjects, are four copies, but most of our patients had fewer C4 copies24, 25. Interestingly, the consistently high serum C4 values were found in P5 and P6, even after 6 months of treatment, and they had the highest C4 copy number among all patients we followed. High serum C3 and C4 are associated with venous thrombosis in postnatal women or in the context of individuals with antiphospholipid antibodies, suggesting the correlation of thrombosis and high C4 concentrations due to CNV in our patients could be significant26, 27. Our findings suggest that genetic variations in the complement genes may account for disease expressivity or variable treatment responsiveness among different CD55 deficient individuals.
Mutant soluble CD55 does not prevent complement hyperactivation
The mutation in patient 12 inserted a stop codon for residue Gly348, which prevented addition of the glycerol phosphate-inositol membrane anchor and allowed CD55 to escape as a soluble serum form (Supplementary Fig. 9). Although typically membrane-bound, CD55 is also secreted as a soluble form that inhibits complement in the fluid phase28. P12 was as sick as the other patients, suggesting this mutant soluble version of CD55 was not protective in the context of GI disease.
Microbiome changes following therapy
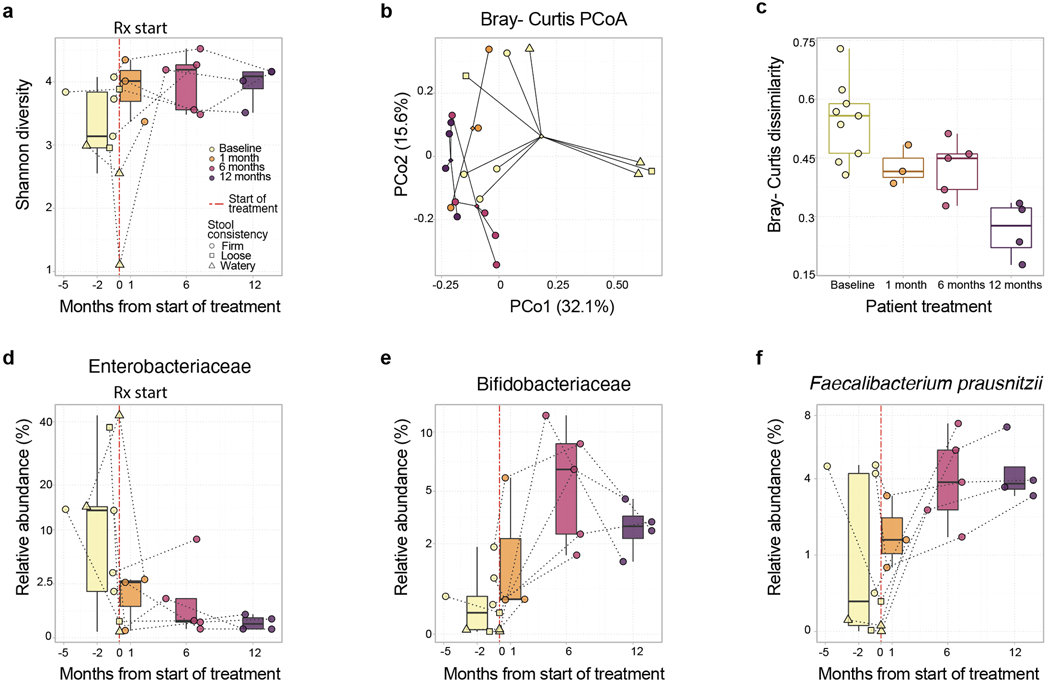
Pathological alterations of the gut microbiota have been observed in inflammatory and metabolic diseases15. Specifically, reductions in alpha diversity and increases in abundance of Enterobacteriaceae taxa occur in chronic inflammatory conditions including inflammatory bowel disease, progressive HIV infection, and necrotizing enterocolitis29, 30, 31, 32, 33. We therefore performed metagenomic profiling of feces from six CHAPLE patients before and after eculizumab. We observed a trend toward increased alpha diversity over 12 months of treatment (P=0.09, linear mixed effects models, Fig. 5a). Pre- and post-treatment bacterial microbiota profiles clustered separately, and baseline microbiota profiles exhibited greater within-group dissimilarity (Fig. 5b,c). A similar profile was evident for fungi but not DNA viruses (Supplementary Fig. 10). Thus, eculizumab treatment causes microbiome restructuring toward a common gut microbial community, possibly by eliminating microbiome stressors, including inflammation, abnormal bowel movements, abnormal food intake, and/or medications, including antibiotics. Moreover, the treatment decreased Enterobacteriaceae (P=0.013, Q value (a p-value that has been adjusted for the False Discovery Rate (FDR)) =0.099, Figure 5d), pro-inflammatory gut pathobionts associated with chronic inflammatory diseases34. Also, the treatment increased Bifidobacteriaceae (P = 0.012, Q = 0.096) and Faecalibacterium prausnitzii (P = 0.019, Q = 0.08), which are important microbiota in healthy infants and are depleted in children with Crohn’s disease (Fig. 5e,f)16, 35. Thus, eculizumab treatment shifted the microbiota composition from an inflammatory profile to an enrichment of taxa comprising a healthy microbiome35.
Fig. 5 ∣. Microbiota composition shifts in eculizumab treated CHAPLE patients.
a, Shannon alpha diversity increases pre- and post-eculizumab treatment are shown. Longitudinal samples from the same patient are connected by dotted lines. A statistical trend toward increased Shannon diversity is seen post-eculizumab treatment (linear mixed effects unadjusted P = 0.09). b, Principal Coordinates Analysis (PCoA) based on Bray-Curtis beta diversity metric of bacterial taxonomic abundances showing clustering by pre- and post-treatment time points (PERMANOVA P = 0.03). Centroids of community distances for each time point group are shown as diamonds. c, Bray-Curtis dispersions (scaled distance from centroid) within each time point group (ANOVA P = 0.00066). d, Enterobacteriaceae (linear mixed effects [LME] P = 0.013, false discovery rate Q=0.099), and e, Bifidobacteriaceae family abundances (LME P = 0.012, Q = 0.096), and f, Faecalibacterium prausnitzii species abundances (LME P = 0.019, Q = 0.08) across time in CHAPLE patients. In a and c-f, the center bar denotes the median, the boxes denote interquartile range, and the whiskers denote the interquartile range × 1.5. The mean of multiple assessments is used for analysis if there are more than one measurement for an individual during a particular time period. ns = not significant. n = 6 patients for all analyses. Q value: a P value that has been adjusted for the False Discovery Rate.
Metabolic response to therapy
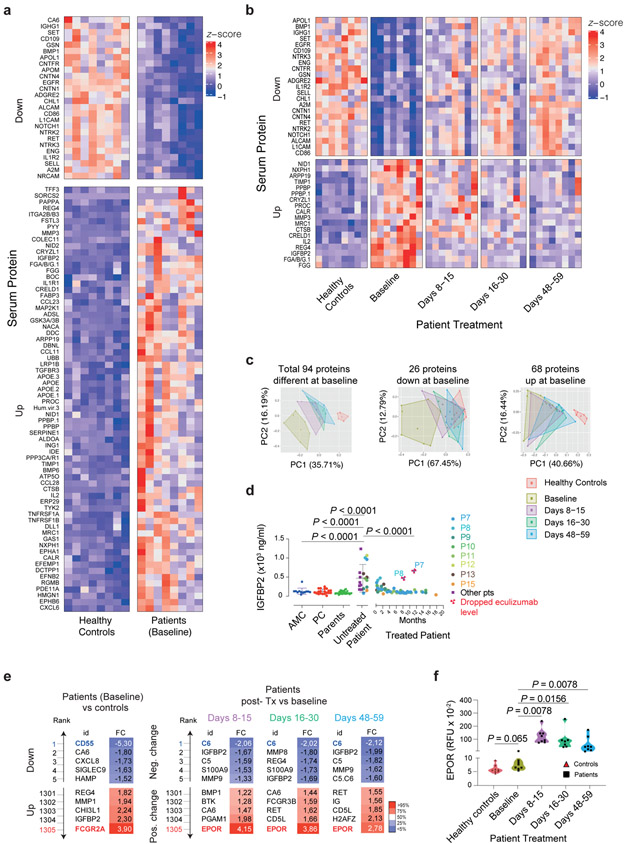
To explore metabolic abnormalities, we profiled 1305 serum proteins in 8 patients using the SOMAlogic aptamer platform. Ninety-four proteins significantly differed between patients and AMCs at baseline with a Q < 0.05)(Fig. 6a and Supplementary Fig. 11). Interestingly, despite PLE, only 26 proteins were reduced (Fig. 6a, top), whereas 68 proteins were increased (Fig. 6a, bottom). Of the 68 upregulated proteins, the greatest increases were in IGFBP2, REG4, ADSL, NACA, APOE, MMP3, PYY, GSK3A & GSK3B, CCL28, and PAPPA. Of the 26 downregulated proteins, those with the greatest decreases were CA6, ADGRE2, BMP1, RET, CNTN4, APOM, NTRK3, EGFR, A2M, and NTRK2, but CD55 was the lowest of any serum protein (Supplementary Fig. 12a,b). After eculizumab, the baseline patient values shifted closer to AMCs, especially for downregulated proteins (Supplementary Fig. 12c). At days 48-59, 23 of 26 downregulated proteins recovered (Fig. 6b, top and Supplementary Fig. 12d), but only 18 upregulated proteins were reduced (Fig. 6b, bottom, and Supplementary Fig. 12d). In an unsupervised principal component (PC) analysis, patient samples at baseline (olive) moved progressively closer towards the controls (red) at 8-15 days (purple), 16-30 days (bright green), and, even more so at 48-59 days (blue) (Fig. 6c). The PC analysis confirms that decreased proteins almost completely recover, whereas the increased proteins reversed but never to normal (Fig. 6c). To validate the SOMAlogic protein changes, we used an ELISA to check IGFBP2, which had the greatest elevation in disease and responded to therapy. The ELISA confirmed elevated IGFBP2 and its correction after treatment (Fig. 6d). Moreover, in P7 and P8, we found that a treatment delay caused an abrupt increase in IGFBP2, suggesting it is a sensitive biomarker of disease activity.
Fig. 6 ∣. Serum proteins altered in patients and response to eculizumab therapy.
a, Compared to healthy age-matched controls (AMC), patients at baseline showed 94 proteins with differences significant at P < 0.05 (two-sided) after correcting for multiple testing of all 1305 proteins measured. 26 of these proteins were downregulated and 68 were upregulated in the patients. b, Comparing baseline to post-treatment timepoints for the patients up to day 59, showed 41 proteins with differences significant at P < 0.05 (two-sided) after correcting for multiple testing of the 94 proteins analyzed due to a difference at baseline. All 41 proteins changed towards the level observed in healthy controls. c, Principal component analysis (PCA) plots using all 94 proteins or separately those down- or up-regulated in patients at baseline. d, Serum IGFBP2 concentrations. Statistics used an ordinary one-way ANOVA with Dunnett’s multiple comparisons correction (Number of samples were: 12 AMC, 20 unrelated processing controls (PC), 36 parent samples, 20 samples from 14 untreated patients, 61 samples from 8 patients post-treatment). Error bars indicate mean and S.D. values. e, Top, and bottom 5 proteins, ranked according to calculated log2 (FC = Fold change) values, either in reference to controls (at baseline), or to pretreatment levels in the patients at the indicated time points (during eculizumab treatment). f, Erythropoietin receptor (EPOR) levels in relative fluorescence units (RFU) are shown. Comparison of healthy controls to baseline was made by Mann-Whitney test and intrapatient comparisons by Wilcoxon matched-pairs signed-rank test (two-sided P value; n = 8 patients, and 8 AMC in a-d and f). The violin plots show the distribution of values across the minimum to maximum range. Protein measurements were performed on the SOMAlogic platform except for IGFBP2 in d, which was determined by ELISA. ECMb: eculizumab.
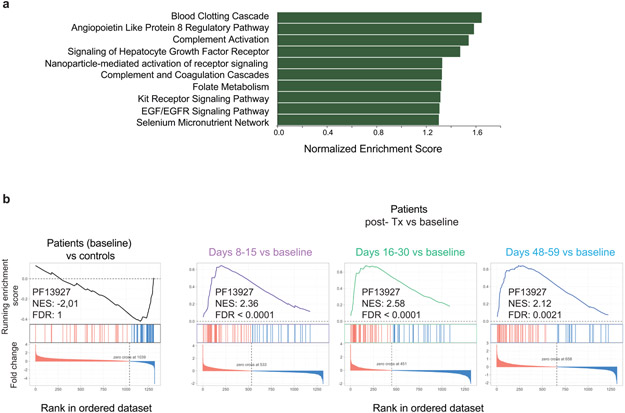
We also observed that certain proteins normal at baseline in CHAPLE patients changed after eculizumab therapy. In a rank analysis, soluble erythropoietin receptor (EPOR), a protein involved in endothelial repair, showed the greatest increase (Fig. 6e,f)36, 37. Among proteomic alterations, complement and coagulation pathways were the top-ranking functional groups, despite an FDR > 0.05 (Extended Data Fig. 6a). CD55 regulates complement, innate immunity, and coagulation, so we selected 53 complement and coagulation proteins measured by SOMAlogic and determined that half (24/53) showed changes at baseline or following therapy (Supplementary Fig. 13).
We also found substantial increases in proteins containing Ig-like domains (Supplementary Fig. 14). A tiny cluster (64 proteins) of such proteins were decreased (Extended Fig. 6b, blue lines). This cluster included several important immunoregulatory proteins, especially cell adhesion molecules, and showed an impressive recovery with treatment (Extended Data Fig. 6b and Supplementary Fig. 15). Interestingly, 12 of the 27 immune-related molecules altered at baseline responded to eculizumab (Supplementary Fig. 16). Thus, inhibiting complement at the C5 level corrects many but not all immune and inflammatory abnormalities.
Discussion
We studied the compassionate use of eculizumab “off label” to treat 16 CHAPLE cases with distinct CD55 gene mutations. We were interested to understand how pharmacologically inhibiting downstream of C5 will affect immune dysregulatory disease caused by CD55 loss affecting upstream control at the C3 convertase. Previously, three members of a single CHAPLE disease family improved with eculizumab treatment3, 13. This success and the global emergence of more CHAPLE cases posed key immunological and metabolic questions that we have now investigated. We demonstrate that eculizumab is broadly effective in CHAPLE patients with different CD55 gene mutations. The recovery from complement damage to gut lymphatics was rapidly achieved in 100% of cases. The loss of Igs, infections, and long-standing functional GI abnormalities were substantially reversed, indicating that the complement-mediated GI inflammation and lymphatic damage is reversible. However, the effects of the drug were temporary. We saw an immediate flare-up of symptoms and serum albumin and Ig loss if the medication was withdrawn. This observation implies that complement and its innate immune and inflammatory effector mechanisms are constantly stimulated, and the patients will require continuous treatment. Thus, eculizumab effectively treats, but does not cure, CHAPLE disease. Nonetheless, despite the high cost of eculizumab, our data support its early and continued use in CHAPLE disease. In addition, the therapeutic potential of an upstream blockade at the C3 level or combinatorial approaches with C3- and C5- blockade at different checkpoints could be considered in future studies, especially in patients with residual findings such as thrombocytosis or thrombosis.
We also explored dosing regimens because eculizumab is extremely expensive, and insurance/health agencies are reluctant to pay for off-label use. The previous study recommended an augmented induction regimen on the theory that PLE would limit drug effectiveness at least early during treatment3, 13. However, we found that this was unnecessary. Our measurements of eculizumab, total C5 (C5 + eculizumab), AH50, and CH50 showed that inhibitory blood concentrations were rapidly achieved, indicating that lymphatic leakage and PLE were surprisingly quickly mended. We also spaced out dosing intervals and found that almost half of the patients only required 30-day maintenance doses, which substantially reduced the cost.
Serum proteomics revealed that GI disease and protein-wasting in CHAPLE patients cause a starvation state, thereby explaining the physiological abnormalities in growth, activity, and maturation. We found that the downregulated proteins were mostly rescued (23 of 26) by treatment, likely due to the prevention of lymph protein loss and anabolic processes. In intestinal lymphangiectasia, albumin and Igs are specifically lost38, 39. We now show that immunoregulatory proteins with structural Ig domains are also selectively lost40. By contrast, only 18 of 68 upregulated proteins showed a significant correction with treatment. We conjecture that because eculizumab blocks at the level of C5, immune activation and inflammation due to upstream C3a anaphylatoxin and C3b opsonization, induces these proteins in a self-perpetuating process despite eculizumab therapy41. Interestingly, eculizumab treatment increased C3 and inactivated C3b. Similar changes occur in PNH following eculizumab, with erythrocytes accumulating C3d on their surface. Opsonized erythrocytes undergo extravascular hemolysis, reducing the treatment benefits42. Interestingly, total C4 concentration was elevated in CHAPLE patients with four or six C4 gene copies. We also found that an SNP encoding the V802I amino acid substitution in C5 protein alters the blood concentration of the eculizumab-bound C5 complex, with potential effects on alternative C5 activation through human neutrophil elastase17. Our data suggests that these genetic factors may modify disease activity and pharmacodynamic properties of eculizumab in CHAPLE disease.
Our proteomics revealed multiple coagulation factor abnormalities. Thus, CHAPLE disease involves the pathological cross-talk of complement abnormalities on coagulation similar to glycosylation disorders that also present with lymphangiectasia and coagulation abnormalities43. Further studies are needed to understand the intersection of complement and coagulation pathways, especially in unexplained lymphangiectasia disorders. Interestingly, we found IGFBP2 as a sensitive biomarker. High IGFBP2 concentrations were reported in various kidney diseases44. It is highly expressed in different cancers and promotes angiogenesis by enhancing VEGF expression45. We did not detect an apparent renal phenotype in CHAPLE; perhaps the biological IGFBP2 activity may be relevant to the circulatory features46. Unexpectedly, we found eculizumab treatment was also associated with increases in certain serum proteins that were normal in untreated disease. The erythropoietin receptor (EPOR) showed the largest increase. Recent studies indicate that the EPOR is expressed in endothelial cells and essential for healthy vasculature and vessel repair36, 37. Thus, EPOR function may contribute to the therapeutic effects of eculizumab in healing damaged GI lymphatic vessels.
Genetic deficiencies of the CD46, CD55, and CD59 complement inhibitors reveal protective roles in different organs. Germline CD55 deficiency in CHAPLE disease, like aHUS, causes abnormalities in coagulation, hematopoietic cells, and endothelium, although has a selective impact on the GI lymphatic vasculature. CD55 is upregulated by diverse stress or danger signals, suggesting it has cytoprotective roles during inflammation, coagulation, and angiogenesis47. The same set of stimuli may not produce an identical response in CD46 or CD59, implying the dominant inhibitor is context-dependent48. For example, CD59 is expressed by the blood vascular endothelial cells but absent in lymphatic endothelium49. PNH involves a somatic mutation in erythrocytes, so presumably, CD55 and CD59 are intact in the gut and prevent GI manifestations9.
Gut microbiota may provide the constant stimulus to complement activation in CHAPLE disease. Our metagenomic profiling showed that CHAPLE patients have pathological microbiota such as inflammationassociated Enterobacteriaceae and reduced Shannon diversity. These are signs of a diseased GI tract and might provide the impetus for ongoing local complement hyperactivation. We found greater inter-patient dissimilarity of microbiota before treatment and homogenization afterwards. This is consistent with the Anna Karenina principle whereby GI diseases cause distinct abnormal microbiota profiles in different patients while microbiota configurations in healthy people converge toward a more homogeneous structure50. Eculizumab treatment removes microbiota stressors and rebalances a healthy microbiome.
Online Methods
Clinical Study Information
This manuscript reports an observational clinical study reporting on the outcome of eculizumab therapy in CHAPLE patients treated in a non-interventional setting in which we collected data on exploratory biomedical parameters. IRB approval was obtained from the Marmara University Medical Faculty Institutional Review Board for Clinical Researches. We recruited verified CD55-deficient patients who were assigned to receive eculizumab by their physicians for enrollment in this study. All patients or their parents for minor subjects provided written informed consent for the study in accordance with the Declaration of Helsinki. Exclusion criteria included: (1) presence of a concomitant disease that leads to hypoproteinemia at the time of starting eculizumab, (2) presence of a concomitant disease that leads to secondary intestinal lymphangiectasia, and (3) unstable clinical condition that precludes allowing blood draw (for biomarker studies). Fifteen patients enrolled in the study through Marmara University and all are reported on in this manuscript. Patient 16 was treated in Netherlands and included into the analyses. We collected clinical data to assess: (1) reversal of protein-losing enteropathy (PLE); (2) reversal of patient-specific major symptoms; (3) reversal of other systemic components of the disease; (4) correction of previous biochemical and radiological abnormalities; and (5) cessation of previous medications.
Dosing regimen
Patients were given an infusion of the complement C5 blocker eculizumab under the brand name Soliris according to the following dosing regimen. Patients older than 18 years of age were given 900 mg weekly for the first 4 weeks and maintained with 1200 mg every other week. For patients younger than 18 years of age, eculizumab was administered on a weight dependent schedule as shown in in Supplementary Table 1. eculizumab was administered within 2 days of the recommended dosage regimen time point. Three patients (P14, P15 and P16) received a modified regimen during the induction phase as proposed by Kurolap et al.13: First 4 eculizumab doses given as 2 injections/week for 2 consecutive weeks (treatment days 1, 4, 8, and 11), followed by three weekly doses (treatment days 15, 22, and 29); thereafter, maintenance infusions on a biweekly basis.
Clinical Markers
Clinical measurements of patient serum were taken at baseline and before each infusion. Total protein, albumin, and immunoglobulin (Ig) G, IgM, IgA, and IgE were measured alongside complete blood counts and the safety lab tests. Additional analyses included the coagulation markers activated partial thromboplastin time (aPTT), D-dimer, and fibrinogen, the additional safety markers total and direct bilirubin, the complement proteins C3, C4, C5, and CH50, and the malabsorption parameters vitamins B12 and D, folate, ferritin, iron, iron binding capacity, calcium, magnesium, and blood lipids.
Symptom Diaries
We collected self-reported daily symptom diaries which tracked abdominal pain, nausea, vomiting, bowel movements, stool formation, facial edema, and inability to feed. Each symptom was scored and recorded daily according to scales described in Supplementary Fig. 5. The patients (adolescent and adults) or parents on behalf children (or youngsters) were asked to complete symptom diaries on daily basis. Data collection was started during the screening visit when patients were evaluated for eligibility for the study and only if a decision to initiate eculizumab was made. Prospectively collected symptom diaries were returned to their physicians on a monthly basis. Patient- reported symptom diaries were collected throughout the induction phase (4 weeks;) plus the subsequent 10 weeks during the maintenance. Some patients continued filling the diaries for longer duration. Symptom scores were entered into a spreadsheet by the primary physician or the principal investigator and analyzed.
Quality of life assessment
The Turkish version “KINDL” questionnaire was used to assess changes in quality of life over the course of the trial, as was previously done (https://www.kindl.org/english/language-versions/turkish/)57. The questionnaire consists of 24 Likert-scaled items grouped into 6 subscales measuring specific aspects of quality of life (QoL), including physical well-being, emotional well-being, self-esteem, family, friends, and school. An additional “Disease” module for patient’s self-identifying as hospitalized or having a long-term illness was used. The presence of the symptom is evaluated based on a quintile Likert scale; “never,” “rarely,” “sometimes,” “frequently,” and “always” options are scored by “1,” “2,” “3,” “4,” and “5”, respectively. Subscale scores are produced by combining the item ratings for each of the 6 subscales and converting each subscale score to a scale of 0–100, with higher scores representing better QoL. Similarly, a total score is produced by combining the item ratings across all 6 subscales (not including the Disease module). The SPSS syntax files provided by the KİNDL-R site were used to enter and analyze the KINDLR data using IBM SPSS Statistics trial version software package for MAC OS. The KINDL parent form was used for 8 to 16-year-old children.
Cells
Peripheral blood mononuclear cells (PBMCs) were isolated from blood by Ficoll gradient density centrifugation. In short, blood was diluted 1:1 in phosphate buffered saline (PBS, Quality Biological). One volume of diluted blood was added to one volume of Ficoll-Paque PLUS density gradient media (GE Healthcare) and centrifuged at 1200 rpm for 20 min with brakes off. PBMCs were isolated from the interface of Ficoll and plasma and underwent ACK lysis (Quality Biological) to remove erythrocytes. PBMCs were aliquoted and cryopreserved in liquid nitrogen in fetal bovine serum (FBS) containing 10 % dimethyl sulfoxide (DMSO).
Flow cytometry
PBMCs were incubated with human Fc receptor blocking solution (BioLegend) in phosphate-buffered saline (PBS) at 25° C for 15 minutes. Cells were stained in ice cold FACS buffer (PBS with 2 % FCS and 1 % sodium azide) with the following antibodies on ice for 30 min at specified dilutions: Purified mouse APC-conjugated anti-human CD3 antibody (BD, cat:347340, clone SK7, dilution 1/10), purified mouse PERCP-conjugated anti-human CD19 antibody (BD, cat: 347540, clone 4G7, dilution 1/10), and purified mouse PE-conjugated anti-human CD55 antibody (BioLegend, cat:311302, clone JS11, dilution 1/100). Cells were washed in FACS buffer prior to acquisition on FACS Calibur (Becton Dickinson) and analyzed with FlowJo v. X (BD). Anaphylatoxins were measured using a Cytometric Bead Array (CBA) Human Anaphylatoxin Kit (BD, cat: 561418). Analyses were performed on a Navios EX flow cytometer (Beckman Coulter, France). Gating strategy is summarized in Supplementary Fig. 1.
Eculizumab and C5 concentrations
Eculizumab concentrations were measured in serum samples using a liquid chromatography-tandem mass spectrometry (LC-MS/MS) method developed in house on a triple-quadrupole mass spectrometer under selected reaction monitoring (SRM) mode, referred to as SRM-LC-MS/MS. Briefly, total protein in serum samples is first denatured, reduced, and digested with trypsin. The digested peptide mixtures are then analyzed by SRM-LC-MS/MS method. Signature peptide(s) unique to eculizumab detected in the sample digests are monitored and quantitated using an eculizumab protein calibration curve. Lower limit of quantitation (LLOQ) of the assay is 4 μg/mL. Total C5 concentrations in human serum were measured in house using an electrochemiluminescence immunoassay that measures the concentration of total C5. Briefly, serum samples were diluted in mild acetic acid containing a biotinylated human anti-C5 antibody as capture reagent, then added into a streptavidin coated MesoScale Discovery (MSD) plate. The C5 capture in the plate is then detected by a second ruthenium labeled anti-C5 antibody and quantified by a purified human C5 calibration curve. LLOQ of the assay was 7.8 μg/ml.
Complement activity
The CH50 assay tests the functional capability of complement components of the classical pathway using sheep red blood cells (SRBCs) pre-coated with rabbit anti-sheep red blood cell antibody. Briefly, each serum sample is diluted serial diluted, and a fixed volume of optimally sensitized SRBCs is added to each diluted sample. After incubation, the mixture is centrifuged, and the degree of hemolysis is quantified by measuring the absorbance of the hemoglobin released into the supernatant at 415 nm. The dilution of patient’s serum needed to lyse 50% of erythrocytes is then determined as the CH50 value. Similarly, the AH50 assay tests the functional capability of complement components of the alternative pathway using rabbit erythrocytes. a buffer containing EGTA is used to chelate calcium and block the activation of the classical pathway58.
Fecal microbiome metagenomics
DNA was isolated using the MagAttract PowerMicrobiome DNA/RNA EP Kit (Qiagen) in an Eppendorf epMotion 5073 automated liquid handling system. Metagenomic libraries were prepared using 30 μl DNA in a 96-well plate with the Nextera DNA Flex Library Prep Kit (Illumina). The library preparation was initiated with tagmentation and post-tagmentation cleanup followed by amplification of tagmented DNA and cleanup using Sample Purification Beads for both the cleanup steps. Library quality control included quantification of the final library using the Qubit dsDNA High Sensitivity assay and library quality assessment using the Agilent High Sensitivity HS D5000 ScreenTape assay on the Agilent 4200 Tapestation. The individual libraries were diluted and pooled at a concentration of 7 nM, to make the final library pool. This pool was normalized to 1.8 pM, spiked-in with 1% PhiX sequencing control and sequenced on an Illumina NextSeq 500 instrument.
Microbiome analysis
For sequence analysis, read pairs were trimmed for quality and adapter sequences using BBDuk (https://jgi.doe.gov/data-and-tools/bbtools). The pairs were subsequently assembled with the metaSPAdes pipeline from SPAdes59. Taxonomic assignment was done with Kraken2 using Bracken 2.5 for abundance estimation and the maxikraken2 (v_1903_140GB) database60. Sequences matching to the human genome were excluded from all analyses. Functional annotation of the metagenomes was done using Prodigal for computational gene finding, InterProScan 5 for annotation, the KEGG orthology for pathway assignment, and MinPath for pathway abundance inference61, 62, 63. Differential abundance statistics were performed using MaAsLin2 (https://huttenhower.sph.harvard.edu/maaslin) with arc-sin transformation and a linear mixed effects model testing the difference in abundances over time per subject. Alpha diversity and ordination were computed with the vegan Community Ecology R package using the diversity and betadisper functions. Statistical analyses were performed with vegan's adonis and the lmerTest R package64.
Serum
Blood was collected in vacutainer tubes without coagulants. The tube was left undisturbed for 30 min after removal of the cap to allow the blood to clot. Then, the tube was spun at 1000 rpm for 10 min. Serum is the supernatant and was stored at −80° C for later use in laboratory experiments.
Plasma
Blood was collected in ACD tubes to isolate PBMCs and plasma. After Ficoll gradient density centrifugation, plasma samples were collected and stored in ultra-low temperature freezer for later use.
Serum proteomics
Serum proteomic analysis was performed using the aptamer-based SOMAscan 1.3k Assay (SomaLogic). SomaLogic detects 1305 protein analytes and is optimized for analysis of human serum65. Briefly, aptamers are short single-stranded DNA sequences modified to confer specific binding to target proteins and can be highly multiplexed for discovery of biomarker signatures. The proteins quantified include cytokines, hormones, growth factors, receptors, kinases, proteases, protease inhibitors, and structural proteins. A complete list of the analytes measured is at http://somalogic.com/wp-content/uploads/2017/06/SSM-045-Rev-2-SOMAscan-Assay-1.3k-Content.pdf. The assay was performed according to manufacturer specifications and analyzed using web tools as previously described66, 67.
Complement markers
Serum C3 and C4 concentrations were determined in the clinical laboratory on fresh blood samples using a turbidimetric method.
Plasma anaphylatoxins (C3a, C4a, and C5a) were measured using a flow cytometry based cytometric bead array (CBA) Human Anaphylatoxin Kit.
The sC5b-9 levels were measured using MicroVue sC5b-9 Plus EIA kit (Quidel Corporation) according to the manufacture’s protocol and as reported elsewhere68. The plasma samples were diluted to 1:10 using the sample diluent provided with the kit. The sC5b-9 was detected by the detection antibody (horseradish peroxidase-conjugated anti-sC5b-9 complex). The signal was developed by adding a chromogenic enzyme-substrate and measured using a spectrophotometer.
The free C5, C3b, inactivated C3b, C4b, and soluble C5b-6 complex (sC5b-6) assessments were made by the SOMAlogic platform using biobanked serum samples as described elsewhere69. In order to test for the active and inactive forms of C3b the SOMAmers 2683-1 and 4480-59 were used. Per the manufacturer’s specifications, SOMAmer 2683-1 was selected against iC3b isolated from human serum, with a similar binding to C3 and C3b. SOMAmer 4480-59 was selected against C3b isolated from human serum; a binding to C3 and iC3b was observed, but it was at least 10× weaker affinity. There was no binding with the C3d or C3a fragments. The characterization of the SOMAmers is presented Supplementary Table 2.
IGFBP2 measurement in serum samples
IGFBP2 concentration was measured in serum samples using a Duoset ELISA kit (R&D Systems, Catalogue # DY674) according to the manufacture’s protocol and as described elsewhere70. Briefly, the 96-well microplates were coated with the capture antibody (mouse anti-human IGFBP2) overnight at 25° C. The serum samples were diluted to 1:250 using the assay buffer provided with the kit. The standards or samples were loaded onto the microplate in duplicates and incubated for 2 h at 25° C. IGFBP2 was detected by the detection antibody (biotinylated goat anti-human IGFBP2) and streptavidin-horseradish peroxidase conjugate.
Whole-Genome Sequencing
Genomic DNA samples were quantified using a fluorescence dye-based assay (PicoGreen dsDNA reagent) before normalization to 20 ng/μL and 55 μL of volume. Samples were added into a Covaris 96 microTUBE plate at 1000 ng gDNA input and sheared using the Covaris LE220 Focused-ultrasonicator targeting a peak size of 410bp. Sequencing libraries were generated from fragmented DNA using the Illumina TruSeq DNA PCR-Free HT Library Preparation Kit and IDT for Illumina TruSeq DNA UD Indexes (96 Indexes, 96 Samples) adapters with minor modifications for automation (Hamilton STAR Liquid Handling System). Library size distribution and absence of adapter dimers was assessed by automated capillary gel-electrophoresis (Advanced Analytical Fragment Analyzer). Library yield was determined by qPCR quantitation using the KAPA qPCR Quantification Kit (Roche Light Cycler 480 Instrument II). Sequencing libraries were normalized, pooled into a 24-plex and quantified as above before dilution to 2.7 nM and sequenced on an Illumina NovaSeq 6000 using a S4 Reagent Kit (300 cycles) using 151+8+8+151 cycle run parameters. Primary sequencing data was demuxed using the Illumina HAS2.2 pipeline and sample-level quality control for base quality, coverage, duplicates and contamination was conducted. The sequenced DNA reads were then mapped to hg19 or hg38 human genome reference using the Burrows-Wheeler Aligner with default parameters. Variant calling was performed using the Genome Analysis Toolkit version 3.4 (the Broad Institute, http://www.broadinstitute.org/gatk/).
For Complement C4 gene copy number, the coverage of the whole C4 regions, the unique C4 long form regions and control regions without CNV changes were calculated by bedtools after removing the duplicated reads and low-quality mapping reads. The coverage of C4 regions were then normalized by the control regions coverage and the rough copy number of C4 genes were estimated for diploid genome. The C4 long form copy numbers were determined by the coverage of the unique C4 intronic region in the C4 long form.
Structural Modeling of Complement C5 and Eculizumab Interactions
To model the interactions and variant impacts of complement C5 and eculizumab, complement C5 protein complexed with eculizumab heavy chain variable domains (PDB ID: 5I5K) was loaded into protein modeling software package ChimeraX19, 55. Variant V802I was created using the rotamerization component in ChimeraX, and associated intermolecular and intramolecular steric hindrances (“clashes”) were calculated based on the predominant rotamer predicted by the software.
Statistics
Unless otherwise stated, all statistical analysis was conducted in Prism (GraphPad) version 8.2.1 with a p-value < 0.05 was considered significant. Statistical analysis was conducted to assess patients’ responses to eculizumab therapy over several different lengths of treatment, as specified in the text. Kaplan-Meier curves were fitted to show cumulative frequency of various parameters as a function of age. To compare patients’ serum albumin, total protein, serum immunoglobulins, total C3 and C4 concentrations, and serum IgG a mixed effects model with Tukey’s multiple comparisons correction was used. We compared plasma C3a values between the 3 groups using an Ordinary one-way ANOVA and the Tukey’s multiple comparisons test. Between group comparisons for plasma C5a and sC5b-9 were made by the Mann Whitney test. The Wilcoxon matched-pairs signed-rank test was used to assess changes in KINDL quality of life (QoL) and platelet counts. Changes in symptom frequency were analyzed using the Friedman test with Dunn’s multiple comparisons correction. Changes in blood triglycerides were assessed using a mixed effects model with Dunnett’s multiple comparisons correction. An ordinary one-way ANOVA with Dunnett’s multiple comparisons correction was used to analyze levels of eculizumab, total C5 (free + eculizumab bound), and CH50 and IGFBP2 levels. Mann-Whitney test and Wilcoxon matched-pairs signed-rank test compared free C5, inactivated C3b, C4b, the soluble C5b-6 complex, CFH, and EPOR levels. The statistics used for the Microbiome changes following therapy section are described in the Microbiome analysis section. For the proteomic analyses, changes in proteins between patients and age-matched healthy controls (AMC) were evaluated by Mann-Whitney U-test, and longitudinal changes within patient by Wilcoxon signed-rank paired test, with p-values corrected for multiple comparison by Benjamini-Hochberg method. There was no blinding in the clinical data acquisition as this is not a placebo-controlled trial. The researchers who carried pharmacokinetic and pharmacodynamic examinations were blinded to the study subjects since each tube containing a patient sample was labeled with a code that included no reference to treatment status of the subject. For the proteomic and metagenomic studies the technicians who analyzed the samples were blinded to the group allocation information. The bioinformaticians carried the computational analyses by matching the sample metadata with the experimental test results, each provided by independent researchers. The bioinformatician who analzed whole genome sequencing data was blinded to serum C4 values when calculating estimated C4 gene copy numbers, but not blinded to patient allocation when searching for C5 gene variants, since the latter analysis required appropriate filtering strategies.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
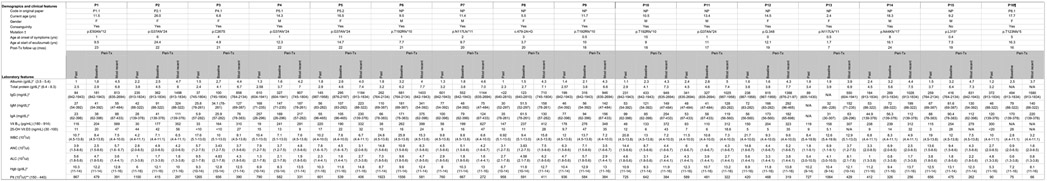
Extended Data Fig. 1. Demographic, clinical and laboratory characteristics of the patients with CHAPLE disease enrolled in the study.
Reference ranges are indicated in parenthesis. NA: Not assessed, NP: New patient, WBC: White blood count, ANC: Absolute neutrophil count, ALC: Absolute lymphocyte count lymphocyte count, Hgb: Hemoglobin, Plt: Platelet count.
† CD55 NCBI Reference Sequence: NM_000574.
* the lowest values in the past (more than 1 year before therapy) are indicated in the "past" columns.
** the highest values in the past (more than 1 year before therapy) are indicated in the "past" columns.
¶ The serum immunoglobulins compared to age-matched serum immunoglobulin reference ranges of Turkish children (Ref 51). Peri-Tx: peri-treatment, referring to the past 12 months before the start of eculizumab until the end of the observation period
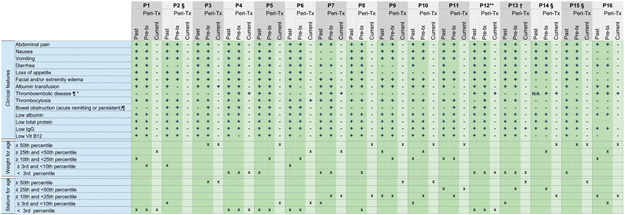
Extended Data Fig. 2. Clinical symptoms, laboratory findings and growth percentiles of CHAPLE patients prior to and on eculizumab treatment.
Past: In the lifetime of the patient. Peri-Tx: Peri-treatment, referring to the past 12 months before the start of eculizumab until the end of the observation period. Current refers to the most recent measurement or the observation period when a particular patient has been on regular eculizumab Tx.
+ present, − absent, N/A: Not assessed
* We detected a recanalization of the narrowed segment in the thrombotic vessel in P5 during the follow up exam on eculizumab. However, there was no documented recovery of any of the thromboembolic complications in other patients during the indicated periods of follow up.
** P12 had ischemic gliotic foci in brain imaging.
¶ As identified by appropriate imaging including abdominal ultrasonography (USG) and/or doppler USG, and/or vascular imaging using a computed tomography or magnetic resonance imaging.
§ Growth percentiles not presented in certain columns; P2 was an adult during the peri-Tx period, and data not available in P14 and P15 for respective periods.
† Abdominal pain cannot be assessed in P13 due to verbal immaturity and corresponding assessment was made for discomfort and abnormal crying behavior.
Extended Data Fig. 3. Medical interventions received by CHAPLE patients prior to and during eculizumab treatment.
Past: In the lifetime of the patient. Peri-Tx: Peri-treatment, referring to the past 12 months before the start of eculizumab until the end of the observation period. Current refers to the most recent measurement or the presence of a finding during the observation period when a particular patient has been on regular eculizumab Tx.
*A repeat bowel surgery had been planned in P8 prior to eculizumab. Likewise, P3 had been planned to undergo bowel surgery before eculizumab. P15 underwent 2 separate bowel resection surgeries and received 39 days of intensive care support during the postoperative period, including mechanical ventilation.
** "Other invasive intervention" includes placement of a central access device in P8 to deliver frequent albumin transfusions; intracardiac blood clot removal surgery, chest tube placement to drain pneumothorax and a cranial surgery to alleviate brain hemorrhage in P4; a percutaneous endoscopic gastrostomy port to support enteral nutrition in P12; a transjugular intrahepatic portosystemic shunt (TIPS) to relieve portal hypertension in P16.
† P8 received intravenous (IV) antibiotics for sepsis and metronidazole for Giardia lamblia enteritis in the past. P4 and P12 experienced cardiovascular problems during the induction period of eculizumab, leading to dose skipping or termination of treatment, respectively.
Eculizumab Tx was restarted in P12 after a period off-therapy, with no recurrence of similar manifestations. Fecal assessment during diarrheal episodes revealed Cryptosporidium parvum in P1 and P13 prior to eculizumab, who were treated with nitazoxanide uneventfully.
¶ The column for past medication before 1 year prior to eculizumab is blank for P13 due to young age and onset of symptoms during the past year.
§ Respiratory support refers to mechanical ventilation and/or oxygen treatment.
PLE: Protein-losing enteropathy.
Extended Data Fig. 4. The effect of dose spacing between injections on albumin, trough eculizumab concentration, and complement markers during the maintenance therapy.
Blood samples were obtained prior to each injection. The horizontal line on left y axis shows lower range for albumin. Calculated mean ± S.D. values for dose intervals during the indicated time spans, or between two consecutive injections are presented in days. ECMb: eculizumab.
Extended Data Fig. 5. Variable dose spacing or interrupted therapy.
a, Normal serum protein levels sustained when switching from biweekly doses to monthly eculizumab injections in P1. Timing of eculizumab doses in relation to serum albumin and total protein concentrations for P1 in top panel, and immunoglobulin isotypes IgG, IgM and IgG in the bottom panel. b. Timeline plotting the incidence and severity of the indicated symptoms in P4 in relation to timing of eculizumab injection at various dosing intervals. Serum albumin levels plotted at the top of the figure with the horizontal bar showing normal range. Each arrow above the x-axis shows an eculizumab injection with the accompanying numbers illustrating days from the previous dose. The top line shows inability to feed (F) ranging from normal appetite (white) to intake completely abandoned (dark purple). The second line shows the presence (purple) or absence (white) of facial edema (E) in purple whereas the third line shows stool formation (S) on a scale from firm (white) to watery (dark grey). The fourth line quantifies the number of bowel movements (M) from 0-1 (white) to >11 (black). The fifth and sixth lines show the incidences of vomiting (V) from none (white) to >3 times per day (dark blue) and severity of nausea (N) from none (white) to severe (green), respectively. The last line shows severity of abdominal pain (P), ranging from none (white) to severe (dark brown). c. Serum albumin, total protein (T. protein), immunoglobulin G (IgG), and vitamin B12 concentrations for P3 before and after treatment at t = 0. Horizontal yellow bars for indicated parameters show normal range. Vertical dashed lines show eculizumab injections and purple bars dose skipping periods with the indicated numbers showing intervals between two consecutive injections.
Extended Data Fig. 6. Biological significance of proteomic changes in serum.
a, Top ranking pathways (Wikipathways) plotted against normalized enrichment score (Ref52, 53). Note, the enrichment of the pathways did not reach adjusted statistical significance level of false discovery rate (FDR) <0.05. b, Enrichment plots from pre-ranked analysis of log2 fold changes (FC) in protein abundances of patients with respect to age matched controls, showing rank-based ordering of proteins belonging to immunoglobulin Pfam domain PF13927 at baseline. Similar plots are generated for indicated Post-treatment (Post-Tx) time points using log2 FC in protein abundances calculated with respect to patient baseline values. PF13927, found to be enriched in the STRING analysis (Ref 54), was added to the Wikipathways gene set to compute the normalized enrichment score (NES) and FDR.
Supplementary Material
Acknowledgements
This work was supported in part by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH, BCBB Support Services Contract HHSN316201300006W/HHSN27200002, and The Marmara University, Scientific Research Projects Committee (BAPKO, grant number SAG-C-TUP-230119-0018). We thank the Turkish National Society of Allergy and Clinical Immunology (TNSACI) for supporting travel expenses for the screening studies. We thank C. Kemper of the National Heart, Lung, and Blood Institute, for thoughtful editing of the final manuscript. We thank M. Quiñones, the Center for Human Immunology and the NIAID Microbiome Program, NIAID, NIH for research support. We also thank A. Kiykim for patient care, A. Dalga, and I. Tatli for technical assistance, H. Su and X.He for advice and assistance; D. Comrie, S. Kubo, and J. Ravell for a critical reading of the manuscript; and R. Kissinger for artwork. We thank important colleagues at Regeneron: A. N. Thomas for sample processing; C. Huang for biomarker analysis; H. Qiu, and S.E for eculizumab analysis; and C. H. Lai, L. DeStefano, and K. Donohue for total C5 analysis. Molecular graphics and analyses performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases.
Footnotes
Competing Interests Statement
The authors M.J.L. and A.O. have a pending patent on C5 inhibitor treatment of CHAPLE. B.S. is a former SomaLogic, Inc. (Boulder, CO, USA) employee and a company shareholder. O.H., Y.G.N., M.D.W., and K.D.N. are employees of Regeneron Pharmaceuticals Inc., a biopharma company. There are no conflicts of interest to report for the remaining coauthors.
Data Availability
The raw data including the adat file used to generate proteomic analyses and the data analysis codes used for microbiome analyses are available upon request from the corresponding authors. The microbiome sequencing data is linked to the NCBI BioProject ID PRJNA629392. The raw data including the adat file used to generate proteomic analyses, the data analysis codes used for microbiome analyses, and all other data that support the findings of this study are available from the corresponding authors upon request to the extent allowed by all laws and institutional policies regarding confidentiality of patient clinical information. Datasets used in the study: Peditools; string db; maxikraken2 (v_1903_140GB) database; gnomAD v2.1.1; and 1000genome.
References
- 1.Waldmann TA, Steinfeld JL, Dutcher TF, Davidson JD & Gordon RS Jr. The role of the gastrointestinal system in "idiopathic hypoproteinemia". Gastroenterology 41, 197–207 (1961). [PubMed] [Google Scholar]
- 2.Ozen A et al. CD55 Deficiency, Early-Onset Protein-Losing Enteropathy, and Thrombosis. N Engl J Med 377, 52–61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurolap A et al. Loss of CD55 in Eculizumab-Responsive Protein-Losing Enteropathy. N Engl J Med 377, 87–89 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Ozen A CHAPLE syndrome uncovers the primary role of complement in a familial form of Waldmann's disease. Immunol Rev 287, 20–32 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Mevorach D Clearance of dying cells and systemic lupus erythematosus: the role of C1q and the complement system. Apoptosis 15, 1114–1123 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Holers VM Complement and its receptors: new insights into human disease. Annu Rev Immunol 32, 433–459 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Kwan WH, van der Touw W & Heeger PS Complement regulation of T cell immunity. Immunol Res 54, 247–253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elvington M, Liszewski MK & Atkinson JP Evolution of the complement system: from defense of the single cell to guardian of the intravascular space. Immunol Rev 274, 9–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zipfel PF & Skerka C Complement regulators and inhibitory proteins. Nat Rev Immunol 9, 729–740 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Rother RP, Rollins SA, Mojcik CF, Brodsky RA & Bell L Discovery and development of the complement inhibitor eculizumab for the treatment of paroxysmal nocturnal hemoglobinuria. Nat Biotechnol 25, 1256–1264 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Wong EK, Goodship TH & Kavanagh D Complement therapy in atypical haemolytic uraemic syndrome (aHUS). Mol Immunol 56, 199–212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris CL, Pouw RB, Kavanagh D, Sun R & Ricklin D Developments in anti-complement therapy; from disease to clinical trial. Mol Immunol 102, 89–119 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Kurolap A et al. Eculizumab Is Safe and Effective as a Long-term Treatment for Protein-losing Enteropathy Due to CD55 Deficiency. J Pediatr Gastroenterol Nutr 68, 325–333 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Di Narzo AF et al. High-Throughput Identification of the Plasma Proteomic Signature of Inflammatory Bowel Disease. J Crohns Colitis 13, 462–471 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pickard JM, Zeng MY, Caruso R & Nunez G Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev 279, 70–89 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwiertz A et al. Microbiota in pediatric inflammatory bowel disease. J Pediatr 157, 240–244 e241 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Giles JL, Choy E, van den Berg C, Morgan BP & Harris CL Functional analysis of a complement polymorphism (rs17611) associated with rheumatoid arthritis. J Immunol 194, 3029–3034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hillebrandt S et al. Complement factor 5 is a quantitative trait gene that modifies liver fibrogenesis in mice and humans. Nat Genet 37, 835–843 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Schatz-Jakobsen JA et al. Structural Basis for Eculizumab-Mediated Inhibition of the Complement Terminal Pathway. J Immunol 197, 337–344 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Fukuzawa T et al. Long lasting neutralization of C5 by SKY59, a novel recycling antibody, is a potential therapy for complement-mediated diseases. Sci Rep 7, 1080 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zelek WM, Taylor PR & Morgan BP Development and characterization of novel anti-C5 monoclonal antibodies capable of inhibiting complement in multiple species. Immunology 157, 283–295 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ardissino G et al. Complement functional tests for monitoring eculizumab treatment in patients with atypical hemolytic uremic syndrome: an update. Pediatr Nephrol 33, 457–461 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Lintner KE et al. Early Components of the Complement Classical Activation Pathway in Human Systemic Autoimmune Diseases. Front Immunol 7, 36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet 80, 1037–1054 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saxena K et al. Great genotypic and phenotypic diversities associated with copy-number variations of complement C4 and RP-C4-CYP21-TNX (RCCX) modules: a comparison of Asian-Indian and European American populations. Mol Immunol 46, 1289–1303 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dahm AEA et al. Elevated Complement C3 and C4 Levels are Associated with Postnatal Pregnancy-Related Venous Thrombosis. Thromb Haemost 119, 1481–1488 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Savelli SL et al. Opposite Profiles of Complement in Antiphospholipid Syndrome (APS) and Systemic Lupus Erythematosus (SLE) Among Patients With Antiphospholipid Antibodies (aPL). Front Immunol 10, 885 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medof ME, Walter EI, Rutgers JL, Knowles DM & Nussenzweig V Identification of the complement decay-accelerating factor (DAF) on epithelium and glandular cells and in body fluids. J Exp Med 165, 848–864 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kostic AD et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22, 292–298 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arthur JC et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lloyd-Price J et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vujkovic-Cvijin I et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med 5, 193ra191 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olm MR et al. Necrotizing enterocolitis is preceded by increased gut bacterial replication, Klebsiella, and fimbriae-encoding bacteria. Sci Adv 5, eaax5727 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin NR, Whon TW & Bae JW Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol 33, 496–503 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Blanton LV et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351, aad3311–aad3311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heeschen C et al. Erythropoietin is a potent physiologic stimulus for endothelial progenitor cell mobilization. Blood 102, 1340–1346 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Ribatti D et al. Human erythropoietin induces a pro-angiogenic phenotype in cultured endothelial cells and stimulates neovascularization in vivo. Blood 93, 2627–2636 (1999). [PubMed] [Google Scholar]
- 38.Homburger F & Petermann ML Studies on hypoproteinemia; familial idiopathic dysproteinemia. Blood 4, 1085–1108 (1949). [PubMed] [Google Scholar]
- 39.Parfitt AM Familial neonatal hypoproteinaemia with exudative enteropathy and intestinal lymphangiectasis. Arch Dis Child 41, 54–62 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srinivasan M & Roeske RW Immunomodulatory peptides from IgSF proteins: a review. Curr Protein Pept Sci 6, 185–196 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Sauter RJ et al. Functional Relevance of the Anaphylatoxin Receptor C3aR for Platelet Function and Arterial Thrombus Formation Marks an Intersection Point Between Innate Immunity and Thrombosis. Circulation 138, 1720–1735 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Notaro R & Sica M C3-mediated extravascular hemolysis in PNH on eculizumab: Mechanism and clinical implications. Semin Hematol 55, 130–135 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Brucker WJ et al. An emerging role for endothelial barrier support therapy for congenital disorders of glycosylation. J Inherit Metab Dis 43, 880–890 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Ding H, Kharboutli M, Saxena R & Wu T Insulin-like growth factor binding protein-2 as a novel biomarker for disease activity and renal pathology changes in lupus nephritis. Clin Exp Immunol 184, 11–18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Azar WJ et al. IGFBP-2 enhances VEGF gene promoter activity and consequent promotion of angiogenesis by neuroblastoma cells. Endocrinology 152, 3332–3342 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Ozen A, Comrie WA & Lenardo MJ CD55 Deficiency and Protein-Losing Enteropathy. N Engl J Med 377, 1499–1500 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Mason JC, Lidington EA, Ahmad SR & Haskard DO bFGF and VEGF synergistically enhance endothelial cytoprotection via decay-accelerating factor induction. Am J Physiol Cell Physiol 282, C578–587 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Mason JC et al. Induction of decay-accelerating factor by cytokines or the membrane-attack complex protects vascular endothelial cells against complement deposition. Blood 94, 1673–1682 (1999). [PubMed] [Google Scholar]
- 49.Park SM et al. Mapping the distinctive populations of lymphatic endothelial cells in different zones of human lymph nodes. PLoS One 9, e94781 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zaneveld JR, McMinds R & Vega Thurber R Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat Microbiol 2, 17121 (2017). [DOI] [PubMed] [Google Scholar]
Methods-only References
- 51.Tezcan I, Berkel AI, Ersoy F & Sanal O Sağlıklı Türk çocukları ve erişkinlerde turbidometrik yöntemle bakılan serum immunoglobulin düzeyleri. Çocuk Sağlığı ve Hastalıkları Dergisi 32, 649–656 (1996). [Google Scholar]
- 52.Slenter DN et al. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Research 46, D661–D667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramanian A et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szklarczyk D et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic acids research 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goddard TD et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shapovalov MV & Dunbrack RL Jr. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure 19, 844–858 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eser E et al. [The psychometric properties of the new Turkish generic health-related quality of life questionnaire for children (Kid-KINDL)]. Turk Psikiyatri Derg 19, 409–417 (2008). [PubMed] [Google Scholar]
- 58.Jaskowski TD, Martins TB, Litwin CM & Hill HR Comparison of three different methods for measuring classical pathway complement activity. Clin Diagn Lab Immunol 6, 137–139 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bankevich A et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wood DE, Lu J & Langmead B Improved metagenomic analysis with Kraken 2. Genome Biol 20, 257 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye Y & Doak TG A parsimony approach to biological pathway reconstruction/inference for genomes and metagenomes. PLoS Comput Biol 5, e1000465 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kanehisa M, Sato Y, Furumichi M, Morishima K & Tanabe M New approach for understanding genome variations in KEGG. Nucleic Acids Res 47, D590–D595 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hyatt D et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuznetsova A, Brockhoff P & Christensen R lmerTest Package: Tests in Linear Mixed Effects Models. Journal of Statistical Software 82, 1–26 (2017). [Google Scholar]
- 65.Gold L et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 5, e15004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Candia J et al. Assessment of Variability in the SOMAscan Assay. Sci Rep 7, 14248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheung F et al. Web Tool for Navigating and Plotting SomaLogic ADAT Files. J Open Res Softw 5, 20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Noris M et al. Dynamics of complement activation in aHUS and how to monitor eculizumab therapy. Blood 124, 1715–1726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emilsson V et al. Co-regulatory networks of human serum proteins link genetics to disease. Science 361, 769–773 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eiseman JL et al. Evaluation of plasma insulin-like growth factor binding protein 2 and Her-2 extracellular domain as biomarkers for 17-allylamino-17-demethoxygeldanamycin treatment of adult patients with advanced solid tumors. Clin Cancer Res 13, 2121–2127 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data including the adat file used to generate proteomic analyses and the data analysis codes used for microbiome analyses are available upon request from the corresponding authors. The microbiome sequencing data is linked to the NCBI BioProject ID PRJNA629392. The raw data including the adat file used to generate proteomic analyses, the data analysis codes used for microbiome analyses, and all other data that support the findings of this study are available from the corresponding authors upon request to the extent allowed by all laws and institutional policies regarding confidentiality of patient clinical information. Datasets used in the study: Peditools; string db; maxikraken2 (v_1903_140GB) database; gnomAD v2.1.1; and 1000genome.