Abstract
Introduction:
Essential tremor (ET) is one of the most common movement disorders. Despite its high prevalence and heritability, its genetic aetiology remains elusive with only a few susceptibility genes identified and poorly replicated. Our aim was to find novel candidate genes involved in ET predisposition through whole exome sequencing.
Methods:
We studied eight multigenerational families (N=40 individuals) with an autosomaldominant inheritance using a comprehensive strategy combining whole exome sequencing, followed by case-control association testing of prioritized variants in a separate cohort comprising 521 ET cases and 596 controls. We further performed gene-based burden analyses in an additional dataset comprising 789 ET patients and 770 healthy individuals to investigate whether there was an enrichment of rare deleterious variants within our candidate genes.
Results:
Fifteen variants co-segregated with disease status in at least one of the families, among which rs749875462 in CCDC183, rs535864157 in MMP10 and rs114285050 in GPR151 showed a nominal association with ET. However, we found no significant enrichment of rare variants within these genes in cases compared with controls. Interestingly, MMP10 protein is involved in the inflammatory response to neuronal damage and has been previously associated with other neurological disorders.
Conclusions:
We prioritized a set of promising genes, especially MMP10, for further genetic and functional studies in ET. Our study suggests that rare deleterious coding variants that markedly increase susceptibility to ET are likely to be found in many genes. Future studies are needed to replicate and further infer biological mechanisms and potential disease causality for our identified genes.
Keywords: essential tremor, genetic risk, WES, rare variants, MMP10
1. Introduction
Essential tremor (ET) is one of the most frequent movement disorders with an estimated prevalence of 0.9% in the general population and up to 4.6% in individuals at age 65 or older [1]. ET subjects show action tremors affecting primarily the upper limbs, although tremors can be present in other body regions. Owing to its clinical heterogeneity, ET is now considered a syndrome that may have multiple genetic and environmental etiologic factors [2].
Many studies have reported a high frequency of familial aggregation of tremor among patients with ET [3], suggesting a high heritability for this disorder [4,5]. Historically, most studies have assumed a Mendelian pattern of inheritance; however, it remains unclear whether such strong familial aggregation is either due to a single highly penetrant rare variant, many low penetrant common variants, or a combination of both.
Despite its high prevalence and frequent familial aggregation, the genetic basis of ET remains poorly understood with only a handful of susceptibility genes identified and not consistently replicated (Table 1). The first studies pointed out three distinct susceptibility loci under autosomal dominant (AD) inheritance models [6–8], in which no candidate gene has been found yet. Under the ‘common disease-common variant’ hypothesis, one could postulate that the genetic component of ET is likely due to a large number of common low-risk alleles [9]. However, genome-wide association studies (GWAS) have only reported five common, low-penetrance genetic variants associated with risk for sporadic ET with low disease odds ratios [10–12]. As in other complex diseases, the identified GWAS hits do not account for the total ET genetic variance [13], suggesting that rare penetrant variants with larger effects may also play a substantial role in the disease etiology, particularly in familial cases [14]. Indeed, whole exome sequencing (WES) has proven to be fruitful in identifying putative causal genes in ET families – such as FUS [15], HTRA2 [16], SORT1 [17], TENM4 [18], SCN4 [19], NOS3, KCNS2, HAPLN4, and USP46 [20] (Table 1) – which have provided clues to disease pathogenesis. For instance, targeted expression of FUS p.Gln290* in both dopaminergic and serotonergic neurons in transgenic Drosophila resulted in motor dysfunction together with impairment in the GABAergic pathway [21], which has been extensively linked to ET pathogenesis [22]. Similarly, Tenm4 knockout mice model showed severe action tremor phenotype and hypomyelination of small-diameter axons in the central nervous system [23]. However, further genetic evidence is needed to consider these genes as causative for ET. We hypothesized that a family-based exome sequencing study would help to identify rare genetic determinants for hereditary ET.
Table 1.
Summary of genes and loci that have been previously associated with essential tremor.
| Genes or loci (chr position) | Ethnic or geographic distribution | Study sample size (cases/controls) | Methodology used | References |
|---|---|---|---|---|
| ETM1 (3q13) | Icelandic | 16 families | Genome-wide scan | Gulcher et al., 1997 [6] |
| ETM2 (2p24.1) | American of Czech ancestry | 1 family | Genome-wide scan | Higgins et al., 1997 [7] |
| ETM3 (6p23) | North American | 7 families | Genome-wide scan | Shatunov et al., 2006 [8] |
| HS1-BP3 | North American | 2 families | Fine mapping studies | Higgins et al., 2005 [*] |
| DRD3 | French | 30 families | Candidate-gene approach | Lucotte et al., 2006 [†] |
| LINGO1 | Icelandic, European and American | 752/15,797 | GWAS | Stefansson et al., 2009 [10] |
| SLC1A2 | European | 658/1,490 | GWAS | Thier et al., 2012 [11] |
| FUS | French-Canadian | 1 family and 270 cases | Exome sequencing | Merner et al., 2012 [15] |
| HTRA2 | Turkish | 1 family | Exome sequencing | Unal Gulsuner et al., 2014 [16] |
| SORT1 | Spanish | 1 family and 151 cases | Exome sequencing | Sanchez et al., 2015 [17] |
| TENM4 | Spanish | 1 family and 299 familial cases | Exome sequencing and targeted resequencing | Hor et al., 2015 [18] |
| SCN4A | Spanish | 1 family, 76 sporadic and 47 familial cases | Exome sequencing | Bergareche et al., 2015 [19] |
| NOS3, KCNS2, HAPLN4, USP46 | North American | 37 families | Exome sequencing | Liu et al., 2016 [20] |
| STK32B, PPARGC1A, CTNNA3 | Northern European and North American | 2,807/6,441 | GWAS | Müller et al., 2016 [12] |
Key: chr, chromosomal; GWAS, Genome-wide association study.
J.J. Higgins, R.Q. Lombardi, J. Pucilowska, J. Jankovic, E.K. Tan, J.P. Rooney, A variant in the HS1-BP3 gene is associated with familial essential tremor, Neurology. 64 (2005) 417–421.
G. Lucotte, J.P. Lagarde, B. Funalot, P. Sokoloff, Linkage with the Ser9Gly DRD3 polymorphism in essential tremor families, Clin. Genet. 69 (2006) 437–440.
We performed WES in 8 families (n=40 individuals) with an AD inherited form of ET, followed by a targeted association testing of prioritized variants, in an attempt to identify novel candidate genes involved in ET.
2. Materials and methods
2.1. Study participants
Eight families of Spanish origin with strong ET aggregation, defined as having at least three affected relatives, were included in the study. Families were selected based on the following criteria: (1) the proband had a diagnosis of definite ET; (2) there were at least two first-degree affected relatives in the family in addition to the proband; and (3) the families had affected members in different generations. The final sample selected for WES included a total of 40 individuals, comprising 29 affected, 9 unaffected relatives, and two individuals with unclear ET diagnosis. The pedigrees for the eight families, indicating the family members that underwent WES, are detailed in Figure 1.
Figure 1. Pedigrees for the families that underwent WES, asterisk indicates the family members sequenced.
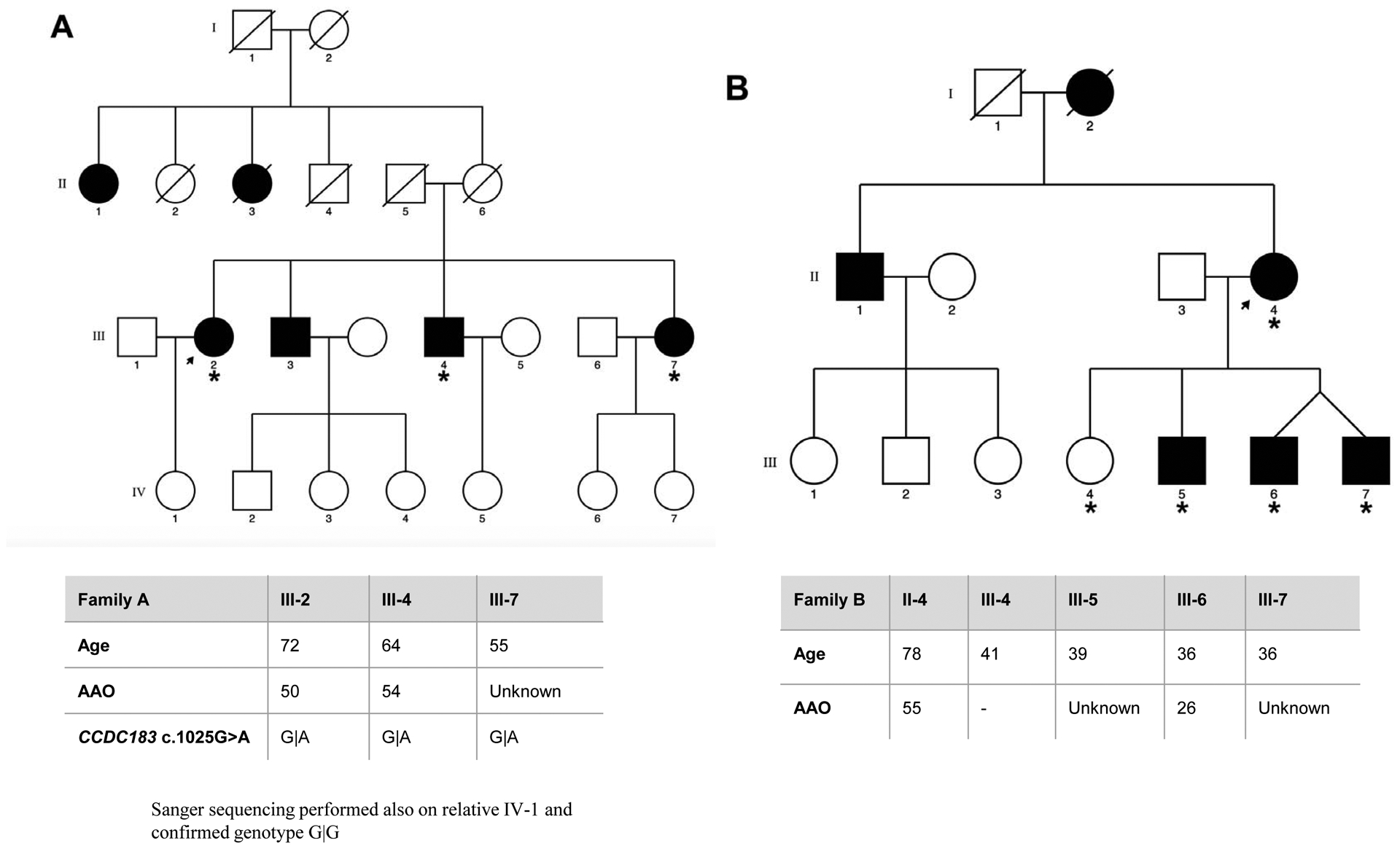
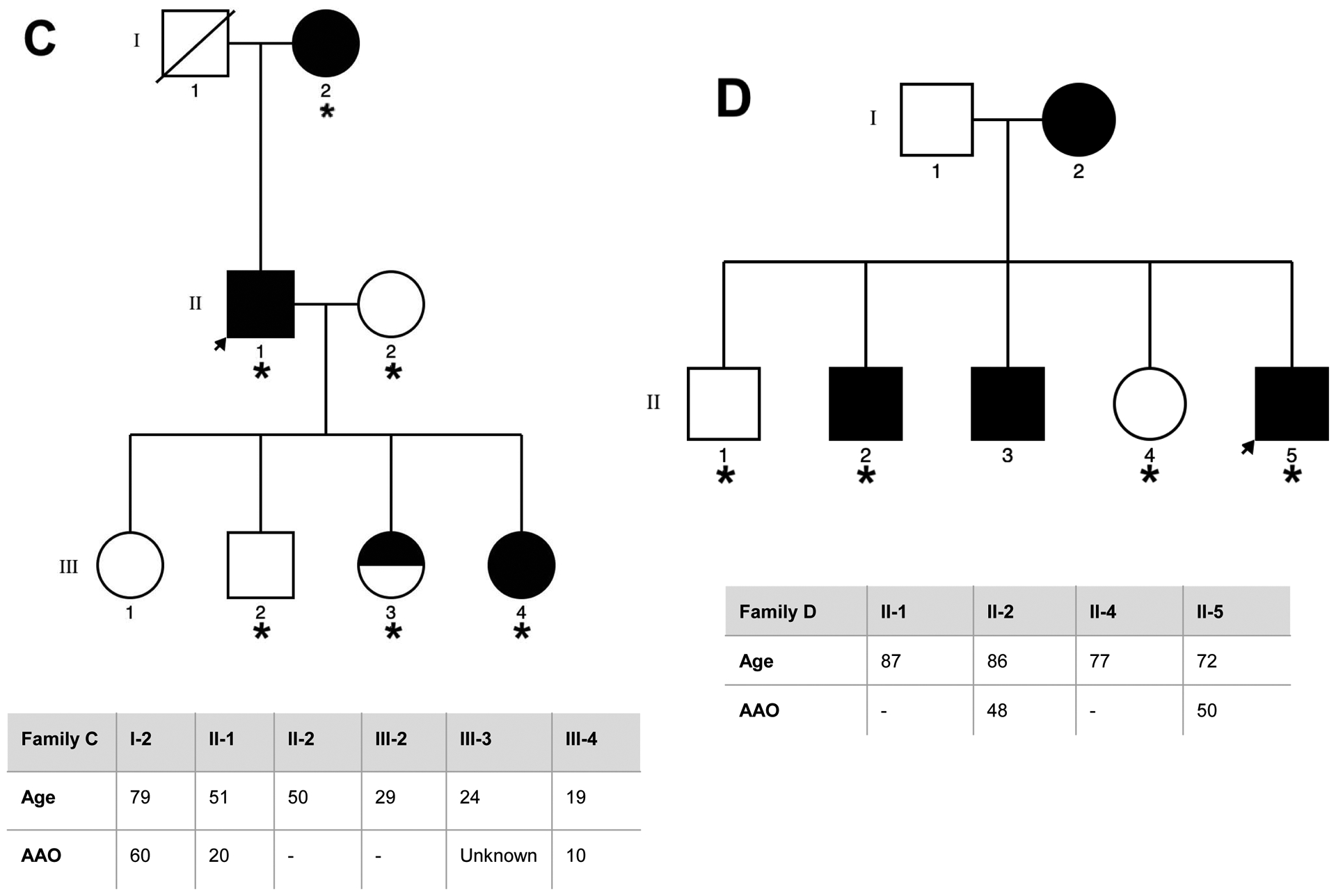
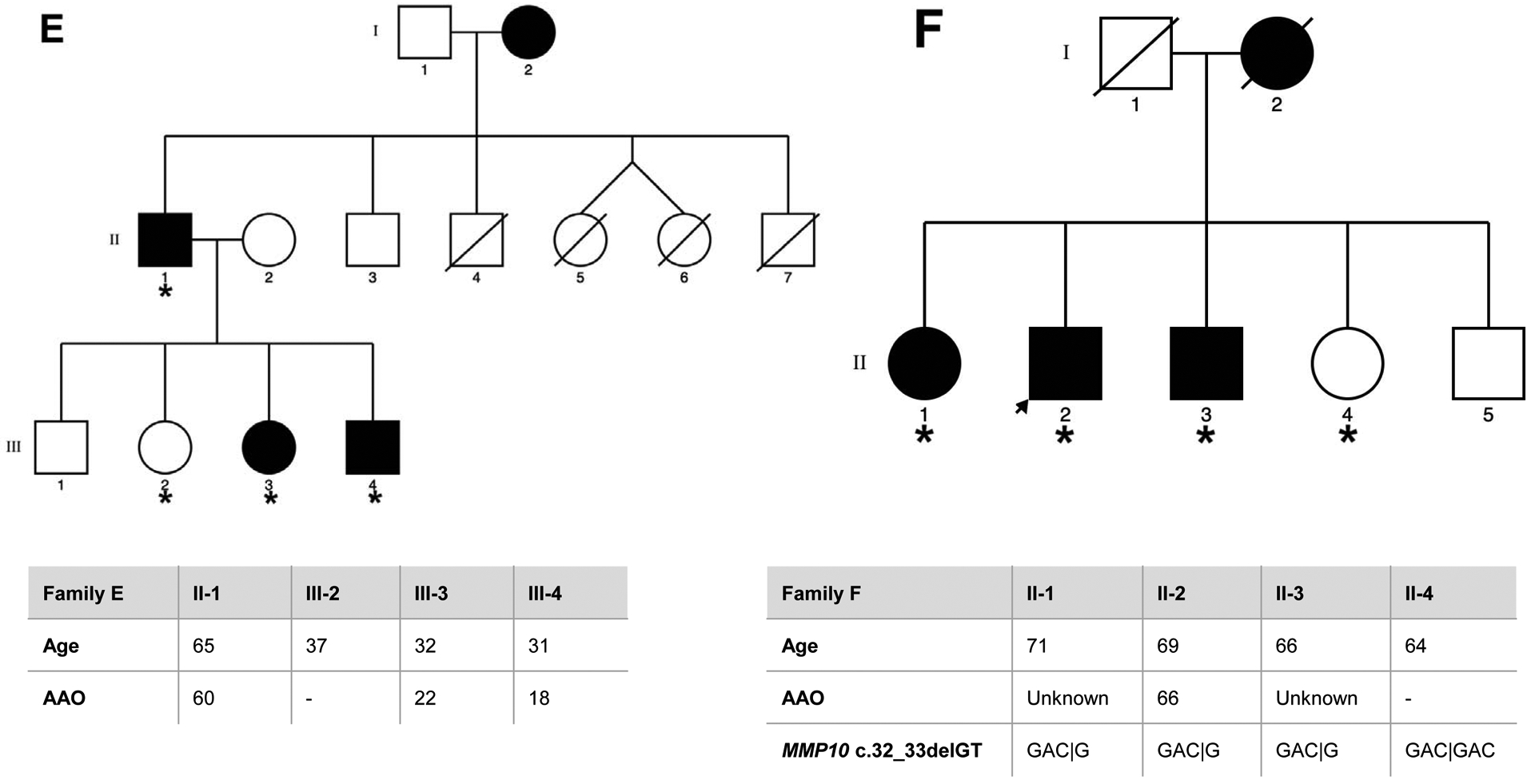
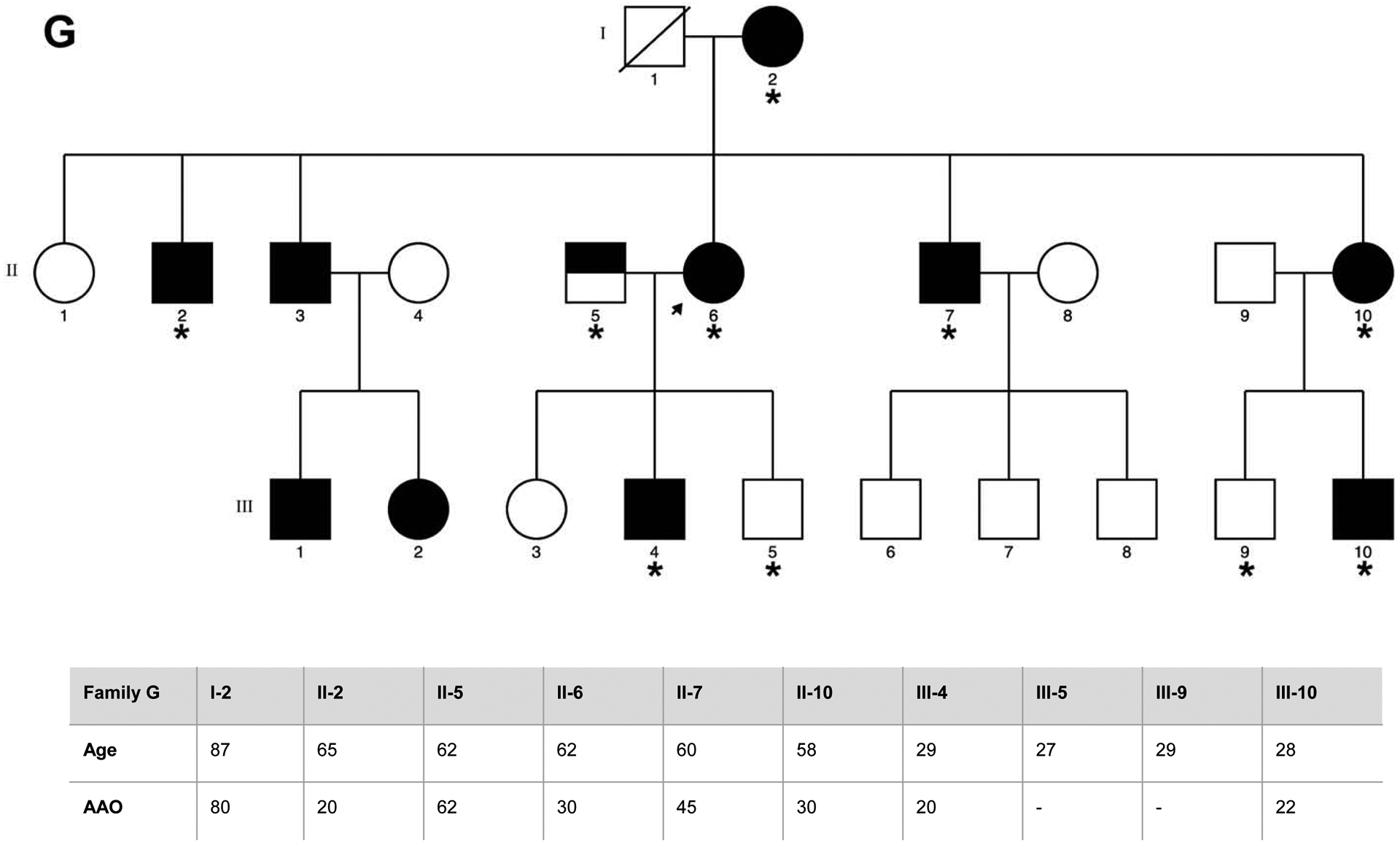
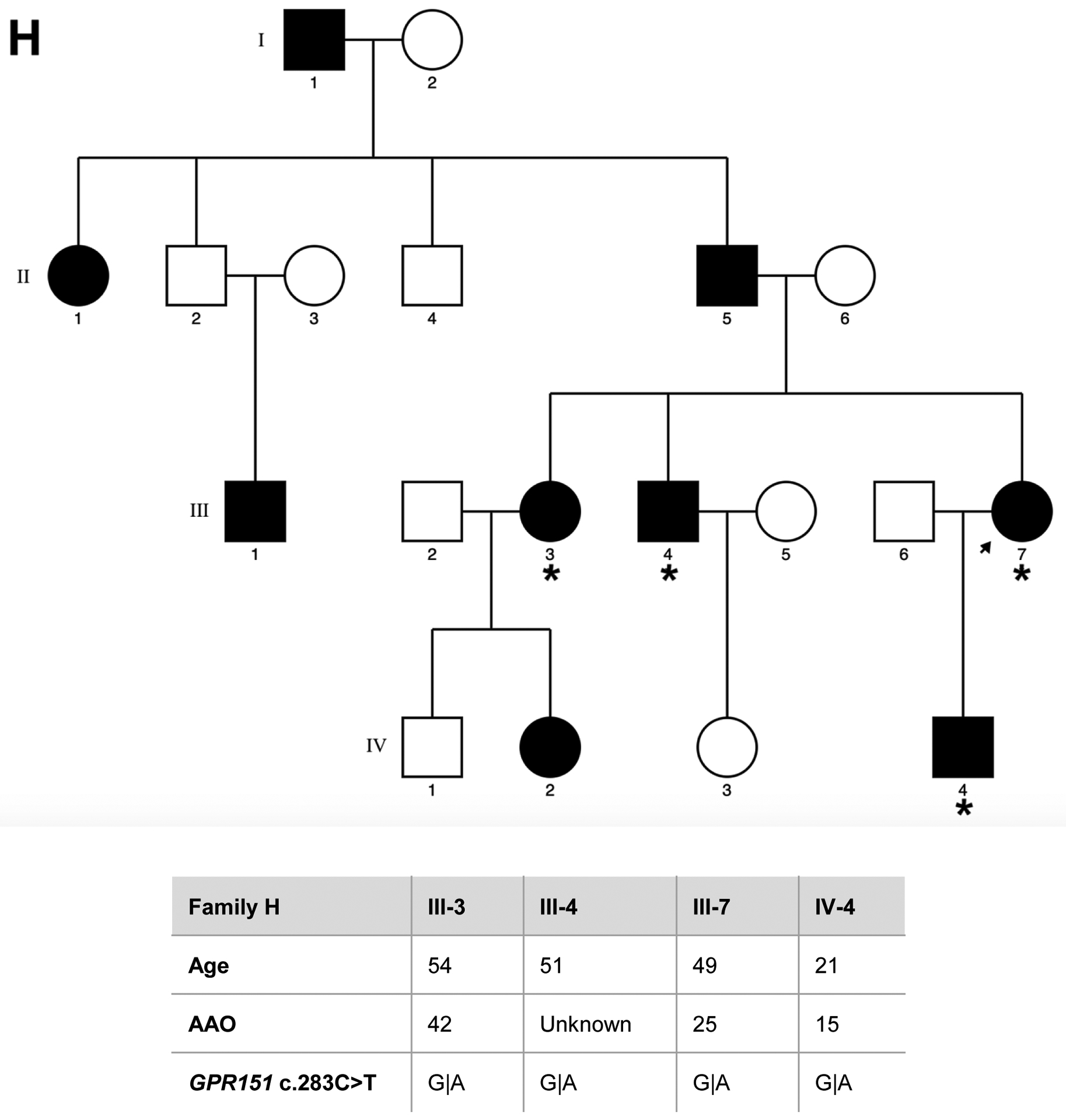
Genetic pedigrees plots of the eight families (A-H) included in the study are shown. The generation in each pedigree is indicated by roman numerals. The proband is indicated by an arrow. Red asterisk indicates those subjects that were exome sequenced (N=40). Below each pedigree, age at enrollment, age at tremor onset (for cases, if known), tremor region and severity are shown in a table. Tremor region (localization of the tremor): Head (H), Tongue (T), Chin (C), Upper limbs (UL), Lower limbs (LL). Tremor severity corresponds to scores in the Part A of the more affected body region according to the Fahn-Tolosa-Marin Clinical Rating Scale for Tremor (FTM) [41]. Shading legend: Symbols completely black indicate individuals with clinical definite ET diagnosis; Symbols half vertical black fill indicate individuals with unclear ET diagnosis; Symbols completely blank indicate unaffected individuals; Symbols with a diagonal line indicated deceased relatives. Segregation within families for the three candidate variants (rs749875462 in CCDC183, rs535864157 in MMP10 and rs114285050) is also shown in families A, F and H.
All participants were examined by at least one movement disorders specialist. Neurological exam included the assessment of rest (assessed with the hands lying on the lap), postural (assessed with the arms extended), and kinetic tremor (assessed with the finger to nose & finger to finger tests), and presence/absence of rigidity and bradykinesia. ET diagnosis was made according to the TRIG classification of essential tremor not following the inclusion criterion “duration of tremor more than 5 years” [24]. Other entities such as dystonia, Parkinson’s disease (PD) and drug-induced tremor were specifically excluded. Enhanced physiological tremor was excluded according to the expert guidelines [25]. All subjects fulfilled the criteria for definite ET, except for individuals II:1 and II.2 from family F who were labeled as having possible ET, since they showed mild parkinsonian signs. Further clinical and demographic details are included in Figure 1. Two of the families included (D and F) had been previously reported (as family 11 and 4 respectively) elsewhere [26]. The study protocol was approved by the Ethics Committee of University Hospital Mutua de Terrassa and University of Navarra as well as the local ethics committees at the different centers. Written informed consent was obtained from all enrollees.
2.2. Whole exome sequencing, variant calling and annotation
Genomic DNA from all individuals was isolated from whole blood using standard procedures. Whole exome sequencing for the 40 individuals was performed at the University of Washington Center for Mendelian Genomics (Seattle, WA, USA). In brief, exomes were captured using the Nimblegen SeqCap EZ Human Exome Library v2.0 (~44 Mb target) (Roche NimbleGen, Inc., Madison, WI, USA). The captured DNA was sequenced with 75bp paired-end reads on an Illumina HiSeq 4000 sequencing platform according to manufacturer’s instructions (Illumina, Inc., San Diego, CA, USA). Exome-captured sequencing reads were aligned to the human reference genome assembly GRCh37 using BWA-MEM (v.0.7.12). PCR duplicates were removed using Picard-tools v1.137 (http://picard.sourceforge.net) and local realignment and base quality score recalibration using Genome Analysis Toolkit (GATK) v3.4–46. Finally, single-nucleotide variants (SNVs) and insertions/deletions (indels) were called using GATK HaplotypeCaller. Additional hard filters applied at variant, genotype and individual levels are detailed in Supplementary Material.
Variants were annotated using hg19, SnpEff and SnpSift. Functional impact was calculated according to Combined Annotation Dependent Depletion (CADD); pathogenicity predictions were calculated according to MutationAssessor, MutationTaster, Polyphen-2 and SIFT algorithms; and population allele frequencies were based on non-Finnish European (NFE) exome samples from the Genome Aggregation Database (gnomAD v.2.1.1) (Supplementary Material).
2.3. Segregation analysis and variant prioritization
Annotated variants were assessed for segregation within the families. Using an AD model of inheritance, only perfectly-segregating variants (present in affected individuals and absent in unaffected relatives) within each family were selected. In addition, only coding, non-synonymous variants with a minor allele frequency (MAF) below 1% and with predicted high impact on protein function, were selected for further analyses. A flow chart of our multi-step filtering strategy is shown in Supplementary Figure 1 and detailed in Supplementary Material.
We obtained a list of “candidate” variants that were exclusively shared by affected ET individuals within a family (Supplementary Table 2). These variants were visually inspected through the Integrative Genomics Viewer and discarded when any sequencing artifact due to strand bias was detected. All variants of interest were confirmed by Sanger sequencing when DNA was available and selected for further analyses (Supplementary Material).
In addition, we checked for presence and segregation of rare variants in the genes previously reported in association with ET in both Mendelian forms of disease and GWAS (Table 1). No rare exonic deleterious variants in ET related genes were found to cosegregate with disease within the families.
2.4. Genetic association analysis with ET status
Next, we performed targeted genotyping for the candidate variants and case-control association testing in a Spanish cohort (Dataset 1) comprising 521 unrelated ET cases and 596 age-matched healthy individuals (Supplementary Materials, Supplementary Table 1). Follow-up genotyping was performed using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry with Sequenom iPLEX Gold custom assays designed using MassARRAY assay design software (Sequenom, San Diego, CA, USA). For variants that could not be genotyped by Sequenom due to specific biochemistry of DNA sequence boundaries where the variant is located, we used the microfluidic-based BioMark™ HD array platform (Fluidigm Corporation, CA, USA). Details of the genotyping procedures are provided in Supplementary Material. After genotyping, we performed an allelic Fisher’s Exact Test in R (v1.2). All reported p-values are two-tailed and the significant threshold was set after Bonferroni correction.
2.5. Gene burden analysis
In order to investigate whether there was an enrichment of rare deleterious variants within our candidate genes in cases compared to controls, we performed gene-based burden analyses for disease trait in a separate cohort (Dataset 2) comprising 789 ET cases and 770 controls of European and North American ancestry (Supplementary Materials, Supplementary Table 1). Molecular inversion probes (MIPs) were used to capture all exons and exon/intron boundaries (5bp flanking) of the genes harboring variants nominally-associated in the case-control association analysis or seen in cases but absent in controls. MIPs were designed, targeted and amplified as previously described [27]. Briefly, the MIPs library was sequenced using Illumina HiSeq 4000 platform at the McGill University and Genome Québec Innovation Centre. Sequencing processing was done using BWA-MEM, Varscan (v2.3.9; samtools v1.9) for variant calling, and ANNOVAR for annotation (Supplementary Material). Quality control was performed as described in Supplementary Material. We then performed fixed threshold aggregation methods (BurdenBT tests) including only exonic, non-synonymous variants with a MAF < 1% as registered in gnomAD using Variant Association Tools (http://varianttools.sourceforge.net/Association/Aggregation).
3. Results
Basic demographic characteristics of the family members that were exome sequenced are shown in Supplementary Table 1. The average age at onset of tremor in ET patients was 38.8 years (±19.5) and average duration of disease was 16.6 years (±11.8). The average age for healthy relatives was 49 years (±22.2). The number of affected individuals enrolled per family ranged from two to seven (mean=3.6).
All 40 samples sequenced passed quality control filters and the total number of SNVs/indels numbered 177,970. After annotating for functional impact, pathogenicity and allele frequencies, we assessed variants for segregation in ET pedigrees. Using an AD model of inheritance, 47,286 variants in 29,089 genes segregated with ET affection status in at least one pedigree, of which 10,726 (22.7%) were seen in more than one family. Following our multi-step filtering strategy, we identified a total of 15 rare variants predicted to affect protein function that segregated perfectly with the disease in seven out of the eight families (Supplementary Table 2). Among these variants, 13 were private (only seen in one family) and two were present in two different families (A and H), and included five stopgained SNVs, three insertions and seven deletions. Twelve variants (80%) were predicted as having a high functional impact (CADD score > 20) and five (33.3%) as being disease causing. A total of five genes contained segregating novel variants.
We then performed single-variant association analysis using an allelic Fisher’s Exact Test. Of note, one variant (rs11288615 in OR5K4) could not be genotyped due to DNA sequence boundaries, being excluded from the analysis. We found no significant results after Bonferroni correction (significance threshold = 0.0036). However, we identified three variants nominally associated (p-value < 0.05) with ET, which included rs749875462 in CCDC183 (p-value = 0.0087), rs535864157 in MMP10 (p-value= 0.0220) and rs114285050 in GPR151 (p-value = 0.0425) (Table 2). Family A. The variant identified in CCDC183 gene (c.1025G>A; rs749875462) results in a nonsense amino-acid substitution (p.Trp342*) predicted as disease-causing. Sanger sequencing in an additional unaffected relative (IV1) confirmed that the variant was only present in affected individuals within the family. We also found this variant in heterozygous state in 10 independent ET cases and in homozygous state in two ET patients, and in heterozygous state in four controls (Table 2). Although the CCDC183 variant identified is at a higher frequency in the African population (0.07%), our ancestry analysis showed no African admixture (Supplementary Figure 1). Family F. We found a possible pathogenic frameshift variant (c.32_33delGT; rs535864157) in MMP10 present in all three affected individuals but absent in one healthy relative. This variant was also present in heterozygous state in additional 5 ET patients but absent in 596 controls (Table 2). The MMP10 rs535864157 variant is situated in the N-terminal signal peptide of the protein, which directs the protein to the secretory pathway and therefore it may impact in its release into the extracellular space. Family H. A SNV (c.283C>T; rs114285050) in GPR151 was found in all four affected individuals and resulted in a nonsense amino-acid substitution (p.Arg95*) predicted as disease-causing. In addition, we found this variant in heterozygous state in 10 unrelated ET cases and 6 controls, and in homozygous state in 2 ET patients (Table 2). The rs114285050 is located in the transmembrane domain, one of the most conserved parts of the protein. G-protein coupled receptors have seven hydrophobic regions, each of which most probably spans the membrane. The N-terminus is located on the extracellular side of the membrane and is often glycosylated, while the C-terminus is cytoplasmic and generally phosphorylated. Three extracellular loops alternate with three intracellular loops to link the seven transmembrane regions.
Table 2.
Single-variant case-control association analysis in 521 ET cases and 596 healthy individuals.
| Variant (HGVS) | Gene | rs ID | Reference allele | Alternate allele | ET [n = 521] (AF) | Controls [n = 596] (AF) | P-value* (OR) [95% CI] |
|---|---|---|---|---|---|---|---|
| NM_001145710.1: c.906delC | FAM228B | Novel | C | DEL | 35 (0.034) | 26 (0.022) | 0.0922 (1.56) [0.91–2.72] |
| NM_003241.3: c.806G>A | TGM4 | rs139860990 | G | A | 18 (0.017) | 16 (0.013) | 0.4918 (1.29) [0.62–2.72] |
| NM_001005516.1: c.904delA | OR5K3 | rs79045298 | A | DEL | 0 (0) | 0 (0) | 1 (nan) |
| NM_183375.2: c.127_131delGTCAG | PRSS48 | Novel | GTCAG | DEL | 0 (0) | 0 (0) | 1 (nan) |
| NM_194251.2: c.283C>T | GPR151 | rs114285050 | G | A | 14 (0.013) | 6 (0.005) | 0.0425 (2.69) [0.97–8.58] |
| NM_015465.4: c.2519_2522delTCAA | GEMIN5 | rs769616395 | TTGA | DEL | 2 (0.002) | 0 (0) | 0.2174 (nan) |
| NM_004999.3: c.2751insA | MYO6 | rs551348450 | DEL | A | 0 (0) | 0 (0) | 1 (nan) |
| NM_001039374.4: c.1025G>A | CCDC183 | rs749875462 | G | A | 14 (0.013) | 4 (0.003) | 0.0087 (4.04) [1.26–16.91] |
| NM_002425.2: c.32_33delGT | MMP10 | rs535864157 | AC | DEL | 5 (0.005) | 0 (0) | 0.0220 (nan) |
| NM_001261828.1: c.167_168insTT | MS4A14 | Novel | DEL | TT | 2 (0.002) | 0 (0) | 0.2174 (nan) |
| NM_144666.2: c.2839G>T | DNHD1 | rs180918289 | G | T | 27 (0.026) | 47 (0.039) | 0.0768 (0.65) [0.39–1.07] |
| NM_001005182.1: c.24delA | OR6C1 | Novel | A | DEL | 0 (0) | 0 (0) | 1 (nan) |
| NM_181536.1: c.3691_3694delCAAA | PKD1L3 | Novel | TTTG | DEL | 0 (0) | 0 (0) | 1 (nan) |
| NM_032423.2: c.1282C>T | ZNF528 | rs150257846 | C | T | 2 (0.002) | 2 (0.002) | 1 (1.14) [0.08–15.81] |
Key: AF, allele frequency; CI, confidence interval; DEL, deletion; HGVS, Human Genome Variation Society; nan, not a number/not available; OR, odds ratio.
P-values derived from Fisher’s Exact test. All p-values were two-tailed and significant threshold was set at 0.0036 after Bonferroni correction (0.05/14 variants). Nominal associations (p < 0.05) are highlighted in bold.
CCDC183, GPR151 and MMP10 genes have observed/expected (o/e) scores of 0.66 (90%CI 0.460.98), 0.93 (90%CI 0.6–1.5) and 0.99 (90%CI 0.71–1.41), indicating that they are highly tolerant to loss-of-function (LOF) variants, which are unlikely to be pathogenic (Supplementary Materials). We also used GeneMANIA software (https://genemania.org/) to identify how these genes may impact tremor circuits leading to tremor but found no potential interactions with proteins involved in ET brain circuits such as GABAergic associated proteins, voltage-gated calcium channels or LINGO1 [28].
Next, we performed gene-based analyses. MIPs sequencing and subsequent variant calling identified 184 SNVs within GPR151, CCDC183, MMP10, GEMIN5, and MS4A14 genes. After filtering for only exonic, non-synonymous variants with a MAF < 1%, 88 rare protein-altering variants remained and were included in gene-based burden analyses (Supplementary Materials). We did not find a significant enrichment of rare deleterious variants within the candidate genes in cases compared to controls (Table 3). However, we found additional rare and potential deleterious variants in those genes in other unrelated ET patients (Supplementary Table 7).
Table 3.
Gene-based burden association tests of candidate genes in 789 ET cases and 770 healthy individuals.
| Gene name | Sample size | N of variants* | Total MAC† | Beta | P-value | Wald statistic |
|---|---|---|---|---|---|---|
| CCDC183 | 1559 | 15 | 48 | −0.2842 | 0.8599 | −1.0798 |
| MMP10 | 1559 | 13 | 37 | 0.1209 | 0.3589 | 0.3615 |
| GPR151 | 1559 | 13 | 19 | −0.1075 | 0.5964 | −0.2442 |
| MS4A14 | 1559 | 16 | 43 | 0.2237 | 0.2317 | 0.7332 |
| GEMIN5 | 1559 | 31 | 138 | −0.2798 | 0.9512 | −1.6564 |
Key: ET, Essential tremor; MAC, minor allele count; N, number.
Only exonic deleterious variants (missense, non-sense SNVs) with a minor allele frequency below 1% in gnomAD non-Finnish Europeans exomes were used for the analysis.
Bonferroni corrected p-value threshold for MAF<1% is 0.01.
The number of variants in each group is adjusted for specified MAF; in this case, below 0.01.
The total minor allele count (MAC) in a group adjusted for mode of inheritance.
4. Discussion
In this study, we sought to identify novel genetic candidates for ET using a comprehensive strategy combining WES, followed by case-control association testing and gene burden analyses. Our stringent filtering approach for only protein-altering variants and subsequent targeted genotyping resulted in a small number of candidate genes nominally-associated with ET risk, which include CCDC183 (family A), MMP10 (family F) and GPR151 (family H). The absence of genes with rare functional variants segregating in more than two families evidences the genetic heterogeneity of ET.
It is of special interest the putative association between MMP10 and ET. MMP10, also called stromelysin-2, belongs to the matrix metalloproteinases (MMPs) family, which are zinc-dependent endoproteases involved in tissue remodeling and degradation of components of the extracellular matrix. MMP10 is also known to play an important role in cell proliferation, migration, differentiation and angiogenesis, as well as vascular integrity [29]. MMP10 is induced by inflammation, known to be highly expressed by activated microglia, and may be involved in the inflammatory response to neuronal damage [30]. Interestingly, some studies have suggested a potential involvement of MMP10 in Alzheimer’s disease pathophysiology. Higher MMP10 levels are found in cerebrospinal fluid (CSF) of Alzheimer’s disease patients compared to both controls and patients with vascular dementia, which indicates a brain-specificity of this protein. MMP10 expression was also increased in neurons of the ischemic brain but not in healthy areas in humans [31] and mice [32], suggesting that its expression is linked to brain injury and to the subsequent inflammatory response. CSF MMP10 levels correlate with both CSF total tau and phosphorylated tau [33] and with cognitive performance [34]. Similarly, a recent study found a positive correlation of CSF MMP10 levels with PD progression [35]. Also, MMP10 is the only metalloproteinase that directly cleaves the huntingtin protein and specific MMP10 inhibitors may have a therapeutic benefit in Huntington’s disease (HD), thus pointing MMP10 as a potential genetic modifier for HD [36]. However, in order to increase the evidence supporting this gene as a causative gene for ET, further replication and functional studies are needed.
We are aware that our association and gene-based analyses are likely underpowered, as rare variant analyses require thousands of individuals to reach acceptable statistical power [37]. A reasonable replication strategy is to select a few genes based on strength of association and perform sequencing and rare-variant associations on new samples, using multiple-test correction threshold applied only to the smaller set of candidate genes [37], as we did in this study. However, we did not identify new genes consistently significant across all of our analyses. Recent sequencing of thousands of exomes from across the world has revealed that the vast majority of genetic variation is rare and highly population-specific [14,38], meaning that our lack of replication might also be explained by genetic differences across populations. In addition, familial clustering in some ET cases may not be due to a single highly-penetrant variant with large effects but a high genetic load of common risk variants. It would have been very useful to perform complementary mapping gene strategies such as linkage analysis – especially in pedigrees G and H – but for most individuals there was not enough DNA available.
Our study has several limitations. First, we did not account for the potential contribution of common variants to the etiology of ET; although it could be argued that common variants may have a reduced impact in multiplex families with strong aggregation of a disease. Second, WES does not cover noncoding or structural variants, which are believed to play a major role in complex trait genetics despite having small effect sizes. Indeed, structural variants have been recently reported to be associated with familial ET [39]. Future long-sequencing analyses might unravel disease-causing structural variants associated with disease, and whole genome sequencing (WGS) may help reveal intronic variants with potential regulatory effects. Indeed, WGS recently identified candidate genes for ET in a subset of families in which WES analysis previously failed to identify the causative variant due to incomplete coverage of the entire coding region of the genome [40]. Third, the power of segregation analysis of individual rare variants in small families is limited and presence of phenocopies within a family could have an impact on the interpretation of apparently non-segregating but pathogenic variants. We cannot exclude the contribution of pathogenic variants found that partially segregate with the disease and that were not further studied. Fourth, some unaffected subjects within the families were young and we could not rule out the possibility that they would develop the disease in the future. Due to the high clinical heterogeneity in ET, we are aware that collecting more samples from additional affected and unaffected relatives would have been very helpful to confirm the causative variants. We argue that more detailed phenotypic data could also help to identify subjects who may have varying amounts of genetic risk.
In summary, here we studied a unique cohort of pedigrees with strong disease aggregation to identify rare variants of high penetrance contributing to ET susceptibility. Our analysis prioritized a set of promising genes, especially MMP10, for further genetic and functional studies in ET, but also suggests that rare deleterious coding variants that markedly increase susceptibility to ET are likely to be found in many genes with low effect size. Future genetic studies including a more detailed clinical characterization of ET patients and larger scale GWAS are needed to better understand the underlying etiology of such a common disease.
Supplementary Material
Highlights.
The genetic aetiology of essential tremor remains elusive.
Whole exome sequencing was performed in 8 multigenerational Spanish ET families.
CCDC183, MMP10 and GPR151 genes were nominally associated with ET.
MMP10 has been associated with other neurological disorders such as AD and PD.
ET is likely to be caused by variants in different genes with small effect sizes.
Acknowledgments
The authors thank the subjects and their families whose help and participation made this work possible. This study was funded by the Spanish Ministry of Science and Innovation to P.P. [SAF201347939-R (2013-2018)]. M.F. was funded by María de Maeztu programme (grant # MDM-2017-0729). Sequencing was provided by the University of Washington Center for Mendelian Genomics (UWCMG) and was funded by NHGRI and NHLBI grants UM1 HG006493 and U24 HG008956. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors thank the technical and human support provided by the Sequencing and Genotyping Unit of Genomics Facility (SGIker) of UPV/EHU and European funding (ERDF and ESF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors declare that they have no conflict of interest.
References
- [1].Louis ED, Ferreira JJ, How common is the most common adult movement disorder? Update on the worldwide prevalence of essential tremor, Movement Disorders. 25 (2010) 534–541. 10.1002/mds.22838. [DOI] [PubMed] [Google Scholar]
- [2].Bhatia KP, Bain P, Bajaj N, Elble RJ, Hallett M, Louis ED, Raethjen J, Stamelou M, Testa CM, Deuschl G, Tremor Task Force of the International Parkinson and Movement Disorder Society, Consensus Statement on the classification of tremors. from the task force on tremor of the International Parkinson and Movement Disorder Society, Mov. Disord 33 (2018) 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Louis ED, Clark L, Ottman R, Familial Aggregation and Co-Aggregation of Essential Tremor and Parkinson’s Disease, Neuroepidemiology. 46 (2016) 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tanner CM, Goldman SM, Lyons KE, Aston DA, Tetrud JW, Welsh MD, Langston JW, Koller WC, Essential tremor in twins: an assessment of genetic vs environmental determinants of etiology, Neurology. 57 (2001) 1389–1391. [DOI] [PubMed] [Google Scholar]
- [5].Lorenz D, Frederiksen H, Moises H, Kopper F, Deuschl G, Christensen K, High concordance for essential tremor in monozygotic twins of old age, Neurology. 62 (2004) 208–211. [DOI] [PubMed] [Google Scholar]
- [6].Gulcher JR, Jónsson P, Kong A, Kristjánsson K, Frigge ML, Kárason A, Einarsdóttir IE, Stefánsson H, Einarsdóttir AS, Sigurthoardóttir S, Baldursson S, Björnsdóttir S, Hrafnkelsdóttir SM, Jakobsson F, Benedickz J, Stefánsson K, Mapping of a familial essential tremor gene, FET1, to chromosome 3q13, Nat. Genet 17 (1997) 84–87. [DOI] [PubMed] [Google Scholar]
- [7].Higgins JJ, Pho LT, Nee LE, A gene (ETM) for essential tremor maps to chromosome 2p22p25, Mov. Disord 12 (1997) 859–864. [DOI] [PubMed] [Google Scholar]
- [8].Shatunov A, Sambuughin N, Jankovic J, Elble R, Lee HS, Singleton AB, Dagvadorj A, Ji J, Zhang Y, Kimonis VE, Hardy J, Hallett M, Goldfarb LG, Genomewide scans in North American families reveal genetic linkage of essential tremor to a region on chromosome 6p23, Brain. 129 (2006) 2318–2331. [DOI] [PubMed] [Google Scholar]
- [9].Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN, Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease, Nat. Genet 33 (2003) 177–182. [DOI] [PubMed] [Google Scholar]
- [10].Stefansson H, Steinberg S, Petursson H, Gustafsson O, Gudjonsdottir IH, Jonsdottir GA, Palsson ST, Jonsson T, Saemundsdottir J, Bjornsdottir G, Böttcher Y, Thorlacius T, Haubenberger D, Zimprich A, Auff E, Hotzy C, Testa CM, Miyatake LA, Rosen AR, Kristleifsson K, Rye D, Asmus F, Schöls L, Dichgans M, Jakobsson F, Benedikz J, Thorsteinsdottir U, Gulcher J, Kong A, Stefansson K, Variant in the sequence of the LINGO1 gene confers risk of essential tremor, Nat. Genet 41 (2009) 277–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Thier S, Lorenz D, Nothnagel M, Poremba C, Papengut F, Appenzeller S, Paschen S, Hofschulte F, Hussl A-C, Hering S, Poewe W, Asmus F, Gasser T, Schöls L, Christensen K, Nebel A, Schreiber S, Klebe S, Deuschl G, Kuhlenbäumer G, Polymorphisms in the glial glutamate transporter SLC1A2 are associated with essential tremor, Neurology. 79 (2012) 243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Müller SH, Girard SL, Hopfner F, Merner ND, Bourassa CV, Lorenz D, Clark LN, Tittmann L, Soto-Ortolaza AI, Klebe S, Hallett M, Schneider SA, Hodgkinson CA, Lieb W, Wszolek ZK, Pendziwiat M, Lorenzo-Betancor O, Poewe W, Ortega-Cubero S, Seppi K, Rajput A, Hussl A, Rajput AH, Berg D, Dion PA, Wurster I, Shulman JM, Srulijes K, Haubenberger D, Pastor P, Vilariño-Güell C, Postuma RB, Bernard G, Ladwig K-H, Dupré N, Jankovic J, Strauch K, Panisset M, Winkelmann J, Testa CM, Reischl E, Zeuner KE, Ross OA, Arzberger T, Chouinard S, Deuschl G, Louis ED, Kuhlenbäumer G, Rouleau GA, Genome-wide association study in essential tremor identifies three new loci, Brain. 139 (2016) 3163–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Diez-Fairen M, Bandres-Ciga S, Houle G, Nalls MA, Girard SL, Dion PA, Blauwendraat C, Singleton AB, Rouleau GA, Pastor P, Genome-wide estimates of heritability and genetic correlations in essential tremor, Parkinsonism Relat. Disord 64 (2019) 262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bomba L, Walter K, Soranzo N, The impact of rare and low-frequency genetic variants in common disease, Genome Biol. 18 (2017) 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Merner ND, Girard SL, Catoire H, Bourassa CV, Belzil VV, Rivière J-B, Hince P, Levert A, Dionne-Laporte A, Spiegelman D, Noreau A, Diab S, Szuto A, Fournier H, Raelson J, Belouchi M, Panisset M, Cossette P, Dupré N, Bernard G, Chouinard S, Dion PA, Rouleau GA, Exome sequencing identifies FUS mutations as a cause of essential tremor, Am. J. Hum. Genet 91 (2012) 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Unal Gulsuner H, Gulsuner S, Mercan FN, Onat OE, Walsh T, Shahin H, Lee MK, Dogu O, Kansu T, Topaloglu H, Elibol B, Akbostanci C, King M-C, Ozcelik T, Tekinay AB, Mitochondrial serine protease HTRA2 p.G399S in a kindred with essential tremor and Parkinson disease, Proc. Natl. Acad. Sci. U. S. A 111 (2014) 18285–18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sánchez E, Bergareche A, Krebs CE, Gorostidi A, Makarov V, Ruiz-Martinez J, Chorny A, Lopez de Munain A, Marti-Masso JF, Paisán-Ruiz C, SORT1 Mutation Resulting in Sortilin Deficiency and p75(NTR) Upregulation in a Family With Essential Tremor, ASN Neuro. 7 (2015). 10.1177/1759091415598290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hor H, Francescatto L, Bartesaghi L, Ortega-Cubero S, Kousi M, Lorenzo-Betancor O, Jiménez-Jiménez FJ, Gironell A, Clarimón J, Drechsel O, Agúndez JAG, Kenzelmann Broz D, Chiquet-Ehrismann R, Lleó A, Coria F, García-Martin E, Alonso-Navarro H, Martí MJ, Kulisevsky J, Hor CN, Ossowski S, Chrast R, Katsanis N, Pastor P, Estivill X, Missense mutations in TENM4, a regulator of axon guidance and central myelination, cause essential tremor, Hum. Mol. Genet 24 (2015) 5677–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bergareche A, Bednarz M, Sánchez E, Krebs CE, Ruiz-Martinez J, De La Riva P, Makarov V, Gorostidi A, Jurkat-Rott K, Marti-Masso JF, Paisán-Ruiz C, SCN4A pore mutation pathogenetically contributes to autosomal dominant essential tremor and may increase susceptibility to epilepsy, Hum. Mol. Genet 24 (2015) 7111–7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liu X, Hernandez N, Kisselev S, Floratos A, Sawle A, Ionita-Laza I, Ottman R, Louis ED, Clark LN, Identification of candidate genes for familial early-onset essential tremor, Eur. J. Hum. Genet 24 (2016) 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tio M, Wen R, Lim YL, Wang H, Ling S-C, Zhao Y, Tan E-K, FUS-linked essential tremor associated with motor dysfunction in Drosophila, Hum. Genet 135 (2016) 1223–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Louis ED, Faust PL, Essential tremor pathology: neurodegeneration and reorganization of neuronal connections, Nat. Rev. Neurol 16 (2020) 69–83. [DOI] [PubMed] [Google Scholar]
- [23].Suzuki N, Fukushi M, Kosaki K, Doyle AD, de Vega S, Yoshizaki K, Akazawa C, Arikawa-Hirasawa E, Yamada Y, Teneurin-4 is a novel regulator of oligodendrocyte differentiation and myelination of small-diameter axons in the CNS, J. Neurosci 32 (2012) 11586–11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Deuschl G, Zimmermann R, Genger H, Lucking CH, Physiologic classification of essential tremor In: Findley LJ, Koller WC, eds. Handbook of Tremor Disorders. New York, NY: Marcel Dekker, 1995:195–208. [Google Scholar]
- [25].Deuschl G, Bain P, Brin M, Consensus statement of the Movement Disorder Society on Tremor. Ad Hoc Scientific Committee, Mov. Disord 13 Suppl 3 (1998) 2–23. [DOI] [PubMed] [Google Scholar]
- [26].Jiménez-Jiménez FJ, García-Martín E, Alonso-Navarro H, Lorenzo-Betancor O, Ortega-Cubero S, Pastor P, Calleja M, Agúndez JAG, A family study of DRD3 rs6280, SLC1A2 rs3794087 and MAPT rs1052553 variants in essential tremor, Neurol. Res 38 (2016) 10:880–7. [DOI] [PubMed] [Google Scholar]
- [27].Ross JP, Dupre N, Dauvilliers Y, Strong S, Ambalavanan A, Spiegelman D, Dionne-Laporte A, Pourcher E, Langlois M, Boivin M, Leblond CS, Dion PA, Rouleau GA, GanOr Z, Analysis of DNAJC13 mutations in French-Canadian/French cohort of Parkinson’s disease, Neurobiology of Aging. 45 (2016) 212.e13–212.e17. [DOI] [PubMed] [Google Scholar]
- [28].Schaefer SM, Rodriguez AV, Louis ED, Brain circuits and neurochemical systems in essential tremor: insights into current and future pharmacotherapeutic approaches, Expert Rev Neurother 18 (2018) 101–110. [DOI] [PubMed] [Google Scholar]
- [29].Brkic M, Balusu S, Libert C, Vandenbroucke RE, Friends or Foes: Matrix Metalloproteinases and Their Multifaceted Roles in Neurodegenerative Diseases, Mediators Inflamm. 2015 (2015) 620581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Murray MY, Birkland TP, Howe JD, Rowan AD, Fidock M, Parks WC, Gavrilovic J, Macrophage migration and invasion is regulated by MMP10 expression, PLoS One. 8 (2013) e63555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cuadrado E, Rosell A, Penalba A, Slevin M, Alvarez-Sabín J, Ortega-Aznar A, Montaner J, Vascular MMP-9/TIMP-2 and neuronal MMP-10 up-regulation in human brain after stroke: a combined laser microdissection and protein array study, J Proteome Res. 8 (2009) 3191–7. [DOI] [PubMed] [Google Scholar]
- [32].Roncal C, Martinez de Lizarrondo S, Salicio A, Chevilley A, Rodriguez JA, Rosell A, Couraud P, Weksler B, Montaner J, Vivien D, Páramo JA, Orbe J, New thrombolytic strategy providing neuroprotection in experimental ischemic stroke: MMP10 alone or in combination with tissue-type plasminogen activator, Cardiovascular Research, 113 (2017) 1219–1229. [DOI] [PubMed] [Google Scholar]
- [33].Craig-Schapiro R, Kuhn M, Xiong C, Pickering EH, Liu J, Misko TP, Perrin RJ, Bales KR, Soares H, Fagan AM, Holtzman DM, Multiplexed immunoassay panel identifies novel CSF biomarkers for Alzheimer’s disease diagnosis and prognosis, PLoS One. 6 (2011) e18850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Whelan CD, Mattsson N, Nagle MW, Vijayaraghavan S, Hyde C, Janelidze S, Stomrud E, Lee J, Fitz L, Samad TA, Ramaswamy G, Margolin RA, Malarstig A, Hansson O, Multiplex proteomics identifies novel CSF and plasma biomarkers of early Alzheimer’s disease, Acta Neuropathol Commun. 7 (2019) 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Santaella A, Kuiperij HB, van Rumund A, Esselink RAJ, van Gool AJ, Bloem BR, Verbeek MM, Inflammation biomarker discovery in Parkinson’s disease and atypical parkinsonisms, BMC Neurol. 20 (2020) 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Miller JP, Holcomb J, Al-Ramahi I, de Haro M, Gafni J, Zhang N, Kim E, Sanhueza M, Torcassi C, Kwak S, Botas J, Hughes RE, Ellerby LM, Matrix metalloproteinases are modifiers of huntingtin proteolysis and toxicity in Huntington’s disease, Neuron. 67 (2010) 199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kiezun A, Garimella K, Do R, Stitziel NO, Neale BM, McLaren PJ, Gupta N, Sklar P, Sullivan PF, Moran JL, Hultman CM, Lichtenstein P, Magnusson P, Lehner T, Shugart YY, Price AL, de Bakker PIW, Purcell SM, Sunyaev SR, Exome sequencing and the genetic basis of complex traits, Nat. Genet 44 (2012) 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gravel S, Henn BM, Gutenkunst RN, Indap AR, Marth GT, Clark AG, Yu F, Gibbs RA, 1000 Genomes Project, C.D. Bustamante, Demographic history and rare allele sharing among human populations, Proc. Natl. Acad. Sci. U. S. A 108 (2011) 11983–11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sun Q-Y, Xu Q, Tian Y, Hu Z-M, Qin L-X, Yang J-X, Huang W, Xue J, Li J-C, Zeng S, Wang Y, Min H-X, Chen X-Y, Wang J-P, Xie B, Liang F, Zhang H-N, Wang C-Y, Lei L-F, Yan X-X, Xu H-W, Duan R-H, Xia K, Liu J-Y, Jiang H, Shen L, Guo J-F, Tang B-S, Expansion of GGC repeat in the human-specific NOTCH2NLC gene is associated with essential tremor, Brain. 143 (2020) 222–233. [DOI] [PubMed] [Google Scholar]
- [40].Odgerel Z, Sonti S, Hernandez N, Park J, Ottman R, Louis ED, Clark L, Whole genome sequencing and rare variant analysis in essential tremor families, PLoS One 14 (2019) e0220512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Fahn S, Tolosa E, Marín C, Clinical rating scale for tremor In: Jankovic J, Tolosa E, eds. Parkinson’s Disease and Movement Disorders. Baltimore, MD: Williams and Wilkins; 1993:271–280. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


