Abstract
Growing evidence implicates histone H3 lysine 9 methylation in tumorigenesis. The SUV family of H3K9 methyltransferases, which include G9a, GLP, SETDB1, SETDB2, SUV39H1 and SUV39H2 deposit H3K9me1/2/3 marks at euchromatic and heterochromatic regions, catalyzed by their conserved SET domain. In cancer, this family of enzymes can be deregulated by genomic alterations and transcriptional mis-expression leading to alteration of transcriptional programs. In solid and hematological malignancies, studies have uncovered pro-oncogenic roles for several H3K9 methyltransferases and accordingly, small molecule inhibitors are being tested as potential therapies. However, emerging evidence demonstrate onco-suppressive roles for these enzymes in cancer development as well. Here, we review the role H3K9 methyltransferases play in tumorigenesis focusing on gene targets and biological pathways affected due to misregulation of these enzymes. We also discuss molecular mechanisms regulating H3K9 methyltransferases and their influence on cancer. Finally, we describe the impact of H3K9 methylation on therapy induced resistance in carcinoma. Converging evidence point to multi-faceted roles for H3K9 methyltransferases in development and cancer that encourages a deeper understanding of these enzymes to inform novel therapy.
Keywords: H3K9methylation; G9a/GLP, SETDB1/2; SUV39H1/2; carcinoma; leukemia; therapy induced resistance; epithelial-to-mesenchymal transition; hypoxia; small molecule inhibitors
1. Introduction
Organismal development requires intricate regulation of gene expression. Post-translational modifications (PTMs) on histones influence gene regulation independent of underlying DNA sequence. Amino acids on histone tails are modified by epigenetic enzymes that deposit or remove different types of chemical moieties which influence transcriptional states. Though di- and tri-methylation of histone H3 at lysine 9 (H3K9me2, H3K9me3) is thought to participate in gene silencing [1,2] in concert with other transcriptional repressors [3], H3K9me1 has been associated with active transcription [4]. Densely packed and transcriptionally silent heterochromatin is broadly enriched with stable H3K9me2/3. H3K9me2/3 is also present at more loosely packed and transcriptionally accessible euchromatin where it plays a role in repressing active genes to dynamically modulate gene expression [2,3]. Chromatin compaction and gene silencing is conserved across eukaryotes, though different modalities have evolved in different organisms. For example, the fission yeast Schizosaccharomyces pombe uses a H3K9methylation-based mechanism for gene silencing and heterochromatin spreading [5], whereas the budding yeast Saccharomyces cerevisiae uses a mechanism analogous to Sir-based heterochromatin formation in metazoans [6,7], which may reflect differential heterochromatin features in these organisms. DNA methylation is also intrinsically linked to heterochromatin formation, where the cooperative action of DNA methyltransferases, histone H3K9 methyltransferases and HP1 protein facilitate heterochromatin formation [8,9]. It has become increasingly clear that deregulation of H3K9 methylation machinery is a common pathogenic feature of solid tumors and hematological malignancies. In this review, we discuss our current understanding of H3K9 methyltransferase function in the progression and pathogenesis of human cancers.
2. Writers of H3K9 methylation
Enzymatic machinery involved in the formation of repressive chromatin is conserved from yeast to humans. Suppressor of variegation (Su(var)3–9) was first discovered in Drosophila melanogaster in a mutagenic screen to identify proteins involved in suppressing position effect variegation or heterochromatin formation [10]. Subsequent studies led to the identification of other enzymes involved in gene silencing. Two mammalian homologues of Drosophila Su(VAR)3–9 (SUV39H1 and SUV39H2) were shown to be expressed in a tissue specific manner [11,12]. G9a and GLP were discovered to have substrate specificity similar to SUV39H depositing H3K9me2 modifications on chromatin [13]. Through a biochemical analysis of KAP-1 binding proteins, SETDB1 was discovered to catalyze H3K9 trimethylation. Homology studies discovered the SETDB2 H3K9 methyltransferase is expressed in a tissue specific manner [14,15]. Thus, early screening assays in Drosophila to identify proteins involved in heterochromatin formation laid the foundation for discovering a number of H3K9 methyltransferases and understanding the role of H3K9 methylation in gene regulation in development and disease.
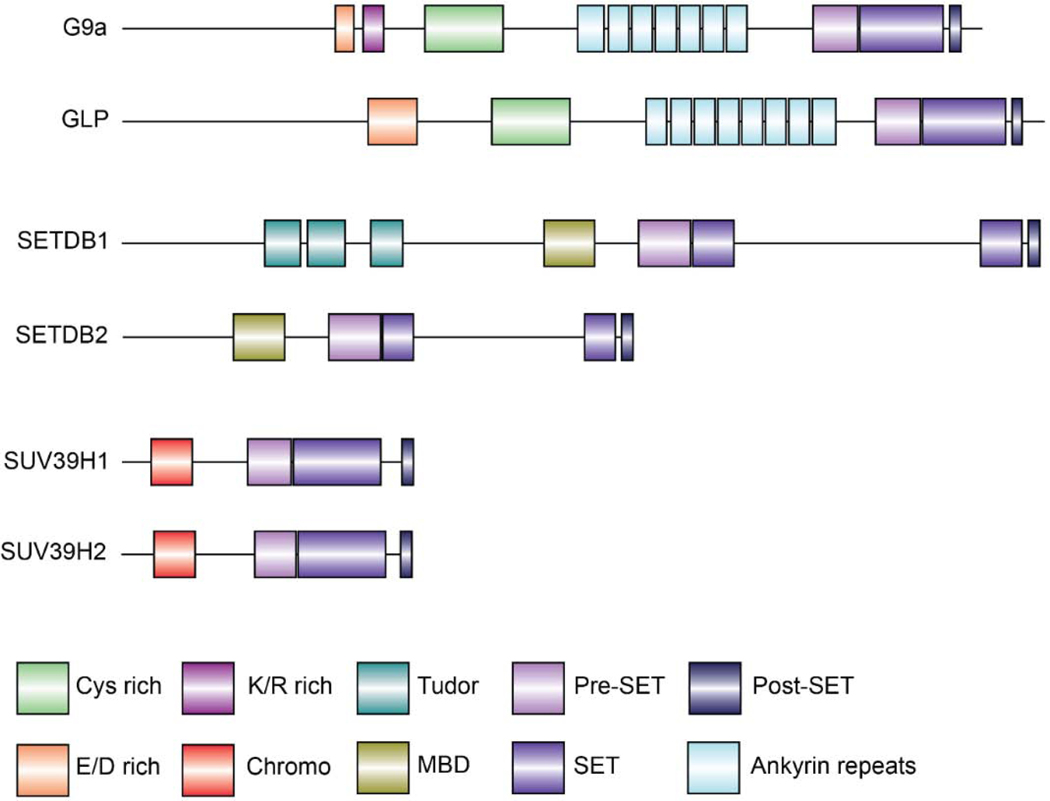
The SUV3–9 family of enzymes comprised of G9a (EHMT2;KMT1C), GLP (EHMT1;KMT1D), SETDB1 (ESET;KMT1E), SETDB2 (KMT1F), SUV39H1 (KMT1A) and SUV39H2 (KMT1B) contain a highly conserved SET domain responsible for methylation of histone H3 at lysine 9, which is associated with gene silencing and heterochromatin formation. Graphical representation of these protein structures is shown in Figure 1.
Figure 1.
Graphical illustration depicting human H3K9 methyltransferase protein structures. G9a; 1210 amino acids (aa) Uniprot: Q96KQ7, GLP; 1298 aa Uniprot: Q9H9B1, SETDB1; 1291 aa Uniprot: Q15047, SETDB2; 719 aa Uniprot: Q96T68, SUV39H1; 412 aa Uniprot: O43463, SUV39H2; 410 aa Uniprot: Q9H5I1. Protein structures are drawn using Dog2.0 tool [223].
G9a and the related GLP are highly homologous proteins and possess identical substrate specificities on histones [13,16]. G9a and GLP form a heteromeric complex and contribute to mono and di-methylation of H3K9 [17]. A function of G9a is to silence euchromatic genes by depositing di-methylation marks at lysine 9 of histone H3. Structurally, G9a and GLP proteins harbor ankyrin repeats which are required for the recognition of histone H3 substrate and interaction with the de novo DNA methyltransferase DNMT3A [18–20]. The catalytic SET domain, flanked by Pre-SET and Post-SET motifs, deposits methyl groups on histone H3K9. In addition, G9a/GLP contains a nuclear localization signal at the N-terminus and a cysteine rich (Cys) domain [21]. A recent report indicated that G9a interacts with CyclinD1 via the Cys domain, suggesting this motif might act as an interaction platform for protein interactions [22]. G9a-deficiency in mouse embryo abolishes H3K9me2 by E9.5 and leads to early embryonic death indicating G9a is critical for development [23]. In addition, G9a/GLP plays a significant role in stem cell commitment to lineage specific cells. During early embryogenesis, lineage commitment and specification requires G9a to suppress pluripotent genes, such as OCT3/4, in co-operation with DNMT3A [18,24]. G9a is also involved in lineage commitment in the hematopoietic system, as well as skeletal myogenic and adipocyte differentiation [21]. A number of studies have demonstrated a role for G9a in cell cycle progression and DNA damage repair [25–28].
SETDB1 deposits H3K9me2/3 at euchromatic regions of the genome and is thought to be involved in silencing active genes as well as telomeric heterochromatin formation [14,29,30]. In addition, SETDB1 has been shown to be involved in establishment of H3K4me3/H3K9me3, H3K14Ac/H3K9me3 bivalent domains, thus indicating a dynamic role in chromatin modification and gene regulation [31–33]. Unlike domain (MBD), a domain involved in protein-protein interaction and DNA methyl-CpG binding [34]. SETDB1/2 contain a SET domain split into two domains leading to their nomenclature, SET bifurcated 1 (SETDB1) and SET bifurcated 2 (SETDB2). SETDB1 possesses triple tudor domains at its N-terminus, which recognize H3K14Ac/H3K9me modifications and facilitate recruitment of SETDB1 complexes to active chromatin sites for gene silencing [32,35]. SETDB1 associates with a variety of co-repressor complexes such as the HUSH complex and PML bodies associated with gene silencing [36,37]. SETDB1 is essential for development as SETDB1-knockout causes early embryonic lethality with defects in neural and hematopoietic development [38–41]. SETDB1 plays a role in suppressing endogenous retroviruses, meiotic progression, and maintenance of methylation homeostasis [42–45] thereby helping to safeguard the genomic integrity of cells. SETDB2 is also important for embryonic development and chromosomal segregation [15,46,47].
SUV39H1/2 are paralogous nuclear proteins that also participate in the formation of constitutive heterochromatin, maintenance of genomic stability, and cell cycle progression [48]. SUV39H1/2 catalyze H3K9 trimethylation through a conserved C-terminal SET domain [49]. H3K9me3 is bound by HP1 at pericentric satellite repeats and contributes to a transcriptional repressive heterochromatin state [5]. They also possess an N-terminal chromo domain that binds H3K9me2/3 and is essential for catalytic activity [50,51].other members of the SUV family of proteins, SETDB1 and SETDB2 contain a methyl CpG binding In mouse, SUV39H1 is detected throughout development from early embryogenesis to adulthood, while SUV39H2 expression is limited to early embryogenesis and adult testes [11,12]. SUV39H1 and SUV39H2 appear to have overlapping functions as either protein is sufficient for embryonic development [52]. Interestingly, deletion of both SUV39H1 and 2 results in a hematopoietic defect [53].
3. Implications of H3K9methyl transferases in carcinogenesis
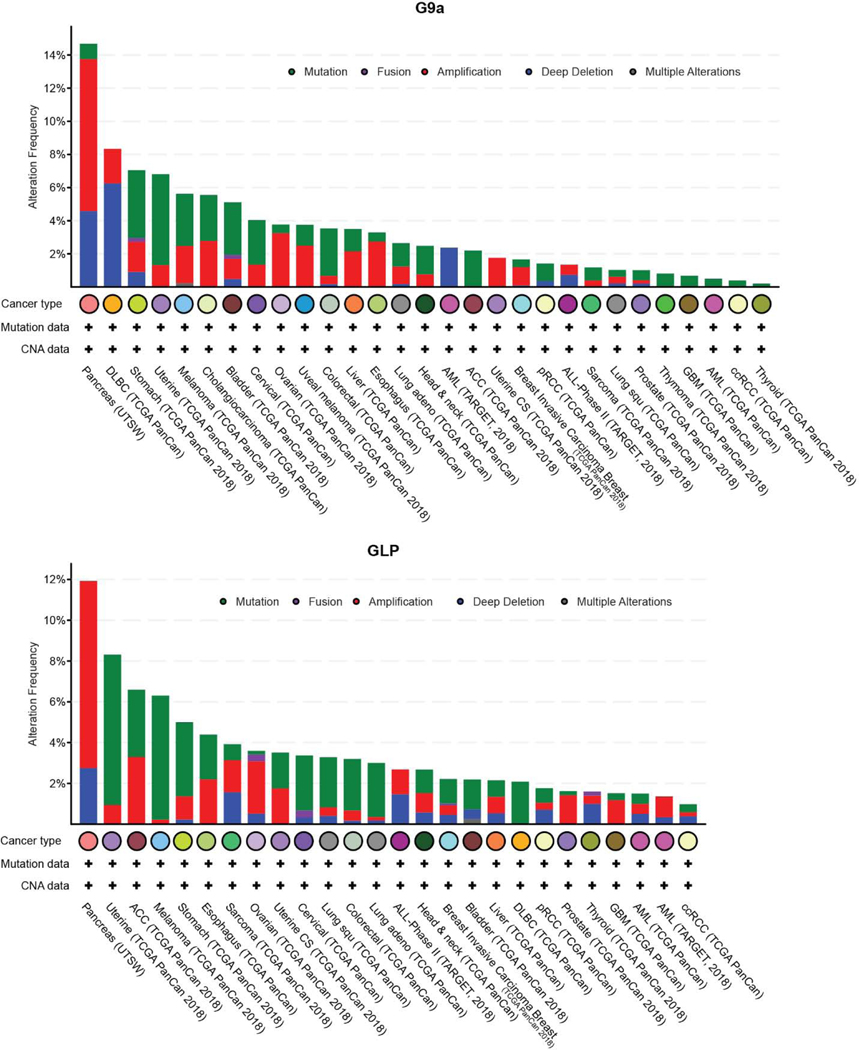
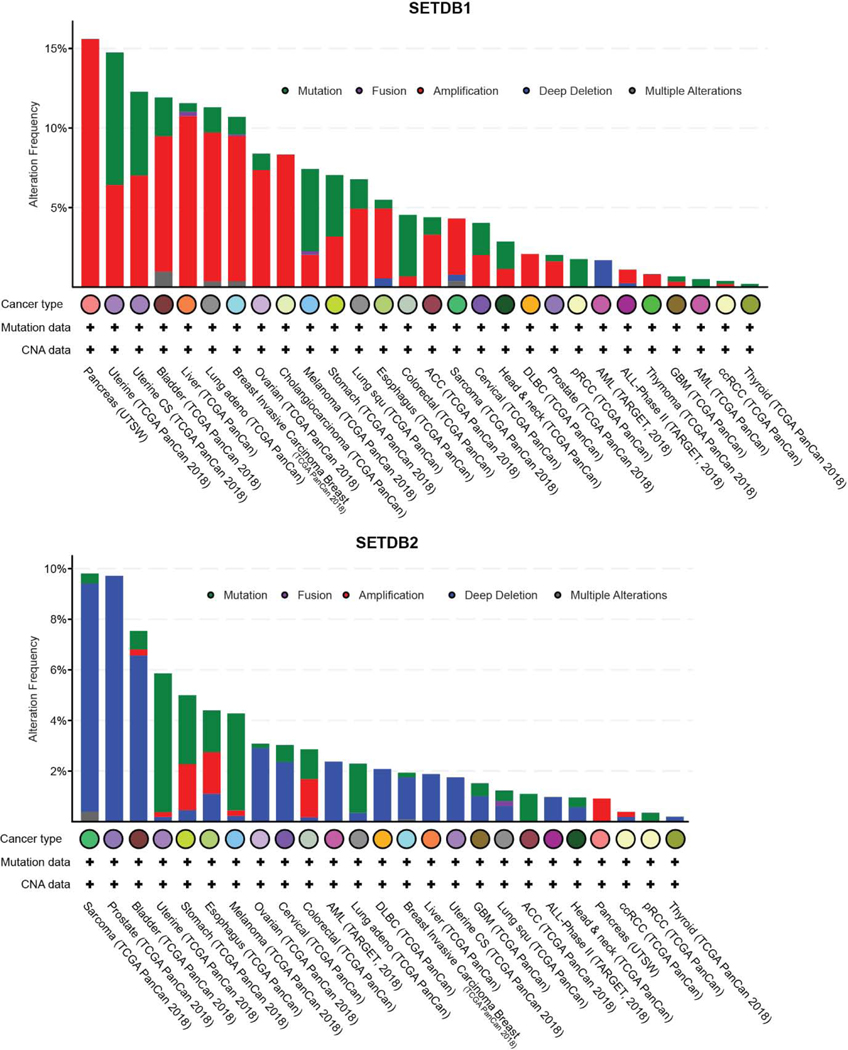
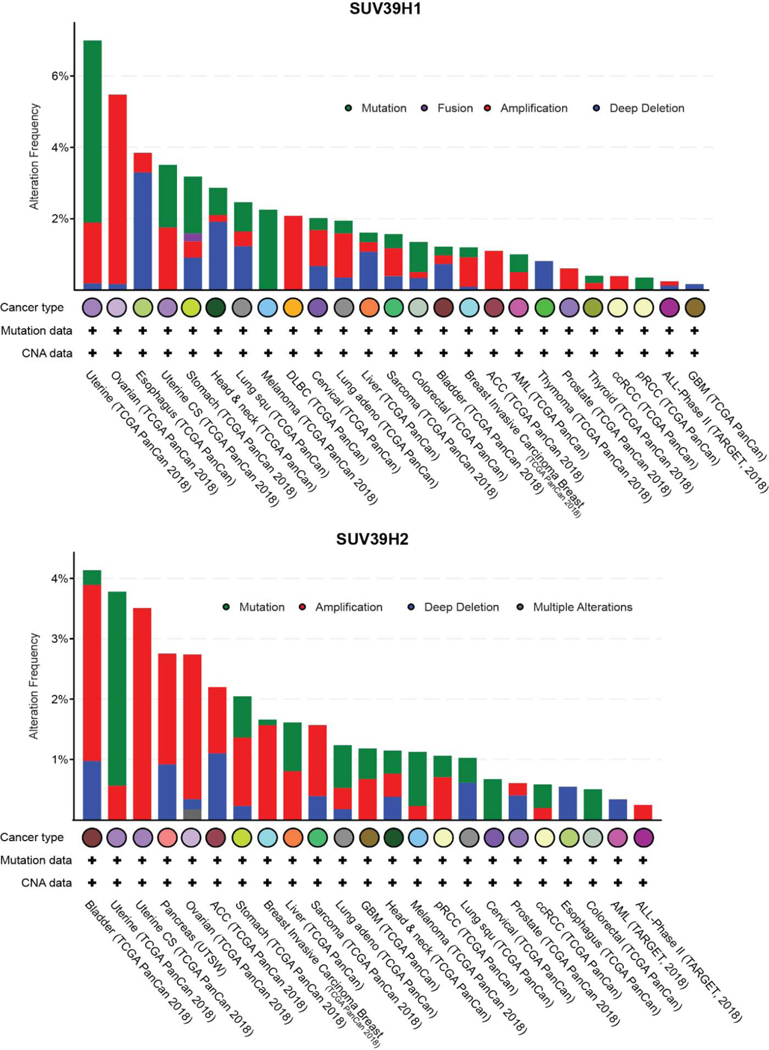
Epigenetic abnormalities are common during neoplastic transformation and result in altered chromatin landscapes and transcriptional deregulation. Altered DNA methylation and histone PTMs are hallmarks of tumorigenesis [54–56]. Aberrant DNA methylation is common in several types of malignancies and recent work in ovarian and lung carcinoma suggest DNA methylation and histone H3K9 methylation are closely linked in the pathogenesis of malignancies [57–59]. The reversible nature of these epigenetic changes, their altered signatures in cancer cells and plasticity employed to avoid drug sensitivity makes them an attractive avenue for therapeutic intervention [60,61]. Wen et al., described the presence of large organized chromatin K9 modifications (LOCKs), which are silent chromatin domains marked by H3K9me2 and associated with developmentally regulated genes. Notably, LOCKs are reduced in cancer cells, suggestive of a role in phenotype plasticity [62,63]. Abnormalities in H3K9methylation arise from mis-expression or regulation of the epigenetic factors that contribute to this modification (H3K9 methyl transferases and demethylases). Genomic alterations to H3K9 methyltransferase genes are observed in different kinds of solid tumors and hematological malignancies (Figure 2, 3 and 4). The different types of gene lesions detected in different cancers may imply non-overlapping roles for H3K9 methyltransferases. Here, we describe the mis-regulation of all H3K9 methyltransferases of the SUV3–9 family and how this affects different types of malignancies. The differential function of H3K9 methyltransferases in carcinogenesis is summarized in Table 1 and 2.
Figure 2.
Genomic alterations in G9a and GLP observed in different types of cancer. Data collected and plotted in c-Bioportal.org
Figure 3.
Genomic alterations in SETDB1 and SETDB2 observed in different types of cancer. Data collected and plotted in c-Bioportal.org
Figure 4.
Genomic alterations in SUV39H1 and SUV39H2 observed in different types of cancer. Data collected and plotted in c-Bioportal.org
Table 1.
Pro-tumorigenic functions of H3K9 methyl transferases
| Pro-tumorigenic activity H3K9 methyl transferase | Cancer type | Biological process affected |
|---|---|---|
| G9a | Lung carcinoma [84,85] | Enhanced EMT [82,85] Activation of WNT signaling [84] Maintenance of stemness [83] |
| Breast carcinoma [64,66] | Enhanced EMT [72] Suppression of autophagy [89] |
|
| Colorectal carcinoma [90] | Enhanced DNA double strand break repair [90] | |
| Gastric cancer [91] | Suppression of autophagy [91] Suppression of tumor suppressors [92] |
|
| Hepatocellular carcinoma [93,94] | Enhanced EMT [95] Suppression of tumor suppressors [94] |
|
| Cholangio carcinoma [96] | Suppression of tumor suppressor [96] | |
| Bladder carcinoma [97] | Suppression of autophagy [97] | |
| Melanoma [98] | Activation of WNT signaling [98] Activation of PI3K/AKT pathway [99] |
|
| Ovarian and cervical carcinoma [100] | Promotes metastasis [100] | |
| Glioma [101] | Suppression of apoptosis [101] | |
| Neuroblastoma cells [102] | Suppression of autophagy [102] | |
| Oral squamous cell carcinoma [103] | Suppression of autophagy [103] | |
| Head and neck squamous cell carcinoma [104] | Supports EMT [104] | |
| G9a/GLP | Hematological malignancies [105] | Suppression of cell cycle inhibitors [106] |
| SETDB1 | Lung carcinoma [107,108] | Activation of WNT signaling [108] |
| Breast carcinoma [76,77] | Promotes EMT [78,79] | |
| Colorectal carcinoma [109,110] | Suppression of tumor suppressors [109,110] | |
| Hepatocellular carcinoma [111,112] | Enhanced metastasis [112,113] | |
| Melanoma [114] | Enhanced metastasis [114] Suppression of cell cycle inhibitors [115] |
|
| Hematological malignancies [116,117] | Suppression of endogenous retroviral elements [116] | |
| Prostate Cancer [118] | - | |
| SETDB2 | Gastric cancer [119] | Suppression of tumor suppressors [119] |
| Hematological malignancies [120] | Suppression of cell cycle inhibitor [121] | |
| SUV39H1 | Colorectal carcinoma [122] | - |
| Gastric cancer [123] | Suppression of tumor suppressors [124] | |
| Hepatocellular carcinoma [125] | Enhanced metastasis [125] | |
| Breast carcinoma [73] | Enhanced EMT [73] | |
| Melanoma [126] | - | |
| Prostate Cancer [127] | - | |
| SUV39H2 | Lung carcinoma [87] | - |
| Colorectal carcinoma [128] | Promotes cell proliferation and metastases [128] | |
| Gastric cancer [129] | Suppression of tumor suppressors [129] | |
| - Further evaluation required | ||
Table 2:
Onco-suppressive functions of H3K9 methyl
| Anti-Tumor activity | ||
|---|---|---|
| H3K9 methyl transferase | Cancer type | Biological process affected |
| G9a | Lung carcinoma [86] | - |
| G9a/GLP | Glioma [130] | Suppression of self-renewal genes [130] |
| Hematological malignancies [117] | - | |
| SETDB1 | Lung carcinoma [131,132] | Suppressor of metastasis [133] |
| Breast carcinoma [80] | Suppressor of metastasis [80] | |
| Hematological malignancies [117,134] | Induced differentiation and suppression of self-renewal genes [134] | |
| SUV39H1 | Breast carcinoma [75] | Suppression of cell cycle gene [75] |
| Ovarian and cervical carcinoma [135] | - | |
| Parathyroid carcinoma [136] | Suppression of cell cycle gene [136] | |
| Hematological malignancies [52,137] Rhabdomyosarcoma [138] |
Induced apoptosis[137] | |
| SUV39H2 | Lung carcinoma [139] | Induced cell cycle arrest [139] |
| - Further evaluation required | ||
3.1. Breast Cancer
Meta-analysis of breast cancer patient specimens display amplification of G9a, which is associated with poor survival [64,65]. Mechanistically, G9a is required for breast cancer tumor growth and metastasis and promotes cell proliferation [64,66]. Multiple gene regulatory networks have been associated with G9a. A recent study reported G9a interacts and represses MYC target genes through deposition of H3K9me2 and is required for MYC dependent breast tumorigenesis [67]. In addition, G9a plays a role in hypoxia, which promotes migration and insensitivity to chemotherapy [68]. Indeed, hypoxia results in enrichment of global H3K9me2 through increased expression of G9a [69,70], which may suppress activity of hypoxia-induced tumor suppressors [69].
H3K9 methyltransferases also play a role in epithelial to mesenchymal transition (EMT) in breast cancer [71]. For example, G9a and SUV39H1 associate with the SNAIL transcription factor to repress anti-EMT genes such as E-cadherin through H3K9me2/3 deposition at its promoter [72,73]. Accordingly, loss of function studies suggest G9a and SUV39H1 promote breast cancer metastasis. Inhibition of SUV39H1 in breast cancer cell lines restores E-cadherin expression through depletion of H3K9me3 at the promoter region [72–74]. Thus, G9a/SUV39H1 and associated H3K9me2/me3 act as important mediators of EMT in breast cancer pathology.
Interestingly, Khanal et al., reported that SUV39H1 suppresses oncogenic transformation in breast cancer cells overexpressing PIN1 [75]. Thus, potential H3K9 methyltransferase therapeutics for breast cancer treatment may require knowledge of underlying genetics.
Recent studies implicate SETDB1 as an oncogenic driver of breast cancer where it is amplified compared to normal tissue [76–78]. SETDB1 depletion blocks tumor cell growth [76,77] consistent with an oncogenic role in breast cancer. Recently, SETDB1 was associated with upregulation of SNAIL1 and SMAD7 expression aiding in EMT, which may promote metastasis [78,79]. The mechanism, however, of SETDB1-mediated activation of SNAIL1 and SMAD7 remains unclear. Paradoxically, Du et al., demonstrated reduced SETDB1 fosters breast cancer metastasis. They found SETDB1 inhibits TGF-β induced EMT and SETDB1-deficiency increased cancer migration in a zebra fish model [80]. These data suggest multiple functions for SETDB1 in breast cancer transformation and EMT that warrants further investigation.
3.2. Lung Carcinoma
Studies in the last decade have implicated gene silencing machinery in lung carcinogenesis and metastasis, including oncogenic roles for G9a in lung carcinoma. Amongst H3K9 methyltransferases, G9a displays the highest expression in several lung cancer cell lines [81]. In a cohort of lung cancer patient samples comprised of lung adenocarcinoma and lung squamous cell carcinoma, G9a expression is elevated compared to normal tissue and correlated with poorer survival [82–85]. Functional studies showed G9a is required for tumor development and enhances dissemination potential of lung carcinoma [84,85]. G9a silences expression of anti-EMT genes, such as EPCAM and CASP1, thus promoting EMT and tumor activity [82,85]. G9a is also involved in activation of WNT signaling by repressing WNT signaling inhibitors such as APC2, DKK1, and WIFI, thus facilitating cell proliferation [84]. Importantly, G9a oncogenic function is dependent on its H3K9 dimethylation activity [82,84,85]. Interestingly, G9a appears to induce expression of OCT4, NANOG and CD133 to maintain stemness in lung cancer cells, though the mechanism remains unclear [83]. In contrast to these oncogenic functions, Rowbotham et al., demonstrated increased proliferation of primitive lung cancer cells upon depletion of G9a suggesting a tumor suppressive role at the initiation stage of transformation. Loss of G9a in tumor propagating cells resulted in a global reduction in H3K9me2, expansion of tumor cells and increased lung cancer dissemination. Clinically, early stage lung cancer patients expressing elevated levels of G9a show better survival. These data suggest a critical balance of H3K9me2 by G9a for cellular homeostasis [86].
A report by Watanabe et al. reported high expression of SUV39 in human lung cancer cell lines, which is important for the growth of transformed human bronchial epithelia [81]. Transcriptomic and immunohistochemistry data demonstrate Suv39H2 is elevated in lung adenocarcinoma patients relative to normal tissue and higher SUV39H2 expression correlates with a poorer survival [87]. A study testing a newly synthesized SUV39H2 inhibitor suggested blocking SUV39H2 activity suppresses the growth of lung cancer cells [88]. SUV39H2 is required for repression of Optoneurin, a gene associated with adenocarcinoma [87], suggesting SUV39H2 catalytic activity contributes to oncogenesis. Lung cancer cell lines treated with the H3K9me inhibitor, Chaetocin, display increased apoptosis and reduced H3K9me3 [140]. While, these studies point to an oncogenic role for SUV39H2 in lung carcinoma, Kim et al. reported SUV39H2 inhibits proliferation and induced cell cycle arrest of the A549 lung adenocarcinoma cell line. Furthermore, clinical data showed a better survival for lung cancer patients with higher SUV39H2 levels [139].
Clinicopathological data indicate SETDB1 expression is elevated in lung cancer and associated with a poorer prognostic outcome [81,107,108,141,142]. Functional analyses indicate SETDB1 exerts pro-proliferative effects on human lung cancer cell lines, [108,143]. Mechanistically, the oncogenic functions of SETDB1 have been attributed to hyperactivation of WNT signaling and suppression of TP53 expression [108]. Interestingly, SETDB1 can also suppress lung cancer migration. Expression of SETDB1 is downregulated in highly metastatic cell lines. In a Zebra fish model, SETDB1 inhibits lung cancer metastasis due to SETDB1-SMAD2/3 cooperative deposition of H3K9me3 and subsequent repression of ANXA2 [133]. Furthermore, inactivating mutations in SETDB1 were found in malignant pleural mesothelioma (MPM), a rare but aggressive form of lung cancer [131,132]. As with other H3K9 methyltransferases, data point to a complex role for SETDB1 in lung carcinoma.
Smoking is a major contributor to lung cancer and is attributed with altering chromatin landscapes and accelerated cell growth and transformation. In normal lung development, Gong et al., found that expression of the tumor suppressor PPAR-γ is reduced upon exposure to nicotine with concomitant increased H3K9me3 and H3K27me3 at its promoter [144,145]. Finally, SETDB1 expression was found to be upregulated in lung cancer patients with a history of smoking compared non-smokers [107].
3.3. Colorectal Carcinoma
Colorectal carcinoma (CRC) is also influenced by H3K9 methyltransferases. Elevated expression of G9a in CRC specimens and functional studies have pointed to an oncogenic role in colorectal carcinoma [90,146–148]. G9a protects against DNA double stranded breaks and may facilitate cancer cell survival. This may occur through reduced surveillance of cell cycle checkpoints caused by G9a-mediated epigenetic repression of PP2A, a phosphatase required for the G2/M checkpoint and cell cycle arrest [90,149,150]. Loss of G9a in vivo attenuated tumor growth and increased sensitivity to radiation treatment and DSB inducing cytotoxic drugs [149]. Modification of non-histone proteins by G9a is also associated with progression of colorectal carcinoma. For instance, G9a-mediated methylation of the transcription factor FOXO1, a tumor suppressor in CRC, reduces protein stability [151] enabling cancer progression.
SUV39H1 and SUV39H2 display oncogenic characteristics and are elevated in CRC. Functional assays indicate SUV39H1/2 promote cell proliferation in vitro and facilitate tumor growth and metastasis in vivo [122,128]. SUV39H1-mediated repression of FAS expression may protect cancer cells against apoptosis as downregulation of FAS is prevalent in CRC [152,153]. Small molecule inhibition of SUV39H1 in CRC cell lines results in derepression of FAS, cell cycle arrest and apoptosis [122,154].
SETDB1 expression is also elevated in CRC patients and associated with a poor prognostic outcome [109,155–158]. Molecular and biochemical characterization indicate SETDB1 functions as an oncogenic driver in colorectal carcinoma. Functionally, SETDB1 is required for cell proliferation, colony formation and cell migration in vitro and accelerates tumor growth in vivo [109,155–158]. Notably, depletion of SETDB1 leads to downregulation of transcription factors that govern cell cycle and stemness including MYC [157]. In addition, SETDB1 suppresses the cell cycle inhibitor, p21 [110]. CRC cells also display increased sensitivity to cancer therapeutic drugs, such as 5-Fluorouracil (5-FU), and cetuximab upon depletion of SETDB1 [109,158], suggesting SETDB1 may be a potential therapeutic target in CRC.
3.4. Gastric Cancer
Recent studies have demonstrated the efficacy of treating gastric cancer (GC) by targeting H3K9 methyltransferases. Molecular analyses of clinical samples indicate G9a is amplified in gastric carcinoma and correlated to increased metastasis and poor survival [91,159,160]. Indeed, depletion of G9a reduces tumor growth and metastasis in vivo, establishing a requisite role for G9a in gastric cancer progression [91,159,161]. Recent studies reveal multiple biological processes are affected by G9a to facilitate transformation. Inhibition of G9a induces apoptosis and an autophagic response through downregulation of the mTOR pathway [91,159,161]. Importantly, G9a was found to activate mTOR by modulating H3K9 monomethylation, suggesting elevated G9a in gastric cancer induces mTOR and suppresses autophagy [91]. Further, kaempferol, a flavonoid present in fruits and vegetables, inhibits G9a and induces autophagic cell death in gastric cancer cell lines [161]. Notably, combination treatment of kaempferol and a G9a inhibitor augments autophagy mediated cell death [161]. Supporting an oncogenic role, in hypoxic conditions G9a delivers H3K9me to the RUNX3 tumor suppressor gene [92]. Finally, ITGB3, an important factor for promoting GC metastasis, was found to be activated by G9a in an enzyme independent manner, suggesting multiple functions in promoting tumorigenesis [160].
Roles for SUV39H1/H2 and SETDB2 in GC stem from research linking these proteins playing a role in silencing tumor suppressors Gastrokine1 and Cdkn2A [123,124,129]. Nishikawaji et al, showed that SETDB2 is overexpressed in GC patient samples, which is correlated with a poor prognosis. SETDB2 also enzymatically silences tumor suppressor genes such as Wwox, and Cadm1 [119].
3.5. Melanoma
A large percentage of melanoma patients present with activating mutations in the serine-threonine kinase BRAF, thereby hyperactivating the MAPK signaling pathway [162]. In addition to BRAF mutations, amplification of chromosome 1q21 (chr1: 147.2–149.2megabases) harboring SETDB1 has been identified in melanoma tumor samples [163,164]. Notably, highly metastatic tumors display elevated global H3K9me3 [165]. Increased SETDB1 copy number and elevated expression are observed in melanoma patient samples relative to benign nevi samples. Further, high SETDB1 expression corresponds to worse prognoses [166]. SETDB1 accelerates cellular migration resulting in more aggressive disease development [114,166]. SETDB1 may suppress the cell cycle inhibitor p16 to promote cellular proliferation [115] Repression of developmental genes, such as HoxA by SETDB1 may also promote melanoma [114]. SUV39H1 also promotes melanoma tumorigenesis and is upregulated in melanoma samples. Functionally, SUV39H1 promotes melanoma in vivo [126]. Targeting the catalytic activity of either SETDB1 or SUV39H1 by genetic manipulation or chemical inhibitor suppresses melanoma tumorigenesis [126,166].
Studies also point to pro-oncogenic roles for G9a in melanoma. Activating G9a mutations (G1069 mutated to L/W) and amplified G9a expression are observed in melanoma cancer samples [98,99]. Further, high G9a expression correlates to poor patient survival. Forced G9a expression promotes melanoma tumorigenesis [98,99]. G9a expression leads to upregulation of Microthalmia-Associated Transcription Factor (MITF), a master regulator of melanocyte survival and melanoma oncogene through activation of the PI3K/AKT pathway [99,167]. G9a enzymatic activity is required to suppress the WNT signaling antagonist DKK1, resulting in activation of WNT signaling and MITF expression [98]. EMT is associated with elevated G9a [99], which may be related to abnormal WNT and PI3K/AKT pathway activity.
3.6. Hepatocellular Carcinoma
H3K9 methyltransferases play a role in hepatocellular carcinoma (HCC) and are being pursued as potential drug targets. Studies demonstrate copy number gains and amplified expression for G9a expression in HCC samples. Clinically, high G9a expression negatively correlates with HCC patient survival [93,94,168]. Testing with genetic models and chemical inhibitors revealed that the enzymatic activity of G9a is required for HCC tumorigenesis [93,94]. G9a suppresses the expression of the tumor suppressor RARRES3 through H3K9me2 deposition at its promoter [94,169]. G9a is also involved in HCC tumor metastasis [95,168]. Notably, G9a forms a complex with EMT regulators SNAIL2 and HDAC1 to repress key anti-EMT genes such as E-Cadherin [95,168].
SETDB1 expression is elevated in HCC compared to non-tumor control and elevated SETDB1 leads to increased cell proliferation and enhanced HCC tumorigenicity and metastasis [111–113,170]. The function of SETDB1 in HCC is related to p53 mutation status. Fei et al., found gain of function mutations in p53 (R249S) conferred sensitivity to Setdb1 deletion [111]. In addition, SETDB1 represses p53 target genes such as Cdkn1a, Puma and Gadd45 and protects HCC cells against irradiation [170]. Similar to SETDB1, SUV39H1 is also upregulated in HCC patient samples and associated with aggressive carcinoma [125,171]. Biochemical characterization indicate SUV39H1 facilitates migration and proliferation of HCC cells [125] and SUV39H1 depletion causes induction of autophagy and increased apoptosis [125,171].
3.7. Hematological Malignancy
Epigenetic dysregulation is a common feature in hematological malignancies [172,173] and H3K9 methyltransferases have become an attractive target for therapeutic intervention [174]. However, studies have described roles for H3K9 methyltransferases in both promoting and suppressing hematologic malignances. For example, G9a was shown to be essential for acute myeloid leukemia (AML) progression by promoting HOXA9/MEIS1 transcriptional activity. Importantly, G9a deletion has no deleterious effect on normal hematopoiesis, suggesting G9a could be a potential target in AML [105]. Small molecules inhibiting G9a/GLP have shown promise in treating AML [175–178]. In acute T-lymphocytic leukemia (T-ALL) G9a/GLP facilitates cell proliferation by repressing cell cycle inhibitor genes such as Cdkn1a and Cdkn2b [106]. Additionally, G9a/GLP is implicated in the development of chronic lymphocytic leukemia (CLL) where G9a/GLP inhibition induces cell death [179]. Recently, G9a has been shown to contribute to multiple myeloma [180]. G9a/GLP inhibitors induces apoptosis of leukemic cells while minimally effecting differentiation [106,181]. These data point to an essential function for G9a/GLP in leukemia cell survival. SETDB1 is also essential in mouse models of leukemia but is also essential for normal hematopoiesis [38]. In this context, SETDB1 suppresses endogenous retroviral elements and silences interferon response in leukemic cells [116]. SETDB2 also displays pro-oncogenic functions in leukemia potentially through suppression of Cdkn1a [120,121]. Interestingly, SETDB2 is also deleted in CLL, which may point to context specific functions [182]. Indeed, treatment with the H3K9 methyltransferases inhibitor chaetocin relieves the differentiation blockade in leukemic cells with a concomitant increase in apoptosis [181,183–185].
Homeobox proteins, including HOXA9 and its co factor MEIS1, are pro-leukemic transcription factors that block differentiation and promote AML and ALL. The posterior HOXA gene cluster is exquisitely regulated by H3K79 methylation, which is deposited by DOT1L [186]. Genetic or small molecule inactivation of DOT1L impairs HOXA9 and MEIS1 expression and leukemia progression [186,187]. Importantly, H3K79 methylation inhibits SUV39H mediated H3K9me3 deposition at the HOXA gene cluster and points to a potential suppressive role for H3K9me in leukemogenesis [188]. Indeed, SETDB1 expression also catalyzes H3K9me3 at the HOXA gene cluster including HOXA9, its co-factor MEIS1 and others [117,134]. Consistent with a suppressive role, forced SETDB1 expression extends AML disease latency in mouse models. Importantly, AML patients with higher SETDB1 expression display a significantly better prognostic outcome than patients with reduced SETDB1 expression [117]. Similar suppressive roles for G9a have been reported in leukemia. For example, G9a/GLP-specific inhibitor treatment of hematopoietic stem and progenitor cells preserves cellular stemness [189,190]. Moreover, overexpression of G9a in leukemic cells impairs proliferation and induces differentiation [117]. A similar tumor suppressive role for SUV39H1 in AML was recently reported. SUV39H1 overexpression induces apoptosis and increased disease latency in AML mouse models. Notably, SUV39H1 expression led to downregulation of HOXB13 in leukemic cells [137]. Together, these data suggest H3K9 methyltransferase-mediated repression of HOX genes is key regulatory mechanism of leukemia inhibition. Forced differentiation of leukemic cells is associated with an increase in G9a-induced H3K9me2 [191,192]. Also consistent with a suppressive role, knockout of SUV39H1 predisposes mice to the development of B-cell lymphoma [52].
Together, these studies point to a potential context and cell specific function for H3K9 methyltransferases that may result in requisite and suppressive roles in hematologic malignancies. These also suggest a fine-tuned molecular mechanism to tightly regulate expression of H3K9 methyltransferases in the hematopoietic system, whose disruption may contribute to leukemogenesis.
3.8. In other types of cancers
Beside the malignancies described above, studies have revealed roles for H3K9 methyltransferases in several other types of cancer. H3K9 methyltransferases expression is altered in cancers of human reproductive organs. In ovarian and cervical carcinoma, G9a and H3K9me2 display cancer promoting functions [100,193]. G9a enhances peritoneal dissemination of ovarian cancer, however, is not required for cell proliferation. Conversely, in cervical cancer, SUV39H1 impedes cancer metastasis and low SUV39H1 levels correlate with poor patient survival [135]. SETDB1 and SUV39H1 are upregulated in prostate cancer cells, which stimulate cell proliferation [118,127]. Recently, G9a was implicated in driving oncogenesis in highly aggressive cholangio carcinoma [96]. G9a also enhances head and neck squamous cells carcinoma metastasis [104]. In glioma, a role for G9a is not well understood. One study reports G9a inhibits cancer stemness by repressing self-renewing genes [130], while others show a cell proliferation defect and increased apoptosis in glioma cells upon treatment with G9a inhibitor [101,194]. SUV39H1 and SETDB1 have also been implicated in glioma [195]. A recent report demonstrates that SETDB1 promotes glioblastoma growth and metastasis through activation of the AKT/mTOR signaling pathway [196]. G9a suppresses autophagy by repressing autophagy promoting genes [197,198], and targeting of G9a leads to induction of autophagy dependent cell death or cellular differentiation in bladder carcinoma, oral squamous cell carcinoma, gastric carcinoma, breast cancer cells, and neuroblastoma cells [91,97,102,103,161,199,200]. In parathyroid carcinoma, SUV39H1 likely has tumor suppressive function as it associates with CDC73 to repress transcription of cell cycle genes such as Cyclin D [136]. These studies point to important functions for H3K9 methylation in tumorigenesis, which require further study.
4. Regulation of H3K9 methyl transferases and implications in tumorigenesis
Several cis and trans acting mechanisms can influence H3K9 methyltransferase activity during oncogenesis. In addition to cis-genomic regulatory regions and transcription factor deregulation that influence KMT expression, microRNAs, PTMs and others impart catalytic regulation during neoplastic development. Here we review our understanding of molecular regulatory mechanisms governing KMT function as well as how these might inform development of therapeutics.
microRNAs (miRNA), such as miRNA-29 and miRNA-621, directly target SETDB1 transcription, which can impede HCC tumorigenesis [112,170]. Reduced expression of these microRNAs in HCC clinical samples with elevated SETDB1 may reflect a mechanism to maintain SETDB1 expression. Though correlational, data suggests miRNA-621 might block WNT mediated cell proliferation by regulating SETDB1 expression [108,201]. SETDB1 is also a target of miRNA-7. Mechanistically, miRNA-7 suppresses EMT in breast cancer by reducing SETDB1 [202]. Another study found miRNA-1 targets G9a and loss of miRNA-1 promotes HCC by increasing G9a activity [94]. Thus, microRNA regulation of H3K9 methyltransferases may safeguard cells from neoplastic transformation and maintain cellular homeostasis.
PTMs on H3K9 methyltransferases can affect catalytic activity and cellular processes. PTMs on G9a/GLP can switch activity from transcriptional repressor to activator. For example, SUMOylation of G9a (K79, K152, K256, and K799) facilitates PCAF mediated activation of E2F1 target genes and promotes myoblast proliferation [25,203]. Interestingly, automethylation of G9a/GLP (G9a; lysine185/GLP; lysine205) facilitate interaction with HP1γ. G9a/GLP, HP1γ, and glucocorticoid receptor form a ternary complex on chromatin to activate glucocorticoid target genes, which block cell migration in lung cancer cells and enhance cell death in B-ALL cells. Conversely, phosphorylation of G9a/GLP (G9a; T186, GLP; T206) by Aurora kinase B compromises co-activator function by destabilizing binding with HP1γ [204,205]. Finally, ubiquitination at lysine867 on SETDB1 by the UBE2E family of E2 enzymes enhances the methyltransferase activity of SETDB1 [206,207]. Thus, multiple mechanisms regulate H3K9 methyltransferase catalytic activity and a more complete understanding is needed to determine the impact on tumorigenesis.
5. H3K9 methyltransferases in therapy induced resistance
Resistance to chemo-therapeutic drugs and radiation therapy are significant and serious obstacles to successful cancer treatment. Sub-populations of induced drug tolerant cancer cells (IDTC) or drug-tolerant persisters (DTPs) [61,208] emerge, in part, through epigenetic mechanisms. Substantial enrichment of repressive H3K9me2/3 and depletion of activation modifications is observed in DTP cancer cells following treatment with chemotherapeutic agents [193,208,209]. Thus, H3K9 methylation may impact chemotherapy resistance and a greater understanding is needed for future therapeutic design.
The role of G9a in resistance to chemotherapies has been explored in several cancer models. G9a is implicated in chemo and radiation therapy resistance in malignant glioma and colorectal cancer. G9a acts as a radioprotector in glioma cell lines by promoting DNA double stranded break repair [194]. In addition, G9a inhibition induces apoptosis in glioma cells by enhancing sensitivity to chemo agents like Temozolomide [210]. Radioresistant colorectal cancer cells display elevated G9a expression and are re-sensitized following G9a depletion. G9a-led protection of CRC cells occurs through heightened DNA repair [149]. G9a/GLP facilitates DSB repair and is observed in high-grade serous cell ovarian carcinoma (HGSOC) resistant to PARP inhibitors (PARPi). G9a/GLP expression and H3K9me2 are elevated in PARPi resistant HGSOC cells. Importantly, disrupting G9a/GLP activity sensitizes resistant cells to PARPi, which leads to increased DNA damage [193]. Gemcitabine (GEM) is a widely administered for several types of malignancies including pancreatic, lung, bladder, cervical, breast, ovarian and head and neck cancers [211]. In pancreatic cancer, expression of G9a is increased upon GEM administration and G9a inhibition sensitizes cells to GEM [212]. Additionally, a correlative study implicates G9a in GEM resistance in cervical cancer [213]. In head and neck squamous cell carcinoma (HNSCC) patients, high G9a expression is correlated with a poor response to chemotherapy. Elevated G9a levels also confer protection of HNSCC cells to cisplatin [214]. From a therapeutic standpoint, treatment of colorectal carcinoma with the H3K9m2/3 inhibitor Verticillin A increased sensitivity to 5-FU [147]. Given that many chemo agents potentiate DNA damage [215], cancer cells may utilize H3K9 methyltransferases to facilitate DNA damage repair and escape the cytotoxic effect of chemo drugs.
Transposable elements are also thought to be implicated in cancer progression. H3K9 methylation plays a role in normal cell homeostasis by silencing transposable elements in the genome [2,216]. LINE-1 elements are the most common form of Long Interspersed Nuclear Elements (LINEs) spanning the human genome. Transposition of non-LTR retrotransposon elements require expression of LINE-1 encoded endonuclease, which generates staggered cuts in DNA triggering DNA damage response [217]. SETDB1 catalytic activity is essential for repression of such endogenous retroviral elements, including LINE-1 [218]. Chemo-resistant cancer cells express elevated levels of SETDB1/2 with simultaneous enrichment of repressive H3K9me3 over LINE-1 genomic loci. Drug sensitivity is restored upon de-repression of LINE-1 elements following inactivation of SETDB1/2 activity [208,209]. Overall, H3K9 methylation machinery is linked with chemoresistance warranting further investigation into H3K9 methylation and cancer.
Cancer Stem cells, or tumor propagating/initiating cells, are a subpopulation of self-renewing cells with the capacity to give rise to bulk tumors and thought to play a key role in the development of resistance to chemotherapeutic treatments. Epigenetic alterations modulate signaling pathways important to cancer stem cells, including WNT and Notch signaling [219]. Because H3K9 methyltransferases contribute to the regulation of signaling pathways like WNT, H3K9 methylation likely plays a role in maintenance of cancer stemness. Indeed, G9a was shown to regulate stem cell programs in neurons [220]. Further, G9a was recently reported to regulate stemness in NSCLC through modulation of DNA methylation. G9a is highly expressed in CD133+ cancer stem cells and inhibition of G9a suppresses CD133 expression and tumor growth in xenograft models [57]. On the contrary, G9a/GLP has been shown to suppress lung adenocarcinoma tumor propagating cells. Inhibition of G9a led to increased cell surface expression of stem cell antigen-1 (Sca-1) and increased disease pathogenicity in vivo [86]. Notably, we have shown that SETDB1 overexpression leads to downregulation of the self-renewal genes HOXA9 and MEIS1 in AML [134]. Though our current knowledge in cancer stem cells is incomplete, the above studies implicate H3K9 methyltransferases in the regulation and/or maintenance of cancer stem cells and evading chemotherapeutics. Future studies will be critical to elucidating the functions of these enzymes in regulating stem cell features in different cancer models.
6. Conclusions and Future Directions
Repressive H3K9me2/3 is important in development cellular transformation. Deregulated H3K9me2/3 results in widespread abnormalities in various cellular functions such as cell proliferation, hypoxia, inflammation, cellular senescence, autophagy, and apoptosis, which can contribute to tumorigenesis. Elevated H3K9 methyltransferases can lead to hyperactivation of prosurvival signaling pathways, (PI3K/AKT, Notch, WNT) and repression of tumor suppressors (p21, p16). EMT is tightly regulated by transcriptional factors and signaling molecules and is a hallmark of cancer metastasis in solid tumors. Several studies implicate H3K9 methyltransferases in tumor migration, indicating these factors play a central role in solid tumor metastasis by suppressing key anti-EMT genes. These findings point to pro-oncogenic functions for H3K9 methyltransferases. Consequently, several promising small molecule inhibitors have been developed targeting H3K9 methyltransferases. For example, the fungal mycotoxin, chaetocin was identified in 2005 as the first H3K9 methyltransferase inhibitor, however this was later found to be not selective. Subsequent studies have identified several new small molecules capable of inhibiting G9a/GLP, including the non-SAM competitive G9a inhibitors BIX-01294, UNC0638 and others (reviewed in [221]). While molecules like UNC0638 have shown efficacy in delaying cancer in mouse models [105], no H3K9 methyltransferase inhibitors are currently in clinical trials. However, as we have noted, studies have also revealed tumor suppressive roles for H3K9 methyltransferases in various cancers and the multi-faceted role of H3K9 methyltransferases is summarized in Figure 5. The dual nature of these proteins suggests tight regulation of H3K9 methyltransferase expression in different cellular and temporal contexts. This also raises the possibility of cell specific roles for H3K9 methyltransferases in different tissues that may reflect their pro- and anti-tumorigenic functions in cancer. As such, care must be taken when targeting H3K9 methyltransferases in tumors to eliminate the possibility of promoting carcinogenesis in other tissues. Thus, a thorough understanding of the role of H3K9 methyltransferases in different tumor environments is essential before small molecule inhibition is pursued as a cancer therapy. This is reinforced by a recent publication showing an early response of melanoma cells to G9a inhibitors only to result in cancer stem cell enrichment and aggressive carcinoma with prolonged latency [222]. Besides tumorigenic and tumor suppressive roles, H3K9 methyltransferases also impact resistance to chemo and radiation therapy. Given our understanding of pro- and anti-tumorigenic roles in cancer, it will be imperative to investigate the optimal levels of H3K9 methylation in disease states. Further, it will be interesting to understand the molecular mechanisms (transcriptional and/or post-transcriptional) utilized by cancer stem cells to maintain (or modulate) H3K9 methylation to promote self-renewal or evade drug treatment. Finally, thorough testing of H3K9 methyltransferase inhibitors in different disease models will be necessary to fully explore the long term effects of H3K9 methylation inhibition on stem and progenitor cell populations. Thus, H3K9 methylation function spans large modalities of cancer biology, which await further investigation.
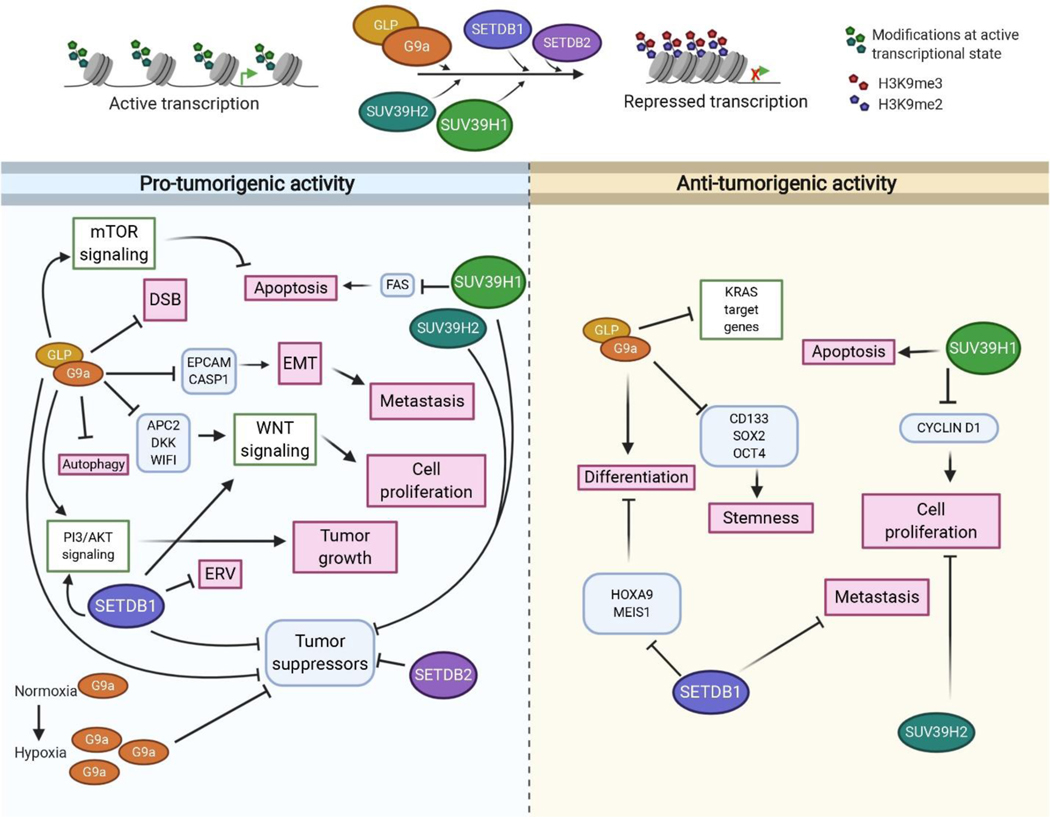
Figure 5.
Graphical illustration summarizing the oncogenic and tumor-suppressive functions of H3K9 methyltransferases. Signaling pathways are shown in green boxes; Genes regulated by H3K9 methyltransferases are shown in blue boxes and affected processes are shown in pink boxes. The bubbles representing H3K9 methyltransferases are not to scale. Created with BioRender.com
Acknowledgements
This study was supported by grants awarded by National Institutes of Health to A.G.M (R01-HL-136420). We thank Dr. James Ropa for critical reading of the manuscript.
Abbreviations
- CRC
Colorectal Carcinoma
- 5-FU
5-Fluorouracil
- GC
Gastric carcinoma
- MITF
Micro thalmia associated transcription factor
- EMT
Epithelial to meshencymal transition
- HCC
Hepatocellular carcinoma
- AML
Acute myeloid leukemia
- CLL
Chronic lymphocytic leukemia
- DSB
DNA double stranded breaks
- ALL
Acute lymphocytic leukemia
- IDTC
Induced Drug tolerance cells
- DTP
Drug tolerant persisters
- HGSOC
High-grade serous cell ovarian carcinoma
- PARPi
PARP inhibitors
- PTM
Post-translational modification
- GEM
Gemcitabine
- HNSCC
Head and neck squamous cell carcinoma
- LINE
Long Interspersed Nuclear Elements
- LOCKS
Large organized chromatin K9 modifications
- Cys
Cysteine rich
- MBD
Methy-CpG binding domain
Footnotes
Declaration of Competing Interest
Authors declare no competing interest
Declaration of Competing Interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
☐ The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Eissenberg JC, Elgin SCR. HP1a: a structural chromosomal protein regulating transcription. Trends Genet. 2014;30:103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ninova M, Fejes Tóth K, Aravin AA. The control of gene expression and cell identity by H3K9 trimethylation. Development. 2019;146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mozzetta C, Boyarchuk E, Pontis J, Ait-Si-Ali S. Sound of silence: the properties and functions of repressive Lys methyltransferases. Nat. Rev. Mol. Cell Biol. 2015;16:499–513. [DOI] [PubMed] [Google Scholar]
- 4.Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. [DOI] [PubMed] [Google Scholar]
- 5.Grewal SIS, Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. [DOI] [PubMed] [Google Scholar]
- 6.Wellinger RJ, Zakian VA. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics. 2012;191:1073–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellahi A, Thurtle DM, Rine J. The Chromatin and Transcriptional Landscape of Native Saccharomy cescerevisiae Telomeres and Subtelomeric Domains. Genetics. 2015;200:505–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehnertz B, Ueda Y, Derijck AAHA, Braunschweig U, Perez-Burgos L, Kubicek S, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. [DOI] [PubMed] [Google Scholar]
- 9.Smallwood A, Estève P-O, Pradhan S, Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 2007;21:1169–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, Reuter G. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3–9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994;13:3822–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Carroll D, Scherthan H, Peters AH, Opravil S, Haynes AR, Laible G, et al. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol. Cell. Biol. 2000;20:9423–9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3–9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev.2011;25:781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu P-F, Zhu K-Y, Jin Y, Chen Y, Sun X-J, Deng M, et al. Setdb2 restricts dorsal organizer territory and regulates left-right asymmetry through suppressing fgf8 activity. Proc. Natl. Acad. Sci. USA. 2010;107:2521–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang Y, Zhang X, Horton JR, Upadhyay AK, Spannhoff A, Liu J, et al. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nat. Struct. Mol. Biol. 2009;16:312–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bittencourt D, Lee BH, Gao L, Gerke DS, Stallcup MR. Role of distinct surfaces of the G9a ankyrin repeat domain in histone and DNA methylation during embryonic stem cell self-renewal and differentiation. Epigenetics Chromatin. 2014;7:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milner CM, Campbell RD. The G9a gene in the human major histocompatibility complex encodes a novel protein containing ankyrin-like repeats. Biochem. J. 1993;290 ( Pt 3):811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 2001;276:25309–25317. [DOI] [PubMed] [Google Scholar]
- 21.Shankar SR, Bahirvani AG, Rao VK, Bharathy N, Ow JR, Taneja R. G9a, a multipotent regulator of gene expression. Epigenetics. 2013;8:16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Jiao X, Di Sante G, Ertel A, Casimiro MC, Wang M, et al. Cyclin D1 integrates G9a-mediated histone methylation. Oncogene. 2019;38:4232–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, et al. G9a-mediated irreversible epigeneticin activation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 2006;8:188–194. [DOI] [PubMed] [Google Scholar]
- 25.Rao VK, Ow JR, Shankar SR, Bharathy N, Manikandan J, Wang Y, et al. G9a promotes proliferation and inhibits cell cycle exit during myogenic differentiation. Nucleic Acids Res. 2016;44:8129–8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W, Wang H-Y, Zhao X, Duan H, Cheng B, Liu Y, et al. A methylation-phosphorylation switch determines Plk1 kinase activity and function in DNA damage repair. Sci. Adv. 2019;5:eaau7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ginjala V, Rodriguez-Colon L, Ganguly B, Gangidi P, Gallina P, Al-Hraishawi H, et al. Protein-lysine methyltransferases G9a and GLP1 promote responses to DNA damage. Sci. Rep. 2017;7:16613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Q, Zhu Q, Lu X, Du Y, Cao L, Shen C, et al. G9a coordinates with the RPA complex to promote DNA damage repair and cell survival. Proc. Natl. Acad. Sci. USA. 2017;114:E6054–E6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gauchier M, Kan S, Barral A, Sauzet S, Agirre E, Bonnell E, et al. SETDB1-dependent heterochromatin stimulates alternative lengthening of telomeres. Sci. Adv. 2019;5:eaav3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torrano J, Al Emran A, Hammerlindl H, Schaider H. Emerging roles of H3K9me3, SETDB1 and SETDB2 in therapy-induced cellular reprogramming. Clin. Epigenetics. 2019;11:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsumura Y, Nakaki R, Inagaki T, Yoshida A, Kano Y, Kimura H, et al. H3K4/H3K9me3 Bivalent Chromatin Domains Targeted by Lineage-Specific DNA Methylation Pauses Adipocyte Differentiation. Mol. Cell. 2015;60:584–596. [DOI] [PubMed] [Google Scholar]
- 32.Jurkowska RZ, Qin S, Kungulovski G, Tempel W, Liu Y, Bashtrykov P, et al. H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1. Nat. Commun. 2017;8:2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price AJ, Manjegowda MC, Kain J, Anandh S, Bochkis IM. Hdac3, Setdb1, and Kap1 mark H3K9me3/H3K14ac bivalent regions in young and aged liver. Aging Cell. 2020;19:e13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roloff TC, Ropers HH, Nuber UA. Comparative study of methyl-CpG-binding domain proteins. BMC Genomics. 2003;4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mader P, Mendoza-Sanchez R, Iqbal A, Dong A, Dobrovetsky E, Corless VB, et al. Identification and characterization of the first fragment hits for SETDB1 Tudor domain. Bioorg. Med. Chem. 2019;27:3866–3878. [DOI] [PubMed] [Google Scholar]
- 36.Robbez-Masson L, Tie CHC, Conde L, Tunbak H, Husovsky C, Tchasovnikarova IA, et al. The HUSH complex cooperates with TRIM28 to repress young retrotransposons and new genes. Genome Res. 2018;28:836–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho S, Park JS, Kang Y-K. Dual functions of histone-lysine N-methyltransferase Setdb1 protein at promyelocytic leukemia-nuclear body (PML-NB): maintaining PML-NB structure and regulating the expression of its associated genes. J. Biol. Chem. 2011;286:41115–41124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koide S, Oshima M, Takubo K, Yamazaki S, Nitta E, Saraya A, et al. Setdb1 maintains hematopoietic stem and progenitor cells by restricting the ectopic activation of nonhematopoietic genes. Blood. 2016;128:638–649. [DOI] [PubMed] [Google Scholar]
- 39.Dodge JE, Kang Y-K, Beppu H, Lei H, Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell. Biol. 2004;24:2478–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan L, Nishi M, Ohtsuka S-T, Matsui T, Takemoto K, Kamio-Miura A, et al. Essential roles of the histone methyltransferase ESET in the epigenetic control of neural progenitor cells during development. Development. 2012;139:3806–3816. [DOI] [PubMed] [Google Scholar]
- 41.Liu S, Brind’Amour J, Karimi MM, Shirane K, Bogutz A, Lefebvre L, et al. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev. 2014;28:2041–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leung D, Du T, Wagner U, Xie W, Lee AY, Goyal P, et al. Regulation of DNA methylation turnover at LTR retrotransposons and imprinted loci by the histone methyltransferase Setdb1. Proc. Natl. Acad. Sci. USA. 2014;111:6690–6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eymery A, Liu Z, Ozonov EA, Stadler MB, Peters AHFM. The methyltransferase Setdb1 is essential for meiosis and mitosis in mouse oocytes and early embryos. Development. 2016;143:2767–2779. [DOI] [PubMed] [Google Scholar]
- 44.Kim J, Zhao H, Dan J, Kim S, Hardikar S, Hollowell D, et al. Maternal setdb1 is required for meiotic progression and preimplantation development in mouse. PLoS Genet. 2016;12:e1005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. [DOI] [PubMed] [Google Scholar]
- 46.Du T-T, Xu P-F, Dong Z-W, Fan H-B, Jin Y, Dong M, et al. Setdb2 controls convergence and extension movements during zebrafish gastrulation by transcriptional regulation of dvr1. Dev. Biol. 2014;392:233–244. [DOI] [PubMed] [Google Scholar]
- 47.Falandry C, Fourel G, Galy V, Ristriani T, Horard B, Bensimon E, et al. CLLD8/KMT1F is a lysine methyltransferase that is important for chromosome segregation. J. Biol. Chem. 2010;285:20234–20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rao VK, Pal A, Taneja R. A drive in SUVs: From development to disease. Epigenetics. 2017;12:177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peters AHFM, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AAHA, Perez-Burgos L, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell. 2003;12:1577–1589. [DOI] [PubMed] [Google Scholar]
- 50.Chin HG, Patnaik D, Estève P-O, Jacobsen SE, Pradhan S. Catalytic properties and kinetic mechanism of human recombinant Lys-9 histone H3 methyltransferase SUV39H1: participation of the chromodomain in enzymatic catalysis. Biochemistry. 2006;45:3272–3284. [DOI] [PubMed] [Google Scholar]
- 51.Wang T, Xu C, Liu Y, Fan K, Li Z, Sun X, et al. Crystal structure of the human SUV39H1 chromodomain and its recognition of histone H3K9me2/3. PLoS One. 2012;7:e52977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. [DOI] [PubMed] [Google Scholar]
- 53.Keenan CR, Iannarella N, Naselli G, Bediaga NG, Johanson TM, Harrison LC, et al. Extreme disruption of heterochromatin is required for accelerated haematopoietic aging. Blood. 2020; [DOI] [PubMed] [Google Scholar]
- 54.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suvà ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. [DOI] [PubMed] [Google Scholar]
- 57.Pangeni RP, Yang L, Zhang K, Wang J, Li W, Guo C, et al. G9a regulates tumorigenicity and stemness through genome-wide DNA methylation reprogramming in non-small cell lung cancer. Clin. Epigenetics. 2020;12:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones PA, Issa J-PJ, Baylin S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet.2016;17:630–641. [DOI] [PubMed] [Google Scholar]
- 59.Liu M, Thomas SL, DeWitt AK, Zhou W, Madaj ZB, Ohtani H, et al. Dual inhibition of DNA and histone methyltransferases increases viral mimicry in ovarian cancer cells. Cancer Res. 2018;78:5754–5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 2009;41:246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat. Rev. Cancer. 2013;13:497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho JC, Abdullah LN, Pang QY, Jha S, Chow EK-H, Yang H, et al. Inhibition of the H3K9 methyltransferase G9A attenuates oncogenicity and activates the hypoxia signaling pathway. PLoS One. 2017;12:e0188051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Casciello F, Windloch K, Gannon F, Lee JS. Functional role of g9a histone methyltransferase in cancer. Front. Immunol. 2015;6:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu X-R, Zhou L-H, Hu J-X, Liu L-M, Wan H-P, Zhang X-Q. UNC0638, a G9a inhibitor, suppresses epithelial mesenchymal transition mediated cellular migration and invasion in triple negative breast cancer. Mol. Med. Rep. 2018;17:2239–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tu WB, Shiah Y-J, Lourenco C, Mullen PJ, Dingar D, Redel C, et al. MYC Interacts with the G9a Histone Methyltransferase to Drive Transcriptional Repression and Tumorigenesis. Cancer Cell. 2018;34:579–595.e8. [DOI] [PubMed] [Google Scholar]
- 68.Camuzi D, de Amorim ÍSS, Ribeiro Pinto LF, Oliveira Trivilin L, Mencalha AL, Soares Lima SC. Regulation Is in the Air: The Relationship between Hypoxia and Epigenetics in Cancer. Cells. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen H, Yan Y, Davidson TL, Shinkai Y, Costa M. Hypoxic stress induces dimethylated histone H3 lysine 9 through histone methyltransferase G9a in mammalian cells. Cancer Res. 2006;66:9009–9016. [DOI] [PubMed] [Google Scholar]
- 70.Casciello F, Al-Ejeh F, Kelly G, Brennan DJ, Ngiow SF, Young A, et al. G9a drives hypoxia-mediated gene repression for breast cancer cell survival and tumorigenesis. Proc. Natl. Acad. Sci. USA. 2017;114:7077–7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun L, Fang J. Epigenetic regulation of epithelial-mesenchymal transition. Cell Mol. Life Sci. 2016;73:4493–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dong C, Wu Y, Yao J, Wang Y, Yu Y, Rychahou PG, et al. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J. Clin. Invest. 2012;122:1469–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dong C, Wu Y, Wang Y, Wang C, Kang T, Rychahou PG, et al. Interaction with Suv39H1 is critical for Snail-mediated E-cadherin repression in breast cancer. Oncogene. 2013;32:1351–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yokoyama Y, Hieda M, Nishioka Y, Matsumoto A, Higashi S, Kimura H, et al. Cancer-associated upregulation of histone H3 lysine 9 trimethylation promotes cell motility in vitro and drives tumor formation in vivo. Cancer Sci. 2013;104:889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khanal P, Kim G, Lim S-C, Yun H-J, Lee KY, Choi H-K, et al. Prolyl isomerase Pin1 negatively regulates the stability of SUV39H1 to promote tumorigenesis in breast cancer. FASEB J. 2013;27:4606–4618. [DOI] [PubMed] [Google Scholar]
- 76.Wu M, Fan B, Guo Q, Li Y, Chen R, Lv N, et al. Knockdown of SETDB1 inhibits breast cancer progression by miR-381–3p-related regulation. Biol. Res. 2018;51:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiao J-F, Sun Q-Y, Ding L-W, Chien W, Liu X-Y, Mayakonda A, et al. The c-MYC-BMI1 axis is essential for SETDB1-mediated breast tumourigenesis. J. Pathol. 2018;246:89–102. [DOI] [PubMed] [Google Scholar]
- 78.Ryu TY, Kim K, Kim S-K, Oh J-H, Min J-K, Jung C-R, et al. SETDB1 regulates SMAD7 expression for breast cancer metastasis. BMB Rep. 2019;52:139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang W, Su Y, Hou C, Chen L, Zhou D, Ren K, et al. SETDB1 induces epithelial mesenchymal transition in breast carcinoma by directly binding with Snail promoter. Oncol. Rep. 2019;41:1284–1292. [DOI] [PubMed] [Google Scholar]
- 80.Du D, Katsuno Y, Meyer D, Budi EH, Chen S-H, Koeppen H, et al. Smad3-mediated recruitment of the methyltransferase SETDB1/ESET controls Snail1 expression and epithelial-mesenchymal transition. EMBO Rep. 2018;19:135–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Watanabe H, Soejima K, Yasuda H, Kawada I, Nakachi I, Yoda S, et al. Deregulation of histone lysine methyltransferases contributes to oncogenic transformation of human bronchoepithelial cells. Cancer Cell Int. 2008;8:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen M-W, Hua K-T, Kao H-J, Chi C-C, Wei L-H, Johansson G, et al. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res. 2010;70:7830–7840. [DOI] [PubMed] [Google Scholar]
- 83.Cheng C-C, Chang J, Huang SC-C, Lin H-C, Ho A-S, Lim K-H, et al. YM155 as an inhibitor of cancer stemness simultaneously inhibits autophosphorylation of epidermal growth factor receptor and G9amediated stemness in lung cancer cells. PLoS One. 2017;12:e0182149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang K, Wang J, Yang L, Yuan Y-C, Tong TR, Wu J, et al. Targeting histone methyltransferase G9a-inhibits growth and Wnt signaling pathway by epigenetically regulating HP1α and APC2 gene expression in non-small cell lung cancer. Mol. Cancer. 2018;17:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang T, Zhang P, Li W, Zhao T, Zhang Z, Chen S, et al. G9A promotes tumor cell growth and invasion by silencing CASP1 in non-small-cell lung cancer cells. Cell Death Dis. 2017;8:e2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rowbotham SP, Li F, Dost AFM, Louie SM, Marsh BP, Pessina P, et al. H3K9 methyltransferases and demethylases control lung tumor-propagating cells and lung cancer progression. Nat. Commun. 2018;9:4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zheng Y, Li B, Wang J, Xiong Y, Wang K, Qi Y, et al. Identification of SUV39H2 as a potential oncogene in lung adenocarcinoma. Clin. Epigenetics. 2018;10:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vougiouklakis T, Saloura V, Park J-H, Takamatsu N, Miyamoto T, Nakamura Y, et al. Development of novel SUV39H2 inhibitors that exhibit growth suppressive effects in mouse xenograft models and regulate the phosphorylation of H2AX. Oncotarget. 2018;9:31820–31831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim SY, Hong M, Heo S-H, Park S, Kwon TK, Sung YH, et al. Inhibition of euchromatin histone-lysineN-methyltransferase 2 sensitizes breast cancer cells to tumor necrosis factor-related apoptosis-inducing ligand through reactive oxygen species-mediated activating transcription factor 4-C/EBP homologous protein-death receptor 5 pathway activation. Mol. Carcinog. 2018;57:1492–1506. [DOI] [PubMed] [Google Scholar]
- 90.Zhang J, He P, Xi Y, Geng M, Chen Y, Ding J. Down-regulation of G9a triggers DNA damage response and inhibits colorectal cancer cells proliferation. Oncotarget. 2015;6:2917–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yin C, Ke X, Zhang R, Hou J, Dong Z, Wang F, et al. G9a promotes cell proliferation and suppresses autophagy in gastric cancer by directly activating mTOR. FASEB J. 2019;33:14036–14050. [DOI] [PubMed] [Google Scholar]
- 92.Lee SH, Kim J, Kim WH, Lee YM. Hypoxic silencing of tumor suppressor RUNX3 by histone modification in gastric cancer cells. Oncogene. 2009;28:184–194. [DOI] [PubMed] [Google Scholar]
- 93.Yokoyama M, Chiba T, Zen Y, Oshima M, Kusakabe Y, Noguchi Y, et al. Histone lysine methyltransferase G9a is a novel epigenetic target for the treatment of hepatocellular carcinoma. Oncotarget. 2017;8:21315–21326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wei L, Chiu DK-C, Tsang FH-C, Law C-T, Cheng CL-H, Au SL-K, et al. Histone methyltransferase G9a promotes liver cancer development by epigenetic silencing of tumor suppressor gene RARRES3. J. Hepatol. 2017;67:758–769. [DOI] [PubMed] [Google Scholar]
- 95.Hu Y, Zheng Y, Dai M, Wang X, Wu J, Yu B, et al. G9a and histone deacetylases are crucial for Snail2-mediated E-cadherin repression and metastasis in hepatocellular carcinoma. Cancer Sci. 2019;110:3442–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ma W, Han C, Zhang J, Song K, Chen W, Kwon H, et al. The Histone Methyltransferase G9a Promotes Cholangiocarcinogenesis through Regulation of the Hippo Pathway Kinase LATS2 and YAP Signaling Pathway. Hepatology. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cao Y-P, Sun J-Y, Li M-Q, Dong Y, Zhang Y-H, Yan J, et al. Inhibition of G9a by a small molecule inhibitor, UNC0642, induces apoptosis of human bladder cancer cells. Acta Pharmacol Sin. 2019;40:1076–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kato S, Weng QY, Insco ML, Chen KY, Muralidhar S, Pozniak J, et al. Gain-of-function genetic alterations of G9a drive oncogenesis. Cancer Discov. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dang N-N, Jiao J, Meng X, An Y, Han C, Huang S. Abnormal overexpression of G9a in melanoma cells promotes cancer progression via upregulation of the Notch1 signaling pathway. Aging (Albany, NY). 2020;12:2393–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hua K-T, Wang M-Y, Chen M-W, Wei L-H, Chen C-K, Ko C-H, et al. The H3K9 methyltransferase G9a is a marker of aggressive ovarian cancer that promotes peritoneal metastasis. Mol. Cancer. 2014;13:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guo A-S, Huang Y-Q, Ma X-D, Lin R-S. Mechanism of G9a inhibitor BIX 01294 acting on U251 glioma cells. Mol. Med. Rep. 2016;14:4613–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ke X-X, Zhang D, Zhu S, Xia Q, Xiang Z, Cui H. Inhibition of H3K9 methyltransferase G9a repressed cell proliferation and induced autophagy in neuroblastoma cells. PLoS One. 2014;9:e106962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ren A, Qiu Y, Cui H, Fu G. Inhibition of H3K9 methyltransferase G9a induces autophagy and apoptosis in oral squamous cell carcinoma. Biochem. Biophys. Res. Commun. 2015;459:10–17. [DOI] [PubMed] [Google Scholar]
- 104.Liu S, Ye D, Guo W, Yu W, He Y, Hu J, et al. G9a is essential for EMT-mediated metastasis and maintenance of cancer stem cell-like characters in head and neck squamous cell carcinoma. Oncotarget. 2015;6:6887–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lehnertz B, Pabst C, Su L, Miller M, Liu F, Yi L, et al. The methyltransferase G9a regulates HoxA9-dependent transcription in AML. Genes Dev. 2014;28:317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huang Y, Zou Y, Lin L, Ma X, Huang X. Effect of BIX-01294 on proliferation, apoptosis and histone methylation of acute T lymphoblastic leukemia cells. Leuk. Res. 2017;62:34–39. [DOI] [PubMed] [Google Scholar]
- 107.Cruz-Tapias P, Zakharova V, Perez-Fernandez OM, Mantilla W, RamÍRez-Clavijo S, Ait-Si-Ali S. Expression of the Major and Pro-Oncogenic H3K9 Lysine Methyltransferase SETDB1 in Non-Small Cell Lung Cancer. Cancers (Basel). 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sun Q-Y, Ding L-W, Xiao J-F, Chien W, Lim S-L, Hattori N, et al. SETDB1 accelerates tumourigenesis by regulating the WNT signalling pathway. J. Pathol. 2015;235:559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen K, Zhang F, Ding J, Liang Y, Zhan Z, Zhan Y, et al. Histone methyltransferase SETDB1 promotes the progression of colorectal cancer by inhibiting the expression of TP53. J. Cancer. 2017;8:3318–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cao N, Yu Y, Zhu H, Chen M, Chen P, Zhuo M, et al. SETDB1 promotes the progression of colorectal cancer via epigenetically silencing p21 expression. Cell Death Dis. 2020;11:351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fei Q, Shang K, Zhang J, Chuai S, Kong D, Zhou T, et al. Histone methyltransferase SETDB1 regulates liver cancer cell growth through methylation of p53. Nat. Commun. 2015;6:8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wong C-M, Wei L, Law C-T, Ho DW-H, Tsang FH-C, Au SL-K, et al. Up-regulation of histone methyltransferase SETDB1 by multiple mechanisms in hepatocellular carcinoma promotes cancer metastasis. Hepatology. 2016;63:474–487. [DOI] [PubMed] [Google Scholar]
- 113.Zhang Y, Huang J, Li Q, Chen K, Liang Y, Zhan Z, et al. Histone methyltransferase SETDB1 promotes cells proliferation and migration by interacting withTiam1 in hepatocellular carcinoma. BMC Cancer. 2018;18:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kostaki M, Manona AD, Stavraka I, Korkolopoulou P, Levidou G, Trigka E-A, et al. High-frequency p16(INK) (4A) promoter methylation is associated with histone methyltransferase SETDB1 expression in sporadic cutaneous melanoma. Exp. Dermatol. 2014;23:332–338. [DOI] [PubMed] [Google Scholar]
- 116.Cuellar TL, Herzner A-M, Zhang X, Goyal Y, Watanabe C, Friedman BA, et al. Silencing of retrotransposons by SETDB1 inhibits the interferon response in acute myeloid leukemia. J. Cell Biol. 2017;216:3535–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ropa J, Saha N, Hu H, Peterson LF, Talpaz M, Muntean AG. SETDB1 mediated histone H3 lysine 9 methylation suppresses MLL-fusion target expression and leukemic transformation. Haematologica. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sun Y, Wei M, Ren S-C, Chen R, Xu W-D, Wang F-B, et al. Histone methyltransferase SETDB1 is required for prostate cancer cell proliferation, migration and invasion. Asian J Androl. 2014;16:319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nishikawaji T, Akiyama Y, Shimada S, Kojima K, Kawano T, Eishi Y, et al. Oncogenic roles of the SETDB2 histone methyltransferase in gastric cancer. Oncotarget. 2016;7:67251–67265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mu G, Chen F. Oncogenic Roles Of A Histone Methyltransferase SETDB2 In AML1-ETO Positive AML. Cancer Manag Res. 2020;12:783–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lin C-H, Wong SH-K, Kurzer JH, Schneidawind C, Wei MC, Duque-Afonso J, et al. SETDB2 Links E2A-PBX1 to Cell-Cycle Dysregulation in Acute Leukemia through CDKN2C Repression. Cell Rep. 2018;23:1166–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lu C, Klement JD, Yang D, Albers T, Lebedyeva IO, Waller JL, et al. SUV39H1 regulates human colon carcinoma apoptosis and cell cycle to promote tumor growth. Cancer Lett. 2020;476:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cai L, Ma X, Huang Y, Zou Y, Chen X. Aberrant histone methylation and the effect of Suv39H1 siRNA on gastric carcinoma. Oncol. Rep. 2014;31:2593–2600. [DOI] [PubMed] [Google Scholar]
- 124.Altieri F, Di Stadio CS, Federico A, Miselli G, De Palma M, Rippa E, et al. Epigenetic alterations of gastrokine 1 gene expression in gastric cancer. Oncotarget. 2017;8:16899–16911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fan DN-Y, Tsang FH-C, Tam AH-K, Au SL-K, Wong CC-L, Wei L, et al. Histone lysine methyltransferase, suppressor of variegation 3–9 homolog 1, promotes hepatocellular carcinoma progression and is negatively regulated by microRNA-125b. Hepatology. 2013;57:637–647. [DOI] [PubMed] [Google Scholar]
- 126.Kim G, Kim J-Y, Lim S-C, Lee KY, Kim O, Choi HS. SUV39H1/DNMT3A-dependent methylation of the RB1 promoter stimulates PIN1 expression and melanoma development. FASEB J. 2018;32:5647–5660. [DOI] [PubMed] [Google Scholar]
- 127.Yu T, Wang C, Yang J, Guo Y, Wu Y, Li X. Metformin inhibits SUV39H1-mediated migration of prostate cancer cells. Oncogenesis. 2017;6:e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shuai W, Wu J, Chen S, Liu R, Ye Z, Kuang C, et al. SUV39H2 promotes colorectal cancer proliferation and metastasis via tri-methylation of the SLIT1 promoter. Cancer Lett. 2018;422:56–69. [DOI] [PubMed] [Google Scholar]
- 129.Li Q, Wang X, Lu Z, Zhang B, Guan Z, Liu Z, et al. Polycomb CBX7 directly controls trimethylation of histone H3 at lysine 9 at the p16 locus. PLoS One. 2010;5:e13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tao H, Li H, Su Y, Feng D, Wang X, Zhang C, et al. Histone methyltransferase G9a and H3K9 dimethylation inhibit the self-renewal of glioma cancer stem cells. Mol. Cell. Biochem. 2014;394:23–30. [DOI] [PubMed] [Google Scholar]
- 131.Kang HC, Kim HK, Lee S, Mendez P, Kim JW, Woodard G, et al. Whole exome and targeted deep sequencing identify genome-wide allelic loss and frequent SETDB1 mutations in malignant pleural mesotheliomas. Oncotarget. 2016;7:8321–8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hmeljak J, Sanchez-Vega F, Hoadley KA, Shih J, Stewart C, Heiman D, et al. Integrative molecular characterization of malignant pleural mesothelioma. Cancer Discov. 2018;8:1548–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wu P-C, Lu J-W, Yang J-Y, Lin I-H, Ou D-L, Lin Y-H, et al. H3K9 histone methyltransferase, KMT1E/SETDB1, cooperates with the SMAD2/3 pathway to suppress lung cancer metastasis. Cancer Res. 2014;74:7333–7343. [DOI] [PubMed] [Google Scholar]
- 134.Ropa J, Saha N, Chen Z, Serio J, Chen W, Mellacheruvu D, et al. PAF1 complex interactions with SETDB1 mediate promoter H3K9 methylation and transcriptional repression of Hoxa9 and Meis1 in acute myeloid leukemia. Oncotarget. 2018;9:22123–22136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rodrigues C, Pattabiraman C, Vijaykumar A, Arora R, Narayana SM, Kumar RV, et al. A SUV39H1-low chromatin state characterises and promotes migratory properties of cervical cancer cells. Exp. Cell Res. 2019;378:206–216. [DOI] [PubMed] [Google Scholar]
- 136.Yang Y-J, Han J-W, Youn H-D, Cho E-J. The tumor suppressor, parafibromin, mediates histone H3 K9 methylation for cyclin D1 repression. Nucleic Acids Res. 2010;38:382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chu Y, Chen Y, Guo H, Li M, Wang B, Shi D, et al. SUV39H1 regulates the progression of MLL-AF9-induced acute myeloid leukemia. Oncogene. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Albacker CE, Storer NY, Langdon EM, Dibiase A, Zhou Y, Langenau DM, et al. The histone methyltransferase SUV39H1 suppresses embryonal rhabdomyosarcoma formation in zebrafish. PLoS One. 2013;8:e64969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim A-R, Sung JY, Rho SB, Kim Y-N, Yoon K. Suppressor of Variegation 3–9 Homolog 2, a Novel Binding Protein of Translationally Controlled Tumor Protein, Regulates Cancer Cell Proliferation. Biomol. Ther. (Seoul). 2019;27:231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu X, Guo S, Liu X, Su L. Chaetocin induces endoplasmic reticulum stress response and leads to death receptor 5-dependent apoptosis in human non-small cell lung cancer cells. Apoptosis. 2015;20:1499–1507. [DOI] [PubMed] [Google Scholar]
- 141.Lafuente-Sanchis A, Zúñiga Á, Galbis JM, Cremades A, Estors M, Martínez-Hernández NJ, et al. Prognostic value of ERCC1, RRM1, BRCA1 and SETDB1 in early stage of non-small cell lung cancer. Clin Transl Oncol. 2016;18:798–804. [DOI] [PubMed] [Google Scholar]
- 142.Inoue Y, Matsuura S, Kurabe N, Kahyo T, Mori H, Kawase A, et al. Clinicopathological and Survival Analysis of Japanese Patients with Resected Non-Small-Cell Lung Cancer Harboring NKX2–1, SETDB1, MET, HER2, SOX2, FGFR1, or PIK3CA Gene Amplification. J. Thorac. Oncol. 2015;10:1590–1600. [DOI] [PubMed] [Google Scholar]
- 143.Rodriguez-Paredes M, Martinez de Paz A, Simó-Riudalbas L, Sayols S, Moutinho C, Moran S, et al. Gene amplification of the histone methyltransferase SETDB1 contributes to human lung tumorigenesis. Oncogene. 2014;33:2807–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gong M, Liu J, Sakurai R, Corre A, Anthony S, Rehan VK. Perinatal nicotine exposure suppresses PPARγ epigenetically in lung alveolar interstitial fibroblasts. Mol. Genet. Metab. 2015;114:604–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Reddy AT, Lakshmi SP, Reddy RC. Pparγ as a novel therapeutic target in lung cancer. PPAR Res. 2016;2016:8972570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Qin J, Zeng Z, Luo T, Li Q, Hao Y, Chen L. Clinicopathological significance of G9A expression in colorectal carcinoma. Oncol. Lett. 2018;15:8611–8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Paschall AV, Yang D, Lu C, Choi J-H, Li X, Liu F, et al. H3K9 Trimethylation Silences Fas Expression To Confer Colon Carcinoma Immune Escape and 5-Fluorouracil Chemoresistance. J. Immunol. 2015;195:1868–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang J, Wang Y, Shen Y, He P, Ding J, Chen Y. G9a stimulates CRC growth by inducing p53 Lys373 dimethylation-dependent activation of Plk1. Theranostics. 2018;8:2884–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Luo C-W, Wang J-Y, Hung W-C, Peng G, Tsai Y-L, Chang T-M, et al. G9a governs colon cancer stem cell phenotype and chemoradioresistance through PP2A-RPA axis-mediated DNA damage response. Radiother. Oncol. 2017;0. [DOI] [PubMed] [Google Scholar]
- 150.Yan Y, Cao PT, Greer PM, Nagengast ES, Kolb RH, Mumby MC, et al. Protein phosphatase 2A has an essential role in the activation of gamma-irradiation-induced G2/M checkpoint response. Oncogene. 2010;29:4317–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chae Y-C, Kim J-Y, Park JW, Kim K-B, Oh H, Lee K-H, et al. FOXO1 degradation via G9a-mediated methylation promotes cell proliferation in colon cancer. Nucleic Acids Res. 2019;47:1692–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sträter J, Hinz U, Hasel C, Bhanot U, Mechtersheimer G, Lehnert T, et al. Impaired CD95 expression predisposes for recurrence in curatively resected colon carcinoma: clinical evidence for immunoselection and CD95L mediated control of minimal residual disease. Gut. 2005;54:661–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Owen-Schaub LB. Fas function and tumor progression: use it and lose it. Cancer Cell. 2002;2:95–96. [DOI] [PubMed] [Google Scholar]
- 154.Lu C, Yang D, Klement JD, Oh IK, Savage NM, Waller JL, et al. SUV39H1 Represses the Expression of Cytotoxic T-Lymphocyte Effector Genes to Promote Colon Tumor Immune Evasion. Cancer Immunol Res. 2019;7:414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ho Y-J, Lin Y-M, Huang Y-C, Chang J, Yeh K-T, Lin L-I, et al. Significance of histone methyltransferase SETDB1 expression in colon adenocarcinoma. APMIS. 2017;125:985–995. [DOI] [PubMed] [Google Scholar]
- 156.Lu Y, Feng Y, Yi-Yi L, Yi-Zhi Z, Yang L, Hong-Mei Y, et al. Histone Methyltransferase SETDB1 Promotes Colorectal Cancer Proliferation through the STAT1-CCND1/CDK6 Axis. Carcinogenesis. 2019; [DOI] [PubMed] [Google Scholar]
- 157.Lee S, Lee C, Hwang CY, Kim D, Han Y, Hong SN, et al. Network Inference Analysis Identifies SETDB1 as a Key Regulator for Reverting Colorectal Cancer Cells into Differentiated Normal-Like Cells. Mol. Cancer Res. 2020;18:118–129. [DOI] [PubMed] [Google Scholar]
- 158.Hou Z, Sun L, Xu F, Hu F, Lan J, Song D, et al. Blocking histone methyltransferase SETDB1 inhibits tumorigenesis and enhances cetuximab sensitivity in colorectal cancer. Cancer Lett. 2020; [DOI] [PubMed] [Google Scholar]
- 159.Lin X, Huang Y, Zou Y, Chen X, Ma X. Depletion of G9a gene induces cell apoptosis in human gastric carcinoma. Oncol. Rep. 2016;35:3041–3049. [DOI] [PubMed] [Google Scholar]
- 160.Hu L, Zang M, Wang H-X, Zhang B-G, Wang Z-Q, Fan Z-Y, et al. G9A promotes gastric cancer metastasis by upregulating ITGB3 in a SET domain-independent manner. Cell Death Dis. 2018;9:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kim TW, Lee SY, Kim M, Cheon C, Ko S-G. Kaempferol induces autophagic cell death via IRE1-JNK-CHOP pathway and inhibition of G9a in gastric cancer cells. Cell Death Dis. 2018;9:875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Inamdar GS, Madhunapantula SV, Robertson GP. Targeting the MAPK pathway in melanoma: why some approaches succeed and other fail. Biochem. Pharmacol. 2010;80:624–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, et al. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 2005;353:2135–2147. [DOI] [PubMed] [Google Scholar]
- 164.Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68:664–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shi X, Tasdogan A, Huang F, Hu Z, Morrison SJ, DeBerardinis RJ. The abundance of metabolites related to protein methylation correlates with the metastatic capacity of human melanoma xenografts. Sci. Adv. 2017;3:eaao5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Orouji E, Federico A, Larribère L, Novak D, Lipka DB, Assenov Y, et al. Histone methyltransferase SETDB1 contributes to melanoma tumorigenesis and serves as a new potential therapeutic target. Int. J. Cancer. 2019;145:3462–3477. [DOI] [PubMed] [Google Scholar]
- 167.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. [DOI] [PubMed] [Google Scholar]
- 168.Qin J, Li Q, Zeng Z, Wu P, Jiang Y, Luo T, et al. Increased expression of G9A contributes to carcinogenesis and indicates poor prognosis in hepatocellular carcinoma. Oncol. Lett. 2018;15:9757–9765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.DiSepio D, Ghosn C, Eckert RL, Deucher A, Robinson N, Duvic M, et al. Identification and characterization of a retinoid-induced class II tumor suppressor/growth regulatory gene. Proc. Natl. Acad. Sci. USA. 1998;95:14811–14815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Shao Y, Song X, Jiang W, Chen Y, Ning Z, Gu W, et al. MicroRNA-621 Acts as a Tumor Radiosensitizer by Directly Targeting SETDB1 in Hepatocellular Carcinoma. Mol. Ther. 2019;27:355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chiba T, Saito T, Yuki K, Zen Y, Koide S, Kanogawa N, et al. Histone lysine methyltransferase SUV39H1 is a potent target for epigenetic therapy of hepatocellular carcinoma. Int. J. Cancer. 2015;136:289–298. [DOI] [PubMed] [Google Scholar]
- 172.Ntziachristos P, Abdel-Wahab O, Aifantis I. Emerging concepts of epigenetic dysregulation in hematological malignancies. Nat. Immunol. 2016;17:1016–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Abdel-Wahab O, Levine RL. Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia. Blood. 2013;121:3563–3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gallipoli P, Huntly BJP. Novel epigenetic therapies in hematological malignancies: Current status and beyond. Semin. Cancer Biol. 2018;51:198–210. [DOI] [PubMed] [Google Scholar]
- 175.Kondengaden SM, Luo L-F, Huang K, Zhu M, Zang L, Bataba E, et al. Discovery of novel small molecule inhibitors of lysine methyltransferase G9a and their mechanism in leukemia cell lines. Eur. J. Med. Chem. 2016;122:382–393. [DOI] [PubMed] [Google Scholar]
- 176.Pappano WN, Guo J, He Y, Ferguson D, Jagadeeswaran S, Osterling DJ, et al. The Histone Methyltransferase Inhibitor A-366 Uncovers a Role for G9a/GLP in the Epigenetics of Leukemia. PLoS One. 2015;10:e0131716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.San José-Enériz E, Agirre X, Rabal O, Vilas-Zornoza A, Sanchez-Arias JA, Miranda E, et al. Discovery of first-in-class reversible dual small molecule inhibitors against G9a and DNMTs in hematological malignancies. Nat. Commun. 2017;8:15424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Jang JE, Eom J-I, Jeung H-K, Chung H, Kim YR, Kim JS, et al. PERK/NRF2 and autophagy form a resistance mechanism against G9a inhibition in leukemia stem cells. J Exp Clin Cancer Res. 2020;39:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Alves-Silva JC, de Carvalho JL, Rabello DA, Serejo TRT, Rego EM, Neves FAR, et al. GLP overexpression is associated with poor prognosis in Chronic Lymphocytic Leukemia and its inhibition induces leukemic cell death. Invest. New Drugs. 2018;36:955–960. [DOI] [PubMed] [Google Scholar]
- 180.Zhang XY, Rajagopalan D, Chung T-H, Hooi L, Toh TB, Tian JS, et al. Frequent upregulation of G9a promotes RelB-dependent proliferation and survival in multiple myeloma. Exp Hematol Oncol. 2020;9:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lai YS, Chen JY, Tsai HJ, Chen TY, Hung WC. The SUV39H1 inhibitor chaetocin induces differentiation and shows synergistic cytotoxicity with other epigenetic drugs in acute myeloid leukemia cells. Blood Cancer J. 2015;5:e313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Parker H, Rose-Zerilli MJJ, Parker A, Chaplin T, Wade R, Gardiner A, et al. 13q deletion anatomy and disease progression in patients with chronic lymphocytic leukemia. Leukemia. 2011;25:489–497. [DOI] [PubMed] [Google Scholar]
- 183.Tran HTT, Kim HN, Lee I-K, Nguyen-Pham T-N, Ahn J-S, Kim Y-K, et al. Improved therapeutic effect against leukemia by a combination of the histone methyltransferase inhibitor chaetocin and the histone deacetylase inhibitor trichostatin A. J. Korean Med. Sci. 2013;28:237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chaib H, Nebbioso A, Prebet T, Castellano R, Garbit S, Restouin A, et al. Anti-leukemia activity of chaetocin via death receptor-dependent apoptosis and dual modulation of the histone methyl-transferase SUV39H1. Leukemia. 2012;26:662–674. [DOI] [PubMed] [Google Scholar]
- 185.Lakshmikuttyamma A, Scott SA, DeCoteau JF, Geyer CR. Reexpression of epigenetically silenced AML tumor suppressor genes by SUV39H1 inhibition. Oncogene. 2010;29:576–588. [DOI] [PubMed] [Google Scholar]
- 186.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chen C-W, Koche RP, Sinha AU, Deshpande AJ, Zhu N, Eng R, et al. DOT1L inhibits SIRT1-mediated epigenetic silencing to maintain leukemic gene expression in MLL-rearranged leukemia. Nat. Med. 2015;21:335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Chen X, Skutt-Kakaria K, Davison J, Ou Y-L, Choi E, Malik P, et al. G9a/GLP-dependent histone H3K9me2 patterning during human hematopoietic stem cell lineage commitment. Genes Dev. 2012;26:2499–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ugarte F, Sousae R, Cinquin B, Martin EW, Krietsch J, Sanchez G, et al. Progressive chromatin condensation and H3K9 methylation regulate the differentiation of embryonic and hematopoietic stem cells. Stem Cell Rep. 2015;5:728–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Son H-J, Kim J-Y, Hahn Y, Seo S-B. Negative regulation of JAK2 by H3K9 methyltransferase G9a in leukemia. Mol. Cell. Biol. 2012;32:3681–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kim K-B, Son H-J, Choi S, Hahm JY, Jung H, Baek HJ, et al. H3K9 methyltransferase G9a negatively regulates UHRF1 transcription during leukemia cell differentiation. Nucleic Acids Res. 2015;43:3509–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Watson ZL, Yamamoto TM, McMellen A, Kim H, Hughes CJ, Wheeler LJ, et al. Histone methyltransferases EHMT1 and EHMT2 (GLP/G9A) maintain PARP inhibitor resistance in high-grade serous ovarian carcinoma. Clin. Epigenetics. 2019;11:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Gursoy-Yuzugullu O, Carman C, Serafim RB, Myronakis M, Valente V, Price BD. Epigenetic therapy with inhibitors of histone methylation suppresses DNA damage signaling and increases glioma cell radiosensitivity. Oncotarget. 2017;8:24518–24532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Spyropoulou A, Gargalionis A, Dalagiorgou G, Adamopoulos C, Papavassiliou KA, Lea RW, et al. Role of histone lysine methyltransferases SUV39H1 and SETDB1 in gliomagenesis: modulation of cell proliferation, migration, and colony formation. Neuromolecul. Med. 2014;16:70–82. [DOI] [PubMed] [Google Scholar]
- 196.Han S, Zhen W, Guo T, Zou J, Li F. SETDB1 promotes glioblastoma growth via CSF-1-dependent macrophage recruitment by activating the AKT/mTOR signaling pathway. J Exp Clin Cancer Res. 2020;39:218. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 197.Artal-Martinez de Narvajas A, Gomez TS, Zhang J-S, Mann AO, Taoda Y, Gorman JA, et al. Epigenetic regulation of autophagy by the methyltransferase G9a. Mol. Cell. Biol. 2013;33:3983–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Park SE, Yi HJ, Suh N, Park Y-Y, Koh J-Y, Jeong S-Y, et al. Inhibition of EHMT2/G9a epigenetically increases the transcription of Beclin-1 via an increase in ROS and activation of NF-κB. Oncotarget. 2016;7:39796–39808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Li F, Zeng J, Gao Y, Guan Z, Ma Z, Shi Q, et al. G9a inhibition induces autophagic cell death via ampk/mtor pathway in bladder transitional cell carcinoma. PLoS One. 2015;10:e0138390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhang J, Yao D, Jiang Y, Huang J, Yang S, Wang J. Synthesis and biological evaluation of benzimidazole derivatives as the G9a Histone Methyltransferase inhibitors that induce autophagy and apoptosis of breast cancer cells. Bioorg Chem. 2017;72:168–181. [DOI] [PubMed] [Google Scholar]
- 201.Tian H, Wang X, Lu J, Tian W, Chen P. MicroRNA-621 inhibits cell proliferation and metastasis in bladder cancer by suppressing Wnt/β-catenin signaling. Chem. Biol. Interact. 2019;308:244–251. [DOI] [PubMed] [Google Scholar]
- 202.Zhang H, Cai K, Wang J, Wang X, Cheng K, Shi F, et al. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells. 2014;32:2858–2868. [DOI] [PubMed] [Google Scholar]
- 203.Srinivasan S, Shankar SR, Wang Y, Taneja R. SUMOylation of G9a regulates its function as an activator of myoblast proliferation. Cell Death Dis. 2019;10:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Poulard C, Baulu E, Lee BH, Pufall MA, Stallcup MR. Increasing G9a automethylation sensitizes B acute lymphoblastic leukemia cells to glucocorticoid-induced death. Cell Death Dis. 2018;9:1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Poulard C, Bittencourt D, Wu D-Y, Hu Y, Gerke DS, Stallcup MR. A post-translational modification switch controls coactivator function of histone methyltransferases G9a and GLP. EMBO Rep. 2017;18:1442–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ishimoto K, Kawamata N, Uchihara Y, Okubo M, Fujimoto R, Gotoh E, et al. Ubiquitination of lysine 867 of the human SETDB1 protein upregulates its histone H3 lysine 9 (H3K9) methyltransferase activity. PLoS One. 2016;11:e0165766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sun L, Fang J. E3-Independent Constitutive Monoubiquitination Complements Histone Methyltransferase Activity of SETDB1. Mol. Cell. 2016;62:958–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Al Emran A, Marzese DM, Menon DR, Stark MS, Torrano J, Hammerlindl H, et al. Distinct histone modifications denote early stress-induced drug tolerance in cancer. Oncotarget. 2018;9:8206–8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Guler GD, Tindell CA, Pitti R, Wilson C, Nichols K, KaiWai Cheung T, et al. Repression of Stress-Induced LINE-1 Expression Protects Cancer Cell Subpopulations from Lethal Drug Exposure. Cancer Cell. 2017;32:221–237.e13. [DOI] [PubMed] [Google Scholar]
- 210.Ciechomska IA, Marciniak MP, Jackl J, Kaminska B. Pre-treatment or Post-treatment of Human Glioma Cells With BIX01294, the Inhibitor of Histone Methyltransferase G9a, Sensitizes Cells to Temozolomide. Front. Pharmacol. 2018;9:1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Mini E, Nobili S, Caciagli B, Landini I, Mazzei T. Cellular pharmacology of gemcitabine. Ann. Oncol. 2006;17 Suppl 5:v7–12. [DOI] [PubMed] [Google Scholar]
- 212.Pan M-R, Hsu M-C, Luo C-W, Chen L-T, Shan Y-S, Hung W-C. The histone methyltransferase G9a as a therapeutic target to override gemcitabine resistance in pancreatic cancer. Oncotarget. 2016;7:61136–61151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Candelaria M, de la Cruz-Hernandez E, Taja-Chayeb L, Perez-Cardenas E, Trejo-Becerril C, Gonzalez-Fierro A, et al. DNA methylation-independent reversion of gemcitabine resistance by hydralazine in cervical cancer cells. PLoS One. 2012;7:e29181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Liu C-W, Hua K-T, Li K-C, Kao H-F, Hong R-L, Ko J-Y, et al. Histone Methyltransferase G9a Drives Chemotherapy Resistance by Regulating the Glutamate-Cysteine Ligase Catalytic Subunit in Head and Neck Squamous Cell Carcinoma. Mol. Cancer Ther. 2017;16:1421–1434. [DOI] [PubMed] [Google Scholar]
- 215.Trenner A, Sartori AA. Harnessing DNA Double-Strand Break Repair for Cancer Treatment. Front. Oncol. 2019;9:1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Wood JG, Helfand SL. Chromatin structure and transposable elements in organismal aging. Front. Genet. 2013;4:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Belgnaoui SM, Gosden RG, Semmes OJ, Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 2006;6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Groh S, Schotta G. Silencing of endogenous retroviruses by heterochromatin. Cell Mol. Life Sci. 2017;74:2055–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Toh TB, Lim JJ, Chow EK-H. Epigenetics in cancer stem cells. Mol. Cancer. 2017;16:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ma DK, Chiang C-HJ, Ponnusamy K, Ming G-L, Song H. G9a and Jhdm2a regulate embryonic stem cell fusion-induced reprogramming of adult neural stem cells. Stem Cells. 2008;26:2131–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kaniskan HÜ, Martini ML, Jin J. Inhibitors of protein methyltransferases and demethylases. Chem. Rev. 2018;118:989–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Avgustinova A, Symeonidi A, Castellanos A, Urdiroz-Urricelqui U, Solé-Boldo L, Martín M, et al. Loss of G9a preserves mutation patterns but increases chromatin accessibility, genomic instability and aggressiveness in skin tumours. Nat. Cell Biol. 2018;20:1400–1409. [DOI] [PubMed] [Google Scholar]
- 223.Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. DOG 1.0: illustrator of protein domain structures. Cell Res. 2009;19:271–273. [DOI] [PubMed] [Google Scholar]