Abstract
Exosomes are a unique subpopulation of naturally occurring extracellular vesicles which are smaller intracellular membrane nanoparticle vesicles. Exosomes have proven to be excellent nanocarriers for carrying lipids, proteins, mRNAs, non-coding RNAs, and DNAs, and disseminating long-distance intercellular communications in various biological processes. Among various cell-line or biological fluid derived exosomes, milk exosomes are abundant in nature and exhibit many nanocarrier characteristics favorable for theranostic applications. To be an effective delivery carrier for their clinical translation, exosomes must inbuilt loading, release, targeting, and imaging/tracking characteristics. Considering the unmet gaps of milk exosomes in theranostic technology it is essential to focus the current review on drug delivery and imaging applications. This review delineates the efficiency of exosomes to load therapeutic or imaging agents and their successful delivery approaches. It is emphasized on possible modifications of exosomes towards increasing the stability and delivery of agents. This article also summarizes the specific applications and the process of developing milk exosomes as a future pharmaceutical drug delivery vehicle.
Keywords: Milk exosomes, Extracellular vesicles, Drug delivery, Imaging agents, Theranostic applications
Graphical abstract
Highlights
-
•
Milk exosomes are nature's abundant drug delivery carriers.
-
•
These exosomes exhibit superior stability, biocompatibility, half-life, and very low immunogenicity.
-
•
This work describes isolation methods, drug/imaging agents loading, and therapeutic implications of milk exosomes.
-
•
Engineered milk exosomes offer superior theranostic applications.
1. Introduction
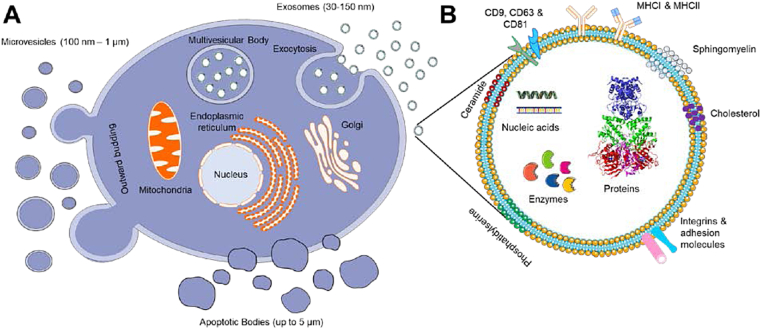
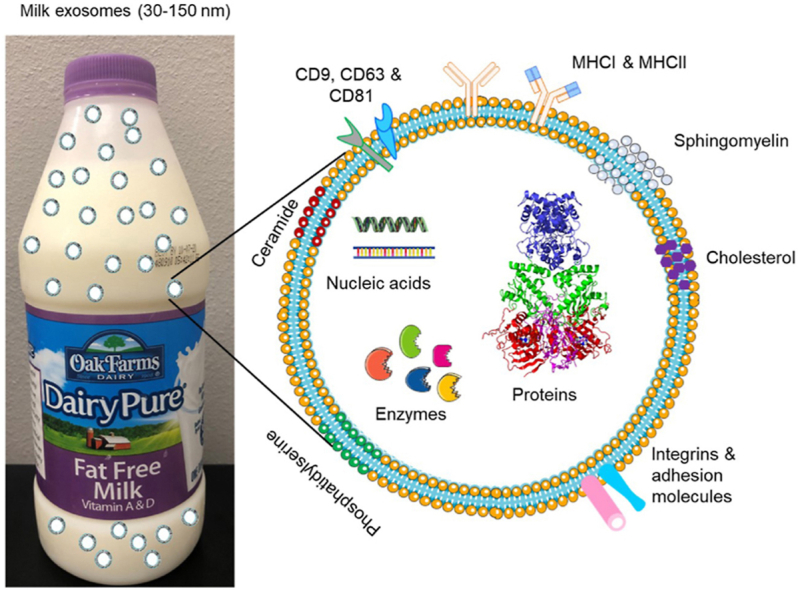
Extracellular vesicles (EVs) are a class of secreted biomolecular lipid-bilayer nanostructures. The prime function of EVs is to transport various types of cellular components such as proteins, lipids, mRNAs, microRNAs, receptors, and effector molecules to the recipient cells (Fig. 1). The EVs were first discovered over three decades ago. Their connection to human metabolism, immunology, and physiology is widely established. Based on the process of intracellular origin, excretion, biogenesis, size, and their function, EVs have been classified into three categories such as exosomes, microvesicles, and apoptotic bodies. Exosomes (Exo) are a unique subpopulation of nanosized spherical membrane vesicles with particle size between 30 and 150 nm (Fig. 1A and B). This term was invented by Dr. Rose Johnstone while identifying the biologic process with maturing reticulocytes [[1], [2], [3]]. His team observed that the cytoplasm of reticulocytes is comprised of large sacs filled with small membrane-enclosed structures of uniform size which are extruded outside the cells. Exosomes are often created by multivesicular bodies which are exported out via fusion with the cell plasma membrane (Figure 1A). Since the process of secretion is opposed to endocytosis, these extruded vesicles were named as “exosomes”. Whereas cellular membrane vesicles in a size range between 50 nm and 1 μm are considered as “microvesicles” (Fig. 1A) [11]. The biogenesis of microvesicles is distinct from exosomes, which occurs due to a straightforward outward blossoming and breaking up of the plasma membrane. Since there is some overlap in the certain size ranges of exosomes and microvesicles, it is important to distinguish based on their mechanism of biogenesis. On the other hand, both exosomes and microvesicles consist of a variety of fragments from the parent cells, including microRNAs, messenger RNAs, lipids, and proteins. Apoptotic bodies often shed cellular contents (deoxyribonucleic acid, ribonucleic acid, and histone proteins) up to 5 μm larger particles (Fig. 1A). Variations among these extracellular vesicles (exosomes, microvesicles, and apoptotic bodies) can be visualized through transmission electron microscopic images [4]. A more detailed explanation of the biogenesis and constitution of exosomes, microvesicles, apoptotic bodies, and other types of extracellular vesicles (components) can be referred to the latest documents [5].
Fig. 1.
A) A schematic representation of cellular vesicle production (exosomes, microvesicles, and apoptotic bodies) from cells including milk. Distinct variation in the biogenesis and excretion from cell to yield different size range excretes. B) An illustration of exosome structure with rich source for protein, nucleic acid, enzyme, lipids, and cargos.
Among all types of EVs, exosomes have received much attention due to their therapeutic, theranostic, and diagnostic applications. EVs are released by numerous cells and their presence of abundance can be seen in several body fluids, such as plasma, urine, saliva, bronchoalveolar lavage fluid, synovial fluids, amniotic fluid, malignant ascites, etc. Wang et al. [6] documented a broad view of statistical analysis of the state of exosomal research based on 4960 publications (from all over the world, 25 countries: USA-highest and Iran-lowest). The co-occurrence analysis mapping confirms that in vitro and in vivo studies were commonly documented, but “exosomal mechanism related research” is extensively reported. A comprehensive survey on the role of exosomes and other extracellular vehicles as diagnostic and therapeutic modalities was delineated by the number of publications, patents, intellectual property, and government funding [7]. Other global scientific trends on exosomal research (2007–2016, from 1850 publications) have been divided into three major clusters: “tumor related”, “stem cell”, and “drug resistance” [8]. It was also observed that “Michigan Cancer Foundation-7” was highly studied due to a breast cancer cell line. The overall publication trends demonstrated that exosomes were highly applied for cancer detection applications [9]. Based on extensive utility in several fields of medicine several US companies are focused on producing and implementing exosomal research including, Aethlon Medical, Inc., Exosome Diagnostics Inc., NanoSomix Inc., ThermoFisher Scientific Inc., YMIR Genomics LLC., and System Biosciences Inc.
2. Milk exosomes
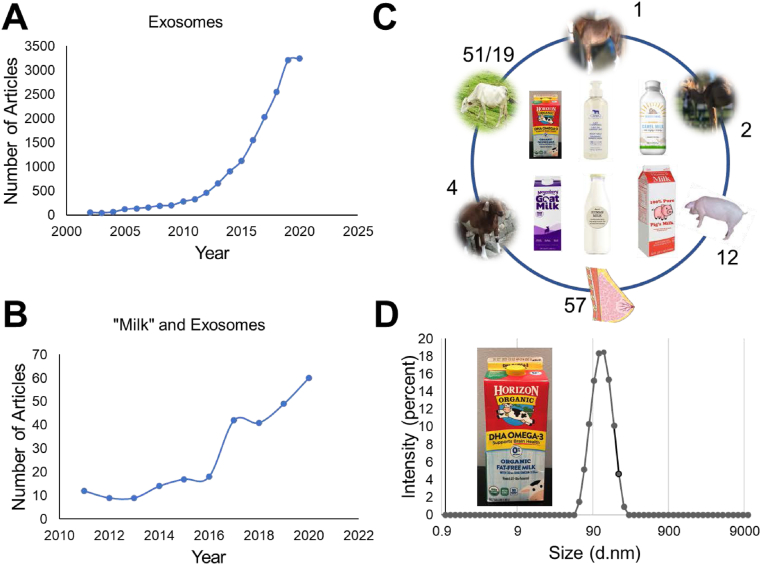
The PubMed search criteria in the title and abstract with the keyword “exosome(s)” led over 15,780 articles (Data accessed on Oct 15, 2020, Fig. 2A). This information suggests that exosomes are highly studied in cancer and drug delivery. The PubMed search revealed that scientific peer-reviewed publications started in early 1966 [1966 (1), 1984 (1), 1986 (1), and 1996 (1)]. The first milk-derived exosome-based research was published in 2007 [10]. There are about 256 publications on milk exosomes (Fig. 2B) in general out of which 57, 51/19, 4, 2, 12, and 1 reports belong to human breast milk, bovine/cow milk, goat milk, camel milk, porcine milk, and horse milk, respectively (Fig. 2C) [11]. These sources can produce liters of milk per day which can yield huge amounts of exosomes compared to cell lines or other biological fluid-based exosomes. These exosomes produce a uniform particle size of ~100 nm (Fig. 2D) which is highly recommended for therapeutic applications.
Fig. 2.
A-C) Peer-reviewed scientific reports and review articles covering various aspects of exosomes recorded using PubMed website https://www.ncbi.nlm.nih.gov/. All data was retrieved on Oct 12, 2020. A) Number of articles found that are published in the literature in each year since 2001 to 2020. B) Number of articles noticed in each year with key word of “Milk” and exosomes, C) Articles found in search with key words of “human milk” and exosome, “bovine/cow milk” and exosome, “horse milk” and exosome, “camel milk” and exosome, “goat milk” and exosome, and “porcine milk” and exosome. D) An example of size distribution of bovine milk exosomes derived from fat free Horizon organic milk using acetic acid precipitation method. It shows a uniform particle size of ~100 nm.
However, up to date, the use of milk exosomes in research and medical sciences is low. Milk is a primary source of postnatal nutrition for all newborn mammals [12]. Breast milk feeding supports the newborn with the growth and development of intestinal microbiota and immunity [13]. Literature covers various investigations on milk exosomes, for their isolation processes, drug delivery, imaging, and therapeutic applications. Table 1, summarizes several review articles comprised of milk exosomes that are specific to i) metabolic regulation, ii) association with human diseases, iii) abundance of microRNAs and other biological molecules, iv) biological influence in infant and adults, and v) isolation methods. A detailed breakdown of individual review reports and their major focus is presented (Table 1). A portion of the article also covered the anticancer drug delivery aspect using milk exosomes, but this article used very limited examples. Altogether, there is no focused review article that is specifically devoted to milk exosomes, for isolation protocols, characterization, their significance in delivery of small molecules, imaging agents, and biomolecules, and developing as hybrid/engineered nanocarrier system for possible therapeutic implications. Therefore, this article aimed to describe conclusive information towards developing milk exosomes as unique nanocarriers for both imaging and therapeutic application. The additional section also covers the possible future directions and translational potential of this nanocarrier system.
Table 1.
Peer-reviewed research review articles and book chapters discussing milk-derived exosomes and their possible in in vitro, in vivo, biological, and medical applications.
| Title of the article | Various topics covered under each article |
|---|---|
| MicroRNA Milk Exosomes: From Cellular Regulator to Genomic Marker [14] | This review documented the possible applications of exosomes in metabolism and health, and genomic markers. |
| Milk-Derived Exosomes and Metabolic Regulation [15] | A detailed review of endogenous exosomes (exosome cargos and cell-to-cell communication), bioavailability and distribution of exosomes and their cargos, and interactions of milk exosomes with the gut microbiome. This study also documents information on phenotypes of dietary depletion of exosomes and cargo, and their influence on immune system. |
| Exosome: An Emerging Source of Biomarkers for Human Diseases [16] | This article reports a cumulative study of exosomes (including breast milk) serving as biomarkers for disease diagnosis and prognosis. This article also highlights the significance of exosomes for possible translational medicine. |
| Milk: an epigenetic amplifier of FTO-mediated transcription? Implications for Western diseases [17] | This review brings awareness on milk exosomes which may transfer dairy cow mammary epithelial cells-derived miRNA-29s and fat mass and obesity-associated mRNA to the milk consumers. The amplifying fat mass and obesity-associated gene expression which may introduce obesity, T2DM, prostate and breast cancer, and neurodegenerative diseases. |
| "Exosomics"-A Review of Biophysics, Biology and Biochemistry of Exosomes With a Focus on Human Breast Milk [18] | This work cites a number of works associated to i) exosomes in health and disease, ii) exosomes in maternal health, and iii) proteomics and micronutrient profiling of human breast milk exosomes. |
| Biological Activities of Extracellular Vesicles and Their Cargos from Bovine and Human Milk in Humans and Implications for Infants [19] | This study confirms the evidence that noncanonical pathways dietary microRNAs may alter gene expression at low concentrations. Breastfed infants have a higher Mental Developmental Index, Psychomotor Development Index, and Preschool Language Scale-3 scores compared to those fed various formulas. |
| Milk is not just food but most likely a genetic transfection system activating mTORC1 signaling for postnatal growth [20] | Milk exosomes often contain miRNA-21 which involves enhancing mTORC1-driven metabolic processes. Therefore, breastfeeding is ideal for infants permitting suitable postnatal growth and species-specific metabolic programming, However, high cow's milk consumption may promote such signaling and its driven diseases of civilization. |
| Exosomes of pasteurized milk: potential pathogens of Western diseases [21] | This review offers an analysis of a number of articles documenting milk consumption, milk-associated miRNA hallmark in western diet, and possible risk developing chronic diseases. |
| The Therapeutic Potential of Breast Milk-Derived Extracellular Vesicles [22] | This article cites a number of therapeutic benefits of breast milk-derived exosomes and their remedial effect in the gastrointestinal disease necrotizing enterocolitis. |
| Breast milk microRNAs harsh journey towards potential effects in infant development and maturation. Lipid encapsulation can help [23] | This work delineates miRNAs of breast milk-derived exosomes and their important roles in infants' development, maturation, and potential biological effects. This work also extends possible implications in pharmacological and pharma-nutritional consequences. |
| Milk's Role as an Epigenetic Regulator in Health and Disease [24] | This review article gives an overview of miRNAs and gene expression via milk-derived miRNAs to consumers. Further, this article also discusses how the commercial interest may further influence the epigenetic miRNA burden for the milk consumer. |
| Milk Exosomes: Isolation, Biochemistry, Morphology, and Perspectives of Use [25] | This book chapter presents i) methods of exosome isolation, purification, and analysis, ii) biochemistry of milk exosomes, and iii) brief overview of its use. |
| Milk Exosomes: Perspective Agents for Anticancer Drug Delivery [26] | Extensively discusses bioactive compounds of milk exosomes: proteins, nucleic acids, and lipids in milk exosomes. It also provides drug delivery applications of milk exosomes in cancer therapy. |
| Milk exosomes: A biogenic nanocarrier for small molecules and macromolecules to combat cancer [27] | This article emphasized the various isolation methods and the evaluation of physicochemical, biodistribution, and delivery characteristics of exosomes to combat cancer. |
3. Milk exosome isolation
A detailed overview of the isolation methods of exosomes from various resources has been well documented in the literature [28]. Ultracentrifugation, size exclusion chromatography, micro-/nano-filtration, immunological, and polymer-based precipitation are some of the widely used methods to separate or isolate exosomes. Exosomal contents (proteins, lipids, and luminal molecules) and population size differ between each type of isolation method and the source of exosomes. Few articles have been specifically devoted to educate about the use of various types of isolation methods, their suitability, possible advantages, and disadvantages [29,30]. Various commercial isolation reagent kits are also available for quick isolation of exosomes using various principles. Among them, ThermoFisher, Sigma, FUJIFILM Wako Pure Chemical Corporation, IZON, Qiagen, Biocompare, System Biosciences, Takara Bio, Miltenyi Biotec, Nano View Biosciences, BioVision Inc., BioCat GmbH, etc are well-known resources. However, many of these kits are specifically applied to cell culture and human biological fluid-derived exosomes. Therefore, in this section, the commonly applied isolation methods are documented for milk exosomes.
There is no one standard method that is followed for the isolation of exosomes from milk. Successful isolation of milk-derived exosomes itself turns out to be a valuable area of research. There is an unmet need to develop a consistent and reproducible exosome isolation method. Isolation or separation of non-vesicular entities from exosomes is not completely accomplished by simple protocols (centrifugation or commercial kits or polymer-based methods). In this regard, the International Society for Extracellular Vesicles (ISEV) established appropriate protocols, recommendations, and requirements for achieving highly purified exosomes with intact morphology and components [31]. These standards are often updated with numerous amendments.
Table 2 presents the commonly used techniques to isolate the milk exosomes and their principles, advantages, disadvantages, yield, and purity [27,32,33]. However, it is important to note that milk-derived exosomes are inexpensive unlike other cell or biological fluids, thus ultracentrifugation is highly preferred [34,35]. All other methods are associated with high cost.
Table 2.
Commonly used techniques to separate exosomes from milk, cells, and biological fluids.
| Technique | Principle | Purity | Yield | Advantage | Disadvantage |
|---|---|---|---|---|---|
| ExoQuick precipitation | Polymer based precipitation | Low | 1 mL | Easy Method | High cost |
| Ultracentrifugation | Differential centrifugation | High to low | 100–500 mL | High yield | Time consuming |
| Density gradient ultracentrifugation | Differential centrifugation | High | 10 mL | High purity | Time consuming |
| Isoelectric precipitation | Precipitation at isoelectric point | Low | 10–20 mL | Low cost | Low purity |
| Size exclusion chromatography | Porous columns filled with polymers | High | 10–100 mL | High yield of exosomal proteins | Complex Method |
The differential ultracentrifugation method is highly suitable for isolating exosomes from bovine and human milk. This method provides the purest form of exosomes with highly intact bio-factories. For example, bovine milk exosomes can be produced with this method by various groups within the range of 30–150 nm [14,[35], [36], [37], [38], [39]]. Ultracentrifugation (UC) is also employed to obtain milk-derived exosomes from different species such as goat (122 nm) [40], horse (~100 nm) [41], camel (30–100 nm) [42], and porcine (50–100 nm) [43]. Both yak milk and cow milk-derived exosomes have a similar particle size distribution of 131.1 ± 53.25 nm and 131.5 ± 52.39 nm, respectively in dynamic light scattering-based measurements [44].
Recent evidences support that acidification is helpful in the quick isolation of milk exosomes [45,46]. Acidification with hydrochloric acid (HCl) and acetic acid (AA) yields exosomes with size ~124 nm and 132 nm, respectively which are close to 126 nm exosomes isolated by UC method [45]. On the other hand, the particle concentration of exosomes was obtained as 2.6 × 109, 1.7 × 109, and 4.8 × 108 particles/mL with AA, HCl, and UC methods, respectively. In addition, the morphology and concentration of protein are very similar to UC derived exosomes. This suggests that the acid treatments yielded significantly higher amounts of exosomes relative to UC. Another comparative method of exosomes isolated from raw milk using ultracentrifugation (70,000 rcf) and isoelectric precipitation (IP) (adjusting with pH of milk to 4.6 with HCl) resulted in a very similar size population of 120 nm [46]. Additionally, IP method resulted in a higher number of exosomes (1.7 × 109 particles/ml) compared to ultracentrifugation (4.8 × 108 particles/ml). However, the ultracentrifugation method provided smooth surface constituted exosomes. Horse milk exosomes with 40–100 nm size range population were obtained for the first time by Sedykh et al. [41]. Further, its purification with gel filtration produced exosomes containing small number of major proteins.
Yamada et al. [35], reported a comparative study for isolating exosomes from bovine milk. Their study concludes that ultracentrifugation with ExoQuick precipitation is a useful method for rapid isolation and high exosome recovery. The ultracentrifugation with a density gradient centrifugation method allows for an efficient and purified form of exosomes with native morphology. These methods are superior compared to ExoQuick precipitation alone. A recent systematic comparison of methods was implemented for the extraction of exosomes isolated from bovine and human milk [34]. In support of this study, a recent investigation with three independent isolation methods suggests that ExoQuick precipitation method is highly suited for milk exosomal isolation than ultracentrifugation [34].
Vaswani et al. [47], validated a superior isolation method for the extraction of extracellular vesicles (30–200 nm) from milk using size exclusion chromatography column (qEV) over ultracentrifugation. The 12–16 fractions of these vesicles that were tested provide over 1 × 108 particles/mL with qEV while ultracentrifugation method resulted in only ~2.2 × 08 particles/mL. Bovine milk exosomes (176.9 nm) yield ~12 × 1013 particles/mL which is significantly higher than human milk exosomes (188.8 nm, ~4.0 × 1012 particles/mL) [48]. A sequential centrifugation along with the size exclusion chromatography isolation method was used to achieve exosomes from bovine and human milk [49]. This study confirms that the concentration of exosomes is higher in bovine milk than human milk, i.e., 1.34 × 1012 to 1.50 × 1012 (bovine milk, centrifugation) compared to 4.88 × 1011 to 1.00 × 1012 (human milk, centrifugation). These studies support the feasibility of scalable isolation of exosomes from the large volumes of milk. Recently, Kaddour et al. [50], developed a high-resolution size guided turbidimetry based particle purification liquid chromatography for achieving superior quality exosomes from various types of resources including cow milk.
Ultracentrifugation uses 100,000–200,000 g (1–20 h) for successful separation/depletion of exosomes from a larger volume often for industrial/commercial usage. This method is highly time-consuming and difficult to standardize. A new solid-phase extraction method based on molecular imprinted polyester capillary-channeled polymer fiber stationary phase in a spin-down tip format is developed to isolate the exosomal population from aqueous medium, urine, milk, and serum [51]. This method can recover the exosomal population fraction from the aqueous, urine, and milk samples ~1.5 × 1010 and ~1.3–1.4 × 109 at 1/100 and 1/1000 dilutions, respectively. As a result, no significant difference was noticed among all types of sources in achieving exosomal population. This further motivated to develop polycarbonate or nitrocellulose-based membrane filters to isolate cells and large EVs in biological samples by combining with ultracentrifugation [52]. Such protocols are highly implemented to generate exosomes from serum or media and biological fluids.
It is imperative to characterize the isolated milk exosomes for their successful use in research, development, and clinical applications. Physicochemical properties of exosomes play an important role in the nanoplatform for drug delivery and theranostic applications. Dynamic light scattering (DLS) is a method of choice to measure size, dispersion, mobility, surface charge, and concentration of exosomes [53]. Several other standard techniques such as transmission electron microscopy [34], scanning electron microscopy [54,55], and atomic force microscopy [55] are also used to examine the shape, morphology, and size of exosomes. In addition to these mentioned techniques, Western blotting [38], antibody based exosomal array [56,57] and flow cytometry [58,59] are some of the methods that are used to evaluate the surface markers of exosomes.
Upon their successful characterization, the biodistribution, pharmacokinetic, and pharmacodynamic profiles of exosomes play a significant role in establishing them as a delivery platform. The biodistribution of exosomes primarily depends on the route of administration. A comprehensive study reports that milk exosomes primarily accumulated in liver, spleen, and brain upon administration (suckling, oral gavage, and intravenous) in mice and pigs [60]. Another study suggests that oral administration of exosomes (DiR, a near infrared dye, loaded) showed a uniform distribution in liver, kidney, lung, spleen, pancreas, colon, brain, and ovaries while intravenously administered exosomes were mostly found in the liver [55]. A bioavailability and biodistribution study of cow milk exosomes after intravenous and oral administration concluded that the milk exosomes were bioavailable in mice but accumulated in liver and spleen [61]. However, only a fraction of orally administered exosomes escaped and reached peripheral tissues after re‐packaged in the intestinal mucosa. Literature revealed that chemical modifications of exosomes also influence the biodistribution. For example, functionalization of exosomes with folic acid promoted the accumulation of exosomes predominately in tumors than other organs of lung cancer mice [62].
4. Milk exosomes: Nature's unique nanocarriers
Many drug molecules and biological agents are unstable in in vivo environments which is a major concern in developing them as therapeutic agents. To solve this issue, various nanoparticulate delivery systems have been developed [[63], [64], [65], [66]]. Among these nanocarriers, polymeric nanoparticles and liposomes are two important nanocarriers for drug delivery applications. Polymeric nanoparticles are composed of biodegradable or biocompatible polymer(s) with stabilizers and possess extensive abilities for entrapment, encapsulation, or conjugation of therapeutic molecules. Whereas liposomes are spherical nanovesicles composed of lipid bilayers of natural or synthetic phospholipids, home to several approved therapeutics. These two nanoplatforms have the ability to enhance the efficacy of therapeutic agents through prolonged circulation half-life, sustained release, and targeted delivery of agents. A number of US Food and Drug Administration approved agents based on polymer and liposomes have been in clinical use and many more are under various stages of clinical development. Liposomes are highly biocompatible but less stable systemically, while polymeric nanoparticles exhibit superior stability but relatively less biocompatible material.
Polymeric nanoparticles are often synthesized by biocompatible polymers and are known to deliver cargos in a sustained manner. One of the biggest advantages of polymeric nanoparticles is, they provide a variety of cargo loading including drugs, nucleic acids, proteins/antibodies, and peptides in order to obtain the targeted therapy [67]. Despite having these many advantages, polymeric nanoparticles have various limitations too such as toxicity of preparation solvents and degradation byproducts, biphasic drug release, batch to batch reproducibility, and large-scale production [68]. On the other hand, due to the presence of both hydrophobic and hydrophilic sites, liposomes allow to load a variety of hydrophobic and hydrophilic drugs and dyes [69]. The average size range for liposomes is 100–250 nm [70]. Liposomes offer enhanced half-life, biodistribution and permeability [71]. However, the use of liposomes has several limitations, mainly, their low loading efficiency, aggregation, poor oral administration, poor stability, toxicity, non-specific protein binding and batch to batch reproducibility [70].
Exosomes mimic liposomal or lipid based-nanocarriers in terms of the outer layer structure. Exosomes are “nature's delivery systems” which facilitate the delivery of various types of in situ encapsulated small-to macro-molecules. Exosomes have the capability of crossing physiological barriers including blood-brain barrier. Exosome-based human clinical trials show encouraging outcomes in terms of biocompatibility, feasibility, and safety. Literature reveals that exosomes can be efficient drug delivery carrier systems due to i) superior circulation capacity and bioavailability (negative zeta potential), ii) their smaller size with deep tissue penetration capacity, iii) similarity with cellular membranes, and iv) avoidance of degradation and quick elimination. Since exosomes are often built up by cellular products and able to evade phagocytosis by macrophages, which offer extended circulation time in the body. Additionally, exosomes also prevent endo- and lyso-somal degradation and facilitate therapeutic payloads into the cytoplasm. Such property improves the delivery or transfection of molecules more efficiently into the cells. Therefore, understanding of loading, release, and stability characteristics of exosomes for drug delivery is highly sought.
Exosomes are natural carriers filled with various biological molecules. These molecules often hamper the external incorporation of therapeutic agents into exosomes. To achieve a maximum loading capacity of exosomes, it is advised to remove the contents that are already present in exosomes. However, there are no efforts or technologies exist to offload agents from exosomes. Selective removal of these molecules may alter the membrane integrity of exosomes. Collective literature indicates the passive and active loading or encapsulation methods used to load therapeutic agents into exosomes. This process is facilitated by three specific routes: endogenous loading, exogenous loading, and fusion methods. The endogenous method of loading represents loading of agents during exosomal biogenesis, while the exogenous loading method is responsible for incorporation after isolation of endosomes. On the other hand, the fusion method leads to the generation of a composite exosomal carrier that is combined with exosomes and other nanoliposomes. Drug loading into milk exosomes is often employed by using the exogenous method.
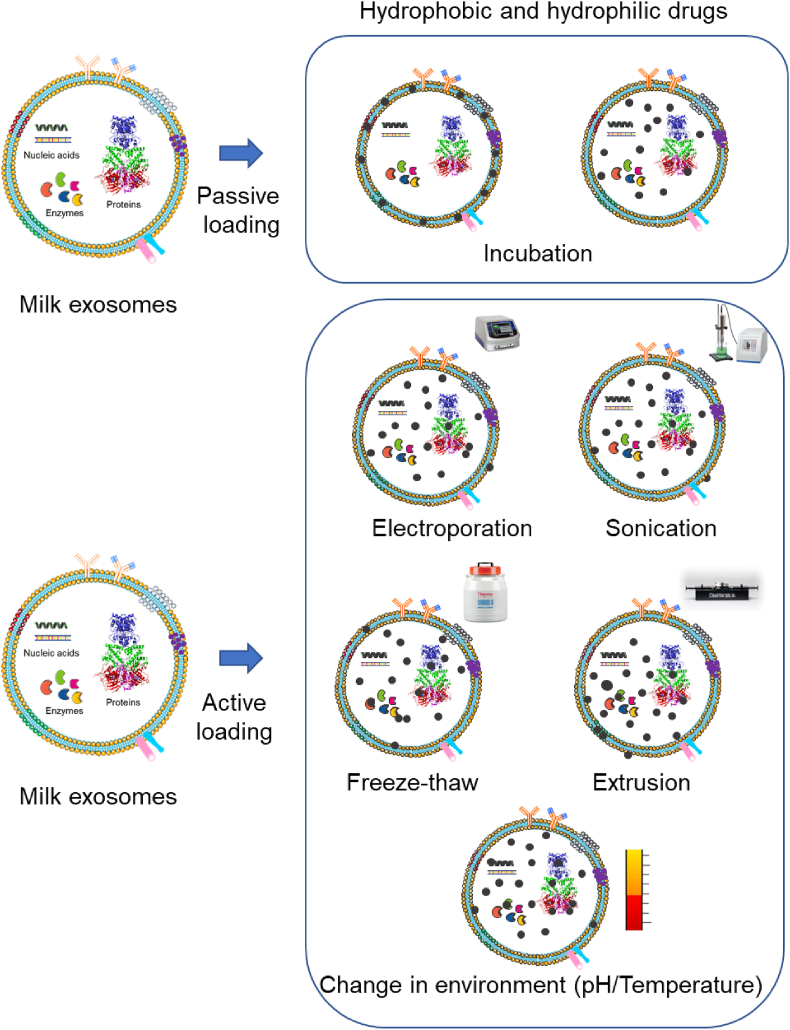
The drug delivery approaches depend on the loading ability of drug molecules into nanocarriers. Drug loading strategies into exosomes relies on passive (incubation) or active stimuli (sonication, electroporation, freeze/thaw, extrusion, or hypotonic dialysis) [72,73] (Fig. 3).
Fig. 3.
The schematic illustration of drug loading or encapsulation in exosomes. A) A simple post representation incubation strategy for passive diffusion into exosome structures. B) An illustrative presentation of stimuli or active drug loading methods in exosomes with the help of electroporation (transfection), sonication, freeze-thaw, extrusion, and temperature/pH gradiation.
Until now, a majority of studies confirm that small chemicals, dyes, macromolecules, and biological agents are loaded into milk exosomes via incubation method. The following section is the comparison of different (passive and active) methods of loading therapeutic cargos/molecules into exosomes (Fig. 3) [74,75]. Table 3 illustrates various methods applied to load therapeutic agents in exosomes, principles involved in drug loading, and efficiency of loading.
Table 3.
Descriptive information of various methods applied in therapeutic loading into exosomes and their associated benefits and perspectives.
| Therapeutic Cargo | Loading Method | Technique/Principle | Comments |
|---|---|---|---|
| Drugs, Nucleic acids, Proteins, and Peptides | Incubation | Diffusion in exosome membrane | Method is easy and simple |
| Proteins, Peptides, and Nucleic acids | Transfection | Gene editing | Cargo stability and loading efficiency |
| Drugs, Nanomaterials, Proteins, and Peptides | Sonication | External mechanical force to create micro pores | High loading efficiency |
| Drugs, Nanomaterials, Nucleic acids, Proteins, and Peptides | Electroporation | External electric force/field to create micro pores | High loading efficiency |
| Protein and Peptides | Freeze-thaw | Membrane fusion with liposomes | Moderate loading efficiency |
| Protein, Peptides, and Nanomaterials | Surfactant treatment | Active agents use to create micro pores | High loading efficiency |
| Drugs and Nucleic acids | Dialysis | pH dependent cargo loading | Method is easy and simple |
| Nanomaterials | In situ assembly and synthesis | Chemical reaction | Exosomes stability |
Incubation: Incubation of milk exosomes or exosome secreting cells with cargo and therapeutic molecule is the easiest process to operate and load cargoes [55,[76], [77], [78]]. It represents a passive loading method. Since exosomes have hydrophobic membranes and some hydrophilic cores, this makes exosomes suitable for both hydrophobic and hydrophilic cargoes. However, low loading efficiency due to physicochemical properties of exosomes and exosomal toxicity through cargos, remain the major challenges with incubation method.
Transfection: To load proteins, nucleic acids, and peptides onto exosomes, transfection technique is used [79,80]. This method is capable to load large molecules such as siRNA, miRNA, and other biomacromolecules. The Exo-Fect method, a chemical transfection (Gene Pulser Xcell) was implemented by transfecting siRNA into bovine exosomes [81]. Exo-Fect successfully inserted 30% of siRNA while electroporation was able to introduce only <5% of siRNA. Without an active loading method, there was almost no siRNA able to be loaded. Apart from commercial methods (lipofection), a layer-by-layer decoration of agents on exosomes is created for efficient delivery of siRNA [82]. However, it is a time consuming process and difficult to quantitate but it provides stable and high loading efficiency.
Sonication and electroporation: Sonication (external shear force) [83] and Electroporation (Extra electrical field) [[84], [85], [86]] are two ways to create micro pores on the exosomes which allows the loading of cargo. Both methods ensure high loading capacity but due to the intensity of the external physical pressure and integrity of exosomes is often compromised. These methods offer potential deformation of membrane and are not highly efficient for hydrophobic drug(s) encapsulation.
Extrusion: Extrusion method allows drug loading by mixing exosomes with drug(s) and the mixture is subjected to extrusion through membranes with varied porous size (100–400 nm) [87,88]. This process leads to disruption of membranes of exosomes which allows the drug molecules inside the exosome compartment. The extrusion method provides high levels of loading compared to sonication and freeze-thaw methods. The draw-back of this method is that deformation of membrane occurs during the loading process.
Freeze-Thaw method: The freeze-thaw method is a straightforward approach widely used to load drugs into exosomes. In this process, exosomes and drugs are mixed and incubated at room temperature, subsequently frozen (−80 °C or in liquid nitrogen or dry ice), and re-thawed. The repetition of these freeze-thaw cycles enables drug encapsulation in exosomes [72,89,90]. A moderate drug loading capacity (lower than that of sonication or extrusion) can be achieved with this method. Repeated freeze–thaw cycles may degrade the proteins and alter the structural changes of EVs [54]. This method may form aggregations during drug loading process.
Surfactant treatment: The surfactant treatment method uses specific surfactants (saponin and triton) which creates micro pores onto the exosomal surface to load the cargo [91]. These active detergents cause the exosome to disintegrate but show high loading capacity due to the increased surface permeation. Moreover, surface treatment induces pores in exosomes which leads to higher toxicity and hemolysis concerns.
Dialysis: This method is a less commonly applied to load desired drug cargos into exosomes [92,93]. In this method, both exosomes and drugs are placed into dialysis tubes or bags and then dialyzed by stirring to achieve drug-loaded exosomes. The hypotonic environment enables the higher drug loading efficiency compared to passive incubation method.
In situ assembly and synthesis: In situ assembly and synthesis is a chemical reaction or deposition of drug molecules into or on the surfaces of exosomes [94,95]. This method is only limited to growing nanoparticles on the surface of exosomes. The prime advantage of this method is no change in membrane integrity of exosome entirety.
5. Milk exosomes for theranostic applications
The milk exosomes derived from various mammals (human, cows, goats, camels, horses, and porcine) are an abundant source compared to cell culture-based fluids and blood plasma. Therefore, it is highly recommended for developing milk exosomes as cargos for the delivery of small molecules, biomacromolecules, nucleic acids, and or contrast imaging agents. Many therapeutic agents are not absorbed by the gastrointestinal tract for inducing their intended therapeutic benefit. This resulted in the implementation of intravenous infusion of these agents which requires multiple rounds of administration and demands huge costs associated with hospital stay and caretaker facilities. Multiple rounds of infusions are also a major risk factor for microbial infection in elderly patients. Therefore, attaining a therapeutic regimen that can be implemented at home results in greater patient compliance while minimizing the associated costs. On the other hand, it is also advisable to minimize the number of infusions by enhancing the therapeutic effects using an exosomal approach. Therefore, milk exosomes may be a suitable carrier for the delivery of drugs and imaging agents.
5.1. Drug delivery
Chemical drugs often exhibit poor water solubility or degradation before reaching the target resulting in severe systemic side effects. The lipid bilayer of exosomes not only improves the solubility of drugs but also protects their chemical contents from degradation [73]. An extensive research effort has been paid towards developing exosomes as drug delivery carriers. They can offer superior benefits as a delivery platform due to their nanostructure composition with cellular membranes and adhesive proteins. Such composition exhibits lower toxicity, unique stability, and overcomes the blood–brain barrier issues, and improve tumor/other organ homing capacity. Exosomes have proven capability of carrying a substantial number of therapeutic agents. It obeys the primary criteria of encapsulation of agents and intracellular delivery of those agents by crossing cellular membranes to induce their therapeutic actions. All these advised that exosomes may be one of the best natural carriers for delivering drug molecules. However, milk exosomal use as a delivery agent is still under development. This section provides systematic documentation of various reports that involve milk exosomes as a drug delivery carrier.
In general, exosomes are considered as a low immune prototype and do not induce major side effects. The unique construction of exosomes with lipid bilayer on the surfaces preserves their internal contents for efficient in vivo delivery [73]. There is a huge amount of literature that represents exosomes as nanodevices for effective delivery of therapeutics [96]. It is important that identifying a suitable, biocompatible, safe, scalable, and cost-effective preparation of exosomes is warranted. Since exosomes are composed of both hydrophilic and hydrophobic components in their structural layer, they can serve as depots for both hydrophilic and lipophilic small drug molecules. The first report by Munagala et al. [55] proposed bovine milk exosomes for successful drug delivery applications. This work confirms the dose and time dependent internalization of milk exosomes in human lung cancer H1299 cells. It was noticed these biological nanocarriers are bioavailable in various organs (liver > spleen > kidneys > lung > pancreas > brain ≥ colon > ovary, in intraperitoneal and intravenous injection of 60 mg/kg mice) and highly safe without inducing any immunological response. Apart from these, the encapsulated paclitaxel and withaferin A exosomes demonstrated superior action against breast (T47D and MDA-MB-231) and lung (A549 and H1299) cancer cells. More importantly, these drug-exosomal formulations promoted the anticancer actions in mice bearing xenografts of A549 lung tumors. Their subsequent investigation applied exosomes to deliver a berry bioactive molecule, anthocyanidins. This Anthos-exosome composition was tested in multiple (A549, H1299; PC3, DU145; HCT116; MCF7, MDA-MB-231; Panc1, MiaPaca2; and OVCa 432; lung, prostate, colon, breast, pancreatic, and ovarian) cancer cell lines [97]. The loading of various chemopreventive and chemotherapeutic agents (withaferin A, curcumin, paclitaxel, docetaxel) was successfully achieved by mixing in ethanol or 1:1 mixture of ethanol and acetonitrile solution with exosome suspension. These studies also confirm there was no effect on exosomal structure and stability identified with the use of organic solvent in the loading process.
Curcumin, a natural molecule exhibiting a number of medicinal properties. Almost all types of nanocarriers have been used to generate curcumin nanoparticles [98,99] for improving its activity while overcoming poor pharmacokinetics, degradation, and elimination. Upon loading curcumin in bovine milk exosomes, exosomal curcumin exhibited enhanced stability over free curcumin and, at the same time, a 3-fold increased transport of curcumin was noticed in transepithelial across Caco2 cell monolayer [100]. Berry anthocyanidins (natural extracts) have enhanced cisplatin-induced anticancer activity against both drug-sensitive and drug-resistant phenotype ovarian cancer cells. Considering this, anthocyanidins were loaded into exosomes and termed as ExoAnthos which significantly reduced the tumor growth of ovarian cancer mouse model [101]. The same approach was extended to another important anticancer molecule, paclitaxel [101]. This study reports that oral exosomal drug delivery exhibited similar therapeutic efficiency as free drug delivery through the intraperitoneal route [101].
Zhang et al. [37], reported a unique milk exosome based system with pH and light sensitive actions by introducing doxorubicin (by hydrolysis) and anthracene endoperoxide and chlorin e6 (EPT1 Ce6, ultrasound sonication). This study exhibited superior anti-tumor activity of this construct against oral squamous cell carcinomas (HSC-3, SCC-9 and CAL-27 cell lines). This activity was noticed as: free Dox (57.28% HSC-3 cells, 48.9% SCC-9 cells and 48.1% CAL-27), Exosome with components (71.32% HSC-3 cells, 59.9% SCC-9 and 65% CAL-27), and treatment with 808 nm laser irradiation (48.02% HSC-3 cells, 37.43% SCC-9 and 36.57% CAL-27). A similar enhanced therapeutic benefit was achieved in vivo. The synergistic photochemistry activity was noticed with NIR contained milk exosomes in regulating tumor growth, i.e., free Dox: ~1.26 cm3 Exosomes with drug: ~0.86 cm3, exosomes with drug and EPT1 Ce6: ~0.05 cm3 [37].
Gene silencing using siRNAs has proven to be a great promise to treat a number of diseases. However, there is no clinically approved method existed to deliver siRNAs. Often, oligonucleotide delivery vectors are positively charged for successful transport into cells. This poses a threat of toxicity issues associated with vectors. Aqil et al. [81], examined the usefulness of bovine milk exosomes as delivery vectors for siRNA delivery to silence specific genes of VEGF, EGFR, AKT, MAPK, and KRAS. This investigation demonstrates that exosomes with siKRAS showed superior internalization in A549 cells and H1299 non-small cells lung cancer cells. Additionally, siKRAS exhibits dose dependent (0–100 pmol/mL) effect on A549 (KRASG12S mutant) but not on A1299 (KRAS wild type) cells. Moreover, this siKRAS exosomal formulation exhibited controlling tumor growth to only 327 ± 181 mm3 while control group tumors grew up to 709 ± 409 mm3. This is a significant reduction (54%).
5.2. Imaging
Imaging technology is widely used to ascertain cellular localization/distribution and tissue accumulation/biodistribution and pharmacokinetics of molecules or agents. Efficient imaging or tracking of exosomes in vitro describes their intra-cellular mechanism of actions and delivery to various compartments. Besides, in vivo tracking of exosomes aids in determining serum availability (half-life), organ-specific delivery, and clearance of exosomes. The common imaging methods to delineate the image characteristics of exosomes (with fluorescence, metal/metal oxide, radiotracer labeling) includes in vitro (microscopy and flow cytometry), in vivo and ex vivo (immunohistochemistry, luciferase activity, fluorescence/radiation/CT-SPEC imaging system). This pre-requisite information directs the specific use of exosomes for proposed diseases and if required, the exosomes can be engineered according to the needs of applicability.
Milk exosomes are often exogenously labeled for the imaging studies. This can be achieved by a two-step process, exosomes are manufactured from various milk sources and then subsequently decorated with a fluorescent membrane dye or radio tracer or other types of contrast agent. Exosomes derived from raw cow milk labeled with NIR dye demonstrated a superior serum availability upon oral administration [102]. A high signal intensity was detected in the blood for 30, 60, 120, 240, and 360 min. The comparative liposome group showed a lower intensity even at 30, 60, and 120 min whereas no signal was noticed for 240 and 360 min. This indicates that milk exosomes are a better choice for the delivery of agents orally. These exosomes efficiently reached major vital organs such as liver, heart, lung, kidney, and spleen and the extent of accumulation was significant over free dye or liposomal-dye groups. More importantly, oral exosomal delivery to the tumor site was ~7500 IR units/protein compared to intravenous delivery ~9000 IR units/protein. However, the tumor delivery capacity is increased to 5.5-folds when these exosomes are surface tagged with iRGD peptides. Another bovine milk exosomal-based nanosystem exhibited feasibility of uniform in vivo distribution [37]. Through monitoring in fluorescent imaging at different time points (1, 4, 8, 24, 48, and 72 h) advised major signals in liver and kidney apart from the tumor. The order of accumulation was noticed as tumor > kidney > liver > spleen > lung ≥ heart. More importantly, these signals are higher compared to free ICG administered group. Goat milk-derived exosomes tagged with a radiotracer, 99mTc for evaluating pharmacokinetics and biodistribution properties [40]. The pharmacokinetic properties of these exosomes were evaluated by administering intravenous, intraperitoneal, and intranasal methods. Intraperitoneal injection showed half-life of 15.97 min which is far higher than intravenous (3.84 min) and intranasal (0.77 min) administration. However, intravenous injections show higher retention (%ID/g) of exosomes in liver (36.6 ± 7.5), spleen (31.1 ± 11.6), and stomach (2.1 ± 0.9). In a recent study, it was demonstrated that bovine milk exosomes can significantly alter their bioavailability and targetability to liver and pancreas with a short-term sonication [103]. This can be detected using a fluorescence dye-labeled exosomes. Goat milk exosomes are biocompatible with glioma cells and showed a strong binding potential [104]. Goat exosomes containing radionuclide 68Ga were capable to target T7 peptide on gliomas in an orthotopic glioma mouse model. A significant accumulation was observed under PET/CT imaging with 4 h after injection. This suggests that exosomes can be used for successful delivery of agents in different organs and can be extended for therapy and non-invasive imaging technique.
6. Engineered milk exosomes
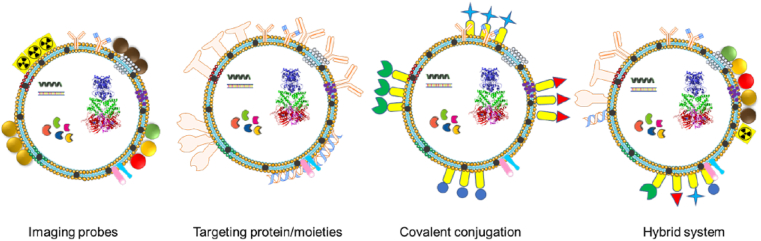
Exosomes have numerous benefits in the treatment and diagnosis of various diseases. Therefore, engineering of the surface of exosomes with an antibody, ligand, aptamer, and other biomacromolecules for organ or tumor-specific delivery/binding is sought (Fig. 4) [[105], [106], [107]]. A simple way to introduce such additional components on the surface of exosomes is through hydrophobic interaction of liposomes which already functionalized with these targeting moieties in exosome membranes either by fusion or freeze-thaw method [108,109]. Other techniques are to establish a i) click chemistry to modify surfaces of exosomes via 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide -N-hydroxy succinimide and azido group conjugation, ii) multi-valent electrostatic interactions, i.e., positive charged moieties (cationic pullulan, lipids, and polymers) insertion onto exosomal (negatively charged) surface membranes, and iii) surface decoration with conjugation or self-assemblies with transferrin, folic acid, hyaluronic acid, etc. [73,[110], [111], [112]]. Additionally, the introduction of stimuli responsive moieties, such as temperature, pH, extracellular matrix, redox-sensitive in exosomal membrane leads to a unique on-demand release characteristic for exosomes [37].
Fig. 4.
Different strategies explored for engineering of exosome surface with imaging probe (radioisotope/radiotracer, magnetic nanoparticles, gold, quantum dots, and labelling fluorescent probes), targeting moieties (monoclonal/polyclonal antibodies, aptamer, fragmented antibodies, and other biomacromolecules), conjugating small, biological, macro-/bio-macromolecules (reporter agents and other molecules), and hybrid nanosystems. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
However, the engineering of milk exosome investigation is limited in the literature. Bovine milk exosomes with doxorubicin (Exo-Dox) self-assembled with DSPE-PEG2000-HA lead HA decorated Exo-Dox (HA-Exo-Dox, ~136 nm) [113]. HA-Exo-Dox exhibited a superior binding with breast (MDA-MB-231 and MCF-7) and lung (HEK293 and A549) cancer cells. This was confirmed by visual evaluation by microscopic and quantitative flow cytometry analysis. A study demonstrated the importance of galactose, N-acetylgalactose, mannose, fructose, and neuraminate molecules insertion on bovine milk exosomes for improved human intestinal cells as well as mice [114]. Among all, N-acetylgalactose and galactose modification resulted in subsequent extensive transport and distribution with minimal accumulation in the liver and pancreas. Another study explored natural ligand-receptor on milk exosomes for efficient binding of siRNAs [82]. This approach uses lactoferrin-poly (l-lysine) mediated layer-by-layer to load siRNAs. This surface modification enables specific internalization in Caco2 and HepG2 cells. The colocalization capacity was up to 41.70 ± 2.39. Together, this method is not only cost-effective but non-toxic and chemical-free compared to commercial agents.
Exosomes comprised of theranostic agents i.e., both chemotherapeutic agent (therapy) and contrast/test agent (imaging/diagnostic agent) are under development for cancer [80]. Along with chemotherapeutic agent(s), the addition of superparamagnetic iron oxide nanoparticles (called as fexosomes for MRI imaging) and gold nanoparticles (called nanosomes for CT/PET imaging) [81]. These agent's inclusion in milk exosomes can be easily adopted.
7. Conclusions and future directions
Exosomes are abundant nanocarriers that can offer a new chapter and a promise for clinical applications. Milk exosomes have superior biocompatibility, no systemic toxicity, and low adverse immune and inflammatory responses. Exosomes are promising for use as vectors for clinical application, owing to their strong biocompatibility. It is important to consider that apart from human, goat, camel, and other mammalian milk, bovine milk can provide a superior scalable source of exosomes. Bovine milk exosomes are feasible to incorporate required specific payloads of chemical, biological, and macromolecules for successful and sustained delivery. In addition, milk exosomes are found to improve safety, bioavailability, and efficacy of therapeutic payload. Cumulative evidence suggests that exosomes with tumor targeting ligands enhance tumor targeted delivery and imaging technology [109,115].
Mounting evidence confirms that milk exosomes are highly useful nanocarriers for drug delivery but there are few challenges noticed. The primary issue is accomplishing uniform large-scale production capability, purity, and standard characterization for their effective translation [116]. Although the source of exosomes is the same, the diet and condition of the source can yield a slight district population of exosomes with difference in shape, size, and cargo contents. The next immediate quest is to develop rapid isolation and purification methods [117]. The other downside of exosomes is related to their low exosomal yield, poor encapsulation/loading efficiency of therapeutic agents. In addition, milk exosomes always carry their exogenous biological cargo that naturally exists inside of the exosomes. To offload such un-wanted endogenous contents altering exosomal structural integrity is needed.
Apart from these, exosomes extend their use as delivery carriers by avoiding unwanted accumulation or homing to the liver which circumvents metabolic issues [60]. These natural carriers are capable of long circulation time and carry therapeutic loads which can cross even blood-brain barrier for implicating some critical therapeutic issues. It is important to report source, variability (batch of source), the concentration of exosomes (number of particles, protein, and other contents), in vitro and in vivo information for efficient clinical translation. More attention is needed to load therapeutic agents without altering much integral membrane stability of exosomes. A standard loading and release characterization method needs to be followed for effective comparisons across multiple investigations.
Overall, exosomes offer an attractive and promising therapeutic tool for the treatment and imaging of various types of diseases. Future directions of exosomes research are aimed to advance multi-dimensional use. These novel therapeutic concept-based implications can certainly enhance the therapeutic ability of exosomes as a universal carrier. Apart from all these, Roche has recognized the importance of milk exosomes in the drug delivery field, a technology deal was made with PureTech to use the exosome platform for delivering small molecules to nucleic acid/biologics for effective oral delivery purpose [118]. Given the extent of uniqueness, exosomes may be the solution to implement as a clinically viable nanocarrier. It is anticipated that the potential of utilization of these carriers as clinically relevant theranostic applications in a decade.
Milk exosomes are particularly explored as delivery carriers rather than biological agent(s). They are in the size range of 50–200 nm which is a good size range to follow EPR mechanism for passive targeting of tumors or extravasation in blood vessels, while other nanosized materials can readily cleared by the kidney (1–30 nm) or the liver or spleen (larger particles) [59]. However, more studies such as toxicity, immune tolerance, tissue distribution, and potency of therapeutic agents loaded exosomes are needed to assess the clinical translation.
Credit author statement
Conceptualization, M.M.Y.; Software, B.A., N.M.C., N.C., M.J., S.C.C., Y.M.M.; Methodology, B.A., N.M.C., N.C., M.M.Y; Resources (Selection and review of literature papers), B.A., N.M.C., Y.M.M.; Writing - Original draft preparation, M.M.Y.; Writing – Review & Editing B.A., N.M.C., N.C., M.J., S.C.C., Y.M.M.; Supervision, M.M.Y.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors acknowledge the support from Department of Immunology and Microbiology, School of Medicine, University of Texas Rio Grande Valley to MMY, MJ, and SCC. This work is partially supported by NIH grants (R01 CA210192, R01 CA206069 and R01 CA204552).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Johnstone R.M., Adam M., Hammond J.R., Orr L., Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J. Biol. Chem. 1987;262(19):9412–9420. [PubMed] [Google Scholar]
- 2.Johnstone R.M. Revisiting the road to the discovery of exosomes. Blood Cell Mol. Dis. 2005;34(3):214–219. doi: 10.1016/j.bcmd.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Johnstone R.M., Mathew A., Mason A.B., Teng K. Exosome formation during maturation of mammalian and avian reticulocytes: evidence that exosome release is a major route for externalization of obsolete membrane. Proteins. 1991;147(1):27–36. doi: 10.1002/jcp.1041470105. [DOI] [PubMed] [Google Scholar]
- 4.Lázaro-Ibáñez E., Sanz-Garcia A., Visakorpi T., Escobedo-Lucea C., Siljander P., Ayuso-Sacido A., Yliperttula M. Different gDNA content in the subpopulations of prostate cancer extracellular vesicles: apoptotic bodies, microvesicles, and exosomes. Prostate. 2014;74(14):1379–1390. doi: 10.1002/pros.22853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akers J.C., Gonda D., Kim R., Carter B.S., Chen C.C. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. Journal of neuro-oncology. 2013;113(1):1–11. doi: 10.1007/s11060-013-1084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang B., Xing D., Zhu Y., Dong S., Zhao B. The state of exosomes research: a global visualized analysis. BioMed Res. Int. 2019;2019:1495130. doi: 10.1155/2019/1495130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roy S., Hochberg F.H., Jones P.S. Extracellular vesicles: the growth as diagnostics and therapeutics; a survey. J. Extracell. Vesicles. 2018;7(1):1438720. doi: 10.1080/20013078.2018.1438720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y., Wang Q., Wei X., Shao J., Zhao J., Zhang Z., Chen Z., Bai Y., Wang N., Wang Y., Li M., Zhai X. Global scientific trends on exosome research during 2007-2016: a bibliometric analysis. Oncotarget. 2017;8(29):48460–48470. doi: 10.18632/oncotarget.17223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ale Ebrahim S., Ashtari A., Zamani Pedram M., Ale Ebrahim N., Sanati-Nezhad A. Publication trends in exosomes nanoparticles for cancer detection. Int. J. Nanomed. 2020;15:4453–4470. doi: 10.2147/IJN.S247210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Admyre C., Johansson S.M., Qazi K.R., Filén J.-J., Lahesmaa R., Norman M., Neve E.P.A., Scheynius A., Gabrielsson S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007;179(3):1969–1978. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- 11.2020. https://pubmed.ncbi.nlm.nih.gov/?term=%22+human+milk%22+and+exosome&sort=datehttps://pubmed.ncbi.nlm.nih.gov/?term=%22+bovine+milk%22+and+exosome&sort=datehttps://pubmed.ncbi.nlm.nih.gov/?term=%22+cow+milk%22+and+exosome&sort=datehttps://pubmed.ncbi.nlm.nih.gov/?term=%22goat+milk%22+and+exosome&sort=datehttps://pubmed.ncbi.nlm.nih.gov/?term=%22camel+milk%22+and+exosome&sort=datehttps://pubmed.ncbi.nlm.nih.gov/?term=%22porcine+milk%22+and+exosome&sort=date PubMed search.
- 12.Haug A., Høstmark A.T., Harstad O.M. Bovine milk in human nutrition – a review. Lipids Health Dis. 2007;6(1):25. doi: 10.1186/1476-511X-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torregrosa Paredes P., Gutzeit C., Johansson S., Admyre C., Stenius F., Alm J., Scheynius A., Gabrielsson S. Differences in exosome populations in human breast milk in relation to allergic sensitization and lifestyle. Allergy. 2014;69(4):463–471. doi: 10.1111/all.12357. [DOI] [PubMed] [Google Scholar]
- 14.Cintio M., Polacchini G., Scarsella E., Montanari T., Stefanon B., Colitti M. MicroRNA milk exosomes: from cellular regulator to genomic marker, animals : an open access. journal from MDPI. 2020;10(7) doi: 10.3390/ani10071126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zempleni J., Sukreet S., Zhou F., Wu D., Mutai E. Milk-derived exosomes and metabolic regulation. Annual review of animal biosciences. 2019;7:245–262. doi: 10.1146/annurev-animal-020518-115300. [DOI] [PubMed] [Google Scholar]
- 16.Xu L., Wu L.F., Deng F.Y. Exosome: an emerging source of biomarkers for human diseases. Curr. Mol. Med. 2019;19(6):387–394. doi: 10.2174/1566524019666190429144310. [DOI] [PubMed] [Google Scholar]
- 17.Melnik B.C. Milk: an epigenetic amplifier of FTO-mediated transcription? Implications for Western diseases. J. Transl. Med. 2015;13:385. doi: 10.1186/s12967-015-0746-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de la Torre Gomez C., Goreham R.V., Bech Serra J.J., Nann T., Kussmann M. Exosomics"-A review of biophysics, biology and biochemistry of exosomes with a focus on human breast milk. Front. Genet. 2018;9:92. doi: 10.3389/fgene.2018.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zempleni J., Aguilar-Lozano A., Sadri M., Sukreet S., Manca S., Wu D., Zhou F., Mutai E. Biological activities of extracellular vesicles and their cargos from bovine and human milk in humans and implications for infants. J. Nutr. 2017;147(1):3–10. doi: 10.3945/jn.116.238949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melnik B.C., John S.M., Schmitz G. Milk is not just food but most likely a genetic transfection system activating mTORC1 signaling for postnatal growth. Nutr. J. 2013;12:103. doi: 10.1186/1475-2891-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melnik B.C., Schmitz G. Exosomes of pasteurized milk: potential pathogens of Western diseases. J. Transl. Med. 2019;17(1):3. doi: 10.1186/s12967-018-1760-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galley J.D., Besner G.E. The therapeutic potential of breast milk-derived extracellular vesicles. Nutrients. 2020;12(3) doi: 10.3390/nu12030745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomé-Carneiro J., Fernández-Alonso N., Tomás-Zapico C., Visioli F., Iglesias-Gutierrez E., Dávalos A. Breast milk microRNAs harsh journey towards potential effects in infant development and maturation. Lipid encapsulation can help. Pharmacol. Res. 2018;132:21–32. doi: 10.1016/j.phrs.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 24.Melnik B.C., Schmitz G. Milk's role as an epigenetic regulator in health and disease. Diseases. 2017;5(1) doi: 10.3390/diseases5010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sedykh S.E., Burkova E.E., Purvinsh L.V., Klemeshova D.A., Ryabchikova E.I., Nevinsky G.A. Milk exosomes: isolation, biochemistry, morphology, and perspectives of use. In: Bon A.G.D., Reales-Calderson J.A., editors. Extracellular Vesicles and Their Importance in Human Health. IntechOpen Limited; London, UK: 2018. pp. 1–28. [Google Scholar]
- 26.Sedykh S., Kuleshova A., Nevinsky G. Milk exosomes: perspective agents for anticancer drug delivery. Int. J. Mol. Sci. 2020;21(18) doi: 10.3390/ijms21186646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kandimalla R., Aqil F., Tyagi N., Gupta R. American journal of reproductive immunology; New York, N.Y.: 1989. Milk Exosomes: A Biogenic Nanocarrier for Small Molecules and Macromolecules to Combat Cancer. 2020. [DOI] [PubMed] [Google Scholar]
- 28.Li P., Kaslan M., Lee S.H., Yao J., Gao Z. Progress in Exosome Isolation Techniques, Theranostics. 2017;7(3):789–804. doi: 10.7150/thno.18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yakimchuk K. Exosomes: isolation and characterization methods and specific markers. Mater Methods. 2015;5:1450. [Google Scholar]
- 30.Doyle L.M., Wang M.Z. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. 2019;8(7) doi: 10.3390/cells8070727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lötvall J., Hill A.F., Hochberg F., Buzás E.I., Di Vizio D., Gardiner C., Gho Y.S., Kurochkin I.V., Mathivanan S., Quesenberry P., Sahoo S., Tahara H., Wauben M.H., Witwer K.W., Théry C. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sidhom K., Obi P.O., Saleem A. A review of exosomal isolation methods: is size exclusion chromatography the best option? Int. J. Mol. Sci. 2020;21(18) doi: 10.3390/ijms21186466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X. Isolation of extracellular vesicles from breast milk. Methods Mol. Biol. 2017;1660:351–353. doi: 10.1007/978-1-4939-7253-1_28. [DOI] [PubMed] [Google Scholar]
- 34.Wijenayake S., Eisha S., Tawhidi Z., Pitino M.A., Steele M.A., Fleming A.S., McGowan P.O. BioRxiv; 2020. Comparison of Methods for Milk Pre-processing, Exosome Isolation, and RNA Extraction in Bovine and Human Milk; p. 2020. 08.14.251629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamada T., Inoshima Y., Matsuda T., Ishiguro N. Comparison of methods for isolating exosomes from bovine milk. J. Vet. Med. Sci. 2012;74(11):1523–1525. doi: 10.1292/jvms.12-0032. [DOI] [PubMed] [Google Scholar]
- 36.Samuel M., Chisanga D., Liem M., Keerthikumar S., Anand S., Ang C.-S., Adda C.G., Versteegen E., Jois M., Mathivanan S. Bovine milk-derived exosomes from colostrum are enriched with proteins implicated in immune response and growth. Sci. Rep. 2017;7(1):5933. doi: 10.1038/s41598-017-06288-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Q., Xiao Q., Yin H., Xia C., Pu Y., He Z., Hu Q., Wang J., Wang Y. Milk-exosome based pH/light sensitive drug system to enhance anticancer activity against oral squamous cell carcinoma. RSC Adv. 2020;10(47):28314–28323. doi: 10.1039/d0ra05630h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaswani K., Koh Y.Q., Almughlliq F.B., Peiris H.N., Mitchell M.D. A method for the isolation and enrichment of purified bovine milk exosomes. Reprod. Biol. 2017;17(4):341–348. doi: 10.1016/j.repbio.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 39.Li B., Hock A., Wu R.Y., Minich A., Botts S.R., Lee C., Antounians L., Miyake H., Koike Y., Chen Y., Zani A., Sherman P.M., Pierro A. Bovine milk-derived exosomes enhance goblet cell activity and prevent the development of experimental necrotizing enterocolitis. PloS One. 2019;14(1) doi: 10.1371/journal.pone.0211431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.González M.I., Martín-Duque P., Desco M., Salinas B. Radioactive labeling of milk-derived exosomes with (99m)Tc and in vivo tracking by SPECT imaging. Nanomaterials. 2020;10(6) doi: 10.3390/nano10061062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sedykh S.E., Purvinish L.V., Monogarov A.S., Burkova E.E., Grigor'eva A.E., Bulgakov D.V., Dmitrenok P.S., Vlassov V.V., Ryabchikova E.I., Nevinsky G.A. Purified horse milk exosomes contain an unpredictable small number of major proteins. Biochimie Open. 2017;4:61–72. doi: 10.1016/j.biopen.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Badawy A.A., El-Magd M.A., AlSadrah S.A. Therapeutic effect of camel milk and its exosomes on MCF7 cells in vitro and in vivo. Integr. Canc. Ther. 2018;17(4):1235–1246. doi: 10.1177/1534735418786000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen T., Xi Q.-Y., Ye R.-S., Cheng X., Qi Q.-E., Wang S.-B., Shu G., Wang L.-N., Zhu X.-T., Jiang Q.-Y., Zhang Y.-L. Exploration of microRNAs in porcine milk exosomes. BMC Genom. 2014;15(1):100. doi: 10.1186/1471-2164-15-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao H.N., Guo H.Y., Zhang H., Xie X.L., Wen P.C., Ren F.Z. Yak-milk-derived exosomes promote proliferation of intestinal epithelial cells in an hypoxic environment. J. Dairy Sci. 2019;102(2):985–996. doi: 10.3168/jds.2018-14946. [DOI] [PubMed] [Google Scholar]
- 45.Rahman M.M., Shimizu K., Yamauchi M., Takase H., Ugawa S., Okada A., Inoshima Y. Acidification effects on isolation of extracellular vesicles from bovine milk. PloS One. 2019;14(9) doi: 10.1371/journal.pone.0222613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamauchi M., Shimizu K., Rahman M., Ishikawa H., Takase H., Ugawa S., Okada A., Inoshima Y. Efficient method for isolation of exosomes from raw bovine milk. Drug Dev. Ind. Pharm. 2019;45(3):359–364. doi: 10.1080/03639045.2018.1539743. [DOI] [PubMed] [Google Scholar]
- 47.Vaswani K., Koh Y.Q., Almughlliq F.B., Peiris H.N., Mitchell M.D. A method for the isolation and enrichment of purified bovine milk exosomes. Reprod. Biol. 2017;17(4):341–348. doi: 10.1016/j.repbio.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 48.Blans K., Hansen M.S., Sørensen L.V., Hvam M.L., Howard K.A., Möller A., Wiking L., Larsen L.B., Rasmussen J.T. Pellet-free isolation of human and bovine milk extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles. 2017;6(1):1294340. doi: 10.1080/20013078.2017.1294340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaswani K., Mitchell M.D., Holland O.J., Qin Koh Y., Hill R.J., Harb T., Davies P.S.W., Peiris H. A method for the isolation of exosomes from human and bovine milk. Journal of Nutrition and Metabolism. 2019;2019:5764740. doi: 10.1155/2019/5764740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaddour H., Lyu Y., Shouman N., Mohan M., Okeoma C.M. Development of novel high-resolution size-guided turbidimetry-enabled particle purification liquid chromatography (PPLC): extracellular vesicles and membraneless condensates in focus. Int. J. Mol. Sci. 2020;21(15) doi: 10.3390/ijms21155361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackson K.K., Powell R.R., Bruce T.F., Marcus R.K. Solid-phase extraction of exosomes from diverse matrices via a polyester capillary-channeled polymer (C-CP) fiber stationary phase in a spin-down tip format. Anal. Bioanal. Chem. 2020;412(19):4713–4724. doi: 10.1007/s00216-020-02728-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kornilov R., Puhka M., Mannerström B., Hiidenmaa H., Peltoniemi H., Siljander P., Seppänen-Kaijansinkko R., Kaur S. Efficient ultrafiltration-based protocol to deplete extracellular vesicles from fetal bovine serum. J. Extracell. Vesicles. 2018;7(1):1422674. doi: 10.1080/20013078.2017.1422674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szatanek R., Baj-Krzyworzeka M., Zimoch J., Lekka M., Siedlar M., Baran J. The methods of choice for extracellular vesicles (EVs) characterization. Int. J. Mol. Sci. 2017;18(6) doi: 10.3390/ijms18061153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bickmore D.C., Miklavcic J.J. Characterization of extracellular vesicles isolated from human milk using a precipitation-based method. Frontiers in nutrition. 2020;7:22. doi: 10.3389/fnut.2020.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Munagala R., Aqil F., Jeyabalan J., Gupta R.C. Bovine milk-derived exosomes for drug delivery. Canc. Lett. 2016;371(1):48–61. doi: 10.1016/j.canlet.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daaboul G.G., Gagni P., Benussi L., Bettotti P., Ciani M., Cretich M., Freedman D.S., Ghidoni R., Ozkumur A.Y., Piotto C., Prosperi D., Santini B., Ünlü M.S., Chiari M. Digital detection of exosomes by interferometric imaging. Sci. Rep. 2016;6(1):37246. doi: 10.1038/srep37246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jørgensen M., Bæk R., Pedersen S., Søndergaard E.K., Kristensen S.R., Varming K. Extracellular Vesicle (EV) Array: microarray capturing of exosomes and other extracellular vesicles for multiplexed phenotyping. J. Extracell. Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ender F., Zamzow P., Bubnoff N.V., Gieseler F. Detection and quantification of extracellular vesicles via FACS: membrane labeling matters! Int. J. Mol. Sci. 2019;21(1) doi: 10.3390/ijms21010291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jara‐Acevedo R., Campos‐Silva C., del Yáñez‐Mó María M., Vales‐Gómez M. Immunostep; 2020. Exosome Detection and Characterization Based on Flow Cytometry; p. 8. [Google Scholar]
- 60.Manca S., Upadhyaya B., Mutai E., Desaulniers A.T., Cederberg R.A., White B.R., Zempleni J. Milk exosomes are bioavailable and distinct microRNA cargos have unique tissue distribution patterns. Sci. Rep. 2018;8(1):11321. doi: 10.1038/s41598-018-29780-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manca S., Giraud D., Zempleni J. vol. 30. 2016. (Bioavailability and Biodistribution of Fluorophore-Labeled Exosomes from Cow's Milk after Intravenous and Oral Administration in C57Bl/6J Mice). S1, 690.8-690.8. [Google Scholar]
- 62.Aqil F., Munagala R., Jeyabalan J., Agrawal A.K., Gupta R. Exosomes for the enhanced tissue bioavailability and efficacy of curcumin. AAPS J. 2017;19(6):1691–1702. doi: 10.1208/s12248-017-0154-9. [DOI] [PubMed] [Google Scholar]
- 63.Yallapu M.M., Jaggi M., Chauhan S.C. Curcumin nanoformulations: a future nanomedicine for cancer. Drug Discov. Today. 2012;17(1–2):71–80. doi: 10.1016/j.drudis.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ganju A., Yallapu M.M., Khan S., Behrman S.W., Chauhan S.C., Jaggi M. Nanoways to overcome docetaxel resistance in prostate cancer. Drug Resist. Updates : reviews and commentaries in antimicrobial and anticancer chemotherapy. 2014;17(1–2):13–23. doi: 10.1016/j.drup.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chowdhury P., Roberts A.M., Khan S., Hafeez B.B., Chauhan S.C., Jaggi M., Yallapu M.M. Magnetic nanoformulations for prostate cancer. Drug Discov. Today. 2017;22(8):1233–1241. doi: 10.1016/j.drudis.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dan N., Setua S., Kashyap V.K., Khan S., Jaggi M., Yallapu M.M., Chauhan S.C. Antibody-drug conjugates for cancer therapy: chemistry to clinical implications. Pharmaceuticals. 2018;11(2) doi: 10.3390/ph11020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Palazzolo S., Bayda S., Hadla M., Caligiuri I., Corona G., Toffoli G., Rizzolio F. The clinical translation of organic nanomaterials for cancer therapy: a focus on polymeric nanoparticles, micelles, liposomes and exosomes. Curr. Med. Chem. 2018;25(34):4224–4268. doi: 10.2174/0929867324666170830113755. [DOI] [PubMed] [Google Scholar]
- 68.Fernandez-Fernandez A., Manchanda R., McGoron A.J. Theranostic applications of nanomaterials in cancer: drug delivery, image-guided therapy, and multifunctional platforms. Appl. Biochem. Biotechnol. 2011;165(7–8):1628–1651. doi: 10.1007/s12010-011-9383-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turjeman K., Bavli Y., Kizelsztein P., Schilt Y., Allon N., Katzir T.B., Sasson E., Raviv U., Ovadia H., Barenholz Y. Nano-drugs based on nano sterically stabilized liposomes for the treatment of inflammatory neurodegenerative diseases. PloS One. 2015;10(7) doi: 10.1371/journal.pone.0130442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jesorka A., Orwar O. Liposomes: technologies and analytical applications, Annual review of analytical chemistry. Palo Alto, Calif.) 2008;1:801–832. doi: 10.1146/annurev.anchem.1.031207.112747. [DOI] [PubMed] [Google Scholar]
- 71.Allen T.M. Liposomal drug formulations. Rationale for development and what we can expect for the future. Drugs. 1998;56(5):747–756. doi: 10.2165/00003495-199856050-00001. [DOI] [PubMed] [Google Scholar]
- 72.Walker S., Busatto S., Pham A., Tian M., Suh A., Carson K., Quintero A., Lafrence M., Malik H., Santana M.X., Wolfram J. Extracellular vesicle-based drug delivery systems for cancer treatment. Theranostics. 2019;9(26):8001–8017. doi: 10.7150/thno.37097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao X., Wu D., Ma X., Wang J., Hou W., Zhang W. Exosomes as drug carriers for cancer therapy and challenges regarding exosome uptake. Biomed. Pharmacother. 2020;128:110237. doi: 10.1016/j.biopha.2020.110237. [DOI] [PubMed] [Google Scholar]
- 74.Fu S., Wang Y., Xia X., Zheng J.C. Exosome engineering: current progress in cargo loading and targeted delivery. NanoImpact. 2020;20:100261. [Google Scholar]
- 75.Luan X., Sansanaphongpricha K., Myers I., Chen H., Yuan H., Sun D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol. Sin. 2017;38(6):754–763. doi: 10.1038/aps.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun D., Zhuang X., Xiang X., Liu Y., Zhang S., Liu C., Barnes S., Grizzle W., Miller D., Zhang H.G. A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes, Molecular therapy. the journal of the American Society of Gene Therapy. 2010;18(9):1606–1614. doi: 10.1038/mt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yano J., Hirabayashi K., Nakagawa S., Yamaguchi T., Nogawa M., Kashimori I., Naito H., Kitagawa H., Ishiyama K., Ohgi T., Irimura T. Antitumor activity of small interfering RNA/cationic liposome complex in mouse models of cancer. Clin. Canc. Res. : an official journal of the American Association for Cancer Research. 2004;10(22):7721–7726. doi: 10.1158/1078-0432.CCR-04-1049. [DOI] [PubMed] [Google Scholar]
- 78.Pascucci L., Coccè V., Bonomi A., Ami D., Ceccarelli P., Ciusani E., Viganò L., Locatelli A., Sisto F., Doglia S.M., Parati E., Bernardo M.E., Muraca M., Alessandri G., Bondiolotti G., Pessina A. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: a new approach for drug delivery. J. Contr. Release : official journal of the Controlled Release Society. 2014;192:262–270. doi: 10.1016/j.jconrel.2014.07.042. [DOI] [PubMed] [Google Scholar]
- 79.Lu M., Zhao X., Xing H., Xun Z., Zhu S., Lang L., Yang T., Cai C., Wang D., Ding P. Comparison of exosome-mimicking liposomes with conventional liposomes for intracellular delivery of siRNA. Int. J. Pharm. 2018;550(1–2):100–113. doi: 10.1016/j.ijpharm.2018.08.040. [DOI] [PubMed] [Google Scholar]
- 80.Shtam T.A., Kovalev R.A., Varfolomeeva E.Y., Makarov E.M., Kil Y.V., Filatov M.V. Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun. Signal. 2013;11(1):88. doi: 10.1186/1478-811X-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aqil F., Munagala R., Jeyabalan J., Agrawal A.K., Kyakulaga A.-H., Wilcher S.A., Gupta R.C. Milk exosomes - natural nanoparticles for siRNA delivery. Canc. Lett. 2019;449:186–195. doi: 10.1016/j.canlet.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 82.Shandilya S., Rani P., Onteru S.K., Singh D. Natural ligand-receptor mediated loading of siRNA in milk derived exosomes. J. Biotechnol. 2020;318:1–9. doi: 10.1016/j.jbiotec.2020.04.015. [DOI] [PubMed] [Google Scholar]
- 83.Kim M.S., Haney M.J., Zhao Y., Mahajan V., Deygen I., Klyachko N.L., Inskoe E., Piroyan A., Sokolsky M., Okolie O., Hingtgen S.D., Kabanov A.V., Batrakova E.V. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomed. Nanotechnol. Biol. Med. 2016;12(3):655–664. doi: 10.1016/j.nano.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wahlgren J., De L.K.T., Brisslert M., Vaziri Sani F., Telemo E., Sunnerhagen P., Valadi H. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012;40(17):e130. doi: 10.1093/nar/gks463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ohno S., Takanashi M., Sudo K., Ueda S., Ishikawa A., Matsuyama N., Fujita K., Mizutani T., Ohgi T., Ochiya T., Gotoh N., Kuroda M. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells, Molecular therapy. the journal of the American Society of Gene Therapy. 2013;21(1):185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tian Y., Li S., Song J., Ji T., Zhu M., Anderson G.J., Wei J., Nie G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35(7):2383–2390. doi: 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- 87.Jang S.C., Kim O.Y., Yoon C.M., Choi D.-S., Roh T.-Y., Park J., Nilsson J., Lötvall J., Kim Y.-K., Gho Y.S. Bioinspired exosome-mimetic nanovesicles for targeted delivery of chemotherapeutics to malignant tumors. ACS Nano. 2013;7(9):7698–7710. doi: 10.1021/nn402232g. [DOI] [PubMed] [Google Scholar]
- 88.Haney M.J., Klyachko N.L., Zhao Y., Gupta R., Plotnikova E.G., He Z., Patel T., Piroyan A., Sokolsky M., Kabanov A.V., Batrakova E.V. Exosomes as drug delivery vehicles for Parkinson's disease therapy. J. Contr. Release. 2015;207:18–30. doi: 10.1016/j.jconrel.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sato Y.T., Umezaki K., Sawada S., Mukai S.A., Sasaki Y., Harada N., Shiku H., Akiyoshi K. Engineering hybrid exosomes by membrane fusion with liposomes. Sci. Rep. 2016;6:21933. doi: 10.1038/srep21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fuhrmann G., Serio A., Mazo M., Nair R., Stevens M.M. Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. J. Contr. Release : official journal of the Controlled Release Society. 2015;205:35–44. doi: 10.1016/j.jconrel.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 91.Fuhrmann G., Serio A., Mazo M., Nair R., Stevens M.M. Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. J. Contr. Release. 2015;205:35–44. doi: 10.1016/j.jconrel.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 92.Jeyaram A., Lamichhane T.N., Wang S., Zou L., Dahal E., Kronstadt S.M., Levy D., Parajuli B., Knudsen D.R., Chao W., Jay S.M. Enhanced loading of functional miRNA cargo via pH gradient modification of extracellular vesicles. Mol. Ther. 2020;28(3):975–985. doi: 10.1016/j.ymthe.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wei H., Chen J., Wang S., Fu F., Zhu X., Wu C., Liu Z., Zhong G., Lin J. A nanodrug consisting of doxorubicin and exosome derived from mesenchymal stem cells for osteosarcoma treatment in vitro. Int. J. Nanomed. 2019;14:8603–8610. doi: 10.2147/IJN.S218988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sancho-Albero M., Rubio-Ruiz B., Pérez-López A.M., Sebastián V., Martín-Duque P., Arruebo M., Santamaría J., Unciti-Broceta A. Cancer-derived exosomes loaded with ultrathin palladium nanosheets for targeted bioorthogonal catalysis. Nature Catalysis. 2019;2(10):864–872. doi: 10.1038/s41929-019-0333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang D., Qin X., Wu T., Qiao Q., Song Q., Zhang Z. Extracellular vesicles based self-grown gold nanopopcorn for combinatorial chemo-photothermal therapy. Biomaterials. 2019;197:220–228. doi: 10.1016/j.biomaterials.2019.01.024. [DOI] [PubMed] [Google Scholar]
- 96.Dai J., Su Y., Zhong S., Cong L., Liu B., Yang J., Tao Y., He Z., Chen C., Jiang Y. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduction and Targeted Therapy. 2020;5(1):145. doi: 10.1038/s41392-020-00261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Munagala R., Aqil F., Jeyabalan J., Agrawal A.K., Mudd A.M., Kyakulaga A.H., Singh I.P., Vadhanam M.V., Gupta R.C. Exosomal formulation of anthocyanidins against multiple cancer types. Canc. Lett. 2017;393:94–102. doi: 10.1016/j.canlet.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yallapu M.M., Jaggi M., Chauhan S.C. Curcumin nanomedicine: a road to cancer therapeutics. Curr. Pharmaceut. Des. 2013;19(11):1994–2010. doi: 10.2174/138161213805289219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yallapu M.M., Nagesh P.K., Jaggi M., Chauhan S.C. Therapeutic applications of curcumin nanoformulations. AAPS J. 2015;17(6):1341–1356. doi: 10.1208/s12248-015-9811-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vashisht M., Rani P., Onteru S.K., Singh D. Curcumin encapsulated in milk exosomes resists human digestion and possesses enhanced intestinal permeability in vitro. Appl. Biochem. Biotechnol. 2017;183(3):993–1007. doi: 10.1007/s12010-017-2478-4. [DOI] [PubMed] [Google Scholar]
- 101.Agrawal A.K., Aqil F., Jeyabalan J., Spencer W.A., Beck J., Gachuki B.W., Alhakeem S.S., Oben K., Munagala R., Bondada S., Gupta R.C. Milk-derived exosomes for oral delivery of paclitaxel. Nanomed. Nanotechnol. Biol. Med. 2017;13(5):1627–1636. doi: 10.1016/j.nano.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 102.Betker J.L., Angle B.M., Graner M.W., Anchordoquy T.J. The potential of exosomes from cow milk for oral delivery. J. Pharmaceut. Sci. 2019;108(4):1496–1505. doi: 10.1016/j.xphs.2018.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sukreet S., Silva B.V.R.E., Adamec J., Cui J., Zempleni J. Sonication and short-term incubation alter the content of bovine milk exosome cargos and exosome bioavailability (OR26-08-19) Current Developments in Nutrition. 2019;3(1) [Google Scholar]
- 104.Zhu Z., Jing B., Lan X., An R. Goat milk-derived exosomes Endowed with Radioactive and Targeting Properties: potential to provide PET/CT monitoring for exosomes-based drug delivery system in gliomas therapy. J. Nucl. Med. 2020;61(1):1064. [Google Scholar]
- 105.Salunkhe S., Dheeraj, Basak M., Chitkara D., Mittal A. Surface functionalization of exosomes for target-specific delivery and in vivo imaging & tracking: strategies and significance. J. Contr. Release. 2020;326:599–614. doi: 10.1016/j.jconrel.2020.07.042. [DOI] [PubMed] [Google Scholar]
- 106.Sato Y.T., Umezaki K., Sawada S., Mukai S.-a., Sasaki Y., Harada N., Shiku H., Akiyoshi K. Engineering hybrid exosomes by membrane fusion with liposomes. Sci. Rep. 2016;6(1):21933. doi: 10.1038/srep21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kalluri R., LeBleu V.S. The biology and biomedical applications of exosomes. 2020;367(6478) doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.You B., Xu W., Zhang B. Engineering exosomes: a new direction for anticancer treatment. American journal of cancer research. 2018;8(8):1332–1342. [PMC free article] [PubMed] [Google Scholar]
- 109.Zhan Q., Yi K., Qi H., Li S., Li X., Wang Q., Wang Y., Liu C., Qiu M., Yuan X., Zhao J., Hou X., Kang C. Engineering blood exosomes for tumor-targeting efficient gene/chemo combination therapy. Theranostics. 2020;10(17):7889–7905. doi: 10.7150/thno.45028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Smyth T., Petrova K., Payton N.M., Persaud I., Redzic J.S., Graner M.W., Smith-Jones P., Anchordoquy T.J. Surface functionalization of exosomes using click chemistry. Bioconjugate Chem. 2014;25(10):1777–1784. doi: 10.1021/bc500291r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang M., Altinoglu S., Takeda Y.S., Xu Q. Integrating protein engineering and bioorthogonal click conjugation for extracellular vesicle modulation and intracellular delivery. PloS One. 2015;10(11) doi: 10.1371/journal.pone.0141860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Antimisiaris S.G., Mourtas S., Marazioti A. Exosomes and exosome-inspired vesicles for targeted drug delivery. Pharmaceutics. 2018;10(4) doi: 10.3390/pharmaceutics10040218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li D., Yao S., Zhou Z., Shi J., Huang Z., Wu Z. Hyaluronan decoration of milk exosomes directs tumor-specific delivery of doxorubicin. Carbohydr. Res. 2020;493:108032. doi: 10.1016/j.carres.2020.108032. [DOI] [PubMed] [Google Scholar]
- 114.Sukreet S., Silva B.V.R.E., Adamec J., Cui J., Zempleni J. Galactose and sialo-galactose modifications in glycoproteins on the surface of bovine milk exosome are essential for exosome uptake in non-bovine species (OR34-07-19) Current Developments in Nutrition. 2019;3(1) [Google Scholar]
- 115.Shao J., Zaro J., Shen Y. Advances in exosome-based drug delivery and tumor targeting: from tissue distribution to intracellular fate. Int. J. Nanomed. 2020;15:9355–9371. doi: 10.2147/IJN.S281890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.van der Meel R., Fens M.H., Vader P., van Solinge W.W., Eniola-Adefeso O., Schiffelers R.M. Extracellular vesicles as drug delivery systems: lessons from the liposome field. J. Contr. Release : official journal of the Controlled Release Society. 2014;195:72–85. doi: 10.1016/j.jconrel.2014.07.049. [DOI] [PubMed] [Google Scholar]
- 117.Lamparski H.G., Metha-Damani A., Yao J.Y., Patel S., Hsu D.H., Ruegg C., Le Pecq J.B. Production and characterization of clinical grade exosomes derived from dendritic cells. J. Immunol. Methods. 2002;270(2):211–226. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- 118.Hargreaves B. William Reed, William Reed Business Media Ltd 2020.; Broadfield Park, Crawley: 2018. Milk-derived Drug Delivery System Given Roche Backing. [Google Scholar]