Abstract
Objective
To assess the risk factors associated with infections and in-hospital mortality, antimicrobial susceptibility patterns and carbapenem resistance mechanisms in E. anophelis.
Methods
This retrospective case–control study was conducted to reveal the risk factors associated with Elizabethkingia anophelis (E. anophelis) infection and in-hospital mortality in a university tertiary hospital in southwest China, using multivariable logistic-regression analyses. Complete 16S rRNA gene sequencing was used to reconfirm the identity of all isolates. We employed the broth microdilution method to investigate the antimicrobial susceptibility profiles. The presence of resistance genes was confirmed by polymerase chain reaction and DNA sequencing. Full-length resistance genes were cloned into the pET-28a vector for further functional studies.
Results
Our multivariate analysis indicated that coronary artery disease, chronic obstructive pulmonary disease, surgery in the past 6 months, anemia and systemic steroid use were independent risk factors for the acquisition of E. anophelis. Additionally, anemia was the only independent risk factor associated with in-hospital mortality in patients with E. anophelis infections. E. anophelis isolates showed high in-vitro susceptibility towards minocycline (100%) and piperacillin/tazobactam (71.8%), but were resistant to colistin, fosfomycin, ceftazidime/avibactam and aztreonam/avibactam. The PCR revealed the presence of blaGOB and blaBlaB in 37 isolates, and blaCME β-lactamase genes in 36 isolates out of 39 E. anophelis isolates. Additionally, we showed that two metallo-β-lactamases (MBLs) BlaB and GOB, were responsible for carbapenem resistance and the serine-β-lactamase, CME, was functionally involved in resistance to cephalosporins and monobactams. Interestingly, the various putative efflux pumps in E. anophelis were not responsible for resistance.
Conclusion
Our findings will help clinicians to identify high-risk patients and suggests that minocycline should be considered as a therapeutic option for E. anophelis infections. Additionally, carbapenem resistance in E. anophelis is mainly associated with the MBLs, BlaB and GOB, rather than various putative efflux pumps.
Keywords: Elizabethkingia anophelis, risk factors, antimicrobial susceptibility patterns, carbapenem resistance
Introduction
Elizabethkingia anophelis (E. anophelis) is an aerobic, immotile, oxidase-positive, indole-positive, Gram-negative, non-fermenting bacillus, belonging to the genus Elizabethkingia and the family Flavobacteriaceae, which was first isolated from the midgut of the mosquito Anopheles gambiae in 2011.1 The first clinically significant E. anophelis infection was associated with a case of neonatal meningitis in Bangui, Central African Republic in 2011.2
Thereafter, within the genus Elizabethkingia, E. anophelis has emerged as a major nosocomial pathogen, which can cause severe pneumonia, meningitis, infections of the bloodstream, osteomyelitis, endocarditis, endophthalmitis, skin and soft tissue infection, urinary tract infection and abdominal infection.3–10,14,15 Whole genome sequencing (WGS) has recently uncovered a wide range of virulence factors contributing to the pathogenesis of E. anophelis, including products of the capsule, lipopolysaccharides, endopeptidases, lipid biosynthesis and metabolism, magnesium transport proteins, macrophage infectivity, heat shock proteins, catalase, peroxidases, superoxide dismutase, two-component regulatory system, and more.5,18–20
Extensive research has shown that E. anophelis isolates are resistant to most β-lactams, carbapenems and aminoglycosides.4,5,14–17 The emergence of multidrug-resistant (MDR) bacterial pathogens is considered a potential public health hazard, they are widely detected in the environment and their transmission to humans is either by the food chain or via infected animals, poultry, and fish.26–29 Furthermore, MDR bacterial pathogens often pose a therapeutic dilemma for clinicians and are therefore associated with a high mortality rate and poor prognosis.
Recently, the incidence of infections caused by the genus Elizabethkingia has increased continuously worldwide, especially those caused by E. anophelis species. A single-hospital study from South Korea reported that there was an increase in the prevalence of Elizabethkingia genus infections among hospitalized patients from 0.02‰ in 2009 to 0.88‰ in 2017.9 The first recorded outbreak of E. anophelis infection was from Singapore in 2012, in which three out of five patients died of septicemia.3 Furthermore, during 2014–2016, several outbreaks have occurred in the Midwestern United States, including Wisconsin, Illinois, and Michigan, in which the patient fatality rate related to E. anophelis infections ranged from 30.8% to 70%.5–8 Therefore, E. anophelis infection is regarded as profoundly serious and important and should be taken seriously by clinicians.
A recent study indicated that E. anophelis has been continuously misidentified as Elizabethkingia meningoseptica (E. meningoseptica) using conventional methods (API/ID32, Phoenix 100 ID/AST, Vitek 2 and Vitek MS).13 Therefore, most of the previously reported data regarding clinical characteristics, antimicrobial susceptibility patterns and carbapenem resistance mechanisms of E. anopheles, may be incorrect. To date, however, the susceptibility patterns of E. anophelis isolates have not been reported in Mainland China, especially with data collated using the more robust broth dilution method. Genome-wide analysis has revealed that this multidrug-resistant pathogen carries a class A serine-β-lactamase, CME, 2 metallo-β-lactamases, GOB and BlaB, in addition to numerous genes encoding for putative efflux pumps.5,18–20 However, no studies have focused on the function of these putative efflux pumps in E. anophelis isolates. In addition, data reporting on risk factors associated with infection and mortality in E. anophelis infected patients may potentially help clinicians identify high-risk patients and help guide future therapeutic strategies.
The present study was therefore initiated to: (i) identify the risk factors associated with E. anophelis infection and in-hospital mortality, (ii) investigate the antimicrobial susceptibility patterns and carbapenem resistance mechanisms of E. anophelis isolates and (iii) characterize the function of β-lactamases and putative efflux pumps expressed in E. anophelis isolates.
Methods
Isolation and Identification of Bacterial Strains
This study used the clinical microbiology database from a 3200-bed university-affiliated medical center (Chongqing, China) to retrospectively collect those strains that were identified as the genus Elizabethkingia between January 2015 and December 2019. Sampling and isolation of bacterial strains were a part of the routine hospital laboratory procedures and microbial identification was performed in the microbiology laboratory using the VITEK2 compact (bioMérieux, Inc., NC, USA) and the VITEK MS (bioMérieux, MO, USA) systems. All strains from the genus Elizabethkingia were stored at −80°C in 15% glycerol until use. Complete 16S rRNA gene sequencing was used to reconfirm the identity of all isolates. The primers used for amplification and sequencing of the 16S rRNA gene are listed in Table S1 in Additional file 1. The sequences were assembled using SeqMan (DNAStar) and compared with publicly available sequences in the NCBI (http://www.ncbi.nlm.nih.gov) using the BLAST algorithm. Strains were considered to be accurately identified when a strain shared >99.0% 16S rRNA sequence with a type of strain in GenBank. The sequences of 16S rRNA were performed using ClustalW, and the phylogenetic trees were constructed in MEGA7 software using the Neighbor-Joining method.
Data Collection and Clinical Definitions
Electronic medical records of the patients were collected retrospectively and we excluded subjects with the following characteristics: patients with polymicrobial infection and patients admitted for <48 hours. Only the first episode was considered for patients with more than one positive E. anophelis culture. To evaluate the risk factors associated with E. anophelis infection, controls were defined as randomly selected patients with non-E. anophelis infections during the same time (at a 3:1 ratio to the case group). Selected epidemiological, demographic, clinical, laboratory, treatment and outcome data were obtained from the electronic medical records. Empirically administering agents to isolates that were not susceptible was defined as inappropriate empirical antimicrobial therapy. Shock was defined as the coexistence of a systolic pressure of <90 mm Hg and organ dysfunction of the respiratory system, liver, or kidneys. Serum total protein content of <60 g/L or albumin content <25 g/L was the criteria used to define hypoproteinemia. Hypokalemia was diagnosed for a serum potassium level <3.5 mmol/L. Systemic steroid use was defined as oral or intravenous administration of at least 20 mg/day of a steroid (prednisone, hydrocortisone, methylprednisolone, or dexamethasone) within 1 month of infection. We defined anemia as a hemoglobin level of <130 g/L in men and <120 g/L in women according to the World Health Organization (WHO) guidelines. The primary clinical outcome was in-hospital mortality.
Antimicrobial Susceptibility Testing
The reference broth microdilution method was used to evaluate the minimum inhibitory concentrations (MICs) of all antibiotics in E. anophelis and recombinant strains according to the Clinical and Laboratory Standards Institute (CLSI) M07-Ed11 (2019). The criterion suggested by the CLSI for “other non-Enterobacteriaceae” was used to determine the susceptibility of isolates to antibiotics except for ceftazidime/avibactam, aztreonam/avibactam, vancomycin, tigecycline, rifampicin, colistin and fosfomycin. The US Food and Drug Administration (FDA) Enterobacteriaceae criteria were used to interpret isolate susceptibility to tigecycline (resistant MIC ≥ 8 μg/mL, susceptible MIC ≤ 2 μg/mL and intermediate MIC = 4 μg/mL). The MIC breakpoint applied to vancomycin and rifampicin was adapted from the CLSI criteria for Staphylococcus spp. A MIC of ≥16/4 µg/mL was considered resistant for the combination of ceftazidime/avibactam and aztreonam/avibactam. MICs for colistin were interpreted at susceptible breakpoints of ≤2 μg/mL and resistant breakpoints of >2μg/mL according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) Enterobacteriaceae criteria. Likewise, for fosfomycin, we elected to use the susceptible breakpoint of ≤32 μg/mL and resistant breakpoint of >32 μg/mL based on EUCAST Enterobacteriaceae criteria. The reference strains Escherichia coli (ATCC 25922) and Pseudomonas aeruginosa (ATCC 27853) were used as negative controls and quality controls for antibiotic susceptibility testing.
Confirmation and Cloning of Drug Resistance Genes
The presence of the carbapenemase genes (blaKPC, blaIMP, blaVIM, blaNDM, blaBlaB, blaGOB and blaOXA-48-like) and extended-spectrum beta-lactamase (ESBL) genes (blaTEM, blaSHV, blaCME and blaCTX-M) were confirmed by touch-down PCR assays and sequencing. Touch-down PCR amplification was performed using a thermal cycler (Applied Biosystems VeritiPro PCR, CA, USA) under the following cycling conditions: Initial step of 95°C for 3 min, followed by 8 cycles of 92°C for 20 sec, with annealing temperatures starting at 68°C for 20 sec (decreasing 2°C/cycle), and with a final extension at 72°C for 30 sec; this step was followed by 21 cycles of 92°C for 20 sec, 55°C for 20 sec, 72°C for 30 sec, and finally, 72°Cfor 5 min. DNA sequencing was performed using an Applied Biosystems 3730 DNA Analyzer.
We amplified the full-length coding sequences (CDSs) of the genes using specific primers flanked by restriction sites (EcoRI, XhoI or BamHI). These included genes encoding for blaCME, blaBlaB, blaGOB, CzcABC family efflux RND transporter, Efflux ABC transporter (ATP-binding protein), the MATE family of MDR efflux pumps, small multidrug resistance family (SMR) proteins and MFS-type transporter. Amplified PCR fragments were purified and cloned into the corresponding sites within the pET-28a plasmid and then electroporated into BL21 competent E. coli. Positive clones were verified by PCR and sequencing. The sets of primers used for amplification and sequencing of target genes are provided in Table S1 in Additional file 1.
Statistical Analyses
The data were evaluated using SPSS statistical software (version 22.0, IBM). Data are presented as counts (proportions) for categorical variables. Direct comparisons between two groups were determined using the χ2 test or Fisher’s exact test. Means (± standard deviation) were used to express normally distributed continuous variables and the median (± inter-quartile range) was calculated for non-normally distributed variables. Comparisons between two groups were conducted using Student’s t-test for normally distributed variables or a Mann–Whitney U-test for non-normally distributed variables. To evaluate independent risk factors for the infection and in-hospital mortality of E. anophelis isolates, we examined all plausible variables using a univariate analysis. Risk factors with a P value <0.1 as detected by the univariate analysis were included in a multivariate logistic-regression model with the enter method. Odds ratios (ORs) and 95% confidence intervals (CIs) were determined for each risk factor of infection and in-hospital mortality. A two-tailed P value <0.05 was considered statistically significant.
Ethical Considerations
The Institutional Review Board and Ethics Committee of Chongqing Medical University approved this study (approval number: 2020–703). The study was conducted in accordance with the Declaration of Helsinki. The collection of culture isolates and collation of anonymous clinical data was in accordance with the approved clinical practice guidelines. The need to give informed consent for this study was waived by the Institutional Review Board due to the nature of the retrospective analysis with no individual patient identifiers.
Results
Clinical Characteristics
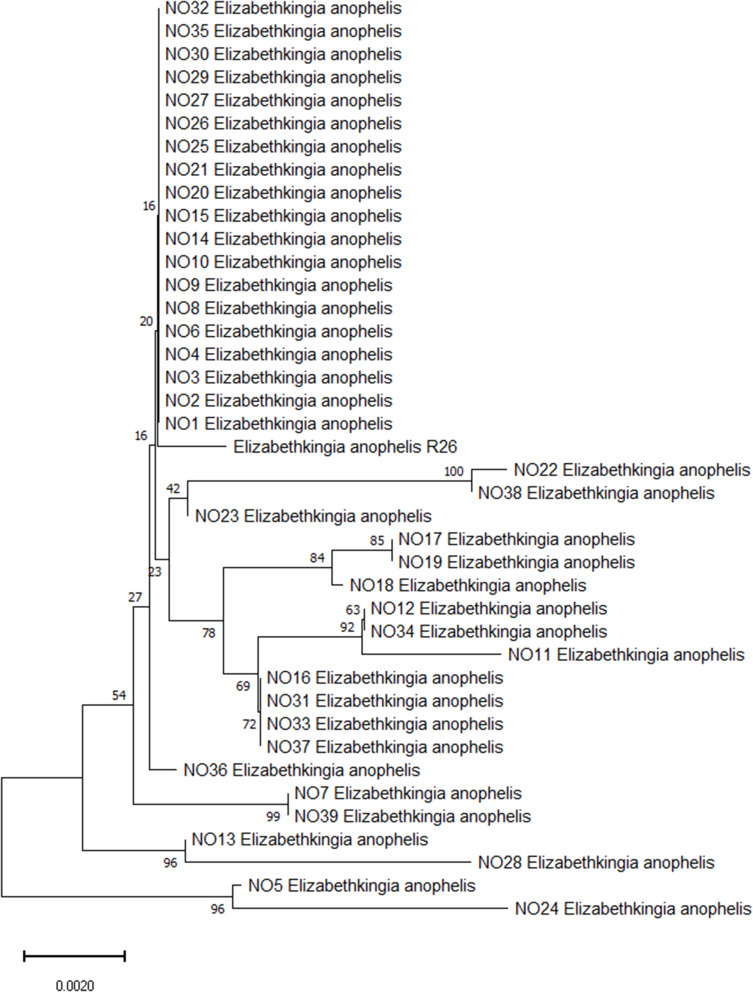
A total of 59 non duplicated Elizabethkingia isolates were collected from the clinical microbiology laboratory for microbial trait investigation. Full 16S rRNA gene sequencing was performed on 59 Elizabethkingia clinical strains. After comparison with available sequences in the NCBI using BLAST, 16S rRNA sequencing showed that 39 (69.6%) isolates were identified as Elizabethkingia anophelis R26. The phylogenetic tree based on 16S rRNA showed the genetic relationship among Elizabethkingia anophelis (Figure 1).
Figure 1.
Phylogenetic tree showing the relationship of the 39 E. anophelis isolates using 16S rRNA gene sequence. Bootstrap support value above branches, the scale bar indicates the number of substitutions per site.
Of the E. anophelis isolates with a documented site of infection, 18 (46.2%) were from the respiratory tract, eight (20.5%) were from the urinary tract, six (15.4%) from blood, three (7.7%) from cerebrospinal fluid, and four were from peritoneal fluid, amniotic fluid, conjunctival sac, and the cornea (Table 5). From January 2015 to December 2019, 39 E. anophelis samples were isolated from 39 consecutive patients who were enrolled in the study. These patients consisted of 21 males (53.8%) and 18 females (46.2%) with a median age of 61.
Table 5.
Empirical Antimicrobial Therapy in Patients Infected with E. anophelis Isolates
| Isolates No. | Specimen | ESBL+MBLs | Empirical Antimicrobial Therapy | Prognosis |
|---|---|---|---|---|
| NO1 | Blood | CME-1+ BlaB-2+ GOB-20 | Ceftazidime+ Levofloxacin+ Teicoplanin | Survival |
| NO2 | Sputum | CME-1+ BlaB-2+ GOB-20 | Cefoperazone-sulbactam+ Minocycline | Non-survival |
| NO3 | Sputum | CME-1+ BlaB-29+ GOB-38 | Cefoperazone-sulbactam+ Teicoplanin | Non-survival |
| NO4 | Urine | CME-1+ BlaB-29+ GOB-38 | Survival | |
| NO5 | Sputum | CME-1+ BlaB-29+ GOB-38 | Cefoperazone-sulbactam+ Minocycline+ Levofloxacin+ Teicoplanin | Non-survival |
| NO6 | Sputum | CME-1+ BlaB-34+ GOB-32 | Piperacillin-tazobactam+ Imipenem | Non-survival |
| NO7 | Urine | CME-1+ BlaB-29+ GOB-38 | Meropenem | Survival |
| NO8 | Urine | CME-1+ BlaB-29+ GOB-38 | Imipenem+ Cefoperazone-sulbactam | Non-survival |
| NO9 | Sputum | CME-1+ BlaB-29+ GOB-38 | Imipenem+ Clindamycin+ Teicoplanin+ Cefepime | Survival |
| NO10 | Blood | CME-1+ BlaB-29+ GOB-38 | Imipenem+ Gentamicin+ Teicoplanin | Non-survival |
| NO11 | Sputum | CME-1+ BlaB-33+ GOB-4 | Cefazolin | Non-survival |
| NO12 | Urine | CME-1+ BlaB-2+ GOB-50 | Teicoplanin | Non-survival |
| NO13 | Sputum | CME-1+ BlaB-29+ GOB-38 | Piperacillin-tazobactam+ ceftriaxone+ Ornidazole | Survival |
| NO14 | Cerebrospinal fluid | Levofloxacin | Survival | |
| NO15 | Blood | CME-1+ BlaB-3+ GOB-45 | Teicoplanin | Survival |
| NO16 | Sputum | CME-1+ BlaB-9+ GOB-32 | Levofloxacin+ Cefazolin+ Amikacin | Non-survival |
| NO17 | Sputum | CME-1+ BlaB-1+ GOB-32 | Imipenem+ Teicoplanin | Non-survival |
| NO18 | Amniotic fluid | CME-1+ BlaB-1+ GOB-38 | Cefuroxime | Survival |
| NO19 | Blood | Ceftriaxone + Vancomycin | Survival | |
| NO20 | Sputum | CME-1+ BlaB-2+ GOB-20 | Cefazolin | Non-survival |
| NO21 | Sputum | CME-1+ BlaB-2+ GOB-50 | Cefoperazone-sulbactam+ Levofloxacin+ Imipenem+ Teicoplanin | Survival |
| NO22 | Sputum | CME-1+ BlaB-17+ GOB-40 | Amikacin | Survival |
| NO23 | Cerebrospinal fluid | CME-1+ BlaB-17+ GOB-20 | Levofloxacin | Survival |
| NO24 | Conjunctival sac | CME-1+ BlaB-29+ GOB-38 | Cefazolin | Survival |
| NO25 | Sputum | CME-2+ BlaB-2+ GOB-39 | Imipenem+ Teicoplanin+ Ornidazole | Survival |
| NO26 | Peritoneal fluid | CME-1+ BlaB-29+ GOB-38 | Meropenem+ Vancomycin | Non-survival |
| NO27 | Urine | CME-1+ BlaB-29+ GOB-38 | Meropenem+ Vancomycin+ Amikacin+ ceftazidime | Non-survival |
| NO28 | Cerebrospinal fluid | CME-1+ BlaB-29+ GOB-38 | Ceftriaxone | Non-survival |
| NO29 | Sputum | CME-1+ BlaB-29+ GOB-38 | Linezolid+ Imipenem | Survival |
| NO30 | Corneal | CME-1+ BlaB-29+ GOB-38 | Amikacin | Survival |
| NO31 | Blood | CME-1+ BlaB-29+ GOB-38 | Imipenem + Levofloxacin+ Amikacin+ Teicoplanin | Non-survival |
| NO32 | Sputum | BlaB-1+ GOB-20 | Cefoperazone-sulbactam+ Minocycline | Non-survival |
| NO33 | Urine | CME-1+ BlaB-1+ GOB-20 | Cefepime+ Cefoperazone-sulbactam | Non-survival |
| NO34 | Sputum | CME-1+ GOB-38 | Imipenem | Survival |
| NO35 | Sputum | CME-1+ BlaB-29+ GOB-38 | Cefoperazone-sulbactam+ Minocycline | Survival |
| NO36 | Urine | CME-1+ BlaB-17+ GOB-50 | Cefoperazone-sulbactam+ Amikacin+ Metronidazole+ Ceftriaxone |
Non-survival |
| NO37 | Blood | CME-2+ BlaB-1+ GOB-20 | Meropenem+ Cefoperazone-sulbactam+ Piperacillin-tazobactam | Non-survival |
| NO38 | Sputum | CME-1+ BlaB-1+ GOB-20 | Imipenem+ Teicoplanin+ Piperacillin-tazobactam | Non-survival |
| NO39 | Urine | CME-1+ BlaB-29+ GOB-38 | Survival |
Abbreviations: ESBL, extended-spectrum beta-lactamase; MBLs, metallo-beta-lactamases; CME, extended-spectrum serine-beta-lactamase CME (class A); BlaB, metallo-beta-lactamase BlaB (subclass B1); GOB, metallo-beta-lactamase GOB (subclass B3).
Notably, chronic obstructive pulmonary disease was the most frequent comorbidity (76.9%), followed by anemia (66.7%). The empirical antibiotics used in the 39 patients included carbapenems (38.5%), β-lactam/lactamase inhibitors (33.3%), β-lactams (30.8%), teicoplanin (25.6%), levofloxacin (17.9%), aminoglycosides (17.9%), minocycline (12.8%) and antibiotics administered either alone or in combination (Table 5), and 84.6% of these empirical antibiotic therapies were deemed as inappropriate antibiotic use. Overall, the in-hospital mortality rate of patients with E. anophelis infection was 51.3%. Furthermore, β-lactam/lactamase inhibitor antibiotics were used significantly more in patients who did not survive (P = 0.041).
Risk Factors and Clinical Outcomes Associated with E. anophelis Infections
Compared to the non-E. anophelis infection controls, the potential risk factors for the acquisition of E. anophelis infections are shown in Tables 1 and 2. Univariate analysis indicated that hypertension, cerebrovascular diseases, chronic obstructive pulmonary disease, renal diseases, surgery in the past 6 months, anemia, hypoproteinemia, and hypokalemia were significantly more frequent in patients with E. anophelis infections (P < 0.05). There was also a positive correlation between patient age and their likelihood of acquiring an E. anophelis infection (P < 0.05). In the multivariate analysis, coronary artery diseases (OR 5.81, 95% CI: 1.09–30.93, P = 0.039), chronic obstructive pulmonary disease (OR 6.71, 95% CI: 1.55–28.99, P = 0.011), surgery in the past 6 months (OR 18.04, 95% CI: 3.29–98.87, P = 0.001), anemia (OR 6.72, 95% CI: 1.12–40.42, P = 0.038) and systemic steroid use (OR 9.87, 95% CI: 1.30–74.94, P = 0.027) were independent risk factors for the acquisition of an E. anophelis infection.
Table 1.
Univariate Analysis of Clinical Features of Patients Infected with E. anophelis Isolates
| Variables | E. anophelis | Controls | Univariable | |
|---|---|---|---|---|
| n=39 (%) | n=117 (%) | OR (95% CI) | P value | |
| Gender (male) | 21 (53.8) | 73 (62.4) | 0.70 (0.34–1.46) | 0.352 |
| Age, median, y (IQR) | 66 (52–76) | 54 (41–67) | – | 0.003 |
| Elderly (≥60 y) | 26 (66.7) | 58 (49.6) | 2.03 (0.95–4.34) | 0.067 |
| ICU admission | 18 (46.2) | 47 (40.2) | 1.28 (0.62–2.65) | 0.575 |
| Underlying diseases and conditions | ||||
| Diabetes | 9 (23.1) | 20 (17.1) | 1.46 (0.60–3.53) | 0.476 |
| Hypertension | 18 (46.2) | 31 (26.5) | 2.38 (1.12–5.04) | 0.029 |
| Shock | 8 (20.5) | 17 (14.5) | 1.52 (0.60–3.86) | 0.450 |
| Solid malignancy | 9 (23.7) | 32 (27.4) | 0.82 (0.35–1.93) | 0.833 |
| Hematologic malignancy | 6 (15.4) | 12 (10.3) | 1.59 (0.55–4.57) | 0.393 |
| Peripheral vascular diseases | 1 (2.6) | 16 (13.7) | 0.17 (0.02–1.30) | 0.073 |
| Coronary artery diseases | 13 (33.3) | 22 (18.8) | 2.16 (0.96–4.86) | 0.076 |
| Cerebrovascular diseases | 15 (38.5) | 20 (17.1) | 3.03 (1.36–6.78) | 0.008 |
| Chronic obstructive pulmonary diseases | 30 (76.9) | 28 (23.9) | 10.60 (4.50–24.98) | 0.000 |
| Gastrointestinal diseases | 14 (35.9) | 26 (22.2) | 1.96 (0.89–4.30) | 0.096 |
| Hepatobiliary diseases | 13 (33.3) | 21 (17.9) | 2.29 (1.01–5.17) | 0.071 |
| Renal diseases | 16 (41.0) | 26 (22.2) | 2.44 (1.12–5.27) | 0.036 |
| Immune system diseases | 0 (0.0) | 14 (12.0) | 0.73 (0.66–0.80) | 0.022 |
| Invasive procedures | ||||
| Mechanical ventilation | 9 (23.1) | 35 (29.9) | 0.70 (0.30–1.63) | 0.538 |
| Central venous catheterization | 14 (35.9) | 27 (23.1) | 1.87 (0.85–4.08) | 0.142 |
| Drainage tube | 11 (28.2) | 35 (29.9) | 0.92 (0.41–2.05) | 1.000 |
| Nasogastric tube insertion | 17 (43.6) | 48 (41.0) | 1.11 (0.53–2.31) | 0.852 |
| Urinary catheter | 16 (41.0) | 59 (50.4) | 0.68 (0.33–1.42) | 0.357 |
| Surgery in the past 6 months | 27 (69.2) | 31 (26.5) | 6.24 (2.82–13.81) | 0.000 |
| Immunosuppressive state | ||||
| Anemia | 26 (66.7) | 14 (12.0) | 14.71 (6.17–35.08) | 0.000 |
| Hypoproteinemia | 36 (92.3) | 60 (51.3) | 11.40 (3.32–39.09) | 0.000 |
| Hypokalemia | 14 (35.9) | 14 (12.0) | 4.12 (1.74–9.74) | 0.002 |
| Chemoradiotherapy | 8 (20.5) | 10 (8.5) | 2.76 (1.00–7.60) | 0.078 |
| Systemic steroid use | 7 (17.9) | 34 (29.1) | 0.53 (0.22–1.33) | 0.210 |
Note: Bold face indicate values that are significant (P < 0.05).
Abbreviations: OR, odds ratio; CI, confidence interval; IQR, interquartile range; ICU, intensive care unit; E. anophelis, Elizabethkingia anophelis.
Table 2.
Multivariate Analysis of Clinical Features of Patients Infected with E. anophelis Isolates
| Variables | Multivariable | |
|---|---|---|
| OR (95% CI) | P value | |
| Coronary artery diseases | 5.81 (1.09–30.93) | 0.039 |
| Chronic obstructive pulmonary diseases | 6.71 (1.55–28.99) | 0.011 |
| Surgery in the past 6 months | 18.04 (3.29–98.87) | 0.001 |
| Anemia | 6.72 (1.12–40.42) | 0.038 |
| Systemic steroid use | 9.87 (1.30–74.94) | 0.027 |
Note: Bold face indicate values that are significant (P < 0.05).
Abbreviations: OR, odds ratio; CI, confidence interval.
Univariate and multivariate logistic-regression analysis results for the factors associated with in-hospital mortality are shown in Table 3. Univariate analysis showed that cerebrovascular disease (P = 0.035), chronic obstructive pulmonary disease (P = 0.020), nasogastric tube insertion (P = 0.008) and anemia (P = 0.002) were associated with a higher mortality rate. Using further multivariate analysis, anemia (OR 86.38, 95% CI: 1.42–5251.29; P = 0.033) was identified as the only independent risk factor for in-hospital mortality in patients with E. anophelis infections.
Table 3.
Risk Factors Associated with In-Hospital Mortality
| Variables | Non-Survival | Survival | Univariable | Multivariable | ||
|---|---|---|---|---|---|---|
| n=20 (%) | n=19 (%) | OR (95% CI) | P value | OR (95% CI) | P value | |
| Gender (female) | 8 (40.0) | 13 (68.4) | 0.31 (0.08–1.15) | 0.080 | ||
| Elderly (≥60 y) | 16 (80.0) | 10 (52.6) | 3.60 (0.87–14.87) | 0.077 | ||
| Drinking history | 2 (10.0) | 7 (36.8) | 0.19 (0.04–1.08) | 0.061 | ||
| ICU admission | 12 (60.0) | 6 (31.6) | 3.25 (0.87–12.14) | 0.080 | ||
| Cerebrovascular disease | 11 (55.0) | 4 (21.1) | 4.58 (1.12–18.80) | 0.035 | ||
| Central nervous system disease | 13 (65.0) | 7 (36.8) | 3.18 (0.86–11.79) | 0.083 | ||
| Chronic obstructive pulmonary disease | 19 (95.0) | 11 (57.9) | 13.82 (1.52–125.65) | 0.020 | ||
| Central venous catheterization | 10 (50.0) | 4 (21.1) | 3.75 (0.92–15.34) | 0.066 | ||
| Nasogastric tube insertion | 13 (65.0) | 4 (21.1) | 6.96 (1.66–29.26) | 0.008 | ||
| Anemia | 19 (95.0) | 7 (36.8) | 32.57 (3.55–298.83) | 0.002 | 86.38 (1.42–5251.29) | 0.033 |
| Systemic steroid use | 5 (25.0) | 2 (10.5) | 2.83 (0.48–16.81) | 0.252 | ||
Note: Bold face indicate values that are significant (P < 0.05).
Abbreviations: OR, odds ratio; CI, confidence interval; ICU, intensive care unit.
Antimicrobial Susceptibility
The susceptibility of the 39 E. anophelis isolates to the antimicrobial agents tested in this study is shown in Table 4. The isolates showed high in-vitro susceptibility towards minocycline (100%), and piperacillin/tazobactam (71.8%) but with lower in-vitro susceptibility towards levofloxacin (38.5%), ciprofloxacin (30.8%), rifampicin (20.5%), piperacillin (17.9%) and tigecycline (10.3%). The MIC50, MIC90 and MIC range for minocycline on the isolates were 0.5, 1, and 0.25–1 μg/mL, respectively, whereas the MIC50, MIC90 and MIC range for piperacillin/tazobactam were 16, 32, and 4–64 μg/mL, respectively. All isolates displayed resistance to vancomycin, ceftazidime, cefepime, aztreonam, ceftazidime/clavulanic acid, cefepime/clavulanic acid, colistin and fosfomycin according to the breakpoints used. It is worth noting that while all isolates produced the MBLs BlaB and GOB, aztreonam/avibactam could not further inhibit growth.
Table 4.
Antimicrobial Susceptibilities of E. Anophelis Isolates Determined by the Broth Microdilution Method
| Antimicrobial Agents | Breakpoint (μg/mL) | MIC (μg/mL) | Susceptibility (%) | |||||
|---|---|---|---|---|---|---|---|---|
| Susceptible | Resistant | Range | 50% | 90% | Susceptible | Intermediate | Resistant | |
| Piperacillin | ≤16 | ≥128 | 16–128 | 32 | 64 | 17.9 | 74.4 | 7.7 |
| Piperacillin-TZB | ≤16 | ≥128 | 4–64 | 16 | 32 | 71.8 | 28.2 | 0 |
| Ceftazidime | ≤8 | ≥32 | 128 to >256 | >256 | >256 | 0 | 0 | 100 |
| Ceftazidime-CLA | ≤8 | ≥32 | 32 to >256 | >256 | >256 | 0 | 0 | 100 |
| Ceftazidime-AVI | ≤8 | ≥16 | 64 to >256 | >256 | >256 | 0 | 0 | 100 |
| Cefepime | ≤8 | ≥32 | 32 to >256 | >256 | >256 | 0 | 0 | 100 |
| Cefepime-CLA | ≤8 | ≥32 | 32 to >256 | 256 | >256 | 0 | 0 | 100 |
| Aztreonam | ≤8 | ≥32 | >256 | >256 | >256 | 0 | 0 | 100 |
| Aztreonam-AVI | ≤8 | ≥16 | >256 | >256 | >256 | 0 | 0 | 100 |
| Imipenem | ≤4 | ≥16 | 16 to >256 | 64 | 128 | 0 | 0 | 100 |
| Meropenem | ≤4 | ≥16 | 16 to >256 | 64 | 256 | 0 | 0 | 100 |
| Levofloxacin | ≤2 | ≥8 | 0.5–32 | 8 | 16 | 38.5 | 2.6 | 60.0 |
| Ciprofloxacin | ≤1 | ≥4 | 0.5 to >256 | 64 | 256 | 30.8 | 2.6 | 66.6 |
| Minocycline | ≤4 | ≥16 | 0.25–1 | 0.5 | 1 | 100 | 0 | 0 |
| Tigecycline | ≤2 | ≥8 | 2–8 | 4 | 4 | 10.3 | 82.1 | 7.6 |
| Fosfomycin | ≤32 | >32 | >256 | >256 | >256 | 0 | 0 | 100 |
| Colistin | ≤2 | >2 | >256 | >256 | >256 | 0 | 0 | 100 |
| Vancomycin | ≤2 | ≥16 | 8–32 | 16 | 32 | 0 | 20.5 | 79.5 |
| Rifampicin | ≤1 | ≥4 | 1–32 | 2 | 16 | 20.5 | 33.3 | 46.2 |
Abbreviations: MIC, minimum inhibitory concentration; TZB, tazobactam at a fixed concentration of 4 μg/mL; AVI, avibactam at a fixed concentration of 4 μg/mL; CLA, clavulanic acid at a fixed concentration of 2 μg/mL.
Identification of β-Lactamases Present in Elizabethkingia anophelis Isolates
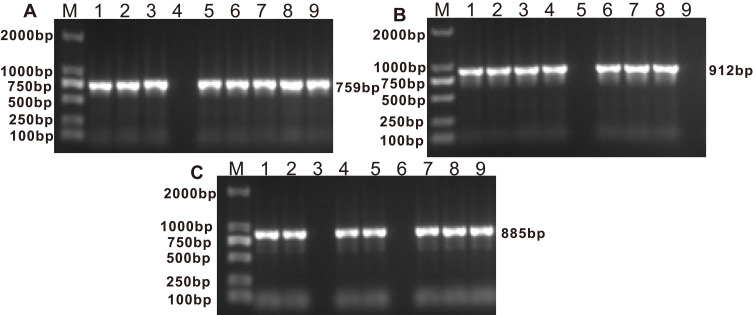
PCR experiments were used to detect the presence of blaGOB and blaBlaB in 37 isolates, and blaCME β-lactamase genes in 36 isolates from the original 39 E. anophelis isolates (Table 5 and Figure 2). Sequence alignments showed that E. anophelis strains harbored two types of the blaCME gene, blaCME-1 (n = 34) and blaCME-2 (n = 2), eight types of the blaBlaB gene, including blaBlaB-29 (17), blaBlaB-2 (7), blaBlaB-1 (5), blaBlaB-17 (3), blaBlaB-3 (1), blaBlaB-9 (1), blaBlaB-33 (1) and blaBlaB-34 (1), and eight types of the blaGOB gene, including blaGOB-38 (19), blaGOB-20 (8), blaGOB-32 (3), blaGOB-50 (3), blaGOB-39 (1), blaGOB-4 (1), blaGOB-40 (1) and blaGOB-45 (1). The most detected combination of β-lactamases was CME-1 + BlaB-29 + GOB-38 (n = 17).
Figure 2.
(A) Electrophoretic pattern of BlaB gene (759 bp); M: 100–2000 bp DNA ladder; Lanes 1, 2, 3, 5, 6, 7, 8, 9: positive E. anophelis strains; Lanes 4: negative E. anophelis strain. (B) Electrophoretic pattern of CME gene (912 bp); M: 100–2000 bp DNA ladder; Lanes 1, 2, 3, 4, 6, 7, 8: positive E. anophelis strains; Lanes 5, 9: negative E. anophelis strains. (C) Electrophoretic pattern of GOB gene (885 bp); M: 100–2000 bp DNA ladder; Lanes 1, 2, 4, 5, 7, 8, 9: positive E. anophelis strains; Lanes 3, 6: negative E. anophelis strains.
We also investigated the presence of other β-lactamase genes, however, none of the 39 E. anophelis isolates harbored carbapenemase genes (blaKPC, blaIMP, blaVIM, blaNDM and blaOXA-48-like) or ESBL genes (blaTEM, blaSHV and blaCTX-M).
MICs of Recombinant Strains
To further evaluate the function of β-lactamases, the most prevalent forms of blaCME, blaBlaB and blaGOB genes from E. anophelis isolates were cloned into a pET28a(+) plasmid vector. We also amplified and cloned genes encoding putative efflux pump proteins including the CzcABC family efflux RND transporter, Efflux ABC transporter (ATP-binding protein), MATE family members of the MDR efflux pump, small multidrug resistance family (SMR) proteins and MFS-type transporter. These plasmids were transformed into BL21 (DE3) E.coli and the MICs of the common antibiotics were tested in the resultant strains. The strain transformed with pET-CME displayed an increased MIC for ampicillin, piperacillin, cefazolin, cefuroxime, ceftazidime, ceftriaxone and aztreonam when compared to the pET28a(+) vector construct. This suggests that the extended-spectrum serine-β-lactamase CME is functionally involved in cephalosporin and monobactam resistance (Table 6). The MIC for imipenem increased 32-fold (from 0.125 to 4 μg/mL) in the presence of the pET-BlaB construct and 16-fold (from 0.125 to 2 μg/mL) in the presence of the pET-GOB construct. This suggests that the MBLs BlaB and GOB, are responsible for increased imipenem resistance (Table 6). Along with the increase in imipenem resistance, the pET-BlaB and pET-GOB constructs also conferred an increased MIC for ampicillin, piperacillin, cefazolin, cefuroxime, and ceftazidime. This indicates that the MBLs BlaB and GOB can also degrade ampicillin, piperacillin, cefazolin, cefuroxime, and ceftazidime (Table 6). All the efflux pump transformants tested, including pET-ABC, pET-MFS, pET-MATE, pET-SMR and pET-RND did not result in increased MICs for any of the antibiotics tested (Table 6).
Table 6.
The Antibiotic Susceptibilities of BL21 (DE3) E. coli Expressing CME, BlaB, GOB, Putative Efflux Proteins or the pET28a(+) Vector
| BL21 with Recombinant Plasmid | MIC (μg/mL) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | PIP | CFZ | CXM | CAZ | CRO | FEP | AZT | IPM | AK | CIP | TGC | |
| pET28a(+) vector | 0.5 | 0.5 | 1 | 1 | 1 | <0.125 | <0.125 | <0.125 | 0.125 | 8 | <0.125 | 4 |
| pET-CME | 32 | 4 | 32 | 32 | 64 | 1 | <0.125 | 8 | 0.25 | 8 | <0.125 | 4 |
| pET-BlaB | 256 | 128 | 2 | 16 | 4 | <0.125 | <0.125 | <0.125 | 4 | 4 | <0.125 | 4 |
| pET-GOB | 4 | 2 | 4 | 8 | 16 | <0.125 | <0.125 | <0.125 | 2 | 4 | <0.125 | 4 |
| pET-ABC | 0.5 | 1 | 1 | 1 | 1 | <0.125 | <0.125 | <0.125 | 0.25 | 8 | <0.125 | 4 |
| pET-MFS | 1 | 0.5 | 2 | 1 | 1 | <0.125 | <0.125 | <0.125 | 0.25 | 8 | <0.125 | 4 |
| pET-MATE | 0.5 | 0.5 | 2 | 1 | 1 | <0.125 | <0.125 | <0.125 | 0.25 | 8 | <0.125 | 4 |
| pET-SMR | 1 | 0.5 | 2 | 1 | 1 | <0.125 | <0.125 | <0.125 | 0.25 | 8 | <0.125 | 4 |
| pET-RND | 0.5 | 0.5 | 2 | 1 | 1 | <0.125 | <0.125 | <0.125 | 0.25 | 8 | <0.125 | 4 |
Note: Bold face indicate MIC values that increase at least 4-fold as compared to the pET28a(+) vector.
Abbreviations: MIC, minimum inhibitory concentration; AMP, ampicillin; PIP, piperacillin; CFZ, cefazolin; CXM, cefuroxime; CAZ, ceftazidime; CRO, ceftriaxone; FEP, cefepime; AZT, aztreonam; IPM, imipenem; AK, amikacin; CIP, ciprofloxacin; TGC, tigecycline; CME, extended-spectrum serine-beta-lactamase CME (class A); BlaB, metallo-beta-lactamase BlaB (subclass B1); GOB, metallo-beta-lactamase GOB (subclass B3); ABC, efflux ABC transporter (ATP-binding protein); MFS, MFS-type transporter; MATE, MATE family of MDR efflux pump; SMR, small multidrug resistance family (SMR) protein; RND, CzcABC family efflux RND transporter.
Discussion
Infection with E. anophelis in humans is increasing in many countries and there have been several reports of E. anophelis outbreaks in the community and nosocomial environment in Singapore and the Midwestern United States, including Wisconsin, Illinois, and Michigan.3–10 However, as suggested previously, E. anophelis is commonly misidentified as E. meningoseptica when using biochemical identification methodologies or automated identification systems in clinical settings. Therefore, data regarding the clinical features, clinical prognosis, and the antimicrobial susceptibility profiles of E. anophelis could be skewed. For these reasons, we have, for the first time identified the risk factors associated with the acquisition of E. anophelis and we have found that anemia is an independent risk factor for in-hospital mortality in patients with E. anophelis infections. Moreover, we demonstrate for the first time that various putative efflux pumps found in E. anophelis do not alter antimicrobial resistance and therefore, do not possess a drug efflux function. β-Lactamases were commonly found in E. anophelis isolates and the MBLs BlaB and GOB, are responsible for carbapenem resistance, whereas the ESBL, CME is functionally involved in resistance to cephalosporins and monobactams.
In the present study, we initially explored risk factors associated with E. anophelis infection and using multivariate analysis, we found that coronary artery diseases, chronic obstructive pulmonary disease, surgery in the past 6 months, anemia and systemic steroid use were independently associated with E. anophelis infection. Previous studies have suggested that patients with E. anophelis infection could have greater underlying comorbidities and this study is the first to provide a statistical analysis to support this hypothesis.3–10 Moreover, surgery in the past 6 months, anemia and systemic steroid use are known to compromise the immune status of the patients. Therefore, these patients are more vulnerable to the acquisition of E. anophelis infection in the same hospital environment.
In previous reports, the case fatality rate of patients with E. anophelis infection ranged from 24% to 60% in different countries.4–14 In line with this, our study showed that the in-hospital mortality rate of patients with an E. anophelis infection was 51.3%. We, therefore, went on to explored the factors influencing this mortality. When compared with the only previous study investigating risk factors for mortality,10 our study demonstrated that anemia was the only independent predictor of mortality in patients infected with E. anophelis, a factor which has not been reported previously and these patients may present as anemic because of the hemolytic activity of E. anophelis. Several studies have reported that this bacterium can lyse erythrocytes to access essential nutrients (such as amino acids) using hemolysins and heme-degrading proteins.20–22 This process may alter the host's physiological status and compromise the immune system, thereby worsening the patient prognosis with E. anophelis. Therefore, anemic patients with a confirmed E. anophelis infection should be considered as higher risk and should be given greater scrutiny and special care. Unexpectedly, inadequate antibiotic therapy was not associated with mortality, this is probably because of the small sample size of infected patients in our study.
Published information concerning the antimicrobial susceptibility patterns of E. anophelis, identified using reliable methods is limited. Studies from Singapore and Taiwan have demonstrated that E. anophelis was resistant to carbapenems, β-lactams, β-lactam/β-lactam inhibitor and aminoglycosides.14–16 However, previous research showed that the susceptibility of E. anophelis to fluoroquinolones, tigecycline, piperacillin, piperacillin-tazobactam and trimethoprim-sulfamethoxazole was variable.4–6,10,17 Using a disk diffusion or agar dilution test, studies from Hong Kong, South Korea and the USA reported the following susceptibilities of E. anophelis to ciprofloxacin (22%–100%), levofloxacin (29%–96%), piperacillin (41.1%–100%), piperacillin-tazobactam (92%), vancomycin (0%–100%) and trimethoprim-sulfamethoxazole (22%–70.6%).4,5,17 However, when using the broth microdilution test, researchers from Taiwan and Singapore showed the following susceptibilities of E. anophelis to ciprofloxacin (1%–21.5%), levofloxacin (16%–78.5%), piperacillin (19.4%), piperacillin-tazobactam (30.6%–92.4%), vancomycin (0%), tigecycline (5.1%–26.4%), minocycline (97.5%–100%) and trimethoprim-sulfamethoxazole (4%–92.4%).14–16 As shown above, there are huge discrepancies in the susceptibility levels of E. anophelis to ciprofloxacin, levofloxacin, piperacillin, and vancomycin when comparing the disk diffusion or agar dilution tests and the broth microdilution test. This suggests that the susceptibility of E. anophelis seen using the disk diffusion test or agar dilution test may be unreliable and inaccurate, as the broth microdilution test represents the gold-standard method for antimicrobial susceptibility testing.
There are also obvious differences in the susceptibility of E. anophelis to antibacterial agents when the standard broth microdilution test is used. A possible explanation for this inconsistency is that natural geographical differences cause variation in the susceptibility patterns observed in previous studies. It is therefore necessary to investigate the antimicrobial susceptibility of E. anophelis in local areas as a guide to antibiotic selection. Our study showed the following susceptibility of E. anophelis to various treatments: minocycline (100%), piperacillin-tazobactam (71.8%), levofloxacin (38.5%), ciprofloxacin (30.8%), piperacillin (17.9%), rifampicin (20.5%) and tigecycline (10.3%). All isolates displayed resistance to ceftazidime, cefepime, aztreonam, ceftazidime/clavulanic acid, cefepime/clavulanic acid, colistin and fosfomycin, according to the breakpoints used. Most of the antimicrobial susceptibility results in this study are consistent with those of previous studies performed using the broth microdilution test. These results suggest that antimicrobial therapy for E. anophelis should prioritize minocycline or piperacillin-tazobactam. However, in our study, patients in the non-survival group were treated with significantly more β-lactam/lactamase inhibitor antibiotics as compared to the survival group (P = 0.041). This observation indicates that piperacillin-tazobactam is not an effective treatment for E. anophelis infections. This study was limited by the fact that antimicrobial susceptibility in-vitro does not equate to in-vivo clinical efficacy. Therefore, large prospective clinical trials are urgently needed to validate therapeutic recommendations.
In this study, we screened for the presence of carbapenemase genes (blaKPC, blaIMP, blaVIM, blaNDM, blaBlaB, blaGOB and blaOXA-48-like) and ESBL genes (blaTEM, blaSHV, blaCME and blaCTX-M) in all 39 E. anophelis isolates. No carbapenemase genes (blaKPC, blaIMP, blaVIM, blaNDM and blaOXA-48-like) or ESBL genes (blaTEM, blaSHV and blaCTX-M) were detected in any isolates. However, we identified 35 (89.7%) isolates co-harboring blaGOB, blaBlaB and blaCME β-lactamase genes. The most detected combination of β-lactamases was CME-1, BlaB-29, and GOB-38 (n = 17).
To further evaluate the function of β-lactamases, recombinant strains harboring either blaCME, blaBlaB or blaGOB were constructed. The transformed strain expressing CMEs displayed an increased MIC for ampicillin, piperacillin, cefazolin, cefuroxime, ceftazidime, ceftriaxone and aztreonam as compared to the pET28a(+) vector construct. This suggests that the ESBL CME is functionally involved in resistance to cephalosporins and monobactams. The MIC for imipenem increased 32-fold (from 0.125 to 4 μg/mL) in the presence of the BlaB-expressing construct and 16-fold (from 0.125 to 2 μg/mL) in the presence of the GOB-expressing construct. This suggests that the metallo-β-lactamases BlaB and GOB are responsible for the observed carbapenem resistance.
Aztreonam/avibactam is a novel class of combinational β-lactamase-inhibitor, designed to treat serious infections of metallo-β-lactamase (MBL)-producing Gram-negative bacteria, which is currently in Phase I clinical trials (NCT01689207). Aztreonam is relatively stable against MBL hydrolysis, however, it is easily inactivated by class A (eg, KPC), class C (eg, AmpC) and certain class D (eg, OXA-48) serine-β-lactamase enzymes.23 Avibactam potently inhibits class A, class C and certain class D serine-β-lactamase enzymes and displays a broader β-lactamase inhibition profile than other β-lactamase inhibitors.23 When in combination, aztreonam/avibactam is effective against isolates co-producing ESBLs and MBLs with porin loss/deficiency.24 However, it was quite unexpected that E. anophelis is resistant to aztreonam/avibactam according to our experimental results.
Genomic annotation of all Elizabethkingia spp. reveals that besides β-lactamases, there are also numerous putative efflux pump proteins including CzcABC family efflux RND transporter, Efflux ABC transporter (ATP-binding protein), MATE family of MDR efflux pumps, small multidrug resistance family (SMR) proteins and MFS-type transporter. Interestingly, however, none of these transporters have been phenotypically characterized.5,18–20 It was, therefore, critical to investigate the function of these putative efflux pumps. Our data showed that all recombinant efflux pump strains including pET-ABC, pET-MFS, pET-MATE, pET-SMR and pET-RND did not result in increased MICs for β-lactam and non-β-lactam antibiotics. These results suggest that the putative efflux pump genes from E. anophelis are not responsible for antimicrobial drug resistance. Similarly, Schindler et al cloned and expressed 21 putative efflux pump genes in Staphylococcus aureus which had no effect on any of the antibiotics tested.25 In summary, we demonstrate for the first time that the various putative efflux pumps found in E. anophelis do not possess antimicrobial drug efflux function.
There were some limitations to our study; however, firstly, the small sample size from the single-center study prevented its translation to the wider population. However, the identification of both a carbapenem resistance mechanism and the susceptibility profile of the drug-resistant E. anophelis are of great clinical importance and warrant an urgent, wider, in-depth study. Secondly, no further investigation into the clonality of these isolates was performed, so that the possibility of infection outbreaks cannot be ruled out.
Conclusions
In conclusion, this study provided a detailed report of risk factors, antimicrobial susceptibility patterns and carbapenem resistance mechanisms in E. anophelis clinical isolates from one medical center in Southwest China. Our data showed that patients with anemia, coronary artery diseases, chronic obstructive pulmonary disease or patients who have received systemic steroids or surgery in the past 6 months are more likely to acquire an E. anophelis infection. Furthermore, patients with anemia have a worse prognosis and therefore require more attention and special care from clinicians. The collected clinical isolates exhibited remarkable multidrug resistance to colistin, fosfomycin, aztreonam/avibactam and tigecycline, which are all regarded as last-resort treatments for carbapenem-resistant Enterobacteriaceae, while minocycline is the most effective antibiotic against E. anophelis in-vitro. Mechanistic analysis revealed that carbapenem resistance is associated with the hydrolytic activity of the MBLs BlaB and GOB and is not associated with various putative efflux pumps expressed in E. anophelis. Future in-vivo and prospective clinical trials are urgently needed to determine optimal antimicrobial agent efficacies based on in-vitro drug susceptibility testing results and resistance mechanisms.
Acknowledgments
This study was supported by the Natural Science Foundation of Chongqing (No.cstc2019jcyj-msxmX0253). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Kampfer P, Matthews H, Glaeser SP, Martin K, Lodders N, Faye I. Elizabethkingia anophelis sp. nov., isolated from the midgut of the mosquito Anopheles gambiae. Int J Syst Evol Microbiol. 2011;61(Pt11):2670–2675. doi: 10.1099/ijs.0.026393-0 [DOI] [PubMed] [Google Scholar]
- 2.Frank T, Gody JC, Nguyen LB, et al. First case of Elizabethkingia anophelis meningitis in the Central African Republic. Lancet. 2013;381(9880):1876. doi: 10.1016/S0140-6736(13)60318-9 [DOI] [PubMed] [Google Scholar]
- 3.Teo J, Tan SY, Tay M, et al. First case of E anophelis outbreak in an intensive-care unit. Lancet. 2013;382(9895):855–856. doi: 10.1016/S0140-6736(13)61858-9 [DOI] [PubMed] [Google Scholar]
- 4.Lau SK, Chow WN, Foo CH, et al. Elizabethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci Rep. 2016;6:26045. doi: 10.1038/srep26045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perrin A, Larsonneur E, Nicholson AC, et al. Evolutionary dynamics and genomic features of the Elizabethkingia anophelis 2015 to 2016 Wisconsin outbreak strain. Nat Commun. 2017;8:15483. doi: 10.1038/ncomms15483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navon L, Clegg WJ, Morgan J, et al. Notes from the field: investigation of Elizabethkingia anophelis cluster - Illinois, 2014–2016. MMWR Morb Mortal Wkly Rep. 2016;65(48):1380–1381. doi: 10.15585/mmwr.mm6548a6 [DOI] [PubMed] [Google Scholar]
- 7.Wisconsin department of health services: Elizabethkingia. Available from: https://www.dhs.wisconsin.gov/disease/elizabethkingia.htm. Accessed April17, 2017.
- 8.CDC. Elizabethkingia, recent outbreaks. Available from: https://www.cdc.gov/elizabethkingia/outbreaks/. Accessed December26, 2016.
- 9.Choi MH, Kim M, Jeong SJ, et al. Risk factors for Elizabethkingia acquisition and clinical characteristics of patients, South Korea. Emerg Infect Dis. 2019;25(1):42–51. doi: 10.3201/eid2501.171985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin JN, Lai CH, Yang CH, Huang YH, Lin HH. Clinical manifestations, molecular characteristics, antimicrobial susceptibility patterns and contributions of target gene mutation to fluoroquinolone resistance in Elizabethkingia anophelis. J Antimicrob Chemother. 2018;73(9):2497–2502. doi: 10.1093/jac/dky197 [DOI] [PubMed] [Google Scholar]
- 11.Janda JM, Lopez DL. Mini review: new pathogen profiles: Elizabethkingia anophelis. Diagn Microbiol Infect Dis. 2017;88(2):201–205. doi: 10.1016/j.diagmicrobio.2017.03.007 [DOI] [PubMed] [Google Scholar]
- 12.Lin JN, Lai CH, Yang CH, Huang YH. Elizabethkingia infections in humans: from genomics to clinics. Microorganisms. 2019;7(9):295. doi: 10.3390/microorganisms7090295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin JN, Lai CH, Yang CH, Huang YH, Lin HF, Lin HH. Comparison of four automated microbiology systems with 16S rRNA gene sequencing for identification of Chryseobacterium and Elizabethkingia species. Sci Rep. 2017;7(1):13824. doi: 10.1038/s41598-017-14244-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin JN, Lai CH, Yang CH, Huang YH. Comparison of clinical manifestations, antimicrobial susceptibility patterns, and mutations of fluoroquinolone target genes between Elizabethkingia meningoseptica and Elizabethkingia anophelis isolated in Taiwan. J Clin Med. 2018;7(12):538. doi: 10.3390/jcm7120538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chew KL, Cheng B, Lin RTP, Teo JWP. Elizabethkingia anophelis is the dominant Elizabethkingia species found in blood cultures in Singapore. J Clin Microbiol. 2018;56(3). doi: 10.1128/JCM.01445-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng YH, Perng CL, Jian MJ, et al. Multicentre study evaluating matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of clinically isolated Elizabethkingia species and analysis of antimicrobial susceptibility. Clin Microbiol Infect. 2019;25(3):340–345. doi: 10.1016/j.cmi.2018.04.015 [DOI] [PubMed] [Google Scholar]
- 17.Han MS, Kim H, Lee Y, et al. Relative prevalence and antimicrobial susceptibility of clinical isolates of Elizabethkingia species based on 16S rRNA gene sequencing. J Clin Microbiol. 2017;55(1):274–280. doi: 10.1128/JCM.01637-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin JN, Lai CH, Yang CH, Huang YH, Lin HH. Genomic features, phylogenetic relationships, and comparative genomics of Elizabethkingia anophelis strain EM361-97 isolated in Taiwan. Sci Rep. 2017;7(1):14317. doi: 10.1038/s41598-017-14841-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breurec S, Criscuolo A, Diancourt L, et al. Genomic epidemiology and global diversity of the emerging bacterial pathogen Elizabethkingia anophelis. Sci Rep. 2016;6:30379. doi: 10.1038/srep30379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Liu Y, Chew SC, et al. Complete genome sequence and transcriptomic analysis of the novel pathogen Elizabethkingia anophelis in response to oxidative stress. Genome Biol Evol. 2015;7(6):1676–1685. doi: 10.1093/gbe/evv101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen S, Bagdasarian M, Walker ED. Elizabethkingia anophelis: molecular manipulation and interactions with mosquito hosts. Appl Environ Microbiol. 2015;81(6):2233–2243. doi: 10.1128/AEM.03733-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kukutla P, Lindberg BG, Pei D, et al. Insights from the genome annotation of Elizabethkingia anophelis from the malaria vector Anopheles gambiae. PLoS One. 2014;9(5):e97715. doi: 10.1371/journal.pone.0097715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong D, van Duin D. Novel beta-lactamase inhibitors: unlocking their potential in therapy. Drugs. 2017;77(6):615–628. doi: 10.1007/s40265-017-0725-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou H, Xiong SJ, Lin QX, Wu ML, Niu SQ, Huang SF. CP-CRE/non-CP-CRE stratification and CRE resistance mechanism determination help in better managing CRE bacteremia using ceftazidime-avibactam and aztreonam-avibactam. Infect Drug Resist. 2019;12:3017–3027. doi: 10.2147/IDR.S219635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schindler BD, Frempong-Manso E, DeMarco CE, et al. Analyses of multidrug efflux pump-like proteins encoded on the Staphylococcus aureus chromosome. Antimicrob Agents Chemother. 2015;59(1):747–748. doi: 10.1128/AAC.04678-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makharita RR, El-Kholy I, Hetta HF, et al. Antibiogram and genetic characterization of carbapenem-resistant gram-negative pathogens incriminated in healthcare-associated infections. Infect Drug Resist. 2020;13:3991–4002. doi: 10.2147/IDR.S276975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Algammal AM, Mabrok M, Sivaramasamy E, et al. Emerging MDR-Pseudomonas aeruginosa in fish commonly harbor oprL and toxA virulence genes and blaTEM, blaCTX-M, and tetA antibiotic-resistance genes. Sci Rep. 2020;10(1):15961. doi: 10.1038/s41598-020-72264-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Algammal AM, Enany ME, El-Tarabili RM, Ghobashy MOI, Helmy YA. Prevalence, antimicrobial resistance profiles, virulence and enterotoxins-determinant genes of MRSA isolated from subclinical bovine mastitis in Egypt. Pathogens. 2020;9(5):362. doi: 10.3390/pathogens9050362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enany ME, Algammal AM, Nasef SA, et al. The occurrence of the multidrug resistance (MDR) and the prevalence of virulence genes and QACs resistance genes in E. coli isolated from environmental and avian sources. AMB Express. 2019;9(1):192. doi: 10.1186/s13568-019-0920-4 [DOI] [PMC free article] [PubMed] [Google Scholar]