Abstract
Purpose
Long noncoding RNAs growth arrest-specific 5 (GAS5) exerts important functions in modulating various tumor behaviors. However, the role of lncRNA GAS5 in laryngeal squamous cell carcinoma (LSCC) remains unknown.
Materials and Methods
Cell viability and apoptosis were, respectively, detected by cell counting kit-8 and flow cytometry, DIANA-LncBase V, Starbase, TargetScan and a dual-luciferase reporter gene assay were employed to assess the relationship among GAS5, miR-26a-5p and uncoordinated 51-like kinase 1 (ULK2), and quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and Western blot were performed to detect the expression of autophagy-relative factors.
Results
The expression level of GAS5 was frequently decreased in LSCC cell lines, and up-regulated GAS5 inhibited AMC-HN-8 cells viability and induced apoptosis. More importantly, we found that GAS5 activated autophagy, with enhanced autophagy-related proteins after GAS5 overexpression. While down-regulated GAS5 had opposite results in Tu 177 cells, GAS5 was found to act as a microRNA sponge in a pathway to regulate miR-26a-5p and its target gene ULK2. MiR-26a-5p mimics inhibited apoptosis and autophagy, which were reversed by GAS5 and siGAS5 in AMC-HN-8 cells and Tu 177 cells, as well as ULK2 in AMC-HN-8 cells. Meanwhile, the concomitant downregulation of ULK2 and miRNA-26a-5p inhibitor decreased the miRNA-26a-5p inhibitor-induced apoptosis and autophagy.
Conclusion
This is the first report of LncRNA GAS5 acting as a tumor suppressor in LSCC by regulating the miR-26a-5p/ULK2 axis, and it could be a new target for gene therapy in LSCC.
Keywords: long noncoding RNAs growth arrest-specific 5, laryngeal squamous cell carcinoma, miR-26a-5p, apoptosis, autophagy
Introduction
Laryngeal cancer is a common head and neck cancer. In many areas, it is second only to nasopharyngeal carcinoma in head and neck tumors. Clinically, laryngeal cancer is mainly characterized by hoarseness, dyspnea, and neck mass. The incidence rate of laryngeal cancer in men is higher than in women, with about 10:1 ratio of male to female incidence in the world.1 Although treatment of laryngeal cancer has been developed for over one hundred years, the related pathogenesis and treatment mechanism of basic have been in progress, the treatment effect is still not satisfactory, postoperative health conditions, particularly the survival rate remains to be an improvement. Thus, it is of important significance to explore a new therapeutic strategy for the treatment of the cancer.
The mammalian genome-wide report shows that about 70–80% of DNAs in genes are transcribed, but protein-coding genes actually account for only 1–2% of the genome.2 The other RNA, which is transcribed but does not have the protein-coding ability, is called non-coding RNA.3,4 In the past, non-coding RNAs have been considered as transcriptional noise without any biological function. With the in-depth study of epigenetics and genomics, despite being unable to be translated into proteins, the non-coding RNAs have been found to participate in various important regulatory processes such as epigenetics, regulation of gene expression at transcription and post-transcription levels, and are closely related to the occurrence and development of human diseases.5
GAS5 gene is located on chromosome 1q25, including 12 exons, and cannot encode protein GAS5.6,7 GAS5 was found in NIH3T3 cells by subtractive hybridization, and its expression was up-regulated when cell growth was inhibited, hence the name8,9. The transcription of GAS5 is widely expressed in tissues, but its expression level is not stable.10 In recent years, a large number of studies have found that the expression level of lncRNA GAS5 is significantly different in various human tumor tissues and normal tissues.11–13 Although this change in the expression level cannot be criticized as a possible secondary change caused by the tumor, studies have also confirmed that GAS5 can play an important inhibitory role in the process of cell malignant transformation by regulating a variety of cell signaling pathways.14–16 However, the mechanism of lncRNA GAS5 on autophagy of laryngeal squamous cell carcinoma cells has not been reported.
Autophagy, also known as type 2 cell death, is a form of programmed cell death characterized by the aggregation of autophagosomes.17 Autophagy is closely related to tumors.18 On the one hand, autophagy could not only maintain the survival of tumor cells in the case of hypoxia and nutritional restriction but also protect cancer cells against injury when they are subjected to chemotherapy and radiotherapy, reducing the effect of chemotherapy and radiotherapy.19 On the other hand, excessive autophagy could also lead to autophagy death or apoptosis.20 This suggests that autophagy may play different roles in different types of tumors or in different stages of the same tumor. Therefore, exploring the autophagy mechanism is extremely important for the study of cancer, including laryngeal cancer.
Based on the above background, this study explored the role of lncRNA GAS5 in the pathogenesis of laryngeal squamous cell carcinoma cell lines, and studied the relationship between lncRNA GAS5 and tumor autophagy, for the clinical treatment of laryngeal squamous cell carcinoma to provide theoretical and experimental basis.
Materials and Methods
Cells and Cultivation
Human normal immortalized epidermal cell line HaCaT and human LSCC cell line AMC-HN-8 were sourced from Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China), Tu 177 was sourced from Shanghai Baili Biotechnology Co., Ltd. (Shanghai, China), SNU-46, SNU-899 and SNU-1076 were sourced from the Korean cell bank. The cells were plated in a 60 mm cell culture dish and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA) containing 100 μg/mL streptomycin, 100 U/mL penicillin (HyClone, Logan, UT, USA) and 10% fetal bovine serum (FBS; HyClone), in a saturated humidity environment with 5% CO2 at 37°C. After culture, the relevant molecular biology test was performed after the cells were grown to about 80% confluence.
Cell Transfection
AMC-HN-8 cells were used to transfect lncRNA-GAS5 and ULK2 plasmid, and plasmid transfection was carried out using Lipofectamine 2000 kit (cat. no. 11668–019; Invitrogen). The pcDNA3.1 plasmids were synthesized from GenePharma (Shanghai, China). Briefly, two 1.5 mL DEPC-treated RNase FREE sterile EP tubes were prepared, 250 μL OPTI-MEM medium was added to each tube, 4 μg of plasmid was added to one tube, and 10 μL of Lipofectamine 2000 transfection reagent was added to the other tube. The two tubes were mixed together, centrifuged carefully, kept to stand for 20 min at room temperature, and add to the corresponding six-well plates.
AMC-HN-8 cells and Tu 177 cells were used to transfect GAS5 overexpression vector, GAS5-siRNA, miR-26a-5p mimic (M), mimic control (MC), inhibitor (I), inhibitor control (IC) and ULK2-siRNA, respectively, two 1.5 mL DEPC-treated RNASE FREE sterile EP tubes were prepared, 250 μL OPTI-MEM medium and 100 μL of serum-free medium were added to each tube, per 150 ng of the siRNA was added to the EP tube, and 12 μL of Lipofectamine 2000 transfection reagent was added to each tube. The mixture was mixed by inversion for 10 seconds, carefully mixed, and allowed to stand at room temperature for 10 min to be added to the corresponding 6-well plate. After 24 h of transfection, biological operations such as qRT-PCR were performed.
CCK-8 Detects Cell Viability
After 24 h of transfection, the cells were laid into 96-well plates, and then cells were digested with trypsin, counted, and then resuspended in complete medium. The cell density was adjusted to 5×104 cells/mL, and 100 μL medium was added to each well. After 24 h of culture, 100 μL of complete medium containing 10% CCK-8 was added to the cells and incubated at 37°C for 2 h, and the absorbance at OD 450 nm was measured. Cell viability was performed according to the instructions of the CCK-8 kit (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany).
Reverse Transcription Quantitative Polymerase Chain Reaction
Cell lysis: Cells in logarithmic growth phase were taken out and washed twice with PBS, and 700 μL of TRIZOL was added to each tube, and allowed to stand at room temperature for 5 min. Three-phase separation: 150 μL of chloroform was added to cells and centrifuged at 12,000 × g for 15 min at 4°C. After centrifugation, the mixture will be divided into three layers. RNA precipitation: The supernatant after centrifugation was carefully pipetted into a new 1.5 mL RNase FREE centrifuge tube, 400 μL of isopropanol was added, mixed, and allowed to stand at room temperature for 15 min, and then centrifuged at 12,000 × g for 10 min at 4°C. RNA cleaning: The supernatant after centrifugation was carefully pipetted into a new 1.5 mL RNase FREE centrifuge tube, and 750 μL of 0.1% DEPC water in 70% ethanol was added, mixed, and centrifuged at 8000 × g for 5 min at 4°C. RNA solubilization: The supernatant was discarded, and the precipitate in the tube was placed on a clean bench for air drying. After air drying, 15 μL DEPC water was added to dissolve the RNA. RNA concentration test: RNA concentration was measured using a NanoDrop™ 2000 spectrophotometer (Thermo Fisher Scientific, Inc.) with a standard of 260/280 ratio between 1.8 and 2.0. Reverse transcription: The oligo-dT or stem-loop reverse transcriptase primers (Takara Bio, Inc., Otsu, Japan) were used to obtain cDNA and the reaction conditions were as follows: 42°C for 60 min, 70°C for 5 min, and 4°C for preservation. Amplification: qPCR was performed by the SYBR® Premix Ex Taq™ II (Takara Bio Inc.) using real-time PCR Detection System (ABI 7500, Life technology, United States). PCR reaction conditions were as follows: pretreatment at 95°C for 10 min; followed by 40 cycles of 94°C for 15 s, 60°C for 1 min, finally at 60°C for 1 min and at 4°C for preservation. The 2−ΔΔCq method was used to process the data. The primers used in this experiment as Table 1.
Table 1.
Primers for RT-qPCR
| Genes | Forward (5ʹ-3ʹ) | Reverse (5ʹ-3ʹ) |
|---|---|---|
| GAS5 | CACACAGGCATTAGACAGA | GCTCCACACAGTGTAGTCA |
| miR-26a-5p | TTGGATCCGTCAGAAATTCTCTCCC GAGG | GGTCTAGATGTGAACTCTGGTGTTG GTGC |
| LC3I | TTACACCCATATCAGATTCTTG | ATTGGAAGGTGTGGGTCA |
| LC3II | AGTGAAGTGTAGCAGGATGA | AAGCCTTGTGAAGGAGAT |
| p62 | GCACACCAAGCTCGCATTC | ACCCGAAGTGTCCGTGTTTC |
| Beclin1 | AAGACAGAGCGATGGTAG | CTGGGCTGTGGTAAGTAA |
| GAPDH | AGAAGGCTGGGGCTCATTTG | AGGGGCCATCCACAGTCTTC |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
Western Blot
The medium in the 6-well plate was discarded 24 h after cell transfection, the cells were washed twice with pre-cooled PBS, and 150 μL of cell lysate containing protease inhibitor and phosphorylase inhibitor was rapidly added to each well, BCA kit is used to detect protein concentration and the protein concentration was adjusted to 5 μg/μL. 30 ug samples were separated by 10% SDS-PAGE gels and transferred to polyvinylidene fluoride (PVDF, Bio-Rad Laboratories, Inc., Hercules, CA, USA). The PVDF membrane was blocked in 5% nonfat milk at room temperature for 1 h, and the target primary antibody was incubated in the corresponding protein band at 4°C overnight. After washing the membrane 3 times with PBST buffer, the target band was incubated with a secondary antibody for 1 h at room temperature. The developer is developed, and the gray value of the strips was analyzed and counted by imageJ (version 5.0; Bio-Rad, Hercules, CA, USA). The primary antibody as followed: anti-LC3I (19 kDa; rabbit; 1:1000; ab48394; abcam), anti-LC3II (17 kDa; rabbit; 1:1000; ab48394; abcam), anti-p62 (51 kDa; mouse; 1:1000; ab56416; abcam), anti-Beclin1 (52 kDa; rabbit; 1:1000; ab62557; abcam), anti-ULK2 (113 kDa; rabbit; 1:1000; ab97695; Abcam) and anti-GAPDH (36 kDa; mouse; 1:1000; ab8245; Abcam). The secondary antibody was horseradish peroxidase (HRP)-conjugated goat anti-mouse/rabbit IgG, 1:2000; sc-516102/sc-2357 (Santa Cruz Biotechnology, Inc. Dallas, TX, USA).
Annexin-V FITC/PI Double Staining for Detection of Apoptosis
After 24 h of transfection, the cells were trypsinized, centrifuged, and the supernatant was discarded, and 500 μL of 1×Annexin-V binding solution prepared in advance was added to prepare a cell suspension at a final concentration of 1×106 cells/mL. Cells were stained with 5 μL of Annexin-V at room temperature for 10 minutes in the dark, and 10 μL of PI. Flow cytometry (version 10.0, FlowJo, FACS CaliburTM, BD, Franklin Lakes, NJ, USA) was used for detection; briefly, Annexin-V FITC was detected by FL1 channel, PI was detected by FL2 channel, and data were analyzed by flow software. Annexin-V+/PI+ and Annexin-V+/PI- are the total number of apoptosis.
Bioinformatic Analysis
The miRNAs that may bind with LncRNA GAS5 were identified using DIANA-LncBase V2 and starBasev2.0 according to the high binding potency, and the target genes of miR-26a-5p were identified using the TargetScan 7.2 according to cumulative weighted context scores.
Luciferase Reporter Assay
Luciferase reporter assay was performed between LncRNA GAS5 and miR-26a-5p, AMC-HN-8 cells were seeded into 24-well plates. After 24 h incubation, 6 ng of pmirGLO report vector carrying wild type (WT) or mutated (Mut) GAS5 was co-transfected with 100 nM miR-26a-5p mimic or 100 nM miR-26a-5p mimic control into the AMC-HN-8 cells. Luciferase reporter assay was also carried out between miR-26a-5p and ULK2, AMC-HN-8 and Tu77 cell lines were seeded into 24-well plates. After 24 h incubation, 6 ng of pmirGLO report vector carrying WT or mutated Mut ULK2 was co-transfected with 100 nM miR-26a-5p mimic or 100 nM inhibitor into the AMC-HN-8 cells and Tu77 cell lines, respectively. Luciferase activities were examined with a dual-luciferase Reporter System (Promega, Madison, Wisconsin, USA).
Statistical Analysis
GraphPad Prism 7 was used to analyze the correlative differences. The value was presented as the mean ± S.E.M. Student’s t test and analysis of variance (ANOVA) were used to analyze the statistical significance for the comparisons of two groups and multiple groups. Statistical significance was set at P < 0.05.
Results
GAS5 Was Low Expressed in LSCC Cell Lines, and Overexpression of GAS5 Suppressed Proliferation and Induced Apoptosis of LSCC Cells
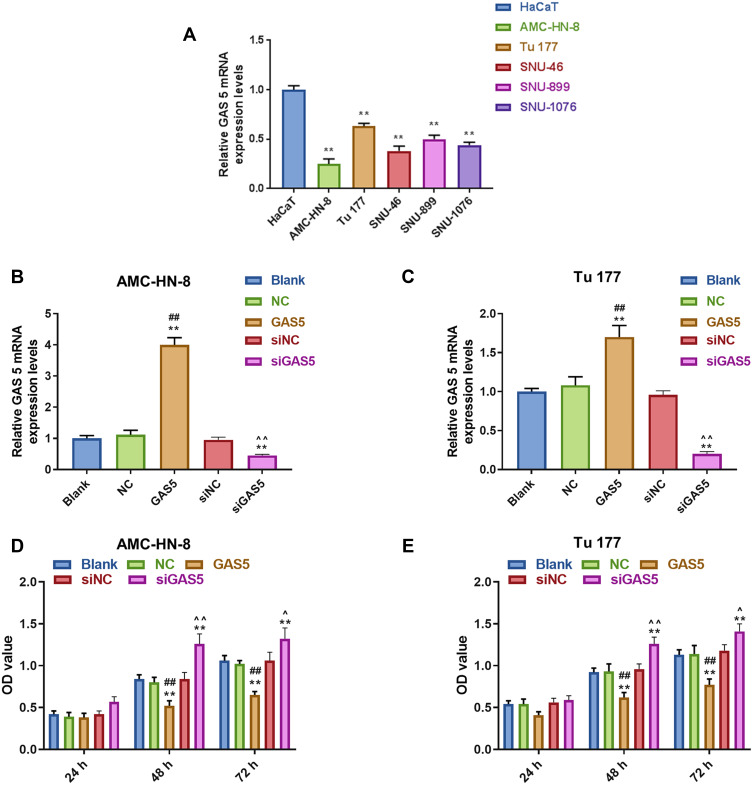
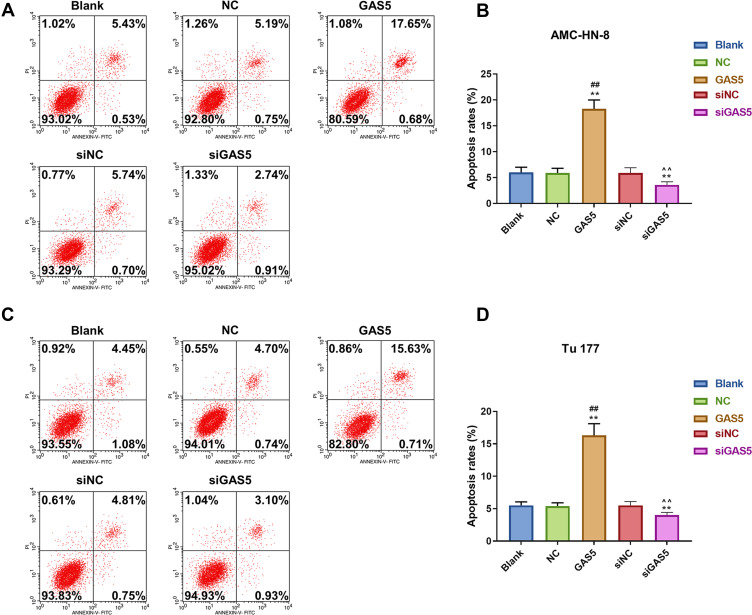
To verify the abnormal expression of GAS5, we performed qRT-PCR assay to determine its expression in 5 LSCC cell lines and normal human immortalized epidermal cell line HaCaT. Compared with HaCaT cells, the expression of GAS5 in the 5 LSCC cell lines, AMC-HN-8, Tu 177, SNU-46, SNU-899 and SNU-1076, had a marked down-regulation, especially in AMC-HN-8 cells, while a comparative lower reduction was shown in Tu 177 cells (Figure 1A). To better reflect the biological function of GAS5 in LSCC, GAS5 and siGAS5 were transfected into AMC-HN-8 cells and Tu177 cells, respectively, to overexpress or knockdown the expression of GAS5, and the transfection efficiency was confirmed by qRT-PCR, as shown in Figure 1B and C, after GAS5 overexpression was transfected to AMC-HN-8 cells, the GAS5 expression was significantly increased in cells, while siGAS5 was transfected to Tu177 cell, the GAS5 expression was significantly inhibited in cell.In addition, qRT-PCR was used to detect the transfection rate of GAS5 in AMC-HN-8 cells transfected with siGAS5. The results showed that GAS5 was significantly lower in AMC-HN-8 cells transfected with siGAS5 (Figure 1B). Moreover, GAS5 was significantly overexpressed in Tu177 cells transfected with GAS5 (Figure 1C). The effects of GAS5 on cell proliferation and apoptosis were investigated by CCK-8 assay and flow cytometry, respectively. The two cells were transfected with above plasmid to culture 24, 48 and 72 h, CCK-8 was used to detect the activity of AMC-HN-8 cells transfected with siGAS5, and the results showed that GAS5 low expression in AMC-HN-8 cell lines increased cell viability (Figure 1D), and high expression of GAS5 in Tu177 cell lines reduces cell viability (Figure 1E), and data from CCK-8 assay showed that up-regulation of GAS5 inhibited AMC-HN-8 cells viability at 48 and 72 h (Figure 1D), while silencing of GAS5 increased Tu177 cells viability at 48 and 72 h (Figure 1E). Results from flow cytometry indicated that an obvious increase of apoptosis rate in AMC-HN-8 cells was observed in GSA5 group (Figure 2A and B), while the low expression of GAS5 in AMC-HN-8 cell lines reduced the apoptosis (Figure 2A and B). Moreover, an opposite tendency of that in Tu177 cells was observed in siGSA5 group (Figure 2C and D).
Figure 1.
Expression of LncRNA GAS5 was downregulated in LSCC. (A) The expression of GAS5 in 5 LSCC cell lines (AMC-HN-8, Tu 177, SNU-46, SNU-899 and SNU-1076,) and a normal human immortalized epidermal cell line (HaCaT) was detected by RT-qPCR. (B) The transfection efficiency of GAS5 in GAS5 or siGAS5 transfected AMC-HN-8 cells was confirmed by RT-qPCR. (C) The transfection efficiency of GAS5 in GAS5 or siGAS5 transfected Tu 177 cells was confirmed by RT-qPCR. (D) Viability of AMC-HN-8 cells was detected by CCK-8 assay. (E) Viability of Tu 177 cells was detected by CCK-8 assay. **P<0.01 vs HaCaT group or Blank group, ##P<0.01 vs NC or siNC, ^P<0.05,^^P<0.01 vs GAS5.
Abbreviations: siNC, silenced negative control; LSCC, laryngeal squamous cell carcinoma; RT-qPCR, quantitative reverse transcription-polymerase chain reaction; GAS5, Long noncoding RNAs growth arrest-specific 5; CCK-8, Cell counting kit-8; OD, optical density; NC, negative control.
Figure 2.
GAS5 induced AMC-HN-8 and Tu 177 cells apoptosis, while siGAS5 inhibited AMC-HN-8 and Tu 177 cells apoptosis. (A) AMC-HN-8 cells apoptosis was induced by GAS5. (B) AMC-HN-8 cells apoptosis rate was quantified. (C) Tu 177 cells apoptosis was inhibited by siGAS5. (D) Tu 177 cells apoptosis rate was quantified. **P<0.01 vs Blank group, ##P<0.01 vs NC or siNC
Abbreviations: siNC, silenced negative control; PI, propidium iodide; FITC, fluorescein isothiocyanate; GAS5, long noncoding RNAs growth arrest-specific 5; siGAS5, silenced Long noncoding RNAs growth arrest-specific 5; NC, negative control.
Autophagy Markers Were Activated by GAS5
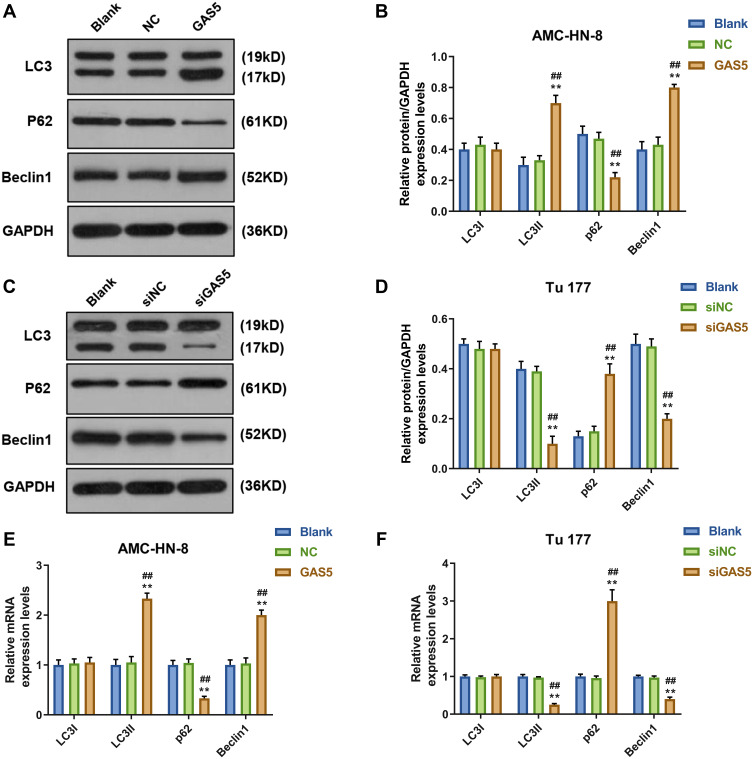
To demonstrate whether GAS5 may affect the expression levels of cellular autophagy-associated regulators, Western blot and qRT-PCR were used to determine the levels of LC3I, LC3II, p62 and Beclin1. Western blot data showed that the levels of LC3II and Beclin1 were significantly increased, but p62 level was decreased in GAS5-treated AMC-HN-8 cells (Figure 3A and B), whereas siGAS5-stimulated Tu177 cells had opposite results (Figure 3C and D). At mRNA level, LC3II and Beclin1 were obviously up-regulated, but p62 was down-regulated in GAS5-treated AMC-HN-8 cells (Figure 3E), whereas siGAS5-stimulated Tu177 cells had opposite results (Figure 3F).
Figure 3.
Autophagy was activated by GAS5 in AMC-HN-8 cells. The protein expressions of LC3I, LC3II, Beclin1 and p62 in AMC-HN-8 cells were determined (A) and quantified (B) by Western blot. The protein expressions of LC3I, LC3II, Beclin1 and p62 in Tu 177 cells were determined (C) and quantified (D) by Western blot. (E) The mRNA levels of LC3I, LC3II, Beclin1 and p62 in AMC-HN-8 cells were determined by qRT-PCR. (F) The mRNA levels of LC3I, LC3II, Beclin1 and p62 in Tu 177 cells were determined by qRT-PCR. **P<0.01 vs Blank group, ##P<0.01 vs NC .
Abbreviations: siNC, silenced negative control; NC, negative control; GAS5, long noncoding RNAs growth arrest-specific 5; siGAS5, silenced Long noncoding RNAs growth arrest-specific 5; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
GAS5 Negatively Regulated the Expression of MiR-26a-5p
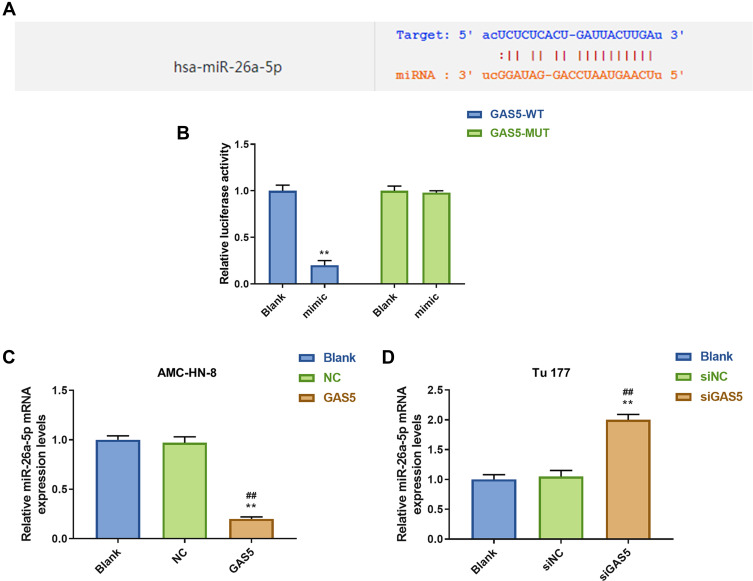
Increasing evidences have revealed that lncRNAs contain sequences that are complementary to miRNAs and could directly or indirectly regulate the expression and activity of miRNAs. DIANA-LncBase V and starbase were used to predict the target gene of GAS5, and the results demonstrated that GAS5 has a position that can bind to miR-26a-5p (Figure 4A). Furthermore, luciferase reporters containing a WT or mutant target site from lnc-GAS5 were also constructed, and miR-26a-5p mimic significantly reduced luciferase activity for the WT lnc- GAS5 reporter (Figure 4B). Meanwhile, we discovered that miR-26a-5p level was lower expressed in GAS5 group than that in NC or Blank group in AMC-HN-8 cells (Figure 4C), while miR-26a-5p level was increased in siGAS5-treated Tu177 cells (Figure 4D).
Figure 4.
MiR-26a-5p was a target gene of GAS5. (A) The target gene of GAS5 was predicted by DIANA-LncBase V2 and starbase. (B) The target gene of GAS5 was confirmed by Luciferase reporter assay. (C) The expression of miR-26a-5p was declined by GAS5 in AMC-HN-8 cells using qRT-PCR. (D) The expression of miR-26a-5p was enhanced by siGAS5 in Tu 177 cells using qRT-PCR. **P<0.01 vs Blank group, ##P<0.01 vs NC or siNC.
Abbreviations: siNC, silenced negative control; NC, negative control; MUT, mutant; WT, wild type; GAS5, long noncoding RNAs growth arrest-specific 5; siGAS5, silenced long noncoding RNAs growth arrest-specific 5.
MiR-26a-5p Mimic Promoted LSCC Proliferation, Which Were Reversed by GAS5
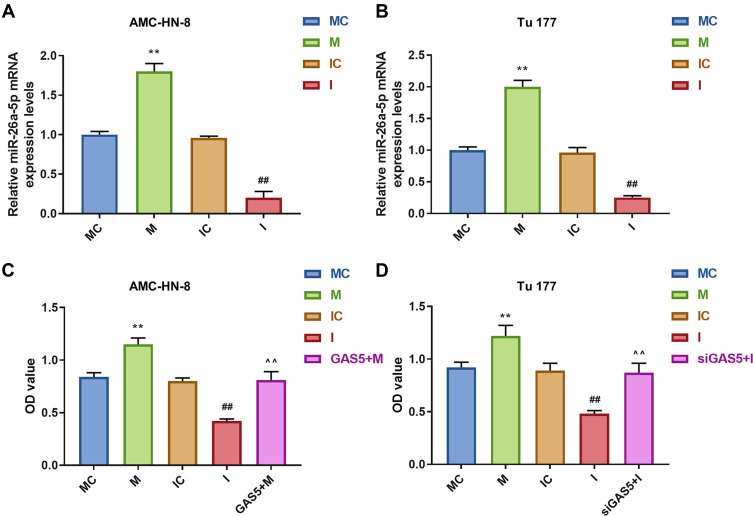
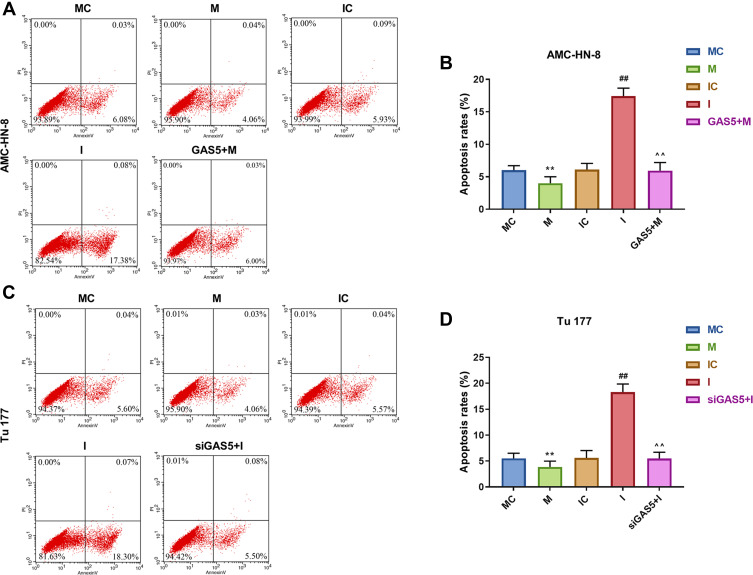
To further evaluate the regulatory relationship between GAS5 and miR-26a-5p, AMC-HN-8 cells and Tu177 cells were transfected with miR-26a-5p mimic, inhibitor sequences and matched controls. Transfection efficiency was confirmed by qRT-PCR (Figure 5A and B). CCK-8 assay indicated that miR-26a-5p mimic enhanced cells viability and miR-26a-5p inhibitor suppressed cells viability both in AMC-HN-8 cells and Tu177 cells, the promotion effect of miR-26a-5p mimic in AMC-HN-8 cells viability was inhibited by overexpression GAS5, and inhibitory effect of miR-26a-5p inhibitor in Tu177 cells viability were increased by siGAS5 (Figure 5C and D). Results from flow cytometry suggested that miR-26a-5p mimic suppressed cells apoptosis and miR-26a-5p inhibitor enhanced cells apoptosis both in AMC-HN-8 cells (Figure 6A and B) and Tu177 cells (Figure 6C and D), the inhibitory effect of miR-26a-5p mimic on AMC-HN-8 cells apoptosis was reversed by overexpression GAS5, and promotion effect of miR-26a-5p inhibitor in Tu177 cells apoptosis was decreased by siGAS5 (Figure 6).
Figure 5.
AMC-HN-8 cells viability was increased by miR-26a-5p mimic, which was reversed by GAS5. Transfection efficiency of miR-26a-5p mimic, inhibitor and negative control in AMC-HN-8 (A) and Tu 177 cells (B) were confirmed by qRT-PCR. (C) AMC-HN-8 cells viability was detected by CCK-8 assay. (D) Tu 177 cells viability was detected by CCK-8 assay. **P<0.01 vs MC, ##P<0.01 vs IC, ^^P<0.01 vs M or I.
Abbreviations: M, mimic; MC, mimic control; I, inhibitor; IC, inhibitor control; OD, optical density; GAS5, long noncoding RNAs growth arrest-specific 5; siGAS5, silenced Long noncoding RNAs growth arrest-specific 5.
Figure 6.
Apoptosis was suppressed by mimic, which were reversed by GAS5 in AMC-HN-8 cells. AMC-HN-8 cells apoptosis was determined (A) and quantified (B) by flow cytometry. Tu 177 cells apoptosis was determined (C) and quantified (D) by flow cytometry. **P<0.01 vs MC, ##P<0.01 vs IC, ^^P<0.01 vs M or I.
Abbreviations: M, mimic; MC, mimic control; I, inhibitor; IC, inhibitor control; PI, propidium iodide; FITC, fluorescein isothiocyanate; GAS5, long noncoding RNAs growth arrest-specific 5; siGAS5, silenced long noncoding RNAs growth arrest-specific 5
MiR-26a-5p Mimic Inhibited Autophagy, Which Were Reversed by GAS5
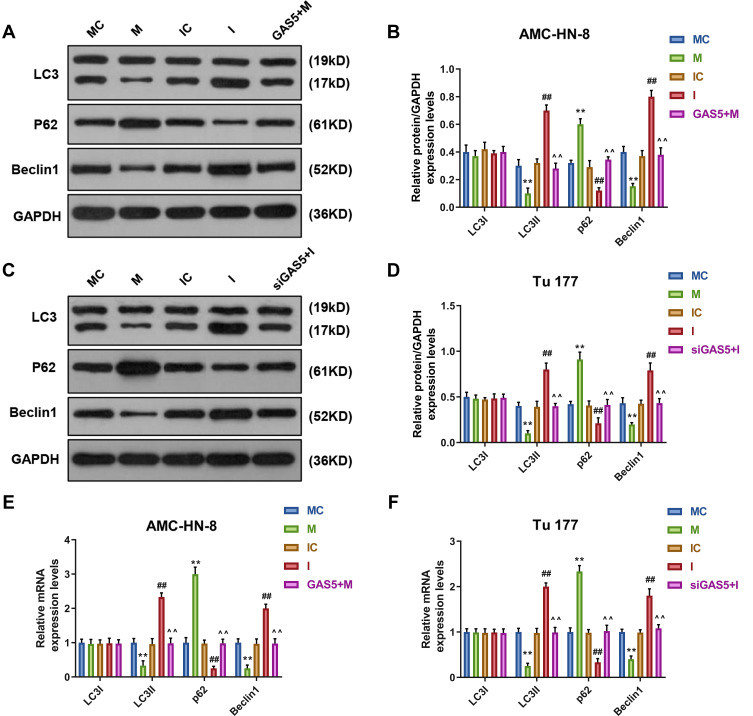
To demonstrate the correlation between the effects of GAS5 on the expression level of autophagy-associated regulatory factors and miR-26a-5p, Western blot and qRT-PCR were used to determine the levels of LC3I, LC3II, p62 and Beclin1. Western blot data showed that, after miR-26a-5p mimic treatment, the levels of LC3II and Beclin1 were significantly decreased, but p62 level was increased in AMC-HN-8 cells and Tu177 cells, while miR-26a-5p inhibitor had opposite results, GAS5 and siGAS5 respectively reversed miR-26a-5p mimic and inhibitor in AMC-HN-8 cells (Figure 7A and B) and Tu177 cells (Figure 7C and D). The qRT-PCR detection showed that results of LC3I, LC3II, p62 and Beclin1 mRNA levels had a similar trend to Western blot data (Figure 7E and F).
Figure 7.
MiR-26a-5p mimic inhibited autophagy, which were reversed by GAS5. The protein expressions of LC3I, LC3II, Beclin1 and p62 in AMC-HN-8 cells were determined (A) and quantified (B) by Western blot. The protein expressions of LC3I, LC3II, Beclin1 and p62 in Tu 177 cells were determined (C) and quantified (D) by Western blot. (E) The mRNA levels of LC3I, LC3II, Beclin1 and p62 in AMC-HN-8 cells were determined by qRT-PCR. (F) The mRNA levels of LC3I, LC3II, Beclin1 and p62 in Tu 177 cells were determined by qRT-PCR. **P<0.01 vs MC, ##P<0.01 vs IC, ^^P<0.01 vs M or I.
Abbreviations: M, mimic; MC, mimic control; I, inhibitor; IC, inhibitor control.
ULK2 Was a Target Gene of MiR-26a-5p
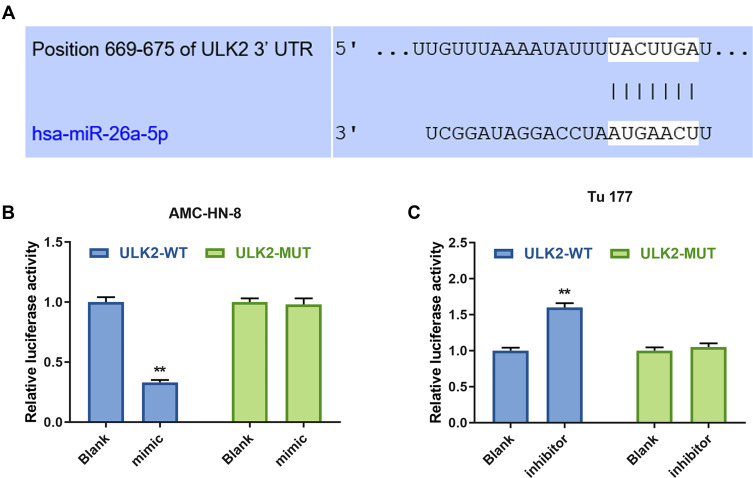
To explore the underlying mechanism of miR-26a-5p in LSCC cell lines, TargetScan 7.2 was used to predict the target gene of miR-26a-5p. The data indicated that ULK2 3ʹ-UTR contained miR-26a-5p binding sites (Figure 8A). Furthermore, luciferase reporters containing a WT or mutant target site from ULK2 were also constructed, and miR-26a mimic significantly reduced luciferase activity for the WT ULK2 reporter in AMC-HN-8 cells (Figure 8B), and miR-26a inhibitor improved luciferase activity for the WT ULK2 reporter in Tu177 cells (Figure 8C).
Figure 8.
ULK2 was a target gene of miR-26a-5p. (A) The target gene of GAS5 was predicted by DIANA-LncBase V2 and starbase. The target gene of miR-26a-5p was confirmed by Luciferase reporter assay in AMC-HN-8 cells (B) and Tu 177 cells (C). **P<0.01 vs Blank group.
Abbreviations: MUT, mutant; WT, wild type; GAS5, long noncoding RNAs growth arrest-specific 5; siGAS5, silenced Long noncoding RNAs growth arrest-specific 5; NC, negative control.
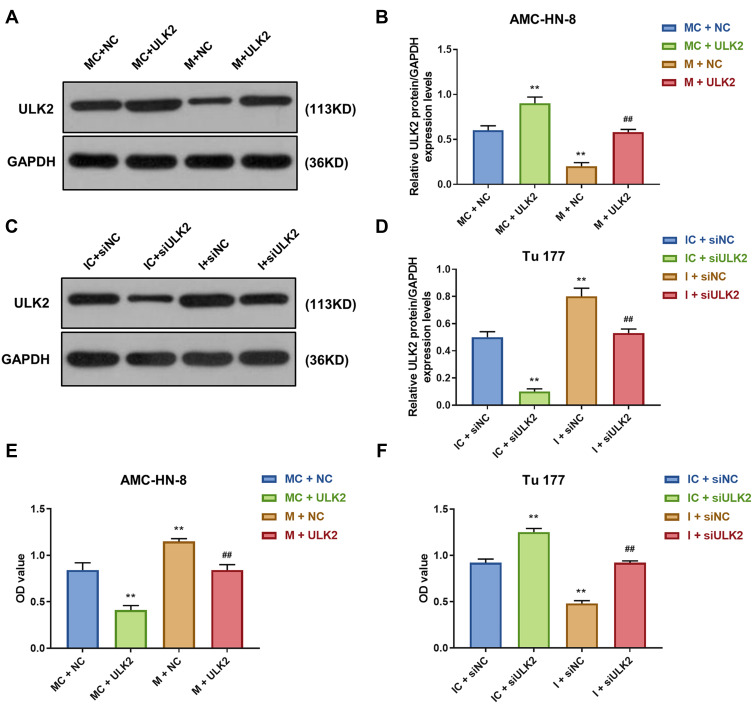
Overexpression of ULK2 Rescued the Effects of MiR-26a-5p Mimic on AMC-HN-8 Cells Proliferation and Apoptosis
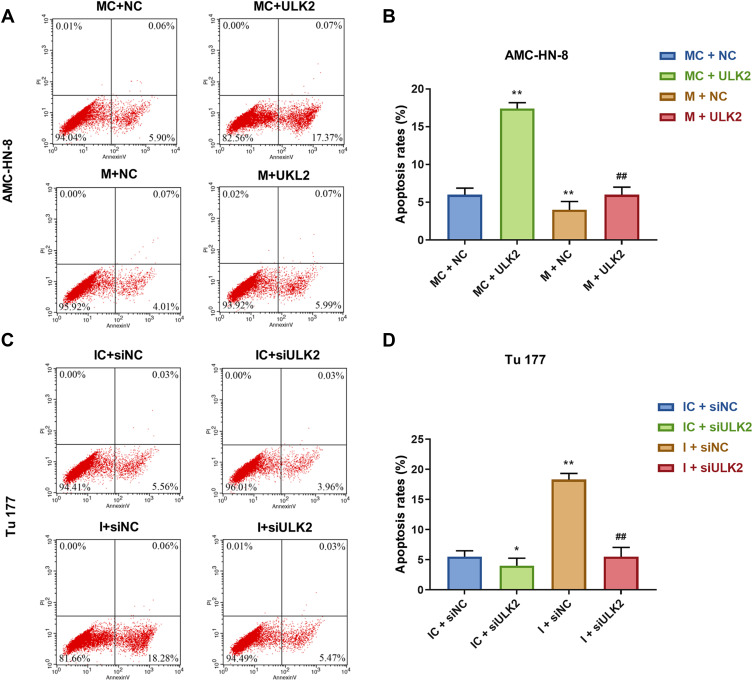
It was investigated whether ULK2 was involved in miR-26a-5p-mediated LSCC cell proliferation and apoptosis, AMC-HN-8 cells were co-transfected with a miR-26a-5p mimic and ULK2 plasmid, and Tu177 cells were co-transfected with a miR-26a-5p inhibitor and ULK2 siRNA. The transfection efficiency was confirmed by Western blot (Figure 9A-D). Data from CCK-8 assay showed that ULK2 inhibited AMC-HN-8 cells viability, mimic promoted cell viability, and co-treated mimic and ULK2 reduced cell viability (Figure 9E). Silencing of ULK2 increased Tu177 cells viability, inhibitor inhibited cell viability, and co-treated inhibitor and siULK2 improved cell viability (Figure 9F). Results from flow cytometry indicated that ULK2 induced AMC-HN-8 cells apoptosis, mimic inhibited cell apoptosis, and co-treated mimic and ULK2 improved cell apoptosis (Figure 10A-B). Silencing of ULK2 decreased Tu177 cells apoptosis, inhibitor increased cell apoptosis, and co-treated inhibitor and siULK2 suppressed cell apoptosis (Figure 10C and D).
Figure 9.
Overexpression of ULK2 rescued the inhibitory effects of miR-26a-5p mimic on AMC-HN-8 cells proliferation. Transfection efficiency of ULK2 in AMC-HN-8 cells was confirmed (A) and quantified (B) by Western blot. Transfection efficiency of ULK2 in Tu177 cells was confirmed (C) and quantified (D) by Western blot. (E) AMC-HN-8 cells viability was tested by CCK-8 assay. (F) Tu177 cells viability was tested by CCK-8 assay. **P<0.01 vs MC+NC or IC+siNC, ##P<0.01 vs M+NC or I+siNC.
Abbreviations: M, mimic; MC, mimic control; I, inhibitor; IC, inhibitor control; siNC, silenced negative control; NC, negative control; OD, optical density; GAS5, long noncoding RNAs growth arrest-specific 5; siGAS5, silenced Long noncoding RNAs growth arrest-specific 5.
Figure 10.
Overexpression of ULK2 rescued the inhibitory effects of miR-26a-5p mimic on AMC-HN-8 cells apoptosis. AMC-HN-8 cells apoptosis was determined (A) and quantified (B) by flow cytometry. Tu 177 cells apoptosis was determined (C) and quantified (D) by flow cytometry. *P<0.05, **P<0.01 vs MC+NC or IC+siNC, ##P<0.01 vs M+NC or I+siNC.
Abbreviations: M, mimic; MC, mimic control; I, inhibitor; IC, inhibitor control; siNC, silenced negative control; PI, propidium iodide; FITC, fluorescein isothiocyanate; NC, negative control; ULK2, uncoordinated 51-like kinase 1.
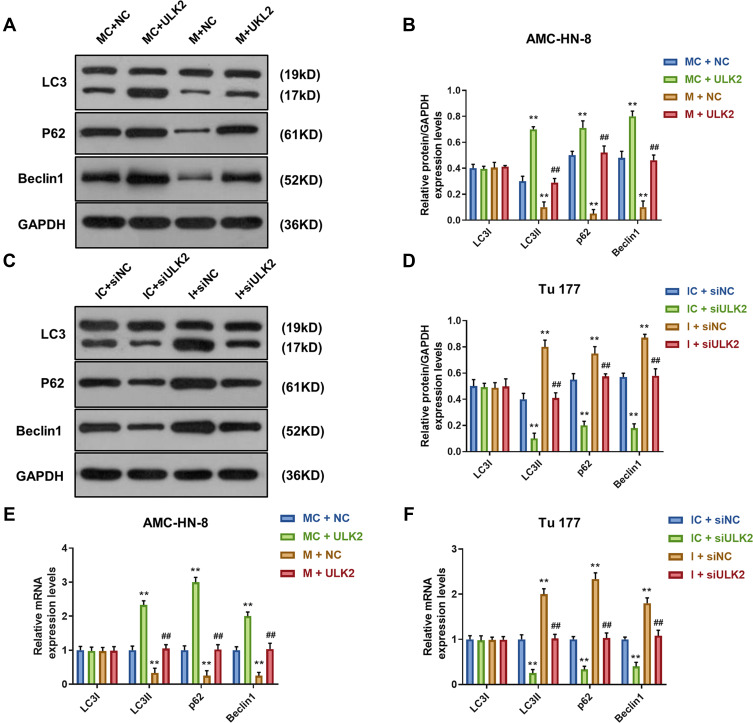
Overexpression of ULK2 Rescued the Inhibitory Effects of MiR-26a-5p Mimic on AMC-HN-8 Cells Autophagy
To demonstrate the correlation between the effects of miR-26a-5p on the expression level of autophagy-associated regulatory factors and ULK2, Western blot and qRT-PCR were used to determine the levels of LC3I, LC3II, p62 and Beclin1. Western blot data showed that, after ULK2 treatment, the levels of LC3II and Beclin1 were significantly increased, while p62 level was decreased in AMC-HN-8 cells, which were reversed by mimic (Figure 11A and B). However, siULK2 had opposite results in Tu177 cells (Figure 11C and D). The qRT-PCR results of LC3I, LC3II, p62 and Beclin1 mRNA levels had a similar trend to Western blot data (Figure 11E and F).
Figure 11.
Overexpression of ULK2 rescued the inhibitory effects of miR-26a-5p mimic on AMC-HN-8 cells autophagy. The protein expressions of LC3I, LC3II, Beclin1 and p62 in AMC-HN-8 cells were determined (A) and quantified (B) by Western blot. The protein expressions of LC3I, LC3II, Beclin1 and p62 in Tu 177 cells were determined (C) and quantified (D) by Western blot. (E) The mRNA levels of LC3I, LC3II, Beclin1 and p62 in AMC-HN-8 cells were determined by qRT-PCR. (F) The mRNA levels of LC3I, LC3II, Beclin1 and p62 in Tu 177 cells were determined by qRT-PCR. **P<0.01 vs MC+NC or IC+siNC, ##P<0.01 vs M+NC or I+siNC.
Abbreviations: M, mimic; MC, mimic control; I, inhibitor; IC, inhibitor control; siNC, silenced negative control; NC, negative control; ULK2, uncoordinated 51-like kinase 1.
Discussion
Laryngeal squamous cell carcinoma (LSCC) accounts for more than 95% of laryngeal cancer.20 Surgery and chemoradiotherapy are the main treatments for LSCC. However, no improvement has been achieved in 5-year survival and quality of life in recent years, especially for advanced patients.21 Thus, there is an urgent need to find new diagnostic biomarkers or new treatments that target LSCC.
One study found that 657 lncRNA transcripts were deregulated in 11 matched laryngeal squamous cell carcinoma (LSCC) matched samples.22 GAS5 was first obtained by Schneider et al from the tumor candidate suppressor gene cDNA subtracted library, which enriched cells in the state of growth stagnation.23 Earlier, Kino et al found that mature LncRNA GAS5 is up-regulated in a state of growth inhibition induced by serum starvation or lack of growth factors.24 Studies found that GAS5 is down-regulated in various tumor tissues such as breast cancer, kidney cancer, prostate cancer and non-small cell carcinoma.25–28 GAS expression induces growth arrest and apoptosis in prostate cancer cells and breast cancer cells.25 These findings have shown that GAS5 functions as a tumor suppressor.28 Moreover, studies found that the decrease of GAS5 expression might be related to the pathogenesis of oral squamous cell carcinoma and is a potential target for tumor therapy.29 In our study, the expression of GAS5 was down-regulated in LSCC cell lines, overexpression GAS5 inhibited AMC-HN-8 cells proliferation, and siGAS5 promoted Tu 177 cells proliferation. These data implied that GAS5 was a tumor candidate suppressor gene in LSCC.
It has been reported that competitive endogenous RNA (ceRNA) forms a large-scale regulatory network in the transcriptome.30 As a sponge of GAS5, miR-26a also regulated both palmitic acid‑induced myocardial inflammatory injury and oxidized low-density lipoprotein-induced impaired autophagy flux as ceRNA manner.31,32 Wu et al found that miR-26a level was down-regulated in LSCC tissues and cell lines, and miR-26a inhibited LSCC tumor growth in vitro and in vivo.33 In this research, we clarified the ceRNA mechanism between LncRNA GAS5 and miR-26a. MiR-26a was confirmed as a direct target of LncRNA GAS5. MiR-26a inhibitor had inhibitory effects in LSCC. Functionally, GAS5 was found to be involved in the development of LSCC through repressing miR-26a function. Thus, GAS5 was a tumor candidate suppressor gene in LSCC through regulating miR-26a.
As a key oncogenic signaling pathway, autophagy plays a pivotal role in various cancers, including hepatocellular carcinoma, breast cancer34 and non-small cell lung cancer. As one of the autophagy genes, ULK2 overexpression significantly inhibited the proliferation of non-small cell lung cancer.35 MiR-26b inhibited autophagy in prostate cancer cells, through down-regulation of ULK2 expression.36 Downregulation of miRNA-26b-induced autophagy and apoptosis in laryngeal carcinoma were reversed by downregulation of ULK2.37 Based on our work, ULK2 was the target gene of miR-26a. Autophagy was induced by GAS5. MiR-26a mimic could inhibit autophagy, which was enhanced by GAS5 and ULK2, respectively. These results further revealed that GAS5/miR-26a/ULK2 regulatory crosstalk in LSCC progression through autophagy pathway, which offered a promising therapy target for LSCC patients.
There are some limitations to our study. The function of GAS5/miR-26a-5p/ULK2 regulatory crosstalk in LSCC progression was only supported by in vitro experiments. Additionally, the observed regulation of autophagy in LSCC is unclear and requires further investigation.
Conclusion
In summary, our present research provides convincing evidence that GAS5/miR-26a/ULK2 axis in LSCC. LSCC evolution is still a multifactorial event and this study provides promising biomarkers for LSCC diagnosis and therapeutic application.
Funding Statement
This work was supported by the Chinese Academy of Medical Sciences (CAMS) Initiative for Innovative Medicine [2017-I2M-1-005].
Disclosure
The authors declare no conflicts of interest.
References
- 1.Dubal PM, Svider PF, Kanumuri VV, Patel AA, Baredes S, Eloy JA. Laryngeal chondrosarcoma: a population-based analysis. The Laryngoscope. 2014;124(8):1877–1881. doi: 10.1002/lary.24618 [DOI] [PubMed] [Google Scholar]
- 2.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- 3.Dey BK, Mueller AC, Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5(4):e944014. doi: 10.4161/21541272.2014.944014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9(6):e1003569. doi: 10.1371/journal.pgen.1003569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 6.Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5ʹ-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18(12):6897–6909. doi: 10.1128/MCB.18.12.6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleming JV, Fontanier N, Harries DN, Rees WD. The growth arrest genes gas5, gas6, and CHOP-10 (gadd153) are expressed in the mouse preimplantation embryo. Mol Reprod Dev. 1997;48(3):310–316. doi: [DOI] [PubMed] [Google Scholar]
- 8.Coccia EM, Cicala C, Charlesworth A, et al. Regulation and expression of a growth arrest-specific gene (gas5) during growth, differentiation, and development. Mol Cell Biol. 1992;12(8):3514–3521. doi: 10.1128/MCB.12.8.3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller AJ, Chatterjee S, Teresky A, Levine AJ. The gas5 gene is disrupted by a frameshift mutation within its longest open reading frame in several inbred mouse strains and maps to murine chromosome 1. Mammalian Genome. 1998;9(9):773–774. doi: 10.1007/s003359900862 [DOI] [PubMed] [Google Scholar]
- 10.Shi X, Sun M, Liu H, et al. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog. 2015;54(Suppl 1):E1–E12. doi: 10.1002/mc.22120 [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Wang Y, Zhang L, et al. ZBTB7A, a miR-663a target gene, protects osteosarcoma from endoplasmic reticulum stress-induced apoptosis by suppressing LncRNA GAS5 expression. Cancer Lett. 2019. [DOI] [PubMed] [Google Scholar]
- 12.Wang G, Sun J, Zhao H, Li H. Long non-coding RNA (lncRNA) Growth Arrest Specific 5 (GAS5) suppresses esophageal squamous cell carcinoma cell proliferation and migration by inactivating Phosphatidylinositol 3-kinase (PI3K)/AKT/Mammalian Target of Rapamycin (mTOR) signaling pathway. Med Sci Monitor. 2018;24:7689–7696. doi: 10.12659/MSM.910867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao H, Yu H, Zheng J, et al. Lowly-expressed lncRNA GAS5 facilitates progression of ovarian cancer through targeting miR-196-5p and thereby regulating HOXA5. Gynecol Oncol. 2018;151(2):345–355. doi: 10.1016/j.ygyno.2018.08.032 [DOI] [PubMed] [Google Scholar]
- 14.Mansoori Y, Tabei MB, Askari A, et al. Expression levels of breast cancer-related GAS5 and LSINCT5 lncRNAs in cancer-free breast tissue: molecular associations with age at menarche and obesity. Breast J. 2018;24(6):876–882. doi: 10.1111/tbj.13067 [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Jing W, Ma W, Liang C, Chai H, Tu J. Down-regulation of long non-coding RNA GAS5-AS1 and its prognostic and diagnostic significance in hepatocellular carcinoma. Cancer Biomarkers. 2018;22(2):227–236. doi: 10.3233/CBM-170781 [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Gu J, The LH. GAS5/miR-222 axis regulates proliferation of gastric cancer cells through the PTEN/Akt/mTOR pathway. Dig Dis Sci. 2017;62(12):3426–3437. doi: 10.1007/s10620-017-4831-4 [DOI] [PubMed] [Google Scholar]
- 17.Tasdemir E, Galluzzi L, Maiuri MC, et al. Methods for assessing autophagy and autophagic cell death. Methods Mol Biol. 2008;445:29–76. [DOI] [PubMed] [Google Scholar]
- 18.Islam Khan MZ, Tam SY, Law HKW. Autophagy-modulating Long Non-coding RNAs (LncRNAs) and their molecular events in cancer. Front Genet. 2018;9:750. doi: 10.3389/fgene.2018.00750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebrahimi S, Hashemy SI. MicroRNA-mediated redox regulation modulates therapy resistance in cancer cells: clinical perspectives. Cell Oncol. 2019;42(2):131–141. doi: 10.1007/s13402-018-00421-z [DOI] [PubMed] [Google Scholar]
- 20.Marioni G, Marchese-Ragona R, Cartei G, Marchese F, Staffieri A. Current opinion in diagnosis and treatment of laryngeal carcinoma. Cancer Treat Rev. 2006;32(7):504–515. doi: 10.1016/j.ctrv.2006.07.002 [DOI] [PubMed] [Google Scholar]
- 21.Steuer CE, El-Deiry M, Parks JR, Higgins KA, Saba NF. An update on larynx cancer. CA Cancer J Clin. 2017;67(1):31–50. doi: 10.3322/caac.21386 [DOI] [PubMed] [Google Scholar]
- 22.Zou AE, Ku J, Honda TK, et al. Transcriptome sequencing uncovers novel long noncoding and small nucleolar RNAs dysregulated in head and neck squamous cell carcinoma. RNA (New York, NY). 2015;21(6):1122–1134. doi: 10.1261/rna.049262.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54(6):787–793. doi: 10.1016/S0092-8674(88)91065-3 [DOI] [PubMed] [Google Scholar]
- 24.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3(107):ra8. doi: 10.1126/scisignal.2000568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28(2):195–208. doi: 10.1038/onc.2008.373 [DOI] [PubMed] [Google Scholar]
- 26.Qiao HP, Gao WS, Huo JX, Yang ZS. Long non-coding RNA GAS5 functions as a tumor suppressor in renal cell carcinoma. Asian Pac j Cancer Prev. 2013;14(2):1077–1082. doi: 10.7314/APJCP.2013.14.2.1077 [DOI] [PubMed] [Google Scholar]
- 27.Pickard MR, Mourtada-Maarabouni M, Williams GT. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta. 2013;1832(10):1613–1623. doi: 10.1016/j.bbadis.2013.05.005 [DOI] [PubMed] [Google Scholar]
- 28.Ma C, Shi X, Zhu Q, et al. The growth arrest-specific transcript 5 (GAS5): a pivotal tumor suppressor long noncoding RNA in human cancers. Tumour Biol. 2016;37(2):1437–1444. [DOI] [PubMed] [Google Scholar]
- 29.Yang M, Xiong X, Chen L, Yang L, Li X. Identification and validation long non-coding RNAs of oral squamous cell carcinoma by bioinformatics method. Oncotarget. 2017;8(64):107469–107476. doi: 10.18632/oncotarget.18178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu B, Pan S, Xiao Y, Liu Q, Xu J, Jia L. LINC01296/miR-26a/GALNT3 axis contributes to colorectal cancer progression by regulating O-glycosylated MUC1 via PI3K/AKT pathway. J Exp Clin Cancer Res. 2018;37(1):316. doi: 10.1186/s13046-018-0994-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang W, Fan T, Liu L, Zhang L. Knockdown of growth-arrest specific transcript 5 restores oxidized low-density lipoprotein-induced impaired autophagy flux via upregulating miR-26a in human endothelial cells. Eur J Pharmacol. 2019;843:154–161. doi: 10.1016/j.ejphar.2018.11.005 [DOI] [PubMed] [Google Scholar]
- 32.Yue Q, Zhao C, Wang Y, et al. Downregulation of growth arrest specific transcript 5 alleviates palmitic acid induced myocardial inflammatory injury through the miR26a/HMGB1/NFkappaB axis. Mol Med Rep. 2018;18(6):5742–5750. doi: 10.3892/mmr.2018.9593 [DOI] [PubMed] [Google Scholar]
- 33.Wu Z, Lu B, Li X, et al. MicroRNA-26a inhibits proliferation and tumorigenesis via targeting CKS2 in laryngeal squamous cell carcinoma. Clin Exp Pharmacol Physiol. 2018;45(5):444–451. doi: 10.1111/1440-1681.12890 [DOI] [PubMed] [Google Scholar]
- 34.Abdel-Mohsen MA, Abdel Malak CA, El-Shafey ES. Influence of copper (I) nicotinate complex and autophagy modulation on doxorubicin-induced cytotoxicity in HCC1806 breast cancer cells. Adv Med Sci. 2019;64(1):202–209. doi: 10.1016/j.advms.2018.08.014 [DOI] [PubMed] [Google Scholar]
- 35.Cheng H, Yang ZT, Bai YQ, Cai YF, Zhao JP. Overexpression of Ulk2 inhibits proliferation and enhances chemosensitivity to cisplatin in non-small cell lung cancer. Oncol Lett. 2019;17(1):79–86. doi: 10.3892/ol.2018.9604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.John Clotaire DZ, Zhang B, Wei N, et al. MiR-26b inhibits autophagy by targeting ULK2 in prostate cancer cells. Biochem Biophys Res Commun. 2016;472(1):194–200. doi: 10.1016/j.bbrc.2016.02.093 [DOI] [PubMed] [Google Scholar]
- 37.Wang S, Guo D, Li C. Downregulation of miRNA-26b inhibits cancer proliferation of laryngeal carcinoma through autophagy by targeting ULK2 and inactivation of the PTEN/AKT pathway. Oncol Rep. 2017;38(3):1679–1687. doi: 10.3892/or.2017.5804 [DOI] [PubMed] [Google Scholar]