Abstract
Background:
Endophytic bacteria reside inside healthy plant tissues and provide several benefits to their host, and help them to tolerate various stresses. Aminocyclopropane-1-carboxylate deaminase (ACCD) production is one of the mechanisms by which these bacteria help the plant to survive under ethylene stress
Objectives:
The main focus of this study was to isolate endophytic bacteria and effectively screen them for ACCD production. The selected isolate was identified and assessed for plant growth-promoting potential under pot conditions.
Materials and Methods:
Endophytic bacteria were isolated from root nodules of Pisum sativum plants, grown in northern India (Haryana state). ACCD activity was initially screened on DF minimal salt medium with ACC as a sole nitrogen source. To narrow down the number of the isolates, another screening method was adopted using a modified medium containing indicator dyes along with ACC. The strain producing ACCD as well as a significant amount of Indole 3 acetic acid (IAA) was identified using 16S rDNA gene sequencing and amplification of acdS gene. Its ability to promote plant growth was evaluated under pot culture conditions.
Results:
Twenty-six endophytic bacteria were isolated from nodules of P. sativum plants. Sixteen isolates showed growth on DF minimal salts medium supplemented with ACC along with negative control. On the modified medium containing indicator dyes, two isolates, PJN13 and PJN17, showed zones of the color gradient. The ACC deaminase activity was further confirmed by enzymatic assay. The strains PJN13 and PJN17 produced 160 and 130 µM of α-ketobutyrate m.g-1 protein h-1, respectively. The IAA production in the strain PJN13 (79.04 ± 0.78 µg.mL -1) was significantly more than that in the strain PJN17 (38.36 ± 1.89 µg.mL-1). It could enhance pea plant growth parameters, including root and shoot length and fresh and dry weight from 1 to 4 times compare to the control (untreated pea plants) under pot conditions. The results of 16S rDNA amplification and sequencing showed that PJN13 has maximum similarity to Bacillus mojavensis, and the sequence submitted to GenBank under accession number MH298523. Also, a band about 800 bp was amplified for the acdS gene.
Conclusions:
Though Bacillus is known as a predominant non-rhizobial endophytic genus, however in the present study, a B. mojavensisBacillus mojavensis PRN2 (MH298523) was reported for the first time as an endophyte from the nodules of pea plants. The isolated strain possesses ACC deaminase activity along with IAA production capability, and high potentials as PGPE (Plant growth-promoting endophyte) for plant growth, so it has potential to be used as biofertilizers in pea fields.
Keywords: ACC deaminase, Bacillus mojavensis, Endophyte, Pisum sativum, Plant growth promotion
1. Background
Leguminous plants are known to be colonized by a wide array of microorganisms and develop a beneficial association with them. Such beneficial microbial association includes bacteria that interact with plants with different relationships that can be beneficial, harmful or neutral ( 1 - 2 ). These bacteria can either be either free-living, present loosely on the surface of roots or invade the various plant tissues and termed as endophytes ( 1 ). Almost all the plant species present on earth are known to be harbor by endophytes. Among endophytic bacteria present in various plant parts, those
which form nodules in the roots of leguminous plants have been widely studied ( 1 ).
The endophytic microbiome of plants plays an imperative role in their growth and development either directly or indirectly ( 1 ). Directly, they can promote plant growth by producing various phytohormones like auxins, enzymes, facilitating nutrient uptake by solubilizing phosphate, fixing atmospheric nitrogen. Indirectly, endophytes can decrease or prevent some lethal effects of the phytopathogenic organism by producing HCN, siderophores and by reducing the levels of stress hormone ethylene ( 4 , 5 ).
Plants synthesize gaseous hormone ethylene from the precursor 1-aminocyclopropane 1- carboxylic acid, which plays multiple roles in development and physiology of plants, such as tissue differentiation, development of lateral bud, the emergence of seedling, shedding of leaves, senescence, root hair elongation, the ripening of fruits, and production of various volatile compounds which leads to fruit aroma ( 3 ). The plant produces the desired amount of ethylene under normal conditions, but its production increases under abiotic and biotic stresses. When the level of ethylene increases from its threshold value, it leads to an unfavorable proliferation of growth parameters and untimely senescence in plants ( 2 ). Endophytic bacteria are known to produce an enzyme 1-aminocyclopropane- 1-carboxylate deaminase (ACCD) which cleaves 1-aminocyclopropane-1-carboxylate (ACC), an immediate precursor of ethylene. It has been reported that the inoculation of plants with ACC deaminase producing Bacteria reduces the amount of ethylene ( 6 ).
Pisum sativum (Pea or garden pea), family Fabaceae, is an important cool-season crop generally cultivated for its green pods in India and is important due to fiber content, protein, starch and many phytochemicals ( 7 ). The flowers and pods of pea are severely affected due to environmental stresses and production declines. Therefore to meet the increasing demand, it is crucial to rescue its pods. Instead of using chemical treatments to reduce the levels of stress ethylene, a better understanding of plant growth-promoting endophytic bacteria is essential to develop effective biological methods.
2. Objectives
The present study was planned to effectively screen ACC deaminase producing bacterial endophytes from pea nodules using a modified method, check the ability of the potent isolate to promote plant growth under pot conditions.
3. Materials and Methods
3.1. Isolation of Endophytic Bacteria
P. sativum plants were collected from fields of Jhajjar (28.7° N: 76.71° E) district, Haryana. The healthy roots and nodules were removed, and surface sterilized. The isolation of endophytic bacteria was done on Tryptone soya agar (TSA) plates. The uncrushed sterilized nodules and water drops of final rinse were also placed on TSA plates to ensure the efficacy of surface sterilization. Purified single colonies were picked up from the plates, maintained on TSA slants and stored at 4 °C in a refrigerator for further studies.
3.2. Screening of ACC Deaminase Producing Endophytes
The endophytic bacteria were screened for ACC deaminase activity. The cultures were freshly grown in broth and centrifuged for 8 min at 8,000 X g, and the pellets were washed twice using sterilized 0.1 M Tris–HCl (pH 7.5) to remove the media. The cell pellets were spotted on modified Dworkin and Foster (DF) ( 8 ) minimal salts agar medium containing trace elements, FeSO4.7H2O and three mM ACC as the sole nitrogen source ( 9 ). For positive control, DF minimal medium supplemented with 0.2% ammonium sulfate, and for the negative control, the DF medium devoid of any nitrogen source was used. Growth, of isolates on DF medium supplemented with ACC, was assessed after incubation for 72 hrs at 30°C and compared to control. The ACCD producers were selected based on growth on ACC amended DF plates ( 10 ). The isolates growing on DF minimal medium were also plated on nitrogen-free media and growth was observed after 48-72 h ( 11 ).
To increase the efficacy of screening, these isolates were again screened on the minimal media supplemented with 3 mM ACC along with bromothymol blue (0.005%) and phenol red (0.008%) as indicator dyes ( 12 ). ACC deaminase catalysis was readily detected by color change of media (due to change in pH) as a result of the breakdown of ACC into ammonia and α-ketobutyrate. The selected isolates were spotted on these plates and incubated at 30 °C for 24 h, observed for the change in color.
3.3. ACC Deaminase Activity Assay
To measure the ACCD activity, cells were firstly induced ( 9 ), collected by centrifugation at 16,000 X g for 5 min. Then the cells were washed using 0.1 M Tris-HCl (pH7.6), resuspended in 600 μL of 0.1 M Tris-HCl (pH 8.5). After washing, 30 μL toluene was added to labilize the cells, mixed by vortexing for 30 s. Two hundred microliters of toluenized cell suspension were incubated with 20 μL of 0.5 M ACC at 30 °C for 15 min. After incubation, 1mL of 0.56 N HCl was added, mixed by vortexing and cell debris was removed by centrifugation at 16000 rpm for 5 min. One mL of culture supernatant was blended with 800 μL of HCl (0.56N) and freshly prepared 300 μL of DNPH reagent (0.1 g 2,4- dinitrophenyl hydrazine in 100mL of 2 N HCl), vortexed and incubated at 30 °C for 30 min. After incubation, 2mL of NaOH (2N) was added, and absorbance was measured at 540nm using UV-Vis spectrophotometer (Thermo scientific GENESYS 10S UV-VIS, Germany). The number of μmoles of α-ketobutyrate produced was determined by comparing with standard ranging between 0.1 to 1mM ( 9 ).
3.4. Protein Concentration Determination
Toluenized cells were used for protein concentration determination by the Bradford method ( 13 ). One hundred microliters aliquots of toluenized cell suspension was mixed with 100 μL of 0.1 N NaOH and incubated for 10 min at 100 °C in a water bath. After cooling the mixture, 50 μL of the sample mixture was transferred to the test tube, and the volume was adjusted to 100 μL with 0.1 M Tris-HCl (pH 8.5). 1 mL of Bradford reagent was added to the sample mixture and incubated at room temperature for 10 min. Absorbance was measured following incubation at 540 nm and compared with the standard bovine serum albumin (BSA) of known concentration.
3.5. IAA Production
IAA production was determined in the culture broth using a standard colorimetric assay ( 14 ). The isolates were inoculated in 25 mL of sterilized YEM (Yeast Extract Mannitol) broth augmented with 0.1 g. L-1 L-tryptophan in Erlenmeyer flasks (150 mL) and incubated in a rotary shaker at 150 rpm at 28±2 ° C for five days. The culture broth was centrifuged at 3000 x g for 3 min after incubation, and the supernatant was mixed with an equal volume of Salkowski (1 mL 0.5 M FeCl3 in 50 mL of 35% HClO4) reagent. After 30 min incubation in the dark, the absorbance was measured at 530 nm, and the IAA present in each sample was calculated using the calibration curve of known IAA concentrations (10 –200 μg. mL-1) ( 15 ). As a negative control, 2 mL of uninoculated YEM broth with equivalent Salkowski reagent volume was taken. All experiments in this study were conducted in triplicates, and the average values were used.
3.6. Identification of Endophytic Bacterial Isolates
The genomic DNA was extracted from freshly grown culture by a modified CTAB method ( 16 ). The universal primers 8F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1541R (5’-AAGGAGGTGATCCAGCCGCA-3’) were used for 16S rDNA amplification ( 17 , 18 ). Amplification of 16S rDNA sequence was done by polymerase chain reaction (PCR) at a volume of 30 μL containing 50 ng DNA template, Taq polymerase buffer (1X), primer (0.25 pmoL.μL-1 each), dNTP’s (0.2 mM each), MgCl2 (1.5mM) and Taq polymerase (1U) (Promega Corp., USA). The PCR amplification was done in a thermocycler (Bio-Rad, T100, USA) with the following program: initial denaturation at 94 °C for 3 min, followed by 30 cycles at 94 °C for 45 s, 58 °C for 45 s, 72 °C for 2 min with final extension of 72 °C for 7 min. The PCR product was resolved for 40 min stained with ethidium bromide on 1.2 percent agarose gel in a 0.5X TBE buffer at 60 V. The gel was visualized under Azure c150 gel documentation system (Azure Biosystems, Dublin, CA 94568 USA). The AgriGenome Labs Private Limited, Kochi, India provided the sequencing facilities and sequence obtained was analyzed for similarity analysis using BLASTN (Nucleotide Specific Local Alignment Search Tool) program available on NCBI website (www.ncbi. nlm.gov/BLAST). Sequence alignment performed using CLUSTAL W, and 1000 bootstrap replicates were used to establish the phylogenetic relationship of the isolate by MEGA 7 program (neighbour-joining (NJ) method), ( 19 )
3.7. Amplification of acdS Gene
The acdS gene was amplified by PCR using universal primers ( 20 ) Sequences of the primers were: (F) 5’-GGCAAGGTCGACATCTATGC-3’ and (R) 5’-GGCTTGCCATTCAGCTATG-3’. PCR reaction was carried out in a volume of 20 μL reaction mixture containing 50 ng of genomic DNA, reaction buffer (1X), dNTP mixture (0.2mM), primers (0.3 μM each) and Taq polymerase (1.5U). The PCR amplification was done with the following program: initial denaturation at 94 °C for 3 min followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 62 °C for 1 min and extension of primers at 72 °C for 3 min, final extension at 72 °C for 5 min in a thermal cycler (Bio-Rad, T100, USA). The PCR product was resolved on 1.2 percent agarose gel in 0.5X TBE buffer at 60 V for 40 min and visualized under Azure c150 gel documentation system (Azure Biosystems, Dublin, CA 94568 USA).
3.8. Pot Experiment
The river sand was washed with acid and sterilized at 180 °C in a hot air oven. Sterilized sand-filled paper cups were autoclaved at 15 lbinch-2 for 1 h. The seeds of P. sativum were surface sterilized and incubated in the overnight grown bacterial suspension. Five seeds per cup were sown, and the experiment was performed in triplicates. For control, sterilized seeds incubated in uninoculated broth were used. These cups were kept in a greenhouse and watered with the Sloger’s nitrogen-free mineral salt solution daily ( 21 , 22 ). The plants were uprooted and observed after 60 days for root and shoot length, the number of lateral roots, fresh and dry roots and shoots weight.
3.9. Statistical Analysis
All the results were reported as mean ± standard errors of three replicates. The data were checked by the Student’s t-test, and significant differences were tested at p<0.001, 0.01 and 0.05.
4. Results
4.1. Isolation of Endophytic Bacteria and Screening for Production of IAA
A total of 26 morphologically distinct endophytic bacteria were isolated from surface-sterilized nodules of P. sativum plants. Out of the total, 73% of the isolates were Gram-positive rods. No bacterial growth was observed on control plates with sterilized nodules and drops of final rinsed water. This ensures proper sterilization and endophytic origin of the isolates.
4.2. Screening of 1-aminocyclopropane-1-carboxylate Deaminase (ACCD) Producing Endophytic Bacterial Isolates
All the isolates were screened for ACC deaminase activity in DF minimal medium supplemented with 1-aminocyclopropane-1-carboxylate (ACC) as the sole nitrogen source (Table 1). Out of the total, 16 isolates showed growth on DF minimal salts medium supplemented with ACC, ammonium sulfate as well as on negative control (i.e. DF minimal salts medium without any nitrogen source) (Fig. 1). The isolates growing on DF minimal medium were also able to grow on nitrogen-free media. Therefore to confirm their ability to produce ACCD, screening was done on modified media.
Table 1.
Screening of ACC deaminase by nodule endophytic bacteria isolated from Pisum sativum@S.No.
| S.No. | Isolates | Growth on DF medium supplemented with | ||
|---|---|---|---|---|
| (NH4)2 SO4+ (Positive control) | ACC- (NH4)2 SO4- (Negative control) | ACC as sole N2 source | ||
| 1 | PJN1 | + | + | - |
| 2 | PJN2 | + | + | + |
| 3 | PJN3 | + | + | + |
| 4 | PJN4 | + | + | ++ |
| 5 | PJN5 | + | + | ++ |
| 6 | PJN6 | + | + | + |
| 7 | PJN7 | + | + | - |
| 8 | PJN8 | + | - | - |
| 9 | PJN9 | + | + | + |
| 10 | PJN10 | + | + | - |
| 11 | PJN11 | + | - | + |
| 12 | PJN12 | + | + | ++ |
| 13 | PJN13 | + | + | +++ |
| 14 | PJN14 | + | - | - |
| 15 | PJN15 | + | + | ++ |
| 16 | PJN16 | + | + | - |
| 17 | PJN17 | + | + | ++ |
| 18 | PJN18 | + | + | ++ |
| 19 | PJN19 | + | + | + |
| 20 | PJN20 | + | + | ++ |
| 21 | PJN21 | + | - | - |
| 22 | PJN22 | + | + | ++ |
| 23 | PJN23 | + | + | - |
| 24 | PJN24 | + | + | ++ |
| 25 | PJN25 | + | + | - |
| 26 | PJN26 | + | - | - |
Figure 1.

Screening of endophytic bacteria on DF media supplemented with (A) ammonium sulphate (B) ACC (C) without nitrogen source
4.3. Modified ACC Medium with 0.005% Bromothymol Blue (BTB) and 0.005% Phenol Red
For effective and rapid screening of endophytic bacterial isolates for ACCD, modified ACC medium supplemented with 0.005% bromothymol blue (BTB) and 0.005% phenol red were used (Fig. 2). A total of 10 isolates (PJN4, PJN5, PJN12, PJN13, PJN15, PJN17, PJN18, PJN20, PJN22, and PJN24) were selected based on their growth on DF media supplemented with ACC and screened on minimal media with indicator dyes (0.005 % bromothymol blue and 0.008% phenol red). Two isolates PJN13 and PJN17 showed positive growth on both indicator plates with different color gradient and zones (Table 2). To confirm the ACC deaminase production by PJN 13 and PJN 17, the enzyme activity was assessed.
Figure 2.

Screening of endophytic bacteria on minimal media with (A) ACC+ Phenol red+ (B) ACC+ Bromothymol blue+
Table 2.
Screening of ACC deaminase on plates with indicator dyes using ACC as sole nitrogen source
| No colony | Colored colony without zone | Colored colony with zone | Colony with intense colored zone |
|---|---|---|---|
| PJN12,PJN15,PJN18, PJN24 | PJN2, PJN4, PJN20,PJN22 | PJN 17 | PJN 13 |
4.4. ACC Deaminase activity
Two endophytic bacterial isolates (PJN13 and PJN17) with maximum zones on indicator dye plates, were assayed for enzymatic activity and quantified by the release of the product (α-ketobutyrate) by deamination of substrate ACC using ACC deaminase. ACC deaminase activity of 160 μM of α-ketobutyrate per mg per protein hour was shown by isolate PJN13 and 130 μM of α-ketobutyrate per mg per protein hour by PJN17.
4.5. IAA Production
PJN13 and PJN17 were assessed for IAA production using Salkowski reagent and formation of pink color, which indicated IAA production. A significant amount of IAA (79.04 ± 0.78 μg.mL-1) was produced by PJN13 in comparison of PJN17 (38.36 ± 1.89 μg.mL-1).
4.6. PCR Amplification of 16S rRNA Gene and Identification of the Isolate PJN13
16S rDNA of PJN13 was amplified using a universal set of primers, which confirmed by obtaining a single band of ~1400bp on 1.2% (w/v) agarose gel. Nucleotide sequence and BLAST analysis revealed 100% similarity with Bacillus mojavensis strain ifo 15718 (accession no. NR118290). The sequence submitted to NCBI under accession number MH298523. Also, the phylogenetic analysis was done with MEGA7 using the neighbour-joining (NJ) method with 1,000 bootstrap replicates (Fig. 3).
Figure 3.

Phylogenetic tree showing relationship of isolate PJN13 with sequences from NCBI using MEGA 7 by the neighbour-joining method
4.7. Amplification of acdS Gene
The ACC deaminase (acdS) gene from PJN13 was amplified using universal primers specific for Bacillus. A specific band about ~800 bp was amplified, that confirming the ACC deaminase activity by the isolate.
4.8. Effect of PJN13 on the Growth of P. sativum
As PJN13 was producing both IAA and ACCD, its efficiency was evaluated under pot culture conditions (Fig. 4, 5). Pisum sativum seeds treated with PJN13 resulted in an increase in the root (4.14 folds) and shoot (1.79 folds) length, root (2.7 folds) and shoot (3 folds) fresh weight, root (1.5 folds) and shoot (2 folds) dry weight, over the uninoculated control plants, which was statistically significant at p<0.05 (Table 3). This increase in root length might be due to IAA production along with ACC deaminase activity of the isolate PJN13, that helping the plant to counteract stresses and better absorption of nutrients from the soil by elongation of roots.
Figure 4.

Effect of PJN 13 (Bacillus mojavensis PRN2) on Pisum sativum plants after 60 days of inoculation
Figure 5.
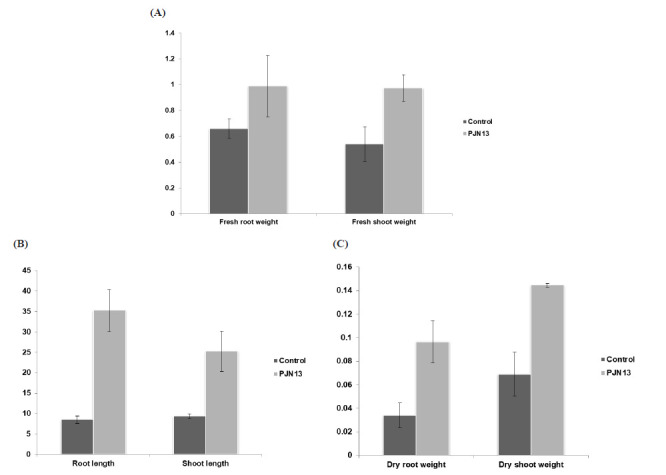
Effect of isolate PJN 13 (Bacillus mojavensis PRN2) on (A) Root and shoot length (B) Fresh root and shoot dry weight (C) Dry root and shoot weight on Pisum sativum plants after 60 days of inoculation along with untreated control
Table 3.
Screening of ACC deaminase on plates with indicator dyes using ACC as sole nitrogen source
| Isolate | Root length (cm) | Shoot length (cm) | Fresh root weight (g) | Fresh shoot weight (g) | Dry root weight (g) | Dry shoot weight (g) |
|---|---|---|---|---|---|---|
| Control | 8.5±0.86 | 9.33±0.57 | 0.65±0.07 | 0.54±0.13 | 0.03±0.01 | 0.07±0.02 |
| PJN13 | 35.25±5.12*** | 25.25±4.92*** | 0.98±0.23* | 0.97±0.10** | 0.09±0.04* | 0.14±0.02*** |
Results shown are the mean values where n=5, ± S.D indicates standard deviation
Statistically significant at P≤0.05
Statistically significant at P ≤ 0.01
Statistically significant at P ≤ 0.001
5. Discussion
Endophytic bacterial microbiome, including both rhizobial as well as non-rhizobial populations, is well known to promote the plant health by the production of various metabolites ( 23 , 24 ). These substances are required for their interaction with plants, nodulation, competition and defence. Plants are in constant exposure to abiotic and biotic stresses, which affects the overall growth of plants and level of stress hormone ethylene increases in response to these factors. This increased level of ethylene was reported to inhibit nodule formation in leguminous plants ( 25 ). Several rhizospheric and endophytic bacterial genera are known to influence the plant ethylene levels either by synthesizing rhizobitoxine or by producing ACC deaminase. Rhizobitoxine is a competitive inhibitor of enzyme ACC synthase while ACC deaminase degrades ACC into ammonia and alpha-ketobutyrate ( 26 ). The presence of ACC deaminase activity has been studied in several endophytic bacterial genera isolated from plants such as Ralstonia, Azospirillum, Rhizobium, Enterobacter, Agrobacterium, Pseudomonas, Achromobacter and Burkholderia ( 27 ).
In our study, we isolated and characterized 26 endophytic bacteria from nodules of Pisum sativum plants for their ability to produce ACC deaminase. Majority of the isolates screened for the production of ACCD were able to grow on DF minimal medium as well as on medium supplemented with ACC as a sole nitrogen source. These isolates were also able to grow on a nitrogen-free medium, and this indicates their ability to fix atmospheric nitrogen ( 28 ). Their nitrogen-fixing ability can be further confirmed by amplification of the nifH gene. But to increase screening spectrum of ACCD producing endophytes, the pH indicator dyes in the medium were used. This requires the efficient method of screening to specify most potential endophytes at a primary level of selection and thus tapered down many isolates in the successive stages ( 12 ). The colonies which grow on basal ACC medium without any indicator dyes were colorless. However, hen a microorganism produces ACCD, it could break down the ACC and consequently ammonia is released. Due to the production of ammonia, the color of medium plates with indicator dyes changes, which forms the basis of this screening method. Ten isolates on the basis on growth on DF medium supplemented with ACC were further screened on minimal medium containing pH indicator dyes (bromothymol blue and phenol red). Following the earlier report, we also observed three types of colonies: colorless without the zone, colonies having gradation in the intensity of zone while colored colonies without colored zone ( 12 ). Out of 10, two isolates PJN13 and PJN17 grew well on both indicator plates and showed the presence of colored zones around them. The zone intensity was high in case of PJN13 as compared to PJN17, which was also confirmed by high enzymatic activity. Further, an amplified product of around 800bp was observed upon amplification of acdS gene using the same primer as reported earlier ( 5 , 20 ).
A large number of species of genus Bacillus has been reported as nodule endophytes ( 29–31 ). The B. mojavensis, closely related to B. subtilis, type and its other strains have been isolated from soil samples of Mojave Desert. It has been reported to endophytically colonize maize and known to promote plant growth directly as well as indirectly by inhibiting the fungal pathogen ( 32 , 33 ). The present study also demonstrates the efficiency of ACC producing isolate PJN13 under pot culture conditions. As it had the capacity to produce both IAA and ACC deaminase it promoted the root, shoot length, fresh and dry weight of root and shoot to a more significant extent. IAA, an active phytohormone, well known to be produced by many endophytic bacteria, influences cell division, differentiation and cell elongation in plants ( 34 ). It is probable that IAA and ACC deaminase stimulate root growth in a coordinated fashion ( 2 ). Significant increase in shoot and root length was also observed upon co-inoculation of soybean seeds with B. megaterium and Bradyrhizobium japonicum ( 35 ). These studies indicated the importance of this genus in terms of plant growth promotion and various other reports supports its effective colonization in plant tissue as an endophyte ( 36 , 37 ).
6. Conclusion
Endophytic bacteria represent an innovative area of research which deals with a wide range of benefits to host plants. The present study dealt with isolation of endophytic bacteria from root nodules of the pea plant and screened them for ACC deaminase (ACCD) activity. For more effective screening of ACCD, we adopted another method using indicator dyes. This method was found to be more effective as compared to the earlier reported method. The results of this study indicate that PJN13 (B. mojavensis (Bacillus mojavensis PRN2)) exhibits high ACC deaminase producing capability along with IAA production. This isolate also resulted in a significant increase in plant growth parameters under pot culture conditions. Though this strain has been isolated as endophyte from other plants, this is the first report on the occurrence of B. mojavensis in nodules of P. sativum plants. Further its putative plant growth-promoting potential still needs to be explored under field conditions.
Acknowledgement
The authors are grateful to the Department of Science & Technology (DST), Govt. of India, New Delhi for financial support to undertake these investigations.
Conflict of Interest: The authors declare no conflict of interest.
References
- 1.Dudeja SS, Giri R, Saini R, Suneja-Madan P, Kothe E. Interaction of endophytic microbes with legumes. J Basic Microbiol. 2012;52(3):248–260. doi: 10.1002/jobm.2011000633. [DOI] [PubMed] [Google Scholar]
- 2.Glick BR. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res. 2014;169(1):30–39. doi: 10.1016/j.micres.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Khosravi H, Alikhani HA, Yakhchali B, Kharkhane AA. Isolation, Cloning and Sequence Analysis of 1-Aminocyclopropane- 1-Carboxylate Deaminase Gene from Native Sinorhizobium meliloti. Iran J Biotechnol. 2014 ;12 (3 ): 50–56 . doi: 10.15171/ijb.1000. [DOI] [Google Scholar]
- 4.Glick BR. Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol Lett. 2005;251( 1):1 –7 . doi: 10.1016/j.femsle.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 5.Singh RP, Shelke GM, Kumar A, Jha PN. Biochemistry and genetics of ACC deaminase: A weapon to “stress ethylene” produced in plants. Front Microbiol. 2015;6(SEP): 1–14 . doi: 10.3389/fmicb.2015.00937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bal HB, Nayak L, Das S, Adhya TK. Isolation of ACC deaminase producing PGPR from rice rhizosphere and evaluating their plant growth, promoting activity under salt stress. Plant Soil. 2013;366(1-): 93– 105. doi: 10.1007/s11104-012-1402-5. [DOI] [Google Scholar]
- 7.Rungruangmaitree R, Jiraungkoorskul W. Pea, Pisum sativum, and its anticancer activity. Pharmacogn Rev. 2017; 11( 21): 39– 42. doi: 10.4103/phrev.phrev_57_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dworkin M, Foster JW. Experiments with some microorganisms which utilize ethane and hydrogen. J Bacteriol. 1958;75:592– 603. doi: 10.1128/jb.75.5.592-603.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Penrose DM, Glick BR. Methods for isolating and characterizing ACC deaminase-containing plant growth- promoting rhizobacteria. Physiol Plant. 2003; 118( 1):10–15. doi: 10.1034/j.1399-3054.2003.00086.x. [DOI] [PubMed] [Google Scholar]
- 10.Ali SZ, Sandhya V, Rao LV. Isolation and characterization of drought-tolerant ACC deaminase and exopolysaccharide- producing fluorescent Pseudomonas sp. Ann Microbiol. 2014;64(2):493–502. doi: 10.1007/s13213-013-0680-3. [DOI] [Google Scholar]
- 11.Liaqat F, Eltem R. Identification and characterization of endophytic bacteria isolated from in vitro cultures of peach and pear rootstocks. 3 Biotech. 2016;6(2):2–9. doi: 10.1007/s13205-016-0442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patil C, Suryawanshi R, Koli S, Patil S. Improved method for effective screening of ACC (1-aminocyclopropane-1- carboxylate) deaminase producing microorganisms. J Microbiol Methods. 2016;131:102 – 104. doi: 10.1016/j.mimet.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Bradford MM, Dong Y, Xu L, Liu S, Bai X, Bradford M, et al. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1-2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 14.Tang YW, Bonner J. The enzymatic inactivation of Indole acetic acid; the physiology of the. Am J Bot. 1948;35(9):570–578. [PubMed] [Google Scholar]
- 15.Gordon SA, Weber RP. Colorimetric estimation of Indoleacetic acid. 1951;26(1):192–5. doi: 10.1104/pp.26.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology, 1995 Ch. 2.4. Wiley, New York. [Google Scholar]
- 17.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suneja P, Piplani S, Dahiya P, Dudeja SS. Molecular characterization of rhizobia from revertants of non-nodulating cultivar and normal cultivar of chickpea. J Agric Sci Technol. 2016 ;18 (3):763–773. [Google Scholar]
- 19.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan J, Müller KM, Charles TC, Vesely S, Glick BR. 1- Aminocyclopropane-1-carboxylate (ACC) deaminase genes in rhizobia from southern saskatchewan. Microb Ecol. 2009;57(3): 423– 436. doi: 10.1007/s00248-008-9407-6. [DOI] [PubMed] [Google Scholar]
- 21.Sloger C. Symbiotic Effectiveness and N2 Fixation in Nodulated Soybean. Plant Physiol. 1969 ; 44( 12):1666 – 1668. doi: 10.1104/pp.44.12.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suneja P, Kumar A, Dahiya P. Characterization of epiphytic bacteria isolated from chickpea (Cicer arietinum L.) nodules. African J Microbiol Res. 2014;8(12):1302–1309. [Google Scholar]
- 23.Brader G, Compant S, Mitter B, Trognitz F, Sessitsch A. Metabolic potential of endophytic bacteria. Current Opinion in Biotechnology. 2014;27:30–37. doi: 10.1016/j.copbio.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suneja P, Duhan JS, Bhutani N, Dudeja SS. Recent Biotechnological Approaches to Study Taxonomy of Legume Nodule Forming Rhizobia. In: Gahlawat SK, Salar RK, Siwach P, Duhan JS, Kumar S, Kaur P, editors. Plant Biotechnology: Recent Advancements and Developments: Springer Singapore. 2017. pp. 101–24. [DOI] [Google Scholar]
- 25.Nukui N, Ezura H, Yuhashi K, Yasuta T, Minamisawa K. Effects of ethylene precursor and inhibitors for ethylene biosynthesis and perception on nodulation in Lotus japonicus and Macroptilium atropurpureum. Plant Cell Physiol. 2000; 41( 7): 893– 897. doi: 10.1093/pcp/pcd011. [DOI] [PubMed] [Google Scholar]
- 26.Van de Poel B, Van DerStraeten D. 1-aminocyclopropane-1- carboxylic acid (ACC) in plants: more than just the precursor of ethylene. Front Plant Sci. 2014; 5( November): 1– 11. doi: 10.3389/fpls.2014.00640/abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blaha D, Prigent-Combaret C, Mirza MS, Moënne-Loccoz Y. Phylogeny of the 1-aminocyclopropane-1-carboxylic acid deaminase-encoding gene acdS in phytobeneficial and pathogenic Proteobacteria and relation with strain biogeography. FEMS Microbiol Ecol. 2006 ;56 ( 3): 455– 470. doi: 10.1111/j.1574-6941.2006.00082.x. [DOI] [PubMed] [Google Scholar]
- 28.Kuklinsky-Sobral J, Araújo WL, Mendes R, Pizzirani-Kleiner AA, Azevedo JL. Isolation and characterization of endophytic bacteria from soybean (Glycine max) grown in soil treated with glyphosate herbicide. Plant Soil. 2005;273(1–2):91–99. doi: 10.1007/s11104-004-6894-1. [DOI] [Google Scholar]
- 29.Palaniappan P, Chauhan PS, Saravanan VS, Anandham R, Sa T. Isolation and characterization of plant growth promoting endophytic bacterial isolates from root nodule of Lespedeza sp. Biol Fertil Soils. 2010;46(8):807–816. doi: 10.1007/s13199-017-0481-8. [DOI] [Google Scholar]
- 30.Pandya M, Naresh Kumar G, Rajkumar S. Invasion of rhizobial infection thread by non-rhizobia for colonization of Vigna radiata root nodules. FEMS Microbiol Lett. 2013;348(1):58–65. doi: 10.1111/1574-6968.12245. [DOI] [PubMed] [Google Scholar]
- 31.Kumar V, Kumar A, Pandey KD, Roy BK. Isolation and characterization of bacterial endophytes from the roots of Cassia tora L. Ann Microbiol. 2015;65(3):1391–1399. doi: 10.1134/S0026261715010105. [DOI] [Google Scholar]
- 32.Bacon CW, Hinton DM. Endophytic and biological control potential of Bacillus mojavensis and related species. Biol Control. 2002;23(3):274–284. doi: 10.1006/bcon.2001.1016. [DOI] [Google Scholar]
- 33.Pyo JS, Shrestha S, Park SH, Kang JS. Biological control of plant growth using the plant growth-promoting rhizobacterium Bacillus mojavensis KJ-s3. J Life Sci. 2014;24(12):1308–1315. doi: 10.5352/JLS.2015.25.8.910. [DOI] [Google Scholar]
- 34.Bhutani N, Maheshwari R, Negi M, Suneja, P. Optimization of IAA production by endophytic Bacillus spp. from Vigna radiata for their potential use as plant growth promoters. Isr. J Plant Sci. 2018;65(01-02):83–96. doi: 10.1163/22238980-00001025. [DOI] [Google Scholar]
- 35.Gopalakrishnan S, Vadlamudi S, Samineni S, Sameer Kumar C V. Plant growth-promotion and biofortification of chickpea and pigeon pea through inoculation of biocontrol potential bacteria. Springerplus. 2016;5(1) doi: 10.1186/s40064-016-3590-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bai Y, D’Aoust F, Smith DL, Driscoll BT. Isolation of plant- growth-promoting Bacillus strains from soybean root nodules. Can J Microbiol. 2002; 48(October 2016):230–238. doi: 10.1139/W02-014. [DOI] [PubMed] [Google Scholar]
- 37.Muresu R, Polone E, Sulas L, Baldan B, Tondello A, Delogu G, et al. Coexistence of predominantly nonculturable rhizobia with diverse, endophytic bacterial taxa within nodules of wild legumes. FEMS Microbiol Ecol. 2008;63(3):383–400. doi: 10.1111/j.1574-6941.2007.00424.x. [DOI] [PubMed] [Google Scholar]


