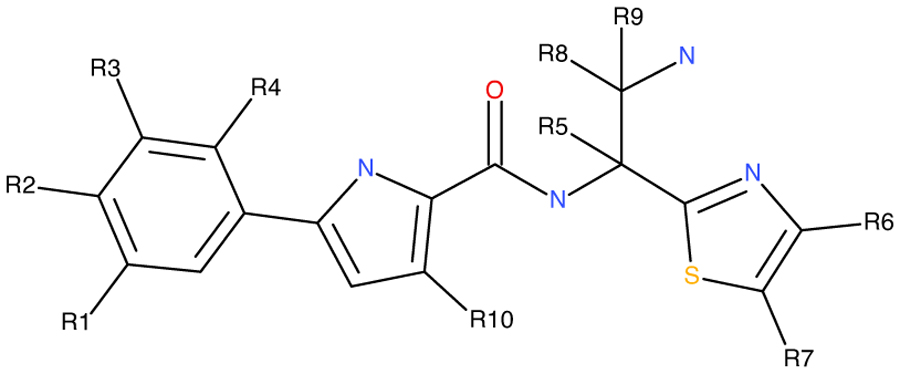
Abstract
We presented our continuing stride to optimize the second-generation NBD entry antagonist targeted to the Phe43 cavity of HIV-1 gp120. We have synthesized thirty-eight new and novel analogs of NBD-14136, earlier designed based on a CH2OH “positional switch” hypothesis, and derived a comprehensive SAR. The antiviral data confirmed that the linear alcohol towards the “N” (C4) of the thiazole ring yielded more active inhibitors than those towards the “S” (C5) of the thiazole ring. The best inhibitor, NBD-14273 (compound 13), showed both improved antiviral activity and selectivity index (SI) against HIV-1HXB2 compared to NBD-14136. We also tested NBD-14273 against a large panel of 50 HIV-1 Env-pseudotyped viruses representing clinical isolates of diverse subtypes. The overall mean data indicate that antiviral potency against these isolates improved by ~3-fold, and SI also improved ~3-fold compared to NBD-14136. This new and novel inhibitor is expected to pave the way for further optimization to a more potent and clinically relevant inhibitor against HIV-1.
Keywords: HIV-1, ENV-pseudovirus, gp120 entry-antagonist, cytotoxicity, broad-spectrum, structure-activity relationship (SAR), selectivity index (SI)
Graphical Abstract
1. Introduction
Human immunodeficiency virus type-1 (HIV-1) is the causative of the acquired immunodeficiency syndrome (AIDS). Based on the 2020 Global HIV Statistics from UNAIDS, 38 million people are living with HIV globally, with about 1.7 million new infections in 2019. Since the start of the epidemic, about 76 million people have become infected with this deadly disease, and more than 32 million have lost their lives due to AIDS-related illnesses.
However, advances in available therapeutics, particularly combination antiretroviral therapy (cART), significantly improved the treatment of HIV infection and facilitated the shift from high morbidity and mortality to a manageable chronic disease. Despite this remarkable success, current treatments suffer from several limitations, including (1) reliance on daily adherence, (2) long-term use resulting in long-term toxicity, (3) limited treatment options due to the development of drug resistance, (4) high cost, and finally, (5) the non-curative nature of current treatments. Further, despite the tremendous effort and investment for over a decade, the availability of an effective vaccine or microbicide is not on the near horizon, and significant hurdles must be overcome to achieve an effective cure. Thus, the continued development of small-molecule drugs with high potency against novel targets, but minimal side effects is imperative. The development of novel therapeutics will aid in increasing the number of new drugs available and will extend the scope of combination therapy.
Viral attachment and fusion to the host cell membrane are critical to the ability of HIV-1 to enter host cells and initiate its life-cycle by utilizing host cell machinery1. Therefore, viral attachment and fusion (often collectively termed “viral entry”) have been targets of new drug discovery for years1–4. Only two drugs currently on the market that target the HIV-1 entry pathway. FUZEON® (enfuvirtide) targets the envelope glycoprotein gp415, 6, and SELZENTRY® (maraviroc) targets the host cell receptor CCR5 to prevent viral entry7–9.
Even though HIV-1 gp120 is critical for viral entry and has been the target of drug discovery efforts, no drugs that target gp120 have been approved by the US FDA. BMS-663068, the most advanced inhibitor in this class, is a prodrug of BMS-626529. The safety and efficacy of BMS-663068 were recently demonstrated in a Phase 2b clinical trial, and the compound is currently undergoing Phase III clinical trials.
Our laboratory had focused on developing novel inhibitors targeted to the Phe43 cavity of gp120 since 2005 when we first reported the discovery of NBD-556, the NBD-series CD4-mimic10. Unfortunately, NBD-556 was shown to enhance HIV-1 infection in CD4−-CCR5+ cells and thus behaved as an HIV-1 entry agonist11, 12. Since this finding, we and others have endeavored to design CD4 mimics with not only higher potency and lower toxicity but also that are devoid of this unwanted agonist property and serve as HIV-1 entry antagonists12–22.
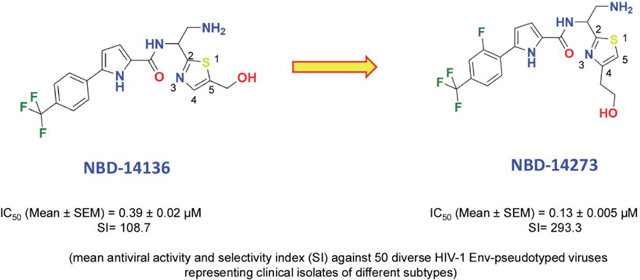
Since the discovery of the small molecule CD4-mimic, NBD-556, we systematically showed by x-ray crystallography that it binds to the Phe43 cavity of the HIV-1 gp12023. The major transformation of CD4-mimic came through the design of NBD-11021. Based on the x-ray structure of NBD-1102112, we further modified the molecules by replacing the piperazine ring with a simple amine, such as NBD-14010. The x-ray structure of NBD-14010 provided critical information on modifying the thiazole ring substituents to achieve robust interactions with the target protein24. One such essential details lead us to postulate the “Positional switch” hypothesis that resulted in more potent antivirals, such as NBD-14136 (IC50=0.27 μM), NBD-14168 (IC50=0.28 μM), and NBD-14189 (IC50=0.089 μM) that we reported earlier (Figure 1)25. Based on this success, we wanted to expand further the structure-activity relationship (SAR) of these series of compounds with significant emphasis on compounds with the p-CF3 substituents, which yielded the most active CD4-mimics as antivirals. In this report, we presented our effort to expand the SAR studies of an extensive series of small molecule CD4-mimics to optimize further this class of HIV-1 entry inhibitors that target the Phe43 cavity of envelope glycoprotein gp120 to improve its antiviral activity and selectivity index (SI).
Figure 1.
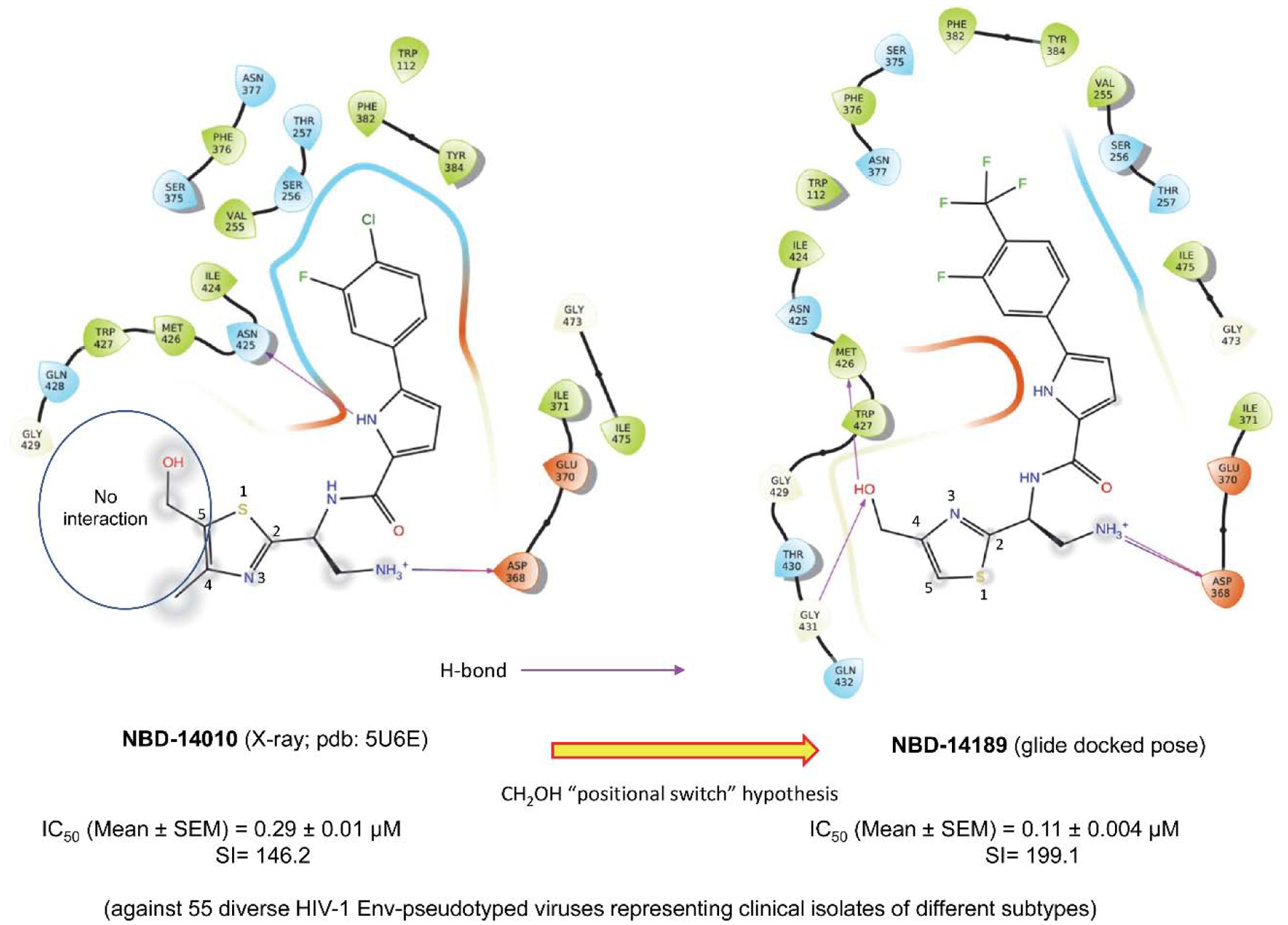
“Positional switch” hypothesis indicating no interaction of the CH2OH group at position 5 in the thiazole ring in the x-ray structure of NBD-14010 bound to HIV-1 gp120. When the CH2OH was switched to position 4 in the thiazole ring in NBD-14189, it forms H-bond (indicated by violet arrows using a glide docking pose) with Met426 and Gly43125.
2. Results and Discussions
2.1. Design, anti-HIV-1 screening, and structure-activity relationships (SARs)
In this study, we focused on one of the best lead NBD-14136 containing a trifluoromethyl (CF3) group at the para position, which showed an IC50 of 0.27 μM and CC50 of 42.4 μM resulting in an SI (where SI = CC50/IC50) = 157. We synthesized a large set of compounds to improve the antiviral activity and SI and derived a comprehensive SAR based on the lead NBD-14136. In most cases, we kept the CF3 substituent at the para position in the phenyl ring. We also decided to minimally alter the substituents in the phenyl ring and make changes in the thiazole moiety that showed improvement in antiviral activity and selectivity before. Due to the presence of one chiral center, all compounds have two stereoisomers (defined as R and S in Table 1). We observed in our earlier studies that in the majority of the cases, the S isomer had better antiviral potency. Therefore, in the SAR, we will mostly focus on comparing the S isomers. We have initially tested all the synthesized compounds against HIV-1HXBR2 in TZM-bl cells (Table 1) to identify the best active molecule with higher SI to further test it against a large panel of HIV-1 ENV-pseudotyped reference viruses from clinical isolates.
Table 1.
Anti-HIV-1 activity (IC50) of NBD compounds in a single-cycle assay against HIV-1HXB2
| New No | Code (enantiomer) | R1 | R2 | R3 | R4 | R5 | R6 | R7 | R9 | R9 | R10 | IC50b | CC50b | SI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NBD-14136 (R)a | H | CF3 | H | H | H | H | CH2OH | H | H | H | 0.27±02 | 42.4±1 | 157 | |
| 1 | NBD-14250 (R) | H | CF3 | H | F | H | CH2OH | H | H | H | H | 0.61±15 | 26.8±0.25 | 43.9 |
| 2 | NBD-14251 (S) | H | CF3 | H | F | H | CH2OH | H | H | H | H | 0.187±0.01 | 25.7±1.2 | 137 |
| 3 | NBD-14260 (R) | H | CN | H | H | H | CH2OH | H | H | H | H | >16 | >82 | - |
| 4 | NBD-14261 (S) | H | CN | H | H | H | CH2OH | H | H | H | H | >16 | >82 | - |
| 5 | NBD-14313 (S) | H | CF3 | H | CF3 | H | CH2OH | H | H | H | H | 4.2±0.4 | 33.4±2 | 7.9 |
| 6 | NBD-14314 (R) | H | CF3 | H | CF3 | H | CH2OH | H | H | H | H | 6.3±1 | 37.6±3.6 | 5.9 |
| 7 | NBD-14278-rac | F | CF3 | H | H | CH3 | CH2OH | H | H | H | H | 1.6±0.08 | 45.3±1 | 28 |
| 8 | NBD-14280-rac | F | Cl | F | H | CH3 | CH2OH | H | H | H | H | 2.6±0.4 | 47.7±1.3 | 18.3 |
| 9 | NBD-14242 (S) | F | Cl | F | H | H | CH2CH2OH | H | H | H | 0.15±0.001 | 32.8±1 | 218 | |
| 10 | NBD-14243 (R) | F | Cl | F | H | H | CH2CH2OH | H | H | H | H | 0.98±0.01 | 32.8±0.5 | 33.5 |
| 11 | NBD-14258 (R) | F | CF3 | H | H | H | CH2CH2OH | H | H | H | H | 0.51±0.05 | 24.2±0.8 | 47.4 |
| 12 | NBD-14259 (S) | F | CF3 | H | H | H | CH2CH2OH | H | H | H | H | 0.2±0.02 | 24.4±0.7 | 122 |
| 13 | NBD-14273 (S) | H | CF3 | H | F | H | CH2CH2OH | H | H | H | H | 0.18±0.02 | 39.6±2 | 220 |
| 14 | NBD-14274 (R) | H | CF3 | H | F | H | CH2CH2OH | H | H | H | H | 0.15±0.01 | 23.4±0.9 | 156 |
| 15 | NBD-14287 (S) | F | CF3 | H | H | H | CH(OH)CH2OH | H | H | H | H | 0.55±0.05 | 72.4±3.1 | 131.6 |
| 16 | NBD-14288 (R) | F | CF3 | H | H | H | CH(OH)CH2OH | H | H | H | H | 2.4±0.1 | 56.3±8.8 | 23.4 |
| 17 | NBD-14289 (S) | F | CF3 | H | H | H | CH2CH2CH2OH | H | H | H | H | 0.25±0.01 | 22.6±2.5 | 90.4 |
| 18 | NBD-14290 (R) | F | CF3 | H | H | H | CH2CH2CH2OH | H | H | H | H | 3.5±0.2 | 38±1.3 | 10.8 |
| 19 | NBD-14303 (S) | F | CF3 | H | H | H | H | CH(OH)CH2OH | H | H | H | 6.2±0.9 | 98±3.5 | 15.8 |
| 20 | NBD-14304 (R) | F | CF3 | H | H | H | H | CH(OH)CH2OH | H | H | H | 0.59±0.1 | 70.5±4.5 | 119.4 |
| 21 | NBD-14271 (S) | F | CF3 | H | H | H | COOH | H | H | H | H | Not soluble | - | - |
| 22 | NBD-14272 (R) | F | CF3 | H | H | H | COOH | H | H | H | H | Not soluble | - | - |
| 23 | NBD-14275 (S) | F | CF3 | H | H | H | COOCH3 | H | H | H | H | 2.9±0.4 | 41.6±1.3 | 14.3 |
| 24 | NBD-14276 (R) | F | CF3 | H | H | H | COOCH3 | H | H | H | H | 4.9±0.9 | 23.4±0.5 | 4.8 |
| 25 | NBD-14267 (R) | F | CF3 | H | H | H | CONH2 | H | H | H | H | 2.3±0.1 | 25.9±0.3 | 11.3 |
| 26 | NBD-14268 (S) | F | CF3 | H | H | H | CONH2 | H | H | H | H | >10 | >52 | |
| 27 | NBD-14325 (S) | F | CF3 | H | H | H | CH2OH | H | H | H | CH3 | 0.54±0.1 | 18.5±0.2 | 34.2 |
| 28 | NBD-14324 (R) | F | CF3 | H | H | H | CH2OH | H | H | H | CH3 | 0.57±0.1 | 14.1±1.6 | 24.7 |
| 29 | NBD-14329 (S) | F | CF3 | H | H | H | H | CH2OH | H | H | CH3 | 0.15±0.02 | 10.3±0.5 | 20.6 |
| 30 | NBD-14328 (R) | F | CF3 | H | H | H | H | CH2OH | H | H | CH3 | 0.21±0.04 | 10.4±0.4 | 49.5 |
| 31 | NBD-14246 (S) | F | Cl | F | H | H | CH2OH | H | CH3 | CH3 | H | 0.12±0.002 | 16±1 | 133 |
| 32 | NBD-14247 (R) | F | Cl | F | H | H | CH2OH | H | CH3 | CH3 | H | 0.15±0.004 | 15.8±0.5 | 105 |
| 33 | NBD-14248 (S) | H | CF3 | H | H | H | CH2OH | H | CH3 | CH3 | H | 0.13±0.005 | 17.5±1 | 134.6 |
| 34 | NBD-14249 (R) | H | CF3 | H | H | H | CH2OH | H | CH3 | CH3 | H | 1.3±0.03 | 19.9±0.5 | 15.3 |
| 35 | NBD-14262 (R) | H | CF3 | H | H | H | H | CH2OH | CH3 | CH3 | H | 0.68±0.12 | 23.1±0.5 | 34 |
| 36 | NBD-14263 (S) | H | CF3 | H | H | H | H | CH2OH | CH3 | CH3 | H | 0.23±0.04 | 25.1±1 | 109 |
| 37 | NBD-14225 (S) | H | CF3 | H | H | H | H | CH2OCH2CH(OH)CH2OH | H | H | H | >13 | >61.9 | - |
| 38 | NBD-14229 (R) | H | CF3 | H | H | H | H | CH2OCH2CH(OH)CH2OH | H | H | H | 2.1±0.2 | ~80 | ~38 |
Data from Reference25
The reported IC50 and CC50 values represent the mean ± standard deviation (SD), n=3
Compound 2 with a CF3 at R2 and a fluoro (F) at R4 position with CH2OH at R6 positions yielded one of the most active compounds in this series (IC50=0.19 μM and SI=137). However, when we kept all groups the same except changing CH2OH to CH2CH2OH, the cytotoxicity improved by about 1.7-fold, providing compound 13 with similar anti-HIV-1 activity but much improved SI of 220. Moving the F to R1 in compound 12 had no apparent effect on both antiviral activity and cytotoxicity when compared to compound 2. We have explored a large number of compounds keeping the substituents the same as in compound 12 by increasing the complexity of the primary alcohol substituent at R6. In general, the cytotoxicity of the majority of the compounds improved, but the activity varied considerably. When the chain length was increased in compound 17, the activity and cytotoxicity remained similar to 12. The antiviral activity dropped considerably when R6 had branched primary alcohols (compounds 15 and 19). When we replaced the primary alcohol with an acid, the compounds became insoluble; therefore, we could not perform the antiviral or cytotoxicity assays. We also observed a considerable drop in antiviral activity when R6 had an amide or ester substituents. Interestingly, when we moved the branched alcohol substituent to the R7 position, antiviral activity dropped substantially (compound 19). It appears that bulkier alcohol substituent in either R6 or R7 position was not well tolerated. Interestingly, we also observed that when we substituted the R2 position with a hydrophilic substituent (CN), we virtually lost the antiviral potency (Compound 4). It was, however, well-established that the Phe43 pocket in that region is hydrophobic. Therefore, the result was not surprising. A chloro (Cl) substituent at R2 and F in both R1 and R3 with CH2CH2OH at R6 (Compound 9) yielded one of the most active inhibitors with an IC50 of 0.15 μM and SI of 218. Interestingly, when we added a methyl group at position R10 on the pyrrole ring, antiviral activity improved remarkably (compounds 27–30). Unfortunately, the cytotoxicity also was higher, making the SI values low. We also observed that when the primary amine had two methyl substituents at R8 and R9 (compounds 31–34) and the CH2OH group was at position R6, the antiviral activities were also improved substantially, but again these additional methyl groups also made those molecules more cytotoxic (CC50). However, when the CH2OH group was moved to R7 (Compound 36), the antiviral activity dropped by about 2-fold, but cytotoxicity improved to make SI >100. In the same series, when we substituted R7 with branched alcohol (compounds 37 and 38), the antiviral potency dropped substantially, but the cytotoxicity improved, although SI values did not improve. Overall, the SAR analyses confirmed that the antiviral activity considerably improved by moving the primary alcohol groups to R6 from R7 (as in NBD-14136). Both p-Cl and p-CF3 containing compounds (compounds 9 and 13) showed high potency with high SI values. The data in Table 1 demonstrated that NBD-14242 and NBD-14273 showed very similar antiviral activity and SI (IC50 of 0.15 and 0.18 μM; and 218 and 220, respectively). We selected NBD-14273 for further study.
2.2. NBD-14273 showed entry antagonist features
NBD-55610, our first-generation gp120-targeted compound, mimic CD4 by inducing a conformational change in gp120 and facilitating HIV infection into CD4-negative cells that expressed the coreceptor CCR513, 26. Since then, we validated our new generation inhibitors by confirming that they do not possess this undesirable trait and work as entry antagonists27–30. We selected NBD-14273, which exhibited the best anti-HIV-1 activity and a higher Selectivity Index value (SI = 220). CD4-negative Cf2TH-CCR5 cells were infected with the recombinant CD4-dependant HIV-1ADA virus in the presence of escalating concentrations of NBD-14273 and NBD-556, which was used as a control. While NBD-556 significantly supported the infection of the Cf2Th-CCR5 cells, NBD-14273 did not, indicating that this compound is an HIV-1 entry antagonist (Figure 2). This data assured us of selecting this inhibitor for further evaluation against a large and diverse panel of HIV-1 ENV-pseudotyped clinical isolates.
Figure 2. Infectivity of CD4-negative Cf2Th-CCR5 cells by CD4-dependent HIV-1ADA.

CCf2Th-CCR5 cells were infected with CD4-dependent HIV-1ADA in the presence of NBD-14273 and NBD-556, which was used as a control. The Relative virus infectivity indicates the ratio of the amount of infection detected in the presence and absence of the compounds. Three independent experiments were performed in triplicate, and the graph is representative of one experiment. The toxicity of the compounds against these cells was evaluated to calculate the CC50 values: for NBD-556, the CC50 was > 60, and for NBD-14273, it was 31.2±1.4. All the values represent the mean ± standard deviation.
2.3. Antiviral activities of NBD-14273 against a large and diverse panel of HIV-1 Env-pseudotyped clinical isolates
Since we reported the identification of NBD-55610 as the first example of HIV-1 entry-inhibitor, we have shown consistent improvement in the anti-HIV-1 activity of our new generations of gp120 entry-antagonists against the control lab-adapted virus HIV-1HXB2 and a large panel of diverse HIV-1 Env-pseudotyped clinical isolates12, 15, 25, 28, 31. In this study, the anti-HIV-1 activity of NBD-14273 was evaluated against a collection of 50 HIV-1 clinical isolates of diverse subtypes, including primary, transmitted, and early founder HIV-1 isolates and a selection of 12 recombinant HIV-1 clones (Table 2). All HIV-1 clones tested use the CCR5 coreceptor for entry except for two dual-tropic (CCR5/CXCR4) clones (NIH # 11563 and 11578). We compared the antiviral activity of this compound with that of the previously described inhibitor NBD-1413628, 32. NBD-14273 exhibited a broad-spectrum antiviral activity, as shown by its activity against all the clinical isolates tested, regardless of the viral subtype or coreceptor usage. The anti-HIV-1 activity of NBD-14273 was about 2.4–3.3-fold higher than the anti-viral activity detected for NBD-14136, as demonstrated by their overall mean IC50 values and their antiviral activities against the different HIV subtypes (Table 2). The overall mean IC50 value for NBD-14136 was 0.39 ± 0.02 μM (with the SI of 108.7 and the IC50 values ranged from 0.19–0.79 μM), whereas the overall mean IC50 value determined for NBD-14273 was 0.135 ± 0.005 μM (with the SI of 293.3 and the IC50 values ranged from 0.11–0.22 μM). It is noteworthy to mention that our earlier reported inhibitor NBD-14189, which showed the best activity profile (as low as 63 nM compared to 94 nM for NBD-14273), but NBD-14273 demonstrated better SI (199.1 for NBD-14189 vs. 293 for NBD-14273)25. In this study, NBD-14273 showed better antiviral activity against all the viruses of different subtypes tested in this work. In contrast, this compound exhibited a slightly better cytotoxicity profile (CC50 of 39.6 ± 2 μM) than that for NBD-14136 (CC50 of 42.4 ± 1.0 μM), which resulted in an improvement of the NBD-14273 SI values of 2.2–3-fold with respect to NBD-14136. Moreover, NBD-14273 was poorly active against the pseudovirus VSV-G, which was used here as a control, suggesting that the inhibitory activities of these compounds are specific to HIV-1. Additionally, the toxicity in the U87-CD4-CXCR4 cell line was similar to what we detected in the other cell lines.
Table 2.
Neutralization Activity of gp120 entry-antagonists against a Panel of HIV-1 Env Pseudoviruses
| IC50 μMa | ||||
|---|---|---|---|---|
| Subtype | NIH # | ENVs | NBD-14136c | NBD-14273 |
| A | 11887 | Q259ENV.W6 | 0.2±0.01 | |
| 11888 | QB726.70M.ENV.C4 | 0.11±0.005 | ||
| 11890 | QF495.23M.ENV.A1 | 0.21±0.02 | ||
| 11891 | QF495.23M.ENV.A3 | 0.11±0.01 | ||
| BG505-T332N | 0.41±0.03 | 0.118±0.003 | ||
| KNH1144 | 0.62±0.08 | 0.11±0.02 | ||
| A/D | 11901 | QA790.204I.ENV.A4 | 0.12±0.004 | |
| 11904 | QA790.204I.ENV.E2 | 0.11±0.02 | ||
| A2/D | 11905 | QG393.60M.ENV.A1 | 0.34±0.02 | 0.13±0.003 |
| 11906 | QG393.60M.ENV.B7 | 0.6±0.005 | 0.127±0.005 | |
| A/G | 11591 | CRF02_AG Clone 211 | 0.58±0.05 | 0.22±0.04 |
| 11594 | CRF02_AG clone 250 | 0.13±0.005 | ||
| 11595 | CRF02_AG clone 251 | 0.11±0.01 | ||
| 11598 | CRF02_AG clone 255 | 0.13±0.01 | ||
| 11599 | CRF02_AG clone 257 | 0.39±0.01 | 0.13±0.006 | |
| 11600 | CRF13_cpx clone 258 | 0.52±0.07 | 0.175±0.03 | |
| 11602 | CRF02_AG clone 266 | 0.51±0.06 | 0.16±0.02 | |
| AE | 11603 | CRF01_AE clone 269 | 0.52±0.02 | 0.13±0.01 |
| B | B41 | 0.139±0.001 | ||
| 11018 | QH0692, clone 42 | 0.13±0.01 | ||
| 11022 | PVO, clone 4 | 0.094±0.007 | ||
| 11023 | TRO, clone 11 | 0.135±0.001 | ||
| 11036 | RHPA4259 clone 7 | 0.13±0.003 | ||
| 11037 | THRO4156 clone 18 | 0.13±0.005 | ||
| 11038 | CAAN5342 clone A2 | 0.12±0.002 | ||
| 11058 | SC422661.8 | 0.32±0.08 | 0.12±0.02 | |
| 11560 | 1006_11.C3.1601 | 0.58±0.06 | 0.135±0.002 | |
| 11561 | 1054.TC4.1499 | 0.25±0.02 | 0.14±0.01 | |
| 11562 | 1056.TA11.1826 | 0.58±0.01 | 0.13±0.01 | |
| 11563 | 1058 11.B11.1550b | 0.17±0.01 | ||
| 11572 | 9021_14.B2.4571 | 0.36±0.08 | 0.13±0.006 | |
| 11578 | WEAUd15.410.5017b | 0.76±0.04 | 0.13±0.004 | |
| C | 11307 | Du172, clone 17 | 0.3±0.03 | 0.12±0.001 |
| 11308 | Du422, clone 1 | 0.63±0.09 | 0.22±0.02 | |
| 11309 | ZM197M.PB7, SVPC6 | 0.27±0.01 | 0.12±0.01 | |
| 11310 | ZM214M.PL15, SVPC7 | 0.24±0.007 | 0.12±0.003 | |
| 11312 | ZM249M.PL1, SVPC10 | 0.55±0.07 | 0.17±0.01 | |
| 11313 | ZM53M.PB12, SVPC11 | 0.54±0.03 | 0.136±0.002 | |
| 11314 | ZM109F.PB4 | 0.12±0.004 | ||
| 11317 | CAP210.2.00.E8, SVPC17 | 0.11±0.003 | ||
| 11502 | HIV-16055–2, clone 3 | 0.123±0.01 | ||
| 11504 | HIV-16936–2, clone 21 | 0.24±0.04 | ||
| 11506 | HIV-25711–2, clone 4 | 0.12±0.01 | ||
| 11507 | HIV-225925–2, clone 22 | 0.11±0.005 | ||
| 11908 | QB099.391M.ENV.B1 | 0.113±0.006 | ||
| D | 11911 | QA013.70I.ENV.H1 | 0.12±0.01 | |
| 11912 | QA013.70I.ENV.M12 | 0.11±0.006 | ||
| 11916 | QD435.100M.ENV.B5 | 0.3±0.01 | 0.11±0.01 | |
| 11918 | QD435.100M.ENV.E1 | 0.19±0.04 | 0.113±0.002 | |
| G | 11596 | CRF02_G clone 252 | 0.26±0.03 | 0.11±0.01 |
|
Overall (n=50) SI |
0.39±0.02 108.7 |
0.135±0.005 293.3 |
||
|
Subtype A (n=6) SI |
0.42±0.05 101 |
0.143±0.02 277 |
||
|
Subtype Arec (n=12) SI |
0.44±0.03 96.4 |
0.139±0.009 285 |
||
|
Subtype B (n=14) SI |
0.39±0.04 108.7 |
0.131±0.004 302.3 |
||
|
Subtype C (n=13) SI |
0.38±0.04 111.6 |
0.14±0.01 282.9 |
||
|
Subtype D (n=4) SI |
0.26±0.03 163.1 |
0.11±0.002 360 |
||
| Control | VSV-Gd | IC50 | >20 | 11.9±0.5 |
| CC50 | 46.3±10.6 | 33.5±0.6 | ||
| IC50 μM (Color Code) | ≤0.2 | >0.2 ≤0.5 | >0.5 | |
The reported IC50 values represent the means ± standard deviations (n = 3).
R5X4-tropic virus all the rest are CCR5-tropic viruses.
Data previously published32
VSV-G was tested in U87-CD4-CCR5 cells
Furthermore, we evaluated the anti-HIV-1 activity of NBD-14273 against a panel of HIV-1 clones, which included a group of paired infant and maternal clones. These HIV clones were isolated from chronically infected mothers and their respective infected infants and belonged to subtypes A and D/A33. it has been shown that the vertically transmitted HIV-1 infant variants were more challenging to neutralize using combinations of broadly neutralizing antibodies (bNAbs) 2G12, biz, 2F5, and 4E1033. In this study, we found that NBD14273, as previously shown for NBD-1413632, equally neutralized both the infant and maternal HIV-1 variants (Table 3), as shown by their overall mean of the IC50 values and the IC50 means for infant and maternal viruses. Of note, NBD-14273 was about 4-fold more effective than NBD-14136 against both viral clones of the infant and maternal panel. These findings suggest that NBD-14273 can neutralize both infant and maternal HIV-1 variants.
Table 3.
Neutralization activity of NBD-14273 against an HIV-1 panel of Paired Infant (B) and Maternal (M) Env Molecular Clones.
| IC50 (μM)a | ||||
|---|---|---|---|---|
| Subtype | NIH # | ENV | NBD-14136b | NBD-14273 |
| A A |
11518-B 11528-M |
BG505.W6M.ENV.C2 MG505.W0M.ENV.A2 |
0.92±0.16 0.93±0.07 |
0.22±0.02 0.17±0.05 |
| A A |
11519-B 11531-M |
B1206.W6P.ENV.A1A MI206.W0M.ENV.D1 |
0.75±0.06 0.7±0.02 |
0.18±0.06 0.22±0.07 |
| A A |
11521-B 11535-M |
BJ613.W6M.ENV.E1 MJ613.W0M.ENV.A2 |
0.8±0.07 0.5±0.05 |
0.13±0.01 0.136±0.02 |
| D/A D/A |
11524-B 11538-M |
BL035.W6M.ENV.C1 ML035.W0M.ENV.G2 |
0.52±0.16 0.59±0.08 |
0.24±0.03 0.18±0.03 |
| A A |
11525-B 11540-M |
BL274.W6M.ENV.A3 ML274.W0M.ENV.B1 |
0.63±0.12 0.66±0.1 |
0.15±0.020.136±0.01 |
| Overall (n=10) | 0.7±0.05 | 0.176±0.01 | ||
| Infant (B) (n=5) | 0.72±0.07 | 0.184±0.02 | ||
| Mother (M) (n=5) | 0.68±0.07 | 0.168±0.016 | ||
The reported IC50 values represent the means ± standard deviations (n = 3).
Data previously published32
2.4. Inhibitory activity of NBD-14273 against a large panel of FDA-approved-drug-resistant viruses
We further evaluated the anti-viral activity of NBD-14273 against a panel of drug-resistant viruses, consisting of 5 HIV-1 clones resistant to the entry-inhibitor Enfuvirtide (T-20); 7 HIV-1 multi-drug-resistant clones specifically, these are non-nucleoside reverse transcriptase inhibitor (NNRTI)-resistant viruses, which carry mutations that confer resistance to both NNRTIs and nucleoside reverse transcriptase inhibitors (NRTIs); 3 HIV-1 clones resistant to the integrase inhibitor Raltegravir; and 9 HIV-1 clones resistant to multiple protease inhibitors (Table 4). As a reference, the activity of NBD-14273 was evaluated against the wild type (WT) HIV-1NL4–3 clone, which was used to obtain the drug-resistant viruses. For this assay, the TZM-bl cells were infected with the WT HIV-1NL4–3 control virus or the drug-resistant viruses pretreated for 30 min with escalating concentrations of NBD-14273. NBD-14273 inhibited the WT HIV-1NL4–3, with an IC50 value of 0.21 μM. All the T-20-resistant viruses were sensitive to NBD-14273; the IC50 values we detected were similar to those determined for the control virus, WT HIV-1NL4–3. Regarding the NNRTI-resistant viruses, also all the viral clones were sensitive to NBD-14273. In some cases, we observed that the drug-resistant viruses appeared to be more sensitive to the gp120 entry-antagonist than the WT virus. The only exceptions were the HIV-1 clone NIH # 12243, which showed a 6.7-fold increase of the IC50 with respect to the IC50 determined for the WT HIV-1NL4–3 control virus. NIH #12233 and NIH #12239 clones showed a 3.38- and 3.85-fold increase of the IC50, respectively. The Raltegravir-resistant viruses and the protease inhibitor-resistant viruses were also sensitive to NBD-14273, as demonstrated by their low IC50 values. In conclusion, NBD-14273 was effective against all the drug-resistant viruses tested in this work, indicating that this compound could potentially be used in combination with other antiviral agents. To explain the low sensitivity of the HIV-1 clones, NIH # 12243, NIH #12233, and NIH #12239 to NBD-14273 will require further investigation.
Table 4.
Inhibitory activity of NBD-14273 against a large panel of FDA approved drug-resistant viruses
| NIH catalog# | Major mutationsa | NBD-14273 IC50 (μM)b | Fold increase/Se nsitive | |
|---|---|---|---|---|
| NL4–3 WT (wild-type) | #114 | - | 0.21±0.04 | - |
| ENTRY (Enfuvirtide)-resistant | #9498 | V38A, N42T | 0.4±0.08 | 1.9 |
| #9490 | V38A | 0.44±0.02 | 2.09 | |
| #9496 | V38E, N42S | 0.31±0.03 | 1.48 | |
| #9491 | N42T, N43K | 0.62±0.07 | 2.95 | |
| #9489 | D36G | 0.34±0.06 | 1.62 | |
| Multi-drug (NNRTI)- resistant | #12227 | K101P, K103N | 0.11±0.006 | Sensitive |
| #12229 | L100I, K103N | 0.39±0.4 | 1.86 | |
| #12231 | K103N, Y181C | 0.12±0.006 | Sensitive | |
| #12233 | K101E, Y181V | 0.71±0.02 | 3.38 | |
| #12237 | Y181C, G190A | 0.3±0.02 | 1.4 | |
| #12239 | K101E, E138K, Y181C | 0.81±0.08 | 3.85 | |
| #12241 | K101E, G190S | 0.26±0.01 | Sensitive | |
| #12243 | L100I, M230L | 1.4±0.05 | 6.7 | |
| Integrase (Raltegravir-resistant) | #11847 | G140S, Q148H | 0.17±0.02 | Sensitive |
| #11850 | E92Q, N155H | 0.11±0.1 | Sensitive | |
| #11851 | N155H | 0.12±0.01 | Sensitive | |
| #11800 | 11I, 32I, 33F, 46I, 47V, 54M, 58E, 73S, 84V, 89V, 90M | 0.1±0.01 | Sensitive | |
| #11801 | 10F, 33F, 43T, 46L, 54V, 82A, 84V, 90M | 0.12±0.02 | Sensitive | |
| #11803 | 33F, 43T, 46I, 48V, 50V, 54S, 82A | 0.38±0.1 | 1.8 | |
| #11804 | 32I, 46I, 47V, 84V | 0.24±0.02 | Sensitive | |
| #11805 | 48V, 53L, 54V, 82A, 90M | 0.16±0.05 | Sensitive | |
| #11807 | 32I, 33F, 47A, 82A, 90M | 0.12±0.03 | Sensitive | |
| #11808 | 10F, 11I, 33F, 43T, 46L, 54V, 73S, 82A, 84V, 89V, 90M | 0.17±0.03 | Sensitive | |
| #12465 | 46I, 54V, 58E, 74P, 82L, 90M | 0.13±0.02 | Sensitive | |
| #12466 | 32I, 33F, 43T, 46I, 47V, 54M, 73S, 82A, 89V, 90M | 0.11±0.02 | Sensitive | |
| #12467 | 0.15±0.07 | Sensitive |
Mutants were reported here as per the data obtained from https://www.aidsreagent.org/ and associated references indicated in the Experimental Section.
The reported IC50 values represent the means ± standard deviations (n = 3).
2.5. gp120 entry-antagonists inhibited cell-to-cell HIV-1 transmission
Our earlier generation of gp120 entry-antagonists displays another critical feature: the ability to inhibit cell-to-cell HIV-1 transmission25, 32. Cell-to-cell HIV-1 infection has been reported to be more efficient than HIV-1 cell-free infection34, 35 as multiple viral particles can be transmitted at the same time. Additionally, this type of infection is resistant to some potent broadly neutralizing antibodies (bNAbs), including CD4 binding site (CD4bs) antibodies36–38 and NRTIs, but not to other antiretrovirals, including entry inhibitors, NNRTIs, and protease inhibitors39. Here, we evaluated the activity of NBD-14273 in the cell-to-cell HIV-1 transmission setting and compared its activity to that of NBD-14136, which has previously been reported to inhibit cell-to-cell HIV-1 transmission25, 32. We used TZM-bl cells as acceptor cells and H9 cells chronically infected with HIV-1IIIB (CXCR4-tropic), and MOLT-4 cells chronically infected with HIV-1ADA (CCR5-tropic) as donor cells. BMS-626529 was used as a control treatment drug. Our results (Table 5) indicated that all the compounds tested in this assay, including BMS-626529, showed better activity against the CXCR4-tropic virus HIV-1IIIB than against the CCR5-tropic virus HIV-1ADA. Furthermore, NBD-14273 had slightly improved activity with respect to that we detected for NBD-14136. Of note, the IC50 value for NBD-14273 calculated against HIV-1ADA in the CCR5-tropic assay was 1-fold lower than the IC50 values calculated for NBD-14136. When we compared the cell-cell HIV-1 transmission inhibition of NBD-14273 with our earlier reported most active inhibitor NBD-14189, it showed somewhat lower activity25.
Table 5.
Inhibitory activity against Cell-to-Cell HIV transmission by NBD-14273.
| Compound | IC50 (μM)a | |
|---|---|---|
| TZM-bl/H9-HIV-1IIIB | TZM-bl/Molt-HIV-1ADA | |
| NBD-14136b | 0.46±0.2 | 0.89±0.14 |
| NBD-14273 | 0.39±0.02 | 0.45±0.01 |
| BMS-626529 | 0.02±0.005 | ~0.2 |
The reported IC50 values represent the means ± standard deviation (SD), n=3.
Data previously published32
3. Conclusion
In summary, we have expanded the SAR of NBD-14136 generated through the “CH2OH” positional switch hypothesis reported earlier. A systematic medicinal chemistry optimization and SAR study yielded several new compounds with improved antiviral potency and SI. We evaluated one of the best inhibitors, NBD-14273, against a large panel of 50 HIV-1 Env-pseudotyped viruses representing clinical isolates of diverse subtypes. The data against this set of clinical isolates indicated an improvement of antiviral potency and selectivity index by ~3-fold. NBD-14273 also showed antiviral activity against a large panel of FDA-approved-drug-resistant HIV viruses. The detailed data is expected to pave the way to optimize this series further to a clinically relevant candidate.
4. Experiment
4.1. Cells and viruses
TZM-bl cells41, U87CD4+CXCR4+ cells42, HIV-1 IIIB, infected H9 Cells43, and MOLT-4 CCR5+ Cells44 were obtained through the NIH ARP. HEK 293T cells were purchased from ATCC. CD4-negative Cf2Th-CCR5+ cells and Env expression vector pSVIIIenv-ADA were kindly provided by Dr. J. G. Sodroski45. HIV-1 Env molecular clone expression vector pHXB2-env (X4) DNA was also obtained through the NIH ARP46. HIV-1 Env molecular clones of gp160 genes for HIV-1 Env pseudovirus production were obtained as follows: clones representing the standard panels A (Q259ENV.W6, QB726.70M.ENV.C4, QF495.23M.ENV.A1, QF495.23M.ENV.A3), A/D, A2/D, D, and C (QB099.391M.Env.B1) were obtained through the NIH ARP from Dr. J. Overbaugh47, 48. The HIV-1 Env molecular clones panel of subtype A/G, A/E, and G Env clones were obtained through the NIH ARP from Drs. D. Ellenberger, B. Li, M. Callahan, and S. Butera49. The HIV-1 Env panel of standard reference subtype B Env clones was obtained through the NIH ARP from Drs. D. Montefiori, F. Gao and M. Li (PVO, clone 4 (SVPB11) TRO, Clone 11 (SVPB12), QH0692, clone 42 (SVPB6), SC422661, clone B (SVPB8)); from Drs. B. H. Hahn and J. F. Salazar-Gonzalez (pRHPA4259, clone 7 (SVPB14)); from Drs. B. H. Hahn and D. L. Kothe (pTHRO4156 clone 18 (SVPB15), pCAAN5342 clone A2 (SVPB19))50, 51. The subtype B clones pWEAUd15.410.5017, p1058_11.B11.1550, p1054.TC4.1499, p1006_11.C3.1601, p1056.TA11.1826 and p9021_14.B2.4571 were obtained through the NIH ARP from Drs. B. H. Hahn, B. F. Keele, and G. M. Shaw52. The subtype C HIV-1 reference panel of Env clones were also obtained through the NIH ARP from Drs. D. Montefiori, F. Gao, S. A. Karim, and G. Ramjee (Du172.17); from Drs. D. Montefiori, F. Gao, C. Williamson, and S. A. Karim (Du422.1), from Drs. B. H. Hahn, Y. Li, and J. F. Salazar-Gonzalez (ZM197M.PB7; ZM214M.PL15, ZM249M.PL1); from Drs. E. Hunter and C. Derdeyn (ZM53M.PB12; ZM109F.PB4); from Drs. L. Morris, K. Mlisana and D. Montefiori, (CAP210.2.00.E8)53–55. The HIV-1 Subtype C Panel of Indian gp160 Env Clones HIV-16055–2 clone 3, HIV-16936–2 clone 21, HIV-25711–2 clone 4, and HIV-225925–2 clone 22 were obtained through the NIH ARP from Drs. R. Paranjape, S. Kulkarni and D. Montefiori49. The panel of paired Infant and maternal HIV-1 Env Molecular Clones were obtained through the NIH ARP from Dr. J. Overbaugh56. The Env pseudotyped genes of BG505.T332N, KNH1144, and B41 were kindly provided by Dr. J. P. Moore of the Weil Cornell Medical College, NY.
The Env-deleted proviral backbone plasmids pNL4–3.Luc.R-.E-DNA (from Dr. N. Landau)57, 58, the pSG3Δenv DNA (from Drs. J. C. Kappes and X. Wu)41, 51, and the pNL4–3 (from Dr. Malcolm Martin)59 were obtained through the NIH ARP Division of AIDS, NIAID, NIH. The following molecular clones were also obtained through the NIH ARP, Division of AIDS, NIAID, NIH: the panel of Enfuvirtide (T-20) resistant viruses from Trimeris, Inc.59, 60; the panel Multi-Drug Resistant NNRTI Infectious Clones from Dr. R. Shafer61; the Raltegravir-resistant infectious molecular clones from Dr. R. Shafer and E. Reuman, M.S62 and the multi-drug Protease inhibitor-resistant infectious clones from Dr. R. Shafer63.
MLV gag-pol-expressing vector pVPack-GP, Env-expressing vector pVPack-VSV-G, and a pFB-Luc vector were obtained from Stratagene (La Jolla, CA).
4.2. Pseudovirus preparation
Pseudoviruses capable of single-cycle infection were prepared as previously described13, 15. Briefly, 5 × 106 HEK293T cells were transfected with an HIV-1 Env-expression plasmid and either the HIV-1 Env-deleted pro-viral backbone plasmid pSG3Δenv or the pNL4–3.Luc.R-.E-DNA, by using FuGENE6 (Promega). The control VSV-G pseudovirus was prepared by transfecting the HEK293T cells with a combination of the Env-expressing plasmid pVPack-VSV-G, the MLV gag-pol-expressing plasmid pVPack-GP, the pFB-Luc plasmid by using FuGENE6. Pseudovirus-containing supernatants were collected two days after transfection, filtered, tittered, and stored in aliquots at −80 °C.
4.3. Measurement of antiviral activity
4.3.1. Single-cycle infection assay in TZM-bl cells
The antiviral activity of the new generation of gp120 entry-antagonists was evaluated in a single-cycle infection assay by infecting TZM-bl cells with HIV-1 pseudotyped with the Env from the lab-adapted CXCR4-tropic HIV-1HXB-2. Additionally, NBD-14136 and NBD-14273 were evaluated against a large group of HIV-1 pseudotyped with the Env from the panel of clinical isolates as previously described13, 15. Briefly, TZM-bl cells were plated at 1 × 104 / well in a 96-well tissue culture plate. Following overnight incubation, the cells were infected with aliquots of HIV-1 pseudovirus pre-treated with graded concentrations of the small molecules for 30 min. On the third day post-infection, the cells were washed and lysed with 50 μl of lysis buffer (Promega). 20 μl of the lysates were transferred to a white plate and mixed with the luciferase assay reagent (Promega). The luciferase activity was measured immediately with a Tecan Spark reader, and the percent inhibition by the compounds and the IC50 (the half-maximal inhibitory concentration) values were calculated using the GraphPad Prism software.
4.3.2. Single-cycle infection assay in U87-CD4-CXCR4 cells
The antiviral activity of NBD-14136 and NBD-14273 was tested against the control pseudovirus VSV-G in U87-CD4-CXCR4 cells. Briefly, U87-CD4-CXCR4 cells were plated in a 96-well tissue culture plate at 1 × 104/well and cultured at 37 °C. Following overnight incubation, the cells were infected with aliquots of pseudovirus pre-treated with graded concentrations of the small molecules for 30 min. On the third day, post-infection, the cells were washed and lysed with 40 μl of lysis buffer. The lysates were then transferred to a white plate and mixed with the luciferase assay reagent. The luciferase activity was immediately measured to calculate the percent of inhibition and IC50 values by using the GraphPad Prism software.
4.3.3. Assay in Cf2Th-CCR5 cells
CD4-negative Cf2Th-CCR5 cells were plated at 6 × 103 cells/well in a 96-well tissue culture plate and incubated at 37 °C. The following day, the cells were infected with the recombinant CD4-dependent pseudovirus HIV-1ADA as previously described11. Briefly, aliquots of HIV-1ADA pseudovirus pre-treated with graded concentrations of NBD-14273 and NBD-556 for 30 min were added to the cells and cultured for 48 h. The cells were then washed with PBS and lysed with 40 μl of cell lysis reagent. The lysates were mixed with the luciferase assay reagent, and the luciferase activity was measured. For each compound, we calculated the relative infection compared to the untreated control. The Relative virus infectivity indicates the ratio of the amount of infection detected in the presence of the compounds and the amount of infection detected in the absence of the compounds.
4.3.4. Measurement of antiviral activity against drug-resistant viruses in TZM-bl cells
We evaluated the antiviral activity of NBD-14273 against a panel of drug-resistant viruses by infecting TZM-bl cells. Briefly, TZM-bl cells were plated at 104/well in a 96 well plate and cultured overnight. On the following day, the HIV-1 drug-resistant viruses were pre-treated with graded concentrations of the small molecules for 30 min and added to the cells. Following 48 h incubation, the cells were washed and lysed. The cellular lysates were mixed with the luciferase substrate, and the luciferase activity was measured to calculate the percent inhibition by the compounds and the IC50 values using the GraphPad Prism software, as reported above.
4.4. Evaluation of cytotoxicity
4.4.1. TZM-bl cells and U87-CD4-CXCR4 cells
The cytotoxicity of the gp120 entry-inhibitors in TZM-bl and U87-CD4-CXCR4 cells was measured by using the colorimetric CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) (Promega) following the manufacturer’s instructions. Briefly, the cells were seeded in a 96-well plate and cultured overnight at 37 °C. The following day the cells were cultured with 100 μl of the compounds at graded concentrations and incubated for three days. The MTS reagent was added to the cells and incubated for 4 h at 37 °C. The absorbance was recorded at 490 nm. The percent of cytotoxicity and the CC50 (the concentration for 50 % cytotoxicity) values were calculated as above.
4.4.2. Cf2Th-CCR5 cells
The cytotoxicity of the small molecules in CD4-negative Cf2Th-CCR5 cells was also measured with the colorimetric assay, as above. Briefly, Cf2Th-CCR5 cells were plated in a 96-well plate and cultured at 37 °C. Following overnight incubation, the cells were cultured with 100 μl of the compounds at graded concentrations and cultured for 48 h. The MTS reagent was added to the cells, and 4 h later, the absorbance was recorded at 490 nm. The percent of cytotoxicity and the CC50 values were calculated as above.
4.5. Cell-to-Cell HIV-1 Transmission
The cell-to-cell HIV-1 transmission inhibition assay was performed as previously described64, 65, with minor modifications. Briefly, the indicator TZM-bl cells (used as ‘acceptor’ cells) were plated at 104/well in a 96 well plate 24 h before the assay. For the CXCR4-tropic assay, as ‘transmitting’ cells, we used H9 cells chronically infected with HIV-1IIIB at 2 × 103 cells/well. For the CCR5-tropic assay, we used MOLT-4/CCR5 cells chronically infected with HIV-1ADA at 2 × 103 cells/well. The transmitting cells were pre-treated with 200 μg/mL mitomycin C (Sigma) for 1 h at 37 °C, washed and incubated with the TZM-bl cells in the presence of escalating concentrations of compounds for 24 h. Then, the cells were washed and lysed. The lysates were mixed with the luciferase substrate, and the luciferase activity was measured to calculate the percent of inhibition and IC50 values by using the GraphPad Prism software.
4.6. Chemistry
For this study, all commercial reagents and solvents were used without further purification. Unless otherwise stated, all the described reactions were performed in an air atmosphere. Reactions were monitored by thin-layer chromatography (TLC) carried out on Merck TLC Silica gel plates (60 F254), using a UV light for visualization and basic aqueous potassium permanganate or iodine fumes as a developing agent. 1H and 13C NMR spectra were recorded on a Bruker Avance 400 instrument with an operating frequency of 400 and 100 MHz, respectively, and calibrated using residual not-deuterated chloroform (δH = 7.28 ppm) and CDCl3 (δC = 77.16 ppm) or not-deuterated DMSO (δH = 2.50 ppm) and DMSO-d6 (δC = 39.51 ppm) as internal references. NMR data spectra abbreviations: s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, br = broad. The purity of compounds was determined via LCMS (Shimadzu LCMS-2010A) using three types of detection systems such as EDAD, ELSD, and UV with purity cut-off at ≥95%.
General procedure A: for Suzuki coupling
To a solution containing appropriate bromide (50 mmol, 1 equiv.), (1-(tert-butoxycarbonyl)-1H-pyrrol-2-yl)boronic acid (50 mmol, 1 equiv.) in THF-H2O (1:1, 100 mL), Na2CO3 (100 mmol, 2 equiv.) and Pd(Ph3P)Cl2 (1 mol. %) were added under a nitrogen atmosphere. The mixture was stirred at reflux for 8–15 h (TLC-control). After cooling to room temperature, water (50 mL) and CH2Cl2 (50 mL) were added. The organic layer was separated; the aqueous layer was extracted with CH2Cl2 (3×50 mL). The combined organic layers were dried over Na2SO4, filtered, and concentrated. Purification by flash chromatography using hexane-EtOAc mixture as eluent afforded the desired compound. Compound S1a–c were obtained following the general procedure A (Scheme 1).
Scheme 1.
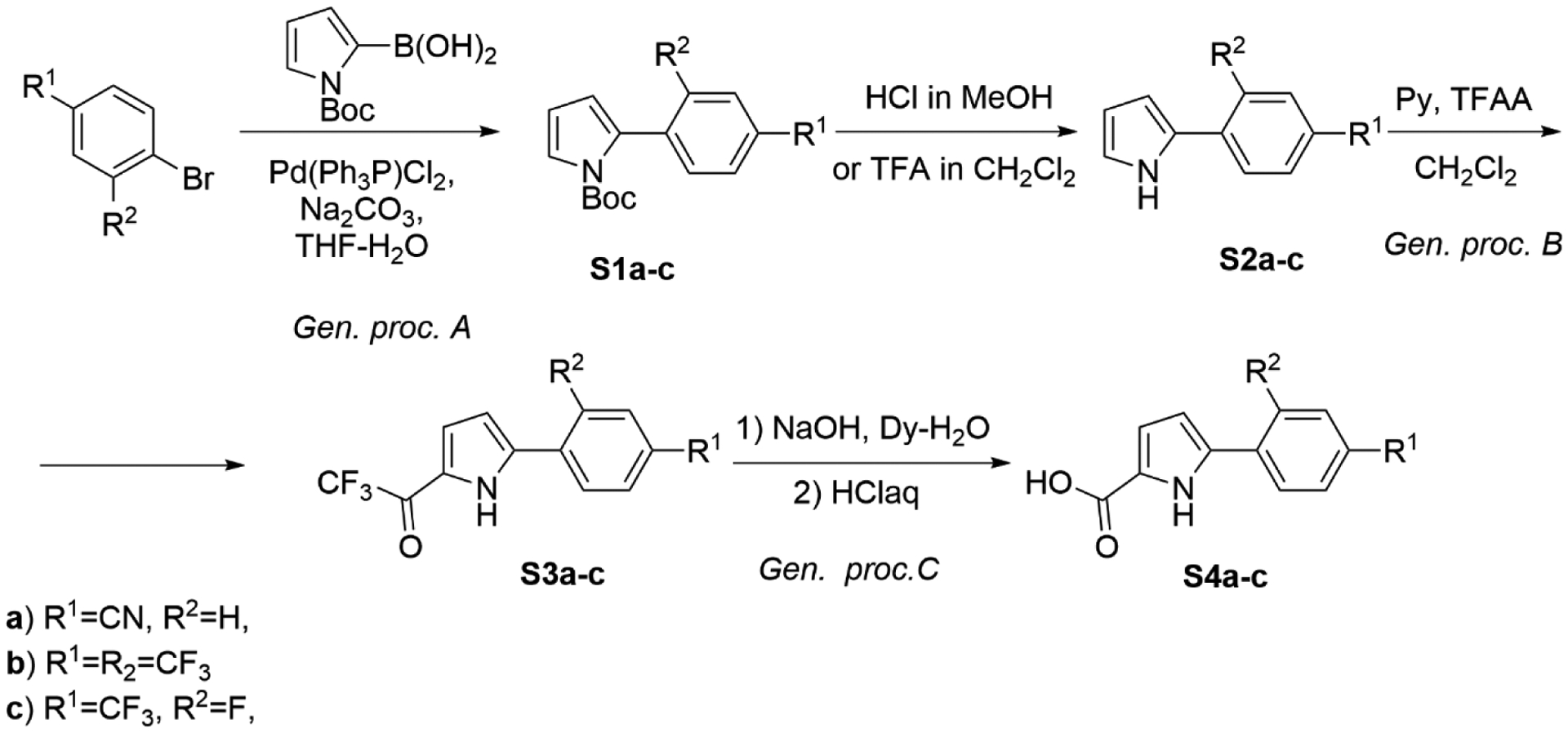
Synthesis of S4a–c.
Tert-butyl 2-(4-cyanophenyl)-1H-pyrrole-1-carboxylate (S1a)
Eluent: Hexane-EtOAc (from 20:1 to 10:1), Rf=0.3 (10:1, Hexane-EtOAc). Yield = 53%.
1H NMR (CDCl3, 400 MHz): δ = 1.43 (s, 9 H), 6.25 – 6.30 (m, 2 H), 7.40 (dd, J=3.2, 1.9 Hz, 1 H), 7.45 – 7.48 (m, 2 H), 7.62 – 7.66 (m, 2 H).
13C NMR (CDCl3, 100 MHz): δ = 27.6 (3 C), 84.3, 110.4, 111.0, 116.1, 118.9, 123.9, 129.5 (2 C), 131.3 (2 C), 133.0, 138.7, 148.9.
Tert-butyl 2-(2,4-bis(trifluoromethyl)phenyl)-1H-pyrrole-1-carboxylate (S1b)
Eluent: Hexane-EtOAc (from 1:0 to 20:1), Rf=0.3–0.4 (20:1, Hexane-EtOAc). Yield = 56%.
1H NMR (CDCl3, 400 MHz): δ = 1.25 (s, 9 H), 6.20 – 6.24 (m, 1 H), 6.28 (t, J=3.3 Hz, 1 H), 7.43 (dd, J=3.4, 1.8 Hz, 1 H), 7.53 (d, J=7.9 Hz, 1 H), 7.81 (d, J=7.4 Hz, 1 H), 7.97 (s, 1 H).
13C NMR (CDCl3, 100 MHz): δ = 27.4 (3 C), 84.0, 110.7, 115.9, 122.4, 123.3 (q, J=274.0 Hz), 123.1 (m), 123.6 (q, J=272.2 Hz), 127.8 (q, J=3.7 Hz), 127.8, 130.6 (q, J=33.5 Hz), 130.8 (q, J=30.6 Hz), 133.6, 138.0, 148.8.
Tert-butyl 2-(2-fluoro-4-(trifluoromethyl)phenyl)-1H-pyrrole-1-carboxylate (S1c)
Eluent: Hexane-EtOAc (from 30:1 to 20:1), Rf=0.5 (20:1, Hexane-EtOAc). Yield = 70%.
1H NMR (CDCl3, 400 MHz): δ = 1.41 (s, 9 H), 6.28 – 6.33 (m, 2 H), 7.30 – 7.38 (m, 1 H), 7.42 – 7.47 (m, 2 H), 7.46 – 7.52 (m, 1 H).
13C NMR (CDCl3, 100 MHz): δ = 27.6, 84.3, 111.0, 112.5 (dq, J=25.5, 3.7 Hz), 116.2, 120.8 (dq, J=7.6, 3.7 Hz), 123.5 (dq, J=272.2, 2.6 Hz), 123.6, 126.8 (d, J=14.9 Hz), 126.8, 131.3 (d, J=3.3 Hz), 131.4 (qd, J=33.2, 7.9 Hz), 149.0, 160.0 (d, J=249.2 Hz).
4-(1H-Pyrrol-2-yl)benzonitrile (S2a)
To a solution containing S1a (10 g, 1 equiv.) in MeOH (20 mL), 1M HCl solution in MeOH (100 ml) was added in one portion. The mixture was stirred at room temperature for 7–8 h, and then the solvent was evaporated. Aqueous K2CO3 (10% solution, 100 mL) was added carefully (CO2 evolution), and the mixture was extracted with CH2Cl2 (3×50 mL). The combined organic layers were dried over Na2SO4, filtered, and concentrated. The crude product was used in the next step without purification. M = 6.01 g. Yield = 96%.
1H NMR (CDCl3, 400 MHz): δ = 6.36 (dt, J=3.5, 2.6 Hz, 1 H), 6.69 (ddd, J=J=3.7, 2.6, 1.4 Hz, 1 H), 6.97 (td, J=2.7, 1.5 Hz, 1 H), 7.53 – 7.57 (m, 2 H), 7.61 – 7.65 (m, 2 H), 8.71 (br. s., 1 H).
13C NMR (CDCl3, 100 MHz): δ = 108.7, 108.9, 111.1, 119.4, 121.1, 123.8 (2 C), 130.2, 132.9 (2 C), 136.9.
2-(2,4-Bis(trifluoromethyl)phenyl)-1H-pyrrole (S2b)
To a solution containing S1b (10 g, 1 equiv.) in CH2Cl2 (130 mL), TFA (10.1 mL, 15.03 g, 132 mmol, 5 equiv.) was added dropwise on the water bath. The mixture was stirred at room temperature overnight, and then the solvent was evaporated. Aqueous K2CO3 (10% solution, 100 mL) was added carefully (CO2 evolution), and the mixture was extracted with CH2Cl2 (3×50 mL). The combined organic layers were dried over Na2SO4, filtered, and concentrated. Purification by flash chromatography using hexane-EtOAc mixture (from 30:1 to 10:1) as eluent (Rf = 0.2 in hexane-EtOAc 10:1). M = 6.34 g. Yield = 86%
1H NMR (CDCl3, 400 MHz): δ = 6.38 – 6.42 (m, 1 H), 6.57 – 6.61 (m, 1 H), 6.99 – 7.04 (m, 1 H), 7.73 (d, J=8.2 Hz, 1 H), 7.83 (d, J=8.3 Hz, 1 H), 8.04 (s, 1 H), 8.62 (br. s., 1 H).
13C NMR (CDCl3, 100 MHz): δ = 110.1, 111.7, 121.0, 123.7 (q, J=272.0 Hz), 123.9 (q, J=273.3 Hz), 124.1 (m), 126.7 (q, J=31.1 Hz), 127.7, 128.8, (q, J=4.0 Hz) 128.9 (q, J=33.5 Hz), 131.9, 136.0.
2-(2-Fluoro-4-(trifluoromethyl)phenyl)-1H-pyrrole (S2c)
S2c was synthesized by the same experimental procedures of S2b.
Eluent: Hexane-EtOAc (from 30:1 to 20:1), Rf=0.4 (20:1, Hexane-EtOAc). Yield = 93%.
1H NMR (DMSO-d6, 400 MHz): δ = 6.18 – 6.27 (m, 1 H), 6.72 (dd, J=2.29, 1.25 Hz, 1 H), 6.96 – 7.06 (m, 1 H), 7.56 (d, J=8.25 Hz, 1 H), 7.63 (d, J=11.86 Hz, 1 H), 7.95 (t, J=8.04 Hz, 1 H), 11.54 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 109.7 (d, J=1.7 Hz), 111.4 (d, J=10.0 Hz), 113.6 (dq, J=26.2, 3.9 Hz), 121.4 (d, J=0.9 Hz), 121.6 (dq, J=7.4, 3.7 Hz), 123.5 (d, J=2.8 Hz), 123.6 (qd, J=271.6, 2.4 Hz), 124.7 (d, J=12.0 Hz), 126.5 (qd, J=33.0, 8.3 Hz), 126.6 (d, J=4.4 Hz), 157.3 (d, J=249.0 Hz).
General procedure B: for acylation
Crude pyrrole from the previous step (1 equiv.) was dissolved in CH2Cl2 (0.5 M solution), and pyridine (1.2 equiv.) was added, followed by the dropwise addition of TFAA (1.2 equiv.). After completion of the addition, the mixture was stirred for 2 h, and the solvent was evaporated. The product was triturated in water, and the precipitate was filtered, washed with water twice, and dried on a filter. Compound S3a–c were obtained following the general procedure B (Scheme 1).
4-(5-(2,2,2-Trifluoroacetyl)-1H-pyrrol-2-yl)benzonitrile (S3a)
Yield = 79%.
1H NMR (DMSO-d6, 400 MHz): δ = 7.04 (dd, J=4.2, 2.4 Hz, 1 H), 7.18 – 7.34 (m, 1 H), 7.87 (d, J=8.4 Hz, 2 H), 8.14 (d, J=8.4 Hz, 2 H), 13.06 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 111.0, 112.3, 117.0 (q, J=289.9 Hz), 118.7, 123.0 (q, J=3.7 Hz), 126.8 (2 C), 126.9, 132.8 (2 C), 134.0, 141.1, 168.3 (q, J=35.2 Hz).
1-(5-(2,4-Bis(trifluoromethyl)phenyl)-1H-pyrrol-2-yl)-2,2,2-trifluoroethanone (S3b)
Yield = 85%.
1H NMR (DMSO-d6, 400 MHz): δ = 6.56 – 6.62 (m, 1 H), 7.31 (dt, J=4.1, 2.1 Hz, 1 H), 7.91 (d, J=8.6 Hz, 1 H), 8.14 – 8.26 (m, 2 H), 13.17 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 114.1 (q, J=2.4 Hz), 116.9 (q, J=289.9 Hz), 121.7 (q, J=3.3 Hz), 123.0 (q, J=273.9 Hz), 123.3 (qq, J=5.9, 4.0 Hz), 123.3 (q, J=272.6 Hz), 126.1, 128.5 (q, J=31.1 Hz), 129.2 (q, J=3.0 Hz), 130.0 (q, J=33.2 Hz), 134.0, 134.1, 138.4, 168.8 (q, J=35.2 Hz).
2,2,2-Trifluoro-1-(5-(2-fluoro-4-(trifluoromethyl)phenyl)-1H-pyrrol-2-yl)ethanone (S3c)
Yield = 92%.
1H NMR (DMSO-d6, 400 MHz): δ = 6.82 (t, J=3.66 Hz, 1 H), 7.13 – 7.36 (m, 1 H), 7.64 (d, J=8.27 Hz, 1 H), 7.73 (d, J=11.13 Hz, 1 H), 8.21 (t, J=7.95 Hz, 1 H), 13.05 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 113.6 (dq, J=26.0, 3.8 Hz), 114.4 (d, J=10.7 Hz), 117.0 (q, J=289.9 Hz), 121.5 (dq, J=7.2, 3.5 Hz), 122.0 (d, J=11.6 Hz), 122.3 (q, J=3.0 Hz), 123.3 (qd, J=277.2, 2.4 Hz), 126.5, 130.1 (d, J=2.6 Hz), 130.7 (qd, J=33.4, 8.5 Hz), 135.4, (d, J=1.8 Hz), 159.0 (d, J=253.0 Hz), 168.7 (q, J=35.2 Hz).
General procedure C: for haloform reaction
Appropriate trifluoroethanone (1 equiv.) was added to a solution of NaOH (5 equiv.) in dioxane-H2O mixture (1:1, 0.5M solution). The resulting reaction mixture was refluxed for 20 h and cooled to room temperature. A concentrated aqueous HCl solution (~12 M, 5 equiv.) was added dropwise. The resulting precipitate was filtered, washed with H2O, and dried on a filter. Compound S4a-c were obtained following the general procedure D (Scheme 1).
5-(4-Cyanophenyl)-1H-pyrrole-2-carboxylic acid (S4a)
Yield = 69%.
1H NMR (DMSO-d6, 400 MHz): δ = 6.75 (dd, J=3.7, 2.5 Hz, 1 H), 6.84 (dd, J=3.8, 2.2 Hz, 1 H), 7.92 (d, J=8.6 Hz, 2 H), 7.97 (d, J=8.6 Hz, 2 H), 12.18 (br. s., 1 H), 12.72 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 109.3, 116.6, 124.9 (2 C), 125.4, 128.9, 129.9 (2 C), 135.4, 135.6, 161.9, 167.2.
5-(2,4-Bis(trifluoromethyl)phenyl)-1H-pyrrole-2-carboxylic acid (S4b)
Yield = 71%.
1H NMR (DMSO-d6, 400 MHz): δ = 6.32 – 6.40 (m, 1 H), 6.83 (dd, J=3.7, 2.4 Hz, 1 H), 7.83 (d, J=8.6 Hz, 1 H), 8.04 – 8.17 (m, 2 H), 12.22 (br. s., 1 H), 12.55 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 111.9 (q, J=3.0 Hz), 115.5, 123.2 (m), 123.4 (q, J=273.9 Hz), 123.6 (q, J=272.4 Hz), 125.3, 127.8 (q, J=30.8 Hz), 128.7 (q, J=33.0 Hz), 128.9, 131.4, 134.0, 135.7, 162.0.
5-(2-Fluoro-4-(trifluoromethyl)phenyl)-1H-pyrrole-2-carboxylic acid (S4c)
Yield = 83%.
1H NMR (DMSO-d6, 400 MHz): δ = 6.65 – 6.72 (m, 1 H), 6.80 – 6.92 (m, 1 H), 7.61 (d, J=8.27 Hz, 1 H), 7.73 (d, J=11.44 Hz, 1 H), 8.20 (t, J=7.95 Hz, 1 H), 12.19 (br. s., 1 H), 12.64 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 112.6 (d, J=10.6 Hz), 113.6 (dq, J=26.2, 3.7 Hz), 116.2, 121.5 (dq, J=6.9, 3.2 Hz), 123.5 (qd, J=272.2, 2.8 Hz), 123.6 (d, J=11.5 Hz), 125.7, 128.7 (d, J=2.3 Hz), 128.7 (qd, J=33.1, 8.7 Hz), 129.1 (d, J=3.2 Hz), 158.4 (d, J=250.9 Hz), 161.9.
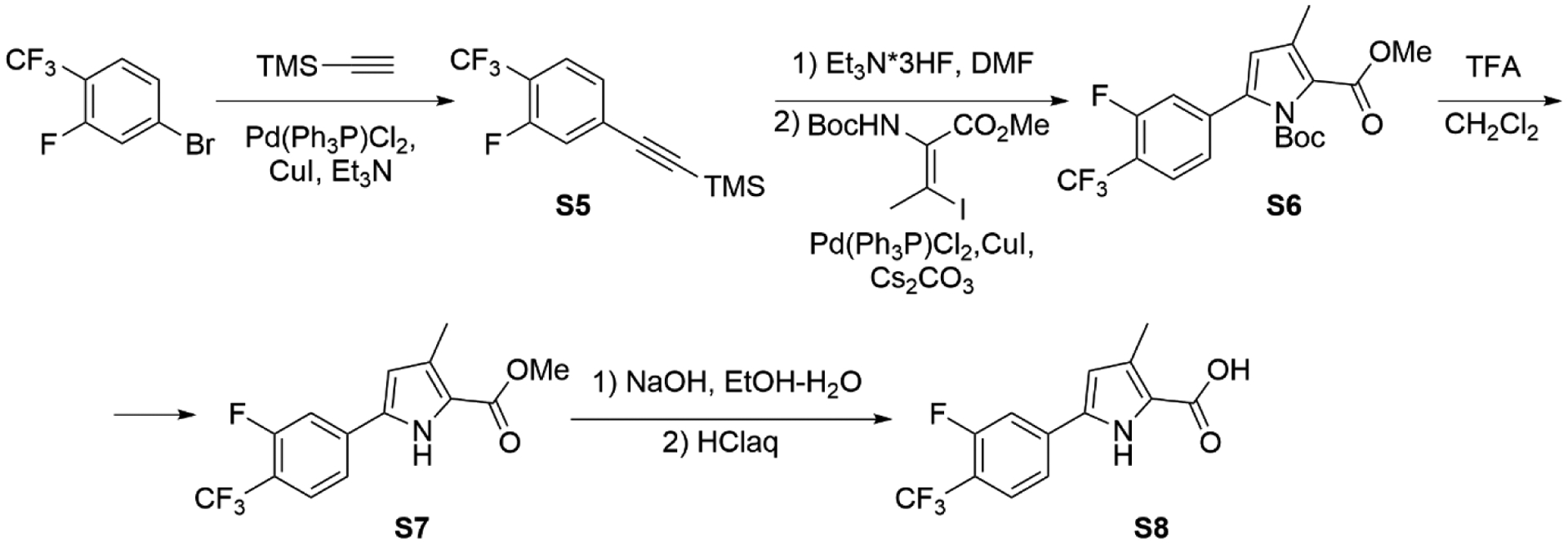
((3-Fluoro-4-(trifluoromethyl)phenyl)ethynyl)trimethylsilane (S5)
We synthesized S5–S8 based on Scheme 2. In a pressurized vessel equipped with magnetic stirring bar containing solution of 4-bromo-2-fluoro-1-(trifluoromethyl)benzene (30 g; 123 mmol, 1 equiv.) in Et3N (250 mL), TMS-acetylene (24.25 g, 34.2 mL, 247 mmol, 2 equiv.), Pd(Ph3P)Cl2 (0.87 g; 1 mol. %), CuI (0.48 g; 2 mol. %) were added under argon atmosphere. It was heated to 50–60 °C, and the mixture was stirred at this temperature for 6–8 h (TLC-control). Water (500 mL) was added and extracted with hexane (3×150 mL). Combined extracts were washed with water and dried with anhydrous sodium sulfate. The solvent was removed by rotary evaporation, and the residue was purified using flash chromatography (eluent: from pure hexane to hexane-EtOAc, 30:1, Rf = 0.6 in hexane). M = 30.1 g. Yield = 94%.
Scheme 2.
Synthesis of S5–S8.
1H NMR (CDCl3, 400 MHz): δ = 0.27 (s, 9 H), 7.28 (d, J=10.8 Hz, 1 H), 7.32 (d, J=8.4 Hz, 1 H), 7.53 (t, J=7.7 Hz, 1 H).
13C NMR (CDCl3, 100 MHz): δ = −0.2, 99.0, 102.1 (d, J=2.6 Hz), 118.5 (qd, J=33.2, 12.5 Hz), 120.1 (d, J=22.1 Hz), 122.5 (q, J=272.7 Hz), 127.2 (qd, J=4.6, 2.2 Hz), 127.8 (d, J=3.7 Hz), 129.4 (d, J=9.8 Hz), 159.5 (dq, J=256.7, 2.0 Hz).
1-tert-Butyl 2-methyl 5-(3-fluoro-4-(trifluoromethyl)phenyl)-3-methyl-1H-pyrrole-1,2-dicarboxylate (S6)
To a solution of S5 (20 g, 77 mmol, 1 equiv.) in DMF (400 mL), triethylamine trihydrofluoride (3.72 g, 3.76 mL, 23 mmol, 0.3 equiv.) was added, and the mixture was stirred for 1 h under argon atmosphere. Then methyl (E)-2-(tert-butoxycarbonylamino)-3-iodo-but-2-enoate (26.2 g, 77 mmol, 1 equiv.), Pd(Ph3P)Cl2 (2.7 g; 5 mol. %), CuI (1.46 g; 10 mol. %) and Cs2CO3 (50.06 g; 154 mmol, 2 equiv.) were added under argon atmosphere. It was heated to 70–80 °C, and the mixture was stirred at this temperature for 10–15 h (TLC-control). Water (800 mL) was added and extracted with Et2O (3×200 mL). Combined extracts were washed with water and brine, dried over anhydrous Na2SO4, filtered, and concentrated. Purification by flash chromatography (eluent: hexane-EtOAc from 20:1 to 5:1, Rf = 0.6 in hexane-EtOAc 5:1). M = 12.73 g, Yield = 41%.
1H NMR (CDCl3, 400 MHz): δ = 1.46 (s, 9 H), 2.30 (s, 3 H), 3.89 (s, 3 H), 6.14 (s, 1 H), 7.25 – 7.33 (m, 2 H), 7.61 (t, J=7.7 Hz, 1 H).
13C NMR (CDCl3, 100 MHz): δ = 12.7, 27.4 (3 C), 51.8, 85.8, 115.2, 117.3 (d, J=21.9 Hz), 117.9 (qd, J=33.3, 12.5 Hz), 122.6 (q, J=272.2 Hz), 123.1, 124.6 (d, J=3.7 Hz), 126.9 (qd, J=4.6, 2.0 Hz), 130.0, 135.2 (d, J=1.8 Hz), 138.1 (d, J=8.9 Hz), 149.3, 159.3 (dq, J=256.3, 2.0 Hz), 161.7.
Methyl 5-(3-fluoro-4-(trifluoromethyl)phenyl)-3-methyl-1H-pyrrole-2-carboxylate (S7)
To a solution of S6 (12.58 g, 31 mmol, 1 equiv.) in CH2Cl2 (150 mL), TFA (17.87 g, 12 mL, 157 mmol, 5 equiv.) was added in one portion, and the mixture was stirred overnight. Then the solvent was evaporated, and 10% aqueous K2CO3 was added, and the mixture was extracted with CH2Cl2 (3×100 mL), dried over anhydrous Na2SO4, filtered, and concentrated. The product was used without further purification. M = 9.28 g. Yield = 98 %.
1H NMR (DMSO-d6, 400 MHz): δ = 2.27 (s, 3 H), 3.81 (s, 3 H), 6.72 (d, J=2.6 Hz, 1 H), 7.70 (t, J=8.1 Hz, 1 H), 7.82 (d, J=8.3 Hz, 1 H), 8.02 (d, J=12.9 Hz, 1 H), 11.99 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 12.8, 51.1, 112.5, 112.8 (d, J=22.7 Hz), 114.2 (qd, J= 32.4, 12.4 Hz), 121.0, 121.3, 122.8 (q, J=271.5 Hz), 127.5 (q, J=3.7 Hz), 128.4, 132.3, 137.9 (d, J=9.0 Hz), 159.4 (d, J=251.4 Hz), 161.2.
5-(3-Fluoro-4-(trifluoromethyl)phenyl)-3-methyl-1H-pyrrole-2-carboxylic acid (S8)
To a solution of S7 (9.19 g, 31 mmol, 1 equiv.) in a mixture of EtOH-H2O (1:1, 120 mL), NaOH (2.44 g, 61 mmol, 2 equiv.) was added in one portion, and the reaction mixture was stirred at reflux for 10–12 h (TLC-control). Then the sodium-salt solution was acidified by the addition of an equivalent amount of HCl (12M, 5.08 mL, 2 equiv.), water (100 mL) was added, and the precipitate was filtered. M = 8.4 g. Yield = 96 %.
1H NMR (DMSO-d6, 400 MHz): δ = 2.28 (s, 3 H), 6.72 (s, 1 H), 7.70 (t, J=8.0 Hz, 1 H), 7.83 (d, J=8.1 Hz, 1 H), 8.03 (d, J=12.9 Hz, 1 H), 11.91 (br. s., 1 H), 12.53 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 12.8, 112.5, 112.6 (d, J=22.7 Hz), 114.0 (qd, J=32.1, 12.7 Hz), 120.9 (d, J=3.0 Hz), 122.4, 122.9 (q, J=270.1 Hz), 127.6 (q, J=3.5 Hz), 127.8, 131.6 (d, J=1.8 Hz), 138.2 (d, J=9.4 Hz), 159.4 (dq, J=249.2, 2.0 Hz), 162.5.
Compounds S9–S14 were synthesized as per Scheme 3.
Scheme 3.
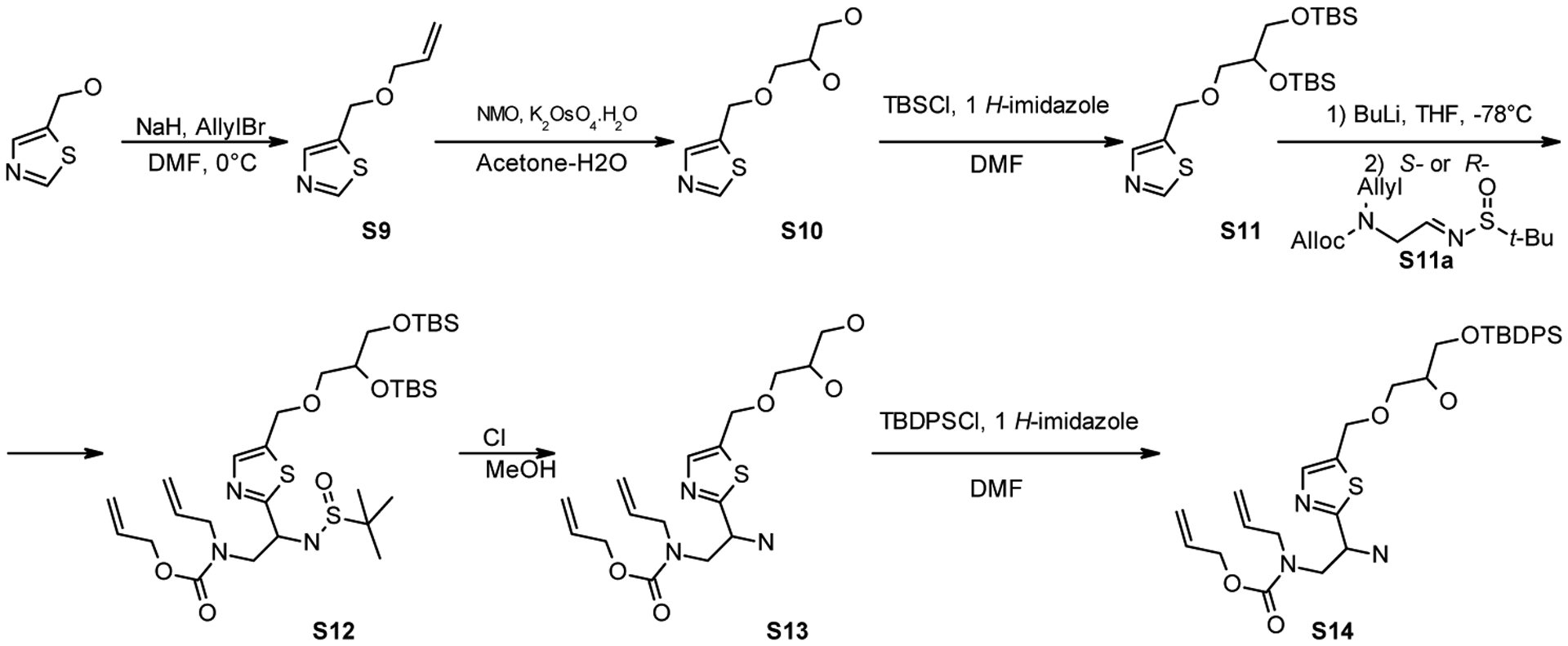
Synthesis of S9–S14 and S21.
5-((Allyloxy)methyl)thiazole (S9)
To a solution of appropriate alcohol (26.1 g, 227 mmol, 1 equiv.) in DMF (226 mL), NaH (40% in mineral oil, 9.97 g, 249 mmol, 1.1 equiv.) was added in several portions at 0 °C. The mixture was stirred until gas evolution stopped. Then allyl bromide (23.5 mL, 32.91 g, 272 mmol, 1.2 equiv.) was added dropwise. The reaction mixture was stirred overnight at room temperature. Water (500 mL) was added and extracted with EtOAc (3×150 mL). Combined extracts were washed with water and brine, dried over Na2SO4, filtered, and concentrated. Purification by distillation under reduced pressure (bp=110–120 °C, 10 torr). M = 28.34 g. Yield = 81%.
1H NMR (CDCl3, 400 MHz): δ = 4.01 (d, J=5.7 Hz, 2 H), 4.71 (s, 2 H), 5.21 (d, J=10.5 Hz, 1 H), 5.29 (dd, J=17.2, 1.6 Hz, 1 H), 5.83 – 5.96 (m, 1 H), 7.77 (s, 1 H), 8.78 (s, 1 H).
13C NMR (CDCl3, 100 MHz): δ = 63.7, 71.0, 117.9, 134.0, 135.6, 142.0, 153.9.
3-(Thiazol-5-ylmethoxy)propane-1,2-diol (S10)
To a solution of alkene S9 (28.34 g, 183 mmol, 1 equiv.) in acetone-water (10:1, 360 mL), NMO monohydrate (37.02 g, 274 mmol, 1.5 equiv.) and potassium osmate (VI) dihydrate (0.67 g, 1 mol. %) were added in that sequence. The mixture was stirred at room temperature for 2–3 h (TLC-control), and then the solvent was evaporated. Crude product was purified by flash chromatography (eluent: from pure EtOAc to CH2Cl2-MeOH 10:1, Rf = 0.3 in EtOAc). M = 32.2 g, Yield = 93%.
1H NMR (CDCl3, 400 MHz): δ = 3.40 – 3.53 (m, 3 H), 3.58 (dd, J=11.4, 3.8 Hz, 1 H), 3.76 – 3.84 (m, 1 H), 4.09 (br. s., 2 H), 4.67 (s, 2 H), 7.70 (s, 1 H), 8.74 (s, 1 H).
13C NMR (CDCl3, 100 MHz): δ = 63.6, 65.0, 70.7, 71.5, 135.4, 141.8, 154.3.
5-((2,3-Bis((tert-butyldimethylsilyl)oxy)propoxy)methyl)thiazole (S11)
To a solution of appropriate diol S10 (32.2 g, 170 mmol, 1 equiv.) DMF (340 mL), imidazole (46.34 g, 681 mmol, 4 equiv.) was added in one portion, followed by portionwise addition of TBSCl (102.57 g, 681 mmol, 4 equiv.). The reaction mixture was stirred overnight at 60–80 °C, cooled to the room temperature, diluted with water (680 mL), and extracted with EtOAc (3 × 200 mL). The combined organic phases were washed with water (200 mL), brine (200 mL), and dried over anhydrous Na2SO4, filtered and evaporated to give an oil, which was purified by flash chromatography (eluent: hexane-EtOAc from 20:1 to 10:1, Rf = 0.4 in hexane-EtOAc 20:1). M = 59.95 g. Yield = 84%.
1H NMR (CDCl3, 400 MHz): δ = 0.03 – 0.08 (m, 12 H), 0.88 (s, 18 H), 3.46 (dd, J=9.9, 5.7 Hz, 1 H), 3.53 – 3.61 (m, 3 H), 3.79 – 3.86 (m, 1 H), 4.77 (s, 2 H), 7.81 (s, 1 H), 8.91 (s, 1 H).
13C NMR (CDCl3, 100 MHz): δ = −5.3, −5.3, −4.6, −4.5, 18.3, 18.4, 25.9 (3 C), 26.0 (3 C), 64.9, 65.4, 72.3, 72.7, 135.9, 141.9, 153.8.
Allyl allyl(2-amino-2-(5-((3-((tert-butyldiphenylsilyl)oxy)-2-hydroxypropoxy)methyl)thiazol-2-yl)ethyl)carbamate (S14)
Experimental procedures are given for the synthesis of R-enantiomer (R):
The thiazole S11 (10.9 g, 26 mmol, 1 equiv.) was dissolved in THF (26 mL) and cooled to −78 °C. At this temperature, n-BuLi (2.5 M in hexane, 10.96 mL, 1.05 equiv.) was added dropwise under a nitrogen atmosphere. The reaction mixture was stirred for 20 min at −78 °C, and appropriate R-imine (S11a: 7.47 g, 26 mmol, 1 equiv.) was added dropwise as a solution in THF (1M, 26 mL). The reaction mixture was slowly (~1 h) warmed to 0 °C and poured into water (150 mL). The organic layer was separated, and water was extracted with CH2Cl2 (3 × 100 mL). Combined organic phases were dried over anhydrous Na2SO4, filtered, and evaporated to give a brown oil which was purified by column chromatography (eluent: hexane-EtOAc, gradient from 5:1 to 0:1, Rf = 0.4 in hexane-EtOAc 1:1). This product (S12) was used without further purification and analysis.
To a solution of protected thiazole from the previous step in MeOH (50 ml), 1M solution of HCl in MeOH (100 ml) was added in one portion. The mixture was stirred for 2–3 h, and the solvent was evaporated. The residue was dissolved in water (50 mL) and extracted with CH2Cl2 (2 × 50 mL). After that, solid K2CO3 was added carefully (CO2 evolution!) to the water phase at pH 10–12. Product was extracted from water with CH2Cl2 (4 × 50 mL), combined organic layers were dried over anhydrous Na2SO4, filtered, and evaporated to give diol S13. M = 3.02 g.
1H NMR (CDCl3, 400 MHz): δ = 2.26 (br. s., 4 H), 3.49 – 3.72 (m, 6 H), 3.77 – 3.98 (m, 3 H), 4.46 (t, J=6.5 Hz, 1 H), 4.60 (d, J=5.3 Hz, 2 H), 4.70 (s, 2 H), 5.08 – 5.24 (m, 3 H), 5.30 (d, J=17.2 Hz, 1 H), 5.70 – 5.97 (m, 2 H), 7.58 (s, 1 H).
Diol S13 (3.02 g, 8 mmol, 1 equiv.) was dissolved in CH2Cl2 (81 mL), and imidazole (0.66 g, 10 mmol, 1.2 equiv.) was added in one portion, followed by dropwise addition of TBDPSCl (2.33 mL, 2.46 g, 9 mmol, 1.1 equiv.). The reaction mixture was stirred overnight, diluted with water (50 mL), and extracted with CH2Cl2 (3 × 50 mL). The combined organic phases were washed with brine and dried over anhydrous Na2SO4. It was then filtered and evaporated to give an oil, which was purified by flash chromatography (eluent: hexane-EtOAc, from 1:1 to 0:1 and then CH2Cl2-MeOH 10:1, Rf = 0.3 in EtOAc). M = 3.75 g. Yield (over three steps) = 24%.
S-enantiomer (S) was synthesized by the same experimental procedures. Yield (over three steps) = 17%.
1H NMR (CDCl3, 400 MHz): δ = 1.06 (s, 9 H), 1.84 (br. s., 2 H), 2.52 (br. s., 1 H), 3.51 – 3.72 (m, 6 H), 3.77 – 4.01 (m, 3 H), 4.45 (t, J=6.4 Hz, 1 H), 4.61 (d, J=5.3 Hz, 2 H), 4.67 (s, 2 H), 5.08 – 5.19 (m, 2 H), 5.21 (dd, J=10.4, 1.2 Hz, 1 H), 5.31 (dd, J=17.2, 1.3 Hz, 1 H), 5.71 – 5.98 (m, 2 H), 7.36 – 7.47 (m, 6 H), 7.57 (s, 1 H), 7.65 (d, J=6.6 Hz, 4 H).
13C NMR (CDCl3, 100 MHz): δ = 19.1, 26.7 (3 C), (50.4, 50.6), (53.1, 53.2), 64.6, 65.3, 66.1, 70.6, 70.9, (116.8, 117.2), (117.3, 117.7), 127.7 (4 C), 129.7 (2 C), 132.7, 133.0 (2 C), 133.1, 135.1, 135.4 (4 C), 141.4, (155.8, 156.7), (175.5, 176.3).
Compounds S15, S16, and S21 were synthesized as per Scheme 4.
Scheme 4.
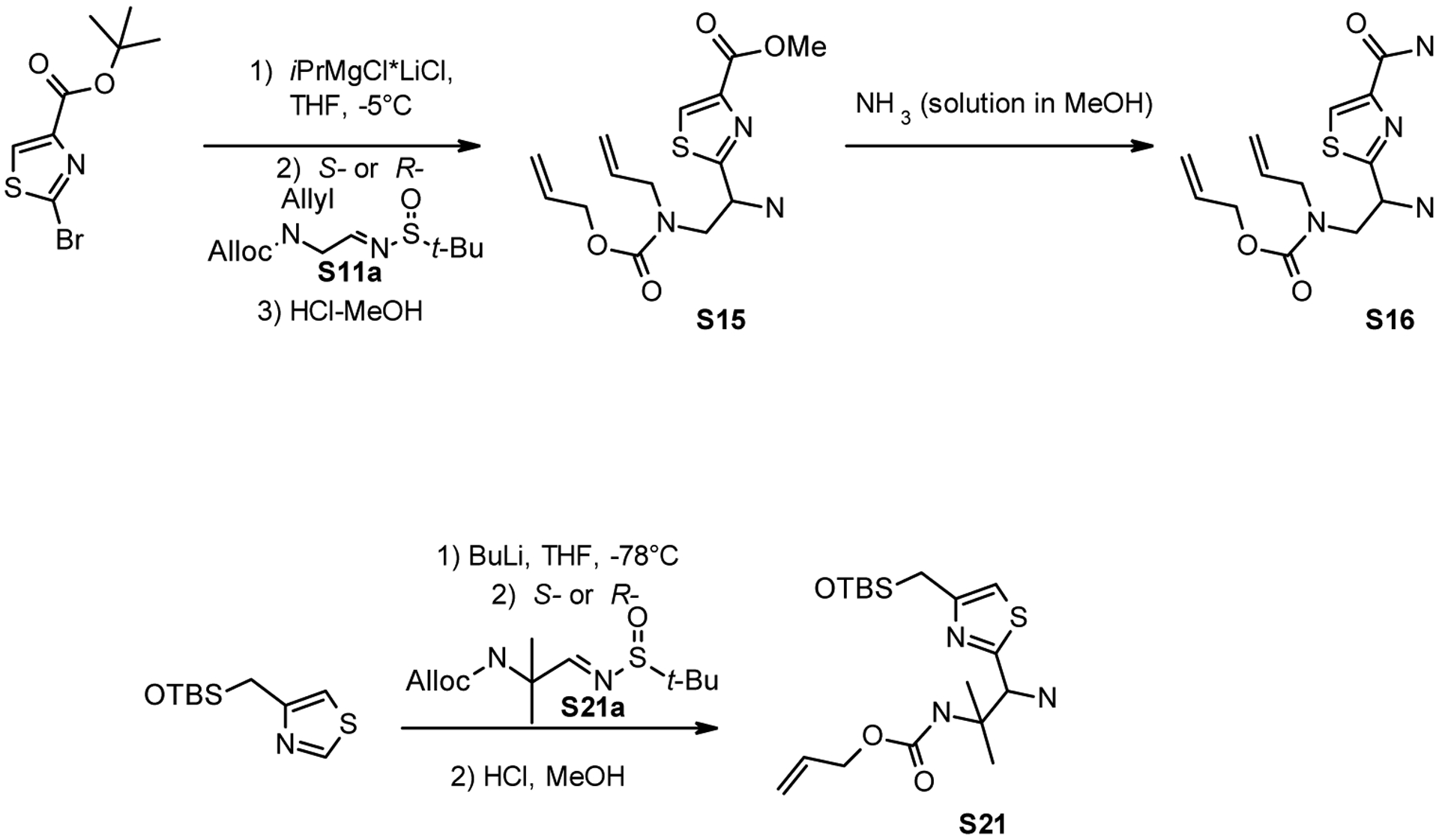
Synthesis of S15, S16 and S21.
Methyl 2-(2-(allyl((allyloxy)carbonyl)amino)-1-aminoethyl)thiazole-4-carboxylate (S15)
A solution of tert-butyl 2-bromothiazole-4-carboxylate (10.0 g, 37.9 mmol, 1 equiv.) and S-imine (S11a) (21.7 g, 75.6 mmol, 2 equiv.) in THF (38 mL) was cooled to 0°C and i-PrMgCl*LiCl solution (1M in tetrahydrofuran, 78 mL, 78 mmol, 2 equiv.) was added maintaining the temperature between 0 and +5 °C. The reaction mixture was then stirred at 0°C for 1h and allowed to warm to room temperature. The reaction was quenched by the addition of aqueous ammonium chloride solution (20% w/w, 0.5 L) and extracted with Et2O (3 × 100 mL). Combined organic layers were washed with brine (3 × 50 mL) dried over Na2SO4 and evaporated to dryness. The residue was dissolved in excess of 1M HCl n methanol (400 mL) and left for 2 days. Methanol was evaporated, and saturated NaHCO3 (100 mL) was added to the residue. The product was extracted with CH2Cl2 (3 × 50 mL). The combined organic layer was dried over anhydrous Na2SO4 and evaporated to dryness. The residue was purified using column chromatography (eluent: hexane-EtOAc from 1:1 to 0:1, and then CH2Cl2-MeOH 10:1, Rf = 0.3–0.4 in EtOAc). M = 7.74 g. Yield = 63%.
Reaction with R-imine (S11a) was performed in the same way. M = 6.22 g. Yield = 51%.
1H NMR (DMSO-d6, 400 MHz): δ = 2.48 (br. s., 2 H), 3.36 – 3.41 (m, 1 H), 3.55 – 3.68 (m, 1 H), 3.81 (s, 3 H), 3.87 (d, J=5.2 Hz, 2 H), 4.32 – 4.54 (m, 3 H), 5.04 – 5.18 (m, 3 H), 5.25 (d, J=17.3 Hz, 1 H), 5.70 – 5.95 (m, 2 H), 8.43 (s, 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 50.0, 51.9, 52.4, (52.7, 53.2), 65.3, (116.0, 116.7), (116.5, 116.9), 129.2, 133.3, (133.8, 134.0), 145.7, (155.1, 155.8), 161.4, (177.9, 178.2).
Allyl allyl(2-amino-2-(4-carbamoylthiazol-2-yl)ethyl)carbamate (S16)
Thiazole S15 (1.5 g, 4.6 mmol) was dissolved in MeOH saturated with NH3 (~7M, 10 mL). It was heated to 70–80 °C in a pressure vessel on an oil bath, and the mixture was stirred at this temperature for 25–30 h (TLC-control). Then it was cooled to the room temperature and evaporated. The product was used without further purification.
1H NMR (CDCl3, 400 MHz): δ = 1.94 (br. s., 2 H), 3.49 – 3.71 (m, 2 H), 3.75 – 3.98 (m, 2 H), 4.42 – 4.63 (m, 3 H), 5.05 – 5.33 (m, 4 H), 5.66 – 5.97 (m, 2 H), 6.28 (br. s., 1 H), 7.15 (br. s., 1 H), 8.07 (s, 1 H).
13C NMR (CDCl3, 100 MHz): δ = 50.3, (52.3, 52.5), (53.4, 52.6), 65.9, (116.6, 117.0), (117.1, 117.4), 123.9, 132.2, 132.7, 149.3, (155.6, 156.4), 163.1, (174.9, 175.3).
We prepared S17–S20 and S22–S28 were prepared by following the methods reported previously25, 27, 32, 67.
Allyl (1-amino-1-(4-(hydroxymethyl)thiazol-2-yl)-2-methylpropan-2-yl)carbamate (S21)
Experimental procedures for the synthesis of R-enantiomer (R):
The 4-(((tert-Butyldimethylsilyl)oxy)methyl)thiazole (20.02 g, 87 mmol, 2.5 equiv.) was dissolved in THF (87 mL) and cooled to −78 °C. At this temperature, n-BuLi (2.5 M in hexane, 34.92 mL, 2.5 equiv.) was added dropwise under a nitrogen atmosphere. The reaction mixture was stirred for 20 min at −78 °C, and appropriate R-imine (S21a) (10.0 g, 35 mmol, 1 equiv.) was added dropwise as a solution in THF (1M, 35 mL). The reaction mixture was slowly (~1 h) warmed to 0 °C and poured into water (200 mL). The organic layer was separated, and water was extracted with CH2Cl2 (4 × 100 mL). Combined organic phases were dried over anhydrous Na2SO4, filtered, and evaporated to give a brown oil which was purified by column chromatography (eluent: hexane-EtOAc, gradient from 5:1 to 0:1, Rf = 0.4 in hexane-EtOAc 1:1). This product was used without analysis.
To a solution of protected thiazole from the previous step in MeOH (50 ml), 1M solution of HCl in MeOH (100 ml) was added in one portion. The mixture was stirred for 2–3 h, and the solvent was evaporated. The residue was dissolved in water (50 mL) and extracted with CH2Cl2 (2 × 50 mL). After that, solid K2CO3 was added carefully (CO2 evolution) to the water phase at pH 10–12. Product was extracted from water with CH2Cl2 (4 × 50 mL), combined organic layers were dried over anhydrous Na2SO4, filtered, and evaporated to give pure amine. M = 3.2 g. Yield = 32%
S-enantiomer (S) was synthesized by the same experimental procedures. Yield = 29%.
1H NMR (CDCl3, 400 MHz): δ = 1.32 (s, 3 H), 1.34 (s, 3 H), 2.49 (br. s., 3 H), 4.45 – 4.56 (m, 3 H), 4.68 (s, 2 H), 5.14 – 5.32 (m, 2 H), 5.59 (s, 1 H), 5.89 (dddd, J=16.8, 11.0, 5.7, 5.4 Hz, 1 H), 7.10 (s, 1 H).
13C NMR (CDCl3, 100 MHz): δ = 23.5, 23.7, 56.0, 59.6, 60.5, 65.1, 114.5, 117.5, 132.9, 155.0, 156.3, 172.6.
General Procedure D: for Amide Coupling
DIPEA (1 equiv.) was added to an appropriate acid (1 equiv.) followed by DMF (10 mL per 1 g of acid) and then HBTU (1 equiv.). The resulting solution was stirred for 10 min and added to a solution of appropriate amine (1 equiv.) in DMF (10 mL per 1 g of amine) in several portions. The reaction mixture was stirred overnight; DMF was evaporated, and the residue was dissolved in CH2Cl2 (50 mL per 1 g of crude product) and successively washed with 5% aqueous NaOH and 10% tartaric acid solutions (25 mL per 1 g of crude product). The organic layer was dried over anhydrous Na2SO4, filtered, evaporated, and dry loaded on silica. Eluting with hexane-EtOAc (1:1, then pure EtOAc) gave the target compounds. Products were used in the next step without analysis.
General Procedure E: for Allyl- and Alloc- Deprotection
To a solution containing protected compound (1 equiv.) and N,N-dimethyl barbituric acid (NDMBA, 3 equiv.) in MeOH (0.1M solution), PPh3 (10 mol %) was added under a nitrogen atmosphere followed by Pd(dba)2 (5 mol %). The mixture was stirred for 1 day under reflux. After cooling, 50 mL of CH2Cl2 was added, and the organic phase was shaken with 10% aqueous K2CO3 (50 mL) to remove the unreacted NDMBA. The organic layer was separated, and the aqueous layer was extracted with CH2Cl2-EtOH (~4:1, (3–5) × 50 mL). Combined organic layers were dried over anhydrous Na2SO4, filtered, and concentrated. Purification by flash chromatography (twice) (eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1 and 5:1, eluent 2: CH2Cl2-MeOH, 4:1, 2:1, 1:1) afforded amine as a slightly yellow or white solid.
General procedure F: for TBDPS- deprotection
TBDPS cleavage: To a solution of TBDPS-protected compound (1 equiv.) in THF (0.1M), solution of TBAF trihydrate (1.1 equiv.) in THF (0.1M) was added in a one portion. The mixture was stirred for 1–2 h at room temperature (TLC-control) and concentrated. Purification by flash chromatography (twice) (eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1, 5:1, 3:1, eluent 2: CH2Cl2-MeOH, 4:1, 3:1, 2:1, 1:1) afforded amine as a slightly yellow or white solid.
Compounds 1–38 were synthesized as per Scheme 5.
Scheme 5.
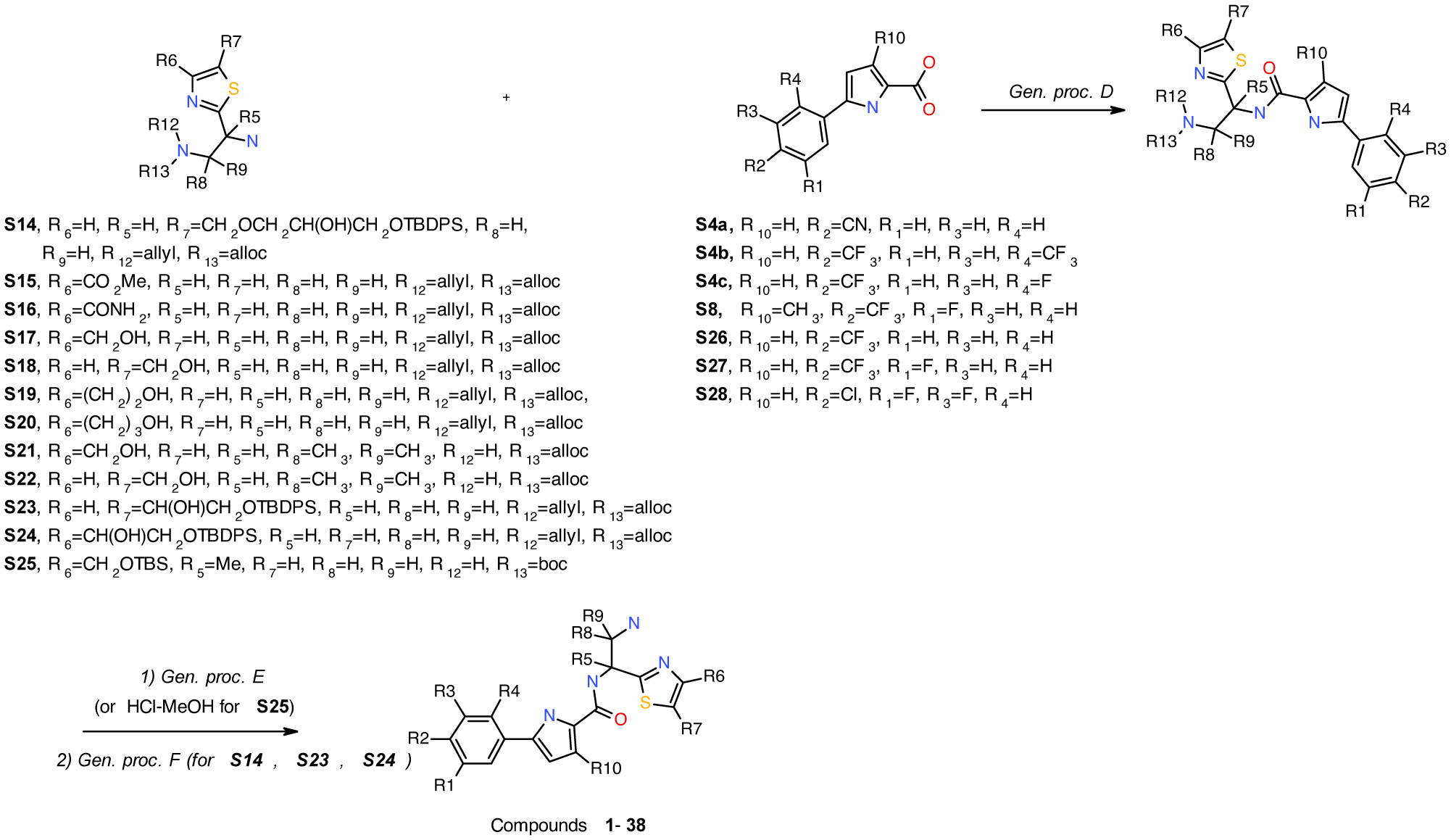
Synthesis of compounds 1–38.
N-(2-amino-1-(4-(hydroxymethyl)thiazol-2-yl)ethyl)-5-(2-fluoro-4-(trifluoromethyl)phenyl)-1H-pyrrole-2-carboxamide
Compounds 1 (NBD-14250) and 2 (NBD-14251) were obtained following the general procedure D, and E from amine S17 and acid S4c Compounds were purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1 and 5:1, Eluent 2: CH2Cl2-MeOH, 4:1 and 2:1.
1 (NBD-14250); (R): M = 501 mg. Yield = 29% (over two steps). rt = 1.317 min. Purity = 100%. LC-MS: m/z [M+H]+ = 429 Da.
2 (NBD-14251); (S): M = 432 mg. Yield = 25% (over two steps). rt = 1.325 min. Purity = 100%. LC-MS: m/z [M+H]+ = 429 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 1.77 (br. s., 2 H), 3.01 (dd, J=13.0, 7.9 Hz, 1 H), 3.14 (dd, J=13.0, 5.1 Hz, 1 H), 4.54 (s, 2 H), 5.18 – 5.27 (m, 1 H), 5.31 (br. s., 1 H), 6.73 (t, J=3.7 Hz, 1 H), 7.09 (d, J=3.8 Hz, 1 H), 7.29 (s, 1 H), 7.62 (d, J=7.6 Hz, 1 H), 7.74 (d, J=11.4 Hz, 1 H), 8.17 (t, J=7.9 Hz, 1 H), 8.73 (d, J=7.6 Hz, 1 H), 11.95 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 45.7, 54.4, 59.8, 112.2 (d, J=10.1 Hz), 112.9, 113.7 (dq, J=26.7, 3.2 Hz), 114.1, 121.53 (qd, J=7.4, 3.7 Hz), 123.5 (qd, J=272.1, 2.8 Hz), 123.7 (d, J=11.5 Hz), 127.1 (d, J=2.8 Hz), 128.1 (qd, J=33.1, 8.3 Hz), 128.4, 128.6 (d, J=3.7 Hz), 157.6, 158.1 (d, J=250.5 Hz), 160.3, 172.1.
HRMS (ESI) calcd for C18H15F4N4O2S [M - H]− 427.0857, found 427.0858.
N-(2-Amino-1-(4-(hydroxymethyl)thiazol-2-yl)ethyl)-5-(4-cyanophenyl)-1H-pyrrole-2-carboxamide
Compounds 3 (NBD-14260) and 4 (NBD-14261) were obtained following the general procedure D and E from amine S17 and acid S4a. Compounds were purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1 and 5:1, Eluent 2: CH2Cl2-MeOH, 4:1 and 2:1.
3 (NBD-14260); (R): M = 207 mg. Yield = 17% (over two steps). rt = 1.077 min. Purity = 91%. LC-MS: m/z [M+H]+ = 368 Da.
4 (NBD-14261); (S): M = 288 mg. Yield = 21% (over two steps). rt = 1.217 min. Purity = 100%. LC-MS: m/z [M+H]+ = 368 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 1.75 (br. s., 2 H), 3.01 (dd, J=13.1, 7.8 Hz, 1 H), 3.14 (dd, J=13.2, 5.3 Hz, 1 H), 4.54 (s, 2 H), 5.17 – 5.26 (m, 1 H), 5.31 (br. s., 1 H), 6.84 (d, J=3.9 Hz, 1 H), 7.05 (d, J=3.9 Hz, 1 H), 7.29 (d, J=0.9 Hz, 1 H), 7.81 (d, J=8.0 Hz, 2 H), 8.01 (d, J=8.3 Hz, 2 H), 8.66 (d, J=7.8 Hz, 1 H), 12.04 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 45.7, 54.4, 59.8, 108.5, 109.6, 113.2, 114.1, 119.1, 125.1 (2 C), 128.8, 132.7 (2 C), 133.1, 136.1, 157.6, 160.4, 172.2.
HRMS (ESI) calcd for C18H16N5O2S [M - H]− 366.1030, found 366.1033.
N-(2-Amino-1-(4-(hydroxymethyl)thiazol-2-yl)ethyl)-5-(2,4-bis(trifluoromethyl)phenyl)-1H-pyrrole-2-carboxamide
Compounds 5 (NBD-14313) and 6 (NBD-14314) were obtained following the general procedure D and E from amine S17 and acid S4b. Compounds were purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1 and 5:1, Eluent 2: CH2Cl2-MeOH, 4:1 and 2:1.
5 (NBD-14313); (S): M = 614 mg. Yield = 35% (over two steps). rt = 1.378 min. Purity = 100%. LC-MS: m/z [M+H]+ = 479 Da.
6 (NBD-14314); (R): M = 662 mg. Yield = 37% (over two steps). rt = 1.365 min. Purity = 100%. LC-MS: m/z [M+H]+ = 479 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 1.89 (br. s., 2 H), 3.00 (dd, J=13.1, 7.9 Hz, 1 H), 3.14 (dd, J=13.2, 5.3 Hz, 1 H), 4.54 (s, 2 H), 5.18 – 5.26 (m, 1 H), 5.31 (br. s., 1 H), 6.39 (d, J=3.2 Hz, 1 H), 7.06 (d, J=3.9 Hz, 1 H), 7.29 (s, 1 H), 7.84 (d, J=8.7 Hz, 1 H), 8.07 – 8.13 (m, 2 H), 8.67 (d, J=8.0 Hz, 1 H), 12.04 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 45.4, 54.0, 59.8, 111.6 (q, J=3.3 Hz), 112.2, 114.2, 123.3 (br.), 123.4 (q, J=273.9 Hz), 123.5 (q, J=272.4 Hz), 127.3 (q, J=30.8 Hz), 128.1, 128.2 (q, J=33.0 Hz), 128.9 (q, J=3.0 Hz), 129.9, 133.7, 135.8, 157.7, 160.5, 172.1.
HRMS (ESI) calcd for C19H17F6N4O2S [M +H]+ 479.0971, found 479.0971.
N-(1-amino-2-(4-(hydroxymethyl)thiazol-2-yl)propan-2-yl)-5-(3-fluoro-4-(trifluoromethyl)phenyl)-1H-pyrrole-2-carboxamide
Compound 7 (NBD-14278) was obtained following the general procedure D and Boc- and TBS-deprotection from amine racemic amine S25 and acid S27.
Deprotection:
To a solution of protected thiazole from the previous step in MeOH (10 ml), 1M solution of HCl in MeOH (50 ml) was added in one portion. The mixture was stirred for 3–4 h, and the solvent was evaporated. After that, 10% aqueous K2CO3 (50 mL) was added, and it was extracted with CH2Cl2-EtOH (~4:1, 4 × 50 mL). Combined organic layers were dried over Na2SO4, filtered, and concentrated. The compound was purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1 and 5:1, Eluent 2: CH2Cl2-MeOH, 5:1 and 2:1.
7 (NBD-14278): M = 451 mg. Yield = 32% (over two steps). rt = 1.350 min. Purity = 100%. LC-MS: m/z [M+H]+ = 443 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 1.77 (s, 3 H), 1.82 (br. s., 2 H), 2.93 (d, J=13.2 Hz, 1 H), 3.12 (d, J=13.1 Hz, 1 H), 4.51 (d, J=2.4 Hz, 2 H), 5.28 (br. s., 1 H), 6.88 (d, J=3.9 Hz, 1 H), 7.01 (d, J=3.9 Hz, 1 H), 7.23 (s, 1 H), 7.73 (t, J=8.1 Hz, 1 H), 7.84 (d, J=8.4 Hz, 1 H), 8.00 (d, J=13.0 Hz, 1 H), 8.15 (s, 1 H), 12.04 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 23.5, 51.8, 59.9, 60.4, 109.9, 112.3 (d, J=22.5 Hz), 112.7, 113.7 (qd, J=32.4, 12.5 Hz), 113.8, 120.6 (d, J=3.0 Hz), 122.9 (q, J=271.5 Hz), 127.6 (q, J=4.4 Hz), 129.6, 131.9 (d, J=2.2 Hz), 138.5 (d, J=9.0 Hz), 156.9, 159.4 (dq, J=253.4, 2.2 Hz), 160.1, 176.4.
HRMS (ESI) calcd for C19H17F4N4O2S [M - H]− 441.1014, found 441.1014.
N-(1-Amino-2-(4-(hydroxymethyl)thiazol-2-yl)propan-2-yl)-5-(4-chloro-3,5-difluorophenyl)-1H-pyrrole-2-carboxamide
Compound 8 (NBD-14280) was obtained following the general procedure D and Boc- and TBS-deprotection from amine racemic amine S25 and acid S28.
Deprotection:
To a solution of protected thiazole from the previous step in MeOH (10 ml), 1M solution of HCl in MeOH (50 mL) was added in one portion. The mixture was stirred for 3–4 h, and the solvent was evaporated. After that, 10% aqueous K2CO3 (50 mL) was added, and it was extracted with CH2Cl2-EtOH (~4:1, 4 × 50 mL). Combined organic layers were dried over Na2SO4, filtered, and concentrated. The compound was purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1 and 5:1, Eluent 2: CH2Cl2-MeOH, 5:1 and 2:1.
8 (NBD-14280): M = 471 mg. Yield = 35% (over two steps). rt = 1.318 min. Purity = 100%. LC-MS: m/z [M+H]+ = 427 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 1.77 (s, 3 H), 1.86 (br. s., 2 H), 2.93 (d, J=13.2 Hz, 1 H), 3.12 (d, J=13.2 Hz, 1 H), 4.52 (s, 2 H), 5.29 (br. s., 1 H), 6.84 (d, J=3.9 Hz, 1 H), 6.99 (d, J=3.9 Hz, 1 H), 7.23 (s, 1 H), 7.84 (d, J=9.1 Hz, 2 H), 8.12 (s, 1 H), 11.93 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 23.5, 51.7, 59.9, 60.3, 105.3 (t, J=21.3 Hz), 108.3 (d, J=23.2 Hz. 2 C), 109.4, 112.5, 113.8, 129.1, 131.6 (t, J=2.8 Hz), 132.8 (t, J=10.3 Hz), 156.9, 158.3 (dd, J=245.8, 4.4 Hz, 2 C), 160.1, 176.4.
HRMS (ESI) calcd for C18H18ClF2N4O2S [M + H]+ 427.0802, found 427.0811.
N-(2-Amino-1-(4-(2-hydroxyethyl)thiazol-2-yl)ethyl)-5-(4-chloro-3,5-difluorophenyl)-1H-pyrrole-2-carboxamide
Compounds 9 (NBD-14242) and 10 (NBD-14243) were obtained following the general procedure D and E from amine S19 and acid S28. Compounds were purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1 and 5:1, Eluent 2: CH2Cl2-MeOH, 4:1 and 2:1.
9 (NBD-14242); (S): M = 292 mg. Yield = 22% (over two steps). rt = 1.284 min. Purity = 95%. LC-MS: m/z [M+H]+ = 427 Da.
10 (NBD-14243); (R): M = 321 mg. Yield = 24% (over two steps). rt = 1.268 min. Purity = 96%. LC-MS: m/z [M+H]+ = 427 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 1.86 (br. s., 2 H), 2.84 (t, J=6.8 Hz, 2 H), 3.00 (dd, J=13.1, 7.9 Hz, 1 H), 3.15 (dd, J=13.1, 5.3 Hz, 1 H), 3.70 (t, J=6.9 Hz, 2 H), 4.68 (br. s., 1 H), 5.17 – 5.26 (m, 1 H), 6.85 (d, J=3.9 Hz, 1 H), 7.04 (d, J=3.9 Hz, 1 H), 7.17 (s, 1 H), 7.86 (d, J=9.0 Hz, 2 H), 8.64 (d, J=7.9 Hz, 1 H), 11.96 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 34.9, 45.8, 54.5, 60.2, 105.4 (t, J=21.5 Hz), 108.4 (d, J=25.7 Hz, 2 C), 109.5, 112.7, 114.1, 128.6, 131.9 (t, J=2.8 Hz), 132.8 (t, J=10.1 Hz), 154.0, 158.4 (dd, J=245.9, 4.2 Hz, 2 C), 160.4, 171.5.
HRMS (ESI) calcd for C18H16ClF2N4O2S [M - H]− 425.0656, found 425.0659.
N-(2-Amino-1-(4-(2-hydroxyethyl)thiazol-2-yl)ethyl)-5-(3-fluoro-4-(trifluoromethyl)phenyl)-1H-pyrrole-2-carboxamide
Compounds 11 (NBD-14258) and 12 (NBD-14259) were obtained following the general procedure D and E from amine S19 and acid S27. Compounds were purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1 and 5:1, Eluent 2: CH2Cl2-MeOH, 4:1 and 2:1.
11 (NBD-14258); (R): M = 582 mg. Yield = 31% (over two steps). rt = 1.333 min. Purity = 98%. LC-MS: m/z [M+H]+ = 443 Da.
12 (NBD-14259); (S): M = 506 mg. Yield = 27% (over two steps). rt = 1.338 min. Purity = 96%. LC-MS: m/z [M+H]+ = 443 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 1.79 (br. s., 2 H), 2.84 (t, J=6.8 Hz, 2 H), 3.00 (dd, J=13.2, 7.9 Hz, 1 H), 3.15 (dd, J=13.2, 5.3 Hz, 1 H), 3.70 (t, J=6.9 Hz, 2 H), 4.67 (br. s., 1 H), 5.18 – 5.26 (m, 1 H), 6.89 (d, J=3.9 Hz, 1 H), 7.06 (d, J=3.9 Hz, 1 H), 7.18 (s, 1 H), 7.73 (t, J=8.1 Hz, 1 H), 7.85 (d, J=8.4 Hz, 1 H), 8.03 (d, J=13.0 Hz, 1 H), 8.67 (d, J=7.9 Hz, 1 H), 12.06 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 34.9, 45.8, 54.5, 60.3, 110.0, 112.4 (d, J=22.5 Hz), 113.0, 113.3 (qd, J=32.6, 12.7 Hz), 114.1, 120.6 (d, J=2.6 Hz), 122.9 (q, J=271.3 Hz), 127.6 (q, J=3.1 Hz), 129.0, 132.1 (d, J=1.8 Hz), 138.5 (d, J=9.2 Hz), 154.1, 159.5 (dq, J=251.3, 2.2 Hz), 160.4, 171.5.
HRMS (ESI) calcd for C19H17F4N4O2S [M - H]− 441.1015, found 441.1015.
N-(2-Amino-1-(4-(2-hydroxyethyl)thiazol-2-yl)ethyl)-5-(2-fluoro-4-(trifluoromethyl)phenyl)-1H-pyrrole-2-carboxamide
Compounds 13 (NBD-14273) and 14 (NBD-14274) were obtained following the general procedure D and E from amine S19 and acid S4c. Compounds were purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1 and 5:1, Eluent 2: CH2Cl2-MeOH, 4:1 and 2:1.
13 (NBD-14273); (S): M = 581 mg. Yield = 37% (over two steps). rt = 1.313 min. Purity = 100%. LC-MS: m/z [M+H]+ = 443 Da.
14 (NBD-14274); (R): M = 548 mg. Yield = 35% (over two steps). rt = 1.308 min. Purity = 100%. LC-MS: m/z [M+H]+ = 443 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 1.80 (br. s., 2 H), 2.84 (t, J=6.9 Hz, 2 H), 3.00 (dd, J=13.2, 7.9 Hz, 1 H), 3.14 (dd, J=13.1, 5.1 Hz, 1 H), 3.70 (t, J=6.9 Hz, 2 H), 4.68 (br. s., 1 H), 5.18 – 5.26 (m, 1 H), 6.73 (t, J=3.7 Hz, 1 H), 7.09 (d, J=3.9 Hz, 1 H), 7.18 (s, 1 H), 7.62 (d, J=8.2 Hz, 1 H), 7.74 (d, J=11.5 Hz, 1 H), 8.17 (t, J=8.0 Hz, 1 H), 8.72 (d, J=7.9 Hz, 1 H), 12.01 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 34.8, 45.8, 54.5, 60.2, 112.2 (d, J=10.3 Hz), 112.9, 113.7 (dq, J=26.4, 3.7 Hz), 114.1, 121.5 (qd, J=7.2, 3.5 Hz), 123.5 (qd, J=271.8, 2.2 Hz), 123.7 (d, J=11.8 Hz), 127.1 (d, J=2.2 Hz), 128.1 (qd, J=33.2, 8.3 Hz), 128.4, 128.6 (d, J=3.7 Hz), 154.0, 158.1 (d, J=250.3 Hz), 160.3, 171.5.
HRMS (ESI) calcd for C19H17F4N4O2S [M - H]− 441.1015, found 441.1014.
N-(2-Amino-1-(4-(1,2-dihydroxyethyl)thiazol-2-yl)ethyl)-5-(3-fluoro-4-(trifluoromethyl)phenyl)-1H-pyrrole-2-carboxamide
Compounds 15 (NBD-14287) and 16 (NBD-14288) were obtained following the general procedure D and E and F from amine S24 and acid S27. Compounds were purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1, 5:1 and 3:1, Eluent 2: CH2Cl2-MeOH, 2:1 and 1:1.
Note that compounds 15 & 16 were obtained as a diastereomeric mixture of 2 single compounds having the absolute configuration of the chiral carbon an as R for 16 and S for 15.
15 (NBD-14287); (S): M = 365 mg. Yield = 19% (over two steps). rt = 1.239 min. Purity = 96%. LC-MS: m/z [M+H]+ = 459 Da.
16 (NBD-14288); (R): M = 402 mg. Yield = 21% (over two steps). rt = 1.304 min. Purity = 95%. LC-MS: m/z [M+H]+ = 459 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 1.80 (br. s., 2 H), 3.01 (ddd, J=13.2, 7.7, 1.7 Hz, 1 H), 3.14 (dt, J=13.2, 4.7 Hz, 1 H), 3.44 – 3.52 (m, 1 H), 3.71 (dt, J=10.9, 3.9 Hz, 1 H), 4.65 (dd, J=6.7, 4.2 Hz, 1 H), 4.71 (br. s., 1 H), 5.17 – 5.26 (m, 1 H), 5.35 (br. s., 1 H), 6.89 (d, J=3.9 Hz, 1 H), 7.06 (d, J=3.9 Hz, 1 H), 7.30 (s, 1 H), 7.74 (t, J=8.1 Hz, 1 H), 7.85 (d, J=8.4 Hz, 1 H), 8.03 (d, J=12.9 Hz, 1 H), 8.68 (d, J=7.8 Hz, 1 H), 12.14 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 45.7, (54.5, 54.5), (65.8, 65.8), (71.3, 71.4), 110.0, 112.4 (d, J=22.5 Hz), 112.9, 113.7 (qd, J=32.6, 12.5 Hz), (114.3, 114.4), 120.6 (d, J=3.0 Hz), 122.9 (q, J=271.5 Hz), 127.6 (q, J=3.0 Hz), 128.9, 132.0 (d, J=1.8 Hz), 138.5 (d, J=9.4 Hz), (158.3, 158.4), 159.4 (dq, J=251.2, 2.2 Hz), (160.3, 160.3), (171.7, 171.8).
HRMS (ESI) calcd for C19H17F4N4O3S [M - H]− 457.0963, found 457.0956.
N-(2-Amino-1-(4-(3-hydroxypropyl)thiazol-2-yl)ethyl)-5-(3-fluoro-4-(trifluoromethyl)phenyl)-1H-pyrrole-2-carboxamide
Compounds 17 (NBD-14289) and 18 (NBD-142990) were obtained following the general procedure D and E from amine S20 and acid S27. Compounds were purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1 and 5:1, Eluent 2: CH2Cl2-MeOH, 4:1 and 2:1.
17 (NBD-14289); (S): M = 519 mg. Yield = 27% (over two steps). rt = 1.348 min. Purity = 95%. LC-MS: m/z [M+H]+ = 457 Da.
18 (NBD-14290); (R): M = 392 mg. Yield = 26% (over two steps). rt = 1.370 min. Purity = 100%. LC-MS: m/z [M+H]+ = 457 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 1.66 (br. s., 2 H), 1.74 – 1.83 (m, 2 H), 2.71 (t, J=7.6 Hz, 2 H), 3.00 (dd, J=13.2, 7.9 Hz, 1 H), 3.15 (dd, J=13.1, 5.3 Hz, 1 H), 3.44 (t, J=6.4 Hz, 2 H), 4.49 (br. s., 1 H), 5.17 – 5.25 (m, 1 H), 6.89 (d, J=3.9 Hz, 1 H), 7.06 (d, J=3.9 Hz, 1 H), 7.13 (s, 1 H), 7.74 (t, J=8.1 Hz, 1 H), 7.85 (d, J=8.4 Hz, 1 H), 8.03 (d, J=12.9 Hz, 1 H), 8.67 (d, J=7.9 Hz, 1 H), 12.13 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 27.6, 32.1, 45.8, 54.5, 60.2, 110.0, 112.4 (d, J=22.5 Hz), 112.9, 113.1, 113.7 (qd, J=32.4, 12.5 Hz), 120.6 (d, J=3.0 Hz), 122.9 (q, J=271.3 Hz), 127.6 (q, J=3.3 Hz), 129.0, 132.1 (d, J=1.8 Hz), 138.5 (d, J=9.4 Hz), 156.6, 159.4 (dq, J=251.2, 2.0 Hz), 160.4, 171.6.
HRMS (ESI) calcd for C20H19F4N4O2S [M - H]− 455.1170, found 455.1170.
N-(2-Amino-1-(5-(1,2-dihydroxyethyl)thiazol-2-yl)ethyl)-5-(3-fluoro-4-(trifluoromethyl)phenyl)-1H-pyrrole-2-carboxamide
Compounds 19 (NBD-14303) and 20 (NBD-14304) were obtained following the general procedure D and E and F from amine S23 and acid S27. Compounds were purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1, 5:1 and 3:1, Eluent 2: CH2Cl2-MeOH, 2:1 and 1:1.
Note that compounds 19 & 20 were obtained as a diastereomeric mixture of 2 single compounds having the absolute configuration of the chiral carbon an as R for 20 and S for 19.
19 (NBD-14303); (S): M = 382 mg. Yield = 31% (over three steps). rt = 1.316 min. Purity = 100%. LC-MS: m/z [M+H]+ = 459 Da.
20 (NBD-14304); (R): M = 216 mg. Yield = 27% (over three steps). rt = 1.230 min. Purity = 100%. LC-MS: m/z [M+H]+ = 459 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 1.68 (br. s., 2 H), 3.00 (dd, J=13.2, 7.9 Hz, 1 H), 3.14 (dd, J=13.2, 5.3 Hz, 1 H), 3.40 – 3.47 (m, 1 H), 3.51 (dd, J=10.8, 6.2 Hz, 1 H), 4.75 (t, J=5.7 Hz, 1 H), 4.94 (br. s., 1 H), 5.15 – 5.25 (m, 1 H), 5.68 (br. s., 1 H), 6.89 (d, J=3.9 Hz, 1 H), 7.06 (d, J=3.9 Hz, 1 H), 7.56 (s, 1 H), 7.74 (t, J=8.1 Hz, 1 H), 7.85 (d, J=8.3 Hz, 1 H), 8.03 (d, J=13.0 Hz, 1 H), 8.66 (d, J=7.9 Hz, 1 H), 12.08 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = (45.6, 45.7), 54.5, 66.7, 68.1, 110.0, 112.4 (d, J=22.6 Hz), 112.9, 113.7 (qd, J=32.4, 12.5 Hz), 120.6 (d, J=2.9 Hz), 122.9 (q, J=271.3 Hz), 127.6 (q, J=3.5 Hz), 129.0, 132.0 (d, J=2.1 Hz), 138.5 (q, J=2.3 Hz), 141.3, 141.4, 159.4 (dq, J=251.3, 1.9 Hz), 160.3, (171.2, 171.2).
HRMS (ESI) calcd for C19H19F4N4O3S [M +H]+ 459.1109, found 459.1109.
2-[2-Amino-1-[[5-[3-fluoro-4-(trifluoromethyl)phenyl]-1H-pyrrole-2-carbonyl]amino]ethyl]thiazole-4-carboxylic acid
Compounds 21 (NBD-14271) and 22 (NBD-14272) were obtained from direct NaOH hydrolysis of 100mg of the corresponding methyl esters 23 (NBD-14275) and 24 (NBD-14276).
21 (NBD-14271); (S): M = 51 mg. LC-MS: m/z [M+H]+ = 443 Da.
22 (NBD-14272); (R): M = 60 mg. LC-MS: m/z [M+H]+ = 443 Da.
Due to broad NMR signals and since the corresponding crudes were not active, no further characterization and purification were performed.
Methyl 2-(2-amino-1-(5-(3-fluoro-4-(trifluoromethyl)phenyl)-1H-pyrrole-2-carboxamido)ethyl)thiazole-4-carboxylate
Compounds 23 (NBD-14275) and 24 (NBD-14276) were obtained following the general procedure D and E from amine S15 and acid S27. Compounds were purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1 and 5:1, Eluent 2: CH2Cl2-MeOH, 4:1 and 2:1.
23 (NBD-14275); (S): M = 344 mg. Yield = 21% (over two steps). rt = 1.377 min. Purity = 100%. LC-MS: m/z [M+H]+ = 457 Da.
24 (NBD-14276); (R): M = 421 mg. Yield = 26% (over two steps). rt = 1.394 min. Purity = 85%. LC-MS: m/z [M+H]+ = 457 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 2.04 (br. s., 2 H), 3.06 (dd, J=13.2, 7.7 Hz, 1 H), 3.18 (dd, J=13.2, 5.4 Hz, 1 H), 3.82 (s, 3 H), 5.22 – 5.29 (m, 1 H), 6.90 (d, J=3.9 Hz, 1 H), 7.07 (d, J=3.9 Hz, 1 H), 7.74 (t, J=8.1 Hz, 1 H), 7.86 (d, J=8.4 Hz, 1 H), 8.04 (d, J=12.9 Hz, 1 H), 8.44 (s, 1 H), 8.82 (d, J=6.4 Hz, 1 H), 12.12 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 45.4, 52.0, 54.5, 110.0, 112.4 (d, J=22.5 Hz), 113.1, 113.8 (qd, J=32.4, 12.5 Hz), 120.7 (d, J=2.8 Hz), 122.9 (q, J=271.3 Hz), 127.6 (q, J=3.3 Hz), 128.8, 129.1, 132.3 (d, J=1.8 Hz), 138.5 (d, J=9.2 Hz), 145.5, 159.5 (dq, J=251.2, 2.2 Hz), 160.5, 161.3, 173.4.
HRMS (ESI) calcd for C19H15F4N4O3S [M - H]− 455.0806, found 455.0808.
2-(2-Amino-1-(5-(3-fluoro-4-(trifluoromethyl)phenyl)-1H-pyrrole-2-carboxamido)ethyl)thiazole-4-carboxamide
Compounds 25 (NBD-14267) and 26 (NBD-14268) were obtained following the general procedure D and E from amine S16 and acid S27. Compounds were purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1 and 5:1, Eluent 2: CH2Cl2-MeOH, 3:1 and 1:1.
25 (NBD-14267); (R): M = 471 mg. Yield = 35% (over two steps). rt = 1.289 min. Purity = 97%. LC-MS: m/z [M+H]+ = 442 Da.
26 (NBD-14268); (S): M = 493 mg. Yield = 36% (over two steps). rt = 1.285 min. Purity = 100%. LC-MS: m/z [M+H]+ = 442 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 1.99 (br. s., 2 H), 3.06 (dd, J=13.3, 7.5 Hz, 1 H), 3.22 (dd, J=13.2, 5.4 Hz, 1 H), 5.20 – 5.30 (m, 1 H), 6.90 (d, J=3.9 Hz, 1 H), 7.08 (d, J=3.9 Hz, 1 H), 7.59 (br. s., 1 H), 7.70 – 7.78 (m, 2 H), 7.86 (d, J=8.4 Hz, 1 H), 8.03 (d, J=13.0 Hz, 1 H), 8.14 (s, 1 H), 8.75 (d, J=6.0 Hz, 1 H), 11.95 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 45.5, 54.5, 110.1, 112.5 (d, J=22.5 Hz), 113.2, 113.9 (qd, J=32.4, 12.5 Hz), 120.7 (d, J=2.8 Hz), 122.9 (q, J=271.5 Hz), 124.0, 127.6 (q, J=3.3 Hz), 128.8, 132.3 (d, J=2.0 Hz), 138.5 (d, J=9.2 Hz), 150.1, 159.5 (dq, J=251.4, 2.0 Hz), 160.6, 162.5, 173.0.
HRMS (ESI) calcd for C18H14F4N5O2S [M - H]− 440.0810, found 440.0811.
N-(2-Amino-1-(4-(hydroxymethyl)thiazol-2-yl)ethyl)-5-(3-fluoro-4-(trifluoromethyl)phenyl)-3-methyl-1H-pyrrole-2-carboxamide
Compounds 27 (NBD-14325) and 28 (NBD-14324) were obtained following the general procedure D and E from amine S17 and acid S8. Compounds were purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1 and 5:1, Eluent 2: CH2Cl2-MeOH, 3:1 and 2:1.
27 (NBD-14325); (S): M = 454 mg. Yield = 27% (over two steps). rt = 1.327 min. Purity = 100%. LC-MS: m/z [M+H]+ = 443 Da.
28 (NBD-14324); (R): M = 571 mg. Yield = 34% (over two steps). rt = 1.336 min. Purity = 100%. LC-MS: m/z [M+H]+ = 443 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 1.81 (br. s., 2 H), 2.32 (s, 3 H), 3.04 (dd, J=13.1, 7.2 Hz, 1 H), 3.12 (dd, J=13.1, 5.3 Hz, 1 H), 4.55 (s, 2 H), 5.20 – 5.27 (m, 1 H), 5.31 (br. s., 1 H), 6.74 (s, 1 H), 7.31 (s, 1 H), 7.72 – 7.82 (m, 2 H), 7.87 (d, J=12.7 Hz, 1 H), 8.28 (d, J=7.0 Hz, 1 H), 11.60 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 12.9, 45.9, 54.0, 59.8, 112.0 (d, J=22.1 Hz), 112.3, 113.8 (qd, J=32.4, 12.4 Hz), 114.2, 120.3 (d, J=2.8 Hz), 122.9 (q, J=271.5 Hz), 124.7, 125.8, 127.8 (q, J=4.1 Hz), 129.9 (d, J=1.8 Hz), 138.4 (d, J=9.2 Hz), 157.7, 159.6 (dq, J=251.6, 2.2 Hz), 160.8, 171.9.
HRMS (ESI) calcd for C19H17F4N4O2S [M-H]− 441.1014, found 441.1013.
N-(2-Amino-1-(5-(hydroxymethyl)thiazol-2-yl)ethyl)-5-(3-fluoro-4-(trifluoromethyl)phenyl)-3-methyl-1H-pyrrole-2-carboxamide
Compounds 29 (NBD-14329) and 30 (NBD-14328) were obtained following the general procedure D and E from amine S18 and acid S8. Compounds were purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1 and 5:1, Eluent 2: CH2Cl2-MeOH, 3:1 and 2:1.
29 (NBD-14329); (S): M = 420 mg. Yield = 24% (over two steps). rt = 1.326 min. Purity = 100%. LC-MS: m/z [M+H]+ = 443 Da.
30 (NBD-14328); (R): M = 620 mg. Yield = 35% (over two steps). rt = 1.318 min. Purity = 100%. LC-MS: m/z [M+H]+ = 443 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 2.08 (br. s., 2 H), 2.33 (s, 3 H), 3.05 (dd, J=13.1, 7.3 Hz, 1 H), 3.14 (dd, J=13.1, 5.1 Hz, 1 H), 4.62 (s, 2 H), 5.18 – 5.25 (m, 1 H), 5.50 (br. s., 1 H), 6.75 (s, 1 H), 7.57 (s, 1 H), 7.73 – 7.82 (m, 2 H), 7.89 (d, J=12.5 Hz, 1 H), 8.32 (d, J=6.8 Hz, 1 H), 11.64 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 12.9, 45.7, 54.0, 55.8, 112.0 (d, J=22.3 Hz), 112.3, 113.7 (qd, J=32.6, 12.5 Hz), 120.3 (d, J=2.8 Hz), 122.8 (q, J=271.3 Hz), 124.6, 125.7, 127.8 (q, J=3.5 Hz), 129.9 (d, J=2.0 Hz), 138.4 (d, J=9.2 Hz), 139.0, 140.2, 159.5 (dq, J=251.7, 2.4 Hz), 160.8, 171.5.
HRMS (ESI) calcd for C19H17F4N4O2S [M-H]− 441.1014, found 441.1014.
N-(2-Amino-1-(4-(hydroxymethyl)thiazol-2-yl)-2-methylpropyl)-5-(4-chloro-3,5-difluorophenyl)-1H-pyrrole-2-carboxamide
Compounds 31 (NBD-14246) and 32 (NBD-14247) were obtained following the general procedure D and E from amine S21 and acid S28. Compounds were purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1 and 5:1, Eluent 2: CH2Cl2-MeOH, 4:1 and 2:1.
31 (NBD-14246); (S): M = 364 mg. Yield = 28% (over two steps). rt = 1.362 min. Purity = 95%. LC-MS: m/z [M+H]+ = 441 Da.
32 (NBD-14247); (R): M = 315 mg. Yield = 24% (over two steps). rt = 1.377 min. Purity = 96%. LC-MS: m/z [M+H]+ = 441 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 1.08 (s, 3 H), 1.13 (s, 3 H), 1.97 (br. s., 2 H), 4.57 (s, 2 H), 5.29 (d, J=7.6 Hz, 1 H), 5.33 (br. s., 1 H), 6.84 (d, J=3.9 Hz, 1 H), 7.08 (d, J=3.9 Hz, 1 H), 7.32 (s, 1 H), 7.83 (d, J=8.9 Hz, 2 H), 8.26 (d, J=8.4 Hz, 1 H), 12.02 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 27.6, 28.5, 52.7, 59.2, 59.8, 105.4 (t, J=21.3 Hz), 108.4 (d, J=25.4 Hz, 2 C), 109.5, 113.2, 114.4, 128.4, 132.0 (t, J=2.8 Hz), 132.8 (t, J=10.1 Hz), 157.3, 158.4 (dd, J=246.4, 4.1 Hz, 2 C), 160.0, 170.2.
HRMS (ESI) calcd for C19H18ClF2N4O2S [M - H]− 439.0813, found 439.0813.
N-(2-Amino-1-(4-(hydroxymethyl)thiazol-2-yl)-2-methylpropyl)-5-(4-(trifluoromethyl)phenyl)-1H-pyrrole-2-carboxamide
Compounds 33 (NBD-14248) and 34 (NBD-14249) were obtained following the general procedure D and E from amine S21 and acid S26. Compounds were purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1 and 5:1, Eluent 2: CH2Cl2-MeOH, 4:1 and 2:1.
33 (NBD-14248); (S): M = 435 mg. Yield = 27% (over two steps). rt = 1.354 min. Purity = 97%. LC-MS: m/z [M+H]+ = 439 Da.
34 (NBD-14249); (R): M = 501 mg. Yield = 31% (over two steps). rt = 1.360 min. Purity = 99%. LC-MS: m/z [M+H]+ = 439 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 1.09 (s, 3 H), 1.14 (s, 3 H), 1.95 (br. s., 2 H), 4.58 (s, 2 H), 5.30 (d, J=8.2 Hz, 1 H), 5.38 (br. s., 1 H), 6.78 (d, J=3.8 Hz, 1 H), 7.08 (d, J=3.8 Hz, 1 H), 7.32 (s, 1 H), 7.71 (d, J=8.4 Hz, 2 H), 8.02 (d, J=8.2 Hz, 2 H), 8.28 (d, J=8.8 Hz, 1 H), 12.05 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 27.6, 28.5, 52.7, 59.2, 59.8, 108.8, 113.5, 114.4, 124.4 (q, J=272.1 Hz), 125.1 (2 C), 125.6 (q, J=4.1 Hz, 2 C), 126.7 (q, J=31.7 Hz), 128.2, 133.4, 135.6, 157.3, 160.0, 170.2.
HRMS (ESI) calcd for C20H20F3N4O2S [M - H]− 437.1265, found 437.1269.
N-(2-Amino-1-(5-(hydroxymethyl)thiazol-2-yl)-2-methylpropyl)-5-(4-(trifluoromethyl)phenyl)-1H-pyrrole-2-carboxamide
Compounds 35 (NBD-14262) and 36 (NBD-14263) were obtained following the general procedure D and E from amine S22 and acid S26. Compounds were purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1 and 5:1, Eluent 2: CH2Cl2-MeOH, 4:1 and 2:1.
35 (NBD-14262); (R): M = 438 mg. Yield = 27% (over two steps). rt = 1.458 min. Purity = 100%. LC-MS: m/z [M+H]+ = 439 Da.
36 (NBD-14263); (S): M = 77 mg. Yield = 4% (over two steps). rt = 1.306 min. Purity = 94%. LC-MS: m/z [M+H]+ = 439 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 1.10 (s, 3 H), 1.14 (s, 3 H), 2.21 (br. s., 2 H), 4.63 (s, 2 H), 5.27 (d, J=8.4 Hz, 1 H), 5.51 (br. s., 1 H), 6.78 (d, J=3.9 Hz, 1 H), 7.06 (d, J=3.9 Hz, 1 H), 7.58 (s, 1 H), 7.72 (d, J=8.4 Hz, 2 H), 8.02 (d, J=8.2 Hz, 2 H), 8.29 (d, J=8.9 Hz, 1 H), 12.07 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 27.5, 28.4, 52.8, 55.8, 59.3, 108.8, 113.6, 124.4 (q, J=271.6 Hz), 125.1 (2 C), 125.6 (q, J=3.7 Hz, 2 C), 126.7 (q, J=31.7 Hz), 128.2, 133.4, 135.6, 138.7, 140.3, 160.0, 170.1.
HRMS (ESI) calcd for C20H20F3N4O2S [M +H]+ 437.1265, found 437.1269.
N-(2-amino-1-(5-((2,3-dihydroxypropoxy)methyl)thiazol-2-yl)ethyl)-5-(4-(trifluoromethyl)phenyl)-1H-pyrrole-2-carboxamide
Compounds 37 (NBD-14225) and 38 (NBD-14229) were obtained following the general procedure D and E and F from amine S14 and acid S26. Compounds were purified using column chromatography on silica gel (twice). Eluent 1: CHCl3-MeOH (saturated with NH3~7M), 10:1, 5:1 and 3:1, Eluent 2: CH2Cl2-MeOH, 2:1 and 1:1.
Note that compounds 37 & 38 were obtained as a diastereomeric mixture of 2 single compounds having the absolute configuration of the chiral carbon as R for 38 and S for 37.
37 (NBD-14225); (S): M = 342 mg. Yield = 17% (over two steps). rt = 1.342 min. Purity = 100%. LC-MS: m/z [M+H]+ = 485 Da.
38 (NBD-14229); (R): M = 381 mg. Yield = 19% (over two steps). rt = 1.456 min. Purity = 95%. LC-MS: m/z [M+H]+ = 485 Da.
1H NMR (DMSO-d6, 400 MHz): δ = 1.77 (br. s., 2 H), 3.04 (dd, J=13.1, 7.8 Hz, 1 H), 3.17 (dd, J=13.3, 5.4 Hz, 1 H), 3.29 – 3.36 (m, 3 H), 3.45 (dd, J=9.9, 4.6 Hz, 1 H), 3.54 – 3.61 (m, 1 H), 4.65 (s, 2 H), 4.72 (br. s., 2 H), 5.19 – 5.28 (m, 1 H), 6.79 (d, J=3.9 Hz, 1 H), 7.06 (d, J=3.8 Hz, 1 H), 7.66 (s, 1 H), 7.71 (d, J=8.4 Hz, 2 H), 8.04 (d, J=8.2 Hz, 2 H), 8.67 (d, J=7.8 Hz, 1 H), 12.00 (br. s., 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 45.6, 54.6, 63.0, 64.5, 70.5, 71.8, 108.9, 113.1, 124.4 (q, J=271.6 Hz), 125.1 (2 C), 125.6 (q, J=3.7 Hz, 2 C), 126.6 (q, J=32.2 Hz), 128.3, 133.3, 135.4, 135.6, 141.0, 160.5, 173.1.
HRMS (ESI): Calcd for C21H24F3N4O4S [M +H]+ 485.1465, found 485.1466.
Acknowledgments
This study was supported by funds from NIH Grant R01 AI104416 (AKD), and the New York Blood Center (AKD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this manuscript.
References
- 1.Caffrey M, HIV envelope: challenges and opportunities for development of entry inhibitors. Trends Microbiol. 2011, 19, 191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Didigu CA; Doms RW, Novel Approaches to Inhibit HIV Entry. Viruses 2012, 4, 309–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu F; Lu L; Du L; Zhu X; Debnath AK; Jiang S, Approaches for Identification of HIV-1 Entry Inhibitors Targeting gp41 Pocket. Viruses 2013, 5, 127–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu L; Yu F; Cai L; Debnath AK; Jiang S, Development of Small-molecule HIV Entry Inhibitors Specifically Targeting gp120 or gp41. Curr. Top. Med. Chem 2016, 16, 1074–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duffalo ML; James CW, Enfuvirtide: a novel agent for the treatment of HIV-1 infection. Ann. Pharmacother 2003, 37 (10), 1448–1456. [DOI] [PubMed] [Google Scholar]
- 6.Hardy H; Skolnik PR, Enfuvirtide, a new fusion inhibitor for therapy of human immunodeficiency virus infection. Pharmacotherapy 2004, 24 (2), 198–211. [DOI] [PubMed] [Google Scholar]
- 7.Dorr P; Westby M; Dobbs S; Griffin P; Irvine B; Macartney M; Mori J; Rickett G; Smith-Burchnell C; Napier C; Webster R; Armour D; Price D; Stammen B; Wood A; Perros M, Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother 2005, 49 (11), 4721–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunt JS; Romanelli F, Maraviroc, a CCR5 coreceptor antagonist that blocks entry of human immunodeficiency virus type 1. Pharmacotherapy 2009, 29 (3), 295–304. [DOI] [PubMed] [Google Scholar]
- 9.Meanwell NA; Kadow JF, Maraviroc, a chemokine CCR5 receptor antagonist for the treatment of HIV infection and AIDS. Curr. Opin. Investig. Drugs 2007, 8 (8), 669–681. [PubMed] [Google Scholar]
- 10.Zhao Q; Ma L; Jiang S; Lu H; Liu S; He Y; Strick N; Neamati N; Debnath AK, Identification of N-phenyl-N’-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamides as a new class of HIV-1 entry inhibitors that prevent gp120 binding to CD4. Virology 2005, 339 (2), 213–225. [DOI] [PubMed] [Google Scholar]
- 11.Si Z; Madani N; Cox JM; Chruma JJ; Klein JC; Schon A; Phan N; Wang L; Biorn AC; Cocklin S; Chaiken I; Freire E; Smith AB III; Sodroski JG, Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc. Natl. Acad. Sci. U S A 2004, 101 (14), 5036–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curreli F; Kwon YD; Zhang H; Scacalossi D; Belov DS; Tikhonov AA; Andreev IA; Altieri A; Kurkin AV; Kwong PD; Debnath AK, Structure-based design of a small molecule CD4-antagonist with broad spectrum anti-HIV-1 activity. J. Med. Chem 2015, 58 (17), 6909–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curreli F; Kwon YD; Zhang H; Yang Y; Scacalossi D; Kwong PD; Debnath AK, Binding mode characterization of NBD series CD4-mimetic HIV-1 entry inhibitors by X-ray structure and resistance study. Antimicrob. Agents Chemother 2014, 58, 5478–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curreli F; Choudhury S; Pyatkin I; Zagorodnikov VP; Bulay AK; Altieri A; Kwon YD; Kwong PD; Debnath AK, Design, synthesis and antiviral activity of entry inhibitors that target the CD4-binding site of HIV-1. J. Med. Chem 2012, 55, 4764–4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curreli F; Belov DS; Ramesh RR; Patel N; Altieri A; Kurkin AV; Debnath AK, Design, synthesis and evaluation of small molecule CD4-mimics as entry inhibitors possessing broad spectrum anti-HIV-1 activity. Bioorg. Med. Chem 2016, 24, 5988–6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Courter JR; Madani N; Sodroski J; Schon A; Freire E; Kwong PD; Hendrickson WA; Chaiken IM; Lalonde JM; Smith AB III, Structure-based design, synthesis and validation of CD4-mimetic small molecule inhibitors of HIV-1 entry: conversion of a viral entry agonist to an antagonist. Acc. Chem. Res 2014, 47, 1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lalonde JM; Elban MA; Courter JR; Sugawara A; Soeta T; Madani N; Princiotto AM; Kwon YD; Kwong PD; Schon A; Freire E; Sodroski J; Smith AB III, Design, synthesis and biological evaluation of small molecule inhibitors of CD4-gp120 binding based on virtual screening. Bioorg. Med. Chem 2011, 19 (1), 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lalonde JM; Le-Khac M; Jones DM; Courter JR; Park J; Schon A; Princiotto AM; Wu X; Mascola JR; Freire E; Sodroski J; Madani N; Hendrickson WA; Smith AB III, Structure-based design and synthesis of an HIV-1 entry inhibitor exploiting X-ray and thermodynamic characterization. ACS Med. Chem. Lett 2013, 4 (3), 338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melillo B; Liang S; Park J; Schon A; Courter JR; Lalonde JM; Wendler DJ; Princiotto AM; Seaman MS; Freire E; Sodroski J; Madani N; Hendrickson WA; Smith AB III, Small-molecule CD4-mimics: structure-based optimization of HIV-1 entry inhibition. ACS Med. Chem. Lett 2016, 7 (3), 330–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashimoto C; Narumi T; Otsuki H; Hirota Y; Arai H; Yoshimura K; Harada S; Ohashi N; Nomura W; Miura T; Igarashi T; Matsushita S; Tamamura H, A CD4 mimic as an HIV entry inhibitor: Pharmacokinetics. Bioorg. Med. Chem 2013, 21 (24), 7884–7889. [DOI] [PubMed] [Google Scholar]
- 21.Narumi T; Arai H; Yoshimura K; Harada S; Hirota Y; Ohashi N; Hashimoto C; Nomura W; Matsushita S; Tamamura H, CD4 mimics as HIV entry inhibitors: lead optimization studies of the aromatic substituents. Bioorg. Med. Chem 2013, 21 (9), 2518–2526. [DOI] [PubMed] [Google Scholar]
- 22.Narumi T; Arai H; Yoshimura K; Harada S; Nomura W; Matsushita S; Tamamura H, Small molecular CD4 mimics as HIV entry inhibitors. Bioorg. Med. Chem 2011, 19 (22), 6735–6742. [DOI] [PubMed] [Google Scholar]
- 23.Kwon YD; Finzi A; Wu X; Dogo-Isonagie C; Lee LK; Moore LR; Schmidt SD; Stuckey J; Yang Y; Zhou T; Zhu J; Vicic DA; Debnath AK; Shapiro L; Bewley CA; Mascola JR; Sodroski JG; Kwong PD, Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 5663–5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curreli F; Kwon YD; Belov DS; Ramesh RR; Kurkin AV; Altieri A; Kwong PD; Debnath AK, Synthesis, antiviral potency, in vitro ADMET and X-ray structure of potent CD4-mimics as entry inhibitors that target the Phe43 cavity of HIV-1 gp120. J. Med. Chem 2017, 60, 3124–3153. [DOI] [PubMed] [Google Scholar]
- 25.Curreli F B. DS; Kwon YD; Ramesh R; Furimsky AM; O’Loughlin K; Byrge PC; Iyer LV; Mirsalis JC; Kurkin AV; Altieri A Debnath AK, Structure-based lead optimization to improve antiviral potency and ADMET properties of phenyl-1H-pyrrole-carboxamide entry inhibitors targeted to HIV-1 gp120. Eur. J. Med. Chem 2018. 154, 367–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schon A; Madani N; Klein JC; Hubicki A; Ng D; Yang X; Smith AB III; Sodroski J; Freire E, Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry 2006, 45 (36), 10973–10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curreli F; Belov DS; Ahmed S; Ramesh RR; Kurkin AV; Altieri A; Debnath AK, Synthesis, antiviral activity, and structure-activity relationship of 1,3-benzodioxolyl pyrrole-based entry inhibitors targeting the Phe43 cavity in HIV-1 gp120. ChemMedChem 2018, 13 (21), 2332–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curreli F; Belov DS; Kwon YD; Ramesh R; Furimsky AM; O’Loughlin K; Byrge PC; Iyer LV; Mirsalis JC; Kurkin AV; Altieri A; Debnath AK, Structure-based lead optimization to improve antiviral potency and ADMET properties of phenyl-1H-pyrrole-carboxamide entry inhibitors targeted to HIV-1 gp120. Eur J Med Chem 2018, 154, 367–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Curreli F; Belov DS; Ramesh RR; Patel N; Altieri A; Kurkin AV; Debnath AK, Design, synthesis and evaluation of small molecule CD4-mimics as entry inhibitors possessing broad spectrum anti-HIV-1 activity. Bioorg Med Chem 2016, 24 (22), 5988–6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curreli F; Kwon YD; Belov DS; Ramesh RR; Kurkin AV; Altieri A; Kwong PD; Debnath AK, Synthesis, Antiviral Potency, in Vitro ADMET, and X-ray Structure of Potent CD4 Mimics as Entry Inhibitors That Target the Phe43 Cavity of HIV-1 gp120. J Med Chem 2017. [DOI] [PubMed] [Google Scholar]
- 31.Curreli F; Kwon YD; Belov DS; Ramesh RR; Kurkin AV; Altieri A; Kwong PD; Debnath AK, Synthesis, antiviral potency, in vitro ADMET and X-ray structure of potent CD4-mimics as entry inhibitors that target the Phe43 cavity of HIV-1 gp120. J Med Chem 2017, 60, 3124 – 3153. [DOI] [PubMed] [Google Scholar]
- 32.Curreli F; Ahmed S; Benedict Victor SM; Iusupov IR; Belov DS; Markov PO; Kurkin AV; Altieri A; Debnath AK, Preclinical Optimization of gp120 Entry Antagonists as anti-HIV-1 Agents with Improved Cytotoxicity and ADME Properties through Rational Design, Synthesis, and Antiviral Evaluation. J Med Chem 2020, 63 (4), 1724–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X; Parast AB; Richardson BA; Nduati R; John-Stewart G; Mbori-Ngacha D; Rainwater SM; Overbaugh J, Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J.Virol 2006, 80 (2), 835–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abela IA; Berlinger L; Schanz M; Reynell L; Gunthard HF; Rusert P; Trkola A, Cell-Cell Transmission Enables HIV-1 to Evade Inhibition by Potent CD4bs Directed Antibodies. PLoS Pathog 2012, 8, e1002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agosto LM; Uchil PD; Mothes W, HIV cell-to-cell transmission: effects on pathogenesis and antiretroviral therapy. Trends Microbiol 2015, 23 (5), 289–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abela IA; Berlinger L; Schanz M; Reynell L; Gunthard HF; Rusert P; Trkola A, Cell-cell transmission enables HIV-1 to evade inhibition by potent CD4bs directed antibodies. PLoS Pathog. 2012, 8 (4), e1002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durham ND; Yewdall AW; Chen P; Lee R; Zony C; Robinson JE; Chen BK, Neutralization resistance of virological synapse-mediated HIV-1 Infection is regulated by the gp41 cytoplasmic tail. J Virol 2012, 86 (14), 7484–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H; Zony C; Chen P; Chen BK, Reduced Potency and Incomplete Neutralization of Broadly Neutralizing Antibodies against Cell-to-Cell Transmission of HIV-1 with Transmitted Founder Envs. J Virol 2017, 91 (9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agosto LM; Zhong P; Munro J; Mothes W, Highly active antiretroviral therapies are effective against HIV-1 cell-to-cell transmission. PLoS.Pathog 2014, 10 (2), e1003982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curreli F; Kwon YD; Zhang H; Scacalossi D; Belov DS; Tikhonov AA; Andreev IA; Altieri A; Kurkin AV; Kwong PD; Debnath AK, Structure-Based Design of a Small Molecule CD4-Antagonist with Broad Spectrum Anti-HIV-1 Activity. J Med Chem 2015, 58 (17), 6909–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei X; Decker JM; Liu H; Zhang Z; Arani RB; Kilby JM; Saag MS; Wu X; Shaw GM; Kappes JC, Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother 2002, 46 (6), 1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bjorndal A; Deng H; Jansson M; Fiore JR; Colognesi C; Karlsson A; Albert J; Scarlatti G; Littman DR; Fenyo EM, Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol 1997, 71 (10), 7478–7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Popovic M; Sarngadharan MG; Read E; Gallo RC, Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science 1984, 224 (4648), 497–500. [DOI] [PubMed] [Google Scholar]
- 44.Baba M; Miyake H; Okamoto M; Iizawa Y; Okonogi K, Establishment of a CCR5-expressing T-lymphoblastoid cell line highly susceptible to R5 HIV type 1. AIDS Res.Hum.Retroviruses 2000, 16 (10), 935–941. [DOI] [PubMed] [Google Scholar]
- 45.Kolchinsky P; Mirzabekov T; Farzan M; Kiprilov E; Cayabyab M; Mooney LJ; Choe H; Sodroski J, Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J. Virol 1999, 73 (10), 8120–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Page KA; Landau NR; Littman DR, Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J. Virol 1990, 5270–5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blish CA; Jalalian-Lechak Z; Rainwater S; Nguyen MA; Dogan OC; Overbaugh J, Cross-subtype neutralization sensitivity despite monoclonal antibody resistance among early subtype A, C, and D envelope variants of human immunodeficiency virus type 1. J. Virol 2009, 83 (15), 7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Long EM; Rainwater SM; Lavreys L; Mandaliya K; Overbaugh J, HIV type 1 variants transmitted to women in Kenya require the CCR5 coreceptor for entry, regardless of the genetic complexity of the infecting virus. AIDS Res. Hum. Retrov 2002, 18 (8), 567–576. [DOI] [PubMed] [Google Scholar]
- 49.Kulkarni SS; Lapedes A; Tang H; Gnanakaran S; Daniels MG; Zhang M; Bhattacharya T; Li M; Polonis VR; McCutchan FE; Morris L; Ellenberger D; Butera ST; Bollinger RC; Korber BT; Paranjape RS; Montefiori DC, Highly complex neutralization determinants on a monophyletic lineage of newly transmitted subtype C HIV-1 Env clones from India. Virology 2009, 385 (2), 505–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li M; Gao F; Mascola JR; Stamatatos L; Polonis VR; Koutsoukos M; Voss G; Goepfert P; Gilbert P; Greene KM; Bilska M; Kothe DL; Salazar-Gonzalez JF; Wei X; Decker JM; Hahn BH; Montefiori DC, Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol 2005, 79 (16), 10108–10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei X; Decker JM; Wang S; Hui H; Kappes JC; Wu X; Salazar-Gonzalez JF; Salazar MG; Kilby JM; Saag MS; Komarova NL; Nowak MA; Hahn BH; Kwong PD; Shaw GM, Antibody neutralization and escape by HIV-1. Nature 2003, 422 (6929), 307–312. [DOI] [PubMed] [Google Scholar]
- 52.Keele BF; Giorgi EE; Salazar-Gonzalez JF; Decker JM; Pham KT; Salazar MG; Sun C; Grayson T; Wang S; Li H; Wei X; Jiang C; Kirchherr JL; Gao F; Anderson JA; Ping LH; Swanstrom R; Tomaras GD; Blattner WA; Goepfert PA; Kilby JM; Saag MS; Delwart EL; Busch MP; Cohen MS; Montefiori DC; Haynes BF; Gaschen B; Athreya GS; Lee HY; Wood N; Seoighe C; Perelson AS; Bhattacharya T; Korber BT; Hahn BH; Shaw GM, Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A 2008, 105 (21), 7552–7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Derdeyn CA; Decker JM; Bibollet-Ruche F; Mokili JL; Muldoon M; Denham SA; Heil ML; Kasolo F; Musonda R; Hahn BH; Shaw GM; Korber BT; Allen S; Hunter E, Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 2004, 303 (5666), 2019–2022. [DOI] [PubMed] [Google Scholar]
- 54.Li M; Salazar-Gonzalez JF; Derdeyn CA; Morris L; Williamson C; Robinson JE; Decker JM; Li Y; Salazar MG; Polonis VR; Mlisana K; Karim SA; Hong K; Greene KM; Bilska M; Zhou J; Allen S; Chomba E; Mulenga J; Vwalika C; Gao F; Zhang M; Korber BT; Hunter E; Hahn BH; Montefiori DC, Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J. Virol 2006, 80 (23), 11776–11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williamson C; Morris L; Maughan MF; Ping LH; Dryga SA; Thomas R; Reap EA; Cilliers T; van HJ; Pascual A; Ramjee G; Gray G; Johnston R; Karim SA; Swanstrom R, Characterization and selection of HIV-1 subtype C isolates for use in vaccine development. AIDS Res. Hum. Retrov 2003, 19 (2), 133–144. [DOI] [PubMed] [Google Scholar]
- 56.Wu X; Parast AB; Richardson BA; Nduati R; John-Stewart G; Mbori-Ngacha D; Rainwater SM; Overbaugh J, Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J. Virol 2006, 80 (2), 835–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Connor RI; Chen BK; Choe S; Landau NR, Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virol 1995, 206 (2), 935–944. [DOI] [PubMed] [Google Scholar]
- 58.He J; Choe S; Walker R; di MP; Morgan DO; Landau NR, Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J.Virol 1995, 69 (11), 6705–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adachi A; Gendelman HE; Koenig S; Folks T; Willey R; Rabson A; Martin MA, Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J.Virol 1986, 59 (2), 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rimsky LT; Shugars DC; Matthews TJ, Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J Virol 1998, 72 (2), 986–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Balamane M; Varghese V; Melikian GL; Fessel WJ; Katzenstein DA; Shafer RW, Panel of prototypical recombinant infectious molecular clones resistant to nevirapine, efavirenz, etravirine, and rilpivirine. Antimicrob Agents Chemother 2012, 56 (8), 4522–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reuman EC; Bachmann MH; Varghese V; Fessel WJ; Shafer RW, Panel of prototypical raltegravir-resistant infectious molecular clones in a novel integrase-deleted cloning vector. Antimicrob Agents Chemother 2010, 54 (2), 934–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Varghese V; Mitsuya Y; Fessel WJ; Liu TF; Melikian GL; Katzenstein DA; Schiffer CA; Holmes SP; Shafer RW, Prototypical Recombinant Multi-Protease-Inhibitor-Resistant Infectious Molecular Clones of Human Immunodeficiency Virus Type 1. Antimicrob Agents Chemother 2013, 57 (9), 4290–4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta P; Lackman-Smith C; Snyder B; Ratner D; Rohan LC; Patton D; Ramratnam B; Cole AM, Antiviral activity of retrocyclin RC-101, a candidate microbicide against cell-associated HIV-1. AIDS Res. 2013, 29 (2), 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lackman-Smith C; Osterling C; Luckenbaugh K; Mankowski M; Snyder B; Lewis G; Paull J; Profy A; Ptak RG; Buckheit RW Jr.; Watson KM; Cummins JE Jr.; Sanders-Beer BE, Development of a comprehensive human immunodeficiency virus type 1 screening algorithm for discovery and preclinical testing of topical microbicides. Antimicrob. Agents Chemother 2008, 52 (5), 1768–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao Q; Ma L; Jiang S; Lu H; Liu S; He Y; Strick N; Neamati N; Debnath AK, Identification of N-phenyl-N’-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamides as a new class of HIV-1 entry inhibitors that prevent gp120 binding to CD4. Virol 2005, 339 (2), 213–225. [DOI] [PubMed] [Google Scholar]
- 67.Belov DS; Ivanov VN; Curreli F; Kurkin AV; Altieri A; Debnath AK, Synthesis of 5-arylpyrrole-2-carboxylic acids as key intermediates for NBD series HIV-1 entry inhibitors. Synthesis 2017, 49 (16), 3692–3699. [Google Scholar]