Abstract
Hepatocellular carcinoma (HCC) is one of the most common types of malignant tumor, and is the second highest cause of cancer-associated mortality, behind lung carcinoma. It is urgent to identify novel genes that can be used to confirm the diagnosis and prognosis of patients with HCC. The present study aimed to investigate the expression pattern of phosphatidylinositol glycan anchor biosynthesis class C (PIGC) in HCC and assess its clinical prognostic significance. Bioinformatics analyses were used to investigate PIGC mRNA expression levels in HCC and adjacent non-cancerous tissue samples. Furthermore, the present study detected the expression levels of PIGC protein in HCC and matched normal tissue samples via immunohistochemistry, and evaluated the prognostic significance of PIGC protein in HCC. The levels of PIGC mRNA and protein were found to be significantly higher in tissue from patients with HCC compared with non-cancerous liver tissue. The survival analysis showed that the expression levels of PIGC mRNA or protein were associated with the survival of patients with HCC. PIGC protein expression was significantly associated with Tumor-Node-Metastasis stage. A negative correlation between PIGC DNA methylation and mRNA expression was observed (Spearman r=−0.453). PIGC is an oncogene that is negatively regulated by DNA methylation, and high levels of PIGC mRNA or protein may predict an unfavorable prognosis in patients with HCC.
Keywords: phosphatidylinositol glycan anchor biosynthesis class C, hepatocellular carcinoma, immunohistochemistry prognosis
Introduction
Primary hepatocellular carcinoma (HCC) is the second most common cause of cancer-associated mortality worldwide (1), with exceedingly high rates in South Asia and sub-Saharan Africa (2). Infection with hepatitis B and C virus (3,4), long-term alcoholism, non-alcoholic fatty liver disease and certain hereditary metabolic diseases contribute to the high incidence rates of HCC (5). The risk of metastasis, which leads to a high incidence of early postoperative recurrence, makes HCC one of the most lethal types of cancer. Despite the fact that several advanced techniques have been successfully applied in the clinical management of patients with HCC (6), the long-term prognosis of patients with HCC remains unsatisfactory. Therefore, it is essential to investigate novel genes or molecules with the aim of developing new therapeutic techniques, and improving the survival rate of patients with HCC.
Phosphatidylinositol glycan anchor biosynthesis class C (PIGC) encodes an endoplasmic reticulum (ER)-associated protein that is vital for the biosynthesis of glycosylphosphatidylinositol (GPI) (7). GPI is utilized to anchor a variety of membrane proteins for expression on the surface of eukaryotic cells. GPI anchoring is a process involved in post-translational modification, which occurs in the ER (1). Mutations in the PIGC gene result in aberrant GPI anchoring, which contributes to seizures, mental retardation and obesity (8–10). Recent studies have demonstrated that dysregulation of PIGC may be implicated in the pathogenesis of several malignant tumor types. Yang et al (11) detected PIGC mutations in the pancreatic ductal adenocarcinoma cell line AsPC-1 and observed that the mutations of PIGC enhanced the motility of cancer cells. Furthermore, Kim et al (12) used Helicobacter pylori-released proteins (G27 strain) to stimulate AGS gastric cancer cells. PIGC was found to be upregulated and may be associated with the transferase activity of cancer cells (12). However, to the best of our knowledge, the expression profile of PIGC in HCC has not yet been investigated and the mechanism underlying its dysregulation in cancer remains unclear. The present study investigated the associations between expression of PIGC and the clinical features of HCC. Furthermore, the present study also aimed to gain an improved understanding of the way in which PIGC is associated with the clinical prognosis of patients with HCC.
Materials and methods
Bioinformatics analysis
PIGC mRNA expression and clinical information data were downloaded from The Cancer Genome Atlas-Liver Hepatocellular Carcinoma database (TCGA-LIHC, http://tcga-data.nci.nih.gov/tcga/) and Cbioportal (http://www.cbioportal.org/). Combined with the two databases, a total of 370 cases of HCC with information on the clinical characteristics and PIGC expression were included in the present study. The expression levels of PIGC mRNA in liver cancer and normal tissues were investigated using Oncomine gene expression array datasets (https://www.oncomine.org). Furthermore, the methylation status of PIGC in HCC tissues and adjacent normal tissues was evaluated using the methHC database (http://methHC.mbc.nctu.edu.tw/php/index.php). The prognostic value of PIGC mRNA in HCC was further verified using Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/). Additionally, in order to investigate the potential roles of PIGC in the pathogenesis of cancer, the present study performed a protein-protein interaction (PPI) analysis of PIGC using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://string-db.org/cgi/input.pl) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of PPI proteins closely associated with PIGC.
Patients and follow-up
Liver cancer and matched adjacent tissue samples were obtained from 97 patients with HCC who received surgical resection at the Surgery Department of Renmin Hospital of Wuhan University (Wuhan, China) between January 2013 and December 2015. All patients provided written informed consent. Out of the 97 patients, three passed away within 1 month after surgery, two patients underwent radiotherapy and four patients received chemotherapy prior to surgery. Thus, these nine patients were excluded from the study. Clinicopathological features, including age, sex, Tumor-Node-Metastasis (TNM) stage, grade (G) stage and American Joint Committee on Cancer stage, were obtained from the electronic record system. The follow-up initiated on the date of surgical resection and ended in March 2018. Follow-up was performed every 3 months during the first 3 years, and twice per year after 3 years once cases became outpatients or received telephone follow-up. Overall survival (OS) was defined as the period from confirmed diagnosis to the end of follow-up or death. The present study was approved by the Institutional Review Board of Renmin Hospital of Wuhan University.
Construction of tissue microarray and analysis of immunohistochemistry (IHC)
Paraffin-embedded HCC tissues and matched non-cancerous tissues were obtained from the Department of Pathology at Renmin Hospital, Wuhan University. The most characteristic liver cancerous and non-cancerous tissue samples were selected to construct a tissue microarray (TMA). The TMA was constructed in line with a previous study (13); a 1.5-mm diameter core for each sample was punched into the TMA. In total, the TMA consisted of 88 HCC and corresponding adjacent tissue samples. The expression of PIGC was assessed using a semi-quantitation method according to the intensity of staining (IS) and the area of positivity (AP) (14). IS was classified as follows: 0, negative; 1, weak; 2, moderate; and 3, strong. AP depended on the percentage of positive-stained cells as follows: 0, 0%; 1, <50% PIGC+ cells; 2, 50–75% PIGC+ cells; and 3, >75% PIGC+ cells. The H-score was calculated by multiplying the IS with the AP. X-tile 3.6.1 software (15) (Yale University, New Haven, CT, USA) was used to determine the optimal cut-off values for the expression of PIGC.
IHC assay
The process of IHC staining was performed as previously described (16). Deparaffinized TMA was washed in 3% H2O2, then subjected to antigen retrieval with citric acid (pH, 6.0). After an overnight incubation at 4°C with primary antibody PIGC (1:50; cat. no. HPA036663; Sigma Aldrich; Merck KGaA), the TMA was incubated for 15 min at room temperature with horseradish peroxidase-labeled polymer conjugated goat anti-rabbit IgG (1:50; cat. no. KIT-5005; Maxvision Biosciences, Inc.). After incubation for 1 min with 3,3′-diaminobenzidine chromogen (Fuzhou Maixin Biotech Co., Ltd.) the TMA was counterstained with hematoxylin. In the negative control group, PBS was used as the primary antibody.
Statistical analysis
SPSS (version 20.0; IBM Corp.) software was used to perform all statistical analyses. Student's t-tests were used to assess the differential expression levels of PIGC and the clinical characteristics. Pearson's χ2 tests were utilized to evaluate the difference between two groups stratified by median PIGC mRNA expression. Kaplan-Meier estimates (log-rank test) were applied to investigate the association between PIGC expression and clinical outcomes in patients with HCC using median PIGC expression or methylation. Cox regression analysis was performed to assess whether the overexpression of PIGC was an independent risk factor for survival in patients with HCC. P<0.05 was considered to indicate a statistically significant difference.
Results
PIGC transcriptional levels of PIGC are higher in liver cancer compared with normal tissue
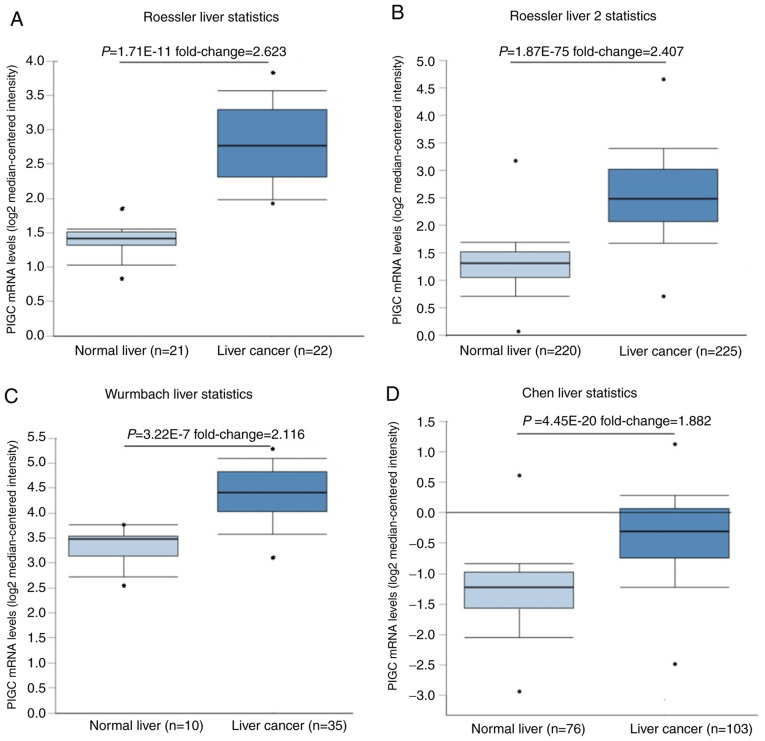
After investigating the transcriptional levels in different types of cancer using the Oncomine database, it was revealed that PIGC expression levels were higher in liver and brain cancer compared with their healthy counterparts, whilst this gene expression was downregulated in HCC compared with normal tissues. The present study exclusively analyzed the levels of PIGC mRNA in HCC in four datasets from the Oncomine database. Using the Roessler Liver Data (P<0.0001; fold change, 2.623), Roessler Liver 2 Data (P<0.0001; fold change, 2.407), Wurmbach Liver Data (P<0.0001; Fold change, 2.116) and Chen Liver Data (P<0.0001; fold change, 1.882), the levels of PIGC expression were observed to be significantly higher in liver cancer compared with that in healthy tissue (Fig. 1), indicating that PIGC is upregulated during the progression of HCC.
Figure 1.
Higher levels of PIGC mRNA in liver cancer than normal tissues. (A) Roessler Liver (B) Roessler Liver 2 (C) Wurmbach Liver and (D) Chen Liver datasets. PIGC, phosphatidylinositol glycan anchor biosynthesis class C.
Associations between PIGC mRNA and clinical characteristics
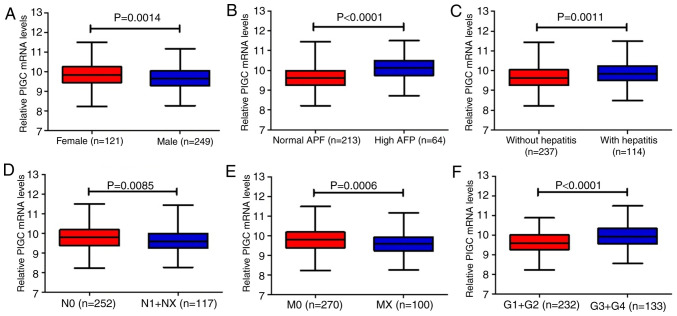
Using data mining from the TCGA (TCGA-LIHC) and the Cbioportal website, the present study included a total of 370 cases of patients with liver cancer with clinical follow-up information and PIGC mRNA expression. By setting the median PIGC mRNA value as the cut-off, it was revealed that the expression of PIGC was significantly associated with sex (χ2=6.496; P=0.011), AFP level (χ2=22.544; P<0.0001), hepatitis (χ2=5.716; P=0.017), G stage (χ2=14.978; P<0.0001), N stage (χ2=5.345; P=0.021) and M stage (χ2=9.264; P=0.002) (Fig. 2). However, no significant associations between the levels of PIGC mRNA and age, alcohol consumption or TNM stage were observed (Table I).
Figure 2.
Associations between PIGC mRNA expression and clinical characteristics. (A) Sex, (B) AFP level, (C) hepatitis, (D) N stage, (E) M stage and (F) G stage data were examined using a Student's t-test. PIGC, phosphatidylinositol glycan anchor biosynthesis class C; AFP, α-feroprotein.
Table I.
Clinical characteristics of HCC stratified by PIGC mRNA expression.
| PIGC mRNA expression | ||||
|---|---|---|---|---|
| Clinical parameter | Low | High | χ2 value | P-value |
| Age, years | 0.302 | 0.583 | ||
| ≤55 | 60 | 65 | ||
| >55 | 125 | 120 | ||
| Sex | 6.496 | 0.011a | ||
| Male | 136 | 113 | ||
| Female | 49 | 72 | ||
| AFP level | 22.544 | <0.001a | ||
| Normal | 122 | 91 | ||
| High | 15 | 49 | ||
| Hepatitis | 5.716 | 0.017a | ||
| No | 130 | 107 | ||
| Yes | 47 | 67 | ||
| Drinking | 3.612 | 0.057 | ||
| No | 118 | 135 | ||
| Yes | 67 | 50 | ||
| T stage | 1.793 | 0.181 | ||
| T1+T2 | 145 | 134 | ||
| T3+T4 | 39 | 50 | ||
| N stage | 5.354 | 0.021a | ||
| N0 | 116 | 136 | ||
| N1+NX | 69 | 48 | ||
| M stage | 9.264 | 0.002a | ||
| M0 | 63 | 37 | ||
| M1+MX | 122 | 148 | ||
| G stage | 14.978 | <0.001a | ||
| G1+G2 | 134 | 98 | ||
| G3+G4 | 51 | 87 | ||
| TNM stage | 1.502 | 0.22 | ||
| I+II | 133 | 123 | ||
| III+IV | 40 | 50 | ||
P<0.05. HCC, hepatocellular carcinoma; PIGC, phosphatidylinositol glycan anchor biosynthesis class C; TNM, tumor node metastasis; AFP, α-feroprotein; G, grade.
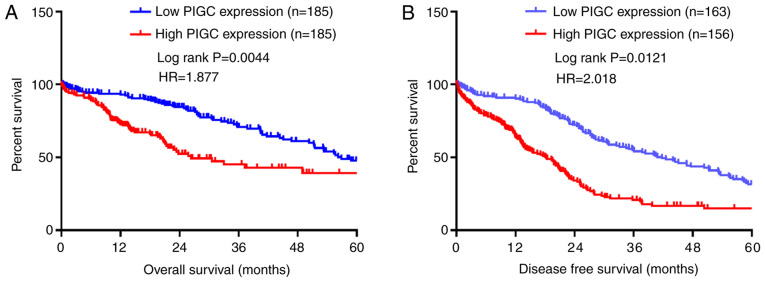
High expression levels of PIGC mRNA predicts worse survival in patients with HCC
Based on the Kaplan-Meier plots (log-rank test), it was demonstrated that a high expression level of PIGC was significantly associated with an unfavorable OS [hazard ratio (HR)=1.877; P=0.0044]. Similarly, it was revealed that the overexpression of PIGC in HCC was significantly associated with decreased disease-free survival (DFS) (HR=2.018; P=0.0121 (Fig. 3A and B). Following the multivariate analysis with Cox regression, the results showed that high expression of PIGC was an independent prognostic factor for poor OS [HR=2.203; 95% confidence interval (CI)=1.476–3.287; P<0.0001] and DFS (HR=1.525; 95% CI=1.114–2.087; P=0.008) in patients with HCC (Tables II and III), suggesting that PIGC may be a novel marker with prognostic significance for HCC. Taken together, the aforementioned results suggested that PIGC mRNA may be a potential independent prognostic marker for patients with HCC.
Figure 3.
High level of PIGC mRNA predicts an unfavorable survival in patients with HCC. (A) High level of PIGC mRNA may predict worse OS. (B) High level of PIGC mRNA may predict shorter DFS. PIGC, phosphatidylinositol glycan anchor biosynthesis class C; OS, overall survival; DFS, disease-free survival.
Table II.
Cox regression analyses of PIGC mRNA level and OS.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Parameter | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age, years | 1.152 (0.795–1.67) | 0.455 | – | – |
| ≤55 | ||||
| >55 | ||||
| Sex | 0.816 (0.573–1.163) | 0.260 | – | – |
| Male | ||||
| Female | ||||
| AFP level | 1.056 (0.646–1.727) | 0.828 | – | – |
| Normal | ||||
| High | ||||
| Hepatitis | 0.562 (0.37–0.816) | 0.007a | 0.665 (0.411–1.075) | 0.096 |
| No | ||||
| Yes | ||||
| Drinking | 1.026 (0.704–1.479) | 0.892 | – | – |
| No | ||||
| Yes | ||||
| T stage | 2.489 (1.75–3.54) | <0.0001a | 1.375 (0.396–4.778) | 0.616 |
| T1+T2 | ||||
| T3+T4 | ||||
| N stage | 1.152 (1.049–2.179) | 0.027a | 1.14 (0.663–1.961) | 0.635 |
| N0 | ||||
| N1+NX | ||||
| M stage | 1.563 (1.0792.264) | 0.018a | 1.686 (0.972–2.926) | 0.063 |
| M0 | ||||
| M1+MX | ||||
| G stage | 1.004 (0.772–1.306) | 0.977 | – | – |
| G1+G2 | ||||
| G3+G4 | ||||
| TNM stage | 2.488 (1.689–3.548) | <0.0001a | 1.74 (0.506–5.985) | 0.380 |
| I+II | ||||
| III+IV | ||||
| PIGC mRNA | 1.654 (1.166–2.347) | 0.005a | 2.203 (1.476–3.287) | <0.0001a |
| Low | ||||
| High | ||||
P<0.05. HCC, hepatocellular carcinoma; PIGC, phosphatidylinositol glycan anchor biosynthesis class C; TNM, tumor node metastasis; AFP, α-feroprotein; OS, overall survival; HR, hazard ratio; CI, confidence interval; G, grade.
Table III.
Cox regression analysis of PIGC mRNA level and DFS.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Parameter | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age, years | 0.966 (0.707–1.321) | 0.828 | – | – |
| ≤55 | ||||
| >55 | ||||
| Sex | 0.885 (0.645–1.124) | 0.45 | – | – |
| Male | ||||
| Female | ||||
| AFP level | 1.108 (0.734–1.673) | 0.625 | – | – |
| Normal | ||||
| High | ||||
| Hepatitis | 0.889 (0.643–1.228) | 0.475 | – | – |
| No | ||||
| Yes | ||||
| Drinking | 1.011 (0.732–1.395) | 0.948 | – | – |
| No | ||||
| Yes | ||||
| T stage | 2.270 (1.643–3.135) | <0.001a | 1.088 (0.432–2.738) | 0.857 |
| T1+T2 | ||||
| T3+T4 | ||||
| N stage | 1.249 (0.904–1.724) | 0.178 | – | – |
| N0 | ||||
| N1+NX | ||||
| M stage | 1.151 (0.826–1.602) | 0.406 | – | – |
| M0 | ||||
| M1+MX | ||||
| G stage | 1.105 (0.811–1.507) | 0.526 | – | – |
| G1+G2 | ||||
| G3+G4 | ||||
| TNM stage | 2.354 (1.692–3.275) | <0.001a | 2.156 (0.874–5.317) | 0.095 |
| I+II | ||||
| III+IV | ||||
| PIGC mRNA | 1.493 (1.107–2.012) | 0.009a | 1.525 (1.114–2.087) | 0.008a |
| Low | ||||
| High | ||||
P<0.05. HCC, hepatocellular carcinoma; PIGC, phosphatidylinositol glycan anchor biosynthesis class C; TNM, tumor node metastasis; AFP, α-feroprotein; DFS, disease-free survival; HR, hazard ratio; CI, confidence interval; G, grade.
PIGC expression is negatively regulated by DNA methylation
By using methHC, the present study generated a contrastive figure showing that the levels of DNA methylation were significantly higher in liver tumor tissues compared with that in healthy tissues (transcript:NM_002642; P<0.005; Fig. S1A). In addition, by investigating the association between PIGC expression and DNA methylation, the present study observed a negative correlation (Spearman's rho=−0.453), indicating that PIGC expression gradually decreased with an increase in PIGC promoter DNA methylation (Fig. S1B). Furthermore, data on PIGC promoter methylation was downloaded from the Cbioportal website and upon analysis, the results matched the follow-up information with TCGA-LIHC data. The present study also investigated whether PIGC methylation status was associated with survival outcomes of patients with HCC. In the two-group analysis stratified by the median status of PIGC DNA methylation, no associations between PIGC methylation and OS or DFS were observed (Fig. S2).
Expression of PIGC protein is upregulated in HCC
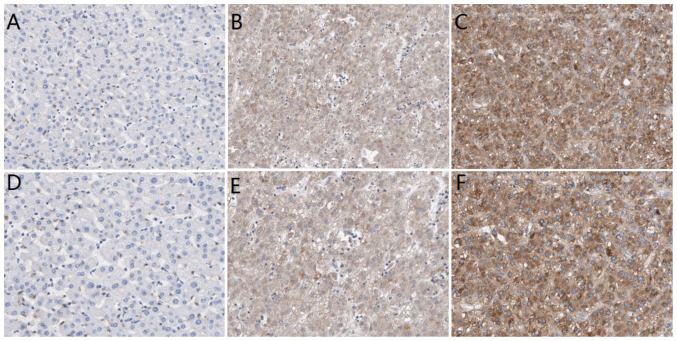
In order to analyze the associations between PIGC protein expression and clinicopathological parameters, the present study set the cut-off value of H-score as 5 points according to Kaplan-Meier curves. If the H-score was <5 points, then the result was defined as low expression; if the H-score was >5 points, then the result was regarded as high expression. The IHC results revealed that PIGC protein was expressed in liver cancerous and non-cancerous tissues, as demonstrated by the brown particles localized in the cytoplasmic membranous with no staining of nucleus (Fig. 4). However, the staining intensity in HCC and non-cancerous tissues was markedly different. In 88 cases of liver cancerous tissues, 63 cases (71.59%) showed high PIGC expression and 25 cases (28.41%) showed low PIGC expression. However, 88 cases of matched tissues exhibited low expression levels of PIGC protein without any case of adjacent non-cancerous tissue showing strong staining. These results suggest that the PIGC protein is highly expressed in HCC tissue and expressed at low levels in corresponding non-cancerous tissues. Thus, PIGC protein may be a useful biomarker for the diagnosis of liver cancer. The χ2 analysis showed that PIGC was significantly associated with N stage (χ2=5.099; P=0.024), M stage (χ2=6.974; P=0.008) and TNM stage (χ2=4.985; P=0.026), but there were no significant associations observed with G stage, T stage, age or sex (Table IV).
Figure 4.
Expression of PIGC protein in liver cancer and normal tissues. Low PIGC protein expression in (A) healthy tissue (B) and HCC tissue. (C) High PIGC protein expression in HCC tissue (×200 magnification). Low PIGC protein expression in (D) healthy tissue (E) and HCC tissue. (F) High PIGC protein expression in HCC tissue (×400 magnification). PIGC, phosphatidylinositol glycan anchor biosynthesis class C; HCC, hepatocellular carcinoma.
Table IV.
Associations between PIGC protein expression and clinical features.
| PIGC expression | ||||
|---|---|---|---|---|
| Factor | High (n=63) | Low (n=25) | χ2 value | P-value |
| Sex | 0.880 | 0.348 | ||
| Male | 42 | 14 | ||
| Female | 21 | 11 | ||
| Age, years | 0.503 | 0.478 | ||
| <60 | 33 | 11 | ||
| ≥60 | 30 | 14 | ||
| G stage | 1.267 | 0.260 | ||
| G1+G2 | 23 | 6 | ||
| G3+G4 | 40 | 19 | ||
| T stage | 0.329 | 0.566 | ||
| T1-T2 | 14 | 7 | ||
| T3-T4 | 49 | 18 | ||
| N stage | 5.099 | 0.024a | ||
| N0 | 19 | 14 | ||
| N1-N2 | 44 | 11 | ||
| M stage | 6.974 | 0.008a | ||
| M0 | 44 | 24 | ||
| M1 | 19 | 1 | ||
| TNM stage | 4.985 | 0.026a | ||
| I–II | 17 | 13 | ||
| III–IV | 46 | 12 | ||
P<0.05. PIGC, phosphatidylinositol glycan anchor biosynthesis class C; TNM, tumor node metastasis; G, grade.
Prognostic value of PIGC protein and Cox regression analysis
Kaplan-Meier analysis was performed to investigate the association between PIGC protein expression and OS in patients with HCC. The Kaplan-Meier survival curve revealed that patients with HCC with high expression levels of PIGC exhibited a worse OS compared with those with low PIGC expression (P=0.0324; Fig. 5). The univariate Cox regression analysis model revealed that PIGC protein expression (HR=3.311; 95% CI=1.156–9.486; P=0.026) and M stage (HR=2.618; 95% CI=1.247–5.496; P=0.011) were prognostic factors for patients with HCC. However, the results showed that PIGC protein expression was not an independent prognostic factor in patients with HCC following the multivariate Cox regression analysis (HR=2.697; 95% CI=0.909–8.01; P=0.074 (Table V).
Figure 5.
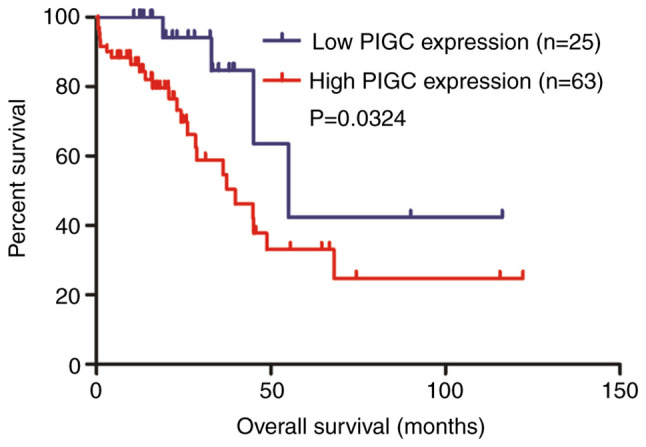
High PIGC expression predicts a worse OS in patients with HCC. PIGC, phosphatidylinositol glycan anchor biosynthesis class C; OS, overall survival; HCC, hepatocellular carcinoma.
Table V.
Cox regression analysis for OS in 88 hepatocellular carcinoma cases.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Factor | HR (95%CI) | P-value | HR (95%CI) | P-value |
| Sex | 0.746 (0.366–0.746) | 0.420 | ||
| Age | 1.04 (0.512–2.11) | 0.914 | ||
| G stage | 0.878 (0.424–1.818) | 0.726 | ||
| T stage | 1.569 (0.598–4.118) | 0.361 | ||
| N stage | 1.261 (0.593–2.682) | 0.547 | ||
| M stage | 2.618 (1.247–5.496) | 0.011 | 2.206 (0.942–4.356) | 0.071 |
| TNM stage | 1.204 (0.566–2.56) | 0.629 | ||
| PIGC expression | 3.311 (1.156–9.486) | 0.026 | 2.697 (0.909–8.01) | 0.074 |
PIGC, phosphatidylinositol glycan anchor biosynthesis class C; TNM, tumor node metastasis; OS, overall survival; HR, hazard ratio; CI, confidence interval; G, grade.
PPI network analysis
The STRING database was utilized to reveal the PPI information about PIGC. As presented in Fig. S3, it was revealed that the PPI network of PIGC consisted of 21 nodes and 104 edges (PPI enrichment, P<1.0×10−16). Furthermore, KEGG pathway enrichment analysis was performed in the present study. According to five KEGG pathways (Table VI), it was revealed that PIGC is primarily involved in ‘GPI-anchor biosynthesis’ [false discovery rate (FDR)=1.72×10−32]. It was also observed that 3 genes (CDIPT, CDS1 and CDS2) may be involved in the phosphatidylinositol signaling pathway (FDR=0.00484), which is highly associated with the proliferation and metastasis of malignant tumors. The KEGG pathway enrichment analysis indicated that PIGC may participate in the occurrence and progression of HCC.
Table VI.
KEGG pathway enrichment analysis of PIGC from STRING.
| Pathway ID | Pathway description | Observed gene count | False discovery rate | Matching proteins in network |
|---|---|---|---|---|
| 563 | Glycosylphosphatidylinositol-anchor biosynthesis | 13 | 1.72×10−32 | DPM2, GPAA1, PIGA, PIGB, PIGC, PIGF, PIGG, PIGH, PIGL, PIGO, PIGP, PIGQ, PIGY |
| 1100 | Metabolic pathways | 19 | 4.98×10−20 | ALG1, CDIPT, CDS1, CDS2, DPAGT1, DPM1, DPM2, DPM3, GPAA1, PIGA, PIGB, PIGC, PIGF, PIGH, PIGL, PIGO, PIGP, PIGQ, PIGY |
| 510 | N-Glycan biosynthesis | 5 | 1.09×10−07 | ALG1, DPAGT1, DPM1, DPM2, DPM3 |
| 4070 | Phosphatidylinositol signaling system | 3 | 4.84×10−03 | CDIPT, CDS1, CDS2 |
| 564 | Glycerophospholipid metabolism | 3 | 6.13×10−03 | CDIPT, CDS1, CDS2 |
PIGC, phosphatidylinositol glycan anchor biosynthesis class C; STRING, Search Tool for the Retrieval of Interacting Genes/Proteins; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Discussion
To the best of our knowledge, the present study is the first to demonstrate the association between the expression of PIGC and prognosis significance of patients with HCC. The present study revealed that PIGC was highly expressed in HCC, both in terms of mRNA and protein levels, and the upregulation was significantly associated with unfavorable prognosis in patients with HCC. Subsequently, a negative association between DNA methylation and PIGC expression was observed according to the Cbioportal website, implying that PIGC expression is negatively regulated by DNA methylation. Based on the aforementioned results, the present study concluded that PIGC is an oncogene regulated by DNA methylation, and may be utilized as a potential therapeutic target for the treatment of HCC.
It has been well established that there are 10 genes encoding enzymes in the process of GPI synthesis that have already been discovered, and PIGC is one of these important genes (7). The transfer of N-acetylglucosamine from UDP-GlcNAc to GPI is the first step in the biosynthesis of GPI, and this crucial step is regulated by PIGC (17). GPI anchoring is essential for embryogenesis, neurogenesis and immune responses. Studies have revealed that mutations of PIGC can lead to the death of embryos and the disruption of PIGC is lethal in yeasts (18,19). Furthermore, GPI anchors are bound to proteins expressed on the surface of eukaryotic cells (20). Over 150 functionally diverse proteins are GPI-anchored, including enzymes, proteins involved in endocytosis (21), adhesion molecules, receptors and complement regulators (20). The overexpression of Glypican 3 (GPC3), a member of GPI-anchored proteoglycans, aids in suppressing hepatocyte proliferation and liver regeneration in transgenic mice (22). Grozdanov et al (23) reported that GPC3 is upregulated in the majority of liver cancer types, but not expressed in normal hepatocytes and benign liver disease. They concluded that GPC3 contributes to the proliferation and metastasis of HCC by activating Wnt signaling. Phosphatidylinositol-glycan biosynthesis class X protein (PIGX) is also an important gene in the biosynthetic pathway of GPI anchoring (23). A recent study reported that PIGX is highly expressed in breast cancer (24). C4.4A, a glycolipid-anchored membrane protein, is overexpressed in patients with esophageal cancer and associated with a poor prognosis (25).
According to the KEGG pathway enrichment analysis, the upregulation of PIGC was associated with the activation of biological processes involved in tumor progression, including the phosphatidylinositol signaling pathway. Phosphatidylinositol signaling, a complicated network of phospholipid messengers and enzymes, represents one of the most substantial signaling systems involved in a variety of lesions, including cancer (26). Previous studies have revealed that the phosphorylation of PI (3–5) P3 mediated by PI3K leads to the recruitment of Akt, causing the activation of downstream signaling cascades (PI3K/Akt) (27,28). PI3K/Akt signaling is an important proliferative pathway involving various cytokines, growth factors and receptors (29). Furthermore, the PI3K/Akt signaling pathway occupies a central position in the regulation of cell invasion and metastasis, and its activation is a crucial feature of the epithelial-mesenchymal transition in the progression of cancer (30). Hence, it is plausible that PIGC may participate in the generation and progression of HCC mediated by phosphatidylinositol signaling (PI3K/Akt pathway), highlighting novel potential targets for therapeutic interventions. Further biochemical, animal and even clinical studies are required in order to clarify the potential mechanism of PIGC involved in the tumorigenesis of HCC.
From the TCGA-LIHC data, the present study observed that PIGC mRNA expression was significantly associated with M and N stages, which was in line with the results from the IHC assay. Although the Cox regression analysis showed that PIGC mRNA overexpression was an independent risk factor for shorter OS, similar results in PIGC protein levels were not observed. The different results between PIGC mRNA and protein may be explained by the following reasons. Firstly, compared with the TCGA-LIHC data, the number of patients with HCC included in the present study from our hospital were relatively small. Secondly, this discrepancy may be due to ethnic differences. Finally, there may be some unknown regulation processes that occur during the translation process of PIGC.
Several limitations exist in the present study. Firstly, as only 88 cases of specimens stained by IHC were included in the present study, the results should be interpreted with caution. Secondly, as only the OS data were recorded and the DFS data were neglected during follow-up, it is not possible to determine the association between PIGC protein expression and the prognosis of patients with HCC. Thirdly, the potential correlations between PIGC methylation and prognostic factors in HCC were not investigated. Finally, the genes co-expressed alongside PIGC were not investigated. Hence, the potential mechanism of PIGC dysregulation in the pathogenesis and development of HCC needs to be further investigated through medical experiments.
In conclusion, PIGC is a novel oncogene regulated by DNA methylation. An elevated expression of PIGC was found to be significantly correlated with an unfavorable survival. PIGC may be utilized as a new prognostic biomarker and is a potential target for personalized medicine in the future.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was funded by Hubei Municipal Health and Family Planning Commission (grant no. WJ2017Q034).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XP and CL contributed to the design of the present study, developed the methodology, collected the bioinformatics data, performed the experiments, analyzed the results and wrote the manuscript. AH, RL and YC contributed to the collection of patient data. XP and WD contributed to the design of the study, critically revised the manuscript and approved the final version to be published. All authors agreed to be accountable for all aspects of the study.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of The Renmin Hospital of Wuhan University (Wuhan, China). Written patient consent was obtained.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, Zhu AX, Murad MH, Marrero JA. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67:358–380. doi: 10.1002/hep.29086. [DOI] [PubMed] [Google Scholar]
- 2.Bertuccio P, Turati F, Carioli G, Rodriguez T, La Vecchia C, Malvezzi M, Negri E. Global trends and predictions in hepatocellular carcinoma mortality. J Hepatol. 2017;67:302–309. doi: 10.1016/j.jhep.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Maucort Boulch D, De Martel C, Franceschi S, Plummer M. Fraction and incidence of liver cancer attributable to hepatitis B and C viruses worldwide. Int J Cancer. 2018;142:2471–2477. doi: 10.1002/ijc.31280. [DOI] [PubMed] [Google Scholar]
- 4.Choi J, Han S, Kim N, Lim YS. Increasing burden of liver cancer despite extensive use of antiviral agents in a hepatitis B virus-endemic population. Hepatology. 2017;66:1454–1463. doi: 10.1002/hep.29321. [DOI] [PubMed] [Google Scholar]
- 5.El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, Yang C, Lu W, Zeng Y. Prognostic significance of glypican-3 expression in hepatocellular carcinoma. Medicine (Baltimore) 2018;97:e9702. doi: 10.1097/MD.0000000000009702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watanabe R, Inoue N, Westfall B, Taron CH, Orlean P, Takeda J, Kinoshita T. The first step of glycosylphosphatidylinositol biosynthesis is mediated by a complex of PIG-A, PIG-H, PIG-C and GPI1. EMBO J. 1998;17:877–885. doi: 10.1093/emboj/17.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edvardson S, Murakami Y, Nguyen TT, Shahrour M, St-Denis A, Shaag A, Damseh N, Le Deist F, Bryceson Y, Abu-Libdeh B, et al. Mutations in the phosphatidylinositol glycan C (PIGC) gene are associated with epilepsy and intellectual disability. J Med Genet. 2017;54:196–201. doi: 10.1136/jmedgenet-2016-104202. [DOI] [PubMed] [Google Scholar]
- 9.Jaeken J, Péanne R. What is new in CDG? J Inherit Metab Dis. 2017;40:569–586. doi: 10.1007/s10545-017-0068-9. [DOI] [PubMed] [Google Scholar]
- 10.Schleinitz D, Klöting N, Lindgren CM, Breitfeld J, Dietrich A, Schön MR, Lohmann T, Dreßler M, Stumvoll M, Mccarthy MI, et al. Fat depot-specific mRNA expression of novel loci associated with waist-hip ratio. Int J Obes (Lond) 2014;38:120–125. doi: 10.1038/ijo.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L, Gao Z, Hu L, Wu G, Yang X, Zhang L, Zhu Y, Wong BS, Xin W, Sy MS, Li C. Glycosylphosphatidylinositol anchor modification machinery deficiency is responsible for the formation of pro-prion protein (PrP) in BxPC-3 protein and increases cancer cell motility. J Biol Chem. 2016;291:3905–3917. doi: 10.1074/jbc.A115.705830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim N, Park WY, Kim JM, Park JH, Kim JS, Jung HC, Song IS. Gene expression of AGS cells stimulated with released proteins by Helicobacter pylori. J Gastroen Hepatol. 2008;23:643–651. doi: 10.1111/j.1440-1746.2007.05241.x. [DOI] [PubMed] [Google Scholar]
- 13.Kawakami F, Sircar K, Rodriguez-Canales J, Fellman BM, Urbauer DL, Tamboli P, Tannir NM, Jonasch E, Wistuba II, Wood CG, et al. Programmed cell death ligand 1 and tumor-infiltrating lymphocyte status in patients with renal cell carcinoma and sarcomatoid dedifferentiation. Cancer. 2017;123:4823–4831. doi: 10.1002/cncr.30937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Y, Song J, Chen J, Xiao J, Ni J, Wu C. Overexpression of NEK3 is associated with poor prognosis in patients with gastric cancer. Medicine (Baltimore) 2018;97:e9630. doi: 10.1097/MD.0000000000009630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 16.Tai CJ, Su TC, Jiang MC, Chen HC, Shen SC, Lee WR, Liao CF, Chen YC, Lin SH, Li LT, et al. Correlations between cytoplasmic CSE1L in neoplastic colorectal glands and depth of tumor penetration and cancer stage. J Transl Med. 2013;11:29. doi: 10.1186/1479-5876-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue N, Watanabe R, Takeda J, Kinoshita T. PIG-C, one of the three human genes involved in the first step of glycosylphosphatidylinositol biosynthesis is a homologue of Saccharomyces cerevisiae GPI2. Biochem Biophys Res Commun. 1996;226:193–199. doi: 10.1006/bbrc.1996.1332. [DOI] [PubMed] [Google Scholar]
- 18.Shamseldin HE, Tulbah M, Kurdi W, Nemer M, Alsahan N, Al Mardawi E, Khalifa O, Hashem A, Kurdi A, Babay Z, et al. Identification of embryonic lethal genes in humans by autozygosity mapping and exome sequencing in consanguineous families. Genome Biol. 2015;16:116. doi: 10.1186/s13059-015-0681-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leidich SD, Kostova Z, Latek RR, Costello LC, Drapp DA, Gray W, Fassler JS, Orlean P. Temperature-sensitive yeast GPI anchoring mutants gpi2 and gpi3 are defective in the synthesis of N-acetylglucosaminyl phosphatidylinositol. Cloning of the GPI2 gene. J Biol Chem. 1995;270:13029–13035. doi: 10.1074/jbc.270.22.13029. [DOI] [PubMed] [Google Scholar]
- 20.Kinoshita T, Fujita M. Biosynthesis of GPI-anchored proteins: Special emphasis on GPI lipid remodeling. J Lipid Res. 2016;57:6–24. doi: 10.1194/jlr.R063313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakhan SE, Sabharanjak S, De A. Endocytosis of glycosylphosphatidylinositol-anchored proteins. J Biomed Sci. 2009;16:93. doi: 10.1186/1423-0127-16-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu B, Bell AW, Paranjpe S, Bowen WC, Khillan JS, Luo JH, Mars WM, Michalopoulos GK. Suppression of liver regeneration and hepatocyte proliferation in hepatocyte-targeted glypican 3 transgenic mice. Hepatology. 2010;52:1060–1067. doi: 10.1002/hep.23794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grozdanov PN, Yovchev MI, Dabeva MD. The oncofetal protein glypican-3 is a novel marker of hepatic progenitor/oval cells. Lab Invest. 2006;86:1272–1284. doi: 10.1038/labinvest.3700479. [DOI] [PubMed] [Google Scholar]
- 24.Nakakido M, Tamura K, Chung S, Ueda K, Fujii R, Kiyotani K, Nakamura Y. Phosphatidylinositol glycan anchor biosynthesis, class X containing complex promotes cancer cell proliferation through suppression of EHD2 and ZIC1, putative tumor suppressors. Int J Oncol. 2016;49:868–876. doi: 10.3892/ijo.2016.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohtsuka M, Yamamoto H, Masuzawa T, Takahashi H, Uemura M, Haraguchi N, Nishimura J, Hata T, Yamasaki M, Miyata H, et al. C4.4A expression is associated with a poor prognosis of esophageal squamous cell carcinoma. Ann Surg Oncol. 2013;20:2699–2705. doi: 10.1245/s10434-013-2900-2. [DOI] [PubMed] [Google Scholar]
- 26.Thapa N, Tan X, Choi S, Lambert PF, Rapraeger AC, Anderson RA. The hidden conundrum of phosphoinositide signaling in cancer. Trends Cancer. 2016;2:378–390. doi: 10.1016/j.trecan.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: Divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2014;15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vanhaesebroeck B, Stephens L, Hawkins P. PI3K signalling: The path to discovery and understanding. Nat Rev Mol Cell Biol. 2012;13:195–203. doi: 10.1038/nrm3290. [DOI] [PubMed] [Google Scholar]
- 29.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song LB, Li J, Liao WT, Feng Y, Yu CP, Hu LJ, Kong QL, Xu LH, Zhang X, Liu WL, et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J Clin Invest. 2009;119:3626–3636. doi: 10.1172/JCI39374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.