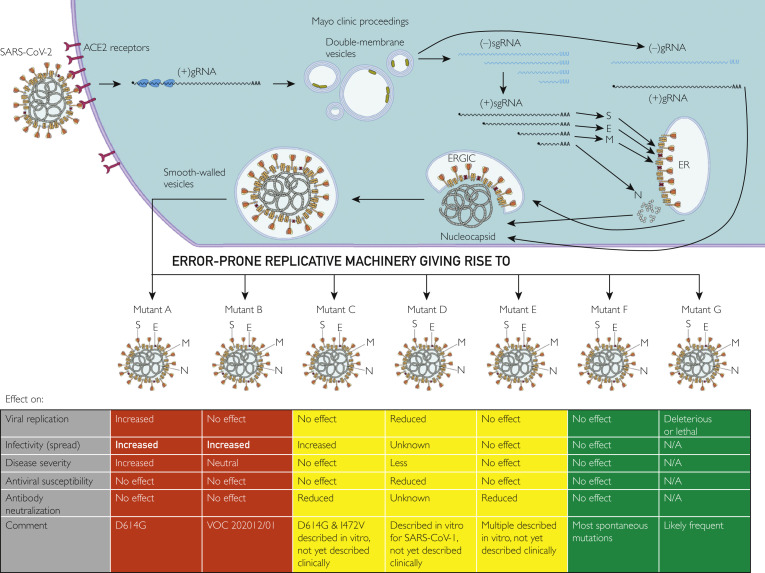
RNA viruses, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), are subject to mutation because of error-prone replication machinery, allowing for potential massive diversity of viral quasispecies. Multiplying by more than 85 million cases suggests that virtually every possible mutation has, or will, eventually be seen. Although “viral mutation” invokes cataclysmic visions of disease even worse than the current ones, reality lies in the principles of viral population evolution, shaped by natural selection, with mutations that are beneficial to the virus being positively selected over time and those that are lethal or deleterious being removed from the population.
The life cycle of any virus involves attachment, penetration, uncoating, replication, assembly, and release. As with all biological processes, each step requires high-affinity protein-protein and protein-RNA interactions. Mutation of any of the proteins can affect the efficiency and kinetics of these interactions, which is then reflected by impaired or enhanced replicative fitness, or the ability of the virus to make more of itself. These mutations can also affect the susceptibility of viral variants to antiviral agents such as remdesivir or therapeutic neutralizing antibodies. Therefore, when evaluating the effect of given viral mutation(s) on human disease, it is essential to interpret mutations in terms of replicative fitness as well as the effect on drug and antibody therapeutic efficacy (Figure ).
Figure.
Cell entry of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) occurs through the interaction of the receptor-binding domain of the spike protein with angiotensin-converting enzyme 2 (ACE2) receptor, which causes cleavage/activation of the spike protein by cellular proteases. After fusion of the viral envelope with the cellular membrane, the viral positive-strand RNA is translated into a polyprotein and cleaved by viral proteases, forming the replicase-transcriptase complex (RTC) within double-membrane vesicles. The RTC that contains error-prone RNA-dependent RNA polymerase then mediates viral RNA replication, which in turn leads to viral protein synthesis and to production of more viral RNA. Viral proteins translocate to the endoplasmic reticulum (ER), in which they assemble in the ER-Golgi intermediate compartment (ERGIC), and daughter virions are released by exocytosis. These daughter virions often contain mutations due to the error-prone nature of the replication cycle, and these mutants may alter different aspects of coronavirus disease 2019 pathophysiology, which can be good (green), bad (red), or indifferent (yellow) to the human host. E, envelope; M, membrane; N, nucleocaspsid; N/A, not applicable; S, spike; (+)gRNA, viral positive strand genomic RNA; (−)gRNA, viral negative strand genomic RNA; (+)sgRNA, positive strand (+) subgenomic RNA; (−)sgRNA, negative strand subgenomic RNA.
One important SARS-CoV-2 viral mutation is the spike protein aspartic acid to glycine substitution at position 614 (D614G). Using in vitro infection models and animal models of disease, this variant is associated with enhanced competitive fitness in human cells and it transmits faster than wild-type virus.1 Consequently, the D614G isolate has become a dominant clinical strain with up to 70% of clinical isolates containing the signature mutation.2
It is important to consider the effect of SARS-CoV-2 mutations not only in terms of viral fitness but also with regard to susceptibility to antibody neutralization—whether or not a virus strain is susceptible to being neutrealized by antibodies, there is great therapeutic importance especially for therapies that rely on antibody neutralization (eg, vaccination, monoclonal antibodies, and plasma). A recent study has evaluated the relationship between viral fitness and antibody neutralization of a diverse panel of SARS-CoV-2 mutants.3 An important finding of this study is that SARS-CoV-2 isolates containing the D614G mutation are equally inhibited by convalescent plasma as wild-type strains,3 a finding that has since been reproduced.1
More recently, attention has focused on SARS-CoV-2 strains B.1.1.7, which is now the dominant viral quasispecies in the United Kingdom,4 with 98% of viral isolates containing the signature mutations,5 and B.1.351 isolated first in South Africa, and both of these appear to have enhanced spread.4 Each virus contains multiple mutations, but they share the N501Y mutation in the receptor-binding domain, which thankfully maintains susceptibility to neutralizing monoclonal antibodies in vitro.6
Reassuringly, of the 106 different mutations that they tested, only 10 variants had decreased sensitivity to convalescent plasma or neutralizing antibodies, and only 1 (D614G plus I472V) exhibited the worrisome combination of both enhanced replicative fitness and decreased susceptibility to antibody neutralization.3 Separately, a study of SARS-CoV-2 escape from therapeutic monoclonal antibodies identified 3 variants that escaped bamlanivimab but 1 mutant (E406W) that escaped the dual antibody therapy with casirivimab and imdevimab,7 further supporting the contention that antibody escape is likely a rare event. The replicative fitness of those variants has not yet been determined.
What about the possibility of selecting for remdesivir resistance, because treating viral infections with a single antiviral agent will often select for drug-resistant variants? Serial passaging of Ebola virus–infected cultured under pressure with remdesivir results in an F548S substitution, which confers resistance to remdesivir,8 and similar experiments in SARS-CoV-1–infected cultures select mutations F476L and V553L, which, when engineered into Middle East respiratory syndrome-related coronavirus, confers remdesivir resistance as well.9 This very concerning result is tempered by the observations that F480L and V557L SARS-CoV-1 variants have reduced viral fitness in cell culture and attenuated SARS-CoV-1 pathogenesis in a mouse model, altogether indicating that the evolutionary cost of remdesivir resistance is a less pathogenic virus.9 Consistent with those observations, a recent survey of more than 90,000 circulating human SARS-CoV-2 isolates identified mutations associated with remdesivir resistance in only 2 cases (0.002%).10
The most effective way to both control the SARS-CoV-2 pandemic and limit the emergence of viral variants is either to prevent infection (eg, with vaccines and public health interventions) or to diagnose early and treat those who become infected with agents that block the viral life cycle (eg, antibody-based approach or antiviral agents). Our current understanding of SARS-CoV-2 mutation, evolution, and the interrelated attributes of fitness, spread, and susceptibility to antibody neutralization/antiviral agents suggests that when antibody or antiviral escape mutants arise, most often they do so at a cost to replicative fitness. Currently, it appears that the virus is spreading faster than it evolves. However, one must temper this observation with the fact that 13 months represents a relatively short time in evolutionary terms. It is possible that if the pandemic persists for a much longer period, we may begin to see additional adaptive mutations that limit the efficacy of available therapies.
Footnotes
Potential Competing Interests: Dr Badley is supported by grants from the National Institute of Allergy and Infectious Diseases (nos. AI110173 and AI120698), amFAR (#109593), and Mayo Clinic (HH Shieck Khalifa Bib Zayed Al-Nahyan Named Professorship of Infectious Diseases). He is a paid consultant for AbbVie and Flambeau Diagnostics, is a paid member of the DSMB for Corvus Pharmaceuticals and Equilium, owns equity for scientific advisory work in Zentalis and nference, and is founder and President of Splissen Therapeutics.
Supplemental Online Material
References
- 1.Hou Y.J., Chiba S., Halfmann P. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370(6523):1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L., Jackson C.B., Mou H. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun. 2020;11(1):6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q., Wu J., Nie J. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.COVID-19: emerging SARS-CoV-2 variants. Centers for Disease Control and Prevention website. https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html Accessed January 27, 2021. [PubMed]
- 5.Volz E., Mishra S., Chand M. Transmission of SARS-CoV-2 lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. https://doi.org/10.1101/2020.12.30.20249034 [published online ahead of print January 2, 2021]. medRxiv.
- 6.Weisblum Y., Schmidt F., Zhang F. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Starr T.N., Greaney A.J., Addetia A. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. https://doi.org/10.1101/2020.11.30.405472 [published online ahead of print December 1, 2020]. bioRxiv. [DOI] [PMC free article] [PubMed]
- 8.Lo M.K., Albariño C.G., Perry J.K. Remdesivir targets a structurally analogous region of the Ebola virus and SARS-CoV-2 polymerases. Proc Natl Acad Sci U S A. 2020;117(43):26946–26954. doi: 10.1073/pnas.2012294117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agostini M.L., Andres E.L., Sims A.C. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9(2) doi: 10.1128/mBio.00221-18. e00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin R., Perry J., Cihlar T., Mo H., Porter D.P., Svarovskaia E.S. Genetic conservation of SARS-CoV-2 RNA replication complex in globally circulating isolates from humans and minks predicts minimal pre-existing resistance to remdesivir. https://doi.org/10.1101/2020.12.19.423600 [published online ahead of print December 19, 2020]. bioRxiv. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.