Abstract
Diamond Blackfan anemia (DBA) is a ribosomopathy of variable expressivity and penetrance characterized by red cell aplasia, congenital anomalies, and predisposition to certain cancers, including early onset colorectal cancer (CRC). DBA is primarily caused by a dominant mutation of a ribosomal protein (RP) gene, although approximately 20% of patients remain genetically uncharacterized despite exome sequencing and copy number analysis. While somatic loss-of-function mutations in RP genes have been reported in sporadic cancers, with the exceptions of 5q- myelodysplastic syndrome (RPS14) and microsatellite unstable CRC (RPL22), these cancers are not enriched in DBA. Conversely, pathogenic variants in RPS20 were previously implicated in familial CRC; however, none of the reported individuals had classical DBA features. We describe two unrelated children with DBA lacking variants in known DBA genes who were found by exome sequencing to have de novo novel missense variants in RPS20. The variants affect the same amino acid but result in different substitutions and reduce RPS20 protein level. Yeast models with mutation of the cognate residue resulted in defects in growth, ribosome biogenesis, and polysome formation. These findings expand the phenotypic spectrum of RPS20 mutation beyond familial CRC to include DBA, which itself is associated with increased risk of CRC.
Keywords: Diamond Blackfan anemia, RPS20, ribosomopathy, familial colorectal cancer, yeast model
INTRODUCTION
Diamond Blackfan anemia (DBA) is a genetic disorder characterized by red cell aplasia and macrocytic anemia typically presenting in the first year of life (Da Costa, Narla, & Mohandas, 2018). Additional associated features include craniofacial, thumb, renal, and cardiac anomalies, and short stature. Underlying these phenotypes is a disorder of ribosomes caused primarily by heterozygous germline pathogenic variants in a small or large ribosomal subunit protein gene (RPS or RPL, respectively). To date, 19 ribosomal protein (RP) genes have been implicated in the pathogenesis of DBA, 10 which encode components of the small, 40S ribosomal subunit, and 9, which encode components of the large, 60S ribosomal subunit (see Supp. Table S1 for genes and MIM#s). The incidence of specific gene involvement varies; however, only 9 genes (RPS19, RPL5, RPS26, RPL11, RPL35A, RPL15, RPS24, RPS17, and RPS10) are mutated in one percent or more of large cohorts of patients with DBA (Quarello et al., 2020; Ulirsch et al., 2018). In addition, rare individuals with DBA have been found to have pathogenic variants in the X-linked genes GATA1 and TSR2, which encode a hematopoietic master transcription factor required for normal erythropoiesis and an RPS26 chaperone protein, respectively (Khajuria et al., 2018; Ludwig et al., 2014). Lastly, some patients who carry an initial diagnosis of DBA have been found to have biallelic pathogenic variants in ADA2 (formerly known as CECR1) (Supp. Table S1), which encodes adenosine deaminase 2, although the mechanism by which this leads to a DBA-like disease is unknown (Ben-Ami et al., 2016). While single nucleotide variants are most common, approximately 10–20% patients harbor large deletions or insertions that disrupt RP gene expression (Farrar et al., 2011).
A recent report described the results of exome sequencing with copy number variant analysis of 472 individuals with DBA (Ulirsch et al., 2018). Rare and predicted deleterious variants in one of the known DBA genes were detected in 78% of cases. Notably, nine mutations in seven previously unreported RP genes (RPL3, RPL34, RPL10, RPL10A, RPL19, RPLP0, and RPS11) were also identified, however, these were not further evaluated at the time of the report. Finally, an additional 13 individuals in that cohort were found to have variants causative of other rare genetic disorders associated with anemia. Thus, even with comprehensive diagnostics, approximately 20% of patients with red cell aplasia and a clinical diagnosis of DBA remain uncharacterized genetically.
Germline pathogenic variants in DBA-associated RP genes are also associated with cancer predisposition. Registry data indicate that individuals with DBA have an increased risk of myelodysplastic syndrome (MDS), acute myeloid leukemia, osteosarcoma, and colorectal cancer (CRC) (Vlachos et al., 2018). The distribution of RP genes mutated in those individuals with MDS or cancer was comparable to that of those mutated in the general DBA population. Although somatic loss-of-function (LOF) mutations in RP genes have been reported in a variety of sporadic cancers, in most cases, the strongest associations have been in cancer types that have not been found to be significantly enriched in DBA [e.g., acute T-cell lymphoblastic leukemia (RPL5, RPL10, and RPL22) (De Keersmaecker et al., 2013; Rao et al., 2012); aggressive chronic lymphocytic leukemia (RPL15) (Landau et al., 2015); and glioblastoma multiforme, uterine corpus endometrial carcinoma, melanoma, and breast cancer (RPL5) (Fancello, Kampen, Hofman, Verbeeck, & De Keersmaecker, 2017)]. 5q-MDS and microsatellite unstable CRC are exceptions to this with somatic mutation in DBA-genes RPS14 and RPL22, respectively (Ebert et al., 2008; Ferreira et al., 2014).
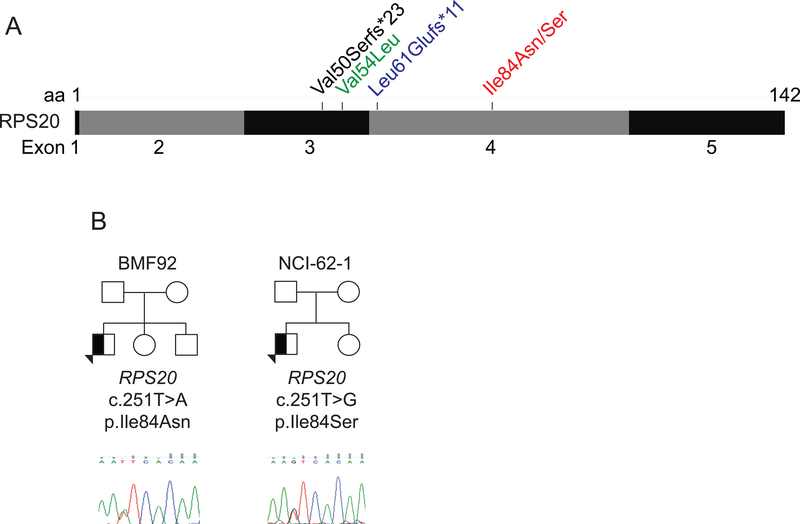
Interestingly, a four-generation kindred with Familial CRC Type X was reported to harbor a germline RPS20 LOF variant [NM_001023.3: c.147dupA, p.(Val50Serfs*23)] co-segregating with CRC (Figure 1A) (Nieminen et al., 2014). The development of CRC was highly penetrant in that family with eight of eight confirmed or obligate RPS20 mutation carriers developing CRC as early as age 24 years (median 52) and two carriers developing multiple primary CRCs. Further supporting RPS20 as a CRC susceptibility gene was the identification of a novel p.(Leu61Glufs*11) LOF variant in a young adult with metachronous CRC, and a rare missense variant, p.Val54Leu, predicted to be deleterious, in a young adult who was likely to have Lynch syndrome based on Amsterdam-criteria (Figure 1A) (Broderick et al., 2017). Notably, none of the individuals with an RPS20 variant was reported to have anemia, short stature, or congenital anomalies, and, heretofore, mutations in RPS20 have not been reported in DBA. However, given the known variable expressivity, incomplete penetrance, spontaneous remissions of anemia, and increased risk of CRC in DBA, the possibility that RPS20 may represent a DBA gene has remained an open question. Here we report novel, de novo RPS20 variants in two unrelated patients with DBA, and evidence to support expansion of the RPS20 phenotype to include DBA.
Figure 1.
Previously reported and two novel RPS20 germline variants. (A) Positions of germline RPS20 variants previously reported in familial colorectal cancer (black), a young adult with metachronous CRC (blue), and a young adult predicted to have Lynch syndrome (green) (Broderick et al., 2017; Nieminen et al., 2014), as well as those identified in BMF92 and NCI-62–1 (red). (B) BMF92 and NCI-62–1 family trees and sequence chromatograms of RPS20 genomic DNA from buccal swab (BMF92) or LCL (NCI-62–1). A half-filled symbol indicates the presence of one variant.
MATERIALS AND METHODS
Ethical Policies and Ethical Considerations
The procedures followed were in accordance with the Helsinki Declaration of 1975, as revised in 2008. Written informed consent was obtained via research protocol H-17698 (BMF# IDs) or H-21708 (RQ4–115) approved by the Baylor College of Medicine Institutional Review Board (IRB), or research protocol NCI 02-C-0052 (NCI-62–1 family) (ClinicalTrials.gov Identifier: NCT00027274) approved by the National Cancer Institute IRB.
Subjects
BMF92 family medical information was obtained and clinical evaluations carried out as part of clinical care provided within Texas Children’s Hospital. Detailed family history and medical history questionnaires were completed by NCI-62–1 and his parents and detailed medical record review, comprehensive questionnaires, and thorough clinical evaluations were conducted at the NIH Clinical Center.
Exome sequencing and variant prioritization
Clinical exome sequencing was performed for BMF92 on peripheral blood DNA prior to hematopoietic cell transplantation (HCT) at the CLIA-certified Baylor Genetics Laboratory, Houston, TX, as previously described (Yang et al., 2014). Variant classification and interpretation were conducted according to the American College of Medical Genetics and Genomics variant interpretation guidelines (Richards et al., 2015).
Exome sequencing on NCI-62–1, his parents, and his unaffected sibling were conducted at the NCI’s Cancer Genomics Research Laboratory as previously described (Ballew et al., 2013; Mirabello et al., 2014) using DNA extracted from whole blood or buccal cells. See Supplemental Methods for description of in silico analyses.
Structure analyses
See Supplemental Methods.
Analysis of RPS20 protein levels
Lymphoblastoid cell line (LCL) protein lysates were prepared in RIPA buffer [50mM Tris pH 8.0, 150mM NaCl, 1% IGEPAL, 0.5% Na deoxycholate, 1% SDS, 5mM EDTA, 1mM PMSF, with protease inhibitors (Millipore 539134)], fractionated by SDS-polyacrylamide gel electrophoresis and analyzed by western blotting using α-β-actin (Sigma A5441), α-RPS20 (Abcam ab133776), and α-neomycin (Millipore AC113) antibodies. For transient overexpression assays, RPS20 p.Ile84Asn and p.Ile84Ser mutations were introduced into myc-RPS20/pCDNA3, kindly provided by Carol Prives, Ph.D. (Columbia University). One or 3 micrograms of plasmid were transfected into 293T cells and lysates prepared using RIPA buffer. Proteins were analyzed as above.
LCL polysome fractionation
Polysome profiling was adapted from the method previously described (Pereboom et al., 2014). In brief, 10 million lymphoblastoid cells (6 OD 260 units after lysis) were resuspended in lysis buffer (110 mM KAc, 20 mM MgAc, 10 mM Tris-HCl, pH 7.6, 100 mM KCl, 10 mM MgCl2, 0.1% NP-40, and freshly added 2 mM DTT, 200 U/ml RNAsin, and 100 μg/ml cycloheximide) and homogenized for 30 strokes in an ice cold Dounce homogenizer. Samples were pre-cleared at 1,300 rpm for 10 minutes at 4°C before loading onto a sucrose gradient (17–50% sucrose in 110 mM KAc, 20 mM MgAc, and 10 mM Tris-HCl, pH 7.6). Samples were spun 38,000 rpm for 2.5 hours at 4°C in a Beckman SW41Ti rotor and fractionated with an ISCO fractionator with UV monitoring at 254 nm. Data collection was accomplished using the Logger Pro software suite.
Yeast growth assays
See Supplemental Methods for construction of yeast plasmids and strains. pRS315 plasmids carrying wild type (pFJZ1072) or mutant (pFJZ1073–8) RPS20 genes were transformed into YAB814 MATa rps20Δ::kanMX his3Δ1 leu2Δ ura3Δ0/pAB913 (CEN URA3 RPS20). Strains were grown on media containing 5-fluoroacetic acid (5-FOA) to select for cells that passively lost the RPS20 covering plasmid (pAB913) and obtain strains bearing the pRS315 plasmids alone. Logarithmically growing yeast cultures were plated at 1:10 serial dilutions on synthetic complete (SC)-Leu media and grown at the indicated temperatures and for the indicated times.
Yeast polysome profiling
Yeast strains carrying wild type (pFJZ1072) or mutant (pFJZ1073–8) RPS20 alleles were cultured in SC-Leu medium at 16°C to OD260 ∼1; cycloheximide was added to 50 μg/mL for 10 min prior to harvesting. Whole cell extracts (WCEs) were prepared in breaking buffer (20 mM Tris-HCl at pH 7.5, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, 200 μg/mL heparin, 50 μg/mL cycloheximide, and 1 Complete EDTA-free Protease Inhibitor Tablet [Roche]/50 mL buffer). Twenty-five OD260 units of WCEs were separated by velocity sedimentation on a 4.5%–45% sucrose gradient by centrifugation at 39,000 rpm for 3 hours at 4°C in an SW41Ti rotor (Beckman). Gradient fractions were scanned at 254 nm to visualize ribosomal species.
Ribosomal RNA analyses
Total RNA was isolated from LCLs using Trizol (Invitrogen). RNA separated by capillary electrophoresis was analyzed by Experion Automated Electrophoresis (Bio-Rad) or by Agilent 2100 Bioanalyzer (by the Baylor College of Medicine Genome and RNA Profiling Core). Precursor ribosomal RNAs were analyzed by northern blotting as previously described using an ITS1 probe (Rouquette, Choesmel, & Gleizes, 2005). Ratios of various pre-rRNA species were determined by quantification of ethidium bromide stained 28S bands and northern blot 21S and 18S-E bands using Image Studio V5.2.5 software (Li-Cor) and ImageQuant software (GE Healthcare Life Sciences).
Statistical methods
For analysis of the 21S/28S and 21S/18S-E ratios, a one way analysis of variance with Dunnet’s comparisons was performed on log transformed data for the data sets having a lognormal rather than normal distribution. Dunnet’s post-hoc test was performed and p values adjusted for multiple comparisons. For the 28S/18S ratio, a paired t test was performed to determine significant difference between BMF92-F and BMF92.
RESULTS
Two unrelated white/Hispanic males with genetically uncharacterized DBA (BMF92 and NCI-62–1) presented with transfusion-dependent anemia during the first month of life (Table 1). Bone marrow evaluations revealed marked erythroid hypoplasia and stabilization of p53 (in the case of BMF92; NCI-62–1 p53 was not analyzed), as characteristic for DBA (Dutt et al., 2011) (Supp. Figure S1). Proband BMF92 experienced a spontaneous remission of his anemia toward the end of the first year of life, possibly related to corticosteroids administered for bronchiolitis. Upon assessment approximately 1 year following his initial remission, his red blood cell (RBC) mean corpuscular volume (MCV) was within normal range and his hemoglobin F was at the upper limits of normal range (2.9%). His erythrocyte adenosine deaminase (eADA) level, however, was elevated [10.5 units (mol/min/gm hg), reference range 0.42–3.5 units (Baylor Genetics, Houston)]. BMF92’s hypoplastic anemia recurred at age 2 ½ years and responded only transiently to corticosteroids on multiple trials. He was treated with an empiric course of leucine 100 mg/kg for 9 months without improvement in his transfusion requirements (Pospisilova, Cmejlova, Hak, Adam, & Cmejla, 2007). He ultimately underwent hematopoietic cell transplantation (HCT) at age 6 years from his histocompatibility locus antigen (HLA)-matched sibling. He is now 3 years status post HCT and is fully engrafted.
Table 1.
Clinical features of RPS20-mutated study patients
| Clinical feature | BMF92 | NCI-62–1 |
|---|---|---|
| Current age/Race/Ethnicity | 8 y/ White/ Hispanic | 18 y/ White/ Hispanic |
| Age at presentation | First month of life | First month of life |
| Mean corpuscular volume | Normal range | Not available† |
| Erythrocyte adenosine deaminase | 10.5 mol/min/gm/hgb (normal range 0.42–3.5) | Not available† |
| Hemoglobin F % | 2.9 (normal range 0–3) | Not available† |
| Congenital anomalies | None | None |
| Stature | Normal (43 %tile) | Normal |
| Gastrointestinal disease | Eosinophilic colitis | Colitis, NOS |
| Family history for anemia or colitis | Negative | Negative |
Red blood cell transfusion-dependent from birth
Hgb, hemoglobin; NOS, not otherwise specified
Proband NCI-62–1 was born anemic with respiratory distress and transfused at birth. Bone marrow biopsy at 4 weeks of age was normocellular showing erythroid hypoplasia consistent with a diagnosis of DBA (Table 1). His anemia was unresponsive to steroid treatment that he received until 14 months of age, and refractory to immunosuppressive therapy with cyclosporine and mycophenolate mofetil. He was RBC transfusion-dependent from birth until age 17 years, which precluded determination of eADA levels. He underwent a 10/10 HLA-matched unrelated donor HCT at age 17 years, after which he developed grade I gastrointestinal graft-versus-host disease but otherwise is doing well at 23 months post-HCT.
BMF92 and NCI-62–1 are of normal stature and have no documented congenital anomalies. Notably, both experienced chronic colitis, which is not a reported characteristic of DBA. BMF92 initially presented with eosinophilic colitis and was later (post-HCT) diagnosed with eosinophilic esophagitis, gastritis, and duodenitis. NCI-62–1 was found to have chronic colitis of unclear etiology within the first year of life. He had multiple colonic biopsies that showed nonspecific colitis. A biopsy at the age of 2 years showed excessive apoptosis in the colonic mucosa. In both cases, family history for DBA was negative and parental eADA levels were within normal limits.
Exome sequencing of peripheral blood DNA did not identify a variant in any of the known DBA genes; however, it did identify heterozygous variants of unknown significance (VUS) in the RP gene RPS20 in both BMF92 and NCI-62–1 (Figure 1B). Parental studies, which confirmed the expected parentage, revealed that the RPS20 VUS were de novo. The variants were at the same genomic position but resulted in different amino acid substitutions in RPS20 [NM_001023.3: c.251T>A, p.Ile84Asn in BMF92 and c.251T>G, p.Ile84Ser in NCI-62–1]. Sanger sequencing of DNA from a buccal swab (BMF92) or LCL (NCI-62–1) obtained prior to HCT confirmed the presence of the variant (Figure 1B), consistent with a germline defect.
Both variants were novel based on inspection of several databases, including the Genome Aggregation Database (gnomAD), dbSNP, Kaviar, Human Gene Mutation Database, ClinVar, and the COSMIC database, which reports somatic mutations in cancer (see Web Resources). In silico analyses were consistent with the variants being damaging and disease causing across numerous prediction tools (including PolyPhen-2, SIFT, Mutation Taster, and VEST4) and ensemble methods (including REVEL), affecting a highly evolutionarily conserved residue (GERP, PhyloP, and PhastCons) (Supp. Table S2). The combined annotation dependent depletion (CADD) phred scores were 31 and 27.4 for RPS20.pIle84Asn and RPS20.pIle84Ser, respectively, predicting the variants to be in the <0.1% and <1% of the most deleterious substitutions possible in the human genome, respectively (Rentzsch, Witten, Cooper, Shendure, & Kircher, 2019).
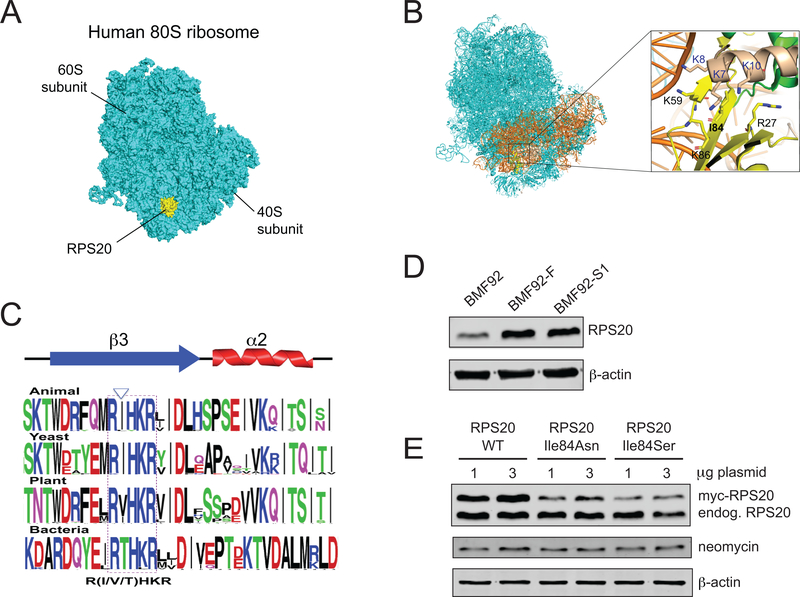
The high resolution cryo-electron microscopy structure of the human 80S ribosome (PDB ID 6EK0) (Natchiar, Myasnikov, Kratzat, Hazemann, & Klaholz, 2017) revealed RPS20 interacts with the 18S ribosomal RNA, RPS3, and RPS29 within the 40S subunit (Figures 2A,B). Whereas portions of RPS20 localize to the ribosome surface (Figure 2A), Ile84 is positioned internally within a conserved motif on β−sheet 3 close to the RPS20 and RPS3 interface (Figures 2B,C). Additionally, it is surrounded by highly conserved lysine residues that form a positively charged pocket (Figure 2B). The mutation observed in BMF92 or NCI-62–1 changes the hydrophobic isoleucine residue to a hydrophilic asparagine or serine residue, respectively, suggesting that the mutant proteins are unable to establish a hydrophobic interaction at position 84. Moreover, additional in silico analyses (MUpro and I-Mutant2.0) predicted that the variants decreased the stability of the protein structure (Supp. Table S2). Consistent with this prediction, RPS20 protein in the BMF92 proband LCL was reduced compared to his father, BMF92-F, and unaffected sibling, BMF92-S1 (Figure 2D). In addition, we found reduced steady state protein levels of the RPS20 mutants compared to wild type following transient over-expression in 293T cells (Figure 2E). Increasing the amount of plasmid transfected from 1 to 3 μg did not increase the mutant protein expression to the level of the wild type protein, possibly indicating an intolerance for the mutant proteins.
Figure 2.
The structure of RPS20 in the human 80S ribosome and reduced expression of mutant RPS20 proteins (A) The localization of RPS20 in the human 80S ribosome (PDB ID: 6EK0) (Garreau de Loubresse et al., 2014; Natchiar et al., 2017). The 80S ribosome and RPS20 are colored cyan and yellow, respectively. (B) Close-up view of RPS20 p.Ile84 in the human 80S ribosome. 18S ribosomal RNA, RPS3, and RPS29 are colored orange, wheat and green, respectively. The amino acid residues located around p.Ile84 are depicted by stick models, and the corresponding amino acid residues are labeled. (C) Graphical representation of the conserved motif within which human p.Ile84 resides [generated by WebLogo (Crooks, Hon, Chandonia, & Brenner, 2004)]; p.Ile84 is indicated by the arrowhead. (D) Representative western blot of RPS20 protein in LCL whole cell lysates from proband BMF92 compared to his unaffected father, BMF92-F, or sibling BMF92-S1. β-actin is a loading control. (E) Representative western blot of wild type, p.Ile84Asn-, or p.Ile84Ser-mutant RPS20 protein in 293T whole cell lysates following transient transfection with the designated quantity of myc-RPS20 plasmid. Neomycin and β-actin are transfection and loading controls, respectively.
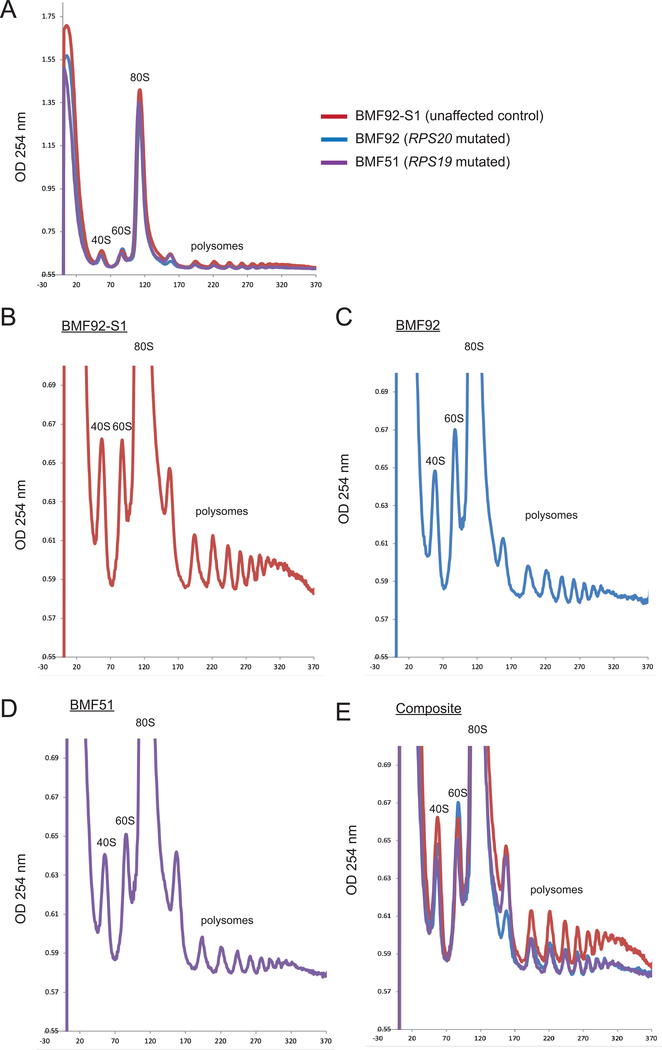
Mutations in DBA-associated RP genes have also been shown to alter ribosome distribution in polysome profiling (Cmejlova et al., 2006). Compared with the unaffected control LCL (BMF92-S1), we observed a reduction in the 80S peak and polysomes (Figure 3A), and an increase in the 60S peak relative to the 40S peak in the BMF92 LCL, similar to the RPS19 mutated-BMF51 LCL (Figures 3B–E). This result was consistent with an RPS gene defect in the RPS20 mutant cells.
Figure 3.
RPS20 p.Ile84Asn missense variant LCL has altered ribosome and polysome profiles. Polysome profile analysis of (A) RPS20 unaffected sibling control (BMF92-S1), BMF92 (RPS20 mutated), and RPS19 mutant (BMF51). Close-up views of (B) BMF92-S1, (C) BMF92, and (D) BMF51 LCL polysome profile analyses. (E) Composite of graphs B-D. Positions of the 40S, 60S, 80S and polysome peaks are shown.
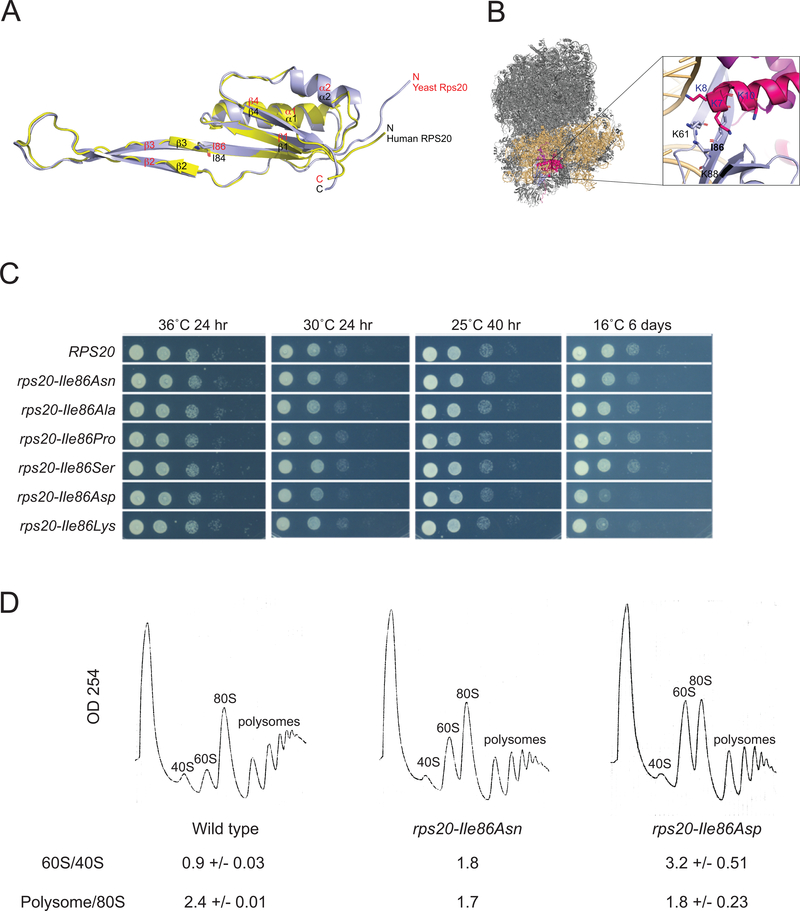
To directly test the impact of the RPS20 mutations on the ribosome, we turned to Saccharomyces cerevisiae, a tractable model system which has proven valuable in the study of ribosome maturation defects in DBA (Leger-Silvestre et al., 2005; Moore, Farrar, Arceci, Liu, & Ellis, 2010). S. cerevisiae harbors a single RPS20 gene, which is essential. Alignment of human and S. cerevisiae proteins identified S. cerevisiae Rps20 p.Ile86 as cognate to human RPS20 p.Ile84 (Supp. Figure S2). Superimposition of S. cerevisiae Rps20 onto human RPS20 gave average root mean square deviations of 0.74 Å for 89 common Cα atoms, demonstrating high similarity between the human and yeast proteins (Figure 4A). Lastly, similar to human RPS20 p.Ile84 (Figure 2B), yeast Rps20 p.Ile86 is surrounded by lysine residues that form a positively charged pocket (Figure 4B). Together, these analyses support the use of the yeast model system for the study of the functional impact of mutation of human RPS20 p.Ile84 on cells.
Figure 4.
Mutation of yeast Rps20 p.Ile86 can affect growth, ribosome biogenesis, and polysome profiles. (A) Superposition of human RPS20 (yellow) and S. cerevisiae Rps20 (light blue) structures. Human Ile84/S. cerevisiae Ile86 are depicted by stick models. (B) Close-up view of Rps20 p.Ile86 interactions in the S. cerevisiae (yeast) 80S ribosome (PDB ID: 4U4R) (Garreau de Loubresse et al., 2014; Natchiar et al., 2017); 18S ribosomal RNA, Rps3, Rps29 and Rps20 are colored light yellow, pink, magenta, and light blue, respectively. (C) S. cerevisiae expressing wild type and mutant Rps20 proteins plated at 1:10 serial dilutions and grown at the designated temperatures for the designated times. (D) Polysome profiling of yeast bearing RPS20 (wild type), rps20-Ile86Asn, or rps20-Ile86Asp alleles. Area under the curve 60S/40S and polysome/80S ratios are shown. The rps20-Ile86Asn strain was analyzed using one transformant, whereas the wild type and rps20-Ile86Asp strains were analyzed using two transformants, with the averages and standard deviations shown.
Plasmids expressing the cognate patient mutations rps20-Ile86Asn or rps20-Ile86Ser, or one of four other amino acid substitutions -Ile86Ala, -Ile86Pro, -Ile86Asp, or -Ile86Lys, were introduced into a yeast strain bearing a deletion of the endogenous RPS20 gene, which was maintained by expression of wild type RPS20 from a centromeric plasmid. Cells expressing rps20-Ile86Asn, rps20-IleI86Asp, and rps20-Ile86Lys had impaired growth at low temperature (16°C) following eviction of the wild type plasmid (Figure 4C). Analysis of polysome profiles of the cells expressing rps20-Ile86Asn and rps20-Ile86Asp showed that these rps20 mutants had markedly increased 60S/40S ratios and slightly reduced polysome/80S monosome ratios (Figure 4D). Together, these data support the critical role of S. cerevisiae Rps20 p.Ile86, and, by extension, human RPS20 p.Ile84, in normal ribosome biogenesis, polysome formation and, thus, potentially translation.
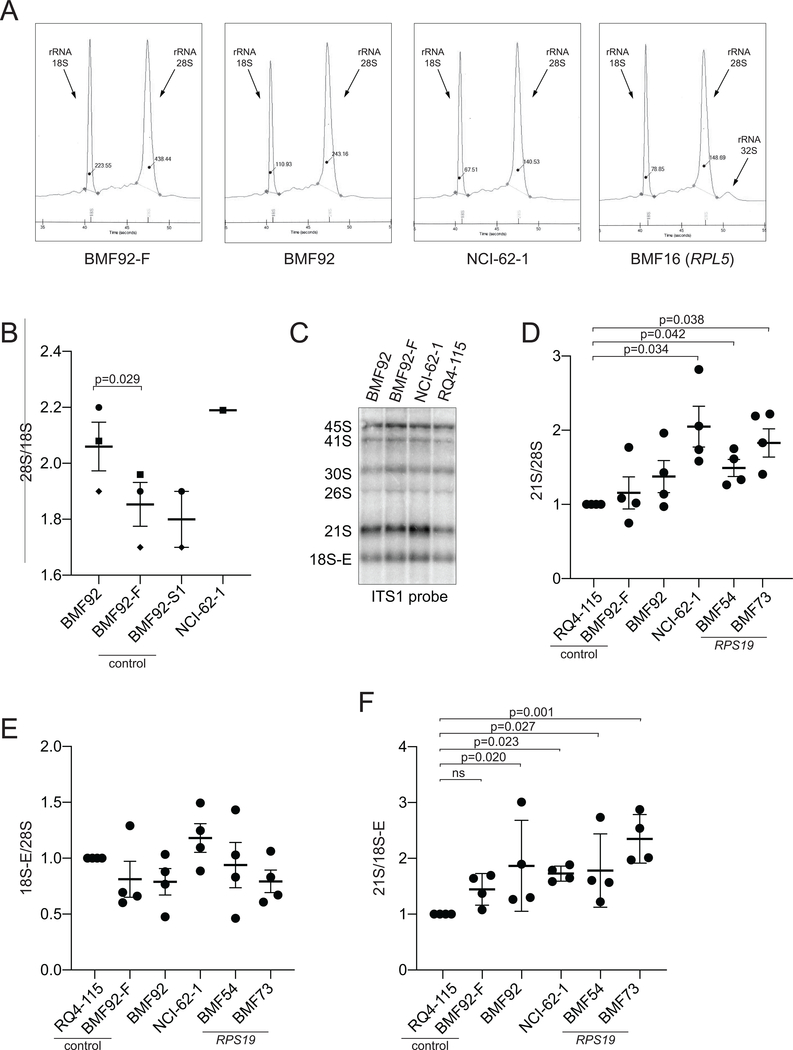
In addition to altering the ribosome and polysome profiles, mutations in DBA-associated RP genes have been shown to impact the processing of precursor rRNAs (pre-rRNAs) and level of mature rRNAs (Choesmel et al., 2007; Farrar et al., 2008; Flygare et al., 2007; Gazda et al., 2008). Mutations in RPS versus RPL genes can be distinguished by changes in the relative abundance of pre-rRNAs (Farrar et al., 2014) or by changes in ratios of 28S and 18S rRNAs (Quarello et al., 2016). Therefore, we compared pre-rRNA processing in LCLs from BMF92 and NCI-62–1 to processing in healthy control LCLs as well as in LCLs from individuals with DBA-causative pathogenic variants in RPL5 (BMF16) and RPS19 (BMF54 and BMF73) (Supp. Table S3).
Amounts of 28S and 18S rRNAs were quantified as peaks resolved by capillary electrophoresis (Figure 5A). BMF92 and NCI-62–1 both showed a higher 28S/18S ratio relative to controls BMF92-F and BMF92-S1 (Figure 5B), as previously observed in cells carrying other DBA-associated RPS gene mutations (Quarello et al., 2016). Also, BMF92 and NCI-62–1 LCLs lacked the elevated 32S pre-rRNA seen in BMF16 (Figure 5A); this small but noticeable 32S peak is a consistent feature of cells with an RPL mutation, but is not seen with RPS mutations (Farrar et al., 2008; Gazda et al., 2008).
Figure 5.
RPS20 p.Ile84 missense variant LCLs have variably altered pre-rRNA processing. (A) Representative electropherograms from LCL total RNA. The 18S and 28S rRNA peaks are indicated, as well as a minor 32S pre-rRNA peak in the BMF16 sample. (B) Comparison of 28S/18S rRNA ratios as measured by electropherograms; similar symbols came from the same experiment. P values were not calculated for BMF92-S1 nor NCI-62–1 as fewer than three biological replicates were performed. Shown is the mean and standard error of the mean (sem). (C) Representative northern blot analysis of RNA isolated from the designated LCLs (listed below) using an ITS1 probe. (D) Quantification of 21S pre-rRNA/28S rRNA ratios from four independent blots as in C. (E) Quantification of 18S-E pre-rRNA/28S rRNA ratios from four independent blots as in C. (F) Quantification of 21S/18S-E pre-rRNA ratios from four independent blots as in C. LCLs tested: BMF92 and NCI-62–1: RPS20 missense variants; BMF16: RPL5 missense variant; BMF92-F: unaffected father of BMF92; BMF92-S1: unaffected sibling of BMF92; RQ4–115: healthy/unaffected control; BMF54 and BMF73: RPS19 missense variants.
Small interfering RNA (siRNA) knockdown of RPS20 in HeLa cells has been shown to increase the amounts of 26S, 21S, and 18S-E pre-rRNAs relative to 28S rRNA two- to eight-fold (O’Donohue, Choesmel, Faubladier, Fichant, & Gleizes, 2010). We therefore examined the relative abundance of these species in LCLs derived from BMF92 and NCI-62–1 or controls (Figure 5C). We found that, compared with control LCL RQ4–115, the 21S/28S ratio was significantly increased in the LCL from NCI-62–1 (Figure 5D), similar to the increase in LCLs BMF54 and BMF73, which were derived from patients with DBA and RPS19 pathogenic variants. The ratios in BMF92 or BMF92-F LCLs were not significantly different from RQ4–115 (Figure 5D). Notably, the 18S-E to 28S ratio showed no statistically significant difference in any of the same samples compared the RQ4–115 control (Figure 5E), however, we did find statistically significant increases in the 21S to 18S-E ratios for the mutant but not the BMF92-F LCLs (Figure 5F). The low abundance of the 26S on the northern blots precluded rigorous assessment (Figure 5C).
DISCUSSION
In summary, we provide evidence that the described variants in RPS20 at the p.Ile84 locus are causative of DBA. At a gene level, the gnomAD database observed a lower than expected number of missense variants in RPS20 [observed/expected ratio 0.66 (90% confidence interval 0.53 – 0.84)], yet novel, de novo variants in RPS20 were identified in these two children afflicted with a rare disorder, DBA, which has an incidence of 1 in 100,000 to 200,000 live births (Boria et al., 2010). At a protein level, RPS20 is a component of the ribosome, which is central in the pathogenesis of DBA. Residue p.Ile84 displays evolutionary conservation and is located within a highly evolutionarily conserved motif [R(I/V/T)HKR] and within a positively charged pocket (Figures 2B,C and Supp. Figure S2). The Ile84Asn and Ile84Ser mutations destabilize RPS20 protein, with markedly reduced levels of RPS20 protein in the proband BMF92 LCL (Figure 2D) and upon transient over-expression. (Figure 2E). While we were unable to test whether transgenic RPS20 expression could rescue the erythropoietic defect in BMF92 or NCI-62–1 CD34+ cells due to a lack of or insufficient cells pre-HCT, respectively, we have presented functional data to support an impact of the mutant RPS20 proteins on ribosomes, mimicking the defects observed in other RPS-mutant DBA lines. We found lymphoblastoid cells bearing the RPS20 p.Ile84Asn variant exhibited an abnormal ribosome and polysome profile (Figure 3B,D) and increased 28S/18S rRNA ratio (Figure 5B), consistent with an RPS20 deficiency (O’Donohue et al., 2010). Lymphoblastoid cells bearing the RPS20 p.Ile84Ser variant exhibited an increase in the 28S/18S, 21S/28S, and 21S/18S-E ratios (Figures 5B–E), similarly consistent with a RPS20 protein defect. Lastly, yeast bearing mutations of the residue cognate to human RPS20 p.Ile84 showed abnormal ribosome biogenesis as well as polysome profiles, indicating an impairment of translation (Figure 4D). While the results of the pre-rRNA analyses of the LCLs do not fully mirror those of RPS20 siRNA-treated HeLa cells (O’Donohue et al., 2010), collectively the data are consistent with a negative impact of the RPS20 variants on ribosome function, which is central to DBA pathophysiology. Lastly, application of the American College of Medical Genetics standards and guidelines for the interpretation of sequence variants in genes that cause Mendelian disorders (Richards et al., 2015) renders both variants as pathogenic (Supp. Table S4).
The absence of additional individuals with an RPS20 variant in a 472 person DBA cohort or in other reports of genetically uncharacterized cases of DBA in which RPS20 was among several RP genes sequenced is notable (Doherty et al., 2010; Gerrard et al., 2013; Ulirsch et al., 2018). Given this and the finding that the same position, p.Ile84, of the 142 amino acid RPS20 was mutated in both patients described in this report, it is likely that only a limited set of mutations in this gene may result in impaired erythropoiesis characteristic of DBA. Thus, while we observed a reduction in RPS20 mutant protein (Figures 2D,E), it may be a combination of reduced and mutant protein that is required to elicit an impact on erythropoiesis.
The possibility of such a genotype-phenotype correlation is supported by the reports of a family with an RPS20 LOF variant [p.(Val50Serfs*23)] segregating with mismatch repair-proficient nonpolyposis CRC and of a young adult bearing a germline RPS20 LOF p.(Leu61Glufs*11) variant with metachronous CRC. Anemia or other DBA-related features were not reported in these studies (Broderick et al., 2017; Nieminen et al., 2014). This suggests that perhaps mutation of RPS20 p.Ile84 does not simply result in reduce protein expression but additionally elicits either a gain-of-function or dominant negative effect. The potential for a genotype-phenotype correlation may underlie the occurrence of RPS20 as the first RP gene in which germline variants are associated with an infant-onset erythroid phenotype characteristic of DBA, as observed in BMF92 and NCI-62–1, as well as a highly penetrant and in some cases early onset CRC phenotype, as observed in the family reported by Nieminen and colleagues (Nieminen et al., 2014). It remains unknown whether the features of eosinophilic colitis manifest in BMF92 and the chronic colitis with excessive apoptosis in the colonic mucosa in NCI-62–1, neither of which are typical of DBA, relate specifically to their RPS20 mutations or potential risk of CRC. Evidence-based guidelines for earlier or potentially more frequent CRC surveillance for individuals with DBA have not been established, but may be of particular benefit for BMF92 and NCI-62–1.
RPS20 is an integral component of the 40S ribosome, where it plays a critical role in the processing of the pre-rRNA, the maturation of the pre-40S ribosomal subunit, and ribosome-mediated quality control (Matsuo et al., 2017; Mitterer et al., 2019; O’Donohue et al., 2010). In addition to this, and similar to several other ribosomal proteins, such as RPL5 and RPL11 (Lohrum, Ludwig, Kubbutat, Hanlon, & Vousden, 2003; Marechal, Elenbaas, Piette, Nicolas, & Levine, 1994), RPS20 has also been shown to interact with the p53 regulator MDM2 and inhibit its E3 ubiquitin ligase activity, leading to p53 stabilization (Daftuar, Zhu, Jacq, & Prives, 2013). The extent to which one or more of the molecular effects of RPS20 mutation impacts the development of CRC remains to be determined.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the patients and their families for their willingness to participate in our study. We thank members of the Bertuch lab for critical reading of the manuscript. Lisa Leathwood, RN, Maureen Risch, RN, and Ann Carr, MS, CGC provided study support through contract HHSN261201100018C with Westat, Inc. (Rockville, MD). Trial registration: ClinicalTrials.gov Identifier: NCT00027274
Grant numbers: This work was supported by Texas Children’s Cancer and Hematology Centers at Texas Children’s Hospital, Medical Genetics Research Fellowship Program (2T32GM007526, to SB, Principal Investigator Brendan Lee, Baylor College of Medicine), and the intramural research program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors report no competing interests.
WEB RESOURCES
Bioinformatic databases and tools utilized for exome interpretation
CADD (Rentzsch et al., 2019) https://cadd.gs.washington.edu/
ClinVar (Landrum et al., 2018) https://www.ncbi.nlm.nih.gov/clinvar/
COSMIC (Tate et al., 2019) https://cancer.sanger.ac.uk/cosmic
dbSNP (Sherry et al., 2001) https://www.ncbi.nlm.nih.gov/snp/
GERP (Davydov et al., 2010) http://mendel.stanford.edu/sidowlab/downloads/gerp/index.html
GnomAD Browser (Lek et al., 2016) http://gnomad.broadinstitute.org/
Human Mutation Database (Stenson et al., 2017) http://www.hgmd.cf.ac.uk/ac/index.php
I-Mutant2.0 (Capriotti, Fariselli, & Casadio, 2005) http://folding.biofold.org/i-mutant/i-mutant2.0.html
Kaviar (Glusman, Caballero, Mauldin, Hood, & Roach, 2011) http://db.systemsbiology.net/kaviar/
MUpro (Cheng, Randall, & Baldi, 2006) http://mupro.proteomics.ics.uci.edu/
Mutation Taster (Schwarz, Rodelsperger, Schuelke, & Seelow, 2010) http://www.mutationtaster.org/
PhyloP (Pollard, Hubisz, Rosenbloom, & Siepel, 2010) http://compgen.cshl.edu/phast/
PolyPhen-2 (Adzhubei et al., 2010) http://genetics.bwh.harvard.edu/pph2
SIFT (Kumar, Henikoff, & Ng, 2009) http://sift.bii.a-star.edu.sg/
Sitewise likelihood-ratio (Massingham & Goldman, 2005) https://www.ebi.ac.uk/goldman-srv/SLR/
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, & Sunyaev SR (2010). A method and server for predicting damaging missense mutations. Nature Methods, 7(4), 248–249. doi: 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballew BJ, Yeager M, Jacobs K, Giri N, Boland J, Burdett L, Alter BP, & Savage SA (2013). Germline mutations of regulator of telomere elongation helicase 1, RTEL1, in Dyskeratosis congenita. Hum Genet, 132(4), 473–480. doi: 10.1007/s00439-013-1265-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ami T, Revel-Vilk S, Brooks R, Shaag A, Hershfield MS, Kelly SJ, Ganson NJ, Kfir-Erenfeld S, Weintraub M, Elpeleg O, Berkun Y, & Stepensky P (2016). Extending the Clinical Phenotype of Adenosine Deaminase 2 Deficiency. Journal of Pediatrics, 177, 316–320. doi: 10.1016/j.jpeds.2016.06.058 [DOI] [PubMed] [Google Scholar]
- Boria I, Garelli E, Gazda HT, Aspesi A, Quarello P, Pavesi E, Ferrante D, Meerpohl JJ, Kartal M, Da Costa L, Proust A, Leblanc T, Simansour M, Dahl N, Frojmark AS, Pospisilova D, Cmejla R, Beggs AH, Sheen MR, Landowski M, Buros CM, Clinton CM, Dobson LJ, Vlachos A, Atsidaftos E, Lipton JM, Ellis SR, Ramenghi U, & Dianzani I (2010). The ribosomal basis of Diamond-Blackfan Anemia: mutation and database update. Human Mutation, 31(12), 1269–1279. doi: 10.1002/humu.21383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick P, Dobbins SE, Chubb D, Kinnersley B, Dunlop MG, Tomlinson I, & Houlston RS (2017). Validation of Recently Proposed Colorectal Cancer Susceptibility Gene Variants in an Analysis of Families and Patients-a Systematic Review. Gastroenterology, 152(1), 75–77 e74. doi: 10.1053/j.gastro.2016.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E, Fariselli P, & Casadio R (2005). I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Research, 33(Web Server issue), W306–310. doi: 10.1093/nar/gki375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Randall A, & Baldi P (2006). Prediction of protein stability changes for single-site mutations using support vector machines. Proteins, 62(4), 1125–1132. doi: 10.1002/prot.20810 [DOI] [PubMed] [Google Scholar]
- Choesmel V, Bacqueville D, Rouquette J, Noaillac-Depeyre J, Fribourg S, Cretien A, Leblanc T, Tchernia G, Da Costa L, & Gleizes PE (2007). Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood, 109(3), 1275–1283. doi: 10.1182/blood-2006-07-038372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmejlova J, Dolezalova L, Pospisilova D, Petrtylova K, Petrak J, & Cmejla R (2006). Translational efficiency in patients with Diamond-Blackfan anemia. Haematologica, 91(11), 1456–1464. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/17082006 [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, & Brenner SE (2004). WebLogo: a sequence logo generator. Genome Research, 14(6), 1188–1190. doi: 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa L, Narla A, & Mohandas N (2018). An update on the pathogenesis and diagnosis of Diamond-Blackfan anemia. F1000Research, 7. doi: 10.12688/f1000research.15542.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daftuar L, Zhu Y, Jacq X, & Prives C (2013). Ribosomal proteins RPL37, RPS15 and RPS20 regulate the Mdm2-p53-MdmX network. PLoS One, 8(7), e68667. doi: 10.1371/journal.pone.0068667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, & Batzoglou S (2010). Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Computational Biology, 6(12), e1001025. doi: 10.1371/journal.pcbi.1001025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keersmaecker K, Atak ZK, Li N, Vicente C, Patchett S, Girardi T, Gianfelici V, Geerdens E, Clappier E, Porcu M, Lahortiga I, Luca R, Yan J, Hulselmans G, Vranckx H, Vandepoel R, Sweron B, Jacobs K, Mentens N, Wlodarska I, Cauwelier B, Cloos J, Soulier J, Uyttebroeck A, Bagni C, Hassan BA, Vandenberghe P, Johnson AW, Aerts S, & Cools J (2013). Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nature Genetics, 45(2), 186–190. doi: 10.1038/ng.2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty L, Sheen MR, Vlachos A, Choesmel V, O’Donohue MF, Clinton C, Schneider HE, Sieff CA, Newburger PE, Ball SE, Niewiadomska E, Matysiak M, Glader B, Arceci RJ, Farrar JE, Atsidaftos E, Lipton JM, Gleizes PE, & Gazda HT (2010). Ribosomal protein genes RPS10 and RPS26 are commonly mutated in Diamond-Blackfan anemia. American Journal of Human Genetics, 86(2), 222–228. doi: 10.1016/j.ajhg.2009.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt S, Narla A, Lin K, Mullally A, Abayasekara N, Megerdichian C, Wilson FH, Currie T, Khanna-Gupta A, Berliner N, Kutok JL, & Ebert BL (2011). Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood, 117(9), 2567–2576. doi: 10.1182/blood-2010-07-295238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, & Golub TR (2008). Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature, 451(7176), 335–339. doi: 10.1038/nature06494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancello L, Kampen KR, Hofman IJ, Verbeeck J, & De Keersmaecker K (2017). The ribosomal protein gene RPL5 is a haploinsufficient tumor suppressor in multiple cancer types. Oncotarget, 8(9), 14462–14478. doi: 10.18632/oncotarget.14895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar JE, Nater M, Caywood E, McDevitt MA, Kowalski J, Takemoto CM, Talbot CC Jr., Meltzer P, Esposito D, Beggs AH, Schneider HE, Grabowska A, Ball SE, Niewiadomska E, Sieff CA, Vlachos A, Atsidaftos E, Ellis SR, Lipton JM, Gazda HT, & Arceci RJ (2008). Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood, 112(5), 1582–1592. doi: 10.1182/blood-2008-02-140012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar JE, Quarello P, Fisher R, O’Brien KA, Aspesi A, Parrella S, Henson AL, Seidel NE, Atsidaftos E, Prakash S, Bari S, Garelli E, Arceci RJ, Dianzani I, Ramenghi U, Vlachos A, Lipton JM, Bodine DM, & Ellis SR (2014). Exploiting pre-rRNA processing in Diamond Blackfan anemia gene discovery and diagnosis. American Journal of Hematology, 89(10), 985–991. doi: 10.1002/ajh.23807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar JE, Vlachos A, Atsidaftos E, Carlson-Donohoe H, Markello TC, Arceci RJ, Ellis SR, Lipton JM, & Bodine DM (2011). Ribosomal protein gene deletions in Diamond-Blackfan anemia. Blood, 118(26), 6943–6951. doi: 10.1182/blood-2011-08-375170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira AM, Tuominen I, van Dijk-Bos K, Sanjabi B, van der Sluis T, van der Zee AG, Hollema H, Zazula M, Sijmons RH, Aaltonen LA, Westers H, & Hofstra RM (2014). High frequency of RPL22 mutations in microsatellite-unstable colorectal and endometrial tumors. Human Mutation, 35(12), 1442–1445. doi: 10.1002/humu.22686 [DOI] [PubMed] [Google Scholar]
- Flygare J, Aspesi A, Bailey JC, Miyake K, Caffrey JM, Karlsson S, & Ellis SR (2007). Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood, 109(3), 980–986. doi: 10.1182/blood-2006-07-038232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garreau de Loubresse N, Prokhorova I, Holtkamp W, Rodnina MV, Yusupova G, & Yusupov M (2014). Structural basis for the inhibition of the eukaryotic ribosome. Nature, 513(7519), 517–522. doi: 10.1038/nature13737 [DOI] [PubMed] [Google Scholar]
- Gazda HT, Sheen MR, Vlachos A, Choesmel V, O’Donohue MF, Schneider H, Darras N, Hasman C, Sieff CA, Newburger PE, Ball SE, Niewiadomska E, Matysiak M, Zaucha JM, Glader B, Niemeyer C, Meerpohl JJ, Atsidaftos E, Lipton JM, Gleizes PE, & Beggs AH (2008). Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. American Journal of Human Genetics, 83(6), 769–780. doi: 10.1016/j.ajhg.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard G, Valganon M, Foong HE, Kasperaviciute D, Iskander D, Game L, Muller M, Aitman TJ, Roberts I, de la Fuente J, Foroni L, & Karadimitris A (2013). Target enrichment and high-throughput sequencing of 80 ribosomal protein genes to identify mutations associated with Diamond-Blackfan anaemia. British Journal of Haematology, 162(4), 530–536. doi: 10.1111/bjh.12397 [DOI] [PubMed] [Google Scholar]
- Glusman G, Caballero J, Mauldin DE, Hood L, & Roach JC (2011). Kaviar: an accessible system for testing SNV novelty. Bioinformatics, 27(22), 3216–3217. doi: 10.1093/bioinformatics/btr540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajuria RK, Munschauer M, Ulirsch JC, Fiorini C, Ludwig LS, McFarland SK, Abdulhay NJ, Specht H, Keshishian H, Mani DR, Jovanovic M, Ellis SR, Fulco CP, Engreitz JM, Schutz S, Lian J, Gripp KW, Weinberg OK, Pinkus GS, Gehrke L, Regev A, Lander ES, Gazda HT, Lee WY, Panse VG, Carr SA, & Sankaran VG (2018). Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis. Cell, 173(1), 90–103 e119. doi: 10.1016/j.cell.2018.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, & Ng PC (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols, 4(7), 1073–1081. doi: 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, Kluth S, Bozic I, Lawrence M, Bottcher S, Carter SL, Cibulskis K, Mertens D, Sougnez CL, Rosenberg M, Hess JM, Edelmann J, Kless S, Kneba M, Ritgen M, Fink A, Fischer K, Gabriel S, Lander ES, Nowak MA, Dohner H, Hallek M, Neuberg D, Getz G, Stilgenbauer S, & Wu CJ (2015). Mutations driving CLL and their evolution in progression and relapse. Nature, 526(7574), 525–530. doi: 10.1038/nature15395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, Karapetyan K, Katz K, Liu C, Maddipatla Z, Malheiro A, McDaniel K, Ovetsky M, Riley G, Zhou G, Holmes JB, Kattman BL, & Maglott DR (2018). ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Research, 46(D1), D1062–D1067. doi: 10.1093/nar/gkx1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger-Silvestre I, Caffrey JM, Dawaliby R, Alvarez-Arias DA, Gas N, Bertolone SJ, Gleizes PE, & Ellis SR (2005). Specific Role for Yeast Homologs of the Diamond Blackfan Anemia-associated Rps19 Protein in Ribosome Synthesis. The Journal of Biological Chemistry, 280(46), 38177–38185. doi: 10.1074/jbc.M506916200 [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, & Exome Aggregation C (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature, 536(7616), 285–291. doi: 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, & Vousden KH (2003). Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell, 3(6), 577–587. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/12842086 [DOI] [PubMed] [Google Scholar]
- Ludwig LS, Gazda HT, Eng JC, Eichhorn SW, Thiru P, Ghazvinian R, George TI, Gotlib JR, Beggs AH, Sieff CA, Lodish HF, Lander ES, & Sankaran VG (2014). Altered translation of GATA1 in Diamond-Blackfan anemia. Nature Medicine, 20(7), 748–753. doi: 10.1038/nm.3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal V, Elenbaas B, Piette J, Nicolas JC, & Levine AJ (1994). The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Molecular and Cellular Biology, 14(11), 7414–7420. doi: 10.1128/mcb.14.11.7414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massingham T, & Goldman N (2005). Detecting amino acid sites under positive selection and purifying selection. Genetics, 169(3), 1753–1762. doi: 10.1534/genetics.104.032144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Ikeuchi K, Saeki Y, Iwasaki S, Schmidt C, Udagawa T, Sato F, Tsuchiya H, Becker T, Tanaka K, Ingolia NT, Beckmann R, & Inada T (2017). Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nature Communications, 8(1), 159. doi: 10.1038/s41467-017-00188-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabello L, Macari ER, Jessop L, Ellis SR, Myers T, Giri N, Taylor AM, McGrath KE, Humphries JM, Ballew BJ, Yeager M, Boland JF, He J, Hicks BD, Burdett L, Alter BP, Zon L, & Savage SA (2014). Whole-exome sequencing and functional studies identify RPS29 as a novel gene mutated in multicase Diamond-Blackfan anemia families. Blood, 124(1), 24–32. doi: 10.1182/blood-2013-11-540278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitterer V, Shayan R, Ferreira-Cerca S, Murat G, Enne T, Rinaldi D, Weigl S, Omanic H, Gleizes PE, Kressler D, Plisson-Chastang C, & Pertschy B (2019). Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism. Nature Communications, 10(1), 2754. doi: 10.1038/s41467-019-10678-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. B. t., Farrar JE, Arceci RJ, Liu JM, & Ellis SR (2010). Distinct ribosome maturation defects in yeast models of Diamond-Blackfan anemia and Shwachman-Diamond syndrome. Haematologica, 95(1), 57–64. doi: 10.3324/haematol.2009.012450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natchiar SK, Myasnikov AG, Kratzat H, Hazemann I, & Klaholz BP (2017). Visualization of chemical modifications in the human 80S ribosome structure. Nature, 551(7681), 472–477. doi: 10.1038/nature24482 [DOI] [PubMed] [Google Scholar]
- Nieminen TT, O’Donohue MF, Wu Y, Lohi H, Scherer SW, Paterson AD, Ellonen P, Abdel-Rahman WM, Valo S, Mecklin JP, Jarvinen HJ, Gleizes PE, & Peltomaki P (2014). Germline mutation of RPS20, encoding a ribosomal protein, causes predisposition to hereditary nonpolyposis colorectal carcinoma without DNA mismatch repair deficiency. Gastroenterology, 147(3), 595–598 e595. doi: 10.1053/j.gastro.2014.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donohue MF, Choesmel V, Faubladier M, Fichant G, & Gleizes PE (2010). Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. Journal of Cell Biology, 190(5), 853–866. doi: 10.1083/jcb.201005117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereboom TC, Bondt A, Pallaki P, Klasson TD, Goos YJ, Essers PB, Groot Koerkamp MJ, Gazda HT, Holstege FC, Costa LD, & MacInnes AW (2014). Translation of branched-chain aminotransferase-1 transcripts is impaired in cells haploinsufficient for ribosomal protein genes. Exp Hematol, 42(5), 394–403 e394. doi: 10.1016/j.exphem.2013.12.010 [DOI] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, & Siepel A (2010). Detection of nonneutral substitution rates on mammalian phylogenies. Genome Research, 20(1), 110–121. doi: 10.1101/gr.097857.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospisilova D, Cmejlova J, Hak J, Adam T, & Cmejla R (2007). Successful treatment of a Diamond-Blackfan anemia patient with amino acid leucine. Haematologica, 92(5), e66–67. doi: 10.3324/haematol.11498 [DOI] [PubMed] [Google Scholar]
- Quarello P, Garelli E, Carando A, Cillario R, Brusco A, Giorgio E, Ferrante D, Corti P, Zecca M, Luciani M, Pierri F, Putti MC, Cantarini ME, Farruggia P, Barone A, Cesaro S, Russo G, Fagioli F, Dianzani I, Ramenghi U, & Anaemia A. w. g. o. D. B. (2020). A 20-year long term experience of the Italian Diamond-Blackfan Anaemia Registry: RPS and RPL genes, different faces of the same disease? British Journal of Haematology. doi: 10.1111/bjh.16508 [DOI] [PubMed] [Google Scholar]
- Quarello P, Garelli E, Carando A, Mancini C, Foglia L, Botto C, Farruggia P, De Keersmaecker K, Aspesi A, Ellis SR, Dianzani I, & Ramenghi U (2016). Ribosomal RNA analysis in the diagnosis of Diamond-Blackfan Anaemia. British Journal of Haematology, 172(5), 782–785. doi: 10.1111/bjh.13880 [DOI] [PubMed] [Google Scholar]
- Rao S, Lee SY, Gutierrez A, Perrigoue J, Thapa RJ, Tu Z, Jeffers JR, Rhodes M, Anderson S, Oravecz T, Hunger SP, Timakhov RA, Zhang R, Balachandran S, Zambetti GP, Testa JR, Look AT, & Wiest DL (2012). Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood, 120(18), 3764–3773. doi: 10.1182/blood-2012-03-415349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch P, Witten D, Cooper GM, Shendure J, & Kircher M (2019). CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Research, 47(D1), D886–D894. doi: 10.1093/nar/gky1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, & Committee ALQA (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine, 17(5), 405–424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquette J, Choesmel V, & Gleizes PE (2005). Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J, 24(16), 2862–2872. doi: 10.1038/sj.emboj.7600752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, & Seelow D (2010). MutationTaster evaluates disease-causing potential of sequence alterations. Nature Methods, 7(8), 575–576. doi: 10.1038/nmeth0810-575 [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, & Sirotkin K (2001). dbSNP: the NCBI database of genetic variation. Nucleic Acids Research, 29(1), 308–311. doi: 10.1093/nar/29.1.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, Hussain M, Phillips AD, & Cooper DN (2017). The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet, 136(6), 665–677. doi: 10.1007/s00439-017-1779-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, Fish P, Harsha B, Hathaway C, Jupe SC, Kok CY, Noble K, Ponting L, Ramshaw CC, Rye CE, Speedy HE, Stefancsik R, Thompson SL, Wang S, Ward S, Campbell PJ, & Forbes SA (2019). COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Research, 47(D1), D941–D947. doi: 10.1093/nar/gky1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulirsch JC, Verboon JM, Kazerounian S, Guo MH, Yuan D, Ludwig LS, Handsaker RE, Abdulhay NJ, Fiorini C, Genovese G, Lim ET, Cheng A, Cummings BB, Chao KR, Beggs AH, Genetti CA, Sieff CA, Newburger PE, Niewiadomska E, Matysiak M, Vlachos A, Lipton JM, Atsidaftos E, Glader B, Narla A, Gleizes PE, O’Donohue MF, Montel-Lehry N, Amor DJ, McCarroll SA, O’Donnell-Luria AH, Gupta N, Gabriel SB, MacArthur DG, Lander ES, Lek M, Da Costa L, Nathan DG, Korostelev AA, Do R, Sankaran VG, & Gazda HT (2018). The Genetic Landscape of Diamond-Blackfan Anemia. Am J Hum Genet, 103(6), 930–947. doi: 10.1016/j.ajhg.2018.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos A, Rosenberg PS, Atsidaftos E, Kang J, Onel K, Sharaf RN, Alter BP, & Lipton JM (2018). Increased risk of colon cancer and osteogenic sarcoma in Diamond-Blackfan anemia. Blood, 132(20), 2205–2208. doi: 10.1182/blood-2018-05-848937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, Veeraraghavan N, Hawes A, Chiang T, Leduc M, Beuten J, Zhang J, He W, Scull J, Willis A, Landsverk M, Craigen WJ, Bekheirnia MR, Stray-Pedersen A, Liu P, Wen S, Alcaraz W, Cui H, Walkiewicz M, Reid J, Bainbridge M, Patel A, Boerwinkle E, Beaudet AL, Lupski JR, Plon SE, Gibbs RA, & Eng CM (2014). Molecular findings among patients referred for clinical whole-exome sequencing. JAMA, 312(18), 1870–1879. doi: 10.1001/jama.2014.14601 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.