Graphical abstract
Keywords: Coronaviruses, SARS-CoV-2, RNA-dependent RNA polymerase (RdRp), 3C-like protease (3CLpro), Papain-like protease (PLpro), Transmembrane serine protease (TMPRSS2)
Abstract
The SARS-CoV-2 outbreak and pandemic that began near the end of 2019 has posed a challenge to global health. At present, many candidate small-molecule therapeutics have been developed that can inhibit both the infection and replication of SARS-CoV-2 and even potentially relieve cytokine storms and other related complications. Meanwhile, host-targeted drugs that inhibit cellular transmembrane serine protease (TMPRSS2) can prevent SARS-CoV-2 from entering cells, and its combination with chloroquine and dihydroorotate dehydrogenase (DHODH) inhibitors can limit the spread of SARS-CoV-2 and reduce the morbidity and mortality of patients with COVID-19. The present article provides an overview of these small-molecule therapeutics based on insights from medicinal chemistry research and focuses on RNA-dependent RNA polymerase (RdRp) inhibitors, such as the nucleoside analogues remdesivir, favipiravir and ribavirin. This review also covers inhibitors of 3C-like protease (3CLpro), papain-like protease (PLpro) and other potentially innovative active ingredient molecules, describing their potential targets, activities, clinical status and side effects.
1. Introduction
The 2019 coronavirus disease (COVID-19) pandemic is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). At present, 90 million people have been infected and more than 1.9 million people have died from this virus. Furthermore, the pandemic has caused socioeconomic chaos on a global scale. Along with SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV-2 is a member of the Betacoronaviridae family; it is an enveloped virus with a positive non-segmented single-stranded RNA genome [1]. Epidemiological studies have shown that SARS-CoV-2 is more infectious than SARS-CoV or MERS-CoV [[2], [3], [4]]. Due to the higher infection rate of SARS-CoV-2 and the lack of effective treatment methods, improved strategies for treating and preventing COVID-19 are urgently needed.
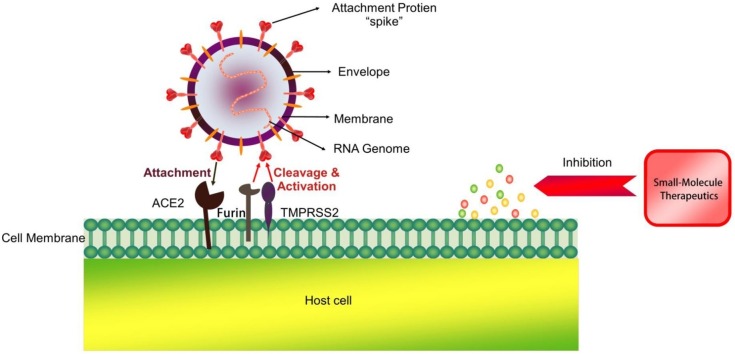
The main structural proteins encoded by SARS-CoV-2 include spike (S), membrane (M), envelope (E) and nucleocapsid (N) proteins. The binding of the S protein to host cell receptor angiotensin-converting enzyme 2 (ACE2) [5,6] is the key event for the entry of SARS-CoV-2 into host cells. Furthermore, the binding of the S protein to the cellular transmembrane serine protease (TMPRSS2) might promote the entry of the virus into the host cells. Therefore, both ACE2 and TMPRSS2 might be pharmacological targets that could restrict the entry of SARS-CoV-2 into cells. The non-structural proteins (nsps) encoded by SARS-CoV-2, such as 3C-like protease (3CLpro), papain-like protease (PLpro) and other nsps, which play key roles in virus replication, are also important targets for drug development [7,8]. To date, some small-molecule drugs, including lopinavir/ritonavir, chloroquine (CQ), hydroxychloroquine (HCQ), arbidol, remdesivir, favipiravir, and ribavirin, have shown beneficial effects in clinical and preclinical trials. In this review, we collected and reviewed the chemical structures, targets, pharmacological effects and adverse reactions of the currently available small-molecule drugs against SARS-CoV-2.
2. RdRp inhibitors
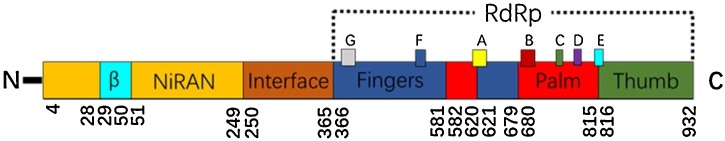
ORF1a and ORF1b, located at the 5′-terminus of the coronavirus (CoV) genome, encode polyproteins 1a and 1b. These two proteins can be cleaved into 16 types of nsps that play essential roles in virus replication and transcription [9]. Therefore, nsps are considered potential virulence factors and CoV targets. Among these nsps, RNA-dependent RNA polymerase (RdRp) is non-structural protein 12 (nsp12) [10,11], which is an enzyme that catalyses the synthesis of RNA strands complementary to a given RNA template and can extend the homopolymerized primer-template substrate by dozens of nucleotides in vitro. The protein structure of RdRp of different RNA viruses is conserved; hence, using RdRp as an anti-RNA virus target could reduce the potential of drug resistance and is essential for the development of a broad-spectrum antiviral drug. Viral/prokaryotic RNA-directed RNA polymerases employ a fold whose organization has been likened to the shape of a right hand with three subdomains, termed “fingers”, “palm” and “thumb” (Fig. 1 ). The palm region of all RdRps consists a three-strand anti-parallel beta sheet as the core and three α-helices on the side and is considered the most conserved structure among all known nucleotide polymerases. In this area, five sequences and domains (A–E) have been identified [12]: A and B have the functions of nucleotide recognition and connection, A and C have the function of phosphate transfer, D comprises the complete structure of the palm region, E is responsible for the junction of the nucleotide primers, and A, B, and C are highly conserved [13].
Fig. 1.
Genome of RdRp.
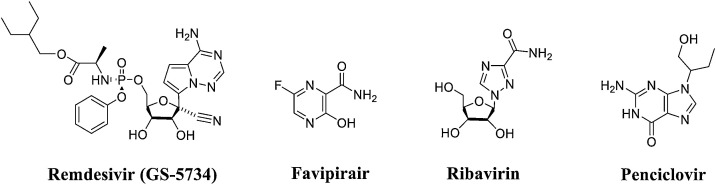
There are two types of RdRp inhibitors: nucleoside drugs and non-nucleoside drugs. Non-nucleoside inhibitors are susceptible to drug resistance and exhibit an inability to act on other subtypes, which limits their development. In contrast, nucleoside drugs have the advantages of directly acting on highly conserved active pockets and acting as catalytic substrates for RdRp. At present, the primary representative nucleoside RdRp inhibitors for COVID-19 are remdesivir (GS-5734), favipiravir, ribavirin, and penciclovir [17,18]. Although these inhibitors have different indications, their mechanisms of action are generally the same. These inhibitors are all prodrugs that are converted into the corresponding active structure (triphosphate structure) in the host cell and are incorporated into the RNA chain by RdRp. This incorporation causes a lethal mutation or terminates immature RNA chain synthesis (Fig. 2 ).
Fig. 2.
Structures of representative nucleoside RdRp inhibitors.
2.1. Remdesivir (GS-5734)
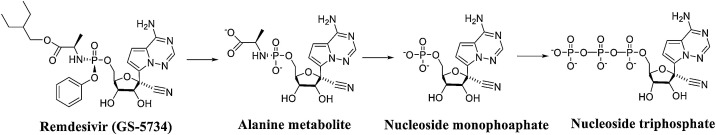
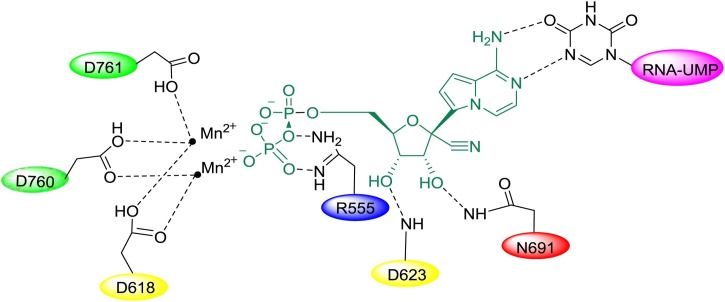
Remdesivir (GS-5734) is a novel nucleotide analogue antiviral prodrug that was developed by Gilead Sciences as a treatment for Ebola virus (EBOV) and Marburg virus infections [14,15], but it also exhibits reasonable antiviral activity against both SARS-CoV and MERS-CoV. Recent studies found that remdesivir shows activity against other coronaviruses, including SARS-CoV-2. An in vitro analysis of the anti-SARS-CoV-2 activity of remdesivir showed that the drug effectively blocked SARS-CoV-2 infection at low micromolar concentrations (EC50 = 0.77 μM) and exhibited low cytotoxicity (CC50>100 μM) with a high selection index (SI > 129.87) [16]. Remdesivir is hydrolysed into alanine metabolites by breaking the ester bond. These metabolites are converted into adenosine monophosphate and then further into adenosine triphosphate, ultimately becoming the active agent targeting the RNA virus polymerase and preventing viral replication (Fig. 3 ) [17,18]. Remdesivir maintains an intact ribose group and uses a hydrogen bond network as its native substrate (Fig. 4 ). Additionally, T680 in SARS-CoV-2 nsp12 is also likely to form hydrogen bonds with the 2′ hydroxyl of remdesivir.
Fig. 3.
Process of remdesivir-metabolized transformation to adenosine triphosphate.
Fig. 4.
Model of the incorporation of remdesivir into SARS-CoV-2 nsp12.
On May 1, 2020, the US Food and Drug Administration (FDA) issued an emergency use authorization (EUA) for treating patients with suspected or confirmed COVID-19 (including adults and children) with the antiviral drug remdesivir. Since then, remdesivir has also been approved for the treatment of COVID-19 patients in Japan, Taiwan, India, Singapore, the United Arab Emirates and the European Union. In other regions, remdesivir remains under investigation and has not been approved. A recent study tested the preventive and therapeutic effects of remdesivir against MERS-CoV in rhesus monkeys [19]. Prophylactic treatment with remdesivir initiated 24 h prior to inoculation completely prevented the clinical disease caused by MERS-CoV by strongly inhibiting the replication of MERS-CoV in the respiratory system and preventing the formation of lung lesions. Therapeutic treatment with remdesivir started 12 h after vaccination provided significant clinical benefits, including reduced clinical signs, decreased viral replication in the lungs and reductions in the number and severity of lung lesions. These findings support the effectiveness of remdesivir in the treatment of COVID-19 and indicate that the drug should be started as soon as possible after symptom onset to maximize its therapeutic effect.
On October 8, 2020, the National Institute of Allergy and Infectious Diseases released the results of the remdesivir trial (Adaptive COVID-19 Treatment Trial, ACTT-1, NCT04280705). Researchers pointed out that the average recovery time of the remdesivir group was 10 days, while that of the placebo group was 15 days. Although there was no significant difference in mortality between the two groups of patients, there was a significant difference in recovery time (p < 0.001). This study demonstrated that remdesivir is superior to placebo in reducing recovery time and can be used to treat COVID-19 [20]. However, on October 15, the World Health Organization (WHO) announced the release of the “Solidarity trial”. The study conducted clinical trials involving 11,266 patients from 405 hospitals in 30 countries. There were 301 deaths among 2743 critically ill patients in the remdesivir group and 303 deaths among 2708 critically ill patients in the control group, for mortality rates of 11 % and 11.2 %, respectively. As there was almost no difference in mortality, the WHO announced that remdesivir has only a rare effect in reducing the mortality of hospitalized patients [21]. By contrast, on October 22, the US FDA officially approved remdesivir as the first and only COVID-19 treatment drug in the country. It can be used for COVID-19 inpatient treatment of adult patients and children over 12 years and weighing over 40 kg. Compared with the Solidarity trial, the design of ACTT-1 (which was randomized, placebo-controlled, and double-blind) was more suitable for rigorously assessing the recovery time endpoint, which is the essence of the effectiveness and safety required for new drug approval. The main goal of the Solidarity trial was to evaluate the impact of treatment intervention on hospital mortality, and the results did not refute the effectiveness of remdesivir, because the effectiveness of remdesivir may be closely related to the shortened recovery time. Meanwhile, it has been reported that the parent nucleotide of remdesivir, GS-441,524, can effectively inhibit the replication of SARS-CoV-2 in Vero E6 cells (IC50 = 0.70 μM). In a rat pharmacokinetics study, it showed a good plasma distribution and a long half-life (t1/2 = 4.8 h). Considering that GS-441,524 is the main metabolite of remdesivir in plasma, it is speculated that GS-441,524 contributes to the anti−COVID-19 effect of remdesivir [22]. GS-441,524 may become a promising cheap drug candidate for the treatment of COVID-19 and future emerging CoV diseases.
2.2. Favipiravir
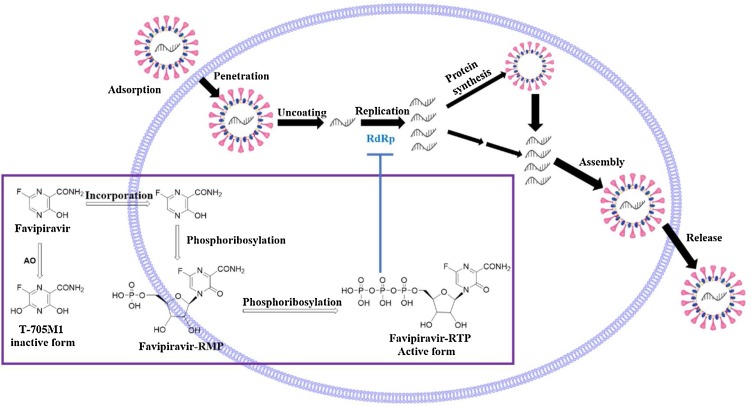
The antiviral drug favipiravir (T-705 or Avignan) is a pyrazinecarboxamide derivative developed by Toyama Chemical Company in Japan that exhibits activity against various RNA viruses [23]. Favipiravir shows activity against influenza virus, West Nile virus, yellow fever virus, foot-and-mouth disease virus, flaviviruses, arenaviruses, bunyaviruses, alphaviruses [24], enterovirus [25] and Rift Valley fever virus [26]. Favipiravir is also a prodrug that is first converted to the intermediate favipiravir-ribofuranosyl-5′-monophosphate through phosphoribosylation, and this intermediate is then further metabolized into its active form [27], favipiravir-ribofuranosyl-5′-triphosphate, which inhibits the activity of RdRp (Fig. 5 ). This drug is available in both oral and intravenous formulations. Clinical trials investigating the use of favipiravir in the treatment of COVID-19 have been performed in many countries (Table 1 ). Favipiravir exhibits significant anti-SARS-CoV-2 activity in Vero E6 cells, with an EC50 value of 67 μM. Studies have confirmed that favipiravir exhibits 100 % efficiency in protecting mice from EBOV [16]. In March 2020, favipiravir was approved by China’s State Drug Administration as China’s first anti−COVID-19 drug because the clinical trial showed that the drug exhibited high efficacy and few side effects. Chen et al. conducted a prospective, randomized, controlled, open-label multicentre trial involving adult COVID-19 patients (ChiCTR2000030254) [28], and elevated serum uric acid levels was the most frequently observed adverse event associated with favipiravir. The study proved that favipiravir is more effective than the antiviral drug Arbidol based on the average antipyretic time and time to achieve cough relief. Yi Shi et al. recently determined the cryo-EM structure of favipiravir bound to the replicating polymerase complex of SARS-CoV-2 in the pre-catalytic state; this structure provides a missing snapshot for visualizing the catalytic dynamics of coronavirus polymerase, facilitating extrapolation of the dynamic catalytic cycle for RNA nucleotide polymerization. These findings shed light on the mechanism of coronavirus polymerase catalysis and provide a rational basis for developing antiviral drugs to treat SARS-CoV-2 infection [29].
Fig. 5.
Mechanism of favipiravir-mediated inhibition of coronavirus replication in host cells (the purple wireframe and arrows represent the intracellular triphosphorylation of favipiravir and the inhibition of RdRp to terminate viral RNA replication). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Table 1.
Clinical trials of favipiravir.
| Study name | Study design | Intervention type | Condition | Identifier (phase) | Population number |
|---|---|---|---|---|---|
| Favipiravir in COVID-19 | Open | Drug therapy | Intensive care, intubation, endotracheal, SARS-CoV-2 infection (COVID-19) | / | 7 |
| Favipiravir in COVID-19: the VIRCO and NCT04445467 studies | Placebo-controlled, randomized |
Drug therapy | SARS-CoV-2 infection (COVID-19) | NCT04445467 | 190 |
| Favipiravir in COVID-19: the NCT04346628 study | Open, randomized |
Drug therapy | SARS-CoV-2 infection (COVID-19) | NCT04346628 | 120 |
| Favipiravir in COVID-19: the NCT04351295 study | Open, placebo-controlled, randomized |
Drug therapy | SARS-CoV-2 infection (COVID-19) | NCT04351295 | 40 |
| Favipiravir in COVID-19: the NCT04333589 study | Open, randomized |
Drug therapy | SARS-CoV-2 infection (COVID-19) | NCT04333589 | 210 |
| Favipiravir in COVID-19/viral pneumonia: the NCT04336904 study | Double-blind, placebo-controlled, randomized |
Drug therapy | Pneumonia, viral, SARS-CoV-2 infection (COVID-19) |
NCT04336904 | 100 |
| Favipiravir in SARS-CoV-2 infection (COVID-19): the NCT04402203 study | Double-blind, placebo-controlled, randomized | Drug therapy | SARS-CoV-2 infection (COVID-19) | NCT04402203 | 50 |
| Favipiravir in COVID-19: the Avi-Mild and NCT04464408 studies | Double-blind, multicentre, placebo-controlled, randomized |
Drug therapy | SARS-CoV-2 infection (COVID-19) | NCT04464408 | 576 |
| Favipiravir in SARS-CoV-2 infection (COVID-19): the NCT04358549 study | Open, randomized |
Drug therapy | SARS-CoV-2 infection (COVID-19) | NCT04358549 | 50 |
| Favipiravir in COVID-19: the NCT04425460 study | Double-blind, placebo-controlled, randomized |
Drug therapy | SARS-CoV-2 infection (COVID-19) | NCT04425460 | 256 |
| Favipiravir in SARS-CoV-2 infection (COVID-19)/respiratory tract infection prevention: the NCT04448119 study CONTROL-COVID | Double-blind, placebo-controlled, randomized | Prevention | SARS-CoV-2 infection (COVID-19) | NCT04448119 | 760 |
| Favipiravir or lopinavir/ritonavir in SARS-CoV-2 infection (COVID-19) | Open | Drug therapy | SARS-CoV-2 infection (COVID-19) | / | 80 |
| Favipiravir vs. hydroxychloroquine in COVID-19: the NCT04387760 study | Comparative, open | Drug therapy | SARS-CoV-2 infection (COVID-19) | NCT04387760 | 150 |
2.3. Ribavirin
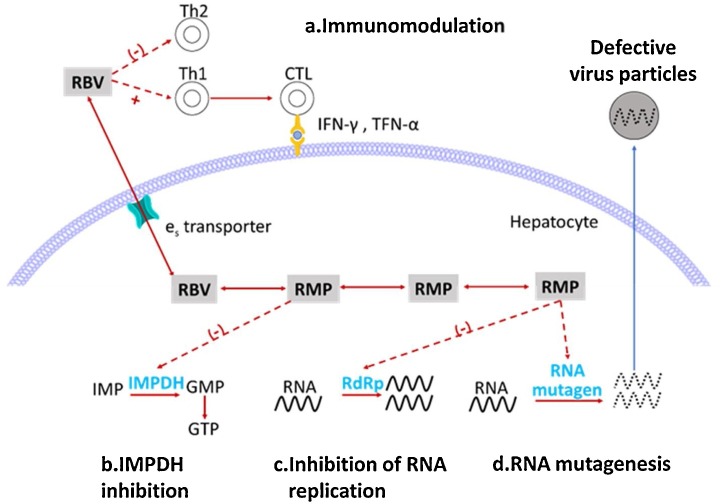
Ribavirin (RBV), also known as tribavirin, is a broad-spectrum antiviral medication used to treat respiratory syncytial virus infection, hepatitis C virus and some viral haemorrhagic fevers. RBV exhibits relatively mild anti-SARS-CoV-2 activity, with an EC50 value of 109.5 μM and a CC50 value higher than 400 μM in Vero E6 cells [16]. RBV is also a prodrug that is converted intracellularly into RBV monophosphate (RMP) by adenosine kinase and further into RBV diphosphate (RDP) and ribavirin triphosphate through nucleoside monophosphate and diphosphate kinases. The antiviral mechanism of RBV is complicated because it exerts its antiviral effects in various ways [30] (Fig. 6 ). Its effects include both indirect and direct actions. The indirect mechanism includes the following: (a) RBV can regulate the immune system by initiating the induction of a shift from the Th2 (T-helper cell 2) to Th1 (T-helper cell 1) immune response, thereby enhancing the host’s cytotoxic T lymphocyte-mediated antiviral immunity and (b) RMP can inhibit inosine monophosphate dehydrogenase (IMPDH) to induce the depletion of guanosine triphosphate (GTP), thereby hindering RNA virus replication. The direct mechanism includes the following: (c) RDP can directly inhibit RNA replication by inhibiting RdRp and (d) RDP can also be used as an RNA mutagen to accelerate the error mutation of rapidly mutating RNA viruses.
Fig. 6.
Mechanism of ribavirin against RNA viruses. The mechanism includes the following: a) induction of a shift from a Th2 to a Th1 immune response, b) inhibition of IMPDH to induce GTP depletion, c) direct inhibition of RdRp, and (d) induction of mutagenesis to trigger the production of defective viral particles.
3. Coronavirus non-structural protein inhibitors
3.1. 3CLpro inhibitors
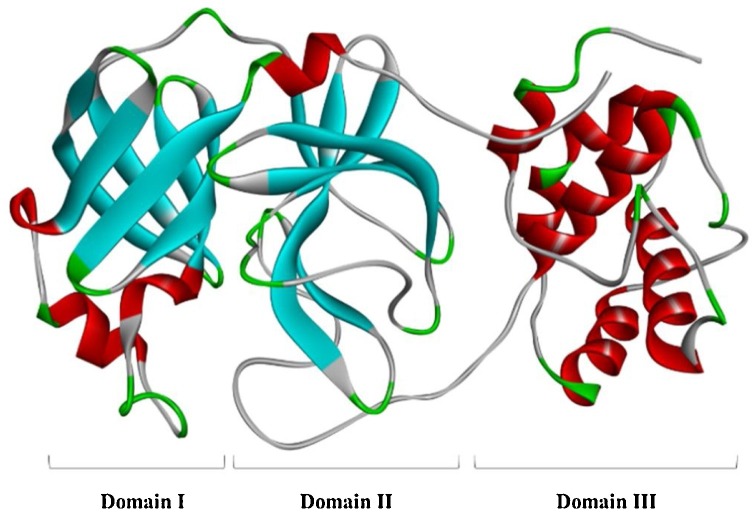
3CLpro, also known as the main protease (Mpro), is a three-domain (domains I to III) cysteine protease composed of 306 amino acids (Fig. 7 ). Domain I (residues 8–101) and domain II (residues 102–184) have anti-parallel β-barrel structures, and domain III (residues 201–303) contains 5 α-helices arranged in one substantially antiparallel globular cluster that is connected to domain II through a long loop region (residues 185–200) [31,32]. SARS-CoV-2 3CLpro has a Cys-His catalytic dyad (Cys145 and His41) in the gap between domains I and II that can specifically recognize 11 cleavage sites of nsp4∼nsp16 to induce the release of additional coronavirus nsps. The nsp4-nsp16 released by 3CLpro hydrolysis and shearing is a carrier of viral genome replication and transcription and is responsible for processes such as protein cleavage and modification and nucleic acid synthesis after translation [33]. Additionally, by comparing the amino acid sequences of SARS-CoV-2, SARS-CoV and MERS-CoV 3CLpro, the 3CLpro of the above three viruses was found to have a high degree of similarity, with significant conservation in the polyprotein cleavage site. The similarity between SARS-CoV 3CLpro and SARS-CoV-2 3CLpro is as high as 96.00 % [34]. Hence, this enzyme is considered one of the most attractive targets for the treatment of COVID-19. A series of 3CLpro inhibitors have been developed, and these can be divided into peptidomimetic and nonpeptidic inhibitors according to their structural type.
Fig. 7.
The crystal structure of SARS-CoV-2 3CLpro (PDB code: 6LU7).
3.1.1. Peptidomimetic 3CLpro inhibitors
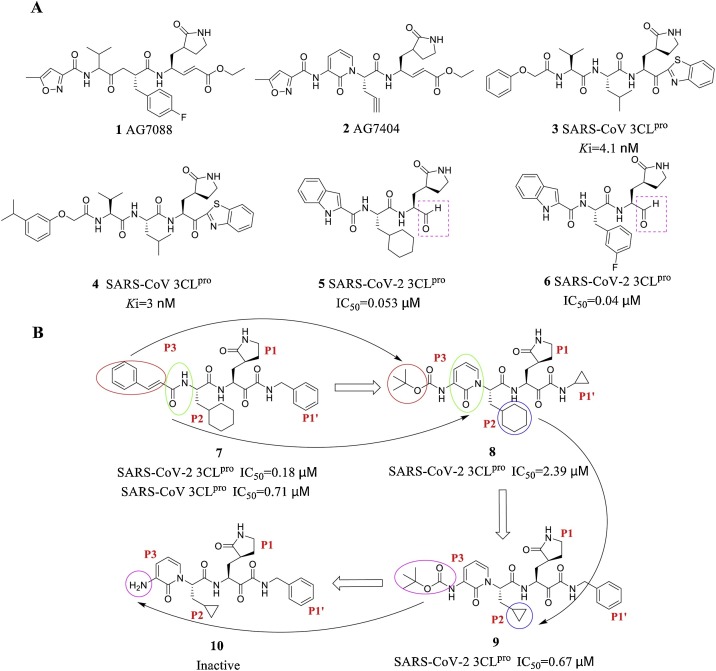
The mechanism of action of peptidomimetic inhibitors includes two steps. First, peptidomimetics that mimic natural peptide substrates bind to 3CLpro and form a noncovalent complex with the warhead group, which is spatially very close to the catalytic residue of the target protein. Second, these inhibitors carry out nucleophilic attack to covalently bind to Cys145 in the 3CLpro S1′ pocket to exert an inhibitory effect [35,36]. AG7088 (1) was the first peptidomimetic irreversible inhibitor and was developed by Pfizer [37]. Due to its off-target effects, research on this molecule was terminated based on results from phase II clinical trials. Optimization of the AG7088 structure yielded the compound AG7404 (2), which exhibited better oral availability and tolerability and has entered phase I clinical trials. Hayashi et al. [38] designed and synthesized the peptidomimetic compounds 3 and 4 using benzothiazolone as the warhead group, and the Ki values reached nanomolar levels. Liu Hong et al. [39] recently designed and synthesized a new class of peptidomimetic inhibitors with aldehyde groups as warheads for SARS-CoV-2 3CLpro. Compounds 5 and 6 exhibited outstanding inhibitory activity against SARS-CoV-2 3CLpro (5: IC50 = 0.053 ± 0.005 μM, 6: IC50 = 0.040 ± 0.002 μM). in vitro antiviral activity assays showed that these two compounds can effectively serve as anti-SARS-CoV-2 drugs. Further pharmacokinetic and safety studies have indicated that both 5 and 6 exhibit good in vivo pharmacokinetic properties with acceptable preliminary safety (5: EC50 = 0.053 μM, 6: EC50 = 0.72 μM) and have the potential to become a new anti-SARS-CoV-2 clinical drug candidates for further research. The research team in Germany guided by Rolf Hilgenfeld at Lübecker University, in cooperation with Chinese colleagues Zhang et al. [40], optimized the previously discovered peptidomimetic α-ketoamide inhibitor 7. The amide bond at the P2-P3 position of compound 7 was hidden on the pyridone ring (Fig. 8 , green circle) and is expected to prevent off-target effects and hydrolysis of the bond by other proteases, which would increase the half-life of the compound in plasma. In addition, the hydrophobic cinnamyl group was replaced by a low-hydrophobic Boc group to increase the solubility of 7 in plasma. The obtained compound 8 was further optimized, and the cyclohexyl group at position P2 was replaced by a smaller cyclopropyl group to yield 9, which exhibited stronger antiviral activity against both SARS-CoV-2 and SARS-CoV. However, compound 10, which was obtained by removing the Boc group of 9, was inactive, which indicates that the hydrophobic group is necessary for penetrating the cell membrane and interacting with the virus 3CLpro. COVID-19 and other coronaviruses mainly affect lung tissue, and pharmacokinetic studies have shown that the concentration of 9 in lung tissue 4 h after its subcutaneous administration is approximately 13 ng/g. Therefore, the lung tropism of 9 is a favourable target organ feature. A mouse lung drug inhalation model showed that 9 is well tolerated without adverse reactions; hence, lung inhalation is the appropriate method for the administration of 9 [41].
Fig. 8.
(A) Representative peptidomimetic 3CLpro inhibitors. (B) Chemical structures of α-ketoamide inhibitors 7-10. The coloured circles and arrows show the specific modifications at each development step.
3.1.2. Nonpeptidic 3CLpro inhibitors
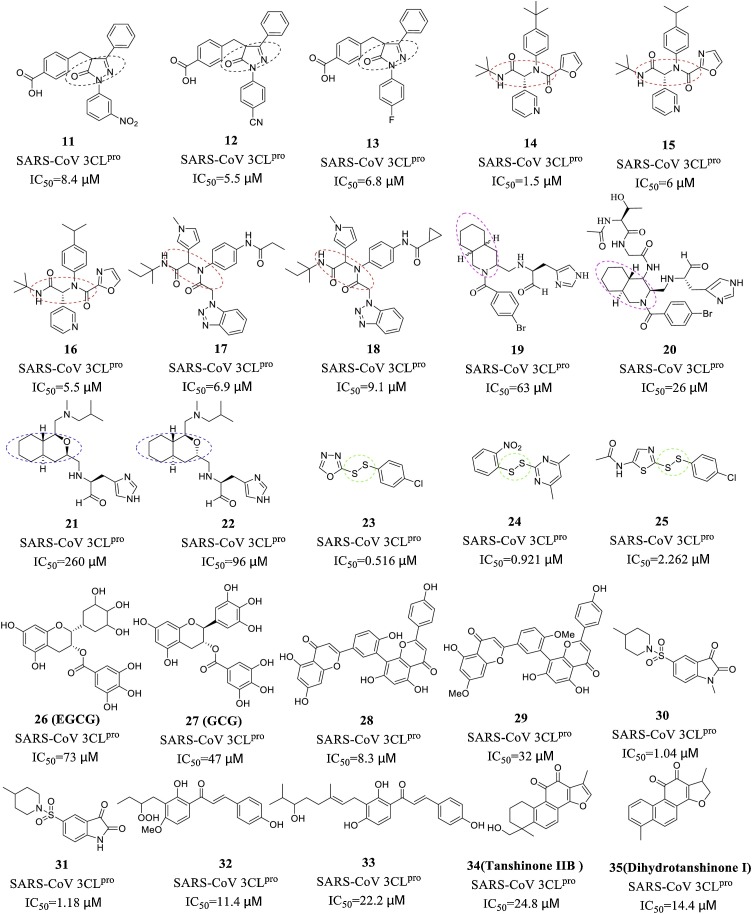
The mechanism of action of nonpeptidic inhibitors mainly involves the formation of hydrogen bonds with the amino acid residues of the S1, S2 and S4 pockets (including Cys145, which has catalytic activity in the S1′ pocket). Nonpeptidic inhibitors are currently an area of intense research due to their diverse structural core and active functional groups. The Ramajayam research team [42] designed and synthesized a series of pyrazolone core compounds (11-13) that exhibited good inhibitory activity against SARS-CoV 3CLpro and extremely low cytotoxicity. Additionally, Jacobs et al. designed and synthesized a series of SARS-CoV 3CLpro inhibitors with diamide scaffolds (14-18) [36,43]. Structure-activity relationship (SAR) studies have shown that pyridyl or triazole moieties are necessary for inhibitory activity, and this finding provides a new feature for the design of nonpeptidic inhibitors. In addition, 3CLpro nonpeptidic inhibitors with decahydroisoquinoline (19-20) [44,45], octahydro-isochromene (21-22) [46], and unsymmetrical aromatic disulphides (23-25) [47] as the core group show good inhibitory activity and can be further studied and modified. Natural product derivatives, such as flavonoids (26-27) [48], bioflavonoids (28-29) [49], isatin (30-31) [50], chalcones (32-33) [51] and terpenoids (compounds 34-35) [52], are also a class of 3CLpro nonpeptidic inhibitors with strong inhibitory activity.
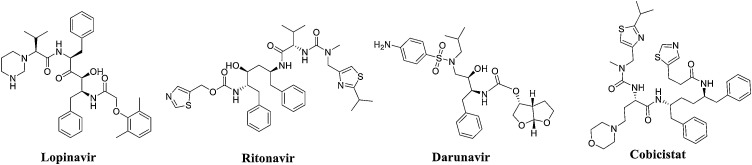
In addition to the abovementioned representative nonpeptidic inhibitors of SARS-CoV 3CLpro, a batch of HIV protease inhibitors with inhibition against SARS-CoV-2 3CLpro was obtained through a virtual screening of known compounds, such as lopinavir, ritonavir, darunavir, and cobicistat, based on the SARS-CoV-2 3CLpro crystal structure presented by the Rao Zihe team (PDB code: 6LU7) (See Fig. 9 and Table 2 ), but further experiments and clinical verification are needed [53]. Lopinavir/ritonavir (LPV/r), which is a combination protease inhibitor used to treat HIV-1 infection, has been included in China’s 2019 Coronavirus Diagnosis and Treatment Guidelines (Seventh Edition) for the treatment of COVID-19. Lopinavir, which is the active ingredient of this drug combination, can prevent the division of Gag-Pol polyprotein, thereby halting the production of immature virus particles and ultimately terminating the infection [54]. In Vero E6 cells, lopinavir exerts a significant inhibitory effect on SARS-CoV-2, with an EC50 of 26.63 μM [55]. Pharmacokinetic studies have shown that lopinavir is mainly metabolized by CYP3A4, and when used alone, this compound produces a lower systemic blood concentration than when used in combination because ritonavir can inhibit the metabolism of lopinavir. A clinical trial conducted at Ruian People’s Hospital included 47 COVID-19 patients admitted from January 22 to January 29, 2020. These patients were divided into an experimental group and a control group based on whether they received LPV/r treatment during their hospitalization. The experimental group was given LPV/r combined with adjuvant drug therapy, and the control group was administered adjuvant drug therapy alone. Changes in body temperature, routine blood tests, and blood biochemistry were observed between the two groups. The results showed that compared with single-use adjuvant drug therapy, LPV/r exerted more obvious effects in reducing the body temperature and restoring normal physiological mechanisms without side effects [56]. In addition, an emergency randomized clinical trial (ChiCTR2000029308) on the efficacy of LPV/r for COVID-19 patients was carried out in Wuhan Jinyintan Hospital. The experimental subjects were 199 adult inpatients with severe COVID-19, of which 100 were assigned to the conventional treatment group and 99 were assigned to the LPV/r group. The LPV/r group received conventional treatment with the addition of LPV/r (400 mg lopinavir, 100 mg ritonavir, twice a day, for 14 days). The results showed that for patients with severe COVID-19, compared with conventional supportive treatment alone, the addition of LPV/r did not improve clinical conditions or reduce mortality [57]. In this study, the main possible factor for why LPV/r did not achieve the expected results was that the research team chose a particularly challenging population. The patients who participated in the study had relatively advanced infections and had considerable tissue damage. Previous studies have shown that even highly active antibacterial agents have limited efficacy against advanced bacterial pneumonia. Based on the above research, the application of LPV/r combined with related adjuvant drugs is recommended for the clinical treatment of early COVID-19 patients.
Fig. 9.
Structure of representative nonpeptidic inhibitors.
Table 2.
Summary of HIV protease inhibitors against human coronaviruses.
| Infectious disease | Drug target | Drugs | Reported mechanism of action | Status |
|---|---|---|---|---|
| 2019-nCoV, MERS-CoV, ARS-CoV, HCoV-229E, HIV, HPV |
3CLpro | Lopinavir | Inhibits 3CLpro | Approved for HIV, in phase 3 trials of 2019-nCoV (NCT04252274, NCT04251871, NCT04255017, ChiCTR2000029539), in phase 2/3 trials for MERS (NCT02845843) |
| 2019-nCoV, MERS-CoV |
3CLpro | Ritonavir | Inhibits 3CLpro | Approved for HIV, in phase 3 trials for 2019-nCoV (NCT04251871, NCT04255017, NCT04261270), in phase 2/3 trials for MERS (NCT02845843) |
| 2019-nCoV | 3CLpro | Darunavir and cobicistat | Inhibits 3CLpro | Approved for HIV, in phase 3 trials for 2019-nCoV (NCT04252274) |
| 2019-nCoV | 3CLpro | ASC09 F (HIV protease inhibitor) | Inhibits 3CLpro | In phase 3 trials for 2019-nCoV in combination with oseltamivir (NCT0426270) |
3.2. PLpro inhibitors
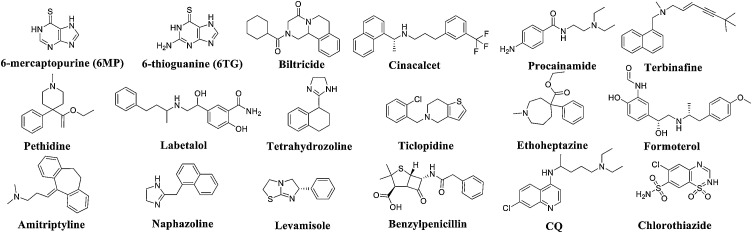
PLpro is a multifunctional protein with protease and phosphatase activity that participates in viral replication, regulates immune responses and antagonizes interferon (IFN) molecules [58,59]. SARS-CoV-2 PLpro is divided into four subdomains: an N-terminal ubiquitin-like domain, an α-helical thumb domain, a β-stranded finger domain and a palm domain. The geometric configuration of the active site of SARS-CoV-2 PLpro is similar to that of SARS-CoV PLpro. All catalytically important residues are unchanged, and the catalytic triad includes residues C111, H272 and D286 [60]. The PLpro cleavable protein causes the release of nsp1, nsp2, and nsp3 from viral polyproteins, and this process is necessary for viral replication [61,62]. Furthermore, PLpro is a deubiquitinating enzyme that can destroy the ubiquitination of host cells, which is not conducive to the innate immune response because ubiquitination is a key factor in regulating the this response [63]. PLpro is also a potential IFN inhibitory protein. After virus infection, the host will immediately activate its natural immune mechanism to produce IFN, which will place the body into an antiviral state and thus limit the replication and spread of the virus. PLpro exerts a significant inhibitory effect on the IFNβ pathway and the interferon regulatory factor 3 pathway. Although the PLpro sequences of SARS-CoV-2 and SARS-CoV exhibit only 83 % similarity, the high-level structure of the active site of PLpro is unchanged [64]. PLpro can be an important target for the development of anti-SARS-CoV-2 drugs. Chouyuan Chou et al. [65] determined that 6-mercaptopurine (6 M P, IC50 = 21.6 μM) and 6-thioguanine (6 TG, IC50 = 5 μM) are widely clinically used for the treatment of childhood acute lymphoblastic leukaemia and myeloid leukaemia and that this combination of SARS-CoV PLpro inhibitors is reversible (Fig. 11); their study constitutes the first investigation of a small-molecule reversible inhibitor of PLpro. Kinetic tests and computer docking studies have revealed that these two compounds are competitive, selective, and reversible PLpro inhibitors, with Ki values ranging from 10 to 20 μM. A SAR study found that the thiocarbonyl moiety of 6 M P or 6 TG is the active pharmacophore for these inhibitory effects. 6 M P or 6 TG might form a hydrogen bond with Cys1651 in the active site of the enzyme through the thiol moiety of the thiocarbonyl group. Hydrogen bonding with Cys1651 blocks the essential sulfhydryl group, and this blockage prevents acylation and inhibits the activity of the enzyme. Although their anti-PLpro activity is in the μM range, these compounds are excellent lead compounds that can be further optimized to develop better antiviral drugs. Many research teams have identified PLpro inhibitors through virtual screening. For example, disulfiram is a potential PLpro inhibitor that acts on the active site of coronavirus PLpro and forms a covalent adduct with Cys112 [66]. Mukesh Kumar et al. used the SARS-CoV PLpro crystal structure (PDB code: 3E9S) as a template to virtually screen potential anti-SARS-CoV-2 PLpro inhibitors and found that 16 FDA-approved drugs exhibit good affinity for SARS-CoV-2 PLpro, which indicates that these drugs can be used as candidates for further research (see Fig. 10 and Table 3 ) [67].
Fig. 11.
Structure of representative PLpro inhibitors.
Fig. 10.
Structure of HIV protease inhibitors with inhibitory activity against SARS-CoV-2 3CLpro.
Table 3.
Sixteen FDA-approved drugs showing the highest affinity for SARS-CoV-2 PLpro.
| Drug | Binding affinity | Current application |
|---|---|---|
| Biltricide | 8 nM-8 μM | Anthelmintic |
| Cinacalcet | 26 nM-3 μM | Calcimimetic, treatment of hyperparathyroidism |
| Procainamide | 30 nM-3 μM | Antiarrhythmic |
| Terbinafine | 33 nM-3 μM | Antifungal |
| Pethidine | 53 nM-5 μM | Narcotic analgesic |
| Labetalol | 113 nM-11 μM | Treatment of hypertension |
| Tetrahydrozoline | 137 nM-14 μM | Over-the-counter eye drops and nasal spray |
| Ticlopidine | 160 nM-16 μM | Inhibitor of platelet aggregation |
| Ethoheptazine | 163 nM-16 μM | Opioid analgesic |
| Formoterol | 716 nM-71 μM | Management of COPD and asthma |
| Amitriptyline | 466 nM-46 μM | Antidepressant with analgesic properties |
| Naphazoline | 697 nM-69 μM | Decongestant in over-the-counter eye drops and nasal preparations |
| Levamisole | 259 nM-26 μM | Antihelminthic used for parasitic, viral, and bacterial infections |
| Benzylpenicillin | 718 nM-71 μM | Narrow-spectrum antibiotic |
| CQ | 858 nM-85 μM | Antimalarial agent |
| Chlorothiazide | 939 nM-93 μM | Diuretic |
Wioletta Rut et al. [68] developed a hybrid combinatorial substrate library containing a wide variety of nonproteinogenic amino acids for SARS-CoV-2 PLpro to analyse the broad substrate specificity of proteolytic enzymes. Their results revealed high substrate specificity for the S2 pocket of SARS-CoV-2 PLpro and that only glycine can be accepted. The S3 pocket exhibits a wide range of substrate specificity that can not only tolerate positively charged residues, such as Phe (Guan), Dap, dab, Arg, Lys, Orn, and hArg, but can also tolerate hydrophobic amino acids, such as hTyr, Phe(F5), Cha, Met, Met(O), Met(O)2, and d-hPhe. However, the S4 pocket can only accommodate hydrophobic residues. Positional scanning technology can be used to design a map of the best fluorescent substrates (Fig. 12 ) of irreversible inhibitors (VIR250 and VIR251) on the best-hit scaffold. Analyses of the crystal structures of the complexes of SARS-CoV-2-PLpro with VIR250 and VIR251 showed that the catalytic Cys111 of SARS-CoV-2-PLpro engages in Michael addition to the b-carbon of the vinyl group of the VME warheads of VIR250 and VIR251, resulting in the formation of a covalent thioether linkage. Moreover, VIR250 and VIR251 both occupy the S4-S1 pocket attached to the active site and form hydrogen bonds with the residues in the pocket. These two inhibitors were found to be active and selectively inhibited SARS-CoV and SARS-CoV-2 PLpro, respectively, but exhibited markedly weaker activity towards MERS-PLpro and no activity towards human UCH-L3. Therefore, these findings provide important information for further research aiming to identify peptide antiviral compounds that target this enzyme.
Fig. 12.
(A) Structures of the tetrapeptide substrate. (B) X-ray crystal structure of VIR250 and VIR251 bound to SARS-CoV-2 PLpro. The residues are shown as sticks; the red, blue and yellow colours indicate oxygen, nitrogen, and sulphur atoms, respectively, and hydrogen bonds are indicated by dashed lines (left: VI2R50, PDB code: 6WUU; right: VIR251, PDB code: 6WX4). (C) VIR250 and VIR251 bound to the binding pocket of SARS-CoV PLpro. VIR250 and VIR251 are shown as sticks, and the P2-P4 positions are labelled. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
4. Host-targeted inhibitors
There are currently two main strategies for designing broad-spectrum antiviral drugs (BSAs): the use of nucleoside or nucleotide analogues and the use of host-targeting antivirals (HTAs). Nucleosides and nucleoside analogues are direct-acting antiviral agents (DAAs), which usually cause drug resistance and toxicity. Owing to viral specificity, the development of new DAAs takes a considerable amount of time when a new virus breaks out. It is necessary to block viral replication to overcome potential viral mutagenesis [69]. Therefore, HTAs are generally more advantageous against viruses.
4.1. Dihydroorotate dehydrogenase (DHODH) inhibitors
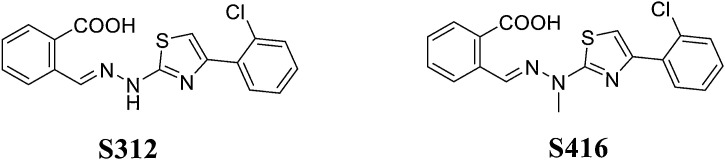
DHODH is an important rate-limiting enzyme in the de novo synthesis of pyrimidine, which catalyses the conversion of dihydroorotate to orotate and provides nucleotides for the synthesis of RNA/DNA [70,71]. Ke Xu [72] confirmed that DHODH can be used as an antiviral drug target by verifying its high antiviral efficacy and low viral replication in DHODH-knockout cells through in vivo experiments. Two effective DHODH inhibitors with good pharmacokinetic characteristics, S312 and S416 (Fig. 13 ), can act against a variety of RNA viruses, including influenza A virus (H1N1, H3N2, and H9N2), Zika virus and Ebola virus, and they exert a strong inhibitory effect on SARS-CoV-2. Notably, S416 exhibits an EC50 value of 17 nM and an SI value of 1,050.88 in SARS-CoV-2-infected cells. Furthermore, S312/S416 and the older drugs (leflunomide/tefnomide) exert dual antiviral and immune regulatory effects. Thus, mutations in SARS-CoV-2 or other RNA viruses circulating around the world are not a concern because they exhibit clinical potential to cure these viral infections.
Fig. 13.
Structures of S312 and S416.
4.2. S-phase kinase-associated protein 2 (SKP2) inhibitors
SARS-CoV-2 infection restricts autophagy by interfering with multiple metabolic pathways. In vitro targeted autophagy-induced compounds can reduce the release of SARS-CoV-2. An in-depth analysis of autophagy signals and metabolomics showed reductions in glycolysis and protein translation by limiting the activation of AMP-activated protein kinase (AMPK) and mTORC1 [73] with Beclin1 (BECN1) as its key regulatory factor. SKP2 inhibitors can reduce BECN1 ubiquitination, which reduces BECN1 degradation and enhances autophagy. The SKP2-BECN1 connection constitutes a promising target for host-directed antiviral drugs (including SARS-CoV-2) and other autophagy-sensitive conditions.
Niclosamide is a drug used to kill tapeworms and has previously been identified as a SKP2 inhibitor [74,75]. Importantly, Theo Rein et al. found that niclosamide reduces the replication of MERS-CoV by 28,000-fold [76]. The maximal inhibition of infectious virus at nontoxic concentrations was identified to be 1.24 μM for niclosamide (>99 %). Mingyi Zhao et al. reported that niclosamide was able to bind to Met124 of ACE. These findings indicate niclosamide as a potential anti-SARS-CoV-2 drug [77,78].
4.3. Potential drugs for the prevention or treatment of cytokine storm
Cytokine storm can cause serious illness in patients and targets host cells. Cytokine storm is characterized by an uncontrolled elevation of proinflammatory cytokines, an excessive recruitment of immune cells, and systemic inflammation leading to tissue and organ damage [79,80]. Therefore, inhibiting the inflammatory storm caused by the release of cytokines has been proposed as a means to limit the severity of COVID-19 [[81], [82], [83]].
4.3.1. Thalidomide
Thalidomide exerts the dual effects of immunosuppression and immunostimulation and might play a role in COVID-19 immune reconstruction, anti-inflammation, lung injury reduction, anti-pulmonary fibrosis and suppression of cytokine storm. Experiments have shown that thalidomide is a promising therapeutic drug for the treatment of COVID-19 [84,85]. A patient with COVID-19 in China received thalidomide combined with low-dose glucocorticoid and antiviral therapy. The patient was administered 100 mg of thalidomide and a small dose of methylprednisolone. The results showed a rapid increase in the oxygen index, the suppression of anxiety, nausea and vomiting, and no reported side effects. The anti-inflammatory and immunomodulatory effects of thalidomide are related to the reduction of inflammatory cytokines (IL-6, IL-10, and IFN-γ) and recovery of the lymphocyte count [86]. Two registered phase II clinical trials have evaluated the efficacy of thalidomide as an immunomodulatory drug for the treatment of SARS-CoV-2 infection. The first clinical trial (NCT04273581) explored the efficacy and safety of the drug combined with low-dose hormones in the treatment of severe COVID-19. The second trial (NCT04273529) investigated the effectiveness and safety in the treatment of pneumonia. Therefore, thalidomide might be a potential drug when it is difficult to achieve short-term antiviral effects.
4.3.2. Pentoxifylline and oxypurinol
Gerardo Lopez-Rodas et al. proved that the administration of pentoxifylline alone or in combination with oxypurinol for the early treatment of patients with COVID-19 can prevent potentially fatal acute respiratory distress. Pentoxifylline is a TNF-α inhibitor, whereas oxypurinol is a xanthine oxidase inhibitor. TNF-α can activate other inflammatory-related genes, such as Nos2, ICAM and IL-6, which regulate the migration and infiltration of neutrophils into the lung interstitial tissue and thereby cause damage to the lung parenchyma. In acute pancreatitis, the anti-inflammatory effect of pentoxifylline might be mediated by preventing the rapid and temporary loss of protein phosphatase 2A activity, which might also occur during the stage of SARS-CoV-2 infection characterized by the release of inflammatory cytokines [87].
4.4. TMPRSS2 inhibitors and furin inhibitors
The coronavirus S protein promotes viral entry into the target cell and depends on the S1 subunit on the surface of the S protein. This subunit helps the virus attach to the surface of the target cell. The entry of S protein needs to be initiated by cellular proteases, and this process requires cleavage of the S protein at the S1/S2 and S2′ sites, which then allows fusion of the virus membrane and cell membrane. The fusion process is driven by the S2 subunit [73]. SARS-CoV-2 binds to ACE2 to enter the host cell, and the S protein on the surface of SARS-CoV-2 mediates the interaction with ACE2. S protein activation is crucial for the entry of SARS-CoV-2 and depends on TMPRSS2 and furin [88,89]. TMPRSS2 and furin cleave S at different sites – furin at the S1/S2 site and TMPRSS2 at the S2′ site. Therefore, TMPRSS2 and furin can be used as a combination strategy of small peptidomimetic inhibitors to synergize S cleavage. This was shown to result in a synergistic blockage of SARS-CoV-2 propagation in cell culture, indicating an eventual therapeutic application.
4.4.1. TMPRSS2 inhibitors
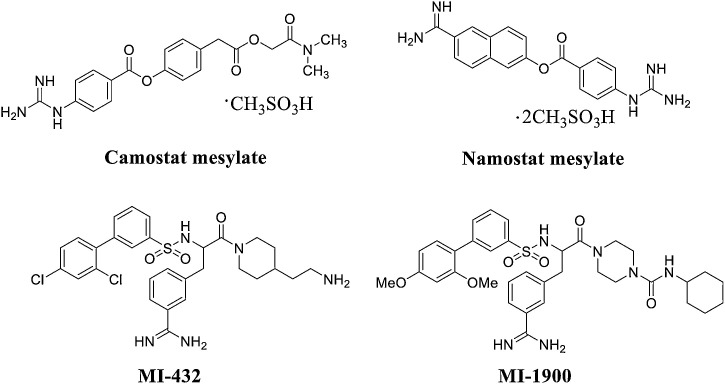
Camostat mesylate is a potent TMPRSS2 inhibitor that blocks SARS-CoV and HCoV-NL63 infection in HeLa cells expressing ACE2 and TMPRSS2 (Fig. 14 ) [90]. Hoffmann and colleagues recently confirmed that this drug effectively blocks the entry of SARS-CoV-2 into lung cells [91]. Another TMPRSS2 inhibitor for the treatment of COVID-19 is nafamostat mesylate, which is an FDA-approved drug for indications not related to coronavirus. It is significantly more effective than camostat mesylate in blocking SARS-CoV-2 infection of human lung cells [92]. Interestingly, Dorothea Bestle et al. [93] found that the peptide mimetic TMPRSS2 inhibitors MI-432 and MI-1900 strongly prevented the proliferation and strong cytopathic effect (CPE) of SARS-CoV-2 in CALU-3 cells in a dose-dependent manner. When MI-432 and MI-1900 were individually applied at 20 and 50 μM, only small foci of infection were visible (Fig. 14). At 10 μM, the diffusion and CPE of SARS-CoV-2 in MI-432- and MI1900-treated cells was reduced compared to that of control cells (no protease inhibitor, 0.5 % DMSO-treated cells). TMPRSS2 is essential for SARS-CoV-2 infection and appears to be a promising target for COVID-19 therapeutics [94].
Fig. 14.
Structures of TMPRSS2 inhibitors.
4.4.2. Furin inhibitors
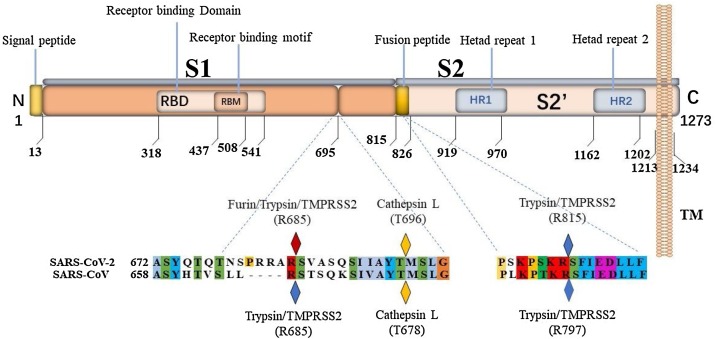
Furin is a pro-protein convertase (PC) located in the trans-Golgi network (TGN) that is activated by an acidic pH environment [95]. This enzyme cleaves precursor proteins with specific motifs to produce mature proteins with biological activity [96]. Furin has key roles in embryonic development and cell homeostasis [97]. Similar to other enveloped viruses that rely on the surface S protein for binding and fusion, the S protein of coronaviruses is cleaved by proteases during virus particle biosynthesis and entry into target cells [98]. A comparison between the SARS-CoV-2 S protein sequence and the highly homologous SARS-CoV sequence revealed that most features of SARS-CoV-2 S protein are similar to those of SARS-CoV. SARS-CoV-2 S protein has an N-terminal signal peptide that is divided into two parts, S1 and S2. S1 contains the N-terminal domain and receptor binding region, and S2 is mainly involved in membrane fusion. The C-terminal region of S2 is S2′, which contains a fusion peptide, heptad repeat 1, heptad repeat 2, and a transmembrane domain (Fig. 15 ) [99]. Unlike SARS-CoV, SARS-CoV-2 S protein contains a PRRA insertion that provides a furin cleavage motif of PRRAR|S at the S1/S2 site (|: cleavage site) [100], which might explain why SARS-CoV-2 infection is more severe than SARS-CoV. In addition, homology alignments and phylogenetic analysis of the SARS-CoV-2 sequence revealed that the PRRA insert did not appear in other close relatives of SARS-CoV-2, indicating that this insert is novel. The existence of this motif might allow the cleavage of S protein into S1 and S2 by furin protease prior to maturation; this effect would provide S1 with the flexibility to change its conformation to better adapt to the host receptor [101,102].
Fig. 15.
Sequence analysis of the SARS-CoV-2 S protein. The amino acid sequence positions of each domain are underneath. The cleavage sites of the SARS-CoV and SARS-CoV-2 S proteins are marked with diamonds.
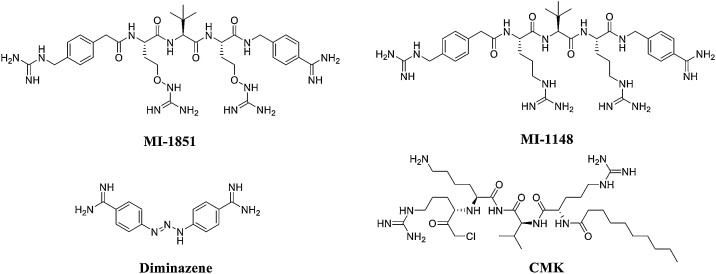
Importantly, furin inhibitors can reduce the activity of the SARS-CoV-2 S protein and reduce the infectivity of SARS-CoV-2. Therefore, furin inhibitors are very likely to be potential SARS-CoV-2-specific targeted therapy drugs. Dorothea Bestle et al. [97] found that a synthetic furin protease inhibitor (MI-1851, Fig. 16 ) strongly inhibited the multicycle replication of SARS-CoV-2 in CALU-3 human airway cells, even at the lowest concentration of 10 μM, producing a 30- to 190-fold reduction in virus titres. This indicates that the inhibitor MI-1851 is a promising compound for further drug development. Interestingly, the authors also found that the combination of TMPRSS2 inhibitors and furin inhibitors had a synergistic antiviral effect: the combination of 10 μM MI-1851 and 10 μM MI-432 showed enhanced antiviral activity against SARS-CoV-2 and reduced virus titres by 10- to 30-fold compared with 10 μM of each inhibitor alone. 4-Guanidinomethyl-phenylacteyl-Arg-Tle-Arg-4-amidinobenzylamide (MI-1148) is a novel peptidomimetic furin inhibitor containing unnatural amino acid residues in the P3 position and has a significant inhibitory effect on furin, with a Ki value of 5.5 pm. MI-1148 has a strong inhibitory effect on the highly pathogenic H5N1 and H7N1 avian influenza viruses and on the reproduction of canine distemper virus. MI-1148 also has the potential to inhibit furin activity in SARS-CoV-2 [103]. Thus, it is a promising compound for the treatment of acute infectious diseases. Hua Li et al. [104] used structure-based virtual screening to screen a library of 4000 compounds (including approved drugs and natural products) for their ability to inhibit furin. The authors then analysed the identified compounds for their inhibitory effects. The antiparasitic drug diminazene showed a high inhibitory effect on furin, with an IC50 of 5.42 ± 0.11 μM; therefore, it should be investigated as a potential COVID-19 therapeutic.
Fig. 16.
Structures of furin inhibitors.
The combined administration of different proteases or other target inhibitors and furin inhibitors might constitute an effective anti-SARS-CoV-2 treatment strategy. Yawen Cheng et al. [105] validated cleavage at a putative furin substrate motif of the SARS-CoV-2 S protein by expressing the protein in Vero E6 cells and found prominent syncytium formation. They found that treatment with the furin protease inhibitor decanoyl-RVKR-chloromethylketone (CMK) and naphthofluorescein prevented cleavage and syncytia. CMK and naphthofluorescein reduced virus production and had cytopathic effects in SARS-CoV-2-infected cells. Further analysis showed that CMK inhibited the virus by preventing viral entry and also inhibited the lysis of the S protein and syncytia, while naphthofluorescein mainly acted by inhibiting viral RNA transcription.
4.5. Repurposing of antimalarial drugs
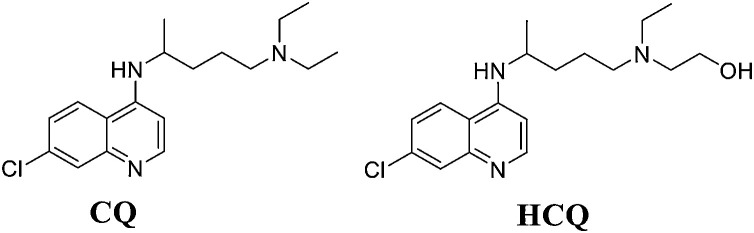
CQ and HCQ are traditional, weak antimalarial drugs (Fig. 17 ). After entering the cell, these drugs preferentially accumulate in acidic organelles, endolysosomes (ELs), and the Golgi and neutralize the lumen of acidic organelles. Malarial parasites rely on acidic digestive vacuoles to survive, which is the main reason for the antimalarial efficacy of CQ and HCQ. SARS-CoV-2 needs an acidic environment to enter the host cell [106]. Therefore, the deacidification effect of CQ and HCQ on ELs and the Golgi might prevent SARS-CoV-2 from being integrated into host cells, thereby exerting an antiviral effect against SARS-CoV-2. CQ can effectively block virus infection at low micromolar concentrations and exhibits a high SI (EC50 = 1.13 μM; CC50>100 μM, SI > 88.50), while the anti-SARS-CoV-2 activity of HCQ appears to be weaker than that of CQ [16]. CQ can inhibit virus replication by reducing ACE2 terminal glycosylation on the surface of host cells and interfering with the binding of S protein to ACE2 [107]. The Hansen group found that HCQ can destroy lipid domains, including monosialotetrahexosylganglioside 1 (GM1) lipid rafts and phosphatidylinositol 4,5-bisphosphate (PIP2). These lipid domains can transform SARS-CoV-2 receptors on the surfaces of blood vessels. ACE2 is recruited to the entry point of endocytosis. HCQ also blocks the ability of GM1 lipid rafts and PIP2 domains to attract and accumulate ACE2 and thus reduces both ACE2 recruitment and virus entry [108]. Both CQ and HCQ can block the transport of SARS-CoV-2 from early endosomes to ELs [109].
Fig. 17.
Structure of CQ and HCQ.
4.6. S protein inhibitor (Arbidol)
Arbidol is a broad-spectrum antiviral drug mainly used to treat upper respiratory tract infections caused by influenza A and B viruses. In recent years, many studies have proven its inhibitory action against SARS-CoV and MERS-CoV [110,111]. Arbidol works by binding to the haemagglutinin (HA) protein. Protein sequence analysis has shown that a short segment (aa947-aa1027) of the trimerization domain (S2) of the SARS-CoV-2 S protein exhibits similarity with the H3N2 HA sequence. The length of the SARS-CoV-2 S protein coding gene sequence is 3822 bp, and the coding product consists of 1273 amino acid residues. It includes four functional domains: a signal peptide, an extracellular domain, a transmembrane domain and a cytoplasmic domain. The S protein forms a corolla-like structure in the form of trimers, which is cleaved into two subunits (S1 and S2) under the action of host cell proteases. The S1 subunit is responsible for binding to the host cell receptor, and the S2 subunit is responsible for fusion of the virus and host cell membranes [112]. The SARS-CoV-2 S protein, located in the outer membrane, is necessary for the adhering to human ACE2 and CD26 receptors on the host cell, and its trimerization is necessary for fusion with the host membrane. Arbidol can effectively block or prevent the trimerization of SARS-CoV-2 S protein, which is key to cell adhesion and entry [113]. Yuan Xue et al. found that Arbidol monotherapy is more effective than lopinavir/ritonavir in the treatment of COVID-19 [110]. However, further clinical research is needed to determine the efficacy and safety of Arbidol when used to treat SARS-CoV-2 infection.
4.7. Corticosteroids
Corticosteroid drugs have been included in the Diagnosis and Treatment Programs of 2019 New Coronavirus Pneumonia (Eighth edition) formulated by the NHC of China. Ruijin Hospital, which is affiliated with the Shanghai Jiaotong University School of Medicine, quickly initiated drug screening and verification experiments for ACE2 activators and found that corticosteroids could improve severe and critical COVID-19 by activating ACE2 and inhibiting IL-6 levels [114]. ACE2 is a protective enzyme that maintains the balance of the respiratory system, cardiovascular system and vital organ functions under physiological conditions. When SARS-CoV-2 invades the body, ACE2 is rapidly depleted, which disrupts the body’s respiratory system, cardiovascular system and other important organs. When this occurs, patients can quickly deteriorate and experience multi-organ failure. Therefore, administering corticosteroids may become an option for COVID-19 treatment. Notably, to varying degrees, commonly used corticosteroids significantly activated the expression of ACE2 and inhibited the IL-6 produced by macrophages. Among these drugs, hydrocortisone was the strongest, followed by prednisolone, dexamethasone and methylprednisolone.
On June 16, 2020, a large-scale clinical trial (RECOVERY project) in the United Kingdom announced that dexamethasone reduced the mortality rate by one-third in severely ill COVID-19 patients. The authors recruited more than 11,000 patients from 175 hospitals in the British National Medical Service System. Overall, 2100 participants recruited from March to June were given low and medium doses (6 mg) of dexamethasone every day for ten consecutive days. The treatment results were compared with those of 4300 people receiving standard care for SARS-CoV-2 infections and showed that dexamethasone works best for critically ill patients who require ventilators, and the fatality rate was reduced by one-third. Patients who needed oxygen but did not use a ventilator also showed improvement after dexamethasone treatment, and the risk of death in this group was reduced by 20 % [115]. These experimental data indicate that the benefits of using low and medium doses of corticosteroid therapy may exceed the potential harm.
5. Discussion and perspectives
Since the emergence of the COVID-19 pandemic, scientists worldwide have been committed to the research and development of SARS-CoV-2 therapeutic drugs. Potential SARS-CoV-2 inhibitors have been discovered through the use of computer simulation techniques, such as molecular docking and free energy calculations. Based on the highly conserved replicated gene sequence of SARS-CoV-2, the development of anti-SARS-CoV-2 or broad-spectrum antiviral drugs targeting RdRp, 3CLpro, PLpro, TMPRSS2, S protein and ACE2 might constitute a future research direction [[116], [117], [118], [119], [120]]. These drugs include inhibitors of TMPRSS2, ACE2, proteases and virus/host cell membrane fusion, as well as existing antimalarial drugs. SARS-CoV-2 uses ACE2 to enter cells; TMPRSS2 is used for the activation of S protein; and furin can accumulate and participate in cell invasion by SARS-CoV-2 together with TMPRSS2 and cathepsin; however, furin does not directly participate in cell invasion. Furin pre-activates the S protein to reduce the dependence of SARS-CoV-2 on the invading target cells, especially cells with low expression levels of TMPRSS2 and/or cathepsin. Therefore, furin protease and TMPRSS2 inhibitors can synergistically block SARS-CoV-2 from entering host cells. This discovery is of great significance for pharmacological and pharmacodynamic studies of drug combinations. Phytochemicals, such as flavonoids, isatins, and terpenoids, which exert recognized curative effects against SARS-CoV-2 infection, might be used in many people owing to their safety, low pharmacological side effects and availability [121,122]. However, the targets and mechanisms of action of several drug candidates remain unclear. Further structural and biophysical studies are needed to determine how these drugs bind to and affect SARS-CoV-2. In addition to the prevention strategies adopted by many countries against SARS-CoV-2 infection, such as the isolation of confirmed cases of infection, the development and provision of vaccines is urgently needed to completely halt the pandemic [123,124].
Existing antiviral medications that have been used for multiple diseases, such as SARS, MERS, HIV/AIDS and malaria, are currently being tested in clinical trials [125]. The pathogenic mechanisms underlying the morbidity and mortality of COVID-19 are diverse, and immuno-inflammatory contributions appear to be a key player [126]. The mechanism of action of the anti-HIV protease inhibitor LPV/r may involve the inhibition of 3CLpro. LPV/r was specifically developed for the aspartate-classified HIV-1 encoded protease, which contains a characteristic Asp-Thr-Gly (Asp25, Thr26 and Gly27) catalytic triad. The SARS-CoV-2 protease contains a cysteine-histidine catalytic site, and the catalytically active sites of the two are completely different. However, whether LPV/r will be an effective treatment for COVID-19 remains to be seen and requires additional clinical observation. Based on previous research, LPV/r is currently the preferred antiviral therapy. CQ is an antimalarial drug. Because it can inhibit the spread of SARS-CoV-2 in kidney-derived Vero cells, it is used for treating COVID-19. However, a recent study showed that CQ could not prevent SARS-CoV-2 from infecting the TMPRSS2-positive lung cell line CALU-3. This means that the virus activation pathway targeted by CQ is not present in lung cells, and CQ is unlikely to prevent SARS-CoV-2 from infecting and spreading within and between individuals. Relevant research on the potential for CQ to treat COVID-19 is increasing; however, further studies are needed [127].
Various treatment agents have emerged from different medical areas. These agents include pharmacological agents [128], corticosteroids [129] and immunological agents and antibodies from the serum of patients who were infected with other coronaviruses or from those who have recovered from COVID-19 [130]. Docking analysis studies have proposed ribavirin, remdesivir, sofosbuvir, galidesivir and tenofovir as potent drugs for treating SARS-CoV-2 infection. Additionally, in silico analyses have confirmed that zanamivir, indinavir, saquinavir and remdesivir can be used as SARS-CoV-2 3CLpro inhibitors [131]. Among the SARS-CoV-2 macromolecules, large polyproteins encode the cysteine protease 3CLpro, which is essential for the viral lifecycle of coronaviruses [132]. This enzyme plays a crucial role in processing viral polyproteins, which are indispensable for viral maturation and infectivity [133].
Hence, viral proteins are considered potential targets for the development of antiviral compounds. 3CLpro is first routinely cleaved from polyproteins to produce mature enzymes, and it subsequently further cleaves downstream nsps at 11 cleavage sites to release nsp4−16 [134]. Cleavage by 3CLpro occurs at the glutamine residue in the P1 position of the substrate through the Cys-His catalytic dyad in which the cysteine thiol functions as the nucleophile in the proteolytic process. Thus, inhibiting the activity of this enzyme arrests the viral replication of SARS-CoV-2 [135]. PLpro is also an important potential target for the treatment of COVID-19. Naphthalene-based PLpro inhibitors can effectively halt SARS-CoV-2 PLpro activity and viral replication and provide a potential strategy for the rapid development of PLpro-targeted therapeutics for use against SARS-CoV-2.
Among the many therapeutic drugs for COVID-19, RdRp inhibitors (e.g. remdesivir) have generally been the most often used [136]. Cell culture and animal model studies have confirmed that remdesivir inhibits coronaviruses, including SARS-CoV and MERS-CoV [137]. Coronaviruses usually exhibit some proofreading ability to detect and correct the incorporation of incorrect nucleoside analogues, but remdesivir outpaces this protective barrier to exert its antiviral activity [138]. In a compassionate use trial, the majority of patients with severe coronavirus symptoms treated with remdesivir exhibited clinical improvement, but the safety and effectiveness of the drug remain controversial. The most common adverse events were elevated liver enzymes, diarrhoea, skin rash, renal impairment, and hypotension. Therefore, further research and verification, including in vitro cell experiments, animal model experiments and drug clinical trials, are needed [139]. One study found that large doses of remdesivir caused insulin pill toxicity and led to sperm parameter deterioration in mice. Therefore, additional studies on the reproductive toxicity of the drug are needed [140]. Nevertheless, remdesivir was approved by the FDA as the first COVID-19 treatment drug in the US. Advances in the structural analysis of SARS-CoV-2 RdRp will also be critical for the design of new drug candidates. Structural information could be used to clarify the inhibition mechanisms of remdesivir and favipiravir and could contribute to the discovery of new drugs. Indeed, a recent study elucidated the cryostructure of the SARS-CoV-2 full-length nsp12 monomer and those of its complexes with nsp7 and nsp8. Ultimately, a combination therapy of RdRp inhibitors with clinical-stage drug candidates that target other viral proteins might provide better therapeutic efficacy for the treatment of COVID-19.
Declaration of Competing Interest
The authors declared that they have no conflicts of interest to this work.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81602967 and 81803784), the China Postdoctoral Science Foundation (Grant Nos. 2016M592898XB and 2019M663921XB), the Basic Research Program of Natural Science of Shaanxi Province (Grant Nos. 2019JQ-779, 2020CGXNG-044, 19JC006, 2019JQ-484 and 2019JQ-252), the Basic Research Plan of the Education Department of Shaanxi Province (Grant No. 19JC006) and the College Students’ Innovative Entrepreneurial Training Program (Grant Nos. 201510708172, 201610708019 and 2019107080827).
References
- 1.Kaul D. An overview of coronaviruses including the SARS-2 coronavirus - Molecular biology, epidemiology and clinical implications. Curr. Med. Res. Pract. 2020;10(2):54–64. doi: 10.1016/j.cmrp.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nature Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., et al. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 4.Ruan Y.J., Wei C.L., Ee A.L., Vega V.B., Thoreau H., Su S.T., Chia J.M., Ng P., Chiu K.P., Lim L., Zhang T., Peng C.K., Lin E.O., Lee N.M., Yee S.L., Ng L.F., Chee R.E., Stanton L.W., Long P.M., Liu E.T. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet (London, England) 2003;361(9371):1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vankadari N. Structure of furin protease binding to SARS-CoV-2 spike glycoprotein and implications for potential targets and virulence. J. Phys. Chem. Lett. 2020;11(16):6655–6663. doi: 10.1021/acs.jpclett.0c01698. [DOI] [PubMed] [Google Scholar]
- 6.Torre-Fuentes L., Matías-Guiu J., Hernández-Lorenzo L., Montero-Escribano P., Pytel V., Porta-Etessam J., Gómez-Pinedo U., Matías-Guiu J.A. ACE2, TMPRSS2, and Furin variants and SARS-CoV-2 infection in Madrid, Spain. J. Med. Virol. 2020 doi: 10.1002/jmv.26319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elhusseiny K.M., Abd-Elhay F.A., Kamel M.G. Possible therapeutic agents for COVID-19: a comprehensive review. Expert Rev. Anti. Ther. 2020:1–15. doi: 10.1080/14787210.2020.1782742. [DOI] [PubMed] [Google Scholar]
- 8.Nittari G., Pallotta G., Amenta F., Tayebati S.K. Current pharmacological treatments for SARS-COV-2: a narrative review. Eur. J. Pharmacol. 2020;882 doi: 10.1016/j.ejphar.2020.173328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimizu H., Saito A., Mikuni J., Nakayama E.E., Koyama H., Honma T., Shirouzu M., Sekine S.I., Shioda T. Discovery of a small molecule inhibitor targeting dengue virus NS5 RNA-dependent RNA polymerase. PLoS Negl. Trop. Dis. 2019;13(11) doi: 10.1371/journal.pntd.0007894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koonin E.V., Gorbalenya A.E., Chumakov K.M. Tentative identification of RNA-dependent RNA polymerases of dsRNA viruses and their relationship to positive strand RNA viral polymerases. FEBS Lett. 1989;252(1–2):42–46. doi: 10.1016/0014-5793(89)80886-5. [DOI] [PubMed] [Google Scholar]
- 11.Ruan Z., Liu C., Guo Y., He Z., Huang X., Jia X., Yang T. SARS-CoV-2 and SARS-CoV: Virtual screening of potential inhibitors targeting RNA-dependent RNA polymerase activity (NSP12) J. Med. Virol. 2020 doi: 10.1002/jmv.26222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10(1):2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., Ray A.S., Cihlar T., Siegel D., Mackman R.L., Clarke M.O., Baric R.S., Denison M.R. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9(2):e00221–18. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wise J. Covid-19: remdesivir is recommended for authorisation by European Medicines Agency. BMJ. 2020;369:m2610. doi: 10.1136/bmj.m2610. [DOI] [PubMed] [Google Scholar]
- 16.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295(20):6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. Mechanism of inhibition of ebola virus RNA-Dependent RNA polymerase by remdesivir. Viruses. 2019;11(4):326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. U.S.A. 2020;117(12):6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.D., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Organization W.H. 2020. Big Global Study Finds Remdesivir Doesn’t Help Covid-19 Patients.https://edition.cnn.com/2020/10/15/health/remdesivir-covid-who-trial-mortality/index.html [Google Scholar]
- 22.Li Y., Cao L., Li G., Cong F., Li Y., Sun J., Luo Y., Chen G., Li G., Wang P., Xing F., Ji Y., Zhao J., Zhang Y., Guo D., Zhang X. Remdesivir metabolite GS-441524 efficiently inhibits SARS-CoV-2 infection in mouse model. bioRxiv. 2020 doi: 10.1021/acs.jmedchem.0c01929. 2020.10.26.353300. [DOI] [PubMed] [Google Scholar]
- 23.Du Y.X., Chen X.P. Favipiravir: Pharmacokinetics and concerns about clinical trials for 2019-nCoV infection. Clin. Pharmacol. Ther. 2020;108(2):242–247. doi: 10.1002/cpt.1844. [DOI] [PubMed] [Google Scholar]
- 24.Furuta Y., Takahashi K., Shiraki K., Sakamoto K., Smee D.F., Barnard D.L., Gowen B.B., Julander J.G., Morrey J.D. T-705 (favipiravir) and related compounds: novel broad-spectrum inhibitors of RNA viral infections. Antiviral Res. 2009;82(3):95–102. doi: 10.1016/j.antiviral.2009.02.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furuta Y., Gowen B.B., Takahashi K., Shiraki K., Smee D.F., Barnard D.L. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Res. 2014;100(2):446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caroline A.L., Powell D.S., Bethel L.M., Oury T.D., Reed D.S., Hartman A.L. Broad spectrum antiviral activity of favipiravir (T-705): protection from highly lethal inhalational rift valley fever. PLoS Negl. Trop. Dis. 2014;8(4) doi: 10.1371/journal.pntd.0002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baranovich S.-S.W.Tatiana, Armstrong Jianling, Marjuki Henju, Webby Richard J., Webster Robert G., Govorkova E.A. T-705 (Favipiravir) induces lethal mutagenesis in influenza a H1N1 viruses in vitro. J. Virol. 2013;87(7):3741. doi: 10.1128/JVI.02346-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C., Zhang Y., Huang J., Yin P., Cheng Z., Wu J., Chen S., Zhang Y., Chen B., Lu M., Luo Y., Ju L., Zhang J., Wang X. Favipiravir versus arbidol for COVID-19: a randomized clinical trial. medRxiv. 2020 [Google Scholar]
- 29.Peng Q., Peng R., Yuan B., Wang M., Zhao J., Fu L., Qi J., Shi Y. Structural basis of SARS-CoV-2 polymerase inhibition by Favipiravir. bioRxiv. 2020 doi: 10.1016/j.xinn.2021.100080. 2020.10.19.345470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Te H.S., Randall G., Jensen D.M. Mechanism of action of ribavirin in the treatment of chronic hepatitis C. Gastroenterol. Hepatol. (N Y) 2011;3(3):218–225. [PMC free article] [PubMed] [Google Scholar]
- 31.Macchiagodena M., Pagliai M., Procacci P. Identification of potential binders of the main protease 3CL(pro) of the COVID-19 via structure-based ligand design and molecular modeling. Chem. Phys. Lett. 2020;750 doi: 10.1016/j.cplett.2020.137489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 33.Berry M., Fielding B.C., Gamieldien J. Potential broad spectrum inhibitors of the Coronavirus 3CLpro: a virtual screening and structure-based drug design study. Viruses. 2015;7(12):6642–6660. doi: 10.3390/v7122963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdelrheem D.A., Ahmed S.A., Abd El-Mageed H.R., Mohamed H.S., Rahman A.A., Elsayed K.N.M., Ahmed S.A. The inhibitory effect of some natural bioactive compounds against SARS-CoV-2 main protease: insights from molecular docking analysis and molecular dynamic simulation. J. Environ. Sci. Health. Part A, Toxic/Hazard. Subst. Environ. Eng. 2020;55(11):1373–1386. doi: 10.1080/10934529.2020.1826192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei P., Fan K., Chen H., Ma L., Huang C., Tan L., Xi D., Li C., Liu Y., Cao A., Lai L. The N-terminal octapeptide acts as a dimerization inhibitor of SARS coronavirus 3C-like proteinase. Biochem. Biophys. Res. Commun. 2006;339(3):865–872. doi: 10.1016/j.bbrc.2005.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobs J., Grum-Tokars V., Zhou Y., Turlington M., Saldanha S.A., Chase P., Eggler A., Dawson E.S., Baez-Santos Y.M., Tomar S., Mielech A.M., Baker S.C., Lindsley C.W., Hodder P., Mesecar A., Stauffer S.R. Discovery, synthesis, and structure-based optimization of a series of N-(tert-butyl)-2-(N-arylamido)-2-(pyridin-3-yl) acetamides (ML188) as potent noncovalent small molecule inhibitors of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL protease. J. Med. Chem. 2013;56(2):534–546. doi: 10.1021/jm301580n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thibaut H.J., De Palma A.M., Neyts J. Combating enterovirus replication: state-of-the-art on antiviral research. Biochem. Pharmacol. 2012;83(2):185–192. doi: 10.1016/j.bcp.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 38.Konno S., Thanigaimalai P., Yamamoto T., Nakada K., Kakiuchi R., Takayama K., Yamazaki Y., Yakushiji F., Akaji K., Kiso Y., Kawasaki Y., Chen S.-E., Freire E., Hayashi Y. Design and synthesis of new tripeptide-type SARS-CoV 3CL protease inhibitors containing an electrophilic arylketone moiety. Bioorg. Med. Chem. 2012;21(2):412–424. doi: 10.1016/j.bmc.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai W., Zhang B., Jiang X.-M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Chen X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.-K., Xu Y., Yang H., Liu H. Structure-Based Design, Synthesis and Biological Evaluation of Peptidomimetic Aldehydes as a Novel Series of Antiviral Drug Candidates Targeting the SARS-CoV-2 Main Protease. bioRxiv. 2020 doi: 10.1126/science.abb4489. 2020.03.25.996348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L., Lin D., Kusov Y., Nian Y., Ma Q., Wang J., von Brunn A., Leyssen P., Lanko K., Neyts J., de Wilde A., Snijder E.J., Liu H., Hilgenfeld R. Α-ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J. Med. Chem. 2020;63(9):4562–4578. doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y., Liang C., Xin L., Ren X., Tian L., Ju X., Li H., Wang Y., Zhao Q., Liu H., Cao W., Xie X., Zhang D., Wang Y., Jian Y. The development of coronavirus 3C-Like protease (3CL(pro)) inhibitors from 2010 to 2020. Eur. J. Med. Chem. 2020;206 doi: 10.1016/j.ejmech.2020.112711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramajayam R., Tan K.-P., Liu H.-G., Liang P.-H. Synthesis and evaluation of pyrazolone compounds as SARS-coronavirus 3C-like protease inhibitors. Bioorg. Med. Chem. 2010;18(22):7849–7854. doi: 10.1016/j.bmc.2010.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turlington M., Chun A., Tomar S., Eggler A., Grum-Tokars V., Jacobs J., Daniels J.S., Dawson E., Saldanha A., Chase P., Baez-Santos Y.M., Lindsley C.W., Hodder P., Mesecar A.D., Stauffer S.R. Discovery of N-(benzo[1,2,3]triazol-1-yl)-N-(benzyl)acetamido)phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: identification of ML300 and noncovalent nanomolar inhibitors with an induced-fit binding. Bioorg. Med. Chem. Lett. 2013;23(22):6172–6177. doi: 10.1016/j.bmcl.2013.08.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimamoto Y., Hattori Y., Kobayashi K., Teruya K., Sanjoh A., Nakagawa A., Yamashita E., Akaji K. Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors. Bioorg. Med. Chem. 2015;23(4):876–890. doi: 10.1016/j.bmc.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohnishi K., Hattori Y., Kobayashi K., Akaji K. Evaluation of a non-prime site substituent and warheads combined with a decahydroisoquinolin scaffold as a SARS 3CL protease inhibitor. Bioorg. Med. Chem. 2019;27(2):425–435. doi: 10.1016/j.bmc.2018.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshizawa S., Hattori Y., Kobayashi K., Akaji K. Evaluation of an octahydroisochromene scaffold used as a novel SARS 3CL protease inhibitor. Bioorg. Med. Chem. 2020;28(4) doi: 10.1016/j.bmc.2019.115273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L., Bao B., Song G., Chen C., Zhang X., Lu W., Wang Z., Cai Y., Li S., Fu S. Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: chemical synthesis, biological evaluation, molecular docking and 3D-QSAR study. Eur. J. Med. Chem. 2017;137:450–461. doi: 10.1016/j.ejmech.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen T.T.H., Woo H., Kang H., Nguyen V.D., Kim Y., Kim D.W., Ahn S., Xia Y., Kim D. Flavonoid-mediated inhibition of SARS coronavirus 3C-like protease expressed in Pichia pastoris. Biotechnol. Lett. 2012;34(5):831–838. doi: 10.1007/s10529-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryu Y.B., Jeong H.J., Kim J.H., Kim Y., Park J., Kim D., Naguyen T.T.H., Park S., Chang J.S., Park K.H. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition. Bioorg. Med. Chem. 2010;18(22):7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu W., Zhu H.-M., Niu G.-J., Shi E.-Z., Chen J., Sun B., Chen W.-Q., Zhou H.-G., Yang C. Synthesis, modification and docking studies of 5-sulfonyl isatin derivatives as SARS-CoV 3C-like protease inhibitors. Bioorg. Med. Chem. $V. 2013;22(1):292–302. doi: 10.1016/j.bmc.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park J., Ko J., Kim D.W., Kim Y.M., Kwon H., Jeong H.J., Kim C.Y., Park K.H., Lee W.S., Ryu Y.B. Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J. Enzyme Inhib. Med. Chem. 2016;31(1):23–30. doi: 10.3109/14756366.2014.1003215. [DOI] [PubMed] [Google Scholar]
- 52.Park J., Kim J.H., Kim Y., Jeong H.J., Kim D.W., Park K.H., Kwon H., Park S., Lee W.S., Ryu Y.B. Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorg. Med. Chem. 2012;20(19):5928–5935. doi: 10.1016/j.bmc.2012.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV), Nature reviews. Drug discovery. 2020;19(3):149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 54.Zheng L., Zhang L., Huang J., Nandakumar K.S., Liu S., Cheng K. Potential treatment methods targeting 2019-nCoV infection. Eur. J. Med. Chem. 2020;205 doi: 10.1016/j.ejmech.2020.112687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choy K.T., Wong A.Y., Kaewpreedee P., Sia S.F., Chen D., Hui K.P.Y., Chu D.K.W., Chan M.C.W., Cheung P.P., Huang X., Peiris M., Yen H.L. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye X.T., Luo Y.L., Xia S.C., Sun Q.F., Ding J.G., Zhou Y., Chen W., Wang X.F., Zhang W.W., Du W.J., Ruan Z.W., Hong L. Clinical efficacy of lopinavir/ritonavir in the treatment of Coronavirus disease 2019. Eur. Rev. Med. Pharmacol. Sci. 2020;24(6):3390–3396. doi: 10.26355/eurrev_202003_20706. [DOI] [PubMed] [Google Scholar]
- 57.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C. A trial of Lopinavir-Ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Báez-Santos Y.M., St John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klemm T., Ebert G., Calleja D.J., Allison C.C., Richardson L.W., Bernardini J.P., Lu B.G., Kuchel N.W., Grohmann C., Shibata Y., Gan Z.Y., Cooney J.P., Doerflinger M., Au A.E., Blackmore T.R., van der Heden van Noort G.J., Geurink P.P., Ovaa H., Newman J., Riboldi-Tunnicliffe A., Czabotar P.E., Mitchell J.P., Feltham R., Lechtenberg B.C., Lowes K.N., Dewson G., Pellegrini M., Lessene G., Komander D. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020 doi: 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao X., Qin B., Chen P., Zhu K., Hou P., Wojdyla J.A., Wang M., Cui S. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S.J., El Oualid F., Huang T.T., Bekes M., Drag M., Olsen S.K. Activity profiling and structures of inhibitor-bound SARS-CoV-2-PLpro protease provides a framework for anti-COVID-19 drug design. bioRxiv. 2020 doi: 10.1126/sciadv.abd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M., Mehdipour A.R., Tascher G., Geurink P.P., Wilhelm A., van der Heden van Noort G.J., Ovaa H., Müller S., Knobeloch K.P., Rajalingam K., Schulman B.A., Cinatl J., Hummer G., Ciesek S., Dikic I. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020 doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Freitas B.T., Durie I.A., Murray J., Longo J.E., Miller H.C., Crich D., Hogan R.J., Tripp R.A., Pegan S.D. Characterization and Noncovalent Inhibition of the Deubiquitinase and deISGylase Activity of SARS-CoV-2 Papain-Like Protease. ACS Infect. Dis. 2020;6(8):2099–2109. doi: 10.1021/acsinfecdis.0c00168. [DOI] [PubMed] [Google Scholar]
- 64.Morse J.S., Lalonde T., Xu S., Liu W. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. ChemRxiv : Prepr. Server Chem. 2020:1. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chou C.Y., Chien C.H., Han Y.S., Prebanda M.T., Hsieh H.P., Turk B., Chang G.G., Chen X. Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biochem. Pharmacol. 2008;75(8):1601–1609. doi: 10.1016/j.bcp.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin M.-H., Moses D.C., Hsieh C.-H., Cheng S.-C., Chen Y.-H., Sun C.-Y., Chou C.-Y. Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes. Antiviral Res. 2017;150:155–163. doi: 10.1016/j.antiviral.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arya R.D., Prashar V., Kumar M. Potential inhibitors a papain-like protease of novel coronavirus (SARS-CoV-2) from FDA approved drugs. chemRxiv. 2020 [Google Scholar]
- 68.Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S.J., El Oualid F., Huang T.T., Bekes M., Drag M., Olsen S.K. Activity profiling and structures of inhibitor-bound SARS-CoV-2-PLpro protease provides a framework for anti-COVID-19 drug design. bioRxiv. 2020 doi: 10.1126/sciadv.abd4596. 2020.04.29.068890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Adalja A., Inglesby T. Broad-spectrum antiviral agents: a crucial pandemic tool. Expert Rev. Anti. Ther. 2019;17(7):467–470. doi: 10.1080/14787210.2019.1635009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mei-Jiao G., Shi-Fang L., Yan-Yan C., Jun-Jun S., Yue-Feng S., Ting-Ting R., Yong-Guang Z., Hui-Yun C. Antiviral effects of selected IMPDH and DHODH inhibitors against foot and mouth disease virus. Biomed. Pharmacother. 2019;118 doi: 10.1016/j.biopha.2019.109305. [DOI] [PubMed] [Google Scholar]
- 71.Chen S., Ding S., Yin Y., Xu L., Li P., Peppelenbosch M.P., Pan Q., Wang W. Suppression of pyrimidine biosynthesis by targeting DHODH enzyme robustly inhibits rotavirus replication. Antiviral Res. 2019;167:35–44. doi: 10.1016/j.antiviral.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 72.Xiong R., Zhang L., Li S., Sun Y., Ding M., Wang Y., Zhao Y., Wu Y., Shang W., Jiang X., Shan J., Shen Z., Tong Y., Xu L., Chen Y., Liu Y., Zou G., Lavillete D., Zhao Z., Wang R., Zhu L., Xiao G., Lan K., Li H., Xu K. Novel and potent inhibitors targeting DHODH are broad-spectrum antivirals against RNA viruses including newly-emerged coronavirus SARS-CoV-2. "Protein & Cell" OnlineFirst. 2020:1–17. doi: 10.1007/s13238-020-00768-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2) doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu J., Shi P.Y., Li H., Zhou J. Broad spectrum antiviral agent niclosamide and its therapeutic potential. ACS Infect. Dis. 2020;6(5):909–915. doi: 10.1021/acsinfecdis.0c00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pindiprolu S., Pindiprolu S.H. Plausible mechanisms of Niclosamide as an antiviral agent against COVID-19. Med. Hypotheses. 2020;140 doi: 10.1016/j.mehy.2020.109765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gassen N.C., Niemeyer D., Muth D., Corman V.M., Martinelli S., Gassen A., Hafner K., Papies J., Mösbauer K., Zellner A., Zannas A.S., Herrmann A., Holsboer F., Brack-Werner R., Boshart M., Müller-Myhsok B., Drosten C., Müller M.A., Rein T. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat. Commun. 2019;10:5770. doi: 10.1038/s41467-019-13659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang X.Y., Huang H.J., Zhuang D.L., Nasser M.I., Yang M.H., Zhu P., Zhao M.Y. Biological, clinical and epidemiological features of COVID-19, SARS and MERS and AutoDock simulation of ACE2. Infect. Dis. Poverty. 2020;9(1):99. doi: 10.1186/s40249-020-00691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gassen N.C., Papies J., Bajaj T., Dethloff F., Emanuel J., Weckmann K., Heinz D.E., Heinemann N., Lennarz M., Richter A., Niemeyer D., Corman V.M., Giavalisco P., Drosten C., Müller M.A. Analysis of SARS-CoV-2-controlled autophagy reveals spermidine, MK-2206, and niclosamide as putative antiviral therapeutics. bioRxiv. 2020 2020.04.15.997254. [Google Scholar]
- 79.Hu B., Huang S., Yin L. The cytokine storm and COVID-19. J. Med. Virol. 2020 doi: 10.1002/jmv.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang C., Wu Z., Li J.W., Zhao H., Wang G.Q. Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int. J. Antimicrob. Agents. 2020;55(5) doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song P., Li W., Xie J., Hou Y., You C. Cytokine storm induced by SARS-CoV-2. Clin. Chim. Acta. 2020;509:280–287. doi: 10.1016/j.cca.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mahmudpour M., Roozbeh J., Keshavarz M., Farrokhi S., Nabipour I. COVID-19 cytokine storm: the anger of inflammation. Cytokine. 2020;133 doi: 10.1016/j.cyto.2020.155151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang X., Zhang Y., Qiao W., Zhang J., Qi Z. Baricitinib, a drug with potential effect to prevent SARS-COV-2 from entering target cells and control cytokine storm induced by COVID-19. Int. Immunopharmacol. 2020;86 doi: 10.1016/j.intimp.2020.106749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Khalil A., Kamar A., Nemer G. Thalidomide-revisited: are COVID-19 patients going to be the latest victims of yet another theoretical drug-repurposing? Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.01248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Atrah H.I. Alternative management of Covid-19 infection. Scott. Med. J. 2020;65(3):72–75. doi: 10.1177/0036933020941497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen C., Qi F., Shi K., Li Y., Li J., Chen Y., Pan J., Zhou T., Lin X., Zhang J., Luo Y., Li X., Xia J. Thalidomide combined with low-dose short-term glucocorticoid in the treatment of critical coronavirus disease 2019. Clin. Transl. Med. 2020;10(2):e35. doi: 10.1002/ctm2.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.López-Iranzo F.J., López-Rodas A.M., Franco L., López-Rodas G. Pentoxifylline and Oxypurinol: potential drugs to prevent the “cytokine release (storm) syndrome” caused by SARS-CoV-2? Curr. Pharm. Des. 2020;26(35):4515–4521. doi: 10.2174/1381612826666200811180232. [DOI] [PubMed] [Google Scholar]
- 88.McKee D.L., Sternberg A., Stange U., Laufer S., Naujokat C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol. Res. 2020;157 doi: 10.1016/j.phrs.2020.104859. 104859-104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stopsack K.H., Mucci L.A., Antonarakis E.S., Nelson P.S., Kantoff P.W. TMPRSS2 and COVID-19: serendipity or opportunity for intervention? Cancer Discov. 2020;10(6):779–782. doi: 10.1158/2159-8290.CD-20-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2) doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ragia G., Manolopoulos V.G. Inhibition of SARS-CoV-2 entry through the ACE2/TMPRSS2 pathway: a promising approach for uncovering early COVID-19 drug therapies. Eur. J. Clin. Pharmacol. 2020:1–8. doi: 10.1007/s00228-020-02963-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hoffmann M., Schroeder S., Kleine-Weber H., Müller M.A., Drosten C., Pöhlmann S. Nafamostat mesylate blocks activation of SARS-CoV-2: new treatment option for COVID-19. Antimicrob. Agents Chemother. 2020;64(6):e00754–20. doi: 10.1128/AAC.00754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bestle D., Heindl M.R., Limburg H., Van Lam van T., Pilgram O., Moulton H., Stein D.A., Hardes K., Eickmann M., Dolnik O., Rohde C., Klenk H.D., Garten W., Steinmetzer T., Böttcher-Friebertshäuser E. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance. 2020;3(9) doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hirano T., Murakami M. COVID-19: a new virus, but a familiar receptor and cytokine release syndrome. Immunity. 2020;52(5):731–733. doi: 10.1016/j.immuni.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Couture F., Kwiatkowska A., Dory Y.L., Day R. Therapeutic uses of furin and its inhibitors: a patent review. Expert Opin. Ther. Pat. 2015;25(4):379–396. doi: 10.1517/13543776.2014.1000303. [DOI] [PubMed] [Google Scholar]
- 96.Jin X., Xu K., Jiang P., Lian J., Hao S., Yao H., Jia H., Zhang Y., Zheng L., Zheng N., Chen D., Yao J., Hu J., Gao J., Wen L., Shen J., Ren Y., Yu G., Wang X., Lu Y., Yu X., Yu L., Xiang D., Wu N., Lu X., Cheng L., Liu F., Wu H., Jin C., Yang X., Qian P., Qiu Y., Sheng J., Liang T., Li L., Yang Y. Virus strain from a mild COVID-19 patient in Hangzhou represents a new trend in SARS-CoV-2 evolution potentially related to Furin cleavage site. Emerg. Microbes Infect. 2020;9(1):1474–1488. doi: 10.1080/22221751.2020.1781551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ivanova T., Hardes K., Kallis S., Dahms S.O., Than M.E., Künzel S., Böttcher-Friebertshäuser E., Lindberg I., Jiao G.S., Bartenschlager R., Steinmetzer T. Optimization of substrate-analogue furin inhibitors. ChemMedChem. 2017;12(23):1953–1968. doi: 10.1002/cmdc.201700596. [DOI] [PubMed] [Google Scholar]
- 98.Izaguirre G. The proteolytic regulation of virus cell entry by furin and other proprotein convertases. Viruses. 2019;11(9):837. doi: 10.3390/v11090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wong Y.C., Lau S.Y., Wang To K.K., Mok B.W.Y., Li X., Wang P., Deng S., Woo K.F., Du Z., Li C., Zhou J., Woo Chan J.F., Yuen K.Y., Chen H., Chen Z. Natural transmission of bat-like SARS-CoV-2 ΔPRRA variants in COVID-19 patients. Clin. Infect. Dis. 2020:1. doi: 10.1093/cid/ciaa953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Madapusi Balaji T., Varadarajan S., Rao U.S.V., Raj A.T., Patil S., Arakeri G., Brennan P.A. Oral cancer and periodontal disease increase the risk of COVID 19? A mechanism mediated through furin and cathepsin overexpression. Med. Hypotheses. 2020;144 doi: 10.1016/j.mehy.2020.109936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nguyen H.T., Zhang S., Wang Q., Anang S., Wang J., Ding H., Kappes J.C., Sodroski J.G. Spike glycoprotein and host cell determinants of SARS-CoV-2 entry and cytopathic effects. bioRxiv. 2020 doi: 10.1128/JVI.02304-20. 2020.10.22.351569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hardes K., Becker G.L., Lu Y., Dahms S.O., Köhler S., Beyer W., Sandvig K., Yamamoto H., Lindberg I., Walz L., von Messling V., Than M.E., Garten W., Steinmetzer T. Novel furin inhibitors with potent anti-infectious activity. ChemMedChem. 2015;10(7):1218–1231. doi: 10.1002/cmdc.201500103. [DOI] [PubMed] [Google Scholar]
- 104.Wu C., Zheng M., Yang Y., Gu X., Yang K., Li M., Liu Y., Zhang Q., Zhang P., Wang Y., Wang Q., Xu Y., Zhou Y., Zhang Y., Chen L., Li H. Furin: a potential therapeutic target for COVID-19. iScience. 2020;23(10) doi: 10.1016/j.isci.2020.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cheng Y.W., Chao T.L., Li C.L., Chiu M.F., Kao H.C., Wang S.H., Pang Y.H., Lin C.H., Tsai Y.M., Lee W.H., Tao M.H., Ho T.C., Wu P.Y., Jang L.T., Chen P.J., Chang S.Y., Yeh S.H. Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep. 2020;33(2) doi: 10.1016/j.celrep.2020.108254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen X., Geiger J.D. Janus sword actions of chloroquine and hydroxychloroquine against COVID-19. Cell. Signal. 2020;73 doi: 10.1016/j.cellsig.2020.109706. 109706-109706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yuan Z., Pavel M.A., Wang H., Hansen S.B. Hydroxychloroquine: mechanism of action inhibiting SARS-CoV2 entry. bioRxiv. 2020 2020.08.13.250217. [Google Scholar]
- 109.Amin S.A., Jha T. Fight against novel coronavirus: a perspective of medicinal chemists. Eur. J. Med. Chem. 2020;201 doi: 10.1016/j.ejmech.2020.112559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhu Z., Lu Z., Xu T., Chen C., Yang G., Zha T., Lu J., Xue Y. Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID-19. J. Infect. 2020;81(1):e21–e23. doi: 10.1016/j.jinf.2020.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rosa S.G.V., Santos W.C. Clinical trials on drug repositioning for COVID-19 treatment. Rev. Panam. Salud Publica. 2020;44:e40. doi: 10.26633/RPSP.2020.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vankadari N. Arbidol: a potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. Int. J. Antimicrob. Agents. 2020;56(2) doi: 10.1016/j.ijantimicag.2020.105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xiang Z., Liu J., Shi D., Chen W., Li J., Yan R., Bi Y., Hu W., Zhu Z., Yu Y., Yang Z. Glucocorticoids improve severe or critical COVID-19 by activating ACE2 and reducing IL-6 levels. Int. J. Biol. Sci. 2020;16(13):2382–2391. doi: 10.7150/ijbs.47652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ledford H. Coronavirus breakthrough: dexamethasone is first drug shown to save lives. Nature. 2020;582(7813):469. doi: 10.1038/d41586-020-01824-5. [DOI] [PubMed] [Google Scholar]
- 116.Amin S.A., Ghosh K., Gayen S., Jha T. Chemical-informatics approach to COVID-19 drug discovery: Monte Carlo based QSAR, virtual screening and molecular docking study of some in-house molecules as papain-like protease (PLpro) inhibitors. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1780946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Naidoo D., Roy A., Kar P., Mutanda T., Anandraj A. Cyanobacterial metabolites as promising drug leads against the M(pro) and PL(pro) of SARS-CoV-2: an in silico analysis. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1794972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.AlAjmi M.F., Azhar A., Owais M., Rashid S., Hasan S., Hussain A., Rehman M.T. Antiviral potential of some novel structural analogs of standard drugs repurposed for the treatment of COVID-19. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1799865. [DOI] [PubMed] [Google Scholar]
- 119.Sisay M. 3CL(pro) inhibitors as a potential therapeutic option for COVID-19: Available evidence and ongoing clinical trials. Pharmacol. Res. 2020;156 doi: 10.1016/j.phrs.2020.104779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu P., Liu H., Sun Q., Liang H., Li C., Deng X., Liu Y., Lai L. Potent inhibitors of SARS-CoV-2 3C-like protease derived from N-substituted isatin compounds. Eur. J. Med. Chem. 2020;206 doi: 10.1016/j.ejmech.2020.112702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Muhseen Z.T., Hameed A.R., Al-Hasani H.M.H., Tahir Ul Qamar M., Li G. Promising terpenes as SARS-CoV-2 spike receptor-binding domain (RBD) attachment inhibitors to the human ACE2 receptor: integrated computational approach. J. Mol. Liq. 2020;320 doi: 10.1016/j.molliq.2020.114493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Forman R., Anderson M., Jit M., Mossialos E. Ensuring access and affordability through COVID-19 vaccine research and development investments: a proposal for the options market for vaccines. Vaccine. 2020;38(39):6075–6077. doi: 10.1016/j.vaccine.2020.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pregelj L., Hine D.C., Oyola-Lozada M.G., Munro T.P. Working hard or hardly working? Regulatory bottlenecks in developing a COVID-19 vaccine. Trends Biotechnol. 2020;38(9):943–947. doi: 10.1016/j.tibtech.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.De Savi C., Hughes D.L., Kvaerno L. Quest for a COVID-19 cure by repurposing small-molecule drugs: mechanism of action, clinical development, synthesis at scale, and outlook for supply. Org. Process Res. Dev. 2020;24(6):940–976. doi: 10.1021/acs.oprd.0c00233. [DOI] [PubMed] [Google Scholar]
- 126.Wang Z., Wang Y., Vilekar P., Yang S.P., Gupta M., Oh M.I., Meek A., Doyle L., Villar L., Brennecke A., Liyanage I., Reed M., Barden C., Weaver D.F. Small molecule therapeutics for COVID-19: repurposing of inhaled furosemide. PeerJ. 2020;8:e9533. doi: 10.7717/peerj.9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hoffmann M., Mösbauer K., Hofmann-Winkler H., Kaul A., Kleine-Weber H., Krüger N., Gassen N.C., Müller M.A., Drosten C., Pöhlmann S. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature. 2020;585:588–590. doi: 10.1038/s41586-020-2575-3. [DOI] [PubMed] [Google Scholar]
- 128.Elfiky A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Russell C.D., Millar J.E., Baillie J.K. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet (London, England) 2020;395(10223):473–475. doi: 10.1016/S0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y., Peng C., Yuan M., Huang J., Wang Z., Yu J., Gao X., Wang D., Yu X., Li L., Zhang J., Wu X., Li B., Xu Y., Chen W., Peng Y., Hu Y., Lin L., Liu X., Huang S., Zhou Z., Zhang L., Wang Y., Zhang Z., Deng K., Xia Z., Gong Q., Zhang W., Zheng X., Liu Y., Yang H., Zhou D., Yu D., Hou J., Shi Z., Chen S., Chen Z., Zhang X., Yang X. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. U.S.A. 2020;117(17):9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hall D.C., Jr., Ji H.F. A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Travel Med. Infect. Dis. 2020;35 doi: 10.1016/j.tmaid.2020.101646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Khan R.J., Jha R.K., Amera G.M., Jain M., Singh E., Pathak A., Singh R.P., Muthukumaran J., Singh A.K. Targeting SARS-CoV-2: a systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2’-O-ribose methyltransferase. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1753577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen L.R., Wang Y.C., Lin Y.W., Chou S.Y., Chen S.F., Liu L.T., Wu Y.T., Kuo C.J., Chen T.S., Juang S.H. Synthesis and evaluation of isatin derivatives as effective SARS coronavirus 3CL protease inhibitors. Bioorg. Med. Chem. Lett. 2005;15(12):3058–3062. doi: 10.1016/j.bmcl.2005.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shannon A., Le N.T., Selisko B., Eydoux C., Alvarez K., Guillemot J.C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I., Bannister R., Park Y., Babusis D., Clarke M.O., Mackman R.L., Spahn J.E., Palmiotti C.A., Siegel D., Ray A.S., Cihlar T., Jordan R., Denison M.R., Baric R.S. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9(396) doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Amirian E.S., Levy J.K. Current knowledge about the antivirals remdesivir (GS-5734) and GS-441524 as therapeutic options for coronaviruses. One Health (Amsterdam, Netherlands) 2020;9 doi: 10.1016/j.onehlt.2020.100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liang C., Tian L., Liu Y., Hui N., Qiao G., Li H., Shi Z., Tang Y., Zhang D., Xie X., Zhao X. A promising antiviral candidate drug for the COVID-19 pandemic: a mini-review of remdesivir. Eur. J. Med. Chem. 2020;201 doi: 10.1016/j.ejmech.2020.112527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fan J., Luo J., Zhao D., Deng T., Weng Y., Sun Y., Li X. A preliminary study on the reproductive toxicity of GS-5734 on male mice. bioRxiv. 2020 2020.04.21.050104. [Google Scholar]