Abstract
All the plants and their secondary metabolites used in the present study were obtained from Ayurveda, with historical roots in the Indian subcontinent. The selected secondary metabolites have been experimentally validated and reported as potent antiviral agents against genetically-close human viruses. The plants have also been used as a folk medicine to treat cold, cough, asthma, bronchitis, and severe acute respiratory syndrome in India and across the globe since time immemorial. The present study aimed to assess the repurposing possibility of potent antiviral compounds with SARS-CoV-2 target proteins and also with host-specific receptor and activator protease that facilitates the viral entry into the host body. Molecular docking (MDc) was performed to study molecular affinities of antiviral compounds with aforesaid target proteins. The top-scoring conformations identified through docking analysis were further validated by 100 ns molecular dynamic (MD) simulation run. The stability of the conformation was studied in detail by investigating the binding free energy using MM-PBSA method. Finally, the binding affinities of all the compounds were also compared with a reference ligand, remdesivir, against the target protein RdRp. Additionally, pharmacophore features, 3D structure alignment of potent compounds and Bayesian machine learning model were also used to support the MDc and MD simulation. Overall, the study emphasized that curcumin possesses a strong binding ability with host-specific receptors, furin and ACE2. In contrast, gingerol has shown strong interactions with spike protein, and RdRp and quercetin with main protease (Mpro) of SARS-CoV-2. In fact, all these target proteins play an essential role in mediating viral replication, and therefore, compounds targeting aforesaid target proteins are expected to block the viral replication and transcription. Overall, gingerol, curcumin and quercetin own multitarget binding ability that can be used alone or in combination to enhance therapeutic efficacy against COVID-19. The obtained results encourage further in vitro and in vivo investigations and also support the traditional use of antiviral plants preventively.
Keywords: ACE-2; Coronavirus; COVID-19; Furin; Main protease; RdRp, spike protein RBD
1. Introduction
Beginning on 17 November 2019 (media reports on unpublished Chinese government data), a novel coronavirus from Hubei's capital Wuhan, China, designated as SARS-CoV-2, has caused an international outbreak of serious and infectious respiratory illness termed COVID-19 [1]. Until September, 30th, 2020, the pandemic virus [2] has infected at least 33.8 million people, and killed 1.01 million worldwide. The COVID-19 manifests from mild cough, fever, self-limiting respiratory tract illness to severe progressive pneumonia in both lungs which can lead to multi-organ failure, and ultimately death occurs [3], [4].
The absence of explicit treatment for COVID-19 leads the population over many regions in the world to use medicinal herbs known in ethnopharmacology as antiviral. Teams around the world are working hard to develop active substances against SARS-CoV-2. This paper reports the state of-the-art antiviral choices against COVID-19, with a specific focus on the recent advances of Ayurvedic antiviral medication repurposing procedures and their potential therapeutic suggestion [5]. As per modern Ayurvedic literature, the root of Ayurveda can be traced back to around 6000 BCE when it originated as an oral tradition known as the “Mother of All Healing” [6]. Several clinical drugs with limited or no observable side effects have been developed and practiced from Ayurveda since ancient times to modern practice as ‘tradition to trend’ [7]. Until today, the capability of Ayurvedic prescriptions has not been completely researched and needs further investigation with modern scientific validation approaches for better therapeutic leads.
The selected bioactive compounds from various plants recorded in Ayurveda with potent antiviral activity against different types of viruses [8], [9], [10], [11] are detailed in Table 1. Several researchers also reported the traditional use and research findings of all the plants related to antiviral activities; Allium sativum [12], [13], Allium cepa [14], Zingiber officinale [15], [16], Syzygium aromaticum [17], Mentha piperita [18], [19], Foeniculum vulgare [20], Ocimum sanctum [21], Origanum vulgare [22], [23], Curcuma longa [21], Tinospora cordifolia [24], [25], Cinnamomum cassia [26] and Cordyceps militaris [27].
Table 1.
Antiviral Ayurvedic medicinal plants with their bioactive compounds tested against closely related human viruses with the reported mode of action (NCBI-Pubmed literature).
| Sl. nos. | Plants | Bioactive principles | Antiviral activity tested on | IC50 | Mechanism of action | References |
|---|---|---|---|---|---|---|
| 1. | Allium sativum L. (Garlic) | Allicin, diallyl trisulfide, ajoene and alliin | Infectious bronchitis virus (Coronavirus), HIV-1, Influenza B, Human cytomegalovirus (HCMV), Parainfluenza virus type 3, Herpes Simplex type 1 and 2, Vaccinia virus, Dengue virus (DENV-2), Vesicular stomatitis virus and Human rhinovirus type 2 | 10–34 μM (HIV-1) | TNF-α inhibitor, Immunomodulatory and anti-inflammatory. | [8], [9], [10], [11] |
| 2. | Allium cepa L.(Onion) | Quercetin, zalcitabine, allicin and ribavirin | Poliovirus and Dengue virus type -2 (DENV-2) | 35.7 μg/mL (DENV-2) | RNAs (-) cannot be transcribed to generate more copies of positive polarity. | [14], [84], [85] |
| 3. | Zingiber officinale Roscoe(Ginger) | Gingerol and zingerone | H1N1 Flu (Swine flu), Influenza, Anti-avian influenza virus H9N2 and Human respiratory syncytial virus | 73.3 mg/mL (HRSV) | Enhance the secretion of IFN-β. | [86], [87], [88] |
| 4. | Syzygium aromaticum L. (Clove) | Eugenol, E-cinnamaldehyde, carvacrol and thymol | Hepatitis C viruses and Herpes simplex virus (HSV-1 and HSV-2) | 25.6 μg/mL (HSV-1) and 16.2 μg/mL (HSV-2) | Inhibition of viral replication, damage to viral envelopes of freshly formed virions. | [89], [90], [91] |
| 5. | Mentha piperita L. (Peppermint) | Menthol and rosmarinic acid | Herpes, Influenza A, Newcastle disease virus, herpes simplex virus (HSV-1) and Vaccinia virus | 0.25 μg/mL (HSV-1) | Inhibits specifically the viral DNA polymerase during the replication cycle. | [80], [92] |
| 6. | Foeniculum vulgare Mill. (Fennel) | Trans-anethole | Herpes simplex virus (HSV-1) and Parainfluenza type-3 (PI-3) | 0.025 μg/mL (HSV-1) | Not reported | [20], [80], [93] |
| 7. | Ocimum sanctum L. (Holy Basil or Tulsi) | Apigenin | Herpes simplex viruses (HSV), adenoviruses (ADV), Hepatitis B virus, Coxsackievirus B1 (CVB1), Enterovirus 71 (EV71) and H1N1Flu virus | 39–55 μg/mL (H1N1) | Increase in the levels of IFN-γ, IL-4 and percentages of T-helper and NK-cells. | [94], [95] |
| 8. | Origanum vulgare L. (Oregano) | Carvacrol | Acyclovir-resistant herpes simplex virus type 1 (ACVR-HHV-1), Acyclovir-sensitive HHV-1, Human respiratory syncytial virus (HRSV), Bovine herpesvirus type 2 (BoHV-2), Bovine viral diarrhea virus (BVDV), Herpes simplex virus type 1 (HSV-1), murine norovirus (MNV), Canine distemper virus (CDV), Canine adenovirus (CAV), Canine coronavirus (CCoV), Respiratory syncytial virus (RSV) and Coxsackievirus B3 (CVB3) | 23.1 μM (RSV long strain) and 81.7 μM (RSV A strain) | Capsid protein disintegration. | [96], [97], [98] |
| 9. | Curcuma longa L. (Turmeric) | Curcumin | Parainfluenza virus type 3 (PIV-3), Feline infectious peritonitis virus (FIPV), Vesicular stomatitis virus (VSV), Flock house virus (FHV), Kaposi's sarcoma virus, Respiratory syncytial virus (RSV), HIV-1, HIV-2, Influenza viruses PR8, H1N1, H6N1, Herpes simplex virus type 1 (HSV-1), HPV-16, HPV-18, Human T-cell leukemia virus type 1 and Japanese encephalitis virus (JEV) | 100 μM (HIV-1) and 250 μM (HIV-2) | Inhibits herpes virus replication, inhibits HIV-1 LTR-directed gene expression, and inhibits transcription of HPV-18. | [99], [100], [101] |
| 10. | Tinospora cordifolia (Willd.) Miers (Giloy or Guduchi) | Tinosporin | H1N1 flu, HIV-1, Herpes simplex virus (HSV-1) and Hepatitis-A virus | 40–50 μg/mL (Hepatitis-A Virus) | Increases IgG antibodies, inhibit HIV-1 protease enzyme activity. | [102], [103], [104] |
| 11. | Cinnamomum cassia (L.) J. Presl (Cinnamon bark) | E-cinnamaldehyde | Human respiratory syncytial virus (HRSV), H1N1 avian influenza virus subtype H7N3, Human immunodeficiency virus (HIV-1) and Herpes simplex virus type-1 | 125 μg/mL (H7N3 influenza A virus) | Cleavage of viral RNA and prevents its translation to the viral protein, inhibiting viral attachment to host cells. | [105], [106], [107] |
| 12. | Cordyceps militaris L. (Caterpillar fungus or Kida jari) | Cordycepin | Human immunodeficiency virus type 1 (HIV-1), Influenza virus, Murine leukemia virus, and Epstein-Barr virus (EBV) | 125 μM (EBV in SNU719 cells) | HIV-1 reverse transcriptase inhibitor and selectively inhibits influenza viral genome replication. | [27], [108], [109], [110] |
Due to the gravity of the situation and worldwide rapid spread of COVID-19, an urgent and complementary effort from researchers is necessary to find therapeutic agents and new preventive methods. Drugs like hydroxychloroquine, lopinavir, ritonavir, arbidol, remdesivir, and favipiravir are currently undergoing clinical trials to test their effectiveness and safety in the treatment of coronavirus disease 2019, and some promising results have been achieved thus far [28].
During COVID-19, spike protein, main protease, human angiotensin-converting enzyme-2, RdRp and furin play vital roles during SARS-CoV-2 infection [29], replication [30] and multiplication [31] in the host body [32]. Therefore, in the present study, these proteins have been targeted with selected medicinal plants to identify potential compounds through in silico methods. In the first phase, virtual screening of compounds was performed using molecular docking and pharmacophore model generation. In the second phase, molecular dynamics (MD) simulation study was performed to further validate the molecular docking results. The binding free energy for these selected protein-ligand complexes was also calculated through the molecular mechanics-Poisson-Boltzmann surface area (MM-PBSA) method. Structure alignment based on 3D descriptors of potent compounds and machine learning algorithms were also used to validate intermolecular interactions. The information obtained through computational approaches provides promising results that reveal the repurposing potential of bioactive compounds from Ayurvedic plants against SARS-CoV-2.
2. Materials and methods
2.1. Selection of antiviral plants
The candidate plants with antiviral activities were selected based on related literature in Ayurveda with historical roots in the Indian subcontinent and the available literature in PubMed-NCBI and Google (References against each candidate plants have been shown in Table 1). The priority of selection was based on one or more criteria:
-
1)
Claimed efficacies in treating various lung diseases (asthma, bronchitis, pneumonia and severe acute respiratory syndrome) in Ayurveda and traditional medicine.
-
2)
Availability of valid experimental proofs (in vitro or in vivo) on lungs protective potentials.
-
3)
Published antiviral activities against genetically-close human viruses, such as influenza (A and B), respiratory syncytial virus, coronaviruses, rhinoviruses, parainfluenza viruses, adenoviruses, herpes simplex, measles and chickenpox [33].
A systematic review of scientific studies, following guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA statement), was carried out [34], [35] on the selected medicinal plants. Furthermore, the PubMed-NCBI database (http://www.ncbi.nlm.nih.gov/pubmed) was systematically searched for experimental trials and related bioassays.
2.2. Acquisition of chemical compound information
As shown in Table 1, the potential compounds, which showed strong antiviral activity against genetically-close human viruses, were identified from the PubMed-NCBI literature. The structures of the natural compounds were downloaded in SDF/Mol2 file format embedded with 3D properties from the PubChem Compound [36] database (http://pubchem.ncbi.nlm.nih.gov/), DrugBank (https://www.drugbank.ca/) [37] and ZINC database (https://zinc.docking.org/) [38]. The molecular arrangement and geometry of all the compounds were fully optimized using the semiempirical quantum chemistry method (PM3) [39]. Finally, the optimized structure of all the ligands was exported in Mol2 format and used for molecular docking study. Fig. 1 shows the chemical structure of potent antiviral compounds identified from Ayurvedic antiviral medicinal plants.
Fig. 1.
Chemical structures of potent antiviral compounds identified from Ayurvedic plant sources.
2.3. Target selection
The SARS-CoV-2 associated target proteins, namely spike protein receptor-binding domain (ID: 6VW1) [40], main protease (6LU7) [41], angiotensin-converting enzyme-2 (ACE2) (ID: 1R42) [42], human furin (ID: 4RYD) [43] and RdRp (ID: 6M71) were used in the present study. The crystal structures of all the target proteins were downloaded from the RCSB Protein Data Bank (PDB) (Source: http://www.rcsb.org/). All the aforesaid target proteins (viral and host) have been reported to intercede SARS-CoV-2 infection, replication, survival and multiplication in the host body [44], [45].
2.4. Molecular docking
The molecular docking between potent antiviral compounds and SARS-CoV-2 associated target proteins were studied using Molegro Virtual Docker (MVD, Molexus IVS, Denmark) 2010.4.0 software for Windows [46], [47]. The molecular docking was performed using a 15 Å grid radius; the number of runs: 10; maximum interactions covered: 1500; maximum population size generated: 50; maximum steps followed: 300; neighbor distance factor: 1.00; the maximum number of poses generated: 5 to cover the ligand-binding site of the target proteins structure [48]. As per MVD algorithm, the docking score is presented as an arbitrary unit; the lower the score is the better the binding affinity [48]. The protein-ligand complex and atomic level chemical interactions were further analyzed and visualized by Chimera software [49] and Discovery Studio [50]. The output data of molecular interaction was shown in radar display and scatter plot using Microsoft Excel-2007 program (Supplementary figures). During graph plot (both radar and scatter plot) X-axis was set as the name of ligands and Y-axis was the MVD docking scores (mean of five poses).
2.5. Molecular dynamics simulations
Molecular dynamics (MD) simulations have been carried out by using Groningen Machine for Chemical Simulations (GROMACS 5.1.5) software [51] to study the dynamic behavior of the top-ranked protein-ligand complexes. The topology parameters of protein and ligands were generated by CHARMM27 [52] using an all-atom force field in GROMACS and SwissParam [53] respectively. Simulations were carried out with the TIP3P water model in dodecahedron boxes. The charged system was neutralized with NaCl solution. The processed setup was then energy minimized with the steepest descent algorithm by running 50,000 steps to remove bad contacts. Before the simulation run, each system was equilibrated for NVT (constant number of atoms, volume and temperature) using V-rescale thermostat [54] for 500 ps at 300 K temperature and NPT (constant number of atoms, pressure and temperature) was performed at 1.0 bar by Parrinello-Rahman barostat [55] for 1000 ps. The protein backbone was restrained while the solvent and counter ions were allowed to move during the equilibration phase. The LINCS algorithm [56] was used for all bound constraints. Particle mesh Ewald (PME) method was used for long-range electrostatics. During simulation run position restraints were removed. Finally, systems were subjected to 100 ns simulation run each under periodic boundary conditions at 300 K temperature and 1.0 bar pressure.
2.6. Binding free energy calculations
Protein-ligand binding affinity was analyzed using the classical simulation method molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) [57]. For current study, the g_mmpbsa tool [58] of GROMACS was employed to calculate the binding affinity of the simulated protein-ligand complexes by taking 50 snapshots from last 50 ns MD trajectories at regular intervals.
The total binding energy (ΔGbinding) of the complex in solvent can be defined as [59]:
| (1) |
Here, Gcomplex is the total energy of the protein-ligand complex and Greceptor and Gligand are individual energies of respective molecules. Further, free energy for each component is defined as [60]:
| (2) |
In the above equation, x denotes the ligand, protein or protein-ligand complex. EMM and Gsolvation are the average molecular mechanics potential energy in the vacuum and free energy of solvation, respectively. EMM can be calculated by using the following equation [61]:
| (3) |
where, Ebonded represents bonded interactions composed of the bond, angle, dihedral and improper interactions. Enon-bonded comprises van der Waals (Evdw) and electrostatic (Eelec) interactions. Also, Gsolvation or free energy of solvation is the combination of Gpolar and Gnon-polar contribution and in MM-PBSA method, implicit solvent model was used to calculate free energy of solvation [62]:
| (4) |
The electrostatic component, Gpolar is calculated by solving the Poisson–Boltzmann (PB) equation [63] and non-electrostatic component, Gnon-polar was calculated with solvent accessible surface area (SASA) model equation [60]:
| (5) |
Here, γ is a coefficient related to the surface tension of the solvent; b is a fitting parameter.
The final ΔGbind values for protein-ligand complexes were the average values from the 50–100 ns of MD simulation trajectories.
2.7. Pharmacophore modeling
Pharmacophore features of top-ranked compounds (higher affinity with target proteins) were determined using the Ligandscout platform, which also reveals possible structural activity relationship (SAR) [64] and optimal molecular interactions with a specific biological target(s). The fully optimized 3D chemical structure of all the compounds in Mol2 format was loaded into Ligandscout software and key pharmacophore features were identified based on 3D descriptors such as H–bond donor, H–bond acceptor, hydrophobic, aromatic, halogen bond donor, positively and negatively ionizable groups [65].
2.8. Validation based on structural superimposition
Molecular shape similarity analysis is based on the similarity of small molecules as the index for determination of minimum common structure from a set of compound libraries. Like pharmacophore theory, the molecular shape similarity index can effectively utilize the overall structural features of integrated compounds [66]. All the selected ligands in Mol2 format were zipped and loaded into PharmaGist web server [67]. The current study emphasizes more to obtain potent compound(s) considering the rule that molecules with similar structures may have similar or the same biological activity [68].
2.9. Validation based on machine learning algorithms
Bayesian machine learning models (FDA-approved drug screening) from Assay Central platform (https://assaycentral.github.io/#) was used to identify the applicability score of potent compounds that may work against SARS-CoV-2. Assay Central is a tool used for building machine learning models (Bayesian, Random Forest and Deep Neural Networks, etc.) that in turn can be used to filter and score target-specific lead compounds prior to testing [69]. It is a collection of predictive Bayesian and Random Forest models in a self-contained executable for non-experts to evaluate the likelihood of activity in a target of interest. The output includes a prediction score and applicability score (AS) (percentage of molecular fragments present in the model input). The prediction score (Pm) is the summation of fingerprint i and contributions, Ci. Contribution is based on the number of active compounds with the fingerprint, Ai, out of the total compounds with the fingerprint, Ti, and the total number of active compounds in the dataset, R [69].
.
.
2.10. Statistical analysis
The experimental results were expressed as mean ± S.D., n = 3. The data analysis was performed by an Analysis of Variance (ANOVA) followed by Tukey's test considering *P ≤ 0.05 as the statically significant values.
3. Results and discussion
Thus far, there are no specific therapeutic agents for controlling coronavirus infections. Drug repurposing is the most efficient alternative in a situation like a pandemic to develop a new cure quickly [30]. A lot of safety data, doses, pharmacokinetics, and bioassays results are already available for the molecule, which allows for a substantially rapid drug development process. Therefore, the present study was undertaken to determine potential therapeutic agents against COVID-19 from the available Ayurvedic antiviral drug candidates by performing a target-based molecular interaction study followed by molecular dynamic and MM-PBSA binding energy calculation.
3.1. Molecular docking
The molecular docking method is useful for modeling the interaction between a small molecule and target proteins at the atomic level, so that the behavior of small molecules at the binding site of target proteins and fundamental biochemical processes can be elucidated [46], [47]. The docking process includes two basic steps: determination of the conformation of the ligand, its position and orientation (pose) and assessment of its binding affinity [70]. These two parameters have been successfully determined in the present study during sampling and scoring functions and are discussed in detail in forthcoming sections.
3.1.1. Docking analysis of angiotensin-converting enzyme-2 (ACE2)
Wan et al. [71] demonstrated that residue 394 (glutamine) in the SARS-CoV-2 receptor-binding domain (RBD) can be recognized by critical lysine 31 on the human ACE2 receptor [72]. Moreover, SARS-CoV-2 binds to ACE2 ten times more strongly than other coronaviruses, making SARS-CoV-2 more infectious than others [73]. So the spike protein of SARS-CoV-2 was predicted to have a strong binding affinity to human ACE2. Thus, target angiotensin-converting enzyme-2 (ACE2) receptor with small molecules may be a viable solution that may restrict the coronavirus from entering into the cells [30]. In this context, molecular docking was performed and the result showed that curcumin has strong binding affinity (−88.9) for ACE2 followed by rosmarinic acid (−88.1) and gingerol (−74.3) (Table S1 and Fig. S1). To have a better understanding, the comparative docking scores relative to the center point of all the twenty antiviral compounds with target proteins are represented in radar display (Fig. S1a). Alternatively, a self-explanatory graphical representation in the form of a scatter plot has been shown, where the individual mean docking scores (poses) are presented in a matrix with top ranked ligands (yellow oval area) (Fig. S1b). In another study, it has been reported that inhibition of myocardial fibrosis by curcumin is associated with modulating expression of angiotensin II (Ang II) receptors and angiotensin-converting enzyme 2 (ACE2) in male Sprague Dawley rats [74].
3.1.2. Docking analysis of furin
For achieving a successful fusion of SARS-CoV-2 with host target cells, the spike protein needs to be cleaved by proteases; this step is critical, as it allows the fusion sequences to be exposed and is generally, mediated by the host furin (α protein convertase) [32]. RRAR, a special furin-like cleavage site (FCS) in SARS-CoV-2 spike protein (S) that is absent from other lineage B βCoVs, such as SARS-CoV, has been reported to be responsible for its high infectivity and transmissibility [75]. Furin is found to be expressed in significant concentrations in the lungs [76]. Thus, this enzyme can be used by viruses in the respiratory tract to transform and activate their own surface glycoproteins [75], [77] that ultimately facilitate interaction with the cell surface receptor ACE2 [78]. This makes their role in viral protein processing noteworthy [75]. Furins are also known to regulate avian influenza A virus infection in which HA glycoprotein cleavage is needed to enter into the host cell [77]. Therefore, furin becomes an additional drug target in terms of inhibiting viral fusion with host cells [32]. Molecular docking study revealed that rosmarinic acid (−89.17) bound tightly to furin followed by curcumin (−87.36) and quercetin (−79.60) (Table S1 and Fig. S2). Several studies indicated that rosmarinic acid has anti-inflammatory and antiviral activity in mice infected with the Japanese encephalitis virus [79], [80]. In another study, it has been mentioned that the rosmarinic acid derivative has shown anti- HIV-1 activity by inhibiting HIV-1 integrase [81].
3.1.3. Docking analysis of SARS-CoV-2 spike protein
The large transmembrane spike glycoprotein (type I) of SARS-CoV-2 accounts for notable features; it is heavily-glycosylated, which is thought to mediate viral entry into the host body [29]. Therefore, its inhibition may be associated with decreasing the viral multiplication. The spike glycoprotein has two functional domains, designated as S1 and S2, both of which are necessary for a coronavirus to enter into a cell [29] successfully. The molecular docking study revealed that curcumin has a strong binding affinity (−109.3) with SARS-CoV-2 RBD domain of spike protein (Table S1) followed by gingerol (−89.59) and ribavirin (−85.92) (Fig. S3). Curcumin formed chemical interactions at RBD domain-human ACE2 interface with Tyr505, Ala387, Asp38, Gln493, Glu 35, His34, Glu 37 and Arg393 in the active site. It is worth mentioning that all these amino acids are localized in the interface region of spike glycoprotein and host receptors, ultimately facilitates receptor-mediated endocytosis [82] during primary infection. In a recent study, it has been reported that pan-coronavirus fusion inhibitor (EK1C4), targeting spike protein successfully restricted (IC50 range: 1.3–15.8 nM) viral entry into the host body against different types of coronaviruses such as SARS-CoV, MERS-CoV, SARS-CoV-2, HCoV-OC43 and SARSr-CoVs [83].
3.1.4. Docking analysis of SARS-CoV-2 main protease
Among coronaviruses, an alluring drug target is the ~ 306 amino acids long main protease (Mpro, 3CLpro), to forestall the spread of disease by restraining the cleavage of the viral polyprotein. Mpro is essential for processing the polyprotein that led to the proteolytic activation of the viral functional proteins [31]. An in silico analysis showed that the curcumin (−115.69) binds strongly with Mpro followed by rosmarinic acid (−105.43) and quercetin (−96.04) (Table S1 and Fig. S4). The Mpro active site amino acids such as Thr190, Pro168, Met165, Glu166 and Cys145 are predicted to play a major role during chemical interactions with curcumin.
3.1.5. Docking analysis of SARS-CoV-2 RNA dependent RNA polymerase (RdRp)
The 3D crystal structure of a SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp) has recently been solved by X-ray crystallography. The single-chain of core RdRp relies on virus-encoded Non-structural protein cofactors such as nsp7 and two units of nsp8 for its optimum function. The crystal structure of RdRp was downloaded from PDB (ID 6M71) with a reported resolution of 2.90 Å. RNA dependent RNA polymerase (RdRp) is involved in the replication and transcription of the SARS-CoV-2 genome. It is the cleavage product of the polyproteins 1a and 1ab from ORF1a and ORF1ab [111]. The minimum complex necessary for its proper functioning is completed by attachment of three additional protein peptides (nsp7-nsp8, and one additional nsp8) to the core polymerase which is chain A and contains 851 amino acids residues. The relative binding free energy between remdesivir and ATP was calculated to be −2.80 ± 0.84 kcal/mol, where remdesivir bound much stronger to SARS-CoV-2 RdRp than the natural substrate ATP [112]. The ∼ 100-fold improvement in the Kd from remdesivir over ATP indicates an effective replacement of ATP in blocking of the RdRp preinsertion site. Key residues Asp618, Ser549, and Arg555 are found to be the contributors to the binding affinity of remdesivir [113]. These findings suggest that remdesivir can potentially act as a SARS-CoV-2 RNA-chain terminator, effectively stopping its RNA replication, and hence remdesivir was used in the present study as a reference ligand to compare with other phytochemicals affinity with target proteins. An in silico result showed that the gingerol (−112) binds strongly with RdRp followed by curcumin (−97) and quercetin (−88) (Tables S1, S3 and Fig. S5). The RdRp active site amino acids such as Asn691, Asp623 and Arg624 are predicted to play a major role during chemical interactions with gingerol. Interestingly, the docking score of gingerol (−112) is found to be comparable with reference ligand, remdesivir (−134).
3.2. Molecular dynamic simulations
Among the different compounds screened through molecular docking studies, curcumin, gingerol, rosmarinic acid, ribavirin and quercetin showed strong intermolecular interaction with the target proteins with high binding scores (−74 to −115) (Table S1). Therefore, these compounds were selected as the potential inhibitors and used for further analysis through MD simulation. A 100 ns simulation was performed for each system to get deep insights into the stability and more reliable binding mode of the ligand-protein complex. The overall stability of each complex was determined through the backbone root mean square deviation (RMSD) of proteins. Further, these compounds were filtered by the MM-PBSA method, which is a reliable method for binding free energy calculations. In last, the binding mode for the selected potential inhibitors from MD simulations and binding free energy analysis was done.
3.2.1. MD simulation analysis for ACE2
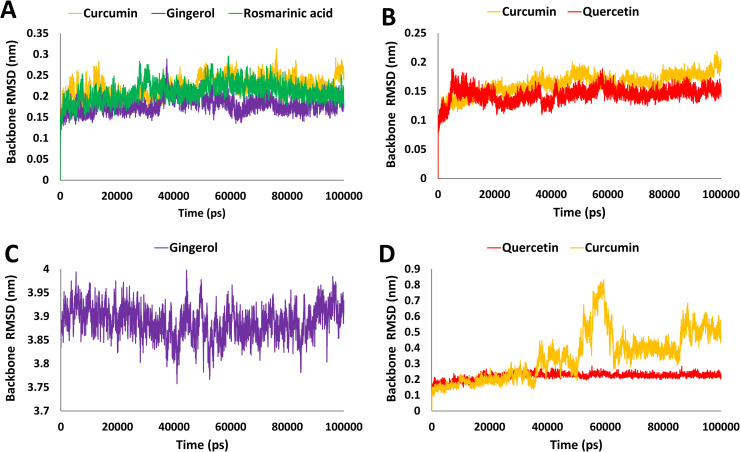
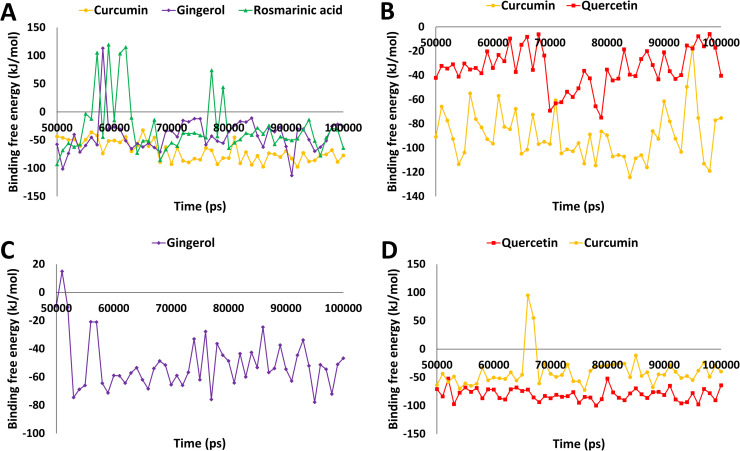
The target protein angiotensin-converting enzyme-2 (ACE2), which was found to have desirable interactions with curcumin, rosmarinic acid and gingerol during molecular docking study were subjected to 100 ns simulation run with respective ligands. After the MD simulation run, the stability of each simulated complex was determined by analyzing the RMSD of protein backbone atoms. The RMSD values for three protein-ligand systems curcumin, rosmarinic acid and gingerol were 0.17, 0.20 and 0.22 nm respectively ( Fig. 2A). The analysis revealed that all three systems obtained a steady-state after convergence and RMSD values were observed within the acceptable value of < 0.3 nm. Further, binding free energy calculations were performed through the MM-PBSA method using the last 50 ns stable RMSD trajectories; a total of 50 snapshots were obtained at regular intervals. The different types of energies which contributed to binding such as van der Waals, electrostatic, polar solvation and SASA energy were calculated; the average values of these energies are depicted in Table S2. The overall average values of the binding energy for curcumin, gingerol and rosmarinic acid were −69.51, −41.62 and −29.18 kJ/mol (Table S2 and Fig. 3A). Our analysis revealed that curcumin displayed stable behavior with ACE2 protein throughout the simulation (100 ns) and displayed the lowest binding free energy and therefore could be considered for further study.
Fig. 2.
Analysis of molecular dynamics simulation results. A) Root mean square deviation (RMSD) of backbone atoms of ACE2 in complex with curcumin, gingerol and rosmarinic acid, B) RMSD of backbone atoms of furin in complex with curcumin and quercetin, C) RMSD of backbone atoms of spike protein RBD-ACE2 in complex with gingerol, D) RMSD of backbone atoms of Mpro in complex with quercetin and curcumin.
Fig. 3.
Binding free energy analysis from MM-PBSA method over the last 50 ns trajectories. A) The ACE2 complexes with curcumin (orange), gingerol (purple), and rosmarinic acid (green) are represented in different color schemes, B) Furin protein complexes with curcumin (orange) and quercetin (red) are represented in different color schemes, C) Spike RBD-ACE2 in complex with gingerol represented here in purple color, D) The Mpro complexes with quercetin (red) and curcumin (orange) are represented in different color schemes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2.2. MD simulation analysis for furin
The third target protein furin was found to have better interaction with rosmarinic acid, curcumin and quercetin during molecular docking analysis. To further validate these results, all three protein-ligand complexes were simulated for 100 ns independently. The MD simulation revealed that furin protein backbone atoms showed RMSD for bound ligands 0.16 (rosmarinic acid), 0.16 (curcumin) and 0.14 (quercetin) nm respectively. The RMSD values were found within the acceptable value of < 0.3 nm. In last, binding free energy calculation using the MM-PBSA tool was performed similarly as stated for other proteins. The different types of energies which contribute to binding such as van der Waals, electrostatic, polar solvation and SASA energy were calculated and the average values of these are depicted in (Table S2). The analysis revealed that curcumin and quercetin displayed the average binding free energy −89.72 and −34.12 kJ/mol (Fig. 3B and Table S2). Unfortunately, rosmarinic acid displayed a positive binding energy value (1.97 kJ/mol), which confirmed its weak affinity towards furin protein and hence was removed from further analysis. Detailed analyses of curcumin and quercetin have been shown in (Figs. 2B and 3B).
3.2.3. MD simulation analysis for spike protein
The second target protein in the present study, large transmembrane spike glycoprotein (type I) of SARS-CoV-2 in complex with humans ACE2 protein, was found to have better interaction with curcumin, gingerol and ribavirin during molecular docking studies. Each protein-ligand complex was further subjected to 100 ns MD simulation independently. The system's stability was then analyzed by RMSD values of protein backbone atoms. The RMSD values of protein-ligand complexes were 3.61, 3.88 and 2.61 nm respectively. Higher values of RMSD were observed but found to be stable. Subsequently, binding free energy calculations were performed through MM-PBSA method. For calculation last 50 ns trajectories of stable RMSD values were selected and a total of 50 frames were generated at an equal interval. The different types of energies which contribute to binding such as van der Waals, electrostatic, polar solvation and SASA energy were calculated, the average values of these energies are depicted in Table S2. Binding free energy calculations revealed that gingerol had an average binding free energy value of −50.59 kJ/mol, which is better than other tested ligands. Rosmarinic acid displayed a positive value of average binding free energy, which is unfavorable for binding. Unfortunately, the curcumin bound complex was not able to give binding free energy values due to instability of the complex and hence discontinued from further analysis. Based on dynamic characteristics and binding free energy score, gingerol displayed better results than other tested ligands. Therefore, only gingerol was selected for detailed analysis (Figs. 2C and 3C). In a recent study, a reference drug remdesivir was studied against spike RBD-ACE2 protein, and recorded average binding free energy −71.88 kJ/mol from the MM-GBSA method [114]. Our hit compound displayed comparatively high binding energy but still under acceptable limit [115] and hence can be considered for further analysis.
3.2.4. MD simulation analysis for Mpro
The fourth targeted protein in this study is Mpro, which was found to have better interaction with curcumin, rosmarinic acid and quercetin during molecular docking analysis. To further validate these results, a 100 ns simulation run was performed and the stability of the protein-ligand complexes were analyzed by protein backbone atoms RMSD. The protein backbone RMSD was found to be 0.34, 0.27 and 0.22 nm for curcumin, rosmarinic acid and quercetin. Curcumin displayed RMSD values slightly greater than 0.3 nm during the simulation run, but was observed to become stable during the later stage of simulation (Fig. 2D). Binding free energy through MM-PBSA was calculated following a similar protocol as mentioned before for Mpro bound ligands. The different types of energies which contribute to binding such as van der Waals, electrostatic, polar solvation and SASA energy were calculated, the average values of these energies are depicted in Table S2. The average binding free energy for potential inhibitors was −80.05 kJ/mol, −40.41 kJ/mol for quercetin and curcumin respectively (Fig. 3D). Unfortunately, rosmarinic acid displayed a positive average binding free energy value (1.37 kJ/mol), which is not favorable for binding and hence, it has been removed from further analysis. In a recent study, the binding free energy for COVID-19 main protease (PBD: 6LU7) bound inhibitor N3 was found to be −51.07 kJ/mol [115]. In contrast, our hit compound quercetin showed very low binding free energy (−80.05 kJ/mol). This observation strongly suggests that quercetin may bind with Mpro more efficiently than reference inhibitor N3 and therefore considered for further analysis.
3.2.5. MD simulation analysis for RdRp
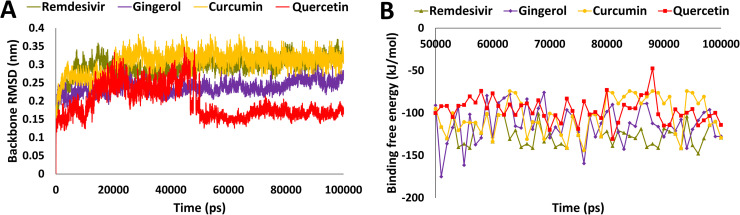
RdRp, which was found to have better interaction with gingerol, curcumin and quercetin during molecular docking analysis. To further validate these results, a 100 ns simulation run was performed and the stability of the protein-ligand complexes were analyzed by protein backbone atoms RMSD. The protein backbone RMSD was found to be 0.29, 0.24, 0.31 and 0.19 nm for remdesivir, gingerol, curcumin and quercetin ( Fig. 4A). Binding free energy through MM-PBSA was calculated following a similar protocol as mentioned above for other receptors. The different types of energies which contribute to binding such as van der Waals, electrostatic, polar solvation and SASA energy were calculated, the average values of these energies are depicted in Table S2. The average binding free energy of potential hit compounds was −123.22 kJ/mol, −114.4 kJ/mol, −105.5 kJ/mol and −96.3 kJ/mol for remdesivir, gingerol, curcumin and quercetin respectively (Fig. 4B and Table S2). In a present study, the binding free energy for SARS-CoV-2 RNA dependent RNA polymerase (RdRp) with reference inhibitor, remdesivir was found to be −123.2 kJ/mol. Interestingly, our hit compound gingerol showed very close binding free energy with reference inhibitor, remdesivir. This observation strongly suggests that gingerol may bind with RdRp efficiently and therefore considered for further analysis.
Fig. 4.
Analysis of molecular dynamics simulation results. A) Root mean square deviation (RMSD) of backbone atoms of RdRp in complex with remdesivir, gingerol, curcumin, and quercetin B) Binding free energy analysis from MM-PBSA method over the last 50 ns trajectories of RdRp. The complexes are represented in different color schemes with remdesivir (olive), gingerol (purple), curcumin (orange), and quercetin (red). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Binding mode analysis
Molecular dynamics simulation is an attractive approach to study molecular interactions and binding mode analysis [116]. In the present study, binding mode analysis was performed by calculating an average structure from the last 10 ns MD trajectories for all four target proteins complexed with respective ligands. Only those ligands were subjected to binding mode analysis, which displayed an acceptable amount of average binding free energy (Table S3). Interestingly, when ligands were superimposed, it was found that only the best-bound inhibitor from MM-PBSA was able to maintain its interaction inside the active site of the protein, and the rest of the inhibitors lost their interactions or showed binding at different sites. Therefore, the molecules displaying the lowest binding free energy were considered and further analyzed for intermolecular interactions inside the respective target protein’s active site.
3.3.1. Binding mode for ACE2 protein
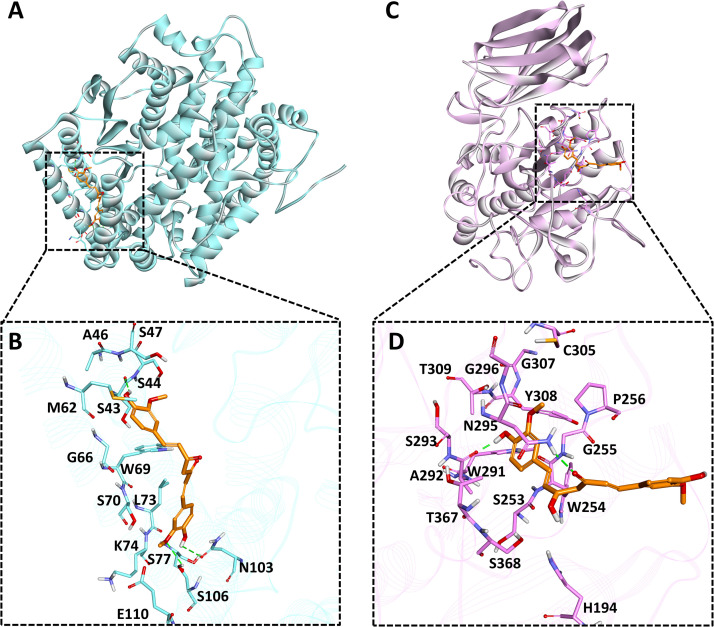
To assess the binding mode of selected compounds from detailed MD simulations and binding free energy calculations, the average structure was calculated from the last 10 ns MD trajectories and compounds were superimposed. Unfortunately, gingerol was not able to show interactions with desirable active site residues and rosmarinic acid found bound outside the active site which is not important for inhibition of protein. Hence, these two compounds were excluded from further analysis ( Fig. 5A). The selected compound, curcumin was found to make three hydrogen bonds with ACE2 active site residues Ser43, Asn103 and Ser106 (Fig. 5B and Table S3). Our investigation also revealed that curcumin makes two types of non-polar interactions with active site residues. The first one is hydrophobic interactions involving Ser44, Ala46, Ser47, Gly66, Trp69, Ser70, Lys74, Ser77, Glu110. The second type of interaction is pi-alkyl, which was observed with Met62 and Leu73 of ACE2 (Fig. S6A and Table S3). A similar pattern of hydrophobic interactions with other inhibitors of ACE2 has been reported recently [117]. Our analysis showed that curcumin shows better binding affinity and molecular interactions such as hydrogen bonds, hydrophobic interactions, and van der Waals interactions with ACE2 active site residues throughout the simulation. Hence, it can be used for further studies against ACE2 protein.
Fig. 5.
Binding mode analysis for human ACE2 and furin proteins. A) ACE2 (cyan color) shown in solid ribbon representation in complex with curcumin (orange color), B) The close depiction of active site of ACE2 with curcumin, here active site residues are shown in cyan sticks C) Furin protein (light pink) shown in solid ribbon representation in complex with curcumin (orange color), D) The close depiction of furin bound curcumin complex, background is shown with protein in line ribbon (pink color). Active site interacting residues are shown in pink sticks. Hydrogen bonds are indicated with green dashed lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3.2. Binding mode for furin
Compounds such as curcumin, quercetin and rosmarinic acid were found to have good interaction during molecular docking but during binding free energy rosmarinic acid displayed unfavorable binding free energy values and therefore, excluded from further analysis. Consequently, last 10 ns MD trajectories data was used to calculate average structure for both the ligands, i.e., curcumin and quercetin. During superimposition of structures, it was observed that quercetin moved outside the active site, so forth only curcumin was considered for binding mode analysis (Fig. 5C). During analysis, it was observed that curcumin formed two hydrogen bonds with active site residues Ala292 and Asn295 of furin protein (Fig. 5D and Table S3). Similar residues were also targeted in other studies providing a strong potentiality to curcumin against furin protein[118]. Apart from hydrogen bonds, various hydrophobic interactions were also observed. Curcumin forms van der Waals interaction with residues His194, Ser253, Gly255, Pro256, Trp291, Ser293, Cys305, Thr309, Thr367, Ser368 and pi-pi interaction with Trp254 (Fig. S6B and Table S3).
3.3.3. Binding mode for spike protein
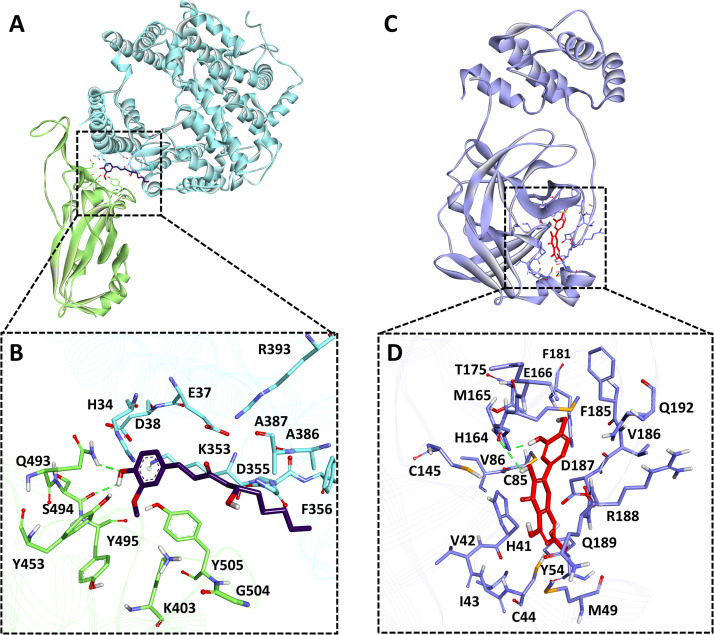
A similar type of analysis was performed for this Spike-ACE2-RBD of SARS-CoV-2. The results obtained after MD simulations and binding free energy calculation revealed that out of three tested ligands, gingerol performs better than other compounds (Table S3). Hence, we further selected gingerol for binding mode analysis. The average structure for the gingerol bound protein was calculated from the last 10 ns of MD simulation trajectories and further analyzed ( Fig. 6A). The analysis revealed that gingerol forms two hydrogen bonds with spike protein RBD domain key residues Ser494 and Gln493 (Fig. 6B and Table S3). A recent study reported Gln493 and Ser494 of spike protein RBD domain interacted with hotspot residues of ACE2 protein, i.e., Lys31 and Lys353 [115], [117]. Similar interactions of gingerol with these residues provide a support to current findings. Gingerol also makes hydrophobic interactions with both the interacting proteins. Hydrophobic interactions with spike RBD were observed with Lys403, Tyr453, Tyr495 and Tyr505; similarly, hydrophobic interactions for ACE2 protein were of two types van der Waals interaction with Glu37, Asp38, Phe356, Ala387, Arg393 and pi-pi interactions with His34, Lys353, Ala386 (Fig. S6C and Table S3). The interaction pattern of gingerol with key residues strongly suggests its inhibitory effect against the Spike-ACE2-RBD complex.
Fig. 6.
Binding mode analysis for SARS-CoV-2 spike and Mpro protein. A) ACE2 protein is shown in cyan and RBD domain of SARS-CoV-2 is shown in green color solid ribbon representation. B) The close depiction of protein complex displays gingerol (purple color) in stick representation inside the spike protein RBD active site. Protein in the background is shown as ribbon. Active site interacting residues of ACE2 protein are shown in cyan sticks whereas, RBD domain active site residues are shown in green sticks. C) Mpro (light purple color) shown in solid ribbon representation bound with quercetin, D) The close depiction of protein displays quercetin (red color) in stick representation inside the Mpro active site. Protein in the background is shown as ribbon (light purple color). Active site interacting residues are shown in purple sticks. Hydrogen bonds are indicated with green dashed lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3.4. Binding mode for Mpro
Binding free energy calculation revealed that quercetin and curcumin showed the highest binding affinity against Mpro. So quercetin and curcumin binding mode was assessed using average structures calculated from the last 10 ns MD trajectories. Superimposition of Mpro complexes with quercetin and curcumin displayed that only quercetin was found bound inside the active site of Mpro (Fig. 6C). Unfortunately, curcumin was bound outside the active site of protein hence excluded from further analysis. Quercetin forms a hydrogen bond interaction with His164, which is one of the key residues reported for the inhibition of Mpro (Fig. 6D). Quercetin also forms van der Waals interactions with the active site residues Cys44, Tyr54, Cys85, Cys145, Gly174, Thr175, Phe181, Phe185, Val186, Asp187, Gln189, Gln192 and pi-pi interaction His41, Met49, Met165, Arg188 (Fig. S6D and Table S3). A similar type of binding pattern for Mpro protein inhibitors was recently noticed [119], [120]. Hence, our findings confirmed that quercetin binds tightly to Mpro and can inhibit its activity through several molecular interactions.
3.3.5. Binding mode for RdRp
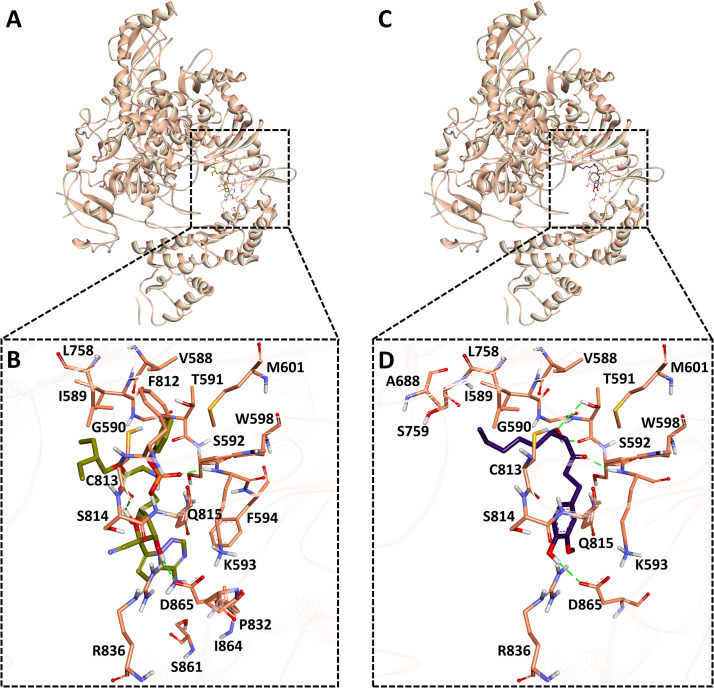
Binding free energy data revealed that gingerol, curcumin and quercetin (Table S2) showed the strong binding affinity against RdRp. Therefore average structure was calculated from the last 10 ns MD trajectories for all mentioned ligands along with remdesivir. It was observed that gingerol formed similar binding mode with RdRp as remdesivir does. Gingerol forms hydrogen bond interactions with Thr591, Lys593 and Asp865 which are one of the key residues reported for the inhibition of RdRp ( Fig. 7, Table S3). Gingerol also forms van der Waals interactions with the active site residues Val588, Gly590, Ser592, Trg598, Met601, Ser759, Cys813, Ser814, Gln815, Arg836 and pi-pi interaction Ile589, Ala688, Leu758 (Fig. S6F and Table S3). A reference inhibitor, remdesivir showed H-bond interactions with key residues Lys593, Cys813, and Asp865 with RdRp (Figs. 7 and S6E). A similar type of binding pattern for RdRp protein inhibitors was recently noticed [112], [113]. Our findings confirmed that gingerol binds tightly to RdRp and can inhibit its activity through several molecular interactions.
Fig. 7.
Binding mode analysis for SARS-CoV-2 RdRp protein. A) RdRp protein is shown in peach color solid ribbon representation. B) The close depiction of protein complex displays remdesivir (olive color) in stick representation inside the RdRp active site C) RdRp protein is shown in peach color solid ribbon representation. D) The close depiction of protein complex displays gingerol (purple color) in stick representation inside the RdRp active site. Protein in the background is shown as ribbon. Active site interacting residues of RdRp protein are shown in peach sticks whereas. Hydrogen bonds are indicated with green dashed lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Pharmacophore modeling
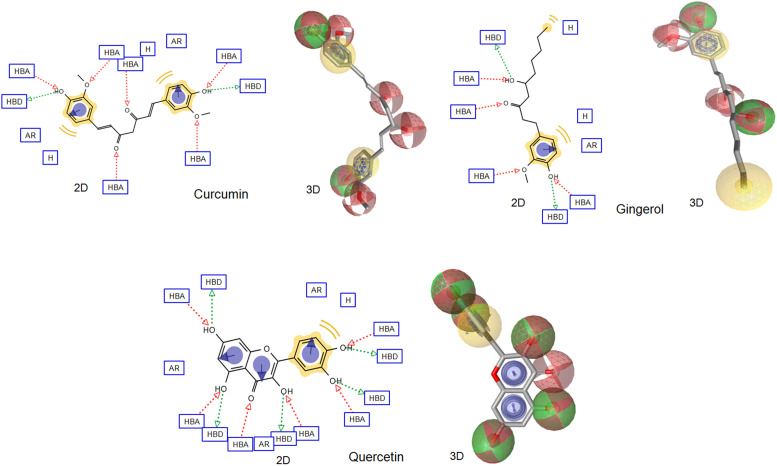
A pharmacophore model of a bioactive compound is a collection of steric and electronic descriptors that is needed to ensure the stability of the ligand-receptor complex and associated biological response [121]. Pharmacophore features also led to the development of more stable and targeted drugs with no or negligible side effects in the host, as they are supposed to be more accurately targeted [122]. The top-ranked ligands (based on MD result), which showed high binding affinity with all the receptors were further subjected to predict pharmacophore features. The 2D and 3D pharmacophore features of top-ranked compounds were determined by LigandScout 4.1 and shown in Fig. 8. The inspected pharmacophoric features of all the compounds include hydrogen bond donors and acceptors as directed vectors, positive and negative ionizable regions, and lipophilic areas illustrated by spheres. All these above-reported pharmacophore features are essential for binding with receptors. These features would enable repurposing COVID-19 drugs candidates at the same time open up new avenues to design new and more potent SARS-CoV-2 inhibitor(s) as prospective antiviral agents.
Fig. 8.
Pharmacophore features (2D and 3D) of top-ranked compounds (based on MD simulation) predicted using Ligandscout software. The pharmacophore color code is yellow for hydrophobic regions, red for hydrogen acceptors and green for hydrogen donors. 2D Pharmacophore features represent HBA: hydrogen bond acceptor, HBD: hydrogen bond donor, AR: aryl, NI: negative ionizable and H: hydrophobic center. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. Validation based on structural superimposition
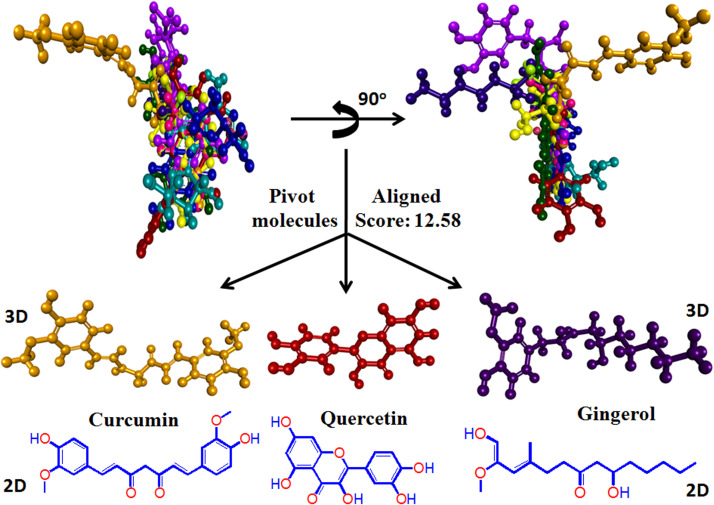
Structural superimposition plays a major role in determining the structural template with a common structure. In the present study, the PharmaGist web server has been used to align all the aforesaid antiviral compounds in 3D orientation, followed by the determination of the pivot molecule(s) [123]. The output file consists of a set of aligned structures with scores based on the common descriptors computed by multiple flexible alignments of the input ligands [67]. After superimposition, the obtained result showed that gingerol, quercetin and curcumin represent pivot molecules and share most of the pharmacophore features of submitted ligands ( Fig. 9) and hence strong intermolecular interactions were observed during docking and MD simulation.
Fig. 9.
PharmaGist based ligands superimposition and determination of pivot molecule (key structure). Curcumin, gingerol and quercetin showed structural similarity with high align score and hence may be able to bind SARS-CoV-2 associated target proteins more efficiently than others.
3.6. Validation based on machine learning algorithms
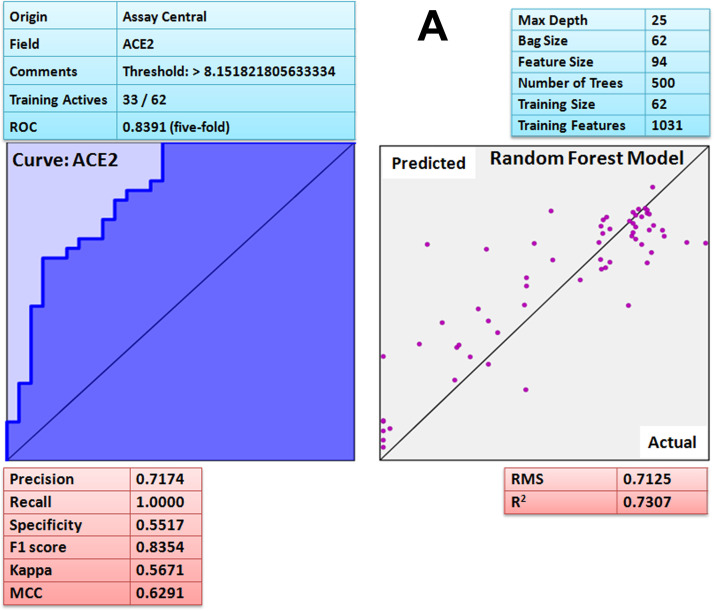
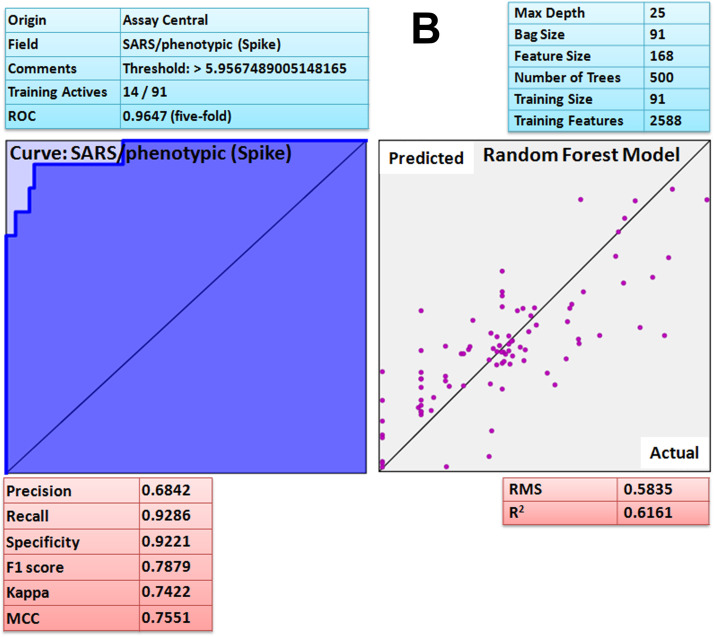
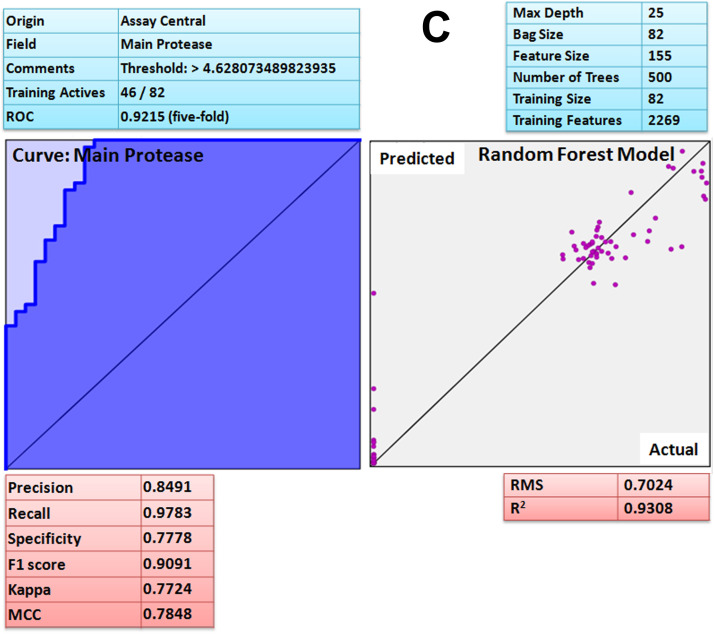
Assay Central (AC) is a stand-alone and user friendly set of predictive Bayesian and Random Forest models for non-experts to determine the likelihood of action of bioactive compounds against the target of interest. After developing the model, each molecule in the selected 'project' receives a relative score known as applicability number that indicates fraction of structural features shared with the training set [69]. Additionally, the prediction score for each compound is also generated that ranges from 0 to 1; a higher score indicates that the compound is more likely to be active at the modeled target. The current threshold value used to consider a compound as active is 0.5 or higher. Furthermore, the applicability score refers to the percentage of fragments of the expected molecule present in the model. The lower applicability score (≤ 0.3) implies that the prediction is not effective because the molecules comprising the model do not have adequate structural similarity to the predicted compound. Such machine learning models could help during prioritization of compounds for testing in vitro and in vivo. The attribute of the ROC or the receiver operator can be interpreted as a percent probability of correctly predicting compound activity against target. Additional metrics included the F1 Score, Cohen's Kappa and Matthew's Correlation Constant (MCC) ranging from 0 to 1 and the value closer to 1 are considered as the most viable model [69]. The Assay Central Bayesian model [69] for coronavirus disease (COVID-19) associated ACE2 receptor has a 5-fold cross-validation receiver operating characteristic (ROC) of 0.84, precision 0.72, recall 1, specificity 0.55, F1-score 0.84, Cohen’s kappa (CK) 0.57, Matthews correlation coefficient (MCC) 0.63 and Random Forest square of the correlation (R2) 0.73 ( Fig. 10A). For ACE2 protein, prediction score (Pm) and applicability score (AS) of potent compound, curcumin was 0.80 and 0.72 respectively. SARS spike protein has a 5-fold cross-validation receiver operating characteristic (ROC) of 0.97, precision 0.68, recall 0.93, specificity 0.92, F1-score 0.79, Cohen’s kappa (CK) 0.74, Matthews correlation coefficient (MCC) 0.76 and Random Forest square of the correlation (R2) 0.62 (Fig. 10B). For spike protein, prediction score (Pm) and applicability score (AS) of potent compound, gingerol was 0.84 and 0.78 respectively. The SARS main protease model has a 5-fold cross-validation ROC of 0.92, precision 0.85, recall 0.98, specificity 0.78, F1-score 0.90, CK 0.77, MCC 0.79 and Random Forest square of the correlation (R2) 0.93 (Fig. 10C). For SARS-CoV-2 main protease prediction score (Pm) and applicability score (AS) of potent compound, quercetin was found to be 0.70 and 0.77 respectively. Moreover, the Assay Central platform lacks data for furin and RdRp therefore; it was not validated by the same method. Hence, based on the prediction (Pm) and applicability score (AS) of Assay Central Bayesian model (Fig. 10A–C) it is revealed that curcumin, gingerol and quercetin has strong affinity for ACE2, spike protein and main protease respectively, interestingly; the result further corroborate the molecular dynamic study. The results were further corroborated by comparing reported IC50 with interaction scores. Interestingly, the compound with low IC50 (reported) has shown strong chemical interactions with target proteins and similar was the case with curcumin, gingerol and quercetin. Thus, based on the machine learning algorithm [69] it can be suggested that aforesaid compounds (curcumin, gingerol and quercetin) possess strong binding affinity against COVID-19 associated target proteins.
Fig. 10.
A Assay Central Visualization. A) Machine learning model performance summary of a Bayesian (left) and Random Forest (right) algorithms. Bayesian machine learning models using ECFP6 fingerprint was used for scoring and selecting compounds having potent binding affinity with ACE-2 target protein. B Assay Central Visualization. A) Machine learning model performance summary of a Bayesian (left) and Random Forest (right) algorithms. Bayesian machine learning models using ECFP6 fingerprint was used for scoring and selecting compounds having potent binding affinity with SARS-CoV-2 spike protein. C Assay Central Visualization. A) Machine learning model performance summary of a Bayesian (left) and Random Forest (right) algorithms. Bayesian machine learning models using ECFP6 fingerprint was used for scoring and selecting compounds having potent binding affinity with SARS-CoV-2 main protease.
Though synthetic antiviral drugs are significant in treatment of viral infections, there are innumerable side effects with alterations due to progressive viral complications. Hence, medicinal plants are considered excellent alternative sources of antiviral compounds [124]. Curcuma longa and Zingiber officinale are reported to have strong antioxidant, and antiviral properties and various reports suggested their use in inhibiting SARS-CoV-2 viral replication and pathogenesis [124]. Various biological effects of curcumin have been documented including anti-tumor, anti-hypertensive and antiviral activities [125], [126]. Curcumin mediated antiviral activity was observed against various viruses, including hepatitis viruses, Zika virus, Chikungunya virus, human papillomavirus, human immunodeficiency virus, herpes simplex virus-2 and respiratory influenza virus [127]. Previously, curcumin has been shown to hinder the replication of SARS-coronavirus as well [128] and identified as a constructive inhibitor of protease. There are ample experimental research findings that support the efficiency of curcumin, gingerol and quercetin against the human viruses including SARS-CoV-2 pathogenesis [84], [85], [129]. Gingerol and curcumin extract demonstrated 45% and 40% of protease inhibition at 0.2 mg/mL concentration in an in vitro protease inhibitory assay and the result was further corroborated by pharmacokinetic properties [129]. SARS-CoV-2 genomic resemblance with SARS-coronavirus (> 80%) and Middle East Respiratory Syndrome-coronavirus (50%) collectively suggests the possibility of curcumin efficacy against COVID-19 [130]. Experimental evidences of curcumin in therapeutic and prophylactic respiratory infection management as well as pathophysiology have been identified and support the implementation of curcumin in COVID-19 [131]. Recent experimental evidence from computational approach predicted the direct binding of curcumin with receptors mediating entry of the virus into host cells as well modulation of associated inflammatory events [132], [133], [134]. The reported ability of curcumin to modulate cellular and physiological consequences [127], [128], [135] associated with SARS-CoV-2 infection along with recent in-silico predictions [132], [133], [134] further support its potential. Synergistic antiviral effects of quercetin and ascorbic acid have been effective in preventing viral entry and replication [136]. Experimental and computational evidence augment the efficacy of curcumin, gingerol and quercetin as anti-protease activity [86], [87], [88], [129]. The present study demonstrated the repurposing possibilities of curcumin, gingerol and quercetin based on their binding ability with multiple COVID-19 targets. Furthermore, their inhibitory activity using molecular dynamic simulations, pharmacophore features and Bayesian machine learning models supported their ability to function as SARS-CoV-2 inhibitors.
Since the emergence of COVID-19, SARS-CoV-2 virus has spread enormously across the globe causing a pandemic. Vaccination or the development of herd immunity seems to be the only way to break the chain of virus spread; however, the appearance of similar mutated strain(s) and failure of initially recommended drugs raised concerns in the near future. Therefore, effective therapies are desperately required to alleviate the burden of this pandemic and reduce mortality rates. Various repurposable drugs were proposed to effectively combat COVID-19. In March 2020, emergency use authorization of hydroxychloroquine was approved by the US food and drug administration (FDA), due to its therapeutic immunomodulatory effects [137]. In a study published in May 2020, remdesivir, an inhibitor of viral RNA dependent RNA polymerase (RdRp) that was used in the treatment of ebola in 2004, was widely considered against COVID-19 [138]. A study by [139] showed the significance inhibition of SARS CoV-2 replication by remdesivir and lopinavir in Vero E6 cells with an EC50 of 23.15 and 26.63 μM respectively. Though, lopinavir-ritonavir, reduced the duration of ICU stay in severely affected patients, they also showed severe gastrointestinal side effects [140]. Early interferon therapy was found more promising by reducing viral counts and lessening symptoms but did not reduce fatality [141]. However, the solidarity trial of treatment evaluation published by WHO in October declared that the four drugs remdesivir, hydroxychloroquine, lopinavir/ritonavir and interferon had no significant effect on covid-19 (www.who.int). It is imperative to understand the mandate of extensive research that focuses on repurposing of drugs and the current study has evaluated a wide range of Ayurvedic components with good efficacy to halt viral replication, attachment and entry. Thus, the repurposing of existing drugs is the fastest and cost effective approach to mitigate the COVID-19 burden for the near future. It remains important to identify medications that can be safely used during the early stage of infection to avoid progression to serious illnesses and deaths.
4. Conclusion
The present study aimed to identify molecules from medicinal plants reported in ancient literature, Ayurveda that may inhibit SARS-CoV-2 by acting on the major target proteins such as spike protein, main protease, angiotensin-converting enzyme-2, furin and RdRp. The obtained results by molecular docking, molecular dynamic simulation, binding free energy calculations, pharmacophore model and structure-based alignment methods showed that curcumin, gingerol and quercetin served as pivot molecules and may work against COVID-19. Furthermore, the binding affinities of all the compounds were also compared with a reference ligand, remdesivir against RdRp target. In addition, we validated molecular docking and molecular dynamic findings using a Bayesian machine learning models using ECFP6 fingerprints and interestingly, prediction score (Pm) and applicability score (AS) of potent compound, gingerol against spike RBD was found to be 0.84 and 0.78 respectively. Furthermore, prediction score (Pm) and applicability score (AS) of curcumin against ACE2 protein was found to be 0.80 and 0.72. Conversely, the prediction score (Pm) and applicability score (AS) of potent compound quercetin against Mpro was turned out to be 0.70 and 0.77. Thus, the results of machine-learning algorithms further corroborate the findings of molecular dynamic simulation and open up new avenues for preclinical investigations of aforesaid compounds in suitable in vivo and in vitro model(s). This study also supports the traditional use of antiviral plants as preventive measures. Hence, it is suggested that the world community should undertake this study further to repurpose clinical studies to test efficacy and safety in the treatment of COVID-19, which is an urgent need of the hour. However, further research is needed to materialize their dosage and safety.
Author contributions
A.K.V. designed the study, performed research, analyzed the data and wrote the paper. V.K. performed molecular dynamic simulation and binding free energy calculations from MM-PBSA and also contributed to writing their parts. S.S., A.S. and I.C. are involved in receiver operating characteristic curve (ROC) validation and analysis, machine learning model validation, data analysis and writing their parts. Professor B.C.G. & K.W.L. contributed in data interpretation and analyzed results. All the authors read, agreed and approved the final manuscript for submission.
Declaration of Competing Interest
There is no conflict of interest for the publication of this article.
Acknowledgments
The Bio and Medical Technology Development Program of the National Research Foundation (NRF), the Korean government is highly acknowledged for using resources of an ongoing project (No. NRF-2018M3A9A70-57263).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.biopha.2021.111356.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Zu Z.Y., Jiang M.D., Xu P.P., Chen W., Ni Q.Q., Lu G.M., Zhang L.J. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020;296:15–25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.W.H. Organization, Coronavirus Disease 2019 (COVID-19): Situation Report, 67, 2020, pp. 1–18.
- 3.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect. Dis. 2020;20:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kui L., Fang Y.Y., Deng Y., Liu W., Wang M.F., Ma J.P., Xiao W., Wang Y.N., Zhong M.H., Li C.H. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin. Med. J. 2020;133:1025–1031. doi: 10.1097/CM9.0000000000000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pizzorno A., Padey B., Terrier O., Rosa-Calatrava M. Drug repurposing approaches for the treatment of influenza viral infection: reviving old drugs to fight against a long-lived enemy. Front. Immunol. 2019;10:1–12. doi: 10.3389/fimmu.2019.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parasuraman S., Thing G.S., Dhanaraj S.A. Polyherbal formulation: concept of ayurveda. Pharmacogn. Rev. 2014;8(16):73. doi: 10.4103/0973-7847.134229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukherjee P.K., Rai S., Kumar V., Mukherjee K., Hylands P., Hider R. Plants of Indian origin in drug discovery. Expert Opin. Drug Discov. 2007;2(5):633–657. doi: 10.1517/17460441.2.5.633. [DOI] [PubMed] [Google Scholar]
- 8.Saber Batiha G. El, Beshbishy A. Magdy, Wasef L.G., Elewa Y.H.A., sagan A.A. Al, Abd El-Hack M.E., Taha A.E., Abd-Elhakim Y.M., Devkota H. Prasad. Chemical constituents and pharmacological activities of garlic (Allium sativum L.): a review. Nutrients. 2020;12:1–21. doi: 10.3390/nu12030872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohajer Shojai T., Ghalyanchi Langeroudi A., Karimi V., Barin A., Sadri N. The effect of Allium sativum (Garlic) extract on infectious bronchitis virus in specific pathogen free embryonic egg. Avicenna J. Phytomed. 2016;6:458–467. [PMC free article] [PubMed] [Google Scholar]
- 10.Shoji S., Furuishi K., Yanase R., Miyazaka T., Kino M. Allyl compounds selectively killed human immunodeficiency virus (type 1)-infected cells. Biochem. Biophys. Res. Commun. 1993;194:610–621. doi: 10.1006/bbrc.1993.1865. [DOI] [PubMed] [Google Scholar]
- 11.Hall A., Troupin A., Londono-Renteria B., Colpitts T.M. Garlic organosulfur compounds reduce inflammation and oxidative stress during dengue virus infection. Viruses. 2017;9:1–10. doi: 10.3390/v9070159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bayan L., Koulivand P.H., Gorji A. Garlic: a review of potential therapeutic effects. Avicenna J. Phytomed. 2014;4:1–14. [PMC free article] [PubMed] [Google Scholar]
- 13.Papu S., Jaivir S., Sweta S., Singh B. Medicinal values of garlic (Allium sativum L.) in human life: an overview. Greener J. Agric. Sci. 2014;4:265–280. [Google Scholar]
- 14.Teshika J.D., Zakariyyah A.M., Zaynab T., Zengin G., Rengasamy K.R., Pandian S.K., Fawzi M.M. Traditional and modern uses of onion bulb (Allium cepa L.): a systematic review. Crit. Rev. Food Sci. Nutr. 2019;59(Suppl. 1):S39–S70. doi: 10.1080/10408398.2018.1499074. [DOI] [PubMed] [Google Scholar]
- 15.Kumar K.M.P., Asish G.R., Sabu M., Balachandran I. Significance of gingers (Zingiberaceae) in Indian system of medicine-Ayurveda: an overview. Anc. Sci. Life. 2013;32:253–261. doi: 10.4103/0257-7941.131989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhowmik D., Tripathi K., Chandira M., Kumar K. Zingiber officinale the herbal and traditional medicine and its therapeutically importance. Res. J. Pharmacogn. Phytochem. 2010;2:102–110. [Google Scholar]
- 17.Bhowmik D., Kumar K.S., Yadav A., Srivastava S., Paswan S., Dutta A.S. Recent trends in Indian traditional herbs Syzygium aromaticum and its health benefits. J. Pharmacogn. Phytochem. 2012;1:13–23. [Google Scholar]
- 18.Loolaie M., Moasefi N., Rasouli H., Adibi H. Peppermint and its functionality: a review. Arch. Clin. Microbiol. 2017;8:1–16. [Google Scholar]
- 19.Herrmann E.C., Jr, Kucera L.S. Antiviral substances in plants of the mint family (Labiatae). III. Peppermint (Mentha piperita) and other mint plants. Proc. Soc. Exp. Biol. Med. 1967;124:874–878. doi: 10.3181/00379727-124-31874. [DOI] [PubMed] [Google Scholar]
- 20.Badgujar S.B., Patel V.V., Bandivdekar A.H. Foeniculum vulgare Mill: a review of its botany, phytochemistry, pharmacology, contemporary application, and toxicology. BioMed. Res. Int. 2014;2014:1–32. doi: 10.1155/2014/842674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jadhav P., Kapoor N., Thomas B., Lal H., Kshirsagar N. Antiviral potential of selected Indian medicinal (ayurvedic) plants against herpes simplex virus 1 and 2. N. Am. J. Med. Sci. 2012;4:641–647. doi: 10.4103/1947-2714.104316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blank D.E., de Oliveira Hübner S., Alves G.H., Cardoso C.A.L., Freitag R.A., Cleff M.B. Chemical composition and antiviral effect of extracts of Origanum vulgare. Adv. Biosci. Biotechnol. 2019;10:188–196. [Google Scholar]
- 23.Leyva-López N., Gutiérrez-Grijalva E.P., Vazquez-Olivo G., Heredia J.B. Essential oils of oregano: biological activity beyond their antimicrobial properties. Molecules. 2017;22:1–14. doi: 10.3390/molecules22060989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rawat N., Roushan R. Guduchi; A Potential Drug Ayurveda. 2018. pp. 355–361. [Google Scholar]
- 25.Saha S., Ghosh S. Tinospora cordifolia: one plant, many roles. Anc. Sci. Life. 2012;31:151–159. doi: 10.4103/0257-7941.107344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vangalapati M., Satya S., Prakash S., Avanigadda S., Satya S., Prakash S., Kiran Sree N., Surya D. A review on pharmacological activities and clinical effects of cinnamon species. Res. J. Pharm. Biol. Chem. Sci. 2012;3:653–663. [Google Scholar]
- 27.Lee H.H., Park H., Sung G.H., Lee K., Lee T., Lee I., Park M.S., Jung Y.W., Shin Y.S., Kang H. Anti-influenza effect of Cordyceps militaris through immunomodulation in a DBA/2 mouse model. J. Microbiol. 2014;52:696–701. doi: 10.1007/s12275-014-4300-0. [DOI] [PubMed] [Google Scholar]
- 28.Dong L., Hu S., Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov. Ther. 2020;14:58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 29.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith M., Smith J.C. Repurposing therapeutics for covid-19: Supercomputer-based docking to the sars-cov-2 viral spike protein and viral spike protein-human ace2 interface. ChemRxiv. 2020:1–15. [Google Scholar]
- 31.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q., Qiu Y., Li J.Y., Zhou Z.J., Liao C.H., Ge X.Y. A unique protease cleavage site predicted in the spike protein of the novel pneumonia coronavirus (2019-nCoV) potentially related to viral transmissibility. Virol. Sin. 2020;35:337–339. doi: 10.1007/s12250-020-00212-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arbab A.H., Parvez M.K., Al-Dosari M.S., Al-Rehaily A.J. In vitro evaluation of novel antiviral activities of 60 medicinal plants extracts against hepatitis B virus. Exp. Ther. Med. 2017;14:626–634. doi: 10.3892/etm.2017.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frederico E., Cardoso A., Moreira-Marconi E., de Sa-Caputo D.D.C., Guimaraes C.A.S., Dionello C.D.F., Morel D.S., Paineiras-Domingos L.L., de Souza P.L., Brandao-Sobrinho-Neto S., Carvalho-Lima R.P., Guedes-Aguiar E.O., Costa-Cavalcanti R.G., Kutter C.R., Bernardo-Filho M. Anti-viral effects of medicinal plants in the management of dengue: a systematic review. Afr. J. Tradit. Complement. Altern. Med. 2017;14:33–40. doi: 10.21010/ajtcam.v14i4S.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liberati A., Altman D.G., Tetzlaff J., Mulrow C., Gøtzsche P.C., Ioannidis J.P., Clarke M., Devereaux P.J., Kleijnen J., Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. Ann. Intern. Med. 2009;151:65–94. doi: 10.7326/0003-4819-151-4-200908180-00136. [DOI] [PubMed] [Google Scholar]
- 36.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A.J.Nar. PubChem substance and compound databases. Nucleic Acids Res. 2016;44:1202–1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wishart D.S., Knox C., Guo A.C., Cheng D., Shrivastava S., Tzur D., Gautam B., Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:901–906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irwin J.J., Shoichet B.K. ZINC: a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gogoi A., Dutta D., Verma A.K., Nath H., Frontera A., Guha A.K., Bhattacharyya M.K. Energetically favorable anti-electrostatic hydrogen bonded cationic clusters in Ni (II) 3, 5-dimethylpyrazole complexes: anticancer evaluation and theoretical studies. Polyhedron. 2019;168:113–126. [Google Scholar]
- 40.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis for receptor recognition by the novel coronavirus from Wuhan. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 42.Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hardes K., Becker G.L., Lu Y., Dahms S.O., Köhler S., Beyer W., Sandvig K., Yamamoto H., Lindberg I., Walz L. Novel furin inhibitors with potent anti‐infectious activity. ChemMedChem. 2015;10:1218–1231. doi: 10.1002/cmdc.201500103. [DOI] [PubMed] [Google Scholar]
- 44.Morse J.S., Lalonde T., Xu S., Liu W. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019–nCoV. ChemBioChem. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xi J., Xu K., Jiang P., Lian J., Hao S., Jia H., Yao H., Zhang Y., Zheng R., Chen D. Virus strain of a mild COVID-19 patient in Hangzhou representing a new trend in SARS-CoV-2 evolution related to Furin cleavage site. Emerg. Microbes Infect. 2020;9:1–74. doi: 10.1080/22221751.2020.1781551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bitencourt-Ferreira G., de Azevedo W.F. Docking Screens for Drug Discovery. Humana; New York, NY: 2019. Molegro virtual docker for docking; pp. 149–167. [DOI] [PubMed] [Google Scholar]
- 47.Kusumaningrum S., Budianto E., Kosela S., Sumaryono W., Juniarti F. The molecular docking of 1, 4-naphthoquinone derivatives as inhibitors of polo-like kinase 1 using Molegro virtual docker. J. Appl. Sci. 2014;4:47–53. [Google Scholar]
- 48.Thomsen R., Christensen M.H. MolDock: a new technique for high-accuracy molecular docking. J. Med. Chem. 2006;49:3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 49.Goddard T.D., Huang C.C., Ferrin T.E. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 50.D.S. Biovia, Discovery Studio Visualizer, Release 2017, Dassault Systèmes, 2016, San Diego, 2020, to be found under 〈http://accelrys.com/products/collaborative-science/biovia-discovery-studio/visualization-download.php〉. (Accessed 12 August 2020).
- 51.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E.J.S. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- 52.Bjelkmar P., Larsson P., Cuendet M.A., Hess B., Lindahl E. Implementation of the CHARMM force field in GROMACS: analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J. Chem. Theory Comput. 2010;6:459–466. doi: 10.1021/ct900549r. [DOI] [PubMed] [Google Scholar]
- 53.Zoete V., Cuendet M.A., Grosdidier A., Michielin O. SwissParam: a fast force field generation tool for small organic molecules. J. Comput. Chem. 2011;32:2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- 54.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:1–7. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 55.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 56.Hess B., Bekker H., Berendsen H.J., Fraaije J.G. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 57.Miller I.I.I., McGee B.R., Jr, Swails T.D., Homeyer N., Gohlke H., Roitberg A.E. MMPBSA. py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 58.Kumari R., Kumar R., Open Source Drug Discovery Consortium, Lynn A. g_mmpbsa A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 59.Kollman P.A., Massova I., Reyes C., Kuhn B., Huo S., Chong L., Lee M., Lee T., Duan Y., Wang W., Donini O. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 60.Gohlke H., Kiel C., Case D.A. Insights into protein–protein binding by binding free energy calculation and free energy decomposition for the Ras–Raf and Ras–RalGDS complexes. J. Mol. Biol. 2003;330:891–913. doi: 10.1016/s0022-2836(03)00610-7. [DOI] [PubMed] [Google Scholar]
- 61.Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins Struct. Funct. Bioinform. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gilson M.K., Honig B. Calculation of the total electrostatic energy of a macromolecular system: solvation energies, binding energies, and conformational analysis. Proteins Struct. Funct. Bioinform. 1988;4:7–18. doi: 10.1002/prot.340040104. [DOI] [PubMed] [Google Scholar]
- 63.Srinivasan J., Cheatham T.E., Cieplak P., Kollman P.A., Case D.A. Continuum solvent studies of the stability of DNA, RNA, and phosphoramidate−DNA helices. J. Am. Chem. Soc. 1998;120:9401–9409. [Google Scholar]
- 64.Wolber G., Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model. 2005;45:160–169. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- 65.Wolber G., Kosara R. Pharmacophores from macromolecular complexes with LigandScout. Methods Princ. Med. Chem. 2006;32:131–150. [Google Scholar]
- 66.Schneidman-Duhovny D., Dror O., Inbar Y., Nussinov R., Wolfson H.J. Deterministic pharmacophore detection via multiple flexible alignment of drug-like molecules. J. Comput. Biol. 2008;15:737–754. doi: 10.1089/cmb.2007.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dror O., Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J. Novel approach for efficient pharmacophore-based virtual screening: method and applications. J. Chem. Inf. Model. 2009;49:2333–2343. doi: 10.1021/ci900263d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin Y.C., Kofron J.L., Traphagen L.M. Do structurally similar molecules have similar biological activity? J. Med. Chem. 2002;45:4350–4358. doi: 10.1021/jm020155c. [DOI] [PubMed] [Google Scholar]
- 69.Clark A.M., Dole K., Coulon-Spektor A., McNutt A., Grass G., Freundlich J.S., Reynolds R.C., Ekins S. Open source Bayesian models. 1. Application to ADME/Tox and drug discovery datasets. J. Chem. Inf. Model. 2015;55:1231–1245. doi: 10.1021/acs.jcim.5b00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meng X.Y., Zhang H.X., Mezei M., Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des. 2011;7:146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94:1–9. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu K., Peng G., Wilken M., Geraghty R.J., Li F. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2012;287:8904–8911. doi: 10.1074/jbc.M111.325803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ortega J.T., Serrano M.L., Pujol F.H., Rangel H.R. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: an in silico analysis. EXCLI J. 2020;19:410–417. doi: 10.17179/excli2020-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pang X.-F., Zhang L.-H., Bai F., Wang N.-P., Garner R.E., McKallip R.J., Zhao Z.-Q. Attenuation of myocardial fibrosis with curcumin is mediated by modulating expression of angiotensin II AT1/AT2 receptors and ACE2 in rats. Drug Des. Dev. Ther. 2015;9:6043. doi: 10.2147/DDDT.S95333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lopez de Cicco R., Bassi D., Page R., Klein-Szanto A.J.P. Furin expression in squamous cell carcinomas of the oral cavity and other sites evaluated by tissue microarray technology. Cancer. 2002;18:29–37. [PubMed] [Google Scholar]
- 77.Kido H., Okumura Y., Takahashi E., Pan H.Y., Wang S., Yao D., Yao M., Chida J., Yano M. Role of host cellular proteases in the pathogenesis of influenza and influenza-induced multiple organ failure. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2012;1824:186–194. doi: 10.1016/j.bbapap.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 78.Abassi Z.A., Skorecki K., Heyman S.N., Kinaneh S., Armaly Z. Covid-19 infection and mortality-a physiologist's perspective enlightening clinical features and plausible interventional strategies. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020;318:1020–1022. doi: 10.1152/ajplung.00097.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Swarup V., Ghosh J., Ghosh S., Saxena A., Basu A. Antiviral and anti-inflammatory effects of rosmarinic acid in an experimental murine model of Japanese encephalitis. Antimicrob. Agents Chemother. 2007;51:3367–3370. doi: 10.1128/AAC.00041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Astani A., Reichling J., Schnitzler P. Screening for antiviral activities of isolated compounds from essential oils. Evid. Based Complement. Altern. Med. 2011;2011:1–8. doi: 10.1093/ecam/nep187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dubois M., Bailly F., Mbemba G., Mouscadet J.-F., Debyser Z., Witvrouw M., Cotelle P. Reaction of rosmarinic acid with nitrite ions in acidic conditions: discovery of nitro-and dinitrorosmarinic acids as new anti-HIV-1 agents. J. Med. Chem. 2008;51:2575–2579. doi: 10.1021/jm7011134. [DOI] [PubMed] [Google Scholar]
- 82.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Castrillo J.L., Carrasco L. Action of 3-methylquercetin on poliovirus RNA replication. J. Virol. 1987;61:3319–3321. doi: 10.1128/jvi.61.10.3319-3321.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zandi K., Teoh B.T., Sam S.S., Wong P.F., Mustafa M.R., Abubakar S. Antiviral activity of four types of bioflavonoid against dengue virus type-2. Virol. J. 2011;8:1–11. doi: 10.1186/1743-422X-8-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Arora R., Chawla R., Marwah R., Arora P., Sharma R.K., Kaushik V., Goel R., Kaur A., Silambarasan M., Tripathi R.P., Bhardwaj J.R. Potential of complementary and alternative medicine in preventive management of novel H1N1 flu (swine flu) pandemic: thwarting potential disasters in the bud. Evid. Based Complement. Altern. Med. 2011;2011:1–16. doi: 10.1155/2011/586506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rasool A., Khan M.U., Ali M.A., Anjum A.A., Ahmed I., Aslam A., Mustafa G., Masood S., Ali M.A., Nawaz M. Anti-avian influenza virus H9N2 activity of aqueous extracts of Zingiber officinalis (Ginger) and Allium sativum (Garlic) in chick embryos. Pak. J. Pharm. Sci. 2017;30:1341–1344. [PubMed] [Google Scholar]
- 88.San Chang J., Wang K.C., Yeh C.F., Shieh D.E., Chiang L.C. Fresh ginger (Zingiber officinale) has anti-viral activity against human respiratory syncytial virus in human respiratory tract cell lines. J. Ethnopharmacol. 2013;145:146–151. doi: 10.1016/j.jep.2012.10.043. [DOI] [PubMed] [Google Scholar]
- 89.Tragoolpua Y., Jatisatienr A. Anti‐herpes simplex virus activities of Eugenia caryophyllus (Spreng.) Bullock and SG Harrison and essential oil, eugenol. Phytother. Res. Int. J. Devoted Pharmacol. Toxicol. Eval. Nat. Prod. Deriv. 2007;21:1153–1158. doi: 10.1002/ptr.2226. [DOI] [PubMed] [Google Scholar]
- 90.Chaieb K., Hajlaoui H., Zmantar T., Kahla‐Nakbi A.B., Rouabhia M., Mahdouani K., Bakhrouf A. The chemical composition and biological activity of clove essential oil, Eugenia caryophyllata (Syzigium aromaticum L. Myrtaceae): a short review. Phytother. Res. Int. J. Devoted Pharmacol. Toxicol. Eval. Nat. Prod. Deriv. 2007;21:501–506. doi: 10.1002/ptr.2124. [DOI] [PubMed] [Google Scholar]
- 91.Pramod K., Ansari S.H., Ali J. Eugenol: a natural compound with versatile pharmacological actions. Nat. Prod. Commun. 2010;5:1999–2006. [PubMed] [Google Scholar]
- 92.Li Y., Liu Y., Ma A., Bao Y., Wang M., Sun Z. In vitro antiviral, anti-inflammatory, and antioxidant activities of the ethanol extract of Mentha piperita L. Food Sci. Biotechnol. 2017;26:1675–1683. doi: 10.1007/s10068-017-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee H.S., Kang P., Kim K.Y., Seol G.H. Foeniculum vulgare mill. protects against lipopolysaccharide-induced acute lung injury in mice through ERK-dependent NF-κB activation. Korean J. Physiol. Pharmacol. 2015;19:183–189. doi: 10.4196/kjpp.2015.19.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chiang L.C., Ng L.T., Cheng P.W., Chiang W., Lin C.C. Antiviral activities of extracts and selected pure constituents of Ocimum basilicum. Clin. Exp. Pharmacol. Physiol. 2005;32:811–816. doi: 10.1111/j.1440-1681.2005.04270.x. [DOI] [PubMed] [Google Scholar]
- 95.Mondal S., Varma S., Bamola V.D., Naik S.N., Mirdha B.R., Padhi M.M., Mehta N., Mahapatra S.C. Double-blinded randomized controlled trial for immunomodulatory effects of Tulsi (Ocimum sanctum Linn.) leaf extract on healthy volunteers. J. Ethnopharmacol. 2011;136:452–456. doi: 10.1016/j.jep.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 96.Gilling D.H., Kitajima M., Torrey J.R., Bright K.R. Antiviral efficacy and mechanisms of action of oregano essential oil and its primary component carvacrol against murine norovirus. J. Appl. Microbiol. 2014;116:1149–1163. doi: 10.1111/jam.12453. [DOI] [PubMed] [Google Scholar]
- 97.Pilau M.R., Alves S.H., Weiblen R., Arenhart S., Cueto A.P., Lovato L.T. Antiviral activity of the Lippia graveolens (Mexican oregano) essential oil and its main compound carvacrol against human and animal viruses. Braz. J. Microbiol. 2011;42:1616–1624. doi: 10.1590/S1517-838220110004000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sharifi-Rad J., Salehi B., Schnitzler P., Ayatollahi S., Kobarfard F., Fathi M., Eisazadeh M., Sharifi-Rad M. Susceptibility of herpes simplex virus type 1 to monoterpenes thymol, carvacrol, p-cymene and essential oils of Sinapis arvensis L., Lallemantia roy leana Benth. and Pulicaria vulgaris Gaertn. Cell Mol. Biol. 2017;63:42–47. doi: 10.14715/cmb/2017.63.8.10. (Noisy le Grand) [DOI] [PubMed] [Google Scholar]
- 99.Li H., Zhong C., Wang Q., Chen W., Yuan Y. Curcumin is an APE1 redox inhibitor and exhibits an antiviral activity against KSHV replication and pathogenesis. Antivir. Res. 2019;167:98–103. doi: 10.1016/j.antiviral.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Richart S.M., Li Y.L., Mizushina Y., Chang Y.Y., Chung T.Y., Chen G.H., Tzen J.T., Shia K.S., Hsu W.L. Synergic effect of curcumin and its structural analogue (Monoacetylcurcumin) on anti-influenza virus infection. J. Food Drug Anal. 2018;26:1015–1023. doi: 10.1016/j.jfda.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zorofchian Moghadamtousi S., Abdul Kadir H., Hassandarvish P., Tajik H., Abubakar S., Zandi K. A review on antibacterial, antiviral, and antifungal activity of curcumin. BioMed. Res. Int. 2014;2014:1–12. doi: 10.1155/2014/186864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Akhtar S. Use of Tinospora cordifolia in HIV infection. Indian J. Pharmacol. 2010;42:57. doi: 10.4103/0253-7613.62402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kalikar M.V., Thawani V.R., Varadpande U.K., Sontakke S.D., Singh R.P., Khiyani R.K. Immunomodulatory effect of Tinospora cordifolia extract in human immuno-deficiency virus positive patients. Indian J. Pharmacol. 2008;40:107–110. doi: 10.4103/0253-7613.42302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Singh D., Chaudhuri P.K. Chemistry and pharmacology of Tinospora cordifolia. Nat. Prod. Commun. 2017;12:299–308. [PubMed] [Google Scholar]
- 105.Brochot A., Guilbot A., Haddioui L., Roques C. Antibacterial, antifungal, and antiviral effects of three essential oil blends. MicrobiologyOpen. 2017;6:1–6. doi: 10.1002/mbo3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Goswami A., Rahman A. Antiviral activity of (E)-cinnamaldehyde revisited with nanoscience tools. Nat. Preced. 2010;1:1–4. [Google Scholar]
- 107.Mbaveng A.T., Kuete V. In: Medicinal Spices and Vegetables from Africa: Therapeutic Potential Against Metabolic, Inflammatory, Infectious and Systemic Diseases. Kuete V., editor. Academic Press; Massachusetts, United States: 2017. Cinnamon species; pp. 385–395. [Google Scholar]
- 108.Ryu E., Son M., Lee M., Lee K., Cho J.Y., Cho S., Lee S.K., Lee Y.M., Cho H., Sung G.-H.J.O. Cordycepin is a novel chemical suppressor of Epstein-Barr virus replication. Oncoscience. 2014;1:866–881. doi: 10.18632/oncoscience.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Montefiori D.C., Sobol R.W., Li S.W., Reichenbach N.L., Suhadolnik R.J., Charubala R., Pfleiderer W., Modliszewski A., Robinson W.E., Mitchell W.M. Phosphorothioate and cordycepin analogues of 2′, 5′-oligoadenylate: inhibition of human immunodeficiency virus type 1 reverse transcriptase and infection in vitro. Proc. Natl. Acad. Sci. 1989;86:7191–7194. doi: 10.1073/pnas.86.18.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tuli H.S., Sandhu S.S., Sharma A.J.B. Pharmacological and therapeutic potential of Cordyceps with special reference to Cordycepin. 3 Biotech. 2014;4:1–12. doi: 10.1007/s13205-013-0121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:1–9. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang L., Zhou R. Structural basis of potential binding mechanism of remdesivir to SARS-CoV-2 RNA dependent RNA polymerase. J. Phys. Chem. B. 2020;124:6955–6962. doi: 10.1021/acs.jpcb.0c04198. [DOI] [PubMed] [Google Scholar]
- 113.Ahmad J., Ikram S., Ahmad F., Rehman I.U., Mushtaq M. SARS-CoV-2 RNA dependent RNA polymerase (RdRp)–a drug repurposing study. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e04502. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chikhale R.V., Sinha S.K., Patil R.B., Prasad S.K., Shakya A., Gurav N., Prasad R., Dhaswadikar S.R., Wanjari M., Gurav S.S. In-silico investigation of phytochemicals from Asparagus racemosus as plausible antiviral agent in COVID-19. J. Biomol. Struct. Dyn. 2020:1–15. doi: 10.1080/07391102.2020.1784289. [DOI] [PubMed] [Google Scholar]
- 115.Sang P., Tian S.H., Meng Z.H., Yang L.Q. Anti-HIV drug repurposing against SARS-CoV-2. RSC Adv. 2020;10:15775–15783. doi: 10.1039/d0ra01899f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hollingsworth S.A., Dror R.O. Molecular dynamics simulation for all. Neuron. 2018;99:1129–1143. doi: 10.1016/j.neuron.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Choudhary S., Malik Y.S., Tomar S. Identification of SARS-CoV-2 cell entry inhibitors by drug repurposing using in silico structure-based virtual screening approach. Front. Immunol. 2020;11:1–14. doi: 10.3389/fimmu.2020.01664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Becker G.L., Sielaff F., Than M.E., Lindberg I., Routhier S., Day R., Lu Y., Garten W., Steinmetzer T.J.J.o.m.c. Potent inhibitors of furin and furin-like proprotein convertases containing decarboxylated P1 arginine mimetics. J. Med. Chem. 2010;53:1067–1075. doi: 10.1021/jm9012455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C.J.N. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 120.Joshi T., Sharma P., Joshi T., Pundir H., Mathpal S., Chandra S. Structure-based screening of novel lichen compounds for SARS coronavirus main protease (Mpro) and angiotensin-converting enzyme 2 (ACE2) inhibitory potentials as multi-target inhibitors of COVID-19. Mol. Divers. 2020:1–13. doi: 10.1007/s11030-020-10118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang S.Y. Pharmacophore modeling and applications in drug discovery: challenges and recent advances. Drug Discov. Today. 2010;15:444–450. doi: 10.1016/j.drudis.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 122.Lewis R.A., Meng E.C. In: Molecular Modelling and Drug Design. Topics in Molecular and Structural Biology. Vinter J.G., Gardner M., editors. Palgrave; London: 1994. A discussion of various computational methods for drug design; pp. 170–210. [Google Scholar]
- 123.Schneidman-Duhovny D., Dror O., Inbar Y., Nussinov R., Wolfson H.J. PharmaGist: a webserver for ligand-based pharmacophore detection. Nucleic Acids Res. 2008;36:223–228. doi: 10.1093/nar/gkn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Laksmiani N.P.L., Larasanty L.P.F., Santika A.A.G.J., Prayoga P.A.A., Dewi A.A.I.K., Dewi N.P.A.K. Active compounds activity from the medicinal plants against SARS-CoV-2 using in silico assay. Biomed. Pharmacol. J. 2020;13:873–881. [Google Scholar]
- 125.Vishvakarma N.K., Kumar A., Singh S.M. Role of curcumin-dependent modulation of tumor microenvironment of a murine T cell lymphoma in altered regulation of tumor cell survival. Toxicol. Appl. Pharmacol. 2011;252:298–306. doi: 10.1016/j.taap.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 126.Zahedipour F., Hosseini S.A., Sathyapalan T., Majeed M., Jamialahmadi T., Al‐Rasadi K., Banach M., Sahebkar A. Potential effects of curcumin in the treatment of COVID‐19 infection. Phytother. Res. 2020:1–10. doi: 10.1002/ptr.6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Praditya D., Kirchhoff L., Brüning J., Rachmawati H., Steinmann J., Steinmann E. Anti-infective properties of the golden spice curcumin. Front. Microbiol. 2019;10:1–16. doi: 10.3389/fmicb.2019.00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wen C.C., Kuo Y.H., Jan J.T., Liang P.H., Wang S.Y., Liu H.G., Lee C.K., Chang S.T., Kuo C.J., Lee S.S., Hou C.C. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J. Med. Chem. 2007;50:4087–4095. doi: 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- 129.Oso B.J., Adeoye A.O., Olaoye I.F. Pharmacoinformatics and hypothetical studies on allicin, curcumin, and gingerol as potential candidates against COVID-19-associated proteases. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1813630. [DOI] [PubMed] [Google Scholar]
- 130.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu B. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Soni V.K., Mehta A., Ratre Y.K., Tiwari A.K., Amit A., Singh R.P., Sonkar S.C., Chaturvedi N., Shukla D., Vishvakarma N.K. Curcumin, a traditional spice component, can hold the promise against COVID-19? Eur. J. Pharmacol. 2020;886:1–10. doi: 10.1016/j.ejphar.2020.173551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen L., Hu C., Hood M., Zhang X., Zhang L., Kan J., Du J., Novel A. Combination of vitamin C, curcumin and glycyrrhizic acid potentially regulates immune and inflammatory response associated with coronavirus infections: a perspective from system biology analysis. Nutrients. 2020;12:1–17. doi: 10.3390/nu12041193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kandeel M., Al-Nazawi M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci. 2020;251:1–5. doi: 10.1016/j.lfs.2020.117627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Maurya V.K., Kumar S., Prasad A.K., Bhatt M.L., Saxena S.K. Structure-based drug designing for potential antiviral activity of selected natural products from Ayurveda against SARS-CoV-2 spike glycoprotein and its cellular receptor. Virus Dis. 2020;31:1–15. doi: 10.1007/s13337-020-00598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Barnard D.L., Kumaki Y. Recent developments in anti-severe acute respiratory syndrome coronavirus chemotherapy. Future Virol. 2011;6:615–631. doi: 10.2217/fvl.11.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Colunga Biancatelli R.M.L., Berrill M., Catravas J.D., Marik P.E. Quercetin and vitamin C: an experimental, synergistic therapy for the prevention and treatment of SARS-CoV-2 related disease (COVID-19) Front. Immunol. 2020;11:1–11. doi: 10.3389/fimmu.2020.01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., de Castilla D. Lopez, Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.D., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. Remdesivir for the treatment of Covid-19-final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Choy K.T., Wong A.Y., Kaewpreedee P., Sia S.F., Chen D., Hui K.P., Chu D.K., Chan M.C., Cheung P.P., Huang X., Peiris M. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020;178:1–5. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Abdolvahab M.H., Moradi-Kalbolandi S., Zarei M., Bose D., Majidzadeh A.K., Farahmand L. Potential role of interferons in treating COVID-19 patients. Int. Immunopharmacol. 2021;90:1–15. doi: 10.1016/j.intimp.2020.107171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li X., Wang Y., Agostinis P., Rabson A., Melino G., Carafoli E., Shi Y., Sun E. Is hydroxychloroquine beneficial for COVID-19 patients? Cell Death Dis. 2020;11:1–6. doi: 10.1038/s41419-020-2721-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Owa A.B., Owa O.T. Lopinavir/ritonavir use in Covid-19 infection: is it completely non-beneficial? J. Microbiol. Immunol. Infect. 2020;53:674–675. doi: 10.1016/j.jmii.2020.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material