Graphical abstract
Keywords: Photocatalyst, TiO2, Inactivate, Pathogenic viruses
Abstract
Previous observations have been reported that viruses were inactivated using strong irradiation. Here, new evidence was disclosed by studying the effects of nanosized TiO2 on viral pathogens under a low irradiation condition (0.4 mW/cm2 at UVA band) that mimics the field setting. We showed that photo-activated TiO2 efficiently inhibits hepatitis C virus infection, and weak indoor light with intensity of 0.6 mW/cm2 at broad-spectrum wavelength and around 0.15 mW/cm2 of UVA band also lead to partial inhibition. Mechanistic studies demonstrated that hydroxyl radicals produced by photo-activated TiO2 do not destroy virion structure and contents, but attack viral RNA genome, thus inactivating the virus. Furthermore, we showed that photo-activated TiO2 inactivates a broad range of human viral pathogens, including SARS-CoV-2, a novel coronavirus responsible for the ongoing COVID-19 pandemic. In conclusion, we showed that photo-catalyzed nanosized TiO2 inactivates pathogenic viruses, paving a way to its field application in control of viral infectious diseases.
1. Introduction
In recent years, the outbreak of several viral infectious diseases have not only threatened human being's health but also caused huge economic losses [1], [2], [3]. As of February 1st of 2021, the ongoing COVID-19 pandemic caused by a novel coronavirus SARS-CoV-2 has infected over 100 million people and killed more than 2,000,000 people in the world (http://coronavirusstatistics.org/). Effective and environmentally-friendly strategies to disinfect viruses of board-spectrum in a field setting are urgently needed to mitigate transmission. The photocatalytic oxidation (PCO) technology using long-wave ultraviolet (315–380 nm) or even visible light spectrum (380–740 nm) renders a safe and durable technique in pathogen inactivation [4], [5]. Photocatalysts exhibit superior light-harvesting capability to create electron-hole pairs and generate reactive oxygen species (ROSs), such as superoxide radical (•O2 −) and hydroxyl radicals (•OH) that are known to be harmful for microbes [6]. Most of these PCO studies focused on the inactivation of bacteria (Escherichia coli and Staphylococcus aureus) [7] and bacteriophages [8]. It was found that the light-induced ROSs disrupt bacterial cell membrane, causing leakage and damage of intracellular bacterial proteins and DNA [9]. Other works demonstrated the photocatalytic virucidal effects on MS2 coliphage by material modification or combining photo-sensitized hydrophilic fullerenes[10], [11], [12]. Similar to the mechanism of bacterial inactivation, •O2 − radicals produced in photocatalysis can rupture the capsid shell of MS2 phage, resulting in the leakage and rapid destruction of capsid proteins and RNA[13].
Most viruses are vulnerable in the acellular environment. Their infectivity, defined as an ability to infect host cells, can be easily lost when viral proteins or genomes are subjected to minor damages or modifications. Unlike bacteria and bacteriophages, the effects of PCO on animal viruses have been less extensively studied, and most of the studies were conducted using strong irradiation. C. Zhang et al. reported that waterborne viruses were completely inactivated after 240 min exposure to illumination at light density of 199.80 mW/cm2 [14]. The commercially available THERAFLEX UV-Platelets system using ultraviolet illumination (wavelength at 254 nm) has been shown to efficiently inactivate hepatitis C virus (HCV)[15] and Zika virus (ZIKV) [16]. Although these strong illumination photocatalytic treatments effectively inactivate viruses, their application in the field setting remains challenging. Some previous studies observed the significant loss of viral proteins and nucleic acids upon photocatalytic inactivation[17], [18]. However, it remains elusive whether this degradation of viral components result directly or indirectly from the ROS damage.
Here, the effects of nanosized TiO2 on pathogens under a low irradiation condition (0.4 mW/cm2 at wavelength of 375 nm) were studied, a condition that more resembles natural light source and has an advantage in the real-world field application. We showed that photo-activated TiO2 efficiently inactivates a broad range of pathogens, including SARS-CoV-2, a novel coronavirus that is responsible for ongoing COVID-19 pandemic. Mechanistic studies demonstrated that photo-induced hydroxyl radicals rather than superoxide radicals play a major role in virus inactivation. Importantly, in contrast to the previous finding, we found that photo-induced radicals do not destroy the virion global structure and contents, but instead attack viral genome, leading to inactivation of the virus. In conclusion, our study demonstrated that nanosized TiO2 is an effective and safe photocatalyst to disinfect pathogenic viruses. Our results paved a way to a field application of this photocatalyst in control of viral infectious diseases.
2. Methods
2.1. Cells and viruses
The hepatoma cell line (Huh7.5.1, Huh7), Vero E6, and Madin-Darby canine kidney (MDCK) cells were maintained in complete Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum, 10 mM HEPES buffer, 100 U/ml penicillin, and 100 mg/ml streptomycin as described previously [19]. HCV (JFH1 strain), H1N1 influenza virus (A/PR/8 strain), EV71, ZIKV, VSV and HSV-1were amplified and the titers were determined as described previously [19], [20], [21], [22], [23], [24]. Viruses are purified by sucrose density-gradient ultracentrifugation as described previously [19]. Briefly, virus was centrifuged at 4000 rpm for 5 min to remove cellular debris and then pelleted through a 20% sucrose cushion at 25,000 rpm for 3 h by using an P28 rotor in an ultracentrifuge (Hitachi). Then the pellet was resuspended in 1 ml of PBS buffer and stored in Thermo Scientific Revco ULT. The experiments involving other viruses or bacteria were performed in BSL-2 facility at Institut Pasteur of Shanghai following the regulations.
The SARS-CoV-2 strain nCoV-SH01 (GenBank accession no. MT121215) was isolated from a COVID-19 patient and propagated in Vero E6 cells for use. The experiments about SARS-CoV-2 were performed in the biosafety level 3 (BSL-3) facility of Fudan University following the regulations [25], [26].
2.2. Cell viability and cytotoxicity assays
Cell viability and cytotoxicity are detected by CellTiter-Glo® Luminescent Cell Viability Assay reagent (Promega GS7570, USA). Briefly, cells were seeded at 15,000 cells per well in 96-well microplates and allowed to attach and grow overnight. The next day, the cell-culture mediums with treated with light/TiO2 or mock were added to the cells. After 48 h of incubation, cells were collected and lysed for the cell cytotoxicity assay.
2.3. Preparation of TiO2 nanoparticles
A typical hydrothermal process was used to synthesize the TiO2 nanoparticles, 6.0 g of tetrabutyl titanate (C16H36O4Ti, AR, 98%, Sinapharm Chemical Reagent Co., Ltd) was mixed with 500 ml ethanol (C2H6O, AR, 99.7%, Zhenxing No.1 Chemical Plant). With continuously stirring, 5 ml deionized water was slowly dropped into the solution. After being stirred for 2 h, the white sediment in the solution was obtained by centrifuge and then transferred to a 200 ml hydrothermal reactor with 150 ml deionized water and reacted for 8 h at 180 °C to get crystallized TiO2 powders. The resultant powder was washed 4 times by deionized water and ethanol and then annealed at 500 °C to remove any organics. The characterization data was summarized in Fig S2.
2.4. The illumination device and the experiment conditions
To treat the virus with as-prepared photocatalytic material (TiO2 NPS), an illumination device has been manufactured. This device consists of two 3 W LED chips (wavelength at 375 nm; Epileds Technologies, China), a direct current power supply, and a cooling system. To make the coating of the photocatalytic material, TiO2 NPs were dispersed in absolute ethanol and sonicated for 20 min. After that, 200 μl solution was taken by Eppendorf and drop to every well of a well plate and transfer to a dryer for 6 h at 50 °C. The distance between LED and well plate was adjustable. To develop an accurate and sensitive experimental system, we optimized the experimental temperature using ice bath combined with LED radiator.
To performed the inactivation assay of SARS-CoV-2 in BSL-3 lab, we designed and produced a mini-sized experimental equipment, which is integrated LED light source consists of two 3 W LED chips (wavelength at 375 nm; Epileds Technologies, China) and high capacity battery (10000mAh, Max output = 5 V, 2A). This unit was assembled outside of BSL-3, and then brought into the biosafety cabinet in the BSL-3 lab.
2.5. DMPO spin-trapping experiment
Electron spin-resonance spectroscopy (ESR, JEOL-FA200, Japan) was utilized to study the generation mechanism of hydroxyl radical (•OH) and superoxide radical (•O2 −). In this experiment 50 mg as-prepared powder was dispersed in a 40 nM 5,5-dimethyl-1-pyrroline-N-oxide DMPO solution tank.
2.6. Viral RNA quantification
HCV RNA levels were determined by quantitative reverse transcription-qPCR (RT-qPCR) as described [19], [27]. For H1N1 and EV71, as mentioned previously [20], [21], [27], [28], the RNA levels were analyzed by RT-qPCR with a pair of H1N1 and EV71-specific primers, respectively [20], [21].
2.7. Indirect immunofluorescence
Intracellular immunostaining of HCV- or ZIKV-infected cells was performed as described previously [23], [27]. Briefly, the virus-infected cells were fixed, and stained with a mouse monoclonal anti-HCV NS5A antibody or a rabbit polyclonal anti-ZIKV NS3 antibody. Bound primary antibodies were detected by using Alexa Fluor 555-conjugated secondary antibodies (Molecular Probes, Eugene, OR). Nuclei were stained with Hoechst dye.
2.8. Western blot analysis
Western blot analysis was performed as described previously [27]. Briefly, HCV viral proteins were detected by a mouse monoclonal anti-E2, anti-core protein and anti-HIV-P24 proteins antibodies, respectively. The HRP-conjugated goat anti-mouse secondary antibodies (Promega, Madison, WI) were used. Proteins were visualized by the ECL kit (Millipore).
2.9. Pseudotyped virus infection
HCV pseudotyped particles (HCVpp) were generated as previously described [27], [29]. Briefly, HEK293T cells were co-transfected with plasmids expressing HCV Ε1, Ε2 glycoproteins, retroviral Core packaging component, and luciferase. The medium was refreshed at 6-hour post-transfection. Supernatants were collected 72 h later and filtered through 0.45 μm-pore membranes. For the infection experiment, 8000 Huh7.5.1 cells seeded in 96-well plates and then infected with HCVpp. Three days after infection, the firefly luciferase activity was measured using the Luciferase Assay System following the manufacturer’s instructions (Promega).
2.10. Plaque assay
For HSV-1 and VSV, the viral titer was determined by the plaque assay as described previously [29]. Briefly, approximately 1.5 × 105 Vero E6 or Huh7 cells were seeded into each well of 24-well plates and maintained in the complete growth medium. Before infection, the cell-culture mediums were removed. After adsorption for 1 h, the inoculums were removed, and the cells were overlaid with 1000 µl plaque medium (containing 2% carboxymethylcellulose, CMC, and 2% FBS). After 48 h of incubation, cells were fixed with the fixed buffer as described previously.
For SARS-CoV-2, approximately 1.0 × 104 ffu viruses were treated in the illumination device for 30 min. And then each treated virus was added into each well. After adsorption for 1 h, the inoculums were removed, and the cells were overlaid with 1000 µl plaque medium. After 48 h of incubation, cells were fixed with the fixed buffer. After staining with1% Crystal Violet, the numbers of plaques were counted.
2.11. Statistical analysis
Results are presented as the means ± SD. Data was analyzed using a two-tailed unpaired Student’s t-test. Differences were considered statistically not significant (ns) when the P value was >0.05, and significant when the P value was <0.05 which can be further defined as **, P<0.01; *, P<0.05; NS, P>0.05.
2.12. Data availability
The data that support the findings of this study are available from the corresponding authors upon request.
3. Results
3.1. Photo-activated TiO2 efficiently inhibits HCV infection
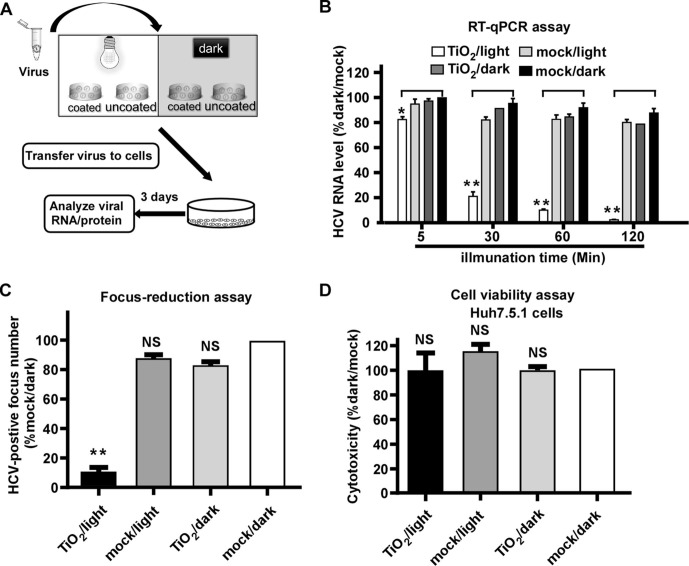
The inactivation assay was carried out in an illumination platform, which comprises an LED light source (wavelength at 375 nm) with light intensity of 0.4 mW/cm2 measured by an illuminometer, and a loading platform sitting in an ice-bath box (Fig. 1 A). First, we tested the effect of photo-activated nanosized TiO2 on HCV, an RNA viral pathogen that has infected 170 million people in the world and is responsible for chronic hepatitis, liver cirrhosis and hepatocellular carcinoma [30]. 1000 focus-forming units (ffu) of HCV (JFH1 strain) [19], [31], [32] was added into nanosized TiO2-coated or uncoated (mock control) 24-well plates, and was light illuminated or kept in dark for 5, 30, 60 and 120 min respectively. After the illumination, HCV was inoculated on Huh7.5.1 cells, a hepatoma-derived cell line that is permissive for HCV infection [19]. At day 3 post-infection, the intracellular HCV RNA levels were determined by RT-qPCR. As shown in Fig. 1B, while the intracellular HCV RNA levels remained unchanged in the group of TiO2/dark, mock/light or mock/dark, they decreased progressively in the TiO2/light group as the illumination time increased. Of note, the 60-min illumination inhibited about 90% HCV infection, therefore, this illumination condition was used for the following experiments.
Fig. 1.
Photo-activated TiO2 efficiently inhibits HCV infection. (A) The schematic of the experiment to test the effect of photo-activated TiO2 on HCV infection. (B) 1000 ffu of HCV (JFH1 strain) was treated with TiO2 or mock in the presence or absence of light illumination for 5, 30, 60 and 120 min respectively, and then inoculated to Huh7.5.1 cells for 3 days. The intracellular HCV RNA levels were determined by RT-qPCR, normalized against cellular GAPDH levels and expressed as the percentage of the mock/dark group. (C) Infectivity titers were determined by immunofluorescence of HCV NS3 proteins after TiO2/light treatment for 60 min. (D) Cytotoxicity of TiO2/light treatment on Huh7.5.1 cells was evaluated under the incubation with the treated medium for 48 h by cell titer assay. The error bars in panels B-D were derived from triplicates. Means and standard deviations for three independent experiments. **, P<0.01; *, P<0.05; NS, P>0.05.
Next we examined the effect of illuminated TiO2 on the HCV infection using a focus-reduction assay [33]. 200 ffu of HCV was subjected to TiO2 or mock with or without illumination for 60 min, and then inoculated on Huh7.5.1 cells for 3-days infection. The number of HCV-positive foci was analyzed by immunofluorescence and quantified. Consistently, TiO2/light significantly inhibited HCV infection (Fig. 1C). Then we assessed the potential cytotoxicity of TiO2. The culture medium was incubated in TiO2-coated or mock-coated 24-well plates with illumination or dark for 60 min, and then transferred to Huh7.5.1 cells. The cells were cultured for 3 days and then subjected to the cell viability assay to measure cell growth. The result demonstrated that the cells grew equally in each of the four groups (Fig. 1D), suggesting that the inhibitory effect of TiO2/light on HCV infection was not due to its cytotoxicity.
3.2. Photo-activated TiO2 targets encapsidated viral RNA
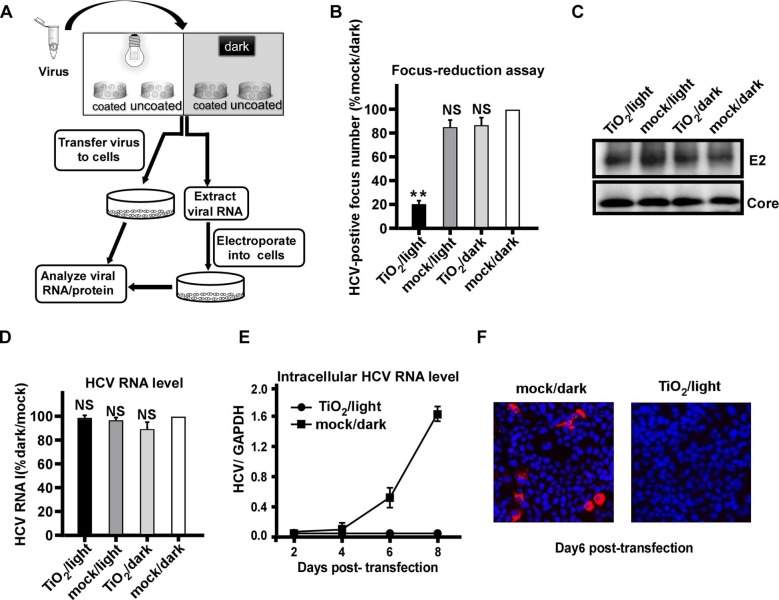
We sought to investigate how illuminated TiO2 inhibits HCV infection. The outline of experiments was shown in Fig. 2 A. After the above described 4 groups of treatment, the quantity of envelope protein E2 and genome RNA of input HCV virions were directly analyzed by Western blot and RT-qPCR respectively. In parallel, after the TiO2/light or mock/dark treatment, the viral RNA of input virions was extracted and then transfected into Huh7.5.1 cells by electroporation, and HCV RNA and protein levels at the different time points after transfection were analyzed by RT-qPCR and immunofluorescence respectively. Consistently, illuminated TiO2 efficiently inhibited HCV infection (Fig. 2B). However, this treatment did not directly reduce the quantity of HCV protein (Fig. 2C) or RNA (Fig. 2D) in the virions, suggesting that illuminated TiO2 do not destroy the virion overall structure and contents. Interestingly, when the viral RNA was extracted from the TiO2/light-treated input virions and then transfected into the cells, it did not leads to viral amplification (Fig. 2E-F), suggesting that although the amount of viral RNA remained unchanged upon the TiO2/light treatment, their biological activity of had been impaired. Altogether, these results suggested that inactivation of HCV virions by illuminated TiO2 was likely due to the damage of the encapsidated viral genome.
Fig. 2.
Photo-activated TiO2 targets encapsidated viral RNA. (A) The schematic of the experiment to test the effect of photo-activated TiO2 on encapsidated viral RNA. (B) HCV (JFH1 strain) was treated with TiO2 or mock in the presence or absence of light illumination for 60 min, and then inoculated to Huh7.5.1 cells for 3 days. The HCV-positive foci were determined by immunofluorescence of HCV NS3 proteins, and expressed as the percentage of the mock/dark group. (C-D) HCV E2 and Core proteins (C) and genome RNA (D) in the input virions after the TiO2/light treatment were directly quantified by Western blotting and RT-qPCR, respectively. The RNA levels were expressed as the percentage of the mock/dark group. (E) The intracellular HCV RNA levels were determined by RT-qPCR on day 2, 4, 6, 8 post-transfection into Huh7.5.1 cells. (F) Immunofluorescence of NS3 proteins (red) in Huh7.5.1 cells transfected with the extracted RNA from TiO2/light treated virions on day 6 post-transfection. Nuclei (blue) were stained with Hoechst dye. The error bars in panels (B), (D) and (E) were derived from triplicates. **, P<0.01; *, P<0.05; NS, P>0.05.
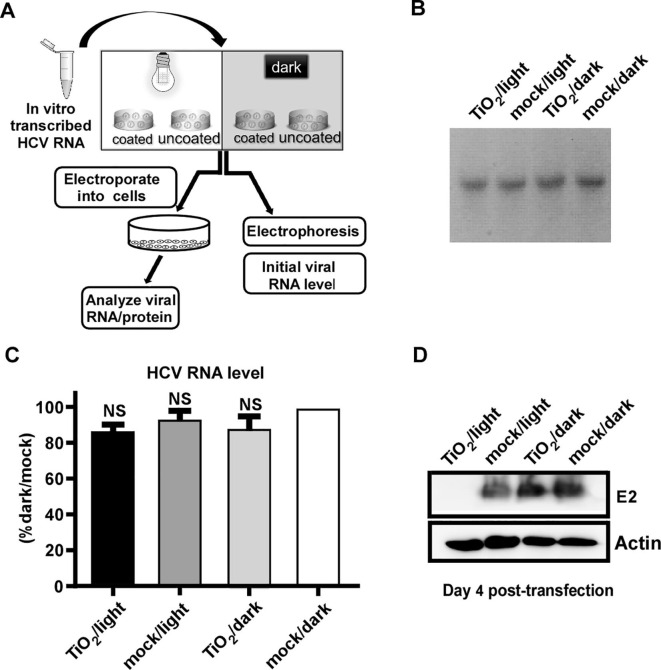
Then, we tested whether the naked HCV RNA can be damaged by the TiO2/light treatment. The in vitro transcribed HCV genomic RNA was subject to the treatment, and then analyzed by gel electrophoresis and RT-qPCR, or transfected into Huh7.5.1 cells by electroporation to analyze the functionality of the RNA (Fig. 3 A). As shown in Fig. 3B-C, the quantity of the naked HCV RNA was not affected by the treatment. However, illuminated TiO2 significantly abolished the ability of the HCV RNA to produce virus in the cells (Fig. 3D).
Fig. 3.
Photo-activated TiO2 inactivates the naked HCV genomic RNA. (A) The schematic of the experiment to test the effect of photo-activated TiO2 on in-vitro-transcribed HCV genomic RNA. (B-C) After the treatment, the viral RNA was analyzed by Electrophoresis (B) and RT-qPCR assay (C). (D) The treated in-vitro-transcribed RNA was electroporated into Huh7.5.1 cells, and viral E2 proteins were analyzed by Western blot analysis at day 4 post-electroporation. The expression level of actin in these transfected cells was detected as control. The error bars in panel C were derived from triplicates. **, P<0.01; *, P<0.05; NS, P>0.05.
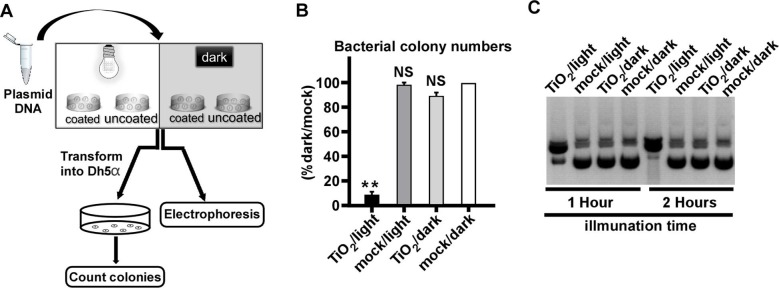
After that, we examined whether the TiO2/light treatment had a similar impact on DNA as on RNA. As shown in the flow chart of the experiment (Fig. 4 A), the plasmid DNA (pUC19-JFH1) that expresses the ampicillin-resistant gene was subjected to the treatment, and then analyzed by gel electrophoresis, or transformed into E. coli (DH5α strain) to analyze its ability to form bacterial colonies upon the ampicillin selection. Transformation of TiO2/light-treated plasmid DNA yielded much fewer bacterial colonies (Fig. 4B). The gel electrophoresis analysis showed that TiO2/light-treated plasmid DNA migrated more slowly (Fig. 4C), likely reflecting a DNA conformational change due to the nick of supercoiled double-stranded plasmid DNA.
Fig. 4.
Photo-activated TiO2 damages bacterial plasmid DNA. (A) The schematic of the experiment to test the effect of photo-activated TiO2 on the plasmid DNA. (B) The DNA plasmid of pUC19 was treated with TiO2/light for 1 h, and then transformed into E.coli (DH5α). The ampicillin-resistant bacterial colony numbers were determined. The error bars in panel C were derived from triplicates. **, P<0.01; *, P<0.05; NS, P>0.05. (C) The TiO2/light-treated plasmid pUC19 for 1 or 2 h were analyzed by electrophoresis.
3.3. Anti-HCV effect of illuminated TiO2 is mediated mainly by the photo-induced hydroxyl radicals
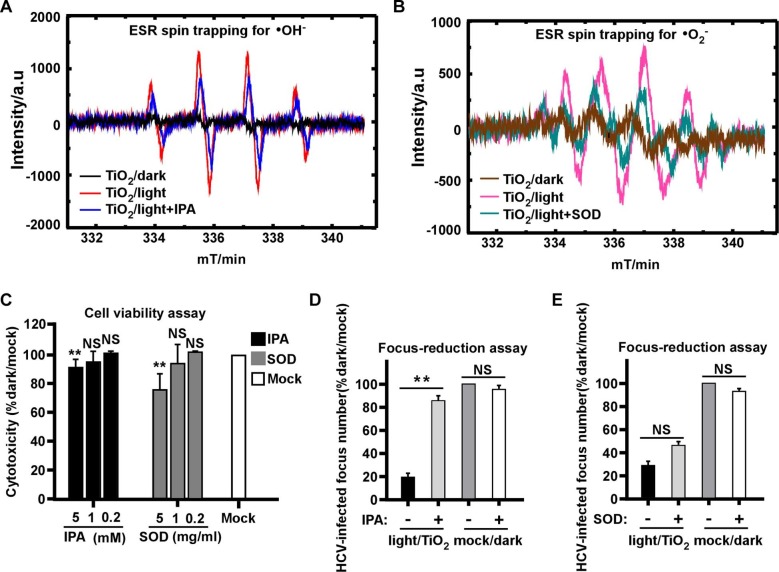
The photocatalytic process involves the production of ROSs, including •OH and •O2 radicals known to harm pathogens [34], [35], thus next we determined the role of different ROSs in the anti-HCV effect of illuminated TiO2. We first analyzed the nature of ROSs produced by TiO2 upon illumination. The production of free radicals was characterized by DMPO spin-trapping ESR measurements. We utilized 1 mmol/L isopropanol (IPA) and Superoxide Dismutase (SOD) as quenching agents for •OH and •O2 radicals, respectively. The results showed both characteristic peaks of •OH and •O2 − radicals reached its highest level with a 10-min illumination dose (0.4 mW/cm2, 375 nm), whereas the peaks of •O2 − and •OH were absent in the dark condition or partially decreased in the presence of the quenching agents (Fig. 5 A-B).
Fig. 5.
The hydroxyl radicals mediate antiviral effect of illuminated TiO2. The radiation intensity was 0.4 mW/cm2 at wavelength of 375 nm for the “light” condition. (A) Hydroxyl radicals (•OH) were detected by DMPO spin trap of TiO2 illuminated for 10 min in the presence or absence of 1 mM isopropanol (IPA) as the quenching agent, or kept in dark. (B) Superoxide radicals (•O2−) were detected by DMPO spin trap of TiO2 illuminated for 10 min in the presence or absence of 1 mg/ml Superoxide Dismutase (SOD) as the quenching agent, or kept in dark. (C) Cytotoxicity of different dose of IPA or SOD that respectively quenches •OH and •O2− radicals in Huh7.5.1 cells for 2 days was evaluated using cell titer assay. (D-E) The effect of 60 min illuminated TiO2 on HCV in the presence of IPA (D) or SOD (E) was determined by the focus-reduction assay. Viral infectivity titers were expressed as a percentage of the value to the mock/dark group. The error bars in panel C were derived from triplicates. **, P<0.01; *, P<0.05; NS, P>0.05.
In order to test the quenching agents in the context of viral infection, we first assessed their potential cytotoxicity. Huh7.5.1 cells were cultured in the presence of 0.2, 1 and 5 mM of IPA or 0.2, 1 and 5 mg/ml SOD for 48 h. The result indicated that the both quenchers at these tested concentrations showed no significant cytotoxicity (Fig. 5C). Then we performed the photocatalytic inactivation experiment in the presence of 1 mM of IPA or 1 mg/ml SOD. The results demonstrated that the anti-HCV effect of illuminated TiO2 was significantly canceled out by the •OH quencher IPA (Fig. 5D), but not by the •O2 − quencher SOD (Fig. 5E), suggesting that the •OH radicals play a more dominant role in the anti-HCV activity.
3.4. Natural indoor sunlight-illuminated TiO2 inactivates HCV
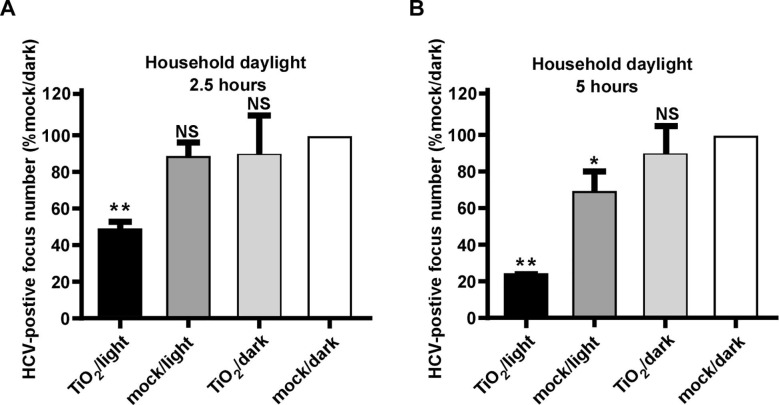
Next, we investigated whether HCV can be inactivated by TiO2 illuminated by natural indoor sunlight, a condition that mimics its application in a real field setting. Unlike the 375-nm illuminating light we used in the above experiments, the natural indoor sunlight consists of broad-spectrum visible lights with a wavelength ranging from 310 to 1000 nm after passing though household windows [36]. The inactivation assay was conducted in a windowed room with broad-spectrum light intensity of 0.6 mW/cm2 measured by a light intensity meter and around 0.15 mW/cm2 at UVA band (Detailed test method included in S3). 200 ffu of HCV was subjected to TiO2 or mock illuminated with indoor sunlight for 2.5 or 5 h or kept in dark respectively. After the treatment, the viruses were inoculated on Huh7.5.1 cells for 3-days infection, and the number of HCV-positive foci was quantified. As shown in Fig. 6 , TiO2 illuminated by natural indoor sunlight inactivated HCV in a time-dependent manner, with 2.5-hour and 5-hour illumination resulting in about 50% and 80% reduction in viral infectivity respectively. Of note, 5-hour illumination by indoor sunlight in the absence of TiO2 slightly decreased HCV infectivity, probably due to the irradiation by residual low-wavelength sunlight that passes through household window.
Fig. 6.
Natural indoor sunlight-illuminated TiO2 inactivates HCV. HCV was subjected to indoor sunlight for 2.5 (A) and 5 h (B), then inoculated to naïve Huh7.5.1 cells. Three days post-infection, the infectivity titers were determined by immunofluorescence of NS3 proteins. Viral infectivity titers were expressed as a percentage of the value in the control treatment. The error bars in panel C were derived from triplicates. **, P<0.01; *, P<0.05; NS, P>0.05.
3.5. Illuminated TiO2 inactivates a wide range of pathogens including SARS-CoV-2
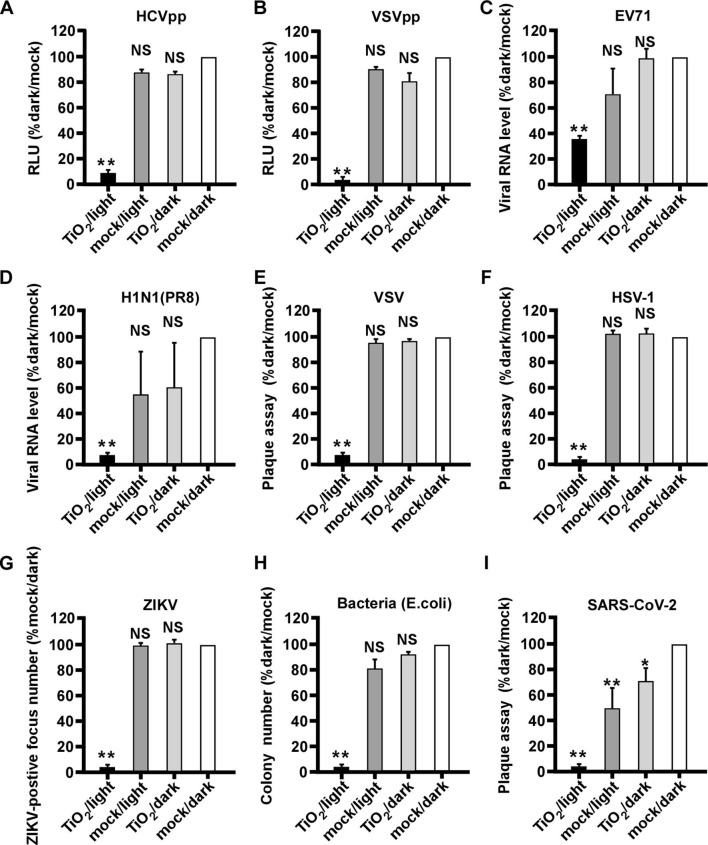
We investigated whether the illuminated nanosized TiO2 can inhibit other viral pathogens. First, we tested its effect on lentivirus-based pseudotype viral particles (chimera) consisting of HCV envelop proteins (HCVpp) or vesicular stomatitis virus glycoproteins (VSVpp) [29]. The both pseudotyped chimera viruses were subjected to the above mentioned four groups of treatment, and their infections in Huh7.5.1 cells were analyzed by the luciferase activity expressed by the lentiviral genome. As shown in Fig. 7 A-B, illuminated TiO2 efficiently inactivated both pseudotyped chimera viruses.
Fig. 7.
Illuminated TiO2 inactivates a wide range of viral pathogens including SARS-CoV-2. Various pathogens were subjected to the TiO2/light treatment, and then inoculated to their corresponding permissive cells to quantify the infection. (A-B) Infection of pseudotype viruses HCVpp (A) and VSVpp (B) in Huh7.5.1 cells for 2 days were analyzed by luciferase assay to measure the luciferase activity expressed by pseudotype viruses. (C) Infection of EV71 in Vero E6 cells for 2 days was analyzed by RT-qPCR to measure the intracellular viral RNA level. (D) Infection of influenza virus H1N1 (PR8 strain) in MDCK cells for 2 days was analyzed by RT-qPCR to measure the intracellular viral RNA level. (E-F) Infection of VSV and HSV-1 in Huh7 cells for 2 days were determined by the plaque assay (G) Infection of ZIKV in Vero E6 cells for 2 days was analyzed by focus-reduction assay. (H) The anti-bacteria efficiency was detected by counting the colony numbers of bacteria (E. coli). (I) Infection of SARS-CoV-2 in Vero E6 cells for 2 days was determined by plaque assay. The error bars in panel C were derived from triplicates. **, P<0.01; *, P<0.05; NS, P>0.05.
Then we tested the effect of illuminated nanosized TiO2 on a range of pathogens, including RNA viruses EV71, influenza virus H1N1 (PR8 strain), vesicular stomatitis virus (VSV), Zika virus (ZIKV), DNA virus herpes simplex virus (HSV-1) as well as bacterium (E. coli). These pathogens were treated as above by TiO2 or mock in the presence or absence of illumination, and then analyzed by RT-qPCR (EV71 and H1N1), plaque assay (VSV and HSV-1), immunofluorescent focus formation assay (ZIKV) or bacterial colony formation assay (E. coli). As shown in Fig. 7C–H, all pathogens can be inactivated by illuminated TiO2, but not by other groups of treatment, suggesting that the photocatalytic TiO2 exhibits a broad-spectrum inactivating potential.
Finally, we investigated whether the illuminated TiO2 can inhibit SARS-CoV-2, a novel coronavirus responsible for the ongoing COVID-19 pandemic. We performed the same inactivation assay, and the SARS-CoV-2 infection was analyzed by a plaque assay. As shown in Fig. 7I, SARS-CoV-2 can be efficiently inactivated by illuminated TiO2, although a low level of decrease in infectivity also can be observed in the mock/light or TiO2/dark treated groups.
4. Discussion
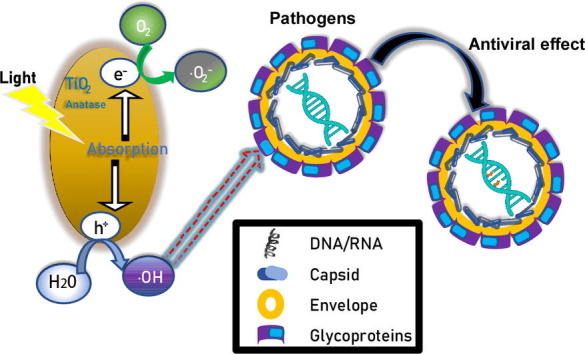
It is imperative to develop an efficient and safe way to disinfect pathogens in a field setting. In this study, we evaluated the effects of nanosized TiO2 thin-film on pathogens under a low irradiation condition (0.4 mW/cm2 at wavelength of 375 nm) as well as in a setting of indoor sunlight. We showed that pathogens can be efficiently inactivated by TiO2 under these illumination conditions that should be readily achievable in a real field setting. Unlike previous studies [13], [17], [18], our mechanistic studies demonstrated that illuminated TiO2 did not decrease the levels of structural components of HCV virions (proteins and RNA), but rather impaired the functions of the viral genome. We proposed a working model for the mechanism of inactivating pathogens by illuminated TiO2. With light illumination, TiO2 nanoparticles can transfer light energy to chemical energy. The resultant •OH radicals damage the HCV RNA genome, leading to the loss of its ability to generate an infectious life cycle. Further experiments will be needed to determine whether this mechanism also applies to the inactivation of other viral pathogens, and how •OH radicals change the structure of nucleic acids of viral genome.
Our results demonstrated that photo-activated TiO2 inactivates a broad range of pathogens, including multiple RNA viruses (H1N1-Flu, VSV, and ZIKV, EV71 and SARS-CoV-2), DNA virus (HSV-1) and bacterium (E. coli). These pathogens include viruses with different genome features (DNA or RNA, genome length, single or multiple segments) or with different virion structural features (envelope and non-envelope). These results are consistent with the proposed antiviral mechanism that •OH radicals induced by photo-activated TiO2 may attack nucleic acids of the pathogen genome, irrespective of their virion structures. It is interesting that the control group of only light treatment seems to inhibit the infectivity of SARS-CoV-2 more than other viruses. The possible explanation is that SARS-CoV-2 has the largest RNA genome (about 30,000 nt) and it may be more sensitive to the photocatalytic inactivation. Furthermore, we showed that TiO2 illuminated by natural indoor sunlight also can inactivate viruses, making it possible to apply it in a field setting to disinfect pathogens. Compared to other conventional disinfecting methods, such as heat, UV, detergents and ethanol [37], [38], photocatalytic TiO2 inactivation possesses several advantages – long lasting effect, easy to use, safe and no secondary pollution to the environment.
5. Conclusions
In summary, we brought forward new evidence of the photocatalytic inactivation properties of nanosized TiO2 on variety of toxic bacteria and viruses under low-irradiation density and of household day light. On top of that, the •OH radicals were considered playing the main role of leading to the inactivation of the pathogens, which confirmed by DMPO spin-trapping experiment. Most importantly, we found that the viral genome was the primary attacking target, instead of virion global structure and contents, thus the virus was inactivated. These results would provide a theoretical basis for photocatalytic materials, which have potential application in building indoor surface paint, air filters and medical devices and eventually help break the chain of transmission, particularly in a field setting of pandemics and epidemics.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We thank Drs. Dongming Zhou and Zhong Huang (Institut Pasteur of Shanghai, Chinese Academy of Sciences) for kindly providing H1N1(PR8) and EV71, Wuhan Institute of Virology, Chinese Academy of Sciences for kindly providing ZIKV.
Author contributions
Yimin Tong, Gansheng Shi, Xiaoyou Hu, Gaowei Hu, Lin Han, Rong Zhang, Xiaofeng Xie and Yongfeng Xu conducted the experiments and analyzed results; Jin Zhong, Jing Sun, Yimin Tong and Gansheng Shi designed experiments and wrote the manuscript. Jin Zhong and Jing Sun supervised the project.
Funding
This study was supported by the grants from National Key Research and Development Program of China (2016YFA0203000) to JS, Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29010205) to JZ, the National Sciences and Technology Major Projects (2018ZX10101004001004) to YT and (2017ZX09101-005-001-005) to YX, National Natural Science Foundation of China (31770189) to YT, NSFC-DFG bilateral organization program (51761135107) to JS. JS and XX acknowledge the technical cooperation with DongPeng Holding Co. Ltd.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cej.2021.128788.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.M.M. Al-Abdallat, D.C. Payne, S. Alqasrawi, B. Rha, R.A. Tohme, G.R. Abedi, M. Al Nsour, I. Iblan, N. Jarour, N.H. Farag, A. Haddadin, T. Al-Sanouri, A. Tamin, J.L. Harcourt, D.T. Kuhar, D.L. Swerdlow, D.D. Erdman, M.A. Pallansch, L.M. Haynes, S.I. Gerber, N. Sabri, M. Al Azhari, H. Khazali, M. Al Maayah, A. Bilbeisi, N. Dawood, B. Al Zubi, J. Meflih, T. Mounds, J. Fitzner, A. Eltom, A. Mafi, C. Miao, H. Caidi, S. Trivedi, S. Kamili, A.J. Hall, A. Curns, J. Moore, H. Pham, C. Zimmerman, E. Farnon, G. Giorgi, R. Gerber, f.t.J.M.-C.I. Team, Clinical Infectious Diseases 59 (2014) 1225-1233.
- 2.Choi A., García-Sastre A. Microbial Forensics. third ed. Academic Press; 2020. pp. 89–104. [Google Scholar]
- 3.Du Toit A. Nat. Rev. Microbiol. 2020 [Google Scholar]
- 4.Sato T., Taya Masahito. Biochem. Eng. J. 2006;28:303–308. [Google Scholar]
- 5.Park G.W., Cho M., Cates E.L., Lee D., Oh B.T., Vinje J., Kim J.H. J. Photochem. Photobiol. B. 2014;140:315–320. doi: 10.1016/j.jphotobiol.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu X., Wang S., Zhang X., Qi A., Qiao X., Liu Z., Wu M., Li L., Wang Z. Nano Energy. 2018;46:29–38. [Google Scholar]
- 7.Yadav H.M., Otari S.V., Koli V.B., Mali S.S., Hong C.K., Pawar S.H., Delekar S.D. J. Photochem. Photobiol. A-Chem. 2014;280:32–38. [Google Scholar]
- 8.Cho M., Chung H., Choi W., Yoon J. Appl. Environ. Microbiol. 2005;71:270–275. doi: 10.1128/AEM.71.1.270-275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi H., Fan J., Zhao Y., Hu X., Zhang X., Tang Z. J. Hazard. Mater. 2020;381 doi: 10.1016/j.jhazmat.2019.121006. [DOI] [PubMed] [Google Scholar]
- 10.Badireddy A.R., Budarz J.F., Chellam S., Wiesner M.R. Environ. Sci. Technol. 2012;46:5963–5970. doi: 10.1021/es300340u. [DOI] [PubMed] [Google Scholar]
- 11.Horovitz I., Avisar D., Luster E., Lozzi L., Luxbacher T., Mamane H. Chem. Eng. J. 2018;354:995–1006. [Google Scholar]
- 12.Park G.W., Cho M., Cates E.L., Lee D., Oh B.-T., Vinjé J., Kim J.-H. J. Photochem. Photobiol., B. 2014;140:315–320. doi: 10.1016/j.jphotobiol.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y., Zhang C., Shuai D., Naraginti S., Wang D., Zhang W. Water Res. 2016;106:249–258. doi: 10.1016/j.watres.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Zhang C., Li Y., Zhang W.L., Wang P.F., Wang C. Chemosphere. 2018;195:551–558. doi: 10.1016/j.chemosphere.2017.12.122. [DOI] [PubMed] [Google Scholar]
- 15.Steinmann E., Gravemann U., Friesland M., Doerrbecker J., Müller T.H., Pietschmann T., Seltsam A. Transfusion. 2013;53:1010–1018. doi: 10.1111/j.1537-2995.2012.03858.x. [DOI] [PubMed] [Google Scholar]
- 16.Fryk J.J., Marks D.C., Hobson-Peters J., Watterson D., Hall R.A., Young P.R., Reichenberg S., Tolksdorf F., Sumian C., Gravemann U., Seltsam A., Faddy H.M. Transfusion. 2017;57:2677–2682. doi: 10.1111/trf.14256. [DOI] [PubMed] [Google Scholar]
- 17.Ishiguro H., Nakano R., Yao Y., Kajioka J., Fujishima A., Sunada K., Minoshima M., Hashimoto K., Kubota Y. Photochem. Photobiol. Sci. 2011;10:1825–1829. doi: 10.1039/c1pp05192j. [DOI] [PubMed] [Google Scholar]
- 18.Kashige N., Kakita Y., Nakashima Y., Miake F., Watanabe K. Curr. Microbiol. 2001;42:184–189. doi: 10.1007/s002840010201. [DOI] [PubMed] [Google Scholar]
- 19.Zhong J., Gastaminza P., Cheng G., Kapadia S., Kato T., Burton D.R., Wieland S.F., Uprichard S.L., Wakita T., Chisari F.V. Proc. Natl. Acad. Sci. U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye X., Ku Z., Liu Q., Wang X., Shi J., Zhang Y., Kong L., Cong Y., Huang Z. J. Virol. 2014;88:72–81. doi: 10.1128/JVI.01848-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song Y., Wang X., Zhang H., Tang X., Li M., Yao J., Jin X., Ertl H.C., Zhou D. J. Virol. 2015;89:7841–7851. doi: 10.1128/JVI.00976-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carmichael J.C., Yokota H., Craven R.C., Schmitt A., Wills J.W. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi Y., Han L., Qi Y., Jin X., Zhang B., Niu J., Zhong J., Xu Y. Antiviral Res. 2020;179 doi: 10.1016/j.antiviral.2020.104813. [DOI] [PubMed] [Google Scholar]
- 24.Elahi S.M., Shen C.F., Gilbert R. J. Biotechnol. 2019;289:144–149. doi: 10.1016/j.jbiotec.2018.11.023. [DOI] [PubMed] [Google Scholar]
- 25.B. Zhao, C. Ni, R. Gao, Y. Wang, L. Yang, J. Wei, T. Lv, J. Liang, Q. Zhang, W. Xu, Y. Xie, X. Wang, Z. Yuan, J. Liang, R. Zhang, X. Lin, Protein Cell (2020). [DOI] [PMC free article] [PubMed]
- 26.Zhang R., et al. J. Microbes Infections. 2020;2020(1):16–21. [Google Scholar]
- 27.Tong Y., Chi X., Yang W., Zhong J. J. Virol. 2017;91 doi: 10.1128/JVI.02468-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brinkhof B., Spee B., Rothuizen J., Penning L.C. Anal. Biochem. 2006;356:36–43. doi: 10.1016/j.ab.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Qi Y., Xiang Y., Wang J., Qi Y., Li J., Niu J., Zhong J. Antiviral Res. 2013;100:392–398. doi: 10.1016/j.antiviral.2013.08.025. [DOI] [PubMed] [Google Scholar]
- 30.Lauer G.M., Walker B.D. N Engl. J. Med. 2001;345:41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 31.Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z., Murthy K., Habermann A., Krausslich H.G., Mizokami M., Bartenschlager R., Liang T.J. Nat. Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindenbach B.D., Evans M.J., Syder A.J., Wolk B., Tellinghuisen T.L., Liu C.C., Maruyama T., Hynes R.O., Burton D.R., McKeating J.A., Rice C.M. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 33.Fournier C., Duverlie G., Francois C., Schnuriger A., Dedeurwaerder S., Brochot E., Capron D., Wychowski C., Thibault V., Castelain S. Virol. J. 2007;4:35. doi: 10.1186/1743-422X-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pham H.N., McDowell T., Wilkins E. J. Environ. Sci. Health Part A-Environ. Sci. Eng. Toxic Hazard. Subt. Control. 1995;30:627–636. [Google Scholar]
- 35.Wainwright M. Int. J. Antimicrob. Agents. 2003;21:510–520. doi: 10.1016/s0924-8579(03)00035-9. [DOI] [PubMed] [Google Scholar]
- 36.M.A. Harry J. Keegan, Belknap, Dorothy J. Cordrey, J. Res. Nat. Bureau Standards 52 (1954) 2505-2508.
- 37.Pfaender S., Brinkmann J., Todt D., Riebesehl N., Steinmann J., Steinmann J., Pietschmann T., Steinmann E. Appl. Environ. Microbiol. 2015;81:1616–1621. doi: 10.1128/AEM.03580-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galasso M., Feld J.J., Watanabe Y., Pipkin M., Summers C., Ali A., Qaqish R., Chen M., Ribeiro R.V.P., Ramadan K., Pires L., Bagnato V.S., Kurachi C., Cherepanov V., Moonen G., Gazzalle A., Waddell T.K., Liu M., Keshavjee S., Wilson B.C., Humar A., Cypel M. Nat. Commun. 2019;10:481. doi: 10.1038/s41467-018-08261-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon request.