Abstract
Pre-implantation embryo development in mammals begins at fertilization with the migration and fusion of the maternal and paternal pro-nuclei, followed by the degradation of inherited factors involved in germ cell specification and the activation of embryonic genes required for subsequent cell divisions, compaction, and blastulation. The majority of studies on early embryogenesis have been conducted in the mouse or non-mammalian species, often requiring extrapolation of the findings to human development. Given both conserved similarities and species-specific differences, however, even comparison between closely related mammalian species may be challenging as certain aspects, including susceptibility to chromosomal aberrations, varies considerably across mammals. Moreover, most human embryo studies are limited to patient samples obtained from in vitro fertilization (IVF) clinics and donated for research, which are generally of poorer quality and produced with germ cells that may be suboptimal. Recent technical advances in genetic, epigenetic, chromosomal, and time-lapse imaging analyses of high quality whole human embryos have greatly improved our understanding of early human embryogenesis, particularly at the single embryo and cell level. This review summarizes the major characteristics of mammalian pre-implantation development from a chromosomal perspective, in addition to discussing the technological achievements that have recently been developed to obtain this data. We also discuss potential translation to clinical applications in reproductive medicine and conclude by examining the broader implications of these findings for the evolution of mammalian species and cancer pathology in somatic cells.
Keywords: Aneuploidy, cancer, chromothripsis, embryo, evolution, fragmentation, IVF, micronuclei, preimplantation
Introduction
Mammalian pre-implantation development encompasses a series of events beginning with the fertilization of a mature oocyte and resulting in the formation of a totipotent embryo capable of implantation into the uterus. Much of our knowledge regarding this critical time in development has been derived from studies using model organisms, including rodents [Whitten and Biggers 1968; Whittingham 1968] and larger agricultural species such as cattle [Brackett et al. 1982]. As in vitro conditions for embryo culture have been expanded and adapted to other mammalian species, a closer examination of higher organisms has provided additional insight into the fundamental aspects of pre-implantation development [Bavister et al. 1984]. Moreover, early studies characterizing human embryos following natural conception [Hertig et al. 1954; Hertig et al. 1956] and more recent studies in the context of in vitro fertilization (IVF) have also significantly contributed to our understanding of human embryogenesis [Edwards et al. 1969; Edwards et al. 1970]. Although mammalian embryos appear morphologically similar at this stage of development, it is clear that while certain features are conserved across species, others are species-specific and may limit extrapolation between different mammals. The susceptibility to chromosomal instability during early embryogenesis is one of these features and the focus of this review.
It is important to note that the mammalian pre-implantation embryo does not develop cell-autonomously and there are numerous extrinsic factors provided by the oviduct in vivo that might not necessarily be emulated by culture media in vitro [Lee and Yeung 2006]. Nevertheless, there is evidence that several of these developmental processes do occur naturally [Buster et al. 1985; Pereda and Croxatto 1978] and recent technological advances in genetic, epigenetic, chromosomal, and time-lapse imaging analyses have greatly assisted in further investigation of the underlying molecules and mechanisms, especially at the single cell level [Chavez et al. 2012; Guo et al. 2014; Hou et al. 2013; Smith et al. 2014; Wong et al. 2010; Xue et al. 2013; Yan et al. 2013]. In this review, we summarize both shared similarities and distinct differences in the major characteristics of pre-implantation development between different mammalian species from a chromosomal point of view as well as the technological achievements that have recently been developed to obtain these findings. We also discuss the potential consequences for the diagnosis and treatment of infertility via assisted reproduction technologies (ARTs), and the broader implications beyond the realm of Reproductive Medicine in species evolution and pathological conditions such as cancer.
A Brief Overview of Pre-Implantation Development Across Mammalian Species
Once a single sperm successfully fertilizes an oocyte by penetrating the outer glycoprotein layer known as the zona pellucida and depositing its genetic material, the fertilized oocyte undergoes second polar body extrusion to remove its remaining set of extra chromosomes. Subsequently, the maternal and paternal pro-nuclei, each containing haploid genomes under normal conditions, appear and migrate towards one another and fuse during a process called syngamy [Payne et al. 1997]. At this stage of development, the 1-cell embryo is termed a zygote; as the parental chromosomes combine, duplicate, and line-up together on the metaphase plate in preparation for mitosis, the embryo remains largely transcriptionally silent. This occurs until embryonic genome activation (EGA), which begins at different times of preimplantation development depending on the mammalian species. The mouse exhibits the earliest transition from maternal to embryonic transcriptional control beginning at the 2-cell stage, but there is minor transcription of a few mRNAs earlier at the 1-cell stage and is often referred to as zygotic gene activation (ZGA) [Flach et al. 1982]. While rat EGA begins at the 2- to 4-cell stage [Zernicka-Goetz 1994] and the 4-cell stage in pigs [Hyttel et al. 2000], human embryos initiate EGA on day 3 at approximately the 4- to 8-cell stage [Braude et al. 1988], which is similar to non-human primates as EGA occurs at the 6- to 8-cell stage in rhesus monkeys [Schramm and Bavister 1999]. Analogous to the mouse, human embryos have also been shown to exhibit minor transcriptional activity of preferential mRNAs prior to day 3 in development [Vassena et al. 2011]. The cow, sheep, and rabbit, initiate EGA later in pre-implantation development at the 8- to 16-cell stage [Brunet-Simon et al. 2001; Crosby et al. 1988; Plante et al. 1994] to likely coincide with delayed implantation timing as compared to other mammalian species. Thus, while all mammalian embryos begin pre-implantation development under relative transcriptional silence, the duration of this quiescence greatly differs between species. It also implies that the mammalian pre-implantation embryo must initially rely on maternally-derived RNAs and proteins, the majority of which are rapidly degraded during the oocyte-to-embryo transition, a process that is largely complete by the 2-cell stage in the mouse and the 8-cell stage in human embryos [Dobson et al. 2004; Evsikov et al. 2006; Galan et al. 2010; Hamatani et al. 2004; Zhang et al. 2009]. However, there must be a certain amount of selectivity in the destruction of maternal mRNAs and proteins to facilitate the successful shift from maternal to embryonic control of development [Alizadeh et al. 2005; Wong et al. 2010]. Additional studies also suggest that a small population of paternal transcripts inherited from sperm is important for sustaining early embryogenesis beginning at the 3- to 4-cell stage in human embryos [Sendler et al. 2013; Taylor et al. 1997] (Figure 1).
Figure 1.
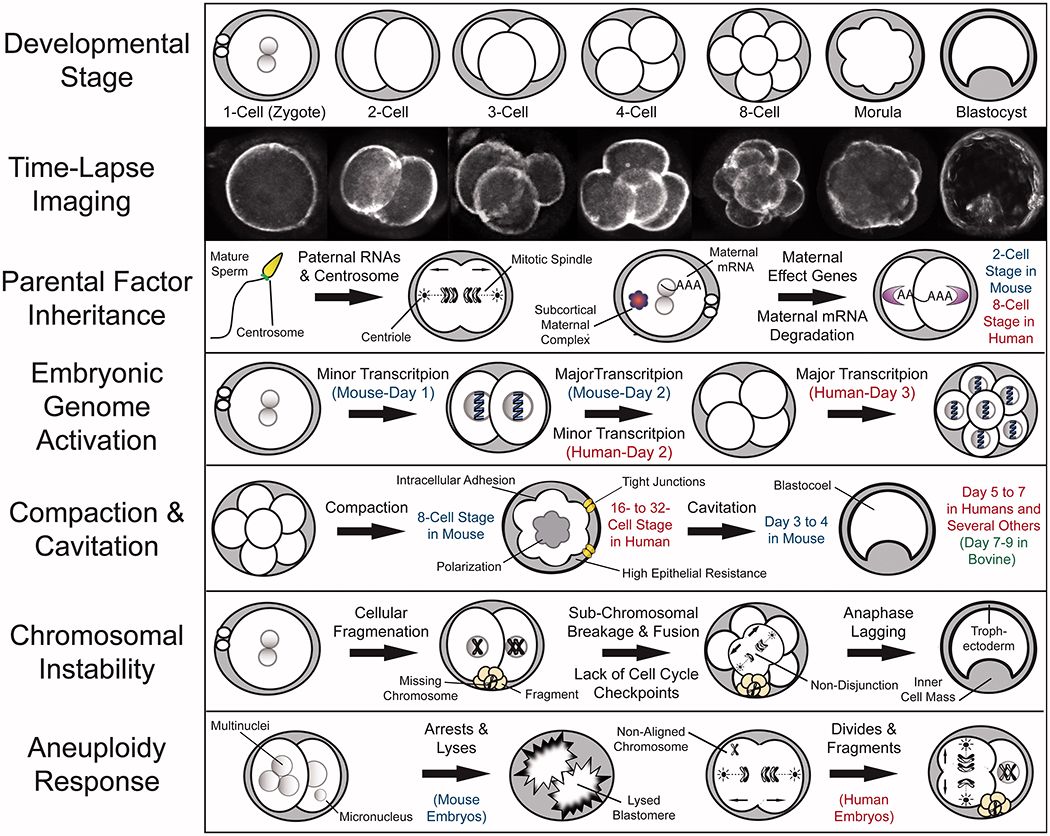
Fundamental aspects of mammalian pre-implantation development from a chromosomal perspective. Recent advances in mammalian embryology, including single cell whole genome/transcriptome analyses and time-lapse imaging, have greatly contributed to our understanding of the key characteristics in each major pre-implantation developmental stage. Time-lapse imaging has provided a non-invasive approach to monitor embryo development as well as the means to identify imaging parameters predictive of developmental success and embryo ploidy status. Embryo chromosomal integrity may be impacted by the lack of and/or inheritance of aberrant parental mRNAs, proteins, or other factors such as paternal contribution of the centrosome, which mediates the first mitotic divisions. A select group of maternal mRNAs termed maternal effect genes are recruited for translation following fertilization, whereas the remaining maternal mRNAs are degraded, a process that is essentially complete by the 2-cell stage in mouse embryos and the 8-cell stage in human embryos. Besides individual maternal proteins, multi-protein complexes, including the subcortical maternal complex (SCMC) is important for embryonic progression beyond the 2-cell stage and potential prevention of mitotic aneuploidy. This oocyte-to-embryo transition is largely dependent upon embryonic genome activation (EGA), the major wave of which occurs in the mouse at the 2-cell stage and begins in humans on day 3 at approximately the 8-cell stage. Following EGA, an embryo undergoes the processes of compaction, intracellular adhesion, and polarization to result in the formation of a morula at the 8-cell and 16- to 32-cell stage in mouse and human embryos, respectively. The majority of mammalian species, including humans, undergo cavitation to form a fluid-filled cavity called a blastocoel between days 5 and 6, whereas mouse embryos begin blastulation earlier between day 3 and 4 and bovine embryos later between day 7 and 8. There are several factors that can contribute to the generation of chromosome instability, particularly in human embryos, including cellular fragmentation, sub-chromosomal breakage and fusion, a lack of cell cycle checkpoints, and chromosomal lagging during anaphase. It appears that unlike human embryos, which respond to chromosomal aberrations by fragmenting and continuing to divide, mouse embryos deal with multi- and micronuclei formation by inducing blastomere lysis.
Following syngamy, the 1-cell zygote undergoes a series of mitotic cell divisions to produce an increasing number of progressively smaller cells called blastomeres without changing the overall size of the embryo [Bell et al. 2008]. This continues until the embryo begins the process of intracellular adhesion known as compaction to form a morula, which is thought to be important for later morphogenetic events such as lineage specification [Kidder and McLachlin 1985; Levy et al. 1986]. Compaction occurs earlier in rodents at the 8-cell stage than in other mammalian species, including humans, non-human primates, cattle, pigs, and sheep, where it begins between the 16- and 32-cell stage [Edwards et al. 1981; Enders et al. 1990; Reima et al. 1993; Steptoe et al. 1971; Van Soom et al. 1997]. In conjunction with compaction, intracellular polarization is also thought to occur, whereby the chromosome-containing nucleus of each blastomere faces basolaterally (inward) and cytoskeletal components such as actin and microtubules accumulate apically (outward) in the embryo [Houliston and Maro 1989; Johnson and Maro 1984; Reeve and Kelly 1983]. Thus, subsequent cell divisions are either symmetric or asymmetric depending on the orientation of the cleavage plane so that this polarity is inherited in the daughter cells. These symmetric versus asymmetric cleavage divisions and/or other previously described factors such as blastomere position within the embryo that has recently regained recognition for their role in cell fate determination [Tarkowski and Wroblewska 1967; Watanabe et al. 2014], generating two distinct cell populations. While the cells on the inside of the embryo will become a part of the inner cell mass (ICM), the cells on the outside will contribute to the trophectoderm (TE) layer [Johnson and Ziomek 1981; Sutherland et al. 1990] (Figure 1).
Regardless of the underlying mechanisms(s), the process of compaction eventually leads to the formation of a totipotent blastocyst that comprises a fluid-filled cavity called a blastocoel. Blastocoel formation is initiated almost immediately after compaction by the assembly of tight junctions and the establishment of high epithelial resistance in TE cells, beginning on day 3 in mouse embryos and later on day 4.5 during human pre-implantation development [Edwards et al. 1981; Sheth et al. 1997]. Because of this delay, human embryos are also likely to undergo at least one additional round of cell division to form an ~256-cell blastocyst, whereas mouse blastocysts typically comprise ~164 cells. However, morphological changes such as cavitation have been shown to be a function of developmental timing rather than cell number per se, at least in the human pre-implantation embryo [Hardy et al. 1989; Niakan et al. 2012]. Analogous to humans, the vast majority of other mammalian species, including non-human primates, sheep, pigs, rabbits, and the rat, form multi-cellular blastocysts on day 5 to 7 [Daniel 1965; Dobrinsky et al. 1996; Gardner et al. 1994; Seshagiri and Hearn 1993; Surani 1975]. Bovine embryos, in comparison, do not exhibit blastocyst formation until day 7 to 9 [Keskintepe et al. 1995] and may be due to differences in DNA damage, metabolic requirements, and/or the extent of placental invasion [Sturmey et al. 2009] (Figure 1). In preparation for implantation into the uterus, the blastocyst then ‘hatches’ from the zona pellucida to allow for increased embryo growth and development as well as TE adhesion to the lining of the uterine wall. Prior to implantation, which occurs on day 7 during human embryonic development, a second lineage decision is made, whereby the ICM of the blastocyst differentiates into either early epiblast or primitive endoderm to form the future fetus and parietal/visceral endoderm, respectively [Cockburn and Rossant 2010].
Incidence and Detection of Aneuploidy During Mammalian Pre-Implantation Development
The formation of a blastocyst is one of first major landmarks in mammalian development and yet, only 30–50% of pre-implantation embryos from the majority of mammals will typically reach this stage when cultured in vitro [Alper et al. 2001]. In contrast, approximately 80% of mouse embryos will form blastocysts in vitro since developmental arrest prior to this stage is far less frequent in this species. Although the cause(s) of embryo arrest may vary between species, an abnormal number of whole chromosomes, or aneuploidy, is thought to be a primary determinant of whether a human embryo will progress in development [Munne et al. 1994]. Previously, the most frequently used method for diagnosing aneuploidy was pre-implantation genetic screening (PGS) of day 3 biopsied blastomeres via DNA florescent in situ hybridization (FISH) [Munne and Weier 1996], which suffers from mosaicism between cells and limited chromosomal analysis [Baart et al. 2006; Kuo et al. 1998; Mastenbroek et al. 2007]. As there is a limitation on the number of cells that can be biopsied from patient embryos, supernumerary cleavage-stage embryos subsequently consented for research have provided an alternative source for aneuploidy assessment, although they are generally of poorer quality.
More recent studies using array-based methods to evaluate all 24 chromosomes in high-quality whole human embryos, demonstrated that 50–80% of human embryos at the cleavage-stage have one or more blastomeres that are indeed aneuploid. In addition, the incidence of aneuploidy in cleaving human embryos appears to be irrespective of fertility status or whether a fresh versus frozen/thawed IVF cycle, since a similar frequency was observed in embryos from fertile couples and following cryopreservation [Chavez et al. 2012; Johnson et al. 2010b; Vanneste et al. 2009]. Furthermore, the current view is that the incidence of aneuploidy in vitro likely reflects the situation in vivo considering that 20-30% of natural human conceptions are estimated to result in a live birth [Macklon et al. 2002; Slama et al. 2002; Zinaman et al. 1996] and chromosomal abnormalities have been reported in the majority of spontaneous miscarriage cases [Benkhalifa et al. 2005; Fritz et al. 2001]. This is in contrast to lower mammalian species, particularly the mouse, which is estimated to exhibit approximately 1% embryonic aneuploidy rates, depending on the strain [Lightfoot et al. 2006]. However, it is important to note that microarrays for the detection of chromosomal copy number variants (CNVs) are far more utilized for humans than other mammalian species due, in large part, to PGS of human embryos in IVF clinics and genetic testing centers. Thus, the overall aneuploidy rates via ‘whole genome’ cytogenetic methods in pre-implantation embryos from non-human species, especially those more closely related to humans, remains to be determined. Aneuploidy analysis via DNA FISH of rhesus macaque embryos produced in vitro [Dupont et al. 2010] as well as frequent observations of abnormal nuclear structure in blastomeres from rhesus embryos as compared to other mammalian species [Chavez et al. 2012] does suggest that the incidence of chromosomal abnormalities in non-human primates is similar to humans (Figure 2).
Figure 2.
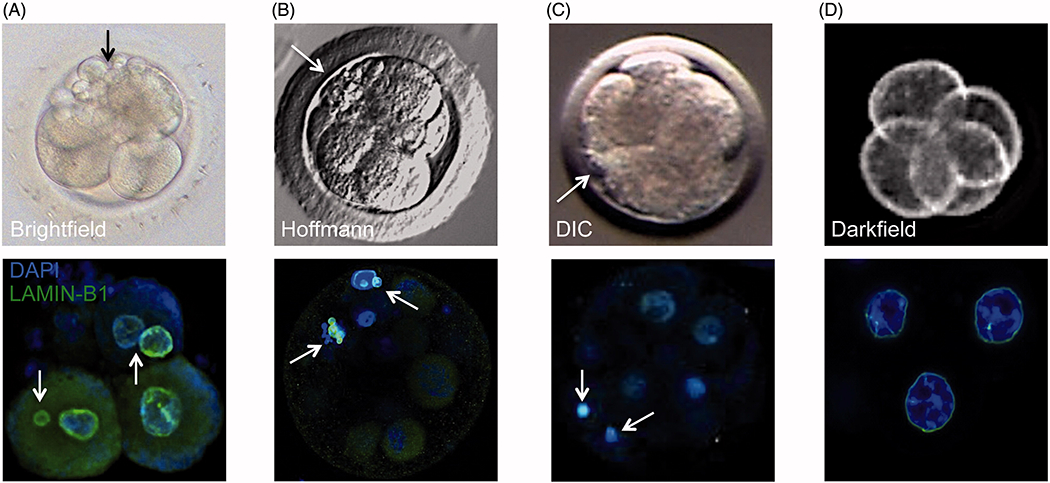
Correlation between cellular fragmentation and multi-/micronuclei formation in cleavage-stage embryos from different mammalian species. Top row: a representative photo of (A) human embryo taken by brightfield imaging, (B) rhesus macaque embryo using Hoffmann modulation contrast, (C) bovine embryo using differential interference contrast (DIC), and (D) mouse embryo taken by darkfield illumination time-lapse imaging. Note the appearance of several cellular fragments (black or white arrows) in cleavage-stage human and non-human primate embryos, but not in mouse embryos, with a lesser extent observed in bovine embryos. Bottom row: the incidence of cellular fragmentation is highly associated with mutli- and micronuclei formation as indicated by confocal microscopy of LAMIN-B1 (green) expression in DAPI-stained (blue) cleavage-stage embryos from the different mammalian species.
Given the high incidence of mosaicism in cleavage-stage human embryos [Baart et al. 2006; Kuo et al. 1998] and potential detriment of blastomere biopsy on embryonic development, alternative approaches such as extended culture of embryos to the blastocyst stage and analysis of chromosomal status via trophectoderm biopsy have recently been implemented to evaluate aneuploidy [Schoolcraft et al. 2010; Scott et al. 2013]. Technological advances in next-generation whole genome sequencing have also provided an additional platform besides array-based methods to comprehensively assess aneuploidy in human embryos, including at the single cell level [Fiorentino et al. 2014; Hou et al. 2013]. While human blastocysts are thought to exhibit less aneuploidy than cleavage-stage embryos [Magli et al. 2000], the presence of two or more cell populations with different chromosomal make-up has been reported at the blastocyst stage and may be more prevalent in embryos from women of advanced maternal age [Fragouli et al. 2011; Liu et al. 2012]. In addition, it is important to differentiate between the proportion of aneuploid cells in an embryo versus the percentage of embryos with mosaicism as the latter increases at the blastocyst stage [van Echten-Arends et al. 2011]. Moreover, there are additional potential risks associated with prolonged embryo culture such as the introduction of epigenetic changes, monozygotic twinning, and other factors that may disrupt embryo integrity in humans and other mammalian species [Fernandez-Gonzalez et al. 2009; Kallen et al. 2010; Katari et al. 2009; Khosla et al. 2001; Peramo et al. 1999]. Nevertheless, extended culture may prevent the selection of cleavage-stage embryos that are destined to arrest, which is why blastocyst transfer with or without the use of PGS has increased [Alper et al. 2001; Munne et al. 1994; Schoolcraft et al. 2010].
Despite higher probability that aneuploid embryos will arrest at the cleavage-stage, embryos with chromosomal abnormalities can still form blastocysts and are often indistinguishable from chromosomally normal (euploid) embryos. Thus, while attempts have been made to correlate morphology with aneuploidy, it is well known that aneuploid embryos can appear normal and suitable for transfer under traditional IVF assessment techniques, especially when obtained by static observations [Baltaci et al. 2006]. More recently, the implementation of time-lapse imaging has provided a non-invasive approach to monitor embryos throughout development and potentially assess embryo viability. Once it was determined that time-lapse monitoring (TLM) was not detrimental to embryo development, several studies began investigating morphological, spatial, and/or temporal correlates between imaging behavior and embryo quality [Cruz et al. 2011; Nakahara et al. 2010; Wong et al. 2010]. To this end, Wong et al. [2010] demonstrated that TLM can be used to predict blastocyst fate prior to EGA by measuring the duration and time between the first three mitotic divisions. Whether the first three mitotic divisions are similarly predictive for other mammalian species besides the human remains to be determined, but an examination of early mitotic timing in murine, bovine, and rhesus monkeys has suggested that this may be the case [Burruel et al. 2014; Pribenszky et al. 2010; Sugimura et al. 2012].
In a follow-up study, Chavez and colleagues determined that the timing of the first three mitotic divisions, in conjunction with assessment of a dynamic process called cellular fragmentation, might also be used to distinguish euploid from aneuploid human embryos at the cleavage-stage [Chavez et al. 2012]. Since this initial report, additional imaging parameters such as the time to fifth cell, initiation of cavitation, and completion of blastulation have been identified that may differentiate between chromosomally normal and abnormal cleaving human embryos as well as blastocysts [Basile et al. 2014; Campbell et al. 2013]. Of course, clinical validation of these findings is required and recent prospective studies of patient embryos have confirmed the importance of certain cell cycle parameters in predicting blastocyst formation and/or aneuploidy risk [Conaghan et al. 2013; Kirkegaard et al. 2013; Yang et al. 2014]. While there currently is some discrepancy as to whether TLM is actually beneficial for embryo selection [Kaser and Racowsky 2014], the publication of the first randomized control trial (RCT), which evaluated implantation rates, ongoing pregnancy, and early pregnancy loss, suggests that TLM is more effective than conventional IVF techniques [Rubio et al. 2014]. It remains to be determined, however, if TLM can also positively impact live birth rates, particularly in cases of single embryo transfers (SET). Nevertheless, it is clear from these studies that (i) standardized nomenclature for time-lapse markers is required as recently described [Kaser and Racowsky 2014], (ii) the method of parameter measurement is important as the use of a common start point such as the time of intra-cytoplasmic sperm injection (ICSI) may have confounding effects on other overlapping parameters, and (iii) additional RCTs investigating SET following TLM and PGS for aneuploidy detection are needed.
Potential Mechanism(s) of Embryonic Aneuploidy Generation and Resolution
It is thought that most chromosomal errors occur during oogenesis as the maternal chromosomes congress and segregate from one another upon division [Nagaoka et al. 2012]. Other errors can occur on the mitotic spindle during embryonic cleavage divisions, which may be perpetuated due to the apparent lack of cell cycle checkpoints in cleaving human embryos [Harrison et al. 2000; Kiessling et al. 2010]. Anaphase lagging, or the failure of one or more chromosomes to connect to the spindle, has also been proposed to contribute to mitotic errors at the blastocyst stage [Coonen et al. 2004]. In comparison to other mammalian species such as the mouse, whereby only 0.05–1% of oocytes are typically aneuploid, estimates of meiotic error rates in women are relatively high at 5–20% of oocytes [Hassold and Hunt 2001]. Therefore, it is not unexpected that maternal meiotic errors are much more frequent than that of paternal origin in human embryos identified as aneuploid [Johnson et al. 2010b]. However, it is important to note that the centrosome, which consists of two centrioles for the generation of spindle microtubules during the first embryonic division(s), are paternally inherited in the majority of mammalian species [Palermo et al. 1994; Sathananthan et al. 1991] (Figure 1). In contrast, it appears that the centrosome is maternally inherited in the mouse embryo [Schatten et al. 1991], the only known exception to date. This may help explain the relatively low aneuploidy rates observed in mouse embryos [Lightfoot et al. 2006] and suggests that paternal contribution to human embryonic aneuploidy is likely greater than thought, thereby invoking consideration of paternal age as maternal age-related aneuploidy is considered [Nagaoka et al. 2012]. Additional studies have supported this idea by determining that mitotic errors are just as, if not more, frequent than meiotic errors in embryos from women of average maternal age [Chavez et al. 2012; Johnson et al. 2010b; Vanneste et al. 2009]. Further evidence is provided by observations of increased mitotic mosaicism in embryos fertilized by testicular sperm extraction (TESE) from men with non-obstructive azoospermia (NOA) to suggest that non-ejaculated spermatozoa may be less effective in organizing the first mitotic spindle [Silber et al. 2003].
Besides the transfer of defective centrosomal components, the lack of and/or inheritance of aberrant parental mRNAs, proteins, or other factors may also influence the chromosomal integrity of embryos [Burruel et al. 2014; Xanthopoulou et al. 2012]. Since sperm are generally considered transcriptionally silent, RNAs detected in the paternal gametes were initially assumed to be either products of transcript degradation or simply contaminants from surrounding cells in the testes or epididymis [Pessot et al. 1989]. More recent studies have shown, however, that sperm actually retain both specific coding as well as non-coding RNAs, some of which have known roles in mitotic progression and the prevention of aneuploidy generation [Dawlaty et al. 2008; Sendler et al. 2013; Taylor et al. 1997].
In contrast to inherited paternal transcripts, the importance of particular maternal RNAs and proteins during the oocyte-to-embryo transition has been more widely characterized. By definition, these factors are termed maternal effect genes as they are transcribed during oogenesis and persist in the embryo, but certain maternal effect genes are not expressed until after EGA to complicate the differentiation between maternal and embryonic effects [Li et al. 2010]. Numerous maternal effect genes have now been identified and include factors involved in maternal mRNA degradation, transcription, chromatin remodelling, and DNA methylation. In addition to individual maternal proteins, multi-protein complexes such as the subcortical maternal complex (SCMC) have also been shown to be important for embryonic progression beyond the 2-cell stage at least in mouse embryos [Li et al. 2008]. One of the first members of this complex to be identified was maternal antigen that embryos require (Mater/Nlrp5) [Tong et al. 2000], and its depletion prevents SCMC formation and results in pre-implantation embryonic lethality in both mice [Li et al. 2008] and rhesus monkeys [Wu 2009]. A similar phenotype was observed in mouse embryos lacking factor located in oocytes permitting embryonic development (Floped/Ooep), another member of the SCMC complex [Tashiro et al. 2010]. The absence of other SCMC proteins such as Filia (RIKEN), in comparison, produces less pronounced phenotypic effects, but a high incidence of mitotic aneuploidy was detected in those embryos that failed to reach the blastocyst stage [Zheng and Dean 2009] (Figure 1). Although humans only possess a Filia ortholog named ES cell-associated transcript 1 (ECAT1/KHDC3L), which was lost in rodents [Pierre et al. 2007], the other SCMC members appear to be conserved across mammalian species [Li et al. 2010]. Thus, it will be important to determine whether these maternal effect proteins play an analogous role in regulating ploidy status during human pre-implantation development.
Even with the existence of parental euploidy-maintaining factors, frequent findings of complex mitotic aneuploid mosaicism, whereby multiple chromosomes are affected in embryos without a single euploid blastomere, indicate that it is unlikely that the majority of chromosomal errors can be corrected through development [Chavez et al. 2012; Vanneste et al. 2009]. Nevertheless, there is some evidence to suggest that chromosomal correction can occasionally occur during pre-implantation development based on PGS and biopsy of an aneuploid blastomere on day 3 followed by euploid TE cells on day 5 [Barbash-Hazan et al. 2009; Munne et al. 2005]. In support of this concept, it was proposed that euploid cells may exhibit advantageous growth over aneuploid cells in cases of diploid-aneuploid mosaicism or perhaps, there is preferential contribution of chromosomally normal cells to the ICM and chromosomally abnormal cells to the TE layer [Barbash-Hazan et al. 2009; Fragouli et al. 2008; Wells and Delhanty 2000]. Recent studies, however, have shown no preferential distribution of chromosomally abnormal cells between the ICM and TE, suggesting that chromosomal asymmetry may not explain how an embryo is able to overcome aneuploidy generation [Capalbo et al. 2013; Johnson et al. 2010a]. Despite this apparent contradiction, the probability that aneuploidy can be resolved during pre-implantation development may instead depend on the type of mosaicism, the number of TE cells biopsied, and/or whether a single or several chromosomes have been impacted [Fragouli et al. 2011; Novik et al. 2014]. There also remains the possibility that alternative mechanisms such as blastomere exclusion or DNA replication in the absence of cell division known as endoreduplication, followed by multi-polar divisions may still play a role in the so-called ‘embryo self-correction’ phenomenon [Capalbo et al. 2013; Johnson et al. 2010a] (Figure 3).
Figure 3.
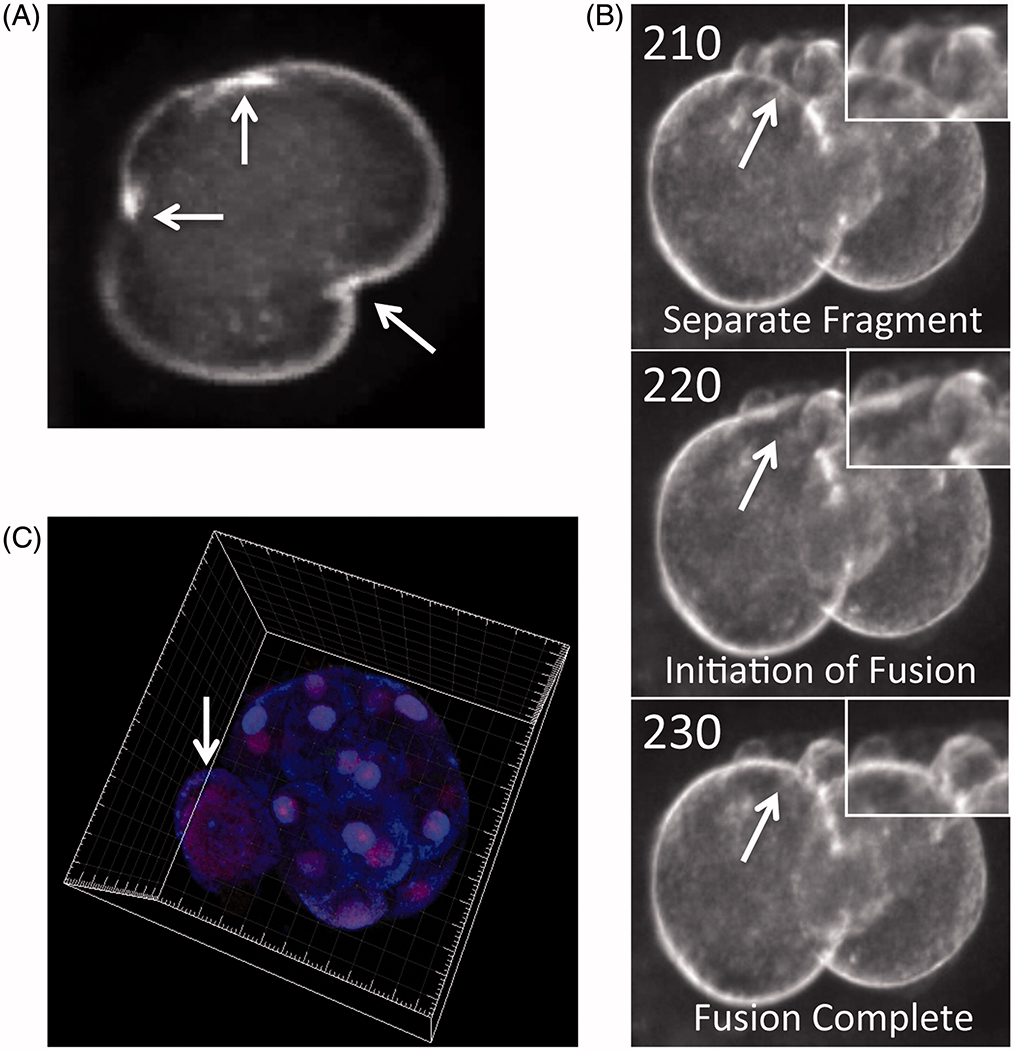
Potential mechanisms of aneuploidy resolution during embryo pre-implantation development. (A) Time-lapse image showing a human zygote with three cleavage furrows as it divides directly from 1-cell to 3-cells. (B) Individual imaging frames of a human embryo with cellular fragmentation demonstrating blastomeric resorption of a fragment. Adapted from Chavez et al. [2012]. (C) Multi-channel confocal analysis of histone modifications in a human morula that is undergoing cavitation to form a blastocyst reveals the presence of a large and likely polyploid blastomere excluded from the embryo. Adapted from Chavez et al. [2014].
As mentioned above, Chavez et al. [2012] demonstrated that dynamic assessment of cell cycle parameters in combination with cellular fragmentation analysis can assist in the differentiation between chromosomally normal and abnormal human embryos. Based on the timing of fragmentation, the authors also determined that embryos with meiotic errors typically exhibited fragmentation at the 1-cell stage, whereas fragmentation was most often detected at the 2-cell stage in embryos with mitotic errors to suggest that the human embryo responds to aneuploidy generation by fragmenting [Chavez et al. 2012]. While the cause(s) of cellular fragmentation has been disputed for several years, greater than 50% of human embryos at the cleavage stage fragment to some degree [Antczak and Van Blerkom 1999] and there is evidence to suggest that fragmentation occurs naturally in vivo, indicating that it is not only a consequence of in vitro embryo culture [Buster et al. 1985; Pereda and Croxatto 1978]. In addition, cellular fragmentation is distinct from the cell death-induced DNA fragmentation that can occur later in mammalian pre-implantation development, most significantly at the morula and blastocyst stages [Hardy 1999; Hardy et al. 2001; Xu et al. 2001]. Although originally thought to represent anucleate cytoplasmic components, Chavez and colleagues demonstrated that cellular fragments can contain nuclear DNA and that the high frequency of human embryonic aneuploidy may have contributions from chromosome-containing embryonic micronuclei, which are likely to be sequestered via fragmentation [Chavez et al. 2012]. It is important to note, however, that not all fragments necessarily enclose mis-segregated chromosomes, and may partially explain how certain fragmented embryos can still implant and result in a successful pregnancy outcome [Alikani et al. 1999; Edwards et al. 1984; Pelinck et al. 2010]. Given this evidence and that fragmentation, which usually arises at the 1- to 2-cell stage [Chavez et al. 2012], well precedes embryo arrest typically at the 8-cell stage, we would suggest that cellular fragmentation is also different from a form of mitotic cell death called chromosome fragmentation [Stevens et al. 2011; Stevens et al. 2007]. Notably, unbalanced partial chromosomal losses and gains have also been observed in fragmented aneuploid embryos and not euploid embryos, to suggest a relationship between sub-chromosomal instability and aneuploidy in the human embryo [Chavez et al. 2012; Vanneste et al. 2009]. Regardless of its origins, cellular fragmentation is observed in pre-implantation embryos from other mammalian species such as non-human primates, cattle, and even mice, although to a lesser extent [Enders et al. 1982; Sugimura et al. 2010] (Figure 2). Thus, it will be important to determine the precise mechanism(s) by which it occurs and how dynamic events such as the timing, degree, and/or resorption of chromosome-containing fragments contributes to the generation and potential correction of embryonic aneuploidy [Chavez et al. 2012; Hardarson et al. 2002; Lemmen et al. 2008; Van Blerkom et al. 2001].
Differential Response to Chromosomal Instability Between Mammalian Species
Unlike human embryos, which appear to respond to aneuploidy generation by fragmenting [Chavez et al. 2012], recent evidence suggests that mouse embryos deal with multi- and micronuclei formation much differently. By microinjecting mouse zygotes with morpholino oligonucleotides (MOs) to Rps6ka4/Msk2, a mitogenic factor known to be involved in the G1 phase of the cell cycle [Bettencourt-Dias et al. 2004], Chavez and colleagues demonstrated that reduced Msk2 expression results in mitotic arrest at the 3- to 8-cell stage [Chavez et al. 2014]. Upon assessment of embryo behavior by time-lapse imaging, the authors also determined that Msk2-MO-injected embryos exhibited an unusual phenotype of increased blastomere movement and lysis following arrest that resembled cellular events described during mitotic catastrophe [Vakifahmetoglu et al. 2008]. As mitotic catastrophe is thought to follow aberrant chromosome segregation, they investigated the nuclear structure of each blastomere and detected the formation of both multi- and micronuclei in Msk2-MO-injected, but not control-MO-injected embryos prior to lysis [Chavez et al. 2014]. This suggests that blastomere lysis may constitute a mechanism for mouse embryos to avoid chromosomal instability and that the low aneuploidy rates observed in mice are the result of a selection process [Lightfoot et al. 2006]. A correlation between MSK2 and human aneuploidy generation was also demonstrated by findings of reduced MSK2 expression, abnormal cell cycle parameter timing and micronuclei formation in aneuploid over euploid human embryos. Besides cell cycle regulators, the deletion of mitotic spindle assembly complex (SAC) components also leads to the formation of micronuclei, chromosome misalignment, aneuploidy, developmental delay, and decreased implantation rates in mice [Wei et al. 2011]. Taken together, this suggests that while human embryos continue to divide and undergo cellular fragmentation in spite of aneuploidy [Chavez et al. 2012], mouse embryos lyse rather than divide when faced with chromosomal instability [Chavez et al. 2014] (Figure 1). Future work will assist in determining if other mammalian species similarly respond to the generation of aneuploidy and whether MSK2 prevents chromosomal aberrations directly or indirectly during preimplantation development.
Clinical Consequences of Embryonic Aneuploidy During Pre-Implantation Development
Despite the progressive increase in the number of IVF cycles performed each year (cdc.gov/art), the percentage of live births has not significantly changed even with the implementation of more advanced IVF techniques, including intracytoplasmic sperm injection (ICSI), PGS, and cryopreservation [Boulet et al. 2015; Harper and Sengupta 2012; Kissin et al. 2014]. Previously, it was thought that low IVF success rates could be explained by infertility and/or advanced maternal age, but both fertile and young women produce embryos that are aneuploid with similar frequency [Vanneste et al. 2009]. By factoring out maternal age, fertility status, and other clinical variables such as whether donor materials were used, a large number of human embryos still arrest before the blastocyst stage and are chromosomally abnormal to suggest that aneuploidy is a common phenomenon among reproductive-age couples [Baart et al. 2006; Franasiak et al. 2014]. It is important to note, however, that the function of sexual reproduction is to reduce the chromosomal instability generated in somatic cells over time. Thus, IVF could bypass this filter, which in turn may potentially increase chromosomal instability in embryos [Horne et al. 2013]. Nevertheless, there is evidence that several of the morphological features observed in embryos following ART procedures, including cellular fragmentation, blastomere asymmetry, and other developmental processes do occur naturally following in vivo conceptions [Buster et al. 1985; Pereda and Croxatto 1978]. Further support for this is provided by the high incidence of mosaicism detected in early spontaneous human abortions [Lebedev et al. 2004; Vorsanova et al. 2005] and similar rates of mosaicism in chorionic villus samples (CVS) between IVF and natural pregnancies later in development [Huang et al. 2009; Mantikou et al. 2012]. Therefore, it will be essential to determine how our increased understanding of chromosomal instability during pre-implantation development may be used to direct efforts toward improved fetal diagnosis and reduced spontaneous loss via cell free DNA or other methods. Lastly, it will also be critical to evaluate how additional recently developed approaches, including time-lapse imaging, DNA-Seq, natural-cycle frozen embryo transfer, either alone or in combination, impacts IVF success rates over the next several years [Evans et al. 2014; Fatemi et al. 2010].
Implications of Chromosomal Instability Beyond Reproductive Medicine: Cancer and Tumor Biology
Chromosomal instability is not only commonly observed during pre-implantation embryo development, but also a quintessential hallmark of cancer cells. The majority of solid tumors contain aneuploid cells as the result of a high frequency of chromosome mis-segregation. In turn, this aneuploidy is often associated with poor prognosis and advanced tumor stage, leading to both metastatic potential and resistance to drugs [Thompson and Compton 2011]. Since its identification almost two decades ago [Lengauer et al. 1997], significant progress has been made to characterize the underlying mechanisms causing chromosomal instability in cancer cells. Among the main culprits, defects in chromatid cohesion, kinetochore-microtubule attachment, spindle assembly, and centrosome copy number appear to play a central role (reviewed in detail by [Thompson et al. 2010]). In addition, numerous proteins involved in cell cycle regulation, including Breast Cancer1 (BRCA1), BRCA2, and tumor protein 53 (TP53), have also been implicated, although their involvement in aneuploidy is less well defined. Merotelic kinetochore orientation, whereby a single kinetochore binds microtubules from both spindle poles, appears to be the most frequent cause for chromosomes to remain at the spindle equator after anaphase (anaphase lagging) in mammalian cells [Cimini et al. 2001]. Importantly, this type of error does not induce mitotic arrest, suggesting that it is not detected during the spindle assembly checkpoint, and most likely the mechanism by which aneuploidy arises in wild type (euploid) cells.
Chromothripsis and Chromosomal Reassembly
Similar to what has been described for aneuploidy generation during pre-implantation development [Chavez et al. 2012], the formation of micronuclei (MN) is also associated with mis-segragation of whole or partial chromosomes in cancer cells and increases with culture and propagation. MN form because mis-segregated chromosomes are able to recruit a nuclear envelope whose structure appears comparable to that surrounding the primary nucleus [Hatch et al. 2013]. However, in vitro experiments with cultured cells have shown that MN display altered nuclear functions, including aberrant DNA replication, transcription and DNA-damage repair. Such studies also proposed a link between the recruitment of chromosomes in MN and a process initially termed genome or karyotype chaos, which is thought to represent a stress response, and is more recently known as chromothripsis [Crasta et al. 2012; Hoffelder et al. 2004; Horne and Heng 2014; Liu et al. 2014; Terradas et al. 2009; Xu et al. 2011]. First described in cancer genomes [Stephens et al. 2011], chromothripsis is a phenomenon through which one or a few chromosomal segments are ‘pulverized’ and randomly reassembled in one unique cellular event [Pellestor et al. 2014]. This is a complex process that includes chromosomal duplications, deletions, translocations, and inversions, and was identified via whole genome sequencing analysis of several cancer genome datasets. A model correlating MN formation with chromosome pulverization was proposed on the basis of in vitro studies in which lagging chromosomes and MN formation were experimentally induced by treating with nocodazole [Crasta et al. 2012] or monastrol [Janssen et al. 2011] and DNA damage was observed in phase G2 entry. DNA breaks appeared to occur in a ‘replication-dependent’ manner as blockage of DNA replication also prevented DNA damage. Interestingly, long-term live cell imaging showed that 97% of MN remain stable during interphase and are not degraded, nor extruded. After breakdown of the nuclear envelope, however, a significant portion (38%) of MN merges with the main nucleus (Figure 4). More detailed analyses of the structural changes occurring in the nuclear envelope of MN in comparison to the primary nucleus showed that DNA damage is a consequence of nuclear envelope collapse in both cultured cells and solid tumors [Hatch et al. 2013]. Destabilization of the nuclear envelope seems to be a consequence of structural defects in the lamina of MN, although the causes of such structural changes are unknown. As a consequence, basic nuclear functions, including DNA repair and replication are altered and genomic stability negatively affected as documented by the accumulation of gamma-H2A histone family, member X (γ-H2AX) foci. This appears to occur irrespective of whether the MN is intact or compromised [Cimini et al. 2001] and has been correlated with impaired DNA replication. However, the precise mechanism underlying DNA damage in MN and how this phenomenon might be linked to the massive accumulation of fragmented chromosomes leading to chromothripsis is still unclear.
Figure 4.
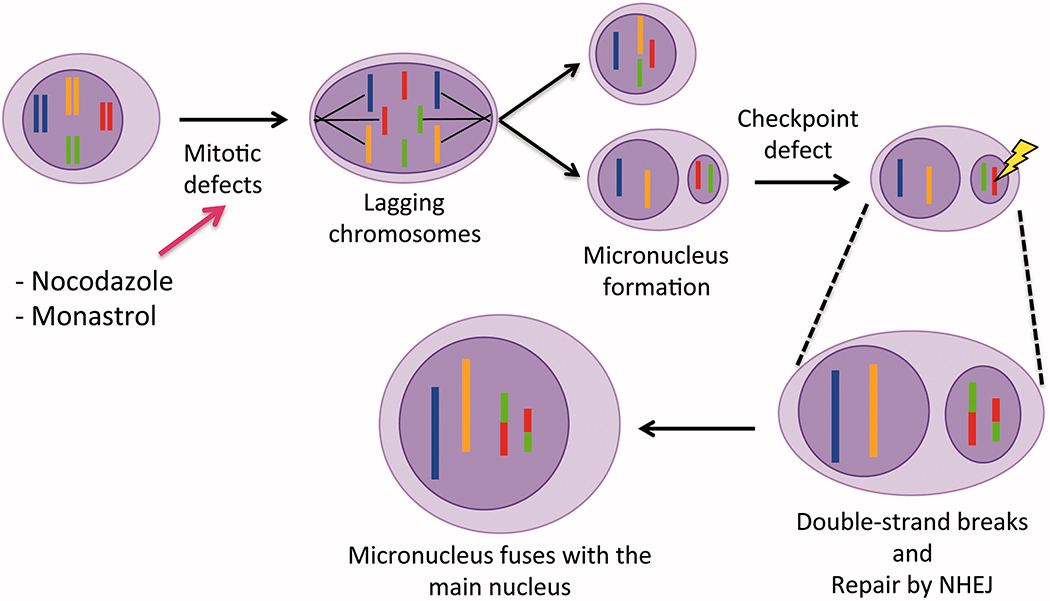
Model describing the association between chromosome mis-segregation, micronuclei, and chromosomal rearrangements. Chromosomes that are lagging during anaphase as a consequence of mitotic defects are encapsulated by nuclear envelope to form a micronucleus (MN). Within the MN, chromosomes tend to sustain frequent double-strand breaks most likely due to changes in chromatin conformation. This induces repair by non-homologous end joining (NHEJ), an error prone mechanism that can produce chromosomal aberrations. As MN may merge back with the main nucleus, this will result in the rearranged chromosomes becoming a part of the genome as described by Janssen et al. [2011].
Regardless of its origins, chromothripsis has also been suggested to occur during pre-implantation development [Pellestor 2014; Pellestor et al. 2014] based on frequent observations of MN formation, cellular fragmentation, and abnormal mitotic divisions in cleavage embryos [Chavez et al. 2012]. Whether aneuploid embryos undergo cellular fragmentation as a survival mechanism or to initiate their demise remains unknown, but we note that cancer cells undergo chromosomal rejoining following fragmentation as a survival strategy in response to cellular stress [Horne and Heng 2014; Liu et al. 2014]. Although a lack of control over the maintenance of genome stability seems contradictory to normal embryo survival, aspects such as heightened cell cycle drivers and diminished checkpoints might make blastomeres more vulnerable to chromosome lagging during early mitotic divisions [Harrison et al. 2000; Kiessling et al. 2010]. Consequently, MN formation may occur and contribute to the high rate of embryonic and fetal aneuploidy observed in vitro and in vivo [Benkhalifa et al. 2005; Chavez et al. 2012; Fritz et al. 2001; Johnson et al. 2010b; Vanneste et al. 2009]. Thus, identification of the genes linked to these functions and the genetic mutations that lead to abnormal cell divisions, chromosome mis-segregation, and MN formation is required for potential therapeutic intervention in both embryology and cancer pathology. In addition, an examination of MN chromosomal content and cellular fate in detail may also assist in determining the similarities and/or differences between embryonic micronuclei [Chavez et al. 2012] and the MN that are prevalent in cancer cells [Crasta et al. 2012; Huang et al. 2012].
Chromosomal Rearrangements and Species Evolution
Although chromosome instability has been extensively examined in the context of cancer biology and reproductive medicine, recent studies have shown that analogous mechanisms might also be behind species evolution. The importance of chromosomal rearrangements in species differentiation was recognized early on by Dobzhansky and Sturtevant in Drosophilid species [Dobzhansky and Sturtevant 1938]. This was later expanded upon by King [1993] and White [1969], who demonstrated that after inversions become fixed in a subpopulation, the hybridization between neighbor populations produced sterile hybrids. As part of the chromosomal speciation theory, it is thought that meiotic recombination between rearranged chromosomes results in unbalanced gametes causing infertility and reproductive isolation within the population. Although cytogenetic studies suggest that most placental mammals have karyotypes similar to the inferred ‘ancestral karyotype’ [Wienberg 2004], comparative genomics has shown that some mutations, including simple point substitutions are relatively frequent. However, other genetic changes such as large-scale chromosomal rearrangements are far less common in the majority of mammalian species. Given that these events are likely to be deleterious, it is not surprising that they tend to be eliminated by natural selection. It is important to note, however, that not all mammals maintain a slow rate of karyotypic evolution since the mouse, dog, and horse are exceptions to this rule. Within primates, gibbons exhibit the most accelerated rate of chromosome evolution, roughly 20 fold higher than in other mammalian species [Carbone et al. 2006]. There are four gibbon genera that separated from each other in only 5 million years and they carry highly diverged [Capozzi et al. 2012] karyotypes, with chromosome numbers ranging from 38 to 52. Interestingly, with 19 recognized species, gibbons are also the most species-rich lineage among the hominoids. Population genetics demonstrated that the radiation of the four gibbon species was almost instantaneous, possibly as the result of a high number of chromosomal rearrangements, in combination with extensive bio-geographical changes occurring in the Sunda shelf around the time of the split [Veeramah et al. 2015]. This rapid divergence, along with extensive lineage sorting experienced by these species, are the main reasons why it has not been possible to reconstruct a consistent phylogenetic tree for the four gibbon genera thus far.
Through sequencing analysis of the gibbon genome reference, a novel gibbon-specific retrotransposon called LAVA [Carbone et al. 2012] was found to preferentially insert into genes whose products are involved in chromosome segregation processes. Retrotransposons, or so-called jumping genes, have been colonizing mammalian genomes for millions of years by creating new genetic copies via production of an RNA intermediate and a copy-and-paste mechanism. The LAVA element is a composite retrotransposon containing portions of repeats commonly found in primate genomes, including, L1, AluS, VNTR, and AluLIKE. Further analysis of the gibbon genome revealed the presence of more than 1,200 full-length LAVA insertions in the Nleu1.0 reference assembly with a preference for intragenic locations [Carbone et al. 2014]. Gene Ontology (GO) term and pathway analyses of genes carrying LAVA insertions also showed functional enrichment for genes whose protein products are involved in chromosome segregation processes, including mitotic spindle integrity/architecture (MAP4 [Samora et al. 2011], CEP164 [Leber et al. 2010], and BUB1B [Baker et al. 2009]), cell division checkpoints, as well as kinetochore assembly and attachment to the spindle (MAD1L1 [Schuyler et al. 2012] and CLASP2 [Maia et al. 2012]). The importance of these genes in several chromosome segregation processes is substantiated by the fact that some are mutated in cancer cells with chromosomal instability [Burum-Auensen et al. 2008]. Given that Carbone et al. [2014] showed that LAVA insertions have the potential to prematurely terminate transcription, it is plausible that LAVA-induced protein truncation in this group of chromosome segregation genes directly influences the elevated frequency of chromosome mis-segregation during meiosis and/or mitosis. This may help explain the high rate of evolutionary chromosomal rearrangements observed in gibbons. Since they are an endangered species, it would be difficult if not impossible to test this hypothesis in gibbons. However, it is possible to extrapolate results from studies in assisted reproduction and cancer biology to the gibbon and potentially uncover the precise mechanisms of increased chromosomal instability that drive the evolution of particular species.
Summary and Conclusions
Human infertility is relatively common and the use of IVF or other ARTs is likely to increase as reproductive-age couples continue to postpone having children [Mascarenhas et al. 2012]. Despite significant advances in treating certain forms of this disorder, IVF success rates still remain only ~30% and one of the main culprits is thought to be chromosomal abnormalities that arise during pre-implantation development (cdc.gov/art). As the tools and technologies necessary for comprehensive chromosomal analysis are further developed and optimized, especially at the single cell level, the direct cause(s) and potential consequence(s) of embryonic chromosomal instability may finally be realized for possible therapeutic intervention. While multiple factors are likely to be involved in aneuploidy generation, micronuclei formation, and cellular fragmentation during early embryogenesis, the findings from these studies may easily translate to cancer risk as well as the shared chromosomal aspects that drive species evolution. Collectively, the ultimate goals of this research and future work is to select and transfer the embryo(s) with the greatest likelihood to result in a normal term pregnancy and enhance our knowledge of the chromosomal requirements of pre-implantation development across different species and the implications for somatic health.
Acknowledgments
We gratefully acknowledge the National Centers for Translational Research in Reproduction and Infertility (NCTRI)/NICHD (P50 HD071836), Howard and Georgeanna Jones Foundation for Reproductive Medicine, Medical Research Foundation of Oregon, and the Collins Medical Trust to SLC for funding.
Footnotes
Declaration of interest
The authors report no conflicts of interest.
References
- Alikani M, Cohen J, Tomkin G, Garrisi GJ, Mack C and Scott RT (1999) Human embryo fragmentation in vitro and its implications for pregnancy and implantation. Fertil Steril 71:836–842. [DOI] [PubMed] [Google Scholar]
- Alizadeh Z, Kageyama S and Aoki F (2005) Degradation of maternal mRNA in mouse embryos: Selective degradation of specific mRNAs after fertilization. Mol Reprod Dev 72:281–290. [DOI] [PubMed] [Google Scholar]
- Alper MM, Brinsden P, Fischer R and Wikland M (2001) To blastocyst or not to blastocyst? That is the question. Hum Reprod 16:617–619. [DOI] [PubMed] [Google Scholar]
- Antczak M and Van Blerkom J (1999) Temporal and spatial aspects of fragmentation in early human embryos: Possible effects on developmental competence and association with the differential elimination of regulatory proteins from polarized domains. Hum Reprod 14:429–447. [DOI] [PubMed] [Google Scholar]
- Baart EB, Martini E, van den Berg I, Macklon NS, Galjaard RJ, Fauser BC, et al. (2006) Preimplantation genetic screening reveals a high incidence of aneuploidy and mosaicism in embryos from young women undergoing IVF. Hum Reprod 21:223–233. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Jin F, Jeganathan KB and van Deursen JM (2009) Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell 16:475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltaci V, Satiroglu H, Kabukcu C, Unsal E, Aydinuraz B, Uner O, et al. (2006) Relationship between embryo quality and aneuploidies. Reprod Biomed Online 12:77–82. [DOI] [PubMed] [Google Scholar]
- Barbash-Hazan S, Frumkin T, Malcov M, Yaron Y, Cohen T, Azem F, et al. (2009) Preimplantation aneuploid embryos undergo self-correction in correlation with their developmental potential. Fertil Steril 92:890–896. [DOI] [PubMed] [Google Scholar]
- Basile N, Nogales Mdel C, Bronet F, Florensa M, Riqueiros M, Rodrigo L, et al. (2014) Increasing the probability of selecting chromosomally normal embryos by time-lapse morphokinetics analysis. Fertil Steril 101:699–704. [DOI] [PubMed] [Google Scholar]
- Bavister BD, Boatman DE, Collins K, Dierschke DJ and Eisele SG (1984) Birth of rhesus monkey infant after in vitro fertilization and nonsurgical embryo transfer. Proc Natl Acad Sci USA 81:2218–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CE, Calder MD and Watson AJ (2008) Genomic RNA profiling and the programme controlling preimplantation mammalian development. Mol Hum Reprod 14:691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkhalifa M, Kasakyan S, Clement P, Baldi M, Tachdjian G, Demirol A, et al. (2005) Array comparative genomic hybridization profiling of first-trimester spontaneous abortions that fail to grow in vitro. Prenat Diagn 25:894–900. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, et al. (2004) Genome-wide survey of protein kinases required for cell cycle progression. Nature 432:980–987. [DOI] [PubMed] [Google Scholar]
- Boulet SL, Mehta A, Kissin DM, Warner L, Kawwass JF and Jamieson DJ (2015) Trends in use of and reproductive outcomes associated with intracytoplasmic sperm injection. J Am Med Assoc 313:255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackett BG, Bousquet D, Boice ML, Donawick WJ, Evans JF and Dressel MA (1982) Normal development following in vitro fertilization in the cow. Biol Reprod 27:147–158. [DOI] [PubMed] [Google Scholar]
- Braude P, Bolton V and Moore S (1988) Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 332:459–461. [DOI] [PubMed] [Google Scholar]
- Brunet-Simon A, Henrion G, Renard JP and Duranthon V (2001) Onset of zygotic transcription and maternal transcript legacy in the rabbit embryo. Mol Reprod Dev 58:127–136. [DOI] [PubMed] [Google Scholar]
- Burruel V, Klooster K, Barker CM, Pera RR and Meyers S (2014) Abnormal early cleavage events predict early embryo demise: Sperm oxidative stress and early abnormal cleavage. Sci Rep 4:6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burum-Auensen E, DeAngelis PM, Schjolberg AR, Roislien J, Mjaland O and Clausen OP (2008) Reduced level of the spindle checkpoint protein BUB1B is associated with aneuploidy in colorectal cancers. Cell Prolif 41:645–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buster JE, Bustillo M, Rodi IA, Cohen SW, Hamilton M, Simon JA, et al. (1985) Biologic and morphologic development of donated human ova recovered by nonsurgical uterine lavage. Am J Obstet Gyn 153:211–217. [DOI] [PubMed] [Google Scholar]
- Campbell A, Fishel S, Bowman N, Duffy S, Sedler M and Hickman CF (2013) Modelling a risk classification of aneuploidy in human embryos using non-invasive morphokinetics. Reprod Biomed Online 26:477–485. [DOI] [PubMed] [Google Scholar]
- Capalbo A, Wright G, Elliott T, Ubaldi FM, Rienzi L and Nagy ZP (2013) FISH reanalysis of inner cell mass and trophectoderm samples of previously array-CGH screened blastocysts shows high accuracy of diagnosis and no major diagnostic impact of mosaicism at the blastocyst stage. Hum Reprod 28:2298–2307. [DOI] [PubMed] [Google Scholar]
- Capozzi O, Carbone L, Stanyon RR, Marra A, Yang F, Whelan CW, et al. (2012) A comprehensive molecular cytogenetic analysis of chromosome rearrangements in gibbons. Genome Res 22:2520–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Harris RA, Gnerre S, Veeramah KR, Lorente-Galdos B, Huddleston J, et al. (2014) Gibbon genome and the fast karyotype evolution of small apes. Nature 513:195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Harris RA, Mootnick AR, Milosavljevic A, Martin DI, Rocchi M, et al. (2012) Centromere remodeling in Hoolock leuconedys (Hylobatidae) by a new transposable element unique to the gibbons. Genome Biol Evol 4:648–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Vessere GM, ten Hallers BF, Zhu B, Osoegawa K, Mootnick A, et al. (2006) A high-resolution map of synteny disruptions in gibbon and human genomes. PLoS Genet 2:e223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez SL, Loewke KE, Han J, Moussavi F, Colls P, Munne S, et al. (2012) Dynamic blastomere behaviour reflects human embryo ploidy by the four-cell stage. Nat Commun 3:1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez SL, McElroy SL, Bossert NL, De Jonge CJ, Rodriguez MV, Leong DE, et al. (2014) Comparison of epigenetic mediator expression and function in mouse and human embryonic blastomeres. Hum Mol Genet 23:4970–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F and Salmon ED (2001) Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol 153:517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn K and Rossant J (2010) Making the blastocyst: Lessons from the mouse. J Clin Invest 120:995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaghan J, Chen AA, Willman SP, Ivani K, Chenette PE, Boostanfar R, et al. (2013) Improving embryo selection using a computer-automated time-lapse image analysis test plus day 3 morphology: Results from a prospective multicenter trial. Fertil Steril 100:412–419 e415. [DOI] [PubMed] [Google Scholar]
- Coonen E, Derhaag JG, Dumoulin JC, van Wissen LC, Bras M, Janssen M, et al. (2004) Anaphase lagging mainly explains chromosomal mosaicism in human preimplantation embryos. Hum Reprod 19:316–324. [DOI] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, et al. (2012) DNA breaks and chromosome pulverization from errors in mitosis. Nature 482:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby IM, Gandolfi F and Moor RM (1988) Control of protein synthesis during early cleavage of sheep embryos. J Reprod Fertil 82:769–775. [DOI] [PubMed] [Google Scholar]
- Cruz M, Gadea B, Garrido N, Pedersen KS, Martinez M, Perez-Cano I, et al. (2011) Embryo quality, blastocyst and ongoing pregnancy rates in oocyte donation patients whose embryos were monitored by time-lapse imaging. J Assist Reprod Genet 28:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JC Jr,. (1965) Studies on the Growth of 5-Day-Old Rabbit Blastocysts in Vitro. J Embryol Exp Morphol 13:83–95. [PubMed] [Google Scholar]
- Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, et al. (2008) Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell 133:103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrinsky JR, Johnson LA and Rath D (1996) Development of a culture medium (BECM-3) for porcine embryos: Effects of bovine serum albumin and fetal bovine serum on embryo development. Biol Reprod 55:1069–1074. [DOI] [PubMed] [Google Scholar]
- Dobson AT, Raja R, Abeyta MJ, Taylor T, Shen S, Haqq C, et al. (2004) The unique transcriptome through day 3 of human preimplantation development. Hum Mol Genet 13:1461–1470. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T and Sturtevant AH (1938) Inversions in the Chromosomes of Drosophila Pseudoobscura. Genetics 23:28–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont C, Segars J, DeCherney A, Bavister BD, Armant DR and Brenner CA (2010) Incidence of chromosomal mosaicism in morphologically normal nonhuman primate preimplantation embryos. Fertil Steril 93:2545–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RG, Bavister BD and Steptoe PC (1969) Early stages of fertilization in vitro of human oocytes matured in vitro. Nature 221:632–635. [DOI] [PubMed] [Google Scholar]
- Edwards RG, Fishel SB, Cohen J, Fehilly CB, Purdy JM, Slater JM, et al. (1984) Factors influencing the success of in vitro fertilization for alleviating human infertility. J In Vitro Fert Embryo Transf 1:3–23. [DOI] [PubMed] [Google Scholar]
- Edwards RG, Purdy JM, Steptoe PC and Walters DE (1981) The growth of human preimplantation embryos in vitro. Am J Obstet Gyn 141:408–416. [DOI] [PubMed] [Google Scholar]
- Edwards RG, Steptoe PC and Purdy JM (1970) Fertilization and cleavage in vitro of preovulator human oocytes. Nature 227:1307–1309. [DOI] [PubMed] [Google Scholar]
- Enders AC, Hendrickx AG and Binkerd PE (1982) Abnormal development of blastocysts and blastomeres in the rhesus monkey. Biol Reprod 26:353–366. [DOI] [PubMed] [Google Scholar]
- Enders AC, Lantz KC and Schlafke S (1990) The morula-blastocyst transition in two Old World primates: The baboon and rhesus monkey. J Med Primatol 19:725–747. [PubMed] [Google Scholar]
- Evans J, Hannan NJ, Edgell TA, Vollenhoven BJ, Lutjen PJ, Osianlis T, et al. (2014) Fresh versus frozen embryo transfer: Backing clinical decisions with scientific and clinical evidence. Hum Reprod Update 20:808–821. [DOI] [PubMed] [Google Scholar]
- Evsikov AV, Graber JH, Brockman JM, Hampl A, Holbrook AE, Singh P, et al. (2006) Cracking the egg: Molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo. Genes Dev 20:2713–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi HM, Kyrou D, Bourgain C, Van den Abbeel E, Griesinger G and Devroey P (2010) Cryopreserved-thawed human embryo transfer: Spontaneous natural cycle is superior to human chorionic gonadotropin-induced natural cycle. Fertil Steril 94:2054–2058. [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, de Dios Hourcade J, Lopez-Vidriero I, Benguria A, De Fonseca FR and Gutierrez-Adan A (2009) Analysis of gene transcription alterations at the blastocyst stage related to the long-term consequences of in vitro culture in mice. Reproduction 137:271–283. [DOI] [PubMed] [Google Scholar]
- Fiorentino F, Bono S, Biricik A, Nuccitelli A, Cotroneo E, Cottone G, et al. (2014) Application of next-generation sequencing technology for comprehensive aneuploidy screening of blastocysts in clinical preimplantation genetic screening cycles. Hum Reprod 29:2802–2813. [DOI] [PubMed] [Google Scholar]
- Flach G, Johnson MH, Braude PR, Taylor RA and Bolton VN (1982) The transition from maternal to embryonic control in the 2-cell mouse embryo. EMBO 1:681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragouli E, Alfarawati S, Daphnis DD, Goodall NN, Mania A, Griffiths T, et al. (2011) Cytogenetic analysis of human blastocysts with the use of FISH, CGH and aCGH: Scientific data and technical evaluation. Hum Reprod 26:480–490. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Lenzi M, Ross R, Katz-Jaffe M, Schoolcraft WB and Wells D (2008) Comprehensive molecular cytogenetic analysis of the human blastocyst stage. Hum Reprod 23:2596–2608. [DOI] [PubMed] [Google Scholar]
- Franasiak JM, Forman EJ, Hong KH, Werner MD, Upham KM, Treff NR, et al. (2014) The nature of aneuploidy with increasing age of the female partner: A review of 15,169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertil Steril 101:656–663 e651. [DOI] [PubMed] [Google Scholar]
- Fritz B, Hallermann C, Olert J, Fuchs B, Bruns M, Aslan M, et al. (2001) Cytogenetic analyses of culture failures by comparative genomic hybridisation (CGH)-Re-evaluation of chromosome aberration rates in early spontaneous abortions. Eur J Hum Genet 9:539–547. [DOI] [PubMed] [Google Scholar]
- Galan A, Montaner D, Poo ME, Valbuena D, Ruiz V, Aguilar C, et al. (2010) Functional genomics of 5- to 8-cell stage human embryos by blastomere single-cell cDNA analysis. PloS One 5:e13615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner DK, Lane M, Spitzer A and Batt PA (1994) Enhanced rates of cleavage and development for sheep zygotes cultured to the blastocyst stage in vitro in the absence of serum and somatic cells: Amino acids, vitamins, and culturing embryos in groups stimulate development. Biol Reprod 50:390–400. [DOI] [PubMed] [Google Scholar]
- Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, et al. (2014) The DNA methylation landscape of human early embryos. Nature 511:606–610. [DOI] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA and Ko MS (2004) Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell 6:117–131. [DOI] [PubMed] [Google Scholar]
- Hardarson T, Lofman C, Coull G, Sjogren A, Hamberger L and Edwards RG (2002) Internalization of cellular fragments in a human embryo: Time-lapse recordings. Reprod Biomed Online 5:36–38. [DOI] [PubMed] [Google Scholar]
- Hardy K (1999) Apoptosis in the human embryo. Rev Reprod 4:125–134. [DOI] [PubMed] [Google Scholar]
- Hardy K, Handyside AH and Winston RM (1989) The human blastocyst: Cell number, death and allocation during late preimplantation development in vitro. Development 107:597–604. [DOI] [PubMed] [Google Scholar]
- Hardy K, Spanos S, Becker D, Iannelli P, Winston RM and Stark J (2001) From cell death to embryo arrest: Mathematical models of human preimplantation embryo development. Proc Natl Acad Sci U S A 98:1655–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JC and Sengupta SB (2012) Preimplantation genetic diagnosis: State of the art 2011. Hum Genet 131:175–186. [DOI] [PubMed] [Google Scholar]
- Harrison RH, Kuo HC, Scriven PN, Handyside AH and Ogilvie CM (2000) Lack of cell cycle checkpoints in human cleavage stage embryos revealed by a clonal pattern of chromosomal mosaicism analysed by sequential multicolour FISH. Zygote 8:217–224. [DOI] [PubMed] [Google Scholar]
- Hassold T and Hunt P (2001) To err (meiotically) is human: The genesis of human aneuploidy. Nat Rev Genet 2:280–291. [DOI] [PubMed] [Google Scholar]
- Hatch EM, Fischer AH, Deerinck TJ and Hetzer MW (2013) Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154:47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertig A, Rock J, Adams E and Mulligan W (1954) On the preimplantation stages of the human ovum: A description of four normal and four abnormal specimens ranging from the second to the fifth day of development. Contrib Embryol Carnegie Instn 35:119–220. [Google Scholar]
- Hertig AT, Rock J and Adams EC (1956) A description of 34 human ova within the first 17 days of development. Am J Anat 98:435–493. [DOI] [PubMed] [Google Scholar]
- Hoffelder DR, Luo L, Burke NA, Watkins SC, Gollin SM and Saunders WS (2004) Resolution of anaphase bridges in cancer cells. Chromosoma 112:389–397. [DOI] [PubMed] [Google Scholar]
- Horne SD, Abdallah BY, Stevens JB, Liu G, Ye KJ, Bremer SW, et al. (2013) Genome constraint through sexual reproduction: Application of 4D-Genomics in reproductive biology. Syst Biol Reprod Med 59:124–130. [DOI] [PubMed] [Google Scholar]
- Horne SD and Heng H (2014) Genome Chaos, Chromothripsis and Cancer Evolution. J Cancer Stud Ther 1:1–6. [Google Scholar]
- Hou Y, Fan W, Yan L, Li R, Lian Y, Huang J, et al. (2013) Genome analyses of single human oocytes. Cell 155:1492–1506. [DOI] [PubMed] [Google Scholar]
- Houliston E and Maro B (1989) Posttranslational modification of distinct microtubule subpopulations during cell polarization and differentiation in the mouse preimplantation embryo. J Cell Biol 108:543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A, Adusumalli J, Patel S, Liem J, Williams J III, and Pisarska MD (2009) Prevalence of chromosomal mosaicism in pregnancies from couples with infertility. Fertil Steril 91:2355–2360. [DOI] [PubMed] [Google Scholar]
- Huang Y, Jiang L, Yi Q, Lv L, Wang Z, Zhao X, et al. (2012) Lagging chromosomes entrapped in micronuclei are not ‘lost’ by cells. Cell Res 22:932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyttel P, Laurincik J, Rosenkranz C, Rath D, Niemann H, Ochs RL, et al. (2000) Nucleolar proteins and ultrastructure in preimplantation porcine embryos developed in vivo. Biol Reprod 63:1848–1856. [DOI] [PubMed] [Google Scholar]
- Janssen A, van der Burg M, Szuhai K, Kops GJ and Medema RH (2011) Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 333:1895–1898. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Cinnioglu C, Ross R, Filby A, Gemelos G, Hill M, et al. (2010a) Comprehensive analysis of karyotypic mosaicism between trophectoderm and inner cell mass. Mol Hum Reprod 16:944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Gemelos G, Baner J, Ryan A, Cinnioglu C, Banjevic M, et al. (2010b) Preclinical validation of a microarray method for full molecular karyotyping of blastomeres in a 24-h protocol. Hum Reprod 25:1066–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MH and Maro B (1984) The distribution of cytoplasmic actin in mouse 8-cell blastomeres. J Embryol Exp Morphol 82:97–117. [PubMed] [Google Scholar]
- Johnson MH and Ziomek CA (1981) The foundation of two distinct cell lineages within the mouse morula. Cell 24:71–80. [DOI] [PubMed] [Google Scholar]
- Kallen B, Finnstrom O, Lindam A, Nilsson E, Nygren KG and Olausson PO (2010) Blastocyst versus cleavage stage transfer in in vitro fertilization: Differences in neonatal outcome? Fertil Steril 94:1680–1683. [DOI] [PubMed] [Google Scholar]
- Kaser DJ and Racowsky C (2014) Clinical outcomes following selection of human preimplantation embryos with time-lapse monitoring: A systematic review. Hum Reprod Update 20:617–631. [DOI] [PubMed] [Google Scholar]
- Katari S, Turan N, Bibikova M, Erinle O, Chalian R, Foster M, et al. (2009) DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet 18:3769–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskintepe L, Burnley CA and Brackett BG (1995) Production of viable bovine blastocysts in defined in vitro conditions. Biol Reprod 52:1410–1417. [DOI] [PubMed] [Google Scholar]
- Khosla S, Dean W, Reik W and Feil R (2001) Culture of preimplantation embryos and its long-term effects on gene expression and phenotype. Hum Reprod Update 7:419–427. [DOI] [PubMed] [Google Scholar]
- Kidder GM and McLachlin JR (1985) Timing of transcription and protein synthesis underlying morphogenesis in preimplantation mouse embryos. Dev Biol 112:265–275. [DOI] [PubMed] [Google Scholar]
- Kiessling AA, Bletsa R, Desmarais B, Mara C, Kallianidis K and Loutradis D (2010) Genome-wide microarray evidence that 8-cell human blastomeres over-express cell cycle drivers and under-express checkpoints. J Assist Reprod Genet 27:265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M (1993) Species Evolution: The Role of Chromosome Change. Cambridge University Press, New York: pp. xxi; 1–336. [Google Scholar]
- Kirkegaard K, Kesmodel US, Hindkjaer JJ and Ingerslev HJ (2013) Time-lapse parameters as predictors of blastocyst development and pregnancy outcome in embryos from good prognosis patients: A prospective cohort study. Hum Reprod 28:2643–2651. [DOI] [PubMed] [Google Scholar]
- Kissin DM, Kawwass JF, Monsour M, Boulet SL, Session DR and Jamieson DJ (2014) Assisted hatching: Trends and pregnancy outcomes, United States, 2000-2010. Fertil Steril 102:795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo HC, Ogilvie CM and Handyside AH (1998) Chromosomal mosaicism in cleavage-stage human embryos and the accuracy of single-cell genetic analysis. J Assist Reprod Genet 15:276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedev IN, Ostroverkhova NV, Nikitina TV, Sukhanova NN and Nazarenko SA (2004) Features of chromosomal abnormalities in spontaneous abortion cell culture failures detected by interphase FISH analysis. Eur J Hum Genet 12:513–520. [DOI] [PubMed] [Google Scholar]
- Leber B, Maier B, Fuchs F, Chi J, Riffel P, Anderhub S, et al. (2010) Proteins required for centrosome clustering in cancer cells. Sci Transl Med 2:33ra38. [DOI] [PubMed] [Google Scholar]
- Lee KF and Yeung WS (2006) Gamete/embryo - oviduct interactions: Implications on in vitro culture. Hum Fertil 9:137–143. [DOI] [PubMed] [Google Scholar]
- Lemmen JG, Agerholm I and Ziebe S (2008) Kinetic markers of human embryo quality using time-lapse recordings of IVF/ICSI-fertilized oocytes. Reprod Biomed Online 17:385–391. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW and Vogelstein B (1997) Genetic instability in colorectal cancers. Nature 386:623–627. [DOI] [PubMed] [Google Scholar]
- Levy JB, Johnson MH, Goodall H and Maro B (1986) The timing of compaction: Control of a major developmental transition in mouse early embryogenesis. J Embryol Exp Morphol 95:213–237. [PubMed] [Google Scholar]
- Li L, Baibakov B and Dean J (2008) A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell 15:416–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zheng P and Dean J (2010) Maternal control of early mouse development. Development 137:859–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot DA, Kouznetsova A, Mahdy E, Wilbertz J and Hoog C (2006) The fate of mosaic aneuploid embryos during mouse development. Dev Biol 289:384–394. [DOI] [PubMed] [Google Scholar]
- Liu G, Stevens JB, Horne SD, Abdallah BY, Ye KJ, Bremer SW, et al. (2014) Genome chaos: Survival strategy during crisis. Cell Cycle 13:528–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang W, Sun X, Liu L, Jin H, Li M, et al. (2012) DNA microarray reveals that high proportions of human blastocysts from women of advanced maternal age are aneuploid and mosaic. Biol Reprod 87:1–9. [DOI] [PubMed] [Google Scholar]
- Macklon NS, Geraedts JP and Fauser BC (2002) Conception to ongoing pregnancy: The ‘black box’ of early pregnancy loss. Hum Reprod Update 8:333–343. [DOI] [PubMed] [Google Scholar]
- Magli MC, Jones GM, Gras L, Gianaroli L, Korman I and Trounson AO (2000) Chromosome mosaicism in day 3 aneuploid embryos that develop to morphologically normal blastocysts in vitro. Hum Reprod 15:1781–1786. [DOI] [PubMed] [Google Scholar]
- Maia AR, Garcia Z, Kabeche L, Barisic M, Maffini S, Macedo-Ribeiro S, et al. (2012) Cdk1 and Plk1 mediate a CLASP2 phospho-switch that stabilizes kinetochore-microtubule attachments. J Cell Biol 199:285–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantikou E, Wong KM, Repping S and Mastenbroek S (2012) Molecular origin of mitotic aneuploidies in preimplantation embryos. Biochim Biophys Acta 1822:1921–1930. [DOI] [PubMed] [Google Scholar]
- Mascarenhas MN, Flaxman SR, Boerma T, Vanderpoel S and Stevens GA (2012) National, regional, and global trends in infertility prevalence since 1990: A systematic analysis of 277 health surveys. PLoS Med 9:e1001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastenbroek S, Twisk M, van Echten-Arends J, Sikkema-Raddatz B, Korevaar JC, Verhoeve HR, et al. (2007) In vitro fertilization with preimplantation genetic screening. N Engl J Med 357:9–17. [DOI] [PubMed] [Google Scholar]
- Munne S, Grifo J, Cohen J and Weier HU (1994) Chromosome abnormalities in human arrested preimplantation embryos: A multiple-probe FISH study. Am J Hum Genet 55:150–159. [PMC free article] [PubMed] [Google Scholar]
- Munne S, Velilla E, Colls P, Garcia Bermudez M, Vemuri MC, Steuerwald N, et al. (2005) Self-correction of chromosomally abnormal embryos in culture and implications for stem cell production. Fertil Steril 84:1328–1334. [DOI] [PubMed] [Google Scholar]
- Munne S and Weier HU (1996) Simultaneous enumeration of chromosomes 13, 18, 21, X, and Y in interphase cells for preimplantation genetic diagnosis of aneuploidy. Cytogenet Cell Genet 75:263–270. [DOI] [PubMed] [Google Scholar]
- Nagaoka SI, Hassold TJ and Hunt PA (2012) Human aneuploidy: Mechanisms and new insights into an age-old problem. Nat Rev Genet 13:493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara T, Iwase A, Goto M, Harata T, Suzuki M, Ienaga M, et al. (2010) Evaluation of the safety of time-lapse observations for human embryos. J Assist Reprod Genet 27:93–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niakan KK, Han J, Pedersen RA, Simon C and Pera RA (2012) Human pre-implantation embryo development. Development 139:829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novik V, Moulton EB, Sisson ME, Shrestha SL, Tran KD, Stern HJ, et al. (2014) The accuracy of chromosomal microarray testing for identification of embryonic mosaicism in human blastocysts. Mol Cytogenet 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo G, Munne S and Cohen J (1994) The human zygote inherits its mitotic potential from the male gamete. Hum Reprod 9:1220–1225. [DOI] [PubMed] [Google Scholar]
- Payne D, Flaherty SP, Barry MF and Matthews CD (1997) Preliminary observations on polar body extrusion and pronuclear formation in human oocytes using time-lapse video cinematography. Hum Reprod 12:532–541. [DOI] [PubMed] [Google Scholar]
- Pelinck MJ, Hoek A, Simons AH, Heineman MJ, van Echten-Arends J and Arts EG (2010) Embryo quality and impact of specific embryo characteristics on ongoing implantation in unselected embryos derived from modified natural cycle in vitro fertilization. Fertil Steril 94:527–534. [DOI] [PubMed] [Google Scholar]
- Pellestor F (2014) Chromothripsis: How does such a catastrophic event impact human reproduction?. Hum Reprod 29:388–393. [DOI] [PubMed] [Google Scholar]
- Pellestor F, Gatinois V, Puechberty J, Genevieve D and Lefort G (2014) Chromothripsis: Potential origin in gametogenesis and preimplantation cell divisions. A review. Fertil Steril 102:1785–1796. [DOI] [PubMed] [Google Scholar]
- Peramo B, Ricciarelli E, Cuadros-Fernandez JM, Huguet E and Hernandez ER (1999) Blastocyst transfer and monozygotic twinning. Fertil Steril 72:1116–1117. [DOI] [PubMed] [Google Scholar]
- Pereda J and Croxatto HB (1978) Ultrastructure of a seven-cell human embryo. Biol Reprod 18:481–489. [DOI] [PubMed] [Google Scholar]
- Pessot CA, Brito M, Figueroa J, Concha II, Yanez A and Burzio LO (1989) Presence of RNA in the sperm nucleus. Biochem Biophys Res Commun 158:272–278. [DOI] [PubMed] [Google Scholar]
- Pierre A, Gautier M, Callebaut I, Bontoux M, Jeanpierre E, Pontarotti P, et al. (2007) Atypical structure and phylogenomic evolution of the new eutherian oocyte- and embryo-expressed KHDC1/DPPA5/ECAT1/OOEP gene family. Genomics 90:583–594. [DOI] [PubMed] [Google Scholar]
- Plante L, Plante C, Shepherd DL and King WA (1994) Cleavage and 3H-uridine incorporation in bovine embryos of high in vitro developmental potential. Mol Reprod Dev 39:375–383. [DOI] [PubMed] [Google Scholar]
- Pribenszky C, Losonczi E, Molnar M, Lang Z, Matyas S, Rajczy K, et al. (2010) Prediction of in-vitro developmental competence of early cleavage-stage mouse embryos with compact time-lapse equipment. Reprod Biomed Online 20:371–379. [DOI] [PubMed] [Google Scholar]
- Reeve WJ and Kelly FP (1983) Nuclear position in the cells of the mouse early embryo. J Embryol Exp Morphol 75:117–139. [PubMed] [Google Scholar]
- Reima I, Lehtonen E, Virtanen I and Flechon JE (1993) The cytoskeleton and associated proteins during cleavage, compaction and blastocyst differentiation in the pig. Differentiation 54:35–45. [DOI] [PubMed] [Google Scholar]
- Rubio I, Galan A, Larreategui Z, Ayerdi F, Bellver J, Herrero J, et al. (2014) Clinical validation of embryo culture and selection by morphokinetic analysis: A randomized, controlled trial of the EmbryoScope. Fertil Steril 102:1287–1294. [DOI] [PubMed] [Google Scholar]
- Samora CP, Mogessie B, Conway L, Ross JL, Straube A and McAinsh AD (2011) MAP4 and CLASP1 operate as a safety mechanism to maintain a stable spindle position in mitosis. Nat Cell Biol 13:1040–1050. [DOI] [PubMed] [Google Scholar]
- Sathananthan AH, Kola I, Osborne J, Trounson A, Ng SC, Bongso A, et al. (1991) Centrioles in the beginning of human development. Proc Natl Acad Sci U S A 88:4806–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatten G, Simerly C and Schatten H (1991) Maternal inheritance of centrosomes in mammals? Studies on parthenogenesis and polyspermy in mice. Proc Natl Acad Sci U S A 88:6785–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoolcraft WB, Fragouli E, Stevens J, Munne S, Katz-Jaffe MG and Wells D (2010) Clinical application of comprehensive chromosomal screening at the blastocyst stage. Fertil Steril 94:1700–1706. [DOI] [PubMed] [Google Scholar]
- Schramm RD and Bavister BD (1999) Onset of nucleolar and extranucleolar transcription and expression of fibrillarin in macaque embryos developing in vitro. Biol Reprod 60:721–728. [DOI] [PubMed] [Google Scholar]
- Schuyler SC, Wu YF and Kuan VJ (2012) The Mad1–Mad2 balancing act–a damaged spindle checkpoint in chromosome instability and cancer. J Cell Sci 125:4197–4206. [DOI] [PubMed] [Google Scholar]
- Scott RT Jr., Upham KM, Forman EJ, Zhao T and Treff NR (2013) Cleavage-stage biopsy significantly impairs human embryonic implantation potential while blastocyst biopsy does not: A randomized and paired clinical trial. Fertil Steril 100:624–630. [DOI] [PubMed] [Google Scholar]
- Sendler E, Johnson GD, Mao SH, Goodrich RJ, Diamond MP, Hauser R, et al. (2013) Stability, delivery and functions of human sperm RNAs at fertilization. Nuc Acids Res 41:4104–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshagiri PB and Hearn JP (1993) In-vitro development of in-vivo produced rhesus monkey morulae and blastocysts to hatched, attached, and post-attached blastocyst stages: Morphology and early secretion of chorionic gonadotrophin. Hum Reprod 8:279–287. [DOI] [PubMed] [Google Scholar]
- Sheth B, Fesenko I, Collins JE, Moran B, Wild AE, Anderson JM, et al. (1997) Tight junction assembly during mouse blastocyst formation is regulated by late expression of ZO-1 alpha+ isoform. Development 124:2027–2037. [DOI] [PubMed] [Google Scholar]
- Silber S, Escudero T, Lenahan K, Abdelhadi I, Kilani Z and Munne S (2003) Chromosomal abnormalities in embryos derived from testicular sperm extraction. Fertil Steril 79:30–38. [DOI] [PubMed] [Google Scholar]
- Slama R, Eustache F, Ducot B, Jensen TK, Jorgensen N, Horte A, et al. (2002) Time to pregnancy and semen parameters: A cross-sectional study among fertile couples from four European cities. Hum Reprod 17:503–515. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A, et al. (2014) DNA methylation dynamics of the human preimplantation embryo. Nature 511:611–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, et al. (2011) Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steptoe PC, Edwards RG and Purdy JM (1971) Human blastocysts grown in culture. Nature 229:132–133. [DOI] [PubMed] [Google Scholar]
- Stevens JB, Abdallah BY, Liu G, Ye CJ, Horne SD, Wang G, et al. (2011) Diverse system stresses: Common mechanisms of chromosome fragmentation. Cell Death Dis 2:e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JB, Liu G, Bremer SW, Ye KJ, Xu W, Xu J, et al. (2007) Mitotic cell death by chromosome fragmentation. Cancer Res 67:7686–7694. [DOI] [PubMed] [Google Scholar]
- Sturmey RG, Hawkhead JA, Barker EA and Leese HJ (2009) DNA damage and metabolic activity in the preimplantation embryo. Hum Reprod 24:81–91. [DOI] [PubMed] [Google Scholar]
- Sugimura S, Akai T, Hashiyada Y, Somfai T, Inaba Y, Hirayama M, et al. (2012) Promising system for selecting healthy in vitro-fertilized embryos in cattle. PLoS One 7:e36627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura S, Akai T, Somfai T, Hirayama M, Aikawa Y, Ohtake M, et al. (2010) Time–lapse cinematography-compatible polystyrene-based microwell culture system: A novel tool for tracking the development of individual bovine embryos. Biol Reprod 83:970–978. [DOI] [PubMed] [Google Scholar]
- Surani MA (1975) Zona pellucida denudation, blastocyst proliferation and attachment in the rat. J Embryol Exp Morphol 33:343–353. [PubMed] [Google Scholar]
- Sutherland AE, Speed TP and Calarco PG (1990) Inner cell allocation in the mouse morula: The role of oriented division during fourth cleavage. Dev Biol 137:13–25. [DOI] [PubMed] [Google Scholar]
- Tarkowski AK and Wroblewska J (1967) Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. J Embryol Exp Morphol 18:155–180. [PubMed] [Google Scholar]
- Tashiro F, Kanai-Azuma M, Miyazaki S, Kato M, Tanaka T, Toyoda S, et al. (2010) Maternal-effect gene Ces5/Ooep/Moep19/Floped is essential for oocyte cytoplasmic lattice formation and embryonic development at the maternal-zygotic stage transition. Genes Cells 15:813–828. [DOI] [PubMed] [Google Scholar]
- Taylor DM, Ray PF, Ao A, Winston RM and Handyside AH (1997) Paternal transcripts for glucose-6-phosphate dehydrogenase and adenosine deaminase are first detectable in the human preimplantation embryo at the three- to four-cell stage. Mol Reprod Dev 48:442–448. [DOI] [PubMed] [Google Scholar]
- Terradas M, Martin M, Tusell L and Genesca A (2009) DNA lesions sequestered in micronuclei induce a local defective-damage response. DNA Repair 8:1225–1234. [DOI] [PubMed] [Google Scholar]
- Thompson SL, Bakhoum SF and Compton DA (2010) Mechanisms of chromosomal instability. Curr Biol 20:R285–R295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL and Compton DA (2011) Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. Proc Natl Acad Sci USA 108:17974–17978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong ZB, Gold L, Pfeifer KE, Dorward H, Lee E, Bondy CA, et al. (2000) Mater, a maternal effect gene required for early embryonic development in mice. Nat Genet 26:267–268. [DOI] [PubMed] [Google Scholar]
- Vakifahmetoglu H, Olsson M and Zhivotovsky B (2008) Death through a tragedy: Mitotic catastrophe. Cell Death Differ 15:1153–1162. [DOI] [PubMed] [Google Scholar]
- Van Blerkom J, Davis P and Alexander S (2001) A microscopic and biochemical study of fragmentation phenotypes in stage-appropriate human embryos. Hum Reprod 16:719–729. [DOI] [PubMed] [Google Scholar]
- van Echten-Arends J, Mastenbroek S, Sikkema-Raddatz B, Korevaar JC, Heineman MJ, van der Veen F, et al. (2011) Chromosomal mosaicism in human preimplantation embryos: A systematic review. Hum Reprod Update 17:620–627. [DOI] [PubMed] [Google Scholar]
- Van Soom A, Boerjan ML, Bols PE, Vanroose G, Lein A, Coryn M, et al. (1997) Timing of compaction and inner cell allocation in bovine embryos produced in vivo after superovulation. Biol Reprod 57:1041–1049. [DOI] [PubMed] [Google Scholar]
- Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, et al. (2009) Chromosome instability is common in human cleavage-stage embryos. Nat Med 15:577–583. [DOI] [PubMed] [Google Scholar]
- Vassena R, Boue S, Gonzalez-Roca E, Aran B, Auer H, Veiga A, et al. (2011) Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development 138:3699–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeramah KR, Woerner AE, Johnstone L, Gut I, Gut M, Marques-Bonet T, et al. (2015) Examining phylogenetic relationships among gibbon genera using whole genome sequence data using an approximate bayesian computation approach. Genetics 200:295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorsanova SG, Kolotii AD, Iourov IY, Monakhov VV, Kirillova EA, Soloviev IV, et al. (2005) Evidence for high frequency of chromosomal mosaicism in spontaneous abortions revealed by interphase FISH analysis. J Histochem Cytochem 53:375–380. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Biggins JS, Tannan NB and Srinivas S (2014) Limited predictive value of blastomere angle of division in trophectoderm and inner cell mass specification. Development 141:2279–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Multi S, Yang CR, Ma J, Zhang QH, Wang ZB, et al. (2011) Spindle assembly checkpoint regulates mitotic cell cycle progression during preimplantation embryo development. PLoS One 6:e21557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells D and Delhanty JD (2000) Comprehensive chromosomal analysis of human preimplantation embryos using whole genome amplification and single cell comparative genomic hybridization. Mol Hum Reprod 6:1055–1062. [DOI] [PubMed] [Google Scholar]
- White MJD (1969) Chromosomal rearrangements and speciation in animals. Annu Rev Genet 3:75–98. [Google Scholar]
- Whitten WK and Biggers JD (1968) Complete development in vitro of the pre-implantation stages of the mouse in a simple chemically defined medium. J Reprod Fertil 17:399–401. [DOI] [PubMed] [Google Scholar]
- Whittingham DG (1968) Fertilization of mouse eggs in vitro. Nature 220:592–593. [DOI] [PubMed] [Google Scholar]
- Wienberg J (2004) The evolution of eutherian chromosomes. Curr Opin Genet Dev 14:657–666. [DOI] [PubMed] [Google Scholar]
- Wong CC, Loewke KE, Bossert NL, Behr B, De Jonge CJ, Baer TM, et al. (2010) Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol 28:1115–1121. [DOI] [PubMed] [Google Scholar]
- Wu X (2009) Maternal depletion of NLRP5 blocks early embryogenesis in rhesus macaque monkeys (Macaca mulatta). Hum Reprod 24:415–424. [DOI] [PubMed] [Google Scholar]
- Xanthopoulou L, Ghevaria H, Mantzouratou A, Serhal P, Doshi A and Delhanty JD (2012) Chromosome breakage in human preimplantation embryos from carriers of structural chromosomal abnormalities in relation to fragile sites, maternal age, and poor sperm factors. Cytogenet Genome Res 136:21–29. [DOI] [PubMed] [Google Scholar]
- Xu B, Sun Z, Liu Z, Guo H, Liu Q, Jiang H, et al. (2011) Replication stress induces micronuclei comprising of aggregated DNA double-strand breaks. PLoS One 6:e18618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Cheung T, Chan ST, Ho P and Yeung WS (2001) The incidence of cytoplasmic fragmentation in mouse embryos in vitro is not affected by inhibition of caspase activity. Fertil Steril 75:986–991. [DOI] [PubMed] [Google Scholar]
- Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, et al. (2013) Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500:593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Yang M, Guo H, Yang L, Wu J, Li R, et al. (2013) Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol 20:1131–1139. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhang J, Salem SA, Liu X, Kuang Y, Salem RD, et al. (2014) Selection of competent blastocysts for transfer by combining time-lapse monitoring and array CGH testing for patients undergoing preimplantation genetic screening: A prospective study with sibling oocytes. BMC Med Genomics 7:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernicka-Goetz M (1994) Activation of embryonic genes during preimplantation rat development. Mol Reprod Dev 38:30–35. [DOI] [PubMed] [Google Scholar]
- Zhang P, Zucchelli M, Bruce S, Hambiliki F, Stavreus-Evers A, Levkov L, et al. (2009) Transcriptome profiling of human preimplantation development. PloS One 4:e7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P and Dean J (2009) Role of Filia, a maternal effect gene, in maintaining euploidy during cleavage-stage mouse embryogenesis. Proc Natl Acad Sci U S A 106:7473–7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinaman MJ, Clegg ED, Brown CC, O’Connor J and Selevan SG (1996) Estimates of human fertility and pregnancy loss. Fertil Steril 65:503–509. [PubMed] [Google Scholar]


