Two-way transmission on mink farms
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a zoonotic virus—one that spilled over from another species to infect and transmit among humans. We know that humans can infect other animals with SARS-CoV-2, such as domestic cats and even tigers in zoos. Oude Munnink et al. used whole-genome sequencing to show that SARS-CoV-2 infections were rife among mink farms in the southeastern Netherlands, all of which are destined to be closed by March 2021 (see the Perspective by Zhou and Shi). Toward the end of June 2020, 68% of mink farm workers tested positive for the virus or had antibodies to SARS-CoV-2. These large clusters of infection were initiated by human COVID-19 cases with viruses that bear the D614G mutation. Sequencing has subsequently shown that mink-to-human transmission also occurred. More work must be done to understand whether there is a risk that mustelids may become a reservoir for SARS-CoV-2.
Science, this issue p. 172; see also p. 120
Genetic evidence indicates that SARS-CoV-2 transmission occurred from humans to mink as well as from mink to humans on farms in the Netherlands.
Abstract
Animal experiments have shown that nonhuman primates, cats, ferrets, hamsters, rabbits, and bats can be infected by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In addition, SARS-CoV-2 RNA has been detected in felids, mink, and dogs in the field. Here, we describe an in-depth investigation using whole-genome sequencing of outbreaks on 16 mink farms and the humans living or working on these farms. We conclude that the virus was initially introduced by humans and has since evolved, most likely reflecting widespread circulation among mink in the beginning of the infection period, several weeks before detection. Despite enhanced biosecurity, early warning surveillance, and immediate culling of animals in affected farms, transmission occurred between mink farms in three large transmission clusters with unknown modes of transmission. Of the tested mink farm residents, employees, and/or individuals with whom they had been in contact, 68% had evidence of SARS-CoV-2 infection. Individuals for which whole genomes were available were shown to have been infected with strains with an animal sequence signature, providing evidence of animal-to-human transmission of SARS-CoV-2 within mink farms.
In late December 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified as the cause of a viral pneumonia outbreak, possibly related to a seafood and live animal market in Wuhan, China (1). Since then, SARS-CoV-2 has spread across the world. By 8 October 2020, more than 36.1 million people had been infected with SARS-CoV-2, resulting in more than 1 million deaths (2). In the Netherlands, more than 155,000 infections have been confirmed, more than 6500 SARS-CoV-2–related deaths have been reported, and nonpharmaceutical interventions have been put into place to prevent further spread of SARS-CoV-2 (3).
In view of the similarities of the new virus and SARS-CoV-1, a zoonotic origin of the outbreak was suspected and linked to a Wuhan fresh market where various animals—including fish, shellfish, poultry, wild birds, and exotic animals—were sold. However, other cases with onset well before the period correlated with the Wuhan market–associated cluster were observed, which suggests the possibility of other sources (4). Although closely related coronaviruses found in bats (5, 6) and pangolins (7, 8) have the greatest sequence identity to SARS-CoV-2, the most likely divergence of SARS-CoV-2 from the most closely related bat sequence is estimated to have occurred between 1948 and 1982 (9). Therefore, the animal reservoir(s) of SARS-CoV-2 is(are) yet to be identified.
Similarly to SARS-CoV-1, SARS-CoV-2 binds to the host angiotensin-converting enzyme 2 (ACE2) receptor. On the basis of ACE2 similarities, a range of different animals have been used as models. Experimental infections in dogs (10), cats (10–13), ferrets (10, 14), hamsters (15, 16), rhesus macaques (17), tree shrews (18), cynomolgus macaques (19), African green monkeys (20), common marmosets (21), rabbits (22), and fruit bats (23) have shown that these species are susceptible to SARS-CoV-2, and experimentally infected cats, tree shrews, hamsters, and ferrets could also transmit the virus. By contrast, experimental infection of pigs and several poultry species with SARS-CoV-2 proved to be unsuccessful (10, 23, 24). SARS-CoV-2 has also sporadically been identified in naturally infected animals. In the United States and Hong Kong, SARS-CoV-2 RNA has been detected in dogs (25). In the Netherlands, France, Hong Kong, Belgium, Spain, and the United States, cats have tested positive for SARS-CoV-2 by reverse transcription polymerase chain reaction (RT-PCR) (26–30). Furthermore, SARS-CoV-2 has been detected in four tigers and three lions in a zoo in New York (31). In Italy, the Netherlands, and Wuhan, China, antibodies to SARS-CoV-2 have been detected in cats (29, 32, 33). Recently, SARS-CoV-2 was detected in farmed mink (Neovison vison), resulting in signs of respiratory disease and increased mortality (29, 34).
In response to the outbreaks in mink farms,the Dutch national response system for zoonotic diseases was activated. Although the public health risk of exposure to animals with SARS-CoV-2 was determined to be low, increased awareness of animals’ possible involvement in the COVID-19 epidemic was needed. Therefore, from 20 May 2020 onward, mink farmers, veterinarians, and laboratories were obliged to report symptoms in mink (family Mustelidae) to the Netherlands Food and Consumer Product Safety Authority (NFCPSA), and an extensive surveillance system was established (35).
Whole-genome sequencing (WGS) can be used to monitor the emergence and spread of pathogens (36–39). As part of the surveillance effort in the Netherlands, more than 1750 SARS-CoV-2 viruses have been sequenced to date from patients from different parts of the country (40). Here, we describe an in-depth investigation into the SARS-CoV-2 outbreak in mink farms and mink farm employees in the Netherlands, combining epidemiological information, surveillance data, and WGS on the human–animal interface.
SARS-CoV-2 was first diagnosed on two mink farms (designated NB1 and NB2) in the Netherlands on 23 and 25 April 2020, respectively. After the initial detection of SARS-CoV-2 on these farms, a thorough investigation was initiated to identify potential transmission routes and to perform an environmental and occupational risk assessment. Here, we describe the results of the investigation of the first 16 SARS-CoV-2–affected mink farms by combining SARS-CoV-2 diagnostics, WGS, and in-depth interviews.
Owners and employees of the 16 mink farms with SARS-CoV-2–positive animals were included in the contact tracing investigation by the Dutch Municipal Health Services and were tested according to national protocol. Ninety-seven individuals were tested by either serological assays and/or RT-PCR. Forty-three of 88 (49%) upper respiratory tract samples tested positive by RT-PCR, whereas 38 of 75 (51%) serum samples tested positive for SARS-CoV-2–specific antibodies. In total, 66 of 97 (68%) tested individuals had evidence for SARS-CoV-2 infection (Table 1).
Table 1. Overview of human sampling on SARS-CoV-2–affected mink farms.
| Farm |
First diagnosis in animals |
Date(s) of sampling of employees and family members |
PCR-positive individuals/tested individuals (%) |
Serology-positive individuals/tested individuals (%) |
Positive employees and family members/tested individuals (PCR and/or serology) |
| NB1 | 23 April 2020 | 28 April 2020 to 11 May 2020 | 5/6 (83%) | 5/5 (100%) | 6/6 (100%) |
| NB2 | 25 April 2020 | 31 March 2020 to 30 April 2020 | 1/2 (50%) | 7/8 (88%) | 7/8 (88%) |
| NB3 | 7 May 2020 | 11 May 2020 to 26 May 2020 | 5/7 (71%) | 0/6 (0%)* | 5/7 (71%) |
| NB4 | 7 May 2020 | 8 May 2020 | 1/3 (33%) | 2/2 (100%) | 2/3 (66%) |
| NB5 | 31 May 2020 | 1 June 2020 | 2/7 (29%) | 3/6 (50%) | 3/7 (43%) |
| NB6 | 31 May 2020 | 1 June 2020 | 1/6 (17%) | 4/6 (66%) | 4/6 (66%) |
| NB7 | 31 May 2020 | 10 June 2020 to 1 July 2020 | 8/10 (80%) | NA† | 8/10 (80%) |
| NB8 | 2 June 2020 | 3 June 2020 | 5/10 (50%) | 5/9 (56%) | 8/10 (80%) |
| NB9 | 4 June 2020 | 7 June 2020 | 1/7 (14%) | 1/7 (14%) | 2/7 (29%) |
| NB10 | 8 June 2020 | 11 June 2020 | 1/8 (13%) | 3/8 (38%) | 4/8 (50%) |
| NB11 | 8 June 2020 | 11 June 2020 | 1/3 (33%) | 0/2 (0%) | 1/3 (33%) |
| NB12 | 9 June 2020 | 11 June 2020 | 6/9 (66%) | 2/8 (25%) | 7/9 (78%) |
| NB13 | 14 June 2020 | 11 June 2020 to 18 June 2020 | 3/3 (100%) | 0/2 (0%) | 3/3 (100%) |
| NB14 | 14 June 2020 | 14 June 2020 | 1/3 (33%) | 5/6 (83%) | 5/6 (83%) |
| NB15 | 21 June 2020 | 10 June 2020 to 30 June 2020 | 2/2 (100%) | NA† | 2/2 (100%) |
| NB16 | 21 June 2020 | 23 June 2020 | 0/2 (0%) | NA† | 0/2 (0%) |
| Total | 43/88 (49%) | 37/75 (49%) | 66/97 (68%) |
*Serology was performed ~1 week before the positive PCR test.
†No serology was performed (NA, not applicable).
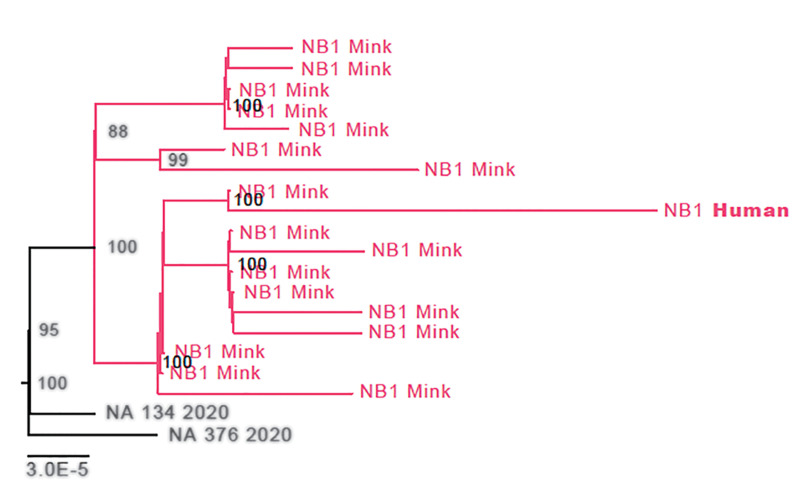
During the interview on 28 April, four of five employees from NB1 reported that they had experienced respiratory symptoms before the outbreak was detected in minks, but none of them had been tested for SARS-CoV-2. Their symptom-onset dates ranged from 1 April to 9 May. For 16 of the mink (sampled on 28 April) and one farm employee (sampled on 4 May), a whole-genome sequence was obtained (hCov-19/Netherlands/NoordBrabant_177/2020). The human sequence clusters within the mink sequences, although it differs from the closest mink sequence by 7 nucleotides (nts) (Fig. 1 and cluster A in Figs. 2 and 3). On farm NB2, SARS-CoV-2 was diagnosed on 25 April. Retrospective analysis showed that one employee from NB2 had been hospitalized with SARS-CoV-2 on 31 March. All samples from the eight employees taken on 30 April were negative by RT-PCR but tested positive for SARS-CoV-2 antibodies. The virus sequence obtained from NB2 animals was distinct from that of NB1 animals, indicating a separate introduction (Figs. 2 and 3, cluster B).
Fig. 1. Phylogenetic analysis of mink farm NB1.
A maximum likelihood analysis was performed using all available SARS-CoV-2 sequences from the Netherlands. Sequences from NB1 are depicted in red, and the employee of NB1 is shown in bold. The two sequences in black at the root of the cluster are the closest-matching human genome sequences from the national SARS-CoV-2 sequence database. The scale bar represents units of substitutions per site.
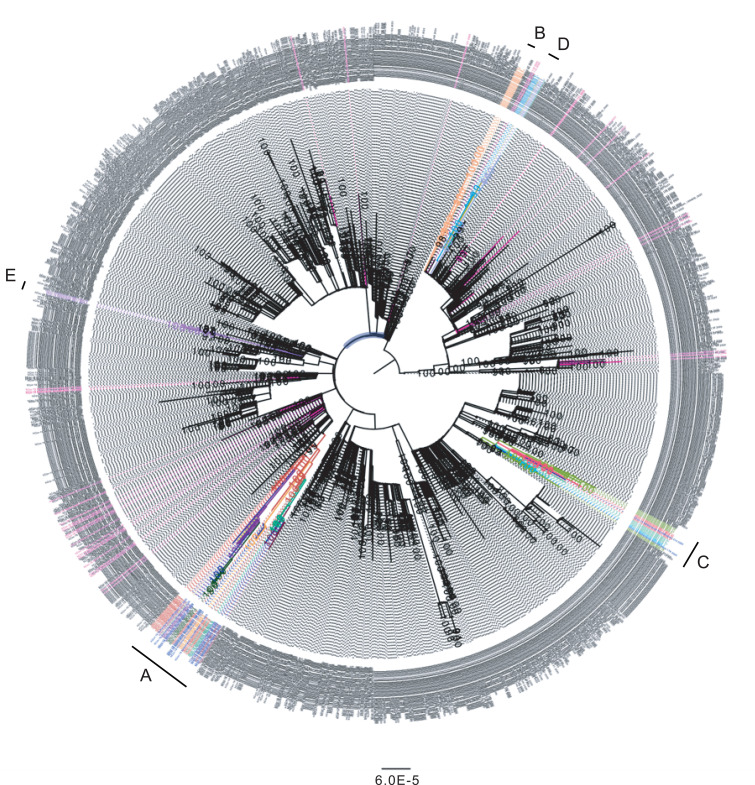
Fig. 2. Maximum likelihood analysis of all SARS-CoV-2 sequences from the Netherlands.
Sequences derived from minks from different farms are indicated with different colors, human sequences related to the mink farms are shown in blue, and samples from similar areas (as determined by four-digit postal code) are shown in magenta. The scale bar represents units of substitutions per site.
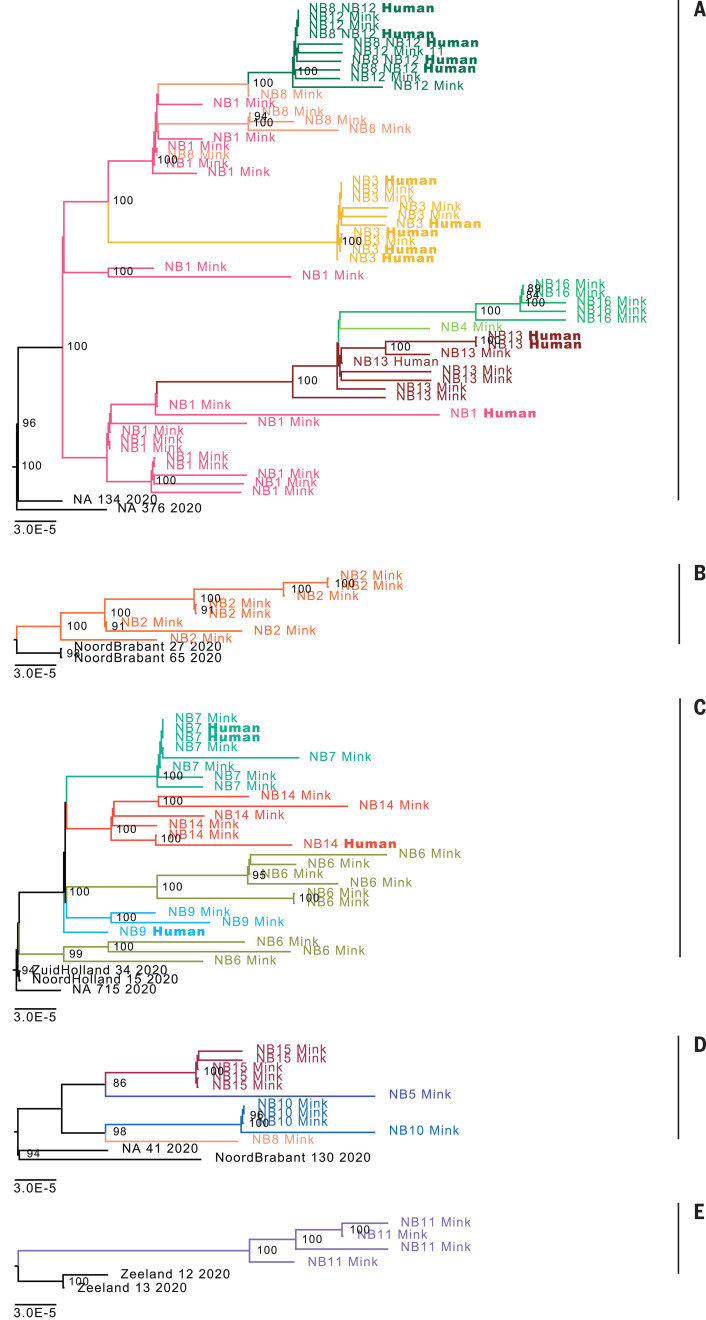
Fig. 3. Phylogenetic analysis of 88 mink and 18 mink-related human SARS-CoV-2 sequences detected in the five mink farm clusters.
Sequences derived from different farms are depicted in different colors. The scale bar represents units of substitutions per site.
On mink farm NB3, SARS-CoV-2 infection was diagnosed on 7 May. Initially, all seven employees tested negative for SARS-CoV-2, but when retested between 19 and 26 May after developing COVID-19–related symptoms, five of seven individuals working or living on the farm tested positive for SARS-CoV-2 RNA. Whole-genome sequences were obtained from these five individuals. The clustering of these sequences with the sequences derived from NB3 mink, together with initial negative test results and subsequent symptom onset, indicate that the employees were infected with SARS-CoV-2 after mink on the farm became infected. An additional infection was identified from contact tracing: An individual who did not visit the farm but had close contact with one of the employees became infected with the SARS-CoV-2 strain found on farm NB3. Animal and human sequences from farm NB3 were close to those from farm NB1, and both were categorized in cluster A.
Similarly, on mink farm NB7, zoonotic transmission from mink to humans likely occurred. On this farm, SARS-CoV-2 infection in mink was diagnosed on 31 May. NB7 employees initially tested negative for SARS-CoV-2, but several of them subsequently began to show symptoms. Between 10 June and 1 July, samples were taken from 10 employees, 8 of which tested positive for SARS-CoV-2 RNA. WGS of two NB7 employee samples showed that their virus sequences clustered with the sequences from the mink at this farm.
The sequences generated from animals and employees of mink farms were compared with ~1775 whole-genome sequences in the national database. To discriminate between community-acquired and mink farm–related infections and to determine the potential risk for people who live near mink farms, WGS was also performed on 34 SARS-CoV-2–positive samples (collected from 4 March until 29 April 2020) from individuals who live in the same geographic area (as determined by four-digit postal code) in which farms NB1 to NB4 are located. These local sequences, sampled in a proxy of ~19 km2, reflected the general diversity of SARS-CoV-2 seen in the Netherlands and were not related to the clusters of mink sequences found on the mink farms, thereby indicating that no spillover to people living in close proximity to mink farms had occurred and that the sequences from SARS-CoV-2–infected animals and farm workers clustered by farm (Fig. 2; sequences from community shown in magenta). The sequences from the mink farm investigation were also compared with sequences from Poland (n = 65) because many of the mink farm workers were seasonal migrants from Poland, but the Polish sequences were more divergent.
Phylogenetic analysis of the mink SARS-CoV-2 genomes showed that mink sequences of 16 farms were grouped into five different clusters (Figs. 2 and 3). Viruses from farms NB1, NB3, NB4, NB8, NB12, NB13, and NB16 belonged to cluster A; sequences from NB2 formed a distinct cluster (cluster B); those from farms NB6, NB7, NB9, and NB14 formed cluster C; those from NB5, NB8, NB10, and NB15 formed cluster D; and those from NB11 were designated as cluster E. On farm NB8, SARS-CoV-2 viruses from clusters A and D were found. A detailed inventory of possible common characteristics—including farm owner, shared personnel, feed supplier, and veterinary service provider—was made. Multiple farms within a cluster shared the same owner; however, in most cases no common factor could be identified for most farms, and clustering could not be explained by geographic distance (Table 2 and Fig. 4).
Table 2. Overview of the clusters detected on the different farms.
Note that veterinarians II and V were from the same veterinary practice. In regard to detection, notification was based on reporting of clinical signs, which was required from 26 April onward. EWS-Ser detection was based on a one-time nationwide compulsory serological screening of all mink farms at the end of May or early June by GD Animal Health. EWS-PM detection was based on the early warning monitoring system (EWS) for which carcasses of animals that died of natural causes were submitted weekly for PCR testing by GD Animal Health from the end of May onward [EWS-PM-1st to -6th postmortem (PM) screening]. NA, not applicable.
| Farm |
Date of diagnosis |
Sequence cluster |
Same owner |
Feed supplier |
Vet |
No. of mink sequences (+no. of human sequences) |
SNP differences (average) |
Mink population size |
Detection |
| NB1 | 24 April 2020 | A | NB1, NB4 | 1 | I | 17 (+1) | 0 to 9 (3.9) | 75,711 | Notification |
| NB2 | 25 April 2020 | B | NA | 1 | II | 8 | 0 to 8 (3.6) | 50,473 | Notification |
| NB3 | 7 May 2020 | A | NA | 2 | III | 5 (+5) | 0 to 2 (0.6) | 12,400 | Notification |
| NB4 | 7 May 2020 | A | NB1, NB4 | 1 | I | 1 | NA | 67,945 | Contact tracing NB1 |
| NB5 | 31 May 2020 | D | NA | 1 | IV | 1 | NA | 38,936 | EWS-Ser+PM-1st |
| NB6 | 31 May 2020 | C | NA | 3 | V | 9 | 0 to 12 (6.8) | 54,515 | EWS-Ser+PM-1st |
| NB7 | 31 May 2020 | C | NB7, NB11, NB15 | 3 | II | 6 (+2) | 0 to 4 (1.4) | 79,355 | EWS-PM-1st |
| NB8 | 2 June 2020 | A and D | NB8, NB12* | 3 | V | 6 (+5) | 0 to 6 (2.6) | 39,144 | EWS-Ser+PM-1st |
| NB9 | 4 June 2020 | C | NA | 2 | V | 2 (+1) | 0 to 3 (1.5) | 32,557 | EWS-Ser+PM-2nd |
| NB10 | 8 June 2020 | D | NA | 3 | II | 4 | 0 to 3 (1.1) | 26,824 | EWS-Ser+PM-2nd |
| NB11 | 8 June 2020 | E | NB7, NB11, NB15 | 3 | II | 4 | 0 to 4 (2.2) | 38,745 | EWS-PM-2nd |
| NB12 | 9 June 2020 | A | NB8, NB12* | 3 | II | 5 | 0 to 3 (1.2) | 55,352 | Notification |
| NB13 | 14 June 2020 | A | NA | 3 | V | 5 (+3) | 0 to 5 (3.2) | 20,366 | EWS-PM-5th |
| NB14 | 14 June 2020 | C | NA | 3 | II | 5 (+1) | 0 to 7 (3.7) | 28,375 | EWS-PM-5th |
| NB15 | 21 June 2020 | D | NB7, NB11, NB15 | 3 | II | 5 | 0 to 2 (0.6) | 35,928 | EWS-PM-6th |
| NB16 | 21 June 2020 | A | NA | 3 | II | 5 | 0 to 4 (1.6) | 66,920 | EWS-PM-6th |
*There was exchange of personnel in these two locations.
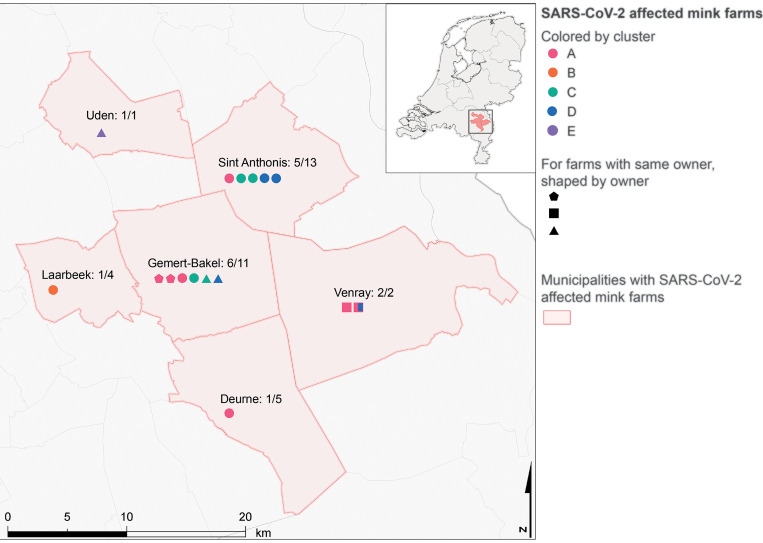
Fig. 4. Geographic overview of mink farms with SARS-CoV-2–positive cases per affected municipality.
The proportion of SARS-CoV-2–affected mink farms over the total number of mink farms (Central Bureau of Statistics database, 2019) is indicated for each area. Symbols for farms with positive cases are colored by cluster; non-circular shapes indicate farms with a single owner.
In total, 18 sequences from mink farm employees or their close contacts were generated from seven different farms. In most cases, these human sequences were nearly identical to the mink sequences from the same farm. For NB1, the situation was different: The human sequence clusters deeply within the sequences derived from mink (Fig. 1), differing from the closest related mink sequence by 7 nts. This was also the case on farm NB14, with a 4-nt difference from the closest related mink sequence. Sequences of employees at mink farm NB8 clustered with those of animals from NB12, likely because personnel were exchanged between these two farms.
SARS-CoV-2 was detected on mink farms NB1 to NB4 after reports of respiratory symptoms and increased mortality in mink. The sequences from farm NB1 showed a difference of 0 to 9 single-nucleotide polymorphisms (SNPs) (average of 3.9 nts) and sequences from NB2 had a difference of 0 to 8 SNPs (average of 3.6 nts), which is more than is generally observed in outbreaks in human settings. In addition, two deletions (one of 12 nts and one of 134 nts) were observed in a single sequence from NB1. The sequences of mink at NB6 had differences of 0 to 12 SNPs, and a deletion of 9 nts was observed in one sequence, whereas diversity was lower for the subsequent farm sequences (Table 2). After the initial detection of SARS-CoV-2, farms were screened weekly. The first, second, fifth, and sixth weekly screening yielded new positive cases.
Several nonsynonymous mutations were identified among the mink sequences when compared with the Wuhan reference sequence NC_045512.2. However, no particular amino acid substitutions were found in all mink samples (fig. S1). Of note, three clusters had the position 614G variant (clusters A, C, and E), and two had the original variant. On the basis of the data available at this stage, there were no obvious disease presentation differences in animals or humans between clusters, but data collection and analysis are ongoing for cases after NB16. This D614G mutation that we observed can also be found in the general human population, and the same mutation was found in human cases related to the mink farms.
Our work shows evidence of ongoing SARS-CoV-2 transmission in mink farms and spillover events to humans. More research in minks and other mustelid species will be needed to determine whether these species are at risk of becoming reservoirs of SARS-CoV-2. After the detection of SARS-CoV-2 on mink farms, 68% of the tested farm workers and/or relatives or contacts would later become or had been infected with SARS-CoV-2, indicating that contact with SARS-CoV-2–infected mink is a risk factor for contracting COVID-19. Recently, an eightfold increase in cytidine-to-uridine (C→U) substitutions compared with uridine-to-cytidine (U→C) substitutions was described, suggestive of host adaptation (41). In the mink sequences, we observed a 3.5-fold increase in C→U compared with U→C substitutions, but the number of substitutions (185) was limited.
A high diversity was observed in the sequences from some mink farms, which is likely explained by multiple generations of viral infections in animals before the increase in mortality was detected. Current estimates indicate that the substitution rate of SARS-CoV-2 is around 1.16 × 10−3 substitutions per site per year in the human population (42), which corresponds to around one mutation per 2 weeks. This could mean that the virus was already circulating in mink farms for some time before it was identified. However, a relatively high sequence diversity was also observed on farms where dead animals tested under the early warning surveillance system tested negative 1 week before a positive test, hinting toward a faster evolutionary rate of the virus in the mink population. Mink farms have large populations of animals, living at high density, which may promote virus transmission. However, the moment of viral introduction was not known, which makes it difficult to draw definite conclusions about the substitution rate in mink farms. The generation interval for SARS-CoV-2 in humans has been estimated to be around 4 to 5 days (43), but with high-dose exposure in a farm with a high number and density of animals, this interval could potentially be shorter.
Further evidence that animals were the most likely source of human infection was provided by the clear phylogenetic separation between mink farm–related human and animal sequences and sequences from human cases within the same geographic area (as determined by four-digit postal code). However, some of the farm-related humans may have been infected within their household, not directly from mink. Spillback into the local community was not observed in our sequence data.
Thus far, the investigation has failed to identify common factors that might explain farm-to-farm spread, possibly via temporary workers who were not included in testing. Since our observations were made, SARS-CoV-2 infections have also been described in mink farms elsewhere (44–46). Additional research efforts are needed, as it is imperative that the fur production and trading sector should not become a reservoir for future spillover of SARS-CoV-2 to humans.
Acknowledgments
We would like to acknowledge R. van Houdt, J. Schinkel, J. Koopsen, and R. Zonneveld (Amsterdam UMC); B. Wintermans (ARDZ); E. Schmid (GGD Goes); B. Rump (GGD Zeeland); J. Filipse and C. Oliveira dos Santos (Isala Hospital, Laboratory of Clinical Microbiology and Infectious Diseases); T. Schuurs (Izore); R. Nijhuis (Meander Medisch Centrum); S. Pas (Microvida); B. de Leeuw (RLM Microbiologie); and R. Jansen, J. Kalpoe, and W. van der Reijden (Streeklab Haarlem) for sample provision and A. van der Linden, M. Boter, and I. Chestakova (Erasmus MC) for technical assistance. Funding: This work has received funding from the European Union’s Horizon 2020 research and innovation program under grants 874735 (VEO), 848096 (SHARP JA), and 101003589 (RECoVER); from ZonMW under grant 10150062010005; and from the Netherlands Ministry of Agriculture, Nature and Food Quality. Author contributions: Conceptualization: B.B.O.M., R.S.S., A.S., and M.P.G.K.; Investigation: B.B.O.M., D.F.N., R.S.S., A.S., M.P.G.K., L.A.M.S., W.H.M.v.d.P., R.J.M., R.J.B., E.M., R.M., A.v.d.S., P.T., A.R., M.B., N.B.-V., F.H., R.H.-v.d.H., M.C.A.W.-B., R.J.B., C.G., A.A.v.d.E., F.C.V., and L.A.M.S.; Supervision: M.P.G.K.; Writing – original draft: B.B.O.M., R.S.S., and M.P.G.K.; Writing – review & editing: all authors. Competing interests: The authors declare no competing interests. Data and material availability: All data, code, and materials used in this work are publicly available. All sequences are publicly available in the GISAID database under accession numbers EPI_ISL_461190 to EPI_ISL_461192, EPI_ISL_461200, EPI_ISL_461202, EPI_ISL_461203, EPI_ISL_522987 to EPI_ISL_523034, EPI_ISL_523040, EPI_ISL_523046, EPI_ISL_523068, EPI_ISL_523070, EPI_ISL_523072, EPI_ISL_523073, EPI_ISL_523075, EPI_ISL_523085, EPI_ISL_523089 to EPI_ISL_523120, EPI_ISL_523282 to EPI_ISL_523286, EPI_ISL_523301, EPI_ISL_523310 to EPI_ISL_523312, EPI_ISL_523333, EPI_ISL_523493, and EPI_ISL_523494. Ethical approval was not required for this study, as anonymous aggregated data were used, and no medical interventions were made on animal or human subjects. This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using such material.
Supplementary Materials
science.sciencemag.org/content/371/6525/172/suppl/DC1
Materials and Methods
Fig. S1
Table S1
MDAR Reproducibility Checklist
References and Notes
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W.; China Novel Coronavirus Investigating and Research Team , A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020). 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E., Du H., Gardner L., An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 20, 533–534 (2020). 10.1016/S1473-3099(20)30120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oude Munnink B. B., Nieuwenhuijse D. F., Stein M., O’Toole Á., Haverkate M., Mollers M., Kamga S. K., Schapendonk C., Pronk M., Lexmond P., van der Linden A., Bestebroer T., Chestakova I., Overmars R. J., van Nieuwkoop S., Molenkamp R., van der Eijk A. A., GeurtsvanKessel C., Vennema H., Meijer A., Rambaut A., van Dissel J., Sikkema R. S., Timen A., Koopmans M.; Dutch-Covid-19 response team , Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in the Netherlands. Nat. Med. 26, 1405–1410 (2020). 10.1038/s41591-020-0997-y [DOI] [PubMed] [Google Scholar]
- 4.Wu F., Zhao S., Yu B., Chen Y. M., Wang W., Song Z. G., Hu Y., Tao Z. W., Tian J. H., Pei Y. Y., Yuan M. L., Zhang Y. L., Dai F. H., Liu Y., Wang Q. M., Zheng J. J., Xu L., Holmes E. C., Zhang Y. Z., A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Wang P., Liu D., Yang J., Holmes E. C., Hughes A. C., Bi Y., Shi W., A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Curr. Biol. 30, 3896 (2020). 10.1016/j.cub.2020.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., Si H. R., Zhu Y., Li B., Huang C. L., Chen H. D., Chen J., Luo Y., Guo H., Jiang R. D., Liu M. Q., Chen Y., Shen X. R., Wang X., Zheng X. S., Zhao K., Chen Q. J., Deng F., Liu L. L., Yan B., Zhan F. X., Wang Y. Y., Xiao G. F., Shi Z. L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lam T. T. Y., Jia N., Zhang Y. W., Shum M. H., Jiang J. F., Zhu H. C., Tong Y. G., Shi Y. X., Ni X. B., Liao Y. S., Li W. J., Jiang B. G., Wei W., Yuan T. T., Zheng K., Cui X. M., Li J., Pei G. Q., Qiang X., Cheung W. Y., Li L. F., Sun F. F., Qin S., Huang J. C., Leung G. M., Holmes E. C., Hu Y. L., Guan Y., Cao W. C., Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583, 282–285 (2020). 10.1038/s41586-020-2169-0 [DOI] [PubMed] [Google Scholar]
- 8.Han G. Z., Pangolins Harbor SARS-CoV-2-Related Coronaviruses. Trends Microbiol. 28, 515–517 (2020). 10.1016/j.tim.2020.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boni M. F., Lemey P., Jiang X., Lam T. T. Y., Perry B. W., Castoe T. A., Rambaut A., Robertson D. L., Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 5, 1408–1417 (2020). 10.1038/s41564-020-0771-4 [DOI] [PubMed] [Google Scholar]
- 10.Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., Zhao Y., Liu P., Liang L., Cui P., Wang J., Zhang X., Guan Y., Tan W., Wu G., Chen H., Bu Z., Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 368, 1016–1020 (2020). 10.1126/science.abb7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halfmann P. J., Hatta M., Chiba S., Maemura T., Fan S., Takeda M., Kinoshita N., Hattori S.-I., Sakai-Tagawa Y., Iwatsuki-Horimoto K., Imai M., Kawaoka Y., Transmission of SARS-CoV-2 in Domestic Cats. N. Engl. J. Med. 383, 592–594 (2020). 10.1056/NEJMc2013400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.GOV.UK, COVID-19 confirmed in pet cat in the UK (2020); www.gov.uk/government/news/covid-19-confirmed-in-pet-cat-in-the-uk.
- 13.Ruiz-Arrondo I., Portillo A., Palomar A. M., Santibáñez S., Santibáñez P., Cervera C., Oteo J. A., Detection of SARS-CoV-2 in pets living with COVID-19 owners diagnosed during the COVID-19 lockdown in Spain: A case of an asymptomatic cat with SARS-CoV-2 in Europe. Transbound. Emerg. Dis. 10.1111/tbed.13803 (2020). 10.1111/tbed.13803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richard M., Kok A., de Meulder D., Bestebroer T. M., Lamers M. M., Okba N. M. A., Fentener van Vlissingen M., Rockx B., Haagmans B. L., Koopmans M. P. G., Fouchier R. A. M., Herfst S., SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat. Commun. 11, 3496 (2020). 10.1038/s41467-020-17367-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sia S. F., Yan L.-M., Chin A. W. H., Fung K., Choy K. T., Wong A. Y. L., Kaewpreedee P., Perera R. A. P. M., Poon L. L. M., Nicholls J. M., Peiris M., Yen H. L., Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 583, 834–838 (2020). 10.1038/s41586-020-2342-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan J. F.-W., Zhang A. J., Yuan S., Poon V. K.-M., Chan C. C.-S., Lee A. C.-Y., Chan W.-M., Fan Z., Tsoi H.-W., Wen L., Liang R., Cao J., Chen Y., Tang K., Luo C., Cai J.-P., Kok K.-H., Chu H., Chan K.-H., Sridhar S., Chen Z., Chen H., To K. K.-W., Yuen K.-Y., Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: Implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 71, 2428–2446 (2020). 10.1093/cid/ciaa325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munster V. J., Feldmann F., Williamson B. N., van Doremalen N., Pérez-Pérez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B., Avanzato V. A., Rosenke R., Hanley P. W., Saturday G., Scott D., Fischer E. R., de Wit E., Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 585, 268–272 (2020). 10.1038/s41586-020-2324-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y., Wang J., Kuang D., Xu J., Yang M., Ma C., Zhao S., Li J., Long H., Ding K., Gao J., Liu J., Wang H., Li H., Yang Y., Yu W., Yang J., Zheng Y., Wu D., Lu S., Liu H., Peng X., Susceptibility of tree shrew to SARS-CoV-2 infection. Sci. Rep. 10, 16007 (2020). 10.1038/s41598-020-72563-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M. M., Oude Munnink B. B., de Meulder D., van Amerongen G., van den Brand J., Okba N. M. A., Schipper D., van Run P., Leijten L., Sikkema R., Verschoor E., Verstrepen B., Bogers W., Langermans J., Drosten C., Fentener van Vlissingen M., Fouchier R., de Swart R., Koopmans M., Haagmans B. L., Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 368, 1012–1015 (2020). 10.1126/science.abb7314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.C. Woolsey, V. Borisevich, A. N. Prasad, K. N. Agans, D. J. Deer, N. S. Dobias, J. C. Heymann, S. L. Foster, C. B. Levine, L. Medina, K. Melody, J. B. Geisbert, K. A. Fenton, T. W. Geisbert, R. W. Cross, Establishment of an African green monkey model for COVID-19. bioRxiv 2020.05.17.100289 [Preprint]. 17 May 2020. 10.1101/2020.05.17.100289. 10.1101/2020.05.17.100289 [DOI]
- 21.Lu S., Zhao Y., Yu W., Yang Y., Gao J., Wang J., Kuang D., Yang M., Yang J., Ma C., Xu J., Qian X., Li H., Zhao S., Li J., Wang H., Long H., Zhou J., Luo F., Ding K., Wu D., Zhang Y., Dong Y., Liu Y., Zheng Y., Lin X., Jiao L., Zheng H., Dai Q., Sun Q., Hu Y., Ke C., Liu H., Peng X., Comparison of nonhuman primates identified the suitable model for COVID-19. Signal Transduct. Target. Ther. 5, 157 (2020). 10.1038/s41392-020-00269-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.B. L. Haagmans, D. Noack, N. M. Okba, W. Li, C. Wang, R. de Vries, S. Herfst, D. de Meulder, P. van Run, B. Rijnders, C. Rokx, F. van Kuppeveld, F. Grosveld, C. GeurtsvanKessel, M. Koopmans, B. Jan Bosch, T. Kuiken, B. Rockx, SARS-CoV-2 neutralizing human antibodies protect against lower respiratory tract disease in a hamster model. bioRxiv 2020.08.24.264630 [Preprint]. 24 August 2020. 10.1101/2020.08.24.264630. 10.1101/2020.08.24.264630 [DOI]
- 23.K. Schlottau, M. Rissmann, A. Graaf, J. Schön, J. Sehl, C. Wylezich, D. Höper, T. C. Mettenleiter, A. Balkema-Buschmann, T. Harder, C. Grund, D. Hoffmann, A. Breithaupt, M. Beer, Experimental Transmission Studies of SARS-CoV-2 in Fruit Bats, Ferrets, Pigs and Chickens. SSRN 3578792. 15 April 2020. 10.2139/ssrn.3578792. 10.2139/ssrn.3578792 [DOI] [PMC free article] [PubMed]
- 24.Suarez D. L., Pantin-Jackwood M. J., Swayne D. E., Lee S. A., DeBlois S. M., Spackman E., Lack of susceptibility to SARS-CoV-2 and MERS-CoV in poultry. Emerg. Infect. Dis. 26, 3074–3076 (2020). 10.3201/eid2612.202989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sit T. H. C., Brackman C. J., Ip S. M., Tam K. W. S., Law P. Y. T., To E. M. W., Yu V. Y. T., Sims L. D., Tsang D. N. C., Chu D. K. W., Perera R. A. P. M., Poon L. L. M., Peiris M., Infection of dogs with SARS-CoV-2. Nature 586, 776–778 (2020). 10.1038/s41586-020-2334-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sailleau C., Dumarest M., Vanhomwegen J., Delaplace M., Caro V., Kwasiborski A., Hourdel V., Chevaillier P., Barbarino A., Comtet L., Pourquier P., Klonjkowski B., Manuguerra J.-C., Zientara S., Le Poder S., First detection and genome sequencing of SARS-CoV-2 in an infected cat in France. Transbound. Emerg. Dis. 67, 2324–2328 (2020). 10.1111/tbed.13659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman A., Smith D., Ghai R. R., Wallace R. M., Torchetti M. K., Loiacono C., Murrell L. S., Carpenter A., Moroff S., Rooney J. A., Barton Behravesh C., First Reported Cases of SARS-CoV-2 Infection in Companion Animals - New York, March-April 2020. MMWR Morb. Mortal. Wkly. Rep. 69, 710–713 (2020). 10.15585/mmwr.mm6923e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ProMED, “COVID-19 update (166): China (Hong Kong) animal, cat, OIE, resolved,” 20200508.7314521 (2020); https://promedmail.org/promed-post/?id=7314521.
- 29.Oreshkova N., Molenaar R. J., Vreman S., Harders F., Oude Munnink B. B., Hakze-van der Honing R. W., Gerhards N., Tolsma P., Bouwstra R., Sikkema R. S., Tacken M. G., de Rooij M. M., Weesendorp E., Engelsma M. Y., Bruschke C. J., Smit L. A., Koopmans M., van der Poel W. H., Stegeman A., SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill. 25, 2001005 (2020). 10.2807/1560-7917.ES.2020.25.23.2001005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segalés J., Puig M., Rodon J., Avila-Nieto C., Carrillo J., Cantero G., Terrón M. T., Cruz S., Parera M., Noguera-Julián M., Izquierdo-Useros N., Guallar V., Vidal E., Valencia A., Blanco I., Blanco J., Clotet B., Vergara-Alert J., Detection of SARS-CoV-2 in a cat owned by a COVID-19-affected patient in Spain. Proc. Natl. Acad. Sci. U.S.A. 117, 24790–24793 (2020). 10.1073/pnas.2010817117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gollakner R., Capua I., Is COVID-19 the first pandemic that evolves into a panzootic? Vet. Ital. 56, 7–8 (2020). 10.12834/VetIt.2246.12523.1 [DOI] [PubMed] [Google Scholar]
- 32.Q. Zhang, H. Zhang, K. Huang, Y. Yang, X. Hui, J. Gao, X. He, C. Li, W. Gong, Y. Zhang, C. Peng, X. Gao, H. Chen, Z. Zou, Z. Shi, M. Jin, SARS-CoV-2 neutralizing serum antibodies in cats: A serological investigation. bioRxiv 2020.04.01.021196 [Preprint]. 3 April 2020. 10.1101/2020.04.01.021196. 10.1101/2020.04.01.021196 [DOI]
- 33.E. I. Patterson, G. Elia, A. Grassi, A. Giordano, C. Desario, M. Medardo, S. L. Smith, E. R. Anderson, T. Prince, G. T. Patterson, E. Lorusso, M. S. Lucente, G. Lanave, S. Lauzi, U. Bonfanti, A. Stranieri, V. Martella, F. S. Basano, V. R. Barrs, A. D. Radford, U. Agrimi, G. L. Hughes, S. Paltrinieri, N. Decaro, Evidence of exposure to SARS-CoV-2 in cats and dogs from households in Italy. bioRxiv 2020.07.21.214346 [Preprint]. 23 July 2020. 10.1101/2020.07.21.214346. 10.1101/2020.07.21.214346 [DOI] [PMC free article] [PubMed]
- 34.Molenaar R. J., Vreman S., Hakze-van der Honing R. W., Zwart R., de Rond J., Weesendorp E., Smit L. A. M., Koopmans M., Bouwstra R., Stegeman A., van der Poel W. H. M., Clinical and Pathological Findings in SARS-CoV-2 Disease Outbreaks in Farmed Mink (Neovison vison). Vet. Pathol. 57, 653–657 (2020). 10.1177/0300985820943535 [DOI] [PubMed] [Google Scholar]
- 35.NVWA, Bedrijfsmatig gehouden dieren en SARS-CoV-2; www.nvwa.nl/nieuws-en-media/actuele-onderwerpen/corona/g/bedrijfsmatig-gehouden-dieren-en-corona.
- 36.Oude Munnink B. B., Münger E., Nieuwenhuijse D. F., Kohl R., van der Linden A., Schapendonk C. M. E., van der Jeugd H., Kik M., Rijks J. M., Reusken C. B. E. M., Koopmans M., Genomic monitoring to understand the emergence and spread of Usutu virus in the Netherlands, 2016-2018. Sci. Rep. 10, 2798 (2020). 10.1038/s41598-020-59692-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arias A., Watson S. J., Asogun D., Tobin E. A., Lu J., Phan M. V. T., Jah U., Wadoum R. E. G., Meredith L., Thorne L., Caddy S., Tarawalie A., Langat P., Dudas G., Faria N. R., Dellicour S., Kamara A., Kargbo B., Kamara B. O., Gevao S., Cooper D., Newport M., Horby P., Dunning J., Sahr F., Brooks T., Simpson A. J. H., Groppelli E., Liu G., Mulakken N., Rhodes K., Akpablie J., Yoti Z., Lamunu M., Vitto E., Otim P., Owilli C., Boateng I., Okoror L., Omomoh E., Oyakhilome J., Omiunu R., Yemisis I., Adomeh D., Ehikhiametalor S., Akhilomen P., Aire C., Kurth A., Cook N., Baumann J., Gabriel M., Wölfel R., Di Caro A., Carroll M. W., Günther S., Redd J., Naidoo D., Pybus O. G., Rambaut A., Kellam P., Goodfellow I., Cotten M., Rapid outbreak sequencing of Ebola virus in Sierra Leone identifies transmission chains linked to sporadic cases. Virus Evol. 2, vew016 (2016). 10.1093/ve/vew016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faria N. R., Kraemer M. U. G., Hill S. C., Goes de Jesus J., Aguiar R. S., Iani F. C. M., Xavier J., Quick J., du Plessis L., Dellicour S., Thézé J., Carvalho R. D. O., Baele G., Wu C.-H., Silveira P. P., Arruda M. B., Pereira M. A., Pereira G. C., Lourenço J., Obolski U., Abade L., Vasylyeva T. I., Giovanetti M., Yi D., Weiss D. J., Wint G. R. W., Shearer F. M., Funk S., Nikolay B., Fonseca V., Adelino T. E. R., Oliveira M. A. A., Silva M. V. F., Sacchetto L., Figueiredo P. O., Rezende I. M., Mello E. M., Said R. F. C., Santos D. A., Ferraz M. L., Brito M. G., Santana L. F., Menezes M. T., Brindeiro R. M., Tanuri A., Dos Santos F. C. P., Cunha M. S., Nogueira J. S., Rocco I. M., da Costa A. C., Komninakis S. C. V., Azevedo V., Chieppe A. O., Araujo E. S. M., Mendonça M. C. L., Dos Santos C. C., Dos Santos C. D., Mares-Guia A. M., Nogueira R. M. R., Sequeira P. C., Abreu R. G., Garcia M. H. O., Abreu A. L., Okumoto O., Kroon E. G., de Albuquerque C. F. C., Lewandowski K., Pullan S. T., Carroll M., de Oliveira T., Sabino E. C., Souza R. P., Suchard M. A., Lemey P., Trindade G. S., Drumond B. P., Filippis A. M. B., Loman N. J., Cauchemez S., Alcantara L. C. J., Pybus O. G., Genomic and epidemiological monitoring of yellow fever virus transmission potential. Science 361, 894–899 (2018). 10.1126/science.aat7115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quick J., Loman N. J., Duraffour S., Simpson J. T., Severi E., Cowley L., Bore J. A., Koundouno R., Dudas G., Mikhail A., Ouédraogo N., Afrough B., Bah A., Baum J. H., Becker-Ziaja B., Boettcher J. P., Cabeza-Cabrerizo M., Camino-Sanchez A., Carter L. L., Doerrbecker J., Enkirch T., Dorival I. G. G., Hetzelt N., Hinzmann J., Holm T., Kafetzopoulou L. E., Koropogui M., Kosgey A., Kuisma E., Logue C. H., Mazzarelli A., Meisel S., Mertens M., Michel J., Ngabo D., Nitzsche K., Pallash E., Patrono L. V., Portmann J., Repits J. G., Rickett N. Y., Sachse A., Singethan K., Vitoriano I., Yemanaberhan R. L., Zekeng E. G., Trina R., Bello A., Sall A. A., Faye O., Faye O., Magassouba N., Williams C. V., Amburgey V., Winona L., Davis E., Gerlach J., Washington F., Monteil V., Jourdain M., Bererd M., Camara A., Somlare H., Camara A., Gerard M., Bado G., Baillet B., Delaune D., Nebie K. Y., Diarra A., Savane Y., Pallawo R. B., Gutierrez G. J., Milhano N., Roger I., Williams C. J., Yattara F., Lewandowski K., Taylor J., Rachwal P., Turner D., Pollakis G., Hiscox J. A., Matthews D. A., O’Shea M. K., Johnston A. M., Wilson D., Hutley E., Smit E., Di Caro A., Woelfel R., Stoecker K., Fleischmann E., Gabriel M., Weller S. A., Koivogui L., Diallo B., Keita S., Rambaut A., Formenty P., Gunther S., Carroll M. W., Real-time, portable genome sequencing for Ebola surveillance. Nature 530, 228–232 (2016). 10.1038/nature16996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sikkema R. S., Pas S. D., Nieuwenhuijse D. F., O’Toole Á., Verweij J., van der Linden A., Chestakova I., Schapendonk C., Pronk M., Lexmond P., Bestebroer T., Overmars R. J., van Nieuwkoop S., van den Bijllaardt W., Bentvelsen R. G., van Rijen M. M. L., Buiting A. G. M., van Oudheusden A. J. G., Diederen B. M., Bergmans A. M. C., van der Eijk A., Molenkamp R., Rambaut A., Timen A., Kluytmans J. A. J. W., Oude Munnink B. B., Kluytmans van den Bergh M. F. Q., Koopmans M. P. G., COVID-19 in health-care workers in three hospitals in the south of the Netherlands: A cross-sectional study. Lancet Infect. Dis. 20, 1273–1280 (2020). 10.1016/S1473-3099(20)30527-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmonds P., Rampant C→U Hypermutation in the Genomes of SARS-CoV-2 and Other Coronaviruses: Causes and Consequences for Their Short- and Long-Term Evolutionary Trajectories. mSphere 5, e00408-20 (2020). 10.1128/mSphere.00408-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Candido D. S., Claro I. M., de Jesus J. G., Souza W. M., Moreira F. R. R., Dellicour S., Mellan T. A., du Plessis L., Pereira R. H. M., Sales F. C. S., Manuli E. R., Thézé J., Almeida L., Menezes M. T., Voloch C. M., Fumagalli M. J., Coletti T. M., da Silva C. A. M., Ramundo M. S., Amorim M. R., Hoeltgebaum H. H., Mishra S., Gill M. S., Carvalho L. M., Buss L. F., Prete C. A. Jr.., Ashworth J., Nakaya H. I., Peixoto P. S., Brady O. J., Nicholls S. M., Tanuri A., Rossi Á. D., Braga C. K. V., Gerber A. L., de C Guimarães A. P., Gaburo N. Jr.., Alencar C. S., Ferreira A. C. S., Lima C. X., Levi J. E., Granato C., Ferreira G. M., Francisco R. S. Jr.., Granja F., Garcia M. T., Moretti M. L., Perroud M. W. Jr.., Castiñeiras T. M. P. P., Lazari C. S., Hill S. C., de Souza Santos A. A., Simeoni C. L., Forato J., Sposito A. C., Schreiber A. Z., Santos M. N. N., de Sá C. Z., Souza R. P., Resende-Moreira L. C., Teixeira M. M., Hubner J., Leme P. A. F., Moreira R. G., Nogueira M. L., Ferguson N. M., Costa S. F., Proenca-Modena J. L., Vasconcelos A. T. R., Bhatt S., Lemey P., Wu C. H., Rambaut A., Loman N. J., Aguiar R. S., Pybus O. G., Sabino E. C., Faria N. R.; Brazil-UK Centre for Arbovirus Discovery, Diagnosis, Genomics and Epidemiology (CADDE) Genomic Network , Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science 369, 1255–1260 (2020). 10.1126/science.abd2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ganyani T., Kremer C., Chen D., Torneri A., Faes C., Wallinga J., Hens N., Estimating the generation interval for coronavirus disease (COVID-19) based on symptom onset data, March 2020. Euro Surveill. 25, 2000257 (2020). 10.2807/1560-7917.ES.2020.25.17.2000257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cahan E., COVID-19 hits U.S. mink farms after ripping through Europe. Science 10.1126/science.abe3870 (2020). 10.1126/science.abe3870 [DOI] [Google Scholar]
- 45.ProMED, “COVID-19 update (319): Spain (AR) animal, farmed mink, 1st rep,” 20200717.7584560 (2020); https://promedmail.org/promed-post/?id=7584560.
- 46.ProMED, “COVID-19 update (266): Denmark (ND) animal, farmed mink, 1st rep,” 20200617.7479510 (2020); https://promedmail.org/promed-post/?id=20200617.7479510.
- 47.RIVM, Signaleringsoverleg zoönosen; www.rivm.nl/surveillance-van-infectieziekten/signalering-infectieziekten/signaleringsoverleg-zoonosen.
- 48.RIVM, Outbreak Management Team (OMT); www.rivm.nl/coronavirus-covid-19/omt.
- 49.Kroneman A., Vennema H., Deforche K., v. d. Avoort H., Peñaranda S., Oberste M. S., Vinjé J., Koopmans M., An automated genotyping tool for enteroviruses and noroviruses. J. Clin. Virol. 51, 121–125 (2011). 10.1016/j.jcv.2011.03.006 [DOI] [PubMed] [Google Scholar]
- 50.Corman V. M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D. K., Bleicker T., Brünink S., Schneider J., Schmidt M. L., Mulders D. G., Haagmans B. L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M. P., Drosten C., Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 25, 2000045 (2020). 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.GeurtsvanKessel C. H., Okba N. M. A., Igloi Z., Bogers S., Embregts C. W. E., Laksono B. M., Leijten L., Rokx C., Rijnders B., Rahamat-Langendoen J., van den Akker J. P. C., van Kampen J. J. A., van der Eijk A. A., van Binnendijk R. S., Haagmans B., Koopmans M., An evaluation of COVID-19 serological assays informs future diagnostics and exposure assessment. Nat. Commun. 11, 3436 (2020). 10.1038/s41467-020-17317-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.R. R. Wick, rrwick/Porechop: adapter trimmer for Oxford Nanopore reads, version 0.2.4, GitHub (2018); https://github.com/rrwick/porechop.
- 53.Oude Munnink B. B., Nieuwenhuijse D. F., Sikkema R. S., Koopmans M., Validating Whole Genome Nanopore Sequencing, using Usutu Virus as an Example. J. Vis. Exp. 10.3791/60906 (2020). 10.3791/60906 [DOI] [PubMed] [Google Scholar]
- 54.T. Seemann, F. Klotzl, A. Page, tseemann/snp-dists: Pairwise SNP distance matrix from a FASTA sequence alignment, version 0.7.0, GitHub (2020); https://github.com/tseemann/snp-dists.
- 55.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.A. Rambaut, FigTree, v1.4.4 (2016); http://tree.bio.ed.ac.uk/software/figtree/.
- 58.Shu Y., McCauley J., GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 22, 30494 (2017). 10.2807/1560-7917.ES.2017.22.13.30494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.H. Wickham, ggplot2: Elegant Graphics for Data Analysis (Springer, 2016). [Google Scholar]
- 60.C.-C. B. voor de Statistiek, Wijk- en buurtkaart 2019; www.cbs.nl/nl-nl/dossier/nederland-regionaal/geografische-data/wijk-en-buurtkaart-2019.
- 61.Pebesma E. J., Bivand R. S., Classes and Methods for Spatial Data: The sp Package. R News 5, 9–13 (2005); https://CRAN.R-project.org/doc/Rnews/. [Google Scholar]
- 62.rhijmans, rspatial/raster: R raster package, version 3.4-7, GitHub (2020); https://github.com/rspatial/raster/.
- 63.rsbivand, cran/rgdal, version 1.5-12, GitHub (2020); https://github.com/cran/rgdal.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
science.sciencemag.org/content/371/6525/172/suppl/DC1
Materials and Methods
Fig. S1
Table S1
MDAR Reproducibility Checklist