The genetics underlying severe COVID-19
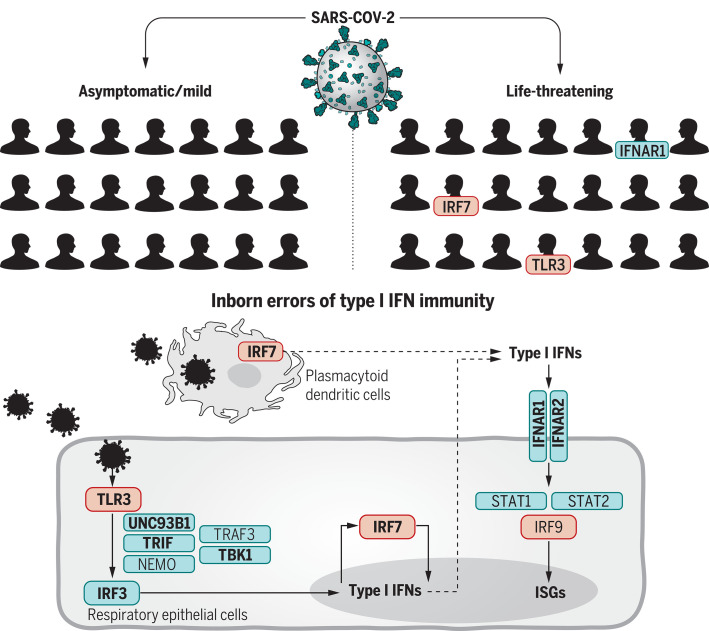
The immune system is complex and involves many genes, including those that encode cytokines known as interferons (IFNs). Individuals that lack specific IFNs can be more susceptible to infectious diseases. Furthermore, the autoantibody system dampens IFN response to prevent damage from pathogen-induced inflammation. Two studies now examine the likelihood that genetics affects the risk of severe coronavirus disease 2019 (COVID-19) through components of this system (see the Perspective by Beck and Aksentijevich). Q. Zhang et al. used a candidate gene approach and identified patients with severe COVID-19 who have mutations in genes involved in the regulation of type I and III IFN immunity. They found enrichment of these genes in patients and conclude that genetics may determine the clinical course of the infection. Bastard et al. identified individuals with high titers of neutralizing autoantibodies against type I IFN-α2 and IFN-ω in about 10% of patients with severe COVID-19 pneumonia. These autoantibodies were not found either in infected people who were asymptomatic or had milder phenotype or in healthy individuals. Together, these studies identify a means by which individuals at highest risk of life-threatening COVID-19 can be identified.
Science, this issue p. eabd4570, p. eabd4585; see also p. 404
A large immunological and genomics study of COVID-19 patients reveals excess mutations in the type I IFN pathway.
INTRODUCTION
Clinical outcomes of human severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection range from silent infection to lethal coronavirus disease 2019 (COVID-19). Epidemiological studies have identified three risk factors for severe disease: being male, being elderly, and having other medical conditions. However, interindividual clinical variability remains huge in each demographic category. Discovering the root cause and detailed molecular, cellular, and tissue- and body-level mechanisms underlying life-threatening COVID-19 is of the utmost biological and medical importance.
RATIONALE
We established the COVID Human Genetic Effort (www.covidhge.com) to test the general hypothesis that life-threatening COVID-19 in some or most patients may be caused by monogenic inborn errors of immunity to SARS-CoV-2 with incomplete or complete penetrance. We sequenced the exome or genome of 659 patients of various ancestries with life-threatening COVID-19 pneumonia and 534 subjects with asymptomatic or benign infection. We tested the specific hypothesis that inborn errors of Toll-like receptor 3 (TLR3)– and interferon regulatory factor 7 (IRF7)–dependent type I interferon (IFN) immunity that underlie life-threatening influenza pneumonia also underlie life-threatening COVID-19 pneumonia. We considered three loci identified as mutated in patients with life-threatening influenza: TLR3, IRF7, and IRF9. We also considered 10 loci mutated in patients with other viral illnesses but directly connected to the three core genes conferring influenza susceptibility: TICAM1/TRIF, UNC93B1, TRAF3, TBK1, IRF3, and NEMO/IKBKG from the TLR3-dependent type I IFN induction pathway, and IFNAR1, IFNAR2, STAT1, and STAT2 from the IRF7- and IRF9-dependent type I IFN amplification pathway. Finally, we considered various modes of inheritance at these 13 loci.
RESULTS
We found an enrichment in variants predicted to be loss-of-function (pLOF), with a minor allele frequency <0.001, at the 13 candidate loci in the 659 patients with life-threatening COVID-19 pneumonia relative to the 534 subjects with asymptomatic or benign infection (P = 0.01). Experimental tests for all 118 rare nonsynonymous variants (including both pLOF and other variants) of these 13 genes found in patients with critical disease identified 23 patients (3.5%), aged 17 to 77 years, carrying 24 deleterious variants of eight genes. These variants underlie autosomal-recessive (AR) deficiencies (IRF7 and IFNAR1) and autosomal-dominant (AD) deficiencies (TLR3, UNC93B1, TICAM1, TBK1, IRF3, IRF7, IFNAR1, and IFNAR2) in four and 19 patients, respectively. These patients had never been hospitalized for other life-threatening viral illness. Plasmacytoid dendritic cells from IRF7-deficient patients produced no type I IFN on infection with SARS-CoV-2, and TLR3−/−, TLR3+/−, IRF7−/−, and IFNAR1−/− fibroblasts were susceptible to SARS-CoV-2 infection in vitro.
CONCLUSION
At least 3.5% of patients with life-threatening COVID-19 pneumonia had known (AR IRF7 and IFNAR1 deficiencies or AD TLR3, TICAM1, TBK1, and IRF3 deficiencies) or new (AD UNC93B1, IRF7, IFNAR1, and IFNAR2 deficiencies) genetic defects at eight of the 13 candidate loci involved in the TLR3- and IRF7-dependent induction and amplification of type I IFNs. This discovery reveals essential roles for both the double-stranded RNA sensor TLR3 and type I IFN cell-intrinsic immunity in the control of SARS-CoV-2 infection. Type I IFN administration may be of therapeutic benefit in selected patients, at least early in the course of SARS-CoV-2 infection.
Inborn errors of TLR3- and IRF7-dependent type I IFN production and amplification underlie life-threatening COVID-19 pneumonia.
Molecules in red are encoded by core genes, deleterious variants of which underlie critical influenza pneumonia with incomplete penetrance, and deleterious variants of genes encoding biochemically related molecules in blue underlie other viral illnesses. Molecules represented in bold are encoded by genes with variants that also underlie critical COVID-19 pneumonia.
Abstract
Clinical outcome upon infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) ranges from silent infection to lethal coronavirus disease 2019 (COVID-19). We have found an enrichment in rare variants predicted to be loss-of-function (LOF) at the 13 human loci known to govern Toll-like receptor 3 (TLR3)– and interferon regulatory factor 7 (IRF7)–dependent type I interferon (IFN) immunity to influenza virus in 659 patients with life-threatening COVID-19 pneumonia relative to 534 subjects with asymptomatic or benign infection. By testing these and other rare variants at these 13 loci, we experimentally defined LOF variants underlying autosomal-recessive or autosomal-dominant deficiencies in 23 patients (3.5%) 17 to 77 years of age. We show that human fibroblasts with mutations affecting this circuit are vulnerable to SARS-CoV-2. Inborn errors of TLR3- and IRF7-dependent type I IFN immunity can underlie life-threatening COVID-19 pneumonia in patients with no prior severe infection.
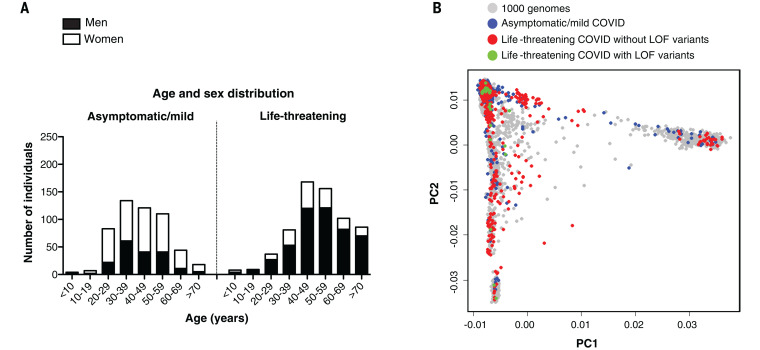
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has already claimed at least 1 million lives, has been detected in at least 20 million people, and has probably infected at least another 200 million. The clinical manifestations range from silent infection to lethal disease, with an infection fatality rate of 0.1 to 0.9%. Three epidemiological factors increase the risk of severity: (i) increasing age, decade by decade, after the age of 50, (ii) being male, and (iii) having various underlying medical conditions (1). However, even taking these factors into account, there is immense interindividual clinical variability in each demographic category considered. Following on from our human genetic studies of other severe infectious diseases (2, 3), we established the COVID Human Genetic Effort (https://www.covidhge.com) to test the general hypothesis that in some patients, life-threatening coronavirus disease 2019 (COVID-19) may be caused by monogenic inborn errors of immunity to SARS-CoV-2 with incomplete or complete penetrance (4). We enrolled 659 patients (74.5% men and 25.5% women, 13.9% of whom died) of various ancestries between 1 month and 99 years of age (Fig. 1A). These patients were hospitalized for life-threatening pneumonia caused by SARS-CoV-2 (critical COVID-19). We sequenced their whole genome (N = 364) or exome (N = 295), and principal component analysis (PCA) on these data confirmed their ancestries (Fig. 1B).
Fig. 1. Demographic and genetic data for the COVID-19 cohort.
(A) Age and sex distribution of patients with life-threatening COVID-19. (B) PCA of patient (with or without LOF variants in the 13 candidate genes) and control cohorts (patients with mild or asymptomatic disease and individuals from the 1000 Genomes Project).
Candidate variants at 13 human loci that govern immunity to influenza virus
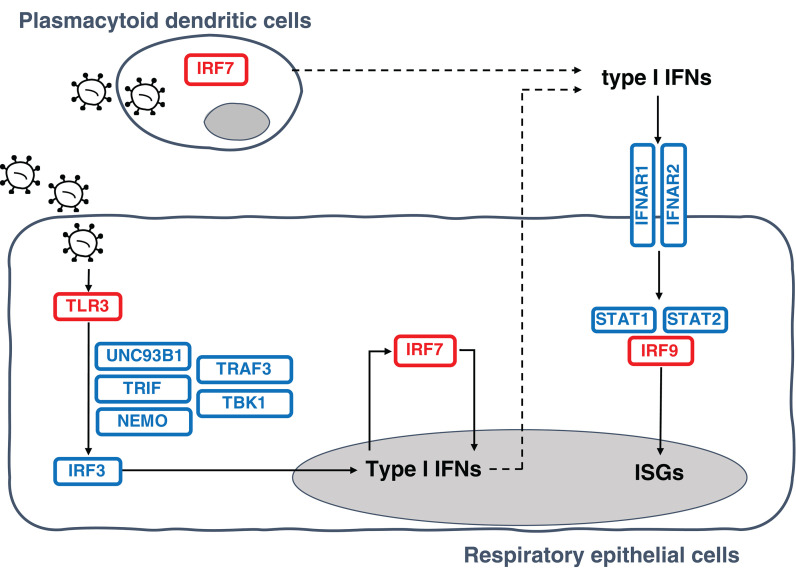
We first tested the specific hypothesis that inborn errors of Toll-like receptor 3 (TLR3)– and interferon regulatory factor 7 (IRF7)–dependent type I interferon (IFN) immunity, which underlie life-threatening influenza pneumonia, may also underlie life-threatening COVID-19 pneumonia (5) (Fig. 2). We considered three loci previously shown to be mutated in patients with critical influenza pneumonia: TLR3 (6), IRF7 (7), and IRF9 (8). We also considered 10 loci mutated in patients with other viral illnesses but directly connected to the three core genes conferring influenza susceptibility: TICAM1/TRIF (9), UNC93B1 (10), TRAF3 (11), TBK1 (12), IRF3 (13), and NEMO/IKBKG (14) in the TLR3-dependent type I IFN induction pathway, and IFNAR1 (15), IFNAR2 (16), STAT1 (17), and STAT2 (18) in the IRF7- and IRF9-dependent type I IFN amplification pathway. We collected both monoallelic and biallelic nonsynonymous variants with a minor allele frequency (MAF) <0.001 at all 13 loci. Twelve of the 13 candidate loci are autosomal, whereas NEMO is X-linked. For the latter gene, we considered only a recessive model (19). Autosomal-dominant (AD) inheritance has not been proven for six of the 12 autosomal loci (UNC93B1, IRF7, IFNAR1, IFNAR2, STAT2, and IRF9). Nevertheless, we considered heterozygous variants because none of the patients enrolled had been hospitalized for critical viral infections before COVID-19, raising the possibility that any underlying genetic defects that they might have display a lower penetrance for influenza and other viral illnesses than for COVID-19, which is triggered by a more virulent virus.
Fig. 2. Illustration of TLR3- and IRF7-dependent type I IFN production and amplification circuit.
Molecules in red are encoded by core genes, deleterious variants of which underlie critical influenza pneumonia with incomplete penetrance; deleterious variants of genes encoding biochemically related molecules in blue underlie other viral illnesses. Type I IFNs also induce themselves. ISGs, interferon-stimulated genes.
Enrichment of variants predicted to be LOF at the influenza susceptibility loci
We found four unrelated patients with biallelic variants of IRF7 or IFNAR1 (Table 1 and table S1). We also found 113 patients carrying 113 monoallelic variants at 12 loci: TLR3 (N = 7 patients/7 variants), UNC93B1 (N = 10/9), TICAM1 (N = 17/15), TRAF3 (N = 6/6), TBK1 (N = 12/11), IRF3 (N = 5/5), IRF7 (N = 20/13), IFNAR1 (N = 14/13), IFNAR2 (N = 17/15), STAT1 (N = 4/4), STAT2 (N = 11/11), and IRF9 (N = 4/4). We detected no copy number variation for these 13 genes. Unexpectedly, one of these variants has been reported in patients with life-threatening influenza pneumonia (TLR3 p.Pro554Ser) (6, 20) and another was shown to be both deleterious and dominant-negative (IFNAR1 p.Pro335del) (21). Nine of the 118 biallelic or monoallelic variants were predicted to be LOF (pLOF), whereas the remaining 109 were missense or in-frame indels (table S1). In a sample of 534 controls with asymptomatic or mild SARS-CoV-2 infection, we found only one heterozygous pLOF variation with a MAF <0.001 at the 13 loci (IRF7 p.Leu99fs). A PCA-adjusted burden test on the 12 autosomal loci revealed significant enrichment in pLOF variants in patients relative to controls [P = 0.01; odds ratio (OR) = 8.28; 95% confidence interval (CI) = 1.04 to 65.64] under an AD mode of inheritance. The same analysis performed on synonymous variants with a MAF <0.001 was not significant (P = 0.19), indicating that our ethnicity-adjusted burden test was well calibrated.
Table 1. Disease-causing variants identified in patients with life-threatening COVID-19.
| Gene | Inheritance | Genetic form | Genotype | Gender | Age [years] | Ancestry/residence | Outcome |
| TLR3 | AD | Known | p.Ser339fs/WT | M | 40 | Spain | Survived |
| TLR3 | AD | Known | p.Pro554Ser/WT | M | 68 | Italy | Survived |
| TLR3 | AD | Known | p.Trp769*/WT | M | 77 | Italy | Survived |
| TLR3 | AD | Known | p.Met870Val/WT | M | 56 | Colombia/Spain | Survived |
| UNC93B1 | AD | New | p.Glu96*/WT | M | 48 | Venezuela/Spain | Survived |
| TICAM1 | AD | Known | p.Thr4Ile/WT | M | 49 | Italy | Survived |
| TICAM1 | AD | Known | p.Ser60Cys/WT | F | 61 | Vietnam/France | Survived |
| TICAM1 | AD | Known | p.Gln392Lys/WT | F | 71 | Italy | Deceased |
| TBK1 | AD | Known | p.Phe24Ser/WT | F | 46 | Venezuela/Spain | Survived |
| TBK1 | AD | Known | p.Arg308*/WT | M | 17 | Turkey | Survived |
| IRF3 | AD | Known | p.Glu49del/WT | F | 23 | Bolivia/Spain | Survived |
| IRF3 | AD | Known | p.Asn146Lys/WT | F | 60 | Italy | Survived |
| IRF7 | AR | Known | p.Pro364fs/p.Pro364fs | F | 49 | Italy/Belgium | Survived |
| IRF7 | AR | Known | p.Met371Val/p.Asp117Asn | M | 50 | Turkey | Survived |
| IRF7 | AD | New | p.Arg7fs/WT | M | 60 | Italy | Survived |
| IRF7 | AD | New | p.Gln185*/WT | M | 44 | France | Survived |
| IRF7 | AD | New | p.Pro246fs/WT | M | 41 | Spain | Survived |
| IRF7 | AD | New | p.Arg369Gln/WT | M | 69 | Italy | Survived |
| IRF7 | AD | New | p.Phe95Ser/WT | M | 37 | Turkey | Survived |
| IFNAR1 | AR | Known | p.Trp73Cys/Trp73Cys | M | 38 | Turkey | Survived |
| IFNAR1 | AR | Known | p.Ser422Arg/Ser422Arg | M | 26 | Pakistan/Saudi Arabia | Deceased |
| IFNAR1 | AD | New | p.Pro335del/WT | F | 23 | China/Italy | Survived |
| IFNAR2 | AD | New | p.Glu140fs/WT | F | 54 | Belgium | Survived |
Experimentally deleterious alleles at the influenza susceptibility loci in 3.5% of patients
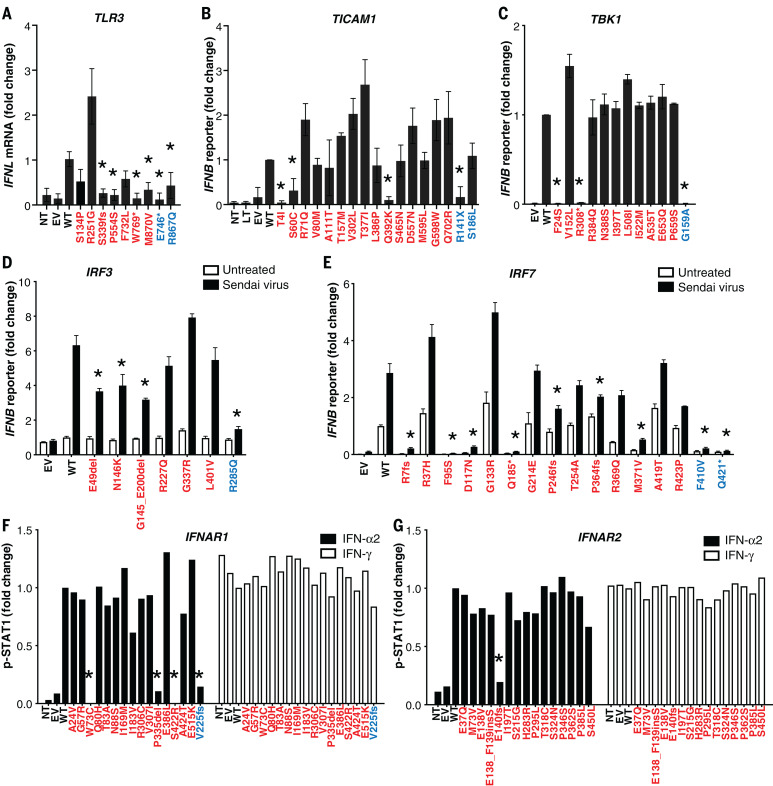
We tested these 118 variants experimentally in ad hoc overexpression systems. We found that 24 variants of eight genes were deleterious (including all the pLOF variants) because they were loss-of-expression, LOF, or severely hypomorphic: TLR3 (N = 4 variants), UNC93B1 (N = 1), TICAM1 (N = 3), TBK1 (N = 2), IRF3 (N = 2), IRF7 (N = 8), IFNAR1 (N = 3), and IFNAR2 (N = 1) (table S1, Fig. 3, and figs. S1 to S8). Consistently, heterozygous LOF variants of IRF3 and IRF7 were reported in single patients with life-threatening influenza pneumonia (22, 23). The remaining 94 variants were biochemically neutral. Twenty-three patients carried these 24 deleterious variants, resulting in four autosomal-recessive (AR) deficiencies (homozygosity or compound heterozygosity for IRF7; homozygosity for IFNAR1) and 19 AD deficiencies. These 23 patients did not carry candidate variants at the other 417 loci known to underlie inborn errors of immunity (table S2) (24–26). These findings suggest that at least 23 (3.5%) unrelated patients of the 659 patients tested suffered from a deficiency at one of eight loci among the 13 tested: four patients with a known AR disorder (IRF7 or IFNAR1) (7, 15), 11 with a known AD disorder (TLR3, TICAM1, TBK1, or IRF3) (6, 9, 12, 13, 20), and eight with a previously unknown AD genetic disorder (UNC93B1, IRF7, IFNAR1, or IFNAR2).
Fig. 3. Impact of TLR3, TICAM1, TBK1, IRF3, IRF7, IFNAR1, and IFNAR2 variants on type I IFN signaling.
(A) TLR3-deficient P2.1 fibrosarcoma cells were stably transfected with plasmids expressing WT or mutant forms of TLR3, and IFNL1 mRNA levels were determined by reverse transcription quantitative PCR. IFNL1 mRNA levels were expressed relative to the housekeeping gene GUS and then normalized. IFNL1 was undetectable in unstimulated cells. The differences between variants and WT were tested using one-way ANOVA (*P < 0.05). (B) TICAM1-deficient SV40-Fib cells were transiently transfected with WT or mutant forms of TICAM1, together with an IFN-β luciferase reporter and a constitutively expressed reporter. Normalized luciferase induction was measured 24 hours after transfection. The differences between variants and WT were tested using one-way ANOVA (*P < 0.05). (C) HEK293T cells were transiently transfected with WT and mutant forms of TBK1, together with an IFN-β luciferase reporter and a constitutively expressed reporter. Normalized luciferase activity was measured 24 hours after transfection. The differences between variants and WT were tested using one-way ANOVA (*P < 0.05). (D) IRF3-deficient HEK293T cells were transiently transfected with WT and mutant forms of IRF3, together with an IFN-β luciferase reporter and a constitutively expressed reporter. Cells were either left untreated or infected with Sendai virus for 24 hours before the normalized measurement of luciferase activity. The differences between variants and WT were evaluated using two-way ANOVA (*P < 0.05). (E) HEK293T cells were transiently transfected with WT and mutant forms of IRF7, together with an IFN-β luciferase reporter and a constitutively expressed reporter. Cells were either left untreated or infected with Sendai virus for 24 hours before the normalized measurement of luciferase activity. The differences between variants and WT were tested using two-way ANOVA (*P < 0.05). (F and G) IFNAR1- or IFNAR2-deficient SV40-Fib cells were transiently transfected with WT or mutant forms of IFNAR1 for 36 hours, and either left untreated or stimulated with IFN-α2 or IFN-γ. Fluorescence-activated cell sorting (FACS) staining with anti-p-STAT1 antibody and the z-score of the MFI were assessed. Asterisks indicate variants with MFI <50% of WT. Variants in red were identified in COVID-19 patients. Variants in blue are known deleterious variants and served as negative controls. EV, empty vector; LT, lipofectamine. Three technical repeats were performed for (A) to (E). Means and SD are shown in the columns and horizontal bars when appropriate.
Impaired TLR3- and IRF7-dependent type I immunity in patient cells in vitro
We tested cells from patients with selected genotypes and showed that PHA-driven T cell blasts (PHA-T cells) from patients with AR or AD IRF7 deficiency had low levels of IRF7 expression (Fig. 4A). We then isolated circulating plasmacytoid dendritic cells (pDCs) from a patient with AR IRF7 deficiency (fig. S9A) (7). These cells were present in normal proportions (fig. S9B), but they did not produce any detectable type I or III IFNs in response to SARS-CoV-2, as analyzed by cytometric bead array (CBA), enzyme-linked immunosorbent assay (ELISA), and RNA sequencing (RNA-seq) (Fig. 4, B and C). We also showed that PHA-T cells from a patient with AR IFN-α/β receptor 1 (IFNAR1) deficiency had impaired IFNAR1 expression and responses to IFN-α2 or IFN-β, and that the patient’s SV40-transformed fibroblast (SV40-Fib) cells did not respond to IFN-α2 or IFN-β (Fig. 5). We then infected TLR3−/−, TLR3+/−, IRF7−/− SV40-Fib cells, and IRF7−/− SV40-Fib cells rescued with wild-type (WT) IRF7; IFNAR1−/− SV40-Fib cells, and IFNAR1−/− SV40-Fib cells rescued with WT IFNAR1, all of which were previously transduced with angiotensin-converting enzyme 2 (ACE2) and transmembrane protease, serine 2 (TMPRSS2). SARS-CoV-2 infection levels were higher in mutant cells than in cells from healthy donors, and transduction of WT IRF7 or IFNAR1 rescued their defects (Fig. 6). Collectively, these findings showed that AR IRF7 deficiency impaired the production of type I IFN by pDCs stimulated with SARS-CoV-2, whereas AR and AD deficiencies of TLR3 or AR deficiency of IFNAR1 impaired fibroblast-intrinsic type I IFN immunity to SARS-CoV2. They also suggest that heterozygosity for LOF variations at the other five mutated loci also underlie life-threatening COVID-19.
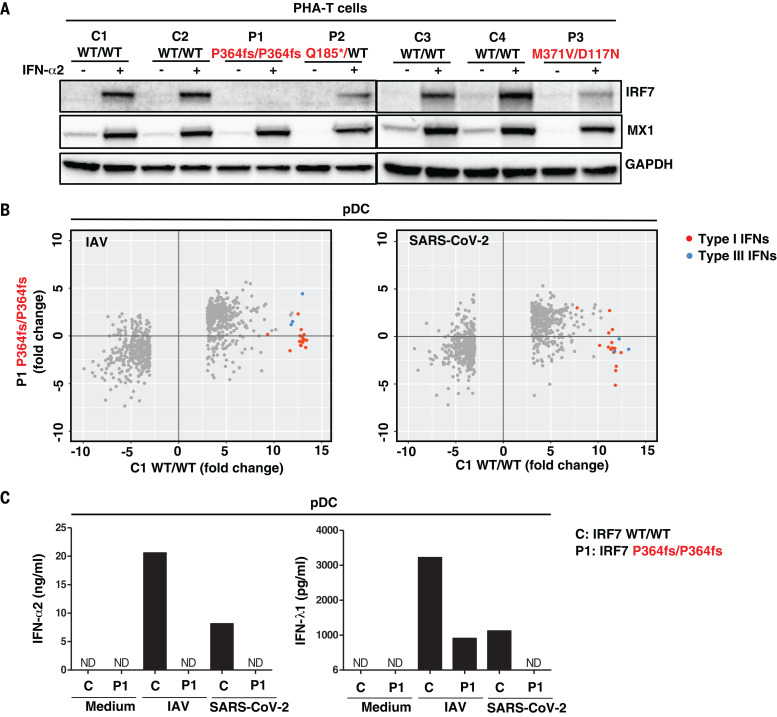
Fig. 4. Type I IFN responses in patient cells defective for IRF7.
(A) Levels of the IRF7 protein in PHA-T cells from two patients with AR IRF7 deficiency (P1 and P3), one patient with AD IRF7 deficiency (P2), and four healthy donors (C1 to C4). Cells were either left untreated or stimulated with IFN-α2 for 24 hours, and protein levels were measured by Western blotting. MX1 was used as a positive control for IFN-α2 treatment. (B) pDCs isolated from an AR IRF7-deficient patient (P1) and a healthy donor (C1) were either left untreated or infected with influenza A virus (IAV) or SARS-CoV-2, and RNA-seq was performed. Genes with expression >2.5-fold higher or lower in C1 after infection are plotted as the fold change in expression. Red dots are type I IFN genes; blue dots are type III IFN genes. (C) pDCs isolated from healthy donor C and IRF7-deficient patient (P1) were either left untreated (Medium) or infected with IAV or SARS-CoV-2, and the production of IFN-α2 and IFN-λ1 was measured by CBA and ELISA, respectively, on the supernatant. ND, not detected.
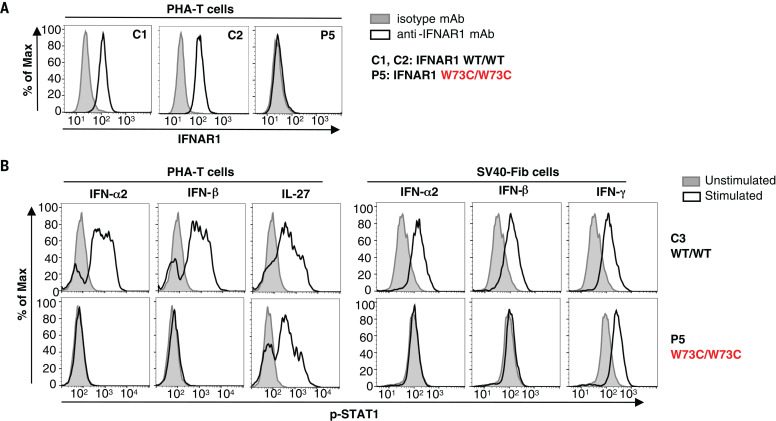
Fig. 5. Type I IFN responses in patient cells defective for IFNAR1.
(A) FACS staining of IFNAR1 on the surface of PHA-T cells from a patient with AR IFNAR1 deficiency (P5) and healthy donors (C1 and C2). (B) PHA-T cells and SV40-Fib from a patient with AR IFNAR1 deficiency (P5) and a healthy donor (C3) were stimulated with IFN-α2 or IFN-β, and p-STAT1 levels were determined by FACS. Interleukin-27 stimulation served as a positive control on PHA-T cells, whereas IFN-γ stimulation served as a positive control on SV40-Fib cells.
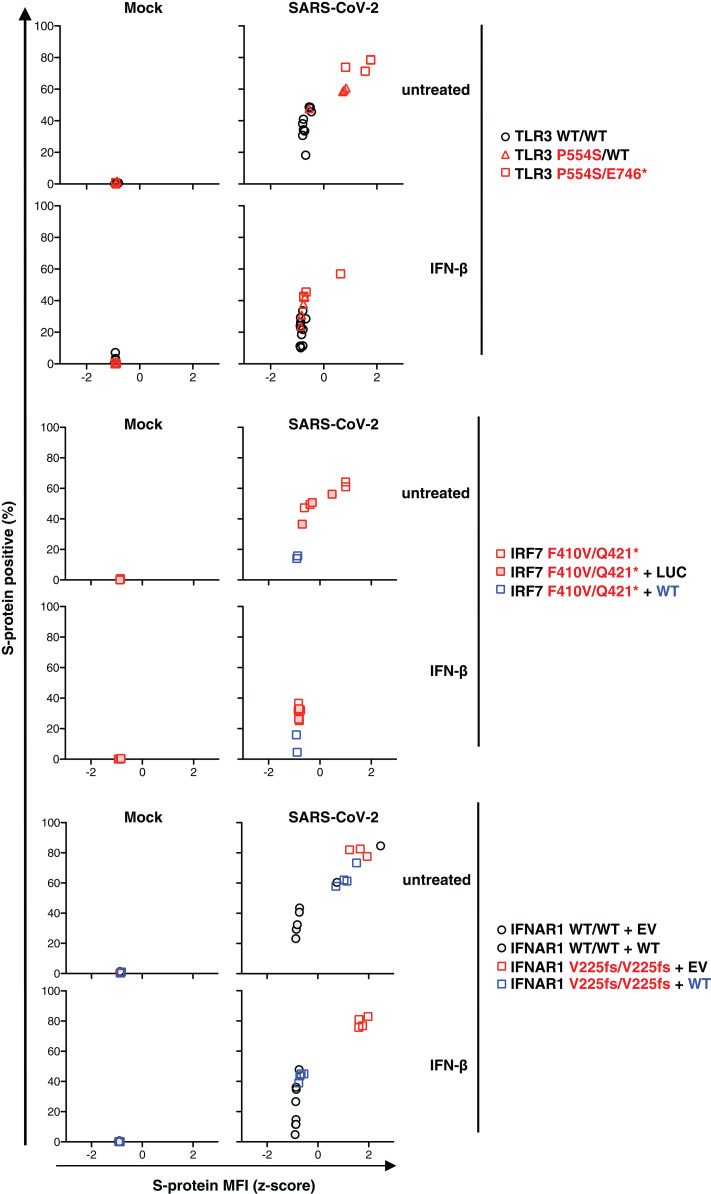
Fig. 6. Cell-intrinsic type I IFN response to SARS-CoV-2.
SV40-Fib cells of TLR3−/−, TLR3+/−, IRF7−/−, and IRF7−/− SV40-Fib cells rescued with WT IRF7; IFNAR1−/− SV40-Fib cells, and IFNAR1−/− SV40-Fib cells rescued with WT IFNAR1 were transduced with ACE2 and TMPRSS2 and then either left untreated or treated with IFN-β for 4 hours. Cells were then infected with SARS-CoV-2 (MOI = 0.5). After staining, ACE2 and viral S-protein levels were measured by high-content microscopy with gating on ACE2+ cells. IRF7-deficient SV40-Fib cells were previously transduced with either WT IRF7 or negative control (Luc). IFNAR1-deficient cells were previously transduced with either WT IFNAR1 or empty vector (EV).
Impaired production of type I IFNs in patients in vivo
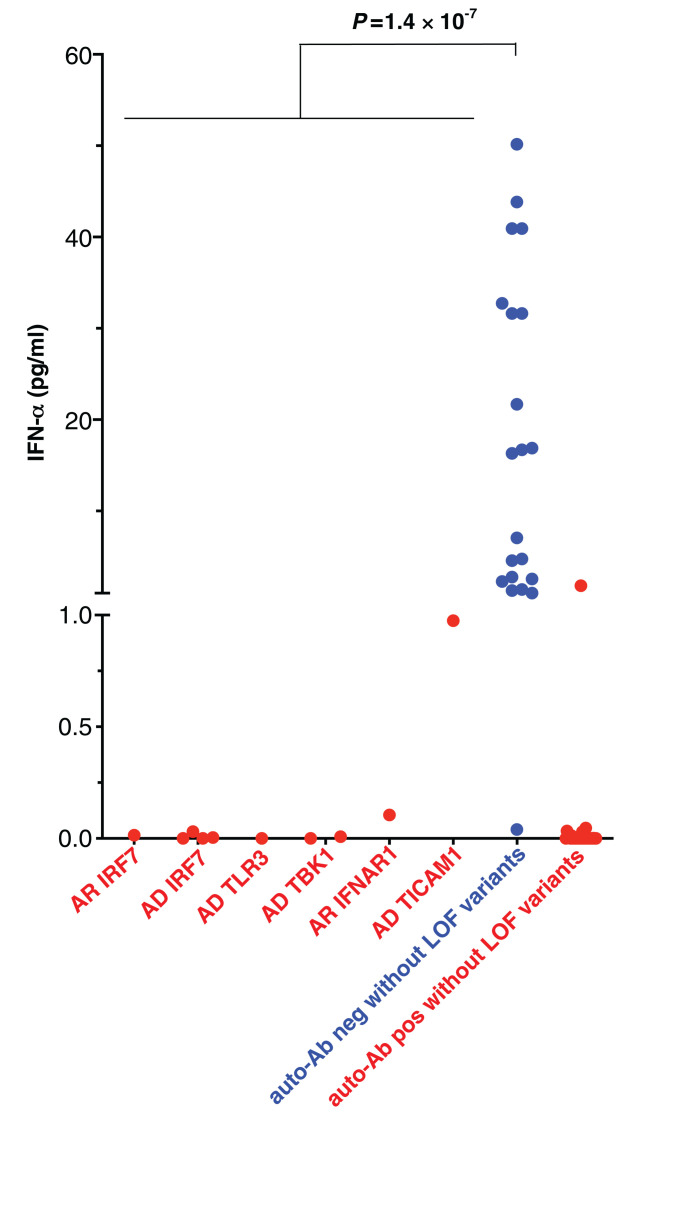
We tested whether these genotypes impaired the production of type I IFN in vivo during the course of SARS-CoV-2 infection. We measured the levels of the 13 types of IFN-α in the blood of patients during the acute phase of COVID-19. We found that 10 of the 23 patients with mutations for whom samples were available (one with AR IRF7 deficiency, four with AD IRF7 deficiency, one with AD TLR3 deficiency, two with AD TBK1 deficiency, one with AR IFNAR1 deficiency, and one with AD TICAM1 deficiency) had serum IFN-α levels <1 pg/ml (Fig. 7). By contrast, previously published cohorts of patients hospitalized with unexplained, severe COVID-19 had various serum IFN-α levels, significantly higher than our 10 patients [one-way analysis of variance (ANOVA), P = 1.4 × 10−7; Fig. 7] (27, 28). Another 29 patients from our cohort displaying auto-antibodies (auto-Abs) against type I IFNs, reported in an accompanying paper, had undetectable levels of serum IFN-α (29). Moreover, none of the 23 patients with LOF mutations of the eight genes had detectable auto-Abs against type I IFNs (29), strongly suggesting that the two mechanisms of disease are similar but independent. Excluding patients with auto-Abs against type I IFN from the burden test of pLOF variants at the 12 autosomal loci strengthened the association signal (P = 0.007; OR = 8.97; 95% CI = 1.13 to 71.09).
Fig. 7. In vivo type I IFN responses to SARS-CoV-2 infections.
Plasma levels of 13 IFN-α were measured by Simoa. Auto-Ab(+) without LOF variants indicates COVID-19 patients with neutralizing anti-IFN-α auto-Abs in our accompanying report (29). P values indicated were evaluated using one-way ANOVA.
Inborn errors of TLR3- and IRF7-dependent type I immunity underlie critical COVID-19
Collectively, our data suggest that at least 23 of the 659 patients with life-threatening COVID-19 pneumonia studied had known (six disorders) or new (four disorders) genetic defects at eight loci involved in the TLR3- and IRF7-dependent induction and amplification of type I IFNs. This discovery reveals the essential role of both the double-stranded RNA sensor TLR3 and type I IFN cell-intrinsic immunity in the control of SARS-CoV-2 infection in the lungs, consistent with their previously documented roles in pulmonary immunity to influenza virus (5–8). These genotypes were silent until infection with SARS-CoV-2. The most thought-provoking examples are the AR deficiencies of IRF7 and IFNAR1. AR IRF7 deficiency was diagnosed in two individuals aged 49 and 50 years, and AR IFNAR1 deficiency was diagnosed in two individuals aged 26 and 38 years, and none of the four patients had a prior history of life-threatening infections (Table 1). One patient with IRF7 deficiency was tested and was seropositive for several common viruses, including various influenza A and B viruses (figs. S10 and S11). These genetic defects therefore display incomplete penetrance for influenza respiratory distress and only manifested clinically upon infection with the more virulent SARS-CoV-2.
Conclusion
The AR form of IFNAR1 deficiency highlights the importance of type I IFN production relative to type III IFN production, which is also impaired by defects of TLR3, IRF7, and IRF9 (5). This conclusion is also supported by our accompanying report of neutralizing auto-Abs against type I IFNs, but not type III IFNs, in other patients with life-threatening COVID-19 pneumonia (29). Inborn errors of TLR3- and IRF7-dependent type I IFN immunity at eight loci were found in as many as 23 patients (3.5%) of various ages (17 to 77 years) and ancestries (various nationalities from Asia, Europe, Latin America, and the Middle East) and in patients of both sexes (Table 1). Our findings suggest that there may be mutations in other type I IFN–related genes in other patients with life-threatening COVID-19 pneumonia. They also suggest that the administration of type I IFN may be of therapeutic benefit in selected patients, at least early in the course of SARS-CoV-2 infection.
Methods
Patients
We included in this study 659 patients with life-threatening COVID-19 pneumonia, defined as patients with pneumonia who developed critical disease, whether pulmonary with mechanical ventilation (CPAP, BIPAP, intubation, hi-flow oxygen), septic shock, or with any other organ damage requiring admission to the intensive care unit. Patients who developed Kawasaki-like syndrome were excluded. The age of the patients ranged from 0.1 to 99 years, with a mean age of 51.8 years (SD 15.9 years), and 25.5% of the patients were female. As controls, we enrolled 534 individuals infected with SARS-CoV-2 based on a positive polymerase chain reaction (PCR) and/or serological test and/or the presence of typical symptoms such as anosmia or ageusia after exposure to a confirmed COVID-19 case, who remained asymptomatic or developed mild, self-healing, ambulatory disease.
Next-generation sequencing
Genomic DNA was extracted from whole blood. For the 1193 patients and controls included, the whole exome (N = 687) or whole genome (N = 506) was sequenced. We used the Genome Analysis Software Kit (GATK) (version 3.4-46 or 4) best-practice pipeline to analyze our whole-exome–sequencing data (30). We aligned the reads obtained with the human reference genome (hg19) using the maximum exact matches algorithm in Burrows–Wheeler Aligner software (31). PCR duplicates were removed with Picard tools (http://broadinstitute.github.io/picard/). The GATK base quality score recalibrator was applied to correct sequencing artifacts.
All of the variants were manually curated using Integrative Genomics Viewer (IGV) and confirmed to affect the main functional protein isoform by checking the protein sequence before inclusion in further analyzes. The main functional protein isoforms were TLR3 (NM_003265), UNC93B1 (NM_030930.4), TICAM1 (NM_182919), TRAF3 (NM_145725.2), TBK1 (NM_013254.4), IRF3 (NM_001571), IRF7 (NM_001572.5), IFNAR1 (NM_000629.3), IFNAR2 (NM_001289125.3), STAT1 (NM_007315.4), STAT2 (NM_005419.4), and IRF9 (NM_006084.5). The analysis of IKBKG was customized to unmask the duplicated region in IKBKG using a specific pipeline previously described (32). We searched the next-generation–sequencing data for deletions in the 13 genes of interest using both the HMZDelFinder (33) and CANOES (34) algorithms.
Statistical analysis
We performed an enrichment analysis on our cohort of 659 patients with life-threatening COVID-19 pneumonia and 534 SARS-CoV2–infected controls, focusing on 12 autosomal IFN-related genes. We considered variants that were pLOF with a MAF <0.001 (gnomAD version 2.1.1) after experimentally demonstrating that all of the pLOF variants seen in the cases were actually LOF. We compared the proportion of individuals carrying at least one pLOF variant of the 12 autosomal genes in cases and controls by means of logistic regression with the likelihood ratio test. We accounted for the ethnic heterogeneity of the cohorts by including the first three principal components of the PCA in the logistic regression model. PC adjustment is a common and efficient strategy for accounting for different ancestries of patients and controls in the study of rare variants (35–38). We checked that our adjusted burden test was well calibrated by also performing an analysis of enrichment in rare (MAF <0.001) synonymous variants of the 12 genes. PCA was performed with Plink version 1.9 software on whole-exome– and whole-genome–sequencing data and the 1000 Genomes (1kG) Project phase 3 public database as a reference, using 27,480 exonic variants with a MAF >0.01 and a call rate >0.99. The OR was also estimated by logistic regression and adjusted for ethnic heterogeneity.
Reporter assays
Cell lines or SV40-Fib cells with known defects were transiently or stably transfected with WT, mutant variants, IFN-β- or ISRE-firefly luciferase reporter, and pRL-TK-Renilla luciferase reporter. Reporter activity was measured with the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Firefly luciferase activity was normalized against Renilla luciferase activity and expressed as a fold change. TRAF3-deficient human embryonic kidney (HEK) 293T cells were kindly provided by M. Romanelli (39).
pDC activation by SARS-CoV-2 and cytokine production
pDCs from an IRF7−/− patient and a healthy donor matched for age and sex were cultured in the presence of medium alone, influenza virus (A/PR/8/34, 2 μg/ml; Charles River Laboratories), or the SARS-CoV-2 primary strain 220_95 (GISAID accession ID: EPI_ISL_469284) at a multiplicity of infection (MOI) of 2. After 12 hours of culture, pDC supernatant was collected for cytokine quantification. IFN-α2 levels were measured using CBA analyzis (BD Biosciences) in accordance with the manufacturer’s protocol using a 20 pg/ml detection limit. IFN-λ1 secretion was measured in an ELISA (R&D Systems, DuoSet DY7246), in accordance with the manufacturer’s instructions.
SARS-CoV-2 infection in patient SV40-Fib
To make patient-derived fibroblasts permissive to SARS-CoV-2 infection, we delivered human ACE2 and TMPRSS2 cDNA to cells by lentivirus transduction using a modified SCRPSY vector (GenBank ID: KT368137.1). SARS-CoV-2 strain USA-WA1/2020 was obtained from BEI Resources. ACE2/TMPRSS2-transduced cells were either left untreated or treated with 500 U/ml IFN-β (11415-1, PBL Assay Science) 4 hours before infection. Cells were infected with SARS-CoV-2 (MOI = 0.5) for 1 hour at 37°C. After 24 hours of infection, cells were fixed and taken out of the BSL3 for staining.
After fixation, cells were stained with SARS-CoV-2 and ACE2 primary antibodies (0.5 and 1 μg/ml, respectively). Primary antibodies were as follows: for SARS-CoV-2, human monoclonal anti-spike-SARS-CoV-2 C121 antibody (40), and for ACE2, mouse monoclonal Alexa Fluor 488–conjugated antibody (FAB9332G-100UG, R&D Systems). Images were acquired with an ImageXpress Micro XLS microscope (Molecular Devices) using the 4× objective. MetaXpress software (Molecular Devices) was used to obtain single-cell mean fluorescence intensity (MFI) values.
Data analysis on single-cell MFI values was done in the R environment (version 4.0.2). ACE2/TMPRSS2-transduced cells were classified as ACE2 positive when the ACE2 log MFI was superior to the log mean MFI of mock-transduced cells plus 2.5 SDs. We excluded all wells with <150 ACE2-positive cells before SARS-CoV-2 scoring. ACE2-expressing cells were classified SARS-CoV-2 positive when the fluorescence intensity value was superior to the MFI of mock-infected cells plus 4 SDs. The median SARS-CoV-2 MFI and percentage SARS-CoV-2–positive cells were calculated for each well (independent infection).
Single-molecule array (Simoa) IFN-α digital ELISA
Serum IFN-α concentrations were determined using Simoa technology, with reagents and procedures obtained from Quanterix Corporation (Quanterix SimoaTM IFNα Reagent Kit, Lexington, MA, USA). According to the manufacturer’s instructions, the working dilutions were 1:2 for all sera in working volumes of 170 μl.
Acknowledgments
We thank the patients, their families, and healthy donors for placing their trust in us; Y. Nemirowskaya, D. Papandrea, M. Woollet, D. Liu, C. Rivalain, and C. Patissier for administrative assistance; A. Adeleye, D. Bacikova, E. McGrath Martinez, A. R. Soltis, K. Dobbs, J. Danielson, H. Matthews, and S. Weber for technical and other assistance; M. M. A. Ata and F. Al Ali for their contribution to VirScan experiments; S. Elledge (Brigham and Women’s Hospital and Harvard Medical School, Boston, MA) for kindly providing the VirScan phage library used in this study; A. W. Ashbrook, the BSL3 manager of the Rice laboratory assistance; M. Lazzaro, Director of Immigration and Academic Appointments, for assistance; W. Chung, K. Kiryluk, S. O'Byrne, D. Pendrick, J. Williamson, C. Andrews, and M. Disco in the J.M. lab for assistance; M. Andreoni (Tor Vergata, Italy) for his clinical contribution; and A. Novelli (Bambino Gesù Hospital, Italy) for his collaboration. We thank the GEN-COVID Multicenter study (https://sites.google.com/dbm.unisi.it/gen-covid). This study used the high-performance computational resources of the National Institutes of Health (NIH) HPC Biowulf cluster (http://hpc.nih.gov) and the Office of Cyber Infrastructure and Computational Biology (OCICB) High Performance Computing (HPC) cluster at the National Institute of Allergy and Infectious Diseases (NIAID), Bethesda, MD. The opinions and assertions expressed herein are those of the authors and are not to be construed as reflecting the views of the Uniformed Services University of the Health Sciences (USUHS) or the U.S. Department of Defense (DoD). Funding: This work was supported by a generous donation from the Fisher Center for Alzheimer’s Research Foundation. The Laboratory of Human Genetics of Infectious Diseases is supported by the Howard Hughes Medical Institute, the Rockefeller University, the St. Giles Foundation, the NIH (R01AI088364), the National Center for Advancing Translational Sciences (NCATS), the NIH Clinical and Translational Science Award (CTSA) program (UL1 TR001866), a Fast Grant from Emergent Ventures, Mercatus Center at George Mason University, the Yale Center for Mendelian Genomics and the GSP Coordinating Center funded by the National Human Genome Research Institute (NHGRI) (UM1HG006504 and U24HG008956), the French National Research Agency (ANR) under the “Investments for the Future” program (ANR-10-IAHU-01), the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence (ANR-10-LABX-62-IBEID), the French Foundation for Medical Research (FRM) (EQU201903007798), the FRM and ANR GENCOVID project, ANRS-COV05, the Square Foundation, Grandir–Fonds de Solidarité pour l’Enfance, the SCOR Corporate Foundation for Science, Institut National de la Santé et de la Recherche Médicale (INSERM), the University of Paris. The French COVID Cohort study group was sponsored by Inserm and supported by the REACTing consortium and by a grant from the French Ministry of Health (PHRC 20-0424). Regione Lombardia, Italy (project “Risposta immune in pazienti con COVID-19 e co-morbidità”), and the Intramural Research Program of the NIAID, NIH. The laboratory of Genomes & Cell Biology of Disease is supported by “Integrative Biology of Emerging Infectious Diseases” (grant no. ANR-10-LABX-62-IBEID), the “Fondation pour la Recherche Medicale” (grant FRM–EQU202003010193), the “Agence Nationale de la Recherche” (ANR FLASH COVID project IDISCOVR cofounded by the “Fondation pour la Recherche Médicale”), University of Paris (“Plan de Soutien Covid-19”: RACPL20FIR01-COVID-SOUL). I.M. is a senior clinical investigator with the FWO Vlaanderen; I.M. and L.M. are supported by FWO G0C8517N – GOB5120N. The VS team was supported by “Agence Nationale de la Recherche” (ANR-17-CE15-0003, ANR-17-CE15-0003-01) and by Université de Paris “PLAN D’URGENCE COVID19”. L.K. was supported by a fellowship from the French Ministry of Research. V.S.-S. is supported by a UKRI Future Leaders Fellowship (MR/S032304/1). S.Z.A.-M. is supported by the Elite Journals Program at King Saud University through grant no. PEJP-16-107. The J.M. laboratory is supported by Columbia University COVID biobank and grant no. UL1TR001873. Work in the Laboratory of Virology and Infectious Disease was supported by NIH grants P01AI138398-S1, 2U19AI111825, and R01AI091707-10S1; a George Mason University Fast Grant; and the G. Harold and Leila Y. Mathers Charitable Foundation. J.L.P. is supported by a European Molecular Biology Organization Long-Term Fellowship (ALTF 380-2018). Work at the Neurometabolic Diseases Laboratory received funding from the European Union’s Horizon 2020 research and innovation program under grant no. 824110 (EasiGenomics grant no. COVID-19/PID12342) to A.P., and Roche and Illumina Covid Match Funds to M.G.. C.R.G. and colleagues are supported by Instituto de Salud Carlos III (COV20_01333 and COV20_01334), Spanish Ministry of Science and Innovation, with the funding of European Regional Development Fund-European Social Fund -FEDER-FSE; (RTC-2017-6471-1; AEI/FEDER, UE), and Cabildo Insular de Tenerife (CGIEU0000219140 and “Apuestas científicas del ITER para colaborar en la lucha contra la COVID-19”). D.C.V. is supported by the Fonds de la recherche en santé du Québec clinician-scientist scholar program. H.S. is adjunct faculty at the University of Pennsylvania. A.-L.N. was supported by the Foundation Bettencourt Schueller. The Amsterdam UMC Covid-19 Biobank was funded by the Netherlands Organization for Health Research and Development (ZonMw, NWO-vici 91819627), The Corona Research Fund (Amsterdam UMC), Dr. J. C. Vaillantfonds, and Amsterdam UMC. Work on COVID-19 at the A.G.-S. laboratory is partly supported by NIH supplements to grants U19AI135972, U19AI142733, and R35 HL135834, and to contract HHSN272201800048C, by a DoD supplement to grant W81XWH-20-1-0270, by DARPA project HR0011-19-2-0020, by CRIP (Center for Research on Influenza Pathogenesis), a NIAID funded Center of Excellence for Influenza Research and Surveillance (CEIRS, contract HHSN272201400008C), by an NIAID funded Collaborative Influenza Vaccine Innovation Center (SEM-CIVIC, contract 75N93019C00051) and by the generous support of the JPB Foundation, the Open Philanthropy Project (research grant 2020-215611(5384)) and anonymous donors. The Virscan analysis presented in fig. S11 was performed with financial support from Sidra Medicine. J.R.H. is supported by Biomedical Advanced Research and Development Authority under Contract (HHSO10201600031C). Author contributions: A.G., A.A., A.A.A., A.L.S., A.-L.N., A.C., A.C., A.P., B.B., B.S.R., C.A., C.M., C.K., C.L., C.M.R., C.L.D., D.D., E.M., E.J., F.A., F.A-M., F.O., F.A., F.K., G.N., G.S., G.G., H.-H.H., H.K.A.S., H.S., I.K.D.S., I.M., J.L.P., J.R., J.E.H., J.C., J.M., J.Y., K.D., K.B., L.A., L.L.-D., L.K., L.M., L.B-M., L.B., L.D.N., M.M-V., M.C., M.O., M.C., M.N., M.F.T., M.S., M.F.A., N.M., N.S., P.B., P.M., Q.Z., Q.Z., Q.P., R.L., R.Y., S.A.T., S.Z.A-M., S.H., S.K., S.H., S.B.-D., T.K., T.M., T.H.M., V.S.-S., V.S., V.B., W.S., X.D., Y.S., and Z.L. either performed or supervised experiments, generated and analyzed data, and contributed to the manuscript. A.S., A.C.U., A.B., A.O., A.P., B.B., D.V.D.B., F.R., G.K., J.M., P.Z., S-Y.Z., T.L.-V., Y.S., and Y.Z. performed computational analysis. A.S., A.N.S., A.M.-N., A.B., C.R., D.M., D.C.V., E.Q.-R., F.H., I.M., I.V., J.B., J.-C.G., L.R.B., L.R., L.I., M.D., P.B., P.S.-P., P.-E.M., R.H., R.C., S.K., S.P., T.O., Y.T.-L., K.K., S.S., J.F., and S.N.K. evaluated and recruited patients to COVID and/or control cohorts. Q.Z. and J.-L.C. wrote the manuscript. All authors edited the manuscript. J.-L.C. supervised the project. Competing interests: The authors declare no competing financial interests. J.-L.C. is listed as an inventor on patent application US63/055,155 filed by The Rockefeller University that encompasses aspects of this publication. R.L. is a non-executive director of Roche and its subsidiary Genentech. Data and materials availability: Plasma, cells, and genomic DNA are available from J.-L.C. or D.V. under a material transfer agreement with Rockfeller University/Research Institute-McGill University Health Centre. pSCRPSY_TMPRSS2-2A-NeoR_ACE2 and Huh-7.5 cells are available upon request from C.R. under a material transfer agreement with The Rockefeller University, or The Rockefeller University and Apath, LLC, respectively. Clinical data, DNA, and other patient samples are available from the Amsterdam UMC Covid-19 Biobank (D.v.d.B.) under a material transfer agreement with Amsterdam UMC. Material and reagents used are almost exclusively commercially available and nonproprietary. Requests for materals derived from human samples may be made available, subject to any underlying restrictions on such samples. J.-L.C. can make material transfer agreements available through The Rockefeller University. Detailed genotype counts for all coding variants in the genes investigated in this manuscript are available at Dryad (41). The whole-genome sequencing datasets used for the analyses, including critical patients and asymptomatic controls described in this manuscript, were deposited in dbGaP under accession number phs002245.v1.p1. All other data are available in the manuscript or the supplementary material. This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using such material.
COVID-STORM Clinicians
Giuseppe Foti1, Giacomo Bellani1, Giuseppe Citerio1, Ernesto Contro1, Alberto Pesci2, Maria Grazia Valsecchi3, Marina Cazzaniga4
1Department of Emergency, Anesthesia and Intensive Care, School of Medicine and Surgery, University of Milano-Bicocca, San Gerardo Hospital, Monza, Italy. 2Department of Pneumology, School of Medicine and Surgery, University of Milano-Bicocca, San Gerardo Hospital, Monza, Italy. 3Center of Bioinformatics and Biostatistics, School of Medicine and Surgery, University of Milano-Bicocca, San Gerardo Hospital, Monza, Italy. 4Phase I Research Center, School of Medicine and Surgery, University of Milano-Bicocca, San Gerardo Hospital, Monza, Italy.
COVID Clinicians
Jorge Abad1, Sergio Aguilera-Albesa2, Ozge Metin Akcan3, Ilad Alavi Darazam4, Juan C. Aldave5, Miquel Alfonso Ramos6, Seyed Alireza Nadji7, Gulsum Alkan8, Jerome Allardet-Servent9, Luis M. Allende10, Laia Alsina11, Marie-Alexandra Alyanakian12, Blanca Amador-Borrero13, Zahir Amoura14, Arnau Antolí15, Sevket Arslan16, Sophie Assant17, Terese Auguet18, Axelle Azot19, Fanny Bajolle20, Aurélie Baldolli21, Maite Ballester22, Hagit Baris Feldman23, Benoit Barrou24, Alexandra Beurton25, Agurtzane Bilbao26, Geraldine Blanchard-Rohner27, Ignacio Blanco1, Adeline Blandinières28, Daniel Blazquez-Gamero29, Marketa Bloomfield30, Mireia Bolivar-Prados31, Raphael Borie32, Cédric Bosteels33, Ahmed A. Bousfiha34, Claire Bouvattier35, Oksana Boyarchuk36, Maria Rita P. Bueno37, Jacinta Bustamante20, Juan José Cáceres Agra38, Semra Calimli39, Ruggero Capra40, Maria Carrabba41, Carlos Casasnovas42, Marion Caseris43, Martin Castelle44, Francesco Castelli45, Martín Castillo de Vera46, Mateus V. Castro37, Emilie Catherinot47, Martin Chalumeau48, Bruno Charbit49, Matthew P. Cheng50, Père Clavé31, Bonaventura Clotet51, Anna Codina52, Fatih Colkesen53, Fatma Çölkesen54, Roger Colobran55, Cloé Comarmond56, David Dalmau57, David Ross Darley58, Nicolas Dauby59, Stéphane Dauger60, Loic de Pontual61, Amin Dehban62, Geoffroy Delplancq63, Alexandre Demoule64, Jean-Luc Diehl65, Stephanie Dobbelaere66, Sophie Durand67, Waleed Eldars68, Mohamed Elgamal69, Marwa H. Elnagdy70, Melike Emiroglu71, Emine Hafize Erdeniz72, Selma Erol Aytekin73, Romain Euvrard74, Recep Evcen75, Giovanna Fabio41, Laurence Faivre76, Antonin Falck43, Muriel Fartoukh77, Morgane Faure78, Miguel Fernandez Arquero79, Carlos Flores80, Bruno Francois81, Victoria Fumadó82, Francesca Fusco83, Blanca Garcia Solis84, Pascale Gaussem85, Juana Gil-Herrera86, Laurent Gilardin87, Monica Girona Alarcon88, Mònica Girona-Alarcón88, Jean-Christophe Goffard89, Funda Gok90, Rafaela González-Montelongo91, Antoine Guerder92, Yahya Gul93, Sukru Nail Guner93, Marta Gut94, Jérôme Hadjadj95, Filomeen Haerynck96, Rabih Halwani97, Lennart Hammarström98, Nevin Hatipoglu99, Elisa Hernandez-Brito100, Cathérine Heijmans101, María Soledad Holanda-Peña102, Juan Pablo Horcajada103, Levi Hoste104, Eric Hoste105, Sami Hraiech106, Linda Humbert107, Alejandro D. Iglesias108, Antonio Íñigo-Campos91, Matthieu Jamme109, María Jesús Arranz110, Iolanda Jordan111, Philippe Jorens112, Fikret Kanat113, Hasan Kapakli114, Iskender Kara115, Adem Karbuz116, Kadriye Kart Yasar117, Sevgi Keles118, Yasemin Kendir Demirkol119, Adam Klocperk120, Zbigniew J. Król121, Paul Kuentz122, Yat Wah M. Kwan123, Jean-Christophe Lagier124, Bart N. Lambrecht33, Yu-Lung Lau125, Fleur Le Bourgeois60, Yee-Sin Leo126, Rafael Leon Lopez127, Daniel Leung125, Michael Levin128, Michael Levy60, Romain Lévy20, Zhi Li49, Agnes Linglart129, Bart Loeys130, José M. Lorenzo-Salazar91, Céline Louapre131, Catherine Lubetzki131, Charles-Edouard Luyt132, David C. Lye133, Davood Mansouri134, Majid Marjani135, Jesus Marquez Pereira136, Andrea Martin137, David Martínez Pueyo138, Javier Martinez-Picado139, Iciar Marzana140, Alexis Mathian14, Larissa R. B. Matos37, Gail V. Matthews141, Julien Mayaux142, Jean-Louis Mège143, Isabelle Melki144, Jean-François Meritet145, Ozge Metin146, Isabelle Meyts147, Mehdi Mezidi148, Isabelle Migeotte149, Maude Millereux150, Tristan Mirault151, Clotilde Mircher67, Mehdi Mirsaeidi152, Abián Montesdeoca Melián153, Antonio Morales Martinez154, Pierre Morange155, Clémence Mordacq107, Guillaume Morelle156, Stéphane Mouly13, Adrián Muñoz-Barrera91, Leslie Naesens157, Cyril Nafati158, João Farela Neves159, Lisa FP. Ng160, Yeray Novoa Medina161, Esmeralda Nuñez Cuadros162, J. Gonzalo Ocejo-Vinyals163, Zerrin Orbak164, Mehdi Oualha20, Tayfun Özçelik165, Qiang Pan-Hammarström166, Christophe Parizot142, Tiffany Pascreau167, Estela Paz-Artal168, Sandra Pellegrini49, Rebeca Pérez de Diego84, Aurélien Philippe169, Quentin Philippot77, Laura Planas-Serra170, Dominique Ploin171, Julien Poissy172, Géraldine Poncelet43, Marie Pouletty173, Paul Quentric142, Didier Raoult143, Anne-Sophie Rebillat67, Ismail Reisli174, Pilar Ricart175, Jean-Christophe Richard176, Nadia Rivet28, Jacques G. Rivière177, Gemma Rocamora Blanch15, Carlos Rodrigo1, Carlos Rodriguez-Gallego178, Agustí Rodríguez-Palmero179, Carolina Soledad Romero180, Anya Rothenbuhler181, Flore Rozenberg182, Maria Yolanda Ruiz del Prado183, Joan Sabater Riera15, Oliver Sanchez184, Silvia Sánchez-Ramón185, Agatha Schluter170, Matthieu Schmidt186, Cyril E. Schweitzer187, Francesco Scolari188, Anna Sediva189, Luis M. Seijo190, Damien Sene13, Sevtap Senoglu117, Mikko R. J. Seppänen191, Alex Serra Ilovich192, Mohammad Shahrooei62, Hans Slabbynck193, David M. Smadja194, Ali Sobh195, Xavier Solanich Moreno15, Jordi Solé-Violán196, Catherine Soler197, Pere Soler-Palacín137, Yuri Stepanovskiy198, Annabelle Stoclin199, Fabio Taccone149, Yacine Tandjaoui-Lambiotte200, Jean-Luc Taupin201, Simon J. Tavernier202, Benjamin Terrier203, Caroline Thumerelle107, Gabriele Tomasoni204, Julie Toubiana48, Josep Trenado Alvarez205, Sophie Trouillet-Assant206, Jesús Troya207, Alessandra Tucci208, Matilde Valeria Ursini83, Yurdagul Uzunhan209, Pierre Vabres210, Juan Valencia-Ramos211, Eva Van Braeckel33, Stijn Van de Velde212, Ana Maria Van Den Rym84, Jens Van Praet213, Isabelle Vandernoot214, Hulya Vatansev215, Valentina Vélez-Santamaria42, Sébastien Viel171, Cédric Vilain216, Marie E. Vilaire67, Audrey Vincent35, Guillaume Voiriot217, Fanny Vuotto107, Alper Yosunkaya90, Barnaby E. Young126, Fatih Yucel218, Faiez Zannad219, Mayana Zatz37, Alexandre Belot220*
1University Hospital and Research Institute “Germans Trias i Pujol,” Badalona, Spain. 2Navarra Health Service Hospital, Pamplona, Spain. 3Division of Pediatric Infectious Diseases, Necmettin Erbakan University, Meram Medical Faculty, Konya, Turkey. 4Department of Infectious Diseases, Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran. 5Hospital Nacional Edgardo Rebagliati Martins, Lima, Peru. 6Parc Sanitari Sant Joan de Déu, Sant Boi de Llobregat, Spain. 7Virology Research Center, National Institutes of Tuberculosis and Lung Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran. 8Division of Pediatric Infectious Diseases, Faculty of Medicine, Selcuk University, Konya, Turkey. 9Intensive Care Unit, Hôpital Européen, Marseille, France. 10Immunology Department, University Hospital 12 de Octubre, Research Institute imas12, and Complutense University, Madrid, Spain. 11Clinical Immuology and Primary Immunodeficiencies Unit, Hospital Sant Joan de Déu, Barcelona, Spain. 12Department of Biological Immunology, Necker Hospital for Sick Children, APHP and INEM, Paris, France. 13Internal Medicine Department, Hôpital Lariboisière, APHP; Université de Paris, Paris, France. 14Internal Medicine Department, Pitié-Salpétrière Hospital, Paris, France. 15Hospital Universitari de Bellvitge, Barcelona, Spain. 16Division of Clinical Immunology and Allergy, Necmettin Erbakan University, Meram Medical Faculty, Konya, Turkey. 17Joint Research Unit, Hospices Civils de Lyon-bio Mérieux, Hospices Civils de Lyon, Lyon Sud Hospital, Lyon, France. 18Hospital U. de Tarragona Joan XXIII, Universitat Rovira i Virgili (URV), IISPV, Tarragona, Spain. 19Private practice, Paris, France. 20Necker Hospital for Sick Children, AP-HP, Paris, France. 21Department of Infectious Diseases, CHU de Caen, Caen, France. 22Consorcio Hospital General Universitario, Valencia, Spain. 23The Genetics Institute, Tel Aviv Sourasky Medical Center and Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel. 24Department of Urology, Nephrology, and Transplantation, APHP-SU, Sorbonne Université, INSERM U 1082, Paris, France. 25Service de Médecine Intensive–Réanimation et Pneumologie, APHP Hôpital Pitié–Salpêtrière, Paris, France. 26Cruces University Hospital, Bizkaia, Spain. 27Paediatric Immunology and Vaccinology Unit, Geneva University Hospitals and Faculty of Medicine, Geneva, Switzerland. 28Hematology, Georges Pompidou Hospital, APHP, Paris, France. 29Pediatric Infectious Diseases Unit, Instituto de Investigación 12 de Octubre imas12, and Hospital Universitario 12 de Octubre, Madrid, Spain. 30Department of Immunology, Motol University Hospital, 2nd Faculty of Medicine, Charles University, Department of Pediatrics, Thomayer’s Hospital, 1st Faculty of Medicine, Charles University, Prague, Czech Republic. 31Centro de Investigación Biomédica en Red de Enfermedades Hepàticas y Digestivas (Ciberehd), Hospital de Mataró, Consorci Sanitari del Maresme, Mataró, Spain. 32Service de Pneumologie, Hopital Bichat, APHP, Paris, France. 33Department of Pulmonology, Ghent University Hospital, Ghent, Belgium. 34Clinical Immunology Unit, Pediatric Infectious Disease Department, Faculty of Medicine and Pharmacy, Averroes University Hospital, LICIA Laboratoire d’Immunologie Clinique, d’Inflammation et d’Allergie, Hassann Ii University, Casablanca, Morocco. 35Endocrinology Unit, APHP Hôpitaux Universitaires Paris-Sud, Le Kremlin-Bicêtre, France. 36Department of Children’s Diseases and Pediatric Surgery, I. Horbachevsky Ternopil National Medical University, Ternopil, Ukraine. 37Human Genome and Stem-Cell Research Center, University of São Paulo, São Paulo, Brazil. 38Hospital Insular, Las Palmas de Gran Canaria, Spain. 39Division of Critical Care Medicine, Department of Anesthesiology and Reanimation, Konya State Hospital, Konya, Turkey. 40MS Center, Spedali Civili, Brescia, Italy. 41Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan, Italy. 42Bellvitge University Hospital, L’Hospitalet de Llobregat, Barcelona, Spain. 43Hopital Robert Debré, Paris, France. 44Pediatric Immuno-hematology Unit, Necker Enfants Malades Hospital, AP-HP, Paris, France. 45Department of Infectious and Tropical Diseases, University of Brescia, ASST Spedali Civili di Brescia, Brescia, Italy. 46Doctoral Health Care Center, Canarian Health System, Las Palmas de Gran Canaria, Spain. 47Hôpital Foch, Suresnes, France. 48Necker Hospital for Sick Children, Paris University, AP-HP, Paris, France. 49Pasteur Institute, Paris, France. 50McGill University Health Centre, Montreal, Canada. 51University Hospital and Research Institute “Germans Trias i Pujol,” IrsiCaixa AIDS Research Institute, UVic-UCC, Badalona, Spain. 52Clinical Biochemistry, Pathology, Paediatric Neurology and Molecular Medicine Departments and Biobank, Institut de Recerca Sant Joan de Déu and CIBERER-ISCIII, Esplugues, Spain. 53Division of Clinical Immunology and Allergy, Department of Internal Medicine, Necmettin Erbakan University, Meram Medical Faculty, Konya, Turkey. 54Department of Infectious Diseases and Clinical Microbiology, Konya Training and Research Hospital, Konya, Turkey. 55Hospital Universitari Vall d’Hebron, Barcelona, Spain. 56Pitié-Salpêtrière Hospital, Paris, France. 57Fundació Docència i Recerca Mútua Terrassa, Barcelona, Spain; Hospital Universitari Mutua Terrassa, Universitat de Barcelona, Terrassa, Catalonia, Spain. 58UNSW Medicine, St. Vincent’s Clinical School, and Department of Thoracic Medicine, St. Vincent’s Hospital Darlinghurst, Sidney, Australia. 59CHU Saint-Pierre, Université Libre de Bruxelles, Brussels, Belgium. 60Pediatric Intensive Care Unit, Robert-Debré University Hospital, APHP, Paris, France. 61Sorbonne Paris Nord, Hôpital Jean Verdier, APHP, Bondy, France. 62Specialized Immunology Laboratory of Dr. Shahrooei, Sina Medical Complex, Ahvaz, Iran. 63Centre de Génétique Humaine, CHU Besançon, Besançon, France. 64Sorbonne Université Médecine and APHP Sorbonne Université Site Pitié-Salpêtrière, Paris, France. 65Intensive Care Unit, Georges Pompidou Hospital, APHP, Paris, France. 66Department of Pneumology, AZ Delta, Roeselare, Belgium. 67Institut Jérôme Lejeune, Paris, France. 68Department of Microbiology and Immunology, Faculty of Medicine, Mansoura University, Mansoura, Egypt. 69Department of Chest, Faculty of Medicine, Mansoura University, Mansoura, Egypt. 70Department of Medical Biochemistry and Molecular Biology, Faculty of Medicine, Mansoura University, Mansoura, Egypt. 71Faculty of Medicine, Division of Pediatric Infectious Diseases, Selcuk University, Konya, Turkey. 72Division of Pediatric Infectious Diseases, Ondokuz Mayıs University, Samsun, Turkey. 73Necmettin Erbakan University, Meram Medical Faculty, Division of Pediatric Allergy and Immunology, Konya, Turkey. 74Centre Hospitalier Fleyriat, Bourg-en-Bresse, France. 75Division of Clinical Immunology and Allergy, Department of Internal Medicine, Necmettin Erbakan University, Meram Medical Faculty, Konya, Turkey. 76Centre de Génétique, CHU Dijon, Dijon, France. 77APHP Tenon Hospital, Paris, France. 78Sorbonne Universités, UPMC University of Paris, Paris, France. 79Department of Clinical Immunology , Hospital Clínico San Carlos, Madrid, Spain. 80Genomics Division, Instituto Tecnológico y de Energías Renovables (ITER), Santa Cruz de Tenerife, Spain; CIBER de Enfermedades Respiratorias, Instituto de Salud Carlos III, Madrid, Spain; Research Unit, Hospital Universitario N.S. de Candelaria, Santa Cruz de Tenerife, Spain; Instituto de Tecnologías Biomédicas (ITB), Universidad de La Laguna, San Cristóbal de La Laguna, Spain. 81CHU Limoges and Inserm CIC 1435 and UMR 1092, Limoges, France. 82Infectious Diseases Unit, Department of Pediatrics, Hospital Sant Joan de Déu, Barcelona, Spain; Institut de Recerca Sant Joan de Déu, Spain; Universitat de Barcelona (UB), Barcelona, Spain. 83Institute of Genetics and Biophysics “Adriano Buzzati-Traverso,” IGB-CNR, Naples, Italy. 84Laboratory of Immunogenetics of Human Diseases, IdiPAZ Institute for Health Research, La Paz Hospital, Madrid, Spain. 85Hematology, APHP, Hopital Européen Georges Pompidou and Inserm UMR-S1140, Paris, France. 86Hospital General Universitario and Instituto de Investigación Sanitaria “Gregorio Marañón,” Madrid, Spain. 87Bégin military Hospital, Bégin, France. 88Pediatric Intensive Care Unit, Hospital Sant Joan de Déu, Barcelona, Spain. 89Department of Internal Medicine, Hôpital Erasme, Université Libre de Bruxelles, Brussels, Belgium. 90Division of Critical Care Medicine, Department of Anesthesiology and Reanimation, Necmettin Erbakan University, Meram Medical Faculty, Konya, Turkey. 91Genomics Division, Instituto Tecnológico y de Energías Renovables (ITER), Santa Cruz de Tenerife, Spain. 92Assistance Publique Hôpitaux de Paris, Paris, France. 93Division of Allergy and Immunology, Necmettin Erbakan University, Meram Medical Faculty, Konya, Turkey. 94CNAG-CRG, Centre for Genomic Regulation (CRG), Barcelona Institute of Science and Technology (BIST); Universitat Pompeu Fabra (UPF), Barcelona, Spain. 95Department of Internal Medicine, National Reference Center for Rare Systemic Autoimmune Diseases, AP-HP, APHP-CUP, Hôpital Cochin, Paris, France. 96Ghent University Hospital, Ghent, Belgium. 97Sharjah Institute of Medical Research, College of Medicine, University of Sharjah, Sharjah, UAE. 98Department of Laboratory Medicine, SE14186, Huddinge, Karolinska Institutet, Stockholm, Sweden. 99Pediatric Infectious Diseases Unit, Bakirkoy Dr. Sadi Konuk Training and Research Hospital, University of Health Sciences, Istanbul, Turkey. 100Department of Immunology, Hospital Universitario de Gran Canaria Dr. Negrín, Canarian Health System, Las Palmas de Gran Canaria, Spain. 101Department of Pediatric Hemato-Oncology, Jolimont Hospital; Department of Pediatric Hemato-Oncology, HUDERF, Brussels, Belgium. 102Intensive Care Unit, Marqués de Valdecilla Hospital, Santander, Spain. 103Hospital del Mar, Parc de Salut Mar, Barcelona, Spain. 104Department of Pediatric Pulmonology and Immunology, Ghent University Hospital, Ghent, Belgium. 105Department of Intensive Care Unit, Ghent University Hospital, Ghent, Belgium. 106Intensive Care Unit, APHM, Marseille, France. 107CHU Lille, Lille, France. 108Department of Pediatrics, Columbia University, New York, NY, USA. 109Centre Hospitalier Intercommunal Poissy Saint Germain en Laye, Poissy, France. 110Fundació Docència i Recerca Mútua Terrassa, Terrassa, Spain. 111Hospital Sant Joan de Déu, Kids Corona Platfform, Barcelona, Spain. 112Department of Intensive Care Unit, University Hospital Antwerp, Antwerp, Belgium. 113Selcuk University, Faculty of Medicine, Chest Diseases Department, Konya, Turkey. 114Division of Allergy and Immunology, Balikesir Ataturk City Hospital, Balikesir, Turkey. 115Division of Critical Care Medicine, Selcuk University, Faculty of Medicine, Konya, Turkey. 116Division of Pediatric Infectious Diseases, Prof. Dr. Cemil Tascıoglu City Hospital, Istanbul, Turkey. 117Departments of Infectious Diseases and Clinical Microbiology, Bakirkoy Dr. Sadi Konuk Training and Research Hospital, University of Health Sciences, Istanbul, Turkey. 118Meram Medical Faculty, Necmettin Erbakan University, Meram Medical Faculty, Konya, Turkey. 119Health Sciences University, Umraniye Education and Research Hospital, Istanbul, Turkey. 120Department of Immunology, 2nd Faculty of Medicine, Charles University and University Hospital in Motol, Prague, Czech Republic. 121Central Clinical Hospital of Ministry of the Interior and Administration in Warsaw, Warsaw, Poland. 122Oncobiologie Génétique Bioinformatique, PC Bio, CHU Besançon, Besançon, France. 123Paediatric Infectious Disease Unit, Hospital Authority Infectious Disease Center, Princess Margaret Hospital, Hong Kong (Special Administrative Region), China. 124Aix Marseille University, IRD, MEPHI, IHU Méditerranée Infection, Marseille, France. 125Department of Paediatrics and Adolescent Medicine, The University of Hong Kong, Hong Kong, China. 126National Centre for Infectious Diseases, Singapore. 127Hospital Universitario Reina Sofía, Cordoba, Spain. 128Imperial College, London, UK. 129Endocrinology and Diabetes for Children, AP-HP, Bicêtre Paris-Saclay Hospital, Le Kremlin-Bicêtre, France. 130Department of Medical Genetics, University Hospital Antwerp, Antwerp, Belgium. 131Neurology Unit, APHP Pitié-Salpêtrière Hospital, Paris University, Paris, France. 132Intensive Care Unit, APHP Pitié-Salpêtrière Hospital, Paris University, Paris, France. 133National Centre for Infectious Diseases; Tan Tock Seng Hospital; Yong Loo Lin School of Medicine; Lee Kong Chian School of Medicine, Singapore. 134Department of Clinical Immunology and Infectious Diseases, National Research Institute of Tuberculosis and Lung Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran. 135Clinical Tuberculosis and Epidemiology Research Center, National Research Institute of Tuberculosis and Lung Diseases (NRITLD), Shahid Beheshti University of Medical Sciences, Tehran, Iran. 136Hospital Sant Joan de Déu and University of Barcelona, Barcelona, Spain. 137Pediatric Infectious Diseases and Immunodeficiencies Unit, Hospital Universitari Vall d’Hebron, Vall d’Hebron Research Institute, Vall d’Hebron Barcelona Hospital Campus. Universitat Autònoma de Barcelona (UAB), Barcelona, Spain. 138Hospital Universitari Mutua de Terrassa, Universitat de Barcelona, Barcelona, Spain. 139IrsiCaixa AIDS Research Institute, ICREA, UVic-UCC, Research Institute “Germans Trias i Pujol,” Badalona, Spain. 140Department of Laboratory, Cruces University Hospital, Barakaldo, Bizkaia, Spain. 141University of New South Wales, New South Wales, Australia. 142APHP Pitié-Salpêtrière Hospital, Paris, France. 143Aix-Marseille University, APHM, Marseille, France. 144Robert Debré Hospital, Paris, France. 145APHP Cohin Hospital, Paris, France. 146Necmettin Erbakan University Meram Faculty of Medicine Department of Pediatric Infectious Diseases, Konya, Turkey. 147University Hospitals Leuven, Leuven, Belgium. 148Hospices Civils de Lyon, Hôpital de la Croix-Rousse, Lyon, France. 149Hôpital Erasme, Brussels, Belgium. 150CH Gonesse, Gonesse, France. 151Vascular Medicine, Georges Pompidou Hospital, APHP, Paris, France. 152Division of Pulmonary and Critical Care, University of Miami, Miami, FL, USA. 153Guanarteme Health Care Center, Canarian Health System, Las Palmas de Gran Canaria, Spain. 154Regional University Hospital of Malaga, Malaga, Spain. 155Aix-Marseille Université, Marseille, France. 156Department of General Paediatrics, Hôpital Bicêtre, AP-HP, University of Paris Saclay, Le Kremlin-Bicêtre, France. 157Department of Internal Medicine, Ghent University Hospital, Ghent, Belgium. 158CHU de La Timone, Marseille, France. 159Centro Hospitalar Universitário de Lisboa Central, Lisbon, Portugal. 160Infectious Diseases Horizontal Technlogy Centre, A*STAR; Singapore Immunology Network, A*STAR, Singapore. 161Department of Pediatrics, Complejo Hospitalario Universitario Insular-Materno Infantil, Canarian Health System, Las Palmas de Gran Canaria, Spain. 162Regional Universitary Hospital of Málaga, Málaga, Spain. 163Hospital Universitario Marqués de Valdecilla, Santander, Spain. 164Faculty of Medicine, Ataturk University, Erzurum, Turkey. 165Department of Molecular Biology and Genetics, Bilkent University, Ankara, Turkey. 166Department of Biosciences and Nutrition, Karolinska Institutet, SE14183, Stockholm, Sweden. 167L’Hôpital Foch, Suresnes, France. 168Department of Immunology, Hospital Universitario 12 de Octubre, Instituto de Investigación Sanitaria Hospital 12 de Octubre imas12, Madrid, Spain. 169APHP Hôpitaux Universitaires Paris-Sud, Le Kremlin-Bicêtre, France. 170Neurometabolic Diseases Laboratory, IDIBELL-Hospital Duran i Reynals, Barcelona; CIBERER U759, ISCiii, Madrid, Spain. 171Hospices Civils de Lyon, Lyon, France. 172Université de Lille, Inserm U1285, CHU Lille, Paris, France. 173Departement of General Pediatrics, University Hospital Robert Debré, APHP, Paris, France. 174Necmettin Erbakan University, Konya, Turkey. 175Germans Trias i Pujol Hospital, Badalona, Spain. 176Medical Intensive Care Unit, Hopital de la Croix-Rousse, Hospices Civils de Lyon, Lyon, France. 177Pediatric Infectious Diseases and Immunodeficiencies Unit, Hospital Universitari Vall d’Hebron, Vall d’Hebron Research Institute, Vall d’Hebron Barcelona Hospital Campus., Barcelona, Spain. 178Department of Immunology, Hospital Universitario de Gran Canaria Dr. Negrín, Canarian Health System, Las Palmas de Gran Canaria, Spain. University Fernando Pessoa Canarias, Las Palmas de Gran Canaria, Spain. 179Neurometabolic Diseases Laboratory, IDIBELL-Hospital Duran i Reynals, Barcelona, Spain. 180Consorcio Hospital General Universitario, Valencia, Spain. 181APHP Hôpitaux Universitaires Paris-Sud, Paris, France. 182Virology Unit, Université de Paris, Cohin Hospital, APHP, Paris, France. 183Hospital San Pedro, Logroño, Spain. 184Respiratory Medicine, Georges Pompidou Hospital, APHP, Paris, France. 185Department of Immunology, Hospital Clínico San Carlos, Madrid, Spain. 186Service de Médecine Intensive Réanimation, Institut de Cardiologie, Hopital Pitié-Salpêtrière, Paris, France. 187CHRU de Nancy, Hôpital d’Enfants, Vandoeuvre, France. 188Chair of Nephrology, University of Brescia, Brescia, Italy. 189Department of Immunology, 2nd Faculty of Medicine, Charles University and Motol University Hospital, Prague, Czech Republic. 190Clínica Universidad de Navarra, Madrid, Spain. 191HUS Helsinki University Hospital, Children and Adolescents, Rare Disease Center, and Inflammation Center, Adult Immunodeficiency Unit, Majakka, Helsinki, Finland. 192Fundació Docència i Recerca Mútua Terrassa, Terrassa, Spain. 193Department of Pulmonology, ZNA Middelheim, Antwerp, Belgium. 194INSERM UMR-S 1140, Biosurgical Research Lab (Carpentier Foundation), Paris University and Hopital Européen Georges Pompidou, Paris, France. 195Department of Pediatrics, Faculty of Medicine, Mansoura University, Mansoura, Egypt. 196Critical Care Unit, Hospital Universitario de Gran Canaria Dr. Negrín, Canarian Health System, Las Palmas de Gran Canaria, Spain. 197CHU de Saint Etienne, Saint-Priest-en-Jarez, France. 198Shupyk National Medical Academy for Postgraduate Education, Kiev, Ukraine. 199Gustave Roussy Cancer Campus, Villejuif, France. 200Intensive Care Unit, Avicenne Hospital, APHP, Bobigny, France. 201Laboratory of Immunology and Histocompatibility, Saint-Louis Hospital, Paris University, Paris, France. 202Department of Internal Diseases and Pediatrics, Primary Immune Deficiency Research Lab, Centre for Primary Immunodeficiency Ghent, Jeffrey Modell Diagnosis and Research Centre, Ghent University Hospital, Ghent, Belgium. 203Department of Internal Medicine, Université de Paris, INSERM, U970, PARCC, F-75015, Paris, France. 204First Division of Anesthesiology and Critical Care Medicine, University of Brescia, ASST Spedali Civili di Brescia, Brescia, Italy. 205Intensive Care Department, Hospital Universitari Mutua Terrassa, Universitat Barcelona, Terrassa, Spain. 206Hospices Civils de Lyon, Lyon Sud Hospital, Lyon, France. 207Infanta Leonor University Hospital, Madrid, Spain. 208Hematology Department, ASST Spedali Civili di Brescia, Brescia, Italy. 209Pneumologie, Hôpital Avicenne, APHP, INSERM U1272, Université Sorbonne Paris Nord, Bobigny, France. 210Dermatology Unit, Laboratoire GAD, INSERM UMR1231 LNC, Université de Bourgogne, Dijon, France. 211University Hospital of Burgos, Burgos, Spain. 212Intensive Care Unit, M. Middelares Ghent, Ghent, Belgium. 213Department of Nephrology and Infectiology, AZ Sint-Jan Brugge-Oostende AV, Bruges, Belgium. 214Center of Human Genetics, Hôpital Erasme, Université Libre de Bruxelles, Brussels, Belgium. 215Department of Chest Diseases, Necmettin Erbakan University, Meram Medical Faculty, Konya, Turkey. 216CHU de Caen, Caen, France. 217Sorbonne Université, Service de Médecine Intensive Réanimation, Hôpital Tenon, Assistance Publique-Hôpitaux de Paris, Paris, France. 218General Intensive Care Unit, Konya Training and Research Hospital, Konya, Turkey. 219CHU de Nancy, Nancy, France. 220University of Lyon, CIRI, INSERM U1111, National Referee Centre RAISE, Pediatric Rheumatology, HFME, Hospices Civils de Lyon, Lyon, France.
*Leader of COVID Clinicians.
Imagine COVID Group
Christine Bole-Feysot, Stanislas Lyonnet*, Cécile Masson, Patrick Nitschke, Aurore Pouliet, Yoann Schmitt, Frederic Tores, Mohammed Zarhrate
Imagine Institute, Université de Paris, INSERM UMR 1163, Paris, France.
*Leader of the Imagine COVID Group.
French COVID Cohort Study Group
Laurent Abel1, Claire Andrejak2, François Angoulvant3, Delphine Bachelet4, Romain Basmaci5, Sylvie Behillil6, Marine Beluze7, Dehbia Benkerrou8, Krishna Bhavsar4, François Bompart9, Lila Bouadma4, Maude Bouscambert10, Mireille Caralp11, Minerva Cervantes-Gonzalez12, Anissa Chair4, Alexandra Coelho13, Camille Couffignal4, Sandrine Couffin-Cadiergues14, Eric D’Ortenzio12, Charlene Da Silveira4, Marie-Pierre Debray4, Dominique Deplanque15, Diane Descamps16, Mathilde Desvallées17, Alpha Diallo18, Alphonsine Diouf13, Céline Dorival8, François Dubos19, Xavier Duval4, Philippine Eloy4, Vincent VE Enouf20, Hélène Esperou21, Marina Esposito-Farese4, Manuel Etienne22, Nadia Ettalhaoui4, Nathalie Gault4, Alexandre Gaymard10, Jade Ghosn4, Tristan Gigante23, Isabelle Gorenne4, Jérémie Guedj24, Alexandre Hoctin13, Isabelle Hoffmann4, Salma Jaafoura21, Ouifiya Kafif4, Florentia Kaguelidou25, Sabina Kali4, Antoine Khalil4, Coralie Khan17, Cédric Laouénan4, Samira Laribi4, Minh Le4, Quentin Le Hingrat4, Soizic Le Mestre18, Hervé Le Nagard24, François-Xavier Lescure4, Yves Lévy26, Claire Levy-Marchal27, Bruno Lina10, Guillaume Lingas24, Jean Christophe Lucet4, Denis Malvy28, Marina Mambert13, France Mentré4, Noémie Mercier18, Amina Meziane8, Hugo Mouquet20, Jimmy Mullaert4, Nadège Neant24, Marion Noret29, Justine Pages30, Aurélie Papadopoulos21, Christelle Paul18, Nathan Peiffer-Smadja4, Ventzislava Petrov-Sanchez18, Gilles Peytavin4, Olivier Picone31, Oriane Puéchal12, Manuel Rosa-Calatrava10, Bénédicte Rossignol23, Patrick Rossignol32, Carine Roy4, Marion Schneider4, Caroline Semaille12, Nassima Si Mohammed4, Lysa Tagherset4, Coralie Tardivon4, Marie-Capucine Tellier4, François Téoulé8, Olivier Terrier10, Jean-François Timsit4, Théo Trioux4, Christelle Tual33, Sarah Tubiana4, Sylvie van der Werf34, Noémie Vanel35, Aurélie Veislinger33, Benoit Visseaux16, Aurélie Wiedemann26, Yazdan Yazdanpanah36
1Inserm UMR 1163, Paris, France. 2CHU Amiens, France. 3Hôpital Necker, Paris, France. 4Hôpital Bichat, Paris, France. 5Hôpital Louis Mourrier, Colombes, France. 6Institut Pasteur, Paris, France. 7F-CRIN Partners Platform, AP-HP, Université de Paris, Paris, France. 8Inserm UMR 1136, Paris, France. 9Drugs for Neglected Diseases Initiative, Geneva, Switzerland. 10Inserm UMR 1111, Lyon, France. 11Inserm Transfert, Paris, France. 12REACTing, Paris, France. 13Inserm UMR 1018, Paris, France. 14Inserm, Pôle Recherche Clinique, Paris, France. 15CIC 1403 Inserm-CHU Lille, Paris, France. 16Université de Paris, IAME, INSERM UMR 1137, AP-HP, University Hospital Bichat Claude Bernard, Virology, Paris, France. 17Inserm UMR 1219, Bordeaux, France. 18ANRS, Paris, France. 19CHU Lille, Lille, France. 20Pasteur Institute, Paris, France. 21Inserm sponsor, Paris, France. 22CHU Rouen–SMIT, Rouen, France. 23FCRIN INI-CRCT, Nancy, France. 24Inserm UMR 1137, Paris, France. 25Centre d’Investigation Clinique, Inserm CIC1426, Hôpital Robert Debré, Paris, France. 26Inserm UMR 955, Créteil, France; Vaccine Research Instiute (VRI), Paris, France. 27F-CRIN INI-CRCT, Paris, France. 28CHU de Bordeaux–SMIT, Bordeaux, France. 29RENARCI, Annecy, France. 30Hôpital Robert Debré, Paris, France. 31Hôpital Louis Mourier–Gynécologie, Colombes, France. 32University of Lorraine, Plurithematic Clinical Investigation Centre Inserm CIC-P; 1433, Inserm U1116, CHRU Nancy Hopitaux de Brabois, F-CRIN INI-CRCT (Cardiovascular and Renal Clinical Trialists), Nancy, France. 33Inserm CIC-1414, Rennes, France. 34Institut Pasteur, UMR 3569 CNRS, Université de Paris, Paris, France. 35Hôpital la Timone, Marseille, France. 36Bichat–SMIT, Paris, France.
CoV-Contact Cohort
Loubna Alavoine1, Karine K. A. Amat2, Sylvie Behillil3, Julia Bielicki4, Patricia Bruijning5, Charles Burdet6, Eric Caumes7, Charlotte Charpentier8, Bruno Coignard9, Yolande Costa1, Sandrine Couffin-Cadiergues10, Florence Damond8, Aline Dechanet11, Christelle Delmas10, Diane Descamps8, Xavier Duval1, Jean-Luc Ecobichon1, Vincent Enouf3, Hélène Espérou10, Wahiba Frezouls1, Nadhira Houhou11, Emila Ilic-Habensus1, Ouifiya Kafif11, John Kikoine11, Quentin Le Hingrat8, David Lebeaux12, Anne Leclercq1, Jonathan Lehacaut1, Sophie Letrou1, Bruno Lina13, Jean-Christophe Lucet14, Denis Malvy15, Pauline Manchon11, Milica Mandic1, Mohamed Meghadecha16, Justina Motiejunaite17, Mariama Nouroudine1, Valentine Piquard11, Andreea Postolache11, Caroline Quintin1, Jade Rexach1, Layidé Roufai10, Zaven Terzian11, Michael Thy18, Sarah Tubiana1, Sylvie van der Werf3, Valérie Vignali1, Benoit Visseaux8, Yazdan Yazdanpanah14
1Centre d’Investigation Clinique, Inserm CIC 1425, Hôpital Bichat Claude Bernard, APHP, Paris, France. 2IMEA Fondation Léon M’Ba, Paris, France. 3Institut Pasteur, UMR 3569 CNRS, Université de Paris, Paris, France. 4University of Basel Children’s Hospital. 5Julius Center for Health Sciences and Primary Care, Utrecht, Netherlands. 6Université de Paris, IAME, Inserm UMR 1137, F-75018, Paris, France, Hôpital Bichat Claude Bernard, APHP, Paris, France. 7Hôpital Pitiè Salpétriere, APHP, Paris. 8Université de Paris, IAME, INSERM UMR 1137, AP-HP, University Hospital Bichat Claude Bernard, Virology, Paris, France. 9Santé Publique France, Saint Maurice, France. 10Pole Recherche Clinique, Inserm, Paris, France. 11Hôpital Bichat Claude Bernard, APHP, Paris, France. 12APHP, Paris, France. 13Virpath Laboratory, International Center of Research in Infectiology, Lyon University, INSERM U1111, CNRS UMR 5308, ENS, UCBL, Lyon, France. 14IAME Inserm UMR 1138, Hôpital Bichat Claude Bernard, APHP, Paris, France. 15Service des Maladies Infectieuses et Tropicales; Groupe Pellegrin-Place Amélie-Raba-Léon, Bordeaux, France. 16Hôpital Hotel Dieu, APHP, Paris, France. 17Service des Explorations Fonctionnelles, Hôpital Bichat–Claude Bernard, APHP, Paris, France. 18Center for Clinical Investigation, Assistance Publique-Hôpitaux de Paris, Bichat-Claude Bernard University Hospital, Paris, France.
Amsterdam UMC Covid-19 Biobank
Michiel van Agtmael1, Anna Geke Algera2, Frank van Baarle2, Diane Bax3, Martijn Beudel4, Harm Jan Bogaard5, Marije Bomers1, Lieuwe Bos2, Michela Botta2, Justin de Brabander6, Godelieve de Bree6, Matthijs C. Brouwer4, Sanne de Bruin2, Marianna Bugiani7, Esther Bulle2, Osoul Chouchane1, Alex Cloherty3, Paul Elbers2, Lucas Fleuren2, Suzanne Geerlings1, Bart Geerts8, Theo Geijtenbeek9, Armand Girbes2, Bram Goorhuis1, Martin P. Grobusch1, Florianne Hafkamp9, Laura Hagens2, Jorg Hamann10, Vanessa Harris1, Robert Hemke11, Sabine M. Hermans1, Leo Heunks2, Markus W. Hollmann8, Janneke Horn2, Joppe W. Hovius1, Menno D. de Jong12, Rutger Koning4, Niels van Mourik2, Jeaninne Nellen1, Frederique Paulus2, Edgar Peters1, Tom van der Poll1, Benedikt Preckel8, Jan M. Prins1, Jorinde Raasveld2, Tom Reijnders1, Michiel Schinkel1, Marcus J. Schultz2, Alex Schuurman13, Kim Sigaloff1, Marry Smit2, Cornelis S. Stijnis1, Willemke Stilma2, Charlotte Teunissen14, Patrick Thoral2, Anissa Tsonas2, Marc van der Valk1, Denise Veelo8, Alexander P.J. Vlaar15, Heder de Vries2, Michèle van Vugt1, W. Joost Wiersinga1, Dorien Wouters16, A. H. (Koos) Zwinderman17, Diederik van de Beek4*
1Department of Infectious Diseases, Amsterdam UMC, Amsterdam, Netherlands. 2Department of Intensive Care, Amsterdam UMC, Amsterdam, Netherlands. 3Experimental Immunology, Amsterdam UMC, Amsterdam, Netherlands. 4Department of Neurology, Amsterdam UMC, Amsterdam Neuroscience, Amsterdam, Netherlands. 5Department of Pulmonology, Amsterdam UMC, Amsterdam, Netherlands. 6Department of Infectious Diseases, Amsterdam UMC, Amsterdam, Netherlands. 7Department of Pathology, Amsterdam UMC, Amsterdam, Netherlands. 8Department of Anesthesiology, Amsterdam UMC, Amsterdam, Netherlands. 9Department of Experimental Immunology, Amsterdam UMC, Amsterdam, Netherlands. 10Amsterdam UMC Biobank Core Facility, Amsterdam UMC, Amsterdam, Netherlands. 11Department of Radiology, Amsterdam UMC, Amsterdam, Netherlands. 12Department of Medical Microbiology, Amsterdam UMC, Amsterdam, Netherlands. 13Department of Internal Medicine, Amsterdam UMC, Amsterdam, Netherlands. 14Neurochemical Laboratory, Amsterdam UMC, Amsterdam, Netherlands. 15Department of Intensive Care, Amsterdam UMC, Amsterdam, Netherlands. 16Department of Clinical Chemistry, Amsterdam UMC, Amsterdam, Netherlands. 17Department of Clinical Epidemiology, Biostatistics and Bioinformatics, Amsterdam UMC, Amsterdam, Netherlands. 18Department of Neurology, Amsterdam UMC, Amsterdam, Netherlands.
*Leader of the AMC Consortium.
COVID Human Genetic Effort
Laurent Abel1, Alessandro Aiuti2, Saleh Al Muhsen3, Fahd Al-Mulla4, Mark S. Anderson5, Andrés Augusto Arias6, Hagit Baris Feldman7, Dusan Bogunovic8, Alexandre Bolze9, Anastasiia Bondarenko10, Ahmed A. Bousfiha11, Petter Brodin12, Yenan Bryceson12, Carlos D. Bustamante13, Manish Butte14, Giorgio Casari15, Samya Chakravorty16, John Christodoulou17, Elizabeth Cirulli9, Antonio Condino-Neto18, Megan A. Cooper19, Clifton L. Dalgard20, Alessia David21, Joseph L. DeRisi22, Murkesh Desai23, Beth A. Drolet24, Sara Espinosa25, Jacques Fellay26, Carlos Flores27, Jose Luis Franco28, Peter K. Gregersen29, Filomeen Haerynck30, David Hagin31, Rabih Halwani32, Jim Heath33, Sarah E. Henrickson34, Elena Hsieh35, Kohsuke Imai36, Yuval Itan8, Timokratis Karamitros37, Kai Kisand38, Cheng-Lung Ku39, Yu-Lung Lau40, Yun Ling41, Carrie L. Lucas42, Tom Maniatis43, Davoud Mansouri44, Laszlo Marodi45, Isabelle Meyts46, Joshua Milner47, Kristina Mironska48, Trine Mogensen49, Tomohiro Morio50, Lisa FP. Ng51, Luigi D. Notarangelo52, Antonio Novelli53, Giuseppe Novelli54, Cliona O’Farrelly55, Satoshi Okada56, Tayfun Ozcelik57, Rebeca Perez de Diego58, Anna M. Planas59, Carolina Prando60, Aurora Pujol61, Lluis Quintana-Murci62, Laurent Renia63, Alessandra Renieri64, Carlos Rodríguez-Gallego65, Vanessa Sancho-Shimizu66, Vijay Sankaran67, Kelly Schiabor Barrett9, Mohammed Shahrooei68, Andrew Snow69, Pere Soler-Palacín70, András N. Spaan71, Stuart Tangye72, Stuart Turvey73, Furkan Uddin74, Mohammed J. Uddin75, Diederik van de Beek76, Sara E. Vazquez77, Donald C. Vinh78, Horst von Bernuth79, Nicole Washington9, Pawel Zawadzki80, Helen C. Su52, Jean-Laurent Casanova81
1INSERM U1163, University of Paris, Imagine Institute, Paris, France. 2San Raffaele Telethon Institute for Gene Therapy, IRCCS Ospedale San Raffaele, Milan, Italy. 3King Saud University, Riyadh, Saudi Arabia. 4Kuwait University, Kuwait City, Kuwait. 5University of California, San Francisco, San Francisco, CA, USA. 6Universidad de Antioquia, Group of Primary Immunodeficiencies, Antioquia, Colombia. 7The Genetics Institute, Tel Aviv Sourasky Medical Center and Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel. 8Icahn School of Medicine at Mount Sinai, New York, NY, USA. 9Helix, San Mateo, CA, USA. 10Shupyk National Medical Academy for Postgraduate Education, Kiev, Ukraine. 11Clinical Immunology Unit, Pediatric Infectious Disease Departement, Faculty of Medicine and Pharmacy, Averroes University Hospital; LICIA Laboratoire d’Immunologie Clinique, d’Inflammation et d’Allergie, Hassann Ii University, Casablanca, Morocco. 12Karolinska Institute, Stockholm, Sweden. 13Stanford University, Stanford, CA, USA. 14University of California, Los Angeles, CA, USA. 15Medical Genetics, IRCCS Ospedale San Raffaele, Milan, Italy. 16Emory University Department of Pediatrics and Children’s Healthcare of Atlanta, Atlanta, GA, USA. 17Murdoch Children’s Research Institute, Victoria, Australia. 18University of São Paulo, São Paulo, Brazil. 19Washington University School of Medicine, St. Louis, MO, USA. 20The American Genome Center; Uniformed Services University of the Health Sciences, Bethesda, MD, USA. 21Centre for Bioinformatics and System Biology, Department of Life Sciences, Imperial College London, South Kensington Campus, London, UK. 22University of California, San Francisco, CA, USA; Chan Zuckerberg Biohub, San Francisco, CA, USA. 23Bai Jerbai Wadia Hospital for Children, Mumbai, India. 24School of Medicine and Public Health, University of Wisconsin, Madison, WI, USA. 25Instituto Nacional de Pediatria (National Institute of Pediatrics), Mexico City, Mexico. 26Swiss Federal Institute of Technology Lausanne, Lausanne, Switzerland. 27Research Unit, Hospital Universitario Nuestra Señora de Candelaria, Canarian Health System, Santa Cruz de Tenerife, Spain. 28University of Antioquia, Medellín, Colombia. 29Feinstein Institute for Medical Research, Northwell Health USA, Manhasset, NY, USA. 30Department of Paediatric Immunology and Pulmonology, Centre for Primary Immunodeficiency Ghent (CPIG), PID Research Lab, Jeffrey Modell Diagnosis and Research Centre, Ghent University Hospital, Edegem, Belgium. 31The Genetics Institute, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel. 32Sharjah Institute of Medical Research, College of Medicine, University of Sharjah, Sharjah, UAE. 33Institute for Systems Biology, Seattle, WA, USA. 34Children’s Hospital of Philadelphia, Philadelphia, PA, USA. 35Anschutz Medical Campus, Aurora, CO, USA. 36Riken, Tokyo, Japan. 37Hellenic Pasteur Institute, Athens, Greece. 38University of Tartu, Tartu, Estonia. 39Chang Gung University, Taoyuan County, Taiwan. 40The University of Hong Kong, Hong Kong, China. 41Shanghai Public Health Clinical Center, Fudan University, Shanghai, China. 42Yale School of Medicine, New Haven, CT, USA. 43New York Genome Center, New York, NY, USA. 44Shahid Beheshti University of Medical Sciences, Tehran, Iran. 45Semmelweis University Budapest, Budapest, Hungary. 46KU Leuven, Department of Immunology, Microbiology and Transplantation, Leuven, Belgium. 47Columbia University Medical Center, New York, NY, USA. 48University Clinic for Children’s Diseases, Skopje, North Macedonia. 49Aarhus University, Aarhus, Denmark. 50Tokyo Medical & Dental University Hospital, Tokyo, Japan. 51Singapore Immunology Network, Singapore. 52National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA. 53Bambino Gesù Children’s Hospital, Rome, Italy. 54Department of Biomedicine and Prevention, University of Rome “Tor Vergata,” Rome, Italy. 55Trinity College, Dublin, Ireland. 56Hiroshima University, Hiroshima, Japan. 57Bilkent University, Ankara, Turkey. 58Laboratory of Immunogenetics of Human Diseases, Innate Immunity Group, IdiPAZ Institute for Health Research, La Paz Hospital, Madrid, Spain. 59IIBB-CSIC, IDIBAPS, Barcelona, Spain. 60Faculdades Pequeno Príncipe e Instituto de Pesquisa Pelé Pequeno Príncipe, Curitiba, Brazil. 61Neurometabolic Diseases Laboratory, IDIBELL–Hospital Duran I Reynals; Catalan Institution for Research and Advanced Studies (ICREA); CIBERER U759, ISCiii Madrid Spain, Barcelona, Spain. 62Institut Pasteur (CNRS UMR2000) and Collège de France, Paris, France. 63Infectious Diseases Horizontal Technology Center and Singapore Immunology Network, Agency for Science Technology (A*STAR), Singapore. 64Medical Genetics, University of Siena, Siena, Italy; Genetica Medica, Azienda Ospedaliero-Universitaria Senese, Italy; GEN-COVID Multicenter Study. 65Hospital Universitario de Gran Canaria Dr. Negrín, Canarian Health System, Canary Islands, Spain. 66Imperial College London, London, UK. 67Boston Children’s Hospital, Harvard Medical School, Boston, MA, USA. 68Saeed Pathobiology and Genetic Lab, Tehran, Iran. 69Uniformed Services University of the Health Sciences, Bethesda, MD, USA. 70Hospital Universitari Vall d’Hebron, Barcelona, Spain. 71University Medical Center Utrecht, Amsterdam, The Netherlands. 72Garvan Institute of Medical Research, Sydney, Australia. 73The University of British Columbia, Vancouver, Canada. 74Holy Family Red Crescent Medical College; Centre for Precision Therapeutics, NeuroGen Children’s Healthcare; Genetics and Genomic Medicine Centre, NeuroGen Children’s Healthcare, Dhaka, Bangladesh. 75Mohammed Bin Rashid University of Medicine and Health Sciences, College of Medicine, Dubai, UAE; The Centre for Applied Genomics, Department of Genetics and Genome Biology, The Hospital for Sick Children, Toronto, Ontario, Canada. 76Amsterdam UMC, University of Amsterdam, Department of Neurology, Amsterdam Neuroscience, Amsterdam, The Netherlands. 77University of California, San Francisco, CA, USA. 78McGill University Health Centre, Montreal, Canada. 79Charité–Berlin University Hospital Center, Berlin, Germany. 80Molecular Biophysics Division, Faculty of Physics, A. Mickiewicz University, Uniwersytetu Poznanskiego 2, Poznań, Poland. 81Rockefeller University, Howard Hughes Medical Institute, Necker Hospital, New York, NY, USA.
*Leaders of the COVID Human Genetic Effort.
NIAID-USUHS/TAGC COVID Immunity Group
Huie Jing1,2, Wesley Tung1,2, Christopher R. Luthers3, Bradly M. Bauman3, Samantha Shafer2,4, Lixin Zheng2,4, Zinan Zhang2,4, Satoshi Kubo2,4, Samuel D. Chauvin2,4, Kazuyuki Meguro1,2, Elana Shaw1,2, Michael Lenardo2,4, Justin Lack5, Eric Karlins6, Daniel M. Hupalo7, John Rosenberger7, Gauthaman Sukumar7, Matthew D. Wilkerson7, Xijun Zhang7
1Laboratory of Clinical Immunology and Microbiology, Division of Intramural Research, NIAID, NIH, Bethesda, MD, USA. 2NIAID Clinical Genomics Program, National Institutes of Health, Bethesda, MD, USA. 3Department of Pharmacology & Molecular Therapeutics, Uniformed Services University of the Health Sciences, Bethesda, MD, USA. 4Laboratory of Immune System Biology, Division of Intramural Research, NIAID, NIH, Bethesda, MD, USA. 5NIAID Collaborative Bioinformatics Resource, Frederick National Laboratory for Cancer Research, Leidos Biomedical Research, Inc., Frederick, MD, USA. 6Bioinformatics and Computational Biosciences Branch, Office of Cyber Infrastructure and Computational Biology, NIAID, NIH, Bethesda, MD, USA. 7The American Genome Center, Uniformed Services University of the Health Sciences, Bethesda, MD, USA.
Supplementary Materials
science.sciencemag.org/content/370/6515/eabd4570/suppl/DC1
Materials and Methods
Figs. S1 to S11
Tables S1 and S2
MDAR Reproducibility Checklist
Contributor Information
Collaborators: Giuseppe Foti, Giacomo Bellani, Giuseppe Citerio, Ernesto Contro, Alberto Pesci, Maria Grazia Valsecchi, Marina Cazzaniga, Jorge Abad, Sergio Aguilera-Albesa, Ozge Metin Akcan, Ilad Alavi Darazam, Juan C. Aldave, Miquel Alfonso Ramos, Seyed Alireza Nadji, Gulsum Alkan, Jerome Allardet-Servent, Luis M. Allende, Laia Alsina, Marie-Alexandra Alyanakian, Blanca Amador-Borrero, Zahir Amoura, Arnau Antolí, Sevket Arslan, Sophie Assant, Terese Auguet, Axelle Azot, Fanny Bajolle, Aurélie Baldolli, Maite Ballester, Hagit Baris Feldman, Benoit Barrou, Alexandra Beurton, Agurtzane Bilbao, Geraldine Blanchard-Rohner, Ignacio Blanco, Adeline Blandinières, Daniel Blazquez-Gamero, Marketa Bloomfield, Mireia Bolivar-Prados, Raphael Borie, Cédric Bosteels, Ahmed A. Bousfiha, Claire Bouvattier, Oksana Boyarchuk, Maria Rita P. Bueno, Jacinta Bustamante, Juan José Cáceres Agra, Semra Calimli, Ruggero Capra, Maria Carrabba, Carlos Casasnovas, Marion Caseris, Martin Castelle, Francesco Castelli, Martín Castillo de Vera, Mateus V. Castro, Emilie Catherinot, Martin Chalumeau, Bruno Charbit, Matthew P. Cheng, Père Clavé, Bonaventura Clotet, Anna Codina, Fatih Colkesen, Fatma Çölkesen, Roger Colobran, Cloé Comarmond, David Dalmau, David Ross Darley, Nicolas Dauby, Stéphane Dauger, Loic de Pontual, Amin Dehban, Geoffroy Delplancq, Alexandre Demoule, Jean-Luc Diehl, Stephanie Dobbelaere, Sophie Durand, Waleed Eldars, Mohamed Elgamal, Marwa H. Elnagdy, Melike Emiroglu, Emine Hafize Erdeniz, Selma Erol Aytekin, Romain Euvrard, Recep Evcen, Giovanna Fabio, Laurence Faivre, Antonin Falck, Muriel Fartoukh, Morgane Faure, Miguel Fernandez Arquero, Carlos Flores, Bruno Francois, Victoria Fumadó, Francesca Fusco, Blanca Garcia Solis, Pascale Gaussem, Juana Gil-Herrera, Laurent Gilardin, Monica Girona Alarcon, Mònica Girona-Alarcón, Jean-Christophe Goffard, Funda Gok, Rafaela González-Montelongo, Antoine Guerder, Yahya Gul, Sukru Nail Guner, Marta Gut, Jérôme Hadjadj, Filomeen Haerynck, Rabih Halwani, Lennart Hammarström, Nevin Hatipoglu, Elisa Hernandez-Brito, Cathérine Heijmans, María Soledad Holanda-Peña, Juan Pablo Horcajada, Levi Hoste, Eric Hoste, Sami Hraiech, Linda Humbert, Alejandro D. Iglesias, Antonio Íñigo-Campos, Matthieu Jamme, María Jesús Arranz, Iolanda Jordan, Philippe Jorens, Fikret Kanat, Hasan Kapakli, Iskender Kara, Adem Karbuz, Kadriye Kart Yasar, Sevgi Keles, Yasemin Kendir Demirkol, Adam Klocperk, Zbigniew J. Król, Paul Kuentz, Yat Wah M. Kwan, Jean-Christophe Lagier, Bart N. Lambrecht, Yu-Lung Lau, Fleur Le Bourgeois, Yee-Sin Leo, Rafael Leon Lopez, Daniel Leung, Michael Levin, Michael Levy, Romain Lévy, Zhi Li, Agnes Linglart, Bart Loeys, José M. Lorenzo-Salazar, Céline Louapre, Catherine Lubetzki, Charles-Edouard Luyt, David C. Lye, Davood Mansouri, Majid Marjani, Jesus Marquez Pereira, Andrea Martin, David Martínez Pueyo, Javier Martinez-Picado, Iciar Marzana, Alexis Mathian, Larissa R. B. Matos, Gail V. Matthews, Julien Mayaux, Jean-Louis Mège, Isabelle Melki, Jean-François Meritet, Ozge Metin, Isabelle Meyts, Mehdi Mezidi, Isabelle Migeotte, Maude Millereux, Tristan Mirault, Clotilde Mircher, Mehdi Mirsaeidi, Abián Montesdeoca Melián, Antonio Morales Martinez, Pierre Morange, Clémence Mordacq, Guillaume Morelle, Stéphane Mouly, Adrián Muñoz-Barrera, Leslie Naesens, Cyril Nafati, João Farela Neves, Lisa F. P. Ng, Yeray Novoa Medina, Esmeralda Nuñez Cuadros, J. Gonzalo Ocejo-Vinyals, Zerrin Orbak, Mehdi Oualha, Tayfun Özçelik, Qiang Pan-Hammarström, Christophe Parizot, Tiffany Pascreau, Estela Paz-Artal, Sandra Pellegrini, Rebeca Pérez de Diego, Aurélien Philippe, Quentin Philippot, Laura Planas-Serra, Dominique Ploin, Julien Poissy, Géraldine Poncelet, Marie Pouletty, Paul Quentric, Didier Raoult, Anne-Sophie Rebillat, Ismail Reisli, Pilar Ricart, Jean-Christophe Richard, Nadia Rivet, Jacques G. Rivière, Gemma Rocamora Blanch, Carlos Rodrigo, Carlos Rodriguez-Gallego, Agustí Rodríguez-Palmero, Carolina Soledad Romero, Anya Rothenbuhler, Flore Rozenberg, Maria Yolanda Ruiz del Prado, Joan Sabater Riera, Oliver Sanchez, Silvia Sánchez-Ramón, Agatha Schluter, Matthieu Schmidt, Cyril E. Schweitzer, Francesco Scolari, Anna Sediva, Luis M. Seijo, Damien Sene, Sevtap Senoglu, Mikko R. J. Seppänen, Alex Serra Ilovich, Mohammad Shahrooei, Hans Slabbynck, David M. Smadja, Ali Sobh, Xavier Solanich Moreno, Jordi Solé-Violán, Catherine Soler, Pere Soler-Palacín, Yuri Stepanovskiy, Annabelle Stoclin, Fabio Taccone, Yacine Tandjaoui-Lambiotte, Jean-Luc Taupin, Simon J. Tavernier, Benjamin Terrier, Caroline Thumerelle, Gabriele Tomasoni, Julie Toubiana, Josep Trenado Alvarez, Sophie Trouillet-Assant, Jesús Troya, Alessandra Tucci, Matilde Valeria Ursini, Yurdagul Uzunhan, Pierre Vabres, Juan Valencia-Ramos, Eva Van Braeckel, Stijn Van de Velde, Ana Maria Van Den Rym, Jens Van Praet, Isabelle Vandernoot, Hulya Vatansev, Valentina Vélez-Santamaria, Sébastien Viel, Cédric Vilain, Marie E. Vilaire, Audrey Vincent, Guillaume Voiriot, Fanny Vuotto, Alper Yosunkaya, Barnaby E. Young, Fatih Yucel, Faiez Zannad, Mayana Zatz, Alexandre Belot, Christine Bole-Feysot, Stanislas Lyonnet, Cécile Masson, Patrick Nitschke, Aurore Pouliet, Yoann Schmitt, Frederic Tores, Mohammed Zarhrate, Laurent Abel, Claire Andrejak, François Angoulvant, Delphine Bachelet, Romain Basmaci, Sylvie Behillil, Marine Beluze, Dehbia Benkerrou, Krishna Bhavsar, François Bompart, Lila Bouadma, Maude Bouscambert, Mireille Caralp, Minerva Cervantes-Gonzalez, Anissa Chair, Alexandra Coelho, Camille Couffignal, Sandrine Couffin-Cadiergues, Eric D’Ortenzio, Charlene Da Silveira, Marie-Pierre Debray, Dominique Deplanque, Diane Descamps, Mathilde Desvallées, Alpha Diallo, Alphonsine Diouf, Céline Dorival, François Dubos, Xavier Duval, Philippine Eloy, Vincent VE Enouf, Hélène Esperou, Marina Esposito-Farese, Manuel Etienne, Nadia Ettalhaoui, Nathalie Gault, Alexandre Gaymard, Jade Ghosn, Tristan Gigante, Isabelle Gorenne, Jérémie Guedj, Alexandre Hoctin, Isabelle Hoffmann, Salma Jaafoura, Ouifiya Kafif, Florentia Kaguelidou, Sabina Kali, Antoine Khalil, Coralie Khan, Cédric Laouénan, Samira Laribi, Minh Le, Quentin Le Hingrat, Soizic Le Mestre, Hervé Le Nagard, François-Xavier Lescure, Yves Lévy, Claire Levy-Marchal, Bruno Lina, Guillaume Lingas, Jean Christophe Lucet, Denis Malvy, Marina Mambert, France Mentré, Noémie Mercier, Amina Meziane, Hugo Mouquet, Jimmy Mullaert, Nadège Neant, Marion Noret, Justine Pages, Aurélie Papadopoulos, Christelle Paul, Nathan Peiffer-Smadja, Ventzislava Petrov-Sanchez, Gilles Peytavin, Olivier Picone, Oriane Puéchal, Manuel Rosa-Calatrava, Bénédicte Rossignol, Patrick Rossignol, Carine Roy, Marion Schneider, Caroline Semaille, Nassima Si Mohammed, Lysa Tagherset, Coralie Tardivon, Marie-Capucine Tellier, François Téoulé, Olivier Terrier, Jean-François Timsit, Théo Trioux, Christelle Tual, Sarah Tubiana, Sylvie van der Werf, Noémie Vanel, Aurélie Veislinger, Benoit Visseaux, Aurélie Wiedemann, Yazdan Yazdanpanah, Loubna Alavoine, Karine K. A. Amat, Sylvie Behillil, Julia Bielicki, Patricia Bruijning, Charles Burdet, Eric Caumes, Charlotte Charpentier, Bruno Coignard, Yolande Costa, Sandrine Couffin-Cadiergues, Florence Damond, Aline Dechanet, Christelle Delmas, Diane Descamps, Xavier Duval, Jean-Luc Ecobichon, Vincent Enouf, Hélène Espérou, Wahiba Frezouls, Nadhira Houhou, Emila Ilic-Habensus, Ouifiya Kafif, John Kikoine, Quentin Le Hingrat, David Lebeaux, Anne Leclercq, Jonathan Lehacaut, Sophie Letrou, Bruno Lina, Jean-Christophe Lucet, Denis Malvy, Pauline Manchon, Milica Mandic, Mohamed Meghadecha, Justina Motiejunaite, Mariama Nouroudine, Valentine Piquard, Andreea Postolache, Caroline Quintin, Jade Rexach, Layidé Roufai, Zaven Terzian, Michael Thy, Sarah Tubiana, Sylvie van der Werf, Valérie Vignali, Benoit Visseaux, Yazdan Yazdanpanah, Michiel van Agtmael, Anna Geke Algera, Frank van Baarle, Diane Bax, Martijn Beudel, Harm Jan Bogaard, Marije Bomers, Lieuwe Bos, Michela Botta, Justin de Brabander, Godelieve de Bree, Matthijs C. Brouwer, Sanne de Bruin, Marianna Bugiani, Esther Bulle, O. Chouchane, Alex Cloherty, Paul Elbers, Lucas Fleuren, Suzanne Geerlings, Bart Geerts, Theo Geijtenbeek, Armand Girbes, Bram Goorhuis, Martin P. Grobusch, Florianne Hafkamp, Laura Hagens, Jorg Hamann, Vanessa Harris, Robert Hemke, Sabine M. Hermans, Leo Heunks, Markus W. Hollmann, Janneke Horn, Joppe W. Hovius, Menno D. de Jong, Rutger Koning, Niels van Mourik, Jeaninne Nellen, Frederique Paulus, Edgar Peters, Tom van der Poll, Benedikt Preckel, Jan M. Prins, Jorinde Raasveld, Tom Reijnders, Michiel Schinkel, Marcus J. Schultz, Alex Schuurman, Kim Sigaloff, Marry Smit, Cornelis S. Stijnis, Willemke Stilma, Charlotte Teunissen, Patrick Thoral, Anissa Tsonas, Marc van der Valk, Denise Veelo, Alexander P.J. Vlaar, Heder de Vries, Michèle van Vugt, W. Joost Wiersinga, Dorien Wouters, A. H. (Koos) Zwinderman, Diederik van de Beek, Laurent Abel, Alessandro Aiuti, Saleh Al Muhsen, Fahd Al-Mulla, Mark S. Anderson, Andrés Augusto Arias, Hagit Baris Feldman, Dusan Bogunovic, Alexandre Bolze, Anastasiia Bondarenko, Ahmed A. Bousfiha, Petter Brodin, Yenan Bryceson, Carlos D. Bustamante, Manish Butte, Giorgio Casari, Samya Chakravorty, John Christodoulou, Elizabeth Cirulli, Antonio Condino-Neto, Megan A. Cooper, Clifton L. Dalgard, Alessia David, Joseph L. DeRisi, Murkesh Desai, Beth A. Drolet, Sara Espinosa, Jacques Fellay, Carlos Flores, Jose Luis Franco, Peter K. Gregersen, Filomeen Haerynck, David Hagin, Rabih Halwani, Jim Heath, Sarah E. Henrickson, Elena Hsieh, Kohsuke Imai, Yuval Itan, Timokratis Karamitros, Kai Kisand, Cheng-Lung Ku, Yu-Lung Lau, Yun Ling, Carrie L. Lucas, Tom Maniatis, Davoud Mansouri, Laszlo Marodi, Isabelle Meyts, Joshua Milner, Kristina Mironska, Trine Mogensen, Tomohiro Morio, Lisa F. P. Ng, Luigi D. Notarangelo, Antonio Novelli, Giuseppe Novelli, Cliona O’Farrelly, Satoshi Okada, Tayfun Ozcelik, Rebeca Perez de Diego, Anna M. Planas, Carolina Prando, Aurora Pujol, Lluis Quintana-Murci, Laurent Renia, Alessandra Renieri, Carlos Rodríguez-Gallego, Vanessa Sancho-Shimizu, Vijay Sankaran, Kelly Schiabor Barrett, Mohammed Shahrooei, Andrew Snow, Pere Soler-Palacín, András N. Spaan, Stuart Tangye, Stuart Turvey, Furkan Uddin, Mohammed J. Uddin, Diederik van de Beek, Sara E. Vazquez, Donald C. Vinh, Horst von Bernuth, Nicole Washington, Pawel Zawadzki, Helen C. Su, Jean-Laurent Casanova, Huie Jing, Wesley Tung, Christopher R. Luthers, Bradly M. Bauman, Samantha Shafer, Lixin Zheng, Zinan Zhang, Satoshi Kubo, Samuel D. Chauvin, Kazuyuki Meguro, Elana Shaw, Michael Lenardo, Justin Lack, Eric Karlins, Daniel M. Hupalo, John Rosenberger, Gauthaman Sukumar, Matthew D. Wilkerson, and Xijun Zhang
References and Notes
- 1.Morens D. M., Fauci A. S., Emerging pandemic diseases: How we got to COVID-19. Cell 182, 1077–1092 (2020). 10.1016/j.cell.2020.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casanova J. L., Abel L., Lethal Infectious Diseases as Inborn Errors of Immunity: Toward a Synthesis of the Germ and Genetic Theories. Annu. Rev. Pathol. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casanova J. L., Abel L., The human genetic determinism of life-threatening infectious diseases: Genetic heterogeneity and physiological homogeneity? Hum. Genet. 139, 681–694 (2020). 10.1007/s00439-020-02184-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casanova J. L., Su H. C.; COVID Human Genetic Effort , A global effort to define the human genetics of protective immunity to SARS-CoV-2 infection. Cell 181, 1194–1199 (2020). 10.1016/j.cell.2020.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q., Human genetics of life-threatening influenza pneumonitis. Hum. Genet. 139, 941–948 (2020). 10.1007/s00439-019-02108-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim H. K., Huang S. X. L., Chen J., Kerner G., Gilliaux O., Bastard P., Dobbs K., Hernandez N., Goudin N., Hasek M. L., García Reino E. J., Lafaille F. G., Lorenzo L., Luthra P., Kochetkov T., Bigio B., Boucherit S., Rozenberg F., Vedrinne C., Keller M. D., Itan Y., García-Sastre A., Celard M., Orange J. S., Ciancanelli M. J., Meyts I., Zhang Q., Abel L., Notarangelo L. D., Snoeck H.-W., Casanova J.-L., Zhang S.-Y., Severe influenza pneumonitis in children with inherited TLR3 deficiency. J. Exp. Med. 216, 2038–2056 (2019). 10.1084/jem.20181621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciancanelli M. J., Huang S. X. L., Luthra P., Garner H., Itan Y., Volpi S., Lafaille F. G., Trouillet C., Schmolke M., Albrecht R. A., Israelsson E., Lim H. K., Casadio M., Hermesh T., Lorenzo L., Leung L. W., Pedergnana V., Boisson B., Okada S., Picard C., Ringuier B., Troussier F., Chaussabel D., Abel L., Pellier I., Notarangelo L. D., García-Sastre A., Basler C. F., Geissmann F., Zhang S.-Y., Snoeck H.-W., Casanova J.-L., Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science 348, 448–453 (2015). 10.1126/science.aaa1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernandez N., Melki I., Jing H., Habib T., Huang S. S. Y., Danielson J., Kula T., Drutman S., Belkaya S., Rattina V., Lorenzo-Diaz L., Boulai A., Rose Y., Kitabayashi N., Rodero M. P., Dumaine C., Blanche S., Lebras M.-N., Leung M. C., Mathew L. S., Boisson B., Zhang S.-Y., Boisson-Dupuis S., Giliani S., Chaussabel D., Notarangelo L. D., Elledge S. J., Ciancanelli M. J., Abel L., Zhang Q., Marr N., Crow Y. J., Su H. C., Casanova J.-L., Life-threatening influenza pneumonitis in a child with inherited IRF9 deficiency. J. Exp. Med. 215, 2567–2585 (2018). 10.1084/jem.20180628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sancho-Shimizu V., Pérez de Diego R., Lorenzo L., Halwani R., Alangari A., Israelsson E., Fabrega S., Cardon A., Maluenda J., Tatematsu M., Mahvelati F., Herman M., Ciancanelli M., Guo Y., AlSum Z., Alkhamis N., Al-Makadma A. S., Ghadiri A., Boucherit S., Plancoulaine S., Picard C., Rozenberg F., Tardieu M., Lebon P., Jouanguy E., Rezaei N., Seya T., Matsumoto M., Chaussabel D., Puel A., Zhang S.-Y., Abel L., Al-Muhsen S., Casanova J.-L., Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J. Clin. Invest. 121, 4889–4902 (2011). 10.1172/JCI59259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casrouge A., Zhang S.-Y., Eidenschenk C., Jouanguy E., Puel A., Yang K., Alcais A., Picard C., Mahfoufi N., Nicolas N., Lorenzo L., Plancoulaine S., Sénéchal B., Geissmann F., Tabeta K., Hoebe K., Du X., Miller R. L., Héron B., Mignot C., de Villemeur T. B., Lebon P., Dulac O., Rozenberg F., Beutler B., Tardieu M., Abel L., Casanova J.-L., Herpes simplex virus encephalitis in human UNC-93B deficiency. Science 314, 308–312 (2006). 10.1126/science.1128346 [DOI] [PubMed] [Google Scholar]
- 11.Pérez de Diego R., Sancho-Shimizu V., Lorenzo L., Puel A., Plancoulaine S., Picard C., Herman M., Cardon A., Durandy A., Bustamante J., Vallabhapurapu S., Bravo J., Warnatz K., Chaix Y., Cascarrigny F., Lebon P., Rozenberg F., Karin M., Tardieu M., Al-Muhsen S., Jouanguy E., Zhang S.-Y., Abel L., Casanova J.-L., Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity 33, 400–411 (2010). 10.1016/j.immuni.2010.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herman M., Ciancanelli M., Ou Y.-H., Lorenzo L., Klaudel-Dreszler M., Pauwels E., Sancho-Shimizu V., Pérez de Diego R., Abhyankar A., Israelsson E., Guo Y., Cardon A., Rozenberg F., Lebon P., Tardieu M., Heropolitańska-Pliszka E., Chaussabel D., White M. A., Abel L., Zhang S.-Y., Casanova J.-L., Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J. Exp. Med. 209, 1567–1582 (2012). 10.1084/jem.20111316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersen L. L., Mørk N., Reinert L. S., Kofod-Olsen E., Narita R., Jørgensen S. E., Skipper K. A., Höning K., Gad H. H., Østergaard L., Ørntoft T. F., Hornung V., Paludan S. R., Mikkelsen J. G., Fujita T., Christiansen M., Hartmann R., Mogensen T. H., Functional IRF3 deficiency in a patient with herpes simplex encephalitis. J. Exp. Med. 212, 1371–1379 (2015). 10.1084/jem.20142274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Audry M., Ciancanelli M., Yang K., Cobat A., Chang H.-H., Sancho-Shimizu V., Lorenzo L., Niehues T., Reichenbach J., Li X.-X., Israel A., Abel L., Casanova J.-L., Zhang S.-Y., Jouanguy E., Puel A., NEMO is a key component of NF-κB- and IRF-3-dependent TLR3-mediated immunity to herpes simplex virus. J. Allergy Clin. Immunol. 128, 610–617.e4 (2011). 10.1016/j.jaci.2011.04.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernandez N., Bucciol G., Moens L., Le Pen J., Shahrooei M., Goudouris E., Shirkani A., Changi-Ashtiani M., Rokni-Zadeh H., Sayar E. H., Reisli I., Lefevre-Utile A., Zijlmans D., Jurado A., Pholien R., Drutman S., Belkaya S., Cobat A., Boudewijns R., Jochmans D., Neyts J., Seeleuthner Y., Lorenzo-Diaz L., Enemchukwu C., Tietjen I., Hoffmann H.-H., Momenilandi M., Pöyhönen L., Siqueira M. M., de Lima S. M. B., de Souza Matos D. C., Homma A., Maia M. L. S., da Costa Barros T. A., de Oliveira P. M. N., Mesquita E. C., Gijsbers R., Zhang S.-Y., Seligman S. J., Abel L., Hertzog P., Marr N., Martins R. M., Meyts I., Zhang Q., MacDonald M. R., Rice C. M., Casanova J.-L., Jouanguy E., Bossuyt X., Inherited IFNAR1 deficiency in otherwise healthy patients with adverse reaction to measles and yellow fever live vaccines. J. Exp. Med. 216, 2057–2070 (2019). 10.1084/jem.20182295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duncan C. J., Mohamad S. M. B., Young D. F., Skelton A. J., Leahy T. R., Munday D. C., Butler K. M., Morfopoulou S., Brown J. R., Hubank M., Connell J., Gavin P. J., McMahon C., Dempsey E., Lynch N. E., Jacques T. S., Valappil M., Cant A. J., Breuer J., Engelhardt K. R., Randall R. E., Hambleton S., Human IFNAR2 deficiency: Lessons for antiviral immunity. Sci. Transl. Med. 7, 307ra154 (2015). 10.1126/scitranslmed.aac4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dupuis S., Jouanguy E., Al-Hajjar S., Fieschi C., Al-Mohsen I. Z., Al-Jumaah S., Yang K., Chapgier A., Eidenschenk C., Eid P., Al Ghonaium A., Tufenkeji H., Frayha H., Al-Gazlan S., Al-Rayes H., Schreiber R. D., Gresser I., Casanova J.-L., Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 33, 388–391 (2003). 10.1038/ng1097 [DOI] [PubMed] [Google Scholar]
- 18.Hambleton S., Goodbourn S., Young D. F., Dickinson P., Mohamad S. M. B., Valappil M., McGovern N., Cant A. J., Hackett S. J., Ghazal P., Morgan N. V., Randall R. E., STAT2 deficiency and susceptibility to viral illness in humans. Proc. Natl. Acad. Sci. U.S.A. 110, 3053–3058 (2013). 10.1073/pnas.1220098110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Döffinger R., Smahi A., Bessia C., Geissmann F., Feinberg J., Durandy A., Bodemer C., Kenwrick S., Dupuis-Girod S., Blanche S., Wood P., Rabia S. H., Headon D. J., Overbeek P. A., Le Deist F., Holland S. M., Belani K., Kumararatne D. S., Fischer A., Shapiro R., Conley M. E., Reimund E., Kalhoff H., Abinun M., Munnich A., Israël A., Courtois G., Casanova J.-L., X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-kappaB signaling. Nat. Genet. 27, 277–285 (2001). 10.1038/85837 [DOI] [PubMed] [Google Scholar]
- 20.Zhang S. Y., Jouanguy E., Ugolini S., Smahi A., Elain G., Romero P., Segal D., Sancho-Shimizu V., Lorenzo L., Puel A., Picard C., Chapgier A., Plancoulaine S., Titeux M., Cognet C., von Bernuth H., Ku C.-L., Casrouge A., Zhang X.-X., Barreiro L., Leonard J., Hamilton C., Lebon P., Héron B., Vallée L., Quintana-Murci L., Hovnanian A., Rozenberg F., Vivier E., Geissmann F., Tardieu M., Abel L., Casanova J.-L., TLR3 deficiency in patients with herpes simplex encephalitis. Science 317, 1522–1527 (2007). 10.1126/science.1139522 [DOI] [PubMed] [Google Scholar]
- 21.Zhang G., deWeerd N. A., Stifter S. A., Liu L., Zhou B., Wang W., Zhou Y., Ying B., Hu X., Matthews A. Y., Ellis M., Triccas J. A., Hertzog P. J., Britton W. J., Chen X., Feng C. G., A proline deletion in IFNAR1 impairs IFN-signaling and underlies increased resistance to tuberculosis in humans. Nat. Commun. 9, 85 (2018). 10.1038/s41467-017-02611-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomsen M. M., Jørgensen S. E., Storgaard M., Kristensen L. S., Gjedsted J., Christiansen M., Gad H. H., Hartmann R., Mogensen T. H., Identification of an IRF3 variant and defective antiviral interferon responses in a patient with severe influenza. Eur. J. Immunol. 49, 2111–2114 (2019). 10.1002/eji.201848083 [DOI] [PubMed] [Google Scholar]
- 23.Thomsen M. M., Jørgensen S. E., Gad H. H., Storgaard M., Gjedsted J., Christiansen M., Hartmann R., Mogensen T. H., Defective interferon priming and impaired antiviral responses in a patient with an IRF7 variant and severe influenza. Med. Microbiol. Immunol. (Berl.) 208, 869–876 (2019). 10.1007/s00430-019-00623-8 [DOI] [PubMed] [Google Scholar]
- 24.Tangye S. G., Al-Herz W., Bousfiha A., Chatila T., Cunningham-Rundles C., Etzioni A., Franco J. L., Holland S. M., Klein C., Morio T., Ochs H. D., Oksenhendler E., Picard C., Puck J., Torgerson T. R., Casanova J.-L., Sullivan K. E., Human inborn errors of immunity: 2019 update on the classification from the International Union of Immunological Societies Expert Committee. J. Clin. Immunol. 40, 24–64 (2020). 10.1007/s10875-019-00737-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bousfiha A., Jeddane L., Picard C., Al-Herz W., Ailal F., Chatila T., Cunningham-Rundles C., Etzioni A., Franco J. L., Holland S. M., Klein C., Morio T., Ochs H. D., Oksenhendler E., Puck J., Torgerson T. R., Casanova J.-L., Sullivan K. E., Tangye S. G., Human Inborn Errors of Immunity: 2019 Update of the IUIS Phenotypical Classification. J. Clin. Immunol. 40, 66–81 (2020). 10.1007/s10875-020-00758-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Notarangelo L. D., Bacchetta R., Casanova J.-L., Su H. C., Human inborn errors of immunity: An expanding universe. Sci. Immunol. 5, eabb1662 (2020). 10.1126/sciimmunol.abb1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pène F., Marin N., Roche N., Szwebel T.-A., Merkling S. H., Treluyer J.-M., Veyer D., Mouthon L., Blanc C., Tharaux P.-L., Rozenberg F., Fischer A., Duffy D., Rieux-Laucat F., Kernéis S., Terrier B., Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369, 718–724 (2020). 10.1126/science.abc6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trouillet-Assant S., Viel S., Gaymard A., Pons S., Richard J.-C., Perret M., Villard M., Brengel-Pesce K., Lina B., Mezidi M., Bitker L., Belot A.; COVID HCL Study Group , Type I IFN immunoprofiling in COVID-19 patients. J. Allergy Clin. Immunol. 146, 206–208.e2 (2020). 10.1016/j.jaci.2020.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bastard P., Rosen L. B., Zhang Q., Michailidis E., Hoffmann H.-H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., Manry J., Shaw E., Haljasmägi L., Peterson P., Lorenzo L., Bizien L., Trouillet-Assant S., Dobbs K., Almeida de Jesus A., Belot A., Kallaste A., Catherinot E., Tandjaoui-Lambiotte Y., Le Pen J., Kerner G., Bigio B., Seeleuthner Y., Yang R., Bolze A., Spaan A. N., Delmonte O. M., Abers M. S., Aiuti A., Casari G., Lampasona V., Piemonti L., Ciceri F., Bilguvar K., Lifton R. P., Vasse M., Smadja D. M., Migaud M., Hadjadj J., Terrier B., Duffy D., Quintana-Murci L., van de Beek D., Roussel L., Vinh D. C., Tangye S. G., Haerynck F., Dalmau D., Martinez-Picado J., Brodin P., Nussenzweig M. C., Boisson-Dupuis S., Rodríguez-Gallego C., Vogt G., Mogensen T. H., Oler A. J., Gu J., Burbelo P. D., Cohen J., Biondi A., Bettini L. R., D'Angio M., Bonfanti P., Rossignol P., Mayaux J., Rieux-Laucat F., Husebye E. S., Fusco F., Ursini M. V., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Castagnoli R., Montagna D., Licari A., Marseglia G. L., Duval X., Ghosn J., HGID Lab, NIAID-USUHS Immune Response to COVID Group, COVID Clinicians, COVID-STORM Clinicians, Imagine COVID Group, French COVID Cohort Study Group, The Milieu Intérieur Consortium, CoV-Contact Cohort, Amsterdam UMC Covid-19 Biobank, COVID Human Genetic Effort, Tsang J. S., Goldbach-Mansky R., Kisand K., Lionakis M. S., Puel A., Zhang S.-Y., Holland S. M., Gorochov G., Jouanguy E., Rice C. M., Cobat A., Notarangelo L. D., Abel L., Su H. C., Casanova J.-L., Auto-antibodies against type I IFNs in patients with life-threatening COVID-19. Science 10.1126/science.abd4585 (2020). [Google Scholar]
- 30.DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., Hartl C., Philippakis A. A., del Angel G., Rivas M. A., Hanna M., McKenna A., Fennell T. J., Kernytsky A. M., Sivachenko A. Y., Cibulskis K., Gabriel S. B., Altshuler D., Daly M. J., A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011). 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boisson B., Honda Y., Ajiro M., Bustamante J., Bendavid M., Gennery A. R., Kawasaki Y., Ichishima J., Osawa M., Nihira H., Shiba T., Tanaka T., Chrabieh M., Bigio B., Hur H., Itan Y., Liang Y., Okada S., Izawa K., Nishikomori R., Ohara O., Heike T., Abel L., Puel A., Saito M. K., Casanova J.-L., Hagiwara M., Yasumi T., Rescue of recurrent deep intronic mutation underlying cell type-dependent quantitative NEMO deficiency. J. Clin. Invest. 129, 583–597 (2019). 10.1172/JCI124011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gambin T., Akdemir Z. C., Yuan B., Gu S., Chiang T., Carvalho C. M. B., Shaw C., Jhangiani S., Boone P. M., Eldomery M. K., Karaca E., Bayram Y., Stray-Pedersen A., Muzny D., Charng W. L., Bahrambeigi V., Belmont J. W., Boerwinkle E., Beaudet A. L., Gibbs R. A., Lupski J. R., Homozygous and hemizygous CNV detection from exome sequencing data in a Mendelian disease cohort. Nucleic Acids Res. 45, 1633–1648 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Backenroth D., Homsy J., Murillo L. R., Glessner J., Lin E., Brueckner M., Lifton R., Goldmuntz E., Chung W. K., Shen Y., CANOES: Detecting rare copy number variants from whole exome sequencing data. Nucleic Acids Res. 42, e97 (2014). 10.1093/nar/gku345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.M. Bouaziz, J. Mullaert, B. Bigio, Y. Seeleuthner, J.-L. Casanova, A. Alcais, L. Abel, A. Cobat, Controlling for human population stratification in rare variant association studies. bioRxiv 969477 [Preprint]. 28 February 2020. . 10.1101/2020.02.28.969477 [DOI] [PMC free article] [PubMed]
- 36.Persyn E., Redon R., Bellanger L., Dina C., The impact of a fine-scale population stratification on rare variant association test results. PLOS ONE 13, e0207677 (2018). 10.1371/journal.pone.0207677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Shen X., Pan W., Adjusting for population stratification in a fine scale with principal components and sequencing data. Genet. Epidemiol. 37, 787–801 (2013). 10.1002/gepi.21764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boisson-Dupuis S., Ramirez-Alejo N., Li Z., Patin E., Rao G., Kerner G., Lim C. K., Krementsov D. N., Hernandez N., Ma C. S., Zhang Q., Markle J., Martinez-Barricarte R., Payne K., Fisch R., Deswarte C., Halpern J., Bouaziz M., Mulwa J., Sivanesan D., Lazarov T., Naves R., Garcia P., Itan Y., Boisson B., Checchi A., Jabot-Hanin F., Cobat A., Guennoun A., Jackson C. C., Pekcan S., Caliskaner Z., Inostroza J., Costa-Carvalho B. T., de Albuquerque J. A. T., Garcia-Ortiz H., Orozco L., Ozcelik T., Abid A., Rhorfi I. A., Souhi H., Amrani H. N., Zegmout A., Geissmann F., Michnick S. W., Muller-Fleckenstein I., Fleckenstein B., Puel A., Ciancanelli M. J., Marr N., Abolhassani H., Balcells M. E., Condino-Neto A., Strickler A., Abarca K., Teuscher C., Ochs H. D., Reisli I., Sayar E. H., El-Baghdadi J., Bustamante J., Hammarström L., Tangye S. G., Pellegrini S., Quintana-Murci L., Abel L., Casanova J.-L., Tuberculosis and impaired IL-23-dependent IFN-γ immunity in humans homozygous for a common TYK2 missense variant. Sci. Immunol. 3, eaau8714 (2018). 10.1126/sciimmunol.aau8714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fochi S., Bergamo E., Serena M., Mutascio S., Journo C., Mahieux R., Ciminale V., Bertazzoni U., Zipeto D., Romanelli M. G., TRAF3 Is Required for NF-κB Pathway Activation Mediated by HTLV Tax Proteins. Front. Microbiol. 10, 1302 (2019). 10.3389/fmicb.2019.01302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robbiani D. F., Gaebler C., Muecksch F., Lorenzi J. C. C., Wang Z., Cho A., Agudelo M., Barnes C. O., Gazumyan A., Finkin S., Hägglöf T., Oliveira T. Y., Viant C., Hurley A., Hoffmann H.-H., Millard K. G., Kost R. G., Cipolla M., Gordon K., Bianchini F., Chen S. T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A. W., Waltari E., Pak J. E., Huey-Tubman K. E., Koranda N., Hoffman P. R., West A. P. Jr.., Rice C. M., Hatziioannou T., Bjorkman P. J., Bieniasz P. D., Caskey M., Nussenzweig M. C., Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584, 437–442 (2020). 10.1038/s41586-020-2456-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Q. Zhang, P. Bastard, Z. Liu, J. Le Pen, M. Moncada-Velez, J. Chen, M. Ogishi, I. K. D. Sabli, S. Hodeib, C. Korol, J. Rosain, K. Bilguvar, J. Ye, A. Bolze, B. Bigio, R. Yang, A. Augusto Arias Sierra, Q. Zhou, Y. Zhang, F. Onodi, S. Korniotis, L. Karpf, Q. Philippot, M. Chbihi, L. Bonnet-Madin, K. Dorgham, N. Smith, W. M. Schneider, B. S. Razooky, H.-H. Hoffmann, E. Michailidis, L. Moens, J. E. Han, L. Lorenzo, L. Bizien, P. Meade, A.-L. Neehus, A. C. Ugurbil, A. Corneau, G. Kerner, P. Zhang, F. Rapaport, Y. Seeleuthner, J. Manry, C. Masson, Y. Schmitt, A. Schlüter, T. Le Voyer, T. Khan, J. Li, J. Fellay, L. Roussel, M. Shahrooei, M. F. Alosaimi, D. Mansouri, H. Al-Saud, F. Al-Mulla, F. Almourfi, S. Z. Al-Muhsen, F. Alsohime, S. Al Turki, R. Hasanato, D. van de Beek, A. Biondi, L. R. Bettini, M. D’Angio, P. Bonfanti, L. Imberti, A. Sottini, S. Paghera, E. Quiros-Roldan, C. Rossi, A. J. Oler, M. F. Tompkins, C. Alba, I. Vandernoot, J.-C. Goffard, G. Smits, I. Migeotte, F. Haerynck, P. Soler-Palacin, A. Martin-Nalda, R. Colobran, P.-E. Morange, S. Keles, F. Çölkesen, T. Ozcelik, K. K. Yasar, S. Senoglu, Ş. N. Karabela, C. Rodríguez-Gallego, G. Novelli, S. Hraiech, Y. Tandjaoui-Lambiotte, X. Duval, C. Laouenan, COVID-STORM Clinicians, COVID Clinicians, Imagine COVID Group, French COVID Cohort Study Group, CoV-Contact Cohort, Amsterdam UMC Covid-19 Biobank, COVID Human Genetic Effort, NIAID-USUHS/TAGC COVID Immunity Group, A. L. Snow, C. L. Dalgard, J. Milner, D. C. Vinh, T. H. Mogensen, N. Marr, A. N. Spaan, B. Boisson, S. Boisson-Dupuis, J. Bustamante, A. Puel, M. Ciancanelli, I. Meyts, T. Maniatis, V. Soumelis, A. Amara, M. Nussenzweig, A. García-Sastre, F. Krammer, A. Pujol, D. Duffy, R. Lifton, S.-Y. Zhang, G. Gorochov, V. Béziat, E. Jouanguy, V. Sancho-Shimizu, C. M. Rice, L. Abel, L. D. Notarangelo, A. Cobat, H. C. Su, J.-L. Casanova, Detailed genotype counts for all coding variants for: Inborn errors of type I IFN immunity in patients with life-threatening COVID-19, Dryad (2020); . 10.5061/dryad.8pk0p2nkk [DOI]
- 42.M. Ogishi, R. Yang, C. Gruber, S. Pelham, A. N. Spaan, J. Rosain, M. Chbihi, J. E. Han, V. K. Rao, L. Kainulainen, J. Bustamante, B. Boisson, D. Bogunovic, S. Boisson-Dupuis, J.-L. Casanova, Multi-batch cytometry data integration for optimal immunophenotyping. bioRxiv 202432 [Preprint]. 15 July 2020. 10.1101/2020.07.14.202432 [DOI]
- 43.Meade P., Kuan G., Strohmeier S., Maier H. E., Amanat F., Balmaseda A., Ito K., Kirkpatrick E., Javier A., Gresh L., Nachbagauer R., Gordon A., Krammer F., Influenza virus infection induces a narrow antibody response in children but a broad recall response in adults. mBio 11, e03243-19 (2020). 10.1128/mBio.03243-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Q. Zhang, P. Bastard, Z. Liu, J. Le Pen, M. Moncada-Velez, J. Chen, M. Ogishi, I. K. D. Sabli, S. Hodeib, C. Korol, J. Rosain, K. Bilguvar, J. Ye, A. Bolze, B. Bigio, R. Yang, A. Augusto Arias Sierra, Q. Zhou, Y. Zhang, F. Onodi, S. Korniotis, L. Karpf, Q. Philippot, M. Chbihi, L. Bonnet-Madin, K. Dorgham, N. Smith, W. M. Schneider, B. S. Razooky, H.-H. Hoffmann, E. Michailidis, L. Moens, J. E. Han, L. Lorenzo, L. Bizien, P. Meade, A.-L. Neehus, A. C. Ugurbil, A. Corneau, G. Kerner, P. Zhang, F. Rapaport, Y. Seeleuthner, J. Manry, C. Masson, Y. Schmitt, A. Schlüter, T. Le Voyer, T. Khan, J. Li, J. Fellay, L. Roussel, M. Shahrooei, M. F. Alosaimi, D. Mansouri, H. Al-Saud, F. Al-Mulla, F. Almourfi, S. Z. Al-Muhsen, F. Alsohime, S. Al Turki, R. Hasanato, D. van de Beek, A. Biondi, L. R. Bettini, M. D’Angio, P. Bonfanti, L. Imberti, A. Sottini, S. Paghera, E. Quiros-Roldan, C. Rossi, A. J. Oler, M. F. Tompkins, C. Alba, I. Vandernoot, J.-C. Goffard, G. Smits, I. Migeotte, F. Haerynck, P. Soler-Palacin, A. Martin-Nalda, R. Colobran, P.-E. Morange, S. Keles, F. Çölkesen, T. Ozcelik, K. K. Yasar, S. Senoglu, Ş. N. Karabela, C. Rodríguez-Gallego, G. Novelli, S. Hraiech, Y. Tandjaoui-Lambiotte, X. Duval, C. Laouenan, COVID-STORM Clinicians, COVID Clinicians, Imagine COVID Group, French COVID Cohort Study Group, CoV-Contact Cohort, Amsterdam UMC Covid-19 Biobank, COVID Human Genetic Effort, NIAID-USUHS/TAGC COVID Immunity Group, A. L. Snow, C. L. Dalgard, J. Milner, D. C. Vinh, T. H. Mogensen, N. Marr, A. N. Spaan, B. Boisson, S. Boisson-Dupuis, J. Bustamante, A. Puel, M. Ciancanelli, I. Meyts, T. Maniatis, V. Soumelis, A. Amara, M. Nussenzweig, A. García-Sastre, F. Krammer, A. Pujol, D. Duffy, R. Lifton, S.-Y. Zhang, G. Gorochov, V. Béziat, E. Jouanguy, V. Sancho-Shimizu, C. M. Rice, L. Abel, L. D. Notarangelo, A. Cobat, H. C. Su, J.-L. Casanova, Detailed genotype counts for all coding variants for: Inborn errors of type I IFN immunity in patients with life-threatening COVID-19, Dryad (2020); . 10.5061/dryad.8pk0p2nkk [DOI]
Supplementary Materials
science.sciencemag.org/content/370/6515/eabd4570/suppl/DC1
Materials and Methods
Figs. S1 to S11
Tables S1 and S2
MDAR Reproducibility Checklist