Abstract
The rumen microbiome ‐ a remarkable example of obligatory symbiosis with high ecological and social relevance
Subject Categories: Digestive System; Ecology; Microbiology, Virology & Host Pathogen Interaction
Ruminants are intimately linked to mankind since their domestication some 8,000 years ago, and their close relationship may have well been one of the main drivers of human civilization (Diamond, 1997). Ruminants—cattle, sheep, goats, deer, gazelles, and so on—also embody the close link between solar energy transformed via photosynthesis and digestion into consumable products, such as meat, milk, leather, or wool, that have sustained humanity for millennia. Throughout this shared history, constant improvements through breeding, husbandry, and industrial livestock farming have greatly increased the production of milk, meat, and other animal‐based products.
Ruminants, more so than any other mammalian group also represent the epitome of mammalian‐microbe symbiosis, as they rely completely on microbial fermentation to sustain their lives. In the rumen, the fermentative organ situated in the upper gastrointestinal tract resides a vast microbial community from all domains of life—bacteria, archaea, and eukarya—that turn indigestible plant feed into food for the animal. The rumen microbiome produces up to 70% of the energy the animal needs for growth and maintenance, and, from mankind's perspective, for the production of food and other consumables.
Ruminants, more so than any other mammalian group, also represent the epitome of mammalian‐microbe symbiosis, as they rely completely on microbial fermentation to sustain their lives.
With growing understanding that these microorganisms are responsible for degrading plant material and supplying nutrients for the animals, a new research discipline emerged along with aspirations to improve the yield of livestock farming. While most research had understandably focused on production efficiency, it also showed that the rumen microbiome is intricately linked to many other phenotypes of the animal. This understanding comes at a time when we increasingly realize that mankind's actions have a detrimental effect on the environment. The microbial fermentation in the rumen produces large amounts of methane, a potent greenhouse gas that has been demonstrated to contribute to global climate change. We therefore need to consider both our increased demand for meat and milk products and aim to mitigate the negative environmental impact of intensive livestock farming. Modulating the microbial community to sustain or further increase productivity while decreasing methane emissions has indeed become a major goal for microbial ecologists studying the rumen microbiome and its interactions with the host animal. In this article, we discuss the driving forces that affect the establishment and composition of the rumen microbiome and its plasticity, and potential avenues for harnessing these forces for a more sustainable production of animal products.
Individuality of the rumen ecosystem across hosts
Although the composition of the rumen microbiome is quite similar across animals as they mature, particularly within the same species, individual ruminants exhibit differences in their microbiome composition. These differences have been strongly implicated in phenotypic outcomes of the host animal, which enables to predict performance based on the composition and function of their rumen microbiome (Nemergut et al, 2013; Fukami, 2015; Shaani et al, 2018; Furman et al, 2020). These studies identified inter‐animal variations in both taxonomic composition and prevalence of specific functions, which may be key to the differences in performance between animals.
This raises several intriguing questions. Where and when does this inter‐individual variability originate? Why do such wide differences exist between different host animals, even in experimental settings with homogeneous management and environmental conditions? Does the initial microbial community influence the later stable state in mature animals? Identifying the sources of these inter‐individual variations and potentially identifying the cause of different phenotypes such as higher methane emission or increased production efficiency requires delving deep into the ecological forces that establish and maintain the microbial community during the animal's life.
As the differences in community composition likely influence metabolic features of the rumen ecosystem and physiological aspects of the host, research, such as the RuminOmics project (http://www.ruminomics.eu/), focuses on the colonization process starting at birth and, more recently, the genetic background of the host animal to find order in the seemingly random variations between animal hosts and their rumen microbiome. The eventual goal is to fine‐tune microbial phenotypes either through external intervention or selective breeding programs. An increased understanding of the ecology of the rumen microbiome could thereby lead to novel approaches to improve its efficiency, resulting in higher yield and lower impact on the environment.
An increased understanding of the ecology of the rumen microbiome could thereby lead to novel approaches to improve its efficiency, resulting in higher yield and lower impact on the environment.
Deeper understanding of the forces driving the assembly and composition of any ecosystem is a key requirement for any attempts to modulate the system. Such forces can be generally divided into two categories: deterministic and stochastic ones. Deterministic forces, such as host genetics, developmental stage, diet, and biotic interactions among the microorganisms, are often tightly linked to microbial composition and can therefore be used to predict and modulate it.
In contrast, stochastic forces are decoupled from environmental conditions and curb our ability to predict microbiome composition. These are unpredictable events, such as the dispersal of specific microorganisms and their invasion into the ecosystem, or random drift in the abundance of specific microbial species (Nemergut et al, 2013). Both deterministic and stochastic forces are not mutually exclusive but entangled in space and time and act in concert to shape the rumen microbiome. Thus, understanding and quantifying their individual contribution to how the microbial community assembles would likely increase our ability to interfere with and modulate the rumen microbiome. The combination of these forces is best exemplified when examining the post‐birth colonization and assembly process.
Stochasticity of the rumen microbiome
During initial colonization, the rumen microbial communities of individual calves exhibit large differences in terms of composition, which is partly due to random invasion of microbial species. These differences are much larger than those observed between adult cows, even with the same diet and other environmental conditions. This observation suggests that initial acquisition of microbes involves a large random component that may define the eventual trajectory of microbiome assembly and its final composition (Furman et al, 2020). In this process, the activity of an early colonizer either preempts colonization of competing organisms (niche preemption) or modifies the local environment so as to create the conditions for subsequent invasion or increased dominance by other organisms (niche modification) (Fukami, 2015). Hence, biotic microbial interactions determine so‐called priority effects which can be seen throughout the life of the animal, both on long‐term and short‐term scales (Shaani et al, 2018).
Priority effects can, for instance, explain functional convergence in the rumen microbial composition across the developmental stages of the host animal. The first steps of colonization are characterized by a rapid invasion of aerobic bacteria followed by the increasing dominance of anaerobic taxa within days. This rapid turnover suggests that niche modification by aerobic bacteria, combined with environmental filtering, invariably leads to the same broad functional niche provided for anaerobic bacteria (Friedman et al, 2017; Furman et al, 2020). Priority effects as a result of stochastic invasion of the initial species pool can also induce different initial environments and consequently set the rumen microbiome on distinct developmental trajectories (Fig 1. Events I, II and IV).
Figure 1. Schematic illustration of the potential compounded effect of stochastic invasion combined with host genetics on the trajectory and mature state of the rumen microbial community.
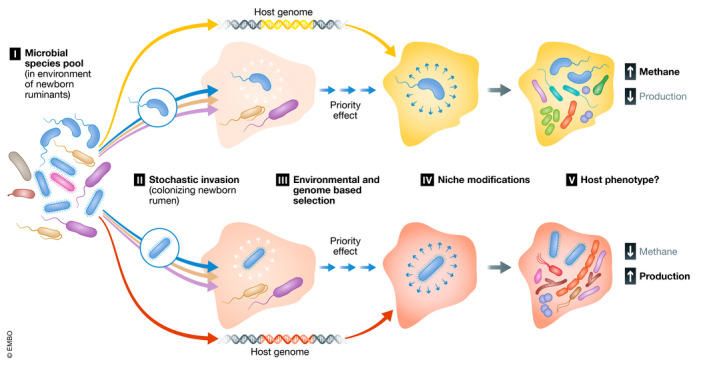
(I) Some of the microorganisms from the common species pool in the environment are able to colonize the newborn's rumen. The shape of the bacteria represents different species, while the color represents their metabolic requirement. In the illustration, the different species of bacteria, depicted in green, can colonize the early‐life rumen along with two other species from the species pool. (II) Stochastic invasion of species in newborn hosts. The two green depicted species with high niche overlap randomly colonize different host, both setting different trajectories of colonization and exclude each other from colonizing their respective hosts. (III) Differential establishment of the microbe community based on the genetic background of the cow. The different shades of the rumen represent the different rumen conditions induced by the host. (IV) Niche modification induced by the different initial invasion. The invading and selected microbes modify the rumen environment, facilitating the arrival of late colonizers. The different rumen shades signify the differences in environmental conditions induced by the niche modification process. (V) Differential phenotypic outcome as a result of the stochastic early colonization combined with the genetic background and management history.
Priority effects […] can also induce different initial environments and consequently set the rumen microbiome on distinct developmental trajectories.
To what extend then does stochasticity in early life affect long‐term composition of the microbiome, despite the observed convergence in taxonomic identity and function? This question was largely examined by interfering with the initial species pool and studying the long‐term effect on the assembly process of the rumen microbiome and its final composition (Furman et al, 2020). Calves born via natural birth—where initial colonization starts in the birth canal—and calves born via C‐section exhibit observable differences throughout the assembly process and the mature stable state of their microbiomes (Furman et al, 2020). These differences can be attributed to the mode of delivery and the initial species pool, as all calves were raised under highly similar conditions and shared a high degree of genetic similarity.
The outcome of the delivery mode could be observed throughout the lifespan of the animal and was mainly defined by different dynamics of colonization along with different patterns of appearance of the colonizing taxa. Thus, early colonizers can steer the rumen assembly process on alternative trajectories, potentially through biotic interactions. Consequently, the history of the individual animal plays a crucial role in the eventual composition of the microbiome. An additional neutral force that could greatly contribute to microbiome assembly and variation is random drift in species representation. Less abundant species are more susceptible to random drift, and changes in their proportion in the community could have a large effect on the whole ecosystem, regardless of their relative low representation.
Rumen microbiome determinants
Nonetheless, stochastic events and the resulting consequences remain framed within the overall constraint of deterministic forces that impose a strong selection based on environmental filtering and that lead to a higher degree of similarity among adult animals. Throughout the animal's life, diet is such a highly deterministic force that has profound effects on the rumen microbiome composition. During the early‐life assembly process and even after the community has reached a stable composition, diet represents the most controllable parameter at our disposal. Indeed, changes in diet across the lifetime of the host are characterized by predictable changes in composition and rumen microbiome function.
During the early‐life assembly process and even after the community has reached a stable composition, diet represents the most controllable parameter at our disposal.
Other deterministic forces can also have a large effect on microbiome composition and may, in some cases, overpower the effect of diet. The developmental stage of an animal is a strong deterministic factor, as it shapes the microbiome composition regardless of other factors, such as diet and the initial species pool. This makes the animal itself—in particular its genetics—a major factor for selection and microbiome composition. Different ruminant species house similar microbiomes, owing to the specific conditions of their habitat that selects for similar groups of microbes out of the available species pool to perform the task of degrading and fermenting plant material. However, microbial communities of different ruminant species—for instance sheep and cows—remain distinct from one another owing to differences in physiology, rumen size, feeding behavior, or pH. Even different breeds within a ruminant species—Norwegian Red and Holstein‐Friesian cows—can exhibit broad physiological differences that affect the composition of the microbial community, even under the same conditions (Roehe et al, 2016; Li et al, 2019). Thus, identifying a connection between host genetics and the microbiome requires a high degree of genetic similarity—and thus physiology—among animals.
Efforts to link microbial composition to host genetics have led to several large‐scale genome‐wide association studies (GWAS) within ruminant breeds. Recent studies (Roehe et al, 2016; Sasson et al, 2017; Li et al, 2019), including the RuminOmics project (Wallace et al, 2019), have shown that a subset of microbes exhibit heritability, that is, correlation between their numerical prevalence in the rumen and the host genome. These heritable microbes were shown to be part of the core community, which is defined as the recurrent microbial species across the majority of cows, some of which were found in a hundred percent of the cows surveyed. This core community was further shown to exhibit a strong consistency in terms of abundance structure: Their ranking in relation to abundance is surprisingly stable across cows, regardless of confounding factors, such as geographical location or management. These heritable core microbes may thus form the functional backbone of the rumen ecosystem (Wallace et al, 2019).
This core community and its heritable fraction is rather small in terms of species number, but it encompasses a large proportion of the community relative abundance, and therefore may have a strong effect on the overall structure of the rest of the microbial community. Furthermore, when looking at the interaction network based on correlations between rumen species, core species that exhibit high heritability are also significantly more connected to other microbes and likely represent keystone species with critical importance and effect on the entire ecosystem. Among these are Ruminococcus flavefaciens and Fibrobacter succinogenes that degrade crystalline cellulose into soluble sugars (Wallace et al, 2019). They are situated at the top of the food chain as two of the three main microbes that degrade this recalcitrant polymer which provides most of the biomass from plant material. These microbes therefore perform a key process that supplies soluble sugar and energy for a great deal of the rumen microbiome and the host animal.
… core species that exhibit high heritability are also significantly more connected to other microbes and likely represent keystone species with critical importance and effect on the entire ecosystem.
Additional evidence about the importance of keystone species can be seen in the strong association between several core heritable microbes with rumen metabolic traits and host physiological traits, including methane production (Sasson et al, 2017; Wallace et al, 2019). It may be therefore possible to modulate their abundance via breeding programs, which could have a tremendous impact on microbiome composition and host phenotype (Fig 1. Events III). Nevertheless, more large‐scale studies and refined experiments are required to adequately address this topic and uncover potential mechanisms by which microbes are selected by the host genotype. Lastly, the importance of these heritable microbes during the assembly process and their effect on microbiome development of the rumen microbiome have yet to be examined.
Can we modulate the rumen microbiome by harnessing ecology?
While the goal of directing the rumen microbial community toward a desired host phenotype remains elusive, recent studies on the topic demonstrate the possibility of disentangling the individual ecological forces and their effect on rumen microbiome trajectory and eventual stable state. The same studies also hint at the possibility of ecology‐guided microbiome modulation by early intervention during the process of colonization. Limiting the degree of stochasticity in the system through post‐birth intervention by various means could increase the predictability of the mature stable state.
Limiting the degree of stochasticity in the system through post‐birth intervention by various means could increase the predictability of the mature stable state.
The recent discoveries point toward the feasibility of those approaches, as events at the beginning of life have a noticeable effect on the dynamics of colonization, and importantly, the eventual stable state of the microbiome. Contingent on further studies establishing how strongly those events can affect host phenotypes, this will provide a framework toward early‐life management, such as optimal dietary strategies, or targeted inoculation of founder communities to prime the rumen for a predictable mature microbiome state. This will also allow a more didactic assessment of the effect of such interventions on the host phenotype, an aspect that has not been elucidated yet.
In this communication, we describe how microbial ecology can assist in resolving some of the biggest challenges mankind is facing today. With an ever‐growing human population and demand for food, and the increasing awareness of our impact on the environment, the recent discoveries about the rumen microbiome may provide novel translational approaches for increased sustainability in agriculture along with insights on the ecology of anaerobic microbial communities in various environments beyond the boundaries of the rumen.
Acknowledgements
The authors want to thank Edward Bayer, Sarah Morais and Tanita Wein for critical reading of this communication and helpful comments. This work was partially funded by the Israeli Science Foundation (grant agreement for Dr. Elie Jami 603/20), European Research Council under the European Union's Horizon 2020 research and innovation program (grant no. 640384) for Itzhak Mizrahi and the Israel Science Foundation (grant no. 1947/19) for Itzhak Mizrahi. We also want to acknowledge the collaborative nature of the work on the rumen microbiome by thanking our community of scientists around the world involved in elucidating the mysteries of the rumen microbiome such as those involved in the RuminOmics project.
EMBO reports (2021) 22: e52269.
References
- Diamond JM (1997) Guns, germs, and steel: the fates of human societies. New York, NY: WW Norton & Company: [Google Scholar]
- Friedman N, Jami E, Mizrahi I (2017) Compositional and functional dynamics of the bovine rumen methanogenic community across different developmental stages. Environ Microbiol 19: 3365–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami T (2015) Historical contingency in community assembly: integrating niches, species pools, and priority effects. Annu Rev Ecol Evol Syst 46: 1–23 [Google Scholar]
- Furman O, Shenhav L, Sasson G, Kokou F, Honig H, Jacoby S, Hertz T, Cordero OX, Halperin E, Mizrahi I (2020) Stochasticity constrained by deterministic effects of diet and age drive rumen microbiome assembly dynamics. Nat Commun 11: 1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Li C, Chen Y, Liu J, Zhang C, Irving B, Fitzsimmons C, Plastow G, Guan LL (2019) Host genetics influence the rumen microbiota and heritable rumen microbial features associated with feed efficiency in cattle. Microbiome 7: 92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut DR, Schmidt SK, Fukami T, O’Neill SP, Bilinski TM, Stanish LF, Knelman JE, Darcy JL, Lynch RC, Wickey P et al (2013) Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev 77: 342–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehe R, Dewhurst RJ, Duthie C‐A, Rooke JA, McKain N, Ross DW, Hyslop JJ, Waterhouse A, Freeman TC, Watson M et al (2016) Bovine host genetic variation influences rumen microbial methane production with best selection criterion for low methane emitting and efficiently feed converting hosts based on metagenomic gene abundance. PLoS Genet 12: e1005846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasson G, Kruger B‐SS, Seroussi E, Doron‐Faigenboim A, Shterzer N, Yaacoby S, Berg MME, White BA, Halperin E, Mizrahi I (2017) Heritable bovine rumen bacteria are phylogenetically related and correlated with the cow’s capacity to harvest energy from its feed. MBio 8: e00703–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaani Y, Zehavi T, Eyal S, Miron J, Mizrahi I (2018) Microbiome niche modification drives diurnal rumen community assembly, overpowering individual variability and diet effects. ISME J 12: 2446–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RJ, Sasson G, Garnsworthy PC, Tapio I, Gregson E, Bani P, Huhtanen P, Bayat AR, Strozzi F, Biscarini F et al (2019) A heritable subset of the core rumen microbiome dictates dairy cow productivity and emissions. Sci Adv 5: eaav8391 [DOI] [PMC free article] [PubMed] [Google Scholar]


