A single-nucleotide change underlies the transition from environmentally induced to genetically encoded matricidal hatching.
Abstract
Genetic assimilation—the evolutionary process by which an environmentally induced phenotype is made constitutive—represents a fundamental concept in evolutionary biology. Thought to reflect adaptive phenotypic plasticity, matricidal hatching in nematodes is triggered by maternal nutrient deprivation to allow for protection or resource provisioning of offspring. Here, we report natural Caenorhabditis elegans populations harboring genetic variants expressing a derived state of near-constitutive matricidal hatching. These variants exhibit a single amino acid change (V530L) in KCNL-1, a small-conductance calcium-activated potassium channel subunit. This gain-of-function mutation causes matricidal hatching by strongly reducing the sensitivity to environmental stimuli triggering egg-laying. We show that reestablishing the canonical KCNL-1 protein in matricidal isolates is sufficient to restore canonical egg-laying. While highly deleterious in constant food environments, KCNL-1 V530L is maintained under fluctuating resource availability. A single point mutation can therefore underlie the genetic assimilation—by either genetic drift or selection—of an ancestrally plastic trait.
INTRODUCTION
A long-standing debate in biology surrounds the question of how an ancestrally plastic, environmentally induced phenotype can evolve into an environmentally insensitive, invariant, and genetically encoded phenotype (1–5). This phenomenon is commonly referred to as genetic assimilation (6–13), a term initially coined by C. H. Waddington (4). While Waddington’s notion of genetic assimilation was tightly linked to the concept of canalization (4, 14–18), this term is much more broadly used today, referring to any scenario in which an environmentally induced trait evolves into a genetically encoded trait, i.e., without necessarily specifying the involved process. Much of the debate surrounding the evolutionary significance of genetic assimilation therefore centers on the nature of the underlying genetic architecture (10, 15, 18–25). Under Waddington’s view, genetic assimilation is the result of selection on “cryptic” genetic variation, genetic variation that is not expressed in the ancestral environment—implicitly a form of nonadditive or epistatic gene interactions (16, 26). Alternatively, fixation of an ancestrally plastic trait may occur without invoking epistasis through fixation of de novo (additive) mutations that mimic the environmentally induced trait.
Here, we identified the molecular basis underlying the evolutionary transition from an ancestral environmentally induced to a derived, genetically encoded trait focusing on matricidal hatching in natural populations of the hermaphroditic nematode Caenorhabditis elegans. Egg-laying behavior of C. elegans is plastic and sensitive to diverse environmental factors (27, 28). Self- and cross-fertilization in C. elegans is internal, with embryos initially developing for 2 to 3 hours in the hermaphrodite uterus, and completing embryogenesis 10 to 12 hours later, under benign environmental conditions (29). Upon starvation, however, hermaphrodites will retain eggs in the uterus where they will hatch into larvae that will continue to grow (“bag of worms” or “facultative viviparity”) and eventually lead to the rapid and premature death of their mother (30, 31). In nutrient-scarce environments, facultative matricidal hatching may be an adaptive strategy because mothers might provide nutritional resources for offspring survival by allowing them, for example, to reach the alternative diapausing dauer larval stage (31, 32). The dauer stage is resistant to nutrient deprivation and will disperse to colonize novel habitats (33). In addition, when facing starvation, early larvae can diapause (34), and so matricidal hatching might also be adaptive because it protects the developing, non-diapausing offspring, which will head-start population growth when conditions become favorable. Hence, matricidal hatching is thought to be an extreme form of offspring provisioning or protection that might be favored by natural selection, even at the cost of maternal reproduction and survival, when populations face fluctuating environments across generations (35, 36).
RESULTS
Natural variation in environmental induction of C. elegans matricidal hatching
Induction of C. elegans matricidal hatching in response to starvation is conserved across genetically divergent wild isolates (Fig. 1A) (32, 37), with the notable exception of the isolate JU751, displaying near-constitutive matricidal hatching irrespective of the environment, in which it was assayed (Fig. 1, A and B). At the phenotypic level, environmentally insensitive matricidal hatching of JU751 thus appears to be derived from an ancestrally plastic state. Although JU751 hermaphrodites laid eggs during early adulthood, they exhibited internally hatched L1 larvae later in life (Fig. 1B and fig. S1, A to F). Comparing JU751 to the wild isolate JU1200 with ancestral C. elegans egg-laying behavior in an ad libitum food environment, we found JU751 to have significantly increased egg retention and correspondingly high matricidal hatching (Fig. 1, C to F, and fig. S1, F to I). Internal hatching led to early maternal death (Fig. 1G and fig. S2, A and B) because larvae moved vigorously inside the mother, causing extensive injury to somatic and gonadal tissues (fig. S1, A to E). Internal larval hatching of JU751 individuals in benign conditions thus seems to carry a considerable fitness cost because maternal death occurred before the end of the reproductive span, prior to exhaustion of hermaphrodite self-sperm (fig. S2, C and D).
Fig. 1. Natural variation in environmental induction of C. elegans matricidal hatching.
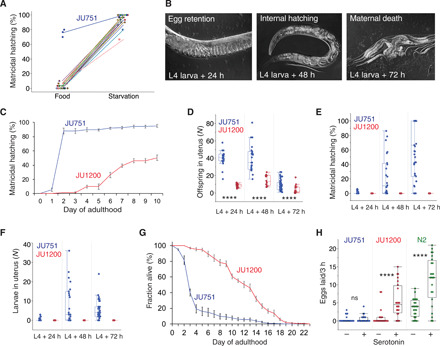
(A) Natural variation in the plasticity of matricidal hatching across genetically divergent C. elegans wild isolates (n = 28) quantified in food (solid) and starvation (liquid) culture. Values of matricidal hatching reflect the percentage of mothers containing one or more internally hatched larvae (n = 30 individuals per strain per environment; except for JU751: n = 3 × 30 individuals). (B) Temporal progression of matricidal hatching of JU751 hermaphrodites in ad libitum food conditions (Nomarski micrographs). (C) Temporal dynamics of matricidal hatching in JU751 versus JU1200: Cumulative percentage of individuals containing one or more internally hatched larvae (n = 5 replicates per strain, n = 29 to 32 individuals per replicate). For further details, see fig. S1 (G to I). (D to F) Egg retention and internal hatching in JU751 and JU1200 in ad libitum food conditions at 20°C (n = 14 to 47 individuals per strain per time point) (see also fig. S1F). (D) Number of total offspring retained in utero (eggs and larvae) during the reproductive span [analysis of variance (ANOVA) performed separately for each time point: ****P < 0.0001]. (E) Percentage of larvae/total offspring retained in utero. (F) Number of internally developing larvae. (G) Survival curves of JU751 and JU1200 hermaphrodites in ad libitum food conditions at 20°C (n = 9 to 15 replicates, n = 9 to 44 individuals per replicate). Matricidal hatching before day 5 of adulthood accounts for >60% of mortality in JU751. For further details, see fig. S2 (A and B). (H) Exogenous application of serotonin [5-hydroxytryptamine (5-HT)] triggered egg-laying in JU1200 and the reference strain N2 but not in JU751 (liquid starvation culture, n = 24 individuals per strain per treatment) (ANOVA performed separately for each strain: ns, not significant; P > 0.05; ****P < 0.0001).
Near-constitutive matricidal hatching in the wild isolate JU751
Multiple observations suggest that the egg-laying apparatus of JU751 is not simply defective. First, JU751 hermaphrodites laid eggs containing advanced embryos during early adulthood (figs. S1F and S2, H to L). Second, egg-laying activity in young adults was the same in JU751 and JU1200, with similar temporal patterns of active and inactive egg-laying phases (fig. S2, H and I), although JU751 laid fewer eggs per active event (fig. S2, J and K). Third, we did not detect any obvious defects in vulval development, feeding (pumping), food sensory behavior (olfaction), or morphology that could be indicative of starvation when fed on the standard Escherichia coli food source. Last, egg retention and matricidal hatching of JU751 were similar and consistently high across different food sources or temperatures (fig. S1, G to I). Therefore, the state of near-constitutive matricidal hatching in JU751 is consistent with a scenario in which the egg-laying circuit exhibits reduced environmental sensitivity. This interpretation is supported by JU751’s lack of response to the neurotransmitter serotonin (Fig. 1H)—known to mimic food stimulation of C. elegans egg-laying (27, 28, 30).
QTL mapping identifies a single large-effect locus explaining variation in matricidal hatching
To characterize the genetic basis of matricidal hatching in JU751, we derived F2 recombinant inbred lines (RILs) from a reciprocal parental cross between isolates JU751 and JU1200 (Fig. 2A and fig. S3A). A total of 144 single-nucleotide polymorphism (SNP)–genotyped RILs were scored for the presence or absence of internally hatched larvae within the first 3 days of adulthood at 15°C (Fig. 2B and fig. S3, B to D). Quantitative trait locus (QTL) mapping uncovered a single significant QTL on chromosome V, spanning approximately 1 Mb (Fig. 2C). The QTL was validated by generating near-isogenic lines (NILs) carrying the QTL region with JU751 genotype backcrossed into the JU1200 background: All NILs with the introgressed QTL region showed strong egg retention and matricidal hatching (fig. S4A). Additional genotyping of the NILs within the QTL region restricted the target interval to 60 kb (fig. S4B) covering three protein-coding genes, which contained a single polymorphism, located in the coding region of the gene kcnl-1 (B0399.1) (Fig. 2D and fig. S4B).
Fig. 2. QTL mapping identifies a single large-effect locus explaining variation in matricidal hatching.
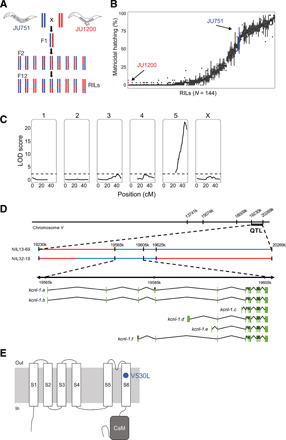
(A) Crossing scheme to generate F2 RILs derived from a parental cross between isolates JU751 and JU1200 (for details on phenotyping and genotyping of RILs, see fig. S3). (B) Phenotypic distribution (cumulative percentage of individuals undergoing matricidal hatching at mid-L4 + 72 hours at 15°C) of all replicates (n = 4 to 5) for each RIL (n = 144). Each boxplot displays all replicate values for a given RIL (x axis). Parental strains are colored in red (JU1200) and blue (JU751). (C) Linkage-mapping analysis of variation in matricidal hatching (proportion at mid-L4 + 72 hours). Each of the 146 SNP markers, separated by chromosome, was examined for correlation with phenotypic variation of the RIL panel (x-axis). The logarithm of the odds (LOD) score is shown on the y-axis. The dashed horizontal line shows the 5% significance threshold calculated with 1000 permutations. Phenotype data from 144 RILs were used for the mapping in R/qtl. (D) From QTL to QTN: Fine-mapping through NILs and additional genotyping of the target region identifies a single polymorphism in the gene kcnl-1, a small-conductance (SK) calcium-activated potassium channel subunit with six predicted isoforms, ranging from 2606 (isoform a) to 1937 (isoform c) amino acids. (E) Predicted structure of the KCNL-1 ion channel subunit (80). The KCNL-1 V530L variant in JU751 affects the predicted S6 transmembrane (TM) domain.
The candidate variant is a single-nucleotide change in the gene kcnl-1, encoding a small-conductance calcium-activated potassium (SK) channel
kcnl-1 encodes a small-conductance calcium-activated potassium (SK) channel subunit with six predicted isoforms (Fig. 2D). KCNL-1 is one of four C. elegans orthologs of the human KCNN SK potassium channel family, characterized by six transmembrane segments and a C-terminal calmodulin (CaM) binding domain (Fig. 2E). The JU751 kcnl-1 variant is a single-nucleotide change (GTG → TTG) causing a valine to leucine substitution (V530L) in the region encoding the S6 transmembrane segment (TM S6) (Fig. 2E). The KCNL-1 protein, including the TM S6 region, shows substantial divergence across distant animal taxa (fig. S4C). The KCNL-1 V530L variant detected in JU751 seems nevertheless exceptional, as the affected TM S6 region is highly conserved not only across all examined species of the genus Caenorhabditis (N > 30) but also across diverse nematode taxa encompassing hundreds of millions of years of species divergence (fig. S4D) (38, 39).
KCNL-1 functions in vulval muscles to regulate egg-laying
To further characterize the physiological function of kcnl-1, we cloned and examined an additional kcnl-1 mutation, previously termed exp-3(n2372) (40). This mutation was initially isolated as a semi-dominant mutation that disrupts the expulsion step of the defecation cycle and increases egg retention (40). The mutation causes a single amino acid change in KCNL-1, A443V, situated in the intracellular loop that connects the S4 and S5 transmembrane segments of KCNL-1 (fig. S5A). Very similar to JU751, the kcnl-1 mutation exp-3(n2372) showed both high egg retention (fig. S5B) and an increased defecation cycle length (fig. S5C). Egg-laying defects of this mutation could be suppressed by RNA interference (RNAi) (fig. S5D), and a suppressor screen uncovered only intragenic loss-of-function mutations. Furthermore, overexpression of mutant kcnl-1(A443V) complementary DNA (cDNA) was sufficient to cause egg retention (fig. S5E), strongly arguing for a gain-of-function effect of the A443V amino acid change. In contrast, and as expected (41), a presumptive loss-of-function (deletion) allele of kcnl-1 (41) was similar to wild type, causing only a slight reduction in egg retention (fig. S5B).
To investigate the kcnl-1 expression pattern, we generated a transcriptional reporter strain using CRISPR-Cas9 gene editing (Fig. 3, A to D). Consistent with the phenotype of kcnl-1 gain-of-function mutants, we observed strong expression in neurons and muscles involved in egg-laying (Fig. 3A) and defecation (Fig. 3B). In addition, kcnl-1 was expressed in body-wall muscles, a few neurons in head and tail ganglia, ventral nerve cord motoneurons, and the PVD mechanosensory neuron (Fig. 3, A to D). To discriminate between a role of KCNL-1 in vulval muscles versus the cholinergic VC neurons that innervate them, and uterine muscles, we restricted expression of the KCNL-1 V530L mutant protein to vulval and uterine muscles by using a kcnl-1 promoter fragment that drives expression in these muscles but not VC neurons. Mosaic analysis showed that animals carrying this transgene in vulval muscles were incapable of laying eggs (n = 497), while only mosaic animals lacking the transgene in vulval muscles could lay eggs normally (n = 34) (fig. S5F). The ability of RNAi, which is inefficient in neurons, to suppress the egg-laying phenotype (fig. S5D) further supported a non-neuronal role for KCNL-1 in C. elegans egg-laying. Matricidal hatching in KCNL-1 gain-of-function variants, V530L (natural variant) and A443V (mutant), thus likely results from a hyperpolarization of vulval muscles (41, 42) that leads to strongly reduced egg-laying activity.
Fig. 3. A single amino acid change in KCNL-1 causes near-constitutive matricidal hatching in JU751.
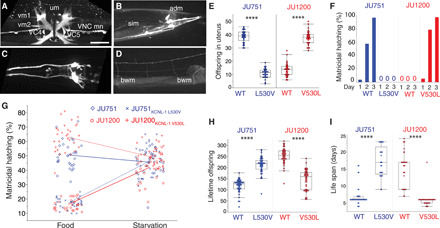
(A to D) Expression analysis of kcnl-1(bln508[kcnl-1::SL2::wrmScarlet]). Scale bar, 20 μm. (A) Strong expression in vulval muscle 1 (vm1) and 2 (vm2), uterine muscle (um), ventral nerve cord motoneurons (VNC mn), and cholinergic neurons VC4 and VC5. (B) Expression in somato-intestinal muscle (sim), anal depressor muscle (adm), and neurons in tail ganglia. (C) Expression in neurons of the head. (D) Weak expression in body-wall muscles (bwm). (E) Offspring number in the uterus (embryos and larvae) of adult hermaphrodites (L4 + 48 hours) in JU751WT, JU751 ARLKCNL-1 L530V and JU1200WT, JU1200 ARLKCNL-1 V530L. n = 30 per strain. (F) Temporal dynamics of matricidal hatching in JU751WT, JU751 ARLKCNL-1 L530V and JU1200WT, JU1200 ARLKCNL-1 V530L across the first 3 days of adulthood (L4 + 24 hours, L4 + 48 hours, L4 + 72 hours). Data show the cumulative percentage of mothers displaying matricidal hatching, i.e., mothers containing one or more internally hatched larva. n = 119 to 140 per strain. (G) Offspring number in uterus (eggs and larvae) of adult hermaphrodites (L4 + 36 hours) in JU751WT, JU751 ARLKCNL-1 L530V and JU1200WT, JU1200 ARLKCNL-1 V530L in food (solid) versus starvation (liquid). n = 27 to 30 per strain (two-way ANOVA, genotype: F3,229 = 176.1, P < 0.0001; environment: F1,229 = 132.4, P < 0.0001; genotype × environment: F3,229 = 124.5, P < 0.0001). (H) Lifetime offspring production of selfing hermaphrodites in JU751WT, JU751 ARLKCNL-1 L530V and JU1200WT, JU1200 ARLKCNL-1 V530L. n = 43 to 47 per strain. (I) Life span of selfing hermaphrodites in JU751WT, JU751 ARLKCNL-1 L530V and JU1200WT, JU1200 ARLKCNL-1 V530L. n = 29 to 45 per strain. (E), (H), and (I): ANOVAs performed separately for each strain background: ****P < 0.0001.
A single amino acid change in KCNL-1 causes matricidal hatching with strongly deleterious fitness effects in constant food environments
We next constructed reciprocal allelic replacement lines (ARLs) in JU751 and JU1200 using CRISPR-Cas9 gene editing by targeting the underlying single-nucleotide variant (GTG → TTG). The resulting amino acid change in the JU1200 ARL (ARLKCNL-1 V530L) led to strong egg retention and highly penetrant matricidal hatching, while the reciprocal nucleotide replacement in JU751 (ARLKCNL-1 L530V) restored fully functional egg-laying and abolished matricidal hatching (Fig. 3, E and F, and fig. S5G). Moreover, restoring the canonical KCNL-1 protein in JU751 also reestablished the ancestral state of strong environment-dependent plasticity of C. elegans matricidal hatching (Fig. 3G). Hence, KCNL-1 V530L represents the central molecular change underlying the evolutionary transition from ancestrally plastic to near-constitutive matricidal hatching in the C. elegans wild isolate JU751. Measuring life history traits in ARLs confirmed the highly deleterious nature of the KCNL-1 V530L variant: Reestablishing the canonical KCNL-1 protein in JU751 nearly doubled its lifetime reproductive output (Fig. 3H) and tripled life span (Fig. 3I).
KCNL-1 V530L has a single evolutionary origin and is maintained in natural populations
The presence of the kcnl-1 gain-of-function allele in JU751 with strongly deleterious effects on reproduction in standard laboratory conditions raised the question of whether this variant is truly of natural origin, or if it reflects a spontaneous mutation acquired after isolation in the laboratory. We therefore characterized independently isolated strains from the JU751 natural habitat, a compost heap in an urban garden close to Paris (France), which was repeatedly sampled during 2004 and 2005 (fig. S6, A and B) (43). Of 36 isolates, 5 additional isolates (isolated on the same day in June of 2005 and sharing the same or similar haplotype as JU751) exhibited the KCNL-1 V530L variant, all of which displayed strong egg retention and matricidal hatching as in JU751 (fig. S6C), consistent with a natural origin of this variant.
Exploring whole-genome sequence variant data of a worldwide collection of C. elegans wild isolates (n ~ 250) (44), we detected kcnl-1 coding sequence variants in additional wild isolates (fig. S7A), including V530L, which was present in two wild isolates collected from garden compost: JU2587 (Haute Loire, France, 2013) and JU2593 (Hauts-de-Seine, France, 2013) (fig. S7, A and B). These two isolates belong to distinct isotypes (genome-wide genotypes) (44) and displayed highly penetrant egg retention and matricidal hatching as observed for JU751 (fig. S7C). Wild isolates with alternative KCNL-1 amino acid variants differed significantly in egg retention; however, none of them displayed high egg retention or matricidal hatching (fig. S7C).
On the basis of whole-genome sequence similarity, JU2587 and JU2593 are closely related to JU751 (fig. S7D) and belong to the same admixture group (EU2) sharing a prominent, globally swept genome-wide haplotype with hundreds of wild isolates (44, 45). Local haplotype analysis showed that the kcnl-1 genomic region is very similar for the three isotypes carrying KCNL-1 V530L, indicative of a likely single evolutionary origin of this variant (fig. S7E). kcnl-1 is located on the right arm of chromosome V, which shows much increased sequence diversity, including a recently identified hyperdivergent region (45); however, this hyperdivergent region does not encompass the kcnl-1 gene (45). On the basis of population genetic analysis using Tajima’s D statistics (46), kcnl-1 and its surrounding genomic region contain an excess of rare alleles (fig. S7F), consistent with a signature of purifying selection, recent select sweeps, or population expansion (45, 46).
Analysis of C. elegans population genomic data thus indicates KCNL-1 V530L to be a rare, derived variant stemming from a single mutational event. The presence of this variant in distinct isolates from different localities, sampled across a span of 8 years, suggests that this variant is being maintained in natural C. elegans populations.
Variable resource availability alleviates negative fitness effects caused by KCNL-1 V530L
How can the KCNL-1 V530L variant be maintained in natural populations given its seemingly strong deleterious effects as assessed in the fairly constant standard laboratory environment with ad libitum food? Given that mutational effects may be strongly dependent on environmental factors, one possibility is that these deleterious effects of KCNL-1 V530L are alleviated in variable and less favorable environments. We mimicked more natural conditions of the C. elegans habitat (33) by exposing JU751, JU1200, and their corresponding kcnl-1 ARLs to starvation (liquid culture) at different maternal ages. Stress exposure experienced at multiple time points during the reproductive span strongly reduced, or eliminated, the negative fitness effects of KCNL-1 V530L (Fig. 4A and fig. S8A). Introducing this variant into JU1200 significantly increased its output of viable offspring when starved as young adults (L4 + 12 hours and L4 + 24 hours) (Fig. 4A). KCNL-1 V530L may thus provide an immediate reproductive advantage depending on the environmental context. In addition, JU1200 ARLKCNL-1 V530L (but not JU751WT) displayed reduced embryonic lethality (Fig. 4B and fig. S8B), indicating that internal hatching may shield embryos from external environmental insults, such as the imposed starvation stress. Environmental heterogeneity can therefore render the evolutionary transition from plastic to near-constitutive matricidal hatching via KCNL-1 V530L potentially beneficial. To confirm that environmental heterogeneity is necessary for the maintenance of KCNL-1 V530L across multiple generations, we competed JU1200WT and JU1200 ARLKCNL-1 V530L against a green fluorescent protein (GFP)–tester strain in a constant food environment. As expected, we found that the V530L variant performed significantly worse under these constant conditions when the variant is introduced at frequencies of either 5% (fig. S8C) or 50% (fig. S8D).
Fig. 4. Variable resource availability alleviates negative fitness effects caused by KCNL-1 V530L.
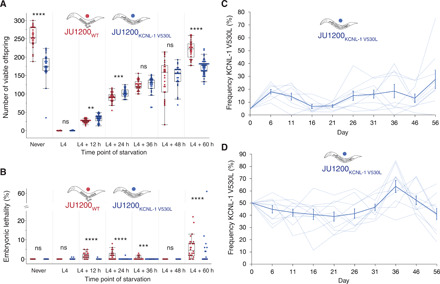
(A) Lifetime production of viable offspring in selfing hermaphrodites in response to starvation encountered at varying maternal age: JU1200WT versus JU1200 ARLKCNL-1 V530L. Age-synchronized populations were transferred from food (solid) to starvation (liquid) environment at different time points of development (n = 24 to 53 per strain per time point). Offspring number reflects combined viable larval offspring produced in nutrient-rich and starvation environments (ANOVA separately performed for each time point: ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (B) Embryonic lethality in response to starvation encountered at varying maternal age: JU1200WT versus JU1200 ARLKCNL-1 V530L [from same experiment as in (A), n = 24 to 53 per strain per time point]. No embryonic lethality was observed in food conditions (ANOVA separately performed for each time point: ns, not significant; P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). (C) Invasive capacity of JU1200 ARLKCNL-1 V530L into a JU1200WT population at a starting frequency of 5% across ~15 generations (n = 9 replicates). Frequencies of the V530L allele are indicated by dashed lines (replicates), and the bold line indicates mean frequencies and SE. (D) Direct competition of JU1200WT versus JU1200 ARLKCNL-1 V530L at an initial 1:1 ratio across ~15 generations (n = 10 replicates). Frequencies of the V530L allele are indicated by dashed lines (replicates), and the bold line indicates mean frequencies and SE.
The KCNL-1 V530L variant is maintained across generations under fluctuating resource availability
Once the derived JU751 variant allele escaped loss by genetic drift after its appearance, it could have been transiently maintained by genetic drift or by balancing selection in fluctuating environment because it seems unconditionally deleterious in constant food environments. We therefore tested whether the V530L allele (JU1200 ARLKCNL-1 V530L) can invade a JU1200WT population when introduced at a starting frequency of 5% [so that it is not immediately lost by genetic drift (47)] in a multiple-generation regime of fluctuating starvation (Fig. 4C). Across ~15 generations, we found that the V530L allele was maintained at a mean frequency of 10 to 20%, and only one of the nine replicate populations went extinct (Fig. 4C); however, the V530L allele was not unconditionally beneficial across the experiment (selection coefficient, s = 0.0139 ± 0.009; χ2 test, P = 0.166). We further quantified the effects of fluctuating starvation by competition of JU1200WT versus JU1200 ARLKCNL-1 V530L at a starting ratio of 1:1 across ~15 generations (Fig. 4D), mimicking a situation where the JU751 allele was already established in the ancestral population. We found that throughout the competition assays, the KCNL-1 V530L allele was maintained at a mean frequency of 40 to 60% (Fig. 4D), again with little evidence that it was unconditionally beneficial across the experiment (s = 0.006 ± 0.004, P = 0.167). These results suggest either that the KCNL-1 V530L allele is neutral or that it is maintained by balancing selection in environments that vary in nutrient availability or other stress factors, similar to the known conditions of the C. elegans natural habitat that exhibit marked boom-and-bust population dynamics under ephemeral food resources (33).
DISCUSSION
The occurrence of near-constitutive C. elegans matricidal hatching due to KCNL-1 V530L in natural populations appears paradoxical given its strong and highly negative effects on fecundity and survival (Fig. 1). Our experimental data and the repeated isolation of KCNL-1 V530L from natural populations suggest that fluctuating environments contribute to the long-term maintenance of such major-effect variants, either by balancing selection or by genetic drift. While KCNL-1 V530L may provide an advantage under a starvation stress (e.g., due to improved protection of embryos that develop internally) (Fig. 4B), this variant could be effectively neutral late in life if reproduction is restricted to early adulthood, a scenario consistent with life history theory on the evolution of aging (48, 49). Our observations suggest that KCNL-1 V530L might indeed be beneficial when the reproductive period is limited to early adulthood due to early starvation onset (Fig. 4A). As in other study systems, major effects of genetic variants uncovered in artificial laboratory conditions may be partly or completely masked in natural populations due to environmental heterogeneity (50).
Physiologically, the KCNL-1 V530L variant does not simply disrupt or abolish egg-laying as hermaphrodites are still capable of laying eggs, albeit at a reduced rate. Practically all individuals will lay eggs early in life, followed by highly penetrant internal hatching late in life. The presence of KCNL-1 V530L therefore does not prevent the functioning of the C. elegans egg-laying system. Rather, this variant strongly dampens environmental sensitivity of the egg-laying circuit: Food signals or neurotransmitters usually triggering egg-laying cannot translate into sufficient or well-coordinated vulval muscle activation, which leads to constitutive egg retention and ultimately to internal hatching and maternal death. Hence, the effects of the KCNL-1 V530L variant are complex, essentially resulting in a combination of reproductive modes, with oviparity early in life and viviparity late in life (Fig. 1F and fig. S1F). Perhaps, it is this novel, mixed reproductive mode, rather than matricidal hatching alone, that could render the KCNL-1 V530L variant beneficial in certain conditions of environmental heterogeneity.
Matricidal hatching in C. elegans caused by KCNL-1 V530L provides a rare example illustrating how a specific single-nucleotide DNA change underlies the evolution of a genetically encoded phenotype, ancestrally only induced in response to the environment. This evolutionary transition can thus be considered a case of genetic assimilation, defined as the evolutionary process whereby an originally environmentally induced phenotype becomes constitutive and environmentally insensitive. As we show, the loss of environmental sensitivity in the case of obligate matricidal hatching in C. elegans reflects the acquisition of a single mutation. This contrasts with Waddington’s scenario of genetic assimilation by means of selection on cryptic genetic variation (5, 14, 15, 51). Our results further indicate that, besides selection, genetic drift can also be responsible for the maintenance of genetic variants underlying assimilated, environmentally insensitive phenotypes. In general, the diverse phenomena attributed to genetic assimilation, including examples of genetic assimilation occurring at macroevolutionary scales (6, 23, 52), likely involve differing genetic bases, which may be shaped by selection and genetic drift. In this regard, recent experimental evidence suggests that, at least in some of Waddington’s Drosophila experiments, stress-induced de novo mutations during laboratory evolution were responsible for genetic assimilation (22). These and other discrepancies surrounding genetic assimilation—and the significance of phenotypic plasticity in the evolutionary process in general—emphasize the need for more work aimed at dissecting the molecular genetic architecture of evolutionary transitions from plastic to invariant phenotype expression.
MATERIALS AND METHODS
Strains and culture conditions
C. elegans strains were maintained on 1.7 or 2.5% agar NGM (nematode growth medium) plates seeded with the E. coli strain OP50 at 15° or 20°C (53, 54). Experiments were performed at 20°C unless noted otherwise. For comparison of the JU751 egg-laying phenotype and QTL mapping, we selected the strain JU1200, a wild isolate showing canonical egg-laying behavior and that is closely related to the C. elegans reference strain N2 (55, 56).The following strains were generated in this study: NIC1627 kcnl-1(cgb1005[L530V]) (ARL in JU751), NIC1642 kcnl-1(cgb1002[V530L]) (ARL in JU1200), JIP1739 kcnl-1(bln488[kcnl-1::d10]), JIP1777 kcnl-1(bln508[kcnl-1::SL2::wrmScarlet]), and JIP1803 blnEx211[pSEM152 Pkcnl-1c::kcnl-1c(V530L)::SL2::GFP]. For a complete list of strains used, including RILs and NILs, see table S1. Strains were thawed to allow for a maximum of four to six generations before phenotypic quantification to avoid accumulation of mutations. Strains were bleached in the second generation after thawing and kept in ad libitum food conditions on NGM plates until phenotypic observations.
Experimental environments
We defined the standard culture condition (NGM agar plate seeded with the E. coli strain OP50) as a benign, favorable environment, in which C. elegans only rarely displays matricidal hatching, and, if so, mostly late in life due to defects in the vulva (57). To assess matricidal hatching under starvation, we used liquid media (M9 or S-Basal) (53) without adding bacterial food, and gentle agitation using a shaker. Starvation was performed in liquid cultures, as C. elegans will leave agar plates without food, thus preventing phenotypic observations. Matricidal hatching is increased in liquid conditions even in the presence of ample food, likely due to increased egg retention caused by changes in oxygen levels and osmolarity, i.e., conditions known to affect egg retention (28, 30, 58). Therefore, the effects of starvation liquid culture on matricidal hatching observed here include other environmental factors in addition to starvation (e.g., mild hypoxia and increased osmolarity). This experimental environment can thus be considered an adverse, unfavorable environment due to multiple stressors. Liquid starvation treatment was imposed using 24-well plates (0.5 ml) or 96-well plates (0.1 ml) or by adding liquid medium directly onto starved NGM agar plates.
Age synchronization and developmental staging
Mixed-age hermaphrodite stock cultures kept at 20°C were hypochlorite-treated to obtain age-synchronized, arrested L1 populations. Hermaphrodites were then picked at the mid-L4 stage based on the morphology of the vulval invagination (59).
Quantification of offspring number in utero and matricidal hatching
Offspring number (eggs and larvae) in utero was measured in randomly picked age-synchronized hermaphrodites, mounted directly on slides, and gently squashed with a coverslip (60). The number of offspring in utero per individual was then counted using a 20× differential interference contrast (DIC) microscope objective. An individual was considered to display matricidal (internal) hatching when one or more L1 larvae were visible in the uterus.
Determination of embryonic stages
Embryonic stages were determined as described earlier (29). The following ranges of embryonic stages were distinguished: early embryo (< 44-cell stage), intermediate embryo [> 44-cell stage < Pretzel (threefold) stage], and late embryo [> Pretzel (threefold) stage] (fig. S1F).
Egg-laying behavioral assays
Egg-laying behavioral assays (fig. S2, H to L) were performed by picking individual animals (L4 + 24 hours) to seeded NGM plates, using a modified, previously published protocol (61). Using a dissecting microscope, plates were scored every 2 min for the presence and number of eggs laid for a total duration of 3 hours. Eggs were immediately removed after egg-laying. Therefore, we only determined time intervals between active and inactive states of egg-laying and not intervals between individual eggs laid within active states, i.e., intracluster intervals (61).
Quantification of hermaphrodite self-sperm number
We quantified the number of self-sperm in synchronized young adult hermaphrodites, i.e., adults containing one to six eggs in utero, as previously described (62). All animals had been isolated at the L4 stage to prevent mating with males. Individuals were collected in S-basal solution [NaCl (5.85 g/liter), K2HPO4 (1 g/liter), KH2PO4 (6 g/liter), cholesterol (5 mg/liter)] and fixed in cold methanol (−20°C) for at least 30 min, washed three times with PBTw (phosphate-buffered saline, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4, containing 0.1% Tween 20), and squashed on a glass slide with Vectashield mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI). Imaging of the anterior spermatheca was performed using an Olympus BX61 microscope with a CoolSNAP HQ2 camera. Images were taken at 60× magnification as Z-sections (1 μm) covering the entire gonad. Sperm number was counted by identifying condensed sperm nuclei on each focal plane using the Fiji plugin Cell Counter. When primary spermatocytes were still present, they were counted as four sperm. We assessed sperm number in a single gonad arm of each individual. Sperm number determined from one gonad arm was multiplied by two to infer the total number of sperm produced.
Quantification of offspring production and embryonic lethality
Lifetime production of viable offspring was analyzed by isolating mid-L4 hermaphrodites on individual NGM plates and transferring them daily to fresh NGM plates until egg-laying ceased. The number of live (viable) larvae and unhatched eggs (embryonic lethality) was counted 24 to 36 hours after each transfer. When mothers died due to matricidal hatching, trapped larvae were allowed to exit the dead mother before being counted. Alternatively, mothers were squashed between slide and coverslip for counting of offspring in the uterus. In liquid culture, offspring production was measured by counting larvae and eggs in wells; eggs that failed to hatch after 24 to 36 hours were considered to be unviable (embryonic lethality). To count offspring in utero, animals in liquid cultures were transferred to a microscope slide using an eyelash. All strains were scored in parallel.
Life-span analysis
Young adult hermaphrodites (L4 + 24 hours) were transferred to NGM plates seeded with fresh E. coli OP50 bacteria. Individuals were transferred daily onto fresh plates until the end of egg-laying and every 3 days after that until the experiment was completed. Animals were scored as dead if they failed to respond to a tip on the head and tail with a platinum wire. For each temperature regime (15° and 20°C), the two strains were scored in parallel.
Egg (embryo) size
Laid eggs of age-synchronized hermaphrodites (at L4 + 24 hours and L4 + 48 hours) were collected using M9 mounted on 4% agar pads on glass slides, and images were obtained with ImageJ software using a 40× DIC microscope objective. The length and width of each egg shell were measured to calculate egg (embryo) volume with the ellipsoid volume function: 4/3 × π × (length/2) × (width/2)2.
Body size
Body size of age-synchronized mid-L4 hermaphrodites was measured using digital images (20× DIC objective). As an estimate of body size, we measured the perimeter of animals by manually tracing the outline of the body in the Fiji software.
Serotonin assays
On the basis of previously described protocols (27, 63, 64), young adults (mid-L4 + 24 hours) were transferred to individual wells of a 96-well microtiter plate containing 100 μl of M9 buffer (without bacterial food) containing serotonin (10 mg/ml; 5-hydroxytryptamine creatinine sulfate complex, Sigma-Aldrich). Egg-laying rates were measured by counting the number of eggs laid after 3 hours in the presence or absence of serotonin at 20°C (Fig. 1H). All strains were scored in parallel.
Defecation assays
Defecation cycle length of singled hermaphrodites was measured at mid-L4 + 24 hours (fig. S5C). Cycle length was determined as the time interval between the first muscular contraction of a defecation event and the first muscular contraction of the next (65). For each individual, five defecation cycles were measured.
Generation of F2 RILs
We generated 144 F2 RILs based on a reciprocal parental cross between wild isolates JU751 (Le Perreux-Sur-Marne, France, isolated from garden compost in 2005) and JU1200 (Scotland, UK, isolated from garden compost in 2007) (fig. S3A) (55). Before crossing of parental isolates, hermaphrodites were depleted of their self-sperm to obtain only cross-progeny in the F1 generation. RILs were derived from 36 randomly picked F2 individuals among F1 cross-progeny obtained per cross direction and parental origin (fig. S3A). A total of 144 F2 individuals were then inbred for 12 generations through selfing by randomly picking a single animal in each generation (fig. S3A).
Genotyping of RILs
DNA from the two parental strains (JU1200 and JU751), the two F1 reciprocals, and 144 RILs was prepared using the Qiagen Blood and Tissue Kit soon after derivation or after thawing from frozen stocks and expansion to at least 103 L1 individuals. Genotyping of 164 biallelic SNPs, uniformly distributed across the six chromosomes according to published genetic distances (66) and parental identities (67), was done with the iPlex Sequenom MALDI-TOF platform (68). For each sample, a multiplex polymerase chain reaction (PCR) targeting between 30 and 35 SNP regions was followed by an SNP allele-specific extension and genotyping by mass spectrometry. A list of the SNPs targeted and the oligonucleotides used for Sequenom analysis can be found in table S2. A total of 164 SNP markers were genotyped, of which 146 SNPs were retained for QTL mapping after quality control. SNP quality control first involved retaining those where either the two parental strains had a different allele or one of the two reciprocal F1s was heterozygous. The recombinant genotypes of RILs are listed in data S1.
Phenotyping of RILs
Across multiple experimental blocks, we scored matricidal (internal) hatching at 15°C. For each RIL (n = 144), 3 to 4 replicates with each 30 mid-L4 were established, and after 24, 48, and 72 hours, the proportion of individuals containing one or more internally hatched larva was scored (Fig. 2B and fig. S3, B to D). Parental isolates were scored in parallel in each block. For QTL mapping, we used the cumulative proportion of animals displaying matricidal hatching on day 3 (mid-L4 + 72 hours).
QTL analysis
QTL mapping was performed using the rQTL package in R (69). The cross data were converted into RILs using the convert2riself function. After visual inspection of the genetic map and recombination fractions as well considering the values from the top.errorlod and droponemarker functions, we removed four markers due to them creating substantial expansions of the genetic map and having anomalous recombination patterns (chromosome 3: III_8052645[C/A], chromosome 5: V_7531256[A/G], V_1297644[C/G], V_3936556[A/G]). The 5% significance threshold for the QTL mapping was calculated using 1000 permutations. The confidence interval for the QTL on chromosome 5 was calculated using the bayesint function in rQTL using a probability of 0.95.
Construction of NILs and fine-mapping
To validate the effect of the uncovered QTL in C. elegans matricidal hatching, we derived NILs from four RILs (NIC613, NIC632, NIC670, and NIC707) displaying constitutive matricidal hatching and different break points within the QTL region (fig. S4B). Corresponding NILs (NIC1566, NIC1570, NIC1573, and NIC1577) were established by eight rounds of backcrossing to the parental JU1200 isolate selecting for individuals with matricidal hatching/markers. NILs were then phenotyped (fig. S4A) and genotyped (fig. S4B).
Molecular biology
pSEM148 was built by assembling the following DNA fragments using Gibson assembly: (i) LHR (left homology arm), (ii) SL2::wrmScarlet cassette, and (iii) RHR (right homology arm). pSEM152 was built similarly by joining (i) the 2.8-kb sequence upstream of the kcnl-1c start codon, (ii) the kcnl-1c locus including the V530L mutation, and (iii) SL2::GFP cassette. pCFJ55 plasmids were generated by three-fragment Gateway assembly from (i) [4-1] Pkcnl-1 isoform C (2.9 kb), (ii) [1-2] kcnl-1 isoform C [A = wt, B = A443V], and (iii) [2-3] GFP unc-54 untranslated region (UTR). See table S2 for detailed reagents.
C. elegans CRISPR-Cas9 gene editing and transgenesis
Gene editing was performed according to previously published protocols (see table S2 for detailed reagents) (70, 71). In brief, bln488 was built by inserting the d10 protospacer sequence at the C terminus of the wild-type kcnl-1 locus using the CRISPR (cr) RNA kcnl-1_Cter and the single-strand oligonucleotide repair template (ssODN-RT) oSEM474. bln508 was generated by using the bln488 d10 locus as a starting point and engineering the SL2::wrmScarlet cassette using the pSEM148 repair template plasmid. The blnEx211([Pkcnl-1c::kcnl-1c(V530L)::SL2::GFP]) transgene was generated by injecting the pSEM152 plasmid and the pCFJ90 (Pmyo-2::mCherry) coinjection marker into the wild-type N2 strain.
The KCNL-1 L530 residue found in the wild isolate JU751 was mutated to V530 (i.e., cg1005) using the crRNA kcnl-1_cg1005 and ssODN-RT kcnl-1_L530, resulting in NIC1627. In brief, JU751 young adult worms were injected with Cas9 protein, kcnl-1 and dpy-10 crRNA, and ssODN-RT. This injection mix was prepared in the following way: (i) combine 3.9 μl of Cas9 protein (Dharmacon; 20 μM), 1.25 μl of trans-activating CRISPR (tracr) RNA (Dharmacon; 0.175 mM), 0.2 μl of dpy-10 crRNA (0.3 mM), and 0.25 μl of kcnl-1 crRNA (kcnl-1_cg1005; 0.6 mM); (ii) incubate for 15 min at 37°C; and (iii) add 0.2 μl of dpy-10 ssODN-RT (AF-ZF-827mod, 0.011 mM) and 0.2 μl of kcnl-1 ssODN-RT (kcnl-1_L530). F1 worms were isolated from P0 plates containing rollers and allowed to produce offspring for 2 days for subsequent use (F2 progeny). The original F1 worms were then individually lysed and screened by PCR using three oligonucleotides: kcnl-1_FWD, 5′ of the SNP; kcnl-1_REV, 3′ of the SNP; and kcnl-1_SNP that carries the modified SNP.
The KCNL-1 V530 residue found in the wild isolate JU1200 was mutated to L530 (i.e., cg1002) using the crRNA kcnl-1_cg1002 and ssODN-RT kcnl-1_V530, resulting in NIC1642. cg1002 animals were detected in the F1 generation based on their egg retention phenotype and verified in the F2 by Sanger sequencing.
Mosaic analysis
We performed kcnl-1 mosaic analysis by correlating the presence/absence of the blnEx211 transgene in vulval and uterine muscles with the egg-laying phenotype (wild-type or egg-laying defective) of individual worms raised at 25°C, over the course of 48 hours after the final larval molt. To discriminate between a role of KCNL-1 in vulval muscles versus the cholinergic VC neurons that innervate them, we restricted expression of the KCNL-1 V530L mutant protein to vulval and uterine muscles by using a kcnl-1 promoter fragment that drives expression in vulval muscles but not VC neurons.
RNA interference
We used an RNAi clone targeting the kcnl-1 gene (clone B0399.1) from the Orfeome RNAi library (72) to test whether RNAi by feeding (73) could suppress the egg-laying defect of the JT6428 [exp-3(n2372)] strain. As a negative control, we cloned a 519–base pair (bp) fragment of enhanced GFP (eGFP) into the pDEST-pL4440 RNAi vector that contains two concordant T7 promoters. We transferred five young adult hermaphrodites to plates with RNAi bacteria and removed all adults after 3 hours. Three days later, we counted the number of adult F1 progeny on the plate. Experiments were performed at room temperature in duplicate on three to five plates.
Overexpression of mutant KCNL-1 protein
To test the causal role of the exp-3(n2372) (A443V) mutation identified in the kcnl-1 gene, we generated wild-type or mutant GFP-tagged cDNAs of the C isoform of kcnl-1. We generated expression constructs using a 2876-bp promoter for the C isoform of kcnl-1 and a 3′UTR from unc-54 by three-fragment Gateway cloning (Thermo Fisher Scientific, USA). See table S2 for details. We generated extra-chromosomal array lines by injection into lin-15(765ts) animals using the Plin-15EK rescue plasmid (74) at 150 ng/μl and the kcnl-1 expression constructs (wild type or mutant) at 10 ng/μl. We tested egg-laying in four independent lines (EG4318, EG4319, EG4320, and EG4321) carrying wild-type or mutant kcnl-1 by allowing five young adult transgenic animals to lay eggs for 3 hours in triplicate and counting the number of adult F1 progeny 3 days later.
Population genetic analyses
Tajima’s D statistical analyses were carried out using the VCF-kit command-line–based collection for performing analysis on Variant Calling Format using the latest hard-filtered SNV VCF release on CeNDR (20200815). Tajima’s D statistics were calculated using a nonoverlapping sliding window of 100 Mb with a step size of 500 bp. Plots were generated using the ggplot2 (version 3.3.2) R package (75). Neighbor-joining trees were established on the basis of the VCF file dataset of CeNDR release 20180527 including 330 isotypes and using a subset of 39 representative isotypes to ease visualization. The subset was extracted using the vcfkeepsamples command-line from the vcflib command-line tools (76) to establish a neighbor-joining network of a region extending 1 Mb on either side of kcnl-1 (fig. S7E). The distance matrix and unrooted neighbor-joining trees were generated using the vk phylo tree nj command-line from the VCF-kit tools (77). Trees were visualized using the software iTOL (v5.6.3) (78).
Reproduction in response to starvation encountered at varying maternal age
Age-synchronized mid-L4 hermaphrodites from either JU1200WT, JU1200 ARLKCNL-1 V530L JU751WT, or JU751 ARLKCNL-1 L530V strain were isolated and cultured on NGM plates seeded with E. coli OP50. Except for the “never” treatment group, which was not exposed to starvation, individuals were transferred to M9 buffer in 12-well plates with gentle shaking at 20°C (1 animal in 1-ml M9 per well, N ~ 30 individuals per treatment group) for all other treatment groups (starvation at 0, 12, 24, 36, 48, and 60 hours). The liquid starvation treatment was applied for 48 hours, during which virtually all animals, except the ones from treatment group “0 hours,” had died within the first 24 hours. For the treatment group 0 hours, individuals were directly transferred to individual wells using an eyelash. For the treatment groups “12 hours” and “24 hours,” individuals were transferred to individual wells 12 or 24 hours after isolation, and the NGM plates of origin were conserved for subsequent counting of the offspring laid by each individual before its transfer to liquid. For the treatment groups “36 hours,” “48 hours,” and “60 hours,” individuals were passaged each day to fresh NGM plates before transfer to the starvation treatment. Plates were conserved for subsequent counting of the offspring before starvation. The treatment group never was maintained on NGM plates until the cessation of reproduction, i.e., L4 + 96 hours), and all individuals were daily transferred to fresh NGM plates. Viable larval offspring and unhatched (dead) embryos produced during the 48-hour starvation treatment were counted using a dissecting microscope. Offspring trapped in dead adult bodies were transferred to slides using an eyelash for microscopy observation using a 20× DIC objective. Larval offspring and unhatched (dead) embryos produced on NGM plates before starvation were counted 24 to 36 hours after transfer of individuals. Total numbers of viable offspring shown in Fig. 4A and fig. S8A include the lifetime production of viable larval offspring produced on NGM plates and liquid starvation. Embryonic lethality in liquid starvation (Fig. 4B and fig. S8B) was inferred from the number of unhatched eggs present in wells after 48 hours of the treatment. No embryonic lethality was observed for any strain on NGM plates.
Competition and invasion experiments
Competition with GFP-tester strain in ad libitum food conditions (fig. S8, C and D)
To evaluate long-term fitness consequences of KCNL-1 V530L, we first performed competition experiments in a favorable, constant ad libitum food environment across multiple generations. We ran competition experiments for both JU1200WT and JU1200 ARLKCNL-1 V530L against a GFP-tester strain (myo-2::gfp; strain PD4790, expressing GFP in the pharynx) with a starting frequency of either 5% (invasion; fig. S8C) or 50% (competition; fig. S8D). To establish experimental populations, age-synchronized arrested L1 larvae of different strains were mixed to obtain a total number of 1000 larvae per NGM plate. For the invasion experiments with a starting ratio of 5:95, we established (i) mixed populations containing 50 L1 larvae of JU1200WT and 950 L1 larvae of myo-2::gfp tester strain (n = 10 replicates) and (ii) mixed populations containing 50 L1 larvae of JU1200 ARLKCNL-1 V530L and 950 L1 larvae of myo-2::gfp tester strain (n = 10 replicates) (fig. S8C). For the competition experiments with a starting ratio of 1:1, we established (i) mixed populations containing 500 L1 larvae of JU1200WT and 500 L1 larvae of myo-2::gfp tester strain (n = 10 replicates) and (ii) mixed populations containing 500 L1 larvae of JU1200 ARLKCNL-1 V530L and 500 L1 larvae of myo-2::gfp tester strain (n = 10 replicates) (fig. S8D). Populations were maintained for 20 days by daily chunking of agar carrying nematodes (approximately one-quarter of the agar) to fresh NGM plates. Every 4 days, genotype frequencies were inferred by quantifying the fraction of GFP-positive individuals among a subpopulation of ~200 to 300 individuals per replicate, using a fluorescence dissecting microscope.
Direct competition in fluctuating starvation environments (Fig. 4, C and D)
We performed direct competition experiments of JU1200 ARLKCNL-1 V530L versus JU1200WT, thus assessing the effects of the single amino acid change (KCNL-1 V530L) in an identical genetic background (JU1200). After initial expansion of the populations, the culturing regime alternated between 72 hours on NGM plates (with food) and 48 hours in liquid (without food). To establish invasion experiments at a 5:95 ratio (Fig. 4C), we mixed 3 mid-L4 JU1200 ARLKCNL-1 V530L and 57 mid-L4 JU1200WT individuals per standard NGM agar plate seeded with E. coli OP50 (n = 9 replicates). We established competition experiments at a 1:1 ratio (Fig. 4D) by mixing 10 mid-L4 JU1200 ARLKCNL-1 V530L and 10 mid-L4 JU1200WT individuals per standard NGM agar plate seeded with E. coli OP50 (n = 10 replicates). After 24 hours, for both invasion and competition experiments, all replicate populations were transferred to the starvation treatment (5-ml M9 liquid culture without bacterial food source in 15-ml Falcon tubes, with gentle shaking) for 48 hours. At this time, we added 0.5 ml of the starved liquid culture to fresh NGM plates seeded with E. coli OP50. Populations were then allowed to grow for 72 hours. The food starvation alternation was then continued for a total of 10 cycles across 56 days. Allele frequencies were determined by phenotyping a random subset (n = 100) of hermaphrodites (at L4 + 48 hours) of each replicate population (days 16, 21, 26, 31, 36, 46, and 56). As validated by PCR genotyping, individuals with >25 eggs in utero could be reliably scored as JU1200 ARLKCNL-1 V530L, whereas JU1200WT individuals contained <20 eggs in utero. Exceptional cases (n = 4) where individuals had 20 - 25 eggs in utero were excluded from analysis. In addition, a random subset of individuals (n = 100) from each replicate population was genotyped at the kcnl-1 locus using PCR primers discriminating between V530 and L530 on days 6 and 46 of the experiment.
Estimation of allele frequency dynamics
kcnl-1 allele frequency dynamics were analyzed using a generalized linear mixed model in R (75), using the lme4 package (79) to estimate the relative fitness of JU751 allele (47). The two competition experiments were analyzed separately as they were done at different starting allele frequencies. kcnl-1 allele counts were modeled as a function of fixed replicate intercept and fixed day of competition while taking the slope of replicate as a random binomial variable. We ignored allele frequencies at setup because they there is a positive correlation between replicate intercept and slope. To test for significance of day of competition—whose estimate is the relative fitness of the JU751 allele—a second model without day as a fixed factor was compared with the first model using a chi-square test.
Statistical analyses
Statistical tests were performed using R and JMP 14.0 software. Data for parametric tests were transformed where necessary to meet the assumptions of analysis of variance (ANOVA) procedures (homogeneity of variances and normal distributions of residuals). For post hoc comparisons, Tukey’s honestly significant difference procedure was used. For data, where ANOVA assumptions could not be met, we used nonparametric tests (e.g., Wilcoxon test).
Box-plots: The median is shown by the horizontal line in the middle of the box, which indicates the 25th to 75th quantiles of the data. The 1.5 interquartile range is indicated by the vertical line.
Acknowledgments
For helpful discussions and pointing out the curious phenotype of JU751, we would like to thank M.-A. Félix. For help with experimental work and data analysis, we thank B. Billard, S. Fausett, N. Poullet, G. Bouzouida, N. Schwartz-Tamoglia, N. Callemeyn-Torre, J.-A. Lepesant, R. Salle, J. Costa, and F. Mallard. For advice and helpful comments on previous versions of the manuscript, we thank E. Abouheif, L. Noble, and three anonymous reviewers. Strains and materials were provided by M.-A. Félix, the C. elegans Natural Diversity Resource (CeNDR), and the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Funding: C.B., C.F., C.G., and P.V. acknowledge financial support by the Centre National de la Recherche Scientifique (CNRS), the Institut national de la santé et de la recherche médicale (Inserm), and the Université Côte d’Azur (UCA). C.B. and H.T. were supported by the Agence Nationale de la Recherche (ANR-17-CE02-0017). H.T. and A.P.-Q. were supported by the European Research Council (FP7/2007-2013/243285) and the Agence Nationale de la Recherche (ANR-14-ACHN-0032-01). J.H. was supported by the Agence Nationale de la Recherche through the “Investments for the Future” LABEX SIGNALIFE (ANR-11-LABX-0028-01). T.B. and S.E.M. were supported by the European Research Council (337702-Kelegans). C.F.-J. was supported by KAUST intramural funding. Author contributions: C.B. conceived and supervised the study and wrote the manuscript. P.V., C.G., and C.F. performed most of the experiments, including construction of RILs, phenotyping, molecular biology, and competition experiments. P.V. and C.G. edited the manuscript. A.V. contributed to construction of RILs and RIL genotyping. A.P.-Q. and H.T. designed and conducted RIL genotyping. H.T. contributed to experimental design, data analysis, and writing of the manuscript. J.H. analyzed RIL genotype and phenotype data and performed the QTL analysis. T.B. and S.E.M. performed kcnl-1 expression and mosaic analysis and edited the manuscript. L.M. performed population genetic analyses. C.F.-J. cloned exp-3, performed RNAi experiments, and edited the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All raw data are provided in data S1. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. All unique biological materials, such as transgenic worm strains, are available from the corresponding author upon request.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/6/eabd9941/DC1
REFERENCES AND NOTES
- 1.Baldwin J. M., A new factor in evolution. Am. Nat. 30, 536–553 (1896). [Google Scholar]
- 2.Simpson G. G., The Baldwin effect. Evolution 7, 110–117 (1953). [Google Scholar]
- 3.Morgan L. C., On modification and variation. Science 4, 733–740 (1896). [DOI] [PubMed] [Google Scholar]
- 4.Waddington C. H., Genetic assimilation of an acquired character. Evolution 7, 118–126 (1953). [Google Scholar]
- 5.Waddington C. H., Canalization of development and the inheritance of acquired characters. Nature 150, 563–565 (1942). [Google Scholar]
- 6.M. J. West-Eberhard, Developmental Plasticity and Evolution (Oxford Univ. Press, 2003). [Google Scholar]
- 7.Price T. D., Qvarnström A., Irwin D. E., The role of phenotypic plasticity in driving genetic evolution. Proc. Biol. Sci. 270, 1433–1440 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pigliucci M., Murren C. J., Schlichting C. D., Phenotypic plasticity and evolution by genetic assimilation. J. Exp. Biol. 209, 2362–2367 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Ghalambor C. K., McKay J. K., Carroll S. P., Reznick D. N., Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Funct. Ecol. 21, 394–407 (2007). [Google Scholar]
- 10.Crispo E., The Baldwin effect and genetic assimilation: Revisiting two mechanisms of evolutionary change mediated by phenotypic plasticity. Evolution 61, 2469–2479 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Ehrenreich I. M., Pfennig D. W., Genetic assimilation: A review of its potential proximate causes and evolutionary consequences. Ann. Bot. 117, 769–779 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheiner S. M., Barfield M., Holt R. D., The genetics of phenotypic plasticity. XV. Genetic assimilation, the Baldwin effect, and evolutionary rescue. Ecol. Evol. 7, 8788–8803 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sommer R. J., Phenotypic plasticity: From theory and genetics to current and future challenges. Genetics 215, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waddington C. H., Canalization of development and genetic assimilation of acquired characters. Nature 183, 1654–1655 (1959). [DOI] [PubMed] [Google Scholar]
- 15.Loison L., Canalization and genetic assimilation: Reassessing the radicality of the Waddingtonian concept of inheritance of acquired characters. Semin. Cell Dev. Biol. 88, 4–13 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Gibson G., Dworkin I., Uncovering cryptic genetic variation. Nat. Rev. Genet. 5, 681–690 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Bateman K. G., The genetic assimilation of four venation phenocopies. J. Genet. 56, 443–474 (1959). [DOI] [PubMed] [Google Scholar]
- 18.Gibson G., Hogness D. S., Effect of polymorphism in the Drosophila regulatory gene Ultrabithorax on homeotic stability. Science 271, 200–203 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Dworkin I., Palsson A., Gibson G., Replication of an Egfr-wing shape association in a wild-caught cohort of Drosophila melanogaster. Genetics 169, 2115–2125 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki Y., Nijhout H. F., Evolution of a polyphenism by genetic accommodation. Science 311, 650–652 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Specchia V., Piacentini L., Tritto P., Fanti L., D’Alessandro R., Palumbo G., Pimpinelli S., Bozzetti M. P., Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature 463, 662–665 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Fanti L., Piacentini L., Cappucci U., Casale A. M., Pimpinelli S., Canalization by selection of de novo induced mutations. Genetics 206, 1995–2006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider R. F., Meyer A., How plasticity, genetic assimilation and cryptic genetic variation may contribute to adaptive radiations. Mol. Ecol. 26, 330–350 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Masel J., Genetic assimilation can occur in the absence of selection for the assimilating phenotype, suggesting a role for the canalization heuristic. J. Evol. Biol. 17, 1106–1110 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Rutherford S. L., Lindquist S., Hsp90 as a capacitor for morphological evolution. Nature 396, 336–342 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Paaby A. B., Rockman M. V., Cryptic genetic variation:evolution’s hidden substrate. Nat. Rev. Genet. 15, 247–258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horvitz H. R., Chalfie M., Trent C., Sulston J. E., Evans P. D., Serotonin and octopamine in the nematode Caenorhabditis elegans. Science 216, 1012–1014 (1982). [DOI] [PubMed] [Google Scholar]
- 28.Schafer W. F., Genetics of egg-laying in worms. Annu. Rev. Genet. 40, 487–509 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Sulston J. E., Schierenberg E., White J. G., Thomson J. N., The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64–119 (1983). [DOI] [PubMed] [Google Scholar]
- 30.C. Trent, “Genetic and behavioral studies of the egg-laying system in Caenorhabditis elegans,” thesis, Massachusetts Institute of Technology (1982); http://dspace.mit.edu/handle/1721.1/15665.
- 31.Chen J., Caswell-Chen E. P., Why Caenorhabditis elegans adults sacrifice their bodies to progeny. Nematology 5, 641–645 (2003). [Google Scholar]
- 32.Chen J., Caswell-Chen E. P., Facultative vivipary is a life-history trait in Caenorhabditis elegans. J. Nematol. 36, 107–113 (2004). [PMC free article] [PubMed] [Google Scholar]
- 33.Frézal L., Félix M.-A., C. elegans outside the Petri dish. eLife 4, e05849 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baugh L. R., To grow or not to grow: Nutritional control of development during Caenorhabditis elegans L1 Arrest. Genetics 194, 539–555 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dey S., Proulx S. R., Teotónio H., Adaptation to temporally fluctuating environments by the evolution of maternal effects. PLOS Biol. 14, e1002388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Proulx S. R., Teotónio H., What kind of maternal effects can be selected for in fluctuating environments? Am. Nat. 189, E118–E137 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Stastna J. J., Snoek L. B., Kammenga J. E., Harvey S. C., Genotype-dependent lifespan effects in peptone deprived Caenorhabditis elegans. Sci. Rep. 5, 16259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiontke K., Fitch D. H. A., The phylogenetic relationships of Caenorhabditis and other rhabditids. WormBook, 1–11 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blaxter M. L., De Ley P., Garey J. R., Llu L. X., Scheldeman P., Vierstraete A., Vanfleteren J. R., Mackey L. Y., Dorrls M., Frisse L. M., Vida J. T., Thomas W. K., A molecular evolutionary framework for the phylum Nematoda. Nature 392, 71–75 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Reiner D. J., Weinshenker D., Thomas J. H., Analysis of dominant mutations affecting muscle excitation in Caenorhabditis elegans. Genetics 141, 961–976 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salkoff L., Wei A. D., Baban B., Butler A., Fawcett G., Ferreira G., Santi C. M., Potassium channels in C. elegans. WormBook 2005, 1–15 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yap F. C., Weber D. S., Taylor M. S., Townsley M. I., Comer B. S., Maylie J., Adelman J. P., Lin M. T., Endothelial SK3 channel-associated Ca2+ microdomains modulate blood pressure. Am. J. Physiol. Hear. Circ. Physiol. 310, H1151–H1163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrière A., Félix M. A., Temporal dynamics and linkage disequilibrium in natural Caenorhabditis elegans populations. Genetics 176, 999–1011 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cook D. E., Zdraljevic S., Roberts J. P., Andersen E. C., CeNDR, the Caenorhabditis elegans natural diversity resource. Nucleic Acids Res. 45, D650–D657 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D. Lee, S. Zdraljevic, L. Stevens, Y. Wang, R. E. Tanny, T. A. Crombie, D. E. Cook, A. K. Webster, R. Chirakar, L. R. Baugh, M. G. Sterken, C. Braendle, M.-A. Félix, M. V. Rockman, E. C. Andersen, Balancing selection maintains ancient genetic diversity in C. elegans bioRxiv 2020.07.23.218420 [Preprint]. 24 July 2020. 10.1101/2020.07.23.218420. [DOI]
- 46.Biswas S., Akey J. M., Genomic insights into positive selection. Trends Genet. 22, 437–446 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Chelo I. M., Nédli J., Gordo I., Teotónio H., An experimental test on the probability of extinction of new genetic variants. Nat. Commun. 4, 2417 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams G. C., Pleiotropy, natural selection, and the evolution of senescence. Evolution 11, 398–411 (1957). [Google Scholar]
- 49.P. B. Medawar, An Unsolved Problem of Biology (H.K. Lewis, 1952). [Google Scholar]
- 50.Taylor M. A., Wilczek A. M., Roe J. L., Welch S. M., Runcie D. E., Cooper M. D., Schmitt J., Large-effect flowering time mutations reveal conditionally adaptive paths through fitness landscapes in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 116, 17890–17899 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braendle C., Flatt T., A role for genetic accommodation in evolution? Bioessays 28, 868–873 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Pfennig D. W., Wund M. A., Snell-Rood E. C., Cruickshank T., Schlichting C. D., Moczek A. P., Phenotypic plasticity’s impacts on diversification and speciation. Trends Ecol. Evol. 25, 459–467 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Stiernagle T., Maintenance of C. elegans. WormBook 2006, 1–11 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brenner S., The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Billard B., Vigne P., Braendle C., A natural mutational event uncovers a life history trade-off via hormonal pleiotropy. Curr. Biol. 30, 4142–4154.e9 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Cook D. E., Zdraljevic S., Tanny R. E., Seo B., Riccardi D. D., Noble L. M., Rockman M. V., Alkema M. J., Braendle C., Kammenga J. E., Wang J., Kruglyak L., Félix M. A., Lee J., Andersen E. C., The genetic basis of natural variation in Caenorhabditis elegans telomere length. Genetics 204, 371–383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pickett C. L., Kornfeld K., Age-related degeneration of the egg-laying system promotes matricidal hatching in Caenorhabditis elegans. Aging Cell 12, 544–553 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang M., Chung S. H., Fang-Yen C., Craig C., Kerr R. A., Suzuki H., Samuel A. D. T., Mazur E., Schafer W. R., A self-regulating feed-forward circuit controlling C. elegans egg-laying behavior. Curr. Biol. 18, 1445–1455 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mok D. Z. L., Sternberg P. W., Inoue T., Morphologically defined sub-stages of C. elegans vulval development in the fourth larval stage. BMC Dev. Biol. 15, 26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poullet N., Vielle A., Gimond C., Carvalho S., Teotónio H., Braendle C., Complex heterochrony underlies the evolution of Caenorhabditis elegans hermaphrodite sex allocation. Evolution 70, 2357–2369 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Waggoner L. E., Zhou G. T., Schafer R. W., Schafer W. R., Control of alternative behavioral states by serotonin in Caenorhabditis elegans. Neuron 21, 203–214 (1998). [DOI] [PubMed] [Google Scholar]
- 62.Gimond C., Vielle A., Silva-Soares N., Zdraljevic S., Mcgrath P. T., Andersen E. C., Braendle C., Natural variation and genetic determinants of Caenorhabditis elegans sperm size. Genetics 213, 615–632 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trent C., Tsuing N., Horvitz H. R., Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics 104, 619–647 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waggoner L. E., Hardaker L. A., Golik S., Schafer W. R., Effect of a neuropeptide gene on behavioral states in Caenorhabditis elegans egg-laying. Genetics 154, 1181–1192 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong A., Boutis P., Hekimi S., Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics 139, 1247–1259 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rockman M. V., Kruglyak L., Recombinational landscape and population genomics of caenorhabditis elegans. PLOS Genet. 5, e1000419 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Andersen E. C., Gerke J. P., Shapiro J. A., Crissman J. R., Ghosh R., Bloom J. S., Félix M. A., Kruglyak L., Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat. Genet. 44, 285–290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bradić M., Costa J., Chelo I. M., Genotyping with Sequenom. Methods Mol. Biol. 772, 193–210 (2012). [DOI] [PubMed] [Google Scholar]
- 69.Broman K. W., Wu H., Sen Ś., Churchill G. A., R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890 (2003). [DOI] [PubMed] [Google Scholar]
- 70.El Mouridi S., Lecroisey C., Tardy P., Mercier M., Leclercq-Blondel A., Zariohi N., Boulin T., Reliable CRISPR/Cas9 genome engineering in Caenorhabditis elegans using a single efficient sgRNA and an easily recognizable phenotype. G3 7, 1429–1437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dickinson D. J., Goldstein B., CRISPR-based methods for Caenorhabditis elegans genome engineering. Genetics 202, 885–901 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rual J. F., Ceron J., Koreth J., Hao T., Nicot A. S., Hirozane-Kishikawa T., Vandenhaute J., Orkin S. H., Hill D. E., van den Heuvel S., Vidal M., Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14, 2162–2168 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Welchman D. P., Zipperien P., Ahringer J., Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 (2003). [DOI] [PubMed] [Google Scholar]
- 74.Clark S. G., Lu X., Horvitz H. R., The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics 137, 987–997 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.R Development Core Team 3.0.1, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2013).
- 76.E. Garrison, Vcflib: A C++ library for parsing and manipulating VCF files (2012).
- 77.Cook D. E., Andersen E. C., VCF-kit: Assorted utilities for the variant call format. Bioinformatics 33, 1581–1582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Letunic I., Bork P., Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bates D., Mächler M., Bolker B. M., Walker S. C., Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 105046 (2015). [Google Scholar]
- 80.Köhler M., Hirschberg B., Bond C. T., Kinzie J. M., Marrion N. V., Maylie J., Adelman J. P., Small-conductance, calcium-activated potassium channels from mammalian brain. Science 273, 1709–1714 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/6/eabd9941/DC1


