All-digital infrared spectroscopic imaging of tumors and their microenvironments predicts colon cancer patient survival.
Abstract
The structure and organization of a tumor and its microenvironment are often associated with cancer outcomes due to spatially varying molecular composition and signaling. A persistent challenge is to use this physical and chemical spatial organization to understand cancer progression. Here, we present a high-definition infrared imaging–based organizational measurement framework (INFORM) that leverages intrinsic chemical contrast of tissue to label unique components of the tumor and its microenvironment. Using objective and automated computational methods, further, we determine organization characteristics important for prediction. We show that the tumor spatial organization assessed with this framework is predictive of overall survival in colon cancer that adds to capability from clinical variables such as stage and grade, approximately doubling the risk of death in high-risk individuals. Our results open an all-digital avenue for measuring and studying the association between tumor spatial organization and disease progression.
INTRODUCTION
A critical gap in predicting tumor behavior lies in determining how the structure and organization of tumor (ranging from well organized to disorganized) affects its ability to interact with the surrounding microenvironment. Access to inflammatory cells that can provide protumorigenic signals (1), access to nutrients (or lack of it, which can make tumors switch their metabolic pathways) (2, 3), changes in pH (4), and interaction of tumor cells with specific types of collagen that can be associated with tumors acquiring a more aggressive mesenchymal phenotype (5) are some of the variables that influence how the tumor progresses. Several of these interactions are dependent on the shape and structure of the tumor (called as tumor and tumor microenvironment spatial organization) that influence the ability of individual tumor cells to interact with the tumor microenvironment. This spatial modulation can affect the availability of signaling factors, nutrients, and oxygen in the tumor microenvironment (6). It can also influence how rapidly prometastatic signals transmit between tumor cells and the tumor microenvironment and subsequently determine how rapidly the tumors progress. The primary limitation in understanding this spatial modulation is the lack of methods to estimate the tumor structure and organization accurately. There is need for high-precision identification of tumor and several of its microenvironment components in tissue images for every pixel to make this estimation feasible.
Pixel-level tissue component identifications by pathologists are labor and time intensive. Such identifications are not feasible for several hundreds of tissue images needed to robustly measure tumor spatial structure and organization. One of the largest annotated image datasets comes from the Cancer Metastases in Lymph Nodes Challenge (CAMELYON), where 400 images were annotated by pathologists identifying only the normal areas and metastatic tumors in lymph node images (7). The tumor microenvironment is much more complex, changing dynamically with tumor development. Estimating tumor structure and organization in relation to this microenvironment would require identification of each unique histological component (HC) in the tissue image (10 or more in colon tissue), as it is difficult to determine a priori which specific tumor microenvironment components play a prognostic role. It is especially important to differentiate between normal stroma, reactive stroma (RS), and immune cell–infiltrated stroma, which have been previously implicated to have prognostic role (8). Hand annotation of these many components on several hundreds of tissue images is infeasible and would also suffer from interobserver variability (9, 10).
Machine learning–based classifiers can greatly aid in accurate pixel-level annotations of tissue images, which can subsequently be used to estimate tumor spatial organization but have not been successfully developed so far. There is a wealth of biochemical signals in tissues, but efforts to segment tissue have been limited to a few components. This limitation is due to the contrast limits of traditionally used methods such as hematoxylin and eosin (H&E), which serve as inputs to the machine learning models. Traditionally used methods for tissue assessment fail to combine detailed biochemical measurements with spatial localization. H&E-stained slide images (11) are poor candidates for machine learning classifiers when more than a few components need to be identified. H&E images provide limited tissue contrast and are affected by experimental and imaging variations that further degrade the performance of machine learning classifiers (12). In addition, such stains may not capture subtle chemical changes that have not yet manifested in gross structural features. Immunostaining can increase contrast in tissue by targeting specific tissue components but is limited to a few markers per slide and impractical due to need for extensive sample processing. Complex workflows also present a barrier to adoption and are an opportunity for error. Mass spectrometric imaging enables multiplexed imaging measurements of the proteome and metabolome and has been shown to be useful in detection and classification of cancers (13–16) and for prognostic measurements (17–19). While mass spectrometric imaging methods provide a detailed overview of the biomolecules in the tissue, they are typically limited in spatial resolution that can be achieved (20), prohibiting their use for studying tumor structure. An ideal assay should instead balance multiplexed content readout with high imaging detail without the need for multiple processing steps. It should also capture the essential features of the tissue structure and organization in a manner simple and robust enough to be routinely adopted.
High-definition Fourier transform infrared (HD-FTIR) spectroscopic imaging is a technique that seeks to measure both the spatial and chemical content of samples without the need for staining (21–23). Specifically, infrared (IR) spectroscopy enables the measurement of abundance classes of molecules, including lipids, proteins, nucleic acids, and carbohydrates. This abundance is read out as the absorbance of IR-absorbing molecular species such as aliphatic groups, amides, phosphates, and ester carbonyl at specific vibrational frequencies dependent on their composition, macromolecule structure, and local environment. The spectrum acts as a bar code of normal or diseased tissues. Previous works have shown that the tissues undergo critical biochemical changes when transforming from normal to cancerous states. For example, changes in lipid composition (24), protein glycosylation (25), and carbohydrate distribution (26) in cancer development have been documented. HD-FTIR combines the optical microscopy capability of providing spatial detail with spectroscopic measurements of the inherent molecular content at every pixel. The molecular content in situ is measured as the increase in absorbance of resonant frequencies corresponding to the characteristic structures of the molecules in the IR range. With recent advancements in HD-FTIR imaging, both the tumor and microenvironment can be measured with subcellular pixel sizes of approximately 1 μm (21), reducing the pixel size by almost sixfold compared with conventional IR imaging instruments (27). This improved definition better localizes spectral signal and provides a higher-quality image, enabling a precise labeling of tumor and microenvironment components on the tissue image and improving the performance of computational algorithms downstream. The spatial-spectral data collected in this manner are highly amenable to machine learning algorithms (27, 28). The unique chemical composition of different histological classes in the tissue gives the contrast necessary for the machine learning–based classification algorithms to perform with high accuracies. High definition of images achieved with this technique further aids in getting accurate estimates of tissue structure and organization.
In this work, we focus on the spatial constraint that is measured by the shape and structure of tumor, which can, in turn, affect how individual tumor cells are able to interact with the tumor microenvironment and specific changes in the tumor stroma (reactive and nonreactive stroma and presence of inflammatory cells). We postulated that with HD-FTIR spectroscopic imaging coupled with machine learning, we can accurately and precisely identify tumor and tumor microenvironment components in tissue images. Building on this pixel-level recognition, we demonstrate that the spatial organization of tumor and its microenvironment is predictive of outcome in patients with cancer (Fig. 1).
Fig. 1. Comparison of artificial intelligence–powered high-definition IR spectroscopic imaging with the conventional pipeline for diagnosis and outcome prediction.
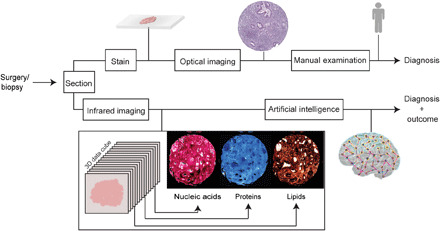
A biopsy or surgical resection is typically (top) stained, imaged using a light microscope and examined by a human for diagnoses. (Bottom) IR spectroscopic imaging does not use dyes or labels but measures the chemical composition of tissue, which can be used by computational algorithms to diagnose disease and its severity.
RESULTS
HD-IR imaging–based segmentation of colon tissue into 10 unique HCs
In this work, we imaged eight colon tissue microarrays (TMAs) with HD-FTIR spectroscopic imaging. This cohort consisted of 604 samples from 320 patients with characteristics described in Table 1. Our first goal was to use the spectral contrast within the tissue to segment the colon tissue into 10 major HCs. These HCs were epithelium (mature), mucin, epithelium (proliferative), necrosis, RS, blood, inflammatory cells, nonreactive stroma, muscle, and loose stroma. To do this 10-class classification (schematically shown in fig. S1A), we acquired ~1 billion spectra with 1506 IR bands per spectrum. One IR spectrum was acquired for every 1 μm × 1 μm pixel from samples approximately 1 mm in diameter. We obtained pathologist annotations on serially acquired H&E images, marking areas that could be identified as an HC with high confidence. These annotations were transferred to HD-IR images to develop ground truth for the classifier. In this manner, we annotated approximately 6900 regions of interest on 419 samples with over 4.3 million spectral pixels.
Table 1. Patient characteristics.
| Number of patients | 320 |
| Median age at surgery (years) | 72.6 (35–94) |
| Median disease-free survival (years) | 4 (0–6.6) |
| Median overall survival (years) | 4.2 (0–6.6) |
| Death | 110 |
| Cancer recurrence | |
| No | 266 |
| Yes | 54 |
| Sex | |
| Male | 162 |
| Female | 158 |
| Site | |
| Colon | 212 |
| Rectum | 108 |
| Stage | |
| 1 | 45 |
| 2 | 133 |
| 3 | 102 |
| 4 | 37 |
| Grade | |
| Well | 8 |
| Moderate | 243 |
| Poor | 69 |
Average spectra from the 10 HCs (Fig. 2A) indicate the chemical constitution differences between these classes. For instance, the most substantial differences were observed in the fingerprint region between 982 and 1480 cm−1, with mucin and mature epithelium showing starkly distinct features in 980 to 1182 cm−1. Mucin is a glycosylated protein that shows a strong absorption at 1038 cm−1 (29). Mature epithelium (goblet cells) containing cytoplasmic mucin also register glycoprotein-associated absorbance but with a distinct protein composition. As seen from the spectra, and the images of adenocarcinomas (Fig. 2B), this functional property of mature epithelium is frequently lost in the malignant or proliferative epithelium. This functional loss results in a difference between the spectra collected from proliferative epithelium and mature, normal-type epithelium. While some of the spectral differences shown in Fig. 2A arise because of differences between histologic components, we also accounted for other sources of variations. Some such sources of variation were experimental variations (array preparation based), patient heterogeneity, and within-patient heterogeneity (fig. S1B). After accounting for these variations, we retained 50 spectral features between 1053 and 1593 cm−1 and between 2939 and 2987 cm−1 that were eventually used for machine learning classifier.
Fig. 2. Pixel-level precise segmentation of tissue components using IR spectroscopic imaging.
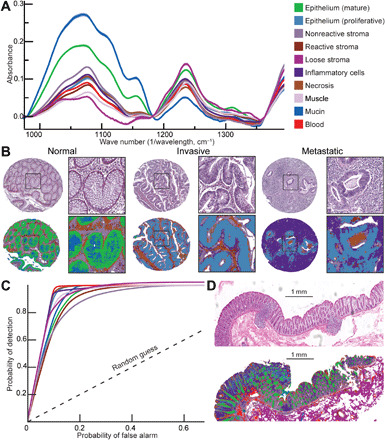
(A) Intrinsic chemical contrast in tissue produces unique IR spectrum for each of the 10 histologic classes. (B) Performance of the artificial intelligence classifier in segmenting normal and invasive tumor containing lymph node tissue cores. (C) ROC curve demonstrating high performance of artificially intelligent classifier in segmenting tissue into 10 unique components as compared with an experienced pathologist. (D) Demonstration of segmentation accuracy on a surgical resection. The color key is common across panels.
Using the spectral signatures isolated from the colon HCs, the HD-IR machine learning classifier demonstrated high classification performance to segment colon tissue. We trained and optimized random forests–based supervised learning algorithm to segment 10 colon HCs from HD-IR spectroscopic imaging data (fig. S1C). This classifier was trained on four arrays (a1 to a4) (fig. S2, A to D) and 126,946 spectra per class, calibrated on two additional arrays (a5 and a6) (fig. S3, A and B), and tested on two independent arrays (a7 and a8) (fig. S4, A and B). For both training and testing, the area under the curve (AUC) of the receiver operating characteristic (ROC) curve was evaluated. High specificity and sensitivity were obtained in both training and independent validation data, yielding an average AUC of 0.94 (fig. S5) and 0.93 (Fig. 2C), respectively.
The cohort used for training in this study comprised paraffin-embedded colon tissue cores, where normal-appearing colorectal mucosa and lymph node metastases were also sampled. Figure 2B compares the H&E images with corresponding HD-IR histologic images for normal colorectal mucosa, invasive tumors, and lymph node metastases. We were also able to identify and localize the presence of malignant cells in samples from lymph node metastases. Prior landmark work (30) had described a deep learning method that was used to perform whole-slide image classification and tumor localization in the lymph node H&E images with AUC of 0.925 and 0.705, respectively. In comparison, we consistently got >0.93 AUC for both whole-slide classification and tumor localization in the lymph node using our classification algorithm, indicating a superior classification performance compared with previously reported H&E benchmarks as measured by the ROC AUC values. We also tested the performance of this classifier on an additional 27 surgical resection samples, which showed good correspondence with the H&E images (Fig. 2D and fig. S6).
Tumor spatial organizational measurements predict survival in the presence of RS
We next focused on characterizing and using the tumor structure and organization for prognostication. To reduce the complexity of the tumor microenvironment to measurable quantities, we focused on three main components of the tumor microenvironment—RS, where the desmoplastic reaction was present; nonreactive stroma, where the desmoplastic response was absent; and lymphocytes. The desmoplastic reaction was identified using previously established criteria, namely, enrichment of “reactive” fibroblasts, the molecular organization of stroma, and presence of other cell types (31, 32). In each case, the pathologist providing the ground truth categorization was blind to all accompanying clinicopathological data, including stage. For this analysis, we selected TMA cores with at least 5% malignant epithelium by area, retaining a total of 245 patients in the study. Of these 245 patients, our model was developed and evaluated on 220 patients, and 25 patients were left out for validation of risk stratification.
We used the annotated images to measure quantitative spatial features that estimate the tumor structure and organization in the context of the microenvironment. For this analysis, we downsampled the image to a third to approximate the pixel size to a single tumor cell and to improve computational time. We defined the tumor structure using the apparent distance between tumor and a nontumor HC (dHC). This distance feature dHC is a measure of the distance between tumor cells and a nontumor HC. Specifically, because we want to understand tumor structure in the context of its microenvironment, dHC varies depending on the presence and proximity of the HC used to calculate dHC. To demonstrate how structure is captured with dHC, we simulated cases where the tumor area was kept constant while varying the tumor shape from cylindrical to spherical (Fig. 3A). On the basis of dHC values, cylindrical tumors have lower dHC as compared with spherical tumors. Although tumor structure is an important component, tumor structure does not capture the abundance of the HC that also influences tumor behavior. Therefore, we defined the microenvironment HC density (NHC) as the density of an HC in a radius R from tumor cells as the center. Therefore, the final spatial organization of tumor and its microenvironment was captured as the interaction product between dHC and NHC (IHC) for each tumor cell (Fig. 3B).
Fig. 3. The tumor spatial organization model and its performance.
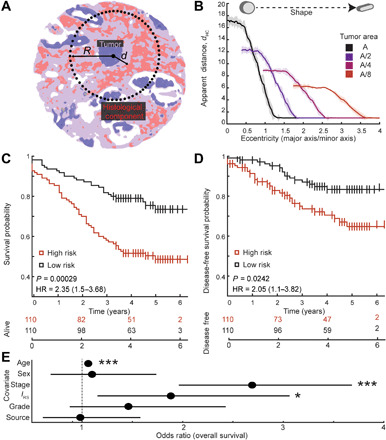
(A) Schematic of tumor spatial organization model. “d” represents the smallest distance between a tumor cell and the RS. R represents the sampling radius to determine the abundance of RS. (B) Relationship between the measured variable “apparent distance” and the tumor shape and structure. As the tumors become rounder, the apparent distance increases, even when area is kept constant. (C) Stratification of overall survival of patients on the basis of dichotomized interaction feature (IRS) with respect to RS as the microenvironment component. (D) Stratification of disease-free survival of patients on the basis of dichotomized interaction feature (IRS) with respect to RS as the microenvironment component. (E) Multivariate Cox regression model shows that interaction feature (IRS) is independently predictive of overall survival, even in presence of other clinical variables such as stage and grade. *P < 0.05, ***P < 0.0005.
To determine whether the quantitative spatial features defined here associated with outcome, we compared the survival of patients against dHC, NHC, and IHC for each of the microenvironment HC, namely, RS, lymphocytes, and normal stroma. We dichotomized each of these candidate features by splitting at the median value, giving us two patient groups per feature per microenvironment component tested. We performed the univariate log-rank test to determine whether there was a statistically significant difference in the overall survival of the two groups (Table 2). From the two tests, only features measured with RS as the microenvironment component showed a statistically significant difference in overall survival. In addition, only the interaction feature (IRS) was significant after correcting for multiple hypothesis test using Bonferroni correction, with P value <0.0005. From this analysis, we prioritized RS as the microenvironment component of interest. In addition, the Kaplan-Meier survival curves for overall survival and disease-free survival showed significantly different outcomes for patients stratified with dichotomized IRS scores (Fig. 3, C and D) with P values of 0.0003 and 0.0274 and at powers 0.93 and 0.40, respectively. Although IRS shows significant patient stratification in determining disease-free survival in univariate analysis, our dataset is not powered for predicting disease-free survival. Given the low power in the current dataset to determine recurrence, it is unclear whether IRS is predictive of disease-free survival. Results of overall survival were validated on an independent set of patients not used in the discovery set, which showed a similar trend in stratifying overall survival (fig. S7).
Table 2. Univariate Cox regression analysis of time to death based on risk features.
NS, normal stroma; L, lymphocytes.
| Microenvironment component | Risk variable | P value (overall survival) |
| RS | dRS | 0.554 |
| NRS | 0.006 | |
| IRS | 0.000 | |
| Normal stroma | dNS | 0.843 |
| NNS | 0.360 | |
| INS | 0.247 | |
| Lymphocytes | dL | 0.580 |
| NL | 0.496 | |
| IL | 0.883 |
Last, to determine whether spatial features add to currently known prognostic markers such as stage and grade, we evaluated multivariate Cox regression model of time to death (Fig. 3E and table S1). We modeled overall survival on the interaction feature IRS, correcting for stage, grade, age, sex, and source of tumor. The interaction feature IRS showed an independent effect on increasing hazard of death for the patients with a P value of 0.011. The P values for three tests testing the fit of the model, the likelihood ratio test, Wald test, and log-rank test were all less than 4 × 10−11, rejecting the null hypothesis that all of the coefficients (β) are equal to 0. In the multivariate Cox regression analysis, the hazard ratio, evaluated as exp.(β), ranged between 1.15 and 3.07, indicating a strong effect size. Other features that showed significant association with risk of death were stage and age, as expected. By contrast, the tumor grade evaluated on whole surgical specimen by pathologists did not show significant association with increased risk when modeled with other covariates.
Aggressive tumors are often detected in later stages, resulting in worse prognosis for patients. We tested whether there was an association between tumor aggression, as measured by the spatial features defined here with stage of the tumor. By performing two-sample t test on continuous interaction feature IRS grouped by stage, we found that low-stage (stage 1 and stage 2) tumors had significantly lower IRS compared with high-stage (stage 3 and stage 4) tumors (Table 3). Two-sample t test on combining these groups had a P value of 0.0012 at 0.90 power. The mean value of IRS was also significantly different between stage 2 and stage 3 at 0.05 significance level (P = 0.00142 and power = 0.9) and between stage 2 and stage 4 (P = 0.00243 and power = 0.87). We did not observe statistically significant difference between stage 1 and stage 4 cases because of the low number of cases in each of these stages. This resulted in low power (0.22) to perform the statistical test. Mann-Whitney test, which does not assume normal distributions and tests whether the patients are sampled from populations with identical distributions, showed similar results.
Table 3. Results of the two-sample t test and Mann-Whitney test to compare values of continuous variable MRS across stages.
| Group | Two-sample t test | Mann-Whitney test | |
| P | Power | P | |
| Stage 1 versus stage 2 | 0.13 | 0.32 | 0.22 |
| Stage 1 versus stage 3 | 0.36 | 0.15 | 0.27 |
| Stage 1 versus stage 4 | 0.23 | 0.22 | 0.21 |
| Stage 2 versus stage 3 | 0.001 | 0.9 | 0.002 |
| Stage 2 versus stage 4 | 0.002 | 0.87 | 0.005 |
| Stage 3 versus stage 4 | 0.51 | 0.1 | 0.61 |
| Low stage (stages 1 and 2) versus high stage (stages 3 and 4) | 0.001 | 0.9 | 9.67 × 10−4 |
Given the predictive power of the interaction feature, we hypothesized that visualizing the continuous risk score on tissue samples can provide a spatial measure of risk that can be easily correlated by the practitioner with conventional stained images. This spatial visualization of risk further protects our method against error introduced by anomalous tissue conditions or isolated errors. When mapping the measured interaction feature score back onto the tissue, we observe a smooth transition of risk in both TMA core samples (Fig. 4A) and large surgical resection (Fig. 4B). While the small TMA punch areas were fairly homogeneous, we observed a gradient of risk score on a much larger surgical resection. It is likely that the risk score is specifically predictive on the invasive fronts of the tumor, since most of the TMA punches were selected from invasive tumor front areas (33). The utility of invasive fronts in cancer study is widely debated. We do not fully understand its role in carcinogenesis and prognosis, although it is clear that the invasive front has some biological relevance. Additional work from surgical resections can shed light on the role of invasive front and possibly develop models that can correct for the invasive front bias in our work.
Fig. 4. Visualization of interaction feature (IRS)–based risk.
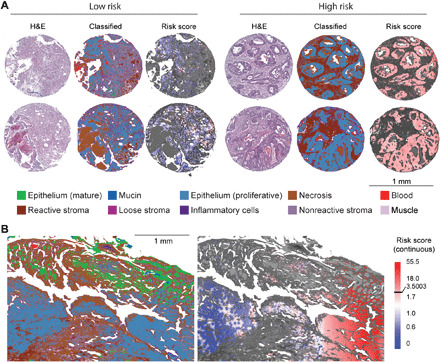
(A) Visualization of interaction feature (IRS)–based risk on TMA cores. Shown are the comparisons with H&E image, HD-IR–classified image, and projection of risk score on tumors. (B) Visualization of interaction feature (IRS)–based risk on large surgical resection shows variation in risk across the tissue. Color keys are common across panels for classification colors and risk scores.
DISCUSSION
The mechanism governing tumor invasion and its interaction with the surrounding stroma and stromal cells is not easy to capture. Understanding this spatial constraint is critical to predict the behavior of tumor. Structural changes in the tumor and its microenvironment at the beginning of metastasis (34) cannot be accounted for in traditional molecular assays. Furthermore, quantitative image analysis is limited by the molecular information of the stain and artifacts introduced at sample preparation, staining, and imaging steps (35). While tissue architecture and morphology can be assessed by pathologists, the determination is largely subjective (36–38). Several outcome-associated objective image assessment criteria, such as nuclear-to-cytoplasmic ratio of molecular markers (39), cellular proportions (40), and measuring immune infiltration (41, 42), have been proposed. These procedures often require additional pathologist and laboratory work and do not measure tumor geometry.
So far, much of the attention has been paid to the molecular aspects of the tumor-tumor microenvironment interaction. Chemical signaling via cytokines and chemokines, presence of cancer-activated fibroblasts, and modifications of other stromal cells have been shown to modulate the behavior of tumors. In addition to chemical signaling, restructuring of the extracellular matrix in tumors is linked to the transformation of tumor to a more aggressive phenotype. Although widely demonstrated, many prognosis-associated features have not found a footing in use due to lack of objective assessment criteria. Unlike some cancers where molecular markers have established a role in determining the outcome and therapy options (43, 44), determining patient outcomes in colon cancers is less precise. For example, characterizing stroma (31) based on the fibrotic stromal response is predictive of 5-year survival rates. Despite the prognostic role of stromal response in colon cancers (45, 46), an objective and reproducible scoring system remains undeveloped, largely due to the concerns arising from lack of concordance (38).
INFORM (INFrared-based ORganizational Measurements) integrates molecular information of the tissue with its spatial context (47), without extensive sample processing or staining. Therefore, INFORM enables us to identify critical biochemical changes in the tissue image. A complex set of factors, at multiple scales, determines a tumor response that can be assessed by INFORM. The association between stage and INFORM-based interaction feature score (IRS) is quite nuanced. In a nutshell, INFORM is able to identify changes in early lesions associated with aggressive behavior that are not currently captured in the clinical approach to staging. Our statistical model that corrected for stage (multivariate Cox regression model of time to death; Fig. 3E) still showed differential survival between patients that have higher INFORM risk score versus patients that do not. This indicates that the INFORM risk score is independent of the stage and potentially adds diagnostic value. However, aggressive tumors that get high INFORM risk score would often be diagnosed at later stages, which we believe may explain an association between stage and INFORM risk score but needs to be tested further. Nevertheless, the novelty of the approach presented here is that a small (<1-mm diameter) biopsy of the tumor contains highly valuable information about the tumor aggression, present as a combination of biochemical and spatial data. Specially in the case of colorectal cancers where diagnosis is typically performed using biopsy taken during colonoscopy and followed up eventually with additional testing for staging, this can prove critical in determining the response urgency, both in terms of further testing and therapy. Notably, from the same biopsy sample, the currently prevalent clinical measure is tumor grade, which is underwhelming in determining prognosis when compared with INFORM risk score, in addition to being subjective and manual. Last, INFORM provides a measure that balances the ease of staging and complexity of the extensive information in the microenvironment.
An easily interpretable, yet automated and objective method, as proposed here, strikes a balance between objective and reproducible scoring and interpretability of results. Here, we were not only able to associate markers of the microenvironment with outcome but also provide guidance to understand how efficacious spatially specific changes in the tumor microenvironment are in predicting patient outcomes. Among the associations between the stromal reaction and patient outcome, the links were much more pronounced in our interaction model. Thus, information from the tumor microenvironment can add new information from an avenue that has never been explored before. This observation holds potential for further discovery and more complex models to characterize the microenvironment. It is important to note that our method does not perturb the tissue in any way and may actually result in reduction of processing steps if technologies such as stainless staining, in which the spectra generate H&E stains computationally (48), are adopted. With the extensive testing on multiple TMAs and on patient surgical resections, the robustness of our approach was established. To our knowledge, this is the most thorough HD-IR imaging study, providing near optical microscopy detail with chemical specificity. Further, we are unaware of any other study that has demonstrated automated, accurate, and precise segmentation of 10 or more HCs in tissues accurate to pixel level in diverse TMAs and also surgical resections. This work presents the largest, most diverse tissue imaging dataset acquired for chemical imaging of any tissue type.
Although INFORM performs very well in stratifying patients, there are many unexplored factors that could potentially stratify patients further. For example, emerging approaches in IR spectroscopic imaging are increasing spatial resolution as well as the level of molecular detail and its fidelity that can be recorded. Finer molecular differences within the RS and cellular heterogeneity in the tumor itself should be probed by combining imaging and molecular assessment modalities. We also anticipate that INFORM can provide key insights into several other cancer types, especially in cases where outcome associated molecular markers are lacking. It is also likely that modern machine learning algorithms can harness the potential of tumor microenvironment much more accurately than the work presented here. Although such methods are powerful, it is difficult to explain where the prediction is coming from. In this work, we have focused on trying to develop easy to interpret measures that can be readily understood, communicated and verified by human inspection if needed.
Last, primary limitations of this study come from the limited sample size and the need to sample the tumor from the invasive fronts. Evidence from this study demonstrates that there is power in the structural determinations of tumor to predict prognosis. However, a rigorous validation in a larger cohort is required. We anticipate that future studies would be able to expand the results from this work further both in terms of sample sizes and the diversity of the tumor microenvironment. In addition, although advanced machine learning models such as deep neural networks have not been applied here because of their limited interpretability and need for large sample sizes, such models can help in advancing this work to clinics. Last, our work demonstrates that combining multiple modalities such as imaging, spectroscopy, and computational modeling can prove beneficial in understanding and predicting tumor behavior. Further integration of modalities that bring complementary information such as genomics and metabolomics can further improve our ability to predict tumor behavior and make it more individualized.
Tumor stage and grade are crucial information for clinical management of cancer. However, these rely only on the characteristics of cancer cells. Prior studies have missed the opportunity to measure the spatial factors that can inhibit or promote tumor progression, controlling the spatial availability of signaling factors and nutrients. We used IR spectroscopic imaging with machine learning algorithms to reliably identify invasive epithelium and RS with pixel-level accuracy in addition to eight other unique histologic types. Next, we measured the tumor structure with respect to specific tumor microenvironment components such as the RS. Last, the interaction feature captured both the structure of tumor and the prevalence of RS. The 6-year probability of survival in “low-risk” patients was 0.73, whereas that in “high-risk” patients was 0.54 at P value of <0.0003. Remarkably, our model was independent of the stage and grade of the tumor in Cox multivariate hazards models. By integrating morphometry and biochemistry to define risk, our work provides a new technique to predict the tumor behavior. We show that while the tumor microenvironment changes are prognostic, an interaction model considering the extent of microenvironment modifications and the tumor morphology is a better predictor of prognosis. This work is especially critical for clinical evaluation of tumors that do not have defined patient outcomes associated with molecular features. Furthermore, our work defines a new dimension to evidence-based pathology and provides interpretable data that can greatly augment clinical practice (49). Recent developments in the field of chemical imaging enable stainless tissue characterization at clinically desirable speeds (50–53). We anticipate routinely applying INFORM in research and clinics, allowing automated diagnosis and outcome predictions that are interpretable and useful.
MATERIALS AND METHODS
Experimental design
The patient cohort used in this study comprised 320 anonymized patients undergoing elective surgery for colorectal carcinoma. Cores of 1-mm diameter were sampled for each patient from representative invasive areas of paraffin-embedded blocks and were used to construct eight TMAs. For some patients, normal colon mucosa samples were also included. This cohort has been used previously (54) and described in detail in the cited reference as cohort II.
Sample preparation
For HD-IR spectroscopic imaging, a 10-μm-thin section of each of the TMAs was obtained on barium fluoride substrate (55). A consecutive section was collected on a glass slide for H&E staining. Because of the IR absorbance of paraffin at 1462 cm−1 (56), before scanning, paraffin was removed by initially dripping the slide with cold hexane followed by complete submersion in continually stirred hexane for 48 hours at 40°C where the solvent was renewed every 3 hours. The disappearance of the representative peak over several locations on the slide confirmed the dissolution of the embedding medium. For supervised classification, data were annotated by labeling histologic classes on H&E images by collaborating pathologists. These annotations were manually copied on the IR spectroscopic images and served as ground truth. For evaluation of surgical resection, paraffin-embedded blocks of colon tissue were obtained from the Carle Foundation Hospital Urbana from anonymized deceased patients who underwent excisional surgery, and processed similarly to the TMA samples.
Imaging
High-magnification images were recorded on an Agilent 870 imaging system in high-magnification mode. This microscope was equipped with 128 × 128 element focal plane array Mercury-Cadmium-Telluride detector. Each pixel of size 1.1 μm was averaged over four scans, and the background spectrum was acquired at 120 scans per pixel on a clean area of the slide. HD-IR spectroscopic images were collected at a spectral resolution of 4 cm−1 and a step size of 2 cm−1 and subsequently truncated to the lower noise spectral range of 900 to 3800 cm−1, providing 1506 data points in the spectrum.
Preprocessing
After stitching the image tiles, noise reduction was performed using minimum noise fraction transform. Savitzky Golay 9 point smoothening, baseline correction, and normalization to amide I peak at 1650 cm−1 were done for each core in the TMA. The spectral data were converted to spectral metrics using methods described previously (56, 57). Briefly, we calculated spectral metrics as ratios of peak heights, peak area and heights, peak areas, and centroid wave number locations of the peaks. In total, we defined 418 spectral metrics.
Feature reduction
In the first stage of feature reduction, we performed analysis of variance (ANOVA) using a nested, random-effects interaction model on three arrays (a1, a3, and a4) using ground truth to label classes, patient, TMA, and patient core numbers. Because specific patients were only found in particular arrays and each patient had multiple cores, the nesting order was array, patient, and patient-specific core. For this analysis, the classes used were malignant epithelium, necrosis, and RS since these classes were most commonly observed in cores. From ANOVA, we determined spectral metrics where interarray, interpatient, and intercore variations were significant at the 0.05 significance level and removed these from the analysis. One hundred sixty-two metrics were retained after this stage. In the second stage, we used minimum redundancy maximum relevance algorithm (mRMR) in R to further remove redundant and irrelevant features. In this model, the feature with maximum mutual information is selected first following the equation (58)
where xi represents ith feature in full dataset X, y represents the output variable (class label), and I represents mutual information given by
The next feature is added to feature vector S by finding the feature with minimum mutual information with respect to all the prior features in S and maximum mutual information with respect to the class label. This is represented as
Applying this using the mRMRe package in R, we found a set of 50 features that were relevant to the classification problem while minimizing the redundancy in the data (table S2).
Supervised classification
In the model learning step, we defined pixel labels for each cell type by duplicating annotations from H&E-stained sections as marked by an expert pathologist onto HD-IR spectroscopic images. Four arrays were used for training (a1 to a4). We used a random forest algorithm for supervised classification in Matlab 2016a. To address fitting issues in the classifier such as high bias or high variance, we used two additional arrays (a5 and a6) to perform optimization of parameters. The parameters that we optimized in the random forest were (i) leaf size, size of the group at which decision tree stops splitting further, and (ii) feature size, number of features sampled by the tree randomly for performing the split. The parameters were optimized by calculating the error in classification for both training and calibration sets for multiple leaf sizes and feature sizes given by
Where e represents error, w represents the weight of the class, n is the number of classes, y is the true class label, ŷ is predicted label, and I is indicator function. Parameters that minimized calibration error and maintained low training error were chosen as optimal. The leaf size and feature size determined for optimal fit were 500 and 7, respectively, with 50 total features and an ensemble of 50 decision trees (fig. S1C). The fully developed supervised model with optimal parameters was validated by two independent arrays (a7 and a8).
Statistical analysis
Classified images of invasive carcinoma cores from all eight TMAs were used for studying tumor-stroma interaction. A total of 245 patients were analyzed using TMA cores containing reactive or nonreactive stroma and at least 5% proliferative epithelium by area. If multiple cores from the same patient were present, then mean over all available cores was calculated. Each risk-associated variable was converted to a dichotomous categorical variable by splitting at the median and evaluated for significance by univariate log-rank test and multivariate Cox regression analysis in R using the package “survival.” Power analysis was performed in R using the package “powerSurvEpi.” Two-sample t test was performed in OriginPro 2017 to test the hypothesis that the means of groups were equal. Mann-Whitney test was performed in OriginPro 2017 as a nonparametric test to test if the distributions were different.
Progression analysis
Three risk-associated features were defined to assess the tumor-stroma interaction. The first risk variable was distance feature (dHC) and measured as the closest encounter distance of the malignant epithelium pixel to the stromal element being probed. The microenvironment HC density (NHC) was measured as the normalized pixel count of the stromal component being probed in a circle of radius R determined experimentally. Empirical work indicated that any radius above ~400 μm captured the spatial characteristics between normal and invasive cores. We set the R for our analysis to be 600 μm. The density factor NHC is the number of HC pixels in area πR2 to accommodate for centers close to the tissue edge. Risk variable interaction feature (IHC) was measured as the interaction of the two features d and N for each pixel. Five hundred random pixels of malignant epithelium were chosen from each of the 30% downsampled classified HD-IR image to calculate the features. Three stromal elements, the RS, lymphocytes, and normal stroma, were evaluated separately for their role in determining outcome.
Acknowledgments
We thank J. Pfister and S. Mittal for help with HD-IR data acquisition. Funding: S.T. was supported by the Beckman Institute Graduate Fellowship and the Nadine Barrie Smith Award. The development of IR imaging technology was funded by NIH grant R01EB009745 to R.B. The funding sources were not involved in study design, analysis and interpretation, writing of the report, and decision to submit the article for publication. Author contributions: S.T. and R.B. conceived the study design and experiments. S.T., A.K.-B., and J.W. performed the experiments and collected the data. S.T., A.K.-B., and R.B. analyzed and interpreted the results. S.T. and R.B. wrote the manuscript. R.B., G.C., S.M.H., and A.K.-B. supervised the work and revised the manuscript. All authors read and approved the final version of the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. HD-IR imaging data, H&E images and annotations, and data analysis are available upon request to the corresponding author.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/6/eabb8292/DC1
REFERENCES AND NOTES
- 1.Grivennikov S. I., Greten F. R., Karin M., Immunity, inflammation, and cancer. Cell 140, 883–899 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boroughs L. K., Deberardinis R. J., Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 17, 351–359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pavlova N. N., Thompson C. B., The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X., Lin Y., Gillies R. J., Tumor pH and its measurement. J. Nucl. Med. 51, 1167–1170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shintani Y., Maeda M., Chaika N., Johnson K. R., Wheelock M. J., Collagen I promotes epithelial-to-mesenchymal transition in lung cancer cells via transforming growth factor-β signaling. Am. J. Respir. Cell Mol. Biol. 38, 95–104 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carmona-Fontaine C., Bucci V., Akkari L., Deforet M., Joyce J. A., Xavier J. B., Emergence of spatial structure in the tumor microenvironment due to the Warburg effect. Proc. Natl. Acad. Sci. U.S.A. 110, 19402–19407 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bándi P., Geessink O., Manson Q., Van Dijk M., Balkenhol M., Hermsen M., Ehteshami Bejnordi B., Lee B., Paeng K., Zhong A., Li Q., Zanjani F. G., Zinger S., Fukuta K., Komura D., Ovtcharov V., Cheng S., Zeng S., Thagaard J., Dahl A. B., Lin H., Chen H., Jacobsson L., Hedlund M., Çetin M., Halici E., Jackson H., Chen R., Both F., Franke J., Kusters-Vandevelde H., Vreuls W., Bult P., Van Ginneken B., Van Der Laak J., Litjens G., From detection of individual metastases to classification of lymph node status at the patient level: The CAMELYON17 challenge. IEEE Trans. Med. Imaging 38, 550–560 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Nissen N. I., Karsdal M., Willumsen N., Collagens and cancer associated fibroblasts in the reactive stroma and its relation to cancer biology. J. Exp. Clin. Cancer Res. 38, 115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Putten P. G., Hol L., Van Dekken H., Han van Krieken J., Van Ballegooijen M., Kuipers E. J., Van Leerdam M. E., Inter-observer variation in the histological diagnosis of polyps in colorectal cancer screening. Histopathology 58, 974–981 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Harris E. I., Lewin D. N., Wang H. L., Lauwers G. Y., Srivastava A., Shyr Y., Shakhtour B., Revetta F., Washington M. K., Lymphovascular invasion in colorectal cancer: An interobserver variability study. Am. J. Surg. Pathol. 32, 1816–1821 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Y. Liu, K. Gadepalli, M. Norouzi, G. E. Dahl, T. Kohlberger, A. Boyko, S. Venugopalan, A. Timofeev, P. Q. Nelson, G. S. Corrado, J. D. Hipp, L. Peng, M. C. Stumpe, Detecting cancer metastases on gigapixel pathology images (2017); http://arxiv.org/abs/1703.02442.
- 12.Otálora S., Atzori M., Andrearczyk V., Khan A., Müller H., Staining invariant features for improving generalization of deep convolutional neural networks in computational pathology. Front. Bioeng. Biotechnol. 7, 198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morse N., Jamaspishvili T., Simon D., Patel P. G., Ren K. Y. M., Wang J., Oleschuk R., Kaufmann M., Gooding R. J., Berman D. M., Reliable identification of prostate cancer using mass spectrometry metabolomic imaging in needle core biopsies. Lab. Invest. 99, 1561–1571 (2019). [DOI] [PubMed] [Google Scholar]
- 14.F. G. Zanjani, A. Panteli, S. Zinger, F. Van Der Sommen, T. Tan, B. Balluff, D. R. N. Vos, S. R. Ellis, R. M. A. Heeren, M. Lucas, H. A. Marquering, I. Jansen, C. D. Savci-Heijink, D. M. de Bruin, P. H. N. de With, in Proceedings - International Symposium on Biomedical Imaging (IEEE Computer Society, 2019), vols. 2019-April, pp. 674–678.
- 15.Cordero Hernandez Y., Boskamp T., Casadonte R., Hauberg-Lotte L., Oetjen J., Lachmund D., Peter A., Trede D., Kriegsmann K., Kriegsmann M., Kriegsmann J., Maass P., Targeted feature extraction in MALDI mass spectrometry imaging to discriminate proteomic profiles of breast and ovarian cancer. Proteomics Clin. Appl. 13, e1700168 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Klein O., Kanter F., Kulbe H., Jank P., Denkert C., Nebrich G., Schmitt W. D., Wu Z., Kunze C. A., Sehouli J., Darb-Esfahani S., Braicu I., Lellmann J., Thiele H., Taube E. T., MALDI-imaging for classification of epithelial ovarian cancer histotypes from a tissue microarray using machine learning methods. Proteomics Clin. Appl. 13, e1700181 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Angel P. M., Bruner E., Bethard J., Clift C. L., Ball L., Drake R. R., Feghali-Bostwick C., Extracellular matrix alterations in low-grade lung adenocarcinoma compared with normal lung tissue by imaging mass spectrometry. J. Mass Spectrom. 55, e4450 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips L., Gill A. J., Baxter R. C., Novel prognostic markers in triple-negative breast cancer discovered by MALDI-mass spectrometry imaging. Front. Oncol. 9, 379 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Randall E. C., Zadra G., Chetta P., Lopez B. G. C., Syamala S., Basu S. S., Agar J. N., Loda M., Tempany C. M., Fennessy F. M., Agar N. Y. R., Molecular characterization of prostate cancer with associated gleason score using mass spectrometry imaging. Mol. Cancer Res. 17, 1155–1165 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchberger A. R., DeLaney K., Johnson J., Li L., Mass spectrometry imaging: A review of emerging advancements and future insights. Anal. Chem. 90, 240–265 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy R. K., Walsh M. J., Schulmerich M. V., Carney P. S., Bhargava R., High-definition infrared spectroscopic imaging. Appl. Spectrosc. 67, 93–105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nallala J., Lloyd G. R., Shepherd N., Stone N., High-resolution FTIR imaging of colon tissues for elucidation of individual cellular and histopathological features. Analyst 141, 630–639 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Nallala J., Lloyd G. R., Hermes M., Shepherd N., Stone N., Enhanced spectral histology in the colon using high-magnification benchtop FTIR imaging. Vib. Spectrosc. 91, 83–91 (2017). [Google Scholar]
- 24.Munir R., Lisec J., Swinnen J. V., Zaidi N., Lipid metabolism in cancer cells under metabolic stress. Br. J. Cancer 120, 1090–1098 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peixoto A., Relvas-Santos M., Azevedo R., Lara Santos L., Ferreira J. A., Protein glycosylation and tumor microenvironment alterations driving cancer hallmarks. Front. Oncol. 9, 380 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J., Liu T., Gao J., Gao L., Zhou L., Cai M., Shi Y., Xiong W., Jiang J., Tong T., Wang H., Variation in carbohydrates between cancer and normal cell membranes revealed by super-resolution fluorescence imaging. Adv. Sci. 3, 1600270 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez D. C., Bhargava R., Hewitt S. M., Levin I. W., Infrared spectroscopic imaging for histopathologic recognition. Nat. Biotechnol. 23, 469–474 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Wald N., Bordry N., Foukas P. G., Speiser D. E., Goormaghtigh E., Identification of melanoma cells and lymphocyte subpopulations in lymph node metastases by FTIR imaging histopathology. Biochim. Biophys. Acta 1862, 202–212 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Patel M. M., Smart J. D., Nevell T. G., Ewen R. J., Eaton P. J., Tsibouklis J., Mucin/poly(acrylic acid) interactions: A spectroscopic investigation of mucoadhesion. Biomacromolecules 4, 1184–1190 (2003). [DOI] [PubMed] [Google Scholar]
- 30.D. Wang, A. Khosla, R. Gargeya, H. Irshad, A. H. Beck, Deep learning for identifying metastatic breast cancer (2016); http://arxiv.org/abs/1606.05718.
- 31.Ueno H., Jones A. M., Wilkinson K. H., Jass J. R., Talbot I. C., Histological categorisation of fibrotic cancer stroma in advanced rectal cancer. Gut 53, 581–586 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ueno H., Jones A., Jass J. R., Talbot I. C., Clinicopathological significance of the ’keloid-like’ collagen and myxoid stroma in advanced rectal cancer. Histopathology 40, 327–334 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Larsson A., Fridberg M., Gaber A., Nodin B., Levéen P., Jönsson G., Uhlén M., Birgisson H., Jirström K., Validation of podocalyxin-like protein as a biomarker of poor prognosis in colorectal cancer. BMC Cancer 12, 282 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thiery J. P., Acloque H., Huang R. Y. J., Nieto M. A., Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Yuan Y., Failmezger H., Rueda O. M., Ali H. R., Gräf S., Chin S.-F., Schwarz R. F., Curtis C., Dunning M. J., Bardwell H., Johnson N., Doyle S., Turashvili G., Provenzano E., Aparicio S., Caldas C., Markowetz F., Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci. Transl. Med. 4, 157ra143 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Santana M. F., Carlos Ferreira L. L., Diagnostic errors in surgical pathology. J. Bras. Patol. Med. Lab. 53, 124–129 (2017). [Google Scholar]
- 37.Gomes D. S., Porto S. S., Balabram D., Gobbi H., Inter-observer variability between general pathologists and a specialist in breast pathology in the diagnosis of lobular neoplasia, columnar cell lesions, atypical ductal hyperplasia and ductal carcinoma in situ of the breast. Diagn. Pathol. 9, 121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okamoto Y., Fujimori T., Ohkura Y., Sugai T., Arai T., Watanabe G., Wada R., Ueno H., Togashi K., Yao T., Kushima R., Mitomi H., Tomita S., Igarashi Y., Ichikawa K., Histological assessment of intra- and inter-institutional reliabilities in detection of desmoplastic reaction in biopsy specimens of early colorectal carcinomas. Pathol. Int. 63, 539–545 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Sung W.-W., Lin Y.-M., Wu P.-R., Yen H.-H., Lai H.-W., Su T.-C., Huang R.-H., Wen C.-K., Chen C.-Y., Chen C.-J., Yeh K.-T., High nuclear/cytoplasmic ratio of Cdk1 expression predicts poor prognosis in colorectal cancer patients. BMC Cancer 14, 951 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West N. P., Dattani M., McShane P., Hutchins G., Grabsch J., Mueller W., Treanor D., Quirke P., Grabsch H., The proportion of tumour cells is an independent predictor for survival in colorectal cancer patients. Br. J. Cancer 102, 1519–1523 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fridman W. H., Pagès F., Sautès-Fridman C., Galon J., The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 12, 298–306 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Galon J., Costes A., Sanchez-Cabo F., Kirilovsky A., Mlecnik B., Lagorce-Pagès C., Tosolini M., Camus M., Berger A., Wind P., Zinzindohoué F., Bruneval P., Cugnenc P.-H., Trajanoski Z., Fridman W.-H., Pagès F., Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313, 1960–1964 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Voduc K. D., Cheang M. C. U., Tyldesley S., Gelmon K., Nielsen T. O., Kennecke H., Breast cancer subtypes and the risk of local and regional relapse. J. Clin. Oncol. 28, 1684–1691 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Partin A. W., Kattan M. W., Subong E. N., Walsh P. C., Wojno K. J., Oesterling J. E., Scardino P. T., Pearson J. D., Combination of prostate-specific antigen, clinical stage, and gleason score to predict pathological stage of localized prostate cancer. JAMA 277, 1445–1451 (1997). [PubMed] [Google Scholar]
- 45.Hirose M., Fukui H., Igarashi Y., Fujimori Y., Katake Y., Sekikawa A., Ichikawa K., Tomita S., Imura J., Ajioka Y., Ueno H., Hase K., Ohkura Y., Kashida H., Togashi K., Nishigami T., Matsui T., Yao T., Wada R., Matsuda K., Watanabe T., Ochiai A., Sugai T., Sugihara K., Fujimori T., Detection of desmoplastic reaction in biopsy specimens is useful for predicting the depth of invasion of early colorectal cancer: A Japanese collaborative study. J. Gastroenterol. 45, 1212–1218 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Kimura R., Fujimori T., Ichikawa K., Ajioka Y., Ueno H., Ohkura Y., Kashida H., Togashi K., Yao T., Wada R., Watanabe T., Ochiai A., Sugai T., Sugihara K., Igarashi Y., Desmoplastic reaction in biopsy specimens of early colorectal cancer: A Japanese prospective multicenter study. Pathol. Int. 62, 525–531 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Bhargava R., Infrared spectroscopic imaging: The next generation. Appl. Spectrosc. 66, 1091–1120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mayerich D., Walsh M. J., Kadjacsy-Balla A., Ray P. S., Hewitt S. M., Bhargava R., Stain-less staining for computed histopathology. Technology 3, 27–31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costa J., Reflections about evidence-based pathology. Int. J. Surg. Pathol. 15, 230–232 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Yeh K., Kenkel S., Liu J., Bhargava R., Fast infrared chemical imaging with a quantum cascade laser. Anal. Chem. 87, 485–493 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tiwari S., Raman J., Reddy V., Ghetler A., Tella R. P., Han Y., Moon C. R., Hoke C. D., Bhargava R., Towards translation of discrete frequency infrared spectroscopic imaging for digital histopathology of clinical biopsy samples. Anal. Chem. 88, 10183–10190 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Mittal S., Yeh K., Leslie L. S., Kenkel S., Kajdacsy-Balla A., Bhargava R., Simultaneous cancer and tumor microenvironment subtyping using confocal infrared microscopy for all-digital molecular histopathology. Proc. Natl. Acad. Sci. U.S.A. 115, E5651–E5660 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pilling M. J., Henderson A., Gardner P., Quantum cascade laser spectral histopathology: Breast Cancer diagnostics using high throughput chemical imaging. Anal. Chem. 89, 7348–7355 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Gaber A., Johansson M., Stenman U.-H., Hotakainen K., Pontén F., Glimelius B., Bjartell A., Jirström K., Birgisson H., High expression of tumour-associated trypsin inhibitor correlates with liver metastasis and poor prognosis in colorectal cancer. Br. J. Cancer 100, 1540–1548 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baker M. J., Trevisan J., Bassan P., Bhargava R., Butler H. J., Dorling K. M., Fielden P. R., Fogarty S. W., Fullwood N. J., Heys K. a., Hughes C., Lasch P., Martin-Hirsch P. L., Obinaju B., Sockalingum G. D., Sulé-Suso J., Strong R. J., Walsh M. J., Wood B. R., Gardner P., Martin F. L., Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 9, 1771–1791 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tiwari S., Bhargava R., Extracting knowledge from chemical imaging data using computational algorithms for digital cancer diagnosis. Yale J. Biol. Med. 88, 131–143 (2015). [PMC free article] [PubMed] [Google Scholar]
- 57.Bhargava R., Fernandez D. C., Hewitt S. M., Levin I. W., High throughput assessment of cells and tissues: Bayesian classification of spectral metrics from infrared vibrational spectroscopic imaging data. Biochim. Biophys. Acta 1758, 830–845 (2006). [DOI] [PubMed] [Google Scholar]
- 58.De Jay N., Papillon-Cavanagh S., Olsen C., El-Hachem N., Bontempi G., Haibe-Kains B., mRMRe: An R package for parallelized mRMR ensemble feature selection. Bioinformatics 29, 2365–2368 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/6/eabb8292/DC1


