Chiral proofreader recruited from Archaea relieves anaerobic stress associated with translation machinery in land plants.
Abstract
Streptophyte algae emerged as a land plant with adaptations that eventually led to terrestrialization. Land plants encounter a range of biotic and abiotic stresses that elicit anaerobic stress responses. Here, we show that acetaldehyde, a toxic metabolite of anaerobic stress, targets and generates ethyl adducts on aminoacyl-tRNA, a central component of the translation machinery. However, elongation factor thermo unstable (EF-Tu) safeguards l-aminoacyl-tRNA, but not d-aminoacyl-tRNA, from being modified by acetaldehyde. We identified a unique activity of archaeal-derived chiral proofreading module, d-aminoacyl-tRNA deacylase 2 (DTD2), that removes N-ethyl adducts formed on d-aminoacyl-tRNAs (NEDATs). Thus, the study provides the molecular basis of ethanol and acetaldehyde hypersensitivity in DTD2 knockout plants. We uncovered an important gene transfer event from methanogenic archaea to the ancestor of land plants. While missing in other algal lineages, DTD2 is conserved from streptophyte algae to land plants, suggesting its role toward the emergence and evolution of land plants.
INTRODUCTION
The root is a critical architectural innovation in land plants, which is essential for anchorage, nutrient, and water uptake (1, 2). Plants being sedentary in their lifestyle, roots have to survive in the oxygen-deprived environment of soil and face multiple stresses, including major anaerobic stress. Thus, roots are the primary sites of acetaldehyde production, an intermediate of ethanol fermentation (3–7). Environmental assaults such as floods, waterlogging, and parasite infection worsen the situation by creating prolonged anoxic conditions, which eventually produce excess acetaldehyde (3, 4, 8). Acetaldehyde is also an ever-present metabolite in plants and involved in multistress response. Moreover, important processes such as seed germination and pollen development are highly dependent on ethanol fermentation. Anomalies in this pathway lead to failure in seed vigor and male fertility (8–12).
Along with anaerobic stress, roots experience tremendous stress from d-amino acids that are found in abundance in the soil. However, the presence of d-amino acids in plants is inevitable because they play vital roles in plant physiology, such in development of pollen tubes, apart from acting as a primary nitrogen source (13). However, d-amino acids also cause cellular toxicity by generating d-aminoacyl-tRNAs (d-aa-tRNAs), which sequester a free tRNA pool and impede translation. Removal of mischarged d-amino acids from d-aa-tRNAs is termed chiral proofreading, which enables the perpetuation of homochirality in the cellular proteome (14, 15). d-amino acid editing modules are universally conserved; d-aa-tRNA deacylase 1 (DTD1) is found in both Bacteria and Eukarya, and DTD2 is present in Archaea and Plantae. Earlier, the presence of DTD2 was noted in all the sequenced genomes of the plant kingdom available at that time. Hence, it was assumed that DTD2 is conserved throughout the plant kingdom (Archaeplastida) (16–18). Plants therefore are unique in having both of the evolutionarily distinct chiral proofreading modules DTD1 and DTD2. Unexpectedly, DTD2 knockout plants, using Arabidopsis as a model system, have been shown to display pronounced sensitivity to both ethanol and acetaldehyde, which are metabolites generated by sustained ethanol fermentation (19, 20).
Acetaldehyde is one of the highly reactive metabolites present inside the cell. It is known to be an unavoidable genotoxic intermediate of anaerobic fermentation. Acetaldehyde forms adducts with DNA and proteins by making an electrophilic attack on free amino groups (21–25). The toxic effects of acetaldehyde are quenched by multiple tiers of detoxifying systems. The first line of defense is provided by acetaldehyde dehydrogenase (ALDH), which converts acetaldehyde to nonreactive acetate. The second level of protection is offered by DNA repair mechanisms such as Fanconi anemia group D2 protein (FANCD2), which maintains chromosomal integrity and stability by repairing acetaldehyde-driven DNA damage. In alcoholic mice and humans, failure of acetaldehyde surveillance mechanisms leads to cancers such as Fanconi anemia, esophageal cancer, and hepatocellular carcinoma (26, 27). Catastrophic effects of acetaldehyde have been reported in various organisms, including plants (28–30). The most intriguing finding among plants is the acetaldehyde hypersensitivity of DTD2 knockouts (17, 19, 20). It has been a long-standing question how a d-amino acid detoxifying system DTD2 confers protection to plants from toxic intermediates of ethanol fermentation, especially from acetaldehyde during anaerobic stress.
Our study aimed at demystifying the link between DTD2 and its role in protecting plants from acetaldehyde-mediated toxicity. The work provides the first empirical evidence that acetaldehyde targets aa-tRNAs, which are highly sensitive compared to nucleic acids and proteins. Acetaldehyde creates ethyl modification of amino acids of aa-tRNA, which makes aa-tRNA more alkali-stable than peptidyl-tRNA. We further identified a unique role of DTD2 as a proofreader of d-aa-tRNA adducts since neither DTD1 nor peptidyl-tRNA hydrolase (PTH) can recycle N-ethyl-aa-tRNAs (NEATs). The unique activity of DTD2 necessitated its recruitment from Archaea to land plants. This is the first evidence of archaeal gene transfer from methanogens to streptophyte algae. Moreover, the distribution of DTD2 is strongly correlated with acetaldehyde biosynthesis. The presence of DTD2 in land plants and their ancestors, starting from early-diverging subaerial streptophyte algae, suggests that acquiring DTD2 from Archaea is one of the key events in the emergence of land plants.
RESULTS
Acetaldehyde creates ethyl modification on aa-tRNA
DTD2 is known to proofread d-aa-tRNAs as a part of translational quality control (16, 17). Therefore, we set out to investigate whether DTD2’s ability to protect plants from acetaldehyde has the involvement of aa-tRNAs. Hence, we treated aa-tRNAs with acetaldehyde, followed by a reducing agent (Fig. 1A). Unexpectedly, this resulted in a differential migration of the spot corresponding to aa-tRNA on thin-layer chromatogram (TLC) when compared with untreated control (Fig. 1B). Additional moiety attached to the α-amino group of aa-tRNA, such as a peptidyl or an acetyl group, is known to increase the stability of the ester linkage between the amino acid and the tRNA and is best assessed by alkali treatment (31). Earlier studies have shown that peptidyl-tRNAs and N-acetyl-l-aa-tRNAs (substrate mimics of peptidyl-tRNAs) are more stable compared to aa-tRNAs and are hydrolyzed by PTH (32). Upon performing the alkali treatment experiment, we found that the ester linkage of acetaldehyde-treated aa-tRNA (modified d-Phe-tRNAPhe; half-life: ~12 min) showed enhanced stability compared to aa-tRNA (d-Phe-tRNAPhe; half-life: ~1 min) and N-acetyl-aa-tRNA (N-acetyl-d-Phe-tRNAPhe; half-life: ~2.5 min) (Fig. 1C). Ultrastability of N-blocked aa-tRNAs makes them more lethal to the cell because they permanently arrest the tRNA pool and prevent it from participating in translation, which is evident in the case of a temperature-sensitive strain of Escherichia coli for PTH (32, 33). Acetaldehyde-treated aa-tRNAs are therefore expected to be more toxic to the cell than N-acetyl-aa-tRNAs and peptidyl-tRNAs because of their high alkali-stable nature. These observations based on TLC and alkali treatment indicated that acetaldehyde could modify aa-tRNA.
Fig. 1. Acetaldehyde targets aa-tRNA and generates ethyl modification.
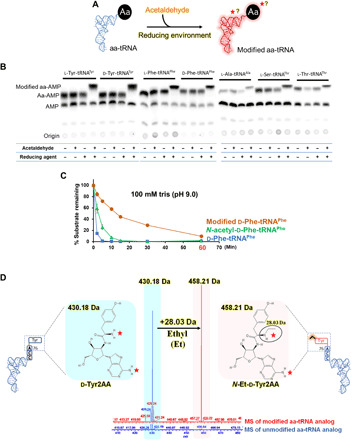
(A) Schematic showing reaction of aa-tRNA and acetaldehyde in the presence of a reducing agent. The red star or question mark indicates that the modification by acetaldehyde could be on amino acid or tRNA or both. (B) TLC is showing differences in the spot positions of aa-AMP corresponding to the unmodified and acetaldehyde-modified l-Tyr-tRNATyr, d-Tyr-tRNATyr, l-Phe-tRNAPhe, d-Phe-tRNAPhe, l-Ala-tRNAAla, l-Ser-tRNAThr, and l-Thr-tRNAThr [for each aa-tRNA, untreated aa-tRNA, aa-tRNA incubated with only acetaldehyde, aa-tRNA incubated with the only reducing agent, and aa-tRNA incubated with both acetaldehyde and reducing agent (in the manuscript, it is referred as “acetaldehyde-treated”) are shown in TLC]. (C) Alkali treatment of d-Phe-tRNAPhe, acetaldehyde-modified d-Phe-tRNAPhe, and N-acetyl-d-Phe-tRNAPhe is showing ultrastability of acetaldehyde-modified d-Phe-tRNAPhe. (D) ESI-MS analysis showing ethyl modification (a clear shift of 28.03 Da) on acetaldehyde-treated substrate analog of d-Tyr-tRNA [d-Tyr2AA (d-tyrosyl-2′-aminoadenosine)] [unmodified d-Tyr2AA with m/z = 430.18 (blue) and modified d-Tyr2AA with m/z = 458.21 (pink)].
To characterize the chemical nature of the modification, electrospray ionization mass spectrometry (ESI-MS) of both acetaldehyde-treated and untreated nonhydrolyzable analogs of aa-tRNAs was performed. These analogs mimic both amino acid and tRNA parts of aa-tRNA, where the amino acid is bound to adenosine (terminal adenosine-76 of tRNA) with nonhydrolyzable amide bond (14). MS data clearly showed that acetaldehyde covalently modifies aa-tRNA with “ethyl” moiety (Fig. 1D and fig. S1, A and B).
Acetaldehyde modifies amino acid of aa-tRNA and is independent of chirality and side chain
Incubation of acetaldehyde with aa-tRNAs (Tyr-tRNATyr, Phe-tRNAPhe, Ala-tRNAAla, Ser-tRNAThr, and Thr-tRNAThr) or with their analogs (Tyr2AA, Ala3AA, Val3AA, and Thr2AA) bearing amino acids having either chirality (l- or d-) and varied side chains revealed that acetaldehyde modifies aa-tRNA irrespective of its chirality and side-chain chemistry (Figs. 1B and 2A and figs. S1, C to J, and S2). Since acetaldehyde is known to target free amino groups, to identify the site of ethyl modification on the aa-tRNA, tandem MS fragmentation (MS2) of ethyl-modified analogs was carried out. Fragmentation experiments (MS2) confirmed that modification happens only on the amino acid part of aa-tRNA (Fig. 2, B to D, and figs. S1 and S2) and not on tRNA. These experiments established the link between acetaldehyde, an anaerobic stress metabolite, and aa-tRNA, a key constituent of the translation machinery (TM). It also prompted us to hypothesize that DTD2 relieves anaerobic stress associated with TM in plants by removing ethyl-modified amino acids from NEATs.
Fig. 2. Acetaldehyde modifies amino acid of aa-tRNA.
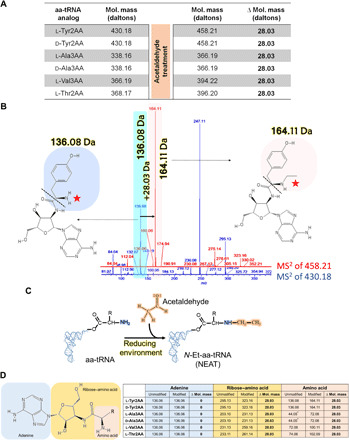
(A) Table showing m/z peaks observed in ESI-MS analysis of acetaldehyde-treated analogs of multiple aa-tRNAs (l-Tyr-tRNA, d-Tyr-tRNA, l-Ala-tRNA, d-Ala-tRNA, l-Val-tRNA, and l-Thr-tRNA). (B) Fragmentation experiments (MS2) showing adenine (136.06 Da) and immonium ion of tyrosine (136.08 Da) peaks in unmodified d-Tyr2AA (blue) and adenine (136.06 Da) and ethyl-modified immonium ion of tyrosine (164.11 Da) peaks in modified d-Tyr2AA (pink). (C) Schematic shows that acetaldehyde creates ethyl modification on the α-NH2 group of aa-tRNA and generates NEAT. (D) Table showing m/z peaks observed during MS2 analysis; adenine and amino acid in unmodified analogs, adenine and ethyl-modified amino acid alone and, along with ribose in modified analogs of various aa-tRNAs [molecular weights of amino acids showed in (B) and (D) correspond to their respective immonium ions. Fragmentation patterns of analogs are mentioned in fig. S1, K and L] (asterisk indicates expected molecular weight of immonium ion of alanine, which was not resolved in our MS experiments. However, this value can be deduced from MS data of ribose–amino acid of l- and d-Ala3AA) (MS data related to this figure are summarized in fig. S2).
DTD2 alleviates anaerobic stress in plants by recycling NEDATs
To explore whether DTD2 deacylates NEATs, in vitro deacylation assays were performed by incubating DTD2 of Arabidopsis thaliana (At DTD2) with both N-ethyl-l-aa-tRNAs [NELATs: N-ethyl-l-Phe-tRNAPhe (At) and N-ethyl-l-Tyr-tRNATyr (Thermus thermophilus) (Tth)] and N-ethyl-d-aa-tRNAs [NEDATs: N-ethyl-d-Phe-tRNAPhe (At) and N-ethyl-d-Tyr-tRNATyr (Tth)]. In line with our hypothesis, At DTD2 readily deacylated NEDATs to N-ethyl-d-amino acids and free tRNAs but not NELATs (Fig. 3, A and B, and fig. S3, A and B). The activity of DTD2 on NEDATs is hereafter referred to as “NEDAT deacylase (ND) activity.” Since plants also have the canonical chiral proofreading module DTD1 alongside DTD2, we carried out deacylation of NELATs and NEDATs with DTD1. Our biochemical data revealed that neither bacterial (E. coli) nor eukaryotic (A. thaliana) DTD1 acts on NELATs or NEDATs (Fig. 3, C and D, and figs. S3, C to J, and S4, A and B). DTD1’s weak activity on NEDATs is corroborated by the earlier structural studies on DTD1 in complex with the d-aa-tRNA analog d-Tyr3AA (14, 34). The cocrystal structure had revealed that the d-amino acid of a d-aa-tRNA fits snugly into the active site of DTD1, thereby sterically excluding the N-ethylated amino group of a d-amino acid (Fig. 3E). PTH, as mentioned before, acts on N-acetyl-l-aa-tRNAs, which are similar to ethylated substrates (Fig. 3F). Moreover, PTH and DTD2 share a similar fold (16); therefore, we expected that PTH could act on NELATs. However, to our surprise, neither bacterial (E. coli)) nor archaeal and the eukaryotic version of PTH (Sulfolobus tokadii) had activity on NELATs (Fig. 3, G to J, and fig. S4, C to L), suggesting their strict chemical selectivity. In light of these findings on the biochemical activities of the editing modules DTD1, PTH, and DTD2, DTD2 stands out with a distinct role as a unique proofreader of d-aa-tRNA adducts.
Fig. 3. NEDATs are readily acted upon by A. thaliana DTD2 but not by DTD1 or PTH.
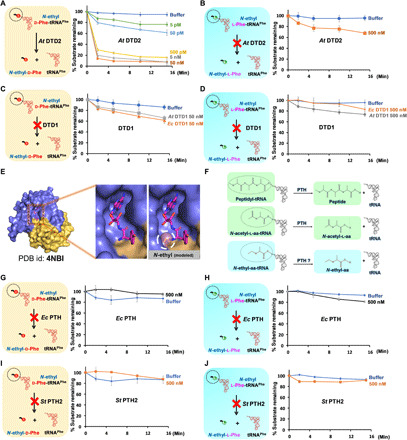
(A) Deacylation of N-ethyl-d-Phe-tRNAPhe (At) with At DTD2. (B) Deacylation of N-ethyl-l-Phe-tRNAPhe (At) with At DTD2. (C) Deacylation of N-ethyl-d-Phe-tRNAPhe with At DTD1 and Ec DTD1. (D) Deacylation of N-ethyl-l-Phe-tRNAPhe with At DTD1 and Ec DTD1. (E) Crystal structure of DTD1 in complex with d-Tyr3AA. Left of the zoomed-in section of the active site shows the binding mode of the snugly fit substrate; right shows a steric clash between the modeled ethyl modification on the α-NH2 group of d-amino acid and active site residues. PDB, Protein Data Bank. (F) Schematics showing subtle chemical differences among peptidyl-tRNA, N-acetyl-aa-tRNA, and NEAT (N-acetyl-aa-tRNAs differ with NEATs only by carbonyl oxygen present on the carbon that is attached to the α-NH2 group of amino acid of aa-tRNA). (G) Deacylation of N-ethyl-d-Phe-tRNAPhe (Ec) with Ec PTH. (H) Deacylation of N-ethyl-l-Phe-tRNAPhe (Ec) with Ec PTH. (I) Deacylation of N-ethyl-d-Phe-tRNAPhe (Pho) with St PTH2. (J) Deacylation of N-ethyl-l-Phe-tRNAPhe (Pho) with St PTH2. In all our biochemical assays, the concentration of substrate used was 0.2 μM. At, A. thaliana; Ec, E. coli; St, S. tokadii.
Elongation factor thermo unstable protects l-aa from acetaldehyde
As mentioned above, the biochemical experiments were performed in vitro, and to show the accumulation of NEDATs in vivo, we used a surrogate (E. coli) system. Initially, E. coli cells were grown in the presence of d-amino acids and acetaldehyde. Later, aa-tRNAs were probed using Northern blotting. As expected, the aa-tRNAs isolated from bacteria exposed to both d-amino acids and acetaldehyde were stable upon alkali treatment. At the same time, the control samples were less stable (Fig. 4, A to D). To further validate the identity of the alkali-stable aa-tRNAs, we performed MS experiments. MS experiments confirmed that these aa-tRNAs were modified to NEDATs by acetaldehyde (Fig. 4E). Similar to in vitro experiments, these in vivo generated NEDATs were deacylated by DTD2 (Fig. 4F). However, acetaldehyde also modified l-aa-tRNAs as efficiently as d-aa-tRNAs in vitro (Figs. 1B and 2A and figs. S1, and S2). Nevertheless, none of the enzymes tested, namely DTD1, DTD2, and PTH, could deacylate the NELATs (Fig. 3, B, D, H, and J, and figs. S3, B and D, and S4D). Therefore, it is important to know how l-aa-tRNAs are protected from acetaldehyde in vivo. Elongation factor thermo unstable (EF-Tu) is one of the most abundant proteins of the cell and is known to bind l-aa-tRNAs with higher affinity compared to d-aa-tRNAs (35). d-aa-tRNAs are discriminated by the EF-Tu and other cellular chiral checkpoints (36). To test EF-Tu’s ability to protect l-aa-tRNAs from freely diffusing acetaldehyde, we performed modification experiments in the presence of EF-Tu. These assays clearly showed that EF-Tu offers protection to l-aa-tRNAs (l-Phe-tRNAPhe and l-Tyr-tRNATyr) but not to d-aa-tRNAs (d-Phe-tRNAPhe and d-Tyr-tRNATyr) from acetaldehyde modification (Fig. 5, A to C). EF-Tu protection experiments led to the identification of its unique role in specifically safeguarding l-aa-tRNAs from reactive metabolites of the cell such as acetaldehyde apart from delivering them to the ribosome. From the crystal structure, it was evident that the α-NH2 group of aa-tRNA is engaged in interactions with EF-Tu, and there is no extra space to accommodate an ethyl group (Fig. 5D) (37). The ability of DTD2 to rescue acetaldehyde toxicity in Arabidopsis in combination with its lack of activity on NELATs highlights that NEDATs are the physiological substrates of DTD2. Together, the above findings suggest that DTD2 alleviates the effect of acetaldehyde by actively clearing NEDATs during anaerobic stress.
Fig. 4. Acetaldehyde generates ethyl modification on d-aa-tRNA in vivo.
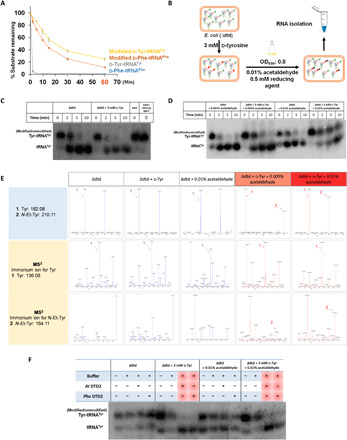
(A) CuSO4 + tris (8.0) treatment of in vitro generated d-Phe-tRNAPhe, d-Tyr-tRNATyr, modified (N-ethyl)-d-Phe-tRNAPhe, and modified (N-ethyl)-d-Tyr-tRNATyr. (B) Schematic showing the method used to generate NEDATs in E. coli. (C) Northern blot analysis showing no accumulation of Tyr-tRNATyr adducts in E. coli cells grown in the presence of only d-tyrosine. (D) Northern blot analysis showing the accumulation of Tyr-tRNATyr adducts in E. coli cells grown in the presence of d-tyrosine and acetaldehyde. (E) Alkali hydrolysis (incubation in 15% of NH4OH at 70°C for 18 hours) of aa-tRNA isolated from Δdtd E. coli, Δdtd E. coli + d-Tyr (grown in the presence of 3 mM d-tyrosine), and Δdtd E. coli + 0.01% acetaldehyde (grown in the presence of 0.01% acetaldehyde) yielded tyrosine peaks. N-ethyl-tyrosine peaks were seen only in Δdtd E. coli + 3 mM d-Tyr + 0.005% acetaldehyde (grown in the presence of 3 mM d-tyrosine and 0.005% acetaldehyde), and Δdtd E. coli + 3 mM d-Tyr + 0.01% acetaldehyde (grown in the presence of 3 mM d-tyrosine and 0.01% acetaldehyde). Fragmentation of the respective peaks also confirms the same. (F) Northern blot analysis showing deacylation of in vivo generated NEDATs by both archaeal DTD2 (Pho DTD2, 50 nM) and plant DTD2 (At DTD2, 50 nM).
Fig. 5. EF-Tu offers protection to l-aa-tRNA from acetaldehyde.
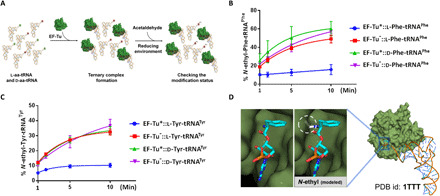
(A) Schematic showing the method that was used for EF-Tu protection assay. (B) Ethyl modification status on l-Phe-tRNAPhe and d-Phe-tRNAPhe by acetaldehyde in the presence of unactivated (denoted as -) and activated EF-Tu (marked as *). (C) Ethyl modification status on l-Tyr-tRNATyr and d-Tyr-tRNATyr by acetaldehyde in the presence of unactivated and activated EF-Tu [this experiment was repeated twice (n = 2)]. (D) The cocrystal structure of EF-Tu and l-Phe-tRNAPhe. Left of the zoomed-in section shows l-phenylalanine in the amino acid binding site; right shows a steric clash between ethyl-modified l-phenylalanine and amino acid binding site.
ND activity is rooted in the Archaea
Apart from plants, DTD2 is also conserved in Archaea (Fig. 6A and fig. S5, A and B) (16, 17), and therefore, we further checked whether archaeal DTD2s have the ND activity. We performed NEAT deacylation experiments with distantly related archaeal DTD2s from the organisms Pyrococcus horikoshii (Pho DTD2), Methanocaldococcus jannaschii (Mj DTD2), and Archaeoglobus fulgidus (Af DTD2). All three archaeal DTD2s acted on NEDATs with similar efficiency as that of plant DTD2s (Fig. 6, B to D, and fig. S5C). Similar to plant DTD2s, archaeal DTD2s also did not act on NELATs, suggesting that not only the ND activity but also chiral selectivity toward adducts is conserved in DTD2s of Archaea (Fig. 6E and fig. S5D). The conservation of DTD2 and its ND activity across Archaea highlights that DTD2’s ability to clear NEDATs is “rooted” in the archaeal branch of life. Therefore, it is clear that plants specifically acquired DTD2 from Archaea during evolution. This is further supported by the complete absence of DTD2 in other major eukaryotic life forms, i.e., opisthokonts, which include fungi and animals.
Fig. 6. DTD2’s ND activity and chiral selectivity are conserved throughout Archaea.
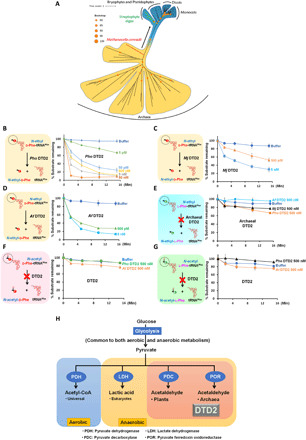
(A) Phylogenetic classification of archaeal and plant DTD2s (generated with scoring matrix LG+R5 (according to the BIC score, of 546 protein models, LG+R5 was chosen as the best amino acid substitution model for the given multiple sequence alignment, which was used to construct a phylogenetic tree). (B) Deacylation of N-ethyl-d-Phe-tRNAPhe (Pho) with Pho DTD2. (C) Deacylation of N-ethyl-d-Phe-tRNAPhe (Pho) with Mj DTD2. (D) Deacylation of N-ethyl-d-Phe-tRNAPhe (Pho) with Af DTD2. (E) Deacylation of N-ethyl-l-Phe-tRNAPhe (Pho) with Pho DTD2, Mj DTD2, and Af DTD2 showing no activity even at 500 nM enzyme concentration. (F) Deacylation of N-acetyl-d-Phe-tRNAPhe with both archaeal (Pho) and plant (At) DTD2s showing no activity even at 500 nM enzyme concentration. (G) Deacylation of N-acetyl-l-Phe-tRNAPhe with both archaeal (Pho) and plant (At) DTD2s showing no activity even at 500 nM enzyme concentration. (H) Co-occurrence of DTD2 with acetaldehyde biosynthesis in Archaea and plants.
DTD2 is specific to ethyl modification and coexists with acetaldehyde biosynthesis
We next interrogated the substrate specificity of DTD2 in the context of modification by performing deacylation with N-acetyl-l-aa-tRNAs and N-acetyl-d-aa-tRNAs. We found that both archaeal (Pho) and plant (At) DTD2s did not hydrolyze acetyl substrates (Fig. 6, F and G, and fig. S5, E and F), suggesting that DTD2 is strictly specific to ethyl modification and has evolved exclusively for ND activity. These findings also indicate that not only DTD2’s ND activity but also its specificity toward ethyl modification is rooted in Archaea. It was therefore intriguing to probe the physiological constraints that could have necessitated the conservation of DTD2 and its ND activity in Archaea and plants. To this end, we probed for the presence of common physiological and biochemical characteristics between Archaea and plants. Notably, we noted that acetaldehyde biosynthesis machinery is conserved exclusively in Archaea and plants (8, 38, 39). In Archaea, pyruvate ferredoxin oxidoreductase converts pyruvate to acetaldehyde. Pyruvate decarboxylase performs the same function in plants as a part of ethanol fermentation, an ancient metabolic pathway (Fig. 6H). In agreement, these machineries are mostly absent in Bacteria and Opisthokonta (40, 41). The above bioinformatic analysis, in combination with the biochemical activity, provides a strong rationale for the requirement of DTD2 in Archaea and plants.
DTD2 distribution is exclusive to land plants and streptophyte algae in the plant kingdom
We next set out to identify when plants acquired the archaeal version of DTD, i.e., DTD2. Extensive bioinformatic analysis of DTD2 distribution in the plant kingdom (Archaeplastida) delineated that DTD2 is strictly confined to land plants (Bryophyta, Pteridophyta, Gymnosperms, and Angiosperms) and streptophyte algae (Chlorokybophyceae, Klebsormidiophyceae, Charophyceae, and Zygnematophyceae). At the same time, DTD2 is entirely absent in different clades of algae, which include glaucophytes, red algae, and green algae. DTD2 is present in the recently sequenced genomes of the streptophyte algae Chlorokybus atmophyticus (42), Klebsormidium nitens (43), Chara braunii (44), and Mesotaenium endlicherianum (45) (Fig. 7A), of which C. atmophyticus is an early-diverging “subaerial/terrestrial” algae (42). Unexpectedly, the early-diverging “aquatic” streptophyte algae Mesostigma viride does not encode DTD2. Structure-based alignment of DTD2 sequences revealed that DTD2 of streptophyte algae have conserved residues, which are essential for catalysis (Fig. 7B). To further validate our sequence analysis, we performed biochemical assays using DTD2 of streptophyte algae [K. nitens DTD2 (Kn DTD2)]. We found that Kn DTD2 not only showed ND activity (Fig. 7C and fig. S5G) but also conserved chiral selectivity (Fig. 7D and fig. S5H) and ethyl specificity on adducts (Fig. 7, E and F, and fig. S5, I and J). These observations suggest that DTD2 may have provided an adaptive advantage to streptophyte algae, allowing them to venture on to the land.
Fig. 7. DTD2 is conserved in land plants and streptophyte algae in the plant kingdom.
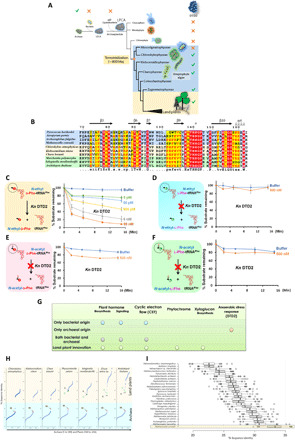
(A) DTD2 distribution is seen only in Archaea, land plants, and streptophyte algae (genome sequences of Coleochaetophyceae members are not available, and hence, DTD2’s status is not depicted in the figure). (B) Structure-based sequence alignment of DTD2 shows conservation of active site residues (16) throughout Archaea, streptophyte algae, and land plants. Deacylation of N-ethyl-d-Phe-tRNAPhe (At) (C), N-ethyl-l-Phe-tRNAPhe (At) (D), N-acetyl-d-Phe-tRNAPhe (At) (E), and N-acetyl-l-Phe-tRNAPhe (At) (F) with K. nitens DTD2. (G) DTD2 is the only gene of exclusive archaeal origin that was recruited for the adaptation of land plants. (H) Sequence identity values between representative plant DTD2s with all the DTD2 sequences (includes both Archaea and plants) [M. conradii (M), C. atmophyticus (C), and K. nitens (K) are highlighted in the figure] [the horizontal axis represents archaeal DTD2 sequences (1 to 149) and plant DTD2 sequences (150 to 234), and the vertical axis represents sequence identity values]. (I) Boxplot displays DTD2 of M. conradii that shares the maximum identity (~33%) with DTD2s of all the plants; the horizontal axis represents sequence identities with all plant DTD2s, and the vertical axis shows the representative archaeal sequences compared with all plants.
Acquisition of DTD2 is one of the first steps on the long road of land plant evolution
Recent evidences have shown that streptophyte algae acquired many land plant–specific characteristics such as phytohormone biosynthesis, phytohormone signaling, xyloglucan biosynthesis, phytochromes, and cyclic electron flow (42–46). These genes are conserved across land plants and are mostly absent in other clades of algae (42, 43). Similarly, the presence of DTD2 from streptophyte algae to land plants suggests that its acquisition is an important land plant–specific adaptation. In addition, the remarkable conservation of DTD2 starting from the early-diverging subaerial/terrestrial streptophyte algae C. atmophyticus indicates that recruitment of DTD2 is one of the key first steps on the long road of land plant evolution.
Moreover, we searched for the origins of the genes mentioned above that are responsible for the adaptations of land plants. Among such genes, DTD2 is the only gene that has an exclusive archaeal origin. In contrast, all the other genes have either only bacterial origin or mixed origins (both bacterial and archaeal) or have appeared de novo as genomic innovations in streptophyte algae (Fig. 7G and fig. S6, A and B). Note that DTD2 was reported to be one of the only two proteins unique to Archaea and plants, while the other protein is topoisomerase VI subunit B (TopVIB) (20). Mutations in TopVIB are known to affect cell proliferation and endoreduplication, which are unique characteristics of the plant kingdom (47–50). While TopVIB is a subunit of archaeal type VI topoisomerase, DTD2 is a stand-alone functional protein. Unlike DTD2, TopVIB is conserved in all the lineages of algae in addition to the land plants (fig. S6C). Therefore, this analysis indicates that DTD2 is the only gene unique to Streptophyta and Archaea.
Recruitment of DTD2 from methanogenic archaea
A thorough phylogenetic analysis of all the known archaeal and plant DTD2s highlighted that among plant DTD2s, DTD2s of streptophyte algae are closer to the archaeal ones (Fig. 6A and fig. S5, A and B). Among archaeal DTD2s, Methanocella conradii shares maximum sequence identity with most of the DTD2s of streptophyte algae and land plants (Fig. 7, H and I). This emphasizes on a single acquisition event of DTD2 from methanogenic archaea to the plant kingdom that happened at the basal radiation of land plants. As mentioned earlier, except streptophyte algae, DTD2 is absent in all the other lineages of algae. This further strengthens that DTD2 was likely acquired from Archaea by streptophyte algae rather than by an endosymbiotic event. Since it is unlikely to be multiple gene loss events in the eukaryotic branch, we propose the likely possibility of a single acquisition event. In agreement, Methanocella is found to be in association with lower plants in bogs. Bogs are the oldest terrestrial ecosystems that are deprived of oxygen and enriched in d-amino acids (51, 52). One of the critical architectural innovations that happened during the evolution of land plants is the “root.” The root is the primary organ that is exposed to higher anaerobic stress and excess d-amino acids in the soil (2, 8, 13, 53, 54). In accordance, DTD2 is highly expressed in roots compared to the other organs (20). The confluence of the two independent stresses, namely d-amino acids and anaerobic stress, aggravates the formation of NEDATs. Therefore, the acquisition of DTD2 from Archaea in the terrestrial ecosystem by streptophyte algae likely benefited them to emerge as land plants during the Neoproterozoic Era (~800 million years ago).
aa-tRNA is the first target to get modified by acetaldehyde
Chronic alcohol consumption in humans causes various cancers (21, 26, 27); the causative agent for the devastating effects is found to be acetaldehyde. Thus, acetaldehyde was demonstrated to be a genotoxic and teratogenic agent in mice and humans (21). Similar to aa-tRNAs, it mounts an electrophilic attack on the free amino groups of nucleic acids and proteins (22–25). To compare relative sensitivities of free amino acids, proteins, and nucleic acids with that of aa-tRNAs, each of these was treated with acetaldehyde. We found that aa-tRNAs are more prone to modification than other biomolecules. Amino acids and peptides are found to be the second preferred targets of acetaldehyde. The requirement of a 1000-fold molar excess or more of acetaldehyde to modify nucleotides suggests that nucleic acids are relatively less prone to modification. However, an equal amount of acetaldehyde is enough to modify aa-tRNAs (Fig. 8 and figs. S7 and S8, A to D). This clearly suggests that during anaerobic stress or acetaldehyde burst, aa-tRNAs are the first cellular components that undergo modification.
Fig. 8. aa-tRNAs are hypersensitive to acetaldehyde.
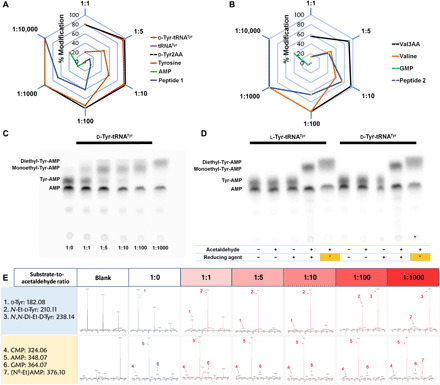
(A and B) Radar charts showing that aa-tRNA (d-Tyr-tRNATyr) and its analogs (d-Tyr2AA and Val3AA) are hypersensitive to acetaldehyde than other biomolecules (each corner represents the ratio of substrate to acetaldehyde, and hexagonal rings depict the percentage of the extent of modification. Related MS data are shown in (E) and figs. S7 and S8D. (C) TLC is showing differential migration of d-Tyr-tRNATyr at the different substrate-to-acetaldehyde ratios. aa-tRNA corresponding spot was observed at 1:0; monoethyl-aa-tRNA corresponding spots were observed at 1:1, 1:5, 1:10, and 1:100; and complete diethyl-aa-tRNA corresponding spot was observed at 1:1000 substrate-to-acetaldehyde ratios [related MS data are shown in (E)]. (D) TLC is showing differences in the spot positions of aa-AMP corresponding to the unmodified, monoethyl, and diethyl-Tyr-tRNATyr [lanes include untreated aa-tRNA, aa-tRNA incubated with only acetaldehyde, aa-tRNA incubated with only reducing agent, aa-tRNA incubated with both acetaldehyde (1:5 substrate-to-acetaldehyde ratio) and reducing agent, and aa-tRNA incubated with both acetaldehyde (1:1000 substrate-to-acetaldehyde ratio and it is indicated as asterisk in the figure) and reducing agent, respectively]. (E) MS data related to (C) (corresponding MS2 data are shown in fig. S8A).
Since aa-tRNAs are the primary targets of acetaldehyde, DTD2 is expected to provide an early response during anaerobic stress. This is further supported by toxicity assays of DTD2 knockout plants that showed acetaldehyde sensitivity within a range of 0.01 to 1% (v/v) of acetaldehyde. Above the 1% (v/v) acetaldehyde, both wild-type and DTD2 knockout plants die (19, 20). This suggests that DTD2 operates to clear the toxic effects of low levels of acetaldehyde that are produced due to the physiological requirement of anaerobic fermentation. An excess acetaldehyde production leads to the modifications on other counterparts such as proteins and nucleic acids (Fig. 8, A, B, and E, and figs. S7 and S8D). In agreement, excess acetaldehyde not only generates ethyl modification on other biomolecules but also creates diethyl modification on aa-tRNA. DTD2 does not deacylate diethyl-d-aa-tRNAs (Fig. 8, C to E, and figs. S7, S8, A to C, and S9). The toxic effects of acetaldehyde are quenched by multiple tiers of detoxifying systems, especially ALDH and FANCD2. ALDH, FANCD2, and other repair mechanisms do not act directly on ethyl modification (21, 26, 27, 55–57), whereas DTD2 is the only enzyme that removes N-ethyl-d-amino acids directly from NEDATs (Fig. 9).
Fig. 9. Acquiring archaeal DTD2 is one of the essential events toward the evolution of land plants to mitigate the toxicity created by the confluence of anaerobic stress and d-amino acids.
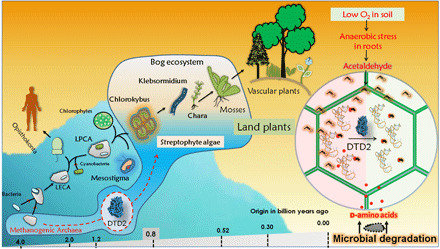
A model showing recruitment of DTD2 in streptophyte algae and its implications toward the land plant evolution (LECA and LPCA are abbreviations of last eukaryotic common ancestor and last plant common ancestor, respectively).
DISCUSSION
Our results show that acetaldehyde attacks a key component of translation, i.e., aa-tRNA, and creates ethyl modification. We also found that aa-tRNAs are hypersensitive to acetaldehyde compared to DNA and proteins. Acetaldehyde is a well-known causative agent of various cancers in alcohol-abused mice and humans (21, 26, 27). A major cause for such diseases is identified as ethyl adducts on DNA and proteins generated by acetaldehyde (22–25). In plants, the anaerobic stress metabolite acetaldehyde is reported to accumulate during multiple environmental stresses that include floods, waterlogging, drought, cold, high salinity, and parasitic infections (3, 4, 8). Enhancing multistress tolerance in crop plants is a key priority in sustainable agriculture. It is therefore worth exploring the possibility of overexpression of DTD2 as a strategy for improving multistress tolerance in crop plants.
Acetaldehyde-modified aa-tRNAs (NEATs) are not acted upon by DTD1 and PTH. Except for plants, most eukaryotes and bacteria do not encode acetaldehyde biosynthesis machinery; hence, the presence of DTD2 is superfluous and is substituted by DTD1 to perform chiral proofreading in these organisms. Distribution of DTD2 is exclusive to Archaea and Streptophyta (including streptophyte algae and land plants), where they encounter the dual problem of acetaldehyde biosynthesis and d-amino acid enrichment. DTD2 and PTH share a common ancestral fold; moreover, PTH is the only known enzyme that acts on N-blocked aa-tRNAs (16). We speculate that during the evolution of Archaea, DTD2 evolved to circumvent early harsh geochemical conditions by stitching an additional domain to the existing basic scaffold of PTH. In addition, it is known that under extreme environmental conditions such as higher temperature, racemization rates of aa-tRNAs are higher and eventually could contribute to the accumulation of d-aa-tRNAs to pernicious levels (58). Notably, PTH does not act on NELATs, which differ subtly with acetyl substrates and therefore delineates PTH’s specificity toward the carbonyl group that is attached to the α-amino group of amino acid. In contrast, DTD2 acts on ethyl modification but not on acetyl that clearly displays its specificity toward methylene group. Mechanistic insights into substrate specificity of DTD2 and PTH may provide the structural basis for their exclusive biochemical selectivity.
Land plant evolution is a major leap in the history of life that occurred during the Neoproterozoic era (~800 million years ago). Terrestrial flora played a pivotal role in changing the global environment and in the diversification of fauna on the land surface (59). Streptophyte algae successfully colonized the land with biochemical, cytological, developmental, and architectural adaptations (42–46). Investigations of the adaptations that occurred in streptophyte algae to be a successful colonizer of land are of extreme importance (42–46). In this regard, our finding of DTD2’s unique role in resolving anaerobic stress exacerbated by d-amino acids brings another dimension to our understanding of the cellular and molecular events that led to the emergence of land plants. Moreover, this also provides the rationale for DTD2’s absence in Opisthokonta, which do not encounter sustained acetaldehyde and d-amino acid stress (40, 41, 52). Notably, DTD2 is the only gene unique to Archaea and land plants and is the only known gene transfer event from Archaea to streptophyte algae. Moreover, the conservation of DTD2 from early-diverging streptophyte algae suggests that DTD2’s recruitment is an early event that likely facilitated the adaptation to terrestrial life (Fig. 9). Overall, our work sheds light on an unexpected cellular target of anaerobic stress and its mitigation, with implications for land plant evolution and multistress tolerance in crop plants.
MATERIALS AND METHODS
Materials
Materials were obtained from Merck unless otherwise mentioned. ESI-MS was performed using a Thermo Fisher Scientific Q-Exactive mass spectrometer. Plasmid pKPY514 coding for different subunits of E. coli phenylalanyl-tRNA synthetase (Ec PheRS) was a gift from D. Tirrell (Addgene plasmid #62598; http://n2t.net/addgene:62598; RRID: Addgene_62598) (60). Superdex 75 and sulfopropyl-Sepharose columns were purchased from GE Healthcare Life Sciences, USA. The nonhydrolyzable analogs of aa-tRNAs were purchased from Jena Biosciences, Germany.
Cloning, expression, and purification
Genes encoding DTD2s of P. horikoshii (Pho), M. jannaschii (Mj), and A. fulgidus (Af) and tyrosyl-tRNA synthetase (TyrRS) of T. thermophilus (Tth) were polymerase chain reaction–amplified from their genomic DNA using appropriate forward and reverse primers. A. thaliana (At) DTD2 gene was amplified from the complementary DNA of A. thaliana. Genes were cloned into the pET28b vector using restriction-free cloning (61). Primers used in this study are listed in table S1. Pho DTD2, Af DTD2, At DTD2, and Tth TyrRS were transformed and overexpressed into E. coli BL21(DE3), Ec PheRS into E. coli M15, and Mj DTD2 into BL21CodonPlus (DE3)-RIL strain of E. coli. Purification of 6× His-tagged proteins (Mj DTD2-C-His, At DTD2-N-His, Tth TyrRS-N-His, and Ec PheRS-N-His) was performed by Ni-NTA (nitrilotriacetic acid) affinity chromatography, followed by size exclusion chromatography (SEC). SEC was performed by using a Superdex 75 column (GE Healthcare Life Sciences, USA). Purification method and buffers for Ni-NTA and SEC were used as described (14). Untagged proteins (Pho DTD2 and Af DTD2) were purified using cation exchange chromatography (CEC) followed by SEC. For CEC, cells were sonicated in a buffer containing 50 mM bis-tris (pH 6.5) and 20 mM NaCl. The lysate was heated at 70°C for 30 min before subjecting to centrifugation (18,000 rpm for 30 min at 4°C). The supernatant was subjected to CEC column, and proteins were eluted with a gradient of NaCl from 50 to 200 mM. Sulfopropyl-Sepharose (GE Healthcare Life Sciences, USA) column was used for CEC. A buffer containing 100 mM tris (pH 8.0), 200 mM NaCl, 5 mM 2-mercaptoethanol (β-ME), and 50% glycerol was used to store all the purified DTD2 proteins.
Generation of α-32P–labeled aa-tRNAs
All the tRNAs used in the study [E. coli (Ec) tRNAPhe, P. horikoshii (Pho) tRNAPhe, A. thaliana (At) tRNAPhe, T. thermophilus (Tth) tRNATyr, and Mus musculus (Mm) tRNAThr (G4:U69)] were generated using the MEGAshortscript T7 Transcription Kit (Thermo Fisher Scientific, USA). tRNAs were end-labeled with [α-32P]ATP (adenosine 5′-triphosphate) (BRIT-Jonaki, India) using CCA-adding enzyme (62). Phenylalanylation of tRNAPhe was done by incubating 1 μM tRNAPhe with 2 μM E. coli PheRS in a buffer containing 100 mM Hepes (pH 7.5), 10 mM KCl, 30 mM MgCl2, 50 μM d-Phe or 50 μM l-Phe, and 2 mM ATP at 37°C for 15 min. Tyrosylation of tRNATyr, alanylation, serylation, and threonylation of tRNAThr (G4:U69) were done as mentioned earlier (14, 63). Aminoacylations were assessed and quantified as discussed below.
Generation of adducts on aa-tRNAs and their analogs
Adducts on aa-tRNAs and their analogs were generated by two different methods, namely single-step and two-step methods (fig. S8E).
Single-step method (probing relative acetaldehyde sensitivities of biomolecules; acetaldehyde titration with aa-tRNA)
A single-step method was used to find relative acetaldehyde sensitivities of aa-tRNAs (acetaldehyde titration with aa-tRNA), aa-tRNA analogs, free amino acids, peptides, and nucleotides. In this method, 200 μM of each of aa-tRNA (Phe-tRNAPhe and Tyr-tRNATyr), nonhydrolyzable analogs (d-Tyr2AA and l-Val3AA), amino acids (d-Tyr and l-Val), peptides (peptides 1 and 2), and nucleotides [adenosine 5′-monophosphate (AMP) and guanosine 5′-monophosphate (GMP)] were incubated with different concentrations of acetaldehyde [200 μM, 1 mM, 2 mM, 20 mM, 200 mM, and 2 M (stocks were prepared in ethanol)] “along with 400 mM NaCNBH3” [stock was prepared in 100 mM potassium acetate (pH 5.4)] at 37°C for 30 min. Except for aa-tRNA, all the other samples were subjected to Eppendorf 5305 Vacufuge plus Concentrator to concentrate the sample. Samples were characterized using MS without any further processing. The method for processing and quantification of modification on aa-tRNA is discussed in detail below.
Two-step method (generation of substrates for deacylation and thin-layer chromatography)
A two-step method was used to generate ethyl modification on the aa-tRNAs that were used for deacylation assays. The final concentration of radiolabeled substrates (modified and unmodified aa-tRNAs) required for deacylation assays was 200 nM. Performing modification reactions with aa-tRNAs at such a low concentration of acetaldehyde (200 nM) was problematic because of its highly volatile nature. On the other hand, raising aa-tRNA concentration to 200 μM (concentrations used in single-step method) or more for each radiolabeled substrate that we used for biochemical assays is resource-intensive, and a large part of it will be unused because the final substrate concentration used for deacylation assays is only 200 nM. On the basis of these considerations, we used a two-step method to generate modification on aa-tRNA that gives maximum modification consistently. Substrates with homogeneous and maximum modification are a prerequisite to perform deacylation assays.
In the two-step method, acetaldehyde and reducing agents were added to aa-tRNAs or aa-tRNA analogs in two discrete steps. In the first step, 2 μM aa-tRNAs or 100 μM aa-tRNA analogs (used as a control) were incubated with 20 mM acetaldehyde at 37°C for 30 min. Before adding the reducing agent, the samples were subjected to Eppendorf 5305 Vacufuge plus Concentrator to “remove excess acetaldehyde” from the reaction. In the second step, the above “mixture was reduced with 400 mM NaCNBH3” by further incubating at 37°C for 30 min. All the reactions were performed on a shaking incubator operated at 300 rpm. Acetaldehyde-treated and untreated analogs were characterized using MS without any further processing. In contrast, acetaldehyde-treated and untreated aa-tRNAs were ethanol-precipitated by overnight incubation at −30°C to remove salts and metal ions. After ethanol precipitation, pellets were resuspended in 5 mM sodium acetate (pH 5.4), and the same pellets were used for deacylation assays.
To check the aminoacylation and modification status, reaction mixtures were assessed before the ethanol precipitation step. One microliter of the reaction mixtures of modified aa-tRNAs or unmodified aa-tRNAs (control) was mixed with 2.5 μl of S1 nuclease (2 U/μl) (Thermo Fisher Scientific, USA) and incubated for 30 min at 22°C. One microliter of S1 nuclease digested samples was spotted on to cellulose F TLC plates (Merck KGaA, Germany). These TLC plates were developed using a mobile phase consisting of 100 mM ammonium chloride and 5% glacial acetic acid; the mobile phase front was allowed to move up to about three-fourth of the vertical length of the TLC plate before air-drying the plate. The developed TLC plates were exposed overnight to imaging plates (Fujifilm, Japan). Typhoon FLA 9000 biomolecular imager (GE Healthcare) was used for phosphorimaging of the exposed image plates. S1 nuclease is an endonuclease that hydrolyzes single-stranded RNA into mononucleotides. S1 digestion of tRNA therefore results in hydrolysis of single-stranded regions such as loops and the 3′-CCA end to free nucleotides. AMP, aa-AMP, and modified aa-AMP were visualized on TLC after phosphorimaging because the 3′-terminal nucleotide A76 was labeled with α-32P. Hence, AMP, aa-AMP, and modified aa-AMP that appear on TLC correspond to free/deacylated tRNA, aa-tRNA, and modified aa-tRNA, respectively (62). Percentages of aminoacylation and modification were assessed using Image Gauge V4.0 software. Protocol for acetaldehyde modification assay was generated and optimized from the insights obtained from the earlier studies (24, 64). N-acetyl-aa-tRNAs were generated and quantified as described (31).
Deacylation assays
Deacylation experiments were performed by incubating different enzymes (PTHs, DTD1s, and DTD2s) of varying concentrations (as mentioned in the figures) with 0.2 μM of different substrates (α-32P–labeled modified or unmodified aa-tRNAs) in a buffer containing 20 mM tris (pH 7.2), 5 mM MgCl2, 5 mM dithiothreitol (DTT), and bovine serum albumin (0.2 mg/ml) at 37°C. One microliter of the reaction mixture was withdrawn at different time points and subjected to S1 digestion (the remaining steps are the same to those discussed in the “Generation of adducts on aa-tRNAs and their analogs” section).
The percentage of aa-AMP or modified aa-AMP present at 0-min time point was considered as 100%. The percentage of aa-AMP or modified aa-AMP remaining after deacylation was plotted against the respective deacylation time points.
Alkali treatment
Aa-tRNAs, acetaldehyde-modified aa-tRNAs, and N-acetyl-aa-tRNAs were initially digested with S1 nuclease as mentioned earlier. S1-digested samples were later subjected to alkali treatment because alkaline conditions are incompatible with S1 digestion. Alkali treatment was carried out by incubating 100 nM S1-digested sample with 100 mM tris (pH 9.0) at 37°C. One microliter of the alkali-treated samples was directly spotted on to a TLC plate at different time intervals, and the latter steps were followed as described above (in the “Generation of adducts on aa-tRNAs and their analogs” section). Half-life values were determined by using GraphPad Prism software by fitting the data points on to the curve according to the first-order exponential decay equation [St] = [S0]e−kt, in which the substrate concentration at time t is denoted as [St], [S0] is the concentration of the substrate at time 0, and k is the first-order decay constant.
Generation and characterization of NEDATs in E. coli
E. coli K12Δdtd::Kan cells were used to establish a surrogate system. Initially, primary cultures were grown at 37°C in 1× minimal salts with 0.2% maltose and kanamycin (50 μg/ml) (minimal medium). One percent of overnight-grown primary culture was inoculated to grow 400-ml secondary cultures (minimal medium with or without 3 mM d-tyrosine). Cultures were grown until OD650 (optical density at 650 nm) reached to 0.8 [it was shown that at 0.8 OD650, of the total Tyr-tRNATyr pool, 40% was d-tyrosylated in the case of d-tyrosine–treated E. coli Δdtd cells (65)]. Two hundred milliliters of each secondary culture was used for RNA isolation, and all the samples were treated with 0.5 mM NaCNBH3 for 30 min at 37°C before RNA isolation. In the case of acetaldehyde-treated samples, before RNA isolation, cultures were additionally treated with either 0.005% (v/v) or 0.01% (v/v) acetaldehyde along with the reducing agent at 37°C for 30 min. Alkali treatment [5 mM CuSO4 + 50 mM tris (pH 8.0)] was used to find out the formation of NEATs in acetaldehyde-treated cells {(Aa-tRNAs and modified aa-tRNAs co-migrate on the acid-urea polyacrylamide gel electrophoresis (PAGE). Hence, CuSO4 + tris (8.0) treatment was carried out to distinguish modified aa-tRNAs from aa-tRNAs. Similar to alkaline treatment [100 mM tris (pH 9.0)], only aa-tRNAs are readily deacylated but not modified aa-tRNAs with CuSO4 + tris (8.0) treatment. Retention (no hydrolysis) was only observed when cultures were grown in the presence of both d-amino acids and acetaldehyde that suggests accumulation of NEDATs}. A similar method was used in the case of N-acetyl-aa-tRNAs and peptidyl-tRNAs (66). Deacylations with both plant and archaeal DTD2s (similar to in vitro deacylation assays) were done to characterize the NEDATs. RNA (0.25 absorbance at 260 nm unit) isolated from the different samples was subjected to alkali treatment or deacylation with DTD2. After deacylation, RNA samples were analyzed by Northern blotting, for which the samples were first fractionated on 6.5% acid-urea polyacrylamide gel (denaturing PAGE analysis) for 20 to 24 hours at 4°C (67). The gel was then subjected to electroblotting on Hybond+ membrane at 16 V, 5 A for 40 min. A 5′-end labeled DNA primer complementary to the anticodon loop of tRNATyrGTA was used as a hybridizing probe (probe sequence: GGCAGATTTACAGTCTGCTC). Radioactive 5′-end labeling of the probe was done with [γ-32P]ATP (BRIT-Jonaki, India) using polynucleotide kinase (NEB) in a reaction as recommended by the manufacturer. The hybridized blot was exposed overnight to image plate followed by phosphorimaging and quantification as mentioned above (in the “Generation of adducts on aa-tRNAs and their analogs” section).
EF-Tu protection assay
EF-Tu from T. thermophilus was activated using a protocol, as mentioned earlier (34). Due to lower activation efficiency (10 to 15%), the effective concentration of activated EF-Tu was assumed to be around 200 to 300 nM in a reaction mix containing 2 μM EF-Tu (68). EF-Tu protection assays were performed at 37°C in a reaction mix containing 2 μM activated EF-Tu or 2 μM unactivated EF-Tu, 0.2 μM l-aa-tRNA or d-aa-tRNA ([32P]-labeled) in 100 mM Hepes (pH 7.2), 2.5 mM DTT with 2 mM acetaldehyde, and 20 mM NaCNBH3. A total of 1.5 μl of the reaction mixture was taken out at various time points and immediately mixed with an equal volume of 250 mM l-cysteine at 37°C to quench the unreacted acetaldehyde. These samples were subjected to S1 nuclease digestion, and the latter steps for TLC were followed as discussed above (in the “Generation of adducts on aa-tRNAs and their analogs” section).
Mass spectrometry
To characterize modification status on aa-tRNAs (titrated with acetaldehyde), modified and unmodified aa-tRNAs (1 mM) were resuspended in 30-μl diethyl pyrocarbonate–treated water and mixed with 60 μl of aqueous ammonia [25% of (v/v) NH4OH] and incubated at 70°C for 18 hours. This treatment resulted in hydrolysis of modified and unmodified amino acids and nucleotides from modified and unmodified aa-tRNAs. Hydrolyzed samples were further concentrated with Eppendorf 5305 Vacufuge plus Concentrator. All the samples (including analogs) were dissolved in water containing 10% methanol and 1% acetic acid. They were subjected to ESI-MS using a Q-Exactive mass spectrometer (Thermo Fisher Scientific) by infusing through heated electrospray ionization source operating at a positive voltage of 3.5 kV. The method of targeted selected ion monitoring (t-SIM) [with an isolation width of 2 m/z (mass/charge ratio) and an inclusion list of theoretical m/z of the MH+ ion species] was used to acquire the mass spectra (at a resolving power of 70,000 at 200 m/z). The high-energy collision-induced MS-MS spectra with a normalized collision energy of 25 of the selected precursor ion species specified in the inclusion list (having the observed m/z value from the earlier t-SIM analysis) were acquired using the method of t-SIM-ddMS2 (at an isolation window of 1 m/z or 0.5 m/z (in cases of AMP and GMP) at a ddMS2 resolving power of 35,000 at 200 m/z).
Bioinformatic analysis
DTD2 protein sequences were retrieved from the National Center for Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov/) (RRID: SCR_006472). The list of plants and archaea, whose genomes had been completely sequenced, were obtained from the KEGG GENOME database (www.genome.jp/kegg/genome.html) (RRID: SCR_012773). Besides, C. atmophyticus, K. nitens, and C. braunii were also included in this study. C. atmophyticus DTD2 gene was predicted using Augustus online server (69) (http://augustus.gobics.de/) with A. thaliana DTD2 reference. Sequence identity values were generated for 80 plant and 140 archaeal DTD2 sequences using Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo/). Box plot was plotted with percentage identity to all plant DTD2 sequences on the horizontal axis and respective archaea on the vertical axis. For control, 290 plant and 500 archaeal sequences of TopVIB were retrieved from UniProt, by blast search using Arabidopsis TopVIB as a query. All TopVIB sequences were used for box plot analysis. T-coffee (http://tcoffee.crg.cat/) (RRID: SCR_011818) server was used to prepare structure-based multiple sequence alignment of DTD2, and the corresponding figure was generated using ESPript 3.0 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). To figure out the origins of genes/processes, which were essential for terrestrialization of plants, we retrieved the protein sequences of the genes involved in various processes (42, 43) from the NCBI (www.ncbi.nlm.nih.gov/) and manually performed protein BLAST-based in silico search against genome sequences of Bacteria and Archaea. We used query coverage (70%) and percentage identity (20%) as a cutoff.
To construct phylogenetic trees, representative DTD2 sequences were selected to maximize the phylogenetic and taxonomic diversity while keeping a minimum number of sequences. These sequences were subjected for multiple sequence alignment using MAFFT server (70) (https://mafft.cbrc.jp/alignment/server/), and alignment was then used to build a phylogenetic tree using IQ-Tree version 2.0.4 (based on maximum likelihood) (71) and MrBayes version 3.2.7 (72). In the IQ-Tree, we have used the default model search option by specifying -m MFP+MERGE -alrt 1000 -bb 1000 -abayes. Here, “MFP+MERGE” looks for the best model by using the Bayesian Information Criterion (BIC). According to the BIC score, of 546 protein models, LG+R5 was chosen as the best amino acid substitution model for the given set multiple sequence alignment, which is used to construct a phylogenetic tree. Approximate likelihood ratio test (aLRT), bootstrap, and abayes were used to validate further and support the phylogenetic tree. MrBayes has ran with default parameters (prset aamodelpr = mixed, mcmc nchains = 1 ngen = 300,000). Clades with a bootstrap value above 90% were well supported, whereas values more than 70% are moderately supported, and the clades with bootstrap values less than 50% were considered unresolved. Web server iTOL (73) (http://itol.embl.de) was used to visualize the phylogenetic tree.
Statistical analysis
All the experiments were performed in triplicates (unless otherwise mentioned). The mean values were used to plot the graphs, and each error bar denotes the SD from the mean value of three independent observations.
Acknowledgments
We acknowledge R. Nagaraj, CSIR-CCMB, for fruitful discussions and providing peptides for MS experiments and S. Raghavan, CSIR-IICT, for insightful discussions on the acetaldehyde modification experiments. Funding: M.M. thanks the Department of Biotechnology, India for research fellowship. R.Si. thanks SERB-NPDF. P.K. thanks CSIR for research fellowship. R.Sa. acknowledges the health care theme project of CSIR, India; J.C. Bose Fellowship of SERB, India; and Centre of Excellence Project of Department of Biotechnology, India. Author contributions: M.M., R.Si., P.K., A.R., S.P.K., and B.R. designed and performed the experiments. R.Sa. conceived and supervised the study. All the authors analyzed the data. M.M., R.Si., and R.Sa. wrote the manuscript with help from P.K., and all the authors reviewed it. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/6/eabe8890/DC1
REFERENCES AND NOTES
- 1.Kenrick P., Crane P. R., The origin and early evolution of plants on land. Nature 389, 33–39 (1997). [Google Scholar]
- 2.Xu B., Ohtani M., Yamaguchi M., Toyooka K., Wakazaki M., Sato M., Kubo M., Nakano Y., Sano R., Hiwatashi Y., Murata T., Kurata T., Yoneda A., Kato K., Hasebe M., Demura T., Contribution of NAC transcription factors to plant adaptation to land. Science 343, 1505–1508 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Drew M. C., Oxygen deficiency and root metabolism: Injury and acclimation under hypoxia and anoxia. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 48, 223–250 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Good A. G., Muench D. G., Long-term anaerobic metabolism in root tissue: Metabolic products of pyruvate metabolism. Plant Physiol. 101, 1163–1168 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kennedy R. A., Rumpho M. E., Fox T. C., Anaerobic metabolism in plants. Plant Physiol. 100, 1–6 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reynoso M. A., Kajala K., Bajic M., West D. A., Pauluzzi G., Yao A. I., Hatch K., Zumstein K., Woodhouse M., Rodriguez-Medina J., Sinha N., Brady S. M., Deal R. B., Bailey-Serres J., Evolutionary flexibility in flooding response circuitry in angiosperms. Science 365, 1291–1295 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreuzwieser J., Scheerer U., Rennenberg H., Metabolic origin of acetaldehyde emitted by poplar (Populus tremula × P. alba) trees. J. Exp. Bot. 50, 757–765 (1999). [Google Scholar]
- 8.Tadege M., Dupuis I., Kuhlemeier C., Ethanolic fermentation: New functions for an old pathway. Trends Plant Sci. 4, 320–325 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Cui X., Wise R. P., Schnable P. S., The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. Science 272, 1334–1336 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Pesis E., Ng T. J., The role of anaerobic respiration in germinating muskmelon seeds: II. Effect of anoxia treatment and alcohol dehydrogenase activity. J. Exp. Bot. 35, 366–372 (1984). [Google Scholar]
- 11.Small J. G. C., Potgieter G. P., Botha F. C., Anoxic seed germination of Erythrina caffra: Ethanol fermentation and response to metabolic inhibitors. J. Exp. Bot. 40, 375–381 (1989). [Google Scholar]
- 12.Tadege M., Bucher M., Stähli W., Suter M., Dupuis I., Kuhlemeier C., Activation of plant defense responses and sugar efflux by expression of pyruvate decarboxylase in potato leaves. Plant J. 16, 661–671 (1998). [Google Scholar]
- 13.Michard E., Lima P. T., Borges F., Catarina Silva A., Teresa Portes M., Carvalho J. E., Gilliham M., Liu L. H., Obermeyer G., Feijó J. A., Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil d-serine. Science 332, 434–437 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Ahmad S., Routh S. B., Kamarthapu V., Chalissery J., Muthukumar S., Hussain T., Kruparani S. P., Deshmukh M. V., Sankaranarayanan R., Mechanism of chiral proofreading during translation of the genetic code. eLife 2, e01519 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calendar R., Berg P., D-tyrosyl RNA: Formation, hydrolysis and utilization for protein synthesis. J. Mol. Biol. 26, 39–54 (1967). [DOI] [PubMed] [Google Scholar]
- 16.Ferri-Fioni M. L., Fromant M., Bouin A. P., Aubard C., Lazennec C., Plateau P., Blanquet S., Identification in archaea of a novel d-Tyr-tRNATyr deacylase. J. Biol. Chem. 281, 27575–27585 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Wydau S., Ferri-Fioni M.-L., Blanquet S., Plateau P., GEK1, a gene product of Arabidopsis thaliana involved in ethanol tolerance, is a d-aminoacyl-tRNA deacylase. Nucleic Acids Res. 35, 930–938 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wydau S., van der Rest G., Aubard C., Plateau P., Blanquet S., Widespread distribution of cell defense against d-aminoacyl-tRNAs. J. Biol. Chem. 284, 14096–14104 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirayama T., Fujishige N., Kunii T., Nishimura N., Iuchi S., Shinozaki K., A novel ethanol-hypersensitive mutant of Arabidopsis. Plant Cell Physiol. 45, 703–711 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Fujishige N., Nishimura N., Iuchi S., Kunii T., Shinozaki K., Hirayama T., A novel Arabidopsis gene required for ethanol tolerance is conserved among plants and Archaea. Plant Cell Physiol. 45, 659–666 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Seitz H. K., Stickel F., Molecular mechanisms of alcohol-mediated carcinogenesis. Nat. Rev. Cancer 7, 599–612 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Fang J., Vaca C., Detection of DNA adducts of acetaldehyde in peripheral white blood cells of alcohol abusers. Carcinogenesis 18, 627–632 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Matsuda T., Terashima I., Matsumoto Y., Yabushita H., Matsui S., Shibutani S., Effective utilization of N2-Ethyl-2′-deoxyguanosine triphosphate during DNA synthesis catalyzed by mammalian replicative DNA polymerases. Biochemistry 38, 929–935 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Fang J. L., Vaca C. E., Development of a 32P-postlabelling method for the analysis of adducts arising through the reaction of acetaldehyde with 2′-deoxyguanosine-3′-monophosphate and DNA. Carcinogenesis 16, 2177–2185 (1995). [DOI] [PubMed] [Google Scholar]
- 25.Carlsson H., Von Stedingk H., Nilsson U., Törnqvist M., LC-MS/MS screening strategy for unknown adducts to N-terminal valine in hemoglobin applied to smokers and nonsmokers. Chem. Res. Toxicol. 27, 2062–2070 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Langevin F., Crossan G. P., Rosado I. V., Arends M. J., Patel K. J., Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475, 53–58 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Garaycoechea J. I., Crossan G. P., Langevin F., Mulderrig L., Louzada S., Yang F., Guilbaud G., Park N., Roerink S., Nik-Zainal S., Stratton M. R., Patel K. J., Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. Nature 553, 171–177 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hadačová V., Plant life in anaerobic environments. Biol. Plant. 21, 480–480 (1979). [Google Scholar]
- 29.Malki E., Waisel Y., Pesis E., Effects of flooding under hydrostatic pressure on the ‘respiratory metabolism of germinated wheat seeds. Physiol. Plant. 77, 519–524 (1989). [Google Scholar]
- 30.Perata P., Alpi A., Ethanol-induced injuries to carrot cells: The role of acetaldehyde. Plant Physiol. 95, 748–752 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker S. E., Fredrick K., Preparation and evaluation of acylated tRNAs. Methods 44, 81–86 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atherly A. G., Peptidyl-transfer RNA hydrolase prevents inhibition of protein synthesis initiation. Nature 275, 769 (1978). [DOI] [PubMed] [Google Scholar]
- 33.Menninger J. R., Accumulation of peptidyl tRNA is lethal to Escherichia coli. J. Bacteriol. 137, 694–696 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Routh S. B., Pawar K. I., Ahmad S., Singh S., Suma K., Kumar M., Kuncha S. K., Yadav K., Kruparani S. P., Sankaranarayanan R., Elongation factor Tu prevents misediting of Gly-tRNA(Gly) caused by the design behind the chiral proofreading site of d-aminoacyl-tRNA deacylase. PLOS Biol. 14, e1002465 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamane T., Miller D. L., Hopfield J. J., Discrimination between d- and l-tyrosyl transfer ribonucleic acids in peptide chain elongation. Biochemistry 20, 7059–7064 (1981). [DOI] [PubMed] [Google Scholar]
- 36.Kuncha S. K., Kruparani S. P., Sankaranarayanan R., Chiral checkpoints during protein biosynthesis. J. Biol. Chem. 294, 16535–16548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nissen P., Kjeldgaard M., Thirup S., Polekhina G., Reshetnikova L., Clark B. F., Nyborg J., Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270, 1464–1472 (1995). [DOI] [PubMed] [Google Scholar]
- 38.Ma K., Hutchins A., Sung S. J. S., Adams M. W. W., Pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon, Pyrococcus furiosus, functions as a CoA-dependent pyruvate decarboxylase. Proc. Natl. Acad. Sci. U.S.A. 94, 9608–9613 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eram M. S., Oduaran E., Ma K., The bifunctional pyruvate decarboxylase/pyruvate ferredoxin oxidoreductase from Thermococcus guaymasensis. Archaea 2014, e349379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Livingstone D. R., Origins and evolution of pathways of anaerobic metabolism in the Animal kingdom. Am. Zool. 31, 522–534 (1991). [Google Scholar]
- 41.Muller M., Mentel M., van Hellemond J. J., Henze K., Woehle C., Gould S. B., Yu R.-Y., van der Giezen M., Tielens A. G. M., Martin W. F., Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 76, 444–495 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S., Li L., Li H., Sahu S. K., Wang H., Xu Y., Xian W., Song B., Liang H., Cheng S., Chang Y., Song Y., Çebi Z., Wittek S., Reder T., Peterson M., Yang H., Wang J., Melkonian B., Van de Peer Y., Xu X., Wong G. K. S., Melkonian M., Liu H., Liu X., Genomes of early-diverging streptophyte algae shed light on plant terrestrialization. Nat. Plants 6, 95–106 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hori K., Maruyama F., Fujisawa T., Togashi T., Yamamoto N., Seo M., Sato S., Yamada T., Mori H., Tajima N., Moriyama T., Ikeuchi M., Watanabe M., Wada H., Kobayashi K., Saito M., Masuda T., Sasaki-Sekimoto Y., Mashiguchi K., Awai K., Shimojima M., Masuda S., Iwai M., Nobusawa T., Narise T., Kondo S., Saito H., Sato R., Murakawa M., Ihara Y., Oshima-Yamada Y., Ohtaka K., Satoh M., Sonobe K., Ishii M., Ohtani R., Kanamori-Sato M., Honoki R., Miyazaki D., Mochizuki H., Umetsu J., Higashi K., Shibata D., Kamiya Y., Sato N., Nakamura Y., Tabata S., Ida S., Kurokawa K., Ohta H., Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 5, 3978 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishiyama T., Sakayama H., de Vries J., Buschmann H., Saint-Marcoux D., Ullrich K. K., Haas F. B., Vanderstraeten L., Becker D., Lang D., Vosolsobě S., Rombauts S., Wilhelmsson P. K. I., Janitza P., Kern R., Heyl A., Rümpler F., Villalobos L. I. A. C., Clay J. M., Skokan R., Toyoda A., Suzuki Y., Kagoshima H., Schijlen E., Tajeshwar N., Catarino B., Hetherington A. J., Saltykova A., Bonnot C., Breuninger H., Symeonidi A., Radhakrishnan G. V., Van Nieuwerburgh F., Deforce D., Chang C., Karol K. G., Hedrich R., Ulvskov P., Glöckner G., Delwiche C. F., Petrášek J., Van de Peer Y., Friml J., Beilby M., Dolan L., Kohara Y., Sugano S., Fujiyama A., Delaux P. M., Quint M., Theißen G., Hagemann M., Harholt J., Dunand C., Zachgo S., Langdale J., Maumus F., Van Der Straeten D., Gould S. B., Rensing S. A., The Chara genome: Secondary complexity and implications for plant terrestrialization. Cell 174, 448–464.e24 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Cheng S., Xian W., Fu Y., Marin B., Keller J., Wu T., Sun W., Li X., Xu Y., Zhang Y., Wittek S., Reder T., Günther G., Gontcharov A., Wang S., Li L., Liu X., Wang J., Yang H., Xu X., Delaux P. M., Melkonian B., Wong G. K. S., Melkonian M., Genomes of subaerial Zygnematophyceae provide insights into land plant evolution. Cell 179, 1057–1067.e14 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Bowman J. L., Kohchi T., Yamato K. T., Jenkins J., Shu S., Ishizaki K., Yamaoka S., Nishihama R., Nakamura Y., Berger F., Adam C., Aki S. S., Althoff F., Araki T., Arteaga-Vazquez M. A., Balasubrmanian S., Barry K., Bauer D., Boehm C. R., Briginshaw L., Caballero-Perez J., Catarino B., Chen F., Chiyoda S., Chovatia M., Davies K. M., Delmans M., Demura T., Dierschke T., Dolan L., Dorantes-Acosta A. E., Eklund D. M., Florent S. N., Flores-Sandoval E., Fujiyama A., Fukuzawa H., Galik B., Grimanelli D., Grimwood J., Grossniklaus U., Hamada T., Haseloff J., Hetherington A. J., Higo A., Hirakawa Y., Hundley H. N., Ikeda Y., Inoue K., ichiro Inoue S., Ishida S., Jia Q., Kakita M., Kanazawa T., Kawai Y., Kawashima T., Kennedy M., Kinose K., Kinoshita T., Kohara Y., Koide E., Komatsu K., Kopischke S., Kubo M., Kyozuka J., Lagercrantz U., Lin S. S., Lindquist E., Lipzen A. M., Lu C. W., De Luna E., Martienssen R. A., Minamino N., Mizutani M., Mizutani M., Mochizuki N., Monte I., Mosher R., Nagasaki H., Nakagami H., Naramoto S., Nishitani K., Ohtani M., Okamoto T., Okumura M., Phillips J., Pollak B., Reinders A., Rövekamp M., Sano R., Sawa S., Schmid M. W., Shirakawa M., Solano R., Spunde A., Suetsugu N., Sugano S., Sugiyama A., Sun R., Suzuki Y., Takenaka M., Takezawa D., Tomogane H., Tsuzuki M., Ueda T., Umeda M., Ward J. M., Watanabe Y., Yazaki K., Yokoyama R., Yoshitake Y., Yotsui I., Zachgo S., Schmutz J., Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304.e15 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Bergerat A., De Massy B., Gadelle D., Varoutas P. C., Nicolas A., Forterre P., An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature 386, 414–417 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Hartung F., Angelis K. J., Meister A., Schubert I., Melzer M., Puchta H., An archaebacterial topoisomerase homolog not present in other eukaryotes is indispensable for cell proliferation of plants. Curr. Biol. 12, 1787–1791 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Hartung F., Puchta H., Molecular characterization of homologues of both subunits A (SPO11) and B of the archaebacterial topoisomerase 6 in plants. Gene 271, 81–86 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Yin Y., Cheong H., Friedrichsen D., Zhao Y., Hu J., Mora-Garcia S., Chory J., A crucial role for the putative Arabidopsis topoisomerase VI in plant growth and development. Proc. Natl. Acad. Sci. U.S.A. 99, 10191–10196 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taffner J., Erlacher A., Bragina A., Berg C., Moissl-Eichinger C., Berg G., What is the role of Archaea in plants? New insights from the vegetation of Alpine bogs. mSphere 3, e00122-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vranova V., Zahradnickova H., Janous D., Skene K. R., Matharu A. S., Rejsek K., Formanek P., The significance of D-amino acids in soil, fate and utilization by microbes and plants: Review and identification of knowledge gaps. Plant and Soil 354, 21–39 (2012). [Google Scholar]
- 53.Steemans P., Le Hérissé A., Melvin J., Miller M. A., Paris F., Verniers J., Wellman C. H., Origin and radiation of the earliest vascular land plants. Science 324, 353 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y., Xiao G., Wang X., Zhang X., Friml J., Evolution of fast root gravitropism in seed plants. Nat. Commun. 10, 3480 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu R. A., Semlow D. R., Kamimae-Lanning A. N., Kochenova O. V., Chistol G., Hodskinson M. R., Amunugama R., Sparks J. L., Wang M., Deng L., Mimoso C. A., Low E., Patel K. J., Walter J. C., TRAIP is a master regulator of DNA interstrand crosslink repair. Nature 567, 267–272 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shakeel S., Rajendra E., Alcón P., O’Reilly F., Chorev D. S., Maslen S., Degliesposti G., Russo C. J., He S., Hill C. H., Skehel J. M., Scheres S. H. W., Patel K. J., Rappsilber J., Robinson C. V., Passmore L. A., Structure of the Fanconi anaemia monoubiquitin ligase complex. Nature 575, 234–237 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hodskinson M. R., Bolner A., Sato K., Kamimae-Lanning A. N., Rooijers K., Witte M., Mahesh M., Silhan J., Petek M., Williams D. M., Kind J., Chin J. W., Patel K. J., Knipscheer P., Alcohol-derived DNA crosslinks are repaired by two distinct mechanisms. Nature 579, 603–608 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dwivedi S., Kruparani S. P., Sankaranarayanan R., A D-amino acid editing module coupled to the translational apparatus in archaea. Nat. Struct. Mol. Biol. 12, 556–557 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Retallack G. J., Early forest soils and their role in Devonian global change. Science 276, 583–585 (1997). [DOI] [PubMed] [Google Scholar]
- 60.Yuet K. P., Doma M. K., Ngo J. T., Sweredoski M. J., Graham R. L. J., Moradian A., Hess S., Schuman E. M., Sternberg P. W., Tirrell D. A., Cell-specific proteomic analysis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 112, 2705–2710 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Den Ent F., Löwe J., RF cloning: A restriction-free method for inserting target genes into plasmids. J. Biochem. Biophys. Methods 67, 67–74 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Ledoux S., Uhlenbeck O. C., [3′-32P]-labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods 44, 74–80 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuncha S. K., Mazeed M., Singh R., Kattula B., Routh S. B., Sankaranarayanan R., A chiral selectivity relaxed paralog of DTD for proofreading tRNA mischarging in Animalia. Nat. Commun. 9, 511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Merryman C., Green R., Transformation of aminoacyl tRNAs for the in vitro selection of “drug-like” molecules. Chem. Biol. 11, 575–582 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Soutourina O., Soutourina J., Blanquet S., Plateau P., Formation of d-tyrosyl-tRNATyr accounts for the toxicity of d-tyrosine toward Escherichia coli. J. Biol. Chem. 279, 42560–42565 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Li Y., Holmes W. B., Appling D. R., RajBhandary U. L., Initiation of protein synthesis in Saccharomyces cerevisiae mitochondria without formylation of the initiator tRNA. J. Bacteriol. 182, 2886–2892 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Varshney U., Lee C. P., RajBhandary U. L., Direct analysis of aminoacylation levels of tRNAs in vivo: Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem. 266, 24712–24718 (1991). [PubMed] [Google Scholar]
- 68.Cvetesic N., Palencia A., Halasz I., Cusack S., Gruic-Sovulj I., The physiological target for Leu RS translational quality control is norvaline. EMBO J. 33, 1639–1653 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stanke M., Steinkamp R., Waack S., Morgenstern B., AUGUSTUS: A web server for gene finding in eukaryotes. Nucleic Acids Res. 32, 309–312 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nguyen L. T., Schmidt H. A., Von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., Larget B., Liu L., Suchard M. A., Huelsenbeck J. P., MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Letunic I., Bork P., Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/6/eabe8890/DC1


