Cancer-associated fibroblast assists cancer cell in building rampart to block T cell infiltration and resist anti–PD-1 drugs.
Abstract
PD-1/PD-L1 blockade therapies provide notable clinical benefits for patients with advanced cancers, but the factors influencing the effectiveness of the treatment remain incompletely cataloged. Here, the up-regulation of laminin γ2 (Ln-γ2) predicted the attenuated efficacy of anti–PD-1 drugs and was associated with unfavorable outcomes in patients with lung cancer or esophageal cancer. Furthermore, Ln-γ2 was transcriptionally activated by transforming growth factor–β1 (TGF-β1) secreted from cancer-associated fibroblasts via JNK/AP1 signaling, which blocked T cell infiltration into the tumor nests by altering the expression of T cell receptors. Coadministration of the TGF-β receptor inhibitor galunisertib and chemotherapy drugs provoked vigorous antitumor activity of anti–PD-1 therapy in mouse tumor models. Therefore, Ln-γ2 may represent a useful biomarker to optimize clinical decisions and predict the response of cancer patients to treatment with anti–PD-1 drugs.
INTRODUCTION
Immune checkpoint blockade therapies have improved the survival outcomes of patients with advanced non–small cell lung cancer (NSCLC) (1) or esophageal squamous cell carcinoma (ESCC) (2). Programmed cell death protein 1 (PD-1) is an inducible receptor expressed on the surface of several immune cells, such as natural killer (NK) cells, T cells, B cells, monocytes, and dendritic cells. Programmed death-ligand 1 (PD-L1) is released by stromal and tumor cells (3, 4). The PD-1/PD-L1 immune checkpoint plays a key role in preventing autoimmunity and provides immune escape for tumor cells by inactivating cytotoxic immunocytes. Because tumor cells are able to evade antitumor immunity by exploiting these immune checkpoints, the administration of anti–PD-1 drugs, such as pembrolizumab and nivolumab, is now the standard therapy for patients with NSCLC (5, 6). However, most patients do not benefit from anti–PD-1 therapy in clinical practice. Therefore, useful clinical biomarkers are needed to predict the response of cancer patients to treatment with anti–PD-1 drugs, and the underlying mechanisms of resistance to anti–PD-1 therapy must be elucidated.
Now, patient selection is based on the expression of PD-1/PD-L1 detected using immunohistochemical staining or the tumor mutational burden tested by nucleic acid sequencing to increase the efficacy of anti–PD-1 immunotherapy (7, 8). Despite their analytical and predictive limitations, PD-1/PD-L1 expression and the tumor mutational burden are currently the main biomarkers used to optimize clinical decisions regarding treatment with immune checkpoint inhibitors. The albumin-globulin ratio was recently identified as a predictive biomarker of the antitumor effect of anti–PD-1 therapy on patients with NSCLC (9). In addition, de Vries et al. (10) distinguished responders and progressors by performing electronic nose analysis of the exhaled breath from patients with NSCLC who were treated with pembrolizumab or nivolumab. Signatures of T cell dysfunction and exclusion might more accurately predict the response of patients with melanoma to treatment of anti–PD-1 or anti-CTLA4 drugs than other biomarkers, such as PD-1/PD-L1 levels and the tumor mutational burden (11). According to a recent study, the lack of response to anti–PD-L1 therapy (atezolizumab) by patients with urothelial cancer is associated with a signature of transforming growth factor–β (TGF-β) signaling in cancer-associated fibroblasts (CAFs) (12). Moreover, increased TGF-β1 level in the tumor microenvironment represents a primary mechanism of immune evasion that promotes T cell exclusion and blocks acquisition of the T helper 1 (TH1) effector phenotype in colon cancer (13). In a mouse model, the therapeutic combination of anti–PD-L1 and anti–TGF-β1 drugs reduced TGF-β signaling in stromal cells, facilitated T cell penetration into the tumor parenchyma, and promoted tumor regression (12–14). On the basis of these findings, TGF-β signaling provides cross-talk between tumor cells and stromal cells and is involved in the acquired ability to resist antitumor immunotherapy, but the mechanisms have not been clearly elucidated.
As a secreted pleiotropic cytokine, TGF-β1 regulates cell growth, proliferation, differentiation, and apoptosis (15). In cancer, TGF-β1 exerts a protumorigenic effect on promoting the epithelial-mesenchymal transition (EMT) of tumor cells, angiogenesis, and metastasis, which are associated with a poor prognosis in patients with many cancers (16–18). In addition, TGF-β1 also plays a critical role in immune suppression within the tumor microenvironment by promoting the expansion of T regulatory (Treg) cells, inhibiting the proliferation and function of effector T cells, antigen-presenting dendritic cells, and NK cells, and regulating the complex behavior of macrophages and neutrophils, thus forming a negative immune regulatory network (18, 19). Recent studies have revealed roles for TGF-β1 in tumor immune evasion and poor responses to cancer immunotherapy (20–22). A high level of TGF-β1 in the tumor microenvironment promotes T cell exclusion from the tumor parenchyma and attenuates the susceptibility of tumors to anti–PD-1/PD-L1 therapies, but the underlying mechanisms require further investigation to improve the efficiency of antitumor immunotherapy (12, 13, 23).
In the present study, CAF-secreted TGF-β1 substantially increased the expression of laminin γ2 (Ln-γ2; encoded by the LAMC2 gene) in NSCLC and ESCC cells via c-Jun N-terminal kinase (JNK)/activating protein 1 (AP1) signaling, which structured a protective barrier of tumor to block T cell penetration into tumor nests. Hence, aberrantly high expression of Ln-γ2 was significantly related to the attenuated response to anti–PD-1 blockade therapy, which was useful for predicting the unfavorable outcomes of patients with NSCLC or ESCC. Therapeutic coadministration of the TGF-β receptor (TGFBR) inhibitor galunisertib (LY2157299) with anti–PD-1 and chemotherapy drugs suppressed tumor progression in tumor-bearing mice, suggesting that treatments targeting Ln-γ2 by reducing TGF-β1 signaling in the tumor microenvironment represent a promising therapeutic strategy for improving the effect of anti–PD-1 therapy.
RESULTS
Cancer patients with high Ln-γ2 display a poor response to anti–PD-1 therapy
Increased TGF-β signaling in the tumor microenvironment has been reported to attenuate the susceptibility of tumor cells to anti–PD-1/PD-L1 therapies, but the mechanisms are unclear (13, 21, 22). RNA sequencing (RNA-seq) was applied to compare differentially expressed genes responsible for reduced susceptibility to the treatment in the human NSCLC cell line A549 and the ESCC cell line KYSE510 treated with or without the human recombinant protein TGF-β1 in vitro (0.5 ng/ml, 24 hours) (fig. S1A and datasheet S1). The Gene Ontology enrichment analysis showed that TGF-β1 signaling primarily mediated extracellular matrix (ECM) remodeling (fig. S1B). The expressions of ECM components, such as laminins, integrins, and collagens, were prominently increased upon TGF-β1 stimulation (fig. S1C). Here, Ln-γ2, a member of the laminin family, was specifically up-regulated by the TGF-β1 treatment in a concentration-dependent manner (fig. S1D). In addition, the up-regulation of Ln-γ2 induced by TGF-β1 could be completely blocked by the TGF-β receptor 1 (TGFBR1) inhibitor galunisertib in A549 and KYSE510 cells (fig. S1E). Moreover, an analysis of The Cancer Genome Atlas (TCGA) database also revealed a positive correlation between the levels of the Ln-γ2 and TGF-β1 mRNAs in human NSCLC and esophageal carcinoma tissues (fig. S2A), as well as in many other cancers (fig. S2B).
Next, the correlation between Ln-γ2 expression and the response to anti–PD-1 therapy was investigated in clinical NSCLC and ESCC specimens. First, analyses of the Gene Expression Omnibus (GEO) public sequencing data indicated a lower level of the Ln-γ2 mRNA in anti–PD-1 responders than in progressors with melanoma (GSE79691 and GSE78220) or urothelial cancer (GSE111636) (fig. S3A). In NSCLC or ESCC samples from patients treated with anti–PD-1 therapies, immunohistochemistry (IHC) staining showed higher expression of the Ln-γ2 in tumor tissues from progressors than in tissues from responders, as evaluated using computed tomography imaging (Fig. 1A and fig. S3B). Conversely, IHC staining with an antibody against CD3 and TUNEL (terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling) staining revealed higher proportions of T cells and apoptotic cells in the tumors from responders than in tumors from progressors treated with anti–PD-1 therapy (Fig. 1, B and C, and fig. S3C). In addition, high expression of Ln-γ2 predicts a longer treatment duration in NSCLC or ESCC patients treated with anti–PD-1 drugs than those with low level of Ln-γ2 (fig. S3D). On the basis of these findings, the level of Ln-γ2 expression in tumor tissues might be a promising biomarker for predicting the response of patients with NSCLC or ESCC to anti–PD-1 therapy.
Fig. 1. High Ln-γ2 expression predicts a diminished response of patients with NSCLC to anti–PD-1 therapy.
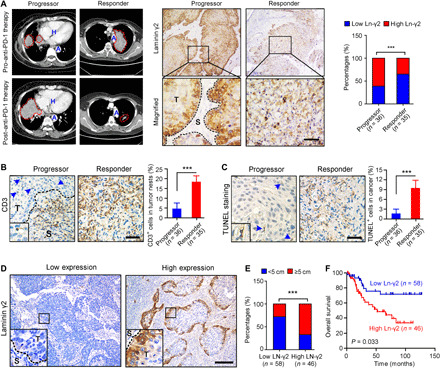
(A) Representative images of computed tomography scans and IHC staining in patients with NSCLC treated with anti–PD-1 (nivolumab) therapy showed significantly higher Ln-γ2 expression in tumor tissues from progressors (n = 36) than in tumor tissues from responders (n = 35). In the left panel, NSCLC tumors are indicated by red dotted lines. H, heart; A, aorta; T, tumor; S, stroma. Scale bar, 100 μm. (B) IHC staining with an antibody against CD3 to detect T cells in NSCLC tissues. Blue arrows indicate CD3+ cells in the tumor. Scale bar, 100 μm. (C) Apoptotic cells in NSCLC tissues from progressors and responders were detected using TUNEL staining. Blue arrows indicate the TUNEL+ cells. Scale bar, 100 μm. (D) Representative images of IHC staining for low or high Ln-γ2 expression in NSCLC tissues. Scale bar, 200 μm. (E) The correlation analysis revealed a positive correlation between high Ln-γ2 expression and the tumor size in patients with NSCLC (total n = 104). (F) Survival curves indicated that high expression of Ln-γ2 (n = 46) predicted shorter survival of patients with NSCLC compared with patients with low expression of Ln-γ2 (n = 58). The data presented in (A) and (E) were analyzed using Pearson’s chi-square (Fisher’s exact) test, statistical analyses of the data presented in (B) and (C) were performed using unpaired two-tailed t test with Welch’s correction, and the data presented in (F) were analyzed with the log-rank (Mantel-Cox) test. In all panels, ***P < 0.001.
High Ln-γ2 predicts shorter survival of patients with NSCLC or ESCC
Next, the associations of Ln-γ2 expression with clinicopathological features, including patient outcomes, were explored in our cohorts of patients with NSCLC (n = 104) and ESCC (n = 260). First, IHC staining revealed significantly higher expression of Ln-γ2 in NSCLC tissues, including LUAD and LUSC, than in corresponding normal lung tissues (fig. S4, A and B). Patients with NSCLC were divided into the low Ln-γ2 group and high Ln-γ2 group based on the median value of IHC staining scores (Fig. 1D). A correlation analysis of the clinicopathological characteristics showed that high Ln-γ2 expression was significantly associated with tumor differentiation (P = 0.003), adjacent organ invasion (P = 0.047), and the tumor size (P < 0.001) (Fig. 1E and table S1). The Kaplan-Meier analysis revealed a significant correlation between high expression of Ln-γ2 and a shorter overall survival of patients with NSCLC (P = 0.033) (Fig. 1F). Moreover, the analysis of TCGA database also indicated high expression of the Ln-γ2 mRNA in LUAD and LUSC tissues, which was associated with poorer overall survival and progression-free survival of patients with NSCLC (fig. S4, C and D).
In addition, IHC staining showed high levels of the Ln-γ2 protein in ESCC tissues, and all normal esophageal epithelial cells were Ln-γ2–negative, consistent with the analysis of TCGA dataset at the mRNA level (fig. S4, E to G). Survival analyses based on the results of IHC staining suggested that ESCC patients with high Ln-γ2 expression had a shorter overall survival than patients with low Ln-γ2 expression (P = 0.017) (fig. S4H). Multivariate Cox proportional regression analyses further revealed that Ln-γ2 expression was an independent prognostic factor for the overall survival of patients with NSCLC (table S2) or ESCC (table S3). Together, high Ln-γ2 promotes NSCLC and ESCC progression.
CAF-derived TGF-β1 up-regulates Ln-γ2 expression
Although TGF-β1 markedly increases the expression of Ln-γ2 in NSCLC and ESCC cells, the primary source of TGF-β1 in the tumor microenvironment remains unknown. An analysis of the GEO database showed that depletion of α–smooth muscle actin–positive (α-SMA+) myofibroblasts decreased the expression of Ln-γ2 in a pancreatic ductal adenocarcinoma transgenic mouse model (GSE52812) (Fig. 2A, left). Conversely, coculture with primary CAFs significantly up-regulated the expression of Ln-γ2 in basal breast cancer cells (GSE41678) (Fig. 2A, right). IHC and immunofluorescence (IF) staining showed that Ln-γ2 was specifically expressed in the periphery of tumor nests, and Ln-γ2+ tumor cells were adjacent to CAFs (α-SMA+) in human NSCLC and ESCC tissues (figs. S5 and S6A). IHC staining of A549- or KYSE510-derived xenografted tumors also indicated near coexpression of α-SMA and Ln-γ2 (fig. S6B), implying that CAFs might up-regulate Ln-γ2 expression via paracrine TGF-β1 signaling.
Fig. 2. CAF-derived TGF-β1 up-regulates Ln-γ2 expression in tumor cells.
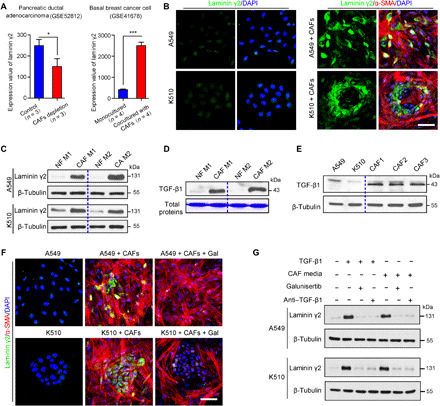
(A) Left: On the basis of an analysis of the GEO database, the depletion of CAFs decreased the expression of Ln-γ2 in a mouse pancreatic ductal adenocarcinoma model (GSE52812, n = 3). Right: Coculture with primary CAFs significantly up-regulated the expression of Ln-γ2 in basal breast cancer cells (GSE41678, n = 4). The statistical analysis was performed using the unpaired two-tailed t test with Welch’s correction. Data are presented as means ± SD. *P < 0.05 and ***P < 0.001. (B) The expression of Ln-γ2 (green) in A549 and KYSE510 cells was determined using double IF staining after coculture with or without CAFs (α-SMA+, red) in vitro. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; blue). Scale bar, 50 μm. (C) Western blotting showed that treatment with CAF-conditioned media (CAF M) significantly increased Ln-γ2 levels compared with treatment with paired NF-conditioned media (NF M; n = 2). (D) Levels of the TGF-β1 protein in CAF M and NF M were analyzed by Western blotting after concentration using an Amicon Ultra centrifugal filter (3 kDa, #UFC200324, Millipore) (n = 2). Total proteins were stained with Coomassie Brilliant Blue R-250. (E) Western blot showing the levels of the Ln-γ2 protein in A549 cells, KYSE510 cells, and three primary CAF lines. (F) IF staining showed that the increase in Ln-γ2 expression observed in A549 and KYSE510 cells after coculture with CAFs was blocked by galunisertib (10 μM, #S2230, Selleck). Scale bar, 50 μm. (G) Western blotting confirmed that galunisertib and the TGF-β1–neutralizing antibody (anti–TGF-β1; 1 μg/ml, #521707, BioLegend) inhibited the increase in the Ln-γ2 levels induced by treatment with CAF-conditioned media. In (C), (E), and (G), β-tubulin was analyzed as a loading control.
A549 and KYSE510 cells were cocultured with primary CAFs or CAF-conditioned media in vitro to verify this hypothesis. IF staining showed the remarkable up-regulation of Ln-γ2 expression in tumor cells cocultured with CAFs (Fig. 2B). Western blotting indicated that treatment with CAF-derived conditioned media increased Ln-γ2 expression in A549 and KYSE510 cells compared with conditioned media from normal fibroblasts (NFs) (Fig. 2C). Fundamentally, TGF-β1 was mainly secreted by CAFs, but not NFs or tumor cells (Fig. 2, D and E). In addition, IF staining and Western blotting showed the increase in the level of Ln-γ2 in tumor cells induced by CAF coculture or CAF-derived conditioned media, which could be blocked by galunisertib and a TGF-β1–neutralizing antibody (anti–TGF-β1) (Fig. 2, F and G, and fig. S6C). These findings suggested that Ln-γ2 expression was specifically increased by CAF-derived TGF-β1 via paracrine signaling in NSCLC and ESCC.
TGF-β1 activates Ln-γ2 expression via JNK/AP1 signaling
We next investigated the mechanism of TGF-β1–mediated Ln-γ2 up-regulation in tumor cells. First, we analyzed the transcription factor binding sites in the promoter region of the Ln-γ2 gene and found three AP1 (mainly c-Jun and c-Fos) binding sites upstream (−92 to −15 nucleotides) of the transcription start site (Fig. 3A). Therefore, chromatin immunoprecipitation coupled with quantitative polymerase chain reaction (ChIP-qPCR) was used to investigate the interaction of activated AP1 [phosphorylated-c-Jun (p-c-Jun) or phosphorylated-c-Fos (p-c-Fos)] and the potential binding sites in A549 and KYSE510 cells treated with TGF-β1 (0.5 ng/ml) or galunisertib (10 μM). TGF-β1 stimulation significantly increased the binding of p-c-Jun and p-c-Fos to the promoter of Ln-γ2, which was effectively inhibited by the galunisertib treatment (Fig. 3B). A luciferase reporter assay also confirmed that Ln-γ2 expression was transcriptionally up-regulated by TGF-β1 through the activation of AP1 in A549 and KYSE510 cells, and these changes were blocked by galunisertib (Fig. 3C).
Fig. 3. Ln-γ2 expression is transcriptionally activated by TGF-β1 via JNK/AP1 signaling.
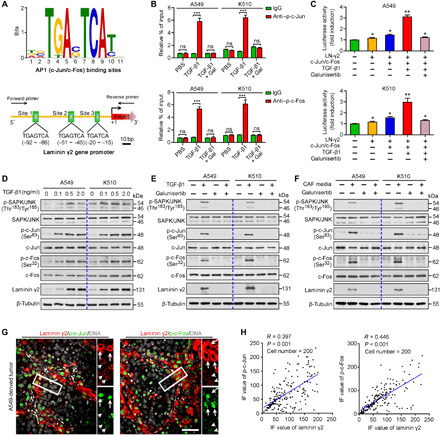
(A) AP1 (including c-Jun/c-Fos) transcriptional binding sites in the Ln-γ2 gene promoter. (B) ChIP-qPCR analysis showing that TGF-β1 stimulation promoted the binding of phosphorylated c-Jun (p-c-Jun) and phosphorylated c-Fos (p-c-Fos) to the Ln-γ2 gene promoter, and these changes were inhibited by galunisertib (10 μM). (C) Luciferase reporter assay showed that TGF-β1 transcriptionally up-regulated Ln-γ2 expression by activating AP1 in A549 and KYSE510 cells. (D) The levels of phosphorylated SAPK/JNK (p-SAPK/JNK), p-c-Jun, and p-c-Fos in tumor cells treated with different concentrations of TGF-β1 were analyzed using Western blotting. (E) Western blotting confirmed that the activation of JNK/AP1 signaling by TGF-β1 (0.5 ng/ml) stimulation was inhibited by galunisertib (10 μM). (F) CAF-conditioned media activated JNK/AP1 signaling in A549 and KYSE510 cells, which was also be blocked by galunisertib (10 μM). (G) Coexpressions of Ln-γ2 (red) with p-c-Jun (green) or p-c-Fos (green) in xenografted tumors derived from A549 cells were shown by double IF staining. Cell nuclei were stained with DAPI (gray). The white long and short arrows indicate the high and low expression of Ln-γ2, respectively. Scale bar, 50 μm. (H) The intensity of IF staining for p-c-Jun, p-c-Fos, and Ln-γ2 in tumor (cell number n = 200) was analyzed with ImageJ software, and a linear regression analysis was performed to test their correlations. In (B) and (C), statistical analyses were performed using unpaired two-tailed t tests with Welch’s correction. Data are presented as means ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001. ns, no significant difference. In (D) to (F), β-tubulin was tested as a loading control.
As AP1 regulates the transcription of the target genes of TGF-β/JNK signaling (24, 25), the levels of phosphorylated stress-activated protein kinase (SAPK)/JNK (p-SAPK/JNK), p-c-Jun, and p-c-Fos were analyzed using Western blotting after tumor cells were treated with different concentrations of TGF-β1 protein. The changes in the levels of Ln-γ2 were consistent with the levels of p-SAPK/JNK, p-c-Jun, and p-c-Fos in A549 or KYSE510 cells (Fig. 3D). Galunisertib and anti–TGF-β1 treatments completely inhibited the up-regulation of Ln-γ2 and the activation of JNK/AP1 signaling induced by TGF-β1 stimulation or CAF-conditioned medium treatment (Fig. 3, E and F, and fig. S7A). Silencing TGFBR1 with two small interfering RNAs (siRNAs) blocked the up-regulation of Ln-γ2 and AP1 activation in tumor cells induced by CAF-derived conditioned media (fig. S7B). In addition, IF staining also showed consistently high levels of Ln-γ2, p-c-Jun, and p-c-Fos in A549 cells treated with TGF-β1 or CAF-conditioned media in the presence or absence of galunisertib (fig. S7C). Coexpression of Ln-γ2 with p-c-Jun or p-c-Fos in xenografted tumors derived from A549 and KYSE510 cells was also confirmed using double IF staining (Fig. 3, G and H, and fig. S7D). Thus, JNK/AP1 signaling was responsible for the TGF-β1–mediated up-regulation of Ln-γ2 in NSCLC and ESCC cells.
Ln-γ2 blocks T cell infiltration into tumor nest
Infiltration of immunocytes into tumors is necessary for the cross-talk between tumor cells and immunocytes, contributing to antigen recognition and tumor destruction. A correlation analysis of gene expression in TCGA dataset did not reveal significant correlations between the expression of Ln-γ2 and other immune-related markers, such as PD-1, PD-L1, CD3, or CD8, in NSCLC and esophageal carcinoma tissues (fig. S8). Double IF staining for Ln-γ2 and CD3, CD4, or CD8 was performed on human NSCLC and ESCC sections to further analyze the correlation between Ln-γ2 expression and the distribution of T cells in tumors. Fewer infiltrating T cells were detected in tumor nests with high levels of Ln-γ2 expression than in tissues with low Ln-γ2 expression (P < 0.001; Fig. 4A and fig. S9A). The TGF-β1 treatment increased the intracellular and extracellular Ln-γ2 levels in NSCLC and ESCC cells (fig. S9, B to D). On the basis of this evidence, secreted Ln-γ2 may block the penetration of T cells into the tumor nests by constructing a protective barrier to prevent the attack of immunocytes, which may subsequently attenuate the effectiveness of anti–PD-1 therapy.
Fig. 4. High Ln-γ2 is responsible for T cell exclusion.
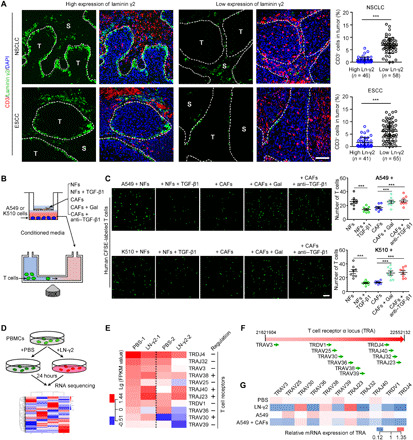
(A) Double IF staining for Ln-γ2 (green) and CD3 (T cells, red) in NSCLC and ESCC tissues. Cell nuclei were counterstained with DAPI (blue). The boundary of tumor and stroma was marked with white dotted lines. Scale bar, 100 μm. The proportions of these T cells in tumor nests were counted in tumor tissues with high or low expression of Ln-γ2. (B) Schematic of the T cell chemotaxis assay. (C) Representative images of migratory T cells (green) labeled with CFSE. Scale bar, 20 μm. The number of migratory T cells was counted in the right panel. (D) RNA-seq was conducted to explore the gene expression in T cells regulated by Ln-γ2. (E) Heat map showing that Ln-γ2 significantly regulated the expression of T cell receptor (TCR) family genes. −, downregulation; +, upregulation. (F) These TCR genes were located in the TCR α chain (TRA). (G) Quantitative real-time PCR (qRT-PCR) confirmed the regulation of TCR gene expression by Ln-γ2 and conditioned media from A549 cells alone or cocultured with CAFs. In (A) and (C), statistical analyses were performed using the unpaired two-tailed t test with Welch’s correction. Data are presented as means ± SEM; ***P < 0.001. ns, no significant difference.
In vitro chemotaxis assays were performed using tumor cell–derived conditioned media after coculture with NFs or CAFs to investigate the regulatory effect of Ln-γ2 on T cell infiltration (Fig. 4B). Conditioned media from tumor cells cocultured with CAFs inhibited T cell chemotaxis, which could be eliminated by galunisertib or anti–TGF-β1 treatment (Fig. 4C), as well as Ln-γ2 silencing in tumor cells (fig. S10, A to C). The activated recombinant Ln-γ2 protein, but not other laminins, directly reduced the chemotaxis of T cells (fig. S10, D to F). In addition, CIBERSORT and TIMER web tools were used to analyze the correlations of Ln-γ2 or TGF-β1 expression levels with immune cell infiltration in NSCLC and ESCA tissues from TCGA datasets. Both Ln-γ2 and TGF-β1 expression levels were negatively correlated with the number of infiltrating lymphocytes (fig. S11). Moreover, the CIBERSORT analysis indicated a significant positive correlation between Ln-γ2 and TGF-β1 levels in 22 immune cells from ESCA and LUSC (fig. S11A).
Next, we performed RNA-seq to analyze the master genes expressed in T cells that were affected by Ln-γ2 (2 μg/ml, 24 hours) (Fig. 4D and datasheet S2). The analysis of differentially expressed genes indicated that Ln-γ2 significantly altered the expression of T cell receptor (TCR) mRNAs in human peripheral blood T cells (Fig. 4E). Notably, all these differentially expressed TCR genes were located in the TCR α chain rather than the β chain (Fig. 4F). Real-time quantitative PCR (qRT-PCR) confirmed the regulatory effect of Ln-γ2 on TCR genes in more T cell samples after treatment with the Ln-γ2 protein or conditioned media from coculture of tumor cells and CAFs (Fig. 4G). Flow cytometry indicated that T cell activation induced by tumor cell–derived conditioned media was inhibited by Ln-γ2 treatment (fig. S12). Therefore, Ln-γ2 inhibited T cell chemotaxis by altering the transcription of TCR genes.
Combination therapies inhibit tumor progression in mice
C57BL/6 mice were subcutaneously inoculated with the mouse Lewis lung cancer cell line LLC or primary ESCC cell line MEC2 after Ln-γ2 silencing to explore the therapeutic potential of targeting Ln-γ2. Knockdown of Ln-γ2 suppressed tumor growth and promoted T cell infiltration into tumor nests (Fig. 5, A and B). As galunisertib and anti–TGF-β1 were very effective in preventing the up-regulation of Ln-γ2 induced by CAFs, a mouse monoclonal antibody against PD-1 [anti–PD-1; 5 mg/kg, intraperitoneally (i.p.)] in combination with galunisertib [100 mg/kg, intragastrically (i.g.)] or anti–TGF-β1 (5 mg/kg, i.p.) was administered to C57BL/6 mice bearing tumors. Carboxymethylcellulose sodium (CMC-Na, 1%, 100 μl, i.g.), the vehicle for galunisertib, and immunoglobulin G2a (IgG2a) isotype control antibody (5 mg/kg, i.p.) were also administered as controls (fig. S13A). The combination treatment of galunisertib or anti–TGF-β1 with anti–PD-1 inhibited tumor growth in vivo (fig. S13B). However, the depletion of CD8+ T cells with a mouse monoclonal neutralizing antibody against CD8a (10 mg/kg, i.p.) attenuated the antitumor effects of the combination treatment with galunisertib and anti–PD-1 (fig. S13, C and D).
Fig. 5. Combination treatments promote tumor regression in mouse models.
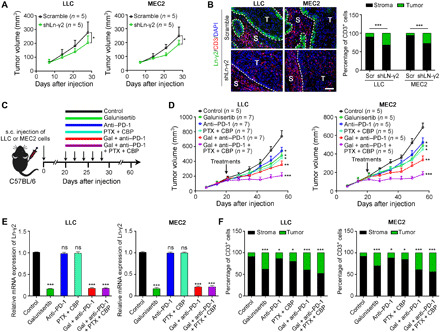
(A) The mouse Lewis lung cancer cell line LLC (2 × 105 cells per mouse) or primary ESCC cell line MEC2 (4 × 106 cells per mouse) in which Ln-γ2 was silenced by a short hairpin RNA was subcutaneously (s.c.) transplanted into C57BL/6 mice (n = 5 mice per group), and tumor volumes were measured weekly for 4 weeks. (B) Double IF staining with antibodies against Ln-γ2 (green) and CD3 (red) was performed on transplanted tumors, and the proportions of CD3+ T cells in the tumor and stroma were summarized in the right panel. Scale bar, 100 μm. (C) Combination therapies, including galunisertib (Gal; 100 mg/kg, i.g.), anti–PD-1 (5 mg/kg, i.p.), and chemotherapeutic drugs, paclitaxel (PTX; 6 mg/kg, i.p.) and carboplatin (CBP; 6 mg/kg, i.p.), were administered to tumor-bearing mice every other day for five treatments. (D) Tumor volumes were measured every week for 8 weeks. (E) The expression of the Ln-γ2 mRNA in tumors from mice subjected to different treatments was analyzed using qRT-PCR. (F) The proportions of tumor- and stroma-infiltrating CD3+ T cells were summarized on the basis of the results of double IF staining. In (A), (D), and (E), statistical analyses were performed using unpaired two-tailed t tests with Welch’s correction, and the data presented in (B) and (F) were analyzed with Pearson’s chi-square (Fisher’s exact) tests. In all panels, data are presented as means ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001; ns, no significant difference.
PD-1/PD-L1 blocking drugs are clinically administered in combination with chemotherapy to patients with NSCLC and ESCC (2, 26), and thus, galunisertib (100 mg/kg, i.g.) and the anti–PD-1 drug (5 mg/kg, i.p.) were administered in combination with the chemotherapeutic drugs paclitaxel (6 mg/kg, i.p.) and carboplatin (6 mg/kg, i.p.) to tumor-bearing mice (Fig. 5C). The combination of carboplatin and paclitaxel significantly increased the antitumor activity of galunisertib and anti–PD-1 treatments (Fig. 5D). In addition, galunisertib alone or in combinations decreased Ln-γ2 expression in tumor cells and promoted T cell penetration into tumor nests (Fig. 5, E and F, and fig. S13, E and F). Together, these data revealed an effective treatment for patients with NSCLC or ESCC by blocking TGF-β and PD-1/PD-L1 signaling pathways in combination with chemotherapy.
DISCUSSION
The administration of PD-1/PD-L1 immune checkpoint blockers is now a promising therapy for advanced cancers (6, 27). However, because of the limitations of PD-1/PD-L1 expression and the tumor mutational burden in predicting the therapeutic response, an unmet need in cancer treatment is to identify useful biomarkers that will accurately predict the response of patients with cancer to anti–PD-1/PD-L1 therapy. Signatures of T cell dysfunction and exclusion have been reported to predict the antitumor immunotherapy response, suggesting the vital role of T cell function during immunotherapy (11). In the present study, CAF-derived TGF-β1 signaling resulted in T cell exclusion by increasing the expression of Ln-γ2 in NSCLC and ESCC cells, which structured a protective barrier for tumors, blocked immune cell penetration into the tumor parenchyma, and attenuated the response to anti–PD-1 therapy. This finding reveals a previously unidentified mechanism of T cell dysfunction that is regulated by the cross-talk between CAFs and tumor cells during antitumor therapy.
Laminin is a major constituent of the ECM that contains α, β, and γ chains. This protein is located outside of cells and plays an important role in cell attachment. Ln-γ2 is a subunit of laminin 332 (Ln-332) that consists of α3, β3, and γ2 chains, is an essential component of epithelial basement membranes, and regulates cell motility and adhesion (28). As part of the specialized hepatocellular carcinoma (HCC) cancer stem cell niche, Ln-332 leads to chemoresistance and quiescence in HCC by maintaining cell stemness (29). In addition, Ln-332 also plays a prominent role in the lymph node metastasis of papillary thyroid carcinoma (30). However, a recent study found that Ln-γ2 was frequently expressed as a monomer in the invasion front of HCC tissues without simultaneous expression of α3 and β3 chains (31). Overexpression of Ln-γ2 promotes tumor cell proliferation, migration, and invasion, and thus, it may be applied to predict the poor prognosis of patients with colorectal cancer and ESCC (32, 33). On the basis of these findings, the monomeric Ln-γ2 chain alone has also physiological activity in regulating cancer progression. In the present study, RNA-seq showed that among the laminin family genes, only Ln-γ2 was significantly up-regulated in NSCLC and ESCC cells treated with TGF-β1. In vitro chemotaxis assays revealed the reduced migration of T cells after supplementation of the monomeric Ln-γ2 chain, which suggested the key role of the Ln-γ2 in regulating T cell exclusion from the tumor nests.
T cell chemotaxis and migration are essential for immune responses. The T cell response is regulated by the ability of cells to precisely arrive at sites of inflammation. For instance, chemokine gradients trigger T cell chemotaxis, and TCR activation induces a migration stop signal in T cells for antigen recognition (34, 35). Therefore, the activation of TCR is an important mechanism regulating T cell migration. In the present study, the RNA-seq analysis revealed that Ln-γ2 impaired T cell infiltration by specifically regulating the expression of the TCR α locus, suggesting that another regulatory mechanism of T cell migration is the control of the levels of TCR genes. High TCR reads are associated with increased T cell infiltration into the tumor nests, which indicated a shorter survival for patients with lower-grade glioma (36). Highly productive TCR α recombination reads, but not the TCR β locus, in renal cell carcinoma correlate with the level of T cell exhaustion in the tumor microenvironment (37). Our qRT-PCR analysis showed that most TCR genes in the α locus were down-regulated by the Ln-γ2 treatment. Ln-γ2 inhibits the activation of T cells induced by tumor cell–derived conditioned media. Therefore, activation and expression of TCR regulated by Ln-γ2 are involved in T cell migration, but the mechanisms require further study.
TGF-β plays an essential role in the regulation of immune responses and immune tolerance. Galunisertib, which was investigated in a phase 2 trial as a treatment for HCC (38), is a small-molecule cancer drug that specifically inhibits TGFBR signaling. Targeting the TGF-β pathway with galunisertib as a monotherapy or in combination with immune checkpoint blockade therapies promotes antitumor immunity. The combination of galunisertib with an anti–PD-L1 drug has been applied to treat colon cancer and has achieved an improved therapeutic effect (14). Galunisertib blocks TGF-β1–mediated suppression of naïve T cell proliferation and enhances the function of T cell and NK cell (39). On the other hand, the antitumor activity of galunisertib in a colon carcinoma model was associated with a modest increase in T cell infiltration in the tumor parenchyma (14). Consistent with the findings of these studies, galunisertib treatment in a mouse model promoted T cell penetration into tumor by breaking the barriers of tumor cells constructed by Ln-γ2 in our study. Moreover, we also confirmed that the therapeutic coadministration of galunisertib, an anti–PD-1 drug, and chemotherapy drugs prominently suppressed tumor growth, which suggested a more efficient therapeutic schedule for patients with NSCLC or ESCC. However, TGFBR1 blockade has been shown to affect multiple immune-related pathways and the EMT of tumor cells (40). TGFBR1 is widely expressed in multiple cells, suggesting potential off-target effects of galunisertib and unavoidable side effects on patients with cancer. Our in vitro and in vivo studies revealed similar effects of galunisertib and anti–TGF-β1 on T cell infiltration. Therefore, treatments directly targeting TGF-β1 itself with anti–TGF-β1 to inhibit Ln-γ2 expression in combination with immune checkpoint inhibitors and chemotherapy may be a promising therapeutic strategy for patients with NSCLC or ESCC. In addition, aberrantly high expression of the Ln-γ2 protein determined using IHC staining was significantly related to the attenuated efficacy of anti–PD-1 blockade therapy and predicted the unfavorable outcomes of patients with NSCLC and ESCC in the present study. Serum monomeric Ln-γ2 levels measured with a chemiluminescent immunoassay represent a promising biomarker for the early detection of HCC among patients with chronic liver diseases (31). Thus, tumor cell–derived Ln-γ2 is potentially released into the bloodstream, and it might also serve as a noninvasive marker to screen patients who would be sensitive to anti–PD-1/PD-L1 therapies.
Consistent with the results of previous studies (41, 42), our immunostaining results showed that Ln-γ2+ cells were located at the periphery and invasion front of tumor clusters. Moreover, high Ln-γ2 expression was associated with poor differentiation and adjacent tissue invasion in patients with NSCLC and ESCC. Several other studies have also reported a role for Ln-γ2 in the invasion and metastasis of tumor cells from different types of carcinoma, such as NSCLC, HCC, and thyroid cancer (43, 44). Therefore, Ln-γ2 likely promotes the metastasis of tumor cells, in addition to the regulatory effect of Ln-γ2 on antitumor immunity. For instance, Ln-γ2 promotes ESCC cell metastasis by interacting with integrin β4 to activate focal adhesion kinase (FAK)–phosphatidylinositol 3-kinase (PI3K)/Akt signaling (33). Elevated Ln-γ2 levels induce the EMT and promote the migration and invasion of lung adenocarcinoma cells by activating the integrin β1 receptor (45). Combination therapeutic strategy with galunisertib, anti–PD-1, and chemotherapeutic drugs may be administered to patients with early NSCLC or ESCC to prevent cancer metastasis. The activation of α2, α4, β1, and β2 integrins is also involved in T cell migration (46–48). Ln-332 interacts with integrin α6β4 to regulate T cell activation (49). Therefore, the multiple activities of Ln-γ2 during cancer progression may be explained by the different expression levels of integrin receptors on the surface of T cells and tumor cells.
Together, CAF-secreted TGF-β1 specifically increased the expression of Ln-γ2 in adjacent tumor cells via JNK/AP1 signaling, which structured a protective barrier for tumors to restrict T cell infiltration into tumor nests. The administration of galunisertib to block TGF-β signaling in combination with chemotherapy increases the response to anti–PD-1 therapy, causing progressors to become responders and suggesting that this approach represents a promising therapeutic strategy for improving the efficacy of antitumor immunotherapy (Fig. 6). Meanwhile, increased Ln-γ2 level was significantly related to the attenuated response to anti–PD-1 blockade therapy and predicted the unfavorable outcomes of patients with NSCLC or ESCC. Hence, Ln-γ2 expression may be useful for distinguishing progressors and responders before immune checkpoint therapy is administered.
Fig. 6. Breaking the Ln-γ2–constructed shield provokes vigorous antitumor immunity.
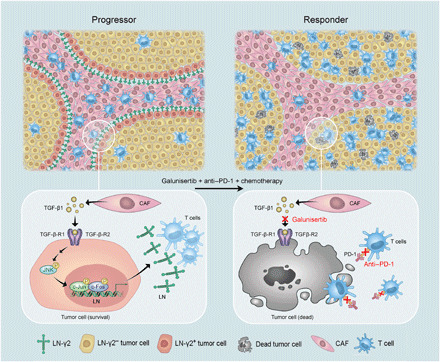
For progressors with NSCLC or ESCC, CAFs up-regulate the expression of Ln-γ2 in adjacent tumor cells via paracrine TGF-β1/JNK/AP1 signaling, which constructs a hermetic shield of tumor cells, impeding T cell penetration into the tumor nests and resisting antitumor immunotherapy. Administration of the inhibitor of TGFBR galunisertib completely inhibits the expression of Ln-γ2, which promotes T cell infiltration from the stroma into the tumor. Administration of galunisertib in combination with chemotherapeutic drugs enhances immunocytotoxicity and improves the response to anti–PD-1 therapy, which may be a promising therapeutic strategy for improving the outcomes of patients with NSCLC and ESCC.
MATERIALS AND METHODS
Clinical samples and cell lines
All clinical specimens, including paraffin-embedded sections, biopsy tissues, blood samples, and prognostic data, were collected from the Sun Yat-sen University Cancer Center (Guangzhou, China). All recruited patients and donors provided written informed consent before samples were collected, and the study protocol was approved by the Committees for Ethical Review of Research at Sun Yat-sen University (Guangzhou, China). NSCLC tumor and nontumor specimens that were surgically removed and embedded in paraffin blocks were obtained from the Department of Thoracic Oncology at Sun Yat-sen University Cancer Center between January 2002 and December 2004. Cancer tissue sections collected from patients with NSCLC or ESCC before PD-1 blockade therapies were obtained from the Sun Yat-sen University Cancer Center, the Affiliated Cancer Hospital of Zhengzhou University, and the First Affiliated Hospital of Zhengzhou University (table S4). Computed tomography imaging was performed to evaluate the response of anti–PD-1 immunotherapy.
The human NSCLC cell lines A549, HCC827, and SK-MES-1; ESCC cell lines KYSE510, KYSE140, and KYSE180; and mouse Lewis lung cancer cell LLC were purchased from the Typical Culture Preservation Commission Cell Bank, Chinese Academy of Sciences (Shanghai, China). Mouse primary ESCC cell line MEC2 was obtained from L. Fu (Shenzhen University, Shenzhen, China). All cell lines were cultured with high-glucose Dulbecco’s Modified Eagle’s Medium (DMEM; Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific) and 1% penicillin/streptomycin (Corning) and routinely cultured at 37°C in a humidified chamber containing 5% CO2.
Isolation of fibroblasts
Primary NFs or CAFs from patients with NSCLC or ESCC were isolated as described previously (50). Briefly, fresh tissues were washed three times with phosphate-buffered saline (PBS) after tumor resection and cut into pieces with surgical scissors in sterile 1.5-ml centrifuge tubes. Tissue digestion was performed using Liberase (0.2 mg/ml; #5401119001, Roche) on a rotary shaker at 37°C for 30 min. Enzymatic digestion was stopped with normal culture medium (DMEM + 10% FBS), and cell suspension was filtered using a sterile cell strainer (40 μm; Corning) to remove tissue pieces that were not completely digested. The single-cell suspension was centrifuged at 1500 rpm for 5 min at 4°C. After three washes with pure DMEM, the cell pellet was resuspended in 3 ml of normal culture medium supplemented with Primocin (100 μg/ml; #ant-pm-1, InvivoGen) to prevent contamination. Primary cells were cultured at 37°C in a humidified chamber containing 5% CO2. In general, the adhesion time and trypsin digestion time for fibroblasts are much shorter than the times required for other cells. Therefore, the nonadherent cells were removed to obtain relatively pure fibroblasts after culture for 30 min at 37°C, and fibroblasts were further purified using short-term digestion with trypsin (approximately 30 s). The digested cells were cultured for fibroblast reisolation. Last, immunostaining with a fibronectin antibody (1:200 dilution; #26836, Cell Signaling Technology) was performed to confirm the purity of the isolated fibroblasts.
Coculture of fibroblasts and cancer cells and preparation of conditioned media
For coculture with direct contact between cells, cancer cells (1 × 104 cells per well) were preseeded in a six-well plate containing a glass coverslip and cultured with normal culture medium at 37°C in a humidified chamber containing 5% CO2. After 2 days, the culture medium was replaced with fresh medium containing primary fibroblasts (1 × 104 cells per well). Double IF staining with antibodies against α-SMA and Ln-γ2 was performed to analyze the regulation of Ln-γ2 expression after 24 hours of coculture. Transwell (pore size, 3 μm; #3415, Corning) was used for noncontact coculture of tumor cells and fibroblasts. Cancer cells (1 × 104 cells per well) were seeded in 24-well plates, and fibroblasts (1 × 104 cells per well) were layered in the upper Transwell chamber. After 24 hours of coculture, conditioned medium was concentrated using an Amicon Ultra centrifugal filter (3 kDa; #UFC900396, Millipore) by centrifugation at 4000g for 10 min at room temperature. The retentate was collected for further experiments.
Isolation of T cells from peripheral blood and chemotaxis assay
First, peripheral blood mononuclear cells (PBMCs) from healthy donors were isolated by density gradient centrifugation using Ficoll-Paque PLUS (#17-1440-02, GE Healthcare) according to the manufacturer’s specifications. T cells were separated from PBMCs using immunomagnetic beads coated with an antibody against CD3 (#130-111-551, Miltenyi Biotec). Then, separated T cells were labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE; 1 μM; #A001, GeneCopoeia) at 37°C for 20 min. Horizontal μ-Slide Chemotaxis chambers (#80326, Ibidi) with or without Matrigel (#356230, Corning Incorporated) were used to investigate the chemotaxis and migration of CFSE-labeled T cells. Matrigel was supplemented with activated or heat-inactivated Ln-γ2 (#MBS2030234, MyBioSource). Tumor cell suspension (1 × 103) in DMEM with 10% FBS was preseeded on the right slide of μ-Slide Chemotaxis chambers. For the migration assay without Matrigel, conditioned media from coculture systems of tumor cells with NFs, CAFs, or CAFs treated with galunisertib (10 μM, #S2230, Selleck) or anti–TGF-β1 (1 μg/ml, #521707, BioLegend) were added to the right slide of chemotaxis chambers. Purified T cells (1 × 103) suspended in RPMI 1640 medium (Thermo Fisher Scientific) with 10% FBS were layered in the left slide of μ-Slide Chemotaxis chambers. The number of migratory T cells was imaged with a fluorescence microscope (OLYMPUS, Japan) after 24 hours of culture at 37°C in a humidified chamber containing 5% CO2.
shRNA- or siRNA-mediated knockdown
Ln-γ2 short hairpin RNA (shRNA) construct (human #sc-35782-SH; mouse #sc-35783-SH, Santa Cruz Biotechnology, Heidelberg, Germany) and control plasmids were cotransfected together with lentivirus packaging vectors (#LT002, GeneCopoeia, Rockville, MD) into 293FT cells (Invitrogen) using the HilyMax transfection reagent (#H357, Dojindo, Japan) according to the manufacturer’s instructions. Lentivirus particles were harvested for the transfection of tumor cells 72 hours after packaging with Polybrene supplement (10 μg/ml; #H9268, Sigma-Aldrich, Burlington, MA) for improving transfection efficiency. Ln-γ2–silenced tumor cells were selected with puromycin treatment (2 μg/ml, #P8833, Sigma, Burlington, MA). For siRNA-mediated knockdown, A549 and KYSE510 cells were plated in six-well plates and transfected with siRNA targeting TGFBR1 (siTGFBR1-1: 5′-GGGUCUGUGACUACAACAUTT-3′; siTGFBR1-2: 5′-GACUGGCAGUAAGACAUGATT-3′) using HilyMax transfection reagent (#H357, Dojindo, Japan) according to the manufacturer’s instructions. Twenty-four hours after transfection, mRNA from tumor cells was harvested, and the knockdown of Ln-γ2 or TGFBR1 in tumor cells was confirmed by qRT-PCR.
IF staining
IF staining was performed as described previously (51, 52). After blocking, the slides were incubated with the primary antibodies to Ln-γ2 (#ab210959, Abcam; dilution ratio, 1:500), pan-cytokeratin (#ab7753, Abcam; dilution ratio, 1:400), α-SMA (#19245, Cell Signaling Technology; dilution ratio, 1:200), CD3 (#ab1669, Abcam; dilution ratio, 1:150 dilution), CD4 (#ab183685, Abcam; dilution ratio, 1:200 dilution), CD8 (#ab217344, Abcam; dilution ratio, 1:200 dilution), EpCAM (#36746, Cell Signaling Technology; dilution ratio, 1:100), p-c-Jun (Ser73) (#3270, Cell Signaling Technology; dilution ratio, 1:800), and p-c-Fos (Ser32) (#5348, Cell Signaling Technology; dilution ratio, 1:200) at 4°C overnight in a moist chamber. After thorough washing, the slides were then incubated with Alexa Fluor 594– or 488–conjugated secondary antibodies (Invitrogen). Last, all slides were mounted with antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI; #S36938, Thermo Fisher Scientific). Images were captured with an OLYMPUS FV2000 fluorescence microscope.
IHC staining and results analysis
IHC staining was performed according to a standard streptavidin-biotin-peroxidase complex method. Briefly, slides with paraffin sections were deparaffinized by xylene, rehydrated with graded ethanol (100, 95, 75, and 50%), and rinsed with deionized water. Hydrogen peroxide (3%) was used to block the endogenous peroxidase activity for 10 min at room temperature. For antigen retrieval, tissue slides were high pressure–treated and boiled in EDTA Antigen Retrieval Solution (pH 8.0; Dako) for 5 min. Nonspecific binding was blocked with 5% bovine serum albumin for 30 min at 37°C. The slides were incubated with Ln-γ2 (#ab210959, Abcam; 1:500 dilution) or CD3 (#ab1669, Abcam; 1:150 dilution) antibodies at 4°C overnight in a humidified chamber. Immunoreactivity was detected using EnVision Detection Systems (#K5007, Dako).
An immunoreactivity score system was applied in the analysis of Ln-γ2 IHC staining. The percentage of Ln-γ2+ cells was scored as follows: 0, <5%; 1, 5 to 25%; 2, 25 to 50%; 3, 50 to 75%; 4, 75 to 100%. The intensity of Ln-γ2+ staining was scored as follows: 0, negative; 1, weak; 2, moderate; 3, strong (fig. S4A). The total score was determined by the following formula: staining index = positive percentage × intensity. The median immunoreactivity score was used as the cut-point to define cases with high or low expression of Ln-γ2. To analyze the proportion of CD3+, CD4+, and CD8+ cells in tumor, five fields in the representative area for each tumor were imaged considering the tumor heterogeneity. Hematoxylin and eosin staining was performed on serial sections that have been performed with IHC staining to evaluate the boundary of tumor and stroma. The number of staining positive cells in tumor was calculated using the counting tool of Photoshop software.
TUNEL staining assay
Apoptotic cells in tissue sides were detected by the In Situ Cell Death Detection Kit (#11-684-817-910, Roche) according to the standard instructions (53). Results of staining were analyzed with an OLYMPUS FV2000 fluorescence microscope. The proportion of TUNEL+ cells in tumor nests was calculated as described for CD3 staining.
Western blotting analysis
Proteins were harvested and lysed with 1× radioimmunoprecipitation assay buffer (#9806, Cell Signaling Technology) supplemented with protease inhibitor (#04693132001, Roche) and phosphatase inhibitor (#4906837001, Roche). Western blot analysis was performed according to the standard procedure (54, 55) with primary antibodies (table S5). β-Tubulin was also used as a loading control.
Quantitative real-time polymerase chain reaction
Total RNA from NSCLC cell A549 and ESCC cell KYSE510 treated with TGF-β1 (0.5 ng/ml, 24 hours) or dimethyl sulfoxide control was extracted using the TRIzol Reagent (#15596018, Thermo Fisher Scientific), and reverse transcription was performed using the PrimeScript Reverse Transcription Kit (#RR047A, Takara). Complementary DNA was subjected to qPCR using FastStart Universal SYBR Green Master (#04913850001, Roche), and the assay was analyzed with a Roche LightCycler 480 detector. ACTB was used as an internal control. The primer sequences of TCR family genes and ACTB were listed in table S6. The relative expression values of target genes (2−ΔΔCt) were normalized to the endogenous ACTB reference (ΔCt).
ChIP-qPCR assay
The ChIP-qPCR assay was performed according to a standard procedure using the Simple ChIP Enzymatic ChIP Kit (#9003, Cell Signaling Technology) with normal rabbit IgG (#2729, Cell Signaling Technology) and primary antibodies against p-c-Fos (Ser32) (#5348, Cell Signaling Technology) or p-c-Jun (Ser73) (#3270, Cell Signaling Technology). qPCR primers (forward: 5′-GCTGGGTCCTCCTTATTCAC-3′; reverse: 5′-TCGGTTGGTGGTTCTCACTC-3′) were designed against the Ln-γ2 promoter (LAMC2, gene ID: 3918), which contains three binding sites for the transcription factors c-Jun and c-Fos. The qPCR results were analyzed using Roche Light Cycler 480 software. Alternatively, ChIP efficiency was manually calculated using the Percent Input Method and the following equation: (i) Input Dilution Factor = (fraction of the input chromatin saved)−1; (ii) ΔCt [normalized ChIP] = (Ct [ChIP] – (Ct [Input] − Log2 (Input Dilution Factor))); (iii) % Input = 2 (−ΔCt [normalized ChIP]).
Luciferase reporter assay
The putative Ln-γ2 promoter (LAMC2, gene ID: 3918) that contains the binding sites for the transcription factors c-Jun/c-Fos was inserted upstream of Gaussia Luciferase (GLuc) coding sequences in the pEZX-PG04 basic reporter plasmid (GeneCopoeia, MD, USA). Human c-Jun and c-Fos coding sequences were cloned into the pEZ-M02 plasmid (GeneCopoeia, MD, USA). A549 and KYSE510 cells were seeded in 96-well plates (2 × 103 cells per well) and then cotransfected with pEZ-c-Jun/c-Fos plasmids and the pEZX-promoter reporter plasmid using Lipofectamine 3000 (#L3000001, Thermo Fisher Scientific). GLuc activity was determined using the Dual-Luciferase Assay System (#E1910, Promega, Madison, WI) according to the technical manual at 48 hours after transfection.
In vivo tumor cell transplantation assay and drug treatments
All animal experiments were approved by the Animal Ethics Committee at Sun Yat-sen University Cancer Center. Five-week-old female C57BL/6 mice with an intact immune system were purchased from the Guangdong Medical Laboratory Animal Center (Foshan, China). The mouse Lewis lung cancer cell line LLC (2 × 105 cells per mouse) or primary ESCC cell line MEC2 (4 × 106 cells per mouse) was resuspended in 100 μl of PBS and injected subcutaneously into the right dorsal flanks of mice. The length (L) and width (W) of tumors were measured every week using calipers for indicated weeks, and tumor volumes were calculated as volume (mm3) = L × W2 × 0.5. If the length of the tumor was larger than 10 mm, the mice were sacrificed by cervical vertebra dislocation after anesthesia.
For drug treatments, the TGFBRI inhibitor galunisertib (#S2230, Selleck; dose, 100 mg/kg) was administered by gavage after dissolution in 1% CMC-Na as the drug vehicle. InVivoMab anti-mouse PD-1 (CD279, #BE0146, Bio X Cell; dose, 5 mg/kg) was used to block PD-1 through an intraperitoneal injection, and InVivoMab rat IgG2a (#BE0089, Bio X Cell; dose, 5 mg/kg) was also administered as an isotype control. The chemotherapeutic drugs paclitaxel (#S1150, Selleck; dose, 6 mg/kg, i.p.) and carboplatin (#S1215, Selleck; 6 mg/kg, i.p.) were intraperitoneally injected in combination with galunisertib and mouse monoclonal antibody anti–PD-1 treatments. Mouse anti–TGF-β1 (#521707, BioLegend; dose, 5 mg/kg, i.p.) in combination with anti–PD-1 (dose, 5 mg/kg, i.p.) was also performed in tumor-bearing mice. Drug therapies were administered every other day for five treatments. Meanwhile, mouse body weights were measured to monitor serious adverse effects.
Statistical analysis
IBM SPSS Statistics 25.0 software (Chicago, IL) was used to analyze all data. Pearson’s χ2 test was used to analyze the categorical variables, and unpaired two-tailed t test with Welch’s correction was used to analyze continuous variables. The prognostic value was calculated using the Kaplan-Meier method, and the P value was calculated with the log-rank test. The results were considered statistically significant if P < 0.05.
Acknowledgments
Funding: This work was supported by grants from the project funded by the National Natural Science Foundation of China (81772554, 81871903, 82072604, and 82072738), the China National Key Sci-Tech Special Project of Infectious Diseases (2018ZX10723204-006-005), the National Science Foundation for Young Scholars of China (81601581 and 81600606), the Basic and Applied Basic Research Foundation of Guangdong Province (2019A1515110660), the National Science Foundation of Guangdong Province (2017A030313764), the Hong Kong Research Grant Council General Research Funds (HKU/7668/11M, 767313, and 17143716), the Hong Kong Theme-based Research Scheme Fund (T12-704/16-R), the Hong Kong Research Grant Council Collaborative Research Funds (C7065-18GF and C7026-18GF), and the Shenzhen Fundamental Research Program (KQDT2015033117210153). X.-Y.G. is Sophie YM Chan Professor in Cancer Research. Author contributions: L.L. conceived the study, coordinated experiments, and wrote the manuscript. J.-R.W. performed RNA-seq and clinicopathological data analyses. J.D., Q.-G.L., H.T., and Y.-X.J. analyzed histopathology and imageology. J.D., Q.-G.L., H.T., Y.-X.J., S.X., and W.T. provided clinical materials. T.-T.Z. assisted animal experiments. Q.-Y.C., Y.-R.Q., Y.-H.Z., and Y.L. provided technical support. X.-Y.G. supervised the study. Competing interests: X.-Y.G. and L.L. are inventors on a pending patent related to this work, and the patent is in the substantive examination stage (patentee: Sun Yat-sen University Cancer Center, 26 August 2020; no. 202010784683.X). The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/6/eabc8346/DC1
REFERENCES AND NOTES
- 1.Doroshow D. B., Herbst R. S., Treatment of advanced non-small cell lung cancer in 2018. JAMA Oncol. 4, 569–570 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Kuo H.-Y., Guo J.-C., Hsu C.-H., Anti-PD-1 immunotherapy in advanced esophageal squamous cell carcinoma: A long-awaited breakthrough finally arrives. J. Formos. Med. Assoc. 119, 565–568 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Tunger A., Sommer U., Wehner R., Kubasch A. S., Grimm M.-O., Bachmann M. P., Platzbecker U., Bornhäuser M., Baretton G., Schmitz M., The evolving landscape of biomarkers for anti-PD-1 or anti-PD-L1 Therapy. J. Clin. Med. 8, 1534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.García-Aranda M., Redondo M., Targeting protein kinases to enhance the response to anti-PD-1/PD-L1 immunotherapy. Int. J. Mol. Sci. 20, 2296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia L., Liu Y., Wang Y., PD-1/PD-L1 blockade therapy in advanced non-small-cell lung cancer: Current status and future directions. Oncologist 24, S31–S41 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tartarone A., Roviello G., Lerose R., Roudi R., Aieta M., Zoppoli P., Anti-PD-1 versus anti-PD-L1 therapy in patients with pretreated advanced non-small-cell lung cancer: A meta-analysis. Future Oncol. 15, 2423–2433 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Hellmann M. D., Ciuleanu T.-E., Pluzanski A., Lee J. S., Otterson G. A., Audigier-Valette C., Minenza E., Linardou H., Burgers S., Salman P., Borghaei H., Ramalingam S. S., Brahmer J., Reck M., O’Byrne K. J., Geese W. J., Green G., Chang H., Szustakowski J., Bhagavatheeswaran P., Healey D., Fu Y., Nathan F., Paz-Ares L., Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N. Engl. J. Med. 378, 2093–2104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reck M., Rodríguez-Abreu D., Robinson A. G., Hui R., Csőszi T., Fülöp A., Gottfried M., Peled N., Tafreshi A., Cuffe S., O’Brien M., Rao S., Hotta K., Leiby M. A., Lubiniecki G. M., Shentu Y., Rangwala R., Brahmer J. R.; KEYNOTE-024 Investigators , Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N. Engl. J. Med. 375, 1823–1833 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Nakanishi Y., Masuda T., Yamaguchi K., Sakamoto S., Horimasu Y., Mimae T., Nakashima T., Miyamoto S., Tsutani Y., Iwamoto H., Fujitaka K., Miyata Y., Hamada H., Okada M., Hattori N., Albumin–globulin ratio is a predictive biomarker of antitumor effect of anti-PD-1 antibody in patients with non-small cell lung cancer. Int. J. Clin. Oncol. 25, 74–81 (2020). [DOI] [PubMed] [Google Scholar]
- 10.de Vries R., Muller M., van der Noort V., Theelen W. S. M. E., Schouten R. D., Hummelink K., Muller S. H., Wolf-Lansdorf M., Dagelet J. W. F., Monkhorst K., Maitland-van der Zee A. H., Baas P., Sterk P. J., van den Heuvel M. M., Prediction of response to anti-PD-1 therapy in patients with non-small-cell lung cancer by electronic nose analysis of exhaled breath. Ann. Oncol. 30, 1660–1666 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Jiang P., Gu S., Pan D., Fu J., Sahu A., Hu X., Li Z., Traugh N., Bu X., Li B., Liu J., Freeman G. J., Brown M. A., Wucherpfennig K. W., Liu X. S., Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 24, 1550–1558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mariathasan S., Turley S. J., Nickles D., Castiglioni A., Yuen K., Wang Y., Kadel E. E. III, Koeppen H., Astarita J. L., Cubas R., Jhunjhunwala S., Banchereau R., Yang Y., Guan Y., Chalouni C., Ziai J., Şenbabaoğlu Y., Santoro S., Sheinson D., Hung J., Giltnane J. M., Pierce A. A., Mesh K., Lianoglou S., Riegler J., Carano R. A. D., Eriksson P., Höglund M., Somarriba L., Halligan D. L., van der Heijden M. S., Loriot Y., Rosenberg J. E., Fong L., Mellman I., Chen D. S., Green M., Derleth C., Fine G. D., Hegde P. S., Bourgon R., Powles T., TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554, 544–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tauriello D. V. F., Palomo-Ponce S., Stork D., Berenguer-Llergo A., Badia-Ramentol J., Iglesias M., Sevillano M., Ibiza S., Cañellas A., Hernando-Momblona X., Byrom D., Matarin J. A., Calon A., Rivas E. I., Nebreda A. R., Riera A., Attolini C. S.-O., Batlle E., TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 554, 538–543 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Holmgaard R. B., Schaer D. A., Li Y., Castaneda S. P., Murphy M. Y., Xu X., Inigo I., Dobkin J., Manro J. R., Iversen P. W., Surguladze D., Hall G. E., Novosiadly R. D., Benhadji K. A., Plowman G. D., Kalos M., Driscoll K. E., Targeting the TGFβ pathway with galunisertib, a TGFβRI small molecule inhibitor, promotes anti-tumor immunity leading to durable, complete responses, as monotherapy and in combination with checkpoint blockade. J. Immunother. Cancer 6, 47 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao S., Mishra L., Targeting transforming growth factor beta signaling in liver cancer. Hepatology 69, 1375–1378 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massague J., TGFβ in cancer. Cell 134, 215–230 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin R.-L., Zhao L.-J., Mechanistic basis and clinical relevance of the role of transforming growth factor-β in cancer. Cancer Biol. Med. 12, 385–393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batlle E., Massagué J., Transforming growth factor-β signaling in immunity and cancer. Immunity 50, 924–940 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanjabi S., Oh S. A., Li M. O., Regulation of the immune response by TGF-β: From conception to autoimmunity and infection. Cold Spring Harb. Perspect. Biol. 9, a022236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flavell R. A., Sanjabi S., Wrzesinski S. H., Licona-Limón P., The polarization of immune cells in the tumour environment by TGFβ. Nat. Rev. Immunol. 10, 554–567 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Principe D. R., Park A., Dorman M. J., Kumar S., Viswakarma N., Rubin J., Torres C., McKinney R., Munshi H. G., Grippo P. J., Rana A., TGFβ blockade augments PD-1 inhibition to promote T-cell–mediated regression of pancreatic cancer. Mol. Cancer Ther. 18, 613–620 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dodagatta-Marri E., Meyer D. S., Reeves M. Q., Paniagua R., To M. D., Binnewies M., Broz M. L., Mori H., Wu D., Adoumie M., Del Rosario R., Li O., Buchmann T., Liang B., Malato J., Arce Vargus F., Sheppard D., Hann B. C., Mirza A., Quezada S. A., Rosenblum M. D., Krummel M. F., Balmain A., Akhurst R. J., α-PD-1 therapy elevates Treg/Th balance and increases tumor cell pSmad3 that are both targeted by α-TGFβ antibody to promote durable rejection and immunity in squamous cell carcinomas. J. Immunother. Cancer 7, 62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta A., Budhu S., Merghoub T., One checkpoint may hide another: Inhibiting the TGFβ signaling pathway enhances immune checkpoint blockade. Hepatobil. Surg. Nutr. 8, 289–294 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rachakonda G., Vu T., Jin L., Samanta D., Datta P. K., Role of TGF-β-induced Claudin-4 expression through c-Jun signaling in non-small cell lung cancer. Cell. Signal. 28, 1537–1544 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Hao Y., Zhu L., Yan L., Liu J., Liu D., Gao N., Tan M., Gao S., Lin B., c-Fos mediates α1, 2-fucosyltransferase 1 and Lewis y expression in response to TGF-β1 in ovarian cancer. Oncol. Rep. 38, 3355–3366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toi Y., Sugawara S., Sugisaka J., Ono H., Kawashima Y., Aiba T., Kawana S., Saito R., Aso M., Tsurumi K., Suzuki K., Shimizu H., Domeki Y., Terayama K., Nakamura A., Yamanda S., Kimura Y., Honda Y., Profiling preexisting antibodies in patients treated with anti-PD-1 therapy for advanced non-small cell lung cancer. JAMA Oncol. 5, 376–383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsoukalas N., Kiakou M., Tsapakidis K., Tolia M., Aravantinou-Fatorou E., Baxevanos P., Kyrgias G., Theocharis S., PD-1 and PD-L1 as immunotherapy targets and biomarkers in non-small cell lung cancer. J. BUON 24, 883–888 (2019). [PubMed] [Google Scholar]
- 28.Marinkovich M. P., Tumour microenvironment: Laminin 332 in squamous-cell carcinoma. Nat. Rev. Cancer 7, 370–380 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Govaere O., Wouters J., Petz M., Vandewynckel Y.-P., Van den Eynde K., Van den Broeck A., Verhulst S., Dollé L., Gremeaux L., Ceulemans A., Nevens F., van Grunsven L. A., Topal B., Vankelecom H., Giannelli G., Van Vlierberghe H., Mikulits W., Komuta M., Roskams T., Laminin-332 sustains chemoresistance and quiescence as part of the human hepatic cancer stem cell niche. J. Hepatol. 64, 609–617 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Oh K. H., Choi J., Woo J.-S., Baek S. K., Jung K. Y., Koh M. J., Kim Y.-S., Kwon S. Y., Role of laminin 332 in lymph node metastasis of papillary thyroid carcinoma. Auris Nasus Larynx 44, 729–734 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Yasuda H., Nakagawa M., Kiyokawa H., Yoshida E., Yoshimura T., Koshikawa N., Itoh F., Seiki M., Unique biological activity and potential role of monomeric laminin-γ2 as a novel biomarker for hepatocellular carcinoma: A review. Int. J. Mol. Sci. 20, 226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang D., Du C., Ji D., Xi J., Gu J., Overexpression of LAMC2 predicts poor prognosis in colorectal cancer patients and promotes cancer cell proliferation, migration, and invasion. Tumour Biol. 39, 1010428317705849 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Liang Y., Chen X., Wu Y., Li J., Zhang S., Wang K., Guan X., Yang K., Bai Y., LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ. 25, 1980–1995 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker C. M., Comrie W. A., Hyun Y.-M., Chung H.-L., Fedorchuk C. A., Lim K., Brakebusch C., McGrath J. L., Waugh R. E., Meier-Schellersheim M., Kim M., Opposing roles for RhoH GTPase during T-cell migration and activation. Proc. Natl. Acad. Sci. U.S.A. 109, 10474–10479 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cernuda-Morollón E., Millán J., Shipman M., Marelli-Berg F. M., Ridley A. J., Rac activation by the T-cell receptor inhibits T cell migration. PLOS ONE 5, e12393 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chobrutskiy B. I., Zaman S., Tong W. L., Diviney A., Blanck G., Recovery of T-cell receptor V(D)J recombination reads from lower grade glioma exome files correlates with reduced survival and advanced cancer grade. J. Neurooncol. 140, 697–704 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Mai A. T., Tong W. L., Tu Y. N., Blanck G., TcR-α recombinations in renal cell carcinoma exome files correlate with an intermediate level of T-cell exhaustion biomarkers. Int. Immunol. 30, 35–40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelley R. K., Gane E., Assenat E., Siebler J., Galle P. R., Merle P., Hourmand I. O., Cleverly A., Zhao Y., Gueorguieva I., Lahn M., Faivre S., Benhadji K. A., Giannelli G., A phase 2 study of galunisertib (TGF-β1 receptor type I inhibitor) and sorafenib in patients with advanced hepatocellular carcinoma. Clin. Transl. Gastroenterol. 10, e00056 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bommireddy R., Saxena V., Ormsby I., Yin M., Boivin G. P., Babcock G. F., Singh R. R., Doetschman T., TGF-β1 regulates lymphocyte homeostasis by preventing activation and subsequent apoptosis of peripheral lymphocytes. J. Immunol. 170, 4612–4622 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Ganesh K., Massagué J., TGF-β inhibition and immunotherapy: Checkmate. Immunity 48, 626–628 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Määttä M., Soini Y., Pääkkö P., Salo S., Tryggvason K., Autio-Harmainen H., Expression of the laminin γ2 chain in different histological types of lung carcinoma. A study by immunohistochemistry and in situ hybridization. J. Pathol. 188, 361–368 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Hamasaki H., Koga K., Hamasaki M., Kiryu H., Nakayama J., Iwasaki H., Nabeshima K., Immunohistochemical analysis of laminin 5-γ2 chain expression for differentiation of basal cell carcinoma from trichoblastoma. Histopathology 59, 159–161 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Garg M., Kanojia D., Okamoto R., Jain S., Madan V., Chien W., Sampath A., Ding L.-W., Xuan M., Said J. W., Doan N. B., Liu L.-Z., Yang H., Gery S., Braunstein G. D., Koeffler H. P., Laminin-5γ-2 (LAMC2) is highly expressed in anaplastic thyroid carcinoma and is associated with tumor progression, migration, and invasion by modulating signaling of EGFR. J. Clin. Endocrinol. Metab. 99, E62–E72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kagesato Y., Mizushima H., Koshikawa N., Kitamura H., Hayashi H., Ogawa N., Tsukuda M., Miyazaki K., Sole expression of laminin gamma 2 chain in invading tumor cells and its association with stromal fibrosis in lung adenocarcinomas. Jpn. J. Cancer Res. 92, 184–192 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moon Y. W., Rao G., Kim J. J., Shim H.-S., Park K.-S., An S. S., Kim B., Steeg P. S., Sarfaraz S., Changwoo Lee L., Voeller D., Choi E. Y., Luo J., Palmieri D., Chung H. C., Kim J.-H., Wang Y., Giaccone G., LAMC2 enhances the metastatic potential of lung adenocarcinoma. Cell Death Differ. 22, 1341–1352 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naci D., Aoudjit F., Alpha2beta1 integrin promotes T cell survival and migration through the concomitant activation of ERK/Mcl-1 and p38 MAPK pathways. Cell. Signal. 26, 2008–2015 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Nordenfelt P., Elliott H. L., Springer T. A., Coordinated integrin activation by actin-dependent force during T-cell migration. Nat. Commun. 7, 13119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han J., Rose D. M., Woodside D. G., Goldfinger L. E., Ginsberg M. H., Integrin α4β1-dependent T cell migration requires both phosphorylation and dephosphorylation of the α4 cytoplasmic domain to regulate the reversible binding of paxillin. J. Biol. Chem. 278, 34845–34853 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Vivinus-Nebot M., Ticchioni M., Mary F., Hofman P., Quaranta V., Rousselle P., Bernard A., Laminin 5 in the human thymus: Control of T cell proliferation via α6β4 integrins. J. Cell Biol. 144, 563–574 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei J.-R., Dong J., Li L., Cancer-associated fibroblasts-derived gamma-glutamyltransferase 5 promotes tumor growth and drug resistance in lung adenocarcinoma. Aging 12, 13220–13233 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li L., Zheng Y.-L., Jiang C., Fang S., Zeng T.-T., Zhu Y.-H., Li Y., Xie D., Guan X.-Y., HN1L-mediated transcriptional axis AP-2γ/METTL13/TCF3-ZEB1 drives tumor growth and metastasis in hepatocellular carcinoma. Cell Death Differ. 26, 2268–2283 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang B., Zhang Z., Li L., Qin Y.-R., Liu H., Jiang C., Zeng T.-T., Li M.-Q., Xie D., Li Y., Guan X.-Y., Zhu Y.-H., TSPAN15 interacts with BTRC to promote oesophageal squamous cell carcinoma metastasis via activating NF-κB signaling. Nat. Commun. 9, 1423 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li L., Li J.-C., Yang H., Zhang X., Liu L.-L., Li Y., Zeng T.-T., Zhu Y.-H., Li X.-D., Li Y., Xie D., Fu L., Guan X.-Y., Expansion of cancer stem cell pool initiates lung cancer recurrence before angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 115, E8948–E8957 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li L., Zeng T.-T., Zhang B.-Z., Li Y., Zhu Y.-H., Guan X.-Y., Overexpression of HN1L promotes cell malignant proliferation in non-small cell lung cancer. Cancer Biol. Ther. 18, 904–915 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng Y.-L., Li L., Jia Y.-X., Zhang B.-Z., Li J.-C., Zhu Y.-H., Li M.-Q., He J.-Z., Zeng T.-T., Ban X.-J., Yuan Y.-F., Li Y., Guan X.-Y., LINC01554-mediated glucose metabolism reprogramming suppresses tumorigenicity in hepatocellular carcinoma via downregulating PKM2 expression and inhibiting Akt/mTOR signaling pathway. Theranostics 9, 796–810 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hamasaki H., Koga K., Aoki M., Hamasaki M., Koshikawa N., Seiki M., Iwasaki H., Nakayama J., Nabeshima K., Expression of laminin 5-γ2 chain in cutaneous squamous cell carcinoma and its role in tumour invasion. Brit. J. Cancer 105, 824–832 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/6/eabc8346/DC1


