Abstract
A major objective of synthetic glycobiology is to re-engineer existing cellular glycosylation pathways from the top-down or construct non-natural ones from the bottom-up for new and useful purposes. Here, we developed a set of orthogonal pathways for eukaryotic O-linked protein glycosylation in Escherichia coli that installed the cancer-associated mucin-type glycans Tn, T, sialyl-Tn and sialyl-T onto serine residues in acceptor motifs derived from different human O-glycoproteins. These same glycoengineered bacteria were used to supply crude cell extracts enriched with glycosylation machinery that permitted cell-free construction of O-glycoproteins in a one-pot reaction. In addition, O-glycosylation-competent bacteria were able to generate an antigenically authentic Tn-MUC1 glycoform that exhibited reactivity with antibody 5E5, which specifically recognizes cancer-associated glycoforms of MUC1. We anticipate that the orthogonal glycoprotein biosynthesis pathways developed here will provide facile access to structurally diverse O-glycoforms for a range of important scientific and therapeutic applications.
Introduction
Protein glycosylation is one of the most abundant and structurally complex post-translational modifications (PTMs)1, 2 and occurs in all domains of life.3 Protein-linked glycans (mono-, oligo- or polysaccharide) play important roles in protein folding, solubility, stability, serum half-life, immunogenicity, and biological function.4 Glycan conjugation is also critical to the development of many biologics, with glycoproteins accounting for more than 70% of current protein-based drugs5 and glycoconjugate vaccines representing one of the safest and most successful vaccination approaches developed over the last 40 years.6 The importance of glycosylation in both nature and the clinic has prompted widespread glycoengineering efforts that seek to: (i) create designer production platforms for controllable glycoprotein synthesis;7–16 and (ii) rationally manipulate glycan structures and their attachment sites as a means to optimize the therapeutic and immunologic properties of proteins.17–21
Genetically engineered eukaryotic expression hosts have provided extensive access to a chemically rich landscape of glycoproteins enabling efforts to generate defined glycoprotein epitopes and engineer proteins with advantageous properties.8, 9, 14–16 However, glycoengineering in eukaryotes is complicated by the fact that glycans are synthesized across several subcellular compartments by the coordinated activities of numerous glycosyltransferases (GTs)22 and that glycosylation is an essential process, with significant alteration of glycosylation pathways often leading to severe fitness defects.23 Glycoengineering in bacteria, on the other hand, is not constrained by these issues due to the non-essential nature of protein glycosylation in bacterial cells and thus has emerged as an attractive alternative that permits customizable glycan construction and protein glycosylation.24 Moreover, some bacteria including laboratory strains of Escherichia coli lack endogenous glycosylation pathways, thereby providing a “clean” chassis for installation of orthogonal glycosylation pathways with little to no interference from endogenous GTs and the potential for more uniformly glycosylated protein products.
Over the last two decades, numerous efforts have collectively endowed E. coli and E. coli-derived cell-free extracts with the catalytic potential to produce diverse N-glycoproteins. Notably, this includes generation of structurally complex glycans, such as the eukaryotic Man3GlcNAc2 structure,7 and their installation at authentic human glycosites.25 In contrast, the analogous construction of O-linked glycosylation pathways in bacteria has received relatively little attention. Two of the earliest examples involved reconstituting the initiating step of vertebrate mucin-type O-glycosylation in E. coli.26, 27 Specifically, human polypeptide N-acetylgalactosaminyl-transferase 2 (GalNAcT2) was used to conjugate GalNAc onto threonine residues of peptides derived from different O-glycoproteins including human mucin 1 (MUC1) or an artificial rat-derived MUC10 in the cytoplasm of E. coli. Most recently, it was shown that the GalNAc installed by GalNAcT2 on threonine residues could be extended by a single galactose (Gal) residue using Campylobacter jejuni β1,3-galactosyltransferase CgtB, yielding acceptor proteins modified with Gal-β1,3-GalNAcα (T antigen or core 1).28 Bacterial protein O-glycosylation pathways have also been successfully reconstituted in E. coli; however, these systems are unlike the processive mechanism used by eukaryotes and instead operate according to an en bloc mechanism that is reminiscent of the canonical N-glycosylation process.24 Here, the glycan structures are assembled on a lipid carrier and subsequently transferred to acceptor proteins by O-oligosaccharyltransferases (O-OSTs) such as PglO from Neisseria gonorrhoeae (NgPglO) and PglL from Neisseria meningitidis (NmPglL). The fact that NmPglL is able to transfer virtually any bacterial glycan from the undecaprenyl-pyrophosphate (Und-PP) carrier29 suggests that bacterial O-OSTs may be useful for a broad range of applications; however, this has not been demonstrated aside from furnishing conjugate vaccines.30
Here, we implemented a synthetic glycobiology approach to engineer E. coli with human-like O-glycosylation pathways based on the bacterial PglL/O paradigm. As proof-of-concept, we created a collection of orthogonal pathways for biosynthesis of proteins decorated with mucin-type O-glycans including Tn, T, sialyl-Tn (STn) and sialyl-T (ST) glycans. Each of these pathways involved cytoplasmic preassembly of desired O-glycan structures on Und-PP by a prescribed set of heterologous GTs expressed in E. coli cells metabolically engineered to produce required nucleotide sugar donors. The addition of heterologous O-OSTs enabled efficient site-directed O-glycosylation of acceptor sequences derived from different human glycoproteins. Glycoengineered E. coli cells were also used to source crude cell extracts selectively enriched with O-glycosylation machinery, enabling a one-pot, cell-free reaction scheme for efficient and site-specific installation of O-glycans on target acceptor proteins. Overall, we anticipate that our glycoengineered bacteria will enable future efforts to produce structurally diverse O-glycoproteins for a variety of applications at the intersection of glycoscience, synthetic biology, and biomedicine.
Results
An engineered pathway for Tn antigen biosynthesis.
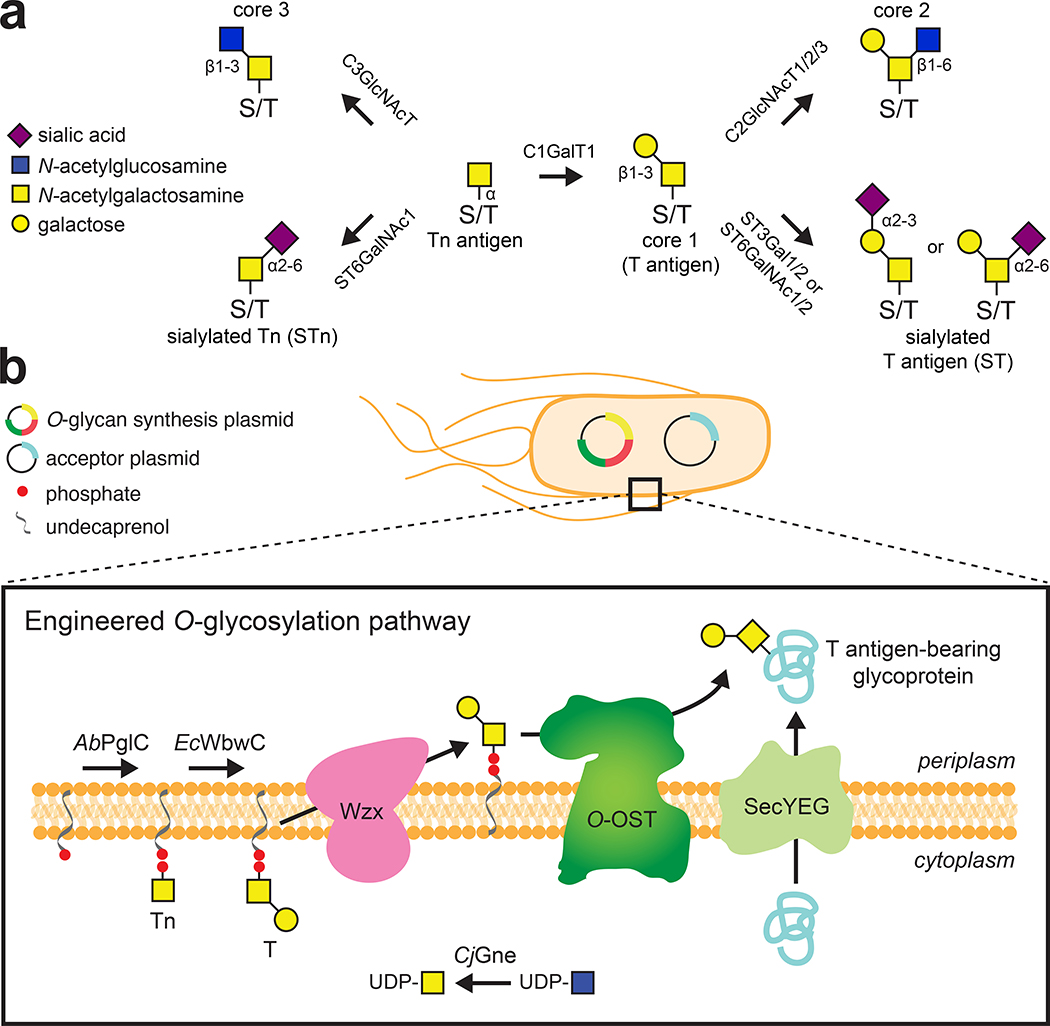
The enable orthogonal O-glycosylation in E. coli required assembling an en bloc pathway for producing the simplest mucin-type O-glycoform, GalNAcα (Tn antigen) (Fig. 1a and b). First, to eliminate formation of Und-PP-GlcNAc, an unwanted precursor in the context of mucin-type O-glycosylation, we deleted the gene encoding the native E. coli phosphoglycosyltransferase WecA from the genome of strain CLM24. This new strain, called CLM25, also lacked the waaL gene encoding the O-antigen ligase, a deletion that makes Und-PP-linked glycans available for the O-OST by preventing their unwanted transfer to lipid A-core.12 Next, we created a plasmid encoding the C. jejuni UDP-Glc(NAc) 4-epimerase (CjGne), which generates the activated sugar donor UDP-GalNAc from UDP-GlcNAc in the cytoplasm. While a number of epimerase homologs were considered, we chose CjGne because of its effectiveness in previous glycoengineering efforts.27, 28, 31 To address the lack of known enzymes that form Und-PP-GalNAc in E. coli, we enlisted PglC from Acinetobacter baumannii ATCC 17978 (AbPglC), which specifically transfers GalNAc to Und-PP in A. baumannii cells.32 Together, the CjGne and AbPglC enzymes comprised a putative pathway for Tn antigen biosynthesis.
Figure 1. Natural and synthetic mucin-type O-glycosylation pathways.
(a) Vertebrate mucin-type O-glycan synthesis originates from the hydroxyl group of a serine or threonine (S/T) amino acid by the addition of GalNAc by GalNAcT2 to form the Tn antigen structure. C1GalT1 adds β1,3-linked Gal to the initial GalNAcα-S/T to generate the T antigen. The Tn and T antigens can be further elaborated with GlcNAc and NeuNAc in a variety of ways, with a few illustrative examples shown here. (b) Representative schematic of engineered pathway for orthogonal O-glycoprotein synthesis in E. coli. CjGne maintains a pool of UDP-GalNAc that serves as the activated nucleotide sugar donor for AbPglC, which catalyzes the formation of Und-PP-linked GalNAc. EcWbwC extends Und-PP-GalNAc by a single Gal residue, yielding lipid-linked Gal-β1,3-GalNAc. Following flipping of the LLO to the periplasmic face of the cytoplasmic membrane by the native E. coli flippase Wzx, the preassembled T antigen glycan is transferred en bloc to a serine amino acid on a Sec pathway-exported acceptor protein by an O-OST such as NgPglO or NmPglL. It should be noted that the absence of EcWbwC enables generation of Tn-modified acceptor proteins while the further elaboration of Gal-β1,3-GalNAc with additional sugars such as NeuNAc followed by transfer to protein is also possible.
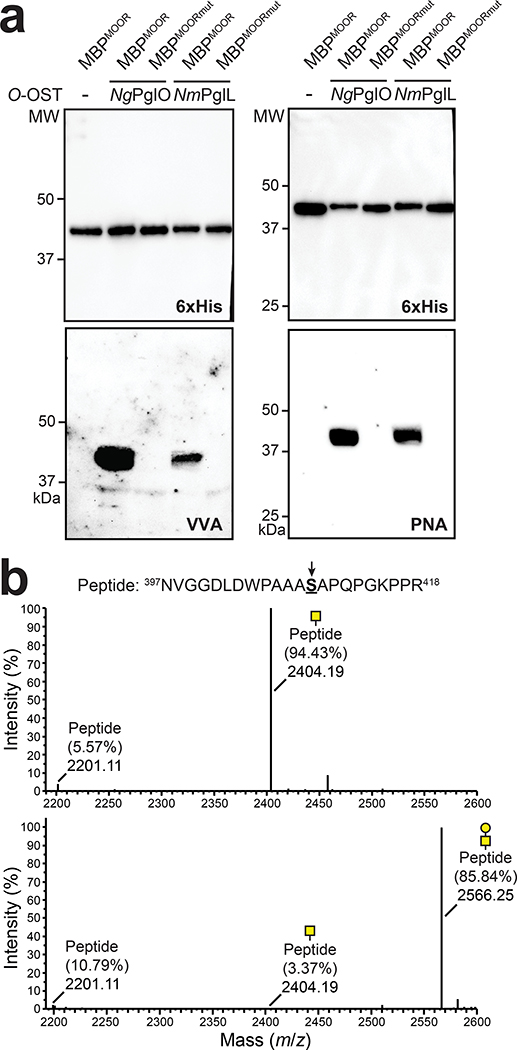
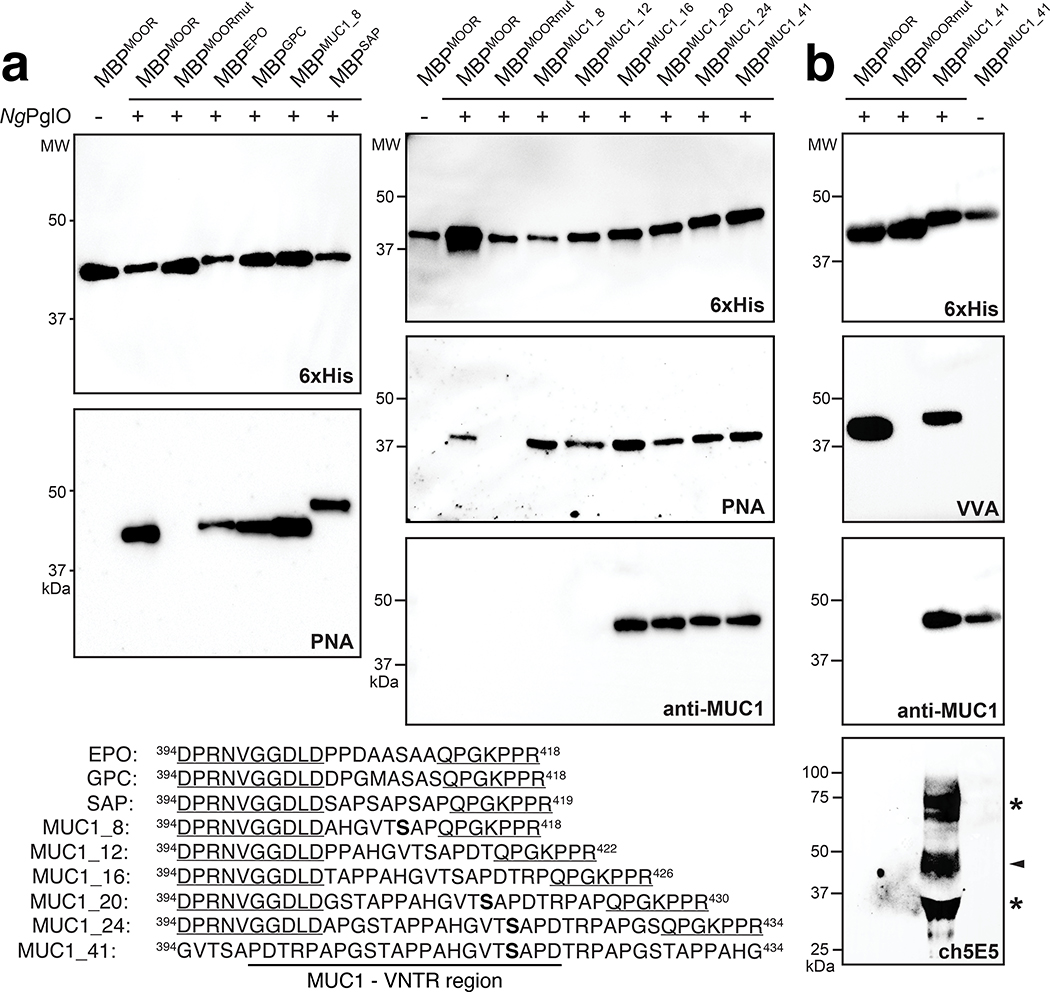
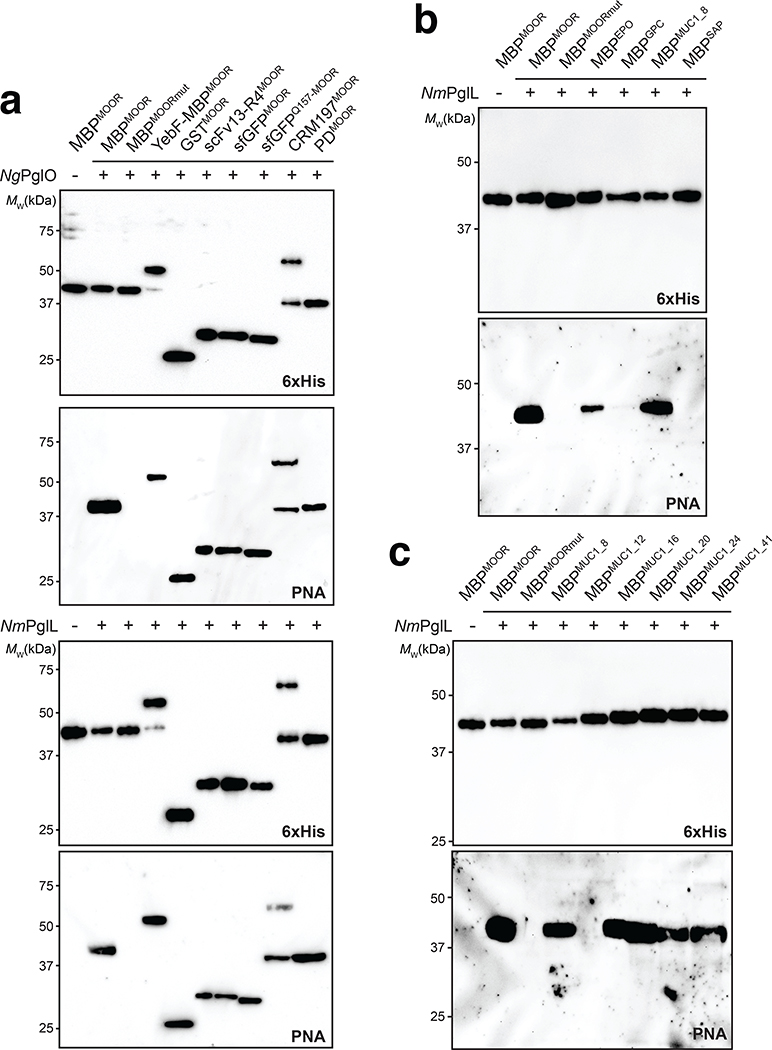
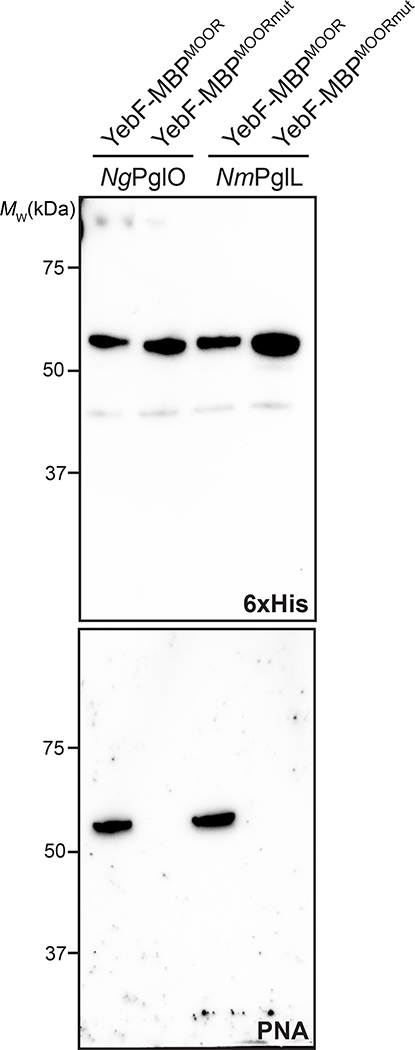
To transfer Und-PP-linked Tn antigen to hydroxylated amino acids in target proteins, we focused on the bacterial O-OST NmPglL and its ortholog NgPglO (95% identity). We hypothesized that these enzymes would recognize preassembled O-glycans on Und-PP and transfer them en bloc to Sec-translocated protein substrates in the periplasm (Fig. 1b). The rationale for this hypothesis was based on earlier findings that NmPglL can be functionally expressed in E. coli, leading to transfer of several structurally diverse glycans assembled on Und-PP.29, 30 To test this hypothesis, an O-OST gene was added to the Tn pathway yielding plasmids pOG-Tn-NmPglL and pOG-Tn-NgPglO. In parallel, we created a pEXT20-based plasmid encoding E. coli maltose-binding protein (MBP) modified at its N-terminus with the periplasmic targeting signal derived from E. coli DsbA33 and at its C-terminus with a MOOR (minimum optimal O-linked recognition) motif that was previously optimized for recognition by NmPglL.30 CLM25 cells co-transformed with these two plasmids produced MBPMOOR that was strongly glycosylated with the Tn antigen as revealed by immunoblots probed with Vicia villosa agglutinin (VVA), a lectin that preferentially binds single αGalNAc residues linked to serine or threonine (Fig. 2a). Importantly, glycosylation was completely undetectable when either O-OST was absent or the serine residue in the MOOR tag was substituted with glycine (MOORmut).
Figure 2. Biosynthesis of O-glycoproteins bearing Tn and T antigens.
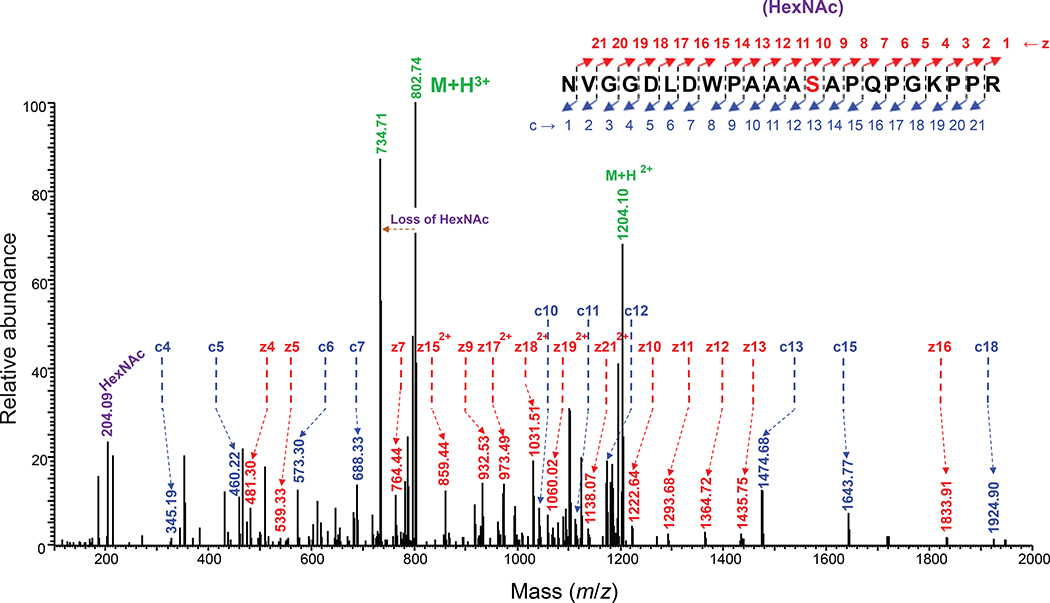
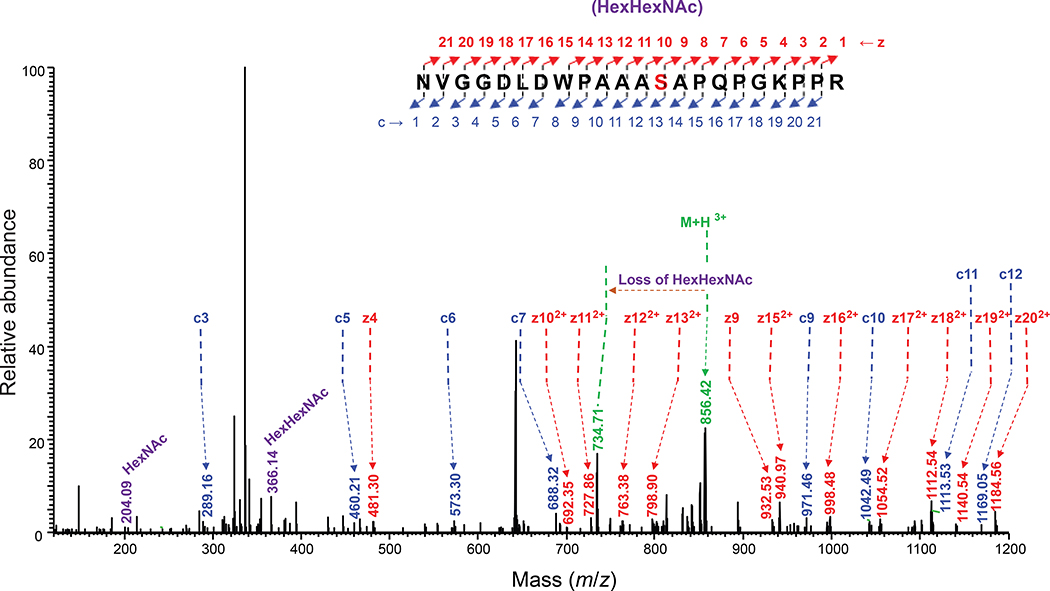
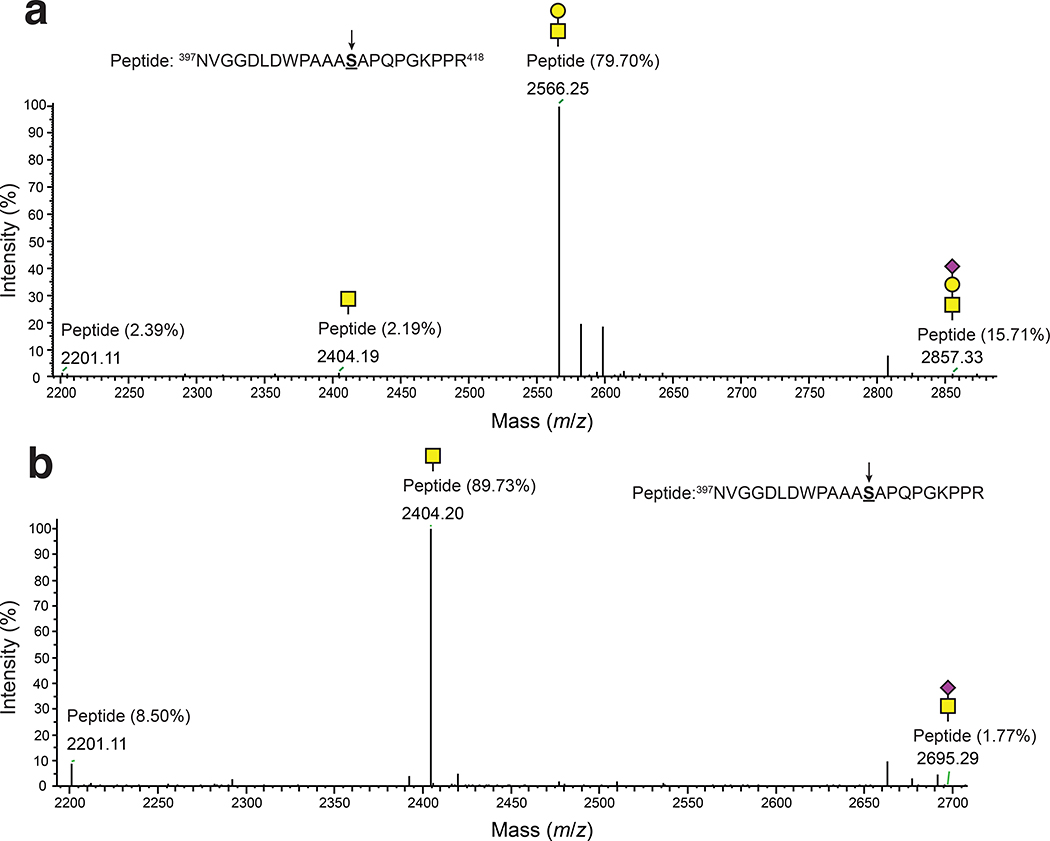
(a) Immunoblot analysis of acceptor proteins purified from CLM25 (W3110 ΔwecA ΔwaaL) cells co-transformed with pOG-Tn (left panels) or pOG-T (right panels) without an O-OST (−), pOG-Tn-NgPglO, or pOG-Tn-NmPglL along with pEXT-spDsbA-MBPMOOR or pEXT-spDsbA-MBPMOORmut as indicated. Absence of O-OST or mutation of acceptor serine to glycine in MBPMOORmut served as controls. Blots were probed with anti-hexa-histidine antibody (6xHis) to detect acceptor proteins and either VVA or PNA lectin to detect the Tn or T antigen, respectively. Molecular weight (MW) markers are indicated on the left. Results are representative of at least three biological replicates. See Supplementary Fig. 1 for uncropped versions of the images. (b) Nano-LC-MS/MS analysis of purified acceptor protein generated by CLM25 cells carrying plasmid pOG-Tn-NgPglO (top spectrum) or pOG-T-NgPglO (bottom spectrum) and pEXT-spDsbA-MBPMOOR. Sequence coverage of 88% and 75% was obtained for glycosylated MBPMOOR with Tn and T antigens, respectively, in the analysis. Spectrum for Tn glycoform reveals a dominant species (94% abundance) corresponding to peptide fragment bearing a single HexNAc and a less abundant (6%) aglycosylated species. Spectrum for T glycoform reveals a dominant species (86% abundance) corresponding to peptide fragment bearing a single HexHexNAc as well as two minor species bearing a single HexNAc and no modification (3% and 11% abundance, respectively). Sequence of detected peptide is shown at top with arrow denoting modified serine (bold underline) as determined by EThcD fragmentation analysis.
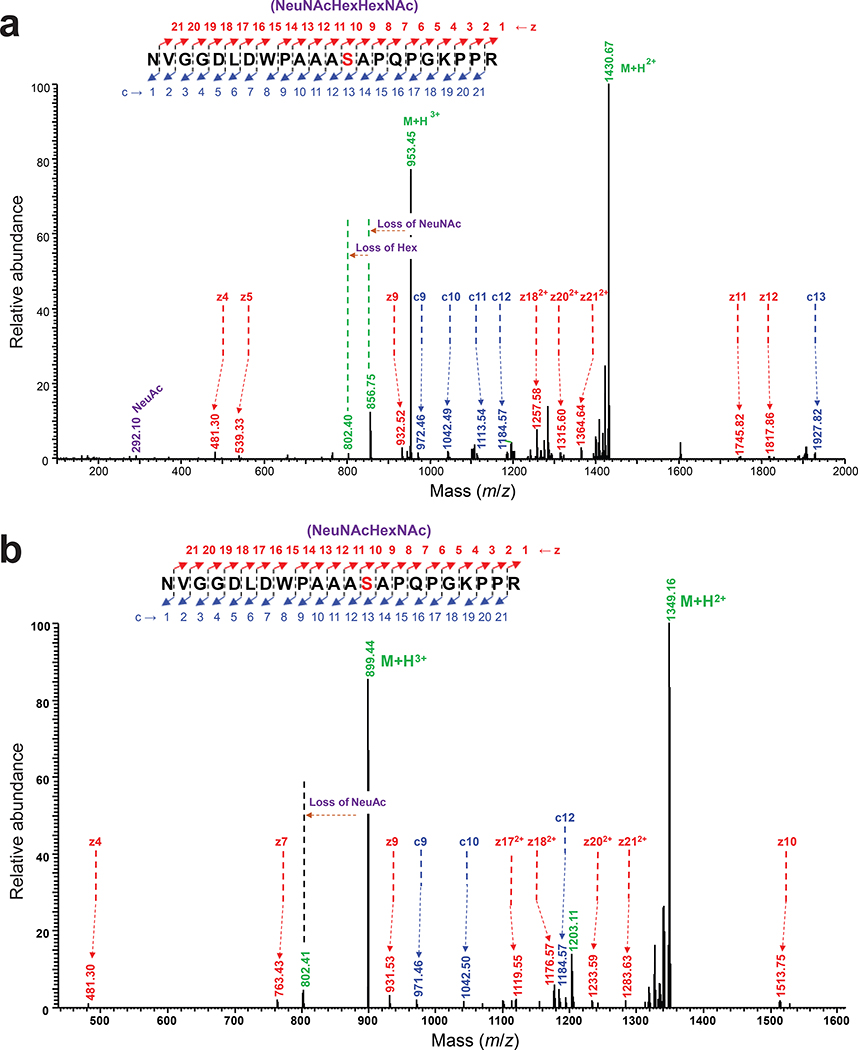
The glycosylated MBPMOOR was further examined by nanoscale liquid chromatography coupled to tandem mass spectrometry (nano-LC-MS/MS) to identify the modification sites. Glycosylation with only HexNAc was identified as the predominant species while a much smaller amount of aglycosylated peptide was also detected (Fig. 2b), consistent with immunoblot analysis. Electron-transfer/higher-energy collision dissociation (EThcD) fragmentation analysis was subsequently performed and unambiguously identified HexNAc modification on S409 within the MOOR sequence of MBPMOOR (Extended Data Fig. 1). Taken together, these results unequivocally established a route for orthogonal biosynthesis of Tn-modified O-glycoproteins.
Pathway extension enables T antigen biosynthesis.
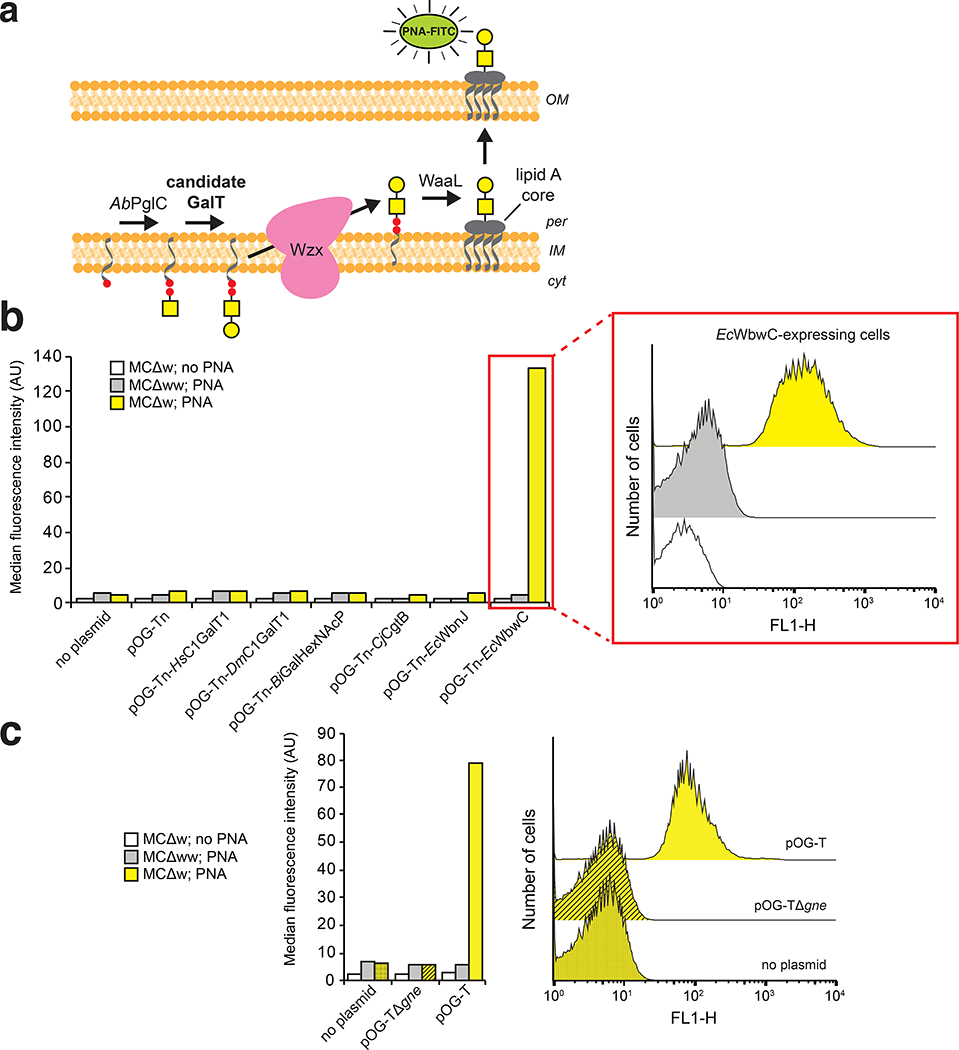
We next attempted biosynthesis of the T antigen (Gal-β1,3-GalNAcα), another mucin-type O-glycan that is absent in most normal tissues but present in many human cancers.34 The challenge here was the fact that Und-PP-GalNAc represents an atypical substrate for eukaryotic Gal transferases (GalT) that prefer GalNAcα-O-S/T. Therefore, we evaluated a panel of GalT enzymes including: core 1 synthase glycoprotein-N-acetylgalactosamine 3-β-galactosyltransferase from Homo sapiens (HsC1GalT1) and Drosophila melanogaster (DmC1GalT1); Bifidobacterium infantis D-galactosyl-β1–3-N-acetyl-D-hexosamine phosphorylase (BiGalHexNAcP); the “S42” mutant of C. jejuni β1–3-galactosyltransferase (CjCgtB) engineered with improved catalytic activity;35 and β−1,3-galactosyltransferases from enteropathogenic E. coli O86 (EcWbnJ) and enterohemorrhagic E. coli O104 (EcWbwC).
To screen GalT activity, we adapted a high-throughput flow cytometric assay previously developed by our group.7, 36 In this assay, Und-PP-linked glycans are flipped into the periplasm by the native E. coli flippase, Wzx, and transferred onto lipid A-core by the O-antigen ligase, WaaL (Extended Data Fig. 2a). Upon shuttling to the outer membrane, lipid A-core displays the attached glycan on the cell surface, where it is readily detected by fluorescently tagged antibodies or lectins. When screened by flow cytometry using FITC-conjugated Arachis hypogaea peanut agglutinin (PNA) lectin, which recognizes T antigen, only cells expressing EcWbwC were observed to transfer galactose to Und-PP-linked GalNAc (Extended Data Fig. 2b); hence, co-expression of CjGne, AbPglC, and EcWbwC from plasmid pOG-T was used for all experiments involving T antigen or derivatives thereof. Importantly, EcWbwC activity was dependent on CjGne, which converts UDP-GlcNAc to UDP-GalNAc (Extended Data Fig. 2c), confirming that the reducing-end monosaccharide was indeed GalNAc.
To transfer T antigen to proteins, O-OST genes were added to the T antigen pathway, yielding plasmids pOG-T-NmPglL and pOG-T-NgPglO. CLM25 cells co-transformed with one of these plasmids along with the plasmid encoding MBPMOOR produced acceptor proteins that were glycosylated with T antigen as revealed by immunoblots probed with PNA (Fig. 2a). As expected, this glycosylation depended on the O-OST and the serine residue in the MOOR tag. Nano-LC-MS/MS analysis revealed glycosylation with HexHexNAc as the predominant species (Fig. 2b), indicating efficient T antigen assembly and transfer to protein by orthogonal pathway enzymes. EThcD fragmentation analysis again confirmed HexHexNAc modification on S409 of MBPMOOR (Extended Data Fig. 3).
Orthogonal biosynthesis of sialylated O-glycoforms.
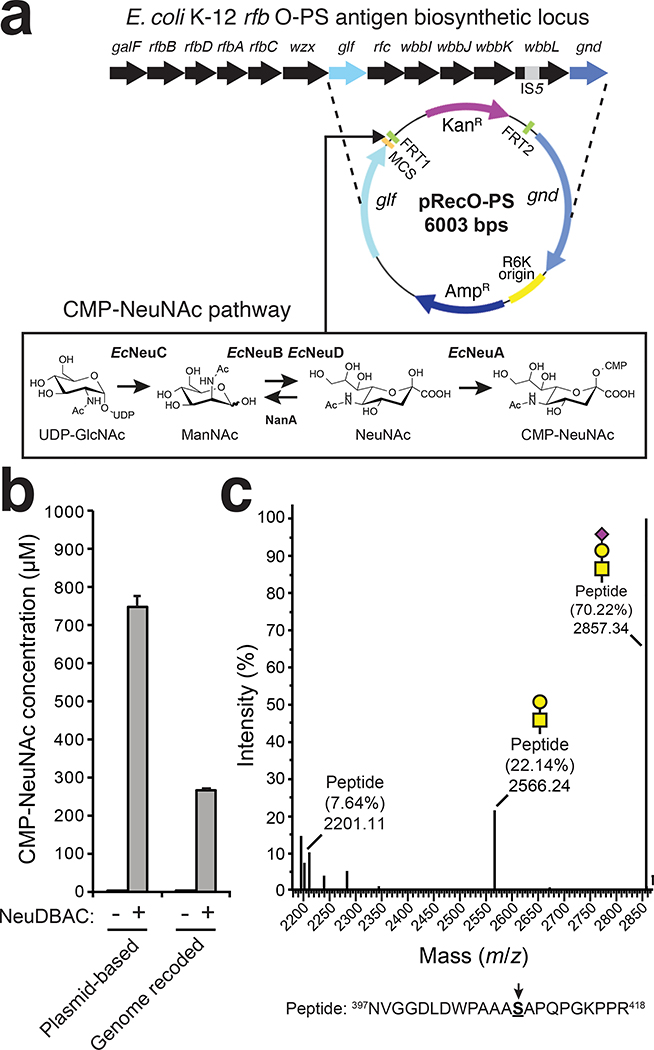
To produce O-glycans bearing sialic acid (NeuNAc), including the STn (NeuNAc-α2,6-GalNAcα) and ST antigens (NeuNAc-α2,3-Gal-β1,3-GalNAcα) (Fig. 1a) that are commonly observed in cancer, required engineering of our host strain to generate CMP-NeuNAc. To this end, we first constructed a plasmid encoding the E. coli K1 neuDBAC genes (Fig. 3a), which enable production of CMP-NeuNAc from UDP-GlcNAc in K-12 strains.31 In addition, the nanA gene encoding N-acetylneuraminate lyase was deleted from the genome of our host strain to avoid catabolism of CMP-NeuNAc. LC-MS analysis confirmed that nanA-deficient cells carrying the CMP-NeuNAc pathway plasmid produced significant levels of CMP-NeuNAc (Fig. 3b). Next, the gene encoding E. coli O104 WbwA (EcWbwA) sialyltransferase, which we predicted would modify Und-PP-linked T antigen with α2,3-linked NeuNAc, was added to the MBPMOOR expression plasmid. When this latter plasmid was added to nanA-deficient cells carrying the CMP-NeuNAc pathway and pOG-T-NgPglO plasmids, glycosylation of MBPMOOR with NeuNAcHexHexNAc was observed (Extended Data Fig. 4a). However, the HexHexNAc-modified glycoform was significantly more abundant, suggesting inefficient extension of T antigens with NeuNAc in this host.
Figure 3. Orthogonal biosynthesis of sialylated O-glycans.
(a) Schematic of glyco-recoding strategy for genomic integration of CMP-NeuNAc biosynthetic pathway in E. coli. Genes encoding E. coli K1 neuDBAC were cloned in shuttle vector pRecO-PS, which was used to insert the neu operon in place of the O-PS pathway between glf and gnd in E. coli MC4100 strain background. (b) LC-MS analysis of lysates derived from glyco-recoded cells, comparing intracellular CMP-NeuNAc levels measured in cells carrying plasmid-encoded copies of neuDBAC genes versus those carrying genomically integrated copy of neuDBAC. Cells lacking the neuDBAC genes served as controls. Data is the average of three biological replicates and error bars represent the standard deviation of the mean. (c) Nano-LC-MS/MS analysis of purified acceptor protein generated by glyco-recoded cells carrying plasmid pOG-T-NgPglO and pEXT-spDsbA-MBPMOOR-EcWbwA. Sequence coverage of 94% was obtained for the MBPMOOR protein in the analysis. Spectrum reveals a dominant species (70% abundance) corresponding to the indicated peptide fragment bearing a single NeuNAcHexHexNAc and two minor species bearing a single HexHexNAc and no modification (22% and 8% abundance, respectively). Sequence of detected peptide is shown at bottom with arrow denoting modified serine (bold underline) as determined by EThcD fragmentation analysis.
We speculated that this low efficiency might be overcome by chromosomal integration of the multi-gene CMP-NeuNAc pathway, a strategy that previously increased glycosylation efficiency of an orthogonal N-linked pathway.37 To test this notion, a glyco-recoding strategy37 was used to integrate the CMP-NeuNAc pathway in place of the non-essential O-polysaccharide (O-PS) antigen biosynthesis pathway in the genome (Fig. 3a). The net effect was a reduction in both the number of required plasmids and the copy number of the neu genes. Following genomic replacement of the O-PS pathway with the CMP-NeuNAc pathway in nanA-deficient cells, appreciable intracellular accumulation of CMP-NeuNAc was again observed (Fig. 3b). While the overall CMP-NeuNAc concentration was lower compared to the plasmid-based system, the amount of sialylated glycan on MBPMOOR was dramatically increased in the glyco-recoded host strain, with this glycan representing the most abundant glycoform (Fig. 3c) and occurring on the expected S409 glycosite (Extended Data Fig. 5a).
A nearly identical strategy for producing STn antigen was carried out using the same glyco-recoded host strain carrying plasmid pOG-Tn-NgPglO in place of pOG-T-NgPglO and the pEXT-based acceptor protein plasmid with α2,6-sialyltransferase from Photobacterium sp. JT-ISH-224 in place of EcWbwA. These cells generated MBPMOOR bearing STn antigen albeit with relatively low sialylation (Extended Data Fig. 4b; Extended Data Fig. 5b). Nonetheless, these results showcase the modularity of the O-glycosylation platform, with the introduction of appropriate GTs providing a direct route to more elaborated glycan structures.
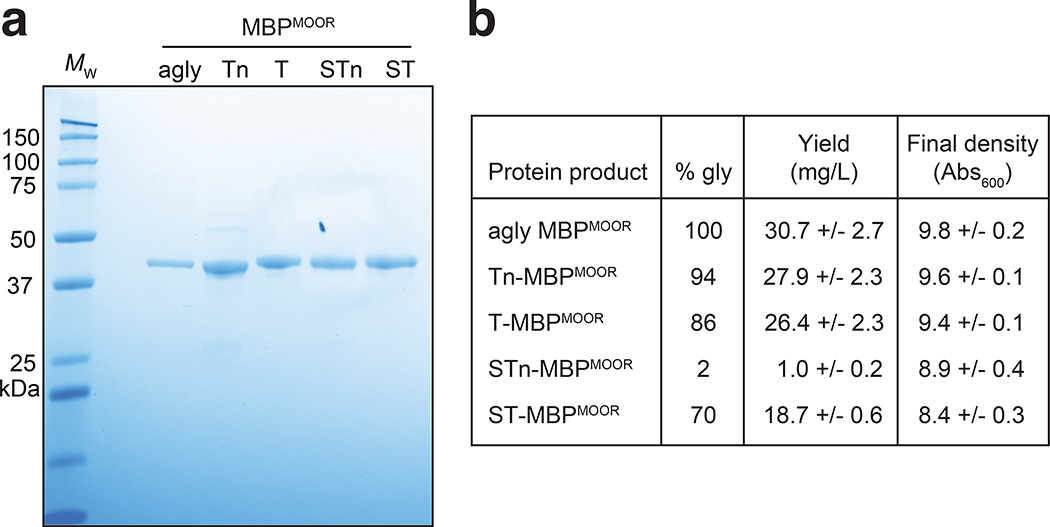
On average, ~30 mg/L of glycosylated MBPMOOR with each of the different O-glycan structures was produced from small-scale cultures (Extended Data Fig. 6a and b). These yields compared favorably to the yields of 60–80 mg/L obtained previously for processive glycosylation of target proteins with T antigen in the E. coli cytoplasm.28 It should also be noted that final culture densities of all glycoprotein-producing strains were comparable to that of the control strain expressing aglycosylated MBPMOOR (Extended Data Fig. 6b).
Cell-free extracts catalyze O-glycosylation.
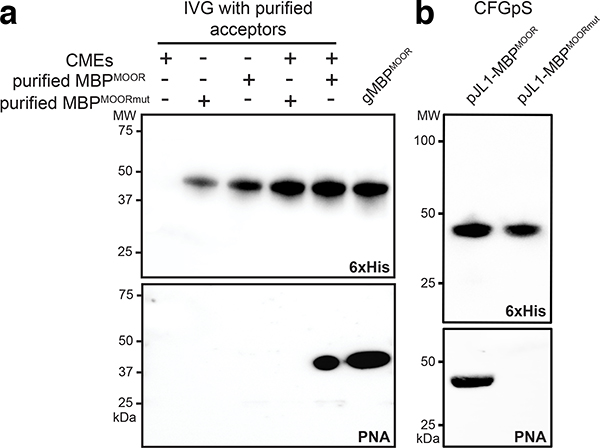
Cell-free modalities are emerging as useful glycoscience tools and for on-demand biomanufacturing of glycoprotein products.10, 11 However, there are currently no cell-free platforms for total biosynthesis of O-glycoproteins. To address this gap, we first evaluated an in vitro glycosylation strategy that combined purified acceptor proteins with partially purified glycosylation machinery. Crude membrane extracts selectively enriched with NgPglO and UndPP-linked T antigen were prepared from CLM25 cells carrying plasmid pOG-T-NgPglO. Upon addition of purified acceptor protein to these “glyco-enriched” extracts, clear glycosylation was observed (Fig. 4a). Next, we attempted a more integrated approach in which cell-free transcription, translation, and glycosylation were carried out together in a single pot. This involved preparing crude S12 extracts from the same CLM25 cells carrying plasmid pOG-T-NgPglO. To initiate cell-free glycoprotein synthesis (CFGpS), the resulting glyco-enriched S12 extracts containing Und-PP-linked T antigen and NgPglO were primed with plasmid DNA encoding the acceptor protein. Following this reaction, clearly detectable MBPMOOR glycosylation was observed, whereas no glycosylation was detected in reactions charged with plasmid DNA encoding MBPMOORmut (Fig. 4b). These results establish that orthogonal O-glycosylation can be functionally reconstituted outside the cell, giving rise to one-pot O-glycoprotein biosynthesis.
Figure 4. Cell-free O-glycosylation using glyco-enriched extracts.
(a) Immunoblot analysis of in vitro glycosylation (IVG) reactions that were performed by incubating purified MBPMOOR or MBPMOORmut acceptor proteins in the presence of crude membrane extracts (CMEs) prepared from CLM25 cells carrying pOG-T-NgPglO (+) or pOG-T without an O-OST (−). Glyco-enriched CMEs alone (lane 1) or glycosylated MBPMOOR (gMBPMOOR) that was previously purified from glycoengineered bacteria (lane 5) served as negative and positive controls, respectively. (b) Immunoblot analysis of acceptor proteins produced by integrated CFGpS in which transcription, translation, and O-glycosylation were performed altogether in a single reaction. Specifically, 1 ml reactions comprised of glyco-enriched S12 extract derived from CLM25 cells carrying pOG-T-NgPglO were primed with plasmid pJL1-MBPMOOR or pJL1-MBPMOORmut as indicated. Blots in (a) and (b) were probed with anti-hexa-histidine antibody (6xHis) to detect the acceptor proteins and PNA to detect the T antigen. Molecular weight (MW) markers are indicated on the left. Results are representative of at least three biological replicates. See Supplementary Fig. 2 for uncropped versions of the images.
O-glycosylation of diverse acceptor protein targets.
To determine the range of glycosylatable acceptor proteins, we grafted the MOOR tag onto the C-terminus of several proteins including: E. coli glutathione-S-transferase (GST); a single-chain Fv antibody fragment specific for β-galactosidase (scFv13-R4); and two conjugate vaccine carrier proteins, namely cross-reacting material 197 (CRM197) and Haemophilus influenzae protein D (PD). We also created a chimera comprised of E. coli secretory protein YebF fused to MBPMOOR as well as two variants of superfolder GFP (sfGFP), one with a C-terminal MOOR tag and the other with the MOOR motif grafted in an internal loop starting at Gln157. It should be noted that scFv13-R4, sfGFP, and YebF have all been N-glycosylated in E. coli previously7, 10, 25, 33 while CRM197 and PD represent carrier proteins used in licensed conjugate vaccines. When expressed in the presence of the T antigen pathway, each protein cross-reacted with PNA (Extended Data Fig. 7a), confirming that O-glycosylation was compatible with different protein contexts including terminal and internal locations. It is also noteworthy that YebF-MBPMOOR and YebF-MBPMOORmut both accumulated in the extracellular culture medium with only YebF-MBPMOOR cross-reacting with PNA (Extended Data Fig. 8), indicating that YebF-mediated secretion is harmonious with en bloc O-glycosylation, as it was for N-glycosylation.33
We further evaluated system modularity by swapping the 8-residue core sequence of the MOOR tag with different human or synthetic O-glycosylation motifs. These included: 8 residues surrounding the S126 O-glycosite in human erythropoietin (EPO);38 8 residues surrounding the S24 O-glycosite in human glycophorin C (GPC), a surface glycoprotein found on red blood cells that marks the Gerbich antigen system;39 8 residues derived from the ectodomain of human mucin 1 (MUC1), which is expressed on the apical surface of glandular epithelial cells at low levels but following oncogenic transformation is expressed at very high levels and with altered glycosylation;34 and synthetic “SAP” motif that was designed de novo based on known glycosite preferences of NmPglL.30 When each construct was expressed in the presence of NgPglO, strong glycosylation with T antigen was observed (Fig. 5a). Interestingly, while NmPglL also robustly glycosylated the EPO- and MUC1-derived sequences, it showed weak glycosylation of the GPC-derived sequence and no detectable activity towards the SAP sequence (Extended Data Fig. 7b), revealing subtle differences in O-OST substrate selectivity. Collectively, these results highlight the ability of our platform to modify O-glycosites in human proteins.
Figure 5. O-linked glycosylation of diverse protein targets.
(a) Immunoblot analysis of acceptor proteins purified from CLM25 cells co-transformed with pOG-T-NgPglO (+) or pOG-T without NgPglO (−) along with pEXT-based plasmid encoding each of the different protein targets as indicated. Absence of NgPglO or mutation of acceptor serine to glycine in MBPMOORmut served as negative controls. Blots were probed with anti-hexa-histidine antibody (6xHis) to detect acceptor proteins and PNA lectin to detect the T antigen. Additional blot for MUC1 variants was probed with murine H23 antibody (anti-MUC1) that is specific for APDTRP motif in human MUC1. Shown at bottom are acceptor sequences derived from human EPO, GPC, and MUC1 as well as synthetic SAP. All acceptor motifs except for MUC1_41 are presented in the context of the hydrophilic flanking regions derived from the MOOR tag (underline). MUC_41 was designed without hydrophilic flanking residues and includes the VNTR region as indicated. Serine amino acids determined to be glycosylated by EThcD fragmentation analysis are shown in bold font. (b) Immunoblot analysis of MUC1_41 expressed in CLM25 cells carrying pOG-Tn-NgPglO (+) or pOG-Tn without NgPglO (−). Also shown is MBPMOOR and MBPMOORmut derived from the same cells. Blots were probed with anti-6xHis antibody to detect acceptor proteins, VVA lectin to detect the Tn antigen, anti-MUC1 to detect MUC1_41, and chimeric 5E5 antibody (ch5E5) to detect Tn-MUC1. Arrow denotes the expected Tn-MUC1 glycoform, while asterisks denote higher and lower molecular weight species that may represent SDS-stable multimers and degradation products, respectively. Molecular weight (MW) markers are indicated on the left of each blot. All immunoblot results are representative of at least three biological replicates. See Supplementary Fig. 3 for uncropped versions of the images.
Biosynthesis of antigenically-relevant MUC1 glycoforms.
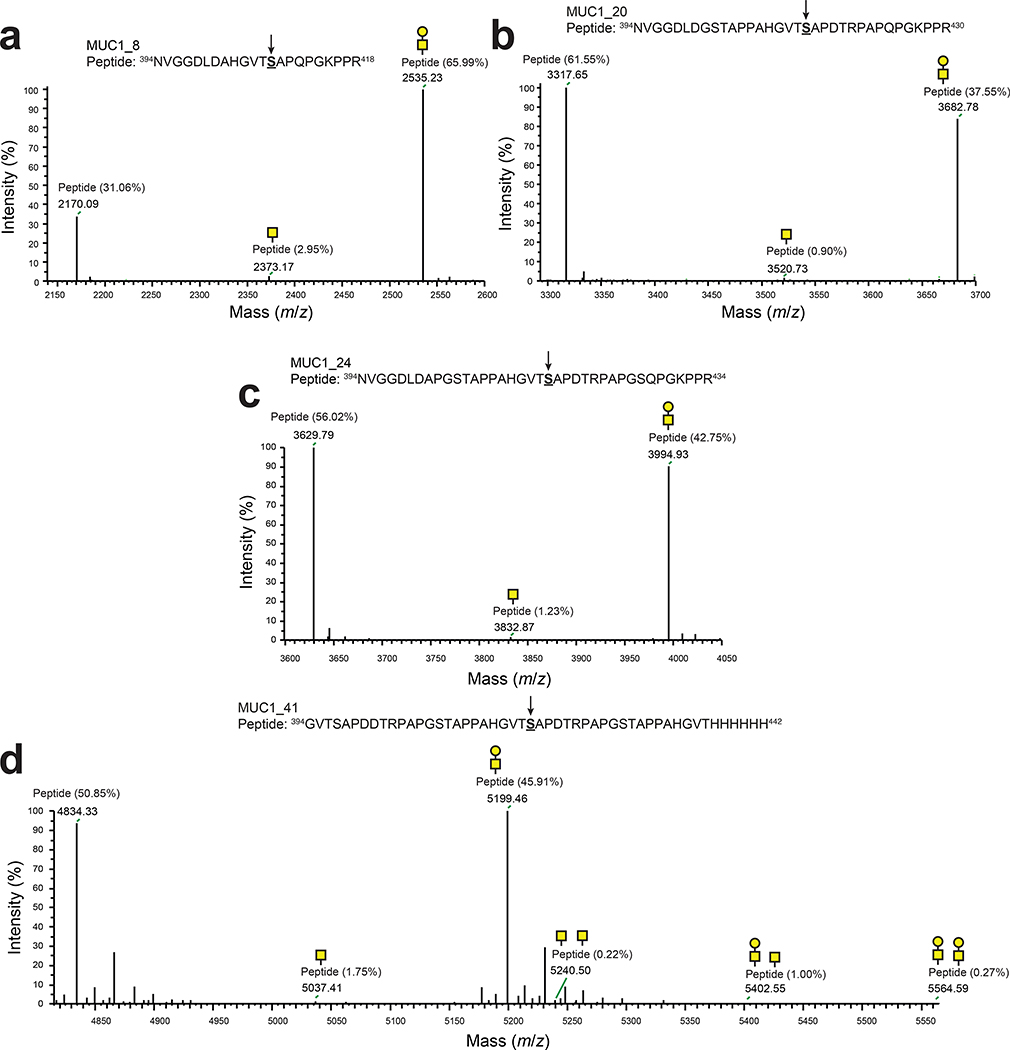
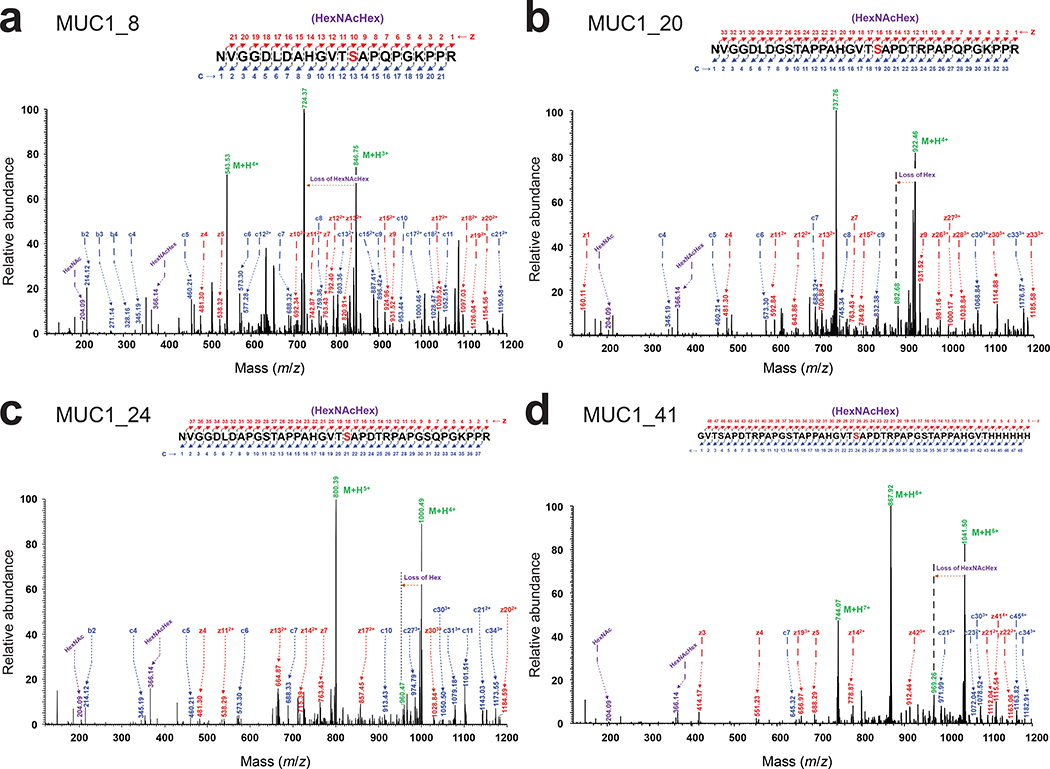
To generate additional MUC1 glycoforms with relevance to human cancer, we focused on the variable number of tandem repeats (VNTRs) of MUC1 that consist of 20–120 repeats of a 20-amino acid sequence (PDTRPAPGSTAPPAHGVTSA) and contain five potential O-glycosylation sites (underlined).40 Here, we created four VNTR-derived sequences by incrementally extending the MUC1_8 motif. Each of these was cloned between the hydrophilic flanking regions of the MOOR motif and subsequently expressed in CLM25 cells carrying either pOG-T-NgPglO or pOG-T-NmPglL. We chose the T antigen-producing host strain because tumor-associated MUC1 is aberrantly glycosylated with truncated O-glycans including T antigen.34 Following expression in bacteria carrying the T antigen pathway, each MUC1 motif was strongly glycosylated by NgPglO (Fig. 5a). NmPglL similarly modified all these motifs except for MUC1_12, which was not detectably glycosylated (Extended Data Fig. 7c) and indicated another subtle difference in O-OST substrate selectivity. It should also be noted that MUC1_16, MUC1_20, and MUC1_24 each cross-reacted with the mouse monoclonal antibody H23 (Fig. 5a), which recognizes the MUC1 APDTRP epitope on the surface of human breast cancer cells41 and confirms the antigenic relevance of these MUC1 peptides. HexHexNAc-modified MUC1_8, MUC1_20, and MUC1_24 were identified as the predominant glycoforms (Extended Data Fig. 9a–c), with the most abundant glycoforms corresponding to HexHexNAc modification at the same serine residue in each construct (Extended Data Fig. 10).
To generate more antigenically authentic glycoforms, we focused on a 41-residue MUC1 sequence containing the 20-residue VNTR flanked with additional stretches of the MUC1 repeat but without the original MOOR flanking residues. Importantly, both NgPglO and NmPglL were able to transfer T antigen to this construct (Fig. 5b; Extended Data Fig. 7c). A single HexHexNAc modification on MUC_41 was the predominant glycoform and was found on the same serine residue identified above (Extended Data Fig. 10). In addition to aglycosylated peptide, other minor T and Tn modifications were also detected (Extended Data Fig. 9d), suggesting multiply glycosylated forms. We attempted targeted HCD and ETD MS/MS analysis to identify and map the location of these minor glycan modifications; however, we were unable to assign the glycosites because of the lower intensities of these glycopeptides and the lack of key fragments on the MS/MS spectrum needed for unambiguous site assignment. Low-resolution ion trap-based detection of ETHcD fragments was also unable to yield conclusive evidence for additional O-glycosylation beyond the S417 modification. Nonetheless, these results demonstrate that authentic human O-glycoprotein epitopes can be generated using our engineered glycosylation system without the need for hydrophilic flanking regions.
As was seen for the other APDTRP-containing MUC1 sequences, T-modified MUC1_41 cross-reacted with H23 (Fig. 5b). While this result confirmed creation of an antigenically-intact MUC1 epitope, H23 binding was not dependent on the O-glycan, consistent with the known specificity of this antibody.41 In contrast, the murine monoclonal antibody 5E5 binds all Tn and STn glycoforms of the MUC1 tandem repeat but does not bind aglycosylated MUC1 peptides.42 To determine whether MUC1 glycoforms could be produced that cross-reacted with this glycoform-specific antibody, we first expressed the MUC1_41 construct in the presence of the Tn pathway, yielding strongly glycosylated MUC1_41 (Fig. 5b). Importantly, the Tn-modified MUC1_41 but not its aglycosylated counterpart was readily detected by the glycoform-specific antibody. This same antibody did not show reactivity for MBPMOOR bearing Tn antigen, consistent with the fact that both glycan and underlying peptide are required for recognition.42 Overall, this glycoform-dependent reactivity provides important validation of our glycoengineered bacteria as a platform for producing glycoprotein epitopes that are antigenically distinct and relevant to cancer immunotherapy.
Discussion
In this work, we engineered orthogonal O-glycoprotein biosynthesis in E. coli by rewiring the cell’s metabolism to provide necessary sugar donors and ectopically expressing specific GTs and OSTs from diverse organisms. The system was highly modular as evidenced by the ability to generate multiple O-glycan structures and post-translationally modify a panel of acceptor protein targets. Unlike previous mucin-type O-glycoengineering in E. coli that focused on processive glycosylation mechanisms,26–28 we took an unconventional approach based on the en bloc O-glycosylation mechanism found natively in some bacteria. Although modeled after this process, the collection of synthetic O-glycosylation pathways described here has no direct biological equivalent and includes the first biosynthetic routes to sialylated mucin-type O-glycosylation in E. coli.
One advantage our strategy is the opportunity to leverage diverse enzymes from all domains of life that naturally operate on lipids as well as proteins. A number of bacteria employ glycomimicry strategies in which endogenous GTs construct human-like oligosaccharides that serve to cloak cell-surface components as a means to evade host immune responses. By enlisting these bacterial GTs, one could further expand the repertoire of O-glycans that can be assembled in E. coli. Moreover, because many human GTs are difficult to functionally express in bacteria, often requiring specialized chaperones or solubility-enhancing fusion partners,43, 44 GTs of microbial origin represent a potential workaround for construction of human-like O-glycans as we demonstrated here.
Another advantage of our strategy is the utilization of bacterial O-OSTs that have an inbuilt ability to transfer glycans onto both serine and threonine residues, whereas human GalNAcT2 used previously is limited to threonine. These enzymes exhibit extreme glycan substrate permissiveness as exemplified by NmPglL.29, 30 Here, we leveraged this promiscuity to show that NmPglL and its NgPglO ortholog can transfer human-like O-glycan structures. The compatibility of acceptor sequences with these enzymes is much less understood. While it has been shown that individual O-OSTs can modify multiple protein substrates,45 there is no clear sequon for glycosylation and the O-glycan attachment sites are in flexible, low-complexity regions, thereby hindering glycoprotein engineering efforts. A breakthrough in this regard was the identification of the MOOR motif that together with two additional hydrophilic flanking sequences could be recognized by NmPglL30 and, as we showed here, NgPglO. Using these hydrophilic flanking sequences, we expanded the list of glycosylatable sequences to include several human and synthetic O-glycosites. The observation that NmPglL and NgPglO could glycosylate varying-length human MUC1 sequences suggested a much greater flexibility than was first reported for these enzymes.30
Most surprising was the site-directed O-glycosylation of MUC1_41 that lacked the flanking sequences, addressing earlier skepticism about the ability of bacterial O-OSTs to discern mammalian O-glycosites.28 The O-glycosylated MUC1_41 produced here was structurally similar to glycopeptides that are reactive towards IgG/IgM antibodies46 and human MHC class I molecules.47 Indeed, recognition of Tn-modified MUC1_41 by a glycoform-specific antibody indicated the creation of an antigenically authentic glycoform. Moreover, the relatively low glycan occupancy on MUC1_41 (~1 or 2 O-glycans per repeat) may bode well for immunotherapeutic discovery given that a synthetic 60-residue MUC1 tandem-repeat peptide, which was extensively glycosylated (5 O-glycans per repeat), elicited only modest antibody responses.42 This weak humoral response results from an inability of antigen-presenting cells to process densely glycosylated MUC1 glycopeptides.48 In contrast, a glycopeptide modified with just a single O-glycan elicited more robust antibody titers and also activated cytotoxic T lymphocytes, which amounted to superior tumor prevention.49
Looking forward, we anticipate that the platform described here could find use in the scalable biosynthesis of O-glycoprotein therapeutics and vaccines. To gain access to greater O-glycoprotein structural space may require additional O-OSTs such as those from Bacteroidetes that modify proteins at a minimal 3-residue motif, D-(S/T)-(A/L/V/I/M/T).50 Directed evolution of GTs to tailor substrate specificity and metabolic engineering to drive pathway performance towards higher conversion could be enabled through a high-throughput screen for O-glycosylation akin to ‘glycoSNAP’, a bacterial colony blot assay for N-linked glycosylation that was used previously to evolve bacterial N-OST variants with greatly relaxed sequon specificity.25 A first important step in this direction was our demonstration that O-glycoproteins can be secreted out of the cell by genetic fusion to the C-terminus of the secretory protein YebF, a feat that is not possible with cytoplasmic O-glycosylation systems. Beyond O-glycoprotein production, the ability of the glycoengineered strains to produce custom glyco-ligands such as O-glycosylated GST and sfGFP could facilitate pulldown assays and cell labeling experiments, respectively, with the potential to uncover and characterize binding partners of structurally defined O-glycoforms. Altogether, our results define a versatile platform for site-directed O-glycosylation of proteins with different mucin-type O-glycans, thereby expanding the bacterial glycoengineering toolkit.
Methods
Bacterial strains and growth conditions.
All strains used in the study are listed in Supplementary Table 1. E. coli strain DH5α and NEB 10-beta were used for cloning and maintenance of plasmids while BL21(DE3) was used to produce purified acceptor proteins for IVG reactions. Unless otherwise noted, strain CLM25 was used for all O-glycoprotein expression and was constructed by deleting wecA from CLM2412 through P1vir phage transduction where strain JW3758–2(Δrfe-735::kan) from the Keio collection51 was used as the donor. MC4100 ΔwecA (MCΔw) and MC4100 ΔwecA ΔwaaL (MCΔΔw) were used as the hosts for flow cytometry screening and glyco-recoding to introduce the CMP-NeuNAc biosynthesis pathway. Strain MCΔw was generated by P1vir phage transduction of strain MC4100 to delete wecA using JW3758–2(Δrfe-735::kan) as the donor. Subsequent P1vir phage transduction of MCΔw to delete waaL using JW3597–1(ΔrfaL734::kan) as donor yielded strain MCΔΔw. In all cases, after each deletion the linked kanamycin resistance (KanR) cassette was removed by transformation with the temperature-sensitive plasmid pCP20 as described in detail elsewhere.52 The E. coli K1 neuDBAC genes encoding the CMP-NeuNAc biosynthesis pathway31 were integrated into the chromosome of MCΔΔw using a previously described glyco-recoding strategy.37 Briefly, the neuDBAC gene cluster was cloned into the pRecO-PS shuttle vector, which is uniquely designed to promote homologous recombination-based insertion of genes-of-interest in place of the existing genomic locus encoding the O-PS biosynthetic pathway between the glf and gnd genes (Fig. 3a). Next, the MCΔΔw strain carrying plasmid pKD46 encoding the λ-red recombinase was rendered electrocompetent and subsequently transformed with a linear PCR product derived from the pRecO-PSneuDBAC shuttle vector, which included the neuDBAC genes, the KanR cassette, and the flanking glf and gnd genes. A kanamycin-resistant chromosomal integrant was then chosen and the KanR marker was removed using the temperature-sensitive pE-FLP plasmid expressing the FLP recombinase, yielding strain MCΔΔw-neuO-PS. Finally, the genomic copy of nanA encoding the N-acetylneuraminate lyase involved in the catabolism of NeuNAc was deleted by P1vir phage transduction using Keio strain JW3194–1 (ΔnanA753::kan) as donor to create strain MCΔΔwΔn-neuO-PS. For extracellular secretion of O-glycoproteins, a secretion-optimized derivative of CLM24 was generated by deleting the yaiW gene53 by P1vir phage transduction using Keio strain JW0369 (ΔyaiW743::kan) as donor.
All cultures were grown at 37°C in Luria-Bertani (LB) media containing D-glucose (0.2% w/v) as well as 20 μg/ml chloramphenicol (Cm), 100 μg/ml trimethoprim (Tmp), and 100 μg/ml ampicillin (Amp) as needed for plasmid maintenance. Induction of protein expression was always performed at mid-log phase (Abs600 ~0.6) with 0.1 mM isopropyl β-D-thiogalactoside (IPTG) and 0.2% (w/v) L-arabinose at 16°C for 16–20 h. For yield determination experiments, cells were grown in 100 ml of Terrific Broth (TB) at 37oC until mid-log phase and then induced with 1 mM IPTG and 0.2% (w/v) L-arabinose at 16°C for 22 h. Following expression, cells were harvested and protein purification was performed as described below.
Plasmid construction.
All plasmids used in the study are listed in Supplementary Table 1. Plasmid construction was performed according to standard cloning protocols using restriction enzymes from New England Biolabs. The pOG backbones were cloned in either the yeast recombineering plasmid pMW07 7 or a modified derivative of pMW07, namely pMW08, in which the yeast origin of replication and URA3 gene were deleted. Plasmid pOG-Tn was generated by the Gibson assembly method. Briefly, the genes encoding CjGne and AbPglC were PCR amplified with overlapping regions, and subsequently cloned into pMW08 using the NEBuilder HiFi DNA Assembly Cloning Kit (New England Biolabs) to generate plasmid pOG-Tn. Each of the candidate GalT enzymes was cloned into pOG-Tn by first obtaining codon-optimized DNA corresponding to each GalT gene synthesized with overlapping regions to facilitate recombination (Twist Biosciences). These genes were then amplified by PCR and cloned into pOG-Tn by Gibson assembly. A similar strategy was followed to generate plasmid pOG-T. Briefly, the genes encoding CjGne, AbPglC, EcWbwC were PCR amplified with overlapping regions, and subsequently cloned into pMW07 using the NEBuilder HiFi DNA Assembly Cloning Kit (New England Biolabs) to generate the pOG-T. Genes encoding NgPglO and NmPglL were added to pOG-Tn and pOG-T as follows. First, codon-optimized DNA encoding the NgPglO and NmPglL genes was synthesized with overlapping regions to facilitate recombination (Twist Biosciences). The synthesized genes were then amplified by PCR to have overlapping ends and recombined with linearized versions of plasmids pOG-Tn and pOG-T using a modified “lazy bones” protocol.54 Briefly, 0.5 ml of an overnight yeast culture was pelleted and washed in sterile TE buffer (10 mM Tris-HCl pH 8.0 and 1 mM EDTA). 0.4 mg of salmon sperm carrier DNA (Sigma), plasmid DNA, and PCR products were added to the pellet along with 0.5 ml lazy bones solution (40% polyethylene glycol MW 3350, 0.1 M lithium acetate, 10 mM Tris-HCl pH 7.5 and 1 mM EDTA). After vortexing for 1 min, the solution was incubated up to 4 d at room temperature. Cells were heat-shocked at 42°C, pelleted and plated on selective medium. Plasmids were isolated from individual transformants and confirmed by DNA sequencing.
All acceptor proteins were cloned in plasmid pEXT20.55 Briefly, the gene encoding E. coli MBP lacking its native 26-residue signal peptide was PCR amplified with primers that introduced the N-terminal signal peptide from E. coli DsbA, which permits periplasmic localization and glycosylation of fused proteins.33 The resulting PCR product was cloned into pEXT20 using restriction cloning between the EcoRI and XbaI sites. The MOOR tag was comprised of an 8-residue core sequence (WPAAASAP) that mimics the S63 glycosite in pilin (PilE), one of the native substrates of NmPglL,30 as well as two hydrophilic flanking sequences (DPRNVGGDLD and QPGKPPR) that are required for glycosylation. This sequence was synthesized as a G block (Integrated DNA Technologies) with a hexa-histidine epitope tag at its C-terminus and cloned between the XbaI and HindIII sites. All other acceptor proteins including GST, scFv13-R4, CRM197, PD, YebF-MBP, sfGFP, and sfGFPQ157 were synthesized as G blocks (Integrated DNA Technologies) and cloned in place of MBP by Gibson assembly using the EcoRI and XbaI sites to linearize the backbone. All additional acceptor peptides including MOORmut, the 8-residue EPO sequence, the 8-residue GPC sequence, the 9-residue SAP sequence, the 8-residue MUC1 sequence (MUC1_8), MUC1_12, MUC1_16, MUC1_20, MUC1_24, and MUC1_41 were synthesized as G blocks (Integrated DNA Technologies) and cloned in place of the MOOR sequence at the C-terminus of MBP by Gibson assembly using the XbaI and HindIII sites to linearize the backbone. The MUC1 sequence designs included motifs based on the most frequent minimal epitopes of natural MUC1 IgG and IgM antibodies including PPAHGVT, PDTRP, and RPAPGS46 and in epitopes that bind to specific human MHC class I molecules including STAPPAHGV, SAPDTRPAP, TSAPDTRPA and APDTRPAPG.56 The sialyltransferase used to produce the ST antigen was cloned adjacent to spDsbA-MBPMOOR in the pEXT20 acceptor plasmid. For sialylation of T antigen, E. coli O104 WbwA was acquired as a codon-optimized G block (Integrated DNA Technologies) and cloned downstream of spDsbA-MBPMOOR in plasmid pEXT20-spDsbA-MBPMOOR using Gibson assembly, yielding plasmid pEXT-spDsbA-MBPMOOR-EcWbwA. For sialylation of Tn antigen, the gene encoding EcWbwA was replaced with α2,6-sialyltransferase from Photobacterium sp. JT-ISH-224, yielding plasmid pEXT-spDsbA-MBPMOOR-PspST6. The plasmid for expression of the neuDBAC genes was constructed by yeast-based recombineering which involved cloning the E. coli K1 neuDBAC genes into plasmid pMLBy, which is a variant of plasmid pMLBAD that contains the yeast origin of replication and URA3 gene. The resulting plasmid was linearized with NheI after which the araC gene and pBAD promoter were replaced with the J23100 constitutive promoter from the Anderson library as described previously.36 The resulting pConNeuDBAC plasmid was used to transform strain ZLKA, a nanA-deficient host used previously for producing CMP-NeuNAc.57 Cell-free expression plasmids were generated by first PCR-amplifying the genes encoding MBPMOOR and MBPMOORmut from pEXT-spDsbA-MBPMOOR and pEXT-spDsbA-MBPMOORmut, respectively. The resulting PCR products were then ligated between NdeI and SalI restriction sites in plasmid pJL1, a pET-based vector used in cell-free glycoprotein synthesis reaction as described previously.10
Finally, a plasmid for expressing chimeric 5E5 antibody was constructed as described previously.58 First, DNA sequences for the VH and VL domains of mouse mAb 5E5 42 were obtained from US Patent US10,189,908 B2 and ordered as genes from GeneArt Gene Synthesis (Thermo Fisher). The 5E5 VH and VL sequences were then swapped with the existing variable region sequences in pVITRO1-Trastuzumab-IgG1/κ (Addgene plasmid #61883) to generate the vector pVITRO1–5E5-IgG1/κ according to previously published method.59 All plasmids were confirmed by DNA sequencing.
Immunoblot analysis.
Glycoprotein expression was carried out in 150-ml cultures for 16–20 h. Cells were pelleted at 10,000 × g for 30 min at 4°C, resuspended in 2 ml of lysis buffer containing 50 mM sodium phosphate, 300 mM sodium chloride, and 10 mM imidazole. Samples were frozen at −80°C overnight. Cells were then thawed, gently agitated at room temperature with 200 μg/ml of lysozyme (Sigma) for 15 min, and lysed by sonication. Lysed samples were then centrifuged at 10,000 × g for 30 min at 4°C and the supernatant was subjected to Ni2+ affinity purification using Ni-NTA spin columns (Qiagen) according to the manufacturer’s protocol. For preparation of extracellular culture supernatants, 10 ml of cells were pelleted by centrifugation at 10,000 x g for 30 min. 5 ml of the cleared supernatant was then transferred to a fresh tube to which 5 ml of 20% chilled trichloroacetic acid was added. The mixture was vortexed and incubated at 4°C without agitation for 16–20 h. The sample was then centrifuged at 21,000 x g for 30 min at 4°C. The supernatant was discarded and the pellet was resuspended in 1 ml of acetone. The sample was again centrifuged at 21,000 x g for 30 min at 4°C, allowed to dry at 37°C for 10 min, and resuspended in 60 μl of PBS.
Purified protein samples were prepared in Bolt LDS Sample Buffer (Thermo Fisher) and resolved on Bolt SDS-PAGE gels (Thermo Fisher). Following electrophoresis, proteins were transferred onto Immobilon-P polyvinylidene difluoride (PVDF) membranes (0.45 μm; Thermo Fisher) according to the manufacturer’s protocol. Antibodies used included: HRP-conjugated anti-hexa-histidine polyclonal antibody (Abcam cat# ab1187; dilution 1:5,000), mouse anti-human MUC1 antibody (BD Biosciences cat # 555925; dilution 1:1,000), biotinylated PNA (Vector labs cat # B-1075; dilution 1:1,000), biotinylated VVA (Vector labs cat # B-1235; dilution 1:500), and chimeric 5E5 antibody (dilution 1:250). The latter antibody was produced in-house using FreeStyle™ 293-F cells (Thermo Fisher) transfected with pVITRO1–5E5-IgG1/κ and purified from cell culture supernatants using Protein A/G agarose (Thermo Fisher) according to the manufacturer’s recommendations. Secondary antibodies included: HRP-conjugated rabbit anti-human IgG (Fc) antibody (Thermo Fisher cat # 31423; 1:2,500 dilution) and HRP-conjugated goat anti-mouse IgG (H&L) antibody (Abcam cat # ab6789; 1:2,500 dilution). Biotinylated lectins were detected using HRP-conjugated Extravidin (Sigma cat # E2886; dilution 1:2,000). Detection of blots was performed using Bio-Rad enhanced chemiluminescent (ECL) substrate. All immunoblots were visualized using a Chemidoc XRS+ system with Image Lab software (Bio-Rad).
Mass spectrometry analysis of protein glycosylation.
All reagents were purchased from Sigma Aldrich unless otherwise mentioned. Proteins were separated on SDS-PAGE gels after which gel pieces containing the glycoprotein bands were excised, cut into small pieces of about 1 mm2, and destained by treatment with 300 μL of a 1:1 mixture of acetonitrile and 50 mM aqueous NH4HCO3 followed by 500 μl of 100% acetonitrile. Since the glycoproteins did not have cysteine residues, reduction and alkylation was not performed. The glycoproteins were directly digested by adding 50 μl of digestion buffer with 12.5 μl of sequencing-grade trypsin (0.4 μg/μl; Promega) to the gel pieces and incubating at 37°C for 12 h. The digested peptides were extracted twice by 5% formic acid in 200 μL of 1:2 water:acetonitrile and filtered through a 0.2-μm filter. The digests were then dried using a SpeedVac, and subsequently re-dissolved in solvent A (0.1% formic acid in water) and stored at −30°C until analysis by nano-LC-MS/MS.
The digests were analyzed on an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher) equipped with a nanospray ion source and connected to a Dionex binary solvent system. Pre-packed nano-LC columns of 15-cm length with 75-μm internal diameter (id), filled with 3-μm C18 material (reverse phase) were used for chromatographic separation of samples. The precursor ion scan was acquired at 120,000 resolution in the Orbitrap analyzer and precursors at a time frame of 3 secs were selected for subsequent MS/MS fragmentation in the Orbitrap analyzer at 15,000 resolution or in ion trap. The threshold for triggering an MS/MS event with either higher-energy collisional dissociation product-triggered electron-transfer dissociation (HCDpdETD) program or electron-transfer dissociation (ETD) was set to 1,000 counts. Charge state screening was enabled, and precursors with unknown charge state or a charge state of +1 were excluded (positive ion mode). Dynamic exclusion was enabled (exclusion duration of 30 secs).
The LC-MS/MS spectra of tryptic digest of glycoproteins were searched against the respective .fasta sequence of mucin fragment using Byonic™ software versions 3.2 and 3.5 with the specific cleavage option enabled, and selecting trypsin as the digestion enzyme. Oxidation of methionine, deamidation of asparagine and glutamine, and O-glycan masses of HexNAc (m/z 203.079), HexHexNAc (m/z 365.132), and NeuNAcHexHexNAc (m/z 656.228) were used as variable modifications. The LC-MS/MS spectra were also analyzed manually for the glycopeptides using Xcalibur 4.2 software. The HCDpdETD and ETD MS2 spectra of glycopeptides were evaluated for the glycan neutral loss pattern, oxonium ions, and the glycopeptide fragmentations to assign the sequence and the presence of glycans in the glycopeptides. The peptide fragments at high resolution from ETD spectra were analyzed for the localization of O-glycosylation sites.
Quantification of in vivo CMP-NeuNAc levels.
For detection and quantification of nucleotide sugars, E. coli cells were pelleted to an equivalent to Abs600 of ~30, resuspended in 1 mL ultrapure water, and lysed by sonication. Following centrifugation at 30,000 × g, the supernatant was collected and analyzed within 4 h. Cleared E. coli lysates were diluted twofold in ultrapure water and injected into an UPLC-ESI-MS system (Waters) for analysis. The autosampler was set at 10°C. Separation was performed on an Acquity BEH C18 Column (1.7 μm, 2.1 mm x 50 mm; Waters). The elution started from 95% mobile phase A (5 mM TBA aqueous solution, adjusted to pH 4.75 with acetic acid) and 5% mobile phase B (5 mM TBA in Acetonitrile), raised to 57% B in 2 min, further raised to 100% B in 0.5 min, and then held at 100% B for 2 min, and returned to initial conditions over 0.1 min and held for 4 min to re-equilibrate the column. The flow rate was set at 0.6 ml/min with an injection volume of 2 μL. The column was preconditioned by pumping the starting mobile phase mixture for 10 min, followed by repeating twice the gradient protocol specified above prior to any injections. LC-ESI-MS chromatograms were acquired in negative ion mode under the following conditions: cpme voltage of 10 V, dry temperature at 520°C, and an acquisition range of m/z 400–900. Selected ion recordings were specified for CMP-NeuNAc. A standard curve was generated using commercial CMP-NeuNAc (CarboSynth).
Flow cytometric analysis.
To analyze the activity of candidate GalT enzymes, a flow cytometry-based screen was adapted from a previous study.36 Briefly, overnight cultures of each strain were grown in LB with relevant antibiotics. Cells were subcultured to an Abs600 of ~0.1 in 10 ml LB and grown for 16–20 h at 30°C. The next day, 1 ml of culture was washed twice with 1 ml PBS and resuspended in 500 μl PBS. All samples were diluted to an Abs600 of ~0.2 in 250 μl PBS. Detection of the disaccharide T antigen was performed with PNA-FITC conjugate (Vector labs cat# FL1071). PNA-FITC was diluted 1:500 in PBS and 250 μl of diluted lectin was added to cells, followed by incubation at 37°C for 30 min. Cells were pelleted at 6,000 × g for 4 min, washed in 1 ml PBS, resuspended in 1 ml PBS, and analyzed by flow cytometry using a FACSCalibur flow cytometer (BD Biosciences). All experiments were performed in triplicate with the resulting data generated through CellQuest Pro 6.0 and analyzed using FlowJo 10.5 software.
Cell-free O-glycosylation reactions.
For IVG reactions, crude membrane extracts enriched with NgPglO and UndPP-linked T antigen was prepared as described previously.10 Briefly, CLM25 cells carrying plasmid pOG-T-NgPglO were grown for 16–20 h at 37°C in LB media. The following day, cells were subcultured into 4 L LB media and allowed to grow at 37°C until mid-log phase (Abs600 ~0.6). Cells were then induced for 20 h at 16°C with 0.2% L-arabinose. Cells were harvested by centrifugation at 10,000 × g for 30 min at 4°C, and then resuspended in buffer containing 50 mM Tris-HCl (pH 8.0) and 25 mM sodium chloride. Cells were lysed by passing the cell suspension through a high-pressure homogenizer (Avestin) five times and the resulting lysate was centrifuged at 15,000 × g for 20 min at 4°C. The supernatant was collected and subjected to ultracentrifugation at 100,000 × g for 2 h at 4°C. The resulting pellet corresponding to the membrane fraction was collected and resuspended in 3 ml of buffer containing 50 mM Tris-HCl (pH 7.0), 25 mM sodium chloride, and 0.1% (w/v) n-dodecyl-β-D-maltoside (DDM). The resuspended pellet was incubated with mild agitation at room temperature for 1 h to enable the solubilization of NgPglO and LLOs. Following incubation, the mixture was centrifuged at 16,000 × g for 1 h at 4°C, and the supernatant was retained as a crude membrane extract. In parallel, acceptor proteins MBPMOOR and MBPMOORmut were purified as described above from a 500-ml culture of BL21(DE3) cells carrying either pEXT-spDsbA-MBPMOOR or pEXT-spDsbA-MBPMOORmut. In vitro glycosylation of purified acceptor proteins was carried out in 1.5-ml reactions containing 50 μg of purified acceptor protein and 1 ml of crude membrane extract in reaction buffer containing 10 mM HEPES (pH 7.5), 10 mM manganese chloride, and 1% (w/v) DDM. The reaction was incubated at 30°C for 16 h with mild tumbling. Upon completion of the reaction, acceptor proteins were purified from the reaction mixture by standard Ni2+ affinity purification using Ni-NTA spin columns (Qiagen) followed by concentration of samples.
For single-pot CFGpS, crude S12 extracts enriched with NgPglO and UndPP-linked T antigen glycans were prepared as described previously10 Briefly, CLM25 cells carrying plasmid pOG-T-NgPglO were grown at 37°C in 2×YTPG (10 g/L yeast extract, 16 g/L tryptone, 5 g/L NaCl, 7 g/L K2HPO4, 3 g/L KH2PO4, 18 g/L glucose, pH 7.2) until the Abs600 reached ~1. The culture was then induced with 0.02% (w/v) L-arabinose and the protein expression was allowed to proceed at 30oC until the Abs600 reached ~3. All subsequent steps were carried out at 4°C unless otherwise stated. Cells were harvested and washed twice using S12 buffer (10 mM tris acetate, 14 mM magnesium acetate, 60 mM potassium acetate, pH 8.2). The pellet was then resuspended in 1 ml per 1 g cells of S12 buffer. The resulting suspension was passed once through a EmulsiFlex-B15 high-pressure homogenizer (Avestin) at 20,000–25,000 psi to lyse cells. The extract was then centrifuged twice at 12,000 × g for 30 min to remove cell debris and the supernatant was collected and incubated at 37oC for 60 min. Following centrifugation at 15,000 x g for 15 min at 4oC, the supernatant was collected, flash-frozen in liquid nitrogen, and stored at −80oC. CFGpS reactions were carried out at 1-ml reaction volumes in a 15-ml conical tube using a modified PANOx-SP system.60 The reaction mixture contained the following components: 0.85 mM each of GTP, UTP, and CTP, 1.2 mM ATP, 34.0 μg/ml folinic acid, 170.0 μg/ml of E. coli tRNA mixture, 130 mM potassium glutamate, 10 mM ammonium glutamate, 12 mM magnesium glutamate, 2 mM each of 20 amino acids, 0.4 mM nicotinamide adenine dinucleotide (NAD), 0.27 mM coenzyme-A (CoA), 1.5 mM spermidine, 1 mM putrescine, 4 mM sodium oxalate, 33 mM phosphoenolpyruvate (PEP), 57 mM HEPES, 6.67 μg/ml plasmid, and 27% (v/v) of cell lysate. Protein synthesis was carried out for 30 min at 30oC, after which protein glycosylation was initiated by the addition of sucrose and tetracycline at the final concentration of 100 mM and 10 μg/ml, respectively, and carried out at 30oC for 16 h. To recover protein products, reaction mixtures were passed through a Ni-NTA spin column (Qiagen) twice, washed, and eluted with 300 mM imidazole. Samples were concentrated and analyzed by SDS-PAGE followed by immunoblotting analysis.
Data availability.
All data generated or analyzed during this study are included in this article (and its supplementary information) or are available from the corresponding authors on reasonable request.
Materials availability.
All unique materials used in this work are available from the authors.
Extended Data
Extended Data Fig. 1 |. MS/MS fragmentation analysis of Tn-modified glycoprotein.
EThcD fragmentation analysis of glycosylated peptide 397NVGGDLDWPAAAS(HexNAc)APQPGKPPR418 derived from MBPMOOR by trypsin digestion. The spectrum Identifies the neutral loss pattern of the single HexNAc monosaccharide, corresponding oxonium ions, and fragments of the glycopeptide (c and z ions), validating the glycosylation and the site of glycosylation at S409 within the 8-resldue WPAAASAP core sequence of MBPMOOR.
Extended Data Fig. 2 |. Flow cytometric screening of Gal transferases for biosynthesis of T antigen.
(a) Schematic of flow cytometric screen to evaluate candidate Gal transferases (GalTs) for their ability to generate lipid-linked T antigen. Once formed, the T antigen is subsequently flipped to periplasm by the native E. coli flippase, Wzx, transferred to lipid A core by the promiscuous O-antlgen llgase WaaL native to E. coli, and ultimately displayed on the cell surface. Cells are labeled with FITC-conjugated PNA that specifically binds the T antigen, (b) Flow cytometric analysis of PNA-labeled E. coli MC4100 ΔwecA (MCΔw) (yellow) or MC4100 ΔwecA ΔwaaL (MCΔww) (gray) carrying no plasmid, plasmid pOG-Tn, or plasmid pOG-Tn modified with one of the candidate GalT enzymes as indicated. (c) Flow cytometric analysis of PNA-labeled MCΔw (yellow) or MCΔww (gray) carrying no plasmid, plasmid pOG-T (producing T antigen glycan with EcWbwC), or plasmid pOG-TΔgne (encoding T antigen pathway but lacking CjGne epimerase). In (b) and (c), unlabeled MCΔw cells (white) were Included as negative controls. Inset histograms show representative flow cytometric data used to generate mean fluorescence Intensity data. See Supplementary Fig. 1 for flow cytometry gating strategy.
Extended Data Fig. 3 |. MS/MS fragmentation analysis of T-modified glycoprotein.
EThcD fragmentation analysis of glycosylated peptide 397NVGGDLDWPAAAS(HexHexNAc)APQPGKPPR418 derived from MBPMOOR by trypsin digestion. The spectrum identifies the neutral loss pattern of the HexHexNAc disaccharide, corresponding oxonium ions, and fragments of the glycopeptide (c and z ions), validating the glycosylation and the site of glycosylation at S409 within the 8-residue WPAAASAP core sequence of MBPMOOR.
Extended Data Fig. 4 |. Orthogonal biosynthesis of sialylated O-glycoforms in E. coli.
(a) Nano-LC-MS/MS analysis of purified acceptor protein generated by nanA-deficient E. coli cells carrying plasmid pConNeuDBAC for CMP-NeuNAc biosynthesis along with pOG-T-NgPglO and pEXT-spDsbA-MBPMOOR-EcWbwA. Sequence coverage of 88% was obtained for the MBPMOOR protein in the analysis. Spectrum reveals a predominant species (80% abundance) corresponding to the indicated peptide fragment bearing a single HexHexNAc modification as well as three less abundant species bearing a single NeuNAcHexHexNAc, a single HexNAc, and no modification (16%, 2%, and 2%, respectively), (b) Same as in (a) but with purified acceptor protein generated by nanA-deficient glyco-recoded cells carrying pOG-Tn-NgPgIO and pEXT-spDsbA-MBPMOOR-PspST6. Sequence coverage of 92% was obtained for MBPMOOR in the analysis. Spectrum reveals a predominant species (90% abundance) corresponding to the indicated peptide fragment bearing a single HexNAc modification as well as two less abundant species bearing a single NeuNAcHexNAc and no modification (2% and 9%, respectively). Arrow denotes modified serine (bold underlined font) as determined by EThcD fragmentation analysis.
Extended Data Fig. 5 |. MS/MS fragmentation analysis of ST- and STn-modified glycoproteins.
EThcD fragmentation analysis of glycosylated peptide 397NVGGDLDWPAAAS(NeuNAcHexHexNAc)APQPGKPPR418 derived from (a) ST-modified MBPMOOR and (b) STn-modified MBPMOOR that were subjected to trypsin digestion. The spectrum identifies the neutral loss pattern of the single NeuNAc and Hex monosaccharides, corresponding oxonium ions, and fragments of the glycopeptide (c and z ions), validating the glycosylation and site of glycosylation at S409 within the 8-residue WPAAASAP core sequence of MBPMOOR.
Extended Data Fig. 6 |. Yield determination for MBPMOOR modified with different O-glycans.
(a) Coomassie-stained SDS-PAGE gel showing MBPMOOR proteins purified from different strains. MBPMOOR bearing Tn or T antigens was produced in CLM25 cells co-transformed with pEXT-based plasmid for acceptor protein and appropriate sialyltransferase expression and either pOG-Tn-NgPglO or pOG-T-NgPglO plasmids, respectively. MBPMOOR bearing STn or ST antigens was produced in glyco-recoded cells carrying the CMP-NeuNAc biosynthesis pathway in the genome and co-transformed with pEXT-based plasmid for acceptor protein expression and either pOG-Tn-NgPglO or pOG-T-NgPglO plasmids, respectively. CLM25 cells co-transformed with only the pEXT-based plasmid for expressing MBPMOOR (agly) and appropriate sialyltransferase served as the control. Molecular weight (Mw) marker included on the left. SDS-PAGE gel is representative of three biological replicates. See Source Data for uncropped version of the image, (b) Yield of each glycoprotein calculated by multiplying the total yield times the percentage glycosylated (% gly), the latter of which was determined from nano-LC-MS/MS analysis of each glycoprotein product. Yield values are the average of three biological replicates and the error is the standard deviation of the mean.
Extended Data Fig. 7 |. O-linked glycosylation of diverse protein targets.
(a) Immunoblot analysis of acceptor proteins purified from CLM25 cells co-transformed with pOG-T-NgPgIO (+, top), pOG-T-NmPgIL (+, bottom), or pOG-T without an O-OST (−) along with pEXT-based plasmid encoding each of the different protein targets as indicated. MBPMOOR and MBPMOORmut derived from the same cells served as positive and negative control, respectively. Blots were probed with anti-hexa-histidine antibody (6xHis) to detect acceptor proteins and PNA lectin to detect the T antigen. Molecular weight (Mw) markers are indicated on the left of each blot. All immunoblot results are representative of at least three biological replicates, (b, c) Same as in (a) with pOG-T-NmPgIL (+) or pOG-T without NmPgIL (−) along with pEXT-based plasmid encoding each of the different protein targets as indicated. See Source Data for uncropped versions of the images.
Extended Data Fig. 8 |. Secretion of O-glycoproteins in the culture supernatant.
Immunoblot analysis of culture supernatants derived from CLM24 ΔyaiW cells co-transformed with pOG-T-NgPglO or pOG-T-NmPgIL along with pEXT-based plasmid encoding YebF-MBPMOOR or YebF-MBPMOORmut as indicated. Mutation of acceptor serine to glycine in YebF-MBPMOORmut served as negative control. Blots were probed with anti-hexa-histidine antibody (6xHis) to detect acceptor proteins and PNA lectin to detect the T antigen. Molecular weight (Mw) markers are indicated on the left of each blot. Immunoblot results are representative of at least three biological replicates. See Source Data for uncropped versions of the images.
Extended Data Fig. 9 |. Orthogonal biosynthesis of different MUC1 O-glycoforms in E. coli.
Nano-LC-MS/MS analysis of purified acceptor protein generated by CLM25 cells carrying plasmid pOG-T-NgPglO along with pEXT-based plasmid for expression of different MUC1 constructs including: (a) MUC1_8; (b) MUC1_20; (c) MUC1_24; and (d) MUC1_41. Sequence coverage of 77% was obtained for MUC1_8, 78% for MUC1.20, 88% for MUC1.24, and 75% for MUC1_41 in the analysis. All spectra reveal a predominant species corresponding to the indicated peptide fragments bearing a single HexHexNAc modification. Additional less abundant species bearing a single HexNAc and no modification were observed in all cases. For MUC1_41, several doubly glycosylated species were also identified as minor species. Arrow denotes modified serine (bold underlined font) as determined by EThcD fragmentation analysis.
Extended Data Fig. 10 |. MS/MS fragmentation analysis of MUC1 O-glycoforms bearing the T antigen.
EThcD fragmentation analysis of glycosylated peptides derived by trypsin digestion. The spectrum identifies the neutral loss pattern of HexHexNAc disaccharide, corresponding oxonium ions, and fragments of the glycopeptide (c and z ions), validating the glycosylation and the sites of glycosylation (S409 in MUC1_8; S415 in MUC1_20; S417 in MUC1_24 and S417 of MUC1_41) within relevant MUC1 peptides as indicated in the inset sequences.
Supplementary Material
Acknowledgements.
The authors would like to thank Robert Lee and Shannon Murphy for their contributions working with GT enzymes, Dr. Laura Yates for helpful discussions with glyco-recoding, Dr. Dominic Mills for helpful discussions regarding O-OSTs, Dr. Matthew Paszek for helpful discussions and provision of reagents, Dr. Mingji Li for technical advice, and Dr. Joshua Wilson, Dr. James Brooks, and Dr. Judith Merritt for help with vector design and yeast-based recombineering. We are also grateful to Dr. Ruchika Bhawal and Dr. Sheng Zhang of the Proteomics and Metabolomics Core Facility in the Cornell Institute of Biotechnology for assistance with LC-MS. This work was supported by the Defense Threat Reduction Agency (GRANT11631647 to M.P.D.), National Science Foundation (grant # CBET-1605242 to M.P.D.), and National Institutes of Health (grant # 1R01GM127578-01 to M.P.D.). Glycomics analysis was supported in part by the National Institutes of Health (grant 1S10OD018530 to P.A.). The work was also supported by seed project funding (to M.P.D.) through the National Institutes of Health-funded Cornell Center on the Physics of Cancer Metabolism (supporting grant 1U54CA210184-01). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. T.J. was supported by a Royal Thai Government Fellowship and also a Cornell Fleming Graduate Scholarship. E.C.C. was supported by a National Institutes of Health Chemical-Biology Interface (CBI) training fellowship (supporting grant T32GM008500).
Footnotes
Competing Interests. M.P.D. has a financial interest in Glycobia, Inc. and Versatope, Inc. M.P.D.’s interests are reviewed and managed by Cornell University in accordance with their conflict of interest policies. All authors declare no other competing interests.
References
- 1.Khoury GA, Baliban RC & Floudas CA Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci Rep 1, 90 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh CT, Garneau-Tsodikova S & Gatto GJ Jr. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl 44, 7342–7372 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Abu-Qarn M, Eichler J & Sharon N Not just for Eukarya anymore: protein glycosylation in Bacteria and Archaea. Curr Opin Struct Biol 18, 544–550 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Varki A Biological roles of glycans. Glycobiology 27, 3–49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sethuraman N & Stadheim TA Challenges in therapeutic glycoprotein production. Curr Opin Biotechnol 17, 341–346 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Rappuoli R Glycoconjugate vaccines: Principles and mechanisms. Sci Transl Med 10, eaat4615 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Valderrama-Rincon JD et al. An engineered eukaryotic protein glycosylation pathway in Escherichia coli. Nat Chem Biol 8, 434–436 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meuris L et al. GlycoDelete engineering of mammalian cells simplifies N-glycosylation of recombinant proteins. Nat Biotechnol 32, 485–489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamilton SR et al. Production of complex human glycoproteins in yeast. Science 301, 1244–1246 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Jaroentomeechai T et al. Single-pot glycoprotein biosynthesis using a cell-free transcription-translation system enriched with glycosylation machinery. Nat Commun 9, 2686 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kightlinger W et al. A cell-free biosynthesis platform for modular construction of protein glycosylation pathways. Nat Commun 10, 5404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldman MF et al. Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc Natl Acad Sci U S A 102, 3016–3021 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tytgat HLP et al. Cytoplasmic glycoengineering enables biosynthesis of nanoscale glycoprotein assemblies. Nat Commun 10, 5403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aumiller JJ, Hollister JR & Jarvis DL A transgenic insect cell line engineered to produce CMP-sialic acid and sialylated glycoproteins. Glycobiology 13, 497–507 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang MM et al. Small-molecule control of antibody N-glycosylation in engineered mammalian cells. Nat Chem Biol 15, 730–736 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Yang Z et al. Engineering mammalian mucin-type O-glycosylation in plants. J Biol Chem 287, 11911–11923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott S et al. Enhancement of therapeutic protein in vivo activities through glycoengineering. Nat Biotechnol 21, 414–421 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Huang W, Giddens J, Fan SQ, Toonstra C & Wang LX Chemoenzymatic glycoengineering of intact IgG antibodies for gain of functions. J Am Chem Soc 134, 12308–12318 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broecker F et al. Multivalent display of minimal Clostridium difficile glycan epitopes mimics antigenic properties of larger glycans. Nat Commun 7, 11224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umana P, Jean-Mairet J, Moudry R, Amstutz H & Bailey JE Engineered glycoforms of an antineuroblastoma IgG1 with optimized antibody-dependent cellular cytotoxic activity. Nat Biotechnol 17, 176–180 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Ilyushin DG et al. Chemical polysialylation of human recombinant butyrylcholinesterase delivers a long-acting bioscavenger for nerve agents in vivo. Proc Natl Acad Sci U S A 110, 1243–1248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarz F & Aebi M Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol 21, 576–582 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Choi BK et al. Use of combinatorial genetic libraries to humanize N-linked glycosylation in the yeast Pichia pastoris. Proc Natl Acad Sci U S A 100, 5022–5027 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Natarajan A, Jaroentomeechai T, Li M, Glasscock CJ & DeLisa MP Metabolic engineering of glycoprotein biosynthesis in bacteria. Emerg Top Life Sci 2, 419–432 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Ollis AA, Zhang S, Fisher AC & DeLisa MP Engineered oligosaccharyltransferases with greatly relaxed acceptor-site specificity. Nat Chem Biol 10, 816–822 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henderson GE, Isett KD & Gerngross TU Site-specific modification of recombinant proteins: a novel platform for modifying glycoproteins expressed in E. coli. Bioconjug Chem 22, 903–912 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Mueller P et al. High level in vivo mucin-type glycosylation in Escherichia coli. Microb Cell Fact 17, 168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du T et al. A bacterial expression platform for production of therapeutic proteins containing human-like O-Linked glycans. Cell Chem Biol 26, 203–212 e205 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Faridmoayer A et al. Extreme substrate promiscuity of the Neisseria oligosaccharyl transferase involved in protein O-glycosylation. J Biol Chem 283, 34596–34604 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan C et al. Biosynthesis of conjugate vaccines using an O-Linked glycosylation system. MBio 7, e00443–00416 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valentine JL et al. Immunization with outer membrane vesicles displaying designer glycotopes yields class-switched, glycan-specific antibodies. Cell Chem Biol 23, 655–665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harding CM, Haurat MF, Vinogradov E & Feldman MF Distinct amino acid residues confer one of three UDP-sugar substrate specificities in Acinetobacter baumannii PglC phosphoglycosyltransferases. Glycobiology 28, 522–533 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Fisher AC et al. Production of secretory and extracellular N-linked glycoproteins in Escherichia coli. Appl Environ Microbiol 77, 871–881 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tarp MA & Clausen H Mucin-type O-glycosylation and its potential use in drug and vaccine development. Biochim Biophys Acta 1780, 546–563 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Yang G et al. Fluorescence activated cell sorting as a general ultra-high-throughput screening method for directed evolution of glycosyltransferases. J Am Chem Soc 132, 10570–10577 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Glasscock CJ et al. A flow cytometric approach to engineering Escherichia coli for improved eukaryotic protein glycosylation. Metab Eng 47, 488–495 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Yates LE et al. Glyco-recoded Escherichia coli: Recombineering-based genome editing of native polysaccharide biosynthesis gene clusters. Metab Eng 53, 59–68 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Lai PH, Everett R, Wang FF, Arakawa T & Goldwasser E Structural characterization of human erythropoietin. J Biol Chem 261, 3116–3121 (1986). [PubMed] [Google Scholar]
- 39.Maier AG et al. Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat Med 9, 87–92 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gendler S, Taylor-Papadimitriou J, Duhig T, Rothbard J & Burchell J A highly immunogenic region of a human polymorphic epithelial mucin expressed by carcinomas is made up of tandem repeats. J Biol Chem 263, 12820–12823 (1988). [PubMed] [Google Scholar]
- 41.Mazor Y, Keydar I & Benhar I Humanization and epitope mapping of the H23 anti-MUC1 monoclonal antibody reveals a dual epitope specificity. Mol Immunol 42, 55–69 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Sorensen AL et al. Chemoenzymatically synthesized multimeric Tn/STn MUC1 glycopeptides elicit cancer-specific anti-MUC1 antibody responses and override tolerance. Glycobiology 16, 96–107 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Ju T & Cummings RD A unique molecular chaperone Cosmc required for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc Natl Acad Sci U S A 99, 16613–16618 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skretas G et al. Expression of active human sialyltransferase ST6GalNAcI in Escherichia coli. Microb Cell Fact 8, 50 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schulz BL et al. Identification of bacterial protein O-oligosaccharyltransferases and their glycoprotein substrates. PLoS One 8, e62768 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Mensdorff-Pouilly S et al. Reactivity of natural and induced human antibodies to MUC1 mucin with MUC1 peptides and n-acetylgalactosamine (GalNAc) peptides. Int J Cancer 86, 702–712 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Apostolopoulos V et al. A glycopeptide in complex with MHC class I uses the GalNAc residue as an anchor. Proc Natl Acad Sci U S A 100, 15029–15034 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ninkovic T & Hanisch FG O-glycosylated human MUC1 repeats are processed in vitro by immunoproteasomes. J Immunol 179, 2380–2388 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Lakshminarayanan V et al. Immune recognition of tumor-associated mucin MUC1 is achieved by a fully synthetic aberrantly glycosylated MUC1 tripartite vaccine. Proc Natl Acad Sci U S A 109, 261–266 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coyne MJ et al. Phylum-wide general protein O-glycosylation system of the Bacteroidetes. Mol Microbiol 88, 772–783 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Methods
- 51.Baba T et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Datsenko KA & Wanner BL One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Natarajan A, Haitjema CH, Lee R, Boock JT & DeLisa MP An engineered survival-selection assay for extracellular protein expression uncovers hypersecretory phenotypes in Escherichia coli. ACS Synth Biol 6, 875–883 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM & O’Toole GA Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72, 5027–5036 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dykxhoorn DM, St Pierre R & Linn T A set of compatible tac promoter expression vectors. Gene 177, 133–136 (1996). [DOI] [PubMed] [Google Scholar]
- 56.Apostolopoulos V, Karanikas V, Haurum JS & McKenzie IF Induction of HLA-A2-restricted CTLs to the mucin 1 human breast cancer antigen. J Immunol 159, 5211–5218 (1997). [PubMed] [Google Scholar]
- 57.Fierfort N & Samain E Genetic engineering of Escherichia coli for the economical production of sialylated oligosaccharides. J Biotechnol 134, 261–265 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Cox EC et al. Antibody-mediated endocytosis of polysialic acid enables intracellular delivery and cytotoxicity of a glycan-directed antibody-drug conjugate. Cancer Res 79, 1810–1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dodev TS et al. A tool kit for rapid cloning and expression of recombinant antibodies. Sci Rep 4, 5885 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jewett MC & Swartz JR Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol Bioeng 86, 19–26 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article (and its supplementary information) or are available from the corresponding authors on reasonable request.