Abstract
Ovomucoid is a major egg white protein which is considered as the most dominant allergen in chicken eggs. Owing to the difficulty of separating ovomucoid from egg whites, researchers have adopted genetic deletion for development of hypoallergenic eggs. Previously, we used CRISPR/Cas9 to establish chickens with ovomucoid gene (OVM) mutations, but it remained unknown whether such hens could produce eggs at maturity. Here, we have reported on eggs laid by OVM-targeted hens. Except for watery egg whites, the eggs had no evident abnormalities. Real-time PCR revealed alternative splicing of OVM mRNA in hens, but their expression was limited. Immunoblotting detected neither mature ovomucoid nor ovomucoid-truncated splicing variants in egg whites. Sixteen chicks hatched from 28 fertilized eggs laid by OVM-targeted hens, and fourteen of the sixteen chicks demonstrated healthy growth. Taken together, our results demonstrated that OVM knockout could almost completely eliminate ovomucoid from eggs, without abolishing fertility. Thus, the eggs developed in this study have potential as a hypoallergenic food source for most patients with egg allergies.
Key words: ovomucoid (OVM)-knockout egg, egg allergen depletion, gene targeting, genome editing, CRISPR/Cas9
Introduction
Chicken eggs are one of the most common allergenic foods, especially in children, in whom allergy prevalence ranges between 0.5 and 2.5% (Rona et al., 2007; Caubet and Wang, 2011). The glycoprotein ovomucoid (OVM) is a major allergenic protein that constitutes 11% of the egg white protein (Kovacs-Nolan et al., 2005). OVM is believed to be the most dominant allergenic protein (Bernhisel-Broadbent et al., 1994; Cooke and Sampson, 1997; Mine and Yang, 2008). Therefore, removal of OVM or elimination of its allergenicity may result in hypoallergenic egg products. Food-processing methods such as proteinase and heat treatments are frequently used to reduce allergenicity of various foods (Verhoeckx et al., 2015). Such methods have been used to reduce egg allergenicity, but their use in egg products is limited because OVM is highly resistant to such treatments (Matsuda et al., 1983; Kovacs-Nolan et al., 2000; Hirose et al., 2004). Physical removal of OVM from egg whites has also been explored, with methods that include solvent extraction and rinsing of boiled egg whites (Urisu et al., 1997; Tanabe et al., 2000). However, physical removal is impractical and difficult to use in food manufacturing because of insufficient OVM elimination, poor cost-efficiency, and destruction of egg white properties such as gelling and foaming (Mine and Zhang, 2001; Chang et al., 2018). In contrast, genetic deletion of OVM from hens implies that OVM protein will not be expressed, thereby potentially generating hypoallergenic OVM-free eggs that reduce immune response in patients with egg allergies (Park et al., 2014; Chojnacka-Puchta and Sawicka, 2020).
We previously used the CRISPR/Cas9 system to produce biallelic OVM knockout (OVM−/−) chickens (Oishi et al., 2016), but we did not examine their egg production capacity. Here, we aimed to determine the egg-laying ability of OVM−/− hens and studied properties of eggs from such hens. To that end, we raised OVM−/− hens to sexual maturity and then evaluated their eggs (hereafter referred to as “OVM-null” eggs) for quality and viability.
Materials and methods
Animal Experiments
All animal experiments were conducted according to protocols approved by the Institutional Animal Care and Use Committees of the National Institute of Advanced Industrial Science and Technology (protocol number 2016-115), the Institute of Livestock and Grassland Science (NILGS), National Agriculture and Food Research Organization (NARO) (protocol number 1611B056), and Cosmo Bio Co., Ltd. (protocol number PMC201703). The OVM mutant and wild-type (WT) chickens were maintained and bred at the animal farm facilities of NILGS (generation (G)0 to G2) and Cosmo Bio Co., Ltd. (G3 and G4). Eggs were allowed to develop in forced-air incubators with tilting at a 60° angle twice each hour at 38.5°C and 60 to 80% relative humidity until hatching. Newly hatched OVM mutant chicks were raised in temperature-controlled, wire-flooring battery brooders. At 4 wk of age, chicks were moved to growing cages and were maintained in the cages until they reached 4 mo of age. Female OVM mutants were then moved to laying cages and male OVM mutants were housed in individual battery cages. Food and water were provided ad libitum throughout the experimental period. After 4 wk of age, the photoperiod for chick housing comprised a 16-hour light/8-hour dark cycle.
Progeny Test of OVM−/− Chickens
Semen samples were collected from every OVM−/− rooster and immediately used for insemination of OVM−/− hens. Eggs were collected 2 to 16 d after insemination and incubated until hatching. The insemination and egg collection-hatching processes were repeated twice in one experiment using the same pair of OVM−/− chickens. The experiments were conducted in triplicate.
DNA Sequencing Analysis
Genomic DNA was extracted from chick feather shafts, using the DNeasy Blood and Tissue kits (Qiagen, Germantown, MD). For genotyping, fragments containing the OVM target site were polymerase chain reaction (PCR)-amplified (MightyAmp DNA Polymerase Ver. 2; Takara Bio, Kusatsu, Japan) with a specific primer set (OVMTg2 target site PCR amplification; Supplementary Table 1). Amplicons were purified using the QIAquick PCR Purification Kit (Qiagen) and sequenced with specific primers (OVMTg2 target site sequencing; Supplementary Table 1) by Eurofins Genomics (Tokyo, Japan). Thermocycling conditions were as follows: 98°C for 2 min and 40 cycles of 98°C for 10 s, 60°C for 10 s, and 72°C for 30 s.
Reverse Transcription PCR
One WT hen and one OVM −/− (Au94) hen were sacrificed for collection of the oviduct magnum. Total RNA was isolated from these samples using the RNeasy plus mini kit (Qiagen). Complementary DNA was synthesized from 500 ng of RNA as per instructions of the SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA). After suspension in 200 μL of 10-mmol Tris-HCl Buffer (pH 7.0), cDNA (1 μL) was PCR-amplified with specific primers corresponding to the 5′ and 3′ untranslated regions of OVM (OVM RT-PCR; Supplementary Table 1). The housekeeping gene GAPDH was also amplified from cDNA with specific primers (GAPDH RT-PCR; Supplementary Table 1). Thermocycling conditions were 98°C for 2 min, followed by 25 cycles (OVM) and 30 cycles (GAPDH) of 98°C for 10 s, 60°C for 10 s, and 72°C for 50 s. Amplicons were purified with a PCR extraction kit (Nippon Genetics, Tokyo, Japan) and then sequenced directly with specific primers (OVM RT-PCR sequencing; Supplementary Table 1). To identify splicing variants of OVM transcripts, purified PCR products were cloned into pGEM-T easy vectors (Promega, Madison, WI) and sequenced with the M13 forward and reverse primers (Supplementary Table 1). Two mutant clones encoding in-frame OVM mutant (with 4-nt insertion/28-nt deletion and exon skipping of exons 3, 4) were used as PCR templates to construct an OVM mutant expression vector (see Expression of OVM Mutations in Cultured Cells).
Quantitative PCR
The reaction mixture (15 μL) contained 1 μL of cDNA (WT and Au 94), 7.5 μL of the Thunderbird SYBR qPCR Mix (Toyobo, Osaka, Japan), and 5 pmol of each primer (OVM exon 1/3 qPCR, OVM exon 6/7 qPCR, and GAPDH qPCR; Supplementary Table 1). The amplification protocol comprised the following conditions: 95°C for 2 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. Relative gene expression was calculated using the ΔΔCT method.
Expression of OVM Mutants in Cultured Cells
Using oviduct cDNA, the 2 in-frame OVM mutants, along with the corresponding WT OVM fragments, were PCR-amplified with specific primers located in exons 1 and 8 of OVM (OVM expression; Supplementary Table 1). These amplicons were cloned into the pcDNA-flag vector (Addgene, Watertown, MA). In addition, cDNA regions from exons 4–5 and exons 6–8 were amplified with specific primer sets (OVM exon 4/5 and OVM exon 6/8, respectively; Supplementary Table 1) and were then cloned into the pEGFP-N1 vector (Takara). All cloning experiments used the In-Fusion HD cloning kit (Takara). Expression vectors were transfected into 293T cells with Lipofectamine 2000 (Thermo Fisher Scientific), following the manufacturer instructions. The cells and culture media were collected 48 h after transfection, lysed, and then used for immunoblotting.
Analysis of OVM Protein in WT and OVM-Null Egg Whites
Sonicated egg whites from WT and OVM-null eggs were suspended in PBS at a ratio of 1:10 for SDS-PAGE. Samples were each mixed with an equal volume of 2 × Laemmli SDS-PAGE buffer (0.125 M Tris-HCl, pH 6.8, 10% (w/v) sucrose, 4% (w/v) SDS, 10% (v/v) 2-mercaptoethanol, and 0.01% bromophenol blue) and subjected to SDS-PAGE gradient (long-life gel 5–20% w/v acrylamide; Oriental Instruments Ltd., Sagamihara, Japan). Each gel was stained with Coomassie Brilliant Blue R-250 (Nacalai, Kyoto, Japan).
For immunoblotting, 50 nL of egg white samples suspended in 10 μL of 1 × Laemmli SDS-PAGE buffer (200-fold dilution) were separated using SDS-PAGE and then transferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Bedford, MA). The membrane was subjected to Western blotting with anti-OVM polyclonal antibodies supplied in a kit for the ovomucoid protein (M2302; Morinaga Institute of Biological Science, Inc., Yokohama, Japan) at manufacturer-recommended 20-fold dilution. Proteins were visualized with horseradish peroxidase-conjugated antirabbit IgG (Jackson ImmunoResearch, West Grove, PA) using an enhanced chemiluminescence system (ImmunoStar reagent; Wako, Osaka, Japan).
To purify OVM, 100 μL of egg white samples from WT and OVM-null eggs were sonicated and diluted with 400 μL of PBS. Sample pH was adjusted to 4.0 with approximately 10 μL of 1M hydrochloric acid solution, and the samples were then heated at 95°C for 15 min. Samples were centrifuged at 9,000 × g for 15 min for collection of both the supernatant and precipitate. Hundred microliters of each fraction was suspended in an equal volume of 2 × Laemmli SDS-PAGE buffer. Each sample was diluted 100-fold with 1 × Laemmli SDS-PAGE buffer and then subjected to SDS-PAGE and immunoblotting.
Statistical Analysis
Data are presented as mean ± SD. Between-species differences in egg weight and albumen height were analyzed with one-way ANOVA, followed by a Tukey-Kramer test for multiple comparisons. Significance was set at P < 0.01.
Results
OVM−/− Chickens
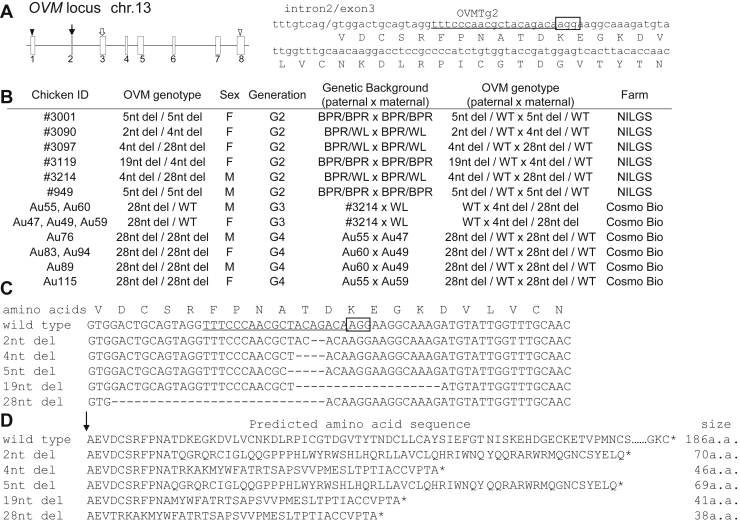
We previously created OVM mutant chickens in which the targeted site was located near the OVM signal sequence in exon 3 (Figure 1A; Oishi et al., 2016). In this study, OVM −/− chickens (G2) were generated as the offspring of OVM +/− chickens (G1). We used the G2 mutants and their offspring for further analysis (G3 and G4; Figure 1B). Our analysis of mutated OVM sequences showed that 2–28 nucleotides around guide RNA-targeted loci were deleted in mutant chickens (Figure 1C). These deletions likely caused frameshift mutations that led to premature stop codons (Figure 1D).
Figure 1.
OVM mutation in gene-targeted chickens. (A) Schematic representation of sgRNA targeting OVM. Left panel, exon-intron organization of OVM. Boxes with numbers indicate OVM exons. Closed and open triangles represent positions of start and stop codons, respectively. A closed arrow indicates the location of the peptide cleavage site region. The open arrow indicates the targeted OVM site. Right panel, OVM target sequence in exon 3. Lowercase and uppercase letters refer to DNA and amino acid sequences, respectively. Underlined and boxed text refer to sgRNA targeting site and adjoining putative protospacer adjacent motif (PAM) sequence, respectively. The slash indicates the boundary of intron 2 and exon 3. (B) OVM genotype of each biallelic mutant chicken in this study. F, female; M, male; WT, wild-type; nt del, nucleotide deletions; BPR, Barred Plymouth Rock; WL, White Leghorn. (C) Sequence analysis of CRISPR/Cas9-mediated deletion mutations in gene-targeted chickens. Wild-type DNA and amino acid sequences are shown in the top rows. The guide RNA targeting site and PAM sequence are underlined and boxed, respectively. The number of deleted nucleotides (2–28) is indicated to the left of each sequence. Deleted nucleotides are shown as dashes. (D) Wild-type amino acid sequences, and amino acid sequences predicted from each mutant DNA sequence. Predicted amino acid size is shown to the right of each sequence. Arrow shows the cleavage site in OVM.
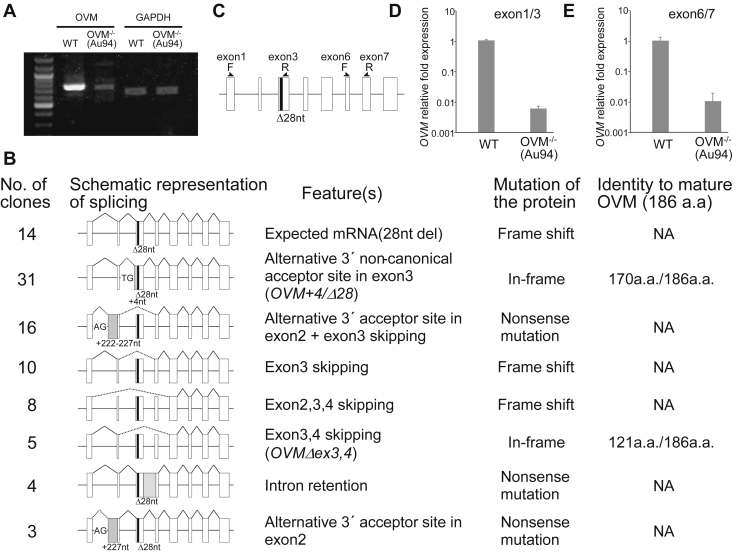
To verify frameshift mutations, we performed RT-PCR. Amplicons were obtained from both WT and OVM −/− (Au94) cDNA templates, but fewer amplicons were obtained from the latter (Figure 2A). Except for a few single-nucleotide polymorphisms, OVM cDNA sequencing traces of exons 1–2 and 4–8 did not differ between OVM −/− and WT (Supplementary Figure 1). However, we observed sequence variation in the region between exons 2 and 4 of the OVM −/− hen (Supplementary Figure 1). To identify aberrant OVM transcripts, we cloned and sequenced 91 OVM cDNA samples from the OVM −/− oviduct and identified 7 alternative splicing variants, in addition to the expected mutant mRNA (Figure 2B). These variants were not observed in WT OVM cDNA (25 clones analyzed). Most splicing variants were unlikely to produce mature protein because of frameshift and nonsense mutations, but 2 transcripts could encode in-frame OVM mutants. One in-frame mutant exhibited an insertion of 4 nucleotides and a deletion of 28 nucleotides (OVM +4/Δ28 nt) in exon 3 (Figure 2B and Supplementary Figure 2). Interestingly, a four-nucleotide insertion was generated via alternative 3′ splicing, using a noncanonical acceptor site (not AG but TG) (Supplementary Figure 2). The other in-frame splice variant showed skipping of exons 3 and 4 (OVM Δ e x 3,4) (Figure 2B).
Figure 2.
Analysis of OVM transcripts expressed in the oviduct of OVM−/− hens. (A) RT-PCR analysis of OVM mRNA in WT and OVM−/− oviduct samples. Primers in exons 1 and 8 of OVM were used (see Materials and Methods). Internal control was GAPDH. (B) Summary of all OVM mRNA variants identified in OVM−/− hens. Schematic represents different types of pre-RNA splicing events. Open boxes represent OVM exons. Solid and dashed lines indicate normal and abnormal splicing, respectively. The black box represents the 28-nucleotide deletion in exon 3; gray boxes indicate aberrant exons. Alternative 3′ acceptor sites (noncanonical TG and canonical AG) are shown. NA, not applicable. (C) Schematic representation of primer positions for quantitative RT-PCR. Two primer sets were designed to quantify OVM transcripts. (D, E) Expression levels of OVM transcripts detected with quantitative RT-PCR. (D) Primers corresponding to exons 1 and 3, and (E) exons 6 and 7 were used. Error bars indicate standard deviation.
Then, we compared OVM mRNA expression in the oviduct of OVM −/− and WT hens using qPCR and 2 primer sets corresponding to the different OVM exons (Figure 2C). The mutant oviduct had significantly lower OVM mRNA expression (<1%) than the WT oviduct (Figures 2D and 2E). These results are consistent with RT-PCR amplicon images (Figure 2A) and suggest that aberrant OVM transcripts, including in-frame mutants, are expressed in OVM −/− hens at significantly lower rates than normal OVM mRNA in WT hens.
We further evaluated the possibility of plasmid integration and off-target mutation in OVM −/− chicken genomes. We did not detect puromycin resistance gene (included in the plasmid used for the CRISPR/Cas9 system in the present study) in the OVM −/− genome, suggesting that unexpected plasmid integration did not occur (Supplementary Figure 3A). To investigate off-target mutations, we amplified 3 potential off-target sites that matched the OVMTg2 seed sequence adjacent to the putative protospacer adjacent motif (Oishi et al., 2016). We did not detect either insertions or deletions in sequence data from 4 mutant hens (Supplementary Figure 3B).
Eggs From OVM −/− Hens
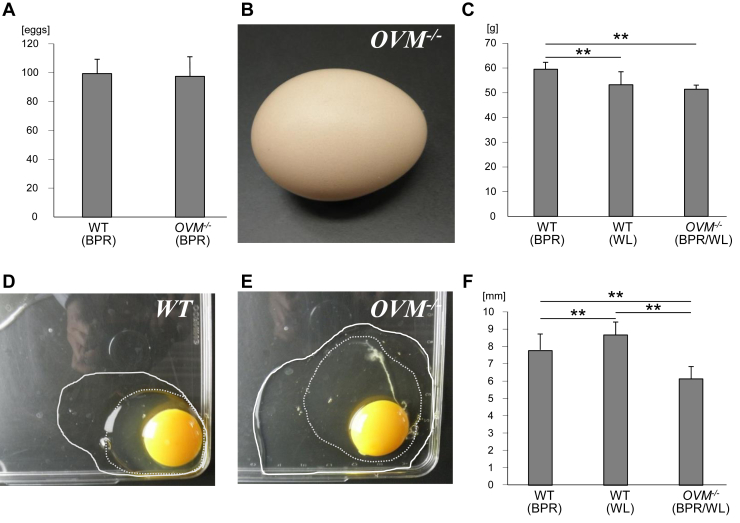
We verified that OVM −/− hens, despite missing a major egg white component, could produce eggs. Average egg production rate within the first 150 d was 98 eggs per hen among sexually mature Barred Plymouth Rock (BPR) OVM −/− hens (n = 2, #3001 and #3119), similar to the rates in WT BPR hens (99 eggs per hen, n = 4) (Figure 3A). All gene-targeted hens produced apparently normal eggs in shape (Figure 3B). Eggs from 10-month-old G4 OVM −/− hens (mixed with BPR and White Leghorn [WL]; Figure 1B) weighed 51.4 ± 1.7 g on average (n = 28), significantly less than WT BPR eggs (59.5 ± 2.7 g, n = 25) but not different from WT WL eggs (53.2 ± 5.3 g, n = 32) (Figure 3C). Distinct from WT eggs, OVM-null eggs tended to have less viscous egg whites (Figures 3D and 3E). Consistent with this observation, fresh egg albumen in OVM-null eggs had lower average height (6.2 ± 0.7 mm, n = 21 from G4 hens) than that in WT eggs (8.7 ± 0.8 mm, n = 26 from WL; 7.8 ± 0.9 mm, n = 25 from BPR) (Figure 3F). Although we did not fully control the genetic background, our results indicated that OVM-null and WT egg whites might have different physical properties.
Figure 3.
Eggs from OVM−/− hens. (A) Average of egg production number of WT (n = 4) and OVM−/− (n = 2) hens within the first 150 d. Genetic backgrounds of hens are indicated. (B) Appearance of an OVM-null egg. (C) Average weight of eggs from WT and OVM−/− hens at 10 mo of age. (D, E) The OVM-null egg had watery whites compared with wild-type. (D) One-week-old WT and (E) OVM-null eggs. Solid and dotted lines indicate the edges bordering thin and thick albumen, respectively. (F) Average height of egg albumen in WT and OVM-null eggs. Error bars indicate standard deviation. ∗∗P < 0.01, one-way ANOVA with the Tukey's test.
OVM Protein in OVM-Null Eggs
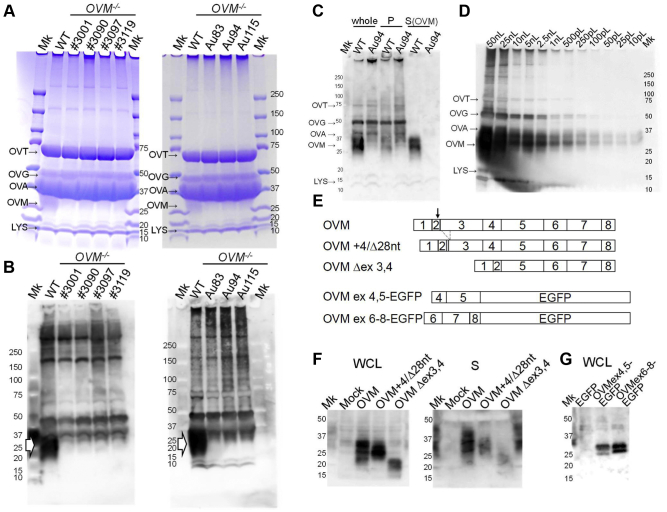
The results of SDS-PAGE and Coomassie Blue staining revealed the presence of major egg white components (i.e., ovalbumin, ovotransferrin, ovoglobulin, and lysozyme) in OVM-null eggs and at expression levels that did not evidently differ from those of WT (Figure 4A). In contrast, OVM protein appeared as a broad band for WT egg whites but was hardly visible for OVM-null egg whites.
Figure 4.
Reduction of OVM protein in OVM-null egg. (A) SDS-PAGE of WT and OVM-null egg whites stained with Coomassie Brilliant Blue. Each lane is labeled with OVM−/- hen IDs. Major egg white components are indicated. OVT, ovotransferrin; OVG, ovoglobulin; OVA, ovalbumin; OVM, ovomucoid; LYS, lysozyme. Per lane, 500 nL of egg white sample was loaded. (B) Immunoblot analysis of WT and OVM-null egg white proteins with anti-OVM antibodies. The arrowheads indicate the position of OVM protein recognized by anti-OVM antibodies. Each lane contained 50 nL of egg white samples. (C) Immunoblots of partially purified WT and OVM-null egg white proteins, using anti-OVM antibodies. Following acid and heat treatment, WT and OVM-null egg white (derived from Au94 OVM−/− hen) were centrifuged to separate precipitate (P) and supernatant (S). Heat-stable OVM protein was prominent in the supernatant and other egg white proteins accumulated in the precipitate. Each lane contained 10 nL of whole egg white samples. (D) Sensitivity tests of anti-OVM antibodies. Each lane (labeled at top) was loaded with different amounts of WT egg white. Immunoblotting shows that anti-OVM antibodies can detect OVM in 10 pL of egg white. (E) Schematic diagram of proteins expressed in cultured cells. Numbers indicate OVM exons. The gray box and dotted lines, respectively, indicate a four-nucleotide insertion and 28-nucleotide deletion in the OVM+4/D28 nt mutant. Arrow shows cleavage site in OVM. (F) Immunoblots of recombinant OVM mutants with anti-OVM antibodies. Samples were whole cell lysate (WCL) and culture S from HEK293 T cells transfected with WT and mutant OVM. Mock: mock transfection with empty plasmid pcDNA3. (G) Immunoblot of the independent OVM region. Whole cell lysate from HEK293 T cells expressing enhanced green fluorescent protein (EGFP) fused with OVM protein (exons 4–5 and exons 6–8) were subjected to immunoblotting using anti-OVM antibodies.
Immunoblotting of anti-OVM antibodies revealed strong signals for OVM (20–37 kDa) in WT but not OVM-null egg whites, thereby indicating that mature OVM was absent in the latter (Figure 4B). As the anti-OVM antibodies partly recognize other egg white components, we observed high background noise. We therefore partially purified OVM protein from egg white samples using acid and heat treatment, followed by further immunoblotting. Supernatant of WT egg whites clearly contained OVM protein (Figure 4C), whereas apparent OVM signals were absent from both the precipitate and supernatant from OVM-null egg whites. Thus, expression of OVM protein, including mutant isoforms, appeared to be lower than the detection limit in OVM-null egg whites. Immunoblotting detected antigen in 10 pL of WT egg white (Figure 4D), but not in 50 nL of OVM-null egg white (Figure 4B), suggesting that OVM-null egg white had <0.02% of OVM protein found in WT egg white.
We evaluated OVM antibodies to determine whether OVM was simply undetectable in OVM-null egg whites because the antibodies did not recognize mutant variants. The precursors of WT OVM and 2 in-frame OVM mutants (OVM +4/Δ28 and OVM Δex3,4, Figure 4D) were expressed in HEK293 T cells and analyzed by immunoblotting. WT and mutant OVM expression were detected in both whole cell lysates and culture supernatant (Figure 4E). This result indicated that OVM antibodies could recognize both WT and mutant OVM, thus reinforcing the explanation that the latter was under the immunoblotting detection limit in OVM-null egg white. Interestingly, OVM mutants had lower relative expression than WT OVM in culture supernatant. Therefore, the 2 in-frame OVM mutants may be difficult to secrete and/or may have unstable mature forms. We further tested whether OVM antibodies could recognize multiple epitopes on OVM. Two different OVM regions, one corresponding to exons 4–5 and the other corresponding to exons 6–8, were fused to EGFP protein (Figure 4E) and expressed in HEK293 T cells. The antibodies recognized both fusion proteins but not EGFP, thereby indicating that they could bind to different OVM regions (Figure 4G). Therefore, these antibodies might detect various in-frame OVM mutants generated by abnormal splicing variants, even if RT-PCR did not identify said mutants. As immunoblotting did not detect evident signals in OVM-null egg whites (Figures 4B and 4C), it could be inferred that the 2 identified in-frame OVM mutants and any unidentified in-frame mutants had low or no expression.
Chicken Development From OVM-Null Eggs
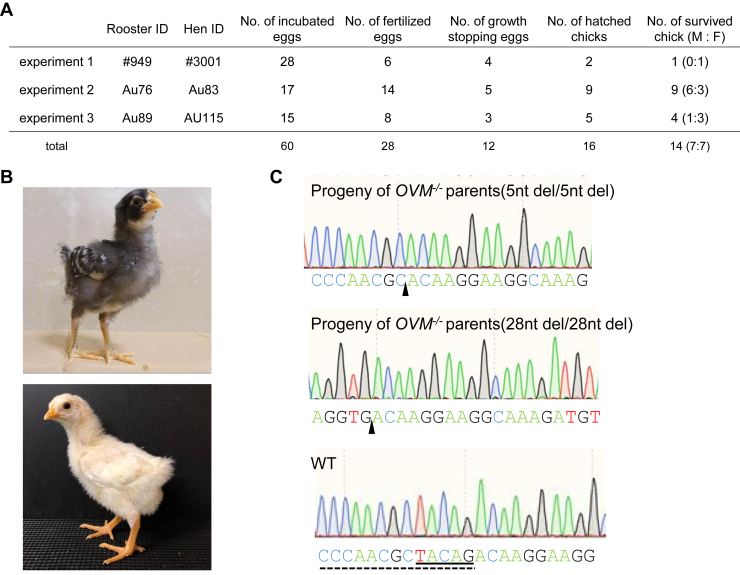
Finally, we examined OVM-null egg fertility and hatchability via crossing between OVM −/− females and OVM −/− males (Figure 5A). We hatched 16 chicks from 28 fertilized eggs, with 14 of the 16 chicks showing healthy growth and development (Figure 5B). Sequencing showed that hatched chicks had the OVM−/− genotype (Figure 5C). These results indicate that OVM−/− hens are fertile and that eggs can be developed successfully even when OVM is almost fully eliminated.
Figure 5.
Progeny of OVM−/− hens. (A) Results of progeny test. Three OVM−/− roosters and hens were crossed. (B) Hatched chicks from OVM-null egg in experiment 1 (top, BPR background) and experiment 2 (bottom, BPR and WL cross). (C) OVM genomic sequences of hatched chicks from experiments 1 (top) and 2 (middle), and from a WT chick (bottom). Progeny genome contained five-base pair (experiment 1) and 28-base pair (experiment 2) homozygous deletions. Closed triangles (top and middle) indicate deleted sequences. Solid and dotted lines (bottom), respectively, indicate deleted sequences in WT.
Discussion
To our knowledge, this is the first study that genetically removed most traces of an egg allergen from egg whites. Not only did our gene-targeted hens harbor biallelic frameshift mutations in the OVM gene, but their eggs also lacked expression of mature OVM protein (Figure 1C). Although OVM −/− hens exhibited 2 abnormal splicing variants encoding in-frame OVM mutants, transcript expression was less than 1% of the WT levels (Figures 2D and 2E). Eggs from OVM −/− hens are likely to be hypoallergenic, with the removal of OVM being especially important because the protein is heat-resistant, differing from other major egg allergens, such as ovalbumin, ovotransferrin, and lysozyme. A food challenge test (Urisu et al., 1997) showed that egg whites heated and rinsed for removal of OVM were significantly less allergenic than heated egg whites containing OVM. Therefore, OVM-null eggs have potential as a safe raw material for heat-processed foods that may be tolerated by patients with egg allergies.
Genome editing–induced frameshift mutations can lead to in-frame exon skipping and the production of short splice variant proteins (Lalonde et al., 2017). Therefore, we evaluated the amount of in-frame OVM mutant proteins in OVM-null eggs. We identified 7 abnormal splicing variants in OVM −/− oviducts, and 2 translated to in-frame OVM mutant proteins. Several allergenic epitopes are present throughout OVM (Mine and Zhang, 2002); short splice variants of OVM may potentially have allergenic activity. Thus, we performed an immunoblotting analysis with commercial polyclonal antibodies and recognized multiple OVM epitopes but did not observe any remarkable expression of shortened OVM splice variants in OVM-null egg whites (Figures 4B and 4C). Therefore, in-frame exon skipping and the production of shortened OVM splice variants were considered to be rare in the OVM-null eggs used in this study. As some patients are allergic to even small amounts of OVM variants, further immunological and clinical studies are required to verify the safety of the modified eggs produced in the present study. However, it is likely that OVM-null egg whites have lower allergenicity than standard egg whites.
Our next experiments focused on OVM-null egg production and egg viability/quality. Reduction of OVM expression in mutant hens did not appear to affect egg production. OVM-null eggs are lighter than WT BPR eggs, but not significantly different in weight from WT WL eggs. Noteworthy, OVM −/− and WT hens had different genetic backgrounds, indicating that comparisons are more difficult. While weight did not differ, egg white viscosity was altered in OVM-null eggs. As OVM constitutes more than 10% of the total egg white proteins, it implies that the characteristics of egg whites differ between OVM-null and WT eggs, but the mechanism by which OVM deletion affects viscosity remains unclear. Other egg white properties, such as foaminess and coagulability, may also have been altered in OVM-null eggs. As such properties may drastically impact the production of processed egg products, further studies are required, especially with respect to the use of OVM-null eggs in the food industry.
In conclusion, our study is the first to demonstrate that eggs with a considerable lack of OVM, a known allergenic protein, laid by genetically engineered hens can develop normally. Such eggs are expected to be hypoallergenic. Although allergen risk assessment remains essential, these eggs are a potential alternative for the millions of people suffering from egg allergies.
Acknowledgements
The authors thank the staff of the Poultry Management Section of the NARO and Cosmo Bio Co. Ltd. for care of the birds and providing fertilized eggs. This work was supported in part by a grant from JSPS KAKENHI (20K21377, 20H03135 to I.O.), and a grant from the Kieikai Research Foundation (to I.O.).
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2020.10.026.
Disclosures
This study was partly funded by Cosmo Bio Co., Ltd., and T. W. is an employee of Cosmo Bio Co., Ltd.
Supplementary data
References
- Bernhisel-Broadbent J., Dintzis H.M., Dintzis R.Z., Sampson H.A. Allergenicity and antigenicity of chicken egg ovomucoid (Gal d III) compared with ovalbumin (Gal d I) in children with egg allergy and in mice. J. Allergy Clin. Immunol. 1994;93:1047–1059. doi: 10.1016/s0091-6749(94)70054-0. [DOI] [PubMed] [Google Scholar]
- Caubet J.C., Wang J. Current understanding of egg allergy. Pediatr. Clin. North Am. 2011;58:427–443. doi: 10.1016/j.pcl.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Lahti T., Tanaka T., Nickerson M.T. Egg proteins: fractionation, bioactive peptides and allergenicity. J. Sci. Food Agric. 2018;98:5547–5558. doi: 10.1002/jsfa.9150. [DOI] [PubMed] [Google Scholar]
- Chojnacka-Puchta L., Sawicka D. CRISPR/Cas9 gene editing in a chicken model: current approaches and applications. J. Appl. Genet. 2020;61:221–229. doi: 10.1007/s13353-020-00537-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke S.K., Sampson H.A. Allergenic properties of ovomucoid in man. J. Immunol. 1997;159:2026–2032. [PubMed] [Google Scholar]
- Hirose J., Kitabatake N., Kimura A., Narita H. Recognition of native and/or thermally induced denatured forms of the major food allergen, ovomucoid, by human IgE and mouse monoclonal IgG antibodies. Biosci. Biotechnol. Biochem. 2004;68:2490–2497. doi: 10.1271/bbb.68.2490. [DOI] [PubMed] [Google Scholar]
- Kovacs-Nolan J., Phillips M., Mine Y. Advances in the value of eggs and egg components for human health. J. Agric. Food Chem. 2005;53:8421–8431. doi: 10.1021/jf050964f. [DOI] [PubMed] [Google Scholar]
- Kovacs-Nolan J., Zhang J.W., Hayakawa S., Mine Y. Immunochemical and structural analysis of pepsin-digested egg white ovomucoid. J. Agric. Food Chem. 2000;48:6261–6266. doi: 10.1021/jf000358e. [DOI] [PubMed] [Google Scholar]
- Lalonde S., Stone O.A., Lessard S., Lavertu A., Desjardins J., Beaudoin M., Rivas M., Stainier D.Y.R., Lettre G. Frameshift indels introduced by genome editing can lead to in-frame exon skipping. PLoS One. 2017;12:e0178700. doi: 10.1371/journal.pone.0178700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T., Watanabe K., Nakamura R. Immunochemical and physical properties of peptic-digested ovomucoid. J. Agric. Food Chem. 1983;31:942–946. doi: 10.1021/jf00119a005. [DOI] [PubMed] [Google Scholar]
- Mine Y., Yang M. Recent advances in the understanding of egg allergens: basic, industrial, and clinical perspectives. J. Agri.C Food Chem. 2008;56:4874–4900. doi: 10.1021/jf8001153. [DOI] [PubMed] [Google Scholar]
- Mine Y., Zhang J.W. The allergenicity of ovomucoid and the effect of its elimination from hen's egg white. J. Sci. Food Agric. 2001;81:1540–1546. [Google Scholar]
- Mine Y., Zhang J.W. Identification and fine mapping of IgG and IgE epitopes in ovomucoid. Biochem. Biophys. Res. Commun. 2002;292:1070–1074. doi: 10.1006/bbrc.2002.6725. [DOI] [PubMed] [Google Scholar]
- Oishi I., Yoshii K., Miyahara D., Kagami H., Tagami T. Targeted mutagenesis in chicken using CRISPR/Cas9 system. Sci. Rep. 2016;6:23980. doi: 10.1038/srep23980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T.S., Lee H.J., Kim K.H., Kim J.S., Han J.Y. Targeted gene knockout in chickens mediated by TALENs. Proc. Natl. Acad. Sci. U.S.A. 2014;111:12716–12721. doi: 10.1073/pnas.1410555111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rona R.J., Keil T., Summers C., Gislason D., Zuidmeer L., Sodergren E., Sigurdardottir S.T., Lindner T., Goldhahn K., Dahlstrom J., McBride D., Madsen C. The prevalence of food allergy: a meta-analysis. J. Allergy Clin. Immunol. 2007;120:638–646. doi: 10.1016/j.jaci.2007.05.026. [DOI] [PubMed] [Google Scholar]
- Tanabe S., Tesaki S., Watanabe M. Producing a low ovomucoid egg white preparation by precipitation with aqueous ethanol. Biosci. Biotechnol. Biochem. 2000;64:2005–2007. doi: 10.1271/bbb.64.2005. [DOI] [PubMed] [Google Scholar]
- Urisu A., Ando H., Morita Y., Wada E., Yasaki T., Yamada K., Komada K., Torii S., Goto M., Wakamatsu T. Allergenic activity of heated and ovomucoid-depleted egg white. J. Allergy Clin. Immunol. 1997;100:171–176. doi: 10.1016/s0091-6749(97)70220-3. [DOI] [PubMed] [Google Scholar]
- Verhoeckx K.C.M., Vissers Y.M., Baumert J.L., Faludi R., Feys M., Flanagan S., Herouet-Guicheney C., Holzhauser T., Shimojo R., van der Bolt N., Wichers H., Kimber I. Food processing and allergenicity. Food Chem. Toxicol. 2015;80:223–240. doi: 10.1016/j.fct.2015.03.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.