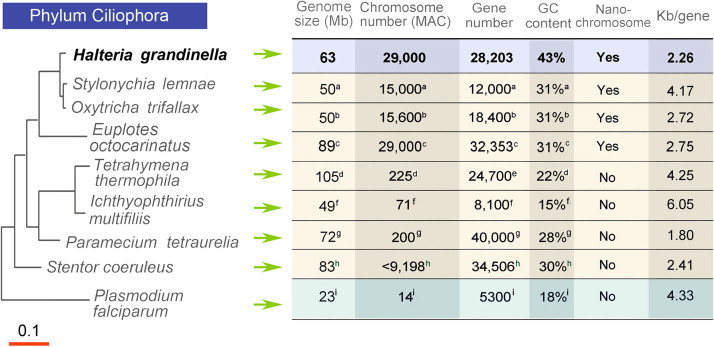
FIG 1.
Comparison of macronuclear genome features. The molecular phylogenetic relationship was constructed by using the maximum likelihood method based on MUSCLE multiple-sequence alignment of 18S rRNA genes (Halteria, MF002432.1; Stylonychia lemnae, AM233915.1; Oxytricha trifallax, FJ545743.1; Euplotes octocarinatus, LT623905.1; Tetrahymena thermophila, M10932.1; Ichthyophthirius multifiliis, U17354.1; Paramecium tetraurelia, AB252009.1; Stentor coeruleus, JQ282899.1; and an outgroup Plasmodium falciparum, MF155937.1). Genome statistics for Halteria were determined in this study. Other statistics were obtained from the following sources: a, reference 25; b, reference 20; c, reference 77; d, reference 18; e, reference 78; f, reference 79; g, reference 19; h, reference 56; i, reference 80. The scale bar corresponds to 0.1 expected substitutions per nucleotide site.