Abstract
In animals, the adaptation to breed at the time of greatest survival of the young is known as seasonal reproduction. This is mainly controlled by the photoperiod, which stimulates the hypothalamic-pituitary-gonadal axis and starts the breeding season. Herein, we have determined the seasonal changes in gene expression patterns of Zhedong white geese pituitary glands under a natural photoperiodism, conducted at autumn equinox (AE), winter solstice (WS), spring equinox (SE), and summer solstice (SS). Pairwise comparisons of WS vs. AE, SE vs. WS, SS vs. SE, and AE vs. SS resulted in 1,139, 33, 704, and 3,503 differently expressed genes, respectively. When compared with SS, AE showed downregulation of genes, such as vasoactive intestinal peptide receptor, prolactin receptor, and thyroid hormone receptor beta, whereas gonadotropin-releasing hormone II receptor was upregulated, indicating that these genes may be responsible for the transition from cessation to egg laying. In addition, the expression levels of 5 transcription factors (POU1F1, Pitx2, NR5A1, NR4A2, and SREBF2) and 6 circadian clock-associated genes (Clock, Per2, ARNTL2, Eya3, Dio2, and NPAS2) also changed seasonally. Gene Ontology term and Kyoto Encyclopedia of Genes and Genomes enrichment analysis revealed that “response to oxidative stress” and steroid biosynthesis pathway also participate in regulating the reproduction seasonality of geese. Overall, these results contribute to the identification of genes involved in seasonal reproduction, enabling a better understanding of the molecular mechanism underlying seasonal reproduction of geese.
Key words: Zhedong white goose, seasonal reproduction, pituitary gland, RNA-Seq analysis
Introduction
Seasonal reproduction is an evolutionary adaptive strategy for animals living in areas with evident seasonal changes in environment, which in particular is important to ensure the survival of the offspring (Ikegami and Yoshimura, 2016; Nishiwaki-Ohkawa and Yoshimura, 2016). Although several factors change seasonally, for example, temperature, precipitation or humidity, the day length (photoperiod) is thought to be responsible for the detection of season variation by many organisms, such as plants (Konstantinova, 1966; Song et al., 2015), insects (Tauber and Kyriacou, 2001; Saunders, 2013), and vertebrates (Hazlerigg and Wagner, 2006; Hazlerigg, 2012). Thus, this phenomenon is called photoperiodism. Based on the relative day length of their reproductive season, seasonal reproduction animals can be classified into long-day (LD) breeders, for example, hamsters and quail, and short-day (SD) breeders, for example, sheep and salmon (Nishiwaki-Ohkawa and Yoshimura, 2016). Long-day breeders develop gonads and display reproductive behaviors during spring when the days become longer and vice versa in SD breeders.
In vertebrates, reproductive process is primarily controlled by the hypothalamic-pituitary-gonadal (HPG) axis. Gonadotropin-releasing hormone (GnRH) produced by the hypothalamus stimulates the biosynthesis and release of luteinizing hormone (LH) and follicle-stimulating hormone (FSH) by the anterior pituitary gland, thus regulating gonadal activation. Prolactin (PRL) and its releasing hormone, vasoactive intestinal peptide (VIP) secreted by the hypothalamus, are also involved in the regulation of annual reproductive activities (Sharp and Blache, 2003; Sharp, 2005). For seasonal breeding animals, the HPG axis is activated at an appropriate time of the year, resulting in a dramatic change in gonadal size. This is particularly evident in birds (Dawson et al., 2001), in which gonadal weight increases more than a hundred-fold during the breeding season (Nicholls et al., 1983). After breeding, the HPG axis is automatically switched off, and the gonads start to regress, although photoperiod still increases. This phenomenon is known as photorefractoriness (Nicholls et al., 1988).
The underlying molecular mechanisms of avian seasonal reproduction have been studied extensively. In contrast to mammals, eyes and pineal gland of birds are not necessary for the regulation of seasonal reproduction because blind ducks and pinealectomized quail still show photostimulation-induced gonadal development (Siopes and Wilson, 1974). Subsequent reports have demonstrated that photoperiodic information is perceived by deep brain photoreceptors, such as OPN5-positive cerebrospinal fluid–contacting neurons, within the mediobasal hypothalamus (Sharp and Follett, 1969; Vigh and Vigh-Teichmann, 1998; Nakane et al., 2010; Yamashita et al., 2010). This information is transmitted to the par tuberalis in the pituitary gland, where the thyroid-stimulating hormone (TSH) is secreted. Long-day–induced TSH acts on the TSH receptor (TSHR) localized in ependymal cells, and upregulating the expression of type 2 deiodinase (DIO2) and downregulating type 3 deiodinase (DIO3) (Yoshimura et al., 2003; Nakao et al., 2008). DIO2 encodes the thyroid hormone (TH)–activating enzyme that converts the prohormone thyroxine (T4) into bioactive triiodothyronine (T3), whereas DIO3 turns both T3 and T4 into inactive metabolites (Yasuo et al., 2005). T3 has been reported to regulate morphological changes in GnRH nerve terminals and the glia, facilitating synthesis and release of GnRH and leading to gonadotropin secretion and gonadal development (Nakane and Yoshimura, 2014). In contrast to long days, the pattern of DIO2 and DIO3 expression during short days depletes T3, and glial cells block the GnRH neuron connection with the basal lamina.
The circadian rhythm is a daily time-keeping mechanism fundamental to a wide range of species. It consists of the transcription-translation feedback of a highly conserved set of genes, collectively known as “clock genes.” It is now commonly accepted that the central core of this gene network consists of “positive” and “negative” elements. The “positive elements” are circadian locomotor output cycles kaput (clock) and the gene that encodes for brain-muscle Arnt-like protein 1 (Arntl or Bmal1). The “negative elements” are genes period 1 to 3 (Per 1-3, but Per1 has not yet been discovered in birds) and cryptochromes cryptochrome 1-2 (Cry1-2). The products of these genes may dynamically interact to trigger rhythmic patterns of transcription, translation, biochemical and physiological processes and ultimately organism behavior (Bell-Pedersen et al., 2005; Eckel-Mahan and Sassone-Corsi, 2013). The heterodimeric protein products of Clock and Bmal1 bind to E-boxes in the promoter regions of Pers and Crys to facilitate their transcription. Following translation, proteins PER and CRY form heterocomplexes that enter the nucleus and inactivate CLOCK and BMAL1 heterodimers, leading to negative feedback, inhibiting their own transcription and eventually shutting-off the loop (Reppert, 2000). Circadian rhythms exist in a wide range of physiological systems that include the reproductive axis. The circadian clock is closely related to the hypothalamus-pituitary regulation of multiple endocrine axes. It is well established that the circadian clock is involved in the photoperiodic response of various vertebrates, including fish (Underwood and Hyde, 1990), birds (Hamner, 1963; Meddle and Follett, 1997), and mammals (Elliott et al., 1972).
The Zhedong white goose (Anser cygnoides), a domestic goose breed in the east of Zhejiang province, is characterized by strict seasonality, high tendency of broodiness, and low egg-laying rate (Zhao et al., 2013). Its regular reproductive season spans from September to May of the following year, that is, from shortening days of early autumn to the longer days of early summer. This behavior results in poor laying performance. The A. cygnoides genome was finally sequenced in 2015, allowing us to perform a system-level analysis of photoperiodic time measurement in goose (Lu et al., 2015). In the present study, we have used high-throughput sequencing technology and bioinformatics to explore the endocrine mechanisms that regulate seasonal breeding in A. cygnoides and to identify genes that are differentially expressed in the pituitary gland at different seasons. Our results provide a better understanding of the molecular regulation that take place in seasonal reproduction.
Materials and methods
Ethics Statement
All animal experimentation was approved by the Institutional Animal Care and Use Committee (IACUC) of Zhejiang University. The methods and protocols were performed in accordance with the Measures for the Administration of Affairs Concerning Experimental Animal of Zhejiang Province, China (approved by the Zhejiang Provincial Government in 2009 and promulgated by Decree No. 263).
Goose Rearing and Sample Collection
Female Zhedong white geese (A. cygnoides) were reared in Xiangshan County, Zhejiang Province, China. They were raised according to common farm practice under the same environmental conditions with natural lighting and temperature. During the experiment, geese were fed ad libitum with rice grain, supplemented with green grass or water plants whenever possible. The feed was given during daytime and geese had free access to drinking water. Twelve healthy Zhedong white geese hatched on the same day in January 2016 were selected for the experiments. They were tested at the autumn equinox (AE) and winter solstice (WS) in 2016 and at the spring equinox (SE) and summer solstice (SS) in 2017, representing different natural photoperiods. Each group consisted of 3 individuals, that is, biological replicates. The geese were anesthetized with sodium pentobarbital and euthanized by exsanguination. The pituitary gland, an oval body located in the sella turcica (hypophysial fossa), was carefully removed from each goose using fine pointed forceps. They were immediately wrapped in freezing tube, snap frozen in liquid nitrogen, and stored at −80°C until RNA extraction.
Total RNA Extraction, Library Preparation, and Illumina Sequencing
Total RNA was extracted from each pituitary gland using TRIzol Reagent (Invitrogen, Carlsbad, CA) followed by treatment with RNase-free DNase I (Thermo Scientific, Waltham, MA) to remove DNA contamination, according to the manufacturer's instructions. Purity, concentration, and integrity of total RNA were measured using a NanoPhotometer spectrophotometer (IMPLEN, Westlake Village, CA), Qubit RNA Assay Kit in Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA), and the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA), respectively. A total amount of 3 μg RNA per sample was used as input material for sequencing library construction. Sequencing libraries were prepared using the NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, Ipswich, MA) following the instruction manual, and index codes were added to attribute sequences to each sample. Briefly, mRNAs were enriched from total RNA using poly-T oligo-attached magnetic beads, which were split into short fragments using divalent cations in NEB-Next First Strand Synthesis Reaction Buffer (5 × ), then reversely transcribed into the first-strand cDNA with random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-). The second-strand cDNA was synthesized using DNA Polymerase I and RNase H. Subsequently, the DNA fragments had their end repaired, with poly(A) added at the 3′ ends. Ligation of NEBNext adaptors for hybridization was followed by purification with AMPure XP system (Beckman Coulter, Beverly, MA) to select cDNA fragments of 150–200 bp. Then PCR was carried out with Phusion High-Fidelity DNA polymerase (NEB, Ipswich, MA), universal PCR primers, and Index (X) primer. After purification of the PCR products, the quality of the cDNA libraries was assessed on an Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA), sequenced on the Illumina HiSeq 4000 platform where 150 bp paired-end reads were generated.
Quantity Control, Reads Mapping, and Gene Annotation
As a part of the quality-control procedure, the clean data were obtained by removing reads containing adapters, reads containing poly-N (N% > 10%), and low-quality reads from raw data. Probabilities Q20 and Q30 (probability of a miscalled base of 1 and 0.1%, respectively) and GC content of the clean data were calculated. The genome assembly and annotation of A. cygnoides were downloaded from GenBank database (accession code AOGC00000000). The filtered, high-quality sequencing reads subsequently were aligned to the assembled reference genome using TopHat (version 2.0.12) (Trapnell et al., 2009).
Gene Quantification and Differential Expression Analysis
HTSeq (version 0.6.1) was used to count the reads numbers mapped to each gene (Anders et al., 2015). Gene expression levels were estimated via FPKM (Fragments Per Kilobase of transcripts per Million fragments mapped) calculated based on the length of the gene and reads count mapped to a gene. Differential expression gene (DEG) analysis of the 4 groups was performed on the DESeq R package (version 1.18.0). To control false discovery rate, P-values were adjusted using the Benjamini and Hochberg's approach, and only genes with an adjust P-value <0.05 were defined as DEG.
GO and KEGG Enrichment Analysis
Gene Ontology (GO) enrichment analysis of all DEG was implemented by the GOseq R package, in which gene length bias was corrected. GO terms with P-value less than 0.01 were considered significantly enriched by DEG.
Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to find pathways in which the DEG were involved. Pathways with a P-value less than 0.05 were defined as significantly enriched pathways. KOBAS software was used to test the statistical enrichment of DEG in KEGG pathways.
Validation of RNA-seq–Based Quantitative RT-PCR
To confirm the DEGs detected by the RNA-seq approach, 15 DEG were selected randomly for qRT-PCR analysis. The qRT-PCR primers were designed using Primer Premier 5.0 software (www.PremierBiosoft.com) and synthesized by Generay biotechnology (Shanghai, China). The primer information is listed in Table 1. Glyceraldehyde-3-phosphate dehydrogenase was used as an internal control. Each 20 μL reaction volume contained 10 μL 2 × ChamQTM Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China), 0.5 μL 10 μM (each) forward and reverse primers, and 1.5 μL cDNA products. The final volume was adjusted using RNA-free water. The qRT-PCR was carried out on LightCycler 960 (Roche) as the following procedure: 1 cycle at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 15 s and annealing at 60°C for 60 s. All reactions were run in triplicate. Relative gene expression levels were calculated using the 2−ΔΔCt method, and the WS group served as calibrator.
Table 1.
Primers used in the real-time quantitative PCR assay of genes.
| Gene name | Forward primer sequence (5′-3′) | Reverse primer sequence (5′-3′) | PCR product (bp) |
|---|---|---|---|
| GAPDH | CTGTCAAGGCTGAGAATG | CAAGAGGCATTGCTGACA | 280 |
| POU1F1 | GAAGCGCAGAACCACCATAA | CCCCTCAGCCATCCTCATAA | 106 |
| VIPR | GGTGCTGGGAGGAAATAAT | GTCCAACGTCTGGGGAATG | 141 |
| TRHR | CTATTTGCATTTCTGTGGAT | TAACTGGCAGGTTTTTTGTC | 224 |
| PPARα | ATGCCAAGGTCTGAGAAGGC | GCCTGCAAGGATGACTCTG | 168 |
| PGR | TGAGGAAGTGCTGTCAAGCC | CTTCTGGCTCAATGCCTCGT | 219 |
| Srebf2 | CCAGCAGCCCGTCATATACC | CCAGAACCTGCTGCTGGATG | 119 |
| IRF2BP2 | GCATTGTCCGCCACCAAAAT | CCTCGTCGGAAGGGATGAAG | 157 |
| CLOCK | TGGCTCCACCCATCATA | CTCGAACCTCCGCATAA | 273 |
| PER2 | CAACCTGAATTAGAGGAAACCACA | AAGCAGGACACTGGACATGG | 198 |
| CGA | TTCTCATGCACGGTTGTCCA | CATTGGAGTGGGATAGGCCC | 119 |
| RELN | CTCATCTGGCTCCTCACT | CCTTCTTCTCTGCTGTCTG | 153 |
| PRL | TGACCTCCTTGCCTATCTG | TGTAATGAAACCCCGACC | 164 |
| GNRHR | CGCACAACTTCACCCAG | AGCGTAGCAGCAGACCAT | 128 |
| SDC1 | TTGAACCTAAAGCCCCTGGC | CATTCCTCCCAGAGTCAGCC | 243 |
| FSH | CCAGCCTGTCCCATTCATACT | ACCTGGGTAAACAAACGTCCTA | 293 |
Results
Overview of RNA-seq Analysis
To identify genes involved in the seasonal reproduction of goose A. cygnoides, we performed transcriptomic sequencing of the pituitary glands of 12 female geese at the 4 seasonal periods described (see Materials and Methods). A total of 704,035,936 raw reads with Q20 > 95% and Q30 > 89% were produced by Illumina HiSeq 4000 from 12 libraries, yielding 678,434,708 clean reads after quality control (see Materials and Methods). The clean reads were aligned to the reference genome of A. cygnoides, and 65.79 to 73.44% of the reads from each library were uniquely mapped, whereas 0.77 to 1.25% were multiple mapped, which was sufficient for subsequent analysis (see details in Table 2). All the sequence data were submitted to the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/sra, NCBI) database (accession number SRP254115).
Table 2.
Data quality and mapping data statistics of RNA-seq libraries.
| Sample name | Raw reads | Clean reads | Q20 (%) | Q30 (%) | Total mapped | Multiple mapped | Uniquely mapped |
|---|---|---|---|---|---|---|---|
| AE1 | 51,733,302 | 50,103,882 | 96.07 | 91.02 | 33,466,593 (66.79%) | 501,793 (1.00%) | 32,964,800 (65.79%) |
| AE2 | 51,334,318 | 49,516,870 | 96.09 | 91.04 | 33,221,268 (67.09%) | 468,132 (0.95%) | 32,753,136 (66.15%) |
| AE3 | 55,244,430 | 53,636,366 | 96.21 | 91.26 | 37,383,365 (69.70%) | 499,916 (0.93%) | 36,883,449 (68.77%) |
| WS1 | 61,937,804 | 59,542,626 | 95.32 | 89.21 | 42,232,194 (70.93%) | 745,463 (1.25%) | 41,486,731 (69.68%) |
| WS2 | 65,701,866 | 63,219,712 | 95.78 | 90.19 | 46,179,485 (73.05%) | 525,540 (0.83%) | 45,653,945 (72.21%) |
| WS3 | 60,739,984 | 58,409,252 | 95.47 | 89.59 | 40,047,979 (68.56%) | 491,402 (0.84%) | 39,556,577 (67.72%) |
| SE1 | 59,318,364 | 57,141,918 | 96.58 | 91.86 | 42,517,522 (74.41%) | 550,699 (0.96%) | 41,966,823 (73.44%) |
| SE2 | 59,493,192 | 57,260,078 | 96.51 | 91.72 | 42,343,912 (73.95%) | 599,498 (1.05%) | 41,744,414 (72.90%) |
| SE3 | 60,894,962 | 58,719,002 | 96.66 | 92.28 | 42,800,229 (72.89%) | 637,200 (1.09%) | 42,163,029 (71.80%) |
| SS1 | 58,078,342 | 55,932,622 | 96.59 | 92.0 | 40,658,843 (72.69%) | 461,450 (0.83%) | 40,197,393 (71.87%) |
| SS2 | 59,175,570 | 57,173,964 | 96.72 | 92.26 | 40,898,203 (71.53%) | 463,165 (0.81%) | 40,435,038 (70.72%) |
| SS3 | 60,383,802 | 57,778,416 | 96.8 | 92.46 | 41,693,939 (72.16%) | 445,167 (0.77%) | 41,248,772 (71.39%) |
AE, WS, SE, and SS represent the autumn equinox, winter solstice, spring equinox, and summer solstice, respectively; 1/2/3 represent 3 biological replicates of each group; Raw reads: all the original data produced by sequencing; Clean reads: the remaining reads after filtering of low quality reads from raw reads; Q20: the proportion of read base whose error rate is less than 1%; Q30: the proportion of read base whose error rate is less than 0.1%; Total mapped: the total reads amount mapped to the reference genome; Multiple mapped: the reads amount mapped to the reference genome more than one site; Uniquely mapped: the reads amount mapped to the reference genome only one site.
Identification of DEGs
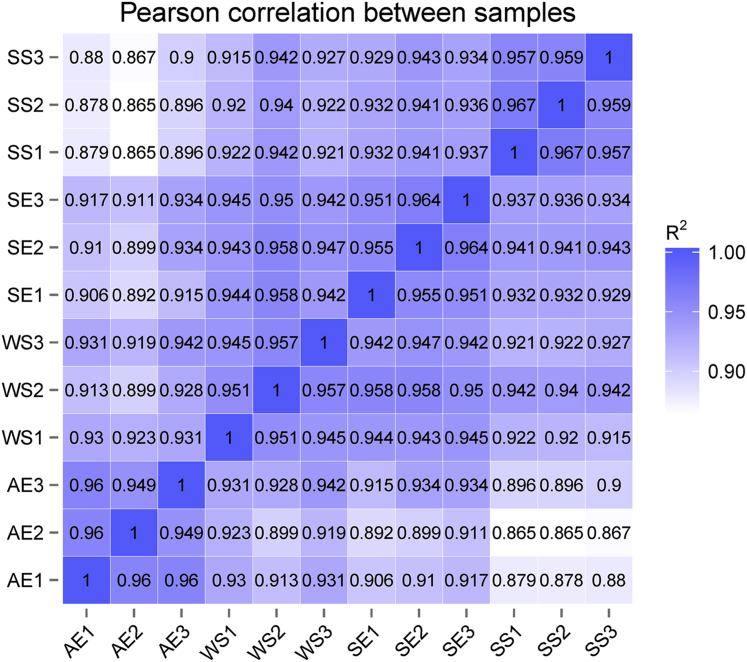
For the 4 groups, the square correlation coefficient (R2 values) between 3 individuals within each group were all above 0.928 (Figure 1), indicating that the three biological replicates were highly repetitive, and the experimental results were reliable and reasonable. Lowest values were found in AE and SS groups, which therefore showed the most variability in expression patterns.
Figure 1.
The Pearson correlation between individuals. Correlation coefficient with R2 value. Abbreviations: AE, autumn equinox; SE, spring equinox; SS, summer solstice; WS, winter solstice. 1, 2, and 3 represent 3 biological replicates of each group.
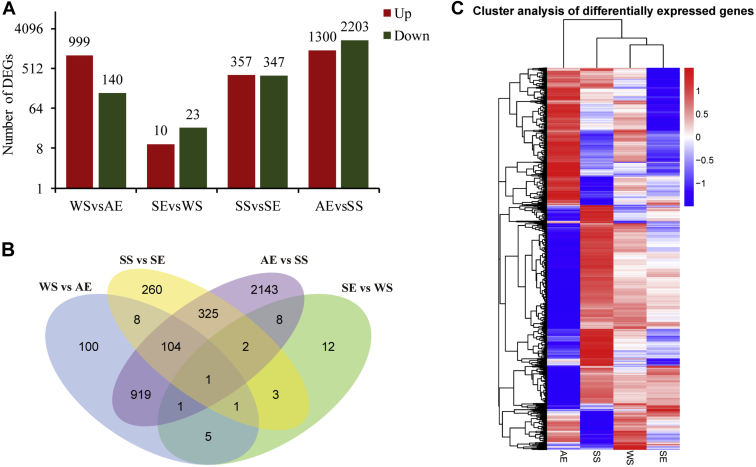
The read count for each sample was standardized by DESeq and used for differential analysis. We identified 1,139, 33, 704, and 3,503 DEGs in the pairwise comparisons WS vs. AE, SE vs. WS, SS vs. SE, and AE vs. SS, respectively (Supplementary Table 1) distributed between upregulated and downregulated (Figure 2A). Only one DEG (ACY_000982, [RELN]) was present in all 4 comparisons (Figure 2B). Differences in gene expression level were visualized via a hierarchical clustering, which clearly displayed the variability in the overall patterns of DEG among the 4 seasons. A large number of genes show stage-specific expression with SS, WS, and SE, clustering away from AE (Figure 2C).
Figure 2.
Statistics of differential expression gene (DEG). (A) Number of upregulated and downregulated DEG in each pairwise comparison. (B) Venn diagrams describing the numbers of unique and shared DEGs between samples. The sum of the numbers in each large circle represents the total number of DEG in the comparison combination, whereas overlapping portions of the circles represent the DEG shared between the combinations. (C) Heatmap comparing expression of the transcriptome at 4 time points. Intensity of color indicates expression levels. Red indicates upregulation and blue indicates downregulation. Similarity between individuals with hierarchical clustering is shown above the heatmap. AE samples were collected at the autumn equinox; WS samples were collected at the winter solstice; SE samples were collected at the spring equinox; and SS samples were collected at the summer solstice.
Hormone-Related Genes
Some DEG were related to three hormone-releasing and hormone receptors, as expected from the known role of the pituitary gland in regulating hormones and seasonal reproduction cycle. For these DEG, 3 different patterns were observed: (1) in AE vs. SS, vasoactive intestinal polypeptide receptor (VIPR) and thyrotropin-releasing hormone-degrading ectoenzyme were downregulated, whereas gonadotropin-releasing hormone II receptor was upregulated significantly; (2) in the comparisons WS vs. AE and AE vs. SS, prolactin receptor (PRLR), glucocorticoid receptor (GR), and thyroid hormone receptor beta were differentially expressed whereas AE had the lowest expression levels; finally (3) estrogen receptor 1 (ER1) was differentially expressed when comparing WS vs. AE, SS vs. SE, and AE vs. SS.
Pituitary Transcription Factors
Several pituitary transcription factors had lower expression levels in the pituitary glands of SS geese. These included pituitary homeobox 2 (Pitx2), Steroidogenic factor 1 (SF-1, also named nuclear receptor subfamily 5 group A member 1, NR5A1), nuclear receptor subfamily 4 group A member 2 (NR4A2), and sterol regulatory element-binding protein 2 (SREBF2). In contrast, level of pituitary-specific positive transcription factor 1 (POU1F1 or Pit-1) was higher, when compared with AE geese.
Clock Genes
We also observed seasonal expression differences of clock genes and proteins. For instance, circadian locomotor output cycles protein kaput (Clock), period circadian protein homolog 2 (Per2), aryl hydrocarbon receptor nuclear translocator-like protein 2 (ARNTL2, also called BMAL2), type II iodothyronine deiodinase (DIO2), aryl hydrocarbon receptor nuclear translocator 2 (ARNT2), CLOCK-interacting pacemaker (CIPC), casein kinase I isoform delta (Csnk1d), pericentrin (PCNT), and nuclear receptor subfamily 1 group D member 2 (NR1D2) were considered to be DEG when comparing AE group with at least one other group. Furthermore, the expression levels of these genes were the highest in the SS group and lowest in the AE group. Except for Per2, its lowest expression level appeared in the WS group, slightly lower than that in the AE group.
Functional Analysis of DEGs
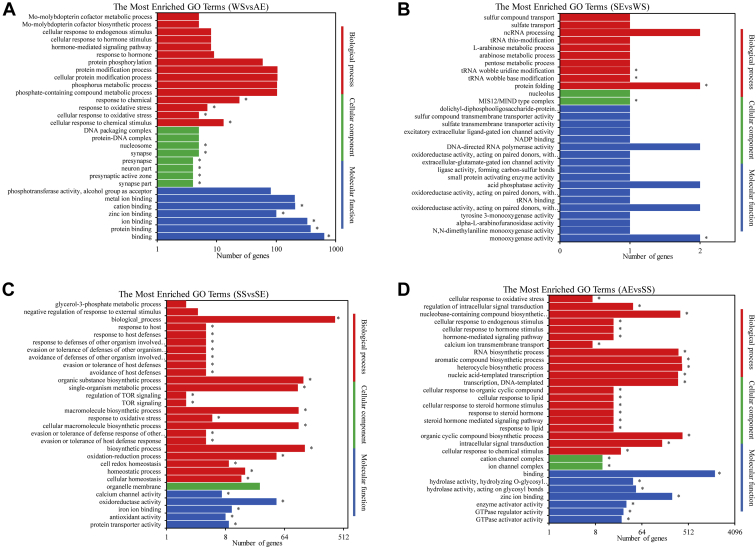
To assign functional categories to the DEG obtained from pairwise comparisons, we performed GO enrichment analysis. GO terms were categorized into 3 main categories, including cellular component, biological process, and molecular function (Supplementary Table 2), and the top 30 highly enriched GO terms were presented in Figure 3. For WS vs. AE (Figure 3A), molecular function such as “binding,” “protein binding” and “ion binding” accounted for the highest proportion of DEG. The significantly enriched cellular components included the “synapse part”, “presynaptic active zone” and “neuron part”. Genes also were involved in biological process, such as “cellular response to oxidative stress”, “response to oxidative stress” and “response to chemical”. Comparison SE vs. WS (Figure 3B) produced only 5 significantly enriched GO terms: “monooxygenase activity”, “MIS12 and MIND type complex”, “protein folding”, “tRNA wobble base modification” and “tRNA wobble uridine modification”. A total of 27 GO terms were significantly enriched of DEG in comparison SS vs. SE (Figure 3C). Among them, for example, “homeostatic process”, “biosynthetic process”, “oxidation-reduction process”, “response to oxidative stress,” and “TOR signaling” were the most important biological process terms. The molecular function category consisted of “protein transporter activity”, “antioxidant activity”, “oxidoreductase activity,” and “calcium channel activity”. For the DEG between AE vs. SS (Figure 3D), most were significantly associated with biological process, including “intracellular signal transduction”, “organic cyclic compound biosynthesis process”, “response to lipid”, “steroid hormone mediated signaling pathway”, “response to steroid hormone”, “transcription, DNA-templated,” and “cellular response to oxidative stress”. The terms belonged to molecular functions, such as “binding”, “GTPase activator activity” and “hydrolase activity, acting on glycosyl bonds” were also significantly enriched. Finally, the term “response to oxidative stress” was enriched in 3 comparisons except SE vs. WS.
Figure 3.
Gene Ontology (GO) functional enrichment of differential expression gene (DEG) between (A) WS vs. AE, (B) SE vs. WS, (C) SS vs. SE, (D) AE vs. SS. The ordinate coordinates represent 3 GO categories under the level of the GO term; the abscissa is the annotation of the term and the number of genes. Three different classifications of GO annotations of 3 basic categories are included (from top to bottom: biological processes, cellular component, and molecular function). The asterisk indicated significantly enriched (P < 0.01). Abbreviations: AE, autumn equinox; SE, spring equinox; SS, summer solstice; WS, winter solstice.
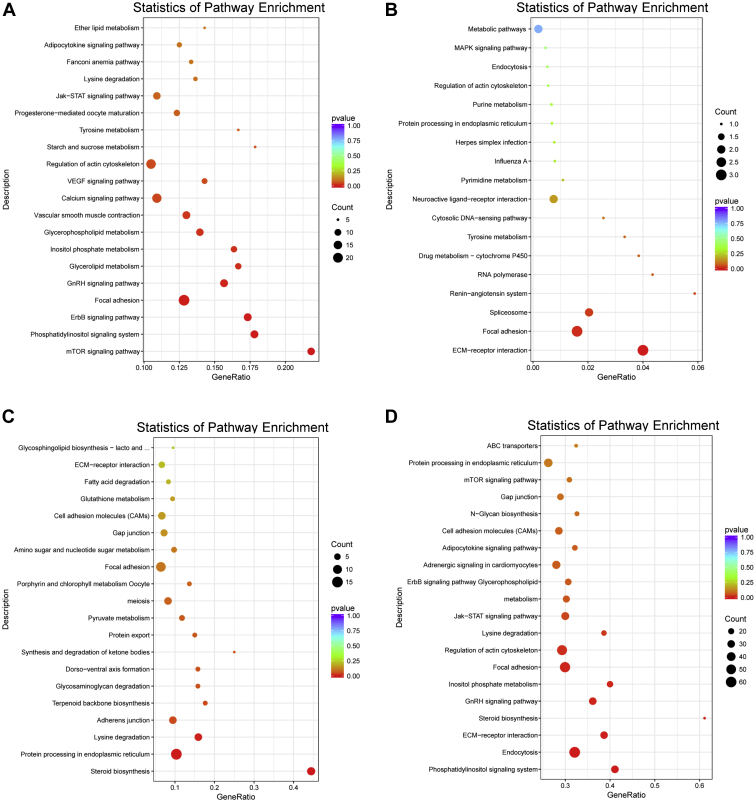
To understand the functional consequences of the expression variations in these 4 pairwise comparisons, the DEG found were mapped to 99, 18, 115, and 147 KEGG pathways, out of which 12, 4, 5, and 10 pathways reached significant levels, respectively. The top 20 pathways with high enrichment significance were displayed in Figure 4. In each comparison, the most significantly enriched pathways were the mTOR signaling pathway (Figure 4A), the extracellular matrix (ECM)-receptor interaction (Figure 4B), steroid biosynthesis (Figure 4C), and phosphatidylinositol signaling system (Figure 4D). Except in SE vs. WS, metabolic pathways accounted for the largest number of DEG (Supplementary Table 3). Eight KEGG pathways were enriched in two or more pairwise comparisons: “Steroid biosynthesis”, “GnRH signaling pathway”, “Lysine degradation”, “Inositol phosphate metabolism”, “Phosphatidylinositol signaling system”, “Focal adhesion”, “ECM-receptor interaction” and “Regulation of acting cytoskeleton”. In the GnRH signaling pathway, expression of GnRH II receptor (gonadotropin-releasing hormone II receptor) increased, while epidermal growth factor receptor and adenylate cyclase type 1 (ADCY1) decreased significantly at AE. Steroid biosynthesis was the most highly represented functional group in SS vs. SE, and was also significantly enriched in AE vs. SS. Genes involved in the steroid biosynthesis pathway, for example, cholesterol side-chain cleavage enzyme (CYP11A1), farnesyl-diphosphate farnesyltransferase 1 (FDFT1), delta (24)-sterol reductase (DHCR24) exhibited decreased expression levels from SE to SS, but were enhanced from SS to AE.
Figure 4.
Pathways enrichment statistical scatter plot of differential expression genes (DEGs) between (A) WS vs. AE, (B) SE vs. WS, (C) SS vs. SE, (D) AE vs. SS. The vertical axis represents the name of pathway; the horizontal axis represents the pathways corresponding rich factor. The gene ratio refers to the ratio of the number of DEG enriched in the pathway and the number of all annotated genes in the pathway. Higher gene ratio indicates greater degrees of enrichment. The 20 pathways with the most significant enrichment were displayed in figure. Abbreviations: AE, autumn equinox; SE, spring equinox; SS, summer solstice; WS, winter solstice.
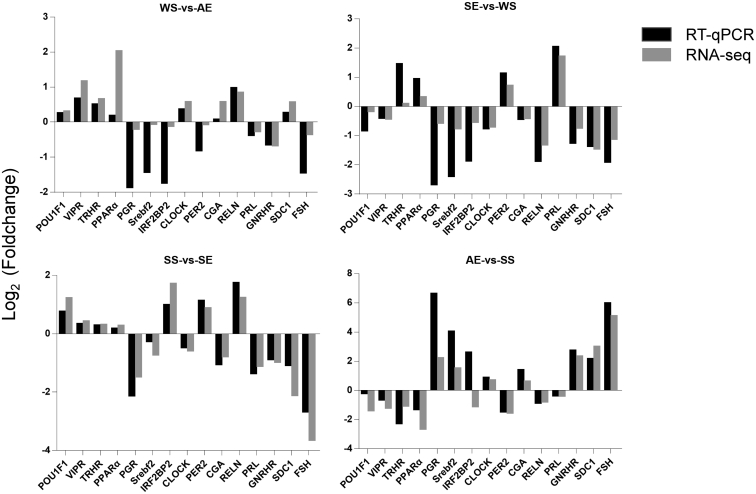
Validation of DEG by qPCR
To evaluate the RNA-seq data, expression levels of 15 randomly selected DEG were analyzed by RT-qPCR, and the log2-fold change was calculated. The trend in expression variation of the 15 genes in intergroup comparisons was consistent with the transcriptome sequencing results (Figure 5). Overall, the RT-qPCR analysis indicated the reliability of the RNA-seq results, which can be further studied and analyzed.
Figure 5.
RT-qPCR validation of differential expression gene (DEG) obtained by RNA-seq. Total RNA extracted from the pituitary glands that were measured by qRT-PCR analysis; relative expression levels were calculated according to the 2−ΔΔCt method using GAPDH as an internal reference gene.
Discussion
To uncover the regulatory mechanisms that synchronize photoperiodic cues and reproduction of A. cygnoides, we have compared transcriptome levels in the pituitary gland, a crucial component of the HPG axis, between different reproductive states seasons. Some DEG found in pairwise comparisons were involved in signaling pathways related to reproductive performance, such as steroid biosynthesis, endocytosis, ECM-receptor interaction, and GnRH signaling pathway.
Genes Associated with Reproductive Hormones and Receptors
Genes known to be associated with reproductive hormones and receptors are candidate gene controlling the reproduction seasonality. In the hypothalamus, photoperiodic information regulates secretions of GnRH and VIP. In the pituitary, these stimulate secretions of LH and PRL, respectively. The latter control annual cycle of gonadal development and breeding activities (Sharp et al., 1998; Sharp and Blache, 2003). In female turkey, increased neuroendocrine activity of the VIP and PRL system suppressed the release of GnRH and LH, and lead to reduction of eggs production (Pitts et al., 1994). Our data show that GnRH receptor (GnRHR) expression level in the SS group was significantly lower than in the AE group, whereas for VIP receptor (VIPR), the results was opposite. GnRH receptor coupled to GnRH, plays an important role in activating the downstream pathway that regulate reproduction. VIP is the avian PRL-releasing factor and its receptor VIPR was strongly suggested to play a prominent role in the regulation of PRL secretion at the pituitary level (Chaiseha et al., 2004). Decreased GnRH receptor expression in SS geese reduces the sensitivity of the pituitary gland to stimulation by GnRH, and upregulated VIPR expression increased sensitivity to VIP and the release of PRL, leading to the cessation of egg production in the SS. We also found transcript expression changes of the PRL receptor (PRLR), which involves in the PRL signal transduction cascade and mediates various biological functions of PRL, including the reproductive processes of seasonally breeding species. PRLR mRNA may be regulated by prolactin in the pituitary (up) and hypothalamus (down) (Zhou et al., 1996). Our results indicate that these genes may participate in regulation of seasonal reproduction and in the reproductive stages transitions.
Circadian Clock-Associated Genes
The circadian rhythmicity of physiological systems provides an organism with the sense of time of day to ensure that physiological and behavioral events coincide (Kennaway, 2005). Circadian clock-associated genes changed significantly during season transitions. The clock protein (CLOCK) and aryl hydrocarbon receptor nuclear translocator-like (BMAL) protein, which can be substituted with Neuronal PAS domain protein 2 (NPAS2) and BMAL2, respectively, paired to form heterodimers CLOCK and BMAL1 or NPAS2 and BMAL2 that activate the transcription of their target genes, such as Period and Cryptochrome (Ikeda et al., 2000; Reick et al., 2001; Oishi et al., 2003; Sasaki et al., 2009). We found no changes in BMAL1 mRNA transcript expression in the pituitary across different seasons. BMAL2, in contrast, changed together with the clock gene, a DEG in comparisons WS vs. AE and AE vs. SS. This is consistent with the active role proposed for BMAL2 in circadian translation (Sasaki et al., 2009).
The circadian clock gene Per2 was highly expressed during long light, consistent with its reported light-inducible expression in different organs of other species (Yoshimura et al., 2000; Dardente et al., 2016). We found a significant enhancement of pituitary gland eyes absent 3 (Eya3) level in SS geese compared with SE and AE. In quail, induction of Eya3 is an early-response gene in par tuberalis activated by long days, where expression patterns are closely matched to that of TSHβ (Nakao et al., 2008). Three conserved E-box are found in Eya3 promoter that suggest regulation by CLOCK and BMAL, which have an additive activation effect on Eya3 promoter constructs (Dardente et al., 2010). Hypothalamic Eya3 expression level increased in photorefractory buntings (Mishra et al., 2017). Overall, this supports that Eya3 plays a role in maintenance of the photoperiodic-induced response (Majumdar et al., 2014; Majumdar et al., 2015) and an involvement of the circadian clock in the regulation of photoperiodism.
Thyroid-Stimulating Hormone
Thyroid-stimulating hormone from the parse tuberalis of the pituitary gland is induced by LD and acts on the ependymal cells to induce DIO2, responsible for converting thyroxine (T4) into triiodothyronine (T3). The enhanced transcription of DIO2 we observed in SS geese may result from the increased day length, consistent with previous observation in Japanese quail (Nicholls et al., 1983) and chicken (Ono et al., 2009). Genes thyroid hormone receptor beta and thyrotropin-releasing hormone-degrading ectoenzyme were upregulated in SS geese, providing a rhythmic gating mechanism for thyroid hormone signaling. Glucocorticoids have been shown to control various physiological functions, notably those involved in development, metabolism, as well as induction of growth hormone expression during pituitary development (Ellestad et al., 2015), and exert most of their effects upon binding to the GR (encoded by NR3A1 gene). In fact, GR is a central receptor and a transcription factor in regulating the HPA axis. These results highlight their potential role in regulating seasonal reproduction via photoperiodicity in goose.
Pituitary Transcription Factors
The present study also identified several pituitary transcription factors or regulators responsible for the reproduction transition between seasons: POU1F1, NR4A2, SF-1, SREBF2, and Pitx2.
POU1F1.
POU1F1 binds and transactivates the promoters of PRL, growth hormone (GH), and TSH beta subunit (TSHβ) genes (Cohen et al., 1996; Miyai et al., 2005) and plays a role in regulating expression of genes for TRH receptor (Matre et al., 1999) and thyroid receptor (Wood et al., 1996). Estrogen responsiveness of the rat prolactin gene requires the presence of both estrogen receptor (ER) and POU1F1, suggesting a synergistic effect in inducing PRL gene transcription, synthesis, and secretion (Day et al., 1990; Nowakowski and Maurer, 1994; Ying and Lin, 2000). POU1F1 also activated PRL gene expression in chicken (Ohkubo et al., 2000) and in domestic turkey (Weatherly et al., 2001).
NR4A2.
NR4A2 works in concert with POU1F1 to enhance PRL expression in cell culture (Peel et al., 2020).
SF-1.
SF-1 regulates expression of the steroidogenic enzyme cytochrome P450 CYP11 A and CYP11 B. In SF-1-deleted mice, expression levels of both LH and FSH were markedly reduced and led to infertility and sexual immaturity (Ingraham et al., 1994; Zhao et al., 2001). SF-1 was claimed to activate the transcription of gonadotrope-specific genes LHβ, GnRH receptor, and neuronal nitric oxide synthase (Fowkes and Burrin, 2003), as well as targeting genes in the HPG axis that are critical for synthesis of progesterone, androgen, estrogens, and steroidogenesis (Jameson, 2004).
SREBF2.
SREBF2, a key transcriptional regulator of cholesterol biosynthesis, was markedly induced by FSH in the granulosa cells (Liu et al., 2009) and expression levels of NR5A1 and SREBF2 strongly associated with the reduced expression levels of steroidogenic genes (Shen et al., 2016).
Pitx2.
Pitx2 is important for pituitary early development, is an upstream regulator of SF-1 gene (Suh et al., 2002) and is implicated in SF-1 gene transcription (Shima et al., 2008). Reduced Pitx2 caused decreased gonadotropes and thyrotropes, also involved in the transcription of TSHβ, GH and GHRHR (Tremblay et al., 2000).
Therefore, we conclude that these 5 transcription factors may target hormone-release genes in goose pituitary gland and are important in the response to changes in photoperiod, triggering the hormonal responses.
Oxidative Stress
GO enrichment analysis of 3 pairwise comparisons show significant enrichment of “response to oxidative stress”, which may play a role in mediating seasonal reproduction transitions. Genes involved in include glutathione peroxidase 3 (Gpx3), sestrin-3 (SESN3), or bromodomain-containing protein 4 (BRD4). These protect against oxidative damage and genotoxic stresses (Budanov et al., 2004; Hussong et al., 2014). The link between reproduction and oxidative stress has been proposed (Metcalfe and Monaghan, 2013) and oxidative stress likely underlie the trade-off between self-maintenance and investment in reproduction (Zimmer and Spencer, 2015). Reproduction is an energy-expensive process for females, with increased resource requirements and metabolism (Foucart et al., 2014). The energetic demands vary at different reproductive stages, and a concomitant change in oxidative stress may be also expected. For instance, song sparrows show increased oxidative stress during the laying period of reproduction as a result of nest predation (Travers et al., 2010). Therefore, our study supports a role for oxidative stress as a reproductive cost during reproductive stages changes.
Steroid Biosynthesis
KEGG enrichment analysis suggests that the involvement of genes enriched in steroid biosynthesis pathway, since expression was reduced sharply during the transition from SE to SS, but increased in AE vs. SS. Production of steroid hormones is associated with steroidogenic enzyme expression, whose changes during follicular development are initiated by gonadotropins (Nett et al., 2002). We observed reduction in mRNA expression levels of seven steroidogenic enzyme genes in the SS group (CYP51A1, MSMO1, NSDHL, FDFT1, Lss, DHCR7, and DHCR24). This might result in the decline of plasma level estrogen in egg-production ceased geese, consistent with results in broody chicken (Shen et al., 2016). Our data suggest that the interaction of steroid biosynthesis pathway and hormone genes, is in part, responsible for the seasonal reproduction in geese.
Conclusions
We have integrated high-throughput sequencing and bioinformatics to compare pituitary mRNA expression profiles among A. cyanoides at 4 season periods, identifying DEGs. Genes of GnRHR, DIO2, POU1F1, Pitx2, NR3C1, Clock, BMAL2, Per2, and NPAS2 have been identified as candidate genes in molecular regulation of reproduction seasonality. GO and KEGG analyses suggest that response to oxidative stress, steroid hormone-mediated signaling pathway, steroid biosynthesis, and GnRH signaling pathway may also play a pivotal role in regulating seasonal reproduction.
Acknowledgments
This work was supported by International S&T Cooperation Program of China (ISTCP) through Grant 2013DFR30980. The authors would like to thank Xiangshan Zhedong White goose breeding farm for raising of goose and help with sample collection.
Discosures
The authors declared that they have no conflicts of interest to this work.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2020.10.049.
Supplementary data
References
- Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D., Cassone V.M., Earnest D.J., Golden S.S., Hardin P.E., Thomas T.L., Zoran M.J. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budanov A.V., Sablina A.A., Feinstein E., Koonin E.V., Chumakov P.M. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- Chaiseha Y., Youngren O.M., El Halawani M.E. Expression of vasoactive intestinal peptide receptor messenger RNA in the hypothalamus and pituitary throughout the Turkey reproductive cycle. Biol. Reprod. 2004;70:593–599. doi: 10.1095/biolreprod.103.022715. [DOI] [PubMed] [Google Scholar]
- Cohen L.E., Wondisford F.E., Radovick S. Role of Pit-1 in the gene expression of growth hormone, prolactin, and thyrotropin. Endocrinol. Metab. Clin. North Am. 1996;25:523–540. doi: 10.1016/s0889-8529(05)70339-x. [DOI] [PubMed] [Google Scholar]
- Dardente H., Wyse C.A., Birnie M.J., Dupre S.M., Loudon A.S., Lincoln G.A., Hazlerigg D.G. A molecular switch for photoperiod responsiveness in mammals. Curr. Biol. 2010;20:2193–2198. doi: 10.1016/j.cub.2010.10.048. [DOI] [PubMed] [Google Scholar]
- Dardente H., Wyse C.A., Lincoln G.A., Wagner G.C., Hazlerigg D.G. Effects of photoperiod Extension on clock gene and Neuropeptide RNA expression in the SCN of the Soay sheep. PLoS One. 2016;11:e0159201. doi: 10.1371/journal.pone.0159201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson A., King V.M., Bentley G.E., Ball G.F. Photoperiodic control of seasonality in birds. J. Biol. Rhythms. 2001;16:365–380. doi: 10.1177/074873001129002079. [DOI] [PubMed] [Google Scholar]
- Day R.N., Koike S., Sakai M., Muramatsu M., Maurer R.A. Both Pit-1 and the estrogen receptor are required for estrogen responsiveness of the rat prolactin gene. Mol. Endocrinol. 1990;4:1964–1971. doi: 10.1210/mend-4-12-1964. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan K., Sassone-Corsi P. Metabolism and the circadian clock converge. Physiol. Rev. 2013;93:107–135. doi: 10.1152/physrev.00016.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellestad L.E., Puckett S.A., Porter T.E. Mechanisms involved in glucocorticoid induction of pituitary GH expression during embryonic development. Endocrinology. 2015;156:1066–1079. doi: 10.1210/en.2014-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott J.A., Stetson M.H., Menaker M. Regulation of testis function in golden hamsters: a circadian clock measures photoperiodic time. Science. 1972;178:771–773. doi: 10.1126/science.178.4062.771. [DOI] [PubMed] [Google Scholar]
- Foucart T., Lourdais O., DeNardo D.F., Heulin B. Influence of reproductive mode on metabolic costs of reproduction: insight from the bimodal lizard Zootoca vivipara. J. Exp. Biol. 2014;217:4049–4056. doi: 10.1242/jeb.104315. [DOI] [PubMed] [Google Scholar]
- Fowkes R.C., Burrin J.M. Steroidogenic factor-1: a key regulator of gonadotroph gene expression. J. Endocrinol. 2003;177:345–350. doi: 10.1677/joe.0.1770345. [DOI] [PubMed] [Google Scholar]
- Hamner W.M. Diurnal rhythm and photoperiodism in Testicular Recrudescence of the House Finch. Science. 1963;142:1294–1295. doi: 10.1126/science.142.3597.1294. [DOI] [PubMed] [Google Scholar]
- Hazlerigg D. The evolutionary physiology of photoperiodism in vertebrates. Prog. Brain Res. 2012;199:413–422. doi: 10.1016/B978-0-444-59427-3.00023-X. [DOI] [PubMed] [Google Scholar]
- Hazlerigg D.G., Wagner G.C. Seasonal photoperiodism in vertebrates: from coincidence to amplitude. Trends Endocrinol. Metab. 2006;17:83–91. doi: 10.1016/j.tem.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Hussong M., Borno S.T., Kerick M., Wunderlich A., Franz A., Sultmann H., Timmermann B., Lehrach H., Hirsch-Kauffmann M., Schweiger M.R. The bromodomain protein BRD4 regulates the KEAP1/NRF2-dependent oxidative stress response. Cell Death Dis. 2014;5:e1195. doi: 10.1038/cddis.2014.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Yu W., Hirai M., Ebisawa T., Honma S., Yoshimura K., Honma K.I., Nomura M. cDNA cloning of a novel bHLH-PAS transcription factor superfamily gene, BMAL2: its mRNA expression, subcellular distribution, and chromosomal localization. Biochem. Biophys. Res. Commun. 2000;275:493–502. doi: 10.1006/bbrc.2000.3248. [DOI] [PubMed] [Google Scholar]
- Ikegami K., Yoshimura T. Comparative analysis reveals the underlying mechanism of vertebrate seasonal reproduction. Gen. Comp. Endocrinol. 2016;227:64–68. doi: 10.1016/j.ygcen.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Ingraham H.A., Lala D.S., Ikeda Y., Luo X., Shen W.H., Nachtigal M.W., Abbud R., Nilson J.H., Parker K.L. The nuclear receptor steroidogenic factor 1 acts at multiple levels of the reproductive axis. Genes Dev. 1994;8:2302–2312. doi: 10.1101/gad.8.19.2302. [DOI] [PubMed] [Google Scholar]
- Jameson J.L. Of mice and men: the tale of steroidogenic factor-1. J. Clin. Endocrinol. Metab. 2004;89:5927–5929. doi: 10.1210/jc.2004-2047. [DOI] [PubMed] [Google Scholar]
- Kennaway D.J. The role of circadian rhythmicity in reproduction. Hum. Reprod. Update. 2005;11:91–101. doi: 10.1093/humupd/dmh054. [DOI] [PubMed] [Google Scholar]
- Konstantinova T.N. Photochemical and dark reactions in photoperiodism in plants. Usp Sovrem Biol. 1966;61:118–131. [PubMed] [Google Scholar]
- Liu Z., Rudd M.D., Hernandez-Gonzalez I., Gonzalez-Robayna I., Fan H.Y., Zeleznik A.J., Richards J.S. FSH and FOXO1 regulate genes in the sterol/steroid and lipid biosynthetic pathways in granulosa cells. Mol. Endocrinol. 2009;23:649–661. doi: 10.1210/me.2008-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Chen Y., Wang Z., Li X., Chen W., Tao Z., Shen J., Tian Y., Wang D., Li G., Chen L., Chen F., Fang D., Yu L., Sun Y., Ma Y., Li J., Wang J. The goose genome sequence leads to insights into the evolution of waterfowl and susceptibility to fatty liver. Genome Biol. 2015;16:89. doi: 10.1186/s13059-015-0652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar G., Rani S., Kumar V. Hypothalamic gene switches control transitions between seasonal life history states in a night-migratory photoperiodic songbird. Mol. Cell Endocrinol. 2015;399:110–121. doi: 10.1016/j.mce.2014.09.020. [DOI] [PubMed] [Google Scholar]
- Majumdar G., Yadav G., Rani S., Kumar V. A photoperiodic molecular response in migratory redheaded bunting exposed to a single long day. Gen. Comp. Endocrinol. 2014;204:104–113. doi: 10.1016/j.ygcen.2014.04.013. [DOI] [PubMed] [Google Scholar]
- Matre V., Hovring P.I., Orstavik S., Frengen E., Rian E., Velickovic Z., Murray-McIntosh R.P., Gautvik K.M. Structural and functional organization of the gene encoding the human thyrotropin-releasing hormone receptor. J. Neurochem. 1999;72:40–50. doi: 10.1046/j.1471-4159.1999.0720040.x. [DOI] [PubMed] [Google Scholar]
- Meddle S.L., Follett B.K. Photoperiodically driven changes in Fos expression within the basal tuberal hypothalamus and median eminence of Japanese quail. J. Neurosci. 1997;17:8909–8918. doi: 10.1523/JNEUROSCI.17-22-08909.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe N.B., Monaghan P. Does reproduction cause oxidative stress? An open question. Trends Ecol. Evol. 2013;28:347–350. doi: 10.1016/j.tree.2013.01.015. [DOI] [PubMed] [Google Scholar]
- Mishra I., Singh D., Kumar V. Seasonal alterations in the daily rhythms in hypothalamic expression of genes involved in the photoperiodic transduction and neurosteroid-dependent processes in migratory blackheaded buntings. J. Neuroendocrinol. 2017;29:1–10. doi: 10.1111/jne.12469. [DOI] [PubMed] [Google Scholar]
- Miyai S., Yoshimura S., Iwasaki Y., Takekoshi S., Lloyd R.V., Osamura R.Y. Induction of GH, PRL, and TSH beta mRNA by transfection of Pit-1 in a human pituitary adenoma-derived cell line. Cell Tissue Res. 2005;322:269–277. doi: 10.1007/s00441-005-0033-z. [DOI] [PubMed] [Google Scholar]
- Nakane Y., Ikegami K., Ono H., Yamamoto N., Yoshida S., Hirunagi K., Ebihara S., Kubo Y., Yoshimura T. A mammalian neural tissue opsin (Opsin 5) is a deep brain photoreceptor in birds. Proc. Natl. Acad. Sci. U S A. 2010;107:15264–15268. doi: 10.1073/pnas.1006393107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane Y., Yoshimura T. Universality and diversity in the signal transduction pathway that regulates seasonal reproduction in vertebrates. Front Neurosci. 2014;8:115. doi: 10.3389/fnins.2014.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao N., Ono H., Yamamura T., Anraku T., Takagi T., Higashi K., Yasuo S., Katou Y., Kageyama S., Uno Y., Kasukawa T., Iigo M., Sharp P.J., Iwasawa A., Suzuki Y., Sugano S., Niimi T., Mizutani M., Namikawa T., Ebihara S., Ueda H.R., Yoshimura T. Thyrotrophin in the pars tuberalis triggers photoperiodic response. Nature. 2008;452:317–322. doi: 10.1038/nature06738. [DOI] [PubMed] [Google Scholar]
- Nett T.M., Turzillo A.M., Baratta M., Rispoli L.A. Pituitary effects of steroid hormones on secretion of follicle-stimulating hormone and luteinizing hormone. Domest. Anim. Endocrinol. 2002;23:33–42. doi: 10.1016/s0739-7240(02)00143-1. [DOI] [PubMed] [Google Scholar]
- Nicholls T.J., Follett B.K., Robinson J.E. A photoperiodic response in gonadectomized Japanese quail exposed to a single long day. J. Endocrinol. 1983;97:121–126. doi: 10.1677/joe.0.0970121. [DOI] [PubMed] [Google Scholar]
- Nicholls T.J., Goldsmith A.R., Dawson A. Photorefractoriness in birds and comparison with mammals. Physiol. Rev. 1988;68:133–176. doi: 10.1152/physrev.1988.68.1.133. [DOI] [PubMed] [Google Scholar]
- Nishiwaki-Ohkawa T., Yoshimura T. Molecular basis for regulating seasonal reproduction in vertebrates. J. Endocrinol. 2016;229:R117–R127. doi: 10.1530/JOE-16-0066. [DOI] [PubMed] [Google Scholar]
- Nowakowski B.E., Maurer R.A. Multiple Pit-1-binding sites facilitate estrogen responsiveness of the prolactin gene. Mol. Endocrinol. 1994;8:1742–1749. doi: 10.1210/mend.8.12.7708061. [DOI] [PubMed] [Google Scholar]
- Ohkubo T., Tanaka M., Nakashima K. Molecular cloning of the chicken prolactin gene and activation by Pit-1 and cAMP-induced factor in GH3 cells. Gen. Comp. Endocrinol. 2000;119:208–216. doi: 10.1006/gcen.2000.7507. [DOI] [PubMed] [Google Scholar]
- Oishi K., Miyazaki K., Kadota K., Kikuno R., Nagase T., Atsumi G., Ohkura N., Azama T., Mesaki M., Yukimasa S., Kobayashi H., Iitaka C., Umehara T., Horikoshi M., Kudo T., Shimizu Y., Yano M., Monden M., Machida K., Matsuda J., Horie S., Todo T., Ishida N. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J. Biol. Chem. 2003;278:41519–41527. doi: 10.1074/jbc.M304564200. [DOI] [PubMed] [Google Scholar]
- Ono H., Nakao N., Yamamura T., Kinoshita K., Mizutani M., Namikawa T., Iigo M., Ebihara S., Yoshimura T. Red jungle fowl (Gallus gallus) as a model for studying the molecular mechanism of seasonal reproduction. Anim. Sci. J. 2009;80:328–332. doi: 10.1111/j.1740-0929.2009.00628.x. [DOI] [PubMed] [Google Scholar]
- Peel M.T., Ho Y., Liebhaber S.A. The transcription factor NR4A2 plays an Essential role in Driving prolactin expression in female pituitary Lactotropes. Endocrinology. 2020;161 doi: 10.1210/endocr/bqaa046. bqaa046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts G.R., Youngren O.M., Silsby J.L., Rozenboim I., Chaiseha Y., Phillips R.E., Foster D.N., el Halawani M.E. Role of vasoactive intestinal peptide in the control of prolactin-induced Turkey incubation behavior. II. Chronic infusion of vasoactive intestinal peptide. Biol. Reprod. 1994;50:1350–1356. doi: 10.1095/biolreprod50.6.1350. [DOI] [PubMed] [Google Scholar]
- Reick M., Garcia J.A., Dudley C., McKnight S.L. NPAS2: an analog of clock operative in the mammalian forebrain. Science. 2001;293:506–509. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- Reppert S.M. Cellular and molecular basis of circadian timing in mammals. Semin. Perinatol. 2000;24:243–246. doi: 10.1053/sper.2000.9122. [DOI] [PubMed] [Google Scholar]
- Sasaki M., Yoshitane H., Du N.H., Okano T., Fukada Y. Preferential inhibition of BMAL2-CLOCK activity by PER2 reemphasizes its negative role and a positive role of BMAL2 in the circadian transcription. J. Biol. Chem. 2009;284:25149–25159. doi: 10.1074/jbc.M109.040758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders D.S. Insect photoperiodism: measuring the night. J. Insect Physiol. 2013;59:1–10. doi: 10.1016/j.jinsphys.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Sharp P.J. Photoperiodic regulation of seasonal breeding in birds. Ann. N. Y. Acad. Sci. 2005;1040:189–199. doi: 10.1196/annals.1327.024. [DOI] [PubMed] [Google Scholar]
- Sharp P.J., Blache D. A neuroendocrine model for prolactin as the key mediator of seasonal breeding in birds under long- and short-day photoperiods. Can J. Physiol. Pharmacol. 2003;81:350–358. doi: 10.1139/y03-025. [DOI] [PubMed] [Google Scholar]
- Sharp P.J., Dawson A., Lea R.W. Control of luteinizing hormone and prolactin secretion in birds. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 1998;119:275–282. doi: 10.1016/s0742-8413(98)00016-4. [DOI] [PubMed] [Google Scholar]
- Sharp P.J., Follett B.K. The effect of hypothalamic lesions on gonadotrophin release in Japanese quail (Coturnix coturnix japonica) Neuroendocrinology. 1969;5:205–218. doi: 10.1159/000121861. [DOI] [PubMed] [Google Scholar]
- Shen X., Bai X., Xu J., Zhou M., Xu H., Nie Q., Lu X., Zhang X. Transcriptome sequencing reveals genetic mechanisms underlying the transition between the laying and brooding phases and gene expression changes associated with divergent reproductive phenotypes in chickens. Mol. Biol. Rep. 2016;43:977–989. doi: 10.1007/s11033-016-4033-8. [DOI] [PubMed] [Google Scholar]
- Shima Y., Zubair M., Komatsu T., Oka S., Yokoyama C., Tachibana T., Hjalt T.A., Drouin J., Morohashi K. Pituitary homeobox 2 regulates adrenal4 binding protein/steroidogenic factor-1 gene transcription in the pituitary gonadotrope through interaction with the intronic enhancer. Mol. Endocrinol. 2008;22:1633–1646. doi: 10.1210/me.2007-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siopes T.D., Wilson W.O. Extraocular modification of photoreception in intact and pinealectomized coturnix. Poult. Sci. 1974;53:2035–2041. doi: 10.3382/ps.0532035. [DOI] [PubMed] [Google Scholar]
- Song Y.H., Shim J.S., Kinmonth-Schultz H.A., Imaizumi T. Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 2015;66:441–464. doi: 10.1146/annurev-arplant-043014-115555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H., Gage P.J., Drouin J., Camper S.A. Pitx2 is required at multiple stages of pituitary organogenesis: pituitary primordium formation and cell specification. Development. 2002;129:329–337. doi: 10.1242/dev.129.2.329. [DOI] [PubMed] [Google Scholar]
- Tauber E., Kyriacou B.P. Insect photoperiodism and circadian clocks: models and mechanisms. J. Biol. Rhythms. 2001;16:381–390. doi: 10.1177/074873001129002088. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers M., Clinchy M., Zanette L., Boonstra R., Williams T.D. Indirect predator effects on clutch size and the cost of egg production. Ecol. Lett. 2010;13:980–988. doi: 10.1111/j.1461-0248.2010.01488.x. [DOI] [PubMed] [Google Scholar]
- Tremblay J.J., Goodyer C.G., Drouin J. Transcriptional properties of Ptx1 and Ptx2 isoforms. Neuroendocrinology. 2000;71:277–286. doi: 10.1159/000054547. [DOI] [PubMed] [Google Scholar]
- Underwood H., Hyde L.L. A circadian clock measures photoperiodic time in the male lizardAnolis carolinensis. J. Comp. Physiol. A. 1990;167:231–243. [Google Scholar]
- Vigh B., Vigh-Teichmann I. Actual problems of the cerebrospinal fluid-contacting neurons. Microsc. Res. Tech. 1998;41:57–83. doi: 10.1002/(SICI)1097-0029(19980401)41:1<57::AID-JEMT6>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Weatherly K.L., Ramesh R., Strange H., Waite K.L., Storrie B., Proudman J.A., Wong E.A. The Turkey transcription factor Pit-1/GHF-1 can activate the Turkey prolactin and growth hormone gene promoters in vitro but is not detectable in lactotrophs in vivo. Gen. Comp. Endocrinol. 2001;123:244–253. doi: 10.1006/gcen.2001.7680. [DOI] [PubMed] [Google Scholar]
- Wood W.M., Dowding J.M., Bright T.M., McDermott M.T., Haugen B.R., Gordon D.F., Ridgway E.C. Thyroid hormone receptor beta2 promoter activity in pituitary cells is regulated by Pit-1. J. Biol. Chem. 1996;271:24213–24220. doi: 10.1074/jbc.271.39.24213. [DOI] [PubMed] [Google Scholar]
- Yamashita T., Ohuchi H., Tomonari S., Ikeda K., Sakai K., Shichida Y. Opn5 is a UV-sensitive bistable pigment that couples with Gi subtype of G protein. Proc. Natl. Acad. Sci. U S A. 2010;107:22084–22089. doi: 10.1073/pnas.1012498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuo S., Watanabe M., Nakao N., Takagi T., Follett B.K., Ebihara S., Yoshimura T. The reciprocal switching of two thyroid hormone-activating and -inactivating enzyme genes is involved in the photoperiodic gonadal response of Japanese quail. Endocrinology. 2005;146:2551–2554. doi: 10.1210/en.2005-0057. [DOI] [PubMed] [Google Scholar]
- Ying C., Lin D.H. Estrogen-modulated estrogen receptor x Pit-1 protein complex formation and prolactin gene activation require novel protein synthesis. J. Biol. Chem. 2000;275:15407–15412. doi: 10.1074/jbc.275.20.15407. [DOI] [PubMed] [Google Scholar]
- Yoshimura T., Suzuki Y., Makino E., Suzuki T., Kuroiwa A., Matsuda Y., Namikawa T., Ebihara S. Molecular analysis of avian circadian clock genes. Brain Res. Mol. Brain Res. 2000;78:207–215. doi: 10.1016/s0169-328x(00)00091-7. [DOI] [PubMed] [Google Scholar]
- Yoshimura T., Yasuo S., Watanabe M., Iigo M., Yamamura T., Hirunagi K., Ebihara S. Light-induced hormone conversion of T4 to T3 regulates photoperiodic response of gonads in birds. Nature. 2003;426:178–181. doi: 10.1038/nature02117. [DOI] [PubMed] [Google Scholar]
- Zhao L., Bakke M., Parker K.L. Pituitary-specific knockout of steroidogenic factor 1. Mol. Cell Endocrinol. 2001;185:27–32. doi: 10.1016/s0303-7207(01)00621-9. [DOI] [PubMed] [Google Scholar]
- Zhao X., Shao T., Wang Y.Q., Lu X.L., Luo J.B., Zhou W.D. The phytoestrogen daidzein may affect reproductive performance of Zhedong White geese by regulating gene mRNA levels in the HPG axis. Br. Poult. Sci. 2013;54:252–258. doi: 10.1080/00071668.2013.767439. [DOI] [PubMed] [Google Scholar]
- Zhou J.F., Zadworny D., Guemene D., Kuhnlein U. Molecular cloning, tissue distribution, and expression of the prolactin receptor during various reproductive states in Meleagris gallopavo. Biol. Reprod. 1996;55:1081–1090. doi: 10.1095/biolreprod55.5.1081. [DOI] [PubMed] [Google Scholar]
- Zimmer C., Spencer K.A. Reduced resistance to oxidative stress during reproduction as a cost of early-life stress. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 2015;183:9–13. doi: 10.1016/j.cbpa.2014.12.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.