Abstract
It is known that nutrition and immunity are connected, but the mechanism is not very clear. Endogenous retroviruses (ERV) account for 8 to 10% of the human and mouse genomes and play an important role in some biological processes of animals. Recent studies indicate that the activation of ERV can affect the expression of the immunity- or inflammation-related genes, and the activities of ERV are subjected to regulation of many factors including nutritional factors. Therefore, we hypothesize that nutritional status can affect the expression of the immunity- or inflammation-related genes via ERV. To verify this hypothesis, the nutritional status of animals was altered by fasting or overfeeding, and the expression of intact ERV (ERVK18P, ERVK25P) and immunity- or inflammation-related genes (DDX41, IFIH1, IFNG, IRF7, STAT3) in the liver was determined by quantitative PCR, followed by overexpressing ERVK25P in goose primary hepatocytes and determining the expression of the immunity- or inflammation-related genes. The data showed that compared with the control group (no fasting), the expression of ERV and the immunity- or inflammation-related genes was increased in the liver of the fasted chickens but decreased in the liver of the fasted geese. Moreover, compared with the control group (routinely fed), the expression of ERV and the immunity- or inflammation-related genes was increased in the liver of the overfed geese. In addition, overexpression of ERVK25P in goose primary hepatocytes can induce the expression of the immunity- or inflammation-related genes. In conclusion, these findings suggest that ERV mediate the effects of fasting and overfeeding on the expression of the immunity- or inflammation-related genes, the mediation varied with poultry species, and ERV and the immunity- or inflammation-related genes may be involved in the development of goose fatty liver. This study provides a potential mechanism for the connection between nutrition and immunity.
Key words: nutrition, immunity, poultry, fatty liver, endogenous retrovirus
Introduction
Endogenous retroviruses (ERV) are considered the remnants of exogenous retroviruses (proviruses). Most of these remained ‘fossil’ sequences contain an amount of mutations that have been accumulated in the process of long-term evolution since their integration into host genomes (Cañadas et al., 2018). They exist in almost all mammalian animals (such as humans, mice, cats, and sheep) and other vertebrates (such as chickens) (Melanie and Nair, 2014; Xu et al., 2014). In the chicken, ERV account for more than 3% of the chicken genome (Huda et al., 2008). Although ERV are abundant in animal genomes, many ERV are not intact. Intact ERV refer to those whose structures are not easily distinguished from exogenous retroviruses. These ERV usually contain 2 long terminal repeats (LTR) that have elements for transcriptional regulation, the coding sequences of viral proteins (group-specific antigen [Gag], reverse transcriptase [Pol], and envelope protein [Env]), polypurine track sequence, and short flanking genomic sequences of their host cells (Jern and Coffin, 2008; Dolei et al., 2015; Küry et al., 2018). So far, there are about 500 relatively intact ERV that are found in the chicken genome (Bolisetty et al., 2012). In addition to the relatively intact ERV, there are other types of ERV, including the ‘slender’ ERV that lack one or more coding genes necessary for self-replication (usually the Env gene) and ‘solo LTR’ ERV. The number of ‘solo LTR’ ERV is about 60 times the number of the relatively intact ERV (Bolisetty et al., 2012). Phylogenetic analysis indicates that avian proretroviruses (i.e., ERV) can be classified into class I (gamma-like), class II (alpha- and beta-like), and class III (distantly spuma-like) proretroviruses. Alpha-like proretroviruses are outnumbered by beta-like, gamma-like, and alphabeta proretroviruses (Bolisetty et al., 2012). Compared with mammalian proretroviruses, the avian proretroviruses are more heterogeneous. The beta-like proretroviruses have undergone an evolutionary transition from beta-like to alphabeta-like and then to alpha-like proretroviruses, with a gradual loss of betaretroviral markers. The alphabeta proretroviruses are the intermediate between alpha-like and beta-like ones, including some earlier recognized avian proretroviruses. The class III proretroviruses appear to be the oldest, followed by the beta-like and gamma-like proretroviruses, whereas the alphabeta and alpha-like proretroviruses appear to be the youngest. Most proretroviruses are integrated in host genes in the sense orientation (Bolisetty et al., 2012).
Like non-LTR transposons (e.g., long or short interspersed nuclear elements), ERV are mobile elements that are able to transpose themselves in the form of DNA sequence from one location to another in the host genome. This transposition is mediated by the RNA intermediate. Although ERV as retrotransposons have strong transposable ability in the early stage of evolution, most of them now have lost this ability (Jern and Coffin, 2008). Moreover, deep sequencing studies indicate that many ERV are generally silent. For instance, only about 20% of ERV are transcribed in chicken embryo fibroblasts, and a subset of these are also transcribed in vivo (Bolisetty et al., 2012). In addition, recent studies show that some silent ERV can be activated and expressed under certain conditions (Crichton et al., 2014), and their expression is affected by many factors, such as cell type or tissue type (especially placenta and germ cells), cell differentiation and aging process, cytokines, the factors that disrupt the normal function of cells, and nutritional factors (Taruscio and Mantovani, 2004; Denner, 2016; Elaheh et al., 2018). In recent decades, the biological functions of ERV have been gradually uncovered: 1) Transposition of ERV may destabilize host genomes, but ERV as an original genetic material allow host animals to increase diversity among and within species, enhance adaptability to environment, and maintain continuous evolution (Zhang et al., 2008); 2) Promoters and enhancers in the LTR regions of ERV can affect the transcription of their adjacent genes and alter the epigenetic status of adjacent regions (such as DNA methylation and histone modification) (Thompson et al., 2016); 3) By binding Env proteins to host receptors, ERV can block the binding of exogenous viruses to the same receptors, thus providing host cells with the ability to resist exogenous viruses (Nadeau et al., 2015); 4) ERV transcripts can activate the innate immune system and induce the production of cytokines such as IFN via the double-stranded RNA-dependent TLR3/MDA5 signaling pathway, thus inhibiting tumors (Chiappinelli et al., 2015); and 5) ERV are also involved in the occurrence and development of some diseases, such as aging, autoimmunity, and degenerative neurological diseases (Mager and Stoye, 2015; Nadeau et al., 2015).
Endogenous retrovirus group K (ERVK) is the most recently endogenized one among the different groups of ERV (ERVW, ERVH, ERVK, and so on). It contains the coding sequence for functional proteins, thus being considered the most intact and biologically active ERV group (Hohn et al., 2013). The upregulated expression of ERVK has been associated with inflammatory disease, neurological disease, autoimmune disease, and so on (Haraguchi et al., 1992; Tolosa et al., 2012). Recent studies show that ERVK activation by the DNA methyltransferase inhibitor, 5-aza-2-deoxycytidine, can enhance cellular innate immunity (Nogues et al., 2018). Compared with human ERVK having many members, avian ERVK has only several members annotated in GenBank. The annotated ERVK members shared by the chicken and goose are just ERVK18P (LOC106029425) and ERVK25P (LOC106046236). At present, the biological or pathological role of avian ERVK remains unknown.
Nutrition and energy statuses are important factors affecting animal growth, reproduction, and immunity. As mentioned previously, nutritional factors may activate the expression of ERV, and ERV may regulate the expression of the immunity- or inflammation-related genes in multiple ways. Therefore, we speculate that the level of nutrition or energy can affect the expression of the immunity- or inflammation-related genes via ERV. To verify this speculation, nutritional status was altered by fasting or overfeeding in experimental chickens or geese, and the expression of ERV and the immunity- or inflammation-related genes in the liver was then determined. In addition, overexpression of ERVK25P in goose primary hepatocytes was also performed to address the relation between ERV and the immunity (or inflammation)-related genes. This study provides a new insight into the mechanism for the connection between nutrition and immunity.
Materials and methods
Experimental Animals
All animal protocols were in accordance with the institutional guidelines on the use of agricultural animals in research and approved by the Animal Care and Use Committee at Yangzhou University in China.
The Jurong Siji goslings from the same hatching batch were reared on the ground under natural lighting and conventional husbandry management at the Mali Experimental Farm (Jurong, Jiangsu, China). At the age of 70 d, 16 healthy geese were randomly divided into 2 groups: fasting group (the geese were fasted for 24 h with free access to water) and control group (no fasting, ad libitum access to feed and water). After 24 h of fasting, all the experimental individuals were sacrificed, and liver samples were collected, snap-frozen in liquid nitrogen, and transferred at −70°C for storage. Similarly, sixteen 20-week-old healthy Rhode Island Red chickens were sacrificed for fasting experiment. In contrast to fasting, sixteen 70-day-old healthy Landes geese (provided by Licheng Animal and Poultry Co., Ltd., Huaian, Jiangsu, China) were randomly and equally divided into the overfeeding (24 d of overfeeding) and the control group (feeding routinely). The protocol for overfeeding was described previously by Geng et al., 2016a. On the 24th day of overfeeding, the liver samples were harvested from both the control and overfeeding groups and stored at −70°C.
Isolation and Culture of Goose Primary Hepatocytes and Overexpression of ERV
The goose primary hepatocytes were isolated and cultured from goose embryos on the 22nd or 23rd day of hatching as described previously by Osman et al., 2016.
The customized overexpression vector of the goose LOC106046236 gene (or ERVK member 25 Pol protein-like, ERVK25P) and empty vector were purchased from Suzhou Jima Gene Co., Ltd. (Suzhou, China). The overexpression vector was constructed using pcDNA3.1 vector containing CMV promoter and the inserted DNA fragment that was the coding sequence of the ERVK25 polymerase gene. The overexpression vector and empty vector were separately transfected into goose primary hepatocytes that had been isolated and cultured for 24 h with Lipofectamine 2000 (cat# 11,668-019, Invitrogen, Co., Ltd., Camarillo). After 32 h of transfection, the cells were collected for gene expression analysis by quantitative fluorescence PCR. The transfection was conducted as previously described by Geng et al., 2013.
RNA Purification and cDNA Synthesis
The total RNA was isolated from liver samples using the TRIzol kit (cat# DP424; Tiangen Biotech (Beijing) Co., Ltd., Beijing, China). The purified RNA samples were reverse transcribed into cDNA using the HiScriptTM Q RTSuperMix reverse transcription kit (cat# R123-01; Vazyme Biotech Co., Ltd., Nanjing, China). Reverse transcription was carried out according to the manufacturer's instructions.
Quantitative PCR Analysis
Based on the reference sequence of each gene in GenBank, quantitative PCR primers for the genes of interest and internal reference gene, GAPDH, were designed using online Primer 3.0 software (Whitehead Institute for Biomedical Research, Cambridge), and the sequence specificity was confirmed using the Primer-BLAST program (National Center for Biotechnology information, Bethesda) on the NCBI website. Primer sequences are listed in Table 1. According to the manufacturer's instructions, quantitative PCR was performed using the Vazyme AceQ qPCR SYBR Green Master Mix kit (cat# Q111-02/03; Vazyme Biotech Co., Ltd., Nanjing, China) and cDNA samples. The relative expression of the genes of interest was calculated using the 2−△△CT method as previously described by Geng et al., 2016b.
Table 1.
List of primer sequences for quantitative PCR.
| Gene name | Primer sequence (5′ to 3′) | Accession number |
|---|---|---|
| GOOSE-ERVK18P | ATTCGCATTCACCCTCCCTG | XM_013170710 |
| TCTGGCAAAGTGTAGGCGAG | ||
| GOOSE-ERVK25P | CTCCCCAGCCCATATTACCT | XM_013197145 |
| CATCAGCAGGGAGAAGTGGA | ||
| GOOSE-DDX41 | AGAAAGCGGAAGCTCGGAAG | XM_013191862 |
| CGGACATGCCCAGGATGTAA | ||
| GOOSE-IFIH1 | GATTGCGGACAAGCTTGGGG | XM_013171142 |
| GGGAACCTGATGGGCAGTTC | ||
| GOOSE-IFNG | CCTTCAGCTGACTGGCTTGAA | XM_013198313 |
| GCATCTCTTTGGAGACTGGCT | ||
| GOOSE-IRF7 | ATCCCCTGGAAGCACAATGC | XM_013174398 |
| GCTGTTCTTGGAGTGGTCCT | ||
| GOOSE-STAT3 | GCTGTGGAACGAAGGGTACA | XM_013199804 |
| CCCATGATGATCTCGGCGAA | ||
| GOOSE-GAPDH | CTGATGCTCCCATGTTCGTG | XM_013199522 |
| CCACGATGCCAAAGTTGTCA | ||
| CHICKEN-ERVK18P | ACTGGAGGCAGGACACATTG | XM_015284763 |
| ACAGCCCGAAGCTCATGCAA | ||
| CHICKEN-ERVK25P | GCCAGACACCCCTTGCAATG | XM_015277371 |
| TGCCGTCAATGCTTCCTCCA | ||
| CHICKEN-DDX41 | TTGCCACTGATGTCGCTTCT | NM_001349708 |
| CGCCCGATACGGTGAACATA | ||
| CHICKEN-IFIH1 | GATTACCAGATGGAAGTTGC | NM_001193638 |
| GGTAATGTAAACAGCCACTC | ||
| CHICKEN-IFNG | CTGACAAGTCAAAGCCGCAC | NM_001193638 |
| TCAAGTCGTTCATCGGGAGC | ||
| CHICKEN-IRF7 | GAGGATCCGGCCAAATGGAA | NM_205372 |
| TGTCATTGGGGACGCCTGAG | ||
| CHICKEN-STAT3 | GTGCTGCTCCGTATCTGAAG | NM_001030931 |
| TCTGCTCCCTCGCTACTGTT | ||
| CHICKEN-GAPDH | GAGAAACCAGCCAAGTATGA | NM_204305 |
| CTGGTCCTCTGTGTATCCTA |
Immunoblotting Analysis
Liver tissue samples were lysed in a buffer containing 50 mmol Tris, pH 7.5, 120 mmol NaCl, 1 mmol EDTA, 15 mmol Na4P2O7, 20 mmol NaF, 1% Nonidet, 0.1% phenylmethyl sulfluoride, and protease inhibitors (0.08 μmol aprotinin, 0.02 μmol leupeptin, 0.04 μmol bestatin, and 15 μmol pepstatin). Protein content in each lysate was determined using the Bio-Rad RC DC protein assay kit (cat no. 500-0119; Bio-Rad, Hercules) according to the manufacturer's instructions. Proteins (10 μg) from tissue lysates were separated by SDS-PAGE and then transferred to nitrocellulose membranes, which were incubated overnight in 5% milk in PBS containing 0.1% Tween 20. The membranes were subsequently incubated with primary antibody overnight at 4°C. The following antibodies were used at 1:1000 dilution in this study: anti-STAT3 (cat no. bs-1141R; Beijing Biosynthesis Biotechnology Co., Ltd., Beijing, China), anti-IFIH1 (cat no. bs-18740R; Beijing Biosynthesis Biotechnology Co., Ltd., Beijing, China), anti-actin (cat no. bsm-33036M; Beijing Biosynthesis Biotechnology Co., Ltd., Beijing, China), and anti-GAPDH (cat no. NB300-221; Novus Biologicals Co., Ltd., CO). Secondary antibodies conjugated with horse radish peroxidase were used at 1:10,000 dilution. Proteins were detected by enhanced chemiluminescence and the Western blotting detection system (Amersham Biosciences, Beijing, China).
Statistical Analysis
The Student t test was used to analyze statistical significance of the difference in gene expression between the treatment and control groups, and P <0.05 was set as the criterion for statistical significance. All data are presented as mean ± SEM.
Results
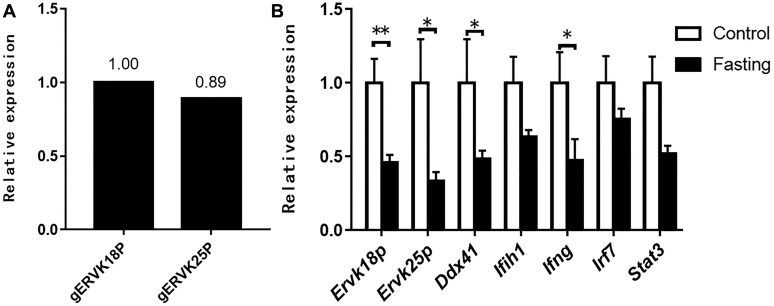
Fasting Suppressed the Expression of ERV and the Immune-Related Genes in the Goose Liver
The quantitative PCR primers for goose ERV genes (ERVK18P or LOC106029425, ERVK25P or LOC106046236) were designed based on the reference sequences in GenBank. Quantitative PCR analysis showed that the expression level of ERVK18P in the goose liver was similar to ERVK25P (Figure 1A). Compared with the control group (no fasting), the expression of ERVK18P and ERVK25P was significantly inhibited in the liver of the geese fasted for 24 h (P < 0.05 or 0.01) (Figure 1B). Accordingly, the mRNA expression of the immunity- or inflammation-related genes (DDX41, IFIH1, IFNG, IRF7, STAT3) was also inhibited, with the difference in mRNA expression of DDX41 and IFNG between the fasted and control geese reaching to a statistically significant level (P < 0.05) (Figure 1B). Immunoblotting analysis showed that the protein level of IFIH1 in the liver of the fasted geese appeared to be lower than that of the control geese (Supplementary Figure 1).
Figure 1.
Expression of ERV and immune-related genes in the goose liver was inhibited by fasting. The relative expression of ERV and immune-related genes was determined by quantitative PCR. (A) The expression of ERVK18P and ERVK25P in the liver of normal adult goose. (B) The expression of ERVK18P, ERVK25P, DDX41, IFIH1, IFNG, IRF7, and STAT3 in the livers of the fasted geese is presented as the fold change over the control (no fasting), n = 6. ∗,∗∗ denote P < 0.05, 0.01 vs. control, respectively. All of the data are shown as mean ± SEM. Abbreviation: ERV, endogenous retrovirus.
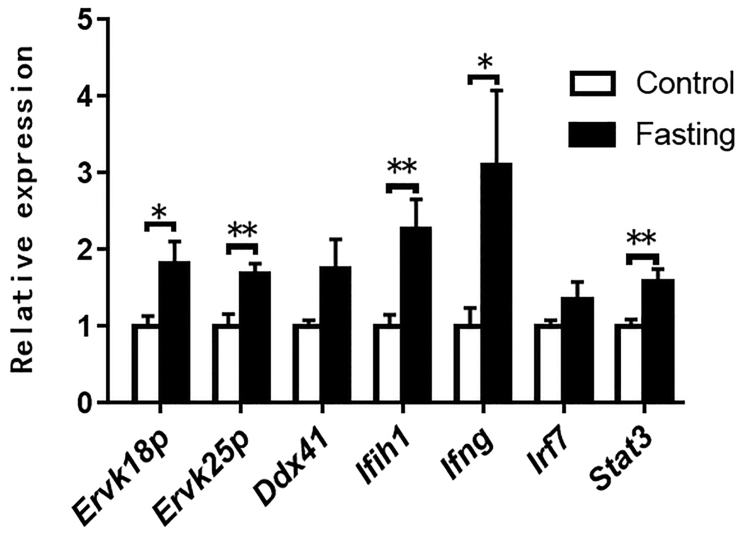
Fasting Induced the Expression of ERV and the Immune-Related Genes in the Chicken Liver
Quantitative PCR analysis showed that compared with the control group (no fasting), fasting induced mRNA expression of ERVK18P and ERVK25P in the chicken liver, and the induction reached a statistically significant level (P < 0.05 or 0.01) (Figure 2). Similarly, fasting also induced mRNA expression of these immunity- or inflammation-related genes, with the difference in mRNA expression of IFIH1, IFNG, IRF7, and STAT3 between the groups reaching to a statistically significant level (P < 0.05 or 0.01) (Figure 2). Immunoblotting analysis showed that the protein level of IFIH1 in the liver of the fasted geese appeared to be higher than that of the control geese (Supplementary Figure 1).
Figure 2.
Expression of ERV and the immunity- or inflammation-related genes in the chicken liver was induced by fasting. The relative expression of ERV and immune-related genes was determined by quantitative PCR. The expression levels of ERVK18P, ERVK25P, DDX41, IFIH1, IFNG, IRF7, and STAT3 in the livers of the fasted chickens is presented as the fold change over the control (no fasting), n = 6. ∗,∗∗ denote P < 0.05, 0.01 vs. control, respectively. All of the data are shown as mean ± SEM. Abbreviation: ERV, endogenous retrovirus.
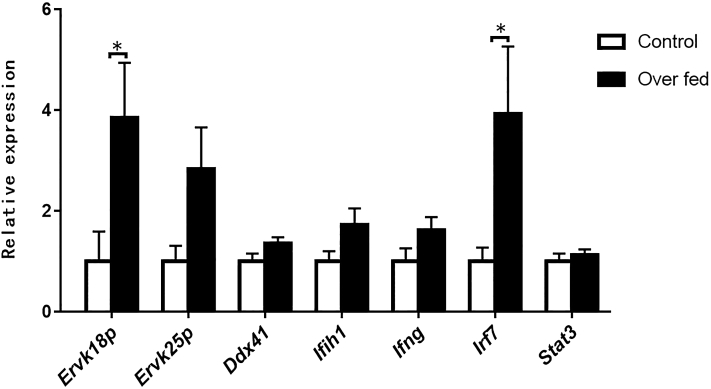
Overfeeding Induced the Expression of ERV and the Immune-Related Genes in the Goose Liver
Quantitative PCR analysis showed that compared with the control group (feeding routinely), the mRNA expression of ERVK18P and ERVK25 P in the liver of geese overfed for 24 d was increased, with the difference in mRNA expression of ERVK18P between the control and overfed geese reaching to a statistically significant level (P < 0.05) (Figure 3). Accordingly, the mRNA expression of the immunity- or inflammation-related genes in the livers of the overfed geese was also increased, with the difference in mRNA expression of IRF7 between the control and overfed geese reaching to a statistically significant level (P < 0.05) (Figure 3). Immunoblotting analysis showed that the protein level of IFIH1 in the liver of the overfed geese appeared to be higher than that of the control geese (Supplementary Figure 1).
Figure 3.
Expression of ERV and the immunity- or inflammation-related genes in the goose liver was induced by overfeeding. The relative expression of ERV and immune-related genes was determined by quantitative PCR. The expression levels of ERVK18P, ERVK25P, DDX41, IFIH1, IFNG, IRF7, and STAT3 in the livers of the overfed geese is presented as the fold change over the control (routine feeding), n = 6. ∗ denotes P < 0.05 vs. control. All of the data are shown as mean ± SEM. Abbreviation: ERV, endogenous retrovirus.
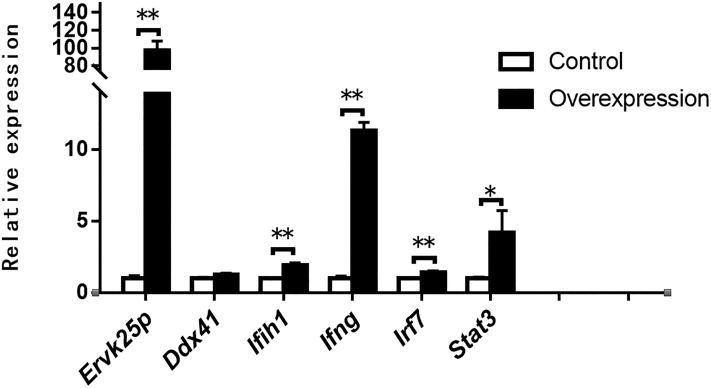
Endogenous Retrovirus Overexpression Induced the Expression of the Immune-Related Genes
After 32 h of transfection of goose primary hepatocytes with the empty vectors or the overexpression vectors containing the coding sequence of the Pol gene of goose ERVK25P, the mRNA expression of the Pol gene in the cells transfected with overexpression vectors was about 97 times more than that in the cells transfected with empty vectors (P < 0.01) (Figure 4). As expected, the expression of the immunity- or inflammation-related genes was induced by ERVK25P overexpression, and the induction of IFIH1, IFNG, IRF7, and STAT3 reached to a statistically significant level (P < 0.05 or 0.01) (Figure 4).
Figure 4.
The expression of the immunity- or inflammation-related genes was induced by ERV overexpression in goose primary hepatocytes. The relative expression of ERV and immune-related genes was determined by quantitative PCR. The expression of ERVK25P, DDX41, IFIH1, IFNG, IRF7, and STAT3 in the goose primary hepatocytes transfected with ERVK25P overexpression vectors is presented as the fold change over the control (the hepatocytes transfected with empty vectors), n = 4. ∗,∗∗ denote P < 0.05, 0.01 vs. control, respectively. All of the data are shown as mean ± SEM. Abbreviation: ERV, endogenous retrovirus.
Discussion
Nutrition and energy levels are important factors affecting animal growth, reproduction, and immunity. Starvation (or fasting) and feeding are 2 typical states affecting nutrition or energy levels, thus often being used as a research model to elucidate the regulation of nutrition or energy on the physiological functions of animals. For example, fasting or feeding can influence the expression of genes related to growth, reproduction, and immunity (Volkoff et al., 2016; Smati et al., 2020). It is however unclear whether fasting or feeding can activate ERV and affect the expression of the immunity- or inflammation-related genes via ERV. In addition, short-term overfeeding (3–4 wk) can lead to the formation of goose fatty liver (commonly known as foie gras), which is similar to nonalcoholic fatty liver disease (NAFLD) in humans and rodents (Nahum et al., 2010; Wang et al., 2019), but it is unknown whether ERV are activated by overfeeding and involved in the development of goose fatty liver. In this study, the expression of ERV and some immunity- or inflammation-related genes in the liver of geese or chickens was determined after altering animal nutrition or energy status by fasting or overfeeding, so that the relationship between nutrition (or energy) status and the expression of ERV and the immunity (or inflammation)-related genes could be clarified. Moreover, ERVK25P overexpression in goose primary hepatocytes was also performed to verify whether the expression of the immunity- or inflammation-related genes was affected by ERV. Indeed, the results provided strong evidence supporting the notion that nutrient or energy status could regulate the expression of the immunity- or inflammation-related genes via ERV in poultry. Interestingly, the effect of fasting on the mRNA expression of ERV and the immunity- or inflammation-related genes in the liver varied with poultry species, that is, the mRNA expression of the genes in the chicken liver was contrary to that in the goose liver. Furthermore, the mRNA expression of the genes in the liver of the fasted geese was contrary to that of the overfed geese. In addition, the data suggest that the immunity- or inflammation-related genes may mediate the regulation of ERV on the development of goose fatty liver (or foie gras).
Previous studies have demonstrated that ERV are generally silenced through DNA methylation in host cells, but ERV can be activated by MER48 (Walsh et al., 1998; Gibb et al., 2015). The induction of ERV is also shown in several diseases, including some metabolic diseases such as multiple sclerosis, wherein immunity- or inflammation-related genes are involved (Perron et al., 2000). Mechanistic studies indicate inflammation plays an important role in the pathogenesis of metabolic diseases usually caused by nutrition or energy oversupply (Eo et al., 2017). Moreover, it is found that fasting or overfeeding can change the DNA methylation level of peroxisome proliferators-activated receptor genes (Jacobsen et al., 2014; Hjort et al., 2017). Based on these findings, it is possible that nutritional change activates the mRNA expression of ERV via epigenetic regulation. On the other hand, nutritional change can induce the mRNA expression of some immunity- or inflammation-related genes (Smati et al., 2020). In line with these findings, this study showed that fasting or overfeeding affected the mRNA expression of ERV and the immunity- or inflammation-related genes and that the mRNA expression of ERV is closely associated with the mRNA expression of the immunity- or inflammation-related genes.
As ERV are abundant in animal genomes, the role of ERV in the regulation of nutrition or energy on immunity or inflammation could be underestimated. This study mainly addressed ERVK18 and ERVK25 mediating the effect of fasting or overfeeding on the mRNA expression of the immunity- or inflammation-related genes. The ERVK genes currently are only the relatively intact ERV annotated in both goose and chicken genomes. Besides these relatively intact ERV, the ‘slender’ ERV and the ‘solo LTR’ ERV could be activated by fasting or overfeeding and thus also contribute to the effect of fasting or overfeeding on the mRNA expression of the immunity- or inflammation-related genes. As ‘slender ERV’ and ‘solo LTR’ ERV usually are located near some host genes (Thompson et al., 2016), fasting or overfeeding may regulate the mRNA expression of the immunity- or inflammation-related genes through cis-effect of the activated ERV. From this viewpoint, the maintenance of immunity at the basic level may be partially due to a small portion of active ERV. Previous studies have shown that some ERV are regularly transcribed in both chicken embryonic fibroblasts (about 20% of ERV) and in vivo (Bolisetty et al., 2012).
The regulation of fasting or overfeeding on the mRNA expression of ERV is most likely through epigenetic modifications (e.g., DNA methylation) as previously mentioned. It has been reported that AZA, an inhibitor of DNA methyltransferase, can significantly induce the mRNA expression of ERV in the cell (Jaenisch et al., 1985; Déborah and Bestor, 2004). The mRNA expression of ERV was differentially regulated in the chicken liver vs. the goose liver by fasting, suggesting that there is a different degree of epigenetic modification to control ERV expression between chickens and geese. This inference is supported by the evidence showing that the mRNA expression of ERV is cell type dependent (Nogues et al., 2018). How fasting or overfeeding affects DNA methylation of ERV, however, remains to be clarified. Moreover, genetic difference between chickens and geese might contribute to the difference in the expression of ERV between the 2 species, such as different response of transcription factors to internal or external stimuli. Indeed, our previous results show that the mRNA expression of many genes in goose fatty liver vs. normal liver is contrary to that in humans with (or mouse) NAFLD vs. normal liver (Liu et al., 2016). In addition, environmental and other factors, such as age, feed, light, and other husbandry conditions, might also be responsible for the different expression of ERV between chickens and geese.
In this study, ERVK25P overexpression increased the expression of the immunity- or inflammation-related genes, which provides strong evidence supporting the notion that fasting or overfeeding regulates the mRNA expression of the immunity- or inflammation-related genes in the liver via ERV. There are 2 potential mechanisms by which the overexpression of the relatively intact ERV increased the mRNA expression of the immunity- or inflammation-related genes: 1) the double-stranded RNA formed within ERV transcripts or by hybridizing with antisense RNAs could activate the NFκB signaling pathway upon the double-stranded RNA binding to its receptors (e.g., TLR3) and in turn induce the expression of the immunity- or inflammation-related genes (Chiappinelli et al., 2015); 2) the relatively intact ERV can also express their encoded proteins or polypeptides, leading to activation of downstream signaling pathways and expression of the immunity- or inflammation-related genes. Although this study could not detect the protein expression of ERVK18P and ERVK25P owing to lack of proper antibodies, previous studies have demonstrated that some ERV, especially ERVK, can synthesize their proteins and induce the expression of the immunity- or inflammation-related genes in neuronal cells (Manghera et al., 2015). These proteins may be sensed by animal receptors such as RIG-1, protein kinase K, and inflammatory body molecule NLRP3 (Mitoma et al., 2013; Mu et al., 2016).
The association between nutrition (or energy) and immunity (or inflammation) has been shown in several nutrition- or energy-related disorders, especially in those obesity-associated diseases including diabetes and NAFLD. Obesity has been considered a chronic inflammation as many inflammation-related genes (e.g., proinflammatory cytokine tumor necrosis factor alpha, MCP1, and IL6) are induced in patients with obesity vs. healthy cohorts (Ferreira et al., 2016). These cytokines can lead to insulin resistance and thus deteriorate obesity-associated metabolic disorders (Li et al., 2019). In this study, data showed that ERV and the immunity- or inflammation-related genes were induced in goose fatty liver vs. normal liver. Liver is an extremely complex organ, which not only plays a central role in nutrient and energy conversions but also has the functions of detoxification and immune regulation. The liver contains a large number of immune cells, such as Kupffer cells (the resident macrophages), natural killer cells, and natural killer T cells. The liver also synthesizes complement components and a large number of other soluble pathogen recognition receptors (Keith et al., 2007). Therefore, the liver is currently regarded as an important immune organ and plays a role in innate immunity (or inflammation) (Xia et al., 2008; Triger, 2010). The ERV and their induced immunity- or inflammation-related genes may be essential to the physiological functions of the liver and serve the link between nutrition metabolism and immunity (or inflammation). In this study, although nutritional (or energy) change induced the mRNA expression of ERV and the immunity- or inflammation-related genes in goose fatty liver vs. normal liver, it is noteworthy that the ERV-induced immunity- or inflammation-related genes may feedback regulate nutrition metabolism (Volkman and Stetson, 2014; Cañadas et al., 2018). Therefore, this study provides some evidence supporting the notion that ERV participate in the development of goose fatty liver via the immunity- or inflammation-related genes.
In this study, we also determined the protein level of IFIH1 in the livers of the fasted geese vs. control geese, the fasted chickens vs. control chickens, and the overfed geese vs. control geese. Although the patterns of the IFIH1 protein level were similar to those of the IFIH1 mRNA level, the difference in the protein level between the treatment and the control groups was not as obvious as that in the mRNA level. The possible explanations include that the protein level of IFIH1 could be regulated post-transcriptionally.
In conclusion, nutrition or energy status affects the expression of some immunity- or inflammation-related genes via ERV, which provides a potential mechanism underlying the association between nutrition (or energy) and immunity (or inflammation). Nutrition or energy status may regulate the expression of ERV via DNA methylation. There is difference in this epigenetic regulation among poultry species, and the specific mechanism needs to be further studied. The immunity- or inflammation-related genes affected by nutrition or energy status may not only participate in innate immune response but also play a role in inflammation, animal growth and apoptosis, and feedback regulation of nutrition or energy metabolism. In addition, this study also revealed for the first time that ERV and their regulated immunity- or inflammation-related genes were involved in the development of goose fatty liver.
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (Beijing, China) to TG (31472086), Jiangsu Province Major Agricultural New Varieties Creation Project (PZCZ201731) to DG, Jiangsu Agriculture Science and Technology Innovation Fund (Jiangsu, China; grant no. CX(18)1004) to DG, Jiangsu Agriculture Industry Technology System to JW, Jiangsu Provincial Qing Lan Project to JW, and the Priority Academic Program Development of Jiangsu Higher Education Institutions to both TG and DG.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2020.11.057.
Disclosures
The authors declare no conflict of interest for this submitted manuscript.
Supplementary data
References
- Bolisetty M., Blomberg J., Benachenhou F., Sperber G., Beemon K. Unexpected diversity and expression of avian endogenous retroviruses. mBio. 2012;3 doi: 10.1128/mBio.00344-12. e00344-00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañadas I., Thummalapalli R., Kim J.W., Kitajima S., Jenkins R.W., Christensen C.L., Campisi M., Kuang Y., Zhang Y., Gjini E., Zhang G., Tian T., Sen D.R., Miao D., Imamura Y., Thai T., Piel B., Terai H., Aref A.R., Hagan T., Koyama S., Watanabe M., Baba H., Adeni A.E., Lydon C.A., Tamayo P., Wei Z., Herlyn M., Barbie T.U., Uppaluri R., Sholl L.M., Sicinska E., Sands J., Rodig S., Wong K.K., Paweletz C.P., Watanabe H., Barbie D.A. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat. Med. 2018;24:1143–1150. doi: 10.1038/s41591-018-0116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli K.B., Strissel P.L., Alexis D., Li H.L., Christine H., Benjamin A., Alexander H., Rote N.S., Cope L.M., Alexandra S., Vladimir M., Sadna B., Dennis J.S., Jedd D.W., Drew M.P., Matthias W.B., Cynthia A.Z., Taha M., Timothy A.C., Stephen B.B., Reiner S. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crichton J.H., Dunican D.S., Marie M., Meehan R.R., Adams I.R. Defending the genome from the enemy within: mechanisms of retrotransposon suppression in the mouse germline. Cell. Mol. Life Sci. 2014;71:1581–1605. doi: 10.1007/s00018-013-1468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déborah B.H., Bestor T.H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Denner J. Expression and function of endogenous retroviruses in the placenta. APMIS. 2016;124:31–43. doi: 10.1111/apm.12474. [DOI] [PubMed] [Google Scholar]
- Dolei A., Uleri E., Ibba G., Caocci M., Piu C., Serra C. The aliens inside human DNA: HERV-W/MSRV/syncytin-1 endogenous retroviruses and neurodegeneration. J. Infect. Dev. Countr. 2015;9:577–587. doi: 10.3855/jidc.6916. [DOI] [PubMed] [Google Scholar]
- Elaheh K., Farzaneh N., Mirshokraei P., Tabatabaeizadeh S.E., Dehghani H. Expression of endogenous retroviruses in pre-implantation stages of bovine embryo. Reprod. Domest. Anim. 2018;53:1405–1414. doi: 10.1111/rda.13269. [DOI] [PubMed] [Google Scholar]
- Eo H., Park J.E., Jeon Y.J., Lim Y. Ameliorative effect of ecklonia cava polyphenol extract on renal inflammation associated with aberrant energy metabolism and oxidative stress in high fat diet-induced obese mice. J. Agric. Food Chem. 2017;65:3811–3818. doi: 10.1021/acs.jafc.7b00357. [DOI] [PubMed] [Google Scholar]
- Ferreira P.S., Spolidorio L.C., Manthey J.A., Cesar T.B. Citrus flavanones prevent systemic inflammation and ameliorate oxidative stress in C57BL/6J mice fed high-fat diet. Food Funct. 2016;7:2675–2681. doi: 10.1039/c5fo01541c. [DOI] [PubMed] [Google Scholar]
- Geng T.Y., Hu W., Broadwater M.H., Snider J.M., Bielawski J., Russo S.B., Schwacke J.H., Ross J., Cowart L.A. Fatty acids differentially regulate insulin resistance through endoplasm reticulum stress-mediated induction of tribbles homologue 3: a potential link between dietary fat composition and the pathophysiological outcomes of obesity. Diabetologia. 2013;56:2078–2087. doi: 10.1007/s00125-013-2973-2. [DOI] [PubMed] [Google Scholar]
- Geng T.Y., Yang B., Li F.Y., Xia L.L., Wang Q., Zhao X., Gong D.Q. Identification of protective components that prevent the exacerbation of goose fatty liver: characterization, expression and regulation of adiponectin receptors. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2016;194:32–38. doi: 10.1016/j.cbpb.2016.01.006. [DOI] [PubMed] [Google Scholar]
- Geng T.Y., Zhao X., Xia L.L., Liu L., Li F.Y., Yang B., Wang Q., Montgomery S., Cui H.M., Gong D.Q. Supplementing dietary sugar promotes endoplasmic reticulum stress-independent insulin resistance and fatty liver in goose. Biochem. Biophys. Res. Commun. 2016;476:665–669. doi: 10.1016/j.bbrc.2016.05.149. [DOI] [PubMed] [Google Scholar]
- Gibb E.A., Warren R.L., Wilson G.W., Brown S.D., Robertson G.A., Morin G.B., Holt R.A. Activation of an endogenous retrovirus-associated long non-coding RNA in human adenocarcinoma. Genome Med. 2015;7:22. doi: 10.1186/s13073-015-0142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi S., Good R.A., Cianciolo G.J., Day N.K. A synthetic peptide homologous to retroviral envelope protein down-regulates TNF-α and IFN-γ mRNA expression. J. Leukoc. Biol. 1992;52:469–472. doi: 10.1002/jlb.52.4.469. [DOI] [PubMed] [Google Scholar]
- Hjort L., Jørgensen S.W., Gillberg L., Hall E., Brøns C., Frystyk J., Vaag A.A., Ling C. 36 h fasting of young men influences adipose tissue DNA methylation of LEP and ADIPOQ in a birth weight-dependent manner. Clin. Epigenetics. 2017;9:40. doi: 10.1186/s13148-017-0340-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn O., Hanke K., Bannert N. HERV-K (HML-2), the best preserved family of HERVs: endogenization, expression, and implications in health and disease. Front. Oncol. 2013;3:246. doi: 10.3389/fonc.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huda A., Polavarapu N., Jordan I.K., McDonald J.F. Endogenous retroviruses of the chicken genome. Biol. Direct. 2008;3:9. doi: 10.1186/1745-6150-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen S.C., Gillberg L., Bork-Jensen J., Ribel-Madsen R., Lara E., Calvanese V., Ling C., Fernandez A.F., Fraga M.F., Poulsen P., Brøns C., Vaag A. Young men with low birthweight exhibit decreased plasticity of genome-wide muscle DNA methylation by high-fat overfeeding. Diabetologia. 2014;57:1154–1158. doi: 10.1007/s00125-014-3198-8. [DOI] [PubMed] [Google Scholar]
- Jaenisch R., Schnieke A., Harbers K. Treatment of mice with 5-azacytidine efficiently activates silent retroviral genomes in different tissues. Proc. Natl. Acad. Sci. U. S. A. 1985;82:1451–1455. doi: 10.1073/pnas.82.5.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jern P., Coffin J.M. Effects of retroviruses on host genome function. Annu. Rev. Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- Keith E.G., Jones R.B., Meiklejohn D.A., Anwar N., Ndhlovu L.C., Chapman J.M., Erickson A.L., Agrawal A., Spotts G., Hecht F.M., Rakoff N.S., Lenz J., Ostrowski M.A., Nixon D.F. T cell responses to human endogenous retroviruses in HIV-1 infection. PLoS Pathog. 2007;3:e165. doi: 10.1371/journal.ppat.0030165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küry P., Nath A., Créange A., Dolei A., Marche P., Gold J., Giovannoni G., Hartung H.P., Perron H. Human endogenous retroviruses in neurological diseases. Trends Mol. Med. 2018;24:379–394. doi: 10.1016/j.molmed.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Y., Wang Y.D., Qi X.Y., Ran L., Hong T., Yang J., Yan B., Liao Z.Z., Liu J.H., Xiao X.H. Serum CCN3 levels are increased in type 2 diabetes mellitus and associated with obesity, insulin resistance and inflammation. Clin. Chim. Acta. 2019;494:52–57. doi: 10.1016/j.cca.2019.03.006. [DOI] [PubMed] [Google Scholar]
- Liu L., Zhao X., Wang Q., Sun X.X., Xia L.L., Wang Q.Q., Yang B., Zhang Y.H., Montgomery S., Meng H., Geng T.Y., Gong D.Q. Prosteatotic and protective components in a unique model of fatty liver: gut microbiota and suppressed complement system. Sci. Rep. 2016;6:1–14. doi: 10.1038/srep31763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager D.L., Stoye J.P. Mammalian endogenous retroviruses. Microbiol. Spectr. 2015;3:1079–1100. doi: 10.1128/microbiolspec.MDNA3-0009-2014. [DOI] [PubMed] [Google Scholar]
- Manghera M., Ferguson J., Douville R. ERVK polyprotein processing and reverse transcriptase expression in human cell line models of neurological disease. Viruses. 2015;7:320–332. doi: 10.3390/v7010320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melanie A.,S., Nair V.K. Prototype endogenous avian retroviruses of the genus Gallus. J. Gen. Virol. 2014;95:2060–2070. doi: 10.1099/vir.0.066852-0. [DOI] [PubMed] [Google Scholar]
- Mitoma H., Hanabuchi S., Kim T., Bao M.S., Zhang Z.Q., Sugimoto N., Liu Y.J. The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity. 2013;39:123–135. doi: 10.1016/j.immuni.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X., Ahmad S., Hur S. Endogenous retroelements and the host innate immune sensors. Adv. Immunol. 2016;132:47–69. doi: 10.1016/bs.ai.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau M.J., Manghera M., Douville R.N. Inside the envelope: endogenous retrovirus-K Env as a Biomarker and Therapeutic Target. Front. Microbiol. 2015;6:1244. doi: 10.3389/fmicb.2015.01244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahum M.S., Marco A., Daniel Z.V., Misael U. Current concepts in the pathogenesis of nonalcoholic fatty liver disease. Liver Int. 2010;27:423–433. doi: 10.1111/j.1478-3231.2007.01483.x. [DOI] [PubMed] [Google Scholar]
- Nogues J.C., Wang Z.M., Smith M., Sotomayor E., Chiappinelli K. The effect of TET mutations on DNA methyltransferase cytotoxicity and ERV induction in B and T cell malignancies. Blood. 2018;132:5157. [Google Scholar]
- Osman R.H., Liu L., Xia L.L., Zhao X., Wang Q.Q., Sun X.X., Zhang Y.H., Yang B., Zheng Y., Gong D.Q., Geng T.Y. Fads1 and 2 are promoted to meet instant need for long-chain polyunsaturated fatty acids in goose fatty liver. Mol. Cell. Biochem. 2016;418:103–117. doi: 10.1007/s11010-016-2737-7. [DOI] [PubMed] [Google Scholar]
- Perron H., Perin J.P., Rieger F., Alliel P.M. Particle-associated retroviral RNA and tandem RGH/HERV-W copies on human chromosome 7q: possible components of achain-reaction'triggered by infectious agents in multiple sclerosis? J. Neurovirol. 2000;6:67–75. [PubMed] [Google Scholar]
- Smati S., Regnier M., Fougeray T., Polizzi A., Fougerat A., Lasserre F., Lukowicz C., Tramunt B., Guillaume M., Burnol A.F., Postic C., Wahli W., Montagner A., Gourdy P., Guillou H. Regulation of hepatokine gene expression in response to fasting and feeding: influence of PPAR-α and insulin-dependent signalling in hepatocytes. Diabetes Metab. 2020;46:129–136. doi: 10.1016/j.diabet.2019.05.005. [DOI] [PubMed] [Google Scholar]
- Taruscio D., Mantovani A. Factors regulating endogenous retroviral sequences in human and mouse. Cytogenet. Genome Res. 2004;105:351–362. doi: 10.1159/000078208. [DOI] [PubMed] [Google Scholar]
- Thompson P., Macfarlan T., Lorincz M. Long terminal repeats: from Parasitic elements to Building blocks of the transcriptional regulatory Repertoire. Mol. Cell. 2016;62:766–776. doi: 10.1016/j.molcel.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolosa J.M., Schjenken J.E., Clifton V.L., Vargas A., Barbeau B., Lowry P., Maiti K., Smith R. The endogenous retroviral envelope protein syncytin-1 inhibits LPS/PHA-stimulated cytokine responses in human blood and is sorted into placental exosomes. Placenta. 2012;33:933–941. doi: 10.1016/j.placenta.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Triger D.R. The liver as an immunological organ. Gastroenterology. 2010;43:54–62. [PubMed] [Google Scholar]
- Volkman H.E., Stetson D.B. The enemy within: endogenous retroelements and autoimmune disease. Nat. Immunol. 2014;15:415. doi: 10.1038/ni.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkoff H., Sabioni R.E., Cyrino J.E.P. Appetite regulating factors in dourado, Salminus brasiliensis: cDNA cloning and effects of fasting and feeding on gene expression. Gen. Comp. Endocrin. 2016;237:34–42. doi: 10.1016/j.ygcen.2016.07.022. [DOI] [PubMed] [Google Scholar]
- Walsh C.P., Chaillet J.R., Bestor T.H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Wang G.S., Jin L., Li Y., Tang Q.Z., Hu S.L., Xu H.Y., Clare A.G., Li M.Z., Wang J. Transcriptomic analysis between Normal and high-intake feeding geese provides insight into adipose deposition and susceptibility to fatty liver in migratory birds. BMC Genomics. 2019;20:372. doi: 10.1186/s12864-019-5765-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Guo Z.H., Xu X.F., Yi H., Wang Q.X., Cao X.T. Hepatic microenvironment programs hematopoietic progenitor differentiation into regulatory dendritic cells, maintaining liver tolerance. Blood. 2008;112:3175–3185. doi: 10.1182/blood-2008-05-159921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.P., Meng F.F., Dong X., Zhao P., Cui Z.Z. Detection and Identification of an endogenous Subgroup E avian Leukosis Virus in a chicken Breeder embryo. Chin. J. Anim. Vet. Sci. 2014;45:1317–1323. [Google Scholar]
- Zhang Y., Maksakova I.A., Gagnier L., van de Lagemaat L.N., Mager D.L. Genome-wide assessments reveal extremely high levels of polymorphism of two active families of mouse endogenous retroviral elements. PLoS Genet. 2008;4:e1000007. doi: 10.1371/journal.pgen.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.