Abstract
Pancreatic ductal adenocarcinoma (PDAC) has a poor 5-year survival rate and lacks effective therapeutics. Therefore, it is of paramount importance to identify new targets. Using multi-plex data from patient tissue, three-dimensional co-culturing in vitro assays, and orthotopic murine models, we identified Netrin G1 (NetG1) as a promoter of PDAC tumorigenesis. We found that NetG1+ cancer-associated fibroblasts (CAFs) support PDAC survival, through a NetG1 mediated effect on glutamate/glutamine metabolism. Also, NetG1+ CAFs are intrinsically immunosuppressive and inhibit NK cell mediated killing of tumor cells. These pro-tumor functions are controlled by a signaling circuit downstream to NetG1, which is comprised of AKT/4E-BP1, p38/FRA1, vesicular glutamate transporter 1, and glutamine synthetase. Finally, blocking NetG1 with a neutralizing antibody stunts in vivo tumorigenesis, suggesting NetG1 as potential target in PDAC.
Keywords: Cancer associated fibroblasts, Netrin G1, NTNG1, pancreatic cancer, PDAC, tumor microenvironment, extracellular matrix, glutamate, glutamine, natural killer (NK) cells, metabolism, immunosuppression, AKT, p38, glutamine synthetase, vesicular glutamate transporter, 4E-BP1, FRA1
Introduction
Pancreatic cancer is projected to become the 2nd leading cause of cancer related deaths by 2030 [1], due to its abysmal 5 year survival rate [2]. The most common form of pancreatic cancer is pancreatic ductal adenocarcinoma (PDAC), to which treatments are often refractory, and where diagnosis is often made at advanced stages of the disease [3], making few patients eligible for surgical interventions [4]. Therefore, there is an urgent need to develop new diagnostic and therapeutic approaches.
PDAC has a unique microenvironment that consists of a fibrous expansion known as desmoplasia, characterized by the deposition of an abundant extracellular matrix (ECM) by cancer associated fibroblasts (CAFs) [5]. Studies have demonstrated that CAFs limit the efficacy of chemotherapies [6–8], promote PDAC progression [9, 10], and correlate with poor prognosis [11]. While there is a clear role for CAFs in facilitating PDAC progression, there is also evidence that CAFs can be tumor restrictive, as complete ablation of fibroblasts from the tumor microenvironment accelerated PDAC progression [12, 13] and provided no added patient benefit [14]. Thus, the role of CAFs in PDAC development and progression is incompletely understood.
An important consequence of desmoplasia is the generation of substantial interstitial pressure that contributes to the collapse of blood vessels [15], resulting in a nutrient deprived [16] and hypoxic microenvironment [17]. Further, Kras driven cancers like PDAC, where ~90% of tumors have mutant Kras, have been suggested to become “addicted” to exogenous sources of glutamine as a main carbon supply [18]. As such, PDAC cells make use of metabolic pathways to promote tumor growth and survival [18–20]. By taking advantage of CAF driven metabolic support [21], CAFs supply PDAC cells with key nutrients [22]. Therefore, further exploration of these mechanisms, aiming to disrupt tumor-stromal crosstalk, is warranted.
In addition to the metabolic roles, CAFs are also at the center of the immunosuppressive PDAC microenvironment [23–26]. CAFs exert immunosuppressive effects through direct modulation of immune cell function via cytokine secretion [27], exclusion of anti-tumor immune cells from the tumor [28], and/or recruitment of immunosuppressive immune cells to the tumor [29]. Intriguingly, recent studies have reported a functional link between metabolism and immune cell function, which has suggested that anti-tumor immune cells rely on a constant supply of nutrients to perform their functions [30, 31]. In the cancer context, key metabolites—glucose, arginine, glutamine, and tryptophan—are depleted, with a concomitant increase of immunosuppressive waste products, such as lactate, inhibiting anti-tumor immune cell function [30, 32, 33]. In PDAC, the immunosuppressive environment created by CAFs, combined with the nutrient poor milieu, build a hostile environment that inhibits anti-tumor immune cell function. Thus, means to simultaneously revert these linked features of the PDAC microenvironment would be therapeutically beneficial.
In this study we sought to clarify the roles of CAFs in PDAC tumorigenesis. We uncovered up-regulation of the glutamatergic pre-synaptic protein, Netrin G1 (NetG1) [34, 35], in CAFs. Of note, we also found expression of Netrin G1 Ligand (NGL-1), the sole known post-synaptic binding partner of NetG1 [36], in PDAC cells. In fact, we saw that NetG1 was particularly overexpressed in human PDAC tissue compared to samples from normal human pancreas and that fibroblastic NetG1 expression correlated inversely with patient survival. We demonstrated that NetG1 in CAFs and NGL-1 in tumor cells enhanced tumorigenesis, as ablation of either protein reduced tumor burden in mice. Functionally, NetG1 was responsible for glutamate, glutamine, and cytokine release by CAFs which allowed PDAC cells to survive in low nutrient conditions and reduced death induced by NK cells. Mechanistically, NetG1 controlled a signaling circuit comprised of glutamine synthetase and vesicular glutamate transporter 1, p38/FRA-1 and AKT/4E-BP1, and systematic inhibition of each component of the pathway revealed their roles in the pro-tumor metabolic and immunosuppressive functions of CAFs. Finally, the therapeutic applicability of targeting NetG1 in vivo was demonstrated, as an anti NetG1 neutralizing monoclonal antibody inhibited tumorigenesis in a murine model of PDAC. Thus, we demonstrate the importance of considering the stroma as a viable therapeutic option in PDAC.
Results
NetG1 is upregulated in CAFs compared to patient matched tumor adjacent fibroblasts
To gain insight into the mechanism by which naive fibroblasts are activated into CAFs, we performed a transcriptomic analysis comparing two sets of patient matched tumor adjacent fibroblasts (TAs) and CAFs (Fig 1A, Sup Table 1) that were cultured in our cell-derived ECM based culturing system that assures maintenance of in vivo-like fibroblastic phenotypes [37, 38]. We identified the most differentially regulated genes between patient matched TAs and CAFs (117 genes, p<0.01, log fold change; Sup Table 1). To characterize the underlying molecular pathways and biological processes, we performed Gene Set Enrichment Analysis (GSEA) and found significant enrichment (Benjamini-Hochberg False Discovery Rate < 0.25) of several pathways previously found to be associated with the transition from normal fibroblasts to CAFs, including upregulation of integrin signaling, focal adhesions, actin cytoskeleton, and ECM (Fig 1B-C, Sup Table 1). Interestingly, results also revealed an upregulation of genes involved with axonal guidance and neurite outgrowth in CAFs compared to TAs. The second most upregulated gene overall and top hit among the neuronal genes was NTNG1, encoding NetG1 [34], a synapse stabilizing protein known for its involvement in axonal communication. We also noticed in CAFs a negative enrichment of pathways associated with antigen presentation (KEGG Systemic Lupus Erythematosus; FDR < 0.25, Normalized Enrichment Score −2.66) and interferon signaling (Reactome Interferon Signaling; FDR < 0.25; NES = −2.66) (Fig 1D, Sup Table 1). Overall, this transcriptomic data confirmed that our ECM cultured CAFs indeed upregulate known pathways previously linked to fibroblast activation in cancer, as well as an association with neuronal gene expression.
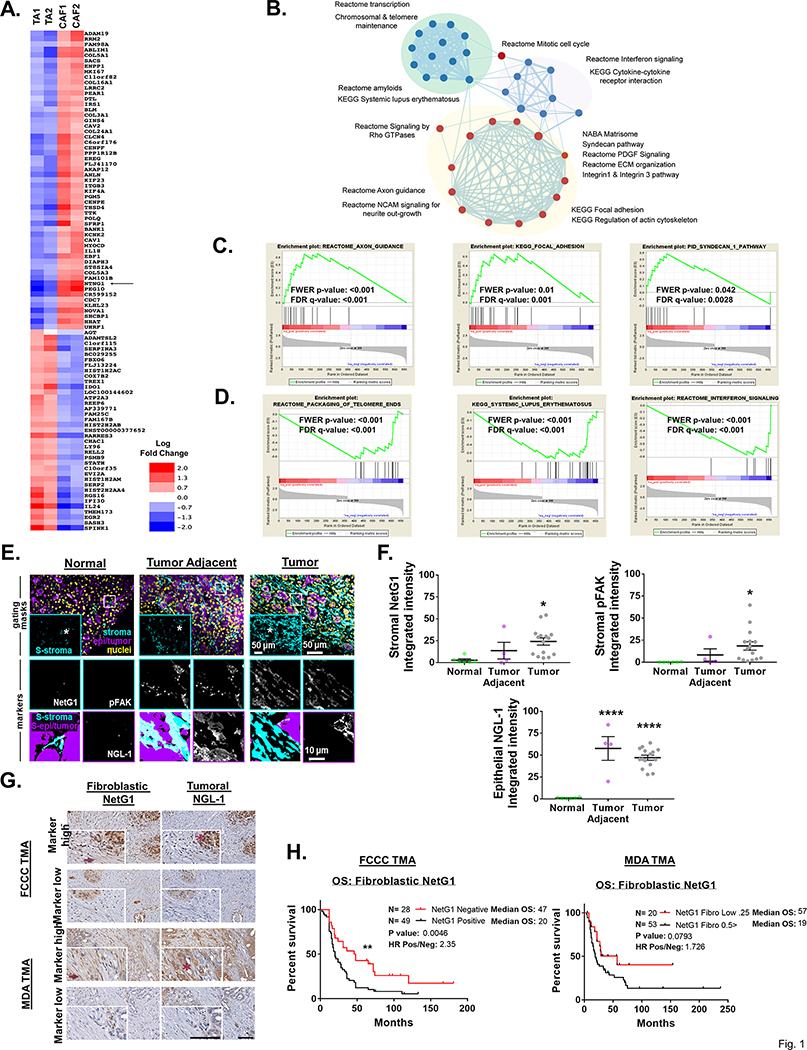
Figure 1. High NetG1 expression in CAFs inversely correlates with patient survival.
A. Heatmap representing increased (red) or decreased (blue) gene expression levels (log fold change) between patient derived tumor adjacent fibroblasts (TA) and CAFs that were maintained in 3D. Position of NetG1 (NTNG1) in the heatmap is highlighted with a black arrow. Top 117 differentially expressed genes were chosen by p-value (<0.01). B. Pathways that are significantly enriched between these two cell types are represented as network map. Each node (circle) represent a pathway while the edges (lines) connecting nodes show shared genes between pathways with thickness of edge corresponds to degree of sharing. Color of node indicate positive (red) or negative (blue) enrichment in CAF cells. Enrichment of select pathways are shown as GSEA enrichment plots with positive (C) and negative (D) enrichment in CAFs. E. Representative images of simultaneous multi-channel immunofluorescent (SMI) approach, performed on formalin fixed paraffin embedded tissue samples corresponding either to normal pancreatic tissue (normal human pancreas), normal tissue adjacent to PDAC tumor (tumor adjacent), or PDAC tumor tissue (tumor) from patient/donor surgical samples. Upper row panels show merged and pseudo-colored images corresponding to the three locations used as “gating masks.” These masks correspond to pan-cytokeratins positive areas in purple demarking epithelial/tumoral compartment, vimentin regions in cyan and DRAQ5 for nuclei in yellow. The SMIA-CUKIE algorithm (described in Material and Methods) was instructed to render an intersection “mask” image corresponding to vimentin positive epithelial/tumoral negative areas (S-stroma in upper panels inserts), whereas epi/tumor positive masks omitted all vimentin positive pixels. The two above-mentioned masks were used as areas to generate the images shown in the medium and lower rows, which are magnified images corresponding to the regions of interest highlighted in upper row images within white squares and asterisks. Medium row show “markers” in stromal areas corresponding to NetG1 and pFAK, while lower row shows NGL-1 at tumor areas. Lower left corner panels indicate the algorithm generated area “masks” used (S-epi/tumor; S/stroma). Representative scale bars are provided for each magnification F. Graphs depict integrated intensities of stromal NetG1, stromal pFAK, or epithelial NGL-1 staining in normal (N= 6), tumor adjacent (N= 4), or tumor (N= 15) pancreatic tissue. *Compared to normal tissue. One-Way ANOVA, Dunnett’s multiple comparison test. *p<0.05, ****p<0.0001. G. Representative images of patient-matched pancreatic cancer tissue evaluated in TMAs obtained from FCCC and MDA. Top row panels show high stromal NG1 and tumoral NGL-1 expression in patients with poor survival, while bottom row images illustrate tissue examples with low levels of mentioned markers, corresponding with patients with extended survival. Inserts in each image show magnified regions with examples of stromal cells pointed by dark red arrows; tumor areas are highlighted by dark red asterisks. Scale bars represent 100 μm. H. Kaplan-Meier plots depicting overall survival (OS) of PDAC patients from two independent TMAs (LEFT: FCCC; N= 80) and (RIGHT: MDA; N=143), stratified by immunohistological scores of fibroblastic NetG1. Log rank test was used to determine statistical significance. **p<0.01.
NetG1 in CAFs, and its binding partner NGL-1 in tumor cells, are overexpressed in PDAC
NetG1 is present on pre-synaptic neurons, and it is known to stabilize excitatory glutamatergic synapses by interacting with its only known receptor on post-synaptic cells, Netrin G1 Ligand (NGL-1) [35, 36]. To assess the clinical relevance of NetG1, we performed Simultaneous Multichannel Immunofluorescence (SMI) [38] on 6 normal, 4 tumor adjacent, and 15 tumor pancreatic tissue samples to discern NetG1 and NGL-1 expression patterns in distinct subsets of cells. NetG1 was upregulated in the stromal compartment (pan-cytokeratin-, vimentin+ areas) in tumor tissue compared to normal pancreatic tissue from individuals without cancer (Fig 1E-F). Moreover, levels of tyrosine 397 phosphorylated focal adhesion kinase (pFAK) followed a similar pattern of increased expression in tumor tissue compared to normal tissue, in line with our established pro-tumor CAF signature [38] (Fig 1E-F). A strong correlation between stromal NetG1 expression and stromal pFAK expression was observed across all tissues (R2 = 0.8046, p<0.0001) (S1A). Further, NGL-1 was upregulated in the epithelial compartment (pan-cytokeratin+ areas) of both tumor adjacent and tumor tissue (Fig 1E-F), with a weak correlation between stromal NetG1 and epithelial NGL-1 expressions across all tissues (R2 = 0.2359, p<0.0001) (S1A). Comparing 4 TAs and 5 CAFs isolated from patient tissue, we confirmed that CAFs upregulated NetG1 protein expression (~2.5 fold increase) (S1B). Additionally, we observed the expression of NGL-1 in 5 patient derived cell lines from PDX models (S1C), as well as upregulated expression in tumor cell lines derived from an isogenic cell line model, hTERT-HPNE E6/E7 (referred to as E6/E7) and hTERT-HPNE E6/E7/K-RasG12D (referred to as PDACc), compared to their immortalized epithelial cell counterpart (hTERT-HPNE; referred to as hTERT) [39] (S1D). Collectively, NetG1 and its binding partner NGL-1 were upregulated in cancer compared to normal tissue in the pancreas, and segregated into distinct cellular compartments (stromal vs. epithelial).
Fibroblastic NetG1 expression correlates with worse overall survival in PDAC patients
Using the cBioportal database [40, 41], we queried both NetG1 (NTNG1) and NGL-1 (LRRC4C) mRNA expression across 31 cancer types and found that PDAC was among the highest expressors of both genes (S2A, arrows). Additionally, we stratified NTNG1 and LRRC4C by clinical subtype according to three independent datasets [42, 43]; (TCGA, [44]) (S2B-E). We found that in two out of three datasets there was a trend towards elevated NTNG1 and LRRC4C in the basal classification, which presents with the worst prognosis of all PDAC subtypes [43].
Moreover, we used the datasets from TCGA and Puleo et al. [42, 44] and stratified patients into 4 groups based on NTNG1 and LRRC4C expression: LRRC4C high/NTNG1 high (UP-UP), (LRRC4C high/NTNG1 low (UP-DN), LRRC4C low/NTNG1 high (DN-UP), and LRRC4C low/NTNG1 low (DN-DN) and examined if these groups correlated with overall survival (OS) (S2F). Although we did not see a significant association between expression and OS, we found a marginal association between relapse free survival (RFS) and co-expression of LRRC4C and NTNG1 (hazard ratio (HR) of 9.45 (1.27–70.27; P = 0.0074; FDR > 0.5) (S2G). These inconclusive results are likely due to the intermingling of stromal and epithelial RNA that is inherent in cancers with a high degree of stromal expansion, such as PDAC [44], highlighting the importance of carefully dissecting the contributions of the stromal and epithelial compartments of the tumor microenvironment.
Thus, to further address the clinical significance of NetG1 and NGL-1, we performed immunohistochemical staining of NetG1 and NGL-1 on two independent tissue microarrays (TMA; Fig 1G, Sup Table 2). Protein expression of fibroblastic NetG1 and tumoral NGL-1 were scored blindly by a pathologist (0–4 scale; Sup Table 3). Strikingly, fibroblastic NetG1 expression correlated inversely with patient overall survival (p<0.01; median OS NetG1+ vs NetG1-: 20 vs 47 months; hazard ratio: 2.35) in the Fox Chase Cancer Center (FCCC) TMA, while there was a similar trend in the MD Anderson (MDA) TMA (p=0.0793; median OS NetG1High vs NetG1Low: 19 vs 57 months; hazard ratio: 1.73) (Fig 1H). Of note, tumoral NGL-1 expression was not predictive of survival (S3). This was likely due to the fact that nearly every patient stained positive for tumoral expression of NGL-1 (72/77 FCCC; 129/135 MDA). Overall, fibroblastic NetG1 expression resulted in worse overall survival for PDAC patients, suggesting that NetG1 could be a useful prognostic marker in PDAC.
RNAseq analysis reveals that ablation of NetG1 in CAFs results in a normalized stromal gene expression signature
CAFs are known to function in an ECM dependent manner and regulate tumorigenesis in a number of ways [5]. Hence, we relied on our well-established 3D cell-derived matrix culturing system, referred to as “3D” for simplicity, which simulates the physiological as well as pathologic tumor microenvironment [37, 38, 45], to dissect the functions of NetG1 in more detail in vitro. To this end, we first generated NetG1 knockout (KO) CAFs through CRISPR/Cas9 (S4A), from one of our patient derived CAF lines that displayed high levels of NetG1 mRNA and protein expression compared to TAs (S4B, S1B). To gain insight into global changes in gene expression, we performed RNAseq analysis comparing control (CON) CAFs and NetG1 KO CAFs (S4C, Sup Table 4). Pathway and gene enrichment analysis identified downregulation of genes associated with many critical pathways, known as being upregulated in CAFs, in our microarray analysis (Fig 1), including: axon guidance, focal adhesion, ECM and TGF-β signaling, and inflammatory responses (S4D-E, Sup Table 4). Thus, taken together, these results suggest a reversion of gene expression profiles linked with known pro-tumor CAF functions upon loss of NetG1 expression.
Ablation of NetG1 does not influence myofibroblastic features of CAFs, but reverses pro-tumorigenic CAF markers
Next, we followed NetG1 expression during ECM production in vitro, as this is a major function of CAFs in the in vivo tumor microenvironment [5]. Importantly, NetG1 protein expression in CAFs was significantly elevated in 3D compared to 2D culturing conditions (S5A). We then assessed the desmoplastic phenotype of CON or NetG1 KO CAFs during ECM production, according to our previous study [38]. Interestingly, NetG1 KO CAFs did not have altered canonical myofibroblastic features, as determined by ECM fiber (fibronectin) alignment and levels/localization of alpha smooth muscle actin (α-SMA) (S5B-C). However, NetG1 KO CAFs displayed reduced levels of active α5β1-integrin and pFAK, indicative of a CAF phenotype that was associated with delayed recurrence following surgery in PDAC patients (S5D-E) [38], consistent with the correlation seen in human tissue samples (S1A). Moreover, we segregated six patient tumor tissues into 2 groups, stromal NetG1 high (N=3) and low (N=3), and questioned if the pan-CAF marker podoplanin (PDPN) [46–48] and the myofibroblastic marker α-SMA correlated with NetG1 expression in vivo. Interestingly, PDPN had a significantly higher stromal IHC score in NetG1 high tissue compared to NetG1 low tissue, while α-SMA levels remained high in both tissues (S5F-G), similarly to the in vitro data (S5B-E). Thus, deletion of NetG1 does not alter canonical myofibroblastic features of CAFs, but attenuates the expression of known pro-tumor CAF markers. Therefore, we hypothesized that deletion of NetG1 would revert pro-tumorigenic functions of CAFs and potentially stunt tumorigenesis in vivo.
Ablation of NetG1 in CAFs and NGL-1 in PDAC cells significantly stunts tumorigenesis in orthotopic murine models of PDAC
To test the impact of NetG1 and NGL-1 in pancreatic cancer in vivo, we performed multiple murine models of PDAC, in which we orthotopically injected CAFs and PDAC cells, and observed tumor progression and burden. In the first model, we co-injected human CON or NetG1 KO CAFs with RFP+ CON or NGL-1 KO PDACc cells (NGL-1 knockout confirmed; S6A), at a CAF:PDAC ratio of 3:1, into the pancreas of severely compromised immunodeficient (SCID) mice and followed tumorigenesis over 1 month. Animals co-injected with CON CAFs and CON PDACc cells had a substantially worse tumor burden, with more obvious macroscopic RFP+ coverage of the pancreas (Fig 2A), as well as greater tumor weight (~2–3 fold) (Fig 2B), pancreas weight (~2–3 fold) (Fig 2C), and tumor area (Fig 2D), than all other conditions. Importantly, mice co-injected with NetG1 KO CAFs and CON or NGL-1 KO PDACc cells developed tumors to a similar extent as mice injected with CON or NGL-1 KO tumor cells alone, suggesting that CAFs lacking NetG1 do not confer any benefit for tumorigenesis. As a control, mice were injected with the CON or NetG1 KO CAF lines alone, and those animals did not develop tumors. Histologically, tumors that developed from CON CAF and CON PDACc co-injections developed significantly more sarcomatous PDAC morphology (~3.5–5 fold more) compared to all other groups (Fig 2E, S6B), which has been associated with an extremely poor patient prognosis [49–51]. We also observed that mice injected with NGL-1 KO PDACc cells alone displayed a tendency for less tumor burden than CON PDACc cells, thus we proceeded to investigate this in more detail.
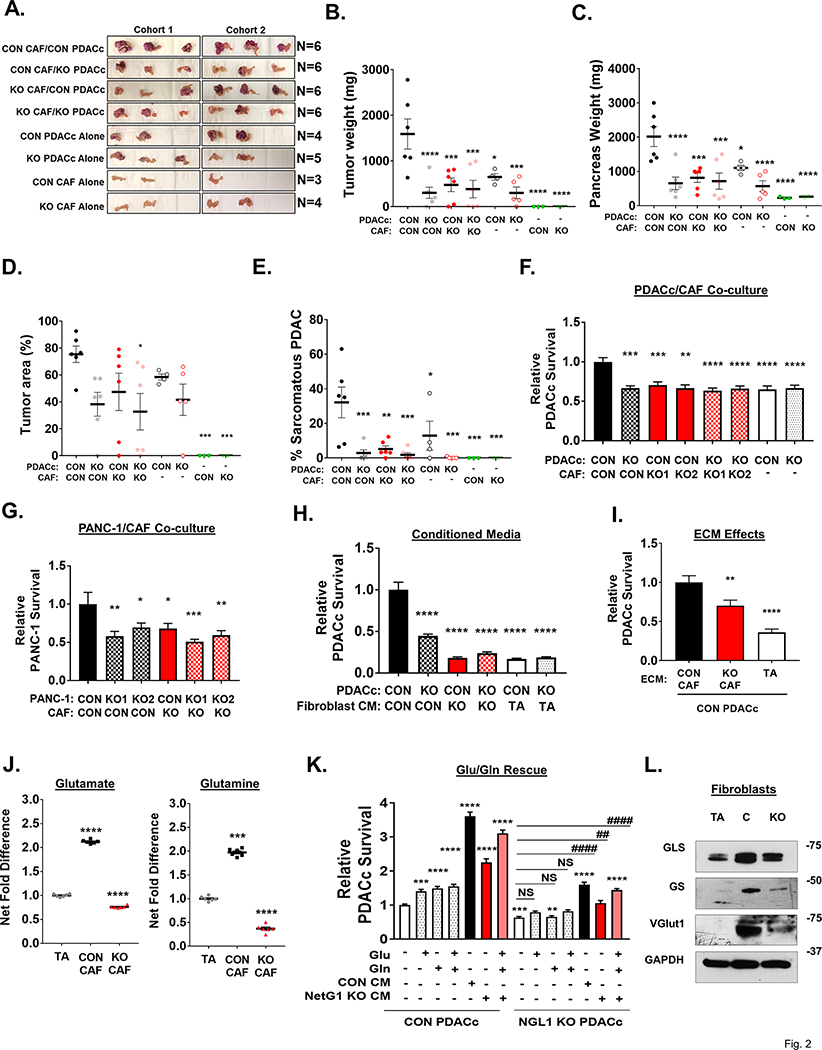
Figure 2. Ablation of NetG1 in CAFs stunts tumorigenesis in vivo and regulates Glu/Gln driven support of nutrient deprived PDAC cells.
7.5×105 CON or NetG1 KO CAFs were orthotopically injected into the pancreas of SCID mice with RFP+ CON or NGL-1 KO PDACc cells (2.5×105), at a 3:1 ratio. CAFs or PDACc cells injected alone served as controls. Mice were sacrificed 1 month after injection and tumorigenesis was assessed. A. Images of the isolated pancreata. B. Graph depicting relative tumor weight from each experimental group. C. Quantification of pancreas weights from each experimental group. D. Graph displaying the quantification of the % area of the pancreas that was classified as a tumor. E. Graph showing quantification of the % area of the tumors that display a sarcomatous phenotype. F. RFP+ CON or NGL-1 KO PDACc (2×104) were co-cultured in 3D with GFP+ CON or NetG1 KO CAFs (2×104) (2 clones, KO1 and KO2) or alone in the absence of serum and Gln for 4 days followed by cell survival assessments. * compared to CON PDACc/CON CAF. G. Same assay as in (F), but with RFP+ CON or NGL-1 KO PANC-1 cells. * compared to CON PANC-1/CON CAF. H. RFP+ CON or NGL-1 KO PDACc (2×104) were co-cultured in 3D with CM from CON, NetG1 KO CAFs, or tumor adjacent fibroblasts (TA) in SF/Gln free media, and PDACc survival was measured after 4 days. * compared to CON PDACc/CON CAF. I. RFP+ CON PDACc (2×104) were grown in 3D in the indicated fibroblast derived ECMs alone in the absence of serum and Gln for 4 days and PDAC cell survival was measured. * compared to CON PDACc/CON CAF. J. Relative glutamate and glutamine levels in the CM of TA, CON or NetG1 KO CAFs. N= 6 biological replicates, all groups were compared to TA condition. K. RFP+ CON or NGL-1 KO PDACc (2×104) were cultured alone in 3D under serum and Gln deprivation (−). Graph depicts relative PDACc survival after exposure to media alone, Glu, Gln, or CAF CM. Treatment groups consisted of Glu (150 μM) and Gln (25 μM) addbacks to determine if those amino acids alone could rescue PDAC cell survival in the absence of CAFs. CM from CON CAFs was used as a positive control for rescue. Note how amino acids alone partially rescue CON PDACc but not NGL-1 KO PDACc, which also benefit from CM media to a lesser extent than their CON counterparts. * compared to CON PDACc alone; # compared to KO PDACc alone. All Graphs: One-Way ANOVA, Dunnett’s multiple comparison test. *p<0.05, **p<0.01, ***p<0.001; ****p<0.0001. L. Representative western blots of glutaminase (GLS), glutamine synthetase (GS) and vesicular glutamate transporter 1 (VGlut1) in TA, CON CAFs (C), and NetG1 KO CAFs (KO). GAPDH was used as a loading control. N= 5.
We first explored NetG1 expression in the stroma of LSL-KrasG12D/+; Pdx-1-Cre (KC) mice, which readily develop precancerous lesions known as PanINs (Pancreatic Intraepithelial Neoplasia) and eventually full blown invasive PDAC [52]. Using SMI, we observed that as KC mice age and develop PanINs, stromal NetG1 expression was detected in pancreatic tissue as early as 12 and 16 weeks, while undetectable in non-tumor bearing mice (S7A). We generated two murine PDAC cell lines, isolated and cloned from the more aggressive LSL-KRASG12D;TP53−/−;PDX1-Cre (KPC) model [53], one with low NGL-1 expression that resulted in an average survival time of 7.5 weeks after orthotopic injection into C57BL/6 mice (KPC4B), and one with high expression of NGL-1 that resulted in an average survival time of 3.5 weeks after orthotopic injection (KPC3) (S7B). These data indicated that, similarly to human PDAC, NetG1/NGL-1 are present in PDAC in mice. Using CRISPR/Cas9, we generated two NGL-1 KO murine PDAC cell lines and one control line from the aggressive KPC3 cells (S7C) and injected them orthotopically into syngeneic C57BL/6 mice (CON, KO1, KO2). Mice injected with CON KPC3 cells (CON group) had a tumor incidence of 100% (7/7), while the groups injected with NGL-1 KO1 and KO2 KPC3 cells (NGL-1 KO group) had an incidence of 22.2% (2/9) and 40% (4/10) with KO tumors displaying significantly less tumor weight (S7D), pancreas weight (S7E) and % tumor area (S7F-G) compared with CON group. In addition, representative MRI images revealed the expansion of the pancreas in CON group over 3 weeks, while the pancreas from the NGL-1 KO group seemingly remained at normal size over that time span (S7H). Moreover, tumors from the KO group had a trend towards being more differentiated than those from the CON group. An average of 79% of the cells from CON tumors were classified as poorly differentiated, compared to 48% in KO tumors (S7I), indicative of less aggressive tumors overall. Consequently, NGL-1 KO tumors also proliferated slower, as measured by Ki67 staining, and had higher rates of apoptosis, as measured by TUNEL staining (S7J). These results were in line with less Ki67 expression in human NGL-1 KO PDACc compared to CON PDACc cultured in 3D in vitro (S7K).
In order to evaluate the impact of the immune system in this model, orthotopic injections of CON or NGL-1 KO KPC3 cells into SCID (lacks T and B cells) and Nod SCID Gamma (NSG; lacks T, B, and NK cells) mice were performed and compared to immunocompetent mice (B6). All animals injected with CON cells presented a greater tumor burden than those injected with NGL-1 KO cells, independently of the immune backgrounds (S7L). This suggests that tumorigenesis, in relation to tumoral NGL-1, is not significantly affected by the immune system. Overall, the results demonstrated that both NetG1 in CAFs, and NGL-1 in tumor cells, promote PDAC tumorigenesis in vivo.
NetG1 increases heterotypic cell-cell interactions between CAFs and PDAC
Because of NetG1’s known role in cell-cell adhesion in the brain, namely through its sole known receptor NGL-1, we tested the potential function of NetG1/NGL-1 in CAF-PDAC cell-cell interactions. Ablation of NetG1 in CAFs reduced cell engagement with control (CON) or NGL-1 KO PDACc by 72% and 79%, respectively (identified by the yellow regions resulted from the interaction between GFP+ CAFs and RFP+ PDACc) (S8A-C). Accordingly, CON or NGL-1 KO PDACc had increased motility (81% and 74%, respectively) when co-cultured with NetG1 KO CAFs in 3D (S8D-E). Unexpectedly, deletion of NGL-1 from PDACc was not sufficient to alter PDACc-CAF cell engagement or PDACc motility, signifying that there may be a redundant mechanism controlling PDAC-CAF engagement in the absence of NGL-1 in tumor cells and that NetG1 in CAFs is the driver of PDAC-CAF heterotypic interactions.
NetG1 in CAFs enhances direct material transfer to PDAC cells, and NGL-1 regulates macropinocytosis in PDAC cells
Because NetG1 in CAFs is needed for PDAC-CAF interactions, we questioned if there was a further functional consequence of the increased heterotypic cell-cell interactions observed during co-culture. Since PDAC cells have been suggested to use numerous extracellular macromolecules as a source of energy [16, 54], and receive nutrients from CAFs [55, 56], we hypothesized that CAFs may provide resources to PDAC cells in a NetG1 dependent manner. To test this, we followed the exchange of GFP from GFP+ CAFs and RFP from RFP+ PDACc. Indeed, after 36 hours of co-culture, GFP was detected intracellularly in PDACc (S9A), demonstrating that PDACc received CAF derived material. Knockout of NetG1 in CAFs, or NGL-1 in PDACc, resulted in a 49%−61% reduction in GFP transfer to PDACc, compared to CON PDACc/CON CAF group (S9B). Interestingly, when performing the same material transfer assay with conditioned media (CM) derived from GFP+ CAFs, instead of direct co-culture, only NGL-1 loss in PDAC cells resulted in a ~55–65% reduction in GFP transfer from either CON or NetG1 KO CM (S9C). This suggested that NGL-1 in PDAC cells could control uptake of material from an extracellular source and thus we sought to understand this uptake mechanism in PDAC cells.
KRAS driven cancers, such as PDAC, have been shown to rely on macropinocytosis to overcome nutrient deprivation [57] and therefore we decided to determine the intrinsic ability of PDACc and PANC-1 cells (NGL-1 knockout confirmed; S9D) to perform macropinocytosis in 3D. First, we performed a dose response curve, using the alamarBlue assay, to find the optimal nontoxic concentration to treat cells with the macropinocytosis inhibitor EIPA (5-(N-ethyl-N-isopropyl) amiloride), which was determined to be ~25μM for all cell lines (S9E). Next, we plated RFP+ CON or NGL-1 KO PDAC cells in 3D in the presence of DMSO or EIPA for 24 hours and tested their ability to uptake FITC-dextran [58]. Both CON PDACc and PANC-1 cells could effectively uptake FITC-dextran, while all EIPA inhibited cells displayed a ~50% decrease in uptake (S9F-G). Strikingly, NGL-1 KO PDACc or PANC-1 cells had a significant impairment in their ability to uptake FITC-dextran (~50% and ~30, respectively), similar to the levels of EIPA treated cells. We next tested whether macropinocytosis was the mechanism PDACc cells used to uptake GFP from CAF CM. Indeed, while CON PDACc cells could efficiently uptake extracellular GFP, NGL-1 KO PDACc had ~50% less uptake of GFP, at similar levels to EIPA treated PDACc cells (S9H).
Therefore, while NGL-1 was dispensable for CAF-PDAC engagement, both NGL-1 and NetG1 were necessary for PDACc to directly receive material from CAFs. Conversely, NGL-1 regulated the macropinocytic ability of PDAC cells to uptake extracellular material, independently of NetG1 expression in CAFs.
NetG1/NGL-1 axis protects PDAC cells from death under nutrient restriction
Since the PDAC microenvironment contains limited nutrients and CAFs can transfer material in a NetG1/NGL-1 dependent manner (S9), we hypothesized that CAFs serve as a critical nutrient conduit and could rescue PDAC cells from nutrient deprivation. To this end, we co-cultured GFP+ CAFs with RFP+ PDACc in 3D under serum free/glutamine (Gln) free conditions and measured the survival of RFP+ PDACc over a 4-day time span. We observed that disruption of NetG1/NGL-1, by deletion of either NetG1 in CAFs or NGL-1 in PDAC cells, resulted in 30–37% decrease in PDACc survival, compared to CON PDACc/CON CAF group, similar to PDACc cultured alone (Fig 2F). These results were replicated using the pancreatic cancer cell line PANC-1 (CON or NGL-1 KO), with a 40–59% reduction in PANC-1 survival compared to CON/CON group (Fig 2G). To determine if CAF secreted factors alone were important for tumor cell survival, PDACc were cultured in 3D with conditioned media (CM) obtained from CON and NetG1 KO CAFs. We found that NetG1 KO CAF CM was 76–82% less efficient at supporting PDACc survival when compared to CON CAF CM, similarly to the CM from TAs (81–83% of CON CM) (Fig 2H). To further assess the effect of NetG1 on the microenvironment generated by CAFs, we performed survival assays using ECMs produced by CON and NetG1 KO CAFs as substrates for cancer cells, and compared them to the ECM produced by TAs, which express less NetG1 than CAFs. Accordingly, the microenvironment generated by NetG1 KO CAFs was less supportive compared to the CON ECM, with a 30% decrease in PDACc survival (Fig 2I). The TA generated ECM was even less supportive, resulting in a decrease in PDACc survival of 65%. For experimental rigor, we also generated NetG1 KD CAFs, using CRISPRi, in two CAF lines (CAF and CAF2) and replicated the survival assay results (S10A-C). Collectively, these results suggest that both direct physical contact with CAFs, and the factors secreted by CAFs, including CM and ECM, support PDAC survival under nutritional deprivation, in a NetG1/NGL-1 dependent fashion.
Alterations in Glutamate/Glutamine generation in NetG1 KO CAFs reduce PDAC cell survival
In an effort to gain insight into key factors that could support PDAC survival under nutritional deprivation, we performed an amino acid screen comparing TAs, CON CAFs, and NetG1 KO CAFs, during ECM production (Sup Tables 5-7). The greatest net change in amino acid secretion between TAs and CAFs was found to be in glutamate (Glu) and Gln, as CAFs secreted 2-fold more of these amino acids (Fig 2J). These results are in line with recent studies that suggest that the catabolism of Glu plays a role in PDAC and other cancers [18, 19, 59, 60]. Strikingly, we found that NetG1 KO CAF CM contained 64% less Glu and 81% less Gln, compared to CON CAFs (Fig 2J), and this was comparable in CAF2 (S10D). To confirm that patient derived CAFs could produce Glu and Gln de novo, we performed an isotope tracing experiment, by providing CAFs with 13C6-glucose and 13C3-pyruvate, and followed labeled carbons of Glu and Gln in CAFs. Indeed, in both 3D and 2D, 70–80% of Glu and ~35% Gln were labeled (S10E), with all M+1–5 populations receiving a fraction of the label (S10F), indicating that CAFs in our system could produce these metabolites de novo.
In order to determine the relative contributions of Glu and Gln to PDAC survival conferred by CON CAFs CM, we performed a Glu/Gln rescue assay, where the amounts of Glu/Gln produced by CON CAFs (150 μM and 25 μM respectively) were added to the depleted media and compared to the rescue provided by CON CAFs CM. Interestingly, while Glu or Gln alone or in combination promoted survival by 40%, 49% or 55% compared to nutrient depleted media alone, CON CAF CM increased CON PDACc survival by 3.5 fold, indicating that there are additional factors in the CM, besides Glu and Gln, contributing to PDAC survival (Fig 2K). Importantly, Glu/Gln were not able to rescue NGL-1 KO PDACc survival, suggesting a potential defect in Glu/Gln utilization in PDAC cells lacking NGL-1. However, supplementing back Glu/Gln at CON CAF CM levels to NetG1 KO CM was able to significantly rescue PDACc survival, although to a lesser degree in NGL-1 KO cells. These results suggest that adding Glu/Gln to factors secreted by NetG1 KO CAFs suffices to mimic CON CAF CM effects in rescuing NGL-1+ PDACc survival under nutrient deprived conditions.
Guided by the clear differences in Glu/Gln levels upon KO of NetG1 in CAFs, we explored if glutaminase (GLS) and glutamine synthetase (GS) protein levels were altered in these cells. We detected expression of GLS and GS in TAs and CAFs (Fig 2L). While GLS levels were generally higher in CON and NetG1 KO CAFs than TAs, GS was significantly reduced in NetG1 KO CAFs, suggesting that NetG1 KO CAFs are compromised in their ability to generate Gln from Glu. Interestingly, NetG1 KO CAFs had reduced levels of vesicular glutamate transporter 1 (VGlut1), a major protein responsible for loading presynaptic vesicles with Glu, assuring synaptic Glu secretion, during neuronal communication [61] (Fig 2L). Thus, while CON and NetG1 KO CAFs expressed similar levels of GLS, NetG1 KO CAFs displayed decreased levels of VGlut1 and GS (also confirmed at the mRNA level, S10G), which could account for the reduced amounts of Gln produced and Glu released by NetG1 KO CAFs. These results were also observed in the NetG1 KD CAFs (S10A) and CAF2 (S11A). As expected from KRAS mutant cells, PDACc expressed lower levels of GS compared to CON CAFs, while maintaining GLS expression (S11B). Interestingly, knockout of NGL-1 in PDACc resulted in a reduction in GLS protein levels, suggesting an even greater metabolic dependence on extracellular nutrients in the KO cells compared to CON PDACc. Additionally, NGL-1 expression in PDAC cells appears to be critical for the utilization of some extracellular factors, such as Gln and Glu, perhaps due to downregulation of glutamate receptor binding pathways, as suggested by TCGA NGL-1 stratification (S12). Collectively, these results suggest that Gln and Glu derived from CAFs help support PDAC survival, under nutrient deprived conditions, and that this support depends on NetG1 expression in CAFs.
Knockout of NetG1 in CAFs abrogates their immunosuppressive phenotype and permits NK cell anti-tumor function, partially through IL-15
Thus far, we noticed that NetG1 expression in CAFs was critical for the ability of CAFs to provide tumor cells with critical nutrients for their survival (Fig 2). Moreover, the RNAseq analysis suggested that KO of NetG1 led to a normalization of CAFs, through the downregulation of pro-tumor fibroblastic pathways, including inflammatory signaling (S4). This led us to question if NetG1 is also a functional mediator of immunosuppression, another key CAF function [5]. Hence, we performed both traditional and multiplex (U-Plex) enzyme-linked immunosorbent assays (ELISAs) with TAs and CAFs cultured in 3D, to identify a cytokine profile that could define the immunosuppressive milieu that CAFs generate. As expected, we found that CON CAFs produce increased levels of GM-CSF, IL-1β, IL-8, CCL20/MIP3α and TGF-β compared to TAs (Fig 3A). Strikingly, ablation of NetG1 in CAFs significantly decreased protein levels of GM-CSF (40.6 fold), IL-6 (1.4 fold), IL-8 (13.3 fold), IL-1β (5.2 fold), CCL20/MIP3α (80 fold), and TGF-β (1.5 fold) compared to CON CAFs (Fig 3A), with a similar downregulation in NetG1 KD CAFs (S13A) and CAF2 (S13B). At the mRNA level, GM-CSF and TGF-β were correspondingly downregulated in NetG1 KO CAFs, with increases in IL-6 and IL-8 mRNA expression (S13C), suggesting post-translational regulation of IL-6 and IL-8. On the other hand, expression of IL-2, IL-12 p70, IL-10, IFN-γ, TNF-α, and IFN-β were all below the limit of detection in all three fibroblastic populations tested (data not shown). Interestingly, we observed that IL-15 protein levels, one of the most potent known NK cell activators, were up-regulated in both CON and NetG1 KO CAFs compared to TAs (4 and 2.9 fold, respectively; Fig 3A). Also, no differences were noted in mRNA expression of IL-15 when comparing CON CAFs to NetG1 KO CAFs (S13C). This suggested that NetG1 KO CAFs may generate a less immunosuppressive microenvironment than the one from CON CAFs, possibly allowing NK cells to more effectively kill PDAC cells, which could represent an important anti-tumor mechanism. Indeed, in our two TMA cohorts, the majority of patients presented with low NK cell infiltrates (represented by IHC of NKp46, S14A) (FCCC: 59/79 or 75%; MDA: 80/137 or 58%). Importantly, similarly to the median OS times observed in stromal NetG1 high patients (Fig 1H), patients with low NK cell infiltrate numbers had a median OS of 20.5 months and 21 months, in the FCCC and MDA TMAs respectively (S14B, Sup Table 3). However, in patients that had more NK cell infiltration, there was a trend for a noticeable increase in survival, 46 and 33 months in the FCCC and MDA TMAs, respectively (p-values: 0.1213, 0.0546; HR: 0.65, 0.68).
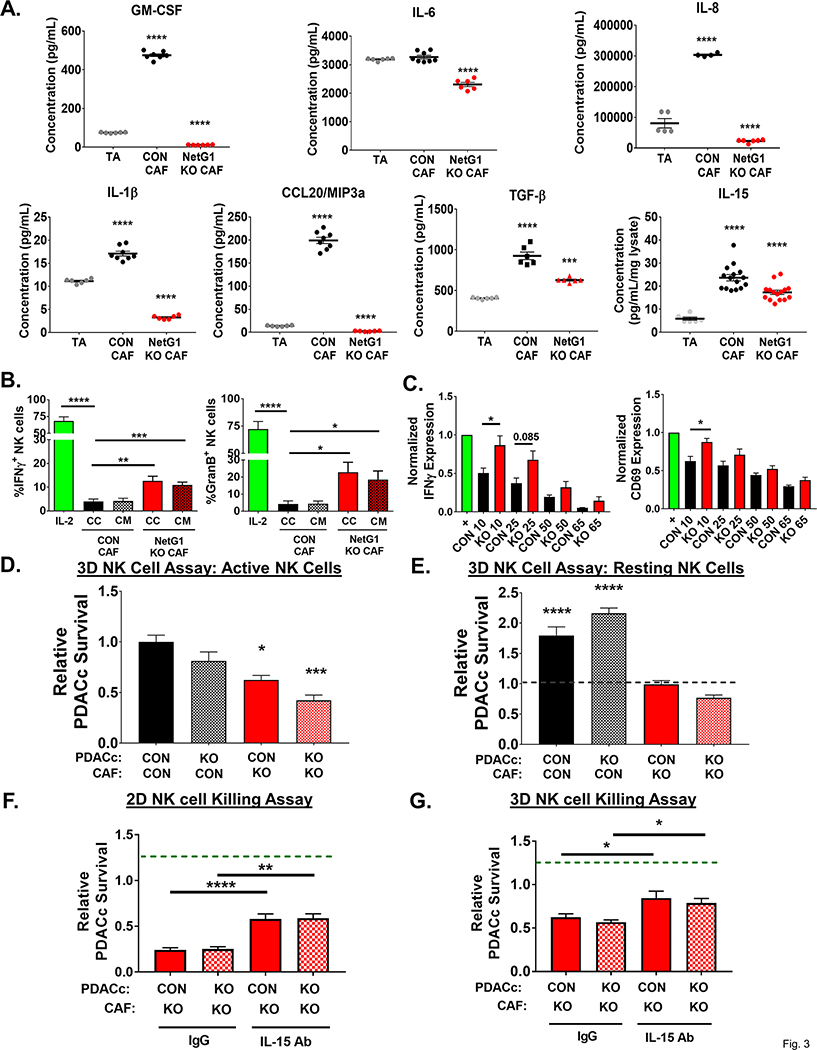
Figure 3. NetG1+ CAFs create an immunosuppressive microenvironment that protects PDAC cells from NK cell induced death.
A. Quantification of U-Plex (multiplex ELISA; GM-CSF, IL-1β, CCL20, IL-6, IL-8) and ELISAs (IL-15, TGF-β) of assorted cytokines with immunomodulatory or immunoattractive potentials, detected in the CM of TA, CON CAFs, or NetG1 KO CAFs, growing in 3D. N= 6 biological replicates. * compared to TA. B. Quantification of the % of NK-92 cells positive for markers of activation (IFNγ and Granzyme B) determined by flow cytometry after IL-2 pre-activated NK-92 cells (8×104) were in direct co-culture (CC) with CON or NetG1 KO CAFs (2×104) or treated with their conditioned media (CM) for 16 hours. * compared to CON CAF CC. C. Primary NK cells (105) were isolated from healthy human donors, pre-activated with IL-2/IL-12, incubated with CM from CON or NetG1 KO CAFs for 16 hours, and their activation status was determined by flow cytometry, using IFNγ and CD69 as markers. Expression of markers was normalized to the positive control (IL-2 alone = 1.0). * comparison between the CON and KO at each % of CM. D-E. RFP+ CON or NGL-1 KO PDACc (2×104) were co-cultured in 3D with GFP+ CON or NetG1 KO CAFs (2×104) and with active (D) or resting (E) NK-92 cells (8×104) for 48 hours and PDAC survival was quantified. Groups were normalized to CON PDACc/CON CAF with active NK cells. Dotted line in the resting graph (RIGHT) denotes PDACc survival with CON PDACc/CON CAF with active NK cells. * compared to CON PDACc/CON CAF with active NK cells. F. NK cell killing assay in 2D, where RFP+ CON or NGL-1 KO PDACc (2×104) were co-cultured with CON or NetG1 KO CAFs (2×104) and 8×104 active NK-92 cells (IL-2 preactivated) for 6 hours in the presence of isotype control IgG or IL-15 neutralizing antibody. Graphs depict PDACc survival, relative to the CON PDACc/CON CAF condition treated with IgG (dotted green line). * compared to IgG treated CON PDACc/CON CAF with NK cells. G. Same assay as in (F), but the co-culture is performed in 3D for 48 hours. * compared to IgG treated CON PDACc/CON CAF with NK cells. One-Way ANOVA, Dunnett’s multiple comparison test (A, B, D, E, F, G) or T-test (C). *p<0.05, **p<0.01, ***p<0.001; ****p<0.0001.
To test the influence of CAF NetG1 in NK cells activation and function, we first assayed the activation of an NK cell line, NK-92, in our in vitro 3D culturing system. After IL-2 treatment alone, 68.5% and 72.3% of the NK cells became positive for IFN-γ and Granzyme B (i.e. activated), respectively (Fig 3B). Conversely, direct co-culture of activated NK cells with CON CAFs or with CM from CON CAFs abolished NK cell activation marker expression, illustrating the potent immunosuppressive potential of CON CAFs. NetG1 KO CAFs, however, were less immunosuppressive, as 12–23% of NK cells maintained IFN-γ or Granzyme B expression. This result is consistent with the decreased levels of immunosuppressive cytokines generated by the NetG1 KO CAFs (Fig 3A). A similar experiment was performed using primary NK cells isolated from healthy human donor blood, with IFN-γ and CD69 as markers of activation. As with NK-92 cells, we observed that while CM from CON CAFs inactivates primary NK cells in a concentration dependent manner (i.e. 10% CM prevented NK activation by ~50%), NetG1 KO CAF CM was significantly less immunosuppressive at the same dilutions (Fig 3C). We next tested if the activation status of NK-92 cells correlated with their functional ability to kill PDAC cells. To this end, we performed an experiment co-culturing resting and active NK cells with CON or NGL-1 KO RFP+ PDACc, using complete media (to avoid stress due to nutrient deprivation) in 3D. Accordingly, active NK cells were twice as effective in killing both CON and NGL-1 KO PDACc than resting NK cells (S15A-B), suggesting that the ability of CAFs to control anti-tumor NK function is independent of PDACc NGL-1 status. To probe how CAFs could directly affect the ability of NK cells to kill PDACc, we co-cultured CON or NetG1 KO CAFs with CON or NGL-1 KO RFP+ PDACc in the presence of resting or active NK cells, again using complete media in 3D. In agreement with the observed immunosuppressive profile of CON CAFs (Fig 3A-B), active NK-92 cells were prevented from killing PDACc in the presence of CON CAFs, and this protection was decreased by 38–39% when cultured with NetG1 KO CAFs (Fig 3D, S15A BOTTOM). These results were effectively replicated when using PANC-1 and CAF2 (S15C). Intriguingly, even when resting NK cells were cultured in the presence of NetG1 KO CAFs, they were 45–65% more effective at eliminating PDACc compared to co-culture with CON CAFs, suggesting that NetG1 KO CAFs could partially activate NK-92 cells (Fig 3E, S15A BOTTOM).
The NK-92 killing assay was replicated in 2D culturing conditions. The KO of NetG1 in CAFs resulted in a significant decrease in CON PDACc or CON PANC-1 survival against NK cell induced death compared to CON CAF conditions (84 and 66% less, respectively) (S15D-F). Interestingly, in 2D conditions, NGL-1 KO cells (KO1 and KO2) were more sensitive to NK cell killing even when co-cultured with CON CAFs (88% and 78% less survival compared to CON PDACc, respectively), suggesting that NetG1/NGL-1 heterotypic cell-cell contacts were important for protection against NK cell induced death in the absence of ECM (S15D-F). Importantly, neither resting nor active NK cells affected CON or NetG1 KO CAFs survival (S15G). To question whether tumor to stromal cell ratios are important for the observed effect, we co-cultured various ratios of PDACc to CAFs (5:1, 3:1, 1:3) and again performed the NK cell killing assay. As the number of CAFs increased, PDACc survival increased as well, indicating a direct effect of CAF numbers on NK cell killing function (S15H).
Overall, NetG1 expression in CAFs creates an immunosuppressive microenvironment that inactivates NK cells and protects PDAC cells from NK cell mediated death. Loss of NetG1 expression in CAFs partially reverts the immunosuppressive capacity of CAFs, allowing NK cells to maintain their activity and eliminate PDAC cells. Moreover, the microenvironment generated by CAFs (i.e. ECM) also plays an important role in the support of PDAC survival, highlighting the differential effects of 2D culture versus 3D culture.
While deletion of NetG1 in CAFs led to a decrease in the expression of immunosuppressive cytokines, levels of IL-15, a key activator of NK cells, remained significantly higher than in TAs (Fig 3A). Thus, we hypothesized that the downregulation of immunosuppressive cytokines, coupled with the maintenance of IL-15 expression, was partially responsible for the observed anti-tumor phenotype of the NetG1 KO CAFs. We repeated the NK cell killing assay in 2D and 3D, co-culturing CON or NetG1 KO CAFs with CON or NGL-1 KO PDACc, in the presence of a neutralizing IL-15 antibody or IgG isotype control. Indeed, neutralization of IL-15 resulted in a ~35% and ~25% (2D and 3D, respectively) increase in PDACc survival compared to IgG treated conditions, independent of PDACc NGL-1 status, suggesting that CAF expressed IL-15 was partially responsible for the anti-tumor microenvironment created by NetG1 KO CAFs (Fig 3F-G). Thus, collectively, the in vitro NK assays and patient data suggest CAFs lacking NetG1 improve NK cell function, which is better for overall survival in PDAC patients, in a mechanism partially dependent on IL-15 expression by CAFs that is accompanied by limiting immunosuppressive cytokine secretion.
p38 and AKT pathways mediate NetG1 dependent pro-tumor CAF functions
Having identified key functions regulated by NetG1, we sought to uncover the downstream mediators of NetG1 that are responsible for pro-tumor functions of CAFs. We identified the downregulation of p-AKT and p-p38 in NetG1 KO CAFs (Fig 4A), which were confirmed in CAF2 (S11A), and postulated that these two pathways were largely mediating the effects of NetG1. Therefore, we employed the use of pharmacological inhibitors of p38 (p38i) and AKT (AKTi) on CAFs in our 3D system and assessed PDAC survival against metabolic stress and NK induced death, as well as Glu/Gln and cytokine production. Interestingly, we observed a greater decrease on PDACc survival under serum/Gln free conditions when CAFs were pre-treated with AKTi (~70%) vs. p38i (~30%) compared to DMSO pre-treated CAFs (Fig 4B). Accordingly, AKTi reduced both Glu/Gln production in CAFs (~30%), while p38i only inhibited Gln production (Fig 4C). Together, these findings suggest that AKT regulated the metabolic parameters of CAFs to a greater degree than p38. On the other hand, p38i CAFs displayed a significant decrease in the secretion of more cytokines than AKTi, with a reduction in GM-CSF, IL-6, and TGF-β levels, and a maintenance of IL-15 (akin to NetG1 deficient CAFs), compared to DMSO treated CAFs (Fig 4D). In contrast, AKTi CAFs had increased GM-CSF production compared to DMSO treated CAFs, with decreases in IL-8, TGF-β, and IL-15 production. Based on these profiles we hypothesized that p38i CAFs would be less immunosuppressive than AKTi CAFs. To test this, we performed the NK killing assay and saw that indeed p38i CAFs lost their immunosuppressive capacity compared to DMSO CAFs, while AKTi CAFs did not (Fig 4E), thus confirming our hypothesis.
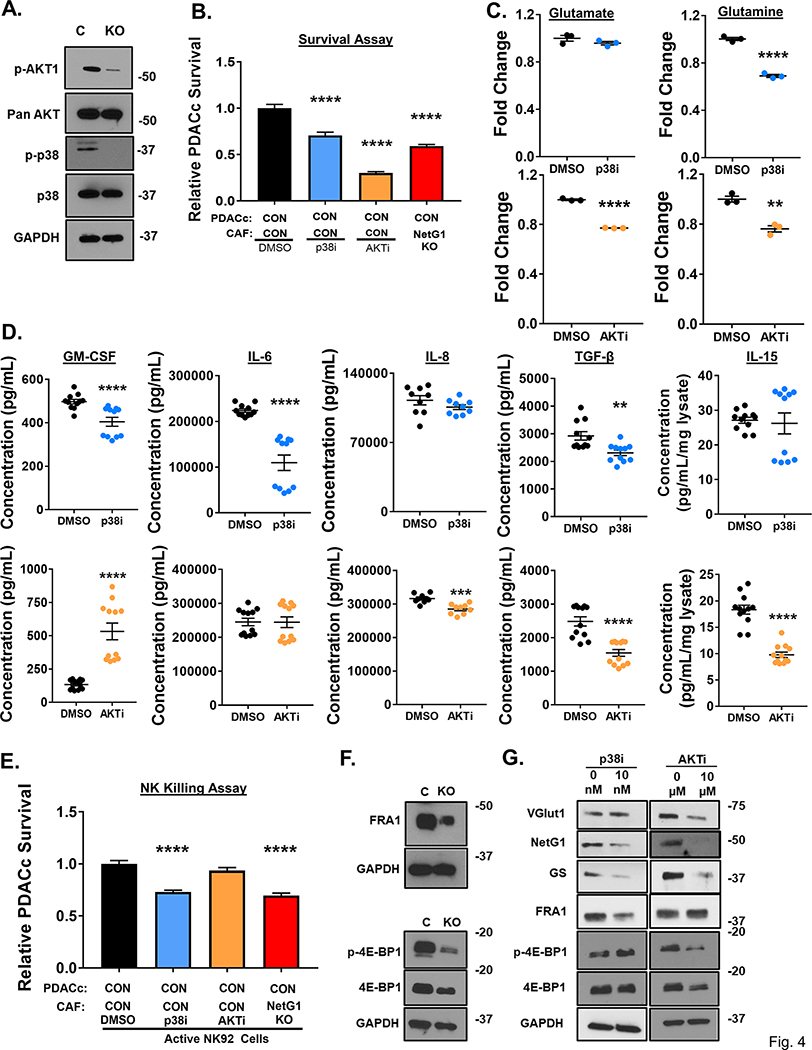
Figure 4. NetG1 functions are mediated through AKT and p38 pathways.
A. Representative western blots demonstrating downregulation of p-AKT and p-p38 in NetG1 KO CAFs compared to CON (N=3). GAPDH was used as a loading control. B. RFP+ PDACc (2×104) were co-cultured with CON CAFs pre-treated for 24 hours with DMSO, 10 nM p38 inhibitor (p38i), or 10 μM AKT inhibitor (AKTi) and PDACc survival was assessed 96 hours later. * compared to DMSO. C. Graphs depicting relative Glu and Gln levels measured from CAFs treated with p38i or AKTi in serum/Gln free media for 48 hours. DMSO was used as a treatment control. * Compared to DMSO. D. ELISAs for GM-CSF, IL-6, IL-8, TGF-β, and IL-15 were performed on CM or lysates of CAFs treated for 48 hours with DMSO or p38i (TOP panel) and DMSO or AKTi (BOTTOM panel). * compared to DMSO. E. Quantification of PDACc survival after co-culture with CON CAFs pre-treated for 24 hours with DMSO, p38i, or AKTi in the presence of active NK cells (8*104). F. Representative western blots demonstrating a reduction in FRA-1, p-4E-BP1, and 4E-BP1 protein expression in NetG1 KO CAFs compared with CON CAFs (N=3). GAPDH was used as a loading control. G. Representative western blots of VGlut1, NetG1, GS, FRA1, p-4E-BP1, and 4E-BP1 from the lysates of DMSO or p38i CAFs (LEFT) and DMSO or AKTi CAFs 48 hour treated CAFs (RIGHT). One-Way ANOVA, Dunnett’s multiple comparison test (B, E) and Student’s T-test (C, D). *p<0.05, **p<0.01, ***p<0.001; ****p<0.0001.
To further dissect the p38 and AKT signaling pathways in CON and NetG1 KO CAFs, we tested the expression of numerous known downstream factors of these two pathways. We found substantial modulations of both pathways in NetG1 KO CAFs compared to CON CAFs, with increased levels of FosB and cFos and reduced levels of c-Jun, JunB and FRA-1 downstream to p38, and reduced protein levels of p-FOXO1, p-FOXO3, p-GSK3β, and p-4E-BP1 downstream to AKT (S16A-B, Fig 4F). To complement our loss of function approaches, we decided to further explore the modulation of the two most downregulated proteins observed in each pathway, FRA-1 and 4E-BP1, due to their previous association with inflammation or fibrosis [62, 63]. The downregulation of FRA1 and 4E-BP1 was also confirmed in CAF2 NetG1 KDs (S11A). Next, we measured the expression of VGlut1, NetG1, GS, as well as FRA1 and 4E-BP1 in p38i and AKTi CAFs to observe how the newly identified signaling circuit changes in response to the kinase inhibitors (Fig 4G). In accordance with Glu/Glu production, p38i CAFs had reduced protein levels of NetG1 and GS, but not VGlut1, while AKTi CAFs exhibited decreases in all three proteins. In addition, FRA1 and 4EBP-1 were only downregulated in response to the corresponding pathway inhibitor (p38 or AKT, respectively). Overall, the data pointed towards a regulation of p38 and AKT downstream to NetG1, with a greater role for p38 in the immunosuppressive functions of CAFs, while AKT regulated metabolic parameters to a greater extent.
FRA1 and 4E-BP1 partially regulate metabolic and immunosuppressive characteristics of CAFs
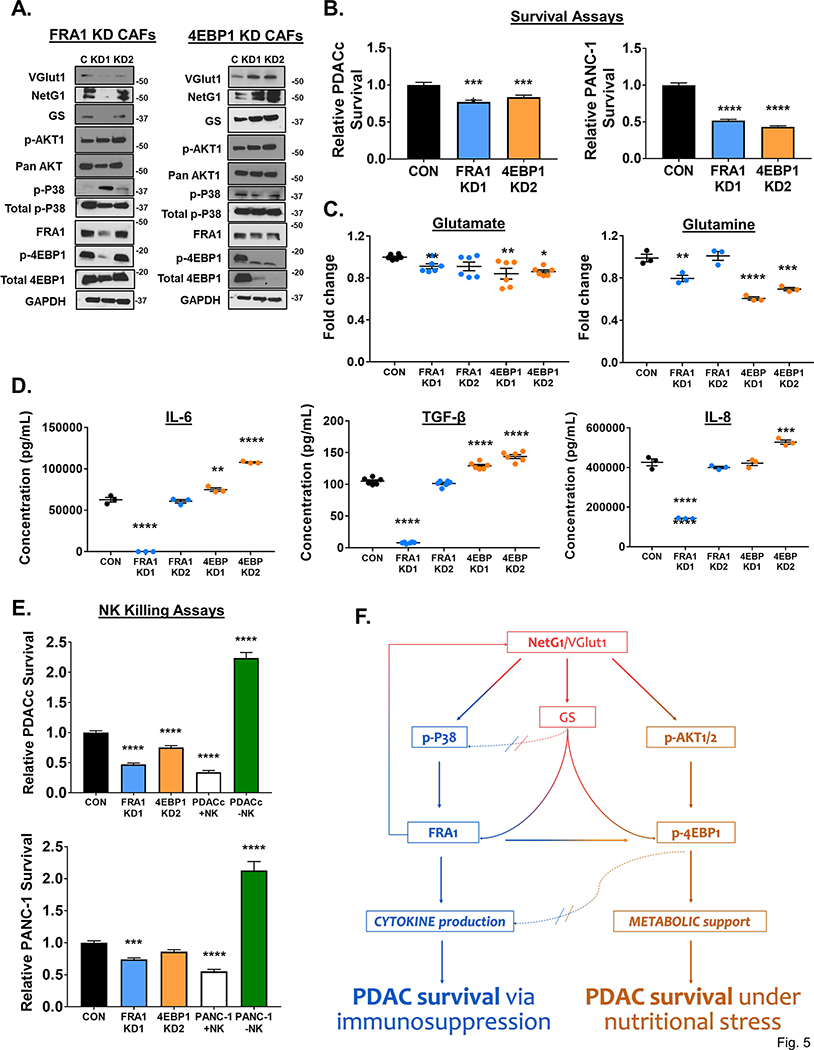
Because there was a large downregulation of FRA1 and 4E-BP1 in NetG1 KO CAFs, we decided to explore the functional roles of these proteins by knocking them down in CAFs, and observe how they fit into the NetG1 signaling circuit. We obtained one effective and one ineffective KD of FRA1, as well as 2 effective KDs for 4E-BP1 (Fig 5A). Intriguingly, the effects of FRA1 and 4E-BP1 KDs in CAFs on the signaling circuit were quite different; FRA1 KD in CAFs resulted in a loss in expression of VGlut1, NetG1, and GS, while 4E-BP1 KD CAFs up-regulated these three proteins. In terms of the upstream kinases, p-AKT remained unchanged in both KDs, while p-p38 increased in FRA1 KD CAFs and remained largely unchanged in 4E-BP1 KD CAFs. Strikingly, while loss of 4E-BP1 in CAFs had no effect on FRA1 expression levels, knockdown of FRA1 resulted in a downregulation of p-4EBP1 in CAFs. These results illustrated a crosstalk between the p38/FRA1 and AKT/4E-BP1 pathways, as well as a potential compensatory feedback loop that upregulated factors upstream in the circuit. Importantly, both FRA1 and 4E-BP1 KD CAFs failed to support PDACc and PANC-1 survival under metabolic stress compared to CON CAFs (Fig 5B). Consequently, Gln levels were lower in the CM of both FRA1 and 4EBP1 KD CAFs compared to CON CAFs, and Glu was lower in 4E-BP1 KD CAFs (Fig 5C), which could account for the decreased PDAC cell survival observed in Figure 5B. On the other hand, when measuring cytokine secretion, FRA1 KD CAFs displayed markedly reduced cytokine levels compared to CON CAFs, and 4E-BP1 KDs had increased cytokine levels, which were in line with the results seen in p38i and AKTi CAFs (Fig 5D). Therefore, we hypothesized that the FRA1 KD CAFs would allow NK cell mediated killing while 4E-BP1 KD CAFs would retain their immunosuppressive capacity. As expected, in the NK killing assay, PDACc and PANC-1 cells co-cultured with FRA1 KD CAFs had significantly reduced survival compared to CON CAFs (Fig 5E). In contrast, more PDACc and PANC-1 cells survived when co-cultured with 4E-BP1 KDs compared to FRA1 KD CAFs, at levels similar to CON CAFs (Fig 5E). Collectively, these results suggest that FRA1 and 4E-BP1 contribute to the metabolic support of PDAC cells by CAFs, while FRA1 more effectively mediates the immunosuppressive function of CAFs. Additionally, FRA1 can modulate the 4E-BP1 side of the signaling circuit, illustrating a crosstalk between the downstream effectors of the p38 and AKT pathways (Fig 5F).
Figure 5. Downstream mediators of p38 and AKT signaling, FRA1 and 4E-BP1, regulate metabolic and immunosuppressive properties of CAFs.
A. Representative blots of VGlut1, NetG1, GS, p-AKT, AKT, p-p38, p38, FRA1, p-4E-BP1, and 4E-BP1 in (LEFT) CON (C) or FRA1 KD1/KD2 (KD) CAFs and (RIGHT) in CON or 4E-BP1 KD CAFs (N=3). GAPDH was used as a loading control. B. Graphs depicting PDAC survival after assays were performed with PDACc cells (TOP) or PANC-1 cells (BOTTOM) co-cultured in 3D, under serum/Gln free conditions, with the most effective KD CAFs (FRA1 KD1 and 4E-BP1 KD2) from (A), *compared with co-culture with CON CAFs. C. Relative Glu and Gln levels detected in the CM of CON, FRA1 KD, or 4E-BP1 KD CAFs after culture for 48 hours in serum/Gln free media in 3D. D. Measurement of indicated cytokine production in the CM of CON, FRA1 or 4E-BP1 KD CAFs after culture for 48 hours in serum/Gln free media in 3D, as determined by ELISA. E. Quantification of PDAC cell survival after co-culture with CON, FRA1 KD1, or 4E-BP1 KD2 KD CAFs in the presence of active NK cells (8*104). PDAC cells cultured alone with or without active NK cells served as controls. LEFT: PDACc; RIGHT: PANC-1. *Compared to CON. One-Way ANOVA, Dunnett’s multiple comparison test. *p<0.05, **p<0.01, ***p<0.001; ****p<0.0001. F. NetG1 Signaling Circuit in CAFs. There are two major arms of the signaling circuit regulated by NetG1, the cytokine arm (LEFT, blue) and the metabolic arm (RIGHT, orange). NetG1 and VGlut1 (see justification for VGlut1 claims in results referring to Figure 6) sit atop the signaling network, and increase levels of GS (see justification for GS claims in results referring to Figures 6 and S17), p-p38, and p-AKT (red arrows). GS, p38, and AKT are further downstream, and regulate FRA1 and 4E-BP1 levels, which begin to diverge in what CAF functions they control. FRA-1 regulates cytokine production in CAFs (blue path), as well as mediates partial effects on CAF generated metabolism (blue to orange arrow), through Gln production. 4E-BP1 has an inhibitory effect on cytokine production (orange to blue dotted / blocked, arrow), while directly controlling CAF generated Glu/Gln (orange path). There is also positive feedback to the top of the circuit (blue to red feedback arrow), and crosstalk to the AKT/4E-BP1 arm, all originating at FRA1 (blue to orange arrow). Thus, NetG1 controls pro-tumor CAF functions, through a complex signaling network, leading to PDAC survival through immunosuppression (blue arm) and metabolic support (orange arm).
VGlut1 and GS inhibition in CAFs modulate metabolic and immunosuppressive functions in CAFs, in a manner similar to NetG1 loss
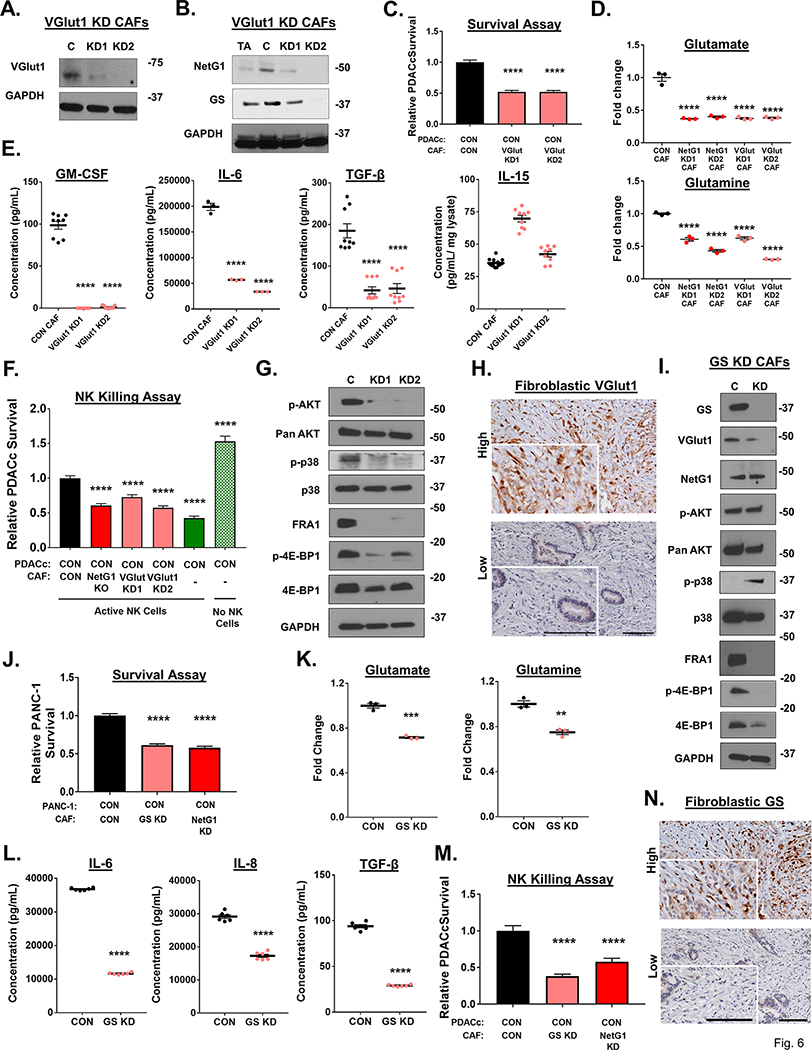
Because of the consistent joint modulation of VGlut1, GS, and NetG1 across nearly all of the genetically and pharmacologically inhibited CAFs, we evaluated if the absence of VGlut1 or GS in CAFs could phenocopy the effects of NetG1 KO in CAFs. First, we produced knockdowns for VGlut1 (VGlut1 KD) using CRISPRi (Fig 6A). Interestingly, VGlut1 KD CAFs lost the expression of NetG1 and GS (Fig 6B), suggesting a co-regulatory mechanism for these proteins. Functionally, VGlut1 KD CAFs had a similar effect on PDACc survival as NetG1 KD CAFs, with PDACc survival reduced by ~50% compared to CON CAFs (Fig 6C). Moreover, VGlut1 KD CAFs produced similar amounts of Glu and Gln, as was observed for NetG1 KD CAFs, both producing significantly less Glu/Gln than CON CAFs (Fig 6D, Sup Table 8). In terms of their cytokine profile, VGlut1 KD CAFs produced significantly less GM-CSF, IL-6, and TGF-β than CON CAFs, while maintaining IL-15 levels (Fig 6E), indicative of less immunosuppressive capacity. Indeed, in the NK killing assay, PDACc cells had ~30–40% decreased survival when co-cultured with VGlut1 KD CAFs compared to CON CAFs (Fig 6F). Related to downstream signaling, VGlut1 KD CAFs displayed reduced levels of p-AKT, p-p38, FRA1, and p-4E-BP1, phenocopying NetG1 KO CAFs (Fig 6G). Lastly, we stained PDAC patient tissue for VGlut1 expression and detected high fibroblastic expression in 4/14 cases, and low expression in 10/14 cases (Fig 6H). In sum, VGlut1 is expressed in human CAFs, and regulates expression of proteins and functions in CAFs involved in supporting PDAC cell survival, phenocopying NetG1 dependent functions (Fig 5F).
Figure 6. VGlut1 and GS regulate pro-tumor CAFs functions, in a manner similar to NetG1.
A. Representative western blots of VGlut1 in CAFs, demonstrating effective knockdown (KD) by CRISPRi (N=3). B. Representative western blot of NetG1 and GS in TA, CON, or VGlut1 KD CAFs. GAPDH was used as a loading control. N=3 C. RFP+ CON PDACc cells (2×104) were cultured in 3D with CON or VGlut1 KD CAFs (2×104) for 96 hours in SF/Gln Free media and PDACc cell survival was assessed. D. Relative Glu and Gln levels in the CM of NetG1 and VGlut1 KD CAFs compared to CON CAF. Note that both NetG1 KD and VGlut1 KD CAFs have a reduction in Glu/Gln levels. * Compared to CON CAF. E. Measurement of indicated cytokine production in the CM of CON or VGlut1 KD CAFs after culture for 48 hours in serum/Gln free media in 3D, as determined by ELISA. * Compared to CON CAF. F. Quantification of PDACc survival after co-culture with CON or VGlut1 KD CAFs in the presence of active NK cells (8*104). One-Way ANOVA, Dunnett’s multiple comparison test. *p<0.05, **p<0.01, ***p<0.001; ****p<0.0001. G. Representative western blots of p-AKT1, AKT, p-p38, p38, FRA1, p-4E-BP1, and 4E-BP1 comparing protein expression in CON and VGlut1 KD CAFs N=2. GAPDH was used as a loading control. H. Representative images of the immunohistochemical staining of VGlut1 in patient tissue, segregated into high or low fibroblastic VGlut1 expression. Inserts correspond to magnified regions, in which is stromal cells are more evident. Scale bars represent 100 μm. I. Representative blots of GS, VGlut1, NetG1, p-AKT, AKT, p-p38, p38, FRA1, p-4E-BP1, and 4E-BP1 in CON (C) or GS KD (KD) CAFs. GAPDH was used as a loading control. N=3. J. RFP+ CON or NGL-1 KO PDACc (2×104) were cultured in 3D with CON or GS KD CAFs (2×104) for 96 hours in SF/Gln Free media and PDACc cell survival was assessed. K. Relative Glu and Gln levels in the CM of GS KD CAFs compared to CON CAF. L. Measurement of indicated cytokine production in the CM of CON or GS KD CAFs after culture for 48 hours in serum/Gln free media in 3D, as determined by ELISA. M. Quantification of PDACc survival after co-culture with CON, GS KD, or NetG1 KD CAFs in the presence of active NK cells (8*104). *Compared to CON. One-Way ANOVA, Dunnett’s multiple comparison test. *p<0.05, **p<0.01, ***p<0.001; ****p<0.0001. N. Representative images of the immunohistochemical staining of GS in patient tissue, segregated into high or low fibroblastic GS expression. Inserts correspond to magnified regions, in which is stromal cells are more evident. Scale bars represent 100 μm.
To address the contribution of GS to the support provided by CAFs for PDACc survival, we genetically and pharmacologically inhibited GS in CAFs, using CRISPRi and increasing concentrations of methionine sulfoximine (MSO), respectively. GS was knocked down with high efficiency (Fig 6I), and MSO effectively inhibited GS expression at concentrations greater than 0.1 mM (S17A). GS KD CAFs exhibited decreased levels of VGlut1, FRA1, and p-4E-BP1, while maintaining expression of NetG1, and p-AKT, and increasing p-p38 (Fig 6I). Transient inhibition with MSO resulted in decreases of VGlut1 and NetG1, as well as downstream mediators p-p38 FRA1, and p-4E-BP1, with p-AKT increasing in expression (S17B). We attribute the differences in signaling molecules to the nature of the treatment (transient vs. permanent), which may have resulted in compensatory up-regulation of p-AKT, for example. Nevertheless, blockade of GS led to an overall downregulation of numerous proteins identified in the novel NetG1 signaling circuit. Therefore, due to the similar profile, we predicted that loss of GS would functionally mimic the loss of NetG1 and VGlut1 in CAFs and reduce PDAC survival in our metabolic stress assay and when challenged with NK cells. Accordingly, PDAC cells that were co-cultured with either GS KD or MSO pre-treated CAFs under serum/Gln free conditions survived significantly less than when cultured with CON CAFs, similarly to co-culture with NetG1 KO CAFs (Fig 6J, S17C). This was likely due to the decreased levels of Glu/Gln detected in the CM of GS KD CAFs (Fig 6K), comparable to other inhibited CAFs in the NetG1 signaling hub that failed to support PDAC survival. The immunosuppressive activity was determined in both GS KD and MSO treated CAFs by measuring cytokine secretion and protection of PDAC cells from NK cell induced death. Both GS KD and MSO pre-treated CAFs showed a decrease in secretion of most cytokines tested (Fig 6L, S17D), and this resulted in a corresponding decrease in PDAC cell survival when co-cultured with GS KD or MSO treated CAFs, similarly to co-culture with NetG1 KO CAFs (Fig 6M, S17E). Finally, we performed IHC on patient PDAC tissue to detect fibroblastic expression of GS and found that 7/15 patients expressed high levels of GS in fibroblasts, and 8/15 had low expression (Fig 6N). Collectively, these data suggested that NetG1, VGlut1, and GS may be all linked in a new signaling circuit in CAFs, driving the metabolic and immunosuppressive functions of CAFs (Fig 5F).
Therapeutic inhibition of NetG1 ablates the pro-tumor functions of CAFs, stunting tumorigenesis in vivo.
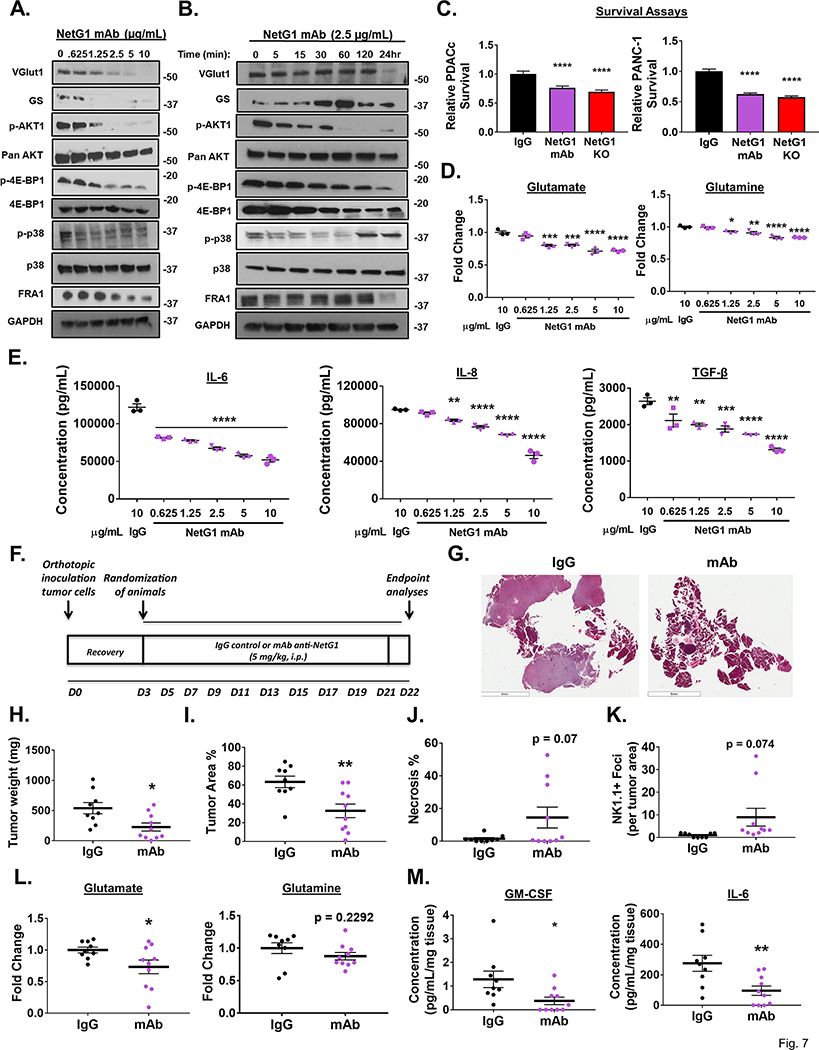
Having established the roles that NetG1 played in driving pro-tumor CAF functions, we next explored the therapeutic potential of targeting NetG1 in vitro and in vivo. We identified a neutralizing monoclonal antibody (mAb) against NetG1, by testing its ability to modulate NetG1 mediated signaling, Glu/Gln/cytokine production, and functions in CAFs in vitro. We measured the changes in expression of the proteins in the signaling hub, by dose and time responses. Anti-NetG1 mAb began to decrease expression of the majority of the signaling circuit at a concentration of 1.25 μg/mL, with maximal efficacy at 10 μg/mL (Fig 7A). We proceeded with the minimal dose that inhibited all proteins (2.5 μg/mL) and explored the changes in protein expression over time. Interestingly, most proteins lost substantial expression by 24 hours of treatment, with only GS having a transient increase in expression between 30–60 minutes of treatment (Fig 7B). p-AKT/p-4E-EP1 remained downregulated after 60 minutes of mAb treatment, while p-p38 levels rapidly decreased starting at 5 minutes, and rebounded by 120 minutes of treatment. These results largely confirm the putative signaling circuit uncovered in CAFs that we observed through the systematic inhibition of each individual component. Functionally, anti-NetG1 mAb pre-treated CAFs co-cultured with PDACc or PANC-1 cells resulted in a ~25–40% reduction in PDAC cell survival under metabolic stress compared to IgG pre-treated CAFs, similar to co-culture with NetG1 KO CAFs (Fig 7C), owed likely to the reduced Glu/Gln production in mAb treated CAFs (Fig 7D). In addition, anti-NetG1 mAb treated CAFs produced less pro-tumor cytokines compared to IgG treated CAFs, showing a dose dependent decrease in IL-6, IL-8, and TGF-β, suggesting less immunosuppressive capacity (Fig 7E). Thus, inhibition of NetG1 by mAb was extremely effective at modulating all pro-tumor signaling and functions of CAFs, serving as the impetus to move the treatment in vivo.
Figure 7. A neutralizing monoclonal antibody targeting NetG1 inhibits pro-tumor properties of CAFs and decreases tumor burden in vivo.
A+B. Representative western blots demonstrating a dose (A) and time (B) dependent decrease in proteins associated with pro-tumorigenic pathways in CAFs after anti-NetG1 monoclonal antibody (mAb) treatment in serum and glutamine free media, with 48 hour treatment for CAFs in (A) and indicated timepoints for CAFs in (B). N=3. C. Quantification of PDAC cell survival (PDACc or PANC-1) under serum and glutamine starvation after 48 hour co-culture with CON (10 μg/mL IgG or mAb treated) or NetG1 KO CAFs, demonstrating the efficacy of NetG1 mAb in a key functional assay. D. Graphs depict amounts of secreted Glu and Gln detected in the conditioned media of CAFs treated with IgG or increasing doses of mAb for 48 hours, as determined by ELISA. E. Graphs depict amounts of secreted pro-tumor cytokines detected in the conditioned media of CAFs treated with IgG or increasing doses of mAb for 48 hours, as determined by ELISA. Note the dose dependent decrease in these key secreted factors in (D) and (E), important for PDAC tumorigenesis. * Compared to IgG. One-way ANOVA: * p <.05, ** p<.01, *** p<.001, **** p<.0001. F. Treatment Strategy: C57/BL6 mice were injected orthotopically with 106 murine pancreatic cancer cells and mice were allowed to heal for 3 days. Starting on day 4, mice were separated into two groups, IgG (n=9) and mAb (n=10), and received either 5mg/kg IgG or mAb against NetG1. Mice were treated 3 times per week until the completion of the model, at day 22. Pancreata were isolated from the mice and H+E sections were developed from a cut of the entire pancreas. G. Representative images of the H+E sections of the pancreas from each treatment group. Scale bar: 6 mm. H. Graph depicting relative tumor weights. I. Quantification of % area of the pancreas that was classified as tumor. J. Quantification of the % area of the pancreas that was classified as necrotic. K. Graph displaying the number of NK1.1 positive Foci, normalized to tumor area per tissue, and these values were subsequently normalized to IgG treated tumors to compute fold changes. L+M. A small piece of tumor tissue (30–60 mg) was excised from the pancreas of each mouse and was cultured overnight in DMEM lacking serum, glutamate (Glu) and glutamine (Gln). The resultant media was collected and the amount of Glu and Gln (L) or cytokines (M) were measured by ELISA and normalized to the weight of tumor tissue cultured. Student’s T-Test was used to determine if samples were significantly different from IgG. * p <.05, ** p<.01, *** p<.001, **** p<.0001. n=9 for IgG, n=10 for mAb.
Next, we decided to test the efficacy of mAb in vivo, relying upon our previously performed syngeneic orthotopic injection model of PDAC in mice. Mice were injected with KPC3 PDAC cells and allowed to recover for 3 days, and were randomized into 2 groups, one receiving 5 mg/kg isotype control IgG and the other receiving 5 mg/kg anti-NetG1 mAb, three times a week for the duration of the experiment (22 days) (Fig 7F). Excitingly, anti-NetG1 mAb treatment was effective at limiting tumorigenesis, evident in the representative images of the pancreata isolated from IgG or mAb treated animals (Fig 7G). Mice treated with anti-NetG1 mAb had less tumor weight, a trend towards reduced pancreas weight, diminished tumor area %, and greater necrosis, all indicative of the efficacy of the therapy (Fig 7H-J, S18A). Importantly, there was greater infiltration of NK cells into the tumors of mAb treated mice compared to IgG (Fig 7K, S18B), and fibroblasts in tumor tissue expressed equal levels of IL-15 (S18C), evident from the IHC staining (S18D). Moreover, analysis of the tumor culture of isolated pieces of the pancreas from each group demonstrated a significant decrease in Glu, GM-CSF, and IL-6 protein levels, with a trend towards less Gln, after mAb treatment, supporting our in vitro findings (Fig 7L-M). Overall, in an extremely aggressive orthotopic model, anti-NetG1 mAb treatment was successful at limiting PDAC tumorigenesis, as well as at reducing key CAF generated metabolites and cytokines, while increasing NK cell infiltrates, highlighting NetG1 as an important druggable target in PDAC.
Discussion
The present study has uncovered a previously unknown target for the potential treatment of pancreatic cancer: the neural glutamatergic synaptic stabilizing protein NetG1. NetG1 (also known as Laminet-1 and not to be confused with Netrin-1) is a unique member of the netrin family of proteins, because it is anchored to the plasma membrane through a glycosylphosphatidylinositol linkage, and is not secreted like the majority of netrin family members [34]. NetG1 is a synapse stabilizing protein, located pre-synaptically on neurons, and its only known receptor is NGL-1, localized on post-synaptic neurons [35, 36], and on microglial cells where it plays a role in neuronal survival [64]. Dysregulation of NetG1 has been associated with Rett syndrome, schizophrenia, bipolar disorder, and anxiety [65–67]. In cancer, NetG1 displays a low mutation rate [68] and NetG1 mRNA expression, methylation, or mutational status has been linked to colon and uterine cancers [68–70]. Nevertheless, no studies have ascribed a functional role to NetG1 in the context of cancer.
PDAC is the most common form of pancreatic cancer, and has a dismal prognosis [2]. CAFs are a major cell type present in the PDAC microenvironment, whose role is incompletely understood. CAFs regulate at least three critical hallmarks of PDAC: 1.) the deposition of desmoplastic ECM; 2.) overcoming nutrient deprivation; and 3.) vast immunosuppression of anti-tumor immune cells [3, 25]. This study aimed to characterize the functions of NetG1 in PDAC, with an emphasis on the contribution of the desmoplastic stroma. Our results confirm that all three hallmarks are interconnected, and that NetG1 expression in CAFs is a central driver of all of them in the PDAC microenvironment.
The PDAC microenvironment is nutrient deprived, due to the collapse of blood vessels enabled by desmoplasia, and this drives PDAC cells to use an alternative nutritional supply [16]. Recent studies have demonstrated that CAFs supply PDAC cells with important metabolites, through exosomes [56] and amino acid secretion [55]. Moreover, the ECM has been shown to be utilized as fuel for cancer cells [54], which potentially positions the scaffolding produced by CAFs in the PDAC microenvironment as a vital source of energy for PDAC cells. Here, we found that CAFs lacking NetG1 were less supportive of PDAC cell survival, in direct co-culture, with their CM, or ECM (Fig 2). These findings prompted us to perform a screen comparing amino acid production and secretion and revealed a NetG1 dependent increase in extracellular Glu/Gln in CAFs.
Interestingly, it has long been recognized that patients with pancreatic cancer have elevated serum levels of Glu [71], with a corresponding depletion of serum Gln levels [72]. PDAC cells have a high capacity to utilize Glu, by catabolizing Gln through the enzyme glutaminase (GLS), in a process known as glutaminolysis [21]. While it is known that inhibition of GLS can decrease cancer cell growth in vitro and in vivo in a number of cancer models [19, 59], a recent study using a potent small molecular inhibitor of GLS displayed little efficacy in preclinical mouse models, in part through up-regulation of compensatory Gln procuring pathways in PDAC cells [73]. Additionally, it has also been demonstrated that the conditions used in cell culture provide an overabundance of nutrients that does not accurately mimic the tumor microenvironment in vivo, and culturing cells in a medium with nutrients at levels more representative of tumors in vivo resulted in tumor cells with a decreased dependence on Gln, due to increased utilization of cystine [74]. Intriguingly, an elegant recent study suggested that in mouse interstitial fluid of PDAC tumors, Gln levels were surprisingly higher than previously thought [75]. It is tempting to speculate that CAFs may be responsible for the maintenance of Gln levels, by utilizing the high levels of Glu produced and secreted by tumor cells. On the other hand, it has been recently shown that PDAC cells can upregulate expression of GS and its tumor specific inhibition stunts tumorigenesis [76]. In this study, the PDAC cells used did not acquire significant expression of GS, while CAFs expressed abundant GS, evident by their direct comparison (S11B). Thus, our findings that CAFs can assist PDAC cells in overcoming nutritional stress by supplying key metabolites, like Glu/Gln, adds to the ways in which cancer cells in PDAC can acquire Gln from the tumor microenvironment.
Our results demonstrated that ablation of NetG1 in CAFs reduced GS (catalyzing conversion of Glu to Gln), but not GLS protein levels (Fig 2). Indeed, GS is up-regulated in CAFs in ovarian cancer compared to their normal counterparts, and inhibition of stromal GS resulted in decreased tumor development [60]. Additionally, we found that deletion of NGL-1 from PDAC cells resulted in a decrease in cancer cell GLS expression (S11), signifying an even greater dependence of these cells on the microenvironment, evident by the lack of rescue during Glu/Gln addition in NGL-1 deficient PDAC cells (Fig 2). An additional interpretation of the data could be that CAF driven metabolism of Glu/Gln is required to first process these amino acids into other metabolites that are then used by PDAC cells. Analysis of TCGA data demonstrated that patients expressing the highest NGL-1 levels have enriched pathway signatures associated with increased glutamate receptor binding genes (S12), suggesting a link between NGL-1 and Glu uptake. Indeed, there are several studies demonstrating the importance of amino acid transporters in pancreatic cancer by which PDAC cells can overcome Gln and nutrient deprivation, such as glutamate/cystine exchanger xCT [74, 77], as well as the neutral amino acid transporters SLC6A14 [78] and SLC38A2 [79]. Pharmacological or genetic inhibition of any of the aforementioned transporters leads to reduced tumor burden in mice, as xCT is required to overcome glutamine deprivation through the utilization of cystine, SLC6A14 for the bulk uptake of amino acids, and SLC38A2 for maintenance of alanine levels, specifically in PDAC cells. Further analysis into the expression and activity of receptors that mediate uptake of extracellular amino acids in PDAC, such as Glu/Gln, as well as TCA cycle metabolites produced by CAFs, will be necessary to gain more insight into the general uptake ability of PDAC cells.
Macropinocytosis is a major nonspecific mechanism by which cancer cells harboring oncogenic KRAS mutations uptake bulk extracellular material (such as amino acids) to overcome nutrient stress [80], and was recently demonstrated to be mediated through sydecan-1 in a large panel of established and primary PDAC cell lines [81]. We observed uptake of CAF derived GFP by NGL-1+ PDAC cells in direct co-culture with CAFs or CAF CM, and direct transfer of GFP was mediated through fibroblastic NetG1/tumoral NGL-1, while only tumoral NGL-1 was required for the uptake of extracellular GFP (S9). Moreover, we found that NGL-1 regulated tumor intrinsic uptake of extracellular material, largely through macropinocytosis (S9). Combined with the survival assays (Fig 2, S10), we believe that while tumor intrinsic bulk uptake of material is an essential PDAC cell survival mechanism, we would also argue that the quality of the content also plays a role in PDAC cell survival, as direct co-culture with NetG1 deficient CAFs or their CM, resulted in a significant decrease in survival for NGL-1+ PDAC cells, despite the maintenance of their bulk uptake ability. Collectively, the data support both PDAC cell autonomous and reciprocal mechanisms to survive nutritional stress.
We also observed a dramatically reduced expression of VGlut1 upon NetG1 loss in CAFs (Fig 2). Interestingly, VGlut1 is present in glutamatergic synapses in neurons, where it loads Glu into pre-synaptic vesicles [61], and VGlut1 expression in neurons was shown to be dependent on NetG1 expression [35]. These studies agree with our data in CAFs, where the KO of NetG1 resulted in downregulation of VGlut1 expression, and provide a potential mechanistic explanation for the decreased levels of Glu in NetG1 KO CAF CM. Interestingly, VGlut1, 2 and 3 have been implicated in pancreatic neuroendocrine tumors, and Glu signaling in these tumor cells drives tumorigenesis [82]. Glu is a key neurotransmitter, but to maintain homeostasis and avoid excitotoxic stress, excess Glu loaded vesicles are then taken up by neighboring astrocytes, which express high levels of GS. Glu is then recycled into Gln, and subsequently secreted back to neurons, where the cycle can continue [83]. Our results suggest that NetG1+ CAFs may serve both as “hyperexcited pre-synaptic neurons” as well as “astrocyte-like cells”, thus providing a constant supply of microenvironmental Gln and Glu, fueling tumorigenesis. Another hallmark of PDAC tumors is an extremely immunosuppressive microenvironment. Numerous studies have confirmed a major role for CAFs in generating factors, such as secreted cytokines and chemokines [26, 27], that contribute to this environment, as well as physically sequestering anti-tumor immune cells [28]. Furthermore, compared to many other tumor types, PDAC has one of the lowest mutational burdens and display of neoantigens [84, 85], which correlates directly with low response rates to immune checkpoint inhibitors, such as PD-1 [86]. Collectively, these findings suggest that even if cytotoxic T-cells could penetrate the tumor and CAF driven immunosuppression was inhibited, the lack of tumor antigens would render them unable to kill PDAC cells effectively. However, the innate cytotoxic immune cell population—natural killer (NK) cells—can destroy PDAC cells independent of neoantigen presentation, and thus represents an attractive anti-tumor cell type for PDAC therapy [87]. Additionally, NK cells play a natural role in the pancreas, which is to remove activated stromal cells during acute fibrosis in wound healing at the end of a process known as nemosis [88]. Importantly, they are unable to accomplish this in the setting of PDAC, due to the overwhelming numbers of CAFs and immunosuppressive signals, resulting in a situation akin to unresolved chronic wound healing [88, 89]. Thus, reverting the immunosuppressive phenotype of CAFs could allow for NK cell stimulating therapies (e.g. IL-15 supplementation) [90] to more efficiently clear pro-tumoral CAFs as well as PDAC cells. In the present work, we confirmed that CAFs produce the immunosuppressive cytokines TGF-β and GM-CSF, which can expand immunosuppressive myeloid cells [91, 92], as well as chemokines known to attract immunosuppressive immune cells (IL-8 and CCL20) (Fig 3). GM-CSF in particular has been demonstrated to play an important role in PDAC tumorigenesis, where mesenchymal cells or tumor cells secrete and respond to this pro-tumor cytokine, and its inhibition results in decreased tumor burden [93–95], in agreement with our in vivo inhibition of NetG1, where less GM-CSF was produced in mAb treated tumor cultures (Fig 7). Intriguingly, we showed that CAFs, compared to TAs, upregulate IL-15, a potent stimulator of NK cell activity. Functionally, however, IL-15 activity is likely overwhelmed by the greater number of immunosuppressive factors secreted by CAFs, especially TGF-β, as CAFs significantly inhibited NK cell activation and function. Our results support this hypothesis, as CAFs lacking NetG1 failed to protect PDAC cells from NK cell induced death, and that IL-15 was responsible for activation of NK cells. Accordingly, IL-15 expression was detected in fibroblasts within tumor tissue treated with mAb, matching our in vitro results. Moreover, NK cell infiltration increased into murine PDAC tumors treated with mAb compared to IgG control, likely contributing to the observed increase in necrosis. Collectively, the data demonstrate that NetG1 in CAFs shapes the immunosuppressive microenvironment in PDAC, and its ablation reprograms CAFs to an anti-tumor phenotype allowing NK cells to eliminate cancer cells.
In recent years, there has been growing interest in linking microenvironmental metabolites with immune cell function [30], and CAFs in the PDAC microenvironment are important players in this connection [21]. Thus, we postulated that CAF metabolism, driven by NetG1, and by extension, VGlut1 and GS, could also be partially responsible for the immunosuppressive phenotype of CAFs. Accordingly, our results demonstrate a link between the metabolic circuit, NetG1-VGlut1-GS, and suppression of anti-tumor immunity. Taking lessons from the brain and synaptic biology, it is known that cytokines, such as TNF-α, IFN-γ, IL-6, TGF-β, and IL1-β regulate glutamatergic synaptic activity and subsequently Glu release and uptake by neurons and astrocytes [96–98]. Our data suggests that a signaling mechanism may exist in CAFs, whereby disruption of synaptic proteins like NetG1 and VGlut1 result in modulation of cytokine secretion from CAFs, and Glu/Gln cycling may also play a role, as GS inhibition also affected cytokine secretion in CAFs. In sum, our findings illustrate that simply modulating NetG1 expression in CAFs can simultaneously “disarm” two major pro-tumorigenic features of CAFs: metabolic support of PDAC cells and protection from immune cell killing (Fig 5F). These data suggest that synaptic biology in CAFs might be a missing link that controls metabolic and immunosuppressive programs in pancreatic cancer. Indeed, PDAC has been found to have a general dysregulation of axonal guidance genes [99], and neo-neurogenesis and perineural invasion have been shown to play roles in promoting tumorigenesis [100]. Thus, PDAC could have a general neural reprograming that is hijacked for the benefit of the tumor.
Mechanistically, little is known about NetG1 mediated signaling. Thus far, studies have shown that NetG1 physically interacts with its receptor, NGL-1, in trans on post-synaptic neurons [36] and microglia [64] and in cis to leukocyte antigen receptor, LAR, a tyrosine protein phosphatase [35], on presynaptic neurons. Additionally, NetG1 has also been shown to modulate expression levels of the potassium channel KATP [101], but no downstream signaling pathways have been associated with any of these findings. Our results revealed that NetG1 controls the activity of the kinases p38 and AKT, as well as known mediators of these pathways (Fig 4–5). In particular, FRA1 and 4E-BP1 were drastically downregulated and, collectively, these pathways could largely account for the pro-tumor functions in CAFs regulated by NetG1. Previously, p38 MAP kinase activity in CAFs has been shown to promote breast and K-Ras driven lung carcinogenesis, by enhancing the secretion of pro-tumor factors (akin to senescence associated secretory proteins) and activating CAFs to produce hyaluronan networks [102, 103]. Additionally, AKT activity in CAFs, mediated through the actin binding protein Girdin, has been shown to enhance lung tumorigenesis [104]. Downstream to p38 is the AP-1 transcription factor complex, which is a dimer that can be comprised of members of the FOS, JUN, ATF, and MAF families, with FRA1 belonging to FOS family [105]. AP-1 has long been associated with cellular responses to stress (activated by MAP kinases such as p38) and inflammatory signals [106]. FRA1 has been linked to activated CAF methylation signatures in PDAC [107], as well as in certain CAF subtypes in breast, as determined by single cell RNAseq [108]. FRA1 is upregulated in a variety of tumors, including pancreatic cancer [105]. Our results demonstrated that FRA1 regulated cytokine production in CAFs and subsequently NK cell induced death of PDAC cells, supporting the notion that FRA1 acts as an AP-1 subunit. We also found that 4E-BP1, a critical regulator of cap dependent translation [109], was highly expressed in CAFs and lost upon ablation of NetG1 (Fig 5). Previous work has shown that 4E-BP1 plays a major role in fibrogenesis [62], and a protein synthesis inhibitor that blocks mTOR/4E-BP1 in pancreatic CAFs, reverts chemotherapeutic resistance in mice, likely by blocking secretory factors from CAFs [110]. This supports our findings that CAFs lacking 4E-BP1 display a less tumorigenic profile, with the downregulation of Glu/Gln and cytokines. Of note, the identification of a neutralizing antibody allowed us to transiently block NetG1, and we were able to further confirm the signaling network downstream to NetG1 (Fig 7). This positions NetG1 as an attractive target, as the blockade of a single protein inhibits multiple pathways known to sustain pro-tumor functions in CAFs. This was further confirmed by the anti-tumor effects seen in vivo, when genetically targeting NetG1 in CAFs (Fig 2) or blocking it with a neutralizing antibody during tumor development (Fig 7). The developing NetG1 signaling circuit is presented in Figure 5F.
Overall, we have identified a new potential desmoplastic target, NetG1, for the treatment of pancreatic cancer, for which there are no effective therapeutic interventions. By targeting NetG1 in CAFs, we can revert tumor promoting CAFs back into tumor restrictive CAFs, ultimately limiting tumorigenesis in vivo. Finally, NetG1 was established as a viable therapeutic target in pre-clinical murine models. It is our hope that this study provides strong rationale to consider effects of the stroma on cancer development and progression, and that specific normalization of the stroma is a reliable therapeutic option.
Methods
Ethical Collection of Human Samples:
The authors declare that they obtained written informed consent from patients that donated samples for research purposes. The studies conducted herein were in accordance with recognized ethical guidelines. Fox Chase Cancer Center, through the Bio Sample Repository Facility, adheres to the International Society for Biological and Environmental Repositories and National Cancer Institute Best Practices for Biospecimen Resources and participate in the NCI Office of Biorepositories and Biospecimen Resources and Biospecimen Research Network. Further, human sample collections were approved by Fox Chase Cancer Center and MD Anderson Institutional Review Boards.
Reproducibility: Key Resources Table
All information (manufacturer/source, RRID, website) regarding key antibodies, mouse models, chemical and biological reagents, software, and instruments has been compiled into a master “Key Resources” Table.
Cell Culture
The PDAC cells used for the majority of the assays, termed “PDACc” in this study [39], were maintained in media containing 1 part M3 Base F (INCELL, San Antonio, TX) to 4 parts Dulbeco Modified Eagle Medium (DMEM) low glucose (1 g/mL) + sodium pyruvate 110 mg/mL + 1% Penicillin-Streptomycin (10,000 U/mL; Thermo Fisher Scientific, Waltham, MA), supplemented with 5% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA). PANC-1 cells were maintained in DMEM supplemented with 5% FBS and 1% penicillin-streptomycin (P/S) (10,000 U/mL). Patient matched tumor adjacent fibroblasts and CAFs [38, 111] were maintained in DMEM containing 15% FBS, 1% P/S, and 4 mM glutamine. For individual experiments, cells were cultured as described in the figure legends or methods, either with the maintenance conditions outlined above, or in DMEM without serum (serum free), or DMEM without serum and without glutamine (−). NK-92 cells were maintained in Alpha Minimum Essential medium without ribonucleosides and deoxyribonucleosides supplemented with the following components: 2 mM L-glutamine, 1.5 g/L sodium bicarbonate, 0.2 mM inositol, 0.1 mM 2-mercaptoethanol, 0.02 mM folic acid, 400 units/mL IL-2, 12.5% horse serum and 12.5% fetal bovine serum. All cell lines used in experiments were within 7–15 passages after being thawed. Cell line authentication was performed by IDEXX BioResearch (Westbrook, ME) and genetic analysis was performed to determine: 1.) Species of Origin; 2.) Genetic Profile based 16 different allelic markers (AMEL, CSF1PO, D13S317, D16S539, D18S51, D21S11, D3S1358, D5S818, D7S820, D8S1179, FGA, Penta_D, Penta_E, TH01, vWA). All cell lines were regularly tested for mycoplasma, as determined by PCR detection methods, and all lines tested negative. Date of last test: 7–20-20.
Isolation and immortalization of patient derived fibroblasts
Fresh patient pancreatic tissue collected after surgery was digested enzymatically, using our established protocol [38, 111]. Briefly, tissue was initially stored in PBS with antibiotics (P/S, Fungizone) Next, tissue was physically minced with a sterile scalpel, and was digested overnight in collagenase (2mg/mL). After a series of filtration and centrifugation steps, cells were plated on gelatin coated plates for subsequent expansion. Cells were characterized as done previously [37, 38], and fibroblasts were confirmed by a lack of cytokeratins, expression of vimentin, cell shape, ability to create substantive ECM, and the presence of lipid droplets (tumor adjacent fibroblasts) or acquisition of lipid droplets after TGF-β signaling inhibition (CAFs). For the immortalization, cells were retrovirally transduced as previously described [38, 111], with the pBABE-neo-hTERT vector, which was a gift from Dr. Robert Weinberg (Addgene plasmid # 1774). In this study, the procedure above generated the following lines: TA (tumor adjacent fibroblasts) and CAFs (CAF1 and CAF2), which were subsequently used in the 3D system (outlined below) or in vivo (CAF1 CON and NetG1 KO).
Generation of Patient Derived PDAC cell lines from patient derived xenographs (PDX)
Procedure was performed with an approved protocol from Fox Chase Cancer Center Institutional Review Board and was done as previously described [112]. Briefly, tumor fragments were obtained after surgery and washed in RPMI media. The washed fragments were then resuspended in 1:1 mixture of RPMI and Matrigel on ice and were implanted subcutaneously into 5–8 week old C-B17.scid mice (Taconic Bioscience). Once tumors developed to a size of at least 150 mm3, but no larger than 1,500 mm3, mice were sacrificed, and the tumors were collected. These tumors were then digested with a solution containing collagenase/hyaluronidase, and dispase (StemCell Technologies, Cambridge, MA). After several washes, dissociated PDX cells were plated in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, and 2 mM glutamine. All of the following reagents were from Thermo Fisher Scientific (Waltham, MA) and supplemented into the media: F12 nutrient mix, 5 μg/mL insulin, 25 μg/mL hydrocortisone, 125 ng/mL EGF, 250 μg/mL fungizone, 10 μg/mL gentamycin,10 mg/mL nystatin, 11.7 mmol/L cholera toxin, and 5 mmol/L Rho kinase inhibitor Y27632 (Sigma-Aldrich, St. Louis, MO). After 1 month of passages, cells were lysed and subjected to western blot analysis for the detection of NGL-1 expression.
3D microenvironment preparation (Cell-derived ECM (CDM) production)
Production of cell-derived ECM (often referred to as CDM) was conducted as previously described [38, 111]. Briefly, CON or NetG1 KO CAFs (1.25×105/mL) were seeded onto crosslinked (1% (v/v) glutaraldehyde then 1M ethanolamine) 0.2% gelatin coated wells of a 24 well or 6 well plate. These cells were grown in DMEM supplemented with 15% FBS, 1% Penicillin-Streptomycin (10,000 U/mL), 4mM glutamine, and 50 μg/mL L-ascorbic acid (Sigma-Aldrich, St. Louis, MO), provided fresh, daily. After 5 days of ECM production, cultures were used to collect secreted factors (described below for conditioned media preparation) or cells were lysed and extracted from ECMs using 0.5% Triton X-100 (Sigma-Aldrich, St. Louis, MO) supplemented with 20 mM Ammonia Hydroxide (Sigma-Aldrich, St. Louis, MO), followed by extensive washes with PBS. The resulting cell-free ECMs were used as the 3D scaffold environment for subsequent functional assays.
Conditioned media (CM) preparation
Fibroblast CM was prepared by growing cells during 3D CDM production (see above) for 5 days and then replacing media with glutamine/serum free (−) media. Cells were allowed to condition the media for 2 days. The CM was then collected and used immediately or aliquoted and frozen at −80°C until use (for up to one month).
mRNA Microarray
RNA was isolated from two sets of patient matched tumor adjacent fibroblasts and tumor associated fibroblasts (CAFs) at the end of CDM production (see above), which were generated in two technical repetitions. The RNA was processed by the Genomics Facility at Fox Chase Cancer Center to render differentially expressed genes between the two sets of matching tumor adjacent fibroblasts and CAFs. Briefly, an Agilent Technologies Gene Chip (~44,000 genes) was used and RNA was hybridized to the chip and run through the Agilent Technologies Instrument (S/N: US22502680). Raw data were processed using the Agilent Feature Extraction 9.5.1.1 Software, outputting processed data into excel spreadsheets. Quantile normalization was applied to microarray data. We applied limma [113] to identify differentially expressed genes between TA and CAFs. Top differentially expressed genes were selected based on p-value < 0.01 and depicted as a heatmap (z-scores). In order to identify pathways that are enriched between these two cell types, we applied GSEA (preranked with default parameters) on significantly differentially expressed genes (p-value < 0.05) with no fold-change cutoff. To depict the resulting enriched pathways (FDR < 0.25) we applied enrichment map method [114]. The full data set, the top hits with p < 0.01, and GSEA list with FDR <0.25, and complete GSEA analysis for positive and negative enrichment, are all available in Supplemental Table 1, organized by tabs. Data were also deposited into the Gene Expression Omnibus database (accession #GSE156962) as part of the superseries “To determine the differentially expressed genes in cancer associated fibroblasts upon ablation of the glutamatergic synapse protein NetrinG1” (accession #GSE156964).
Quantitative Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR)
The Ambion PureLink kit (Life Technologies, Carlsbad, CA) was used, following manufacturer’s instructions, to extract total RNA cells from the various experimental conditions and tested for RNA quality using a Bioanalyzer (Agilent, Santa Clara, CA). Contaminating genomic DNA was removed using Turbo DNA free from Ambion. RNA concentrations were determined with a spectrophotometer (NanoDrop; Thermo Fisher Scientific, Waltham, MA). RNA was reverse transcribed (RT) using Moloney murine leukemia virus reverse transcriptase (Ambion, Life Technologies, Carlsbad, CA) and a mixture of anchored oligo-dT and random decamers (Integrated DNA Technologies, Skokie, Illinois). Two reverse-transcription reactions were performed for each biological replicate using 100 and 25 ng of input RNA. Taqman assays were used in combination with Taqman Universal Master Mix and run on a 7900 HT sequence detection system (Applied Biosystems, Foster City, CA). Cycling conditions were as follows: 95°C, 15 minutes, followed by 40 (two-step) cycles (95°C, 15 s; 60°C, 60 s).
Ct (cycle threshold) values were converted to quantities (in arbitrary units) using a standard curve (four points, four fold dilutions) established with a calibrator sample. Values were averaged per sample and standard deviations were from a minimum of two independent PCRs. Polymerase (RNA) II (DNA directed) polypeptide F (POLR2F) was used as internal control. Identification numbers of commercial assays (Life Technologies, Carlsbad, CA) and sequences of the NTNG1 primers (FW and RV) and probe for the POLR2F assay are as follows:
GGAAATGCAAGAAGAATTATCAGG (forward),
GTTGTCGCAGACATTCGTACC (reverse),
6fam-CCCCATCATCATTCGCCGTTACC-bqh1 (probe)
For other genes, the same RNA extraction was performed, but RT reaction was performed using High Capacity cDNA RT Kit and qRT-PCR reactions were performed in a barcoded 96 well plate using Power SYBR Green PCR master mix, gene specific primers, and template DNA, using the StepOnePlus Real Time-PCR system. All reagents and instruments were from Applied Biosystems/Thermo Fisher Scientific, Waltham MA. All primer sequences were obtained from PrimerBank [115] and are available in the Key Resources Table. Relative mRNA expression for each gene was calculated by the ΔΔCt method, with 18s used as a housekeeping control, and then normalized to CON CAFs to determine fold change differences in gene expression.
SDS-PAGE and Western Blot Analysis
Cell lysates were prepared by adding 150 μL standard RIPA buffer to 6 well plates containing 106 cells. The lysates were homogenized using a sonicator. After centrifugation, the supernatant was collected and 30 μg of lysate (determined by the Bradford Assay) was diluted in 2x loading buffer (Bio-Rad, Hercules, CA) and was loaded onto 4–20% polyacrylamide gels (Bio-Rad, Hercules, CA). The gels were run at 70 volts for 2 hours, and transferred to PVDF membranes (Millipore, Burlington, MA) using the semi-dry transfer system (Bio-Rad, Hercules, CA). Membranes were blocked in 5% nonfat milk in 0.1% Tween in tris buffered saline (TBST) for 1 hour at room temperature (RT), and primary antibodies were incubated with the membranes overnight at 4°C. Next, membranes were washed 5 times with 0.1% TBST and then secondary antibodies linked to HRP were diluted in 5% nonfat milk in 0.1% TBST and were applied for 2 hours at RT. Membranes were washed again as before, and incubated for 5 minutes in Immobilon Western Chemiluminescent HRP substrate (Millipore, Burlington, MA). Membranes were developed using film and scanned to create digital images. Primary and secondary antibodies are listed in the Reagents table.
Key Resources Table.
| Antibodies | ||
| Reagent or Resource | Source | Identifier |
| Netrin G1 Antibody (D-2) (1:100) | Santa Cruz | RRID:AB_10707668 |
| Netrin G1 Antibody (N2C1) (1:100) | Genetex | RRID:AB_10625511 |
| Glutaminase antibody [EP7212] (1:100,000) | Abcam | #ab156876 |
| Glutamine Synthetase antibody (1:50,000) | Abcam | #ab73593 |
| Histone H3 (D1H2) XP® antibody (1:10,000) | Cell Signaling Technology | #4499 |
| EpCam (MOC-31) antibody (1:200) | Novus Biologicals | #NBP2–22388 |
| pan-cytokeratin (AE1/AE3) antibodies (1:200) | DAKO | #M3515 |
| CD-70 antibody (1:200) | Biolegend | RRID:AB_2561429 |
| Vimentin antibody (1:200) | Abcam | #ab92547 |
| GAPDH antibody (1:10,000) | Abcam | RRID:AB_307275 |
| VGlut1 antibody (1:100) | Thermo Fisher Scientific | RRID:AB_2533843 |
| Fibronectin antibody (1:200) | Sigma-Aldrich | RRID:AB_476961 |
| α-SMA antibody (1:300) | Sigma-Aldrich | RRID: AB_476701 |
| SNAKA51 Antibody (45 μg/mL) | M Humphries | N/A |
| p-FAK [Y397] Antibody (1:200) | Thermo Fisher Scientific | RRID:AB_2533701 |
| Ki67 Antibody (1:500) | Abcam | RRID:AB_443209 |
| Granzyme B Antibody (1:50) | ebioscence | RRID:AB_1659718 |
| IFNγ secretion assay detection kit-PE | Miltenyi Biotec | #130–054–20 |
| NKp80-APC | Biolegend | RRID:AB_2044041 |
| CD56-BUV395 | BD Horizon | RRID:AB_2687886 |
| CD69-Pac Blue | Biolegend | RRID:AB_493667 |
| CD3-Cy7APC | BD Pharmigen | RRID:AB_1645475 |
| donkey anti-rabbit secondary antibody- Cy5 conjugation (1:200) | Jackson ImmunoResearch | RRID:AB_2340607 |
| donkey anti-mouse secondary antibody- TRITC conjugation (1:200) | Jackson ImmunoResearch | RRID:AB_2340766 |
| Human IL-15 Antibody (10 μg/mL) | R&D Systems | RRID:AB_2124577 |
| Anti-Mouse IgG (whole molecule)–Peroxidase antibody (1:5,000) | Sigma-Aldrich | RRID:AB_258431 |
| Goat anti-Rabbit IgG (H+L) Secondary Antibody, HRP (1:10,000) | Thermo Fisher Scientific | RRID:AB_2533967 |
| p-p38 (Thr180/Tyr182) (1:250) | Cell Signaling Technology | RRID: AB_2139682 |
| p38 (1:2000) | Cell Signaling Technology | RRID: AB_10999090 |
| p-AKT (Ser473) (1:1000) | Cell Signaling Technology | #4060 |
| Pan AKT (1:2500) | Cell Signaling Technology | RRID: AB_915783 |
| p-4E-BP1 (Thr37/46) (1:1000) | Cell Signaling Technology | RRID: AB_560835 |
| 4E-BP1 (1:2000) | Cell Signaling Technology | RRID: AB_2097841 |
| FRA1 (1:2000) | Cell Signaling Technology | RRID: AB_10557418 |
| FRA2 (1:2000) | Cell Signaling Technology | RRID: AB_2722526 |
| FosB (1:1000) | Cell Signaling Technology | RRID: AB_2106903 |
| cFOS (1:1000) | Cell Signaling Technology | RRID: AB_2247211 |
| c-Jun (1:1000) | Cell Signaling Technology | RRID: AB_2130165 |
| JunB (1:1000) | Cell Signaling Technology | RRID: AB_2130002 |
| p-mTOR (Ser2448) (1:500) | Cell Signaling Technology | #5536 |
| p-FOXO1 (Ser256) (1:1000) | Cell Signaling Technology | RRID: AB_2800035 |
| FOXO1 (1:1000) | Cell Signaling Technology | RRID: AB_2106495 |
| p-FOXO3 (Ser294) (1:1000) | Cell Signaling Technology | #5538 |
| FOXO3 (1:1000) | Cell Signaling Technology | RRID: AB_836876 |
| p-GSK3β (Ser9) (1:500) | Cell Signaling Technology | #9323 |
| Mouse IgG1 | Bio X Cell | #BE-0083 |
| IL-15 (1:50) | Lifespan Biosciences | # LS-B10080 |
| NK1.1 (1:50) | Invitrogen-Thermo Fisher Scientific | # MA1–70100 |
| Podoplanin/PDPN (1:100) | Biolegend | # 916606 |
| Bacterial Strains | ||
| Reagent or Resource | Source | Identifier |
| One Shot Stbl3 Chemically Competent E. coli | Thermo Fisher Scientific | # C737303 |
| Chemicals | ||
| Reagent or Resource | Source | Identifier |
| L-glutamate | Abcam | #ab120049 |
| MK-2206 | Cayman Chemical | #11593 |
| SB 202190 | Tocris Bioscience | #1264 |
| Sulfosalicylic acid | Sigma-Aldrich | #S-7422 |
| L-Ascorbic acid | Sigma-Aldrich | #A92902–100G |
| 25% Glutaraldehyde Solution in water | Sigma-Aldrich | #G6257–1L |
| Ethanolamine | Sigma-Aldrich | #E9508–1L |
| Polybrene | Santa Cruz Biotechnology | # sc-134220 |
| L-Methionine sulfoximine (MSO) | Sigma-Aldrich | #M5379–250MG |
| D-GLUCOSE (U-13C6) | Cambridge Isotope Laboratories | #CLM-1396-PK |
| SODIUM PYRUVATE (13C3) | Cambridge Isotope Laboratories | #CLM-2440-PK |
| MOX reagent | Thermofisher | # TS-45950 |
| N-tertbutyldimethylsilyl-N-methyltrifluoroacetamide with 1% tert-butyldimethylchlorosilane | Sigma-Aldrich | # 375934 |
| Fisherbrand™ Blood Bank Saline 0.85% - 0.9% | Thermo Fisher Scientific | #p-4527840 |
| Cell Culture | ||
| Reagent or Resource | Source | Identifier |
| FBS | Atlanta Biologicals | #S11150 |
| DMEM | Corning | #50–013-PB |
| L-Glutamine | Corning | #25–005CI |
| Penicillin/Streptomycin | Corning | #30–002-CI |
| DMEM low glucose | Corning | #10–014 |
| M3 Base F | INCELL | #M300F-500 |
| Alpha MEM | Corning | # 10–022-CV |
| IL-2 | Fox Chase Cancer Center | N/A |
| IL-12 | Fox Chase Cancer Center | N/A |
| Cell Lines | ||
| Reagent or Resource | Source | Identifier |
| hTERT-HPNE E6/E7/K-RasG12D (KRAS) | ATCC | RRID:CVCL_C469 |
| hTERT-HPNE (hTERT) | ATCC | RRID:CVCL_C466 |
| hTERT-HPNE E6/E7 (E6/E7) | ATCC | RRID:CVCL_C467 |
| KPC3 | This paper | N/A |
| KPC4B | This paper | N/A |
| Panc-1 | ATCC | RRID:CVCL_0480 |
| Cancer associated fibroblasts (patient derived) | This Paper | N/A |
| Tumor adjacent fibroblasts (patient derived) | This Paper | N/A |
| NK-92 cells | ATCC | RRID:CVCL_2142 |
| 293T Cells | ATCC | RRID:CVCL_0063 |
| Phoenix-Amphotropic (φNX) | ATCC | RRID:CVCL_H716 |
| Organisms/Strains (Mice) | ||
| Reagent or Resource | Source | Identifier |
| C57BL/6 | Fox Chase Cancer Center | N/A |
| KPC model: LSL-Kras(G12D/+) ; Trp53(flox/flox) ;Pdx-1-Cre | Kerry Campbell, Igor Astsaturov | Original Mice: PMID: 15894267 |
| KC Model: LSL-Kras(G12D/+); Pdx-1-Cre | Kerry Campbell | Original Mice: PMID: 14706336 |
| C.B17 scid | Taconic Bioscience | #CB17SC |
| Nod SCID Gamma (NSG) | Jackson Laboratories | #005557 |
| Plasmids | ||
| Reagent or Resource | Source | Identifier |
| Plv-Cmv-Puro | Fox Chase Cancer Center | N/A |
| LentiCRISPR v2 | Addgene (Sanjana et al., 2014) | #52961 |
| psPAX2 | Addgene | #12260 |
| pCMV-VSV-G | Addgene | #8454 |
| pBABE-neo-hTERT | Addgene | #1774 |
| CRISPRi-Puro | Modified from Addgene; (Thakore et al., 2015) | #71236 |
| Critical Commercial Reagents | ||
| Reagent or Resource | Source | Identifier |
| PureLink™ RNA Mini Kit | Thermo Fisher Scientific | #12183025 |
| SuperScript III First Strand Synthesis System | Thermo Fisher Scientific | #18080–051 |
| Phusion High-Fidelity PCR Master Mix | Thermo Fisher Scientific | # F-531L |
| 2x Laemmli buffer | BioRad | #1610737 |
| Sytox Blue | Thermo Fisher Scientific | #S11348 |
| Odyssey Blocking Buffer (PBS) | LI-COR | #927–40000 |
| DeadEnd™ Fluorometric TUNEL System | Promega | #G3250 |
| Immobilon Western Chemiluminescent HRP Substrate | EMD Millipore | #WBKLS0500 |
| Ultrafree-MC-GV centrifugal filters | Millipore | #UFC30GVNB |
| Dnase I | Thermo Fisher Scientific | #EN0525 |
| X-tremeGene9 | Sigma-Aldrich | #6365787001 |
| U-PLEX Biomarker Group 1 (hu) Assays, SECTOR (1 PL) | Meso Scale Discoveries | #K15067L-1 |
| DuoSet ELISAs (IL-15, IFN-β, TGF-β, IL-6, IL-8, GM-CSF, mTGF-β, mGM-CSF, mIL-6) | R&D Systems | #DY247–05, #DY814–05, #DY240–05, #DY206–05, #DY208–05, #DY215–05, #DY1679–05, #DY415–05, # DY406–05 |
| DuoSet ELISA Ancillary Reagent Kit 1 | R&D Systems | DY007 |
| DuoSet ELISA Ancillary Reagent Kit 2 | R&D Systems | DY008 |
| Sample Activation Kit 1 | R&D Systems | DY010 |
| Immobilon-P PVDF Membrane | EMD Millipore | IPVH00010 |
| SYBR Green | Invitrogen | S7563 |
| Collagenase/Hyaluronidase | StemCell Technologies | #07912 |
| Dispase | StemCell Technologies | #7446 |
| Magnevist | Bayer | 50419–188–58 |
| SiteClick Qdot labeling Kits (525–655) | Thermo Fisher Scientific | #S10449, S10450, S10451, S10469, S10452, S10453 |
| Truseq stranded mRNA library kit | Illumina, Inc. | # 20020595 |
| SuperscriptII reverse transcriptase | Thermo Fisher Scientific | #18064014 |
| SPRIselect beads | Beckman Coulter | # B23318 |
| HiSeq rapid SBS kit v2 (50 cycle) | Illumina, Inc. | # FC-402–4022 |
| Qubit 1x dsDNA HS assay kit | Thermo Fisher Scientific | #Q33216 |
| High sensitivity DNA kit | Agilent Technologies | # 5067–4626 |
| Glutamate Assay Kit | Abcam | # ab83389 |
| Glutamine ELISA Kit | MyBioSource | # MBS732543 |
| High Capacity cDNA Reverse Transcription Kit | Thermo Fisher Scientific | #4368814 |
| PowerSYBR Green PCR Master Mix | Thermo Fisher Scientific | #4367659 |
| MicroAmp Fast Optical 96 well Reaction Plate with Barcode (0.1 mL) | Thermo Fisher Scientific | #4346906 |
| ProLong™ Gold Antifade Mountant | Thermo Fisher Scientific | #P10144 |
| Dextran, Fluorescein, 40,000 MW, Anionic, Lysine Fixable | Thermo Fisher Scientific | #D1845 |
| 5-(N-Ethyl-N-isopropyl)amiloride [EIPA] | Tocris | # 3378 |
| alamarBlue reagent | Bio-Rad | # BUF012A |
| Envision+ polymer system | Dako | #K4000 (anti-mouse) #K4002 (anti-rabbit) |
| Oligonucleotides | ||
| Reagent or Resource | Source | Identifier |
| GFP Xba FW: TAGGTATCTAGAATGGTGAGCAAGGGCGAGG | This paper | N/A |
| GFP Xho RV: TAGGTACTCGAGCTTGTACAGCTCGTCCATGCC | This paper | N/A |
| mCherry Xba FW: TAGGTATCTAGAatggtgagcaagggcgag | This paper | N/A |
| mCherry Xho RV: TAGGTACTCGAGttacttgtacagctcgtccatgc | This paper | N/A |
| NTNG1 qPCR FW: GGAAATGCAAGAAGAATTATCAGG | This paper | N/A |
| NTNG1 qPCR RV: GTTGTCGCAGACATTCGTACC | This paper | N/A |
| POLR2F qPCR internal control: 6fam-CCCCATCATCATTCGCCGTTACC-bqh1 | This paper | N/A |
| eGFP gRNA 1.1: CACCGCATGTGATCGCGCTTCTCGT | This paper | N/A |
| eGFP gRNA 1.2 : AAACACGAGAAGCGCGATCACATGC | This paper | N/A |
| NetrinG1 gRNA 1.1: CACCGGCGTCCAGACCAAATGATCC | This paper | N/A |
| NetrinG1 gRNA 1.2: AAACGGATCATTTGGTCTGGACGCC | This paper | N/A |
| NetrinG1 gRNA 2.1: CACCGTGATGCGAGTACCCCTGAGC | This paper | N/A |
| NetrinG1 gRNA 2.2: AAACGCTCAGGGGTACTCGCATCAC | This paper | N/A |
| NGL1 gRNA1.1: CACCGAACCTGCGTGAGGTTCCGGA | This paper | N/A |
| NGL1 gRNA1.2: AAACTCCGGAACCTCACGCAGGTTC | This paper | N/A |
| NGL1 gRNA 2.1: CACCGTGCCATCCGGAACCTCACGC | This paper | N/A |
| NGL1 gRNA 2.2: AAACGCGTGAGGTTCCGGATGGCAC | This paper | N/A |
| murine NGL1 gRNA 1.1: CACCGCGATTTCAATGGTTCGAATA | This paper | N/A |
| murine NGL1 gRNA 1.2: AAACTATTCGAACCATTGAAATCGC | This paper | N/A |
| murine NGL1 gRNA 2.1: CACCGAACCTTCGTGAAGTTCCGGA | This paper | N/A |
| murine NGL1 gRNA 2.2: AAACTCCGGAACTTCACGAAGGTTC | This paper | N/A |
| NetrinG1 CRISPRi gRNA 1.1: CACCGGCGCCCCGAGGTCGTGGAG | (Horlbeck et al., 2016 | N/A |
| NetrinG1 CRISPRi gRNA 1.2: AAACCTCCACGACCTCGGGGCGCCC | (Horlbeck et al., 2016 | N/A |
| NetrinG1 CRISPRi gRNA 2.1: CACCGACAGCAACAGCGAGCGGGA | (Horlbeck et al., 2016 | N/A |
| NetrinG1 CRISPRi gRNA 2.2: AAACTCCCGCTCGCTGTTGCTGTCC | (Horlbeck et al., 2016 | N/A |
| NetrinG1 CRISPRi gRNA 3.1: CACCGCGAGAGCCGGAAAGAGGAG | (Horlbeck et al., 2016) | N/A |
| NetrinG1 CRISPRi gRNA 3.2: AAACCTCCTCTTTCCGGCTCTCGCC | (Horlbeck et al., 2016 | N/A |
| VGlut1 CRISPRi gRNA 1.1: CACCGAGAGAGAGTGGAGCCGGGT | (Horlbeck et al., 2016 | N/A |
| VGlut1 CRISPRi gRNA 1.2: AAACACCCGGCTCCACTCTCTCTCC | (Horlbeck et al., 2016) | N/A |
| VGlut1 CRISPRi gRNA 2.1: CACCGGCTCCGCTCGGGGGGAAGG | (Horlbeck et al., 2016) | N/A |
| VGlut1 CRISPRi gRNA 2.2: AAACCCTTCCCCCCGAGCGGAGCCC | (Horlbeck et al., 2016) | N/A |
| VGlut1 CRISPRi gRNA 3.1: CACCGAGAGACCCAGAAGTGGTGG | (Horlbeck et al., 2016) | N/A |
| VGlut1 CRISPRi gRNA 3.2: AAACCCACCACTTCTGGGTCTCTCC | (Horlbeck et al., 2016) | N/A |
| GS CRISPRi 1.1: CACCGGCTCTGCAGAGTCGAGAGT | (Horlbeck et al., 2016) | N/A |
| GS CRISPRi 1.2: AAACACTCTCGACTCTGCAGAGCCC | (Horlbeck et al., 2016) | N/A |
| GS CRISPRi 2.1: CACCGGAGCGTGTGAGCAGTACTG | (Horlbeck et al., 2016) | N/A |
| GS CRISPRi 2.2: AAACCAGTACTGCTCACACGCTCCC | (Horlbeck et al., 2016) | N/A |
| GS CRISPRi 3.1: CACCGGACGGGTCCAAGCCACCAG | (Horlbeck et al., 2016) | N/A |
| GS CRISPRi 3.2: AAACCTGGTGGCTTGGACCCGTCCC | (Horlbeck et al., 2016) | N/A |
| 4E-BP1 CRISPRI 1.1: CACCGCACAGGAGACCATGTCCGG | (Horlbeck et al., 2016) | N/A |
| 4E-BP1 CRISPRI 1.2: AAACCCGGACATGGTCTCCTGTGC | (Horlbeck et al., 2016) | N/A |
| 4E-BP1 CRISPRI 2.1: CACCGAGGGCAGCGAGAGGTTCGC | (Horlbeck et al., 2016) | N/A |
| 4E-BP1 CRISPRI 2.2: AAACGCGAACCTCTCGCTGCCCTC | (Horlbeck et al., 2016) | N/A |
| 4E-BP1 CRISPRI 3.1: CACCGACCGGGTGTCCAGGCTCAA | (Horlbeck et al., 2016) | N/A |
| 4E-BP1 CRISPRI 3.2: AAACTTGAGCCTGGACACCCGGTC | (Horlbeck et al., 2016) | N/A |
| FRA1 CRISPRi 1.1: CACCGAAGTCTCGGAACATGCCCG | (Horlbeck et al., 2016) | N/A |
| FRA1 CRISPRi 1.2: AAACCGGGCATGTTCCGAGACTTC | (Horlbeck et al., 2016) | N/A |
| FRA1 CRISPRi 2.1: CACCGGGCGGCTCTGCGGGGTACA | (Horlbeck et al., 2016) | N/A |
| FRA1 CRISPRi 2.2: AAACTGTACCCCGCAGAGCCGCCC | (Horlbeck et al., 2016) | N/A |
| FRA1 CRISPRi 3.1: CACCGGGCATGTTCCGAGACTTCG | (Horlbeck et al., 2016) | N/A |
| FRA1 CRISPRi 3.2: AAACCGAAGTCTCGGAACATGCCC | (Horlbeck et al., 2016) | N/A |
| VGlut1 qPCR FW: CTGGGGCTACATTGTCACTCA | Primer Bank | N/A |
| VGlut1 qPCR RV: GCAAAGCCGAAAACTCTGTTG | Primer Bank | N/A |
| GS qPCR FW: AAGAGTTGCCTGAGTGGAATTTC | Primer Bank | N/A |
| GS qPCR RV: AGCTTGTTAGGGTCCTTACGG | Primer Bank | N/A |
| IL6 qPCR FW: ACTCACCTCTTCAGAACGAATTG | Primer Bank | N/A |
| IL6 qPCR RV: CCATCTTTGGAAGGTTCAGGTTG | Primer Bank | N/A |
| IL8 qPCR FW: TTTTGCCAAGGAGTGCTAAAGA | Primer Bank | N/A |
| IL8 qPCR RV: AACCCTCTGCACCCAGTTTTC | Primer Bank | N/A |
| IL15 qPCR FW: CAGTGCAGGGCTTCCTAAAAC | Primer Bank | N/A |
| IL15 iso1 qPCR RV: TGGGGTGAACATCACTTTCCG | Primer Bank | N/A |
| TGFB1 qPCR FW: GGCCAGATCCTGTCCAAGC | Primer Bank | N/A |
| TGFB1 qPCR RV: GTGGGTTTCCACCATTAGCAC | Primer Bank | N/A |
| GM-CSF qPCR FW: TCCTGAACCTGAGTAGAGACAC | Primer Bank | N/A |
| GM-CSF qPCR RV: TGCTGCTTGTAGTGGCTGG | Primer Bank | N/A |
| 18s qPCR FW: GGCCCTGTAATTGGAATGAGTC | Primer Bank | N/A |
| 18s qPCR RV: CCAAGATCCAACTACGAGCTT | Primer Bank | N/A |
| Instruments | ||
| Reagent or Resource | Source | Identifier |
| Agilent Technologies Gene Chip | Agilent | http://www.genomics.agilent.com |
| Agilent Technologies Microarray Instrument | Agilent | http://www.genomics.agilent.com |
| Confocal spinning disk Ultraview | Perkin-Elmer | http://www.perkinelmer.com/ |
| NanoDrop 1000 | Thermo Fisher Scientific | http://www.thermofisherscientific.com |
| FlourChem E Imaging System | Protein Simple | https://www.proteinsimple.com/ |
| Biochrom 30 amino acid analyzer | Biochrom | http://www.biochrom.co.uk/ |
| Nikon Eclipse TE2000U | Nikon | http://www.nikon.com/ |
| MSD SECTOR Imager 2400 | Meso Scale Discoveries | https://www.mesoscale.com |
| Spark® Multimode Microplate | Tecan | https://www.tecan.com |
| CoolSNAP HQ CCD Camera | Photometrics | https://www.photometrics.com/ |
| Transblot SD Semi Dry Transfer System | BioRad | www.bio-rad.com |
| Bruker DRX 300 | Bruker | www.bruker.com |
| DB-35MS column | Agilent Technologies | www.agilent.com |
| Agilent 7890A gas chromatograph | Agilent Technologies | www.agilent.com |
| Agilent 5997B mass spectrometer | Agilent Technologies | www.agilent.com |
| HiSeq2500 System | Illumina, Inc. | www.illumina.com |
| Agilent 2100 bioanalyzer | Agilent Technologies | www.agilent.com |
| Qubit 3.0 flurometer | Thermo Fisher Scientific | http://www.thermofisherscientific.com |
| StepOnePlus Real Time-PCR System | Applied Biosystems/Thermo Fisher Scientific | https://www.thermofisher.com/order/catalog/product/4376600#/4376600 |
| Nikon Eclipse Ti2-E Inverted Microscope Imaging System | Nikon | https://www.microscope.healthcare.nikon.com/products/inverted-microscopes/eclipse-ti2-series |
| Nikon A1 camera | Nikon | https://www.microscope.healthcare.nikon.com/products/confocal-microscopes/a1hd25-a1rhd25 |
| Software/Algorithms | ||
| Reagent or Resource | Source | Identifier |
| Agilent Feature Extraction 9.5.1.1 Software | Agilent | http://www.genomics.agilent.com/en/Microarray-Scanner-Processing-Hardware/Feature-Extraction-Software/?cid=AG-PT-144&tabId=AG-PR-1050 (RRID:SCR_014963) |
| ImageJ | NIH | https://imagej.nih.gov/ij/ (RRID:SCR_003070) |
| Metamorph 7.8.1.0 Image Anaylsis Software | Molecular Devices | http://www.moleculardevices.com/Products/Software/Meta-Imaging-Series/MetaMorph.html (RRID:SCR_002368) |
| Volocity 3D Image Analysis Software | Perkin Elmer | http://www.perkinelmer.com/pages/020/cellularimaging/products/volocity.xhtml (RRID:SCR_002668) |
| SMIA-CUKIE | Franco-Barraza et al., 2017 | https://github.com/cukie/SMIA (RRID:SCR_014795) |
| FlowJo | FlowJo LLC | https://www.flowjo.com/solutions/flowjo (RRID:SCR_008520) |
| ParaVision | Bruker | http://www.bruker.com/service/support-upgrades/software-downloads/mri.html (RRID:SCR_001964) |
| OrientationJ plugin for ImageJ | Rezakhaniha et al, 2012 | http://bigwww.epfl.ch/demo/orientation/ (RRID:SCR_014796) |
| MSD Workbench 4.0 software | Meso Scale Discoveries | https://www.mesoscale.com |
| Graph Pad Prism 7.0 software | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ (RRID:SCR_000306) |
| ParaVision | Bruker | https://www.bruker.com (RRID:SCR_001964) |
| Adobe Photoshop CS6 13.0.1 | Adobe | http://www.adobe.com/fr/products/photoshop.html (RRID: SCR_014199) |
| Nuance 3.0.1 | Caliper Life Sciences/Perkin Elmer | https://www.moleculardevices.com/Products/Software/Meta-Imaging-Series/MetaMorph.html (RRID: SCR_002368) |
| Illumina base space | Illumina, Inc. | (https://basespace.illumina.com |
Indirect Immunofluorescence
CAFs were allowed to generate ECM (3D microenvironment) as outlined above (CDM production). After 5 days of ECM production, cells on coverslips were fixed and simultaneously permeabilized for 5 minutes at RT in 4% paraformaldehyde containing 0.5% TritonX-100 and 150 mM sucrose and allowed to continue fixation, using the same preparation lacking triton, for an additional 20 minutes at RT. Fixed/permeabilized cells were then blocked for 1 hour at RT with Odyssey Blocking Buffer (PBS base; Li-Cor, Lincoln, NE). Cells were stained for 1 hour at RT with the following antibodies diluted in blocking buffer: fibronectin (Sigma-Aldrich, St. Louis, MO) and α-smooth muscle actin (α-SMA) (Sigma-Aldrich, St. Louis, MO). After 1 hour incubation, cells were washed 3 times with 0.05% Tween in phosphate buffered saline (PBST), and secondary antibodies diluted in blocking buffer were applied for 1 hour at RT (TRITC for α-SMA; Cy5 for fibronectin, Jackson ImmunoResearch, West Grove, PA). Next, cells were washed 3 times with 0.05% PBST, and nuclei were counter stained with SYBR green (1:10,000; Invitrogen, Carlsbad, CA). Stained coverslips were imaged using Spinning disk confocal microscopy (Perkin Elmer, Waltham, MA), captured by a CoolSNAP CCD Camera (Photometrics, Tucson, AZ) attached to an Eclipse 2000-S inverted microscope (Nikon, Tokyo, Japan). Images were exported from Volocity 3D Image Analysis Software (Perkins Elmer, Waltham, MA). The integrated intensity of α-SMA was quantified using Metamorph Image Analysis Software (Molecular Devices, San Jose, CA). Fibronectin alignment was quantified as described below.
ECM fiber orientation analysis
ECM fiber orientation, indicative of alignment, was performed as previously described [38, 111]. Briefly, confocal images of fibronectin in CAF generated ECM’s were analyzed for fiber orientation using the OrientationJ plugin [116] for ImageJ. Fiber angles generated by the plugin were normalized to fit within the angle range of −90° to +90°. Next, normalized angles were plotted in Excel to generate orientation curves (alignment output at every angle). Finally, the percentage of fibers between −15° to +15° was plotted as a measurement indicative of ECM fiber alignment. In general, tumor adjacent fibroblasts tend to have disorganized −15° to +15° angle fibers (<50%), while CAFs tend to have fibers with angles >50%.
PDAC Cell Proliferation Assay
CON or NGL-1 KO PDACc cells (2×104) were seeded in CAF-derived 3D ECMs and allowed to proliferate overnight. Ki67, a nuclear marker of proliferation, and nuclei (DAPI) were stained by IF (as outlined above). The number of Ki67 positive cells/nuclei per field was calculated using Metamorph image analysis software.
CRISPR/Cas9 Mediated Knockout of NetG1 and NGL-1
Knockout of NetG1 in CAFs, or NGL-1 in PDACc or PANC-1 cells, was achieved by introducing a frameshift mutation in coding regions of each gene, induced by cuts made by CRISPR/Cas9 and gene specific gRNAs introduced into cells by lentiviral transduction.
gRNA design:
Using the MIT Optimized CRISPR Design website: http://crispr.mit.edu/, the first 200 bp of exon 5 for NetG1 (first common exon for all splice variants) and first 220 bp of the NGL-1 gene were used to generate gRNAs. The top two hits for each gene were selected and designed to include BsmBI/Esp3I overhangs (bold, underline, italics), the specific gRNA sequence (plain text). G (italics, underline) was added to the .1 gRNAs (and corresponding C on the .2 gRNA) to optimize its expression for the human U6 promoter. The gRNA sequences for NerinG1 were: 1.1, CACCGGCGTCCAGACCAAATGATCC; 1.2, AAACGGATCATTTGGTCTGGACGCC; 2.1, CACCGTGATGCGAGTACCCCTGAGC; and 2.2, AAACGCTCAGGGGTACTCGCATCAC. The gRNA sequences for NGL-1 were: 1.1, CACCGAACCTGCGTGAGGTTCCGGA; 1.2, AAACTCCGGAACCTCACGCAGGTTC; 2.1 CACCGTGCCATCCGGAACCTCACGC; and 2.2 AAACGCGTGAGGTTCCGGATGGCAC. Empty vector or Nontargeting gRNA against eGFP were used as a control, the eGFP gRNA sequences used were: 2.1, CACCGCATGTGATCGCGCTTCTCGT; and 2.2, AAACACGAGAAGCGCGATCACATGC.
Generation of Lentiviral Vectors:
Designed oligos were ordered from Integrated DNA Technologies (Skokie, Illinois) and cloned into the LentiCRISPR v2 vector, as previously published (a gift from Feng Zhang; Addgene plasmid # 52961; [38, 117]. Briefly, oligos were phosphorylated using T4 PNK (New England Biolabs, Ipswich, MA) and annealed in a thermal cycler. Annealed oligos were cloned into a gel purified (GeneJet Gel Extration Kit; Thermo Fisher Scientific, Waltham, MA), dephosphorylated (Fast AP; Thermo Fisher Scientific, Waltham, MA), and digested vector (Fast Digest Esp3I; Thermo Fisher Scientific, Waltham, MA). Annealed oligos were ligated into the digested vector using quick ligase (New England Biolabs, Ipswich, MA). The ligated vectors were transformed into Stbl3 strain of Escherichia coli (Thermo Fisher Scientific, Waltham, MA) and selected with 100 μg/mL ampicillin on LB/agar plates. The following day, single colonies of bacteria were screened by colony PCR using a forward primer for the U6 promoter (GAGGGCCTATTTCCCATGATT) and the reverse primer sequence (gRNA x.2) for each gene. Positive clones were subjected to sequencing and subsequently expanded for plasmid purification and lentivirus production.
Lentiviral Transduction:
To package the LentiCRISPRv2 plasmid into functional virus particles, 10 μg LentiCRISPRv2, 5 μg psPAX2 (psPAX2 was a gift from Didier Trono; Addgene plasmid # 12260) and 2 μg VSVg were mixed with 30 μL X-tremeGene9 transfection reagent (Sigma-Aldrich, St. Louis, MO) in 1 mL of serum free/antibiotic free DMEM and held at RT for 30 minutes. The transfection mixture was added dropwise to 293T cells containing 5 mL serum free/antibiotic free DMEM and the cells were incubated at 37°C overnight. The next day, the media was replaced with complete DMEM media. On days 2 and 4 post-transfection, the media with viral particles was collected and syringe filtered through a 0.45 μM filter (Millipore, Burlington, MA). The filtered viral supernatant was used immediately to transduce target cells (PDACc/PANC-1 or fibroblasts) with 10 μg/mL polybrene (Santa Cruz Biotechnology, Dallas, Texas) or stored at −80°C for use in the future. Target cells were selected with puromycin (2 μg/mL for CAFs, 5 μg/mL PANC-1, 12 μg/mL KRAS) for 2 weeks. Cells that survived selection were used for future experiments.
Generation of RFP+ and GFP+ CRISPR/Cas9 transduced cells:
eGFP and mCherry (RFP) were PCR amplified using Phusion HF polymerase master mix (Thermo Fisher Scientific, Waltham, MA) to add overhangs compatible with restriction enzymes. Then cloned into XbaI/XhoI (New England Biolabs, Ipswich, MA) digested, Fast AP dephosphorylated pLV-CMV-H4-puro vector (kindly provided by Dr. Alexey Ivanov, West Virginia University School of Medicine, Morgantown, WV, USA). Empty vector transduced (CON) or NetG1 KO CAF were transduced with pLV-CMV-H4-puro-GFP and the CON or NGL-1 KO PDACc were transduced with pLV-CMV-H4-puro-RFP, with 10 μg/mL polybrene. Cells were selected by flow cytometry. The selected cells were used for future experiments.
CRISPR interference (CRISPRi) mediated knockdown of target genes
We generated a lentiviral vector named CRISPRi-Puro (modified from pLV hU6-sgRNA hUbC-dCas9-KRAB-T2a-Puro; Addgene Plasmid #71236; a gift from Charles Gersbach) [118] containing dead Cas9 (dCas9) fused to Krueppel-associated box (KRAB) domain, gRNA cloning site, and puromycin resistance. The gRNA cloning site was modified to contain a “stuffer” (cut from LentiCRISPRv2 plasmid using the restriction enzyme BsmBI and ligated into our vector) that resulted in a higher gRNA cloning efficiency. Upon insertion of gene specific gRNA, dCas9-KRAB mediated knockdown of target genes could be achieved. Three gRNA oligos were selected from the top ten gRNA sequences generated from the study “Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation” [119]. Generation of the vector and transduction of the target cells was performed as described above for the generation and transduction of the LentiCRISPRv2 vector. The best two knockdown cell lines generated were subsequently used for all experiments. Cells transduced with vector alone were used as a control. The NetrinG1 CRISPRi gRNA sequences were: 1.1, CACCGGCGCCCCGAGGTCGTGGAG; 1.2, AAACCTCCACGACCTCGGGGCGCCC; 2.1, CACCGACAGCAACAGCGAGCGGGA; 2.2, AAACTCCCGCTCGCTGTTGCTGTCC; 3.1, CACCGCGAGAGCCGGAAAGAGGAG; and 3.2, AAACCTCCTCTTTCCGGCTCTCGCC. The VGlut1 CRISPRi gRNA sequences were: 1.1, CACCGAGAGAGAGTGGAGCCGGGT; 1.2, AAACACCCGGCTCCACTCTCTCTCC; 2.1, CACCGGCTCCGCTCGGGGGGAAGG; 2.2, AAACCCTTCCCCCCGAGCGGAGCCC; 3.1, CACCGAGAGACCCAGAAGTGGTGG; and 3.2, AAACCCACCACTTCTGGGTCTCTCC. The GS CRISPRi selected gRNA sequences were: 1.1, CACCGGCTCTGCAGAGTCGAGAGT; 1.2, AAACACTCTCGACTCTGCAGAGCCC; 2.1, CACCGGAGCGTGTGAGCAGTACTG; 2.2, AAACCAGTACTGCTCACACGCTCCC; 3.1, CACCGGACGGGTCCAAGCCACCAG; and 3.2, AAACCTGGTGGCTTGGACCCGTCCC. For 4E-BP1 the selected gRNA sequences were: 1.1, CACCGCACAGGAGACCATGTCCGG; 1.2, AAACCCGGACATGGTCTCCTGTGC; 2.1, CACCGAGGGCAGCGAGAGGTTCGC; 2.2, AAACGCGAACCTCTCGCTGCCCTC; 3.1, CACCGACCGGGTGTCCAGGCTCAA; and 3.2, AAACTTGAGCCTGGACACCCGGTC. The FRA1 CRISPRi sequences were: 1.1, CACCGAAGTCTCGGAACATGCCCG; 1.2, AAACCGGGCATGTTCCGAGACTTC; 2.1, CACCGGGCGGCTCTGCGGGGTACA; 2.2, AAACTGTACCCCGCAGAGCCGCCC; 3.1, CACCGGGCATGTTCCGAGACTTCG; and 3.2, AAACCGAAGTCTCGGAACATGCCC.
Cell Engagement/Motility Assays
RFP+ CON or NGL-1 KO PDACc (5×104) were plated with GFP+ CON or NetG1 KO CAFs (5×104) in 3D and allowed to attach for 2 hours. Following cell attachment, 3 regions per well, where there was obvious clustering of red PDACc and green CAFs, were selected to be imaged every 15 minutes for 24 hours, using a motorized XYZ stage (Optical Apparatus Co., Ardmore, PA) controlled by MetaMorph 6.3r7 (Molecular Devices, Downingtown, PA) software. Time-lapse movies were built with images from each corresponding fluorescence channels acquired every 15 min for a period of 24 hours using either 4X or 20X Pan Fluor objectives mounted on a Nikon TE-2000U wide field inverted microscope (Optical Apparatus Co., Ardmore, PA) with a Cool Snap HQ camera rendering 3 time-lapse movies per condition for a total of 9 videos generated over 3 independent experimental repetitions. The resulting time-lapse videos were analyzed to quantify heterotypic cell engagement (yellow interacting areas using both channels simultaneously in merged file stacks) and PDACc cell motility (using only the monochromatic images corresponding to the red channel), as outlined below.
Cell Engagement:
For image analysis MetaMorph offline 7.8.1.0 software was used. To achieve sequence coherence, out of focus or blank images from particular fluorescence channels were removed, together with its fluorescence counterpart image. All movies were normalized (trimmed) to 18 hours data for analysis. Images were scaled and pseudocolored (RFP in red, GFP in green) similarly for all conditions. After merging of fluorescence channels, cell engagement areas (CEA) obtained from regions of fluorescence colocalization (in yellow) were digitally detected, isolated and measured for each condition over time.
PDAC Cell Motility:
Monochromatic channel of RFP+ PDAC cells were tracked using the Track Object function in MetaMorph 7.8.1.0 offline software (Molecular Devices, Downingtown, PA). A minimum of 5 cells per region were chosen to carefully follow. Red PDACc that were touching other red PDACc, moving out of the imaging plane, or that began to round up prior to mitosis were not counted in this experiment. Motility was analyzed according to the total distance (μm) traveled over the timespan of the experiment (24 hr), expressed as velocity (μm/hr).
AlamarBlue Assay
In order to determine the optimal (non-cytotoxic) concentration of 5-(N-ethyl-N-isopropyl) amiloride (EIPA; TOCRIS Bioscience, Bristol, UK) to inhibit macropinocytosis in PDAC cells, 5×103 CON or NGL-1 PDAC cells (PDACc or PANC-1) were plated in a 96 well plate and cultured in 100 μL PDAC media (M3 base, DMEM low glucose, 5%FBS, P/S) overnight in order for cells to attach. The following day, cells were allowed to proliferate for 24 hours in the presence of increasing concentrations of EIPA (0–500μM). DMSO was used as a vehicle control. Next, 10 μL alamarBlue reagent (Bio-Rad, Hercules, CA) was added to the wells, and PDAC cells metabolized the alamarBlue reagent for 6 hours (reduction of alamarBlue reagent). For controls, one set of 6 wells contained only media with no cells or reagent (media blank), while for another set of 6 wells, contained only media and alamarBlue reagent (filter correction factor). The 96 well plate was measured for absorbance at 570nm and 600nm using the Spark® Multimode Microplate Reader (Tecan, Switzerland). The reduction of alamarBlue reagent was calculated according to the following formula:
The % reduction of alamarBlue was normalized to the DMSO control and graphed to determine the highest concentration possible without affecting cell proliferation. This concentration was determined to be 25 μM for all PDAC cells tested and was used in subsequent macropinocytosis assays.
Macropinocytosis Assay
Uptake of the macropinocytosis substrate FITC-Dextran
The macropinocytic activity of PDAC cells was assessed as previously described [58], with minor modifications. To determine the baseline macropinocytosis capacity of PDAC cells cultured in 3D, 2×104 RFP+ CON or NGL-1 KO PDAC cells (PDACc or PANC-1) were plated onto coverslips containing CAF ECM and cultured for 24 hours in glutamine free/serum free DMEM, treated either with DMSO control or EIPA (25 μM) to block macropinocytosis. The following day, lysine fixable FITC-Dextran (D1845; Thermo Fisher Scientific, Waltham, MA) was added to the cells at a final concentration of 1mg/mL and the cells were incubated at 37°C for 30 min. Next, cells were placed on ice and washed 5 times with ice cold PBS. Then cells were fixed in a 4% paraformaldehyde PBS solution for 30 min at room temperature. Cells were then washed 3 times with PBS, and nuclei were counterstained with a DAPI solution (1:10,000) for 15 min. Finally, samples were washed 3 times with PBS and coverslips were mounted onto slides using the ProLong™ Gold Antifade Mountant (P10144; Thermo Fisher Scientific, Waltham, MA). Slides were then imaged and macropinocytotic ability of cells was quantified as outlined below.
Imaging and Quantification
A minimum of 30 confocal images (60x objective; .5 μM slices) per condition were captured by a Nikon A1 camera (Nikon, Melville, NY, USA) linked to a Nikon Eclipse Ti2-E Inverted Microscope Imaging System (Nikon, Melville, NY, USA) for each channel (performed in triplicate, 2 independent experiments; N = 6 coverslips), which included RFP+ PDAC cells (red channel), FITC labeled dextran (green channel), and DAPI labeled nuclei (blue channel). First, each channel of stacked images was merged by max intensity using FIJI/Image J plugin “Z project”. Next, MetaMorph 7.8.1.0 offline software (Molecular Devices, Downingtown, PA) was used to quantify the amount of FITC-dextran taken up by RFP+ PDAC cells. RFP was used to generate a mask and the amount of FITC under that mask was queried (integrated density) and recorded. The integrated density of FITC under RFP was normalized to the DMSO treated CON PDAC cells (PDACc or PANC-1) for each experiment.
Material Transfer Assay
2×104 RFP+ CON or NGL-1 KO PDACc and/or 2×104 GFP+ CON or NetG1 KO CAFs were seeded in 3D and were co-cultured for 36 hours, in serum free DMEM. Spinning disk confocal images (Perkin Elmer, Waltham, MA) were captured by a CoolSNAP CCD Camera (Photometrics, Tucson, AZ) attached to an Eclipse 2000-S inverted microscope (Nikon, Tokyo, Japan). Images were exported from Volocity 3D Image Analysis Software (Perkins Elmer, Waltham, MA) and quantified for the integrated intensity of GFP+ pixels under RFP+ pixels, using Metamorph Image Analysis Software (Molecular Devices, San Jose, CA), which was indicative CAF derived GFP material transfer to RFP+ PDACc.
For the material transfer assays involving CAF CM and EIPA, 2×104 RFP+ PDACc (CON or NGL-1 KO) were seed in 3D onto coverslips and co-cultured for 24 hours with 50% GFP+ CON CAF CM in the presence of DMSO or EIPA (25 μM). Next, coverslips were then washed 3 times with PBS and fixed in 4% paraformaldehyde PBS solution for 30 min at room temperature. Cells were then washed 3 times and nuclei were labeled with DAPI (1:10,000) for 15 min at room temperature. Finally, cells were washed 3 times and mounted onto slides using ProLong™ Gold Antifade Mountant. Imaging and quantification were performed exactly as described above in the macropinocytosis assay, with the amount of GFP+ under RFP+ mask indicative of CAF derived material transfer (GFP+) to PDACc cells (RFP+).
PDAC Survival Assay
2×104 RFP+ CON or NGL-1 KO PDAC cells (PDACc or PANC-1) and/or 2×104 GFP+ CON or NetG1 KO CAFs were seeded in 3D (CON CAF, NetG1 KO, or tumor adjacent fibroblast ECMs, as indicated) in 24 well plates. Cells were co-cultured for 96 hours in glutamine free/serum free DMEM. Next, at least 10 images were taken of red PDAC cells and green CAFs using a Cool Snap 1HQ camera (Roper Scientific, Vianen, Netherlands) on an Eclipse 2000-U inverted microscope (Nikon, Tokyo, Japan). PDAC cell survival was quantified as the number of RFP+ per field of view, using Image J. Dead cells were excluded by Sytox Blue (1 μM; Thermo Fisher Scientific, Waltham, MA) positivity or cell size. Briefly, identical thresholds were established for each color channel for each batch of images per experiment. Then the images were batch processed using a macro that measures cell counts and % area coverage of cells in each image, at a given threshold. The mean RFP+/Sytox blue negative cells (live) from each condition was calculated (counts or % area, depending on the experiment) and normalized to the CON PDAC/CON CAF condition. Cell counts were chosen when cells were clearly isolated (accurate cell counts), and % area was chosen when cells had more overlap (could not accurately determine cell counts). The 96 hour timepoint was determined empirically, when cell death became evident by eye. Other PDAC survival assays were performed with CAF conditioned media (CM), VGlut1, GS, 4E-BP1, and FRA1 KD CAFs, or CAFs pretreated 24 hours with 1mM L-Methionine sulfoximine (glutamine synthetase inhibitor; Sigma-Aldrich, St. Louis, MO), 10 nM SB 202190 (P38 inhibitor; Tocris Bioscience, Bristol, UK), 10 μM MK-2206 (AKT inhibitor; Cayman Chemical, Ann Arbor, MI), or 2.5 μg/mL NetG1 mAb (Santa Cruz, Dallas, TX), as indicated. Note: all small molecule inhibitors were used to pre-treat CAFs overnight and CAFs were washed three times with PBS before co-culture with PDAC cells.
NK-92 cell killing assay
2×104 RFP+ CON or NGL-1 KO PDAC cells and/or 2×104 GFP+ CON or NetG1 KO CAFs were seeded in 3D in 24 well plates and were co-cultured in 500 μL NK-92 media without IL-2 to establish their niche. The following day, wells were seeded with 8×104 resting or active NK-92 cells, in 500 μL of NK-92 media without IL-2. Cells were co-cultured for 48 hours and PDAC survival was assessed identically to the PDAC cell survival assay. To produce active NK-92 cells, cells were maintained with 400 units/mL IL-2 and for resting NK-92 cells, cells were maintained for one week in 100 units/mL IL-2 and switched overnight to NK culture media containing no IL-2 the day before the experiment. For 2D conditions, all experimental conditions were the same, except the endpoint of the experiment was at 6–8 hours, as lack of CAF derived ECM (3D microenvironment) allowed NK-92 cells to kill at a faster rate. For the IL-15 neutralizing experiments, an IL-15 neutralizing antibody (10 μg/mL; R&D Systems, Minneapolis, MN) or Nonspecific IgG isotype control (10 μg/mL; Jackson Immunoresearch, West Grove, PA) were added to the co-cultures where specified. Whenever CON or NetG1 KO CAF CM was used, it was diluted 1:1 in NK-92 media without IL-2. Images were quantified for live PDAC cells as in the “PDAC Survival Assay” section above. Note: all CAFs that were pre-treated with a small molecule inhibitor were washed three times with PBS before subsequent co-culture with PDAC and/or NK-92 cells.
NK-92 cell activation assay
8×104 IL-2 activated NK-92 cells were directly co-cultured (CC) with CAFs or with conditioned media (CM) derived from CAFs for 2 hours. After the co-culture period, NK-92 activation status was determined by staining Granzyme B intracellularly, or by using the IFNγ secretion assay detection kit. Stained cells were subjected to flow cytometry using by LSRII (Beckton Dickinson, San Jose, CA) and analyzed with FlowJo 9.9 software (Tree Star, San Carlos, CA). IL-2 activated NK-92 cells cultured alone were used as the positive control. Percentage of cells that were positive for either Granzyme B or IFNγ were considered activated.
Enzyme-linked immunosorbent assay (ELISA)
ELISAs for IL-15, IFN-β, TGF-β, IL-6, IL-8, GM-CSF, mTGF-β, mGM-CSF, and mIL-6 (R&D Systems, Minneapolis, MN), as well as glutamate (Abcam, Cambridge, UK) and glutamine (MyBioSource, San Diego, CA), were performed as recommended by the manufacturers. Briefly, buffers for the IL-15, IFN-β, IL-6, IL-8, GM-CSF, mGM-CSF, and mIL-6 ELISAs were prepared using the DuoSet ELISA Ancillary Reagent Kit 2, and buffers were prepared using the DuoSet ELISA Ancillary Reagent Kit 1 for the TGF-β and mTGF-β ELISAs (R&D Systems, Minneapolis, MN). Standards, CM or lysates from various fibroblastic lines or murine tumor cultures were incubated for 2 hours on plates pre-coated overnight with capture antibody. For the TGF-β ELISAs, samples first underwent activation using the Sample Activation Kit 1 (R&D Systems, Minneapolis, MN) before sample incubation. Next, detection antibody linked to biotin was added to the wells and incubated for 2 hours. Streptavidin-HRP was then added to the wells and incubated for 20 minutes. Samples were rinsed 3 times with wash buffer in between all steps. Substrate was added for 20 minutes and was neutralized with stop buffer. Plates were read using Spark® Multimode Microplate Reader (Tecan, Switzerland) at 450 nm to measure the specified protein present in sample. Plate imperfections, measured at 570 nm, were subtracted from the readings at 450 nm, to produce corrected readings. Finally, media alone (BLANK) was subtracted from the corrected readings for each sample. To determine the concentration of the indicated protein in each sample, a standard curve was generated using known concentrations of analyte, and the blank corrected values determined for each sample were compared to the standard curve. The extrapolated values were normalized to cell number or total protein content.
U-Plex Assay
U-Plex Assay (multiplex ELISA) was carried out as recommended by the manufacturer (Meso Scale Discoveries, Rockville, MD). Briefly, TA and CON or NetG1 KO CAF CM samples were generated and added to a pre-coated plate containing 10 capture antibodies per well for the detection of the following proteins: IL-1β, IL-2, IFNγ, GM-CSF, IL-6, IL-8, IL-10, IL-12p70, MIP3α, and TNFα. After 3 washes, sample, standards, or calibrator were added to the wells and incubated for 1 hour. Next, after 3 washes, detection antibody was added to the wells and incubated for 1 hour. After a final set of 3 washes, read buffer was added to the wells and the plate was read using the MSD SECTOR Imager 2400 (Meso Scale Discoveries, Rockville, MD) and the data was processed using the MSD Workbench 4.0 software (Meso Scale Discoveries, Rockville, MD).
Amino Acid Screen: Biochrom Method
Media:
20 μL of media was mixed with 80 μL of water and 100 μL of 10% Sulfosalicylic acid (Sigma-Aldrich, St. Louis, MO) followed by incubation for 1 hour at 4°C and then centrifugation at 13,000 rpm (15 min; 4°C). No visible pellet was seen. Supernatant was filtered using Ultrafree-MC-GV centrifugal filters (Millipore, Burlington, MA) and used for amino acid analysis employing Biochrom 30 amino acid analyzer. Results are in terms of μM and were normalized to total protein content.
Cell Lysates:
14.55 μg of cell lysate protein in RIPA buffer was brought to a total volume of 50 μL in water and was mixed with 50 μL of 10% Sulfosalicylic acid (Sigma-Aldrich, St. Louis, MO) followed by incubation for 1 hour at 4°C and then centrifugation at 13,000 rpm (15 minutes; 4°C). Supernatant was filtered using Ultrafree-MC-GV centrifugal filters (Millipore, Burlington, MA) and used for amino acid analysis employing the Biochrom 30 amino acid analyzer (Biochrom US, Holliston, MA). Results were provided in nmol/mg protein units (normalized to total protein content).
These results are available in Supplemental Tables 5-7.
Amino Acid Screen: Mass Spectrometry
Metabolite Extraction and Quantitation
Metabolites were extracted from media and quantitated as described previously [74]. 10μL of media was mixed with 10μL of a mixture of isotopically labeled metabolites and amino acids of known concentrations (Cambridge Isotope Laboratories, Tewksbury, MA) and extracted with 600μL ice-cold HPLC grade methanol. Samples were vortexed for 10 minutes at 4oC, centrifuged at max speed on a tabletop centrifuge for 10 minutes 4oC, and then 450uL of supernatant was dried under nitrogen gas and frozen at −80oC until analyzed by GC-MS.
Gas Chromatograpy-Mass Spectrometry Analysis
Polar metabolites were analyzed by GC-MS as described previously [74]. Dried metabolite extracts were derivatized with 16μL MOX reagent (Thermofisher, Waltham, MA) for 90 minutes. at 37°C, followed by 20 μL of N-tertbutyldimethylsilyl-N-methyltrifluoroacetamide with 1% tert-butyldimethylchlorosilane (Sigma Aldrich, St. Louis, MO) for 60 minutes. at 60°C. After derivatization, samples were analyzed by GC-MS using a DB-35MS column (Agilent Technologies, Santa Clara, CA) installed in an Agilent 7890A gas chromatograph coupled to an Agilent 5997B mass spectrometer. Helium was used as the carrier gas at a flow rate of 1.2 mL/minute. One microliter of sample was injected in split mode (all samples were split 1:1) at 270°C. After injection, the GC oven was held at 100°C for 1 minute. and increased to 300°C at 3.5 °C/minute. The oven was then ramped to 320°C at 20 °C/minute and held for 5 minutes. at this 320°C. The MS system operated under electron impact ionization at 70 eV and the MS source and quadrupole were held at 230°C and 150°C respectively. The detector was used in scanning mode, and the scanned ion range was 100–650 m/z.
All results were normalized to cell number and are available in Supplemental Table 8.
Stable isotope tracing of cultured fibroblasts
To measure steady state incorporation of glucose and pyruvate carbon into polar metabolites, fibroblasts were plated at 200,000 cell/well in 2mL of DMEM media with 10% FBS into six well dishes with or without ECM (termed 2D without fibroblast derived ECM or 3D with fibroblast derived ECM). Cells were then incubated and allowed to adhere overnight. After attachment, the cells were washed twice with 2mL of PBS. Cells were then incubated for 48 hours in 2 mL of Glu/Gln free DMEM supplemented with 10% dialyzed FBS where glucose and pyruvate were replaced with 25mM 13C6-glucose (Cambridge Isotopes Laboratories, Tewksbury, MA; #CLM-1396-PK) and 1mM 13C3-pyruvate (Cambridge Isotopes Laboratories, Tewksbury, MA; #CLM-2440-PK). Following the labeling period, the cells were placed on ice and all subsequent steps took place with the cells on ice. First, the media was aspirated. Each well was then washed rapidly with ~8mL of ice cold blood bank saline (Thermo Fisher Scientific, Waltham, MA; #p-4527840). The saline was then aspirated and 600 μL of ice cold methanol: water (4:1) was added. Cells were then scraped and the resulting extracts were transferred to microcentrifuge tubes. The extracts were then vortexed for 10 min and centrifuged to pellet insoluble material at 21,000 RCF for 10 min. 450 μL of each extract was carefully removed to avoid contamination with insoluble material and transferred to new microcentrifuge tubes and dried under nitrogen gas and stored −80°C until further analysis. GC-MS analysis was then performed on these extracts as described in Gas Chromatography-Mass Spectrometry Analysis.
Mass isotopomer distributions were then determined by integrating appropriate ion fragments for each metabolite [120] using in-house software [121] that corrects for natural abundance using previously described methods [122].
Human NK cell Isolation and Activation Experiments
Isolation of Primary NK cells:
Blood was collected into heparinized tubes from healthy volunteer donors. The authors received written informed consent using HIPAA-compliant procedures approved by the Institutional Review Board of Fox Chase Cancer Center. NK cells were enriched from healthy donor PBMCs using the EasySep NK cell negative selection kit (StemCell Technologies, Cambridge, MA) and confirmed to be >95% purity using flow cytometry.
Primary NK Cell Activation Assay:
Purified NK cells were incubated overnight at 37°C in indicated percentages of CON or NetG1 CAF CM diluted in NK cell media (10%−65%) in the presence of IL-2 (500 units/mL) and IL-12 (10 ng/mL). NK cells activated with IL-2 and IL-12 were used as a positive control. After overnight incubation, the population of IFNγ secreting cells was identified using the IFNγ Secretion assay detection kit (PE) (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s protocol. After a 2-hour incubation with IFNγ catch reagent, cells were washed twice. Next, cells were stained with IFNγ detection antibody and antibodies against NKp80 (anti-NKp80-APC, Biolegend, San Diego, CA), CD3 (anti-CD3-Cy7APC, BD Pharmigen, San Jose, CA), CD56 (anti-CD56-BUV395, BD Horizon, San Jose, CA), CD69 (anti-CD69-Pac Blue, San Diego, CA). Cells were centrifuged and rinsed twice, with propidium iodide in the second wash. NK cells were defined as NKp80+CD56+CD3- cells.
Flow cytometry and data analysis:
Stained cells were analyzed on a BD ARIA II flow cytometer (Becton Dickinson, Franklin Lakes, NJ). Data was collected with BD FACS Diva software (v6) and analyzed with FlowJo (v9.9; Tree Star Inc., Ashland, OR). IFNγ and CD69 expression were identified as the percentage of IFNγ+ cells or mean fluorescence intensity (MFI) of CD69+ cells. The data were normalized to the positive control (NK cells with IL2/IL12 treatment alone).
Mouse Models
Tissue for stromal analysis was obtained from the LSL-KRASG12D, PDX1-Cre (KC) model [52], and KPC4B and KPC3 murine PDAC cell lines were derived from tumors generated from a variant of LSL-KRASG12D, TP53−/−, PDX1-Cre (KPC) mice [53].
PDAC cell orthotopic injections
All animal work was performed under protocols approved by IACUC at Fox Chase Cancer Center. A small incision was made in the abdominal skin of the mice, and gently moved aside the spleen to reveal the pancreas. Next, 106 murine CON or NGL-1 KO #1 or #2 PDAC cells (derived from KPC tumors), were injected (100 μL) orthotopically into the pancreas of syngeneic C57BL/6 mice (source: FCCC, 6–8 weeks old) and tumors were allowed to develop for 3.5 weeks. Mice were monitored by observation, palpation, and weekly MRIs and sacrificed at 3.5 weeks. Pancreas weights were obtained and pancreatic tissue was further analyzed as described below.
To determine the effect of the immune system on tumorigenesis in the orthotopic model, the injections were performed as above, with the following mice used as hosts:
C57BL/B6 (immunocompetent; FCCC), C.B17 Severe Combined Immunodeficient, (SCID; lacks T and B cells; Taconic Bioscience), and NOD.Cg-Prkdcscid IL2rgtm1Wjl/SzJ: NOD scid gamma (NSG; lacks NK, T, and B cells; Jackson Laboratories). Mice were 6–8 weeks old.
For the human tumor cell/CAF co-injections, RFP+ PDACc cells (CON or NGL-1 KO) and CAFs (CON or NetG1 KO) were co-injected at a 1:3 ratio (2.5*105 tumor: 7.5*105 CAF) into SCID mice (6–8 weeks old). CAFs and tumor cells injected alone were used as controls. Tumors were allowed to form for 4 weeks. Subsequently, mice were sacrificed and tissue was analyzed as outline below.
For the NetG1 neutralizing monoclonal antibody (mAb) experiment, after mice (C57BL/B6, 6–8 weeks old) recovered from the tumor cell (106) orthotopic injections for 3 days, mice were divided into two groups (IgG; n=9 or mAb; n=10) and were treated with 5 mg/kg isotype specific IgG (IgG1; West Lebanon, New Hampshire) or mAb (Santa Cruz, Dallas, TX) 3 times per week, until the completion of the model (3.5 weeks, as above).
Magnetic Resonance Imaging (MRI)
Animals were imaged in a 7 Tesla vertical wide-bore magnet, using a Bruker DRX 300 spectrometer (Billerica, MA) with a micro-imaging accessory, which included a micro 2.5 gradient set, a 30 cm radiofrequency coil, and Paravision software (Bruker, Billerica, MA). Animals were anesthetized with a mixture of oxygen and isoflurane (2–3%). An injection of 0.2 mL of 10:1 diluted Magnevist (gadopentetate dimeglumine; Bayer, Whippany, NJ) was made into the shoulder region immediately preceding the scan. Scout scans were performed in the axial and sagittal orientations, permitting us to accurately prescribe an oblique data set in coronal and sagittal orientations that included the organs of interest. A two-dimensional spin echo pulse sequence was employed with echo time 15 msec, repetition time 630 msec, field of view = 2.56 cm, acquisition matrix= 256×256, slice thickness= 0.75 mm, 2 averages, scan time = 5 minutes. Fat suppression (standard on Bruker DRX systems) was used for all scans. Raw data files were converted to Tiff format image files using the Bruker plugin on ImageJ.
Murine tissue preparation and histopathological analysis
Tissue Preparation:
Tissues were collected and fixed in 10% phosphate-buffered formaldehyde (formalin) 24–48 hours, dehydrated and embedded in paraffin. Tissues were processed by dehydration in a series of ethanol followed by xylene (70% ethanol 3 hours, 95% ethanol 2 hours, 100% ethanol 2 hours, 100% ethanol Xylene mixed 1hours, Xylene 3hours, then immersed in Paraffin). Paraffin blocks were cut into 5 μm sections, mounted on microscope slides, and stored at room temperature until used. Sections were stained with hematoxylin and eosin for pathologic evaluation.
All H&E slides were viewed with a Nikon Eclipse 50i microscope and photomicrographs were taken with an attached Nikon DS-Fi1 camera (Melville, NY, USA).
Tumor area analysis:
H&E stained slides were also scanned using Aperio ScanScope CS scanner (Aperio, Vista, CA). Scanned images were then viewed with Aperio’s image viewer software (ImageScope). Selected regions of interest for PDAC, necrotic, and normal tissue were outlined respectively by a pathologist (Dr. Kathy Q. Cai), who was blinded to the treatment groups. The ratio of PDAC, necrotic, and normal tissue to the total pancreatic areas scanned was calculated for each animal.
Differentiation status of tumor cells:
H&E images of pancreata injected with CON or NGL-1 KO cells were scored for the percentage of cells that were well differentiated, moderately differentiated, or poorly differentiated, as determined by a pathologist, who was blinded to the treatment groups. For the co-injection model, sarcomatoid/sarcomatous PDAC was also scored, as it represented a significant tumor cell type in this model.
TUNEL Staining:
Apoptotic cells were quantified using the DeadEnd™ Fluorometric TUNEL System (Promega, Madison, WI), per manufacturer’s instructions. Briefly, tissue was deparaffinized and fixed in 4% formaldehyde for 15 minutes at RT. After 2 PBS washes, tissue was treated with 20 μg/mL proteinase K solution for 10 minutes at RT. After 2 PBS washes, slides were fixed again in 4% formaldehyde for 5 minutes at RT. Tissue was then submerged in Equilibration buffer at RT for 5 minutes. Next, apoptotic cells were labeled with TdT reaction mixture for 1 hour at 37°C. After 1 hour, the reaction was stopped by adding SSC buffer for 15 minutes at RT. Tissue was then washed 3x in PBS and then stained with DAPI (1:10,000) for 15 minutes at RT. Finally, after an additional wash, tissue was mounted and imaged using the Eclipse 2000-U inverted microscope. Metamorph software was used to quantify dead cells (green)/nuclei (blue) in each field of view. At least 10 images (20X) per sample were acquired.
Ki67 staining (proliferation):
IF on tissue sections was done as previously published [38]. Briefly, deparaffinized tissue underwent antigen retrieval by boiling in buffer containing 10 mM Sodium citrate, 0.05% Tween 20, pH 6.0 for 1 hour. Next, tissue was blocked in Odyssey Blocking Buffer (PBS base) (LI-COR Biosciences, Lincoln, NE) for 1 hour at RT. Tissue was then stained with primary antibody against Ki67 (Abcam, Cambridge, UK) overnight at 4°C. The following day, tissue was washed 3 times in 0.05% PBST, and secondary antibody conjugated to TRITC (Jackson ImmunoResearch, West Grove, PA) was applied to the samples for 2 hours at RT. After 3 additional washes, nuclei were stained with DAPI (1:10,000) for 15 minutes at RT. After one final wash, samples were mounted and images (20X) were captured using an Eclipse 2000-U inverted microscope. Proliferating cells (red)/nuclei (blue) in each field of view were quantified using Metamorph Software.
Tissue preparation, histology, and immunohistochemistry for NetG1 mAb mouse model (NK1.1 and IL-15)
Tissue preparation
Mouse tissues were collected and fixed in 10% phosphate-buffered formaldehyde (formalin) for 24–48 hrs, dehydrated and embedded in paraffin. Hematoxylin and eosin (H&E) stained sections were used for morphological evaluation purposes and unstained sections for immunohistochemical (IHC) studies.
Immunohistochemical staining of NK1.1 and IL-15
Immunohistochemical staining was carried out according to standard methods. Briefly, 5- μm formalin-fixed, paraffin embedded sections were deparaffinized and hydrated. Sections were then subjected to heat-induced epitope retrieval with EDTA (pH 9.0) for NK1.1 and with Citrate buffer (pH 6.0) for IL-15. Endogenous peroxidases were quenched by the immersion of slides in 3% hydrogen peroxide solution. The sections were incubated overnight with primary antibodies to anti NK1.1 (Mouse, 1:50, MA1–70100; Invitrogen-Thermo Fisher Scientific, Waltham, MA) and IL-15 (Rabbit, 1:50, LS-B10080; Lifespan Biosciences, Seattle WA) at 4 °C in a humidified slide chamber. Immunodetection was performed using the Dako Envision+ polymer system (#K4000 and #K4002, for mouse and rabbit, respectively) (Dako, Tokyo, Japan) and immunostaining was visualized with the chromogen 3, 3’-diaminobenzidine.
The sections were then washed, counterstained with hematoxylin, dehydrated with ethanol series, cleared in xylene, and mounted.
All slides were viewed with a Nikon Eclipse 50i microscope and photomicrographs were taken with an attached Nikon DS-Fi1 camera (Melville, NY, USA).
NK1.1 and IL-15 IHC scoring
NK1.1 and IL-15 IHC of mouse tissue was scored blindly by a pathologist. For NK1.1, the number of clusters (foci) of NK1.1+ cells were counted. Each focus consisted of at least 20 NK1.1 positive cells. Foci were normalized to tumor area for each mouse, and then normalized to the IgG group to compute fold changes between IgG and mAb treated groups. For IL-15, fibroblastic cells were scored on a 0–4 scale, based on intensity and coverage of the staining.
Supernatant Collection of Tumor Cultures
Small pieces of tissue (30–60 mg) were excised from the pancreata isolated from mice treated with IgG or mAb (described above) and were cultured in serum/Gln free media overnight. The following day, the media was centrifuged for 15 minutes at 15,000 rpm to remove cellular debris. The resultant supernatant was collected and subjected to ELISAs to measure levels of Glu, Gln, mTGF-β, mGM-CSF, and mIL-6. All data were normalized to mg of tissue cultured.
Acquisition and Use of Human Tissue (used in Tissue Microarray)
All human tissues used in this study, for the purpose of generating Tissue Micro Arrays, were collected using HIPAA approved protocols and exemption-approval of the Fox Chase Cancer Center’s Institutional Review Board, after patients signed a written informed consent in which they agreed to donate surgical specimens to research. To protect patient’s identity, samples were classified, coded and distributed by the Institutional Biosample Repository Facility. Normal pancreatic tissue was obtained through US Biomax Inc. (Rockville, MD) following their rigorous ethical standards for collecting tissue from healthy expired donors.
Tissue Microarray (TMA)
Acquisition of the TMAs:
The FCCC tissue microarray was generated by the Biosample Repository at Fox Chase Cancer Center. There were 80 patient samples generated from three independent TMAs. The MDA TMA was generated at the University of Texas MD Anderson Cancer Center. There were 143 patient samples generated from three independent TMAs. The UICC system was used for all of the samples from both TMAs for tumor grading and staging. The clinical parameters for each TMA are available in Supplemental Table 2.
Immunohistochemistry (IHC):
TMA sections were cut from formalin fixed paraffin embedded (FFPE) blocks. After deparaffinizing in xylene, sections were hydrated in graded alcohol and quenched in 3% H2O2. Slides were subjected to heat induced epitope retrieval in citrate buffer (pH 6.0) for 1 hour. After cooling to room temperature, sections were permeabilized in Triton X-100 for 20 minutes, followed by a 1 hour incubation in blocking buffer (1%BSA, 2% Normal Goat Sera, 0.2% Cold water fish skin gelatin in PBS). Antibodies against NGL-1 (GTX 121508), NetG1 (GTX 115637), GS (Abcam), VGlut1 (ThermoFisher), PDPN (Biolegend, 916606), α-SMA (Sigma), and NKp46 (R&D Systems, AF1850) were used at 1:100 dilution and the incubation was carried out overnight at 4° C. The sections were washed and incubated with anti- rabbit HRP (Vector Lab MP-7451) and then treated with DAB (ImmPACT SK-4105) for 2.5 minutes. Sections were counter stained in Mayer’s Hematoxylin to show nuclei, dehydrated in graded alcohol and mounted in Cytoseal-60 after clarifying in toluene.
Blind Pathological Scoring of TMAs:
A pathologist (Dr. Klein-Szanto) blindly scored the 80 and 143 stained TMA cores for fibroblastic NetG1 or tumoral NGL-1 on a scale of 0–4, which is based on staining intensity and coverage. NKp46 was also scored on a 0–4 scale, but with each value representing # of cells staining positive for NKp46, per core. 0 = 0 cells, 1 = 1–10 cells, 2 = 11–20 cells, 3 = 21–30 cells, and 4 = >30 cells. Each patient had two cores stained per marker and these were averaged together to generate the IHC score. These values, along with overall survival data, are available in Supplemental Table 3.
Generation of Kaplan-Meier Plots:
Kaplan-Meier (KM) plots were generated for overall survival correlated with immunohistochemical staining of fibroblastic NetG1, tumoral NGL-1, and NKp46.
Simultaneous Multichannel Immunofluorescence (SMI)
Qdot antibody conjugation
Corresponding antibodies were linked to Qdot molecules using SiteClick Qdot labeling from Molecular Probes-ThermoFisher Scientific. In brief, approximately 100 mg of specific IgG were pre-purified to remove excessive carrier proteins (if needed) and each conjugation was performed following manufacturer’s three-step protocol requiring antibody’s carbohydrate domain modifications to allowing an azide molecule attachment to facilitate linking DIBO-modified nanocrystals to the antibodies.
Formalin fixed and paraffin embedded (FFPE) tissue processing
FFPE sections were deparaffinized using xylene and rehydrated in progressive ethanol to water dilutions. Tissues then were processed through heat-induced epitope retrieval using pH 6.0 Citrate buffer, followed by permeabilization with 0.5% TritonX-100. After treating the specimens with Odyssey (PBS base) blocking buffer (LI-COR Biosciences, Lincoln, NE) samples were labeled by a three-day indirect immunofluorescence scheme. Initially, primary antibodies against mouse monoclonal NetG1 (D-2, Santa Cruz. Dallas, TX) and rabbit polyclonal NGL-1 (N1N3, Genetex Inc., Irvine, CA) were incubated at 4°C overnight, followed by anti-mouse Q625 and anti-rabbit Q655 secondary antibodies, together with primary rabbit polyclonal Y397P-FAK (Genetex Inc., Irvine, CA) pre-linked to Q605, for an overnight incubation (see Q-dots antibody conjugation details above). After washes, specimens were treated with corresponding monovalent Fab IgG fragments (Jackson Immuno Research Inc., West Grove, PA) to avoid cross-reaction during the incubation of the following cocktail of primary antibodies: mouse monoclonal anti-pan-cytokeratin (clones AE1/AE3, DAKO) to detect the epithelial compartment; rabbit monoclonal anti-vimentin (EPR3776, Abcam) antibodies for mesenchymal (stromal) components. Secondary antibodies used were donkey anti-mouse Cy3 and donkey anti-rabbit Cy2 (Jackson Immuno Research Inc., West Grove, PA). Nuclei were labeled by DRAQ5 (Invitrogen-Thermo Fisher Scientific, Waltham, MA). Lastly, sections were dehydrated in progressive ethanol concentrations and clarified in Toluene before mounting using Cytoseal-60 (Thermo Fisher Scientific, Waltham, MA). Slides were cured overnight in dark at RT before the imaging.
Image acquisitions for SMI analysis
Imaging of fluorescently labeled tissue sections was performed utilizing Nuance-FX multispectral imaging system (Caliper LifeSciences, PerkinElmer), using full capabilities of its Tunable Liquid Crystal imaging module, attached to a Nikon Eclipse TE-2000-S epifluorescence microscope. Images were acquired using a Plan Fluor 40X/0.75 objective. A specific wavelength based spectral library for each system was built using control pancreatic tissues (i.e. murine and human), individually labeled for each Qdot and fluorophore used. Unstained specimens were also included to digitally discard specific auto-fluorescence spectra. Each particular spectral library was used for the subsequent image acquisition and analysis. All excitations were achieved via a high-intensity mercury lamp using the following filters for emission-excitation, for Nuance-FX, DAPI (450–720), FITC (500–720), TRITC (580–720), CY5 (680–720). Excitation was conducted from the infrared to ultraviolet spectrum, attempting to avoid energy transfer and excitation of nearby fluorophores. Emission was collected using ‘DAPI’ filter (wavelength range 450–720) for all Qdot labeled markers, while masks used the conventional FITC, TRITC, and CY5 filters.
Collected image cubes (multi-spectral data files) were unmixed to obtain 16-bit (gray scale) individual monochromatic files. Next, the images were processed in bulk for similar levels adjustment and 8-bit monochromatic image conversion for each channel, using Photoshop’s ‘Levels’ and ‘Batch processing’ automated functions. The resulting images were sampled to set identical threshold values for each marker (NetG1, Y397P-FAK and NGL-1) and masks (vimentin and nuclei). Threshold values distinguishing cytokeratin in normal versus tumor samples needed to be independently adjust for each tissue set to assure variations in intensity staining patterns between the two types of epithelium (i.e., normal vs tumoral) were accounted for, as these were needed to be used as “gating masks” identifying epithelial/tumoral area pixels. Processed images were used to analyze distribution, intensity and co-localization of positive-selected pixels using SMIA-CUKIE (SCR_014795) algorithm, available online at https://github.com/cukie/SMIA [38].
SMIA-CUKIE usage and outputs
As described previously [38], SMIA-CUKIE algorithm (SCR_014795) was used to analyze high throughput monochromatic images acquired from SMI-processed FFPE tissue sections. In brief, the algorithm is based on the usage of digital masks, constructed from pixel-positive area distribution, resulting in tissue compartmentalization (i.e. cytokeratin positive/vimentin negative area: epithelium/tumor compartment vs. vimentin positive/cytokeratin negative area: stromal compartment). Next, we queried mean, median, coverage area, integrated intensity of the markers of interest (i.e. NetG1, NGL-1, Y397P-FAK), under each mask. Each mask and marker required a numeric threshold (0–255), indicating the value of pixel intensity considered positive for each channel (corresponding to each mask and marker). These were kept identical for each marker and mask throughout the study. Finally, the analysis generated an Excel file containing numerical values, which were scrutinized for statistical significance as described in the corresponding statistical analysis section. Black lines through a row indicate an unquantifiable image. The algorithm is available at the public domain: https://github.com/cukie/SMIA_CUKIE.
RNA sequencing (RNAseq)
Stranded mRNA-seq library: 1000ng total RNAs from each sample were used to make library according to the product guide of Truseq stranded mRNA library kit (Illumina, Cat# 20020595). In short, mRNAs were enriched twice via poly-T based RNA purification beads, and subjected to fragmentation at 94 degree for 8 minutes via divalent cation method. The 1st strand cDNA was synthesized by Superscript II reverse transcriptase (ThermoFisher, Cat# 18064014) and random primers at 42 degree for 15 minutes, followed by 2nd strand synthesis at 16 degree for 1hour. During second strand synthesis, the dUTP was used to replace dTTP, thereby the second strand was quenched during amplification. A single ‘A’ nucleotide is added to the 3’ ends of the blunt fragments at 37 degree for 30 minutes. Adapters with illuminaP5, P7 sequences as well as indices were ligated to the cDNA fragment at 30 degree for 10 minutes. After SPRIselect beads (Beckman Coulter, Cat# B23318) purification, a 15-cycle PCR reaction was used to enrich the fragments. PCR was set at 98 degree for 10 sec, 60 degree for 30 sec and extended at 72 degree for 30 sec. Libraries were again purified using SPRIselect beads, had a quality check on Agilent 2100 bioanalyzer (serial # DE34903146) using Agilent high sensitivity DNA kit (Cat# 5067–4626), and quantified with Qubit 3.0 flurometer (ThermoFisher Scientific, Cat#Q33216) using Qubit 1x dsDNA HS assay kit (Cat#Q33230). Sample libraries were subsequently pooled and loaded to the Hiseq 2500 (Illumina, serial number SN930). Single end reads at 50bp were generated by using Hiseq rapid SR cluster kit V2 (Illumina, Cat# GD-402–4002) and HiSeq rapid SBS kit v2 50 cycles (Illumina, Cat# FC-402–4022). Faseq files were obtained at Illumina base space (https://basespace.illumina.com) and aligned to Hg38 using Tophat2 [123] and resulting BAM files were used to estimate the gene counts using HT-Seq [124]. Subsequently, to identify differentially expressed genes between CON and NetG1 KO CAFs, we applied DESeq2 [125]. The resulting differentially expressed genes were used as input to GSEA [126] (default parameters with enrichment statistic set as classic). Heatmap depicting differentially expressed genes using mean-centered rlog transformed gene counts were plotted using pheatmap package available in R. Data are organized in tabs (Gene list p<0.001, GSEA list with FDR <0.25, and complete GSEA analysis for positive and negative enrichment), available in Supplemental Table 4. Data were also deposited into the Gene Expression Omnibus database (accession #GSE156963) as part of the superseries “To determine the differentially expressed genes in cancer associated fibroblasts upon ablation of the glutamatergic synapse protein NetrinG1” (accession #GSE156964).
Pancreatic Cancer Datasets
We obtained the gene expression data from [42] (processed data available at ArrayExpress ID: E-MTAB-6134), [43] (processed expression data available at Gene Expression Omnibus ID GSE21501) and TCGA (rsem normalized gene expression data available at GDAC Firehose repository for 76 high tumor purity samples). Clinical outcomes data indicating patient overall survival was obtained from the respective data repositories. In the case of Moffitt and TCGA studies, we used Moffitt et al. [43] classification to stratify PDAC tumors into basal and classical subtypes. In the case of Puleo et al. [42] we applied 5 different classes as described by that study. In order to assess the association between expression of LRRC4C and NTNG1 in PDAC tumors to overall survival (OS), we used expression and clinical data for TCGA and Puleo et al. [42, 44]. We stratified the expression data into lower (DN) and upper quartiles (UP) for both LRRC4C and NTNG1 and categorized them into 4 different groups (DN-DN; DN-UP; UP-DN and UP-UP). We then estimated the distributions of OS and RFS using Kaplan-Meier methods and were compared using log-rank tests. Analyses to compare the overall survival among the basal and classical subtypes were performed by comparing survival curves with log-rank tests. These were calculated using the R ‘survival’ package Therneau, T. M. & Grambsch, P. M. Modeling Survival Data: Extending the Cox Model (Springer-Verlag, 2010). We applied Gene Set Variation Analysis (GSVA) [127] with mx.diff parameter set to true on all the 76 PDAC tumor cases in TCGA using 12,913 gene sets available from MSigDB [128]. To classify NGL-1 into low- and high-expressors, we selected cases on Z-scores (85th CI for mean). Wilcoxon-rank sum test was applied to identify significantly enriched MSigDB datasets (p<0.005). Select enriched pathways that are significantly different between low- and high-expressors of NGL-1 were plotted as waterfall plots with low- (green color) and high-expressors (red). Cases that did not satisfy the Z-score cut-off were plotted as gray bars.
KM Plots Generated with KM Plotter:
To generate KM Plots correlating with overall survival (OS) and relapse free survival (RFS) data with NTNG1 and LRRC4C mRNA expression from the TCGA dataset, we used the online tool KM Plotter (kmplot.com; [129]). We selected the pancreatic adenocarcinoma dataset (N=177) from the Pan-Cancer RNAseq section of the website for overall survival. For the RFS analysis, we selected the RFS dataset, for which there was data available for 69 patients [130]. We used the default auto cutoff algorithm to establish the high and low expressors of NTNG1 and LRRC4C (cutoff = 22) combined. The false discovery rate (FDR) for the cutoff values for NTNG1 and LRRC4C were >0.5. KM plots were generated and Hazard Ratios (HR) and p-values (log rank test) were obtained.
Statistics
For all in vitro and animal experiments, Prism 7.0 (Graph Pad Software, San Diego, California) was used for all statistical analysis. For comparison between two groups, a two tailed Student’s T-Test was performed. For comparisons between more than two groups, a One-Way ANOVA was performed, using either a Dunnett’s multiple comparisons test (compared to control condition) or Tukey’s multiple comparisons test (comparing all conditions to one another), depending on the experiment. Groups were deemed statistically significantly different from one another if the p-value was less than or equal to 0.05. On graphs, significance was defined as follows: * p<0.05; ** p<0.01; *** < 0.001; **** p< 0.0001. For the correlation of the SMI analysis, linear regression was applied to the SMIA generated values of two markers at the designated components, and the R2 value was calculated. All in vitro experiments were performed at least three times (unless explicitly stated in the figure legend) and the orthotopic injections were performed in two cohorts each time, with the following resultant sample sizes:
Key Resources Table
Supplementary Material
Significance.
This study demonstrates the feasibility of targeting a fibroblastic protein, NetG1, which can limit PDAC tumorigenesis in vivo by reverting the pro-tumorigenic properties of CAFs. Moreover, inhibition of metabolic proteins in CAFs altered their immunosuppressive capacity, linking metabolism with immunomodulatory function.
Human co-injection SCID model: N=6 for all co-injections; N=4 and 5 for CON or NGL-1 KO PDAC cells alone; N= 3 and 4 for CON or NetG1 KO CAFs alone.
Murine cancer cell model: N=7 for Control, and N=19 for combined NGL-1 KO injected mice.
Murine immunocompromised mouse models: N=5 for all groups
mAb model: N=9 for IgG group; N =10 for mAb group.
Acknowledgments
This study is dedicated to the memories of the late Dr. Patricia Keely (pioneer in the field of ECM biology and an amazing mentor) and Neelima Shah (a noteworthy co-author on this study, with over 30+ years in research and the most beautiful soul), who continue to inspire our work. We would like to thank Dr. Martin Humphries for the SNAKA 51 antibody. We appreciate the advice from Dr. Alexander Macfarlane on NK cells and flow cytometry. We thank Jim Oesterling for his feedback and technical assistance with flow cytometry. We greatly appreciate the NetG1/NGL-1 discussions with Dr. Shigeyoshi Itohara. We would also like to acknowledge Dr. Stephen M. Sykes, Dr. Daniela Di Marcantonio, and Esteban Martinez for their time and expertise with RT-qPCR. We are grateful for the kind gifts provided by Robert Weinberg (pBABE-neo-hTERT vector; Addgene plasmid # 1774), Dr. Feng Zhang (LentiCRISPRv2; Addgene plasmid # 52961), Dr. Didier Trono (psPAX2; Addgene plasmid # 12260), Charles Gersbach (pLV hU6-sgRNA hUbC-dCas9-KRAB-T2a-Puro; Addgene plasmid #71236), and Dr. Alexey Ivanov (pLV-CMV-H4-puro vector). This work was supported in part by gifts donated to the memory of Judy Costin, funds from the Marianne DiNofrio Pancreatic Research Foundation, funds from the Martin and Concetta Greenberg Pancreatic Cancer Institute, Pennsylvania’s DOH Health Research Formula Funds, the 5th AHEPA Cancer Research Foundation, Inc., a grant by the Worldwide Cancer Research (E.C.), DOD grant WX81XWH-15-1-0170 (E.C./R.F./J.F.B.), as well as NIH/NCI grants R01CA113451-10 (E.C.), T32CA009035 (R.F. and D.B.V.C., independently), R01CA113451-09S1 (J.F.B./E.C.), R21 CA231252 (E.C./I.A.), R01CA232256 (E.C.), R01CA188430 (I.A.), R03CA212949 (I.A.), R01GM116911 (I.A.), R01CA194263 (K.S.C.), and the Core Comprehensive Cancer Center Grant CA06927 in support to Fox Chase Cancer Center’s facilities including: Bio Sample Repository, Light Microscopy, Small Animal Imaging, Biostatistics and Bioinformatics, Flow Cytometry, Laboratory Animal, Cell Culture, DNA sequencing, Genotyping and Real-time PCR, Histopathology, Immune Monitoring Facility, and Talbot Library.. The authors also acknowledge support from F32CA213810 (A.M.), the Damon Runyon Cancer Research Foundation (A.N.L.), R01CA168653, R01CA201276, and P30CA1405141 (M.G.V.H), the Lustgarten Foundation (M.G.V.H.), the MIT Center for Precision Medicine (M.G.V.H.), SU2C (M.G.V.H.), and the Ludwig Center at MIT (M.G.V.H.). M.G.V.H. is a Howard Hughes Medical Institute Faculty Scholar.
Footnotes
Declaration of Interests: M.G.V.H. discloses that he is a consultant and SAB member for Agios Pharmaceuticals, Aeglea Biotherapeutics and Auron Therapeutics. E.C. discloses that she is a consultant and SAB member for Phenomic AI. The rest of the authors declare no competing interests.
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, and Matrisian LM, Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the united states. Cancer Res, 2014; 74: p. 2913–21. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, and Jemal A, Cancer statistics, 2020. CA Cancer J Clin, 2020; 70: p. 7–30. [DOI] [PubMed] [Google Scholar]
- 3.Ryan DP, Hong TS, and Bardeesy N, Pancreatic adenocarcinoma. N Engl J Med, 2014; 371: p. 2140–1. [DOI] [PubMed] [Google Scholar]
- 4.Hidalgo M, Pancreatic cancer. N Engl J Med, 2010; 362: p. 1605–17. [DOI] [PubMed] [Google Scholar]
- 5.Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, et al. , A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer, 2020; 20: p. 174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. , Inhibition of hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science, 2009; 324: p. 1457–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCarroll JA, Naim S, Sharbeen G, Russia N, Lee J, Kavallaris M, et al. , Role of pancreatic stellate cells in chemoresistance in pancreatic cancer. Front Physiol, 2014; 5: p. 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan TS, Hsu CC, Pai VC, Liao WY, Huang SS, Tan KT, et al. , Metronomic chemotherapy prevents therapy-induced stromal activation and induction of tumor-initiating cells. J Exp Med, 2016; 213: p. 2967–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang RF, Moore T, Arumugam T, Ramachandran V, Amos KD, Rivera A, et al. , Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res, 2008; 68: p. 918–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Djurec M, Grana O, Lee A, Troule K, Espinet E, Cabras L, et al. , Saa3 is a key mediator of the protumorigenic properties of cancer-associated fibroblasts in pancreatic tumors. Proc Natl Acad Sci U S A, 2018; 115: p. E1147–E1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erkan M, Michalski CW, Rieder S, Reiser-Erkan C, Abiatari I, Kolb A, et al. , The activated stroma index is a novel and independent prognostic marker in pancreatic ductal adenocarcinoma. Clin Gastroenterol Hepatol, 2008; 6: p. 1155–61. [DOI] [PubMed] [Google Scholar]
- 12.Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, et al. , Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell, 2014; 25: p. 719–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. , Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell, 2014; 25: p. 735–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim EJ, Sahai V, Abel EV, Griffith KA, Greenson JK, Takebe N, et al. , Pilot clinical trial of hedgehog pathway inhibitor gdc-0449 (vismodegib) in combination with gemcitabine in patients with metastatic pancreatic adenocarcinoma. Clin Cancer Res, 2014; 20: p. 5937–5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Provenzano PP, Cuevas C, Chang AE, Goel VK, Von Hoff DD, and Hingorani SR, Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell, 2012; 21: p. 418–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, et al. , Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res, 2015; 75: p. 544–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erkan M, Kurtoglu M, and Kleeff J, The role of hypoxia in pancreatic cancer: A potential therapeutic target? Expert Rev Gastroenterol Hepatol, 2016; 10: p. 301–16. [DOI] [PubMed] [Google Scholar]
- 18.Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, et al. , Glutamine supports pancreatic cancer growth through a kras-regulated metabolic pathway. Nature, 2013; 496: p. 101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin L, Alesi GN, and Kang S, Glutaminolysis as a target for cancer therapy. Oncogene, 2016; 35: p. 3619–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seo JW, Choi J, Lee SY, Sung S, Yoo HJ, Kang MJ, et al. , Autophagy is required for pdac glutamine metabolism. Sci Rep, 2016; 6: p. 37594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyssiotis CA and Kimmelman AC, Metabolic interactions in the tumor microenvironment. Trends Cell Biol, 2017; 27: p. 863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Ahrens D, Bhagat TD, Nagrath D, Maitra A, and Verma A, The role of stromal cancer-associated fibroblasts in pancreatic cancer. J Hematol Oncol, 2017; 10: p. 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basso D, Gnatta E, and Plebani M, Pancreatic cancer fostered immunosuppression privileges tumor growth and progression. Journal of Clinical & Cellular Immunology, 2014: p. 16. [Google Scholar]
- 24.Clark CE, Hingorani SR, Mick R, Combs C, Tuveson DA, and Vonderheide RH, Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res, 2007; 67: p. 9518–27. [DOI] [PubMed] [Google Scholar]
- 25.Fu Y, Liu S, Zeng S, and Shen H, The critical roles of activated stellate cells-mediated paracrine signaling, metabolism and onco-immunology in pancreatic ductal adenocarcinoma. Mol Cancer, 2018; 17: p. 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harper J and Sainson RC, Regulation of the anti-tumour immune response by cancer-associated fibroblasts. Semin Cancer Biol, 2014; 25: p. 69–77. [DOI] [PubMed] [Google Scholar]
- 27.Wu Q, Tian Y, Zhang J, Zhang H, Gu F, Lu Y, et al. , Functions of pancreatic stellate cell-derived soluble factors in the microenvironment of pancreatic ductal carcinoma. Oncotarget, 2017; 8: p. 102721–102738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ene-Obong A, Clear AJ, Watt J, Wang J, Fatah R, Riches JC, et al. , Activated pancreatic stellate cells sequester cd8+ t cells to reduce their infiltration of the juxtatumoral compartment of pancreatic ductal adenocarcinoma. Gastroenterology, 2013; 145: p. 1121–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar V, Donthireddy L, Marvel D, Condamine T, Wang F, Lavilla-Alonso S, et al. , Cancer-associated fibroblasts neutralize the anti-tumor effect of csf1 receptor blockade by inducing pmn-mdsc infiltration of tumors. Cancer Cell, 2017; 32: p. 654–668 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buck MD, Sowell RT, Kaech SM, and Pearce EL, Metabolic instruction of immunity. Cell, 2017; 169: p. 570–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kouidhi S, Ben Ayed F, and Benammar Elgaaied A, Targeting tumor metabolism: A new challenge to improve immunotherapy. Front Immunol, 2018; 9: p. 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renner K, Singer K, Koehl GE, Geissler EK, Peter K, Siska PJ, et al. , Metabolic hallmarks of tumor and immune cells in the tumor microenvironment. Front Immunol, 2017; 8: p. 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardiner CM and Finlay DK, What fuels natural killers? Metabolism and nk cell responses. Front Immunol, 2017; 8: p. 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakashiba T, Ikeda T, Nishimura S, Tashiro K, Honjo T, Culotti JG, et al. , Netrin-g1: A novel glycosyl phosphatidylinositol-linked mammalian netrin that is functionally divergent from classical netrins. J Neurosci, 2000; 20: p. 6540–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song YS, Lee HJ, Prosselkov P, Itohara S, and Kim E, Trans-induced cis interaction in the tripartite ngl-1, netrin-g1 and lar adhesion complex promotes development of excitatory synapses. J Cell Sci, 2013; 126: p. 4926–38. [DOI] [PubMed] [Google Scholar]
- 36.Lin JC, Ho WH, Gurney A, and Rosenthal A, The netrin-g1 ligand ngl-1 promotes the outgrowth of thalamocortical axons. Nat Neurosci, 2003; 6: p. 1270–6. [DOI] [PubMed] [Google Scholar]
- 37.Amatangelo MD, Bassi DE, Klein-Szanto AJ, and Cukierman E, Stroma-derived three-dimensional matrices are necessary and sufficient to promote desmoplastic differentiation of normal fibroblasts. Am J Pathol, 2005; 167: p. 475–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franco-Barraza J, Francescone R, Luong T, Shah N, Madhani R, Cukierman G, et al. , Matrix-regulated integrin alphavbeta5 maintains alpha5beta1-dependent desmoplastic traits prognostic of neoplastic recurrence. Elife, 2017; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell PM, Groehler AL, Lee KM, Ouellette MM, Khazak V, and Der CJ, K-ras promotes growth transformation and invasion of immortalized human pancreatic cells by raf and phosphatidylinositol 3-kinase signaling. Cancer Res, 2007; 67: p. 2098–106. [DOI] [PubMed] [Google Scholar]
- 40.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. , The cbio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov, 2012; 2: p. 401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. , Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci Signal, 2013; 6: p. pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puleo F, Nicolle R, Blum Y, Cros J, Marisa L, Demetter P, et al. , Stratification of pancreatic ductal adenocarcinomas based on tumor and microenvironment features. Gastroenterology, 2018; 155: p. 1999–2013 e3. [DOI] [PubMed] [Google Scholar]
- 43.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, et al. , Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet, 2015; 47: p. 1168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cancer Genome Atlas Research Network. Electronic address aadhe and Cancer Genome Atlas Research N, Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell, 2017; 32: p. 185–203 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cukierman E, Pankov R, Stevens DR, and Yamada KM, Taking cell-matrix adhesions to the third dimension. Science, 2001; 294: p. 1708–12. [DOI] [PubMed] [Google Scholar]
- 46.Friedman G, Levi-Galibov O, David E, Bornstein C, Giladi A, Dadiani M, et al. , Cancer-associated fibroblast compositions change with breast cancer progression linking the ratio of s100a4+ and pdpn+ cafs to clinical outcome. Nature Cancer, 2020; 1: p. 692–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirayama K, Kono H, Nakata Y, Akazawa Y, Wakana H, Fukushima H, et al. , Expression of podoplanin in stromal fibroblasts plays a pivotal role in the prognosis of patients with pancreatic cancer. Surg Today, 2018; 48: p. 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shindo K, Aishima S, Ohuchida K, Fujiwara K, Fujino M, Mizuuchi Y, et al. , Podoplanin expression in cancer-associated fibroblasts enhances tumor progression of invasive ductal carcinoma of the pancreas. Mol Cancer, 2013; 12: p. 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiarelli M, Burati M, Tagliabue F, Terragni S, Guttadauro A, Cioffi G, et al. , Undifferentiated pancreatic carcinoma: Presentation, classification and prognosis. Journal of the Pancreas, 2019; 21: p. 1–6. [Google Scholar]
- 50.Strobel O, Hartwig W, Bergmann F, Hinz U, Hackert T, Grenacher L, et al. , Anaplastic pancreatic cancer: Presentation, surgical management, and outcome. Surgery, 2011; 149: p. 200–8. [DOI] [PubMed] [Google Scholar]
- 51.Xie Y, Xiang Y, Zhang D, Yao X, Sheng J, Yang Y, et al. , Sarcomatoid carcinoma of the pancreas: A case report and review of the literature. Mol Med Rep, 2018; 18: p. 4716–4724. [DOI] [PubMed] [Google Scholar]
- 52.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. , Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell, 2003; 4: p. 437–50. [DOI] [PubMed] [Google Scholar]
- 53.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. , Trp53r172h and krasg12d cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell, 2005; 7: p. 469–83. [DOI] [PubMed] [Google Scholar]
- 54.Muranen T, Iwanicki MP, Curry NL, Hwang J, DuBois CD, Coloff JL, et al. , Starved epithelial cells uptake extracellular matrix for survival. Nat Commun, 2017; 8: p. 13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, et al. , Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature, 2016; 536: p. 479–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao H, Yang L, Baddour J, Achreja A, Bernard V, Moss T, et al. , Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife, 2016; 5: p. e10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Recouvreux MV and Commisso C, Macropinocytosis: A metabolic adaptation to nutrient stress in cancer. Front Endocrinol (Lausanne), 2017; 8: p. 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Commisso C, Flinn RJ, and Bar-Sagi D, Determining the macropinocytic index of cells through a quantitative image-based assay. Nat Protoc, 2014; 9: p. 182–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiang Y, Stine ZE, Xia J, Lu Y, O’Connor RS, Altman BJ, et al. , Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. J Clin Invest, 2015; 125: p. 2293–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang L, Achreja A, Yeung TL, Mangala LS, Jiang D, Han C, et al. , Targeting stromal glutamine synthetase in tumors disrupts tumor microenvironment-regulated cancer cell growth. Cell Metab, 2016; 24: p. 685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liguz-Lecznar M and Skangiel-Kramska J, Vesicular glutamate transporters (vgluts): The three musketeers of glutamatergic system. Acta Neurobiol Exp (Wars), 2007; 67: p. 207–18. [DOI] [PubMed] [Google Scholar]
- 62.Woodcock HV, Eley JD, Guillotin D, Plate M, Nanthakumar CB, Martufi M, et al. , The mtorc1/4e-bp1 axis represents a critical signaling node during fibrogenesis. Nat Commun, 2019; 10: p. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rajasekaran S, Reddy NM, Zhang W, and Reddy SP, Expression profiling of genes regulated by fra-1/ap-1 transcription factor during bleomycin-induced pulmonary fibrosis. BMC Genomics, 2013; 14: p. 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fujita Y, Nakanishi T, Ueno M, Itohara S, and Yamashita T, Netrin-g1 regulates microglial accumulation along axons and supports the survival of layer v neurons in the postnatal mouse brain. Cell Rep, 2020; 31: p. 107580. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Q, Sano C, Masuda A, Ando R, Tanaka M, and Itohara S, Netrin-g1 regulates fear-like and anxiety-like behaviors in dissociable neural circuits. Sci Rep, 2016; 6: p. 28750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eastwood SL and Harrison PJ, Decreased mrna expression of netrin-g1 and netrin-g2 in the temporal lobe in schizophrenia and bipolar disorder. Neuropsychopharmacology, 2008; 33: p. 933–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nectoux J, Girard B, Bahi-Buisson N, Prieur F, Afenjar A, Rosas-Vargas H, et al. , Netrin g1 mutations are an uncommon cause of atypical rett syndrome with or without epilepsy. Pediatr Neurol, 2007; 37: p. 270–4. [DOI] [PubMed] [Google Scholar]
- 68.Hao W, Yu M, Lin J, Liu B, Xing H, Yang J, et al. , The pan-cancer landscape of netrin family reveals potential oncogenic biomarkers. Sci Rep, 2020; 10: p. 5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sho S, Court CM, Winograd P, Russell MM, and Tomlinson JS, A prognostic mutation panel for predicting cancer recurrence in stages ii and iii colorectal cancer. J Surg Oncol, 2017; 116: p. 996–1004. [DOI] [PubMed] [Google Scholar]
- 70.Andrew AS, Baron JA, Butterly LF, Suriawinata AA, Tsongalis GJ, Robinson CM, et al. , Hyper-methylated loci persisting from sessile serrated polyps to serrated cancers. Int J Mol Sci, 2017; 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Castro-Bello F, Ramos F, Vivanco F, and Marina-Fiol C, High serum glutamic acid levels in patients with carcinoma of the pancreas. Digestion, 1976; 14: p. 360–3. [DOI] [PubMed] [Google Scholar]
- 72.Roux C, Riganti C, Borgogno SF, Curto R, Curcio C, Catanzaro V, et al. , Endogenous glutamine decrease is associated with pancreatic cancer progression. Oncotarget, 2017; 8: p. 95361–95376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Biancur DE, Paulo JA, Malachowska B, Quiles Del Rey M, Sousa CM, Wang X, et al. , Compensatory metabolic networks in pancreatic cancers upon perturbation of glutamine metabolism. Nat Commun, 2017; 8: p. 15965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muir A, Danai LV, Gui DY, Waingarten CY, Lewis CA, and Vander Heiden MG, Environmental cystine drives glutamine anaplerosis and sensitizes cancer cells to glutaminase inhibition. Elife, 2017; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sullivan MR, Danai LV, Lewis CA, Chan SH, Gui DY, Kunchok T, et al. , Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability. Elife, 2019; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bott AJ, Shen J, Tonelli C, Zhan L, Sivaram N, Jiang YP, et al. , Glutamine anabolism plays a critical role in pancreatic cancer by coupling carbon and nitrogen metabolism. Cell Rep, 2019; 29: p. 1287–1298 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Daher B, Parks SK, Durivault J, Cormerais Y, Baidarjad H, Tambutte E, et al. , Genetic ablation of the cystine transporter xct in pdac cells inhibits mtorc1, growth, survival, and tumor formation via nutrient and oxidative stresses. Cancer Res, 2019; 79: p. 3877–3890. [DOI] [PubMed] [Google Scholar]
- 78.Coothankandaswamy V, Cao S, Xu Y, Prasad PD, Singh PK, Reynolds CP, et al. , Amino acid transporter slc6a14 is a novel and effective drug target for pancreatic cancer. Br J Pharmacol, 2016; 173: p. 3292–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parker SJ, Amendola CR, Hollinshead KER, Yu Q, Yamamoto K, Encarnacion-Rosado J, et al. , Selective alanine transporter utilization creates a targetable metabolic niche in pancreatic cancer. Cancer Discov, 2020; 10: p. 1018–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, et al. , Macropinocytosis of protein is an amino acid supply route in ras-transformed cells. Nature, 2013; 497: p. 633–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yao W, Rose JL, Wang W, Seth S, Jiang H, Taguchi A, et al. , Syndecan 1 is a critical mediator of macropinocytosis in pancreatic cancer. Nature, 2019; 568: p. 410–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li L and Hanahan D, Hijacking the neuronal nmdar signaling circuit to promote tumor growth and invasion. Cell, 2013; 153: p. 86–100. [DOI] [PubMed] [Google Scholar]
- 83.Popoli M, Yan Z, McEwen BS, and Sanacora G, The stressed synapse: The impact of stress and glucocorticoids on glutamate transmission. Nat Rev Neurosci, 2011; 13: p. 22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, et al. , Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med, 2017; 9: p. 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Evans RA, Diamond MS, Rech AJ, Chao T, Richardson MW, Lin JH, et al. , Lack of immunoediting in murine pancreatic cancer reversed with neoantigen. JCI Insight, 2016; 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yarchoan M, Hopkins A, and Jaffee EM, Tumor mutational burden and response rate to pd-1 inhibition. N Engl J Med, 2017; 377: p. 2500–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Van Audenaerde JRM, Roeyen G, Darcy PK, Kershaw MH, Peeters M, and Smits ELJ, Natural killer cells and their therapeutic role in pancreatic cancer: A systematic review. Pharmacol Ther, 2018; 189: p. 31–44. [DOI] [PubMed] [Google Scholar]
- 88.Vaheri A, Enzerink A, Rasanen K, and Salmenpera P, Nemosis, a novel way of fibroblast activation, in inflammation and cancer. Exp Cell Res, 2009; 315: p. 1633–8. [DOI] [PubMed] [Google Scholar]
- 89.Heneberg P, Paracrine tumor signaling induces transdifferentiation of surrounding fibroblasts. Crit Rev Oncol Hematol, 2016; 97: p. 303–11. [DOI] [PubMed] [Google Scholar]
- 90.Van Audenaerde JRM, De Waele J, Marcq E, Van Loenhout J, Lion E, Van den Bergh JMJ, et al. , Interleukin-15 stimulates natural killer cell-mediated killing of both human pancreatic cancer and stellate cells. Oncotarget, 2017; 8: p. 56968–56979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Trotta R, Dal Col J, Yu J, Ciarlariello D, Thomas B, Zhang X, et al. , Tgf-beta utilizes smad3 to inhibit cd16-mediated ifn-gamma production and antibody-dependent cellular cytotoxicity in human nk cells. J Immunol, 2008; 181: p. 3784–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Takeuchi S, Baghdadi M, Tsuchikawa T, Wada H, Nakamura T, Abe H, et al. , Chemotherapy-derived inflammatory responses accelerate the formation of immunosuppressive myeloid cells in the tissue microenvironment of human pancreatic cancer. Cancer Res, 2015; 75: p. 2629–40. [DOI] [PubMed] [Google Scholar]
- 93.Bayne LJ, Beatty GL, Jhala N, Clark CE, Rhim AD, Stanger BZ, et al. , Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and t cell immunity in pancreatic cancer. Cancer Cell, 2012; 21: p. 822–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, and Bar-Sagi D, Oncogenic kras-induced gm-csf production promotes the development of pancreatic neoplasia. Cancer Cell, 2012; 21: p. 836–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Waghray M, Yalamanchili M, Dziubinski M, Zeinali M, Erkkinen M, Yang H, et al. , Gm-csf mediates mesenchymal-epithelial cross-talk in pancreatic cancer. Cancer Discov, 2016; 6: p. 886–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Galic MA, Riazi K, and Pittman QJ, Cytokines and brain excitability. Front Neuroendocrinol, 2012; 33: p. 116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Haroon E, Miller AH, and Sanacora G, Inflammation, glutamate, and glia: A trio of trouble in mood disorders. Neuropsychopharmacology, 2017; 42: p. 193–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vesce S, Rossi D, Brambilla L, and Volterra A, Glutamate release from astrocytes in physiological conditions and in neurodegenerative disorders characterized by neuroinflammation. Int Rev Neurobiol, 2007; 82: p. 57–71. [DOI] [PubMed] [Google Scholar]
- 99.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. , Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature, 2012; 491: p. 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Demir IE, Friess H, and Ceyhan GO, Neural plasticity in pancreatitis and pancreatic cancer. Nat Rev Gastroenterol Hepatol, 2015; 12: p. 649–59. [DOI] [PubMed] [Google Scholar]
- 101.Long Y, Zhuang K, Ji Z, Han Y, Fei Y, Zheng W, et al. , 2-deoxy-d-glucose exhibits anti-seizure effects by mediating the netrin-g1-katp signaling pathway in epilepsy. Neurochem Res, 2019; 44: p. 994–1004. [DOI] [PubMed] [Google Scholar]
- 102.Brichkina A, Bertero T, Loh HM, Nguyen NT, Emelyanov A, Rigade S, et al. , P38mapk builds a hyaluronan cancer niche to drive lung tumorigenesis. Genes Dev, 2016; 30: p. 2623–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alspach E, Flanagan KC, Luo X, Ruhland MK, Huang H, Pazolli E, et al. , P38mapk plays a crucial role in stromal-mediated tumorigenesis. Cancer Discov, 2014; 4: p. 716–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yamamura Y, Asai N, Enomoto A, Kato T, Mii S, Kondo Y, et al. , Akt-girdin signaling in cancer-associated fibroblasts contributes to tumor progression. Cancer Res, 2015; 75: p. 813–23. [DOI] [PubMed] [Google Scholar]
- 105.Jiang X, Xie H, Dou Y, Yuan J, Zeng D, and Xiao S, Expression and function of fra1 protein in tumors. Mol Biol Rep, 2020; 47: p. 737–752. [DOI] [PubMed] [Google Scholar]
- 106.Atsaves V, Leventaki V, Rassidakis GZ, and Claret FX, Ap-1 transcription factors as regulators of immune responses in cancer. Cancers (Basel), 2019; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bhagat TD, Von Ahrens D, Dawlaty M, Zou Y, Baddour J, Achreja A, et al. , Lactate-mediated epigenetic reprogramming regulates formation of human pancreatic cancer-associated fibroblasts. Elife, 2019; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Busch S, Andersson D, Bom E, Walsh C, Stahlberg A, and Landberg G, Cellular organization and molecular differentiation model of breast cancer-associated fibroblasts. Mol Cancer, 2017; 16: p. 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Qin X, Jiang B, and Zhang Y, 4e-bp1, a multifactor regulated multifunctional protein. Cell Cycle, 2016; 15: p. 781–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Duluc C, Moatassim-Billah S, Chalabi-Dchar M, Perraud A, Samain R, Breibach F, et al. , Pharmacological targeting of the protein synthesis mtor/4e-bp1 pathway in cancer-associated fibroblasts abrogates pancreatic tumour chemoresistance. EMBO Mol Med, 2015; 7: p. 735–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Franco-Barraza J, Beacham DA, Amatangelo MD, and Cukierman E, Preparation of extracellular matrices produced by cultured and primary fibroblasts. Curr Protoc Cell Biol, 2016; 71: p. 10 9 1–10 9 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Beglyarova N, Banina E, Zhou Y, Mukhamadeeva R, Andrianov G, Bobrov E, et al. , Screening of conditionally reprogrammed patient-derived carcinoma cells identifies ercc3-myc interactions as a target in pancreatic cancer. Clin Cancer Res, 2016; 22: p. 6153–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. , Limma powers differential expression analyses for rna-sequencing and microarray studies. Nucleic Acids Res, 2015; 43: p. e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Merico D, Isserlin R, Stueker O, Emili A, and Bader GD, Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS One, 2010; 5: p. e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang X, Spandidos A, Wang H, and Seed B, Primerbank: A pcr primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res, 2012; 40: p. D1144–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rezakhaniha R, Agianniotis A, Schrauwen JT, Griffa A, Sage D, Bouten CV, et al. , Experimental investigation of collagen waviness and orientation in the arterial adventitia using confocal laser scanning microscopy. Biomech Model Mechanobiol, 2012; 11: p. 461–73. [DOI] [PubMed] [Google Scholar]
- 117.Sanjana NE, Shalem O, and Zhang F, Improved vectors and genome-wide libraries for crispr screening. Nat Methods, 2014; 11: p. 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Thakore PI, D’Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, et al. , Highly specific epigenome editing by crispr-cas9 repressors for silencing of distal regulatory elements. Nat Methods, 2015; 12: p. 1143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y, et al. , Compact and highly active next-generation libraries for crispr-mediated gene repression and activation. Elife, 2016; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, et al. , Tracing compartmentalized nadph metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell, 2014; 55: p. 253–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Young JD, Walther JL, Antoniewicz MR, Yoo H, and Stephanopoulos G, An elementary metabolite unit (emu) based method of isotopically nonstationary flux analysis. Biotechnol Bioeng, 2008; 99: p. 686–99. [DOI] [PubMed] [Google Scholar]
- 122.Fernandez CA, Des Rosiers C, Previs SF, David F, and Brunengraber H, Correction of 13c mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom, 1996; 31: p. 255–62. [DOI] [PubMed] [Google Scholar]
- 123.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL, Tophat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol, 2013; 14: p. R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.sAnders S, Pyl PT, and Huber W, Htseq--a python framework to work with high-throughput sequencing data. Bioinformatics, 2015; 31: p. 166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Love MI, Huber W, and Anders S, Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biol, 2014; 15: p. 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A, 2005; 102: p. 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hanzelmann S, Castelo R, and Guinney J, Gsva: Gene set variation analysis for microarray and rna-seq data. BMC Bioinformatics, 2013; 14: p. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, and Tamayo P, The molecular signatures database (msigdb) hallmark gene set collection. Cell Syst, 2015; 1: p. 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nagy A, Lanczky A, Menyhart O, and Gyorffy B, Validation of mirna prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep, 2018; 8: p. 9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. , An integrated tcga pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell, 2018; 173: p. 400–416 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.